Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ag2WO4 nanocatalyst-driven green synthesis of pyrano[2,3-d]pyrimidinones: an integrated experimental, DFT, and cytotoxic investigation
Sathiaseelan Perumalab,
Perumal Muthurajac,
M. Sasikumar b,
R. Hari Krishnad,
Paramasivam Manisankar*a and
Viswanathan Subramanian
b,
R. Hari Krishnad,
Paramasivam Manisankar*a and
Viswanathan Subramanian *a
*a
aDepartment of Industrial Chemistry, Alagappa University, Karaikudi, Tamil Nadu 630 006, India. E-mail: manisankarp@alagappauniversity.ac.in; rsviswa@gmail.com
bDepartment of Chemistry, Bishop Heber College, Tiruchirappalli, Tamil Nadu 620 017, India
cDepartment of Chemistry, Indian Institute of Technology Guwahati, Guwahati 781039, India
dDepartment of Chemistry, M.S. Ramaiah Institute of Technology, Bangalore-560 054, India
First published on 16th January 2026
Abstract
The development of environmentally sustainable routes for synthesizing bioactive heterocycles remains a central objective in contemporary medicinal chemistry. In this work, silver tungstate (Ag2WO4) nanoparticles were synthesized and characterized by X-ray diffraction and scanning electron microscopy, confirming their crystalline structure and surface morphology. The prepared nanoparticles served as an efficient heterogeneous nanocatalyst for the one-pot green synthesis of pyrano[2,3-d]pyrimidinone derivatives (2a–2h) in an ethanol–water medium, affording excellent yields under mild conditions and demonstrating high catalytic efficiency with strong alignment to green chemistry principles. Comprehensive in silico analyses, including density functional theory (DFT), molecular docking, molecular dynamics (MD) simulations, and ADME screening, were performed to elucidate the electronic distribution, binding interactions, and pharmacokinetic profiles of the synthesized scaffolds. Docking studies revealed that compounds 2g (−7.58 kcal mol−1) and 2h (−7.34 kcal mol−1) exhibited binding affinities comparable to the reference drug afatinib (−8.01 kcal mol−1). In vitro cytotoxic evaluation against A549 lung carcinoma cells further identified compound 2h as the most potent derivative (IC50 = 39.29 µM), significantly outperforming the unsubstituted analogue 2a (IC50 = 120.65 µM). Overall, this integrated experimental, computational, and biological study establishes Ag2WO4 as a robust and sustainable nanocatalyst, offering an efficient pathway for the rapid synthesis of pyrano[2,3-d]pyrimidinone scaffolds with promising anticancer potential.
1 Introduction
The pyrano-pyrimidinone moiety is a significant class of fused heterocycles with diverse biological activities, such as antioxidant,1 antimicrobial,2 anti-inflammatory,3 and anticancer4 properties, making them attractive scaffolds for therapeutic development.5 Owing to their structural diversity and biological relevance, the development of efficient and sustainable methods for synthesising these frameworks is a priority in medicinal chemistry.To date, various methodologies have been developed for constructing the pyrano[2,3-d]pyrimidinone core.6–15 Multicomponent reactions (MCRs) are eco-friendly owing to their high atom economy, operational simplicity, and ability to produce complex heterocycles in a single step.16,17 Simultaneously, nanocatalysts have gained popularity in organic synthesis because of their improved surface reactivity and reusability, thus adhering to green chemistry.18,19 However, innovative catalytic systems that offer both high efficiency and environmental friendliness are still required, particularly for pharmaceutically significant MCRs.
In this context, a review of the literature shows that silver tungstate (Ag2WO4) has been studied mainly for its redox activity and photocatalytic efficiency.20 However, reports on its application as a heterogeneous nanocatalyst in organic synthesis, particularly in multicomponent reactions leading to heterocycles, are still limited.21 These unexplored potential positions Ag2WO4 NPs as promising candidates for developing greener synthetic methodologies. Guo et al. demonstrated the bifunctional catalytic role of silver tungstate (Ag2WO4) in the carbonylation of terminal alkynes.22 Motivated by these findings, we synthesised Ag2WO4 nanoparticles (NPs) to further enhance their surface activity and extend the investigation.
This study introduces an environmentally benign one-pot protocol that utilises Ag2WO4 nanoparticles as heterogeneous nanocatalystss for the synthesis of pyrano[2,3-d]pyrimidinone derivatives. Importantly, the protocol proceeds efficiently under mild conditions, delivering excellent yields and purities within short reaction times without requiring chromatographic purification. By reducing energy consumption and waste, this methodology significantly enhances the green credentials of heterocyclic synthesis processes.
Building on this synthetic success, the compounds were systematically assessed to complement the experimental synthesis using a combination of computational and biological methods, such as density functional theory (DFT) calculations, ADME prediction, molecular docking, molecular dynamics (MD) simulations, MM-GBSA, and free energy landscape (FEL) mapping.23 Cytotoxicity was assessed in A549 lung cancer cells using an MTT assay.24 Afatinib, an FDA-approved EGFR tyrosine kinase inhibitor, was selected as the reference drug owing to its cytotoxicity in A549 cells.25 Although Afatinib primarily targets EGFR, its docking with PDB ID: 4ZXT (ERK2 kinase domain) was performed in this study as an exploratory benchmark, enabling a comparative analysis of the binding affinities of the clinically validated drug and the newly synthesised derivatives.
Overall, these integrated synthetic, computational, and biological approaches address the critical gap in the sustainable production of pharmaceutically important heterocycles. This study breaks new ground by utilising Ag2WO4 NPs in a one-pot method to synthesise pyrano[2,3-d]pyrimidinones. This highlights their potential as anticancer agents and supports ongoing initiatives for sustainable drug discovery.
2 Results and discussion
2.1 Chemistry
 | ||
| Fig. 1 X-ray diffraction (XRD) pattern of synthesized Ag2WO4 nanocatalyst, showing characteristic diffraction peaks. | ||
Scanning electron micrographs of Ag2WO4 at different magnifications are shown in Fig. 2a–d. At lower magnification, the surface morphology was found to be a highly agglomerated mass of particle clusters. A closer observation of the micrographs revealed that the agglomerated particles had a mixed morphology consisting of irregular particle aggregates and fused rod-like structures (Fig. 2b). However, at higher magnifications, the SEM images clearly show the dominance of rod-like fused structures with uneven growth. This uneven growth results in a rough surface, and the tailing of rod-like structures can be observed. Furthermore, to evaluate the elemental composition and distribution, EDAX analysis with colour mapping was performed, as shown in Fig. 2. The EDAX and colour mapping results confirmed that, except for Ag, W, and O, no other elements were present in the compound, suggesting simple purity of the sample. Furthermore, colour mapping showed a uniform distribution of elements throughout the product and the homogeneous nature of the product.
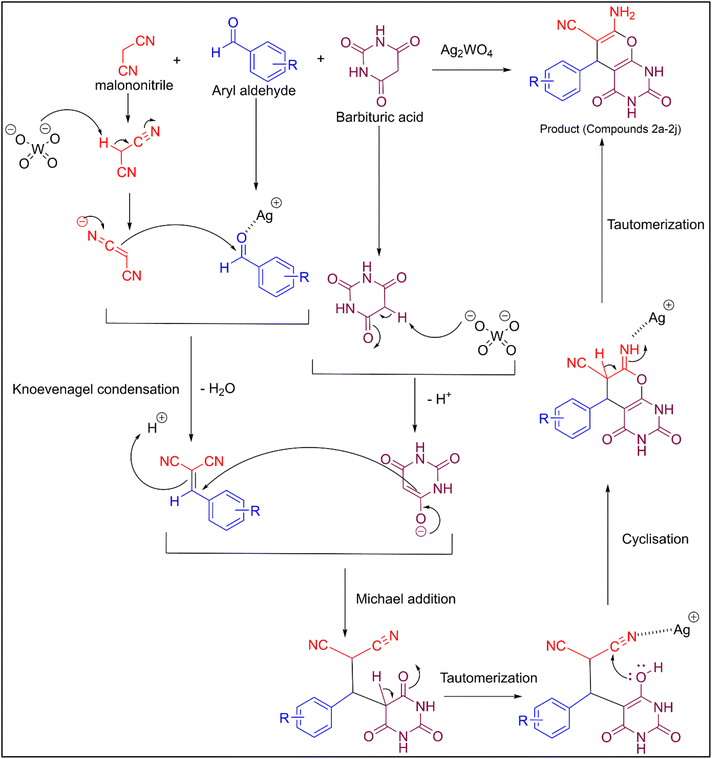
The Ag2WO4 catalyst demonstrated superior performance compared with previously reported catalysts for the synthesis of pyrano[2,3-d]pyrimidinone derivatives. Specifically, Ag2WO4 catalysed the one-pot synthesis of pyrano[2,3-d]pyrimidinones in an EtOH/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solvent mixture, achieving product yields of 95% within a reaction time of 5–8 min at 70 °C. The superior performance of Ag2WO4 can be attributed to the operational simplicity and rapidity of the catalyst and/or its adherence to the principles of green chemistry. Overall, the results demonstrated that Ag2WO4 is a highly active and environmentally compliant catalyst that can match or exceed conventional systems in terms of activity and practicability.
1) solvent mixture, achieving product yields of 95% within a reaction time of 5–8 min at 70 °C. The superior performance of Ag2WO4 can be attributed to the operational simplicity and rapidity of the catalyst and/or its adherence to the principles of green chemistry. Overall, the results demonstrated that Ag2WO4 is a highly active and environmentally compliant catalyst that can match or exceed conventional systems in terms of activity and practicability.
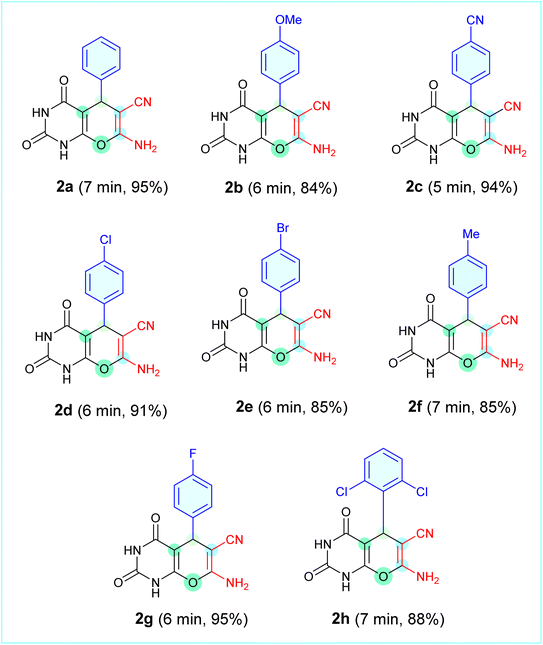
Table 1 lists the generality of the proposed method. All aromatic aldehydes examined, regardless of whether they had an electron-donating or electron-withdrawing group, underwent a clean reaction to produce the corresponding derivatives (2a–2h) in very high yields (84–95%) over a short time period (5–7 min). Thus, the unsubstituted analogue (2a) was produced in 95% yield (7 min), while p-chloro (2d) and p-fluoro (2g) analogues were produced in 91% and 95% yields, respectively, within 6 min. Additionally, the o-dichloro-substituted analogue (2h) was produced in 88% yield within 7 min, demonstrating the wide applicability of the Ag2WO4-NP-based system. In summary, the proposed mechanism and scope demonstrate that Ag2WO4 nanoparticles act as a bifunctional nanocatalyst to enable a green precipitation-assisted protocol for the facile and efficient synthesis of pyrano[2,3-d]pyrimidinones.
 | ||
| Fig. 3 1H NMR spectrum (in DMSO) of compound 2h recorded at 600 MHz, showing the chemical shifts (δ, ppm) with proton assignments corresponding to the molecular structure. | ||
 | ||
| Fig. 4 13C NMR spectrum (in DMSO) of compound 2h recorded at 600 MHz, showing the chemical shifts (δ, ppm) with carbon assignments corresponding to the molecular structure. | ||
Such glutarimide-type deshielding significantly lowers the shielding of the N–H protons, leading to resonances far from downfield. In compound 2h, these signals were assigned to H31 (δ 12.06 ppm, amide N–H) and H30 (δ 11.02 ppm, imide N–H). The –NH2 protons at C2 appeared as a broad signal at δ ∼7.4 ppm, confirming the presence of an exocyclic amino group. The methine proton (H24) attached to C6 of the pyran ring appeared at δ ∼5.2 ppm, serving as a diagnostic marker for ring closure. The aromatic protons (H26–H28) of the phenyl substituent were observed at δ 7.2–7.3 ppm, with minor chemical shift variations across the derivatives reflecting the electronic influence of substituents such as –Cl, –Br, –F, –CH3, –OCH3, and –CN. This overall spectral pattern, such as two downfield NH signals, a mid-field amino group, a methine resonance near δ 4.0–5.2 ppm, and aromatic protons near δ 7.2–7.3 ppm was consistently observed across all derivatives, confirming the integrity of the prano[2,3-d]pyrimidinone framework.
The 13C NMR spectra further substantiated these structural assignments. We observed the most deshielded signals with respect to the sp2 carbon (C2) of the pyran ring, which is connected to O15 and N1, followed by the carbonyl carbons of the pyrimidinone core, resonating in the δ 160–167 ppm range, which is consistent with conjugation and strong electron-withdrawing effects. In compound 2h, the characteristic carbonyl resonances were observed at δ = 158.68 and 149.46 ppm. The cyano carbon signal was consistently detected at δ 118 ppm (δ 118.46 ppm for 2h), serving as a reliable fingerprint of the –CN substitution at the pyran ring. The sp2 hybridised carbon atoms at the ring junction of the pyran ring appeared in the δ 50–90 ppm range (δ 54.09 and 86.07 ppm for 2h), while the sp3 carbon of the system resonated between δ 35–39 ppm (δ 32.09 ppm in 2h). The substituent effects are clearly reflected in the aromatic carbon region; electron-withdrawing groups, such as halogens (Cl, Br, and F), produced deshielding of the ortho and ipso carbons (δ ∼135–140 ppm), whereas electron-donating substituents (–OCH3 and –CH3) induced slight upfield shifts in the phenyl carbons.
In summary, experimental NMR data confirmed the target structure. The reproducible downfield NH resonances, characteristic methine and amino signals in the proton spectra, carbonyl and cyano resonances in the carbon spectra, and substituent-dependent aromatic shifts confirmed the successful synthesis of the scaffolds (2a–2h). To further validate the structural assignments, the experimental 1H and 13C NMR spectra of compound 2h were compared with the theoretical chemical shifts calculated at the B3LYP/def2-TZVP level of theory for the DMSO phase (Table 2). For 1H NMR, the calculated chemical shifts aligned with the experimental trend with good accuracy (RMSD = 2.6 ppm). The deshielded protons H31 (δ_exp = 12.0 ppm) and H30 (δ_exp = 11.0 ppm), arising from the glutarimide-type N–H groups, were predicted theoretically (δ_calc ∼7.0–7.1 ppm), albeit slightly underestimated because of the known limitations of DFT shielding calculations.
| 1H NMR | 13C NMR | ||||
|---|---|---|---|---|---|
| Atom | B3LYP | Exp | Atom | B3LYP | Exp |
| H22 | 5.0 | 7.4 | C6 | 45.2 | 32.1 |
| H23 | 4.5 | 7.4 | C3 | 65.6 | 54.1 |
| H24 | 5.8 | 5.3 | C13 | 100.8 | 86.1 |
| H26 | 7.6 | 7.2 | C4 | 130.9 | 118.5 |
| H27 | 7.5 | 7.3 | C9 | 139.7 | 128.6 |
| H28 | 7.5 | 7.2 | C10 | 139.8 | 129.3 |
| H30 | 7.0 | 11.0 | C11 | 141.7 | 130.3 |
| H31 | 7.1 | 12.0 | C7 | 151.4 | 134.1 |
| RMSD | 2.6 | C12 | 152.7 | 135.7 | |
| C8 | 153.8 | 135.9 | |||
| C17 | 156.9 | 149.5 | |||
| C14 | 167.6 | 153.0 | |||
| C19 | 170.9 | 158.7 | |||
| C2 | 176.4 | 162.1 | |||
| RMSD | 12.7 | ||||
The methine proton H24 (attached to C6) appeared at δ_exp = 5.3 ppm, which closely matched the predicted value of δ_calc = 5.8 ppm. The aromatic protons H26–H28 clustered around δ_exp 7.2–7.4 ppm, also showing good correlation with the theoretical data δ_calc = 7.5–7.6 ppm. The 13C NMR results showed a good match between the experimental and theoretical shifts, with an RMSD of 12.7 ppm, which was acceptable. The carbonyl carbons (C17 and C19) resonated experimentally at δ 149.5 and 158.7 ppm, respectively, and were reproduced theoretically (δ_calc ∼157–171 ppm). The cyano carbon (C4) appeared at δ_exp = 118.5 ppm, in line with its strong electron-withdrawing environment, and was well predicted by DFT (δ_calc ≈ 130.9 ppm). The methine carbon (C6) gave a signal at δ_exp = 32.1 ppm (δ_calc = 45.2 ppm), which is consistent with the sp3-hybridised environment in the fused pyran ring. The most downfield carbon, C2 (δ_exp = 162.1 ppm), corresponds to the sp2-hybridised carbon in the pyran ring conjugated with adjacent heteroatoms and was predicted to be δ_calc = 176.4 ppm. The remaining aromatic carbons (C7–C12) resonated between δ_exp 126–136 ppm, with theoretical values showing good correspondence.
The strong correlation between the theoretical and experimental NMR results strongly supported the structural assignment and confirmed the theoretically predicted electronic environment of the molecules. The similar comparison table is also shown for each of the other derivatives 2a–2h in the SI (Tables S1–S16). This confirmed the assignments based on experimental NMR data for each derivative. In addition to NMR analysis, FT-IR analysis was performed to further validate the key functional groups and vibrational features of the synthesised scaffolds.
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N) was consistently observed as a sharp, medium-intensity band at 2195–2200 cm−1, confirming the presence of nitrile functionality. Two strong bands corresponding to the C
N) was consistently observed as a sharp, medium-intensity band at 2195–2200 cm−1, confirming the presence of nitrile functionality. Two strong bands corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of the pyrimidinone/lactam functional groups appeared in the range of 1720–1670 cm−1, which is characteristic of conjugated carbonyl groups within the fused heteroaromatic system.
O stretching vibrations of the pyrimidinone/lactam functional groups appeared in the range of 1720–1670 cm−1, which is characteristic of conjugated carbonyl groups within the fused heteroaromatic system.Another characteristic band at 1380–1090 cm−1, assigned to the C–O–C stretching of the pyran ring, and absorptions in the fingerprint region (900–700 cm−1) correspond to the aromatic C–H bending modes. Substituent effects were also evident; halogenated derivatives (2a–2h) exhibited stronger bands in the 600–500 cm−1 region corresponding to C–Cl or C–Br stretching, while the methoxy-substituted compound (2b) showed an additional band near 1096 cm−1 characteristic of C–O stretching. The IR spectra of the synthesised derivatives (2a–2h are shown in the SI (Fig. S1–S32), with comprehensive FT-IR data for each derivative detailed in Sections 2.2.1–2.2.8.
The comparative IR analysis of compound 2h, as shown in Fig. 5, demonstrates excellent agreement between the experimental spectrum and the theoretically predicted vibrational profile, thereby validating the optimised geometry and electronic structure calculations. Experimentally, strong absorption bands were observed at ∼3389 and 3213 cm−1, corresponding to primary amino and amide N–H stretching vibrations respectively, which were reproduced in the theoretical spectrum with slight shifts due to anharmonic and solvation effects. The sharp absorption near 2196 cm−1 was assigned to the cyano (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N) stretch, which was consistent with the experimental and theoretical results. In the carbonyl region, distinct stretching bands were observed at approximately 1717 and 1673 cm−1, which correlated well with the theoretically predicted C
N) stretch, which was consistent with the experimental and theoretical results. In the carbonyl region, distinct stretching bands were observed at approximately 1717 and 1673 cm−1, which correlated well with the theoretically predicted C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrations, confirming the integrity of the pyranone and pyrimidinone frameworks. Additionally, a C–O–C stretching band appeared near 1096 cm−1 in both spectra, highlighting the characteristic contribution of the heterocyclic oxygen.
O vibrations, confirming the integrity of the pyranone and pyrimidinone frameworks. Additionally, a C–O–C stretching band appeared near 1096 cm−1 in both spectra, highlighting the characteristic contribution of the heterocyclic oxygen.
The close correspondence between the theoretical and experimental spectra not only supports the structural assignment of 2h but also reinforces the reliability of the DFT-based vibrational analysis for the entire series of derivatives (2a–2h). This similarity provides confidence in linking computational insights to experimental validation. Although 2h is shown as a representative example, comparative experimental and theoretical IR spectra of the derivatives (2a–2h) are presented in the SI (Fig. S25–S32), thus validating the characteristic vibrational features.
2.2 Computational analysis
Following the synthesis and characterisation of pyrano[2,3-d]pyrimidinones, we proceeded with the application of computational methods that would provide additional evidence and support the experimental data and explain the properties of compounds 2a–2h. The use of computational methods for this purpose provided an opportunity to utilise the DFT method to provide HOMO–LUMO, MEP, and other reactivity descriptors to illustrate the electronic distribution and reaction of derivatives 2a–2h. Additionally, docking of derivatives 2a–2h with PDB ID: 4ZXT using afatinib (A549 assay) as a reference drug provided an assessment of the binding affinities of the derivatives with the target protein. Dynamic stability was also investigated through 100 ns molecular dynamics simulations, which included additional analyses such as MM/GBSA,26 PCA and FEL.27,28 Finally, ADME prediction of the pharmacokinetic potential of derivatives 2a–2h was performed, and the results were correlated with MTT cytotoxicity to bridge the gap between computational methods and biological systems.| Code | ELUMO (eV) | EHOMO (eV) | ΔE | (I) | (A) | (n) | (z) | (χ) | (µ) | (ω) | µ (D) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a ELUMO = energy of LUMO, EHOMO = energy of HOMO, ΔE = |EHOMO–ELUMO|, I = ionisation potential, A = electron affinity, n = chemical hardness, z = chemical softness, χ = electronegativity, µ = chemical potential, ω = electrophilicity, µ = dipole moment. | |||||||||||
| 2a | −1.65 | −6.53 | 4.88 | 6.53 | 1.65 | 2.44 | 0.20 | 4.09 | −4.09 | 3.43 | 5.12 |
| 2b | −1.57 | −6.05 | 4.48 | 6.05 | 1.57 | 2.24 | 0.22 | 3.81 | −3.81 | 3.24 | 3.89 |
| 2c | −2.07 | −6.87 | 4.80 | 6.87 | 2.07 | 2.40 | 0.21 | 4.47 | −4.47 | 4.16 | 8.77 |
| 2d | −1.77 | −6.57 | 4.80 | 6.57 | 1.77 | 2.40 | 0.21 | 4.17 | −4.17 | 3.62 | 5.95 |
| 2e | −1.79 | −6.55 | 4.76 | 6.55 | 1.79 | 2.38 | 0.21 | 4.17 | −4.17 | 3.65 | 6.17 |
| 2f | −1.60 | −6.41 | 4.81 | 6.41 | 1.60 | 2.41 | 0.21 | 4.01 | −4.01 | 3.33 | 4.98 |
| 2g | −1.73 | −6.58 | 4.85 | 6.58 | 1.73 | 2.43 | 0.21 | 4.16 | −4.16 | 3.56 | 5.84 |
| 2h | −1.68 | −6.43 | 4.75 | 6.43 | 1.68 | 2.38 | 0.21 | 4.06 | −4.06 | 3.46 | 4.35 |
| Afatinib | −1.81 | −5.72 | 3.91 | 5.72 | 1.81 | 1.96 | 0.26 | 3.77 | −3.77 | 3.63 | 3.76 |
The chemical potential (µ), which reflects the tendency of electrons to escape, was consistently negative (−3.81 to −4.47 eV) across the series. The most negative values (2c, −4.47 eV; 2g, −4.16 eV) suggest a lower tendency to lose electron density, which stabilises these molecules against spontaneous charge transfer. In contrast, 2b (−3.81 eV) and afatinib (−3.77 eV) showed relatively higher µ values, indicating a greater propensity to engage in donor–acceptor interactions within the protein-binding sites. The electrophilicity index (ω) of the scaffolds (2a–2h) ranged from 3.24–4.16 eV. Compounds 2c (4.16 eV) and 2e (3.65 eV) showed the highest electrophilic character, making them favourable candidates for electron-acceptor roles in protein-ligand binding. Compound 2h (ω = 3.46 eV) displayed electrophilicity comparable to that of afatinib (3.63 eV), suggesting a balanced profile of reactivity and selectivity. The results of dipole moment analysis further support these findings. The calculated dipole moments ranged from 3.89 D (2b) to 8.77 D (2c), reflecting the variations in the overall molecular polarity. Highly polar molecules such as 2c (8.77 D) and 2e (6.17 D) are assumed to have stronger solubility in aqueous environments and enhanced orientation within the polar regions of the active site of PDB ID: 4ZXT. In contrast, derivatives with lower dipole moments, including 2b (3.89 D) and 2h (4.35 D), may favour hydrophobic pockets and balanced solubility, contributing to favourable bioavailability. Afatinib exhibited the lowest dipole moment (3.76 D) among the scaffolds (2a–2h), suggesting that moderate polarity, rather than excessive polarity, may be optimal for achieving strong and specific binding interactions.
Therefore, the general trend throughout the series indicates that all electron-withdrawing substituents (CN, Halogen in compounds 2c, 2d, 2e, 2g, and 2h) increased the electrophilicity and dipole moment of the compound, while electron-donating substituents (OCH3 in 2b and OH in 2f) raised the HOMO energy levels and decreased the dipole moment, thereby lowering the nucleophilicity. Compound 2h represents an intermediate with a moderately lowered HOMO energy level (−6.43 eV), low LUMO energy (−1.68 eV), appropriate chemical potential (−4.06 eV), reasonable electrophilic index (3.46 eV), and moderate dipole moment (4.35 D). The above characteristics explain the good IC50 value (39.29 µM) against the A549 cell line, which clearly shows a strong relationship between the predicted results from the computer models and the measured activity in the cell lines.
The MEP results indicate that 2h may interact via two different mechanisms: first, by forming a hydrogen bonding interaction between a polar hydrogen atom from 2h and an acceptor group on the protein; second, by forming a non-polar interaction (π–σ or π–alkyl) between 2h and the protein. The MEP results suggested that this increased flexibility is beneficial for protein binding. Furthermore, the calculated electrophilicity index (ω = 3.46 eV) for 2h was similar to that of afatinib, suggesting a favourable binding profile. Importantly, the results shown in the MEP maps were consistent with the docking of 2h into 4ZXT (Fig. 7D). In this complex, the electronegative regions of 2h were aligned with the donor groups of the protein, and the electropositive sites of 2h complement the acceptor sites (ASP167 and ASP111) on the enzyme. These MEP-derived predictions were validated using the different types of interactions present in the docking complex. MEP plots and the optimised geometries along with the HOMO–LUMO for each of the synthesised compounds (2a–2h) are provided in the SI (Fig. S33)
| Donor | Acceptor | E(2) kcal mol−1)a | ΔE (a.u.)b | F(i, j) (a.u.)c |
|---|---|---|---|---|
| a E2 is the energy of the hyperconjugative interactions.b Energy difference between the donor and acceptor i and j NBO orbitals.c F(i, j) is the Fock matrix element between i and j. NBO orbitals. | ||||
| LP (N18) | π* (C17–O21) | 63.71 | 0.27 | 0.118 |
| LP (N16) | π* (C17–O21) | 53.12 | 0.29 | 0.111 |
| LP (N18) | π* (C19–O20) | 51.48 | 0.29 | 0.109 |
| LP (N16) | π* (C13–C14) | 46.74 | 0.30 | 0.106 |
| LP (N1) | π* (C2–C3) | 44.49 | 0.31 | 0.104 |
| LP (O15) | π* (C13–C14) | 33.18 | 0.38 | 0.100 |
| LP (O21) | σ* (N16–C17) | 30.88 | 0.63 | 0.124 |
| LP (O20) | σ* (N18–C19) | 30.75 | 0.63 | 0.124 |
| LP (O15) | π* (C2–C3) | 30.29 | 0.39 | 0.096 |
| LP (O21) | σ* (C17–N18) | 28.68 | 0.66 | 0.123 |
| π (C13–C14) | π* (C19–O20) | 25.11 | 0.32 | 0.079 |
| π (C2–C3) | π* (C4–N5) | 23.52 | 0.41 | 0.088 |
| π (C8–C9) | π* (C7–C12) | 22.61 | 0.28 | 0.071 |
| LP (O20) | σ* (C13–C19) | 22.08 | 0.68 | 0.110 |
| π (C10–C11) | π* (C7–C12) | 21.81 | 0.27 | 0.068 |
| π (C10–C11) | π* (C8–C9) | 21.27 | 0.27 | 0.068 |
| π (C8–C9) | π* (C10–C11) | 19.93 | 0.29 | 0.068 |
| π (C7–C12) | π* (C10–C11) | 19.03 | 0.30 | 0.067 |
| π (C7–C12) | π* (C8–C9) | 18.72 | 0.29 | 0.065 |
| LP (Cl25) | π* (C8–C9) | 16.39 | 0.32 | 0.065 |
| LP (Cl29) | π* (C7–C12) | 15.96 | 0.33 | 0.065 |
Complementary π → π* delocalizations across the conjugated framework e.g., π(C13–C14) → π*(C19–O20) (∼25.1 kcal mol−1), π(C2–C3) → π*(C4–N5) (∼23.5 kcal mol−1), and π(C8–C9) → π*(C7–C12) (∼22.6 kcal mol−1) extend electron density over the aryl/pyrimidinone surface, explaining the π–σ/π–alkyl contacts with Leu156, Val39, Ala52, and Ile31 observed in the interaction map. Moderate σ → σ* hyperconjugations around C17–O21/N16–C17 (e.g. σ(C17–O21) → σ*(N16–C17) ∼28.7 kcal mol−1) further tune the bond polarisation and help maintain the partially planar conjugated geometry observed in the optimised structure.
This orbital-level rationale was also aligned with the MEP map (positive potential over N–H hydrogens and negative potential over C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and cyano regions), providing a coherent electronic basis for the observed binding mode of 2h.
O and cyano regions), providing a coherent electronic basis for the observed binding mode of 2h.
2.3 ADME analysis
Following the evaluation of the electronic structure and reactivity descriptors (FMO, MEP, and NBO), the pharmacokinetic profiles and drug-likeness of the synthesised scaffolds were assessed.The drug-likeness and pharmacokinetic profiles of the synthesised pyrano[2,3-d]pyrimidinone derivatives (2a–2h) were assessed using SwissADME and MolSoft web servers (Table 5). All compounds complied with Lipinski's rule of five, with no violations, and had molecular weights between 282–361 g mol−1, well below that of afatinib (487.9 g mol−1), indicating favourable oral bioavailability. The predicted bioavailability scores were consistently 0.55, which is consistent with the drug-like behaviour. The topological polar surface area (TPSA) values ranged from 124 to 149 Å2, balancing solubility and permeability. Compounds 2a, 2d, and 2f, with TPSA values of approximately 124 Å2, were well-suited for membrane transport, whereas 2c (148.6 Å2) showed the highest polarity and correspondingly lower gastrointestinal (GI) absorption. All other derivatives were predicted to have high GI absorption, and none was BBB-permeable, which is advantageous for anticancer therapy because it limits CNS side effects. Lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P = 1.1–1.5) suggests moderate hydrophobicity, which is favourable for passive diffusion, while solubility (log
P = 1.1–1.5) suggests moderate hydrophobicity, which is favourable for passive diffusion, while solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = −2.28 to −3.52) is acceptable and higher than that of afatinib (−4.29). The predicted skin permeability (log
S = −2.28 to −3.52) is acceptable and higher than that of afatinib (−4.29). The predicted skin permeability (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp = −7.1 to −7.9 cm s−1) indicated a uniform pharmacokinetic behaviour.
Kp = −7.1 to −7.9 cm s−1) indicated a uniform pharmacokinetic behaviour.
| Code | M.Wt | RB | HBA | HBD | TPSA (Å2) | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) po/wi log po/wi log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S (ESOL) S (ESOL) |
GI | BBB | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp cm s−1 Kp cm s−1 |
Violations | BAS | DLS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
a Parameter definitions: M.Wt, molecular weight; RB, rotatable bonds; HBA, number of hydrogen bond acceptors; HBD-number, hydrogen bond donors; TPSA – topological polar surface area; log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, octanol–water partition coefficient; log P, octanol–water partition coefficient; log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, solubility (ESOL); GI – gastrointestinal absorption; BBB, blood–brain barrier permeability; log S, solubility (ESOL); GI – gastrointestinal absorption; BBB, blood–brain barrier permeability; log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp, skin permeability; BAS, bioavailability score; DLS, drug-likeness score. Kp, skin permeability; BAS, bioavailability score; DLS, drug-likeness score. |
|||||||||||||
| 2a | 282.25 | 1 | 4 | 3 | 124.76 | 1.13 | −2.34 | High | No | −7.58 | 0 | 0.55 | −0.62 |
| 2b | 312.28 | 2 | 5 | 3 | 133.99 | 1.40 | −2.40 | High | No | −7.79 | 0 | 0.55 | −0.43 |
| 2c | 307.26 | 1 | 5 | 3 | 148.55 | 1.35 | −2.28 | Low | No | −7.93 | 0 | 0.55 | −0.67 |
| 2d | 316.70 | 1 | 4 | 3 | 124.76 | 1.40 | −2.93 | High | No | −7.34 | 0 | 0.55 | −0.20 |
| 2e | 361.15 | 1 | 4 | 3 | 124.76 | 1.50 | −3.24 | High | No | −7.57 | 0 | 0.55 | −0.59 |
| 2f | 296.28 | 1 | 4 | 3 | 124.76 | 1.39 | −2.63 | High | No | −7.41 | 0 | 0.55 | −0.53 |
| 2g | 300.24 | 1 | 5 | 3 | 124.76 | 1.22 | −2.49 | High | No | −7.62 | 0 | 0.55 | −0.39 |
| 2h | 351.14 | 1 | 4 | 3 | 124.76 | 1.47 | −3.52 | High | No | −7.11 | 0 | 0.55 | −0.58 |
| Afatinib | 487.95 | 9 | 6 | 3 | 87.22 | 3.95 | −4.29 | High | No | −7.30 | 0 | 0.55 | 1.12 |
Among the series, compound 2h stood out, combining a favourable molecular weight, optimal log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (1.47), balanced polarity (TPSA = 124.8 Å2), high GI absorption, and good solubility, making it a promising lead candidate.
P (1.47), balanced polarity (TPSA = 124.8 Å2), high GI absorption, and good solubility, making it a promising lead candidate.
2.4 Molecular docking analysis
Although ADME Profiling has demonstrated that the pharmacokinetics of the synthesised scaffolds are acceptable, the therapeutic relevance of these synthesised scaffolds will depend on the degree to which they can interact with the biological target. Molecular docking was used to predict the binding affinity and types of interactions of the synthesised scaffolds (2a–2h) within the active site of the protein (PDB ID: 4ZXT). The results of the docking studies, listed in Table 6, indicated that the binding affinities ranged from −6.69 to −7.58 kcal mol−1, whereas the reference drug afatinib exhibited a stronger affinity (−8.01 kcal mol−1). Among the synthesised compounds, 2g (−7.58 kcal mol−1) and 2h (−7.34 kcal mol−1) were identified as the two most potent scaffolds with binding affinities comparable to that of afatinib. The binding mode of compound 2h (Fig. 7) demonstrated the defined positioning of the ligand within the active site of 4ZXT.| Ligand | 2a | 2b | 2c | 2d | 2e | 2f | 2g | 2h | Afatiniba |
|---|---|---|---|---|---|---|---|---|---|
| a The IC50 value for afatinib was adapted from Yuanbiao Tu et al. Discovery of novel quinazoline derivatives bearing semicarbazones. Computational and Structural Biotechnology Journal 16 (2018) 462–478. | |||||||||
| Binding affinity (kcal mol−1) | −7.20 | −7.24 | −6.69 | −6.91 | −6.83 | −6.83 | −7.58 | −7.34 | −8.01 |
The key stabilising interactions between compound 2h and 4ZXT included conventional hydrogen bonds between the amide/amine hydrogens of compound 2h and ASP111 and ASP167, consistent with the electrostatic predictions from the MEP Analysis. Furthermore, the π–cation interactions of compound 2h with LYS114 and the π–sigma/π–alkyl interactions of compound 2h with VAL39, LEU156, and ALA52 also contributed to ligand anchoring. Together, these hydrogen bonding and hydrophobic interactions suggest a dual stabilisation mechanism, consistent with kinase-ligand binding models.
Moreover, NBO analysis of 2h predicted strong donor–acceptor delocalisation between the Amide N–H and heteroaryl π-systems, directly correlating with the observed hydrogen bonding to acidic residues (ASP111/ASP167). Overall, the docking results indicated that 2g and 2h exhibited balanced hydrophilic and hydrophobic interactions in the active site of the protein. These findings provide a robust foundation for subsequent MD simulations to probe dynamic stability and MTT cytotoxic assays to confirm the cellular activity. However, it is important to note that although afatinib is used as a reference standard in MTT assays, it primarily targets EGFR and is considered a negative/organic control for ERK2 kinase domain PDB ID: 4ZXT docking. The steric clashes and donor–donor penalties in the binding site support its off-target profile, indicating that the favourable binding observed for 2g and 2h is more representative of active site engagement.
2.5 Molecular dynamics (MD) simulation
Because docking can provide a very rigid view of how compounds bind, we further evaluated the 2h – 4ZXT complex using molecular dynamics (MD) simulations to determine whether the 2h – 4ZXT complex remains stable in a physiologically relevant environment. MD simulations were used in conjunction with docking analysis to assess the stability of the 2h – 4ZXT complex. Fig. 8A shows the RMSD traces for the 2h – 4ZXT complex. The RMSD traces showed that by the end of the first 10–15 ns of the simulation time, the protein backbone was stabilised within the 2.5–3.0 Å range, indicating a stable protein structure in the presence of 2h under physiological conditions. Furthermore, the ligand RMSD trace was very similar to that of the complex, indicating that during the course of the simulation, 2h did not undergo significant rotation away from the binding pocket or dissociate from the binding pocket.The RMSF traces (Fig. 8B) provide information about localised structural flexibility on a per-residue basis. Consistent with our expectations, the loop and terminal regions showed moderate levels of motion. However, the residues near the active site of the protein showed minimal movement, demonstrating that binding for 2h had a stabilising effect on the active site of the protein. SASA traces (Fig. 8C) fluctuated between 185–200 nm2, suggesting that no large-scale unfolding of the protein occurred. Additionally, the radius of gyration (Fig. 8D) remained at approximately 2.25–2.30 nm, indicating that the protein remained folded around compound 2h. These data collectively demonstrate that compound 2h not only favourably interacts with 4ZXT but also maintains the stability of the 2h – 4ZXT complex over long periods of simulation. Therefore, these studies, when combined with docking and DFT-based descriptors, demonstrated that compound 2h is a chemically robust lead compound that should be assessed for its free binding energy (MM/GBSA) and subsequently validated biologically.
It is also important to note that the persistence of these interactions at various points during the simulation indicated that compound 2h did not experience significant displacement during the course of the simulation, which is consistent with the RMSD and Rg data. Overall, the PLIF data indicate that compound 2h forms a dynamic balance between hydrogen bonding and hydrophobic contacts, which can account for the favourable docking affinity and MD-derived stability of compound 2h.
Additionally, FEL analysis (Fig. 10A), projected onto the first two principal components, demonstrated low-energy basins of motion for the complex, each separated by moderate barriers, and therefore indicated that the majority of the simulation time (i.e. 100 ns) of the complex was spent exploring the most favourable energetics. The fact that the FEL analysis identified specific, well defined minima corresponds to the previously observed stable RMSD and Rg values, and therefore indicates that the 2h maintains the protein in a very constrained conformational space with little to no unfolding of the protein. This conclusion is further supported by the observation of PCA clustering, which showed that the complex converges into a small conformational subspace over time. In addition, trajectory progression (Fig. 10C) clearly shows that the active site residues of 4ZXT are unable to move freely when bound for 2h, and therefore support the conclusions drawn from the SASA fluctuations. Overall, these additional analyses demonstrate that not only does compound 2h binds to 4ZXT with energetically favourable binding properties and holds the kinase in a restrained state of motion, which is a highly desirable property for selective inhibition.
2.6 In vitro cytotoxicity studies by MTT assay
Although computational studies have provided strong evidence for the stability, binding energetics, and pharmacokinetic potential of pyrano[2,3-d]pyrimidinone derivatives, it is essential to validate these predictions in a biological context. Therefore, the cytotoxic potential of the synthesised scaffolds was evaluated in vitro using the A549 lung carcinoma cell line. The cytotoxic effects of the scaffolds (2a–2h) were thoroughly assessed in the A549 human lung carcinoma cell line using theMTT assay. Afatinib served as a positive control in the cytotoxicity tests against A549 cells, enabling direct comparison of the growth-inhibitory effects of the synthesized derivatives within a biologically relevant cellular environment.The IC50 values summarised in Table 7 reveal a clear structure–activity relationship among the derivatives. Compound 2h exhibited the most potent inhibitory effect (IC50 = 39.29 µM), followed by 2d (47.91 µM), 2f (58.15 µM), and 2c (59.13 µM), indicating that halogen and pseudohalogen substitutions at the phenyl ring significantly enhanced antiproliferative activity. In contrast, the derivatives 2a (120.65 µM) and 2e (90.17 µM) displayed weaker activities. Afatinib was considered the reference drug and served two complementary roles in this study: as a biological positive control in MTT assays (EGFR-driven cytotoxicity in A549 cells) and as a docking comparator against the crystal structure 4ZXT, which corresponds to the ERK2 kinase domain. In this context, afatinib was used as a negative control to highlight the potential off-target interactions. This dual consideration allowed us to place the newly synthesised derivatives in perspective, comparing their anticancer activity with that of a clinically validated drug while also probing their putative engagement with the ERK2 signalling framework represented by 4ZXT.
| Code | 2a | 2b | 2c | 2d | 2e | 2f | 2g | 2h | Afatiniba |
|---|---|---|---|---|---|---|---|---|---|
| a The IC50 value of afatinib was adapted from the study by Tsai et al. Afatinib triggers a Ni2+-resistant Ca2+ influx pathway in A549 non-small cell lung cancer cells. Fundamental & Clinical Pharmacology 2022, 37, 253–262. | |||||||||
| IC50 (µM) | 120.65 | 89.09 | 59.13 | 47.91 | 90.17 | 58.15 | 69.97 | 39.29 | 1.4 |
The reference drug afatinib demonstrated superior potency (IC50 = 1.4 µM);29 however, compound 2h exhibited a reasonable cytotoxic profile within the micromolar range, highlighting its potential as a lead scaffold for further optimisation. Microscopic imaging of A549 cells treated for 2h (Fig. 11) provided direct morphological evidence of the cytotoxicity. Untreated control cells exhibited a dense, confluent monolayer with intact morphology, whereas the treated cells displayed progressive dose-dependent changes, including cell shrinkage, rounding, detachment, and loss of adherence at higher concentrations. These visual alterations correlated well with the quantitative viability assay, where cell survival sharply declined at concentrations above 20 µg mL−1, consistent with the IC50 value. Representative cell viability plots and microscopic triplicates for compounds 2a–2h are shown in Fig. S36.
Thus, the MTT assay confirmed that 2h exerted significant antiproliferative effects on A549 cells, with potency aligned with its favourable ADME profile, docking interactions within the active site, and dynamic stability observed in MD simulations. This integrative evidence strengthens the case of 2h as a promising candidate for further preclinical evaluation.
2.7 Structure–activity relationship (SAR) analysis
While docking scores provided the first indication of how well ligands would bind, regression analysis of the MTT cytotoxicity data for compounds 2a–2h in Fig. S35 indicated that there was no correlation between docking energy and cytotoxicity, indicating that substituent-dependent structure–activity relationships (SAR) and other pharmacokinetic properties are better indicators of biological responses than docking energy. The combination of theoretical insights from docking, MD simulations, MMGBSA, and free energy landscape calculations with experimental cytotoxicity data supports this view, where compound 2h was identified as the most promising compound based on both docking energy and cytotoxicity.Compound 2a exhibited very low activity (IC50 = 120.65 µM), establishing the importance of incorporating a phenyl ring into the scaffold to improve its bioactivity. Compouns 2b exhibited a moderately higher activity owing to the addition of an electron-donating methoxy group (IC50 = 89.09 µm). Addition of an electron-withdrawing cyano group to compound 2c resulted in increased polarity and only a moderate increase in cytotoxicity (IC50 = 59.13 µm). Among the halo-substituted analogues, the introduction of a chlorine atom (compound 2d) significantly increased cytotoxicity (IC50 = 47.91 µm); however, the introduction of a larger bromine atom (compound 2e) decreased cytotoxicity, which could be attributed to sterically hindered access to the active site of the protein. Compounds 2f (methoxy) and 2g (fluoro) exhibited moderate cytotoxicity (IC50 = 58.15 and 65.97 µm, respectively). The high level of cytotoxicity observed for compound 2h (IC = 39.29 µm) can be explained by the optimal hydrophobic packing and steric complementarity afforded by the two chlorine atoms present at the ortho position, validating its identification as the preferred lead scaffold.
3 Conclusions
In this study, a green and efficient methodology was developed for the one-pot synthesis of pyrano[2,3-d]pyrimidinone derivatives (2a–2h) using silver tungstate (Ag2WO4) nanoparticles as a heterogeneous catalyst. This approach successfully combined synthetic innovation with computational and biological validation to explore the anticancer potential of the resulting scaffolds in the A549 lung carcinoma cell line. Future studies will extend the cytotoxic evaluation of the synthesized pyrano[2,3-d]pyrimidinones to additional cancer cell lines in order to further validate their anticancer potential and broaden the biological relevance of the present findings.Silver tungstate nanoparticles exhibit a very high catalytic performance and allow for fast and selective transformations at low temperatures and under non-toxic conditions. Compound 2h demonstrated the highest cytotoxic activity among the synthesised compounds, with an IC50 of 39.29 µM, which was significantly higher than that of unsubstituted parent scaffold 2a (IC50 = 120.65). Complementary in silico analyses, including DFT, molecular docking, MD simulations, and ADME screening, provided insights into the structural stability, interactions with biological targets, and pharmacokinetics of the synthesised compounds, thereby revealing the relationship between the predicted theoretical data and experimental results.
This study is the first to report the application of Ag2WO4 nanoparticles as nanocatalysts for the construction of pyrano[2,3-d]pyrimidinone frameworks, establishing a sustainable route for generating bioactive heterocyclic scaffolds with promising anticancer profiles. Although the compounds described herein are foundational scaffolds, further structural modification could improve the selectivity, potency, and drug-like properties to enable the synthesis of clinically viable anticancer drugs derived from the green synthetic platform used in this study.
4 Materials and methods
4.1 Preparation of Ag2WO4 catalyst
Stoichiometric aqueous solutions of sodium tungstate dihydrate (Na2WO4·2H2O, 5 mmol) and silver nitrate (AgNO3, 10 mmol) were prepared. The silver nitrate solution was slowly introduced into the sodium tungstate solution with continuous stirring until complete precipitation occurred. The mixture was then placed in a 100 mL Teflon-lined stainless-steel autoclave and subjected to hydrothermal processing at 180 °C for 16 h. After natural cooling to room temperature, the precipitate was separated by centrifugation, thoroughly rinsed with deionised water and ethanol, and dried at 60 °C overnight.4.2 General procedure for the synthesis of pyrano[2,3-d]pyrimidine-6 carbonitrile
A 100 mL round-bottom flask was loaded with malononitrile (1.1 mmol), aldehyde (1.0 mmol), barbituric acid (1.0 mmol), and Ag2WO4 (2.5 mol%) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ethanol–water mixture. The mixture was stirred at 70 °C for the required duration, and its progress was monitored using thin-layer chromatography (TLC) with ethyl acetate/petroleum ether (3
1 ethanol–water mixture. The mixture was stirred at 70 °C for the required duration, and its progress was monitored using thin-layer chromatography (TLC) with ethyl acetate/petroleum ether (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) as the mobile phase. Once the reaction was complete, the mixture was allowed to cool to room temperature, and the precipitate was separated by vacuum filtration. The crude solid was dissolved in ethanol with a few drops of DMSO to enhance the solubility and filtered again to eliminate any remaining Ag2WO4 catalyst. The resulting filtrate was poured into cold water to re-precipitate the product, which was collected and further purified by recrystallisation from ethanol to yield pure pyrano[2,3-d]pyrimidinone derivatives (2a–2h). The structures of the obtained compounds were characterised using FT-IR, 1H NMR, and 13C NMR spectroscopy.
1) as the mobile phase. Once the reaction was complete, the mixture was allowed to cool to room temperature, and the precipitate was separated by vacuum filtration. The crude solid was dissolved in ethanol with a few drops of DMSO to enhance the solubility and filtered again to eliminate any remaining Ag2WO4 catalyst. The resulting filtrate was poured into cold water to re-precipitate the product, which was collected and further purified by recrystallisation from ethanol to yield pure pyrano[2,3-d]pyrimidinone derivatives (2a–2h). The structures of the obtained compounds were characterised using FT-IR, 1H NMR, and 13C NMR spectroscopy.
4.3 Optimization of reaction
To identify the most suitable eco-friendly solvent system and catalyst loading, a model multicomponent reaction (Scheme 1) was performed using various green solvents and temperature in the presence of Ag2WO4. The optimisation results are listed in Table 8.| Entry | Ag2WO4 (mol%) | Solvent/temperaturea (°C) | Time (min) | Yieldb (%) |
|---|---|---|---|---|
| a Reaction conditions: barbituric acid (1.0 mmol), benzaldehyde (1.0 mmol), and malononitrile (1.1 mmol) were combined with different solvents and catalyst amounts at different temperatures.b Isolated yields. | ||||
| 1 | 2.5 | H2O/RT | 30 | 71 |
| 2 | 2.5 | EtOH/RT | 30 | 75 |
| 3 | 2.5 | EtOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)/RT 1)/RT |
5–8 | 80 |
| 4 | 2.5 | H2O/100 | 15 | 78 |
| 5 | 2.5 | EtOH/80 | 10 | 84 |
| 6 | 2.5 | EtOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)/70 1)/70 |
5–8 | 95 |
| 7 | 2.5 | PEG-400/RT | 30 | 78 |
| 8 | 2.5 | PEG-400/100 | 10 | 81 |
| 9 | 5.0 | EtOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)/70 1)/70 |
5–8 | 95 |
Reactions performed in pure aqueous medium at room temperature afforded only a moderate yield (71% in 30 min), whereas ethanol under identical conditions resulted a marginal improvement (yield = 75%). In contrast, a mixed protic solvent system (EtOH–H2O, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) at 70 °C was highly efficient, delivering the target product within 5–8 min in excellent yield (95%). This remarkable enhancement can be attributed to the synergistic hydrogen-bonding environment of the binary solvent, in which the high surface energy of water promotes proton transfer and facilitates rapid product precipitation, thereby accelerating reaction completion. To evaluate the influence of catalyst loading, the amount of Ag2WO4 was increased from 2.5 mol% to 5.0 mol% in the EtOH–H2O (1
1 v/v) at 70 °C was highly efficient, delivering the target product within 5–8 min in excellent yield (95%). This remarkable enhancement can be attributed to the synergistic hydrogen-bonding environment of the binary solvent, in which the high surface energy of water promotes proton transfer and facilitates rapid product precipitation, thereby accelerating reaction completion. To evaluate the influence of catalyst loading, the amount of Ag2WO4 was increased from 2.5 mol% to 5.0 mol% in the EtOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) system. However, no improvement in yield was observed (remaining at 95%), confirming that 2.5 mol% Ag2WO4 was sufficient to efficiently promote the transformation.
1) system. However, no improvement in yield was observed (remaining at 95%), confirming that 2.5 mol% Ag2WO4 was sufficient to efficiently promote the transformation.
Thus, EtOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 70 °C with 2.5 mol% Ag2WO4 was established as the optimal condition, affording the desired products in excellent yields within a few minutes (Table 8). These findings underscore the critical role of binary protic solvents in enhancing the reactivity and highlight the sustainability advantages of this protocol, including minimal catalyst usage, reduced energy demand, and excellent efficiency.
1) at 70 °C with 2.5 mol% Ag2WO4 was established as the optimal condition, affording the desired products in excellent yields within a few minutes (Table 8). These findings underscore the critical role of binary protic solvents in enhancing the reactivity and highlight the sustainability advantages of this protocol, including minimal catalyst usage, reduced energy demand, and excellent efficiency.
4.4 General experimental procedures
The synthesised pyrano[2,3-d]pyrimidine scaffolds were characterised using spectroscopic techniques. Infrared spectra were obtained using a PerkinElmer Spectrum Two FTIR spectrometer equipped with an attenuated total reflectance (ATR) accessory that included a diamond crystal and a deuterated triglycine sulfate (DTGS) detector. The instrument was operated within the range of 4000–400 cm−1, with four scans per sample and a resolution of 4 cm−1. The spectra were collected in ATR mode without any additional sample preparation. 1H and 13C NMR spectra were recorded in DMSO-d6 using a JEOL 600 MHz spectrometer.4.5 Spectral results
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1683 & 1634 (C
N), 1683 & 1634 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1094 (C–O–C); HRMS (ESI) m/z [M + Na]+ calcd for C14H10N4O3Na+:305.0645, found: 305.0641.
O), 1094 (C–O–C); HRMS (ESI) m/z [M + Na]+ calcd for C14H10N4O3Na+:305.0645, found: 305.0641.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1716 &1674 (C
N), 1716 &1674 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C15H13N4O4+:313.0931, found: 313.0933.
O), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C15H13N4O4+:313.0931, found: 313.0933.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1719 &1670 (C
N), 1719 &1670 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1100 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C15H10N5O3+:308.0778, found: 308.0771.
O), 1100 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C15H10N5O3+:308.0778, found: 308.0771.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1717 &1673 (C
N), 1717 &1673 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H10ClN4O3+:317.0436, found: 317.0430.
O), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H10ClN4O3+:317.0436, found: 317.0430.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1717 &1673 (C
N), 1717 &1673 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1098 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H10BrN4O3+:360.9931, found: 360.9933.
O), 1098 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H10BrN4O3+:360.9931, found: 360.9933.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1717 &1673 (C
N), 1717 &1673 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C15H13N4O3+:297.0982, found: 297.0985.
O), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C15H13N4O3+:297.0982, found: 297.0985.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1721 &1675 (C
N), 1721 &1675 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1099 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H10FN4O3+:301.0731, found: 307.0726.
O), 1099 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H10FN4O3+:301.0731, found: 307.0726.![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N), 1717 &1673 (C
N), 1717 &1673 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O str), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H9Cl2N4O3+:349.9973 found: 349.9971.
O str), 1096 (C–O–C); HRMS (ESI) m/z [M + H]+ calcd for C14H9Cl2N4O3+:349.9973 found: 349.9971.4.6 Computational studies
All ligand structures were first geometry-optimised using ORCA 6.0.1 (B3LYP/def2-TZVP). The optimised geometries were converted into 3D structures and further processed for docking by assigning Gasteiger charges and defining rotatable bonds using Open Babel,48 RDKit,49 and Meeko50 in a Google Colab environment. The final ligand files were prepared in PDBQT format.
Docking simulations were performed using AutoDock Vina.51 A grid box was defined to encompass the active site of 4ZXT, ensuring coverage of all key residues involved in ligand binding. Each ligand was docked independently, and the lowest-energy binding pose was selected based on the binding affinity and favourable interaction profiles. The resulting protein-ligand complexes were analysed using Discovery Studio Visualiser52 and UCSF ChimeraX53 to examine the binding orientation, hydrophobic contacts, and hydrogen bonding within the active site.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps or until a tolerance of 1000 kJ mol−1 nm−1 was achieved. This was followed by position-restrained equilibration under the NVT and NPT ensembles at 298 K and 1 atm pressure, utilising the V-rescale thermostat and Parrinello-Rahman barostat, respectively.57,58 Post-simulation analyses, were primarily performed using the built-in GROMACS utilities, whereas the processed data were further visualised and plotted using Python 3 libraries. Principal Component Analysis (PCA), FEL, and MM/GBSA analyses were performed on the 100 ns MD trajectories to evaluate the essential dynamics, conformational free-energy basins, and binding free energies of the 4ZXT–ligand complexes. The free energy landscape (FEL) and principal component analysis (PCA) were constructed from covariance matrices of the backbone atoms, whereas MM/GBSA calculations were conducted using gmx_MMPBSA to estimate the van der Waals, electrostatic, and solvation energy contributions.
000 steps or until a tolerance of 1000 kJ mol−1 nm−1 was achieved. This was followed by position-restrained equilibration under the NVT and NPT ensembles at 298 K and 1 atm pressure, utilising the V-rescale thermostat and Parrinello-Rahman barostat, respectively.57,58 Post-simulation analyses, were primarily performed using the built-in GROMACS utilities, whereas the processed data were further visualised and plotted using Python 3 libraries. Principal Component Analysis (PCA), FEL, and MM/GBSA analyses were performed on the 100 ns MD trajectories to evaluate the essential dynamics, conformational free-energy basins, and binding free energies of the 4ZXT–ligand complexes. The free energy landscape (FEL) and principal component analysis (PCA) were constructed from covariance matrices of the backbone atoms, whereas MM/GBSA calculations were conducted using gmx_MMPBSA to estimate the van der Waals, electrostatic, and solvation energy contributions.4.7 MTT assay
Author contributions
All authors contributed significantly to the design, execution, and interpretation of experiments. Sathiaseelan Perumal: writing – original draft, methodology, investigation, and data curation. Perumal Muthuraja: resources, writing – review & editing M. Sasikumar: writing – review & editing. R. Hari Krishna: investigation Manisankar Paramasivam: project administration and resources, conceptualization. Viswanathan Subramanian: project administration and resources, writing – review & editing, and supervision.Conflicts of interest
There are no conflicts of interest to declare.Data availability
All data generated or analysed during this study are included in this published article and its supporting information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra08635c.Acknowledgements
PS would like to thank Bishop Heber College for providing the Management Seed funding (F. No. MRP/1022/2021).References
- T. E. Ali, D. A. Bakhotmah and M. A. Assiri, Synth. Commun., 2020, 50, 3314–3325 Search PubMed.
- N. E. Abd El-Sattar, K. El-Adl, M. A. El-Hashash, S. A. Salama and M. M. Elhady, Bioorganic Chem., 2021, 115, 105186 Search PubMed.
