Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
S-Adenosylmethionine: considerations on its role in the origin and evolution of life
Andreas Kirschning
 ab
ab
aInstitute of Organic Chemistry, Leibniz Universität Hannover, Schneiderberg 1B, D-30167 Hannover, Germany. E-mail: andreas.kirschning@oci.uni-hannover.de; Fax: (+49)-(0)511-762 3011; Tel: (+49)-(0)511-762 4614
bUppsala Biomedical Center (BMC), Uppsala University, Husargatan 3, 752 37 Uppsala, Sweden
First published on 19th January 2026
Abstract
Covering: up to 2025
S-Adenosylmethionine (SAM) belongs to the class of group-transferring coenzymes, whereby alkyl group transfers, especially electrophilic methylations, on the one hand, and radical reactions, which are characterised by initial H radical abstractions, on the other hand, are predominant. From an evolutionary point of view, these types of reactions are fundamental e.g. in the modification of nucleobases and fatty acids but also in methionine biosynthesis. At which point of chemical and biological evolution did SAM come into play? Since SAM is closely tied to nucleotide biochemistry both structurally and biosynthetically, a discussion linking it to RNA appears to be reasonably. Apart from general overviews of the early evolutionary role of coenzymes and cofactors, the appearance of SAM on the evolutionary stage has only been dealt with superficially so far. This report attempts to achieve such a classification, both prebiotically and biosynthetically within the RNA world theory.
1. Introduction
As early as 1976, White III proposed that coenzymes are the surviving vestiges of nucleic acid enzymes due to the fact that many coenzymes contain structural elements derived from RNA nucleotides.1 He claimed that their occurrence preceded the evolution of ribosomal protein synthesis. Although this led to some profound reflections in the years that followed,2,3 this earlier insight only recently gained momentum with respect to hypotheses on the origin of life.4 In fact, coenzymes and cofactors must be very ancient, because (a) they occur in all kingdoms of life, (b) the Last Universal Common Ancestor (LUCA) model,5 a theoretical one recently substantiated by phylogenetic and bioinformatics analyses, must have used the majority of coenzymes and cofactors known today6 and (c) the biosynthesis of proteinogenic amino acids and the enzymatic formation of coenzymes represent a classical case of a “chicken and egg” problem.7 From a chemical point of view, they can be considered the most chemical species that nature has developed, since their almost unaltered sole purpose is to act as (co-) catalysts for enzymes, and this was probably also the case in LUCA Fig. 1.5,8S-Adenosylmethionine (SAM, 1)9 falls precisely into this category, as it contains an adenosine group and thus belongs to those small molecules that fit into the concept of the RNA-first theory.10 Other examples of coenzymes containing adenosine are nicotinamide dinucleotide (NAD+), flavin adenine dinucleotide (FAD) and coenzyme A (CoASH). Associated with iron–sulfur clusters, SAM is also involved in radical chemistry. In addition, iron–sulfur complexes are suggested to be the oldest redox systems present when Life emerged on planet earth.11 In view of the close relationship with S-adenosylmethionine in radical SAM biochemistry,12 a discussion on SAM in the context of the origin and evolution of Life seems to be overdue.
2. Coenzymes including SAM and the RNA world theory
The history of Earth is commonly divided into four major phases, each of which is further subdivided. The four geological eons are the Hadean (4.6–4.0 Ga ref. 13), the Archaen (4.0–2.5 Ga), the Proterozoic (2.5 Ga-538.8 Ma), and the current geologic eon called Phanerozoic.Prebiotic evolution began under conditions that existed during the Hadean Earth and provided the first organic molecules and early pre-metabolic networks. Dramatic events separate the Archaen eon from the Proterozoic, and the Proterozoic in turn from the Phanerozoic. Both are associated with the sudden and substantial increase of oxygen concentration in the atmosphere and, in part, in dissolved form in the oceans.14 Several theories on the origin of life are being discussed, including the topic of prebiotic chemistry and, building on this, the question of which of the three central biomacromolecules of life (proteins, DNA, and RNA) appeared first on Earth, supposedly in the Archaen eon. This has led to the emergence of the RNA world theory. Its scenario states that RNA with a defined sequence must have preceded proteins with a defined sequence. This assumption is based on the fact that, unlike proteins, RNA molecules have the ability to self-replicate and also perform the function of transcribing protein structures. Furthermore, RNA exhibits catalytic properties, and so-called ribozymes can perform a limited number of chemical transformations. This is evident, for example, in the ability of RNAs to catalyze their own synthesis from activated monomers, as has been demonstrated in vitro.15 This dual role forms the basis for the RNA-first theory. In essence, RNA combines elements of coding with metabolic properties. Accordingly, it is assumed that RNA played a key role in the evolution of prebiotic replication.16 This assumption has recently found support by chemical approaches to the synthesis of nucleotides under plausible prebiotic conditions.16
However, without metal or co-enzymes, the variety of known ribozyme-promoted reactions is very limited and is based mainly on acid-base-catalysed hydrolysis and a few group transfer reactions of nucleotide and peptide elements. Redox chemistry, alkylations and group transfer chemistry require co-catalysts or coenzymes, including S-adenosylmethionine (1).
If RNA was indeed a precursor to the protein-based enzymes that appeared later and thus played a decisive role in governing pre-metabolism, the reaction repertoire of RNA must had been considerably larger. It can therefore be assumed that small, co-catalytically active molecules ought to have been around that provided functional co-ribozymes through covalent, coordinative or hydrogen bonds to RNA. This brings coenzymes and metallic cofactors into play, especially as some coenzymes contain the adenosine phosphate handle. Several cases have been described in which the weak binding of small molecules to the RNA template has been achieved or demonstrated.17 Alternatively, adenosyl-containing coenzymes can themselves be regarded as nucleotide building blocks which, when covalently bound to the 5′-terminus of a ribozyme, would in fact resemble catalytically active nucleotide building blocks.
Today, the concept of RNA able to bind small molecules is manifested in small mRNAs called riboswitches.18 This includes several coenzymes like thiamine pyrophosphate (TPP),19 flavine mononucleotide (FMN),20 tetrahydrofolate (THF),21 adenosyl cobalamine (AdoCbl),22 SAM23 and others.24 Binding of metabolites occurs directly via electrostatic ineractions or through hydrogen bonding, without be induced by proteins. Upon binding, the secondary RNA structure is altered resulting in a change in production of the proteins encoded by this mRNA.25 This is how SAM biosynthesis is regulated by SAM riboswitches in bacteria.23 SAM itself is biosynthesised by methionine adenosyltransferase from L-methionine (4) and adenosine triphosphate (ATP). After SAM 1, bound in the corresponding protein matrix, has transferred its methyl group to a nucleophilic substrate, S-adenosyl-L-homocysteine (SAH, 2) remains in the enzyme pocket and has to be regenerated to 1 via several enzymatic steps. Most organisms convert homocysteine into methionine using methylfolate or methylcobalamin (MeCbl) as methyl donors.26,27 It is known that several classes of SAM riboswitches act as control points in this “SAM cycle.” (Scheme 1A).28
Assuming that SAM was already present at an early stage of life (a detailed discussion can be found in Section 4), two scenarios are conceivable as to how a ribozyme can become a catalytic system with alkylating properties. On the one hand, SAM can be bound to a protoribozyme via weak interactions, as is currently the case in SAM riboswitches 5. Could there be other possible grounds for the existence of an early and active form of SAM? The “AMP handle” present in some coenzymes and adenosine in SAM point not only structurally but also functionally to the RNA world theory, because adenine can be actively associated with RNA by pairing with the base uracil. It is also noteworthy that nicotinamide (in NADH) forms hydrogen bonds with uracil and cytosine (Scheme 1B) so that nicotinamide was designated as a possible pre-genetic code dehydrogenase.3d–f Indeed, Breaker expressed the idea early on that some riboswitch aptamers, especially those that bind coenzymes, could be derived from early ribozymes.25 Either they were responsible for the synthesis of coenzymes, or they served as RNA-coenzyme complexes for the catalysis of metabolic reactions. In either case, a coenzyme-dependent ribozyme as well as a coenzyme-responsive riboswitch would have to have a coenzyme binding site.
Recently, Höbartner and colleagues described a ribozyme that, by binding SAM, is able to alkylate RNA in a site-specific manner, thus supporting the theory presented here of the close evolutionary entanglement of ribozymes and coenzymes.29
Assuming that SAM was already present at an early stage of life (a detailed discussion can be found in Section 4), two scenarios are conceivable as to how a ribozyme can become a catalytic system with alkylating properties (Scheme 1C1 and C2). On the one hand, SAM can be bound to a protoribozyme via weak interactions, as is currently the case in SAM riboswitches 5. In contrast to riboswitches, this complex acts as a chemically active species for the alkylation of substrates of an early stage of metabolism. Alternatively, ribozymes could bind methionine at the perphosphorylated 5′-terminal end of the ribozyme. The resuting sulfonium species 6 is in essence SAM with an oligonucleotide. In fact, it has been demonstrated that methionine30 reacts spontaneously with ATP, but also with adenosyl monophosphate which is more relevant from a prebiotic perspective.31 However, these approaches have a fundamental weakness if one includes the question of regeneration, i.e., the reloading with a methyl group. At this point, it is necessary to consider plausible prebiotic chemistry.
3. Prebiotic SAM models
3.1 Electrophilic methylations
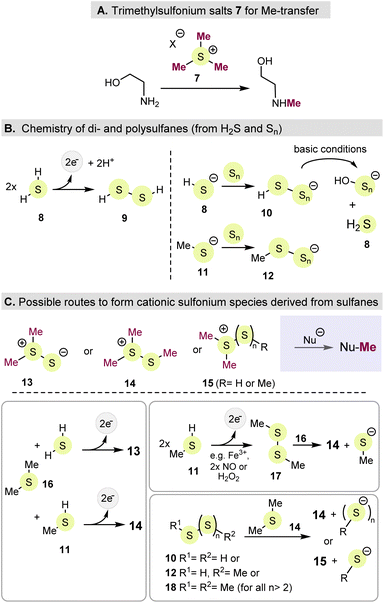
The primary metabolic role of SAM is methyl group transfer onto nucleobases in DNA and tRNA32 as well as onto homocysteine, guanidine acetate and α-amino acids in proteins. Other methyl acceptors are aminoalcohols such aminoethanol and the precursor of epinephrine. Also methylations of unsaturated fatty acids are driven by SAM leading to branched or cyclopropyl fatty acids. In search for simpler, prebiotic models of SAM, first experimental studies on the methylation of ethanol amine set the trimethylsulfonium ion (Me3S+, 7) into focus.33 The chemoselective methylation was successful with 7 conducted under dilute, aqueous conditions (Scheme 2A).However, the probable origin of these cationic species under prebiotic conditions has not yet been clarified, as a chemical route would have to start from dimethyl sulfide 16. While 16, like thiomethanol 11, are products of deep-sea vents, the source of the third methyl group is unclear, which calls into question the legitimacy of 7 as a plausible prebiotic source of an electrophilic methyl group.33 Alternatively, formaldehyde (CH2O)34,35 and diazomethane36 were proposed as ancient methylating agents. The former was used in N-methylations of primary amines via intermediate imine formation, while the latter was formed under presumably prebiotic conditions, but due to its high chemical instability, the relevance of diazomethane on early Earth can be critically dicussed.
3.2 Sulfanes as precursors for mercaptodimethylsulfonium cations – a “Gedankenexperiment”
An alternative to trialkylsulfonium species would be to replace one of the three methyl groups in 7 with a heteroatom-bearing substituent such as sulfur or nitrogen.37 The early world was very likely marked by the presence of sulfur and this element played a key role in prebiotic chemistry as it was supposedly present in multiple forms such H2S 8 and elemental Sn as well as SO2, sulfite, sulfate and thiosulfate.38 In addition, dimethyl sulfide 16 and thiomethanol 11 are found to be present in deep sea vent systems (black smoker) which make them candidates already present in a prebiotic world.39It is proposed here, that the mercaptodimethylsulfonium species 13–15 could be sensible alternatives to the classic trialkylsulfonium species (e.g. 7) as well as in SAM 1. Here, a thio substituent is part of the substitution sphere around the central sulfonium species. But how can such sulfonium species be generated under plausible prebiotic conditions and are they capable of performing methyl transfer chemistry? Polysulfanes 10 or 18 can serve as starting point for conducting such a “Gedankenexperiment” (Scheme 2B).
In analogy to alkanes,40 polysulfides are also called sulfanes or polysulfanes for which H2(S)n 10 and 12 are typical representatives. But investigations on this special sulfur geochemistry are still in their infancy including considerations linked to the origin of Life. Sulfanes are relevant for pyrite formation and metal chelation. They are stable under alkaline conditions, but below a neutral pH value, polysulfides form molecular sulfur S8. In prebiotic times, the S–S bonds in di- (HSSH 9 or MeSSMe 17) and polysulfanes 10, 12 and 18 could have formed from the corresponding thiols (e.g. MeSH),41 but also from H2S by various oxidizing agents such as H2O2, NO, or by trace metals such as Fe(III).42 In this context, H2S 8 plays a central role in the initiation and control of complex sulfane equilibria. Furthermore, polysulfanes 10 and 12 can also form when hydrogen sulfide reacts with elemental sulfur (Sn).43
In theory this dynamic system can serve as initiation point for the formation of sulfonium species 13–15. E.g. if dimethyl sulfide reacted with electrophilic sulfur present in polysulfanes, these type of sulfonium species would be formed and would, in principle, be capable of undergoing methyl transfer reactions in the same way as sulfonium species like 7 or SAM 1.
What evidence would support this hypothesis? Dimethyl(methylthio)sulfonium salts (14 or 15) are well established and are usually obtained by methylation of disulfides or by reactions of thiols with electrophilic thiols (e.g. sulfenyl chlorides); these represent the disulfides or polysulfanes suggested here (Scheme 2B and C).44a,b It has also been shown that these cationic species can act as alkylating agents with various nucleophiles,44c confirming that these salts can in principle be regarded as prebiotic SAM substitutes.
Interestingly, it has been shown that dimethyl(methylthio)sulfonium salts such as 14 form trimethylsulfonium salt 7 under the influence of organic trialkyl or triphenyl phosphites. The corresponding phosphorothioate triester is formed as a by-product.45 Is this the missing link to the simplest prebiotic analogue of SAM, sulfonium cation 7? However, in order to draw this conclusion, prebiotic equivalents of such phosphites must first be found.
An ideal pre-geological location where one could imagine this type of chemistry taking place are hydrothermal fields with periodical “wet-and-dry” cycles. Such oscillating cycles in hydrothermal ponds have been identified as crucial environments for promoting prebiotic chemistry, particularly for condensation and oligomerization processes (such as peptides, RNA) during dry periods. In contrast, colder rainy periods cause reaction partners to dissolve and mix, which in turn enables chemical reactions in solutions that lead from simple building blocks of life to more complex structures and even protocells. Conditions prevailing during the “dry phase” and high local concentration in particular could ensure the presumed formation of sulfonium cations 13–15 as proposed here.46
3.3 Radical SAM models
Iron–sulfur clusters are ancient cofactors47 that play a fundamental role in metabolism and radical SAM enzymes contain iron–sulfur clusters which act by generating radicals as a result of a homolytic cleavage of S-adenosylmethionine in SAM.48 The resulting adenosyl radical induces hydrogen atom abstractions from the substrate, leading to a plethora of subsequent reactions.This kind of chemistry may also have impacted the prebiotic chemistry that led to life. In fact several accounts showed that iron–sulfur clusters can be produced under plausible prebiotic conditions, whereby photochemical excitation of the starting components was one of the activation methods.49 This opens the gate for considering radical SAM chemistry in a prebiotic context. Iron–sulfur clusters are stabilised by a wide range of cysteine-containing peptides, where either prebiotically produced cysteine50 or thiomethanol 11 could have taken on this role before the appearance of complex peptides.
Aqueous FeS clusters, in which FeS clusters are directly hydrated by water, were first observed in seawater by Buffle et al.51a Aqueous FeS clusters are also found in marine sediments51b–d and hydrothermal vents in the deep sea. The stoichiometry of these FeS clusters appears to range from Fe2S2 to Fe150S150. The smaller aqueous clusters Fe2S2 and Fe4S4 show structures similar to those of the cluster form in FeS proteins. Further evidence that iron sulfide was available from geochemical sources during prebiotic times is revealed by the fact that the structure of Fe2S2 resembles the basic structural component of the mineral mackinawite (FeS).51e
For a prebiotic radical SAM system to exist, it would also be necessary to identify a prebiotic form of sulfonium cations for which suggestions are given above. Only in recent years has synthetic chemistry developed an awareness for radical chemistry based on sulfonium salts, whereby photochemical activation has typically been used to achieve homolytic bond cleavage.52a However, biomimetic electron transfer from a metal, has hardly been studied to date. And only a few examples of alkyl radical formation has being pursued so far;52c instead, aryl radical formation dominates the field. Since this chemistry has not yet been investigated under plausible prebiotic reaction conditions e.g., with abiotic iron–sulfur clusters, no statement can be made at this point as to whether radical SAM chemistry played a significant role at such an early stage in Earth's history.
4. SAM – an early player in biological evolution?
4.1 How “simple” is SAM biosynthesis?
The biosynthesis of SAM is based on the building blocks ATP and methionine. The coenzymes pyridoxal phosphate (PLP), NADH and THF are necessary for the regeneration of the sulfonium species, all of which may be regarded to be old with respect to their appearance, due to the simplicity of their biosynthesis with respect to coenzymes required (Scheme 3).8 | ||
| Scheme 3 Proposed timing of coenzyme biosynthesis, taking into account the necessary building blocks and coenzymes. According to ref. 6, all of these biosynthesis already existed in LUCA (R5P = ribose-5-phosphate; GAP = glyceraldehyde-3-phosphate; DHAP = 3-deoxy-D-arabino-heptulosonate-7-phosphate; OAA = oxaloacetate, PRPP = 5-phospho-D-ribose-1-diphosphate, Gly = glycine, Cys = cysteine, GTP = guanosine triphosphate, Glu = glutamic acid, PABA = para-aminobenzoic acid, Met = methionine, Thr = threonine, NAMN = nicotinamide mononucleotide, DMB = dimethyl-benzimidazole). | ||
In fact, the biosynthesis of PLP appears to be the simplest of all coenzymes and cofactors, assuming that nucleotides existed in view of the RNA world theory. Only ATP and building blocks from carbohydrate metabolism are required here. NADH is also easily accessible biosynthetically if the same type of analysis is chosen. Similarly, the biosynthesis of the simplest porphyrin, uroporphyrinogen III, and iron–sulfur clusters can be considered simple too. Next, NADH is required for the biosynthesis of THF, and hence THF is required for the regeneration process of SAM. p-Aminobenzoic acid (PABA), being structural element in THF is produced by the shikimic acid pathway, and its biosynthesis splits off from it at the stage of chorismic acid. The classic shikimate biosynthesis pathway requires a TPP-dependent transketolase step. According to this kind of analysis the biosynthesis of TPP did not necessarily appear at an early stage in evolution when THF became necessary as a coenzyme for methionine regeneration. However, an alternative, TPP-free biosynthetic approach to shikimic acid was recently discovered in Methanocaldococcus jannaschii.53 This shows that in such a case the THF-dependent regeneration system could have been already present at an early stage of a biological proto-life and this would consequently mean that the SAM biochemistry still represents a biosynthetically fairly simple system.
4.2 Tetrahydrofolic acid or cobalamin – which methyl regenerating system is older?
The above analysis demonstrates that THF is a relatively readily accessible coenzyme in terms of biosynthesis. What about cobalamin, which can serve as an alternative to THF? Unlike uroporphyrinogen III, the biosynthesis of the corinoid core in cobalamin requires up to ten equivalents of SAM and one equivalent of FeS/SAM (from uroporphyrinogen III) as well as a series of other coenzymes (Scheme 3). From an evolutionary perspective, cobalamin must therefore have appeared long after the establishment of SAM biosynthesis and consequently the cobalamin-based regeneration system from L-homocysteine 3 to SAM 4 (Scheme 1A) can be considered a “latecomer”. Thus, the tetrahydrofolate-dependent regeneration of SAM represents the older system in evolutionary terms. Another implication of this analysis is that the second adenosyl radical-induced chemistry, which utilises vitamin B12, appeared evolutionarily later than radical SAM, solely due to the complex biosynthesis of the corinoid ligand.4.3 How old might the radical SAM system be?
A similar argument can be made with regard to the occurrence of iron–sulfur clusters. According to the analysis used here, these can be described as “early arrivals.” Consequently, the couple [FeS] clusters and SAM have likely also existed early in the evolution of Life and may have been involved in fundamental biological processes such as formation of selected coenzymes and metal cofactors as well as in radical SAM reactions.54 The analysis also reveals that methanogenesis may have arrived at a later stage in evolution as several cofactors require SAM and radical SAM for the biosynthetic generation.4.4 Impact of SAM on the biosynthesis of other cofactors
S-Adenosylmethionine (1) is not only involved in electrophilic alkylations and sulfur ylide chemistry, but various coenzymes and cofactors are known whose biosyntheses are based on iron–sulfur clusters in combination with SAM (Scheme 4).8aThese are lipoic acid, biotin, thiamine pyrophosphate (TPP; those biosynthetic variants that are found in bacteria and archaea), menaquinone,55 pyrroloquinoline quinone, FeM (M = Mo, V, Fe) cofactors found in different nitrogenases,56 [FeFe]-hydrogenase H-cluster,57 the molybdenum cofactor (Moco) and at least three cofactors involved in methanogenesis, specifically coenzymes F0/F420 and tetrahydro-methanopterin (THMPT).58 This suggests that many fundamental biochemical processes, from an evolutionary perspective, are only conceivable in their current form due to the presence of SAM and its coupling to 1-electron transfer systems. This applies in particular to Wood–Ljungdahl C1 fixation coupled with molecular hydrogen and nitrogen fixation. But archaean electron transfer chains and the diverse redox chemistry mediated by Moco also need to be taken into account.
5. Conclusions
Coenzymes and cofactors have most likely played an important, if not decisive, role in the evolution and origin of life; they may be the key to resolving the often-cited dichotomy between the “RNA world” theory and the “metabolism first” theory. Their chemical role is clearly and predominantly located in metabolism, while several representatives, including S-adenosylmethionine discussed here, bear structural features of nucleotides. In an “RNA world”, such coenzymes and cofactors were partners of RNA rather than of proteins.An analysis based on biosynthetic aspects prescribes SAM biosynthesis at an early stage of biological evolution; the same applies to iron–sulfur clusters, so that not only the classic chemical role of SAM, especially methylations, but also radical SAM reactions can be declared to be ‘ancient’. If we continue the chosen analysis based on this finding, then many fundamental biochemical processes such as methanogenesis, nitrogen fixation and hydrogenation, the latter being embedded in the Wood–Ljungdahl C1 fixation pathway, only appeared in the form we know today with the emergence of SAM. The analysis also shows that corinoids appeared relatively late in biological evolution. Radical chemistry induced by the adenosyl radical emerged twice, with the homolytic cleavage of the sulfonium cation initiated by the iron–sulfur cluster in SAM having the edge in terms of timing.
Clearly this report is speculative, but that applies to the entire field of deciphering evolution and the origin of Life.59 As Albert Eschenmoser put it: “The origin of Life cannot be discovered, it has to be reinvented”.60
6. Conflicts of interest
There are no conflicts of interest to declare.7. Data availability
No supporting data come along with this article.8. Acknowledgements
I thank the taxpayers for funding salaries and providing an excellent scientific environment. This article is dedicated to Harold B. White III (University of Delaware).9. References
- H. B. White III, Coenzymes as fossils of an earlier metabolic state, J. Mol. Evol., 1976, 7, 101–104 CrossRef PubMed.
- (a) M. Yarus, Getting past the RNA world: the initial Darwinian ancestor, Cold Spring Harbor Perspect. Biol., 2011, 3, a003590 Search PubMed; (b) V. R. Jadhav and M. Yarus, Coenzymes as coribozymes, Biochimie, 2002, 84, 877–888 CrossRef CAS PubMed; (c) G. J. Connell and E. L. Christian, Utilization of cofactors expands metabolism in a new RNA world, Orig. Life Evol. Biosph, 1993, 23, 291–297 Search PubMed.
- (a) G. L. Holliday, J. M. Thornton, A. Marquet, A. G. Smith, F. Rebeille, R. Mendel, H. L. Schubert, A. D. Lawrence and M. J. Warren, Nat. Prod. Rep., 2007, 24, 972–987 RSC; (b) A. Eschenmoser and E. Loewenthal, Chem. Sov. Rev., 1992, 21, 1–16 RSC; (c) G. A. M. King, Evolution of the coenzymes, BioSystems, 1980, 13, 2345 CrossRef PubMed; (d) C. M. Visser, Evolutionary roots of catalysis by nicotinamide and flavins in CH oxidoreductases and in photosynthesis, Orig. Life, 1982, 12, 165–179 CrossRef CAS PubMed; (e) C. M. Visser, Evolution of biocatalysis 1. Possible pre-genetic-code RNA catalysts which are their own replicases, Orig.Life, 1984, 14, 291–300 CrossRef CAS PubMed; (f) F. Wenzek, A. Biallas and S. Müller, Nicotinamide riboside: What it takes to incorporate it into RNA, Molecules, 2024, 29, 3788 CrossRef CAS PubMed.
- A. Kirschning, Coenzymes and their role in the evolution of Life, Angew. Chem. Int Ed., 2021, 60, 6242–6269 CrossRef CAS PubMed.
- E. R. R. Moody, S. Álvarez-Carretero, T. A. Mahendrarajah, J. W. Clark, H. C. Betts, N. Dombrowski, L. L. Szánthó, R. A. Boyle, S. Daines, X. Chen, N. Lane, Z. Yang, G. A. Shields, G. J. Szöllősi, A. Spang, D. Pisani, T. A. Williams, T. M. Lenton and P. C. J. Donoghue, The nature of the last universal common ancestor and its impact on the early Earth syste,, Nat. Ecol. Evol., 2024, 8, 1654–1666 CrossRef PubMed.
- M. C. Weiss, F. L. Sousa, N. Mrnjavac, S. Neukirchen, M. Roettger, S. Nelson-Sathi and W. F. Martin, The physiology and habitat of the last universal common ancestor,, Nat. Microbiol., 2016, 16116 CrossRef CAS PubMed.
- A. Kirschning, The coenzyme/protein pair: A neglected "chicken and egg" problem with significance for the origin of life,, Nat. Prod. Rep., 2021, 38, 993–1010 RSC.
- (a) A. Kirschning, On the evolution of coenzyme biosynthesis, Nat. Prod. Rep., 2022, 39, 2175–2199 RSC; (b) A. Kirschning, Why pyridoxal phosphate could be a functional predecessor of thiamine pyrophosphate and speculations on a primordial metabolism, RSC Chem. Biol., 2024, 5, 508–517 RSC.
- G. L. Cantoni, S-Adenosylmethionine; a new intermediate formed enzymatically from L-methionine and ATP, J. Biol. Chem., 1953, 204, 403–416 CrossRef CAS PubMed.
- (a) W. Gilbert, Origin of life: The RNA world,, Nature, 1986, 319, 618 CrossRef; (b) G. F. Joyce, Nature, 2002, 418, 214–221 CrossRef CAS PubMed; (c) M. P. Robertson and G. F. Joyce, The origins of the RNA world,, Cold Spring Harb. Perspect. Biol., 2012, 4, a003608 Search PubMed; (d) D. Penny, An Interpretive Review of the Origin of Life Research, Biol. Philos., 2005, 20, 633–671 CrossRef.
- (a) H. Beinert, Iron–sulfur proteins: ancient structures, still full of surprises, Biol. Philosoph., 2005, 20, 633–671 CrossRef; (b) J. Meyer, Iron–sulfur protein folds, iron–sulfur chemistry, and evolution, J. Biol. Inorg. Chem., 2008, 13, 157–170 CrossRef CAS PubMed.
- (a) A. Marquet, B. T. S. Bui, A. G. Smith and M. J. Warren, Iron–sulfur proteins as initiators of radical chemistry, Nat. Prod. Rep., 2007, 24, 1027–1040 RSC; (b) K. Yokoyama and E. A. Lilla, C–C bond forming radical SAM enzymes involved in the construction of carbon skeletons of cofactors and natural products, Nat. Prod. Rep., 2018, 35, 660–694 RSC.
- GA is a common abbreviation for Gigaannum, meaning one billion years.
- J. Olejarz, Y. Iwasa, A. H. Knoll and M. A. Nowak, The Great Oxygenation Event as a consequence of ecological dynamics modulated by planetary change,, Nat. Commun., 2012, 12, 3985 CrossRef PubMed.
- (a) T. R. Cech and C. Uhlenbeck, Hammerhead nailed down, Nature, 1994, 372, 39–40 Search PubMed; (b) M. J. Fedor and J. R. Williamson, The catalytic diversity of RNAs, Rev. Mol. Cell Biol., 2005, 6, 399–412 CrossRef CAS PubMed.
- C. Anastasi, F. F. Buchet, M. A. Crowe, A. L. Parkes, M. W. Powner, J. M. Smith and J. D. Sutherland, Prebiotic systems chemistry: complexity overcoming clutter, Chem. Biol. Div., 2017, 4, 721–739 Search PubMed.
- (a) S. Tsukiji, S. B. Pattnaik and H. Suga, Reduction of an Aldehyde by a NADH/Zn2+-Dependent Redox Active Ribozyme, J. Am. Chem. Soc., 2004, 126, 5044–5045 CrossRef CAS PubMed; (b) F. Huang, C. W. Bugg and M. Yarus, RNA-Catalyzed CoA, NAD, and FAD Synthesis from Phosphopantetheine, NMN, and FMN, Biochemistry, 2000, 39, 15548–15555 CrossRef CAS PubMed; (c) R. R. Breaker and G. F. Joyce, Self-incorporation of coenzymes by ribozymes, J. Mol. Evol., 1995, 40, 551–558 CrossRef CAS PubMed.
- (a) P. J. McCown, K. A. Corbino, S. Stav, M. E. Sherlock and R. R. Breaker, Riboswitch diversity and distribution, RNA, 2017, 23, 995–1011 CrossRef CAS PubMed; (b) A. V. Sherwood and T. M. Henkin, Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses, Ann. Rev. Microbiol., 2016, 70, 361–374 CrossRef CAS PubMed; (c) A. Serganov and D. J. Patel, Metabolite Recognition Principles and Molecular Mechanisms Underlying Riboswitch Function, Ann. Rev. Biophys, 2012, 41, 343–370 CrossRef CAS PubMed.
- (a) W. Winkler, A. Nahvi and R. R. Breaker, Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression, Nature, 2002, 419, 952–955 CrossRef CAS PubMed; (b) A. Serganov, A. Polonskaia, A. Tuan Phan, R. R. Breaker and D. J. Patel, Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch, Nature, 2006, 441, 1167–1171 CrossRef CAS PubMed; (c) M. T. Croft, M. Moulin, M. E. Webb and A. G. Smith, Thiamine biosynthesis in algae is regulated by riboswitches, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20770–20775 CrossRef CAS PubMed.
- (a) A. Serganov, L. Huang and D. J. Patel, Nature, 2009, 458, 233–237 Search PubMed; (b) D. B. Pedrolli, A. Matern, J. Wang, M. Ester, K. Siedler, R. Breaker and M. Mack, Nucl. Acids Res., 2012, 40, 8662–8673 CrossRef CAS PubMed.
- T. D. Ames, D. A. Rodionov, Z. Weinberg and R. R. Breaker, A eubacterial riboswitch class that senses the coenzyme tetrahydrofolate, Chem. Biol., 2010, 17, 681–685 CrossRef CAS PubMed.
- E. E. Regulski, R. H. Moy, Z. Weinberg, J. E. Barrick, Z. Yao, W. L. Ruzzo and R. R. Breaker, A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism,, Mol. Microbiol., 2008, 68, 918–932 CrossRef CAS PubMed.
- (a) F. J. Grundy and T. M. Henkin, The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in Gram-positive bacteria,, Mol. Microbiol., 1989, 30, 737–749 CrossRef PubMed; (b) W. C. Winkler, A. Nahvi, N. Sudarsan, J. E. Barrick and R. R. Breaker, An mRNA structure that controls gene expression by binding S-adenosylmethionine, Nat. Struct. Biol., 2003, 10, 701–707 CrossRef CAS PubMed.
- A. Nahvi, N. Sudarsan, M. S. Ebert, X. Zou, K. L. Brown and R. R. Breaker, Genetic control by a metabolite binding mRNA, Chem. Biol., 2002, 9, 1043–1049 CrossRef CAS PubMed.
- (a) R. R. Breaker, Riboswitches and the RNA world, Cold Spring Harb. Perspect. Biol., 2012, 4, a003566 Search PubMed; (b) A. R. Ferré-D'Amaré, Use of a coenzyme by the glmS ribozyme-riboswitch suggests primordial expansion of RNA chemistry by small molecules, Philos. Trans R. Soc. Lond. B Biol. Sci., 2011, 366, 2942–2948 CrossRef PubMed.
- M. N. Price, A. M. Deutschbauer and A. P. Arkin, Four families of folate-independent methionine synthases, PLoS Genet., 2021, 17, e1009342 CrossRef CAS PubMed.
- (a) S. W. Ragsdale and E. Pierce, Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation, Biochim. Biophys. Acta, 2008, 1784, 1873–1898 CrossRef CAS PubMed; (b) S. W. Ragsdale, Enzymology of the Wood–Ljungdahl pathway of acetogenesis, Ann. N. Y. Acad. Sci., 2008, 1125, 129–136 CrossRef CAS PubMed.
- (a) J. X. Wang, E. R Lee, D. R. Morales, J. Lim and R. R Breaker, Riboswitches that Sense S-adenosylhomocysteine and Activate Genes Involved in Coenzyme Recycling, Mol. Cell, 2008, 29, 691–702 CrossRef CAS PubMed; (b) D.-J. Tang, X. Du, Q. Shi, J.-L. Zhang, Y.-P. He, Y.-M. Chen, Z. Ming, D. Wang, W.-Y. Zhong, Yu-W. Liang, J.-Y. Liu, J.-M. Huang, Y.-S. Zhong, S.-Q. An, H. Gu and J.-L. Tang, A SAM-I riboswitch with the ability to sense and respond to uncharged initiator tRNA,, Nat. Commun., 2020, 11, 2794 CrossRef CAS PubMed.
- H. A. Chen, T. Okuda, A. K. Lenz, C. P. M. Scheitl, H. chindelin and C. Höbartner, Structure and catalytic activity of the SAM-utilizing ribozyme SAMURI, Nature Chem. Biol., 2025 DOI:10.1038/s41589-024-01808-w.
- E. T. Parker, H. J. Cleaves, M. P. Callahan, J. P. Dworkin, D. P. Glavin, A. Lazcano and J. L. Bada, Prebiotic Synthesis of Methionine and Other Sulfur-Containing Organic Compounds on the Primitive Earth: A Contemporary Reassessment Based on an Unpublished 1958 Stanley Miller Experiment,, Orig, Life Evol Biosph., 2011, 41, 201–212 CrossRef CAS PubMed.
- P. Laurino and D. S. Tawfik, Spontaneous Emergence of S-Adenosylmethionine and the Evolution of Methylation, Angew. Chem., Int. Ed., 2017, 56, 343–345 CrossRef CAS PubMed.
- (a) A. L. Mattei, N. Bailly and A. Meissner, DNA methylation: a historical perspective, Trends Gen., 2022, 38, 676–707 CrossRef CAS PubMed; (b) S. Zaccara, R. J. Ries and S. R. Jaffrey, Reading, writing and erasing mRNA methylation, Nat. Rev. Mol. Cell Biol., 2019, 20, 608–624 CrossRef CAS PubMed.
- B. E. Prieur, A new hypothesis for the origin of life,, J. Biol. Phys., 1994, 20, 301–312 CrossRef CAS.
- H. Sandström and M. Rahm, Crossroads at the Origin of Prebiotic Chemical Complexity: Hydrogen Cyanide Product Diversification, J. Phys. Chem. A, 2023, 127, 4503–4510 CrossRef PubMed.
- G. Waddell, L. L. Eilders, B. P. Patel and M. Sims, Prebiotic methylation and the evolution of methyl transfer reactions in living cells, Orig. Life Evol. Biospheres, 2000, 30, 539–548 CrossRef PubMed.
- C. Schneider, S. Becker, H. Okamura, A. Crisp, T. Amatov, M. Stadlmeier and T. Carell, Noncanonical RNA Nucleosides as Molecular Fossils of an Early Earth—Generation by Prebiotic Methylations and Carbamoylations, Angew. Chem., Int. Ed., 2018, 57, 5943–5946 Search PubMed.
- (a) A. Schröberl, and A. Wagner, in Methoden der Organischen Chemie (Houben-Weyl), E. Müller, Georg Thieme Verlag Stuttgart, 4th edn, 1955, vol. IX, pp 86–92 Search PubMed; (b) K.-D. Gundermann, and K. Hümke in Methoden der Organischen Chemie (Houben-Weyl), E. Klamann, Georg Thieme Verlag, Stuttgart, New York, 4th edn, 1985, Band E11/Teil 1, pp 129–187 Search PubMed.
- (a) S. Ono, Photochemistry of sulfur dioxide and the origin of mass-independent isotope fractionation in Earth's atmosphere, Annu. Rev. Earth Planet. Sci., 2017, 45, 301–329 Search PubMed; (b) S. Ono, J. L. Eigenbrode and A. A. Pavlov, New insights into Archean sulfur cycle from mass-independent sulfur isotope records from the Hamersley Basin, Australia, Earth Planet. Sci. Lett., 2003, 213, 15–30 Search PubMed; (c) S. Ono, N. Beukes and D. Rumble, Origin of two distinct multiple-sulfur isotope compositions of pyrite in the 2.5 Ga Klein Naute Formation, Griqualand West Basin, South Africa. Ono, Precambrian Res., 2009, 169, 48–57 Search PubMed; (d) W. Martin, J. Baross, D. Kelley and M. J. Russell, Hydrothermal vents and the origin of life, Nat. Rev. Microbiol., 2008, 6, 805–814 Search PubMed.
- Y. Xu, M. A. A. Schoonen, D. K. Nordstrom, K. M. Cunningham and J. W. Ball, Sulfur geochemistry of hydrothermal waters in Yellowstone National Park: I. The origin of thiosulfate in hot spring waters,, Geochim. Cosmochim. Acta, 1998, 62, 3729–3743 Search PubMed.
- The term “sulfane” (sometimes polysulfane) usually refers to sulfur atoms that are covalently bonded in chains solely to other sulfur atoms: (a) F. Feher and W. Laue, Beiträge zur Chemie des Schwefels. XXIX. Über die Darstellung von Rohsulfanen, Z. Anorg. Allg. Chem., 1956, 288, 103–112 CrossRef CAS; (b) H. P. Meissner, E. R. Conway and H. S. Mickley, Synthesis of Hydrogen Persulfides, Ind. Eng. Chem., 1956, 48, 1347–1353 CrossRef CAS; (c) W. Giggenbach, Optical spectra and equilibrium distribution of polysulfide ions in aqueous solution at 20.deg, Inorg. Chem., 1972, 11, 1201 CrossRef CAS; (d) J. Boulegue, Equilibria in a sulfide rich water from Enghien-les-Bains, France, Geochim. Cosmochim. Acta, 1997, 41, 1751 CrossRef.
- K. R. Olson, Hydrogen sulfide, reactive sulfur species and coping with reactive oxygen species, Free Radic. Biol. Med., 2019, 140, 74–83 CrossRef CAS PubMed.
- D. Rickard and G. W. Luther, Chem. Rev., 2007, 107, 514–562 CrossRef CAS PubMed.
- (a) Y. Qin, S. Yue, D. Xu, M. Yang and L. Zhang, Formation pathways of hydrogen polysulfides in sulfur-bearing natural gas reservoirs from density functional theory calculations, J. Mol. Model, 2025, 31, 157 CrossRef CAS PubMed; (b) Y. Bai, and Q. Bai, Subsea Corrosion and Scale, Subsea Engineering Handbook, 2010, ch 17, 505–540 Search PubMed; (c) H. B. van der Heijde, in Organic Sulfur Compounds, N. Kharasch, Pergamon Press, Oxford, London, New York, Paris, 1961, vol. 1, ch 2 Search PubMed.
- (a) S. H. Smallcombe and M. C. Caserío, Kinetics of Sulfur-Sulfur Bond Cleavage in Methylated Methyl Disulfide by Nuclear Magnetic Resonance, J. Am. Chem. Soc., 1971, 93, 5826–5833 CrossRef CAS; (b) J. K. Kim and M. C. Caserío, Alkylated Disulfides. A Degenerate Rearrangement,, J. Am. Chem. Soc., 1974, 96, 1930–1932 CrossRef CAS; (c) C. Douat, E. Berni, R. Jacquet, L. Pouységu, D. Deffieux and S. Quideau, Protecting-Group-Free Solid-Phase Anchoring of Polyphenolic C-Glucosidic Ellagitannins and Synthesis of 1-Alkylamino-Vescalagin Derivatives, Eur. J. Org Chem., 2014, 4963–4972 CrossRef CAS.
- H. Minato, T. Miura and M. Kobayashi, Reactions of dimethylmethylsulfonium fluoroborate with some nucleophiles, Chem. Lett., 1975, 701–706 CrossRef CAS.
- B. Damer and D. Deamer, Coupled Phases and Combinatorial Selection in Fluctuating Hydrothermal Pools: A Scenario to Guide Experimental Approaches to the Origin of Cellular Life,, Life, 2015, 5, 872–887 CrossRef CAS PubMed.
- (a) H. Beinert, R. H. Holm and E. Münck, Iron–Sulfur Clusters: Nature's Modular, Multipurpose Structures, Science, 1997, 277, 653–659 CrossRef CAS PubMed; (b) S. Bandyopadhyay, K. Chandramouli and M. K. Johnson, Iron–sulfur cluster biosynthesis, Biochem. Soc. Trans., 2008, 36, 1112–1119 CrossRef CAS PubMed; (c) C. Ayala-Castro, A. Saini and F. W. Outten, Fe-S cluster assembly pathways in bacteria, Microbiol. Mol. Biol. Rev., 2008, 72, 110–125 CrossRef CAS PubMed.
- (a) H. J. Sofia, G. Chen, B. G. Hetzler, J. F. Reyes-Spindola and N. E. Miller, Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods, Nucl. Acids Res., 2001, 29, 1097–1106 CrossRef CAS PubMed; (b) Q. Zhang, W. A. van der Donk and W. Liu, Radical-Mediated Enzymatic Methylation: A Tale of Two SAMS,, Acc. Chem. Res., 2012, 45, 555–564 Search PubMed; (c) P. A. Frey, Radical Mechanisms of Enzymatic Catalysis1, Annu. Rev. Biochem., 2001, 70, 121–148 Search PubMed.
- (a) S. Scintilla, D. Rossetto, M. Clémancey, J. Rendon, A. Ranieri, G. Guella, M. Assfalg, M. Borsari, S. Gambarelli, G. Blondin and S. S. Mansy, Prebiotic synthesis of the major classes of iron–sulfur clusters, Chem. Sci., 2025, 16, 4614–4624 RSC; (b) C. Bonfio, L. Valer, S. Scintilla, S. Shah, D. J. Evans, L. Jin, J. W. Szostak, D. D. Sasselov, J. D. Sutherland and S. S. Mansy, UV-light-driven prebiotic synthesis of iron–sulfur clusters, Nature Chem., 2017, 9, 1229–1234 CrossRef CAS PubMed.
- (a) E. T. Parker, H. J. Cleaves, J. P. Dworkin, D. P. Glavin, M. Callahan, A. Aubrey, A. Lazcano and J. L. Bada, Chemical analysis of a “Miller-type” complex prebiotic broth: Part I: Chemical diversity, oxygen and nitrogen based polymers, Proc. Natl. Acad. Sci. U.S.A., 2011, 108, 5526–5531 CrossRef CAS PubMed; (b) E. T. Parker, H. J. Cleaves, M. P. Callahan, J. P. Dworkin, D. P. Glavin, A. Lazcano and J. L. Bada, Prebiotic Synthesis of Methionine and Other Sulfur-Containing Organic Compounds on the Primitive Earth: A Contemporary Reassessment Based on an Unpublished 1958 Stanley Miller Experiment,, Orig. Life Evol. Biosph., 2011, 41, 201–212 CrossRef CAS PubMed; (c) H. J. Cleaves, J. H. Chalmers, A. Lazcano, S. L. Miller and J. L. Bada, A Reassessment of Prebiotic Organic Synthesis in Neutral Planetary Atmospheres,, Orig. Life Evol. Biosph., 2008, 38, 105–115 Search PubMed; (d) J. L. Bada, New insights into prebiotic chemistry from Stanley Miller's spark discharge experiments, Chem. Soc. Rev., 2013, 7, 2186–2196 RSC.
- (a) J. Buffle, R. R. de Vitre, D. Perret, and G. G. Leppard, Combining field measurements for speciation in non perturbable water samples (application to the iron and sulfide cycles in a eutrophic lake), In Metal Speciation: Theory. Analysis and Application, J. R. Kramer and M. E. Allen, Lewis Publications, Michigan, 1988, ch 5, pp. 99–124 Search PubMed; (b) G. W. Luther III, B. T. Glazer, J. Hohman, I. Popp, M. Taillefert, T. F. Rozan, P. J. Brendel, S. Theberge and D. B. Nuzzio, Sulfur speciation monitored in situ with solid state gold amalgam voltammetric microelectrodes: polysulfides as a special case in sediments, microbial mats and hydrothermal vent waters, J. Environ. Monit., 2001, 3, 61–66 Search PubMed; (c) G. W. Luther III, T. F. Rozan, M. Taillefert, D. B. Nuzzio, C. Di Meo, T. M. Shank, R. A. Lutz and S. C. Cary, Chemical speciation drives hydrothermal vent ecology, Nature, 2001, 410, 813–816 Search PubMed; (d) G. W. Luther III, S. M. Theberge and D. Rickard, Evidence for aqueous clusters as intermediates during zinc sulfide formation, Geochim. Cosmochim. Acta, 1999, 63, 3159–3169 CrossRef; (e) M. Wolthers, S. J. Van der Gaast and D. Rickard, The structure of disordered mackinawite, Am. Mineral., 2003, 88, 2007–2015 CrossRef CAS.
- (a) X. Wu, P. Gao and F. Chen, Synthetic Applications of Sulfonium Salts as Aryl Radical Precursors, Eur. J. Org Chem., 2023, 26, e202300864 Search PubMed; (b) Á. Péter, G. J. P. Perry and D. J. Procter, Radical C–C Bond Formation using Sulfonium Salts and Light, Adv. Synth. Catal., 2020, 362, 2135–2142 CrossRef; (c) C. Chen, Z. J. Wang, H. Lu, Y. Zhao and Z. Shi, Generation of non-stabilized alkyl radicals from thianthrenium salts for C–B and C–C bond formation, Nat. Commun., 2021, 12, 4526 CrossRef CAS PubMed.
- (a) R. H. White, L-Aspartate semialdehyde and a 6-deoxy-5-ketohexose 1-phosphate are the precursors to the aromatic amino acids in Methanocaldococcus jannaschii, Biochemistry, 2004, 43, 7618–7627 CrossRef CAS PubMed; (b) R. H. White and H. Xu, Methylglyoxal is an intermediate in the biosynthesis of 6-deoxy-5-ketofructose-1-phosphate: a precursor for aromatic amino acid biosynthesis in Methanocaldococcus jannaschii, Biochemistry, 2006, 45, 12366–12379 CrossRef CAS PubMed; (c) R. H. White, Biochemical origins of lactaldehyde and hydroxyacetone in Methanocaldococcus jannaschii, Biochemistry, 2008, 47, 5037–5046 CrossRef CAS PubMed; (d) M. K. Gulko, M. Dyall-Smith, O. Gonzalez and D. Oesterhelt, How Do Haloarchaea Synthesize Aromatic Amino Acids, PLoS One, 2014, 9, e107475 Search PubMed.
- B. M. Hoffman, W. E. Broderick and J. B. Broderick, Mechanism of Radical Initiation in the Radical SAM Enzyme Superfamily, Ann. Rev.Biochem., 2023, 92, 333–349 Search PubMed.
- B. Schoepp-Cothenet, C. Lieutaud, F. Baymann, A. Vermeglio, T. Friedrich, D. M. Kramerd and W. Nitschke, Menaquinone as pool quinone in a purple bacterium, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 8549–8554 Search PubMed.
- D. E. Canfield, A. N. Glazer and P. G. Falkowski, The Evolution and Future of Earth's Nitrogen Cycle,, Science, 2010, 330, 192–196 CrossRef CAS PubMed.
- R. D. Britt, G. Rao and L. Tao, Bioassembly of complex iron–sulfur enzymes: hydrogenases and nitrogenases, Nat. Rev. Chem., 2020, 4, 542–549 CrossRef CAS PubMed.
- (a) T. Sato and H. Atomi, Novel metabolic pathways in Archaea, Curr. Opin. Microbiol., 2011, 14, 307–314 CrossRef CAS PubMed; (b) J. M. Wolfe and G. P. Fournier, Horizontal gene transfer constrains the timing of methanogen evolution, Nat. Ecol. Evol., 2018, 2, 897–903 CrossRef PubMed.
- L. L. J. Schoenmakers, T. Reydon and A. Kirschning, Evolution at the Origins of Life?, Life, 2024, 14, 175 CrossRef CAS PubMed.
- A. Eschenmoser, The search for the chemistry of life's origin,, Tetrahedron, 2007, 63, 12821–12844 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |