 Open Access Article
Open Access ArticleBiosynthesis of Pseudomonas aeruginosa mediated silver nanoparticles for remediation of crude oil contaminated water
Agatha Abamhekhelu Ikhumetsea,
Peter Olabisi Abioyeab,
Oluwafemi Adebayo Oyewole *a,
Abdulsalami Sanni Kovoc and
Udeme Joshua Josiah Ijaha
*a,
Abdulsalami Sanni Kovoc and
Udeme Joshua Josiah Ijaha
aDepartment of Microbiology, Federal University of Technology, Minna, Nigeria. E-mail: oa.oyewole@futminna.edu.ng; Tel: +2348140031875
bDepartment of Public Health, Federal University of Technology, Minna, Nigeria
cDepartment of Chemical Engineering, Federal University of Technology, Minna, Nigeria
First published on 2nd January 2026
Abstract
This study was aimed at biosynthesizing silver nanoparticle (AgNP) for remediation of total petroleum hydrocarbon (TPH) from crude oil-contaminated water samples from Nigeria's Niger-Delta ecosystem. The physicochemical and bacteriological analyses of the water samples were ascertained by standard techniques. Screening for hydrocarbon utilization was done for all isolates and the highest degrader was selected for biosynthesis of AgNP and characterized. TPH were adsorbed using the AgNP. The TPH response was designed and optimized using response surface methodology (RSM). The TPH in some of the water samples were above permissible limits in surface water given by Department of Petroleum Resources (DPR) and ranged from 6.55 ± 0.91 to 2324.36 ± 4.23 mg L−1. The Total hydrocarbon utilizing bacterial counts ranged from 0.33 ± 0.01 × 103 cfu mL−1 to 13.67 ± 1.20 × 103 cfu mL−1. The isolate that recorded highest hydrocarbon degradation of 97.80% was identified as Pseudomonas aeruginosa strain AgA (PAE). The AgNP showed a peak at 425 nm, spherical, particle size distribution of 34.60 ± 13.04 nm, and a significant signal in the silver region, crystalline structure with an average mean crystallite size of 18.01504 ± 4.03 nm. The coefficient of determination (R2) had a value of 0.9190, the RSM for design and optimization demonstrated a satisfactory match for the 2FI regression model for the AgNP. At optimal contact time (22.5 min/100 mL), stirring speed (1620 rpm/100 mL), dosage (0.3206 g) and temperature (67.5 °C), the maximum expected TPH removal for PAE was 99.98%, demonstrating that the bacterial was helpful in synthesizing AgNPs to improve effective removal of pollutants from water.
1 Introduction
Concern among the public is rising as a variety of dangerous substances are being unintentionally or intentionally discharged into the environment.1,2 Among these substances are petroleum hydrocarbons, which are continuously discharged in significant quantities into the Niger Delta aquatic environment, which is the backbone of the Nigerian economy through several routes such as pipeline failures, natural seepages and discharges during tank or ship transportation.3,4 Oil spills have become more frequent due to as a result of hydrocarbons building up over time in delicate natural environments due to increased exploration and inadequate environmental management techniques.5–8 These spills can cause several illnesses, such as kidney disease, potential bone marrow destruction and an elevated risk of cancer, because they include harmful carcinogenic and mutagenic substances.9 Furthermore, oil spills have a detrimental effect on the ecosystem, agricultural output, water bodies, bird productivity and the spread of the microbial community across the environment.1,10 Nitric oxide (NO) released from oil spills contributes to atmospheric pollution through oxidation of nitrogen-containing hydrocarbons and oxidative stress in marine environments.11,12 The severe repercussions of environmental oil pollution therefore necessitate the use of effective cleanup strategies.13The physical and chemical approaches used in conventional crude oil remediation are expensive and have the potential to release secondary pollutants into water bodies.8,14 Bacteria and other microbes survive and eventually thrive in this environment, utilizing the hydrocarbons in petroleum products as a source of energy and carbon. The biodegradation of hydrocarbons by a naturally-occurring population of microorganisms is one of the primary techniques for eliminating petroleum contamination from the environment.3 Bioremediation uses a variety of microorganisms, especially naturally-occurring bacteria.15 Even in the face of extreme environmental circumstances, such as those brought on by pollution from crude oil, some bacteria can live. They accomplish this by using a range of adaptation techniques, such as metabolic pathway modifications and spore production permeability.10 Therefore, it is possible to argue that biological techniques to remediation are beneficial, but they are also directly impacted by variables like the time, oil content, nutrients and the availability of suitable microbes.16
Nanoparticles (NPs) are a collection of zero-dimensional atoms in different shapes that are separated into two primary groups: organic and inorganic.17 The physical and chemical characteristics of metal nanoparticles such as silver nanoparticles (AgNPs) differ greatly from those of traditional bulk metal since they are inorganic nanoparticles. Because of their special qualities, including high conductivity, size, form, chemical stability, crystallinity and superior adsorptive qualities, researchers now employ AgNPs more than any other inorganic nanoparticles.18 The synthesis of nanoparticles can be done in a variety of ways, such as chemical, biological and physical.19 Environmental issues arise from the use of hazardous and combustible chemicals as stabilizing and regenerating agents in the synthesis of nanoparticles by chemical processes.17 However, one drawback of physical approaches is that they take a lot of energy to sustain the temperature and pressure needed for reaction stages.18 As a result, biological synthesis using living bacteria, fungi and plant cells is thought to be the most environmentally-friendly process and a potential substitute for physical or chemical manufacturing,20 because it is inexpensive, environmentally friendly, and uses less toxic chemicals.21,22 Furthermore, because biological media contain both reducing and stabilizing agents, biological approaches can synthesize NPs in a single step, as opposed to the two-step process employed by physicochemical methods (for NP reduction and stabilization).17 Previous studies indicated the potential for Pseudomonas aeruginosa in hydrocarbon remediation through biodegradation and the production of biosurfactants. For instance, Muthukumar et al.23 and Rehman et al.24 reported 72% crude oil removal under aerobic conditions using novel P. aeruginosa strains and 65–70% degradation in biosurfactant-enhanced systems, respectively. Ahmadi et al.25 discussed soil remediation with pyrene, emphasizing assisted desorption by biosurfactants. However, none of these studies have elaborated on the use of biologically synthesized silver nanoparticles together with P. aeruginosa for the remediation of aqueous hydrocarbons. Presently, this study reports the biosynthesis of AgNPs using P. aeruginosa, their physicochemical properties, and demonstrates enhanced crude oil removal in water under mild conditions. Thus, it is considered a synergistic approach to microbial degradation, combined with nanoparticle-assisted remediation, which represents a distinct stride from conventional biodegradation methods.
However, biosynthesized AgNPs can still exert biotoxicity, including oxidative stress and growth inhibition in aquatic organisms and reduced enzymatic activity in soil microbes.26,27 Their environmental fate involves aggregation, surface transformation, and gradual dissolution, influencing mobility and persistence. To mitigate ecological risks in remediation, strategies include optimizing dosage, immobilizing AgNPs on supports and incorporating capping agents to reduce ion release. This study involved the identification and screening of bacteria from water samples contaminated by crude oil from communities in the Jones Creek Oil Field, Niger Delta region of Nigeria. The highest degrader was utilized in the biosynthesis of AgNPs, which were then characterized and employed to remediate crude oil contamination in the freshwater environment of the Niger Delta using a response surface methodology (RSM). The findings of this study open a new and uncharted field of nanomaterials for the Niger Delta environmental remediation since they will have a big influence on the creation of better biosynthesized AgNPs products for the nanotechnology and biotechnological sectors.
2 Materials and methods
2.1 Study area and description
The Jones Creek Oil Field communities of Akpata Egbemu and Okerenkoko, which are situated in Warri South West Local Government, Delta State, Nigeria, between latitudes 7° 45′ N and 7° 50′ N and longitudes 4° 15′ E and 4° 50′ E, served as the research sample site (Fig. 1). Multinational firms control several oil and gas facilities in the Jones Creek that have been in operation for more than 50 years. It is an oil-contaminated water body location that is a component of the Niger Delta's western ecology.42.2 Collection and processing of samples
Crude oil-contaminated water samples were aseptically collected from four (4) separate locations, two from each community, spaced fifty meters apart along the river's course. Samples collected from each community were combined to get a composite sample for that community. Water samples were also collected along the Escravos River, which served as control and had no prior history of oil pollution. Four (4) Liter glass bottles that had been sterilized with 70% alcohol 24 hours before the final collection were used to collect water samples for bacteriological examination. Prior to the last collection, the bottles were rinsed three (3) times using water samples. In order to determine the dissolved oxygen (DO), two more sets of water samples were taken using 60 mL Winchester glass stoppered vials. In the field, 0.5 mL of Winkler I and II reagents were applied to one set of DO samples as soon as they were collected. In order to determine the biological oxygen demand (BOD5), the other set was fixed following five days of incubation at 20 °C.28 Water samples for additional physicochemical characteristics were collected in separate 4 L sterile plastic and glass bottles. The bottles were labeled accordingly. The samples were transported in a cool box (4 ± 1 °C) to the Microbiology laboratory of the Federal University of Technology, Minna, Nigeria for physicochemical and bacteriological analyses using standard procedures outlined in the American Public Health Association29 and for screening of bacteria for hydrocarbon utilizing ability.30,31 The crude oil used was the bonny light crude (BLC), which was obtained from the Nigerian Petroleum Development Company (NPDC) in Benin City, Edo State, Nigeria.2.3 Methods
2.3.7.1 Screening of bacterial isolates for potential to utilize crude oil. The broth dilution method and plate diffusion method were used to screen the bacterial isolates for their ability to utilize crude oil and for their possible application in the synthesis of silver nanoparticles.31 The gravimetric method was used to screen the bacterial isolates for hydrocarbon degradation.15
2.3.7.2 Broth dilution method. The bacteria isolates were grown on mineral salt medium (MSM) supplemented with 1% crude oil (oil broth) to screen their ability to degrade crude oil. Here, mineral salt medium supplemented with 1% crude oil was prepared with these compositions (g L−1): Na2HPO4 2.0, KH2PO4 2.0, MgSO4·7H2O 0.01, NaNO3 2.5, NaCl 0.8, CaCl2, 0.2, KCl, 0.8, FeSO4·7H2O, 0.001, using crude oil (1%) as carbon source and 1 mL of a trace element solution. The trace element solution contained (mg L−1): ZnSO4·7H2O, 525; MnSO4·4H2O, 200; CuSO4·5H2O 705; NaMnO4·2H2O, 15; CoCl2·6H2O, 200; H3BO3, 15; NiSO4·6H2O, 27.31 The MSM was filter-sterilized, then 100 mL of the media was dispensed into 250 mL flasks. One milliliter (1 mL) of the 106 cfu mL−1 of cells of the corresponding standardized isolates' inoculum was added to the medium and incubated in an incubator shaker at 35 °C for 28 days. Bacterial growth was shown by the turbidity of the culture. This was measured with a spectrophotometer at 550 nm. The selection of bacteria capable of breaking down crude oil was based on their superior capacity to proliferate when crude oil was used as the only carbon and energy source in the growth media. Growth was seen as a sign of the capacity to degrade hydrocarbons.38 The bacterial isolates were further plated on MSM using plate diffusion method.
2.3.7.3 Plate diffusion method. Bacterial isolates were also screened for crude oil-utilizing ability using the plate diffusion method.30 Positive results from the broth dilution method were serially diluted and further sub-cultured using spread plate method onto the surface of freshly-prepared mineral salt medium (with 1.0% crude oil as carbon and energy source and supplemented with agar agar). The plates were incubated at 35 °C for 7 days. Discrete colonies that developed were counted and transferred onto nutrient agar plates and incubated at 35 °C for 24 hours, after which they were stored in the refrigerator for further use.
2.3.7.4 Secondary screening of bacterial isolates by gravimetric method. The nine bacteria that were isolated from water samples contaminated with crude oil were each incubated at 37 °C for 24 hours in nutrient broth. Fifty milliliters (50 mL) of the mineral salt medium (oil broth) supplemented with 1% crude oil was poured into conical flasks, which were then autoclaved for 15 min at 121 °C to sterilize them, and then allowed to cool. Each MSM medium was then inoculated with 1 mL of the corresponding standardized isolates and the mixture was incubated at 35 °C in an incubator shaker at 150 rpm for 30 days (Remi RS-12R, India). The uninoculated flask (control), was likewise incubated. Samples were drawn every 10 days to determine the residual concentrations of each oil. The content was subsequently transferred to a separating funnel and extracted twice using di-ethyl ether (cold extraction). The solvent (di-ethyl ether) was allowed to evaporate and the amount of each remaining oil was measured gravimetrically.15 The percentage of residual oil (hydrocarbon degradation percentage) was determined using eqn (1):39
 | (1) |
2.3.7.5 Molecular identification of test organism. At the molecular level, three bacterial isolates that exhibited significantly high growth, more colonies, and the highest percentage of degradation were identified; the isolate with the highest potential for degradation was, however, chosen for the synthesis of silver nanoparticles and for further research. Molecular identification of the isolate was carried out via amplification of the 16S ribosomal DNA (16S rDNA) fragment followed by direct sequencing.40 Genomic DNA was extracted using the protocol stated by Trindade et al.41 The 16S rDNA fragment was amplified by polymerase chain reaction (PCR) using 27F (AGAGTTTGA TCMTGGCTCAG3′) and 1525R (5′AAGGAGGTGATCCAGCC3′) primers.42 Amplification products were sequenced. Forward and reverse sequences were processed and manually edited using Bioedit Sequence Alignment Editor Software 7.0. Contigs was constructed using Bioedit and was subjected to Basic Local Alignment Search Tool (BLAST) search via NCBI42 to determine the most likely identity and designation of taxonomic units based on similarities. DNA alignments were done using MAFFT online software43 and Gblocks program44 to cure the poorly aligned regions, and to further select the conserved areas of the multiple sequence alignment.
2.4 Extracellular synthesis of silver nanoparticles using hydrocarbon-utilizing bacterium
The bacterial strain was utilized in the synthesis of the AgNP following the procedure of Saleh and Alwan20 with just slight modifications, especially to the temperature and experiment time. After being inoculated into sterile nutrient broth, the bacterial strain was incubated at 37 °C for 24 hours. The broth's culture supernatant was extracted by centrifuging it at 8000 rpm for 10 min. Next, 0.034 g of silver nitrate (AgNO3) (Sigma Aldrich Chemicals, Germany) was dissolved in 100 mL of distilled water to produce 2 mM AgNO3. Then, in a 500 mL Erlenmeyer flask, 100 mL culture supernatant was added to 100 mL of an aqueous solution containing 2 mM AgNO3. The reaction mixtures were incubated at 37 °C and 150 rpm in an orbital shaker for 30 min in a bright environment. Additionally, two flasks were made to act as controls. Only the supernatant of the bacterial strain under study was present in the first flask while the AgNO3 solution was in the second flask. Both were incubated under similar conditions. In addition to other characterization techniques, the reduction of silver nitrate to silver and the formation of AgNP was visually verified by a colour shift from light yellow to dark-brown. This was further confirmed using a UV-visible spectrophotometer (Shimadzu UV-1800, Germany).202.5 Characterization of the bacterial silver nanoparticle
UV-visible spectroscopy was used to analyze the optical properties of the produced silver nanoparticle and a Malvern nano-zetasizer (nano-zetasizer 3000, UK) was used to measure the particle size distributions of the AgNPs. Using an X-ray diffractometer (D8, Bruker, Germany), the chemical structure and crystalline phase of the AgNP was identified. The morphology and elemental composition of the AgNP was then ascertained by subjecting it to a combination of energy dispersive spectroscopy (EDS) and High-resolution Transmission Electron Microscope (HRTEM)/Selected Area Electron Diffraction (SAED) analysis.45
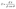 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ = Bragg diffraction angle.
θ = Bragg diffraction angle.2.6 Adsorption experiment using RSM for TPH nanobioremediation
Two liters (2 L) of the sample taken from each community were combined to get a composite sample for TPH removal using AgNPs synthesized from Pseudomonas aeruginosa strain AgA (PAE). A range of 250 mL Erlenmeyer flasks bearing the numbers 1 through 27 were used to dispense 100 mL of the water samples. Using PAE at various nanoadsorbent doses (0.1625–0.3875 g L−1), stirring speeds (875–1625 rpm), temperatures (42.5–67.5 °C) and contact times (22.5–47.5 min), nanobioremediation studies on TPH removal were carried out. Design Expert Software (version 13, Statease, Minneapolis, USA) in conjunction with Box-Behnken Design (BBD) and Response Surface Methodology (RSM) was used to conduct the nanobioremediation experimental design. The study used a BBD with three levels for each independent variable at three center point replicates. With the coded values −1 (minimum), 0 (center point) and +1 (maximal), the independent variables were temperature (°C), adsorbent dosage (g), stirring speed (rpm), and contact duration (mins) (Table 1). To determine the best possible combination, twenty-seven (27) separate runs of the tests were conducted for the removal of TPH. Throughout the experiment, the samples were constantly swirled in a water bath with temperature control. At regular intervals, samples were drawn for examinations. Following a 10 min centrifugation at 1000 rpm to separate the adsorbent and adsorbate solution, the filtrate was examined. Gas chromatography-mass spectrometry (GC-MS) was used to examine changes in the chemical composition of the water samples after residual crude oil was extracted using the previously described gravimetric method. Every adsorption study condition was repeated three times, and the average was noted. The removal efficiency (RE%) of TPH in the crude oil-contaminated water samples using PAE AgNPs was determined using eqn (3):35
 | (3) |
| Variables | Levels | ||
|---|---|---|---|
| Low (−1) | Middle (0) | High (+1) | |
| A: Contact time | 22.5 | 35 | 47.5 |
| B: Stirring speed | 875 | 1250 | 1625 |
| C: Dosage | 0.1625 | 0.275 | 0.3875 |
| D: Temperature | 42.5 | 55 | 67.5 |
2.7 Data analysis
GraphPad Prism version 8.0.1 and the statistical program for social science (SPSS) version 26 were used to analyze the data, which were then shown as Mean ± SEM. Duncan's Multiple Range Test (DMRT) and two-way Analysis of Variance (ANOVA) were used to compare various groups. P < 0.05 were regarded as significant. To determine the relationship between the parameters, correlation analysis was done.3 Results and discussion
3.1 Physicochemical parameters of crude oil-polluted water
Table 2 shows the results of the physicochemical examination of water samples contaminated by crude oil collected from the Jones Creek Oil Field communities. Turbidity, pH, potassium, BOD5, nitrates, sulphates and phosphates of the water samples fell within the acceptable ranges set for surface waters by the World Health Organization,53 the Standard Organization of Nigeria.54 And the Federal Ministry of Environment.55 The mean turbidity ranged from 0.05 ± 0.00 to 0.11 ± 0.00 mg L−1 and the pH ranged from 6.63 ± 0.01 to 6.88 ± 0.01. In contrast, the mean values of potassium (K+), BOD5, nitrates, sulphates and phosphates ranged from 8.21 ± 0.12 to 8.99 ± 0.24, 1.45 ± 0.12 to 9.25 ± 0.61, 0.14 ± 0.00 to 0.16 ± 0.00, 0.49 ± 0.00 to 0.61 ± 0.02, and 0.68 ± 0.02 to 0.97 ± 0.02 mg L−1 respectively. All the water samples examined had electrical conductivity, total hardness, total dissolved solids and chloride levels that were higher than recommended standards. With a mean value of 22700.00 ± 12.78 µS cm−1, the water sample taken from the Escravos River had the highest electrical conductivity while the water sample taken from Akpata Ekpemu had the lowest, measuring 11650.00 ± 50.37 µS cm−1. All water samples that were examined had total hardness and total dissolved solids (TDS) above the WHO and FEPA permissible limits. The water samples from the Escravos River had the highest mean values of total hardness and TDS (2452.00 ± 25.84 and 11350.00 ± 45.66 mg L−1 respectively) while the water samples from Akpata Ekpemu had the lowest mean values (972.00 ± 28.28 and 5750.00 ± 50.00 mg L−1 respectively). The COD in the water sample collected from the Escravos River was within the WHO and FEPA allowed limits while water samples from Akpata Ekpemu and Okerenkoko had mean values of 860.00 ± 60.00 and 620.00 ± 20.11 mg L−1 respectively, which were beyond levels prescribed by WHO and FEPA. The dissolved oxygen (DO) levels were high in Akpata Ekpemu (16.40 ± 0.10 mg L−1) and Okerenkoko (17.75 ± 0.15 mg L−1) but was within the permissible limits for surface water prescribed by WHO for water collected along the Escravos River (12.350 ± 05 mg L−1).| Parameters | Water samples | ||||
|---|---|---|---|---|---|
| A | B | C | WHO (mg L−1) | FEPA/SON (mg L−1) | |
| a Keys: A = River water from Akpata Ekpemu, B = River water from Okerenkoko, C = River water from Escravos River (Control), TDS = Total Dissolved Solids, BOD = Biochemical Oxygen Demand, COD = Chemical Oxygen Demand, DO = Dissolved Oxygen, Nephelometric Turbidity Unit (NTU), EC = Electrical Conductivity, NS = Not stated, HU = Hazen units, FEPA = Federal Environmental Protection Agency, WHO = World Health Organization, SON = Standard Organization of Nigeria. Values are expressed in mean ± standard error of mean of triplicate determination. Values with different superscripts on the same column are significantly different at p < 0.05. | |||||
| pH | 6.83 ± 0.01b | 6.63 ± 0.01a | 6.88 ± 0.01c | 6.5–9.0 | 6.5–8.0 |
| Turbidity (NTU) | 0.09 ± 0.02b | 0.11 ± 0.00b | 0.05 ± 0.00a | 5 | 5 |
| EC (µS/cm) | 11650.00 ± 50.37a | 13000.00 ± 10.65b | 22700.00 ± 12.78c | 1000 | 1000 |
| Total hardness (mg L−1) | 972.00 ± 28.28a | 1132.00 ± 28.56b | 2452.00 ± 25.84c | 200 | 150 |
| TDS (mg L−1) | 5750.00 ± 50.00a | 6450.00 ± 50.62b | 11350.00 ± 45.66c | 1000 | 500 |
| DO (mg L−1) | 16.40 ± 0.10b | 17.75 ± 0.15c | 12.350 ± 05a | 14 | 14 |
| K+ (mg L−1) | 8.21 ± 0.12a | 8.46 ± 0.12a | 8.99 ± 0.24a | 12 | NS |
| Ca+ (mg L−1) | 1.34 ± 0.13a | 1.44 ± 0.03a | 1.80 ± 0.01b | 75 | NS |
| BOD (mg L−1) | 5.10 ± 0.00b | 9.25 ± 0.61c | 1.45 ± 0.12a | 10 | NS |
| COD (mg L−1) | 860.00 ± 60.00b | 620.00 ± 20.11bb | 240.00 ± 12.62a | ˂250 | ˂250 |
| Chlorides (mg L−1) | 466.40 ± 3.12b | 167.43 ± 2.22a | 585.99 ± 8.02c | 250 | 250 |
| Nitrates (mg L−1) | 0.16 ± 0.00b | 0.15 ± 0.01b | 0.14 ± 0.00a | ≤50.0 | 50 |
| Sulphates (mg L−1) | 0.61 ± 0.02a | 0.49 ± 0.00a | 0.53 ± 0.01a | 250 | 100 |
| Phosphates (mg L−1) | 0.97 ± 0.02b | 0.68 ± 0.02a | 0.83 ± 0.01b | 5 | NS |
The number of ionized compounds in water determines its electrical conductivity (EC), which is a measurement of its ability to carry electrical current.56 It evaluates the total amount of dissolved solids in water. Additionally, it is an indicator characteristic that describes the salt and organic content. In its natural condition, pure water conducts electricity.57 The water sample (Table 2) from the Jones Creek communities had mean EC values that were generally high, ranging from 11650.00 µS cm−1 to 22700.00 µS cm−1. These values exceeded the acceptable limits of 500 and 1000 µS cm−1 set by the FEPA,55 SON54 and WHO53 standards, respectively. The rush of seawater brought on by high tide may be the cause of the water samples' elevated conductivity levels. The EC values found in this study are consistent with high EC values of 33.489–33.592 µS cm−1 from the estuary of the Bonny/New Calabar River reported by Onojake et al.58 and 1245–1824 µS cm−1 from the Ogoloma River in Okrika Local Government Area, Rivers State, Nigeria, reported by Efekemo et al.56 Effiong and Etteokon59 observed higher EC values from the tropical mangrove estuarine system in Akwa Ibom State (Niger Delta), Nigeria, ranging from 3472.53 to 39611.68 µS cm−1. The researchers ascribed the high EC values to the regular waste flow from human activity. The elevated levels of total dissolved solids are supported by the high concentration of EC found in this investigation.
The presence of cations in water, especially calcium and magnesium, is what causes water hardness.60 Water hardness has no health-based guidelines, however concentrations over 200 mg L−1 may cause scale buildup on boilers and in distribution systems, poor lather creation that leads to overuse of soap and scum development.61 The total hardness level of the water samples obtained from various communities in the Jones Creek Oil Field is higher than the WHO, SON and FEPA recommended standards for freshwater, indicating poor water quality. The presence of various minerals and heavy metals in the water may be the cause of the observed hardness. The results of this finding support the findings of Sam et al.,62 but it contrasts with the findings of Ajayi and Okeke,61 which indicated total hardness in the range of 19.0 to 38.0 mg L−1 from surface water resources of Kolo Creek and environs, Niger Delta, Nigeria.
Total dissolved solids (TDS) are measurements of inorganic salts, organic matter and other dissolved materials in a typical aquatic environment.5 In general, its concentration is related to the level of pollution.63 The various communities in Jones Creek yielded total dissolved solids readings ranging from 5750.00 to 11350.00 mg L−1. The SON and WHO allowable limits were exceeded by these levels. High concentration of dissolved solids tends to raise the viscosity of water, decrease the solubility of gases in water and alter the osmoregulation of aquatic animals.56 Over an extended period of time, elevated TDS levels in waterways can contribute to eutrophication. Living things have a hard time moving along the water column in an aquatic habitat with a high TDS content. Public health risks from excessive water consumption, which is high in dissolved solids, include effects on the central nervous system, paralysis of the tongue, lips, and face as well as prickliness and vertigo.56
The water samples taken from the Okerenkoko and Akpata Ekpemu communities had dissolved oxygen levels ranging from 16.40 to 17.75 mg L−1 (Table 2), which was greater than what was recommended by the SON and WHO. Dissolved oxygen (DO) is one of the most crucial markers of high-water quality and a vital component for fish and other aquatic species' survival, which plays a vital role in biological processes.62 For example, high DO levels in drinking water may taste better than low DO levels, but they can also harm industrial components, such as water pipe corrosion. The DO levels found in this study exceeded the 14 mg L−1 threshold set by the relevant bodies (SON and WHO). To survive in any aquatic habitat, aquatic species need a minimum concentration of 5.0 to 6.0 mg L−1. The DO levels found in this study were comparable to those found in water from the Bodo64 and Sagbama65 Creeks in the Niger Delta of Nigeria, but they did not support the values reported by Iyeritei et al.28 and Chukwuma et al.66
The water samples under investigation had pH levels that fell between the 6.5 and 9.0 range recommended by the WHO, SON and FEPA for surface water. Water system's pH is a dynamic indicator of water value and quality that shows how polluted the water source is. Low pH values are indicative of acidic waters because they can dissolve harmful substances and components, such as heavy metals, creating toxic habitats that are unfavourable to aquatic plant and animal life.67 The study's pH readings indicated that the water samples had a minor acidity. The increase in acidic precipitate caused by oil spills on nearby water bodies and waste disposal may be the cause of mild acidity.1,2 The pH values that were observed were within the range that other researchers in the Niger Delta environment had recorded.60,68
The water samples had nitrate contents that were within the WHO-established acceptable limit of 50 mg L−1.53 In the aquatic environment, nitrate is less harmful than ammonia and nitrite, two additional types of nitrogen. Surface water typically has a low nitrate concentration, but runoff from agriculture, particularly from fertilizer-contaminated soils, or sewage discharge from households and businesses can raise it to high levels.69 Lakes and slow-moving streams can become eutrophic due to algal blooms brought on by elevated nitrate concentrations over the threshold value.67 The near-complete lack of agricultural activity in the areas under investigation may be the cause of the low levels found in this study. Similarly, in their study on water quality evaluation utilizing a pollution model and water quality index in a few settlements in Gbaramatu Kingdom, Niger Delta, Nigeria, Sam et al.62 found low nitrate concentrations in the range of 0.37 to 0.92 mg L−1. However, compared to other authors67,70 in comparable environments, the observed values in this study were lower.
The water samples had low phosphate levels that were within an acceptable range (Table 2). As a result, the water may be suitable for household usage. The phosphate concentration ranged from 0.68 to 0.97 mg L−1, which is comparable to the findings of Friday et al.67 from Nta-Wogba Stream, which revealed that, in the absence of human interference, phosphate levels typically fall between low and moderate in freshwater media. The main causes of turbidity in water are colloid and finely suspended particles. Additionally, it is linked to the number of pathogenic organisms in water, which can be caused by soil runoff.60 All the tested samples had minimal turbidity (Table 3), falling below the World Health Organization's 2011 recommended threshold of 5 NTU. Similar turbidity readings from surface water near Odeama Community in the Nigerian Niger Delta were observed by Okoro and Diejomaoh.71
| Samples | THBC (cfu mL−1 × 105) | THUBC (cfu mL−1 × 103) |
|---|---|---|
| a Values are expressed in mean ± standard error of mean of triplicate determination. Values with different alphabet on the same row are statistically significant different at p < 0.05. Keys: THBC = Total Heterotrophic Bacterial Counts, THUBC = Total Hydrocarbon Utilizing Bacterial Counts, cfu mL−1 = Colony Forming Units per mL. | ||
| A | 120.33 ± 0.88c | 2.33 ± 0.88a |
| B | 16.67 ± 2.73b | 13.67 ± 1.20b |
| C | 4.33 ± 0.88a | 0.33 ± 0.01a |
Chloride levels in water samples (Table 2) collected from Akpata Ekpemu and the Escravos River (Samples A and C) exceeded the WHO53 recommended limit of 250 mg L−1, however the Okerenkoko water sample was within the range. The high chloride levels in this study were consistent with the findings of Edori et al.,69 who suggested that industrial activity in the vicinity of the study region and storm water surface runoffs to the river could be the cause of the high chloride levels. Sewage, industrial discharges and the re-suspension of sediments contaminated with chloride are the main sources of chloride in water. The presence of chloride in water is a sign of sewage-related pollution.
Leaching from gypsum-containing rocks and other sulfur-derived minerals causes sulphates to naturally occur in water. The concentration of sulphate in water is further increased by atmospheric precipitation, household sewage and waste from industry outputs (such as mining operations, tanneries and paper production enterprises).70 Excessive sulphate levels in water have the potential to lower the pH of the water, which would encourage the growth of bacteria.67 All the sampled communities in Jones Creek Oil Field had sulphate levels below the 200 mg L−1 threshold established by the FEPA and WHO for freshwaters. The sulfate concentrations found in this study were lower than those found in a similar study conducted by Edori5 in Ede Onyima Creek, Okarki-Engenni, Rivers State, Nigeria, and by Akpan et al.70 in the Qua Iboe River, Ikot Ekpene stretch, Akwa Ibom State, Nigeria. Researchers have also found higher sulfate levels in comparable settings. Sam et al.,62 for example, reported sulphate levels ranging from 0.33 to 181 mg L−1 while evaluating the water quality index and pollution model in a few communities in Gbaramatu Kingdom, Niger Delta, Nigeria. They attributed the findings to atmospheric deposition, mineral dissolution and other anthropogenic sources like fertilizer, mining, oil and gas exploration and production that were related to the study area.
3.2 Total petroleum hydrocarbon concentrations of crude oil-polluted water samples
Samples taken from Akpata Ekpemu, Okerenkoko and the composite sample (Samples A, B, and D) had mean concentrations of total petroleum hydrocarbons (TPH) in surface water that were higher than the 10.00 mg L−1 limits set by the Department of Petroleum Resources72 and the FEPA, with mean concentrations of 14.00 ± 1.42 mg L−1, 380.85 ± 0.55 mg L−1 and 2324.36 ± 4.23 mg L−1 respectively. Nonetheless, the water sample collected from the Escravos River had a mean TPH value of 6.55 ± 0.91 mg L−1, which was within permissible limits (Fig. 2).Total petroleum hydrocarbons were not evenly distributed throughout the Jones Creek communities (Fig. 2). There were noticeable differences between stations. This demonstrates unequivocally that human activity was the cause of pollution. Hydrocarbons in freshwater environments are caused by anthropogenic processes, including industrial operations, burning fossil fuels, oil plants and refineries and urban garbage.73 Illegal bunkering in the vicinity and commercial activities (such as speed boat fishing), where the engine may be releasing lubricating or engine oil into the water, may be blame for the elevated TPH levels found in Jones Creek. The greater quantity of TPH in the research location may possibly be explained by inadequate cleanup following an oil spill, as this creek also receives sewage from nearby oil businesses.
Petroleum hydrocarbons will inevitably poison and harm the environment wherever oil exploration and production occur.74 Through bioaccumulation and olfactory cell blockage, petroleum hydrocarbons, whether found in sediments or surface water, can impair aquatic organism life forms.56 The increase in total petroleum hydrocarbons found in this study suggested that Jones Creek's freshwater is unsuitable for household purposes like drinking and cooking because even a small amount of hydrocarbon product in drinking water poses a major risk to public health since it can cause a number of illnesses, including cancer.28,53
3.3 Bacterial counts of the water samples
Table 3 displays the findings of the bacteriological examinations of water samples collected from the Jones creek oil field's communities. In the water samples, the average Total Heterotrophic Bacterial Counts (THBC) varied between 4.33 ± 0.88 cfu mL−1 to 120.33 ± 0.88 × 105 cfu mL−1. However, it was observed that there was a significant difference (P < 0.05) in THBC between the samples. Akpata Ekpemu (Sample A) had the highest mean count (120.33 ± 0.88 × 105 cfu mL−1), whereas water samples taken from the Escravos River (Sample C) had the lowest count (2.33 ± 0.33 × 105 cfu mL−1). The mean Total Hydrocarbon Utilizing Bacteria Counts (THUBC) varied from 0.33 ± 0.01 cfu mL−1 to 13.67 ± 1.20 × 103 cfu mL−1, as shown in Table 3. The highest bacterial counts were found in crude oil-contaminated water samples from Okerenkoko (Sample B), whereas the lowest bacterial counts were found in samples taken along the Escravos River, which also functioned as the control (Sample C) (Table 3).The bacterial community in this investigation did not exhibit any discernible growth pattern, but their numbers were greater than those in the control (Escravos River). The THBC indicates that the overall environmental microbial population may rise because of an increase in nutrient load brought on by pollution.13,75 According to other researchers, the presence of bacterial growth from crude oil-contaminated water samples is also proof that these bacteria can adapt to such an environment. For example, Omoruyi and Amadi76 reported a mean THBC of 5.91 log cfu mL−1 in Egbaoma Flow Station, Delta State, Nigeria and Akpoka et al.9 reported a THBC of 19.3 ± 8.41 × 105 cfu mL−1 for surface water in similar environments. The results of this study are consistent with the findings of these investigations. However, in the surface water of the Kono River in Rivers State, Nigeria, and the Borikiri wetlands, Benibo et al.16 and Osadebe et al.14 showed reduced THUB and THBC numbers respectively. The study's findings showed that the natural microbial populations in the water tainted by crude oil could mineralize environmental contaminants to a level that was safe and acceptable.
Given that the river and the surrounding area are associated with chronic crude oil pollution, the percentage of hydrocarbon (HC) utilizers seen in this study was anticipated. Because the bacterial population present in communities surrounding petroleum products had the capacity to consume hydrocarbon due to recurrent exposure to petroleum products, high abundances of HC-utilizers are expected in locations that experience repeated spills over an extended period of time. In accordance with Galitskaya et al.,77 following a pollution event, the number of HC-utilizers in the environment tends to increase dramatically. As pollutant levels decline over time, their numbers gradually decline but hardly ever reach their pre-pollution levels. This is particularly true for areas like the communities of the Jones Creek Oil Field that are subject to ongoing pollution.
3.4 Identity of bacterial isolates in the crude oil-polluted water samples
The bacterial isolates from crude oil-contaminated water samples in this study were identified as Pseudomonas aeruginosa, Pseudochrobactrum asaccharolyticum, Bacillus megaterium, Micrococcus luteus, Bacillus subtilis, Escherichia coli, Bacillus cereus, Macrococcus caseolyticus and Bacillus thuringiensis. These organisms were identified based on their cultural, morphological and biochemical characteristics. The morphological and biochemical characteristics of the bacterial isolates are presented in Table S1. Other researchers have confirmed that some of the species recovered from this study area (Table S1) are present in Niger Delta soil, ground water and surface water.16,76,78,79 The fact that these organisms were isolated from these habitats demonstrate that they can break down hydrocarbons and have developed adaptation mechanisms to use these pollutants as sources of carbon and energy. Therefore, it is not surprising that these organisms were isolated from contaminated water, and they are shown to be promising candidates for the biodegradation of petroleum hydrocarbons. Potential oil degraders are best isolated from oil-contaminated areas.76Since microorganisms are found throughout nature, their presence in particular habitats may serve as a biomarker for the presence of certain contaminants or as an indication of contamination.76 Therefore, the distribution of bacterial isolates from the different sample locations suggests that metabolically active strains that might use hydrocarbons as a source of carbon and energy are frequently found. Bacteria that use hydrocarbons are incredibly diverse and may easily adapt to live in unfavourable conditions.9,80 Different microbial genera break down petroleum compounds using different paths; some of these pathways take less time to finish while others take more time. Biodegradation processes are also greatly influenced by the kinds of enzymes that microorganisms use and the components of petroleum products.10
Bacteria have been identified as the main and active agents in the breakdown of hydrocarbons among microorganisms.80,81 Hydrocarbon-degrading bacteria are not limited to a small number of taxa but are found throughout nature. The bacteria that were isolated and described in this study, such as Pseudomonas, Bacillus and Micrococcus, are known to be oleophilic in contaminated environments.14,82 Some of the various genera of bacteria that have been reported as hydrocarbon degraders include the Bacillus, Acinetobacter, Pseudomonas, Micrococcus, Gordonia, Rhodococcus, Cobetia, Halomonas, Alcanivorax, Marinobacter, Microbacterium, Corynebacterium, Ochrobactrum, Stenotrophomonas and Hahella.76,78,81
One of the well-known hydrocarbon degraders that use the terminal oxidation pathway is Pseudomonas species.83 Several authors have found Pseudomonas species in a variety of hydrocarbon-contaminated environments.6,16,84 Up to 100% of the C13–C35 hydrocarbons in crude oil can be broken down by Pseudomonas aeruginosa, which was isolated from an oil-contaminated lake wetland.84 The detection of Bacillus species in the water samples used in this investigation may be explained by their capacity to produce spores in situations when nutrients are few and to endure in a variety of conditions, including water contaminated by hydrocarbons.14,16,81 Because they can lower surface and intersurface tension, biosurfactants—a class of surface-active molecules with emulsifying activity—are also produced by several Bacillus species. This characteristic is thought to be a biological tactic to improve the biodegradation rates and raise the bioavailability of carbon sources that are difficult to reach, like hydrocarbons.81
Some of the bacteria that were isolated from the water samples (Table S1) have been connected to water-borne illnesses including diarrhea and are of public health concern.79 These bacterial isolates' presence indicates that the water sources are unfit for human use. Escherichia coli, for example, is a sign that surface water has been contaminated by feces as a result of untreated sewage being dumped directly into the water body.16 Urinary tract infections, diarrhea, nosocomial pneumonia, and dysentery are intestinal illnesses that can be brought on by Escherichia coli. Additionally, some Pseudomonas species, such Pseudomonas aeruginosa, have the potential to produce septicemia and bacteremia.79
Members of the genus Macrococcus have been isolated from people and other animals, and Macrococcus caseolyticus is a non-pathogenic bacterium. Lamb was the original host of Macrococcus caseolyticus.85 In their search for animal-borne antibiotic-resistant bacteria, Baba et al.86 identified this organism from a domestic chicken's skin swab; Karani et al.87 isolated it from semi-dried prawns that had undergone radiation processing. However, Pelletier and Draper88 isolated the same bacterium from four horses' diseased hooves. Furthermore, two species—Macrococcus equipercicus and Macrococcus carouselicus—have been isolated from ponies' and horses' skins, although they haven't been linked to any diseases.89 Macrococcus and Staphylococcus are the two bacterial genera that are most closely related.
It was reclassified to be a separate genus due to its significantly lower genome size than staphylococci, after formerly belonging to the staphylococcal family.86 Staphylococcus epidermidis is oxidase negative, whereas it is oxidase positive. Currently, seven (7) species are included in the genus Macrococcus: Macrococcus bovicus, Macrococcus carouselicus, Macrococcus caseolyticus, Macrococcus equipercicus, Macrococcus brunensis, Macrococcus hajekii and Macrococcus lamae.86,87 In contrast to staphylococcal species, macrococci are usually isolated from animal skin and foods like milk and meat and do not cause infections in humans or other animals. However, very little is known about this organism's physiological characteristics and there aren't many published reports on macrococci. In addition to Staphylococcus, Bacillus species are closely linked to Macrococcus caseolyticus, according to phylogenetic relationship research based on 16S rRNA sequences. This organism has a globular shape; however, its cells are bigger than staphylococci's.86,87 Sharma and Dhingra90 have previously isolated Pseudochrobactrum asaccharolyticum from soil, water and the root nodules of leguminous plants.
3.5 Tolerance of bacterial isolates to hydrocarbon based on turbidity
Fig. 3 displays the primary screening for crude oil-degradation capacity based on turbidity of the isolated bacteria from the different communities of the Jones Creek oil field. The findings indicated that on the 14th day, all the bacterial isolates were at their highest level. Of all the bacterial isolates in the MSM medium, Pseudomonas aeruginosa had the highest optical density (OD) value of 1.86 on the 14th day of incubation. At the end of the incubation period, Escherichia coli had the lowest optical density value of all the bacterial isolates, measuring 0.109. On day 14, Pseudochrobactrum asaccharolyticum reached its maximum OD of 1.73; on day 28, it dropped to 1.53. Bacillus thuringiensis, Bacillus subtilis, Bacillus megaterium, and Bacillus cereus recorded ODs of 0.21, 0.17, 0.18, and 0.14 on Day 28, respectively while Macrococcus caseolyticus had the highest OD of 1.50 on Day 14 and decreased to 1.23 at the end of the study. The 14th day likewise had the maximum OD of 0.51 for Micrococcus luteus (Fig. 3).Mineral Salt Medium (MSM) supplemented with 1% crude oil was used to examine the bacterial isolates' capacity to break down hydrocarbons. Bacillus megaterium, Micrococcus luteus, Bacillus subtilis, Escherichia coli, Bacillus cereus, Macrococcus caseolyticus, Bacillus thuringiensis, Pseudomonas aeruginosa and Pseudochrobactrum asaccharolyticum were the bacterial isolates (Fig. 3). The bacterial growth was assessed by measuring optical density (550 nm) at regular intervals (7 days) after the nine isolates were incubated for 28 days on 1% crude oil as the only source of carbon and energy. This served as an indicator for the utilization of hydrocarbons10,14 After 28 days of incubation, these isolates used the crude oil to varying extents (Fig. 3). After 14 days of incubation, Pseudomonas aeruginosa had the highest optical density value of all bacterial isolate in the MSM medium, measuring 1.855. At the conclusion of the incubation period, Escherichia coli had the lowest optical density value of any bacterial strain, measuring 0.109.
Turbidity measurement is an indirect indicator of microbial cell mass.91 Although the degree of degradation varied across the isolates, the study's findings (Fig. 3) demonstrated that all the bacterial isolates had efficiently used crude oil. For every bacterial isolate, the OD increased linearly between days 7 and 14. Hydrocarbon breakdown was caused by bacterial growth, as evidenced by the steady rise in OD (turbidity).3 While lowering the pollutant content in the environment, the microorganisms that break down crude oil were able to change and mineralize the pollutant for use as an energy source and for other cellular processes.92 The oil layer gradually and visibly disappeared due to the rise in cell densities brought on by hydrocarbon usage and after a long time, the oil film completely vanished. The broth's turbidity gradually decreased from day 21 to day 28, which may indicate a drop in the bacterial population and the breakdown of the hydrocarbon between days 7 and 21. The findings unequivocally demonstrated that the nine (9) bacterial isolates employed in this investigation could utilize, proliferate and break down the crude oil found in the mineral salt medium. The bacterial isolates that used carbon as their only energy source adapted, survived and multiplied, as evidenced by other researchers' reports of an increase in optical density and turbidity of the mineral salt medium of the bioremediation setup that lasted for 28 days.3,82
3.6 Tolerance of bacterial isolates to hydrocarbon based on viable plate counts
Table S2 shows the results of the primary screening for crude oil-degradation capacity of the isolated bacteria from the different communities of the Jones Creek oil field based on viable plate counts. Pseudomonas aeruginosa had the highest viable count of isolates from the water samples during the course degradation experiment, followed by Pseudochrobactrum asaccharolyticum and Macrococcus caseolyticus. Escherichia coli had the lowest capacity to degrade the crude oil used in this investigation. However, it was noted that there was no discernible difference in the overall viable counts of the other five bacterial isolates. From day 1 to day 21, the organisms grew steadily and were able to use the nutrients that were available (Table S2). Additionally, the overall viable counts gradually decreased after day 21, suggesting that the cells had stopped dividing. This might have happened because of nutritional depletion. It's also possible that the buildup of harmful metabolic products the organism made caused growth to stop. It's possible that the presence of harmful waste products altered the culture media and prevented the bacteria from growing.37,823.7 Crude oil biodegradation by bacterial isolates
Fig. S1 displays the rates and overall extent of biodegradation of the bacterial isolates based on secondary screening to establish their capacity to break down crude oil using the gravimetric method. When compared to the percentage of biodegradation in the control flasks without bacterial inoculation, it was found that the bacterial isolates degraded the light crude oil to differing degrees. The flasks inoculated with different bacterial isolates showed a notable loss of oil. Within the first 10 days of incubation, the crude oil biodegraded quickly in the flasks injected with Pseudomonas aeruginosa (36.30%), as opposed to the ˂1% biodegradation seen in the flasks without inoculation. At the end of 30 days, this isolate also had the highest percentage of oil degradation (97.80%), followed by Pseudochrobactrum asaccharolyticum and Macrococcus caseolyticus, which had crude oil degradation rates of 96.20% and 94.30%, respectively. In contrast, the control had 1% crude oil degradation during the same period. Following 30 days, Escherichia coli degraded the light crude oil at the lowest percentage (61.62%).Compared to the other bacterial strains, Pseudomonas aeruginosa displayed a higher capacity to degrade hydrocarbons. Because Pseudomonas aeruginosa can use these hydrocarbons as an energy source, it grew quickly and widely during the hydrocarbon-degradability test. Other researchers' similar reports78,83 demonstrated that Pseudomonas spp. were the most common species in habitats contaminated by hydrocarbons.
The study's findings (Fig. S1) are consistent with those of Jeya et al.,15 who discovered that Pseudomonas aeruginosa broke down crude oil more effectively than other isolates in 30 days. The findings of this investigation, however, contradict those of Abioye et al.,78 who discovered that in 28 days, Micrococcus sp., Pseudomonas sp., and Bacillus sp. degraded crude oil by 77.6%, 73.1%, and 67.7% respectively. When compared to these results, the variations in the percentage of oil biodegradation may be the result of the various types of oil utilized in the research. Another notable bacterial genus that has been shown to breakdown petroleum hydrocarbons is Bacillus.3,14
In this investigation, it was discovered that Escherichia coli could survive on crude oil as the only carbon source (Fig. S1). This suggested that it could be used to clean up crude oil spills. Crude oil degradation by this bacterium has been documented in other investigations. According to Sarfo et al.,93 who employed Oxford nanopore sequencing to isolate and characterize crude-oil-dependent bacteria from the Ghanaian coast, this organism has shown the ability to degrade crude oil. Transformant Escherichia coli was identified in soil contaminated by used motor oil by Musa94 and has also been linked to the breakdown of crude oil (phenanthrene).95
Mono and dioxygenases are found in hydro-carbonoclastic bacteria, and they catalyze the splitting of oxygen molecules into atoms, which are then incorporated into aliphatic and aromatic hydrocarbons.96 When the hydrocarbon is attacked, alkanes are converted to alkanols, which are then oxidized to alkanals and, ultimately, to the corresponding fatty acids. After that, β-oxidation breaks down the fatty acids into units of acetyl-CoA. This intermediate is processed through the TCA cycle to CO2 and H2O for the manufacture of ATP and the cycle's keto acid, which creates amino acids for the synthesis of bacterial cell components. This encourages the bacteria to grow.82 The metabolic pathway mentioned above was probably employed by the bacterial isolates identified in this investigation, which showed optimal development when exposed to crude oil. Only one of the nine (9) bacterial isolates was chosen for additional research, but three (3) that showed good growth and optical density were identified down to the molecular level.
3.8 Molecular identities of selected bacterial isolates
The National Centre for Biotechnology Information (NCBI) databases' Basic Local Alignment Search Tool (BLAST) was used to identify the bacterial 16S rRNA sequences, as shown in Table S3. The top four isolates that degraded hydrocarbons were confirmed to be Pseudomonas aeruginosa (A), Pseudomonas aeruginosa (B), Pseudochrobactrum asaccharolyticum (C) and Macrococcus caseolyticus (D) by genomic analysis (Table S3). Isolates A and B's 16S rRNA gene sequencing revealed 99.43% and 99.78% sequence similarity to Pseudomonas aeruginosa respectively. As a result, isolates A and B were identified as Pseudomonas aeruginosa strain AgA and Pseudomonas aeruginosa strain AgB. Isolate C's physiological and morphological characteristics were similar to those of Pseudochrobactrum asaccharolyticum, which had 99.85% sequence similarity and was identified as Pseudochrobactrum asaccharolyticum strain AgC. However, isolate D had 99.72% identity with Macrococcus caseolyticus and was identified as Macrococcus caseolyticus strain AgD. By sequencing the 16S rRNA, the isolated bacterial strains were molecularly identified. All four isolates had 1500 base pairs (Plate S1). The PCR-amplified 16S rRNA sequences employed for this were used as a sensitive and specific microorganism detection approach.3,97 The three (3) bacterial isolates were identified using a universal primer pair unique to a 16S rRNA gene fragment. All isolates showed positive results with an amplification band corresponding to 1500 bp, confirming that they were all bacterial species (Plate S1). The Pseudomonas aeruginosa sequence found in this investigation matched the similar sequences of a few other known species that break down hydrocarbons. The phylum Proteobacteria typically possesses traits that are strongly linked to aliphatic and aromatic hydrocarbon-degrading organisms, according to the findings from previous investigations by Bekele et al.98 The NCBI database now contains strains. However, Pseudomonas aeruginosa strain AgA was selected for the synthesis of silver nanoparticles and further study.3.9 Extracellular synthesized silver nanoparticles
The culture supernatant of Pseudomonas aeruginosa strain AgA was used to synthesize AgNP. Using a visual indication that could be seen by the reaction's colour shift, the biosynthesis of AgNPs was carried out (Plate S2). The main indicator of the biosynthesis process was the visual monitoring of the bioformation of AgNPs from the AgNO3 combination using bacterial supernatant. The control media (culture supernatant without AgNO3 and AgNO3 solution alone) incubated under the same conditions, however, did not change colour (Plate S3).One of the main indicators of the bioreduction of Ag+ ions to Ag0 is the change in colour from yellow to dark-brown when bacterial supernatant is present (Plates S2–3).99 The control (Plates S2–3) showed no change in colour, indicating that the media's compounds could not influence the Ag+ ion's reduction.100 The colour shift also occurred when light was present, suggesting that sunlight was crucial to the synthesis of AgNPs.101 Various investigations have shown that the application of various reducing agents in the presence of sunlight triggers the biosynthesis of AgNPs. Similar colour changes under illumination were also noted by Hamouda et al.,102 who suggested that AgNO3 may be decreased in the reaction mixture due to electrons moving between energy levels and generating silver nanoparticles. An oscillation of electrons while in resonance with light waves caused this colour shift, which was triggered by the activation of the AgNPs' Surface Plasmon Resonance (SPR).103
3.10 Characteristics of the synthesized silver nanoparticles
The biosynthesized AgNP showed a prominent broad peak at 425 nm in the UV-visible absorption spectrum (Fig. 4). A surface plasmon resonance of the particle has been connected to the detection of such a band.99 The findings of this investigation were in line with prior work by Tariq et al.,104 who noted that all aliquots of the reaction mixture derived from the culture supernatant that was 48 hours old had strong peaks at 426 nm. Similarly, Abeer-Mohammed et al.100 indicated that a strong surface plasmon reverberation placed at 400 nm was exposed when the AgNPs synthesized by Pseudomonas aeruginosa strain display.
According to Nagaraja et al.,105 distinctive AgNPs generally displayed characteristic surface plasmon resonance at wavelengths between 400 and 450 nm. Any shift in the SPR peak might be explained by biomolecules serving as capping and reducing agents.106 The size and form of the metal AgNPs, the dielectric constant and the medium surrounding the metal particles all affected the variation in frequency and the breadth of the SPR absorption band.21 This study also found the same thing. The optical absorption spectra of metal nanoparticles, which change to a longer wavelength as particle size rises, are mostly determined by SPR.107
Abeer-Mohammed et al.100 found that the bacterial cell's metabolic products, including proteins, amino acids and/or extracellular enzymes, might reduce Ag+ ions. In contrast, the control tests' colour remained unchanged, indicating that Pseudomonas aeruginosa's metabolic byproducts—rather than the media's constituents—were in charge of reducing Ag+ during the biosynthesis process. The researchers concluded that the high content of secondary metabolites and enzymes in Pseudomonas aeruginosa cell-free supernatant may be connected to the high production rate of AgNPs at 50%. NADPH was discovered to be the co-factor of the nitrate reductase enzyme, and Giri et al.103 also revealed that silver ions exposed to nitrate reductase produced exceptionally stable silver AgNPs. According to the study's findings, the bioreduction of Ag+ to Ag0 and the subsequent creation of silver nanoparticles may be caused by the bacterial isolates under examination because they are known to secrete the co-factor NADH and NADH-dependent enzymes, particularly nitrate reductase. The biosynthesis of AgNPs by Pseudomonas aeruginosa strain is primarily driven by catalytic proteins—especially nitrate reductase—which donate electrons to reduce Ag+ to Ag0, as reported in Ali et al.108 Enzyme activity assays show elevated nitrate reductase activity correlating with rapid nanoparticle formation. Simultaneously, extracellular polymeric substances, including proteins and phenolic-rich metabolites, cap and stabilize the forming nanoparticles by providing functional groups (–NH, –COOH) that prevent aggregation. This dual action of enzymatic reduction and polymer-mediated stabilization aligns with green antimicrobial AgNP mechanisms described by Mikhailova.109
The dynamic light scattering (DLS) method of particle size characterization relies on the interaction of light with particles and can be used to calculate narrow particle size distributions (Fig. 5), particularly in the 1–100 nm range.110 These results corroborate those of Esmail et al.,17 who generated AgNPs using Bacillus ROM6 culture supernatant and found that they had an average size of 20–70 nm. In their research, Khaleghi et al.111 also demonstrated that Bacillus thuringiensis formed silver nanoparticles with an average particle size of 42 nm. As a result, the findings of this investigation are aligned with those of other researchers.
 | ||
| Fig. 6 (a) High resolution HRSEM (b) low resolution HETEM (c) SAED of and (d) EDS of biosynthesized AgNP. | ||
The presence of capping peptides surrounding each particle, which served to stabilize the nanoparticles, was indicated by the small aggregates. Crystal lattice fringes with an average d-spacing of 1.77 ± 0.13 Å evidence the presence of well-defined atomic planes within synthesized AgNPs, hence confirming their crystalline nature (Fig. 6b). Such fringes are associated with the periodic arrangement of atoms in metallic silver, and the measured d-spacing should relate to interplanar distances corresponding to a particular crystallographic plane. Verma et al.112 reported that such lattice fringes relate to face-centered cubic (fcc) silver and align with the reflections recorded in XRD patterns. The narrow variation (±0.13 Å) around the mean value could point to high structural homogeneity and controlled nucleation during synthesis. The presence of clear and sharp lattice fringes would, therefore, suggest minimal defects and strong crystallinity, which enhance AgNP optical, catalytic, and antimicrobial properties. Nevertheless, there was little aggregation and an uneven distribution of the bacterial AgNP across the surface. For a crystalline material like AgNP, d-spacing displays the inter-atomic spacing. The average d-spacing for Pseudomonas aeruginosa strain AgA was 1.77 ± 0.13 Å. AgNP would be more crystalline, more rigid and more stable with lower d-spacings, and their performance would be superior.
The TEM analysis of Pseudomonas aeruginosa strain AgA was comparable to that of Pseudomonas sp. ef1 bio-AgNPs by John et al.,113 which showed smooth-surfaced poly-dispersed particles that ranged in size from 12.5 to 100 nm and had a smooth surface. However, this was in contrast to the mono-dispersed and smaller (6 to 16 nm) bio-AgNPs from Pseudomonas putida by Gopinath et al.114 while Abeer-Mohammed et al.100 reported that the biosynthesized AgNPs synthesized by Pseudomonas aeruginosa were well dispersed with different shapes, aspherical and square, ranging between 11 and 25 nm in diameter, Singh et al.115 synthesized irregular shapes of the AgNPs from Pseudomonas aeruginosa in size of 10 to 40 nm.
In this study, AgNPs synthesized using the P. aeruginosa strain AgA showed well-defined diffraction rings indicating their polycrystalline nature (Fig. 6c). Five distinct concentric rings corresponding to the lattice planes of (111), (200), (220), (311), and (400) were observed in the SAED patterns. These correspond to the fcc structure of elemental silver. These Bragg reflection planes match very well with the JCPDS card for pure Ag, thereby confirming that the nanoparticles formed because of bacterial reduction of silver ions possess a crystalline metallic silver core rather than silver oxide or other mixed phases. The prominent (111) plane indicates that a favorable orientation or a high-intensity reflection is paramount, as expected for biosynthesized AgNPs due to thermodynamically favorable atomic packing. The presence of multiple rings further confirms that the AgNPs are polycrystalline and are constituted of several small crystallites aggregated within each nanoparticle. The crystallographic features seen in the present study from SAED are further in agreement with the XRD, which also confirmed the formation of fcc silver with similar diffraction peaks. The crystallinity observed in the present study is also in good agreement with other studies, such as Chand et al.116 and Al-Mashud et al.,117 in which SAED was used to confirm the crystalline nature of green- or microbially synthesized AgNPs. Such a ring pattern corresponding to the (111), (200), (220), and (311) planes also appeared, showing that the biological method always produced fcc-structured silver with good polycrystallinity. Moreover, Hassan-Afandy et al.118 pointed out that the crystalline structure of green-synthesized plasmonic AgNPs tends to enhance its antibacterial efficiency through enhanced surface reactivity and electron transfer dynamics. Their SAED study also stressed the existence of intense diffraction rings related to fcc silver, pointing out the central role of crystalline quality in determining the functional performance.
These results are also in agreement with the work of Muddassir et al.,119 who biosynthesized AgNPs for evaluating their antibacterial activity against resistant strains of Pseudomonas aeruginosa. The authors demonstrated that the crystalline nature of AgNPs contributes significantly to their antimicrobial potential, thereby enhancing the interaction with bacterial membranes and facilitating the release of Ag+ ions. They presented diffraction patterns characteristic of polycrystalline fcc AgNPs, which agrees with the structural features described in this work. This is in good agreement with reports indicating that microbial synthesis routes, especially those involving P. aeruginosa, often generate crystalline AgNPs possessing higher biological activities. The polycrystalline nature revealed from the SAED patterns indicates that the biosynthesized AgNPs are made up of multiple crystallographic domains contributing to the overall structural stability and high surface energy of the nanoparticles. The presence of multiple orientation domains also indicates that the nanoparticles possess a high number of active facets, mainly the (111) plane, which has been reported to play a key role in interaction with microbial cell walls.
Silver (Ag), carbon (C), and oxygen (O) atoms were detected in the AgNP produced using the bacterial strain under study by energy dispersive X-ray analysis (Fig. 6d). Ag was found in substantial amounts in the EDS analysis. The atomic percentages of silver (Ag) 87.35%, carbon (C) 10.19%, and oxygen (O) 2.46% were revealed by the EDS results of the AgNP produced utilizing the Pseudomonas aeruginosa strain AgA. Silver had the greatest peak in the bacterial AgNPs' EDS spectra (Fig. 6d), while carbon and oxygen had the weakest peaks. This demonstrates that the chemical composition of AgNP production is comparatively pure. Silver was the predominant element in this study, as evidenced by the prominent peak (signal) of silver that emerged at the 3 keV area due to surface plasma resonance.120 This is consistent with the findings of earlier investigations.121,122 A few or minor peaks (spectra signals) were also found in the EDS analysis, indicating that the nanoparticles synthesized from the investigated bacterial strain contained elements other than silver metal, such as carbon and oxygen. These elements could be explained by the presence of conjugated biomolecules, such as proteins and enzymes, over the surface of AgNPs.8,123
The investigated bacterial strain produced a silver plane, although silver was the primary crystalline phase. Additionally, there were a few extra, unassigned peaks in the XRD examination. The crystalline biological components on the surface of the nanoparticles or the chemical utilized in the synthesis (silver nitrate) may be the cause of these unaccounted-for peaks. Other AgNPs studies conducted elsewhere also show these unidentified peaks in the XRD.103,123 Furthermore, the AgNPs' comparable diffraction profiles showed that the stabilizing and reducing chemicals in the supernatants had fully reduced the silver ions (Ag+) to (Ag0).123 According to the XRD examination, the produced nanoparticle was crystalline and composed of metallic silver. This outcome is consistent with previous researchers' findings.17,99 According to El-Saadony et al.,121 the existence of face-centered cubic (FCC) of the crystalline AgNPs is the cause of these distinctive XRD peaks.
3.11 RSM of TPH removal
The Box Behnken Design (BBD) and their responses for the removal of TPH removal using biosynthesized P. aeruginosa–AgNPs are shown in Table 4. The highest TPH removal of 99.02% using AgNPs synthesized from Pseudomonas aeruginosa strain AgA (PAE) was recorded by Experimental run 17, which was conducted at a temperature of 67.5 °C, a contact time of 35 min, a stirring speed of 1625 rpm and a dosage of 0.275 g. This was higher than the predicted TPH removal of 98.98%. At a contact time of 35 min, stirring speed of 875 rpm, dosage of 0.1625 g and temperature of 55 °C, Run 21 produced the lowest TPH elimination of 90.57%, which was less than the predicted 92.06% using PAE. In Akl et al.,124 the TPH removal for biosynthesized P. aeruginosa–AgNPs was higher compared to chemically synthesized AgNPs, single-microbe degradation, and physical adsorbents; it offered lower cost, enhanced biodegradation, and reduced ecological toxicity. This nanobioremediation approach can boast of a superior catalytic–biological synergy compared to algae-mediated AgNPs and biochar-based stimulation in Saeed et al.125 for faster, cleaner, and more sustainable crude-oil remediation.| Run | Factors | Response (TPH removal) (%) | ||||
|---|---|---|---|---|---|---|
| A: Contact time (mins) | B: Stirring speed (rpm) | C: Dosage (g) | D: Temp (°C) | PAE | ||
| Actual | Predicted | |||||
| a PAE = AgNPs synthesized using Pseudomonas aeruginosa strain AgA. | ||||||
| 1 | 22.5 | 1625 | 0.275 | 55 | 98.82 | 99.44 |
| 2 | 22.5 | 1250 | 0.275 | 67.5 | 97.56 | 97.16 |
| 3 | 35 | 1250 | 0.1625 | 42.5 | 94.82 | 94.14 |
| 4 | 22.5 | 875 | 0.275 | 55 | 93.97 | 94.68 |
| 5 | 35 | 1625 | 0.1625 | 55 | 98.71 | 98.17 |
| 6 | 22.5 | 1250 | 0.1625 | 55 | 95.55 | 95.35 |
| 7 | 35 | 1250 | 0.1625 | 67.5 | 98.97 | 98.09 |
| 8 | 35 | 875 | 0.275 | 67.5 | 94.91 | 94.75 |
| 9 | 35 | 1250 | 0.275 | 55 | 96.95 | 95.73 |
| 10 | 35 | 875 | 0.3875 | 55 | 97.72 | 98.24 |
| 11 | 47.5 | 1250 | 0.1625 | 55 | 96.76 | 96.87 |
| 12 | 35 | 1250 | 0.275 | 55 | 97.05 | 95.73 |
| 13 | 47.5 | 1625 | 0.275 | 55 | 93.67 | 93.20 |
| 14 | 22.5 | 1250 | 0.3875 | 55 | 98.46 | 98.77 |
| 15 | 35 | 1625 | 0.275 | 42.5 | 93.76 | 93.65 |
| 16 | 22.5 | 1250 | 0.275 | 42.5 | 97.49 | 96.96 |
| 17 | 35 | 1625 | 0.275 | 67.5 | 99.02 | 98.98 |
| 18 | 47.5 | 1250 | 0.3875 | 55 | 91.32 | 91.94 |
| 19 | 47.5 | 1250 | 0.275 | 67.5 | 96.21 | 96.58 |
| 20 | 35 | 1250 | 0.3875 | 67.5 | 96.06 | 95.65 |
| 21 | 35 | 875 | 0.1625 | 55 | 90.57 | 92.06 |
| 22 | 47.5 | 875 | 0.275 | 55 | 95.99 | 95.61 |
| 23 | 35 | 1250 | 0.275 | 55 | 95.45 | 95.73 |
| 24 | 35 | 1625 | 0.3875 | 55 | 91.97 | 92.46 |
| 25 | 35 | 1250 | 0.3875 | 42.5 | 95.28 | 95.06 |
| 26 | 47.5 | 1250 | 0.275 | 42.5 | 91.98 | 92.23 |
| 27 | 35 | 875 | 0.275 | 42.5 | 95.77 | 95.54 |
One potential solution that is environmentally-friendly and saves time, energy, and money is nanobioremediation.126 This study examined the efficacy of several nanoadsorbent doses of 0.15625–0.3875, contact time (22.5–47.5 min), stirring speed (875–1625 rpm) and temperature (42.5–67.5 °C) in removing TPH from crude oil-contaminated water samples using synthesized AgNP made with Pseudomonas aeruginosa strain AgA (PAE). The maximum adsorption effectiveness of 99.02% was shown by the results (Table 4). The surface area connected to the accessible binding sites for adsorption may be the reason for this AgNP's significant % elimination of TPH.50 The experimental run's values were within prediction error ± 10%, which is very near to the expected responses. The biogenic AgNPs' greater surface area and smaller size, along with the functional groups affixed to their surface, may contribute to their potential for quicker TPH degrading activity.127,128 These results are consistent with those of Rasheed et al.,128 who employed biologically and chemically produced AgNPs to break down dyes and found that biogenic AgNPs exhibited a quicker degradation trend. Biologically produced AgNPs' organic surface chemicals also serve as capping agents, enhancing the NPs' activity.127 The smaller size of the AgNPs may have contributed to PAE's increased propensity for TPH degradation.
Table S4 provides a summary of the model fit and lack of fit findings. The design chose 2FI as the most suitable response surface model for TPH removal utilizing the AgNP that was under investigation. PAE had an adjusted R2 (0.8683), a predicted R2 of 0.7785, a standard deviation of 0.8705, and an R2 of 0.9190. Using the AgNP being studied, the 2FI model was chosen for TPH degradation. According to Table S4, PAE's standard deviation was 0.8705, its R2 was 0.9190, its adjusted R2 was 0.8683, and its predicted R2 was 0.7785. The obtained determination coefficient (R2) of 0.9190 indicates that 91.90% of the variability caused by the biosynthesized AgNP can be explained by the model. The difference between the adjusted R2 value and the predicted R2 value for AgNP is less than 0.2. The coefficient of determination (R2) is typically used to assess the model's fitness. A higher correlation between the experimental and expected responses is indicated by an R2 value near 1 (Table S4). A decent model's R2 should therefore fall between 0 and 1, with the closer the R2 is to 1, the more fit the model is.129
The signal to noise ratio is measured with adequate precision, and values higher than 4 are typically preferred.130 According to Sarkar et al.,131 the PAE's suitable precision value of 14.818 indicated a sufficient signal, suggesting that this model may be utilized to traverse the design space. The percentage of coefficient of variation (CV) reflected the degree of accuracy of the chosen model that the experiments were related with; a high CV value suggested that the experiment was less reliable. A low CV of 0.9093% for PAE in this investigation suggested that the model experiments were more accurate and reliable.132 These results showed a strong and positive relationship between the experiment and the statistical model. Because the model matched the experimental data, it could be utilized to generate predictions.
Table S5 displays the ANOVA findings for the response surface model. Using PAE, the ANOVA examined the impacts of two factors at a time as well as the combined effects of two factors (temperature, dosage, stirring speed and contact duration) on TPH elimination. While the lack of fit for PAE was 0.6303, the P-value for the AgNP under examination was less than 0.0001. For PAE, a high model F-value of 18.14 was observed. The statistical ANOVA findings for the response showed that the model's F-values (Table S5) for PAE were 18.14, indicating the model's significance and the 0.01% probability that an F-value this large could be caused by noise. Given that P-values less than 0.05 (p < 0.05) indicated that model terms were significant, the AgNP's P-value of < 0.0001 suggested that the model was very significant (Table S5). Lack of fit for PAE yielded a value of 0.6303. This demonstrated that it was not significant in relation to the pure error because the chosen model should have a negligible lack-of-fit.133 This shows that the model is statistically correct, which is desirable. The effectiveness of the AgNP (PAE) in removing TPH from crude oil-contaminated water was taken into consideration when it was utilized independently. The single effect and two-factor interaction revealed that contact time (A), stirring speed (B) and temperature (D) had a linearly significant (p < 0.05) impact on TPH removal by PAE, whereas dosage (D) had no significant (p > 0.05) impact on TPH removal using the AgNP under study. TPH removal using PAE was significantly impacted by the two-factor interaction of AB, AC, AD, BC and BD, whereas CD had no discernible impact. In contrast to the findings of Abubakar et al.,50 who stated that the removal of pollutants increased with an increase in dosage, the result showed that a change in the amount of dosage (C) alone did not significantly affect the percentage of TPH removal by the AgNP. It also showed that the proportion of TPH removed by the AgNPs may not be greatly impacted by the dosage amount or temperature (D).
The impact of a single variable on the elimination of TPH by AgNPs made with Pseudomonas aeruginosa strain AgA (PAE) is shown in Fig. S2. The impact of contact time on TPH removal is depicted in Fig. S2a. Reducing contact duration has a significant (p < 0.0001) impact on the removal of TPH. The TPH removal increases with decreasing contact time. The impact of stirring speed on TPH removal is depicted in Fig. S2b. Stirring speed had a substantial impact on the removal of TPH (p = 0.0334). At 875 rpm, the highest TPH removal (95%) was recorded, and it gradually dropped as the stirring speed decreased TPH removal was not significantly impacted by the dosage (p = 0.1492) (Fig. S2c). With a dosage of 0.1625 g, the highest TPH removal of 96% was achieved. However, as the dosage increased, the amount of TPH removed decreased. Fig. S2d shows how temperature affects the removal of TPH. Temperature had a substantial impact on TPH removal (p = 0.0003), and this effect increased as the temperature rose. At 67 °C, the highest TPH removal rate of 97% was achieved (Fig. S2d).
When the other parameters were at their optimal levels, it was demonstrated that contact duration influenced the removal of TPH for AgNP. Since the adsorbent's active binding sites significantly increased the efficacy of TPH removal in the early stages, optimal TPH removal was observed within the first 23 min for the adsorbent. This could be because the AgNP had more vacant sites with numerous active functional groups. This outcome is comparable to the findings of Mandal et al.134 The functional binding sites of the nanoadsorbents eventually became saturated, leading to a continuous decline in TPH removal over time and making the binding sites less effective. The adsorption process was slowed down towards equilibrium in the early stages of contact time due to the possibility of absorbing a large number of unoccupied surface patches.135 TPH adsorption capacity for PAE was 97% at 23 min of contact time. PAE was found to have rapid adsorption, which may be attributed to its high surface area.50 Above all, because the nanoadsorbents became saturated, the removal of the pollutant decreased as the adsorption process progressed past the equilibrium time.136 In line with the pattern from a prior investigation, a greater concentration of AgNPs did not always lead to the elimination of more TPH.137 This is because a further increase in the number of AgNPs would not facilitate the removal of the hydrocarbons because the active sites on the biomolecules were now completely occupied by silver ions, making it impossible for the extra silver ions to bind to the biomolecules.
The three-dimensional (3D) plots and response surface curves (RSC) of the interactive effects of independent variables on the removal of TPH by AgNPs produced with Pseudomonas aeruginosa strain AgA (PAE) are shown in Fig. 8 and 9 illustrates the 3D Plots of the interactive effects of independent variables on TPH Removal by the synthesized AgNP (PAE). The AgNP was significant (p < 0.0001), the two-factor interaction (2FI) model had a non-significant (0.6303) lack of fit. The model's predicted R2 was 0.7785, adjusted R2 was 0.8683 and R2 was 0.9190. The coefficient of variation (CV%) was 0.9093. Two factors were varied while the others were kept constant. The RSC's increased TPH removal was indicated by the colour gradient, which went from blue to green to yellow to red. With the exception of dosage, all other variables (contact time, stirring speed and temperature) had a significant (p < 0.05) impact on TPH elimination for all interactions (as indicated by the RSC).
The RSC and contour plot of the effect of contact time and stirring speed on the removal of TPH by AgNPs made with Pseudomonas aeruginosa strain AgA (PAE) are shown in Fig. 8a. The relationship between stirring speed and contact time had a substantial (p = 0.0008) impact on the elimination of TPH with a contact time of 25.5 min and a stirring speed of 1525 rpm, the maximum TPH removal of approximately 99% was recorded; with a contact time of 45.5 min and a stirring speed of 1375 rpm, the minimum TPH removal of about 94% was noted. The elimination of TPH by AgNP made with Pseudomonas aeruginosa strain AgA (PAE) was significantly impacted by the two-factor interaction between dosage and contact duration (p = 0.0002) (Fig. 8b). At 24.5 min and 0.3375 g (the red zone), the maximum TPH removal of 98% was achieved, while at 40.5 min and 0.2925 g, the lowest TPH removal of 94% was recorded. The removal of TPH by AgNPs made using Pseudomonas aeruginosa strain AgA (PAE) was significantly impacted by the interaction of temperature and contact time, as seen in Fig. 8c (p = 0.0295). The lowest removal of TPH, 93%, was achieved at 45.5 °C and 43.5 min while the maximum removal, 97%, was achieved at 47.5 °C and 29.5 min of contact time. The interaction between stirring speed and dosage produced the lowest TPH removal (94%) at 1075 rpm and dosage of 0.2325 g, and the highest TPH removal (98%) at 1425 rpm and 0.2125 g (the red shade) (Fig. 8d). This interaction was found to have a significant (p < 0.0001) impact on the removal of TPH. Significant (p = 0.0029) effects on the removal of TPH were observed from the interaction between temperature and stirring speed. At a temperature of 63 °C and a stirring speed of 1425 rpm, the red zone achieved the highest TPH removal of 98%. In contrast, at 1500 rpm and 44.5 °C, the lowest TPH removal of 94% was recorded. TPH removal was improved by raising the temperature and decreasing the stirring speed, and vice versa (Fig. 8e). There was no discernible (p = 0.0708) impact of the temperature-dosage interaction on TPH removal. It was found that while TPH removal reduced with increasing dosage, it increased with increasing temperature. The lowest TPH removal (95%) occurred at a dosage of 0.3725 g and at a temperature of 48 °C, while the highest TPH efficiency (97%) was noted at 0.2525 g and at 60.5 °C (Fig. 8f).
The interaction between temperature and contact duration showed that TPH removal was high when using PAE at a higher temperature and a shorter contact time. The improved TPH removal may have resulted from the AgNP's faster chemical precipitation rate in water at higher temperatures, which occurred in a shorter time.138 Likewise, it may be related to the enhanced mobility of free electrons, which accelerated the rate of reaction. This aligns with the results of earlier studies.139 It was found that TPH removal increased with an increase in adsorbent dosage at a shorter time using PAE, indicating that the contact time–dosage interaction had a significant impact on the adsorption capacity. This may be explained by the adsorbent's larger surface area and increased number of adsorption sites, which enhance the elimination of TPH.140 With an increase in adsorbent dosage for the same quantity of adsorbate molecules, the existence of more adsorption sites may also be responsible for the rise in removal effectiveness.134 The findings have shown that raising the temperature enhanced the adsorption capacity, which, when employing PAE, rose to 97% at 60.5 °C. However, in the temperature-dosage interaction, the AgNP's adsorption capacity decreased as the adsorption dosage increased. The decrease in solution viscosity and the increase in the kinetic energy of the hydrocarbon molecules to efficiently make contact with the active sites of the nanoadsorbents through diffusion to the interior pores and across the boundary layer may be the cause of the observed increase in TPH adsorption with increasing temperature.141
The quantitative prediction result of the reaction from optimal parameters (temperature, adsorbent dosage, stirring speed and contact time) is shown in Fig. S3. The AgNP removed the TPH with the best projected values for the following factors: temperature (67.5 °C), dosage (0.3206 g), stirring speed (1620 rpm/100 mL), and contact time (22.5 min/100 mL). The dependent variable produced a 99.98% response. On the other hand, the entire model achieved a desirability of 99.40%. The ideal experimental conditions for the removal of hydrocarbons by biosynthesized AgNPs were determined to be at a temperature of 67.5 °C, a stirring speed of 1620 rpm, a contact time of 22.5 min, a dosage of 0.3206 g, and seeking 27 starting points in the response surface changes (Fig. S3). The experiment was repeated under ideal conditions to determine the ideal parameters, which is consistent with the findings of Olatunji et al.142 Using AgNPs synthesized by Pseudomonas aeruginosa strain AgA (PAE), the corresponding maximum response (% TPH removal) of the dependent variable was 99.98%. The estimated function's desirability of 0.994 suggested that it might represent the experimental model and the desired conditions. The adsorbent's maximum capacity revealed a large adsorption capacity for the hydrocarbons. In this investigation, the optimized conditions matched the expected response value, as indicated by the desirability value of 0.994 (99.4%) recorded.
The software's recommended predicted value for the response (characteristic) was equivalent to the experimental value obtained, and these values could be utilized to optimize TPH removal, as validated by the optimal formulation for the generated AgNP (Table S6). The actual (experimental) value and the predicted value differed by a very small percentage, with a change of 1.22%. A significant correlation was indicated between the AgNP under study and the response variable by the validation table, which shows the correlation between the experimental and predicted values for TPH degradation by the AgNP. Both values were strongly and positively correlated (Table S6). The predicted model and optimal conditions may be applied to hydrocarbon degradation, as the experimental and predicted values of RSM for TPH removal are identical.
3.12 Chromatogram of the hydrocarbons
| PK | Compound | Formula | RT | Area (%) |
|---|---|---|---|---|
| a PK = Peak number, RT = Retention time. | ||||
| 1 | 1,3,5-Triazabicyclo[3.2.1]octane | C5H11N3 | 3.782 | 0.21 |
| 2 | 1,2-Dimethylpyrrolidine | C6H13N | 4.291 | 0.34 |
| 3 | Pentadecane | C15H32 | 6.177 | 0.92 |
| 4 | 2,6,10-Trimethyltridecane | C16H34 | 7.771 | 0.39 |
| 5 | Tridecane | C13H28 | 8.429 | 1.00 |
| 6 | N-[3-[N-Aziridyl]propylidene]tetra | C10H21N3 | 10.200 | 0.27 |
| 7 | Tetradecane | C14H30 | 10.823 | 0.96 |
| 8 | 1-Octadecanesulphonyl chloride- | C18H37ClO2S | 12.241 | 0.39 |
| 9 | Nonane | C9H20 | 13.247 | 0.28 |
| 10 | N-Methyl-3-piperidinecarboxamide | C7H14N2O | 14.470 | 0.34 |
| 11 | Hexadecane | C16H34 | 15.881 | 0.97 |
| 12 | Methoxyacetic acid | C3H6O3 | 17.367 | 1.10 |
| 13 | Octadecyl ester | C36H72O2 | 17.996 | 0.23 |
| 14 | 2,6,10,14-Tetramethyl pentadecane | C19H40 | 18.899 | 5.64 |
| 15 | 11,13-Dimethyl-12-tetradecen-1-olacetate | C18H34O2 | 19.459 | 0.46 |
| 16 | Triacontyl heptafluorobutyrate | C34H61F7O2 | 19.756 | 0.52 |
| 17 | 2-Dodecen-1-yl(−)succinic anhydride | C16H26O3 | 20.168 | 0.51 |
| 18 | Hexadecane,2,6,10,14-tetramethylHeptadecane | C21H44 | 20.934 | 6.66 |
| 19 | 1-Eicosene 2(1H)-Naphthalenone | C10H8O | 21.454 | 1.84 |
| 20 | Dibutyl phthalate | C16H22O4 | 21.905 | 2.62 |
| 21 | Octadecane, 3-methyl- decane | C19H40 | 22.202 | 3.32 |
| 22 | 2-Methyl-Z-4-tetradecene | C15H30 | 22.391 | 0.37 |
| 23 | 1,7,11-Trimethyl-4-(1-methylethyl) cyclohexane | C20H40 | 22.768 | 1.00 |
| 24 | Disulfide, di-tert-dodecyl | C24H50S2 | 23.40 | 2.15 |
| 25 | n-Nonenylsuccinic anhydride | C13H20O3 | 23.574 | 1.38 |
| 26 | Phosphine sulfide | C8H11PS | 23.980 | 1.03 |
| 27 | Nonadecyl heptafluorobutyrate | C23H39F7O2 | 24.186 | 0.23 |
| 28 | Cyclotetradecane | C14H28 | 24.506 | 2.38 |
| 29 | Tetratetracontane | C44H90 | 24.700 | 2.73 |
| 30 | Triacontyl pentafluoropropionate | C33H61F5O2 | 25.094 | 2.17 |
| 31 | Cyclopropane carboxamide | C4H7NO | 25.272 | 0.22 |
| 32 | 2-Methylallyl undecyl ester | C19H34O4 | 25.494 | 2.14 |
| 33 | 2,5-Furandione | C4H4O3 | 25.712 | 1.11 |
| 34 | 2,6,10,14-Tetramethyl-7-(3-methylpent-4-enylidene) pentadecane | C25H48 | 25.957 | 2.36 |
| 35 | 3-Dodecyl- docosane | C22H46 | 26.140 | 2.32 |
| 36 | Tricosane | C23H48 | 26.906 | 1.87 |
| 37 | 2-Methyl-cis-7,8-epoxynonadecane | C20H40O | 27.089 | 1.05 |
| 38 | 11,13-Dimethyl-12-tetradecen-1-ol | C18H34O2 | 27.089 | 1.20 |
| 39 | 2,6,10,14-Tetramethyl-7-(3-methylp ent-4-enylidene) pentadecane | C25H48 | 27.386 | 1.20 |
| 40 | Cyclohexane, 1-(1,5-dimethylhexyl) -4-(4-methylpentyl) | C20H40 | 27.535 | 7.11 |
| 41 | (1R,2R,8S,8Ar)-8-hydroxy-1-(2-hydr oxyethyl)-1,2,5,5-tetramethyl-cis decalin | C16H30O2 | 28.198 | 1.60 |
| 42 | 2-Methyl-7-hydroxy-8-allyl-isoflavone | C13H12O | 28.564 | 5.13 |
| 43 | 2,6,10,14-Tetramethyl-7-(3-methylp ent-4-enylidene) pentadecane | C25H48 | 29.078 | 1.97 |
| 44 | Heptadecyl propargyl ester | C14H20O | 30.164 | 1.70 |
| 45 | Butanamide, 3-(3-fluorobenzoylhydrazono)-N-(4-fluorobenzyl) | C11H15FN2O | 30.524 | 1.92 |
| 46 | 3,7-Decadiyne, 2,2,5,5,6,6,9,9-oct amethyl | C18H30 | 30.718 | 0.67 |
| 47 | Decahydro-8a-ethyl-1,1,4a,6-tetra methylnaphthalene | C16H30O2 | 31.067 | 1.47 |
| 48 | 3-Ethyl-4,6-dimethyl-2,6-dioxo-4,5,6,7-tetrahydro-1,2,3-triazolo[4,5-d]pyrimidine | C17H10ClF3N2 | 31.496 | 1.00 |
| 49 | D-Homoandrostane, (5.alpha.,13.alp ha.) | C20H34 | 32.004 | 0.88 |
| 50 | 2-(4-Fluoro-phenyl)-4-(3-methyl-benzylidene)-4H-oxazol-5-one | C16H11NO2 | 32.216 | 0.55 |
| 51 | 1-Methyl-4-(1-methylethyl)-3-[1-methyl-1-(4-methylpentyl)-5-methylheptyl]cyclohexene | C14H22O | 32.524 | 1.14 |
| 52 | 5-Butyl-6-hexyloctahydro (2Z,4E)-3,7,11-trimethyl-2,4,10-dodecatriene | C15H26 | 32.679 | 0.47 |
| 53 | Isoxazole, 4-(4-fluorobenzoyl)-5-m ethyl-3-phenyl decahydro-8a-ethyl-1,1,4a,6-tetra methylnaphthalene | C12H9F3N2O2 | 33.050 | 2.00 |
| 54 | 1H-Indene, 5-butyl-6-hexyloctahydro isoxazole, 4-(4-fluorobenzoyl)-5-m ethyl-3-phenyl | C19H36 | 33.387 | 0.45 |
| 55 | 28-Nor-17.alpha.(H)-hopane 17.alfa.,21.beta.-28,30-bisnorhopane | C29H50 | 33.530 | 0.82 |
| 56 | Phenanthrene | C14H10 | 34.130 | 2.55 |
| 57 | Cyclobarbital | C12H16N2O3 | 34.599 | 0.63 |
| 58 | 1,2,5-Oxadiazol-3-amine | C2H3N3O | 34.988 | 1.46 |
| 59 | (E)-2-Bromobutyloxychalcone | C19H19BrO2 | 35.370 | 0.47 |
| 60 | Ethyl 2-(2-chloroacetamido)-3,3,3- trifluoro-2-(4-fluoroanilino) propionate | C13H13ClF4N2O3 | 35.753 | 0.71 |
| 61 | Benzamide | C7H7NO | 35.931 | 2.30 |
| 62 | 5-Methyl-2-phenylindolizine | C15H13N | 36.559 | 0.25 |
| 63 | 2,3-Dihydroxy-6-nitroquinoxaline | C8H5N3O4 | 36.788 | 0.29 |
| 64 | 6-Fluoro-2-trifluoromethylbenzoic acid | C15H6Cl2F4O3 | 37.022 | 1.39 |
| 65 | 2,4,5-Trichlorophenyl ester | C13H7Cl3O2 | 37.274 | 2.14 |
| 66 | Tris(trimethylsilyl) ester | C9H27AsO3Si3 | 38.051 | 0.32 |
| 67 | (E)-2-Bromobutyloxychalcone | C19H19BrO2 | 39.148 | 0.43 |
| 68 | 1,2-Benzisothiazol-3-amine | C7H6N2S | 39.394 | 0.46 |
| 69 | Tris(tert-butyldimethylsilyloxy) arsane | C18H45AsO3Si3 | 40.886 | 0.24 |
| 70 | Benzo[h]quinolone | C13H9N | 41.303 | 0.30 |
The findings indicated that the C30 component was present in the surface water (Fig. S5). Edori et al.74 made a similar observation from the Edagberi River in the Niger Delta, Nigeria, suggesting that the compound's hydrophilic nature may be the cause of its existence. The presence of C10, C11, C22, C28 and C30 in the water may also result from river-related disturbances and flow patterns.74 Similar to the findings of Sharma et al.,143 cyclic chemicals, including derivatives of naphthalene, phenanthrene, phenol and annulene, were also found in the water sample. These substances are extremely harmful, mutagenic and carcinogenic to both people and other living things.144
| PK | Compound | Formula | PT | Area |
|---|---|---|---|---|
| a PK = peak number, RT = retention time. | ||||
| 1 | 2-Butenedioic acid (Z)-, dibutyl ester | C12H20O4 | 8.506 | 8.78 |
| 2-Butenedioic acid (Z)-, dibutyl ester | C12H20O4 | |||
| 1,4-Dioxaspiro [4.6] undec-7-ene | C9H14O2 | |||
| 2 | 1,2-Benzenedicarboxylic acid, bis(2-methylpropyl) ester | C16H22O4 | 12.106 | 77.00 |
| 1,2-Benzenedicarboxylic acid, bis(2-methylpropyl) ester | C16H22O4 | |||
| Phthalic acid, isobutyl octyl ester | C20H30O4 | |||
| 3 | Phthalic acid, 1-tert-butoxyprop-2-yl isobutyl ester | C19H28O4 | 13.072 | 3.73 |
| Phthalic acid, 1-tert-butoxyprop-2-yl butyl ester | C19H28O4 | |||
| Phthalic acid, isobutyl 5-methoxy-3-methylpent-2-yl ester | C18H22O5 | |||
| 4 | Bis(2-ethylhexyl) phthalate | C24H38O4 | 18.027 | 3.75 |
| Diisooctyl phthalate | C24H38O4 | |||
| Bis(2-ethylhexyl) phthalate | C24H38O4 | |||
| 5 | Nonyl tetradecyl ether | C23H48O | 18.479 | 3.21 |
| Dodecyl nonyl ether | C21H44O | |||
| 1-Octadecanesulphonyl chloride | C18H37CLO2S | |||
| 6 | Eicosyl isopropyl ether | C23H48O | 19.228 | 3.53 |
| 2-Methyltetracosane | C25H52 | |||
| Tetratetracontane | C44H90 | |||
The chromatograms (Fig. S5) showed that the samples that the AgNPs worked upon had lower levels of total petroleum hydrocarbon (TPH). This finding validated the biosynthesized AgNP's capacity to degrade several crude oil compounds found in the water that was polluted. Numerous alkanes (C10–C44), including both long chain alkanes [heneicosane (C21), n-docosane (C22), n-heptacosane (C27), hexatriacontane (C36), and tetratetracontane (C44)] and short chain alkanes [n-decane (C10), n-undecane (C11), dodecane (C12), n-hexadecane (C16), heptadecane (C17), and n-nonadecane (C19)] that were present in the untreated sample were broken down to their derivatives or the concentration was attenuated or completely degraded by the bacterially produced AgNP (Tables 5 and 6). Similar to the findings of Ndekhedehe et al.,145 other aromatic hydrocarbons were also present (benzamide, (E)-2-bromobutyloxychalcone, 5-methyl-2-phenylindolizine, and 1-eicosene 2(1H)-naphthalenone), along with some polycyclic aromatic hydrocarbons (phenanthrene, benzo[h]quinolone), which were found to have decreased following remediation. These organic compounds are typically hydrophobic in nature due to the presence of two or more benzene rings, which makes them resistant to structural degradation.146 The success of this study may therefore be attributed to the nanobioremediation technique.92
3.13 Mechanistic degradation of hydrocarbon using biosynthesized AgNP
Biosynthesis of P. aeruginosa-mediated silver nanoparticles for remediation of crude oil-contaminated water follows a coupled biological and physicochemical degradation mechanism. When Pseudomonas aeruginosa-mediated silver nanoparticles are introduced into crude-oil-contaminated water, these biogenic AgNPs enhance degradation through both catalytic and synergistic microbial pathways. These nanoparticles have strong electron-shuttling capabilities to accelerate the oxidative cleavage of long-chain hydrocarbons. Their large specific surface area facilitates adsorption of aliphatic and aromatic hydrocarbons, offering increased contact between the contaminant and degradative enzymes secreted by P. aeruginosa. Simultaneously, AgNPs stimulate bacterial membrane permeability at sub-lethal concentrations and up-regulate hydrocarbon-degrading enzymes. These enzymes oxidize hydrocarbons in a sequential manner, which transforms them into alcohols, aldehydes, and carboxylic acids; these are further mineralized via β-oxidation and the tricarboxylic acid cycle into CO2 and H2O. The biogenic AgNPs further produce low levels of ROS, which stimulate partial oxidation and fragmentation of high-molecular-weight fractions of crude oil, hence transforming recalcitrant compounds into intermediates more amenable to biodegradation. The capping molecules on the surface of AgNP enhance emulsification through reduction of interfacial tension, which favors breakdown of hydrophobic crude oil droplets into smaller micelles accessible to microbial cells. These processes collectively lead to a synergistic catalytic–biodegradation system wherein the nanoparticles accelerate initial physicochemical breakdown while P. aeruginosa accomplishes metabolic mineralization.4 Conclusion
This study demonstrates that P. aeruginosa strain AgA is an efficient hydrocarbon-degrading bacterium capable of mediating the green synthesis of silver nanoparticles for effective crude-oil pollution remediation. Total petroleum hydrocarbons were found to be present in high concentrations in the water samples obtained from the Jones Creek oil Field communities, with mean values ranging from 6.55 ± 0.91 to 2324.36 ± 4.23 mg L−1. The analysis of the physicochemical parameters also showed that the water samples were contaminated. From the water contaminated by crude oil, nine (9) bacterial species were identified, characterized and screened for their ability to degrade crude oil. The microorganisms that degraded bonny light crude oil at different rates were identified as Pseudomonas aeruginosa, Pseudochrobactrum asaccharolyticum, Bacillus megaterium, Micrococcus luteus, Bacillus subtilis, Escherichia coli, Bacillus cereus, Macrococcus caseolyticus and Bacillus thuringiensis. The bacterial isolate that demonstrated high growth and the ability to degrade petroleum was further identified using molecular techniques such as Pseudomonas aeruginosa strain AgA and was used for the extracellular synthesis of AgNPs, which was initially verified by visual observation and characterized by UV-vis spectroscopy, XRD, HRTEM and EDS analyses. The distinctive plasmon absorption peak for AgNPs was visible in the UV-vis spectra at 425 nm. The resulting nanoparticle had a spherical form, was polydispersed and had an average crystallite size of 18.01504 ± 4.03 nm with a particle size distribution of 34.60 ± 13.04 nm. The existence of elemental silver and the crystalline form of the AgNP were confirmed by the strong signals in the silver region of the EDS and XRD spectra. The 2FI model for the AgNP was chosen after TPH removal was designed and optimized using response surface methodology. Based on a P-value of <0.05, the ANOVA analysis demonstrated that both the model and its terms were significant. It was discovered that the following parameters were ideal for the AgNP to remove the most TPH: 22.5 min of contact time, 1620 rpm of stirring speed, 0.3206 g of dosage and 67.5 °C of temperature. AgNP synthesized using Pseudomonas aeruginosa strain AgA (PAE) yielded an optimal TPH removal effectiveness of 99.98% under the optimized conditions. At a desirability value of 0.994 (99.4%), the response value was very similar to the experiment value under the best conditions suggested. The experimental results verified that the model chosen was a promising innovation in lowering hydrocarbons and other harmful pollutants in water samples from the creeks of the Niger Delta and similar ecosystems elsewhere, and that all the factors examined were significant for the variable response.Consent to publish
All the authors agreed to submit the manuscript to this journal for publication.Conflicts of interest
There is no any competing interest in the study.Data availability
The data supporting this article have been included as part of the Supplementary Information (SI). Supplementary information: Tables S1–S6, Fig. S1–S5 and Plates S1–S3. See DOI: https://doi.org/10.1039/d5ra08196c.Acknowledgements
The study was not funded by any organization; the authors contributed their financial resources for the study.References
- A. A. Ikhumetse, O. P. Abioye, U. J. J. Ijah and M. T. Bankole, A critical review of oil spills in the Niger Delta aquatic environment: causes, impacts, and bioremediation assessment, Environ. Monit. Assess., 2002, 194(11), 816 Search PubMed.
- P. O. Olajide and A. O. Adeloye, Hydrocarbon biodegradation by Proteus and Serratia strains isolated from oil-polluted water in Bonny community, Niger Delta, Nigeria, Results Chem., 2023, 5, 1–5 Search PubMed.
- C. O. Udinyiwe, I. Idemudia and F. O. Ekhaise, Biodegradation potential of bacterial isolates from crude oil contaminated soil samples from Gelegele River, Edo State, J. Sci. Technol. Res., 2022, 4(1), 244–257 Search PubMed.
- A. A. Ikhumetse, O. P. Abioye, U. J. J. Ijah and A. S. Kovo, Assessment of physicochemical quality, heavy metals, and total petroleum hydrocarbon concentration in surface water from communities of the Jones creek oil field, Niger Delta, Nigeria, IJRIAS, 2024, 9(5), 75–93 CrossRef.
- E. S. Edori, Physical and chemical characteristics of water from Ede Onyima Creek, Okarki-Engenni, Rivers State, Nigeria, Chem. Res. J., 2022,(3), 144–154 Search PubMed.
- M. P. Wanjala, L. Odokuma, I. Etela, R. Ramkat and B. A. Odogwu, Characterization of culturable microbial community in oil contaminated soils in Greater Port Harcourt area, Nigeria, Afr. J. Microbiol. Res., 2022, 16(1), 32–42 CrossRef.
- S. A. Aransiola, S. S. Leh-Togi Zobeashia, A. A. Ikhumetse, O. I. Musa, O. P. Abioye, U. J. J. Ijah and N. R. Maddela, Niger Delta mangrove ecosystem: biodiversity, past and present pollution, threat and mitigation, Reg. Stud. Mar. Sci., 2024, 75, 103568 Search PubMed.
- A. A. Ikhumetse, O. P. Abioye, U. J. J. Ijah and A. S. Kovo, Biosynthesis of silver nanoparticles using Macrococcus caseolyticus strain AgD isolated from crude oil contaminated water samples and their application in remediation of crude oil contamination in the Niger Delta aquatic environment, Environ. Nanotechnol., Monit. Manage., 2025, 23, 101057 CAS.
- O. A. Akpoka, G. O. Erifeta, O. S. Imade, E. J. Okafor-Elenwo, A. A. Enaigbe and D. S. Abolarin, Isolation and characterization of crude oil degrading bacteria in association with microalgae in saver pit from Egbaoma flow station, Niger Delta, Nigeria, Arch. Ecotoxicol., 2020, 2(2), 12–16 CrossRef.
- C. E. Achife, U. J. J. Ijah, J. D. Bala and S. B. Oyeleke, Microbial population of soil and water around petroleum depot Suleja, Nigeria, and their hydrocarbon utilization, Int. J. Life Sci. Biotechnol., 2021, 4(1), 90–113 CrossRef CAS.
- W. Dai, C. Wang, Y. Wang, J. Sun, H. Ruan, Y. Xue and S. Xiao, Unlocking photocatalytic NO removal potential in an S-type UiO-66-NH2/ZnS (en) 0.5 heterostructure, Interdiscip. Mater., 2024, 3(3), 400–413 CAS.
- J. Guo, H. Ma, H. Shang, W. Wang, R. Yang, S. Wang, Y. Miao, D. L. Phillips, G. Li and S. Xiao, Dual-site Langmuir-Hinshelwood mechanism in ZnCr-LDH/NH2-UIO66 heterojunction for efficient photocatalytic NO oxidation, J. Hazard. Mater., 2025, 492, 138060 CrossRef CAS PubMed.
- M. A. El-Liethy, M. M. El-Noubi, A. L. K. Abia, M. G. El-Malky, A. I. Hashem and G. E. El-Taweel, Eco-friendly bioremediation approach for crude oil-polluted soils using a novel and biostimulated Enterobacter hormaechei ODB H32 strain, Int. J. Environ. Sci. Technol., 2022, 19, 10577–10588 CrossRef CAS.
- A. U. Osadebe, C. J. Ogugbue and G. C. Okpokwasili, Diversity and degradative potency of extant autochthonous crude oil-metabolizing species in a chronically polluted River, Pollution, 2023, 9(2), 795–809 CAS.
- K. R. Jeya, M. Veerapagu, A. Sankaranarayanan and R. Sathyapriya, Isolation and screening of kerosene oil degrading bacteria from contaminated soil of motor mechanic workshop, Int. J. Pharm. Biol. Sci., 2018, 8(2), 683–688 CAS.
- N. Benibo, O. Obire, S. I. Douglas and R. R. Nrior, Hydrocarbonoclastic bacteria and polycyclic aromatic hydrocarbon profile of surface water in Borikiri wetlands, Port Harcourt, Nigeria, Acta Sci. Microbiol., 2023, 6(3), 41–45 CrossRef.
- R. Esmail, A. Afshar, M. Morteza, A. Abolfaz and E. Akhondi, Synthesis of silver nanoparticles with high efficiency and stability by culture supernatant of Bacillus ROM6 isolated from Zarshouran gold mine and evaluating its antibacterial effects, BMC Microbiol., 2022, 22(97), 1–10 Search PubMed.
- M. G. Muñoz-Carrillo, C. Garcidueñas-Piña, R. C. Valerio-García, J. L. Carrazco-Rosales and J. F. Morales-Domínguez, Green synthesis of silver nanoparticles from the Opuntia ficus-indica fruit and its activity against treated wastewater microorganisms, J. Nanomater., 2020, 1–10 Search PubMed.
- S. S. Salem, E. N. Hammad, A. A. Mohamed and W. El-Dougdoug, A comprehensive review of nanomaterials: types, synthesis, characterization and applications, Biointerface Res. Appl. Chem., 2023, 13(1), 1–30 Search PubMed.
- M. N. Saleh and S. K. Alwan, Biosynthesis of silver nanoparticles from bacteria Klebsiella pneumonia: Their characterization and antibacterial studies, J. Phys., 2020, 1664, 1–16 Search PubMed.
- S. Mandal, S. B. Marpu, R. Hughes, M. A. Omary and S. Q. Shi, Green synthesis of silver nanoparticles using Cannabis sativa extracts and their anti-bacterial activity, Green Sustainable Chem., 2021, 11, 38–48 CAS.
- S. Sattar, S. Siddiqui, A. Shahzad, A. Bano, M. Naeem, R. Hussain, N. Khan, B. L. Jan and H. Yasmin, Comparative analysis of microbial consortiums and nnoparticles for rehabilitating petroleum waste contaminated soils, Molecules, 2022, 27, 1945 CrossRef CAS PubMed.
- B. Muthukumar, M. S. Al Salhi, J. Narenkumar, S. Devanesan, T. N. Rao, W. Kim and A. Rajasekar, Characterization of two novel strains of Pseudomonas aeruginosa on biodegradation of crude oil and its enzyme activities, Environ. Pollut., 2022, 304, 119223 CrossRef CAS PubMed.
- R. Rehman, M. I. Ali, N. Ali, M. Badshah, M. Iqbal, A. Jamal and Z. Huang, Crude oil biodegradation potential of biosurfactant-producing Pseudomonas aeruginosa and Meyerozyma sp, J. Hazard. Mater., 2021, 418, 126276 Search PubMed.
- M. Ahmadi, F. Niazi, N. Jaafarzadeh, S. Ghafari and S. Jorfi, Characterization of the biosurfactant produced by Pseudomonas aeruginosa strain R 4 and its application for remediation pyrene-contaminated soils, J. Environ. Health Sci. Eng., 2021, 19(1), 445–456 CrossRef CAS.
- C. A. Ottoni, M. L. Neto, P. Léo, B. D. Ortolan, E. Barbieri and A. O. De Souza, Environmental impact of biogenic silver nanoparticles in soil and aquatic organisms, Chemosphere, 2020, 239, 124698 CrossRef CAS.
- H. Jangid and G. Kumar, Ecotoxicity of fungal-synthesized silver nanoparticles: mechanisms, impacts, and sustainable mitigation strategies, 3 Biotech, 2025, 15(4), 1–31 CrossRef PubMed.
- F. Iyeritei, O. Obire and S. I. Douglas, An evaluation of the physicochemical quality of Lobia creek in the Niger Delta, Int. J. Curr. Microbiol. Appl. Sci., 2022, 11(06), 224–237 Search PubMed.
- APHA, American Public Health Association, Standard Methods for Examination of Water and Waste Water, American Public Health Association, Washington, DC, 2017, 23rd edn, pp. 127–134 Search PubMed.
- H. Liu, J. Yao, Z. Yuan, Y. Shang, H. Chen, F. Wang, K. Masakorala, C. Yu, M. Cai, R. E. Blake and M. M. F. Choi, Isolation and characterization of crude-oil-degrading bacteria from oil-water mixture in Dagang oilfield, China, Int. Biodeterior. Biodegrad., 2014, 87, 52–59 Search PubMed.
- H. S. El-Sheshtawy, N. M. Khalil, W. Ahmed and A. A. Nabila, Enhancement the bioremediation of crude oil by nanoparticle and biosurfactants, Egypt. J. Chem., 2017, 60(5), 835–848 Search PubMed.
- A. M. El-Borai, K. M. Eltayeb, A. R. Mostafa and S. A. El-Assar, Biodegradation of industrial oil-polluted wastewater in Egypt by bacterial consortium immobilized in different types of carriers, J. Environ. Stud., 2016, 25(5), 1901–1909 CAS.
- V. C. Eze, C. E. Onwuakor and F. E. Orok, Microbiological and physicochemical characteristics of soil contaminated with used petroleum products in Umuahia, Abia State, Nigeria, J. Appl. Environ. Microbiol., 2014, 2(6), 281–286 Search PubMed.
- S. A. Raed and R. H. Shimaa, Bacterial biodegradation of crude oil using local isolates, Int. J. Bacteriol., 2014, 2014, 1–8 Search PubMed.
- A. Gupte and S. Sonawdekar, Study of oil degrading bacteria isolated from oil contaminated sites, IJRASET, 2015, 3(2), 345–349 Search PubMed.
- R. S. Al-Wasify and S. R. Hamed, Bacterial biodegradation of crude oil using local isolates, Int. J. Bacteriol., 2014, 1–8 Search PubMed.
- M. Hassanshahian, G. Emtiazi and S. Cappello, Isolation and characterization of crude-oil-degrading bacteria from the Persian Gulf and the Caspian Sea, Mar. Pollut. Bull., 2012, 64, 7–12 CrossRef CAS PubMed.
- M. Bao, L. Wang, P. Sun, L. Cao, J. Zou and Y. Li, Biodegradation of crude oil using an efficient microbial consortium in a simulated marine environment, Mar. Pollut. Bull., 2012, 64, 1177–1185 CrossRef CAS PubMed.
- N. Daxini and K. Mistry, Bio-surfactant assistance in crude oil degradation by halophilic Bacillus cereus ND1, Indian J. Geo-Mar. Sci., 2018, 47(8), 1640–1647 Search PubMed.
- H. Lodish, A. Berk, and P. Matsudaira, Molecular Cell Biology, WH Freeman, New York, NY, 2004, 5th edn, pp. 123–126 Search PubMed.
- L. C. Trindade, E. Marques, D. B. Lopes and M. A. S. V. Ferreira, Development of a molecular method for detection and identification of Xanthomonas campestris pv. Viticola, Summa Phytopathol., 2007, 33(1), 16–23 Search PubMed.
- C. Pisapia, E. Gerard, M. Gerard and B. Meñez, Mineralizing filamentous bacteria from the Porny Bay hydrothermal field give new insights into the functioning of serpentinization-based subsea floor ecosystems, Front. Microbiol., 2017, 8, 1–18 Search PubMed.
- K. Katoh and H. Toh, Recent developments in the MAFFT multiple sequence alignment program, Briefings Bioinf., 2008, 9(4), 286–298 Search PubMed.
- G. Talavera and J. Castresana, Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments, Syst. Biol., 2007, 56(4), 564–577 Search PubMed.
- J. S. Adriano, G. G. Oyong, E. C. Cabrera and J. I. B. Janairo, Screening of silver-tolerant bacteria from a major Philippine landfill as potential bioremediation agents, Ecol. Chem. Eng., S, 2018, 25(3), 469–485 Search PubMed.
- E. Ibrahim, H. Fouad, M. Zhang, Y. Zhang, W. Qiu, C. Yan, B. Li, J. Moc and J. Chen, Biosynthesis of silver nanoparticles using endophytic bacteria and their role in inhibition of rice pathogenic bacteria and plant growth promotion, RSC, 2019, 9, 29293–29299 CAS.
- K. I. Alsamhary, Eco-friendly synthesis of silver nanoparticles by Bacillus subtilis and their antibacterial activity, Saudi J. Biol. Sci., 2020, 27, 2185–2191 Search PubMed.
- A. Salayová, Z. Bedlovicová, N. Daneu, M. Baláž, Z. L. Bujnáková, L. Balážová and L. Tkáˇciková, Green synthesis of silver nanoparticles with antibacterial activity using various medicinal plant extracts: Morphology and antibacterial efficacy, Nanomaterials, 2021, 11, 1–20 CrossRef PubMed.
- J. Sukweenadhi, K. I. Setiawan, C. Avanti, K. Kartini, E. J. Rupa and D. Yang, Scale-up of green synthesis and characterization of silver nanoparticles using ethanol extract of Plantago major L. leaf and its antibacterial potential, S. Afr. J. Chem. Eng., 2021, 38, 1–8 Search PubMed.
- H. L. Abubakar, J. O. Tijani, A. S. Abdulkareem, T. C. Egbosiuba, M. Abdullahi, S. Mustapha and E. A. Ajiboye, Effective removal of malachite green from local dyeing wastewater using zinc-tungstate based materials, Heliyon, 2023, 9, 1–26 CrossRef.
- C. Rodrıguez-Serrano, J. Guzman-Moreno, C. Angeles-Chave, V. Rodrıguez-Gonzalez, J. J. Ortega-Sigala and R. M. Ramırez-Santoyo, Biosynthesis of silver nanoparticles by Fusarium scirpi and its potential as antimicrobial agent against uropathogenic Escherichia coli biofilms, PLoS One, 2020, 15(3), 1–20 CrossRef.
- O. D. Onukwuli, P. C. Nnaji, M. C. Menkiti, V. C. Anadebe, E. O. Oke, C. N. Ude, C. J. Ude and N. A. Okafor, Dual-purpose optimization of dye-polluted wastewater decontamination using bio-coagulants from multiple processing techniques via neural intelligence algorithm and response surface methodology, J. Taiwan Inst. Chem. Eng., 2021, 125, 372–386 CrossRef CAS.
- WHO, World Health Organization, Guidelines for Standard Operating Procedures for Microbiology: in: Bacteriological Examination of Water, World Health Organization Regional Office for South-East Asia, 2012, pp. 115–126 Search PubMed.
- SON, Standards Organization of Nigeria, Nigerian Industrial Standard: Nigerian Standard for Drinking Water Quality, Abuja, 2007, pp. 11–19 Search PubMed.
- Federal Environmental Protection Agency, Guidelines and Standards for Environmental Pollution Control in Nigeria, 2003, p. 420 Search PubMed.
- O. Efekemo, I. C. Davies and C. O. Orororo, Water quality assessment and heavy metal levels in Mudskipper (Periophthalmus papilio), sediments and water of mangrove swamps, Rivers State, Nigeria, AJENSR, 2024, 7(1), 128–145 Search PubMed.
- E. I. Asionye, A. P. Bariweni, J. E. Idome and A. P. Onyena, Assessment of physicochemical and microbiological qualities of potable water sources in Okerenkoko, Delta State, Nigeria, JLBSR, 2023, 4(1), 31–39 CrossRef.
- M. C. Onojake, F. D. Sikoki, O. Omokheyeke and R. U. Akpiri, Surface water and trace metal level of Bonny/New Calabar River Estuary, Niger Delta, Nigeria, Appl. Water Sci., 2017, 7(2), 951–959 CrossRef CAS.
- U. Effiong and S. E. Etteokon, Preliminary study on the diversity of plankton flora and water quality of a tropical mangrove estuarine system, Akwa Ibom State, Niger Delta Area, Nigeria, AJOL, 2020, 3(6), 34–45 Search PubMed.
- P. E. Edeki, E. C. Isaha and N. Mokogwua, Assessment of physicochemical and bacteriological quality of drinking water in Sapele Local Government Area of Delta State, South-South, Nigeria, J. Water Health, 2023, 21(2), 286–298 Search PubMed.
- O. Ajayi and O. C. Okeke, Assessment of quality of surface water resources of Kolo creek and environs, Niger Delta, Nigeria, using water quality index and multivariate statistical analysis, Sci. Technol. Stud., 2024, 3(1), 101–138 Search PubMed.
- K. Sam, A. P. Onyena, O. J. Eriegha and F. Eze, Water quality evaluation using water quality index and pollution model in selected communities in Gbaramatu Kingdom, Niger Delta, Nigeria, Afr. J. Environ. Sci. Technol., 2023, 17(6), 118–134 CrossRef.
- A. I. Israel, I. Etim and O. V. Etim, Effects of gas flaring on surface water in Mkpanak community of Akwa Ibom State, Nigeria, Int. J. Eng. Res. Sci. Technol., 2019, 8(9), 566–573 Search PubMed.
- I. F. Vincent-Akpu, A. N. Tyler, C. Wilson and G. Mackinnnon, Assessment of physico-chemical properties and metal contents of water and sediments of Bodo Creek, Niger Delta, Nigeria, Toxicol. Environ. Chem., 2015, 97(2), 135–144 CrossRef CAS.
- E. I. Seiyaboh, S. C. Izah and S. Oweibi, Assessment of water quality from Sagbama Creek, Niger Delta, Nigeria, Biotechnol. Res., 2017, 3(1), 20–24 CAS.
- A. Chukwuma, J. Ugbebor and E. Ugwoha, Assessment of the physicochemical characteristics and water quality index of Nkisa river in the Niger Delta, Nigeria, IJNRES, 2024, 10(2), 95–105 Search PubMed.
- K. Friday, O. S. Edori and V. C. Nwokanma, Assessment of physicochemical parameters in Nta-Wogba stream in Port Harcourt, Rivers State, Nigeria, J. Sci. Eng. Res., 2020, 7(8), 124–132 CAS.
- N. O. Ahmed, G. J. Udom and A. A. Obafemi, Chemophysical and metallic characterization of surface water and precipitation for environmental quality assessment in Oyigbo LGA, Rivers State, Nigeria, J. Global Ecol. Environ., 2024, 2(1), 28–57 Search PubMed.
- E. S. Edori, W. A. Iyama and M. Q. C. Amadi, Variation of some physicochemical parameters in surface water of Elelenwo River, Rivers State, Niger Delta, Nigeria, IJRSI, 2020, 7(5), 230–235 Search PubMed.
- N. A. Akpan, R. B. Udombeh, M. B. Ukpong and I. E. Udosen, Impacts of human activities on trace metals of Qua Iboe River, Ikot Ekpene stretch, Akwa Ibom State, Nigeria, Int. J. Sci. Res. Arch., 2024, 11(01), 2120–2128 CrossRef.
- D. Okoro and L. C. Diejomaoh, Profiling the surface water around Odeama Community of the Niger Delta area of Nigeria, J. Environ. Chem. Ecotoxicol., 2022, 14(1), 9–25 Search PubMed.
- DPR, Department of Petroleum Resources, Environmental Guidelines and Standards for the Petroleum Industry in Nigeria (EGASPIN), Universal Press, Lagos, Nigeria, Revised Edition, 2011, pp. 276–297 Search PubMed.
- C. A. Dimkpa, I. H. Dimkpa and M. A. Achadu, Analysis of heavy metals and petroleum-based contaminants in surface water and sediment near two illegal refining sites in Niger Delta, Nigeria, PPEJ, 2024, 8(2), 1–9 Search PubMed.
- E. S. Edori, F. Kpee and A. C. Marcus, Total petroleum hydrocarbon fractions (components) in surface water and sediments of Edagberi River, Niger Delta, Nigeria, IJRIAS, 2020, 5(10), 1–8 Search PubMed.
- W. Z. Zhang, P. Gu, W. J. Zhu, N. Wang, M. Q. Jiang and J. He, Phenotype changes of Cyanobacterial and microbial distribution characteristics of surface sediments in different periods of cyanobacterial blooms in Taihu Lake, Aquat. Ecol., 2020, 54, 591–607 Search PubMed.
- I. M. Omoruyi and J. I. Amadi, Impact of petrochemical activities on the physicochemical and microbiological quality of surface water, ground water and soil in Otu-Jeremi, Iwerehkan and Okpare communities, Ughelli South Local Government Area, Delta State, Nigeria, FJS, 2022, 6(3), 1–14 CrossRef.
- P. Galitskaya, L. Biktasheva, S. Blagodatsky and S. Selivanovskaya, Response of bacterial and fungal communities to high petroleum pollution in different soils, Sci. Rep., 2021, 11, 164–183 CrossRef CAS.
- O. P. Abioye, P. O. Amaefule, S. A. Aransiola and E. Stephen, Screening of bacterial consortium isolated from oil contaminated soil for its potential to degrade crude oil, Adv. Sci. Focus, 2013, 1(3), 1–4 Search PubMed.
- E. I. Seiyaboh, F. O. Youkparigha, S. C. Izah and I. D. Daniels, Bacteriological quality of groundwater in Imiringi Town, Bayelsa State, Nigeria, J. Biotechnol. Biomedi. Sci., 2020, 2(2), 34–40 Search PubMed.
- M. Hassanshahian, The effects of crude oil on marine microbial communities in sediments from the Persian Gulf and the Caspian Sea: A microcosm experiment, Int. J. Adv. Biol. Biomed. Res., 2014, 2(1), 1–17 CAS.
- S. D. Limaa, A. F. Oliveiraa, R. Golina, V. C. P. Lopesa, D. S. Caixetaa, Z. M. Limaa and E. B. Moraisa, Isolation and characterization of hydrocarbon-degrading bacteria from gas station leaking-contaminated groundwater in the Southern Amazon, Brazil, Braz. J. Biol., 2020, 80(2), 354–361 CrossRef PubMed.
- E. C. Omenna, K. Omage, E. Ezaka and M. A. Azeke, Tolerance, taxonomic and phylogenetic studies of some bacterial isolates involved in bioremediation of crude oil-polluted soil in the southern region of Nigeria, Heliyon, 2023, 9, 1–14 CrossRef.
- N. Mukjang, T. Chitov, W. Mhuantong, V. Champreda, W. Pathom-aree, P. Sattayawat and S. Bovonsombut, Bacterial communities associated with crude oil bioremediation through composting approaches with indigenous bacterial isolate, Life, 2022, 12(1712), 1–14 Search PubMed.
- H. Liu, G. Yang, H. Jia and B. Sun, Crude oil degradation by a novel strain Pseudomonas aeruginosa AQNU-1 isolated from an oil-contaminated lake wetland, Processes, 2022, 10, 307–322 CrossRef CAS.
- R. De-la-Fuente, G. Suarez, J. A. Ruiz-Santa-Quiteria, H. Meugnier and M. Bes, Identification of coagulase-negative staphylococci isolated from lambs as Staphylococcus caseolyticus, Comp. Immunol., Microbiol. Infect. Dis., 1992, 15, 47–52 CrossRef CAS PubMed.
- T. Baba, K. Kuwahara-Arai, I. Uchiyama, F. Takeuchi, T. Ito and K. Hiramatsu, Complete genome sequence of Macrococcus caseolyticus strain JSCS5402, reflecting the ancestral genome of the human-pathogenic Staphylococci, J. Bacteriol., 2009, 191(4), 1180–1190 CrossRef CAS.
- M. Karani, R. Shashidhar, A. Kakatkar, R. K. Gautam, S. Sukhi, L. Pansare-Godambe and J. Bandekar, Radiation-resistant Macrococcus caseolyticus (A) isolated from radiation-processed semidried prawns, Can. J. Microbiol., 2015, 61, 89–92 Search PubMed.
- M. Pelletier and J. Draper, Characterization and identification of bacterial flora from infected equine hooves, Int. J. Vet. Sci. Res., 2022, 8(2), 050–056 CrossRef.
- W. E. Kloos, D. N. Ballard, C. G. George, J. A. Webster, R. J. Hubner, W. Ludwig, K. H. Schleifer, F. Fiedler and K. Schubert, Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., and Macrococcus bovicus sp. no. and Macrococcus carouselicus sp. Nov, Int. J. Syst. Bacteriol., 1998, 48, 859–877 Search PubMed.
- M. Sharma and H. K. Dhingra, Isolation and culture conditions optimization for PHB production by Pseudochrobactrum asaccharolyticum, IJSR, 2015, 4(10), 1895–1901 Search PubMed.
- L. Edemhanria, A. A. Daodu, E. I. Ebhohimen and C. C. Osubor, Crude oil utilization and degradation potential of microbes isolated from aged crude oil polluted soil in Niger Delta, Nigeria, NRSD, 2020, 10(2), 262–271 Search PubMed.
- Y. M. Polyak, L. G. Bakina, M. V. Chugunova, N. V. Mayachkina, A. O. Gerasimov and V. M. Bure, Effect of remediation strategies on biological activity of oil-contaminated soil- A field study, Int. Biodeterior. Biodegrad., 2018, 126, 57–68 Search PubMed.
- M. K. Sarfo, S. F. Gyasi, A. T. Kabo-Bah, B. Adu, Q. Mohktar, A. S. Appiah and Y. Serfor-Armah, Isolation and characterization of crude-oil-dependent bacteria from the coast of Ghana using oxford nanopore sequencing, Heliyon, 2023, 9, 1–13 CrossRef PubMed.
- S. I. Musa, Isolation and identification of diesel oil-degrading bacteria in used engine oil-contaminated soil, J. Appl. Sci. Environ. Manage., 2019, 23(3), 431–435 CAS.
- X. Jia, Y. He, D. Jiang, C. Liu and W. Lu, Construction and analysis of an engineered Escherichia coli-Pseudomonas aeruginosa co-culture consortium for phenanthrene bioremoval, Biochem. Eng. J., 2019, 148, 214–223 Search PubMed.
- N. Ali, N. Dashti, M. Khanafer, H. Al-Awadhi and S. Radwan, Bioremediation of soils saturated with spilled crude oil, Sci. Rep., 2020, 10, 1116–1125 CrossRef CAS.
- M. M. Aladwan, B. F. Dababneh, H. S. Farah and M. A. H. Abusalah, Identification of oil degrading bacteria from oil-contaminated soil in the Northeastern part of Jordan, J. Ecol. Eng., 2024, 25(5), 306–320 Search PubMed.
- G. K. Bekele, S. A. Gebrie, E. Mekonen, T. T. Fida, A. A. Woldesemayat, E. M. Abda, M. Tafesse and F. Assefa, Isolation and characterization of diesel-degrading bacteria from hydrocarbon-contaminated sites, flower farms and soda lakes, Int. J. Microbiol., 2022, 2022, 1–12 CrossRef.
- M. Amr, S. H. Abu Hussien, R. Ismail, A. Aboubakr, R. Wael, M. Yasser, B. Hemdan, S. M. El Sayed, A. Bakry, N. M. Ebeed, H. Elhariry, A. Galal and B. T. Abd Elhalim, Utilization of biosynthesized silver nanoparticles from Agaricus bisporus extract for food safety application: Synthesis, characterization, antimicrobial efficacy and toxicological assessment, Sci. Rep., 2023, 13(15048), 1–14 Search PubMed.
- A. B. Abeer-Mohammed, M. M. Abd-Elhamid, M. K. M. Khalil, A. S. Ali and R. N. Abbas, The potential activity of biosynthesized silver nanoparticles of Pseudomonas aeruginosa as an antibacterial agent against multidrug-resistant isolates from intensive care unit and anticancer agent, Environ. Sci. Eur., 2022, 34(1), 109–123 Search PubMed.
- A. J. Das, R. Kumar, S. P. Goutam and S. S. Sagar, Sunlight irradiation induced synthesis of silver nanoparticles using glycolipid bio-surfactant and exploring the antibacterial activity, J. Bioeng. Biomed. Sci., 2016, 6(5), 208–213 Search PubMed.
- R. A. Hamouda, M. H. Hussein, R. A. Abo-elmagd and S. S. Bawazir, Synthesis and biological characterization of silver nanoparticles derived from the cyanobacterium Oscillatoria limnetica, Sci. Rep., 2019, 9, 13071–13088 CrossRef PubMed.
- A. K. Giri, B. Jena, B. Biswal, A. K. Pradhan, M. Arakha, S. Acharya and L. Acharya, Green synthesis and characterization of silver nanoparticles using Eugenia roxburghii DC. extract and activity against biofilm-producing bacteria, Sci. Rep., 2022, 12, 8383–8392 CrossRef CAS.
- F. Tariq, N. Ahmed, M. Afzal, M. A. U. Khan and B. Zeshan, Synthesis, characterization and antimicrobial activity of Bacillus subtilis-derived silver nanoparticles against multidrug-resistantbacteria, Jundishapur J. Microbiol., 2020, 13(5), 1–7 Search PubMed.
- S. K. Nagaraja, S. K. Niazi, A. Bepari, R. A. Assiri and S. Nayaka, Leonotis nepetifolia flower bud extract mediated green synthesis of silver nanoparticles, their characterization and in vitro evaluation of biological applications, Materials, 2022, 15(8990), 1–20 Search PubMed.
- F. Jalilian, A. Chahardoli, K. Sadrjavadi, A. Fattahi and Y. Shokoohinia, Green synthesized silver nanoparticle from Allium ampeloprasum aqueous extract: Characterization, antioxidant activities, antibacterial and cytotoxicity effects, Adv. Powder Technol., 2020, 31(3), 1323–1332 Search PubMed.
- S. Gevorgyan, R. Schubert, S. Falke, K. Lorenzen, K. Trchounian and C. Betzel, Structural characterization and antibacterial activity of silver nanoparticles synthesized using a low-molecular-weight royal jelly extract, Sci. Rep., 2022, 12, 14077–14089 Search PubMed.
- J. Ali, A. Hameed, S. Ahmed, M. I. Ali, S. Zainab and N. Ali, Role of catalytic protein and stabilising agents in the transformation of Ag ions to nanoparticles by Pseudomonas aeruginosa, IET Nanobiotechnol., 2016, 10(5), 295–300 CrossRef PubMed.
- E. O. Mikhailova, Green silver nanoparticles: an antibacterial mechanism, Antibiotics, 2024, 14(1), 5 CrossRef PubMed.
- S. A. Ezeamalu, P. C. Aleke and P. F. Uzor, Antimicrobial evaluation and characterization of green synthesized silver nanoparticles using Annona muricata L. leaf extract, SARJNP, 2023, 6(2), 64–73 Search PubMed.
- M. Khaleghi, S. Khorrami and H. Ravan, Identification of Bacillus thuringiensis bacterial strain isolated from the mine soil as a robust agent in the biosynthesis of silver nanoparticles with strong antibacterial and anti-biofilm activities, Biocatal. Agric. Biotechnol., 2019, 18(2019), 101047 CrossRef.
- R. Verma, A. B. Khan, A. K. Amar, M. I. K. Khan and S. Sah, Opto-structural characteristics and biomedical applications of microwave irradiated green synthesised AM-AgNP from Atalantia monophylla (L.) leaf extract, ES Energy Environ., 2022, 17(3), 44–55 CAS.
- M. S. John, J. A. Nagoth, K. P. Ramasamy, A. Mancini, G. Giuli, A. Natalello, P. Ballarini, C. Miceli and S. Pucciarelli, Synthesis of bioactive silver nanoparticles by a Pseudomonas strain associated with the antarctic psychrophilic Protozoon Euplotes focardii, Mar. Drugs, 2020, 18, 38–51 CrossRef CAS PubMed.
- V. Gopinath, S. Priyadarshini, M. FaiLoke, J. Arunkumar, E. Marsili, D. Mubarak and A. J. Vadivelu, Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity., Arab. J. Chem, 2015, 10, 1107–1117 CrossRef.
- H. Singh, J. Du, P. Singh and T. H. Yi, Extracellular synthesis of silver nanoparticles by Pseudomonas sp. THG-LS1.4 and their antimicrobial application, J. Pharm. Anal., 2018, 8, 258–264 Search PubMed.
- K. Chand, C. Jiao, M. N. Lakhan, A. H. Shah, V. Kumar, D. E. Fouad, M. B. Chandio, A. A. Maitlo, M. Ahmed and D. Cao, Green synthesis, characterization and photocatalytic activity of silver nanoparticles synthesized with Nigella Sativa seed extract, Chem. Phys. Lett., 2021, 763(2021), 138218 CrossRef CAS.
- M. A. Al-Mashud, M. Moinuzzaman, M. S. Hossain, S. Ahmed, G. Ahsan, A. Reza, R. B. A. Ratul, M. H. Uddin, M. A. Momin and M. A. H. M. Jamal, Green synthesis of silver nanoparticles using Cinnamomum tamala (Tejpata) leaf and their potential application to control multidrug-resistant Pseudomonas aeruginosa isolated from hospital drainage water, Heliyon, 2022, 8, 1–9 Search PubMed.
- H. Hassan-Afandy, D. K. Sabir and S. B. Aziz, Antibacterial activity of the green synthesized plasmonic silver nanoparticles with crystalline structure against gram-positive and gram-negative bacteria, Nanomaterials, 2023, 13(8), 1327 Search PubMed.
- M. Muddassir, A. Raza, S. Munir, A. Basirat, M. Ahmed, M. S. Butt, O. A. Dar, S. S. Ahmed, S. Shamim and S. Z. H. Naqvi, Antibacterial efficacy of silver nanoparticles (AgNPs) against metallo-β-lactamase and extended spectrum β-lactamase producing clinically procured isolates of Pseudomonas aeruginosa, Sci. Rep., 2022, 12(1), 20685 Search PubMed.
- R. Vijayabharathi, A. Sathya and S. Gopalakrishnan, Extracellular biosynthesis of silver nanoparticles using Streptomyces griseoplanus SAI-25 and its antifungal activity against Macrophomina phaseolina, the charcoal rot pathogen of sorghum, Biocatal. Agric. Biotechnol., 2018, 14, 166–171 CrossRef.
- M. T. El-Saadony, N. A. El-Wafai, H. I. A. El-Fattah and S. A. Mahgoub, Biosynthesis, optimization and characterization of silver nanoparticles using a soil isolate of Bacillus pseudomycoides MT32 and their antifungal activity against some pathogenic fungi, Adv. Anim. Vet. Sci, 2019, 7(4), 238–249 Search PubMed.
- S. M. Alsharif, S. S. Salem, M. A. Abdel-Rahman, A. Fouda, A. Mohamed-Eid, S. E. Hassan, M. A. Awad and A. A. Mohamed, Multifunctional properties of spherical silver nanoparticles fabricated by different microbial taxa, Heliyon, 2020, 6, 1–13 Search PubMed.
- S. K. Nagaraja, S. K. Niazi, A. Bepari, R. A. Assiri and S. Nayaka, Leonotis nepetifolia flower bud extract mediated green synthesis of silver nanoparticles, their characterization and in vitro evaluation of biological applications, Materials, 2022, 15(8990), 1–20 Search PubMed.
- F. M. Akl, M. M. El-Sheekh, S. I. Ahmed and M. E. Makhlof, Bioadsorption of crude petroleum oil from seawater using the marine alga Hormophysa triquetra mediated silver nanoparticles, Mar. Pollut. Bull., 2024, 206, 116763 Search PubMed.
- M. Saeed, N. Ilyas, K. Jayachandran, S. Gaffar, M. Arshad, M. S. Ahmad, F. Bibi, K. Jeddi and K. Hessini, Biostimulation potential of biochar for remediating the crude oil contaminated soil and plant growth, Saudi J. Biol. Sci., 2021, 28(5), 2667–2676 Search PubMed.
- A. S. El-Shehawy, A. Elsayed, O. A. El-Shehaby and E. M. Ali, Potentiality of the green synthesized silver nanoparticles for heavy metal removal using Laurencia papillosa seaweed, Egypt. J. Aquat. Res., 2023, 49, 513–519 CrossRef.
- M. Noman, M. Shahid, T. Ahmed, M. B. K. Niazi, S. Hussain and F. Song, Use of biogenic copper nanoparticles synthesized from a native Escherichia sp. as photocatalysts for azo dye degradation and treatment of textile effluents, Environ. Pollut., 2020, 257, 113514 CrossRef CAS PubMed.
- A. Rasheed, S. Hussain, W. Mushtaq, M. Zubair, K. Siddique, K. Attia, N. Khan, S. Fiaz, F. Azeem and Y. Chen, Application of silver nanoparticles synthesized through varying biogenic and chemical methods for wastewater treatment and health aspects, Environ. Sci. Pollut. Res., 2022 DOI:10.1007/s11356-022-24761-4.
- N. Adibah, B. E. Firdianti and S. Suprapto, Synthesis of silver nanoparticles using response surface methodology, J. Nano- Electron. Phys., 2023, 15(3), 03001–03004 CAS.
- S. Denrah and M. Sarkar, Design of experiment for optimization of nitrophenol reduction by green synthesized silver nanocatalyst, Chem. Eng. Res. Des., 2019, 144, 494–504 CrossRef CAS.
- M. Sarkar, S. Denrah, M. Das and M. Das, Statistical optimization of bio-mediated silver nanoparticles synthesis for use in catalytic degradation of some azo dyes, Chem. Phys. Impact, 2021, 3, 1–12 Search PubMed.
- D. H. Itam, N. U. Udeh and E. Ugwoha, Modelling and optimizing the effect of pH on remediation of crude oil-polluted soil with biochar blend: RSM approach, Adv. Res., 2023, 24(3), 56–73 CrossRef.
- B. O. Ogunsile, O. S. Okohb, I. P. Ejidikeb and O. R. Omolajaa, Biosynthesis and optimization of AgNPs yield from Chromolaena odorata leaf extract using response surface methodology (RSM), Phys. Chem. Res., 2024, 12(1), 21–31 CAS.
- S. Mandal, S. Hwang, S. B. Marpu, M. A. Omary, V. Prybutok and S. Q. Shi, Bioinspired synthesis of silver nanoparticles for the remediation of toxic pollutants and enhanced antibacterial activity, Biomolecules, 2023, 13, 1054–1077 CrossRef CAS PubMed.
- A. El-Shahawy, M. F. Mubarak, M. El-Shafied and H. M. Abdullae, Fe (III) and Cr (VI) ions' removal using AgNPs/GO/chitosan nanocomposite as an adsorbent for wastewater treatment, RSC Adv., 2022, 12(27), 17065–17084 RSC.
- T. C. Egbosiuba, A. S. Abdulkareem, A. S. Kovo, E. A. Afolabi, J. O. Tijani and W. D. Roos, Enhanced adsorption of As (V) and Mn (VII) from industrial wastewater using multi-walled carbon nanotubes and carboxylated multi-walled carbon nanotubes, Chemosphere, 2020, 254(2020), 126780 Search PubMed.
- X. Yang and J. Y. Wu, Synthetic conditions, physical properties and antibacterial activities of silver nanoparticles with exopolysaccharides of a medicinal fungus, Materials, 2022, 15, 5620 CrossRef CAS PubMed.
- A. Ahmadi, S. Heidarzadeh and A. R. Mokhtari, Optimization of heavy metal removal from aqueous solutions by maghemite (g-Fe2O3) nanoparticles using response surface methodology, J. Geochem. Explor., 2014, 147, 151–158 CrossRef CAS.
- M. Ibrahim, J. B. Agboola, A. S. Abdulkareem, O. Adedipe and J. O. Tijani, Optimization of green synthesis of silver nanoparticles using response surface method (RSM). Nanotechnology applications in Africa: Opportunities and constraints, IOP Conf. Ser.:Mater. Sci. Eng., 2020, 805, 1–14 Search PubMed.
- V. Javanbakht and S. M. Ghoreishi, Application of response surface methodology for optimization of lead removal from an aqueous solution by a novel superparamagnetic nanocomposite, Adsorpt. Sci. Technol., 2017, 35(1–2), 241–260 Search PubMed.
- M. E. Mahmoud, R. M. El-Sharkawy and G. A. A. Ibrahim, A novel bionanocomposite from doped lipase enzyme into magnetic graphene oxide-immobilized-cellulose for efficient removal of methylene blue and malachite green dyes, J. Mol. Liq., 2022, 368, 120676 Search PubMed.
- O. M. Olatunji, D. H. Itam, G. E. Akpan and I. T. Horsfall, Predictive modeling coupled with multiple optimization techniques for assessing the effect of various process parameters on oil and pectin extraction from watermelon rind, Process Integr. Optim. Sustain, 2022, 6(3), 765–779 CrossRef.
- K. Sharma, S. Kalita, N. S. Sarma and A. Devi, Treatment of crude oil contaminated wastewater via an electrochemical reaction, RSC Adv., 2020, 10, 1925–1936 RSC.
- M. T. Alnuaimi, T. A. Taher, Z. Z. Aljanabi and M. M. Adel, High-resolution GC/MS study of biodegradation of crude oil by Bacillus megaterium, Res. Crops., 2020, 21(3), 650–657 Search PubMed.
- E. I. Ndekhedehe, E. S. Shaibu, E. U. Itoro and S. N. Essien, Systematic intrinsic biodegradation studies of crude oil-contaminated soil of Bdere community in South-South, Nigeria, UJMR, 2023, 8(2), 40–55 CrossRef.
- O. Anaejionu, P. A. Ahiarammunnah and C. J. Nri-ezedi, Hydrocarbon pollution in the Niger-Delta: Geographies of impacts and appraisal of lapses in extant legal framework, Resour. Policy, 2015, 45, 65–77, DOI:10.1016/j.resourpol.2015.03.012..
| This journal is © The Royal Society of Chemistry 2026 |