- N. P. Veeranna, Y. D. Bodke, M. Basavaraju and P. Krishnamurthy, Nucleosides Nucleotides Nucleic Acids, 2023, 42, 281–295 Search PubMed.
- A. Kumar, K. K. Bhagat, A. K. Singh, H. Singh, T. Angre, A. Verma, H. Khalilullah, M. Jaremko, A.-H. Emwas and P. Kumar, RSC Adv., 2023, 13, 6872–6908 RSC.
- P. Seboletswe, P. Awolade and P. Singh, ChemMedChem, 2021, 16, 2050–2067 CrossRef CAS PubMed.
- R. Karimi-Chayjani, N. Daneshvar, H. Tajik and F. Shirini, ChemistrySelect, 2019, 4, 1205–1213 Search PubMed.
- S. Azad Poshtmakhi, K. Rad-Moghadam, H. Hosseinzadeh and S. Toorchi-Roudsari, ChemistrySelect, 2023, 8, e202300533 CrossRef CAS.
- K. B. Badiger and K. Kamanna, Polycycl. Aromat. Compd., 2023, 43, 5976–5995 Search PubMed.
- N. Seyyedi, F. Shirini and M. S. N. Langarudi, RSC Adv., 2016, 6, 44630–44640 RSC.
- P. K. Paliwal, S. R. Jetti and S. Jain, Med. Chem. Res., 2013, 22, 2984–2990 Search PubMed.
- A. Maleki, A. A. Jafari and S. Yousefi, Carbohydr. Polym., 2017, 175, 409–416 Search PubMed.
- M. A. Zolfigol, R. Ayazi-Nasrabadi and S. Baghery, Appl. Organomet. Chem., 2016, 30, 273–281 CrossRef CAS.
- S. Momeni and R. Ghorbani-Vaghei, ACS Omega, 2024, 9, 10332–10342 CrossRef CAS.
- A. R. Nesaragi, T. Gasti, T. V. Metre, A. Anand, R. R. Kamble, R. B. Chougale and R. S. Keri, ChemistrySelect, 2022, 7, e202200604 CrossRef CAS.
- F. Mohamadpour, Curr. Res. Green Sustain. Chem., 2025, 10, 100444 CrossRef CAS.
- X. Shen, G. Hong and L. Wang, Org. Biomol. Chem., 2025, 23, 2059–2078 RSC.
- M. Tandi, V. Sharma, B. Gopal and S. Sundriyal, RSC Adv., 2025, 15, 1447–1489 Search PubMed.
- S. H. Gebre, Appl. Nanosci., 2023, 13, 15–63 CrossRef CAS.
- R. P. Patil, V. A. Kalantre and K. N. Alasundkar, Res. Chem. Intermed., 2023, 49, 5163–5203 CrossRef CAS.
- S. Zinatloo-Ajabshir, M. Baladi, O. Amiri and M. Salavati-Niasari, Sep. Purif. Technol., 2020, 248, 117062 CrossRef CAS.
- A. A. Almehizia, M. A. Al-Omar, A. M. Naglah, H. A. Ghabbour, A. S. Hassan and T. K. Khatab, Pol. J. Chem. Technol., 2024, 26, 1–7 CrossRef CAS.
- Q.-W. Song, B. Yu, X.-D. Li, R. Ma, Z.-F. Diao, R.-G. Li, W. Li and L.-N. He, Green Chem., 2014, 16, 1633–1638 RSC.
- P. Chunarkar-Patil, M. Kaleem, R. Mishra, S. Ray, A. Ahmad, D. Verma, S. Bhayye, R. Dubey, H. N. Singh and S. Kumar, Biomedicines, 2024, 12, 201 CrossRef CAS.
- S. Thanigachalam and M. Pathak, RSC Adv., 2024, 14, 13062–13082 RSC.
- K. Yonesaka, K. Kudo, S. Nishida, T. Takahama, T. Iwasa, T. Yoshida, K. Tanaka, M. Takeda, H. Kaneda, I. Okamoto, K. Nishio and K. Nakagawa, Oncotarget, 2015, 6, 33602–33611 CrossRef PubMed.
- M. S. Valdés-Tresanco, M. E. Valdés-Tresanco, P. A. Valiente and E. Moreno, J. Chem. Theory Comput., 2021, 17, 6281–6291 CrossRef PubMed.
- A. Amadei, A. B. Linssen and H. J. Berendsen, Proteins Struct. Funct. Bioinforma., 1993, 17, 412–425 Search PubMed.
- P. Sfriso, A. Emperador, L. Orellana, A. Hospital, J. L. Gelpí and M. Orozco, J. Chem. Theory Comput., 2012, 8, 4707–4718 Search PubMed.
- T.-Y. Tsai, C.-Y. Chen, L.-R. Shiao, T.-T. Ou, C.-H. Wu, Y.-M. Leung and L. W. C. Chow, Fundam. Clin. Pharmacol., 2022, 37, 253–262 CrossRef.
- M. D. Hanwell, D. E. Curtis, D. C. Lonie, T. Vandermeersch, E. Zurek and G. R. Hutchison, J. Cheminformatics, 2012, 4, 17 Search PubMed.
- C. Bannwarth, S. Ehlert and S. Grimme, J. Chem. Theory Comput., 2019, 15, 1652–1671 CrossRef CAS.
- F. Neese, WIRES Comput Molec Sci, 2012, 2, 73–78 CrossRef CAS.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 Search PubMed.
- B. Helmich-Paris, B. de Souza, F. Neese and R. Izsák, J. Chem. Phys., 2021, 155, 104109 Search PubMed.
- F. Neese, J Comp Chem, 2003, 24, 1740–1747 CrossRef CAS.
- F. Neese, F. Wennmohs, A. Hansen and U. Becker, Chem. Phys., 2009, 356, 98–109 CrossRef CAS.
- M. Franz, F. Neese and S. Richert, Chem. Sci., 2022, 13, 12358–12366 RSC.
- M. Minteguiaga, E. Dellacassa, M. A. Iramain, C. A. N. Catalán and S. A. Brandán, J. Mol. Struct., 2019, 1177, 499–510 CrossRef CAS.
- I. A. E. Hassani, S. A. Brandán, S. Mortada, S. Arshad, E. Romano, Y. Ramli, J. T. Mague, M. E. A. Faouzi, K. Karrouchi and M. Ansar, J. Mol. Struct., 2024, 1295, 136620 CrossRef CAS.
- T. Tsuneda, J.-W. Song, S. Suzuki and K. Hirao, J. Chem. Phys., 2010, 133, 174101 CrossRef.
- E. Glendening, K. Badenhoop, A. Reed, J. Carpenter, J. Bohmann, C. Morales, P. Karafiloglou, C. Landis and F. Weinhold, J. Comput. Chem., 2012, 33, 580–592 CrossRef PubMed.
- R. Izsák, F. Neese and W. Klopper, J. Chem. Phys., 2013, 139, 094111 Search PubMed.
- F. Neese, Chem. Phys. Lett., 2000, 325, 93–98 Search PubMed.
- G. A. Zhurko and D. A. Zhurko, Acad, Version, 2005, 1, (Academic license) Search PubMed.
- S. H. Shin, M.-H. Lee, M. Malakhova, I. Kurinov, Q. Wu, J. Xu, Y. Jiang, Z. Dong, K. Liu and K. Y. Lee, Oncotarget, 2016, 7, 35001 CrossRef PubMed.
- P. W. Rose, B. Beran, C. Bi, W. F. Bluhm, D. Dimitropoulos, D. S. Goodsell, A. Prlić, M. Quesada, G. B. Quinn and J. D. Westbrook, Nucleic Acids Res., 2010, 39, D392–D401 CrossRef.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, J. Cheminformatics, 2011, 3, 1–14 CrossRef.
- RDKit: Open-source cheminformatics. https://www.rdkit.org, Accessed July 2025.
- Meeko, https://github.com/forlilab/meeko, Accessed July 2025.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS.
- BIOVIA, Dassault Systèmes, Discovery Studio Visualizer.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, E. C. Meng, G. S. Couch, T. I. Croll, J. H. Morris and T. E. Ferrin, Protein Sci., 2021, 30, 70–82 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1, 19–25 CrossRef.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. De Groot, H. Grubmüller and A. D. MacKerell Jr, Nat. Methods, 2017, 14, 71–73 CrossRef CAS.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 Search PubMed.
- G. Bussi, T. Zykova-Timan and M. Parrinello, J. Chem. Phys., 2009, 130, 074101 Search PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- MolSoft drug-likeness calculator, https://molsoft.com/mprop/.
- R. I. Freshney, Man. Basic Tech., Wiley, 1987 Search PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |