Open Access Article
Open Access ArticleA platform for the formation of uniform DNA condensate droplets using vibration-induced local vortices
Zhitai Huanga,
Kanji Kanekoa,
Ryotaro Yoneyamaa,
Tomoya Maruyama bd,
Takeshi Hayakawa
bd,
Takeshi Hayakawa a,
Masahiro Takinoue
a,
Masahiro Takinoue bcde and
Hiroaki Suzuki
bcde and
Hiroaki Suzuki *a
*a
aGraduate School of Science and Engineering, Chuo University, Kasuga 1-13-27, Bunkyo-ku, 112-8551, Japan. E-mail: suzuki@mech.chuo-u.ac.jp
bDepartment of Life Science and Technology, Institute of Science Tokyo, 4259 Nagatsutacho, Midori-ku, Yokohama, Kanagawa 226-8501, Japan
cDepartment of Computer Science, Institute of Science Tokyo, 4259 Nagatsutacho, Midori-ku, Yokohama, Kanagawa 226-8501, Japan
dResearch Center for Autonomous Systems Materialogy, Institute of Integrated Research, Institute of Science Tokyo, 4259 Nagatsutacho, Midori-ku, Yokohama, Kanagawa 226-8501, Japan
eLaboratory for Chemistry and Life Science, Institute of Integrated Research, Institute of Science Tokyo, 4259 Nagatsutacho, Midori-ku, Yokohama, Kanagawa 226-8501, Japan
First published on 25th November 2025
Abstract
DNA condensate droplets (hereafter referred to as DNA condensates), which arise from specific interactions between sticky ends embedded in multi-arm DNA nanostructures, hold significant promise as programmable smart materials. However, from an engineering standpoint, the controlled preparation of DNA condensates with uniform size and a well-defined structure remains a major challenge due to the stochastic nature of the condensation process. This study presents a novel approach that employs vibration-induced local vortices (VILV) within a microfluidic platform to achieve spatial control over DNA condensate dimensions and enable their parallel generation. A key advantage of this platform is its ability to support direct observation and real-time tracking of structural morphology and dynamics. Through flow-field analysis of the VILV system, we demonstrate that uniform microvortices serve as semi-closed compartments, wherein DNA molecules confined within each vortex space rapidly aggregate and relax into uniform spherical condensate droplets. By modulating parameters such as DNA concentration and micropillar dimensions, the VILV platform not only enables systematic control of condensate size but also facilitates the construction of complex, multicomponent “patchy” condensates with consistent morphology. This platform provides a robust and scalable tool for studying liquid–liquid phase separation (LLPS) and offers broad potential for applications in the bottom-up synthesis of condensed molecular systems.
New conceptsDNA condensates hold great promise for various applications because their physical properties and molecular recognition capabilities can be programmed via sequence design. However, standardized and reproducible methods for producing uniform and homogeneous condensates have not yet been established. This work demonstrates the use of an array of stable, vibration-induced local vortices (VILV) as ‘virtual compartments’ for the parallel and controlled self-assembly of sticky DNA nanostructures in a purely aqueous system. This approach fundamentally differs from conventional methods by replacing physical boundaries with designable hydrodynamic traps. This technique brings a new perspective to biomolecular materials science by establishing a ‘hydrodynamic assembly platform’, enabling a predictable and scalable relationship between the macroscopic device geometry and the resulting condensate size. This control is characterized by accelerated Y-motif DNA nanostar aggregation and the ability to observe its relaxation process. This high degree of control is further highlighted by the successful formation of uniform, multicomponent ‘patchy’ DNA condensates with a consistent morphology, showcasing a powerful strategy for providing a clean testbed to study complex phase transition dynamics and fabricating advanced functional condensed biomaterials. |
Introduction
In the fields of life sciences and polymer chemistry, research on molecular condensates formed through liquid–liquid phase separation (LLPS) has become increasingly active.1–4 Cellular granules and membraneless organelles, along with their functions, have attracted significant attention, leading to growing elucidation of their formation mechanisms and properties.5,6 Fundamentally, LLPS occurs either due to the entropic mutual exclusion of polymers or specific associative interactions. Various types of interactions fall under the latter category, and molecular-level understanding of condensate formation continues to advance.Among the various LLPS systems, molecular condensates composed of DNA (DNA droplets or DNA hydrogels) have garnered significant attention in both materials science and information science due to their unique properties and potential applications.7–11 These condensates are particularly valued for their biocompatibility, biodegradability, and capacity to incorporate genetic information. A category of DNA condensates consist of networks of DNA nanostructures, often multi-arm motifs such as Y- or X-shaped, formed through highly specific Watson–Crick interactions.12–15 Because the molecular interactions between DNA nanostructures can be precisely and specifically tuned through structural design and sticky-end (SE) sequences located at the tips of each arm, both physical properties and molecular recognition abilities can be controlled in a highly programmable manner.16–21 Unlike traditional DNA nanostructures such as DNA origami, DNA condensates exist on the microscale, enabling the observation of dynamic changes due to molecular interactions or environmental stimuli using standard optical microscopy.
Despite the rapid advancement of molecular understanding and applications of LLPS droplets, including DNA condensates, there remains no universally effective method for controlling their formation process and size. Molecular condensation is inherently stochastic; as with water droplet formation on a windowpane, condensates form across a broad size range.22 In the microfluidics community, generation methods for emulsion droplets composed of water and oil have been widely studied, and various approaches for producing uniform droplets have been reported.23,24 In such systems, the large interfacial tension between water and oil (on the order of mN m−1) facilitates stable droplet breakup via shear forces.23 However, LLPS droplets exhibit interfacial tensions that are 1/10 to 1/100 of those at oil–water interfaces, making their boundaries more prone to continuous stretching under shear rather than droplet breakup.25
Previous efforts to control the size of LLPS droplets can be broadly categorized into two approaches: microfluidics-based flow-focusing methods and microscale compartmentalization. The former adapts techniques for generating uniform water-in-oil (W/O) or oil-in-water (O/W) droplets for LLPS systems, but thus far, its application has been limited to segregative aqueous two-phase systems (ATPS), typically polyethylene glycol (PEG)/dextran systems.26–29 In these methods, the interface must form immediately upon contact between two immiscible polymer solutions, making them unsuitable for associative polymer systems such as DNA condensates, where condensation occurs gradually via intermolecular interactions. A related method has shown that uniform ATPS droplets can spontaneously form inside glass capillaries,30 although this technique likely produces droplets only comparable in size to the inner diameter of the capillary. The latter approach uses microscale compartments of uniform volume, such as microwells, W/O droplets, or liposomes, to encapsulate associative polymers. Because the amount of polymer within each compartment is defined, the resulting condensate size is also controlled. In an early example, Sokolova et al.31 demonstrated that encapsulating Escherichia coli cell lysate in uniform W/O droplets and inducing osmotic shrinkage led to phase separation and uniform condensate formation. The same group later showed that uniform poly-L-lysine (PLL)/adenosine triphosphate (ATP) coacervates and DNA condensates could be formed within uniform giant unilamellar vesicles (GUVs) or W/O droplets generated via microfluidics.32,33 Our group has also recently demonstrated the formation of uniform DNA condensates within liposomes produced using polydimethylsiloxane (PDMS) microfluidic channels.34 Additionally, Masukawa et al. attempted the formation of uniform DNA droplets within microwell arrays.35 These compartment-based methods are particularly promising for constructing hierarchical artificial cells.36,37 However, from other perspectives, it would be highly beneficial to generate uniform LLPS droplets directly in aqueous solution without compartments—particularly for detailed observation of molecular condensation processes, post-formation perfusion of molecules to study interactions, or for collecting condensates for off-chip applications.
In this context, the group led by S. Mann demonstrated the formation of uniform arrays of coacervate droplets composed of polydiallyldimethylammonium chloride (PDDA)/ATP at nodes of a pressure potential field generated by acoustic standing waves.38 Using this system, they investigated how coacervate droplets respond to environmental concentration gradients of model morphogen molecules.39 These studies controlled LLPS in aqueous environments by introducing virtual compartments via pressure fields, representing a potential groundbreaking tool in LLPS research.
We previously demonstrated that aggregates of nano- and microparticles with specific affinities can be formed using a vibration-induced flow (VIF) system.40–42 We proposed that this system could serve as a simple detection method for biological nanoparticles.43 The system operates by generating a uniform array of microvortices—hereafter referred to as vibration-induced local vortices (VILV)—within the spaces between square-arranged micropillars, through small rotational oscillations applied to the substrate. As aggregates form and grow within these vortices, they rapidly accumulate at the vortex centers (<10 min), resulting in microscale aggregate formation.
In this study, we examined the usefulness of this VILV system for parallel generation of uniform molecular condensates and directly observing their dynamics. As a result, similar to micro- and nanoparticle aggregate formation, Y-motif structures gathered at the center of the vortex in a short time (∼5 min), forming uniform aggregates. The initial aggregates had an amorphous morphology with a shaggy contour due to rapid SE bonding caused by an increase in local concentration, but relaxed into spherical, dense condensates over time. The VILV method allows the size of the condensates to be controlled by changing the Y-motif concentration conditions and the geometry of the micropillars, and subsequent changes can be observed under a microscope. We demonstrated that even complex condensates composed of multiple DNA motifs can be formed and observed with uniform and consistent morphology.
Experimental
PDMS device fabrication
Microfluidic devices were fabricated using standard soft lithography techniques with PDMS, as previously described in detail.41,42 Master molds featuring micropillar arrays and spacer structures (Fig. S1) were fabricated on 2-inch silicon wafers using a deep reactive ion etching process (RIE-400iPB, Samco, Japan). To prevent PDMS adhesion, a fluorinated release agent (Fluoro Surf FG-5084, FluoroTechnology Co., Ltd, Japan) was applied to the mold surface and dried on a hot plate at 120 °C for 20 min. PDMS resin (Sylgard 184, Dow Corning, USA) was mixed with its curing agent at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 weight ratio, degassed, poured over the master mold, and cured on a hot plate at 80 °C for at least 2 h. After curing, PDMS replicas were carefully demolded and cut into individual devices.
1 weight ratio, degassed, poured over the master mold, and cured on a hot plate at 80 °C for at least 2 h. After curing, PDMS replicas were carefully demolded and cut into individual devices.
For the design using 100 µm diameter and 100 µm height micropillars, a 35 × 35 array of cylindrical micropillars was patterned. The center-to-center spacing between adjacent micropillars was set to 200 µm, resulting in a 100 µm edge-to-edge spacing. To define the fluidic channel height and prevent coverslip slippage during experiments, 100 µm-high spacer structures were integrated at the corners and sides of the PDMS layout (Fig. S1).
Devices with other micropillar diameters (75 µm, 50 µm, and 25 µm) were fabricated following a similar design principle, with center-to-center spacings set at twice the micropillar diameter. The array configurations were 46 × 46 for 75 µm pillars, 70 × 70 for 50 µm pillars, and 140 × 140 for 25 µm pillars. The pillar height was 100 µm for the 75 µm devices, and 50 µm for the 50 µm and 25 µm devices.
