Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Design, synthesis, and structural analysis of an inhibitor of the gastric proton pump with a diaza-tricyclic skeleton
Nariyoshi Umekubo a,
Airi Hashizumea,
Haruki Saitob,
Satoru Katoa,
Chisato Kanai
a,
Airi Hashizumea,
Haruki Saitob,
Satoru Katoa,
Chisato Kanai c,
Chai C. Gopalasingam
c,
Chai C. Gopalasingam bd,
Christoph Gerle
bd,
Christoph Gerle d,
Hideki Shigematsu
d,
Hideki Shigematsu e,
Atsushi Yoshimori
e,
Atsushi Yoshimori *f,
Kazuhiro Abe
*f,
Kazuhiro Abe *b and
Satoshi Yokoshima
*b and
Satoshi Yokoshima *a
*a
aGraduate School of Pharmaceutical Sciences, Nagoya University, Nagoya, Aichi 464-8601, Japan. E-mail: yokoshima.satoshi.v7@f.mail.nagoya-u.ac.jp
bDepartment of Chemistry, Faculty of Science, Hokkaido University, Sapporo, Japan. E-mail: kabe@sci.hokudai.ac.jp
cINTAGE Healthcare, Inc., 3-5-7, Kawaramachi Chuo-ku, Osaka 541-0048, Japan
dRIKEN SPring-8 Center, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan
eJapan Synchrotron Radiation Research Institute (JASRI), Spring-8, 1-1-1 Kouto, Sayo, Hyogo 679-5148, Japan
fInstitute for Theoretical Medicine, Inc., 26-1, Muraoka-Higashi 2-chome, Fujisawa, Kanagawa 251-0012, Japan. E-mail: yoshimori@itmol.com
First published on 5th January 2026
Abstract
The development of potent K+-competitive acid blockers (P-CABs) as inhibitors of acid gastric secretion attracts much research attention. In this study, the structure-guided design and enantioselective synthesis of P-CABs yielded a diaza-tricyclic compound with moderate inhibitory activity against the gastric proton pump. The eutomer was experimentally confirmed, consistent with pharmacophore predictions, and its binding mode to the gastric proton pump was elucidated via cryo-electron microscopy.
Introduction
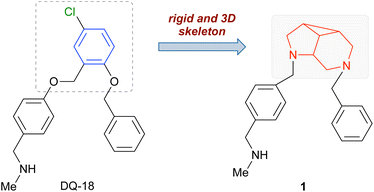
K+-competitive acid blockers (P-CABs, Fig. 1) are potent inhibitors of acid gastric secretion and as such are used for the treatment of acid-related gastric diseases such as peptic ulcers and gastroesophageal reflux disease. After the development of SCH28080 as the prototype of P-CABs,1 several molecules have been investigated, among which tegoprazan,2 revaprazan,3 and vonoprazan4 have reached clinical application in some Asian countries.In 2023, we reported the development of novel P-CABs employing an approach combining cryo-electron microscopy (cryo-EM) for structure analysis, deep generative models for de novo drug design, and organic synthesis.5 Thus, pharmacophores were defined in the drug-bound gastric proton pump structures,6 and compounds satisfying the pharmacophores were designed in silico using our deep generative models “Deep Quartet”. Several compounds were synthesized and evaluated for their inhibition activities in vitro. The binding poses of the compounds were analyzed via cryo-EM and fed back into the compound design. This approach allowed identifying DQ-18 as a potent P-CAB with a Ki value of 47.6 nM (Fig. 2).
DQ-18 has an N-methylamino group common to vonoprazan, which interacts in the gastric proton pump with the cation-binding site where three glutamates (Glu343, Glu795, and Glu820) are located. The benzene ring connecting to the N-methylamino group occupies the hydrophobic conduit of the gastric proton pump. Meanwhile, the benzene ring at the other terminal of DQ-18 occupies another hydrophobic region near the Ala123 residue. These three structural features, i.e., the N-methylamino group and two benzene rings, play important roles in the binding. We hypothesized that a novel P-CAB could be designed by replacing the central benzene ring (shown in blue, Fig. 2) with a different core structure while preserving the three key features. Specifically, we aimed to design a rigid and three-dimensional skeleton because these two characteristics are essential in drug discovery. In particular, three-dimensional structures, which are built with sp3 carbons, exhibit beneficial properties for drug discovery, such as high solubility, low promiscuity, and low CYP inhibitory activity.7 The rigidity of the skeleton is also required because it allows the substituents to be fixed in specific positions.8 Herein, we disclose the development of a novel P-CAB with a rigid and three-dimensional skeleton.
Results and discussion
Design and synthesis of P-CABs
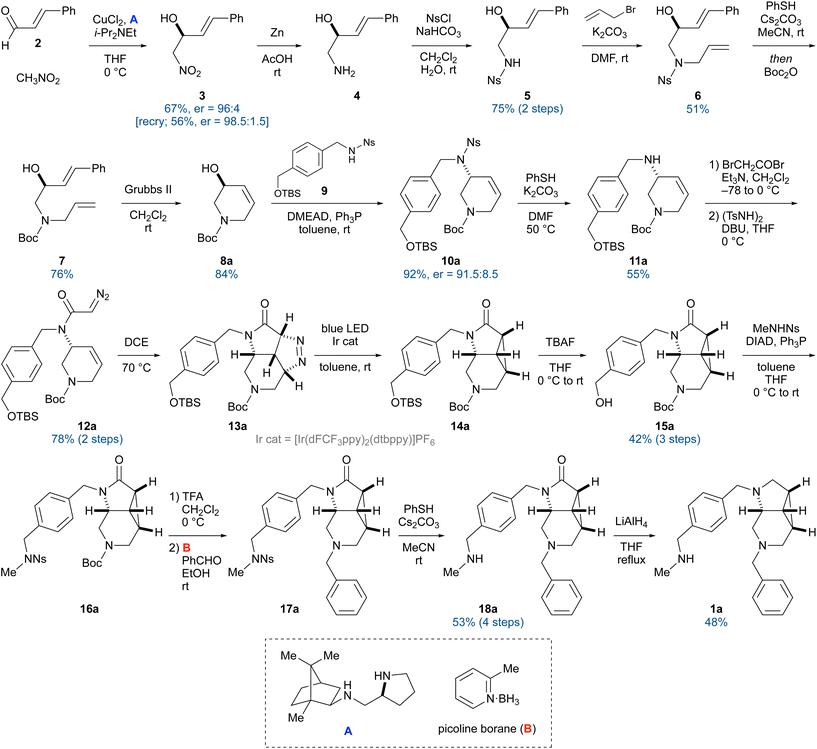
To obtain a novel P-CAB, we designed various rigid and three-dimensional core skeletons by considering the size of the binding pocket and their synthetic accessibility. By attaching two substituents on the skeletons, candidate structures were generated and screened in silico. Among them, compound 1 was identified as a potent molecule with a diaza-tricyclic skeleton consisting of pyrrolidine, piperidine, and cyclopropane rings (Scheme 1).9 To our surprise, the synthesis of the diaza-tricyclic skeleton was not previously reported, except as a partial structure of a pentacyclic aminal.10 In addition, pharmacophore fitting of the enantiomers suggested that both can interact with the gastric proton pump, with a slight preference for one of them (vide infra). To verify this preference, we sought to obtain compound 1 in an optically active form by developing an asymmetric synthetic route for the core scaffold (Scheme 2).The synthesis commenced with the copper-catalyzed enantioselective nitroaldol reaction of cinnamaldehyde (2) using chiral diamine A as the ligand,11 which afforded the known alcohol 3 in 67% yield with an enantiomer ratio (er) of 96![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. Recrystallization from xylene improved the enantiopurity to 98.5
4. Recrystallization from xylene improved the enantiopurity to 98.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5. Reduction of the nitro group in 3 with zinc in acetic acid yielded aminoalcohol 4, onto which a nosyl (Ns, 2-nitrobenzenesulfonyl) group was introduced.12 Allylation of the resulting nosylamide 5 under basic conditions proceeded smoothly to give compound 6. After replacing the Ns group with a tert-butoxycarbonyl (Boc) group, a tetrahydropyridine ring was constructed via ring-closing metathesis, affording 8a.13 A Mitsunobu reaction with nosylamide 9 furnished compound 10a.14 At this stage, the enantiomer ratio was confirmed via chiral HPLC, showing that partial racemization occurred (er = 91.5
1.5. Reduction of the nitro group in 3 with zinc in acetic acid yielded aminoalcohol 4, onto which a nosyl (Ns, 2-nitrobenzenesulfonyl) group was introduced.12 Allylation of the resulting nosylamide 5 under basic conditions proceeded smoothly to give compound 6. After replacing the Ns group with a tert-butoxycarbonyl (Boc) group, a tetrahydropyridine ring was constructed via ring-closing metathesis, affording 8a.13 A Mitsunobu reaction with nosylamide 9 furnished compound 10a.14 At this stage, the enantiomer ratio was confirmed via chiral HPLC, showing that partial racemization occurred (er = 91.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8.5). In general, the Mitsunobu reaction of allylic alcohols proceeds predominantly via an SN2 mechanism. However, allylic migration with anti-addition also occurred, leading to partial racemization.15 The Ns group was then removed under standard conditions. The resulting secondary amine 11a was converted into diazoacetamide 12a in a two-step sequence involving bromoacetylation and a reaction with N,N′-ditosylhydrazine.16 Direct cyclopropanation of 12a via treatment with a rhodium catalyst did not produce the desired compound, most likely because the rhodium carbenoid generated in situ reacted with the benzene ring.17 However, to our delight, heating in 1,2-dichloroethane (DCE) at 70 °C promoted the intramolecular cycloaddition of the diazo moiety with the C–C double bond to form pyrazoline 13a, which was then converted into the requisite cyclopropane 14a by irradiating with blue LED light in the presence of an iridium complex.17a,18 Removal of the tert-butyldimethylsilyl group with tetra-n-butylammonium fluoride, followed by a Mitsunobu reaction with N-methyl-nosylamide, gave compound 16a. After acidic cleavage of the Boc group, a benzyl group was introduced on the resulting secondary amine via reductive alkylation with benzaldehyde. Finally, removal of the Ns group and subsequent reduction of the lactam moiety with lithium aluminum hydride produced compound 1a.
8.5). In general, the Mitsunobu reaction of allylic alcohols proceeds predominantly via an SN2 mechanism. However, allylic migration with anti-addition also occurred, leading to partial racemization.15 The Ns group was then removed under standard conditions. The resulting secondary amine 11a was converted into diazoacetamide 12a in a two-step sequence involving bromoacetylation and a reaction with N,N′-ditosylhydrazine.16 Direct cyclopropanation of 12a via treatment with a rhodium catalyst did not produce the desired compound, most likely because the rhodium carbenoid generated in situ reacted with the benzene ring.17 However, to our delight, heating in 1,2-dichloroethane (DCE) at 70 °C promoted the intramolecular cycloaddition of the diazo moiety with the C–C double bond to form pyrazoline 13a, which was then converted into the requisite cyclopropane 14a by irradiating with blue LED light in the presence of an iridium complex.17a,18 Removal of the tert-butyldimethylsilyl group with tetra-n-butylammonium fluoride, followed by a Mitsunobu reaction with N-methyl-nosylamide, gave compound 16a. After acidic cleavage of the Boc group, a benzyl group was introduced on the resulting secondary amine via reductive alkylation with benzaldehyde. Finally, removal of the Ns group and subsequent reduction of the lactam moiety with lithium aluminum hydride produced compound 1a.
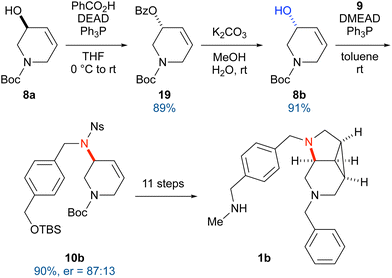
For the preparation of the opposite enantiomer, we attempted the inversion of the configuration of 8a via a Mitsunobu reaction (Scheme 3). Specifically, 8a was treated with diethyl azodicarboxylate (DEAD), triphenylphosphine, and benzoic acid, giving benzoate 19. Methanolysis of 19 under basic conditions afforded 8b. After the Mitsunobu reaction with 9, the enantiomer ratio was confirmed via chiral HPLC, which revealed the occurrence of partial racemization (er = 87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13). Using the same procedure, 10b was converted into 1b.
13). Using the same procedure, 10b was converted into 1b.
Biological evaluation and cryo-EM analysis
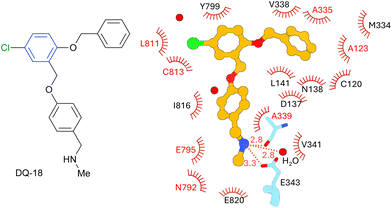
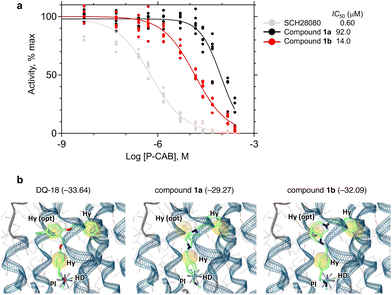
With both enantiomers in hand, we evaluated their potency by measuring the dose-dependence of ATPase activity inhibition using H+,K+-ATPase-enriched membrane fractions, according to previously reported protocols (Fig. 3a).5 Despite exhibiting lower IC50 values than the reference compound SCH28080 (IC50,SCH = 0.60 μM), compounds 1a and 1b inhibit the H+,K+-ATPase activity in a dose-dependent manner with IC50 values of 92.0 and 14.0 μM, respectively. The configuration of the more active enantiomer (eutomer, 1b) is the same as that of the optical isomer that exhibited stronger binding in the pharmacophore fitting (Fig. 3b).We also performed a cryo-EM analysis of H+,K+-ATPase bound to compound 1b to verify its binding mode (Fig. 4).19 The EM map analyzed at an overall resolution of 2.66 Å unambiguously resolved the densities corresponding to compound 1b with surrounding water molecules and amino acids (Fig. 4b). As expected according to the positions of the pharmacophore features set in Deep Quartet and the docking simulations (Fig. 3b), compound 1b is bound to the luminal-facing conduit in the transmembrane region of the gastric proton pump, which connects the cation-binding site (e.g., E343, E795, and E820) to the gastric luminal solution (Fig. 4c). The cationic secondary amine moiety of 1b is located close to the cation-binding site, suggesting a weak electrostatic interaction with the Glu343 side chain (3.9 Å). This characteristic is observed in DQ-related compounds and vonoprazan, faithfully reflecting the pharmacophore feature defined in the Deep Quartet calculation.5 A hydrogen bond with the main chain carbonyl of Ala339 (2.9 Å) is also observed. These polar interactions may contribute to fixing the binding position of 1b in the hydrophobic pocket. Apart from the abovementioned polar interactions, there are many van der Waals interactions with surrounding amino acids, including Val341 (3.4 Å), Glu795 (3.7 Å), Asn792 (3.5 Å), and Glu820 (3.4 Å) (Fig. 4f). The two benzene rings are positioned to satisfy the defined pharmacophore features and thus engage in hydrophobic interactions with the snugly fitted binding pocket (Fig. 4d and e). The core diaza-tricyclic skeleton of 1b is juxtaposed to Tyr799 (Fig. 4c). To our surprise, despite the bulkiness of this scaffold, it fits well within the binding pocket (Fig. 4d and e). However, the π–π interaction that most P-CABs form with Y799 by placing an sp2 functional group at this position is not expected for compound 1b. This may be one of the reasons for its lower apparent affinity compared with DQ18 and other P-CABs. Interestingly, when compared with the DQ18-bound structure, the positions of Y799 and TM2 are displaced (Fig. 4g). Owing to the presence of the bulky diaza-tricyclic skeleton, Y799 moves by 0.9 Å, and TM2, including Asn137 and Asn138, shifts by 0.7 Å, widening the binding pocket. This demonstrates that the relationship between the proton pump binding site and the inhibitor is not a simple lock-and-key model; instead, the compound binding induces small-scale conformational changes. Such induced fit was not observed with SCH28080, whose bicyclic imidazopyridine ring occupies the position of the diaza-tricyclic skeleton, suggesting the characteristic effect of this sp3-rich, nonplanar, and bulky scaffold.
Conclusions
We designed P-CABs based on the cryo-EM structure of the gastric proton pump bound to DQ-18, and pharmacophore fitting allowed identifying a potent candidate with a diaza-tricyclic skeleton consisting of pyrrolidine, piperidine, and cyclopropane rings. We established a synthetic route to the diaza-tricyclic skeleton via an enantioselective nitroaldol reaction, construction of the cyclopropane ring through a 1,3-dipolar cycloaddition of a diazo compound, and subsequent nitrogen extrusion from the resulting pyrazoline intermediate. Stereoinversion via a Mitsunobu reaction enabled the preparation of both enantiomers. Biological evaluation revealed that both enantiomers exhibited moderate inhibitory activity against the gastric proton pump, with compound 1b being the more active enantiomer, as predicted by pharmacophore fitting. The binding features of compound 1b to the gastric proton pump were elucidated via a cryo-EM analysis.Author contributions
S. Y. designed the study with K. A., A. Y., and N. U. C. K. and A. Y. performed in silico screening. N. U., A. H., S. K., and S. Y. synthesized the compounds. Ha. S. performed the ATPase assay. Ha. S. expressed and purified the protein. Ha. S. and K. A. performed the cryo-EM analysis with C. C. G, C. G., and Hi. S. Ha. S., C. C. G., and K. A. performed image processing, model building, and structural interpretations. S. Y., K. A., A. Y., and N. U. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5ob01828e.The structural data generated in this study have been deposited in the Protein Data Bank and EM Data Bank under accession codes 9VVO and EMD-65385.19a,b
Acknowledgements
This work was financially supported by JSPS KAKENHI (Grant Numbers JP23H02602, JP23K14321, JP24H01767, and JP24K01957); by the Research Support Project for Life Science and Drug Discovery [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from the Japan Agency for Medical Research and Development (AMED) under Grant Number JP25ama121044 (support number 4085) and JP25ama121001 to Masaki Yamamoto, C. G., C. C. G., and H. S.; and by JST CREST (Grant number JPMJCR22E4). Cryo-EM data are acquired using EM01CT of SPring-8 with approval of the Japan Synchrotron Radiation Research Institute (proposal number 2024B2519).References
- B. Wallmark, C. Briving, J. Fryklund, K. Munson, R. Jackson, J. Mendlein, E. Rabon and G. Sachs, J. Biol. Chem., 1987, 262, 2077–2084 Search PubMed.
- D. K. Kim, K.-H. Lee, S.-J. Kim, S.-J. Kim, S. J. Lee, C. H. Park, B.-T. Kim, G.-S. Song, B.-S. Moon and S.-Y. Ryu, J. Pharmacol. Exp. Ther., 2019, 369, 318–327, DOI:10.1124/jpet.118.254904 30894456.
- K. S. Han, Y. G. Kim, J. K. Yoo, J. W. Lee and M. G. Lee, Biopharm. Drug Dispos., 1998, 19, 493–500, DOI:10.1002/(sici)1099-081x(1998110)19:8<493::aid-bdd129>3.0.co;2-z.
- K. Otake, Y. Sakurai, H. Nishida, H. Fukui, Y. Tagawa, H. Yamasaki, M. Karashima, K. Otsuka and N. Inatomi, Adv. Ther., 2016, 33, 1140–1157, DOI:10.1007/s12325-016-0345-2.
- K. Abe, M. Ozako, M. Inukai, Y. Matsuyuki, S. Kitayama, C. Kanai, C. Nagai, C. C. Gopalasingam, C. Gerle, H. Shigematsu, N. Umekubo, S. Yokoshima and A. Yoshimori, Commun. Biol., 2023, 6, 956 Search PubMed.
- (a) K. Abe, K. Irie, H. Nakanishi, H. Suzuki and Y. Fujiyoshi, Nature, 2018, 556, 214 Search PubMed; (b) S. Tanaka, M. Morita, T. Yamagishi, H. V. Madapally, K. Hayashida, H. Khandelia, C. Gerle, H. Shigematsu, A. Oshima and K. Abe, J. Med. Chem., 2022, 65, 7843 Search PubMed.
- (a) F. Lovering, J. Bikker and C. Humblet, J. Med. Chem., 2009, 52, 6752 Search PubMed; (b) F. Lovering, MedChemComm, 2013, 4, 515 Search PubMed.
- A. D. G. Lawson, M. MacCoss and J. P. Heer, J. Med. Chem., 2018, 61, 4283 Search PubMed.
- Several skeletons have been identified as likely. Works on other skeletons are currently underway and will be reported in due course.
- O. Tsuge, S. Kanemasa and S. Takenaka, Bull. Chem. Soc. Jpn., 1983, 56, 2073 Search PubMed.
- Y. Zhou, J. Dong, F. Zhang and Y. Gong, J. Org. Chem., 2011, 76, 588 Search PubMed.
- (a) T. Fukuyama, C.-K. Jow and M. Cheung, Tetrahedron Lett., 1995, 36, 6373 Search PubMed; (b) T. Kan and T. Fukuyama, Chem. Commun., 2004, 353 Search PubMed.
- (a) C. Lecourt, S. Dhambri, L. Allievi, Y. Sanogo, N. Zeghbib, R. Ben Othman, M. I. Lannou, G. Sorin and J. Ardisson, Nat. Prod. Rep., 2018, 35, 105 Search PubMed; (b) M. Yu, S. Lou and F. Gonzalez-Bobes, Org. Process Res. Dev., 2018, 22, 918 Search PubMed; (c) E. J. Groso and C. S. Schindler, Synthesis, 2019, 1100 Search PubMed.
- (a) O. Mitsunobu, Synthesis, 1981, 1 Search PubMed; (b) K. C. K. Swamy, N. N. B. Kumar, E. Balaraman and K. V. P. P. Kumar, Chem. Rev., 2009, 109, 2551 Search PubMed; (c) S. Fletcher, Org. Chem. Front., 2015, 2, 739 Search PubMed.
- B. K. Shull, T. Sakai, J. B. Nichols and M. Koreeda, J. Org. Chem., 1997, 62, 8294 Search PubMed.
- T. Toma, J. Shimokawa and T. Fukuyama, Org. Lett., 2007, 9, 3195 Search PubMed.
- (a) Y. Ohfune, T. Demura, S. Iwama, H. Matsuda, K. Namba, K. Shimamoto and T. Shinada, Tetrahedron Lett., 2003, 44, 5431 Search PubMed; (b) T. Miura, Y. Funakoshi and M. Murakami, J. Am. Chem. Soc., 2014, 136, 2272 Search PubMed.
- M. J. R. Richter, F. J. Zécri, K. Briner and S. L. Schreiber, Angew. Chem., Int. Ed., 2022, 61, e202203221 Search PubMed.
- (a) 9VVO: H. Saito and K. Abe, Cryo-EM structure of gastric proton pump bound to YK01, 2026; (b) EMD-65385: H. Saito and K. Abe, Cryo-EM structure of gastric proton pump bound to YK01, 2026.
| This journal is © The Royal Society of Chemistry 2026 |