Open Access Article
Open Access ArticleGreen gold of the Pacific: unlocking compounds from terrestrial flora for antitumor and immunomodulatory drug discovery
Paul Huchedé
 *a,
Vincent Dumontet
b and
Mariko Matsui
a
*a,
Vincent Dumontet
b and
Mariko Matsui
a
aGroup “BIOactivities of NAtural Products and Derivatives” (BIONA), Institut Pasteur of New Caledonia, Member of the Pasteur Network, Noumea, New Caledonia. E-mail: phuchede@pasteur.nc
bUniversité Paris-Saclay, CNRS, Institut de Chimie des Substances Naturelles (ICSN), UPR 2301, 91198 Gif-sur-Yvette, France
First published on 5th January 2026
Abstract
Covering up to 2025
Natural products (NPs) from the terrestrial biodiversity play a key role in oncology drug discovery. While historically identified through bioactivity-guided fractionation, recent advances in high-content screening (HCS) assays, metabolomics, and in silico modeling have significantly enhanced the potential and attractiveness of flora-derived NPs for the development of anticancer therapeutics. This includes immunomodulatory molecules that are able to target the tumor microenvironment to promote immune-mediated clearance of the tumor, thereby improving patient response. This review highlights the untapped potential of molecules extracted from the South Pacific's terrestrial flora in the search for novel antitumor and immunomodulatory compounds. The unique biodiversity of Oceania, including Australia, New Zealand, and Pacific Island Countries and Territories (PICTs) across Micronesia, Melanesia and Polynesia, offers a promising yet largely unexplored reservoir for discovering plant-derived molecules with antitumor and immunomodulatory activities. Herein, we examine the recent pharmacological advances in this field and highlight the need for sustainable and collaborative research. Leveraging cutting-edge technologies could help overcome the challenge of NP-based drug discovery on these geographically isolated islands, unlocking the region's vast potential for plant-derived cancer therapeutics.
1. Introduction: cancer in the Pacific Island Countries and Territories
The latest WHO estimates (2022) put the number of cancer cases worldwide at almost 20 million, with 9.7 million associated deaths.1 It represents the most significant clinical, social, and economic burden in terms of cause-specific disability-adjusted life years (DALY) among all human diseases.2 269![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 088 cases and 73
088 cases and 73![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 776 deaths are attributed to Oceania, including Australia, New Zealand, and Pacific Island Countries and Territories (PICTs) in the regions of Micronesia, Melanesia, and Polynesia.1 This encompasses 22 countries and territories, including American Samoa, Cook Islands, the Federated States of Micronesia, Fiji, French Polynesia, Guam, Kiribati, Marshall Islands, Nauru, New Caledonia, Niue, Northern Mariana Islands, Palau, Papua New Guinea, Pitcairn Islands, Samoa, Solomon Islands, Tokelau, Tonga, Tuvalu, Vanuatu, and Wallis-and-Futuna (Fig. 1). PICT populations face a growing cancer burden, with a profile that includes both cancers linked to poverty and infection and cancers associated with aging population, exposure to tobacco, and changing diets.3,4 Indeed, increased international trade links and variable weather patterns caused by climate change5 have contributed to greater availability of and reliance on imported and heavily processed foods rich in carbohydrate, free sugars, trans fats, and salt among PICT populations.3,5 This nutritional shift, coupled with a decline in physical activity, has significantly elevated obesity rates, with the number of affected individuals exceeding half of the adult population in several PICTs and has been associated with a rising incidence of obesity-related cancers.3,6 Globally, for the referenced PICTs, WHO predicts that cancer incidence will at least double by 2050,7 posing a significant public health challenge in the coming decades. Moreover, specific features of PICTs, including small, geographically dispersed, and isolated populations, with often restricted resources and infrastructures, complicate patient care.3,4 Health services are often overburdened, and cancer surveillance systems are generally weaker than in high-income countries, with patients presenting with advanced cancer stages.3,4 Given these circumstances, it is absolutely necessary to collaborate and share resources at the regional level, create or strengthen partnerships between oncology specialists in high-income countries and health-care professionals in PICTs, improve cancer registration, and support capacity building strategies and regional training.4 There is an urgent need for these countries to counteract the so-called non-communicable disease epidemic by developing sustainable, locally adapted strategies for the management of these diseases, which should also include a therapeutic component.
776 deaths are attributed to Oceania, including Australia, New Zealand, and Pacific Island Countries and Territories (PICTs) in the regions of Micronesia, Melanesia, and Polynesia.1 This encompasses 22 countries and territories, including American Samoa, Cook Islands, the Federated States of Micronesia, Fiji, French Polynesia, Guam, Kiribati, Marshall Islands, Nauru, New Caledonia, Niue, Northern Mariana Islands, Palau, Papua New Guinea, Pitcairn Islands, Samoa, Solomon Islands, Tokelau, Tonga, Tuvalu, Vanuatu, and Wallis-and-Futuna (Fig. 1). PICT populations face a growing cancer burden, with a profile that includes both cancers linked to poverty and infection and cancers associated with aging population, exposure to tobacco, and changing diets.3,4 Indeed, increased international trade links and variable weather patterns caused by climate change5 have contributed to greater availability of and reliance on imported and heavily processed foods rich in carbohydrate, free sugars, trans fats, and salt among PICT populations.3,5 This nutritional shift, coupled with a decline in physical activity, has significantly elevated obesity rates, with the number of affected individuals exceeding half of the adult population in several PICTs and has been associated with a rising incidence of obesity-related cancers.3,6 Globally, for the referenced PICTs, WHO predicts that cancer incidence will at least double by 2050,7 posing a significant public health challenge in the coming decades. Moreover, specific features of PICTs, including small, geographically dispersed, and isolated populations, with often restricted resources and infrastructures, complicate patient care.3,4 Health services are often overburdened, and cancer surveillance systems are generally weaker than in high-income countries, with patients presenting with advanced cancer stages.3,4 Given these circumstances, it is absolutely necessary to collaborate and share resources at the regional level, create or strengthen partnerships between oncology specialists in high-income countries and health-care professionals in PICTs, improve cancer registration, and support capacity building strategies and regional training.4 There is an urgent need for these countries to counteract the so-called non-communicable disease epidemic by developing sustainable, locally adapted strategies for the management of these diseases, which should also include a therapeutic component.
In this context, the use of plants for medicinal purposes remains deeply rooted in cultural healthcare practices, while phytochemistry continues to drive the discovery of new anticancer agents in modern pharmaceutical research. In PICTs, the transmission of ancestral knowledge about medicinal plants remains prevalent, and these traditional practices often form a routine part of healthcare, offering a culturally accepted and locally accessible resource.8–12 Although this review does not address ethnobotanical knowledge, it is important to acknowledge its significant and ongoing relevance to contemporary drug discovery.13,14 When regulated and appropriately supervised, including the determination of the concentrations of active ingredients or potentially toxic secondary compounds, the integration of medicinal plants can provide cost-effective therapeutic options that resonate with local populations. The high cost and limited availability of anticancer therapies, particularly in smaller countries and territories with fragile economies, such as the PICTs, underscore the need for innovative and accessible healthcare solutions. Consequently, the dual perspective of utilizing plant-based therapies both as a cornerstone of local pharmacopeia and as a catalyst for economic development through the isolation of active molecules (notably antitumor and immunomodulatory compounds) holds promise for advancing healthcare outcomes, enhancing patient care, and strengthening regional resilience.9,12,15
This review will first summarize recent advances in the contribution of terrestrial flora to oncology drug discovery, as well as the opportunities that emerging technologies present for shaping the field in the coming years. It will then focus on the exceptional biodiversity of the South Pacific and highlight studies covering up to 2025 that describe the antitumor activities of molecules that are either unique to, or were initially discovered in, endemic species from Micronesia, Melanesia, and Polynesia regions, as well as Australia, emphasizing their structural diversity and singularity. Plant genera and species, as well as their geographical distributions, were obtained by consulting Plants of the World Online (POWO, https://powo.science.kew.org), an online taxonomic database published by the Royal Botanic Gardens, along with the Global Biodiversity Information Facility database (GBIF, https://www.gbif.org). In this review, we will also outline the value of investigating compounds with immunomodulatory properties for the development of new anticancer immunotherapies, a therapeutic avenue that has emerged as groundbreaking over the past decade for developing more effective and specific treatments. Through these topics, we aim to illustrate how the rich terrestrial flora of the South Pacific can make a significant impact on global efforts in oncology drug discovery while demonstrating how its potential can be unlocked through new technologies and analytical methods.
2. Unlocking the potential of terrestrial natural products in drug discovery: where are we headed?
2.1. The enduring relevance of terrestrial natural products in drug development
Natural products (NPs) continue to play a central role in the global drug discovery process. Several inherent difficulties associated with NP-based drug development exist, including securing sufficient biological material in a sustainable manner, isolating and characterizing bioactive compounds, developing efficient synthetic routes, implementing robust dereplication strategies to avoid the rediscovery of known molecules, and elucidating molecular mechanisms and targets. However, recent scientific and technological advances are helping to overcome these obstacles.16 As a result, terrestrial NPs are becoming increasingly attractive candidates.The Dictionary of Natural Products® (DNP), one of the most comprehensive NP databases with 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 NPs, reports that two-thirds (67%) of referenced NPs with documented organism classification originate from plants, predominantly comprising alkaloids, terpenoids, and flavonoids.17 Marine NPs are gaining increasing attention due to their unique structural features and drug-like scaffolds. However, they remain largely underexploited considering their abundance and diversity.18–20 Terrestrial NPs tend to have smaller molecular sizes, greater hydrophilicity, and a higher proportion of compounds meeting Lipinski's Rule of Five.18 This rule, used as a first, albeit insufficient,21 proxy for evaluating drug-likeness through the qualitative estimation of a molecule's absorption and permeability, supports the relevance of terrestrial NPs as promising candidates for small-molecule development.
000 NPs, reports that two-thirds (67%) of referenced NPs with documented organism classification originate from plants, predominantly comprising alkaloids, terpenoids, and flavonoids.17 Marine NPs are gaining increasing attention due to their unique structural features and drug-like scaffolds. However, they remain largely underexploited considering their abundance and diversity.18–20 Terrestrial NPs tend to have smaller molecular sizes, greater hydrophilicity, and a higher proportion of compounds meeting Lipinski's Rule of Five.18 This rule, used as a first, albeit insufficient,21 proxy for evaluating drug-likeness through the qualitative estimation of a molecule's absorption and permeability, supports the relevance of terrestrial NPs as promising candidates for small-molecule development.
When looking specifically at antitumor molecules approved worldwide between 1981 and 2019, 70% of the 247 drugs are considered of natural origin, a significantly higher rate than that for all other types of treatment (56%). This includes biological molecules produced by an organism or a cell line (21%), NPs or semi-synthetic derivatives (25%), and synthetic compounds based on a NP pharmacophore (23%).22 Since the discovery of paclitaxel,23 camptothecin,24 vinblastine and vincristine,25,26 to name but the best-known, still frequently cited to highlight the success of plants as a source of anticancer drugs, research into the discovery of antitumor agents from terrestrial flora has evolved considerably and benefited from major technological advances, giving new life to the field.16
2.2. New frontiers in natural product sourcing
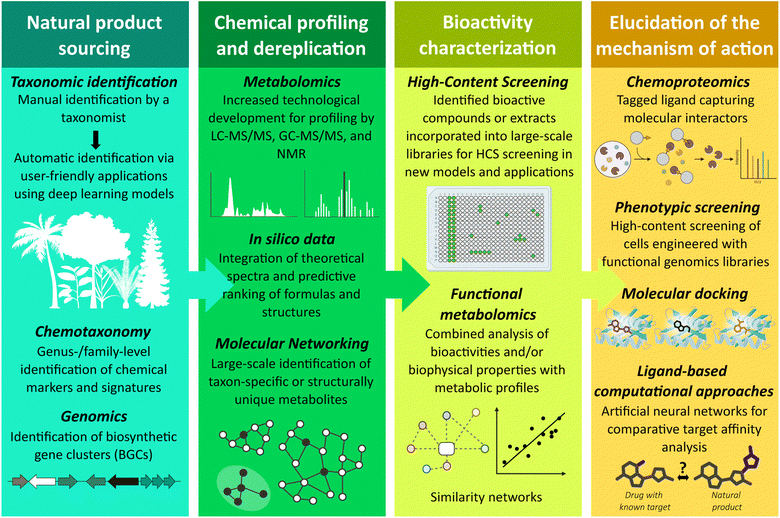
Anticancer NP lead discoveries often begin with the identification of a novel or unique source of cytotoxic compounds from a huge number of plants. Taxonomic identification of unknown or less-studied specimens provides an excellent opportunity to find novel natural sources, traditionally achieved manually by professional taxonomists by assigning identification keys. The incredible growth in computer processing power and the emergence of sophisticated artificial intelligence (AI) models have given way to automatic plant identification techniques for bridging the botanical taxonomic gap, from deep learning models to user-friendly applications, such as Flora Incognita, Pl@ntNet and LeafNet.27–31 Moreover, computer-aided chemotaxonomy has emerged as a valuable tool for the discovery of antitumor analogs, which involves leveraging biological classification data to identify relevant chemical markers27 (Fig. 2). This approach has notably enabled the characterization of metabolic differences between the Fagaceae and Asteraceae families, which correlate with distinct antioxidant and tyrosinase inhibitory activities.32 It has also facilitated the identification of diterpene distribution patterns across Lamiaceae subfamilies,33 compounds known for their cytotoxic activity and, therefore, of particular interest in the development of anticancer agents.34 In addition, genome sequence-based mining is an effective strategy for identifying novel bioactive metabolites through the detection of biosynthetic gene clusters (BGCs).35 Recent omics studies have revealed that BGCs are not only a hallmark of microbes and fungi but also can, albeit infrequently, be found in plants.36–39 In silico prediction tools have emerged to find BGCs, also called metabolic gene clusters (MGCs), in plant genomes.40 Although plant BGC discovery is still in its early stages, it has already led to the identification of several BGCs in rice, which code for the biosynthesis of diterpenoid 5,10-diketo-casbene,41 hydroxycinnamoylputrescine,42 and momilactones.43 The latter compounds exhibit in vitro cytotoxic activity, particularly against human leukemia, lymphoma and colon tumor cells.44–46 Similarly, biosynthetic genes for noscapine, an alkaloid from Papaver somniferum, are organized in a complex gene cluster.47 While noscapine is an alkaloid approved as a cough suppressant, this compound and its derivatives, referred to as noscapinoids, have attracted interest for their anticancer properties48,49 and their ability to act as β-tubulin inhibitors.50,51 This recent approach opens access to a vast reservoir of genetic sequences encoding potential anticancer compounds. In the coming years, the study of plant BGCs could become a valuable complement to botanical and chemotaxonomic strategies both for the reevaluation of known plant genomes and as a prospective tool for the discovery of new NPs from terrestrial flora37,38 (Fig. 2).2.3. Unlocking natural product chemistry: profiling, dereplication, and fractionation
The next step involves in-depth metabolite profiling and dereplication to chemically characterize the NP composition. Advances in analytical instrumentation and separation science, combined with computational tools, have significantly transformed the field, enabling metabolomics to play a central role in the discovery of novel bioactive compounds from plant secondary metabolites.16,27 To avoid the rediscovery of known molecules, the process of dereplication integrates chromatographic and spectroscopic approaches with database searching.52 State-of-the-art ultra-high-performance liquid chromatography coupled with high-resolution mass spectrometry (UHPLC-HRMS) offers high-throughput acquisition of MS and MS/MS spectra with great sensitivity and accuracy,53,54 allowing the direct comparison of acquired data with reference mass spectral libraries, such as MSnLib, FragHub, MassBank, METLIN, and other preprocessed or aggregated databases available through the Global Natural Products Social Molecular Networking (GNPS) platform.55–59 Used in conjunction with gas chromatography (GC)-MS techniques and nuclear magnetic resonance (NMR), the latter providing quantitative information and detailed structural insights, this approach enables comprehensive elucidation of the chemical composition of complex NP extracts, such as those from plants54,60–62 (Fig. 2). Moreover, AI is increasingly being integrated into existing workflows to enable de novo identification of novel compounds from MS data.27,63 This includes advanced molecular database matching using deep neural networks,64 prediction of molecular formulae directly from MS spectra,65 and integration of theoretical spectra.66,67 In parallel, spatial representation through molecular networking (MN) further enhances the identification of metabolite families and the global characterization of complex extracts, and it is now incorporated into modern data analysis pipelines.68 This multi-scale pairwise alignment allows the visualization of structural relationships among molecules and helps highlight original and taxonomically specific bioactive plant metabolites across large extract libraries.69,70 Various MN-based strategies have been developed, reviewed by other colleagues,71–73 and, for some, implemented into the GNPS platform.74–76 Notably, through examining 20 genera of the Euphorbiaceae family, this approach facilitates the identification of the genus Austrobuxus and specifically A. carunculatus, which is endemic to New Caledonia, as a source of previously unreported picrotoxane-type norditerpene dilactones,70 as detailed in Section 4.2.Although classical NP-based drug discovery began with limited biological screening of crude extracts to identify bioactive hits, this in-depth metabolite profiling now allows for the selection of the most promising candidates from larger compound libraries for subsequent in vitro screening steps. Bioactivity-guided isolation, the fractionation process used to isolate active NPs, presents several limitations that have been alleviated by recent technological innovations.16 Crude extracts may first be subjected to prefractionation, generating sub-fractions through chromatographic separation techniques.77 This prefractionation step enhances screening performance by sequestering common nuisance compounds, such as cytotoxins or assay-interfering products, and by concentrating potent active metabolites. The development of prefractionated NP libraries thereby streamlines downstream workflows for isolating bioactive components from both marine78 and terrestrial79 NPs, and the libraries are well suited for integration into automated liquid handling systems for drug discovery.77 In addition, prefractionation methods can be tailored to yield sub-fractions enriched in compounds with drug-like properties, including optimal hydrophilicity, thereby narrowing the pool of candidates less likely to advance to preclinical testing.77
2.4. From bench to scale: high-throughput experiments for natural product bioactivity testing
To scale up the in vitro testing of crude extracts or their subfractions in biological assays, high-throughput screening (HTS) and high-content screening (HCS) have represented a quantum leap in drug discovery. Both rely on microplate-based assays combined with automated operating systems and sensitive, rapid detection instruments (Fig. 2). Compared to HTS, HCS offers the added advantage of high-resolution cellular imaging to capture phenotypic changes at the cellular or subcellular levels. A notable example of HCS application in the discovery of antitumor and immunomodulatory compounds from plants is ingenol mebutate (ingenol-3-angelate), a hydrophobic diterpene ester isolated from Euphorbia peplus (Euphorbiaceae). Initially characterized as a topical chemotherapeutic agent used for the treatment of skin cancer in preclinical settings,80 it was approved by the FDA in 2012 for the treatment of actinic keratosis, a sun-related preneoplastic lesion that can progress to squamous cell carcinoma.81 More recently, HCS has brought this compound back into focus, revealing its capacity to reverse T cell exhaustion and enhance B7-H3 CAR T cell cytolysis activity against osteosarcoma cells, making it a promising lead for the development of next-generation immunotherapies.82,83 HCS has also revitalized interest in homoharringtonine (HHT), also named omacetaxine mepesuccinate, an alkaloid originally discovered in Cephalotaxus harringtonia (Cephalotaxaceae) with antitumor activity against leukemia and lymphoma cells.84–86 Approved by the FDA in 2012 for the treatment of resistant chronic myelogenous leukemia (CML), it is also included or currently explored in several clinical trials for combination regimens in acute myeloid leukemia (AML).87,88 Its subsequent inclusion in large anticancer compound libraries has enabled HCS to reveal selective activity against FMS-like tyrosine kinase-3–internal tandem duplication (FLT3-ITD)-positive AML cells, particularly in synergy with FLT3 inhibitors.89 Furthermore, additional HCS-driven studies have revealed that HHT is also active against solid tumors, including von Hippel-Lindau (VHL)-deficient clear cell renal cell carcinoma (ccRCC) compared to VHL-intact cells in vitro and in vivo.90 Among 291 compounds tested, HHT emerged as the most active compound in fluorouracil-resistant rectal cancer cell lines by impeding mitochondrial function in vivo.91Beyond repurposing known compounds, high-throughput experiments also facilitate the re-evaluation of crude plant extract libraries to uncover new sources of known active molecules. For instance, HTS of over 2000 extract fractions using the NCI-60 human tumor cell line panel from the National Cancer Institute (NCI, USA) identified phyllanthusmin D (lactone) and dichapetalin (triterpenoid) as the major cytotoxic molecules of Flueggea virosa extract (Phyllanthaceae, also known as Securinega virosa).92 These compounds are under investigation for derivative synthesis and preclinical evaluation and are of great interest in antitumor and immunomodulatory drug development.93–95
Interestingly, HTS/HCS have also been expanded into approaches aimed at deciphering the functional roles of metabolites at the scale of the metabolome, referred to as “functional metabolomics”. These strategies focus on linking metabolomic data to biological or biophysical properties. Large untargeted metabolomics datasets from complex NP fractions have been integrated with bioactivity assays or compared with profiles of metabolites and synthetic small molecules of known bioactivity to construct correlation/similarity networks that help infer mechanisms of action or identify active metabolites in complex mixtures (Fig. 2).96 These approaches have, for example, unveiled 12-deoxyphorbols from Bocquillonia nervosa as highly potent inhibitors of the WNT pathway,97 a key signaling cascade in both cancer and immune cells,98,99 as well as (re-) identified several metabolites underpinning anti-hepatocarcinoma activity.100,101 Further details and additional examples of functional metabolomic strategies are presented in recent reviews.102,103
2.5. From bioactivity to mechanism: identifying molecular targets
In the process of identifying NP molecular targets, chemoproteomic approaches remain the gold standard. It involves the use of a tagged ligand to capture molecular interactors, which are subsequently identified by MS.104 For example, chemoproteomic profiling enabled the identification of SLC25A20 as a cellular target of the previously mentioned ingenol mebutate used for the treatment of actinic keratosis.105 Complementary approaches include phenotypic screening of cell lines engineered with functional genomics libraries, including small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs), and the now gold-standard CRISPR/Cas9 libraries, to confirm target involvement based on altered responses following gene knockout or overexpression.106,107 Upon their introduction, these technologies revolutionized the field, enabling, for example, the identification of specific genes involved in non-small-cell lung cancer (NSCLC) progression or lung metastases in vivo in transplanted mouse models.108Although these tools remain widely used in target deconvolution workflows, AI has emerged as a catalyst for the prediction of antitumor and immunomodulatory NP-target interactions, supporting the rational design of analogs and helping to anticipate off-target effects.27,109 AI-driven approaches typically rely on either structure-based or ligand-based computational approaches. Among structure-based approaches, molecular docking remains the most used in silico technique, predicting how and with what affinity a compound binds to a protein based on its 3D structure.27,109 Notably, the 2024 Nobel Prize in Chemistry recognized the developers of AlphaFold, a tool that has significantly advanced drug discovery by accurately predicting 3D protein structures.110 Alongside other platforms,111 this achievement has greatly facilitated molecular docking and small-molecule interaction modeling. Conversely, the ligand-based approach is based on the principle that ligands structurally similar to a known bioactive compound, including NPs, are likely to exhibit similar biological activities towards shared targets.27 These relationships can be identified using artificial neural networks, such as self-organizing maps, which assess drug equivalence and target similarity112 (Fig. 2). For example, a bioinformatic analysis of the NCI 60-cell NP extract screening data identified Phyllanthus engleri (Phyllanthaceae) CH2Cl2 extract as selectively active against renal carcinoma cells. This activity has been linked to the presence of sesquiterpene (−)-englerin A, active in nanomolar concentrations.113 Further work revealed (−)-englerin A as a modulator of voltage-dependent L-type calcium channels,114 a discovery supported by computational target-inference approaches that also confirmed minimal off-target interactions.115
Altogether, recent technological advances are revitalizing the discovery of terrestrial NPs, reaffirming their central role in drug development, particularly for antitumor and immunomodulatory agents. Innovations in AI-driven taxonomy, genome mining, analytical instrumentation, and HTS/HCS are overcoming traditional obstacles in sourcing, characterization, and bioactivity assessment of NPs. When integrated with advanced computational tools for comprehensive metabolite profiling and target identification, these approaches are streamlining and accelerating the journey from crude extracts to therapeutic leads (Fig. 2). As a result, terrestrial flora remains an invaluable and expanding source of novel drug candidates, poised to fuel the development of next-generation therapeutics.
3. The South Pacific: land of exceptional flora
The Pacific region encompasses 9 of the 36 global biodiversity hotspots, territories with exceptionally high rates of endemism that are increasingly threatened. These include two hotspots spanning Pacific and Indian oceans (Sundaland, covering parts of Malaysia and Indonesia, and Wallacea in Indonesia), two in the North Pacific (Japan and the Philippines), and 5 specific to the South Pacific: Polynesia–Micronesia, the East Melanesian Islands, the Forests of Eastern Australia, New Caledonia, and New Zealand116 (Fig. 3). Notably, New Guinea has recently been designated as the island with the world's richest flora, comprising 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 634 species, 68% of which are endemic, distributed across 1742 genera and 264 families, with over 95% of the species in the Ericaceae, Gesneriaceae, and Zingiberaceae families being endemic.117 Kier and colleagues also highlighted that New Caledonia exhibits by far the greatest endemism-scaled richness of vascular plants worldwide, with Polynesia–Micronesia and Eastern Pacific islands ranked in third place.118 Furthermore, the analysis of 4306 islands using a restricted phylogenetic endemism index (PER) highlights that 32 of the 47 (68%) significant island areas of endemism are located in the Pacific region, including 20 (43%) in Micronesia, Melanesia, Polynesia regions, and the nearby state of Tasmania.119 13 of these are shared exclusively among 3 territories: Papua New Guinea (PNG), New Caledonia (NC) and New Zealand (NZ). The associated islands for these are the Isle of Pines, Lifou Island, and the Grande Terre Island in NC; D'Urville Island, Rauhomaumau Island, and South Island in NZ; and Bougainville Island, Dawila, Kairiru Island, Misima, New Britain, New Ireland, Tagula Island, and the island of New Guinea in PNG, which is partly shared with Indonesia.119
634 species, 68% of which are endemic, distributed across 1742 genera and 264 families, with over 95% of the species in the Ericaceae, Gesneriaceae, and Zingiberaceae families being endemic.117 Kier and colleagues also highlighted that New Caledonia exhibits by far the greatest endemism-scaled richness of vascular plants worldwide, with Polynesia–Micronesia and Eastern Pacific islands ranked in third place.118 Furthermore, the analysis of 4306 islands using a restricted phylogenetic endemism index (PER) highlights that 32 of the 47 (68%) significant island areas of endemism are located in the Pacific region, including 20 (43%) in Micronesia, Melanesia, Polynesia regions, and the nearby state of Tasmania.119 13 of these are shared exclusively among 3 territories: Papua New Guinea (PNG), New Caledonia (NC) and New Zealand (NZ). The associated islands for these are the Isle of Pines, Lifou Island, and the Grande Terre Island in NC; D'Urville Island, Rauhomaumau Island, and South Island in NZ; and Bougainville Island, Dawila, Kairiru Island, Misima, New Britain, New Ireland, Tagula Island, and the island of New Guinea in PNG, which is partly shared with Indonesia.119
This remarkable island biodiversity has arisen from lineages that both colonized isolated oceanic islands via long-distance dispersal and persisted on continental fragments or land-bridge islands following geological separation. Subsequent evolution in isolation, combined with dynamic and heterogeneous island environments, has driven diversification.120,121 In this way, endemism richness is 9.5 times higher on islands than in mainland regions.118 Insular territories contain 26.1% of all plant range equivalents—the sum of species' proportional ranges occurring within specific regions—even though they account for only 3.6% of the terrestrial surface considered in this study118 (a figure later evaluated at 6.67% (ref. 122)). Globally, 67% of plant species native to islands are endemics, with 70% of these restricted to a single island.123 However, the insular biota displays several geographic, demographic, and genetic characteristics that have enabled these species to thrive on islands, but now make them disproportionately vulnerable to a range of environmental stresses.120 Of note, the biodiversity of PICTs has been extensively altered, with native vegetation now covering only 17.5% of New Caledonia, 10.7% of the East Melanesian Islands, and 5.2% of the Polynesia–Micronesia hotspot.124 These regions face intense climatic and agroeconomic pressures: for example, projections for the East Melanesian Islands estimate a 47–59% loss of pristine vegetations across all Representative Concentration Pathway (RCP)/Shared Socioeconomic Pathway (SSP) scenarios by 2050, along with a 7–35% endemic plant species loss under the RCP4.5 scenario.116
This notable, though endangered, region, with its high rate of endemic species, harbors exceptional chemodiversity. Indeed, geographical isolation has profoundly influenced the phytochemical landscape, particularly with respect to secondary metabolites, and it may have driven the biosynthesis of novel or rare compounds that are absent in related taxa from the mainland (if there are any). This diversification has been quantified using indicators measuring, notably, phytochemical richness (the number of metabolites) and disparity (the structural dissimilarity among them).125 Richness and disparity are evident across multiple biological scales, from entire ecosystems to specific plant lineages. For example, at a broad level, metabolomic analyses have shown that trees in tropical ecosystems have a higher phenolic and polyphenolic compound diversity compared to their temperate counterparts,126 as well as a high, species-specific diversity of volatile organic compounds.127 Furthermore, at the subfamily scale, research on Amaryllidoideae has demonstrated a significant correlation between phylogeny and the diversity and bioactivity of alkaloids, specifically regarding the inhibition of acetylcholinesterase (AChE) and binding to the serotonin reuptake transporter (SERT) in vitro.128 This pattern also persists at the genus level: in Erysimum, diversity and abundance of glucosinolates and cardenolide compounds highly differ across species, with rare carboxylic or indole glucosinolates and unique cannogenins or acetyl cannogenols found in only a few species.129
Given the high rate of endemism seen in Pacific islands, which encompass both rare ancient (long-branch) and recent (short-branch) plant lineages,119 it is likely that this unique endemism generates a remarkable diversity of metabolites—in terms of both richness and disparity. The extensive review work by Coulerie and Poullain has documented the exceptional chemodiversity of the flora of New Caledonia, highlighting the occurrence of unique compounds.130–132 Notably, the majority of the territory's endemic species have yet to be chemically characterized, underscoring New Caledonia as a highly promising site for NP discovery. Moreover, Meesakul and colleagues recently reviewed the phytochemistry and biological activities of Hawaiian endemic plants, emphasizing their rich chemical diversity.133 Selected examples of promising bioactive compounds are discussed in the following sections.
Thus, the South Pacific region is home to some of the world's most unique and diverse ecosystems and floras, distinguished by a high degree of endemism and representing perhaps the largest reservoir of molecules with such a vast and singular chemodiversity, much of which remains unknown to this day. This underlines the need for collaborative approaches that support sustainable bioprospecting to actively valorize the unique potential of these terrestrial plant resources while emphasizing the importance of their conservation in the face of climate change and increasing anthropogenic pressures.120,134
4. Chemical structures and biological activities of endemic species
In this section, we focus on antitumor and/or immunomodulatory compounds identified exclusively in the endemic terrestrial flora of the South Pacific, including Australia, New Zealand, and the 22 previously mentioned PICTs. We included both compounds that, to date, have only been found in endemics of this region, as well as those first isolated from South Pacific plants and subsequently detected in species from other parts of the world, thereby facilitating their recognition and re-identification in a broader geographical context, with this latter case specified in the relevant examples. In some cases, comparisons were drawn with structurally similar molecules whose antitumor and/or immunomodulatory activities are better documented, so as to emphasize their therapeutic potential.4.1. Alkaloids
Ellipticine, a tetracyclic pyrido[4,3-b]carbazole shown in Fig. 4, was first identified in the small tropical evergreen tree Ochrosia elliptica (Apocynaceae), native to the Gilbert Islands, Nauru, New Caledonia, Norfolk Island, Queensland (Australia), and Vanuatu. Since its discovery in the late 1950s,135 interest in this molecule has steadily grown, and it has since been found in other plants, including other species of the Ochrosia genus, which are predominantly found in Oceania. Many studies have focused on synthesizing ellipticine derivatives for their enhanced antitumor properties, which have now been demonstrated in numerous in vitro and in vivo models.136,137 Several molecular mechanisms have been attributed to its antitumor properties, including DNA intercalation, topoisomerase II inhibition, p53 activation, induction of apoptosis through intrinsic and extrinsic pathways, and PI3K/AKT inhibition. However, its clinical development has not advanced beyond phase II trials, primarily due to its moderate activity and associated toxic side effects.138,139 Nevertheless, 9-hydroxy-N-methylellipticinium acetate (known as elliptinium acetate, Celiptium®), although no longer used today, has shown efficacy for treating patients with metastatic breast cancer resistant to anthracyclines.139,140 A closely related compound, 6-methylellipticine, exhibited sub-micromolar IC50 and was the most active derivative on a panel of 12 solid tumor cell lines in vitro.141 Other recent derivatives, including 9-(dimethylamino)-ethyloxy as well as 11-substituted benzylamide and unsaturated ketone derivatives, exhibit high cytotoxic activity against several myeloma cell lines at sub-micromolar concentrations and against the NCI-60-cell line panel at micromolar concentrations, respectively.142,143 Given the previous clinical advancement of ellipticinium acetate, these efforts to discover more potent and active derivatives hold promise for the potential clinical development of another analogue.Two unprecedented compounds of the indolo[2,3-a]quinolizinium family with an unusual n-propyl group at C-2, namely nukuhivensium and N12-methylnukuhivensium (Fig. 4), have been discovered in the stem bark of Rauvolfia nukuhivensis (Apocynaceae), a species endemic to the Marquesas Islands in French Polynesia,144 whereas the Rauvolfia genus is widely distributed throughout intertropical regions. The total extract of R. nukuhivensis has been shown to block IL-22-induced hyperproliferation via phosphatase and tensin homolog (PTEN) and filaggrin up-regulation, as well as a downregulation of the antiapoptotic Bcl-2-encoding gene, making them interesting candidates for countering both apoptosis-resistance mechanisms and tumor-promoting inflammation in cancer.145 Furthermore, their structural resemblance to two other alkaloids with characterized in vitro and in vivo antitumor properties, flavopereirine146,147 (found in Geissospermum and Strychnos genera)148,149 and sempervirine150,151 (found in Gelsemium sempervirens and G. elegans),152 underscores their therapeutic potential.
Two ajmaline-type alkaloids, sandwicine and isosandwicine, have been discovered in two other Rauvolfia species endemic to Hawaii: R. sandwicensis and R. vomitoria.153,154 Interestingly, a deeper phytochemical analysis of the Marquesan R. nukuhivensis has described 11 other indole alkaloids, including the newly discovered norsandwicine and isonorsandwicine, which feature the loss of the methyl group at N1 of sandwicine and isosandwicine; Nb-methylisosandwicine, with an additional methyl at N4; and three carboxylic acid derivatives: 10-methoxypanarine (a sarpagine derivative), nortueiaoine and tueiaoine (macroline derivatives).155 Norsandwicine, 10-methoxypanarine, and N12-methylnukuhivensium affected the viability of SH-SY5Y neuroblastoma cells but only in high concentrations (maximum observed: 67% reduction with N12-methylnukuhivensium at 100 µM). Interestingly, norsandwicine and 10-methoxypanarine (Fig. 4), alongside nukuhivensium and N12-methylnukuhivensium, have been shown to significantly inhibit the activity of the human Ether-à-go-go Related Gene (hERG), a voltage-dependent potassium channel,155 and should therefore be considered in oncology since these channels are aberrantly expressed in many cancer subtypes and play important roles in cancer progression.156 This recent finding, which sheds light on the potential mechanism of action of these alkaloids associated with both cytotoxic and immunomodulatory activities, underscores the importance of investigating or re-investigating NPs in light of recently identified mechanisms sustaining oncogenic properties (such as hERG channels), thereby enabling their evaluation on new potential targets.
Macrocyclic spermine alkaloid derivatives, named homalium alkaloids (including homaline, hopromine, hoprominol, hoprominone, and hopromalinol (Fig. 4)) have been discovered in Homalium guillainii (Salicaceae). This specific species is endemic to New Caledonia,157 while other Homalium species are distributed throughout intertropical regions. This family of compounds, which possesses original 1,5-diazocan-2-one moieties, has aroused great interest within the scientific community and has prompted the development of new synthetic methods in recent years.158,159 Although the bioactivities of these specific molecules remain unknown, other synthetic derivatives containing 1,5-diazocan-2-one groups have exhibited immunomodulatory properties with the inhibition of IL-1β secretion by THP-1 cells in vitro.160 Additionally, they demonstrated antitumor properties both in vitro and in vivo, suppressing breast and ovarian tumor cell growth at nanomolar concentrations by inducing apoptosis.161,162 Their ability to bind the anti-apoptotic proteins XIAP, cIAP1, and cIAP2, along with a favorable pharmacokinetic profile,161,162 makes them promising candidates for the clinical development of anticancer second mitochondria-derived activator of caspase (SMAC) mimetics with pro-apoptotic properties.
Vatine, a hexameric pyrrolidinoindoline-type alkaloid, together with its hepta- and octa-meric congeners, Vatamine and Vatamidine, have been identified in Psychotria milnei (Rubiaceae, formerly Calycodendron milnei), a species endemic to Fiji and Vanuatu.163 To our knowledge, they have not yet been found in any other species. Other oligomeric cyclotryptamine alkaloids have been identified in several species from the Pacific region, including hodgkinsine; quadrigemines A and B (all originally isolated from Eumachia frutescens, formerly Hodgkinsonia frutescens, native to Queensland, Australia164–166); quadrigemines C, D, and I; isopsychotridines A and C; oleoidine and caledonine (isolated from Eumachia oleoides, formerly Psychotria oleoides, native to New Caledonia167–169); and psychotridine (isolated from Eumachia leptothyrsa var. leptothyrsa, formerly Psychotria beccarioides, native to the Philippines, Indonesia and Papua New Guinea170). This list underscores the importance of the genera Psychotria and Eumachia, abundant sources of tropical plants, in the discovery of innovative compounds, some of which display structural specificities unique to species endemic to the South Pacific region. Notably, several of the above-mentioned compounds have been tested in vitro on non-tumor Vero African green monkey kidney cells and exhibited IC50 values in the micromolar range, with notably better selectivity against proliferative cells compared to their quiescent counterparts.171 Cytotoxic activities have also been reported on rat hepatocarcinoma cells in vitro, with Quadrigemine A outperforming the chemotherapeutic agent vincristine,172 even though this chemotherapeutic is no longer used for the treatment of liver tumors except for pediatric hepatoblastomas.173 Quadrigemine C, presented in Fig. 4, has been shown to be the most active compound in a panel of 14 pyrrolidinoinoline alkaloids on DU145 prostate cancer and A2508 melanoma cell lines in vitro (IC50 = 2.2 and 1.7 µM, respectively).174 Moreover, a recent study highlighted quadrigemine I as an effective antitumor agent both in vitro and in vivo using lymphoma xenograft models. Quadrigemine I induced apoptosis and tumor regression, modulated mitogen-activated protein kinase (MAPK) signaling pathway, and inhibited the production of pro-inflammatory cytokines and nitric oxide (NO).175 Thus, the original and complex structures of oligomeric cyclotryptamine derivatives have attracted considerable interest within the chemistry community, inspiring efforts to develop stereocontrolled synthetic routes to these compounds and new derivatives,174,176,177 including metal-catalysed syntheses.178,179 All these studies highlight the antitumor and immunomodulatory potential of these original alkaloid structures, which should undergo more extensive pharmacokinetic and pharmacodynamic studies to assess their translational potential for clinical application.
4.2. Terpenoids
Nearly one-third of all naturally occurring plant-based compounds with documented antitumor properties belong to the terpenoid family,180 making it the most extensively studied subgroup in antitumor research. These molecules are also abundant in plants endemic to the South Pacific region, with some promising candidates identified exclusively in this area.Thanks to extensive screening of plant ethyl acetate extracts of the Euphorbiaceae family from species mostly endemic to New Caledonia, a series of unprecedented dilactone norditerpene picrotoxanes has been discovered in the fruits of Austrobuxus carunculatus (Picrodendraceae).70 Molecular networks from UHPLC-HRMS2 data highlighted several clusters of ions specific to the genus Austrobuxus, with 13 new tutin derivatives bearing a butyrolactone moiety at C-13 and additional saturated-carbon ester side chains, named austrobuxusins E to M. Among these, austrobuxusins F and L, which have an acyl chain at C-2 (Fig. 5), exhibited in vitro cytotoxicity against A549 lung adenocarcinoma and U-87 MG glioma cells, with IC50 = 5.8 µM and 0.7 µM, respectively.70 Of interest, this nomenclature continues from four previously identified picrotoxane-type compounds, austrobuxusins A–D, and precedes the discovery of austrobuxusin N, all discovered in the Australian endemic plant Austrobuxus swanii.181,182 However, these compounds show little to no cytotoxic activity on colorectal adenocarcinoma (Caco-2) and ovarian cancer (SK-OV-3) cell lines in vitro.182 These findings highlight the potential of the entire Austrobuxus genus, whose species are exclusively endemic to the Pacific region.
Four new limonoids—dysoxylin, dysoxylone (Fig. 5), tigloyldysoxylin, and 6α-acetoxyobacunol acetate—have been found for the first time in the methanolic extract of the leaves of Didymocheton alliaceus (Meliaceae, formerly Dysoxylum richii),183,184 an ethnomedicinal plant native to Fiji, Kiribati, Niue, Samoa, Solomon Islands, Tonga, Vanuatu, and Wallis-and-Futuna islands.185 While the bioactivities of these limonoids remain uncharacterized, they display an unusual dilactonic triterpenoid structure with both a γ-lactone and an ω-lactone, reminiscent of obacunone-type limonoids.186 Dysoxylin and tigloyldysoxylin possess a tetrahydrofuran ring similar to that in limonin,187 with additional ester groups at C-6 (6α-acetoxyobacunol acetate) and C-7 (6α-acetoxyobacunol acetate, dysoxylone, tigloyldysoxylin). These new structural variants are of particular interest due to both the well-documented antitumor activity and inhibition of pro-inflammatory signaling cascades exhibited by obacunone-type limonoids,188–191 compounds commonly found in Citrus and other species of the Rutaceae family, as well as the ongoing clinical development of terpenoid lactones, including those of plant origin.192
Moreover, four dammarane-type triterpenoids—methyl richenoate, richenone (shown in Fig. 5), richenol and richenoic acid—have been isolated from the fruits of D. alliaceus.193 These resemble previously known dammarane triterpenoids, but uniquely possess tetrahydrofuran moieties substituted with propylene in positions where other Dysoxylum/Didymocheton species have hydroxy-substituted variants.194 Additionally, five new apotirucallane triterpenoids, dysorones A–E, have been isolated from the dichloromethane fraction of the leaf methanolic extract of Didymocheton roseus (formerly Dysoxylum roseum), endemic to New Caledonia.195 Dysorone E (Fig. 5), the major compound, has been shown to have a cytotoxic effect on KB cells (IC50 = 7.5 µM).195 There is ongoing interest in the scientific community in synthesizing new derivatives based on these terpenoid scaffolds, as they may exert selective cytotoxicity against several solid tumor cell lines.196 These discoveries highlight the interest in the genus Didymocheton, which is mainly distributed in the South Pacific. Its classification has recently been challenged,197 as it was formerly included within the genus Dysoxylum, underlining the need for chemotaxonomic studies to elucidate the distribution of families of bioactive compounds with therapeutic interest.
Several drimane terpenoids have been discovered in Zygogynum pancheri, Z. acsmithii, and Z. baillonii, species belonging to the Winteraceae family and all endemic to New Caledonia.198,199 For several of them, IC50 values for cytotoxic activity in vitro ranged from 0.1 to 0.5 µM on KB, HL60, and HCT116 tumor cells.198,199 Further studies have identified dialdehyde-substituted compounds, including NA1-115-7 (Fig. 5), as BH3-mimetics that covalently bind to MCL-1 with strong affinity, thereby inducing apoptosis in lymphoma cells in vitro without exhibiting toxicity toward normal blood cells or cardiomyocytes.200,201 Additionally, lipid nano-emulsions of NA1-115-7 exhibited improved solubility and stability, as well as cytotoxic activity, as evidenced by their comparable induction rate of cell death at half the dose.202 This makes NA1-115-7 a promising preclinical candidate for the treatment of lymphomas. Of note, other dialdehyde-substituted drimane-type terpenoids have been found in the Winteraceae family and are considered specific markers of endemic species from New Zealand in the Pseudowintera genus.203 Among them, polygodial is a well-known dialdehyde-substituted drimane terpenoid that has been identified in numerous species widely distributed across the world. The extraction of polygodial and the synthesis of its derivatives have attracted considerable interest in the scientific community204–207 because of their antitumor properties. Notable cytotoxic effects have been observed against taxane-resistant prostate cancer cell lines, while the diastereoisomer 9-epipolygodial and the dimeric unsaturated ester derivatives have demonstrated greater activity than polygodial across a broader panel of tumor cell lines.205–208 These findings could inspire the development of additional derivatives of dialdehyde-substituted drimane terpenoids, including NA1-115-7. Other unprecedented compounds, such as colorata-4(13),8-dienolide isolated from Pseudowintera colorata,209 a species endemic to New Zealand, possess unusual C1 branches at the C-3/C-4 positions in the A ring. Although no biological activities have yet been described for this compound, its total synthesis has been reported.210 Altogether, these findings highlight the potential of the Winteraceae family, particularly the Zygogynum and Pseudowintera genera, which are exclusively present in the South Pacific, as a valuable source of novel molecular scaffolds for future drug discovery efforts.
Sixteen previously undescribed triterpenoid saponins and three norlupane terpenoids were identified in the leaves, stems, and bark extracts of Jaffrea xerocarpa (Rhamnaceae, syn. Alphitonia xerocarpus and A. xerocarpa), also endemic to New Caledonia.211,212 Norlupane compounds exhibited cytotoxic activity against KB cells in vitro, with 58.4% cell death for the newly identified 29-hydroxyceanothenic acid at 10 µg mL−1. This activity is slightly lower than that of ceanothenic acid (78.5%, IC50 = 1.2 µg mL−1),211 a well-characterized triterpene commonly found in the Rhamnaceae family.213 Interestingly, the new 2α-formyl-A(1)norlup-20(29)-en-28-oic acid (Fig. 5) has been identified as the major constituent of J. xerocarpa ethyl acetate bark and stem extracts.212 This compound, along with its 2β isomer also identified in the extract, exhibited cytotoxic activity with IC50 values of 7.9 µM and 7 µM, respectively. Both aldehyde isomers, regardless of stereochemistry, exhibited cytotoxic activity similar to that of their known alcoholic counterpart, alphitolic acid.212 Other discovered triterpenoid saponins showed low (or not tested) cytotoxic properties against KB cells.211,212
Other compounds isolated from Alphitonia species include alphitexolide, an unusual γ-lactone derivative of ceanothic acid discovered in A. excelsa,214 endemic to Australia and New Guinea (Fig. 5); alphitonin, a hydroxybenzyl coumarone found in the wood of both A. petriei, endemic to Queensland and New South Wales (Australia), and A. excelsa;214 and several known steroids identified in A. petriei.215 These compounds demonstrate promising immunomodulatory properties with TNF-α and NO inhibition,215 as well as radical scavenging activity and α-glucosidase inhibition.216
4.3. Flavonoids and coumarins
Flavonoids play a well-established antioxidant role responsible for their various health-protective functions.217 Given the dysregulated oxidative stress balance in tumor cells and its potential pro-oncogenic role, targeting these pathways has emerged as a therapeutic strategy.218 Therefore, natural phenolic derivatives, including flavonoids and coumarins, hold significant promise for the development of novel anticancer treatments.217In addition to the previously mentioned chemical and biological characterization of Alphitonia species, an unprecedented flavonoid derivative, 3-O-(6-E-feruloyl)-β-D-glucopyranosyl-(1→2)–[β-D-xylopyranosyl-(1→2)–]α-L-rhamnopyranosyl-quercetin (Fig. 6), has been found in the n-butanol-soluble fraction of the hydromethanolic extract of the fruits of A. neocaledonica, a species endemic to New Caledonia.219 While the isolated compound exhibited poor anti-tyrosinase activity, like other glycosides present in the extract, the total n-butanol fraction exhibited high antioxidant (93.2% at 200 µg mL−1) and tyrosinase inhibitory (92.1% at 4 mg mL−1) activities. Given the diverse phytochemical profiles of Alphitonia species, which comprise 19 species native to Southeast Asia, Australia, and PICTs, along with the biological activity of the above-mentioned terpenoids, as well as their significant role in traditional medicine,220 we advocate for further investigations into these plants to identify potential antitumor and immunomodulatory compounds.
 | ||
| Fig. 6 Active flavonoids, coumarins, and other hybrid structures identified in the terrestrial flora of the South Pacific, along with reference compounds (rocaglamide and elliptifoline). | ||
Molimau-Samasoni and colleagues elucidated the mechanism of action underlying the traditional use of Psychotria insularum (Rubiaceae), a plant endemic to Niue, Samoa, Society Islands, Tonga, and Wallis-and-Futuna, by integrating genomics and metabolomics approaches.221 Although chemical characterization revealed only the known flavonol glycosides, rutin and nicotiflorin, other bioactive fractions notably contained polymeric flavonoids that could not be fully characterized due to broad and unresolved NMR signals, warranting further investigation. Nevertheless, the leaf juice of P. insularum was found to strongly impact iron homeostasis by inhibiting intracellular iron content and heme synthesis.221 Given the importance of iron metabolism in cancer progression and the growing relevance of targeting this pathway,222,223 these findings further emphasize the potential of NPs to act on novel oncogenic targets. Moreover, the homogenate exhibited immunomodulatory activity, inhibiting the production of inflammatory cytokines, precisely IL-4, IL-12p40, and TNF-α under unstimulated conditions and IL-17α, IL-6, IFN-γ, and IL-1α in concanavalin A- and/or lipopolysaccharide-activated splenocytes in vitro. These inhibition rates exceeded those of both rutin and the reference compound ibuprofen, suggesting the involvement of uncharacterized compound(s) with a stronger anti-inflammatory effect or with the potential to act synergistically with characterized compounds to enhance overall bioactivity. The dual potential as an inducer of cell death via ferroptosis and as an immunomodulatory agent demonstrates the potential of P. insularum leaf juice for use in cancer immunotherapy and illustrates the important role of modern integrative omics approaches in fostering such discoveries in the South Pacific region.
The genus Geijera (Rutaceae), comprising six species endemic to Australia, New Guinea, and New Caledonia, is also of interest due to its unique phytochemical composition and potential biological activities.224 In particular, unprecedented coumarins, including geiparvarin (Fig. 6), 2′-3′-dihydrogeiparvarin, 6-(methoxyl)geiparvarin, 6′-dehydromarmin, (R)-6-O-(4-geranyloxy-2-hydroxy)cinnamoylmarmin, and parvifloranines A and B (Fig. 6), have been discovered in the extracts of G. salicifolia, endemic to New Guinea, Australia, and New Caledonia, as well as in G. parviflora, endemic to Australia.225–228 Interestingly, some of these compounds exhibited both anti-inflammatory and antitumor properties. In particular, several studies have focused on the structural optimization of geiparvarin to enhance its cytotoxic properties.229,230 It has been demonstrated that introducing a methyl group at position 1′ (Fig. 6) significantly enhances cytotoxicity against several tumor cell lines in vitro, with IC50 values in the sub-micromolar range.230 These compounds effectively induced apoptosis in a concentration-dependent manner and exhibited similar or even lower IC50 values against multidrug-resistant breast cancer cells (MCF7-MDR), vinblastine-resistant leukemia cells (CEM-VBL10), and doxorubicin-resistant colon adenocarcinoma cells (LoVo/Doxo).230 Furthermore, geiparvarin has been identified as the most active compound isolated from G. parviflora for inhibiting TNF-α secretion and NO production, with IC50 = 4.1 µM and 3.8 µM, respectively.228 Two naturally occurring alkaloid derivatives of geiparvarin, parvifloranine A, substituted with a 2-pyrrolidinecarboxylic acid moiety (Fig. 6), and parvifloranine B, bearing an asparagine substituent, have also been identified in G. parviflora.226 Parvifloranine A exhibited NO inhibition in lipopolysaccharide-stimulated RAW264.7 macrophages, albeit with lower activity (IC50 = 23.4 µM) compared to that of geiparvarin.226 Given the compelling bioactivities of these coumarin derivatives, along with the presence of a diverse array of bioactive alkaloids,224,231 although not exclusive to the genus, these studies underscore the potential of Geijera species as a promising source for developing drug candidates and overcoming therapeutic resistance, while emphasizing the need for particular care with certain endangered species.232
4.4. Other chemical structures
A study on the dichloromethane extract from the leaves of Melicope barbigera (Rutaceae), endemic to the Hawaiian island of Kauai, highlighted four new acetophenones and 2H-chromenes, namely melibarbinon A and B and melibarbichromen A and B.233 Although these compounds resemble other acetophenones found in Acronychia species (Rutaceae), they are distinguished by a methoxy substitution at C-4′/C-5 rather than the more common hydroxy group, and they represent new regioisomeric forms relative to previously described acronyculatins and acrophenones.234–236 They exhibited cytotoxic activities on A2780 human ovarian cancer cells, with melibarbinon B (Fig. 6) being the most active (IC50 = 30 µM).233 These compounds expand the diverse family of plant-derived acetophenones, predominantly contributed to by the genera Acronychia and Melicope, both distributed across the Indo-Pacific region. Further, they provide new scaffolds for chemical modification, as well as opportunities to investigate structure–activity relationship (SAR) and the influence of regioisomerism on bioactivity.237Aglaia is the largest genus in the Meliaceae family, with over 150 species in the tropical and subtropical forests of Southern Asia, Northern Australia and the Pacific region.238 So far, flavaglines are the most described metabolites in the genus, with a total of 98 compounds, representing 34% of all isolated compounds from the Aglaia species.238 These compounds are characterized by the cycloaddition of a flavonoid nucleus with a cinnamic acid moiety, constituting a cyclopenta[b]benzofuran.239 Among them, two flavaglines, marikarin and 3′-hydroxymarikarin (Fig. 6), and a cyclopenta[bc]benzopyran flavagline, desacetylaglain A, a saturated diastereoisomer of the alkene-containing Elliptifoline (Fig. 6), were discovered in A. gracilis, endemic to Fiji.240 These compounds share structural similarities with other aglains, notably those identified in A. argentea, A. forbesii, A. foveolate, A. odorata, and A. rimosa (syn. A. elliptifolia).241–244 This includes the well-known flavagline rocaglamide,243 while marikarin is further distinguished by a fused pyrazinone and pyrrolidine moiety (Fig. 6), a structural feature also found in other compounds discovered in A. odorata (formerly A. duperreana).245,246 Among these molecules, rocaglamide, elliptifoline, and silvestrol are widely recognized for their potent antitumor properties in vitro and in vivo, as well as their immunomodulatory activity.247–249 In particular, recent studies have demonstrated that rocaglamide treatment can overcome TRAIL-resistance in various tumor cell lines in vitro, as well as in a mouse xenograft model in vivo.250–252 Additionally, rocaglamide enhances the ability of NK cells to mediate non-small cell lung cancer cell killing253,254 and inhibits TNF-α-induced NF-κB activity,255 further supporting its dual role as an antitumor and immunomodulatory agent. Given their therapeutic potential, new flavagline analogs with enhanced activity are being synthesized and screened252,255–257 through various methods, including in silico approaches,258 positioning them as key candidates for the development of anticancer immunotherapies. The discovery of naturally occurring flavaglines in Aglaia species endemic to the Pacific region could further contribute to this effort, providing valuable leads for drug development.
5. Concluding remarks
By showcasing the potential of terrestrial flora from the South Pacific for antitumor and immunomodulatory drug discovery, this review aims to stimulate further research and investment in this promising field. We believe that the convergence of traditional knowledge with the biodiversity of the Pacific region, combined with advances in high-throughput experiments, new metabolomics pipelines, and AI-assisted in silico drug discovery, can drive the development of potent and accessible anticancer therapies. The exceptional biodiversity endemic to the South Pacific produces unprecedented metabolites exhibiting remarkable structural diversity across various NP families (i.e. alkaloids, terpenoids, flavonoids, coumarins, and hybrid structures). These range from compounds featuring key additional chemical groups at precise positions (for example, an n-propyl group at C-2 in nukuhivensiums or a dialdehyde substitution in NA1-115-7) to more original scaffolds (such as oligomeric cyclotryptamine alkaloids from Eumachia species or picrotoxane-type norditerpene lactone derivatives from Austrobuxus carunculatus), displaying notable antitumor or immunomodulatory activities. The dual activity is particularly relevant given the major role that the development of next-generation immunotherapies has played in anticancer therapy over the past decade. Among others, the previously mentioned FDA-approved ingenol mebutate has attracted renewed interest due to its ability to reactivate hypofunctional CD8+ T cells82,259 and enhance CAR T cell function,83 directly potentiating antitumor efficacy and thereby underscoring the significant potential of NPs as relevant anticancer immunotherapies. This paves the way for further investigation of other NPs, including those from the South Pacific, such as Quadrigemine I discovered in Psychotria oleoides or new geiparvarin derivatives from endemic Geijera species, whose dual antitumor and immunomodulatory properties have already been described in the literature in preclinical settings and warrant further attention.175,229,230,260Although not addressed in the present review, metabolites derived from terrestrial fungi could play a complementary role in anticancer and immunomodulatory drug discovery. Although they have been much less studied than their plant and bacterial counterparts, recent technological advances in genomics and metabolomics, particularly in the search for biosynthetic gene clusters (BGCs) and the annotation of high-resolution mass spectra, could potentially lead to the discovery of novel therapeutically relevant metabolites.261–263 Many fungal metabolites have shown promising antitumor activities in preclinical settings, yet none have so far resulted in an approved anticancer drug.263 Instead, the major contribution has been to immunomodulatory drug discovery, most notably through the success of immunosuppressive agents, such as cyclosporin A from Tolypocladium inflatum and mycophenolic acid from Penicillium species.264,265 Endemic fungi from the South Pacific, including endophytic and mycorrhizal species, represent a significant gap in our knowledge, and much remains to be discovered about their potential as a source of novel molecules with therapeutic value.
In many of the cited studies, biological-activity characterization has been limited to preliminary in vitro assays. We advocate for more comprehensive evaluation using advanced cell models that are now widely used, including spheroids, tumoroids, multicellular systems, and tumor-on-a-chip, as well as for studies on whole organisms. Notably, some original compounds that have been discovered have never been tested for their activity, despite displaying structural homology to compounds with recognized antitumor and immunomodulatory activities in preclinical settings. This is probably due to a lack of resources or limited technologies available at the time of their initial discovery, particularly during the surge in NP chemistry research in the second half of the 20th century. By highlighting these studies alongside recent literature, we emphasize the need to reinvestigate the activity of these original NPs, since even small structural modifications can dramatically alter their interactions with oncogenic or immunogenic targets, significantly influencing their therapeutic potential. This structural diversity expands the chemical space available for antitumor and immunomodulatory drug discovery, offering new opportunities to explore novel mechanisms of action and to advance NP drug discovery in innovative and impactful ways.
In another aspect, PICTs are characterized by their geographical dispersion, which presents significant logistical challenges. Difficulties in accessing remote locations and ensuring proper conservation of biological materials during transport can hinder efficient and high-quality sample collection. Moreover, these regions often have small populations and limited resources and infrastructure, further complicating the development of robust research pipelines necessary for the effective discovery of therapeutically relevant molecules from natural sources. To address these challenges, it is crucial to promote regional and international collaborations, share access to high-throughput equipment to enhance screening capacity, and invest in capacity-building initiatives. Establishing such integrated research pipelines is essential to meeting the long-term demands of bioprospecting campaigns and the chemical and biological characterizations of NPs, and to translating this research into tangible successes.
We also call for collaborative efforts to ensure sustainable and ethical bioprospecting practices for these valuable natural resources in the Pacific region, and the need to protect them in the context of climate change and anthropogenic pressures.120,134 Bioprospecting has to be conducted in compliance with current legal regulations, such as the Nagoya Protocol and other Access and Benefit-Sharing frameworks, with efficient implementation and enforcement of these frameworks. Furthermore, bioprospecting and the exploitation of biological resources must follow sustainable practices to ensure responsible valorization of NPs through the establishment of reproducible and viable production platforms. These include sustainable crop cultivation, as well as alternative methods for metabolite production, such as the development of semi- or fully synthetic routes and the application of advanced metabolic engineering to create cell-based or cell-free systems, thereby providing scalable solutions for industrial applications and drug production. Biodiversity conservation is now an integral component of bioprospecting and NP discovery strategies, recognized as an urgent and multidisciplinary global challenge.266–269 Ultimately, by aligning research on terrestrial flora from the South Pacific and worldwide with principles of sustainability and responsibility, we can unlock the full potential of their molecular diversity and singularity to advance oncology drug discovery and address the wider unmet medical needs.
6. Author contributions
Conceptualization, visualization, writing – original draft: P. H. Writing – review & edition: P. H., V. D., M. M. Supervision: M. M. All authors have read and agreed to the published version of the manuscript.7. Conflicts of interest
There are no conflicts to declare.8. Data availability
There are no additional data associated with this article.9. Acknowledgements
P. H. received the Calmette & Yersin and the Pasteur-Roux-Cantarini postdoctoral fellowships from the Pasteur Network and the Institut Pasteur. M. M. is financed by the Government of New Caledonia. We thank the Pacific Community (SPC) for providing the base map of the Pacific Island Countries and Territories.10. References
- WHO, Globocan 2022 Estimates, Cancer Today, https://gco.iarc.who.int/today/, accessed March 1, 2024.
- C. Mattiuzzi and G. Lippi, J. Epidemiol. Global Health, 2019, 9, 217–222 CrossRef PubMed.
- D. Sarfati, R. Dyer, F. A.-L. Sam, M. Barton, F. Bray, E. Buadromo, A. Ekeroma, S. Foliaki, J. Fong, J. Herman, L. Huggins, K. Maoate, I. Meredith, G. Mola, N. Palafox, V. Puloka, H.-R. Shin, J. Skeen, W. Snowdon, M. Tafuna'i, A. Teng, D. Watters and P. Vivili, Lancet Oncol., 2019, 20, e475–e492 CrossRef.
- A. Ekeroma, R. Dyer, N. Palafox, K. Maoate, J. Skeen, S. Foliaki, A. J. Vallely, J. Fong, M. Hibma, G. Mola, M. Reichhardt, L. Taulung, G. Aho, T. Fakakovikaetau, D. Watters, P. J. Toliman, L. Buenconsejo-Lum and D. Sarfati, Lancet Oncol., 2019, 20, e493–e502 CrossRef.
- L. McIver, R. Kim, A. Woodward, S. Hales, J. Spickett, D. Katscherian, M. Hashizume, Y. Honda, H. Kim, S. Iddings, J. Naicker, H. Bambrick, A. J. McMichael and K. L. Ebi, Environ. Health Perspect., 2016, 124, 1707–1714 CrossRef PubMed.
- T. Kessaram, J. McKenzie, N. Girin, A. Roth, P. Vivili, G. Williams and D. Hoy, Aust. N. Z. J. Public Health, 2015, 39, 336–343 CrossRef PubMed.
- WHO, Globocan 2022 Estimates, Cancer Tomorrow, https://gco.iarc.who.int/today/, accessed March 31, 2025.
- R. Fuller, N. Z. J. Botan., 2013, 51, 116–138 CrossRef.
- M. Balick, Planta Med., 2012, 78, IL29 Search PubMed.
- G. Bradacs, J. Heilmann and C. S. Weckerle, J. Ethnopharmacol., 2011, 137, 434–448 CrossRef PubMed.
- F. Chassagne, J.-F. Butaud, F. Torrente, E. Conte, R. Ho and P. Raharivelomanana, J. Ethnopharmacol., 2022, 292, 115186 CrossRef CAS PubMed.
- S. Shah and J. A. Bhat, J. Integr. Med., 2019, 17, 244–249 CrossRef PubMed.
- D. Domingo-Fernández, Y. Gadiya, S. Mubeen, T. J. Bollerman, M. D. Healy, S. Chanana, R. G. Sadovsky, D. Healey and V. Colluru, iScience, 2023, 26(9), 107729 CrossRef PubMed.
- I. Süntar, Phytochem. Rev., 2020, 19, 1199–1209 CrossRef.
- I. Vandebroek, J. Ethnopharmacol., 2013, 148, 746–754 Search PubMed.
- A. G. Atanasov, S. B. Zotchev, V. M. Dirsch and C. T. Supuran, Nat. Rev. Drug Discovery, 2021, 20, 200–216 Search PubMed.
- F. Chassagne, G. Cabanac, G. Hubert, B. David and G. Marti, Phytochem. Rev., 2019, 18, 601–622 CrossRef CAS.
- J. Shang, B. Hu, J. Wang, F. Zhu, Y. Kang, D. Li, H. Sun, D.-X. Kong and T. Hou, J. Chem. Inf. Model., 2018, 58, 1182–1193 Search PubMed.
- A. R. Carroll, B. R. Copp, T. Grkovic, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2024, 41, 162–207 Search PubMed.
- C. Lyu, T. Chen, B. Qiang, N. Liu, H. Wang, L. Zhang and Z. Liu, Nucleic Acids Res., 2021, 49, D509–D515 CrossRef CAS PubMed.
- N. Lohit, A. K. Singh, A. Kumar, H. Singh, J. P. Yadav, K. Singh and P. Kumar, Let. Drug Des. Discovery, 2024, 21, 1334–1358 Search PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 Search PubMed.
- M. C. Wani, H. L. Taylor, M. E. Wall, P. Coggon and A. T. McPhail, J. Am. Chem. Soc., 1971, 93, 2325–2327 CrossRef CAS PubMed.
- M. E. Wall, M. C. Wani, C. E. Cook, K. H. Palmer, A. T. McPhail and G. A. Sim, J. Am. Chem. Soc., 1966, 88, 3888–3890 Search PubMed.
- R. L. Noble, C. T. Beer and J. H. Cutts, Biochem. Pharmacol., 1959, 1, 347–348 CrossRef.
- I. S. Johnson, H. F. Wright and G. H. Svoboda, J. Lab. Clin. Med., 1959, 54, 830 Search PubMed.
- G. Li, P. Lin, K. Wang, C.-C. Gu and S. Kusari, Trends Cancer, 2022, 8, 65–80 CrossRef CAS PubMed.
- P. Mäder, D. Boho, M. Rzanny, M. Seeland, H. C. Wittich, A. Deggelmann and J. Wäldchen, Methods Ecol. Evol., 2021, 12, 1335–1342 CrossRef.
- P. Barré, B. C. Stöver, K. F. Müller and V. Steinhage, Ecol. Inf., 2017, 40, 50–56 CrossRef.
- H. Zhu, Q. Liu, Y. Qi, X. Huang, F. Jiang and S. Zhang, Multimedia Tools Appl., 2018, 77, 29779–29797 CrossRef.
- A. Joly, P. Bonnet, H. Goëau, J. Barbe, S. Selmi, J. Champ, S. Dufour-Kowalski, A. Affouard, J. Carré, J.-F. Molino, N. Boujemaa and D. Barthélémy, Multimedia Syst., 2016, 22, 751–766 Search PubMed.
- S. Lee, D.-G. Oh, S. Lee, G. R. Kim, J. S. Lee, Y. K. Son, C.-H. Bae, J. Yeo and C. H. Lee, Molecules, 2015, 20, 19719–19734 CrossRef CAS PubMed.
- A. Barbosa Silva Cavalcanti, R. P. Costa Barros, V. C. d. O. Costa, M. Sobral da Silva, J. Fechine Tavares, L. Scotti and M. T. Scotti, Molecules, 2019, 24, 3908 Search PubMed.
- D. Cox-Georgian, N. Ramadoss, C. Dona and C. Basu, Med. Plants, 2019, 333–359 Search PubMed.
- B. C. Covington, F. Xu and M. R. Seyedsayamdost, Annu. Rev. Biochem., 2021, 90, 763–788 CrossRef CAS PubMed.
- K. S. Singh, J. J. J. van der Hooft, S. C. M. van Wees and M. H. Medema, Nat. Prod. Rep., 2022, 39, 1876–1896 Search PubMed.
- C. Zhan, S. Shen, C. Yang, Z. Liu, A. R. Fernie, I. A. Graham and J. Luo, Trends Plant Sci., 2022, 27, 981–1001 CrossRef CAS PubMed.
- X. Zhou and Z. Liu, Plant Commun., 2022, 3, 100300 CrossRef CAS.
- H.-W. Nützmann, A. Huang and A. Osbourn, New Phytol., 2016, 211, 771–789 Search PubMed.
- N. Töpfer, L.-M. Fuchs and A. Aharoni, Nucleic Acids Res., 2017, 45, 7049–7063 Search PubMed.
- C. Zhan, L. Lei, Z. Liu, S. Zhou, C. Yang, X. Zhu, H. Guo, F. Zhang, M. Peng, M. Zhang, Y. Li, Z. Yang, Y. Sun, Y. Shi, K. Li, L. Liu, S. Shen, X. Wang, J. Shao, X. Jing, Z. Wang, Y. Li, T. Czechowski, M. Hasegawa, I. Graham, T. Tohge, L. Qu, X. Liu, A. R. Fernie, L.-L. Chen, M. Yuan and J. Luo, Nat. Plants, 2020, 6, 1447–1454 Search PubMed.
- H. Fang, S. Shen, D. Wang, F. Zhang, C. Zhang, Z. Wang, Q. Zhou, R. Wang, H. Tao, F. He, C. Yang, M. Peng, X. Jing, Z. Hao, X. Liu, J. Luo, G.-L. Wang and Y. Ning, Sci. Bull., 2021, 66, 2381–2393 Search PubMed.
- N. Kitaoka, J. Zhang, R. K. Oyagbenro, B. Brown, Y. Wu, B. Yang, Z. Li and R. J. Peters, Plant Cell, 2021, 33, 290–305 CrossRef PubMed.
- S.-J. Kim, H.-R. Park, E. Park and S.-C. Lee, J. Agric. Food Chem., 2007, 55, 1702–1706 CrossRef CAS PubMed.
- C. Park, N. Y. Jeong, G.-Y. Kim, M. H. Han, I.-M. Chung, W.-J. Kim, Y. H. Yoo and Y. H. Choi, Oncol. Rep., 2014, 31, 1653–1660 Search PubMed.
- S. C. Lee, I.-M. Chung, Y. J. Jin, Y. S. Song, S. Y. Seo, B. S. Park, K. H. Cho, K. S. Yoo, T.-H. Kim, S.-B. Yee, Y.-S. Bae and Y. H. Yoo, Nutr. Cancer, 2008, 60, 542–551 CrossRef CAS PubMed.
- T. Winzer, V. Gazda, Z. He, F. Kaminski, M. Kern, T. R. Larson, Y. Li, F. Meade, R. Teodor, F. E. Vaistij, C. Walker, T. A. Bowser and I. A. Graham, Science, 2012, 336, 1704–1708 CrossRef CAS PubMed.
- M. Mahmoudian and P. Rahimi-Moghaddam, Recent Pat. Anti-Cancer Drug Discovery, 2009, 4, 92–97 Search PubMed.
- X. Chen, T.-T. T. Dang and P. J. Facchini, Phytochemistry, 2015, 111, 7–13 Search PubMed.
- F. Nemati, I. Bischoff-Kont, P. Salehi, S. Nejad-Ebrahimi, M. Mohebbi, M. Bararjanian, N. Hadian, Z. Hassanpour, Y. Jung, S. Schaerlaekens, D. Lucena-Agell, M. A. Oliva, R. Fürst and H. R. Nasiri, Bioorg. Chem., 2021, 115, 105135 CrossRef CAS PubMed.
- F. Nemati, P. Salehi, M. Bararjanian, N. Hadian, M. Mohebbi, G. Lauro, D. Ruggiero, S. Terracciano, G. Bifulco and I. Bruno, Bioorg. Med. Chem. Lett., 2020, 30, 127489 CrossRef CAS PubMed.
- J. Hubert, J.-M. Nuzillard and J.-H. Renault, Phytochem. Rev., 2017, 16, 55–95 Search PubMed.
- D. Li and E. Gaquerel, Annu. Rev. Plant Biol., 2021, 72, 867–891 CrossRef CAS PubMed.
- J.-L. Wolfender, J.-M. Nuzillard, J. J. J. van der Hooft, J.-H. Renault and S. Bertrand, Anal. Chem., 2019, 91, 704–742 CrossRef CAS PubMed.
- C. Brungs, R. Schmid, S. Heuckeroth, A. Mazumdar, M. Drexler, P. Šácha, P. C. Dorrestein, D. Petras, L.-F. Nothias, V. Veverka, R. Nencka, Z. Kameník and T. Pluskal, Nat. Methods, 2025, 1–4 Search PubMed.
- A. Dablanc, S. Hennechart, A. Perez, G. Cabanac, Y. Guitton, N. Paulhe, B. Lyan, E. L. Jamin, F. Giacomoni and G. Marti, Anal. Chem., 2024, 96, 12489–12496 CAS.
- R. Tautenhahn, K. Cho, W. Uritboonthai, Z. Zhu, G. J. Patti and G. Siuzdak, Nat. Biotechnol., 2012, 30, 826–828 CrossRef CAS PubMed.
- H. Horai, M. Arita, S. Kanaya, Y. Nihei, T. Ikeda, K. Suwa, Y. Ojima, K. Tanaka, S. Tanaka and K. Aoshima, J. Mass Spectrom., 2010, 45, 703–714 CrossRef CAS PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. B. P, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O'Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P.-M. Allard, P. Phapale, L.-F. Nothias, T. Alexandrov, M. Litaudon, J.-L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D.-T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. O. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- H. Tsugawa, A. Rai, K. Saito and R. Nakabayashi, Nat. Prod. Rep., 2021, 38, 1729–1759 RSC.
- F. M. M. Ocampos, A. J. B. de Souza, G. H. Ribeiro, L. S. Almeida, N. R. B. Cônsolo and L. A. Colnago, Front. Nat. Prod., 2024, 3, 1414506 CrossRef.
- N. Feizi, F. S. Hashemi-Nasab, F. Golpelichi, N. Saburouh and H. Parastar, Trends Anal. Chem., 2021, 138, 116239 CrossRef CAS.
- M. W. Mullowney, K. R. Duncan, S. S. Elsayed, N. Garg, J. J. J. van der Hooft, N. I. Martin, D. Meijer, B. R. Terlouw, F. Biermann, K. Blin, J. Durairaj, M. Gorostiola González, E. J. N. Helfrich, F. Huber, S. Leopold-Messer, K. Rajan, T. de Rond, J. A. van Santen, M. Sorokina, M. J. Balunas, M. A. Beniddir, D. A. van Bergeijk, L. M. Carroll, C. M. Clark, D.-A. Clevert, C. A. Dejong, C. Du, S. Ferrinho, F. Grisoni, A. Hofstetter, W. Jespers, O. V. Kalinina, S. A. Kautsar, H. Kim, T. F. Leao, J. Masschelein, E. R. Rees, R. Reher, D. Reker, P. Schwaller, M. Segler, M. A. Skinnider, A. S. Walker, E. L. Willighagen, B. Zdrazil, N. Ziemert, R. J. M. Goss, P. Guyomard, A. Volkamer, W. H. Gerwick, H. U. Kim, R. Müller, G. P. van Wezel, G. J. P. van Westen, A. K. H. Hirsch, R. G. Linington, S. L. Robinson and M. H. Medema, Nat. Rev. Drug Discovery, 2023, 22, 895–916 CrossRef CAS PubMed.
- H. W. Kim, M. Wang, C. A. Leber, L.-F. Nothias, R. Reher, K. B. Kang, J. J. J. van der Hooft, P. C. Dorrestein, W. H. Gerwick and G. W. Cottrell, J. Nat. Prod., 2021, 84, 2795–2807 CrossRef CAS PubMed.
- F. Wang, J. Liigand, S. Tian, D. Arndt, R. Greiner and D. S. Wishart, Anal. Chem., 2021, 93, 11692–11700 CrossRef CAS PubMed.
- P. L. Bremer, A. Vaniya, T. Kind, S. Wang and O. Fiehn, J. Chem. Inf. Model., 2022, 62, 4049–4056 CrossRef CAS PubMed.
- P.-M. Allard, G. Genta-Jouve and J.-L. Wolfender, Curr. Opin. Chem. Biol., 2017, 36, 40–49 CrossRef CAS PubMed.
- P.-M. Allard, T. Péresse, J. Bisson, K. Gindro, L. Marcourt, V. C. Pham, F. Roussi, M. Litaudon and J.-L. Wolfender, Anal. Chem., 2016, 88, 3317–3323 CrossRef CAS PubMed.
- F. Olivon, S. Remy, G. Grelier, C. Apel, C. Eydoux, J.-C. Guillemot, J. Neyts, L. Delang, D. Touboul, F. Roussi and M. Litaudon, J. Nat. Prod., 2019, 82, 330–340 CrossRef CAS PubMed.
- F. Olivon, P. Retailleau, S. Desrat, D. Touboul, F. Roussi, C. Apel and M. Litaudon, J. Nat. Prod., 2020, 83, 3069–3079 CrossRef CAS PubMed.
- Y. Wang, Y. Wang, Z. Zhang, K. Xu, Q. Fang, X. Wu and S. Ma, J. Pharm. Biomed. Anal., 2025, 259, 116741 CrossRef CAS PubMed.
- M. A. Beniddir, K. Bin Kang, G. Genta-Jouve, F. Huber, S. Rogers and J. J. J. van der Hooft, Nat. Prod. Rep., 2021, 38, 1967–1993 RSC.
- A. E. F. Ramos, L. Evanno, E. Poupon, P. Champy and M. A. Beniddir, Nat. Prod. Rep., 2019, 36, 960–980 RSC.
- A. T. Aron, E. C. Gentry, K. L. McPhail, L.-F. Nothias, M. Nothias-Esposito, A. Bouslimani, D. Petras, J. M. Gauglitz, N. Sikora, F. Vargas, J. J. J. van der Hooft, M. Ernst, K. B. Kang, C. M. Aceves, A. M. Caraballo-Rodríguez, I. Koester, K. C. Weldon, S. Bertrand, C. Roullier, K. Sun, R. M. Tehan, C. A. P. Boya, M. H. Christian, M. Gutiérrez, A. M. Ulloa, J. A. Tejeda Mora, R. Mojica-Flores, J. Lakey-Beitia, V. Vásquez-Chaves, Y. Zhang, A. I. Calderón, N. Tayler, R. A. Keyzers, F. Tugizimana, N. Ndlovu, A. A. Aksenov, A. K. Jarmusch, R. Schmid, A. W. Truman, N. Bandeira, M. Wang and P. C. Dorrestein, Nat. Protoc., 2020, 15, 1954–1991 CrossRef CAS PubMed.
- L.-F. Nothias, D. Petras, R. Schmid, K. Dührkop, J. Rainer, A. Sarvepalli, I. Protsyuk, M. Ernst, H. Tsugawa, M. Fleischauer, F. Aicheler, A. A. Aksenov, O. Alka, P.-M. Allard, A. Barsch, X. Cachet, A. M. Caraballo-Rodriguez, R. R. Da Silva, T. Dang, N. Garg, J. M. Gauglitz, A. Gurevich, G. Isaac, A. K. Jarmusch, Z. Kameník, K. B. Kang, N. Kessler, I. Koester, A. Korf, A. Le Gouellec, M. Ludwig, C. H. Martin, L.-I. McCall, J. McSayles, S. W. Meyer, H. Mohimani, M. Morsy, O. Moyne, S. Neumann, H. Neuweger, N. H. Nguyen, M. Nothias-Esposito, J. Paolini, V. V. Phelan, T. Pluskal, R. A. Quinn, S. Rogers, B. Shrestha, A. Tripathi, J. J. J. van der Hooft, F. Vargas, K. C. Weldon, M. Witting, H. Yang, Z. Zhang, F. Zubeil, O. Kohlbacher, S. Böcker, T. Alexandrov, N. Bandeira, M. Wang and P. C. Dorrestein, Nat. Methods, 2020, 17, 905–908 Search PubMed.
- R. Schmid, D. Petras, L.-F. Nothias, M. Wang, A. T. Aron, A. Jagels, H. Tsugawa, J. Rainer, M. Garcia-Aloy, K. Dührkop, A. Korf, T. Pluskal, Z. Kameník, A. K. Jarmusch, A. M. Caraballo-Rodríguez, K. C. Weldon, M. Nothias-Esposito, A. A. Aksenov, A. Bauermeister, A. Albarracin Orio, C. O. Grundmann, F. Vargas, I. Koester, J. M. Gauglitz, E. C. Gentry, Y. Hövelmann, S. A. Kalinina, M. A. Pendergraft, M. Panitchpakdi, R. Tehan, A. Le Gouellec, G. Aleti, H. Mannochio Russo, B. Arndt, F. Hübner, H. Hayen, H. Zhi, M. Raffatellu, K. A. Prather, L. I. Aluwihare, S. Böcker, K. L. McPhail, H.-U. Humpf, U. Karst and P. C. Dorrestein, Nat. Commun., 2021, 12, 3832 CrossRef CAS PubMed.
- B. A. P. Wilson, C. C. Thornburg, C. J. Henrich, T. Grkovic and B. R. O'Keefe, Nat. Prod. Rep., 2020, 37, 893–918 RSC.
- A. Cutignano, G. Nuzzo, A. Ianora, E. Luongo, G. Romano, C. Gallo, C. Sansone, S. Aprea, F. Mancini, U. D'Oro and A. Fontana, Mar. Drugs, 2015, 13, 5736–5749 CrossRef CAS PubMed.
- C. J. Henrich, L. K. Cartner, J. A. Wilson, R. W. Fuller, A. E. Rizzo, K. M. Reilly, J. B. McMahon and K. R. Gustafson, J. Nat. Prod., 2015, 78, 2776–2781 CrossRef CAS PubMed.
- S. M. Ogbourne, A. Suhrbier, B. Jones, S.-J. Cozzi, G. M. Boyle, M. Morris, D. McAlpine, J. Johns, T. M. Scott, K. P. Sutherland, J. M. Gardner, T. T. T. Le, A. Lenarczyk, J. H. Aylward and P. G. Parsons, Cancer Res., 2004, 64, 2833–2839 CrossRef CAS PubMed.
- M. Lebwohl, N. Swanson, L. L. Anderson, A. Melgaard, Z. Xu and B. Berman, N. Engl. J. Med., 2012, 366, 1010–1019 CrossRef CAS PubMed.
- B. S. Marro, J. Zak, R. B. Zavareh, J. R. Teijaro, L. L. Lairson and M. B. A. Oldstone, Cell Rep., 2019, 29, 3293–3302 CrossRef CAS PubMed.
- H. W. Lee, C. O'Reilly, A. N. Beckett, D. G. Currier, T. Chen and C. DeRenzo, J. Exp. Clin. Cancer Res., 2024, 43, 97 CrossRef CAS PubMed.
- R. G. Powell, D. Weisleder, C. R. Smith and I. A. Wolff, Tetrahedron Lett., 1969, 10, 4081–4084 CrossRef.
- R. G. Powell, D. Weisleder, C. R. Smith and W. K. Rohwedder, Tetrahedron Lett., 1970, 815–818 CrossRef CAS PubMed.
- R. G. Powell, D. Weisleder and C. R. Smith, J. Pharm. Sci., 1972, 61, 1227–1230 CrossRef CAS PubMed.
- J. Jin, J.-X. Wang, F.-F. Chen, D.-P. Wu, J. Hu, J.-F. Zhou, J.-D. Hu, J.-M. Wang, J.-Y. Li, X.-J. Huang, J. Ma, C.-Y. Ji, X.-P. Xu, K. Yu, H.-Y. Ren, Y.-H. Zhou, Y. Tong, Y.-J. Lou, W.-M. Ni, H.-Y. Tong, H.-F. Wang, Y.-C. Mi, X. Du, B.-A. Chen, Y. Shen, Z. Chen and S.-J. Chen, Lancet Oncol., 2013, 14, 599–608 CrossRef CAS PubMed.
- W. Wang, L. He, T. Lin, F. Xiang, Y. Wu, F. Zhou and Y. He, Front. Oncol., 2025, 15, 1522273 CrossRef CAS PubMed.
- S. S. Y. Lam, E. S. K. Ho, B.-L. He, W.-W. Wong, C.-Y. Cher, N. K. L. Ng, C.-H. Man, H. Gill, A. M. S. Cheung, H.-W. Ip, C.-C. So, J. Tamburini, C. W. E. So, D. N. Ho, C.-H. Au, T.-L. Chan, E. S. K. Ma, R. Liang, Y.-L. Kwong and A. Y. H. Leung, Sci. Transl. Med., 2016, 8, 359ra129 Search PubMed.
- N. C. Wolff, A. Pavía-Jiménez, V. T. Tcheuyap, S. Alexander, M. Vishwanath, A. Christie, X.-J. Xie, N. S. Williams, P. Kapur, B. Posner, R. M. McKay and J. Brugarolas, Oncotarget, 2015, 6, 16951–16962 CrossRef PubMed.
- H. Chenghao, L. Xuefeng, P. Junli, W. Ke, L. Haixia, H. Guangyue, L. Qingqin and W. Feng, Biochem. Biophys. Res. Commun., 2025, 743, 151141 CrossRef PubMed.
- C. C. Thornburg, J. R. Britt, J. R. Evans, R. K. Akee, J. A. Whitt, S. K. Trinh, M. J. Harris, J. R. Thompson, T. L. Ewing, S. M. Shipley, P. G. Grothaus, D. J. Newman, J. P. Schneider, T. Grkovic and B. R. O'Keefe, ACS Chem. Biol., 2018, 13, 2484–2497 CrossRef CAS PubMed.
- Y. Deng, J. Chu, Y. Ren, Z. Fan, X. Ji, B. Mundy-Bosse, S. Yuan, T. Hughes, J. Zhang, B. Cheema, A. T. Camardo, Y. Xia, L.-C. Wu, L.-S. Wang, X. He, A. D. Kinghorn, X. Li, M. A. Caligiuri and J. Yu, J. Immunol., 2014, 193, 2994–3002 CrossRef CAS PubMed.
- A. N. Young, D. Herrera, A. C. Huntsman, M. A. Korkmaz, D. D. Lantvit, S. Mazumder, S. Kolli, C. C. Coss, S. King, H. Wang, S. M. Swanson, A. D. Kinghorn, X. Zhang, M. A. Phelps, L. N. Aldrich, J. R. Fuchs and J. E. Burdette, Mol. Cancer Ther., 2018, 17, 2123–2135 CrossRef CAS PubMed.
- I. Addae-Mensah, G. A. Dziwornu, M. A. Chama and D. Osei-Safo, Nat. Prod. Rep., 2024, 41, 1579–1603 RSC.
- S. K. Hight, T. N. Clark, K. L. Kurita, E. A. McMillan, W. Bray, A. F. Shaikh, A. Khadilkar, F. P. J. Haeckl, F. Carnevale-Neto, S. La, A. Lohith, R. M. Vaden, J. Lee, S. Wei, R. S. Lokey, M. A. White, R. G. Linington and J. B. MacMillan, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2208458119 CrossRef CAS PubMed.
- F. Olivon, P.-M. Allard, A. Koval, D. Righi, G. Genta-Jouve, J. Neyts, C. Apel, C. Pannecouque, L.-F. Nothias, X. Cachet, L. Marcourt, F. Roussi, V. L. Katanaev, D. Touboul, J.-L. Wolfender and M. Litaudon, ACS Chem. Biol., 2017, 12, 2644–2651 CrossRef CAS PubMed.
- J. M. Bugter, N. Fenderico and M. M. Maurice, Nat. Rev. Cancer, 2021, 21, 5–21 CrossRef CAS PubMed.
- W.-J. Chae and A. L. M. Bothwell, Trends Immunol., 2018, 39, 830–847 CrossRef CAS PubMed.
- S. S. El-Hawary, R. Mohammed, A. F. Tawfike, S. F. AbouZid, M. A. Taher, U. R. Abdelmohsen and E. Amin, Sci. Rep., 2021, 11, 8405 CrossRef CAS PubMed.
- F. Chassagne, M. Haddad, A. Amiel, C. Phakeovilay, C. Manithip, G. Bourdy, E. Deharo and G. Marti, Fitoterapia, 2018, 127, 226–236 CrossRef CAS PubMed.
- G. A. Vitale, C. Geibel, V. Minda, M. Wang, A. T. Aron and D. Petras, Nat. Prod. Rep., 2024, 41, 885–904 RSC.
- M. M. Rinschen, J. Ivanisevic, M. Giera and G. Siuzdak, Nat. Rev. Mol. Cell Biol., 2019, 20, 353–367 CrossRef CAS PubMed.
- G. Drewes and S. Knapp, Trends Biotechnol., 2018, 36, 1275–1286 CrossRef CAS PubMed.
- C. G. Parker, C. A. Kuttruff, A. Galmozzi, L. Jørgensen, C.-H. Yeh, D. J. Hermanson, Y. Wang, M. Artola, S. J. McKerrall, C. M. Josyln, B. Nørremark, G. Dünstl, J. Felding, E. Saez, P. S. Baran and B. F. Cravatt, ACS Cent. Sci., 2017, 3, 1276–1285 CrossRef CAS PubMed.
- C. Bock, P. Datlinger, F. Chardon, M. A. Coelho, M. B. Dong, K. A. Lawson, T. Lu, L. Maroc, T. M. Norman, B. Song, G. Stanley, S. Chen, M. Garnett, W. Li, J. Moffat, L. S. Qi, R. S. Shapiro, J. Shendure, J. S. Weissman and X. Zhuang, Nat. Rev. Methods Primers, 2022, 2, 9 CrossRef PubMed.
- L. Przybyla and L. A. Gilbert, Nat. Rev. Genet., 2022, 23, 89–103 CrossRef CAS PubMed.
- S. Chen, N. E. Sanjana, K. Zheng, O. Shalem, K. Lee, X. Shi, D. A. Scott, J. Song, J. Q. Pan, R. Weissleder, H. Lee, F. Zhang and P. A. Sharp, Cell, 2015, 160, 1246–1260 CrossRef CAS PubMed.
- T. Rodrigues and G. J. L. Bernardes, Curr. Opin. Chem. Biol., 2020, 56, 16–22 CrossRef CAS PubMed.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritzel, O. Ronneberger, L. Willmore, A. J. Ballard, J. Bambrick, S. W. Bodenstein, D. A. Evans, C.-C. Hung, M. O'Neill, D. Reiman, K. Tunyasuvunakool, Z. Wu, A. Žemgulytė, E. Arvaniti, C. Beattie, O. Bertolli, A. Bridgland, A. Cherepanov, M. Congreve, A. I. Cowen-Rivers, A. Cowie, M. Figurnov, F. B. Fuchs, H. Gladman, R. Jain, Y. A. Khan, C. M. R. Low, K. Perlin, A. Potapenko, P. Savy, S. Singh, A. Stecula, A. Thillaisundaram, C. Tong, S. Yakneen, E. D. Zhong, M. Zielinski, A. Žídek, V. Bapst, P. Kohli, M. Jaderberg, D. Hassabis and J. M. Jumper, Nature, 2024, 630, 493–500 CrossRef PubMed.
- M. Bugnon, U. F. Röhrig, M. Goullieux, M. A. S. Perez, A. Daina, O. Michielin and V. Zoete, Nucleic Acids Res., 2024, 52, W324–W332 CrossRef PubMed.
- G. Schneider and P. Schneider, Expert Opin. Drug Discovery, 2017, 12, 271–277 CrossRef PubMed.
- R. Ratnayake, D. Covell, T. T. Ransom, K. R. Gustafson and J. A. Beutler, Org. Lett., 2009, 11, 57–60 CrossRef PubMed.
- Y. Akbulut, H. J. Gaunt, K. Muraki, M. J. Ludlow, M. S. Amer, A. Bruns, N. S. Vasudev, L. Radtke, M. Willot, S. Hahn, T. Seitz, S. Ziegler, M. Christmann, D. J. Beech and H. Waldmann, Angew Chem. Int. Ed. Engl., 2015, 54, 3787–3791 CrossRef PubMed.
- T. Rodrigues, F. Sieglitz, V. J. Somovilla, P. M. S. D. Cal, A. Galione, F. Corzana and G. J. L. Bernardes, Angew. Chem., Int. Ed., 2016, 55, 11077–11081 CrossRef PubMed.
- J. C. Habel, L. Rasche, U. A. Schneider, J. O. Engler, E. Schmid, D. Rödder, S. T. Meyer, N. Trapp, R. Sos del Diego, H. Eggermont, L. Lens and N. E. Stork, Conserv. Lett., 2019, 12, e12668 CrossRef.
- R. Cámara-Leret, D. G. Frodin, F. Adema, C. Anderson, M. S. Appelhans, G. Argent, S. Arias Guerrero, P. Ashton, W. J. Baker, A. S. Barfod, D. Barrington, R. Borosova, G. L. C. Bramley, M. Briggs, S. Buerki, D. Cahen, M. W. Callmander, M. Cheek, C.-W. Chen, B. J. Conn, M. J. E. Coode, I. Darbyshire, S. Dawson, J. Dransfield, C. Drinkell, B. Duyfjes, A. Ebihara, Z. Ezedin, L.-F. Fu, O. Gideon, D. Girmansyah, R. Govaerts, H. Fortune-Hopkins, G. Hassemer, A. Hay, C. D. Heatubun, D. J. N. Hind, P. Hoch, P. Homot, P. Hovenkamp, M. Hughes, M. Jebb, L. Jennings, T. Jimbo, M. Kessler, R. Kiew, S. Knapp, P. Lamei, M. Lehnert, G. P. Lewis, H. P. Linder, S. Lindsay, Y. W. Low, E. Lucas, J. P. Mancera, A. K. Monro, A. Moore, D. J. Middleton, H. Nagamasu, M. F. Newman, E. Nic Lughadha, P. H. A. Melo, D. J. Ohlsen, C. M. Pannell, B. Parris, L. Pearce, D. S. Penneys, L. R. Perrie, P. Petoe, A. D. Poulsen, G. T. Prance, J. P. Quakenbush, N. Raes, M. Rodda, Z. S. Rogers, A. Schuiteman, P. Schwartsburd, R. W. Scotland, M. P. Simmons, D. A. Simpson, P. Stevens, M. Sundue, W. Testo, A. Trias-Blasi, I. Turner, T. Utteridge, L. Walsingham, B. L. Webber, R. Wei, G. D. Weiblen, M. Weigend, P. Weston, W. de Wilde, P. Wilkie, C. M. Wilmot-Dear, H. P. Wilson, J. R. I. Wood, L.-B. Zhang and P. C. van Welzen, Nature, 2020, 584, 579–583 CrossRef PubMed.
- G. Kier, H. Kreft, T. M. Lee, W. Jetz, P. L. Ibisch, C. Nowicki, J. Mutke and W. Barthlott, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 9322–9327 CrossRef PubMed.
- S. Veron, T. Haevermans, R. Govaerts, M. Mouchet and R. Pellens, Sci. Rep., 2019, 9, 11693 CrossRef PubMed.
- J. M. Fernández-Palacios, H. Kreft, S. D. H. Irl, S. Norder, C. Ah-Peng, P. A. V. Borges, K. C. Burns, L. de Nascimento, J.-Y. Meyer, E. Montes and D. R. Drake, Global Ecol. Conserv., 2021, 31, e01847 CrossRef PubMed.
- B. H. Warren, D. Simberloff, R. E. Ricklefs, R. Aguilée, F. L. Condamine, D. Gravel, H. Morlon, N. Mouquet, J. Rosindell, J. Casquet, E. Conti, J. Cornuault, J. M. Fernández-Palacios, T. Hengl, S. J. Norder, K. F. Rijsdijk, I. Sanmartín, D. Strasberg, K. A. Triantis, L. M. Valente, R. J. Whittaker, R. G. Gillespie, B. C. Emerson and C. Thébaud, Ecol. Lett., 2015, 18, 200–217 CrossRef PubMed.
- R. Sayre, S. Noble, S. Hamann, R. Smith, D. Wright, S. Breyer, K. Butler, K. Van Graafeiland, C. Frye, D. Karagulle, D. Hopkins, D. Stephens, K. Kelly, Z. Basher, D. Burton, J. Cress, K. Atkins, D. P. Van Sistine, B. Friesen, R. Allee, T. Allen, P. Aniello, I. Asaad, M. J. Costello, K. Goodin, P. Harris, M. Kavanaugh, H. Lillis, E. Manca, F. Muller-Karger, B. Nyberg, R. Parsons, J. Saarinen, J. Steiner and A. Reed, J. Oper. Oceanogr., 2019, 12, S47–S56 Search PubMed.
- J. Schrader, P. Weigelt, L. Cai, M. Westoby, J. M. Fernández-Palacios, F. J. Cabezas, G. M. Plunkett, T. A. Ranker, K. A. Triantis, P. Trigas, Y. Kubota and H. Kreft, Nature, 2024, 634, 868–874 CrossRef PubMed.
- S. Sloan, C. N. Jenkins, L. N. Joppa, D. L. A. Gaveau and W. F. Laurance, Biol. Conserv., 2014, 177, 12–24 CrossRef.
- H. Petrén, R. A. Anaia, K. S. Aragam, A. Bräutigam, S. Eckert, R. Heinen, R. Jakobs, L. Ojeda-Prieto, M. Popp, R. Sasidharan, J.-P. Schnitzler, A. Steppuhn, F. M. Thon, S. B. Unsicker, N. M. van Dam, W. W. Weisser, M. J. Wittmann, S. Yepes, D. Ziaja, C. Müller and R. R. Junker, Ecol. Monogr., 2024, 94, e1635 CrossRef.
- G. Peguero, A. Gargallo-Garriga, J. Maspons, K. Klem, O. Urban, J. Sardans and J. Peñuelas, Plants, 2021, 10, 554 CrossRef PubMed.
- E. A. Courtois, C. E. T. Paine, P.-A. Blandinieres, D. Stien, J.-M. Bessiere, E. Houel, C. Baraloto and J. Chave, J. Chem. Ecol., 2009, 35, 1349–1362 CrossRef PubMed.
- N. Rønsted, M. R. E. Symonds, T. Birkholm, S. B. Christensen, A. W. Meerow, M. Molander, P. Mølgaard, G. Petersen, N. Rasmussen, J. van Staden, G. I. Stafford and A. K. Jäger, BMC Evol. Biol., 2012, 12, 182 CrossRef PubMed.
- T. Züst, S. R. Strickler, A. F. Powell, M. E. Mabry, H. An, M. Mirzaei, T. York, C. K. Holland, P. Kumar, M. Erb, G. Petschenka, J.-M. Gómez, F. Perfectti, C. Müller, J. C. Pires, L. A. Mueller and G. Jander, eLife, 2020, 9, e51712 CrossRef PubMed.
- P. Coulerie and C. Poullain, Chem. Biodivers., 2015, 12, 841–858 CrossRef PubMed.
- P. Coulerie and C. Poullain, Chem. Biodivers., 2016, 13, 18–36 CrossRef.
- P. Coulerie and C. Poullain, Chem. Biodivers., 2016, 13, 366–379 CrossRef PubMed.
- P. Meesakul, T. Shea, R. Fenstemacher, S. X. Wong, Y. Kuroki, A. Wada and S. Cao, Int. J. Mol. Sci., 2023, 24, 16323 CrossRef PubMed.
- W. L. Applequist, J. A. Brinckmann, A. B. Cunningham, R. E. Hart, M. Heinrich, D. R. Katerere and T. van Andel, Planta Med., 2020, 86, 10–18 Search PubMed.
- S. Goodwin, A. F. Smith and E. C. Horning, J. Am. Chem. Soc., 1959, 81, 1903–1908 CrossRef.
- W. Hou, X.-L. Xu, L.-J. Huang, Z.-Y. Zhang, Z.-N. Zhou, J.-Y. Wang, X. Ouyang, S.-Y. Xin, Z.-Y. Zhang, Y. Xiong, H. Huang and J.-X. Lan, Chem. Biodivers., 2024, 21, e202400210 CrossRef PubMed.
- C. M. Miller and F. O. McCarthy, RSC Adv., 2012, 2, 8883–8918 RSC.
- C. Paoletti, J. B. Le Pecq, N. Dat-Xuong, P. Juret, H. Garnier, J. L. Amiel and J. Rouesse, Recent Results Cancer Res., 1980, 74, 107–123 CrossRef PubMed.
- J. Rouëssé, M. Spielmann, F. Turpin, T. Le Chevalier, M. Azab and J. M. Mondésir, Eur. J. Cancer, 1993, 29, 856–859 CrossRef PubMed.
- C. Colichi, S. Delaloge, M. Spielman, L. Albiges, A. Goubar, A. Auperin and F. André, Eur. J. Cancer Suppl., 2009, 7, 267 CrossRef.
- F. M. Deane, E. C. O'Sullivan, A. R. Maguire, J. Gilbert, J. A. Sakoff, A. McCluskey and F. O. McCarthy, Org. Biomol. Chem., 2013, 11, 1334–1344 RSC.
- E. Tian, T. H. Landowski, O. W. Stephens, S. Yaccoby, B. Barlogie and J. D. Shaughnessy Jr, Mol. Cancer Ther., 2008, 7, 500–509 CrossRef PubMed.
- C. M. Miller, E. C. O'Sullivan and F. O. McCarthy, Pharmaceuticals, 2019, 12, 90 CrossRef PubMed.
- N. J. Martin, S. Prado, G. Lecellier, O. P. Thomas and P. Raharivelomanana, Molecules, 2012, 17, 12015–12022 CrossRef PubMed.
- F. Abdallah, G. Lecellier, P. Raharivelomanana and C. Pichon, Sci. Rep., 2019, 9, 4132 CrossRef.
- J.-M. Li, Y.-C. Huang, Y.-H. Kuo, C.-C. Cheng, F.-C. Kuan, S.-F. Chang, Y.-R. Lee, C.-C. Chin and C.-S. Shi, Cancers, 2019, 11, 1034 CrossRef PubMed.
- J.-J. Wu, S.-H. Chen, C.-H. Lee, Y.-Z. Li, Y.-W. Hsu, M.-Y. Hsieh and Y.-R. Lee, Am. J. Cancer Res., 2024, 14, 3317–3334 CrossRef PubMed.
- E. Bächli, C. Vamvacas, H. Schmid and P. Karrer, Helv. Chim. Acta, 1957, 40, 1167–1187 CrossRef.
- N. A. Hughes and H. Rapoport, J. Am. Chem. Soc., 1958, 80, 1604–1609 CrossRef.
- R. Yue, H. Liu, Y. Huang, J. Wang, D. Shi, Y. Su, Y. Luo, P. Cai, G. Jin and C. Yu, Front. Pharmacol, 2021, 12, 806091 CrossRef PubMed.
- B. Zhang, W. Wang, Y. Song, H. Chen, X. Lin, J. Chen, Y. Chen, J. Huang, D. Li and S. Wu, Pharmaceuticals, 2024, 17, 1318 CrossRef PubMed.
- R. B. Woodward and B. Witkop, J. Am. Chem. Soc., 1949, 71, 379 Search PubMed.
- M. Gorman, N. Neuss, C. Djerassi, J. P. Kutney and P. J. Scheuer, Tetrahedron, 1957, 1, 328–337 CrossRef.
- F. Ronchetti, G. Russo, E. Bombardelli and A. Bonati, Phytochemistry, 1971, 10, 1385–1388 CrossRef.
- N. J. Martin, S. F. Ferreiro, F. Barbault, M. Nicolas, G. Lecellier, C. Paetz, M. Gaysinski, E. Alonso, O. P. Thomas, L. M. Botana and P. Raharivelomanana, Phytochemistry, 2015, 109, 84–95 CrossRef PubMed.
- S. He, M. T. Moutaoufik, S. Islam, A. Persad, A. Wu, K. A. Aly, H. Fonge, M. Babu and F. S. Cayabyab, Biochim. Biophys. Acta, Rev. Cancer, 2020, 1873, 188355 CrossRef PubMed.
- M. Païs, R. Sarfati, F.-X. Jarreau and R. Goutarel, Tetrahedron, 1973, 29, 1001–1010 CrossRef.
- S. G. Davies and J. E. Thomson, in The Alkaloids: Chemistry and Biology, ed. H.-J. Knölker, Academic Press, 2015, vol. 74, pp. 121–158 Search PubMed.
- J. P. Matheny, P. M. Yamanushkin, P. A. Petillo and M. Rubin, RSC Adv., 2020, 10, 44183–44190 RSC.
- K. A. Oppong, C. D. Ellis, M. C. Laufersweiler, S. V. O'Neil, Y. Wang, D. L. Soper, M. W. Baize, J. A. Wos, B. De, G. K. Bosch, A. N. Fancher, W. Lu, M. K. Suchanek, R. L. Wang and T. P. Demuth, Bioorg. Med. Chem. Lett., 2005, 15, 4291–4294 CrossRef PubMed.
- Y. Peng, H. Sun, Z. Nikolovska-Coleska, S. Qiu, C.-Y. Yang, J. Lu, Q. Cai, H. Yi, S. Kang, D. Yang and S. Wang, J. Med. Chem., 2008, 51, 8158–8162 Search PubMed.
- R. Sheng, H. Sun, L. Liu, J. Lu, D. McEachern, G. Wang, J. Wen, P. Min, Z. Du, H. Lu, S. Kang, M. Guo, D. Yang and S. Wang, J. Med. Chem., 2013, 56, 3969–3979 CrossRef PubMed.
- Y. Adjibadé, H. Saad, T. Sévenet, B. Kuballa, J. C. Quirion and R. Anton, Planta Med., 1990, 56, 212–215 CrossRef.
- E. F. L. J. Anet, G. K. Hughes and E. Ritchie, Aust. J. Chem., 1961, 14, 173–174 CrossRef.
- K. P. Parry, Alkaloids from Hodgkinsonia frutescens: The Structure of the Quadrigemines, The University of Manchester, 1968 Search PubMed.
- K. P. Parry and G. F. Smith, J. Chem. Soc., Perkin Trans. 1, 1978, 1671–1682 RSC.
- F. Libot, C. Miet, N. Kunesch, J. Poisson, J. Pusset and T. Sévenet, J. Nat. Prod., 1987, 50(3), 468–473 Search PubMed.
- F. Guéritte-Voegelein, T. Sévenet, J. Pusset, M.-T. Adeline, B. Gillet, J.-C. Beloeil, D. Guénard, P. Potier, R. Rasolonjanahary and C. Kordon, J. Nat. Prod., 1992, 55, 923–930 CrossRef.
- V. Jannic, F. Guéritte, O. Laprévote, L. Serani, M.-T. Martin, T. Sévenet and P. Potier, J. Nat. Prod., 1999, 62, 838–843 Search PubMed.
- N. Hart, S. Johns, J. Lamberton and R. Summons, Aust. J. Chem., 1974, 27, 639–646 CrossRef CAS.
- H. E. Saad, S. H. el-Sharkawy and W. T. Shier, Planta Med., 1995, 61, 313–316 CrossRef CAS.
- A. Roth, B. Kuballa, C. Bounthanh, P. Cabalion, T. Sévenet, J. P. Beck and R. Anton, Planta Med., 1986, 450–453 CrossRef CAS PubMed.
- H. M. Katzenstein, W. L. Furman, M. H. Malogolowkin, M. D. Krailo, M. B. McCarville, A. J. Towbin, G. M. Tiao, M. J. Finegold, S. Ranganathan, S. P. Dunn, M. R. Langham, E. D. McGahren, C. Rodriguez-Galindo and R. L. Meyers, Cancer, 2017, 123, 2360–2367 Search PubMed.
- S. M. Canham, B. D. Hafensteiner, A. D. Lebsack, T. L. May-Dracka, S. Nam, B. A. Stearns and L. E. Overman, Tetrahedron, 2015, 71, 6424–6436 CrossRef CAS PubMed.
- J. Pu, W. Shi, J. Cui, H. Yang, J. Cao, Y. Liu, S. Xiao and G. Cheng, Int. J. Mol. Sci., 2025, 26, 4848 CrossRef CAS PubMed.
- T. Z. Scott, V. F. Armelin and M. Movassaghi, Org. Lett., 2022, 24, 2160–2164 Search PubMed.
- P. Lindovska and M. Movassaghi, J. Am. Chem. Soc., 2017, 139, 17590–17596 CrossRef CAS PubMed.
- C. R. Jamison, J. J. Badillo, J. M. Lipshultz, R. J. Comito and D. W. C. MacMillan, Nat. Chem., 2017, 9, 1165–1169 CrossRef CAS PubMed.
- R. H. Snell, M. J. Durbin, R. L. Woodward and M. C. Willis, Chem.–Eur. J., 2012, 18, 16754–16764 CrossRef CAS.
- M. Mangal, P. Sagar, H. Singh, G. P. S. Raghava and S. M. Agarwal, Nucleic Acids Res., 2013, 41, D1124–D1129 CrossRef CAS PubMed.
- O. Demirkiran, M. Campitelli, C. Wang and Y. Feng, Tetrahedron, 2016, 72, 8400–8405 CrossRef CAS.
- J. Zhang, D. T. Nguyen, G. K. Pierens, I. Cock and Y. Feng, Nat. Prod. Res., 2022, 36, 5199–5205 CrossRef CAS PubMed.
- M. K. Jogia and R. J. Andersen, Phytochemistry, 1987, 26, 3309–3311 CrossRef CAS.
- M. K. Jogia and R. J. Andersen, Can. J. Chem., 1989, 67, 257–260 CrossRef CAS.
- S. Shah and J. A. Bhat, J. Integr. Med., 2019, 17, 244–249 CrossRef PubMed.
- J. K. Rugutt, N. H. Fischer, M. A. Nauman, T. J. Schmidt and D. K. Berner, Spectrosc. Lett., 1996, 29, 711–726 CrossRef CAS.
- S. Fan, C. Zhang, T. Luo, J. Wang, Y. Tang, Z. Chen and L. Yu, Molecules, 2019, 24, 3679 CrossRef CAS PubMed.
- R. Melong, P. C. Tamokoue Kengne, J. P. Dzoyem, A. A. Fusi, E. Allemann, F. Delie, C. G. Bochet, U. Beifuss and G. D. W. F. Kapche, Nat. Prod. Res., 2022, 36, 2783–2790 CrossRef CAS.
- X. Luo, Z. Yu, B. Yue, J. Ren, J. Zhang, S. Mani, Z. Wang and W. Dou, Pharm. Biol., 2020, 58, 886–897 CrossRef CAS PubMed.
- Y. Gao, R. Hou, F. Liu, H. Liu, Q. Fei, Y. Han, R. Cai, C. Peng and Y. Qi, J. Cell. Biochem., 2018, 119, 837–849 CrossRef CAS PubMed.
- X. Luo, B. Yue, Z. Yu, Y. Ren, J. Zhang, J. Ren, Z. Wang and W. Dou, Front. Microbiol., 2020, 11, 497 CrossRef PubMed.
- Y. Ren and A. D. Kinghorn, J. Med. Chem., 2020, 63, 15410–15448 CrossRef CAS.
- W. Aalbersberg and Y. Singh, Phytochemistry, 1991, 30, 921–926 CrossRef CAS.
- H.-J. Yan, J.-S. Wang and L.-Y. Kong, J. Nat. Prod., 2014, 77, 234–242 CrossRef CAS PubMed.
- S. A. Adesanya, M. Païs, T. Sévenet and J. P. Cosson, J. Nat. Prod., 1991, 54, 1588–1594 CrossRef CAS.
- L. Ma, X. Wang, W. Li, D. Miao, Y. Li, J. Lu and Y. Zhao, Eur. J. Med. Chem., 2020, 187, 111964 CrossRef CAS PubMed.
- L. Holzmeyer, F. Hauenschild, D. J. Mabberley and A. N. Muellner-Riehl, Taxon, 2021, 70, 1248–1272 CrossRef.
- N. Allouche, C. Apel, M.-T. Martin, V. Dumontet, F. Guéritte and M. Litaudon, Phytochemistry, 2009, 70, 546–553 CrossRef CAS PubMed.
- D. Fomekong Fotsop, F. Roussi, C. Le Callonec, H. Bousserouel, M. Litaudon and F. Guéritte, Tetrahedron, 2008, 64, 2192–2197 CrossRef CAS.
- F. Daressy, L. Séguy, L. Favre, S. Corvaisier, C. Apel, A.-C. Groo, M. Litaudon, V. Dumontet, A. Malzert-Fréon, S. Desrat, F. Roussi, A. Robert and J. Wiels, Biomed. Pharmacother., 2022, 154, 113546 CrossRef CAS PubMed.
- F. Daressy, F. Malard, L. Seguy, V. Guérineau, C. Apel, V. Dumontet, A. Robert, A.-C. Groo, M. Litaudon, J. Bignon, S. Desrat, A. Malzert-Fréon, J. Wiels, E. Lescop and F. Roussi, ChemMedChem, 2021, 16, 1789–1798 CrossRef PubMed.
- L. Séguy, F. Daressy, S. Lahlil, S. Corvaisier, V. Dumontet, M. Litaudon, C. Apel, F. Roussi, J. Wiels, A. Robert, A.-C. Groo and A. Malzert-Fréon, Int. J. Pharm., 2023, 630, 122433 CrossRef.
- K. A. Wayman, P. J. de Lange, L. Larsen, C. E. Sansom and N. B. Perry, Phytochemistry, 2010, 71, 766–772 CrossRef CAS PubMed.
- J. Nadia, K. Shahbaz, M. Ismail and M. M. Farid, ACS Sustainable Chem. Eng., 2018, 6, 862–871 CrossRef CAS.
- V. Maslivetc, B. Laguera, S. Chandra, R. Dasari, W. J. Olivier, J. A. Smith, A. C. Bissember, M. Masi, A. Evidente, V. Mathieu and A. Kornienko, Int. J. Mol. Sci., 2021, 22, 11256 CrossRef CAS PubMed.
- R. Dasari, A. De Carvalho, D. C. Medellin, K. N. Middleton, F. Hague, M. N. M. Volmar, L. V. Frolova, M. F. Rossato, J. J. De La Chapa, N. F. Dybdal-Hargreaves, A. Pillai, V. Mathieu, S. Rogelj, C. B. Gonzales, J. B. Calixto, A. Evidente, M. Gautier, G. Munirathinam, R. Glass, P. Burth, S. C. Pelly, W. A. L. van Otterlo, R. Kiss and A. Kornienko, ChemMedChem, 2015, 10, 2014–2026 CrossRef CAS PubMed.
- R. Dasari, A. De Carvalho, D. C. Medellin, K. N. Middleton, F. Hague, M. N. M. Volmar, L. V. Frolova, M. F. Rossato, J. J. De La Chapa, N. F. Dybdal-Hargreaves, A. Pillai, R. E. Kälin, V. Mathieu, S. Rogelj, C. B. Gonzales, J. B. Calixto, A. Evidente, M. Gautier, G. Munirathinam, R. Glass, P. Burth, S. C. Pelly, W. A. L. van Otterlo, R. Kiss and A. Kornienko, Eur. J. Med. Chem., 2015, 103, 226–237 CrossRef CAS PubMed.
- R. Venkatesan, M. A. Hussein, L. Moses, J. S. Liu, S. R. Khetani, A. Kornienko and G. Munirathinam, Cancers, 2022, 14, 5260 CrossRef CAS PubMed.
- R. E. Corbett and T. L. Chee, J. Chem. Soc., Perkin Trans. 1, 1976, 850–857 RSC.
- Ae. de Groot, M. P. Broekhuysen, L. L. Doddema, M. C. Vollering and J. M. M. Westerbeek, Tetrahedron Lett., 1982, 23, 4831–4834 CrossRef CAS.
- D. Muhammad, N. Lalun, H. Bobichon, E. Le Magrex Debar, S. C. Gangloff, M. Nour and L. Voutquenne-Nazabadioko, Phytochemistry, 2016, 129, 45–57 CrossRef CAS PubMed.
- D. Muhammad, N. Lalun, H. Bobichon, E. Le Magrex Debar, S. C. Gangloff, M. Nour and L. Voutquenne-Nazabadioko, Phytochemistry, 2017, 141, 121–130 CrossRef CAS.
- P. L. Julian, J. Pikl and R. Dawson, J. Am. Chem. Soc., 1938, 60, 77–79 CrossRef CAS.
- G. Branch, D. Burgess, P. Dunstan, L. Foo, G. Green, J. G. Mack, E. Ritchie and W. Taylor, Aust. J. Chem., 1972, 25, 2209–2216 CrossRef CAS.
- R. Raju, D. Gunawardena, M. A. Ahktar, M. Low, P. Reddell and G. Münch, Molecules, 2016, 21, 1521 CrossRef PubMed.
- T. H. Kim, J. Lee, H.-J. Kim and C. Jo, J. Agric. Food Chem., 2017, 65, 6929–6935 CrossRef CAS PubMed.
- H. Slika, H. Mansour, N. Wehbe, S. A. Nasser, R. Iratni, G. Nasrallah, A. Shaito, T. Ghaddar, F. Kobeissy and A. H. Eid, Biomed. Pharmacother., 2022, 146, 112442 CrossRef CAS PubMed.
- C. Gorrini, I. S. Harris and T. W. Mak, Nat. Rev. Drug Discovery, 2013, 12, 931–947 CrossRef CAS PubMed.
- D. Muhammad, J. Hubert, N. Lalun, J.-H. Renault, H. Bobichon, M. Nour and L. Voutquenne-Nazabadioko, Phytochem. Anal., 2015, 26, 137–144 CrossRef CAS PubMed.
- R. Al Omar, R. Micklewright, K. Masud, T. Naz, S. Vemulpad and J. Jamie, J. Ethnopharmacol., 2022, 294, 115168 CrossRef PubMed.
- S. Molimau-Samasoni, V. H. Woolner, S. T. Foliga, K. Robichon, V. Patel, S. K. Andreassend, J. P. Sheridan, T. Te Kawa, D. Gresham, D. Miller, D. J. Sinclair, A. C. La Flamme, A. V. Melnik, A. Aron, P. C. Dorrestein, P. H. Atkinson, R. A. Keyzers and A. B. Munkacsi, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2100880118 CrossRef CAS.
- T. Cañeque, L. Baron, S. Müller, A. Carmona, L. Colombeau, A. Versini, S. Solier, C. Gaillet, F. Sindikubwabo, J. L. Sampaio, M. Sabatier, E. Mishima, A. Picard-Bernes, L. Syx, N. Servant, B. Lombard, D. Loew, J. Zheng, B. Proneth, L. K. Thoidingjam, L. Grimaud, C. S. Fraser, K. J. Szylo, E. Der Kazarian, C. Bonnet, E. Charafe-Jauffret, C. Ginestier, P. Santofimia-Castaño, M. Estaras, N. Dusetti, J. L. Iovanna, A. S. Cunha, G. Pittau, P. Hammel, D. Tzanis, S. Bonvalot, S. Watson, V. Gandon, A. Upadhyay, D. A. Pratt, F. P. Freitas, J. P. Friedmann Angeli, B. R. Stockwell, M. Conrad, J. M. Ubellacker and R. Rodriguez, Nature, 2025, 642, 492–500 CrossRef.
- K. Roemhild, F. von Maltzahn, R. Weiskirchen, R. Knüchel, S. von Stillfried and T. Lammers, Trends Pharmacol. Sci., 2021, 42, 640–656 CrossRef CAS.
- D. Dugan, R. J. Bell, R. Brkljača, C. Rix and S. Urban, Metabolites, 2024, 14, 81 CrossRef CAS PubMed.
- D. L. Dreyer and A. Lee, Phytochemistry, 1972, 11, 763–767 CrossRef CAS.
- Q. Shou, L. K. Banbury, D. E. Renshaw, J. E. Smith, X. He, A. Dowell, H. J. Griesser, M. Heinrich and H. Wohlmuth, J. Nat. Prod., 2013, 76, 1384–1387 CrossRef CAS PubMed.
- J. J. Brophy, R. J. Goldsack and P. I. Forster, J. Essent. Oil Res., 2005, 17, 169–174 CrossRef.
- L. K. Banbury, Q. Shou, D. E. Renshaw, E. H. Lambley, H. J. Griesser, H. Mon and H. Wohlmuth, J. Ethnopharmacol., 2015, 163, 251–255 CrossRef CAS PubMed.
- G. Viola, D. Vedaldi, F. dall'Acqua, G. Basso, S. Disarò, M. Spinelli, B. Cosimelli, M. Boccalini and S. Chimichi, Chem. Biodiv., 2004, 1, 1265–1280 CrossRef CAS.
- S. Chimichi, M. Boccalini, A. Salvador, F. Dall'Acqua, G. Basso and G. Viola, ChemMedChem, 2009, 4, 769–779 CrossRef CAS PubMed.
- Q. Shou, L. K. Banbury, A. T. Maccarone, D. E. Renshaw, H. Mon, S. Griesser, H. J. Griesser, S. J. Blanksby, J. E. Smith and H. Wohlmuth, Fitoterapia, 2014, 93, 62–66 CrossRef CAS.
- D. Bruy, G. Lannuzel, G. Gâteblé and J. Munzinger, Phytotaxa, 2023, 578, 228–240 CrossRef.
- K.-T. Le, J. J. Bandolik, M. U. Kassack, K. R. Wood, C. Paetzold, M. S. Appelhans and C. M. Passreiter, Molecules, 2021, 26, 688 CrossRef CAS PubMed.
- K. Miyake, A. Suzuki, C. Morita, M. Goto, D. J. Newman, B. R. O'Keefe, S. L. Morris-Natschke, K.-H. Lee and K. Nakagawa-Goto, J. Nat. Prod., 2016, 79, 2883–2889 CrossRef CAS PubMed.
- C.-R. Su, P.-C. Kuo, M.-L. Wang, M.-J. Liou, A. G. Damu and T.-S. Wu, J. Nat. Prod., 2003, 66, 990–993 CrossRef CAS PubMed.
- C. Ito, M. Hosono, H. Tokuda, T.-S. Wu and M. Itoigawa, Nat. Prod. Commun., 2016, 11, 1934578X1601100929 Search PubMed.
- H. Ahmadpourmir, H. Attar, J. Asili, V. Soheili, S. F. Taghizadeh and A. Shakeri, Chem. Biodivers., 2024, 14, 28 CAS.
- D. Harneti and U. Supratman, Phytochemistry, 2021, 181, 112540 CrossRef CAS PubMed.
- H. Greger, Phytochem. Rev., 2022, 21, 725–764 CrossRef CAS.
- H. Greger, T. Pacher, B. Brem, M. Bacher and O. Hofer, Phytochemistry, 2001, 57, 57–64 CrossRef CAS PubMed.
- V. Dumontet, O. Thoison, O. R. Omobuwajo, M.-T. Martin, G. Perromat, A. Chiaroni, C. Riche, M. Païs, T. Sévenet, A. Hamid and A. Hadi, Tetrahedron, 1996, 52, 6931–6942 CrossRef CAS.
- B. Y. Hwang, B.-N. Su, H. Chai, Q. Mi, L. B. S. Kardono, J. J. Afriastini, S. Riswan, B. D. Santarsiero, A. D. Mesecar, R. Wild, C. R. Fairchild, G. D. Vite, W. C. Rose, N. R. Farnsworth, G. A. Cordell, J. M. Pezzuto, S. M. Swanson and A. D. Kinghorn, J. Org. Chem., 2004, 69, 3350–3358 CrossRef CAS PubMed.
- M. L. King, C.-C. Chiang, H.-C. Ling, E. Fujita, M. Ochiai and A. T. McPhail, J. Chem. Soc., Chem. Commun., 1982, 1150–1151 RSC.
- S.-K. Wang, Y.-J. Cheng and C.-Y. Duh, J. Nat. Prod., 2001, 64, 92–94 CrossRef CAS PubMed.
- J. H. Chaidir, B. W. Nugroho, F. I. Bohnenstengel, V. Wray, L. Witte, P. D. Hung, L. C. Kiet, W. Sumaryono and P. Proksch, Phytochemistry, 1999, 52, 837–842 CrossRef CAS.
- B. W. Nugroho, R. A. Edrada, B. Güssregen, V. Wray, L. Witte and P. Proksch, Phytochemistry, 1997, 44, 1455–1461 CrossRef CAS.
- G. Schulz, C. Victoria, A. Kirschning and E. Steinmann, Nat. Prod. Rep., 2021, 38, 18–23 RSC.
- G. Peron, A. Mastinu, S. I. Peña-Corona, H. Hernández-Parra, G. Leyva-Gómez, D. Calina and J. Sharifi-Rad, Biomed. Pharmacother., 2024, 177, 117047 CrossRef CAS PubMed.
- Q. Yin, G. Chen, J. Hao, B. Lin, Q. Meng, L. Xu, D. Zhou, Y. Hou and N. Li, Phytochemistry, 2025, 229, 114298 CrossRef CAS PubMed.
- Y. Wu, M. Giaisi, R. Köhler, W.-M. Chen, P. H. Krammer and M. Li-Weber, Cancer Lett., 2017, 389, 70–77 CrossRef CAS PubMed.
- Y. Huang, X. Yang, T. Xu, Q. Kong, Y. Zhang, Y. Shen, Y. Wei, G. Wang and K.-J. Chang, Int. J. Oncol., 2016, 49, 153–163 CrossRef CAS PubMed.
- A. D. Nalli, L. E. Brown, C. L. Thomas, T. J. Sayers, J. A. Porco and C. J. Henrich, Sci. Rep., 2018, 8, 17519 CrossRef PubMed.
- C. Yao, Z. Ni, C. Gong, X. Zhu, L. Wang, Z. Xu, C. Zhou, S. Li, W. Zhou, C. Zou and S. Zhu, Autophagy, 2018, 14, 1831–1844 CrossRef CAS PubMed.
- X. Yan, C. Yao, C. Fang, M. Han, C. Gong, D. Hu, W. Shen, L. Wang, S. Li and S. Zhu, Int. J. Biol. Sci., 2022, 18, 585–598 CrossRef CAS PubMed.
- B. Baumann, F. Bohnenstengel, D. Siegmund, H. Wajant, C. Weber, I. Herr, K.-M. Debatin, P. Proksch and T. Wirth, J. Biol. Chem., 2002, 277, 44791–44800 CrossRef CAS PubMed.
- S. D. Stone, N. J. Lajkiewicz, L. Whitesell, A. Hilmy and J. A. Jr. Porco, J. Am. Chem. Soc., 2015, 137, 525–530 CrossRef CAS PubMed.
- L. M. Blair, M. B. Calvert and J. Sperry, in The Alkaloids: Chemistry and Biology, ed. H.-J. Knölker, Academic Press, 2017, vol. 77, pp. 85–115 Search PubMed.
- I. V. F. dos Santos, R. S. Borges, G. M. Silva, L. R. de Lima, R. S. Bastos, R. S. Ramos, L. B. Silva, C. H. T. P. da Silva and C. B. R. dos Santos, Front. Mol. Biosci., 2022, 9, 836572 CrossRef PubMed.
- T. T. T. Le, J. Gardner, D. Hoang-Le, C. W. Schmidt, K. P. MacDonald, E. Lambley, W. A. Schroder, S. M. Ogbourne and A. Suhrbier, Vaccine, 2009, 27, 3053–3062 Search PubMed.
- Q. Shou, L. K. Banbury, D. E. Renshaw, J. E. Smith, X. He, A. Dowell, H. J. Griesser, M. Heinrich and H. Wohlmuth, J. Nat. Prod., 2013, 76, 1384–1387 CrossRef PubMed.
- N. P. Keller, Nat. Rev. Microbiol., 2019, 17, 167–180 CrossRef PubMed.
- A. Evidente, A. Kornienko, A. Cimmino, A. Andolfi, F. Lefranc, V. Mathieu and R. Kiss, Nat. Prod. Rep., 2014, 31, 617–627 Search PubMed.
- A. Kornienko, A. Evidente, M. Vurro, V. Mathieu, A. Cimmino, M. Evidente, W. A. L. van Otterlo, R. Dasari, F. Lefranc and R. Kiss, Med. Res. Rev., 2015, 35, 937–967 CrossRef PubMed.
- J. F. Borel, C. Feurer, H. U. Gubler and H. Stähelin, Agents Actions, 1976, 6, 468–475 CrossRef PubMed.
- R. Bentley, Chem. Rev., 2000, 100, 3801–3826 CrossRef PubMed.
- D. G. I. Kingston, J. Nat. Prod., 2011, 74, 496–511 CrossRef PubMed.
- D. Zouraris, K. Graikou, P. Vasileiou, V. Dimitrov, Z. D. Stevanovic, A. R. Bilia, J. Zivkovic, A. Dias, K. Kasiotis, K. Gardikis, P. Dias, M. Oluški, J. R. M. Montaño, H. Hristova, H. Iliev, G. Petrangolini, A. Afantitis and N. Aligiannis, Comput. Struct. Biotechnol. J., 2025, 29, 85–94 CrossRef PubMed.
- M.-J. R. Howes, C. L. Quave, J. Collemare, E. C. Tatsis, D. Twilley, E. Lulekal, A. Farlow, L. Li, M.-E. Cazar, D. J. Leaman, T. A. K. Prescott, W. Milliken, C. Martin, M. N. De Canha, N. Lall, H. Qin, B. E. Walker, C. Vásquez-Londoño, B. Allkin, M. Rivers, M. S. J. Simmonds, E. Bell, A. Battison, J. Felix, F. Forest, C. Leon, C. Williams and E. Nic Lughadha, Plants People Planet, 2020, 2, 463–481 CrossRef.
- G. Llauradó Maury, D. Méndez Rodríguez, S. Hendrix, J. C. Escalona Arranz, Y. Fung Boix, A. O. Pacheco, J. García Díaz, H. J. Morris-Quevedo, A. Ferrer Dubois, E. I. Aleman, N. Beenaerts, I. E. Méndez-Santos, T. Orberá Ratón, P. Cos and A. Cuypers, Antioxidants, 2020, 9, 1048 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |