 Open Access Article
Open Access ArticleHigh reliability Ag/Ni/NiO nanowire-based SERS for cancer detection: a study on breast cancer
Alondra
Hernandez Cedillo
 *ab,
Javier
Mendez-Lozoya
ab,
Coral
Hernandez Cedillo
ab,
Alfred L. M.
Bothwell
cd and
Miguel Jose
Yacaman
*ab,
Javier
Mendez-Lozoya
ab,
Coral
Hernandez Cedillo
ab,
Alfred L. M.
Bothwell
cd and
Miguel Jose
Yacaman
 ab
ab
aDepartment of Applied Physics and Materials Science, Northern Arizona University, Flagstaff, Arizona 86011, USA. E-mail: ah3526@nau.edu
bCenter for Materials Interfaces in Research and Applications, Northern Arizona University, Flagstaff, Arizona 86011, USA
cDepartment of Pathology, Microbiology, and Immunology, University of Nebraska Medical Center, Omaha, Nebraska 68198, USA
dDepartment of Immunobiology, Yale University School of Medicine, New Haven, Connecticut 06520, USA
First published on 28th January 2026
Abstract
Over the past decade, substantial research has focused on identifying cancer biomarkers using Raman spectroscopy. However, no commercial Raman-based diagnostic tests are currently available. A major barrier is the need to amplify the inherently weak Raman signal, typically achieved through Surface-Enhanced Raman Spectroscopy (SERS). Because SERS relies on nanostructured substrates, its clinical translation has been hindered primarily by poor reproducibility, which undermines data reliability and prevents regulatory approval. In this pilot study, we evaluate an improved SERS substrate based on Ag/Ni/NiO nanowires, previously reported by our group, which significantly enhances sensitivity, specificity, detection limits, and critically substrate reproducibility. We further strengthen analytical performance by incorporating machine learning methods for spectral interpretation. Using this optimized platform, we analyzed SERS spectra from tissue samples of 31 breast cancer patients and compared them with matched healthy controls. Our results demonstrate a clear spectral distinction between cancerous and non-cancerous tissue. More importantly, we showed that SERS combined with machine learning, can differentiate major breast cancer subtypes, including luminal A (LUMA), luminal B (LUMB), HER2-enriched (HER2), and triple-negative breast cancer (TNBC). PCA-LDA modeling yielded exceptional diagnostic metrics, achieving up to 99% accuracy, sensitivity, and specificity across multiple classification tasks. Although this pilot study includes a limited number of samples, the findings demonstrate that improved SERS substrates paired with machine-learning analysis can generate a unique molecular fingerprint of breast cancer and its subtypes. This work opens a pathway for developing assays, potentially using blood or saliva, to enable early detection and subtype identification based on SERS-derived cancer fingerprints.
Introduction
Breast cancer is the most commonly diagnosed cancer among women worldwide, and its impact on public health continues to rise. A recent global study published in Nature Medicine (2025)1 estimates that 1 in 20 women globally will be diagnosed with breast cancer during their lifetime. If current trends persist, the annual number of new cases could reach 3.2 million by 2050, with approximately 1.1 million deaths each year, disproportionately affecting countries with limited healthcare resources.Biomarkers that aid diagnosis, prognosis, and prediction of breast cancer are essential for early detection and effective disease management throughout treatment. Advances in genetic profiling and molecular signalling pathways have led to the identification of various tissue- and blood-based biomarkers (i.e., liquid biopsies) that can assess cancer progression, recurrence risk, treatment response, and drug tolerance.2
Multiple classes of macromolecules have been proposed as key diagnostic biomarkers, including circular RNAs, microRNAs (miRNAs), DNA, proteins, exosomes, and antibodies. However, clinical trials assessing their diagnostic efficacy have not yet produced conclusive results.3 In contrast, the use of biomarkers in drug development and clinical research has shown encouraging progress.4,5 Early detection is critical for improving breast cancer survival rates and treatment outcomes. Mammography is traditionally regarded as the gold standard for screening,6 enabling clinicians to identify abnormalities in breast tissue before symptoms appear. Other imaging techniques, such as ultrasound and magnetic resonance imaging (MRI), are often used alongside mammography, particularly in women with dense breast tissue or elevated risk factors.7
Despite their widespread use, these screening methods have notable limitations. For example, mammography can miss tumors or generate false positives8–11 leading to unnecessary biopsies and potential overdiagnosis.12–15 Its sensitivity is also reduced in younger women or those with dense breast tissue, where lesions may be obscured.16,17 While ultrasound and MRI offer greater sensitivity in some cases, they are more expensive, less accessible in many regions, and also prone to higher false-positive rates.18–20
Surface-Enhanced Raman Spectroscopy (SERS) is one of the most promising techniques for detecting cancer biomarkers and developing early diagnostic tests. This method enables the detection of organic molecules with exceptionally high sensitivity, reaching detection limits close to the single-molecule level, making it particularly well-suited for biomarker identification.21,22 A literature survey reveals approximately 2000 publications over the past two decades reporting cancer biomarker detection using SERS (see, for example, reviews22–27), yet no SERS-based clinical test for early cancer detection has been approved to date.
The primary challenge limiting the clinical translation of SERS lies in the lack of data consistency and reproducibility, which undermines the reliability required for clinical trials.28–30 SERS relies on nanomaterials, such as nanoparticles, nanowires, and nanorods, to enhance the Raman signal, providing a high signal-to-noise ratio and enabling quantitative analysis.31
However, achieving precise control of nanomaterial shape, size, and crystal structure is challenging, resulting in significant batch-to-batch variation in substrate performance and amplification efficiency. Additionally, analyte molecules must adsorb onto the nanostructure surface to generate SERS enhancement. Many biomolecules exhibit low affinity for the substrate, and may instead fall into non-enhancing regions, resulting in a higher-than-desired limit of detection (LOD). Because sensitivity is proportional to approximately three times the noise divided by LOD, a high LOD directly reduces achievable sensitivity.32,33
Specificity is another critical parameter and depends on instrument noise, fluorescence background, and spectral peak overlap among different analytes. In biological samples, biomarker molecules are frequently bound to other molecular species, producing peak shifts that complicate comparison with reference spectra of isolated molecule. This mismatch introduces errors in peak identification. Sensitivity and specificity are largely independent parameters, and successful Raman-SERS assays require high performance in both, coupled with a low LOD and high substrate reproducibility.34
In this study, we address the fundamental issues that have hindered the translation of Raman-based diagnostic tests to the clinical and commercial sectors.
2. The SERS substrate (Ag/Ni/NiO)
2.1 Enhancement modes
Ag/Ni/NiO nanowires are used as the SERS substrate. Nanowires are fabricated using a standard method.35 The nanowires are thoroughly cleaned to remove all organic materials and subsequently coated with a nickel (Ni) layer a few nanometers thick. Upon exposure to air, the top Ni layer forms NiO, which passivates the nanowires. As a result, the nanowires become magnetic and can be easily aligned on the substrate. Fig. 1(a) shows SEM images of a single nanowire and nanowires deposited on an aluminum substrate, as well as nanowires aligned using a magnetic field. The substrate coverage by nanowires is approximately 98%. Fig. 1(b) High-resolution TEM image of an individual Ag/Ni/NiO nanowire; the schematic illustrates the layer structure, and the nanowire exhibits a five-fold symmetry. Fig. 1(c) SEM image of nanowires aligned along a preferred direction after magnetic-field-assisted assembly. Fig. 1(d) STEM image of a single nanowire. Fig. 1(e) STEM-EDS elemental map of Ag (green), corresponding to the nanowire first layer. Fig. 1(f) STEM-EDS elemental map of Ni (red), corresponding to the second layer. Fig. 1(g) Overlay of Ag (green) and Ni (red) elemental maps, confirming the presence and uniformity of the surface coating. Ag/Ni/NiO nanostructures support all major Raman enhancement mechanisms: plasmonic, chemical, and magnetic.2.2 Sensitivity, specificity, and limit of detection (LOD) of the SERS substrates
One of the critical aspects of Raman analysis is achieving optimal sensitivity and specificity. Sensitivity, defined as the ability to detect true positives, is closely linked to the limit of detection (LOD); lower LOD values result in higher sensitivity. The LOD depends on the SERS enhancement factor, system noise, chemical affinity between the substrate and analyte, and substrate coverage. Analyte molecules that fall into uncovered regions of the substrate are not detected. In addition, nanostructure degradation or aggregation over time can increase the LOD. In the proposed substrate, nanowire coverage reaches approximately 98%, and the three-dimensional architecture enhances analyte capture probability. The magnetic field of the Ag/Ni/NiO nanowires and their affinity for proteins further guarantee high sensitivity. Since most analyte molecules possess a dipole moment, the magnetic field attracts them toward the nanowires. Furthermore, the multilayer 3D structure increases the likelihood of analyte attachment. Sensitivity and specificity of the Ag/Ni/NiO nanowires were evaluated using two test molecules: rhodamine and sialic acid (Neu5Ac). Raman peak intensities were measured as a function of concentration, as shown in Fig. 3. The LODs for rhodamine and sialic acid were determined to be approximately 10−14 M and 10−10 M, respectively. Specificity measures the ability to avoid false positives and is not directly related to LOD. Instead, it depends on peak separation, spectral interference, and surface chemistry. Effective separation of peaks from different analytes is critical for achieving high specificity. | ||
| Fig. 3 (a) Pairwise ROC and (b) box plot analyses comparing response of sialic acid analyte at different concentrations and box plot of measured intensities at 940 cm−1. | ||
2.3 Software and fingerprint approach
One of the main challenges of Raman analysis is that real samples contain multiple molecular species, including both biomarkers of interest and background substances. Additionally, each molecule produces multiple Raman peaks, many of which overlap and exhibit frequency shifts due to chemical bonding. Consequently, peak intensities in experimental spectra represent convolutions of contributions from multiple species, which introduces error and limits spectral consistency. To correctly identify an analyte, its complete vibrational fingerprint must be observed rather than relying on individual peaks. A common example in breast cancer research is sialic acid (SA), which is often considered a cancer biomarker. However, SA binds to glycoproteins and Siglecs, and its concentration increases in many cancers as well as inflammatory conditions. Therefore, SA elevation alone cannot be uniquely linked to breast cancer. In this work, we adopt a fingerprint-based approach by analyzing averaged spectra from cancerous tissue. An optical microscope coupled with a motorized stage was used to scan regions containing stromal and tumor tissue. Multiple spectra (27 per sample) were acquired and averaged to generate a representative spectral fingerprint. We propose that this averaged spectrum reflects the molecular fingerprint of the tumor. The resulting SERS data were analyzed using Principal Component Analysis (PCA) and Linear Discriminant Analysis (LDA), which facilitate dimensionality reduction and supervised classification, respectively.34 Breast cancer is a highly heterogeneous disease comprising multiple molecular subtypes, including luminal A (LUMA), luminal B (LUMB), HER2-enriched (HER2), and triple-negative breast cancer (TNBC).35–38 The goal of this study is to determine whether Raman-based spectroscopic techniques can reliably differentiate these subtypes by identifying consistent and reproducible spectral biomarkers, thereby supporting histopathological diagnosis and enabling more targeted clinical decision-making.Results and discussion
The averaged Surface-Enhanced Raman Scattering (SERS) spectra obtained from 31 samples are shown in Fig. 4, representing five distinct breast tissue classifications: normal (blue), Triple-Negative Breast Cancer (TNBC, yellow), HER2-positive (HER2, green), Luminal A (LUMA, red), and Luminal B (LUMB, purple). The spectra reveal distinct vibrational profiles among the groups, reflecting biochemical differences associated with each subtype. These averaged spectra provide a clear overview of spectral trends across breast cancer subtypes and healthy tissue, serving as the foundation for subsequent statistical and machine learning analyses.To further examine intra-group consistency and variability, Fig. 5 and 6 display grouped 2D and 3D average SERS spectra for individual breast tissue subtypes.Fig. 5(a) and 6(a) show the averaged spectra of six normal tissue samples, characterized by consistent peaks indicative of a uniform biochemical composition. Fig. 5(b) and 6(b) present spectra from eight TNBC samples, which exhibit low intra-group variability, suggesting molecular homogeneity commonly associated with this aggressive subtype. Fig. 5(c) and 6(c) include four HER2-positive spectra, showing distinct vibrational features that may be linked to HER2 overexpression. Fig. 5(d) and 6(d) display spectra from ten Luminal A (LUMA) samples, while Fig. 5(e) and 6(e) represent five Luminal B (LUMB) samples.
 | ||
| Fig. 5 3D Grouped average SERS spectra of individual breast tissue subtypes: (a) normal, (b) TNBC, (c) HER 2, (d) LUMA, and (e) LUMB. | ||
 | ||
| Fig. 6 2D Grouped average SERS spectra of individual breast tissue subtypes: (a) normal, (b) TNBC, (c) HER 2, (d) LUMA, and (e) LUMB. | ||
Fig. 7(a) displays 2D and Fig. 7(b) 3D grouped average SERS spectra of breast tissue subtypes: Normal, TNBC, HER 2, LUMA, and LUMB. Collectively, these results highlight both inter- and intra-group spectral diversity captured by SERS, reinforcing its potential as a sensitive tool for biochemical fingerprinting and breast cancer classification. To assess potential contributions from paraffin and tissue processing, SERS spectra were acquired from FFPE sections before and after deparaffinization using ethanol, hexane, and water, revealing no appreciable changes in spectral features or classification-relevant bands (Fig. S1). Pathology-guided tumor-region delineation in FFPE breast cancer tissue has been demonstrated in prior work by our group,39 supporting the feasibility of integrating histological annotation with SERS.
 | ||
| Fig. 7 (a) 2D and (b) 3D grouped average SERS spectra of breast tissue subtypes: normal (blue), TNBC (yellow), HER 2 (green), LUMA (red), and LUMB (purple). | ||
Fig. 8 and Table 1 show a comparative SERS spectral analysis between breast cancer and normal breast tissue, reveal distinct vibrational signatures, indicative of underlying biochemical alterations associated with tumor malignancy, a complete list of SERS peaks and vibrational mode assignments on Table S1. A pronounced peak at 482 cm−1 is observed exclusively in the breast cancer tissue, with no corresponding signal in the normal spectrum. This peak is attributed to glycogen, a biomarker of metabolic adaptation under hypoxia, and its elevated presence in cancer aligns with increased glycolytic activity characteristic of the Warburg effect.40 Similarly, the 865 cm−1 peak, associated with collagen, lipids, saccharides, and proline, is significantly elevated in cancerous samples and absent in normal tissue. This elevation reflects extracellular matrix remodelling and enhanced collagen deposition, hallmarks of tumor progression.
 | ||
| Fig. 8 Differentiation of normal and breast cancer tissue using average SERS spectra and Ag/Ni/NiO NWs substrate. | ||
| Breast cancer tissue (SERS shift cm−1) | Normal tissue (SERS shift cm−1) | Raman assignment | Biochemical assignment | Biological relevance | Cancer context | References |
|---|---|---|---|---|---|---|
| 482 | Glycogen | Metabolic adaptation; glycolytic activity | Elevated in tumors under hypoxia | 40 | ||
| 865 | C–C stretching | Collagen, lipid, saccharides, proline | Breast tissue carcinogenesis | Elevated in tumors | 41 | |
| 890 | 890 | Protein | Structural protein signature | Elevated in cancerous tissue | 42 | |
| 1005 | 1005 | ν(C–C) ring breathing modes | Phenylalanine | Protein synthesis, metabolic regulation | Increased in tumors | 43 |
| 1030 | Collagen | Proliferation of fibroblasts and accumulation of collagen | Increased in tumors | 44 | ||
| 1063 | 1063 | C–C stretch, C–O and C–N stretch, O–P–O stretch | Lipids, proteins, DNA and RNA, and proline (collagen) | Altered lipid metabolism | decrease lipid signals | 45 and 46 |
| 1092 | 1109 | C–C stretch | Protein | Protein vibration | Higher in cancer | 47 |
| 1134 | 1134 | C–C stretch | Tryptophan | Tumor metabolic state | Decreased intensity | 48 |
| 1172 | 1172 | C–H bending | Proteins | Amide III, side-chain vibrations | Elevated in tumors | 49 |
| 1270 | ν(C–N) | Amide III | Lipid composition | Ratio used to differentiate tissue types | 50 | |
| 1296 | 1296 | ν(CN) and ν(NH) | Phospholipids | Cell proliferation | Increased in tumors | 51 |
| 1369 | 1369 | (CH3) stretch | Proteins/Lipid oxidation | Lipid peroxidation; redox balance | Oxidation leads to depletion | 52 |
| 1418 | 1418 | Lipids | Lipogenesis | Increased in tumors | 46, 51 and 53 | |
| 1440 | 1440 | CH2 bending mode, C–O symmetric stretching, CH2/CH3 deformation | Lipids | CH2 bending mode | Lower in cancer; indicator of lipid loss | 49 and 52 |
| 1463 | 1463 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching N stretching |
Lipids | Lipogenesis | Increased in tumors | 41 |
Protein-related features also demonstrate clear differentiation. The 890 cm−1 peak, corresponding to structural proteins, and the 1005 cm−1 peak, associated with phenylalanine, are both higher in cancerous tissue, supporting literature that links increased protein synthesis and metabolic regulation with tumor proliferation.43,42 Furthermore, the 1030 cm−1 collagen band is markedly higher in breast cancer tissue and absent in normal spectra, suggesting increased fibroblast activity and matrix stiffening. The 1063 cm−1 peak, reflecting a complex mixture of lipids, proteins, nucleic acids, and collagen-related proline, appears slightly reduced in cancer tissue, implying disrupted lipid metabolism and altered structural composition, both consistent with cancer-associated metabolic reprogramming.
Additional differences are evident in peaks related to protein folding and structural motifs. The 1109 cm−1 peak, associated with protein vibrations, is stronger in breast cancer tissue, consistent with elevated protein turnover in malignancy.47 The 1134 cm−1 band, linked to tryptophan, shows decreased intensity in cancer, reflecting shifts in tumor metabolic states. The 1172 cm−1 Amide III band, linked to side-chain vibrations, and the 1270 cm−1 peak, specific to cancerous tissue, are both associated with protein secondary structure and are commonly elevated in tumors.49 These features further underscore the SERS platform's sensitivity to subtle conformational changes in biomolecules during tumorigenesis.
Lipid-associated peaks also present distinct trends. The 1296 cm−1 phospholipid band, linked to membrane dynamics and cell proliferation, shows a slightly lower intensity in cancer tissue, potentially due to increased membrane turnover. In contrast, the 1369 cm−1 peak, related to lipid oxidation and redox imbalance, is reduced in cancer tissue, reflecting oxidative stress and depletion of oxidized species.52 The 1418 cm−1 lipid band is higher in normal tissue, possibly indicating lipid loss in malignancy. Interestingly, the 1440 cm−1 and 1463 cm−1 CH2 bending modes are significantly elevated in breast cancer spectra, suggesting lipid accumulation or membrane reorganization within tumor cells. Together, these findings demonstrate that Ag/Ni/NiO nanowire-based SERS not only captures reliable molecular fingerprints of breast cancer but also provides mechanistic insights into tumor biochemistry through label-free vibrational profiling.
The SERS spectra obtained from breast cancer tissue samples exhibited distinctive vibrational fingerprints that varied by molecular subtype. These spectral differences were analyzed to identify key peaks and assign their molecular origins, thereby elucidating biochemical characteristics associated with each subtype.
Triple-negative breast cancer (TNBC)
The TNBC subtype demonstrated unique SERS features characterized by sharp peaks at 892, 1065, 1112, 1135, 1297, and 1463 cm−1 (Table S2).46,48,54–56 These peaks are attributed to protein structural vibrations, C–C skeletal stretching in polysaccharides, and nucleic acid components. Notably, the absence of the 1172 cm−1 band in TNBC supports its distinctive protein conformation compared to other subtypes.46HER2-enriched subtype
HER2-positive spectra revealed prominent peaks at 1062, 1171, and 1461 cm−1 (Table S3).45,46,55 These features correspond to C–C skeletal modes, CH2 bending in proteins, and ring stretching vibrations in amino acids. The HER2 spectra notably lacked the 892 and 1112 cm−1 peaks present in TNBC, suggesting differing structural motifs in the extracellular matrix or membrane-associated proteins.46,54,57Luminal A (LUMA) subtype
LUMA spectra featured a distinct 1172 cm−1 peak, which is assigned to CH2 bending in proteins (Table S4).46 This peak's presence suggests a specific protein configuration not observed in TNBC. Other peaks such as 1062 and 1297 cm−1 were also present but with lower intensity.45,46,56,57 These spectral signatures indicate subtle differences in protein folding and lipid content.Luminal B (LUMB) subtype
LUMB shared the 1172 cm−1 protein bending peak46 with LUMA but also exhibited bands at 1109 and 1461 cm−1 (Table S5), implying a unique vibrational pattern.46,54,55,57 The co-occurrence of these peaks suggests a hybrid molecular environment, potentially reflecting the intermediate clinical behaviour of this subtype.The comprehensive comparison of subtype-specific spectral shifts is summarized in Table S6 and Fig. S2. TNBC and HER2 subtypes displayed the most distinct profiles, while LUMA and LUMB shared some overlapping features. These vibrational differences, particularly in protein- and lipid-associated bands, form the basis for successful classification using PCA and LDA models. The variables contributing most strongly to classification are highlighted in the SI.
Our findings affirm the potential of Ag/Ni/NiO nanowire-based SERS substrates in discriminating between breast cancer subtypes through non-destructive, label-free detection of biochemical changes. These insights are further detailed in Supplementary Tables S1–S6 and Fig. S2
Vibrational band assignments are presented as putative biochemical associations based on prior experimental and DFT-supported validation of the Ag/Ni/NiO nanowire SERS substrate and established Raman literature, rather than as definitive molecular identifications.
Fig. 9 shows a stacked overlay of SERS peak positions across breast cancer subtypes and demonstrates that SERS captures both shared oncogenic features and subtype-specific biochemical alterations across breast cancer phenotypes. These results underscore the power of SERS as a label-free, molecularly sensitive diagnostic approach capable of differentiating breast cancer subtypes based on their unique vibrational fingerprints, with strong potential for early detection, precise classification, and support of personalized therapeutic strategies.
 | ||
| Fig. 9 Stacked overlay of SERS peak positions across breast cancer subtypes (TNBC, HER2-enriched, Luminal A, and Luminal B). | ||
Discriminant model performance and classification metrics
To evaluate the capability of SERS Raman spectroscopy in differentiating normal breast tissue from various molecular subtypes of breast cancer, a classification model based on Principal Component Analysis followed by Linear Discriminant Analysis (PCA-LDA) was implemented. This multivariate approach reduces data dimensionality while preserving relevant spectral information and enables supervised classification to optimize class separation. PCA was initially applied to transform the original, highly correlated spectral variables into orthogonal components, thereby minimizing noise and redundancy. These principal components were then used as input for LDA, which identifies the linear combinations that best separate the predefined classes.The PCA-LDA model was employed to classify Raman spectral data into cancerous and non-cancerous categories, as well as to discriminate among breast cancer molecular subtypes, including Luminal A, Luminal B, HER2-enriched, and Triple-Negative Breast Cancer (TNBC). A total of 31 averaged spectra were analyzed, comprising normal tissue (control, n = 6), Luminal A (n = 8), Luminal B (n = 5), HER2-positive (n = 4), and TNBC (n = 8). To assess the primary distinction between malignant and non-malignant samples, a binary classification was first performed using MATLAB. Although multiple spectra were acquired per tissue section, all statistical analyses were performed at the patient level using averaged spectra to reduce pseudo-replication and limit overfitting.
As shown in Fig. 10(a), the LDA projection onto the first two discriminant axes reveals a clear separation between cancerous and control spectra, with distinct clustering primarily along the LD2 axis. Dimensionality reduction, facilitated by the preceding PCA step, enhanced both visualization and interpretability of class differences. To reduce the risk of overfitting and to provide a more conservative estimate of model performance, classification was evaluated using stratified k-fold cross-validation performed at the patient level, ensuring that all spectra from a given patient were confined to a single fold; detailed validation results, including confusion matrix and ROC curves across repeated cross-validation runs, are provided in Figs. S2 and 3.
The PCA-LDA model exhibited outstanding discriminate capability, achieving and overall classification accuracy of 99% 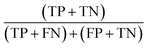 .All cancerous spectra (n = 25) and normal spectra (n = 6) were correctly clasified, yielding a sensitivity
.All cancerous spectra (n = 25) and normal spectra (n = 6) were correctly clasified, yielding a sensitivity 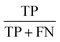 of 99% and specificity
of 99% and specificity 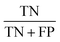 of 99%, where TP denotes true positives, TN true negatives, FP false positives, and FN false negatives.58
of 99%, where TP denotes true positives, TN true negatives, FP false positives, and FN false negatives.58
These findings highlight the robustness of the PCA-LDA approach for reliable classification of complex SERS spectral data and underscore its potential for diagnostic applications in breast cancer.
The PCA-LDA model was also applied to differentiate between luminal (Luminal A and Luminal B) and non-luminal (HER2-enriched and TNBC) breast cancer subtypes. As shown in Fig. 10(b), the linear discriminant projection resulted in a clear separation between the two groups along the LD1 axis, with minimal overlap. Classification performance was evaluated using standard metrics. The model achieved an overall accuracy of 99%, correctly classifying all samples into their respective luminal and non-luminal categories. Sensitivity and specificity were both 99%, indicating perfect classification of luminal cases (true positives) and non-luminal cases (true negatives).
The PCA-LDA model was further applied to distinguish HER2-enriched breast cancer cases from non-HER2 subtypes (Luminal A, Luminal B HER2-, and TNBC). As shown in Fig. 10(c), the projection onto the first two linear discriminants reveals a clear and complete separation between the HER2-positive and non-HER2 groups, primarily along the LD1 axis, with no observed overlap. This distinct separation reflects the unique molecular and biochemical profile of HER2-enriched tumors, particularly the overexpression of ERBB2 and associated signaling pathways. The classification model again achieved an overall accuracy of 99%, correctly identifying all HER2-positive (n = 4) and non-HER2 (n = 21) spectra. Sensitivity and specificity were both 99%, confirming the model's ability to reliably detect HER2 status based on SERS spectral signatures.
Fig. 10(d) shows the projection onto the first two linear discriminants, which reveals a clear and complete separation between the TNBC and non-TNBC groups, primarily along the LD1 axis, with no observed overlap between the two classes. This distinct separation reflects underlying molecular and biochemical differences between TNBC and other breast cancer subtypes. The classification model achieved an overall accuracy of 99%, correctly identifying all TNBC (n = 8) and non-TNBC (n = 18) samples. Sensitivity and specificity were both 99%, confirming the model's robustness in discriminating TNBC based on the input spectral and molecular features.
Multiple platforms were utilized for comparative analysis and visualization, including Python (scikit-learn), MATLAB, and OriginLab.
In Python, LDA was performed using the LinearDiscriminantAnalysis class from the scikit-learn library, reducing the feature space to two linear discriminants (LD1 and LD2). The resulting 2D scatter plot (Fig. 11) illustrates distinct separation among the five breast tissue classes: Normal (blue), TNBC (orange), HER2 (green), LUMA (red), and LUMB (purple). Each point represents a sample projected into the LDA space, with 95% confidence ellipses indicating class-specific distributions. HER2 samples are well separated along LD1, while TNBC and normal samples cluster distinctly in the upper right quadrant. LUMA and LUMB show some overlap along LD2 but remain largely distinguishable, demonstrating LDA's effectiveness in preserving class-specific variance for subtype classification.
As shown in Fig. 11, the projection onto the first two linear discriminants (LD1 and LD2) reveals a well-defined separation among all five groups: Normal, TNBC, HER2, Luminal A (LUMA), and Luminal B (LUMB). The spatial distribution of each class, combined with the non-overlapping confidence ellipses, indicates excellent model performance in distinguishing breast cancer subtypes from normal tissue. The classification model achieved an overall accuracy of 99%, correctly assigning each sample to its respective group. Both sensitivity and specificity reached 99% for all classes, confirming the robustness of the discriminant model in identifying subtle spectral variations and molecular signatures characteristic of each subtype.
The close proximity of LUMA and LUMB in the LDA plot can be explained by their overlapping molecular signatures revealed through SERS analysis. Both subtypes share the prominent 1172 cm−1 peak, attributed to CH2 bending in proteins, indicating a similar protein folding motif absent in TNBC.46 While LUMA spectra also include peaks at 1062 and 1297 cm−1 associated with protein and lipid content these features are relatively subtle and of lower intensity.45,46,56,57 LUMB, meanwhile, exhibits additional peaks at 1109 and 1461 cm−1, suggesting slightly different vibrational characteristics.46,54,56,57 However, the shared presence of key protein-related bands and the partial overlap in lipid-associated vibrations imply a degree of biochemical similarity between these two subtypes.
To validate and enhance the interpretability of the results, MATLAB was used to construct an LDA pipeline based on manually computed within-class and between-class scatter matrices. Fig. 12 presents a 2D LDA scatter plot from this analysis, showing similar class clustering patterns with clearly distinguishable subtypes. This MATLAB implementation also enabled the export of linear discriminant coordinates and class labels for further analysis.
Fig. 13 displays a corresponding LDA plot generated in OriginLab, following PCA-based dimensionality reduction. This visualization likewise highlights clear group separation, with minimal overlap among the five subtypes: HER2 (green), Normal (black), TNBC (red), LUMA (blue), and LUMB (cyan), reinforcing the robustness of the PCA-LDA pipeline in extracting meaningful spectral distinctions.
 | ||
| Fig. 13 2D LDA visualization of spectral signatures from breast cancer tissue samples using OriginLab. | ||
This work represents a pilot study. The cohort size was limited, with smaller numbers in HER2 and Luminal B subtypes, which restricts statistical power and generalizability. Accordingly, the reported classification performance reflects internal feasibility rather than definitive clinical accuracy. The primary contribution of this study is the demonstration that reproducible, subtype-associated biochemical fingerprints can be obtained from FFPE breast tissue using Ag/Ni/NiO nanowire-based SERS. These findings establish a foundation for future studies incorporating larger, prospectively powered cohorts and independent external validation.
The robust classification performance observed in the PCA-LDA models from distinctive Raman spectral features that characterize each breast cancer subtype. For instance, TNBC samples displayed unique and consistently high-intensity peaks at 892, 1065, 1112, 1135, and 1297 cm−1, primarily associated with C–C stretching in polysaccharides and protein backbone vibrations. These spectral markers were either absent or significantly weaker in other subtypes, enabling the model to clearly separate TNBC from HER2-enriched and luminal types. In contrast, HER2-enriched spectra lacked the 892 and 1112 cm−1 peaks, reflecting a reduced presence of certain structural protein and carbohydrate components. The absence of these peaks served as strong negative discriminants, enhancing the classification accuracy of HER2 against TNBC and LUMB. Principal Component Analysis (PCA) followed by Linear Discriminant Analysis (LDA) was performed using a combination of Python, MATLAB, and Origin, ensuring consistency and cross-validation of results across platforms.
Furthermore, subtle but significant differences between luminal subtypes also contributed to the model's high discriminative power. Both LUMA and LUMB exhibited a peak at 1172 cm−1, associated with CH2 bending in proteins, but LUMB showed this peak more prominently, along with moderate contributions from lipid- and nucleic acid-related peaks at 1002 and 1446 cm−1. LUMA, meanwhile, was characterized by sharper and more intense peaks at 750, 1002, and 1446 cm−1, suggesting a more defined biochemical profile with high lipid and nucleic acid content. These nuanced variations were successfully captured by PCA and further separated by LDA into distinct, non-overlapping clusters. The use of multiple analytical platforms allowed for robust multivariate modeling, reproducible spectral interpretation, and high accuracy in breast cancer subtype classification.
Previous studies have successfully demonstrated the potential of SERS combined with silver-based nanostructures for breast cancer detection using clinical samples. For instance, Wang et al. (2024)59 utilized Ag nanoparticles (50–60 nm) to acquire fingerprint SERS spectra from fine needle aspiration (FNA) samples, achieving high diagnostic sensitivity and specificity through machine learning models such as PCA-LDA, PLS-DA, and SVM. Similarly, Lyng et al. (2021)56 compared SERS and conventional Raman spectroscopy using serum samples and reported that SERS provided higher sensitivity, specificity, and area under receiving operating curve values for classifying different breast cancer stages. While these studies highlight the diagnostic power of SERS using silver nanoparticles, our work is the first to employ Ag/Ni/NiO nanowires as the plasmonic substrate. The elongated geometry of nanowires offers a higher surface area and enhanced electromagnetic and improved molecular sensitivity. This novel substrate choice could represent a significant advancement in SERS-based breast cancer diagnostics, especially in distinguishing subtypes at the molecular level. While immunohistochemical and molecular assays remain the clinical gold standard for breast cancer60,61 subtyping, the present study demonstrates that Ag/Ni/NiO nanowire-based SERS can capture subtype-specific biochemical signatures in agreement with pathology-defined classifications, supporting its potential role as a complementary, label-free diagnostic tool.
Experimental
Materials
Sodium chloride (NaCl), ethylene glycol (EG, (CH2OH)2), silver Nitrate (AgNO3), polyvinylpyrrolidone (PVP, Mw ≈ 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), ethanol (C2H6O, EtOH), diethyl hydroxylamine (DEHA, C4H11NO), hydrazine (N2H4), and nickel(II) nitrate hexahydrate (Ni(NO3)2·6H2O) were all purchased from Sigma-Aldrich. Deionized water was used throughout the entire experiments.
000), ethanol (C2H6O, EtOH), diethyl hydroxylamine (DEHA, C4H11NO), hydrazine (N2H4), and nickel(II) nitrate hexahydrate (Ni(NO3)2·6H2O) were all purchased from Sigma-Aldrich. Deionized water was used throughout the entire experiments.
Preparation of silver nanowires (AgNWs) and silver nanowires with nickel coating (Ag/Ni/NiO NWs)
Silver nanowires (AgNWs) were synthesized using a modified polyol reduction method, in which silver nitrate (AgNO3) was reduced by ethylene glycol, serving both as the solvent and reducing agent, in the presence of polyvinylpyrrolidone (PVP) as a stabilizer. The reaction conditions including temperature, stirring rate, and the PVP-to-AgNO3 ratio were optimized to produce uniform, high-aspect-ratio nanowires, as described by Hernandez Cedillo et al.35 Subsequently, Ag/Ni/NiO nanowires were fabricated by coating the as-prepared AgNWs with a thin nickel layer. This approach, as demonstrated by Hernandez Cedillo et al., promotes the formation of a well-defined structure with improved stability and functional properties.Sample collection and preparation
Breast tissue samples were sectioned into 5-micron slices and mounted onto aluminum substrates at room temperature. Tissue sections were collected from 31 patients, including controls (n = 6), luminal A (LUMA, n = 8), luminal B (LUMB, n = 5), HER2-enriched (HER2-, n = 4), and triple-negative breast cancer (TNBC, n = 8) for measurement and analysis. These samples were obtained from the University of Nebraska Medical Center Department of Pathology, Microbiology, and Immunology Tissue Services Facility (TSF) with ethical approval for de-identified samples, as well as from commercial sources.All experimental protocols were reviewed and approved by the respective institutional review boards of the collaborating institutions, in accordance with relevant ethical guidelines and regulations. Biological materials were collected anonymously, as specified in the approved ethics documentation. Informed consent was obtained from all participants under the oversight of the University of Nebraska Medical Center Department of Pathology, Microbiology, and Immunology. This study was designed as a pilot feasibility investigation using retrospectively available FFPE breast tissue specimens to assess the technical performance and discriminatory potential of Ag/Ni/NiO nanowire-based SERS prior to larger, powered validation studies.
For the SERS experiments, 50 µL of Ag/Ni/NiO nanowires were deposited onto an aluminum substrate and allowed to dry at room temperature, ensuring proper adhesion. Once the substrate was prepared, a section of either breast cancer or normal tissue was carefully placed on top of the nanowire-coated surface for analysis.
Raman spectroscopy
Raman spectroscopy was conducted using an XploRA™ PLUS MicroRaman Spectrometer (Horiba), equipped with a laser output of approximately 20 mW and a diffraction grating of 1800 grooves/mm for spectral acquisition. To minimize photodamage to the analyte, a 638 nm laser filtered to 10% intensity was directed through a 50× objective lens, producing a focused beam spot with a diameter of approximately 2 µm. Spectral data were collected in the 400–1800 cm−1 range, using 10 accumulations per measurement with an integration time of 10 seconds per acquisition.For each sample, spectra were obtained from three distinct regions, yielding in nine spectra per sample. A total of 837 SERS spectra were collected and pre-processed using polynomial baseline correction, smoothing, and noise reduction prior to averaging.
Data analysis using statistical methods
To analyze the Raman spectra obtained from breast tissue samples, including normal tissue (NT) and the molecular subtypes Luminal A, Luminal B, HER2-enriched, and Triple-Negative Breast Cancer, a discriminant classification model based on Principal Component Analysis followed by Linear Discriminant Analysis (PCA-LDA) was employed. The PCA-LDA approach offers several advantages for high-dimensional datasets such as Raman spectra, gene expression profiles or medical imaging data. PCA reduces model complexity by removing non-informative variation (noise) while preserving the most relevant variance. This dimensionality reduction is especially beneficial for biological datasets, which typically involve a large number of variables and a limited number of samples, thereby mitigation the risk of overfitting.Moreover, since LDA assumes that input variables are uncorrelated, a condition seldom met in biological data, PCA is first applied to transform the correlated original variables into orthogonal principal components. This preprocessing step enhances the performance and robustness of the LDA classifier. The combined PCA-LDA model also enables two-dimensional visualization of the data, facilitating maximal class separation and aiding in clinical interpretation.62,63
The analysis was conducted in several stages. Initially, a binary classification (cancer vs. normal, luminal vs. Non-Luminal, HER2-enriched vs. Non-HER2-enriched, and TNBC vs. Non-TNBC) was performed to distinguish cancerous tissues (Luminal A, Luminal B, HER2-enriched, and TNBC) from non-cancerous NT samples using MATLAB's discriminant analysis functions. Subsequently, a multiclass classification was applied to the cancerous subtypes to evaluate their biochemical and molecular distinctions. In particular, the luminal group (Luminal A and Luminal B) was compared to the non-luminal group (HER2-enriched and TNBC) to differentiate hormone-dependent tumors from more aggressive, less differentiated forms.64 Further analysis compared HER2-enriched samples with non-HER2-enriched subtypes (including Luminal A, Luminal B, and TNBC), anticipating distinctive molecular and clinical profiles driven by HER2-enriched (ERBB2) overexpression, which is known to correlate with increased proliferation and tumor aggressiveness.65,66
Finally, TNBC was compared with non-TNBC subtypes (Luminal A, Luminal B, and HER2-enriched). TNBC is known to exhibit distinct biochemical and molecular characteristics, including elevated Raman band intensities associated with disordered proteins, nucleic acids (DNA/RNA), and phosphate groups indicative of high mitotic activity. It also displays diminished lipid-associated signals, reflecting reduced cellular differentiation, alongside enhanced features corresponding to proteins, nucleic acids, and active cellular membranes.67
All spectral processing was conducted using Python (v3.13.2), MATLAB (R2023b), and OriginLab (2024) to ensure consistency and robustness across analytical platforms. Principal Component Analysis (PCA) was used for unsupervised dimensionality reduction to identify variance within the spectral data, followed by Linear Discriminant Analysis (LDA) for supervised classification of tissue types.
In Python, PCA and LDA were performed using the scikit-learn library, with pandas and numpy for data handling, and matplotlib and seaborn for data visualization. In MATLAB, the within-class scatter matrix (Sw) and between-class scatter matrix (Sb) were computed, and the generalized eigenvalue problem Sbv = λSwv was solved to obtain discriminant vectors, where v represents the eigenvectors and λ the corresponding eigenvalues. LDA was also performed in OriginLab 2024 using its built-in multivariate analysis tools. A PCA step was included prior to LDA in all platforms to stabilize the classification space and improve model performance.
Conclusions
The present report is a pilot study of breast cancer detection using a combination of a new reliable Ag/Ni/NiO nanowire substrate (previously developed),35 combined with machine learning analysis that overcomes many limitations of SERS analysis. We demonstrated the effectiveness of these Ag/Ni/NiO nanowire substrates in enabling accurate, label-free differentiation of breast cancer subtypes using formalin-fixed tissue samples. We successfully discriminated between luminal A (LUMA), luminal B (LUMB), HER2-enriched, and triple-negative breast cancer (TNBC), as well as from normal tissue, based on their distinct biochemical signatures. In this study, we report sensitivity, specificity, and LOD values combined with high reproducibility that are within the accepted parameters for a successful clinical trial.Distinct spectral differences were observed among the various breast cancer subtypes that provide clues to the understanding of cancer growth mechanisms. Triple-negative breast cancer (TNBC) exhibited unique peaks at 892, 1065, 1112, 1135, and 1297 cm−1, corresponding to C–C stretching in polysaccharides and protein-related vibrations, indicative of its metabolic and structural heterogeneity. HER2-enriched tissues lacked signals at 892 and 1112 cm−1, suggesting a reduced contribution from specific protein and carbohydrate components. Luminal B (LUMB) tissues showed a distinct peak at 1172 cm−1, also present in luminal A (LUMA), associated with CH2 bending in proteins, but differed in the relative intensities of nucleic acid- and lipid-associated peaks (e.g., 750, 1002, and 1446 cm−1), reflecting differences in molecular composition. LUMA spectra were characterized by sharp nucleic acid and lipid peaks, consistent with lower proliferation rates and more stable molecular profiles.
This work shows that by using a reliable SERS substrate and mapping and averaging the spectra, it is possible to eliminate variations in different regions of the sample and obtain a spectral fingerprint of the cancer. We have shown that the spectra of different types of cancer are very distinct. The next logical step will be to look for those fingerprints in samples such as saliva or plasma and advance in the development of a breast cancer test that can be approved by the regulating authorities and make its way to the market. Raman spectroscopy analysis can take very short times, and it is capable of identifying cancer biomarkers. At present, there are no point-of-care cancer tests that will help to save many lives. The road to develop those tests is still long. However, we believe that this study provides a very solid step towards that goal.
Author contributions
A. H. C. conceptualization, methodology, investigation, formal analysis, and writing (original draft); J. M. L. investigation and formal analysis; C. H. C. investigation; A. L. M. B. resources, funding acquisition, and writing (review and editing); and M. J. Y. conceptualization, supervision, funding acquisition, and writing (review and editing).Conflicts of interest
The authors declare no conflict of interest.Data availability
The data supporting the findings of this study are included in the Supplementary Information (SI). These data comprise: (i) a comparison of Surface-Enhanced Raman Scattering (SERS) spectra acquired from formalin-fixed paraffin-embedded (FFPE) breast tissue sections before and after deparaffinization (Fig. S1); (ii) a complete list of SERS peaks and corresponding vibrational mode assignments (Table S1); (iii) distributions of SERS spectral shifts for each breast cancer molecular subtype, including Triple-Negative Breast Cancer (TNBC, Table S2), HER2-enriched (Table S3), Luminal A (LUMA, Table S4), and Luminal B (LUMB, Table S5); and (iv) a consolidated summary of SERS peaks across all subtypes (Table S6). Additional supplementary materials include the confusion matrix for PCA-LDA-based classification of breast cancer subtypes (Fig. S2) and the corresponding receiver operating characteristic (ROC) curves (Fig. S3). No additional datasets or code were generated or deposited in external repositories as part of this study. See DOI: https://doi.org/10.1039/d5na00890e.Acknowledgements
The authors gratefully acknowledge financial support from the U.S. Department of Energy (DOE), Office of Science, under Award No. DE-SC0023343 (Mission DAC: Molecular mechanisms of moisture-driven direct air capture within charged polymers); the Army Research Office (ARO) under Award No. W911NF-23-2-0014 (Cognitive distributed sensing in congested radio-frequency environments: FREEDOM); and the National Science Foundation (NSF) under Grant No. 2025490 and 2427869 (I-CORPs), as well as support from the NSF National Nanotechnology Coordinated Infrastructure (NNCI) program through the Nanotechnology Collaborative Infrastructure Southwest (NCI-SW). Additional funding was provided by the Secretariat of Science, Humanities, Technology and Innovation (SECIHTI) under Grant No. 827567 and 795368. The authors also acknowledge support from the Nebraska Research Initative (NRI); the Fred and Pamela Buffett Cancer Center through the National Cancer Institute (NCI) Cancer Support Grant P30CA036727; the National Institute of General Medical Sciences (NIGMS) under Grant No. P20 GM103427 (NE-INBRE), P30 GM106397 (NCS), and P20 GM130447 (CoNDA); the National Institutes of Health (NIH) under Award No. S10OD030486 and S10RR02730; and the National Cancer Institute of the NIH under Grant No. P30 CA230074. The authors further acknowledge the Center for Materials Interfaces in Research and Applications (¡MIRA!) at the Northern Arizona University for access to its facilities. The authors also gratefully acknowledge Dr J. Jesus Velazquez-Salazar for assitance with Ag/Ni/NiO nanowire synthesis.References
- J. Kim, A. Harper, V. McCormack, H. Sung, N. Houssami, E. Morgan, M. Mutebi, G. Garvey, I. Soerjomataram and M. M. Fidler-Benaoudia, Global patterns and trends in breast cancer incidence and mortality across 185 countries, Nat. Med., 2025, 31(4), 1154–1162, DOI:10.1038/s41591-025-03502-3.
- B. M. Moloney, D. O’Loughlin, S. Abd Elwahab and M. J. Kerin, Breast cancer detection—a synopsis of conventional modalities and the potential role of microwave imaging, Diagnostics, 2020, 10(103), 1–13, DOI:10.3390/diagnostics10020103.
- E. Nalejska, E. Mączyńska and M. A. Lewandowska, Prognostic and predictive biomarkers: Tools in personalized oncology, Mol. Diagn. Ther., 2014, 18(3), 273–284, DOI:10.1007/s40291-013-0077-9.
- J. L. Parker, S. S. Kuzulugil, K. Pereverzev, S. Mac, G. Lopes, Z. Shah, A. Weerasinghe, D. Rubinger, A. Falconi, A. Bener, B. Caglayan, R. Tangri and N. Mitsakakis, Does biomarker use in oncology improve clinical trial failure risk? A large-scale analysis, Cancer Med., 2021, 10(6), 1955–1963, DOI:10.1002/cam4.3732.
- R. A. Beckman, J. Clark and C. Chen, Integrating predictive biomarkers and classifiers into oncology clinical development programmes, Perspectives, 2011, 10, 735–748, DOI:10.1049/pbpo016e_ch7.
- G. Redonda and M. D. Miller, Breast cancer screening can we talk?, J. Gen. Intern. Med., 2004, 16(3), 206–207, DOI:10.1023/a:1005893126451.
- R. A. Reeves and T. Kaufman, In Mammography, StastPearls Publishing, Statpearls, Treasure Island, FL, 2025, (National Center for biotechnology Information) Search PubMed.
- S. Hofvind, A. Ponti, J. Patnick, N. Ascunce, S. Njor, M. Broeders, L. Giordano, A. Frigerio and S. Törnberg, False-Positive Results in Mammographic Screening for Breast Cancer in Europe: A Literature Review and Survey of Service Screening Programmes, J. Med. Screen, 2012, 19(1), 57–66, DOI:10.1258/jms.2012.012083.
- C. L. Christiansen, Predicting the Cumulative Risk of False-Positive Mammograms, J. Natl. Cancer Inst., 2000, 92(20), 1657–1666, DOI:10.1093/jnci/92.20.1657.
- R. A. Hubbard, K. Kerlokowske, C. I. Flowers and W. Zhu, Cumulative probability of false-positive recall or biopsy recommendation after 10 years of screening mammography, Ann. Intern. Med., 2011, 155(8), 481–492, DOI:10.7326/0003-4819-155-8-201110180-00004.
- J. E. Joy, E. E. Penhoet and D. B. Petitti, Saving Women's Lives: Strategies for Improving Breast Cancer Detection and Diagnosis. Washington, D.C.: National Academies Press, 2005, DOI: DOI:10.17226/11016..
- P.-H. Zahl, J. Mæhlen and H. G. Welch, The Natural History of Invasive Breast Cancers Detected by Screening Mammography, Arch. Intern. Med., 2008, 168(21), 2311–2317, DOI:10.1001/archinte.168.21.2311.
- M. Løberg, M. L. Lousdal, M. Bretthauer and M. Kalager, Benefits and harms of mammography screening, Breast Cancer Res., 2015, 17(63), 1–12, DOI:10.1186/s13058-015-0525-z.
- D. Puliti, S. W. Duffy, G. Miccinesi, H. De Koning, E. Lynge, M. Zappa and E. Paci, Overdiagnosis in Mammographic Screening for Breast Cancer in Europe: A Literature Review, J. Med. Screen, 2012, 19(Suppl 1), 42–56, DOI:10.1258/jms.2012.012082.
- M. Kalager, H.-O. Adami, M. Bretthauer and R. M. Tamimi, Overdiagnosis of Invasive Breast Cancer Due to Mammography Screening: Results From the Norwegian Screening Program, Ann. Intern. Med., 2012, 156(7), 491–499, DOI:10.7326/0003-4819-156-7-201204030-00005.
- N. F. Boyd, L. Sun, J. Stone, E. Fishell, R. A. Jong and A. Chiarelli, Mammographic Density and the Risk and Detection of Breast Cancer, N. Engl. J. Med., 2007, 356(3), 227–236, DOI:10.1056/NEJMoa062790.
- E. A. Sickles, The Use of Breast Imaging to Screen Women at High Risk for Cancer, Radiol. Clin. N. Am., 2010, 48(5), 859–878, DOI:10.1016/j.rcl.2010.06.012.
- K. J. Wernli, W. B. DeMartini, L. Ichikawa, C. D. Lehman, T. Onega, K. Kerlikowske, L. M. Henderson, B. M. Geller, M. Hofmann and B. C. Yankaskas, Patterns of Breast Magnetic Resonance Imaging Use in Community Practice, JAMA Intern. Med., 2014, 174(1), 125–132, DOI:10.1001/jamainternmed.2013.11963.
- W. A. Berg, J. D. Blume, A. M. Adams, R. A. Jong, R. G. Barr, D. E. Lehrer, E. D. Pisano, W. P. Evans, M. C. Mahoney, L. Hovanessian Larsen, G. J. Gabrielli and E. B. Mendelson, Reasons Women at Elevated Risk of Breast Cancer Refuse Breast MR Imaging Screening: ACRIN 6666, Radiology, 2010, 254(1), 79–87, DOI:10.1148/radiol.2541090953.
- W. A. Berg, L. Gutierrez, M. S. NessAiver, W. B. Carter, M. Bhargavan, R. S. Lewis and O. B. Ioffe, Diagnostic Accuracy of Mammography, Clinical Examination, US, and MR Imaging in Preoperative Assessment of Breast Cancer, Radiology, 2004, 233(3), 830–849, DOI:10.1148/radiol.2333031484.
- V. Moisoiu, S. D. Iancu, A. Stefancu, T. Moisoiu, B. Pardini, M. P. Dragomir, N. Crisan, L. Avram, D. Crisan, I. Andras, D. Fodor, L. F. Leopold, C. Socaciu, Z. Bálint, C. Tomuleasa, F. Elec and N. Leopold, SERS liquid biopsy: An emerging tool for medical diagnosis, Colloids Surf. B Biointerfaces, 2021, 208(112064), 1–9, DOI:10.1016/j.colsurfb.2021.112064.
- J. Li, Y. Li, P. Li, Y. Zhang, L. Du, Y. Wang, C. Zhang and C. Wang, Exosome detection via surface-enhanced Raman spectroscopy for cancer diagnosis, Acta Biomater., 2022, 144, 1–14, DOI:10.1016/j.actbio.2022.03.036.
- M. Vendrell, K. K. Maiti, K. Dhaliwal and Y.-T. Chang, Surface-enhanced Raman scattering in cancer detection and imaging, Trends Biotechnol., 2013, 31(4), 249–257, DOI:10.1016/j.tibtech.2013.01.013.
- R. Beeram, K. R. Vepa and V. R. Soma, Recent Trends in SERS-Based Plasmonic Sensors for Disease Diagnostics, Biomolecules Detection, and Machine Learning Techniques, Biosensors, 2023, 13(328), 1–37, DOI:10.3390/bios13030328.
- L. Vázquez-Iglesias, G. M. Stanfoca Casagrande, D. García-Lojo, L. Ferro Leal, T. A. Ngo, J. Pérez-Juste, R. M. Reis, K. Kant and I. Pastoriza-Santos, SERS sensing for cancer biomarker: Approaches and directions, Bioact. Mater., 2024, 34, 248–268, DOI:10.1016/j.bioactmat.2023.12.018.
- L. Xu, Y. Xie, J. Lin, A. Wu and T. Jiang, Advancements in SERS-based biological detection and its application and perspectives in pancreatic cancer, View, 2024, 5, 20230070, DOI:10.1002/viw.20230070.
- S. Lee, H. Dang, J.-I. Moon, K. Kim, Y. Joung, S. Park, Q. Yu, J. Chen, M. Lu, L. Chen, S.-W. Joo and J. Choo, SERS-based microdevices for use as in vitro diagnostic biosensors, Chem. Soc. Rev., 2024, 53(11), 5394–5427, 10.1039/d3cs01055d.
- S. E. J. Bell, G. Charron, E. Cortés, J. Kneipp, M. L. De La Chapelle, J. Langer, M. Procházka, V. Tran and S. Schlücker, Towards Reliable and Quantitative Surface-Enhanced Raman Scattering (SERS): From Key Parameters to Good Analytical Practice, Angew. Chem., Int. Ed., 2020, 59(14), 5454–5462, DOI:10.1002/anie.201908154.
- A. I. Pérez-Jiménez, D. Lyu, Z. Lu, G. Liu and B. Ren, Surface-enhanced Raman spectroscopy: benefits, trade-offs and future developments, Chem. Sci., 2020, 11(18), 4563–4577, 10.1039/d0sc00809e.
- R. Panneerselvam, G.-K. Liu, Y.-H. Wang, J.-Y. Liu, S.-Y. Ding, J.-F. Li, D.-Y. Wu and Z.-Q. Tian, Surface-enhanced Raman spectroscopy: bottlenecks and future directions, Chem. Commun., 2018, 54(1), 10–25, 10.1039/c7cc05979e.
- R. D. Norton, H. T. Phan, S. N. Gibbons and A. J. Haes, Quantitative Surface-Enhanced Spectroscopy, Annu. Rev. Phys. Chem., 2022, 73(1), 141–162, DOI:10.1146/annurev-physchem-082720-033751.
- P. Lepucki, A. P. Dioguardi, D. Karnaushenko, O. G. Schmidt and H.-J. Grafe, The normalized limit of detection in NMR spectroscopy, J. Magn. Reson., 2021, 332, 107077, DOI:10.1016/j.jmr.2021.107077.
- S. Toghi Eshghi, X. Li and H. Zhang, Targeted Analyte Detection by Standard Addition Improves Detection Limits in Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry, Anal. Chem., 2012, 84(18), 7626–7632, DOI:10.1021/ac301423f.
- X. Terzapulo, A. Kassenova and R. Bukasov, Immunoassays: Analytical and Clinical Performance, Challenges, and Perspectives of SERS Detection in Comparison with Fluorescent Spectroscopic Detection, Int. J. Mol. Sci., 2024, 25, 2080, DOI:10.3390/ijms25042080.
- A. Hernandez Cedillo, J. J. Velazquez Salazar, J. Mendez Lozoya, J. A. Agyei-Mensah, R. A. Guirado-López, C. Hernandez Cedillo, A. Lehr and M. Jose Yacaman, Ag@Ni–NiO NW Core–Shell Nanowires: A Reliable Surface-Enhanced Raman Scattering (SERS) Substrate, J. Phys. Chem. C, 2025, 129(8), 4113–4125, DOI:10.1021/acs.jpcc.4c08450.
- M. Aeschlimann, J. P. Bange, M. Bauer, U. Bovensiepen, H.-J. Elmers, T. Fauster, L. Gierster, U. Höfer, R. Huber, A. Li, X. Li, S. Mathias, K. Morgenstern, H. Petek, M. Reutzel, K. Rossnagel, G. Schönhense, M. Scholz, B. Stadtmüller, J. Stähler, S. Tan, B. Wang, Z. Wang and M. Weinelt, Time-resolved photoelectron spectroscopy at surfaces, Surf. Sci., 2025, 753, 122631, DOI:10.1016/j.susc.2024.122631.
- G. Lin, M. Wu, R. Tang, B. Wu, Y. Wang, J. Zhu, J. Zhang and W. Wu, A Surface-Enhanced Raman Scattering Substrate with Tunable Localized Surface Plasmon Resonance Absorption Based on AgNPs, Sensors, 2024, 24, 5778, DOI:10.3390/s24175778.
- S. R. Tadi, A. G. Shenoy, A. Bharadwaj, C. Mukhopadhyay, K. Sadani and P. Nag, Recent advances in the design of SERS substrates and sensing systems for biosensing applications: Systems from single cell to single molecule detection, F1000Res., 2025, 13, 670, DOI:10.12688/f1000research.149263.3.
- A. Sivakoses, H. Q. Marcarian, A. M. Arias, A. R. Lam, O. C. Ihedioha, J. A. Santamaria, G. C. Gurtner and A. L. M. Bothwell, Triple Negative Breast Cancer Cells Acquire Lymphocyte Proteins and Genomic DNA During Trogocytosis with T Cells, PeerJ, 2024, 13, e19236, DOI:10.1101/2024.08.09.607029.
- D. Lee, J. Du, R. Yu, Y. Su, J. R. Heath and L. Wei, Visualizing Subcellular Enrichment of Glycogen in Live Cancer Cells by Stimulated Raman Scattering, Anal. Chem., 2020, 92(19), 13182–13191, DOI:10.1021/acs.analchem.0c02348.
- C. Zheng, L. Liang, S. Xu, H. Zhang, C. Hu, L. Bi, Z. Fan, B. Han and W. Xu, The use of Au@SiO2 shell-isolated nanoparticle-enhanced Raman spectroscopy for human breast cancer detection, Anal. Bioanal. Chem., 2014, 406(22), 5425–5432, DOI:10.1007/s00216-014-7967-5.
- R. E. Kast, G. K. Serhatkulu, A. Cao, A. K. Pandya, H. Dai, J. S. Thakur, V. M. Naik, R. Naik, M. D. Klein, G. W. Auner and R. Rabah, Raman spectroscopy can differentiate malignant tumors from normal breast tissue and detect early neoplastic changes in a mouse model, Biopolymers, 2008, 89(3), 235–241, DOI:10.1002/bip.20899.
- T. Bhattacharjee, S. Tawde, R. Hudlikar, M. Mahimkar, G. Maru, A. Ingle and C. Murali Krishna, Ex vivoRaman spectroscopic study of breast metastatic lesions in lungs in animal models, J. Biomed. Opt., 2015, 20(8), 085006, DOI:10.1117/1.jbo.20.8.085006.
- A. S. Haka, K. E. Shafer-Peltier, M. Fitzmaurice, J. Crowe, R. R. Dasari and M. S. Feld, Identifying Microcalcifications in Benign and Malignant Breast Lesions by Probing Differences in Their Chemical Composition Using Raman Spectroscopy, Cancer Res., 2002, 15(62), 5375–5380 Search PubMed.
- W. C. Zúñiga, V. Jones, S. M. Anderson, A. Echevarria, N. L. Miller, C. Stashko, D. Schmolze, P. D. Cha, R. Kothari, Y. Fong and M. C. Storrie-Lombardi, Raman Spectroscopy for Rapid Evaluation of Surgical Margins during Breast Cancer Lumpectomy, Sci. Rep., 2019, 9, 14639, DOI:10.1038/s41598-019-51112-0.
- Y. Zhang, Z. Li, Z. Li, H. Wang, D. Regmi, J. Zhang, J. Feng, S. Yao and J. Xu, Employing Raman Spectroscopy and Machine Learning for the Identification of Breast Cancer, Biol. Proced. Online, 2024, 26, 28, DOI:10.1186/s12575-024-00255-0.
- A. M. Fuentes, A. Narayan, K. Milligan, J. J. Lum, A. G. Brolo, J. L. Andrews and A. Jirasek, Raman spectroscopy and convolutional neural networks for monitoring biochemical radiation response in breast tumour xenografts, Sci. Rep., 2023, 13, 1530, DOI:10.1038/s41598-023-28479-2.
- S. Contorno, R. E. Darienzo and R. Tannenbaum, Evaluation of aromatic amino acids as potential biomarkers in breast cancer by Raman spectroscopy analysis, Sci. Rep., 2021, 11, 1698, DOI:10.1038/s41598-021-81296-3.
- Z. Movasaghi, S. Rehman and I. U. Rehman, Raman Spectroscopy of Biological Tissues, Appl. Spectrosc. Rev., 2007, 42(5), 493–541, DOI:10.1080/05704920701551530.
- Q. Zheng, J. Li, L. Yang, B. Zheng, J. Wang, N. Lv, J. Luo, F. L. Martin, D. Liu and J. He, Raman spectroscopy as a potential diagnostic tool to analyse biochemical alterations in lung cancer, Analyst, 2020, 145(2), 385–392, 10.1039/c9an02175b.
- H. Abramczyk and B. Brozek-Pluska, Raman Imaging in Biochemical and Biomedical Applications. Diagnosis and Treatment of Breast Cancer, Chem. Rev., 2013, 113(8), 5766–5781, DOI:10.1021/cr300147r.
- B. Brożek-Płuska, I. Placek, K. Kurczewski, Z. Morawiec, M. Tazbir and H. Abramczyk, Breast cancer diagnostics by Raman spectroscopy, J. Mol. Liq., 2008, 141(3), 145–148, DOI:10.1016/j.molliq.2008.02.015.
- M. Ma, J. Zhang, Y. Liu, X. Wang and B. Han, Advances in the clinical application of Raman spectroscopy in breast cancer, Appl. Spectrosc. Rev., 2024, 59(10), 1459–1493, DOI:10.1080/05704928.2024.2352519.
- S. K. Koya, M. Brusatori, S. Yurgelevic, C. Huang, C. W. Werner, R. E. Kast, J. Shanley, M. Sherman, K. V. Honn, K. R. Maddipati and G. W. Auner, Accurate identification of breast cancer margins in microenvironments of ex-vivo basal and luminal breast cancer tissues using Raman spectroscopy, Prostaglandins Other Lipid Mediators, 2020, 151, 106475, DOI:10.1016/j.prostaglandins.2020.106475.
- J. De Gelder, K. De Gussem, P. Vandenabeele and L. Moens, Reference database of Raman spectra of biological molecules, J. Raman Spectrosc., 2007, 38(9), 1133–1147, DOI:10.1002/jrs.1734.
- F. M. Lyng, D. Traynor, T. N. Q. Nguyen, A. D. Meade, F. Rakib, R. Al-Saady, E. Goormaghtigh, K. Al-Saad and M. H. Ali, Discrimination of breast cancer from benign tumours using Raman spectroscopy, PLoS One, 2019, 14(2), e0212376, DOI:10.1371/journal.pone.0212376.
- H. F. Nargis, H. Nawaz, H. N. Bhatti, K. Jilani and M. Saleem, Comparison of surface enhanced Raman spectroscopy and Raman spectroscopy for the detection of breast cancer based on serum samples, Spectrochim. Acta, Part A, 2021, 246, 119034, DOI:10.1016/j.saa.2020.119034.
- M. Toğaçar, B. Ergen and Z. Cömert, Application of breast cancer diagnosis based on a combination of convolutional neural networks, ridge regression and linear discriminant analysis using invasive breast cancer images processed with autoencoders, Med. Hypotheses, 2020, 135, 109503, DOI:10.1016/j.mehy.2019.109503.
- M. Wang, K. Zhang, L. Yue, X. Liu, Y. Lai and H. Zhang, Robust Diagnosis of Breast Cancer Based on Silver Nanoparticles by Surface-Enhanced Raman Spectroscopy and Machine Learning, ACS Appl. Nano Mater., 2024, 7(11), 13672–13680, DOI:10.1021/acsanm.4c02191.
- G. R. Bean, C.-Y. Lin, M. Krystel-Whittemore and L. Sun, Catching Up to Increased Complexity in Breast Cancer Molecular Testing, J. Mol. Diagn., 2025, 27(7), 602–604, DOI:10.1016/j.jmoldx.2025.03.007.
- S. Masuda and Y. Nakanishi, Application of Immunohistochemistry in Clinical Practices as a Standardized Assay for Breast Cancer, Acta Histochem. Cytochem., 2023, 56(1), 1–8, DOI:10.1267/ahc.22-00050.
- S. Gardner-Lubbe, Linear discriminant analysis for multiple functional data analysis, J. Appl. Stat., 2021, 48(11), 1917–1933, DOI:10.1080/02664763.2020.1780569.
- M. O. Adebiyi, M. O. Arowolo, M. D. Mshelia and O. O. Olugbara, A Linear Discriminant Analysis and Classification Model for Breast Cancer Diagnosis, Appl. Sci., 2022, 12, 11455, DOI:10.3390/app122211455.
- C. M. Perou, T. Sørlie, M. B. Eisen, M. Van De Rijn, S. S. Jeffrey, C. A. Rees, J. R. Pollack, D. T. Ross, H. Johnsen, L. A. Akslen, Ø. Fluge, A. Pergamenschikov, C. Williams, S. X. Zhu, P. E. Lønning, A.-L. Børresen-Dale, P. O. Brown and D. Botstein, Molecular portraits of human breast tumours, Nature, 2000, 406(6797), 747–752, DOI:10.1038/35021093.
- Y. Yarden and M. X. Sliwkowski, Untangling the ErbB signalling network, Nat. Rev. Mol. Cell Biol., 2001, 2(2), 127–137, DOI:10.1038/35052073.
- N. Iqbal and N. Iqbal, Human Epidermal Growth Factor Receptor 2 (HER2) in Cancers: Overexpression and Therapeutic Implications, Mol. Biol. Int., 2014, 2014, 852748, DOI:10.1155/2014/852748.
- A. S. Melitto, V. E. A. Arias, J. Y. Shida, L. H. Gebrim and L. Silveira, Diagnosing molecular subtypes of breast cancer by means of Raman spectroscopy, Laser Surg. Med., 2022, 54(8), 1143–1156, DOI:10.1002/lsm.23580.
| This journal is © The Royal Society of Chemistry 2026 |






