 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Engineering delivery platforms for CRISPR-Cas and their applications in healthcare, agriculture and beyond
Nisha Bharti ab,
Unnati Modi
ab,
Unnati Modi bc,
Dhiraj Bhatia
bc,
Dhiraj Bhatia *b and
Raghu Solanki
*b and
Raghu Solanki *b
*b
aSchool of Life Sciences, Central University of Gujarat, Kundhela, Taluka-Dabhoi, Vadodara, Gujarat-391107, India
bDepartment of Biological Sciences and Engineering, Indian Institute of Technology Gandhinagar, Palaj, Gandhinagar, Gujarat-382355, India. E-mail: raghu.solanki@iitgn.ac.in; dhiraj.bhatia@iitgn.ac.in
cSchool of Biotechnology & Bioengineering, Institute of Advanced Research, Koba Institutional Area, Near GIFT City Bridge, Gandhinagar, Gujarat-382426, India
First published on 5th January 2026
Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems have transformed genome editing through unprecedented precision, and next-generation variants (base and prime editors) further enhance specificity by enabling targeted nucleotide changes without introducing double-strand DNA breaks. These technologies have unlocked broad applications in therapeutic gene correction, functional genomics, infectious disease management, diagnostics, agricultural engineering, environmental biotechnology, and synthetic biology. However, the targeted delivery of these systems remains a major challenge due to the large and chemically distinct nature of their components, including Cas protein or its base/prime editor fusions, guide RNA, and in some cases, DNA repair templates—which complicate packaging, stability, and cellular uptake. Additional hurdles arise from tissue and cell-type specificity, differential intracellular environments, variable editing efficiencies, and the persistent risk of off-target genome modifications. This review outlines the key challenges in the delivery of CRISPR technologies as well provides a comprehensive overview of both current and emerging delivery strategies, including viral vectors (adenovirus, adeno-associated virus, and lentivirus), non-viral physical approaches (microinjection, electroporation, ultrasound, and hydrodynamic tail-vein injection), and nanoparticle-based modalities (lipid and polymeric nanoparticles, gold nanoparticles, DNA nanostructures, and extracellular vesicles). We also discussed the diverse applications of CRISPR-Cas9 in gene therapy, immune cell engineering for cancer therapies, and agricultural innovation.
1 Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) systems were first identified as adaptive immune mechanisms in bacteria and archaea, providing sequence-specific defense against genetic elements such as bacteriophages, plasmids, and transposons.1–3 Early sequencing studies revealed that these systems capture fragments of invading DNA (known as spacers) and integrate them into CRISPR arrays (a repetitive loci interspersed with unique spacer sequences) that function as “heritable molecular memories” of past infections.4,5 Upon subsequent infections, the CRISPR array is transcribed to generate the CRISPR RNA (crRNA), which dictates target specificity, and the trans-activating CRISPR RNA (tracrRNA), which acts as a scaffold to recruit Cas9 nuclease. Together, the crRNA-tracrRNA duplex forms a ribonucleoprotein complex with Cas9 that scans for complementary foreign DNA, forms an RNA–DNA hybrid, and directs Cas9 to cleave the foreign DNA to eliminate the threat. This discovery established CRISPR-Cas as a programmable, nucleic-acid-guided defense system, in which DNA targeting can be reprogrammed simply by altering the guide RNA sequence.6,7 Because of its inherent modularity and the simplicity of its RNA-protein architecture, CRISPR-Cas9 rapidly emerged as a broadly applicable and powerful platform for precise genome manipulation.8,9Building on this natural defense mechanism, researchers further simplified the CRISPR-Cas9 system by engineering the crRNA and tracrRNA into a single guide RNA (sgRNA) that directs the Streptococcus pyogenes Cas9 nuclease to a chosen genomic locus. Target recognition requires both Watson–Crick complementarity between the sgRNA spacer, the target DNA strand (TS) and the presence of a protospacer adjacent motif (PAM) on the opposite non-target strand (NTS) adjacent to the target site. PAM binding initiates local DNA unwinding and R-loop formation, allowing the sgRNA to pair with the target strand and properly orient Cas9's catalytic domains for DNA cleavage (Fig. 1). The HNH domain cuts the target strand, and the RuvC domain cleaves the non-target strand, generating a site-specific double-strand break (DSB).6,7,10 Cellular repair pathways, primarily non-homologous end joining (NHEJ) or homology-directed repair (HDR), then process the DSBs to produce targeted gene disruption, correction, or insertion.11–15 Together with these mechanistic insights, the demonstration by Emmanuelle Charpentier and Jennifer Doudna of programmable genome editing, and its broad applications, ultimately led to them being awarded the 2020 Nobel Prize in Chemistry.16
Advances in protein and RNA engineering have dramatically expanded the CRISPR toolkit beyond canonical DSB-mediated genome editing. Structure-guided mutagenesis of Cas9's catalytic domains such as mutation of the RuvC nuclease domain (D10A) or the HNH domain (H840A) generates nickase Cas9 (nCas9), which introduces single-strand breaks, while the combination of both mutations (D10A/H840A) yields catalytically inactive Cas9 (dCas9).6,10 These nuclease domain engineered backbones have enabled a broad suite of precision CRISPR technologies developed by David Liu.17 Fusing nCas9 to cytosine or adenine deaminases generated base editors that install targeted C → T or A → G conversions without making double-strand breaks.18,19 Importantly, approximately 60 percent of known pathogenic human single-nucleotide variants fall within these transition classes, underscoring the therapeutic potential of base editing.20 Prime editing further extends this concept by coupling nCas9 to an engineered reverse transcriptase and by redesigning the guide RNA into a prime-editing guide RNA (pegRNA), which both directs Cas9 to the target site and provides the encoded repair template. Prime editing can install all 12 possible base substitutions, including transition, transversion mutations, small insertions, small deletions, and their combinations, making it a versatile genome editing suite.21 In parallel, dCas9 has been adapted for gene regulation by tethering transcriptional activators or repressors to create CRISPRa and CRISPRi systems that modulate endogenous gene expression.22–28 dCas9 can also be fused to fluorescent proteins or paired with fluorescently labeled guide RNAs to allow real-time imaging of chromatin organization, genome movement, and transcriptional activity in living cells.29–31 Collectively, these engineered derivatives position CRISPR-Cas technology as a modular and versatile platform for targeted sequence modification, epigenomic control, and high-resolution visualization of genome function.32,33
The development of CRISPR-Cas9 genome editing has revolutionized both basic and applied scientific research by enabling rapid analysis of previously inaccessible questions. Of all scientific disciplines, medical science has witnessed the swiftest translation of CRISPR technologies.34–37 For example, CASGEVY (exagamglogene autotemcel) is the first FDA-approved CRISPR/Cas9 therapy for severe sickle cell disease and transfusion-dependent β-thalassemia (Table 1). This treatment involves editing the erythroid enhancer region of the BCL11A gene in a patient's own hematopoietic stem cells, thereby reactivating fetal hemoglobin expression.38,39 Numerous other CRISPR-based therapies are currently in clinical development, including NTLA-2001 for transthyretin amyloidosis, EDIT-101 for Leber congenital amaurosis, CTX110 and CTX130 as allogeneic CAR-T cell therapies, VERVE-101 and VERVE-102 for familial hypercholesterolemia, BEAM-101 for sickle cell disease using base editing, and PM359 for inherited chronic granulomatous disease. These ongoing trials underscore the rapid progression of CRISPR-based technologies in a decade from bench to bedside (Table 1).40,41 CRISPR-based diagnostics such as SHERLOCK and DETECTR further extend the translational impact of CRISPR-technologies by enabling rapid, point-of-care detection of pathogens and genetic variants.42–45 These diagnostic technologies used the collateral cleavage activity of Cas enzymes (Cas12 or Cas13) to enable highly sensitive SARS-CoV-2 detection during the COVID-19 pandemic.46,47
| Clinical trial/product (modality) | Target gene(s)/locus | Therapeutic indication | Clinical trial phase | Clinical trial identifier |
|---|---|---|---|---|
| Exagamglogene autotemcel (exa-cel/CTX001/Casgevy) CRISPR-Cas9 (ex vivo) | BCL11A erythroid enhancer | Sickle cell disease β-thalassemia | Phase 2/3 completed; FDA/EMA approved | NCT03745287, NCT03655678, plus long-term follow-ups |
| NTLA-2001 CRISPR-Cas9 (in vivo) | TTR | Hereditary transthyretin amyloidosis | Phase 1; phase 3 ongoing | NCT04601051, NCT06128629 |
| EDIT-101 (AGN-151587) CRISPR-Cas9 (in vivo, subretinal) | CEP290 IVS26 mutation | Leber congenital amaurosis type 10 (LCA10) | Phase 1/2 | NCT03872479 |
| CTX110 CRISPR-Cas9 allogeneic CAR-T | TRAC, B2M knock-out; anti-CD19 CAR insertion | B-cell malignancies (e.g., LBCL) | Phase 1/2 | NCT04035434 |
| CTX130 CRISPR-Cas9 allogeneic CAR-T | CD70 CAR insertion (with edits in donor T cells) | T-cell lymphoma; clear-cell renal carcinoma | Phase 1/2 | NCT04438083, NCT04502446 |
| VERVE-101 in vivo adenine base editor (LNP) | PCSK9 | Familial hypercholesterolemia/ASCVD | Phase 1b | NCT05398029 |
| VERVE-102 in vivo base editor (GalNAc-LNP) | PCSK9 | Familial hypercholesterolemia; premature CAD | Phase 1b | NCT06164730 |
| BEAM-101 ex vivo adenine base-edited autologous HSPCs | HBG1/HBG2 promoter (to derepress HbF) | Sickle cell disease | Phase 1/2 | NCT05456880 |
| PM359 (Prime-0101) ex vivo prime editing | NCF1 (p47phox) | Chronic granulomatous disease (p47phox CGD) | Phase 1/2 | NCT06559176 |
Beyond therapeutics, CRISPR technologies help accelerating the development of disease-resistant, climate-resilient, and high-yield crops.48 For example, editing the wheat susceptibility gene in Mildew resistance locus O (MLO-B1) with a 304 kb deletion produced powdery mildew-resistant wheat plants with the same yield as wild type.49 In livestock science, genome editing is enabling precise enhancement of commercially and biologically important traits including the animals with disease resistant traits.50 For example, editing the CD163 receptor in pigs or CD46 receptors in cows produced animals resistant to porcine reproductive and respiratory syndrome (PRRS) and bovine viral diarrhea virus (BVDV), respectively.51,52 Environmental biotechnology is also rapidly advancing, with CRISPR being applied to engineer microbial systems for improved bioremediation and sustainable bio-manufacturing.53,54 CRISPR-based gene driven strategies are under evaluation for population control and disease-vector suppression in regions burdened by insect-transmitted pathogens such as malaria and Lyme disease.55–58
In this review, we discuss key challenges associated with the delivery of CRISPR technologies and illustrate how advances in viral, non-viral, and physical delivery strategies are addressing these limitations to improve the safety and efficiency of genome editing. Finally, we will highlight the use of delivery technologies in translational applications of CRISPR-based systems across gene therapy, agriculture, and other emerging areas (Fig. 2).
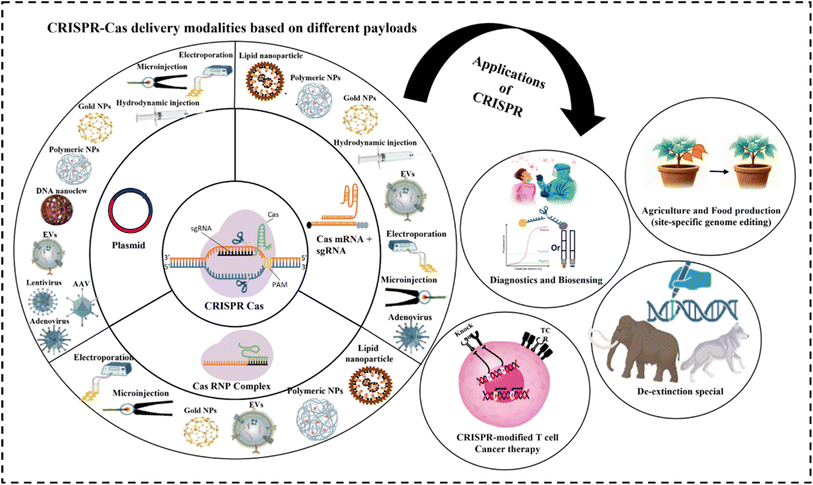 | ||
| Fig. 2 Schematic illustration of delivery platforms for CRISPR-Cas and their applications in healthcare and agriculture. | ||
2 Different modalities for CRISPR-Cas9 delivery
Despite its transformative impact, conventional CRISPR-Cas systems face several fundamental limitations that restrict their broad translational applicability. A major constraint lies in the intrinsic size and structural complexity of CRISPR components. The widely used SpCas9 is a ∼160 kDa nuclease (∼10 nm), and its associated gRNAs add an additional ∼3–5 nm; together these components far exceed the permeability limits of cellular membranes, which typically permit diffusion of small molecules such as glucose (∼1 nm).59 This size mismatch underscores the difficulty of achieving efficient tissue and cell-specific delivery, particularly in vivo, where physiological barriers, immune clearance and extracellular matrix density further impede transport. Moreover, CRISPR knockout (CRISPRko) strategies rely on endogenous DNA repair pathways to resolve Cas9 induced DSBs, often suffer from variable cutting efficiencies and produce heterogeneous populations of cells.60 DSBs can also trigger genotoxic stress, cytotoxicity or apoptosis.61,62 These issues can amplify in cancer cells where excessive DSB formation may generate false-positive phenotypes or selective cell loss. In parallel, off target editing, unintended chromosomal rearrangements and immunogenicity of bacterial Cas proteins remain major safety concerns for clinical translation.63,64Conventional methods to regulate Cas9 activity, including systems that modulate nuclease half-life, are often slow, inefficient and reliant on large (>230 amino acids) regulatory modules or expensive chemical inducers with poor pharmacological profiles, limiting their suitability for in vivo use.65 These biological constraints are compounded by persistent delivery challenges: the field still lacks universal, safe, and efficient vectors capable of achieving precise spatiotemporal control, minimizing immune responses and providing mechanisms such as kill switches for reversible or self-limiting genome editing.
Although viral vectors, non-viral nanocarriers and physical delivery strategies each offer partial solutions (Fig. 3), they also introduce trade-offs in cargo capacity, tissue tropism, toxicity and manufacturability.66–68 For example, adeno-associated virus (AAV) vectors remain among the most reliable systems for in vivo delivery of base editors; however, their prolonged transgene expression increases the risks of off-target editing and vector integration, raising concerns about genotoxicity and carcinogenesis. These limitations highlight the need for next-generation CRISPR platforms that emphasize precision, safety and controllability to fully realize the therapeutic potential of genome editing. For example, the challenges associated with AAV have driven the development of alternative strategies such as direct delivery of base editor ribonucleoproteins (RNPs), which exhibit shorter intracellular halftimes and reduced off-target activity and emerging virus-like particle platforms that can deliver RNPs without introducing a viral genetic material.69 In the following sections, we discuss different delivery modalities explored for CRISPR-based genome engineering with their advantages, disadvantages and case studies.
2.1 Viral delivery systems
Recombinant viral vectors engineered to deliver therapeutic genes to diseased tissues by harnessing the natural ability of many viruses to introduce foreign genetic material into host cells.70 Viral vectors have been in development for over 30 years to deliver genetic treatments like various nucleic acid-based therapeutics; more than 1000 viral gene therapy clinical trials have been conducted and some therapies have even been approved for clinical use. While there are concerns about their safety and the risk of causing unwanted mutations, viral vectors remain one of the most effective methods for getting plasmid-based genetic material into mammalian cells, both in the lab and in living organisms. Because of this, they have become a popular tool for delivering CRISPR-Cas9 gene editing technology into mammalian cells.68 The most used viral vectors are adeno-associated virus (AAV), lentivirus, and adenovirus71 (Table 2).| Vector type | Package limitation | Superiority | Advantages | Disadvantages | Application | References |
|---|---|---|---|---|---|---|
| Adeno-associated virus (AAV) | 4.5 kb | Low immunogenicity, stable transgene expression | High infection efficiency, broad cell tropism | Low packaging capacity, difficulty in production | Treatment of lipoprotein lipase deficiency patients, edit the VEGFR2 gene in retinal endothelial cells within a mouse model, single-cell CRISPR screening in mouse brain | 72–74 |
| Lentivirus (LV) | 8 kb | Large packaging capacity, low cell cycle tendency | High infection efficiency, large packing size, long-term gene expression, persistent gene transfer | Long lasting expression of Cas9, potential for insertional mutagenesis | Epigenome editing targets cytokines (TNF-α, IL-1β) in the intervertebral disc to treat degenerative disc disease and chronic low-back pain, gene editing tool delivery with a gag-only strategy, vaccine development | 75,76 |
| Adenovirus (AV) | >8 kb | Large packaging capacity, no integration to the host genome | Risk of off-target effects, strong immunogenicity, limited cargo capacity for gene constructs, limited reusability, high delivery efficiency | High immunogenicity, difficult scale production | Induces targeted mutations in a wide range of human cells, selective targeting of vascular endothelial cells in vivo, cancer treatment, gene therapy | 77,78 |
To overcome this, AAV have evolved strategies like overlapping open reading frames (ORFs) to increase coding capacity without expanding genome length.82 The European Medicines Agency (EMA) approved Glybera (alipogene tiparvovec) as a first AAV-based gene therapy drug in the European Union in 2012. Glybera is an adeno-associated viral vector engineered to express lipoprotein lipase gene into the muscle for the treatment of lipoprotein lipase deficiency patients.83 Despite its advantages, in vivo genome editing still faces challenges related to efficiency and safety. A targeted approach involves using the endothelial-specific intercellular adhesion molecule 2 (pICAM2) promoter, which drives expression of SpCas9, ensuring targeted expression of the Cas9 enzyme within vascular endothelial cells that are efficiently transduced by AAV1; this system is effective effectively used to edit the VEGFR2 gene in retinal endothelial cells in a mouse model of oxygen-induced retinopathy, showcasing its potential for targeted vascular genome editing.84
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 genes and includes over 64
080 genes and includes over 64![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 unique guide sequences, allows for both positive and negative selection screening in human cells and improving whole-genome screens.86 The feasibility of lentiviral CRISPR epigenome editing as a targeted gene therapy approach to reduce harmful inflammation, particularly by cytokines such as TNF-α and IL-1β within the intervertebral disc (IVD), which drive the Degenerative disc disease (DDD), the leading cause of chronic low-back pain and disability, has been demonstrated. While TNFR1 suppression proved effective (reducing NF-κB activity, cell death, and pro-inflammatory gene expression following cytokine challenge), IL1R1 targeting presents challenges due to feedback mechanisms, as some patient samples showed rebound expression driven by IL-1β. Additionally, RNA sequencing identified two key transcription factors, IRF1 and TFAP2C, as central regulators of inflammatory signaling in IVD cells. Still, this method opens the door for novel therapies that address the root inflammatory drivers of DDD.87
000 unique guide sequences, allows for both positive and negative selection screening in human cells and improving whole-genome screens.86 The feasibility of lentiviral CRISPR epigenome editing as a targeted gene therapy approach to reduce harmful inflammation, particularly by cytokines such as TNF-α and IL-1β within the intervertebral disc (IVD), which drive the Degenerative disc disease (DDD), the leading cause of chronic low-back pain and disability, has been demonstrated. While TNFR1 suppression proved effective (reducing NF-κB activity, cell death, and pro-inflammatory gene expression following cytokine challenge), IL1R1 targeting presents challenges due to feedback mechanisms, as some patient samples showed rebound expression driven by IL-1β. Additionally, RNA sequencing identified two key transcription factors, IRF1 and TFAP2C, as central regulators of inflammatory signaling in IVD cells. Still, this method opens the door for novel therapies that address the root inflammatory drivers of DDD.872.2 Non-viral and physical delivery systems
Viral delivery systems with high issues related to safety and the risk of causing unwanted mutation there is need to use physical delivery system91,92 (e.g. microinjection, ultrasound, hydrodynamic tail-vein injection and electroporation) and non-viral delivery system93–95 (e.g. nanoparticles) are relatively simple to carry out but are primarily suited for in vitro applications (Table 3). Nanotechnology refers to the engineer nanomaterials at the nanoscale, generally within the range of 1–100 nm such as carbon nanotubes, gold nanoparticles and quantum dots.96 In biology and medicine, nanoparticles can be used for drug delivery, biosensing, diagnostic imaging and even gene editing. Their unique properties of nanoparticles, such as their size and reactivity, make them ideal candidates to interact with biological systems of living cells in ways that allow for visualizing, high-resolution imaging and monitoring of cellular processes at the molecular level, and their surface properties can be engineered to target specific cells or tissues. It provides a promising solution to challenges in sensitivity, resolution, real-time tracking and efficiency of delivery. The intersection of CRISPR-Cas9 technology and nanotechnology particularly advanced nanomaterials allows for the engineering of nanoscale materials that enable real-time tracking and high-resolution imaging of the CRISPR-Cas9 gene-editing process. This interdisciplinary approach promises breakthroughs in molecular biology, agriculture, precision medicine, and therapeutic development.97,98| Delivery system | Advantages | Disadvantages | Application | References |
|---|---|---|---|---|
| Microinjection | Dosage controllable, direct delivery, highly specific, suitable for all strategies of CRISPR-Cas9 | Labor-intensive, low throughput, potential for cell damage, time consuming, in vitro only | Animal model generation, plant genome editing, precision editing in oocytes and zygotes | 99 |
| Ultrasound | Non-invasive, potential for targeted and controlled delivery | Limited tissue penetration, potential for tissue damage, requires specialized equipment | Potential for non-invasive gene therapy, cancer treatment, stem cell engineering with minimal damage | 100 and 101 |
| Hydrodynamic tail-vein injection (HTVI) | Easy to operate, low price | Traumatic to tissue, low specificity, can cause hepatocyte membrane disruption and cellular stress, reducing gene delivery efficiency | In vitro and in vivo gene editing efficiency, efficient liver-targeted gene delivery using lentiviral vectors through HTVI with reduced off-target distribution to other organs | 102 and 103 |
| Induced transduction by osmocytosis and propanebetaine (iTOP) | Reliable method for delivery of cas9 protein and sgRNA, virus-free and non-integrative | Unsuitable for in vivo applications, high salt concentrations required | Primarily utilized in ex vivo gene editing applications, highly efficient transduction of native proteins, efficient manipulation of a wide variety of primary cell types | 104 |
| Electroporation | Easy to operate, suitable for any cell type, high transfection efficiency, suitable for all strategies of CRISPR-Cas9 and can be used in vitro and in vivo | Potential for cell damage, affects cell viability, requires specialized equipment, in vitro only and nonspecific transfection | Ex vivo and in vivo delivery (e.g., cancer immunotherapy) | 105 and 106 |
A novel method called Easy Electroporation of Zygotes (EEZ) offers a highly efficient way to generate CRISPR/Cas9 transgenic mice while also reducing the number of animals needed.127 This approach provides a faster and less invasive alternative to traditional pronuclear injections for genome editing in mice. EEZ is particularly well-suited for introducing targeted mutations in C57BL/6 mice by electroporating intact zygotes using a standard electroporator, synthetic CRISPR/Cas9 components, and minimal technical complexity. Unlike earlier protocols that require specialized electroporation equipment or harsh pre-treatment of zygotes often at the expense of embryo viability EEZ minimizes physical damage, resulting in improved embryo development. However, genome editing success has been reported in up to 100% of live-born offspring using this method, making it a practical and accessible tool for researchers.128 Mutant mice can be generated in a single step by directly delivering CRISPR/Cas9 genome editing components into mouse zygotes via electroporation. This approach helps to overcome the limitations of traditional low-throughput methods and enables the efficient recovery of live mice carrying targeted mutations through both nonhomologous end joining (NHEJ) and homology-directed repair (HDR).129
Nucleofection-based CRISPR/Cas9 genetic engineering has been proven as an effective and rapid method for creating mutant CD8+ T cells from both naïve and in vitro-activated primary mouse cells. This technique allows for efficient genetic modification without significantly compromising the cells' in vivo functionality. Experimental testing of their immune responses revealed that nucleofected naïve CD8+ T cells retained antiviral immune activity comparable to that of non-nucleofected cells. While nucleofection of in vitro-activated CD8+ T cells resulted in slightly reduced expansion and survival shortly after adoptive transfer, it also led to a more noticeable contraction phase over time.130 Researchers have showed that in vivo electroporation of morpholino oligonucleotides targeting fgfr1 and msxb proteins offers a practical and effective method for protein knockdown during zebrafish fin regeneration (a widely used model organism for studying tissue regrowth). This technique led to reduced fin outgrowth, with Fgfr1 knockdown mimicking the effects of a known Fgfr1 inhibitor. Additionally, the approach provided direct evidence of msxb's functional role in caudal fin regeneration. Interestingly, while knocking down Fgfr1 disrupted the expression of msxc in the regenerative blastema, msxb knockdown did not indicate that this method can also be used to investigate genetic relationships and epistasis within regeneration pathways. Thus, this convenient reverse genetic tool enables researchers to rapidly assess the role of genes expressed during fin regeneration, screen candidate genes for functional importance and map genes to the molecular pathways that drive regeneration.131 (Table 4). represents the non-viral delivery systems.
| Delivery system | Advantages | Disadvantages | Application | References |
|---|---|---|---|---|
| Lipid nanoparticles (LNPs) | Easy to prepare, low cost, high transfection efficiency, biocompatible, biodegradable, safe and suitable for all strategies of CRISPR-Cas9 | Potential immunogenicity, off-target effects, possible degradation in biological fluid before reaching target, low delivery efficiency, specific cell tropism | In vitro and in vivo gene therapy, (e.g., lung and liver editing, epigenetic editing into solid tumors, cancer therapy, potential for neurodegenerative disease treatment) | 132 and 133 |
| Polymer nanoparticles (PNPs) | Easy to prepare, high stability, controlled release, biocompatible, safe and suitable for all strategies of CRISPR-Cas9 | Lower transfection efficiency, some PNPs may induce cytotoxicity, low delivery efficiency | In vitro and ex vivo stem cell engineering, controlled release editing for prolong gene expression, chronic disease treatment, (e.g., genome-editing for CNS disease, etc) | 134 and 135 |
| Gold nanoparticles (AuNPs) | High delivery efficiency, easy surface functionalization, efficient delivery of CRISPR RNPs, mainly lower cytotoxicity | Limited capacity for delivery large gene-editing payloads, expensive production, potential long-term accumulation and toxicity concerns in vivo at high concentration | Potential for non-invasive CRISPR delivery to brain, cellular imaging and tracking and minimal off-target editing, precise gene editing in cancer therapy, (e.g., breast cancer) | 136 |
| DNA nanostructure | Stable assembly, multiple editing, well histocompatibility, biocompatible, controllable size and architecture | Assembly is complicated, stimuli responsive release, requires modification of template DNA | In vivo as well as in vitro (e.g., cancer treatment, gene therapy, etc) | 137 |
| Extracellular vesicle (EVs) | Biocompatible, low immunogenicity, efficient cellular uptake, target delivery, high delivery capacity, cross biological barriers | Limited loading capacity, potential off-target effects, unpredictable biodistribution as it may accumulate in different organs | Gene therapy, neurological disease treatment, cancer therapy, immune cell engineering, stem cell and regenerative medicine (e.g., gene editing livestock gametes and embryos, cardiac repair) | 138 and 139 |
Researchers have combined two powerful tools: CRISPR/Cas9 gene editing and a targeted nanoparticle-based delivery system, supported by focused ultrasound (FUS) technology to construct lipid-polymer hybrid nanoparticles for delivery of the CRISPR/Cas9 system.142 In combination with focused ultrasound-microbubbles (FUS-MBs), this platform enables the opening of the blood–brain barrier (BBB) and knock down of MGMT (a gene known to drive resistance to temozolomide drug). The MBs-LPHNs-cRGD delivery system holds promise as an effective alternative method for targeted gene therapy in the treatment of glioblastoma (GBM) which is one of the toughest challenges in oncology. It has the potential to treat a wide range of genetic disorders, including cancers, but delivering it safely and effectively, especially within the central nervous system (CNS), remains a significant challenge due to the lack of reliability in vivo targeting systems.
When the nanoclew DNA sequence shared partial complementarity with the sgRNA guide sequence it shows the highest gene editing efficiency, suggesting a useful design principle for optimizing delivery. This method offers a flexible and efficient platform not only for CRISPR-Cas9 but also for transporting other DNA-interacting proteins or functional nucleic acids in a similar fashion.155 For example, Francesca Miceli et al. developed MAIGRET (Molecular Assay based on antibody-Induced Guide-RNA Enzymatic Transcription), a two-step CRISPR-powered immunoassay to detect specific antibodies and antigens with high sensitivity using cell-free RNA transcription. In this process target antibody binds and triggers the transcription of a guide RNA, which activates Cas12a to cleave an optically labelled hairpin DNA reporter (fluorescent reporter), which leads to a measurable fluorescence signal. It can detect even complex samples like antibodies in the low picomolar range and can support antigen detection using a competitive format. Though slightly less sensitive than other methods (like-UCAD), it offers better signal control and broader adaptability with potential applications in diagnostics and could even be used in future gene-based therapies.156
3 Application of CRISPR-Cas9 technology across different areas
3.1 CRISPR-based diagnostics
In the process of CRISPR based diagnosis the guide RNA (gRNA or sgRNA) finds a complementary target sequence in DNA or RNA, then Cas protein cleaves the target recognized by guide RNA and ensures initial detection known as cis-cleavage. After binding the target in cis, many Cas enzymes become activated and begin cutting other nearby non-target nucleic acids indiscriminately, known as trans-cleavage, which is used as a signal amplification tool in which the trans-cleavage behavior can be directed toward specially designed reporter molecules—usually short single-stranded DNA or RNA tagged with a fluorophore and a quencher. When the trans-cleavage happens, the fluorophore is separated from the quencher, leading to a visible fluorescent signal. This engineered system allows researchers to toggle between target-specific (cis) cleavage and non-specific (trans) cleavage activity, by adjusting factors like guide RNA design, Cas protein variants, and reaction conditions which helps scientist to optimize the sensitivity, specificity, and timing of the signal generation. This activity is very important to avoid false positives, enhance signal clarity, and ensure accurate detection even at very low concentrations of the target DNA or RNA.The CRISPR-Cas system has recently gained popularity as a diagnostic tool in various clinical and point-of-care applications due to its ability to specifically target genes and cleave surrounding single-stranded DNA for precise and rapid nucleic acid detection.162,163 Emerging studies suggest that CRISPR-based systems can also be adapted to detect a broader range of targets, including proteins, antibiotics, metal ions, and amino acid metabolites. However, CRISPR-based detection is currently slightly less sensitive than the widely used RT-PCR test.
For example, scientists reported that CRISPR-Cas-based diagnostics offers a safe, non-invasive way to detect conditions like Trisomy 13, 18, and 21 using the cell-free fetal DNA (cffDNA) just extracted from the maternal blood sample during the early gestation period using the kit method. In this process fetal DNA is gently extracted and amplified with fluorescent primers and targeted by specific guide RNAs and Cas enzymes, then detection of 3 targets will be favored by adding reporters for Cas12f, Cas12b, and Cas12a in HEX (hexachlorofluorescein), TEX (TEX615), and FAM channels to help with early detection of any chromosomal issues with high sensitivity and specificity. Then, a clear fluorescent signal generated from the DNA-Cas-sgRNA complex confirms the presence of trisomy. It allows quick, affordable testing and avoids the risks of traditional procedures like amniocentesis.164
A rapid diagnosis of SARS-CoV-2 is essential to enable timely treatment for COVID-19; reverse transcription-polymerase chain reaction (RT-qPCR) assays were developed to detect the novel coronavirus within 4–6 hours and are considered the gold standard for identifying viral nucleic acids due to their high sensitivity and specificity.165 However, these assays require expensive equipment and trained personnel, hindering their accessibility in resource-constrained settings. On the other hand, serology tests are faster and require minimal equipment but are less effective for diagnosing acute SARS-CoV-2 infections, as antibody responses can take several days to weeks to develop. To address the need for instrument-free nucleic acid detection, various isothermal amplification methods—such as Recombinase Polymerase Amplification (RPA) and Loop-mediated Isothermal Amplification (LAMP)—have been developed. These techniques are simpler, faster, and more cost-effective than conventional PCR assays.166 Alternative approaches based on combinations of RPA, LAMP and CRISPR-mediated detection have been developed. These methods enable instrument-free, sensitive, precise and rapid point-of-care testing (POC) for COVID-19.167 In this process, patient samples (usually nasal or throat swabs) are collected and processed to extract viral RNA, which is then amplified using isothermal techniques like RT-LAMP or RPA. CRISPR enzymes (e.g., Cas12 or Cas13), guided by virus-specific RNA sequences, bind the amplified target and activate trans-cleavage, cutting reporter molecules to produce a detectable fluorescent or colorimetric signal.
 | ||
| Fig. 4 CRISPR-based diagnosis system: (a) detection using the HOLMESv2 technique, figure adapted from ref. 172. Copyright© 2020, American Chemical Society. (b) dCas9-based electrochemical detection using a handheld device, figure adapted from ref. 173. Copyright© 2019, Reza Hajian et al. (c) detection using the CASLFA technique, figure adapted from ref. 174. Copyright© 2020, American Chemical Society. (d) Detection using the AIOD-CRISPR technique, figure adapted from ref. 47. Copyright© 2020, James P. Broughton et al. | ||
3.2 CRISPR-based real-time tracking and visualization
Real-time tracking is used to monitor the chromosome dynamics during genome editing. Traditional methods for real-time monitoring have several limitations, including low temporal resolution, destructive sampling techniques, limited sensitivity, and inadequate spatial resolution. Real-time visualization provides a deeper understanding of CRISPR-Cas9 interactions with DNA, its editing efficiency, and potential off-target effects, all of which are crucial for optimizing CRISPR-based genome editing and ensuring its safety in therapeutic applications. Consequently, real-time tracking of CRISPR-mediated genome editing has emerged as a key research goal. In today's world, various CRISPR-based bioimaging techniques are available.175Visualization of chromosome dynamics is crucial for understanding various fundamental intra-nuclear processes. For example, Shaho et al. developed an innovative method utilizing a modified single-guide RNA (sgRNA) within the CRISPR/Cas9 system to enable dual-color chromosomal locus imaging.176 This approach involves structure-guided engineering of the sgRNA by inserting RNA aptamers that specifically bind to fluorescent protein-tagged effectors, thereby optimizing the imaging strategy. Moreover, studies have demonstrated that this sgRNA-based labeling technique exhibits significantly greater resistance to photobleaching compared to Cas9-based labeling, due to the rapid exchange dynamics of the RNA aptamer-binding effectors.
Another method was established by Yang J. et al., employing aptamer-modified DNA origami as a delivery vehicle to dynamically track target loci in a human breast cancer cell line (MCF-7).177 The ribonucleoprotein (RNP) complex, consisting of dCas9-EGFP and cyanine-5 (Cy5)-labelled gRNA, is efficiently encapsulated, protected, and delivered by a barrel-shaped, hollow DNA origami nanostructure known as TUBE. Following a 6 hour incubation with the nanostructures, high-resolution multi-color and labelling of target genes in living cells is achieved without affecting cell viability, proliferation, or chromosome dynamics, demonstrating the rapid and biocompatible nature of this DNA origami-based delivery method.
Furthermore, to investigate the spatiotemporal dynamics and functional role of non-repetitive genes in living cells, Chen B et al. developed a CRISPR-Tag system that utilizes 1 to 4 highly active sgRNAs to specifically label protein-coding genes with a high signal-to-noise ratio for real-time imaging via wide-field fluorescence microscopy.178 Incorporating 2–6 CRISPR-targetable DNA sequence repeats from Caenorhabditis elegans into human genes helps to efficiently label endogenous genes like HIST2H2BE, LMNA, and HSPA8 with fluorescent dCas9. Robust imaging of repetitive sequences in telomeres and coding genes in living cells has also been demonstrated using a structurally optimized single-guide RNA (sgRNA) in combination with an EGFP-tagged, endonuclease-deficient Cas9 (dCas9) protein.
Sun et al. developed a CRISPR-Sunspot imaging system based on SunTag technology to enhance signals from single-molecule mRNA in living cells.179 Using CRISPR-Sunspot, the co-localization of Camk2a mRNA with the regulatory protein Xlr3b in neurons was successfully monitored (Fig. 5A). Liu H. et al. assembled a light-activated CRISPR-Cas12a (LAC12a) system for enhanced detection of miRNA-21 and its imaging in living cells (Fig. 5B).180 This study is significant as it introduces a photoactivated CRISPR-Cas12a platform for improved miRNA detection, leveraging spatiotemporal imaging capabilities that enable single-cell resolution analysis of miRNA expression.
 | ||
| Fig. 5 CRISPR-based bioimaging strategies. (A) Schematic diagram of the SunTag-mediated single-molecule RNA imaging method (CRISPR-Sunspot), figure adapted from ref. 179 Copyright© 2020, Sun et al. (B) diagram illustrating how a response module linked to the CRISPR-Cas12a (LAC12a) system can be light-activated to release cellular protection and enhance miRNA detection, figure adapted from ref. 180 Copyright© 2024, the American Association for the Advancement of Science. (C) Diagram of an MCM-CRISPR/Cas12a system for sensitive and specific imaging of multiple microRNAs within cells, figure adapted from ref. 179 Copyright© 2024, Chen X. et al. (D) schematic illustration of the construction and application of CRISPR nanorobots consisting of an AuNP functionalized with molecular constructs to enable CRISPR activity, figure adapted from ref. 181 Copyright© 2024 American Chemical Society. | ||
Furthermore, Chen X. et al. developed a mini crRNA-mediated CRISPR/Cas12a (MCM-CRISPR/Cas12a) system for sensitive and targeted imaging of miRNA-21, miRNA-155, and miRNA-10b both inside and outside cells (Fig. 5C).182 Their results showed that molecular beacons (MBs) in the MCM-CRISPR/Cas12a system produced significantly higher fluorescence intensity compared to MBs opened directly by the target microRNAs. Yuan A. et al. designed a study to overcome the limitations of traditional methods that require supplying high concentrations of substrates into cells.181 In their approach, high local concentrations of both enzyme and substrate were achieved within a nanorobot, in which crRNA-Cas12a RNP complexes hybridize with thousands of anchor strands and hundreds of substrate strands conjugated to an AuNP scaffold, forming the functional nanorobot (Fig. 5D).
3.3 Medical healthcare (cancer therapy and gene editing)
Gene therapy represents a promising therapeutic approach that involves the modification or replacement of defective genes to correct genetic mutations, thereby offering the potential for curative treatment or prevention for various human genetic disorders caused by gene mutations including point mutations, gene defects or abnormal gene expression.183 This approach holds therapeutic potential for conditions such as cancers (e.g., tumors), monogenic diseases (e.g., sickle cell disease, β-thalassemia), hematological malignancies like leukemia, and viral infections (e.g., hepatitis B virus-induced hepatocellular carcinoma, human papillomavirus-associated cervical cancer, and HIV-induced acquired immunodeficiency).184 Unlike conventional treatment modalities, gene therapy represents a transformative paradigm that targets the underlying genetic etiology, offering the possibility of long-term or permanent cures. The CRISPR-Cas9 genome editing system has emerged as a transformative tool in this domain, owing to its precision, efficiency, and programmability. It has been extensively employed in both preclinical and clinical investigations for therapeutic gene correction.Conventional cancer treatments such as surgery, radiation and chemotherapy can slow down cancer progression and extend patient survival. However, these methods often come with the risk of the cancer coming back or the body becoming resistant to the drugs, that can result in a poor outlook for patients.185 Moreover, the lack of precision in chemotherapy and radiation therapy can cause severe side effects, and can sometimes even be life-threatening. Recently, the US Food and Drug Administration (FDA) approved CAR-T cell therapy for cancer treatment.186 CAR-T cell therapy is a form of immunotherapy that involves modification of T cells (engineered T cells) to express chimeric antigen receptors, known as CAR T cells.187 CARs are specially engineered proteins that help T cells recognize and attack cancer. They are created by linking single-chain variable fragments to transmembrane and intracellular signaling regions, often including one or two co-stimulatory domains. These recombinant antigen receptors can reprogram the specificity and function of T lymphocytes, are often enhanced with additional support signals, and enable a stronger, more targeted anti-tumor response.188
However, there is a continued strong need for innovative cancer therapies, and CRISPR/Cas9 technology holds the potential to significantly transform cancer treatment and could be a game-changer in the way the disease is treated.189 For example, Stadtmauer et al. reported a first-in-human phase 1 clinical trial (clinicaltrials.gov (http://clinicaltrials.gov); trial NCT03399448) where three genes (TRAC, TRBC, and PDCD1) of T cells extracted from the body of three patients, two of them with advanced treatment-resistant myeloma and one with metastatic sarcoma, were targeted and disrupted by the multiplex CRISPR-Cas9 gene editing tool to boost the cells' ability to fight tumors.190 Here, the edits removed endogenous TCRs and PD-1 to enhance T cell function and persistence, while reducing immune exhaustion, and a cancer-specific TCR transgene (NY-ESO-1) was added to redirect T cell specificity toward tumor antigens. Then modified T cells were safely infused back into the patient's body and showed stable engraftment for ≥9 months. As a result, all patients tolerated the therapy well, with no serious side effects linked to the gene edits. The TCR α chain gene (TRAC) showed the most effective editing, and reductions in NY-ESO-1/LAGE-1 antigens were observed in myeloma patients. While travelling to tumor sites, the cells showed a drop in key cancer markers. Off-target effects were rare, and chromosomal translocations occurred in vitro but declined post-infusion. The study confirms that CRISPR-Cas9 editing is clinically feasible with potential for advancing cancer immunotherapy.
Furthermore, CRISPR-Cas9 facilitates the generation of genetically engineered animal models, enabling in-depth studies of disease mechanisms and the development of targeted interventions.191 For example, Oura S. et al. reported an engineered variant of the Streptococcus pyogenes Cas9 nuclease, termed SpCas9-NG, which recognizes a relaxed NGN protospacer adjacent motif (PAM), thereby significantly expanding the range of targetable genomic loci. In their study, they demonstrated the potential of SpCas9-NG-mediated genome editing to correct the pathologically expanded CAG repeat tract characteristic of Huntington's disease (HD). By specifically targeting the boundary of the CAG repeat expansion in embryonic stem (ES) cells derived from an HD mouse model, SpCas9-NG facilitated precise genome editing, leading to a precise contraction of the aberrant repeat region.192
Morever, Wang Y. et al. reported a stimulus-responsive silica nanoparticle (SNP, ∼50 nm) capable of encapsulating biomacromolecules with high loading efficiency and releasing cargo in response to glutathione (GSH) via disulfide crosslinking (Fig. 6A).193 These SNPs demonstrated efficient and biocompatible delivery of nucleic acids (DNA, mRNA) and CRISPR genome editors (Cas9/sgRNA RNPs), enabling effective genome editing. In vivo studies further showed successful mRNA and RNP delivery to murine retinal pigment epithelium (RPE) and liver cells, achieving targeted genome editing. In another study, nanoparticles containing a protein-based CRISPR/dCas9 system targeting Sirt1 were prepared, and their intranasal delivery was evaluated in a mouse model of ischemic stroke (Fig. 6B).194 Intranasal administration of this system (dCas9/CaP/PEI-PEG-bHb nanoparticles) improved survival following permanent middle cerebral artery occlusion and reduced cerebral edema via the upregulation of target gene Sirt1 in the brain.
 | ||
| Fig. 6 Applications of CRISPR-based technologies in gene editing. (A) Schematic illustration of a stimulus-responsive silica nanoparticle (SNP) for intracellular trafficking of CRISPR genome editors (e.g., RNP and RNP + ssODN) and nucleic acids (e.g., DNA and mRNA), figure adapted with permission from ref. 193 Copyright© 2021 Elsevier. (B) Schematic representation of dCas9/CaP/PEI-PEG-bHb administered intranasally for the treatment of ischemic stroke, figure adapted from ref. 194 Copyright© 2024 Jee-Yeon Ryu et al. (C) schematic diagram of NIR-sensitive and reducing agent-sensitive nitrilotriacetic acid-decorated micellar nanoparticles encapsulated with anticancer photosensitizer Ce6 and Nrf2-targeting Cas9/sgRNA for application in photodynamic therapy, figure adapted from ref. 195 Copyright© 2020 Deng S. et al. American Association for the Advancement of Science. | ||
Deng S. et al. designed near-infrared (NIR)- and reducing agent-responsive nanoparticles and achieved controlled release of CRISPR-Cas9 RNPs along with the anticancer photosensitizer chlorin e6 (Ce6) in a mouse animal model (Fig. 6C).195 They validated the synergistic effects of Nrf2 gene editing and Ce6-mediated photodynamic therapy in vivo. These approaches demonstrate the potential of CRISPR-based gene-editing strategies, synergistically integrated with nanotechnology and anticancer therapeutics, for advanced biomedical applications.
3.4 Agricultural and environmental applications
Food security remains a critical challenge as the global population grows, with agricultural productivity needing to increase sustainably to tackle malnutrition and hunger. Projections indicate that global food demand will rise by approximately 100–110% between 2005 and 2050, potentially exacerbating environmental impacts depending on the methods used for agricultural expansion.196 Focusing on efficient farming practices and improved technologies could help meet this demand while minimizing land clearing and reducing greenhouse gas emissions and environmental harm.The CRISPR-Cas9 site-specific genome editing technology is a promising approach towards global food security/safety, crop improvement and livestock engineering.197 As a rapid, efficient and cost-effective technology, CRISPR-Cas9 enables the development of genetically modified organisms (GMOs) with enhanced traits. This approach facilitates the biofortification of major crops with essential vitamins and minerals, thereby improving their nutritional value.198 Furthermore, CRISPR has been successfully employed to enhance crop resilience against various abiotic stresses such as drought, salinity and heat,199 as well as biotic stresses including disease resistance.200 The adoption of next-generation CRISPR platforms—including methods like de novo domestication of plants from scratch, editing genes in specific tissues, using prime editing for greater accuracy and fine-tuning of gene expression within multidisciplinary breeding strategies—holds significant promise for the development of climate-resilient crops, ultimately contributing to sustainable global food security.201,202
CRISPR-Cas technology has emerged as a powerful tool for crop quality enhancement, with a primary focus on improving physical appearance—such as shape, size, color and texture for prolonged shelf life.203–205 Additionally, this technology has been employed to enhance nutritional quality by increasing carotenoid content and GABA levels, modifying fatty acid composition, biofortifying micronutrients and eliminating anti-nutritional factors.206–208 CRISPR has also contributed to enhancing edible quality and palatability, such as improving cooking and eating quality and optimizing flavor, thus facilitating the development of crop varieties with preferred consumer traits.209–211
The CRISPR/Cas toolkit has expanded into the RNA-editing domain with the advent of Cas13—a versatile “Swiss Army knife” for plant researchers.212 This RNA-targeting system offers unprecedented opportunities to manipulate plant transcriptomes. Nuclease-dead versions of Cas13 enable real-time visualization of RNA dynamics in living cells and facilitate precise, programmable RNA editing through base-specific deamination or chemical conjugation.213,214 Cas13 offers several advantages over traditional technologies, including scalability, multiplex RNA targeting, and the ability to target non-coding nuclear transcripts and pre-mRNA when fused with nuclear localization signals.
While CRISPR/Cas9 has made significant developments in plant genome editing, its application in plant pathogen diagnostics remains underdeveloped. Early pathogen detection could help farmers to manage diseases and reduce crop losses. CRISPR-Cas9 enables quicker and more efficient development of disease-resistant crops, offering an alternative to traditional breeding methods which are slow and labor-intensive.167For example, the development of DNA detection systems using Cas12a, in conjunction with highly active crRNAs and lateral flow assays (LFAs), has shown promise for low-cost, efficient, and field-deployable diagnostics in plant disease surveillance and GMO monitoring.215 Moreover, recent studies showed that CRISPR-Cas9-mediated inactivation of the Downy Mildew Resistance 6 (DMR6) gene in Arabidopsis thaliana confers resistance to multiple pathogens. Similarly, targeted mutagenesis of the DMR6 ortholog (SlDMR6-1 orthologue Solyc03g080190) in tomato also leads to broad-spectrum disease resistance against Pseudomonas syringae, P. capsica, Xanthomonas spp, pv. tomato and Phytophthora capsica as this gene is significantly upregulated upon infection.216
The emergence of infectious diseases caused by bacterial pathogens, including foodborne species such as Staphylococcus aureus (S. aureus), represents a persistent global public health concern due to their serious microbiological risks and potential to cause food poisoning and other infectious diseases. Several studies have been reported to develop rapid, efficient, and ultrasensitive methods for detecting S. aureus in food samples. For example, Zhuang J. et al. developed a microfluidic paper-based analytical device (µPAD) integrating recombinase polymerase amplification (RPA) with CRISPR/Cas12a (RPA-Cas12a-µPAD), for ultrasensitive surface-enhanced Raman scattering (SERS) detection217 (Fig. 7A). The platform achieved a dynamic detection range of 1–108 CFU mL−1 with a limit of detection of approximately 3–4 CFU mL−1 for S. typhi in spiked milk and meat samples. Precise detection of foodborne pathogens in dietary samples was accomplished within 45 minutes using the RPA-Cas12a-µPAD. In another study, Yujie Ma et al. developed a magnetic enrichment cascade single-step recombinase polymerase amplification (RPA)-CRISPR/Cas12a assay (Fig. 7D).218 This assay could detect as low as 10 CFU mL−1 of S. aureus and 1 copy per µL (five copies per reaction) of extracted DNA within 40 minutes. Moreover, the assay produced results consistent with qPCR and successfully detected S. aureus in real food samples, including lettuce, pork, orange juice, and lake water.
 | ||
| Fig. 7 Applications of CRISPR/Cas9 in agriculture and environmental sustainability. (A) Schematic design of the proposed CRISPR-Cas12a-based biosensor employing gold nanostar@4-mercaptobenzoic acid@gold nanoshell structures (AuNS@4-MBA@Au) for SERS-based bacterial detection, figure adapted with permission from ref. 217 Copyright© 2022 Elsevier. (B) Schematic illustration of a ‘signal-on’ colorimetric/photothermal biosensor (psG4-CHA/Cas) developed for portable detection of environmental contaminants, utilizing a catalytic hairpin assembly (CHA) with Cas12a for amplification and phosphorothioate-modified G4 as a reporter, figure adapted with permission from ref. 219 Copyright© 2024 Elsevier. (C) Schematic diagram of the RPA-CRISPR/Cas12a environmental DNA (eDNA) assay, figure adapted with permission from ref. 220 Copyright© 2024 Elsevier. (D) Schematic illustration of the RPA-CRISPR/Cas12a-based platform for S. aureus detection, figure adapted with permission from ref. 218 Copyright© 2024 Elsevier. | ||
CRISPR technology has revolutionized molecular biology with broad applications in environmental biotechnology.219,221 It enables the engineering of microorganisms for bioremediation, degradation of pollutants, and clean-up of oil spills and heavy metal contamination.220 For example, Kai Shi et al. designed a “signal-on” colorimetric/photothermal biosensor (psG4-CHA/Cas) that combines catalytic hairpin assembly (CHA) with Cas12a as an amplification strategy and phosphorothioate-modified G4 as a reporter for the portable detection of environmental contaminants (Fig. 7B).219 For the colorimetric detection of Pb2+ and kanamycin, a smartphone was employed. The detection limits for kanamycin and Pb2+ were found to be 100 pM and 50 pM, respectively. Additionally, a portable thermometer was used, achieving detection limits of 10 pM for kanamycin and 8 pM for Pb2+.
CRISPR supports the development of fourth-generation biofuels, control of invasive species, genetic enhancement for ecosystem restoration, and modulation of methane emissions.221 Environmental DNA (eDNA) research enables near real-time global species monitoring, offering enormous potential to advance biodiversity science and conservation efforts. For example, Kim K. et al. developed a two-step RPA-CRISPR-Cas12a eDNA assay, comprising target eDNA amplification followed by a CRISPR-Cas12 reaction, for the detection of seasonal Chrysaora pacifica (Fig. 7C).220 The assay results were validated and compared using visual observations of jellyfish, absolute quantification via dPCR, and eDNA metabarcoding. The RPA-CRISPR-Cas12a assay, with its isothermal operation, high specificity, sensitivity, and precision, demonstrates significant potential for low-cost, on-site eDNA detection of non-indigenous species. Additionally, CRISPR/Cas-based biosensors offer promising tools for monitoring environmental health, while CRISPR diagnostics enable early detection of pathogens and contaminants, such as waterborne microbes.222,223 Furthermore, by integrating with nanotechnology it forms smart nanostructure hybrids that can enhance environmental clean-up and long-term ecological sustainability.224
By inserting carefully edited DNA sequences—representing traits of an extinct species—into the genome of a closely related living species, researchers can generate hybrid organisms. These hybrids are not exact replicas of the extinct species, but rather new organisms that retain essential features of the original ones while incorporating traits from their modern relatives. According to Colossal Biosciences, a Dallas–Texas based bioscience and genetic engineering company focused on species de-extinction,227 they have achieved a groundbreaking milestone: the successful birth of three healthy dire wolves, a species that roamed North America but has been extinct for over 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 years.228 While many dire wolf fossils were discovered in the La Brea Tar Pits in Los Angeles, the tar didn't preserve species DNA.
500 years.228 While many dire wolf fossils were discovered in the La Brea Tar Pits in Los Angeles, the tar didn't preserve species DNA.
Reintroducing revived species (mainly the keystone species) into environments where they can thrive not only adds to biodiversity but also strengthens the resilience of entire ecosystems.229 Colossal Biosciences' pioneering conservation efforts exemplify how advanced genetics can be harnessed not merely to preserve endangered species, but to revive ecosystems. By introducing woolly mammoth traits into Asian elephants, the goal extends beyond species conservation to the restoration of the ancient Arctic grasslands once inhabited by mammoths. Releasing mammoth-elephant hybrids into the tundra could stimulate large-scale ecological recovery. Their movement and behaviour, such as trampling permafrost, toppling low-oxygen trees, and creating open spaces for carbon-sequestering grasses, could help rejuvenate the tundra ecosystem. This transformation has the potential to enhance carbon capture, mitigate greenhouse gas emissions, and contribute meaningfully to global climate regulation.
4 Limitations of CRISPR-Cas9 technology
The development of the CRISPR-Cas9 system has provided transformative potential and widespread applications across many fields, including medical therapeutics, agricultural engineering, and biological research,230,231 as it offers relatively simple, precise, programmable, and versatile tools and an efficient method for altering DNA sequences in living organisms. However, despite their promise, the efficacy of first-generation CRISPR-based gene editing tools is often limited by several challenges such as efficiency of delivery, off-target effects, targeting scope, half-life of CRISPR components, and endogenous DSB repair mechanism efficiency to achieve genomic edits, ethical concerns and the need for real-time monitoring during editing processes at the cellular level. This will enable a deeper understanding of how CRISPR-Cas9 interacts with DNA, which is critical for optimizing the CRISPR editing system and ensuring its safety in therapeutic applications.88Hsu et al. reported that decreasing the content of Cas9 protein in the CRISPR-Cas9 system will reduce or overcome the off-target effect challenges, but it will also cause low efficiency of targeted cutting, which is the main role of CRISPR technology.232 Off-target effects (OTEs) occur when CRISPR-Cas9 binds to and cuts DNA even when mismatches exist between the guide RNA and the target sequence. Although this mechanism helps defend against viruses in nature, in genome editing it can lead to unintended modifications at similar genomic sites, potentially causing mutations or chromosomal alterations. Therefore, scientists are working to improve the accuracy and safety of CRISPR for medical applications, with ongoing efforts focused on enhancing detection techniques such as in silico tools and high-throughput sequencing.233
Other researchers also designed different techniques to resolve these challenges by continuous enhancement or upgradation of the specificity by improvement of guide RNA (gRNA) design, bioinformatic tools and high-fidelity Cas9 enzymes for recognition of sites as well as a series of structural modifications to the Cas9 protein. The modifications of Cas9 protein generally result in prime editing, base editing, transcriptional modulation (gene regulation) and RNA editing (Fig. 1(c)). For example, in 2020, Lin Q. et al. reported prime editing created by fusing a CRISPR-Cas9 nickase (H840A) with reverse transcriptase and guided by pegRNAs to make precise changes in DNA without needing donor templates or causing double-strand breaks. This system was tailored in plants by optimizing codons, promoters, and editing conditions, which successfully produced point mutations, insertions, and deletions in rice and wheat protoplast. In rice, prime-edited plants were regenerated with editing efficiencies reaching up to 21.8%.234 Newby G.A. et al. (2021) demonstrated an alternative approach for modifying Cas proteins to treat sickle cell disease, which is caused by a mutation in the HBB gene. Using the specialized adenine base editor ABE8e-NRCH, they converted the harmful SCD variant HBBS gene to the harmless HBBG (Makassar β-globin) by introducing the editor and guide RNA into blood stem cells from SCD patients, achieving up to 80% correction and long-lasting effects after transplantation in mice. This approach improved blood health (even when only about 20% of the cells were corrected), avoided DNA damage seen in traditional CRISPR, and could offer a safe, one-time treatment for Sickle cell disease (SCD).235
5 Conclusion and future prospects
Genetic modification is quickly shaping our world—from better crops to medicines. The heart of this revolution is CRISPR-Cas9, a powerful tool making gene editing faster, easier, and more precise. For example, CRISPR is used in diagnosis, real-time tracking and visualization, and in advancing cancer immunotherapy. It can also help fortify crops by engineering genes that boost vitamin production, improve yield, and increase resilience to environmental stress, supporting global efforts to end hunger and improve nutrition by 2030.236 CRISPR tools are also helping to detect diseases like Zika,44 especially in remote areas, as outbreaks and reemergence of tropical diseases (TDs) often impact remote and rural communities with limited access to medical diagnostic tools. With support from e-healthcare and telemedicine, we can further boost access to such innovations. To use CRISPR safely and effectively, we need a comprehensive understanding of how its components work and how to deliver it where it's needed. CRISPR has enormous potential, but challenges like safe delivery, accuracy, precision, off-target effects and ethical concerns remain, and overcoming these is key to its safe, widespread use.As technology evolves, its integration with emerging innovations, especially artificial intelligence and nanotechnology, could unlock personalized, powerful breakthroughs in medicine and beyond. Combining CRISPR biosensors with artificial intelligence (AI), cloud tech, the Internet of Things (IoT) and the Industrial Internet of things (IIoT) can make real-time health monitoring a reality.237 But to get there, we still need to improve things like accuracy, data privacy, and portability.168 In the future, simple, all-in-one diagnostic tools and smarter imaging could make CRISPR-based healthcare faster, easier, and more accessible for everyone. Future research may combine CRISPR with cell-based drug delivery combining gene editing with the natural ability of cells for personalized treatments. Scientists are also exploring its use to bring back extinct species, which could help restore ecosystems. With continued research, these innovations could provide a more sustainable future for both our planet and future generations.
Author contributions
Nisha Bharti: literature review and writing – original draft preparation, Unnati Modi: literature review and writing – original draft preparation, Dhiraj Bhatia: conceptualization, writing – review and editing, and supervision. Raghu Solanki: conceptualization, writing – review and editing. All authors confirmed the final version of the manuscript.Conflicts of interest
The authors declare that they have no conflict of interest.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
The authors would like to thank IIT Gandhinagar for providing the necessary facilities. RS thanks the Anusandhan National Research Foundation (ANRF), Govt. of India for an N-PDF fellowship (PDF/2023/000365). We would like to acknowledge the support of Dr Sreekanth Vedagopuram in improving review article by providing valuable feedback. DB thanks SERB-DST, GUJCOST, GSBTM, and STARS-MoES for research grants. The authors also thank the DB lab members for their feedback on the review article.References
- B. Wiedenheft, S. H. Sternberg and J. A. Doudna, Nature, 2012, 482, 331–338 Search PubMed.
- L. A. Marraffini and E. J. Sontheimer, Nat. Rev. Genet., 2010, 11, 181–190 Search PubMed.
- K. S. Makarova, D. H. Haft, R. Barrangou, S. J. J. Brouns, E. Charpentier, P. Horvath, S. Moineau, F. J. M. Mojica, Y. I. Wolf and A. F. Yakunin, Nat. Rev. Microbiol., 2011, 9, 467–477 Search PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709–1712 Search PubMed.
- P. Horvath and R. Barrangou, Science, 2010, 327, 167–170 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 Search PubMed.
- G. Gasiunas, R. Barrangou, P. Horvath and V. Siksnys, Proceedings of the National Academy of Sciences, 2012, 109, E2579–E2586 Search PubMed.
- J. A. Doudna and E. Charpentier, Science, 2014, 346–1258096 Search PubMed.
- P. D. Hsu, E. S. Lander and F. Zhang, Cell, 2014, 157, 1262–1278 Search PubMed.
- H. Nishimasu, F. A. Ran, P. D. Hsu, S. Konermann, S. I. Shehata, N. Dohmae, R. Ishitani, F. Zhang and O. Nureki, Cell, 2014, 156, 935–949 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang and L. A. Marraffini, Science, 2013, 339, 819–823 Search PubMed.
- S. W. Cho, S. Kim, J. M. Kim and J.-S. Kim, Nat. Biotechnol., 2013, 31, 230–232 CrossRef CAS PubMed.
- M. R. Lieber, Annu. Rev. Biochem., 2010, 79, 181–211 CrossRef CAS PubMed.
- P. Mali, L. Yang, K. M. Esvelt, J. Aach, M. Guell, J. E. DiCarlo, J. E. Norville and G. M. Church, Science, 2013, 339, 823–826 CrossRef CAS PubMed.
- W.-D. Heyer, K. T. Ehmsen and J. Liu, Annu. Rev. Genet., 2010, 44, 113–139 Search PubMed.
- H. Ledford and E. Callaway, Nature, 2020, 586, 346–347 Search PubMed.
- A. V Anzalone, L. W. Koblan and D. R. Liu, Nat. Biotechnol., 2020, 38, 824–844 CrossRef PubMed.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Nature, 2016, 533, 420–424 Search PubMed.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2017, 551, 464–471 CrossRef CAS PubMed.
- H. A. Rees and D. R. Liu, Nat. Rev. Genet., 2018, 19, 770–788 CrossRef CAS PubMed.
- A. V Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby and A. Raguram, Nature, 2019, 576, 149–157 Search PubMed.
- M. Nakamura, Y. Gao, A. A. Dominguez and L. S. Qi, Nat. Cell Biol., 2021, 23, 11–22 CrossRef CAS PubMed.
- P. Mali, J. Aach, P. B. Stranges, K. M. Esvelt, M. Moosburner, S. Kosuri, L. Yang and G. M. Church, Nat. Biotechnol., 2013, 31, 833–838 Search PubMed.
- L. A. Gilbert, M. H. Larson, L. Morsut, Z. Liu, G. A. Brar, S. E. Torres, N. Stern-Ginossar, O. Brandman, E. H. Whitehead and J. A. Doudna, Cell, 2013, 154, 442–451 CrossRef CAS PubMed.
- N. C. Yeo, A. Chavez, A. Lance-Byrne, Y. Chan, D. Menn, D. Milanova, C.-C. Kuo, X. Guo, S. Sharma and A. Tung, Nat. Methods, 2018, 15, 611–616 CrossRef CAS PubMed.
- A. Chavez, J. Scheiman, S. Vora, B. W. Pruitt, M. Tuttle, E. PR Iyer, S. Lin, S. Kiani, C. D. Guzman and D. J. Wiegand, Nat. Methods, 2015, 12, 326–328 CrossRef CAS PubMed.
- S. Konermann, M. D. Brigham, A. E. Trevino, J. Joung, O. O. Abudayyeh, C. Barcena, P. D. Hsu, N. Habib, J. S. Gootenberg and H. Nishimasu, Nature, 2015, 517, 583–588 Search PubMed.
- L. S. Qi, M. H. Larson, L. A. Gilbert, J. A. Doudna, J. S. Weissman, A. P. Arkin and W. A. Lim, Cell, 2013, 152, 1173–1183 CrossRef CAS PubMed.
- B. Chen, L. A. Gilbert, B. A. Cimini, J. Schnitzbauer, W. Zhang, G.-W. Li, J. Park, E. H. Blackburn, J. S. Weissman and L. S. Qi, Cell, 2013, 155, 1479–1491 CrossRef CAS PubMed.
- H. Wang, M. Nakamura, T. R. Abbott, D. Zhao, K. Luo, C. Yu, C. M. Nguyen, A. Lo, T. P. Daley and M. La Russa, Science, 2019, 365, 1301–1305 Search PubMed.
- H. Ma, L.-C. Tu, A. Naseri, M. Huisman, S. Zhang, D. Grunwald and T. Pederson, Nat. Biotechnol., 2016, 34, 528–530 CrossRef CAS PubMed.
- M. Adli, Nat. Commun., 2018, 9, 1911 Search PubMed.
- L. Villiger, J. Joung, L. Koblan, J. Weissman, O. O. Abudayyeh and J. S. Gootenberg, Nat. Rev. Mol. Cell Biol., 2024, 25, 464–487 Search PubMed.
- J. Y. Wang and J. A. Doudna, Science, 2023, 379 Search PubMed.
- D. B. T. Cox, R. J. Platt and F. Zhang, Nat. Med., 2015, 21, 121–131 CrossRef CAS PubMed.
- M. H. Porteus, N. Engl. J. Med., 2019, 380, 947–959 Search PubMed.
- J. A. Doudna, Nature, 2020, 578, 229–236 CrossRef CAS PubMed.
- H. Frangoul, D. Altshuler, M. D. Cappellini, Y.-S. Chen, J. Domm, B. K. Eustace, J. Foell, J. de la Fuente, S. Grupp and R. Handgretinger, N. Engl. J. Med., 2021, 384, 252–260 Search PubMed.
- H. Frangoul, F. Locatelli, A. Sharma, M. Bhatia, M. Mapara, L. Molinari, D. Wall, R. I. Liem, P. Telfer and A. J. Shah, N. Engl. J. Med., 2024, 390, 1649–1662 Search PubMed.
- J. D. Gillmore, E. Gane, J. Taubel, J. Kao, M. Fontana, M. L. Maitland, J. Seitzer, D. O'Connell, K. R. Walsh and K. Wood, N. Engl. J. Med., 2021, 385, 493–502 Search PubMed.
- S. Levesque and D. E. Bauer, Nat. Rev. Drug Discov., 2025, 1–19 Search PubMed.
- M. M. Kaminski, O. O. Abudayyeh, J. S. Gootenberg, F. Zhang and J. J. Collins, Nat. Biomed. Eng., 2021, 5, 643–656 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian, J. M. Palefsky and J. A. Doudna, Science, 2018, 360, 436–439 Search PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer and C. A. Freije, Science, 2017, 356, 438–442 Search PubMed.
- M. Patchsung, K. Jantarug, A. Pattama, K. Aphicho, S. Suraritdechachai, P. Meesawat, K. Sappakhaw, N. Leelahakorn, T. Ruenkam and T. Wongsatit, Nat. Biomed. Eng., 2020, 4, 1140–1149 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados and A. Sotomayor-Gonzalez, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed.
- C. Gao, Cell, 2021, 184, 1621–1635 CrossRef CAS PubMed.
- S. Li, D. Lin, Y. Zhang, M. Deng, Y. Chen, B. Lv, B. Li, Y. Lei, Y. Wang and L. Zhao, Nature, 2022, 602, 455–460 CrossRef CAS PubMed.
- C. Proudfoot, S. Lillico and C. Tait-Burkard, Animal Frontiers, 2019, 9, 6–12 Search PubMed.
- K. M. Whitworth, R. R. R. Rowland, C. L. Ewen, B. R. Trible, M. A. Kerrigan, A. G. Cino-Ozuna, M. S. Samuel, J. E. Lightner, D. G. McLaren and A. J. Mileham, Nat. Biotechnol., 2016, 34, 20–22 CrossRef CAS PubMed.
- A. M. Workman, M. P. Heaton, B. L. Vander Ley, D. A. Webster, L. Sherry, J. R. Bostrom, S. Larson, T. S. Kalbfleisch, G. P. Harhay and E. E. Jobman, PNAS nexus, 2023, 2, pgad125 CrossRef PubMed.
- D. J. Lea-Smith, F. Hassard, F. Coulon, N. Partridge, L. Horsfall, K. D. J. Parker, R. D. J. Smith, R. R. McCarthy, B. McKew and T. Gutierrez, Nat. Commun., 2025, 16, 3538 CrossRef CAS PubMed.
- Y. Zhou, L. Lin, H. Wang, Z. Zhang, J. Zhou and N. Jiao, Commun. Biol., 2020, 3, 98 CrossRef CAS PubMed.
- N. Windbichler, M. Menichelli, P. A. Papathanos, S. B. Thyme, H. Li, U. Y. Ulge, B. T. Hovde, D. Baker, R. J. Monnat and A. Burt, Nature, 2011, 473, 212–215 CrossRef CAS PubMed.
- A. Hammond, R. Galizi, K. Kyrou, A. Simoni, C. Siniscalchi, D. Katsanos, M. Gribble, D. Baker, E. Marois and S. Russell, Nat. Biotechnol., 2016, 34, 78–83 Search PubMed.
- J. Buchthal, S. W. Evans, J. Lunshof, S. R. Telford and K. M. Esvelt, Philos. Trans. R. Soc. B, 2019, 374, 20180105 CrossRef PubMed.
- E. Bier, Nat. Rev. Genet., 2022, 23, 5–22 CrossRef CAS PubMed.
- S. K. Alsaiari, B. Eshaghi, B. Du, M. Kanelli, G. Li, X. Wu, L. Zhang, M. Chaddah, A. Lau and X. Yang, Nat. Rev. Mater., 2025, 10, 44–61 CrossRef CAS.
- A. E. Modell, D. Lim, T. M. Nguyen, V. Sreekanth and A. Choudhary, Trends Pharmacol. Sci., 2022, 43, 151–161 CrossRef CAS PubMed.
- J. Boutin, D. Cappellen, J. Rosier, S. Amintas, S. Dabernat, A. Bedel and F. Moreau-Gaudry, CRISPR J., 2022, 5, 19–30 CrossRef CAS PubMed.
- C. Xue and E. C. Greene, Trends Genet., 2021, 37, 639–656 CrossRef CAS PubMed.
- J. M. T. Hunt, C. A. Samson, A. du Rand and H. M. Sheppard, Hum. Genet., 2023, 142, 705–720 CrossRef PubMed.
- N. Kalter, C. Fuster-García, A. Silva, V. Ronco-Díaz, S. Roncelli, G. Turchiano, J. Gorodkin, T. Cathomen, K. Benabdellah and C. Lee, Mol. Ther. Nucleic Acids, 2025, 36(3), 102636 CrossRef CAS PubMed.
- V. Sreekanth, M. Jan, K. T. Zhao, D. Lim, S. Siriwongsup, J. R. Davis, M. McConkey, V. Kovalcik, S. Barkal and B. K. Law, J. Am. Chem. Soc., 2025, 147, 23844–23856 CrossRef CAS PubMed.
- H.-X. Wang, M. Li, C. M. Lee, S. Chakraborty, H.-W. Kim, G. Bao and K. W. Leong, Chem. Rev., 2017, 117, 9874–9906 Search PubMed.
- S. Honrath, M. Burger and J.-C. Leroux, Int. J. Pharm., 2025, 125470 Search PubMed.
- C. Liu, L. Zhang, H. Liu and K. Cheng, J. Contr. Release, 2017, 266, 17–26 CrossRef CAS PubMed.
- S. Banskota, A. Raguram, S. Suh, S. W. Du, J. R. Davis, E. H. Choi, X. Wang, S. C. Nielsen, G. A. Newby and P. B. Randolph, Cell, 2022, 185, 250–265 CrossRef CAS PubMed.
- M. Le Hars, C. Joussain, T. Jégu and A. L. Epstein, Gene Ther., 2025, 32, 173–183 CrossRef CAS PubMed.
- C. E. Nelson and C. A. Gersbach, Annu. Rev. Chem. Biomol. Eng., 2016, 7, 637–662 CrossRef CAS PubMed.
- E. V Pankevich and C. Bock, Nat. Protoc., 2025, 1–2 Search PubMed.
- S. Ylä-Herttuala, Mol. Ther., 2012, 20, 1831–1832 CrossRef PubMed.
- W. Wu, Y. Yang, F. Yao, L. Dong, X. Xia, S. Zhang and H. Lei, Methods, 2021, 194, 12–17 CrossRef CAS PubMed.
- J. Jia, Y. Hao, L. Zhang, X. Cao, L. An, H. Wang, Q. Ma, X. Jin and X. Ma, Eur. J. Med. Res., 2025, 30, 242 CrossRef CAS PubMed.
- E. Fang, G. He, Y. Chang, Q. He, P. Chen and K. Hu, Mol. Biotechnol., 2025, 1–17 Search PubMed.
- S. Tang, X. Chen, X. Tong and L. Zhu, Int. J. Med. Sci., 2025, 22, 3625 CrossRef PubMed.
- K. Singh, D. Jain, P. Sethi, J. K. Gupta, A. Dubey, A. Al Noman, S. Lal, P. D. Sharma and E. M. Abdallah, 3 Biotech, 2025, 15, 196 CrossRef PubMed.
- C. S. Lee, E. S. Bishop, R. Zhang, X. Yu, E. M. Farina, S. Yan, C. Zhao, Z. Zeng, Y. Shu and X. Wu, Genes Dis., 2017, 4, 43–63 Search PubMed.
- T. Gaj, B. E. Epstein and D. V Schaffer, Mol. Ther., 2016, 24, 458–464 CrossRef CAS PubMed.
- J. Saraiva, R. J. Nobre and L. P. de Almeida, J. Controlled Release, 2016, 241, 94–109 CrossRef CAS PubMed.
- F. Sonntag, K. Schmidt and J. A. Kleinschmidt, Proceedings of the National Academy of Sciences, 2010, 107, 10220–10225 Search PubMed.
- S. Ylä-Herttuala, Mol. Ther., 2012, 20, 1831–1832 Search PubMed.
- W. Wu, Y. Yang, F. Yao, L. Dong, X. Xia, S. Zhang and H. Lei, Methods, 2021, 194, 12–17 CrossRef CAS PubMed.
- T. Schnell, P. Foley, M. Wirth, J. Munch and K. Uberla, Hum. Gene Ther., 2000, 11, 439–447 Search PubMed.
- O. Shalem, N. E. Sanjana, E. Hartenian, X. Shi, D. A. Scott, T. S. Mikkelsen, D. Heckl, B. L. Ebert, D. E. Root and J. G. Doench, Science, 2014, 343, 84–87 Search PubMed.
- N. Farhang, M. Ginley-Hidinger, K. C. Berrett, J. Gertz, B. Lawrence and R. D. Bowles, Hum. Gene Ther., 2019, 30, 1161–1175 CrossRef CAS PubMed.
- M. Taghdiri and C. Mussolino, Int. J. Mol. Sci., 2024, 25, 7333 Search PubMed.
- I. Maggio, M. Holkers, J. Liu, J. M. Janssen, X. Chen and M. A. F. V Gonçalves, Sci. Rep., 2014, 4, 5105 Search PubMed.
- R. Lorincz, A. B. Alvarez, C. J. Walkey, S. A. Mendonça, Z. H. Lu, A. E. Martinez, C. Ljungberg, J. D. Heaney, W. R. Lagor and D. T. Curiel, Biomed. Pharmacother., 2023, 158, 114189 Search PubMed.
- Y. Yu, Y. Gao, L. He, B. Fang, W. Ge, P. Yang, Y. Ju, X. Xie and L. Lei, MedComm (Beijing), 2023, 4, e259 Search PubMed.
- X. Du, J. Wang, Q. Zhou, L. Zhang, S. Wang, Z. Zhang and C. Yao, Drug Deliv., 2018, 25, 1516–1525 Search PubMed.
- M. Kim, Y. Hwang, S. Lim, H.-K. Jang and H.-O. Kim, Pharmaceutics, 2024, 16, 1197 Search PubMed.
- H. Lee, W.-Y. Rho, Y.-H. Kim, H. Chang and B.-H. Jun, Molecules, 2025, 30, 542 CrossRef CAS PubMed.
- J. Kim, Y. Eygeris, R. C. Ryals, A. Jozić and G. Sahay, Nat. Nanotechnol., 2024, 19, 428–447 CrossRef CAS PubMed.
- R. Solanki, A. Shankar, U. Modi and S. Patel, Mater. Today Chem., 2023, 101478 CrossRef CAS PubMed.
- G. Kaur, J. Arora, A. S. Sodhi, S. Bhatia and N. Batra, Mol. Biotechnol., 2024, 1–23 Search PubMed.
- K. Khanna, P. Ohri and R. Bhardwaj, Environ. Sci. Pollut. Res., 2023, 30, 118049–118064 Search PubMed.
- H. Wang, H. Yang, T. Li, Y. Chen, J. Chen, X. Zhang, J. Zhang, Y. Zhang, N. Zhang and R. Ma, Int. J. Mol. Sci., 2025, 26, 1065 Search PubMed.
- B. S. Owusu-Yaw, Mol. Ther., 2025, 33, 831–832 Search PubMed.
- T. Yamaguchi, Y. Endo-Takahashi, T. Amano, A. Ihara, T. Sakuma, T. Yamamoto, T. Fukazawa and Y. Negishi, Pharmaceutics, 2025, 17, 1053 CrossRef CAS PubMed.
- T. Dalsgaard, C. R. Cecchi, A. L. Askou, R. O. Bak, P. O. Andersen, D. Hougaard, T. G. Jensen, F. Dagnæs-Hansen, J. G. Mikkelsen, T. J. Corydon and L. Aagaard, Improved lentiviral gene delivery to mouse liver by hydrodynamic vector injection through tail vein, Mol. Ther.–Nucleic Acids, 2018, 12, 672–683 Search PubMed.
- M. G. Sebestyén, V. G. Budker, T. Budker, V. M. Subbotin, G. Zhang, S. D. Monahan, D. L. Lewis, S. C. Wong, J. E. Hagstrom and J. A. Wolff, Mechanism of plasmiddelivery by hydrodynamic tail vein injection. I.Hepatocyte uptake of various molecules, J. Gene Med ., 2006, 8(7), 852–873 Search PubMed.
- D. S. D’Astolfo, R. J. Pagliero, A. Pras, W. R. Karthaus, H. Clevers, V. Prasad, R. J. Lebbink, H. Rehmann and N. Geijsen, Efficient intracellular delivery of native proteins, Cells, 2015, 161(3), 674–690 Search PubMed.
- H. Karp, M. Zoltek, K. Wasko, A. L. Vazquez, J. Brim, W. Ngo, A. Schepartz and J. A. Doudna, Nucleic Acids Res., 2025, 53, gkaf105 CrossRef CAS PubMed.
- Q. Yan, Q. Zhang, H. Yang, Q. Zou, C. Tang, N. Fan and L. Lai, Cell Regen., 2014, 3, 3–12 Search PubMed.
- A. Hruscha and B. Schmid, Neuronal Cell Death: Methods and Protocols, 2015, 341–350 Search PubMed.
- H. Sasaki, K. Yoshida, A. Hozumi and Y. Sasakura, Dev. Growth Differ., 2014, 56, 499–510 Search PubMed.
- X. Chen, F. Xu, C. Zhu, J. Ji, X. Zhou, X. Feng and S. Guang, Sci. Rep., 2014, 4, 7581 Search PubMed.
- S. Basu, A. Aryan, J. M. Overcash, G. H. Samuel, M. A. E. Anderson, T. J. Dahlem, K. M. Myles and Z. N. Adelman, Proceedings of the National Academy of Sciences, 2015, 112, 4038–4043 Search PubMed.
- Y. Nakagawa, T. Sakuma, T. Sakamoto, M. Ohmuraya, N. Nakagata and T. Yamamoto, BMC Biotechnol., 2015, 15, 1–10 Search PubMed.
- Z. Zhang, J. Wang, J. Li, X. Liu, L. Liu, C. Zhao, W. Tao, D. Wang and J. Wei, Biology (Basel), 2023, 12, 336 Search PubMed.
- S. R. Porazinski, H. Wang and M. Furutani-Seiki, J. Vis. Exp., 2010, e1937 Search PubMed.
- H. Yin, L. Sun, Y. Pu, J. Yu, W. Feng, C. Dong, B. Zhou, D. Du, Y. Zhang and Y. Chen, ACS Cent. Sci., 2021, 7, 2049–2062 CrossRef CAS PubMed.
- K. Hazel, D. Singh, S. He, Z. Guertin, M. C. Husser and B. Helfield, Mol. Ther., 2025, 33(3), 986–996 CrossRef CAS PubMed.
- Y. Li, P. Wu, M. Zhu, M. Liang, L. Zhang, Y. Zong and M. Wan, Adv. Healthc. Mater., 2023, 12, 2203082 CrossRef CAS PubMed.
- F. Liu, Y. K. Song and D. Liu, Gene Ther., 1999, 6, 1258–1266 Search PubMed.
- T. Suda, T. Yokoo, T. Kanefuji, K. Kamimura, G. Zhang and D. Liu, Pharmaceutics, 2023, 15, 1111 Search PubMed.
- T. Kanefuji, T. Yokoo, T. Suda, H. Abe, K. Kamimura and D. Liu, Mol. Ther.–Methods Clin. Dev., 2014, 1, 14029 Search PubMed.
- D. L. Lewis and J. A. Wolff, Adv. Drug Deliv. Rev., 2007, 59, 115–123 Search PubMed.
- W. M. Kholosy, M. Visscher, K. Ogink, H. Buttstedt, K. Griffin, A. Beier, J. P. Gerlach, J. J. Molenaar, N. Geijsen and M. de Boer, J. Biotechnol., 2021, 338, 71–80 Search PubMed.
- J. A. Zuris, D. B. Thompson, Y. Shu, J. P. Guilinger, J. L. Bessen, J. H. Hu, M. L. Maeder, J. K. Joung, Z.-Y. Chen and D. R. Liu, Nat. Biotechnol., 2015, 33, 73–80 Search PubMed.
- G.-L. Yang, D.-G. Wang, X. Zhao, D.-L. Hao, Z.-Q. Zhang and D.-P. Liu, Mol. Ther.–Methods Clin. Dev., 2025, 33(3), 101542 Search PubMed.
- F. Liu, R. Su, S. Wang, W. Mu and L. Chang, Nanoscale, 2024, 16, 10500–10521 Search PubMed.
- S.-E. Choi, H. Khoo and S. C. Hur, Chem. Rev., 2022, 122, 11247–11286 Search PubMed.
- M. Abbasi and B. Heath, Drug Deliv. Transl. Res., 2025, 15, 1962–1984 Search PubMed.
- A. J. Modzelewski, S. Chen, B. J. Willis, K. C. K. Lloyd, J. A. Wood and L. He, Nat. Protoc., 2018, 13, 1253–1274 Search PubMed.
- S. E. Tröder, L. K. Ebert, L. Butt, S. Assenmacher, B. Schermer and B. Zevnik, PLoS One, 2018, 13, e0196891 CrossRef PubMed.
- W. Qin, S. L. Dion, P. M. Kutny, Y. Zhang, A. W. Cheng, N. L. Jillette, A. Malhotra, A. M. Geurts, Y.-G. Chen and H. Wang, Genetics, 2015, 200, 423–430 CrossRef CAS PubMed.
- P. Pfenninger, L. Yerly and J. Abe, Front. Immunol., 2022, 13, 777113 Search PubMed.
- R. Thummel, S. Bai, M. P. Sarras, P. Song, J. McDermott, J. Brewer, M. Perry, X. Zhang, D. R. Hyde and A. R. Godwin, Dev. Dyn., 2006, 235, 336–346 Search PubMed.
- K. Chen, H. Han, S. Zhao, B. Xu, B. Yin, A. Lawanprasert, M. Trinidad, B. W. Burgstone, N. Murthy and J. A. Doudna, Nat. Biotechnol., 2024, 1–13 Search PubMed.
- E. A. Woodward, E. Wang, C. Wallis, R. Sharma, A. W. J. Tie, N. Murthy and P. Blancafort, in Epigenome Editing: Methods and Protocols, Springer, 2024, pp. 267–287 Search PubMed.
- R. Khodaei, M. Bayandori, L. M. G. Sarpoli, M. Souri, I. Hasanzade, R. Khodaee, S. Saeedi, J. Kiani and M. Karimi, Adv. Nat. Sci. Nanosci. Nanotechnol., 2024, 15, 045003 Search PubMed.
- S. Iqbal, A. Alexander-Bryant and J. Larsen, in Engineering Biomaterials for Neural Applications, Springer, 2022, pp. 229–258 Search PubMed.
- J. Padayachee and M. Singh, Nanobiomedicine (Rij.), 2020, 7, 1849543520983196 Search PubMed.
- M. A. Rauf, A. Rao, S. S. Sivasoorian and A. K. Iyer, Cells, 2025, 14, 1136 Search PubMed.
- G. A. Ferronato, J. C. Silveira and A. M. M. Ferraz, Biol. Reprod., 2025, 113(6), 1321–1339 CrossRef CAS PubMed.
- S. Xie, N. F. Ilahibaks, Z. Lei and J. P. Sluijter, Harnessing extracellular vesicles to deliver genetic medicine for cardiac repair, Nanomedicine, 2025, 1–3 Search PubMed.
- B. B. Mendes, J. Conniot, A. Avital, D. Yao, X. Jiang, X. Zhou, N. Sharf-Pauker, Y. Xiao, O. Adir and H. Liang, Nat. Rev. Methods Primers, 2022, 2, 24 Search PubMed.
- D. Wilbie, J. Walther and E. Mastrobattista, Acc. Chem. Res., 2019, 52, 1555–1564 CrossRef CAS PubMed.
- Q. Yang, Y. Zhou, J. Chen, N. Huang, Z. Wang and Y. Cheng, Int. J. Nanomed., 2021, 185–199 CrossRef PubMed.
- R. Kumar, C. F. Santa Chalarca, M. R. Bockman, C. Van Bruggen, C. J. Grimme, R. J. Dalal, M. G. Hanson, J. K. Hexum and T. M. Reineke, Chem. Rev., 2021, 121, 11527–11652 Search PubMed.
- L. Peng and E. Wagner, Biomacromolecules, 2019, 20, 3613–3626 CrossRef CAS PubMed.
- Y. Rui, D. R. Wilson, J. Choi, M. Varanasi, K. Sanders, J. Karlsson, M. Lim and J. J. Green, Sci. Adv., 2019, 5, eaay3255 CrossRef CAS PubMed.
- G. A. Kanu, J. B. M. Parambath, R. O. Abu Odeh and A. A. Mohamed, Cancers (Basel), 2022, 14, 5366 Search PubMed.
- J. Zhang, L. Mou and X. Jiang, Chem. Sci., 2020, 11, 923–936 Search PubMed.
- R. Mout, M. Ray, G. Yesilbag Tonga, Y.-W. Lee, T. Tay, K. Sasaki and V. M. Rotello, ACS Nano, 2017, 11, 2452–2458 CrossRef CAS PubMed.
- P. Wang, L. Zhang, W. Zheng, L. Cong, Z. Guo, Y. Xie, L. Wang, R. Tang, Q. Feng and Y. Hamada, Angew. Chem., Int. Ed., 2018, 57, 1491–1496 CrossRef CAS PubMed.
- R. Hu, X. Zhang, Z. Zhao, G. Zhu, T. Chen, T. Fu and W. Tan, Angew. Chem., 2014, 126, 5931–5936 CrossRef.
- N. Singh, A. Sharma, A. Goel, K. Kumar, R. Solanki and D. Bhatia, Small, 2025, 2410440 CrossRef CAS PubMed.
- N. Jain, A. Singh and D. Bhatia, Nanoscale, 2025, 17, 18–52 RSC.
- L. Zhou, Z. Chen, K. Dong, M. Yin, J. Ren and X. Qu, Adv. Mater., 2014, 26, 2424–2430 CrossRef CAS PubMed.
- W. Sun, W. Ji, J. M. Hall, Q. Hu, C. Wang, C. L. Beisel and Z. Gu, Angew. Chem., 2015, 127, 12197–12201 Search PubMed.
- N. Z. Fantoni, T. Brown and A. Kellett, ChemBioChem, 2021, 22, 2184–2205 CrossRef CAS PubMed.
- F. Miceli, S. Bracaglia, D. Sorrentino, A. Porchetta, S. Ranallo and F. Ricci, Nucleic Acids Res., 2025, 53, gkaf238 CrossRef CAS PubMed.
- Y. Liang, Z. Iqbal, J. Wang, L. Xu, X. Xu, K. Ouyang, H. Zhang, J. Lu, L. Duan and J. Xia, Biomater. Sci., 2022, 10, 4095–4106 RSC.
- K. Leandro, D. Rufino-Ramos, K. Breyne, E. Di Ianni, S. M. Lopes, R. J. Nobre, B. P. Kleinstiver, P. R. L. Perdigão, X. O. Breakefield and L. P. de Almeida, Adv. Drug Deliv. Rev., 2024, 115346 Search PubMed.
- Z. Song, Y. Tao, Y. Liu and J. Li, Front. Immunol., 2024, 15, 1444437 Search PubMed.
- L. Duan, L. Xu, X. Xu, Z. Qin, X. Zhou, Y. Xiao, Y. Liang and J. Xia, Nanoscale, 2021, 13, 1387–1397 RSC.
- X. Huang, A. Li, P. Xu, Y. Yu, S. Li, L. Hu and S. Feng, J. Nanobiotechnol., 2023, 21, 184 CrossRef PubMed.
- S. Singh, R. Thakran, A. Kaushal, R. V Saini, A. Saini and S. Datta, Microchem. J., 2024, 110421 CrossRef CAS.
- Y. Xia, R. Rao, M. Xiong, B. He, B. Zheng, Y. Jia, Y. Li and Y. Yang, Anal. Chem., 2024, 96, 8091–8108 Search PubMed.
- D. W. Bianchi, D. Chudova, A. J. Sehnert, S. Bhatt, K. Murray, T. L. Prosen, J. E. Garber, L. Wilkins-Haug, N. L. Vora and S. Warsof, JAMA, 2015, 314, 162–169 CrossRef CAS PubMed.
- S. A. Bustin, Int. J. Mol. Sci., 2024, 25, 9326 CrossRef CAS PubMed.
- M. R. Rahman, M. A. Hossain, M. Mozibullah, F. Al Mujib, A. Afrose, M. Shahed-Al-Mahmud and M. A. I. Apu, Biomed. Pharmacother., 2021, 140, 111772 Search PubMed.
- P. Bhardwaj, R. Kant, S. P. Behera, G. R. Dwivedi and R. Singh, Int. J. Mol. Sci., 2022, 23, 6052 Search PubMed.
- A. U. Ibrahim, F. Al-Turjman, Z. Sa’id and M. Ozsoz, Multimed. Tool. Appl., 2022, 81, 35143–35171 Search PubMed.
- M. J. Kellner, J. G. Koob, J. S. Gootenberg, O. O. Abudayyeh and F. Zhang, Nat. Protoc., 2019, 14, 2986–3012 Search PubMed.
- Y. M. Hassan, A. S. Mohamed, Y. M. Hassan and W. M. El-Sayed, Clin. Exp. Med., 2025, 25, 33 Search PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados and A. Sotomayor-Gonzalez, Nat. Biotechnol., 2020, 38, 870–874 Search PubMed.
- L. Li, S. Li, N. Wu, J. Wu, G. Wang, G. Zhao and J. Wang, ACS Synth. Biol., 2019, 8, 2228–2237 Search PubMed.
- R. Hajian, S. Balderston, T. Tran, T. DeBoer, J. Etienne, M. Sandhu, N. A. Wauford, J.-Y. Chung, J. Nokes and M. Athaiya, Nat. Biomed. Eng., 2019, 3, 427–437 Search PubMed.
- X. Wang, E. Xiong, T. Tian, M. Cheng, W. Lin, H. Wang, G. Zhang, J. Sun and X. Zhou, ACS Nano, 2020, 14, 2497–2508 CrossRef CAS PubMed.
- E. Kolesova, S. Pulone, D. Kostyushev and E. Tasciotti, Adv. Drug Deliv. Rev., 2025, 115619 Search PubMed.
- S. Shao, W. Zhang, H. Hu, B. Xue, J. Qin, C. Sun, Y. Sun, W. Wei and Y. Sun, Nucleic Acids Res., 2016, 44, e86 Search PubMed.
- J. Yang, X. Xu, L. Yang, Y. Tian, J. Wang and D. Han, Small Methods, 2025, 2401559 Search PubMed.
- B. Chen, W. Zou, H. Xu, Y. Liang and B. Huang, Nat. Commun., 2018, 9, 5065 CrossRef PubMed.
- N.-H. Sun, D.-Y. Chen, L.-P. Ye, G. Sheng, J.-J. Gong, B.-H. Chen, Y.-M. Lu and F. Han, Theranostics, 2020, 10, 10993 Search PubMed.
- H. Liu, J. Dong, Z. Duan, F. Xia, I. Willner and F. Huang, Sci. Adv., 2024, 10, eadp6166 Search PubMed.
- A. Yuan, R. Sha, W. Xie, G. Qu, H. Zhang, H. Wang, X. C. Le, G. Jiang and H. Peng, J. Am. Chem. Soc., 2024, 146, 26657–26666 Search PubMed.
- X. Chen, Q. You, Y. Guo, J. Zhang, Q. Hu, C. Huang, Z. Wu, J. Deng, J. Xu and P. Zhao, Chem. Biomed. Imaging., 2025, 3(6), 369–378 CrossRef CAS PubMed.
- M. R. Cring and V. C. Sheffield, Gene Ther., 2022, 29, 3–12 CrossRef CAS PubMed.
- F. Arabi, V. Mansouri and N. Ahmadbeigi, Biomed. Pharmacother., 2022, 153, 113324 CrossRef CAS PubMed.
- B. Liu, H. Zhou, L. Tan, K. T. H. Siu and X.-Y. Guan, Signal Transduct. Targeted Ther., 2024, 9, 175 CrossRef PubMed.
- R. Thirumalaisamy, S. Vasuki, S. M. Sindhu, T. M. Mothilal, V. Srimathi, B. Poornima, M. Bhuvaneswari and M. Hariharan, Mol. Biotechnol., 2025, 67, 469–483 CrossRef CAS PubMed.
- K. K. Patel, M. Tariveranmoshabad, S. Kadu, N. Shobaki and C. June, Mol. Ther. Search PubMed.
- J. Zeng, J. Luo and Y. Zeng, Funct Integr Genomics, 2025, 25, 200 Search PubMed.
- S. C. Selvakumar, K. A. Preethi, K. Ross, D. Tusubira, M. W. A. Khan, P. Mani, T. N. Rao and D. Sekar, Mol. Cancer, 2022, 21, 83 Search PubMed.
- E. A. Stadtmauer, J. A. Fraietta, M. M. Davis, A. D. Cohen, K. L. Weber, E. Lancaster, P. A. Mangan, I. Kulikovskaya, M. Gupta and F. Chen, Science, 2020, 367 Search PubMed.
- B. Zhang, J. Cell. Physiol., 2021, 236, 2459–2481 Search PubMed.
- S. Oura, T. Noda, N. Morimura, S. Hitoshi, H. Nishimasu, Y. Nagai, O. Nureki and M. Ikawa, Commun. Biol., 2021, 4, 771 CrossRef CAS PubMed.
- Y. Wang, P. K. Shahi, X. Wang, R. Xie, Y. Zhao, M. Wu, S. Roge, B. R. Pattnaik and S. Gong, J. Controlled Release, 2021, 336, 296–309 CrossRef CAS PubMed.
- J.-Y. Ryu, C. Cerecedo-Lopez, H. Yang, I. Ryu and R. Du, Theranostics, 2024, 14, 4773 CrossRef CAS PubMed.
- S. Deng, X. Li, S. Liu, J. Chen, M. Li, S. Y. Chew, K. W. Leong and D. Cheng, Sci. Adv., 2020, 6, eabb4005 Search PubMed.
- D. Tilman, C. Balzer, J. Hill and B. L. Befort, Proceedings of the national academy of sciences, 2011, 108, 20260–20264 Search PubMed.
- S. Ahmad, L. Tang, R. Shahzad, A. M. Mawia, G. S. Rao, S. Jamil, C. Wei, Z. Sheng, G. Shao and X. Wei, J. Agric. Food Chem., 2021, 69, 8307–8323 CrossRef CAS PubMed.
- A. Chaudhary, R. K. Singh, T. Bisht, A. Verma, A. Sharma, P. Dahiya, N. Kaushik and R. K. Bhatia, in Plant-microbiome Interactions for Climate-Resilient Agriculture, ed U. Pankaj, P. Babele and A. K. Singh, Springer Nature Singapore, Singapore, 2025, pp. 281–319 Search PubMed.
- K. Chennakesavulu, H. Singh, P. K. Trivedi, M. Jain and S. R. Yadav, Plant Cell Rep., 2021, 1–17 Search PubMed.
- D. Schenke and D. Cai, iScience, 2020, 23(9), 101478 CrossRef CAS PubMed.
- P. Jasrotia, P. Kumari and P. L. Kashyap, in Cutting Edge Technologies for Developing Future Crop Plants, ed A. Mann, N. Kumar, A. Kumar, P. Chandra, S. K. Sanwal and P. Sheoran, Springer Nature Singapore, Singapore, 2025, pp. 219–247 Search PubMed.
- C. Sangh, M. G. Mallikarjuna, M. K. Pandey, T. K. Mondal, T. Radhakrishnan, R. S. Tomar, H. P. Gajera and S. K. Bera, in Breeding Climate Resilient and Future Ready Oilseed Crops, ed M. K. Pandey, M. G. Mallikarjuna, H. C. Lohithaswa, M. S. Aski and S. Gupta, Springer Nature Singapore, Singapore, 2025, pp. 265–301 Search PubMed.
- C. Yuyu, Z. Aike, X. Pao, W. Xiaoxia, C. Yongrun, W. Beifang, Z. Yue, S. Liaqat, C. Shihua and C. Liyong, Rice Sci., 2020, 27, 405–413 Search PubMed.
- Z.-S. Xu, Q.-Q. Yang, K. Feng and A.-S. Xiong, Plant Physiol., 2019, 181, 195–207 Search PubMed.
- D. Wang, N. H. Samsulrizal, C. Yan, N. S. Allcock, J. Craigon, B. Blanco-Ulate, I. Ortega-Salazar, S. E. Marcus, H. M. Bagheri and L. Perez Fons, Plant Physiol., 2019, 179, 544–557 Search PubMed.
- S.-K. Sun, X. Xu, Z. Tang, Z. Tang, X.-Y. Huang, M. Wirtz, R. Hell and F.-J. Zhao, Nat. Commun., 2021, 12, 1392 Search PubMed.
- P. T. Do, C. X. Nguyen, H. T. Bui, L. T. N. Tran, G. Stacey, J. D. Gillman, Z. J. Zhang and M. G. Stacey, BMC Plant Biol., 2019, 19, 1–14 Search PubMed.
- N. Kaur, A. Alok, P. Kumar, N. Kaur, P. Awasthi, S. Chaturvedi, P. Pandey, A. Pandey, A. K. Pandey and S. Tiwari, Metab. Eng., 2020, 59, 76–86 CrossRef CAS PubMed.
- H. Gao, M. J. Gadlage, H. R. Lafitte, B. Lenderts, M. Yang, M. Schroder, J. Farrell, K. Snopek, D. Peterson and L. Feigenbutz, Nat. Biotechnol., 2020, 38, 579–581 Search PubMed.
- Y. Li, W. Zhang, M. Li, X. Ling, D. Guo, Y. Yang, Q. Liu, B. Zhang and J. Wang, Plant Commun., 2025, 6(1), 101141 CrossRef CAS PubMed.
- R. Parveen, A. Mohanty, R. K. Sohane, M. Kumar and T. Ranjan, J. Plant Biochem. Biotechnol., 2025, 1–23 Search PubMed.
- F. Wolter and H. Puchta, Plant J., 2018, 94, 767–775 Search PubMed.
- L.-Z. Yang, Y. Wang, S.-Q. Li, R.-W. Yao, P.-F. Luan, H. Wu, G. G. Carmichael and L.-L. Chen, Mol. Cell, 2019, 76, 981–997 Search PubMed.
- J. Yu, J. Shin, J. Yu, J. Kim, D. Yu and W. Do Heo, Nat. Commun., 2024, 15, 673 Search PubMed.
- Y. Zhang, Y. Zhang and K. Xie, Mol. Breed., 2020, 40, 11 CrossRef.
- L. K. Acharya, S. Manik, M. Pradhan, A. Birah, M. K. Khokhar, D. Majumder and A. Hossain, in Plant Stress Tolerance, CRC Press, pp. , pp. 99–112 Search PubMed.
- J. Zhuang, Z. Zhao, K. Lian, L. Yin, J. Wang, S. Man, G. Liu and L. Ma, Biosens. Bioelectron., 2022, 207, 114167 Search PubMed.
- Y. Ma, H. Wei, Y. Wang, X. Cheng, H. Chen, X. Yang, H. Zhang, Z. Rong and S. Wang, J. Hazard. Mater., 2024, 465, 133494 CrossRef CAS PubMed.
- K. Shi, Y. Tian, S. Liu, W. Luo, K. Liu, L. Zhang, Y. Zhang, J. Chang, J. Zhang and S. Wang, Anal. Chim. Acta, 2024, 1308, 342649 CrossRef CAS PubMed.
- K. Kim, U. J. Maji, K.-Y. Shim, I.-C. Yeo and C.-B. Jeong, Sci. Total Environ., 2024, 955, 176945 CrossRef CAS PubMed.
- J. Verdezoto-Prado, C. Chicaiza-Ortiz, A. B. Mejía-Pérez, C. Freire-Torres, M. Viteri-Yánez, L. Deng, C. Barba-Ostria and L. P. Guamán, Discov. Appl. Sci., 2025, 7, 167 Search PubMed.
- H. Daneshgar, M. Safarkhani, S. Sojdeh, M. Bagherzadeh and N. Rabiee, TrAC, Trends Anal. Chem., 2025, 118174 Search PubMed.
- A. K. Wani, N. Akhtar, C. Chopra, R. Singh, J. C. Hong and U. S. Kadam, Environ. Technol. Innov., 2024, 34, 103625 Search PubMed.
- E. Bahl, A. Jyoti, A. Singh, A. Siddqui, S. K. Upadhyay, D. Jain, M. P. Shah and J. Saxena, Environ. Sci. Pollut. Res., 2024, 31, 67479–67495 CrossRef CAS PubMed.
- L. Evans Ogden, Bioscience, 2014, 64, 469–475 Search PubMed.
- H. T. Greely, Hastings Center Report, 2017, 47, S30–S36 Search PubMed.
- A. Nelson and K. O'Riordan, Sci. Cult., 2025, 1–31 CAS.
- Colossal laboratories and biosciences, https://colossal.com/, accessed 2 May 2025.
- L. P. Thiele, Glob. Environ. Polit., 2020, 20, 9–27 Search PubMed.
- A. Parsaeimehr, R. I. Ebirim and G. Ozbay, Biotechnol. Rep., 2022, 34, e00731 CrossRef CAS PubMed.
- R. Barrangou and J. A. Doudna, Nat. Biotechnol., 2016, 34, 933–941 Search PubMed.
- P. D. Hsu, D. A. Scott, J. A. Weinstein, F. A. Ran, S. Konermann, V. Agarwala, Y. Li, E. J. Fine, X. Wu and O. Shalem, Nat. Biotechnol., 2013, 31, 827–832 Search PubMed.
- C. Guo, X. Ma, F. Gao and Y. Guo, Front. Bioeng. Biotechnol., 2023, 11, 1143157 Search PubMed.
- Q. Lin, Y. Zong, C. Xue, S. Wang, S. Jin, Z. Zhu, Y. Wang, A. V Anzalone, A. Raguram and J. L. Doman, Nat. Biotechnol., 2020, 38, 582–585 CrossRef CAS PubMed.
- G. A. Newby, J. S. Yen, K. J. Woodard, T. Mayuranathan, C. R. Lazzarotto, Y. Li, H. Sheppard-Tillman, S. N. Porter, Y. Yao and K. Mayberry, Nature, 2021, 595, 295–302 Search PubMed.
- D. Kumar, A. Yadav, R. Ahmad, U. N. Dwivedi and K. Yadav, Front. Genet., 2022, 13, 932859 CrossRef CAS PubMed.
- S. Dixit, A. Kumar, K. Srinivasan, P. M. D. R. Vincent and N. Ramu Krishnan, Front. Bioeng. Biotechnol., 2024, 11, 1335901 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |


