 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Leveraging mirror-image glycans in carbohydrate materials
Dominik Wehab and
Martina Delbianco *b
*b
aDepartment of Chemistry and Biochemistry, Freie Universität Berlin, Arnimallee 22, 14195 Berlin, Germany
bDepartement of Biomolecular Systems, Max Planck Institute of Colloids and Interfaces, Am Mühlenberg 1, 14476 Potsdam, Germany. E-mail: martina.delbianco@mpikg.mpg.de
First published on 12th January 2026
Abstract
Chirality has become a fundamental design principle to craft peptide materials. In contrast, the systematic exploitation of chirality to build glycan materials remains largely unexplored, despite the rich chiral diversity of carbohydrates, with both D- and L-configurations readily available in nature. Here, we emphasize the added value of exploring mirror-image glycans to tailor carbohydrate materials. By examining the distinctive chiral features of carbohydrates in comparison to peptides, we demonstrate how these characteristics provide powerful opportunities to modulate and elucidate the rules governing glycan assembly. We discuss examples of carbohydrate materials based on individual enantiomers, the co-assembly of racemic mixtures, and the assembly of heterochiral sequences. Each section is introduced with key insights from peptide materials, serving as inspiration and guidance for the future design of glycan assemblies.
Introduction
Life is based on chiral building blocks such as nucleotides, amino acids, and carbohydrates which assemble to form higher-order structures.1,2 Likewise, synthetic molecular chiral systems have advanced the fields of asymmetric catalysis,3 optics,4,5 and materials science.2,6–8 Chirality dictates the spatial configuration of molecules, which in turn influences their interactions with other molecules. In supramolecular systems, the chiral information encoded in the individual components governs the interactions among them, ultimately leading to the generation of unique materials.9 Chirality shapes the landscape of possible non-covalent interactions, including hydrogen bonding, ionic interactions, π–π stacking, and van der Waals forces, guiding self-assembly across different length scales.10,11Although molecular asymmetry is well understood, the critical features and driving forces that govern chiral propagation from asymmetric molecules towards ordered supramolecular structures remain elusive.3,10–12 To address this, assemblies based on synthetic enantiopure molecules have been studied to grasp the rules of chiral transmission across scales in nature.10 In parallel, systems based on the co-assembly of both enantiomers have been explored. These works revealed that the co-assembly of enantiomeric building blocks can alter the molecular packing within supramolecular structures, opening up new possibility not available in our predominantly homochiral world.
This approach has been extensively explored to modulate the secondary structure and assembly behavior of peptides, ultimately generating novel peptide-based materials.13–26 Co-assemblies of natural L-peptides with their non-natural D-enantiomers have resulted in materials with enhanced mechanical properties.13–18 Similarly, the combination of D- and L-amino acids—whether within the same sequence or as co-assemblies—allowed access to unusual geometries, broadening the portfolio of peptide-based engineered nanomaterials.18,19,27–29 The preferential interactions between enantiomers have been also leveraged to simplify the structural analysis of difficult-to-crystallize peptides and proteins through racemic crystallization.30
Similar to peptides, carbohydrates (i.e. oligosaccharides and polysaccharides, a.k.a. glycans) are versatile building blocks for creating programmable supramolecular structures.31 Their multiple hydroxyl groups and chiral centers offer several options for directional, site-specific functionalization enabling fine-tuning of both structures and functions. However, chiral carbohydrate assemblies have been significantly less explored compared to their peptide counterparts. To date, the chiral characteristics of glycans have primarily been exploited in the context of cellulose and chitin nanocrystal assemblies, which have been extensively reviewed2 and fall outside the scope of this review.
Here, we propose that chirality should be a fundamental design principle in crafting carbohydrate materials. Carbohydrates offer a much bigger chiral pool compared to peptides, with D- and L-monosaccharides – each including multiple stereocenters – readily available in nature.32 Their remarkable chiral features offer a powerful opportunity to modulate the formation of chiral supramolecular structures as well as to elucidate the rules governing glycans assembly in natural systems. By comparing the exceptional chiral features of carbohydrates to those of peptides, we emphasize the added value of exploring mirror-image glycans to enhance the tuning of carbohydrate materials. Our focus will be on the assembly of individual enantiomers, the co-assembly of racemic mixtures, and the assembly of heterochiral sequences (i.e. sequences containing both enantiomers). Each section will be introduced with key highlights from the broad literature on peptide materials, serving as inspiration and guidance for the future design of glycan assemblies.
To classify the systems discussed throughout this review, we will adopt the following nomenclature (Fig. 1). This nomenclature applies to both peptides and glycans:
Glycan's chiral features
A huge natural chiral pool
Glycans exhibit a remarkable structural diversity, arising from more than 100 fundamental building blocks (i.e. monosaccharides) that vary in ring size and hydroxyl group orientations. The glycan pool of biologically relevant monosaccharides includes both D- and L-configurations.32–34 For example, the most common natural form of galactose is D-galactose while fucose primarily exists as L-fucose. Additionally, some monosaccharides like fucose, rhamnose, and xylose can even be found in both D-and L-configurations, even though one enantiomer is significantly less abundant in nature.32–34 For arabinose both D-and L-enantiomers are widespread.32–34 This plasticity in glycan structures contrasts with peptides, where proteinogenic amino acids are almost exclusively found in the L-configuration.Another significant difference lies in the structural complexity of the building blocks. Amino acids are based on a single asymmetric carbon, while each monosaccharide can be viewed as an array of chiral centers. Inverting the stereoconfiguration at a single position of a monosaccharide results in a new compound (i.e. a diastereoisomer) with distinctly different physical properties (Fig. 2A). For example, the inversion of position C2 of D-glucose (D-Glc) yields D-mannose (D-Man), while inverting the C4 position produces D-galactose (D-Gal).35 The presence of multiple stereocenters also results in different monosaccharides with analog three-dimensional arrangements of some hydroxyl groups (Fig. 2A).35–37 For example, the enantiomer of D-Man, L-mannose (L-Man), shares structural similarities with L-rhamnose (L-Rha; 6-deoxy-L-mannose), and L-fucose (L-Fuc; 6-deoxy-L-galactose) resembles L-galactose (L-Gal). In addition, D-Man and L-Fuc present the same three-dimensional arrangement of hydroxyl groups at position C2, C3 and C4 (Fig. 2B) displaying equivalent binding epitopes for some lectins, i.e. glycan binding proteins. For this reason, the C-type lectin DC-SIGN recognizes D-Man and L-Fuc as canonical binders but also binds their close analogs, D-Rha (6-deoxy-D-mannose) and L-Gal (6-hydroxyl-L-fucose).36
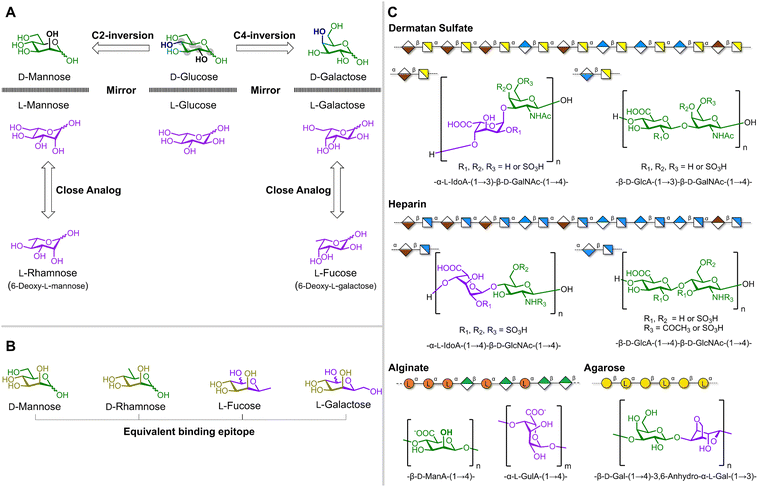 | ||
| Fig. 2 (A) D-Man and D-Gal described as diasteroisomers of D-Glc. Chiral centers in D-Glc are marked with gray dots. Inversion of chiral center at C2 in D-Glc generates D-Man while inversion of the chiral center at C4 yields D-Gal. Mirroring the D-enantiomers (green) produces L-enantiomers (purple) L-Man, L-Glc and L-Gal which possess close 3D-analogs, i.e. L-Rha (6-deoxy-L-mannose) and L-Fuc (6-deoxy-L-galactose). (B) Equivalent binding-epitopes of D-Man, D-Rha (6-deoxy-D-mannose), L-Fuc and L-Gal (6-hydroxyl-L-fucose).36 (C) SNFG38 representation and chemical structure of “pseudo” heterochiral natural polysaccharides. Illustrative SNFG38 representation of dermatan sulfate and chemical structure of disaccharide repeating units. The disaccharide blocks are distributed heterogeneously within the polysaccharide forming domains of only L-iduronic acid (L-IdoA)-containing disaccharide blocks, alternating L-IdoA and D-glucuronic acid (D-GlcA) blocks, or exclusively blocks of D-GlcA.39 Illustrative SNFG38 representation of heparin and chemical structure of disaccharide repeating units. The disaccharide blocks are distributed heterogeneously within the polysaccharide.40 Illustrative SNFG38 representation of alginate and chemical structure of the β-D-mannuronic (β-D-ManA) and α-L-guluronic acid (α-L-GulA) residues. The distribution of monomer blocks within the polysaccharide depends on the source of the algae.41 Illustrative SNFG38 representation of agarose and chemical structure of disaccharide repeating unit.42 | ||
Such rich three-dimensional information encoded within each monosaccharide scaffold, combined with different glycosidic linkages (α vs. β) and ring sizes (pyranose vs. furanose), contributes to a unique structural diversity in glycans.35 Moreover, polysaccharides that contain both D- and L-configurations are common in nature (Fig. 2C). For example, L-iduronic acid (L-IdoA) is a key component in linear, anionic glycosaminoglycans (GAGs), such as dermatan sulfate (in combination with D-glucuronic acid (D-GlcA) and D-N-acetyl galactosamine (D-GalNAc)) or heparin (in combination with D-GlcA and D-N-acetyl glucosamine (D-GlcNAc)). These polymers are involved in a variety of important biological processes, including cell proliferation and signaling, controlling coagulation, and wound healing.39,40,43 Similarly, L-guluronic (L-GulA) and D-mannuronic (D-ManA) acid build up the linear anionic polysaccharide alginate which is a major constituent in brown algae cell walls and produced by some bacteria.44 To date alginate has found applications in biomedical science and tissue engineering.41,45 Agarose, unique for its self-gelling capabilities, is another example of linear polysaccharide composed of repeating D-Gal and 3,6-anhydro-L-galactopyranose.42 Such “pseudo” heterochiral polysaccharides, containing both D- and L-configurations, are utilized by nature to perform function.39–45 Nevertheless, only a few synthetic non-natural heterochiral systems have been reported,46–50 which is surprising considering the abundance of heterochiral glycans in nature.
To date, glycans chiral features have been mainly discussed in the context of glycobiology, particularly as determinants of glycan–protein interactions and immunological responses.35 We argue that glycan chirality could offer an important design tool to modulate secondary structures and assembly behavior of carbohydrates, opening the way to tunable glycomaterials. Heterochiral peptide sequences are frequently used to trigger novel types of assemblies with interesting material properties. In analogy, we speculate that heterochiral, or “pseudo” heterochiral, glycan sequences could present untapped opportunities for the design of synthetic carbohydrate-based materials.
Glycan chirality influences assembly chirality
In materials science, glycans have primarily been used to decorate supramolecular systems tuning their self-assembly51–53 or to generate glycoamphiphiles that form low molecular weight gelators.54,55 In glycan-decorated systems, factors such as the stereochemistry of the glycosidic linkage (α vs. β) or the orientation of the hydroxyl groups determines the capability to engage in directional non-covalent interactions, dramatically affecting aggregation. Variation in these parameters can alter the secondary structure, molecular packing, hierarchical assembly, and morphology of the resulting supramolecular structure.The impact of different glycan decorations on assembly has been systematically documented in recent reviews, which we highlight for a more detailed background.51–53,55 Notable examples include the decoration of benzene-1,3,5-tricarboxamides (BTA) supramolecular monomers with epimers (e.g. D-Glc vs. D-Man) resulting in helical fibers with opposite handedness (Fig. 3).56
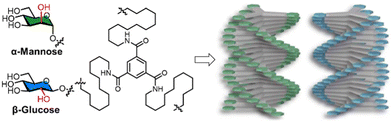 | ||
| Fig. 3 Chemical structures of BTA-D-mannose and BTA-D-glucose which assemble into helical fibers with opposite handedness. Figure was adapted from ref. 52, CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/). | ||
Similarly, the distinct packing modes observed in 4-nitrophenylmethoxycarbonyl (NPmoc) C2-functionalized with epimers D-glucosamine (D-GlcN) and D-galactosamine (D-GalN) resulted in different hydrogen bonding patterns, ultimately yielding either columnar or spherical structures.57 These well-defined synthetic models yielded important insights on the stereochemistry-directed assembly of diastereoisomers, emphasizing that glycans are versatile chiral building blocks capable of creating materials through self-assembly.
In these examples, the assembly is primarily driven by the non-carbohydrate portion of the molecule (i.e. aglycone), while differences in the type of carbohydrate decoration determine the chirality of the system. Most of these systems investigated how the decoration with different diastereoisomers (e.g. D-Glc vs. D-Man) led to different supramolecular outcomes. We believe that a systematic analysis of chiral interactions between glycans could expand the opportunities for carbohydrate assembly. In this review, we will systematically discuss how carbohydrate enantiomers (e.g. D-Glc vs. L-Glc and “pseudo-enantiomers”, e.g. D-Man vs. L-Rha) could be harnessed to tune supramolecular interactions.
Assembly and co-assembly of homochiral molecules
Assembly and co-assembly of homochiral peptides
In the past decades, synthetic chiral peptides have provided critical insights into the stereochemistry-directed co-assembly of opposite enantiomers, navigating the balance between conglomerate (self-sorting) and racemate (co-assembling) formations.13–23,26,30,58–60 In some cases, self-sorting of short β-sheet forming peptide sequences has been observed as a result of preferred homochiral pairing, which allowed for stronger non-covalent interactions compared to heterochiral pairing.58–60 In other instances, a stereocomplex was obtained due to preferential heterochiral pairing of enantiomers.13–23,30 In the following examples, we highlight the heterochiral co-assembly of peptide enantiomers and the underlying driving factors influencing their aggregation. While experimental factors that dictate either kinetic or thermodynamic control during assembly are important considerations, they will not be the primary focus of this review.21 Instead, we will concentrate on how heterochiral co-assembly enhances structural complementarity in the resulting supramolecular structures compared to homochiral assembly.24Rippled β-sheets
To enhance complementarity within β-sheet structures, a rippled architecture was first hypothesized by Pauling and Corey in 1953.61 Their model suggested that equimolar mixtures of D- and L-peptides should assemble in alternated fashion, leading to a more effective side chain distribution across the sheet (Fig. 4A).61 Indeed, experimental results confirmed this hypothesis, showing the formation of rippled β-sheets from aggregating mirror-image peptide strands.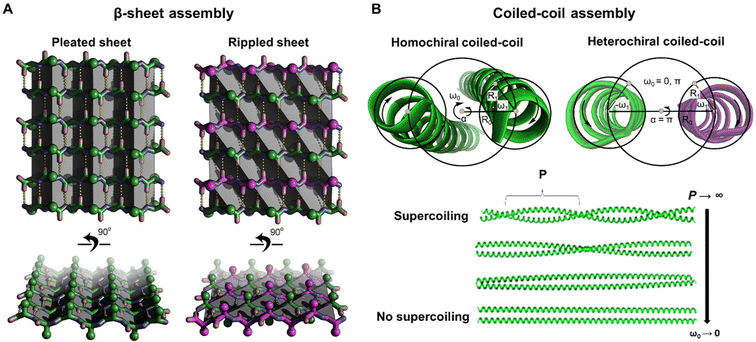 | ||
| Fig. 4 (A) Model comparison between pleated and rippled β-sheet. The pleated sheet is entirely composed of L-amino acids (green), forming uniform zigzag arrangements (pleats, highlighted as gray planes) aligned along the hydrogen-bonding direction (yellow dots). The rippled sheet contains alternating strands of L- (green) and D- (purple) amino acids with ripples (gray planes) running diagonal to the hydrogen bond network. This arrangement leads to a more effective distribution of side chains (green and purple spheres) across the rippled sheet compared to the pleated sheet.23 (B) Conventional Crick coiled-coil parameters shown for an axial view of a homochiral coiled-coil dimer (GCN4, PDB 2ZTA) and heterochiral coiled-coil dimer (M2-TM I42A). Due to opposite helicity of enantiomers no supercoiling occurs in the heterochiral coil-coil dimer. As the pitch, P of a coiled-coil super helix becomes larger, the angular frequency of the superhelix goes to zero (as P → ∞, ω0 → 0).20 Part A was adapted from ref. 23, CC BY-NC 3.0 (https://creativecommons.org/licenses/by-nc/3.0/). Part B was adapted from ref. 20, Copyright 2019, American Chemical Society. | ||
Mirror-image complementarity also affected the mechanical properties of peptide materials. For instance, MAX1, a 20-amino-acid peptide that folds into a β-hairpin, self-assembled into a hydrogel composed of well-defined fibrils. Its strands are based on alternating hydrophobic (valine, V) and hydrophilic (lysine, K) residues and are connected with a central four residue core sequence (VDPPT). The co-assembly of equimolar amounts of L-MAX1 and D-MAX1 lead to a fibrillar hydrogel exhibiting a 4-fold higher rigidity than the homochiral hydrogels.16 The fibrils consisted of a peptide bilayer forming hydrogen bonds along the long axis, with enantiomers arranged in an alternating manner, generating an extended heterochiral rippled sheet. The alternation between D- and L-enantiomers allowed for a staggered arrangement of valine residues, leading to nested hydrophobic interactions not possible in the homochiral hydrogel, thus responsible for the increased rigidity.17
A similar observation was made during the co-assembly of an amphipathic Ac-(FKFE)2-NH2 sequence. A fibrillar hydrogel composed of a flat nanoribbon formed from the racemic mixture, while single enantiomers assembled into helical fibrils. Both types of hydrogels (i.e. heterochiral and homochiral) exhibited a defined β-sheet secondary structure, but, akin to MAX1, the heterochiral co-assembly lead to a rippled sheet structure.13 The heterochiral hydrogels showed enhanced rigidity and proteolytic stability compared to the homochiral hydrogel, readily degraded by common proteases.15
Recently, the first crystal structure of rippled β-sheets was reported, advancing our understanding of how opposite enantiomeric strands are organized within these strcutures.21–23,26 In combination with theoretical calculation, this work indicated that peptide strands tend to assembled in a supramolecular motif that maximizes backbone hydrogen bonding and minimizes steric hindrance between side chains.25
In the above examples, heterochiral co-assembly did not change dramatically the overall morphology of the supramolecular structure; both heterochiral co-assembly and homochiral assembly resulted in fibrillar hydrogels. In contrast, the heterochiral co-assembly of L- and D-KYFIL pentapeptides resulted in a network of platelets, while a fibrillar hydrogel was obtained from the homochiral assembly.18 The network of platelets exhibited a lower stiffness, but increased proteolytic stability, compared to the homochiral hydrogels. This unexpected behavior may be linked to the side chain arrangement in the primary sequence. For instance, the KYFIL peptide sequence contained stretches of four hydrophobic side chains whereas the MAX1 peptides featured alternating hydrophobic and hydrophilic residues.18 This difference might influence the hydrophobic interactions governing supramolecular assembly, leading to different material properties (stiffness) and morphology (platelets instead of fibrils).
Helical coiled-coil complexes
Heterochiral interactions are also prominent in the co-assembly of α-helix enantiomers, affecting the morphology of coiled-coil peptide assemblies (Fig. 4B).20 Canonical homochiral coiled-coils peptide motifs exhibit a seven amino acid repeating pattern (heptad) often labelled abcdefg. In this sequence, positions a and d are hydrophobic residues whereas e and g are ionic. This arrangement facilitates hydrophobic knob-into-hole (KIH) and electrostatic interactions.14 In contrast, heterochiral coiled-coils preferers a 11-residue/hendecad motif (3,4,4 spacing). This preference arises because dimeric heterochiral coiled-coil systems cannot undergo supercoiling due to the opposite helicity of the enantiomers, which precludes any crossing angle other than 180°. In the hendecad motif, supercoiling is not necessary to compensate for the drift in hydrophobic side chains of heptad sequence patterns.20,62 Instead, the hydrophobic side chains in the 3,4,4-spacing pattern (hendecad) are displayed nearly parallel to the helical axis, mitigating the need of supercoiling. As a result, a dimeric heterochiral superhelix can be described as two parallel lines that have phases of either 0 or π-radians.Systematic studies showed that heterochiral complexes have increased stability against enzymatic digestion compared to their homochiral counterpart, as demonstrated for heterochiral co-assembly of glutamic acid/lysine (E/K)-rich coils.14 Additionally, the heterochiral hendecad coiled-coils displayed greater structural stability than their homochiral analogues. These properties makes heterochiral coiled-coil systems highly attractive for biotechnological applications, potentially overcoming the limited in vivo lifetime of natural coiled-coils.14
Assembly and co-assembly of homochiral glycans
The co-assembly of peptide enantiomers has provided access to new morphologies and materials with advantageous properties, such as increased stiffness and proteolytic stability. Inspired by these findings in peptides, the assembly and co-assembly of homochiral glycan samples were investigated.The asymmetric nature of glycans ensured that homochiral assembly of individual enantiomers led to mirroring supramolecular structures, independently of functional group modifications.46,63–65 For instance, glucose equipped stearoyl esters readily assembled into left- (for D-GLc) and right-handed (for L-Glc) cookie-shaped microparticles, sharing the same thermal properties.63
Mirroring supramolecular structures were also observed upon self-assembly of benzyl substituted glucose disaccharides DD and LL (Fig. 5A), forming micrometer long and nanometer wide helical fibers with left- (DD) or right- (LL) handed twists.64
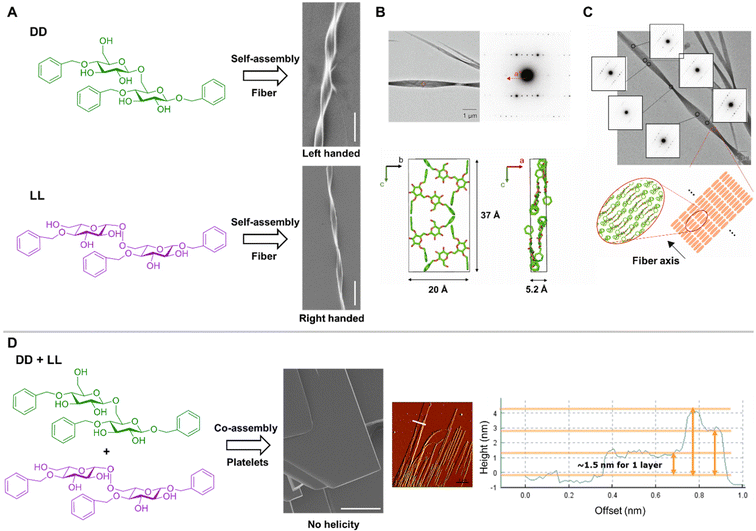 | ||
| Fig. 5 (A) Chemical structures of DD (green) and LL (purple) benzylated disaccharides and SEM images of their supramolecular assembly (scale bar: 10 µm). DD (green) and LL (purple) assemble into helical fibers with opposite chirality.64 (B) MicroED analysis of the assembled DD crystal with electron diffraction diagram obtained from the circled area. This allowed for a tentative molecular packing model of DD in the determined unit cell.64 (C) Sequential electron diffraction experiment along the fiber axis with a schematic image of the molecular packing manner in the fibrillar DD crystal.64 (D) Chemical structure of DD and LL and SEM-image of their supramolecular co-assembly (scale Bar: 10 µm). Co-assembly of the racemic mixture DD + LL generates a flat sheet-like structure. AFM image and cross-sectional analysis indicated a layer-bi-layer assembly.64 Figure was adapted from ref. 64, CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/). | ||
The combination of different techniques including optical microscopy, NMR spectroscopy, and microcrystal electron diffraction (microED) revealed the molecular organization within the assembly (Fig. 5B and C). The fibers exhibited high crystallinity with an orthorhombic unit cell (a = 5.2 Å, b = 20 Å, c = 37 Å) containing four DD molecules. The molecules adopted an overall flat conformation and stacked along the a axis with glucose ring planes oriented roughly in the bc plane and aromatic rings in close proximity to each other. The interactions between the aromatic rings were mostly CH-π type edge-to-face interactions, whereas no face-to-face π–π interactions occurred. The relatively low density of 1.06 g cm−3 implied the presence of water molecules in the crystal lattice, which may be involved in hydrogen bonding between molecules. Alongside the local molecular organization, microED-analysis afforded structural insights into the supramolecular organization. Sequential electron diffraction experiments along the fiber axis dissected the twist origin. In all diffraction patterns, the a*-axis was oriented along the fiber axis of the crystal indicating that stacking of flat molecular sheets occurred parallel to the fiber axis. The crystal therefore twisted along the stacking direction with a half helical pitch twist (180° rotation) of 5 µm, which resembled a rotation of 0.02 degree per unit cell.
In contrast, the co-assembly of DD and LL enantiomers (Fig. 5D) resulted in a different morphology lacking supramolecular chirality. The flat platelets were analyzed by atomic force microscopy (AFM), indicating a layer-bi-layer assembly with sheets height varying from few hundred nanometers to several micrometers. Each layer consisted of around 1.5 nm, which correlated with the dimension of a single disaccharide, suggesting that the enantiomers aligned laterally forming a single layer, as observed for some peptide assemblies.19 These results demonstrated that tuning the supramolecular chirality in carbohydrate systems through heterochiral co-assembly of enantiomers can give rise to assemblies with new morphologies and material properties, as commonly observed in peptide systems.18
A synthetic system to dissect cellulose chirality across scales
Well-defined glycan oligomers and their enantiomers were not only utilized to tune morphology and mechanical properties, but also to gain insights into the molecular basis of chirality transfers across scales in natural glycans (Fig. 6).46 | ||
| Fig. 6 (A) Chemical structures of D6 (green) and L6 (purple) cellulose hexasaccharide oligomers. (B) Cryo-TEM images of self-assembled platelets with no evident chiral features (scale bar: 200 nm) and 3D-model of a platelet composed of cellulose oligomers (D6) arranged in antiparallel fashion according to cellulose-II crystal structure.46 (C) TEM images of assembled bundles with opposite supramolecular chirality (scale bar: 500 nm). The fan-like arrangement of the stacking was interpreted as a rotation between the (001) planes.46 (D) Chemical structure of racemic mixture D6 + L6. Powder XRD profiles for assembled D6 (crystalline) and co-assembled D6 + L6 (amorphous) showing no defined assembly. Recrystallization regenerated platelets as indicated by powder XRD (D6 + L6a) and TEM (scale bar: 500 nm).46 Figure was adapted from ref. 46, CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/). | ||
Hexasaccharides representative of natural (D6) and enantiomeric (L6) cellulose self-assembled in aqueous solution to give thin platelets with thickness matching the length of a single oligomer chain (Fig. 6B). The oligomers arranged antiparallel to each other's (Cellulose-II type) along the thickness direction, as confirmed by electron diffraction. Crystal growth along the hydrophobic [110] direction was faster than along the hydrophilic [1−10] direction, resulting in elongated platelets.66 While platelets itself exhibited no obvious chiral features (e.g. a helical twist), upon drop casting, the platelets came together into twisted bundles (Fig. 6C). A right-handed twist was observed for D6, whereas bundles of L6 showed a left-handed twist. Thus, the bundles displayed an intrinsic chiral feature which directly connected to the monosaccharide composition. This behavior was also observed for synthetic chitin (oligomers of N-acetyl glucosamine) nanocrystals.67
The heterochiral co-assembly of D6 with L6 oligomers produced neither platelets nor bundles, suggesting that non-covalent interactions between the two enantiomers inhibited further assembly (Fig. 6D). A comparable phenomenon was noted in amyloid-like β-sheet peptide sequences, where strong co-aggregation between enantiomers prevented progression toward higher-order structures.68 In this case, non-covalent hydrophobic interactions between peptide enantiomers led to the formation of globular aggregates instead of the helical fibers characteristic of single enantiomers. Recrystallization of D6 + L6 racemic mixtures via a solvent switch method (DMSO to MeOH)46 ultimately allowed for the formation of crystalline platelets, as indicated by X-ray diffraction (XRD) and transmission electron microscopy (TEM). However, no further aggregation into higher-order assemblies with chiral features was observed. Thus, it appears that non-covalent interactions governing enantiomer co-assembly are as critical for carbohydrates as they are for peptides.
The role of chirality in cellulose aggregation was further investigated using CO-tipped Scanning-Tunneling-Microscopy (STM) imaging, a non-destructive imaging method capable of resolving atomic-level molecular structures on surface (Fig. 7).69 Separate deposition of D6 and L6 on an Au(111) surface using Electrospray Ion Beam Deposition (ESIBD) resulted in mirroring assemblies (Fig. 7A). In contrast, when the racemic mixture was deposited, the enantiomers self-sorted to form chiral assemblies (conglomerates), highlighting the enantiospecificity of chiral interactions between glucose oligomer enantiomers (Fig. 7B).
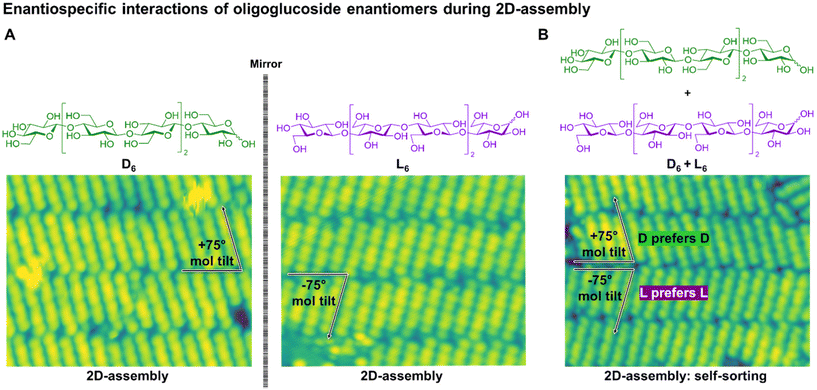 | ||
| Fig. 7 (A) Chemical structures of D6 (green) and L6 (purple). STM imaging of enantiospecific interaction of oligoglucoside enantiomers on Au(111) surface. D6 assembles with a +75° tilt along the assembly propagation axis whereas L6 assembles with a −75° tilt.69 (B) When both D6 and L6 were present on surface the enantiomers self-sorted producing conglomerates, i.e. domains that exclusively contain D6 or L6 due to preferential D–D and L–L interactions.69 Figure was adapted from ref. 69, CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/). | ||
This observation contrasted with the results from the co-assembly of the same oligomers in aqueous solution. If self-sorting had occurred in solution, one would expect the formation of platelets that subsequently organize into bundles with right- (D6) or left- (L6) handed twists. Instead, the formation of amorphous material lacking chiral features indicated that interactions between enantiomers hindered extended homochiral organization. Chiral interactions might therefore differ between 2D and 3D-assembly, influencing the formation of higher-order structures.12 Self-sorting during heterochiral co-assembly has been observed for short peptide sequences as well, both upon deposition on surface and in solution.58–60,70
Heterochiral co-assembly of cyclodextrins
Cyclodextrins (CDs) are cyclic oligosaccharides capable of binding guest molecules within their hydrophobic cavity in aqueous solution. These feature makes CDs attractive compounds in host–guest and supramolecular chemistry. Their inherent chirality paired with Cn-symmetry made cyclodextrins popular for chiral recognition, for separation technologies, as catalysts in chemical reactions, and as building blocks for the creation of chiral materials.65 In the past decades, CDs aggregation was extensively studied in solid-state.65,71–73While naturally occurring CDs offer only one set of enantiomers (i.e. D-Glc), chemical synthesis enriched the CD toolkit with the mirror image L-α-,β-,γ-cyclodextrins.65 Solid-state X-ray structure determination and Induced Electronic Circular Dichroism (IECD) spectroscopy, using host–guest complexes formed from complexation of an achiral guests (i.e. methylorange), confirmed the opposite chirality of D- and L-CDs. The availability of both L- and D-CDs prompted investigations into the behavior of β-CD racemic mixtures in the solid-state (Fig. 8).
 | ||
| Fig. 8 Solid-state structure of β-CD racemic mixture as stick representations. Carbon atoms are shown in green (β-D-CD) and purple (β-L-CD). Oxygen atoms are in shown in red, hydrogen atoms and solvent molecules have been omitted for clarity.65 (A) Solid-state supramolecular structure viewed along the a axis. β-D-CD and β-L-CD self-sort into layered assemblies with two adjacent layers composed of the same enantiomers.65 (B) Supramolecular structure viewed along the c axis. The molecular packing produces open channels associated with β-D and β-L-CDs.65 (C) Side-on view of a discrete stack of β-D-CD and β-L-CD showing an alternating DDLL pattern. Within the enantiomeric pairs, the two CD-molecules align in a head-to-head manner.65 Figure was adapted from ref. 65 Copyright 2024, The Authors (s), under exclusive license to Springer Nature Limited. | ||
Racemate containing equimolar amounts of both enantiomers packed into the centrosymmetric space group C2/c. Further assembly along the c axis was governed by a discrete stack of alternated DDLL pattern, in which the enantiomeric pairs CD molecules were aligned in a head-to-head manner. This organization allowed for favorable hydrogen bonding interactions between the two secondary faces of CDs with identical chirality. Along the a axis, self-sorted assemblies composed of same enantiomers were formed. The stacking of CDs along the c axis lead to open channels, with the D–L interface held together via a hydrogen bond network mediated by multiple H2O molecules. This solid-state structural analysis of racemic β-CD demonstrated that chiral interactions govern the assembly of cyclic oligosaccharides, suggesting opportunities for the creation of supramolecular materials with tailored networks and patterns.74–76
Racemic crystallization
The preferential interaction of racemic molecules was not only beneficial for generating new assemblies, but also for structural analysis via X-ray crystallography. In general, obtaining an X-ray crystal structure from a single enantiomer of complex molecules, such as proteins, presents challenges due to the difficulty of achieving: (i) a highly ordered protein crystal and (ii) the phase information necessary to solve its structure. Access to the unnatural D-proteins enabled the analysis of racemic mixtures, which facilitated crystallization (as predicted by Wukovitz and Yeates in 1995)77 and simplified the phase problem (as postulated by Mackay in 1989).78Protein racemic mixtures can crystallize in centrosymmetric space groups, such as P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , which restricts all possible phases to 0 or 180° degrees. In contrast, single enantiomers cannot form centrosymmetric crystals and can only crystallize in chiral space groups, allowing phases to take on any values from 0 to 360°. For a more detailed background on protein crystallography we recommend the detailed review by Yeates and Kent.79
, which restricts all possible phases to 0 or 180° degrees. In contrast, single enantiomers cannot form centrosymmetric crystals and can only crystallize in chiral space groups, allowing phases to take on any values from 0 to 360°. For a more detailed background on protein crystallography we recommend the detailed review by Yeates and Kent.79
Racemic crystallization has enabled structure elucidation of various difficult-to-crystallize proteins.30 This approach can also be applied to enantiomorphs, which are near mirror-image molecules, but not true enantiomers. This principle is commonly referred to as quasi-racemic crystallization, and offered a powerful tool for elucidating the structures of series of L-protein analogues that exhibit small structural differences, utilizing a single D-protein.30
Quasi-racemic crystallization was employed to obtain the crystal structure of the glycosylated chemokine L-Ser-CCL1 with a non-glycosylated D-Ser-CCL1 protein (Fig. 9).80 Glycosylation, one of the most frequent post-translational modification, plays a crucial role in the folding and function of proteins. However, structural determination of glycoproteins is particularly challenging due to their inherent structural complexity, heterogenicity and flexibility of the conjugated glycan moieties, which can hinder crystallization.81
 | ||
| Fig. 9 Quasi racemic crystallography of glycosylated L-Ser-CCL1 and D-Ser-CCL1. (A) Amino acid sequence of chemokine Ser-CCL1 and chemical structure of N-linked glycan attached at residue Asn29.80 (B) Unit cell of the quasi-racemate in space group P1 with protein molecules in ribbon representation.80 (C) Ribbon representation of glycosylated L-Se-CCL1 (purple) and non-glycosylated D-Ser-CCL1 (green). The inset shows 2 Fo – Fc electron density map of the glycosylated Asn29-Ser31 consensus sequence (contoured at a σ level of 1.0).80 Figure was adapted from ref. 80. Copyright 2014, Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
While crystals were not obtained from the glycosylated L-Ser-CCL1, they readily formed through equimolar crystallization with the non-glycosylated D-Ser-CCL1. With crystals in hand, the structure was solved via X-ray diffraction at a resolution of 2.6–2.1 Å, revealing a space group P1 forming a pseudo-centrosymmetrical arrangement and with all structural features of Ser-CCL1 identified. The interactions between glycosylated L-protein and non-glycosylated D-protein were similar to those observed in the previously resolved racemic X-ray crystal structure of non-glycosylated L- and D-protein, suggesting that, in this particular example, glycosylation did not affect the structure of the protein.80 In this experiment, the complete conformation of the glycan was not fully resolved due to its flexibility. The authors hypothesized that racemic crystallization of the natural D-glycan-L-protein and the unnatural L-glycan-D-protein might facilitate a comprehensive structure determination of the glycoprotein.
While racemic crystallization has achieved substantial success in the structural analysis of peptides and proteins, it has yet to be effectively applied to glycans. Studies on simple racemic mixtures of monosaccharides demonstrated the formation of stable racemic crystals.82 However, progress in this area has likely been prevented by several bottlenecks. Glycan synthesis is complex and time-consuming, especially when involving non-natural enantiomers. Automated synthetic platforms are not as available as those for peptide synthesis. Furthermore, glycan crystallography is less developed compared to protein crystallography. The intrinsic flexibility of certain glycans complicates their crystallization. Moreover, the radiation sensitivity of glycans, along with a limited understanding of their crystallization processes, has often hindered in-depth investigations of carbohydrates through crystallographic methods.83
Nevertheless, significant practical knowledge has been accumulated over the past decades, improving the structural analysis of glycans.83 As demonstrated in the previous section, preferential interactions between enantiomeric glycans are indeed possible. With the advent of new synthetic platforms that standardize the synthesis of complex non-natural glycans,84,85 we speculate that racemic crystallization could emerge as an attractive technique for enhancing our understanding of glycan structures.
Assembly of heterochiral molecules
Assembly of heterochiral peptides
The construction of higher-order structures is highly dependent on the availability and directionality of non-covalent interactions. As discussed in the previous chapter, well-defined chiral synthetic peptide model systems have provided valuable insights into the stereochemistry-directed co-assembly of enantiomers, underscoring the importance of geometric complementarity.13–24,26,30,58,59 Fundamentally, the self-sorting or co-assembling of peptide strands is driven by the need to maximize backbone hydrogen bonding while minimizing steric hindrance between side chains.25 Opposite chirality can enhance steric complementarity between sequences, thereby affecting the directionality of non-covalent interactions that govern self-assembly. For example, the co-assembly of opposing peptide enantiomers promoted the formation of a rippled β-sheet structure due to the alternating packing of these enantiomers. This structural motif resembles a conventional pleated β-sheet structure, as both are stabilized through either parallel or antiparallel backbone hydrogen bonding. However, in the rippled structure, the side chains of neighboring strands align in a staggered arrangement, leading to the formation of morphologically flat fibril-like assemblies instead of left- or right-handed helical fibrils.13,16,24,26 This configuration provides additional stability to the motif through minimized steric hindrance between side chains and favorable hydrophobic interactions.13,16,24,26An alternative approach to modulate these interactions is to incorporate both enantiomers into the same primary sequence.24,27–29,86–92 This strategy allows for the tuning of steric complementarity and non-covalent interactions, thereby enabling better control over the assembly process.
The impact of block chirality on peptide self-assembly was investigated on the extensively studied amphiphatic Ac-(FKFE)2-NH2 peptide (Fig. 10A).27 As previously mentioned, the co-assembly of amphiphatic L- and D-Ac-(KEFE)2-NH2 peptides triggered the formation of a rippled β-sheet further aggregating into a fibrillar hydrogel with enhanced material properties.13,15 Substituting the FKFE block by its D-enantiomer gave the heterochiral analogues Ac-FKFEfkfe-NH2 (LD) and Ac-fkfeFKFE-NH2 (DL). TEM indicated that, upon heterochiral assembly, both peptides assembled into supramolecular helices with average width and pitch values of ≈108 ± 55 nm (width), ≈900–1200 nm (pitch). This morphology starkly contrasted with the observed helical fibrils formed upon homochiral assembly (LL: ≈8.1 ± 0.3 nm width, ≈19 nm pitch; DD: ≈7.9 ± 0.3 nm width), or the flat nanoribbons formed during the heterochiral co-assembly of homochiral peptides (LL + DD:≈3.0 ± 0.4 nm width).13,27 The resulting hydrogels exhibited distinct rheological properties associated with their morphological differences at both the molecular and supramolecular levels.
 | ||
| Fig. 10 (A) Top: Stick representation of homochiral Ac-(FKFEFKFE)-NH2 peptide (capital letter indicates L-configuration). Self-assembled helical fibrils composed of a bilayer of antiparallel β-sheets. Bottom: Stick representation of heterochiral Ac-(FKFEfkfe)-NH2 peptide (capital letter indicates L-configuration; lowercase letter indicates D-configuration). Self-assembled helical tapes composed of a monolayer of antiparallel β-sheets. Negative-stain TEM images of self-assembled peptides Ac-(FKFEFKFE)-NH2 and Ac-(FKFEfkfe)-NH2 at 0.25 mM in water (scale bar: 250 nm).27 (B) Stick representation of heterochiral Ac-(LlLlK)-NH2 peptide (capital letter indicates L-configuration; lowercase letter indicates D-configuration). Self-assembled nanotubes composed of a monolayer of α-sheets. Cryo-TEM images of assembled Ac-(LlLlK)-NH2 (scale bar: 200 nm).28 (C) Relationship between conformation of a β-pleated strand and that of a β-helix.93 Top to bottom: (1) homochiral idealized pleated conformation with Φ = −120° and Ψ = 120° viewed parallel to the plane of the backbone.93 (2) Inversion of chirality of the marked purple residue from L to D. Inverting chirality of a single amino acid in a pleated strand causes a change of local geometry (Φ = 120° and Ψ = −120°) introducing a kink in the peptide backbone.93 (3) Continuing this process every second residue causes a continuous bend, introducing a pleated helical conformation.93 (4) Stick and ribbon representation of solid-state NMR-derived single-stranded helical conformation of gramicidin A (HCO-Val-Gly-Ala-leu-Ala-val-Val-Val-Trp-leu-Trp-leu-Trp-leu-Trp-NHCH2CH2OH) in hydrated lipid bilayers, stabilized through β-sheet type hydrogen bonding. D-Amino acids are colored purple, L-amino acids green (PDB 1MAG).94 (D) Chemical structure of cyclic peptide CPx (capital letter and green color indicates L-configuration; lowercase letter and purple color indicates D-configuration). Self-assembled amphiphilic nanotubes that further undergo 2D-assembly into nanosheets composed of nanotube bilayers with hydrophobic cores, held together by Leu zippers (yellow) and Trp stacks (gray). Polar residues on are placed on the surface.29 Part A was adapted from ref. 27. This is an unofficial adaptation of an article that appeared in an ACS publication. ACS has not endorsed the content of this adaptation or the context of its use. Copyright 2020, American Chemical Society.27 Part B was adapted from ref. 28. Copyright 2022, American Chemical Society.28 Part D was adapted from ref. 29. Copyright 2020, American Chemical Society.29 | ||
Spectroscopic and scattering measurements (CD, FT-IR, SAXS and WAXS), combined with theoretical simulations (MD), suggested that the heterochiral peptide strands formed a monolayer of β-sheets stabilized by antiparallel hydrogen bonding. In contrast, the homochiral peptide strands formed a putative bilayer of antiparallel β-sheets, due to the nonpolar phenylalanine side chains that defined a hydrophobic surface.13 The two enantiomeric blocks within the heterochiral peptide caused a structural rearrangement of the peptide backbone and the side chains, causing a different assembly behavior compared to homochiral sequences.27 The influence of block chirality was further investigated increasing the sequence length from 8 to 12 amino acids of (Ac-(FKFE)n-NH2).89
Similarly, the self- and co-assembly behavior of aromatic FFFK (F3K) peptides, including F3K (LL), f3k (DD), F3k (LD), f3K (DL), provided valuable insights on the interplay of aromatic π–π-stacking and steric complementarity.24,91 Overall, these examples confirmed that patterns of L- and D-enantiomers into peptide primary sequences gave rise to unique intermolecular interactions that direct assembly towards novel higher-order structures.
Heterochiral peptides to access unusual secondary structures
The strategic positioning of D- and L-amino acids within a peptide sequence has enabled the formation of unusual secondary structures, such as π-helices and α-pleated sheets. These structures are less stable compared to conventional secondary structures (e.g., β-pleated sheets and α-helices), making them much rarer in native proteins and peptides. They are often regarded as transient conformations that occur during the folding processes of proteins and peptides.28The α-pleated sheet was first predicted by Pauling and Corey, showing that, similar to β-sheets, this secondary structure is stabilized by regular hydrogen bonding between adjacent strands in the “α-extended chain” conformation.95 In this arrangement, each residue is defined as locally helical in the right-handed (αR) and left-handed regions (αL); however, the alternation causes the formation of an extended chain.96
To facilitate the formation of α-pleated sheet secondary structures, the alternation of L- and D-amino acids within a single sequence has been explored.97–99 Two engineered heterochiral amphiphilic peptide sequences Ac-LlLlK-NH2 and Ac-lLlLk-NH2 organized into an α-pleated sheet secondary structure that served as a foundation for helical ribbons and nanotubes, with widths of 150–200 nm and a monolayer thickness of 2.1 nm (Fig. 10B).28 When compared to homochiral analogs (Ac-LLLLK-NH2), the heterochiral pentapeptides exhibited a greater degree of molecular ordering within their supramolecular structures, attributable to enhanced steric complementarity.
Linear peptides composed of alternating D- and L-amino acids were also utilized to access β-helical conformations. In this arrangement, helix stabilization occurs through regular hydrogen bonding between adjacent strands, resembling the pleated β-sheet secondary structure (Fig. 10C). In an idealized homochiral pleated β-strand, the main chain dihedral angles between adjacent amino acids result in a sinusoidal backbone geometry that positions α-carbons alternately above and below the backbone plane. Since enantiomers exhibit opposite but equivalent conformational properties, inverting the chirality of a single amino acid in a pleated strand causes a change of local geometry, introducing a bend in the peptide backbone. Thus, the D-enantiomer reinforces the bending direction of the adjacent L-enantiomers. Continuing this process for every second residue causes a continuous bend, generating the pleated helical conformation, stabilized through β-sheet-type hydrogen bonding. In this arrangement, the side chains radiate toward the outside, minimizing unfavorable steric interactions. Thus, the helix displays a screw symmetry axis with a dipeptide containing both enantiomers as repeating unit.93 This design has facilitated the creation of transmembrane pores, with hydrophilic interior acting as a cation channel and hydrophobic side chains helping to anchor the channel in the lipid membrane.100–102
Heterochiral cyclic peptides
Cyclic peptide sequences with an even number of alternating D- and L-amino acids effectively stack through β-type intermolecular hydrogen bonding.103 The resulting cylindrical supramolecular structure is conceptually related to the β-helix, but allows for higher control and predictability in assembly. Furthermore, the hollow tubular structure exhibits a constant cavity diameter that can be readily tuned with the size of the peptide ring.93Cyclic peptide nanotubes were created from the assembly of the octapeptide cyclo[-(ala-Glu-ala-Gln)2], exploiting the ionization state of glutamic acid side chains to trigger self-assembly in aqueous solution. Under basic conditions, the negatively charged carboxylate groups resulted in strong intermolecular electrostatic repulsion, which inhibited stacking and thus maintained solubility. Controlled acidification decreased this repulsion, facilitating ordered self-assembly into nanotubes.92
Hierarchical 1D-to-2D self-assembly was achieved by encoding hydrophilic (Glu-his-Gln-his-Glu) and hydrophobic (leu-Trp-leu) domains within the cyclic octapeptide CPx (Fig. 10D).29 Upon annealing, the cyclic CPx stacked via parallel β-sheet type hydrogen bonding and π–π stacking (between tryptophan units) to give amphiphilic nanotubes (1D). The subsequent assembly process towards defined 2D-nanosheets was the result of attractive (H-bonds, hydrophobic interactions) and repulsive (electrostatic) forces modulated by the presence of glutamic acids.
Self-assembled cyclic peptide nanotubes and their higher order assemblies have shown potential in various fields including size-selective ion channels, molecular switches, tubular composites, antibiotics, charge transport systems, and drug delivery systems.29
Assembly of heterochiral glycans
In analogy to peptides, modulating the pattern of L- and D-monosaccharides within a glycan sequence offers an important design mode to direct assembly.46–50As discussed in the previous chapter, hexasaccharides representative of natural (D6) and enantiomeric (L6) cellulose self-assembled in aqueous solution to give 2D-like crystalline platelets. These platelets further stacked via the (001) surface into helical bundles with a right- (D6) and left- (L6) handed twist. Since the individual platelet did not display evident chiral features, these results suggested that the chirality of the monosaccharide exposed on the (001) surface determined the direction of the supramolecular twist. To verify this hypothesis, well-defined heterochiral sequences with different chiral patterns were prepared. Five oligomers were synthesized to selectively modulate the chirality at the termini of each sequence and investigate the determinants of chirality across scales.46 Two groups of compounds were designed based on the known antiparallel arrangement of oligosaccharide sequences within the crystals. Group 1 (LD6, L2D4, L3D3) displayed alternating D- and L-Glc residues at the (001) surface, while Group 2 (LD5L and LD6L) displayed only L-Glc residues at the (001) surface, preserving the crystal core composed of D-Glc units.
LD6 preserved the cellulose II type of assembly, yielding thin crystalline platelets further ordering into bundles. However, the introduction of L-Glc resulted in lower twisting tendency and no clear handedness (Fig. 11A). With increasing number of L-Glc units, such as in L2D4, L3D3, crystallinity was lost and solubility drastically increased. When L-Glc was positioned at both termini of the oligosaccharide sequence, such as in LD5L and LD6L (Group 2), crystalline platelets further aggregating into bundles were detected (Fig. 11B). As observed for LD6, the bundles exhibited attenuated chiral features and, in some cases, left-handed handedness. These experiments demonstrated that perturbing the molecular nature of the (001) surface of cellulose II platelets drastically affects the assembly behavior, weakening the twisting tendency. However, bundles of LD6L-platelets (displaying only L-Glc on their surface) did not display complete inversion of chirality, indicating that the core sequence also contributes to the overall supramolecular twist.
 | ||
| Fig. 11 (A) Chemical structure of LD6 (D-Glc in green, L-Glc in purple). Simplified model of LD6 platelets arranged in antiparallel fashion according to the cellulose-II type of packing and top view of the (001) face. TEM images of LD6 bundles (aqueous suspension) showing a left-handed twisted morphology (red arrows, scale bars 500 nm).46 (B) Simplified model of LD6L platelets arranged in antiparallel fashion according to the cellulose-II type of packing and top view of the (001) face. TEM images of LD6L bundles (MeOH suspension) showing a twisted morphology (red arrows, scale bars 500 nm).46 (C) Chemical structure of D6. Schematic representation of a 2D-crystal composed of D6 showing the cellulose II type of alignment within the crystal. Cryo-TEM analysis of D6 assemblies (scale bar: 200 nm).47 (D) Chemical structure of D3LD4. Schematic representation of a 3D crystal composed of D3LD4 and the cellulose IVII type of alignment within the crystal. SEM and TEM analysis of D3LD4 assemblies (scale bar: 500 nm, 1 µm).47 Part A, B and C was adapted from ref. 46 CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Part C and D was adapted from ref. 47, CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/). | ||
To further examine the self-assembly behavior of heterochiral cellulose-like sequences, a single L-Glc residue was placed within a D-Glc oligomer (D3LD4). This modification triggered the assembly into square-like platelets based on the rare cellulose IVII allomorph (Fig. 11D).47,104 The electron diffraction-pattern of D3LD4 confirmed the antiparallel arrangement of oligosaccharides and larger intermolecular spacing compared to the most common cellulose II allomorph. A higher resolution, approaching 1 Å, surpassed previous electron diffraction patterns obtained from natural cellulose IVII crystals, indicating improved crystalline ordering of molecules resulting from the insertion of L-Glc.47 The crystallinity was further enhanced upon enzyme triggered assembly of the same heterochiral sequence, achieving resolutions beyond 1 Å.105 These results suggest that engineered heterochiral glycan sequences could lead to the formation of rare structures, similar to what observed for peptides. This approach opens new avenues for the structural investigation of lesser-known cellulose allomorphs and, more broadly, glycan structures at atomic resolution.
The on surface self-assembly behavior of the heterochiral cellulose sequences was also investigated using CO-tipped Scanning-Tunneling-microscopy (STM)-Imaging.69 On surface, conglomerate formation was observed for the racemic mixture of D6 and L6 indicating enantiospecific interactions between glucose enantiomers on surface (Fig. 7). These highly enantiospecific D–D and L–L interactions were also detected upon deposition of the heterochiral L2D4-sequence (Fig. 12). Two sets of glucose subunits were identified within the assembly: the L-Glc domains preferentially bound to L-Glc subunits of neighboring molecules, while the D-Glc domains exhibited a similar preference for D-Glc subunits. This specific interaction, not observed in solution phase, led to the formation of patterned chiral surfaces and could inspire the generation of well-defined chiral surfaces.
 | ||
| Fig. 12 Chemical structure of L2D4 (D-Glc in green, L-Glc in purple). STM imaging of enantiospecific interactions of glucose enantiomers on Au(111) surface. The preferential D–D (green) and L–L (purple) interactions was observed for heterochiral sequences.69 Figure was adapted from ref. 69, CC-BY 4.0 (https://creativecommons.org/licenses/by/4.0/). | ||
Overall, the discussed examples of assemblies of well-defined heterochiral glycan sequences suggest that, similar to peptides, a careful combination of D- and L-monosaccharides serves as a valuable design tool for accessing glycan materials with intriguing and unusual morphologies.
Heterochiral cyclic glycans
Similar to peptides, the alternation of D- and L-residues facilitates the generation of cyclic structures. Heterochiral cyclic oligomers composed of alternating D- and L-monosaccharides have been utilized to craft cyclodextrin analogues which effectively stack forming nanotubes with potential applications in host–guest chemistry.48–50 This motif closely resembles the nanotubes formed by cyclic peptide systems composed of alternating L- and D-amino acids. A polycondensation/cyclization approach using disaccharide precursors enabled the synthesis of a range of highly symmetric α-1,4-linked cyclic oligosaccharides composed of alternating units of L-Rha and D-Man (rM), L- and D-Rha (rR) and L- and D-Man (mM). NMR spectroscopy confirmed the formation of highly symmetric macrocycles with identical faces.X-ray crystallographic analysis of alternated rR-octasaccharide revealed a C2-symmetric macrocycle in solid state, departing from ideal S8 symmetry (Fig. 13A). No intramolecular hydrogen bonding was found, a feature known to stabilize the toroidal conformation in natural CDs. The absence of a molecular rotation-reflection symmetry (Sn) and the slight folding of the macrocycle plane resulted in two diastereotopic faces. Consequently, the molecules ordered in a head-to-tail manner, forming discrete stacks. In contrast to cyclic peptide nanotubes, no intermolecular hydrogen bonding within the stacks was observed; instead, adjacent stacks were stabilized by hydrogen bonds. Each stack of molecules formed large open channels.50
 | ||
| Fig. 13 (A) Chemical structure of D-Rha and L-Rha. Solid-state structure of C2-symmetric cyclic octasaccharide rR in stick representation (CCDC 1221815). Solid-state supramolecular structure viewed along the b axis. Cyclic octasaccharide rR order in a near close-packed hexagonal array. Interstack hydrogen bonds are omitted. The yellow and red cross symbols show the respective polarities of the stacks. Molecules order head-to-tail within the stacks.50 (B) Chemical structure of D-Man and L-Man. Solid-state structure of Ci-symmetric cyclic hexasaccharide mM (CCDC 1221816). Solid-state supramolecular structure of cyclic hexasaccharide mM. Molecules pack parquet-like. Intra- and intermolecular hydrogen bonds are omitted.50 | ||
Such hollow tubular structures were also observed for the alternated rR-decasaccharide and rM-octasaccharide. In contrast, the mM-hexasaccharide macrocycle crystallized into an interlocked parquet-like pattern, forming no channels (Fig. 13B). The system was stabilized through intramolecular and intermolecular hydrogen bonding.50 These findings demonstrate that the assembly behavior of cyclic heterochiral oligosaccharides is highly dependent on monosaccharide composition. Manipulating the macrocycle composition could lead to the development of useful glycan-based supramolecular architectures.
Conclusions and outlook
The strategic combination of D- and L-amino acids – whether incorporated within the same peptide sequence or organized as co-assemblies of homochiral peptides – allowed access to unusual architectures, difficult to achieve with homochiral sequences. These structures exhibit intriguing properties, including enhanced stability in biological environments and superior mechanical performances.The few examples of chiral carbohydrate assemblies formed by the integration of D- and L-monosaccharides described here indicate a similar potential for crafting glycan-based programmable supramolecular structures.46–50,64,65 We argue that this approach could reveal many more exciting opportunities, especially as new sequence designs come into play. Still, the systematic exploitation of chirality in glycan material development remains largely untapped, likely due to the complexity of glycan synthesis and structural analysis. Recent advancements in automated glycan synthesis84,85 and controlled polymerization techniques106 are enabling access to a broader variety of glycans than ever before. Relatively long sequences (up to 1000 units)107,108 can now be accessed routinely, maintaining a full control over the primary sequence and allowing of rare and non-canonical monosaccharide units. Currently, most examples of heterochiral glycan sequences and assemblies are based on combinations of D- and L-Glc. While access to non-natural enantiomers, like L-Glc, is usually associated with larger financial costs, a few synthetic methods exist for the synthesis of L-Glc and progress in this area remains crucial for larger scale usage.109,110 The systematic screening of glycan sequences based on other monosaccharides and their enantiomers could dramatically expand the geometrical diversity of glycan architectures. In this context, rare non-natural enantiomers like L-Man or L-Gal could be replaced with structurally similar, naturally occurring enantiomers such as L-Rha or L-Fuc to create innovative frameworks and reduce costs.
Analytical techniques – including scanning tunneling microscopy (STM),69 non-contact atomic force microscopy (nc-AFM),111 and cryo-electron microscopy (cryo-EM)83 – are emerging, enabling deeper insights into structural details of glycan assemblies.
The creation of tunable chiral glycans could open up numerous opportunities. For example, glycan chirality could be exploited for chiral recognition. Recently, D- and L-Glc were effectively utilized as chiral selectors in high-performance liquid chromatography (HPLC), inverting the elution order of chiral substrates.112 New heterochiral polymers could be engineered as matrices for separation technologies. Naturally occurring CDs and their derivatives are highly versatile compounds that found applications in host–guest and supramolecular chemistry, formulation and catalysis.74–76,113–119 The recent synthesis of L-α,β and γ-cyclodextrins allow access to systems with opposite chirality;65 heterochiral CD analogues, made from both D- and L-monosaccharides with varying lengths and geometries, could significantly broaden these portfolios, expanding options for stereoselective binding and encapsulation.
Heterochiral polysaccharides (e.g. GAGs or alginates) are involved in a variety of biological processes. However, the synthesis of well-defined, naturally occurring heterochiral polysaccharides remains complex.120 Heterochiral sequences that mimic some structural features of these natural glycans could inspire a new class of biopolymers that could complement existing natural examples.
Beyond material design, the preferential interaction between enantiomers may be exploited to expand our understanding of glycan structures. Racemic crystallization has obtained considerable success in elucidating the structure of difficult-to-crystallize peptides and proteins. A similar technique could reveal details of glycans secondary structure and aggregation, paving the way for further discoveries in the glycoscience.
Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting the findings of this review are derived from previously published literature and are openly available. All relevant data, figures, and references cited in this manuscript can be accessed through the respective journals and databases. No primary data from the authors’ experiments are included in this review.Acknowledgements
We thank the Max Planck Society, the German Federal Ministry of Education and Research (BMBF, grant number 13XP5114), and the European Research Council (ERC) under the Horizon Europe research and innovation programme (Project 101075357—GLYCOFOLD) for generous financial support. Open Access funding provided by the Max Planck Society.References
- C. J. C. Edwards-Gayle and I. W. Hamley, Org. Biomol. Chem., 2017, 15, 5867–5876 Search PubMed.
- B. Frka-Petesic, T. G. Parton, C. Honorato-Rios, A. Narkevicius, K. Ballu, Q. Shen, Z. Lu, Y. Ogawa, J. S. Haataja, B. E. Droguet, R. M. Parker and S. Vignolini, Chem. Rev., 2023, 123, 12595–12756 Search PubMed.
- R. Noyori, Angew. Chem., Int. Ed., 2002, 41, 2008–2022 Search PubMed.
- Z. A. VanOrman, W. R. Kitzmann, A.-P. M. Reponen, T. Deshpande, H. J. Jöbsis and S. Feldmann, Nat. Rev. Chem., 2025, 9, 208–223 Search PubMed.
- J. R. Brandt, F. Salerno and M. J. Fuchter, Nat. Rev. Chem., 2017, 1, 0045 Search PubMed.
- X. Zhang, Y. Xu, C. Valenzuela, X. Zhang, L. Wang, W. Feng and Q. Li, Light:Sci. Appl., 2022, 11, 223 Search PubMed.
- B. Pokroy, S. H. Kang, L. Mahadevan and J. Aizenberg, Science, 2009, 323, 237–240 Search PubMed.
- Z. Yang, S. K. Yorke, T. P. J. Knowles and M. J. Buehler, Sci. Adv., 2025, 11, eadv1971 Search PubMed.
- G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 Search PubMed.
- S. M. Morrow, A. J. Bissette and S. P. Fletcher, Nat. Nanotechnol., 2017, 12, 410–419 Search PubMed.
- E. Grelet and M. M. C. Tortora, Nat. Mater., 2024, 23, 1276–1282 Search PubMed.
- S. Dutta and A. J. Gellman, Chem. Soc. Rev., 2017, 46, 7787–7839 Search PubMed.
- R. J. Swanekamp, J. T. M. DiMaio, C. J. Bowerman and B. L. Nilsson, J. Am. Chem. Soc., 2012, 134, 5556–5559 Search PubMed.
- V. P. Gray and R. A. Letteri, Biomacromolecules, 2024, 25, 5273–5280 Search PubMed.
- R. J. Swanekamp, J. J. Welch and B. L. Nilsson, Chem. Commun., 2014, 50, 10133–10136 Search PubMed.
- K. J. Nagy, M. C. Giano, A. Jin, D. J. Pochan and J. P. Schneider, J. Am. Chem. Soc., 2011, 133, 14975–14977 Search PubMed.
- K. Nagy-Smith, P. J. Beltramo, E. Moore, R. Tycko, E. M. Furst and J. P. Schneider, ACS Cent. Sci., 2017, 3, 586–597 Search PubMed.
- I. J. Duti, J. R. Florian, A. R. Kittel, C. D. Amelung, V. P. Gray, K. J. Lampe and R. A. Letteri, J. Am. Chem. Soc., 2023, 145, 18468–18476 Search PubMed.
- F. Xu, I. J. Khan, K. McGuinness, A. S. Parmar, T. Silva, N. S. Murthy and V. Nanda, J. Am. Chem. Soc., 2013, 135, 18762–18765 Search PubMed.
- D. F. Kreitler, Z. Yao, J. D. Steinkruger, D. E. Mortenson, L. Huang, R. Mittal, B. R. Travis, K. T. Forest and S. H. Gellman, J. Am. Chem. Soc., 2019, 141, 1583–1592 Search PubMed.
- A. J. Kuhn, B. Ehlke, T. C. Johnstone, S. R. J. Oliver and J. A. Raskatov, Chem. Sci., 2022, 13, 671–680 Search PubMed.
- A. Hazari, M. R. Sawaya, M. Sajimon, N. Vlahakis, J. Rodriguez, D. Eisenberg and J. A. Raskatov, J. Am. Chem. Soc., 2023, 145, 25917–25926 Search PubMed.
- A. Hazari, M. R. Sawaya, H. Lee, M. Sajimon, H. Kim, W. A. Goddard Iii, D. Eisenberg and J. A. Raskatov, Chem. Sci., 2025, 16, 5907–5917 Search PubMed.
- H. Qi, K. Qi, Y. Wang, X. Ju, Y. Han, S. M. King, S. E. Rogers, Y. Wang, F. Zhou, J. R. Lu, H. Xu and X. Hu, J. Am. Chem. Soc., 2025, 147, 14231–14243 Search PubMed.
- H. Lee, A. Hazari, J. A. Raskatov, H. Kim and W. A. Goddard, III, J. Am. Chem. Soc., 2025, 147, 17642–17650 Search PubMed.
- T. D. Samdin, J. Lubkowski, C. F. Anderson and J. P. Schneider, J. Am. Chem. Soc., 2025, 147, 14377–14387 Search PubMed.
- T. M. Clover, C. L. O'Neill, R. Appavu, G. Lokhande, A. K. Gaharwar, A. E. Posey, M. A. White and J. S. Rudra, J. Am. Chem. Soc., 2020, 142, 19809–19813 Search PubMed.
- P. Zhou, X. Hu, J. Li, Y. Wang, H. Yu, Z. Chen, D. Wang, Y. Zhao, S. M. King, S. E. Rogers, J. Wang, J. R. Lu and H. Xu, J. Am. Chem. Soc., 2022, 144, 21544–21554 Search PubMed.
- I. Insua and J. Montenegro, J. Am. Chem. Soc., 2020, 142, 300–307 Search PubMed.
- K. Harrison, A. S. Mackay, L. Kambanis, J. W. C. Maxwell and R. J. Payne, Nat. Rev. Chem., 2023, 7, 383–404 Search PubMed.
- S. Djalali, N. Yadav and M. Delbianco, Nat. Rev. Mater., 2024, 9, 190–201 Search PubMed.
- A. Varki, R. D. Cummings, J. D. Esko, P. Stanley, G. W. Hart, M. Aebi, D. Mohnen, T. Kinoshita, N. H. Packer, J. H. Prestegard, R. L. Schnaar and P. H. Seeberger, Essentials of Glycobiology, Cold Spring Harbor Laboratory Press, Cold Spring Harbor (NY), 2022 Search PubMed.
- F. Di Lorenzo, K. A. Duda, R. Lanzetta, A. Silipo, C. De Castro and A. Molinaro, Chem. Rev., 2022, 122, 15767–15821 Search PubMed.
- A. Adibekian, P. Stallforth, M.-L. Hecht, D. B. Werz, P. Gagneux and P. H. Seeberger, Chem. Sci., 2011, 2, 337–344 Search PubMed.
- R. Derda, K. F. Aoki-Kinoshita, C. S. Bennett, C. R. Bertozzi, D. Bojar, C. De Castro, T. Feizi, E. D. Goddard-Borger, A. Imperty, B. Imperiali, J. Jiménez-Barbero, D. L. Kasper, L. L. Kiessling, R. A. Laine, N. E. Lewis, F. Lisacek, Y. Liu, T. L. Lowary, M. S. Macauley, L. K. Mahal, A. Molinaro, N. H. Packer, J. C. Paulson, R. J. Payne, M. R. Pratt, C. Rademacher, P. H. Seeberger, S. J. Williams, R. J. Woods and I. Yamada, Science, 2024, 386, 1351–1353 Search PubMed.
- J. D. Martínez, R. Núñez-Franco, P. Valverde, S. Delgado, A. Ardá, J. Jiménez-Barbero, G. Jiménez-Oses and F. J. Cañada, Chem. – Eur. J., 2025, 31, e202501420 Search PubMed.
- E. J. Carpenter, C. Peng, S.-K. Wang, R. Greiner and R. Derda, bioRxiv, 2025, preprint, DOI:10.1101/2025.01.21.633632.
- A. Varki, R. D. Cummings, M. Aebi, N. H. Packer, P. H. Seeberger, J. D. Esko, P. Stanley, G. Hart, A. Darvill, T. Kinoshita, J. J. Prestegard, R. L. Schnaar, H. H. Freeze, J. D. Marth, C. R. Bertozzi, M. E. Etzler, M. Frank, J. F. Vliegenthart, T. Lütteke, S. Perez, E. Bolton, P. Rudd, J. Paulson, M. Kanehisa, P. Toukach, K. F. Aoki-Kinoshita, A. Dell, H. Narimatsu, W. York, N. Taniguchi and S. Kornfeld, Glycobiology, 2015, 25, 1323–1324 Search PubMed.
- M. A. Thelin, B. Bartolini, J. Axelsson, R. Gustafsson, E. Tykesson, E. Pera, Å. Oldberg, M. Maccarana and A. Malmstrom, FEBS J., 2013, 280, 2431–2446 Search PubMed.
- A. Onishi, K. S. Ange, J. S. Dordick and R. J. Linhardt, Front. Biosci.-Landmark, 2016, 21, 1372–1392 Search PubMed.
- K. Y. Lee and D. J. Mooney, Prog. Polym. Sci., 2012, 37, 106–126 Search PubMed.
- M. Lahaye and C. Rochas, Hydrobiologia, 1991, 221, 137–148 Search PubMed.
- J. M. Trowbridge and R. L. Gallo, Glycobiology, 2002, 12, 117R–125R Search PubMed.
- H. Ertesvåg, Front. Microbiol., 2015, 6, 523 Search PubMed.
- D. Ji, J. M. Park, M. S. Oh, T. L. Nguyen, H. Shin, J. S. Kim, D. Kim, H. S. Park and J. Kim, Nat. Commun., 2022, 13, 3019 Search PubMed.
- G. Fittolani, D. Vargová, P. H. Seeberger, Y. Ogawa and M. Delbianco, J. Am. Chem. Soc., 2022, 144, 12469–12475 Search PubMed.
- N. Hribernik, D. Vargová, M. C. S. Dal Colle, J. H. Lim, G. Fittolani, Y. Yu, J. Fujihara, K. Ludwig, P. H. Seeberger, Y. Ogawa and M. Delbianco, Angew. Chem., Int. Ed., 2023, 62, e202310357 Search PubMed.
- P. R. Ashton, C. L. Brown, S. Menzer, S. A. Nepogodiev, J. F. Stoddart and D. J. Williams, Chem. – Eur. J., 1996, 2, 580–591 Search PubMed.
- G. Gattuso, S. Menzer, S. A. Nepogodiev, J. F. Stoddart and D. J. Williams, Angew. Chem., Int. Ed. Engl., 1997, 36, 1451–1454 Search PubMed.
- P. R. Ashton, S. J. Cantrill, G. Gattuso, S. Menzer, S. A. Nepogodiev, A. N. Shipway, J. F. Stoddart and D. J. Williams, Chem. – Eur. J., 1997, 3, 1299–1314 Search PubMed.
- Y. Yao, X. Meng, C. Li, K. V. Bernaerts and K. Zhang, Small, 2023, 19, 2208286 Search PubMed.
- L. Su, S. I. S. Hendrikse and E. W. Meijer, Curr. Opin. Chem. Biol., 2022, 69, 102171 Search PubMed.
- M. Delbianco, P. Bharate, S. Varela-Aramburu and P. H. Seeberger, Chem. Rev., 2016, 116, 1693–1752 Search PubMed.
- A. Brito, S. Kassem, R. L. Reis, R. V. Ulijn, R. A. Pires and I. Pashkuleva, Chem, 2021, 7, 2943–2964 Search PubMed.
- V. I. B. Castro, R. L. Reis, R. A. Pires and I. Pashkuleva, Chem. Soc. Rev., 2025, 54, 8325–8344 Search PubMed.
- S. I. S. Hendrikse, L. Su, T. P. Hogervorst, R. P. M. Lafleur, X. Lou, G. A. van der Marel, J. D. C. Codee and E. W. Meijer, J. Am. Chem. Soc., 2019, 141, 13877–13886 Search PubMed.
- S. L. Higashi and M. Ikeda, JACS Au, 2021, 1, 1639–1646 Search PubMed.
- D. M. Chung and J. S. Nowick, J. Am. Chem. Soc., 2004, 126, 3062–3063 Search PubMed.
- S. Basak, I. Singh, A. Ferranco, J. Syed and H.-B. Kraatz, Angew. Chem., Int. Ed., 2017, 56, 13288–13292 Search PubMed.
- X. Li, S. E. Rios and J. S. Nowick, Chem. Sci., 2022, 13, 7739–7746 Search PubMed.
- L. Pauling and R. B. Corey, Proc. Natl. Acad. Sci. U. S. A., 1953, 39, 253–256 Search PubMed.
- D. E. Mortenson, J. D. Steinkruger, D. F. Kreitler, D. V. Perroni, G. P. Sorenson, L. Huang, R. Mittal, H. G. Yun, B. R. Travis, M. K. Mahanthappa, K. T. Forest and S. H. Gellman, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 13144–13149 Search PubMed.
- Y. Yao, Q. Tang, S. Rosenfeldt, M. Krüsmann, M. Karg and K. Zhang, Small, 2021, 17, 2102938 Search PubMed.
- S. Gim, G. Fittolani, Y. Nishiyama, P. H. Seeberger, Y. Ogawa and M. Delbianco, Angew. Chem., Int. Ed., 2020, 59, 22577–22583 Search PubMed.
- Y. Wu, S. Aslani, H. Han, C. Tang, G. Wu, X. Li, H. Wu, C. L. Stern, Q.-H. Guo, Y. Qiu, A. X. Y. Chen, Y. Jiao, R. Zhang, A. H. G. David, D. W. Armstrong and J. Fraser Stoddart, Nat. Synth., 2024, 3, 698–706 Search PubMed.
- M. Wada, S. Wakiya, K. Kobayashi, S. Kimura, M. Kitaoka, R. Kusumi, F. Kimura and T. Kimura, Cellulose, 2021, 28, 6757–6765 Search PubMed.
- S. Djalali, Y. Jing, Y. Ogawa and M. Delbianco, Chem. Sci., 2025, 16, 1390–1395 Search PubMed.
- T. Koga, M. Matsuoka and N. Higashi, J. Am. Chem. Soc., 2005, 127, 17596–17597 Search PubMed.
- J. Seibel, G. Fittolani, H. Mirhosseini, X. Wu, S. Rauschenbach, K. Anggara, P. H. Seeberger, M. Delbianco, T. D. Kühne, U. Schlickum and K. Kern, Angew. Chem., Int. Ed., 2023, 62, e202305733 Search PubMed.
- M. Lingenfelder, G. Tomba, G. Costantini, L. Colombi Ciacchi, A. De Vita and K. Kern, Angew. Chem., Int. Ed., 2007, 46, 4492–4495 Search PubMed.
- K. Lindner and W. Saenger, Biochem. Biophys. Res. Commun., 1980, 92, 933–938 Search PubMed.
- B. Hingerty and W. Saenger, J. Am. Chem. Soc., 1976, 98, 3357–3365 Search PubMed.
- K. Lindner and W. Saenger, Angew. Chem., Int. Ed. Engl., 1978, 17, 694–695 Search PubMed.
- I. Roy and J. F. Stoddart, Acc. Chem. Res., 2021, 54, 1440–1453 Search PubMed.
- L. Hu, X. Zhu, C. Yang and M. Liu, Angew. Chem., Int. Ed., 2022, 61, e202114759 Search PubMed.
- D. Colesnic, P. J. Hernando, L.-M. Chamoreau, L. Bouteiller, M. Ménand and M. Sollogoub, Chem. – Eur. J., 2023, 29, e202300150 Search PubMed.
- S. W. Wukovitz and T. O. Yeates, Nat. Struct. Biol., 1995, 2, 1062–1067 Search PubMed.
- A. L. Mackay, Nature, 1989, 342, 133–133 Search PubMed.
- T. O. Yeates and S. B. H. Kent, Annu. Rev. Biophys., 2012, 41, 41–61 Search PubMed.
- R. Okamoto, K. Mandal, M. R. Sawaya, Y. Kajihara, T. O. Yeates and S. B. H. Kent, Angew. Chem., Int. Ed., 2014, 53, 5194–5198 Search PubMed.
- R. A. Dwek, Chem. Rev., 1996, 96, 683–720 Search PubMed.
- B. Tyson, C. M. Pask, N. George and E. Simone, Cryst. Growth Des., 2022, 22, 1371–1383 Search PubMed.
- Y. Ogawa and J.-L. Putaux, Front. Chem., 2022, 10, 835663 Search PubMed.
- J.-Y. Huang and M. Delbianco, Synthesis, 2022, 1337–1354 Search PubMed.
- W. Yao and X.-S. Ye, Acc. Chem. Res., 2024, 57, 1577–1594 Search PubMed.
- S. Marchesan, C. D. Easton, F. Kushkaki, L. Waddington and P. G. Hartley, Chem. Commun., 2012, 48, 2195–2197 Search PubMed.
- A. M. Garcia, D. Iglesias, E. Parisi, K. E. Styan, L. J. Waddington, C. Deganutti, R. De Zorzi, M. Grassi, M. Melchionna, A. V. Vargiu and S. Marchesan, Chem, 2018, 4, 1862–1876 Search PubMed.
- A. M. Garcia, M. Melchionna, O. Bellotto, S. Kralj, S. Semeraro, E. Parisi, D. Iglesias, P. D'Andrea, R. De Zorzi, A. V. Vargiu and S. Marchesan, ACS Nano, 2021, 15, 3015–3025 Search PubMed.
- C. L. O'Neill, J. L. Fascetti, Z. Clapacs, L. K. Kaplita, C.-Y. Liu, D. Kim, M. A. White and J. S. Rudra, Chem. – Eur. J., 2025, 31, e202404603 Search PubMed.
- H. Qi, K. Qi, J. Li, C. He, M. Liao, X. Hu, Y. Zhao, Y. Ke, C. Zhang, J. Zhang, J. Wang, J. R. Lu and H. Xu, Nano Res., 2023, 16, 12230–12237 Search PubMed.
- K. Qi, H. Qi, M. Wang, X. Ma, Y. Wang, Q. Yao, W. Liu, Y. Zhao, J. Wang, Y. Wang, W. Qi, J. Zhang, J. R. Lu and H. Xu, Nat. Commun., 2024, 15, 6186 Search PubMed.
- M. R. Ghadiri, J. R. Granja, R. A. Milligan, D. E. McRee and N. Khazanovich, Nature, 1993, 366, 324–327 Search PubMed.
- D. T. Bong, T. D. Clark, J. R. Granja and M. R. Ghadiri, Angew. Chem., Int. Ed., 2001, 40, 988–1011 Search PubMed.
- R. R. Ketchem, K. C. Lee, S. Huo and T. A. Cross, J. Biomol. NMR, 1996, 8, 1–14 Search PubMed.
- L. Pauling and R. B. Corey, Proc. Natl. Acad. Sci. U. S. A., 1951, 37, 251–256 Search PubMed.
- V. Daggett, Acc. Chem. Res., 2006, 39, 594–602 Search PubMed.
- F. Heitz, G. Detriche, F. Vovelle and G. Spach, Macromolecules, 1981, 14, 47–50 Search PubMed.
- D. Shea, C.-C. Hsu, T. M. Bi, N. Paranjapye, M. C. Childers, J. Cochran, C. P. Tomberlin, L. Wang, D. Paris, J. Zonderman, G. Varani, C. D. Link, M. Mullan and V. Daggett, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 8895–8900 Search PubMed.
- G. Hopping, J. Kellock, R. P. Barnwal, P. Law, J. Bryers, G. Varani, B. Caughey and V. Daggett, eLife, 2014, 3, e01681 Search PubMed.
- F. Kovacs, J. Quine and T. A. Cross, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 7910–7915 Search PubMed.
- D. W. Urry, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 1610–1614 Search PubMed.
- R. R. Ketchem, W. Hu and T. A. Cross, Science, 1993, 261, 1457–1460 Search PubMed.
- P. De Santis, S. Morosetti and R. Rizzo, Macromolecules, 1974, 7, 52–58 Search PubMed.
- A. Buleon and H. Chanzy, J. Polym. Sci., Polym. Phys. Ed., 1980, 18, 1209–1217 Search PubMed.
- J. P. van Trijp, N. Hribernik, J. H. Lim, M. C. S. Dal Colle, Y. V. Mena, Y. Ogawa and M. Delbianco, Angew. Chem., Int. Ed., 2024, 63, e202410634 Search PubMed.
- L. Wu, Z. Zhou, D. Sathe, J. Zhou, S. Dym, Z. Zhao, J. Wang and J. Niu, Nat. Chem., 2023, 15, 1276–1284 Search PubMed.
- W. Yao, D.-C. Xiong, Y. Yang, C. Geng, Z. Cong, F. Li, B.-H. Li, X. Qin, L.-N. Wang, W.-Y. Xue, N. Yu, H. Zhang, X. Wu, M. Liu and X.-S. Ye, Nat. Synth., 2022, 1, 854–863 Search PubMed.
- A. A. Joseph, A. Pardo-Vargas and P. H. Seeberger, J. Am. Chem. Soc., 2020, 142, 8561–8564 Search PubMed.
- W. A. Szarek, G. W. Hay, D. M. Vyas, E. R. Ison and L. J. J. Hronowski, Can. J. Chem., 1984, 62, 671–674 Search PubMed.
- T.-Y. Xia, Y.-B. Li, Z.-J. Yin, X.-B. Meng, S.-C. Li and Z.-J. Li, Chin. Chem. Lett., 2014, 25, 1220–1224 Search PubMed.
- M. Grabarics, B. Mallada, S. Edalatmanesh, A. Jiménez-Martín, M. Pykal, M. Ondráček, P. Kührová, W. B. Struwe, P. Banáš, S. Rauschenbach, P. Jelínek and B. de la Torre, Nat. Commun., 2024, 15, 9482 Search PubMed.
- A. F. Lehrhofer, M. Bacher, I. Melikhov, I. Sulaeva, W. Lindner, M. Kohout, H. Kamitakahara, A. Potthast, T. Rosenau and H. Hettegger, Carbohydr. Polym., 2025, 367, 123896 Search PubMed.
- Y. Chen and Y. Liu, Chem. Soc. Rev., 2010, 39, 495–505 Search PubMed.
- G. Chen and M. Jiang, Chem. Soc. Rev., 2011, 40, 2254–2266 Search PubMed.
- G. Crini, Chem. Rev., 2014, 114, 10940–10975 Search PubMed.
- K. Takahashi, Chem. Rev., 1998, 98, 2013–2034 Search PubMed.
- G. Xu, S. Leloux, P. Zhang, J. Meijide Suárez, Y. Zhang, E. Derat, M. Ménand, O. Bistri-Aslanoff, S. Roland, T. Leyssens, O. Riant and M. Sollogoub, Angew. Chem., Int. Ed., 2020, 59, 7591–7597 Search PubMed.
- J. Ji, W. Wu, W. Liang, G. Cheng, R. Matsushita, Z. Yan, X. Wei, M. Rao, D.-Q. Yuan, G. Fukuhara, T. Mori, Y. Inoue and C. Yang, J. Am. Chem. Soc., 2019, 141, 9225–9238 Search PubMed.
- M. E. Davis and M. E. Brewster, Nat. Rev. Drug Discovery, 2004, 3, 1023–1035 Search PubMed.
- S. Perez, O. Makshakova, J. Angulo, E. Bedini, A. Bisio, J. L. de Paz, E. Fadda, M. Guerrini, M. Hricovini, M. Hricovini, F. Lisacek, P. M. Nieto, K. Pagel, G. Paiardi, R. Richter, S. A. Samsonov, R. R. Vivès, D. Nikitovic and S. Ricard Blum, JACS Au, 2023, 3, 628–656 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |

