Recent advances in chemoenzymatic synthesis of oligosaccharides and polysaccharides
Tomonari
Tanaka

Department of Biobased Materials Science, Graduate School of Science and Technology, Kyoto Institute of Technology, Matsugasaki, Sakyo-ku, Kyoto 606-8585, Japan. E-mail: t-tanaka@kit.ac.jp
First published on 14th November 2025
Abstract
Chemoenzymatic methods are essential tools for synthesising a wide range of saccharides and their derivatives, including oligosaccharides, polysaccharides, and glycoconjugates, in the fields of glycotechnology and polymer chemistry. This review summarises recent progress and provides an overview of research on chemoenzymatic synthesis of oligosaccharides and polysaccharides through the combined use of chemical and enzymatic reactions. The methodologies are discussed separately for each class of enzyme: glycosyltransferases, glycan phosphorylases, and glycosyl hydrolases.
Introduction
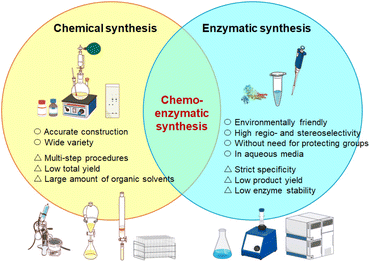
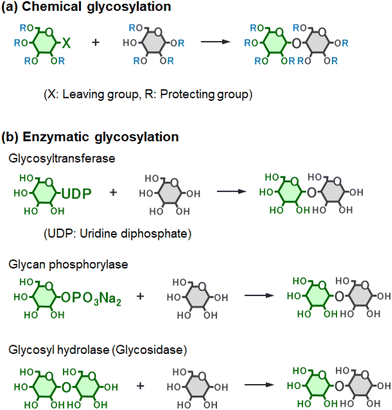
Carbohydrates, along with nucleotides and amino acids, are fundamental biomolecules. In particular, saccharide chains such as oligosaccharides and polysaccharides are often referred to as the ‘third class of biomolecular chains’, following nucleic acids and proteins, and are widely expressed across diverse living organisms. Oligosaccharides generally consist of two to several dozen monosaccharide units, whereas longer chains are classified as polysaccharides; however, these terms are not strictly defined by molecular weight or chain length. A key distinction between carbohydrates and nucleotides or proteins is the current inability to modify them through genetic engineering. Therefore, the synthesis of oligosaccharides, polysaccharides, and their conjugates—including glycosylated peptides and glycoproteins—remains a critical area requiring continued methodological development.The synthetic methodologies for oligo- and polysaccharides can be broadly divided into two categories: chemical synthesis and enzymatic synthesis (Fig. 1). Chemical synthesis enables the accurate construction of a wide variety of oligo- and polysaccharides; however, it typically requires complex multi-step procedures involving extensive protection and deprotection of hydroxy, carboxy, amino, and other functional groups on saccharide residues (Fig. 2a). Moreover, large amounts of organic solvents are often necessary for both reaction and purification processes. Since the development of Fischer glycosylation in the 1980s, chemical synthesis of oligosaccharides has continued to advance, with recent developments showing remarkable progress. The key step in chemical synthesis is glycosylation, in which monosaccharides are covalently linked either to one another or to other molecules. These advances have led to remarkable improvements in stereoselectivity at the anomeric position of saccharide residues. Numerous reviews have comprehensively summarised strategies for chemical glycosylation.1–7 Furthermore, the development of automated chemical glycosylation has greatly expanded the field, enabling the synthesis of polysaccharides containing thousands of monosaccharide units.8–11
Enzymes are essential biocatalysts and environmentally friendly alternatives for glycosylation. Enzymatic glycosylation is generally categorised by enzyme type: glycosyltransferases, glycan phosphorylases and glycosyl hydrolases (glycosidases) (Fig. 2b).12 Glycosyltransferases (EC 2.4) are categorised into two types: Leloir-type and non-Leloir-type. Leloir-type glycosyltransferases are the key enzymes for the formation of glycosidic bonds in nature and use sugar nucleotides as activated donor substrates.13 Chemical and enzymatic synthesis of sugar nucleotides has been reported.14–16 Glycan phosphorylases are categorised into non-Leloir-type glycosyltransferases. In this paper, the terms ‘glycosyltransferase’ and ‘phosphorylase’ are used to refer to Leloir-type and non-Leloir-type, respectively. Owing to their high regio- and stereoselectivity, these enzymes enable accurate synthesis without the need for protecting groups, and the reactions naturally proceed in aqueous media. The application of enzyme-catalysed glycosylation has expanded considerably with advances in genetic engineering, which have facilitated the expression and ready availability of diverse enzymes. The challenge is that product yields are often lower than those obtained by chemical synthesis. Details of each enzyme are provided in the respective sections below.
The combination of chemical and enzymatic approaches, known as the chemoenzymatic method, is a powerful strategy that leverages the advantages of both systems. In general, two modes of combination are possible: one in which an enzymatic reaction follows chemical synthesis, and another in which the product of an enzymatic reaction serves as a substrate for chemical modification. Regardless of the sequence, both approaches are referred to as chemoenzymatic synthesis. Enzymatic methods are often well-suited for integration with chemical strategies because chemical systems can compensate for challenges such as the complexity of enzyme systems, low yields, strict specificity, or the absence of suitable catalysts. Chemoenzymatic methods are valuable not only in carbohydrate synthetic chemistry but also in the synthesis of a broad range of other compounds. Several review articles have described the applications of chemoenzymatic synthesis in natural products,17–20 polypeptides,21,22 proteins,23 and polymeric materials.24 In the field of carbohydrate synthetic chemistry, chemoenzymatic approaches have been actively explored and continue to evolve. This review focuses on recent advances and provides an overview of chemoenzymatic strategies for the synthesis of oligo- and polysaccharides through the combined use of chemical and enzymatic reactions. The methodologies are discussed separately for each class of enzyme: glycosyltransferases, glycan phosphorylases and glycosyl hydrolases.
Chemoenzymatic methods using glycosyltransferases
Glycosyltransferases are enzymes that catalyse the transfer of saccharide moieties from nucleotide sugars—such as uridine diphosphate (UDP) and cytidine-5′-monophosphate (CMP) saccharides—to specific acceptor molecules, thereby forming glycosidic bonds (Scheme 1). In vivo, they typically function in metabolic glycosylation processes. The carbohydrate-active enzymes (CAZy) database (https://www.cazy.org/) provides an up-to-date classification of glycosyltransferases, grouping enzymes that use saccharide nucleotide diphosphates, saccharide nucleotide monophosphates, saccharide phosphates, and related donor substrates into distinct sequence-based families. This section provides an overview of the chemoenzymatic synthesis of oligo- and polysaccharides via glycosyltransferase-catalysed glycosylation.Around the year 2000, Nishimura and co-workers developed an automated oligosaccharide synthesiser using glycosyltransferases, known as Golgi™.25–27 More recently, automated enzymatic glycan synthesis using glycosyltransferases has continued to attract considerable attention and is in and ongoing development (Fig. 3).28,29
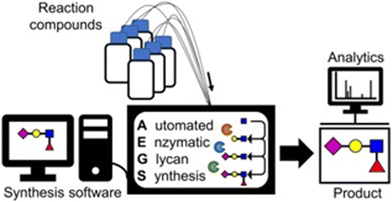 | ||
| Fig. 3 Automated enzymatic glycan synthesis (reproduced with permission from Elsevier (Biotechnol. Adv., 2023)).29 | ||
Although hundreds of thousands of glycosyltransferases have been identified, those successfully used in the chemoenzymatic synthesis of human glycans are primarily transferases of D-glucose (Glc), D-galactose (Gal), D-mannose (Man), L-fucose (Fuc), N-acetyl-D-glucosamine (GlcNAc), N-acetyl-D-galactosamine (GalNAc), and sialic acids such as N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc). Ideal glycosyltransferases for chemoenzymatic synthesis should exhibit high expression levels, stability during storage and use, high catalytic activity, and accurate regio- and stereoselectivity. Because wild-type enzymes do not always possess all of these properties, mutant glycosyltransferases with improved synthetic performance have been developed using enzyme engineering techniques.
Enzymatic glycosylation onto chemically synthesised saccharides
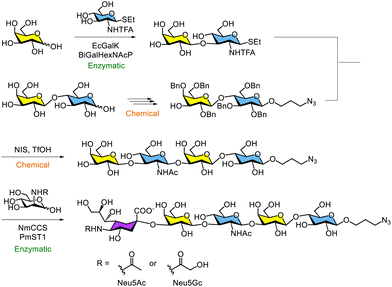
In the chemoenzymatic synthesis of oligosaccharides and their conjugates, such as glycopeptides, glycosyltransferase-catalysed reactions are commonly used to construct oligosaccharide moieties. Recently, numerous enzymatic syntheses of human milk oligosaccharides catalysed by glycosyltransferases have been reported. Since the mid-20th century, human milk oligosaccharides —composed of Glc, Gal, GlcNAc, Fuc and Neu5Ac—have been recognised as important contributors to the growth of bifidobacteria.30–34 These oligosaccharides are present in breast milk alongside lactose and include trisaccharides as well as higher-order oligosaccharides. Moreover, human milk oligosaccharides play critical roles in various biological processes, including protection of the gastrointestinal tract against toxins, inhibition of viral and bacterial adhesion, modulation of the immune response, and support of infant brain development.35–37In 2015, Cao and co-workers reported the chemoenzymatic synthesis of derivatives of lacto-N-tetraose (Galβ1-3GalNAcβ1-3Galβ1-4Glc), a major component and one of the most abundant core structures of human milk oligosaccharides.38 This method combines multiple chemical and enzymatic glycosylation steps (Scheme 2). A thioglycoside derivative of a disaccharide, enzymatically synthesised using galactokinase from E. coli K-12 (EcGalK) and D-galactosyl-β1,3-N-acetyl-D-hexosamine phosphorylase from Bifidobacterium infantis (BiGalHexNAcP), was reacted with a chemically synthesised disaccharide derivative containing an azide group. The lacto-N-tetraose derivative was obtained via chemical glycosylation using the thioglycoside as a glycosyl donor, with N-iodosuccinimide (NIS) and trifluoromethanesulfonic acid (TfOH) as activators. Similar one-pot multienzyme syntheses of UDP-monosaccharides, catalysed by kinases and hexosamine phosphorylases, have also been reported.39–43 In addition, the enzymatic synthesis of sialyllacto-N-tetraose containing two different sialic acid forms, Neu5Ac and Neu5Gc, has been achieved using a one-pot, three-enzyme system. In this approach, the sialic acid precursors N-acetyl-D-mannosamine and N-glycolyl-D-mannosamine were converted into Neu5Ac and Neu5Gc, respectively, in the presence of sialic acid aldolase from E. coli K-12. These sialic acids were then transformed into CMP-Neu5Ac and CMP-Neu5Gc using cytidine 5′-triphosphate and CMP-sialic acid synthetase from Neisseria meningitidis (NmCSS). Finally, the resulting glycosyl donors, CMP-Neu5Ac and CMP-Neu5Gc, were used in the reaction catalysed by α2,3-sialyltransferase 1 from Pasteurella multocida (PmST1), yielding α2,3-sialyllacto-N-tetraose via the one-pot multienzyme system.
Wang and co-workers reported the chemoenzymatic synthesis of a library of 31 human milk oligosaccharides (Fig. 4a).44 This was achieved through enzymatic terminal modification of chemically synthesised tetra- and pentaoligosaccharides with Gal, Fuc and Neu5Ac moieties. Yu and co-workers reported the enzymatic synthesis of fucosylated human milk oligosaccharides, including Lewis X, Lewis A, and sialyl Lewis X antigen structures, catalysed by α-1,3/1,4-fucosyltransferase from Helicobacter pylori DSM 6709.45,46 Boons's group also demonstrated the systematic enzymatic synthesis of asymmetrical multiantennary human milk oligosaccharides using a limited set of human glycosyltransferases (Fig. 4b).47 A lactose derivative bearing a hydrophobic coumarin moiety was converted into a linear tetrasaccharide, lacto-N-neotetraose, catalysed by β1,3-N-acetylglucosaminyltransferase 2 (B3GNT2) and human β1,4-galactosyltransferase (B4GalT1) in the presence of UDP-GlcNAc and UDP-Gal, respectively. Lacto-N-neotetraose was then selectively modified by the I-branching enzyme, N-acetyllactosaminide β1,6-N-acetylglucosaminyltransferase (GCNT2), introducing a branched β1,6-linked GlcNAc moiety and yielding an asymmetric pentasaccharide. The galactose residue at the nonreducing end was then sialylated using α2,6-sialyltransferase 1 (ST6GAL1) in the presence of CMP-Neu5Ac, producing a sialylhexasaccharide. Subsequent modifications of the β6 antenna of this sialylhexasaccharide, catalysed by B4GalT1, B3GNT2, galactoside α2-fucosyltransferase 1 (FUT1), and β1,3-galactosyltransferase 5 (B3GALT5), enabled the synthesis of oligosaccharides ranging from octa- to dodecasaccharides. More recently, Fang and co-workers reported an enzymatic modular strategy for generating a library of branched human milk oligosaccharides using β1,6-N-acetylglucosaminyltransferase 2 expressed in Pichia pastoris.48,49 Additionally, a recent review summarised practical and scalable approaches for automated enzymatic production of tailored human milk oligosaccharides.50
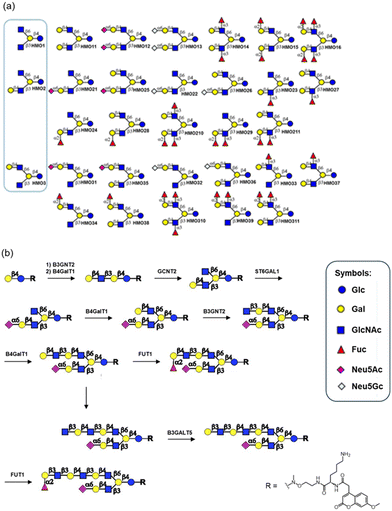 | ||
| Fig. 4 Chemoenzymatic synthesis of human milk oligosaccharides with various glycosyltransferases (reproduced with permission from American Chemical Society (J. Org. Chem., 2016) and National Academy of Sciences of the USA (Proc. Natl. Acad. Sci. U. S. A., 2017)).44,47 | ||
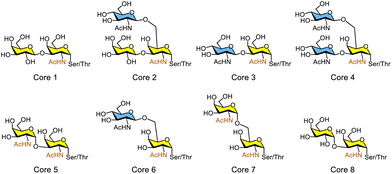
O-Linked glycans (O-glycans), also referred to as mucin-type glycans, are viscous biomolecules secreted by living organisms into various fluids. Their most common structural feature is an α-O-linked GalNAc moiety attached to serine or threonine residues. O-Glycan core structures are classified based on their chemical composition, typically designated as cores 1–8, which contain GalNAc, Gal, and GlcNAc residues (Fig. 5). Chen and co-workers reported a general regioselective strategy using glycosyltransferases to access diverse O-glycan core 2 oligosaccharides, which can be further extended to synthesise other complex branched glycans.51 A chemically synthesised core 2 trisaccharide derivative bearing a p-methoxyphenyl (PMP) group was modified through β1,4-galactosylation using β1,4-galactosyltransferase from Neisseria meningitidis (NmLgtB), α2,3-sialylation using the α2,3-sialyltransferase 1 M144D mutant from Pasteurella multocida (PmST1 M144D), α2,3-sialylation using α2,3-sialyltransferase 3 from Pasteurella multocida (PmST3), and α2,6-sialylation using the α2,6-sialyltransferase A366G mutant from Photobacterium species (Psp2,6ST A366G), yielding sialylated core 2 glycans (Fig. 6a). Wen and Li's group reported the chemoenzymatic synthesis of both sulfated and non-sulfated core 2 oligosaccharides via glycosyltransferase-catalysed reactions, generating 36 structurally well-defined O-glycans (Fig. 6b).52 Other O-glycan core oligosaccharides have been synthesised using a robust chemoenzymatic modular assembly strategy.53 The key to this approach is the convergent assembly of O-glycan cores 1–4 and 6 from three chemical building blocks, followed by enzymatic diversification using 13 tailored enzyme modules, resulting in a total of 83 distinct O-glycan oligosaccharides.
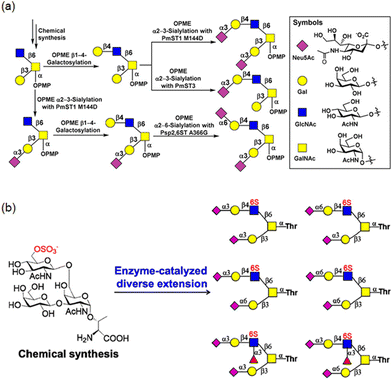 | ||
| Fig. 6 Chemoenzymatic synthesis of core 2 oligosaccharides (reproduced with permission from American Chemical Society (ACS Catal., 2019 and J. Org. Chem., 2021)).51,52 | ||
Recently, Li and co-workers reported the chemical synthesis of three α-linked O-glycan core saccharides—cores 5, 7, and 8—containing 1,2-cis-linked Gal and GalNAc residues at the nonreducing end. Their chemically synthesised α-linked cores were subsequently converted into sialylated forms using α2,6-sialyltransferase from Photobacterium damselae (Scheme 3).54 Boon and co-workers also reported the chemoenzymatic synthesis of a sulfonate isostere of 6-sulfo-sialyl Lewis X via multiple glycosylation steps catalysed by glycosyltransferases.55 Recently, Wen and co-workers reported the chemoenzymatic preparation of oligosaccharide libraries, including sulfated and sialylated glycans as well as N-glycans, using glycosyltransferases.56–58
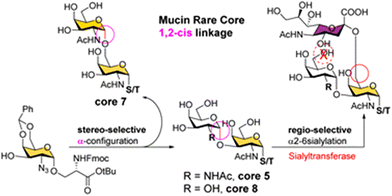 | ||
| Scheme 3 Chemoenzymatic synthesis of O-glycan cores 5, 7, and 8 (reproduced with permission from Royal Society of Chemistry (Chem. Sci., 2023)).54 | ||
Chemoenzymatic synthesis of glycosaminoglycans
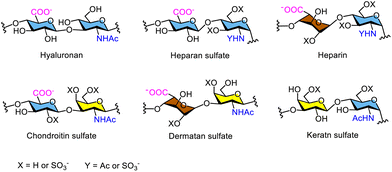
Glycosaminoglycans are linear heteropolysaccharides composed of repeating disaccharide units consisting of alternating hexosamine and uronic acid or galactose residues. Examples include hyaluronic acid (hyaluronan), heparan sulfate and heparin, chondroitin sulfate, dermatan sulfate, and keratan sulfate (Fig. 7).59,60 Glycosaminoglycans serve as structural components and play key regulatory roles in numerous biological functions of proteins, cells, and tissues throughout the human body. Therefore, glycosaminoglycans are used extensively in different medical applications.61 Glycosaminoglycans exhibit heterogeneous chemical structures; for example, their modification patterns, including sulfation and epimerization, are influenced by seasonal variations, environmental factors, animal species, tissue source, age, breed, and health status when isolated from animal sources. Therefore, the synthesis of structurally well-defined glycosaminoglycans is essential and in high demand because they are challenging to obtain in pure and consistent form from natural sources.Chemoenzymatic synthesis of glycosaminoglycans has been comprehensively reviewed elsewhere.62,63 A typical approach combines enzymatic glycosylation, catalysed by glycosyltransferases, with chemical modification of the resulting oligosaccharides. Liu and co-workers reported the chemoenzymatic synthesis of ultralow molecular weight (ULMW) heparins, with molecular weights ranging from 1500 to 3000 Da, corresponding to 5–10 saccharide units. These sulfated oligosaccharides are used clinically for the treatment of thrombotic disorders (Scheme 4).64,65 The synthesis began with a disaccharide precursor, which was elongated to a tetrasaccharide using two bacterial glycosyltransferases: N-acetylglucosaminyltransferase from Escherichia coli K5 and heparosan synthase-2 from Pasteurella multocida. The tetrasaccharide was elongated to a heptasaccharide in three steps and subsequently converted into ULMW heparins through modification of the N-trifluoroacetyl glucosamine residue to an N-sulfonated glucosamine, followed by epimerization and sulfation at the 2- and 6-positions of the iduronic acid residues. Similarly, another heptasaccharide with a GlcNAc residue sulfated at the 2- and 6-positions at the nonreducing end was synthesised. These ULMW heparins exhibit excellent in vitro anticoagulant activity and display pharmacokinetic properties comparable to Arixtra™, a clinically used low molecular weight heparin-like anticoagulant.
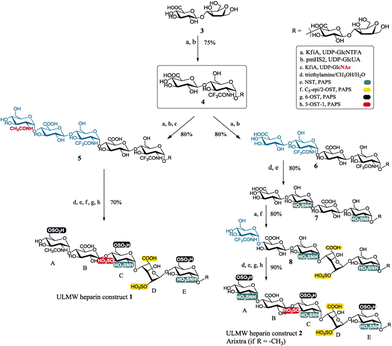 | ||
| Scheme 4 Chemoenzymatic synthesis of ULMW heparins (reproduced with permission from Science (Science, 2011)).64 | ||
Chemical sulfation of glycosaminoglycans is typically performed using sulfur trioxide complexes, such as sulfur trioxide–pyridine, sulfur trioxide–N,N-dimethylformamide (DMF), or sulfur trioxide–triethylamine, in organic solvents such as DMF.66–72N-Sulfation of suitably N-deacylated glycosaminoglycans can also be achieved in aqueous media using hydrazine sulfate in the presence of anhydrous hydrazine, yielding sulfonated derivatives.73,74 Regioselective enzymatic sulfation using heparan sulfate sulfotransferases from Kluyveromyces lactis has been reported to produce heparan sulfates, although its application remains limited by the small scale of reactions and the restricted availability of sulfotransferases and the cofactor 3′-phosphoadenosine 5′-phosphosulfate.75 Additional reviews on the enzymatic synthesis of glycosaminoglycans using glycosyltransferases have also been published.76,77
Chemoenzymatic methods using glycan phosphorylases
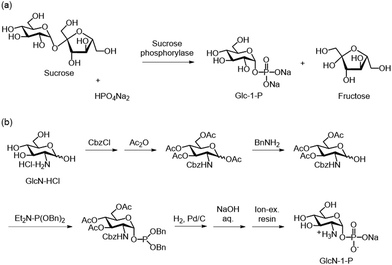
Glycan phosphorylases are classified as non-Leloir-type glycosyltransferases in the hexosyltransferase and glycoside hydrolase families according to the CAZy database, based on sequence similarity. Details on their types, structures, mechanisms, roles, and related aspects are summarised in other review papers.78–84 Glycan phosphorylases catalyse the transfer of a phosphate group from inorganic phosphate to a glycosyl acceptor (Scheme 5). Most glycan phosphorylases catalyse phosphorolysis at the nonreducing end of glycosidic linkages. Importantly, these reactions are reversible, allowing glycan phosphorylases to catalyse both the phosphorolysis of nonreducing end glycosidic bonds in the presence of inorganic phosphate and the formation of glycosidic bond in the presence of excess monosaccharide 1-phosphates.Glycan phosphorylases use accessible and relatively stable monosaccharide 1-phosphates for the regio- and stereospecific synthesis of glycosidic bonds. Their substrate specificity is generally strict; therefore, different glycan phosphorylases are used for the synthesis of oligo- and polysaccharides with defined glycosidic linkages.85–87 α-Glucose 1-phosphate (Glc-1-P) is commercially available and serves as a glycosyl donor in glycan phosphorylase-catalysed glycosylation. It can also be enzymatically synthesised from sucrose by sucrose phosphorylase (SCP) (Scheme 6a).88,89 Furthermore, the enzymatic synthesis of Glc-1-P from cellobiose catalysed by cellobiose phosphorylase (CBP) has also been reported.90 Other non-commercial monosaccharide 1-phosphates, such as glucosamine 1-phosphate (GlcN-1-P), are typically synthesised via multistep chemical processes involving protection of hydroxy and other functional groups, introduction of a phosphate group at the anomeric position, and subsequent deprotection (Scheme 6b).91 This section provides an overview of the chemoenzymatic synthesis of oligo- and polysaccharides through enzymatic glycosylation catalysed by glycan phosphorylases.
Enzymatic synthesis of α-glucans and their derivatives catalysed by α-glucan phosphorylases
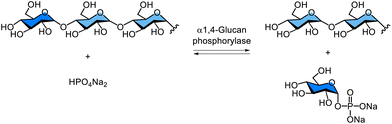
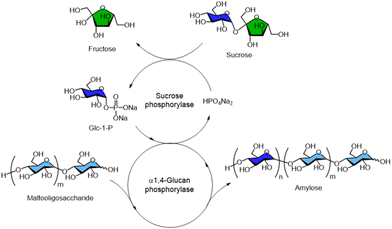
α-Glucan phosphorylases are essential in both the degradation and synthesis of α1,4-glucosidic linkages, particularly in the metabolism of starch and glycogen. Therefore, numerous studies have reported the synthesis of α-linked oligo- and polysaccharides catalysed by α-glucan phosphorylases.For example, the enzymatic synthesis of amylose—a linear glucose polymer with α1,4-glycosidic linkages and a major component of starch—has been achieved using two phosphorylases starting from sucrose, a disaccharide composed of glucose and fructose (Fig. 8). Glc-1-P was first enzymatically generated from sucrose in the presence of phosphoric acid using SCP, and was subsequently polymerised by α1,4-glucan phosphorylase to yield amylose. A combination of thermostable SCP and potato-derived L-type α1,4-glucan phosphorylase has been reported by Ezaki Glico Co., Ltd for the production of amylose from sucrose.92,93 Enzymatic synthesis of α-linked oligo- and polysaccharides using sucrose phosphorylases and related enzymes belonging to glycoside hydrolase family has also been reported.94,95
Amylose-containing copolymers can be synthesised using α1,4-glucan phosphorylases. Loos and co-workers reported the enzymatic synthesis of rod–coil block copolymers composed of amylose and synthetic polymers, such as polystyrene and poly(2-vinyl pyridine), catalysed by potato phosphorylase (Scheme 7).96–99 The group also demonstrated the synthesis of hyperbranched glycoconjugates by combining potato phosphorylase with the glycogen branching enzyme from Deinococcus geothermalis.100 Furthermore, Loos's group reported the production of amylose-modified silica particles through potato phosphorylase-catalysed polymerisation, as well as densely packed α1,4- and α1,6-linked hyperbranched polysaccharide brush coatings using the phosphorylase in combination with the microbial branching enzyme from Deinococcus geothermalis.101,102
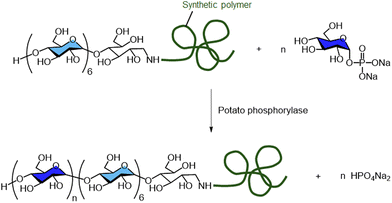 | ||
| Scheme 7 Phosphorylase-catalysed enzymatic synthesis of rod–coil block copolymers composed of amylose and a synthetic polymer. | ||
Because phosphorylases exhibit relatively relaxed substrate specificity, various monosaccharide 1-phosphates have been used as glycosyl donors in phosphorylase-catalysed enzymatic glycosylation to produce unnatural oligo- and polysaccharides.103 Enzymatic synthesis of such unnatural polysaccharides using α1,4-glucan phosphorylases has been demonstrated. For example, Thiem's group and Kadokawa's group independently reported that phosphorylases, such as potato phosphorylase and thermostable phosphorylase from Aquifex aeolicus VF5 catalysed α1,4-glycosylation of Man104 2-deoxy-D-glucose (2dGlc),105D-xylose (Xyl),106D-glucosamine (GlcN),91 and N-formyl D-glucosamine (GlcNF),107D-glucuronic acid (GlcA)108 at the nonreducing end of maltooligosaccharide acceptor using the corresponding α-monosaccharide 1-phosphates as a glycosyl donor in aqueous buffer conditions (Scheme 8).
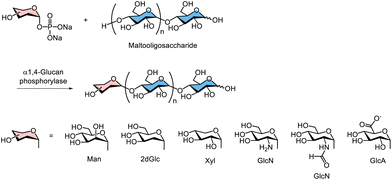 | ||
| Scheme 8 Phosphorylase-catalysed glycosylation using different monosaccharide-1-phosphates as glycosyl donors. | ||
Kadokawa and co-workers also applied phosphorylase-catalysed polymerisation for the synthesis of amylose-grafted functional heteropolysaccharide materials. Maltooligosaccharide-bearing polysaccharides, including chitosan, chitin, cellulose, alginate, xanthan gum, and carboxymethyl cellulose, were first chemically prepared and subsequently converted into amylose-grafted heteropolysaccharides via phosphorylase-catalysed polymerisation (Fig. 9a).109–115 This approach has also been extended to the modification of glycogen, a water-soluble, high-molecular-weight natural polysaccharide composed of linear α1,4-glucose chains, by grafting amylose chains onto its structure. The resulting solution of amylose-elongated glycogen was completely converted into a hydrogel in aqueous media through cross-link formation arising from the double-helix conformation of the elongated amylose chains among the glycogen molecules (Fig. 9b).116 Using the abovementioned phosphorylase-catalysed glucuronylation and glucosaminylation reactions with the analogue glycosyl donor substrates GlcA-1-P and GlcN-1-P, highly branched amphoteric glycogens containing both acidic GlcA and basic GlcN residues can also be synthesised.117–119 Pich's group reported phosphorylase-catalysed grafting-from polymerisation of Glc-1-P, catalysed by rabbit muscle phosphorylase, onto poly(N-vinylcaprolactam) (PVCL)-based maltoheptaose-modified microgels, resulting in the formation of amylose-coated PVCL microgels (Fig. 9c).120
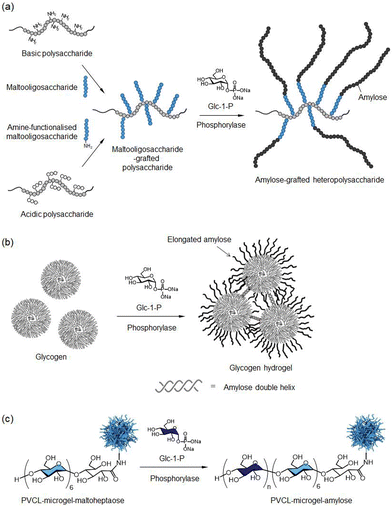 | ||
| Fig. 9 Chemoenzymatic synthesis of (a) amylose-grafted heteropolysaccharides, (b) glycogen hydrogel, and (c) amylose-covered PVCL microgels by phosphorylase-catalysed polymerization (reproduced from Polymers, 2016).115 | ||
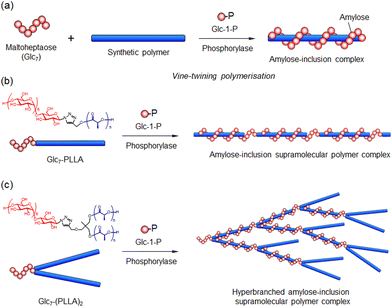
Furthermore, Kadokawa and co-workers developed a novel phosphorylase-catalysed polymerisation technique, termed vine-twining polymerisation, which uses a maltooligosaccharide such as maltoheptaose (Glc7) as a primer, a synthetic polymer as a guest, and Glc-1-P as a monomer to synthesise amylose–polymer inclusion complexes (Fig. 10a).121–123 This enzymatic polymerisation system for forming amylose–synthetic polymer inclusion complexes resembles the way plant vines twine around a rod; this methodology is named ‘vine-twining polymerisation’. The group reported the synthesis of various amylose–polymer inclusion complexes with hydrophobic polymers such as poly(tetrahydrofuran) (PTHF) and poly(oxetane) (POXT);124,125 polyesters including poly(δ-valerolactone) (PVL), poly(ε-caprolactone) (PCL), poly(glycolic acid-co-ε-caprolactone), and poly(ester-ether)s; poly-L-lactide (PLLA);126–129 polycarbonates such as poly(tetramethylene carbonate); and an amphiphilic triblock copolymer, poly(2-methyl-2-oxazoline-block-tetrahydrofuran-block-2-methyl-2-oxazoline),130,131 as guest polymers via vine-twining polymerisation. Vine-twining polymerisation has also been applied to the selective inclusion of amylose with polymers of defined structures, molecular weight distributions, and chirality.129,132–134
Kadokawa's group and our group jointly applied vine-twining polymerisation to synthesise amylose-inclusion supramolecular polymer complexes composed of amylose and guest polymers, such as PLLA and PTHF, using Glc7-bearing polymers as primer–guest polymer conjugates (Fig. 10b).135–137 Interestingly, when Glc7-bearing poly-D-lactide (PDLA) was used as the primer–guest polymer conjugate, no amylose-inclusion supramolecular polymer complex formed. Instead, an amylose–PDLA block copolymer was obtained, which was attributed to the chirality-specific recognition of polylactic acid by amylose, preventing inclusion. Additionally, hyperbranched amylose-inclusion supramolecular polymer complexes were enzymatically synthesised via vine-twining polymerisation using a branched Glc7–PLLA conjugate (Glc7–(PLLA)2) and Glc-1-P (Fig. 10c).138 Amylose supramolecular network hydrogels were fabricated via the formation of amylose–polymer inclusion complexes through vine-twining polymerisation. Grafted copolymers such as PVL-grafted poly(acrylic acid sodium salt), PCL-grafted carboxymethyl cellulose, and PCL-grafted poly(γ-glutamic acid) were used as guest polymers to produce the hydrogels (Fig. 11).139–141
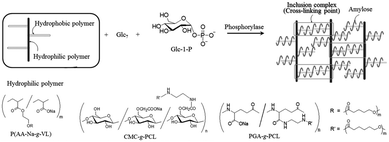 | ||
| Fig. 11 Preparation of amylose–inclusion supramolecular network materials by vine-twining polymerization using grafted copolymers (reproduced from Polymers, 2016).115 | ||
Enzymatic synthesis of β-glucans catalysed by β-glucan phosphorylases
Bottom-up enzymatic synthesis of β-linked oligo- and polysaccharides catalysed by β-glucan phosphorylases has attracted considerable attention. This approach enables the production of well-defined compounds essential for fundamental studies of β-glucan properties and for the development of functional materials because β-glucans are important and renewable biomacromolecules in materials science. Since the discovery of β1,3-oligoglucan phosphorylase in 1963,142 additional β-glucan phosphorylases have been identified, differing in the type of glycosidic linkage they form and/or in the degree of polymerisation (DP) of the products.143–145Kitaoka and Nishimoto's group reported the enzymatic synthesis of the human milk disaccharide lacto-N-biose I (LNB, Galβ1-3GlcNAc) from sucrose. The reaction used a galacto/lacto-N-biose phosphorylase and galactose 1-phosphate, combined with SCP, UDP-glucose–hexose-1-phosphate uridylyltransferase, and UDP-glucose 4-epimerase (Fig. 12).146,147 They further demonstrated practical LNB production using crude extracts from bifidobacterial cells, which contained the four enzymes naturally produced as intracellular proteins in various Bifidobacterium strains.148
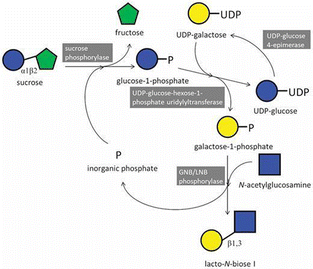 | ||
| Fig. 12 Enzymatic synthesis of lacto-N-biose I catalysed by phosphorylases (reproduced with permission from Oxford University Press (Biosci. Biotechnol. Biochem., 2020)).146 | ||
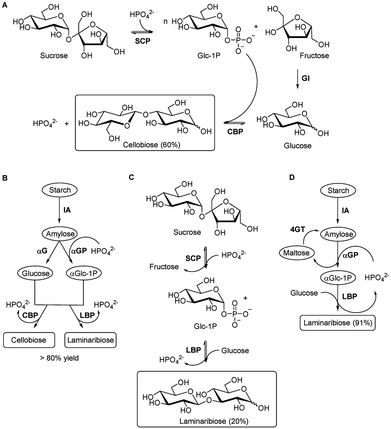
Cellobiose, a β1,4-linked disaccharide of glucose (Glcβ1-4Glc) and a component of cellulose, is valuable as a zero-calorie sweetener and potential food additive, making it commercially attractive. Enzymatic synthesis of cellobiose from sucrose has been achieved for industrial production by combining SCP and CBP. Wei and co-workers developed a multi-enzyme, one-pot synthesis of cellobiose using three enzymes. The process begins with phosphorolysis of sucrose by an SCP from Thermoanaerobacterium thermosaccharolyticum, generating Glc-1-P and fructose (Fig. 13A).149 The resulting fructose is subsequently isomerised to glucose by a glucose isomerase (GI) from Streptomyces murinus. Cellobiose is produced via the reaction catalysed by CBP from Clostridium thermocellum using Glc-1-P. You and co-workers developed an in vitro enzymatic platform for the synthesis of disaccharides from starch (Fig. 13B).150 This multi-enzyme system operates in parallel: α-glucosidase (αG) converts amylose, which is generated from starch by isoamylase (IA), into glucose, while α-glucan phosphorylase (αGP) converts amylose into Glc-1-P. These intermediates, glucose and Glc-1-P, are then transformed into the disaccharides cellobiose and laminaribiose (Glcβ1-3Glc) by CBP and laminaribiose phosphorylase (LBP), respectively, achieving yields greater than 80%. Other valuable disaccharides, including sophorose (Glcβ1-2Glc) and trehalose (Glcα1-1αGlc), were also synthesised from starch using similar enzymatic strategies. Jördening and co-workers reported the enzymatic synthesis of laminaribiose from sucrose and glucose in a one-pot reaction using individually immobilised SCP and Euglena gracilis extract enriched in LBP activity (Fig. 13C).151 An improved two-enzyme system was subsequently implemented in a packed-bed reactor for continuous laminaribiose production, achieving yields above 46 g L−1 while maintaining stable biocatalyst half-lives.152,153 You's group developed an alternative multi-enzyme approach starting from starch, a cheaper substrate than sucrose (Fig. 13D).154 Using four enzymes, laminaribiose was synthesised from starch and glucose with a yield of 91%. The process involves the following steps: (1) debranching starch using IA to generate amylose, (2) converting amylose into Glc-1-P, and (3) catalysing the final reaction with LBP in the presence of external glucose. The byproduct maltose is recycled into maltooligosaccharides via 4-α-glucanotransferase (4GT). Isono and co-workers reported the one-pot enzymatic synthesis of sophorose (Glcβ1-2Glc) from sucrose and glucose using three enzymes, sucrose phosphorylase from Leuconostoc mesenteroides, 1,2-β-oligoglucan phosphorylase from Enterococcus italicus and exo-β-1,2-glucooligosaccharide sophorohydrolase from Parabacteroides distasonis.155
 | ||
| Fig. 13 Enzymatic synthesis of cellobiose and laminaribiose catalysed by phosphorylases (reproduced with permission from Elsevier (Carbohydr. Res., 2021)).144 | ||
The efficient synthesis of well-defined cellooligosaccharides using wild-type phosphorylases remains challenging. Cellodextrin phosphorylase (CDP), a β1,4-glucan phosphorylase belonging to glycoside hydrolase family 94 (GH94), reversibly phosphorolyzes cellooligosaccharides with a DP ≥3 to produce Glc-1-P.156–159 Desmet and co-workers investigated engineered CDP from Clostridium cellulosi and CBP from Cellulomonas uda as potential biocatalysts for the enhanced production of cellotriose from Glc-1-P and cellobiose.160 However, CDP displayed rapid elongation towards higher DP, even after extensive mutagenesis, which complicates the selective synthesis of defined oligosaccharides. In contrast, an optimised CBP variant efficiently converted cellobiose to cellotriose with a molar yield of 73%. Cellotriose accounted for 82% of the final soluble cellodextrin mixture (DP = 2–5), achieving the highest reported purity to date. Loos's group reported the enzymatic synthesis of cellooligosaccharides from cellobiose and Glc-1-P using CDPs from Clostridium stercorarium and Clostridium thermocellum.161 An average DP of ∼14 was obtained for the synthesised cellooligosaccharides.
Serizawa and co-workers investigated the effects of β-glucosyl derivatives bearing hydrophilic and hydrophobic non-reactive substituents on cellulose self-assembly when used as primers for CDP from Clostridium thermocellum. They reported that oligoethylene glycol chains as primers yielded cellulose hydrogels with crystalline nanoribbon networks during enzymatic polymerisation.162 The length of the oligoethylene glycol chain influenced both the DP of the cellulose and the morphology of the nanoribbons, indicating a role in colloidal stability. Similarly, different alkyl β-glucoside primers with varying alkyl chain lengths produced distinct nanostructures, including nanoribbons, helical nanorods, and distorted nanosheets. Shorter alkyl chains favoured the formation of cellulose II-like nanoribbons and hydrogels, whereas longer chains led to dispersions with cellulose I-like structures, suggesting that alkyl chain length modulates intermolecular interactions and self-assembly (Fig. 14).163
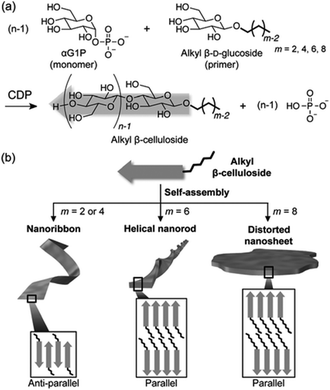 | ||
| Fig. 14 (a) Enzymatic synthesis of alkyl β-cellulosides and (b) their self-assembled structures with different alkyl chain lengths (reproduced with permission from American Chemical Society (Langmuir, 2016)).163 | ||
Nidetzky and co-workers reported the enzymatic synthesis of a reducing-end thiol-modified cellulose oligomer from 1-thio-β-D-glucose, catalysed by CBP and CDP via 1-thio-β-cellobiose (Fig. 15).164 The resulting thiol-modified cellulose oligomer self-assembled into a highly ordered crystalline material (cellulose allomorph II), forming long, thin nanosheets with micrometre-scale length and a thickness of 5–7 nm. These thiol-containing cellulose II nanosheets were subsequently loaded with silver nanoparticles, which were well-dispersed and selectively bound to the surface. The resulting hybrid material exhibited excellent antibacterial activity against Escherichia coli and Staphylococcus aureus. Wada and co-workers reported the enzymatic synthesis of plate-like cellulose II nanosheets using 13C-enriched glucose acceptors and Glc-1-P, catalysed by CDP. Solid-state nuclear magnetic resonance (NMR) spectroscopy revealed that the reducing-end units located on the nanosheet surface predominantly adopted the β-anomeric configuration, which is sterically more stable than the α-anomeric configuration.165
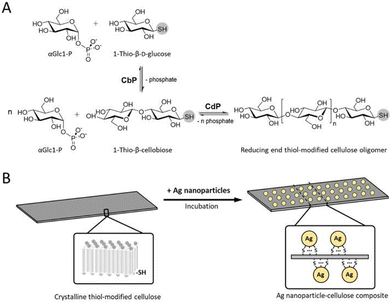 | ||
| Fig. 15 (A) Enzymatic synthesis of reducing end thiol-modified cellulose oligomer and (B) the templated assembly of silver nanoparticles onto the surface of thiol-modified cellulose (reproduced with permission from Elsevier (Carbohydr. Polym., 2021)).164 | ||
Wada and Kobayashi's group also reported the enzymatic synthesis of cellulose oligomers catalysed by cellodextrin phosphorylase.166 They investigated the effects of various cellooligosaccharide primers on the structural characteristics of the resulting products. The yield of the synthesised products correlated positively with the enzyme's activity towards the primer, whereas the DP and molecular weight distribution exhibited an inverse correlation. Similarly, Field and co-workers examined the properties of enzymatically synthesised cellulose nanosheets using fluorine-modified donors catalysed by CDP. The incorporation of multiple 6-deoxy-6-fluoro-glucose units led to shorter cellulose nanosheets with a previously unobserved crystalline allomorph, a higher average DP of approximately 10, and chains extending up to DP 15.167 The formation of highly ordered, hierarchical nanostructures through in situ self-assembly of insoluble enzymatically synthesised cellulose oligomers is influenced not only by chemical functionalisation but also by reaction conditions and the choice of natural substrates. Furthermore, Serizawa and co-workers reported that the self-assembly of cellooligosaccharides was enhanced in the presence of concentrated water-soluble polymers, such as dextran, poly(N-vinylpyrrolidone), poly(ethylene glycol), and the highly branched, high-molecular-weight neutral hydrophilic polysaccharide Ficoll.168,169 Stable hydrogels composed of well-developed crystalline nanoribbon networks of cellulose II were obtained under in vitro macromolecular crowding conditions, independent of the polymer species used. In contrast, conventional rectangular, sheet-like precipitates formed in the absence of these polymers. Furthermore, a robust double-network hydrogel was prepared through pH-triggered self-assembly of cellulose oligomers and gelatin. Insoluble cellooligosaccharides with a DP of approximately 10 dissolved in an alkaline solution and self-assembled upon lowering the pH from 7.4 in the presence of warm acidic gelatin solution, resulting in a hydrogel with enhanced stiffness.170 This strategy also enabled the formation of physically cross-linked cellulose nanoribbon hydrogels with anti-biofouling properties when applied to more complex mixtures, such as serum-containing cell culture media.171 These bottom-up approaches have considerably expanded the range of applications for functionalised, self-assembled nanocellulose materials.
Chemoenzymatic methods using glycoside hydrolases
Glycoside hydrolases (EC 3.2.1), also known as glycosidases, are enzymes that catalyse the hydrolysis of glycosidic bonds in carbohydrates and their derivatives.172–175 These ubiquitous enzymes perform diverse roles in nature, including the degradation of polysaccharide biomass, antibacterial defence, pathogenesis, and normal cellular functions such as glycoprotein biosynthesis and quality control through glycan trimming. Enzymatic glycosylation, also referred to as glycosidase-catalysed transglycosylation, has been extensively studied and applied since the 1970s. Zeuner and co-workers recently reported the enzymatic elongation of lacto-N-biose in human milk oligosaccharides using GH136 lacto-N-biosidase (LnbX) from Bifidobacterium longum subsp. infantis. This reaction involves disaccharide transglycosylation, with lacto-N-tetraose serving as the glycosyl donor and lacto-N-neotetraose as the acceptor, yielding para-lacto-N-hexaose (pLNH) (Fig. 16).176 This section provides an overview of glycosidase-catalysed glycosylation strategies for the synthesis of oligo- and polysaccharides.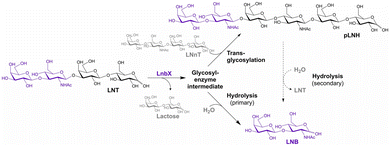 | ||
| Fig. 16 Enzymatic synthesis of para-lacto-N-hexaose (pLNH) from lacto-N-tetraose (LNT) and lacto-N-neotetraose (LNnT) (reproduced with permission from Elsevier (Enzyme Microb. Technol., 2025)).176 | ||
Chemoenzymatic synthesis of polysaccharides via glycosidase-catalysed polycondensation
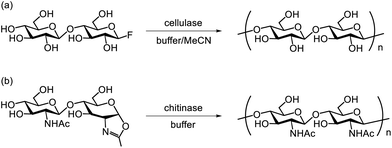
The door to chemoenzymatic synthesis of polysaccharides catalysed by glycosidases was widely opened by the cellulose synthesis reported by Kobayashi and Shoda's group in 1991. Artificial cellulose was synthesised via enzymatic polycondensation catalysed by cellulase using β-cellobiosyl fluoride as an activated monomer substrate in an acetonitrile-containing buffer (Scheme 9a).177 The use of activated substrates, such as glycosyl fluorides, is important for achieving efficient glycosidase-catalysed glycosylation.178 In general, glycosyl fluorides are chemically synthesised in four steps from unprotected saccharides: (1) acetyl protection of hydroxy groups, (2) bromination at the anomeric position, (3) introduction of fluorine at the anomeric position, and (4) deprotection of the acetyl groups. Glycosyl fluorides are moderately stable under aqueous enzymatic reaction conditions at near-neutral pH. The fluorine atom at the anomeric position serves as an efficient leaving group, facilitating both glycosidase-catalysed and chemical glycosylation. Prior to the use of glycosyl fluorides, p-nitrophenyl (pNP) glycosides were commonly used as glycosyl donors in glycosidase-catalysed reactions to synthesise various oligosaccharides and polysaccharides.179,180 This glycosidase–glycosyl fluoride methodology has also been applied to the synthesis of other β-linked polysaccharides, including cellulose derivatives and xylan.181,182Furthermore, in 1996, Kobayashi and Shoda's group reported the enzymatic synthesis of artificial chitin catalysed by chitinase, using a 1,2-oxazoline derivative of chitobiose as an activated monomer under basic buffer conditions (Scheme 9b).183 This oxazoline derivative was chemically synthesised from an unprotected saccharide in three steps: (1) acetyl chlorination, (2) formation of the oxazoline ring, and (3) deprotection of acetyl groups. The use of transition-state analogue substrates lowered the activation energy required for forming the oxazolinium ion intermediate compared to pNP-glycosides, enabling the enzymatic reaction to proceed efficiently (Fig. 17). Furthermore, glycosylation was favoured under basic conditions owing to the decreased hydrolytic activity of the enzyme. Although chitinase exhibits optimal activity near neutral pH, the glycosylation reaction was performed at pH 10. Oxazoline-mediated glycosidase-catalysed glycosylation has been applied to various glycosidases that hydrolyse the glycosidic bond of 2-acetamido-2-deoxy saccharides via a substrate-assisted catalytic mechanism.184 While the Michaelis constants (Km) of these enzymes for oxazoline derivatives are higher than for pNP-glycosides, the first-order rate constants (kcat) for oxazoline derivatives are considerably higher, facilitating efficient enzymatic glycosylation. This leads to reduced substrate recognition but an increased turnover number, enabling oxazoline-mediated glycosidase-catalysed glycosylation to efficiently produce glycosylation products. Following this approach, Kobayashi and co-workers reported the chemoenzymatic synthesis of various artificial β-linked polysaccharides, including hyaluronan,185 chondroitin and its derivatives,186 and chondroitin sulfates,187 catalysed by hyaluronidase using oxazoline derivatives of saccharides as activated monomers (Fig. 18). These methodologies have been summarised in a review article.188 Additionally, keratanase II-catalysed glycosylation has been used to synthesise keratan sulfate,189,190 sulfated type 2 carbohydrate chains,191 and sulfo-sialyloligosaccharides.192 Wang and co-workers reported the enzymatic synthesis of glycopeptides catalysed by endo-β-N-acetylglucosaminidase using an oxazoline derivative of high-mannose-type oligosaccharide as the glycosyl donor.193 They further demonstrated the chemoenzymatic synthesis of glycoproteins using sugar oxazoline derivatives, particularly via glycosidase-catalysed transglycosylation of N-glycans by endo-β-N-acetylglucosaminidases from Arthrobacter protophormiae (endo-A), Mucor hiemalis (endo-M), and other sources.194,195 These sugar oxazoline derivatives were chemically synthesised through multistep procedures involving the protection and deprotection of hydroxy groups on the saccharide moieties, as previously mentioned.
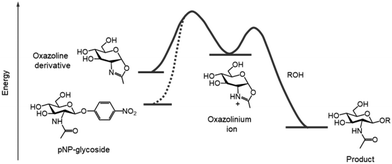 | ||
| Fig. 17 Energy profile of glycosidase-catalysed glycosylation using oxazoline derivatives as activated substrates. | ||
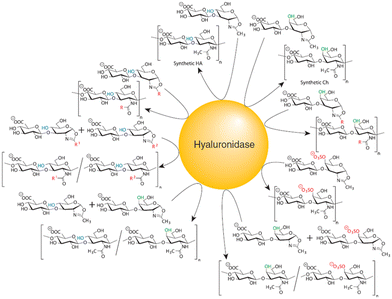 | ||
| Fig. 18 Hyaluronidase-catalysed polysaccharide synthesis using oxazoline derivatives (reproduced from Proc. Jpn. Acad., Ser. B, 2007).188 | ||
Glycosidase-catalysed glycosylation using glycosyl fluorides as donors has been extended to the synthesis of various oligosaccharides. For example, the enzymatic synthesis of oligosaccharides containing a galactose unit at the nonreducing end has been reported. β-Lactosyl fluoride was used as the glycosyl donor for enzymatic lactosylation catalysed by cellulase from Trichoderma viride (Scheme 10a).196,197 This glycosylation proceeds with complete regio- and stereoselectivity, yielding products exclusively with β1,4-glycosidic bonds. Similarly, the formation of β1,4-glycosidic bonds of GlcNAc has been achieved using chitinase from Bacillus sp. and the oxazoline derivative of N-acetyllactosamine (Scheme 10b).198 Glycosidase-catalysed synthesis of human milk oligosaccharides has been summarised by Meyer and co-workers for sialidase, α-L-fucosidase, and β-galactosidase.199 To date, numerous glycosidase-catalysed syntheses of human milk oligosaccharide analogues have been reported by the groups of Usui, Meyer, and others.200–208
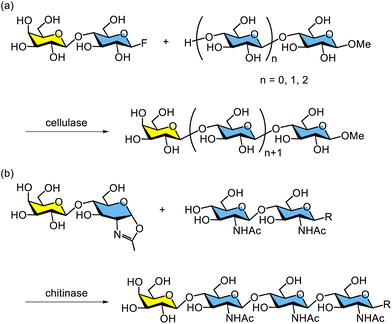 | ||
| Scheme 10 Enzymatic glycosylation using (a) cellulase with a glycosyl fluoride and (b) chitinase with an oxazoline derivative. | ||
Chemoenzymatic synthesis using mutant glycosidases
Glycosidases are generally classified as either retaining or inverting enzymes. Retaining glycosidases hydrolyse glycosidic bonds while retaining the anomeric configuration of the substrate. Wild-type retaining glycosidases are also capable of catalysing transglycosylation. In 1998, Withers and co-workers reported mutant glycosidases, termed glycosynthases, which retained the ability to catalyse glycosylation but lacked hydrolytic activity. For example, mutation of the catalytic glutamate residue to alanine in β-glucosidases from Agrobacterium sp. produces a glycosynthase capable of glycosylation without hydrolysis.209 This approach has been widely applied in the enzymatic glycosylation of peptides, proteins, and antibodies.210–214 The combination of activated glycosyl donors, such as glycosyl fluorides or sugar oxazoline derivatives, with glycosynthases enables highly efficient enzymatic synthesis of oligosaccharides.In contrast, reports of mutant inverting glycosidases used for enzymatic glycosylation are relatively scarce compared to retaining glycosidases. In 2006, Honda and Kitaoka's groups reported the first glycosynthase derived from an inverting glycosidase.215 To generate this glycosynthase, the catalytic base residue, Asp263, in the xylose-releasing exo-β-oligoxylanase was mutated. Among nine Asp263 variants—Gly, Ala, Val, Thr, Leu, Asn, Cys, Pro, and Ser—each exhibited glycosynthase activity, catalysing the formation of xylotriose from α-xylobiosyl fluoride and xylose. The mutantation of Asp263 to Cys (D263C) displayed the highest xylotriose production.
Glycosidase-catalysed glycosylation using triazinyl glycosides
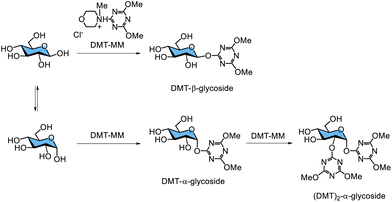
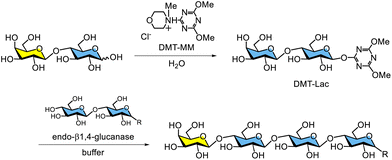
The chemical synthesis of activated glycosyl donors for glycosidase-catalysed glycosylation typically requires multistep procedures, including the protection and deprotection of hydroxy groups on saccharide moieties. In 2008, the author, then a member of Shoda's group, reported the direct synthesis of a novel triazinyl glycoside, 4,6-dimethoxy-1,3,5-triazin-2-yl glycoside (DMT-glycoside), as an activated glycosyl donor for glycosidase-catalysed glycosylation.216 DMT-glycosides can be synthesised directly from free saccharides without hydroxy group protection using the water-soluble dehydrative condensing agent 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM)217,218 under basic aqueous conditions at room temperature. Free aldoses, which are saccharides containing a hemiacetal group, can react with DMT-MM in water to form DMT-glycosides (Fig. 19). These glycosides are obtained as the main products because the anomeric hydroxy group preferentially attacks the 2-position of DMT-MM owing to its higher acidity compared with other hydroxy groups on the saccharide. The pKa value of the hemiacetal group at the anomeric position is 12.2, whereas the pKa values of primary and secondary hydroxy groups are approximately 16. The pKa value of water is 15.7, lying between these values. Aldohexoses with an equatorial hydroxy group at the 2-position, such as glucose and galactose, predominantly yield DMT-β-glycosides. Simultaneously, an α-glycoside with DMT disubstitution at the 1- and 2-positions of the saccharide moiety is formed as a byproduct. Early in the reaction, a DMT-monosubstituted α-glycoside is produced, which subsequently converts into the DMT-α-disubstituted product. The nucleophilicity of the hydroxy group at the 2-position in the DMT-α-glycoside may be enhanced intramolecularly by the lone pair on the nitrogen atom of the triazine ring at the anomeric position. In the case of mannose, which has an axial hydroxy group at the 2-position, the main product is a DMT-α-monosubstituted glycoside, while a DMT-β-disubstituted glycoside is formed as a minor byproduct.The author reported that DMT-β-lactoside (DMT-Lac) was directly synthesised from free lactose using DMT-MM (Scheme 11).216 It was recognised as a glycosyl donor by cellulase, specifically endo-β1,4-glucanase from Trichoderma reesei, yielding enzymatic glycosylation products. Additionally, several dialkoxytriazine-type glycosyl donors were directly synthesised by reacting free saccharides with 2-chloro-4,6-dimethoxy-1,3,5-triazine in water. These substrates were also successfully applied in enzymatic glycosylation catalysed by endo-β1,4-glucanase.219
Cellulase-catalysed polycondensation using DMT-glycosides as monomer substrates enables the production of artificial polysaccharides. DMT-glycosides of oligoxyloglucans, namely the cellotetraose-backboned heptasaccharide (XXXG) and nonasaccharide (XLLG) (DMT-XXXG and DMT-XLLG, respectively), can be used to synthesise non-natural xyloglucan polysaccharides with defined repeating structures via polycondensation catalysed by endo-β1,4-glucanase from Trichoderma reesei (Scheme 12).220,221
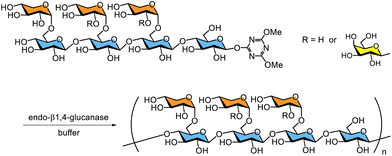 | ||
| Scheme 12 Synthesis of xyloglucans by enzymatic polycondensation using DMT-glycosides as monomer substrates. | ||
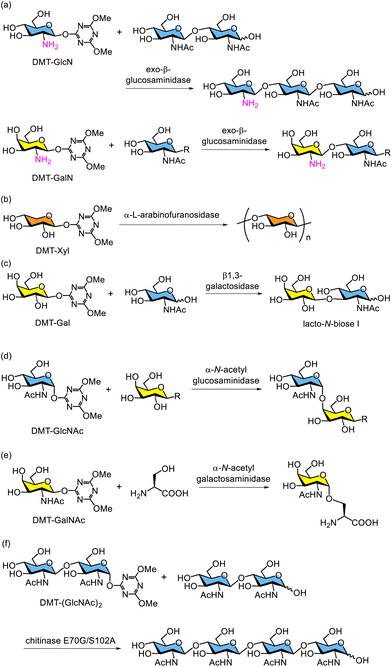
Various glycosidases have been reported to catalyse glycosylation using DMT-glycosides as glycosyl donors. For example, GH2 exo-β-D-glucosaminidase from Amycolatopsis orientalis catalyses enzymatic glucosamination and galactosamination using DMT-β-D-glucopyranosaminide (DMT-GlcN) and DMT-β-D-galactopyranosaminide (DMT-GalN), respectively (Scheme 13a).222 Enzymatic synthesis of oligoxylans using DMT-β-D-xylopyranoside (DMT-Xyl) catalysed by GH51 α-L-arabinofuranosidase from Thermotoga maritima has also been reported (Scheme 13b).223 Additionally, lacto-N-biose I was synthesised using DMT-β-D-galactopyranoside (DMT-Gal) catalysed by GH23 β1,3-galactosidase from Bacillus circulans (Scheme 13c).224 This enzymatic methodology using DMT-glycosides can be applied for the formation of 1,2-cis-glycosides of 2-amino sugars. Chemical α-glycosylation of GlcNAc is generally challenging because the predominant product is the undesired 1,2-trans isomer, the β-glycoside, owing to participation of the neighbouring 2-acetamido group in GlcNAc. However, the reaction of unprotected N-acetyl-2-amino sugars, such as GlcNAc and GalNAc, with DMT-MM in water produces DMT-α-glycosides as the main products. These DMT-α-glycosides are recognised as glycosyl donors by the corresponding glycosidases. For example, the enzymatic synthesis of oligosaccharides containing α-linked GlcNAc was reported using DMT-α-N-acetylglucopyranoside (DMT-GlcNAc) as a glycosyl donor catalysed by α-N-acetylglucosaminidase from Bacteroides thetaiotaomicron VPI5482 (Scheme 13d).225 Similarly, DMT-α-N-acetylgalactopyranoside (DMT-GalNAc) was recognised as a glycosyl donor by GH129 α-N-acetylgalactosaminidase from Bifidobacterium bifidum. This approach was subsequently used to form an α-glycosidic bond between GalNAc and L-serine, a common structure at the reducing end of O-glycans (Scheme 13e).226 A major advantage of using DMT-glycosides is the ability to increase the substrate concentration. For example, N,N′,N″,N‴-tetraacetylchitotetraose ((GlcNAc)4) was enzymatically synthesised using DMT-α-N,N′-diacetylchitobioside (DMT-(GlcNAc)2) as the glycosyl donor and chitobiose ((GlcNAc)2) as the glycosyl acceptor, catalysed by a mutant GH19 inverting chitinase from Bryum coronatum (BcChiA) (Scheme 13f).227 The double mutant E70G/S102A of BcChiA successfully produced (GlcNAc)4, where E70 acts as the catalytic base and S102 stabilises a nucleophilic water molecule. The single mutants of BcChiA, E70G and S102A, have been reported to exhibit glycosynthase activity, enabling the synthesis of (GlcNAc)4 using α-(GlcNAc)2 fluoride as a glycosyl donor.228 However, these single mutants did not yield the desired product when DMT-(GlcNAc)2 was used as the glycosyl donor for BcChiA-catalysed (GlcNAc)4 synthesis. Mutation of serine 102 to alanine creates sufficient space at the catalytic centre to accommodate the DMT moiety of the glycosyl donor. This is the first report of enzymatic glycosylation using a glycosynthase with a DMT-glycoside as the glycosyl donor. For reference, chemical glycosylation using DMT-glycosides and their derivatives as glycosyl donors has been previously reported.229–233 Based on the broad substrate recognition by corresponding glycosidases, further glycosidase-catalysed glycosylation has been explored.
Chemoenzymatic synthesis of oligosaccharides via direct synthesis of sugar oxazoline derivatives
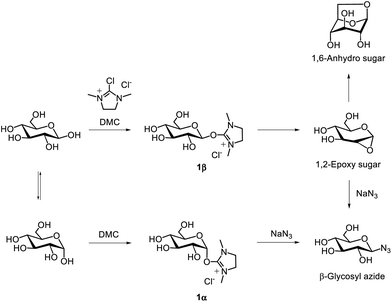
The chemical synthesis of sugar oxazoline derivatives generally requires multiple steps, including the protection and deprotection of hydroxy groups on the saccharide moieties in organic solvents. While the chemical synthesis of oxazoline derivatives of mono- and disaccharides is relatively straightforward, the preparation of oxazoline derivatives of oligosaccharides—such as N-glycan oligosaccharides with a GlcNAc moiety at the reducing end—requires a laborious procedure and accurate control of reaction conditions. In particular, even more careful handling is necessary owing to the labile α-sialyl glycosidic bond in sialyloligosaccharides.In 2009, Shoda's group developed a method for the direct synthesis of 1,2-oxazoline derivatives of N-acetyl-2-amino sugars with high yields. This approach uses a water-soluble dehydration condensing agent, 2-chloro-1,3-dimethylimidazolinium chloride (DMC),234 and does not require protection of the hydroxy groups, allowing the reaction to be performed directly in water (Fig. 20).235 This methodology for direct anomeric activation of unprotected sugars using DMC is referred to as ‘Shoda's activation’, and the reagent itself is commonly called ‘Shoda's reagent’ in carbohydrate chemistry.236,237 The reactions proceed efficiently in the presence of a base, such as triethylamine or N,N-diisopropylethylamine. This direct oxazoline formation can be applied to a variety of unprotected mono- and oligosaccharides containing N-acetyl-2-amino sugars at the reducing end, including GlcNAc, N-acetyllactosamine, chitooligosaccharides ((GlcNAc)n, n = 2–6), N-acetylglucosamine 6-sulfate (GlcNAc-6S), N-acetylglucosamine 6-phosphate (GlcNAc-6P), high-mannose-type N-glycans, and complex-type N-glycans (Fig. 20c). Additionally, Shoda's group developed 2-chloro-1,3-dimethyl-1H-benzimidazol-3-ium chloride (CDMBI) as a stable derivative of DMC, which improves stability and reduces hygroscopicity (Fig. 20b). CDMBI efficiently converts free saccharides into the corresponding oxazoline derivatives in the presence of an inorganic base, such as trisodium phosphate.238 The hydrolysate of CDMBI can be readily removed from the reaction mixture by filtration owing to its hydrophobicity and water insolubility.
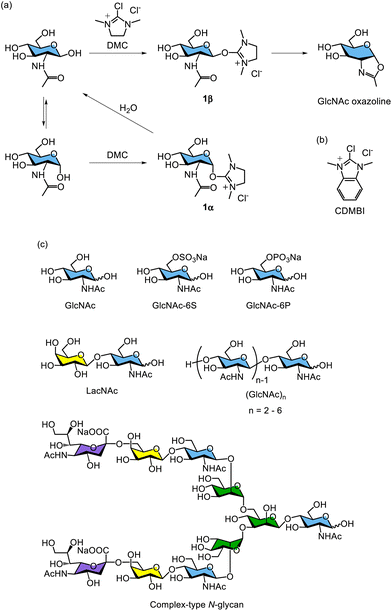 | ||
| Fig. 20 (a) Reaction mechanism of synthesizing sugar oxazoline derivatives using DMC. (b) Chemical structure of CDMBI. (c) Chemical structures of unprotected sugar as starting materials. | ||
The reaction mechanism using DMC is similar to that of DMT-MM in DMT-glycoside synthesis. The β-type hemiacetal hydroxy group at the anomeric position of the free saccharide reacts with DMC to form a β-configuration intermediate (1β) (Fig. 20a). This intermediate undergoes an intramolecular attack by the carbonyl oxygen of the 2-acetamido group, followed by deprotonation of the amide proton by a base, yielding the corresponding oxazoline derivative. In contrast, the α-type hemiacetal hydroxy group, which is in equilibrium with the β-form, reacts with DMC to form an α-configuration intermediate (1α). However, the α-type intermediate is not converted into an oxazoline derivative. Instead, it is hydrolysed by water and reverted to the original free saccharide because intramolecular oxazoline ring formation from the α-type intermediate is sterically hindered.
Recently, Shoda's activation has been widely applied in the chemoenzymatic synthesis of glycoconjugates, including glycopeptides and glycoproteins (Fig. 21).239–244 This strategy is particularly relevant in the context of mutant glycosidases for chemoenzymatic synthesis. Research on the enzymatic modification of oligosaccharides onto antibodies and the synthesis of glycosylated antibodies using sugar oxazoline derivatives has advanced rapidly.
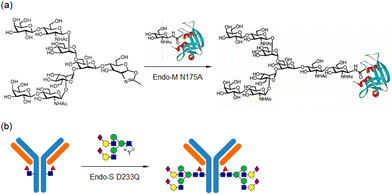 | ||
| Fig. 21 Glycosynthase-catalysed (a) enzymatic synthesis of glycoproteins and (b) remodelling of antibodies (reproduced with permission from Elsevier (Biochim. Biophys. Acta., Gen. Subj., 2010) and American Chemical Society (ACS Chem. Biol., 2016)).212,240 | ||
Additionally, the Fairbanks and co-workers reported a nonenzymatic glycosylation method using sugar oxazoline derivatives via Shoda's activation with DMC, enabling the synthesis of β1,6-linked disaccharides of 2-acetamido sugars.245
One-pot and protecting-group-free chemoenzymatic synthesis of oligosaccharides and glycopolymers in water
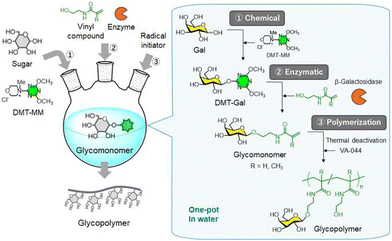
The one-pot synthesis method is widely used not only in chemical synthesis but also in chemoenzymatic synthesis, including synthetic carbohydrate chemistry.246–251 Its advantage lies in using water-soluble dehydration condensing agents—such as DMT-MM, DMC, and their derivatives—in aqueous conditions for the direct synthesis of DMT-glycosides and sugar oxazoline derivatives. This approach enables a one-pot chemoenzymatic process without the need to isolate intermediates. Upon adding a glycosyl acceptor and glycosidase, enzymatic glycosylation proceeds to yield oligosaccharides—such as lactosyl oligosaccharides, chitoheptoaose, and N-glycans—, eliminating the isolation of glycosyl intermediate donors (Scheme 14).216,252,253 Additionally, the author described the one-pot chemoenzymatic synthesis of glycopolymers—biologically functional polymers featuring a synthetic polymer backbone with attached saccharides. Glycomonomers containing a Gal residue were synthesised via β-galactosidase-catalysed reaction using DMT-Gal as the glycosyl donor and vinyl compounds as acceptor substrates, without isolating DMT-Gal. After thermally deactivating the enzyme, a water-soluble radical initiator, 2,2′-azobis[2-(2-imidazolin-2-yl)propane] dihydrochloride (VA-044), was added, and radical polymerisation was performed to produce glycopolymers bearing Gal moieties. These glycopolymers were successfully synthesised from the unprotected sugars using a one-pot procedure in aqueous conditions (Fig. 22).254,255 | ||
| Scheme 14 One-pot chemoenzymatic glycosylation via direct synthesis of the sugar oxazoline derivative. | ||
1,2-Epoxy sugar as an intermediate of glycosidases
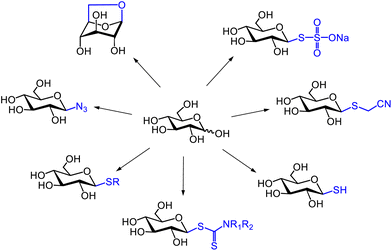
1,2-Anhydro sugars, also called 1,2-epoxy sugars, feature an epoxide ring where an oxygen atom is bonded to the 1- and 2-positions of a pyranose ring. Protected 1,2-epoxy sugars are versatile saccharide derivatives widely used as glycosyl donors in chemical glycosylation.256–259 In contrast, unprotected 1,2-epoxy sugars are known to be unstable and are thus not used as substrates in glycosylation reactions.Shoda's activation of unprotected sugars using DMC enables the direct synthesis of a variety of saccharide derivatives in water. For instance, 1,6-anhydro sugars,260 glycosyl azides,261 thioglycosides,262–264 glycosyl dithiocarbamates,265 glycosyl thiols,266 cyanomethyl thioglycosides,267 and glycosyl Bunte salts268 can be directly synthesised from free saccharides using DMC (Fig. 23). Additionally, sugar nucleoside diphosphates were synthesised using DMC and imidazole, forming 2-imidazolyl-1,3-dimethylimidazolinium chloride (ImIm) in situ in water.269,270 This compound can activate nucleoside 5′-monophosphates in water to produce a reactive phosphorimidazolide intermediate. Coupling this intermediate with sugar-1-phosphates in water afforded sugar nucleoside diphosphates in good yields. Additionally, α-glycosyl chlorides were synthesised from protected saccharides bearing a hydroxy group at the anomeric position, using DMC in an organic solvent.271 In studies on the direct synthesis of various saccharide derivatives from unprotected glucose, 1,2-epoxy sugars were proposed as a second intermediate following the β-glycosyl intermediate 1β, illustrating the reaction mechanism (Fig. 24). However, these 1,2-epoxy sugars were not detected at that time. It was only in 2017 that Shoda's group successfully detected unprotected 1,2-epoxy sugars in an acetonitrile-containing aqueous reaction mixture using low-temperature NMR measurements.272,273
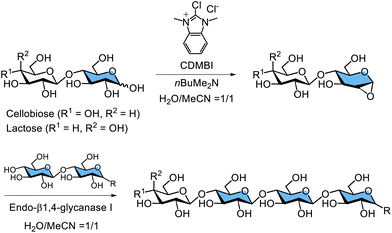
The notion that 1,2-epoxy sugars serve as reaction intermediates in glycosidase-catalysed hydrolysis is controversial. The hydrolysis mechanism of family GH99 endo-α1,2-mannosidase suggests that the 1,2-epoxy sugar acts as an intermediate, as evidenced by X-ray crystallography.274 Additionally, 1,2-epoxy sugar has recently been proposed as an intermediate in endo-α1,2-mannosidase-catalysed hydrolysis owing to neighbouring-group participation by the 2-position hydroxy group on mannose.275 Furthermore, Shoda's group has pioneered new approaches in glycosidase-catalysed glycosylation using 1,2-epoxy sugars as glycosyl donors. They explored the enzymatic glycosylation activity of family GH12 endo-β1,4-glucanase I from Aspergillus aculeatus and demonstrated glycosylation of 1,2-epoxy sugars derived from free cellobiose and lactose, using CDMBI as the glycosyl donor under acetonitrile–water conditions (Scheme 15).276 Additionally, the glycosylation activity of family GH5 endo-β1,4-glucanase II from Aspergillus aculeatus was shown to transfer 1,2-anhydro cellobiose, an important finding in this field. Moreover, endo-β1,4-glucanases from Trichoderma reesei, belonging to the GH5 and GH12 families, exhibited similar glycosylation capabilities.277–279 Family GH16 xyloglucan endo-transglycosylase/hydrolase from Arabidopsis thaliana, which catalyses the hydrolysis and glycosylation of β1,4-glycosidic bonds in xyloglucan, recognised the 1,2-anhydro sugar of the xyloglucan heptasaccharide (XXXG) and generated a xyloglucan 14-sugar product (XXXG)2. Based on these experimental findings and previous crystal structure data of the substrate complex, it has been hypothesised that the hydrolysis mechanism of certain retaining endo-β-glucanases involves a 1,2-anhydro sugar intermediate. This intermediate forms through neighbouring-group participation by the 2-position hydroxy group during glycosidic bond cleavage. Since the 1990s, the crystal structures of the covalent intermediates of anomer-retaining enzymes, including the endo-β1,4-glucanases belonging to the GH5 and GH12 families, have been analysed. The Koshland mechanism, which involves glycosyl ester intermediates, is widely accepted.280–283 However, Shoda argues that experimental evidence supporting 1,2-anhydro sugar intermediates does not necessarily contradict the Koshland mechanism, although this remains a matter of debate.276 Therefore, there is still potential to improve glycosidase-catalysed glycosylation using 1,2-anhydro sugars as glycosyl donors in other glycosidases.
Conclusions
The recent shift towards a circular economy and carbon neutrality has been strongly driven by combining accurate chemical synthesis with environmentally friendly enzymatic reactions. In glycoscience and glycotechnology, chemoenzymatic methods play a crucial role in synthesising oligosaccharides, polysaccharides, and glycosoconjugates. These methods advance the understanding of carbohydrate chemistry and the enzymology of carbohydrate-active enzymes.This review summarised chemoenzymatic synthesis techniques, organised by enzyme types: glycosyltransferases, glycan phosphorylases, and glycosyl hydrolases. Each enzyme's unique properties were leveraged to synthesise a variety of oligosaccharides, polysaccharides, and their derivatives. As new enzymes are discovered and their functions leveraged, chemoenzymatic synthesis will continue to evolve and expand in glycotechnology and polymer chemistry.
Author contributions
This manuscript was conceptualized and originally written by the author.Conflicts of interest
There are no conflicts to declare.Data availability
Since this is a review article, there is no research data available that would have to be disclosed.Acknowledgements
This review article is dedicated to Professor Christine Luscombe, of the Okinawa Institute of Science and Technology in Japan, to celebrate her appointment as the editor-in-chief of Polymer Chemistry. The author would like to thank all co-workers and laboratory members for their contributions to my research presented in this paper.References
- X. Zhu and R. R. Schmidt, Angew. Chem., Int. Ed., 2009, 48, 1900–1934 CrossRef CAS PubMed.
- S. S. Nigudkar and A. V. Demchenko, Chem. Sci., 2015, 6, 2687–2704 RSC.
- R. Das and B. Mukhopadhyay, ChemistryOpen, 2016, 5, 401–433 CrossRef CAS PubMed.
- S. Shoda, Proc. Jpn. Acad., Ser. B, 2017, 93, 125–145 CrossRef CAS.
- P. O. Adero, H. Amarasekara, P. Wen, L. Bohé and D. Crich, Chem. Rev., 2018, 118, 8242–8284 CrossRef CAS.
- A. M. Vibhute, N. Komura, H. Tanaka, A. Imamura and H. Ando, Chem. Rec., 2021, 21, 3194–3223 CrossRef CAS.
- Y. Singh, S. A. Geringer and A. V. Demchenko, Chem. Rev., 2022, 122, 11701–11758 CrossRef CAS.
- M. Panza, S. G. Pistorio, K. J. Stine and A. V. Demchenko, Chem. Rev., 2018, 118, 8105–8150 CrossRef CAS.
- M. Guberman and P. H. Seeberger, J. Am. Chem. Soc., 2019, 141, 5581–5592 CrossRef CAS PubMed.
- W. Yao, D.-C. Xiong, Y. Yang, C. Geng, Z. Cong, F. Li, B.-H. Li, X. Qin, L.-N. Wang, W.-Y. Xue, N. Yu, H. Zhang, X. Wu, M. Liu and X.-S. Ye, Nat. Synth., 2022, 1, 854–863 CrossRef CAS.
- J. Suri and R. Gilmour, Angew. Chem., Int. Ed., 2025, 64, e202422766 CrossRef CAS PubMed.
- S. Shoda, H. Uyama, J. Kadokawa, S. Kimura and S. Kobayashi, Chem. Rev., 2016, 116, 2307–2413 CrossRef CAS PubMed.
- L. Na, R. Li and X. Chen, Curr. Opin. Chem. Biol., 2021, 61, 81–95 CrossRef CAS PubMed.
- G. K. Wagner, T. Pesnota and R. A. Field, Nat. Prod. Rep., 2009, 26, 1172–1194 RSC.
- T. Bülter and L. Elling, Glycoconjugate J., 1999, 16, 147–159 CrossRef.
- B. Roy, A. Depaix, C. Périgaud and S. Peyrottes, Chem. Rev., 2016, 116, 7854–7897 CrossRef CAS.
- J. Li, A. Amatuni and H. Renata, Curr. Opin. Chem. Biol., 2020, 55, 111–118 CrossRef CAS PubMed.
- F. Kaspar and A. Schallmey, Curr. Opin. Biotechnol., 2022, 77, 102759 CrossRef CAS.
- C. N. Stout, N. M. Wasfy, F. Chen and H. Renata, J. Am. Chem. Soc., 2023, 145, 18161–18181 CrossRef CAS.
- M. Zheng, Z. Lin, S. Lin and X. Qu, Eur. J. Org. Chem., 2024, e202301066 CrossRef CAS.
- K. Yazawa and K. Numata, Molecules, 2014, 19, 13755–13774 CrossRef PubMed.
- K. Tsuchiya and K. Numata, Macromol. Biosci., 2017, 17, 1700177 CrossRef.
- R. E. Thompson and T. W. Muir, Chem. Rev., 2020, 120, 3051–3126 CrossRef CAS PubMed.
- Y. Yang, J. Zhang, D. Wu, Z. Xing, Y. Zhou, W. Shi and Q. Li, Biotechnol. Adv., 2014, 32, 642–651 CrossRef CAS PubMed.
- S. Nishimura and K. Yamada, J. Am. Chem. Soc., 1997, 119, 10555–10556 CrossRef CAS.
- M. Fumoto, H. Hinou, T. Matsushita, M. Kurogochi, T. Ohta, T. Ito, K. Yamada, A. Takimoto, H. Kondo, T. Inazu and S. Nishimura, Angew. Chem., Int. Ed., 2005, 44, 2534–2537 CrossRef CAS.
- M. Fumoto, H. Hinou, T. Ohta, T. Ito, K. Yamada, A. Takimoto, H. Kondo, H. Shimizu, T. Inazu, Y. Nakahara and S. Nishimura, J. Am. Chem. Soc., 2005, 127, 11804–11818 CrossRef CAS.
- L. Wen, G. Edmunds, C. Gibbons, J. Zhang, M. R. Gadi, H. Zhu, J. Fang, X. Liu, Y. Kong and P. G. Wang, Chem. Rev., 2018, 118, 8151–8187 CrossRef CAS PubMed.
- K. P. Hussnaetter, P. Palm, A. Pich, M. Franzreb, E. Rapp and L. Elling, Biotechnol. Adv., 2023, 67, 108208 CrossRef CAS PubMed.
- M. Kitaoka, Adv. Nutr., 2012, 3, 422S–429S CrossRef CAS.
- D. Garrido, S. Ruiz-Moyano and D. A. Mills, Anaerobe, 2012, 18, 430–435 CrossRef CAS PubMed.
- T. Urashima, J. Hirabayashi, S. Sato and A. Kobata, Trends Glycosci. Glycotechnol., 2018, 30, SE51–SE65 CrossRef.
- J. Zheng, H. Xu, J. Fang and X. Zhang, Carbohydr. Polym., 2022, 291, 119564 CrossRef CAS PubMed.
- M. Hu, M. Miao, K. Li, Q. Luan, G. Sun and T. Zhang, Carbohydr. Polym., 2023, 316, 121067 CrossRef CAS PubMed.
- L. Bode, Glycobiology, 2012, 22, 1147–1162 CrossRef CAS.
- X. Chen, Adv. Carbohydr. Chem. Biochem., 2015, 72, 113–190 CrossRef.
- G. Georgi, N. Bartke, F. Wiens and B. Stahl, Am. J. Clin. Nutr., 2013, 98, 578S–585S CrossRef CAS PubMed.
- W. Yao, J. Yan, X. Chen, F. Wang and H. Cao, Carbohydr. Res., 2015, 401, 5–10 CrossRef CAS.
- X. Ma and J. Stöckigt, Carbohydr. Res., 2001, 333, 159–163 CrossRef CAS PubMed.
- M. M. Muthana, J. Qu, Y. Li, L. Zhang, H. Yu, L. Ding, H. Malekan and X. Chen, Chem. Commun., 2012, 48, 2728 RSC.
- C. Ou, C. Li, R. Zhang, Q. Yang, G. Zong, Y. Dai, R. L. Francis, S. Bournazos, J. V. Ravetch and L.-X. Wang, Bioconjugate Chem., 2021, 32, 1888–1897 CrossRef CAS.
- H. Yu and X. Chen, Org. Biomol. Chem., 2016, 14, 2809–2818 RSC.
- Y. Li, Q. Chen, S. Liu, L. Deng, S. Li and R. Gao, J. Agric. Food Chem., 2024, 72, 3644–3653 CrossRef CAS PubMed.
- Z. Xiao, Y. Guo, Y. Liu, L. Li, Q. Zhang, L. Wen, X. Wang, S. M. Kondengaden, Z. Wu, J. Zhou, X. Cao, X. Li, C. Ma and P. G. Wang, J. Org. Chem., 2016, 81, 5851–5865 CrossRef CAS.
- T.-W. Tsai, J.-L. Fang, C.-Y. Liang, C.-J. Wang, Y.-T. Huang, Y.-J. Wang, J.-Y. Li and C.-C. Yu, ACS Catal., 2019, 9, 10712–10720 CrossRef CAS.
- Y.-T. Huang, Y.-C. Su, H.-R. Wu, H.-H. Huang, E. C. Lin, T.-W. Tsai, H.-W. Tseng, J.-L. Fang and C.-C. Yu, ACS Catal., 2021, 11, 2631–2643 CrossRef CAS.
- A. R. Prudden, L. Liu, C. J. Capicciotti, M. A. Wolfert, S. Wang, Z. Gao, L. Meng, K. W. Moremen and G.-J. Boons, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6954–6959 CrossRef CAS.
- Y. Li, Y. Li, Y. Guo, C. Chen, L. Yang, Q. Jiang, P. Ling, S. Wang, L. Li and J. Fang, Carbohydr. Polym., 2024, 333, 121908 CrossRef CAS.
- G. M. Vos, Y. Wu, R. van der Woude, R. P. de Vries and G. Boons, Chem. – Eur. J., 2024, 30, e202302877 CrossRef CAS.
- B. Petschacher and B. Nidetzky, J. Biotechnol., 2016, 235, 61–83 CrossRef CAS PubMed.
- A. Santra, Y. Li, T. Ghosh, R. Li, H. Yu and X. Chen, ACS Catal., 2019, 9, 211–215 CrossRef CAS PubMed.
- Z. Xu, Y. Deng, Z. Zhang, W. Ma, W. Li, L. Wen and T. Li, J. Org. Chem., 2021, 86, 10819–10828 CrossRef CAS PubMed.
- S. Wang, C. Chen, M. R. Gadi, V. Saikam, D. Liu, H. Zhu, R. Bollag, K. Liu, X. Chen, F. Wang, P. G. Wang, P. Ling, W. Guan and L. Li, Nat. Commun., 2021, 12, 3573 CrossRef CAS PubMed.
- M. R. Gadi, C. Chen, S. Bao, S. Wang, Y. Guo, J. Han, W. Xiao and L. Li, Chem. Sci., 2023, 14, 1837–1843 RSC.
- Y. Wu, J. Weber, A. L. M. Kimpel, L. Unione, E. Uslu, R. P. de Vries and G.-J. Boons, JACS Au, 2025, 5, 3328–3338 CrossRef PubMed.
- S. Ma, P. Zhang, J. Ye, Y. Tian, X. Tian, J. Jung, M. S. Macauley, J. Zhang, P. Wu and L. Wen, J. Am. Chem. Soc., 2024, 146, 29469–29480 CrossRef CAS PubMed.
- S. Ma, J. Zhang, F. Wei, X. Tian, Y. Tian and L. Wen, J. Am. Chem. Soc., 2025, 147, 35042–35054 CrossRef CAS PubMed.
- F. Wei, L. Zang, P. Zhang, J. Zhang and L. Wen, Chem, 2024, 10, 2844–2860 CAS.
- R. L. Jackson, S. J. Busch and A. D. Cardin, Physiol. Rev., 1991, 71, 481–539 CrossRef CAS PubMed.
- M. Mende, C. Bednarek, M. Wawryszyn, P. Sauter, M. B. Biskup, U. Schepers and S. Bräse, Chem. Rev., 2016, 116, 8193–8255 CrossRef CAS PubMed.
- A. Köwitsch, G. Zhou and T. Groth, J. Tissue Eng. Regener. Med., 2018, 12, e23–e41 CrossRef PubMed.
- P. L. DeAngelis, J. Liu and R. J. Linhardt, Glycobiology, 2013, 23, 764–777 CrossRef CAS PubMed.
- X. Zhang, L. Lin, H. Huang and R. J. Linhardt, Acc. Chem. Res., 2020, 53, 335–346 CrossRef CAS PubMed.
- Y. Xu, S. Masuko, M. Takieddin, H. Xu, R. Liu, J. Jing, S. A. Mousa, R. J. Linhardt and J. Liu, Science, 2011, 334, 498–501 CrossRef CAS.
- T. Wang, L. Liu and J. Voglmeir, Biochim. Biophys. Acta, Proteins Proteomics, 2020, 1868, 140301 CrossRef CAS.
- K. Nagasawa, H. Uchiyama and N. Wajima, Carbohydr. Res., 1986, 158, 183–190 CrossRef CAS.
- K. Sugahara, Y. Tanaka, S. Yamada, N. Seno, H. Kitagawa, S. M. Haslam, H. R. Morris and A. Dell, J. Biol. Chem., 1996, 271, 26745–26754 CrossRef CAS PubMed.
- C. Cai, K. Solakyildirim, B. Yang, J. M. Beaudet, A. Weyers, R. J. Linhardt and F. Zhang, Carbohydr. Polym., 2012, 87, 822–829 CrossRef CAS PubMed.
- M. Roriguez, B. Jann and K. Jann, Eur. J. Biochem., 1988, 177, 117–124 Search PubMed.
- M. Manzoni, S. Bergomi, F. Molinari and V. Cavazzoni, Biotechnol. Lett., 1996, 18, 383–386 CrossRef CAS.
- D. J. Mans, H. Ye, J. D. Dunn, R. E. Kolinski, D. S. Long, N. L. Phatak, H. Ghasriani, L. F. Buhse, J. F. Kauffman and D. A. Keire, Anal. Biochem., 2015, 490, 52–54 CrossRef CAS PubMed.
- E. Bedini, A. Laezza and A. Iadonisi, Eur. J. Org. Chem., 2016, 3018–3042 CrossRef CAS.
- V. D. Nadkarni, T. Toida, C. L. Van Gorp, R. L. Schubert, J. M. Weiler, K. P. Hansen, E. E. O. Caldwell and R. J. Linhardt, Carbohydr. Res., 1996, 290, 87–96 CrossRef CAS PubMed.
- G. Li, C. Cai, L. Li, L. Fu, Y. Chang, F. Zhang, T. Toida, C. Xue and R. J. Linhardt, Anal. Chem., 2014, 86, 326–330 CrossRef CAS PubMed.
- X. Zhou, K. Chandarajoti, T. Q. Pham, R. Liu and J. Liu, Glycobiology, 2011, 21, 771–780 CrossRef CAS PubMed.
- K. Sugahara and H. Kitagawa, Curr. Opin. Struct. Biol., 2000, 10, 518–527 CrossRef CAS.
- J. Gottschalk and L. Elling, Curr. Opin. Struct. Biol., 2021, 61, 71–80 CrossRef CAS.
- M. Kitaoka and K. Hayashi, Trends Glycosci. Glycotechnol., 2002, 14, 35–50 CrossRef CAS.
- R. S. Rathore, N. Garg, S. Garg and A. Kumar, Crit. Rev. Biotechnol., 2009, 29, 214–224 CrossRef CAS PubMed.
- H. Nakai, M. Kitaoka, B. Svensson and K. Ohtsubo, Curr. Opin. Chem. Biol., 2013, 17, 301–309 CrossRef CAS PubMed.
- V. Puchart, Biotechnol. Adv., 2015, 33, 261–276 CrossRef CAS PubMed.
- M. S. Gentry, M. K. Brewer and C. W. Vander Kooi, Curr. Opin. Struct. Biol., 2016, 40, 62–69 CrossRef CAS PubMed.
- G. Pergolizzi, S. Kuhaudomlarp, E. Kalita and R. A. Field, Protein Pept. Lett., 2017, 24, 696–709 CrossRef CAS PubMed.
- Z. Ubiparip, K. Beerens, J. Franceus, R. Vercauteren and T. Desmet, Appl. Microbiol. Biotechnol., 2018, 102, 8187–8202 CrossRef CAS PubMed.
- E. C. O'Neill and R. A. Field, Carbohydr. Res., 2015, 403, 23–37 CrossRef PubMed.
- F. N. Awad, Biocatal. Agric. Biotechnol., 2021, 31, 101886 CrossRef CAS.
- K. Loos and J. Kadokawa, in Enzymatic Polymerization towards Green Polymer Chemistry, ed. S. Kobayashi, H. Uyama and J. Kadokawa, Springer, 2019, pp. 47–87 Search PubMed.
- S. J. Reid and V. R. Abratt, Appl. Microbiol. Biotechnol., 2005, 67, 312–321 CrossRef CAS.
- C. Goedl, T. Sawangwan, P. Wildberger and B. Nidetzky, Biocatal. Biotransform., 2010, 28, 10–21 CrossRef CAS.
- Y.-K. Kim, M. Kitaoka, M. Krishnareddy, Y. Mori and K. Hayashi, J. Biochem., 2002, 132, 197–203 CrossRef CAS PubMed.
- M. Nawaji, H. Izawa, Y. Kaneko and J. Kadokawa, Carbohydr. Res., 2008, 343, 2692–2696 CrossRef CAS PubMed.
- K. Fujii, M. Iiboshi, M. Yanase, T. Takaha and T. Kuriki, J. Appl. Glycosci., 2006, 53, 91–97 CrossRef CAS.
- M. Yanase, H. Takata, K. Fujii, T. Takaha and T. Kuriki, Appl. Environ. Microbiol., 2005, 71, 5433–5439 CrossRef CAS PubMed.
- C. Luley-Goedl and B. Nidetzky, Biotechnol. J., 2010, 5, 1324–1338 CrossRef CAS.
- J. Franceus and T. Desmet, Int. J. Mol. Sci., 2020, 21, 2526 CrossRef CAS PubMed.
- K. Loos and R. Stadler, Macromolecules, 1997, 30, 7641–7643 CrossRef CAS.
- K. Loos and A. H. E. Müller, Biomacromolecules, 2002, 3, 368–373 CrossRef CAS.
- K. Loos, A. Böker, H. Zettl, M. Zhang, G. Krausch and A. H. E. Müller, Macromolecules, 2005, 38, 873–879 CrossRef CAS.
- K. Kumar, A. J. J. Woortman and K. Loos, Macromol. Rapid Commun., 2015, 36, 2097–2101 CrossRef CAS.
- J. van der Vlist, M. Faber, L. Loen, T. J. Dijkman, L. A. T. W. Asri and K. Loos, Polymers, 2012, 4, 674–690 CrossRef.
- K. Loos, V. von Braunmühl, R. Stadler, K. Landfester and H. W. Spiess, Macromol. Rapid Commun., 1997, 18, 927–938 CrossRef CAS.
- J. van der Vlist, I. Schönen and K. Loos, Biomacromolecules, 2011, 12, 3728–3732 CrossRef CAS PubMed.
- J. Kadokawa, Trends Glycosci. Glycotechnol., 2013, 25, 57–69 CrossRef CAS.
- B. Evers and J. Thiem, Bioorg. Med. Chem., 1997, 5, 857–863 CrossRef CAS PubMed.
- B. Evers, P. Mischnick and J. Thiem, Carbohydr. Res., 1994, 262, 335–341 CrossRef CAS PubMed.
- M. Nawaji, H. Izawa, Y. Kaneko and J. Kadokawa, J. Carbohydr. Chem., 2008, 27, 214–222 CrossRef CAS.
- S. Kawazoe, H. Izawa, M. Nawaji, Y. Kaneko and J. Kadokawa, Carbohydr. Res., 2010, 345, 631–636 CrossRef CAS PubMed.
- Y. Umegatani, H. Izawa, M. Nawaji, K. Yamamoto, A. Kubo, M. Yanase, T. Takaha and J. Kadokawa, Carbohydr. Res., 2012, 350, 81–85 CrossRef CAS.
- S. Matsuda, Y. Kaneko and J. Kadokawa, Macromol. Rapid Commun., 2007, 28, 863–867 CrossRef CAS.
- Y. Omagari, S. Matsuda, Y. Kaneko and J. Kadokawa, Macromol. Biosci., 2009, 9, 450–455 CrossRef CAS PubMed.
- Y. Omagari, Y. Kaneko and J. Kadokawa, Carbohydr. Polym., 2010, 82, 394–400 CrossRef CAS.
- T. Arimura, Y. Omagari, K. Yamamoto and J. Kadokawa, Int. J. Biol. Macromol., 2011, 49, 498–503 CrossRef CAS PubMed.
- J. Kadokawa, T. Arimura, Y. Takemoto and K. Yamamoto, Carbohydr. Polym., 2012, 90, 1371–1377 CrossRef CAS PubMed.
- D. Hatanaka, Y. Takemoto, K. Yamamoto and J. Kadokawa, Fibers, 2014, 2, 34–44 CrossRef.
- J. Kadokawa, Polymers, 2016, 8, 138 CrossRef PubMed.
- H. Izawa, M. Nawaji, Y. Kaneko and J. Kadokawa, Macromol. Biosci., 2009, 9, 1098–1104 CrossRef CAS.
- Y. Takemoto, H. Izawa, Y. Umegatani, K. Yamamoto, A. Kubo, M. Yanase, T. Takaha and J. Kadokawa, Carbohydr. Res., 2013, 366, 38–44 CrossRef CAS.
- Y. Takata, R. Shimohigoshi, K. Yamamoto and J. Kadokawa, Macromol. Biosci., 2014, 14, 1437–1443 CrossRef CAS PubMed.
- Y. Takata, K. Yamamoto and J. Kadokawa, Macromol. Chem. Phys., 2015, 216, 1415–1420 CrossRef CAS.
- E. Gau, F. Flecken, T. Belthle, M. Ambarwati, K. Loos and A. Pich, Macromol. Rapid Commun., 2019, 40, 1900144 CrossRef.
- Y. Kaneko and J. Kadokawa, Chem. Rec., 2005, 5, 36–46 CrossRef CAS PubMed.
- J. Kadokawa, Polymers, 2012, 4, 116–133 CrossRef CAS.
- J. Kadokawa, Biomolecules, 2013, 3, 369–385 CrossRef.
- J. Kadokawa, Y. Kaneko, S. Nagase, T. Takahashi and H. Tagaya, Chem. – Eur. J., 2002, 8, 3321–3326 CrossRef CAS.
- J. Kadokawa, Y. Kaneko, H. Tagaya and K. Chiba, Chem. Commun., 2001, 449–450 RSC.
- J. Kadokawa, Y. Kaneko, A. Nakaya and H. Tagaya, Macromolecules, 2001, 34, 6536–6538 CrossRef CAS.
- J. Kadokawa, A. Nakaya, Y. Kaneko and H. Tagaya, Macromol. Chem. Phys., 2003, 204, 1451–1457 CrossRef CAS.
- S. Nomura, T. Kyutoku, N. Shimomura, Y. Kaneko and J. Kadokawa, Polym. J., 2011, 43, 971–977 CrossRef CAS.
- Y. Kaneko, K. Ueno, T. Yui, K. Nakahara and J. Kadokawa, Macromol. Biosci., 2011, 11, 1407–1415 CrossRef CAS PubMed.
- Y. Kaneko, K. Beppu and J. I. Kadokawa, Macromol. Chem. Phys., 2008, 209, 1037–1042 CrossRef CAS.
- J. Kadokawa, K. Yano, S. Orio and K. Yamamoto, ACS Omega, 2019, 4, 6331–6338 CrossRef CAS PubMed.
- Y. Kaneko, K. Beppu and J. Kadokawa, Biomacromolecules, 2007, 8, 2983–2985 CrossRef CAS PubMed.
- Y. Kaneko, K. Beppu, T. Kyutoku and J. Kadokawa, Polym. J., 2009, 41, 279–286 CrossRef CAS.
- Y. Kaneko, K. Beppu and J. Kadokawa, Polym. J., 2009, 41, 792–796 CrossRef CAS.
- T. Tanaka, S. Sasayama, S. Nomura, K. Yamamoto, Y. Kimura and J. Kadokawa, Macromol. Chem. Phys., 2013, 214, 2829–2834 CrossRef CAS.
- T. Tanaka, S. Sasayama, K. Yamamoto, Y. Kimura and J. Kadokawa, Macromol. Chem. Phys., 2015, 216, 794–800 CrossRef CAS.
- T. Tanaka, A. Tsutsui, R. Gotanda, S. Sasayama, K. Yamamoto and J. Kadokawa, J. Appl. Glycosci., 2015, 62, 135–141 CrossRef CAS.
- T. Tanaka, R. Gotanda, A. Tsutsui, S. Sasayama, K. Yamamoto, Y. Kimura and J. Kadokawa, Polymer, 2015, 73, 9–16 CrossRef CAS.
- Y. Kaneko, K. Fujisaki, T. Kyutoku, H. Furukawa and J. Kadokawa, Chem. – Asian J., 2010, 5, 1627–1633 CrossRef CAS PubMed.
- J. Kadokawa, S. Nomura, D. Hatanaka and K. Yamamoto, Carbohydr. Polym., 2013, 98, 611–617 CrossRef CAS PubMed.
- J. Kadokawa, K. Tanaka, D. Hatanaka and K. Yamamoto, Polym. Chem., 2015, 6, 6402–6408 RSC.
- L. R. Maréchal and S. H. Goldemberg, Biochem. Biophys. Res. Commun., 1963, 13, 106–109 CrossRef.
- Z. Ubiparip, M. De Doncker, K. Beerens, J. Franceus and T. Desmet, Appl. Microbiol. Biotechnol., 2021, 105, 4073–4087 CrossRef CAS PubMed.
- G. S. Bulmer, P. de Andrade, R. A. Field and J. M. van Munster, Carbohydr. Res., 2021, 508, 108411 CrossRef CAS PubMed.
- M. Nakajima, S. Motouchi, N. Tanaka and T. Masaike, Appl. Microbiol. Biotechnol., 2025, 109, 49 CrossRef CAS PubMed.
- M. Nishimoto and M. Kitaoka, Biosci. Biotechnol. Biochem., 2007, 71, 2101–2104 CrossRef CAS PubMed.
- M. Nishimoto, Biosci. Biotechnol. Biochem., 2020, 84, 17–24 CrossRef CAS PubMed.
- S. Machida, K. Saito, M. Nishimoto and M. Kitaoka, J. Appl. Glycosci., 2022, 69, 15–22 CrossRef CAS PubMed.
- C. Zhong, P. Wei and Y.-H. P. Zhang, Chem. Eng. Sci., 2017, 161, 159–166 CrossRef CAS.
- S. Sun, X. Wei, X. Zhou and C. You, J. Agric. Food Chem., 2021, 69, 302–314 CrossRef CAS PubMed.
- C. Müller, T. Ortmann, A. Abi, D. Hartig, S. Scholl and H.-J. Jördening, Appl. Biochem. Biotechnol., 2017, 182, 197–215 CrossRef PubMed.
- A. Abi, C. Müller and H.-J. Jördening, Biotechnol. Bioprocess Eng., 2017, 22, 272–280 CrossRef CAS.
- A. Abi, A. Wang and H.-J. Jördening, Appl. Biochem. Biotechnol., 2018, 186, 861–876 CrossRef CAS PubMed.
- S. Sun, X. Wei and C. You, Biotechnol. J., 2019, 14, 1800493 CrossRef PubMed.
- Y. Tatebe, Y. Yamamoto and N. Isono, J. Appl. Glycosci., 2025, 72, 7201201 CAS.
- M. L. Lee and K. H. Muench, J. Biol. Chem., 1969, 244, 223–230 CrossRef CAS PubMed.
- M. Arai, K. Tanaka and T. Kawaguchi, J. Ferment. Bioeng., 1994, 77, 239–242 CrossRef CAS.
- M. Reichenbecher, F. Lottspeich and K. Bronnenmeier, Eur. J. Biochem., 1997, 247, 262–267 CrossRef CAS.
- M. Krishnareddy, Y.-K. Kim, M. Kitaoka, Y. Mori and K. Hayashi, J. Appl. Glycosci., 2002, 49, 1–8 CrossRef CAS.
- Z. Ubiparip, D. S. Moreno, K. Beerens and T. Desmet, Appl. Microbiol. Biotechnol., 2020, 104, 8327–8337 CrossRef CAS PubMed.
- D. M. Petrović, I. Kok, A. J. J. Woortman, J. Ćirić and K. Loos, Anal. Chem., 2015, 87, 9639–9646 CrossRef.
- T. Nohara, T. Sawada, H. Tanaka and T. Serizawa, Langmuir, 2016, 32, 12520–12526 CrossRef CAS.
- Y. Yataka, T. Sawada and T. Serizawa, Langmuir, 2016, 32, 10120–10125 CrossRef CAS PubMed.
- C. Zhong, K. Zajki-Zechmeister and B. Nidetzky, Carbohydr. Polym., 2021, 260, 117772 CrossRef CAS PubMed.
- Y. Kita, R. Kusumi, T. Kimura, M. Kitaoka, Y. Nishiyama and M. Wada, Cellulose, 2020, 27, 1899–1907 CrossRef CAS.
- T. Konishi, A. Sasaki, R. Kusumi, M. Wada and K. Kobayashi, Carbohydr. Polym. Technol. Appl., 2025, 9, 100731 CAS.
- P. de Andrade, J. C. Muñoz-García, G. Pergolizzi, V. Gabrielli, S. A. Nepogodiev, D. Iuga, L. Fábián, R. Nigmatullin, M. A. Johns, R. Harniman, S. J. Eichhorn, J. Angulo, Y. Z. Khimyak and R. A. Field, Chem. – Eur. J., 2021, 27, 1374–1382 CrossRef CAS.
- Y. Hata, T. Kojima, T. Koizumi, H. Okura, T. Sakai, T. Sawada and T. Serizawa, ACS Macro Lett., 2017, 6, 165–170 CrossRef CAS.
- Y. Hata, T. Sawada and T. Serizawa, Polym. J., 2017, 49, 575–581 CrossRef CAS.
- Y. Hata, T. Kojima, T. Maeda, T. Sawada and T. Serizawa, Macromol. Biosci., 2020, 20, 202000187 CrossRef.
- T. Serizawa, T. Maeda and T. Sawada, ACS Macro Lett., 2020, 9, 301–305 Search PubMed.
- S. Withers, Carbohydr. Polym., 2001, 44, 325–337 CrossRef CAS.
- Y. Bourne and B. Henrissat, Curr. Opin. Struct. Biol., 2001, 11, 593–600 Search PubMed.
- P. Bojarová and V. Křen, Trends Biotechnol., 2009, 27, 199–209 CrossRef.
- T. V. Vuong and D. B. Wilson, Biotechnol. Bioeng., 2010, 107, 195–205 CrossRef CAS PubMed.
- M. Vuillemin, M. Lengyel, J. Muschiol, M. Matwiejuk, Y. Zhang, N. Cannac, D. Molnar-Gabor, A. S. Meyer and B. Zeuner, Enzyme Microb. Technol., 2025, 189, 110660 CrossRef CAS.
- S. Kobayashi, K. Kashiwa, T. Kawasaki and S. Shoda, J. Am. Chem. Soc., 1991, 113, 3079–3084 CrossRef CAS.
- S. J. Williams and S. G. Withers, Carbohydr. Res., 2000, 327, 27–46 CrossRef CAS PubMed.
- S. Kobayashi and S. Shoda, Int. J. Biol. Macromol., 1995, 17, 373–379 Search PubMed.
- T. Murata and T. Usui, Trends Glycosci. Glycotechnol., 2000, 12, 161–174 CrossRef CAS.
- E. Okamoto, T. Kiyosada, S. Shoda and S. Kobayashi, Cellulose, 1997, 4, 161–172 CrossRef CAS.
- K. Kobayashi and S. Kamiya, Macromolecules, 1996, 29, 8670–8676 CrossRef CAS.
- S. Kobayashi, T. Kiyosada and S. Shoda, J. Am. Chem. Soc., 1996, 118, 13113–13114 CrossRef CAS.
- Q. Wang and S. G. Withers, J. Am. Chem. Soc., 1995, 117, 10137–10138 CrossRef CAS.
- S. Kobayashi, H. Morii, R. Itoh, S. Kimura and M. Ohmae, J. Am. Chem. Soc., 2001, 123, 11825–11826 CrossRef CAS.
- S. Kobayashi, S. Fujikawa and M. Ohmae, J. Am. Chem. Soc., 2003, 125, 14357–14369 CrossRef CAS.
- S. Fujikawa, M. Ohmae and S. Kobayashi, Biomacromolecules, 2005, 6, 2935–2942 CrossRef CAS PubMed.
- S. Kobayashi, Proc. Jpn. Acad., Ser. B, 2007, 83, 215–247 CrossRef CAS PubMed.
- M. Ohmae, K. Sakaguchi, T. Kaneto, S. Fujikawa and S. Kobayashi, ChemBioChem, 2007, 8, 1710–1720 CrossRef CAS PubMed.
- Y. Yamazaki, S. Kimura and M. Ohmae, Carbohydr. Res., 2018, 456, 61–68 CrossRef CAS PubMed.
- Y. Yamazaki, K. Sezukuri, J. Takada, S. Kimura and M. Ohmae, ChemBioChem, 2016, 17, 1879–1886 CrossRef CAS PubMed.
- S. Yuge, A. Tateishi, K. Numata and M. Ohmae, Biomacromolecules, 2022, 23, 316–325 CrossRef CAS PubMed.
- B. Li, Y. Zeng, S. Hauser, H. Song and L. X. Wang, J. Am. Chem. Soc., 2005, 127, 9692–9693 CrossRef CAS PubMed.
- H. Ochiai, W. Huang and L.-X. Wang, J. Am. Chem. Soc., 2008, 130, 13790–13803 CrossRef CAS PubMed.
- L.-X. Wang, Carbohydr. Res., 2008, 343, 1509–1522 CrossRef CAS PubMed.
- S. Kobayashi, T. Kawasaki, K. Obata and S. Shoda, Chem. Lett., 1993, 22, 685–686 CrossRef.
- O. Karthaus, S. Shoda, H. Takano, K. Obata and S. Kobayashi, J. Chem. Soc., Perkin Trans. 1, 1994, 1851 RSC.
- S. Shoda, M. Fujita, C. Lohavisavapanichi, Y. Misawa, K. Ushizaki, Y. Tawata, M. Kuriyama, M. Kohri, H. Kuwata and T. Watanabe, Helv. Chim. Acta, 2002, 85, 3919–3936 CrossRef CAS.
- B. Zeuner, C. Jers, J. D. Mikkelsen and A. S. Meyer, J. Agric. Food Chem., 2014, 62, 9615–9631 CrossRef CAS.
- T. Murata, T. Inukai, M. Suzuki, M. Yamagishi and T. Usui, Glycoconjugate J., 1999, 16, 189–195 CrossRef CAS PubMed.
- T. Murata, S. Morimoto, X. Zeng, S. Watanabe and T. Usui, Carbohydr. Res., 1999, 320, 192–199 CrossRef CAS.
- T. Murata and T. Usui, Biotechnol. Bioprocess Eng., 2002, 7, 263–267 CrossRef CAS.
- T. Murata and T. Usui, Biosci. Biotechnol. Biochem., 2006, 70, 1049–1059 CrossRef CAS PubMed.
- B. Zeuner, C. Nyffenegger, J. D. Mikkelsen and A. S. Meyer, New Biotechnol., 2016, 33, 355–360 CrossRef CAS PubMed.
- L. Guo, X. Chen, L. Xu, M. Xiao and L. Lu, Appl. Environ. Microbiol., 2018, 84, e00071–e00018 CAS.
- B. Zeuner, D. Teze, J. Muschiol and A. S. Meyer, Molecules, 2019, 24, 2033 CrossRef CAS PubMed.
- B. Zeuner and A. S. Meyer, Carbohydr. Res., 2020, 493, 108029 CrossRef CAS.
- P. Nekvasilová, M. Hovorková, Z. Mészáros, L. Petrásková, H. Pelantová, V. Křen, K. Slámová and P. Bojarová, Int. J. Mol. Sci., 2022, 23, 4106 CrossRef.
- L. F. Mackenzie, Q. Wang, R. A. J. Warren and S. G. Withers, J. Am. Chem. Soc., 1998, 120, 5583–5584 CrossRef CAS.
- J. R. Rich and S. G. Withers, Nat. Chem. Biol., 2009, 5, 206–215 CrossRef CAS PubMed.
- W. Huang, C. Li, B. Li, M. Umekawa, K. Yamamoto, X. Zhang and L. X. Wang, J. Am. Chem. Soc., 2009, 131, 2214–2223 CrossRef CAS.
- P. M. Danby and S. G. Withers, ACS Chem. Biol., 2016, 11, 1784–1794 CrossRef CAS PubMed.
- M. Hayes and J. Pietruszka, Molecules, 2017, 22, 1434 CrossRef.
- T. Katoh and K. Yamamoto, J. Appl. Glycosci., 2021, 68, 1–9 CrossRef CAS PubMed.
- Y. Honda and M. Kitaoka, J. Biol. Chem., 2006, 281, 1426–1431 CrossRef CAS.
- T. Tanaka, M. Noguchi, A. Kobayashi and S. Shoda, Chem. Commun., 2008, 2016–2018 RSC.
- M. Kunishima, C. Kawachi, J. Morita, K. Terao, F. Iwasaki and S. Tani, Tetrahedron, 1999, 55, 13159–13170 CrossRef CAS.
- M. Kunishima, C. Kawachi, K. Hioki, K. Terao and S. Tani, Tetrahedron, 2001, 57, 1551–1558 CrossRef CAS.
- T. Tanaka, M. Noguchi, K. Watanabe, T. Misawa, M. Ishihara, A. Kobayashi and S. Shoda, Org. Biomol. Chem., 2010, 8, 5126–5132 RSC.
- T. Tanaka, M. Noguchi, M. Ishihara, A. Kobayashi and S. Shoda, Macromol. Symp., 2010, 297, 200–209 CrossRef CAS.
- A. Kobayashi, T. Tanaka, K. Watanabe, M. Ishihara, M. Noguchi, H. Okada, Y. Morikawa and S. Shoda, Bioorg. Med. Chem. Lett., 2010, 20, 3588–3591 CrossRef CAS PubMed.
- T. Tanaka, T. Wada, M. Noguchi, M. Ishihara, A. Kobayashi, T. Ohnuma, T. Fukamizo, R. Brzezinski and S. Shoda, J. Carbohydr. Chem., 2012, 31, 634–646 CrossRef CAS.
- D. H. Im, K. Kimura, F. Hayasaka, T. Tanaka, M. Noguchi, A. Kobayashi, S. Shoda, K. Miyazaki, T. Wakagi and S. Fushinobu, Biosci. Biotechnol. Biochem., 2012, 76, 423–428 CrossRef CAS PubMed.
- T. Ohnuma, T. Taku, T. Nagatani, A. Horii, S. Imaoka and T. Tanaka, Biosci. Biotechnol. Biochem., 2021, 85, 1716–1719 CrossRef PubMed.
- M. Noguchi, M. Nakamura, A. Ohno, T. Tanaka, A. Kobayashi, M. Ishihara, M. Fujita, A. Tsuchida, M. Mizuno and S. Shoda, Chem. Commun., 2012, 48, 5560–5562 RSC.
- M. Kiyohara, T. Nakatomi, S. Kurihara, S. Fushinobu, H. Suzuki, T. Tanaka, S. Shoda, M. Kitaoka, T. Katayama, K. Yamamoto and H. Ashida, J. Biol. Chem., 2012, 287, 693–700 CrossRef CAS PubMed.
- T. Ohnuma, T. Tanaka, A. Urasaki, S. Dozen and T. Fukamizo, J. Biochem., 2019, 165, 497–503 CrossRef CAS PubMed.
- T. Ohnuma, T. Fukuda, S. Dozen, Y. Honda, M. Kitaoka and T. Fukamizo, Biochem. J., 2012, 444, 437–443 CrossRef CAS PubMed.
- U. Huchel, C. Schmidt and R. R. Schmidt, Eur. J. Org. Chem., 1998, 1353–1360 CrossRef CAS.
- Y. Fujimoto, K. Mitsunobe, S. Fujiwara, M. Mori, M. Hashimoto, Y. Suda, S. Kusumoto and K. Fukase, Org. Biomol. Chem., 2013, 11, 5034 RSC.
- M. Ishihara, Y. Takagi, G. Li, M. Noguchi and S. Shoda, Chem. Lett., 2013, 42, 1235–1237 CrossRef CAS.
- T. Tanaka, N. Kikuta, Y. Kimura and S. Shoda, Chem. Lett., 2015, 44, 846–848 CrossRef CAS.
- G. Li, M. Noguchi, M. Ishihara, Y. Takagi, M. Nagaki, S. Saito, M. Saito, X. Ye and S. Shoda, Carbohydr. Res., 2023, 534, 108940 CrossRef CAS PubMed.
- T. Isobe and T. Ishikawa, J. Org. Chem., 1999, 64, 6984–6988 CrossRef CAS.
- M. Noguchi, T. Tanaka, H. Gyakushi, A. Kobayashi and S. Shoda, J. Org. Chem., 2009, 74, 2210–2212 CrossRef CAS.
- A. Novoa, S. Barluenga, C. Serba and N. Winssinger, Chem. Commun., 2013, 49, 7608–7610 RSC.
- A. J. Fairbanks, Carbohydr. Res., 2021, 499, 108197 CrossRef CAS PubMed.
- M. Noguchi, T. Fujieda, W. C. Huang, M. Ishihara, A. Kobayashi and S. Shoda, Helv. Chim. Acta, 2012, 95, 1928–1936 CrossRef CAS.
- W. Huang, Q. Yang, M. Umekawa, K. Yamamoto and L. Wang, ChemBioChem, 2010, 11, 1350–1355 CrossRef CAS PubMed.
- M. Umekawa, T. Higashiyama, Y. Koga, T. Tanaka, M. Noguchi, A. Kobayashi, S. Shoda, W. Huang, L. X. Wang, H. Ashida and K. Yamamoto, Biochim. Biophys. Acta, Gen. Subj., 2010, 1800, 1203–1209 CrossRef CAS.
- W. Huang, J. Giddens, S. Q. Fan, C. Toonstra and L. X. Wang, J. Am. Chem. Soc., 2012, 134, 12308–12318 CrossRef CAS.
- Y. Higuchi, Y. Eshima, Y. Huang, T. Kinoshita, W. Sumiyoshi, S. Nakakita and K. Takegawa, Biotechnol. Lett., 2017, 39, 157–162 CrossRef CAS.
- A. J. Fairbanks, Chem. Soc. Rev., 2017, 46, 5128–5146 RSC.
- X. Zhang, C. Ou, H. Liu and L.-X. Wang, Bioconjugate Chem., 2022, 33, 1179–1191 CrossRef CAS.
- X. Qiu, A. L. Garden and A. J. Fairbanks, Chem. Sci., 2022, 13, 4122–4130 RSC.
- K. M. Koeller and C.-H. Wong, Chem. Rev., 2000, 100, 4465–4494 CrossRef CAS.
- C.-C. Wang, M. M. L. Zulueta and S.-C. Hung, Chimia, 2011, 65, 54 CrossRef CAS.
- S. S. Kulkarni, C.-C. Wang, N. M. Sabbavarapu, A. R. Podilapu, P.-H. Liao and S.-C. Hung, Chem. Rev., 2018, 118, 8025–8104 CrossRef CAS PubMed.
- C.-W. Cheng, C.-Y. Wu, W.-L. Hsu and C.-H. Wong, Biochemistry, 2020, 59, 3078–3088 CrossRef CAS PubMed.
- L. Cai, L. Meng, J. Zeng and Q. Wan, Org. Chem. Front., 2021, 8, 3150–3165 RSC.
- H. Xu, B. Shen, M. Qiao, R. J. Linhardt and X. Zhang, Carbohydr. Polym., 2021, 258, 117672 CrossRef CAS PubMed.
- N. Yoshida, T. Tanaka, M. Noguchi, A. Kobayashi, K. Ishikura, T. Ikenuma, H. Seno, T. Watanabe, M. Kohri and S. Shoda, Chem. Lett., 2012, 41, 689–690 CrossRef CAS.
- M. Noguchi, T. Fujieda, W. C. Huang, M. Ishihara, A. Kobayashi and S. Shoda, Helv. Chim. Acta, 2012, 95, 1928–1936 CrossRef CAS.
- T. Tanaka, A. Matsuura, Y. Aso and H. Ohara, Chem. Commun., 2020, 56, 10321–10324 RSC.
- T. Tanaka, A. Matsuura, Y. Aso and H. Ohara, J. Appl. Glycosci., 2020, 67, 119–127 CrossRef CAS.
- F. H. Newth, Q. Rev., Chem. Soc., 1959, 13, 30 RSC.
- K. Chow and S. Danishefsky, J. Org. Chem., 1990, 55, 4211–4214 CrossRef CAS.
- S. Wagschal, J. Guilbaud, P. Rabet, V. Farina and S. Lemaire, J. Org. Chem., 2015, 80, 9328–9335 CrossRef CAS PubMed.
- M. Tanaka, A. Nakagawa, N. Nishi, K. Iijima, R. Sawa, D. Takahashi and K. Toshima, J. Am. Chem. Soc., 2018, 140, 3644–3651 CrossRef CAS PubMed.
- T. Tanaka, W. C. Huang, M. Noguchi, A. Kobayashi and S. Shoda, Tetrahedron Lett., 2009, 50, 2154–2157 CrossRef CAS.
- T. Tanaka, H. Nagai, M. Noguchi, A. Kobayashi and S. Shoda, Chem. Commun., 2009, 3378–3379 RSC.
- T. Tanaka, T. Matsumoto, M. Noguchi, A. Kobayashi and S. Shoda, Chem. Lett., 2009, 38, 458–459 CrossRef CAS.
- N. Yoshida, M. Noguchi, T. Tanaka, T. Matsumoto, N. Aida, M. Ishihara, A. Kobayashi and S. Shoda, Chem. – Asian J., 2011, 6, 1876–1885 CrossRef CAS.
- N. Yoshida, T. Fujieda, A. Kobayashi, M. Ishihara, M. Noguchi and S. Shoda, Chem. Lett., 2013, 42, 1038–1039 CrossRef CAS.
- G. Li, M. Noguchi, H. Kashiwagura, Y. Tanaka, K. Serizawa and S. Shoda, Tetrahedron Lett., 2016, 57, 3529–3531 CrossRef CAS.
- S. Köhling, M. P. Exner, S. Nojoumi, J. Schiller, N. Budisa and J. Rademann, Angew. Chem., Int. Ed., 2016, 55, 15510–15514 CrossRef PubMed.
- S. R. Alexander and A. J. Fairbanks, Org. Biomol. Chem., 2016, 14, 6679–6682 RSC.
- Y. Meguro, M. Noguchi, G. Li and S. Shoda, Org. Lett., 2018, 20, 76–79 CrossRef CAS.
- H. Tanaka, Y. Yoshimura, M. R. Jørgensen, J. A. Cuesta-Seijo and O. Hindsgaul, Angew. Chem., Int. Ed., 2012, 51, 11531–11534 CrossRef CAS.
- K. Fujihara, H. Yamamura and A. Miyagawa, Synth. Commun., 2024, 54, 1245–1251 CrossRef CAS.
- M. B. Tatina, D. T. Khong and Z. M. A. Judeh, Eur. J. Org. Chem., 2018, 2208–2213 CrossRef CAS.
- K. Serizawa, M. Noguchi, G. Li and S. Shoda, Chem. Lett., 2017, 46, 1024–1026 CrossRef CAS.
- G. Li, M. Noguchi, K. Serizawa and S. Shoda, Chimia, 2018, 72, 874 CrossRef CAS.
- A. J. Thompson, R. J. Williams, Z. Hakki, D. S. Alonzi, T. Wennekes, T. M. Gloster, K. Songsrirote, J. E. Thomas-Oates, T. M. Wrodnigg, J. Spreitz, A. E. Stütz, T. D. Butters, S. J. Williams and G. J. Davies, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 781–786 CrossRef CAS.
- L. F. Sobala, G. Speciale, S. Zhu, L. Raich, N. Sannikova, A. J. Thompson, Z. Hakki, D. Lu, S. Shamsi Kazem Abadi, A. R. Lewis, V. Rojas-Cervellera, G. Bernardo-Seisdedos, Y. Zhang, O. Millet, J. Jiménez-Barbero, A. J. Bennet, M. Sollogoub, C. Rovira, G. J. Davies and S. J. Williams, ACS Cent. Sci., 2020, 6, 760–770 CrossRef CAS PubMed.
- S. Shoda, M. Noguchi, G. Li and K. Serizawa, Bull. Appl. Glycosci., 2019, 9, 83–89 CrossRef CAS.
- D.-D. Meng, X. Liu, S. Dong, Y.-F. Wang, X.-Q. Ma, H. Zhou, X. Wang, L.-S. Yao, Y. Feng and F.-L. Li, Biochem. J., 2017, 474, 3373–3389 CrossRef CAS.
- A. A. Telke, N. Zhuang, S. S. Ghatge, S.-H. Lee, A. Ali Shah, H. Khan, Y. Um, H.-D. Shin, Y. R. Chung, K. H. Lee and S.-W. Kim, PLoS One, 2013, 8, e65727 CrossRef CAS PubMed.
- M. Sandgren, G. I. Berglund, A. Shaw, J. Ståhlberg, L. Kenne, T. Desmet and C. Mitchinson, J. Mol. Biol., 2004, 342, 1505–1517 CrossRef CAS PubMed.
- G. J. Davies, L. Mackenzie, A. Varrot, M. Dauter, A. M. Brzozowski, M. Schülein and S. G. Withers, Biochemistry, 1998, 37, 11707–11713 CrossRef CAS.
- G. Sulzenbacher, L. F. Mackenzie, K. S. Wilson, S. G. Withers, C. Dupont and G. J. Davies, Biochemistry, 1999, 38, 4826–4833 CrossRef CAS.
- E. Sabini, G. Sulzenbacher, M. Dauter, Z. Dauter, P. L. Jørgensen, M. Schülein, C. Dupont, G. J. Davies and K. S. Wilson, Chem. Biol., 1999, 6, 483–492 CrossRef CAS PubMed.
- S. R. Andrews, S. J. Charnock, J. H. Lakey, G. J. Davies, M. Claeyssens, W. Nerinckx, M. Underwood, M. L. Sinnott, R. A. J. Warren and H. J. Gilbert, J. Biol. Chem., 2000, 275, 23027–23033 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |