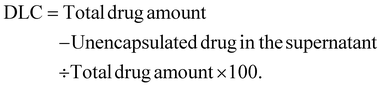Open Access Article
Open Access ArticleFormulation and evaluation of a Tridax procumbens-loaded phospholipid complex (phytosome) gel for wound healing and antimicrobial activities
Priyanka S.
Jadhav
 ,
Priyanka H.
Shinde
,
Priyanka H.
Shinde
 and
Jameel Ahmed S.
Mulla
and
Jameel Ahmed S.
Mulla
 *
*
Department of Pharmaceutics, Shree Santkrupa College of Pharmacy, Ghogaon-Karad, Maharashtra-415111, India. E-mail: jameelahmed5@gmail.com; Tel: +919845463472
First published on 18th November 2025
Abstract
Tridax procumbens is a widely recognized medicinal plant known for its remarkable antimicrobial and wound-healing effects. Nevertheless, its therapeutic efficacy is often restricted due to low bioavailability. This study aimed to formulate and evaluate a Tridax procumbens-loaded phospholipid complex (phytosome) gel to enhance its transdermal delivery and therapeutic efficacy. The optimization of the phytosomal gel was conducted using a central composite design, where soy lecithin and cholesterol were the primary independent variables affecting particle size and entrapment efficiency. The ideal formulation showed a particle diameter of 460.2 nm and a loading capacity of 93.14%, ensuring improved permeation and prolonged drug release. Antimicrobial studies demonstrated improved efficacy against E. coli, with the phospholipid complex gel exhibiting a zone of inhibition (ZOI) of 32 mm, compared to 28 mm for the ethanolic extract and 30 mm for the standard drug amikacin. In vitro wound healing studies using L929 fibroblast cells showed that the phospholipid complex gel achieved 54.34% wound closure after 48 hours, compared to 41.57% for the ethanolic extract and 87.53% for the standard drug cipladine. These results suggest that the phospholipid complex (phytosome) system significantly enhances the bioavailability and therapeutic potential of Tridax procumbens for wound healing and antimicrobial applications.
1. Introduction
Tridax procumbens, often called “coat buttons”, is a renowned medicinal plant extensively utilized in Ayurveda, especially for its antimicrobial and wound-healing benefits.1 It is a herbaceous weed that grows widely in India, especially in the states of Maharashtra, Madhya Pradesh, and Chhattisgarh.2 The medicinal properties of Tridax procumbens are attributed to its rich content of secondary metabolites, including flavonoids, alkaloids, tannins, carotenoids, and saponins, which significantly contribute to its antimicrobial, anti-inflammatory, and antioxidant properties.3 The leaves of Tridax procumbens are specifically known for their wound-healing capabilities, with traditional applications in stopping bleeding and accelerating tissue repair.4Although Tridax procumbens offers significant therapeutic benefits, its clinical application is restricted due to the poor bioavailability of its phytoconstituents, mainly resulting from low lipid solubility and complex polyphenolic structures.5,6 This inherent low solubility limits their absorption across the lipid-rich biological membranes of the skin. Phytosome technology has been introduced as an innovative novel drug delivery system developed specifically to enhance the penetration and bioavailability of herbal extracts.7 Phospholipid complexes (phytosomes) are advanced vesicular systems in which plant bioactive compounds form stable complexes with phospholipids, thereby significantly improving pharmacokinetic and pharmacodynamic properties.8 By converting Tridax procumbens leaf extract into a phospholipid complex (phytosome), its wound healing and antimicrobial efficacy can be substantially enhanced.9,10
The complexation with phospholipids facilitates better permeation through biological membranes, ensuring improved systemic absorption and therapeutic effectiveness.11,12 This novel system not only enhances bioavailability but also maximizes the plant's inherent healing potential, making it a promising approach for advanced wound care therapeutics.13
Despite its rich pharmacological profile, Tridax procumbens suffers from limited clinical translation due to its poor solubility, instability under physiological conditions, and restricted skin permeability. The phytosome approach not only enhances bioavailability but also stabilizes phenolic and flavonoid compounds against enzymatic degradation, ensuring targeted dermal delivery. This lipid-compatible complex facilitates stronger interaction with cell membranes, thereby improving therapeutic retention at the wound site and enabling dual antimicrobial–healing action. Thus, a phytosomal formulation provides a scientifically justified route to maximize Tridax procumbens’ pharmacodynamic potential for wound management.
2. Materials and methods
2.1 Materials
This study utilized the following materials: Tridax procumbens extract, soy lecithin, cholesterol, ethanol, dichloromethane, methanol, and phosphate buffer for the preparation of phytosomes. For the formulation of the phytosomal gel, Carbopol 934, triethanolamine, glycerin, propylene glycol, methyl 4-hydroxybenzoate, propyl 4-hydroxybenzoate and distilled water were used.14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in a 250 ml iodine flask. The mixture is left to macerate for 72 hours, during which it is occasionally stirred or shaken to enhance the extraction process. After the maceration period, the liquid extract is separated from the plant residues through filtration. The filtrate is then concentrated by evaporating the solvent using a rotary evaporator or allowed to air dry, yielding the crude extract. This extract can be further dried if necessary and stored in an airtight container.15
10 in a 250 ml iodine flask. The mixture is left to macerate for 72 hours, during which it is occasionally stirred or shaken to enhance the extraction process. After the maceration period, the liquid extract is separated from the plant residues through filtration. The filtrate is then concentrated by evaporating the solvent using a rotary evaporator or allowed to air dry, yielding the crude extract. This extract can be further dried if necessary and stored in an airtight container.15
2.2 Preparation of a phospholipid complex (phytosome)
The active plant extract (50 mg) is dissolved in a dichloromethane and methanol mixture in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
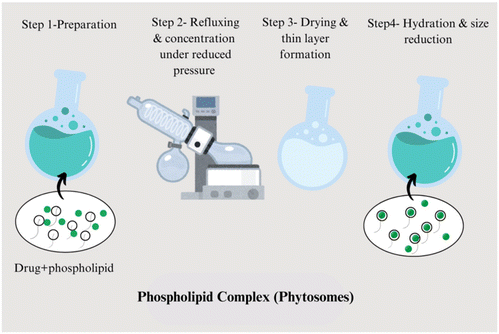
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. Soy lecithin and cholesterol are also dissolved in the same solvent. These components are mixed thoroughly to form a homogeneous solution. The mixture is then subjected to evaporation under vacuum using a rotary evaporator, which removes the solvent and leaves behind a thin lipid film containing the plant extract, lecithin, and cholesterol. This film is hydrated using a phosphate buffer, leading to the formation of a phospholipid complex (phytosome). The resulting suspension can be sonicated to reduce the particle size, ensuring uniform vesicle formation (Fig. 1).16
1 ratio. Soy lecithin and cholesterol are also dissolved in the same solvent. These components are mixed thoroughly to form a homogeneous solution. The mixture is then subjected to evaporation under vacuum using a rotary evaporator, which removes the solvent and leaves behind a thin lipid film containing the plant extract, lecithin, and cholesterol. This film is hydrated using a phosphate buffer, leading to the formation of a phospholipid complex (phytosome). The resulting suspension can be sonicated to reduce the particle size, ensuring uniform vesicle formation (Fig. 1).16
| Encoded data tier | Independent variables | |
|---|---|---|
| X1 soy lecithin (mg) | X2 cholesterol (mg) | |
| −1 | 50 | 5.55 |
| 0 | 100 | 11.11 |
| +1 | 150 | 16.66 |
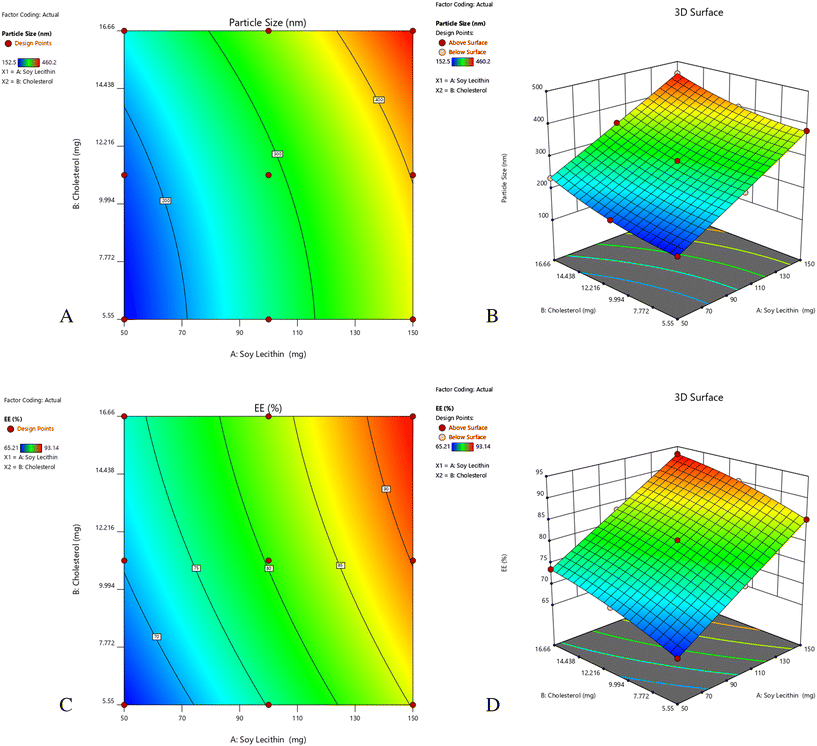
The central composite design (CCD) employed a two-factor, three-level factorial design comprising 9 experimental runs to assess the effects of soy lecithin (X1: 50–150 mg) and cholesterol (X2: 5.55–16.66 mg) on particle size and entrapment efficiency. The model included four factorial points, four axial points, and one center point to ensure rotatability and predict curvature effects.
2.2.1.1 Particle size. Particle size analysis via DLS was used to measure the vesicle size and polydispersity index (PDI) with a Horiba Nanopartica SZ-100 particle size analyzer. The sample was diluted with distilled water as the solvent and measured in a quartz cuvette at a 90-degree scattering angle. All the batches were analyzed in triplicate, and the mean and SD were calculated. The vesicle size (in nm) and polydispersity index (PDI) were determined to assess uniformity and dispersion stability.19,20
2.2.1.2 Charge-dependent mobility. Charge-dependent mobility was measured based on the surface charge of Tridax procumbens-loaded phytosomes using a Horiba Nanopartica model SZ-100 particle size analyzer. The sample was diluted (20 folds) using distilled water and then analysis was performed at 24.9 °C and 3.4 volts. The charge-dependent mobility of the phytosomes was calculated. The obtained zeta potential (mV) indicated the surface charge and colloidal stability, with values above ±30 mV suggesting good stability and reduced aggregation.21
2.2.1.3 Drug loading capacity. The drug loading capacity (DLC) of the formulated phospholipid complex (phytosome) was evaluated through the ultracentrifugation method. The formulation solutions the samples were spun at 15
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 minutes. After centrifugation, the supernatant was decanted, and the concentration of the unencapsulated drug was determined at 257 nm using a UV spectrophotometer (Shimadzu). The entrapment efficiency was then determined using a specific mathematical formula.22,23
000 rpm for 30 minutes. After centrifugation, the supernatant was decanted, and the concentration of the unencapsulated drug was determined at 257 nm using a UV spectrophotometer (Shimadzu). The entrapment efficiency was then determined using a specific mathematical formula.22,232.3 Preparation of the Tridax procumbens loaded phospholipid complex (phytosome) gel
An accurately measured amount of Carbopol 934 was added to a beaker that contained 100 mL of distilled water and allowed to hydrate for 30 minutes. Separately, methylparaben and propylene glycol were dissolved in water. The swollen Carbopol 934 was stirred continuously until a smooth, lump-free mixture was obtained. Subsequently, 5–6 drops of triethanolamine, along with methylparaben, propyl paraben, and glycerin were incorporated into the Carbopol 934 solution followed by continuous mixing until a clear, translucent gel was obtained. Finally, the prepared gel was combined with the phospholipid complex (phytosome) containing Tridax procumbens extract and gently stirred to obtain the phospholipid complex (phytosome) gel (Table 2).| Sl no. | Ingredients | Formulation |
|---|---|---|
| 1 | Tridax procumbens extract | 0.10 g |
| 2 | Carbopol 934 | 1.5 g |
| 3 | Propylene glycol | 10 ml |
| 4 | Methyl 4-hydroxybenzoate | 200 mg |
| 5 | Propyl 4-hydroxybenzoate | 100 mg |
| 6 | Trolamine | qs |
| 7 | Glycerine | 1.2 ml |
| 8 | Distilled water | 100 ml |
2.4 pH determination
The pH of each formulation was assessed using a pre-calibrated pH meter. The calibration was performed using standard buffer solutions with pH values of 4, 7, and 9. For measurement, one gram of emulgel was mixed with 100 ml of distilled water and allowed to rest for 2 hours before recording the pH.24,252.5 Spreadability
Two glass slides, each measuring 2.5 cm width × 7 cm length, were selected for the spreadability test. A specific amount of the phospholipid complex (phytosome) gel was placed between the two glass slides, and a 20 g weight was applied to the upper slide to ensure uniform spreading and create an even layer. The time required for the slides to separate was recorded using a stopwatch.26The spreadability (S) was calculated using the equation:
| S = M × L/T |
2.6 Consistency
The viscosity of the phospholipid complex (phytosome) gel was measured by using a Brookfield viscometer with spindle number S64. 30 gm of the prepared gel was kept in a 50 ml beaker, set at room temperature and spindled at 5, 10, 20, 30 & 50.272.7 Uniformity
All the gel formulations that were prepared were visually inspected for uniformity after being set in their containers. The assessment focused on detecting the presence and appearance of any aggregates.282.8 FTIR
FTIR spectroscopy was performed to identify the functional groups present in the composition, pure extract, phytosomes, and the phytosomal gel. The spectra were obtained using a Bruker FTIR device over a wavelength range of 4000–400 cm−1. The samples were prepared by mixing with potassium bromide (KBr) and compressed into pellets before scanning. The obtained spectra were analyzed to identify characteristic peaks corresponding to various functional groups and to confirm the successful formation of the phospholipid complex (phytosome) and phytosomal gel.292.9 Antimicrobial efficacy
The pathogen suppression ability of the test samples was evaluated employing the agar well diffusion assay against E. coli. Muller Hinton Agar (MHA) plates were prepared and sterilized, and the test microorganism was inoculated using the spread plate technique. Three different samples were evaluated: amikacin (used as a positive control), an extract of Tridax procumbens leaves, and the phospholipid complex (phytosome) gel formulated with Tridax procumbens extract. A sterile cork borer was utilized to punch 10 mm diameter wells in the agar. The plates were kept at 37 °C for 24 hours of incubation, and the zone of inhibition (ZOI) was measured afterward using a Vernier calliper to assess antimicrobial efficacy.30–322.10 Assessment of in vitro wound healing activity
The in vitro wound healing potential of the Tridax procumbens extract and its gel formulation was evaluated using the scratch wound healing assay on fibroblast cells. Wound closure rates were measured at various time points by observing the movement of cells into the wound area. Both the extract and gel demonstrated substantial wound closure compared to the control, indicating an increase in cell proliferation and migration. These results indicate that the phytoconstituents in Tridax procumbens play a key role in its wound-healing activity, positioning it as a promising candidate for topical applications.332.11 Scratch assay
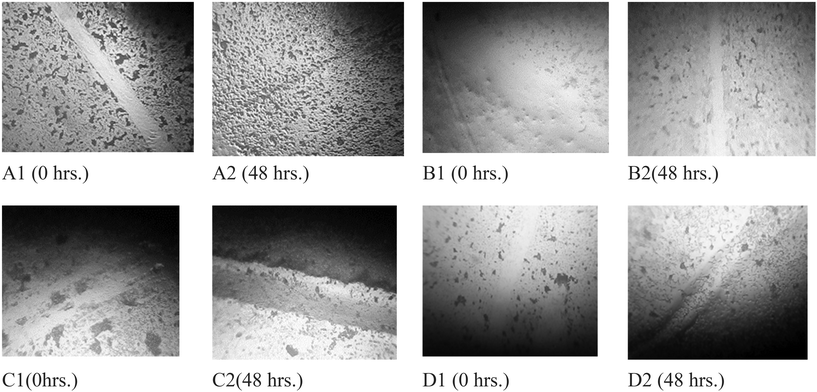
An in vitro migration experiment was employed to analyze the regenerative potential of sample J12 with L929 fibroblasts. A cell mixture at a density of 2 × 105 cells per mL was introduced into 6-well plates and allowed to incubate overnight. The following day, a scratch assay was performed with a sterile 200 µL pipette tip after washing the cells in DPBS. Detached cells and particles were eliminated by flushing with DPBS. Subsequently, the cells were treated with 100 µL of sample J12, while a 5 µg mL−1 concentration of cipladine, a known wound-healing agent, served as the active control. Cells not exposed to treatment functioned as the placebo control. Following a 24-hour incubation, cell movement and morphological alterations were monitored through images captured using a reverse microscope equipped with a digital camera. The experiment was conducted three times (n = 3), and wound closure and scratch width were analyzed at different time points (48 hours) using SAGLO software.3. Results
3.1 Enhanced experimental configuration
The result of the central composite design was statistically evaluated using Design Expert software. Soy lecithin (X1) and cholesterol (X2) were investigated as independent variables and their impact on particle diameter and drug loading capacity were examined as dependent variables. The 32 factorial results in 9 runs are shown in Table 3.| Formulation | Variable 1 | Variable 2 | Response 1 | Response 2 |
|---|---|---|---|---|
| A: soy lecithin (mg) | B: cholesterol (mg) | Particle size (nm) | ee (%) | |
| 1 | 50 | 50 | 152.5 | 65.21 |
| 2 | 50 | 100 | 177.1 | 70.23 |
| 3 | 50 | 150 | 233.72 | 73.59 |
| 4 | 100 | 50 | 260.2 | 75.11 |
| 5 | 100 | 100 | 288.3 | 80.45 |
| 6 | 100 | 150 | 348.9 | 83.22 |
| 7 | 150 | 50 | 380.7 | 85.24 |
| 8 | 150 | 100 | 400.7 | 90.11 |
| 9 | 150 | 150 | 460.2 | 93.14 |
3.1.1.1 Fit statistics. The statistical analysis showed a standard deviation of 3.11 and a mean of 300.26, with a low C.V. % of 1.04%, indicating minimal variability. The model exhibited strong correlation and predictive accuracy, with R2 = 0.9997, adjusted R2 = 0.9991, and predicted R2 = 0.9960. An adequate accuracy of 121.7530 confirmed a high signal-to-noise ratio, ensuring the model's reliability for optimization. The predicted R2 value of 0.9960 aligns well with the modified R-squared value of 0.9991, as the deviation remains below 0.2 (Table 4).
| Source | Total squared variance | df | Average square | F-Value | p-Value | |
|---|---|---|---|---|---|---|
| The factorial coding is designated as coded. The total squared variance follows the type III partial analysis approach. The F-ratio of the model of 1811.73 indicates statistical significance. P-Values below 0.0500 suggest that the model terms are significant. In this case, A, B, and B2 are identified as significant model terms. | ||||||
| Model | 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 653.23 653.23 |
5 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530.65 530.65 |
1811.73 | <0.0001 | Significant |
| A-soy lecithin | 76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 677.29 677.29 |
1 | 76![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 677.29 677.29 |
7924.32 | <0.0001 | |
| B-cholesterol | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 368.39 368.39 |
1 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 368.39 368.39 |
1071.54 | <0.0001 | |
| AB | 0.7396 | 1 | 0.7396 | 0.0764 | 0.8001 | |
| A2 | 5.69 | 1 | 5.69 | 0.5880 | 0.4990 | |
| B2 | 601.12 | 1 | 601.12 | 62.12 | 0.0043 | |
| Residual | 29.03 | 3 | 9.68 | |||
| Cor total | 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 682.26 682.26 |
8 | ||||
Adequate precision assesses the clarity-to-distortion ratio, with a value above 4 being preferable. In this context, a proportion of 121.753 signifies a strong signal, confirming the model's reliability. Thus, this model is suitable for exploring the optimization space.
3.1.2.1 Fit statistics. The statistical analysis showed a standard deviation of 0.1609 and a mean of 79.59, with a low C.V. % of 0.2022, indicating minimal variability. The model exhibited excellent correlation and predictive accuracy, with R2 = 0.9999, modified R-squared = 0.9997, and predictive R2 = 0.9991 (Table 5). An adequate accuracy of 212.6897 confirmed a strong clarity-to-distortion ratio, ensuring the model's reliability for optimization. The predictive R2 value of 0.9991 closely aligns with the modified R-squared value of 0.9997, as the difference remains below 0.2. Adequate accuracy evaluates the clarity-to-distortion ratio, with a value exceeding 4 being preferable. In this case, a ratio of 212.690 confirms a strong signal, demonstrating the model's reliability. Therefore, this model is suitable for exploring the design space (Fig. 2).
| Source | Total squared variance | df | Average square | F-Stat | Probability value | |
|---|---|---|---|---|---|---|
| The factorial coding is set as coded. The total squared variance follows the type III partial analysis approach. The model's F-stat of 5331.27 signifies its statistical significance. Probability values below 0.0500 indicate that certain model terms are significant. In this case, A, B, and B2 are identified as significant factors in the model. | ||||||
| Model | 690.50 | 5 | 138.10 | 5331.27 | <0.0001 | Significant |
| A-soy lecithin | 589.25 | 1 | 589.25 | 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 747.66 747.66 |
<0.0001 | |
| B-cholesterol | 99.15 | 1 | 99.15 | 3827.46 | <0.0001 | |
| AB | 0.0576 | 1 | 0.0576 | 2.22 | 0.2327 | |
| A2 | 0.0001 | 1 | 0.0001 | 0.0034 | 0.9570 | |
| B2 | 2.05 | 1 | 2.05 | 79.02 | 0.0030 | |
| Residual | 0.0777 | 3 | 0.0259 | |||
| Cor total | 690.58 | 8 | ||||
3.2 Zeta potential
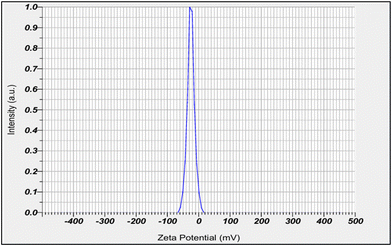
The optimized Tridax procumbens-loaded phospholipid complex (phytosome) was assessed for zeta potential at a temperature of 24.9 °C, the velocity of dispersion was 0.897 mPa s, and the conductivity was 0.205 mS cm−1. The electrode voltage difference was set at 3.4 V. The zeta potential measured was −25.0 mV, with an electrophoretic velocity of −0.000194 cm2 V−1 s−1, as shown in the peak in Fig. 3.3.3 Physical properties of the Tridax procumbens gel
The physical characteristics of a topical formulation play a crucial role in patient compliance and therapeutic efficacy. The formulated Tridax procumbens phospholipid complex (phytosome) gel was evaluated for its appearance, color, and odor to ensure its suitability for topical application.3.4 pH determination
The pH of the prepared formulation Tridax procumbens phospholipid complex (phytosome) gel was determined to be between 6 and 7.5, which falls within the typical pH range of the skin. Specifically, the pH was found to be 7.35. Hence, the formulated Tridax procumbens phospholipid complex (phytosome) gel is within the skin pH range.3.5 Determination of spreadability
Spreadability is a key factor in patient adherence, as it ensures even distribution of the Tridax procumbens phytosomal gel on the skin. The spreading coefficient values for the formulated gel indicate that it spreads easily on the skin's surface. The spreadability test reveals a value of 27.5, which indicates excellent spreading properties.3.6 Determination of viscosity
The viscosity of the Tridax procumbens phospholipid complex (phytosome) gel was found to be 7100 cps at 5 rpm, indicating that it has a semi-solid texture, which is appropriate for topical use. The high viscosity at low shear rates suggests shear-thinning behavior, which is essential for ease of application and better skin retention.3.7 FTIR analysis
FTIR analysis was performed using a Bruker-Alpha II. The range of wavenumbers used to capture the spectrum was 4000 to 500 cm−1. The FTIR spectra of the Tridax procumbens extract, Tridax procumbens phospholipid complex (phytosome) and Tridax procumbens phytosomal gel are shown, along with the values of their principal peaks, in Fig. 4.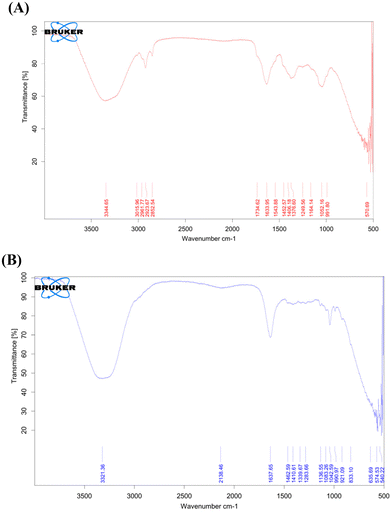 | ||
| Fig. 4 (A) FTIR spectrum of the Tridax procumbens extract. (B) FTIR spectrum of the Tridax procumbens phospholipid complex (phytosome) gel. | ||
The FTIR spectrum analysis identified key functional groups in the sample. A wide peak at 3344.65 cm−1 indicates O–H stretching (hydroxyl groups). Peaks at 2915.59 cm−1 and 2852.54 cm−1 are associated with C–H stretching in alkanes. The strong peak at 1744.62 cm−1 confirms C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching (carbonyl groups), while 1633.95 cm−1 denotes C
O stretching (carbonyl groups), while 1633.95 cm−1 denotes C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching in alkenes or aromatic rings. The peak at 1543.88 cm−1 is related to N–O asymmetric stretching in nitro compounds. C–O and C–O–C stretching at 1269.56 cm−1 and 1091.26 cm−1 suggest the presence of esters, ethers, or polysaccharides. Finally,
C stretching in alkenes or aromatic rings. The peak at 1543.88 cm−1 is related to N–O asymmetric stretching in nitro compounds. C–O and C–O–C stretching at 1269.56 cm−1 and 1091.26 cm−1 suggest the presence of esters, ethers, or polysaccharides. Finally, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H bending at 991.80 cm−1 confirms alkenes/aromatic rings.
C–H bending at 991.80 cm−1 confirms alkenes/aromatic rings.
The FTIR spectrum analysis confirmed key functional groups in the sample. A wide peak at 3321.36 cm−1 indicates O–H stretching (hydroxyl groups). The peak at 2138.46 cm−1 suggests C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C or C
C or C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N stretching (alkynes or nitriles). C
N stretching (alkynes or nitriles). C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching at 1637.65 cm−1 confirms alkenes or aromatic rings, while peaks at 1410.61 cm−1 and 1002.97 cm−1 indicate aromatic compounds. C–O and C–O–C stretching at 1283.67 cm−1, 1183.66 cm−1, and 1038.25 cm−1 suggest phenols, esters, or ethers. Peaks at 654.69 cm−1 and 546.22 cm−1 confirm the presence of alkyl halides.
C stretching at 1637.65 cm−1 confirms alkenes or aromatic rings, while peaks at 1410.61 cm−1 and 1002.97 cm−1 indicate aromatic compounds. C–O and C–O–C stretching at 1283.67 cm−1, 1183.66 cm−1, and 1038.25 cm−1 suggest phenols, esters, or ethers. Peaks at 654.69 cm−1 and 546.22 cm−1 confirm the presence of alkyl halides.
3.8 Determination of antimicrobial activity
This study investigates the antimicrobial efficacy of Tridax procumbens extract and its gel formulation in comparison with amikacin. The extract is prepared using suitable solvents, and the gel is formulated for topical application. The antimicrobial activity is assessed using methods such as the disk diffusion test against selected bacterial strains (Fig. 5).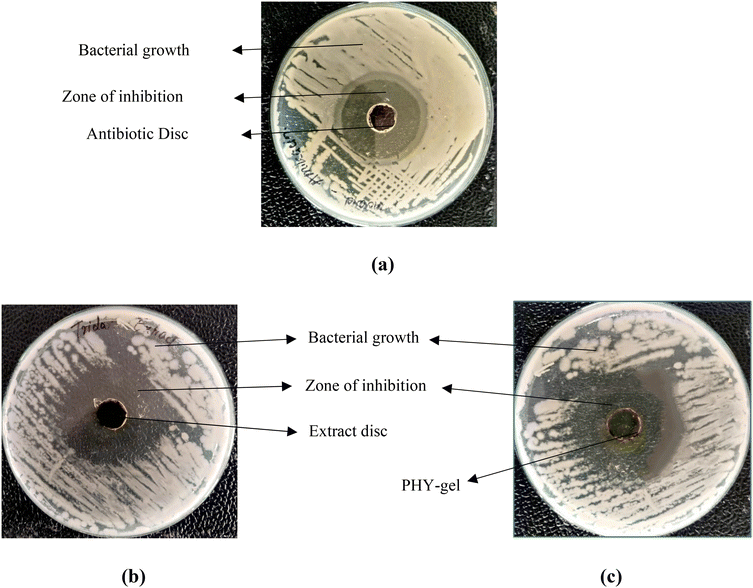 | ||
| Fig. 5 Antimicrobial activity indicating (a) the standard drug amikacin, (b) Tridax procumbens leaf extract, and (c) the Tridax procumbens phospholipid complex (phytosome) gel. | ||
The findings of the antimicrobial performance study are presented in Table 6. The total ZOI, including the well diameter (10 mm), was found to be 40 mm for amikacin, 38 mm for Tridax procumbens extract, and 42 mm for the phospholipid complex (phytosome) gel formulation. After subtracting the well diameter, the net ZOI values were calculated to be 30 mm, 28 mm, and 32 mm, respectively (Table 7).
| Physical evaluation | Results |
|---|---|
| Appearance | Semi-solid |
| Colour | Light greenish colour |
| Odour | Pleasant odour |
| Sample | Total ZOI (mm) | Well diameter (mm) | Net ZOI (mm) |
|---|---|---|---|
| Amikacin (standard) (30 µg) | 40 | 10 | 30 |
| Tridax procumbens extract (40 mg) | 38 | 10 | 28 |
| Tridax procumbens phospholipid complex (phytosome) gel (40 mg) | 42 | 10 | 32 |
3.9 Determination of in vitro wound-healing activity
The scratch assay results showed considerable wound healing potential of the phospholipid complex (phytosome) gel formulation (PHY-Gel) compared to the ethanolic extract (Ex) and control (Fig. 6).• Control group: minimal wound closure was observed, with an average wound width reduction of 9.79% over 48 hours.
• Standard (cipladine, 5 µg mL−1): showed the highest wound closure efficiency with 87.53% reduction.
• Ex (5 µg mL−1): showed a moderate wound healing response, with 41.57% wound closure.
• PHY-Gel (5 µg mL−1): showed enhanced wound healing with 54.34% wound closure (Table 8).
| Reading 0 hours (mm) | Mean | Reading 48 hours (mm) | Mean | Percentage of cell reduction | |
|---|---|---|---|---|---|
| Control | 10.00 | 10.01 | 09.50 | 9.79 | |
| 09.98 | 09.48 | ||||
| 10.05 | 10.00 | ||||
| Standard – cipladine 5 µg ml−1 | 10.00 | 10.01 | 1.04 | 1.22 | 87.53 |
| 09.98 | 1.20 | ||||
| 10.05 | 1.42 | ||||
| Sample – Ex | 10.00 | 10.01 | 5.90 | 5.72 | 41.57 |
| 09.98 | 5.64 | ||||
| 10.05 | 5.63 | ||||
| Sample – PHY-Gel | 10.00 | 10.01 | 4.52 | 4.47 | 54.34 |
| 09.98 | 4.30 | ||||
| 10.05 | 4.61 |
4. Discussion
The statistical analysis revealed significant model terms, confirming the suitability of the design for optimizing phospholipid complex (phytosome) formulations. The ANOVA results indicated that both soy lecithin and cholesterol significantly affected particle size and entrapment efficiency. The high R2 values (0.9997 for particle size and 0.9999 for ee), low coefficient of variation (C.V.) and adequate precision values greater than 4 confirm a strong signal-to-noise ratio, ensuring the model's reliability and suitability for formulation prediction and optimization. These results confirm that the central composite design was effective in optimizing the phospholipid complex (phytosome) gel formulation by fine-tuning the soy lecithin and cholesterol concentrations.The negative charge on the phospholipid complex (phytosome) is attributed to the phospholipid content, which helps in electrostatic repulsion, preventing aggregation and ensuring dispersion stability. The electrophoretic mobility (−0.000194 cm2 V−1 s−1) and dispersion viscosity (0.897 mPa s) further support the stability of the developed phospholipid complex (phytosome) system. It suggests that the formulation is stable under normal conditions.
These physical characteristics ensure that the gel is aesthetically acceptable and suitable for patient use.
A neutral pH is particularly beneficial for wound healing applications, as it prevents excessive acidity or alkalinity that could interfere with skin repair mechanisms. These findings confirm that the pH of the formulated gel is suitable for dermatological use.
The formulation's consistency allowed it to be easily applied without excessive fluidity or thickness. The observed spreadability confirms that the gel has a balanced texture for user-friendly application.
This rheological property ensures sustained drug release and enhances the formulation's stability. The reduction in viscosity with an increasing shear rate further supports its pseudoplastic nature, making it ideal for spreading and absorption. The high viscosity ensures sustained release of the active ingredient, reducing the need for frequent reapplication.
FTIR was used to validate the presence of functional groups in the Tridax procumbens extract, phospholipid complex (phytosome), and the phytosomal gel. The shifts in characteristic peaks between the extract and phospholipid complex (phytosome) confirmed successful complexation of the bioactive components with phospholipids. FTIR results validated the successful formation of the phospholipid complex (phytosome) system and retention of key phytoconstituents.
The phospholipid complex (phytosome) gel showed the largest zone of inhibition, indicating enhanced antimicrobial activity compared to the crude extract. The improved antimicrobial effect is likely due to the maximum amount of extract and phytosomal gel being used as compared to the standard drug. It facilitates enhanced penetration and bioavailability of phytochemicals in the phytosomal delivery system.
Phospholipid complex (phytosome) gel: 54.34% wound closure, which was significantly higher than the crude extract. The phytosomal system enhanced fibroblast migration and proliferation, confirming its potential to accelerate wound healing. The improved efficacy of the PHY-Gel formulation suggests that the phytosomal system facilitates enhanced bioavailability and sustained delivery at the wound site.
Although the phytosomal gel demonstrated superior wound closure compared to the ethanolic extract, its healing rate remained below that of the standard drug (cipladine). This discrepancy could be due to the absence of potent antiseptic agents in the phytosomal system or limited penetration depth of the herbal actives. Nonetheless, the results validate significant biological activity enhancement after phytosomal complexation. Future optimization can incorporate synergistic agents or permeation enhancers to bridge the efficacy gap with the standard drug.
5. Conclusion
The formulated Tridax procumbens-loaded phospholipid complex (phytosome) gel exhibited improved antimicrobial and wound healing activities in comparison with the ethanolic extract, owing to improved bioavailability and sustained release. The higher entrapment efficiency (93.14%) and enhanced wound closure (54.34%) highlight the advantages of the phytosomal system in topical drug delivery. The formulation demonstrated better antimicrobial efficacy than the ethanolic extract and was comparable to the standard drug amikacin. These findings indicate that phospholipid complex (phytosome) gel technology can be a promising approach for developing effective herbal-based wound healing treatments. Further in vivo and clinical studies are required to confirm its therapeutic potential.While the in vitro results demonstrate promising wound-healing and antimicrobial efficacy, further in vivo investigations on animal models are warranted to confirm biocompatibility, dermal safety, and therapeutic consistency. Long-term toxicological assessments and stability studies are essential before advancing toward clinical translation.
Author contributions
PSJ and PHS carried out the investigation, analysis, methodology development and data visualisation. JASM was responsible for conceptualisation of the research, project administration and supervision. PSJ and PHS wrote the original draft of the manuscript and PSJ, PHS and JASM were responsible for review and editing.Conflicts of interest
The authors do not have any conflict of interest.Data availability
All data generated during this study supporting its findings are available within the manuscript. All data are available from the corresponding author upon reasonable request.Acknowledgements
The authors are thankful to the Management and Principal of Shree Santkrupa College of Pharmacy, Ghogaon, for providing necessary facilities to carry out this research work.References
- V. V. Ingole, P. C. Mhaske and S. R. Katade, Phytomed. Plus, 2022, 100199, DOI:10.1016/j.phyplu.2021.100199.
- S. Agrawal and D. Mohale, Med. Plants – Int. J. Phytomed. Relat. Ind., 2010, 2, 73–78, DOI:10.5958/j.0975-4261.2.2.012.
- S. Beck, H. Mathison, T. Todorov, E. Calderón-Juárez and O. Kopp, J. Plant Stud., 2018, 7, 19–35, DOI:10.5539/jps.v7n1p19.
- Y. Park, S. Park, M. Valan Arasu, N. Al-Dhabi, H. Ahn, J. Kim and S. Park, Molecules, 2018, 22, 313, DOI:10.3390/molecules22020313.
- J. Bhise, O. Bhusnure, S. Jagtap, S. Gholve and R. Wale, J. Drug Delivery Ther., 2019, 9, 924–930, DOI:10.22270/jddt.v9i3-s.2863.
- Z. Teng, C. Yuan, F. Zhang, M. Huan, W. Cao and K. Li, et al. , PLoS One, 2012, 7, e29647, DOI:10.1371/journal.pone.0029647.
- M. Kapse and J. Mulla, Acta Mater. Med., 2024, 3, 509–520 CAS.
- V. Kheradkar and J. Mulla, World J. Pharmacogn. Phytochem., 2023, 1, 1–8 Search PubMed.
- M. Barani, E. Sangiovanni and M. Angarano, Int. J. Nanomed., 2021, 16, 6983–7022, DOI:10.2147/IJN.S318416.
- M. Lu, Q. Qiu and X. Luo, Asian J. Pharm. Sci., 2019, 14, 265–274, DOI:10.1016/j.ajps.2018.05.011.
- N. Dewan, D. Dasgupta, S. Pandit and P. Ahmed, J. Pharmacogn. Phytochem., 2016, 5, 104–108 CAS.
- P. Kidd and K. Head, Altern. Med. Rev., 2005, 10, 193–203 Search PubMed , https://pubmed.ncbi.nlm.nih.gov/16164374/.
- R. Singh, H. Gangadharappa and K. Mruthunjaya, J. Drug Delivery Sci. Technol., 2017, 39, 166–179, DOI:10.1016/j.jddst.2017.03.027.
- Z. Hooresfand, S. Ghanbarzadeh and H. Hamishehkar, Pharm. Sci., 2015, 21, 145–151, DOI:10.15171/PS.2015.29.
- A. Khade, Y. Kutal and P. Gorde, Int. J. Creat. Res. Thoughts, 2023, 11, 2320–2882 Search PubMed.
- A. Anthrayose and N. George, World J. Pharm. Res., 2018, 7, 1026–1041, DOI:10.32604/or.2023.042228.
- J. Mulla, P. Chalke, S. Londhe, M. Patil, S. Nalawade and R. Sawant, Indian J. Novel Drug Delivery, 2023, 15, 189–199 Search PubMed.
- R. Dubal, J. Mulla and M. Kapse, J. Pharm. Biol. Sci., 2024, 12, 144–150, DOI:10.18231/j.jpbs.2024.021.
- J. Mulla, N. Shetty, S. Panchamukhi and I. Khazi, Int. J. Res. Ayurveda Pharm., 2010, 1, 192–200 CrossRef CAS PubMed.
- M. Mabrouk, D. Chejara, J. Mulla, R. Badhe, Y. Choonara, P. Kumar, L. Du Toit and V. Pillay, Int. J. Pharm., 2015, 490, 429–437, DOI:10.1016/j.ijpharm.2015.05.082.
- J. Mulla, M. Mabrouk, Y. Choonara, P. Kumar, D. Chejara, L. C. du Toit and V. Pillay, J. Drug Delivery Sci. Technol., 2017, 41, 13–19, DOI:10.1016/j.jddst.2017.06.017.
- J. Mulla, V. Aralelimath, O. Tipugade, S. Shinde, N. Tetgure, A. Mulla and D. Gavali, Indian J. Novel Drug Delivery, 2020, 12, 222–227 Search PubMed.
- S. Patil and J. Mulla, Indian J. Novel Drug Delivery, 2024, 16, 104–112 Search PubMed.
- A. Naikawadi and J. Mulla, World J. Drug Delivery, 2023, 1, 28–33 Search PubMed.
- V. Sabale and S. Vora, Int. J. Pharm. Invest., 2012, 2, 140–149, DOI:10.4103/2230-973X.104397.
- A. Gosavi, P. Thorat and J. Mulla, J. Drug Delivery Ther., 2024, 14, 122–130, DOI:10.22270/jddt.v14i9.6786.
- S. Chakorkar and J. Mulla, Indian J. Pharm. Educ. Res., 2024, 28, s502–s514, DOI:10.5530/ijper.58.2s.52.
- M. H. Song, Y. Yan, B. Chen, L. Gong, L. Chen and J. Feng, et al. , Pharmaceutics., 2025, 17, 1118, DOI:10.3390/pharmaceutics17091118.
- S. Jacob, A. B. Nair, S. H. Boddu, B. Gorain, N. Sreeharsha and J. Shah, Pharmaceutics, 2021, 13, 1206, DOI:10.3390/pharmaceutics13081206.
- S. Panchamukhi, J. Mulla, N. Shetty, M. Khazi, A. Khan, M. Kalashetti and I. Khazi, Arch. Pharmacal, 2011, 344, 358–365, DOI:10.22270/jddt.v14i9.6786.
- P. Rajalakshmi, M. Sakthivel, S. Mohammed Halith, L. Aslam, S. Lenin, J. Manimegalai, P. Manoj and J. Matheshwaran, Int. J. Pharm. Res. Appl., 2023, 79, 176–180, DOI:10.47583/ijpsrr.2023.v79i02.028.
- N. Badiger, J. Mulla and I. Khazi, Pharm. Chem. J., 2013, 46, 667–671, DOI:10.1007/s11094-013-0866-9.
- S. R. Bolla, A. M. Al-Subaie, R. Y. Al-Jindan, J. P. Balakrishna, P. K. Ravi, V. P. Veeraraghavan, A. A. Pillai, S. S. R. Gollapalli, J. P. Joseph and K. M. Surapaneni, Heliyon, 2019, 5, e01648, DOI:10.1016/j.heliyon.2019.e01648.
| This journal is © The Royal Society of Chemistry 2026 |