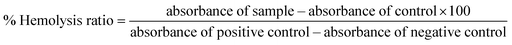Design, synthesis, and biological evaluation of sulfonamide-functionalized pyridine carbothioamides as potent tubulin-targeting anticancer agents
Fatima
Younas
 a,
Jahan Zaib
Arshad
a,
Jahan Zaib
Arshad
 *a,
Waqas Ali
Shah
*a,
Waqas Ali
Shah
 *b,
Sundas
Arshad
c,
Adnan
Ashraf
d,
Syed Shoaib Ahmad
Shah
*b,
Sundas
Arshad
c,
Adnan
Ashraf
d,
Syed Shoaib Ahmad
Shah
 *e,
Muhammad Asam
Raza
*e,
Muhammad Asam
Raza
 f,
Amara
Mumtaz
f,
Amara
Mumtaz
 g,
Nasir
Shahzad
g,
Nasir
Shahzad
 h and
Tariq
Javed
h and
Tariq
Javed
 i
i
aDepartment of Chemistry, Government College Women University Sialkot, Sialkot, 51310, Pakistan. E-mail: jahanzaib.arshad@gcwus.edu.pk
bSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang, 453007, China. E-mail: waqasalishah1@hotmail.com
cInternational Research Institute for Steel Technology, Collaborative Innovation Center for Advanced Steels, Wuhan University of Science and Technology, Wuhan 430081, China
dDepartment of Chemistry, The University of Lahore, Lahore, Pakistan
eDepartment of Chemistry, School of Natural Sciences, National University of Sciences and Technology, Islamabad, 44000, Pakistan. E-mail: shoaib03ahmad@outlook.com
fDepartment of Chemistry, Hafiz Hayat Campus, University of Gujrat, Gujrat, Pakistan
gDepartment of Chemistry, COMSATS University Islamabad, Abbottabad Campus, 22060, Abbottabad, Pakistan
hGovernment Postgraduate College No.1 Abbottabad, Abbottabad, Pakistan
iDepartment of Chemistry, University of Sahiwal, Sahiwal, Pakistan
First published on 24th October 2025
Abstract
Pyridine carbothioamides (PCAs) are recognized for their gastric mucosal protective effects and low in vivo toxicity, making them attractive scaffolds for anticancer drug development. In this study, a series of N-phenyl 4-substituted and 2,4-disubstituted PCAs (1–8) incorporating a sulfonamide pharmacophore were synthesized, fully characterized, and evaluated as tubulin polymerization inhibitors. The compounds were tested against four cancer cell lines (A549, MCF-7, PC-3, HepG2) with colchicine and doxorubicin as reference drugs. Among them, compounds 3 and 5 exhibited potent cytotoxicity, being 2–6-fold more active than colchicine and up to 2.5-fold stronger than doxorubicin in PC-3 cells. Importantly, both showed ∼4-fold lower toxicity toward normal HLMEC cells and displayed higher selectivity towards tested cancer cells than doxorubicin. Tubulin polymerization assays confirmed their activity, with IC50 values of 1.1 μM (3) and 1.4 μM (5), outperforming colchicine (10.6 μM) and CA-4 (2.96 μM). Molecular docking revealed strong binding at the colchicine site, supported by favorable inhibition constants and free binding energies. In silico ADME predictions indicated that the most lipophilic compounds 3 and 5 demonstrated favorable drug-likeness, as expected from computational studies, along with excellent gastrointestinal absorption, favorable bioavailability, and low hemolytic activity. Collectively, these findings highlight compounds 3 and 5 as promising lead candidates for the development of orally active anticancer and antimitotic agents.
Introduction
Cancer is a major contributor to global mortality, causing about 1 in every 6 deaths, with 20 million new cases and 9.7 million deaths reported in 2022. By 2050, the burden is expected to rise by 77%.1,2 Cancer is a complex and life-threatening disease arising from uncontrolled cell proliferation. Currently, anticancer agents often lack selectivity, damaging both healthy and malignant cells, and are further limited by intrinsic or acquired resistance mechanisms.3,4 These limitations underscore the need for new, targeted chemotherapeutics with high potency and reduced toxicity.Microtubules, integral cytoskeletal filaments, are indispensable in eukaryotic cells.5–8 They play an important role in the maintenance of cell shape and mitotic cell division and ensuring the precise segregation of duplicated chromosomes into two identical sets prior to cleavage of the cell into two daughter cells. The pivotal role of microtubules in mitosis and cell division renders them a critical target for anticancer drugs.9,10 Microtubule targeting agents not only disrupt the mitotic process but also induce apoptotic cell death during interphase,11 exerting their effects through interactions with tubulin at three primary binding sites: the taxane/epothilone, the vinca alkaloid, and the colchicine site. For taxanes and vinca alkaloids, there are many drugs developed. However, these agents are prone to multidrug resistance through P-glycoprotein efflux. In contrast, colchicine-site inhibitors (CSIs) are less susceptible to MDR and thus attractive alternatives.12–14 Combretastatin A-4 (CA-4) and its prodrug fosbretabulin illustrate the promise of CSIs, though issues of solubility and stability have limited their development.15–17
Sulfonamides represent an important class of pharmacologically active agents, with drugs containing this pharmacophore widely employed for the treatment of numerous diseases. Sulfonamide containing compounds have been used in clinical trials for years owing to their various pharmacological activities including anti-bacterial,18,19 anti-inflammatory,20 anti-diabetic,21,22 antimalarial,23,24 antiviral,25,26 carbonic anhydrase inhibitors,27,28 anti-tubercular,29 antioxidant,30,31 and anticancer.32 Over 150 FDA-approved drugs incorporate sulfonyl groups, including celecoxib, meloxicam, piroxicam, and sulfasalazine.33,34 Sulfonamides are versatile pharmacophores present in many FDA-approved drugs with diverse activities, including anticancer effects. Clinically relevant examples include belinostat (HDAC inhibitor for T-cell lymphoma),35,36 amsacrine (topoisomerase II inhibitor for leukemia and lymphomas),37 and dabrafenib (BRAF inhibitor for melanoma)38 (Fig. 1). In addition, several sulfonamide derivatives such as ABT-751 (E7010), batabulin, and T900607 have advanced to clinical trials as colchicine-site tubulin inhibitors, though their progress has been limited by toxicity, solubility, or resistance issues (Fig. 2).39–42
 | ||
| Fig. 1 Clinically approved anticancer drugs containing a sulfonamide pharmacophore.35,38,49–51 | ||
 | ||
| Fig. 2 The lead tubulin polymerization inhibitors bearing the sulfonamide pharmacophore.39,52 | ||
Pyridine-2-carbothioamides (PCAs) are low-toxicity ligands that exhibit potent antiproliferative activity, influenced by phenyl-ring substitutions. Lipophilic derivatives proved strong cytotoxic agents with enhanced drug-likeness.43–45 and a hydroxamic acid-functionalized PCA (JAZZ-90) demonstrated superior cytotoxicity, HDAC inhibition, and anti-angiogenic activity, highlighting their potential as multitargeted anticancer agents.46
Building on these findings and guided by the need for more potent anticancer agents, we hypothesized that functionalizing bioactive pyridine-2-carbothioamides (PCAs) with aryl sulfonamide moieties could yield novel colchicine-site tubulin inhibitors with enhanced potency, selectivity, bioavailability, and drug-like properties. This design was guided by the pharmacophore model for colchicine-site binders, which typically requires a heterocyclic ring (as in ABT-751/E7010-type structures) to improve pharmacological targeting, a substituted aromatic unit with electron-rich or polar features to fit tightly within the binding pocket, and a linker to optimally orient these groups within the binding site.47,48 The synthesized PCA scaffold contributes a pyridine ring and thioamide linker capable of hydrogen-bonding and electronic interactions, while the sulfonamide-substituted phenyl ring introduces additional polarity, cytotoxic potential, and drug-likeness. Notably, these features structurally align with the clinically investigated sulfonamide-based colchicine-site inhibitor ABT-751 (E7010) reinforcing the strategic depth of our design. Inspired by the previously conducted SAR studies of PCAs,44 small substituents (methyl, dimethyl, fluoro, methoxy, and hydroxy) were introduced at the N-phenyl 2- and 4-positions to systematically evaluate the influence of steric hindrance, lipophilicity, and electronic effects on tubulin binding and anticancer activity. The resulting derivatives were systematically evaluated for cytotoxicity against multiple cancer and normal cell lines, tubulin polymerization inhibition, and molecular docking interactions, along with correlations to physicochemical/ADME and hemolytic profiles, to identify promising candidates for orally active anticancer therapy.
Results and discussion
Pyridine carbothioamides (PCAs) are versatile N,S-bidentate bioactive ligands with multitargeted nature towards biomolecules and also famous for strong coordination with metal ions resulting in the formation of a library of organometallic and coordination compounds.44–46,53–55 PCAs were previously reported as gastric mucosal protectants with reduced acute toxicity in vivo,43 and as strong cytotoxic agents with potential to inhibit proteins such as histone deacetylase involved in tumor formation or progression.44,46 We synthesized novel derivatives of N-phenyl 4-substituted or 2,4-disubstituted PCAs 2–8 functionalized with a sulfonamide moiety, a common pharmacophore that interacts with the active sites of various proteins or enzymes.56–58 The new PCAs 2–8 containing the sulfonamide motif were synthesized by a method from the literature that was previously utilized to prepare N-(4-sulfamoylphenyl)pyridine-2-carbothioamide 1.53 In short, the sulfanilamide derivatives (1 eq.) were refluxed for 72 hours with 2-picoline (2 eq.) and sulfur (2.5 eq.) in the presence of catalytic amounts of sodium sulfide nonahydrate (0.5%) to afford the synthesis of PCA derivatives 1–8 functionalized with the sulfonamide motif (Scheme 1). After work up, recrystallization from acetonitrile has been used to purify and yield the PCAs from 69% to 85% yield which is comparable to what has been previously reported for related compounds and their melting points were determined in the range of 131 to 158 °C.The synthesized PCAs 1–8 were characterized by FTIR, NMR, ESI-MS and elemental analysis. The FTIR spectra of the synthesized compounds 1–8 were obtained in the 4000–400 cm−1 range. The FTIR spectra confirmed the presence of a thioamide group in compounds 1–8, with significant C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S stretching vibrations from 1690–1720 cm−1 and modest N–H sharp stretching vibrations in the range of 3355–3439 cm−1. The synthesis of sulfonamide-substituted PCAs 1–8 was also confirmed by the two strong symmetric and asymmetric stretching vibrations of the SO2 group in the range of 1115–1152 cm−1 and 1302–1325 cm−1, respectively. The stretching vibrations of aromatic C–H groups were observed in the 3084–3439 cm−1 range. In compounds 1 and 4–8, the existence of two symmetric and asymmetric vibrations in the range of 3215–3238 cm−1 and 3248–3291 cm−1 revealed the presence of the NH2 group (Fig. 3) while these peaks are not observed in compounds 2 and 3 due to replacement of primary amine hydrogen by methyl groups. In compound 7, the intense broad peak at 3361 cm−1 was assigned to the –OH group while in 8 the two strong symmetric and asymmetric vibrations at 1148 cm−1 and 1320 cm−1 respectively, were assigned to the –NO2 group.
S stretching vibrations from 1690–1720 cm−1 and modest N–H sharp stretching vibrations in the range of 3355–3439 cm−1. The synthesis of sulfonamide-substituted PCAs 1–8 was also confirmed by the two strong symmetric and asymmetric stretching vibrations of the SO2 group in the range of 1115–1152 cm−1 and 1302–1325 cm−1, respectively. The stretching vibrations of aromatic C–H groups were observed in the 3084–3439 cm−1 range. In compounds 1 and 4–8, the existence of two symmetric and asymmetric vibrations in the range of 3215–3238 cm−1 and 3248–3291 cm−1 revealed the presence of the NH2 group (Fig. 3) while these peaks are not observed in compounds 2 and 3 due to replacement of primary amine hydrogen by methyl groups. In compound 7, the intense broad peak at 3361 cm−1 was assigned to the –OH group while in 8 the two strong symmetric and asymmetric vibrations at 1148 cm−1 and 1320 cm−1 respectively, were assigned to the –NO2 group.
In the 1H NMR spectra of compounds 2–8 in DMSO-d6 (Fig. S1–S7), the presence of thioamide protons resonating at approximately 12 ppm confirmed the formation of compounds. In compounds 4–8 sulfonamide protons were observed as a singlet at about 7 ppm while in compound 2 containing an N-methyl substituted sulfamoyl group [–SO2NH(CH3)] the secondary amide proton was shielded due to the presence of a methyl group and detected at 5.30 ppm. Furthermore, in compound 3 containing an N,N-dimethyl substituted sulfamoyl group [–SO2N(CH3)2], methyl groups protons were observed as a singlet at 2.94 ppm. The pyridine ring protons (H1–H4) were detected in the range of 7.50–8.80 ppm, whereas the aromatic phenyl (H8–12) protons exhibited signals between 6.60–8.03 ppm. In compounds 5–7, the presence of methyl, methoxy and hydroxyl proton(s) were observed as singlets at 3.32 ppm (Fig. 4), 3.79 ppm and 5.59 ppm, respectively.
In the 13C NMR spectra of newly synthesized compounds 2–8 (Fig. S8–S14), the presence of quaternary carbon (C6) confirmed the presence of the thioamide group in the range of 197.2–199.2 ppm. The quaternary carbon of the pyridine ring (C5) was detected at ∼148 ppm whereas in the pyridine ring the quaternary carbon (C10) containing a sulfonamide or N-substituted sulfamoyl group exhibited signals between 134.5–135.5 ppm. In compounds 5–7 the quaternary carbon (C8) containing the electron donating groups was resonating at ∼128 ppm. Similarly, the presence of electron withdrawing substituents in compounds 4 and 8 resulted in downfield shift of quaternary carbon (C8) by ∼25 ppm and was observed in the range of approximately 152.1–153.3 ppm. The chemical shifts of the individual carbon atoms of the pyridine ring were observed at 122.2–149.1 ppm while those attributed to the aromatic phenyl carbons appeared in the range of 116.0–153.3 ppm. In compounds 2 and 3N-methyl substituents were resonating at 29.3 ppm and 37.9 ppm, respectively. Similarly, in compounds 5 and 6, the carbon atoms of methyl and methoxy substituents at the C8 position appeared at 20.9 ppm (Fig. 5) and 56.2 ppm respectively.
The ESI-mass spectra of compounds 2–8 are supported by the formation of a molecular ion with a proton at m/z 308.0541, 322.0662, 312.0254, 308.0531 (Fig. 6), 324.0502, 310.0334 and 339.0239, respectively, which is in close agreement with the calculated values (Fig. S15–S21). The elemental analysis confirmed the formation and purity of the compounds by providing data in close agreement with the theoretical values.
In vitro anticancer studies
Previously, we have reported that pyridine carbothioamides exhibited strong anticancer activities in the low micromolar range against human colorectal carcinoma (HCT116), non-small cell lung carcinoma (NCI-H460), cervical carcinoma (SiHa) and colon carcinoma (SW480) cells in the sulforhodamine B (SRB) assay.44 Recently, we have synthesized sulfonamide-substituted PCAs 1–8, and evaluated their antiproliferative activity in comparison with doxorubicin and colchicine against human non-small lung carcinoma cells (A549), breast carcinoma cells (MCF-7), prostate cancer cells (PC3) and liver cancer cells (HepG2) using the MTT assay, with doxorubicin and colchicine as reference drugs (Table 1).| Compounds | IC50 value (μM) cancer cells | |||
|---|---|---|---|---|
| A549 | MCF-7 | PC3 | HepG2 | |
| 1 | 63 ± 2.5 | >200 | >200 | 71 ± 1 |
| 2 | 6.7 ± 1.3 | 9.1 ± 0.9 | 7.3 ± 0.3 | 5.9 ± 0.3 |
| 3 | 1.3 ± 0.2 | 4.9 ± 0.2 | 2.3 ± 0.4 | 1.2 ± 0.1 |
| 4 | 7.7 ± 1.3 | 12 ± 2.1 | 13 ± 1.8 | 9.1 ± 1.5 |
| 5 | 2.3 ± 0.9 | 5.9 ± 1.1 | 2.9 ± 0.6 | 3.7 ± 0.5 |
| 6 | 14 ± 2.4 | 36 ± 3 | 31 ± 2.7 | 21 ± 3.1 |
| 7 | >200 | >200 | >200 | >200 |
| 8 | >200 | >200 | >200 | >200 |
| Doxorubicin | 0.69 ± 0.02 | 1.78 ± 0.05 | 7.97 ± 0.09 | 1.63 ± 0.03 |
| Colchicine | 0.043 ± 0.01 | 10.44 ± 0.15 | 4.36 ± 0.11 | 7.38 ± 0.15 |
Among all the synthesized PCAs bearing sulfonamide, compounds 2, 3 and 5 exhibited potent cytotoxicity against all investigated cancer cell lines with IC50 values in the low micro molar range of 1.2–9.1 μM. However, compound 4 containing the o-fluoro substituent showed slightly reduced anticancer activity (7.7–13 μM) compared with the most active compounds 2, 3 and 5. A moderate antiproliferative behaviour was exhibited by compound 6 bearing the o-methoxy substituent with an IC50 value in the range of 14–36 μM. On the other hand, the already reported sulfonamide-substituted PCA 1 showed moderate cytotoxicity against A549 (IC50 = 63 μM) and HepG2 (IC50 = 71 μM), Interestingly, in our previous study the mono-substituted PCA containing the sulfonamide moiety 1 showed moderate cytotoxicity to inactivity against HCT116, NCI-H460, SiHa and SW480 cancer cells while the addition of a second substituent at the N-phenyl 2-position in compounds 4 and 5 or the replacement of the hydrogen atoms of the NH2 group of sulfonamides with methyl groups in 2 and 3 resulted in a marked increase in anticancer activity. However, compounds 7 and 8 bearing –OH and –NO2 groups as second substituents proved inactive, respectively.
In comparative analysis, compound 3 proved as the most cytotoxic agent (1.2–4.9 μM) against all investigated cancer cells and displayed 2-, 3.5- and 6-fold higher potency against MCF-7, PC3 and HePG2, respectively than reference drug colchicine. Similarly, in comparison to colchicine, the second most active compound 5 revealed ∼1.5 times higher cytotoxicity against PC3 and 2 times higher anticancer activity against MCF-7 and HepG2 cancer cell lines. On the other hand, in comparison with standard anticancer drug doxorubicin, compounds 3 and 5 exhibited ∼2-fold and 2.7-fold higher antiproliferative activity only against PC3 cells, respectively. In general the synthesized compounds observed the following trends in terms of cytotoxicity 3 > 5 > 2 > 4 > 6 > 1 > 7 = 8 (Table 1). Consequently, compounds 3 and 5 demonstrated potent anticancer activity, suggesting their potential as promising candidates for further development.
Additionally, the cytotoxicity of the anticancer-active PCAs (2–6), along with the reference drug doxorubicin, was evaluated against a normal human cell line, human lung microvascular endothelial cells (HLMEC), using the MTT assay (Table 2). Among these, the most potent anticancer compounds, 3 and 5, exhibited minimal cytotoxicity toward HLMEC cells, with IC50 values of 63.29 μM and 62.11 μM, respectively. In contrast, the moderately active PCAs 2, 4, and 6 showed moderate cytotoxicity toward HLMEC cells, with IC50 values of 48.53 μM, 35.71 μM, and 29.45 μM, respectively. Comparative analysis revealed that compounds 3 and 5 demonstrated approximately 4-fold lower toxicity against HLMEC cells than doxorubicin (IC50 = 15.91 μM). However, compound 6 was notably more cytotoxic towards normal cells, exhibiting nearly 2-fold higher toxicity than doxorubicin. Overall, the relative cytotoxicity of the synthesized PCAs (2–6) against HLMEC cells, compared with doxorubicin, followed the order: 6 > doxorubicin > 4 > 2 > 5 > 3.
| Compounds | IC50 value (μM) normal cells |
|---|---|
| HLMEC | |
| 2 | 48.53 ± 1.3 |
| 3 | 63.29 ± 1.9 |
| 4 | 35.71 ± 2.2 |
| 5 | 62.11 ± 1.5 |
| 6 | 9.45 ± 1.2 |
| Doxorubicin | 15.91 ± 1.8 |
Selectivity index (SI)
The drug's selectivity was assessed using the selectivity index (SI), defined as the ratio of its cytotoxic activity in normal cell lines to that in cancer cell lines. Specifically, the SI was calculated as the IC50 value in a normal cell line (HLMEC) divided by the IC50 value in the corresponding tumor cell line, according to the following equation:An SI value above 1.0 indicates preferential cytotoxicity toward cancer cells over normal cells.59 The SI values of the tested compounds (2–6) and doxorubicin are summarized in Table 3. Among the anticancer-active sulfonamide-substituted PCAs (2–6), compounds 2–5 displayed selectivity against all tested cancer cell lines, whereas compound 6 showed only weak cytotoxicity. Within the series, compounds 3 and 5 emerged as the most cytotoxic and exhibited the highest selectivity indices across all cancer cell lines. Comparative analysis revealed that compounds 3 and 5, bearing N,N-dimethyl and methyl substituents, respectively, were more selective than doxorubicin. Notably, compounds 2 and 4 demonstrated 1.5- and 3.5-fold higher selectivity than doxorubicin against PC-3 cells. Furthermore, compound 3, the most potent anticancer agent in the series, exhibited 2-, 1.5-, 14-, and 5.5-fold greater selectivity than doxorubicin against A549, MCF-7, PC-3, and HepG2 cells, respectively. Overall, the selectivity trend among the tested compounds was: 3 > 5 > 2 > 4 > 6 (Table 3).
| Compounds | Selectivity index (SI) | |||
|---|---|---|---|---|
| A549 | MCF-7 | PC3 | HepG2 | |
| 2 | 7.24 | 5.33 | 6.66 | 8.22 |
| 3 | 48.68 | 12.91 | 27.51 | 52.74 |
| 4 | 4.63 | 2.97 | 2.74 | 3.92 |
| 5 | 27.0 | 10.52 | 21.41 | 16.78 |
| 6 | 0.67 | 0.26 | 0.30 | 0.45 |
| Doxorubicin | 23.05 | 8.9 | 1.99 | 9.76 |
In vitro tubulin polymerization inhibition studies
To determine whether the antiproliferative activity of the synthesized compounds was linked to tubulin inhibition, an in vitro tubulin polymerization inhibition experiment was conducted in comparison with the reference drugs colchicine and combretastatin (CA-4) (Table 4 and Fig. 7). Among the synthesized PCAs bearing the sulfonamide moiety, compounds 3, 4, 5 and 8 showed the highest inhibition of tubulin polymerization in the range of 1.1–7.3 μM, while compounds 2, 6 and 7 displayed moderate inhibitory activity with IC50 in the range of 29.7–59.6 μM. Interestingly, compounds 3–5 which proved as potent inhibitors of tubulin polymerization also showed potent anticancer activity. Surprisingly the anticancer inactive sulfonamide-substituted PCA bearing the –NO2 group 8 showed effective inhibition capacity towards microtubules but proved inactive in the cytotoxic assay. This discrepancy may occur because tubulin inhibition alone is not always sufficient to induce cell death. Additional factors, such as cellular uptake, efflux mechanisms, metabolic stability, and intracellular target accessibility, can significantly influence the overall cytotoxic outcome. Similar inconsistencies between tubulin inhibition and cytotoxicity have also been reported in the literature, where limited permeability or pharmacokinetic properties restricted cellular efficacy despite measurable tubulin interaction.17,60,61 Moreover, compound 1 was found inactive to inhibit the polymerization of tubules. In general, the following trend was observed within the synthesized compounds in terms of tubulin polymerization inhibition: 3 > 5 > 4 > 8 > 2 > 6 > 7 > 1 (Table 4).| Compounds | IC50 value (μM) |
|---|---|
| Tubulin polymerization inhibition | |
| 1 | >100 |
| 2 | 29.7 ± 2.3 |
| 3 | 1.1 ± 0.1 |
| 4 | 2.7 ± 0.1 |
| 5 | 1.4 ± 0.09 |
| 6 | 45.2 ± 2.1 |
| 7 | 59.6 ± 3.3 |
| 8 | 7.3 ± 0.2 |
| CA-4 | 2.93 ± 0.02 |
| Colchicine | 10.5 ± 0.03 |
 | ||
| Fig. 7 Graphical representation of tubulin polymerization inhibitory activity IC50 (μM) of PCAs 1–8 and reference drugs CA-4 and colchicine. | ||
In comparison, as the most potent inhibitor of microtubules 3 manifested ∼2.5 and ∼9.5 times higher inhibitory activity than reference drugs CA-4 and colchicine, respectively. Similarly, compounds 5 exhibited 2- and ∼7.5-fold higher tubulin inhibitory potential than CA-4 and colchicine, respectively, while compound 4 displayed ∼4-fold antimitotic capacity than colchicine. On the other hand, compound 8 demonstrated ∼1.5 times higher microtubule inhibition than colchicine. The results suggest that tubulin inhibition may contribute to the observed cytotoxic activity; however, additional studies such as apoptosis assays, cell-cycle analysis, and microtubule imaging will be necessary to fully elucidate the underlying mechanism of action. In future work, we plan to conduct these studies to determine the precise mechanism of action of these compounds. Nonetheless, the present findings provide a strong foundation for the further development and optimization of sulfonamide-functionalized PCAs, particularly the lead molecules 3 and 5, as promising anticancer agents.
Molecular docking studies
Molecular docking is a valuable tool in drug discovery due to its capability to predict the conformation of ligands or molecules within the target binding site.62 In the present stud, molecular docking of the synthesized PCAs 1–8 and CA-4 was performed at the colchicine binding site of tubulin to evaluate their inhibition constants, binding energies, and binding interactions, and to compare these parameters with the standard drug CA-4 (Tables 5 and S1). All the synthesized compounds 1–8 displayed binding energies and inhibition constants in the range of −8.66 to −14.91 kcal mol−1 and 1.26 to 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 nM, respectively. Compounds 3, 4 and 5 showed strong binding potentials towards tubulin with inhibition constants in the low nanomolar range of 1.26–9.58 nM and revealed slightly stronger binding energies (−13.57 to −14.91 kcal mol−1) than that of CA-4 with inhibition constant = 11.92 nM and binding energy −13.39 kcal mol−1 which is consistent with the previously reported value of −13.42 kcal mol−1.63 However, the rest of compounds 2, 6, 7 and 8 with moderate inhibition constants (18.35–67.13 μM) showed lower free binding energies (−12.55 to −9.79 kcal mol−1) than that of CA-4, while compound 1 exhibited the lowest binding energy of −8.66 kcal mol−1 and proved inactive (inhibition constant = 83
700 nM, respectively. Compounds 3, 4 and 5 showed strong binding potentials towards tubulin with inhibition constants in the low nanomolar range of 1.26–9.58 nM and revealed slightly stronger binding energies (−13.57 to −14.91 kcal mol−1) than that of CA-4 with inhibition constant = 11.92 nM and binding energy −13.39 kcal mol−1 which is consistent with the previously reported value of −13.42 kcal mol−1.63 However, the rest of compounds 2, 6, 7 and 8 with moderate inhibition constants (18.35–67.13 μM) showed lower free binding energies (−12.55 to −9.79 kcal mol−1) than that of CA-4, while compound 1 exhibited the lowest binding energy of −8.66 kcal mol−1 and proved inactive (inhibition constant = 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 nM) towards tubulin enzyme. Notably, lead compounds 3 and 5 demonstrated outstanding binding energies and inhibition constants along with excellent tubulin inhibitory and cytotoxic potential, and could be developed as potent anticancer agents targeting tubulin polymerization.
700 nM) towards tubulin enzyme. Notably, lead compounds 3 and 5 demonstrated outstanding binding energies and inhibition constants along with excellent tubulin inhibitory and cytotoxic potential, and could be developed as potent anticancer agents targeting tubulin polymerization.
| Compounds | Inhibition constant (nM) | Binding energy (kcal per mole) |
|---|---|---|
| 1 | 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 700 |
−8.66 |
| 2 | 37.6 | −11.36 |
| 3 | 1.26 | −14.91 |
| 4 | 9.58 | −13.57 |
| 5 | 4.30 | −14.05 |
| 6 | 52.95 | −9.93 |
| 7 | 67.13 | −9.79 |
| 8 | 18.35 | −12.55 |
| CA-4 | 11.92 | −13.39 |
In 3D molecular docking analysis, the synthesized PCAs 1–8 exhibited strong hydrogen bonding and several lipophilic interactions with amino acid residues within the colchicine binding pocket of the tubulin enzyme (Fig. 8). Moreover, to perform a comparative analysis, we docked the reference drug CA-4 within the colchicine binding site of tubulin. CA-4 exhibited one hydrogen bond with Thr353(B) via one of the methoxy groups of the trimethoxylated phenol moiety, along with five carbon–hydrogen bond interactions with Val315(B), Asn350(B), Asn258(B), Ala317(B), and Thr179(A) through the methoxy groups of both phenolic moieties. In addition, CA-4 exhibited a π–σ interaction with Leu248(B) through the phenolic moiety containing 3,4,5-trimethoxy groups and showed six π–alkyl interactions with Ala354(B), Ala316(B), Ala180(A), Val181(A), Lys352(B), and Met259(B) through both phenyl rings and their attached methoxy groups. These interactions contributed to the stability of the ligand–protein complex, with a binding energy of −13.39 kcal mol−1.
In comparative analysis, the most potent inhibitor of tubulin 3 showed two H-bonds with Asn101(A) and Lys254(B) through one oxygen atom of the sulfonyl group and two C–H bonds with Ser178(A) and Glu183(A) through the amine group of the sulfonamide moiety. Moreover, the pyridine and phenyl ring of compounds 3 demonstrated seven pi–alkyl interactions with Ala180(A), Ala250(B), Ala316(B), Lys352(B), Met259(B), Val181(A), Leu248(B) improving significantly the stability of the ligand/receptor complex to score the best free binding energy of −14.91 kcal mol−1 among the docked compounds. Compound 5, the second most potent tubulin inhibitor in the series, formed one hydrogen bond with Val315(B) via the oxygen atom of the SO2 group. Additionally, Thr314(B) and Asp251(B) established two C–H interactions with the sulfonamide moiety and the pyridine ring, respectively. Moreover, the pyridine and phenyl ring formed eight lipophilic interactions involving one pi–sulfur with Met259(B), two pi–sigma interactions with Ala250(B) and Leu255(B), and five pi–alkyl interactions with Ala180(B), Ala181(B), Ala316(B), Leu248(B), and Lys352(B), which augmented the ligand/receptor complex stability with an excellent free binding energy of −14.05 kcal mol−1. Compound 4, ranked as the third highest inhibitor of tubulin, formed two hydrogen bonds with Ala317(B) and Lys352(B) through –NH2 of the sulfonamide moiety. Also, it formed two additional hydrogen bonds with Val238(B) and Cys241(B) through –NH of thioamide and the o-fluoro substituent in the phenyl ring, respectively. Moreover, the additional lipophilic interactions involved two pi–sigma, one pi–sulfur and five pi–alkyl interactions with the pyridine and phenyl ring resulting in an increased stability of the ligand/receptor complex with a free binding energy of −13.57 kcal mol−1. Surprisingly, the PCA derivative 8 bearing the nitro substituent proved inactive in the cytotoxic assay but ranked fourth both in tubulin inhibition and docking studies and demonstrated a significant inhibition constant (18.35 nM) with a free binding energy of −12.55 kcal mol−1. In compound 8, the amine group of sulfonamide formed two H-bonds with Val315(B) and Asn350(B), and one H-bond with Asn258(B) through the nitro substituent in the phenyl ring. Furthermore, one additional C–H bond with Thr314(B) and several lipophilic interactions with Leu255(B), Met259(B), Ala250(B), Ala316(B), and Cys241(B) may contributed its enhanced interaction towards tubulin enzyme.
The PCA derivatives 2, 6 and 7, which showed moderate tubulin inhibition activity among the synthesized compounds, showed moderate binding affinities of −11.36 kcal mol−1, −9.93 kcal mol−1 and −9.79 kcal mol−1 respectively which are comparatively lower than 3, 4, 5 and CA-4.
In further investigation, the oxygen atoms and amine group of the sulfonamide moiety of compounds 2 and 6 demonstrated three H-bonds with Val238(B), Leu242(B), Cys241(B) and Val315(B), Met259(B), Asn350(B), respectively. Unfortunately, compound 7 established the highest four H-bonds among the docked compounds with Val315(B), Lys254(B), Asn258(B), and Leu255(B) through the oxygen and amine of sulfonamide, –NH of thioamide and hydroxyl group of the phenyl ring but showed moderate binding affinity towards tubulin enzyme. Likewise other compounds, ligands 2, 6 and 7 formed several lipophilic interactions with the pyridine and phenyl ring. However, in the series only compound 1 established one H-bond with Val315(B) through the amine of the sulfonamide moiety, hence resulting in a very weak binding affinity of −8.66 kcal mol−1, and also proved inactive in tubulin inhibition assay. These in silico observations revealed that 3 and 5 containing strong electron donating N,N-dimethyl and methyl substituents manifested the highest binding interactions, affinities and inhibition constants as compared to reference drug CA-4, indicating their potential as lead structures for further exploration as tubulin assembly inhibitors.
While molecular docking in this study provides useful preliminary insights into binding orientations and relative affinities, its static nature and simplified scoring functions limit accuracy in capturing protein flexibility and solvent effects. More advanced computational methods, such as molecular dynamics (MD) simulations and free energy calculations (FEP/MM-PBSA), could offer deeper and more reliable insights into protein–ligand interactions. These approaches were beyond the scope of the present study due to resource and expertise constraints but will be considered in future work to further validate the findings.
Hemolysis studies
To ensure safe usage, therapeutic agents must be tested in vitro for blood compatibility. The hemolytic assay is a standard test for determining the bioactive compounds' initial hemocompatibility.64 In view of this, the hemolytic activity of the most potent anticancer and antimitotic agents 3 and 5 in comparison with reference anticancer drug doxorubicin in mouse red blood cells was assessed to gain further insight into their toxicity (Fig. 9). At a low concentration of 20 μg mL−1 both compounds 3 and 5 displayed hemolytic activity in the range of ∼2.5–3% while doxorubicin reported 21.8% toxicity.65 When the concentration is increased to 30 and 40 μg mL−1, the PCAs 3 and 5 showed about ∼6–7% hemolytic activity, which was significantly lower than that of doxorubicin, which displayed 36.4% and 49.5% hemolytic activity at the same concentrations, respectively.65 In general, at concentrations of 20, 30 and 40 μg mL−1, the most potent compound 3 in the series displayed 8.5-, 6- and 8-fold lower toxicity, respectively compared to the standard anticancer drug doxorubicin. | ||
| Fig. 9 Percentage of haemolysis of mouse blood cells induced by sulfonamide-substituted PCAs 3 and 5 at various concentrations in comparison to doxorubicin.65 | ||
Lipophilicity
Lipophilicity is associated with enhanced anticancer efficacy due to its impact on cellular absorption and oral bioavailability of anticancer drugs.66,67 Higher lipophilicity is accounted as beneficial for compounds to move across cellular membranes68 and is generally indicated as the octanol/water partition coefficient (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P). The octanol/water partition coefficients (clog
P). The octanol/water partition coefficients (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) of the sulfonamide-substituted PCAs 1–8 were evaluated using ChemDraw 12.0, Molinspiration (https://www.molinspiration.com), and ALOGPS 2.1 and compared with standard anticancer drugs doxorubicin and colchicine (Tables S2 and 6). The Lipinski rule-of-five suggested that compounds with a log
P) of the sulfonamide-substituted PCAs 1–8 were evaluated using ChemDraw 12.0, Molinspiration (https://www.molinspiration.com), and ALOGPS 2.1 and compared with standard anticancer drugs doxorubicin and colchicine (Tables S2 and 6). The Lipinski rule-of-five suggested that compounds with a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value between 0 and 3 are ideal for oral administration due to their balance of solubility and permeability.69,70 In this investigation, compounds 1–8 reflected optimum solubility in the range of 1.13–2.67, indicating a favourable balance of solubility and permeability. Moreover, compounds 1–5 exhibited higher lipophilicity (1.52–2.05) than the reference dugs doxorubicin and colchicine i.e. 1.41 and 1.50, respectively. However, the following trend was observed within the synthesized compounds in terms of lipophilicity 3 > 5 > 2 > 4 > 1 > 6 > 8 > 7. Generally, in line with previously reported PCAs,44 the cytotoxicity of compounds 1–8 increases with the increase in lipophilicity. Interestingly, compounds 3 and 5 (containing strong electron donating N,N-dimethyl and methyl substituents) with the highest log
P value between 0 and 3 are ideal for oral administration due to their balance of solubility and permeability.69,70 In this investigation, compounds 1–8 reflected optimum solubility in the range of 1.13–2.67, indicating a favourable balance of solubility and permeability. Moreover, compounds 1–5 exhibited higher lipophilicity (1.52–2.05) than the reference dugs doxorubicin and colchicine i.e. 1.41 and 1.50, respectively. However, the following trend was observed within the synthesized compounds in terms of lipophilicity 3 > 5 > 2 > 4 > 1 > 6 > 8 > 7. Generally, in line with previously reported PCAs,44 the cytotoxicity of compounds 1–8 increases with the increase in lipophilicity. Interestingly, compounds 3 and 5 (containing strong electron donating N,N-dimethyl and methyl substituents) with the highest log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of 2.05 and 2.0 respectively, revealed their highest tendency towards gastrointestinal and cellular absorption which is also reflected in their potential as strong cytotoxic agents with potent inhibition of microtubules.
P value of 2.05 and 2.0 respectively, revealed their highest tendency towards gastrointestinal and cellular absorption which is also reflected in their potential as strong cytotoxic agents with potent inhibition of microtubules.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and druglikeness QEDmow values of PCAs 1–8
P and druglikeness QEDmow values of PCAs 1–8
| Compounds | clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
QEDmow |
|---|---|---|
| 1 | 1.52 | 0.840 |
| 2 | 1.87 | 0.813 |
| 3 | 2.05 | 0.855 |
| 4 | 1.67 | 0.847 |
| 5 | 2.00 | 0.845 |
| 6 | 1.39 | 0.819 |
| 7 | 1.13 | 0.819 |
| 8 | 1.27 | 0.636 |
| Doxorubicin | 1.41 | 0.234 |
| Colchicine | 1.50 | 0.732 |
Quantitative estimates of drug-likeness
The sulfonamide-bearing PCAs were designed to achieve oral bioavailability, and a quantitative measure of drug-likeness was used to forecast their potential as orally active anticancer drugs. The weighted quantitative estimate of drug-likeness of the compounds based on the maximum information content (QEDmow) was computed for compounds 1–8 and the reference anticancer drugs doxorubicin and colchicine (Tables 6 and S3). The QED varies from 0 to 1 and indicates the percentage of drug-likeness. The synthesized compounds 1–8 displayed effective drug-likeness with QEDmow ranging from 0.63–0.86 and greater than the reference anticancer drug doxorubicin (0.234). Compounds 1–7 showed higher drug-likeness of 0.81–0.86 than the reference drug colchicine (0.73) except compound 8 (0.63). Among all the compounds, compounds 1–7 exhibited druglikeness of more than 0.8 or higher than 80% showing ability to appear as a drug. Interestingly, compound 3 exhibited drug-likeness with the highest QEDmow value of 0.86 along with strong cytotoxicity and tubulin inhibitory potential suggesting its promise as a lead candidate requiring further clinical evaluation as an orally active anticancer and antimitotic agent.Bioavailability radar
Bioavailability radar is a tool used in drug discovery to quickly estimate drug-likeness and bioavailability of molecules represented in a form of distorted hexagonal model. Bioavailability radar evaluates six physicochemical qualities abbreviated as: LIPO (lipophilicity), SIZE (size), POLAR (polarity), INSOLU (insolubility), FLEX (flexibility), and INSATU (unsaturation). The pink area in the radar represents the optimal range for all parameters, including lipophilicity (XLOGP3 = 0.7 to +5.0), solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S = 6), polarity (TPSA = 20 to 130 Å), saturation (sp3 hybridised carbon = 0.25 percent), and flexibility (less than 9 rotatable bonds).71 The drug-like region is often represented by a pink area on the graph, and compounds with physiochemical properties lying inside this range are regarded more promising for drug development. The bioavailability radar of synthesized compounds 1–8 and the reference drugs doxorubicin 9 and colchicine 10 exhibited ideal bioavailability patterns (Fig. 10) with all the parameters found in the compatible range suggesting good drug-likeness and potential for oral bioavailability. However, the synthesized compounds 1–8 and doxorubicin 9 revealed one offshoot value i.e. higher unsaturation and increase polarity respectively, but all of the other parameters were found to be within the suitable range, indicating drug-likeness and oral bioavailability.
S = 6), polarity (TPSA = 20 to 130 Å), saturation (sp3 hybridised carbon = 0.25 percent), and flexibility (less than 9 rotatable bonds).71 The drug-like region is often represented by a pink area on the graph, and compounds with physiochemical properties lying inside this range are regarded more promising for drug development. The bioavailability radar of synthesized compounds 1–8 and the reference drugs doxorubicin 9 and colchicine 10 exhibited ideal bioavailability patterns (Fig. 10) with all the parameters found in the compatible range suggesting good drug-likeness and potential for oral bioavailability. However, the synthesized compounds 1–8 and doxorubicin 9 revealed one offshoot value i.e. higher unsaturation and increase polarity respectively, but all of the other parameters were found to be within the suitable range, indicating drug-likeness and oral bioavailability.
BOILED-egg
The BOILED-egg model is a computational tool for predicting the gastrointestinal absorption and brain penetration of small compounds, notably in drug development. It employs a graphical representation based on a molecule's polarity (TPSA) and lipophilicity (WLOGP), shown as a “BOILED-egg”. The model evaluates a molecule's capacity for passive absorption through the gastrointestinal system (white region) and its ability to cross the blood–brain barrier (BBB) (yellow area or yolk). Furthermore, it determines if a molecule is a substrate for P-glycoprotein (P-gp), a protein that actively transports chemicals out of the brain. In view of this, the BOILED-egg model of the synthesized compounds 1–8 and the standard drug colchicine 9 was assessed utilizing SMILE strings of compounds (Fig. 11).72 The BOILED-egg model revealed that compounds 1–6 and reference drugs doxorubicin and colchicine 9 are located in the white region, indicating a likelihood of greater gastrointestinal absorption, which may contribute to their potential oral bioavailability. Compounds 7 and 8 were found both outside the yolk and white region implicating poor pharmacokinetic properties which is also reflected in their inactive nature in cytotoxic assays.P-glycoprotein (PGP) acts as an active efflux pump, removing xenobiotics and numerous toxins from cells while also playing a key role in drug extraction and absorption. In the BOILED-egg image, the blue (PGP+) and red (PGP−) dots indicate whether the chemical can or cannot be effluxed from the central nervous system (CNS), respectively.73 In the current study, compounds 1–8 are indicated as red dots, implicating that these molecules do not bind to p-glycoprotein, indicating a negative PGP impact while the colchicine 9 showed PGP positive behavior (blue dots) reflecting that it can be actively effluxed from the central nervous system.
It should be noted that these drug-likeness, bioavailability radar, and BOILED-egg predictions are based solely on in silico models and therefore provide only preliminary indications of absorption and bioavailability; experimental validation through in vitro permeability assays (e.g., PAMPA, Caco-2) was not performed in the present study due to resource constraints, but will be considered in our future work to strengthen these findings.
Conclusion
PCAs act as gastro mucosal protectants and are famous for their promising anticancer and HDAC enzyme inhibition activities with low toxicity in an in vivo mice model. Herein, we have synthesized new analogues of our previously reported sulfonamide-substituted PCA 1 by introducing new electron-withdrawing and -donating substituents at N-phenyl 2 or 4 positions to study the effect of substitutions on the physicochemical properties, hemolytic, anticancer and tubulin inhibition activities. The anticancer studies revealed that compounds 3 and 5 demonstrated stronger cytotoxicity than reference drug colchicine against MCF-7, PC3 and HePG2 cancer cells while they showed higher activity than doxorubicin only against the PC3 cell line. In the active PCAs (2–6), compounds 3 and 5 showed ∼4-fold reduced cytotoxicity in normal HLMEC cells, resulting in significantly higher selectivity indices compared to doxorubicin, underscoring their improved therapeutic window. Tubulin polymerization assays confirmed potent inhibitory activity, with IC50 values of 1.1 μM (3) and 1.4 μM (5), surpassing both colchicine and CA-4. Their favorable physicochemical profiles, high lipophilicity, drug-likeness, bioavailability, and gastrointestinal absorption correlated strongly with their tubulin inhibitory potential and anticancer activities, while hemolysis assays revealed markedly lower toxicity (∼6–7%) than doxorubicin (49.5%). Docking analyses substantiated these observations, demonstrating that compounds 3 and 5 form robust interactions with tubulin active sites, characterized by low micromolar inhibition constants and superior free binding energies (−14.91 and −14.05 kcal mol−1) compared to CA-4 (−13.42 kcal mol−1). Overall, these findings highlight compounds 3 and 5 as promising lead scaffolds for the rational development of next-generation tubulin polymerization inhibitors. The newly synthesized PCAs (3 and 5) exhibit distinct advantages over standard reference drugs, including enhanced anticancer potency, improved selectivity with lower toxicity, potent tubulin inhibition, and favorable physicochemical and ADMET properties, underscoring their potential as safer and more effective anticancer agents. Nevertheless, pharmacological and mechanistic studies are warranted to validate their therapeutic promise and clinical applicability.Experimental
Materials and methods
Commercially available chemicals and solvents of analytical quality were used exactly as supplied. Sulfanilamide, sulfur, sodium sulfide nonahydrate, 2-picoline, sodium hydroxide, conc. HCl, silica gel, 4-amino-N-methylbenzenesulfonamide, 4-amino-N,N-dimethylbenzenesulfonamide, 4-amino-3-fluorobenzenesulfonamide, 4-amino-3-hydroxy benzenesulfonamide, dichloromethane and acetonitrile were from Sigma-Aldrich, 4-amino-3-methylbenzenesulfonamide was from Fischer Scientific, 4-amino-3-methoxy benzenesulfonamide was from AK Scientific, and 4-amino-3-nitrobenzene sulfonamide was from molport. N-(4-Sulfamoylphenyl)pyridine-2-carbothioamide was synthesized by adopting standard procedures.53A Bruker Avance AVIII 400 MHz NMR spectrometer at the COMSATS University Islamabd, Abbottabad Campus, Abbottabad was used to record the NMR spectra at room temperature at 400.13 MHz (1H) or 100.61 MHz (13C) using DMSO-d6 as the solvent. SpinWorks (3.1) was used for visualization and basic processing of NMR data. A Shimadzu FT-IR Prestige-21 infrared spectrophotometer at the Government College Women University Sialkot, Sialkot, Pakistan was used to record the FTIR spectra using KBr plates in the 4000–400 cm−1 range. The FTIR graph was processed and plotted using OriginPro software. High resolution mass spectra were recorded in the positive electron spray ionization mode on a MaXis II-ESI-QTOF at the HEJ Research Institute of Chemistry, University of Karachi, Karachi, Pakistan. The elemental analyses were carried out using LECO's CHNS-932 Elemental Analyzer at the University of Sargodha, Sargodha, Pakistan.
General procedure for synthesis of pyridine carbothioamides functionalized with sulfonamide
The pyridine carbothioamides functionalized with sulfonamide 1–8 were synthesized by refluxing a mixture of sulfanilamide derivatives (12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.5 mol%) and 2-picoline (25 mmol) for 72 h. The reaction mixture was cooled down at room temperature followed by addition of 2 M aqueous solution of sodium hydroxide (2 × 75 mL) and the resulting suspension was filtered. Concentrated hydrochloric acid was added dropwise into the desired filtrate solution to acidify it to pH 5 and the resulting precipitate was filtered and washed with 100 mL of water.74 The crude product was dissolved in dichloromethane and filtered through a bed of silica gel. Recrystallization from acetonitrile resulted in production of the pure crystalline product which was dried.53N-(4-Sulfamoylphenyl)pyridine-2-carbothioamide (1)
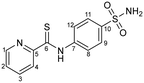 Compound 1 was synthesized and characterized by NMR, mass spectrometry and elemental analysis by using the reported method.53
Compound 1 was synthesized and characterized by NMR, mass spectrometry and elemental analysis by using the reported method.53
m.p. 146 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3423 (m) (–NH), 3277, 3228 (m) (–NH2), 3122 (m) (CHarom), 1726 (s) (C
= 3423 (m) (–NH), 3277, 3228 (m) (–NH2), 3122 (m) (CHarom), 1726 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1322, 1143 (s) (–SO2).
S), 1322, 1143 (s) (–SO2).
N-(4-(N-Methylsulfamoyl)phenyl)pyridine-2-carbothioamide (2)
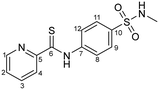 Compound 2 was synthesized by following the general procedure using 4-amino-N-methylbenzenesulfonamide (2.327 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 2.84 g (73%); m.p. 152 °C. IR (KBr, cm−1):
Compound 2 was synthesized by following the general procedure using 4-amino-N-methylbenzenesulfonamide (2.327 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 2.84 g (73%); m.p. 152 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3411 (m) (–NH), 3115 (m) (CHarom), 1708 (s) (C
= 3411 (m) (–NH), 3115 (m) (CHarom), 1708 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1309, 1123 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 11.99 (s, 1H, –NH), 8.64 (d, 3J = 5 Hz, 1H, H-4), 8.52 (d, 3J = 8 Hz, 1H, H-1), 8.02 (td, 3J = 8 Hz, 4J = 1 Hz, 1H, H-3), 7.71 (d, 3J = 9 Hz 2H, H-9/H-11), 7.61 (m, 1H, H-2), 6.60 (d, 3J = 9 Hz, 2H, H-8/H-12), 5.30 (s, 1H, –SO2NH), 2.94 (s, 3H, –SO2NCH3) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 198.2 (C-6), 148.7 (C-1), 148.4 (C-5), 136.8 (C-3), 135.6 (C-10), 132.0 (C-7), 127.8 (C-9/C-11), 124.7 (C-2), 122.3 (C-4), 118.6 (C-8/C-12), 29.3 (–SO2N(CH3)) ppm. MS (ESI+): m/z 308.0541 [2 + H]+ (m/zcalc 308.0527). Anal. found: C, 50.92; H, 3.97; N, 13.93; calc. for C16H14N2O3: C, 50.80; H, 4.26; N, 13.67.
S), 1309, 1123 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 11.99 (s, 1H, –NH), 8.64 (d, 3J = 5 Hz, 1H, H-4), 8.52 (d, 3J = 8 Hz, 1H, H-1), 8.02 (td, 3J = 8 Hz, 4J = 1 Hz, 1H, H-3), 7.71 (d, 3J = 9 Hz 2H, H-9/H-11), 7.61 (m, 1H, H-2), 6.60 (d, 3J = 9 Hz, 2H, H-8/H-12), 5.30 (s, 1H, –SO2NH), 2.94 (s, 3H, –SO2NCH3) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 198.2 (C-6), 148.7 (C-1), 148.4 (C-5), 136.8 (C-3), 135.6 (C-10), 132.0 (C-7), 127.8 (C-9/C-11), 124.7 (C-2), 122.3 (C-4), 118.6 (C-8/C-12), 29.3 (–SO2N(CH3)) ppm. MS (ESI+): m/z 308.0541 [2 + H]+ (m/zcalc 308.0527). Anal. found: C, 50.92; H, 3.97; N, 13.93; calc. for C16H14N2O3: C, 50.80; H, 4.26; N, 13.67.
N-(4-(N,N-Dimethylsulfamoyl)phenyl)pyridine-2-carbothioamide (3)
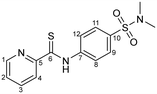 Compound 3 was synthesized by following the general procedure using 4-amino-N,N-dimethylbenzenesulfonamide (2.503 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.17 g (78%); m.p. 158 °C. IR (KBr, cm−1):
Compound 3 was synthesized by following the general procedure using 4-amino-N,N-dimethylbenzenesulfonamide (2.503 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.17 g (78%); m.p. 158 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3397 (m) (–NH), 3095 (m) (CHarom), 1697 (s) (C
= 3397 (m) (–NH), 3095 (m) (CHarom), 1697 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1302, 1115 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.11 (s, 1H, –NH), 8.66 (d, 3J = 7 Hz, 1H, H-4), 8.55 (d, 3J = 8 Hz, 1H, H-1), 8.04 (td, 3J = 7 Hz, 4J = 1 Hz, 1H, H-3), 7.92 (m, 2H, H-8/H-12), 7.64 (ddd, 3J = 7 Hz, 4J = 1 Hz, 1H, H-2), 6.78 (d, 3J = 9 Hz, 2H, H-9/H-11), 2.95 (s, 6H, –SO2N(CH3)2) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 198.1 (C-6), 148.8 (C-1), 148.3 (C-5), 136.7 (C-3), 134.5 (C-10), 132.2 (C-7), 127.9 (C-9/C-11), 124.8 (C-2), 122.4 (C-4), 118.8 (C-8/C-12), 37.9 (–SO2N(CH3)2) ppm. MS (ESI+): m/z 322.0662 [3 + H]+ (m/zcalc 322.0684). Anal. found: C, 52.05; H, 4.59; N, 12.87; calc. for C16H14N2O3: C, 52.32; H, 4.70; N, 13.07.
S), 1302, 1115 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.11 (s, 1H, –NH), 8.66 (d, 3J = 7 Hz, 1H, H-4), 8.55 (d, 3J = 8 Hz, 1H, H-1), 8.04 (td, 3J = 7 Hz, 4J = 1 Hz, 1H, H-3), 7.92 (m, 2H, H-8/H-12), 7.64 (ddd, 3J = 7 Hz, 4J = 1 Hz, 1H, H-2), 6.78 (d, 3J = 9 Hz, 2H, H-9/H-11), 2.95 (s, 6H, –SO2N(CH3)2) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 198.1 (C-6), 148.8 (C-1), 148.3 (C-5), 136.7 (C-3), 134.5 (C-10), 132.2 (C-7), 127.9 (C-9/C-11), 124.8 (C-2), 122.4 (C-4), 118.8 (C-8/C-12), 37.9 (–SO2N(CH3)2) ppm. MS (ESI+): m/z 322.0662 [3 + H]+ (m/zcalc 322.0684). Anal. found: C, 52.05; H, 4.59; N, 12.87; calc. for C16H14N2O3: C, 52.32; H, 4.70; N, 13.07.
N-(2-Fluoro-4-sulfamoylphenyl)pyridine-2-carbothioamide (4)
 Compound 4 was synthesized by following the general procedure using 4-amino-3-fluorobenzenesulfonamide (2.377 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.27 g (83%); m.p. 138 °C. IR (KBr, cm−1):
Compound 4 was synthesized by following the general procedure using 4-amino-3-fluorobenzenesulfonamide (2.377 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.27 g (83%); m.p. 138 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3439 (m) (–NH), 3291, 3238 (m) (–NH2), 3129 (m) (CHarom), 1735 (s) (C
= 3439 (m) (–NH), 3291, 3238 (m) (–NH2), 3129 (m) (CHarom), 1735 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1325, 1152 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.01 (s, 1H, –NH), 8.80 (d, 3J = 7 Hz, 1H, H-4), 8.56 (d, 3J = 8 Hz, 1H, H-1), 8.03 (m, 2H, H-9/H-11), 7.90 (td, 3J = 7 Hz, 4J = 1 Hz, 1H, H-3), 7.50 (ddd, 3J = 7 Hz, 4J = 1 Hz, 1H, H-2), 7.15 (d, 3J = 9 Hz, 1H, H-12), 7.42 (s, 2H, –SO2NH2) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 197.9 (C-6), 153.3 (C-8), 148.9 (C-1), 148.1 (C-5), 136.8 (C-7), 136.3 (C-10), 127.8 (C-11), 125.5 (C-3), 124.7 (C-2), 122.3 (C-4), 120.7 (C-12), 116.0 (C-9) ppm. MS (ESI+): m/z 312.0254 [4 + H]+ (m/zcalc 312.0277). Anal. found: C, 45.96; H, 3.08; N, 13.62; calc. for C16H14N2O3: C, 46.29; H, 3.24; N, 13.50.
S), 1325, 1152 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.01 (s, 1H, –NH), 8.80 (d, 3J = 7 Hz, 1H, H-4), 8.56 (d, 3J = 8 Hz, 1H, H-1), 8.03 (m, 2H, H-9/H-11), 7.90 (td, 3J = 7 Hz, 4J = 1 Hz, 1H, H-3), 7.50 (ddd, 3J = 7 Hz, 4J = 1 Hz, 1H, H-2), 7.15 (d, 3J = 9 Hz, 1H, H-12), 7.42 (s, 2H, –SO2NH2) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 197.9 (C-6), 153.3 (C-8), 148.9 (C-1), 148.1 (C-5), 136.8 (C-7), 136.3 (C-10), 127.8 (C-11), 125.5 (C-3), 124.7 (C-2), 122.3 (C-4), 120.7 (C-12), 116.0 (C-9) ppm. MS (ESI+): m/z 312.0254 [4 + H]+ (m/zcalc 312.0277). Anal. found: C, 45.96; H, 3.08; N, 13.62; calc. for C16H14N2O3: C, 46.29; H, 3.24; N, 13.50.
N-(2-Methyl-4-sulfamoylphenyl)pyridine-2-carbothioamide (5)
 Compound 5 was synthesized by following the general procedure using 4-amino-3-methylbenzenesulfonamide (2.340 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 2.69 g (69%); m.p. 154 °C. IR (KBr, cm−1):
Compound 5 was synthesized by following the general procedure using 4-amino-3-methylbenzenesulfonamide (2.340 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 2.69 g (69%); m.p. 154 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3355 (m) (–NH), 3248, 3215 (m) (–NH2), 3095 (m) (CHarom), 1705 (s) (C
= 3355 (m) (–NH), 3248, 3215 (m) (–NH2), 3095 (m) (CHarom), 1705 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1311, 1119 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.21 (s, 1H, –NH), 8.67 (d, 3J = 6 Hz, 1H, H-4), 8.53 (d, 3J = 8 Hz, 1H, H-1), 8.04 (td, 3J = 8 Hz, 4J = 1 Hz, 2H, H-3), 7.85 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.65 (m, 1H, H-2), 7.26 (d, 3J = 8 Hz, 1H, H-12), 7.32 (s, 2H, –SO2NH2), 3.32 (s, 3H, –CH3) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 197.2 (C-6), 148.9 (C-1), 148.5 (C-5), 136.9 (C-3), 136.5 (C-10), 133.8 (C-7), 128.4 (C-8), 128.1 (C-9), 127.8 (C-11), 124.6 (C-2), 122.2 (C-4), 120.8 (C-12), 20.9 (–CH3) ppm. MS (ESI+): m/z 308.0531 [5 + H]+ (m/zcalc 308.0527). Anal. found: C, 50.64; H, 4.51; N, 13.83; calc. for C16H14N2O3: C, 50.80; H, 4.26; N, 13.67.
S), 1311, 1119 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.21 (s, 1H, –NH), 8.67 (d, 3J = 6 Hz, 1H, H-4), 8.53 (d, 3J = 8 Hz, 1H, H-1), 8.04 (td, 3J = 8 Hz, 4J = 1 Hz, 2H, H-3), 7.85 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.65 (m, 1H, H-2), 7.26 (d, 3J = 8 Hz, 1H, H-12), 7.32 (s, 2H, –SO2NH2), 3.32 (s, 3H, –CH3) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 197.2 (C-6), 148.9 (C-1), 148.5 (C-5), 136.9 (C-3), 136.5 (C-10), 133.8 (C-7), 128.4 (C-8), 128.1 (C-9), 127.8 (C-11), 124.6 (C-2), 122.2 (C-4), 120.8 (C-12), 20.9 (–CH3) ppm. MS (ESI+): m/z 308.0531 [5 + H]+ (m/zcalc 308.0527). Anal. found: C, 50.64; H, 4.51; N, 13.83; calc. for C16H14N2O3: C, 50.80; H, 4.26; N, 13.67.
N-(2-Methoxy-4-sulfamoylphenyl)pyridine-2-carbothioamide (6)
 Compound 6 was synthesized by following the general procedure using 4-amino-3-methoxybenzenesulfonamide (2.527 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 2.94 g (72%); m.p. 149 °C. IR (KBr, cm−1):
Compound 6 was synthesized by following the general procedure using 4-amino-3-methoxybenzenesulfonamide (2.527 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 2.94 g (72%); m.p. 149 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3384 (m) (–NH), 3256, 3218 (m) (–NH2), 3084 (m) (CHarom), 1712 (s) (C
= 3384 (m) (–NH), 3256, 3218 (m) (–NH2), 3084 (m) (CHarom), 1712 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1316, 1128 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.19 (s, 1H, –NH), 8.66 (d, 3J = 5 Hz, 1H, H-4), 8.54 (d, 3J = 9 Hz, 1H, H-1), 8.03 (td, 3J = 8 Hz, 4J = 2 Hz, 2H, H-3), 7.89 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.64 (m, 1H, H-2), 7.01 (d, 3J = 9 Hz, 1H, H-12), 7.19 (s, 2H, –SO2NH2), 3.79 (s, 3H, –OCH3) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 198.6 (C-6), 149.1 (C-1), 148.7 (C-5), 144.8 (C-11), 136.8 (C-3), 133.3 (C-7), 127.8 (C-8), 127.5 (C-3), 124.7 (C-2), 122.3 (C-4), 120.7 (C-12), 116.0 (C-7), 56.2 (–OCH3) ppm. MS (ESI+): m/z 324.0502 [6 + H]+ (m/zcalc 324.0477). Anal. found: C, 48.06; H, 3.95; N, 12.91; calc. for C16H14N2O3: C, 48.28; H, 4.05; N, 12.99.
S), 1316, 1128 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.19 (s, 1H, –NH), 8.66 (d, 3J = 5 Hz, 1H, H-4), 8.54 (d, 3J = 9 Hz, 1H, H-1), 8.03 (td, 3J = 8 Hz, 4J = 2 Hz, 2H, H-3), 7.89 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.64 (m, 1H, H-2), 7.01 (d, 3J = 9 Hz, 1H, H-12), 7.19 (s, 2H, –SO2NH2), 3.79 (s, 3H, –OCH3) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 198.6 (C-6), 149.1 (C-1), 148.7 (C-5), 144.8 (C-11), 136.8 (C-3), 133.3 (C-7), 127.8 (C-8), 127.5 (C-3), 124.7 (C-2), 122.3 (C-4), 120.7 (C-12), 116.0 (C-7), 56.2 (–OCH3) ppm. MS (ESI+): m/z 324.0502 [6 + H]+ (m/zcalc 324.0477). Anal. found: C, 48.06; H, 3.95; N, 12.91; calc. for C16H14N2O3: C, 48.28; H, 4.05; N, 12.99.
N-(2-Hydroxy-4-sulfamoylphenyl)pyridine-2-carbothioamide (7)
 Compound 7 was synthesized by following the general procedure using 4-amino-3-hydroxybenzenesulfonamide (2.352 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.56 g (91%); m.p. 135 °C. IR (KBr, cm−1):
Compound 7 was synthesized by following the general procedure using 4-amino-3-hydroxybenzenesulfonamide (2.352 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.56 g (91%); m.p. 135 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3405 (m) (–NH), 3361 (s) (OH), 3283, 3225 (m) (–NH2), 3110 (m) (CHarom), 1709 (s) (C
= 3405 (m) (–NH), 3361 (s) (OH), 3283, 3225 (m) (–NH2), 3110 (m) (CHarom), 1709 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1315, 1124 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.35 (s, 1H, –NH), 8.69 (d, 3J = 5 Hz, 1H, H-4), 8.52 (d, 3J = 7 Hz, 1H, H-1), 8.04 (td, 3J = 8 Hz, 4J = 2 Hz, 1H, H-3), 7.99 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.66 (m, 1H, H-2), 7.51 (d, 3J = 9 Hz, 1H, H-12), 7.33 (s, 2H, –SO2NH2), 5.59 (s, 1H, –OH) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): 197.3 (C-6), 148.6 (C-1), 148.6 (C-5), 136.7 (C-3), 136.3 (C-10), 133.2 (C-7), 128.7 (C-8), 128.3 (C-9), 127.6 (C-11), 124.4 (C-2), 122.4 (C-4), 120.5 (C-12) ppm. MS (ESI+): m/z 310.0334 [7 + H]+ (m/zcalc 310.0320). Anal. found: C, 46.72; H, 3.49; N, 13.36; calc. for C16H14N2O3: C, 46.59; H, 3.58; N, 13.58.
S), 1315, 1124 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.35 (s, 1H, –NH), 8.69 (d, 3J = 5 Hz, 1H, H-4), 8.52 (d, 3J = 7 Hz, 1H, H-1), 8.04 (td, 3J = 8 Hz, 4J = 2 Hz, 1H, H-3), 7.99 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.66 (m, 1H, H-2), 7.51 (d, 3J = 9 Hz, 1H, H-12), 7.33 (s, 2H, –SO2NH2), 5.59 (s, 1H, –OH) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): 197.3 (C-6), 148.6 (C-1), 148.6 (C-5), 136.7 (C-3), 136.3 (C-10), 133.2 (C-7), 128.7 (C-8), 128.3 (C-9), 127.6 (C-11), 124.4 (C-2), 122.4 (C-4), 120.5 (C-12) ppm. MS (ESI+): m/z 310.0334 [7 + H]+ (m/zcalc 310.0320). Anal. found: C, 46.72; H, 3.49; N, 13.36; calc. for C16H14N2O3: C, 46.59; H, 3.58; N, 13.58.
N-(2-Nitro-4-sulfamoylphenyl)pyridine-2-carbothioamide (8)
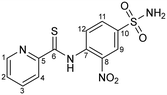 Compound 8 was synthesized by following the general procedure using 4-amino-3-nitrobenzenesulfonamide (2.715 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.63 g (85%); m.p. 131 °C. IR (KBr, cm−1):
Compound 8 was synthesized by following the general procedure using 4-amino-3-nitrobenzenesulfonamide (2.715 g, 12.5 mmol), sulfur (0.96 g, 30 mmol), sodium sulfide nonahydrate (0.15 g, 5 mol%) and 2-picoline (2.46 mL, 25 mmol). Yield: 3.63 g (85%); m.p. 131 °C. IR (KBr, cm−1): ![[small nu, Greek, tilde]](https://www.rsc.org/images/entities/i_char_e0e1.gif) = 3431 (m) (–NH), 3288, 3230 (m) (–NH2), 3126 (m) (CHarom), 1732 (s) (C
= 3431 (m) (–NH), 3288, 3230 (m) (–NH2), 3126 (m) (CHarom), 1732 (s) (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 1542 (s), (NO2), 1320, 1148 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.45 (s, 1H, –NH), 8.70 (d, 3J = 5 Hz, 1H, H-4), 8.55 (d, 3J = 9 Hz, 1H, H-1), 8.06 (td, 3J = 8 Hz, 4J = 2 Hz, 1H, H-3), 7.94 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.65 (m, 2H, H-2/H-12), 7.43 (s, 2H, –SO2NH2) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 199.2 (C-6), 152.1 (C-8), 149.0 (C-1), 148.7 (C-5), 142.2 (C-3), 132.9 (C-7), 136.5 (C-10), 129.2 (C-11), 127.8 (C-11), 126.8 (C-9), 124.7 (C-2), 122.3 (C-4), 120.7 (C-12) ppm. MS (ESI+): m/z 339.0239 [7 + H]+ (m/zcalc 339.0222). Anal. found: C, 42.94; H, 3.12; N, 16.81; calc. for C16H14N2O3: C, 42.60; H, 2.98; N, 16.56.
S), 1542 (s), (NO2), 1320, 1148 (s) (–SO2). 1H NMR (400.13 MHz, DMSO-d6, 25 °C) δ = 12.45 (s, 1H, –NH), 8.70 (d, 3J = 5 Hz, 1H, H-4), 8.55 (d, 3J = 9 Hz, 1H, H-1), 8.06 (td, 3J = 8 Hz, 4J = 2 Hz, 1H, H-3), 7.94 (d, 3J = 9 Hz, 2H, H-9/H-11), 7.65 (m, 2H, H-2/H-12), 7.43 (s, 2H, –SO2NH2) ppm. 13C{1H}NMR (100.61 MHz, DMSO-d6, 25 °C): δ = 199.2 (C-6), 152.1 (C-8), 149.0 (C-1), 148.7 (C-5), 142.2 (C-3), 132.9 (C-7), 136.5 (C-10), 129.2 (C-11), 127.8 (C-11), 126.8 (C-9), 124.7 (C-2), 122.3 (C-4), 120.7 (C-12) ppm. MS (ESI+): m/z 339.0239 [7 + H]+ (m/zcalc 339.0222). Anal. found: C, 42.94; H, 3.12; N, 16.81; calc. for C16H14N2O3: C, 42.60; H, 2.98; N, 16.56.
MTT assay
The cytotoxicity of pyridine carbothioamides 1–8, doxorubicin and colchicine against human cancer cell lines (A549, MCF-7, PC3 and HepG2) and human normal cancer cells (HLMEC) was concluded using the MTT [3 (4,5-dimethylthiazolyl)-2,5-diphenyl tetrazolium bromide] assay (from Sigma Aldrich, China) conducted at the Institute of Pharmaceutical Process, School of Medicine, Wuhan University of Science and Technology, Wuhan, China.75 Concisely, prior to drug interaction, in 96-well plates at a density of 2000 cells per well, the cells were seeded in 100 μL of growth medium and pre-incubated for 24 hours. In DMSO, 1–8, colchicine and doxorubicin stock solutions were arranged, whereas in PBS, the cDDP stock solution was prepared. The stock solutions were diluted in a complete medium and thereafter added into 100 μL aliquots from each well (DMSO concentration <0.5%). Exposure time of 72 h was chosen to allow sufficient interaction of the compounds with the cancer cells and to capture both early and delayed cytotoxic effects and then cells were administrated with MTT (20 μL, 5 mg mL−1 in PBS). After the removal of medium DMSO (200 μL) was used to dissolve purple formazan crystals. After shaking the plates for 10 minutes, the solution's absorbance at 570 nm was measured using a Varioskan flash multimode reader (Tokyo, Japan). The presented IC50 values are the mean of at least 3 independent experiments.In vitro tubulin polymerization inhibition assay
The synthesized compounds 1–8, CA-4 and colchicine were selected to evaluate their potential as in vitro tubulin polymerization inhibitors (conducted at the Institute of Pharmaceutical Process, School of Medicine, Wuhan University of Science and Technology, Wuhan, China). In this assay, the polymerization protocol was followed to carry out the standard tubulin polymerization reactions followed by incubation of the synthesized compounds (minus tubulin ligands).63 In short, the standard polymerization reaction consists of 100 μL volume of 4 mg mL−1 tubulin in 80 mM PIPES, pH 6.9, 0.5 mM EGTA, 2 mM MgCl2, and 1 mM GTP. After incubation, the tubulin polymerization reaction started at 37 °C, followed by monitoring the absorption at 340 nm. The polymerization reached maximal optimal density at 340 nm (OD340) between 0.15 and 0.25 within 30 min. Under these conditions, the tubulin polymerization reaction processed through three stages involved: I (nucleation), II (growth), and III (steady state). In this assay, (100 μL volume in a spectrophotometer with a 0.5 cm path length), an OD340 of 0.1 corresponds to 1 mg mL−1 of polymer mass. This standard protocol resulted in approximately 40% tubulin polymerization, allowing the evaluation of compounds as potential enhancers or inhibitors of polymerization. The IC50 values were expressed as mean ± SD from 3 independent experiments.Molecular docking assay
To elucidate the binding interactions of pyridine carbothioamide derivatives 1–8 and CA-4 within the binding pocket of tubulin (colchicine-binding site), molecular docking simulations were conducted using Autodock 4.0.76–78 The 3D structure of tubulin (PDB ID: 4O2B) was retrieved from the RCSB Protein Data Bank (https://www.rcsb.org/pdb/home/home.do). The 3D structure of all the compounds 1–8 and CA-4 was generated using Chem3D 21.0 and subsequently converted into PDB format using OpenBabel GUI for docking compatibility. The tubulin protein structure was prepared using MGL Tools, involving the removal of crystallographic water molecules, addition of polar hydrogen, and assignment of Gasteiger partial charges to optimize the structure for molecular docking. Docking simulations were performed using the Autodock Tools 1.5.7 package, with the search grid centered on the colchicine-binding site (x = 17.336, y = 63.576, z = 43.586) and encompassing a 60 × 60 × 60 Å cubic space to ensure complete coverage of the binding pocket. An exhaustiveness value of 10 was implemented to ensure the perfect position of compounds 1–8 and CA-4 within the colchicine-binding pocket of tubulin. Post-docking analysis was carried out using Discovery Studio Visualizer to examine protein–ligand interactions.Hemolysis assay
The hemolytic activity of the most active anticancer PCAs 3 and 5 in comparison with doxorubicin65 was evaluated at the Institute of Pharmaceutical Process, School of Medicine, Wuhan University of Science and Technology, Wuhan, China, through previously reported procedures.79,80 Here, 5 mL of freshly drawn rabbit blood cells (collected from Xinglong Laboratory Animal Breeding Plant, China) was mixed with 5 mL of PBS (pH = 7.4), centrifuged for 15 minutes at 3000 rpm, and then red blood cells (RBCs) precipitate after the clear supernatant is discarded. To wash and filter the RBCs, this procedure was carried out three times. Following purification, the RBCs were diluted with PBS until they reached a final concentration of 2% (v/v). On the other hand, different concentrations of PCAs (3 and 5) and doxorubicin were prepared at 10, 20, 30, and 40 μg mL−1. After thoroughly mixing 500 mL of the tested substance with 500 mL of PBS, 500 mL of deionised water, and 500 mL of the suspension of RBCs, the mixture was incubated for 4 hours at 37 °C. After centrifuging the mixtures for 15 minutes at 3000 rpm, each group was arranged in increasing order of concentration in a UV-visible spectrophotometer. After that the clear supernatants were collected and the absorbance value of released hemoglobin was measured at 540 nm. For negative and positive controls, the tested compounds or samples were replaced with PBS and deionized water, respectively. The test was replicated three times under identical conditions for investigated samples, as well as negative and positive control groups. The hemolysis ratio was computed based on the following equation mentioned below:Statistical analysis
All data are expressed as mean values with standard errors. For the in vitro antitumor and tubulin inhibitory activities and hemolytic assays, one-way analysis of variance (ANOVA) was performed to analyze the results. Student's t-test or Duncan's multiple comparison test was used to determine statistical significance, with differences considered significant at P < 0.05.Calculated logarithmic octanol/water partition coefficient (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) P)
P)
ChemBioDraw Ultra 12.0, Molinspiration (https://www.molinspiration.com) and ALOGPS2.1 were used to assess the lipophilicity of compounds 1–8, doxorubicin and colchicine using the logarithmic octanol/water partition coefficient (clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P).44,75,81
P).44,75,81
Quantitative estimate of drug-likeness
The QED for 1–8, doxorubicin and colchicine was calculated using previously mentioned methods.44,75,81SwissADME studies
The ADME (absorption, distribution, metabolism, and excretion) of pyridine carbothioamides 1–8, doxorubicin 9 and colchicine 10 was evaluated using SwissADME.75,81 SwissADME is a web-based software platform (https://www.swissadme.ch) developed and maintained by the Swiss Institute of Bioinformatics (SIB). The physicochemical properties and medicinal chemistry profiles of the synthesized compounds were assessed using a range of metrics, including the gastrointestinal absorption, molar refractivity, solubility, molecular weight, bioavailability radar map and BOILED-egg model.71,82Author contributions
Fatima Younas: methodology; writing – original draft. Jahan Zaib Arshad: project administration; supervision; methodology; writing – original draft; writing – review, editing, validation. Sundas Arshad: software and data curation. Waqas Ali Shah: supervision, resources and formal analysis. Adnan Ashraf: writing—review and editing. Syed Shoaib Ahmed Shah: supervision; resources; formal analysis. Muhammad Asam Raza: software; formal analysis. Amara Mumtaz: resources; formal analysis. Nasir Shahzad: visualization; data curation. Tariq Javed: writing—review and editing.Conflicts of interest
All the authors have declared no conflict of interest.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary information (SI). Supplementary information: which includes NMR and ESI-mass spectra of compounds 2–7, a molecular docking interactions table for compounds 1–8 and CA-4 with tubulin amino acids, and lipophilicity dataand quantitative estimates of drug-likeness for compounds 1–8, doxorubicin, and colchicine. See DOI: https://doi.org/10.1039/d5md00693g.Acknowledgements
We thank the Higher Education Commission of Pakistan for IPFP Fellowship (Ref. No. 187/IPFP-II(Batch-I)/HRD/HEC/2020/61) and SRGP Project (Ref. No. 187/IPFP-II(Batch-I)/SRGP/NAHE/HEC/2020/34) awarded to Dr. J. Z. Arshad. Moreover, the Table of Contents image was prepared for this article using original illustrations and software tools (ChemBio3D Ultra 12.0, ChemDraw, Microsoft PowerPoint and https://BioRender.com).References
- H. M. Bizuayehu, K. Y. Ahmed, G. D. Kibret, A. F. Dadi, S. A. Belachew, T. Bagade, T. K. Tegegne, R. L. Venchiarutti, K. T. Kibret and A. H. Hailegebireal, JAMA Netw. Open, 2024, 7, e2443198 CrossRef.
- I. Soerjomataram and F. Bray, Nat. Rev. Clin. Oncol., 2021, 18, 663–672 CrossRef PubMed.
- C. Holohan, S. Van Schaeybroeck, D. B. Longley and P. G. Johnston, Nat. Rev. Cancer, 2013, 13, 714–726 CrossRef CAS PubMed.
- H. Zahreddine and K. L. Borden, Front. Pharmacol., 2013, 4, 28 Search PubMed.
- M. Burute and L. C. Kapitein, Annu. Rev. Cell Dev. Biol., 2019, 35, 29–54 CrossRef CAS PubMed.
- A. L. Parker, M. Kavallaris and J. A. McCarroll, Front. Oncol., 2014, 4, 153 Search PubMed.
- R. H. Wade, Mol. Biotechnol., 2009, 43, 177–191 CrossRef CAS PubMed.
- C. M. Logan and A. S. Menko, Exp. Biol. Med., 2019, 244, 1240–1254 CrossRef CAS.
- H. C. Assunção, P. M. Silva, H. Bousbaa and H. Cidade, Molecules, 2025, 30, 3314 CrossRef.
- J. Sebastian and K. Rathinasamy, Curr. Drug Targets, 2023, 24, 889–918 CrossRef CAS.
- R. Kaul, A. L. Risinger and S. L. Mooberry, J. Nat. Prod., 2019, 82, 680–685 CrossRef CAS PubMed.
- A. Kumar, P. R. Sharma and D. M. Mondhe, Anti-Cancer Drugs, 2017, 28, 250–262 CrossRef CAS PubMed.
- E. C. McLoughlin and N. M. O'Boyle, Pharmaceuticals, 2020, 13, 8 CrossRef CAS PubMed.
- X. Wang, B. Gigant, X. Zheng and Q. Chen, MedComm: Oncol., 2023, 2, e46 CAS.
- M. Gao, D. Zhang, Q. Jin, C. Jiang, C. Wang, J. Li, F. Peng, D. Huang, J. Zhang and S. Song, Oncotarget, 2016, 7, 58133 CrossRef PubMed.
- M. Zweifel, G. C. Jayson, N. Reed, R. Osborne, B. Hassan, J. Ledermann, G. Shreeves, L. Poupard, S.-P. Lu and J. Balkissoon, Ann. Oncol., 2011, 22, 2036–2041 CrossRef CAS.
- M. Hawash, Biomolecules, 2022, 12, 1843 CrossRef CAS PubMed.
- H. Azevedo-Barbosa, D. F. Dias, L. L. Franco, J. A. Hawkes and D. T. Carvalho, Mini-Rev. Med. Chem., 2020, 20, 2052–2066 CrossRef CAS.
- R. C. Ochu, U. C. Okoro, J. Conradie and D. I. Ugwu, Eur. J. Med. Chem. Rep., 2024, 10, 100136 CAS.
- F. A. Khan, S. Mushtaq, S. Naz, U. Farooq, A. Zaidi, S. M. Bukhari, A. Rauf and M. S. Mubarak, Curr. Org. Chem., 2018, 22, 818–830 CrossRef CAS.
- S. D. Durgapal and S. S. Soman, Synth. Commun., 2019, 49, 2869–2883 CAS.
- M. S. Ayoup, N. Khaled, H. Abdel-Hamid, D. A. Ghareeb, S. A. Nasr, A. Omer, A. Sonousi, A. E. Kassab and A. S. Eltaweil, RSC Adv., 2024, 14, 7664–7675 RSC.
- L. C. Dantas, V. R. Campos, J. C. Borges and L. C. Pinheiro, Mini-Rev. Med. Chem., 2023, 23, 2073–2088 CrossRef CAS.
- D. P. Mishra, R. Sahu, P. K. Sahu, S. Ghadai, A. K. Padhan, T. K. Sahu, S. Naik and B. Acharya, Future Med. Chem., 2025, 1–21 Search PubMed.
- M. Y. Moskalik, Molecules, 2022, 28, 51 CrossRef PubMed.
- D. Iwan, K. Kamińska, M. Denel-Bobrowska, A. B. Olejniczak and E. Wojaczyńska, Biomed. Pharmacother., 2022, 153, 113473 CrossRef CAS PubMed.
- C. T. Supuran, Clin. Sci., 2021, 135, 1233–1249 CrossRef CAS.
- C. T. Supuran, Future Med. Chem., 2021, 13(22), 1935–1937 CrossRef PubMed.
- P. Shekharagouda, G. Mamatha, G. Nagaraju, C. Krishnamurthy, V. P. Sajjan, B. Ali Al-Asbahi, N. Mohammed Al-Hada, B. G. Gowda and L. Naik, Mol. Phys., 2025, 123, e2356189 CrossRef.
- K. Abha Mishra, N. Kumari, F. Carta, G. Renzi, C. T. Supuran and K. K. Sethi, J. Biochem. Mol. Toxicol., 2025, 39, e70135 CrossRef CAS PubMed.
- D. Alhyari, N. A. Qinna, H. M. Sheldrake, S. Kantamneni, B. Y. Ghanem and K. J. Paluch, Antioxidants, 2025, 14, 374 CrossRef CAS PubMed.
- S. H. Al-Ziyadi, A. H. Halawa, A. Belal, N. K. A. Albezrah, A. J. Obaidullah, N. S. Abdelshafi, H. F. Al-Shareef, A. B. Mehany, S. M. Hassan and W. E. Elgammal, ACS Omega, 2025, 10(35), 39772–39790 CrossRef CAS PubMed.
- R. Nandakishore, P. R. Yalavarthi, Y. R. Kiran and M. Rajapranathi, Curr. Drug Discovery Technol., 2014, 11, 127–132 CrossRef CAS PubMed.
- C. Willette, in Pharmacology in Veterinary Anesthesia and Analgesia, John Wiley & Sons, Inc., 2024, pp. 138–163 Search PubMed.
- R. Parveen, D. Harihar and B. P. Chatterji, Cancer, 2023, 129, 3372–3380 CrossRef CAS PubMed.
- K. A. Elsayad, G. F. Elmasry, S. T. Mahmoud and F. M. Awadallah, Bioorg. Chem., 2024, 147, 107409 CrossRef CAS.
- S. Mastrangelo, G. Attina, S. Triarico, A. Romano, P. Maurizi and A. Ruggiero, Biomed. Pharmacol. J., 2022, 15, 553–562 CAS.
- J. J. Grob, M. M. Amonkar, B. Karaszewska, J. Schachter, R. Dummer, A. Mackiewicz, D. Stroyakovskiy, K. Drucis, F. Grange, V. Chiarion-Sileni, P. Rutkowski, M. Lichinitser, E. Levchenko, P. Wolter, A. Hauschild, G. V. Long, P. Nathan, A. Ribas, K. Flaherty, P. Sun, J. J. Legos, D. O. McDowell, B. Mookerjee, D. Schadendorf and C. Robert, Lancet Oncol., 2015, 16, 1389–1398 CrossRef CAS PubMed.
- A. Vicente-Blázquez, M. González, R. Álvarez, S. del Mazo, M. Medarde and R. Peláez, Med. Res. Rev., 2019, 39, 775–830 CrossRef PubMed.
- T. M. Mahgoub, E. J. Jordan, A. F. Mahdi, V. Oettl, S. Huefner, N. O'Donovan, J. Crown and D. M. Collins, Cancer Chemother. Pharmacol., 2024, 93, 427–437 CrossRef CAS.
- A. Sargsyan, H. Sahakyan and K. Nazaryan, ACS Omega, 2023, 8, 29448–29454 CrossRef CAS PubMed.
- J. Wang, D. D. Miller and W. Li, Drug Discovery Today, 2022, 27, 759–776 CrossRef CAS PubMed.
- W. A. Kinney, N. E. Lee, R. M. Blank, C. A. Demerson, C. S. Sarnella, N. T. Scherer, G. N. Mir, L. E. Borella, J. F. DiJoseph and C. Wells, J. Med. Chem., 1990, 33, 327–336 CrossRef CAS.
- J. Arshad, M. Hanif, S. Movassaghi, M. Kubanik, A. Waseem, T. Söhnel, S. M. F. Jamieson and C. G. Hartinger, J. Inorg. Biochem., 2017, 177, 395–401 CrossRef CAS PubMed.
- S. M. Meier, M. Hanif, Z. Adhireksan, V. Pichler, M. Novak, E. Jirkovsky, M. A. Jakupec, V. B. Arion, C. A. Davey and B. K. Keppler, Chem. Sci., 2013, 4, 1837–1846 RSC.
- M. Hanif, J. Arshad, J. W. Astin, Z. Rana, A. Zafar, S. Movassaghi, E. Leung, K. Patel, T. Söhnel, J. Reynisson, V. Sarojini, R. J. Rosengren, S. M. F. Jamieson and C. G. Hartinger, Angew. Chem., 2020, 59, 14609–14614 CrossRef CAS.
- E. C. McLoughlin and N. M. O'Boyle, Pharmaceuticals, 2020, 13(1), 8 CrossRef CAS PubMed.
- W. Li, H. Sun, S. Xu, Z. Zhu and J. Xu, Future Med. Chem., 2017, 9, 1765–1794 CrossRef CAS PubMed.
- D. Chan, Y. Zheng, J. W. Tyner, W. J. Chng, W. W. Chien, S. Gery, G. Leong, G. D. Braunstein and H. P. Koeffler, J. Cancer Res. Clin. Oncol., 2013, 139, 1507–1514 CrossRef PubMed.
- K. Yamamoto, T. Kokubun, K. Sato, T. Akaishi, A. Shimazaki, M. Nakamura, Y. Shiga, S. Tsuda, K. Omodaka, H. Saya and T. Nakazawa, Sci. Rep., 2019, 9, 19288 CrossRef CAS.
- M. P. Economides, D. McCue, G. Borthakur and N. Pemmaraju, Expert Opin. Pharmacother., 2019, 20, 1637–1644 CrossRef CAS PubMed.
- M. Dong, F. Liu, H. Zhou, S. Zhai and B. Yan, Molecules, 2016, 21(10), 1375 CrossRef PubMed.
- J. Arshad, M. Hanif, A. Zafar, S. Movassaghi, K. K. H. Tong, J. Reynisson, M. Kubanik, A. Waseem, T. Söhnel, S. M. F. Jamieson and C. G. Hartinger, ChemPlusChem, 2018, 83, 612–619 CrossRef CAS.
- J. Arshad, K. K. H. Tong, S. Movassaghi, T. Söhnel, S. M. F. Jamieson, M. Hanif and C. G. Hartinger, Molecules, 2021, 26, 833 CrossRef CAS.
- Z. Riaz, B. Y. Lee, J. Stjärnhage, S. Movassaghi, T. Söhnel, S. M. Jamieson, M. A. Shaheen, M. Hanif and C. G. Hartinger, J. Inorg. Biochem., 2023, 241, 112115 CrossRef CAS PubMed.
- C. T. Supuran, A. Casini and A. Scozzafava, Med. Res. Rev., 2003, 23, 535–558 CrossRef CAS.
- F. Carta, A. Scozzafava and C. T. Supuran, Expert Opin. Ther. Pat., 2012, 22, 747–758 CrossRef CAS PubMed.
- J.-Y. Winum, A. Scozzafava, J.-L. Montero and C. T. Supuran, Med. Res. Rev., 2006, 26, 767–792 CrossRef CAS.
- K. Klimek, K. Tyśkiewicz, M. Miazga-Karska, A. Dębczak, E. Rój and G. Ginalska, Molecules, 2021, 26, 2366 CrossRef CAS PubMed.
- C. Dumontet and M. A. Jordan, Nat. Rev. Drug Discovery, 2010, 9, 790–803 CrossRef CAS PubMed.
- Y. Lu, J. Chen, M. Xiao, W. Li and D. D. Miller, Pharm. Res., 2012, 29, 2943–2971 CrossRef CAS.
- X.-Y. Meng, H.-X. Zhang, M. Mezei and M. Cui, Curr. Comput.-Aided Drug Des., 2011, 7, 146–157 CrossRef CAS PubMed.
- A. O. El-Abd, S. M. Bayomi, A. K. El-Damasy, B. Mansour, N. I. Abdel-Aziz and M. A. El-Sherbeny, ACS Omega, 2022, 7, 33599–33613 CrossRef CAS PubMed.
- P. Urbán, N. J. Liptrott and S. Bremer, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2019, 11, e1546 Search PubMed.
- A. Ansary, A. Osman and M. E. El-Khouly, RSC Adv., 2025, 15, 6457–6473 RSC.
- M. Bajda, S. Boryczka, J. Wietrzyk and B. Malawska, Biomed. Chromatogr., 2007, 21, 123–131 CrossRef CAS PubMed.
- F. E. Stuurman, B. Nuijen, J. H. Beijnen and J. H. Schellens, Clin. Pharmacokinet., 2013, 52, 399–414 CrossRef CAS PubMed.
- X. Liu, B. Testa and A. Fahr, Pharm. Res., 2011, 28, 962–977 CrossRef CAS PubMed.
- Z. Mandic, Physico Chemical Methods in Drug Discovery and Development, IAPC Publishing, Zagreb, Croatia, 2012 Search PubMed.
- V. Ivanović, M. Rančić, B. Arsić and A. Pavlović, Popul. Sci. Article, 2020, 3, 171–177 Search PubMed.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 42717 CrossRef PubMed.
- A. Daina and V. Zoete, ChemMedChem, 2016, 11, 1117–1121 CrossRef CAS PubMed.
- S. Mollazadeh, A. Sahebkar, F. Hadizadeh, J. Behravan and S. Arabzadeh, Life Sci., 2018, 214, 118–123 CrossRef CAS PubMed.
- M. H. Klingele and S. Brooker, Eur. J. Org. Chem., 2004, 2004, 3422–3434 CrossRef.
- J. Z. Arshad, S. Tabassum, M. S. Kiani, S. Arshad, M. A. Hashmi, I. Majeed, H. Ali and S. S. A. Shah, Chem. – Asian J., 2023, 18, e202300804 CrossRef CAS PubMed.
- A. E. Prota, F. Danel, F. Bachmann, K. Bargsten, R. M. Buey, J. Pohlmann, S. Reinelt, H. Lane and M. O. Steinmetz, J. Mol. Biol., 2014, 426, 1848–1860 CrossRef CAS PubMed.
- G. Wang, Z. Peng, J. Zhang, J. Qiu, Z. Xie and Z. Gong, Bioorg. Chem., 2018, 78, 332–340 CrossRef CAS PubMed.
- G. Wang, J. Qiu, X. Xiao, A. Cao and F. Zhou, Bioorg. Chem., 2018, 76, 249–257 CrossRef CAS PubMed.
- M. A. Khan, A. N. Aljarbou, Y. H. Aldebasi, M. S. Alorainy and A. Khan, Int. J. Nanomed., 2015, 6331–6338 CrossRef CAS PubMed.
- H. Cheng, Y. Zhao, Y. Wang, Y. Hou, R. Zhang, M. Zong, L. Sun, Y. Liu, J. Qi and X. Wu, Int. J. Nanomed., 2023, 6813–6828 CrossRef CAS PubMed.
- S. Shaheen, J. Z. Arshad, M. Haider, A. Ashraf, M. M. Ahmad, M. Ashfaq, M. A. Ismail, T. Najam and S. S. A. Shah, New J. Chem., 2024, 48, 19427–19440 RSC.
- B. Bakchi, A. D. Krishna, E. Sreecharan, V. B. J. Ganesh, M. Niharika, S. Maharshi, S. B. Puttagunta, D. K. Sigalapalli, R. R. Bhandare and A. B. Shaik, J. Mol. Struct., 2022, 1259, 132712 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |