Open Access Article
Open Access ArticleDevelopment of substituted 2-(4-(sulfonyl)piperazin-1-yl)quinazoline molecular hybrids as a new class of antimalarials
Hari Madhav†
 ab,
Ashutosh Panda†b,
Dipanwita Dattab,
Kashfa Shaheena,
Abdur Rahmanc,
Azhar Tariq Khana,
Souvik Bhattacharjeec,
Dhiraj Kumarb,
Pawan Malhotra*b and
Nasimul Hoda
ab,
Ashutosh Panda†b,
Dipanwita Dattab,
Kashfa Shaheena,
Abdur Rahmanc,
Azhar Tariq Khana,
Souvik Bhattacharjeec,
Dhiraj Kumarb,
Pawan Malhotra*b and
Nasimul Hoda *a
*a
aDrug Design and Synthesis Laboratory, Department of Chemistry, Jamia Millia Islamia, New Delhi-110025, India. E-mail: nhoda@jmi.ac.in
bInternational Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi-110067, India. E-mail: pawanmal@gmail.com; pawanm@icgeb.res.in
cSpecial Centre for Molecular Medicine, Jawaharlal Nehru University, New Delhi-110067, India
First published on 2nd January 2026
Abstract
The rapid emergence of drug resistance makes malaria elimination a global challenge despite the prevalence of artemisinin-based combination therapies (ACTs), thus highlighting the urgent need for the development of new antimalarials with novel modes of action. The present study aimed to develop new quinazoline hybrid antimalarials using bioactive small building blocks. The antimalarial activity results revealed that most molecular hybrids have IC50 values below 10 µM for the drug-sensitive Pf3D7 strain. The study identified molecular hybrids 19, N-(2-chloro-4-((4-(4-(((tetrahydrofuran-2-yl)methyl)amino)quinazolin-2-yl)piperazin-1-yl)sulfonyl)phenyl)acetamide and 27, 2-(4-((2-nitrophenyl)sulfonyl)piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline as potent antimalarials with an IC50 value of 3.4 µM and 2.9 µM against Pf3D7, respectively. The cytotoxicity investigation against mammalian A549 cells and activated macrophages derived from THP1 monocytes revealed that the compounds were relatively non-cytotoxic, and their antimalarial activity was not associated with cytotoxicity. In silico studies were conducted to predict plausible drug targets of the compounds, and the results suggested that the antimalarial activity of the compounds may be due to the inhibition of zinc metalloprotease PfFLN, with concurrent inhibition of cysteine proteases PfFP2 and PfFP3. The MM-GBSA analysis revealed that the binding free energies of 19 and 27 with PfFLN were −50.3223 and −51.5066 kcal mol−1, respectively. The predicted ADME properties of the compounds fall within the Schrödinger range, which encompasses 95% of all known medications. The study thus emphasised the significance of the molecular hybridisation approach and highlighted compounds 19 and 27 as potent hit molecules that could be further optimised for the development of new antimalarials.
1. Introduction
Malaria is a life-threatening illness that contributed to 597![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities worldwide in 2023, with an increase of 10% over the previous three years.1 Despite the artemisinin-based combination therapy's (ACTs) effectiveness, the rapid emergence of drug resistance to its constituent parts poses a serious challenge to the management and eradication of malaria.2 Although there are new antimalarial medications in the clinical pipeline, it won't be long before they're widely accessible.3 A clinical trial (NCT 04546633) disclosed that one of the candidate drugs, KAF-156, was markedly potent against artemisinin-resistant parasites in adults and is currently being investigated in Phase 2 studies in children when combined with the partner medication lumefantrine.4 Likewise, to challenge artemisinin resistance, the use of three ACTs was also investigated and found effective in clinical trials.5,6 Consequently, there is an urgent need for the development of new antimalarials with a novel mode of action to address the drug resistance towards mainline therapy ACTs.
000 fatalities worldwide in 2023, with an increase of 10% over the previous three years.1 Despite the artemisinin-based combination therapy's (ACTs) effectiveness, the rapid emergence of drug resistance to its constituent parts poses a serious challenge to the management and eradication of malaria.2 Although there are new antimalarial medications in the clinical pipeline, it won't be long before they're widely accessible.3 A clinical trial (NCT 04546633) disclosed that one of the candidate drugs, KAF-156, was markedly potent against artemisinin-resistant parasites in adults and is currently being investigated in Phase 2 studies in children when combined with the partner medication lumefantrine.4 Likewise, to challenge artemisinin resistance, the use of three ACTs was also investigated and found effective in clinical trials.5,6 Consequently, there is an urgent need for the development of new antimalarials with a novel mode of action to address the drug resistance towards mainline therapy ACTs.
A number of versatile scaffolds have been identified in medicinal chemistry that exhibit varied pharmacological activities against multiple pathogens. Quinazoline is one such scaffold that has been shown to be active against malaria,7 bacterial infections,8,9 cancer,10 hypertension and viral infections,11 and tuberculosis.12 For antimalarial development, Gilson, et al.13 disclosed 2-anilino 4-amino substituted quinazolines in nanomolar activity against Pf3D7, PfW2, and potent oral in vivo activity. A study by Bouchut, et al.14 highlighted in vitro and in vivo antimalarial potential of quinazolines as DNA methyltransferase (DNMT3a) inhibitors. Similarly, Mizukawa, et al.11 disclosed the nanomolar antimalarial 6,7-dimethoxyquinazoline-2,4-diamines up to 6 nM against P. falciparum malaria. Artemisinin-quinazoline derivatives were also shown to be effective antimalarials by the study of Fröhlich, et al.15
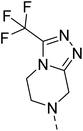
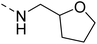
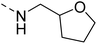
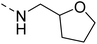
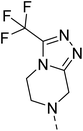
Likewise, the sulphonamide functional group is frequently explored in medicinal chemistry, which has led to the development of efficient antibacterial, antiviral, and antifungal agents.16 The anticancer,17 antidepressant,18 antimalarials,19 antiproliferative,20 and antitubercular21 potential of sulphonamides was also accounted for. Recently, Pingaew, et al.22 explored indole-sulphonamide hybrids as dual-acting agents for treating cancer and malaria. Similarly, Karpina, et al.23 reported a series of 1,2,4 triazolo[4,3-a]pyridine sulfonamides as effective antimalarials and proposed them as inhibitors of PfFP2. Furthermore, the moiety 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine is highlighted as an important pharmacophore of Sitagliptin 24, a marketed drug for the treatment of type-II diabetes. The 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine was used in developing inhibitors of bromodomain-containing protein 4 (BRD-4),25,26 poly (ADP-ribose) polymerase-1 (PARP1), and anaplastic lymphoma kinase (ALK)27 inhibitors for anti-cancer and cellular antiproliferative activity. Similarly, this group has also been reported as an important building block for developing antimicrobial28 and potential anticonvulsant agents.29 Tetrahydrofurfurylamine has proven to be a versatile and valuable building block in the design of new therapeutic agents. Shan, et al.30 developed tetrahydrofurfurylamino derivatives of geldanamycin that were able to block hepatitis C virus (HCV) replication. Lawrence, et al.31 used the same building block to develop new anticancer molecules that inhibit the ACK1 enzyme. Elbadawi, et al.32 designed tetrahydrofurfurylamine-hybridised 2-arylquinolines that target both EGFR and FAK, offering another promising approach to cancer treatment. Similarly, Balam et al.33 developed compounds combining tetrahydrofurfurylamine with N-(3-picolyl) benzoxazaphosphinamides, showing potential for treating metabolic disorders such as heart disease, obesity, and diabetes by inhibiting the 11β-HSD1 enzyme. Collectively, these studies highlight the significant role of the tetrahydrofurfurylamine motif in tuning biological activity and enabling the development of therapeutically relevant compounds.
The polypharmacological properties of quinazolines, sulphonamides, tetrahydrofurfurylamine, and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine thus strongly highlight the biological importance of these small building blocks. Here, we designed a series of hybrid molecules using the above-mentioned biologically active building blocks and studied their antimalarial potential. The study thus advocates designing unique trifunctional hybrids through integration for the treatment of malaria and investigating their potential for the development of distinct antimalarials.
1.1. Design strategy and hypothesis
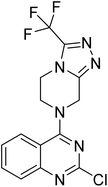
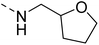
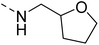
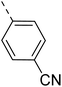
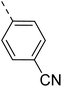
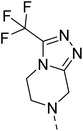
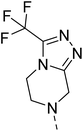
The molecular hybridisation approach was employed to design proposed quinazoline-sulphonamide hybrids by combining three biologically important small building blocks: quinazoline as a core scaffold, substituted triazolo[4,3-a]pyrazine or tetrahydrofurfurylamine at the 4th position of quinazoline, and substituted sulphonamides at the 2nd position of the quinazoline. The significance of quinazolines for the development of new antimalarials could be underlined by the study of Gilson, et al.13 which disclosed that substitution at the 2nd and 4th positions of quinazolines plays a substantial role in tuning the antimalarial efficacy of the compounds. The compound A, 4-((4-(((tetrahydrofuran-2-yl)methyl)amino)quinazolin-2-yl)amino)benzamide of the study highlighted that the tetrahydrofurfuryl amine at the 4th position of the quinazoline ring was fruitful to afford potent antimalarials with an EC50 value of 0.691 µM against the Pf3D7 strain, Fig. 1. | ||
| Fig. 1 Design of new tetrahydrofurfurylamine/triazolo[4,3-a]pyrazine-quinazoline-sulphonamide hybrids. | ||
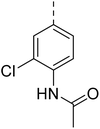
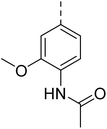
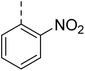
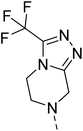
Similarly, Bouchut, et al.14 reported molecule B, N-(1-benzylpiperidin-4-yl)-2-(4-phenylpiperazin-1-yl)quinazolin-4-amine and underlined the significance of the quinazoline core for the development of new antimalarials. Moreover, our recent study revealed compound C, ethyl 7-methoxy-4-(((tetrahydrofuran-2-yl)methyl)amino)quinoline-3-carboxylate, which utilised tetrahydrofurfurylamine, displayed significant efficacy against the Pf3D7 strain with an IC50 value of 3.96 µM.34 Furthermore, the substituted triazolo[4,3-a]pyrazine was also explored for the development of disease-modifying small molecules, such as Hu, et al.35 reported compound D, (S)–N-(3-(1H-indol-3-yl)-1-oxo-1-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)propan-2-yl)benzenesulfonamide with an MIC of 32 and 16 µg ml−1 against S. aureus and E. coli, which highlighted the antibacterial potential of triazolo[4,3-a]pyrazine and aryl sulphonamide hybrids. Similarly, Raveesha, et al.36 developed triazolo[4,3-a]pyrazine analogues and highlighted compound F, N-(2-fluoro-4-(trifluoromethyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazine-7(8H)-carboxamide as promising anticancer agents with an IC50 of 8.18 and 15.23 µM against human colon cancer cell lines HT-29, and HCT-116, respectively. Hence, this building block was also utilised at the 4th position of the quinazoline core to explore its potential application in the identification of new antimalarials. Like compound D, the significance of sulphonamides was accounted for in several studies for the development of new antimalarial agents. Ezugwu, et al.19 reported compound E, (2S)-4-methyl-N-(3-methyl-1-oxo-1-(phenylamino)butan-2-yl)-2-((4-nitrophenyl) sulfonamido)pentanamide as a sulphonamide-based dipeptide antimalarial with 64.7% inhibition of parasitaemia in P. berghei-infected mice.19 As a result, sulphonamides' variable structure makes them great candidates for the development of new multi-target therapeutic molecules in the developing discipline of polypharmacology.
The emerging resistance to current mainline antimalarial drugs underscores the need for the development of new antimalarials with a novel mechanism of action. To combat multidrug-resistant parasite strains, the concept of hybridisation of two or more active pharmacophores into a single chemical entity displayed potential advantages over artemisinin-based combination therapy (ACT).37 Therefore, keeping in mind the antimalarial potential of quinazoline,13,14 tetrahydrofurfurylamine,13,34 triazolo[4,3-a]pyrazine,38 and sulphonamides19,23 through different drug targets, the current idea was conceived to design a new hybrid antimalarial for the treatment of malaria. Combining them via a molecular hybridisation strategy at the quinazoline's C2 and C4 positions was expected to afford enhanced antimalarial activity through synergistic polypharmacology with a probable novel mechanism of action. This scaffold fills gaps in the antimalarial chemical space by introducing a trifunctional hybrid that has not been observed in literature.
Efforts were made to establish the structure–activity relationship (SAR) for antimalarial efficacy. The hybrids were screened against drug-sensitive and artemisinin-resistant strains of P. falciparum. The cytotoxicity of the hybrids was assessed against lung cancer A549 cells and activated macrophages from THP1 monocytes. Additionally, the physicochemical characteristics and drug-likenepolyss of the synthesised hybrids were also predicted.
2. Results and discussion
2.1. Chemistry
The multi-step synthetic protocol was followed to achieve target molecular hybrids as displayed in Schemes 1–6. | ||
| Scheme 1 Synthesis of substituted 4-acetamidobenzene-1-sulfonyl chlorides 5 (a–b). Reagents and conditions: (i) Et3N, DCM, 0 °C – RT, 1.5 h; (ii) 0–60 °C, 3 h. | ||
 | ||
| Scheme 2 Synthesis of 3-acetylbenzenesulfonyl chloride 7. Reagents and conditions: (i) 0–90 °C, 12 h. | ||
 | ||
| Scheme 3 Synthesis of 2,4-dichloroquinazoline 11. Reagents and conditions: (i) 180–210 °C, 3 h; (ii) POCl3, DIPEA, RT – 90 °C, 6 h. | ||
 | ||
| Scheme 4 Synthesis of substituted N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine 14 (a–b). Reagents and conditions: (i) Et3N, THF, 60 °C, 1 h; (ii) K2CO3, DMF, morpholine/piperazine, 95 °C, 12 h. | ||
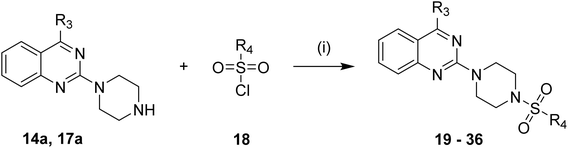 | ||
| Scheme 6 Synthesis of substituted quinazoline-sulphonamides 19–36. Reagents and conditions: (i) DMF, K2CO3, 70 °C, 3.5 h. | ||
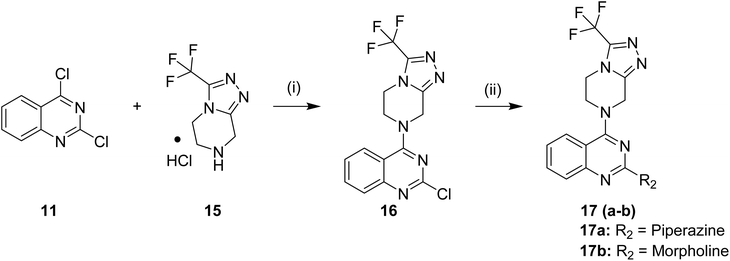
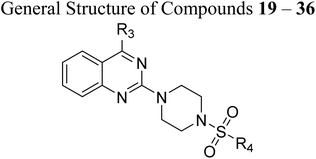
Substituted 4-acetamidobenzene-1-sulfonyl chlorides 5 (a–b), Scheme 1, were afforded by the acylation of different anilines in DCM in the presence of Et3N, followed by reaction with chlorosulphonic acid at different temperatures. Likewise, 3-acetylbenzenesulfonyl chloride 7 was afforded by treating chlorosulphonic acid with acetophenone, as shown in Scheme 2. On the other hand, as displayed in Scheme 3, 2,4-dichloroquinazoline 11 was synthesised by reacting urea with methyl anthranilate at high temperature, followed by chlorination with phosphorus oxychloride. Synthesised 2,4-dichloroquinazoline 11 was further reacted with tetrahydrofurfurylamine and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine, HCl to achieve respective intermediates 13 and 16. The intermediates 13 and 16 were further reacted with piperazine and morpholine to afford intermediates 14 (a–b) and 17 (a–b), as shown in Schemes 4 and 5. Finally, target quinazoline-sulphonamides 19–36 were obtained by reacting intermediates 14a and 17a with respective sulphonyl chlorides as shown in Scheme 6. The final structures and antimalarial properties of compounds 19–36 are shown in Table 1.
The synthesised intermediates and target compounds were purified by column chromatography using silica gel (200–400) as the stationary phase. The chemical structures and molecular weight of the compounds were confirmed by NMR spectroscopy (Fig. S1–S48) and high-resolution mass spectrometry (HRMS) (Fig. S49–S72). The purified compounds were stored in a cool, dark place and used for biological studies.
2.2. Antimalarial study of the compounds
The antimalarial efficacy of the compounds was investigated against the drug-sensitive 3D7 strain of the P. falciparum (Pf3D7) parasite. The study used DMSO and chloroquine (CQ) as a control. The half-maximal inhibitory concentration (IC50) of the compounds is shown in Table 1 and Fig. S73 and S74.Results indicate that most of the compounds were able to inhibit the growth of drug-sensitive parasites with an IC50 below 25 µM. A total of four compounds, 19, 21, 25, and 27, displayed an IC50 value below 5 µM, while ten compounds, 14a, 14b, 17a, 20, 24, 28, 30, 31, 33, and 34, displayed an IC50 value below 25 µM. The structure–activity relationship (SAR) study disclosed a compelling correlation between the structure of the compounds and their antimalarial activity. Substitution at 4th position of 2,4-dichloroquinazoline with tetrahydrofurfurylamine and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine resulted as compounds 13 and 16, respectively. Both compounds remained inactive against drug-sensitive P. falciparum, but further substitution with piperazine at the 2nd position of both compounds resulted in 14a and 17a, making both compounds active against Pf3D7 with an IC50 value of 8.26 µM and 13.53 µM, respectively. The results of a subsequent SAR study showed that the bioactivity of 14b did not change significantly when the piperazine ring of 14a was replaced with morpholine, but it was lost in the case of 17b, indicating that the NH group of piperazine with triazolo[4,3-a]pyrazine at the 4th position of quinazoline was helpful in identifying the antimalarial potency of the compounds. In order to study the antimalarial potential of quinazoline-sulphonamide hybrids, intermediates 14a and 17a were hybridised with the substituted aromatic sulphonamides. The multiple substitutions were performed at the aromatic ring of sulphonamides to fine-tune the antimalarial efficacy of the compounds. In this order, compound 14a was hybridised with 4-acetamido-3-chlorobenzenesulfonyl chloride, and the low micromolar antimalarial potency against Pf3D7 strain was obtained and identified as compound 19, with an IC50 value of 3.43 µM. Further substitution in the compound 19 with an electron-donating methoxy group at the 3rd position of the phenyl ring of the sulphonamide group resulted in 20, which displayed ∼6.31-fold activity loss with an IC50 of 21.64 µM. This might be a result of the ability of the chloro group of 19 to grab electrons towards itself, which decreased the density and increased the positive charge of the π-electron cloud and thus facilitated its ability to interact with the stacked aromatic rings of the potential target protein 39.
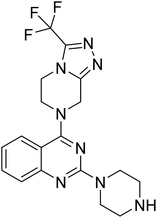
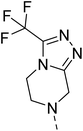
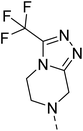
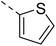
Hybridisation of 4-acetamido-3-methoxybenzenesulfonyl chloride with 17a resulted in 21, which displayed ∼3-fold improved antimalarial efficacy. The compounds 20 and 21 have similar benzene sulphonamide groups but have a structural difference at the 4th position of the quinazoline ring, resulting in a ∼5-fold activity difference. This indicated that the 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine plays a significant role in defining the antimalarial efficacy of the compounds. A similar activity pattern was followed for compounds 25 and 28, which have identical sulphonamide groups but a structural difference at the 4th position of the quinazoline ring. The antimalarial efficacy of 25 with tetrahydrofurfurylamine group was ∼2-fold better than 28 with 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine. Likewise, compound 27 with 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine was around 3-fold more potent than its analogue 24 with a tetrahydrofurfurylamine group at the 4th position. Another example of compound 34 revealed that 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine significantly improved the antimalarial efficacy, with an IC50 value of 4.83 µM, as its analogue 33 with a tetrahydrofurfurylamine group displayed almost 3.8-fold lower antimalarial efficacy. Compounds 33 and 34 had an electron-withdrawing chloro group substituted on the thiophene ring of the sulphonamide group and showed micromolar activity against drug-sensitive malaria; however, the absence of the chloro group made both compounds inactive. Likewise, the significance of the positional substitution of NO2 at the phenyl ring of the sulphonamide group related to the antimalarial efficacy was also noted. The substitution of the NO2 group at the 2nd position of the benzene ring was found to be more fruitful than the substitution at the 4th position. Compounds 24 and 27 with the NO2 group at the 2nd position of the phenyl ring displayed IC50 values of 8.80 µM and 2.90 µM, respectively, while their analogues with the NO2 group at the 4th position were either inactive or less potent against the Pf3D7 strain. Thus, the SAR study emphasised that 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine played a significant role in the development of new antimalarials, and it was found fruitful when hybridised with quinazoline-sulphonamides.
The detailed SAR investigation revealed that the positional substitution of different electron-withdrawing groups on the phenyl ring of the sulphonamides was found to be significant to fine-tune the antiplasmodial efficacy of the compounds. Finally, our study highlights compounds 19 and 27 as potential leads for the development of a new class of antimalarials.
2.3. Effect of the lead compounds on the development and morphology of the parasites
Post-IC90 estimation, the selected compounds were tested against various asexual stages of Plasmodium falciparum to assess their effects on parasite morphology and development. Dimethyl sulfoxide (DMSO) was used as the solvent control throughout the experiment. Synchronised ring-stage parasites (6–16 hours post-invasion) were exposed to the compounds for 64 hours, and Giemsa-stained blood smears were examined microscopically at intervals of 6–16, 30–34, 44–48 hours, and again at 6–16 hours (Fig. 2). | ||
| Fig. 2 Representative images of the morphology of test compounds treated parasites at different time intervals. | ||
In the DMSO-treated control group, parasites progressed normally through the intraerythrocytic developmental cycle, maturing from rings to trophozoites, then schizonts, and producing new ring-stage progeny. The images at different time intervals clearly indicate the formation of hemozoin crystals at a 30–34 h time interval, which is also observed later at 44–48 h. In the case of the test compounds treated parasites, no visible crystal was observed at any stage of the parasitic development. This phenomenon suggested that the compounds may target the parasite's hemozoin formation pathway, haeme detoxification pathway, and/or haemoglobin digestion pathway to show their antimalarial efficacy. In contrast, treatment with compound 14b notably inhibited the transition from trophozoites to schizonts, leading to developmental arrest at the schizont stage, with a swollen effect on the parasite's food vacuole. Parasites exposed to compounds 14a, 19, and 25 exhibited marked shrinkage within erythrocytes after 34 h of treatment, with complete developmental arrest at the ring stage and no further progression observed. Notably, compounds 21 and 27 induced developmental arrest at the trophozoite stage, characterised by a distinct swelling of the parasite's food vacuole, suggesting a potential disruption of vacuolar function. Literature suggests that inhibitors of parasitic aspartic proteases and cysteine proteases result in swollen food vacuoles.40,41 Therefore, the swelling effect on the parasite's food vacuole may correspond to the binding of the test compounds to aspartic proteases and/or cysteine proteases.
Collectively, these findings demonstrate that the six lead compounds exert stage-specific inhibitory effects on the asexual blood stages of P. falciparum. Further mechanistic studies and structural optimisation of these compounds hold promise for the development of potent, low-nanomolar antimalarial agents with diverse modes of action.
2.4. Cytotoxicity of the compounds
A549 lung carcinoma cell cultures were adopted to investigate the viability of the most potent compounds against the Pf3D7 strain. The obtained results are reported in Table 2, which indicates that compound 27 is non-cytotoxic to the cancerous cell A549 and more selective towards parasitic cells, with a selective index of 66.72. Furthermore, compounds 19 and 21 were found to be toxic to some extent in cancerous cells and were selective towards parasitic cells, with a selective index of 19.45 and 17.26, respectively. Compound 14b was found to be toxic to the cancerous cells with an IC50 value of 28.66 µM and may be further optimised for the development of new anticancer agents.| Compounds | A549 cells IC50 (µM) | Selective index |
|---|---|---|
| a The results indicated are the averages of triplicate measurements. Selective index: IC50 value of cytotoxicity assay/IC50 value of antimalarial assay. | ||
| 14b | 28.66 ± 2.06 | 2.88 |
| 19 | 66.72 ± 2.03 | 19.45 |
| 21 | 76.12 ± 3.49 | 17.26 |
| 25 | 57.02 ± 4.60 | 16.25 |
| 27 | 193.5 ± 1.73 | 66.72 |
Furthermore, cytotoxicity was also assessed using an MTT assay on activated macrophages derived from THP1 monocytes at 100 and 500 µM concentrations of the compounds. The obtained results in Fig. 3 revealed that untreated cells grew normally.
 | ||
| Fig. 3 Effect of the test compounds on the growth of the activated macrophages from THP1 monocytes (the results were the average of the five independent experiments). | ||
The compound 19 displayed cell growth of 88.76% and 81.54% at 100 µM and 500 µM concentration, respectively, and compound 21 showed cell growth of 97.75% and 96.79% at 100 µM and 500 µM concentration, respectively. The results indicated that the test compounds displayed cell growth of more than 80% and may be considered non-toxic at selected concentrations. Therefore, the antimalarial activity of the compounds was specific and not associated with cytotoxicity.
2.5. In silico studies
Falcipain II and III are hemoglobinases of the cysteine protease family,44 which share a common catalytic mechanism involving a nucleophilic cysteine thiol in their catalytic triad to hydrolyse the haemoglobin at multiple sites in the food vacuole.45,46 Fig. 4 disclosed that 19 and 27 were tightly bound with PfFP2 with docking scores of −5.2 and −5.7, respectively. Fig. 4(a) revealed that ligand 19 interacted with PfFP2 through a halogen bond between Cl and HIS174 with a bond length of 2.58 Å, and two aromatic H-bonds between the hydrogens of the quinazoline ring and ASP234 with a bond length of 2.50 and 2.69 Å. Similarly, ligand 27 was involved in making an H-bond acceptor between the N of the triazole group and HIS174 with a bond length of 2.79 Å, and a salt bridge between the NO2 group of the ligand ASP234 with a bond length of 4.68 Å, Fig. 4(b). Furthermore, 19 and 27 were docked with PfFP3 with docking scores of −5.3 and −6.1, respectively (Fig. 5). The protein ligand interactions with PfFP3 revealed that the NH group and the quinazoline ring of 19 were involved through a hydrogen bond and an aromatic hydrogen bond with GLY92, with the bond length of 1.85 Å and 2.30 Å, respectively, Fig. 5(a). There was no visible interaction observed in Maestro for 27 with PfFP3, Fig. 5(b).
Falcilysin (FLN) is a zinc-dependent metalloprotease belonging to the M16C family, playing a central role in haemoglobin breakdown within the parasite's digestive vacuole. It acts downstream of primary hemoglobinases such as falcipains, plasmepsins, and HAP, processing the short peptide fragments they generate. The enzyme functions through a dynamic catalytic cycle: it initially opens to permit substrate entry into its spacious active site, then shifts into a closed conformation that facilitates peptide cleavage, and finally reopens to release the processed products.47 Recent studies revealed that FLN is the target for the candidate drug MK-4815.48 The ligands were docked at the MK-4815 binding site (allosteric site of FLN) and visualised to investigate the protein ligand interactions, represented in Fig. 6.
Fig. 6 revealed that 19 and 27 were deeply seated in the groove of the PfFLN and strongly bound with the target protein. Ligand 19 was involved in making an H-bond through the NH group with ASP526, with a bond length of 2.08 Å, Fig. 6(a). Another H-bond was also observed between the oxygen of the acetamide group and ASN420, with the bond length of 2.30 Å. Additionally, two aromatic H-bonds were also observed between hydrogens of quinazoline ring and ASP526 and ASP451 with the bond length of 2.63 Å and 2.45 Å, respectively.
Similarly, ligand 27 was bound to the target protein with multiple interactions. Fig. 6(b) revealed that the quinazoline ring interacted with PHE545 through π–π stacking with the bond length of 5.16 Å. Furthermore, the SO2 and NO2 groups of 27 were interacted with ASN420 and ASN421 through H-bond with the bond length of 2.11 Å and 2.33 Å, respectively. The π–cation type of interaction was observed between the N+ of the NO2 group and HIE425, with the bond length of 5.68 Å. Additionally, an aromatic H-bond was also observed between the hydrogens of the quinazoline ring and ASP526, with a bond length of 2.40 Å. The ligand binding results were consistent with the earlier studies of Eagon, et al.49 and Kahlon, et al..50
The docking studies revealed that the synthesised hybrid molecules may bind with the PfFP2, PfFP3, and PfFLN through significant protein–ligand interactions. In the case of docking, only the ligand is flexible, while the receptor is rigid; however, as the temperature changes, both become flexible. Therefore, the accuracy of the docking remains very limited; hence, to investigate the stability of these complexes, a molecular dynamics simulation study was performed on Desmond through Maestro of Schrödinger.
2.5.2.1 Root-mean square deviation (RMSD). The RMSD trajectory of the apo form of PfFP2 displayed initial stabilisation at ∼1.2 Å and then a gradual increase toward 2.5 Å over the simulation time, suggesting conformational changes as the protein explores its dynamic states, Fig. S82(a). The same can also be observed in the simulation movie, Movie M1.
The RMSD trajectory of the PfFP2-19 complex displayed initial conformational changes upon binding with the ligand, but was observed to be comparatively tight and stable, while the ligand RMSD trajectory fluctuates for a long stretch with two sharp spikes ∼100 ns and ∼150 ns, Fig. S82(b). This event indicated that ligand 19 keeps the protein more stable than the apo state, but the ligand itself explores multiple poses, higher–deviation transitions, which reveal more dynamic binding.51 Moreover, the protein RMSD trajectory of the PfFP2-27 complex rises steadily and stays high and peaks at ∼4.0 Å late in the run, but the ligand RMSD remains lower compared to the 19, with occasional spikes, indicating a persistent pose with moderate jitter. The RMSD trajectory of the ligand in PfFP2-27 complex, Fig. S82(c), revealed that 27 binds to the protein in a more self-consistent pose, but it induced larger global rearrangements of the protein as compared to the apo form or complex with 19. Hence, ligand 19 develops a complex with good stability, while ligand 27 increases backbone deviation in the protein, suggesting broader conformational adaptation. The confirmations of PfFP2-19 and PfFP2-27 complexes and ligand–protein interaction events during the simulation may also be observed in simulation movies, Movies M2 and M3, respectively.
The RMSD trajectory for the apo form of PfFP3, Fig. S83(a), displayed a quick rise in the earliest frames to ∼1.6–1.9 Å by ∼20–40 ns, then drifts upward slowly and stabilises roughly between ∼1.8–2.4 Å up to 200 ns. This trend indicates gradual relaxation from the starting (crystal) conformation to a slightly shifted but compact ensemble. The RMSD trajectory of PfFP3-19 complex in Fig. S83(b) displayed that the protein initially follows a similar pattern as the apo form, but exhibits slightly higher average values, ∼2.0–2.7 Å, between ∼40–140 ns and stabilises throughout the simulation. The increase in RMSD may be due to the ligand-induced conformational changes in the protein. The RMSD trajectory of the ligand 19 displayed low-to-moderate fluctuations up to ∼80 ns, then stabilised for the rest of the simulation with a sharp spike at ∼120 ns. This trajectory pattern suggests the ligand reorientation events and partial displacement from the initial pose. These confirmational changes and ligand behaviour of the apo protein and in complex with 19 during the simulation may also be observed from the simulation movies, Movies M4 and M5, respectively. The RMSD trajectory pattern of PfFP3-27 complex revealed that the protein RMSD displayed a quicker rise at ∼20 ns than at ∼50 ns, which was overall larger than the apo form or in the complex with 19, Fig. S83(c). That indicates a more pronounced backbone rearrangement during the run.52 The ligand RMSD for 27 remains relatively low early on, but after ∼100 ns, it increases substantially and becomes highly variable, reaching very large values (double-digit Å in late frames), which strongly suggests the ligand underwent a major displacement and partially relocated in the original binding site, also confirmed by the simulation movie, Movie M6. The RMSD results of apo PfFP3 and its complexes revealed that the experimental crystal structure relaxes to a stable solution ensemble with modest conformational drift over 200 ns. For complexes, 19 is likely a moderately stable binder with multiple binding microstates, which keeps the protein closer to its apo state compared to 27. This may be due to the fewer tight contacts that remain associated with the protein. The RMSD results for ligand 27 suggest that (i) its interactions may require larger protein rearrangement (induced fit) but ultimately are not stable or lead to ligand escape, (ii) the ligand initially finds an alternate pocket that perturbs backbone geometry, and later the complex relaxes to an alternative state with the ligand shifted away.34,53 Both cases imply strong local dynamics and a binding mode that is not rigidly conserved over 200 ns.
Furthermore, the RMSD trajectory of the apo form of PfFLN displayed a stable RMSD during the first 50 ns, followed by a progressive rise, reaching values of ∼2.5–3.5 Å after ∼100 ns and maintaining fluctuations around this range until the end of the simulation, Fig. S84(a) and Movie M7. These fluctuations suggest that PfFLN undergoes significant conformational rearrangements in the apo state, likely linked to its intrinsic flexibility as a metalloprotease.54,55 The RMSD trajectory of protein in complex with 19 displayed an early rise and reached equilibrium at ∼30 ns, which remains lower than the apo trajectory, with average backbone RMSD in the ∼1.8–2.2 Å range after equilibration, Fig. S84(b). The ligand RMSD was also relatively small and well-behaved (∼1.3–1.8 Å), indicating that ligand 19 retains a consistent binding pose for most of the simulation, Movie M8. In Fig. S84(c) of complex PfFLN-27, the protein RMSD was intermediate to high, typically ∼1.8–2.5 Å, but with several transient excursions beyond 2.5 Å later in the run. The ligand 27 exhibits wider fluctuations in RMSD, ranging from ∼1.2 to 3.5 Å, including episodes of larger deviation late in the trajectory. The combined behaviour suggests a period of induced conformational adjustment followed by increased ligand mobility. The 19 complex reaches a plateau within ∼30 ns and remains near that plateau, consistent with rapid local relaxation and a stable ligand–protein complex. The 27 complex displays a two-phase behaviour: an initial relaxation and induced fit (0–100 ns) accompanied by modest protein rearrangements, followed by increased ligand mobility and several ligand deviation events after ∼120 ns. These deviations are temporally coincident with transient increases in protein RMSD, suggesting coupled ligand–protein rearrangements. These events can also be confirmed by the simulation movie, Movie M9.
The comparative analysis demonstrates that both ligands exhibit strong binding affinity toward the target proteins, with greater structural stability observed in the complex with PfFLN relative to PfFP2 and PfFP3. The RMSD profiles predicted that PfFLN may be the principal target implicated in the antimalarial potential of these ligands, while the concurrent inhibition of PfFP2 and PfFP3 may provide a complementary or synergistic contribution to their overall efficacy.
2.5.2.2 Root mean square fluctuation (RMSF). Root mean square fluctuation (RMSF) is useful for characterising local changes along the protein chain. The RMSF plot of the apo proteins and their complexes provides clear insights into the protein's conformational flexibility, ligand effects, and implications for drug design.
The RMSF plot of apo PfFP2 in Fig. S85(a) shows higher peaks and relatively higher residue fluctuation, especially in loop regions, compared to both ligand–bound complexes. Fig. S85(b) and S85(c) revealed that the binding of ligands 19 and 27 affected the protein's residue fluctuations and resulted in a decrease in fluctuations, which indicates the stabilisation of the protein by ligand binding. The plot highlighted that 27 induced stronger stabilisation, as confirmed by the lower RMSF in most of the regions, while 19 retained moderate flexibility, which may correspond to exposed or mobile loop areas. In both cases, the profile for Cα and backbone atoms was very similar, supporting that stabilisation was global, but most pronounced near the ligand-binding regions. The vertical green bars indicate ligand contact with protein residues during the simulation. The RMSF results for the apo form of PfFP3 and its complexes with 19 and 27 were shown in Fig. S86.
Fig. S86(a) revealed that apo falcipain-3 exhibited higher RMSF values at both N- and C-terminal ends and several loop regions, indicating greater intrinsic flexibility where no ligand is bound. When ligand 19 was bound to the falcipain-3, Fig. S86(b), the residue stretches were greatly affected. The vertical green bar indicated that the residues of these regions were involved in making non-covalent interactions, and fluctuations highlighted that the protein–ligand complex is changing conformational state to get a lower energy state. Fig. S86(c) indicates that ligand 27 exhibits fewer protein–ligand contacts compared to ligand 19, but stabilises more residual fluctuation (residues 75–90) than the apo form complex with ligand 19. Residues outside the ligand contact zones retain their flexibility, like the apo protein. Differences between the ligands (19 vs. 27) in the extent and location of stabilisation may be attributed to their distinct binding modes, chemical structures, and interaction networks within the flacipain-3 active or allosteric sites. Ligand 19, showing a broader interaction footprint than ligand 27, potentially translates to enhanced inhibitory potential. The RMSF analysis results of falcilysin in apo form and in complex with ligands 19 and 27 were presented in Fig. S87.
The RMSF plot of the apo protein, Fig. S87(a), displayed several distinct peaks with higher flexibility in multiple residue regions. Peaks likely correspond to loops or surface regions, which are generally more mobile in the absence of ligands. The average RMSF is higher across the range compared to the complexes. Fig. S87(b) and S87(c) indicate that the binding of ligands 19 and 27 noticeably reduces the RMSF values in multiple regions, especially at the ligand binding sites, highlighted using green vertical bars. The RMSF data highlighted that interacting residues are tightly associated with the ligand, resulting in noticeable stabilisation and lower atomic displacement at and near the ligand binding sites. Few peaks remain in the complex plots, but their intensities have been reduced compared to the apo form, which suggests effective ligand-induced rigidification.
The RMSF data for the apo and complex forms of all three target proteins revealed that both ligands induced local stabilisation at their respective binding sites, reducing RMSF values of associated amino acid residues, which is consistent with classical MD simulation interpretations where ligand binding restricts atomic displacements and conformational freedom locally.56,57 The higher fluctuation profile of the apo form of the proteins highlights larger conformation sampling and flexibility, which is also needed for substrate recognition, which decreases upon binding with the ligands.58,59 Both ligands, 19 and 27, display similar stabilising trends, though the specific residues affected and the degree of rigidity differ, which may reflect differing binding poses, interaction strengths, or allosteric effects. The lower RMSF fluctuations in complexes are generally defined as evidence of stable binding and limited flexibility at interacting residues, while higher flexibility refers to the inherent protein flexibility not modulated by ligand attachment.60,61 Hence, the RMSF trends were in correlation with the RMSD data and supported our hypothesis that the antimalarial activity of the ligands may be due to the inhibition of cysteine proteases and falcilysin.
2.5.2.3 Protein–ligand contacts study. The type and number of interactions between the target protein and the ligand can be monitored throughout the MD simulation. These interactions can be divided into different categories, such as hydrogen bonds, hydrophobic, ionic and water bridges, etc. Hydrogen bonds are further characterised into four sub-parts, i.e. backbone acceptor, backbone donor, side-chain acceptor, and side-chain donor. Similarly, halogen atoms can form attractive interactions by functioning as both electron donor and acceptor sites and can play an important role in ligand specificity. Likewise, hydrophobic interactions can be categorised into three sub-categories, i.e. π–cation, π–π interaction, and other non-specific interactions. Traditionally, these types of interactions are formed between a hydrophobic amino acid and an aromatic or aliphatic group on the ligand. Moreover, ionic interactions are formed between two oppositely charged atoms, and water bridges are considered in MD simulations as hydrogen-bonded protein–ligand interactions mediated by a water molecule.
Fig. 7 revealed that most of the ligands exhibited significant protein–ligand interactions during the course of simulation. Fig. 7(a) and S88 disclosed that ligand 19 was involved with PfFP2 mainly through hydrophobic interaction via LEU84 for more than 35% of simulation time, and water bridge, hydrogen bond via LEU172 and ASN173. Similarly, ligand 19 was involved with PfFP3 through multiple hydrogen bonds and hydrophobic interactions. Fig. 7(b) and S90 displayed that TYR93 was involved with the ligand through π–π interaction and hydrogen bond for 34% and 50% of simulation time, respectively. ASN86 also displayed a hydrogen bond with the ligand 19 for 44% of simulation time. Protein–ligand contact analysis displayed strong binding between PfFLN and 19, as shown in Fig. 7(c) and S92. The amino acid residues ASN420 and ASN421 were involved in making a hydrogen bond for 98% and 90% of the simulation time, respectively. TYR413 and GLU530 displayed interaction through a water bridge for 44% and 63% of simulation time, respectively.
 | ||
| Fig. 7 2D representation of protein–ligand contacts during 200 ns MD simulations. (a) PfFP2-19, (b) PfFP3-19, (c) PfFLN-19, (d) PfFP2-27, (e) PfFP3-27, and (f) PfFLN-27 complexes. | ||
The study of protein–ligand interaction of ligand 27 disclosed multiple interactions with the target protein throughout the simulation. It was involved with PfFP2 mainly through three hydrogen bonds with GLN36, CYS42, and ILE85 for 64%, 81%, and 88% of simulation time, respectively, as shown in Fig. 7(d) and S89. Ligand 27 was contacted with PfFP3 mainly through TRP215 via π–π interaction for 53% of simulation time, as indicated in Fig. 7(e) and S91. Similar to ligand 19, ligand 27 was also strongly bound to PfFLN through hydrogen bonds and hydrophobic interactions. Fig. 7(f) and S93 disclosed that residues ASN420 and ASN421 of PfFLN were involved in making hydrogen bonds with 86% and 43%, respectively. Furthermore, PHE529 was involved in making π–π interactions with 27 for 39% of simulation time, along with other hydrophobic interactions. The higher length of bars in the protein–ligand histogram for ASN420, ASN421, and PHE529 is due to the multiple interactions of a single type of residue with the same ligand.
The results of the protein–ligand contact study for all six complexes showed close proximity, as indicated by RMSD and RMSF values. In silico studies predicted that all ligands were tightly bound to PfFLN, as well as to PfFP2 and PfFP3. Hence, it suggests that the associated antimalarial efficacy may be due to the inhibition of these proteins.
| Complex | ΔG average (kcal mol−1) | Standard deviation |
|---|---|---|
| PfFP2-19 | −48.2992 | ±6.10 |
| PfFP2-27 | −59.7697 | ±5.39 |
| PfFP3-19 | −54.7571 | ±10.17 |
| PfFP3-27 | −56.8884 | ±5.70 |
| PfFLN-19 | −50.3223 | ±4.55 |
| PfFLN-27 | −51.5066 | ±4.73 |
The post-MD MM-GBSA analysis confirmed stable binding of both ligands with their respective target proteins. Among the complexes, PfFLN exhibited the lowest standard deviations in binding free energy, suggesting that ligand interactions with PfFLN were more consistent throughout the simulation when compared to PfFP2 and PfFP3. Taken together, these in silico findings predict that the observed antimalarial activity of ligands 19 and 27 may be mediated through the plausible inhibition of PfFLN in addition to PfFP2 and PfFP3. To validate the in silico predictions, future in vitro enzymatic assays would be essential to provide deeper and clearer mechanistic insight into their mode of action. The ADME predictions and physicochemical properties of the synthesised compounds have been discussed in the SI and Table S1.
3. Conclusion
To combat the resistance to the current antimalarials, there is a challenge and an urgent need for the development of new antimalarials with a novel mode of action. The present study aimed to develop quinazoline-based trifunctional hybrid antimalarials with potential antimalarial activity. Bioactive small pharmacophores such as 2,4-dichloroquinazoline, tetrahydrofurfurylamine, substituted sulfonamides, and triazolo[4,3-a]pyrazine were utilised to construct a new class of antimalarials. Results revealed that most quinazoline sulphonamide hybrids were shown to be potential antimalarials, with an IC50 value below 10 µM for the drug-sensitive Pf3D7 strain. The results revealed that molecular hybrid 27, with 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine at the 4th position and substituted sulphonamides at the 2nd position of quinazoline, was the most potent against the Pf3D7 strain with an IC50 value of 2.9 µM. Moreover, hybrid 19, with tetrahydrofurfurylamine at the 4th position and substituted sulphonamides at the 2nd position of quinazoline, was the second most potent compound against the Pf3D7 strain with an IC50 value of 3.4 µM. The MTT assay was performed on A549 lung cancer cells and activated macrophages derived from THP-1 monocytes to assess the associated cytotoxicity of the compounds. The results against A549 cells disclosed that 19 and 27 were more selective towards parasitic cells, with the selective index of 19.45 and 66.72, respectively. The compounds 14b, 24, and 28 were moderately cytotoxic to the cancerous cells and may be further developed as potential anticancer agents. The results against THP1 cells revealed that the cell growth was more than 90% on treatment with 100 µM concentration of the test compounds, except 19, which displayed 88.76% cell growth. Therefore, the results indicated that the antimalarial activity of the compounds was specific and not associated with cytotoxicity. The in silico studies, i.e. docking followed by MD simulation and MM-GBSA analysis, were conducted to computationally predict the plausible mechanism of action of the potent molecules. The in silico predictions suggested that the antimalarial efficacy of the compounds may be attributed to the plausible inhibition of zinc metalloprotease PfFLN, with concurrent inhibition of cysteine proteases PfFP2 and PfFP3, which require further validation through enzymatic assays. The calculated and predicted physicochemical parameters of the compounds fall within the Schrödinger range, which is relied on by 95% of all known medications. The study addressed the significance of the molecular hybridisation approach for the identification of an entirely distinct class of antimalarials. Compounds 19 and 27 were thus highlighted in the study as striking molecules that could be further optimised for the development of new antimalarials.4. Method and materials
4.1. Chemistry
All chemicals and reagents were purchased in AR grade from commercial suppliers and used without further purification. Substituted anilines, acyl chloride, chlorosulphonic acid, anhydrous piperazine, morpholine, and acetophenone were procured from Spectrochem Pvt. Ltd, India. Hünig's base N,N-diisopropylethylamine (DIPEA) was procured from Sisco Research Laboratories Pvt. Ltd, India. Methyl anthranilate and tetrahydrofurfurylamine were procured from Alfa-Aesar, USA. Urea and EtOH were obtained from Merck Millipore, India, and 3-(trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine, HCl was obtained from BLD Pharmatech (India) Pvt. Ltd, India. Different sulphonyl chlorides were procured from TCI Chemicals (India) Pvt. Ltd, India. Solvents such as N,N-dimethylformamide (DMF), dichloromethane (DCM), and tetrahydrofuran (THF) were anhydrous and procured from Spectrochem Pvt. Ltd, India. The 1H & 13C NMR spectral data were recorded with a Bruker Avance Neo 400 NMR spectrometer. Tetramethylsilane (TMS) was used as an internal standard, and the observed chemical shift (δ) value was marked in ppm. The multiplicity of 1H NMR signals was labelled as s (singlet), d (doublet), t (triplet), m (multiplets), q (quartet), p (pentet), dd (double doublets), td (triplet of doublets), dt (doublet of triplets), dq (doublet of quartets), qd (quartet of doublets), tt (triplet of triplets), qt (quartet od triplets), dtd (doublet of triplet of doublets), ddd (doublet of doublet of doublets). HRMS spectra of the compounds were calculated with an Agilent G6530AA (LC-HRMS-Q-TOF). Silica gel-coated aluminium plates, TLC Silica gel 60 F254 of Merck Millipore, were used for conducting analytical thin-layer chromatography (TLC) experiments and visualised under a UV-chamber equipped with long-wavelength (365 nm) and short-wavelength (254 nm). The silica gel of 200–400 mesh was procured from Sisco Research Laboratories Pvt. Ltd, India, and used for column chromatography. Chloroquine-resistant P. falciparum 3D7 strain was obtained from the Malaria Research and Reference Reagent Resource Center (MR4). Human blood was used from P. falciparum culture in this study. Donor blood was obtained from Rotary Blood Bank (RBB), New Delhi, India. RBB is an ISO 9001:2008 certified blood bank established in 2002. It follows stringent screening procedures, careful documentation, and good laboratory practices for collecting, processing, and testing blood. Albumax was obtained from Gibco, New Zealand and RPMI media from Invitrogen, India. Chloroquine, Molecular Biology Grade DMSO, tetrazolium salt [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide], sorbitol, and Phorbol 12-Myristate 13-Acetate (PMA) were obtained from Sigma-Aldrich. SYBR Green dye was obtained from Invitrogen, India. Lung cancer A549 epithelial cells and THP1 monocytes were obtained from ATCC. Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were obtained from Gibco, USA. RPMI-1640 medium was obtained from Cellclone, India.Chlorosulfonic acid (15.756 mmol) was added in a dropwise fashion to the respective N-phenylacetamide (3a–3b) (3.03 mmol) in a round-bottom flask equipped with a magnetic bar at 0 °C, followed by stirring for 0.25 h. The resulting mixture was heated to the optimal temperature and stirred by almost all gas evolution was completed (10 °C/0.5 h for 5a; 60 °C/4 h for 5b). After the reaction was complete, the mixture was cooled and quenched with crushed ice in a beaker while being vigorously stirred to free any associated sulfonyl chlorides from the beaker's walls. The precipitate was filtered and washed with ice-cold water and purified using column chromatography to afford 4-acetamidobenzene-1-sulfonyl chlorides (5a–5b) in 75–85% yield. A similar protocol was followed to afford 3-acetylbenzenesulfonyl chloride (7) with some modifications, Scheme 2. On addition of sulphonyl chloride to acetophenone, the resulting mixture was heated at 90 °C for 12 h, followed by quenching and purification. Purified products 5a–5b and 7 were used for the next steps.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (80
EtOAc (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) as the mobile phase. The solvent was removed under reduced pressure, and 2,4-dichloroquinazoline (11) was obtained as a white solid. Yield = ∼90%.
20) as the mobile phase. The solvent was removed under reduced pressure, and 2,4-dichloroquinazoline (11) was obtained as a white solid. Yield = ∼90%.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (40
EtOAc (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
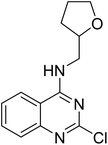
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) as the mobile phase. The solvent was evaporated under vacuum to afford product 2-chloro-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (13) as a white solid. Yield = 99%. 1H NMR (400 MHz, chloroform-d) δ 7.66–7.57 (m, 3H), 7.32 (ddd, J = 13.2, 8.4, 4.8 Hz, 1H), 6.60 (t, J = 5.2 Hz, 1H), 4.22–4.11 (m, 1H), 3.97 (ddd, J = 16.8, 6.4, 3.2 Hz, 1H), 3.88 (dt, J = 8.4, 6.8 Hz, 1H), 3.82–3.72 (m, 1H), 3.39 (ddd, J = 13.6, 8.4, 4.4 Hz, 1H), 2.10–2.00 (m, 1H), 1.95–1.86 (m, 2H), 1.59 (dq, J = 12.0, 7.2 Hz, 1H). 13C NMR (101 MHz, CDCl3) δ 161.0, 157.5, 150.7, 133.5, 127.6, 126.1, 121.0, 113.2, 77.2, 68.2, 45.3, 28.9, 25.9. ESI HRMS, found 264.0898 (C13H14ClN3O, [M + H]+ requires 264.0898).
60) as the mobile phase. The solvent was evaporated under vacuum to afford product 2-chloro-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (13) as a white solid. Yield = 99%. 1H NMR (400 MHz, chloroform-d) δ 7.66–7.57 (m, 3H), 7.32 (ddd, J = 13.2, 8.4, 4.8 Hz, 1H), 6.60 (t, J = 5.2 Hz, 1H), 4.22–4.11 (m, 1H), 3.97 (ddd, J = 16.8, 6.4, 3.2 Hz, 1H), 3.88 (dt, J = 8.4, 6.8 Hz, 1H), 3.82–3.72 (m, 1H), 3.39 (ddd, J = 13.6, 8.4, 4.4 Hz, 1H), 2.10–2.00 (m, 1H), 1.95–1.86 (m, 2H), 1.59 (dq, J = 12.0, 7.2 Hz, 1H). 13C NMR (101 MHz, CDCl3) δ 161.0, 157.5, 150.7, 133.5, 127.6, 126.1, 121.0, 113.2, 77.2, 68.2, 45.3, 28.9, 25.9. ESI HRMS, found 264.0898 (C13H14ClN3O, [M + H]+ requires 264.0898).Similar protocol was followed to synthesize 2-chloro-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (16). 2,4-dichloroquinazoline (11, 5.03 mmol) was added to the solution of 3-(Trifluoromethyl)-5,6,7,8-tetrahydro-[1,2,4]triazolo[4,3-a]pyrazine, HCl (5.28 mmol) and KOH (11.61 mmol) in anhydrous THF (25 ml) and stirred for 4 h at 40 °C, Scheme 5. The desired product 16 was afforded as an off-white solid. Yield = 95%. 1H NMR (400 MHz, chloroform-d) δ 7.92 (ddd, J = 8.4, 5.6, 1.2 Hz, 2H), 7.88–7.81 (m, 1H), 7.59 (ddd, J = 10.0, 7.2, 1.6 Hz, 1H), 5.33 (s, 2H), 4.52 (t, J = 5.6 Hz, 2H), 4.31 (t, J = 5.2 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 165.0, 155.8, 153.5, 143.9 (q, J = 38.88 Hz, 1C), 149.88, 134.43, 128.64, 126.85, 124.07, 118.3 (q, J = 270.28 Hz, 1C), 114.41, 47.39, 45.15, 42.63. ESI HRMS, found 355.0678 (C14H10ClF3N6, [M + H]+ requires 355.0681).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (80
MeOH (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) as the mobile phase. The solvent was evaporated under a vacuum to afford product 14a in an off-white molten state, and it solidified at room temperature. The piperazine was replaced with morpholine (3.80 mmol) by following a similar protocol to afford the desired product 14b as an off-white solid product, Scheme 4.
20) as the mobile phase. The solvent was evaporated under a vacuum to afford product 14a in an off-white molten state, and it solidified at room temperature. The piperazine was replaced with morpholine (3.80 mmol) by following a similar protocol to afford the desired product 14b as an off-white solid product, Scheme 4.
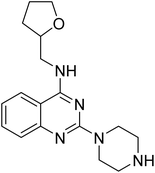
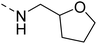
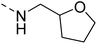
4.1.4.1 2-(Piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a). Yield = ∼95%. 1H NMR (400 MHz, chloroform-d) δ 7.61–7.50 (m, 2H), 7.47 (dd, J = 8.4, 1.2 Hz, 1H), 7.12 (ddd, J = 9.6, 6.8, 1.6 Hz, 1H), 6.14 (t, J = 5.6 Hz, 1H), 4.17 (qd, J = 7.2, 3.6 Hz, 1H), 4.11 (t, J = 5.2 Hz, 4H), 3.98–3.88 (m, 2H), 3.87–3.77 (m, 1H), 3.52 (ddd, J = 16.0, 8.8, 4.0 Hz, 1H), 3.16 (t, J = 5.2 Hz, 4H), 2.10–2.02 (m, 1H), 2.00 (t, J = 5.2 Hz, 2H), 1.94 (q, J = 8.0 Hz, 1H), 1.75–1.60 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 177.66, 160.19, 158.24, 151.55, 132.79, 125.82, 121.75, 121.01, 110.69, 68.22, 44.71, 44.05, 43.69, 42.12, 28.93, 25.92, 23.15. ESI HRMS, found 314.1975 (C17H23N5O, [M + H]+ requires 314.1976).
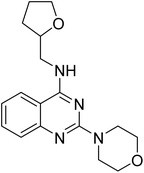
4.1.4.2 2-Morpholino-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14b). Yield = ∼95%. 1H NMR (400 MHz, chloroform-d) δ 7.58–7.52 (m, 1H), 7.51–7.40 (m, 2H), 7.03 (ddd, J = 10.0, 6.4, 2.0 Hz, 1H), 6.44 (s, 1H), 4.11 (qd, J = 7.2, 3.6 Hz, 1H), 3.90–3.84 (m, 1H), 3.82 (dd, J = 6.4, 4.8 Hz, 4H), 3.79–3.74 (m, 2H), 3.74–3.70 (m, 4H), 3.48 (ddd, J = 13.6, 7.2, 4.8 Hz, 1H), 2.02–1.93 (m, 1H), 1.92–1.82 (m, 2H), 1.67–1.54 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 159.91, 157.75, 132.92 (2C), 124.57, 121.81, 121.34, 110.48, 77.37, 68.22, 67.00 (2C), 44.79, 44.73 (2C), 28.98, 25.88. ESI HRMS, found 315.1817 (C17H22N4O2, [M + H]+ requires 315.1816).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (50
MeOH (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) as the mobile phase. The solvent was evaporated under a vacuum to afford product 17a as a white solid. The piperazine was replaced with morpholine (3.80 mmol) by following a similar protocol to afford the desired product 17b as a white solid.
50) as the mobile phase. The solvent was evaporated under a vacuum to afford product 17a as a white solid. The piperazine was replaced with morpholine (3.80 mmol) by following a similar protocol to afford the desired product 17b as a white solid.
4.1.5.1 2-(Piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a). Yield = ∼98%. 1H NMR (400 MHz, chloroform-d) δ 7.62 (dd, J = 8.0, 1.2 Hz, 1H), 7.59–7.47 (m, 2H), 7.11 (ddd, J = 10.0, 6.4, 1.6 Hz, 1H), 4.98 (s, 2H), 4.32 (t, J = 5.6 Hz, 2H), 4.04 (t, J = 5.2 Hz, 2H), 3.79 (dd, J = 6.0, 4.4 Hz, 4H), 3.71 (dd, J = 6.0, 2.8 Hz, 4H), 1.43 (d, J = 202.4 Hz, 1H). 13C NMR (101 MHz, CDCl3) δ 164.71, 157.78, 154.48, 151.22, 143.6 (q, J = 39.19 Hz, 1C), 133.52, 126.83, 123.90, 121.95, 118.5 (q, J = 266.135 Hz, 1C), 111.45, 66.90 (2C), 46.90, 46.18, 44.50 (2C), 43.13. ESI HRMS, found 405.1758 (C18H19F3N8, [M + H]+ requires 405.1758).
4.1.5.2 4-(4-(3-(Trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazolin-2-yl)morpholine (17b). Yield = ∼93%. 1H NMR (400 MHz, chloroform-d) δ 7.63 (dd, J = 8.4, 1.6 Hz, 1H), 7.58–7.47 (m, 2H), 7.12 (ddd, J = 9.6, 6.4, 1.2 Hz, 1H), 4.99 (s, 2H), 4.33 (t, J = 5.2 Hz, 2H), 4.05 (t, J = 5.6 Hz, 2H), 3.94 (t, J = 5.2 Hz, 4H), 3.02 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, CDCl3) δ 177.37, 164.79, 157.39, 154.39, 151.18, 143.6 (q, J = 34.04 Hz, 1C), 133.59, 126.87, 123.89, 122.15, 118.35 (q, J = 264.11 Hz, 1C), 111.44, 63.69, 46.88, 46.19, 44.26, 43.10, 22.86. ESI HRMS, found 406.1602 (C18H18F3N7O, [M + H]+ requires 406.1598).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc (40
EtOAc (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) as the mobile phase. The solvent was evaporated under a vacuum to afford the final product as a solid.
60) as the mobile phase. The solvent was evaporated under a vacuum to afford the final product as a solid.
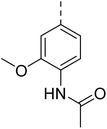
4.1.6.1 N-(2-Chloro-4-((4-(4-(((tetrahydrofuran-2-yl)methyl)amino)quinazolin-2-yl)piperazin-1-yl)sulfonyl)phenyl)acetamide (19). 4-Acetamido-3-chlorobenzenesulfonyl chloride (5b) was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 19 as a white solid. Yield = 90%. 1H NMR (400 MHz, chloroform-d) δ 8.53 (d, J = 8.8 Hz, 1H), 7.74–7.68 (m, 2H), 7.57 (dd, J = 8.8, 2.0 Hz, 1H), 7.49–7.40 (m, 2H), 7.36 (d, J = 8.4 Hz, 1H), 7.01 (ddd, J = 9.2, 6.8, 1.2 Hz, 1H), 6.06 (s, 1H), 4.08 (qd, J = 7.2, 3.2 Hz, 1H), 3.95 (dd, J = 6.4, 4.4 Hz, 3H), 3.85 (dt, J = 8.4, 6.4 Hz, 2H), 3.80–3.69 (m, 1H), 3.66 (s, 1H), 3.42 (ddd, J = 13.6, 7.2, 4.4 Hz, 1H), 3.01 (t, J = 5.2 Hz, 4H), 2.19 (s, 3H), 2.02–1.92 (m, 1H), 1.87 (p, J = 6.4 Hz, 2H), 1.65–1.51 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 175.41, 168.54, 160.09, 157.96, 151.35, 138.62, 132.82, 130.77, 128.43, 127.58, 125.53, 122.45, 121.65, 121.00, 120.84, 110.55, 68.22, 63.70, 46.13, 44.67, 43.27, 28.91, 25.90, 25.04, 20.92. ESI HRMS, found 545.1730 (C25H29ClN6O4S, [M + H]+ requires 545.1733).
4.1.6.2 N-(2-Methoxy-4-((4-(4-(((tetrahydrofuran-2-yl)methyl)amino)quinazolin-2-yl)piperazin-1-yl)sulfonyl)phenyl)acetamide (20). 4-Acetamido-3-methoxybenzenesulfonyl chloride (5a) was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 20 as a white solid. Yield = 90%. 1H NMR (400 MHz, chloroform-d) δ 8.80 (d, J = 2.0 Hz, 1H), 7.77 (s, 1H), 7.49 (ddd, J = 9.6, 3.2, 2.0 Hz, 4H), 7.10–7.02 (m, 1H), 6.92 (d, J = 8.8 Hz, 1H), 5.96 (s, 1H), 4.15 (qd, J = 7.2, 3.2 Hz, 1H), 4.01 (t, J = 5.2 Hz, 4H), 3.93 (s, 3H), 3.91–3.76 (m, 3H), 3.46 (ddd, J = 13.6, 7.6, 4.4 Hz, 1H), 3.10 (t, J = 5.2 Hz, 4H), 2.20 (s, 3H), 2.05 (dt, J = 13.6, 6.8 Hz, 1H), 1.95 (p, J = 7.2 Hz, 2H), 1.72–1.60 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 168.32, 159.98, 150.71, 134.56, 132.69, 128.13, 127.59, 125.79, 124.00, 121.38, 120.88 (2C), 118.78, 110.47, 109.53, 77.47, 68.20, 56.16 (2C), 46.30 (2C), 44.71, 43.23, 28.90, 25.94, 24.89. ESI HRMS, found 541.2225 (C26H32N6O5S, [M + H]+ requires 541.2228).
4.1.6.3 N-(2-Methoxy-4-((4-(4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazolin-2-yl)piperazin-1-yl)sulfonyl)phenyl)acetamide (21). 4-Acetamido-3-methoxybenzenesulfonyl chloride (5a) was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 21 as a white solid. Yield = 92%. 1H NMR (400 MHz, chloroform-d) δ 8.77 (d, J = 2.4 Hz, 1H), 8.01 (s, 1H), 7.78 (s, 1H), 7.66 (dd, J = 8.0, 1.2 Hz, 1H), 7.59 (ddd, J = 10.0, 6.8, 1.6 Hz, 1H), 7.54–7.48 (m, 1H), 7.16 (ddd, J = 9.6, 6.8, 1.2 Hz, 1H), 6.95 (d, J = 8.8 Hz, 1H), 5.03 (s, 2H), 4.39 (t, J = 5.2 Hz, 2H), 4.09 (t, J = 5.6 Hz, 2H), 3.99 (dd, J = 8.0, 4.4 Hz, 4H), 3.94 (s, 3H), 3.11 (d, J = 7.6 Hz, 4H), 2.20 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 168.40, 164.72, 162.57, 157.08, 154.33, 151.07, 143.6 (q, J = 39.89 Hz, 1C), 133.53, 128.12, 127.50, 126.79, 124.07, 123.90, 122.03, 118.85, 118.3 (q, J = 258.56 Hz, 1C) 111.30, 109.62, 56.17, 46.39, 46.10, 45.93, 43.30, 43.05, 36.52, 31.45, 24.84. ESI HRMS, found 632.2013 (C27H28F3N9O4S, [M + H]+ requires 632.2010).
4.1.6.4 1-(3-((4-(4-(3-(Trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazolin-2-yl)piperazin-1-yl)sulfonyl)phenyl)ethan-1-one (22). 3-Acetylbenzenesulfonyl chloride (7) was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 22 as an off-white solid. Yield = 82%. 1H NMR (400 MHz, chloroform-d) δ 8.04 (dd, J = 8.0, 1.6 Hz, 1H), 7.68–7.60 (m, 2H), 7.55 (ddd, J = 13.2, 7.6, 1.2 Hz, 2H), 7.49 (dd, J = 8.8, 1.6 Hz, 1H), 7.32 (dd, J = 7.2, 1.2 Hz, 1H), 7.11 (ddd, J = 10.0, 6.8, 1.6 Hz, 1H), 5.03–4.91 (m, 2H), 4.31 (t, J = 5.6 Hz, 2H), 4.06–3.98 (m, 4H), 3.97–3.79 (m, 4H), 3.71–3.59 (m, 2H), 3.20 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 168.25, 164.78, 157.52, 151.14, 146.37, 143.6 (q, J = 40.20 Hz, 1C), 137.85, 136.68, 133.94, 133.57, 129.83, 129.74, 127.32, 126.87, 123.89, 122.09, 118.42 (q, J = 258.76 Hz, 1C), 111.43, 47.36, 46.84, 46.23, 45.56, 43.51, 43.40, 43.09, 41.90. ESI HRMS, found 587.1798 (C26H25F3N8O3S, [M + H]+ requires 587.1795).
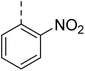
4.1.6.5 2-(4-((4-Nitrophenyl)sulfonyl)piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (23). 4-Nitrobenzenesulfonyl chloride was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 23 as a white solid. Yield = 95%. 1H NMR (400 MHz, chloroform-d) δ 8.38–8.33 (m, 2H), 8.02 (s, 1H), 7.97–7.92 (m, 2H), 7.56–7.48 (m, 1H), 7.42 (dd, J = 8.4, 1.2 Hz, 1H), 7.09 (ddd, J = 9.2, 6.8, 1.2 Hz, 1H), 6.17 (t, J = 5.6 Hz, 1H), 4.07–4.00 (m, 4H), 3.92 (dt, J = 8.4, 6.8 Hz, 1H), 3.85–3.77 (m, 1H), 3.49 (ddd, J = 13.6, 7.6, 4.8 Hz, 1H), 3.13 (t, J = 5.2 Hz, 4H), 2.97 (s, 1H), 2.89 (d, J = 0.8 Hz, 1H), 2.07 (s, 1H), 1.94 (p, J = 6.4 Hz, 2H), 1.64 (dq, J = 20.0, 7.6 Hz, 1H). 13C NMR (101 MHz, CDCl3) δ 162.79, 160.13, 157.93, 151.25, 150.22, 141.69, 132.88, 128.93, 125.51, 124.37, 121.80, 121.04, 110.58, 68.21, 63.72, 46.03, 44.68, 43.34, 36.61, 31.53, 28.91, 25.88, 20.97. ESI HRMS, found 499.1756 (C23H26N6O5S, [M + H]+ requires 499.1758).
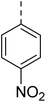
4.1.6.6 2-(4-((2-Nitrophenyl)sulfonyl)piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (24). 3-Nitrobenzenesulfonyl chloride was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 24 as a white solid. Yield = 97%. 1H NMR (400 MHz, chloroform-d) δ 8.01–7.94 (m, 1H), 7.73–7.64 (m, 2H), 7.62–7.56 (m, 1H), 7.53 (ddd, J = 9.2, 3.6, 2.4 Hz, 2H), 7.44 (d, J = 7.6 Hz, 1H), 7.14–7.06 (m, 1H), 6.09 (t, J = 5.2 Hz, 1H), 4.17 (qd, J = 7.2, 3.6 Hz, 1H), 4.04–3.99 (m, 4H), 3.93 (dt, J = 8.4, 6.8 Hz, 1H), 3.89–3.78 (m, 2H), 3.51 (ddd, J = 13.6, 7.6, 4.8 Hz, 1H), 3.37 (t, J = 5.2 Hz, 4H), 2.08 (s, 1H), 1.95 (p, J = 6.4 Hz, 2H), 1.71–1.62 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 160.12, 158.28, 151.59, 148.56, 133.78, 132.80, 131.49, 130.98, 130.88, 125.70, 124.07, 121.64, 120.99, 110.62, 77.55, 68.23, 63.70, 46.06, 44.69, 43.68, 28.91, 25.92, 20.88. ESI HRMS, found 499.1759 (C23H26N6O5S, [M + H]+ requires 499.1758).
4.1.6.7 2-(4-(Naphthalen-2-ylsulfonyl)piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (25). Naphthalene-2-sulfonyl chloride was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 25 as a yellow solid. Yield = 97%. 1H NMR (400 MHz, chloroform-d) δ 8.27 (d, J = 1.6 Hz, 1H), 7.90–7.84 (m, 2H), 7.83–7.78 (m, 1H), 7.68 (dd, J = 8.4, 1.6 Hz, 1H), 7.58–7.49 (m, 2H), 7.40 (dtd, J = 7.2, 3.6, 1.2 Hz, 2H), 7.31 (dd, J = 8.8, 1.2 Hz, 1H), 6.97 (ddd, J = 9.6, 6.8, 1.2 Hz, 1H), 5.88 (t, J = 5.6 Hz, 1H), 4.04 (qd, J = 7.2, 3.6 Hz, 1H), 3.94 (dd, J = 6.0, 4.0 Hz, 4H), 3.83 (dt, J = 8.4, 6.8 Hz, 1H), 3.76–3.67 (m, 2H), 3.39 (ddd, J = 13.6, 7.6, 4.8 Hz, 1H), 3.07 (t, J = 4.8 Hz, 4H), 2.00–1.90 (m, 1H), 1.84 (p, J = 6.4 Hz, 2H), 1.60–1.50 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 160.12, 158.25, 151.82, 135.00, 132.77, 132.67, 132.28, 129.33, 129.29, 129.23, 128.96, 128.01, 127.68, 125.84, 123.10, 121.52, 120.97, 110.60, 68.27, 46.35, 44.67, 43.34, 36.63, 31.54, 28.95, 25.98, 20.91. ESI HRMS, found 504.2068 (C27H29N5O3S, [M + H]+ requires 504.2064).
4.1.6.8 2-(4-((4-Nitrophenyl)sulfonyl)piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (26). 4-Nitrobenzenesulfonyl chloride was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 26 as white solid. Yield = 95%. 1H NMR (400 MHz, chloroform-d) δ 8.31–8.25 (m, 2H), 7.90–7.85 (m, 2H), 7.60 (dd, J = 8.0, 1.2 Hz, 1H), 7.53 (ddd, J = 9.6, 6.4, 1.2 Hz, 1H), 7.45 (dd, J = 8.4, 1.2 Hz, 1H), 7.11 (ddd, J = 9.6, 6.8, 1.2 Hz, 1H), 4.92 (s, 1H), 4.27 (t, J = 5.6 Hz, 2H), 4.02 (t, J = 5.6 Hz, 2H), 3.94 (dd, J = 6.0, 4.4 Hz, 4H), 3.67 (s, 1H), 3.05 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, CDCl3) δ 164.84, 162.61, 156.94, 154.22, 151.16, 150.27, 146.19, 143.6 (q, J = 35.75 Hz, 1C), 141.58, 133.72, 128.93, 126.85, 124.42, 123.86, 122.39, 118.34 (q, J = 266.64 Hz, 1C), 111.46, 63.73, 47.10, 46.02, 45.91, 43.31, 43.11, 36.54. ESI HRMS, found 590.1545 (C24H22F3N9O4S, [M + H]+ requires 590.1541).
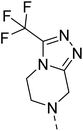
4.1.6.9 2-(4-((2-Nitrophenyl)sulfonyl)piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (27). 3-Nitrobenzenesulfonyl chloride was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 27 as a white solid. Yield = 97%. 1H NMR (400 MHz, chloroform-d) δ 8.03–7.96 (m, 1H), 7.75–7.66 (m, 3H), 7.65–7.58 (m, 2H), 7.56 (dd, J = 8.8, 1.6 Hz, 1H), 7.19 (ddd, J = 10.0, 6.8, 1.6 Hz, 1H), 5.05 (s, 2H), 4.39 (t, J = 5.6 Hz, 2H), 4.12 (t, J = 5.2 Hz, 2H), 4.00 (t, J = 5.2 Hz, 4H), 3.37 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, CDCl3) δ 164.83, 157.17, 154.32, 151.11, 148.48, 143.6 (q, J = 36.68 Hz, 1C), 133.91, 133.65, 131.60, 131.04, 130.88, 126.87, 124.14, 123.92, 122.27, 118.3 (q, J = 248.86 Hz, 1C), 111.45, 46.77, 46.26, 45.86 (2C), 43.71 (2C), 43.08. ESI HRMS, found 590.1542 (C24H22F3N9O4S, [M + H]+ requires 590.1541).
4.1.6.10 2-(4-(Naphthalen-2-ylsulfonyl)piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (28). Naphthalene-2-sulfonyl chloride was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 28 as a yellow solid. Yield = 97%. 1H NMR (400 MHz, chloroform-d) δ 8.34 (d, J = 2.0 Hz, 1H), 7.96 (t, J = 7.6 Hz, 2H), 7.91–7.86 (m, 1H), 7.75 (dd, J = 8.8, 2.0 Hz, 1H), 7.62 (dtd, J = 8.8, 4.4, 1.6 Hz, 3H), 7.57 (ddd, J = 6.8, 2.4, 1.6 Hz, 1H), 7.49 (dd, J = 8.4, 1.2 Hz, 1H), 7.14 (ddd, J = 9.6, 6.8, 1.2 Hz, 1H), 4.98 (s, 2H), 4.34 (t, J = 5.6 Hz, 2H), 4.05 (t, J = 5.2 Hz, 2H), 4.00 (dd, J = 6.0, 4.4 Hz, 4H), 3.13 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, CDCl3) δ 164.75, 157.00, 154.30, 151.12, 143.6 (q, J = 36.16 Hz, 1C), 134.93, 133.57, 132.80, 132.44, 132.19, 129.32, 129.26, 129.16, 128.96, 127.91, 127.67, 126.79, 123.83, 122.97, 122.12, 118.4 (q, J = 256.64 Hz, 1C), 111.36, 46.90, 46.10 (2C), 43.33 (2C), 43.11. ESI HRMS, found 595.1845 (C28H25F3N8O2S, [M + H]+ requires 595.1846).
4.1.6.11 4-((4-(4-(((Tetrahydrofuran-2-yl)methyl)amino)quinazolin-2-yl)piperazin-1-yl)sulfonyl)benzonitrile (29). 4-Cyanobenzenesulfonyl chloride was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 29 as an off-white solid. Yield = 98%. 1H NMR (400 MHz, chloroform-d) δ 7.82–7.72 (m, 4H), 7.44 (dd, J = 6.8, 1.2 Hz, 2H), 7.33 (dd, J = 9.2, 1.2 Hz, 1H), 7.02 (td, J = 14.8, 6.8, 1.2 Hz, 1H), 5.86 (t, J = 5.2 Hz, 1H), 4.06 (qd, J = 7.2, 3.6 Hz, 1H), 3.95 (dd, J = 6.4, 4.4 Hz, 4H), 3.85 (dt, J = 8.4, 6.4 Hz, 1H), 3.74 (tt, J = 13.6, 6.4, 3.6 Hz, 2H), 3.40 (ddd, J = 13.6, 7.6, 4.8 Hz, 1H), 3.03 (t, J = 5.2 Hz, 4H), 2.00–1.91 (m, 1H), 1.87 (p, J = 7.2 Hz, 2H), 1.62–1.52 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 160.11, 158.19, 151.84, 140.20, 132.90 (2C), 132.77, 128.33 (2C), 125.96, 121.62, 120.87, 117.20, 116.65, 110.60, 77.37, 68.22, 46.07 (2C), 44.68, 43.23 (2C), 28.92, 25.93. ESI HRMS, found 479.1862 (C24H26N6O3S, [M + H]+ requires 479.1860).
4.1.6.12 4-((4-(4-(3-(Trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazolin-2-yl)piperazin-1-yl)sulfonyl)benzonitrile (30). 4-Cyanobenzenesulfonyl chloride was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 30 as a white solid. Yield = 98%. 1H NMR (400 MHz, chloroform-d) δ 7.90–7.81 (m, 4H), 7.68 (dd, J = 8.4, 1.6 Hz, 1H), 7.61 (ddd, J = 10.0, 6.8, 1.2 Hz, 1H), 7.53 (dd, J = 8.4, 1.2 Hz, 1H), 7.19 (ddd, J = 9.6, 6.8, 1.2 Hz, 1H), 5.00 (s, 2H), 4.35 (t, J = 5.2 Hz, 2H), 4.09 (t, J = 5.6 Hz, 2H), 4.01 (t, J = 5.2 Hz, 4H), 3.10 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, CDCl3) δ 164.85, 156.96, 154.25, 151.16, 143.6 (q, J = 39.29 Hz, 1C), 140.03, 133.72, 132.97 (2C), 128.33 (2C), 126.86, 123.86, 122.38, 118.4 (q, J = 271.89 Hz, 1C), 117.15, 116.76, 111.47, 47.12, 46.03, 45.91 (2C), 43.30 (2C), 43.10. ESI HRMS, found 570.1646 (C25H22F3N9O2S, [M + H]+ requires 570.1642).
4.1.6.13 2-(4-(Pyridin-3-ylsulfonyl)piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (31). Pyridine-3-sulfonyl chloride was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 31 as an off-white solid. Yield = 95%. 1H NMR (400 MHz, chloroform-d) δ 8.93 (dd, J = 2.0, 0.8 Hz, 1H), 8.73 (dd, J = 5.2, 1.6 Hz, 1H), 7.98 (ddd, J = 8.0, 2.4, 1.6 Hz, 1H), 7.48–7.42 (m, 2H), 7.42–7.33 (m, 2H), 7.06–6.98 (m, 1H), 5.95 (s, 1H), 4.07 (qd, J = 7.2, 3.6 Hz, 1H), 3.97 (t, J = 5.2 Hz, 4H), 3.85 (dt, J = 8.4, 6.8 Hz, 1H), 3.80–3.70 (m, 2H), 3.41 (ddd, J = 13.6, 7.6, 4.8 Hz, 1H), 3.06 (t, J = 4.8 Hz, 4H), 2.02–1.92 (m, 1H), 1.87 (p, J = 6.4 Hz, 2H), 1.63–1.54 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 162.62, 160.11, 153.46, 148.51, 135.45, 132.80, 132.51, 125.85, 123.78 (2C), 121.67, 120.92, 110.58, 68.21, 63.72, 46.03, 44.69, 43.28, 36.52, 31.47, 28.92, 25.91. ESI HRMS, found 455.1856 (C22H26N6O3S, [M + H]+ requires 455.1860).
4.1.6.14 N-((Tetrahydrofuran-2-yl)methyl)-2-(4-(thiophen-2-ylsulfonyl)piperazin-1-yl)quinazolin-4-amine (32). Thiophene-2-sulfonyl chloride was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 32 as a white solid. Yield = 94%. 1H NMR (400 MHz, chloroform-d) δ 7.59 (dd, J = 4.8, 1.2 Hz, 1H), 7.54 (dd, J = 3.6, 1.2 Hz, 1H), 7.53–7.48 (m, 2H), 7.42 (dd, J = 8.4, 1.2 Hz, 1H), 7.14–7.05 (m, 2H), 5.93 (s, 1H), 4.15 (qd, J = 7.2, 3.6 Hz, 1H), 4.04 (t, J = 4.8 Hz, 4H), 3.92 (dt, J = 8.4, 6.8 Hz, 1H), 3.88–3.77 (m, 2H), 3.49 (ddd, J = 14.0, 7.6, 4.8 Hz, 1H), 3.14 (t, J = 4.8 Hz, 4H), 2.04 (ddd, J = 18.0, 12.0, 6.8 Hz, 1H), 1.94 (p, J = 6.8 Hz, 2H), 1.70–1.60 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 160.10, 158.32, 151.93, 135.84, 132.72, 132.55, 132.22, 127.70, 125.96, 121.49, 120.86, 110.59, 68.22, 46.22 (2C), 44.67, 43.11 (2C), 30.95, 28.91, 25.94. ESI HRMS, found 460.1474 (C21H25N5O3S2, [M + H]+ requires 460.1472).
4.1.6.15 2-(4-((5-Chlorothiophen-2-yl)sulfonyl)piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (33). 5-Chlorothiophene-2-sulfonyl chloride was treated with 2-(piperazin-1-yl)-N-((tetrahydrofuran-2-yl)methyl)quinazolin-4-amine (14a) and K2CO3 in anhydrous DMF to afford 33 as a white solid. Yield = 96%. 1H NMR (400 MHz, chloroform-d) δ 7.57–7.48 (m, 2H), 7.43 (dd, J = 8.4, 1.2 Hz, 1H), 7.31 (d, J = 4.0 Hz, 1H), 7.13–7.05 (m, 1H), 6.95 (d, J = 4.0 Hz, 1H), 5.95 (t, J = 5.2 Hz, 1H), 4.15 (qt, J = 7.2, 3.6 Hz, 1H), 4.05 (dd, J = 6.0, 4.4 Hz, 4H), 3.93 (dt, J = 8.0, 6.8 Hz, 1H), 3.89–3.76 (m, 2H), 3.50 (ddd, J = 13.6, 7.6, 4.8 Hz, 1H), 3.14 (t, J = 5.2 Hz, 4H), 2.09–2.00 (m, 1H), 1.95 (p, J = 6.4 Hz, 2H), 1.71–1.61 (m, 1H). 13C NMR (101 MHz, CDCl3) δ 160.13, 158.25, 151.89, 137.57, 133.84, 132.76, 131.92, 127.16 (2C), 125.95, 121.56, 120.90, 110.61, 68.23, 46.16 (2C), 44.68, 43.08 (2C), 28.92, 25.94. ESI HRMS, found 494.1082 (C21H24ClN5O3S2, [M + H]+ requires 494.1082).
4.1.6.16 2-(4-((5-Chlorothiophen-2-yl)sulfonyl)piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (34). 5-Chlorothiophene-2-sulfonyl chloride was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 34 as a white solid. Yield = 97%. 1H NMR (400 MHz, chloroform-d) δ 7.69 (dd, J = 8.0, 1.2 Hz, 1H), 7.62 (ddd, J = 10.0, 6.8, 1.6 Hz, 1H), 7.55 (dd, J = 8.4, 1.2 Hz, 1H), 7.33 (d, J = 4.0 Hz, 1H), 7.19 (ddd, J = 9.6, 6.8, 1.2 Hz, 1H), 6.96 (d, J = 4.0 Hz, 1H), 5.03 (s, 2H), 4.38 (t, J = 5.6 Hz, 2H), 4.11 (t, J = 5.6 Hz, 2H), 4.04 (dd, J = 6.0, 4.0 Hz, 4H), 3.14 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, CDCl3) δ 164.86, 157.04, 154.33, 151.12, 143.6 (q, J = 39.89 Hz, 1C), 137.74, 133.74, 133.67, 132.05, 127.20, 126.89, 123.88, 122.28, 118.34 (q, J = 251.79 Hz, 1C), 111.47, 46.94, 46.19, 45.98 (2C), 43.18 (2C), 43.12. ESI HRMS, found 585.0865 (C22H20ClF3N8O2S2, [M + H]+ requires 585.0864).
4.1.6.17 2-(4-(Pyridin-3-ylsulfonyl)piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (35). Pyridine-3-sulfonyl chloride was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 35 as an off-white solid. Yield = 94%. 1H NMR (400 MHz, chloroform-d) δ 8.92 (dd, J = 2.4, 0.8 Hz, 1H), 8.74 (dd, J = 4.8, 1.6 Hz, 1H), 8.03–7.97 (m, 1H), 7.60 (dd, J = 8.0, 1.2 Hz, 1H), 7.54 (ddd, J = 10.0, 6.8, 1.2 Hz, 1H), 7.47 (dd, J = 8.4, 1.2 Hz, 1H), 7.42 (ddd, J = 8.0, 4.8, 0.8 Hz, 1H), 7.11 (ddd, J = 9.6, 6.8, 1.6 Hz, 1H), 4.94 (s, 2H), 4.30 (t, J = 5.6 Hz, 2H), 4.02 (t, J = 5.2 Hz, 2H), 3.95 (dd, J = 6.0, 4.4 Hz, 4H), 3.06 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, DMSO) δ 164.87, 157.09, 154.21, 153.72, 152.14, 147.98, 143.1 (q, J = 31.81 Hz, 1C), 136.27, 133.94, 132.00, 126.07, 125.33, 125.10, 122.40, 118.7 (q, J = 274.21 Hz, 1C), 111.53, 63.02, 46.72, 45.98 (2C), 45.51, 43.26 (2C). ESI HRMS, found 546.1644 (C23H22F3N9O2S, [M + H]+ requires 546.1642).
4.1.6.18 2-(4-(Thiophen-2-ylsulfonyl)piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (36). Thiophene-2-sulfonyl chloride was treated with 2-(piperazin-1-yl)-4-(3-(trifluoromethyl)-5,6-dihydro-[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl)quinazoline (17a) and K2CO3 in anhydrous DMF to afford 36 as a white solid. Yield = 96%. 1H NMR (400 MHz, chloroform-d) δ 7.68 (dd, J = 8.0, 1.2 Hz, 1H), 7.63–7.59 (m, 2H), 7.56–7.53 (m, 2H), 7.18 (ddd, J = 9.6, 6.8, 1.2 Hz, 1H), 7.13 (dd, J = 5.2, 4.0 Hz, 1H), 5.03 (s, 2H), 4.38 (t, J = 5.2 Hz, 2H), 4.10 (t, J = 5.6 Hz, 2H), 4.03 (dd, J = 6.0, 4.0 Hz, 4H), 3.13 (t, J = 5.2 Hz, 4H). 13C NMR (101 MHz, CDCl3) δ 164.82, 157.09, 154.32, 151.12, 143.6 (q, J = 38.88 Hz, 1C), 135.64, 133.65, 132.69, 132.40, 127.79, 126.84, 123.89, 122.23, 118.3 (q, J = 246.64 Hz, 1C), 111.42, 46.89, 46.19, 46.04 (2C), 43.20 (2C), 43.12. ESI HRMS, found 551.1250 (C22H21F3N8O2S2, [M + H]+ requires 551.1254).
4.2. P. falciparum culture and inhibition assay
The P. falciparum 3D7 strain of malaria parasite was cultured following a well-described protocol, as previously outlined.64 Human erythrocytes (4% hematocrit) were supplemented with 0.5% albumax and 4% hematocrit in RPMI media, and they were synchronised by frequent sorbitol treatment. The growth inhibition assay was performed in triplicate and repeated twice. The desired concentration (0–100 µM) was achieved by adding 4 µL of the test compound to each well of the 96-well plate. Parasite culture (96 µL) at 1% parasitemia (3 to 10 h p.i.) and 2% hematocrit was added to each well of the 96-well plate and was incubated for the next 48 h. As a control experiment, along with test compounds, 4 µM chloroquine and 0.4% DMSO were used. As described by Smilkstein, et al.,65 a DNA fluorescent dye-binding assay using SYBR green was carried out after incubation for 48 h to assess the parasite's development in each well.4.3. Cytotoxicity assay
The associated cytotoxicity was investigated against the cancerous cell line A549 by employing the reported protocol66 and THP1 monocytes. The MTT assay was used to measure the cell viability, which is mostly reliant on the mitochondrial activity of the cell. It is based on the reduction of a tetrazolium salt [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] to a formazan. Dulbecco's modified Eagle's medium, augmented with 10% fetal bovine serum, was used to grow the A549 cells. The cells at a density of 1 × 104 cells per well in a 160 µL medium were incubated at 37 °C for 24 h in a 96-well plate to seed, followed by the addition of the test compounds in different concentrations. The media was discarded after incubation for 24 h, followed by the addition of 25 µL of 4 mmgg mmLL-1-1 of MTT, and further incubated for 4 h. After discarding the media, the formazan crystals were liquified by the addition of 200 µL of DMSO. To calculate the IC50 values, each experiment was independently performed in triplicate, and the cell viability was obtained by comparing control wells. The IC50 values were calculated using the nonlinear regression analysis method of GraphPad Prism, US.Moreover, THP1 monocytes were maintained in RPMI-1640 medium. Cells were cultured at 37 °C in a 5% CO2 environment. All media were supplemented with 10% fetal bovine serum (FBS). The cells were stimulated by administering Phorbol 12-Myristate 13-Acetate (PMA). The activated macrophages were seeded into a 96-well microtiter plate (2 × 104 cells per well) in five replicates for each condition and treated with compounds at concentrations of 100 µM and 500 µM. After 24 h, 100 µl of MTT stock solution (1 mg ml−1) was added to each well. The cells were then incubated at 37 °C for 45 minutes. The coloured formazan crystals were dissolved in 100 µl of DMSO. The OD values of the solutions were measured at 595 nm using a plate reader.
4.4. In silico studies
To calculate the binding free energy, the MM-GBSA calculations were carried out post-MD simulations using ‘thermal_mmgbsa.py’, a Python script of the Schrödinger.69 This script calculates the binding free energies of the ligand–protein complexes, an essential aspect of understanding the interaction dynamics. The general structure of the command used for running the script was “run thermal_mmgbsa.py -j [name_of_job] -o [output_file_name.csv] -step_size [step] -HOST [host_name] -NJOBS 10 -lig_asl [ligand_name] [input_.cms_file]”. In this study, the script was executed with a step size of 10 to analyse each frame from 0 to 200 ns.
Author contributions
HM: design, methodology, chemical synthesis, characterisation, rigorous computational studies, evaluation of the results, and writing the original draft of the manuscript. AP: designing and performing antimalarial assays, compiling and analysing the antimalarial activity results. AR and SB: designing and performing cytotoxicity assays on A549 cells. KS: synthesis of intermediates. DD and DK: design and supervision of cytotoxicity assays of THP1 cells, analysis of the results, and reviewing and editing the original draft. ATK: reviewing the original draft. PM: design and supervision of antimalarial assays, analysis of the results, and reviewing and editing the original draft. NH: conceptualisation, methodology, study supervision, resources, investigation of the results, reviewing and editing the original draft, and arrangement of funds.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra08515b.Acknowledgements
The author HM would like to acknowledge the Department of Biotechnology and Indian Council of Medical Research, Government of India, New Delhi, India, for providing financial support during the study as the M K Bhan-Young Researcher Fellowship Program (File No. HRD-16016/17/2025-HRD-DBT) and a Senior Research Fellowship (Award No. 45/02/2020-Nan/BMS), respectively. The PM acknowledges the DBT-Wellcome Team Science Grant (WTA/24/006) and the JC Bose Grant (DST/20/015) for providing financial support during the study. The NH acknowledges the Indian Council for Medical Research (ICMR), New Delhi, India (BMI/Adhoc/36/2022–23) for providing financial support during the study. We also thank the Rotary Blood Bank, New Delhi, India, for providing human red blood cells.References
- World Health Organisation, World Malaria Report 2024: Addressing Inequity in the Global Malaria Response, Geneva, 2024 Search PubMed.
- B. Hanboonkunupakarn and N. J. White, Advances and roadblocks in the treatment of malaria, Br. J. Clin. Pharmacol., 2022, 88, 374–382, DOI:10.1111/bcp.14474.
- H. Madhav and N. Hoda, An insight into the recent development of the clinical candidates for the treatment of malaria and their target proteins, Eur. J. Med. Chem., 2021, 210, 112955, DOI:10.1016/j.ejmech.2020.112955.
- P. Jagannathan and A. Kakuru, Malaria in 2022: Increasing challenges, cautious optimism, Nat. Commun., 2022, 13, 2678, DOI:10.1038/s41467-022-30133-w.
- R. W. van der Pluijm, R. Tripura, R. M. Hoglund, A. Pyae Phyo, D. Lek, A. ul Islam, A. R. Anvikar, P. Satpathi, S. Satpathi, P. K. Behera, A. Tripura, S. Baidya, M. Onyamboko, N. H. Chau, Y. Sovann, S. Suon, S. Sreng, S. Mao, S. Oun, S. Yen, C. Amaratunga, K. Chutasmit, C. Saelow, R. Runcharern, W. Kaewmok, N. T. Hoa, N. V. Thanh, B. Hanboonkunupakarn, J. J. Callery, A. K. Mohanty, J. Heaton, M. Thant, K. Gantait, T. Ghosh, R. Amato, R. D. Pearson, C. G. Jacob, S. Gonçalves, M. Mukaka, N. Waithira, C. J. Woodrow, M. P. Grobusch, M. van Vugt, R. M. Fairhurst, P. Y. Cheah, T. J. Peto, L. von Seidlein, M. Dhorda, R. J. Maude, M. Winterberg, N. T. Thuy-Nhien, D. P. Kwiatkowski, M. Imwong, P. Jittamala, K. Lin, T. M. Hlaing, K. Chotivanich, R. Huy, C. Fanello, E. Ashley, M. Mayxay, P. N. Newton, T. T. Hien, N. Valecha, F. Smithuis, S. Pukrittayakamee, A. Faiz, O. Miotto, J. Tarning, N. P. J. Day, N. J. White, A. M. Dondorp, R. W. van der Pluijm, R. Tripura, R. M. Hoglund, A. P. Phyo, D. Lek, A. ul Islam, A. R. Anvikar, P. Satpathi, S. Satpathi, P. K. Behera, A. Tripura, S. Baidya, M. Onyamboko, N. H. Chau, Y. Sovann, S. Suon, S. Sreng, S. Mao, S. Oun, S. Yen, C. Amaratunga, K. Chutasmit, C. Saelow, R. Runcharern, W. Kaewmok, N. T. Hoa, N. V. Thanh, B. Hanboonkunupakarn, J. J. Callery, A. K. Mohanty, J. Heaton, M. Thant, K. Gantait, T. Ghosh, R. Amato, R. D. Pearson, C. G. Jacob, S. Gonçalves, M. Mukaka, N. Waithira, C. J. Woodrow, M. P. Grobusch, M. van Vugt, R. M. Fairhurst, P. Y. Cheah, T. J. Peto, L. von Seidlein, M. Dhorda, R. J. Maude, M. Winterberg, N. T. Thuy-Nhien, D. P. Kwiatkowski, M. Imwong, P. Jittamala, K. Lin, T. M. Hlaing, K. Chotivanich, R. Huy, C. Fanello, E. Ashley, M. Mayxay, P. N. Newton, T. T. Hien, N. Valeche, F. Smithuis, S. Pukrittayakamee, A. Faiz, O. Miotto, J. Tarning, N. P. Day, N. J. White and A. M. Dondorp, Triple artemisinin-based combination therapies versus artemisinin-based combination therapies for uncomplicated Plasmodium falciparum malaria: a multicentre, open-label, randomised clinical trial, Lancet, 2020, 395, 1345–1360, DOI:10.1016/S0140-6736(20)30552-3.
- T. J. Peto, R. Tripura, J. J. Callery, D. Lek, H. D. T. Nghia, C. Nguon, N. T. H. Thuong, R. W. van der Pluijm, N. T. P. Dung, M. Sokha, V. Van Luong, L. T. Long, Y. Sovann, J. Duanguppama, N. Waithira, R. M. Hoglund, P. Chotsiri, N. H. Chau, A. Ruecker, C. Amaratunga, M. Dhorda, O. Miotto, R. J. Maude, H. Rekol, K. Chotivanich, J. Tarning, L. von Seidlein, M. Imwong, M. Mukaka, N. P. J. Day, T. T. Hien, N. J. White and A. M. Dondorp, Triple therapy with artemether–lumefantrine plus amodiaquine versus artemether–lumefantrine alone for artemisinin-resistant, uncomplicated falciparum malaria: an open-label, randomised, multicentre trial, Lancet Infect. Dis., 2022, 22, 867–878, DOI:10.1016/S1473-3099(21)00692-7.
- T. D. Ashton, A. Ngo, P. Favuzza, H. E. Bullen, M. R. Gancheva, O. Romeo, M. Parkyn Schneider, N. Nguyen, R. W. J. Steel, S. Duffy, K. N. Lowes, H. J. Sabroux, V. M. Avery, J. A. Boddey, D. W. Wilson, A. F. Cowman, P. R. Gilson and B. E. Sleebs, Property activity refinement of 2-anilino 4-amino substituted quinazolines as antimalarials with fast acting asexual parasite activity, Bioorg. Chem., 2021, 117, 105359, DOI:10.1016/j.bioorg.2021.105359.
- S. Malasala, M. N. Ahmad, R. Akunuri, M. Shukla, G. Kaul, A. Dasgupta, Y. V. Madhavi, S. Chopra and S. Nanduri, Synthesis and evaluation of new quinazoline-benzimidazole hybrids as potent anti-microbial agents against multidrug resistant Staphylococcus aureus and Mycobacterium tuberculosis, Eur. J. Med. Chem., 2021, 212, 112996, DOI:10.1016/j.ejmech.2020.112996.
- S. S. Al Neyadi, A. G. Al Blooshi, H. L. Nguyen and M. A. Alnaqbi, UiO-66-NH 2 as an effective solid support for quinazoline derivatives for antibacterial agents against Gram-negative bacteria, New J. Chem., 2021, 45, 20386–20395, 10.1039/D1NJ03749H.
- S. Shagufta and I. Ahmad, An insight into the therapeutic potential of quinazoline derivatives as anticancer agents, Medchemcomm, 2017, 8, 871–885, 10.1039/C7MD00097A.
- Y. Mizukawa, M. Ikegami-Kawai, M. Horiuchi, M. Kaiser, M. Kojima, S. Sakanoue, S. Miyagi, C. Nanga Chick, H. Togashi, M. Tsubuki, M. Ihara, T. Usuki and I. Itoh, Quest for a potent antimalarial drug lead: Synthesis and evaluation of 6,7-dimethoxyquinazoline-2,4-diamines, Bioorg. Med. Chem., 2021, 33, 116018, DOI:10.1016/j.bmc.2021.116018.
- A. Dutta, P. Trivedi, P. S. Gehlot, D. Gogoi, R. Hazarika, P. Chetia, A. Kumar, A. K. Chaliha, V. Chaturvedi and D. Sarma, Design and Synthesis of Quinazolinone-Triazole Hybrids as Potent Anti-Tubercular Agents, ACS Appl. Bio Mater., 2022, 5(9), 4413–4424, DOI:10.1021/acsabm.2c00562.
- P. R. Gilson, C. Tan, K. E. Jarman, K. N. Lowes, J. M. Curtis, W. Nguyen, A. E. Di Rago, H. E. Bullen, B. Prinz, S. Duffy, J. B. Baell, C. A. Hutton, H. Jousset Subroux, B. S. Crabb, V. M. Avery, A. F. Cowman and B. E. Sleebs, Optimization of 2-Anilino 4-Amino Substituted Quinazolines into Potent Antimalarial Agents with Oral in Vivo Activity, J. Med. Chem., 2017, 60, 1171–1188, DOI:10.1021/acs.jmedchem.6b01673.
- A. Bouchut, D. Rotili, C. Pierrot, S. Valente, S. Lafitte, J. Schultz, U. Hoglund, R. Mazzone, A. Lucidi, G. Fabrizi, D. Pechalrieu, P. B. Arimondo, T. S. Skinner-Adams, M. J. Chua, K. T. Andrews, A. Mai and J. Khalife, Identification of novel quinazoline derivatives as potent antiplasmodial agents, Eur. J. Med. Chem., 2019, 161, 277–291, DOI:10.1016/j.ejmech.2018.10.041.
- T. Fröhlich, C. Reiter, M. M. Ibrahim, J. Beutel, C. Hutterer, I. Zeitträger, H. Bahsi, M. Leidenberger, O. Friedrich, B. Kappes, T. Efferth, M. Marschall and S. B. Tsogoeva, Synthesis of Novel Hybrids of Quinazoline and Artemisinin with High Activities against Plasmodium falciparum , Human Cytomegalovirus, and Leukemia Cells, ACS Omega, 2017, 2, 2422–2431, DOI:10.1021/acsomega.7b00310.
- A. H. Shntaif, S. Khan, G. Tapadiya, A. Chettupalli, S. Saboo, M. S. Shaikh, F. Siddiqui and R. R. Amara, Rational drug design, synthesis, and biological evaluation of novel N-(2-arylaminophenyl)-2,3-diphenylquinoxaline-6-sulfonamides as potential antimalarial, antifungal, and antibacterial agents, Digit. Chinese Med., 2021, 4, 290–304, DOI:10.1016/j.dcmed.2021.12.004.
- M. N. Peerzada, P. Khan, K. Ahmad, M. I. Hassan and A. Azam, Synthesis, characterization and biological evaluation of tertiary sulfonamide derivatives of pyridyl-indole based heteroaryl chalcone as potential carbonic anhydrase IX inhibitors and anticancer agents, Eur. J. Med. Chem., 2018, 155, 13–23, DOI:10.1016/j.ejmech.2018.05.034.
- K. N. de Oliveira, P. Costa, J. R. Santin, L. Mazzambani, C. Bürger, C. Mora, R. J. Nunes and M. M. de Souza, Synthesis and antidepressant-like activity evaluation of sulphonamides and sulphonyl-hydrazones, Bioorg. Med. Chem., 2011, 19, 4295–4306, DOI:10.1016/j.bmc.2011.05.056.
- J. A. Ezugwu, U. C. Okoro, M. A. Ezeokonkwo, K. S. Hariprasad, M. Rudrapal, D. I. Ugwu, N. Gogoi, D. Chetia, I. Celik and O. C. Ekoh, Design, Synthesis, Molecular Docking, Molecular Dynamics and In Vivo Antimalarial Activity of New Dipeptide-Sulfonamides, ChemistrySelect, 2022, 7(5), e202103908, DOI:10.1002/slct.202103908.
- A. Sabt, O. M. Abdelhafez, R. S. El-Haggar, H. M. F. Madkour, W. M. Eldehna, E. E.-D. A. El-Khrisy, M. A. Abdel-Rahman and L. A. Rashed, Novel coumarin-6-sulfonamides as apoptotic anti-proliferative agents: synthesis, in vitro biological evaluation, and QSAR studies, J. Enzyme Inhib. Med. Chem., 2018, 33, 1095–1107, DOI:10.1080/14756366.2018.1477137.
- M. V. Papadopoulou, W. D. Bloomer, H. S. Rosenzweig, A. Arena, F. Arrieta, J. C. J. Rebolledo and D. K. Smith, Nitrotriazole- and Imidazole-Based Amides and Sulfonamides as Antitubercular Agents, Antimicrob. Agents Chemother., 2014, 58, 6828–6836, DOI:10.1128/AAC.03644-14.
- R. Pingaew, P. Mandi, V. Prachayasittikul, A. Thongnum, S. Prachayasittikul, S. Ruchirawat and V. Prachayasittikul, Investigations on Anticancer and Antimalarial Activities of Indole-Sulfonamide Derivatives and In Silico Studies, ACS Omega, 2021, 6, 31854–31868, DOI:10.1021/acsomega.1c04552.
- V. R. Karpina, S. S. Kovalenko, S. M. Kovalenko, O. G. Drushlyak, N. D. Bunyatyan, V. A. Georgiyants, V. V. Ivanov, T. Langer and L. Maes, A Novel Series of [1,2,4]Triazolo[4,3-a]Pyridine Sulfonamides as Potential Antimalarial Agents: In Silico Studies, Synthesis and In Vitro Evaluation, Molecules, 2020, 25, 4485, DOI:10.3390/molecules25194485.
- S. Dhillon, Sitagliptin, Drugs, 2010, 70, 489–512, DOI:10.2165/11203790-000000000-00000.
- J. Xing, R. Zhang, X. Jiang, T. Hu, X. Wang, G. Qiao, J. Wang, F. Yang, X. Luo, K. Chen, J. Shen, C. Luo, H. Jiang and M. Zheng, Rational design of 5-((1H-imidazol-1-yl)methyl)quinolin-8-ol derivatives as novel bromodomain-containing protein 4 inhibitors, Eur. J. Med. Chem., 2019, 163, 281–294, DOI:10.1016/j.ejmech.2018.11.018.
- X. Chang, D. Sun, D. Shi, G. Wang, Y. Chen, K. Zhang, H. Tan, J. Liu, B. Liu and L. Ouyang, Design, synthesis, and biological evaluation of quinazolin-4(3H)-one derivatives co-targeting poly(ADP-ribose) polymerase-1 and bromodomain containing protein 4 for breast cancer therapy, Acta Pharm. Sin. B, 2021, 11, 156–180, DOI:10.1016/j.apsb.2020.06.003.
- X. Jiang, J. Zhou, J. Ai, Z. Song, X. Peng, L. Xing, Y. Xi, J. Guo, Q. Yao, J. Ding, M. Geng and A. Zhang, Novel tetracyclic benzo[b]carbazolones as highly potent and orally bioavailable ALK inhibitors: Design, synthesis, and structure—activity relationship study, Eur. J. Med. Chem., 2015, 105, 39–56, DOI:10.1016/j.ejmech.2015.10.005.
- M. Patil, A. Noonikara-Poyil, S. D. Joshi, S. A. Patil, S. A. Patil, A. M. Lewis and A. Bugarin, Synthesis, molecular docking studies, and in vitro antimicrobial evaluation of piperazine and triazolo-pyrazine derivatives, Mol. Divers., 2022, 26, 827–841, DOI:10.1007/s11030-021-10190-x.
- R. Raveesha, K. Y. Kumar, M. S. Raghu, S. B. B. Prasad, A. Alsalme, P. Krishnaiah and M. K. Prashanth, Synthesis, in silico ADME, toxicity prediction and molecular docking studies of N-substituted [1,2,4]triazolo[4,3-a]pyrazine derivatives as potential anticonvulsant agents, J. Mol. Struct., 2022, 1255, 132407, DOI:10.1016/j.molstruc.2022.132407.
- G. Shan, Z. Peng, Y. Li, D. Li, Y. Li, S. Meng, L. Gao, J. Jiang and Z. Li, A novel class of geldanamycin derivatives as HCV replication inhibitors targeting on Hsp90: synthesis, structure–activity relationships and anti-HCV activity in GS4.3 replicon cells, J. Antibiot., 2011, 64, 177–182, DOI:10.1038/ja.2010.161.
- H. R. Lawrence, K. Mahajan, Y. Luo, D. Zhang, N. Tindall, M. Huseyin, H. Gevariya, S. Kazi, S. Ozcan, N. P. Mahajan and N. J. Lawrence, Development of Novel ACK1/TNK2 Inhibitors Using a Fragment-Based Approach, J. Med. Chem., 2015, 58, 2746–2763, DOI:10.1021/jm501929n.
- M. M. Elbadawi, W. M. Eldehna, A. A. Abd El-Hafeez, W. R. Somaa, A. Albohy, S. T. Al-Rashood, K. K. Agama, E. B. Elkaeed, P. Ghosh, Y. Pommier and M. Abe, 2-Arylquinolines as novel anticancer agents with dual EGFR/FAK kinase inhibitory activity: synthesis, biological evaluation, and molecular modelling insights, J. Enzyme Inhib. Med. Chem., 2022, 37, 355–378, DOI:10.1080/14756366.2021.2015344.
- S. K. Balam, S. K. Krishnammagari, J. Soora Harinath, S. P. Sthanikam, S. S. Chereddy, V. R. Pasupuleti, N. K. Yellapu, V. G. R. Peddiahgari and S. R. Cirandur, Synthesis of N-(3-picolyl)-based 1,3,2λ5-benzoxazaphosphinamides as potential 11β-HSD1 enzyme inhibitors, Med. Chem. Res., 2015, 24, 1119–1135, DOI:10.1007/s00044-014-1194-7.
- E. Jameel, H. Madhav, P. Agrawal, M. K. Raza, S. Ahmedi, A. Rahman, N. Shahid, K. Shaheen, C. H. Gajra, A. Khan, M. Z. Malik, M. A. Imam, M. Kalamuddin, J. Kumar, D. Gupta, S. M. Nayeem, N. Manzoor, A. Mohammad, P. Malhotra and N. Hoda, Identification of new oxospiro chromane quinoline-carboxylate antimalarials that arrest parasite growth at ring stage, J. Biomol. Struct. Dyn., 2023, 1–22, DOI:10.1080/07391102.2023.2188959.
- Z. Hu, H. Dong, Z. Si, Y. Zhao and Y. Liang, Synthesis and Antibacterial Activity of Novel Triazolo[4,3-a]pyrazine Derivatives, Molecules, 2023, 28, 7876, DOI:10.3390/molecules28237876.
- R. Raveesha, M. G. D. Kumar and S. B. B. Prasad, Synthesis of 3-Trifluoromethyl-5,6-dihydro-[1,2,4]triazolo Pyrazine Derivatives and Their Anti-Cancer Studies, Molbank, 2020, 2020, M1173, DOI:10.3390/M1173.
- D. Agarwal, R. D. Gupta and S. K. Awasthi, Are Antimalarial Hybrid Molecules a Close Reality or a Distant Dream?, Antimicrob. Agents Chemother., 2017, 61 DOI:10.1128/AAC.00249-17.
- D. Johnson, I. Jenkins, C. Huxley, M. Coster, K. Lum, J. White, V. Avery and R. Davis, Synthesis of New Triazolopyrazine Antimalarial Compounds, Molecules, 2021, 26, 2421, DOI:10.3390/molecules26092421.
- P. Mignon, Influence of the - interaction on the hydrogen bonding capacity of stacked DNA/RNA bases, Nucleic Acids Res., 2005, 33, 1779–1789, DOI:10.1093/nar/gki317.
- P. S. Sijwali and P. J. Rosenthal, Gene disruption confirms a critical role for the cysteine protease falcipain-2 in hemoglobin hydrolysis by Plasmodium falciparum, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4384–4389, DOI:10.1073/pnas.0307720101.
- P. A. Moura, J. B. Dame and D. A. Fidock, Role of Plasmodium falciparum Digestive Vacuole Plasmepsins in the Specificity and Antimalarial Mode of Action of Cysteine and Aspartic Protease Inhibitors, Antimicrob. Agents Chemother., 2009, 53, 4968–4978, DOI:10.1128/AAC.00882-09.
- J. M. Matz, Plasmodium's bottomless pit: properties and functions of the malaria parasite's digestive vacuole, Trends Parasitol., 2022, 38, 525–543, DOI:10.1016/j.pt.2022.02.010.
- D. Jani, R. Nagarkatti, W. Beatty, R. Angel, C. Slebodnick, J. Andersen, S. Kumar and D. Rathore, HDP—A Novel Heme Detoxification Protein from the Malaria Parasite, PLoS Pathog., 2008, 4, e1000053, DOI:10.1371/journal.ppat.1000053.
- R. Ettari, S. Previti, C. Di Chio and M. Zappalà, Falcipain-2 and Falcipain-3 Inhibitors as Promising Antimalarial Agents, Curr. Med. Chem., 2021, 28, 3010–3031, DOI:10.2174/0929867327666200730215316.
- P. S. Sijwali, B. R. Shenai and P. J. Rosenthal, Folding of the Plasmodium falciparum Cysteine Protease Falcipain-2 Is Mediated by a Chaperone-like Peptide and Not the Prodomain, J. Biol. Chem., 2002, 277, 14910–14915, DOI:10.1074/jbc.M109680200.
- M. Mishra, V. Singh and S. Singh, Structural Insights Into Key Plasmodium Proteases as Therapeutic Drug Targets, Front. Microbiol., 2019, 10, 394, DOI:10.3389/fmicb.2019.00394.
- J. Lin, X. Yan, Z. Chung, C. W. Liew, A. El Sahili, E. V. Pechnikova, P. R. Preiser, Z. Bozdech, Y.-G. Gao and J. Lescar, Inhibition of falcilysin from Plasmodium falciparum by interference with its closed-to-open dynamic transition, Commun. Biol., 2024, 7, 1070, DOI:10.1038/s42003-024-06774-6.
- G. Wirjanata, J. Lin, J. M. Dziekan, A. El Sahili, Z. Chung, S. Tjia, N. E. Binte Zulkifli, J. Boentoro, R. Tham, L. S. Jia, K. D. Go, H. Yu, A. Partridge, D. Olsen, N. Prabhu, R. M. Sobota, P. Nordlund, J. Lescar and Z. Bozdech, Identification of an inhibitory pocket in falcilysin provides a new avenue for malaria drug development, Cell Chem. Biol., 2024, 31, 743–759, DOI:10.1016/j.chembiol.2024.03.002.
- S. Eagon, M. Howland, M. Heying, E. Callant, N. Brar, E. Pompa and J. P. Mallari, Identification of Plasmodium falciparum falcilysin inhibitors by a virtual screen, Bioorg. Med. Chem. Lett., 2021, 52, 128394, DOI:10.1016/j.bmcl.2021.128394.
- G. Kahlon, R. Lira, N. Masvlov, E. Pompa, N. Brar, S. Eagon, M. O. Anderson, A. Andaya, J. P. Chance, H. Fejzic, A. Keniston, N. Huynh, N. Celis, B. Vidal, N. Trieu, P. Rodriguez and J. P. Mallari, Structure guided development of potent piperazine-derived hydroxamic acid inhibitors targeting falcilysin, Bioorg. Med. Chem. Lett., 2021, 32, 127683, DOI:10.1016/j.bmcl.2020.127683.
- H. Guterres and W. Im, Improving Protein–Ligand Docking Results with High-Throughput Molecular Dynamics Simulations, J. Chem. Inf. Model., 2020, 60, 2189–2198, DOI:10.1021/acs.jcim.0c00057.
- L. Fusani, D. S. Palmer, D. O. Somers and I. D. Wall, Exploring Ligand Stability in Protein Crystal Structures Using Binding Pose Metadynamics, J. Chem. Inf. Model., 2020, 60, 1528–1539, DOI:10.1021/acs.jcim.9b00843.
- H. Madhav, S. A. Abdel-Rahman, M. A. Hashmi, M. A. Rahman, M. Rehan, K. Pal, S. M. Nayeem, M. T. Gabr and N. Hoda, Multicomponent Petasis reaction for the identification of pyrazine based multi-target directed anti-Alzheimer’s agents: In-silico design, synthesis, and characterization, Eur. J. Med. Chem., 2023, 254, 115354, DOI:10.1016/j.ejmech.2023.115354.
- K. Teilum, J. G. Olsen and B. B. Kragelund, Functional aspects of protein flexibility, Cell. Mol. Life Sci., 2009, 66, 2231–2247, DOI:10.1007/s00018-009-0014-6.
- S. Fatima, D. G. Boggs, N. Ali, P. J. Thompson, M. C. Thielges, J. Bridwell-Rabb and L. Olshansky, Engineering a Conformationally Switchable Artificial Metalloprotein, J. Am. Chem. Soc., 2022, 144, 21606–21616, DOI:10.1021/jacs.2c08885.
- X. Song, L. Bao, C. Feng, Q. Huang, F. Zhang, X. Gao and R. Han, Accurate Prediction of Protein Structural Flexibility by Deep Learning Integrating Intricate Atomic Structures and Cryo-EM Density Information, Nat. Commun., 2024, 15, 5538, DOI:10.1038/s41467-024-49858-x.
- J. F. Fatriansyah, A. G. Boanerges, S. R. Kurnianto, A. F. Pradana, Fadilah and S. N. Surip, Molecular Dynamics Simulation of Ligands from Anredera cordifolia (Binahong) to the Main Protease (Mpro) of SARS-CoV-2, J. Trop. Med., 2022, 2022, 1–13, DOI:10.1155/2022/1178228.
- S. Ghahremanian, M. M. Rashidi, K. Raeisi and D. Toghraie, Molecular dynamics simulation approach for discovering potential inhibitors against SARS-CoV-2: A structural review, J. Mol. Liq., 2022, 354, 118901, DOI:10.1016/j.molliq.2022.118901.
- I. Yasuda, K. Endo, E. Yamamoto, Y. Hirano and K. Yasuoka, Differences in ligand-induced protein dynamics extracted from an unsupervised deep learning approach correlate with protein–ligand binding affinities, Commun. Biol., 2022, 5, 481, DOI:10.1038/s42003-022-03416-7.
- T. Yu, N. Sudhakar and C. D. Okafor, Illuminating ligand-induced dynamics in nuclear receptors through MD simulations, Biochim. Biophys. Acta - Gene Regul. Mech., 2024, 1867, 195025, DOI:10.1016/j.bbagrm.2024.195025.
- S. De Vita, M. G. Chini, G. Bifulco and G. Lauro, Insights into the Ligand Binding to Bromodomain-Containing Protein 9 (BRD9): A Guide to the Selection of Potential Binders by Computational Methods, Molecules, 2021, 26, 7192, DOI:10.3390/molecules26237192.
- S. Manzoor, S. K. Prajapati, S. Majumdar, K. Raza, M. T. Gabr, S. Kumar, K. Pal, H. Rashid, S. Kumar, S. Krishnamurthy and N. Hoda, Discovery of new phenyl sulfonyl-pyrimidine carboxylate derivatives as the potential multi-target drugs with effective anti-Alzheimer’s action: Design, synthesis, crystal structure and in-vitro biological evaluation, Eur. J. Med. Chem., 2021, 215, 113224, DOI:10.1016/j.ejmech.2021.113224.
- H. Hu, Y. Dong, M. Li, R. Wang, X. Zhang, P. Gong and Y. Zhao, Design, synthesis and biological evaluation of novel thieno[3,2-d]pyrimidine and quinazoline derivatives as potent antitumor agents, Bioorg. Chem., 2019, 90, 103086, DOI:10.1016/j.bioorg.2019.103086.
- A. Singh, M. Kalamuddin, M. Maqbool, A. Mohmmed, P. Malhotra and N. Hoda, Quinoline carboxamide core moiety-based compounds inhibit P. falciparumfalcipain-2: Design, synthesis and antimalarial efficacy studies, Bioorg. Chem., 2020, 104514, DOI:10.1016/j.bioorg.2020.104514.
- M. Smilkstein, N. Sriwilaijaroen, J. X. Kelly, P. Wilairat and M. Riscoe, Simple and Inexpensive Fluorescence-Based Technique for High-Throughput Antimalarial Drug Screening, Antimicrob. Agents Chemother., 2004, 48, 1803–1806, DOI:10.1128/AAC.48.5.1803-1806.2004.
- K. Pal, M. K. Raza, J. Legac, A. Rahman, S. Manzoor, S. Bhattacharjee, P. J. Rosenthal and N. Hoda, Identification, in-vitro anti-plasmodial assessment and docking studies of series of tetrahydrobenzothieno[2,3-d]pyrimidine-acetamide molecular hybrids as potential antimalarial agents, Eur. J. Med. Chem., 2023, 248, 115055, DOI:10.1016/j.ejmech.2022.115055.
- T. A. Halgren, Identifying and Characterizing Binding Sites and Assessing Druggability, J. Chem. Inf. Model., 2009, 49, 377–389, DOI:10.1021/ci800324m.
- R. A. Friesner, R. B. Murphy, M. P. Repasky, L. L. Frye, J. R. Greenwood, T. A. Halgren, P. C. Sanschagrin and D. T. Mainz, Extra Precision Glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein–Ligand Complexes, J. Med. Chem., 2006, 49, 6177–6196, DOI:10.1021/jm051256o.
- V. Pandya, P. Rao, J. Prajapati, R. M. Rawal and D. Goswami, Pinpointing top inhibitors for GSK3β from pool of indirubin derivatives using rigorous computational workflow and their validation using molecular dynamics (MD) simulations, Sci. Rep., 2024, 14, 49, DOI:10.1038/s41598-023-50992-7.
Footnote |
| † These authors contributed equally to the article (Joint First Authors). |
| This journal is © The Royal Society of Chemistry 2026 |