Open Access Article
Open Access ArticleNanoarchaeosomes loaded with tumor antigens elicit antigen-presenting cell activation and T cell response for cervical cancer immunotherapy
Abirami
Seetharaman
a,
Parimalanandhini
Duraisamy
a,
Subastri
Ariraman
a,
Priya
Ramanathan
b and
Swathi
Sudhakar
 *a
*a
aDepartment of Applied Mechanics and Biomedical Engineering, Indian Institute of Technology Madras, Chennai, India. E-mail: swathi.s@iitm.ac.in
bDepartment of Molecular Oncology, Cancer Institute (WIA), Adyar, Chennai, India
First published on 27th November 2025
Abstract
Cancer remains one of the leading causes of mortality worldwide, necessitating the development of innovative therapeutic strategies such as cancer immunotherapy, which harnesses the body's immune system to recognize and eliminate tumor cells. Despite the promise of nanoparticle-based cancer immunotherapy, current platforms often suffer from poor thermal stability and rapid loss of bioactivity, limiting their clinical efficacy. To address these shortcomings, in this study, we developed a protein-based therapeutic nanovaccine targeting cervical cancer by incorporating cervical tumor antigens into nanoarchaeosomes (NACDPs). The formulated NACDPs were characterized using SEM, DLS, and zeta potential analyses, confirming optimal particle size, surface charge, and stability. NACDPs exhibit high thermostability and enhanced protein loading efficiency. When tested on isolated PBMCs, monocytes and dendritic cells showed high internalization efficiency of NACDPs. Moreover, treatment of dendritic cells and macrophages with 500 µg mL−1 of NACDPs led to significant upregulation of MHC II (HLA-DR) and co-stimulatory surface markers (CD86, CD80, and CD83), indicating maturation of antigen-presenting cells (APCs). Fluorescence imaging revealed that NACDP-treated APCs demonstrated high cytoplasmic uptake of DIO-labeled nanoarchaeosomes compared to control formulations. Co-culture of these APCs with T cells (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio) showed cellular interactions within 30 minutes. This was accompanied by enhanced T cell proliferation, elevated IFN-γ levels, and increased expression of key T cell markers CD4, CD8, and CD20. Notably, T cells primed with NACDPs induced approximately 80% lysis of target cancer cells at a 1
10 ratio) showed cellular interactions within 30 minutes. This was accompanied by enhanced T cell proliferation, elevated IFN-γ levels, and increased expression of key T cell markers CD4, CD8, and CD20. Notably, T cells primed with NACDPs induced approximately 80% lysis of target cancer cells at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 effector-to-target ratio. Based on the various salient outcomes, it is noted that tumor protein incorporated with nanoarchaeosomes unveils a significant role as a vaccine targeting tumor cells in immunotherapy.
10 effector-to-target ratio. Based on the various salient outcomes, it is noted that tumor protein incorporated with nanoarchaeosomes unveils a significant role as a vaccine targeting tumor cells in immunotherapy.
1. Introduction
Cervical cancer is the fourth most prevalent cancer among women worldwide, primarily caused by persistent infection with human papillomavirus (HPV), and poses a global health challenge.1 A global assessment across 185 countries revealed a higher incidence of cervical cancer among younger women under the age of 45, with high mortality rates in regions with limited socioeconomic resources and low levels of human development.2,3 Currently, preventive strategies such as regular screening and HPV vaccination (e.g., Cervarix, Gardasil, Gardasil-9, Heplisav-B, and Bacillus Calmette–Guérin [BCG]) have been implemented in many developed countries.4While these vaccines have demonstrated preventive potential, they are often associated with a broad spectrum of side effects and, in some cases, provoke an unintended immune response, where healthy cells are misdirected to express cancer-like antigens.4 Recently, therapeutic cancer vaccines based on whole tumor proteins or cell lysates have gained interest due to their ability to present a wide array of tumor antigens, thereby mimicking the heterogeneity of tumor cells. However, these protein-based vaccines often suffer from poor immunogenicity, rapid degradation, and challenges related to stability and targeted delivery. These limitations restrict their effectiveness in generating a robust and durable anti-tumor immune response. Current strategies, such as immune checkpoint inhibitors (e.g., PD-1, PD-L1, CTLA-4),5 oncolytic viruses (e.g., adenoviruses, HSV), and CAR-based approaches (CAR-T, CAR-NK, CAR-M),6 have shown efficacy in certain cancers. However, their application in solid tumors like cervical cancer remains limited due to poor antigen presentation, immune evasion, and the presence of a highly immunosuppressive tumor microenvironment. Similarly, therapeutic vaccines under clinical trials for HPV-associated cervical cancer, including VGX-3100 and ADXS11-001,7,8 though promising, are often constrained by low immunogenicity and the secretion of immunosuppressive cytokines.9 Furthermore, vaccination often induces systemic side effects such as anorexia, fatigue, chills, fever, headache, myalgia, and neuralgia.10
In addition to immunological barriers, logistical challenges such as storage and transportation of vaccines under refrigerated conditions (typically 2–8 °C) hinder widespread deployment, particularly in resource-limited or remote settings. Interruptions in the cold chain can severely compromise vaccine stability and efficacy.
To address these limitations, various nanocarriers—such as lipid nanoparticles, polymeric nanoparticles, and inorganic nanoparticles—have been explored for vaccine delivery.11 However, many of these systems still suffer from poor thermal stability and loss of bioactivity over time. In contrast, thermostable nanoarchaeosomes have emerged as promising candidates due to their exceptional rigidity, low permeability, unique stereochemistry, and heterogeneous lipid composition, which together contribute to improved physicochemical stability and biological performance compared to conventional liposomes.12–14
Furthermore, our previous studies demonstrated that the synthesised nanoarchaeosomes (NAs) exhibited excellent pH stability, maintained structural integrity at room temperature, and showed high biocompatibility and efficient entrapment of cancer drugs such as doxorubicin.15 In this context, the current in vitro study investigates the potential of nanoarchaeosomes loaded with cervical cancer cell-derived proteins (NACDPs) as a cervical cancer nanovaccine. The NACDPs demonstrated high internalisation efficiency in PBMC-derived dendritic cells and macrophages, promoting effective antigen presentation and activation of antigen-presenting cells (APCs). Moreover, co-culture assays revealed significant stimulation of T lymphocytes, resulting in a robust immune response against cancer cells. These findings highlight the potential of tumor protein-loaded nanoarchaeosomes as a stable and effective cervical cancer vaccine platform and pave the way for developing personalised nanovaccines with improved storage, delivery, and immunological outcomes.
2. Materials and methods
Fine chemicals, including 1-stearoyl-2-oleoyl-sn-glycero-3-phosphocholine (SOPC) and 1,2-di-O-phytanyl-sn-glycero-3-phosphocholine (archaeal lipids), were purchased from Avanti Polar Lipids, 3,3′-dioctadecyloxacarbocyanine perchlorate (DIO) was purchased from Thermo Fisher Scientific, the Bradford reagent was purchased from SRL, cell staining dyes, including CFSE dye (carboxyfluorescein succinimidyl ester) and CMPTX (Cell Tracker red), were acquired from Thermo Fisher Scientific and CD4, CD8, CD20, CD14, CD80, CD86, CD83, HLA DR, CD11B, and CD163 antibodies were procured from Beckman Coulter.2.1. Procurement and maintenance of the cell line
The human cervical cancer (squamous cell carcinoma) cell line SiHa was obtained from American Type Culture Collection, VA, USA. The cells were cultured and maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1) at 37 °C in an incubator with a 5% CO2 supply.
000 U mL−1) at 37 °C in an incubator with a 5% CO2 supply.
2.2. Extraction of tumour cell proteins from SiHa cells
The SiHa cells were cultured in a T75 flask with complete DMEM medium and observed for 80% confluency, at which the cells were rinsed twice with PBS (phosphate-buffered saline) (pH 7.2) and scraped gently using a cell scraper in 1 mL of PBS suspension. Then, the cellular suspension was collected and then subjected to 5 cycles of freeze–thaw using liquid nitrogen and a dry bath (55 °C), followed by five cycles of homogenisation using a syringe and needle (20-gauge) for complete lysis of cells. Given its downstream processing, the lysis buffer was not used for cell lysis. The lysed cells were then centrifuged at 3200 × g for 10 min at 4 °C to remove debris, and the whole supernatant was collected and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min at 4 °C. The supernatant containing cell line-derived proteins (CDP) was obtained and quantified using the Bradford assay. The assay was performed using the commercial Bradford reagent. 10 µL of the protein sample and 190 µL of the Bradford reagent were mixed and incubated for 10 min at room temperature, and the optical density (OD values) of the test samples was measured at 595 nm using a plate reader (Biotek, Synergy H1). Furthermore, the aliquots of CDP were stored at −80 °C for further use.
000 rpm for 30 min at 4 °C. The supernatant containing cell line-derived proteins (CDP) was obtained and quantified using the Bradford assay. The assay was performed using the commercial Bradford reagent. 10 µL of the protein sample and 190 µL of the Bradford reagent were mixed and incubated for 10 min at room temperature, and the optical density (OD values) of the test samples was measured at 595 nm using a plate reader (Biotek, Synergy H1). Furthermore, the aliquots of CDP were stored at −80 °C for further use.
2.3. Preparation of nanoarchaeosomes (NAs) and loading of tumour cell protein in nanoarchaeosomes (NACDPs)
NAs were prepared following the previously established protocol with slight modifications.16 Briefly, a dry thin film of SOPC (1 mg) was rehydrated using cell-derived protein (1 mL). The mixture was then gently mixed and briefly vortexed to ensure proper dispersion. Separately, a dry film of archaeal lipid (1 mg) was rehydrated with 1 mL of Milli-Q water and vortexed thoroughly. The NA formulation was prepared by combining 800 µL of SOPC rehydrated with cell-derived protein and 200 µL of archaeal lipid rehydrated in Milli-Q water, resulting in an 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (v/v) suspension. This suspension was sonicated for about 30 minutes at 45 °C and subjected to five cycles of freeze–thaw between −80 °C and 37 °C to promote efficient entrapment of the tumour cell proteins into the nanoarchaeosomes. The final nanoformulation was centrifuged at 15
20 (v/v) suspension. This suspension was sonicated for about 30 minutes at 45 °C and subjected to five cycles of freeze–thaw between −80 °C and 37 °C to promote efficient entrapment of the tumour cell proteins into the nanoarchaeosomes. The final nanoformulation was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 10 minutes. The pellet containing the nanoarchaeosomes loaded with tumor cell proteins (NACDPs) was aliquoted and stored at −80 °C until further experimental use. The entrapped NACDPs were quantified by measuring the free, unbound protein in the supernatant using the Bradford assay. The NA samples were stored at 4 °C.
000 rpm at 4 °C for 10 minutes. The pellet containing the nanoarchaeosomes loaded with tumor cell proteins (NACDPs) was aliquoted and stored at −80 °C until further experimental use. The entrapped NACDPs were quantified by measuring the free, unbound protein in the supernatant using the Bradford assay. The NA samples were stored at 4 °C.
2.4. Characterisation of NAs and NACDPs through SEM, DLS and zeta potential analyses
The particle size and surface charge are an integral part of the characterization of nanoformulations. The size and zeta potential of the nanoarchaeosomal formulations (NAs and NACDPs) were measured using scanning electron microscopy (SEM) (S-4800, Hitachi), dynamic light scattering (DLS) (Horiba Scientific DLS) and zeta potential analysis (Horiba Scientific Zeta Analyzer). Subsequently, to evaluate the colloidal stability of NACDPs and the liposomes loaded with CDP at different temperatures, the size of the particles was determined through DLS analyses at 24 °C, 37 °C, and 60 °C. The samples for the various analyses were prepared by subjecting them to ultrasonication for 10 min at 37 °C (BR Biochem Life Sciences) to achieve uniform dispersion of nanoarchaeosomal formulations.2.5. Quantification of total cell-derived protein loaded into nanoarchaeosomes.
The loading efficiency of cell-derived tumour protein in NAs was determined using the Bradford assay. The assay was performed by adding 10 µL of the test samples to 190 µL of the Bradford reagent and incubating for 10 min at room temperature. Post incubation, the optical density (OD) of the test samples was measured at 595 nm. The loading efficiency was further calculated by using the formula given below. Subsequently, to determine the stability of proteins encapsulated in NAs, the NACDPs were stored at room temperature for 14 days. The protein concentration was estimated using the Bradford assay as mentioned in Section 2.5. The protein concentrations were estimated on days 0, 7, and 14.2.6. Qualitative analysis of cell-derived protein loaded into nanoarchaeosomes
Subsequently, the molecular weight and protein retention in NACDPs were analysed qualitatively through electrophoretic separation utilizing SDS–PAGE. About 100 µg mL−1 of the protein samples were mixed with equal volumes of SDS and β-2-mercaptoethanol-containing sample buffer and heated in a dry bath at 95 °C for denaturation. The prepared sample was loaded with the protein marker in a 5% stacking and 12% resolving gel of SDS–PAGE and electrophoresis was performed at 50 V in a stacking gel and the voltage was increased to 100 V for the resolving gel. After electrophoresis, the gel was stained with Coomassie brilliant blue dye for 4–5 h and subsequently destained with the destaining solution containing methanol and glacial acetic acid and then observed for separated protein bands.2.7. Isolation of PBMCs from human whole blood
The peripheral blood mononuclear cells (PBMCs) were derived from the peripheral blood of healthy volunteers (n = 5). Around 10 mL of blood was collected in an EDTA-coated tube. Consequently, the collected blood samples were subjected to gradient centrifugation at 1580 rpm for 30 min at 15 °C using Ficoll Paque Premium (GMP-grade, density: 1.073 g mL−1; GE Healthcare, Chalfont St Giles, UK) as per the manufacturer's instructions. After centrifugation, the plasma and the buffy coat containing the mononuclear cells were collected and washed twice with PBS at 1580 rpm for 10 min at 15 °C. Furthermore, the cell pellet was resuspended in complete RPMI medium with 10% FBS, and viable cells were counted by trypan blue exclusion.2.8. FACS analysis to determine leucocyte population in PBMCs
