Open Access Article
Open Access ArticlePiperidine-catalyzed synthesis of (E)-2-cyanoacrylamides: crystallographic, antiproliferative, and docking studies
Camilo Serrano-Sterlinga,
Isabel Iriepa bc,
Mario A. Macías
bc,
Mario A. Macías d,
Juan-Carlos Castillo
d,
Juan-Carlos Castillo *a and
Diana Becerra*a
*a and
Diana Becerra*a
aEscuela de Ciencias Químicas, Universidad Pedagógica y Tecnológica de Colombia, Avenida Central del Norte 39–115, Tunja 150003, Colombia. E-mail: juan.castillo06@uptc.edu.co; diana.becerra08@uptc.edu.co
bDepartamento de Química Orgánica y Química Inorgánica, Universidad de Alcalá, Ctra. Madrid-Barcelona, km. 33, 6, 28871 Madrid, Spain
cInstitute of Chemical Research Andrés M. del Río, Universidad de Alcalá, Alcalá de Henares, 28805 Madrid, Spain
dCristalografía y Química de Materiales, Departamento de Química, Universidad de los Andes, Carrera 1 No. 18A–10, Bogotá 111711, Colombia
First published on 2nd January 2026
Abstract
A piperidine-catalyzed Knoevenagel condensation between (hetero)aromatic aldehydes 1a–i and 2-cyanoacetamide 2a was developed to afford (E)-2-cyano-3-(het)arylacrylamides 3a–i in 40–95% yields under mild and environmentally friendly conditions. The methodology was further extended to other methylene active compounds, including malonamide 2b and ethyl cyanoacetate 2c, providing the corresponding adducts 3m–o in 54–81% yields. The (E)-stereochemistry of 3-arylacrylamide 3i was unambiguously confirmed by single-crystal X-ray diffraction analysis. The antiproliferative activity of compounds 3a–o was evaluated across the NCI-60 human cancer cell line panel. Compounds 3f and 3o exhibited the highest potency against the CAKI-1 renal cancer cell line, with GI50 values of 0.287 µM and 0.336 µM, respectively, while compound 3n showed its strongest activity against the RPMI-8226 melanoma cell line (GI50 = 0.367 µM). These values are comparable to or lower than those of the reference drug Osimertinib (GI50 = 0.343 µM for CAKI-1 and 1.95 µM for RPMI-8226 cell lines). In all active cases, LC50 to GI50 ratios equal to or greater than 100 indicated selective growth inhibition rather than nonspecific cytotoxicity. Compound 3n exhibited a slightly higher cytotoxic response compared with its structural analogues. To rationalize this behavior, toxicity profiling revealed a coordinated activation of Nrf2-mediated oxidative stress, p53-dependent DNA damage response, and androgen receptor (AR-LBD) signaling pathways. Molecular docking studies further demonstrated favorable binding interactions of compounds 3f, 3n, and 3o within the tyrosine kinase domain of the epidermal growth factor receptor (EGFR), with predicted affinities surpassing that of Erlotinib.
1 Introduction
Covalent fragment-based drug discovery has become a central strategy in modern drug development.1–5 Targeted covalent inhibitors (TCIs) combine a noncovalent scaffold, which ensures selective target recognition, with a reactive warhead capable of forming reversible or irreversible covalent bonds with specific amino acid residues.6,7 Approximately 36% of these warheads are designed to target cysteine due to its low abundance in the proteome and high nucleophilic reactivity.8 TICs offer several advantages, including enhanced potency and selectivity, prolonged duration of action, lower dosing frequency, and a lower risk of drug resistance.1,4 However, potential drawbacks include off-target effects, immunogenicity, and dose-limiting systemic toxicity.9,10 Advances in understanding their mechanisms of action and warhead reactivity have driven the development of multiple drug candidates and several FDA-approved therapies.5,11–13Since 2001, the US FDA has approved 45 drugs that form irreversible covalent bonds with their target proteins,14 including 11 tyrosine kinase inhibitors (TKIs) for cancer treatment (Fig. 1).11,12 Of these, 10 feature an acrylamide moiety as the warhead, enabling covalent bond formation with target enzymes via a Michael addition.15 Representative examples include Afatinib,16 Osimertinib,17 Dacomitinib,18 Mobocertinib,19 and Lazertinib,20 which covalently bind to the thiol group of Cys797, thereby inhibiting the mutant epidermal growth factor receptor (EGFR) in non-small cell lung cancer (NSCLC).12,13 Neratinib targets human epidermal growth factor receptor 2 (HER2) via covalent binding to Cys805 and is approved for HER2-positive breast cancer.21
Other clinically relevant TCIs include Ibrutinib, Acalabrutinib, and Zanubrutinib, which inhibit Bruton's Tyrosine Kinase (BTK) and are indicated for mantle cell lymphoma, chronic lymphocytic leukemia (CLL), small lymphocytic lymphoma (SLL), and marginal zone lymphoma.22–24 Futibatinib targets fibroblast growth factor receptors 1–4 (FGFR1–4) in patients with advanced intrahepatic cholangiocarcinoma harboring FGFR2 gene fusions or rearrangements,25 while Ritlecitinib selectively inhibits Janus kinase 3 (JAK3) for the treatment of alopecia areata.26 Finally, Lazertinib is also indicated for locally advanced or metastatic NSCLC harboring an EGFR exon 19 deletion or an exon 21 L858R substitution mutation.12
Based on their α-substituents, acrylamide-based warheads are categorized into α-unsubstituted, α-fluoro, and α-cyano derivatives (Fig. 2a).15 Introduced in the 1990s, α-unsubstituted acrylamides can enhance inhibitor activity and overcome resistance but carry significant risks, including severe off-target effects and irreversible inhibition.15 In contrast, α-cyanoacrylamides offer notable advantages, such reduced reactivity and the ability to form reversible covalent bonds with cysteine, thereby improving target selectivity and minimizing adverse effects.27 In 2022, Owens and colleagues reported PRN473 and PRN1008 as reversible covalent BTK inhibitors, with IC50 values of 1.8 nM and 1.3 nM, respectively (Fig. 2b).28 In 2016, Forster and co-workers described FM381 as a reversible covalent JAK3 inhibitor, exhibiting an IC50 value of 0.154 nM (Fig. 2b).29
 | ||
| Fig. 2 (a) Classification of acrylamide-based warheads, and (b) applications of α-cyanoacrylamides in BTK inhibitors. | ||
Notably, PRN1008 and FM381 form reversible covalent bonds with Cys481 and Cys909, respectively, through their α-cyanoacrylamide warheads. These findings underscore that incorporating a α-cyanoacrylamide-based warhead can facilitate the development of novel TKIs.
Traditional synthetic routes to α-cyanoacrylamides typically involve a Knoevenagel condensation between an aldehyde and 2-cyanoacetamide in the presence of a Brønsted base.30 In recent decades, both homogenous and heterogenous catalytic strategies have been successfully developed to improve this transformation.30,31 Among them, metal-free catalysis has been extensively explored using readily available Brønsted bases such as triethylamine,32 4-methylpiperidine,33 piperazine,33 1-methylpiperazine,34 ammonium acetate,35 and sodium hydroxide.36 The use of piperidine under reflux in ethanol has also been documented.37,38 Moreover, innovative approaches employing 1-methyl-3-butyl imidazolium bicarbonate as an ionic liquid and microwave-assisted heating have emerged as notable alternatives.36,39 In our previous work, we reported the application of biogenic carbonates and functionalized hydrotalcites as efficient heterogenous catalysts for the solvent-free synthesis of α-cyanoacrylamides.40,41 Despite these advances, current methodologies still suffer from the use of hazardous catalysts or solvents, complex catalyst preparation, high catalyst loadings, and labor-intensive purification, which limit scalability and sustainability.
This study overcomes these limitations by developing a water-promoted synthesis of α-cyanoacrylamides via a piperidine-catalyzed Knoevenagel condensation between (het)aromatic aldehydes and 2-cyanoacetamide at room temperature. Subsequent in vitro anticancer assays and molecular docking studies demonstrated their potential as promising EGFR inhibitors in medicinal chemistry.
2 Results and discussion
2.1 Chemistry
We initiated the investigation of an efficient and straightforward approach to synthesize (E)-2-cyano-3-arylacrylamide 3a through a model Knoevenagel reaction between 4-chlorobenzaldehyde 1a and 2-cyanoacetamide 2a (Table 1). Due to the significant acidity of 2-cyanoacetamide (pKa = 2.96 and pKb = 11.0), our study began with the optimization of the base. Initially, we stirred a stoichiometric mixture of 4-chlorobenzaldehyde 1a and 2-cyanoacetamide 2a in ethanol at room temperature for 3 h without adding a base (Entry 1, Table 1). TLC analysis showed no formation of product 3a, highlighting the essential role of a base in this reaction. We then screened several bases using a 10 mol% catalyst loading under the same conditions. Inorganic bases, such as NaOH and KOAc, produced product 3a in 52% and 30% yields, respectively (Entries 2 and 3, Table 1). Additionally, various organic amine bases, including triethylamine (Et3N, pKb = 3.2), piperidine (PP, pKb = 3.0), and 1,8-diazabicyclo(5.4.0)undec-7-ene (DBU, pKb = 2.0), were tested by stirring in ethanol at room temperature for 3 h (Entries 4–6, Table 1). Remarkably, piperidine delivered the most favorable result, yielding 76% of product 3a. We attributed this higher yield to piperidine's balanced nucleophilicity, steric accessibility, and appropriate basicity, which likely contributed to its superior performance compared to Et3N and DBU. Following this, we optimized the reaction conditions by adjusting both the reaction time and solvent. Extending the reaction time to 6 h and using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of EtOH/H2O increased the yield of product 3a to 85% and 89%, respectively (Entries 7 and 8, Table 1). Replacing ethanol with water alone significantly improved the yield of 3a (93%) after stirring in water at room temperature for 6 h, using piperidine (10 mol%) as an organocatalyst (Entry 9, Table 1). However, reducing the catalyst loading to 5 mol% and extending the reaction time to 12 h resulted in lower yields of 3a (Entries 10 and 12, Table 1).
1 mixture of EtOH/H2O increased the yield of product 3a to 85% and 89%, respectively (Entries 7 and 8, Table 1). Replacing ethanol with water alone significantly improved the yield of 3a (93%) after stirring in water at room temperature for 6 h, using piperidine (10 mol%) as an organocatalyst (Entry 9, Table 1). However, reducing the catalyst loading to 5 mol% and extending the reaction time to 12 h resulted in lower yields of 3a (Entries 10 and 12, Table 1).
| Entrya | Base | mol% | Solvent | Time (h) | Yield 3a (%) |
|---|---|---|---|---|---|
a Reaction conditions: 1a (1 mmol), 2a (1 mmol), base (5–10 mol%), solvent (3 mL), room temperature. The precipitate was filtered, washed with a cold EtOH/H2O mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), and dried.b 1 1, v/v), and dried.b 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v). 1 (v/v). |
|||||
| 1 | — | — | EtOH | 3 | — |
| 2 | NaOH | 10 | EtOH | 3 | 52 |
| 3 | KOAc | 10 | EtOH | 3 | 30 |
| 4 | Et3N | 10 | EtOH | 3 | 67 |
| 5 | Piperidine | 10 | EtOH | 3 | 76 |
| 6 | DBU | 10 | EtOH | 3 | 72 |
| 7 | Piperidine | 10 | EtOH | 6 | 85 |
| 8b | Piperidine | 10 | EtOH/H2O | 6 | 89 |
| 9 | Piperidine | 10 | H2O | 6 | 93 |
| 10 | Piperidine | 5 | H2O | 6 | 78 |
| 11 | Piperidine | 10 | H2O | 12 | 91 |
| 12 | Piperidine | 5 | H2O | 12 | 80 |
With the optimized reaction conditions established, the piperidine-catalyzed Knoevenagel condensation was applied to structurally diverse (hetero)aromatic aldehydes 1a–l and 2-cyanoacetamide 2a to synthesize (E)-2-cyano-3-(het)arylacrylamides (Scheme 1). Aromatic aldehydes bearing electron-withdrawing substituents, such as 4-bromo and 4-nitro, furnished compounds 3b and 3c in 87% and 67% yields, respectively, after reaction times of 6 h and 24 h. Likewise, substrates containing 2,4-dichlorophenyl and 4-chloro-3-nitrophenyl moieties provided products 3d and 3e in good yields within 6 h. In contrast, aldehydes bearing electron-donating groups, such as 4-diphenylamino, 4-methyl, 4-methoxy, 4-hydroxyl, and 4-dimethylamino, required longer reaction times (6–24 h) and generally gave lower yields (54–88%) of products 3f–j. This outcome can be attributed to the diminished electrophilicity of the carbonyl carbon, which reduces its susceptibility to nucleophilic attack during the Knoevenagel condensation. Notably, 9-anthraldehyde, a substrate containing an extended polycyclic aromatic system, afforded compound 3k in 85% yield after 24 h, suggesting the tolerance of the reaction toward bulky aromatic frameworks. Finally, the heteroaromatic substrate 5-(hydroxymethyl)furfural delivered compound 3l in 40% yield after 12 h. In all cases, the (E)-2-cyano-3-(het)arylacrylamides were efficiently purified by simple filtration followed by washing with a cold EtOH/H2O mixture. The (E)-stereochemistry of compound 3i was unambiguously confirmed by single-crystal X-ray diffraction analysis.
Subsequently, the piperidine-catalyzed Knoevenagel protocol was extended to other methylene active compounds, such as malonamide 2b and ethyl cyanoacetate 2c (Scheme 2). Preliminary experiments revealed that water was unsuitable as the reaction medium due to the limited solubility of the precursors, which resulted in reduced reactivity. Consequently, ethanol was selected as a sustainable alternative. The condensation of 4-chlorobenzaldehyde 1a with malonamide 2b in ethanol for 6 h furnished compound 3m in 62% yield after simple filtration. Next, the protocol was applied to the less reactive 4-(diphenylamino)benzaldehyde 1f with methylene active compounds 2b and 2c for 24 h, affording products 3n and 3o in 54% and 81% yields, respectively. Due to the high solubility of these products in ethanol, purification was performed by column chromatography on silica gel.
Finally, we performed functionalization reactions to obtain 3-arylacrylamide derivatives, given their relevance in both medicinal chemistry and chemical sensing.42,43 Accordingly, selective O-alkylation of the hydroxyl group in 3-arylacrylamide 3i was performed using propargyl bromide 4a and cesium carbonate in DMF at room temperature for 3 h, affording the terminal alkyne 5 in 89% yield (Scheme 3). Subsequently, a copper-catalyzed azide–alkyne cycloaddition (CuAAC) was performed using O-propargylated 3-arylacrylamide 5, benzyl bromide 4b, and sodium azide in the presence of CuI (10 mol%) as the catalyst in a mixture of ethanol and water at 60 °C for 12 h, affording the 1,4-disubstituted 1,2,3-triazole 6 in 79% yield. Notably, this multicomponent transformation enabled the formation of three C–N bonds in a single step, employing an eco-friendly solvent and proceeding under ligand-free conditions.
2.2 FT-IR and NMR spectroscopy studies
The solid-state FT-IR spectra of the Knoevenagel adducts 3a–o exhibited characteristic absorption bands consistent with the functional groups anticipated in their proposed molecular structures (Fig. S2–S7 in SI). For compounds 3a–n, the asymmetric and symmetric N–H stretching vibrations of non-hydrogen-bonded groups were observed in the ranges 3362–3455 cm−1 and 3302–3373 cm−1, respectively. Additional absorption bands at 3236–3288 cm−1 and 3151–3193 cm−1 were assigned to the asymmetric and symmetric hydrogen-bonded N–H stretching vibrations. The amide C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration appeared in the range 1652–1715 cm−1 for compounds 3a–n, while compound 3o exhibited an ester C
O stretching vibration appeared in the range 1652–1715 cm−1 for compounds 3a–n, while compound 3o exhibited an ester C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration at 1711 cm−1. Finally, the C
O stretching vibration at 1711 cm−1. Finally, the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretching vibration was detected in the range of 2200–2230 cm−1 in compounds 3a–l and 3o.
N stretching vibration was detected in the range of 2200–2230 cm−1 in compounds 3a–l and 3o.
Further structural elucidation was performed using 1H and 13C NMR spectroscopy in DMSO-d6 and CDCl3 (Fig. S8–S24 in SI). The (E)-2-cyano-3-(het)arylacrylamides 3a–l and 3o exhibited a characteristic singlet in the range of δ 7.93–9.07 ppm, attributable to the H-3 proton. The 13C NMR spectra confirmed the presence of the C-3 carbon (δ 135.9–150.5 ppm) and the carbonyl carbon of the acrylamide moiety (δ 161.6–164.0 ppm). In the Knoevenagel adducts 3m–o, the β-proton and β-carbon appeared in the ranges of δ 7.28–8.09 ppm and δ 132.8–154.1 ppm, respectively, reflecting the electronic effects of the substituents on the conjugated aryl system. Moreover, the ester carbonyl in compound 3o was observed at δ 163.8 ppm, while compounds 3m and 3n exhibited two distinct amide carbonyl signals in the range of δ 165.6–169.6 ppm, indicative of non-equivalent amide environments.
2.3 X-ray crystallographic studies
The crystal structure of compound 3i was determined by single-crystal X-ray diffraction (Table S18 shows the crystallographic data). Fig. 3a shows the ORTEP representation of 3i, illustrating its nearly planar conformation with only a slight distortion at the carbonyl group, most likely resulting from packing effects.The electrostatic potentials (EPSs) mapped on the Hirshfeld surface (calculated at the B3LYP level with the 6-31G(d,p) basis set) indicate that the most electronegative and electropositive regions are localized on the carbonyl/cyano and hydroxyl/amino groups, respectively (Fig. 3b). Consistently, the strongest hydrogen bond is formed between the hydroxyl group (donor) and the carbonyl group (acceptor) (Fig. 3c). These hydrogen bonds are complemented by short N–H⋯N and N–H⋯O non-covalent interactions involving the amino (donor) and hydroxyl (acceptor) groups, leading to the formation of molecular layers stabilized by π⋯π stacking interactions and van der Waals forces (Fig. 3d). The resulting supramolecular structure displays layered architecture, with strong interactions holding the sheets together and weaker interactions operating in the interlayer zone. Detailed crystallographic data (Table S18), hydrogen-bonding parameters (Table S19), bond lengths (Table S22), bond/valence angles (Table S23), and torsion/dihedral angles (Table S24) are provided in the SI.
After extensive attempts to optimize the crystallization conditions, low-quality crystals of compound 3n were obtained and subjected to X-ray data collection. Although the diffraction data yielded suboptimal statistical parameters, they were sufficient to confirm the molecular structure (Table S20). Due to low-quality data, the CIF file was not deposited in the CCDC; however, the file, including the embedded HKL data, has been provided in the SI. According to these data, compound 3n crystallizes in the triclinic space group P-1 with unit cell parameters a = 9.1161(13) Å, b = 11.3530(12) Å, c = 21.0258(17) Å, α = 99.481(8)°, β = 95.989(10)°, γ = 102.881(11)°, and Z = 2 with Z′ = 1. In the supramolecular structure, the 2-methylenemalonamide fragment promotes short C–H⋯O and N–H⋯N hydrogen bonds that assemble into molecular sheets stacked along the c axis, further connected through the N,N-diphenylaniline fragment by C–H⋯π contacts. Detailed crystallographic data (Table S20), hydrogen-bond parameters (Table S21), the ORTEP representation, and the crystal packing (Fig. S1) are provided in the SI.
2.4 Physicochemical and drug-likeness profiling
To determine whether the Knoevenagel adducts 3a–o comply with Lipinski's Rule of Five (Ro5), their physicochemical properties were analyzed using Molinspiration (Table 2). The Ro5 predicts oral bioavailability by evaluating key parameters related to aqueous solubility and membrane permeability, including molecular weight (≤500 Da), lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ≤ 5), hydrogen bond donors (≤5), and hydrogen bond acceptors (≤10).44 Among the series, only compound 3o exceeded the recommended log
P ≤ 5), hydrogen bond donors (≤5), and hydrogen bond acceptors (≤10).44 Among the series, only compound 3o exceeded the recommended log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, with a value of 6.12. This deviation results from the replacement of the primary amide (–CONH2) in the α-cyanoacrylamide core with an ethyl ester (–COOEt), eliminating a hydrogen-bond donor, combined with the presence of an N,N-diphenylamino group that extends the aromatic surface. These features reduce polarity, increase hydrophobic bulk, and promote partitioning into nonpolar phases, explaining the elevated log
P, with a value of 6.12. This deviation results from the replacement of the primary amide (–CONH2) in the α-cyanoacrylamide core with an ethyl ester (–COOEt), eliminating a hydrogen-bond donor, combined with the presence of an N,N-diphenylamino group that extends the aromatic surface. These features reduce polarity, increase hydrophobic bulk, and promote partitioning into nonpolar phases, explaining the elevated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P.
P.
| Compound | %ABSb | TPSA (Å2)a | nHBA (ON)a | nHBD (OHNH)a | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pa Pa |
MW (Da)a | No violationsa | Fsp3 c | Mean %Gd |
|---|---|---|---|---|---|---|---|---|---|
| a Values obtained from https://www.molinspiration.com.b %ABS = 109 − (0.345 × TPSA).c Fraction of sp3 carbon atoms (Fsp3).d Values obtained from the National Cancer Institute (NCI, USA). | |||||||||
| 3a | 85.92 | 66.89 | 3 | 2 | 1.80 | 206.63 | 0 | 0 | 103.28 |
| 3b | 85.92 | 66.89 | 3 | 2 | 1.93 | 251.08 | 0 | 0 | 103.74 |
| 3c | 70.12 | 112.71 | 6 | 2 | 1.08 | 217.18 | 0 | 0 | 103.48 |
| 3d | 85.92 | 66.89 | 3 | 2 | 2.22 | 241.08 | 0 | 0 | 100.21 |
| 3e | 70.12 | 112.71 | 6 | 2 | 1.68 | 251.63 | 0 | 0 | 94.54 |
| 3f | 84.81 | 70.12 | 4 | 2 | 4.62 | 339.40 | 0 | 0 | 40.97 |
| 3g | 85.92 | 66.89 | 3 | 2 | 1.57 | 186.21 | 0 | 0.091 | 103.65 |
| 3h | 82.74 | 76.12 | 4 | 2 | 1.18 | 202.21 | 0 | 0.091 | 103.84 |
| 3i | 78.95 | 87.11 | 4 | 3 | 0.64 | 188.19 | 0 | 0 | 103.04 |
| 3j | 84.81 | 70.12 | 4 | 2 | 1.22 | 215.26 | 0 | 0.17 | 99.42 |
| 3k | 85.92 | 66.89 | 3 | 2 | 3.23 | 272.31 | 0 | 0 | 90.54 |
| 3l | 74.41 | 100.25 | 5 | 3 | −0.23 | 192.17 | 0 | 0.11 | 105.20 |
| 3m | 79.26 | 86.19 | 4 | 4 | 0.74 | 224.65 | 0 | 0 | 64.54 |
| 3n | 78.15 | 89.43 | 5 | 4 | 3.56 | 357.41 | 0 | 0 | −63.43 |
| 3o | 90.60 | 53.34 | 4 | 0 | 6.12 | 368.44 | 1 | 0.083 | 36.80 |
Topological polar surface area (TPSA), a critical descriptor for predicting intestinal absorption and blood brain barrier (BBB) penetration, generally indicates that values above 140 Å2 correlate with poor absorption.45 All Knoevenagel adducts 3a–o presented TPSA values between 53.34 Å2 and 112.71 Å2, remaining within the optimal range for effective absorption. Notably, compounds 3a, 3b, 3d, 3g, 3k, and 3o exhibited the lowest values (≤66.89 Å2), with 3o showing the minimum (53.34 Å2), in line with its reduced hydrogen-bonding capacity and increased hydrophobicity.
Consistent with the TPSA trends, the estimated percentage of absorption (%ABS), a key pharmacokinetic parameter reflecting a compound's ability to permeate biological membranes,46 ranged from 70.12% to 90.60% for compounds 3a–o. Notably, compounds 3a, 3b, 3d, 3f, 3g, 3h, 3j, 3k, and 3o exhibited %ABS values exceeding 80%, a feature attributable to their low TPSA (≤76.12 Å2), which facilitates passive membrane permeability. Among these, compound 3o achieved the highest absorption (90.60 Å2), in line with its minimal TPSA (53.34 Å2) and elevated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (6.12).
P (6.12).
The fraction of sp3-hybridized carbons (Fsp3) was analyzed, as this parameter is directly related to molecular three-dimensionality and can significantly influence solubility, conformational flexibility, and pharmacokinetic properties.47 The Knoevenagel adducts 3a–o exhibited low Fsp3 values, not exceeding 0.17, consistent with predominantly unsaturated molecular frameworks. Within the series, compounds 3g, 3h, 3j, 3l and 3o showed slightly higher Fsp3 values (>0.08). Among these, 3j reached the highest Fsp3 (0.17), attributable to the N,N-dimethylamino substituent, which increases the proportion of sp3-hybridized carbons.
2.5 Antiproliferative activity
The in vitro antiproliferative activity of Knoevenagel adducts 3a–o was evaluated by the National Cancer Institute (NCI, USA). All compounds were initially subjected to a single-dose screening at 10 µM against 60 human cancer cell lines representing nine cancer panels (leukemia, melanoma, lung, colon, central nervous system, ovarian, renal, prostate, and breast) using the sulforhodamine B (SBR) assay. Results, expressed as NCI mean graphs, indicated that lower growth percentage (%G) correspond to higher growth inhibition percentage (%GI = 100 − %G), while negative %G values denote lethality (Fig. S25–S43 in SI). The mean growth percent (mean %G) represents the average of %G values across the panel of cancer cell lines; thus, mean %G ≤ 50 were considered indicative of significant active. According to this criterion, compounds 3f, 3n, and 3o exhibited the highest antiproliferative activity with the mean %G of 40.97, −63.43, and 36.80, respectively, among the tested series (Table 2).48,49The physicochemical and drug-likeness profiling revealed a clear correlation between antiproliferative activity and lipophilicity (Table 2). The most active compounds 3f, 3n, and 3o showed high log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of 4.62, 3.56, and 6.12, respectively, indicating that increased hydrophobicity enhances activity. This relationship can be rationalized by the presence of the N,N-diphenylamino substituent, which increases the aromatic surface, reduces molecular polarity, and facilitates membrane partitioning into nonpolar environments. Conversely, no consistent correlation was observed between mean %G and either topological polar surface area (TPSA) or the predicted absorption (%ABS). The structure–activity relationship (SAR) analysis derived from %GI, lethality, and mean %G data revealed two key trends: (i) incorporation of the R1 = NPh2 substituent within the α,β-unsaturated carbonyl framework of compounds 3f, 3n, and 3o markedly enhances antiproliferative activity, and (ii) substitution at R3 = CONH2 in 3n further increases activity relative to the corresponding nitrile analogues (R3 = CN) in 3f and 3o.
P values of 4.62, 3.56, and 6.12, respectively, indicating that increased hydrophobicity enhances activity. This relationship can be rationalized by the presence of the N,N-diphenylamino substituent, which increases the aromatic surface, reduces molecular polarity, and facilitates membrane partitioning into nonpolar environments. Conversely, no consistent correlation was observed between mean %G and either topological polar surface area (TPSA) or the predicted absorption (%ABS). The structure–activity relationship (SAR) analysis derived from %GI, lethality, and mean %G data revealed two key trends: (i) incorporation of the R1 = NPh2 substituent within the α,β-unsaturated carbonyl framework of compounds 3f, 3n, and 3o markedly enhances antiproliferative activity, and (ii) substitution at R3 = CONH2 in 3n further increases activity relative to the corresponding nitrile analogues (R3 = CN) in 3f and 3o.
Compounds 3f, 3n, and 3o met the NCI selection threshold of mean %G ≤ 50% and were advanced to the five-dose screening at 0.01, 0.1, 1.0, 10, and 100 µM. This assay enabled the determination of GI50 (growth inhibitory concentration) and LC50 (lethal concentration) values. Compounds were considered active when they achieved a selectivity index LC50/GI50 ≥ 100, ensuring that their antiproliferative effects reflected selective growth inhibition rather than nonspecific cytotoxicity.48,49 Although 3f, 3n, and 3o frequently exhibited very low GI50 values, the separation from their LC50 values was often insufficient to meet this threshold. Consequently, part of their apparent activity could not be unequivocally distinguished from nonspecific cytotoxic effects.
In Table 3, results highlighted in blue denote selective antiproliferative activity, whereas those in orange indicate cases where the LC50/GI50 ratio fell below the required threshold. Accordingly, the subsequent discussion will focus on the blue results obtained for 3f, 3n, and 3o, particularly those showing activity at concentrations close to 1 µM, in comparison with Osimertinib as the reference drug.
| a GI50 corresponds to the concentration required to reduce net protein content by 50% in control cells, as determined by the SRB assay, after exposure to five concentrations (0.01, 0.1, 1.0, 10, and 100 µM).b LC50 represents the concentration needed to induce 50% cell death.c Not determined.d Activity data for Osimertinib in the NCI-60 cancer cell line panel were obtained from the NCI database: https://dtp.cancer.gov/dtpstandard/cancerscreeningdata/index.jsp. |
|---|
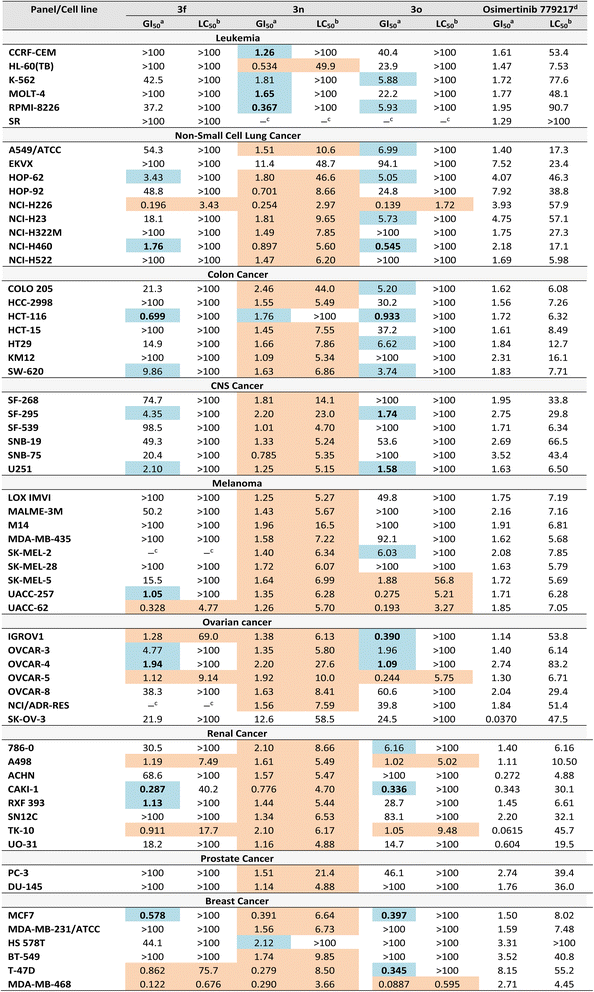 |
Compound 3f exhibited the strongest antiproliferative effect against the CAKI-1 renal cancer cell line (GI50 = 0.287 µM). It also showed potent activity against NCI-H460 (non-small cell lung cancer, GI50 = 1.76 µM), HCT-116 (colon cancer, GI50 = 0.699 µM), UACC-257 (melanoma, GI50 = 1.05 µM), OVCAR-4 (ovarian cancer, GI50 = 1.94 µM), RXF 393 (renal cancer, GI50 = 1.13 µM), and MCF7 (breast cancer, GI50 = 0.578 µM). Remarkably, compound 3f displayed higher activity than Osimertinib across all these cell lines (Table 3, bold entries).
For compound 3n, although it consistently yielded low GI50 values across multiple cancer cell lines, it failed to achieve the required 100-fold separation from LC50, suggesting that nonspecific cytotoxicity may partly account for its activity profile. Nevertheless, compound 3n displayed potent and selective antiproliferative activity against hematological malignancies, including CCRF-CEM, K-562, MOLT-4, and RPMI-8226, with GI50 values ranging from 0.367 to 1.81 µM. Remarkably, 3n was 1.3-, 1.1-, and 5.3-fold more potent than Osimertinib against CCRF-CEM, MOLT-4, and RPMI-8226, respectively (Table 3, bold entries). In addition, 3n showed notable activity against the HCT-116 colon cancer cell line (GI50 = 1.76 µM), comparable to Osimertinib (GI50 = 1.72 µM).
Compound 3o exhibited its highest antiproliferative activity against the CAKI-1 renal cancer cell line (GI50 = 0.336 µM), showing comparable potency to Osimertinib (GI50 = 0.343 µM). It also demonstrated strong activity against NCI-H460 (non-small cell lung cancer, GI50 = 0.545 µM), HCT-116 (colon cancer, GI50 = 0.933 µM), SF-295 (CNS cancer, GI50 = 1.74 µM), and U251 (CNS cancer, GI50 = 1.58 µM), surpassing Osimertinib in these cell lines (Table 3, bold entries). Moreover, 3o displayed notable potency against ovarian cancer cell lines IGROV1, OVCAR-4, and OVCAR-3, with GI50 values of 0.390 µM, 1.09 µM, and 1.96 µM, respectively, being 2.9- and 2.5-fold more potent than Osimertinib against IGROV1 and OVCAR-4. Finally, 3o exerted pronounced antiproliferative effects against the breast cancer cell lines T47D and MCF7, with GI50 values of 0.345 µM and 0.397 µM, respectively, corresponding to 23.6- and 3.8-fold greater potency than Osimertinib.
Taken together, compounds 3f and 3o incorporating a primary amide (–CONH2) and an ethyl ester (–COOEt), respectively, exhibited potent and selective antiproliferative activity at concentrations close to 1 µM across multiple cancer types, including non-small cell lung (NCI-H460), colon (HCT-116), ovarian (OVCAR-4), renal (CAKI-1), and breast (MCF7), with GI50 values ranging from 0.287 µM to 1.94 µM (LC50/GI50 ≥ 100 for all cases). Notably, both compounds exhibited their strongest activity against the CAKI-1 renal cancer cell line, with GI50 values of 0.287 µM for 3f and 0.336 µM for 3o, closely comparable to Osimertinib (GI50 = 0.343 µM).
In contrast, compound 3n, bearing two primary amide groups and lacking the cyano substituent, exhibited both low GI50 and LC50 values, suggesting that replacement of –CN with –CONH2 may promote nonspecific cytotoxicity (GI50 ≈ LC50). Despite this, 3n maintained selective antiproliferative activity against hematological cell lines CCRF-CEM, K-562, MOLT-4, and RPMI-8226 (GI50 = 0.367–1.81 µM), where it proved more potent than Osimertinib in most cases (GI50 = 1.61–1.95 µM).
Importantly, compounds 3f, 3o, and 3n also exhibited reproducible antiproliferative activity against the HCT-116 colon cancer cell line, with GI50 values of 0.699 µM, 0.933 µM, and 1.76 µM, respectively (LC50/GI50 ≥ 100 for all cases). Notably, compounds 3f and 3o were 2.5- and 1.8-fold more potent than Osimertinib, whereas 3n showed comparable potency to the reference drug.
Overall, these findings underscore that the Knoevenagel adducts 3f, 3n, and 3o, featuring an N,N-diphenylamino substituent, represent highly promising molecular scaffolds for the rational design and development of new anticancer agents.
Additionally, the mean GI50 values for the cancer panels of compounds 3f, 3n, and 3o were compared with those of the reference drug Osimertinib (Fig. 4). Among them, compound 3n exhibited consistently higher antiproliferative activity than 3f and 3o across all cancer types. Remarkably, 3n was more potent than Osimertinib in leukemia, non-small cell lung, colon, CNS, melanoma, prostate, and breast cancer panels, with fold increases of 1.5, 1.6, 1.1, 1.7, 1.2, 1.7, and 3.3, respectively. These findings suggest that 3n may serve as a promising lead molecule for anticancer drug discovery; however, its activity cannot be clearly differentiated from nonspecific cytotoxic effects (GI50 ≈ LC50), a limitation also observed for Osimertinib across multiple cancer cell lines (Table 3).
 | ||
| Fig. 4 Mean GI50 values per panel for compounds 3f, 3n, and 3o compared with the reference drug Osimertinib across the NCI-60 human cancer cell line panel. | ||
2.6 Mechanistic toxicological profiling
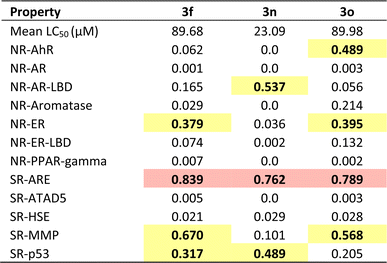
To complement the experimental cytotoxicity data (LC50 values, Table 3) and to elucidate potential molecular mechanisms underlying the observed nonspecific cytotoxicity, a Tox21 pathway analysis was performed using ADMETlab 3.0. This computational tool predicts the likelihood of activation or inhibition of nuclear receptor (NR) and stress response (SR) pathways, with output values ranging from 0 (inactive) to 1 (active). According to ADMETlab's classification, predictions are categorized as excellent/low risk (green, <0.3), moderate (yellow, 0.3–0.7), and poor/high risk (red, >0.7). The analyzed endpoints included NR-AhR (oxidative stress and xenobiotic metabolism), NR-AR and NR-AR-LBD (androgen receptor and its ligand-binding domain; endocrine signaling), NR-Aromatase (estrogen biosynthesis and endocrine disruption), NR-ER and NR-ER-LBD (estrogen receptor and its ligand-binding domain; hormonal regulation), NR-PPAR-gamma (lipid metabolism and inflammatory response), SR-ARE (Nrf2-mediated oxidative stress response), SR-ATAD5 (genomic integrity maintenance), SR-HSE (cellular stress induced by protein misfolding), SR-MMP (mitochondrial dysfunction and apoptosis), and SR-p53 (DNA damage response and apoptosis). As shown in Table 4, compounds 3f, 3n, and 3o exhibited mean LC50 values of 89.68, 23.09, and 89.98 µM, respectively. All compounds exhibited moderate probabilities of activation for the Nrf2-mediated oxidative stress pathway (SR-ARE = 0.762–0.839), suggesting potential induction of adaptive antioxidant mechanisms. Notably, compound 3n displayed additional moderate activity toward androgen receptor binding (NR-AR-LBD = 0.537) and p53-dependent DNA damage response (SR-p53 = 0.489), which may be associated with its higher cytotoxic effect. These results indicate that the nonspecific cytotoxicity of 3n likely arises from the combined influence of oxidative stress, hormonal signaling, and apoptotic mechanisms. Enhanced Nrf2 (SR-ARE) activity may be associated with increased levels of reactive oxygen species, while interaction with the androgen receptor (NR-AR-LBD) could disrupt redox homeostasis and promote oxidative damage that activates the p53-mediated apoptotic pathway.50,512.7 Molecular docking studies
EGFR was selected as the molecular target for docking studies due to its well-established role as a driver in cancer progression and its validation as the clinical target of FDA-approved acrylamide-based inhibitors, such as Osimertinib. The incorporation of the acrylamide moiety not only enables irreversible binding to the EGFR kinase domain but has also been shown to enhance drug-like properties in EGFR inhibitor design.42,52 Given that compounds 3f, 3n, and 3o demonstrated notable antiproliferative activity comparable or superior to Osimertinib across multiple NCI-60 cancer cell lines, targeting EGFR via docking provided a rational framework to correlate their biological activity with their predicted binding conformations.53Molecular docking studies were conducted to evaluate the interactions of the Knoevenagel adducts 3f, 3n, and 3o with the EGFR active site (PDB ID: 1M17). To ensure accuracy and energetic plausibility of the predicted binding poses, the docking protocol was validated by redocking the co-crystallized ligands into its binding site. This validation confirmed that AutoDock Vina reliably reproduced the experimental orientation and position of the ligand,54 with redocking yielded RMSD values below 2.0 Å, thereby verifying the robustness of the docking parameters. Self-docking of the co-crystallized ligand Erlotinib resulted in a docking score of −6.6 kcal mol−1 and an RMSD of 1.31 Å, validating the protocol. These results confirmed the reliability and accuracy of the molecular docking simulations applied in this study.
To further investigate the binding interactions of (E)-2-cyano-3-arylacrylamide 3f within the EGFR active site, molecular docking simulations revealed that its acrylamide moiety occupied the region typically filled by the quinazoline ring of Erlotinib, forming two key hydrogen bonds with Met769 and an additional hydrogen bond with Pro770 (Fig. 5 and 6). The para-disubstituted benzene ring of 3f was oriented toward the hydrophobic side chains of Val702 and Leu820. In this conformation, one benzene ring overlapped with the region commonly occupied by the anilino moiety of Erlotinib, while the second benzene ring extended toward the opposite side of the binding pocket, engaging in several non-covalent interactions, including a π–alkyl interaction with Val702, a π–cation interaction with Lys721, and π–anion and π–σ interactions with Asp831 and Phe699, respectively. Additional van der Waals contacts further stabilized the complex. Overall, compound 3f engaged six key side-chain residues critical for EGFR inhibition (Leu694, Leu820, Ala719, Met769, Lys721, and Val702), along with three additional active-site residues (Phe699, Pro770, and Asp831) that are not commonly targeted by reference ligands. The remarkable binding affinity of compound 3f (−8.7 kcal mol−1), surpassing that of Erlotinib (−6.6 kcal mol−1), highlights its strong interaction with the EGFR binding site and supports its potential as an effective inhibitor.
 | ||
| Fig. 5 Overlay of the docked conformations of erlotinib (magenta), compound 3f (blue), compound 3n (yellow) and compound 3o (green) within the EGFR binding pocket. | ||
 | ||
| Fig. 6 2D representation of the binding interactions of compound 3f with key amino acid residues in the EGFR active site. | ||
Following the same docking protocol, compound 3n was found to occupy the EGFR active site in a binding mode distinct from that of compound 3f. The acrylamide moiety of 3n was positioned within the region typically occupied by the anilino moiety of Erlotinib, while one of its benzene rings aligned with the area corresponding to the 2-methoxyethoxy substituent of Erlotinib (Fig. 5). Binding 3n to the EGFR pocket involved conventional hydrogen bonds with Ala719, Glu738, and Thr830; π–alkyl interactions with Leu694, Val702, and Lys721; along with a π–anion interaction with Asp831. Importantly, compound 3n preserved five key interactions observed in the reference ligand, involving residues Leu694, Val702, Ala719, Lys721, and Leu820, which are essential for defining the EGFR binding site and its catalytic mechanism. In addition, compound 3n established novel hydrogen bonds with Glu738 and Thr830, further stabilizing its binding conformation (Fig. 7). The favorable binding affinity of compound 3n (−7.8 kcal mol−1), surpassing that of Erlotinib (−6.6 kcal mol−1), underscores its effective engagement with critical residues in the EGFR active site and supports its potential as a promising inhibitor.
 | ||
| Fig. 7 2D representation of the binding interactions of compound 3n with key amino acid residues in the EGFR active site. | ||
Compound 3o, with a binding energy of −7.9 kcal mol−1, displayed interaction patterns with key amino acid residues in the EGFR active site closely resembling those of compound 3f. Remarkably, the three aromatic rings of compound 3f overlap with those of 3o, engaging the same residues (Phe699, Val702, Ala719, Lys721, Met742, Thr766, Asn818, Leu820, Asp831, and Met769) (Fig. 5 and 8). The acrylamide moiety of compound 3o, although adopting slightly different orientation, forms interactions with several of the same residues (Leu768, Met769, Pro770, and Gly772). The most distinctive feature between the two compounds, and one of mechanistic significance, is the additional interaction with Cys773, a residue that plays a pivotal role in ligand recognition, binding affinity, and selectivity within the EGFR catalytic domain.
 | ||
| Fig. 8 2D representation of the binding interactions between compound 3o and key amino acid residues within the EGFR active site. | ||
3 Conclusions
In summary, we developed a piperidine-catalyzed synthesis of (E)-2-cyano-3-(het)arylacrylamides 3a–i in 40–95% yields via a Knoevenagel condensation between (hetero)aromatic aldehydes 1a–i and 2-cyanoacetamide 2a under mild conditions. The methodology was successfully extended to other methylene active compounds, such as malonamide 2b and ethyl cyanoacetate 2c, affording Knoevenagel adducts 3m–o in 54–81% yields. This strategy is distinguished by its operational simplicity, use of green solvents (water or ethanol), low reaction temperatures, minimal catalyst loading, broad synthetic scope, and straightforward purification by simple filtration in most cases. Additionally, functionalization of the propargyl moiety in α-cyanoacrylamide 5 via a CuAAC reaction enabled the synthesis of the 1,4-disubstituted 1,2,3-triazole 6 in 79% yield.X-ray diffraction analysis of compound 3i reveals a planar molecular conformation, with the highest electronegative and electropositive potentials (from ESP maps) localized on the carbonyl/cyano and hydroxyl/amino groups, respectively. This distribution of electrostatic potentials, combined with the molecular planarity, accounts for the formation of molecular sheets interconnected through π–π stacking interactions and van der Waals forces.
The antiproliferative activity of Knoevenagel adducts 3a–o was assessed against the NCI panel of 60 human cancer cell lines using the SRB assay. In the single-dose screening at 10 µM, compounds 3f, 3n, and 3o, each bearing an N,N-diphenylamino substituent, exhibited the strongest activity within the series, with mean %G values of 40.97, −63.43, and 36.80, respectively.
In the subsequent five-dose screening at 100, 10, 1.0, 0.1, and 0.01 µM, compounds 3f (bearing a primary amide, –CONH2) and 3o (bearing an ethyl ester, –COOEt) demonstrated potent and selective antiproliferative activity, with GI50 values ranging from 0.287 µM to 1.94 µM and LC50/GI50 ≥ 100 across multiple cancer types, including non-small cell lung (NCI-H460), colon (HCT-116), ovarian (OVCAR-4), renal (CAKI-1), and breast (MCF7). Remarkably, both compounds displayed their strongest activity against the CAKI-1 renal cancer, with GI50 values of 0.287 µM for 3f and 0.336 µM for 3o, closely comparable to Osimertinib (GI50 = 0.343 µM). In contrast, compound 3n characterized by the presence of two –CONH2 groups and the absence of the cyano substituent, exhibited a cytotoxic profile consistent with nonspecific activity (GI50 ≈ LC50). Despite this limitation, 3n retained notable selective antiproliferative activity against hematological cancer cell lines CCRF-CEM, K-562, MOLT-4, and RPMI-8226 (GI50 = 0.367–1.81 µM), demonstrating superior potency to Osimertinib in most cases (GI50 = 1.61–1.95 µM).
Overall, compounds 3f, 3o, and 3n consistently demonstrated antiproliferative activity against the HCT-116 colon cancer cell line, with GI50 values of 0.699 µM, 0.933 µM, and 1.76 µM, respectively (LC50/GI50 ≥ 100 in all three cases). Notably, 3f and 3o were 2.5- and 1.8-fold more potent than Osimertinib, while 3n exhibited efficacy comparable to the reference drug. Importantly, molecular docking studies elucidated the binding modes of compounds 3f, 3n, and 3o within the tyrosine kinase domain of the epidermal growth factor receptor (EGFR). These results suggest that the concurrent incorporation of an N,N-diphenylamino group into α,β-unsaturated carbonyl scaffolds could provide a privileged structural framework for the rational design and future development of novel anticancer agents.
4 Experimental
4.1 Materials and methods
The progress of all reactions was monitored by thin-layer chromatography (TLC) using Merck Kieselgel 60 F254 precoated silica gel plates, with visualization under UV light at 254 and 365 nm. Unless otherwise stated, solvents and reagents were purchased from Sigma-Aldrich and used without further purification. Infrared spectra were recorded at room temperature using a PerkinElmer Polymer ID Analyzer equipped with an ATR accessory, operating over a wavenumber range of 400–4000 cm−1, with a spectral resolution of 4 cm−1 and an acquisition of 64 scans per sample. 1H and 13C NMR spectra were obtained using a Bruker Avance 400 MHz spectrometer with DMSO-d6 as the deuterated solvent. Chemical shifts (δ) are reported in parts per million (ppm) relative to residual solvent signals (δ = 2.50 ppm for 1H and 39.52 ppm for 13C) and coupling constants (J) are expressed in hertz (Hz). Multiplicity is denoted as s (singlet), d (doublet), t (triplet), and m (multiplet). High-resolution mass spectrometry (HRMS) was performed using a Q-TOF 6520 spectrometer with electrospray ionization (ESI, 4000 V).4.2 Knoevenagel synthesis of (E)-2-cyano-3-(het)arylacrylamides 3
A mixture of the (hetero)aromatic aldehyde 1a–l (1 mmol), active methylene compound 2a–c (1 mmol), and piperidine (0.1 mol, 9.9 µL) in distilled water or absolute ethanol (3 mL) was stirred at room temperature for 6–24 h. The resulting precipitate was filtered, washed with a cold EtOH/H2O mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v), and dried to afford the (E)-2-cyano-3-(het)arylacrylamides 3a–o. Crystals of compounds 3i and 3n suitable for single-crystal X-ray diffraction analysis were obtained by slow evaporation of a solution of the compound in a mixture of N,N-dimethylformamide and methanol (1
1, v/v), and dried to afford the (E)-2-cyano-3-(het)arylacrylamides 3a–o. Crystals of compounds 3i and 3n suitable for single-crystal X-ray diffraction analysis were obtained by slow evaporation of a solution of the compound in a mixture of N,N-dimethylformamide and methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v, 2 mL) over the course of one month under ambient temperature and pressure conditions. The identity of the products was confirmed by comparison of their NMR data with values reported in the literature.
1 v/v, 2 mL) over the course of one month under ambient temperature and pressure conditions. The identity of the products was confirmed by comparison of their NMR data with values reported in the literature.
![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1704 (νC
N), 1704 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (νC
O), 1602 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1587 (νC
C), 1587 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1486, 1381, 1207, 1092, 1008, 957, 826, 784, 707 (νC–Cl), 577, 458 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.65 (d, J = 8.8 Hz, 2H), 7.82 (br s, 1H, NHa), 7.90–7.97 (m, 3H), 8.18 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 107.3 (C), 116.3 (C), 129.4 (2CH), 130.8 (C), 131.7 (2CH), 136.9 (C), 149.3 (CH, C-3), 162.5 (C, CE001O) ppm. These NMR data matched previously reported data.55
C), 1486, 1381, 1207, 1092, 1008, 957, 826, 784, 707 (νC–Cl), 577, 458 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.65 (d, J = 8.8 Hz, 2H), 7.82 (br s, 1H, NHa), 7.90–7.97 (m, 3H), 8.18 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 107.3 (C), 116.3 (C), 129.4 (2CH), 130.8 (C), 131.7 (2CH), 136.9 (C), 149.3 (CH, C-3), 162.5 (C, CE001O) ppm. These NMR data matched previously reported data.55![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1701 (νC
N), 1701 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (νC
O), 1602 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1489, 1378, 1187, 1074, 1008, 829, 811, 697, 580 (νC–Br), 470 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.78 (d, J = 8.4 Hz, 2H), 7.82 (br s, 1H, NHa), 7.86 (d, J = 8.4 Hz, 2H), 7.95 (br s, 1H, NHb), 8.16 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 107.4 (C), 116.3 (C), 126.0 (C), 131.2 (C), 131.8 (2CH), 132.4 (2CH), 149.4 (CH, C-3), 162.5 (C, C
C), 1489, 1378, 1187, 1074, 1008, 829, 811, 697, 580 (νC–Br), 470 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.78 (d, J = 8.4 Hz, 2H), 7.82 (br s, 1H, NHa), 7.86 (d, J = 8.4 Hz, 2H), 7.95 (br s, 1H, NHb), 8.16 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 107.4 (C), 116.3 (C), 126.0 (C), 131.2 (C), 131.8 (2CH), 132.4 (2CH), 149.4 (CH, C-3), 162.5 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.56
O) ppm. These NMR data matched previously reported data.56![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1690 (νC
N), 1690 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (νC
O), 1602 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1509, 1380, 1344, 1203, 1106, 855, 782, 769, 748, 683, 568, 531, 474, 443 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.93 (br s, 1H, NHa), 8.06 (br s, 1H, NHb), 8.12 (d, J = 8.8 Hz, 2H), 8.30 (s, 1H, H-3), 8.37 (d, J = 8.4 Hz, 2H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 110.6 (C), 115.8 (C), 124.2 (2CH), 131.0 (2CH), 138.1 (C), 148.3 (CH, C-3), 148.8 (C), 162.1 (C, C
C), 1509, 1380, 1344, 1203, 1106, 855, 782, 769, 748, 683, 568, 531, 474, 443 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.93 (br s, 1H, NHa), 8.06 (br s, 1H, NHb), 8.12 (d, J = 8.8 Hz, 2H), 8.30 (s, 1H, H-3), 8.37 (d, J = 8.4 Hz, 2H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 110.6 (C), 115.8 (C), 124.2 (2CH), 131.0 (2CH), 138.1 (C), 148.3 (CH, C-3), 148.8 (C), 162.1 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.56
O) ppm. These NMR data matched previously reported data.56![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1715 (νC
N), 1715 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1608 (νC
O), 1608 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1585 (νC
C), 1585 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1470, 1393, 1144, 1110, 1051, 924, 842, 826, 792, 765, 592, 559, 532, 446 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.65 (dd, J = 2.0, 8.8 Hz, 1H), 7.86 (d, J = 2.0 Hz, 1H), 7.94 (br s, 1H, NHa), 8.01 (d, J = 8.4 Hz, 1H), 8.08 (br s, 1H, NHb), 8.30 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 111.5 (C), 115.4 (C), 128.2 (CH), 129.5 (C), 129.7 (CH), 130.9 (CH), 135.1 (C), 137.0 (C), 146.0 (CH, C-3), 161.6 (C, C
C), 1470, 1393, 1144, 1110, 1051, 924, 842, 826, 792, 765, 592, 559, 532, 446 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.65 (dd, J = 2.0, 8.8 Hz, 1H), 7.86 (d, J = 2.0 Hz, 1H), 7.94 (br s, 1H, NHa), 8.01 (d, J = 8.4 Hz, 1H), 8.08 (br s, 1H, NHb), 8.30 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 111.5 (C), 115.4 (C), 128.2 (CH), 129.5 (C), 129.7 (CH), 130.9 (CH), 135.1 (C), 137.0 (C), 146.0 (CH, C-3), 161.6 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.56
O) ppm. These NMR data matched previously reported data.56![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1707 (νC
N), 1707 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1607 (νC
O), 1607 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1538, 1478, 1392, 1352, 1213, 1112, 1052, 941, 824, 789, 670, 581, 524, 488, 451 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.92 (br s, 1H, NHa), 8.00 (d, J = 8.4 Hz, 1H), 8.01 (br s, 1H, NHb), 8.21 (dd, J = 2.2, 8.6 Hz, 1H), 8.25 (s, 1H, H-3), 8.56 (d, J = 2.0 Hz, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 109.9 (C), 115.7 (C), 127.0 (CH), 128.2 (C), 132.3 (C), 132.7 (CH), 134.2 (CH), 147.3 (CH, C-3), 147.6 (C), 161.9 (C, C
C), 1538, 1478, 1392, 1352, 1213, 1112, 1052, 941, 824, 789, 670, 581, 524, 488, 451 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.92 (br s, 1H, NHa), 8.00 (d, J = 8.4 Hz, 1H), 8.01 (br s, 1H, NHb), 8.21 (dd, J = 2.2, 8.6 Hz, 1H), 8.25 (s, 1H, H-3), 8.56 (d, J = 2.0 Hz, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 109.9 (C), 115.7 (C), 127.0 (CH), 128.2 (C), 132.3 (C), 132.7 (CH), 134.2 (CH), 147.3 (CH, C-3), 147.6 (C), 161.9 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. HRMS (ESI+): m/z calculated for C10H735ClN3O3+ 252.0170 [M + H]+; found 252.0176.
O) ppm. HRMS (ESI+): m/z calculated for C10H735ClN3O3+ 252.0170 [M + H]+; found 252.0176.![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1652 (νC
N), 1652 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1607 (νC
O), 1607 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1576 (νC
C), 1576 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1490, 1368, 1326, 1301, 1193, 1179, 1075, 926, 834, 826, 756, 7256.89 (d, J = 8.8 Hz, 2H), 7.16–7.25 (m, 6H), 7.41 (dd, J = 7.6, 7.6 Hz, 4H), 7.62 (br s, 1H, NHa), 7.75 (br s, 1H, NHb), 7.82 (d, J = 8.8 Hz, 2H), 8.02 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 101.4 (C), 117.4 (C), 118.7 (2CH), 123.5 (C), 125.4 (2CH), 126.2 (4CH), 130.0 (4CH), 132.1 (2CH), 145.5 (2C), 149.8 (CH, C-3), 151.1 (C), 163.3 (C, C
C), 1490, 1368, 1326, 1301, 1193, 1179, 1075, 926, 834, 826, 756, 7256.89 (d, J = 8.8 Hz, 2H), 7.16–7.25 (m, 6H), 7.41 (dd, J = 7.6, 7.6 Hz, 4H), 7.62 (br s, 1H, NHa), 7.75 (br s, 1H, NHb), 7.82 (d, J = 8.8 Hz, 2H), 8.02 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 101.4 (C), 117.4 (C), 118.7 (2CH), 123.5 (C), 125.4 (2CH), 126.2 (4CH), 130.0 (4CH), 132.1 (2CH), 145.5 (2C), 149.8 (CH, C-3), 151.1 (C), 163.3 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.43
O) ppm. These NMR data matched previously reported data.43![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1692 (νC
N), 1692 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1589 (νC
O), 1589 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1508, 1366, 1212, 1182, 1104, 822, 790, 670, 600, 511, 488, 458 cm−1. 1H NMR (400 MHz, DMSO-d6): 2.38 (s, 3H), 7.38 (d, J = 8.0 Hz, 2H), 7.74 (br s, 1H, NHa), 7.85 (d, J = 8.0 Hz, 2H), 7.89 (br s, 1H, NHb), 8.13 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 21.3 (CH3), 105.3 (C), 116.7 (C), 129.2 (C), 129.9 (2CH), 130.2 (2CH), 143.0 (C), 150.5 (CH, C-3), 162.9 (C, C
C), 1508, 1366, 1212, 1182, 1104, 822, 790, 670, 600, 511, 488, 458 cm−1. 1H NMR (400 MHz, DMSO-d6): 2.38 (s, 3H), 7.38 (d, J = 8.0 Hz, 2H), 7.74 (br s, 1H, NHa), 7.85 (d, J = 8.0 Hz, 2H), 7.89 (br s, 1H, NHb), 8.13 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 21.3 (CH3), 105.3 (C), 116.7 (C), 129.2 (C), 129.9 (2CH), 130.2 (2CH), 143.0 (C), 150.5 (CH, C-3), 162.9 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.55
O) ppm. These NMR data matched previously reported data.55![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1694 (νC
N), 1694 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1581 (νC
O), 1581 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1384, 1364, 1310, 1260, 1177, 1025, 961, 825, 672, 578, 550, 530, 472, 455 cm−1. 1H NMR (400 MHz, DMSO-d6): 3.85 (s, 3H), 7.13 (d, J = 8.8 Hz, 2H), 7.68 (br s, 1H, NHa), 7.81 (br s, 1H, NHb), 7.96 (d, J = 8.8 Hz, 2H), 8.11 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 55.6 (CH3), 102.9 (C), 114.8 (2CH), 117.1 (C), 124.4 (C), 132.5 (2CH), 150.2 (CH, C-3), 162.6 (C), 163.1 (C, C
C), 1384, 1364, 1310, 1260, 1177, 1025, 961, 825, 672, 578, 550, 530, 472, 455 cm−1. 1H NMR (400 MHz, DMSO-d6): 3.85 (s, 3H), 7.13 (d, J = 8.8 Hz, 2H), 7.68 (br s, 1H, NHa), 7.81 (br s, 1H, NHb), 7.96 (d, J = 8.8 Hz, 2H), 8.11 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 55.6 (CH3), 102.9 (C), 114.8 (2CH), 117.1 (C), 124.4 (C), 132.5 (2CH), 150.2 (CH, C-3), 162.6 (C), 163.1 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.56
O) ppm. These NMR data matched previously reported data.56![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1652 (νC
N), 1652 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1599 (νC
O), 1599 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1570 (νC
C), 1570 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1511, 1411, 1374, 1288, 1229, 1180, 956, 930, 671, 568, 543, 525, 503, 488, 431 cm−1. 1H NMR (400 MHz, DMSO-d6): 6.93 (d, J = 8.8 Hz, 2H), 7.63 (br s, 1H, NHa), 7.76 (br s, 1H, NHb), 7.87 (d, J = 8.8 Hz, 2H), 8.05 (s, 1H, H-3), 10.59 (br s, 1H, OH) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 101.5 (C), 116.2 (2CH), 117.4 (C), 123.0 (C), 132.9 (2CH), 150.5 (CH, C-3), 161.8 (C), 163.4 (C, C
C), 1511, 1411, 1374, 1288, 1229, 1180, 956, 930, 671, 568, 543, 525, 503, 488, 431 cm−1. 1H NMR (400 MHz, DMSO-d6): 6.93 (d, J = 8.8 Hz, 2H), 7.63 (br s, 1H, NHa), 7.76 (br s, 1H, NHb), 7.87 (d, J = 8.8 Hz, 2H), 8.05 (s, 1H, H-3), 10.59 (br s, 1H, OH) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 101.5 (C), 116.2 (2CH), 117.4 (C), 123.0 (C), 132.9 (2CH), 150.5 (CH, C-3), 161.8 (C), 163.4 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.56
O) ppm. These NMR data matched previously reported data.56![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1686 (νC
N), 1686 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1610 (νC
O), 1610 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1562 (νC
C), 1562 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1522, 1440, 1362, 1323, 1238, 1190, 1169, 943, 810, 663, 590, 525, 501, 461 cm−1. 1H NMR (400 MHz, DMSO-d6): 3.04 (s, 6H), 6.81 (d, J = 8.8 Hz, 2H), 7.48 (br s, 1H, NHa), 7.58 (br s, 1H, NHb), 7.85 (d, J = 8.8 Hz, 2H), 7.97 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 39.6 (CH3), 97.2 (C), 111.6 (2CH), 118.3 (C), 118.7 (C), 132.7 (2CH), 150.5 (CH, C-3), 153.0 (C), 164.0 (C, C
C), 1522, 1440, 1362, 1323, 1238, 1190, 1169, 943, 810, 663, 590, 525, 501, 461 cm−1. 1H NMR (400 MHz, DMSO-d6): 3.04 (s, 6H), 6.81 (d, J = 8.8 Hz, 2H), 7.48 (br s, 1H, NHa), 7.58 (br s, 1H, NHb), 7.85 (d, J = 8.8 Hz, 2H), 7.97 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 39.6 (CH3), 97.2 (C), 111.6 (2CH), 118.3 (C), 118.7 (C), 132.7 (2CH), 150.5 (CH, C-3), 153.0 (C), 164.0 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.57
O) ppm. These NMR data matched previously reported data.57![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.43
O) ppm. These NMR data matched previously reported data.43![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1686 (νC
N), 1686 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1594 (νC
O), 1594 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1569 (νC
C), 1569 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1512, 1390, 1356, 1204, 1024, 973, 939, 799, 741, 631, 615, 536, 453 cm−1. 1H NMR (400 MHz, DMSO-d6): 4.50 (d, J = 6.0 Hz, 2H), 5.55 (t, J = 6.0 Hz, 1H, OH), 6.64 (d, J = 4.4 Hz, 1H), 7.33 (d, J = 4.4 Hz, 1H), 7.68 (br s, 1H, NHa), 7.79 (br s, 1H, NHb), 7.93 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 56.0 (CH2), 100.4 (C), 110.9 (CH), 116.4 (C), 122.4 (CH), 135.9 (CH, C-3), 147.6 (C), 161.6 (C), 162.8 (C, C
C), 1512, 1390, 1356, 1204, 1024, 973, 939, 799, 741, 631, 615, 536, 453 cm−1. 1H NMR (400 MHz, DMSO-d6): 4.50 (d, J = 6.0 Hz, 2H), 5.55 (t, J = 6.0 Hz, 1H, OH), 6.64 (d, J = 4.4 Hz, 1H), 7.33 (d, J = 4.4 Hz, 1H), 7.68 (br s, 1H, NHa), 7.79 (br s, 1H, NHb), 7.93 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 56.0 (CH2), 100.4 (C), 110.9 (CH), 116.4 (C), 122.4 (CH), 135.9 (CH, C-3), 147.6 (C), 161.6 (C), 162.8 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.40
O) ppm. These NMR data matched previously reported data.40![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1602 (νC
O), 1602 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1492, 1429, 1380, 1331, 1277, 1090, 1014, 827, 679, 646, 625, 617, 500, 430 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.24 (br s, 1H), 7.28 (s, 1H), 7.37 (br s, 1H), 7.47 (d, J = 8.8 Hz, 2H), 7.52 (br s, 1H), 7.56 (d, J = 8.8 Hz, 2H), 7.86 (br s, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 128.7 (2CH), 130.9 (2CH), 132.8 (CH), 133.0 (C), 133.8 (C), 134.1 (C), 165.6 (C, C
C), 1492, 1429, 1380, 1331, 1277, 1090, 1014, 827, 679, 646, 625, 617, 500, 430 cm−1. 1H NMR (400 MHz, DMSO-d6): 7.24 (br s, 1H), 7.28 (s, 1H), 7.37 (br s, 1H), 7.47 (d, J = 8.8 Hz, 2H), 7.52 (br s, 1H), 7.56 (d, J = 8.8 Hz, 2H), 7.86 (br s, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 128.7 (2CH), 130.9 (2CH), 132.8 (CH), 133.0 (C), 133.8 (C), 134.1 (C), 165.6 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 168.9 (C, C
O), 168.9 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.58
O) ppm. These NMR data matched previously reported data.58![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1579 (νC
O), 1579 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1486, 1326, 1282, 1178, 1073, 922, 832, 759, 693, 654, 606, 593, 529, 507, 487, 450 m cm−1. 1H NMR (400 MHz, DMSO-d6): 6.88 (d, J = 8.8 Hz, 2H), 7.03–7.15 (m, 7H), 7.22 (s, 2H), 7.34 (dd, J = 8.0, 8.0 Hz, 4H), 7.43–7.49 (m, 3H), 7.83 (s, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 121.1 (2CH), 124.0 (2CH), 124.9 (4CH), 127.0 (C), 129.7 (4CH), 130.8 (2CH), 130.9 (C), 133.8 (CH), 146.5 (2C), 148.2 (C), 165.9 (C, C
C), 1486, 1326, 1282, 1178, 1073, 922, 832, 759, 693, 654, 606, 593, 529, 507, 487, 450 m cm−1. 1H NMR (400 MHz, DMSO-d6): 6.88 (d, J = 8.8 Hz, 2H), 7.03–7.15 (m, 7H), 7.22 (s, 2H), 7.34 (dd, J = 8.0, 8.0 Hz, 4H), 7.43–7.49 (m, 3H), 7.83 (s, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 121.1 (2CH), 124.0 (2CH), 124.9 (4CH), 127.0 (C), 129.7 (4CH), 130.8 (2CH), 130.9 (C), 133.8 (CH), 146.5 (2C), 148.2 (C), 165.9 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 169.6 (C, C
O), 169.6 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. HRMS (ESI+): m/z calculated for C22H18N3O2+ 356.1394 [M − H2 + H]+; found 356.1398.
O) ppm. HRMS (ESI+): m/z calculated for C22H18N3O2+ 356.1394 [M − H2 + H]+; found 356.1398.![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1711 (νC
N), 1711 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1569 (νC
O), 1569 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1503, 1484, 1336, 1319, 1265, 1215, 1176, 1091, 1022, 964, 830, 758, 696, 635, 618, 604, 583, 507, 455 cm−1. 1H NMR (400 MHz, CDCl3): 1.38 (t, J = 7.0 Hz, 3H), 4.35 (q, J = 7.0 Hz, 2H), 6.98 (d, J = 8.8 Hz, 2H), 7.15–7.22 (m, 6H), 7.35 (t, J = 7.8 Hz, 4H), 7.85 (d, J = 8.8 Hz, 2H), 8.09 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, CDCl3): 14.4 (CH3), 62.3 (CH2), 97.4 (C), 116.9 (C), 119.2 (2CH), 123.5 (C), 125.6 (2CH), 126.6 (4CH), 129.9 (4CH), 133.3 (2CH), 145.8 (2C), 152.6 (C), 154.1 (CH, C-3), 163.8 (C, C
C), 1503, 1484, 1336, 1319, 1265, 1215, 1176, 1091, 1022, 964, 830, 758, 696, 635, 618, 604, 583, 507, 455 cm−1. 1H NMR (400 MHz, CDCl3): 1.38 (t, J = 7.0 Hz, 3H), 4.35 (q, J = 7.0 Hz, 2H), 6.98 (d, J = 8.8 Hz, 2H), 7.15–7.22 (m, 6H), 7.35 (t, J = 7.8 Hz, 4H), 7.85 (d, J = 8.8 Hz, 2H), 8.09 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, CDCl3): 14.4 (CH3), 62.3 (CH2), 97.4 (C), 116.9 (C), 119.2 (2CH), 123.5 (C), 125.6 (2CH), 126.6 (4CH), 129.9 (4CH), 133.3 (2CH), 145.8 (2C), 152.6 (C), 154.1 (CH, C-3), 163.8 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. These NMR data matched previously reported data.59
O) ppm. These NMR data matched previously reported data.594.3 Synthesis of (E)-2-cyano-3-(4-(prop-2-yn-1-yloxy)phenyl)acrylamide 5
A mixture of (E)-2-cyano-3-(4-hydroxyphenyl)acrylamide 3i (188 mg, 1.0 mmol), propargyl bromide solution (80 wt% in toluene, 112 µL, 1.0 mmol) 4a, and cesium carbonate (326 mg, 1.0 mmol) in N,N-dimethylformamide (2.0 mL) was stirred at room temperature for 3 h. Upon completion, the reaction mixture was filtered, and the solid residue was washed with ethyl acetate (10 mL). The filtrate was treated with 1.0 M NaOH (2 mL) and extracted with ethyl acetate (3 × 5 mL). The organic layers were combined, washed with brine (2 × 10 mL), dried over anhydrous Na2SO4, filtered, and concentrated under reduced pressure to obtain the crude product. Purification by flash chromatography on silica gel afforded compound 5 as a white solid (201 mg, 89%). M.p. 165–166 °C (amorphous). FTIR (ATR): 3450 (νas NH2 non-hydrogen bonded), 3399 (νas NH2 hydrogen bonded), 3310 (νs NH2 non-hydrogen bonded), 3182 (νs NH2 hydrogen bonded), 3090, 2230 (νC![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1715 (νC
N), 1715 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1608 (νC
O), 1608 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1585 (νC
C), 1585 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1470, 1393, 1144, 1110, 1051, 924, 842, 826, 792, 765, 592, 559, 532, 446 cm−1. 1H NMR (400 MHz, DMSO-d6): 3.65 (t, J = 2.4 Hz, 1H), 4.92 (d, J = 2.4 Hz, 2H), 7.18 (d, J = 8.8 Hz, 2H), 7.71 (br s, 1H, NHa), 7.84 (br s, 1H, NHb), 7.97 (d, J = 8.8 Hz, 2H), 8.12 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 55.8 (CH2), 78.7 (C), 78.9 (CH), 103.5 (C), 115.6 (2CH), 117.1 (C), 125.1 (C), 123.4 (2CH), 150.1 (CH, C-3), 160.4 (C), 163.1 (C, C
C), 1470, 1393, 1144, 1110, 1051, 924, 842, 826, 792, 765, 592, 559, 532, 446 cm−1. 1H NMR (400 MHz, DMSO-d6): 3.65 (t, J = 2.4 Hz, 1H), 4.92 (d, J = 2.4 Hz, 2H), 7.18 (d, J = 8.8 Hz, 2H), 7.71 (br s, 1H, NHa), 7.84 (br s, 1H, NHb), 7.97 (d, J = 8.8 Hz, 2H), 8.12 (s, 1H, H-3) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 55.8 (CH2), 78.7 (C), 78.9 (CH), 103.5 (C), 115.6 (2CH), 117.1 (C), 125.1 (C), 123.4 (2CH), 150.1 (CH, C-3), 160.4 (C), 163.1 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. HRMS (ESI+): m/z calculated for C13H11N2O2+ 227.0815 [M + H]+; found 227.0814.
O) ppm. HRMS (ESI+): m/z calculated for C13H11N2O2+ 227.0815 [M + H]+; found 227.0814.
4.4 Synthesis of (E)-3-(4-((1-benzyl-1H-1,2,3-triazol-4-yl)methoxy)phenyl)-2-cyanoacrylamide 6
A mixture of (E)-2-cyano-3-(4-(prop-2-yn-1-yloxy)phenyl)acrylamide 5 (113 mg, 0.50 mmol), benzyl bromide 4b (59 µL, 0.50 mmol), sodium azide (32 mg, 0.50 mmol), and CuI (9.5 mg, 10 mol%) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v mixture of distilled water and ethanol (2.0 mL) was stirred at 60 °C for 12 h. Upon completion, the resulting solid was filtered, washed with a cold 1
1 v/v mixture of distilled water and ethanol (2.0 mL) was stirred at 60 °C for 12 h. Upon completion, the resulting solid was filtered, washed with a cold 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v mixture of distilled water and ethanol (5.0 mL), and dried to afford the 1,4-disubstituted 1,2,3-triazole 6 as a pale yellow solid (142 mg, 79%). M.p. 145–147 °C (amorphous). FTIR (ATR): 3450 (νas NH2 non-hydrogen bonded), 3399 (νas NH2 hydrogen bonded), 3310 (νs NH2 non-hydrogen bonded), 3182 (νs NH2 hydrogen bonded), 3090, 2230 (νC
1 v/v mixture of distilled water and ethanol (5.0 mL), and dried to afford the 1,4-disubstituted 1,2,3-triazole 6 as a pale yellow solid (142 mg, 79%). M.p. 145–147 °C (amorphous). FTIR (ATR): 3450 (νas NH2 non-hydrogen bonded), 3399 (νas NH2 hydrogen bonded), 3310 (νs NH2 non-hydrogen bonded), 3182 (νs NH2 hydrogen bonded), 3090, 2230 (νC![[triple bond, length half m-dash]](https://www.rsc.org/images/entities/char_e007.gif) N), 1715 (νC
N), 1715 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1608 (νC
O), 1608 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1585 (νC
C), 1585 (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1470, 1393, 1144, 1110, 1051, 924, 842, 826, 792, 765, 592, 559, 532, 446 cm−1. 1H NMR (400 MHz, DMSO-d6): 5.26 (s, 2H), 5.62 (s, 2H), 7.23 (d, J = 8.4 Hz, 2H), 7.30–7.40 (m, 5H), 7.71 (br s, 1H, NHa), 7.83 (br s, 1H, NHb), 7.96 (d, J = 8.4 Hz, 2H), 8.11 (s, 1H), 8.35 (s, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 52.9 (CH2), 61.4 (CH2), 103.1 (C), 115.5 (2CH), 117.1 (C), 124.7 (C), 125.1 (CH), 125.1 (C), 128.0 (2CH), 128.2 (CH), 128.8 (2CH), 132.4 (2CH), 136.0 (C), 150.1 (CH, C-3), 161.3 (C), 163.1 (C, C
C), 1470, 1393, 1144, 1110, 1051, 924, 842, 826, 792, 765, 592, 559, 532, 446 cm−1. 1H NMR (400 MHz, DMSO-d6): 5.26 (s, 2H), 5.62 (s, 2H), 7.23 (d, J = 8.4 Hz, 2H), 7.30–7.40 (m, 5H), 7.71 (br s, 1H, NHa), 7.83 (br s, 1H, NHb), 7.96 (d, J = 8.4 Hz, 2H), 8.11 (s, 1H), 8.35 (s, 1H) ppm. 13C{1H} NMR (101 MHz, DMSO-d6): 52.9 (CH2), 61.4 (CH2), 103.1 (C), 115.5 (2CH), 117.1 (C), 124.7 (C), 125.1 (CH), 125.1 (C), 128.0 (2CH), 128.2 (CH), 128.8 (2CH), 132.4 (2CH), 136.0 (C), 150.1 (CH, C-3), 161.3 (C), 163.1 (C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. HRMS (ESI+): m/z calculated for C20H18N5O2+ 360.1455 [M + H]+; found 360.1452.
O) ppm. HRMS (ESI+): m/z calculated for C20H18N5O2+ 360.1455 [M + H]+; found 360.1452.
4.5 Single-crystal X-ray diffraction analysis
X-ray crystallographic analysis was performed at ambient temperature (298 K) using CuKα radiation (λ = 1.54184 Å) and ω-scan measurements on an Agilent SuperNova diffractometer (Dual source, Cu at Zero configuration) equipped with an Atlas four-circle goniometer and CCD detector. Diffraction frames were processed and integrated with the CrysAlis PRO software suite,60 and empirical absorption corrections were applied using the SCALE3 ABSPACK scaling algorithm implemented in the same package. The molecular structures of compounds 3i and 3n were solved iteratively and completed by Fourier difference mapping.61 Subsequent refinement of the crystal structure was performed with SHELXL2018/3,62 and molecular as well as supramolecular graphics were generated with Mercury.63 Electrostatic potential mapping over the Hirshfeld surface was performed using the CrystalExplorer program.64 Crystallographic data for compound 3i have been deposited in the Cambridge Crystallographic Data Center under deposition number CCDC-2483802. Crystallographic data for compound 3n has been included in the SI due to the low quality of the data.4.6 Anticancer activity
All biological assays, including the handling of reagents and human cancer cell lines, were conducted externally by the National Cancer Institute (NCI, USA) as part of the NCI-60 human tumor cell screening program.65 All compounds submitted to this program are initially evaluated at a single concentration (10 µM) across the entire panel. The one-dose experiments provide the relative growth of treated cells compared with untreated controls and the initial cell population at time zero. Compounds that display significant growth inhibition in the one-dose screen are subsequently tested at five concentrations in the NCI-60 cell panel.65The human tumor cell lines included in the screening panel are cultured in RPMI-1640 medium supplemented with 5% fetal bovine serum and 2 mM l-glutamine. For each assay, cells are inoculated into 96-well microtiter plates (100 µL per well) at plating densities ranging from 5000 to 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well, depending on the doubling time of each cell line. Following inoculation, the plates are incubated for 24 h at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air prior to the addition of the experimental drugs. After 24 h, two plates of each cell line are fixed in situ with trichloroacetic acid (TCA) to determine the cell population at the time of drug addiction (Tz). The samples are dissolved in dimethylsulfoxide (DMSO) at 400 times the desired final maximum test concentration and stored frozen until use. At the time of sample addition, an aliquot of the frozen concentrate is thawed and diluted to twice the intended final concentration using complete medium supplemented with 50 µg mL−1 gentamicin. An additional four 10-fold or 1/2 log serial dilutions were made to provide a total of five drug concentrations plus the control. Aliquots of 100 µL from each dilution are then added to the appropriate wells already containing 100 µL of medium, resulting in the required final sample concentrations. After the tested compounds were added, the plates are incubated for an additional 48 h at 37 °C in a humidified atmosphere of 5% CO2, 95% air, and 100% relative humidity. For adherent cells, the assay is terminated by the addition of cold trichloroacetic acid (TCA). Cells are fixed in situ by the gentle addition of 50 µL of cold 50% (w/v) TCA (final concentration, 10% TCA) and incubated for 60 min at 4 °C. The supernatant is discarded, and the plates are washed five times with tap water and air dried. Sulforhodamine B (SRB) solution (100 µL) at 0.4% (w/v) in 1% acetic acid is added to each well, and the plates are incubated for 10 min at room temperature. After staining, the unbound dye is removed by washing five times with 1% acetic acid, and the plates are air dried. The bound stain is subsequently solubilized with 10 mM trizma base, and the absorbance is measured using an automated plate reader at a wavelength of 515 nm. For suspension cells, the methodology is identical, except that the assay is terminated by fixing the settled cells at the bottom of the wells through the gentle addition of 50 µL of 80% TCA (final concentration, 16% TCA). Using seven absorbance measurements [time zero (Tz), control growth (C), and test growth in the presence of drug at the five concentration levels (Ti)], the percentage growth is calculated at each of the drug concentrations levels. Growth percentage is calculated as: [(Ti − Tz)/(C − Tz)] × 100 for concentrations where Ti ≥ Tz, and [(Ti − Tz)/Tz] × 100 for concentrations where Ti < Tz. Three dose response parameters are calculated for each experimental compound. The 50% growth inhibition (GI50) value is determined from the equation [(Ti − Tz)/(C − Tz)] × 100 = 50, corresponding to the drug concentration that produces a 50% reduction in the net protein increase (as measured by SRB staining) relative to the untreated control cells during incubation. The total growth inhibition (TGI) value is defined as the drug concentration at which Ti = Tz, indicating complete cessation of cell growth. The 50% lethal concentration (LC50) represents the drug concentration that causes a 50% reduction in total cellular protein at the end of treatment compared to the initial value, calculated using the equation [(Ti − Tz)/Tz] × 100 = 50, and reflects a net loss of viable cells. These three parameters are calculated whenever the corresponding activity levels are achieved; if not, or if they exceed the tested range, the results are reported as greater or less than the maximum or minimum concentration evaluated.65
000 cells per well, depending on the doubling time of each cell line. Following inoculation, the plates are incubated for 24 h at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air prior to the addition of the experimental drugs. After 24 h, two plates of each cell line are fixed in situ with trichloroacetic acid (TCA) to determine the cell population at the time of drug addiction (Tz). The samples are dissolved in dimethylsulfoxide (DMSO) at 400 times the desired final maximum test concentration and stored frozen until use. At the time of sample addition, an aliquot of the frozen concentrate is thawed and diluted to twice the intended final concentration using complete medium supplemented with 50 µg mL−1 gentamicin. An additional four 10-fold or 1/2 log serial dilutions were made to provide a total of five drug concentrations plus the control. Aliquots of 100 µL from each dilution are then added to the appropriate wells already containing 100 µL of medium, resulting in the required final sample concentrations. After the tested compounds were added, the plates are incubated for an additional 48 h at 37 °C in a humidified atmosphere of 5% CO2, 95% air, and 100% relative humidity. For adherent cells, the assay is terminated by the addition of cold trichloroacetic acid (TCA). Cells are fixed in situ by the gentle addition of 50 µL of cold 50% (w/v) TCA (final concentration, 10% TCA) and incubated for 60 min at 4 °C. The supernatant is discarded, and the plates are washed five times with tap water and air dried. Sulforhodamine B (SRB) solution (100 µL) at 0.4% (w/v) in 1% acetic acid is added to each well, and the plates are incubated for 10 min at room temperature. After staining, the unbound dye is removed by washing five times with 1% acetic acid, and the plates are air dried. The bound stain is subsequently solubilized with 10 mM trizma base, and the absorbance is measured using an automated plate reader at a wavelength of 515 nm. For suspension cells, the methodology is identical, except that the assay is terminated by fixing the settled cells at the bottom of the wells through the gentle addition of 50 µL of 80% TCA (final concentration, 16% TCA). Using seven absorbance measurements [time zero (Tz), control growth (C), and test growth in the presence of drug at the five concentration levels (Ti)], the percentage growth is calculated at each of the drug concentrations levels. Growth percentage is calculated as: [(Ti − Tz)/(C − Tz)] × 100 for concentrations where Ti ≥ Tz, and [(Ti − Tz)/Tz] × 100 for concentrations where Ti < Tz. Three dose response parameters are calculated for each experimental compound. The 50% growth inhibition (GI50) value is determined from the equation [(Ti − Tz)/(C − Tz)] × 100 = 50, corresponding to the drug concentration that produces a 50% reduction in the net protein increase (as measured by SRB staining) relative to the untreated control cells during incubation. The total growth inhibition (TGI) value is defined as the drug concentration at which Ti = Tz, indicating complete cessation of cell growth. The 50% lethal concentration (LC50) represents the drug concentration that causes a 50% reduction in total cellular protein at the end of treatment compared to the initial value, calculated using the equation [(Ti − Tz)/Tz] × 100 = 50, and reflects a net loss of viable cells. These three parameters are calculated whenever the corresponding activity levels are achieved; if not, or if they exceed the tested range, the results are reported as greater or less than the maximum or minimum concentration evaluated.65
4.7 Molecular docking
The Knoevenagel adducts 3f, 3n, and 3o were modelled in Discovery Studio 2022 (DS 2022) using standard bond lengths and angles. Their molecular geometries were optimized by energy minimization with the adopted-based Newton-Rapson algorithm until an RMS gradient below 0.01 kcal mol−1 Å−2 was achieved. The 3D crystal structure of EGFR in complex with Erlotinib (PDB ID: 1M17) was obtained from the RCSB Protein Data Bank. Protein preparation in DS 2022 involved removal of water molecules, heteroatoms, co-crystallized solvents, and ligands. Subsequently, hydrogens and partial charges were added to proteins and ligands using AutoDockTools (ADT version 1.5.7), with Gasteiger charges assigned. Ligands were allowed to adopt flexible conformations during docking. The docking box was generated with ADT, centered within the protein, and defined dimensions of 62 Å × 64 Å × 50 Å and a resolution of 1 Å, centered at coordinates x = 24.500, y = 6.900, and z = 58.487. Docking calculations were performed with AutoDock Vina,52 and the top-ranked binding pose of the ligands within the EGFR active site were selected for further analysis in Discovery Study (DS).Author contributions
CS-S: investigation, methodology; II: computational analysis, writing – original draft; MAM: X-ray crystallographic analysis, writing – original draft; J-CC: conceptualization, methodology, writing – original draft; DB: conceptualization, methodology, writing – original draft.Conflicts of interest
There are no conflicts to declare.Abbreviations
| TCIs | Targeted covalent inhibitors |
| TKIs | Tyrosine kinase inhibitors |
| EGFR | Epidermal growth factor receptor |
| NSCLC | Non-small cell lung cancer |
| HER2 | Human epidermal growth factor receptor 2 |
| BTK | Bruton's tyrosine kinase |
| CLL | Chronic lymphocytic leukemia |
| SLL | Small lymphocytic lymphoma |
| FGFR | Fibroblast growth factor receptor |
| JAK3 | Janus kinase 3 |
| PP | Piperidine |
| DBU | 1,8-Diazabicyclo(5.4.0)undec-7-ene |
| BBB | Blood brain barrier |
| TPSA | Topological polar surface area |
| %ABS | Percentage of absorption |
| NCI | National Cancer Institute |
| SRB | Sulforhodamine B |
Data availability
The data supporting this article are provided in the supplementary information (SI). Supplementary information: IR, NMR, and HRMS spectra, as well as X-ray crystallographic data for compounds reported in this study. See DOI: https://doi.org/10.1039/d5ra07121f.CCDC 2483802 (3i) contains the supplementary crystallographic data for this paper.66
Acknowledgements
The authors acknowledge the financial support provided by the Dirección de Investigaciones at the Universidad Pedagógica y Tecnológica de Colombia (Projects SGI-3724 and SGI-3927). I. I. acknowledges support from the Universidad de Alcalá. M. A. A. gratefully acknowledges support from the Facultad de Ciencias at the Universidad de los Andes. We also extend our sincere gratitude to the National Cancer Institute (NCI) for performing the in vitro anticancer assays.References
- F. Sutanto, M. Konstantinidou and A. Dömling, RSC Med. Chem., 2020, 11, 876–884 RSC.
- W. Lu, M. Kostic, T. Zhang, J. Che, M. P. Patricelli, L. H. Jones, E. T. Chouchaniae and N. S. Gray, RSC Chem. Biol., 2021, 2, 354–367 Search PubMed.
- L. Boike, N. J. Henning and D. K. Nomura, Nat. Rev. Drug Discovery, 2022, 21, 881–898 CrossRef CAS.
- K. McAulay, A. Bilsland and M. Bon, Pharmaceuticals, 2022, 15, 1366 CrossRef CAS PubMed.
- L. Arce-Ramos, J.-C. Castillo and D. Becerra, Pharmaceuticals, 2023, 16, 1265 Search PubMed.
- M. Gehringer and S. A. Laufer, J. Med. Chem., 2019, 62, 5673–5724 Search PubMed.
- N. V. Mehta and M. S. Degani, Drug Discovery Today, 2023, 28, 103799 Search PubMed.
- P. Ábrányi-Balogh and G. M. Keseru, in Advances in Chemical Proteomics, ed. X. Yao, Elsevier, Amsterdam, 2022, ch. 2, pp. 47–73 Search PubMed.
- T. A. Baillie, Expert Opin. Drug Discovery, 2020, 16, 275–287 CrossRef.
- L. Hillebrand, X. J. Liang, R. A. M. Serafim and M. Gehringer, J. Med. Chem., 2024, 67, 7668–7758 Search PubMed.
- R. Roskoski Jr., Pharmacol. Res., 2024, 200, 107059 CrossRef PubMed.
- R. Roskoski Jr., Pharmacol. Res., 2025, 216, 107723 CrossRef PubMed.
- T. Tang, J. Luo, D. Zhang, Y. Lu, W. Liao and J. Zhang, Eur. J. Med. Chem., 2025, 284, 117202 Search PubMed.
- S. E. Dalton, O. Di Pietro and E. Hennessy, J. Med. Chem., 2025, 13, 2307–2313 CrossRef PubMed.
- L. Liang, Z. Zhang, Q. You and X. Guo, Bioorg. Med. Chem., 2024, 112, 117902 CrossRef CAS PubMed.
- Y. Jiang, X. Fang, Y. Xiang, T. Fang, J. Liu and K. Lu, Curr. Oncol., 2023, 30, 5337–5349 CrossRef.
- Y. N. Lamb, Targeted Oncol., 2021, 16, 687–695 Search PubMed.
- J. E. Shin, H. A. Jung, S. Park, J.-M. Sun, S.-H. Lee, J. S. Ahn, M.-J. Ahn and B. Y. Shim, Sci. Rep., 2025, 15, 4593 CrossRef CAS.
- N. Gupta, A. Largajolli, H. Witjes, P. M. Diderichsen, S. Zhang, M. J. Hanley, J. Lin and M. Mehta, Clin. Pharmacol. Ther., 2022, 112, 327–334 Search PubMed.
- H.-L. Jeon, M. Kwak, S. Kim, H.-Y. Yu, J.-Y. Shin and H. A. Jung, Sci. Rep., 2024, 14, 14659 Search PubMed.
- E. D. Deeks, Drugs, 2017, 77, 1695–1704 CrossRef CAS PubMed.
- K. J. Maddocks, A. S. Ruppert, G. Lozanski, N. A Heerema, W. Zhao, L. Abruzzo, A. Lozanski, M. Davis, A. Gordon, L. L. Smith, R. Mantel, J. A. Jones, J. M. Flynn, S. M. Jaglowski, L. A. Andritsos, F. Awan, K. A. Blum, M. R. Grever, A. J. Johnson, J. C. Byrd and J. A. Woyach, JAMA Oncol., 2015, 1, 80–87 Search PubMed.
- A. Markham and S. Dhillon, Drugs, 2018, 78, 139–145 Search PubMed.
- C. S. Tam, J. L. Muñoz, J. F. Seymour and S. Opat, Blood Cancer J., 2023, 13, 141 Search PubMed.
- M. Javle, G. King, K. Spencer and M. J. Borad, Oncologist, 2023, 28, 928–943 Search PubMed.
- B. King, J. Soung, C. Tziotzios, L. Rudnicka, P. Joly, M. Gooderham, R. Sinclair, N. A. Mesinkovska, C. Paul, Y. Gong, S. D. Anway, H. Tran, R. Wolk, S. H. Zwillich and A. Lejeune, Am. J. Clin. Dermatol., 2024, 25, 299–314 Search PubMed.
- I. M Serafimova, M. A. Pufall, S. Krishnan, K. Duda, M. S. Cohen, R. L. Maglathlin, J. M. McFarland, R. M. Miller, M. Frödin and J. Taunton, Nat. Chem. Biol., 2012, 8, 471–476 Search PubMed.
- T. D. Owens, K. A. Brameld, E. J. Verner, T. Ton, X. Li, J. Zhu, M. R. Masjedizadeh, J. M. Bradshaw, R. J. Hill, D. Tam, A. Bisconte, E. O. Kim, M. Francesco, Y. Xing, J. Shu, D. Karr, J. LaStant, D. Finkle, N. Loewenstein, H. Haberstock-Debic, M. J. Taylor, P. Nunn, C. L. Langrish and D. M. Goldstein, J. Med. Chem., 2022, 65, 5300–5316 CrossRef CAS.
- M. Forster, A. Chaikuad, S. M. Bauer, J. Holstein, M. B. Robers, C. R. Corona, M. Gehringer, E. Pfaffenrot, K. Ghoreschi, S. Knapp and S. A. Laufer, Cell Chem. Biol., 2016, 23, 1335–1340 Search PubMed.
- S. Johari, M. R. Johan and N. G. Khaligh, Org. Biomol. Chem., 2022, 20, 2164–2186 RSC.
- J. N. Appaturi, R. Ratti, B. L. Phoon, S. M. Batagarawa, I. U. Din, M. Selvaraj and R. J. Ramalingam, Dalton Trans., 2021, 50, 4445–4469 Search PubMed.
- C. Serrano-Sterling, D. Becerra, J. Portilla, H. Rojas, M. Macías and J.-C. Castillo, J. Mol. Struct., 2021, 1244, 130944 CrossRef CAS.
- J. Stec and W. H. Witola, Results Chem., 2023, 6, 101212 CrossRef CAS.
- C. Nitsche, C. Steuer and C. D. Klein, Bioorg. Med. Chem., 2011, 19, 7318–7337 CrossRef CAS.
- K. M. Uddin, M. H. Meem, M. Akter, S. Rahman, M. A. Al-Gawati, N. Alarifi, H. Albrithen, A. Alodhayb, R. A. Poirier and M. H. Bhuiyan, MethodsX, 2024, 12, 102691 CrossRef CAS.
- D. E. Quintero Jimenez, L. Lima Zanin, L. Farinelli Diniz, J. Ellena and A. L. Meleiro Porto, Curr. Microwave Chem., 2019, 6, 54–60 CrossRef CAS.
- T. R. Burke, B. Lim, V. E. Marquez, Z.-H. Li, J. B. Bolen, I. Stefanova and I. D. Horak, J. Med. Chem., 1993, 36, 425–432 CrossRef CAS.
- S. Son, H. Kim, H. Y. Yun, D. H. Kim, S. Ullah, S. J. Kim, Y.-J. Kim, M.-S. Kim, J.-W. Yoo, P. Chun and H. R. Moon, Bioorg. Med. Chem., 2015, 23, 7728–7734 CrossRef CAS.
- F. Pandolfi, M. Feroci and I. Chiarotto, ChemistrySelect, 2018, 3, 4745–4749 CrossRef CAS.
- E. Y. Mesa Castro, A. F. Monroy Ramírez, J. J. Martínez, J.-C. Castillo and G. A. Caicedo Pineda, Catalysts, 2024, 14, 927 CrossRef CAS.
- S. Mancipe, J.-C. Castillo, M. H. Brijaldo, V. P. López, H. Rojas, M. A. Macías, J. Portilla, G. P. Romanelli, J. J. Martínez and R. Luque, ACS Sustainable Chem. Eng., 2022, 10, 12602–12612 CrossRef CAS.
- K.-D. Wu, G. S. Chen, J.-R. Liu, C.-E. Hsieh and J.-W. Chern, ACS Med. Chem. Lett., 2019, 10, 22–26 CrossRef CAS.
- A. Tigreros, M.-C. Ríos, C. Serrano-Sterling, D. Becerra, J.-C. Castillo and J. Portilla, J. Mol. Struct., 2025, 1321, 139926 CrossRef CAS.
- A. C. Lipinski, Drug Discovery Today: Technol., 2004, 1, 337–341 CrossRef PubMed.
- S. Prasanna and R. J. Doerksen, Curr. Med. Chem., 2009, 16, 21–41 CrossRef CAS.
- M. Bitew, T. Desalegn, T. B. Demissie, A. Belayneh, M. Endale and R. Eswaramoorthy, PLoS One, 2021, 16, e0260853 CrossRef CAS PubMed.
- W. Wei, S. Cherukupalli, L. Jing, X. Liu and P. Zhan, Drug Discovery Today, 2020, 25, 1839–1845 CrossRef CAS PubMed.
- National Cancer Institute, DCTD Division of Cancer Treatment & Diagnosis, available online: https://dtp.cancer.gov/discovery_development/nci-60/methodology.htm, accessed on 15 July 2025 Search PubMed.
- L. M. Moreno, J. Quiroga, R. Abonia, A. Lauria, A. Martorana, H. Insuasty and B. Insuasty, RSC Adv., 2020, 10, 34114 RSC.
- N. Khurana and S. C. Sikka, Cancers, 2018, 10, 352 CrossRef CAS PubMed.
- F. A. Verza, G. C. Da Silva and F. G. Nishimura, Oncol. Res., 2025, 33, 1819–1834 Search PubMed.
- P. A. Morese, A. Ahmad, M. P. Martin, R. A. Noble, S. Pintar, L. Z. Wang, S. Xu, A. Lister, R. A. Ward, A. K. Bronowska, M. E. M. Noble, H. L. Stewart and M. J. Waring, Commun. Chem., 2025, 8, 111 CrossRef CAS PubMed.
- D. N. do Amaral, J. Lategahn, H. H. Fokoue, E. M. B. da Silva, C. M. R. Sant'Anna, D. Rauh, E. J. Barreiro, S. Laufer and L. M. Lima, Sci. Rep., 2019, 9, 14 CrossRef.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- R. Gunasekar, P. Thamaraiselvi, R. S. Rathore, K. I. Sathiyanarayanan and S. Easwaramoorthi, J. Org. Chem., 2015, 80, 12351–12358 Search PubMed.
- B. D. Rupanwar, S. S. Chavan, A. M. Shelkea and G. M. Suryavanshi, New J. Chem., 2018, 42, 6433–6440 Search PubMed.
- M. Curini, F. Epifano, M. C. Marcotullio, O. Rosati and A. Tsadjout, Synth. Commun., 2002, 32, 355–362 CrossRef CAS.
- C. N. O'Callaghan and T. B. H. McMurry, J. Chem. Res., 1989, 6, 1219–1231 Search PubMed.
- P. S. Hariharan, V. K. Prasad, S. Nandi, A. Anoop, D. Moon and S. P. Anthony, Cryst. Growth Des., 2017, 17, 146–155 CrossRef CAS.
- CrysAlisPro 1.171.39.46e, Rigaku Oxford Diffraction, 2018 Search PubMed.
- L. Palatinus and G. Chapuis, J. Appl. Crystallogr., 2007, 40, 786–790 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C:Struct. Chem., 2015, 71, 3–8 Search PubMed.
- C. F. Macrae, I. J. Bruno, J. A. Chisholm, P. R. Edgington, P. McCabe, E. Pidcock, L. Rodriguez-Monge, R. Taylor, J. van de Streek and P. A. Wood, J. Appl. Crystallogr., 2008, 41, 466–470 CrossRef CAS.
- M. J. Turner, J. J. McKinnon, S. K. Wolff, D. J. Grimwood, P. R. Spackman, D. Jayatilaka and M. A. Spackman, CrystalExplorer17, University of Western Australia, 2017 Search PubMed.
- NCI-60 Screening Methodology, https://dctd.cancer.gov/drug-discovery-development/assays/high-throughput-screening-services/nci60/submitting-compounds/screening-methodology.pdf, accessed November 4, 2025 Search PubMed.
- CCDC 2483802: Experimental Crystal Structure Determination, 2025, DOI:10.5517/ccdc.csd.cc2pclnb.
| This journal is © The Royal Society of Chemistry 2026 |


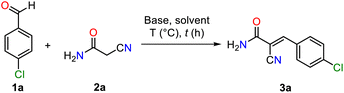
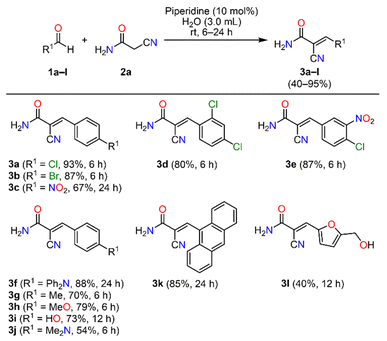
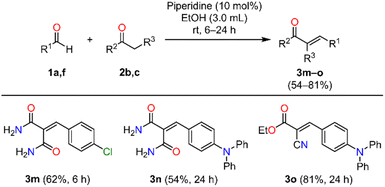
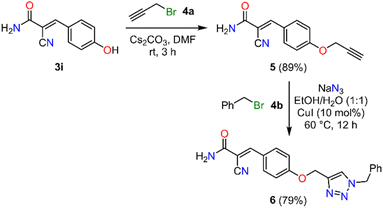

![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , with unit cell parameters
, with unit cell parameters