Open Access Article
Open Access ArticlePlasmon-enhanced photocatalytic nanoreactors for selective inactivation of murine leukemia virus (MLV)
Tianhong
Ouyang
a,
Koustav
Kundu
 a,
James
Hood
b,
Baichuan
Cheng
c,
Yixin
Mei
d,
Ainsley
Gray
a,
Suryaram
Gummuluru
b and
Björn M.
Reinhard
a,
James
Hood
b,
Baichuan
Cheng
c,
Yixin
Mei
d,
Ainsley
Gray
a,
Suryaram
Gummuluru
b and
Björn M.
Reinhard
 *a
*a
aDepartment of Chemistry and the Photonics Center, Boston University, Boston, MA 02215, USA. E-mail: bmr@bu.edu
bDepartment of Virology, Immunology and Microbiology, Boston University Chobanian & Avedisian School of Medicine, Boston, MA 02118, USA
cDepartment of Material Engineering, Boston University, Boston, MA 02215, USA
dDepartment of Biomedical Engineering, Boston University, Boston, MA 02215, USA
First published on 16th December 2025
Abstract
The lack of effective virucides that can eradicate viruses under mild conditions that do not harm mammalian cells or high value biologics poses risks for the food, health care, and pharmaceutical industries. Here, we examine plasmonic nanoreactors that contain the photocatalyst [Ru(bpy)3]2+ localized in the evanescent electric (E-) field of a silver nanoparticle (AgNP) as a selective virucide. The AgNP is passivated by a lipid coating and functionalized with annexin V to target and bind enveloped viruses with surface-exposed phosphatidylserine and localize the light-driven reactivity of the plasmonic nanoreactor virucide (PNV) in the proximity of the virus to enhance inactivation efficacy and minimize collateral damage. The lipid coating prevents premature Ag+ release under “dark” conditions and minimizes cytotoxicity. Upon illumination at 470 nm, plasmon-enhanced excitation of [Ru(bpy)3]2+ induces photoreactivity and generates reactive oxygen species (ROS) that damage the bound virus and increase the permeability of the lipid coating around the AgNP, facilitating the release of Ag+ ions. Using murine leukemia virus (MLV) as a model, annexin V-functionalized PNVs achieved over 85% viral inactivation after 30 minutes of illumination with 470 nm light (65 mW cm−2) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 virus
1 virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNV ratio, with no measurable cytotoxicity in mammalian host cells. These results demonstrate that PNVs combine light-activated reactivity with targeting to achieve potent, selective virucidal activity under mild conditions, paving a path to safeguarding biologics and cell cultures against viral contamination.
PNV ratio, with no measurable cytotoxicity in mammalian host cells. These results demonstrate that PNVs combine light-activated reactivity with targeting to achieve potent, selective virucidal activity under mild conditions, paving a path to safeguarding biologics and cell cultures against viral contamination.
Introduction
Viral contaminations of food, biologics, or growth media pose significant health and economic risks in the food industry, health care settings and pharmaceutical manufacturing and motivate the development of effective virucides.1–4 Although effective “broadband” chemical and physical antiviral strategies have been established, many of these strategies apply harsh conditions that not only inactivate virus contaminants but also deteriorate sensitive samples.5–7 For instance, monoclonal antibodies are sensitive towards treatment with heat, irradiation, or reactive chemicals and require currently sterilization via ultrafiltration, which is expensive and prone to induce aggregation.8–10 There has been a growing interest in the field of nanotechnology in recent years to engineer nanomaterials for applications at the molecular level with the goal to target and inactivate cell-free virus particles highly selectively with minimal collateral damage to cells or valuable biologics.11–13 Different strategies for nanoparticle (NP)-mediated inactivation of virus particles have been explored,14,15 including perturbation of viral envelope proteins or capsid structures through binding interactions with NPs,16–18 inhibition of viral replication,19 photochemical degradation through semiconductor NPs20–23 or photosensitizer loaded NPs,24 and denaturation of viral components through photothermal and photodynamic effects25 associated with plasmonic nanomaterials.26,27 In some cases, a combination of AgNPs and wide-bandgap semiconductor nanostructures has been found to be beneficial for achieving viral photoinactivation under visible light illumination.28 These previous studies demonstrated efficacy against enveloped viruses, including SARS-CoV-2,21,28 Zika,19 West Nile,19 dengue,19 HIV,23 and influenza20 virus. In this study, we describe the development of discrete hybrid nanostructures of noble metal nanoparticles and molecular photocatalysts that generate increased chemical reactivity upon light illumination. Furthermore, selective targeting of these light-activated plasmonic nanoreactors to the virus particle using annexin V – phosphatidylserine (PS) interactions maximizes the reactivity-based virucidal effect while simultaneously minimizing collateral damage.The plasmonic nanoreactor design is based on silver nanoparticles (AgNPs) encapsulated in a lipid coating that binds tris(bipyridine)ruthenium(II) [Ru(bpy)3]2+ and positions the photocatalyst in the evanescent field of the AgNPs. AgNPs support a localized surface plasmon resonance (LSPR) that overlaps with the metal-to-ligand-charge-transfer (MLCT) band in [Ru(bpy)3]2+ in the range between 450 nm to 480 nm.29,30 Resonant excitation of the AgNP core of the plasmonic nanoreactors facilitates a plasmon-enhanced excitation of the molecular photocatalyst, which results in increased photoreactivity.31 In its excited, long-lived triplet state, [Ru(bpy)3]2+ is simultaneously a good oxidizer and reducer and can also generate reactive oxygen species (ROS) including single O2, superoxide anions and hydroxyl radicals.29,32–35 If ROS are generated selectively in the proximity of virus particles, virus particles are preferentially affected by the generated chemical reactivity while the effect on bystander cells is limited due to the short diffusion lengths of ROS that lie between 5 nm in the case of the hydroxyl radical and 300 nm for the superoxide anion.36,37
We implement in this study plasmonic nanoreactor virucides (PNVs) that target murine leukemia virus (MLV) as a model system for enveloped viruses, to examine the antiviral effect of these nanostructures with and without targeting functionality and characterize the mechanisms underlying viral inactivation. We chose MLV as a model to represent potential contaminations of the bioreactor feed material by viruses shed by rodents, which is a problem in the fabrication of biologics. In the absence of light illumination, the plasmonic nanoreactors show a high degree of cytocompatibility, but light illumination triggers the generation of ROS by [Ru(bpy)3]2+ that facilitates the release of virucidal Ag+ ions38,39 from the NP core through increased permeability of NP lipid coating and promotes damage to the targeted virus particles via multiple mechanisms. The PNVs demonstrate >85% virus inactivation after 30 min of 470 nm illumination under benign conditions for a virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNV ratio of 1
PNV ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which confirms the potential of targeted plasmonic nanoreactors for the selective inactivation of enveloped viruses in sensitive applications at relatively low concentrations.
1, which confirms the potential of targeted plasmonic nanoreactors for the selective inactivation of enveloped viruses in sensitive applications at relatively low concentrations.
Results and discussion
Assembly and characterization of plasmonic nanoreactor virucides (PNVs)
The structure of the investigated PNVs (Fig. 1A) is similar to that of plasmonic nanoreactors previously introduced as antibacterials.29,31 The main difference is that we included annexin V within the lipid coating surrounding the NPs as a specific targeting functionality for enveloped viruses. The lipid coating around the 60 nm AgNP in this design serves three main purposes: (1) it is designed to provide cytocompatibility, passivate the AgNPs, and minimize the leakage of Ag+ ions under dark conditions (i.e. no light illumination). (2) It provides a matrix for [Ru(bpy)3]2+, which has been predicted to enrich at the lipid/water interface around the NPs.29 (3) Functionalized lipids in the coating allow the integration of viral targeting functionalities, such as annexin V, into the NP surface. Annexin V binds to phosphatidylserine (PS),40,41 a phospholipid that is localized to the inner leaflet of healthy mammalian cells but is exposed in the outer leaflet of virus particles and virus-infected host cells.42,43 The presentation of PS in the outer leaflet of enveloped viruses provides the basis of a selective targeting by annexin V-functionalized NPs.PNVs are assembled in a “one-pot” synthesis by combining colloidal 60 nm Ag NPs with an aqueous mix of [Ru(bpy)3]2+ and lipids in the presence of 1-octadecanethiol.31 The lipids used in this work were comprised of DPPC (dipalmitoyl phosphatidylcholine), cholesterol, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PA (1,2-dioleoyl-sn-glycero-3-phosphate), and DSPE-PEG(2000)-azide (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[azido(polyethylene glycol)-2000]) in a molar ratio of 29
1 PA (1,2-dioleoyl-sn-glycero-3-phosphate), and DSPE-PEG(2000)-azide (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[azido(polyethylene glycol)-2000]) in a molar ratio of 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The main ingredient of the lipid coating, DPPC, is a zwitterionic saturated lipid that prefers rigid packing, but the addition of cholesterol increases the fluidity of the membrane-like coating. 18
1. The main ingredient of the lipid coating, DPPC, is a zwitterionic saturated lipid that prefers rigid packing, but the addition of cholesterol increases the fluidity of the membrane-like coating. 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PA is a negatively charged unsaturated lipid that improves the colloidal stability of the NPs but also increases the susceptibility of the lipid coating to ROS. Peroxidation of the unsaturated lipids is expected to destabilize the lipid coating44–46 and enhance the permeability for Ag+ ions. DSPE-PEG(2000)-azide was included to facilitate the covalent attachment of annexin V (see below).
1 PA is a negatively charged unsaturated lipid that improves the colloidal stability of the NPs but also increases the susceptibility of the lipid coating to ROS. Peroxidation of the unsaturated lipids is expected to destabilize the lipid coating44–46 and enhance the permeability for Ag+ ions. DSPE-PEG(2000)-azide was included to facilitate the covalent attachment of annexin V (see below).
High-resolution transmission electron microscopy (HRTEM) images of individual PNVs and energy dispersive X-ray spectroscopy (EDX) element maps recorded in scanning TEM (STEM) mode (Fig. 1B) confirm that the AgNPs are encapsulated by a membrane-like lipid coating and contain [Ru(bpy)3]2+. Microwave Plasma Atomic Emission Spectroscopy (MP-AES) yielded average concentrations of Ag and Ru of 937.1 ± 156.2 ppb and 10.0 ± 1.8 ppb, respectively, for the hybrid material. We also measured UV-visible (UV-vis) spectra of the hybrids, which confirmed that the plasmon resonance of the AgNPs and the MLCT of [Ru(bpy)3]2+ overlap (Fig. 1C). Table 1 summarizes peak plasmon resonances, hydrodynamic diameters, and ζ-potentials for all investigated experimental conditions, controls, and relevant intermediate structures.
| Sample | Resonance peak (nm) | Hydrodynamic diameter (nm) | Zeta potential (meV) |
|---|---|---|---|
| a Citrate capped AgNP. b Lipid-coated AgNP. c Lipid-coated AgNP mixed with annexin V (ANXV). d Lipid-coated AgNP containing azide ligands. e Lipid-coated AgNP covalently functionalized with annexin V. f Lipid-coated AgNP with bound [Ru(bpy)3]2+. g Lipid-coated AgNP with bound [Ru(bpy)3]2+ mixed with annexin V. h Lipid-coated AgNP with bound [Ru(bpy)3]2+ and containing azide ligands. i PNVs with covalently bound annexin V. | |||
| AgNP a | 448 | 60.5 ± 0.4 | −24.3 ± 2.5 |
| L-AgNP b | 455 | 62.5 ± 1.6 | −29.6 ± 7.1 |
| L-AgNP + ANXV c | 449 | 92.5 ± 2.0 | −17.1 ± 1.3 |
| L-AgNP-azide d | 447 | 76.0 ± 0.4 | −14.9 ± 0.3 |
| L-AgNP-azide @ ANXV e | 450 | 89.2 ± 2.0 | −11.6 ± 0.3 |
| L-AgNP-Ru f | 449 | 68.4 ± 1.1 | −9.6 ± 2.1 |
| L-AgNP-Ru + ANXV g | 454 | 111.5 ± 2.4 | −11.1 ± 2.9 |
| PNVs h | 450 | 73.9 ± 0.8 | −9.1 ± 0.9 |
| PNVs @ ANXV i | 452 | 97.7 ± 2.9 | −12.6 ± 0.3 |
Introducing targeting functionalities into PNVs
PNVs were assembled containing DSPE-PEG(2000)-azide in the lipid coating for the covalent attachment of DBCO-functionalized annexin V via strain-promoted azide–alkyne cycloaddition (SPAAC).50,51 The scheme of the reaction is presented in Fig. 2A. Annexin V has a total of 22 lysine residues,47–49,52 allowing an easy incorporation of DBCO through sulfo-NHS ester chemistry. Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) validated the successful introduction of DBCO into annexin V (Fig. 2B). The mass increases from 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 746 m/z for annexin V to 36
746 m/z for annexin V to 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 m/z, which indicates that most annexin V molecules react with two DBCO-sulfo-NHS ester molecules (532.5 g mol−1). The tail of the product peak at higher masses reflects a sub-population of annexin V with more than 2 DBCO-sulfo-NHS molecules bound. The DBCO-functionalized annexin V was combined with PNVs to achieve cross-linking by SPAAC. The resulting annexin V-functionalized PNVs had a hydrodynamic diameter of 97.7 ± 2.9 nm, a ζ-potential of −12.6 ± 0.3 meV, and a peak plasmon resonance of 452 nm.
800 m/z, which indicates that most annexin V molecules react with two DBCO-sulfo-NHS ester molecules (532.5 g mol−1). The tail of the product peak at higher masses reflects a sub-population of annexin V with more than 2 DBCO-sulfo-NHS molecules bound. The DBCO-functionalized annexin V was combined with PNVs to achieve cross-linking by SPAAC. The resulting annexin V-functionalized PNVs had a hydrodynamic diameter of 97.7 ± 2.9 nm, a ζ-potential of −12.6 ± 0.3 meV, and a peak plasmon resonance of 452 nm.
 | ||
| Fig. 2 (A) Functionalization of the PNV (PNV@ANXV) for annexin V-mediated targeting of MLV. Structure of annexin V (UniProt ID: P08758) was obtained from the AlphaFold Protein Structure Database.47–49 (B) MALDI-MS of the annexin-V (red) and the DBCO-functionalized annexin-V (Blue). (C) Number of annexin V/NP determined by ELISA for (left to right) L-AgNP mixed with annexin V, PNVs that lack [Ru(bpy)3]2+, PNVs that lack azide groups, and complete PNVs. (D) TEM image of MLV. (E) TEM image of MLV containing two bound PNV@ANXV particles. Samples in D and E were treated with a 2% uranyl acetate solution. Scale bars are 10 nm. | ||
We validated the successful crosslinking of the DBCO-functionalized annexin V with the DSPE-PEG(2000)-azide on the metal NPs through enzyme-linked immunosorbent assay (ELISA) for PNVs as well as for lipid-coated AgNP (L-AgNP) that lacked [Ru(bpy)3]2+ (Fig. 2C). In both cases, the cross-linking results in a large increase in detected annexin V, confirming a successful introduction of the targeting functionality into the NP surface with no apparent negative effect associated with the presence of [Ru(bpy)3]2+.
Finally, the binding of annexin V-functionalized PNVs to MLVs was verified by transmission electron microscopy (Fig. 2D, E and S1). Only for annexin V-functionalized PNVs a strong localization of AgNPs on MLVs or membrane debris from the host cell is observed, while PNVs that lack annexin V show a random distribution with no preferential association with MLVs or membrane debris.
Switching between cytocompatibility and reactivity through resonant excitation
For many applications, it is desirable that the nanoreactors lack reactivity and do not create ROS or release substantial concentrations of cytotoxic Ag+ ions unless they are bound to their viral target and activated by optical excitation to maximize the antiviral effect and minimize any Ag+ cytotoxicity related side effects.53–56 One intended function of the lipid coating around the AgNP of the PNV is to insulate the NP core, reduce its cytotoxicity, and increase its overall cytocompatibility. This was experimentally tested by comparing the viability of Rat-2 cells after incubation with citrate-stabilized AgNPs and lipid coated PNVs (Fig. S2). The lipid coating profoundly reduced the number of dead cells when compared with AgNPs, suggesting that the lipid coating passivates and stabilizes the AgNP core in PNVs.Illumination at the plasmon resonance is expected to trigger the generation of ROS through excitation of [Ru(bpy)3]2+ and to activate PNVs for virucidal applications. To validate this concept, we quantified ROS generation for L-AgNP without [Ru(bpy)3]2+ and PNVs as a function of 470 nm illumination time using dihydrorhodamine 123 (DHR123) which is converted by ROS into the fluorescent rhodamine 123 (Fig. 3A).57,58 The PNV sample exhibited a substantial increase in the rhodamine 123 fluorescence signal over the L-AgNP baseline within the first 10 min of illumination, confirming that the [Ru(bpy)3]2+ in the PNV is a source of ROS generation. A change in the AgNP core in PNVs with AuNPs resulted in a decrease in rhodamine 123 generation by ∼50% after 5 min of illumination (Fig. S3A). L-AgNP and L-AuNP that lack [Ru(bpy)3]2+ show low ROS generation both under 470 nm illumination and in the dark. PNVs (with a AgNP core) illuminated at 420, 450, or 470 nm demonstrated the strongest ROS generation at 470 nm (Fig. S3B).
ROS generation may also initiate lipid peroxidation and destabilize the lipid coating of PNVs.44,46 A degradation of the passivating PNV lipid coating would expose the AgNP core and could lead to increased Ag+ ion release.29 We evaluated the stability of the PNV lipid coating before and after illumination with 470 nm light with an irradiance of 65 mW cm−2 using a AgNP dissolution assay. Sodium thiosulfate can dissolve citrate-stabilized AgNPs in the presence of oxygen,59–61 and this process can be monitored by UV-vis spectroscopy (Fig. 3B). Importantly, L-AgNP that lack [Ru(bpy)3]2+ were stable after addition of sodium thiosulfate when illuminated at 470 nm for 60 min, indicating that the lipid coating protects the AgNP core (Fig. 3C). Likewise, PNVs that have not been illuminated at 470 nm were also stable against treatment with sodium thiosulfate. However, upon photoexcitation at 470 nm a dissolution of PNVs is detected by UV-vis spectroscopy (Fig. 3C). Overall, these observations indicate a light-induced ROS generation and subsequent degradation of the lipid coating. The induced lipid damage increases the permeability of the protective coating of PNVs and results in overall reduced protection against AgNP dissolution.
Viral inactivation through PNVs
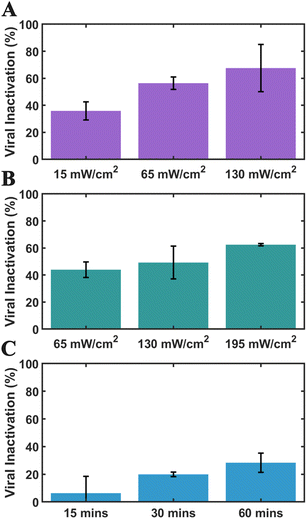
We examined the virucidal effect of PNVs by quantifying the reduction in MLV infectivity under different experimental conditions. The absorbance spectrum of PNVs shows a broad resonance that peaks at 452 nm with a width of approximately 100 nm. We first evaluated the baseline virucidal effect of different LED lamps with emission wavelengths between 420 and 470 nm that are available to excite the PNVs (Fig. 4). These experiments were performed at room temperature. After 15 minutes of illumination with 65 mW cm−2, 420 and 450 nm emitting LEDs achieved a viral inactivation of 53% and 44%, respectively (Fig. 4A and B). Even with a reduced fluence of 15 mW cm−2 the 420 nm LED still achieves a viral inactivation of 36%. In contrast, 30 minutes of illumination with a 470 nm LED at 65 mW cm−2 provides a baseline viral inactivation of less than 20% (Fig. 4C). Since our interest in this work is based on characterizing the effect of PNVs, we chose this condition for our experiments to minimize the contribution from light-driven viral inactivation.Avoiding excitation wavelengths far in the blue spectrum is also beneficial for avoiding photodamage and related collateral effects in future applications of PNVs. Noble metal NPs have large optical cross-sections, and dissipation in the NPs can result in thermal effects. The calculated62 temperature increase for an individual NP under the continuous wave (CW) illumination conditions applied here is, however, very low. Even for a solution of the highest NP concentration used in this work, 5 × 109 particles/175 µL, the available heating rate is insufficient to produce a significant increase above ambient temperature when heat transfer is taken into account. We therefore do not consider photothermal effects to be relevant for virus inactivation under the chosen experimental conditions.
The virucidal effect of annexin V-functionalized PNVs and selected controls under 470 nm LED illumination is summarized in Fig. 5A. We included the following samples in these experiments: virus control, virus + L-AgNPs (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio), and virus + PNVs at two different virus
1 ratio), and virus + PNVs at two different virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNV ratios (5
PNV ratios (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). For all samples viral inactivation was evaluated without illumination (dark) and with 470 nm illumination. Both the virus control and virus + lipid-coated AgNPs show no measurable inactivation in the absence of illumination after an incubation time of 30 min (Fig. 5A). The PNVs show a moderate inactivation of 13% after 30 min in the dark. After 30 min of 470 nm illumination, the virus and virus + L-AgNP controls show an inactivation of 14% and 15%, respectively, which results from the 470 nm light effect discussed above. Importantly, the illuminated PNV samples achieve a significantly higher (p < 0.01) viral inactivation of 59% and 86% for virus
1). For all samples viral inactivation was evaluated without illumination (dark) and with 470 nm illumination. Both the virus control and virus + lipid-coated AgNPs show no measurable inactivation in the absence of illumination after an incubation time of 30 min (Fig. 5A). The PNVs show a moderate inactivation of 13% after 30 min in the dark. After 30 min of 470 nm illumination, the virus and virus + L-AgNP controls show an inactivation of 14% and 15%, respectively, which results from the 470 nm light effect discussed above. Importantly, the illuminated PNV samples achieve a significantly higher (p < 0.01) viral inactivation of 59% and 86% for virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNV ratios of 5
PNV ratios of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Considering a net light effect of ∼13%, the targeted PNVs achieve a gain in viral inactivation of approx. 45% and 72%. These increases in virus inactivation are substantial when one considers the applied virus
1. Considering a net light effect of ∼13%, the targeted PNVs achieve a gain in viral inactivation of approx. 45% and 72%. These increases in virus inactivation are substantial when one considers the applied virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNV ratios and may indicate that the annexin V-functionalized PNV binds to multiple virus particles and thus enhance their effect. PNVs that were not functionalized with annexin V only achieved an inactivation of 13%. The difference in the much higher (p < 0.001) inactivation levels for annexin V functionalized PNVs underlines the importance of virus targeting for the efficacy of the PNVs.
PNV ratios and may indicate that the annexin V-functionalized PNV binds to multiple virus particles and thus enhance their effect. PNVs that were not functionalized with annexin V only achieved an inactivation of 13%. The difference in the much higher (p < 0.001) inactivation levels for annexin V functionalized PNVs underlines the importance of virus targeting for the efficacy of the PNVs.
Cell viability under the previously defined virus and virus + PNV infection conditions was simultaneously evaluated through lactate dehydrogenase (LDH) assays (Fig. 5B). For the virus control (no treatment) and L-AgNP incubation, infection results in approximately 50% cell death in the absence of 470 nm illumination. For the PNV samples the cell death rate increases to 57 and 63% at 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 virus
1 virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNV ratios. Upon illumination with 470 nm LED light, the cell death drastically decreases to 23% and 5%, for PNV samples with virus
PNV ratios. Upon illumination with 470 nm LED light, the cell death drastically decreases to 23% and 5%, for PNV samples with virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNV ratios of 5
PNV ratios of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. These changes had a statistical significance of p < 0.001. In contrast, the cell viability decreases by < 6% for the virus control, and it remains nearly unchanged for the L-AgNP sample with illumination. These striking differences exclude a decrease in host cell viability as a cause for the observed drop in viral load observed with PNV treatment and provide further evidence of a viral inactivation induced by targeted PNV under resonant illumination.
1, respectively. These changes had a statistical significance of p < 0.001. In contrast, the cell viability decreases by < 6% for the virus control, and it remains nearly unchanged for the L-AgNP sample with illumination. These striking differences exclude a decrease in host cell viability as a cause for the observed drop in viral load observed with PNV treatment and provide further evidence of a viral inactivation induced by targeted PNV under resonant illumination.
Addition of the singlet O2 scavenger NaN3 (Fig. 5C) reduced the viral inactivation obtained with PNVs under resonant illumination, confirming that the light-driven generation of ROS is an important factor in the observed virus inactivation.63,64 The baseline level of virus inactivation was increased in the presence of NaN3 (see virus + NaN3 condition), which is attributed to the cytotoxicity of the azide.
Conclusion
We have examined the virucidal effect of plasmonic nanoreactors that localize their light-driven reactivity to enveloped viruses by targeting of PS in the viral membrane and demonstrated a viral inactivation of up to 86% with MLV as the test case. The PNV design is based on a hybrid nanostructure design that contains a passivating lipid coating around a AgNP core. The lipid coating provides a matrix for the localization of the photocatalyst ([Ru(bpy)3]2+) in the evanescent field of the AgNP to achieve plasmon-enhanced, light-driven ROS generation. Furthermore, the AgNP also allows a combination of photocatalyst and virus-targeting recognition elements. In this work annexin V was used to target PS, which is present in the outer leaflet of enveloped viruses but restricted to the inner leaflet of healthy mammalian cells. PNVs targeting viral membranes through annexin V can affect MLVs through multiple channels. The ROS generated in direct proximity of the virus can damage viral functions. The ROS can also act on the lipid coating of the PNV itself that contains unsaturated lipids and trigger damage that subsequently increases the permeability of the membrane-like coating. The “local” release of Ag+ ions from AgNPs bound to the virus target constitutes an additional virucide that further enhances the antiviral effect. Due to the passivating surface properties of the lipid coating of the AgNPs, the cytotoxicity of the plasmonic nanoreactors to healthy host cells was overall low. Targeting of PNVs to enveloped viruses provides a potential path for their selective inactivation even in the presence of mammalian cells. This approach is particularly relevant for biopharmaceutical processes that utilize cell-based bioreactors, which are susceptible to virus contamination.Experimental section
Preparation of the DBCO-annexin V crosslinker
The Tag-free human annexin A5 reconstitute of protein (annexin V, Acro biosystems) was added in 100 mmol of sodium bicarbonate (NaHCO3, Sigma Aldrich) to reach a final concentration of 17.4 µmol. 20 µL of annexin A5 solution was then mixed with 4 µL of 0.83 mM dibenzocyclooctyne-sulfo-N-hydroxysuccinimidyl ester (DBCO-sulfo-NHS ester, Sigma Aldrich) solution in 0.5× PBS. The mixture was incubated for 24 hours at 4 °C. After the incubation, the mixture underwent dialysis in 0.5× PBS for 48 hours.Preparation of PNVs
PNVs were prepared through a modified one-pot self-assembly method. A lipid mixture containing chloroform solutions of 58 mol% DPPC, 10 mol% 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PA, 30 mol% cholesterol, and 2 mol% of DSPE-PEG(400)-azide(Avanti Polar Lipids) was mixed with a 2 mol% methanol solution of [Ru(bpy)3]Cl2 (Sigma Aldrich). The mixture was then rotary evaporated at 32 °C and desiccated for 3 hours. 2 mL of water was added to a round-bottom flask containing the dehydrated lipid film, followed by tip sonication for 5 min to form liposomes that contained [Ru(bpy)3]2+. The liposomes were next mixed with 160 µL of 2 mg mL−1 octadecanethiol (ODT, Sigma-Aldrich) ethanol solution, followed by mixing with 1 mL of citrate stabilized 60 nm AgNPs (1010 particles/mL) (nanoComposix, NanoXact Silver Nanospheres). The solution was incubated overnight to allow the formation of functionalized lipid-coated AgNP containing [Ru(bpy)3]2+ and DSPE-PEG-azide. Finally, the AgNP solution was washed by centrifugation (3500 g), followed by storage at 4 °C.
1 PA, 30 mol% cholesterol, and 2 mol% of DSPE-PEG(400)-azide(Avanti Polar Lipids) was mixed with a 2 mol% methanol solution of [Ru(bpy)3]Cl2 (Sigma Aldrich). The mixture was then rotary evaporated at 32 °C and desiccated for 3 hours. 2 mL of water was added to a round-bottom flask containing the dehydrated lipid film, followed by tip sonication for 5 min to form liposomes that contained [Ru(bpy)3]2+. The liposomes were next mixed with 160 µL of 2 mg mL−1 octadecanethiol (ODT, Sigma-Aldrich) ethanol solution, followed by mixing with 1 mL of citrate stabilized 60 nm AgNPs (1010 particles/mL) (nanoComposix, NanoXact Silver Nanospheres). The solution was incubated overnight to allow the formation of functionalized lipid-coated AgNP containing [Ru(bpy)3]2+ and DSPE-PEG-azide. Finally, the AgNP solution was washed by centrifugation (3500 g), followed by storage at 4 °C.
Mass spectroscopy
The measurement was conducted on a Bruker Autoflex Speed MALDI-TOF. The sample preparation started with dissolving 10 mg of sinapic acid in 1 mL of ethanol as the matrix. Then, the protein sample was mixed with the matrix in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 2 µL of the mixture was dropcast onto the sample plate.
1, and 2 µL of the mixture was dropcast onto the sample plate.
ELISA
The experiment was performed with an Invitrogen Human annexin V ELISA kit (Fisher Thermo). The kit comes with all the chemicals used in this section. The 8-well strips coated with monoclonal annexin V antibody were washed twice with PBS with 1% Tween 20. A serial dilution of the human annexin V standard (from 50 to 0.78 ng mL−1) and a blank sample were prepared for the comparison of counts of annexin V incorporated into the lipid membrane. The four experimental samples consisted of L-NP-annexin V, L-NP-azide-annexin V, L-NP-Ru-annexin V, and L-NP-Ru-azide-annexin V, as well as the supernatants from the second washing. Each of the conditions contains 108 of the particles. Each experimental well was composed of 50% particles and 50% diluent by volume. 50 µL of Biotin-conjugate anti-human annexin V antibody was added to all wells and incubated at room temperature for 2 hours. The wells were emptied and washed four times with PBS with 1% Tween 20. Then, 100 µL streptavidin-HRP was added to all wells and incubated at room temperature for one hour. The wells were emptied and washed four times with PBS with 1% Tween 20 before a tetramethyl-benzidine substrate solution was added. The reaction was stopped after 10 minutes by the addition of 1 M phosphoric acid stop solution. Optical densities at 450 nm and 620 nm were measured using a microplate reader to determine the concentration of annexin V.Characterization of PNVs
Hydrodynamic size and ζ-potential measurements were performed by using a Zetasizer Nano ZS90 (Malvern). In the hydrodynamic size measurements, AgNPs were diluted to 100 µL of 1010 particles per mL with Milli-Q water. Respectively, NPs were diluted to 1 mL of 10 mM NaCl solution for the ζ-potential measurement.For the Microwave Plasma Atomic Emission Spectroscopy (MP-AES) measurement, the particles were washed twice with Milli-Q water and redispersed in 200 µL of Milli-Q water. In the 12-well plate, NPs were mixed with aqua regia at 60 °C overnight. 1 mL of 2% HNO3 was added to the 12-well plate to redissolve the ions. The measurement was performed in an Agilent 4210 MP-AES. Standards of Ag and Ru were made in 2% HNO3 from 10 ppb to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ppb.
000 ppb.
Transmission Electron Microscopy (TEM) was performed on a JEOL F200 TEM at 80 kV. Energy Dispersive X-ray spectroscopy (EDX) was performed with a high-angle annular dark-field detector in Scanning Transmission Electron Microscopy (STEM) mode and a JEOL EDAX detector with Oxford instruments Aztec software. Samples were prepared on cooper/carbon TEM grids (Tedpella Inc.). 20 µL of functionalized lipid-coated AgNPs were dropcast onto the grids and incubated for 30 minutes in a water-saturated environment followed by removal with a Kimwipe.
Sodium thiosulfate dissolution assay
1 M dilution of sodium thiosulfate solution (Na2S2O3, Sigma Aldrich) was prepared in Milli-Q water. After functionalized lipid-coated AgNPs were illuminated by the respective LED, 100 µL of sodium thiosulfate solution was added to 1 mL of functionalized lipid-coated AgNPs. UV-vis spectroscopy was performed using a Cary-5000 in the range of 300 nm to 600 nm. The data were automatically generated by taking a scan of 30 seconds every 2 minutes.ROS measurement
A 50 µL sample of nanoparticles was mixed with 30 µM dihydrorhodamine 123 (Med Chem Express) solution in DMSO and diluted to a total volume of 150 µL with Milli-Q water. The mixture was then illuminated by using a 470 nm LED with a power density of 15 mW cm−2 20 µL of liquid was taken out at each time point and diluted to 400 µL. Fluorescence was measured by using a Horiba Jobin Yvon FluoroMax 3 Fluorimeter (Ex: 485 nm, Em: 523 nm).Virus preparation
Single round luciferase reporter murine leukemia virus (MLV) was53 prepared with a pCL-Eco packaging plasmid and pLNC-luc MLV reporter provirus construct plasmid. Virus-containing cell supernatants were harvested 2 days post-transfection, cleared of cell debris by passing through 0.45 µm filters, and purified and concentrated by ultracentrifugation on a 20% sucrose cushion (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 2 h with a SW32Ti or SW28 rotor (Beckman Coulter)). The virus pellets were resuspended in PBS, aliquoted, and stored at −80 °C until use.
000 rpm and 4 °C for 2 h with a SW32Ti or SW28 rotor (Beckman Coulter)). The virus pellets were resuspended in PBS, aliquoted, and stored at −80 °C until use.
Virus titering
Concentrated viral stocks were titered using the QuickTiter™ MuLV Core Antigen ELISA Kit (Cell Biolabs, Inc Catalog ID 130-050-201). The viral particles per µL was calculated by multiplying the ELISA p30 ng µL−1 value by the constant: 1 ng p30 = 9.009 × 10 × 106 viral MLV particles.Nanoparticle-mediated viral inactivation and infection assays
1 × 104 Rat-2 cells (ATCC CRL-1764) were seeded in 100 µL cell growth media (DMEM+10% fetal bovine serum+5% penicillin/streptomycin) 24 hours prior to MLV infection. For a virus![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) particle ratio of 5
particle ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1 × 109 particles of PNVs or L-AgNP controls in 50 µL were mixed with 2.25 µL of MLV with a concentration of 2.1 × 109 viral particles per µL and 0.5× PBS, bringing the final volume of the mixture to 175 µL. The mixture was incubated at 4 °C for 1 hour. Next, the illumination was performed at room temperature using a 470 nm LED with 65 mW cm−2 for 30 minutes unless otherwise stated. Following inactivation, cell media was replaced with 50 µL of 20 µg mL−1 polybrene (Millipore) followed by 50 µL of a sample mixture. The cells were spinoculated with a virus/nanoparticle mixture at 2300 rpm for 1 hour at room temperature and then moved to a cell culture incubator for 2 hours before replacing all experimental conditions with growth media. Cells were lysed at 48 hours post-infection, and cell lysates were used for quantification of luciferase activity using a Promega Bright-Glo Luciferase Assay System (Catalog ID PR-E2620) as a measure of MLV infection establishment.
1, 1 × 109 particles of PNVs or L-AgNP controls in 50 µL were mixed with 2.25 µL of MLV with a concentration of 2.1 × 109 viral particles per µL and 0.5× PBS, bringing the final volume of the mixture to 175 µL. The mixture was incubated at 4 °C for 1 hour. Next, the illumination was performed at room temperature using a 470 nm LED with 65 mW cm−2 for 30 minutes unless otherwise stated. Following inactivation, cell media was replaced with 50 µL of 20 µg mL−1 polybrene (Millipore) followed by 50 µL of a sample mixture. The cells were spinoculated with a virus/nanoparticle mixture at 2300 rpm for 1 hour at room temperature and then moved to a cell culture incubator for 2 hours before replacing all experimental conditions with growth media. Cells were lysed at 48 hours post-infection, and cell lysates were used for quantification of luciferase activity using a Promega Bright-Glo Luciferase Assay System (Catalog ID PR-E2620) as a measure of MLV infection establishment.
Cytotoxicity assay
Cell cytotoxicity was assayed by measuring LDH release in cell supernatants and MTT metabolization in cells. LDH release was assayed using the CytoTox 96® non-radioactive cytotoxicity assay (catalog ID PR-G1780). MTT metabolization was assayed by removing cell growth media and adding 100 µL MTT working solution (5 mg mL−1, Sigma Catalog ID M5655-500 MG) in phosphate-buffered saline (PBS). Cells were incubated for 3–4 hours at 37 °C in a cell culture incubator. MTT working solution was then replaced with 100 µL solubilization solution consisting of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volumetric ratio of ethanol and DMSO. Absorbance was read with a plate reader at 550–570 nm with a background reading at 650 nm.
1 volumetric ratio of ethanol and DMSO. Absorbance was read with a plate reader at 550–570 nm with a background reading at 650 nm.
The viral inactivation (%) was determined based on the luciferase counts. All inactivation experiments were performed in triplicate. The inactivation is defined as percentage reduction relative to the control: (1 – a/b) × 100. In this expression a is the luciferase count for a specific condition and b is the count for the virus only control without any treatment.
Author contributions
TO and KK contributed equally to this manuscript. TO, KK, SG, and BMR conceived the study and designed the experiments. TO and KK performed the experiments, analyzed the data, and generated the figures assisted by BC and AG. TO and JH performed the viral infection experiments. The manuscript was written through the contributions of all authors. All authors have approved the final version of the manuscript.Conflicts of interest
BMR and SG hold patents for lipid-coated metal nanoparticles.Data availability
Raw data for this article, including optical spectra, inactivation results, and nanoparticle characterization data, are available at Zenodo under the digital objective identifier (DOI): https://doi.org/10.5281/zenodo.16746910.Supplementary information (SI): Fig. S1–S3: larger scale TEM images for the PNVs@ANV binding, cell cytotoxicity test with different conditions of AgNPs, and additional controls on ROS generation. See DOI: https://doi.org/10.1039/d5na00784d.
Acknowledgements
The authors acknowledge support from the National Institutes of Health through grants R01GM142012 (BMR, SG) and R01AI175068 (SG). Parts of this work were performed at the Harvard University Center for Nanoscale Systems (CNS); a member of the National Nanotechnology Coordinated Infrastructure Network (NNCI), which is supported by the National Science Foundation under NSF award no. ECCS-2025158.References
- Z. Lou, Y. Sun and Z. Rao, Current Progress in Antiviral Strategies, Trends Pharmacol. Sci., 2014, 35(2), 86–102, DOI:10.1016/j.tips.2013.11.006
.
- B. C. Gonçalves, M. G. Lopes Barbosa, A. P. Silva Olak, N. Belebecha Terezo, L. Nishi, M. A. Watanabe, P. Marinello, D. Zendrini Rechenchoski, S. P. Dejato Rocha and L. C. Faccin-Galhardi, Antiviral Therapies: Advances and Perspectives, Fundam. Clin. Pharmacol., 2021, 35(2), 305–320, DOI:10.1111/fcp.12609
.
- H.-J. Henzler and K. Kaiser, Avoiding Viral Contamination in Biotechnological and Pharmaceutical Processes, Nat. Biotechnol., 1998, 16(11), 1077–1079, DOI:10.1038/3538
.
- P. W. Barone, M. E. Wiebe, J. C. Leung, I. T. M. Hussein, F. J. Keumurian, J. Bouressa, A. Brussel, D. Chen, M. Chong, H. Dehghani, L. Gerentes, J. Gilbert, D. Gold, R. Kiss, T. R. Kreil, R. Labatut, Y. Li, J. Müllberg, L. Mallet, C. Menzel, M. Moody, S. Monpoeho, M. Murphy, M. Plavsic, N. J. Roth, D. Roush, M. Ruffing, R. Schicho, R. Snyder, D. Stark, C. Zhang, J. Wolfrum, A. J. Sinskey and S. L. Springs, Viral Contamination in Biologic Manufacture and Implications for Emerging Therapies, Nat. Biotechnol., 2020, 38(5), 563–572, DOI:10.1038/s41587-020-0507-2
.
- F. Vigant, N. C. Santos and B. Lee, Broad-Spectrum Antivirals against Viral Fusion, Nat. Rev. Microbiol., 2015, 13(7), 426–437, DOI:10.1038/nrmicro3475
.
- M. Karim, C.-W. Lo and S. Einav, Preparing for the next Viral Threat with Broad-Spectrum Antivirals, J. Clin. Invest., 2023, 133(11), e170236, DOI:10.1172/JCI170236
.
- A. Brai, R. Fazi, C. Tintori, C. Zamperini, F. Bugli, M. Sanguinetti, E. Stigliano, J. Esté, R. Badia, S. Franco, M. A. Martinez, J. P. Martinez, A. Meyerhans, F. Saladini, M. Zazzi, A. Garbelli, G. Maga and M. Botta, Human DDX3 Protein Is a Valuable Target to Develop Broad Spectrum Antiviral Agents, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(19), 5388–5393, DOI:10.1073/pnas.1522987113
.
- H. K. Meng, K. T. Pang, C. Wan, Z. Y. Zheng, Q. Beiying, Y. Yang, W. Zhang, Y. S. Ho, I. Walsh and S. Chia, Thermal and pH Stress Dictate Distinct Mechanisms of Monoclonal Antibody Aggregation, Int. J. Biol. Macromol., 2024, 282, 136601, DOI:10.1016/j.ijbiomac.2024.136601
.
- B. Fongaro, V. Cian, F. Gabaldo, G. De Paoli, G. Miolo and P. Polverino De Laureto, Managing Antibody Stability: Effects of Stressors on Ipilimumab from the Commercial Formulation to Diluted Solutions, Eur. J. Pharm. Biopharm., 2022, 176, 54–74, DOI:10.1016/j.ejpb.2022.05.005
.
- N. Whitaker, S. E. Pace, K. Merritt, M. Tadros, M. Khossravi, S. Deshmukh, Y. Cheng, S. B. Joshi, D. B. Volkin and P. Dhar, Developability Assessments of Monoclonal Antibody Candidates to Minimize Aggregation During Large-Scale Ultrafiltration and Diafiltration (UF-DF) Processing, J. Pharm. Sci., 2022, 111(11), 2998–3008, DOI:10.1016/j.xphs.2022.08.001
.
- E. Mosidze, G. Franci, F. Dell'Annunziata, N. Capuano, M. Colella, F. Salzano, M. Galdiero, A. Bakuridze and V. Folliero, Silver Nanoparticle-Mediated Antiviral Efficacy against Enveloped Viruses: A Comprehensive Review, Global Chall., 2025, 2400380, DOI:10.1002/gch2.202400380
.
- A. Bhatti and R. K. DeLong, Nanoscale Interaction Mechanisms of Antiviral Activity, ACS Pharmacol. Transl. Sci., 2023, 6(2), 220–228, DOI:10.1021/acsptsci.2c00195
.
- J. J. Broglie, B. Alston, C. Yang, L. Ma, A. F. Adcock, W. Chen and L. Yang, Antiviral Activity of Gold/Copper Sulfide Core/Shell Nanoparticles against Human Norovirus Virus-Like Particles, PLoS One, 2015, 10(10), e0141050, DOI:10.1371/journal.pone.0141050
.
- N. Lin, D. Verma, N. Saini, R. Arbi, M. Munir, M. Jovic and A. Turak, Antiviral Nanoparticles for Sanitizing Surfaces: A Roadmap to Self-Sterilizing against COVID-19, Nano Today, 2021, 40, 101267, DOI:10.1016/j.nantod.2021.101267
.
- A. Warghane, R. Saini, M. Shri, I. Andankar, D. K. Ghosh and B. A. Chopade, Application of Nanoparticles for Management of Plant Viral Pathogen: Current Status and Future Prospects, Virology, 2024, 592, 109998, DOI:10.1016/j.virol.2024.109998
.
- E. Kolanthai, C. J. Neal, U. Kumar, Y. Fu and S. Seal, Antiviral Nanopharmaceuticals: Engineered Surface Interactions and Virus-selective Activity, Wiley Interdiscip. Rev.:Nanomed. Nanobiotechnol., 2022, 14(5), e1823, DOI:10.1002/wnan.1823
.
- V. Cagno, P. Andreozzi, M. D'Alicarnasso, P. Jacob Silva, M. Mueller, M. Galloux, R. Le Goffic, S. T. Jones, M. Vallino, J. Hodek, J. Weber, S. Sen, E.-R. Janeček, A. Bekdemir, B. Sanavio, C. Martinelli, M. Donalisio, M.-A. Rameix Welti, J.-F. Eleouet, Y. Han, L. Kaiser, L. Vukovic, C. Tapparel, P. Král, S. Krol, D. Lembo and F. Stellacci, Broad-Spectrum Non-Toxic Antiviral Nanoparticles with a Virucidal Inhibition Mechanism, Nat. Mater., 2018, 17(2), 195–203, DOI:10.1038/nmat5053
.
- J. M. de Souza e Silva, T. D. M. Hanchuk, M. I. Santos, J. Kobarg, M. C. Bajgelman and M. B. Cardoso, Viral Inhibition Mechanism Mediated by Surface-Modified Silica Nanoparticles, ACS Appl. Mater. Interfaces, 2016, 8(26), 16564–16572, DOI:10.1021/acsami.6b03342
.
- B. Schroeder, P. Demirel, C. Fischer, E. Masri, S. Kallis, L. Redl, T. Rudolf, S. Bergemann, C. Arkona, C. Nitsche, R. Bartenschlager and J. Rademann, Nanoparticular Inhibitors of Flavivirus Proteases from Zika, West Nile and Dengue Virus Are Cell-Permeable Antivirals, ACS Med. Chem. Lett., 2021, 12(12), 1955–1961, DOI:10.1021/acsmedchemlett.1c00515
.
- R. Nakano, H. Ishiguro, Y. Yao, J. Kajioka, A. Fujishima, K. Sunada, M. Minoshima, K. Hashimoto and Y. Kubota, Photocatalytic Inactivation of Influenza Virus by Titanium Dioxide Thin Film, Photochem. Photobiol. Sci., 2012, 11(8), 1293–1298, 10.1039/c2pp05414k
.
- S. Khaiboullina, T. Uppal, N. Dhabarde, V. R. Subramanian and S. C. Verma, Inactivation of Human Coronavirus by Titania Nanoparticle Coatings and UVC Radiation: Throwing Light on SARS-CoV-2, Viruses, 2020, 13(1), 19, DOI:10.3390/v13010019
.
- I. De Pasquale, C. Lo Porto, M. Dell'Edera, M. L. Curri and R. Comparelli, TiO2-Based Nanomaterials Assisted Photocatalytic Treatment for Virus Inactivation: Perspectives and Applications, Curr. Opin. Chem. Eng., 2021, 34, 100716, DOI:10.1016/j.coche.2021.100716
.
- K. Yamaguchi, T. Sugiyama, S. Kato, Y. Kondo, N. Ageyama, M. Kanekiyo, M. Iwata, Y. Koyanagi, N. Yamamoto and M. Honda, A Novel CD4-conjugated Ultraviolet Light-activated Photocatalyst Inactivates HIV-1 and SIV Efficiently, J. Med. Virol., 2008, 80(8), 1322–1331, DOI:10.1002/jmv.21235
.
- G. Zhang, L. Kuang, Y. Liu, C. Jiang, R. Yang, Q. Lv, K. Sun, X. Liang and K. Tao, Efficient Inactivation of Enveloped Viruses Using a Nanoparticle-Based Photodynamic Method, ACS Appl. Nano Mater., 2023, 6(19), 17647–17656, DOI:10.1021/acsanm.3c02880
.
- G. Zhang, J. Zhou, Q. Lv, R. Yang, Y. Zhang, J. Chu, H. Zhang, Y. Han, K. Sun, C. Yuan and K. Tao, Rapid Virus Inactivation by Nanoparticles-Embedded Photodynamic Surfaces, J. Colloid Interface Sci., 2025, 679, 609–618, DOI:10.1016/j.jcis.2024.10.114
.
- N. Chen, Y. Zheng, J. Yin, X. Li and C. Zheng, Inhibitory Effects of Silver Nanoparticles against Adenovirus Type 3 in Vitro, J. Virol. Methods, 2013, 193(2), 470–477, DOI:10.1016/j.jviromet.2013.07.020
.
- H. H. Lara, E. N. Garza-Treviño, L. Ixtepan-Turrent and D. K. Singh, Silver Nanoparticles Are Broad-Spectrum Bactericidal and Virucidal Compounds, J. Nanobiotechnol., 2011, 9(1), 30, DOI:10.1186/1477-3155-9-30
.
- R. Djellabi, N. Basilico, S. Delbue, S. D'Alessandro, S. Parapini, G. Cerrato, E. Laurenti, E. Falletta and C. Bianchi, Oxidative Inactivation of SARS-CoV-2 on Photoactive AgNPs@TiO2 Ceramic Tiles, Int. J. Mol. Sci., 2021, 22(16), 8836, DOI:10.3390/ijms22168836
.
- X. An, N. Naowarojna, P. Liu and B. M. Reinhard, Hybrid Plasmonic Photoreactors as Visible Light-Mediated Bactericides, ACS Appl. Mater. Interfaces, 2020, 12(1), 106–116, DOI:10.1021/acsami.9b14834
.
- K. Mori, M. Kawashima, M. Che and H. Yamashita, Enhancement of the Photoinduced Oxidation Activity of a Ruthenium(II) Complex Anchored on Silica-Coated Silver Nanoparticles by Localized Surface Plasmon Resonance, Angew. Chem., Int. Ed., 2010, 49(46), 8598–8601, DOI:10.1002/anie.201004942
.
- X. An, D. Stelter, T. Keyes and B. M. Reinhard, Plasmonic Photocatalysis of Urea Oxidation and Visible-Light Fuel Cells, Chem, 2019, 5(8), 2228–2242, DOI:10.1016/j.chempr.2019.06.014
.
- P. Parakh, S. Gokulakrishnan and H. Prakash, Visible Light Water Disinfection Using [Ru(Bpy)2(Phendione)](PF6)2·2H2O and [Ru(Phendione)3]Cl2·2H2O Complexes and Their Effective Adsorption onto Activated Carbon, Sep. Purif. Technol., 2013, 109, 9–17, DOI:10.1016/j.seppur.2013.02.022
.
- F. D. Abreu, I. C. N. Diógenes, L. G. de F. Lopes, E. H. S. Sousa and I. M. M. de Carvalho, Ruthenium(II) Bipyridine Complexes with Pendant Anthracenyl and Naphthyl Moieties: A Strategy for a ROS Generator with DNA Binding Selectivity, Inorg. Chim. Acta, 2016, 439, 92–99, DOI:10.1016/j.ica.2015.09.034
.
- Z. Ye, B. Song, Y. Yin, R. Zhang and J. Yuan, Development of Singlet Oxygen-Responsive Phosphorescent Ruthenium(Ii) Complexes, Dalton Trans., 2013, 42(40), 14380, 10.1039/c3dt52020j
.
- R. S. da Silva, M. S. P. Marchesi, A. C. Tedesco, A. Mikhailovsky and P. C. Ford, Generation of Reactive Oxygen Species by Photolysis of the Ruthenium(Ii) Complex Ru(NH3)5(Pyrazine)2+ in Oxygenated Solution, Photochem. Photobiol. Sci., 2007, 6(5), 515–518, 10.1039/b617350k
.
- H. Li, A. Jacque, F. Wang and R. W. Byrnes, Diffusion Distances of Known Iron Complexes in Model Systems, Free Radical Biol. Med., 1999, 26(1), 61–72, DOI:10.1016/S0891-5849(98)00166-X
.
- M. Saran and W. Bors, Signalling by O2−˙ and NO˙: How Far Can Either Radical, or Any Specific Reaction Product, Transmit a Message under in Vivo Conditions?, Chem.-Biol. Interact., 1994, 90(1), 35–45, DOI:10.1016/0009-2797(94)90109-0
.
- S. Galdiero, A. Falanga, M. Vitiello, M. Cantisani, V. Marra and M. Galdiero, Silver Nanoparticles as Potential Antiviral Agents, Molecules, 2011, 16(10), 8894–8918, DOI:10.3390/molecules16108894
.
- H. H. Lara, N. V. Ayala-Nuñez, L. Ixtepan-Turrent and C. Rodriguez-Padilla, Mode of Antiviral Action of Silver Nanoparticles against HIV-1, J. Nanobiotechnol., 2010, 8(1), 1, DOI:10.1186/1477-3155-8-1
.
- T. R. Hawthorne, R. Bürgi, H. Grossenbacher and J. Heim, Isolation and Characterization of Recombinant Annexin V Expressed in Saccharomyces Cerevisiae, J. Biotechnol., 1994, 36(2), 129–143, DOI:10.1016/0168-1656(94)90049-3
.
- H. Montón, C. Parolo, A. Aranda-Ramos, A. Merkoçi and C. Nogués, Annexin-V/Quantum Dot Probes for Multimodal Apoptosis Monitoring in Living Cells: Improving Bioanalysis Using Electrochemistry, Nanoscale, 2015, 7(9), 4097–4104, 10.1039/C4NR07191C
.
- F. K. Yoshimura, T. Wang and S. Nanua, Mink Cell Focus-Forming Murine Leukemia Virus Killing of Mink Cells Involves Apoptosis and Superinfection, J. Virol., 2001, 75(13), 6007–6015, DOI:10.1128/JVI.75.13.6007-6015.2001
.
- E. Hamard-Peron, F. Juillard, J. S. Saad, C. Roy, P. Roingeard, M. F. Summers, J.-L. Darlix, C. Picart and D. Muriaux, Targeting of Murine Leukemia Virus Gag to the Plasma Membrane Is Mediated by PI(4,5)P2/PS and a Polybasic Region in the Matrix, J. Virol., 2010, 84(1), 503–515, DOI:10.1128/JVI.01134-09
.
- H. Yin, L. Xu and N. A. Porter, Free Radical Lipid Peroxidation: Mechanisms and Analysis, Chem. Rev., 2011, 111(10), 5944–5972, DOI:10.1021/cr200084z
.
- I. O. L. Bacellar, M. C. Oliveira, L. S. Dantas, E. B. Costa, H. C. Junqueira, W. K. Martins, A. M. Durantini, G. Cosa, P. Di Mascio, M. Wainwright, R. Miotto, R. M. Cordeiro, S. Miyamoto and M. S. Baptista, Photosensitized Membrane Permeabilization Requires Contact-Dependent Reactions between Photosensitizer and Lipids, J. Am. Chem. Soc., 2018, 140(30), 9606–9615, DOI:10.1021/jacs.8b05014
.
- E. E. Farmer and M. J. Mueller, ROS-Mediated Lipid Peroxidation and RES-Activated Signaling, Annu. Rev. Plant Biol., 2013, 64(1), 429–450, DOI:10.1146/annurev-arplant-050312-120132
.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly Accurate Protein Structure Prediction with AlphaFold, Nature, 2021, 596(7873), 583–589, DOI:10.1038/s41586-021-03819-2
.
- M. Varadi, S. Anyango, M. Deshpande, S. Nair, C. Natassia, G. Yordanova, D. Yuan, O. Stroe, G. Wood, A. Laydon, A. Žídek, T. Green, K. Tunyasuvunakool, S. Petersen, J. Jumper, E. Clancy, R. Green, A. Vora, M. Lutfi, M. Figurnov, A. Cowie, N. Hobbs, P. Kohli, G. Kleywegt, E. Birney, D. Hassabis and S. Velankar, AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models, Nucleic Acids Res., 2022, 50(D1), D439–D444, DOI:10.1093/nar/gkab1061
.
- M. Varadi, D. Bertoni, P. Magana, U. Paramval, I. Pidruchna, M. Radhakrishnan, M. Tsenkov, S. Nair, M. Mirdita, J. Yeo, O. Kovalevskiy, K. Tunyasuvunakool, A. Laydon, A. Žídek, H. Tomlinson, D. Hariharan, J. Abrahamson, T. Green, J. Jumper, E. Birney, M. Steinegger, D. Hassabis and S. Velankar, AlphaFold Protein Structure Database in 2024: Providing Structure Coverage for over 214 Million Protein Sequences, Nucleic Acids Res., 2024, 52(D1), D368–D375, DOI:10.1093/nar/gkad1011
.
- N. J. Agard, J. M. Baskin, J. A. Prescher, A. Lo and C. R. Bertozzi, A Comparative Study of Bioorthogonal Reactions with Azides, ACS Chem. Biol., 2006, 1(10), 644–648, DOI:10.1021/cb6003228
.
- N. A. Danilkina, A. I. Govdi, A. F. Khlebnikov, A. O. Tikhomirov, V. V. Sharoyko, A. A. Shtyrov, M. N. Ryazantsev, S. Bräse and I. A. Balova, Heterocycloalkynes Fused to a Heterocyclic Core: Searching for an Island with Optimal Stability-Reactivity Balance, J. Am. Chem. Soc., 2021, 143(40), 16519–16537, DOI:10.1021/jacs.1c06041
.
- M. J. Matos, B. L. Oliveira, N. Martínez-Sáez, A. Guerreiro, P. M. S. D. Cal, J. Bertoldo, M. Maneiro, E. Perkins, J. Howard, M. J. Deery, J. M. Chalker, F. Corzana, G. Jiménez-Osés and G. J. L. Bernardes, Chemo- and Regioselective Lysine Modification on Native Proteins, J. Am. Chem. Soc., 2018, 140(11), 4004–4017, DOI:10.1021/jacs.7b12874
.
- L. M. Stabryla, K. A. Johnston, J. E. Millstone and L. M. Gilbertson, Emerging Investigator Series: It's Not All about the Ion: Support for Particle-Specific Contributions to Silver Nanoparticle Antimicrobial Activity, Environ. Sci.:Nano, 2018, 5(9), 2047–2068, 10.1039/C8EN00429C
.
- Z. A. Ratan, F. R. Mashrur, A. P. Chhoan, S. M. Shahriar, M. F. Haidere, N. J. Runa, S. Kim, D.-H. Kweon, H. Hosseinzadeh and J. Y. Cho, Silver Nanoparticles as Potential Antiviral Agents, Pharmaceutics, 2021, 13(12), 2034, DOI:10.3390/pharmaceutics13122034
.
- S. Galdiero, A. Falanga, M. Vitiello, M. Cantisani, V. Marra and M. Galdiero, Silver Nanoparticles as Potential Antiviral Agents, Molecules, 2011, 16(10), 8894–8918, DOI:10.3390/molecules16108894
.
- M. Rai, A. Yadav and A. Gade, Silver Nanoparticles as a New Generation of Antimicrobials, Biotechnol. Adv., 2009, 27(1), 76–83, DOI:10.1016/j.biotechadv.2008.09.002
.
- C. Szabó, A. L. Salzman and H. Ischiropoulos, Peroxynitrite-Mediated Oxidation of Dihydrorhodamine 123 Occurs in Early Stages of Endotoxic and Hemorrhagic Shock and Ischemia-Reperfusion Injury, FEBS Lett., 1995, 372(2–3), 229–232, DOI:10.1016/0014-5793(95)00984-H
.
- E. Yu, E. Ban, M. K. In and Y. S. Yoo, Development for the Analysis of Reactive Oxygen Species Using Capillary Electrophoresis with Laser-Induced Fluorescence Detection, J. Microcolumn Sep., 2001, 13(8), 327–331, DOI:10.1002/mcs.10014
.
- B. Liu, Z. Ma and K. Li, Sculpturing Effect of Sodium Thiosulfate in Shape Transformation of Silver Nanoparticles from Triangular Nanoprisms to Hexagonal Nanoplates, J. Nanosci. Nanotechnol., 2011, 11(6), 5001–5006, DOI:10.1166/jnn.2011.4115
.
- R. Yu. Bek and O. N. Shevtsova, Kinetics of Silver Anodic Dissolution in Thiosulfate Electrolytes, Russ. J. Electrochem., 2011, 47(3), 248–255, DOI:10.1134/S1023193511030037
.
- Z. Adamczyk, M. Oćwieja, H. Mrowiec, S. Walas and D. Lupa, Oxidative Dissolution of Silver Nanoparticles: A New Theoretical Approach, J. Colloid Interface Sci., 2016, 469, 355–364, DOI:10.1016/j.jcis.2015.12.051
.
- G. Baffou and R. Quidant, Thermo-plasmonics: Using Metallic Nanostructures as Nano-sources of Heat, Laser Photonics Rev., 2013, 7(2), 171–187, DOI:10.1002/lpor.201200003
.
- N. Chakraborty and B. C. Tripathy, Involvement of Singlet Oxygen in 5-Aminolevulinic Acid-Induced Photodynamic Damage of Cucumber (Cucumis Sativus L.) Chloroplasts 1, Plant Physiol., 1992, 98(1), 7–11, DOI:10.1104/pp.98.1.7
.
- L. Huang, T. G. St. Denis, Y. Xuan, Y.-Y. Huang, M. Tanaka, A. Zadlo, T. Sarna and M. R. Hamblin, Paradoxical Potentiation of Methylene Blue-Mediated Antimicrobial Photodynamic Inactivation by Sodium Azide: Role of Ambient Oxygen and Azide Radicals, Free Radical Biol. Med., 2012, 53(11), 2062–2071, DOI:10.1016/j.freeradbiomed.2012.09.006
.
| This journal is © The Royal Society of Chemistry 2026 |