The collected PBMCs were analysed for various leucocyte populations using flow cytometry analysis through the expression of surface markers like CD14 (monocytes), CD20 (lymphocytes) and CD3 (B cells). The collected PBMCs were washed with PBS at 1500 rpm at 4 °C. Furthermore, the cell pellets were resuspended in PBS, and 1 × 106 cells were used for the analysis. Live/dead fixable violet dye (1 µL mL−1) was used to exclude dead cells. The cells to be stained were blocked using 5% FBS for 15 min at room temperature to block high levels of Fc receptor, which could cause nonspecific binding of antibodies. Subsequently, surface staining was carried out with optimal antibody concentration as per the manufacturer's instructions. The cell surface marker antibodies were added to the cells and incubated for 30 min in the dark at 4 °C. Post incubation, the cells were washed with PBS to remove unbound antibodies by centrifuging at 1500 rpm for 5 min. The cell pellet was further fixed with 2% paraformaldehyde solution and acquired in a flow cytometer (Beckman Coulter CytoFLEX). The results were analysed using CytExpert software version 5.1.2.9. Internalisation efficiency analysis of NAs and NACDPs by leucocyte population
The PBMCs were analysed for their internalization efficiency by incubating the synthesized and characterized NAs and NACDPs with PBMCs. The NAs and NACDPs were labelled with DIO fluorescent dye (1 mM) for the analysis. The cells after 30 minutes of incubation with NAs and NACDPs were collected and washed with PBS. Around 100 µL of cell suspension was aliquoted in a tube and stained for surface antibodies (CD14, CD3 and CD20) for 30 min in the dark at 4 °C. Post incubation, the cells were washed with 200 µL of wash buffer and centrifuged at 1500 rpm for 5 min. The cells were then resuspended in 200 µL of PBS and immediately analysed using a flow cytometer.2.10. Generation of antigen presenting cells (APCs—macrophages and dendritic cells) from PBMCs
Macrophages and dendritic cells were obtained using well-developed methods with minor modifications.17 Briefly, the isolated PBMCs as mentioned above were suspended in RPMI medium, plated on 6-well plates at 2.5 × 106 cells per well and allowed to adhere for 2 hours at 37 °C in a humidified 5% CO2 atmosphere. Monocytes can attach to the plastic surfaces; hence, the cells (other than monocytes) in suspension/floating were collected and stored for coculture assays, whereas the adhered cells were considered as enriched monocytes. Fresh RPMI 1640 medium supplemented with granulocyte macropahge colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4) (600 IU and 100 IU per mL) was added to the adhered cells and cultured for 5 days to differentiate monocytes in to dendritic cells, similarly macropahge colony stimulating factor (M-CSF) (1 µg mL−1) and IL-4 (1 µg mL−1) were added to the adherent monocytic cells for macrophage differentiation. The differentiated cells (dendritic cells and macrophages) were harvested on day 5 using a non-enzymatic cell dissociation solution (Sigma-Aldrich).2.11. Determination of macrophage activation and dendritic cell maturation through phenotypic analysis using FACS
The macrophages and dendritic cells incubated with growth factors and cytokines, as mentioned above, were removed and replaced with a fresh medium on day 6 for activation with the antigen (NACDPs). The cells were incubated with NACDPs at a concentration of 500 µg mL−1. The positive control cells were obtained by incubating the cells with 100 ng mL−1 lipopolysaccharide (LPS) and 20 ng mL−1 IFN-γ, while cells without NACDP stimulation were considered unstimulated negative control UDCs. Subsequently, before co-incubation with cells, the NACDP suspensions were all sterilised with 0.22 µm filters and incubated for 72 h with macrophages or DCs and analysed for the expression of cell surface markers such as CD80, CD86, CD83, HLADR, CD11B and CD163 using flow cytometry. The cells were stained with the respective antibodies and incubated for 30 min in the dark at room temperature. The unbound dye antibodies were removed by washing the cells in PBS at 1500 rpm for 5 min, followed by 2% paraformaldehyde fixation of the cell pellet for later acquisition in FACS. Additionally, a fluorescence microscopy-based imaging system was also used to confirm the internalisation of NAs and NACDPs by labelling the nanoformulation with DIO fluorescent dye, which has a hydrophilic nature. The fluorescence of DIO is greatly enhanced upon interaction with cell membranes bound to lipophilic biomolecules. The dendritic cells and macrophages were counterstained with CMPTX red (viable cell tracker dye) at a final concentration of 5 µM, and images were acquired using a confocal microscope (Leica TCS SP8).2.12. Morphological characterisation by scanning electron microscopy
The morphologies of the differentiated dendritic cells and macrophages were analysed using a scanning electron microscope (S-4800, Hitachi). The synthesised and harvested cell pellets of DCs and macrophages were resuspended at a dilution of 1 mL of PBS, and the cell suspensions were placed on a pre-cleaned glass slide and incubated at room temperature for 30 min. Furthermore, the cells were fixed with 2.5% glutaraldehyde and subjected to gold sputter coating at 2.5 kV and 20 mA at a rate of 10 nm per min (Polaron SC7640 gold sputter). The morphologies of cells were characterised at a voltage of 1–5 kV.2.13. Analysis of interferon-gamma secretion in T cells after co-culture with APCs
Autologous mixed lymphocytes were isolated from healthy donor blood (n = 5). The mixed lymphocytes were labelled with CFSE (carboxyl fluorescein succinimidyl ester) dye (Life Technologies). Briefly, the cells were resuspended in 1 mL of PBS, stained with a final concentration of 5 µM of Cell Trace CFSE dye, and incubated for 20 min in a 37 °C water bath. Post incubation, the unbound CFSE was removed by diluting the cell pellet in FBS containing RPMI medium and incubating further for 5 min at 37 °C. The cells were then centrifuged at 1500 rpm for 5 min to pellet down the cells. The cells were again resuspended in RPMI medium and distributed to perform a coculture assay. The co-culture of stimulated DCs and macrophages with mixed lymphocytes was performed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratio of differentiated cells with mixed lymphocytes. The cells without any stimulation by DCs or macrophages were considered unstimulated or a negative control. After seven days of T cell proliferation, IFN-γ secretion was assessed by flow cytometry. To assess the proliferating B cells (CD20) and T cells, the CD4 (helper T cell) and CD8 (cytotoxic T cell) markers were used along with CFSE. The procedure was carried out following standard surface staining procedures as mentioned above. The divided and undivided populations of cells were determined using a CFSE baseline control. The covalently bound CFSE divides equally between daughter cells, allowing discrimination of successive rounds of cell division; as the intensity of the dye decreases with an increase in cell proliferation, a maximum of eight generations of cell division can be analysed. To determine the intracellular secretion of IFNγ, the collected cells were initially fixed with 4% paraformaldehyde (PFA) for 15 min and to remove excess PFA the cells were washed with wash stain buffer consisting of PBS with 2% Tween 20, 10% sodium azide and 5% saponin. Furthermore, the cells were permeabilised with 100 µL of 0.25% saponin solution and incubated at 4 °C for 15 min. Subsequently, the cells were washed with wash buffer, and the fluorochrome-conjugated IFN-γ antibody was added to 100 µL of cell suspension and incubated for 15 min at 4 °C. Then the cells were washed twice with wash buffer and resuspended with 2% PFA before flow acquisition.
50 ratio of differentiated cells with mixed lymphocytes. The cells without any stimulation by DCs or macrophages were considered unstimulated or a negative control. After seven days of T cell proliferation, IFN-γ secretion was assessed by flow cytometry. To assess the proliferating B cells (CD20) and T cells, the CD4 (helper T cell) and CD8 (cytotoxic T cell) markers were used along with CFSE. The procedure was carried out following standard surface staining procedures as mentioned above. The divided and undivided populations of cells were determined using a CFSE baseline control. The covalently bound CFSE divides equally between daughter cells, allowing discrimination of successive rounds of cell division; as the intensity of the dye decreases with an increase in cell proliferation, a maximum of eight generations of cell division can be analysed. To determine the intracellular secretion of IFNγ, the collected cells were initially fixed with 4% paraformaldehyde (PFA) for 15 min and to remove excess PFA the cells were washed with wash stain buffer consisting of PBS with 2% Tween 20, 10% sodium azide and 5% saponin. Furthermore, the cells were permeabilised with 100 µL of 0.25% saponin solution and incubated at 4 °C for 15 min. Subsequently, the cells were washed with wash buffer, and the fluorochrome-conjugated IFN-γ antibody was added to 100 µL of cell suspension and incubated for 15 min at 4 °C. Then the cells were washed twice with wash buffer and resuspended with 2% PFA before flow acquisition.
2.14. Interaction of DCs and macrophages with lymphocytes
Dendritic cells and macrophages were stained with CMPTX red fluorescent viable cell tracker dye, which helps in monitoring the movement of cells and proper tracking. To distinguish the cells from APCs and to visualize the distinct interaction, the T cells were labelled with CFSE fluorescent dye. After an hour of co-culture of labelled APCs and CFSE, the cells were fixed with 4% paraformaldehyde, incubated for 5 min, and washed to remove excess fixative. The interactions were visualised using a confocal microscope (Leica TCS SP8).2.15. Assessment of tumour cell cytotoxicity in vitro
To assess the cytotoxicity of tumor cells, the activated lymphocytes were co-cultured with CFSE-labelled SiHa cancer cells for 24 h at different ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10–1
10–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50). The culture medium was prepared by mixing half DMEM medium and half complete RPMI 1640 medium to maintain the cells. Post incubation, the co-cultured cells were collected and resuspended in 100 µL of PBS. Subsequently, the cells were stained with 5–10 µL of propidium iodide (PI), which is a membrane-impermeant dye, followed by incubation in the dark for 1 min, and analysed using a flow cytometer. Initially, the data were acquired for unstained and single-colour positive controls, and subsequently, the stained cells were analysed for viable and dead cells through a dot-plot of forward scatter versus PI stain. With the acquired data, the percentage of tumour cell death was analysed, and double-positive cells (both CFSE and PI) were considered dead tumor cells.
50). The culture medium was prepared by mixing half DMEM medium and half complete RPMI 1640 medium to maintain the cells. Post incubation, the co-cultured cells were collected and resuspended in 100 µL of PBS. Subsequently, the cells were stained with 5–10 µL of propidium iodide (PI), which is a membrane-impermeant dye, followed by incubation in the dark for 1 min, and analysed using a flow cytometer. Initially, the data were acquired for unstained and single-colour positive controls, and subsequently, the stained cells were analysed for viable and dead cells through a dot-plot of forward scatter versus PI stain. With the acquired data, the percentage of tumour cell death was analysed, and double-positive cells (both CFSE and PI) were considered dead tumor cells.
2.16. Statistical analysis
Data are mean ± S.D. and n = 5 unless indicated. Statistical significance was determined by one-way ANOVA (*P < 0.05, **P < 0.01).3. Results and discussion
3.1. Synthesis and characterisation of NAs and NAs loaded with cell-derived proteins (NACDPs)
Nanoarchaeosomes (NAs) were synthesized by sonication of archaeal total polar lipids in phosphate-buffered saline, following previously established protocols.15 For protein loading, crude tumor protein was extracted from SiHa cervical cancer cells at ∼90% confluence using phosphate-buffered saline (PBS). The extracted protein was then encapsulated into NAs using a freeze–thaw method. Both NAs and NAs loaded with cell-derived proteins (NACDPs) were characterized to evaluate their physicochemical properties. The synthesized NAs and NACDPs were initially characterized to determine the size through SEM and DLS analyses, as shown in Fig. 1. The SEM results revealed a uniform spherical vesicle with an average size of 52.5 ± 3 nm and 57 ± 3 nm for NAs and NACDPs, respectively. Consistent with the SEM analysis, the results obtained from DLS also suggested an average size of 53 ± 5 and 58 ± 4 nm and a polydispersity index (PDI) of 0.3. The charge densities determined using a zeta analyser were −52.5 ± 2.83 mV and −42 ± 4.5 mV for NAs and NACDPs, respectively (Fig. S1a & b). The reduction in the surface charge indicates successful encapsulation of cell-derived proteins and protein–lipid interactions. Additionally, elemental analysis by energy dispersive X-ray (EDX) spectroscopy showed strong peaks for carbon and oxygen in NAs, while NACDPs displayed spectral signatures for nine elements (Tables S1–S3), with increased elemental weight confirming successful protein encapsulation within the NA matrix. The quantitative validation yielded a loading efficiency of approximately 94 ± 5%, as determined by UV-vis spectrophotometric analysis at 480 nm. Additionally, the protein stability was analysed qualitatively using the Bradford assay and the data are presented in SI Section S1.2.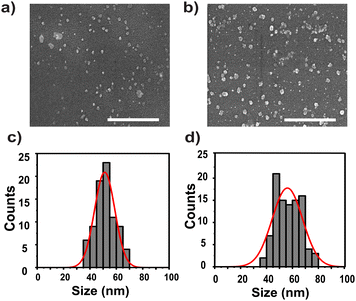 | ||
| Fig. 1 SEM images of (a) NAs and (b) NAs loaded with cancer cell derived proteins (NACDPs); DLS measurements of (c) NAs and (d) NACDPs. Scale: 500 nm. | ||
To qualitatively validate protein encapsulation, the preliminary analysis of SDS–PAGE was performed on both the total tumor protein and NACDPs. The gel electrophoresis results showed clear protein bands in NACDPs (lane II) comparable to the original tumor protein (lane I), confirming the successful incorporation of cell-derived proteins into the NAs (Fig. S1c).
NAs possess a unique bilayer structure composed of archaeal lipids with ether linkages and isoprenoid chains, which enhances membrane rigidity and stability, facilitating high encapsulation efficiency. The freeze–thaw method promotes the formation of large internal aqueous compartments, allowing efficient entrapment of hydrophilic tumor proteins.18 Additionally, it is speculated that strong protein–lipid interactions due to the heterogeneous lipid composition of NAs further support stable protein incorporation.
Furthermore, we tested the stability of the NACDPs and conventional liposomes loaded with CDP by measuring their average size at various temperatures using DLS. The results suggested that, for NACDPs, no significant size increase is observed with an increase in temperature, confirming their colloidal stability. However, for the liposomes loaded with CDP, the average size increased from 55.5 ± 2.3 to 274 ± 3.7 nm with an increase in temperature (Fig. S2a and b).
The stability of nanocarriers during storage is an important factor that has to be considered for their therapeutic applications. The developed formulations should have the capability to remain unaltered concerning their chemical and physical properties. From the obtained results, it is evident that the NACDPs maintained their size under various temperature conditions compared to conventional liposomes.
3.2. Determination of leucocyte population in PBMCs using FACS
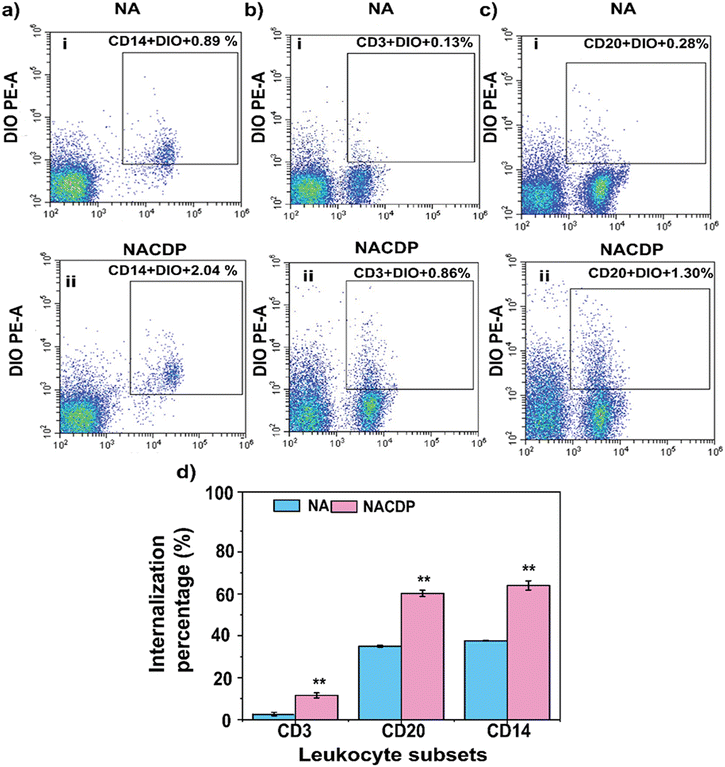
Peripheral blood mononuclear cells (PBMCs) isolated from whole blood were analyzed using flow cytometry (FACS) to identify various leukocyte subtypes. Distinct leukocyte populations were gated based on specific cell surface markers: CD14 for monocytes, CD3 for T lymphocytes, and CD20 for B cells. As shown in Fig. S3, FACS analysis revealed that CD14 monocytes constituted approximately 11.6 ± 2.3% of the total leukocytes. CD3 T cells accounted for 47.1 ± 3.2%, while CD20 B cells represented around 3.1 ± 0.1 7%. These results indicate a relatively higher proportion of T cells compared to monocytes and B cells within the PBMC population.3.3. Internalization efficiency of NAs and NACDPs by leucocytes
We further quantified the internalization efficiency and cytotoxicity of DIO-labelled NAs and NACDPs across different leukocyte populations. There is no significant decrease in the viability of the NA- and NACDP-treated leukocyte populations compared to control cells, confirming the biocompatibility of NAs and NACDPs. Among all cell types analyzed, monocytes demonstrated the highest uptake, with internalization efficiencies of 37.6 ± 14.9% for NAs and 64 ± 13.2% for NACDPs, as shown in Fig. 2a (i & ii) and d. In contrast, both T and B lymphocytes exhibited lower uptake levels: 2.6 ± 0.3% and 11.5 ± 2.4% for NAs, and 35 ± 11% and 60 ± 12% for NACDPs (Fig. 2b-c (i & ii) and d, respectively. These results confirm that monocytes possess superior internalization capacity, making them the preferred population for further experimental analyses. The enhanced internalization of NACDPs across all leukocyte subsets, particularly monocytes and B cells, may be attributed to the presence of cell-derived proteins. These proteins likely mimic endogenous antigens or damage-associated molecular patterns (DAMPs), enhancing recognition by immune receptors and promoting uptake.19Additionally, the unique membrane composition of archaeosomes—rich in ether lipids—facilitates strong interactions with immune cells, especially phagocytic cells, thereby increasing internalization efficiency.20 Monocytes were prioritized for subsequent studies due to their pronounced phagocytic activity and innate immune response. Leukocytes are generally activated upon exposure to foreign substances, and monocytes, macrophages, and dendritic cells express a diverse repertoire of surface proteins essential for phagocyte function. Prior research has shown that monocytes have a higher capacity (up to 12%) to engulf 200 nm sized nanoparticles compared to other immune cells.21 Importantly, previous studies have demonstrated that archaeosomes composed of total polar lipids from Methanococcus voltae, Methanosarcina mazei, and Methanobrevibacter smithii, particularly when combined with conventional phospholipids, exhibit significantly higher uptake by phagocytic cells than standard liposomes.22 These findings highlight the potential of NAs as robust delivery vehicles for antigen presentation and immune modulation.
3.4. Morphological and phenotypic determination of dendritic cells and macrophages through microscopic and FACS analyses
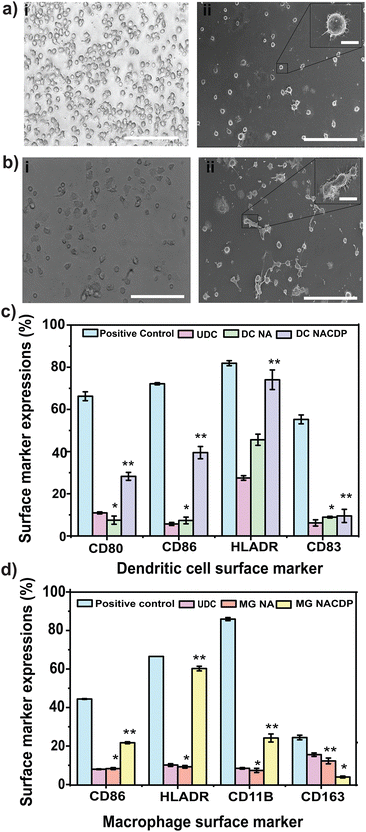
The monocytes from PBMCs with higher internalisation efficiency were further differentiated into dendritic cells and macrophages, and their morphologies were analysed using light microscopy and scanning electron microscopy (SEM). As shown in Fig. 3a(i), DCs displayed an adherent phenotype with a spread-out cytoplasm and prominent nuclei under light microscopy, while SEM images (Fig. 3a(ii)) revealed the presence of ruffled membranes and extended dendrites indicative of activated morphology. Similarly, differentiated macrophages exhibited distinct morphological features under light microscopy (Fig. 3b(i)), and SEM analysis confirmed well-spread structures and membrane projections typical of macrophage activation (Fig. 3b(ii)).To further evaluate the maturation status, flow cytometry analysis was performed to quantify the expression of characteristic surface markers on DCs and macrophages. Fig. 3c shows the expression of co-stimulatory molecules (CD80, CD86, CD83 and HLA-DR) of the unprimed dendritic cells (UDCs) and DCs treated with lipopolysaccharide (LPS) (positive control), NAs (DC NAs) and NACDPs (DC NACDPs). The DC NACDPs exhibited significantly elevated levels of the co-stimulatory molecules CD80 (28 ± 1.8%) and CD86 (39 ± 2.9%), as well as CD83 (9.49 ± 3%), a key marker of dendritic cell maturation, compared to the DC NAs and UDCs. Additionally, HLA-DR expression, critical for antigen presentation, was observed at 78 ± 3% in DC NACDPs. UDCs showed minimal expression of all DC-specific markers, serving as a negative control. No significant differences were observed between DC NAs and UDCs, confirming that NAs alone do not induce dendritic cell maturation. The positive control (PC), treated with LPS, showed the highest expression across all markers, as expected for fully mature DCs.
Macrophage analysis (Fig. 3d) revealed that the MG NACDP group showed significantly higher expression levels of CD86 (21 ± 0.5%), HLA-DR (60 ± 1.2%), CD11B (24 ± 2.1%), and CD163 (4 ± 0.5%) in comparison to MG NA and UDC groups, which showed minimal marker expression. These results suggest successful phenotypic polarization and functional priming of macrophages following NACDP treatment. LPS stimulated macrophages (PC) demonstrated the highest expression of these markers.
To evaluate the activation status of macrophages, flow cytometry analysis was also carried out to detect surface markers relevant to macrophage polarization and function. The MG-NACDP group showed a significant increase in the surface expression of CD86 (21 ± 2%), HLA-DR (60 ± 4%), and CD11B (24 ± 2%), indicating a shift toward an activated and antigen-presenting phenotype. A modest increase in CD163 (4 ± 1%), a marker commonly associated with the M2-like regulatory phenotype, was also observed. In contrast, the UDC and MG NA groups displayed minimal expression of all tested markers, suggesting that NAs alone do not significantly influence macrophage activation. The highest levels of all markers were again observed in the LPS-treated macrophages (PC), confirming successful polarization and activation. These results suggest that NACDPs induce functional activation of macrophages, enhancing their potential for antigen presentation and immune modulation.
The high expression of CD86 and HLA-DR in both DCs and MGs indicates enhanced T cell co-stimulation and antigen presentation, respectively. In DCs, the increased CD83 expression—a classical marker of mature dendritic cells—further confirms that NACDPs drive phenotypic maturation, potentially through endosomal Toll-like receptor (TLR) engagement or inflammasome activation. The encapsulated cancer cell derived proteins (CDPs) likely contribute additional antigen-specific signaling, facilitating MHC class II loading and adaptive immune priming. In macrophages, the upregulation of CD11B and CD86 suggests a transition toward an M1-like phenotype, while the simultaneous expression of CD163, typically linked with M2 macrophages, points to a mixed or hybrid phenotype.
This dual expression pattern may reflect a plastic activation state that balances pro-inflammatory responses with tissue remodeling and resolution signals—a hallmark of functional antigen-presenting macrophages. It is plausible that NACDPs trigger both TLR-mediated pro-inflammatory signaling and a feedback loop involving IL-10 or TGF-β, thereby inducing partial CD163 expression.23 Moreover, the lack of significant surface marker induction in the DC-NA and MG-NA groups reinforces that NAs alone do not induce immune activation in the absence of antigenic cargo, thereby highlighting the critical role of antigen-specific immune modulation in NACDPs. Collectively, these findings suggest that NACDPs not only ensure efficient delivery and presentation of CDP but also act as an immune adjuvant, capable of priming both innate and adaptive immune responses.
3.5. NACDPs’ internalization and T cell activation by antigen-presenting cells
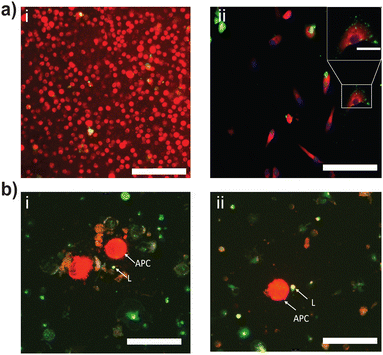
Next, the internalization efficiency of DCs and macrophages was assessed using fluorescence microscopy. NACDPs were labeled with DIO (green fluorescence), while dendritic cells and macrophages were counterstained with CMTPX (red fluorescence), as shown in Fig. 4. These APCs displayed their characteristic phagocytic behaviour by actively engulfing the NACDPs. The internalized DIO-labelled NACDPs appeared as punctate fluorescent signals within the cytoplasmic region of both dendritic cells and macrophages, as shown in Fig. 4a(i & ii), confirming successful uptake.Following this step, the primed dendritic cells and macrophages were co-cultured with T cells for the antigen presentation process. The fluorescence microscopy observation results showed that 30 min of co-incubation of APCs with T cells exhibited an explicit cell-to-cell interaction and demonstrated effective immune function by triggering T cell activation as shown in Fig. 4b(i & ii). The results further demonstrate clustering and a vivid interaction of T cells (DIO-labelled) with APCs (CMPTEX dye); moreover, this observation ensures that a minimum interaction time of APCs with T cells could effectively stimulate signals for immune activity. Dendritic cells and macrophages share similarities in their developmental origin, surface phenotype, and immunological roles. Both exhibit high phagocytic activity and are central to the activation of T cells via antigen presentation.24 Under immunosuppressive conditions, such as in cancer, T cells often become dysfunctional or remain in a dormant state.25 In such cases, conventional dendritic cells and macrophages play a pivotal role in reactivating anti-tumor immune responses.26 However, APCs can also influence the immune balance by stimulating regulatory T cells (Tregs), which subsequently modulate effector T cell responses.27
In the present study, NACDP-treated dendritic cells and macrophages successfully internalized the nanocarriers and localized them in the cytoplasmic region. Following internalization, the NACDP-primed APCs engaged in active antigen presentation, leading to the activation of naïve T cells into effector T cells—an essential step for mounting a robust immune response.
3.6. Proliferation and IFN-γ production in T cells
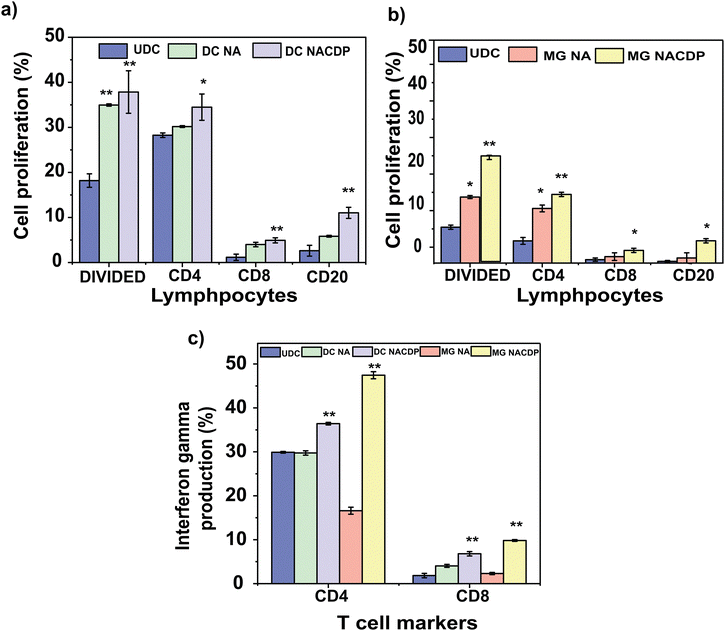
T cell proliferation and production of interferon-gamma is an essential process to mount an effective immune response against tumor cells under cancer conditions. Interferon-gamma is a soluble cytokine produced by T lymphocytes that plays a vital role in innate and adaptive immunity. The present study assessed the proliferative capacity and IFN-γ production of effector T cells through FACS analysis. The proliferation rate of specific lymphocytes among the divided cells was tracked with CFSE dye (Fig. S4 a and b). The analysis results suggested that the mature dendritic cells (DCs) treated with NACDPs (DC NACDPs) effectively enhanced the population of CD4 (T helper cell marker) at a percentage of 34.48 ± 2.9%, CD8 (cytotoxic lymphocytes) at 4.92 ± 0.5% and CD 20+ (B cell marker) at a percentage of 11.03 ± 1.2%, whereas the DC cells treated with NAs (DC-NA) alone expressed 30.18 ± 0.2% of CD4, 3.99 ± 0.5% of CD8 and 5.83 ± 0.2% of CD20+ markers as shown in Fig. 5a. Similarly, the mature macrophage (MG) cells treated with NACDPs (MG NACDPs) upon interaction with T cells significantly boosted the expression of CD4, CD8 and CD20 at a percentage of 16.09 ± 0.5%, 2.97 ± 0.5% and 5.2 ± 0.5%, respectively, whereas the MG cells treated with NA (MG NA) stimulated T cell marker expression at a percentage rate of 12.8 ± 0.8% (CD4), 1.5 ± 0.9% (CD8) and 1.2 ± 1.2% (CD20) (Fig. 5b). Subsequently, the production of IFN-γ by T cells was estimated at the end of 5 days of co-culture with primed APCs (Fig. 5c and Fig. S4c and d).The results revealed that the DC NACDPs stimulated T cells (CD4) to produce a significant amount of IFN-γ at a percentage rate of 36 ± 0.3% when compared to that of DC NAs alone. Likewise, MG NACDPs contributed to a significant increase in IFN-γ production with 47.44 ± 1% in T cells (CD4). It is also observed that T cells with the CD8 marker treated with DC NACDPs and MG NACDPs also produced 7 ± 1% and 10 ± 0.2% of IFN-γ, which is significantly higher than that of DC NA, MG NA and control samples. The results from the present analysis revealed that the APCs primed with NACDPs stimulated the T cells to produce a significantly increased level of IFN-γ when compared to that of APCs primed with NAs alone and the control, as shown in Fig. 5c.
The proliferation of T cells and cytokine production are crucial factors involved in the activation and development of immune responses against any tumour conditions. Among the various secreted mediators, interferon-gamma is a critical mediator of tumour immunity. Activated lymphocytes like CD4 and CD8 are primarily involved in the production of IFN-γ.28 Moreover, CD4 and CD8 T cells play a major part in the cancer-immune cycle and exert a significant influence on clinical outcomes.29 With the obtained results, the NACDPs significantly influenced the populations of CD4 and CD8 T cells and enhanced the immune function.
3.7. Cytotoxic effect of T cells on tumor cells (SiHa cells)
To evaluate the tumor-killing potential of effector T cells activated via NACDP-primed antigen-presenting cells (APCs), various effector-to-target (E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) T) cell ratios were tested in co-culture with SiHa cervical cancer cells. Notably, effector T cells generated by NACDP-primed dendritic cells exhibited robust cytotoxicity, achieving an 87 ± 1.5% kill rate at a minimum ratio of 1
T) cell ratios were tested in co-culture with SiHa cervical cancer cells. Notably, effector T cells generated by NACDP-primed dendritic cells exhibited robust cytotoxicity, achieving an 87 ± 1.5% kill rate at a minimum ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (E
20 (E![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) T). In comparison, effector T cells stimulated by NACDP-primed macrophages demonstrated a significant, albeit slightly lower, cytotoxic effect of 67.49 ± 3.2%. Subsequently, the dendritic cells and macrophages primed with NAs demonstrated a cytotoxic effect of 41 ± 1% and 51 ± 1.2% on SiHa cells at the same ratio, as illustrated in Fig. 6 and Fig. S5.
T). In comparison, effector T cells stimulated by NACDP-primed macrophages demonstrated a significant, albeit slightly lower, cytotoxic effect of 67.49 ± 3.2%. Subsequently, the dendritic cells and macrophages primed with NAs demonstrated a cytotoxic effect of 41 ± 1% and 51 ± 1.2% on SiHa cells at the same ratio, as illustrated in Fig. 6 and Fig. S5.
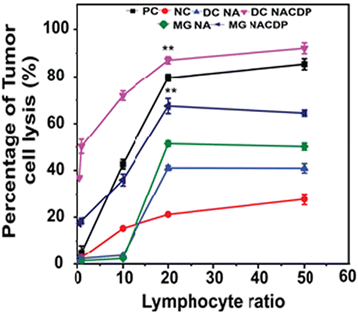 | ||
| Fig. 6 Quantitative determination of cell lysis in SiHa cells incubated with various ratios of effector T cells (PC—positive control; NC—negative control). | ||
These findings indicate a dose-dependent relationship between the number of effector T cells and the observed cytotoxic response. This enhanced tumoricidal activity confirms that both dendritic cells and macrophages primed with NACDPs effectively induce functional effector T cells with strong cytotoxic properties. The results underscore the pivotal role of both innate and adaptive immune components in eliciting a robust anticancer response. Importantly, cytotoxicity in the tumor microenvironment is predominantly mediated by activated CD4 and CD8 T lymphocytes, which are also major producers of IFN-γ.CD4 T cells contribute to tumor cell killing through the secretion of pro-inflammatory cytokines, cytotoxic granzymes (GZM), and perforin (PRF1),30 whileCD8 T cells serve as the primary cytotoxic effectors with potent antitumor activity. In alignment with these established mechanisms, our study confirms that NACDPs efficiently activate CD4 and CD8 T cells, equipping them with strong effector functions to combat tumor cells. These observations further validate the therapeutic potential of NACDPs in modulating immune responses within the tumor microenvironment.
4. Conclusions
The findings from this in vitro study present a compelling immunotherapeutic strategy for cervical cancer by harnessing tumor-derived proteins encapsulated within lipid-based nanoarchaeosomes. This innovative neoantigen-loaded nanovaccine platform demonstrates significant potential to reshape the landscape of cancer immunotherapy by enabling efficient and targeted activation of the immune system. Notably, the nanoarchaeosomal formulation effectively facilitates antigen delivery to primary antigen-presenting cells—dendritic cells and macrophages—without the requirement for high concentrations of tumor protein, thereby promoting their maturation and polarization. Furthermore, this versatile nano-delivery system can be engineered to carry either crude tumor antigens or tumor-specific neopeptides, allowing tailored immunotherapy across various cancer types. An important future direction for NACDPs lies in their application to personalized cancer immunotherapy. The archaeosomal platform, owing to its exceptional thermostability and ability to preserve antigen integrity, offers a promising avenue for the incorporation of patient-specific tumor antigens. In principle, tumor-associated proteins or neoantigens isolated from a patient's biopsy or circulating tumor material could be directly loaded into NACDPs, generating a personalized nanovaccine tailored to the individual's tumor antigenic landscape. This strategy has the potential to overcome tumor heterogeneity and enhance immune recognition of unique patient-specific antigens, thereby improving therapeutic efficacy compared to “one-size-fits-all” vaccines. The feasibility of such an approach is supported by the demonstrated stability of NACDPs under both elevated temperatures and prolonged storage, which ensures that labile patient-derived proteins could remain structurally intact during formulation and administration. Moreover, the nanoscale size and archaeosome-based membrane composition may facilitate efficient delivery to antigen-presenting cells, a key requirement for robust immune activation. However, several challenges must be considered for clinical translation. The reliable and rapid isolation of tumor proteins or peptides from patient samples remains technically demanding and may require the integration of high-throughput proteomic platforms. Standardization of antigen quality, yield, and purity would also be critical to ensure reproducibility across patients. Additionally, regulatory and manufacturing pathways for patient-specific formulations are more complex compared to off-the-shelf vaccines, necessitating specialized infrastructure for individualized production. Despite these challenges, the flexibility of the NACDP platform, combined with advances in tumor antigen discovery, makes it highly adaptable to personalized medicine strategies. Future work focusing on integrating NACDPs with patient-derived antigens could establish a new paradigm in individualized cancer immunotherapy, bridging the gap between robust nanocarrier design and precision oncology.Author contributions
Abirami Seetharaman: investigation, validation of experimental methodologies, experiments, preparation of the original draft, formal analysis and data curation. Parimalanandhini Duraisamy: writing of the original draft, formal analysis, data interpretation and editing of the original draft. Priya Ramanathan: instrumental facility. Subastri Ariraman: methodology validation and review and editing of the original draft. Swathi Sudhakar: supervision, resources, project administration, methodology, investigation, funding acquisition, formal analysis and conceptualization.Conflicts of interest
The authors declare no conflicts of interest.Data availability
The data supporting the findings of this study are available within the article. Further inquiries regarding the data can be directed to the corresponding author.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5ma01014d.
Acknowledgements
The authors duly acknowledge the funding support from the Indian Institute of Technology Madras (IITM) and, through the Ministry of Education, Goverment of India (Project No: SP24250430AMMOEX009000), Indo-French Centre for the Promotion of Advanced Research (Project No: SP24251136AMCEFI009000), Department of Biotechnology, Goverment of India (Project No: SP25261072AMDBTX009000). The authors are also grateful to Dr Priya Ramanathan (Adyar Cancer Institute) and Dr Saumendra K Bajpai (Applied Mechanics and Biomedical Engineering) for providing instrumentation facilities.References
- F. Bray, J. Ferlay, I. Soerjomataram, R. L. Siegel, L. A. Torre and A. Jemal, Ca-Cancer J. Clin., 2018, 68, 394–424 CrossRef.
- M. Arbyn, E. Weiderpass, L. Bruni, S. de Sanjosé, M. Saraiya, J. Ferlay and F. Bray, Lancet Global Health, 2020, 8, e191–e203 CrossRef PubMed.
- D. Singh, J. Vignat, V. Lorenzoni, M. Eslahi, O. Ginsburg, B. Lauby-Secretan, M. Arbyn, P. Basu, F. Bray and S. Vaccarella, Lancet Global Health, 2023, 11, e197–e206 CrossRef CAS.
- N. Abd-Aziz and C. L. Poh, J. Oncol., 2022, 2022, 9749363 CrossRef.
- M. D. Hellmann, R. Nanda and J. R. Bauman, Nat. Rev. Clin. Oncol., 2021, 18, 477–489 Search PubMed.
- S. Tahmasebi, A. Nunez and Y. Zhi, J. Clin. Oncol., 2024, 42, 450–464 Search PubMed.
- P. K. Bhuyan, M. Dallas, K. Kraynyak, T. Herring, M. Morrow, J. Boyer, S. Duff, J. Kim and D. B. Weiner, Hum. Vaccines Immunother., 2021, 17, 1288–1293 CrossRef CAS.
- B. A. Miles, B. J. Monk and H. P. Safran, Gynecol. Oncol. Res. Pract., 2017, 4, 9 CrossRef.
- A. Kamath, Cancer Immunol. Res, 2021, 9, 324–336 CrossRef.
- G. P. Dunn, Cancer Research Institute, available online: https://www.cancerresearch.org/cancer-vaccines, accessed April 18, 2025.
- J. Chiodini, Nurs. Stand., 2014, 28, 45–52 CrossRef.
- G. B. Patel and G. D. Sprott, Crit. Rev. Biotechnol., 1999, 19, 317–357 CrossRef CAS.
- M. J. Altube, M. M. B. Martínez, B. Malheiros, P. C. Maffía, L. R. S. Barbosa, M. J. Morilla and E. L. Romero, Mol. Pharm., 2019, 17, 70–83 CrossRef.
- N. Charó, H. Jerez, S. Tatti, E. L. Romero and M. Schattner, Pharmaceutics, 2022, 14, 736 CrossRef.
- K. V. Babunagappan, A. Seetharaman, S. Ariraman, P. B. Santhosh, J. Genova, N. P. Ulrih and S. Sudhakar, Nanoscale Adv., 2024, 6, 2026–2037 RSC.
- S. Ariraman, A. Seetharaman, K. V. Babunagappan and S. Sudhakar, Mater. Adv., 2024, 5, 6944–6956 RSC.
- G. Cechim and J. A. Chies, An. Acad. Bras. Cienc., 2019, 91, e20190310 CrossRef.
- P. B. Santhosh and J. Genova, ACS Omega, 2022, 8, 1–9 CrossRef PubMed.
- F. Liang and K. Loré, Clin. Transl. Immunol., 2016, 5, e74 CrossRef.
- M. Bhandari, R. Kumar, S. Gangwar, A. Singh, D. U. Dhakrey, A. Patel, N. Wadhwani and R. E. Jesudasan, Rev. Article (journal unspecified, please confirm for proper formatting) Search PubMed.
- R. Mohammapdour and H. Ghandehari, Adv. Drug Delivery Rev., 2022, 180, 114022 CrossRef CAS PubMed.
- D. L. Tolson, R. K. Latta, G. B. Patel and G. D. Sprott, J. Liposome Res., 1996, 6, 755–776 CrossRef CAS.
- A. B. Alves-Januzzi, M. K. Brunialti and R. Salomao, Cytometry, Part B, 2017, 92, 192–198 CrossRef CAS PubMed.
- I. Mellman and R. M. Steinman, Cell, 2001, 106, 255–258 CrossRef CAS.
- D. S. Thommen and T. N. Schumacher, Cancer Cell, 2018, 33, 547–562 CrossRef CAS.
- D. Nagorsen, S. Voigt, E. Berg, H. Stein, E. Thiel and C. Loddenkemper, J. Transl. Med., 2007, 5, 1 CrossRef.
- A. Visperas, J. Do and B. Min, J. Immunol. Res., 2014, 2014, 750374 Search PubMed.
- J. D. Burke and H. A. Young, Semin. Immunol., 2019, 43, 101280 CrossRef CAS.
- S. Hadrup, M. Donia and P. T. Straten, Cancer Microenviron., 2013, 6, 123–133 CrossRef CAS PubMed.
- L. Xie, J. Fang, J. Yu, W. Zhang, Z. He, L. Ye and H. Wang, MedComm, 2023, 4, e390 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |