Open Access Article
Open Access ArticleDesign, synthesis and evaluation of new pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidines as tacrine-like acetylcholinesterase inhibitors
Liubov V. Muzychka *,
Oksana V. Muzychka,
Oleksandr L. Kobzar
*,
Oksana V. Muzychka,
Oleksandr L. Kobzar ,
Andriy I. Vovk and
Oleg B. Smolii
,
Andriy I. Vovk and
Oleg B. Smolii
V.P. Kukhar Institute of Bioorganic Chemistry and Petrochemistry, NAS of Ukraine, 1 Academic Kukhar Str., Kyiv 02094, Ukraine. E-mail: liubovmuzychka@gmail.com
First published on 12th January 2026
Abstract
The development of acetylcholinesterase (AChE) inhibitors remains a promising research direction in drug discovery for Alzheimer's disease. A series of eighteen pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives was synthesized as novel tacrine-like AChE inhibitors. Sixteen compounds inhibited AChE in the micromolar range. Among them, 4-(dimethylamino)-7,8-dimethylpyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidin-6(7H)-one (22) exhibited the highest inhibitory activity against the enzyme with an IC50 value of 0.22 ± 0.02 µM, showing mixed-type inhibition. In silico studies showed that 22 occupies the catalytic anionic site of hAChE and forms strong π–π stacking interactions with Trp86, similar to those of tacrine. This study demonstrates the potential use of methyl-substituted pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidines in the development of potent AChE inhibitors.
Introduction
Alzheimer's disease (AD) is a progressive neurodegenerative brain disorder that causes memory loss, cognitive decline, and abnormal behaviour.1–3 Acetylcholine deficiency is a key contributor to the cognitive symptoms of AD. Cholinesterase inhibitors are used to alleviate the symptoms of neurological diseases and improve cognitive function in patients with AD.4,5 Currently, there are four FDA-approved acetylcholinesterase (AChE) inhibitors, donepezil, rivastigmine, galantamine, and benzgalantamine, that lead to increased acetylcholine levels in the synaptic cleft. Together with memantine, the N-methyl-D-aspartate antagonist, they constitute all the approved small-molecule drugs for the treatment of AD.2,6–9 However, these drugs have suboptimal pharmacokinetics and pharmacodynamics and may cause gastrointestinal tract disorders.10–12 Due to concerns about their toxicity and side effects, there is a growing interest in developing better treatments for AD through the structural optimisation of existing drug scaffolds.6Heterocyclic compounds have played a significant role in the discovery of potent cholinesterase inhibitors.13–18 Tacrine was the first AChE inhibitor approved for the treatment of AD, but it was withdrawn from the market due to its hepatotoxicity and poor therapeutic efficacy.19 Despite its limitations, the tacrine scaffold remains an attractive starting point for the development of new AChE inhibitors with improved pharmacological profiles.20–22 Indeed, most tacrine derivatives exhibit high AChE inhibitory activity and lower toxicity.23–26 Many tacrine-like tricyclic compounds have been designed and tested against AChE. In particular, thiazolo[5′,4′:4,5]pyrimido[1,2-a]azepinones, tetrahydropyridothienopyrimidinones, cyclohepta[b]thieno-oxazines, 1,2,3-triazolo[4,5-b]aminoquinolines, pyrrolo[2,1-b]quinazolin-9(1H)-ones, and pyrido[1,2-a]thiazolo[5,4-d]pyrimidinones inhibited AChE in the micromolar range.27–32 Notably, pyrazino[1,2-a]indol-1(2H)-one, tetrahydrobenzo[1,8-b]naphthyridine, and pyrazolo[1,2-b]phthalazine derivatives showed nanomolar IC50 values.33–35 Overall, replacing the benzene ring in the tacrine structure with various heterocyclic systems contributed to high anticholinesterase activity and selectivity.6,27,30–35
We recently reported the synthesis and evaluation of 4,7-substituted 8,9-dihydropyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives. The studied compounds containing different alkyl, dimethylaminoalkyl, or methoxy substituents at positions 4 and 7 demonstrated promising inhibitory activity against AChE.36 In addition, the tricyclic thienopyrimidinone28 and indol-1(2H)-one33 derivatives showed effective inhibition of AChE, enhanced by non-polar substituents at positions 7 and 8. Inspired by these results, we present the design and synthesis of eighteen new tacrine-like AChE inhibitors with a pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine core (Fig. 1). We explored two series with a dihydropyrazine (structure A) or a pyrazine ring (structure B) and introduced methyl substituents at positions 4, 7, and 8 to study the structure–activity relationship. We hypothesized that the addition of methyl groups would provide additional hydrophobic interactions, while modification of the azatricyclic system with amino groups would lead to more efficient hydrogen bonding with key tyrosine residues at the AChE active site. The synthesized compounds were evaluated in vitro for their inhibitory activity against AChE. The binding of the most potent compound was further investigated by kinetic analysis, molecular docking, and molecular dynamics simulations. Its cytotoxicity was also studied in a hepatocellular carcinoma cell line HepG2. Additionally, ADMET properties of the synthesized compounds were predicted in silico to assess their drug-likeness.
Results and discussion
Chemistry
The synthesis of 8,9-dihydropyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidines 5, 6, 8–10, and 13 was completed in three or four steps, depending on the substitution at positions 3 and 7 (Scheme 1). Methyl 4-chloro-7H-pyrrolo[2,3-d]pyrimidine-6-carboxylate 1 was used as the starting material to synthesize the intermediates 3, 4, and 7 in two steps, as previously reported.36 3-Substituted derivative 12 was prepared from 2 via reflux in 6 M hydrochloric acid, followed by methylation with methyl iodide in dimethylformamide in the presence of K2CO3.36 The formation of the dihydropyrazine ring was the last step of the synthesis of the target compounds 5, 6, 8, 9, and 13. It was accomplished by treating compounds 3, 4, 7, and 12 with excess aqueous ammonia or methylamine at room temperature or by refluxing 7 with methylamine solution in MeOH. Heating of 9 with ammonium acetate gave the target 4-amino-substituted derivative 10. | ||
| Scheme 1 Synthesis of the target 8,9-dihydropyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidines 5, 6, 8–10, and 13. | ||
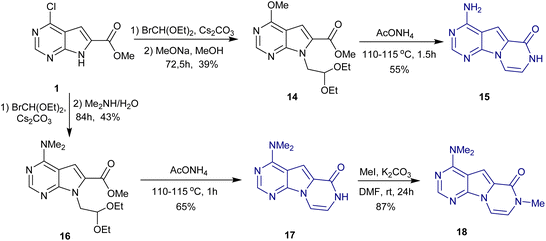
Analogs 15, 17, and 18 containing a pyrazine ring were also synthesized from carboxylate 1 (Scheme 2). Alkylation of 1 and subsequent nucleophilic substitution with sodium methoxide or dimethylamine gave key intermediates 14 and 16. Then, 15 and 17 were prepared via treatment with ammonium acetate, which mediated pyrazine ring formation and the conversion of the methyl ether of 15 to the amino group at position 4. Methylation of 17 afforded the target compound 18.
A new efficient synthetic approach was developed to introduce a methyl group at position 8 and to prepare derivatives 22 and 25 (Scheme 3). The key intermediate 19 was obtained by the alkylation of 1 with chloroacetone. Then, 22 was synthesized from 19 via three steps: nucleophilic substitution, cyclization, and methylation. Alternatively, 19 was treated with sodium methoxide in methanol to convert the chloride into the ether 23. The reaction of 23 with ammonium acetate yielded both 24 and 25, depending on the reaction time (Scheme 3). Functionalization of 24 afforded three more derivatives 26, 27, and 28 (Scheme 4).
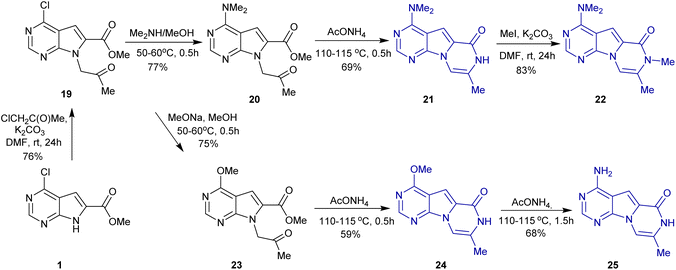 | ||
| Scheme 3 Synthesis of the target pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives 21, 22, 24, and 25. | ||
Additionally, 3-methyl substituted pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine 33 was prepared in four steps (Scheme 5). The starting material iodomethylpyrimido[5′,4′:4,5]pyrrolo[2,1-c][1,4]oxazine 29 was obtained according to the previously reported procedures.37,38 Methylation of 29 and iodine elimination with sodium acetate gave the alkene intermediate 31. Upon treatment with ammonium acetate, the oxazine ring was converted into a pyrazine, and methylation of 32 gave 33 in a high yield.
The structures of the synthesized compounds were confirmed by the elemental analysis, 1H and 13C NMR spectroscopy, and mass spectrometry.
Acetylcholinesterase inhibition
The inhibitory activity of pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives against AChE from electric eel was tested in vitro using a modified Ellman's method.39 The studied compounds inhibited AChE with IC50 values ranging from >100 to 0.22 µM (Table 1).| Compound | IC50, µM |
|---|---|
| a IC50 values are shown as the average value ± standard deviation.b Reference compound. | |
| 5 | 15.86 ± 1.58 |
| 6 | 7.17 ± 1.01 |
| 8 | >100 |
| 9 | 78.93 ± 12.57 |
| 10 | 77.87 ± 3.02 |
| 13 | 14.92 ± 0.51 |
| 15 | >100 |
| 17 | 12.03 ± 0.91 |
| 18 | 2.43 ± 0.26 |
| 21 | 1.03 ± 0.08 |
| 22 | 0.22 ± 0.02 |
| 24 | 6.23 ± 1.53 |
| 25 | 15.84 ± 1.26 |
| 26 | 0.61 ± 0.14 |
| 27 | 0.46 ± 0.02 |
| 28 | 0.61 ± 0.11 |
| 32 | 14.26 ± 1.73 |
| 33 | 1.24 ± 0.31 |
| Donepezilb | 0.016 ± 0.001 |
| Tacrineb | 0.054 ± 0.001 |
A structure–activity relationship study indicates that the presence of a pyrazine moiety in the inhibitor structures increases their potency compared to the compounds with a dihydropyrazine fragment (5, 6, 8–10, and 13). The methyl group at position 7 of the pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine core also played an important role in the enzyme inhibition. Compounds 9, 18, 22, 26, 27, and 33 exhibited higher inhibitory activity than their respective unsubstituted analogs 8, 17, 21, 24, 25, and 32. Similarly, methyl substitution at position 8 led to a significant improvement in the potency of 21, 22, and 25 compared to their unsubstituted derivatives 17, 18, and 15. Overall, a dimethylamino group at position 4 was more favorable for AChE inhibition than (methyl)amino, methoxy, or methylamide moieties, as is evident when comparing IC50 values of compounds 22, 28, 27, 26, and 33. These results demonstrate that methyl substituents can enhance inhibitor binding affinity to AChE.
Among the synthesized compounds, 4-(dimethylamino)-7,8-dimethylpyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidin-6(7H)-one (22) was the most potent AChE inhibitor, showing an IC50 value of 0.22 ± 0.02 µM. Kinetic studies were performed to elucidate the mechanism of the enzyme inhibition. A Lineweaver–Burk plot (Fig. 2) showed mixed-type inhibition, indicating that the inhibitor can bind to both the free enzyme and the enzyme–substrate complex. The calculated Ki and  values are 0.207 ± 0.05 µM and 0.343 ± 0.07 µM, respectively.
values are 0.207 ± 0.05 µM and 0.343 ± 0.07 µM, respectively.
 | ||
| Fig. 2 Lineweaver–Burk plot for inhibition of AChE by compound 22. The concentrations of the inhibitor were 0 (○), 70 nM (Δ), 140 nM (□), and 280 nM (◊). | ||
All tested compounds were also evaluated in vitro for their activity against butyrylcholinesterase from equine serum. However, no inhibitory effect was observed at a concentration of 10 µM.
Molecular docking
To simulate interactions between the enzyme and the inhibitor, compound 22 was docked to human acetylcholinesterase (hAChE) using AutoDock Vina software.40 The human enzyme shares conserved amino acid residues of the active site and has remarkable homology with the AChE obtained from the electric eel.41 Molecular docking, which sheds light on intermolecular interactions between the inhibitor and the enzyme, is considered a pivotal technique in the rational design of AChE inhibitors.42 The crystal structure with PDB code 7XN1 ref. 43 was used for docking. The tacrine molecule in the anionic subsite of the enzyme's catalytic site (CAS) was removed before calculations. First, a blind docking calculation to the entire surface of hAChE was performed to identify the inhibitor binding sites. The results showed that compound 22 is capable of occupying the CAS as well as the peripheral anionic site (PAS), located at the entrance to the catalytic gorge. Although ligand positioning at the PAS was observed more frequently, the CAS location was approximately 1 kcal mol−1 more favorable. The docking was repeated 18 times, and the binding poses with the lowest docking energies were analyzed.Based on these data, compound 22 was specifically docked into the anionic subsite of the CAS. When occupying the anionic subsite (Fig. 3; calculated docking energy was −8.5 kcal mol−1), the pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine scaffold of the inhibitor forms multiple π–π stacking contacts with the indole ring of Trp86, similar to those of tacrine in the crystal structure PDB 7XN1. The pyrazinone and pyrimidine rings of the ligand have hydrogen bonds with Tyr133 and Tyr337 through the keto group and nitrogen atom, respectively. The dimethylamino group of the inhibitor is directed to Tyr72. Additionally, the binding pose of compound 22 within the CAS of hAChE is stabilized by electrostatic and van der Waals interactions with surrounding amino acid residues.
 | ||
| Fig. 3 Binding mode of compound 22 in the CAS of hAChE predicted by molecular docking calculations. Hydrogen atoms of the amino acid residues are hidden. | ||
Molecular dynamics simulations
Molecular dynamics (MD) simulations were performed to better understand the intermolecular interactions in the hAChE-inhibitor complex. Studies of the hAChE-tacrine complex (co-crystalized ligand) were carried out to validate the molecular dynamics calculation protocols and to compare these results with those obtained for compound 22. As shown in Fig. 4A, the time-dependent changes in the root mean square deviation (RMSD) of the protein backbone atoms for studied complexes were similar and have average values ± standard deviation of 1.235 ± 0.132 Å for free hAChE, 1.402 ± 0.216 Å for hAChE with compound 22 located in the CAS, and 1.271 ± 0.154 Å for hAChE with tacrine located in the CAS. Despite the slight RMSD perturbation observed for the hAChE-22 complex, the studied model systems reach equilibration approximately after 10 ns of the calculation. The root mean square fluctuation (RMSF) analysis shown in Fig. 4B demonstrates that the bound ligands only slightly change the fluctuations of the surrounding amino acid residues. The time-dependent changes in the radius of gyration (Rg), shown in Fig. 4C, indicate that the overall structural compactness of the free hAChE and its complexes with inhibitors was maintained throughout the simulation. The solvent-accessible surface area (SASA) values (Fig. 4D) showed moderate fluctuations, reflecting minor conformational flexibility of the protein while preserving its overall structural integrity. These combined results supported the conclusion that the model systems remained stable throughout the molecular dynamics simulation and were suitable for further analysis.The binding mode of compound 22 in the CAS of hAChE, obtained at the last frame of MD simulations, is shown in Fig. 5. As can be seen, the inhibitor maintains its position relative to the docking results. Notably, the minor non-planarity of the pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine scaffold is caused by the nitrogen atom at position 10 being parameterized as a pyramidal trigonal nitrogen, which adopts a pyramidal rather than planar configuration under the applied force field during the MD simulation. The dimethylamino group adopts a planar geometry during the MD simulation, in contrast to its pyramidal shape observed in the docking calculation. This is attributed to the difference in the force fields used in docking and MD simulations.
The MM/GBSA method was employed to estimate the binding free energies and their contributing components for the enzyme–inhibitor complexes. Calculated data shown in Table 2 suggest that van der Waals (VDWAALS) and electrostatic (EEL) energies contribute to stabilization of the enzyme–inhibitor complexes, while polar solvation energy (EGB) leads to destabilization. The ΔGtotal values indicate favorable binding in complexes.
| MM/GBSA component | Compound 22 | Tacrine |
|---|---|---|
| a VDWAALS is a van der Waals energy describing non-covalent dispersion interactions between the ligand and the enzyme in the gas phase; EEL is the electrostatic energy showing coulombic interactions between the ligand and enzyme in the gas phase; EGB polar solvation energy represents the desolation penalty upon complex formation, estimated using the Generalized Born (GB) model; ESURF is nonpolar solvation energy approximated based on the solvent-accessible surface area (SASA)), accounting for hydrophobic contributions; ΔGgas is total interaction energy in the gas phase; ΔGsolv is total solvation energy (polar and nonpolar contributions); ΔGtotal is the free binding energy of the complex formation. | ||
| VDWAALS | −42.46 ± 3.08 | −31.58 ± 2.34 |
| EEL | −26.58 ± 2.81 | −194.59 ± 8.41 |
| EGB | 39.10 ± 1.93 | 202.16 ± 6.70 |
| ESURF | −4.14 ± 0.10 | −3.51 ± 0.12 |
| ΔGgas | −69.04 ± 3.19 | −226.18 ± 8.42 |
| ΔGsolv | 35.85 ± 1.91 | 198.65 ± 6.68 |
| ΔGtotal | −33.18 ± 2.21 | −27.52 ± 3.00 |
The per-residue decomposition of the binding free energies for amino acid residues located within 4 Å of the ligands in the complexes, calculated using the MM/GBSA method, is presented in Fig. 6. According to the results, the interactions of compound 22 with Trp86, Tyr133, and Tyr337, as well as those of tacrine with Trp86 and His447, which were identified in the docking binding modes, remained stable throughout the MD simulation. These interactions were among the strongest observed and are likely responsible for stabilizing the enzyme–inhibitor complexes.
In silico ADMET prediction
The drug-like properties of the pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives were evaluated based on the in silico ADMET predictions. The physicochemical and ADMET characteristics of the compounds were calculated using the SwissADME web tool and the Deep-PK.44,45 It was found that all the pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine inhibitors of AChE fully comply with Lipinski's rule of five.46 Compared to tacrine, new derivatives have higher molecular weight, lower log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values, a slightly larger number of H-bond acceptors, and greater topological polar surface area (TPSA) (Table 3). The TPSA values are below 90 Å, suggesting that the compounds may be able to cross the blood–brain barrier.47 This assumption is further supported by the categorical model results obtained from the Deep-PK (Table 3). However, the calculated log PS values indicate only moderate penetration into the central nervous system. According to log
P values, a slightly larger number of H-bond acceptors, and greater topological polar surface area (TPSA) (Table 3). The TPSA values are below 90 Å, suggesting that the compounds may be able to cross the blood–brain barrier.47 This assumption is further supported by the categorical model results obtained from the Deep-PK (Table 3). However, the calculated log PS values indicate only moderate penetration into the central nervous system. According to log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PS values, as well as the BOILED-Egg model obtained from the SwissADME server, compound 22, which was the most potent AChE inhibitor, has a higher probability of blood–brain barrier penetration compared to the other tested pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidines, except for compound 6. The calculated log
PS values, as well as the BOILED-Egg model obtained from the SwissADME server, compound 22, which was the most potent AChE inhibitor, has a higher probability of blood–brain barrier penetration compared to the other tested pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidines, except for compound 6. The calculated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Paap values predict that all AChE inhibitors exhibit high Caco-2 cell permeability, suggesting good intestinal mucosal absorption. Similar to tacrine, the studied pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives are predicted to have a risk of drug-induced liver injury. Overall, the in silico ADMET profiles of these compounds are comparable to those of tacrine.
Paap values predict that all AChE inhibitors exhibit high Caco-2 cell permeability, suggesting good intestinal mucosal absorption. Similar to tacrine, the studied pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives are predicted to have a risk of drug-induced liver injury. Overall, the in silico ADMET profiles of these compounds are comparable to those of tacrine.
| Compound | MW, g mol−1 | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P (WLOGP) P (WLOGP) |
H-bond acceptors | H-bond donors | TPSA, Å | Caco-2 permeability, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Paap (cm s−1) Paap (cm s−1) |
Blood–brain barrier (CNS), log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PS PS |
Blood–brain barrier | Liver injury I | Liver injury II |
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 231.25 | −0.01 | 3 | 1 | 63.05 | −4.54 | −2.47 | Penetrable | Toxic | Toxic |
| 6 | 245.28 | 0.20 | 3 | 0 | 54.26 | −4.58 | −2.01 | Penetrable | Toxic | Toxic |
| 8 | 218.21 | −0.20 | 4 | 1 | 69.04 | −4.8 | −3.07 | Penetrable | Toxic | Toxic |
| 9 | 202.21 | 0.14 | 3 | 0 | 51.02 | −4.58 | −2.38 | Penetrable | Toxic | Toxic |
| 10 | 217.23 | −0.27 | 3 | 1 | 77.04 | −4.82 | −2.42 | Penetrable | Toxic | Toxic |
| 13 | 232.24 | −0.56 | 3 | 0 | 60.13 | −4.77 | −2.52 | Penetrable | Toxic | Toxic |
| 15 | 203.20 | −0.62 | 3 | 2 | 85.83 | −4.99 | −2.51 | Penetrable | Toxic | Toxic |
| 17 | 229.24 | 0.64 | 3 | 1 | 66.29 | −4.82 | −2.53 | Penetrable | Safe | Toxic |
| 18 | 243.26 | 0.65 | 3 | 0 | 55.43 | −4.66 | −2.31 | Penetrable | Safe | Toxic |
| 21 | 243.26 | 0.95 | 3 | 1 | 66.29 | −4.82 | −2.37 | Penetrable | Safe | Toxic |
| 22 | 257.29 | 0.96 | 3 | 0 | 55.43 | −4.73 | −2.10 | Penetrable | Safe | Toxic |
| 24 | 230.22 | 0.89 | 4 | 1 | 72.28 | −4.83 | −3.17 | Penetrable | Safe | Toxic |
| 25 | 215.21 | 0.47 | 3 | 2 | 89.07 | −5.06 | −2.58 | Penetrable | Safe | Toxic |
| 26 | 244.25 | 0.90 | 4 | 0 | 61.42 | −4.79 | −2.85 | Penetrable | Safe | Toxic |
| 27 | 229.24 | 0.48 | 3 | 1 | 78.21 | −4.88 | −2.34 | Penetrable | Safe | Toxic |
| 28 | 243.26 | 0.74 | 3 | 1 | 64.22 | −4.81 | −2.35 | Penetrable | Safe | Toxic |
| 32 | 232.24 | −0.51 | 3 | 1 | 68.92 | −4.96 | −2.71 | Penetrable | Toxic | Toxic |
| 33 | 244.25 | 0.19 | 3 | 0 | 61.30 | −4.83 | −2.57 | Penetrable | Toxic | Toxic |
| Tacrine | 198.26 | 2.70 | 1 | 1 | 38.91 | −4.71 | −1.97 | Penetrable | Safe | Toxic |
Evaluation of cytotoxicity of 22 on the HepG2 cell line
To evaluate the hepatotoxicity of 22 as the most active compound in the series, we tested its effect on the cell viability of the human hepatocellular carcinoma cell line HepG2. HepG2 cells were exposed to the compound for 48 hours, followed by the resazurin assay to determine cell viability. The IC50 for compound 22 was calculated from the top plateau at 56.4% inhibition of cell viability. The obtained IC50 value was 34.5 µM, indicating moderate liver cytotoxicity (1 µM < IC50 < 100 µM).Experimental
Chemistry
1H and 13C NMR spectra were obtained on Varian Unity INOVA 400 or Bruker Avance DRX-500 spectrometers in CDCl3 [residual CHCl3 (δH = 7.26 ppm) or CDCl3 (δC = 77.16 ppm) as internal standard] or DMSO-d6 [residual SO(CD3)(CD2H) (δH = 2.50 ppm) or SO(CD3)2 (δC = 39.52 ppm) as internal standard]. LCMS spectra were performed on Agilent 1260 LC equipped with a G6140 MSD detector (ESI mode). Elemental analysis was performed at the Analytical Laboratory of the V.P. Kukhar Institute of Bioorganic Chemistry and Petrochemistry of the NAS of Ukraine. Melting points were determined on the Boetius hot stage apparatus. Reaction progress was monitored by thin-layer chromatography.Characteristics and spectral data of the synthesized compounds are provided in the SI.
Synthesis of compounds 1, 2, 4, 11, and 12 was described previously.36
Methyl 7-(2-bromoethyl)-4-(methylamino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxylate (3) was synthesized according to the procedure described for compound 4.36
General procedure for the synthesis of compounds 5, 6, and 13
A mixture of a pyrrolo[2,3-d]pyrimidine-6-carboxylate 3, 4, or 12 (1 mmol) and methylamine (40% aqueous solution) (4 mmol) in acetonitrile (1 mL) was stirred at room temperature for 12 h. The precipitate was filtered and recrystallized from methanol.Methyl 7-(2-bromoethyl)-4-methoxy-7H-pyrrolo[2,3-d]pyrimidine-6-carboxylate (7)
A mixture of pyrrolo[2,3-d]pyrimidine-6-carboxylate 2 (5 mmol) and sodium methylate (5 mmol) in methanol (15 mL) was heated at 50–60 °C for 0.5 h. The reaction mixture was left at room temperature for 1 h. The resulting precipitate was filtered and recrystallized from methanol.4-Methoxy-8,9-dihydropyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidin-6(7H)-one (8)
A mixture of compound 7 (2 mmol) and ammonium hydroxide solution (25%, 10 mmol) in acetonitrile (2 mL) was stirred at room temperature for 72 h. The reaction mixture was extracted with ethyl acetate, and the solvent was evaporated under reduced pressure. The resulting product was purified by crystallization from methanol.4-Methoxy-7-methyl-8,9-dihydropyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidin-6(7H)-one (9)
A mixture of pyrrolo[2,3-d]pyrimidine-6-carboxylate 7 (3 mmol) and methylamine (2 M in methanol) (20 mL) was refluxed for 1 h. The solvent was evaporated under reduced pressure. The resulting product was purified by crystallization from methanol.General procedure for the synthesis of compounds 10, 15, 17, 21, 24, 25, 27, and 32
A mixture of the appropriate starting material 9, 14, 16, 20, 23, 24, 26, or 31 (3 mmol) and ammonium acetate (15 mmol) was heated at 110–115 °C for 0.5–2 h (monitored by TLC). The reaction mixture was cooled and diluted with water (10 mL). The precipitate was filtered, washed with water, and recrystallized from the appropriate solvent.General procedure for the synthesis of compounds 14 and 16
A mixture of pyrrolo[2,3-d]pyrimidine 1 (10 mmol), bromoacetal (40 mmol), and Cs2CO3 (40 mmol) in acetonitrile (25 mL) was refluxed for 72 h. The precipitate was filtered, and the solvent was evaporated under reduced pressure. The obtained residue was treated with boiling hexane. The solvent was evaporated, and dimethylamine (40% aqueous solution, 3 mL) was added to the residue. The reaction mixture was stirred at room temperature for 12 h. The product 16 was filtered and recrystallized from 2-propanol. To obtain compound 14, the residue was heated for 0.5 h at 50–60 °C with sodium methylate in methanol.General procedure for N-methylation
A mixture of the appropriate starting material 1, 17, 21, 24, 29 or 32 (1 mmol), methyl iodide or chloroacetone (in case of compound 1) (2 mmol), and K2CO3 (2 mmol) in dimethylformamide (10 mL) was stirred at room temperature for 24 h. Water (20 mL) was added to the reaction mixture. The precipitate was filtered, washed with water, and dried. The obtained compounds 18, 19, 22, 26, 30, and 33 did not require further purification.General procedure for the synthesis of compounds 20 and 23
A mixture of pyrrolo[2,3-d]pyrimidine-6-carboxylate 19 (3 mmol) and dimethylamine (2 M in methanol, 20 mL) or sodium methylate (3 mmol) in methanol (10 mL) was heated at 50–60 °C for 0.5 h. The reaction mixture was left at room temperature for 0.5 h. The resulting precipitate was filtered and recrystallized from methanol.7,8-Dimethyl-4-(methylamino)pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidin-6(7H)-one (28)
A mixture of compound 26 (1 mmol) and dimethylamine (40% aqueous solution, 5 mmol) in acetonitrile (2 mL) was stirred at room temperature for 36 h. The precipitate was filtered and recrystallized from a methanol/DMF mixture.3-Methyl-8-methylene-8,9-dihydro-4H-pyrimido[5′,4′:4,5]pyrrolo[2,1-c][1,4]oxazine-4,6(3H)-dione (31)
A mixture of compound 30 (3 mmol) and anhydrous sodium acetate (6 mmol) in dimethylformamide (10 mL) was heated at 90 °C for 1 h. The reaction mixture was cooled and diluted with water (10 mL) after 1 h. The precipitate was filtered, washed with water, and recrystallized from a methanol/DMF mixture.Acetylcholinesterase inhibition assay
Acetylcholinesterase from Electrophorus electricus (electric eel) was purchased from Sigma-Aldrich. The synthesized compounds were evaluated for their AChE inhibitory activity using modified Ellman's method.39 The tested compounds were dissolved in DMSO. The total volume of the reaction mixture was 2 mL and contained 25 mM phosphate buffer (pH 7.4), inhibitor, enzyme, and 1% DMSO. The solution was thermostated for 5 min at 25 °C. Then DTNB was added for the detection of AChE activity. The reaction was started by adding S-acetylthiocholine iodide. The concentrations of DTNB and the substrate in the final volume of the reaction mixture were 1 mM and 0.1 mM, respectively. The activity of AChE was monitored spectrophotometrically at 412 nm by measuring the formation of 5-thio-2-nitrobenzoate after reaction of DTNB with thiocholine generated from the S-acetylthiocholine iodide by the enzyme. Donepezil and tacrine were used as reference compounds. The IC50 values represent the concentrations of compounds that reduce enzyme activity by 50% at the substrate concentration of 0.1 mM. The IC50 values were determined from dose-dependent curves obtained by plotting the percentage of remaining enzyme activity versus the logarithm of inhibitor concentration. The results were presented as mean ± standard deviation of three independent series of experiments.Molecular docking
The crystal structure of hAChE with co-crystalized tacrine molecule at the anionic subsite of the catalytic site with PDB code 7XN1 43 was downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/).48 Before performing docking calculations, the ligands, water molecules, and subunit B were removed from the downloaded file. The file containing subunit A was opened in AutoDockTools 1.5.6,49 hydrogens were added, Gasteiger charges were assigned, and the structure was saved in PDBQT format. The structure of compound 22 was drawn using Marvin Sketch and saved in MOL2 format. The geometry was then optimized with the MMFF94 s force field in Avogadro.50 Subsequently, the MOl2 file was opened in AutoDockTools 1.5.6 and converted to PDBQT format, preserving the assigned charges. Docking calculations were performed using AutoDock Vina.40 Blind docking calculation was carried out using the whole surface of the subunit A. The grid box size was set to 80 with x, y, and z grid center coordinates of 53.992, −32.624, and −28.740, respectively. The grid box size was set to 15 with x, y, and z grid center coordinates of 48.178, −40.077, and −30.751 for ligand docking to the catalytic site. The redocking of the tacrine molecule into the catalytic site was carried out for validation according to the docking protocol. The RMSD of the heavy atoms between the re-docked pose and the co-crystallized ligand was 0.57 Å. Discovery Studio 3.5 visualizer (Accelrys, San Diego, USA) was used for analysis and visualization of the docking and molecular dynamics results.Molecular dynamics
To investigate the obtained binding modes of compound 22 and tacrine at the catalytic site, molecular dynamics simulations were performed using the NAMD 2.14 software package.51 To add the missing loop (Glu491-Ala497) to the protein structure of the complexes, a homology model was built with the help of the SWISS-MODEL server,52 using the amino acid sequence of human acetylcholinesterase (UniProt53 entry P22303) and the PDB crystal structure with code 7XN1 as a template. The PROPKA method,54 implemented in the PDB2PQR software,55 was used to verify and correct the protonation states of amino acid residues at pH 7.4, which agree with in vitro study conditions. According to these calculations, Glu202 and His447 were represented in the MD simulations as Glh202 and Hid447, respectively.The structure of hAChE was parameterized using the Amber ff14SB force field,56 while parameterizations of the compound 22 and tacrine were carried out by ACPYPE software57 using the generalized AMBER force field (GAFF)58 with AM1-BCC charge method. The enzyme–inhibitor complexes were prepared by the LEaP module of AmberTools22. The enzyme–ligand complexes were solvated using the TIP3P water model in a box having dimensions 92.9 Å × 81.3 Å × 104.4 Å (total van der Waals box size), with a minimum distance of 12 Å between the amino acids and the edge of the water box. The sodium ions were added to the model systems to neutralize.
Before starting the production MD simulation, the model systems were energy minimized, heated, and equilibrated. Minimization was performed for 100 ps, keeping heavy atoms fixed to enable water molecules to spread correctly throughout the system. Periodic boundary conditions were employed alongside the particle mesh Ewald (PME) method, using a PME grid spacing of 1 Å and a tolerance of 1 × 10−6. The simulations were conducted with a time step of 1 fs. Then, minimization for 100 ps was carried out without any atomic restraints. Subsequently, the system was heated from 0 K to 298.15 K over 50 ps at a rate of 0.007 K per step within the NVT ensemble. Equilibration was performed for 100 ps under NVT conditions and for 1 ns under NPT conditions. Temperature and pressure were controlled at 298.15 K and 1 atm (≈1.01325 bar) using Langevin thermostat and barostat methods, with damping coefficient, piston period, and piston decay set to 1 ps−1, 50 fs, and 25 fs, respectively. The production molecular dynamics simulations were performed in the NPT ensemble for 20 ns, saving frames every 250 steps, resulting in a total of 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames. The trajectory files were subsequently down-sampled to 2000 frames using VMD version 1.9.3 REF. 59 and then analysed for root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration, and solvent-accessible surface area (SASA).
000 frames. The trajectory files were subsequently down-sampled to 2000 frames using VMD version 1.9.3 REF. 59 and then analysed for root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration, and solvent-accessible surface area (SASA).
All the calculations and preparations described above, except the MD simulation, were performed in the conda environment.
MM/GBSA calculation
Free binding energies of compound 22 and tacrine at the CAS were calculated using MM-PBSA.py60 available in AmberTools22. The calculation was performed using the last 1000 frames of the 2000-frame trajectory for each complex, with a sampling interval of 5 frames. The ionic strength was set to 0, and other parameters were set to default.Drug-likeness evaluation
Assessment of physicochemical parameters, as well as pharmacokinetic and toxicity properties, was conducted by SwissADME (https://www.swissadme.ch/) and Deep-PK (https://biosig.lab.uq.edu.au/deeppk/). Canonical SMILES of the pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives were generated and entered into the list of SMILES section of SwissADME or saved in a txt format and loaded into Deep-PK. In the obtained results, particular attention was given to the analysis of compliance of the physicochemical parameters of the compounds with Lipinski's rule of five.46 In the case of the ADMET properties, the intestinal absorption, blood–brain barrier penetration, and hepatotoxicity were selected.Cytotoxicity assay on HepG2 cell line
HepG2 (ACC 180) cell line was provided by DSMZ, Germany. Cells were cultured in DMEM Hg medium containing 10% fetal bovine serum, 2 mM GlutaMax, 100 units per mL penicillin, and 100 µg mL−1 streptomycin. Cells were maintained in the logarithmic growth phase at 37 °C in a humidified atmosphere containing 5% CO2. Before plating, the cells were washed with DPBS and trypsinized with TrypLE solution. The appropriate volume of culture medium was placed in a flask to stop trypsinization. Cells were centrifuged, resuspended, stained with Trypan Blue, and counted using a counting chamber. The appropriate volumes of the cell suspensions were placed in the falcon tube containing the assay medium. HepG2 cells at 5000 cells per well density were seeded in sterile 384-well plates (Greiner Bio-One, Cat# 781091) in 60 µl/well and covered with sterile transparent seals. Plates were left overnight in a humidified atmosphere at 37 °C and 5% CO2 for adaptation and adherence. The appropriate cell concentrations correspond to 30–35% of confluence before incubation with drugs. Cytotoxicity for the test compound was assessed at the final concentrations ranging from 0.005 µM to 100.0 µM (10 points, 3-fold serial dilutions). Incubation was performed for 48 hours in a humidified atmosphere at 37 °C and 5% CO2. The final DMSO concentration in the assay was 0.5%. Then resazurin (50 µM final concentration) was added and incubated for 3 hours in a humidified atmosphere at 37 °C and 5% CO2. The presence of resorufin was quantified by measuring fluorescence Ex – 555 nm, Em – 585 nm, cut-off 570, bottom read. The concentration–response relationship was analyzed using a four-parameter logistic (4 PL) model.Conclusions
In summary, eighteen pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidines were designed and synthesized as potential tacrine-like AChE inhibitors. A new method for the synthesis of 8-methyl-substituted pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine derivatives was developed. In vitro evaluation showed that sixteen target compounds inhibited AChE in the micromolar range. Replacement of a dihydropyrazine ring with a pyrazine one and introduction of methyl groups at positions 7 and 8 of the pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine core resulted in a significant improvement in the inhibitory activity. Among the tested compounds, 4-(dimethylamino)-7,8-dimethylpyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidin-6(7H)-one (22) was the most potent AChE inhibitor with an IC50 value of 0.22 ± 0.02 µM. Kinetic studies of compound 22 confirmed mixed-type inhibition. Like tacrine, the inhibitor showed strong π–π stacking interactions with Trp86 in the CAS of hAChE. In silico analysis suggested that 22 has an ADMET profile similar to that of tacrine. In addition, 22 demonstrates moderate cytotoxicity against the HepG2 cell line. Overall, the obtained results highlight pyrazino[1′,2′:1,5]pyrrolo[2,3-d]pyrimidine as a promising scaffold for the development of novel AChE inhibitors.Author contributions
Conceptualization, L. V. Muzychka, O. B. Smolii, A. I. Vovk; synthesis of compounds, L. V. Muzychka, O. B. Smolii; bioactivity studies, O. V. Muzychka, O. L. Kobzar; writing – review and editing, L. V. Muzychka, O. V. Muzychka, O. L. Kobzar, A. I. Vovk, O. B. Smolii.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ra06700f.Acknowledgements
This work was supported by the National Academy of Sciences of Ukraine (projects 0125U000356 and 0125U000355). We are grateful to Liana Dmytrovska and Dr Petro Borysko (Enamine) for evaluating the in vitro cytotoxicity in HepG2 cells and to Inna Sokolenko for her help with manuscript preparation. The authors also thank the brave defenders of Ukraine, whose service allowed us to continue our scientific work and made this publication possible.Notes and references
- A. E. Abdallah, RSC Adv., 2024, 14, 11057–11088 RSC
.
- A. Sharma, S. Rudrawar, S. B. Bharate and H. R. Jadhav, RSC Med. Chem., 2024, 16, 652–693 RSC
.
- H. Xing, S. Yue, R. Qin, X. Du, Y. Wu, D. Zhangsun and S. Luo, Int. J. Mol. Sci., 2025, 26, 3905 CrossRef CAS PubMed
.
- J. Marco-Contelles and M. J. Oset-Gasque, Chem.-Biol. Interact., 2025, 413, 111497 CrossRef CAS PubMed
.
- L. C. Llanes, I. Kuehlewein, I. V. França, L. V. da Silva and J. W. da Cruz Junior, Curr. Med. Chem., 2023, 30, 701–724 CrossRef CAS PubMed
.
- A. Sobha, A. Ganapathy, S. Mohan, N. Madhusoodanan, A. D. Babysulochana, K. Alaganandan and S. B. Somappa, Eur. J. Med. Chem. Rep., 2024, 12, 100237 CAS
.
- P. Arora, S. R. Swati, S. Jha, S. Gupta and S. Kumar, Mol. Divers., 2025, 29, 5367–5396 CrossRef CAS
.
- S.-C. Peitzika and E. Pontiki, Molecules, 2023, 28, 1084 CrossRef CAS PubMed
.
- N. V. Mehta, A. Kapadia, M. Khambete and A. Abhyankar, ChemBioChem, 2025, 26, e202500053 Search PubMed
.
- S. M. Gupta, A. Behera, N. K. Jain, A. Tripathi, D. Rishipathak, S. Singh, N. Ahemad, M. Erol and D. Kumar, RSC Adv., 2023, 13, 26344–26356 Search PubMed
.
- H. D. Nguyen and M. S. Kim, Comput. Biol. Chem., 2023, 104, 107872 CrossRef CAS
.
- H. D. Nguyen and M. S. Kim, J. Biomol. Struct. Dyn., 2024, 42, 7128–7149 CrossRef CAS PubMed
.
- M. A. Kale and M. Faisal, Curr. Drug Res. Rev., 2025, 17, 266–281 Search PubMed
.
- F. Tok, Chem. Biodivers., 2025, 22, e202402837 CrossRef CAS
.
- R. J. Obaid, N. Naeem, E. U. Mughal, M. M. Al-Rooqi, A. Sadiq, R. S. Jassas, Z. Moussa and S. A. Ahmed, RSC Adv., 2022, 12, 19764–19855 RSC
.
- O. M. Waly, K. M. Saad, H. I. El-Subbagh, S. M. Bayomi and M. A. Ghaly, Eur. J. Med. Chem., 2022, 231, 114152 CrossRef CAS
.
- A. Dorababu, Eur. J. Pharmacol., 2022, 920, 174847 CrossRef CAS PubMed
.
- N. Puranik and M. Song, Neurol. Int., 2025, 17, 26 CrossRef CAS PubMed
.
- S. Mitra, M. Muni, N. J. Shawon, R. Das, T. B. Emran, R. Sharma, D. Chandran, F. Islam, M. J. Hossain, S. Z. Safi and S. H. Sweilam, Oxid. Med. Cell Longev., 2022, 2022, 7252882 Search PubMed
.
- B. Sameem, M. Saeedi, M. Mahdavi and A. Shafiee, Eur. J. Med. Chem., 2017, 128, 332–345 CrossRef CAS
.
- M. Przybyłowska, S. Kowalski, K. Dzierzbicka and I. Inkielewicz-Stepniak, Curr. Neuropharmacol., 2019, 17, 472–490 Search PubMed
.
- I. I. Jevtić, R. V. Suručić, G. Tovilović-Kovačević, N. Zogović, S. V. Kostić-Rajačić, D. B. Andrić and J. Z. Penjišević, Bioorg. Med. Chem., 2024, 101, 117649 CrossRef
.
- A. Babu, R. Chandran, S. Meleveetil, D. Vijayan, S. Kenchaiah, A. Khade, G. Zyryanov and N. J. Muthipeedika, J. Mol. Struct., 2025, 1330, 141479 CrossRef CAS
.
- S. Fares, W. M. El Husseiny, K. B. Selim and M. A. M. Massoud, ACS Omega, 2023, 8, 26012–26034 CrossRef CAS
.
- S. Šegan, M. Mosić, V. Šukalović and I. Jevtić, J. Chromatogr. B, 2025, 1253, 124481 CrossRef
.
- B. Svobodova, Z. Moravcova, A. Misiachna, G. Novakova, A. Marek, V. Finger and J. Korabecny, Eur. J. Med. Chem., 2025, 292, 117678 CrossRef CAS
.
- Y. Zeng, L. Nie, L. Liu, K. Bozorov and J. Zhao, J. Heterocycl. Chem., 2024, 61, 1542–1553 Search PubMed
.
- J. Zhang, Y. Li, J. D. Shao, G. Wei, N. Y. Zhang, K. K. Zhu, K. M. Wang, C. S. Jiang and J. H. Wang, Mol. Divers., 2025 DOI:10.1007/s11030-025-11354-9
.
- E. A. Fayed, S. A. El-Sebaey, M. A. Ebrahim, K. Abu-Elfotuh, R. El-Sayed Mansour, E. K. Mohamed, A. M. E. Hamdan, F. T. Al-Subaie, G. S. Albalawi, T. M. Albalawi, A. M. Hamdan, A. A. Mohammed and T. M. Ramsis, Eur. J. Med. Chem., 2025, 284, 117201 CrossRef CAS
.
- Y. G. Kappenberg, P. A. Nogara, F. S. Stefanello, C. P. Delgado, J. B. T. Rocha, N. Zanatta, M. A. P. Martins and H. G. Bonacorso, Bioorg. Chem., 2023, 139, 106704 CrossRef CAS
.
- D. Turgunov, L. Nie, A. Nasrullaev, Z. Murtazaeva, B. Wang, D. Kholmurodova, R. Kuryazov, J. Zhao, K. Bozorov and H. A. Aisa, Molecules, 2025, 30, 2791 CrossRef CAS
.
- Y. Zeng, Z. Chen, Z. Yang, F. Yuan, L. Nie and C. Niu, Mol. Divers., 2025, 29, 3915–3931 CrossRef CAS
.
- B. Kuzu and Y. Demir, Arch. Biochem. Biophys., 2025, 771, 110504 CrossRef CAS PubMed
.
- A. Samadi, C. Valderas, C. de los Ríos, A. Bastida, M. Chioua, L. González-Lafuente, I. Colmena, L. Gandía, A. Romero, L. Del Barrio, M. D. Martín-de-Saavedra, M. G. López, M. Villarroya and J. Marco-Contelles, Bioorg. Med. Chem., 2011, 19, 122–133 CrossRef CAS PubMed
.
- L. Jalili-Baleh, H. Nadri, A. Moradi, S. N. A. Bukhari, M. Shakibaie, M. Jafari, M. Golshani, F. Homayouni Moghadam, L. Firoozpour, A. Asadipour, S. Emami, M. Khoobi and A. Foroumadi, Eur. J. Med. Chem., 2017, 139, 280–289 CrossRef CAS
.
- L. Muzychka, O. Muzychka and O. Smolii, Chem. Biodivers., 2025, 22, e202401874 Search PubMed
.
- I. O. Yaremchuk, L. V. Muzychka, O. B. Smolii, O. V. Kucher and S. V. Shishkina, Tetrahedron Lett., 2018, 59, 442–444 Search PubMed
.
- L. V. Muzychka, I. O. Yaremchuk, E. V. Verves and O. B. Smolii, Chem. Heterocycl. Comp., 2019, 55, 397–400 Search PubMed
.
- G. L. Ellman, K. D. Courtney, V. Andress and R. M. Featherstone, Biochem. Pharmacol., 1961, 7, 88–95 Search PubMed
.
- O. Trott and A. J. Olson, J. Comp. Chem., 2010, 31, 455–461 Search PubMed
.
- A. Ganeshpurkar, R. Singh, S. Shivhare, Divya, D. Kumar, G. Gutti, R. Singh, A. Kumar and S. K. Singh, Mol. Divers., 2022, 26, 1455–1479 CrossRef CAS PubMed
.
- M. F. Reynoso-Garcia, D. E. Nicolas-Alvarez, A. Y. Tenorio-Barajas and A. Reyes-Chaparro, Int. J. Mol. Sci., 2025, 26, 3781 Search PubMed
.
- K. V. Dileep, I. Kentaro, M.-T. Chiemi, K.-N. Mutsuko, Y. Mayumi, H. Kazuharu, S. Mikako and K. Y. J. Zhang, Int. J. Biol. Macromol., 2022, 15, 172–181 CrossRef
.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef
.
- Y. Myung, A. G. de Sá and D. B. Ascher, Nucleic Acids Res., 2024, 52, W469–W475 Search PubMed
.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Adv. Drug Deliv. Rev., 1997, 23, 3–25 Search PubMed
.
- D. E. Clark, Drug Discov. Today, 2003, 8, 927–933 CrossRef CAS PubMed
.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed
.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comp. Chem., 2009, 30, 2785–2791 Search PubMed
.
- M. D. Hanwell, D. E. Curtis and D. C. Lonie, J. Cheminformatics, 2012, 4, 4–17 Search PubMed
.
- J. C. Phillips, D. J. Hardy, J. D. C. Maia, J. E. Stone, J. V. Ribeiro, R. C. Bernardi, R. Buch, G. Fiorin, J. Henin, W. Jiang, R. McGreevy, M. C. R. Melo, B. K. Radak, R. D. Skeel, A. Singharoy, Y. Wang, B. Roux, A. Aksimentiev, Z. Luthey-Schulten, L. V. Kale, K. Schulten, C. Chipot and E. Tajkhorshid, J. Chem. Phys., 2020, 153, 044130 CrossRef CAS
.
- A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. T. Heer, T. A. P. de Beer, C. Rempfer, L. Bordoli, R. Lepore and T. Schwede, Nucleic Acids Res., 2018, 46, W296–W303 Search PubMed
.
- The Uniprot Consortium, UniProt: the universal Protein knowledgebase in 2025, Nucleic Acids Res., 2025, 53, D609–D617 CrossRef PubMed
.
- H. Li, A. D. Robertson and J. H. Jensen, Proteins, 2005, 61, 704–721 Search PubMed
.
- T. Dolinsky, P. Czodrowski, H. Li, J. E. Nielsen, J. H. Jensen, G. Klebe and N. A. Baker, Nucleic Acids Res., 2007, 35, W522–W525 CrossRef
.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed
.
- A. W. Sousa da Silva and W. F. Vranken, BMC Res. Notes., 2012, 5, 367 CrossRef
.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 Search PubMed
.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics., 1996, 14, 33–38 CrossRef CAS
.
- B. R. Miller, T. D. McGee, J. M. Swails, N. Homeyer, H. Gohlke and A. E. Roitberg, J. Chem. Theory Comput., 2012, 8, 3314–3321 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2026 |