Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
iRGD-engineered exosomes mediate siMYC delivery for effective tumor suppression in triple-negative breast cancer
Hui Li†
a,
Weiguang Yuan†bc,
Jialin Liudef,
Yingjie Wangdef,
Fang Fangdef,
Yuanyuan Yudef,
Jianxun Houdef,
Mengru Jindef,
Siwei Lidef,
Siyu Liudef,
Yajie Gongdef,
Yijun Chudef,
Xingda Zhangdef,
Shilu Zhaodef,
Wenhui Haodef,
Xuquan Qindef,
Qinchen Fandef,
Xi Cheng,
Youxue Zhang*def,
Da Pang*def and
Xianyu Zhang‡
*def
aDepartment of Ultrasound, The First Affiliated Hospital of Harbin Medical University, Harbin, China
bInstitute of Cancer Prevention and Treatment, Harbin Medical University, Harbin, China
cInstitute of Cancer Prevention and Treatment, Heilongjiang Academy of Medical Sciences, Harbin, China
dHeilongjiang Clinical Research Center for Breast Cancer, Harbin Medical University Cancer Hospital, Harbin, China. E-mail: zhangxianyu@ems.hrbmu.edu.cn; pangda@ems.hrbmu.edu.cn; alxazyx@163.com
eDepartment of Breast Surgery, Harbin Medical University Cancer Hospital, Harbin, China
fTranslational Medicine Research and Cooperation Center of Northern China, Heilongjiang Academy of Medical Sciences, Harbin, China
gDepartment of Breast Surgery, The Second Affiliated Hospital of Harbin Medical University, Harbin, China
First published on 12th December 2025
Abstract
Triple-negative breast cancer (TNBC) is an aggressive cancer with a poor prognosis. MYC overexpression drives tumor progression, but the lack of an efficient siMYC delivery system remains a major challenge. Exosomes (Exos), as biocompatible nanocarriers, offer a solution. Here, we engineered internalizing RGD peptide (iRGD)-Exos to enhance siMYC delivery and improve therapeutic efficacy. iRGD-Exos were generated by transfecting FreeStyle™ 293-F (293-F) cells with an iRGD-Flag-Lamp2b plasmid, followed by ultracentrifugation and isolation. siMYC was loaded via electroporation. Exosomes were characterized, and their uptake efficiency was measured. CCK-8 assays and flow cytometry were conducted to analyze the effects of exosomes on the proliferation and apoptosis of TNBC cells. Apoptosis staining was also conducted on patient-derived organoids (PDOs). In a TNBC xenograft mouse model, fluorescence imaging, tumor volume measurement, and histological analysis were conducted to assess tumor targeting and therapeutic effects of engineered exosomes. Systemic toxicity was evaluated based on hematological, biochemical, and histopathological analyses. The iRGD modification significantly enhanced the uptake efficiency of exosomes by αvβ3 integrin-positive Hs578T TNBC cells. Following siMYC loading via electroporation, iRGD-Exos-siMYC markedly suppressed the proliferation of TNBC cells and induced their apoptosis. Additionally, it promoted apoptosis in PDOs, further supporting its antitumor potential. In vivo, iRGD-Exos-siMYC exhibited superior tumor-targeting capability, effectively inhibiting tumor growth and significantly downregulating MYC expression. Moreover, biosafety evaluations confirmed that iRGD-Exos-siMYC possesses good biosafety. This study demonstrated that iRGD-modified exosomes can effectively deliver siMYC to TNBC cells, enhancing gene silencing and antitumor efficacy. The targeted exosomal drug delivery system showed high tumor selectivity and minimal systemic toxicity. These findings provide new insights into exosome-based gene therapy and highlight the value of iRGD-Exos-siMYC as a novel treatment strategy for TNBC.
Introduction
Triple-negative breast cancer (TNBC) is a highly aggressive subtype of breast cancer, characterized by the absence of estrogen receptors, progesterone receptors, and human epidermal growth factor receptor 2 (HER2).1–3 This lack of molecular targets makes TNBC difficult to treat, leading to poor prognosis and high metastasis rates.4,5 Currently, chemotherapy is the primary treatment option for TNBC, but it comes with several limitations, including drug resistance, toxic side effects, and the lack of specificity in targeting tumor cells.6,7 These challenges have spurred the exploration of alternative therapeutic strategies, such as small-interfering RNA (siRNA)-mediated gene therapy, which holds great potential for selectively silencing oncogenes and overcoming the limitations of traditional treatments.8MYC is a key molecular player in TNBC progression. It is a transcription factor that regulates various cellular processes, including cell cycle progression, apoptosis, and metabolism.9–11 MYC is often overexpressed in TNBC and promotes tumor growth and metastasis.9,10 Targeting MYC through siRNA offers a promising strategy for inhibiting tumor progression and improving the outcomes of patients with TNBC. However, the efficient delivery of siRNA to the tumor site remains a major challenge in cancer treatment.
The complex tumor microenvironment makes it more difficult to effectively deliver siRNA to tumor cells, presenting physical and biological barriers to drug penetration.12,13 Conventional drug delivery systems often struggle to achieve the desired therapeutic concentration at the tumor site.14 Nanocarrier-based drug delivery systems have shown considerable advancement in overcoming these challenges, offering improved drug stability, controlled release, and enhanced bioavailability.15–18 However, for successful translation of these systems into clinical practice, several hurdles must be addressed, including the need for active cellular targeting and overcoming biological barriers, such as the tumor extracellular matrix.19,20
Exosome-based nanocarriers are emerging as a particularly promising solution to these challenges. Exosomes are natural lipid bilayer vesicles secreted by cells that possess intrinsic advantages as drug delivery vehicles. They mimic the cell membrane composition and offer biocompatibility and low immunogenicity.21–23 Moreover, exosomes can cross biological barriers, such as the blood–brain barrier, and deliver therapeutic agents directly to tumor cells.24–26 To enhance their targeting capabilities, exosomes can be engineered to carry ligands or peptides that specifically recognize tumor-associated receptors.27,28 One of such receptors, integrin αvβ3, is a member of the integrin family that plays a crucial role in tumor angiogenesis and metastasis. Integrin αvβ3 is normally expressed at low levels in normal tissues but is significantly overexpressed in tumor cells and endothelial cells of intratumoral blood vessels.29–31 Targeting integrin αvβ3 using peptides, such as iRGD, which binds specifically to the receptor, can improve the targeting ability of exosome-based nanocarriers.32,33 iRGD peptide can enhance the penetration of exosomes into tumors and improve their therapeutic efficacy by binding to integrin αvβ3 on the surface of tumor endothelial cells and cancer cells.34,35
In this study, we designed and engineered an iRGD-modified exosome to enhance the targeting capability of exosomes. As a gene delivery vehicle, iRGD-Exos encapsulated siMYC via electroporation. The targeted iRGD-Exos-siMYC was applied to TNBC cells to evaluate its targeting efficiency and therapeutic effects. In vitro and in vivo results indicated that the newly developed targeted drug delivery system effectively targeted TNBC cells and downregulated the expression of MYC, thereby exhibiting robust antitumor efficacy. This innovative platform holds promising translational potential and offers exciting clinical prospects, providing novel insights into the treatment of TNBC.
Experimental
Cell line and culture
TNBC cell lines (MDA-MB-231, MDA-MB-436, MDA-MB-468, BT549, Hs578T, and HCC1937) were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in the recommended medium containing 10% fetal bovine serum (FBS), 1% penicillin, and 1% streptomycin. The cells were incubated in a 5% CO2 incubator at 37 °C. All cell lines were validated to be free of mycoplasma contamination. FreeStyle™ 293-F (293-F) cells (Thermo Fisher Scientific, R79007, USA) were cultured in FreeStyle™ 293 Expression Culture (Thermo Fisher Scientific, 12338018, USA) in a shaker incubator (105008, NEST, China) with 120 rpm, at 37 °C, and 5% CO2.RNA isolation and qRT-PCR
Total RNA was isolated from the cells using Trizol reagent (Invitrogen, #269201, USA). Subsequently, reverse transcription was conducted with the FastKing gDNA Dispelling RT SuperMix (TIANGEN, GKR118, China) to synthesize cDNA. Real-time PCR was conducted on a 7500 Fast Real-Time PCR system (Applied Biosystems, USA) using FastStart Universal SYBR Green Master (ROX) (Roche, #04913914001, Switzerland). Data were analyzed using the 2−ΔΔCt method, and β-actin was considered the housekeeping gene. ITGAV, forward: 5′-AGGAGAAGGTGCCTACGAAGCT-3′; reverse: 5′-GCACAGGAAAGTCTTGCTAAGGC-3′. ITGB3, forward: 5′-CATGGATTCCAGCAATGTCCTCC-3′; reverse: 5′-TTGAGGCAGGTGGCATTGAAGG-3′. MYC, forward: 5′-GGCTCCTGGCAAAAGGTCA-3′; reverse: 5′-CTGCGTAGTTGTGCTGATGT-3′.Protein extraction and western blotting
Cells and tissues were lysed using RIPA buffer (Solarbio, R0010, China) containing 1% PMSF for 20 minutes at 4 °C. The total protein concentration of cell lysates, tissue lysates, and exosomal lysates was determined using a BCA assay kit (Beyotime, P0010, China). For each sample, 40 µg of total protein from cells, tissues, and exosomes was loaded per lane. Proteins were separated using 10% or 12.5% SDS-PAGE gels based on the target molecular weight (10% for high molecular weight proteins and 12.5% for small proteins) using a two-step voltage protocol (70 V for 15 minutes followed by 110 V for approximately 90 minutes). The separated proteins were subsequently transferred onto 0.2 μm PVDF membranes (Millipore, 03010040001, USA) at 200 mA for approximately 120 minutes in a cold transfer buffer. After blocking with 5% skimmed milk at room temperature for 90 minutes, the membranes were incubated with primary antibodies at 4 °C overnight. The membranes were washed three times with TBST (0.1% Tween-20), followed by incubation with HRP-conjugated secondary antibodies at room temperature for 1 hour. Next, the membranes were washed three times with TBST. Finally, the protein bands were visualized using enhanced chemiluminescence (Meilunbio, MA0186, China), and images were captured using the BIO-RAD ChemiDoc XRS + System. The following antibodies were used for western blotting: integrin αv (Cell Signaling Technology, #4711, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), integrin β3 (Cell Signaling Technology, #13166, 1
1000), integrin β3 (Cell Signaling Technology, #13166, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), β-actin (ZSGB-BIO, TA-09, 1
1000), β-actin (ZSGB-BIO, TA-09, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), c-Myc (Cell Signaling Technology, #5605, 1
1000), c-Myc (Cell Signaling Technology, #5605, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), Lamp2b (Abcam, ab18529, 1
1000), Lamp2b (Abcam, ab18529, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), Flag (Sigma, F1804, 1
1000), Flag (Sigma, F1804, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), CD63 (Abcam, ab134045, 1
1000), CD63 (Abcam, ab134045, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), ALIX (Abcam, ab186429, 1
1000), ALIX (Abcam, ab186429, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), TSG101 (Abcam, ab125011, 1
1000), TSG101 (Abcam, ab125011, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), Calnexin (Abcam, ab92573, 1
1000), Calnexin (Abcam, ab92573, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000), goat anti-mouse (Proteintech, No. SA00001-1, 1
2000), goat anti-mouse (Proteintech, No. SA00001-1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), and goat anti-rabbit (Proteintech, No. SA00001-2, 1
000), and goat anti-rabbit (Proteintech, No. SA00001-2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000).
000).
siRNA transfection
First, 2 × 105 cells were seeded in six-well culture plates for siRNA transfection. At a density of nearly 70%, the cells were transfected using JetPrime (Polyplus, #114–15, Germany). Briefly, 10 µL of siRNA was diluted in 200 µL of the jetPRIME® buffer and mixed thoroughly. Then, 4 µL of the jetPRIME® reagent was added to the mixture and mixed gently. The solution was incubated at room temperature for 10 minutes to allow complex formation. Subsequently, 200 µL of the transfection mixture was added dropwise to each well containing 2 mL of culture medium. Three siRNAs targeting human MYC were synthesized by Saiwen Innovation (Beijing) Biotechnology Co., Ltd (Beijing, China). Knockdown cell lines were identified using qRT-PCR or western blotting to validate transfection efficiency. The sequences of siRNAs were as follows: siMYC 1#: 5′-GAGGAGACAUGGUGAACCA-3′; siMYC 2#: 5′-GGGUCAAGUUGGACAGUGU-3′; siMYC 3#: 5′-CGACGAGACCUUCAUCAAA-3′.Plasmid construction and transfection
The iRGD-Flag-Lamp2b plasmid was synthesized by Genechem (Shanghai, China). Human embryonic kidney 293-F cells were employed for suspension culture and transient transfection. The cell density was adjusted to 6–7 × 105 cells per mL the day before transfection to prepare for transfection on Day 2. On the transfection day, cell viability was measured using trypan blue exclusion to ensure a cell viability of more than 90%. For each 30 mL transfection, 3 × 107 cells were required. In total, 30 μg of plasmid DNA was diluted in 1 mL of Opti-MEM™ I (31985062, Thermo Fisher Scientific, USA) and mixed gently. Separately, 60 μL of 293fectin™ (Thermo Fisher Scientific, 12347019, USA) was diluted in 1 mL of Opti-MEM™ I, mixed gently, and incubated at room temperature for 5 minutes. After incubation, diluted DNA was added to the diluted 293fectin™, reaching a total volume of 2 mL. They were incubated at room temperature for 30 minutes to form the DNA-reagent complex. Meanwhile, 28 mL of the pre-warmed FreeStyle™ 293 expression medium was added to a sterile 125 mL conical flask. After incubation, the 2 mL DNA–reagent complex was added to the conical flask, resulting in a total culture volume of 30 mL and a cell density of approximately 1 × 106 cells per mL. The flasks were incubated at 37 °C with 5% CO2 on a shaking platform at 120 rpm. For downstream analyses, the transfected cells and their conditioned medium were harvested 48 hours after transfection.Isolation of exosomes
Exosomes were isolated from culture supernatants using differential centrifugation. First, the culture medium was centrifuged at 300g for 10 minutes to remove cells, followed by centrifugation at 2000g for 10 minutes to remove dead cells. The supernatant was then centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 minutes to eliminate cell debris. To pellet the exosomes, the supernatant was centrifuged at 100
000g for 30 minutes to eliminate cell debris. To pellet the exosomes, the supernatant was centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 minutes. The exosome pellet was resuspended in PBS and subjected to another round of centrifugation at 100
000g for 70 minutes. The exosome pellet was resuspended in PBS and subjected to another round of centrifugation at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 minutes to wash and remove residual proteins and other contaminants. Finally, the exosome pellet was resuspended in PBS and stored at −80 °C for subsequent assays. All centrifugation steps were conducted at 4 °C.
000g for 70 minutes to wash and remove residual proteins and other contaminants. Finally, the exosome pellet was resuspended in PBS and stored at −80 °C for subsequent assays. All centrifugation steps were conducted at 4 °C.
Characterization of exosomes
For transmission electron microscopy (TEM), 15 μL of the exosome sample was placed on a copper grid and allowed to sit for 1 minute. Then, a filter paper was used to blot the exosome sample from the copper grid. Next, 15 μL of 2% uranyl acetate staining solution was added using a pipette and incubated at room temperature for 1 minute. Afterward, the exosome sample was blotted on the copper grid with filter paper again. The stained sample was placed under a lamp and baked for 10 minutes. Finally, images were observed and captured using a TEM (FEI, Tecnai G2 spititi, USA), and the pictures were saved. A nanoparticle tracking analysis (NTA) instrument (Malvern, NanoSight NS300, Britain) was employed to assess the size and concentration of exosomes.Exosome labeling
Exosomes were labeled with the fluorescent dye Dil (Beyotime, C1991, China) for 30 minutes, and then centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 minutes to remove any unbound dye. The labeled exosomes were then washed twice with PBS at 100
000g for 70 minutes to remove any unbound dye. The labeled exosomes were then washed twice with PBS at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 minutes each time. Finally, the exosomes were resuspended in PBS.
000g for 70 minutes each time. Finally, the exosomes were resuspended in PBS.
Exosomes loading and loading efficiency
To load siMYC into the exosomes, 24 μg of exosomes and 2500 nM of siMYC were gently mixed in 400 μL Gene Pulser Electroporation Buffer (BioRad, 1652677, USA) at 4 °C. Thereafter, the mixture was electroporated in a 4 mm cuvette (BioRad, 1652081, USA) using a Gene Pulser II Electroporator (BioRad, 1652660, USA) at 400 V and 125 μF. Next, the supernatant was centrifuged at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 70 min to remove free siMYC.
000g for 70 min to remove free siMYC.
To evaluate siRNA loading efficiency, total RNA was extracted from siMYC-loaded exosomes using the miRcute miRNA Isolation Kit (TIANGEN, DP501, China) following the manufacturer's protocol. The exosomes were lysed in Buffer MZ containing guanidinium salts and β-mercaptoethanol. Small RNAs were purified on silica-based spin columns and eluted in RNase-free ddH2O. cDNA synthesis was conducted using the miRcute Plus miRNA First-Strand cDNA Kit (TIANGEN, KR211, China), which combines poly(A)-tailing and reverse transcription with Oligo(dT)-Universal Tag primers. The 20 μL reaction mixture was incubated at 42 °C for 60 min and then at 95 °C for 3 min. The resulting cDNA was used for qRT-PCR. qRT-PCR was conducted using the miRcute Plus miRNA qPCR Kit (SYBR Green) (TIANGEN, FP411, China) on an ABI PRISM 7500 system. The reaction mixture (20 μL) contained 10 μL of 2× miRcute Plus miRNA PreMix (SYBR & ROX), 0.4 μL of forward (siMYC 2#: 5′-GGCGGGTCAAGTTGGACAGTGTA-3′) and 0.4 μL of reverse primers (10 μM), 2 μL of cDNA, 1.6 μL of 50× ROX dye, and nuclease-free water. The cycling conditions were 95 °C for 15 min, followed by 40 cycles of 94 °C for 20 s, and 60 °C for 34 s. The amount of siMYC was determined based on qRT-PCR using siMYC-specific primers. The siRNA-loading efficiency (%) was calculated as the ratio of the amount of siMYC detected in exosomes to the total amount of siMYC initially added during electroporation.
Cellular uptake study in vitro
In total, 2 × 104 Hs578T cells were seeded into confocal dishes and incubated overnight. Subsequently, 30 μg of Dil-blank-Exos or Dil-iRGD-Exos were added to the Hs578T cell culture medium and incubated at 37 °C for 2, 4, or 6 hours. The cells were washed three times with PBS and fixed with 4% paraformaldehyde for 30 minutes. Following fixation, the cells were stained with DAPI for 20 minutes. Finally, cell images were captured using a confocal microscope (Olympus, Japan).Flow cytometry was performed to quantify cellular uptake efficiency. Briefly, 2 × 105 Hs578T cells were seeded into 6-well plates and incubated overnight. Then, 30 μg of Dil-blank-Exos or Dil-iRGD-Exos were added to the Hs578T cell culture medium and incubated for 3 or 6 hours. Subsequently, the cells were washed three times with PBS. After collection, the cells were resuspended in PBS, transferred to BD Falcon™ round-bottom polystyrene tubes (BD Biosciences, Cat. No. 352054, USA), and placed on ice. Flow cytometry was conducted using a BD Biosciences flow cytometer. At least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample were acquired for analysis.
000 events per sample were acquired for analysis.
siRNA uptake assay
Hs578T cells were seeded into confocal dishes at a density of 2 × 104 cells per dish and incubated overnight to evaluate the cellular internalization efficiency of siRNA-loaded exosomes. Cy3-labeled siMYC2# (Saiwen, China) was loaded into blank-Exos or iRGD-Exos via electroporation as described previously. The labeled exosomes (30 μg per dish, corresponding to 0.221 nmol siMYC) were then co-cultured with Hs578T cells at 37 °C for 6 hours. After incubation, the cells were washed three times with PBS and fixed with 4% paraformaldehyde for 30 minutes, followed by nuclear staining with DAPI for 20 minutes. The intracellular localization of Cy3-siMYC was visualized using a confocal fluorescence microscope (Olympus, Japan). The fluorescence intensity of Cy3 was quantified using ImageJ software.Cell proliferation assays
Hs578T cells were seeded into a 96-well plate at a density of 2000 cells per well for 24 hours. Subsequently, the old culture medium in each well was replaced with fresh complete medium containing 1 μg per well (corresponding to 7.4 pmol siRNA per well) of blank-Exos-siNC, blank-Exos-siMYC, iRGD-Exos-siNC, or iRGD-Exos-siMYC. After incubation for 6 hours, the medium was replaced with exosome-free complete medium, and the cells were incubated for 48 hours. Thereafter, 10 μL of the CCK-8 solution was added to each well, and the plate was incubated at 37 °C for 1 hour. Cell viability in each well was determined by reading the absorbance at 450 nm using a microplate reader (BioTek, USA).Apoptosis assay
First, 2 × 105 Hs578T cells were inoculated into a 6-well plate for 24 hours. Subsequently, the old culture medium in each well was replaced with fresh complete medium containing 30 μg per well (corresponding to 0.221 nmol siMYC per well) of blank-Exos-siNC, blank-Exos-siMYC, iRGD-Exos-siNC, or iRGD-Exos-siMYC. After 6 hours of incubation, the cells were gently washed twice with PBS and cultured in exosome-free complete medium for 48 hours. Cell apoptosis was detected using the Annexin V-FITC Apoptosis Detection Kit (Beyotime, C1062, China) following the manufacturer's instructions. The cells were digested with EDTA-free trypsin and centrifuged at 1000g for 5 minutes. Thereafter, the supernatant was discarded. The cells were then gently resuspended in PBS and counted. In total, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–100
000–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were collected and centrifuged at 1000g for 5 minutes. After removing the supernatant, the cell pellet was gently resuspended in 195 μL of Annexin V-FITC binding buffer. Then, 5 μL of Annexin V-FITC and 10 μL of propidium iodide (PI) staining solution were added, mixed gently, and dark incubated at room temperature for 15 minutes. After staining, the samples were transferred to BD Falcon™ round-bottom polystyrene tubes and placed on ice. Flow cytometry was conducted on a BD Biosciences flow cytometer, and at least 10
000 cells were collected and centrifuged at 1000g for 5 minutes. After removing the supernatant, the cell pellet was gently resuspended in 195 μL of Annexin V-FITC binding buffer. Then, 5 μL of Annexin V-FITC and 10 μL of propidium iodide (PI) staining solution were added, mixed gently, and dark incubated at room temperature for 15 minutes. After staining, the samples were transferred to BD Falcon™ round-bottom polystyrene tubes and placed on ice. Flow cytometry was conducted on a BD Biosciences flow cytometer, and at least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events per sample were acquired for analysis.
000 events per sample were acquired for analysis.
Organoid preparation
In this study, clinical samples were obtained from patients undergoing routine surgical procedures at Harbin Medical University Cancer Hospital. The use of these samples for research purposes was approved by the Ethics Committee of Harbin Medical University Cancer Hospital (Approval no. KY2020-09). Informed consent was obtained from all patients before sample collection and use. Breast tumor tissue samples obtained from patients were placed in pre-cooled PBS, minced into small fragments, and enzymatically digested. The cell suspension was centrifuged at 300g at 4 °C for 5 minutes, and the resulting pellet was kept on ice. The cell pellet was resuspended in matrix gel, and 40 μL of the mixture was seeded into each well of a 24-well plate. Subsequently, 500 μL of breast cancer organoid culture medium was added to each well. The cultures were incubated in a CO2 incubator at 37 °C, and the medium was refreshed every two days.Tumor targeting in vivo
Animal experiments were approved by the Medical Experimental Animal Care Committee of the Second Affiliated Hospital of Harbin Medical University (Approval no. SYDW2025-018) and were conducted in accordance with the guidelines defined by the National Institutes of Health (NIH) for animal care and ethical standards. Female BALB/c nude mice, aged 6–8 weeks, were obtained from Beijing Weitong Lihua Experimental Animal Technology Co., Ltd. All animals were housed in a clean-grade barrier facility at the Medical Experimental Animal Center of the Second Affiliated Hospital of Harbin Medical University. The animals had free access to food and water under constant temperature. After anesthesia with avertin (0.020 mL g−1 body weight, intraperitoneal injection), 5 × 106 Hs578T cells were suspended in 200 μL of serum-free medium and injected directly into the right mammary fat pad. Six mice were used in total (n = three mice per group). One week later, 150 μg per mouse (corresponding to 1.10 nmol siMYC per mouse) of Dil-labeled exosomes (Dil-blank-Exos and Dil-iRGD-Exos) were intravenously administered via the tail vein. After 24 hours, mice were euthanized in a CO2 environment, followed by cervical dislocation to ensure death and minimize suffering. Thereafter, the mice were carefully dissected to harvest the tumor, heart, liver, spleen, lung, and kidney tissues. After rinsing the tissues with PBS, fluorescence images were captured using the IVIS Spectrum Imaging System.Anti-tumor efficacy and biosafety in vivo
After anesthesia with avertin (0.020 mL g−1 body weight, intraperitoneal injection), 5 × 106 Hs578T cells were suspended in 200 μL of serum-free medium and injected directly into the right mammary fat pad. A mouse model bearing Hs578T tumor was initially established using 25 female BALB/c nude mice (6–8 weeks old). One week later, tumor-bearing mice were randomly divided into five groups (n = 5 per group): control, blank-Exos-siNC, blank-Exos-siMYC, iRGD-Exos-siNC, iRGD-Exos-siMYC. Each mouse intravenously received the corresponding treatment via the tail vein every three days at a siRNA dose of 0.6 mg kg−1 (corresponding to approximately 12 µg siRNA or 126 µg of engineered exosomes per 20 g mouse). Mouse body weight was recorded every three days using a digital scale. Tumor volume was measured every three days using a caliper. Tumor volume was calculated using the following formula: V = 1/2(length × width2).After treatment and anesthesia with avertin (0.020 mL g−1 body weight, intraperitoneal injection), approximately 600–800 µL of blood was immediately collected from the retro-orbital sinus. Subsequently, mice were euthanized in a CO2 chamber, followed by cervical dislocation to ensure death and minimize suffering. The tumors and the heart, lungs, liver, spleen, kidneys, and other major organs were carefully dissected. Blood samples and major organs were harvested for hematological and histochemical analyses. The serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), blood urea nitrogen (BUN), and creatinine (CRE) were measured. White blood cells (WBC), red blood cells (RBC), hemoglobin (HGB), hematocrit (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), and platelet (PLT) counts were also recorded. The heart, lungs, liver, spleen, kidneys, and other major organs were fixed in formaldehyde for 48 hours and then embedded in paraffin. Tissue sections were cut into slices and subjected to routine hematoxylin and eosin (H&E) staining.
An additional in vivo experiment was conducted using tumor-bearing BALB/c nude mice to evaluate the superiority of iRGD-Exos-siMYC over free siRNA. Mice were divided into the following groups (n = 5 per group): control, free siMYC 2#, iRGD-Exos-siMYC 2#, and iRGD-Exos-siMYC 3#. Each mouse received a tail-vein injection of Free siMYC or iRGD-Exos-siMYC at a siRNA dose of 0.6 mg kg−1 (equivalent to approximately 12 µg siRNA or 126 µg of exosomal protein per 20 g mouse), every three days. Tumor size was measured every 3 days, and tumor volumes were calculated as described above. After the final treatment, the tumors were excised, photographed, and weighed to measure therapeutic efficacy.
Immunohistochemical (IHC) staining
Tumor tissues were fixed, embedded in paraffin, and sectioned to a thickness of 3 μm. Following standard protocols, paraffin-embedded sections were subjected to deparaffinization, antigen retrieval, background blocking, and subsequent detection using the specified antibodies. Detection was conducted using liquid DAB+ and counterstained with Carazzi's hematoxylin. Two pathologists blinded to the grouping measured gene expression. Briefly, the percentage of tumor cells with positive staining was scored as follows: 0 (no positive staining), 1 (≤10% positive), 2 (10% < positive ≤ 25%), 3 (25% < positive ≤ 50%), 4 (50% < positive ≤ 75%), and 5 (>75% positive). The staining intensity was scored as follows: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The histochemical score (H-score) was determined by multiplying the proportion of positively stained tumor cells by the staining intensity.Statistical analysis
All data are presented as mean ± standard deviation. Student's t-test was used to compare the experimental and control groups. One-way ANOVA was employed for multiple group comparisons. Statistical analyses were conducted using GraphPad Prism 10.1. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 were deemed statistically significant.Results
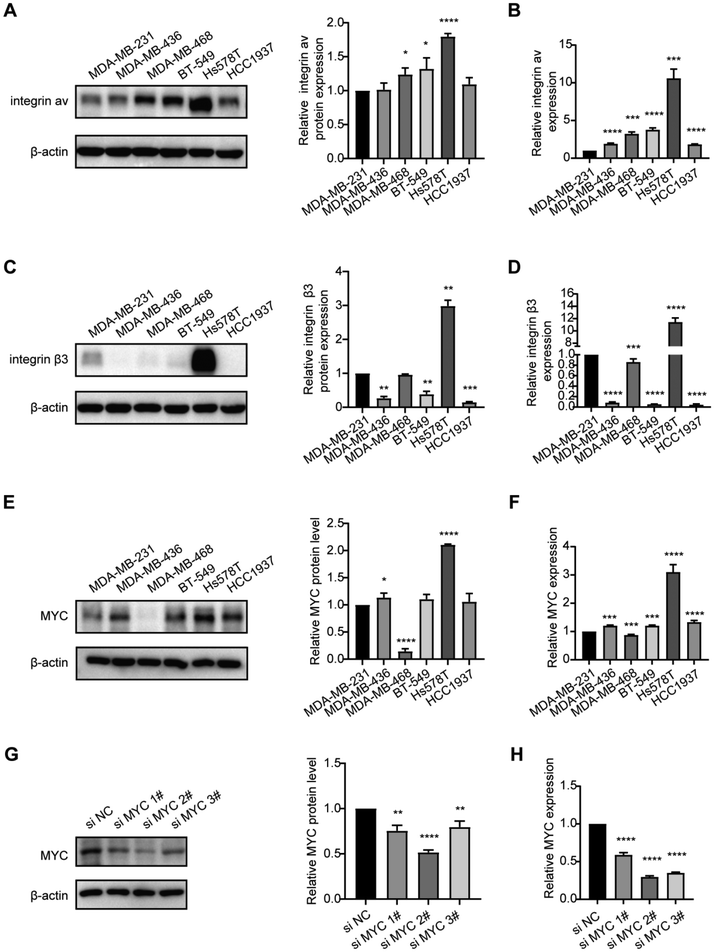
Expression of integrin αvβ3 and MYC in TNBC cell lines
qRT-PCR and western blotting were utilized to assess the mRNA and protein levels of integrin αvβ3 in TNBC cell lines, including MDA-MB-231, MDA-MB-436, MDA-MB-468, BT549, Hs578T, and HCC1937. Among TNBC cell lines, Hs578T exhibited the highest mRNA and protein levels of integrin αvβ3 (Fig. 1A–D). Additionally, both the mRNA and protein levels of MYC were also highest in Hs578T cells (Fig. 1E and F). Therefore, Hs578T cells were selected for subsequent experiments. Since siMYC 2# led to a greater decrease in MYC expression in Hs578T cells compared to siMYC 1# or siMYC 3# (Fig. 1G and H), siMYC 2# was used in the following experiments.Isolation and characterization of exosomes
We transfected 293-F cells with the iRGD-Flag-Lamp2b plasmid. western blotting revealed that, compared to 293-F cells transfected with the empty vector, the transfected 293-F cells exhibited high protein levels of Lamp2b and Flag (Fig. 2A). Exosomes were then isolated from the culture supernatants of untransfected 293-F cells (ctrl-Exos), 293-F cells transfected with the empty vector (blank-Exos), and 293-F cells transfected with the overexpression plasmid (iRGD-Exos). ctrl-Exos, blank-Exos, and iRGD-Exos were characterized using TEM, NTA, and western blotting. Exosomal protein markers CD63, ALIX, and TSG101 were positively expressed in ctrl-Exos, blank-Exos, and iRGD-Exos, while calnexin was absent in these vesicles (Fig. 2B). Furthermore, a flag-positive band was detected exclusively in iRGD-Exos, accompanied by a markedly increased Lamp2b protein signal compared to blank-Exos, suggesting that the engineered exosomes successfully carried and expressed the iRGD peptide on their surface (SI Fig. S1A). In TEM and NTA analyses, ctrl-Exos, blank-Exos, and iRGD-Exos all exhibited a typical cup-shaped structure, with diameters of 138, 134, and 119 nm (Fig. 2C and D), respectively, which is consistent with the size of typical exosomes. These findings suggest the successful isolation of ctrl-Exos, blank-Exos, and iRGD-Exos.Targeting of iRGD-Exos in vitro
Next, we investigated the in vitro tumor-targeting capability of iRGD-Exos and evaluated whether iRGD peptide modification can enhance exosome binding to Hs578T cells. Dil-labeled blank-Exos or iRGD-Exos were incubated with Hs578T cells for 2, 4, and 6 hours. Both types of exosomes were successfully internalized by recipient cells (Fig. 3A). Notably, the Dil fluorescence signal in the iRGD-Exos group was significantly higher than that in the blank-Exos group. Furthermore, flow cytometry revealed that the cellular uptake of iRGD-Exos was significantly greater than that of blank-Exos, consistent with the in vitro results in confocal microscopy (Fig. 3B). These findings suggest that iRGD peptide modification significantly enhanced the targeting capability of exosomes toward Hs578T cells.The antitumor effect of iRGD-Exos-siMYC in vitro
We measured the feasibility of loading siMYC2# into iRGD-Exos using an electroporation method. qRT-PCR quantification exhibited that the siRNA loading efficiency was approximately 17.65%, confirming the successful encapsulation of siMYC2# into exosomes. The highest loading efficiency was achieved when the concentration of siMYC reached 2500 nM (SI Fig. S2A). Subsequently, TEM and NTA analyses revealed no significant changes in the shape and size of exosomes after loading siMYC2# into blank-Exos or iRGD-Exos via electroporation (Fig. 4A and B). These results indicate that electroporation did not affect the physical properties of the exosomes.To determine whether iRGD-Exos loaded with siMYC2# can specifically transfer siMYC2# into Hs578T cells in vitro, Cy3-labeled siMYC2# was loaded into blank-Exos or iRGD-Exos via electroporation, followed by co-culturing with Hs578T cells for 6 hours. The Cy3 fluorescence signal in the iRGD-Exos-siMYC group was significantly stronger than that in the blank-Exos-siMYC group (Fig. 4C and D). qRT-PCR and western blotting showed that, compared to the control group or the blank-Exos-siMYC group, the mRNA and protein levels of MYC were significantly downregulated in Hs578T cells treated with iRGD-Exos-siMYC (Fig. 4E and F). These data indicate that iRGD-Exos loaded with siMYC2# specifically delivered siMYC2# to Hs578T cells in vitro.
Next, we analyzed the effect of iRGD-Exos-siMYC on the proliferation of Hs578T cells. Four types of exosomes were constructed, including blank-Exos-siNC, blank-Exos-siMYC, iRGD-Exos-siNC, and iRGD-Exos-siMYC, and their targeting ability was validated by western blotting (Fig. 5A). Hs578T cells were treated with control medium, blank-Exos-siNC, blank-Exos-siMYC, iRGD-Exos-siNC, or iRGD-Exos-siMYC for 48 hours. We found that the iRGD-Exos-siMYC group exhibited the strongest inhibitory effect on cell proliferation compared to other groups (Fig. 5B). Meanwhile, flow cytometry analysis revealed that the iRGD-Exos-siMYC group had the highest proportion of apoptotic cells compared to other groups (Fig. 5C and D).
We also evaluated the growth-inhibitory effect of iRGD-Exos-siMYC in patient-derived organoids (PDOs) from patients with TNBC. Apoptosis staining showed that treatment with iRGD-Exos-siMYC significantly induced apoptosis in PDOs (Fig. 5E).
In summary, iRGD-Exos-siMYC effectively suppressed tumor growth by inhibiting proliferation and inducing apoptosis in TNBC models, including both the Hs578T cell line and PDOs.
Targeting of iRGD-Exos in vivo
The tumor-targeting capability of iRGD-Exos was validated in a mouse model bearing Hs578T tumors. 24 hours after tail vein injection of Dil-labeled blank-Exos or iRGD-Exos, tumor tissues, as well as the heart, liver, spleen, lungs, and kidneys, were harvested. In vivo fluorescence imaging was conducted to monitor the fluorescence signal. Fluorescence imaging showed strong fluorescence accumulation in the tumor region, liver, and kidneys (SI Fig. S3A). Compared to the blank-Exos group, the iRGD-Exos group exhibited a stronger Dil fluorescence signal in the tumor. These results suggest that iRGD-Exos possess tumor-targeting capabilities in vivo.Antitumor effect of iRGD-Exos-siMYC in vivo
Next, we evaluated the antitumor efficacy of iRGD-Exos-siMYC in vivo. A xenograft Hs578T mouse model was established. One week after tumor inoculation, the mice were randomly divided into five groups, including control, blank-Exos-siNC, blank-Exos-siMYC, iRGD-Exos-siNC, and iRGD-Exos-siMYC, and treated via tail vein injection (Fig. 6A). The iRGD-Exos-siMYC group exhibited the slowest tumor growth (Fig. 6B and C). Additionally, at the end of the observation period, tumors were excised from each mouse and weighed. The mice treated with iRGD-Exos-siMYC had the lowest tumor weight (Fig. 6D).We assessed the protein expression of MYC in tumor tissues using western blotting and found that MYC expression was lowest in the iRGD-Exos-siMYC group (Fig. 6E). Additionally, we conducted immunofluorescence and immunohistochemical staining to detect MYC expression. The results were consistent with those of western blotting (Fig. 6F, G and SI Fig. S4A). Furthermore, we measured the expression of Ki67 and cleaved caspase-3 in tumor tissues. The results showed that treatment with iRGD-Exos-siMYC downregulated Ki67 expression and upregulated cleaved caspase-3 expression in tumor tissues (Fig. 6H and I).
An additional in vivo experiment was conducted in Hs578T tumor-bearing nude mice to verify the therapeutic superiority and reproducibility of iRGD-Exos-mediated siMYC delivery. Mice were randomly divided into four groups: control, free siMYC 2#, iRGD-Exos-siMYC 2#, and iRGD-Exos-siMYC 3#. Both iRGD-Exos-siMYC 2# and iRGD-Exos-siMYC 3# led to visible tumor shrinkage compared to the free siMYC and control groups (SI Fig. S5A). Tumor growth curves showed consistently slower tumor progression in both iRGD-Exos-siMYC groups (SI Fig. S5B). The final tumor volumes and weights were significantly reduced compared to free siMYC (SI Fig. S5C). Importantly, the comparable outcomes observed between iRGD-Exos-siMYC 2# and iRGD-Exos-siMYC 3# groups confirmed the reproducibility and robustness of the iRGD-Exos-mediated siRNA delivery strategy. Moreover, both iRGD-Exos-siMYC formulations exhibited markedly stronger tumor growth inhibition than free siMYC, highlighting the superior therapeutic efficacy achieved through iRGD-mediated targeted delivery. These results suggest that iRGD modification can effectively enhance siMYC accumulation in tumor tissues and substantially improve the overall antitumor potency compared with free siRNA.
Safety evaluation
Firstly, we measured the in vitro viability of Hs578T cells treated with different concentrations of iRGD-Exos (0–25 μg mL−1). Cell viability remained more than 95% across all tested concentrations, indicating the excellent biocompatibility of iRGD-Exos with minimal cytotoxicity to normal cells (SI Fig. S6A).In addition to therapeutic efficacy, biosafety is another critical parameter for evaluating delivery systems. At the end of the experiment, there were no significant differences in the average body weight of mice across groups, suggesting that all treatment regimens were well-tolerated (Fig. 7A). Furthermore, blood biochemistry and hematological analyses were conducted at the end of the observation period. Biochemical parameters, including liver function markers, such as ALT and AST, and kidney function markers, such as UREA and CRE, were also assessed. All of these parameters were consistent with the control group (Fig. 7B), suggesting that iRGD-Exos-siMYC did not lead to significant hepatic or renal toxicity within the administered dose range. Regarding hematological analysis, WBC, RBC, HGB, HCT, MCV, MCH, MCHC, and PLT in the iRGD-Exos-siMYC treatment group were comparable to those in the control group (Fig. 7C). H&E staining was conducted to measure histological changes in major organs, including the heart, liver, spleen, lungs, and kidneys. There were no significant changes in the H&E staining images of the major organs in all five groups compared to the control group (Fig. 7D). This indicates that none of the treatments led to noticeable side effects and all exhibited good biosafety. These results suggest that iRGD-Exos-siMYC did not lead to hematological toxic effects nor adversely affect major organs in mice.
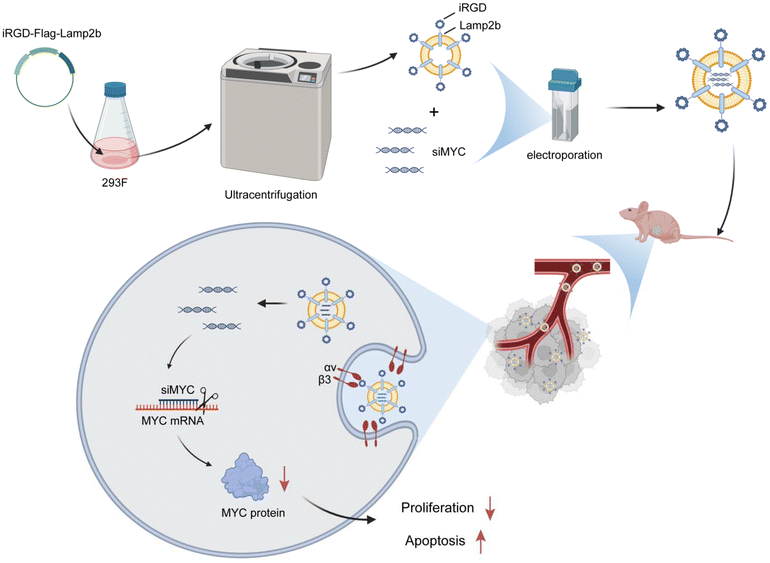
Discussion
Currently, targeted therapy has become a crucial approach in cancer treatment, as it targets specific molecular markers on cancer cells, effectively inhibiting tumor growth and metastasis.36 Common targeted therapies include monoclonal antibodies and small-molecule inhibitors.37 These treatment options are highly specific, minimize damage to normal cells, and offer fewer side effects compared to traditional chemotherapy.37 However, several challenges remain. Tumor cells may develop resistance through mechanisms, such as genetic mutations and activation of alternative signaling pathways.38–41 Additionally, the efficacy of targeted drugs is limited by the selectivity of the target and the narrow therapeutic window. Some tumor cells may not express the common targets, rendering these treatments ineffective in certain types of cancer. In this context, RNA interference (RNAi) technology has emerged as a new and promising strategy for gene therapy. RNAi utilizes small RNA molecules, such as siRNA, to specifically degrade the mRNA of target genes, thereby inhibiting their expression.42,43 Compared to traditional targeted therapies, RNAi provides a more precise regulation of the expression of tumor-related genes, offering more personalized and targeted treatment options.42 However, the clinical application of RNAi still faces challenges, including issues with effective delivery, stability, and off-target effects.44,45 Overcoming these challenges remains a critical area of current research to enhance the therapeutic efficacy of RNAi. In this study, we established an exosome-based delivery system to deliver siRNA to tumor cells for targeted therapy (Fig. 8).Exosomes, which were previously considered cellular “waste” carriers, actually possess powerful drug and gene delivery capabilities, effectively transporting bioactive molecules to target cells.46,47 The binding affinity of exosomes to specific receptors on tumor cells can be significantly enhanced by modifying exosomes with targeting peptides.28,48 Targeting peptides, such as iRGD, have shown tremendous potential in enhancing the delivery of therapeutic agents to tumor cells. Researchers have reported that exosomes secreted by genetically engineered donor cells, modified with the iRGD peptide, can efficiently target integrin-positive glioblastoma multiforme (GBM),49 anaplastic thyroid carcinoma (ATC),50 breast cancer,35 nasopharyngeal carcinoma (NPC),51,52 gastric cancer,53 lung adenocarcinoma,54 and diffuse large B-cell lymphoma (DLBCL) cells.55 Additionally, Han et al. reported that genetic modification with the iRGD peptide can enhance the targeting ability and uptake efficiency of exosomes from human cord blood mesenchymal stem cells (cbMSCs) in tumor cells expressing the NRP-1 receptor, significantly improving tumor targeting capabilities.56 In this study, we employed iRGD-modified exosomes to target breast cancer cells, particularly Hs578T cells. iRGD modification significantly enhanced the uptake of exosomes by tumor cells, more efficiently targeting tumor cells.
Exosomes, as natural nanoscale carriers, possess an intrinsic nanospace that enables them to encapsulate and efficiently load various bioactive molecules. Previous studies have reported that active loading strategies, such as electroporation, sonication, and freeze–thaw, generally achieve higher loading efficiency compared to passive incubation.57 Among these methods, electroporation has proven to be an effective method. This technique utilizes an electrical field to transiently disrupt the lipid bilayer of exosomes, allowing the incorporation of cargo molecules. As a notable advantage, electroporation allows exosomes to efficiently load a diverse range of bioactive compounds without significantly compromising their structural integrity or biological function. This method is relatively simple and reproducible.58–60 Moreover, electroporation is widely recognized as one of the most commonly used and highly efficient physical methods for loading nucleic acids, particularly siRNA.61,62 For example, MAPK1 siRNA has been successfully loaded into exosomes using electroporation, whereas no siRNA signal was observed in the non-electroporated control samples.63 Likewise, an siRNA loading efficiency of approximately 20% has been achieved using electroporation, further supporting its effectiveness for small RNA delivery.64 In this study, electroporation-mediated loading of siMYC into exosomes achieved a loading efficiency of 17.65%, without significantly altering the size or structural integrity of exosomes, as confirmed by TEM and NTA. This process preserved the stability and functionality of the exosomes. Furthermore, the electroporated exosomes successfully loaded siMYC and efficiently delivered it to Hs578T cells. siMYC delivered via electroporation significantly downregulated MYC expression at both the mRNA and protein levels, indicating effective gene transfer and silencing. In vivo, the iRGD-Exos-siMYC group exhibited the lowest MYC expression level in tumor tissues. These findings highlight the potential of electroporation as a reliable and non-destructive method for drug loading into exosomes, particularly for targeted gene silencing.
Through surface modification with specific ligands or peptides and by loading therapeutic cargo molecules, engineered exosomes can exhibit enhanced targeting capability and therapeutic potential. As one of the effective anti-tumor drugs, the side effects of doxorubicin (DOX) can be minimized by using exosomes as delivery carriers.65 Exosomes modified by iRGD-Lamp2b or LAMP2b-DARPin were loaded with DOX through electroporation, targeting MDA-MB-231 cells and HER2-positive SKBR3 cells highly expressing αv integrin. Both studies verified the therapeutic effects of engineered exosomes on breast cancer through animal experiments.35,66 HEK293T cells were transfected with pLEX-LAMP-DARP in lentivirus to produce exosomes that can bind to HER2-positive breast cancer cells. Through the targeting effect of exosomes, siRNA TPD52 was transferred to SKBR3 cells and the expression of the TPD52 gene was silencd.67 These findings suggest that engineered exosomes carrying different miRNAs, such as miR-588, LNA-anti-miR-142-3p, and miR-145, can effectively target breast cancer cells, inhibit tumor growth, and suppress proliferation and metastasis by modulating the expression of tumor suppressor genes.67–69 Moreover, immunostimulatory exosomes combined with dual-targeted Coxsackievirus B3 (miR-CVB3) therapy were shown to reshape the immunosuppressive tumor microenvironment, enhance the anti-tumor immune response, and significantly suppress tumor growth in breast cancer.70 It was found that engineered exosomes modified with the GE11 peptide can target EGFR-expressing breast cancer cells and deliver miRNA let-7a, which possesses anti-tumor properties, thereby effectively inhibiting tumor growth in a breast cancer xenograft model.71 Wang et al. reported the same experimental results. Although extracellular vesicles (EVs) were loaded with let-7a, EVs were conjugated with the aptamer AS1411, which binds to nucleolar protein through chemical modification.72 Wan et al. have produced PTX-loaded AS1411-ENVs via a chemical conjugation method. AS1411-ENVs exhibited anti-tumor effects.73 Engineered exosomes modified with CD62L and OX40L (ExoSmart) significantly enhanced the anti-tumor immune response by activating effector T cells and suppressing Tregs, thereby effectively inhibiting the growth of metastatic breast cancer.74 Additionally, SMART-Exos or GEMINI-Exos, displaying specific monoclonal antibodies on their surface, such as those targeting CD3 and HER2 or those targeting CD3 and EGFR, were shown to significantly enhance the anti-tumor immune response and inhibit tumor growth in breast cancer, demonstrating immense potential in targeted therapy and immunotherapy.75,76 Engineered exosomes not only improve the targeting and intracellular delivery efficiency of the drug but also significantly enhance the therapeutic efficacy of breast cancer through sonodynamic therapy (SDT).77,78 In this study, iRGD-Exos-siMYC effectively inhibited cell proliferation and induced apoptosis not only in TNBC cell lines but also in patient-derived organoids, suggesting its potent antitumor activity in vitro. In vivo, treatment with iRGD-Exos-siMYC significantly suppressed tumor growth, demonstrating its potent therapeutic efficacy.
Engineered exosomes have garnered significant attention in drug delivery due to their inherent biocompatibility. As natural lipid bilayer vesicles, exosomes usually do not provoke immune responses compared to synthetic nanoparticles, making them an ideal choice for therapeutic applications. Moreover, the ability of exosomes to mimic the cellular membrane structure enhances their stability and safety.79–81 In this study, treatment with iRGD-Exos-siMYC did not significantly change body weight, organ function, or hematological parameters, indicating the biosafety of iRGD-Exos-siMYC.
There are some limitations to this study. First, the use of human-derived cells in immunodeficient mice did not fully replicate the tumor microenvironment. Particularly, it did not reproduce the effect of the immune system on treatment. Therefore, future studies could introduce spontaneous tumor models or humanized mouse models to more comprehensively evaluate the therapeutic effects. Additionally, some exosomes still accumulate in the liver and kidneys, and their in vivo biodistribution needs further optimization to reduce non-specific uptake and enhance tumor-targeting efficiency. Third, this study did not directly compare exosomes with other commonly used RNA delivery systems, such as lipid nanoparticles (LNPs) or viral vectors. Hence, future studies should systematically compare the differences in siRNA-loading efficiency, in vivo biodistribution, gene silencing effects, and biosafety across various delivery systems, providing more reliable evidence for optimizing siRNA delivery strategies.
Conclusion
iRGD-modified exosomes loaded with siMYC effectively targeted Hs578T cells, inhibited MYC expression, and significantly suppressed tumor growth both in vitro and in vivo, with favorable biosafety. These findings underscore the potential of iRGD-Exos-siMYC as a promising therapeutic strategy for breast cancer.Author contributions
Hui Li: writing – original draft, investigation. Weiguang Yuan: data curation. Jialin Liu: investigation. Yingjie Wang: data curation. Fang Fang: methodology. Yuanyuan Yu: investigation. Jianxun Hou: investigation. Mengru Jin: data curation. Siwei Li: methodology. Siyu Liu: data curation. Yajie Gong: investigation. Yijun Chu: methodology. Xingda Zhang: investigation. Shilu Zhao: methodology. Wenhui Hao: investigation. Xuquan Qin: methodology. Xi Chen: investigation. Youxue Zhang: conceptualization. Da Pang: conceptualization. Xianyu Zhang: conceptualization, supervision.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information. Supplementary information: Supplementary figure S1–S6. See DOI: https://doi.org/10.1039/d5nr04841a.Acknowledgements
This work is supported by the National Natural Science Foundation of China (Grant Numbers: 82073410, 82272623 and 82173235); Key Special Projects of Heilongjiang Province Key Research and Development Program (Grant Numbers: 2023ZX06C10 and JD22C004); Nn10 Program of Harbin Medical University Cancer Hospital (Grant Numbers: 2017-02); Spring Goose Support Program of Heilongjiang Province (Grant Numbers: CYQN24010). The authors would like to express their gratitude to EditSprings (https://www.editsprings.com) for the expert linguistic services provided.References
- P. Zagami and L. A. Carey, npj Breast Cancer, 2022, 8, 95 CrossRef CAS PubMed.
- U. Mehraj, U. Mushtaq, M. A. Mir, A. Saleem, M. A. Macha, M. N. Lone, A. Hamid, M. A. Zargar, S. M. Ahmad and N. A. Wani, Semin. Cancer Biol., 2022, 86, 769–783 CrossRef CAS PubMed.
- W. D. Foulkes, I. E. Smith and J. S. Reis-Filho, N. Engl. J. Med., 2010, 363, 1938–1948 CrossRef CAS PubMed.
- R. Dent, M. Trudeau, K. I. Pritchard, W. M. Hanna, H. K. Kahn, C. A. Sawka, L. A. Lickley, E. Rawlinson, P. Sun and S. A. Narod, Clin. Cancer Res., 2007, 13, 4429–4434 CrossRef PubMed.
- H. Kennecke, R. Yerushalmi, R. Woods, M. C. Cheang, D. Voduc, C. H. Speers, T. O. Nielsen and K. Gelmon, J. Clin. Oncol., 2010, 28, 3271–3277 CrossRef PubMed.
- R. K. Landow, Dermatol. Clin., 1988, 6, 575–584 CrossRef CAS.
- C. Kim, R. Gao, E. Sei, R. Brandt, J. Hartman, T. Hatschek, N. Crosetto, T. Foukakis and N. E. Navin, Cell, 2018, 173, 879–893.e13 CrossRef CAS PubMed.
- A. Afghahi and G. W. Sledge Jr., Cancer J., 2015, 21, 294–298 CrossRef CAS PubMed.
- Y. Fallah, J. Brundage, P. Allegakoen and A. N. Shajahan-Haq, Biomolecules, 2017, 7, 53 CrossRef PubMed.
- M. C. Stipp and A. Acco, Gene, 2025, 941, 149209 CrossRef CAS PubMed.
- D. J. Liao and R. B. Dickson, Endocr.-Relat. Cancer, 2000, 7, 143–164 CAS.
- M. R. Thomas, A. K. Badekila, V. Pai, N. S. Y. Bhandary, A. Rai and S. Kini, Adv. Healthc. Mater., 2025, e2403107, DOI:10.1002/adhm.202403107.
- S. Rivera and F. Yuan, Curr. Pharm. Biotechnol., 2012, 13, 1279–1291 CAS.
- D. Haussecker, J. Controlled Release, 2014, 195, 49–54 CrossRef CAS PubMed.
- S. Peng, X. Yuan, H. Li, Y. Wei, B. Zhou, G. Ding and J. Bai, Eur. J. Med. Chem., 2023, 252, 115259 CrossRef CAS PubMed.
- J. K. Patra, G. Das, L. F. Fraceto, E. V. R. Campos, M. D. P. Rodriguez-Torres, L. S. Acosta-Torres, L. A. Diaz-Torres, R. Grillo, M. K. Swamy, S. Sharma, S. Habtemariam and H. S. Shin, J. Nanobiotechnol., 2018, 16, 71 CrossRef PubMed.
- P. Dong, K. P. Rakesh, H. M. Manukumar, Y. H. E. Mohammed, C. S. Karthik, S. Sumathi, P. Mallu and H. L. Qin, Bioorg. Chem., 2019, 85, 325–336 CrossRef CAS PubMed.
- Z. Li, S. Tan, S. Li, Q. Shen and K. Wang, Oncol. Rep., 2017, 38, 611–624 CrossRef CAS.
- D. Rosenblum, N. Joshi, W. Tao, J. M. Karp and D. Peer, Nat. Commun., 2018, 9, 1410 CrossRef PubMed.
- A. Nasir, A. Khan, J. Li, M. Naeem, A. A. K. Khalil, K. Khan and M. Qasim, Curr. Top. Med. Chem., 2021, 21, 1360–1376 CrossRef CAS PubMed.
- H. Zeng, S. Guo, X. Ren, Z. Wu, S. Liu and X. Yao, Cells, 2023, 12, 1416 CrossRef CAS PubMed.
- L. Zhu, H. T. Sun, S. Wang, S. L. Huang, Y. Zheng, C. Q. Wang, B. Y. Hu, W. Qin, T. T. Zou, Y. Fu, X. T. Shen, W. W. Zhu, Y. Geng, L. Lu, H. L. Jia, L. X. Qin and Q. Z. Dong, J. Hematol. Oncol., 2020, 13, 152 CrossRef CAS PubMed.
- Y. Zhang, Q. Liu, X. Zhang, H. Huang, S. Tang, Y. Chai, Z. Xu, M. Li, X. Chen, J. Liu and C. Yang, J. Nanobiotechnol., 2022, 20, 279 CrossRef CAS PubMed.
- P. Vader, E. A. Mol, G. Pasterkamp and R. M. Schiffelers, Adv. Drug Delivery Rev., 2016, 106, 148–156 CrossRef CAS PubMed.
- F. U. Rehman, Y. Liu, M. Zheng and B. Shi, Biomaterials, 2023, 293, 121949 CrossRef CAS PubMed.
- R. Pandit, L. Chen and J. Gotz, Adv. Drug Delivery Rev., 2020, 165–166, 1–14 CrossRef CAS PubMed.
- Y. Guo, Z. Wang, X. Shi and M. Shen, Exploration, 2022, 2, 20210171 CrossRef CAS PubMed.
- S. Salunkhe, D. Dheeraj, M. Basak, D. Chitkara and A. Mittal, J. Controlled Release, 2020, 326, 599–614 CrossRef CAS.
- J. S. Desgrosellier and D. A. Cheresh, Nat. Rev. Cancer, 2010, 10, 9–22 CrossRef CAS PubMed.
- F. Debordeaux, L. Chansel-Debordeaux, J. B. Pinaquy, P. Fernandez and J. Schulz, Nucl. Med. Biol., 2018, 62–63, 31–46 CrossRef CAS PubMed.
- H. Hamidi and J. Ivaska, Nat. Rev. Cancer, 2018, 18, 533–548 CrossRef CAS PubMed.
- V. M. D'Amore, G. Donati, E. Lenci, B. S. Ludwig, S. Kossatz, M. Baiula, A. Trabocchi, H. Kessler, F. S. Di Leva and L. Marinelli, J. Chem. Inf. Model., 2023, 63, 6302–6315 CrossRef PubMed.
- C. Mantis, I. Kandela, F. Aird and Reproducibility Project: Cancer Biology, Elife, 2017, 6, e17584 CrossRef PubMed.
- H. Zuo, J. Oncol., 2019, 2019, 9367845 Search PubMed.
- Y. Tian, S. Li, J. Song, T. Ji, M. Zhu, G. J. Anderson, J. Wei and G. Nie, Biomaterials, 2014, 35, 2383–2390 CrossRef CAS PubMed.
- Y. T. Lee, Y. J. Tan and C. E. Oon, Eur. J. Pharmacol., 2018, 834, 188–196 CrossRef CAS PubMed.
- R. Kalyn, J. Oncol. Pharm. Pract., 2007, 13, 199–205 CrossRef CAS PubMed.
- M. Martini, L. Vecchione, S. Siena, S. Tejpar and A. Bardelli, Nat. Rev. Clin. Oncol., 2011, 9, 87–97 CrossRef PubMed.
- J. Jin, X. Wu, J. Yin, M. Li, J. Shen, J. Li, Y. Zhao, Q. Zhao, J. Wu, Q. Wen, C. H. Cho, T. Yi, Z. Xiao and L. Qu, Front. Oncol., 2019, 9, 263 CrossRef PubMed.
- Y. M. Thu, K. Suzawa, S. Tomida, K. Ochi, S. Tsudaka, F. Takatsu, K. Date, N. Matsuda, K. Iwata, K. Nakata, K. Shien, H. Yamamoto, M. Okazaki, S. Sugimoto and S. Toyooka, PLoS One, 2024, 19, e0300644 CrossRef CAS PubMed.
- H. Zhong, Z. Zhou, H. Wang, R. Wang, K. Shen, R. Huang and Z. Wang, Int. J. Mol. Sci., 2024, 25, 13376 CrossRef CAS PubMed.
- Y. Zhou, C. Zhang and W. Liang, J. Controlled Release, 2014, 193, 270–281 CrossRef CAS PubMed.
- Y. L. Chiu and T. M. Rana, RNA, 2003, 9, 1034–1048 CrossRef CAS PubMed.
- Y. J. Ren and Y. Zhang, Expert Opin. Biol. Ther., 2014, 14, 1581–1592 CrossRef CAS PubMed.
- D. Bumcrot, M. Manoharan, V. Koteliansky and D. W. Sah, Nat. Chem. Biol., 2006, 2, 711–719 CrossRef CAS PubMed.
- Y. Couch, E. I. Buzas, D. Di Vizio, Y. S. Gho, P. Harrison, A. F. Hill, J. Lotvall, G. Raposo, P. D. Stahl, C. Thery, K. W. Witwer and D. R. F. Carter, J. Extracell. Vesicles, 2021, 10, e12144 CrossRef CAS PubMed.
- N. P. Hessvik and A. Llorente, Cell. Mol. Life Sci., 2018, 75, 193–208 CrossRef CAS PubMed.
- Y. Wu, Y. Cao, L. Chen, X. Lai, S. Zhang and S. Wang, Biol. Proced. Online, 2024, 26, 15 CrossRef PubMed.
- D. Gecys, A. Kazlauskas, E. Gecyte, N. Pauziene, D. Kulakauskiene, I. Lukminaite and A. Jekabsone, Biology, 2022, 11, 1483 CrossRef CAS PubMed.
- C. Wang, N. Li, Y. Li, S. Hou, W. Zhang, Z. Meng, S. Wang, Q. Jia, J. Tan, R. Wang and R. Zhang, J. Nanobiotechnol., 2022, 20, 247 CrossRef CAS PubMed.
- J. Wang, Y. Liu, Y. Zhang, X. Li, M. Fang and D. Qian, Cancer Med., 2023, 12, 12608–12621 CrossRef CAS PubMed.
- J. Wang, Q. Jiang, O. D. Faleti, C. M. Tsang, M. Zhao, G. Wu, S. W. Tsao, M. Fu, Y. Chen, T. Ding, T. Chong, Y. Long, X. Yang, Y. Zhang, Y. Cai, H. Li, M. Peng, X. Lyu and X. Li, Mol. Ther. – Nucleic Acids, 2020, 22, 153–165 CrossRef CAS PubMed.
- L. Xin, Y. W. Yuan, C. Liu, L. Q. Zhou, L. Liu, Q. Zhou and S. H. Li, Dig. Dis. Sci., 2021, 66, 1045–1053 CrossRef CAS PubMed.
- J. Chen, F. Cao, Y. Cao, S. Wei, X. Zhu and W. Xing, J. Biomed. Nanotechnol., 2022, 18, 1075–1083 CrossRef CAS PubMed.
- Q. Liu, G. Dai, Y. Wu, M. Zhang, M. Yang, X. Wang, M. Song, X. Li, R. Xia and Z. Wu, Front. Oncol., 2022, 12, 822805 CrossRef CAS PubMed.
- S. Han, G. Li, M. Jia, Y. Zhao, C. He, M. Huang, L. Jiang, M. Wu, J. Yang, X. Ji, X. Liu, C. Chen and X. Chu, Front. Mol. Biosci., 2021, 8, 743013 CrossRef CAS PubMed.
- H. I. Kim, J. Park, Y. Zhu, X. Wang, Y. Han and D. Zhang, Exp. Mol. Med., 2024, 56, 836–849 CrossRef CAS PubMed.
- J. Gehl, Acta Physiol. Scand., 2003, 177, 437–447 CrossRef CAS PubMed.
- M. A. C. Pomatto, F. Negro and G. Camussi, Methods Mol. Biol., 2022, 2504, 219–230 CrossRef CAS PubMed.
- X. M. Xi, S. J. Xia and R. Lu, Pharmazie, 2021, 76, 61–67 CAS.
- Y. Lu, W. Huang, M. Li and A. Zheng, Pharmaceutics, 2023, 15, 598 CrossRef CAS PubMed.
- Y. Han, T. W. Jones, S. Dutta, Y. Zhu, X. Wang, S. P. Narayanan, S. C. Fagan and D. Zhang, Processes, 2021, 9, 356 CrossRef CAS PubMed.
- J. Wahlgren, L. K. T. De, M. Brisslert, F. Vaziri Sani, E. Telemo, P. Sunnerhagen and H. Valadi, Nucleic Acids Res., 2012, 40, e130 CrossRef CAS PubMed.
- L. Alvarez-Erviti, Y. Seow, H. Yin, C. Betts, S. Lakhal and M. J. Wood, Nat. Biotechnol., 2011, 29, 341–345 CrossRef CAS PubMed.
- Z. S. Hashemi, M. Ghavami, F. Mohammadi, M. Shokrollahi Barough, F. Shokati, S. Asghari, S. Khalili, M. Akbari Yekta, A. Ghavamzadeh and R. Sarrami Forooshani, Iran. J. Basic Med. Sci., 2024, 27, 1604–1615 Search PubMed.
- H. Gomari, M. Forouzandeh Moghadam, M. Soleimani, M. Ghavami and S. Khodashenas, Int. J. Nanomed., 2019, 14, 5679–5690 CrossRef CAS PubMed.
- S. K. Limoni, M. F. Moghadam, S. M. Moazzeni, H. Gomari and F. Salimi, Appl. Biochem. Biotechnol., 2019, 187, 352–364 CrossRef CAS PubMed.
- Z. Zhang, X. Luo, X. Xue, M. Pang, X. Wang, L. Yu, J. Qian, X. Li, M. Tian, A. Lu, C. Lu and Y. Liu, Int. J. Nanomed., 2024, 19, 743–758 CrossRef CAS PubMed.
- Z. Naseri, R. K. Oskuee, M. R. Jaafari and M. Forouzandeh Moghadam, Int. J. Nanomed., 2018, 13, 7727–7747 CrossRef CAS PubMed.
- T. G. Nguyen Cao, J. H. Kang, J. Y. You, H. C. Kang, W. J. Rhee, Y. T. Ko and M. S. Shim, ACS Appl. Mater. Interfaces, 2021, 13, 25575–25588 CrossRef CAS PubMed.
- S. Ohno, M. Takanashi, K. Sudo, S. Ueda, A. Ishikawa, N. Matsuyama, K. Fujita, T. Mizutani, T. Ohgi, T. Ochiya, N. Gotoh and M. Kuroda, Mol. Ther., 2013, 21, 185–191 CrossRef CAS PubMed.
- Y. Wang, X. Chen, B. Tian, J. Liu, L. Yang, L. Zeng, T. Chen, A. Hong and X. Wang, Theranostics, 2017, 7, 1360–1372 CrossRef CAS PubMed.
- Y. Wan, L. Wang, C. Zhu, Q. Zheng, G. Wang, J. Tong, Y. Fang, Y. Xia, G. Cheng, X. He and S. Y. Zheng, Cancer Res., 2018, 78, 798–808 CrossRef CAS PubMed.
- M. Wu, Z. Zhang, D. Li, X. Ruan, J. Yang, S. Chen, X. Li and W. Ling, Theranostics, 2025, 15, 68–85 CrossRef CAS PubMed.
- M. Sheykhhasan, N. Kalhor, A. Sheikholeslami, M. Dolati, E. Amini and H. Fazaeli, BioMed Res. Int., 2021, 2021, 5516078 CrossRef PubMed.
- Q. Cheng, Z. Dai, G. Smbatyan, A. L. Epstein, H. J. Lenz and Y. Zhang, Mol. Ther., 2022, 30, 3066–3077 CrossRef CAS PubMed.
- X. Shi, Q. Cheng, T. Hou, M. Han, G. Smbatyan, J. E. Lang, A. L. Epstein, H. J. Lenz and Y. Zhang, Mol. Ther., 2020, 28, 536–547 CrossRef CAS PubMed.
- Z. Guo, G. Li, L. Shen, J. Pan, D. Dou, Y. Gong, W. Shi, Y. Sun, Y. Zhang, K. Ma, C. Cui, W. Li, Q. Liu and X. Zhu, Int. J. Nanomed., 2025, 20, 1147–1169 CrossRef CAS PubMed.
- M. D. Hade, C. N. Suire and Z. Suo, Cells, 2021, 10, 1959 CrossRef CAS PubMed.
- A. Alptekin, M. Parvin, H. I. Chowdhury, M. H. Rashid and A. S. Arbab, Immunol. Rev., 2022, 312, 76–102 CrossRef CAS PubMed.
- J. Radler, D. Gupta, A. Zickler and S. E. Andaloussi, Mol. Ther., 2023, 31, 1231–1250 CrossRef PubMed.
Footnotes |
| † These authors contributed equally. |
| ‡ Lead contact. |
| This journal is © The Royal Society of Chemistry 2026 |