Open Access Article
Open Access ArticleHijacking exosome biogenesis: viral glycoproteins as modular scaffolds for engineering functionalized extracellular vesicles
Daniel Levy†
a,
David Wang† bcd,
Haseeb Afzalia,
Mai Anh Doa,
Jiayi Zhanga,
Renceh Flojoa,
Joy Kua,
Kyle Asanoa,
David Diebolda,
Aijun Wang
bcd,
Haseeb Afzalia,
Mai Anh Doa,
Jiayi Zhanga,
Renceh Flojoa,
Joy Kua,
Kyle Asanoa,
David Diebolda,
Aijun Wang *bcd and
Biao Lu
*bcd and
Biao Lu *a
*a
aDepartment of Bioengineering, School of Engineering, Santa Clara University, 500 El Camino Real, Santa Clara, CA 95053, USA. E-mail: dhlevy@alumni.scu.edu; hafzali@scu.edu; mdo@alumni.scu.edu; jzhang10@alumni.scu.edu; rflojo@alumni.scu.edu; jku@alumni.scu.edu; kasano@alumni.scu.edu; ddiebold@alumni.scu.edu; blu2@scu.edu
bDepartment of Surgery, University of California Davis, Sacramento, CA 95817, USA
cDepartment of Biomedical Engineering, University of California Davis, Sacramento, CA 95616, USA
dInstitute for Pediatric Regenerative Medicine, Shriners Hospitals for Children, Sacramento, CA 95817, USA. E-mail: dtwang@ucdavis.edu; aawang@health.ucdavis.edu
First published on 2nd January 2026
Abstract
Small extracellular vesicles (sEVs) are emerging as versatile, biologically derived nanocarriers for precision drug delivery, yet strategies to enhance their targeting efficiency remain limited. Inspired by viral tropism, this study investigates whether viral envelope glycoproteins (GPs) can co-opt native biogenesis pathways to functionalize sEV membranes. Diverse viral GPs from both DNA and RNA viruses—including VSVG, HSV-gpB, SARS-CoV-1 spike, and RD114A—are shown to efficiently incorporate into sEV membranes in human cells, independent of viral assembly. Live-cell confocal imaging and co-localization with canonical sEV markers (CD63, XPACK) reveals that these GPs hijacked endosomal trafficking routes to access the sEV biogenesis machinery. Strikingly, truncation of the ectodomain does not impede sEV sorting, indicating the transmembrane and cytoplasmic domains as primary determinants of incorporation. Functionally, sEV bearing VSVG exhibit over a 2-fold increase in uptake by recipient cells compared to unmodified sEVs. These findings uncover a conserved mechanism by which viral GPs exploit host sEV pathways and establish a modular strategy for sEV surface engineering. This work paves the way for the rational design of targeted, virus-inspired sEV therapeutics for cancer, neurological disease, and gene delivery applications.
Introduction
Extracellular vesicles (EVs) are a diverse group of membrane-enclosed nanoparticles involved in numerous physiological and pathological processes, including immune regulation,1,2 inflammation,3 infection,4 cancer progression,5–7 and tissue regeneration.8–10 In humans, nearly all cell types secrete EVs, which are generally categorized into two major subtypes based on their biogenesis: exosomes and microvesicles. Exosomes are small vesicles, typically 30–100 nm in diameter, formed within the endosomal system.11,12 They originate from the inward budding of the limiting membrane of late endosomes, resulting in the formation of intraluminal vesicles (ILVs) within multivesicular bodies (MVBs). Upon fusion of the MVB with the plasma membrane, ILVs are released into the extracellular space as exosomes.13,14 Given their heterogeneous composition and physical properties, exosomes are commonly referred to as small EVs (sEV) in accordance with the MISEV2023 guidelines. In contrast, microvesicles—ranging from 100–1000 nm—are generated by the outward budding and shedding of the plasma membrane.15,16 Due to their nanoscale size, intrinsic stability, low immunogenicity, and ability to cross biological barriers, EVs—especially sEVs—have emerged as promising platforms for targeted drug delivery and precision therapeutics.17–21 Nevertheless, strategies to improve their targeting specificity and cellular uptake remain a significant challenge in the field.Viral envelope glycoproteins (GPs), evolved to mediate virus–host cell interactions, offer a compelling solution.22–25 These transmembrane proteins determine viral tropism by engaging specific host receptors. Emerging evidence suggests they can also integrate into sEV membranes.26 For example, spike (S) proteins from SARS-CoV-1 and SARS-CoV-2 have been detected on sEVs, enabling ACE2-dependent targeting and immune activation.27–31 Other GPs, including HIV-1 Env and Epstein–Barr virus gp350, similarly modulate sEV biodistribution and cellular responses, highlighting their potential for sEV surface engineering.32,33 Viral GPs offer unique advantages for EV engineering as they have naturally evolved to mediate efficient membrane anchoring, vesicle trafficking, and highly specific interactions with target cell receptors. Compared to current methods such as synthetic modifications or peptide tags, GPs have the potential to enhance EV stability, tropism, and fusion capabilities.
Among viral GPs, vesicular stomatitis virus glycoprotein (VSVG) has been widely used to pseudotype viral vectors, and more recently sEVs to improve delivery of therapeutic enzymes and gene editing tools.34–37 Yet despite growing interest, the molecular mechanisms governing GP incorporation into sEVs—particularly in the absence of viral replication—remain poorly understood.38–41 Whether this process is conserved across viral families, and which domains are required for sEV targeting, are fundamental questions that have not been resolved. This gap in knowledge serves as one of several obstacles in their translation to therapeutic applications via off-target interactions and variability in incorporation efficiency across different EV types. To highlight these considerations, we provide a concise comparison of representative GPs that offer unique advantages and limitations to demonstrate the feasibility of GP integration into sEV membranes.
In this study, we systematically investigated the integration of several viral GPs into sEV membranes by expressing fluorescently tagged GPs from both RNA and DNA viruses—including VSVG, HSV-gpB, SARS-CoV-1 spike, and RD114A—in human cells.42–45 For example, VSVG enhances broad uptake but may carry immunogenic concerns. Alternatively, SARS-CoV-1 spike enables receptor-specific targeting but can be restricted by expression. Using live-cell confocal microscopy, co-localization with exosome and sEV markers, and functional uptake assays, we demonstrate that these GPs hijack endogenous exosome biogenesis pathways for membrane incorporation. Notably, we show that truncation of the ectodomain does not impair sEV sorting, implicating the transmembrane and cytoplasmic regions as key determinants. Moreover, sEVs bearing VSVG exhibited dramatically enhanced uptake in recipient cells, underscoring their potential for functionalization.
Materials and methods
Materials
Human 293T cells were obtained from Alstem Inc. (Richmond, CA). High-glucose DMEM, GlutaMax, penicillin–streptomycin, and puromycin were obtained from Gibco (Billings, MT). UltraCulture serum-free media was obtained from Lonza (Hayward, CA), while fetal bovine serum (FBS) was from HyClone Laboratories (Logan, UT). Transfection reagents—Lipofectamine 2000, FuGene6, and polyethyleneimine (PEI)—were obtained from ThermoFisher Scientific (Fremont, CA), Promega (Madison, WI), and Sigma-Aldrich (St Louis, MO), respectively. The exosome isolation reagent ExoQuick TC, ExoGlow-RNA exosome labeling kit, and exosome mark constructs (CD63 and XPACK) were obtained from System Biosciences (Palo Alto, CA). Hoechst 33342 dye was obtained from ThermoFisher Scientific. SARS-CoV-1/-2 spike-GFP fusion constructs in pcDNA3.1 vector (SARS-Linker_pcDNA3.1(+)-C-eGFP and 2019-nCov-Linker_pcDNA3.1(+)-C-eGFP) were obtained from Genscript (Piscataway, NJ).Plasmid construction
VSVG-GFP and VSVG-RFP were constructed as described previously and modified to include a puromycin resistance gene.22 RD114A glycoprotein coding sequences (pLREES114A, plasmid #17576) were obtain from Addgene (Watertown, MA), the RD114A fusion with reporter genes, such as Gaussian luciferase (gLuc), GFP, and/or RFP were created using seamless fusion techniques.46,47 The HSV-gpB-GFP fusion gene and biocistronic VSVG-RFP and CD63-GFP constructs with P2A linkers were synthesized and cloned into pcDNA3 vector via a serve by Genscript All constructs were sequence-verified and are detailed in SI1.Cell culture, transient transfection, and stable cell line generation
Human 293T cells were cultured in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM GlutaMax, and 100 U mL−1 penicillin–streptomycin, and maintained at 37 °C in a humidified atmosphere containing 5% CO2. For transient transfection, cells were seeded in 6-well plates and transfected with 1–2 µg of plasmid DNA per well using transfection reagents such as Lipofectamine 2000, FuGene6, or polyethyleneimine (PEI), as previously described.48,49 Protein expression was monitored up to 72 hours post-transfection. For stable cell line generation, transfected cells were switched to a selection medium containing 3–5 µg mL−1 puromycin 48 hours after transfection. Cells that exhibited puromycin resistance over 20 passages were deemed to be stably transformed.Nuclei staining, confocal imaging, and co-localization analysis
Nuclei were stained with Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution) for 10 minutes at 37 °C. Confocal imaging was performed using either Olympus or Leica TCS SP8 microscopes, with Hoechst fluorescence detected using DAPI excitation at 461 nm. For co-localization studies, cells were co-transfected with glycoprotein (GP) constructs and exosomal markers (e.g., CD63 or XPACK). Fluorescent signals from GFP and RFP channels were acquired under identical imaging settings. ImageJ software was used to quantify co-localization by calculating Pearson's correlation coefficient across three independent image sets. Image brightness and contrast were uniformly adjusted across each full frame to ensure consistency. Data are presented as mean ± standard deviation (SD) of the Pearson's coefficients.
1000 dilution) for 10 minutes at 37 °C. Confocal imaging was performed using either Olympus or Leica TCS SP8 microscopes, with Hoechst fluorescence detected using DAPI excitation at 461 nm. For co-localization studies, cells were co-transfected with glycoprotein (GP) constructs and exosomal markers (e.g., CD63 or XPACK). Fluorescent signals from GFP and RFP channels were acquired under identical imaging settings. ImageJ software was used to quantify co-localization by calculating Pearson's correlation coefficient across three independent image sets. Image brightness and contrast were uniformly adjusted across each full frame to ensure consistency. Data are presented as mean ± standard deviation (SD) of the Pearson's coefficients.
Small extracellular vesicle isolation
sEVs were isolated from the conditioned medium of transfected 293T cells, following a previously established protocol.50 Briefly, 293T cells were transfected with GP-expression plasmid DNAs. After 24 hours, the transfected cells were switched to serum-free UltraCulture medium to promote the secretion of GP-modified sEVs into fresh medium. The conditioned medium was collected after an additional 48-hour incubation. The collected medium was first centrifuged at 1500g for 10 minutes to remove cellular debris and then filtered through a 0.22 µm membrane to eliminate larger particles. The filtered medium was combined with ExoQuick-TC solution at a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (medium:ExoQuick-TC) and incubated overnight at 4 °C to precipitate sEVs. The sEV-enriched fraction was pelleted by centrifugation at 3000g for 90 minutes, resuspended in phosphate-buffered saline (PBS), and stored at −80 °C for subsequent analysis. Protein concentrations of sEVs were determined by BCA assay as described previously.52
1 ratio (medium:ExoQuick-TC) and incubated overnight at 4 °C to precipitate sEVs. The sEV-enriched fraction was pelleted by centrifugation at 3000g for 90 minutes, resuspended in phosphate-buffered saline (PBS), and stored at −80 °C for subsequent analysis. Protein concentrations of sEVs were determined by BCA assay as described previously.52
Nanoparticle tracking analysis (NTA)
sEV size and concentration were determined using NTA as described in prior studies.22,51 Briefly, 1 mL of diluted sEV samples was analyzed with a NanoSight LM10 instrument equipped with a 405 nm violet laser. The samples were diluted to an appropriate concentration to ensure optimal particle visibility and minimize overlapping trajectories. For each sample, three 60-second videos were recorded under consistent flow conditions to capture particle movement. The data were analyzed using NanoSight NTA software (version 3.30; Malvern P Analytical, Malvern, UK), which tracks Brownian motion of the particles to determine size distribution and concentration.Dot-blot detection of sEV markers
To detect sEV markers, ∼100 µg of extracellular vesicle (EV) isolates was applied to pre-printed dot-blot membranes embedded with antibodies specific to established sEV markers. These included CD63, CD81, ALIX, FLOT1, ICAM1, EpCAM, ANXA5, and TSG101. The immune binding and signal detection steps were carried out according to the protocol provided by the membrane manufacturer.52 This ensured specificity and reliability in identifying the targeted sEV markers.sEV uptake assays
Recipient 293T cells were cultured in serum-free UltraCulture medium and incubated with sEVs for up to 72 hours. For confocal imaging, ells were treated with GFP-tagged sEVs, and internalization was monitored at designated time points using confocal microscopy. For flow cytometry analysis, sEVs from both unmodified controls and GP-GFP-modified samples were fluorescently labeled using the ExoGlow-RNA exosome labeling kit (System Biosciences, SBI) following the manufacturer's protocol. Labeled sEVs were added to recipient cells at a final concentration of 30 μg mL−1 and incubated for 0, 5, 24, 48, or 72 hours. After incubation, cells were washed with PBS, trypsinized, and resuspended in PBS buffer. Cellular uptake of sEVs was quantified using an Accuri C6 Plus flow cytometer (BD Biosciences).50 Untreated cells were included as negative controls to account for background autofluorescence. Data were analyzed using CFlow Plus software, and results were reported as mean ± SD of fluorescence intensity from triplicate experiments.Results
Fusion gene design and molecular trafficking strategy
To investigate whether viral envelope glycoproteins (GPs) can be used to functionalize sEV membranes, we engineered a panel of expression constructs encoding full-length GPs fused to fluorescent or luminescent reporters. We selected four viral GPs representing diverse tropisms: VSVG (broad tropism),53 RD114A (hematopoietic targeting),54 SARS-CoV-1/-2 spike (epithelial targeting),55 and HSV-gpB (neuronal targeting).56 Reporter genes including Gaussia luciferase (gLuc), GFP, RFP, and puromycin resistance were fused to the C-terminus of each GP (Fig. 1A).Domain analysis revealed that all GPs shared a conserved topology, consisting of an N-terminal signal peptide, a large extracellular ectodomain, a single transmembrane helix (TMH), and a short cytoplasmic tail (Fig. 1B). This architecture orients the ectodomain on the sEV surface and positions the fused reporter internally within the sEV lumen (Fig. 1C). All constructs were designed to preserve membrane trafficking signals while enabling real-time visualization of expression, localization, and incorporation. These fusion designs provide a modular framework to study GP-mediated trafficking and facilitate sEV surface engineering with customizable functional payloads.
Viral GPs are efficiently incorporated into sEVs in transient transfected cells
To determine whether viral envelope glycoproteins (GPs) can be incorporated into sEV membranes independently of viral particle assembly, we transiently transfected 293T cells with plasmids encoding GP-reporter fusion constructs. These included VSVG-GFP-Puro, RD114A-Gluc-GFP, SARS-CoV-1 spike-GFP, and HSV-gpB-GFP. Time-course confocal imaging of cells transfected with VSVG-GFP-Puro or RD114A-Gluc-GFP revealed that both fusion proteins localized primarily to cytoplasmic puncta (arrows), with only weak fluorescence observed at the plasma membrane (arrowheads), suggesting trafficking through endosomal and exosomal compartments (Fig. 2A and B). Based on image quality and localization patterns, the 48 hours timepoint post transfection was selected for subsequent imaging of SARS-CoV-1 spike-GFP and HSV-gpB-GFP. As shown in Fig. 2C and D, both constructs displayed similar intracellular vesicular distribution.To further validate the localization of viral glycoproteins (GPs) to exosome biogenesis compartments, we co-transfected 293T cells with VSVG-RFP-Puro and either CD63-GFP or XPACK-GFP, two well-established exosomal markers. Confocal microscopy revealed strong co-localization of VSVG with both markers within intracellular vesicular structures (Fig. 2E and F), as quantified by Pearson's correlation coefficients ranging from 0.75 to 0.88 (SI Fig. S1 and S2). In a separate experiment, VSVG-RFP and CD63-GFP were co-expressed from a bicistronic cassette to normalize expression levels. Under this condition, fluorescence signals again exhibited strong co-localization, with a Pearson's coefficient of 0.876 (SI Fig. S3). These results indicate that viral GPs, such as VSVG, can effectively engage host intracellular trafficking machinery and be incorporated into sEV membranes, independent of viral particle assembly.
Stable expression of viral GPs in 293T cells confirms sEV localization
To minimize variability from transient transfection, we generated stable 293T cell lines expressing either VSVG-GFP/RFP or RD114A-Gluc-RFP using puromycin selection (Fig. 3A). Stable expression was maintained over 20 passages. Confocal imaging of these cell lines confirmed that both VSVG and RD114A predominantly localized to punctate intracellular vesicles with minimal surface expression (Fig. 3B and C), consistent with their sorting into endosomal compartments.To further validate sEV targeting, RD114A-Gluc-RFP cells were transiently co-transfected with CD63-GFP or XPACK-GFP. Co-localization analyses revealed overlapping signals within vesicular structures (Fig. 3D and E), confirming that stably expressed GPs are directed to exosome-producing compartments. These results indicate that the stably expressed viral GPs are selectively sorted into exosomal compartments, validating the use of these engineered cell lines for downstream sEV-based applications.
sEV incorporation is independent of the ectodomain of viral GPs and enables functional display
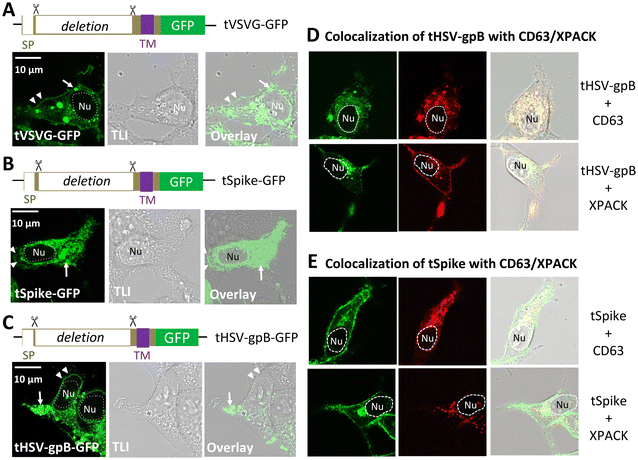
To determine whether the ectodomain is necessary for sEV targeting, we engineered truncated versions of three viral GPs: HSV-gpB, SARS-CoV-1 spike, and VSVG, by deleting their ectodomains while retaining the signal peptide (SP), transmembrane helix (TMH), and cytoplasmic domains (Fig. 4). Each of these constructs (tHSV-gpB-GFP, tSpike-GFP, and tVSVG-GFP) was transiently transfected into 293T cells. Confocal microscopy revealed that all three truncated GPs exhibited punctate vesicular localization (arrows), with limited plasma membrane signal (Fig. 4A–C, arrowheads), similar to their full-length counterparts.Similarly, co-transfection of these truncated GPs with CD63-RFP or XPACK-RFP confirmed their localization within exosome-associated compartments (Fig. 4D and E). Furthermore, a modified construct in which the VSVG ectodomain was replaced by a gLuc-Hinge fusion (gLuc-tVSVG-GFP) retained similar subcellular distribution and high colocalization with CD63-RFP and XPACK-RFP (Pearson's r = 0.911 and 0.722, respectively; Fig. S4C). These findings demonstrate that the transmembrane and cytoplasmic regions are sufficient for sEV targeting and support the use of truncated GPs as modular scaffolds for functional surface display. These results suggest that ectodomain-deleted viral GPs can function as modular scaffolds to display non-native functional proteins (e.g., reporters, ligands) on sEV membranes, expanding their utility for engineering sEV-based delivery systems.
Viral GP-modified sEVs are released and maintain molecular and structural integrity
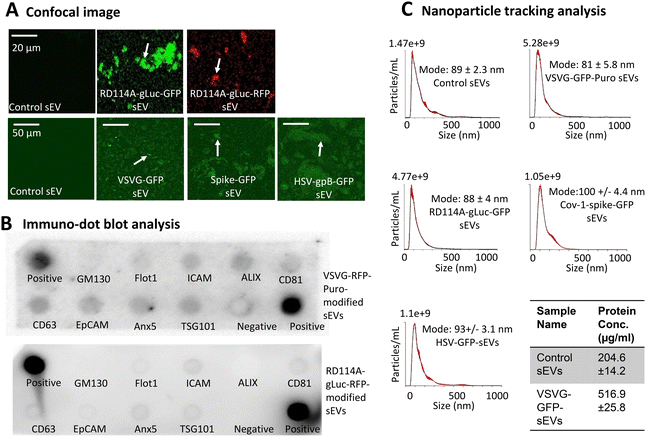
To evaluate the release and integrity of sEVs containing viral GPs, we isolated EVs from the conditioned media of 293T cells transiently expressing GP-GFP or GP-RFP constructs. Using serum-free, chemically defined medium for sEV production and a combination of centrifugation and ExoQuick precipitation for isolation, we minimized potential contamination by protein aggregates and lipoproteins from serum.34,52 Confocal imaging of the sEV preparations revealed strong fluorescence signals from RD114A-gluc-GFP/RFP, VSVG-GFP, SARS-CoV-1 spike-GFP and HSV-gpB-GFP, confirming the incorporation of viral GPs into sEV membranes (Fig. 5A). Next, dot blot analyses were performed using either VSVG-RFP-Puro- or RD114A-gLuc-RFP-moidifed sEVs. The results demonstrated the presence of established sEV markers, including CD63, CD81, ALIX, ICAM1, EpCAM, Flotillin-1, Anx5, and TSG101, and the absence of Golgi marker GM130, supporting sEV identity (Fig. 5B). Cryogenic electron microscopy (cryo-EM) of isolated sEVs showed morphological features consistent with EVs, including an intact lipid bilayer membrane and circular 2-dimensional structure (Fig. S5A). Presence of a lipid bilayer membrane was also visualized by super-resolution microscopy using a lipophilic membrane dye MemGlow700. Quantification of these images revealed that over 70% of control sEVs and viral GP-modified sEVs were lipid-positive, confirming bona fide lipid vesicles (Fig. S5B). Canonical marker positivity using dot-blot analysis is consistent with our previous report by using chip-based single-vesicle assays and also in line with recent super-resolution microscopy using detection pan-antibodies against CD9, CD63, and CD81 tetraspanin proteins. Quantification of these images revealed over 90% of sEVs to be tetraspanin positive, indicating bona fide sEVs following isolation (Fig. S5C). To further verify particle size and distribution, we carried out NTA as previously describe.52 All sEV samples exhibited a modal particle diameter in the range of 81–94 nm, consistent with the expected size distribution of sEVs (Fig. 5C). The sEV protein concentrations were 204.6 µg ml−1 and 516.9 µg ml−1 for control and VSVG-modified samples respectively, matching that of the roughly 3× sEV sample concentration difference between VSVG-GFP and control samples. This confirmed no significant changes in sEV protein-to-particle ratios following glycoprotein incorporation, indicating a maintenance of sEV purity. Together, these findings demonstrate that viral GP-modified sEVs are successfully released and retain structural integrity and molecular identity characteristics of sEVs.VSVG-modified sEVs exhibit enhanced uptake by recipient cells
To evaluate whether GP-modified sEVs retain functional activity, we assessed their uptake and intracellular routing in recipient cells. Confocal imaging revealed robust cytoplasmic accumulation of RD114A-Gluc-GFP and RD114A-Gluc-RFP in 293T cells, confirming efficient internalization (Fig. 6A). To examine intracellular trafficking, recipient cells treated with VSVG-GFP-modified sEVs were co-stained with EndoTracker-RFP and LysoTracker-RFP. Strong co-localization indicated that internalized sEVs entered the endosomal and lysosomal compartments, consistent with canonical vesicular processing (Fig. 6B).To quantitatively compare uptake efficiency, we performed flow cytometry on 293T cells incubated with either unmodified controls or VSV-G-modified sEVs for 0, 5, 24, 48, and 72 hours. Both groups showed time-dependent increases in fluorescein intensity, reaching peak levels at 48–72 hours. However, VSVG-modified sEVs exhibited significantly greater uptake at all time points, with a 14-fold increase at 5 hours and more than a 40-fold increase by 48–72 hours (Fig. 7A and B). In contrast, unmodified control sEVs reached a maximum of 18-fold enhancement (Fig. 7C and D). Comparing across experimental groups, VSVG-modified sEVs demonstrated a ∼2.33-fold increase in uptake at the 48 hours and 72 hours timepoints compared to the unmodified sEV control group. The increased cellular uptake of VSVG-modified sEVs is likely mediated by the VSVG ectodomain, as its deletion significantly reduces vesicle internalization.22 These results confirm that VSVG incorporation not only promotes cellular uptake but also facilitates efficient trafficking through endolysosomal pathways, highlighting its potential to enhance functional delivery in sEV-based applications.
Discussion
Our study reveals key molecular principles governing the integration of viral envelope glycoproteins (GPs) into sEV membranes and highlights their utility in engineering functionalized sEVs for targeted delivery. We demonstrate that envelope GPs from diverse DNA and RNA viruses—including HSV, VSV, FeLV, and SARS-CoV-1/-2—are efficiently incorporated into sEV membranes. This finding supports a shared exploitation of endogenous exosome biogenesis pathways by these viral proteins, particularly through trafficking via endosomal compartments and co-localization with exosomal markers such as CD63 and XPACK. These insights shed light on the mechanistic convergence between viral particle assembly and sEV formation.Functionally, GP-modified sEVs exhibited enhanced cellular uptake, as shown by increased internalization of VSV-G-bearing sEVs in recipient cells. Notably, deletion of the ectodomain did not disrupt sEV localization, implicating the transmembrane and cytoplasmic domains as key determinants for membrane incorporation. These findings align with our observation that the truncated VSV-G variant retained high colocalization with exosomal markers (CD63/XPACK) and enabled the surface display of imaging moieties such as gLuc. This suggests that viral GPs, even when stripped of their receptor-binding ectodomains, serve as effective scaffolds to load functional molecules onto the sEV surface.
Our results also emphasize the translational potential of viral GP-decorated sEVs. The capacity to selectively incorporate GPs from distinct viruses enables tailored cellular tropism, leveraging native viral targeting strategies—such as ACE2 recognition by SARS-CoV spike proteins or CD34+ targeting by RD114—to direct sEV delivery.57,58 This specificity is critical for applications requiring tissue-selective delivery, including cancer therapeutics, gene therapy, and treatments for neurological disorders.59–63
Moreover, the use of ectodomain-deleted GPs for sEV surface engineering enables modular design strategies, facilitating the attachment of customized functional moieties while minimizing off-target interactions. This approach broadens the therapeutic potential of sEVs not only as delivery vehicles but also as platforms for non-invasive imaging, immune modulation, and diagnostic applications.64–66 Our findings further contribute to the growing understanding of sEV-virus interplay. The efficient trafficking of viral GPs through exosomal biogenesis pathways offers a framework to study viral mimicry and may inform future antiviral strategies that selectively target this shared pathway. Engineered sEVs could also serve as tractable tools to model viral pathogenesis or to deliver therapeutic agents that interfere with viral entry mechanisms.67,68 Future studies will extend evaluation of GP-modified sEVs to primary cells and in vivo models to further assess tissue-targeting and therapeutic potential.
In conclusion, this study establishes a robust platform for sEV engineering using viral envelope glycoproteins as versatile membrane anchors. By exploiting viral tropism and the modular nature of GP domains, we provide a scalable strategy for functionalizing sEVs for diverse biomedical applications. These findings bridge virology and nanomedicine and lay the groundwork for the development of next-generation sEV-based therapeutics. Future efforts should prioritize the evaluation of targeting efficiency, immunogenicity, and safety of GP-functionalized sEVs in vivo to facilitate their clinical translation.
Author contributions
Dr. Biao Lu conceived the study, designed the experiments, coordinated the project, analyzed the data, and wrote the manuscript. Daniel Levy and Mai Anh Do established stable cell lines and performed confocal imaging experiments. Haseeb Afzali, David Wang, Jiayi Zhang, Renceh Flojo, and Joy Ku, carried out sEV isolation, imaging acquisition, and data analysis. Kyle Asano and David Diebold conducted exosome uptake assays and performed flow cytometry analysis. Daniel Levy, David Wang, and Aijun Wang contributed to data interpretation and manuscript revision. Dr. Aijun Wang co-led the study with Dr. Lu and provided conceptual insights, critical perspectives and funding support throughout the study.Conflicts of interest
All authors declare no competing conflict of interest.Data availability
All datasets generated and/or analyzed during this study have been deposited in the Dryad Digital Repository and are publicly available at: https://doi.org/10.5061/dryad.x0k6djhz1. The data include full plasmid amino acid sequences. These materials are provided to ensure transparency, reproducibility, and facilitate reuse in future research.Supplementary information (SI) is available. These data include co-localization images by confocal microscopy, cryo-TEM images, and super-resolution images of engineered sEVs. See DOI: https://doi.org/10.1039/d5nr03704b.
Acknowledgements
We thank Dr Yan Jiang for her thoughtful review and editorial assistance with the manuscript. We also acknowledge student researchers Zach Ehlinger, Anja Beard and Hanzhe Chen for their outstanding technical support in establishing stable cell lines, performing imaging acquisition and analysis, and conducting flow cytometry experiments. This work was supported by the School of Engineering at Santa Clara University and the National Institutes of Health, National Institute on Aging under Award Number R15AG092927-01 and National Institute of Neurological Disorders and Stroke under Award Number 1R01NS131538-01A1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.References
- P. Gangadaran, H. Madhyastha, R. Madhyastha, R. L. Rajendran, Y. Nakajima, N. Watanabe, A. K. G. Velikkakath, C. M. Hong, R. V. Gopi and G. K. Muthukalianan, et al., The emerging role of exosomes in innate immunity, diagnosis and therapy, Front. Immunol., 2022, 13, 1085057 Search PubMed.
- E. I. Buzas, The roles of extracellular vesicles in the immune system, Nat. Rev. Immunol., 2023, 23(4), 236–250 Search PubMed.
- B. D. Chan, W. Y. Wong, M. M. Lee, W. C. Cho, B. K. Yee, Y. W. Kwan and W. C. Tai, Exosomes in Inflammation and Inflammatory Disease, Proteomics, 2019, 19(8), e1800149 Search PubMed.
- H. M. Hosseini, A. A. Fooladi, M. R. Nourani and F. Ghanezadeh, The role of exosomes in infectious diseases, Inflammation Allergy:Drug Targets, 2013, 12(1), 29–37 Search PubMed.
- R. Kalluri and K. M. McAndrews, The role of extracellular vesicles in cancer, Cell, 2023, 186(8), 1610–1626 Search PubMed.
- M. Majood, S. Rawat and S. Mohanty, Delineating the role of extracellular vesicles in cancer metastasis: A comprehensive review, Front. Immunol., 2022, 13, 966661 Search PubMed.
- M. D. A. Paskeh, M. Entezari, S. Mirzaei, A. Zabolian, H. Saleki, M. J. Naghdi, S. Sabet, M. A. Khoshbakht, M. Hashemi and K. Hushmandi, et al., Emerging role of exosomes in cancer progression and tumor microenvironment remodeling, J. Hematol. Oncol., 2022, 15(1), 83 Search PubMed.
- J. Basu and J. W. Ludlow, Exosomes for repair, regeneration and rejuvenation, Expert Opin. Biol. Ther., 2016, 16(4), 489–506 Search PubMed.
- R. Long and S. Wang, Exosomes from preconditioned mesenchymal stem cells: Tissue repair and regeneration, Regener. Ther., 2024, 25, 355–366 Search PubMed.
- Y. An, S. Lin, X. Tan, S. Zhu, F. Nie, Y. Zhen, L. Gu, C. Zhang, B. Wang and W. Wei, et al., Exosomes from adipose-derived stem cells and application to skin wound healing, Cell Proliferation, 2021, 54(3), e12993 Search PubMed.
- J. A. Welsh, D. C. I. Goberdhan, L. O'Driscoll, E. I. Buzas, C. Blenkiron, B. Bussolati, H. Cai, D. Di Vizio, T. A. P. Driedonks and U. Erdbrugger, et al., Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches, J. Extracell. Vesicles, 2024, 13(2), e12404 Search PubMed.
- D. M. Pegtel and S. J. Gould, Exosomes, Annu. Rev. Biochem., 2019, 88, 487–514 Search PubMed.
- M. Xu, J. Ji, D. Jin, Y. Wu, T. Wu, R. Lin, S. Zhu, F. Jiang, Y. Ji and B. Bao, et al., The biogenesis and secretion of exosomes and multivesicular bodies (MVBs): Intercellular shuttles and implications in human diseases, Genes Dis., 2023, 10(5), 1894–1907 Search PubMed.
- Q. F. Han, W. J. Li, K. S. Hu, J. Gao, W. L. Zhai, J. H. Yang and S. J. Zhang, Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer, Mol. Cancer, 2022, 21(1), 207 Search PubMed.
- J. C. Akers, D. Gonda, R. Kim, B. S. Carter and C. C. Chen, Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus-like vesicles, and apoptotic bodies, J. Neurooncol., 2013, 113(1), 1–11 Search PubMed.
- J. W. Clancy, M. Schmidtmann and C. D'Souza-Schorey, The ins and outs of microvesicles, FASEB BioAdv., 2021, 3(6), 399–406 Search PubMed.
- R. Kalluri and V. S. LeBleu, The biology, function, and biomedical applications of exosomes, Science, 2020, 367(6478), 6977 Search PubMed.
- J. Rezaie, M. Feghhi and T. Etemadi, A review on exosomes application in clinical trials: perspective, questions, and challenges, Cell Commun. Signaling, 2022, 20(1), 145 Search PubMed.
- H. I. Kim, J. Park, Y. Zhu, X. Wang, Y. Han and D. Zhang, Recent advances in extracellular vesicles for therapeutic cargo delivery, Exp. Mol. Med., 2024, 56(4), 836–849 Search PubMed.
- F. U. Rehman, Y. Liu, M. Zheng and B. Shi, Exosomes based strategies for brain drug delivery, Biomaterials, 2023, 293, 121949 Search PubMed.
- O. M. Elsharkasy, J. Z. Nordin, D. W. Hagey, O. G. de Jong, R. M. Schiffelers, S. E. Andaloussi and P. Vader, Extracellular vesicles as drug delivery systems: Why and how?, Adv. Drug Delivery Rev., 2020, 159, 332–343 Search PubMed.
- C. Meyer, J. Losacco, Z. Stickney, L. Li, G. Marriott and B. Lu, Pseudotyping exosomes for enhanced protein delivery in mammalian cells, Int. J. Nanomed., 2017, 12, 3153–3170 Search PubMed.
- I. K. Herrmann, M. J. A. Wood and G. Fuhrmann, Extracellular vesicles as a next-generation drug delivery platform, Nat. Nanotechnol., 2021, 16(7), 748–759 Search PubMed.
- S. Hong, S. Ruan, Z. Greenberg, M. He and J. L. McGill, Development of surface engineered antigenic exosomes as vaccines for respiratory syncytial virus, Sci. Rep., 2021, 11(1), 21358 Search PubMed.
- M. Richter, P. Vader and G. Fuhrmann, Approaches to surface engineering of extracellular vesicles, Adv. Drug Delivery Rev., 2021, 173, 416–426 Search PubMed.
- S. Bansal, S. Perincheri, T. Fleming, C. Poulson, B. Tiffany, R. M. Bremner and T. Mohanakumar, Cutting Edge: Circulating Exosomes with COVID Spike Protein Are Induced by BNT162b2 (Pfizer-BioNTech) Vaccination prior to Development of Antibodies: A Novel Mechanism for Immune Activation by mRNA Vaccines, J. Immunol., 2021, 207(10), 2405–2410 Search PubMed.
- E. Pesce, N. Manfrini, C. Cordiglieri, S. Santi, A. Bandera, A. Gobbini, P. Gruarin, A. Favalli, M. Bombaci and A. Cuomo, et al., Exosomes Recovered From the Plasma of COVID-19 Patients Expose SARS-CoV-2 Spike-Derived Fragments and Contribute to the Adaptive Immune Response, Front. Immunol., 2021, 12, 785941 Search PubMed.
- S. K. Tey, H. Lam, S. W. K. Wong, H. Zhao, K. K. To and J. W. P. Yam, ACE2-enriched extracellular vesicles enhance infectivity of live SARS-CoV-2 virus, J. Extracell. Vesicles, 2022, 11(5), e12231 Search PubMed.
- E. Barberis, V. V. Vanella, M. Falasca, V. Caneapero, G. Cappellano, D. Raineri, M. Ghirimoldi, V. De Giorgis, C. Puricelli and R. Vaschetto, et al., Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection, Front. Mol. Biosci., 2021, 8, 632290 Search PubMed.
- L. Li, Z. Yang and J. Li, Exosomes and SARS-CoV-2 infection, Front. Immunol., 2024, 15, 1467109 Search PubMed.
- Y. Fu and S. Xiong, Tagged extracellular vesicles with the RBD of the viral spike protein for delivery of antiviral agents against SARS-COV-2 infection, J. Controlled Release, 2021, 335, 584–595 Search PubMed.
- A. Arakelyan, W. Fitzgerald, S. Zicari, C. Vanpouille and L. Margolis, Extracellular Vesicles Carry HIV Env and Facilitate Hiv Infection of Human Lymphoid Tissue, Sci. Rep., 2017, 7(1), 1695 Search PubMed.
- H. Vallhov, C. Gutzeit, S. M. Johansson, N. Nagy, M. Paul, Q. Li, S. Friend, T. C. George, E. Klein and A. Scheynius, et al., Exosomes containing glycoprotein 350 released by EBV-transformed B cells selectively target B cells through CD21 and block EBV infection in vitro, J. Immunol., 2011, 186(1), 73–82 Search PubMed.
- M. A. Do, D. Levy, A. Brown, G. Marriott and B. Lu, Targeted delivery of lysosomal enzymes to the endocytic compartment in human cells using engineered extracellular vesicles, Sci. Rep., 2019, 9(1), 17274 Search PubMed.
- P. Gee, M. S. Y. Lung, Y. Okuzaki, N. Sasakawa, T. Iguchi, Y. Makita, H. Hozumi, Y. Miura, L. F. Yang and M. Iwasaki, et al., Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping, Nat. Commun., 2020, 11(1), 1334 Search PubMed.
- X. Zhang, Q. Xu, Z. Zi, Z. Liu, C. Wan, L. Crisman, J. Shen and X. Liu, Programmable Extracellular Vesicles for Macromolecule Delivery and Genome Modifications, Dev. Cell, 2020, 55(6), 784–801 Search PubMed.
- J. A. Whitley and H. Cai, Engineering extracellular vesicles to deliver CRISPR ribonucleoprotein for gene editing, J. Extracell. Vesicles, 2023, 12(9), e12343 Search PubMed.
- D. Cortes-Galvez, J. A. Dangerfield and C. Metzner, Extracellular Vesicles and Their Membranes: Exosomes vs. Virus-Related Particles, Membranes, 2023, 13(4), 397 Search PubMed.
- P. Chaudhari, V. Ghate, M. Nampoothiri and S. Lewis, Multifunctional role of exosomes in viral diseases: From transmission to diagnosis and therapy, Cell Signalling, 2022, 94, 110325 Search PubMed.
- E. Nolte-'t Hoen, T. Cremer, R. C. Gallo and L. B. Margolis, Extracellular vesicles and viruses: Are they close relatives?, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(33), 9155–9161 Search PubMed.
- M. H. Saad, R. Badierah and E. M. Redwan, El-Fakharany EM: A Comprehensive Insight into the Role of Exosomes in Viral Infection: Dual Faces Bearing Different Functions, Pharmaceutics, 2021, 13(9), 1405 Search PubMed.
- G. J. Letchworth and L. L. Rodriguez, Del cbarrera J: Vesicular stomatitis, Vet. J., 1999, 157(3), 239–260 Search PubMed.
- K. Madavaraju, R. Koganti, I. Volety, T. Yadavalli and D. Shukla, Herpes Simplex Virus Cell Entry Mechanisms: An Update, Front. Cell. Infect. Microbiol., 2020, 10, 617578 Search PubMed.
- V. C. Cheng, S. K. Lau, P. C. Woo and K. Y. Yuen, Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection, Clin. Microbiol. Rev., 2007, 20(4), 660–694 Search PubMed.
- E. A. Hoover and J. I. Mullins, Feline leukemia virus infection and diseases, J. Am. Vet. Med. Assoc., 1991, 199(10), 1287–1297 Search PubMed.
- C. Uhde-Stone, N. Sarkar, T. Antes, N. Otoc, Y. Kim, Y. J. Jiang and B. Lu, A TALEN-based strategy for efficient bi-allelic miRNA ablation in human cells, RNA, 2014, 20(6), 948–955 Search PubMed.
- N. Curley, D. Levy, M. A. Do, A. Brown, Z. Stickney, G. Marriott and B. Lu, Sequential deletion of CD63 identifies topologically distinct scaffolds for surface engineering of exosomes in living human cells, Nanoscale, 2020, 12(22), 12014–12026 Search PubMed.
- C. Olson, P. Zhang, J. Ku, R. Flojo, D. Boyes and B. Lu, A Novel Dual-Reporter System Reveals Distinct Characteristics of Exosome-Mediated Protein Secretion in Human Cells, Biol. Proced. Online, 2023, 25(1), 25 Search PubMed.
- D. Levy, M. A. Do, J. Zhang, A. Brown and B. Lu, Orchestrating Extracellular Vesicle With Dual Reporters for Imaging and Capturing in Mammalian Cell Culture, Front. Mol. Biosci., 2021, 8, 680580 Search PubMed.
- J. Zhang, A. Brown, B. Johnson, D. Diebold, K. Asano, G. Marriott and B. Lu, Genetically Engineered Extracellular Vesicles Harboring Transmembrane Scaffolds Exhibit Differences in Their Size, Expression Levels of Specific Surface Markers and Cell-Uptake, Pharmaceutics, 2022, 14(12), 2564 Search PubMed.
- C. Olson, K. Ivanov, D. Boyes, D. Bengford, J. Ku, R. Flojo, P. Zhang and B. Lu, Dual-Omics Approach Unveils Novel Perspective on the Quality Control of Genetically Engineered Exosomes, Pharmaceutics, 2024, 16(6), 824 Search PubMed.
- N. Duong, K. Curley, A. Brown, A. Campanelli, M. A. Do, D. Levy, A. Tantry, G. Marriott and B. Lu, Decoy exosomes as a novel biologic reagent to antagonize inflammation, Int. J. Nanomed., 2019, 14, 3413–3425 Search PubMed.
- E. Hastie, M. Cataldi, I. Marriott and V. Z. Grdzelishvili, Understanding and altering cell tropism of vesicular stomatitis virus, Virus Res., 2013, 176(1–2), 16–32 Search PubMed.
- A. J. Bell Jr., D. Fegen, M. Ward and A. Bank, RD114 envelope proteins provide an effective and versatile approach to pseudotype lentiviral vectors, Exp. Biol. Med., 2010, 235(10), 1269–1276 Search PubMed.
- R. Peng, L. A. Wu, Q. Wang, J. Qi and G. F. Gao, Cell entry by SARS-CoV-2, Trends Biochem. Sci., 2021, 46(10), 848–860 Search PubMed.
- T. Suenaga, T. Satoh, P. Somboonthum, Y. Kawaguchi, Y. Mori and H. Arase, Myelin-associated glycoprotein mediates membrane fusion and entry of neurotropic herpesviruses, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(2), 866–871 Search PubMed.
- A. G. Wrobel, Mechanism and evolution of human ACE2 binding by SARS-CoV-2 spike, Curr. Opin. Struct. Biol., 2023, 81, 102619 Search PubMed.
- V. Sandrin, B. Boson, P. Salmon, W. Gay, D. Negre, R. Le Grand, D. Trono and F. L. Cosset, Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34 + cells derived from human and nonhuman primates, Blood, 2002, 100(3), 823–832 Search PubMed.
- R. Cecchin, Z. Troyer, K. Witwer and K. V. Morris, Extracellular vesicles: The next generation in gene therapy delivery, Mol. Ther., 2023, 31(5), 1225–1230 Search PubMed.
- Z. Xu, S. Zeng, Z. Gong and Y. Yan, Exosome-based immunotherapy: a promising approach for cancer treatment, Mol. Cancer, 2020, 19(1), 160 Search PubMed.
- D. J. Beetler, D. N. Di Florio, K. A. Bruno, T. Ikezu, K. L. March, L. T. Cooper Jr., J. Wolfram and D. Fairweather, Extracellular vesicles as personalized medicine, Mol. Aspects Med., 2023, 91, 101155 Search PubMed.
- F. Tan, X. Li, Z. Wang, J. Li, K. Shahzad and J. Zheng, Clinical applications of stem cell-derived exosomes, Signal Transduction Targeted Ther., 2024, 9(1), 17 Search PubMed.
- B. Lu, J. Ku, R. Flojo, C. Olson, D. Bengford and G. Marriott, Exosome- and extracellular vesicle-based approaches for the treatment of lysosomal storage disorders, Adv. Drug Delivery Rev., 2022, 188, 114465 Search PubMed.
- M. Sancho-Albero, N. Ayaz, V. Sebastian, C. Chirizzi, M. Encinas-Gimenez, G. Neri, L. Chaabane, L. Lujan, P. Martin-Duque and P. Metrangolo, et al., Superfluorinated Extracellular Vesicles for In Vivo Imaging by (19)F-MRI, ACS Appl. Mater. Interfaces, 2023, 15(7), 8974–8985 Search PubMed.
- A. T. Gharavi, N. A. Hanjani, E. Movahed and M. Doroudian, The role of macrophage subtypes and exosomes in immunomodulation, Cell. Mol. Biol. Lett., 2022, 27(1), 83 Search PubMed.
- B. Zhou, K. Xu, X. Zheng, T. Chen, J. Wang, Y. Song, Y. Shao and S. Zheng, Application of exosomes as liquid biopsy in clinical diagnosis, Signal Transduction Targeted Ther., 2020, 5(1), 144 Search PubMed.
- M. Jamalkhah, Y. Asaadi, M. Azangou-Khyavy, J. Khanali, M. Soleimani, J. Kiani and E. Arefian, MSC-derived exosomes carrying a cocktail of exogenous interfering RNAs an unprecedented therapy in era of COVID-19 outbreak, J. Transl. Med., 2021, 19(1), 164 Search PubMed.
- D. El Safadi, A. Mokhtari, M. Krejbich, A. Lagrave, U. Hirigoyen, G. Lebeau, W. Viranaicken and P. Krejbich-Trotot, Exosome-Mediated Antigen Delivery: Unveiling Novel Strategies in Viral Infection Control and Vaccine Design, Vaccines, 2024, 12(3), 280 Search PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2026 |