 Open Access Article
Open Access ArticleMetal-based triazoles as a medical marvel of the modern era: a comprehensive review
Kashif Haseeb
a,
Muhammad Hasnain Mustafa
a,
Wardha Zafar
a,
Abrar Ul Hassan
 b,
Zahid Hussain Chohan
c and
Sajjad Hussain Sumrra
b,
Zahid Hussain Chohan
c and
Sajjad Hussain Sumrra
 *a
*a
aDepartment of Chemistry, University of Gujrat, Gujrat 50700, Punjab, Pakistan. E-mail: sajjadchemist@uog.edu.pk; sajjadchemist@gmail.com
bDepartment of Chemistry, Lunen Institute of Technology, Tanghzhou 37299, China
cDepartment of Chemistry, University of Southern Punjab, Multan, Punjab, Pakistan
First published on 5th January 2026
Abstract
Antimicrobial resistance is raising serious health concerns across the globe, and its adverse effects are elevating day by day. Researchers are making efforts to find new and more efficient pharmaceutical agents to overcome this growing challenge. Metal-based drugs are very useful in this regard, and hence, they are gaining more attention from researchers. This review systematically examines metal-based triazole Schiff base compounds, delving into their synthetic methodologies, structural characterization and a range of bioactivities. With a precise emphasis on antimicrobial properties along with cytotoxicity effects, DNA interactions and anti-cancerous and enzymatic applications, researches explores that these compounds are innovative solutions to the growing crisis of antimicrobial resistance. The synergistic combination of metal ions and organic ligands within these complexes often results in enhanced antimicrobial efficacy compared to traditional organic antimicrobials. This review provides a comprehensive overview of triazole-based metal complexes under research from 2006 to 2024, which can be used as antibacterial, antifungal, cytotoxic, anticancer, DNA interaction and enzyme inhibition agents.
1 Introduction
1.1 Antimicrobial resistance: a silent pandemic
One of the most important developments that has improved human wellness is the discovery of medications to prevent potentially fatal infections caused by bacteria. On the other hand, mortality rates, safe alimentary products and therapeutic advances are all hampered by antibiotic resistance.1 The phenomenon of antimicrobial resistance has become more prominent in recent years. Antimicrobial resistance occurs when microorganisms like fungi and bacteria do not respond to the antibacterial drugs or when the effect of these microbes is elevated. Global estimates place the amount of money lost due to antibiotic resistance around the range of $300 billion and $1 trillion until 2050.2 The Organization for Economic Cooperation and Development (OECD) estimates that the combined GDP loss for its member countries will be between twenty dollars and 35 trillion dollars by 2050. Furthermore, an analysis by the World Bank indicated that antibiotic resistance would raise the poverty plateau and mostly affect nations that are most impoverished. AMR-related high health care costs are often incurred mostly for the need of additional staffing and healthcare services; this issue is particularly noticeable in developing countries. This is mostly because these nations depend more heavily on labor revenues and have greater rates of contagious infections.3Antimicrobial resistance is causing serious health issues all around the world and increasing the death rates. According to the CDC (Center for Disease Control and Prevention), around two million people suffer from infections and 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people die every year due to antimicrobial resistance.4 AMR also increased exponentially during Covid-19. Taking an excess dose of antimicrobial medicines is the major cause of enhanced antimicrobial resistance.5 The exponentially elevating death rates due to antimicrobial resistance indicate that it might overtake all other causes of death in the coming years.6
000 people die every year due to antimicrobial resistance.4 AMR also increased exponentially during Covid-19. Taking an excess dose of antimicrobial medicines is the major cause of enhanced antimicrobial resistance.5 The exponentially elevating death rates due to antimicrobial resistance indicate that it might overtake all other causes of death in the coming years.6
The main causes of antimicrobial resistance are antibiotic overuse and misuse. Inappropriate antibiotic prescriptions frequently result in the selection of resistant strains of viruses, such as when the drugs are used ineffectively to treat viral illnesses.7 Resistance is also fostered by self-medication and the use of leftover antibiotics, which lead to inadequate treatment regimens.8 One major factor for the emergence of antimicrobial resistance in the agricultural field is the use of antibiotics in cattle to stimulate growth and prevent diseases (Fig. 1). Antibiotics administered to animals have the potential to favor the development of resistant microorganisms, which can be passed on to humans via environmental routes, contaminated meat intake or direct contact.9 The emergence of resistant microorganisms in hospital settings is also significantly influenced by subpar infection prevention and control procedures. Inadequate hand washing techniques and the reuse of medical equipment are two examples of poor hygiene habits that make it easier for the resistant strains to spread between patients.10 Antimicrobial resistance is made worse by the dearth of newly developed antibiotics. The current antibiotic arsenal loses effectiveness as bacteria evolve and develop resistance, and there are fewer treatment choices due to the sluggish rate of new antibiotic discoveries.11
1.2 Metals to combat antimicrobial resistance
The ongoing quest for novel, potent antimicrobial agents is gaining momentum due to the fact that the existing therapeutic cure is not up to the task of combating the rising incidence and dissemination of resistant strains of bacteria.12 The majority of the substances that medical chemists worldwide have created since the 1920s, when Alexander Fleming discovered antibiotics, were entirely organic. Even though metals and their compounds have been used for a very long time, they were mostly utilized as substances or catalysts because of their poisonous qualities to some extent.13 However, the first successful therapy for syphilis is the use of an arsenic-containing organometallic complex discovered in the early twentieth century, which led to the widespread application of metallic compounds with specified structures in pharmacology (Salvarsan).14 Innovative medications and diagnostic instruments can be designed using metal ions or metal ion–interacting components. Metal ions are key components of life. In biological solutions, because they easily release electrons and produce positively charged ions, metal atoms are soluble in liquid media.15 They may easily engage with biological molecules that are rich in electrons, such as DNA or proteins, as they lack electrons. As a result, they can take part in catalysis or in the stabilizing or recognition of tertiary or quaternary structural changes in these molecules.16Modifications and upgradation of existing antimicrobial drugs or finding alternative drugs to replace the existing pharmaceuticals are needed. Metals are essential components for the body, and they perform vital functions within the body.17 Metal-based drugs are making a huge impact on medicinal chemistry due to their high bioactivity and excellent efficiency. Many metals play a crucial role in the metabolism occurring in the body.18 Iron is the most important metal for the body. Other metals like “K, I, Zn, Mg, and Mn” are also vital to human metabolism. Metal-based drugs are considered better than conventional organic drugs because metal complexes can change the geometric configuration, oxidation states and coordination number.19 An additional advantage of metal-based drugs is the formation of chelates with organic drugs that enhance their activity. Metal-based pharmaceutical agents can also play a crucial role as diagnostic agents.20 Numerous metalloproteins have the function of anti-cancer, antimalarial and antidiabetic agents. The overtones concept indicates that when ligands are coordinated with metal ions, they exhibit enhanced chemical and physical properties compared to their uncoordinated counterparts.21
The use of metals in the pharmaceutical industry is not new. The Chinese, Greeks and Indian scientists were amazed to find their excellent bioactive attributes.22 During ancient times, copper, iron and silver therapies were very much in practice for the treatment of many diseases.23 By comparing metal-based drugs to traditional organic antibacterial medications, it was found that there are diverse mechanisms of action. Furthermore, when metals are added to organic antibiotics, new and different modes of action are made available that would not be possible with only the organic medication.24 Because these metal-based composites have these unique and extra mechanisms of action, they may prove to be a promising therapy for bacterial illnesses that are resistant to antibiotics, either when used alone or in combination.25
1.3 Half life and stabilization of metal complexes
The stability and half-life of metal-triazole compounds under physiological conditions are important factors to take into account when assessing their potential as therapeutic options. The nitrogen donors of 1,2,4-triazole ligands frequently form strong coordination bonds with transition metals such as Cu(II), Zn(II), Co(II), Ni(II), and VO(IV), which increases the stability of the complexes.26 The effective half-life and bioavailability of these complexes may be decreased by ligand exchange, partial dissociation, or metal release caused by factors like biomolecules, pH variations, and redox-active species in in vivo environments.27 Research shows that chelating and multidentate triazole ligands frequently improve kinetic stability, despite the lack of experimental data on precise half-lives.28 With half-lives of about 15 minutes at physiological pH, mixed-ligand copper(II) complexes containing triazole Schiff bases exhibit strong catalytic action in the detoxification of organophosphorus pesticides.29 Lanthanide-based complexes such as those that incorporate cerium(III) into a macrocyclic triazole Schiff base ligand function as powerful synthetic phosphotriesterases with half-lives of approximately five minutes.30 With a half-life of roughly ten minutes, the zinc(II) analogue hydrolyzes 4-nitrophenyl phosphate considerably more quickly than its nickel(II) or cobalt(II) counterparts.311.4 Schiff base compounds
Therapeutic applications of Schiff bases are studied worldwide because of their versatility, especially when they behave as metal complexing agents. The condensation reaction of carbonyl compounds with a primary amine can be used to prepare Schiff bases.32 The creation of new molecules with strong biological activity relies heavily on Schiff bases. Hugo Schiff originally disclosed the existence of Schiff bases in 1864.33 Schiff bases are condensation products when primary amines are allowed to react with carbonyl compounds. The azomethine functional group found in these scaffolds has the general formula RHC = N − R1, where R and R1 are cycloalkyl, heterocyclic, alkyl, or aryl groups that can be substituted in many ways.34 These substances also go by the names azomethines, imines, and anils. The presence of only one pair of electrons in the sp2 hybridized orbital of the nitrogen atom belonging to the azomethine group has been demonstrated to have significant chemical and biological significances.35 Organic processes use Schiff bases as intermediates, which are further examined in light of their usefulness. Scientists are examining a variety of approaches for the creation of novel, green technologies with an emphasis on Schiff bases and their derivatives.36 Schiff bases are now gaining interest from researchers in the medical field because of their use in chemotherapy.37 These formed complexes can be utilized in the synthesis of many antimicrobial agents, which are more effective than conventional drugs. In addition, they exhibit a variety of pharmaceutical and biological features. The substances in question exhibit several biological features, mostly because of the imine group present in them.38The molecules that comprise azomethines are additionally used as polymeric stabilizing agents, corrosion inhibitors, coloring agents, catalysts, antioxidants, antibacterial agents, anticancer agents, etc. Every metal ion is likely to be chelated by Schiff base complexes due to their multi dentate ligands.39 These ligands are powerful for the fascinating new therapeutic strategy to improve our understanding of illnesses and treat them.40 Schiff bases containing heterocyclic compounds are considered an important class of organic compounds. These compounds are considered very important constituents of the drugs for the treatment of many diseases. They are used largely due to their antiviral, antifungal, antibacterial and antirheumatic properties.41 Triazoles are a significant category of heterocyclic substances. Major antimicrobial agents contain a core skeleton of triazole, which has more effectiveness and less toxicity.42
After complexing with metals, Schiff base ligands exhibit dramatically increased bioactivity because of many synergistic effects. First, metal complexation strengthens Schiff base ligands, increasing their stability and resistance to hydrolysis and degradation. In biological systems, this increased stability is essential for preserving bioactivity over extended periods.43 Due to their immense biological potential, Schiff base metal complexes have emerged as versatile therapeutic candidates. Their anticancer activities are mediated through various mechanisms such as oxidative stress, disruption of mitochondria, and induction of apoptosis.44 Other important enzymatic inhibitions, especially against targets like the proteasome, further contribute toward the death of cancerous cells.45
Besides these properties, many of these complexes interact strongly with DNA, often by intercalation or groove binding, leading to structural damage and interference with replication processes. These combined properties translate into notable cytotoxic effects across various cancer cell lines, positioning Schiff base metal complexes as promising multifunctional agents in medicinal chemistry. The electronic distribution of Schiff base ligands is changed by the coordination of metal ions with them, which improves the ligand's capacity to interact with biological macromolecules, including DNA, proteins and enzymes. The increased bioactivity exhibited by metal complexes, which has been well documented in several studies, is especially significant for this change in electronic characteristics.46,47
1.5 Chemistry of triazoles
Triazoles belong to the class of heterocyclic compounds that are considered the fundamental moieties in bioinorganic chemistry. They are five-membered solid compounds containing 3 nitrogen atoms, and their molecular formula is C2H3N3.48 Bladin came up with the term triazole in 1885. An alternative name for triazole was introduced in 1889 by Andrococei as “pyrrodiazole”. The discovery of triazole opened new ways of finding different pathways for drug synthesis.49 Triazoles are cyclic compounds that contain a 5-membered ring. The heterocyclic ring has a couple of carbon atoms and three nitrogen atoms in it. Triazole shows two isomeric forms, namely, 1,2,3-triazole and 1,2,4-triazole (Fig. 2).1,2,3-Triazole is present in the form of hygroscopic crystals, which have a sweet taste, no color, a melting point of 24 °C and a boiling point of 209 °C. These crystals can dissolve in water. 1,2,4-Triazole is present in the form of colorless crystals with a melting point 121 °C and a boiling point of 260 °C, and they also dissolve in water. The planner 1,2,4-triazole is considered to be more significant due to its fused heterocyclic structure and its wide bioactivity.50,51 Nitrogen substitutes in the ring exhibit more prominent pharmacological characteristics, according to the structure–activity relationship (SAR) analysis of triazole drugs. Because substitutes come in a wide range of forms and can specifically alter the chemistry of triazoles, the biological impact of substituted triazoles is increased. Numerous triazole ring locations that can be used in pharmaceutical applications have been identified by an in-depth study of the chemistry of 1,2,4-triazole and its numerous conjugates.52
1.6 Comparison of 1,2,3- and 1,2,4-triazoles and tautomer clarification
Due to the abundance of available data, this study focuses on metal-coordinated 1,2,4-triazole complexes; however, 1,2,3-triazole systems and even their metal chelates are worth mentioning because they represent a developing, complementary area in biomedical chemistry. The main difference lies in the arrangement of nitrogen atoms: 1,2,4-triazole places the nitrogen atoms at positions 1, 2, and 4, while 1,2,3-triazole carries three nearby nitrogen atoms at positions 1, 2, and 3. By altering the N-donor topology, this has a substantial impact on chelation geometry and complex stability. It is actually challenging to synthetically coordinate 1,2,3-triazolylidene (trz) ligands to first-row transition metals due to orbital mismatch and ligand–metal electronic incompatibility.28 Nevertheless, recent studies have indicated that metal-1,2,3-triazole chelates, especially mixed-ligand or hybrid systems, may exhibit promising pharmacological properties like cytotoxic and antibacterial activity.26,53 Despite the lack of available data, we choose to focus on 1,2,4-triazole metal complexes because they have repeatable coordination, well-characterized structures, and consistent biological evaluations. However, we highlight 1,2,3-triazoles as a promising but understudied avenue for future investigation.Finally, in terms of tautomerism in 1,2,4-triazoles, the 1H-1,2,4-triazole tautomer is thermodynamically preferred over the 4H form under ambient conditions, and the 1H form is used as a ligand precursor in nearly all reported metal coordination studies, either explicitly or implicitly. Thus, throughout this review, “triazole” refers to 1H-1,2,4-triazole unless otherwise specified.
1.7 Triazoles as medicinal agents
Triazoles are gaining more attention from researchers due to their exceptional binding attributes.54 They possess a pi conjugated system and three nitrogen atoms that can coordinate with metal ions, which contributes to their wide range of biological activities. These include antihypertensive, antimicrobial, antiviral, anticancer, anti-inflammatory, analgesic and antidiabetic effects. In addition, their ability to form hydrogen bonds further strengthens their potential as promising pharmaceutical agents.55 Moreover, the heterocyclic structure of triazole is capable of forming nonbonding relation with the enzyme receptors found in biological systems. This property has enabled them to form chromophores, which are vital in medicinal chemistry.56 SAR studies of triazole disclosed that the substituent attached to nitrogen is responsible for the biomedical properties. These properties can be enhanced or swapped by altering these substituted groups.57 There are enormous drugs containing triazoles, which can be used as antifungal, anti-estrogen, anti-viral, anti-cancer and many other various pharmaceutical agents (Fig. 3).Triazole compounds provide a route for glucose transfer in gene transcription, making them a highly effective anti-diabetic medicine. Triazole derivatives can act as ACE inhibitors to prevent the conversion of angiotensin, exhibiting antihypertensive properties.58 Urea hydrolysis results in ammonia, which is the major cause of urinary tract stones. The urease enzyme's activity can be inhibited by Schiff bases that contain a triazole moiety. Additionally, many enzymes are inhibited by triazole compounds. These include tyrosinase, which is largely related to Parkinson's disease and other neurodegenerative ailments; alpha-amylase, alpha-glucosidase and the enzyme known as bacterial DNA gyrase are involved in the replication of bacteria.59 Moreover, the alpha-amylase enzyme has exceptional hypo-glycemic pharmaceutical attributes. Triazole-containing substances have strong anti-leishmanial and anti-parasitic properties.60 Pre- and postharvest treatments of triazoles are utilized to manage a range of fungal infections in vegetables, legumes, fruits and cereal crops. Because they impede ergosterol generation, they interfere with the construction of fungal cell walls, which is the biochemical mechanism underlying their antifungal activity.61
First-generation triazoles, like fluconazole and itraconazole, and second-generation triazoles, like voriconazole, posaconazole and isavuconazole, are the two major categories of triazoles. These medications can be used in various therapeutic settings due to their distinct pharmacokinetic characteristics and activity spectra.62 Fig. 4 shows the structures of some commercially available triazole derivatives.
To treat a variety of superficial and systemic fungal diseases, fluconazole is a frequently prescribed medication. Furthermore, itraconazole works well against a wider variety of fungi such as the species of Aspergillus. An effective second-generation triazole that works well against a range of fungi is voriconazole.63 Triazoles have been investigated for possible anticancer effects. For instance, several triazole compounds function as aromatase inhibitors, preventing the synthesis of estrogen and therefore impeding the development of breast tumors that are estrogen-dependent. Another triazole of the second generation, posaconazole, has a wider range of activities, including the ability to combat some strains of fluconazole-resistant Candida species. The latest drug in the triazole family, isavuconazole, has been licensed for the treatment of invasive mucormycosis and aspergillosis.64
1.8 Triazole Schiff bases
Because of their unique characteristics, Schiff base ligands incorporating heterocycles have long been the subject of great study attention. In addition, extensive research was conducted on Schiff base complexes of the heterocyclic kind, which are formed involving amino acids that contain nitrogen and oxygen donor atoms and carbonyl derivatives.65 The chemistry of Schiff base ligands is very important, particularly for the formation of Schiff base complexes, and many of these compounds exhibit outstanding catalytic properties in a variety of reactions at high temperatures and under the influence of water. Schiff bases can form stable coordination compounds when allowed to react with metal.66 Triazole Schiff bases are considered as the most efficient biologically active Schiff base. The presence of extra N-donor atoms that generate mono-, bi- or tridentate locations for binding has led to an increase in the durability of 1,2,4-triazole-derived Schiff base compounds as ligands over the past decade. This is because of the chelate effect. Triazole-derived ligands have also been used to create a variety of metal scaffolds with intriguing features due to their flexible interaction patterns.671.9 Metal-based triazole Schiff bases
According to antibiotic studies, metal chelates have more antibacterial activity than unchelated substrates. Tweedy's idea of chelation and Overtone's notion can be used to explain this. According to Overtone's theory of cell permeability, only lipophilic substances are able to cross the lipid bilayer that envelops the microbial cell.68 Thus, liposolubility is a crucial factor in regulating the antibacterial action. Due to the ligand orbitals' overlap and the metal ion's partial positive charge sharing with the donating site of the ligand, the polar nature of metals is greatly lowered after chelation.69Metal-based triazoles are very well recognized for their extraordinary biochemical activity. They have a large number of biological properties like antibacterial, anticancer, analgesic, antiviral, antihypertensive and anti-inflammatory functions.70 Overall, metal-induced triazoles have exhibited development in various fields including medicine and chemistry. Their versatile coordination attributes and wide biological activities make them attractive candidates for more research and development.71 The structures of some biologically active triazole Schiff bases are shown in Fig. 5.
Triazole ligands and metal ions interact in triazole Schiff base metal complexes, leading to a variety of mechanisms of action. The coordination between the metal ions and the nitrogen atoms in the triazole ring and the Schiff base alters the electronic characteristics of the metal and the ligand. Complex's capacity to block certain enzymes is strengthened by this interaction, upsetting vital cellular metabolic processes.72 Reactive oxygen species (ROS) can also be produced by these metal complexes when they come into contact with biological components. Oxidative stress and cell death can result from reactive oxygen species (ROS), causing oxidative damage to macromolecules in the cell, including proteins, lipids and nucleic acids.73 By engaging with cellular receptors or transport mechanisms, the metal ions in the complexes may promote greater cellular absorption, improving the bioavailability and efficacy of the triazole Schiff base.74
1.10 Mechanistic and structural comparison of metal-triazole complexes
The biological properties of metal-triazole complexes vary depending on the metal ion and its coordination environment. Cu(II) complexes, which are typically redox-active and generate ROS, facilitate strong antibacterial and cytotoxic reactions through oxidative DNA breakage.75 Zn(II) complexes often have potent antibacterial and enzyme-inhibitory profiles with decreased toxicity because of their redox inactivity, which mainly operates through Lewis acid-mediated biomolecular activation.76 Ni(II) and Co(II)/Co(III) complexes typically maintain octahedral geometries that promote DNA binding and enzyme inhibition. Vanadyl (VO2+) complexes exhibit insulin-mimetic and anticancer effects due to their variable oxidation states and involvement with phosphate-dependent signaling pathways.772 General procedure for the synthesis of triazole Schiff base ligand and its metal complexes
The ligand and its related metal complexes can be synthesized by the usual approach of reflux condensation78 (Fig. 6). Depending on the solubility of the reactants required for the synthesis of ligands, the solvent is selected. Similarly, the solvent chosen is one in which the ligand and metallic salts are highly soluble. After their full dissolution in the solvent, the reactants are combined in a round-bottom flask. Furthermore, in order to reflux the mixture, magnetic stirring and heat are applied using a hotplate. Every synthetic reaction is carried out in an oil bath, which ensures even heating. By utilizing multiple mobile phases in varied ratios, comparative thin layer chromatography (TLC) can be used to continually assess the product formation.79 Using the hit-and-trial procedure, the appropriate solvents are identified as the functional mobile phase. After the reaction is completed, the product is extracted by filtering the product. In case of a clear solution, the rotary evaporation method can be adopted to obtain the desired product. Similar refluxation processes using metallic salt solutions and the triazole Schiff base ligand are used to prepare the metal complexes of the synthesized azomethine ligand. The metal salts are used at suitable ligand-to-metal salt ratios.3 Biological efficacy of metal-embedded triazoles
3.1 Anti-bacterial activity
Public health is severely hampered by the existence and pathogenicity of bacteria in healthcare facilities, which have an array of negative effects on healthcare systems and communities. Bacterial diseases can spread via food, drink, air, water or live vectors.80 Many governmental, nonprofit and international groups have acknowledged the need for novel antibacterial medications to treat multidrug-resistant (MDR) bacterial infections, which is a serious global health concern.81 It is crucial to create new classes of antibiotics with cutting-edge mechanisms. The exploration of novel targets includes peptides with potential antibiotic characteristics identified by artificial intelligence on the human proteome, as well as reflux pumps and biofilms or combination medicines that target both important bacterial activities and resistance-causing components.82 Triazole Schiff base complexes have proven to be very efficient against bacterial strains, and they can be used as antibacterial agents. A lot of triazole-based antibacterial drugs have been discovered and utilized in the pharmaceutical field to overcome the increasing pathogenic infections. The following possible mechanisms are explained by the researchers, through which these drugs can act as antibacterial drugs (Fig. 7). | ||
| Fig. 7 Mechanism of the antibacterial activity of metal complexes, adapted from ref. 83. | ||
Al-Hassani et al., 2023 stated the creation of a novel triazole ligand L1, which is formed through the condensation of an amino-substituted triazole and an aldehyde derivative (Scheme 1). Metal complexes (1a–1f) were formed by coordinating the bivalent metal ions to the ligand. Analytical and spectrometric approaches were used to corroborate the geometry of the novel ligand and its complexes. The anti-bacterial action was examined through bacterial cultures, and the MIC (minimum inhibitory concentration) values were equated to the conventional medicines. The MIC value comparison proved that these complexes have a greater response against the bacterial species. Anti-fungal properties also showed that these complexes were bioactive against several fungal strains.84
 | ||
| Scheme 1 Synthesis of the Schiff base L1 and metal complexes 1a–1f, redrawn from ref. 84. | ||
At present, metal-based triazoles are the most widely used pharmaceutical agents due to their high bioactivity. They can be used as antibacterial and antifungal agents. Metal ions are coordinated to the Schiff bases to enhance their action against the bacterial and fungal species. Sumrra and his coworkers published the preparation of triazole metal complexes (2a–2h). Schiff base 2-[(1Z)-N-(1H-1,2,4-triazol-3-yl)-etanimidoyl]phenol (L2) was synthesized by reacting an amine-substituted triazole and an aldehyde. The solution was refluxed for 8 hours, and the formation of the product was observed through TLC. Metal ions of transition metals were added to the ligand in a stoichiometric ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to form complexes, which were the desired product (Scheme 2). The ligand- and metal-coordinated complexes were studied through analytical techniques like UV-vis spectroscopy, IR spectroscopy, NMR spectroscopy, elemental studies and conductance examination. The antibacterial action of the prepared complex was observed against a few species of bacteria. The experimental results showed that metal complexes were more bio-active than the parent triazole ligand.85
1 to form complexes, which were the desired product (Scheme 2). The ligand- and metal-coordinated complexes were studied through analytical techniques like UV-vis spectroscopy, IR spectroscopy, NMR spectroscopy, elemental studies and conductance examination. The antibacterial action of the prepared complex was observed against a few species of bacteria. The experimental results showed that metal complexes were more bio-active than the parent triazole ligand.85
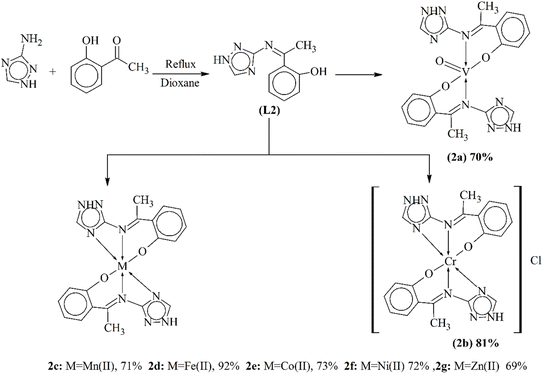 | ||
| Scheme 2 Synthetic route for metal complexes 2a–2g of the ligand L2, redrawn from ref. 85. | ||
Trivalent lanthanide complexes (3a–3l) were synthesized by reacting anhydrous lanthanide(III) chloride with Schiff bases L3a–L3c derived from 3-(phenyl/substitutedphenyl)-4-amino-5-mercapto-1,2,4-triazole with diacetyl/benzil in methanol, as shown in Scheme 3. The complexes were characterized using elemental analysis, electrical conductance, magnetic moment and various spectroscopic techniques (IR, 1H, 13C-NMR, and UV-vis), as well as XRD. Spectral data indicated that the Schiff base ligands acted as dibasic tetradentate chelating agents with coordination sites at two thiol sulfur atoms and two azomethine nitrogen atoms. The presence of coordinated water in the metal complexes was confirmed by thermal and IR analyses. The antibacterial and antifungal activities of the complexes and their ligands were evaluated against several strains. Lanthanide complexes of 1,2-diketones are known for their optical properties and have various applications in electronics, NMR and biomedical fields. The metal complexes displayed improved antimicrobial activity when compared to the free ligands, suggesting their potential as bacteriostatic agents. This increased activity is caused by factors such as cell permeability and dipole moment, which are dependent on the presence of the metal ion.86
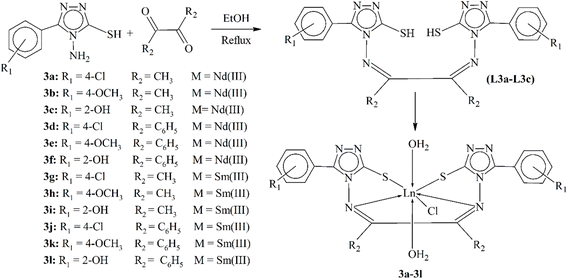 | ||
| Scheme 3 Synthesis of ligands L3a–L3c and Ln complexes 3a–3l, redrawn from ref. 86. | ||
A series of Mn(II), Fe(III) and Zn(II) complexes (4a–4l) were synthesized using Schiff bases L4a–L4d resulting from isatin and 3-substituted-4-amino-5-mercapto-1,2,4-triazole (Scheme 4). Characterization included elemental, spectroscopic (IR, NMR, UV-vis, fluorescence, and redox properties) and magnetic moment studies, suggesting an octahedral shape for the synthesized complexes. Both the ligands and their metal complexes exhibited fluorescence phenomenon. Antimicrobial studies against bacterial (Bacillus subtilis, Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa) and fungal (Aspergillus niger and Penicillium chrysogenum) species revealed stronger antimicrobial activities of the complexes than those of the ligands. Additionally, the property of metal ions to cleave inside the DNA strand indicated the significant role of the metal ions in biological systems. The complexes were found to be stable, non-hygroscopic, and insoluble in common organic solvents but soluble in DMF and DMSO. Elemental analyses indicated that the Mn(II) and Zn(II) complexes had a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry of the ML2 type, while Fe(III) complexes possessed a 1
2 stoichiometry of the ML2 type, while Fe(III) complexes possessed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio of the type [ML2]Cl. The molar conductance values of Mn(II) and Zn(II) complexes suggested a non-electrolytic nature, whereas the high molar conductance values of Fe(III) complexes indicated the electrolytic behavior in DMF.87
2 molar ratio of the type [ML2]Cl. The molar conductance values of Mn(II) and Zn(II) complexes suggested a non-electrolytic nature, whereas the high molar conductance values of Fe(III) complexes indicated the electrolytic behavior in DMF.87
 | ||
| Scheme 4 Synthetic scheme of ligands and metal complexes 4a–4l, redrawn from ref. 87. | ||
Sharma and coworkers produced two novel ligand L5 bearing azomethine linkage having 1,2,4-triazole moieties (Scheme 5) and their corresponding oxovanadium(IV) complexes (5a–5b). They characterized these compounds using various spectroscopic and analytical techniques such as UV-vis, FTIR, NMR, EPR, XRD, CV and elemental analysis. The oxovanadium(IV) complexes were non-electrolytic and had particle sizes of 47.53 nm and 26.28 nm. The compounds were assessed for their antibiotic property against four bacterial pathogens. The oxovanadium(IV) complexes (5a–5b) exhibited higher antibacterial activity as compared to their precursor Schiff bases. Molecular docking studies revealed that both the ligands and complexes had significant binding affinity to bacterial proteins. Molecular dynamics simulations confirmed the stability of the protein–ligand interactions and indicated the spontaneity of the binding process.88
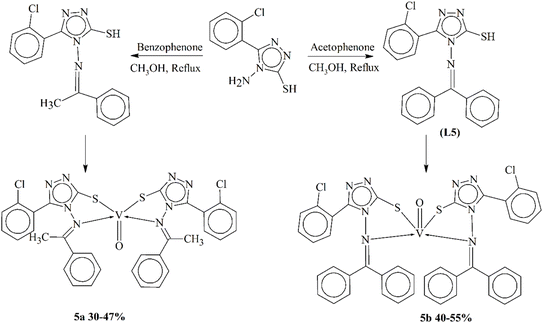 | ||
| Scheme 5 Synthesis of oxovanadium(IV) complexes 5a–5b, redrawn from ref. 88. | ||
A set of metal complexes (6a–6j) of Co(II), Ni(II), Cu(II), Zn(II), and Pd(II) with the ligand L6 4-((4-isopropoxybenzylidene)amino)-5-methyl-4H-1,2,4-triazole-3-thiol (Scheme 6) were prepared and subsequently characterized using various analytical techniques by Siwach and Singh. IR, 1H, 13C-NMR and mass spectrometry were used for the analysis of the Schiff base ligand, while FT-IR, proton NMR, elemental analysis, ESR and electronic spectral studies were used to analyze the metal complexes. Magnetic moment and electrochemical behavior studies were carried out by CV. TGA displayed the thermal stability of the complexes. Fluorescence spectra of the ligand and metal complexes were also analyzed in different solvents. Octahedral geometry was proposed for Co, Ni and Zn complexes, while square planar geometries were proposed for Cu and Pd complexes, according to the findings. Biological screening against various bacterial and fungal strains showed that the complexes were found to possess antibacterial activity comparable with standard drugs against E. coli. The potential antimalarial activity of the complexes was also screened. The molar conductance values indicated the non-electrolytic nature of the complexes. The synthesized metal complexes show great potential to be further used therapeutically.89
 | ||
| Scheme 6 Synthetic scheme of Schiff base L6 and complexes 6a–6j, redrawn from ref. 89. | ||
Chohan and Hanif explored the potential of novel triazole Schiff bases L7a–L7c and their corresponding Zn(II) complexes (7a–7c) as broad-spectrum antimicrobial agents (Scheme 7). Triazole-containing molecules have gained noteworthy interest due to diversity in bioactivities, and the authors leverage this established knowledge to design these new compounds. The researchers evaluated the efficacy of ligands and their Zn(II) complexes against various bacterial and fungal strains. The results revealed a marked increase in antimicrobial activity for the metal complexes compared to the free ligands. The authors proposed that chelation with Zn(II) reduces the polarity of the metal ion, leading to increased lipophilicity and potentially enhanced penetration through the microbial cell membrane. This mechanism could explain the observed improvement in activity.90
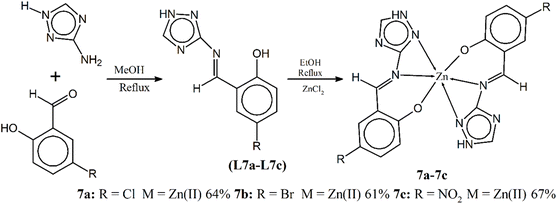 | ||
| Scheme 7 Scheme of the synthesis of ligands and metal complexes 7a–7c, redrawn from ref. 90. | ||
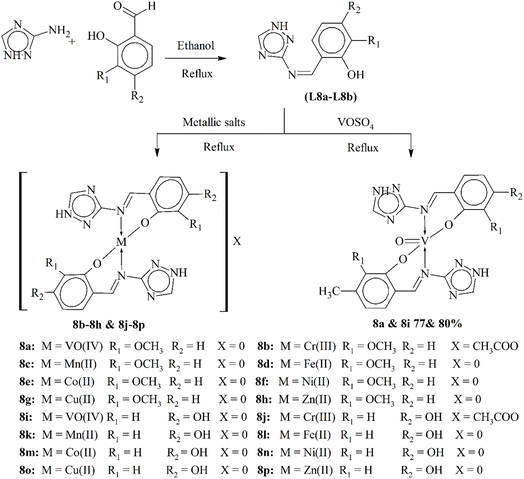
Metal coordinates are the trending pharmacophores due to their vast biological attributes. Metals can be utilized to enhance the bioactivity of several compounds through forming interactions with the Schiff bases. Sumrra and his coworkers described triazole ligands L8a–L8b via a condensation reaction of a triazole moiety (Scheme 8). Metallic salts were introduced to the ligand to form metal scaffolds (8a–8p). Divalent transition metal salts were incorporated, which showed enhanced biological functionality in a chemical ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (L
1 (L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M). The geometrical and physical traits of these metal scaffolds were examined through several analytical approaches. The antibacterial functionality of these compounds was tested against several bacterial strains. The complexes 8a and 8b showed increased biochemical function when compared to the parent ligand.91
M). The geometrical and physical traits of these metal scaffolds were examined through several analytical approaches. The antibacterial functionality of these compounds was tested against several bacterial strains. The complexes 8a and 8b showed increased biochemical function when compared to the parent ligand.91
 | ||
| Scheme 8 Synthetic route for metal complexes 8a–8p, redrawn from ref. 91. | ||
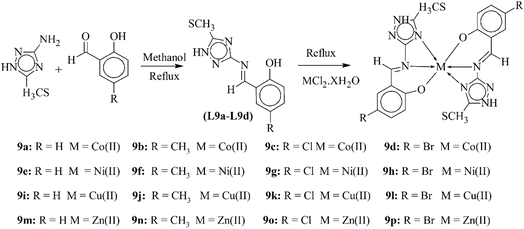
Chohan and his coworkers (2013) published a new sequence of triazole compounds, which act as tridentate ligands L9a–L9d (Scheme 9). The bonding and geometry was studied by IR, NMR, electronic and magnetic studies. The biological testing showed that metallic compounds of transition metals possessed great antibacterial and antifungal bioactivities. It was noticed that metal-infused complexes had more bioactivity than the simple Schiff base ligands. Zinc-based complexes (9m–9p) showed more antibacterial activity while cobalt complexes (9a–9d) showed more antifungal bioactivity than other metal complexes.92
 | ||
| Scheme 9 Scheme for metal complexes 9a–9p of ligands L9a–L9d, redrawn from ref. 92. | ||
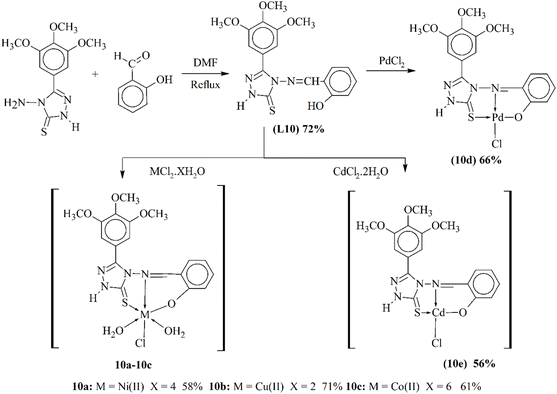
Al-Jibouri and his coworkers synthesized a new Schiff base ligand. This ligand L10 was used to synthesize metal-based complexes (10a–10c) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with divalent transition metals (Scheme 10). These complexes were characterized using analytical techniques like 1HNMR, CNMR, atomic absorption flame, FTIR and elemental analysis. The antibiotic action of the metal-based compounds was observed against two G(−ve) bacteria E. coli and Burkholderia and G(+ve) bacteria B. subtilis and S. aureus and fungi C. albicans. The metal complexes of bivalent metals showed higher anti-microbial activity while 10a, 10b and 10e showed mild bioactivity against these microbes.93
1 ratio with divalent transition metals (Scheme 10). These complexes were characterized using analytical techniques like 1HNMR, CNMR, atomic absorption flame, FTIR and elemental analysis. The antibiotic action of the metal-based compounds was observed against two G(−ve) bacteria E. coli and Burkholderia and G(+ve) bacteria B. subtilis and S. aureus and fungi C. albicans. The metal complexes of bivalent metals showed higher anti-microbial activity while 10a, 10b and 10e showed mild bioactivity against these microbes.93
 | ||
| Scheme 10 Synthesis of the solid metal complexes 10a–10c, redrawn from ref. 93. | ||
Mahmoud et al., (2021) reported the creation of new triazole ligand L11 and metallic compounds (Scheme 11) of iron, copper and zinc (11a–11c). All the prepared metal complexes were observed through thermal study, FTIR spectroscopy, conductivity measurements and elemental analysis. Conductivity measurements showed that metal complexes of 11a and 11b have an electrolytic nature, while the 11c complex was a nonelectrolyte. These compounds were examined for the anti-microbial properties against G(+ve) bacteria B. subtilis and S. aureus and G(−ve) bacteria E. coli and P. aeruginosa. They were also inspected against two fungal strains: C. albicans and A. flavus. The complexes showed enhanced bioactivity as compared to the ligands.94
 | ||
| Scheme 11 Proposed structures of metal complexes 11a–11c, redrawn from ref. 94. | ||
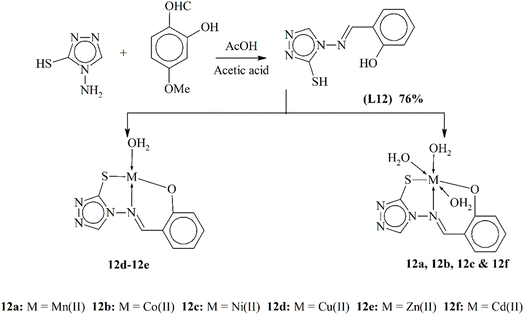
A novel nitrogen-containing ligand L12 was synthesized and subsequently employed to generate a series of transition metal complexes (12a–12f). Spectroscopic data exposed that the ligand functions as a tridentate donor, coordinating to metal ions through sulfur, nitrogen and oxygen atoms. The geometric configuration of the complexes varied, with 12a, 12b, 12c and 12f displaying octahedral structures, while 12d formed a square planar complex, and 12e adopted a tetrahedral arrangement (Scheme 12). Antimicrobial assessments indicated that the metal complexes have improved antimicrobial properties compared to their ligand, signifying their potential as antimicrobial agents.95
 | ||
| Scheme 12 Proposed structure of metal complexes 12a–12f, redrawn from ref. 95. | ||
Chohan and coworkers investigated a novel series of oxovanadium(IV) complexes (13a–13d) formed with Schiff bases L13a–L13d bearing triazole L13a–L13d. These ligands were synthesized by the condensation of 3,5-diamino-1,2,4-triazole with various aldehydes (Scheme 13). Comprehensive characterization techniques including IR, NMR and mass spectrometry confirmed the geometry of both the Schiff bases and their square pyramidal VO(IV) complexes. The complexes exhibited a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio for the metal and ligand. Furthermore, the study evaluated the bioactivity of both the ligands and their VO(IV) complexes. The results revealed antibacterial, antifungal and cytotoxic properties for both sets of compounds. However, coordination with vanadium(IV) generally enhanced the antimicrobial and antifungal efficacy against various bacterial and fungal strains.96 Fig. 8 shows the comparison of the antibacterial activities of metal complexes.
2 molar ratio for the metal and ligand. Furthermore, the study evaluated the bioactivity of both the ligands and their VO(IV) complexes. The results revealed antibacterial, antifungal and cytotoxic properties for both sets of compounds. However, coordination with vanadium(IV) generally enhanced the antimicrobial and antifungal efficacy against various bacterial and fungal strains.96 Fig. 8 shows the comparison of the antibacterial activities of metal complexes.
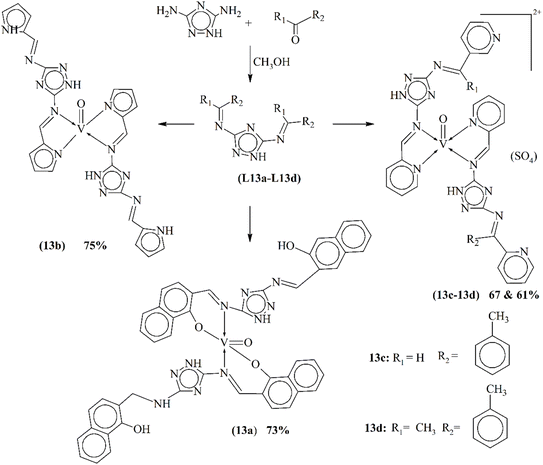 | ||
| Scheme 13 Synthesis of Schiff base ligands L13a–L13d and metal coordinates 13a–13d, redrawn from ref. 96. | ||
3.2 Anti-fungal activity
Fungal infections (FIs), which cause over 1.7 million fatalities yearly, are one of the undervalued emerging illnesses.97 Taking these data into consideration, it is expected that malaria grounds nearly 405![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mortalities per year and TB 1.5 million deaths annually.98 However, the effects of FIs on medicine go well beyond these horrifying death rates. Every year, FIs impact over one billion individuals, with over 150 million instances including severe and potentially fatal FIs. Metal-based drugs are playing a very crucial role in tackling the fungal infection as well as the AMR.99 Triazole Schiff base metal coordinates can be employed as antifungal agents. Triazole metal scaffolds act as antifungal agents through several mechanisms. Major types of mechanisms are presented in Fig. 9.
000 mortalities per year and TB 1.5 million deaths annually.98 However, the effects of FIs on medicine go well beyond these horrifying death rates. Every year, FIs impact over one billion individuals, with over 150 million instances including severe and potentially fatal FIs. Metal-based drugs are playing a very crucial role in tackling the fungal infection as well as the AMR.99 Triazole Schiff base metal coordinates can be employed as antifungal agents. Triazole metal scaffolds act as antifungal agents through several mechanisms. Major types of mechanisms are presented in Fig. 9.
 | ||
| Fig. 9 Mechanism of the antifungal activity of metal complexes, adapted from ref. 100–103. | ||
In the vast expanse of bioinorganic chemistry, a novel series of Schiff base ligands L14a–L14e and their coordination complexes with oxovanadium(IV) as in 14a–14e have been synthesized and examined by Chohan and Sumrra. These Schiff bases, which are derivatives of triazole, form a bond with oxovanadium(IV) to form complexes that exhibit a unique geometric structure (Scheme 14). These complexes have been put to the test against different microbes, demonstrating a promising range of antimicrobial activity. Particularly, the complexes 14d and 14e have shown significant efficiency, offering a beacon of hope in the face of drug-resilient straining, prevalent in different pharmacological practices. The study also underscores the role of vanadium in bioinorganic chemistry, highlighting its potential in various biological activities.104
 | ||
| Scheme 14 Synthesis of triazole Schiff bases and their complexes 14a–14e, redrawn from ref. 104. | ||
A novel series of oxovanadium(IV) complexes were synthesized by allowing vanadyl sulfate to react with ligands L15a–L15c derived from 4-amino-5-(substitutedphenoxyacetic acid)-1,2,4-triazole-3-thiol and benzyl as Schiff bases (Scheme 15). These complexes (15a–15c) showed solubility in dimethylformamide (DMF) and dimethylsulphoxide (DMSO), and their low molar conductance values indicated non-electrolytic behavior in the solution. The complexes were characterized using elemental analysis, spectral techniques (UV-vis, IR, EPR and XRD) and magnetic moment measurements. EPR spectra indicated that the free electron was in the dxy orbital. The activity against fungi Aspergillus niger, Colletotrichum falcatum, and Colletotrichum pallescence, as well as antibacterial activity against Escherichia coli and Salmonella typhi and Staphylococcus aureus and Bacillus subtilis bacterial strains, was determined for both the ligands and the synthesized complexes. The results showed increased activity upon complexation. This study demonstrates the potential of these oxovanadium(IV) complexes as antibacterial and antifungal agents against resistant strains.105
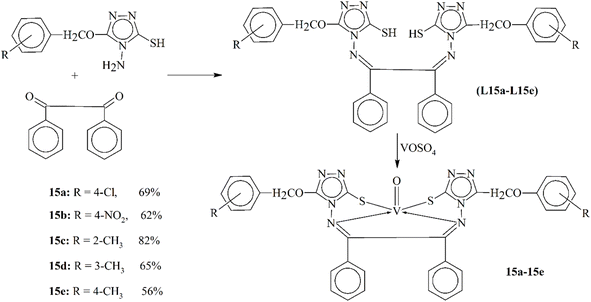 | ||
| Scheme 15 Synthetic diagram of ligands L15a–L15c and vanadyl complexes 15a–15e, redrawn from ref. 105. | ||
Chohan and Hanif studied a novel series of metal-based triazoles derived from Schiff base ligands L16a–L16c. The ligands were produced by condensing 3-amino-1H-1,2,4-triazole with furan-2-carboxaldehydes substituted with methyl-, chloro- and nitro- groups followed by complexation with Co(II), Ni(II), Cu(II) and Zn(II) metals in order to form complexes (16a–16l), as shown in Scheme 16. Various physical, analytical and spectroscopic methods were used for characterizing and gaining insights into the synthesized compounds. The antibacterial and antifungal activities of these compounds were evaluated against several bacterial and fungal strains. The complexes showed superior antimicrobial actions as compared to their parent ligands, indicating a potential enhancement in bioactivity upon coordination with metal ions. This enhanced activity could be attributed to the tridentate coordination of the ligands to the metal ions, involving the azomethine-N, triazole ring-N and furanyl-O atoms.106
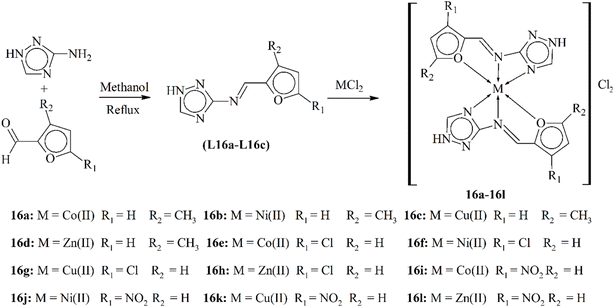 | ||
| Scheme 16 Synthetic scheme of the Schiff bases L16a–L16c and their complexes 16a–16l, redrawn from ref. 106. | ||
Biologically active iron and manganese metal complexes were prepared by Kumar and his coworkers. The compounds (17a–17c) were synthesized by using triazole Schiff base ligand L17 and then coordinating the metal ions to the bidentate ligand (Scheme 17). A triazole derivative and an aldehyde were refluxed using ethanol as the solvent for three hours. Cream color crystals of the ligand were separated and dried. TLC was utilized to confirm the synthesis of ligands. Metal salts were added to the ligand in methanol, and they were refluxed to form the metal complexes. The structural properties of the complexes were examined using nuclear magnetic resonance, infrared and UV-vis spectroscopic techniques and other microanalytical approaches.107
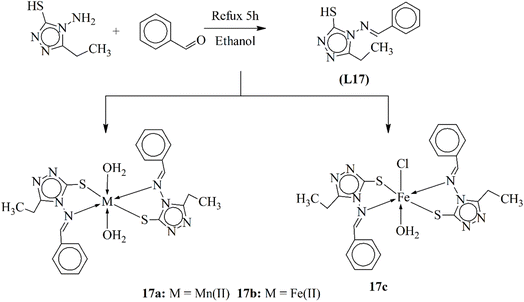 | ||
| Scheme 17 Synthetic scheme of metal complexes 17a–17b, redrawn from ref. 107. | ||
Joshi et al. 2020 introduced a new series of complex scaffolds (18a–18b) by incorporating tetravalent organotin compounds in a triazole ligand L18, which contains a carbon atom attached to the nitrogen atom through a double bond (Scheme 18). The structural detailing was performed using various approaches such as FT-IR, NMR and spectrometric approaches. Analysis based on DFT was implemented to evaluate the geometrical attributes of novel complex coordinates. The nature of organotin bonding was examined through an NBO approach. The antifungal action of the complex was tested against several fungal strains, which elucidated that the complexes were showing better antifungal characteristics than the parent ligand.108
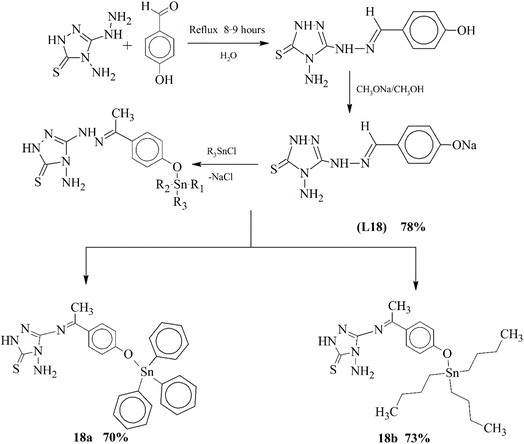 | ||
| Scheme 18 Structure of metal complexes 18a–18b of ligand L18a, redrawn from ref. 108. | ||
Metal complexes are the most vital pharmacological agents of modern era. They attracted the researchers all around the world due to their higher bioactivity and lesser antimicrobial resistance. Sumrra and coworkers published their work on a modern triazole ligand L19 Schiff base formed using a triazole moiety and an aldehyde in equal molar ratios (Scheme 19). Physical approaches, spectral techniques, elemental analysis and computational investigations were used to describe the obtained ligand. The computational study of the ligand deeply elaborated the structure, composition and stability. The synthesized ligand was complexed with the bivalent metal ions of transition metals. Spectroscopic and physical techniques were utilized to infer the structural properties of the formed complexes (19a–19h). The synthesized ligand and complexes were assessed for bioactivity against numerous bacteria. Complex 19g was found to be more active against bacteria than the other complexes.109
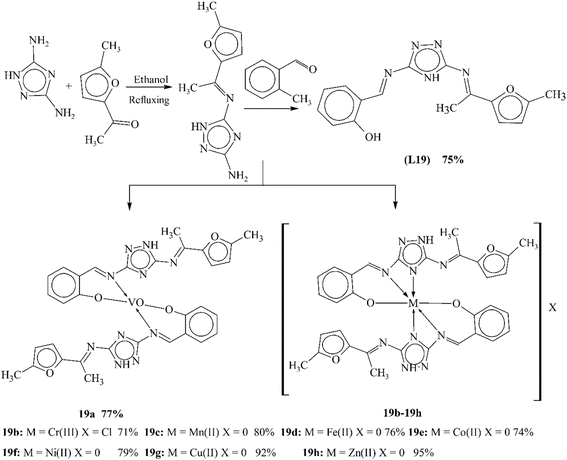 | ||
| Scheme 19 Synthetic route for metal complexes 19a–19f, redrawn from ref. 109. | ||
A new series of triazole ligands were synthesized by Munawar and his coworkers via a condensation reaction of an aldehyde derivative and an amino-substituted triazole compound (Scheme 20). Metal ions of vanadium oxide were incorporated into the ligands L20a–L20d to form the complex compounds 20a–20d. The produced compounds were examined using FT-IR spectroscopy, elemental analysis, NMR spectroscopy, molar susceptibility and conduction strategies, thermo-gravimetry and the melting point method. Bonding details were identified through red- and blue-shifts of UV-visible spectroscopy. The bioactivity of the VO(IV) complexes was found to be much higher than the parent ligand.110 A comparison of the antifungal activities of metal complexes is shown in Fig. 10. Table 1 shows the comparison of the antibacterial and antifungal activity values of some metal complexes.
 | ||
| Scheme 20 Synthetic route for metal complexes 20a–20d of ligands L20a–L20d, redrawn from ref. 110. | ||
| Comp. | Anti-bacterial activity | Anti-fungal activity | Ref. | Comp. | Anti-bacterial activity | Anti-fungal activity | Ref. |
|---|---|---|---|---|---|---|---|
| 1f | IZD = 14 mm (E. coli) | — | 84 | 16j | — | PI = 65% (F. solani) | 106 |
| 1b | — | IZD = 29 mm (Penicillium spp.) | 84 | 18a | — | IZD = 37 mm (A. pullulans) | 108 |
| 2g | IZD = 17 mm (E. coli) | — | 85 | 19b | IZD = 10 mm (N. gonorrhea) | — | 109 |
| 2f | IZD = 24 mm (S. typhi) | — | 85 | 20c | IZD = 35 mm (E. coli) | IZD = 62 mm (P. notatum) | 110 |
| 3d | PI = 82% (S. aureus) | — | 86 | 21b | — | PI = 73% (C. glabrata) | 111 |
| 4b | IZD = 82% (S. aureus) | — | 87 | 21f | IZD = 24 mm (B. subtilis) | — | 111 |
| 4k | — | IZD = 17 mm (A. niger) | 87 | 22e | IZD = 26 mm (E. coli) | — | 112 |
| 5b | IZD = 24 mm (S. typhi) | — | 88 | 23g | IZD = 25 mm (P. aeruginosa) | — | 113 |
| 6f | MIC = 500 µg mL−1 (P. aeruginosa) | — | 89 | 23q | — | PI = 74% (F. solani) | 113 |
| 7c | IZD = 27 mm (P. aeruginosa) | — | 90 | 24d | IZD = 30 mm (P. aeruginosa) | PI = 60% (T. longifusus) | 114 |
| 8h | — | PI = 82% (C. glabra90ta) | 91 | 25a | IZD = 20 mm (S. aureus) | — | 115 |
| 8m | IZD = 25 mm (H. salina) | — | 91 | 26a | IZD = 15 mm (P. aeruginosa) | — | 116 |
| 9h | IZD = 24 mm (B. subtilis) | — | 92 | 26c | IZD = 13 mm (S. aureus) | — | 116 |
| 9o | IZD = 21 mm (E. coli) | — | 92 | 27b | IZD = 13 mm (S. aureus) | — | 117 |
| 10e | IZD = 33 mm (E. coli) | — | 93 | 31a | IZD = 25 mm (P. aeruginosa) | — | 118 |
| 11a | — | IZD = 13 mm (A. flavus) | 94 | 31f | — | PI = 82% (C. glabrata) | 118 |
| 11c | IZD = 18 mm (P. aeruginosa) | — | 94 | 32c | MIC = 3.94 µg mL−1 (E. coli) | MIC = 31.25 µg mL−1 (E. coli) | 119 |
| 12f | IZD = 20 mm (S. aureus) | — | 95 | 33d | IZD = 16 mm (S. typhimurium) | IZD = 11 mm (C. albicans) | 120 |
| 13d | IZD = 25 mm (P. aeruginosa) | — | 96 | 35a | IZD = 7 mm (B. subtilis) | — | 121 |
| 14c | — | PI = 78% (C. glabrata) | 104 | 43d | IZD = 18 mm (E. coli, B. subtilis) | — | 122 |
| 15a | — | PI = 87% (A. niger) | 105 | 44f | IZD = 13 mm (E. coli) | IZD = 19 mm (A. alternate) | 123 |
| 45b | MIC = 3.5 µM (E. coli) | — | 124 |
3.3 Cytotoxic activity
Metal complexes have special properties because of their metal center, which can affect their way of interaction with biological molecules and ultimately influence their cytotoxicity. In medicinal chemistry, the cytotoxic activity of metal complexes is an important area of research, especially for the development of anticancer drugs and antimicrobial agents.125 Here is an overview of how these complexes work as cytotoxic agents (Fig. 11). | ||
| Fig. 11 Mechanism of the cytotoxic activity of metal complexes, adapted from ref. 126. | ||
In 2012, Hanif and Chohan presented the synthesis of triazole ligands L21a–L21c by reacting 3-amino-1H-1,2,4-triazole with pyrrole-2-carboxaldehyde, 5-iodo-2-hydroxy benzaldehyde and 4-bromo-thiophene2-carboxaldehyde (Scheme 21). The above compounds were utilized to generate metallic coordinates (21a–21l) by reacting them with the metal salts in a stoichiometric ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (L
1 (L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M). The structural studies have confirmed that copper complexes showed a distorted octahedral geometry, while other metal complexes showed an octahedral structure. Biological studies revealed that the prepared metal compounds presented bioactivity against bacterial and fungal strains. Cytotoxic properties were also examined by in vitro brine shrimp bioassay. The observed data revealed that the bioactivity of the metallic compounds is greater than the normal ligand. The reason for increased antimicrobial activity is the chelation phenomenon.111
M). The structural studies have confirmed that copper complexes showed a distorted octahedral geometry, while other metal complexes showed an octahedral structure. Biological studies revealed that the prepared metal compounds presented bioactivity against bacterial and fungal strains. Cytotoxic properties were also examined by in vitro brine shrimp bioassay. The observed data revealed that the bioactivity of the metallic compounds is greater than the normal ligand. The reason for increased antimicrobial activity is the chelation phenomenon.111
 | ||
| Scheme 21 Synthesis of ligands and their metal complexes 21a–21h, redrawn from ref. 111. | ||
Sumrra and Chohan provided an in-depth look into the creation and medicinal importance of Schiff base ligands and their oxovanadium(IV) complexes (22a–22f). The ligands L22a–L22f were synthesized by reacting 3,5-diamino-1,2,4-triazole with various substituted 2-hydroxybenzaldehydes (Scheme 22). These bases then reacted with oxovanadium(IV) sulphate to form oxovanadium(IV) complexes in a stoichiometric ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (M
2 (M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L). Synthesized complexes, exhibiting a square-pyramidal geometry, were then tested for their bioactivity using antimicrobial and brine shrimp bioassay tests, with complexes showing better action than the original Schiff bases. The study also highlights the role of vanadium chemistry in bioinorganic chemistry, noting its antimicrobial, anti-tumor, anti-leukemic, spermicidal, anti-amoebic, antioxidant and osteogenic activity. Particularly, the oxovanadium(IV) complexes are known as potential inhibitors of various enzymes and have insulin-mimetic activity, which could be beneficial in treating diabetes mellitus.112
L). Synthesized complexes, exhibiting a square-pyramidal geometry, were then tested for their bioactivity using antimicrobial and brine shrimp bioassay tests, with complexes showing better action than the original Schiff bases. The study also highlights the role of vanadium chemistry in bioinorganic chemistry, noting its antimicrobial, anti-tumor, anti-leukemic, spermicidal, anti-amoebic, antioxidant and osteogenic activity. Particularly, the oxovanadium(IV) complexes are known as potential inhibitors of various enzymes and have insulin-mimetic activity, which could be beneficial in treating diabetes mellitus.112
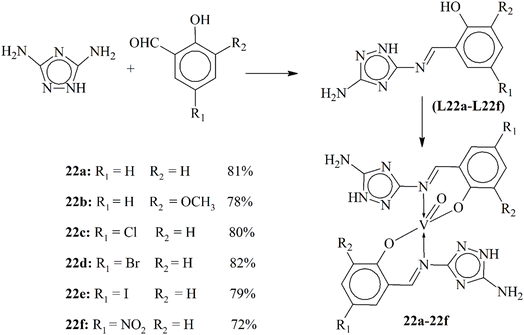 | ||
| Scheme 22 Synthetic scheme of oxovanadium metal complexes 22a–22f, redrawn from ref. 112. | ||
Chohan and Hanif explored a novel class of potential bioactive agents based on triazole derivatives. They successfully synthesized five Schiff base ligands L23a–L23e via condensation reactions between 3-amino-1,2,4-triazole and various substituted thiophene-2-carboxaldehydes (Scheme 23). A comprehensive array of techniques including IR spectroscopy, NMR, magnetic susceptibility and elemental analysis confirmed the structures of the ligands and their metal complexes (23a–23t) with cobalt, copper, nickel and zinc. Notably, X-ray diffraction analysis provided further structural validation for each ligand. The study revealed that all metal complexes exhibited an octahedral geometry, with the exception of the copper complexes, which adopted a distorted octahedral arrangement. To assess their potential therapeutic value, researchers evaluated the antibacterial activity of these compounds. The findings demonstrated that all ligands and their metal complexes displayed moderate to significant antibacterial action against various bacterial strains. Importantly, the metal complexes displayed superior antibacterial efficacy compared to their free ligand counterparts. This enhanced activity is attributed to chelation, a process that increases the lipophilicity of the complexes, potentially facilitating their penetration through bacterial cell membranes.113
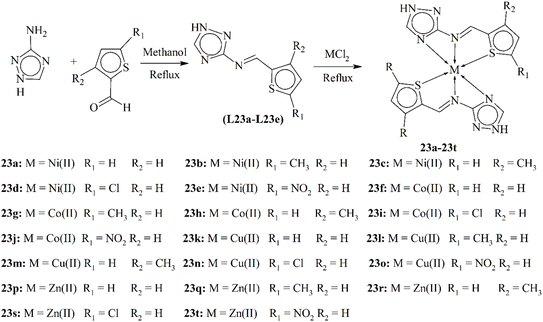 | ||
| Scheme 23 Synthetic scheme of triazole ligands L23a–L23c and metal complexes 23a–23t, redrawn from ref. 113. | ||
In the intriguing domain of bioinorganic chemistry, the work was done on the synthesis and characterization of Schiff base ligands L24a–L24e and their oxovanadium(IV) complexes (24a–24e) by Chohan and Sumrra. These Schiff base bidentate ligands form a bond with an oxovanadium moiety through azomethine, resulting in vanadyl complexes with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (M
2 (M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L) stoichiometry and a square-pyramidal shape (Scheme 24). They further delved the study into the in vitro antimicrobial actions of these complexes against a number of bacteria and fungi. The outcomes indicate that all the synthesized ligands and their complexes exhibit moderate to higher activity against bacterial strains, but better antifungal actions against different strains. In addition, a brine shrimp bioassay was conducted to assess the cytotoxicity of these compounds. This research thus provides valuable insights into the potency of these oxovanadium(IV) compounds as antimicrobial drugs, underscoring the importance of vanadium chemistry in medicinal applications.114
L) stoichiometry and a square-pyramidal shape (Scheme 24). They further delved the study into the in vitro antimicrobial actions of these complexes against a number of bacteria and fungi. The outcomes indicate that all the synthesized ligands and their complexes exhibit moderate to higher activity against bacterial strains, but better antifungal actions against different strains. In addition, a brine shrimp bioassay was conducted to assess the cytotoxicity of these compounds. This research thus provides valuable insights into the potency of these oxovanadium(IV) compounds as antimicrobial drugs, underscoring the importance of vanadium chemistry in medicinal applications.114
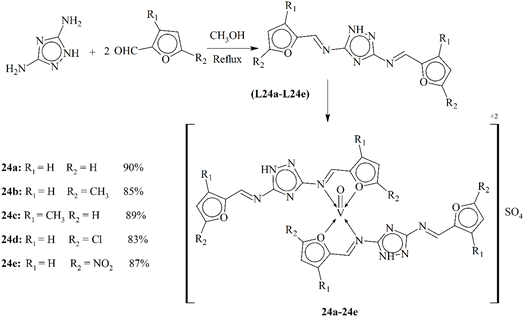 | ||
| Scheme 24 Proposed scheme of oxovanadium metal complexes 24a–24e, redrawn from ref. 114. | ||
3.4 Anti-cancer activity
One out of six deaths in the world is due to cancer. In 2020, around 19.3 million new cases and about 10 million losses of lives occurred in the world.127 Cancer is a lethal disease that grows over time and causes disorder in the growth of cells.128 For several years, the only options available to cancer patients were radiation therapy, chemotherapy and surgery, either independently or combined.129 The tremendous capability of metal complexes to treat a variety of cancers has drawn the interest of researchers all over the world. The special characteristics of metal complexes such as kinetics (such as ligand exchange rates) and thermodynamics (such as metal–ligand bond strengths and redox potential) can be adjusted through modification of the central metal ion, atom or their oxidation states.130 Their exceptional biomedical attributes make them a great option to cure many diseases.131 For example, platinum-incorporated anti-cancer agents like cisplatin, carboplatin and oxaliplatin were prepared.132Triazole Schiff base metal scaffolds are used as anticancer agents. They have been found to be highly effective against cancer cells. Following are the types of mechanisms through which triazole metal scaffolds can act as anticancer agents (Fig. 12).
 | ||
| Fig. 12 Anticancer activity of triazole metal complexes, adapted from ref. 136. | ||
1. Redox modulation and oxidative stress133
2. DNA binding and cleavage134
3. Metal ion chelation135
In 2021, Deodware et al. published the synthesis of modern triazole ligands L25a–L25d, 4-(2′/3′/4′-nitrobenzelideneimino)-3-methyl/ethyl-5-mercapto-1,2,4-triazole and its bivalent cobalt complexes (25a–25d). The ligand and its metal complexes were studied using NMR analysis, thermal, electronic and magnetic moment calculation studies. The structural observation elucidated that all the cobalt complexes were present in octahedral geometry (Scheme 25). Gravimetric studies showed that the coordinated metal also contained two water molecules, and the complex was also found to be highly bioactive against fungal strains. Another important aspect of these complexes was their ability to work as anticancer agents, and these complexes were found to be active against blood, lung, ovary and prostate cancers.115
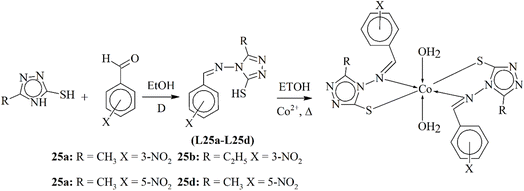 | ||
| Scheme 25 Synthetic scheme for the ligands and Co(II) complexes 25a–25d, redrawn from ref. 115. | ||
Deghadi and his coworkers published the synthesis of unique metal complexes (26a–26c) of uranium dioxide, erbium and lanthanum metals (Scheme 26). The Schiff base ligand L26 was synthesized through a condensation process. The ligand and the complexes formed were observed through physical and spectrometric approaches to examine the structural and physical aspects of the scaffolds. The bonding properties and octahedral geometry were also observed through computational and TGA studies. The antibacterial assay was studied against four bacterial strains. The experimental results proved that the complexes were highly active against these bacteria. It was found that these complexes were biologically active against cancer cells. This phenomenon approved their anti-cancer function. The anticancer activity was also confirmed through molecular docking.116
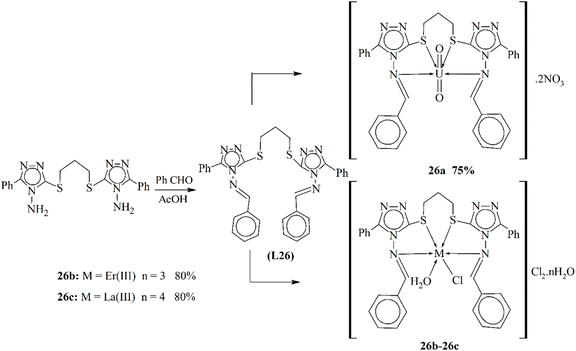 | ||
| Scheme 26 Scheme of metal coordinates 26a–26c, redrawn from ref. 116. | ||
Metal coordinates of triazole-incorporated ligands can be utilized for the treatment of tumors. These metal scaffolds and ligands can be used as antioxidant agents. The above-mentioned biological assays were investigated in 2014 by Gaber and his coworkers. They published the synthesis of a Schiff base L27 having 3-amino-1,2,4-trizole (Scheme 27). The ligand was prepared using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 concentration of an aldehyde and triazole. The reflux process resulted in the formation of a crystalline ligand, which was separated by washing with methanol. Metallic chlorides were used to form metallic compounds (27a–27c) chelated to the synthesized ligand. Several physical and analytical techniques were used to study the structural and binding properties of the compound. The studies revealed that 27b and 27c complexes have a square planar geometry, while 27a complex has an octahedral geometry. The anti-bacterial and anti-fungal actions of ligand and metal scaffolds were also tested using G-(+ve) and G-(−ve) bacteria. The synthesized compound also presented anti-cancer and anti-oxidant properties. Comparative studies revealed that ligand was more efficient than the metal complexes.117
1 concentration of an aldehyde and triazole. The reflux process resulted in the formation of a crystalline ligand, which was separated by washing with methanol. Metallic chlorides were used to form metallic compounds (27a–27c) chelated to the synthesized ligand. Several physical and analytical techniques were used to study the structural and binding properties of the compound. The studies revealed that 27b and 27c complexes have a square planar geometry, while 27a complex has an octahedral geometry. The anti-bacterial and anti-fungal actions of ligand and metal scaffolds were also tested using G-(+ve) and G-(−ve) bacteria. The synthesized compound also presented anti-cancer and anti-oxidant properties. Comparative studies revealed that ligand was more efficient than the metal complexes.117
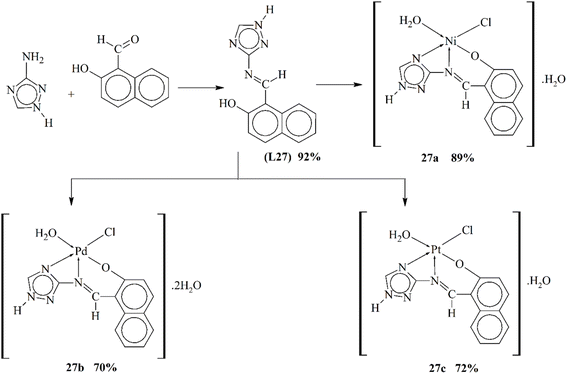 | ||
| Scheme 27 Proposed structures of metal complexes 27a–27c, redrawn from ref. 117. | ||
Two Schiff base ligands L28a–L28b, which were found to be biologically active, were synthesized by reacting 4-amino-5-(pyridin-4-yl)-4H-1,2,4-triazole-3-thiol with thiophene-2-carbaldehyde and furan-2-carbaldehyde, respectively, in equimolar ratios (Scheme 28). The structures of these ligands were confirmed using various analytical methods. The Schiff bases exhibited solubility in DMF, DMSO and methanol upon heating. The Schiff bases were then complexed with Co(II), Ni(II) and Cu(II) ions, forming crystalline solids that were stable in air but decomposed above 280 °C. Characterization was performed using 1H-NMR spectroscopy, UV-vis spectroscopy, TGA, IR spectroscopy, mass spectrometry and molar conductivity studies, with DFT studies showing an octahedral geometry for the metallic coordinates. Cytotoxicity studies against MCF-7 and HEPG-2 cell lines showed moderate to significant cytotoxicity for all metal scaffolds, indicating their effectiveness as anticancer agents.137
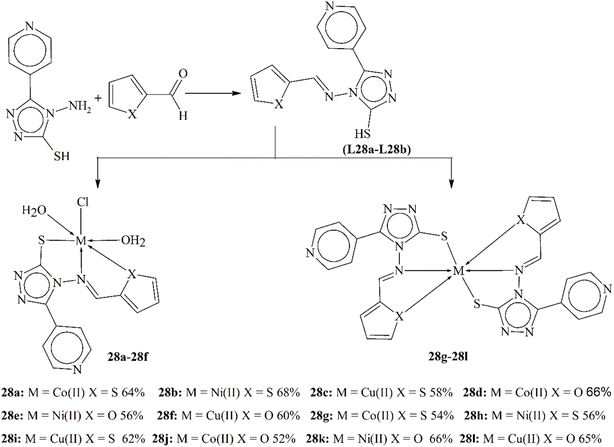 | ||
| Scheme 28 Structure of the ligands and metal complexes 28a–28l, redrawn from ref. 137. | ||
Two fresh Schiff base ligands were obtained by condensing an amine derivative of the 1,2,4-triazole moiety with 2-hydroxy-4-methoxybenzaldehyde. These ligands L29a–L29b were then coordinated with Co(II), Ni(II), Cu(II) and Zn(II) ions (Scheme 29). The structural characterization of both the Schiff bases and the resulting metal coordinates (29a–29h) was performed using various techniques including NMR spectroscopy, UV-vis spectroscopy, IR spectroscopy, mass spectrometry and molar conductivity. The fluorescence mechanism of the metal complexes revealed interesting binding behavior, with Zn(II) and Cu(II) complexes showing stronger affinity toward BSA (bovine serum albumin). Spectral data indicated that the ligands behave as tridentate ligands. Based on the spectral analyses, the metal complexes exhibited distinct geometries: octahedral for Co(II) and Ni(II), square planar shape for Cu(II), and tetrahedral geometry for Zn(II) complexes. Notably, the metal complexes demonstrated increased cytotoxicity in cell proliferation assays compared to the free ligand. The observed inhibition of cell proliferation may be attributed to the presence of the azomethine linkage and other heteroatoms within these compounds.138
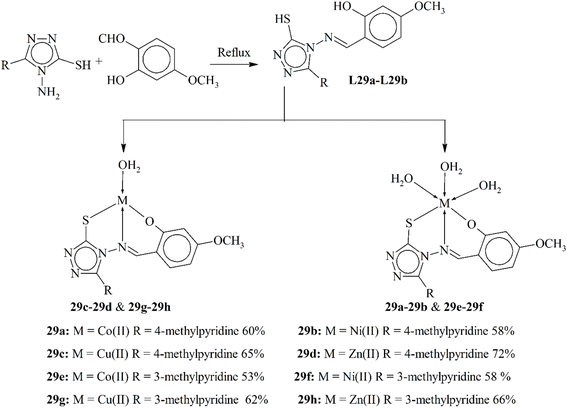 | ||
| Scheme 29 Preparation of ligands L29a–L29b and their respective complexes 29a–29h redrawn from ref. 138. | ||
Eno and his coworker prepared the Schiff base L30 and its metal scaffolds (30a–30b) through refluxing the mixture of 2-hydroxy-1-naphtahledehyde and a triazole moiety (Scheme 30). The Schiff base in the form of crystals was filtered and dried. The metal salt of cadmium was used to prepare metal complexes (30a–30b). The bioactive assay revealed that the synthesized metal coordinates were found to be active against plasmodium. NBO and FMO analyses were performed to study the structural and bonding attributes of the metal compound. Detailed specification of the metal coordinates were investigated through FT-IR spectroscopy, NMR spectroscopy and XRD analysis.139
 | ||
| Scheme 30 Synthetic presentation of cadmium complexes 30a–30b, redrawn from ref. 139. | ||
Sumrra et al. (2015) presented the synthesis of two triazole ligands L31a–L31c. Metal complexes (31a–31d) of vanadium(IV) metal ions were synthesized (Scheme 31). They were observed by using elemental, spectral and physical data. The prepared complexes were investigated for their antibiotic action opposing six bacterial types S. flexneri, P. aeruginosa, E. coli, S. aureus, S. typhi and B. subtilis. The experimental studies showed that metal complexes of vanadium(IV) were found to be bioactive against more than one fungal and bacterial type. The cytotoxicity of this metal complex was also examined through the brine shrimp bioassay.118 Table 2 elaborates the anticancer and cytotoxic activity values of some metal complexes.
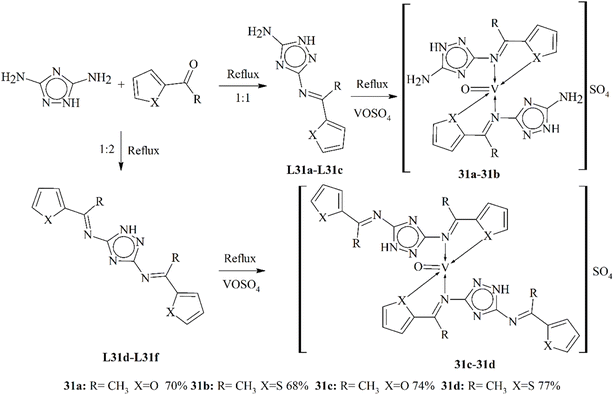 | ||
| Scheme 31 Scheme of mono- and di-substituted ligands and metal complexes 31a–31d, redrawn from ref. 118. | ||
| Comp. | Cytotoxic activity | Anti-cancer activity | Ref. | Comp. | Cytotoxic activity | Anti-cancer activity | Ref. |
|---|---|---|---|---|---|---|---|
| 7a | LD50 = 5.76 × 10−4 M mL−1 | — | 111 | 26a | — | IC50 = 50 µM (MCF-7) | 116 |
| 14b | LD50 = 2.154 × 10−4 M mL−1 (A. salina) | — | 104 | 27b | — | IC50 = 19.7 µg L−1 (HEPG 2) | 117 |
| 16b | LD50 = 9.48 × 10−4 M mL−1 | — | 106 | 28k | — | GI50(10 µM) = 38% (MCF-7) | 137 |
| 21d | LD50 = 1.15 × 10−3 M mL−1 | — | 111 | 29g | — | GI50(10 µM) = 39% (HEPG 2) | 138 |
| 22d | LD50 = 2.236 × 10−2 M mL−1 | — | 112 | 31a | LD50 = 6.11 × 10−3 M mL−1 | — | 118 |
| 23h | LD50 = 9.44 × 10−4 M mL−1 | — | 113 | 35b | — | IC50 = 200 g mL−1 (MCF-7) | 121 |
| 24e | LD50 = 6.819 × 10−3 M mL−1 | — | 114 | 37i | IC50 = 0.32 µM (HEP-2) | 140 | |
| 25b | — | PG = 85% (OVCAR-3) | 115 | 43b | — | GI = 135% (SPC-3) | 122 |
3.5 Enzyme inhibition activity
Because of the implications for drug design and therapeutic uses, the study of enzyme inhibitory activity of metal complexes is an important field in biochemistry and pharmacology. Enzymatic activity can be either promoted or inhibited by metal ions depending on their interaction with the enzymes. Gaining insights into these relationships is essential for creating potent inhibitors, particularly when it comes to diseases like cancer.141 Triazole metal complexes are also very important is this aspect as they can act as enzyme inhibitors for the treatment of many diseases. Possible mechanisms of these complexes are given in Fig. 13. | ||
| Fig. 13 Possible mechanism of enzyme inhibitory activity of metal complexes, adapted from ref. 142–145 | ||
Vinush et al. (2020) prepared a new triazole ligand, 5-[(3,4-dimethoxybenzylidene)amino]-(4H-1,2,4-triazole)-3-thiol. Bivalent transition metal complexes were prepared with a metal-to-ligand ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (M
2 (M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) L). The prepared scaffolds were characterized through NMR spectroscopy, UV-visible spectroscopy, TGA and IR spectroscopy. The antimicrobial activity of these compounds was assessed against nine food pathogens. It was observed through in vitro α-amylase inhibitory analysis that metal-based compounds were more efficient than simple triazole Schiff base ligands.146
L). The prepared scaffolds were characterized through NMR spectroscopy, UV-visible spectroscopy, TGA and IR spectroscopy. The antimicrobial activity of these compounds was assessed against nine food pathogens. It was observed through in vitro α-amylase inhibitory analysis that metal-based compounds were more efficient than simple triazole Schiff base ligands.146
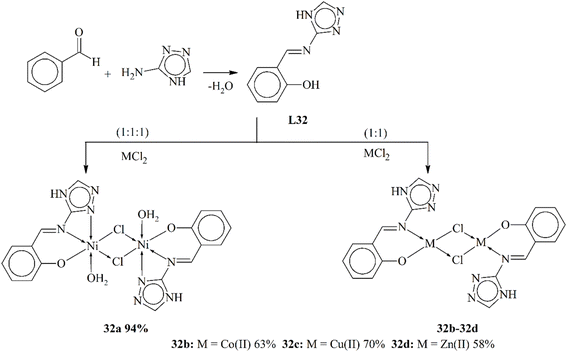
Calu and his coworkers (2015) published the synthesis of unique metal complexes (32a–32d) of transition metals (Scheme 32). The Schiff base ligand L32 was synthesized through a condensation process. The amine-containing triazole and aldehyde were refluxed in ethanol as a solvent. The refluxation process was continued for 5 hours at 50 °C. The ligand and the complexes formed were observed through specific physical and spectrometric approaches to examine the structural and physical aspects of the scaffolds. The bonding properties and octahedral geometry was also observed through computational and TGA studies. The present literature was used to confirm the successful synthesis of the ligand. Complex 32a showed an octahedral geometry, while divalent 32c showed a square planar geometry. CV (cyclic voltammetry) was used to study the redox characteristic of the metal coordinates. The anti-bacterial assay was observed against few bacterial strains. The experimental results proved that the complexes were highly active against these bacteria. Another important aspect of these complexes was their ability to work as anticancer agents.119
 | ||
| Scheme 32 Synthesis of the triazole ligand and its metal coordinates 32a–32d, redrawn from ref. 119. | ||
Hassan et al. (2019) published the preparation of two ligands N-(furan-2-ylmethylene)-[1,2,4-triazole-3-amine] and N-(thiophene-2-ylmethylene)-1H-1,2,4-triazole-3-amine (L33) and their metallic compounds (33a–33d) of cobalt, nickel, copper and zinc (Scheme 33). The structural and bonding studies of their metal complexes were conducted using UV-visible spectroscopy, FT-IR spectroscopy, mass spectrometry, nuclear magnetic resonance spectroscopy, electron spin resonance and thermal activity. Zinc complexes of these generated compounds were studied for their antimicrobial activity.120
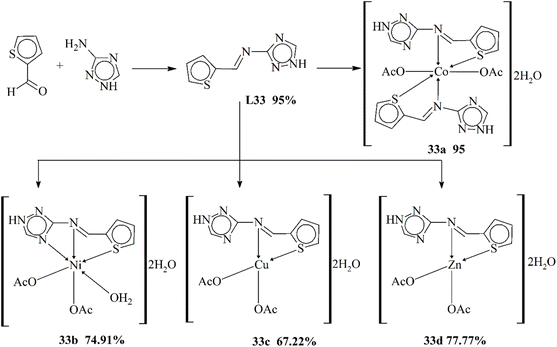 | ||
| Scheme 33 Synthesis of the triazole-based ligand and its metal scaffolds 33a–33d, redrawn from ref. 120. | ||
In 2009, Al-Masoudi et al. described the synthesis of novel Schiff base triazole-coordinated ligands L34a–L34d from 5-amino-4-phenyl-4H-1,2,4-triazole-3-thiol and substituted benzaldehydes, along with a new benzothiazole derivative (Scheme 34). These compounds were characterized using spectral analysis. Their metal coordinates (34a–34f) with Cu(II), Fe(II), Au(III) and Mn(II) were also synthesized and checked for anti-HIV-1 and HIV-2 activities using MT-4 cells. Karl-Fischer titration showed the presence of water molecules in these complexes, supported by a band around 3390–3520 cm−1 related to water molecules (ν(OH)) associated with the metallic compounds. The IR spectrum of the ligand showed a band at ∼1625 cm−1 related to ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) of azomethine, which shifted to a lower wave number by around 25–30 cm−1 upon chelation of the ligand with a metallic ion, particularly evident in the Cu(II) complexes (34a–34b).147
N) of azomethine, which shifted to a lower wave number by around 25–30 cm−1 upon chelation of the ligand with a metallic ion, particularly evident in the Cu(II) complexes (34a–34b).147
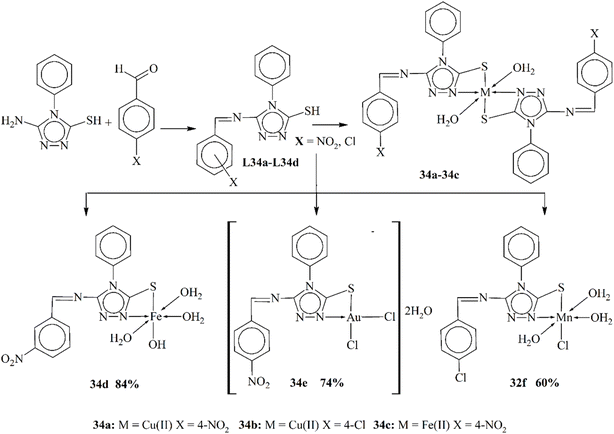 | ||
| Scheme 34 Synthesis of ligands and their metal scaffolds 34a–34c, redrawn from ref. 147. | ||
3.6 DNA-binding and cleavage activity
Novel DNA interaction that affects the biological activity can result from the covalent bonding of the organic moiety to transition metal coordinates, resulting in metallo-intercalators.148 Metal complexes in which aromatic scaffolds are linked through σ bonds can show dual functionality, allowing them to interact with DNA both through metal chelation and by intercalation of the aromatic ligand. These aromatic moieties provide novel approaches to DNA binding that entail interactions between functional groups that are kept close to one another.149 Fig. 14 shows some examples of mechanism through which triazole metal complexes interact with DNA strands. | ||
| Fig. 14 Mechanism of DNA binding and cleavage activity of metal complexes, adapted from ref. 150–154. | ||
Triazole-based compounds can be used as bioactive entities for the treatment of breast cancer as well as for fungal infections. Triazole Schiff base L35 and its metal scaffolds were analyzed by Sangeetha and coworkers. Triazole Schiff base was synthesized by mixing a triazole entity with an aldehyde derivative in 10 mL equimolar ratio of ethanol and acetonitrile. The resulting ligand was then utilized to synthesize the metal complexes (35a–35b) through refluxing it with metallic salts of manganese chloride (Scheme 35). Physical and structural analyses were performed through FT-IR spectroscopy, CNMR EPR and electronic techniques. The catalase bioactivity assay was performed against Gram-negative and Gram-positive bacteria. It was observed that the bioactivity was increased 21 percent as compared to the original ligand. DNA docking analysis also supported the above-mentioned bioactive property of the prepared compounds.121
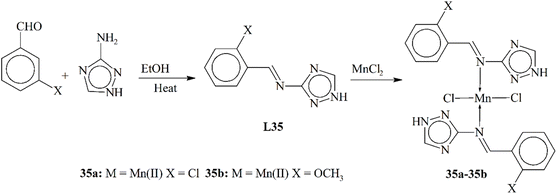 | ||
| Scheme 35 Synthetic route of ligand and metal complexes 35a–35b, redrawn from ref. 121. | ||
Bheemarasetti with his fellows investigated a brand new Schiff base ligand, L36, obtained from a chromone and a triazole and its metallic scaffolds (36a–36e) with Co(II), Ni(II), Cu(II), Zn(II) and Pd(II). The ligand was found to coordinate to the metallic ions in a bidentate pattern, forming complexes with different geometries (Scheme 36). Most complexes adopted tetrahedral structures, while Cu(II) and Pd(II) formed square-planar complexes. These compounds exhibited strong binding affinity to DNA and were capable of cleaving DNA strands, suggesting their potential for applications in gene therapy or as anticancer agents. Furthermore, the complexes demonstrated promising biological activities, including antimicrobial and antioxidant properties. The antimicrobial capability was checked against both Gram-positive and Gram-negative bacteria, as well as fungi. The antioxidant activity was attributed to the complexes' ability to scavenge reactive oxygen species, which are implicated in various diseases. These studies urged the potential of Schiff base complexes derived from this ligand for development as therapeutic agents or materials with specialized properties.155
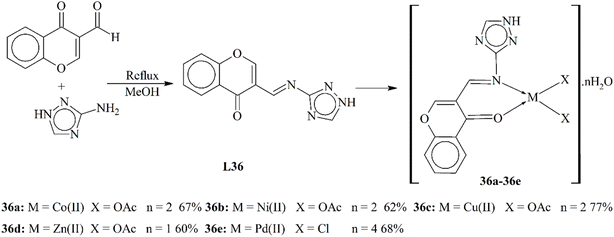 | ||
| Scheme 36 Synthetic scheme of ligand and metal complexes 36a–36e, redrawn from ref. 155. | ||
The development of novel metallodrugs with enhanced biological actions and cytotoxicity while maintaining less toxicity is crucial in the fight against cancer. Utthra and coworkers presented a series of octahedral metal complexes (37a–37l) containing a triazole-induced Schiff base scaffold L37 (Scheme 37). These complex coordinates were characterized using various techniques. Their interactions with DNA, antimicrobial properties and electrochemical behavior were investigated. While all complexes showed activity, complex 37i exhibited exceptional DNA binding, cleavage and antimicrobial efficacy. Additionally, complexes 37a, 37e and 37f demonstrated antiproliferative activity against human tumor cell lines with less toxicity to the normal cell.140
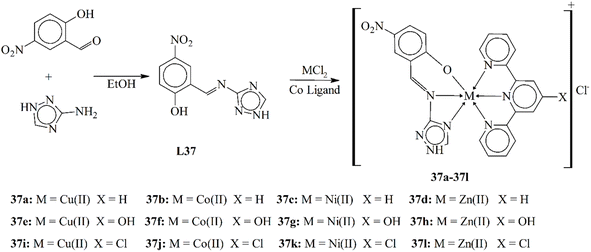 | ||
| Scheme 37 Schematic of metal complexes 37a–37l synthesis, redrawn from ref. 140. | ||
Kulkarni and coworkers described the chemical synthesis and profiling of cobalt(II), nickel(II) and copper(II) scaffolds (38a–38f) obtained from Schiff bases L38a–L38b formed via the reaction of 3-substituted-4-amino-5-mercapto-1,2,4-triazole and fluvastatin. The Schiff bases are chelated to the metal ions by the azomethine nitrogen and thiolate sulfur atoms (Scheme 38). Based on analytical, spectral, magnetic and thermal studies, the square planar structure was proposed for all the metallic adducts. The chemotherapeutic activities of the Schiff bases and their metal complexes were evaluated against assorted bacterial and fungal species using the minimum inhibitory concentration approach. The metallic entities demonstrated superior microbicidal activity compared to the Schiff bases. Additionally, the cobalt(II), nickel(II) and copper(II) chelates were found to cleave DNA isolated from Aspergillus niger.156
 | ||
| Scheme 38 Synthesis of imine ligands and transition metal scaffolds 38a–38f, redrawn from ref. 156. | ||
Chaurasia and fellows presented the synthesis and characterization of a novel Schiff base ligand L39 derived from 2-hydroxy-4-methoxybenzaldehyde and 3-amino-1,2,4-triazole (Scheme 39). Metal complexes (39a–39d) of the ligand with Co(II), Ni(II), Cu(II) and Zn(II) were characterized using various spectroscopic techniques. The ligand was found to act as a tridentate ligand, attached to the metal ions by means of phenolic oxygen, imine nitrogen and triazole nitrogen atoms. Computational studies using Gaussian 09W revealed octahedral geometry for 39a, tetrahedral for 39b and 39d and tetragonal for 39c. DNA binding studies of the metal complexes using UV absorbance, fluorescence and CD spectroscopy demonstrated significant binding to calf thymus DNA, suggesting their potential for biological applications.157
 | ||
| Scheme 39 Synthesis of the ligand and metallic chelates 39a–39d, redrawn from ref. 157. | ||
The world of coordination chemistry, particularly focused on macro cyclic complexes, has seen a remarkable transformation thanks to the development of innovative synthetic techniques. Template reactions have proven to be a powerful tool in creating a wide range of macro cyclic complexes, especially those containing nitrogen atoms. Bagihalli and Patil have explored the synthesis of metal complexes (40a–40p) using cobalt, nickel, copper and zinc, combined with newly discovered biologically active ligands L40a–L40d derived from 3-substituted-4-amino-5-hydrazino-1,2,4-triazole and orthophthalaldehyde (Scheme 40). By analyzing the elements present and conducting various spectroscopic tests, researchers have been able to propose likely structures for these complexes. Electrochemical studies have revealed that these complexes are not electrolytic in specific solvents like N,N-dimethylformamide and DMSO. Moreover, the Schiff bases and their metal adducts have been evaluated for their effectiveness against various microorganisms and their ability to cleave DNA.158 Table 3 presents the enzyme inhibition and DNA interaction activity values of some metal coordinates.
 | ||
| Scheme 40 Synthesis of ligands and metal complexes 40a–40d, redrawn from ref. 158. | ||
| Comp. | Enzyme inhibition activity | DNA-binding activity | Ref. | Comp. | Enzyme inhibition activity | DNA-binding activity | Ref. |
|---|---|---|---|---|---|---|---|
| 20d | PI = 100% (ALP) | — | 110 | 36c | — | Binding kb = 4.8 × 104 M−1 | 155 |
| 32d | GI = 67.80% (HeLa cells) | — | 119 | 37i | — | Binding kb = 3.5 × 105 M−1 | 140 |
| 34e | C50 = 2.11 µg mL−1 (MT-4, HIV) | — | 147 | 39b | — | Binding Abs = 1 × 10−5 M (etDNA) | 157 |
| 35a | — | BI = 5.9 kJ mole−1 (DNA-hexamer) | 121 | ||||
3.7 Anti-oxidant activity
Antioxidants are substances that neutralize the effects of reactive species such as free radicals and protect the body against oxidative damage-related diseases and aging.159 The ability of a synthetic chemical to scavenge and trap free radicals is a useful tool for measuring the antioxidant activity. These free radicals have the ability to oxidize biological molecules such as proteins, lipids, DNA, nucleic acids and tissue damage.160 They can also start degenerative illnesses. Oxidative damage is a major pathogenic factor in a number of human diseases including cancer, cirrhosis, emphysema, atherosclerosis and arthritis.161 Triazole metal complexes act as antioxidant agents through the following mechanism (Fig. 15). | ||
| Fig. 15 Possible mechanisms of the antioxidant activity of metal complexes, adapted from ref. 162–165. | ||
Sumrra and coworkers investigated a new class of potential antimicrobial agents designed to fight the growing threat of bacterial biofilms, and the study meticulously characterized ligands using various techniques. The ligand L41 readily formed complexes (41a–41f) with transition metals, which were then subjected to rigorous analysis including advanced theoretical calculations (Scheme 41). These calculations revealed the ligand's enhanced stability compared to the complexes. The most exciting finding lies in the biological activity. The metallic adducts significantly outperformed the free ligand in the case of antibacterial and antifungal activities, with the complex 41c excelling against E. coli and the complex 41d demonstrating potency against Aspergillus niger. Furthermore, all compounds exhibited promising antioxidant properties. This synergy between theoretical predictions and experimental results suggests that these triazole-based compounds hold immense potential as future antibiotics.166
 | ||
| Scheme 41 Schematic pathway of the ligand and metal complexes 41a–41f, redrawn from ref. 166. | ||
Singh and his coworkers published the synthesis of new transition metal chelates (42a–42f) through attaching metal ions to triazole ligand L42. This ligand was synthesized through condensation of a carbonyl derivative and amine-substituted triazole (Scheme 42). The Schiff base ligand was formed, which was confirmed through a TLC study. Metal ions were added to that ligand and refluxed for 2–3 hours. The structural characteristics of the generated ligand were studied using FT-IR spectroscopy, NMR spectroscopy and elemental studies. All the complexes showed an octahedral geometry except copper scaffolds that presented a tetrahedral structure. The bioactive parameters were studied against different kinds of bacteria and fungi. These studies elaborated that metal coordinates showed increased activity against microorganisms as compared to the parent ligand. The anti-oxidant activity of the complexes was also examined using a 1,1-diphenyl-2-picrylhydrazy assay.167
 | ||
| Scheme 42 Synthetic scheme of ligand L42 and its metallic scaffolds 42a–42f, redrawn from ref. 167. | ||
Dhale et al. (2023) reported a sequence of triazole ligands L43a–L43c and the metal complexes (43a–43f) of divalent nickel and cobalt metals (Scheme 43). The structure and geometry were examined through Fourier transform infrared spectroscopy, nuclear magnetic resonance, mass spectrometry and TGA studies. The above-mentioned techniques elucidated that these complexes have an octahedral geometry. These synthesized compounds displayed greater anti-tubercular activity than the conventional drugs. The antimicrobial activity was tested against G(−ve) E. coli bacteria and G(+ve) S. aureus bacteria. The synthesized complexes also showed antioxidant properties. Biological studies revealed that these complexes can be used as potent drugs.122
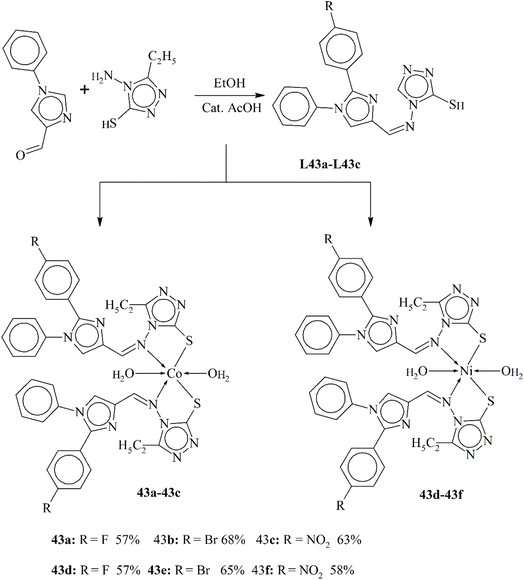 | ||
| Scheme 43 Possible structures of the complexes 43a–43f, redrawn from ref. 122. | ||
In 2020, Sumrra et al. unveiled a new class of potential therapeutic agents, which are a triazole-based ligand L44 and its related metallic scaffolds (44a–44f). The researchers meticulously designed and characterized the ligand, confirming its structure. They then successfully coordinated the ligand with various metal ions, resulting in a series of complexes with specific structures (Scheme 44). Interestingly, advanced calculations revealed the ligand's exceptional stability, a key property for potential drugs. The most exciting finding lies in the biological activity. Both ligand and its metal chelates displayed a range of therapeutic benefits including fighting bacteria and fungi, acting as antioxidants and inhibiting the harmful process of glycation. Notably, these activities were significantly amplified upon complexation with metals, suggesting that metal binding plays a crucial role in boosting the ligand's therapeutic potential.168
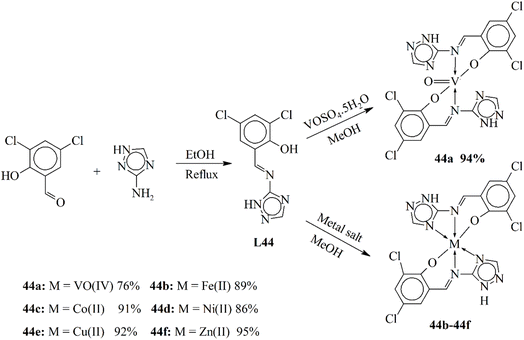 | ||
| Scheme 44 Structure of the ligand and its metal complexes 44a–44f, redrawn from ref. 168. | ||
A series of Fe(III) complexes (45a–45d) were synthesized using ferric nitrate and a Schiff base L45 (Scheme 45). The metallic scaffolds were soluble in both DMF and DMSO, showing low-level molar conductance values implying non-electrolytic behavior. Geometric and spectroscopic analyses using FT-IR as well as 1H and 13C-NMR, electronic spectra and FAB mass spectra revealed that the ligand existed in the tautomeric enol form with intramolecular hydrogen bonding. The complexes were synthesized by refluxing the ligand with Fe(NO3)3·9H2O and sodium acetate, filtering the precipitate, washing and drying. Magnetic and spectral studies indicated an octahedral structure for the complexes. The complexes exhibited antioxidant activity as determined by the FRAP assay, with compounds 45b and 45c showing high activity compared to 45a and 45d.124 Table 4 shows the antioxidant activity of some metal scaffolds.
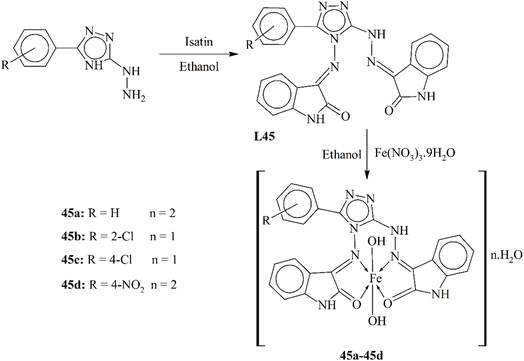 | ||
| Scheme 45 Synthesis and structure of the ligand and metal complexes 45a–45d, redrawn from ref. 124. | ||
| Comp. | Anti-oxidant activity | Ref. | Comp. | Anti-oxidant activity | Ref. |
|---|---|---|---|---|---|
| a IZD = inhibition zone diameter, MIC = minimum inhibitory concentration, PI = percentage inhibition, IC50 = half maximal inhibitory concentration, GI50 = concentration causing 50% cell growth inhibition, FRP = ferric reducing power, TPC = total phenolic content. | |||||
| 35b | PI = 83% (FRP) | 121 | 44c | PI = 17.53% (DPPH), Abs = 0.79 nm (FRP) | 123 |
| 41b | PI = 79% (TPC) | 166 | 44e | Abs = 0.79 nm (TPC) | 123 |
| PI = 72% (DPPH) | |||||
| 42e | PI = 22% (DPPH) | 167 | 45a | PI = 87.42% (RPA) | 124 |
| 43a | PI = 65.55% (DPPH) | 122 | 45b | PI = 82.76% (FRP) | 124 |
4 Core metal triazole systems and their predominant biological activity profiles
For each particular biological activity covered in this review, the activity values of all reported complexes are displayed in the related tables. While the article contains several metal-triazole complexes, the metals included in Table 5 are the main and most significant contributors to the biological patterns that have been identified. In the context of this review, these particular metals need special attention as they offer the most reliable structure–activity connections.| Metal | Representative complex number | Main biological activity |
|---|---|---|
| VO(IV) | 5b, 14b, 14c, 20d, 24e | Antibacterial, cytotoxic, antifungal, enzyme inhibition |
| Fe(II/III) | 11a, 4lb | Antifungal, antioxidant |
| Co(II) | 6f, 8m, 23g, 23h, 25b | Antibacterial, cytotoxic, anti-cancer |
| Ni(II) | 2f, 9h, 28k, 39b | Antibacterial, anticancer, DNA binding |
| Cu(II) | 2g, 32c, 36c | Antibacterial, antifungal, DNA binding |
| Zn(II) | 1f, 2h, 7c, 16j, 32d | Antibacterial, antifungal, enzyme inhibition |
5 Challenges and limitations of metal-triazole complexes
The development of triazole-Schiff base metal complexes as successful therapeutic drugs is hampered by a complex web of scientific and translational challenges, despite their encouraging in vitro bioactivity. A significant physicochemical barrier is their inherent low water solubility, which is directly caused by the hydrophobic and aromatic nature of the organic framework. This greatly lowers bioavailability and necessitates the use of often dangerous organic co-solvents for in vivo research, which complicates the interpretation of pharmacological data.169 Furthermore, even though the dynamic covalent nature of the imine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) link is advantageous for synthesis, it presents a serious risk under physiological conditions (pH ∼7.4), where it is susceptible to hydrolysis by acid catalysis. The intended synergistic effect of the coordination may be negated if the complex prematurely dissociates in vivo, releasing free ligand and metal ions before they reach the cellular target.170 The layer of structure–activity relationship (SAR) is even more complex. Rather than the metal ion or ligand alone, bioactivity is determined by the complex interaction of the triazole substituents, the Schiff base backbone, the coordination geometry of the metal ion, and the total charge of the complex. This makes rational drug design a great challenge because even minor structural changes can result in unexpected and disproportionate changes in target specificity, membrane permeability, and potency.171
N) link is advantageous for synthesis, it presents a serious risk under physiological conditions (pH ∼7.4), where it is susceptible to hydrolysis by acid catalysis. The intended synergistic effect of the coordination may be negated if the complex prematurely dissociates in vivo, releasing free ligand and metal ions before they reach the cellular target.170 The layer of structure–activity relationship (SAR) is even more complex. Rather than the metal ion or ligand alone, bioactivity is determined by the complex interaction of the triazole substituents, the Schiff base backbone, the coordination geometry of the metal ion, and the total charge of the complex. This makes rational drug design a great challenge because even minor structural changes can result in unexpected and disproportionate changes in target specificity, membrane permeability, and potency.171
Perhaps the largest barrier to clinical translation is the complicated issue of toxicity. This covers a variety of distinct but related problems:
5.1 Inherent metal toxicity
Bioactive metal ions like Cu(II), Ni(II), and Co(II) can have two sides. They can catalyze the Fenton and Haber–Weiss reactions, which result in cytotoxic reactive oxygen species (ROS) that cause non-selective damage to lipids, proteins, and DNA, causing off-target toxicity in healthy tissues. Even essential metals like copper and zinc require homeostasis, and exogenous delivery can disrupt intricate metal-ion regulation circuits, leading to systemic toxicity.1725.2 Ligand-mediated toxicity
The biological effects of complex breakup on the Schiff base ligand are rarely investigated. Unusual toxicity that cannot be connected to the metal complex may result from these organic compounds' production of reactive metabolites or their own unidentified toxicophores.5.3 Narrow therapeutic window
Despite the fact that a number of studies demonstrate potent in vitro cytotoxicity against cancer cells, they are unable to demonstrate a significant selectivity index over non-malignant cell lines (like HEK-293 and MRC-5). This lack of selective toxicity indicates a widespread rather than a targeted cytotoxic effect, which poses a serious risk to therapeutic use.173Triazole-metal complex limitations can be reduced by modifying ligand structures to increase stability and solubility.67 Nanocarrier-based delivery methods improve bioavailability and lower toxicity.174 Computational optimization can be used to further modify metal–ligand geometry and pharmacokinetic behavior.175 Furthermore, off-target toxicity is decreased without compromising activity by employing biocompatible metals such as Zn(II) or Fe(III).176
6 Conclusion
Triazole Schiff bases based on metals have become a very promising family of molecules with a wide range of powerful biological activities. Their increased pharmacological characteristics are largely due to their unique structural framework, which is the result of the fusion of triazole and Schiff base moieties with transition metals. Their diverse activities as anticancer, antibacterial, antifungal, cytotoxic, DNA-binding, enzyme-inhibitory and antioxidant compounds have been emphasized in the review, illustrating the wide range of their therapeutic potential. The coordination of metals with triazole Schiff bases modifies the complexes' interactions with biological targets such DNA, enzymes, and cellular membranes in addition to increasing the complexes' stability and lipophilicity. The capacity of certain metal complexes to suppress harmful bacterial and fungal strains, trigger apoptosis in cancer cells, and stop oxidative stress all of which are essential in contemporary medication development is especially noteworthy. More thorough research is necessary to completely determine their pharmacokinetic characteristics, mechanism of action, and safety profiles, even with the encouraging in vitro and in vivo outcomes.7 Future prospects
Although metal-based triazole Schiff bases have demonstrated robust biological activity, they have not yet attained their full potential. To reduce the side effects, future research should focus on developing compounds showing greater selectivity toward certain targets including tumor cells or resistant microbes. These complexes may provide better therapeutic results when combined with conventional medications, especially when there is drug resistance. Their stability, controlled release and transport to certain tissues can all be enhanced by incorporating them into nanocarriers such as polymeric nanoparticles or liposomes. To fully comprehend their precise modes of action, including how they interact with enzymes, DNA or biological pathways, more research is required. This can be supported by computational methods that anticipate how these chemicals attach to biological targets, such as dynamics simulations and molecular docking. The assessment of biological screening's wider applicability can be made easier by adding resistant bacterial strains, virus models or neglected illnesses.Author contributions
Kashif Haseeb: writing – original draft; Muhammad Hasnain Mustafa: writing – original draft; Wardha Zafar: writing – review & editing, visualization; Abrar Ul Hassan: data curation; Zahid Hussain Chohan: validation; Sajjad Hussain Sumrra: conceptualization, supervision, project administration.Conflicts of interest
The authors declare no conflict of interest regarding the publication of this review paper.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.References
- T. Pulingam, T. Parumasivam, A. M. Gazzali, A. M. Sulaiman, J. Y. Chee, M. Lakshmanan, C. F. Chin and K. Sudesh, Eur. J. Pharm. Sci., 2022, 170, 106103 CrossRef CAS PubMed.
- A. Kapoor, S. B. Mudaliar, V. G. Bhat, I. Chakraborty, A. S. B. Prasad and N. Mazumder, 3 Biotech, 2024, 14, 256 CrossRef.
- C. L. Ventola, Pharm. Ther., 2015, 40, 277 Search PubMed.
- F. Prestinaci, P. Pezzotti and A. Pantosti, Pathog. Global Health, 2015, 109, 309–318 CrossRef.
- M. S. Butler, V. Gigante, H. Sati, S. Paulin, L. Al-Sulaiman, J. H. Rex, P. Fernandes, C. A. Arias, M. Paul and G. E. Thwaites, Antimicrob. Agents Chemother., 2022, 66, 1–20 CAS.
- B. J. Langford, M. So, M. Simeonova, V. Leung, J. Lo, T. Kan, S. Raybardhan, M. E. Sapin, K. Mponponsuo and A. Farrell, Lancet Microbe, 2023, 4, 179–191 CrossRef.
- T. U. Berendonk, C. M. Manaia, C. Merlin, D. Fatta-Kassinos, E. Cytryn, F. Walsh, H. Bürgmann, H. Sørum, M. Norström and M. N. Pons, Nat. Rev. Microbiol., 2015, 13, 310–317 CrossRef CAS.
- C. Llor and L. Bjerrum, Ther. Adv. Drug Saf., 2014, 5, 229–241 CrossRef PubMed.
- T. P. Boeckel, E. E. Glennon, D. Chen, M. Gilbert, T. P. Robinson, B. T. Grenfell, S. A. Levin, S. Bonhoeffer and R. Laxminarayan, Science, 2017, 357, 1350–1352 Search PubMed.
- P. W. Schreiber, H. Sax, A. Wolfensberger, L. Clack and S. P. Kuster, Infect. Control Hosp. Epidemiol., 2018, 39, 1277–1295 CrossRef PubMed.
- E. Tacconelli, E. Carrara, A. Savoldi, S. Harbarth, M. Mendelson, D. L. Monnet, C. Pulcini, G. Kahlmeter, J. Kluytmans and Y. Carmeli, Lancet Infect. Dis., 2018, 18, 318–327 CrossRef.
- A. Frei, A. D. Verderosa, A. G. Elliott, J. Zuegg and M. A. Blaskovich, Nat. Rev. Chem., 2023, 7, 202–224 Search PubMed.
- E. Altun, M. O. Aydogdu, E. Chung, G. Ren, S. Homer-Vanniasinkam and M. Edirisinghe, Appl. Phys. Rev., 2021, 8, 041303 Search PubMed.
- A. Lance-Byrne, B. Lindquist-Kleissler and T. C. Johnstone, Eur. J. Inorg. Chem., 2024, 27, e202300717 Search PubMed.
- A. Kumar Singh, A. Kumar, H. Singh, P. Sonawane, P. Pathak, M. Grishina, J. Pal Yadav, A. Verma and P. Kumar, Chem. Biodiversity, 2023, 20, 202300061 CrossRef.
- M. Claudel, J. V. Schwarte and K. M. Fromm, Chemistry, 2020, 2, 849–899 CrossRef CAS.
- J. E. Waters, L. Stevens-Cullinane, L. Siebenmann and J. Hess, Curr. Opin. Microbiol., 2023, 75, 102347 CrossRef CAS.
- I. Kostova, Inorganics, 2023, 11, 56 Search PubMed.
- A. Wang, M. Walden, R. Ettlinger, F. Kiessling, J. J. Gassensmith, T. Lammers, S. Wuttke and Q. Peña, Adv. Funct. Mater., 2023, 2308589 Search PubMed.
- C. N. Njoku, T. U. Maduoma, W. Emori, R. E. Odey, B. M. Unimke, E. Yakubu, C. C. Anorondu, D. I. Udunwa, O. C. Njoku and K. B. Oyoh, Natural and synthetic drugs as eco-friendly and sustainable corrosion inhibitors for metals: a review, Pigm. Resin Technol., 2023, 1074–1087 Search PubMed.
- W. Zafar, M. Ashfaq and S. H. Sumrra, J. Mol. Struct., 2023, 135744 Search PubMed.
- M. S. Capper, H. Packman and M. Rehkämper, ChemBioChem, 2020, 21, 2111–2115 CrossRef CAS.
- K. H. Thompson and C. Orvig, Dalton Trans., 2006, 6, 761–764 RSC.
- E. Sánchez-López, D. Gomes, G. Esteruelas, L. Bonilla, A. L. Lopez-Machado, R. Galindo, A. Cano, M. Espina, M. Ettcheto and A. Camins, Nanomaterials, 2020, 10, 292 CrossRef PubMed.
- A. Evans and K. A. Kavanagh, J. Med. Microbiol., 2021, 70, 001363 CrossRef CAS PubMed.
- H. A. Qasem, Arabian J. Chem., 2025, 18, 1622024 CrossRef.
- A. Aljuhani, M. S. Nafie, N. R. Albujuq, M. Alsehli, S. K. Bardaweel, K. M. Darwish, S. Y. Alraqa, M. R. Aouad and N. Rezki, RSC Adv., 2025, 15, 3570–3591 RSC.
- W. Stroek and M. Albrecht, Chem. Soc. Rev., 2024, 53, 6322–6344 RSC.
- X. Li, D. Zhang, Z. Liu, Y. Xu and D. Wang, Inorg. Chim. Acta, 2017, 471, 280–289 CrossRef.
- X. Li, X. Chen, J.-W. Yuan, Y. Liu, P. Li, L. Qu and Y. Zhao, Phosphorus Sulfur Silicon Relat. Elem., 2014, 190, 277–291 Search PubMed.
- N. Behera, T. Behera, J. Rout and S. Moharana, Significant Aspects of Heterocyclic Schiff Bases and Their Metal Complexes, 2024 Search PubMed.
- D. Chaturvedi and M. Kamboj, J. Chem. Sci., 2016, 7, 114 Search PubMed.
- E. Raczuk, B. Dmochowska, J. Samaszko-Fiertek and J. Madaj, Molecules, 2022, 27, 787 CrossRef CAS PubMed.
- M. S. Hossain, P. K. Roy, C. Zakaria and M. Kudrat-E-Zahan, Int. J. Chem. Stud, 2018, 6, 19–31 CAS.
- M. A. Ashraf, K. Mahmood, A. Wajid, M. J. Maah and I. Yusoff, Int. Proc. Chem., Biol. Environ. Eng., 2011, 10, 185 Search PubMed.
- N. K. Bhattacharyya, D. Dutta and J. Biswas, Indian J. Chem., Sect. B: Org. Chem. Incl. Med. Chem., 2021, 60, 1478–1489 Search PubMed.
- A. A. Alezzy, H. Alnahari and S. A. Al-horibi, J. Chem. Nutr. Biochem., 2022, 3, 44–57 Search PubMed.
- P. Ghanghas, A. Choudhary, D. Kumar and K. Poonia, Inorg. Chem. Commun., 2021, 130, 108710 CrossRef CAS.
- Z. Zhang, Q. Song, Y. Jin, Y. Feng, J. Li and K. Zhang, Metals, 2023, 13, 386 CrossRef CAS.
- S. Arulmurugan, H. P. Kavitha and B. Venkatraman, Rasayan J. Chem., 2010, 3, 385–410 Search PubMed.
- N. R. Joshi, S. G. Mule, V. A. Gore, R. D. Suryawanshi, G. T. Pawar, S. R. Bembalkar and R. P. Pawar, J. Explor. Res. Pharmacol., 2022, 7, 202–207 Search PubMed.
- J. Dai, S. Tian, X. Yang and Z. Liu, Front. Chem., 2022, 10, 891484 CrossRef CAS.
- R. Çakmak, B. Ay, E. Çınar, E. Başaran, S. Akkoç, M. Boğa and E. Taş, J. Mol. Struct., 2023, 1292, 136225 CrossRef.
- L. Lv, T. Zheng, L. Tang, Z. Wang and W. Liu, Coord. Chem. Rev., 2025, 525, 216327 CrossRef CAS.
- T. M. Dhanya, K. J. Rajimon, N. K. Mohanan, A. Anilkumar, S. S. Pillai and P. V. Mohanan, Discov. Chem., 2024, 1, 48 CrossRef.
- E. Basaran, H. G. Sogukomerogullari, R. Cakmak, S. Akkoc, T. Taskin-Tok and A. Köse, Bioorg. Chem., 2022, 129, 106176 CrossRef CAS.
- S. Bhandarkar, P. Pathare and B. Khobragade, Mater. Today: Proc., 2023, 92, 807–816 CAS.
- M. V. Gil, M. J. Arévalo and O. Lopez, Synthesis, 2007, 2007, 1589–1620 CrossRef.
- R. Kharb, P. C. Sharma and M. S. Yar, J. Enzyme Inhib. Med. Chem., 2011, 26, 1–21 CrossRef CAS.
- A. Kashyap and O. Silakari, Triazoles: multidimensional 5-membered mucleus for designing multitargeting agents, in Key Heterocycle Cores for Designing Multitargeting Molecules, Elsevier, 2018, pp. 323–342 Search PubMed.
- N. Sahu, J. K. Sahu and A. Kaushik, Curr. Res. Pharm. Sci., 2013, 108–113 Search PubMed.
- B. S. Creaven, M. Devereux, A. Foltyn, S. McClean, G. Rosair, V. R. Thangella and M. Walsh, Polyhedron, 2010, 29, 813–822 CrossRef CAS.
- L. Todorov and I. Kostova, Front. Chem., 2023, 11, 1247805 CrossRef CAS.
- I. Pibiri and S. Buscemi, Curr. Bioact. Compd., 2010, 6, 208–242 CrossRef CAS.
- S. M. A. Rahman, J. S. Bhatti, S. Thareja and V. Monga, Eur. J. Med. Chem., 2023, 5, 115699 CrossRef.
- L. da S. M. Forezi, C. G. S. Lima, A. A. P. Amaral, P. G. Ferreira, M. C. B. V. de Souza, F. de C. da Silva and V. F. Ferreira, Chem. Rec., 2021, 21, 2782–2807 CrossRef.
- S. H. Sumrra, U. Habiba, W. Zafar, M. Imran and Z. H. Chohan, J. Coord. Chem., 2020, 73, 2838–2877 CrossRef CAS.
- E. Bonandi, M. S. Christodoulou, G. Fumagalli, D. Perdicchia, G. Rastelli and D. Passarella, Drug Discov. Today., 2017, 22, 1572–1581 CrossRef CAS PubMed.
- M. Xu, Y. Peng, L. Zhu, S. Wang, J. Ji and K. Rakesh, Eur. J. M. Chem., 2019, 180, 656–672 CrossRef CAS.
- T. Akhtar, W. Zafar and S. Sumrra, South. J. Res., 2022, 2, 189–203 Search PubMed.
- S. Sathish Kumar and P. Kavitha, Mini-Rev. Org. Chem., 2013, 10, 40–65 CrossRef.
- H. Zhou and C. Y. Wang, Curr. Med. Chem., 2012, 19, 239–280 CrossRef.
- J. Khan, A. Rani, M. Aslam, R. Maharia, G. Pandey and B. Nand, FDA Approvals, and Clinical Trial Updates-A Comprehensive Review, Tetrahedron, 2024, 162, 134122 CrossRef CAS.
- F. Bongomin, S. Gago, R. O. Oladele and D. W. Denning, J. Fungi, 2017, 3, 57 CrossRef PubMed.
- T. R. Arun, H. P. Kumar and S. Kamalesu, Inorg. Chem. Commun., 2024, 170, 113251 CrossRef.
- C. Boulechfar, H. Ferkous, A. Delimi, A. Djedouani, A. Kahlouche, A. Boublia, A. S. Darwish, T. Lemaoui, R. Verma and Y. Benguerba, Inorg. Chem. Commun., 2023, 150, 110451 CrossRef CAS.
- A. Kumar, S. Devi, S. Kumar and K. Lal, Coord. Chem. Rev., 2025, 536, 216675 CrossRef CAS.
- S. Thakur, A. Jaryal and A. Bhalla, Results Chem., 2024, 7, 101350 CrossRef CAS.
- V. K. Juyal, A. Pathak, M. Panwar, S. C. Thakuri, O. Prakash, A. Agrwal and V. Nand, J. Organomet. Chem., 2023, 999, 122825 CrossRef CAS.
- R. Khandelwal, M. Vasava, R. Abhirami and M. Karsharma, Bioorg. Med. Chem. Lett., 2024, 15, 129927 CrossRef.
- M. M. Matin, P. Matin, M. R. Rahman, T. Ben Hadda, F. A. Almalki, S. Mahmud and S. Alshehri, Front. Mol. Biosci., 2022, 9, 864286 CrossRef CAS.
- M. U. Rehman and S. Long, Future Med. Chem., 2024, 16, 2451–2453 CrossRef CAS.
- C. A. Juan, J. M. Pérez de la Lastra, F. J. Plou and E. Pérez-Lebeña, Int. J. Mol. Sci., 2021, 22, 4642 CrossRef CAS.
- B. Hu, H. Zhao, Z. Chen, C. Xu, J. Zhao and W. Zhao, Molecules, 2018, 23, 709 CrossRef PubMed.
- M. Arshad, A. Rashid, F. Mahmood, S. Saeed and A. Ahmed, Eur. J. Chem., 2023, 14, 155–164 CrossRef CAS.
- S. Hong, H. Lu, D. Tian, Y. Chang, Q. Lu and F. Gao, Front. Chem., 2025, 13, 1545259 CrossRef CAS.
- S. de Oliveira Pinheiro, J. L. Braga, W. M. B. da Silva, G. H. M. Do Nascimento, R. A. Montes, D. R. Alves, D. S. Rodrigues, A. C. Leitão, V. P. de Farias Cabral and L. E. A. Moreira, Future Med. Chem., 2025, 17, 767–778 CrossRef CAS PubMed.
- S. H. Sumrra, S. Kausar, M. A. Raza, M. Zubair, M. N. Zafar, M. A. Nadeem, E. U. Mughal, Z. H. Chohan, F. Mushtaq and U. Rashid, J. Mol. Struct., 2018, 1168, 202–211 CrossRef CAS.
- P. Mahadevi, S. Sumathi, S. Dasgupta and A. Noor, Inorg. Chem. Commun., 2024, 167, 112800 Search PubMed.
- J. Soni, S. Sinha and R. Pandey, Front. Microbiol., 2024, 15, 1370818 Search PubMed.
- P. Biegański, Ł. Szczupak, M. Arruebo and K. Kowalski, RSC Chem. Biol., 2021, 2, 368–386 Search PubMed.
- D. G. Moriel, D. Piccioli, M. M. Raso and M. Pizza, Semin. Immunopathol., 2024, 45, 481–491 CrossRef PubMed.
- W. Zafar, S. H. Sumrra and Z. H. Chohan, Eur. J. Med. Chem., 2021, 222, 113602 CrossRef CAS PubMed.
- R. A. M. Al Hassani, A. A. Balakit and H. Khuder, Revista Bionatura, 2023, 8, 63 Search PubMed.
- S. Sumrra, S. Ramzan, G. Mustafa, M. Ibrahim, E. Mughal, M. Nadeem, Z. Chohan and M. Khalid, Russ. J. Gen. Chem., 2018, 88, 1707–1711 Search PubMed.
- Q. Ain, S. Pandey, O. Pandey and S. Sengupta, Spectrochim. Acta, Part A, 2015, 140, 27–34 CrossRef CAS.
- S. A. Patil, M. Manjunatha, A. D. Kulkarni and P. S. Badami, Complex Met., 2014, 1, 128–137 CrossRef.
- B. P. Sharma, J. A. Subin, B. P. Marasini, R. Adhikari, S. K. Pandey and M. L. Sharma, Heliyon, 2023, 9, e15239 CrossRef CAS.
- K. Singh and P. Siwach, Chem. Biol. Lett., 2022, 9, 407 Search PubMed.
- Z. H. Chohan and M. Hanif, Phenol, Appl. Organomet. Chem., 2011, 2, 4 Search PubMed.
- S. Sumrra, A. Suleman, Z. Chohan, M. Zafar, M. Raza and T. Iqbal, Russ. J. Gen. Chem., 2017, 87, 1281–1287 Search PubMed.
- Z. H. Chohan and M. Hanif, J. Enzyme Inhib. Med. Chem., 2013, 28, 944–953 CrossRef CAS.
- M. N. Al-jibouri, W. A. Jawad, A. A. Balakit and M. Obies, Egypt. J. Chem., 2021, 64, 5227–5239 Search PubMed.
- N. F. Mahmoud, A. A. Abbas and G. G. Mohamed, Appl. Organomet. Chem., 2021, 35, 6219 Search PubMed.
- S. P. Bhale, A. R. Yadav, P. G. Pathare, S. U. Tekale, F. P. Franguelli, L. Kótai and R. P. Pawar, Eur. Chem. Bull., 2020, 9, 4 Search PubMed.
- Z. H. Chohan, S. H. Sumrra, M. H. Youssoufi and T. B. Hadda, Eur. J. Med. Chem., 2010, 45, 2739–2747 Search PubMed.
- W. Fang, J. Wu, M. Cheng, X. Zhu, M. Du, C. Chen, W. Liao, K. Zhi and W. Pan, J. Biomed. Sci., 2023, 30, 42 CrossRef CAS PubMed.
- J. Houšť, J. Spížek and V. Havlíček, Metabolites, 2020, 10, 106 CrossRef PubMed.
- Y. Lin, H. Betts, S. Keller, K. Cariou and G. Gasser, Chem. Soc. Rev., 2021, 50, 10346–10402 RSC.
- K. Bajaj, R. M. Buchanan and C. A. Grapperhaus, J. Inorg. Biochem., 2021, 225, 111620 CrossRef CAS PubMed.
- N. Raman, J. Joseph, A. S. Kumara Velan and C. Pothiraj, Mycobiology, 2006, 34, 214–218 CrossRef CAS.
- S. Gopalakrishnan and J. Joseph, Mycobiology, 2009, 37, 141–146 CrossRef CAS.
- A. Frei, A. G. Elliott, A. Kan, H. Dinh, S. Bräse, A. E. Bruce, M. R. Bruce, F. Chen, D. Humaidy and N. Jung, JACS Au, 2022, 2, 2277–2294 CrossRef CAS PubMed.
- Z. H. Chohan and S. H. Sumrra, J. Enzyme Inhib. Med. Chem., 2012, 27, 187–193 CrossRef CAS PubMed.
- M. Sahani, S. Pandey, O. Pandey and S. Sengupta, J. Mol. Struct., 2014, 1074, 401–407 CrossRef CAS.
- M. Hanif and Z. H. Chohan, Appl. Organomet. Chem., 2013, 27, 36–44 CrossRef CAS.
- A. Kumar, V. Kumar, P. Kumar, P. K. Yadav and L. K. Mishra, Int. J. Res. Appl. Sci. Eng. Technol., 2023, 11, 1585–1590 CrossRef.
- R. Joshi, A. Kumari, K. Singh, H. Mishra and S. Pokharia, J. Mol. Struct., 2020, 1206, 127639 CrossRef CAS.
- S. H. Sumrra, I. Sahrish, M. A. Raza, Z. Ahmad, M. N. Zafar, Z. H. Chohan, M. Khalid and S. Ahmed, Monatsh. Chem., 2020, 151, 549–557 CrossRef CAS.
- K. Munawar, S. Ali, M. Tahir, N. Khalid, Q. Abbas, I. Qureshi and S. Shahzadi, Russ. J. Gen. Chem., 2015, 85, 2183–2197 Search PubMed.
- M. Hanif and Z. H. Chohan, Spectrochim. Acta, Part A, 2013, 104, 468–476 Search PubMed.
- S. H. Sumrra and Z. H. Chohan, J. Enzyme Inhib. Med. Chem., 2013, 28, 1291–1299 Search PubMed.
- Z. H. Chohan and M. Hanif, J. Enzyme Inhib. Med. Chem., 2010, 25, 737–749 Search PubMed.
- Z. H. Chohan and S. H. Sumrra, J. Enzyme Inhib. Med. Chem., 2010, 25, 599–607 CrossRef CAS PubMed.
- S. A. Deodware, U. B. Barache, U. B. Chanshetti, D. Sathe, U. P. Ashok, S. H. Gaikwad and S. P. Kollur, Results Chem., 2021, 3, 100162 CrossRef CAS.
- R. G. Deghadi, A. A. Abbas and G. G. Mohamed, Appl. Organomet. Chem., 2021, 35, 6292 Search PubMed.
- M. Gaber, H. El-Ghamry and S. Fathalla, Spectrochim. Acta, Part A, 2015, 139, 396–404 CrossRef CAS.
- S. H. Sumrra, M. Hanif and Z. H. Chohan, J. Enzyme Inhib. Med. Chem., 2015, 30, 800–808 CrossRef CAS.
- L. Calu, M. Badea, M. C. Chifiriuc, C. Bleotu, G. I. David, G. Ioniţă, L. Măruţescu, V. Lazăr, N. Stanică and I. Soponaru, J. Therm. Anal. Calorim., 2015, 120, 375–386 CrossRef CAS.
- A. Hassan, B. H. Heakal, A. Younis, M. A. E. M. Bedair and M. M. A. Mohamed, Egypt. J. Chem., 2019, 62, 1603–1624 Search PubMed.
- T. Sangeetha and S. Mohanapriya, Indian J. Chem., Sect. A, 2021, 60, 797–805 Search PubMed.
- P. C. Dhale, P. A. Ubale, K. D. Sonawane, N. M. Naik, M. Afzal, L. A. Ghule, S. A. Deodware, K. D. Gaikwad, U. B. Barache and S. H. Gaikwad, Results Chem., 2023, 6, 101155 Search PubMed.
- S. H. Sumrra, M. Anees, A. Asif, M. N. Zafar, K. Mahmood, M. F. Nazar, M. Khalid, M. A. Nadeem and M. U. Khan, Bull. Chem. Soc. Ethiop., 2020, 34, 335–351 CrossRef CAS.
- G. Kharadi, Spectrochim. Acta, Part A, 2013, 110, 311–316 Search PubMed.
- S. K. Raju, A. Karunakaran, S. Kumar, P. Sekar, M. Murugesan and M. Karthikeyan, Ger. J. Pharm. Biomater., 2022, 1, 6–28 Search PubMed.
- S. Kowalski, D. Wyrzykowski and I. Inkielewicz-Stępniak, Molecules, 2020, 25, 1757 Search PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef.
- S. W. D. Merriel, S. M. Ingle, M. T. May and R. M. Martin, BMJ Open, 2021, 11, 044420 CrossRef.
- D. T. Debela, S. G. Muzazu, K. D. Heraro, M. T. Ndalama, B. W. Mesele, D. C. Haile, S. K. Kitui and T. Manyazewal, SAGE Open Med., 2021, 9, 20503121211034366 CrossRef.
- I. Romero-Canelon and P. J. Sadler, Inorg. Chem., 2013, 52, 12276–12291 CrossRef CAS.
- A. G. Weidmann, A. C. Komor and J. K. Barton, Comments Inorg. Chem., 2014, 34, 114–123 CrossRef CAS PubMed.
- P. Jia, R. Ouyang, P. Cao, X. Tong, X. Zhou, T. Lei, Y. Zhao, N. Guo, H. Chang and Y. Miao, J. Coord. Chem., 2017, 70, 2175–2201 CrossRef CAS.
- U. Jungwirth, C. R. Kowol, B. K. Keppler, C. G. Hartinger, W. Berger and P. Heffeter, Antioxid. Redox Signaling, 2011, 15, 1085–1127 Search PubMed.
- D. Chen, V. Milacic, M. Frezza and Q. P. Dou, Curr. Pharm. Des., 2009, 15, 777–791 Search PubMed.
- K. Gaur, A. M. Vázquez-Salgado, G. Duran-Camacho, I. Dominguez-Martinez, J. A. Benjamín-Rivera, L. Fernández-Vega, L. Carmona Sarabia, A. Cruz García, F. Pérez-Deliz and J. A. Méndez Román, Inorganics, 2018, 6, 126 CrossRef CAS PubMed.
- L. A. Alfonso-Herrera, D. Hernández-Romero, J. A. Cruz-Navarro, Á. Ramos-Ligonio, A. López-Monteon, J. M. Rivera-Villanueva, D. Morales-Morales and R. Colorado-Peralta, Coord. Chem. Rev., 2024, 505, 215698 Search PubMed.
- P. Tyagi, S. Chandra, B. Saraswat and D. Yadav, Spectrochim. Acta - A: Mol. Biomol. Spectrosc., 2015, 145, 155–164 CrossRef CAS.
- P. Tyagi, M. Tyagi, S. Agrawal, S. Chandra, H. Ojha and M. Pathak, Spectrochim. Acta, Part A, 2017, 171, 246–257 Search PubMed.
- E. A. Eno, E. E. Okon, H. Louis, N. U. Obot, S. A. Adalikwu, T. O. Unimuke, T. E. Gber, U. G. Chukwu, P. Ekoja and J. Ogar, Vietnam J. Chem., 2023, 61, 187–203 CrossRef CAS.
- P. Ponya Utthra, G. Kumaravel, R. Senthilkumar and N. Raman, Appl. Organomet. Chem., 2017, 31, e3629 CrossRef.
- K. J. Kilpin and P. J. Dyson, Chem. Sci., 2013, 4, 1410–1419 Search PubMed.
- T. Ezedom and S. Asagba, Toxicol Rep, 2016, 25, 708–715 CrossRef PubMed.
- B. Şahin and S. Çenesiz, SU URUNLERI DERGISI, J. Fish. Aquat. Sci., 2021, 38, 479–48638 Search PubMed.
- Â. de Fátima, C. de Paula Pereira, C. R. S. D. G. Olímpio, B. G. de Freitas Oliveira, L. L. Franco and P. H. C. da Silva, J. Adv. Res., 2018, 13, 113–126 Search PubMed.
- M. Sohrabi, M. R. Binaeizadeh, A. Iraji, B. Larijani, M. Saeedi and M. Mahdavi, RSC Adv., 2022, 12, 12011–12052 RSC.
- H. Vinusha, S. P. Kollur, R. Ramu, P. S. Shirahatti, N. Prasad and M. Begum, Lett. Appl. NanoBioScience, 2020, 9, 1372–1388 Search PubMed.
- N. A. Al-Masoudi, N. M. Aziz and A. T. Mohammed, Phosphorus, Sulfur Silicon Relat. Elem., 2009, 184, 2891–2901 Search PubMed.
- N. Kumar, R. Kaushal and P. Awasthi, J. Mol. Struct., 2023, 1288, 135751 Search PubMed.
- H. K. Liu and P. J. Sadler, Acc. Chem. Res., 2011, 44, 349–359 Search PubMed.
- H. L. Lu, J. J. Liang, Z. Z. Zeng, P. X. Xi, X. H. Liu, F. J. Chen and Z. H. Xu, Transition Met. Chem., 2007, 32, 564–569 CrossRef CAS.
- K. Cheng, J. J. Yan and Y. Qiu, Asian J. Chem., 2013, 25, 1152 Search PubMed.
- R. B. Dixit, T. S. Patel, S. F. Vanparia, A. P. Kunjadiya, H. R. Keharia and B. C. Dixit, Sci. Pharm., 2011, 79, 293 CrossRef CAS PubMed.
- L. Andrezálová and Z. Országhová, J. Inorg. Biochem., 2021, 225, 111624 Search PubMed.
- F. M. Ibrahim and S. M. Abdalhadi, Al-Nahrain J. Sci., 2021, 24, 1–10 CrossRef.
- M. Bheemarasetti, K. Palakuri, S. Raj, P. Saudagar, D. Gandamalla, N. R. Yellu and L. R. Kotha, J. Iran. Chem. Soc., 2018, 15, 1377–1389 CrossRef CAS.
- A. D. Kulkarni, S. A. Patil, V. H. Naik and P. S. Badami, Med. Chem. Res., 2011, 20, 346–354 CrossRef CAS.
- M. Chaurasia, D. Tomar and S. Chandra, J. Mol. Struct., 2019, 1179, 431–442 CrossRef CAS.
- G. B. Bagihalli and S. A. Patil, J. Coord. Chem., 2009, 62, 1690–1700 CrossRef CAS.
- S. Ivanova, S. Balkanski, P. Atanasov, M. Chaneva, D. Obreshkova, V. Dimitrov, K. Kazalukova, L. Peikova and O. Markov, Pharmacia, 2023, 70, 375–382 CrossRef CAS.
- H. M. A. El-Lateef, T. El-Dabea, M. M. Khalaf and A. M. Abu-Dief, Antioxidants, 2023, 12, 213 CrossRef CAS PubMed.
- A. Choudhary, R. Sharma, M. Nagar, M. Mohsin and H. S. Meena, J. Chil. Chem. Soc., 2011, 56, 911–917 CrossRef CAS.
- N. R. Perron and J. L. Brumaghim, Cell Biochem. Biophys., 2009, 53, 75–100 CrossRef CAS PubMed.
- J. Nagaj, K. Stokowa-Sołtys, I. Zawisza, M. Jeżowska-Bojczuk, A. Bonna and W. Bal, J. Inorg. Biochem., 2013, 119, 85–89 CrossRef CAS.
- E. Rodríguez-Arce and M. Saldías, Biomed. Pharmacother., 2021, 143, 112236 Search PubMed.
- V. A. Timoshnikov, O. Y. Selyutina, N. E. Polyakov, V. Didichenko and G. J. Kontoghiorghes, Int. J. Mol. Sci., 2022, 23, 1247 CrossRef CAS PubMed.
- S. H. Sumrra, W. Zafar, H. Javed, M. Zafar, M. Z. Hussain, M. Imran and M. A. Nadeem, Biometals, 2021, 34, 1329–1351 CrossRef CAS PubMed.
- K. Singh, B. Kumari and A. Sharma, Spectrosc. Lett., 2021, 54, 742–762 CrossRef CAS.
- R. Hussain, S. L. Rubab, A. Maryam, T. Ashraf, M. Arshad, K. Lal, S. H. Sumrra, S. Ashraf and B. Ali, Co(II), and Ni(II) complexes: experimental and theoretical studies, ACS Omega, 2023, 8, 42598–42609 CrossRef CAS.
- S. P. Vaidya, S. Gadre, R. T. Kamisetti and M. Patra, Biosci. Rep., 2022, 42, BSR20212160 CrossRef CAS.
- A. Bhatnagar, R. P. Pakhariya and G. Pemawat, Top. Curr. Chem., 2025, 383, 27 CrossRef CAS.
- N. Naeem, E. U. Mughal, A. Sadiq, G. A. Othman and B. Shakoor, Arch. Pharm., 2025, 358, e70059 CrossRef CAS PubMed.
- R. kamoon, A. Talib, Z. Sadiq and S. Abdulhadi, Sci. Rev. Chem. Commun., 2025, 7, 1–24 CAS.
- E. J. Anthony, E. M. Bolitho, H. E. Bridgewater, O. W. L. Carter, J. M. Donnelly, C. Imberti, E. C. Lant, F. Lermyte, R. J. Needham, M. Palau, P. J. Sadler, H. Shi, F. X. Wang, W. Y. Zhang and Z. Zhang, Chem Sci, 2020, 11, 12888–12917 RSC.
- K. Singh, J. K. Gupta, G. Lakhchora, D. Jain, A. Bhatt, M. C. Sharma, M. Chaitanya and M. Tabish, Curr. Alzheimer Res., 2025, 22, 327–343 CrossRef CAS PubMed.
- K. El-Baradie, Y. S. El-Sayed, N. El-Wakiel, B. M. Salem and A. El-Nagar, Chem. Biol. Technol. Agric., 2025, 12, 58 CrossRef CAS.
- S. A. Unnisa, R. Ervaguda, A. Rasool, F. Sri and H. Krismastuti, Environ. Rep., 2025, 7, 84–103 CrossRef.
| This journal is © The Royal Society of Chemistry 2026 |