To prevent condensate adhesion, the PDMS surfaces were passivated with a polymer brush. The surface was first activated by a 10-s oxygen plasma treatment (20 mL min−1 O2 flow, 75 W power; Compact Etcher FA-1, SAMCO, Japan) to enhance polymer solution spreading. Immediately afterward, 200 µL of a 5% (w/v) polyvinyl alcohol (PVA; Mw 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–23
000–23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, 87–89% hydrolyzed, Sigma-Aldrich, USA) solution was applied to the micropillar side and left to stand for 1 h. Subsequently, the devices were rinsed with deionized water and dried on a hot plate at 80 °C for 30 min to evaporate residual water. A second 10-s oxygen plasma treatment was applied prior to the VIF experiments to further enhance wettability, minimize air bubble entrapment, and promote uniform solution distribution.
000, 87–89% hydrolyzed, Sigma-Aldrich, USA) solution was applied to the micropillar side and left to stand for 1 h. Subsequently, the devices were rinsed with deionized water and dried on a hot plate at 80 °C for 30 min to evaporate residual water. A second 10-s oxygen plasma treatment was applied prior to the VIF experiments to further enhance wettability, minimize air bubble entrapment, and promote uniform solution distribution.
Preparation of DNA condensate solutions
Single-stranded DNA (ssDNA) oligonucleotides (custom-synthesized by Eurofins Genomics; sequences in the SI, Table S1), designated Y1, Y2, and Y3, were initially dissolved in nuclease-free water to prepare 100 µM stock solutions, stored at −20 °C. Before use, stock solutions were thawed and equilibrated to room temperature for at least 30 min.The standard composition for experiments was 10 µM of each ssDNA (Y1, Y2, and Y3), 50 mM NaCl, and 20 mM Tris-HCl (pH 8.0). To examine the effect of DNA concentration on condensate formation, four different solutions containing 2.5, 5, 10, or 20 µM of each ssDNA were prepared (see the SI, Table S2(b)). All solutions included a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution of YOYO™-1 Iodide (Y3601, Invitrogen, CA, USA) for DNA staining.
100 dilution of YOYO™-1 Iodide (Y3601, Invitrogen, CA, USA) for DNA staining.
To ensure stable Y-motif assembly, DNA mixtures (containing Y1, Y2, and Y3 in buffer) were heated to 70 °C for 10 min, ensuring full dissociation and availability for correct hybridization into Y-motifs. The solution was then cooled to the working temperature for the experiments.
Preparation of patchy DNA condensate solution
For patchy DNA condensates, two orthogonal 4-nt Y-motifs were prepared (see Table S3 for details). The first type was formed using strands Y1, Y2, and Y3 as described above. For fluorescence visualization, 10 molar% of Y2 was replaced with 6-FAM-labeled Y2 (ex/em = 493/517 nm), resulting in 5 µM of each Y1, Y2/Y2-FAM, and Y3. The second type was formed using three different ssDNA strands designated as orthY1, orthY2, and orthY3 (sequences in the SI, Table S1), with 10 molar% of orthY2 replaced by orthY2 labeled with hexachlorofluorescein (HEX, ex/em = 533/549 nm). Thus, orthY1, orthY2/orthY2-HEX, and orthY3 were each at a final concentration of 5 µM to form this second Y-motif. Additionally, a linker DNA nanostructure (S-motif), composed of six ssDNA strands (S1 to S6; sequences in the SI, Table S1), was added at 0.5 µM to bridge the two orthogonal Y-motifs. All components for the patchy DNA condensate formation were prepared according to the protocol described in the previous subsection, omitting YOYO™-1 iodide DNA staining dye.Operation of the VIF system
Immediately after the Y-motif formation step, 14 µL of the mixture solution was pipetted onto the micropillar face of the PDMS device for vibrational manipulation experiments. A coverslip (18 × 18 mm2) was gently placed over the micropillar device. For 4nt Y-motif systems, an on-chip heating step was included: devices were placed on a 70 °C hot plate (SS-100, BLAST, Japan) for 3 min to dissolve spontaneously formed initial condensates in solution, ensuring a consistent monomer starting state prior to the commencement of vibration. This step was not required for 6nt systems due to slower condensation kinetics. Vibration was applied using a sinusoidal signal with a 90° phase shift delivered to the xy-piezo stage (PK2H130-030U(-N), THK Precision Co., LTD, Japan), mounted on the microscope. The waveform generator (AG 1022F, OWON, China) produced two sinusoidal waves in 2.0 Vpp amplitude at 600 Hz with a 90° phase difference. This signal was amplified 15-fold using a power amplifier (PH301, THK Precision Co., LTD, Japan), resulting in a 30 Vpp output that drove the stage in a circular vibration with a fixed physical amplitude of 3 µm. This frequency was determined to be optimal for stable vortex formation and reproducible condensation based on systematic screening of five different frequencies (Fig. S2).Velocity field measurement
Flow around micropillars under circular vibration was characterized using micro-particle image velocimetry (µ-PIV). Fluorescent tracer particles (1 µm diameter, Polysciences, Cat#17154-10) were suspended 4.6 × 108 particles per mL. Sequential images were captured with a high-speed camera (HAS-EX, DITECT, Japan), using either a 20× objective (LUCPlanFLN 20×/0.70, Olympus, Japan) or a 10× objective (A-Plan 10×/0.25, Carl Zeiss, Germany), depending on the required field of view. Images were acquired at 120 or 200 fps. Streamlines were generated by superimposing particle images.The average velocity field was determined by cross-correlation of particle images captured at similar phases of oscillation. Analysis was performed using PIV software (Flow Expert, KATO Koken, Japan). Velocity vectors were calculated using a directional cross-correlation with a 32 × 32-pixel (11.9 × 11.9 µm2) interrogation window, with a 12-pixel (4.5 µm) search range in all directions.
Fluorescence microscopy imaging
DNA condensates were imaged using an upright fluorescence microscope (Axio Scope A1, Carl Zeiss, Germany) with a Colibri 7 LED light source (Carl Zeiss, Germany) using excitation wavelengths of 469 nm (for YOYO-1 and FAM) and 556 nm (for HEX). Images were acquired using a cooled charge-coupled camera (Axiocam 503 mono, Carl Zeiss AG, Germany). For general observations and area measurements, an A-Plan 10×/0.25 objective lens (Carl Zeiss, Germany) was used, providing a 1.4 µm per pixel resolution.Fluorescence intensity was maintained during image acquisition. Brightness and contrast were uniformly adjusted using Zeiss ZEN software. Images were exported as uncompressed TIFF files for analysis.
Condensate cross-sectional areas were measured using ImageJ's “Analyze Particles” function (NIH, USA) after exclusion of micropillar regions and binarization. To minimize the inclusion of background noise or very small, non-specific aggregates, particles smaller than 50 µm2, were excluded from the analysis. From the measured maximum cross-sectional area, an equivalent diameter or volume was estimated assuming spherical or circular profiles for simplicity.
Results and discussion
Characterization of the vibration-induced flow
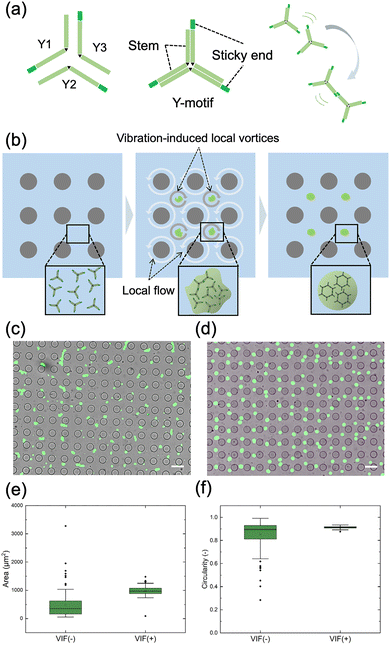
We first characterized the flow generated by applying small circular vibration to the micropillar array device (Fig. 1a). Under 600 Hz frequency and 3 µm amplitude, streamline diagrams from 200 superimposed fluorescent-tracer images (Fig. 1b and Movie S1) revealed the emergence of stable, highly ordered VILV located at the centers of four-pillar junctions. Corresponding PIV analysis (Fig. 1c) confirmed uniform VILV patterns, with peak peripheral velocities approximately 300 µm s−1. This stable vortex formation was consistently observed across devices with different micropillar geometries (D = 25 µm, 75 µm, and 100 µm), as shown in Fig. S3. The physical mechanism is attributed to momentum transfer from the oscillating pillar surfaces to the fluid via the Stokes boundary layer. Notably, our results indicate that while the vortical pattern remains consistent, the area of the vortex increases as the pillar dimensions increase. This result demonstrates that local hydrodynamic environments can be systematically controlled by tuning micropillar geometry.Effect of VILV on the DNA condensate formation
We next investigated the condensation behavior of Y-motif DNA structures in the VILV field. The Y-motif DNA nanostructure, illustrated in Fig. 2a, consists of a 16-nt stem and 4-nt SEs at the terminal of each arm. Condensation occurs via base pairing between SEs, influenced by both the Y-motif and salt concentrations. We expected that, when the solution containing the Y-motif was introduced into the VILV field, Y-motif molecules within each vortex region gathered to the center and formed uniform condensates (Fig. 2b).Experiments were performed with and without VIF in a micropillar array device with a pillar diameter and height of 50 µm. Typically, the Y-motif with 4-nt SEs formed small condensates visible under the microscope immediately after removal of heat. In the absence of VIF, these small condensates, dispersed within the fluid volume between micropillars, fused over time to form larger condensates with irregular positions and morphologies. As time progressed, the amorphous condensates gradually relaxed into rounded shapes due to surface tension, but even after 4 h, they remained heterogeneous in size and not fully spherical (Fig. 2c). Conversely, VIF application led to the rapid accumulation of Y-motifs at the centers of vortices, producing uniform condensates with shaggy outlines within 10 min (time-lapse images during VIF application are shown in Fig. S4 for the D = 100 µm pillar array; see also the SI, Movie S2). After stopping the vibration, the shaggy contours relaxed into smooth, spherical shapes within 3 h (Fig. 2d). Image analysis quantitatively confirmed that the initial VIF application significantly reduced variability in condensate size and shape. Specifically, the box plots in Fig. 2e revealed that without VIF, condensate areas were broadly distributed between 160 and 628 µm2 (as determined by the interquartile range, IQR, a distance between the 3rd and 1st quartiles), with numerous outliers exceeding 1000 µm2. In contrast, with VIF, the areas were tightly clustered around 981 µm2 (IQR: 886–1078 µm2), with few outliers. Regarding circularity (Fig. 2f), defined as 4π × area per perimeter2, condensates formed without VIF exhibited a wide range (0.81–0.93), whereas those formed with VIF exhibited a much narrower distribution (0.91–0.92). These results demonstrate that the VILV flow field significantly enhances centralization of condensate formation, shape regularity, and size uniformity.
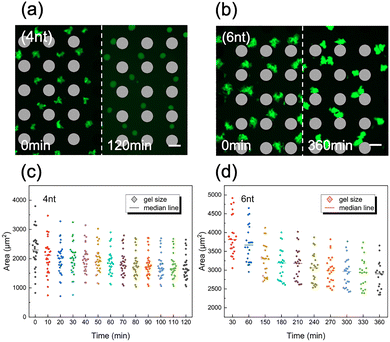
Effect of sticky-end length on DNA condensation dynamics
Using the VILV platform, we compared the condensation behavior of Y-motifs with 4nt-SE and 6nt-SE. Representative fluorescence images are shown in Fig. 3a (4nt-SE) and Fig. 3b (6nt-SE). After 10 min of VILV application (defined as t = 0 min), both constructs formed uniform but non-spherical, dendrimer-like aggregates. Vibration was then stopped, and the condensates were observed under static (no flow) conditions to monitor morphological change. During time-lapse imaging, 4nt-SE Y-motif aggregates contracted into uniform, nearly spherical condensates within 2 h (Fig. 3a, also see the SI, Movie S3). In contrast, 6nt-SE Y-motif aggregates retained fuzzy, irregular shapes and did not achieve spherical morphology even after 6 h (Fig. 3b). Scatter plots of projected condensate area over time (Fig. 3c for 4nt and Fig. 3d for 6nt) further revealed that 4nt-SE Y-motif condensates reached a stable size in 80 min, whereas the size of 6nt-SE Y-motif condensates continued to decrease even at 6 h. Furthermore, throughout the entire shrinkage process, condensates formed from the 6nt-SE Y-motif were consistently larger than those from the 4nt-SE Y-motif, despite identical Y-motif and NaCl concentrations. These observations suggest that the SE length significantly affects the kinetics, morphology and the molecular density of DNA condensates.44 First, shorter SEs confer greater fluidity, promoting faster transition into spherical shapes. Second, condensates with shorter SEs exhibit higher molecular density.These differences can be attributable to the varying thermodynamic stabilities of the SE interactions: the shorter 4nt SEs have lower binding free energy (−6.73 kcal mol−1 in 50 mM NaCl at 25 °C, calculated using the nucleic acid package (NUPACK)45) and a lower proportion of bound SEs, resulting in greater fluidity at room temperature. In contrast, the longer 6nt-SEs form more stable bonds (−7.48 kcal mol−1) immediately upon contact, producing less fluid and spatially diffuse aggregates following VIF application. Subsequently, condensates formed from 6nt-SE Y-motifs exhibited much slower relaxation into spherical condensates. It is worth noting that random migration and fusion may occur after cessation of vibration, particularly during prolonged observation, thereby compromising monodispersity (Fig. 3b). However, this can be effectively prevented by continuing low-frequency vibration (e.g., 200 Hz), which maintains droplet position over extended periods of time (Fig. S5).
Based on these differences in structural dynamics, we selected the 4nt-SE Y-motif as the standard building unit in subsequent experiments to reduce the time required for evaluating condensate uniformity.
Controlling DNA condensate size by varying micropillar-array dimensions
To investigate the effect of structural parameters on condensate size, we designed and fabricated microfluidic chips with four distinct micropillar diameters (D): 25 µm, 50 µm, 75 µm, and 100 µm (Fig. S1). Due to limitations in the deep etching process, the corresponding pillar heights were 50 µm for D = 25 µm and 50 µm, and 100 µm for D = 75 µm and 100 µm.The results from an initial 5-min VIF application to a solution containing Y-motif DNA at a fixed concentration of 10 µM demonstrated that shaggy aggregates of nearly identical size formed at vortex centers (Fig. S3) and relaxed into rounded shapes within 4 h across all micropillar dimensions. The final condensates exhibited good uniformity in size (Fig. 4a–d) compared to the control conditions without VIF application (Fig. S6), and the results were reproducible across independent experiments (Fig. S7). Under the D = 100 µm condition, the condensates remained non-circular after 4 h (Fig. 4d), likely due to the longer time required for larger condensates to fully relax in shape. This is attributable to the slower relaxation (shrinkage) process in larger condensates. Image analysis (Fig. 4e) revealed a clear positive correlation between condensate size and micropillar diameter: the median diameters were 15.0 µm for D = 25 µm (N = 136), 21.0 µm for D = 50 µm (N = 36), 40.3 µm for D = 75 µm (N = 62), and 47.6 µm for D = 100 µm pillars. The IQRs were 2.18, 1.6, 4.4, and 8.9 µm, respectively, indicating consistent data dispersion across conditions. Note that the coefficient of variation in fluorescent intensity among these condensates was calculated to be ∼6.4%, indicating these condensates were highly uniform in terms of the Y-motif density. Also, we noted from Fig. 4(e) that the standard deviation of the condensate size appears to trend to be larger for D = 25 µm and 100 µm devices. Indeed, we consistently observed better monodispersity with D = 50 µm and 75 µm devices. It is because, in the small micropillar device, the net induced velocity magnitude becomes smaller. Thus, when initial condensates in neighbouring vortices came into contact, the shear force was too weak to separate them. In contrast, in the large micropillar device, initially formed small aggregates merge and fuse to grow to form large condensates. In this process, initial small aggregates tended to travel across the neighbouring vortices stochastically, resulting in larger variation. The maximum velocity around the pillar under conditions typically yielding monodisperse results is approximately 300 µm s−1. These observations suggest that an appropriate pillar size should be chosen to obtain the molecular condensates in small variation depending on the concentration of building blocks.
Additionally, a significant step change in condensate size was observed between the 50 and 75 µm pillar diameter conditions. This is primarily attributed to the increase in channel height from 50 µm to 100 µm, which expands the available volume for condensate formation. To quantify this relationship, we calculated the volumetric condensation factor, defined as the ratio of the final condensate volume to the fluid volume available in a unit cell formed by four pillars. Across all geometric conditions, this factor remained remarkably consistent at 0.06 ± 0.01, indicating that the Y-motif DNA initially at 10 µM was concentrated approximately 17-fold. We compared this factor to our previous work, where Y-motif condensation occurred within the confined space of liposomes.34 The 5 µM Y-motif in the liposome was condensed to 6.6 times, which is nearly half of the value in the present system. Considering the initial Y-motif concentration was also half, the ratio of Y-motif molecules to the condensate size agrees between two distinct systems. This observation supports that our VILV platform efficiently captures and concentrates nearly all DNA molecules within the vortex. These findings suggest that the VILV field functions as virtual, uniform fluidic compartments, within which Y-motif DNA molecules accumulate to form condensates with well-defined volume.
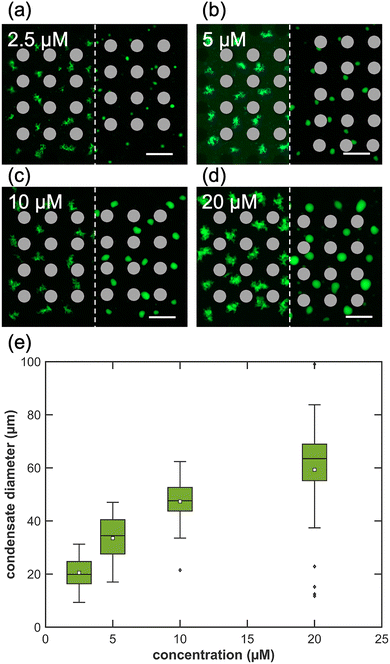
Controlling DNA condensate size by varying Y-motif concentration
To further explore control over DNA condensate size, we performed VILV-based condensate formation using four different concentrations of the 4nt-SE Y-motif: 2.5 µM, 5 µM, 10 µM, and 20 µM. All experiments were conducted within the same microstructures (D = 100 µm, 100 µm height) under identical vibration conditions.Representative images of condensates (Fig. 5a–d) demonstrate the transition from uniform shaggy aggregates to rounded shapes after 10 min of VIF application. The final condensate size at 240 min increased with higher initial DNA concentrations. During VIF application, we observed that larger condensates, occupying a greater fraction of the vortex volume, remained stationary, whereas smaller condensates formed at lower concentrations tended to circulate along the vortex periphery and occasionally migrated between neighbouring vortices. The box plot analysis shown in Fig. 5e provides a more detailed statistical perspective on the diameter distributions. It clearly illustrates a positive correlation, with median diameters systematically increasing with DNA concentration: 19.9 µm at 2.5 µM (N = 47), 34.5 µm at 5 µM (N = 35), 47.6 µm at 10 µM (N = 31), and 63.5 µm at 20 µM (N = 33). The IQRs were relatively consistent—8.4, 12.9, 8.9, and 13.8 µm, respectively—indicating that condensate size uniformity was well-maintained despite increasing average sizes. The distribution of condensate volumes for each condition, detailed in Fig. S8, further confirmed a linear relationship between Y-motif concentration and final condensate volume.
These results underscore that, under a stable flow field, DNA concentration serves as an effective and programmable parameter for modulating condensate size. Interestingly, we also observed a trade-off between condensate size and final morphology: larger condensates formed at higher concentrations exhibited less smooth surface contours compared to the smaller condensates formed at lower concentrations.
Construction of uniform patchy DNA condensates
We further explored the formation of more complex DNA condensates composed of different multi-arm nanostar motifs. It has been previously demonstrated that by introducing distinct SE sequences, Y-motif molecules with identical SEs will coalesce into condensates, while those with different SEs remain immiscible and form segregated condensates. The interaction between these orthogonal (immiscible) DNA condensates can be modulated by introducing a “linker” motif that includes both SE types within a single structure.17,21 Previously, we demonstrated that the morphology of such complex DNA condensates can be precisely tuned within monodisperse giant vesicles.46 Here, we tested a similar degree of control can be achieved in an aqueous environment using the VILV platform.We prepared two types of orthogonal 4nt Y-motifs, featuring either (5′-GCGC-3′) or (5′-GGCC-3′, referred to as orthY-motif) SEs. Additionally, a 6-arm linker DNA strand containing three of each SE type was designed to link immiscible condensates at their interfaces. The concentration ratio of each Y-motif to linker motif was set at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (see the SI, Table S3, for details). This solution was added to the D = 100 µm micropillar device and subjected to 20 min of vibration.
1 (see the SI, Table S3, for details). This solution was added to the D = 100 µm micropillar device and subjected to 20 min of vibration.
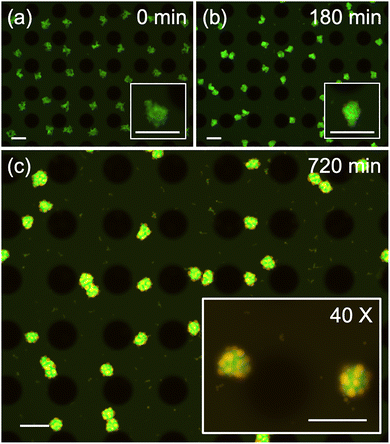
Time-lapse fluorescent imaging (Fig. 6; see also the SI, Movie S4) revealed that, despite identical length and GC content of the SEs, large Y-motif aggregates (depicted in green) appeared first after 5 min of VIF application (Fig. 6a), similar to the results shown in Fig. 6d. Subsequently, during the relaxation period without VIF, these shaggy condensates gradually become more compact. Around 3 h post-VIF, many small orange dots, representing the condensed orthY-motif, appeared on the surface of the green condensates (Fig. 6b). By 12 h, uniform patchy condensates were observed, consisting of green condensates (D = 42.3 ± 1.7 µm, N = 37) surrounded by numerous smaller orange condensates (D = 10.7 ± 4.1 µm, N = 300) (Fig. 6c). This transition resembles a typical phase separation process previously reported17 when the linker motif in a fully mixed condensate was enzymatically digested. We presume that during the initial formation of shaggy condensates, both orthY-motif and linker motifs were incorporated into the large Y-motif structures. As the condensates relaxed at room temperature, phase separation of the orthY-motif progressed on the surface of the Y-motif condensates. Over time, fusion among the small orthY-motif condensates led to their growth while remaining tethered to the parent Y-motif condensates through the linker. Notably, the resulting patchy condensates exhibited high uniformity and morphological consistency.
To confirm the role of the connecting motif in directing this specific morphology, we conducted a control experiment with reduced linker concentration (Fig. S9). When the concentration ratio of each Y-motif to the linker motif was reduced to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, two distinct condensates formed independently yet remained in contact with each other due to the weaker interaction. These results unequivocally demonstrate that the linker motif is the key component determining the morphology of the complex multi-component DNA condensates. A more comprehensive study on the generation of various structures through sequence design is currently underway.
1, two distinct condensates formed independently yet remained in contact with each other due to the weaker interaction. These results unequivocally demonstrate that the linker motif is the key component determining the morphology of the complex multi-component DNA condensates. A more comprehensive study on the generation of various structures through sequence design is currently underway.
Conclusions
In this study, we successfully developed a versatile microfluidic platform that utilizes VILV to precisely control the formation process of DNA condensates. Our results demonstrate that by employing micropillar arrays with diameters ranging from 25 to 100 µm, we can microscopically regulate and observe the condensation behavior of sticky Y-motif DNA nanostructures. The VILV generated in this system not only accelerates initial aggregate formation but also provides spatial control over condensate positioning within the aqueous environment. Further structural control can be achieved by adjusting key parameters such as ssDNA concentration and micropillar geometry. These findings highlight the platform's significant advantages for investigating LLPS and DNA condensation mechanisms. The ability of the VILV system to promote the formation of large, defined DNA condensates in an aqueous medium is especially valuable for applications requiring hierarchical condensate formation and responsiveness to external stimuli.Furthermore, this micropillar-based VIF platform offers distinct benefits. It significantly reduces the material costs and simplifies observation compared to conventional flow-based microfluidic systems that rely on external pumps. The platform also facilitates direct, real-time monitoring of condensate dynamics and can potentially integrate temperature or environmental controls. Furthermore, this system can be readily scaled up to obtain a large number of homogeneous condensates by simply enlarging the micropillar array substrate. Consequently, this system provides a simplified yet powerful tool for studying a broad range of biomolecular interactions and biomechanical processes, including those relevant to molecular computing.
This platform is expected to improve the reproducibility and quantitative understanding of the reaction kinetics in complex, DNA-based condensate systems and logic operations.18,47 We also envision its adaption for generating other types of monodisperse, phase-separated condensates (e.g., peptide/protein or synthetic-polymer coacervates) as reproducible and controllable models for artificial cells and microscale chemical reactors.48 The methodologies developed in this work are expected to significantly advance research in these fields.
Author contributions
HS supervised the project. ZH and KK designed the experimental system. ZH performed the fabrication, conducted the measurements, and analyzed the data. TH developed the VIF technology and performed PIV measurement. RY contributed to the refinement of the experiments. TM and MT provided valuable suggestions and modifications. ZH and HS drafted the manuscript. All authors have reviewed and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5mh01304f.Acknowledgements
This work was supported in part by JSPS grants (24K01320, 24H01155, 23K22725, 20H05935, and 24K08214), Japan Keirin Autorace foundation (JKA), and the Institute of Science and Engineering at the Chuo University. We thank Naoki ASAI in our group for performing additional experiments.References
- Y. Shin and C. P. Brangwynne, Science, 2017, 357, eaaf4382 CrossRef PubMed.
- C. D. Crowe and C. D. Keating, Interface Focus, 2018, 8, 20180032 CrossRef PubMed.
- N. A. Yewdall, A. A. M. André, T. Lu and E. Spruijt, Curr. Opin. Colloid Interface Sci., 2021, 52, 101416 CrossRef CAS.
- C. E. Sing and S. L. Perry, Soft Matter, 2020, 16, 2885–2914 RSC.
- D. Bracha, M. T. Walls and C. P. Brangwynne, Nat. Biotechnol., 2019, 37, 1435–1445 CrossRef CAS PubMed.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- E. Lattuada, M. Leo, D. Caprara, L. Salvatori, A. Stoppacciaro, F. Sciortino and P. Filetici, Front. Pharmacol., 2020, 11, 01345 CrossRef CAS PubMed.
- H. Udono, J. Gong, Y. Sato and M. Takinoue, Adv. Biol., 2023, 7, 2200180 CrossRef CAS PubMed.
- M. Takinoue, Interface Focus, 2023, 13, 20230021 CrossRef PubMed.
- Y. Sato and M. Takinoue, Biophysics, 2024, 21, e210010 CAS.
- D. Wang, P. Liu and D. Luo, Angew. Chem., Int. Ed., 2022, 61, e202110666 CrossRef CAS PubMed.
- K. Matsuura, K. Masumoto, Y. Igami, T. Fujioka and N. Kimizuka, Biomacromolecules, 2007, 8, 2726–2732 CrossRef CAS PubMed.
- S. Biffi, R. Cerbino, F. Bomboi, E. M. Paraboschi, R. Asselta, F. Sciortino and T. Bellini, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15633–15637 CrossRef CAS PubMed.
- N. Conrad, T. Kennedy, D. K. Fygenson and O. A. Saleh, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 7238–7243 CrossRef CAS PubMed.
- S. H. Um, J. B. Lee, N. Park, S. Y. Kwon, C. C. Umbach and D. Luo, Nat. Mater., 2006, 5, 797–801 CrossRef CAS PubMed.
- D. T. Nguyen, B. Jeon, G. R. Abraham and O. A. Saleh, Langmuir, 2019, 35, 14849–14854 CrossRef CAS PubMed.
- Y. Sato, T. Sakamoto and M. Takinoue, Sci. Adv., 2020, 6, eaba3471 CrossRef CAS PubMed.
- J. Gong, N. Tsumura, Y. Sato and M. Takinoue, Adv. Funct. Mater., 2022, 32, 2202322 CrossRef CAS.
- H. Udono, M. Fan, Y. Saito, H. Ohno, S. M. Nomura, Y. Shimizu, H. Saito and M. Takinoue, ACS Nano, 2024, 18, 15477–15486 CrossRef CAS PubMed.
- D. B. McIntosh, G. Duggan, Q. Gouil and O. A. Saleh, Biophys. J., 2014, 106, 659–666 CrossRef CAS PubMed.
- B. Jeon, D. T. Nguyen and O. A. Saleh, J. Phys. Chem. B, 2020, 124, 8888–8895 CrossRef CAS PubMed.
- C. Ma, L. Chen, L. Wang, W. Tong, C. Chu, Z. Yuan, C. Lv and Q. Zheng, Nat. Commun., 2022, 13, 5381 CrossRef CAS PubMed.
- S. Sadasivan, S. Pradeep, J. C. Ramachandran, J. Narayan and M. J. Gęca, Microfluid. Nanofluid., 2025, 29, 17 CrossRef CAS.
- P. Zhu and L. Wang, Lab Chip, 2017, 17, 34–75 RSC.
- N. A. Yewdall, A. A. M. André, T. Lu and E. Spruijt, Curr. Opin. Colloid Interface Sci., 2021, 52, 101416 CrossRef CAS.
- I. Ziemecka, V. Van Steijn, G. J. M. Koper, M. Rosso, A. M. Brizard, J. H. Van Esch and M. T. Kreutzer, Lab Chip, 2011, 11, 620–624 RSC.
- I. Ziemecka, V. Van Steijn, G. J. M. Koper, M. T. Kreutzer and J. H. Van Esch, Soft Matter, 2011, 7, 9878 RSC.
- D. van Swaay, T.-Y. D. Tang, S. Mann and A. de Mello, Angew. Chem., Int. Ed., 2015, 54, 8398–8401 CrossRef CAS PubMed.
- B.-U. Moon, N. Abbasi, S. G. Jones, D. K. Hwang and S. S. H. Tsai, Anal. Chem., 2016, 88, 3982–3989 CrossRef CAS PubMed.
- M. Shono, R. Ito, F. Fujita, H. Sakuta and K. Yoshikawa, Sci. Rep., 2021, 11, 23570 CrossRef CAS PubMed.
- E. Sokolova, E. Spruijt, M. M. K. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. S. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS PubMed.
- N. Deng and W. T. S. Huck, Angew. Chem., Int. Ed., 2017, 56, 9736–9740 CrossRef CAS PubMed.
- Q. Zhao, F. Cao, Z. Luo, W. T. S. Huck and N. Deng, Angew. Chem., Int. Ed., 2022, 61, e202117500 CrossRef CAS PubMed.
- R. Yoneyama, N. Morikawa, R. Ushiyama, T. Maruyama, R. Sato, M. Tsugane, M. Takinoue and H. Suzuki, JACS Au, 2025, 5, 3533–3544 CrossRef PubMed.
- M. K. Masukawa, Y. Okuda and M. Takinoue, Front. Genet., 2021, 12, 705022 CrossRef CAS PubMed.
- Z. Lin, T. Beneyton, J. Baret and N. Martin, Small Methods, 2023, 7, 2300496 CrossRef CAS PubMed.
- A. D. Slootbeek, M. H. I. Van Haren, I. B. A. Smokers and E. Spruijt, Chem. Commun., 2022, 58, 11183–11200 RSC.
- L. Tian, N. Martin, P. G. Bassindale, A. J. Patil, M. Li, A. Barnes, B. W. Drinkwater and S. Mann, Nat. Commun., 2016, 7, 13068 CrossRef CAS PubMed.
- L. Tian, M. Li, A. J. Patil, B. W. Drinkwater and S. Mann, Nat. Commun., 2019, 10, 3321 CrossRef PubMed.
- T. Hayakawa, S. Sakuma, T. Fukuhara, Y. Yokoyama and F. Arai, Micromachines, 2014, 5, 681–696 CrossRef.
- K. Kaneko, T. Osawa, Y. Kametani, T. Hayakawa, Y. Hasegawa and H. Suzuki, Micromachines, 2018, 9, 668 CrossRef PubMed.
- K. Kaneko, Y. Hasegawa, T. Hayakawa and H. Suzuki, J. Appl. Phys., 2024, 135, 154702 CrossRef CAS.
- K. Kaneko, M. Tsugane, T. Sato, T. Hayakawa, Y. Hasegawa and H. Suzuki, Sens. Actuators, B, 2025, 430, 137312 CrossRef CAS.
- Y. Sato and M. Takinoue, Nanoscale Adv., 2023, 5, 1919–1925 RSC.
- J. N. Zadeh, C. D. Steenberg, J. S. Bois, B. R. Wolfe, M. B. Pierce, A. R. Khan, R. M. Dirks and N. A. Pierce, J. Comput. Chem., 2011, 32, 170–173 CrossRef CAS PubMed.
- R. Yoneyama, R. Ushiyama, T. Maruyama, R. Sato, M. Tsugane, M. Takinoue and H. Suzuki, RSC Appl. Interfaces, 2025, 2, 1599–1606 RSC.
- T. Maruyama, J. Gong and M. Takinoue, Nat. Commun., 2024, 15, 7397 CrossRef CAS PubMed.
- A. J. Lin, A. Z. Sihorwala and B. Belardi, ACS Synth. Biol., 2023, 12, 1889–1907 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |