Open Access Article
Open Access ArticleEngineering tunable GTP/TPi-responsive liposomes through liposomal membrane modulation using a bis-triphenylphosphonium lipid switch
Brooke E.
Smith
 ,
Caleb G.
Russell
,
Mayesha B.
Mustafa
and
Michael D.
Best
,
Caleb G.
Russell
,
Mayesha B.
Mustafa
and
Michael D.
Best
 *
*
Department of Chemistry, University of Tennessee, Knoxville, TN 37996, USA. E-mail: mdbest@utk.edu
First published on 4th December 2025
Abstract
Liposomes are effective nanocarriers for targeted therapeutic delivery, yet challenges regarding the extent and specificity of cargo release persist. Many disease conditions result in metabolite concentration dysregulation, increasing the appeal to harness overly abundant metabolite concentrations as triggers for targeted delivery and cargo release. Here, we introduce a novel stimuli-responsive liposomal platform with a tunable response to either guanosine triphosphate (GTP) or tripolyphosphate (TPi) that was achieved through incorporation of a novel bis-phosphonium-based lipid switch (BPLS) and strategic manipulation of liposome composition. This platform enables selective cargo release triggered by GTP, a metabolite upregulated in many fast-growing cancer cells. Fine-tuning of liposome composition also allows for TPi triggered release, a model phosphate compound to illustrate the dual response of this system. Hydrophobic and hydrophilic dye release assays, dynamic light scattering, transmission electron microscopy, and kinetic cargo release studies confirmed metabolite-responsive membrane perturbation driven by BPLS, inciting controlled release of both polar and non-polar cargo. By fine-tuning liposome composition to control metabolite selectivity and release kinetics, this platform offers a versatile framework for addressing complex metabolite profiles in diseased cells, expanding the stimuli-responsive liposome toolbox toward the potential of customized drug delivery.
Introduction
Liposomes are versatile nanocarriers that are capable of encapsulating a wide range of molecular cargo and are especially useful for applications in therapeutic drug delivery.1,2 Therefore, liposomes have found widespread utility in clinical therapeutic applications. While conventional liposomes offer improvements to therapeutic delivery, such as increased solubility and favorable pharmacokinetics, they also exhibit minimal cargo release and lack delivery specificity in the absence of a triggering mechanism.1 These particular challenges have propelled recent advances in liposomal modifications to improve these properties, particularly through the development of stimuli-responsive liposomes. Liposome platforms utilizing both active (i.e. heat, ultrasound, light)3–5 and passive (redox, pH, and enzyme expression)6–8 release systems have been developed, but challenges remain for the clinical translation of each of these. For example, external stimuli that are commonly harnessed by active release systems can be toxic to healthy cells, while passive, internal release systems have historically targeted stimuli that only exhibit slight differences between healthy and diseased cells.9,10Further expansion of this field has led to the recent development of metabolite-responsive liposomes, which exploit the benefits of passive release stimuli while addressing the challenges of diseased versus healthy cell differentiation due to the significant upregulation of metabolite concentrations that are common in diseased cells. These liposomes leverage chemical species specifically upregulated within disease states, such as zinc,11 calcium,12 guanosine triphosphate (GTP),13 and adenosine triphosphate (ATP),14 as internal triggers to selectively incite cargo release in the presence of the target stimulus.15 This is achieved by incorporating synthetic lipid switches that undergo significant conformational changes upon metabolite binding, thereby disrupting liposome membrane packing to release encapsulated contents.16 While such innovations have the potential to enhance liposome delivery capabilities, fine-tuning these carriers to respond to multiple metabolites or specific triggers by modulating liposome composition remains underexplored, limiting the ability to tailor delivery platforms to diverse disease profiles.
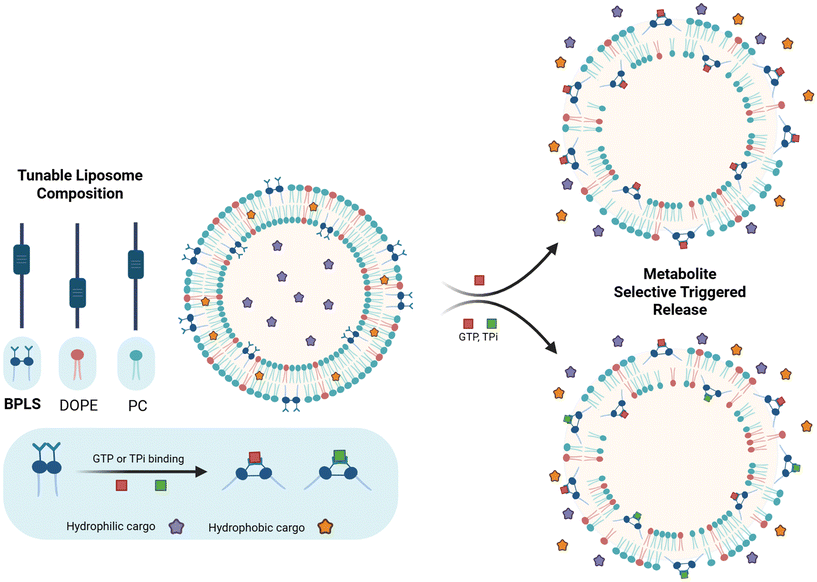
Here, we report a novel liposomal platform that achieves tunable responsiveness to phosphorylated metabolites including GTP and tripolyphosphate (TPi) by varying the liposome composition (Fig. 1). GTP is a metabolite of interest due to its involvement in a wide range of intracellular enzymatic reactions and its support of protein synthesis.17–19 Rapid tumor cell growth necessitates an abundance of GTP since protein synthesis requires two molecules of GTP per amino acid addition, and its concentration within various cancerous cell lines has been documented at 800 to 2400 µM, elevated from healthy cell concentrations.20 TPi, while not biologically relevant, serves as a proof-of-principle model phosphate, allowing us to assess the adaptability of this platform to diverse phosphorylated metabolites. The tunable responsiveness relies on varying the ratio of lipids within the liposome formulation, for which we utilized lipids including 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), L-α-phosphatidylcholine (PC), L-α-phosphatidic acid (PA), and our bis-phosphonium lipid switch (BPLS). The lipid mixture modulates the responsiveness of the liposome by varying membrane self-assembly properties. Serendipitously discovered during synthetic efforts, BPLS provides an alternative option to a copper(II) dipicolyamine (CuDPA) lipid we reported previously that is responsive toward GTP,13 but instead demonstrates tunable responses to GTP and TPi. Furthermore, the two copper chelates included within the structure of our original GTP-responsive lipid switch may limit its application in biological systems.
Our studies described herein show that liposomes formulated in a 40/35/25 BPLS/DOPE/PC ratio exhibit dual responsiveness to both GTP and TPi, whereas adjustment of this ratio to 45/20/35 yields GTP-selective hydrophobic cargo release over other structurally similar nucleotide triphosphates. Dynamic light scattering (DLS) and transmission electron microscopy (TEM) experiments confirmed morphology changes consistent with membrane disruption due to metabolite interactions with our lipid switch. Fluorescent dye release assays using both hydrophobic and hydrophilic cargo provide insight into the release capabilities of our liposome systems, highlighting the tunable release response between different metabolites. By demonstrating liposome composition-dependent metabolite selectivity, this platform expands the toolbox for metabolite-responsive liposomes and offers the potential for a versatile approach to eventually customize drug delivery for diseases with dysregulated metabolite profiles.
Results and discussion
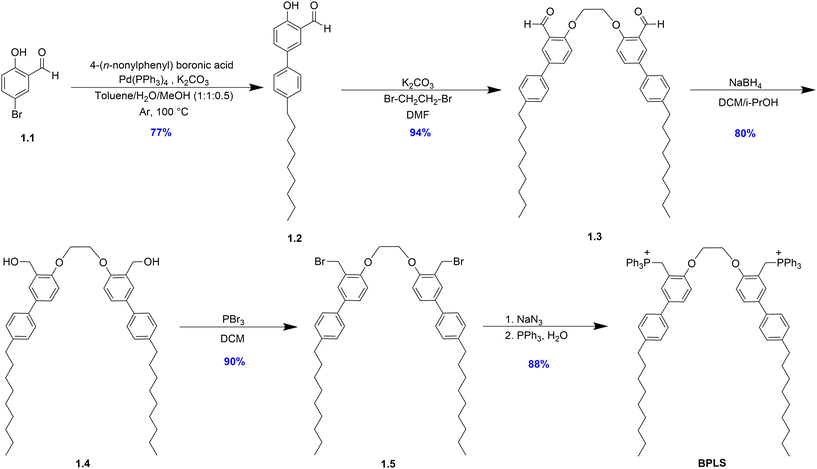
To advance the tunable metabolite-responsive lipid switch toolbox, we integrated structural insights from a previously reported ATP-responsive lipid switch14 through novel headgroup modification, leading to the incorporation of triphenylphosphonium (TPP) headgroups serendipitously discovered during synthesis. The synthetic route to BPLS (Scheme 1) adapts the previously reported work, beginning with a Suzuki coupling between 5-bromo-2-hydroxybenzaldehyde as the core of the headgroup structure and 4-(n-nonylphenyl) boronic acid, which introduces the hydrophobic lipid tail to produce 1.2. This was followed by dimerization with a 1,2-dibromoethane linker for 1.3 generation. Headgroup modification continued via reduction to 1.4 and bromination to 1.5. We next intended to perform azide substitution followed by a Staudinger reduction to produce a bis-amino product, but instead observed addition of the two TPP units of BPLS. Presumably, this occurred through substitution of either the benzylic bromide or azide groups by triphenylphosphine nucleophiles. It should be noted that the TPP groups of BPLS could drive mitochondrial targeting, and established use of alternative TPP-conjugated lipids points towards potential low cytotoxicity and biodegradability of BPLS, although further testing would be required to confirm these possibilities.21–24After producing BPLS as an unexpected product containing positively charged groups that could engage in complexation, we set out to evaluate this structure as a lipid switch for metabolite-triggered release from liposomes. Utilizing a Nile red (NR) dye release assay,22 where NR fluorescence is activated within the hydrophobic interior of the liposomal membrane but deactivated upon exposure to the aqueous buffer environment, we measured the extent of cargo release driven by bilayer perturbation through changes in fluorescence. Liposomes for these release experiments were prepared through standard thin-film hydration at 2 mM concentration and extruded through 200 nm membranes, followed by DLS analysis to confirm stable liposome formation.23
Initially, metabolite-responsive liposome selectivity was investigated using liposomes containing 35/40/25 percentages of BPLS, DOPE, and PC lipids, respectively. Liposomes were incubated for 16 hours with 1, 5 or 10 mM concentrations of common phosphate- or carboxylate-containing anionic metabolites (Fig. 2), including sodium phosphate (Pi), sodium pyrophosphate (PPi), adenosine 5′-diphosphate (ADP), adenosine 5′-triphosphate (ATP), D-fructose-6-phosphate (FP), D-fructose-1,6-bisphosphate (FBP), TPi, cytidine 5′-triphosphate (CTP), GTP, uridine 5′-triphosphate (UTP), adenosine 5′-monophosphate (AMP), malic acid (Mal), fumaric acid (Fum), oxaloacetic acid (Oxa), and succinic acid (Suc). This metabolite selectivity screen revealed that BPLS liposomes exhibited significant cargo release in response to GTP and TPi (∼70% and 72% NR fluorescence decrease, respectively), prompting our investigation into this lipid's capabilities and subsequent findings of tunable response. Some moderate metabolite driven release was also observed with ATP and UTP treated liposomes, although only at higher metabolite concentrations and resulting in less extensive cargo release. Conversely, CTP, an additional nucleotide triphosphate, yielded minimal release despite structural similarities to GTP. However, CTP has been noted as a weaker electron donor due to its pyrimidone functionality, which may account for its decreased binding affinity with BPLS.24 While data show some overlap in triggering by other nucleotide triphosphates, this is not expected to be problematic since increased concentrations of all nucleotide triphosphates are generally observed in cancer cells with rapid cell growth environments.25 Several other anionic metabolites showed virtually no NR release under these conditions, showcasing partial selectivity with this liposomal triggered release platform.
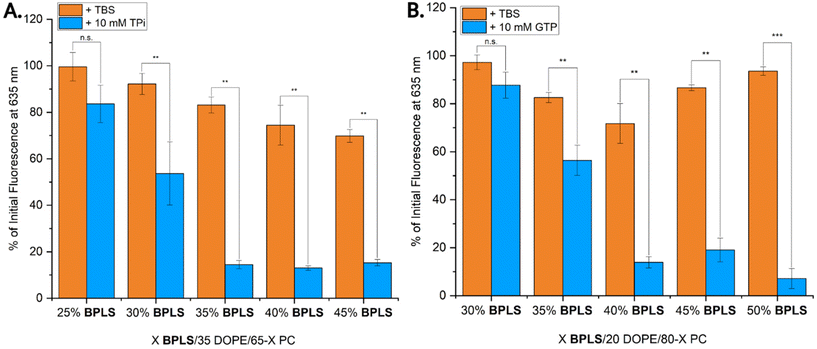
After the initial metabolite selectivity screen revealed responsiveness of BPLS liposomes to GTP and TPi, a wider range of liposome formulations were tested with the same NR release assay, specifically focusing on GTP as the triggering metabolite (summarized in Table S1). Ultimately, two key liposome formulations exhibiting favorable GTP-triggered release were identified as either 35% DOPE or 20% DOPE, with the remaining percentage shared in different ratios of BPLS and PC. TPi and GTP treatment of these two liposome formulation types yielded an interesting response, where 40/35/25 BPLS/DOPE/PC liposomes (Fig. S1A) exhibited triggered cargo release with both metabolites, but lipid ratio adjustment to 45/20/35 BPLS/DOPE/PC (Fig. S1B) yielded enhanced cargo release selectivity by GTP. This suggests that the liposome composition plays a key role in lipid switch selectivity by modulating the membrane environment in a way that can be fine-tuned to promote cargo release response to a specific metabolite.
To further investigate this tunable metabolite selectivity of BPLS and observe how lipid switch percentage and cargo release correlate, end-point NR release assays were next completed where DOPE percentages were held constant at 35 or 20% while BPLS and PC percentages were varied (Fig. 3). The inclusion of DOPE within liposome formulations aids in liposome destabilization,26 priming liposomes for triggering and subsequent cargo release. However, with DOPE inclusion, inherent liposome leakiness can also arise due to excessive destabilization. While the two previous formulations exhibited tunable release, we expected that liposome leakiness could be improved from ∼25% passive cargo release (Fig. S1A). For liposomes with 35% DOPE treated with TPi, our findings indicated a linear relationship between NR release and BPLS percentage (Fig. 3A), plateauing at ∼82% release with 35–45% BPLS. However, destabilization of these liposomes to promote triggered cargo release also resulted in increased passive cargo leakage. Of this less-selective liposome type, formulations with 35/35/30 BPLS/DOPE/PC percentages maintained the maximum cargo release while decreasing passive cargo release to ∼15%, balancing stability while maintaining metabolite triggered release capabilities.
For GTP-selective liposomes containing 20% DOPE, an initial linear trend in liposome destabilization was observed up to 30–40% BPLS (Fig. 3B). However, with higher concentrations of BPLS, a re-stabilization effect appeared, where passive cargo leakage decreased while maximum cargo release was maintained. The optimal formulation of this type was determined as 50/20/30 BPLS/DOPE/PC liposomes, which exhibited ∼5% passive leakage and ∼92% cargo release when treated with GTP.
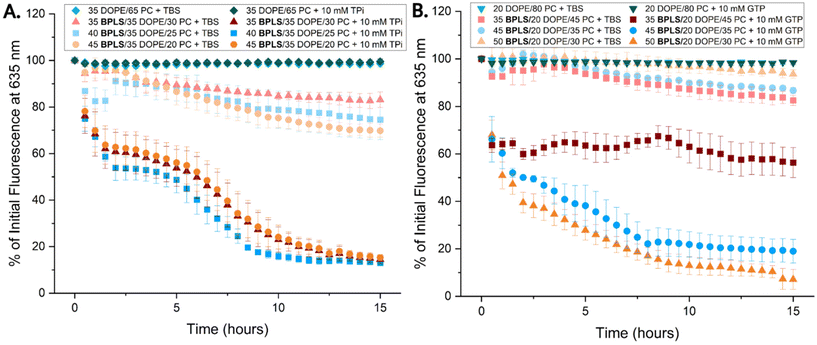
We next explored an enhanced perspective of metabolite-driven release from BPLS liposomes by studying the kinetics of NR release (Fig. 4). Here, we selected three formulations of each liposome type from the previous endpoint release assay (Fig. 3) that exhibited cargo release while minimizing passive leakage. Liposomes were prepared at 2 mM and treated with either 10 mM TPi (X/35/65-X BPLS/DOPE/PC) or 10 mM GTP (X/20/80-X BPLS/DOPE/PC). The three liposome formulations treated with TPi (Fig. 4A) showed similar kinetic release curves for NR release, plateauing at similar extents of cargo release and exhibiting ∼40–50% cargo release within 2.5 hours. The GTP treated liposomes differed in kinetic release curves for the three formulations tested, increasing in release extent as BPLS was increased. Cargo release within the first 2.5 hours ranged from ∼40–60%. NR release of the BPLS liposomes was also measured in more complex environments, where liposomes hydrated in 1x PBS still exhibited minimal cargo leakage in a phosphate-rich environment (Fig. S29) and GTP-selective release was still observed for 50/20/30 BPLS/DOPE/PC liposomes treated with 6 mM of competing metabolites (similar to the metabolites in Fig. 2) and 6 mM GTP (Fig. S30). GTP-selective liposomes were then also measured in media (Fig. S31), where GTP-selective cargo release was also observed for 50/20/30 BPLS/DOPE/PC liposomes. Overall cargo release was diminished compared to the same liposomes in buffer, but still exhibited significant release of ∼44% of NR. Buffer treated controls indicated higher overall liposome stability with decreasing passive leakage as BPLS was increased. Control liposomes without BPLS exhibited minimal NR release after metabolite treatment, indicating NR release was facilitated through lipid switch and metabolite interaction. Rate constants and t1/2 were calculated for the GTP/TPi treated samples (Fig. S12–S17).
A final NR release study was then completed (Fig. 5) to reinvestigate the metabolite selectivity of the two optimized BPLS formulations and compare to the original screen (Fig. 2), focusing specifically on the nucleotide triphosphates GTP, ATP, CTP, and UTP along with TPi. The chosen formulations represent the optimized liposome composition for response to either TPi or GTP, based on the previous kinetic release curves. NR encapsulation efficiencies for these specific formulations were found to be (19.2 ± 0.4) and (63.3 ± 0.8)% for 35/35/30 and 50/20/30 BPLS/DOPE/PC, respectively, using a calibration curve (Fig. S22). For the less-selective TPi/GTP responsive formulation, 35/35/30 BPLS/DOPE/PC liposomes exhibited triggered cargo release resulting from both TPi and GTP treatment as expected (Fig. 5), along with some moderate response with UTP treatment.
Raw DLS data indicated a split population for the GTP treated liposomes, with some particles retaining the original size and others shifted to larger diameters (Fig. S2). This can be clearly contrasted with more uniformly large aggregates from TPi-triggered liposomes in the same study. While these liposomes release cargo in the presence of either metabolite, the liposomal response and reorganization differed, highlighting how membrane dynamics may play a role in our responsive BPLS liposomes. Additionally, zeta potential studies (Fig. S3 and S4) indicated an overall negative charge flip for 35% BPLS liposomes treated with TPi versus GTP, which may be another factor responsible for this formulation's decreased selectivity even with a higher Kd determined for BPLS-TPi (Fig. S5 and S6). Both zeta potential and fluorescence binding data provide validation of BPLS binding to GTP or TPi.
Triggered release from ATP and UTP incubation for these liposomes was also reduced compared to the initial selectivity screening, confirming this formulation's increased selectivity towards GTP and TPi. This contrasted with the GTP-selective formulation, where 50/20/30 BPLS/DOPE/PC liposomes exhibited significant cargo release after GTP incubation (Fig. 5 and S7), with minimal release from alternate metabolite incubation, verifying the tunable selectivity of BPLS against similar metabolites. Both liposome formulations also exhibited minimal changes in fluorescence over a longer, untreated 7-day period, indicating minimal passive leakage and thus shelf-stability of both liposome types (Fig. S8). Both liposome formulation types were additionally loaded with Doxorubicin, where their loading capacities were determined to be (5.93 ± 0.4)% and (5.12 ± 0.4)% for 35/35/30 and 50/20/30 BPLS/DOPE/PC liposomes, respectively. Partially selective 35% BPLS liposomes indicated Dox release with both 10 mM TPi and GTP treatment whereas the more selective 50% BPLS liposomes exhibited significant Dox release with GTP treatment only (Fig. S32).
We expected that triggered release from BPLS liposomes would be accompanied by changes in liposome size and morphology due to membrane perturbations we attribute to lipid switch conformational changes upon metabolite binding. To probe potential changes in liposome morphology, we next monitored particle size changes through DLS experiments before and after TPi and GTP treatment. The samples showed uniform particle sizes of ∼200 nm before treatment (Fig. S9A and B), indicating stable liposomes were initially formed. Upon incubation with 10 mM GTP or TPi, no notable changes were observed for 0% BPLS (negative control) liposomes, indicating a lack of non-specific response. In an additional negative control, BPLS liposomes treated with buffer exhibited no significant size changes, indicating that NR release from tris-buffered saline (TBS) treated samples in the previous release experiments likely resulted from passive cargo leakage. However, liposomes containing BPLS showed significant size increases after treatment with GTP (Fig. S9A and B) or TPi (Fig. S9A), confirming that metabolite responsiveness was the source of cargo release. The less selective formulations with 35% DOPE exhibited significant size changes after treatment with both triggers. When DOPE is decreased to 20% and BPLS ranges from 30–50%, selectivity increases such that significant size changes were only observed after GTP treatment.
GTP/TPi driven membrane perturbations with BPLS binding were further supported by TEM imaging studies (Fig. 6 and S10). The dual-metabolite responsive liposomes (35/35/30 BPLS/DOPE/PC) were imaged using a negative staining technique before and after GTP or TPi treatment. While certain untreated liposomes had difficulty surviving stain treatment, we were able to image several examples of unilamellar, uniform liposomes (Fig. 6A and B). TEM images of the liposomes after one minute of incubation with 10 mM GTP (Fig. 6C and D), 30 mM GTP (Fig. 6E and F), 10 mM TPi (Fig. 6G) or 30 mM TPi (Fig. 6H) highlighted drastic morphology changes within the liposomes. Multivesicular aggregates were noted for all metabolite treatment conditions, but an interesting shift in aggregate morphology from 10 mM to 30 mM treatment was noted. Liposomes treated with 10 mM GTP or TPi showed multivesicular aggregates with more spherical vesicles identifiable within the larger aggregate. Increasing metabolite treatment to 30 mM shifted the captured morphology where individual spherical vesicles were much more difficult to identify, indicating more extensive membrane perturbations resulting from BPLS triggering.
While the prior studies primarily investigated NR hydrophobic dye release using BPLS, the unique bilayer structure of liposomes also allows for encapsulation of hydrophilic cargo within the aqueous liposome core for delivery. Release of polar, hydrophilic cargo is of particular interest due to the recent rise in RNA treatment strategies including CRISPR/Cas 9 gene editing or mRNA sequence delivery.27–29 Eliciting polar cargo release from liposomes can present additional challenges since this cargo must escape through the non-polar membrane bilayer. This may require more extreme membrane perturbation, necessitating a fine balance of liposome stability before and after encountering the stimulus. Therefore, inclusion of additional membrane-destabilizing lipids such as DOPE and PA can enhance triggered release, but requires formulation testing to ensure that passive cargo leakage remains low.
For hydrophilic cargo release studies, we utilized sulforhodamine B (SRB) dye as a model for polar cargo. SRB encapsulated at high concentrations within liposomes exhibits self-quenching due to collisional effects, which is counteracted through dye release into solution that can be tracked through fluorescence increases.30 Non-specific dye encapsulation within liposomes was performed by hydrating liposomes with a SRB-containing buffer, and unencapsulated dye was removed by size-exclusion chromatography (SEC) (Fig. S11). Encapsulation efficiencies and dye to lipid ratios were calculated to be 0.201 ± 0.005% (12.1 ± 0.3 nmol SRB per µmol lipid) and 0.075 ± 0.004% (4.51 ± 0.2 nmol SRB per µmol lipid) for 37.5/32.5/2/28 and 37.5/36.5/2/24 BPLS/DOPE/PA/PC liposomes, respectively. Polar dye encapsulation is randomly trapped during liposome formation, usually leading to low efficiencies.31 Osmolarities of the Tris–HCl buffer and SRB were corrected to be isotonic with physiological environments. SRB release results were calibrated as a percentage of total release of encapsulated dye, which was determined by treating liposomes with Triton X-100 detergent at the end of each assay.
After liposome formulation screening with various percentages of BPLS, DOPE, PA, and PC, two liposome formulations were identified for promising polar cargo release while minimizing passive cargo leakage (Fig. 7). BPLS percentages were kept constant within the two formulations at 37.5%, with changes in the DOPE/PA/PC ratios altering metabolite selectivity and overall cargo release. Liposomes composed of 37.5/32.5/2/28 BPLS/DOPE/PA/PC released ∼15% of SRB cargo upon 10 mM GTP incubation and ∼20% cargo with 10 mM TPi incubation. Conversely, liposomes with 37.5/36.5/2/24 BPLS/DOPE/PA/PC percentages released ∼10% of cargo with 10 mM GTP incubation versus 25% cargo with 10 mM TPi incubation, again indicating tunability even with liposomes that were purposely destabilized for polar cargo release. As hydrophilic cargo release requires polar molecules to move through the entire membrane, SRB release was not as extensive NR release, which allows for future optimization. However, passive cargo leakage was minimized below 5% for both liposome formulations, indicating balanced stability preventing passive leakage while still possessing sufficient liposome destabilization capabilities to facilitate polar cargo release. Rate constants and t1/2 were calculated for the GTP/TPi treated samples (Fig. S18–S21).
Conclusions
We have developed a novel liposomal platform that exhibits tunable responsiveness to GTP and TPi through strategic modulation of lipid compositions. By incorporating our TPP-based BPLS lipid switch, we achieved tunable, semi-selective and controlled cargo release with both hydrophobic and hydrophilic dye cargos. These results were confirmed using NR release assays, kinetic cargo release curves, DLS, metabolite selectivity screening, TEM imaging, and SRB kinetic release assays. Further development of this work is necessary for translation into clinical application, including optimization of trigger concentration sensitivity and release kinetics. Pharmacological evaluations completed through studies such as biodistribution, biodegradability, and toxicity analysis will additionally be necessary for further translation of the system. However, our chemical proof-of-concept results do highlight the critical role of liposome composition in fine-tuning metabolite triggered selectivity and release kinetics. While the structure of the lipid switch is important for metabolite selectivity and subsequent switch triggering, the percentage of the lipid switch in relation to other bulk lipids within liposomes also plays a key role in triggered release activity. By leveraging GTP upregulation in diseased states and exploring TPi as a model phosphate to investigate liposome platform tunability, this work provides the foundation towards a versatile approach for complex metabolic profiles in diseased cells. With this adaptable framework, a modular toolbox of lipid switches could be envisaged to eventually target a wide range of disease conditions with various upregulated metabolites. Instead of utilizing one unique lipid switch per metabolite of interest, strategically modifying liposome composition may allow a smaller number of unique switches to selectively respond to a wider range of metabolites through tuning. In the long term, liposomes that respond to targeted metabolites show promise for enhancing the selectivity of delivery to diseased cells in which those particular metabolites are upregulated. Toward that end, this work expands the stimuli-responsive liposome toolbox and paves the way for exploration of multi-metabolite responsive systems.Author contributions
B.E.S.: conceptualization, data curation, formal analysis, investigation, methodology, project administration, validation, visualization, writing – original draft, writing – review & editing; C.G.R.: investigation, methodology, writing – review & editing; M.B.M.: investigation, writing-review & editing; M.D.B.: conceptualization, funding acquisition, project administration, resources, supervision, writing – review.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). The SI includes synthesis and characterization of compounds, experimental procedures, DLS, zeta potential, binding isotherms, stability studies, additional TEM images, rate constant and t1/2 fittings, fluorescent cargo calibration curves, and additional dye and drug release assays. See DOI: https://doi.org/10.1039/d5pm00206k.Additional information is available upon request from the authors.
Acknowledgements
This material is based upon work supported by the National Science Foundation under grant DMR-1807689 (to M.D.B.) and through the Graduate Research Fellowship Program under award 2439861 (to B.E.S.). We thank Jaydeep Kolape at the University of Tennessee Advanced Microscopy and Imaging Center for assistance in obtaining TEM images and Dr. Jinchao Lou for helpful discussions.References
- B. S. Pattni, V. V. Chupin and V. P. Torchilin, Chem. Rev., 2015, 115, 10938–10966 CrossRef CAS PubMed.
- A. Akbarzadeh, R. Rezaei-Sadabady, S. Davaran, S. W. Joo, N. Zarghami, Y. Hanifehpour, M. Samiei, M. Kouhi and K. Nejati-Koshki, Nanoscale Res. Lett., 2013, 8, 102 CrossRef PubMed.
- E. Yuba, J. Mater. Chem. B, 2020, 8, 1093–1107 RSC.
- Y. Lee and D. h. Thompson, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9, e1450 Search PubMed.
- Y.-S. Kim, M. J. Ko, H. Moon, W. Sim, A. S. Cho, G. Gil and H. R. Kim, Pharmaceutics, 2022, 14, 1314 CrossRef CAS PubMed.
- W. Ong, Y. Yang, A. C. Cruciano and R. L. McCarley, J. Am. Chem. Soc., 2008, 130, 14739–14744 CrossRef CAS.
- N. M. AlSawaftah, N. S. Awad, W. G. Pitt and G. A. Husseini, Polymers, 2022, 14, 936 CrossRef CAS.
- F. Fouladi, K. J. Steffen and S. Mallik, Bioconjugate Chem., 2017, 28, 857–868 CrossRef CAS PubMed.
- B. A. Webb, M. Chimenti, M. P. Jacobson and D. L. Barber, Nat. Rev. Cancer, 2011, 11, 671–677 CrossRef CAS PubMed.
- G. J. Fisher, Z. Wang, S. Datta, J. Varani, S. Kang and J. Voorhees, N. Engl. J. Med., 1997, 337, 1419–1429 CrossRef CAS PubMed.
- R. Sagar, J. Lou, A. J. Watson and M. D. Best, Bioconjugate Chem., 2021, 32, 2485–2496 CrossRef CAS PubMed.
- J. Lou, A. J. Carr, A. J. Watson, S. I. Mattern-Schain and M. D. Best, Chem. – Eur. J., 2018, 24, 3599–3607 CrossRef CAS.
- J. Lou, M. M. Hudson, C. F. Ancajas and M. D. Best, Chem. Commun., 2023, 59, 3285–3288 RSC.
- J. Lou, J. A. Schuster, F. N. Barrera and M. D. Best, J. Am. Chem. Soc., 2022, 144, 3746–3756 CrossRef CAS.
- J. Lou, R. Sagar and M. D. Best, Acc. Chem. Res., 2022, 55, 2882–2891 CrossRef CAS.
- V. A. Frolov, A. V. Shnyrova and J. Zimmerberg, Cold Spring Harbor Perspect. Biol., 2011, 3, a004747 Search PubMed.
- K. Sumita, Y.-H. Lo, K. Takeuchi, M. Senda, S. Kofuji, Y. Ikeda, J. Terakawa, M. Sasaki, H. Yoshino, N. Majd, Y. Zheng, E. R. Kahoud, T. Yokota, B. M. Emerling, J. M. Asara, T. Ishida, J. W. Locasale, T. Daikoku, D. Anastasiou, T. Senda and A. T. Sasaki, Mol. Cell, 2016, 61, 187–198 CrossRef CAS PubMed.
- D. W. Wolff, A. Bianchi-Smiraglia and M. A. Nikiforov, Trends Mol. Med., 2022, 28, 758–769 CrossRef CAS PubMed.
- N. Mrnjavac and W. F. Martin, Biochim. Biophys. Acta, Bioenerg., 2024, 1866, 149514 CrossRef PubMed.
- K. Kopra, R. Mahran, T. Yli-Hollo, S. Tabata, E. Vuorinen, Y. Fujii, I. Vuorinen, A. Ogawa-Iio, A. Hirayama, T. Soga, A. T. Sasaki and H. Härmä, Anal. Bioanal. Chem., 2023, 415, 6689–6700 CrossRef CAS.
- M. F. Ross, G. F. Kelso, F. H. Blaikie, A. M. James, H. M. Cochemé, A. Filipovska, T. Da Ros, T. R. Hurd, R. a. J. Smith and M. P. Murphy, Biochem. – Biokhimiia, 2005, 70, 222–230 CrossRef CAS.
- P. Greenspan and S. D. Fowler, J. Lipid Res., 1985, 26, 781–789 CrossRef CAS.
- H. Zhang, in Liposomes: Methods and Protocols, ed. G. G. M. D'Souza, Springer, New York, NY, 2017, pp. 17–22 Search PubMed.
- D. A. Jose, S. Mishra, A. Ghosh, A. Shrivastav, S. K. Mishra and A. Das, Org. Lett., 2007, 9, 1979–1982 CrossRef CAS.
- N. J. Mullen and P. K. Singh, Nat. Rev. Cancer, 2023, 23, 275–294 CrossRef CAS PubMed.
- M. Johnsson and K. Edwards, Biophys. J., 2001, 80, 313–323 CrossRef CAS PubMed.
- B. N. Aldosari, I. M. Alfagih and A. S. Almurshedi, Pharmaceutics, 2021, 13, 206 CrossRef CAS.
- R. Tenchov, R. Bird, A. E. Curtze and Q. Zhou, ACS Nano, 2021, 15, 16982–17015 CrossRef CAS.
- S. Zhen and X. Li, Cancer Gene Ther., 2020, 27, 515–527 CrossRef CAS PubMed.
- R. I. MacDonald, J. Biol. Chem., 1990, 265, 13533–13539 CrossRef CAS PubMed.
- J. O. Eloy, M. Claro de Souza, R. Petrilli, J. P. A. Barcellos, R. J. Lee and J. M. Marchetti, Colloids Surf., B, 2014, 123, 345–363 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |