Open Access Article
Open Access ArticleMannose-6-phosphate functionalized liposomal nanocarrier for lysosome-specific delivery of β-glucuronidase in a Drosophila model of MPS VII
Abinash
Padhy†
 a,
Apurba
Das†
b,
Keya
Mondal
a,
Basudeb
Mondal
a,
Apurba
Das†
b,
Keya
Mondal
a,
Basudeb
Mondal
 a,
Rupak
Datta
*b and
Sayam
Sen Gupta
a,
Rupak
Datta
*b and
Sayam
Sen Gupta
 *a
*a
aDepartment of Chemical Sciences, Indian Institute of Science Education and Research Kolkata, Nadia, Mohanpur, West Bengal 741246, India. E-mail: sayam.sengupta@iiserkol.ac.in
bDepartment of Biological Sciences, Indian Institute of Science Education and Research Kolkata, Nadia, Mohanpur, West Bengal 741246, India. E-mail: rupak.datta@iiserkol.ac.in
First published on 15th December 2025
Abstract
Lysosomal dysfunction leads to critical lysosomal storage disorder (LSD) conditions, and among all, Mucopolysaccharidosis VII (MPS VII), or Sly syndrome, is a rare LSD caused by the deficiency of the β-glucuronidase (β-GUS) enzyme, leading to the accumulation of glycosaminoglycans (GAGs) in tissues and organs. Since enzyme replacement therapy (ERT) lacks target-specific delivery of recombinant enzymes, targeted enzyme delivery approaches are desired to enhance the success of existing ERT methods. The effective targeted delivery of functional enzymes to lysosomes remains a significant therapeutic challenge. In this study, we report the design and development of a mannose-6-phosphate (M6P) functionalized liposomal nanocarrier for targeted lysosomal delivery of β-GUS. The liposomal nanocarriers were formulated with surface-decorated M6P ligand functionality to exploit the previously established M6P receptor-mediated endocytosis pathway, ensuring high specificity and enhanced intracellular trafficking to lysosomes. Comprehensive physicochemical characterization confirmed the stability, size uniformity, and successful surface functionalization of the liposomes. Internalization studies using HEK293 cells and hemocytes derived from the Drosophila model of MPS VII demonstrated significantly improved cellular internalization and colocalization with lysosomes. The time- and dose-dependent restoration of β-GUS activity was also studied. Furthermore, the nanocarrier exhibited minimal cytotoxicity, indicating its potential for safe and effective ERT. This targeted liposomal system represents a promising platform for lysosome-specific delivery of therapeutic enzymes and could be broadly applied to other lysosomal storage disorders.
Introduction
The cellular waste disposal system, known as lysosomes, uses over 50 hydrolytic enzymes, classified as proteases, lipases, sulfatases, glycosidases, nucleases, phosphatases, transferases, and reductases, to degrade macromolecules.1 Lysosomal enzymes break down unwanted proteins, lipids, carbohydrates, nucleic acids, damaged organelles, and pathogens, helping maintain cellular homeostasis. Enzyme deficiencies and dysregulated lysosomal activity can severely impact cell health, leading to several critical diseases like cancer, neurological disorders, and infectious diseases.2–8 Additionally, lysosomal storage disorders (LSDs) are genetic disorders caused by the deficiency of lysosomal glycosidases, membrane proteins, transporters, and proteases.9,10 In LSDs, the substrates of the respective enzymes, such as glycosaminoglycans (GAGs), sphingolipids, glycogens, glycoproteins, proteins, etc., remain undegraded or partially degraded, leading to cellular and organismal dysfunction due to macromolecule accumulation.11 In addition to common symptoms like neurological defects, muscle weakness, developmental delay, presence of engorged lysosomes, and storage bodies, the severity becomes fatal in some cases, resulting in premature death.12Mucopolysaccharidosis (MPS) (seven different types) constitutes a special class of LSD caused by a defect in GAG turnover. MPS VII, also known as Sly syndrome, is caused by a mutation in the gene encoding β-glucuronidase (β-GUS).13 β-GUS (EC 3.2.1.31) catalyses the degradation of mucopolysaccharides by removing β-glucuronosyl from the non-reducing end of the GAGs.14 The deficiency or malfunction of β-GUS leads to the accumulation of GAGs, namely dermatan sulfate, keratan sulfate, heparan sulfate, and chondroitin sulfate. It spans a spectrum of disease severity, having systemic and neurological effects, with symptoms including hepatosplenomegaly, musculoskeletal deformities, developmental delay, slow cognition, cardiac valve disease, and occasionally hydrops fetalis in newborns, ultimately leading to premature death.13,15 Most MPS disorders, as well as other LSDs, primarily rely on enzyme replacement therapy (ERT), offering a lifeline, delivering recombinant enzymes via intravenous infusion.16,17 The FDA has approved the recombinant human enzyme therapies for LSDs like Gaucher's disease,18 Pompe disease,19 Fabry disease,20 MPS I,21 MPS II,22 MPS IVA,23 MPS VI,24 CLN2,25 and Wolman disease.26 The FDA-approved ERT for MPS VII is currently the only available treatment (Ultragenyx Pharmaceutical Inc.), Mepsevii™ (vestronidase-α).27 ERT has been effective in reducing accumulated GAG levels, improving muscle strength, pulmonary function, and fatigue reduction, but is unable to correct neurological defects.28–30 The shortcoming is that these enzymes lack precision, struggling to reach the required tissues and organelles, including the blood–brain barrier (BBB) to treat neurological symptoms.29,31,32 On top of this, concerns regarding enzyme stability in the blood, the inefficient distribution in tissues, the staggering costs and lifelong dependency on ERT emphasize the need for alternative enzyme delivery strategies.33 The future lies in using smarter and efficient targeted enzyme delivery systems, which may potentially change the existing therapeutic landscapes for combating LSD.
To combat MPS VII, researchers have explored delivering native β-GUS for ERT in murine models; however, cytosolic delivery via protein transduction has proven ineffective in human trials.34–36 Davidson and colleagues found that a non-denatured β-GUS-Tat fusion protein, expressed via an adenoviral vector, achieved broader distribution than native β-GUS when administered intravenously or directly into the brain.37 Meanwhile, Sly and co-workers mapped the pathway for Tat-mediated β-GUS delivery in murine models.38 However, such approaches require recombinant β-GUS (prepared by protein engineering), and the modality lacks specificity for targeting the recombinant GUS to lysosomes, the organelle that houses these enzymes. Lysosomal enzymes are trafficked to lysosomes via mannose-6-phosphate (M6P) receptors after undergoing post-translational modifications, resulting in M6P-tagged enzymes, which are crucial for their facile transport within the cells.39,40 Inspired by this cellular pathway, researchers, including our group, have employed a similar strategy to deliver drugs into lysosomes.41,42 We have demonstrated that M6P-functionalized macromolecular architectures43–45 and nanocarriers46–48 can be designed to achieve lysosome-specific cargo delivery.42 We have recently showcased the successful delivery of the β-glucosidase enzyme into lysosomes as a potential alternative treatment for Gaucher's disease, utilizing advanced protein engineering and bioconjugation techniques.49 However, the complexity of macromolecule synthesis poses a challenge for the widespread application of these nanocarriers. Additionally, chemical modifications to formulate hybrid conjugates may compromise the enzyme's native function.
To address these limitations, an effective approach would be the design of small amphiphilic lipid molecules capable of forming liposomes and encapsulating proteins of varying molecular weights, thereby ensuring targeted cell- or organelle-specific delivery. Liposomes are highly effective nanocarriers in nanomedicine, offering stability, biocompatibility, and targeted drug delivery.50 Small liposomes (100–200 nm) are optimal for intravenous use, as they avoid rapid kidney clearance and prolong circulation.51 Currently, 15 liposomal-based nanomedicines have been approved for clinical trials, with many more under various stages of development.52–54 Liposome-encapsulated enzyme and gene delivery have been explored for the treatment of LSDs like Gaucher's disease,55 Pompe disease,56 Niemann-Pick,57etc. Additionally, it can diffuse through the BBB and thus has emerged as a potential treatment for brain-related diseases.58 Thus, we hypothesize that packaging β-GUS inside efficient lysosome-targeting liposomal nanocarriers may be rewarding towards enhancing the efficiency of ERT and addressing the challenges associated with direct chemical manipulation.
In this study, we have designed and synthesized a novel M6P-functionalized glycolipid (M6PGL) with a non-ester backbone, offering a streamlined and scalable approach. We prepared M6P-functionalized liposomal nanocarriers (LN) using M6PGL as the targeting lipid, along with commercially available 1,2-di-O-hexadecyl-sn-glycero-phosphocholine (DHGPC) and cholesterol (Chl) as co-lipids for targeted lysosomal delivery of the large-sized β-GUS (270 kDa tetramer) enzyme. To validate its efficacy, we first demonstrated lysosome-specific delivery using the model dye calcein and the model protein fluorescently labelled bovine serum albumin (Fl_BSA). Next, we showed the lysosomal delivery of fluorescently labeled β-GUS (Fl_GUS) in the mammalian cells by the LN using confocal microscopy. Finally, through confocal imaging and in vitro assays, we demonstrated that primary hemocyte cells extracted from β-GUS-knockout Drosophila (MPS VII fly models) exhibit enhanced β-GUS enzyme activity compared to free-GUS following LN-mediated delivery. The novel lysosome-targeting LN described here open a new avenue for the treatment of MPS VII and other LSDs.
Experimental section
Materials and methods
Details of chemicals, instruments, synthesis of all components, and characterization methods have been provided in the SI.Synthesis of Protected M6P glycolipid (4)
M6P-carboxylic acid (624 mg, 1 mmol) was taken in a 100 mL two-neck RB and solubilized in dry dichloromethane (DCM) (5 mL) and dry N,N-dimethylformamide (DMF) (1 mL). Solid EDC·HCl (240 mg, 1.25 mmol), NHS (144 mg, 1.25 mmol), and a catalytic amount of DMAP were added sequentially to the ice-cold and stirred solution under an argon atmosphere. After half an hour, N-6-aminohexyl-N,N-di-n-octadecylamine (682 mg, 1.1 mmol, prepared as explained in the SI) dissolved in 4 mL dry DCM was added to the reaction mixture at 0 °C. The resulting solution was left stirring at room temperature overnight, then diluted with excess DCM and washed sequentially with saturated sodium bicarbonate (3 × 50 mL), water (2 × 50 mL), and brine (50 mL). The organic layer was dried over anhydrous sodium sulphate, filtered, and the solvent from the filtrate was removed by rotary evaporation. The residue obtained after column chromatographic purification with 100–200 mesh silica gel, using 70% EtOAc and 30% hexane as the eluent, afforded 0.61 g (50% yield) of the pure product as a light-yellow, sticky solid.1H NMR (500 MHz, CDCl3) δ 0.87 (t, 6H), 1.15–1.45 (m, 68H), 1.58 (s, 6H), 1.84 (d, 3H), 1.98 (d, 3H), 2.03 (s, 2H), 2.06 (s, 3H), 2.13 (d, 2H), 3.80–3.92 (m, 3H), 4.07 (dd, 2H), 4.26 (dd, 1H), 4.32 (d, 1H), 4.88–4.97 (m, 2H), 4.98–5.08 (m, 3H), 5.22–5.44 (m, 2H), 7.33 (d, 10H), 7.33–7.34 (–NH, amide).
31P NMR (203 MHz, CDCl3) δ −1.91.
HRMS (ESI-TOF): m/z [M + H]+: calculated – 1227.8528, experimental – 1227.8322.
Deprotection of M6P glycolipid (5)
(i)Step 1 – For benzyl deprotection, the protected M6P lipid was employed for hydrogenation using 10% Pd/C in methanol/DCM (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) under hydrogen balloon pressure for 24 h. After completion, the crude mixture was filtered over a Celite pad, and the benzyl-deprotected lipid was obtained quantitatively after removing the solvent from the filtrate. This lipid was used for the next deprotection step without further purification.
1) under hydrogen balloon pressure for 24 h. After completion, the crude mixture was filtered over a Celite pad, and the benzyl-deprotected lipid was obtained quantitatively after removing the solvent from the filtrate. This lipid was used for the next deprotection step without further purification.
(ii)Step 2 – For acetyl deprotection, the intermediate lipid from step 1 was exposed to hydrazine monohydrate (5 eq.), which was added to the lipid solution in methanol (10 mg mL−1), followed by stirring for 12 h at room temperature. After deprotection, the reaction mixture was quenched by adding acetone, and the solvent was removed under reduced pressure. Furthermore, it was purified by washing with pentane repeatedly (3–4 times) to isolate a pure, deprotected M6P lipid (90% yield) as a light-yellow powder.
1H NMR (500 MHz, CDCl3) δ 0.88 (t, 6H), 1.25 (s, 64H), 1.60 (d, 4H), 1.81 (d, 6H), 2.71 (s, 2H), 2.95 (s, 4H), 3.40 (s, 3H), 3.65 (s, 2H), 3.74 (t, 4H), 5.60 (s, 1H), 6.42–6.63 (m, 2H), 6.97–7.14 (m, 3H).
31P NMR (203 MHz, CDCl3) δ −0.61.
HRMS (ESI-TOF): m/z [M + H]+: calculated – 921.7272, experimental – 921.7286.
Formulation of the liposomal nanocarrier
The liposomes were formulated using the thin-film hydration method.59,60 A stock solution of M6PGL, 1,2-di-O-hexadecyl-sn-glycero-phosphocholine (DHGPC), and cholesterol (Chl) was made in chloroform. The required amount of M6PGL, DHGPC, and Chl was transferred to a glass/plastic vial in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio to make a total lipid of 8 µmoles (5 mg of total lipid = 1.5 mg M6PGL, 2.25 mg DHGPC, and 1.25 mg Chl). Chloroform was evaporated using slow evaporation with a nitrogen flow to form a thin film on the surface of the vial. The organic volatiles were completely removed by applying a high vacuum for 7–8 h. Next, the lipid film was kept hydrated overnight by adding 5 mL of Milli-Q water to swell the lipid film. The next day, the solution was agitated using a vortex (30 sec) and bath sonication (30 sec) to fully remove the film from the bottom of the vial, and the solution turned cloudy due to the formation of multi-lamellar vesicles (MLVs). Further, to form uni-lamellar vesicles, the hazy solution was probe sonicated for 25 cycles under an ice bath (using a Ti-probe sonicator applying pulse ON for 10 sec and pulse OFF for 50 sec), after which the solution turned clear. The clear solution was filtered through a 0.22 µm syringe filter to afford monodisperse liposomal nanocarriers (LNs), which were further characterized using DLS, TEM, and AFM.
2 molar ratio to make a total lipid of 8 µmoles (5 mg of total lipid = 1.5 mg M6PGL, 2.25 mg DHGPC, and 1.25 mg Chl). Chloroform was evaporated using slow evaporation with a nitrogen flow to form a thin film on the surface of the vial. The organic volatiles were completely removed by applying a high vacuum for 7–8 h. Next, the lipid film was kept hydrated overnight by adding 5 mL of Milli-Q water to swell the lipid film. The next day, the solution was agitated using a vortex (30 sec) and bath sonication (30 sec) to fully remove the film from the bottom of the vial, and the solution turned cloudy due to the formation of multi-lamellar vesicles (MLVs). Further, to form uni-lamellar vesicles, the hazy solution was probe sonicated for 25 cycles under an ice bath (using a Ti-probe sonicator applying pulse ON for 10 sec and pulse OFF for 50 sec), after which the solution turned clear. The clear solution was filtered through a 0.22 µm syringe filter to afford monodisperse liposomal nanocarriers (LNs), which were further characterized using DLS, TEM, and AFM.
Dye encapsulation in the liposomal nanocarriers
For hydrophobic dye (rhodamine B octadecyl ester (RBOE) in this case) encapsulation, 150 µL of RBOE (stock solution of 1 mg mL−1) in acetone was added to the lipids in chloroform and was evaporated to remove the organic solvents fully, after which the liposomes were formulated using the protocol mentioned above. To remove the unentrapped dye (RBOE), the solution was filtered using a 0.22 µm syringe filter, affording RBOE-loaded liposomal nanocarrier (R-LN).For hydrophilic dye (Calcein) encapsulation, 250 µL of dye (stock solution of 1 mg mL−1) in 5 mL of water was added to the dried film for hydration. To remove the unentrapped dye (Calcein), dialysis against Milli-Q water for 24 h was performed using a 3.5 kDa molecular weight cut-off dialysis membrane, with the water changed six times, to afford Calcein-loaded liposomal nanocarrier (C-LN).
The dual dye Calcein and RBOE-loaded liposomal nanocarrier are referred to as CR-LN.
Dye entrapment efficiency (DEE) was determined by using the following formula:
| DEE% = (weight of encapsulated dye)/(weight of dye in feed) × 100 |
| Calcein entrapment efficiency of C-LN = ∼58% |
Protein encapsulation in liposomal nanocarriers
The thin film was prepared as mentioned in the earlier written protocol. Then, 5 mL of water containing 500 µL of Fl_BSA (stock 1 mg mL−1) or 5 mL of 20 mM phosphate buffer containing 600 µL of β-glucuronidase (Fl_GUS) (stock 5 mg mL−1) was added to the dried film for hydration and was kept at 4 °C overnight. The next day, the solution was agitated (using a vortex) to form the MLVs, and then extrusion (10 times) was performed using a 0.1 µm polycarbonate membrane from Whatman. The solution was employed for five freeze–thaw cycles (freeze using liquid N2 and thawed using a water bath at 50 °C). Furthermore, the solution was loaded onto a Sephadex-G25 (for BSA) and Sepharose CL-6B (for GUS) column, and the unentrapped BSA/GUS was separated from the BSA/GUS encapsulated in liposomal nanocarriers. The loading of BSA/GUS was confirmed by fluorescence spectroscopy and DLS. The BSA-loaded liposomal nanocarrier are named BSA-LN, and the GUS-loaded liposomal nanocarrier are named GUS-LN.Enzyme entrapment efficiency (EEE) was determined by using the following formula:
| ELE% = (weight of encapsulated enzyme)/(weight of enzyme in feed) × 100 |
| EEE for BSA-LN = ∼42% |
| EEE for GUS-LN = ∼37% |
In vitro cytotoxicity assay
The HEK293 cell viabilities upon liposomal nanocarrier (LN) treatment were quantified by the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) assay as described.61 HEK293 cells were seeded in 6-well plates in MEM (Gibco) complete media (supplemented with 2 mM L−1 of glutamine, 100 U ml−1 penicillin, 100 µg mL−1 of streptomycin, and 10% FBS), incubated at 37 °C with 5% CO2. After 12 hours, the LN was added to achieve final concentrations of 5, 10, 20, 40, 50, 75, 100, and 200 µg mL−1 (weight of LN after formulation), respectively, in MEM containing 10% FBS. The cells were incubated for 48 h at 37 °C with 5% CO2. An equal number of untreated and treated cells (1 × 104) were seeded in a 96-well plate. They were incubated with 0.5 mg mL−1 MTT for 3 h at 37 °C with 5% CO2 conditions to allow the formation of purple formazan. Next, the cells in 96-well plates were centrifuged at 4000 rpm for 10 min, followed by a wash with PBS. 100 µL of DMSO was added to rupture the cells and dissolve the formazan. The OD at 595 nm was recorded for the solubilised purple formazan using a microplate reader (BioTek Cytation 5). The formazan (OD of which was measured) is produced by the action of mitochondrial dehydrogenases of metabolically active cells, correlating with the number of viable cells. The percentage of viability upon LN treatment was calculated using the formula (ODtreated/ODcontrol × 100). As stated differently, cell viability was represented as a percentage relative to the untreated control cells. The average OD of the three replicates for each sample (mean ± SEM) was plotted in GraphPad.Confocal microscopy of the dye/protein-loaded liposomal nanocarrier in HEK293 cells
HEK293 cells were seeded in a 35 mm glass-bottom dish in MEM complete media, and the plate was incubated at 37 °C with 5% CO2 for 12 h. Then, the incubation medium was replaced with the dye/protein-loaded liposomal nanocarrier (Calcein-loaded, fluorescein labelled-BSA-loaded, fluorescein labelled-GUS-loaded liposomal nanocarrier: C-LN, BSA-LN, and GUS-LN) (equivalent to 200 µg mL−1 of LN containing different amounts of Calcein, Fl_BSA, and Fl_β-GUS) treatment. It was incubated for 4 h at 37 °C with 5% CO2. As a control, no treatment was given to the other dish. After a 4 h incubation and a brief wash, Lysotracker Red DND-99 (Invitrogen) at a concentration of 75 nM was added to stain the lysosomes in complete media. The cells were then incubated for 30 minutes at 37 °C with 5% CO2. The cells were washed twice with PBS and then incubated in HBSS buffer containing glucose during live imaging. Images were captured using a Leica SP8 confocal microscope with a 63X oil immersion objective. Images were analysed using LASx software.Isolation of the primary hemocytes from Drosophila larvae
The wildtype (W1118) and CG2135−/− knockout flies were used to study the isolation of the hemocytes, following the report by Goodman and co-workers.62 3rd instar larvae were used for the isolation of hemocytes. In brief, to obtain 3rd instar larvae, an equal number of embryos were reared in a fly food-containing vial (corn flour 75 g L−1, sugar 80 g L−1, agar 10 g L−1, dry yeast 15 g L−1, and malt extract 30 g L−1). After 5 days, the hatched larva enters the 3rd instar or wandering stage. During this time, they leave the media and seek dry spots to transform into pupae. A total of 30–40 3rd instar larvae were collected using a paintbrush and washed twice in distilled water to clean the media from their body. They are incubated in 70% ethanol for 5 min to disinfect the larvae. After this, the larvae were transferred to fresh distilled water. 30 larvae were bled into 200 µL of S2 (Gibco) complete media supplemented with 10% FBS, 1% pen-strep, and 25 µM phenylthiourea (PTU). PTU is added to prevent the formation of crystal cells, which cause melanisation of the hemocytes. For dissection, the larva was torn apart to allow leakage of the hemolymph into the media. After bleeding, the cells were allowed to attach to the surface of the 35 mm dish for 5 min. Then, the carcass was discarded and left at room temperature for 5 min. Finally, washed with sterile PBS and incubated in complete S2 media in a 25 °C incubator.Confocal microscopy of the dye/protein-loaded liposomal nanocarrier in hemocytes
Hemocyte cells were isolated from dissected 3rd instar larvae and seeded in a 35 mm glass-bottom dish in S2 complete media (composition described above). Then, the incubation medium was replaced with the protein-loaded liposomal nanocarrier (fluorescein labelled-GUS-loaded liposomal nanocarrier; GUS-LN) (equivalent to 200 µg mL−1 of LN) treatment and incubated for 4 hours at 25 °C. As a control, no treatment was given to another dish. After incubation, Lysotracker Red DND-99 (Invitrogen) at a concentration of 75 nM was added to stain the lysosomes in S2 complete media, and the cells were incubated for 30 min at 25 °C in the incubator. The cells were washed twice with PBS and then incubated in PBS during live imaging. Images were captured using a Leica SP8 confocal microscope with a 63X oil immersion objective. Images were analysed using LASx software, and Pearson's coefficient was quantified using the Coloc-2 module in ImageJ Fiji.In vitro activity assay of internalized β-GUS
β-GUS activity of the internalized GUS-loaded liposomal nanocarrier (GUS-LN) was measured fluorometrically using 4-methylumbelliferyl β-D-glucuronide (4-MUG) as a substrate.63 The hemocytes were incubated with GUS-LN (effective concentration of GUS in GUS-LN is 100 or 200 µg mL−1) or free-GUS (200 µg mL−1) for 2 or 4 h at 25 °C, and untreated cells were kept as controls. After incubation, the cells were washed twice with PBS to remove the uninternalized liposome-containing media. Then the cells were scraped, collected in an Eppendorf tube, and centrifuged at 5000 rpm for 5 minutes at room temperature. The pellet was resuspended in 100 µL lysis buffer (1× PBS + 1× PIC). To lyse the cells, sonication was used (10 sec pulse, 20 sec pauses, 3 cycles). Next, the lysate (at least 0.5 µg µL−1) was incubated with the substrate solution containing 10 mM of 4-MUG and 1 mg mL−1 BSA in 0.1 M acetate buffer, pH 4.8. The solution was incubated for 4 h at 37 °C. A stop buffer of glycine-carbonate, pH 10.5, was added to stop the reaction, and the GUS-catalyzed product (4-MU) was measured using a BioTek plate reader at an excitation wavelength of 365 nm and an emission wavelength of 445 nm. The amount of 4-MU produced was quantified using the standard curve of known 4-MU concentrations. The activity was normalized to protein concentration, as determined by Lowry's method.64Results and discussion
Synthesis and characterization of amphiphilic M6P glycolipid
The self-assembly of amphiphilic lipids is a widely used method for the formulation of liposomal nanocarriers. Short amphiphilic lipids offer advantages over macromolecular architectures due to their simpler synthesis, making them more industrially viable. Previously, a dual enzyme-responsive (esterase and alkaline phosphatase) mannose-6-phosphate (M6P)-functionalized lipid was developed using palmitic and stearic acid for lysosomal cargo delivery.46 To enable targeted enzyme delivery, the delivery system must ensure extended stability in circulation while avoiding rapid degradation by blood plasma esterases.65Initially, M6P-functionalized carboxylic acid (1g) was synthesized over 10 steps following our previous report,43 and subsequently, an amine-functionalized hydrophobic tail (3b) was synthesized starting from hexamethylenediamine. The hydrophobic tail consists of two octadecyl (18-carbon-long) chains linked to a tertiary nitrogen group. Furthermore, the acid-amine coupling was performed using EDC and NHS coupling to afford the protected M6P glycolipid in 50% yield (the scheme is given in Fig. 1). The protected glycolipid was characterized using 1H, 31P and 13C NMR (see NMR section in the SI) and HR-MS (Fig. S1 in the SI). In 1H NMR, the peaks at 0.87, 1.25, and 1.58 ppm corresponded to the hydrophobic part, and the peaks at 2.0, 3.8–5.4, and 7.3 ppm corresponded to the protected sugar (M6P) part (NMR section in the SI). The peak at 1.91 ppm in 31P NMR confirmed the presence of the phosphate group (NMR section in the SI).
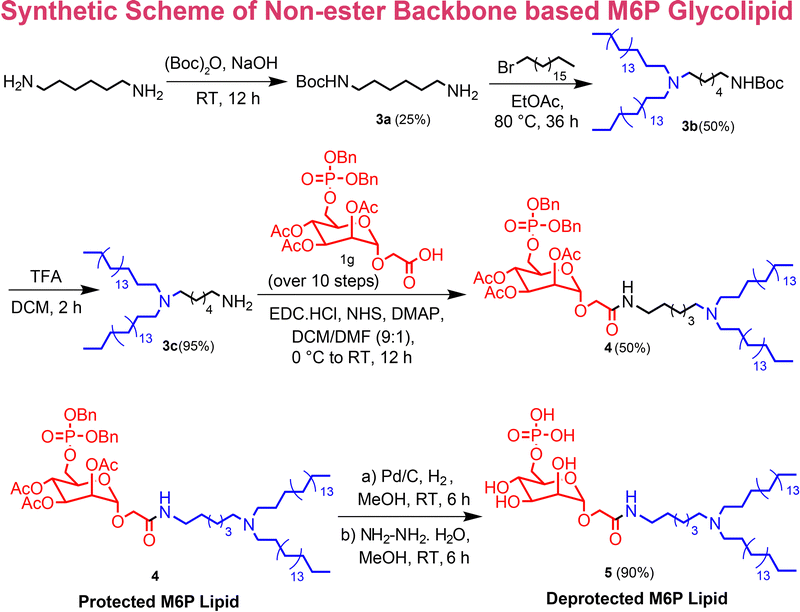 | ||
| Fig. 1 Synthetic scheme for the synthesis of M6P functionalized nonester backbone-based glycolipid (M6PGL). | ||
Furthermore, the deprotection of the benzyl and acetyl groups on the M6P group was performed to afford the fully deprotected amphiphile (M6PGL). Firstly, benzyl deprotection was done by hydrogenolysis using Pd/C/H2, followed by acetyl group deprotection using hydrazine hydrate in methanol (the scheme is given in Fig. 1). After purification, these compounds were characterized using 1H, 31P, 13C NMR, and HR-MS. The absence of 2.0 and 7.3 ppm peaks in the 1H NMR (Fig. 2A) confirmed the deprotection of the acetyl and benzyl groups, respectively, which was further confirmed by 13C NMR (Fig. 2B) and HR-MS (Fig. S2 in the SI). The peak at 0.61 ppm in 31P NMR confirmed the presence of the phosphate group (NMR section in the SI).
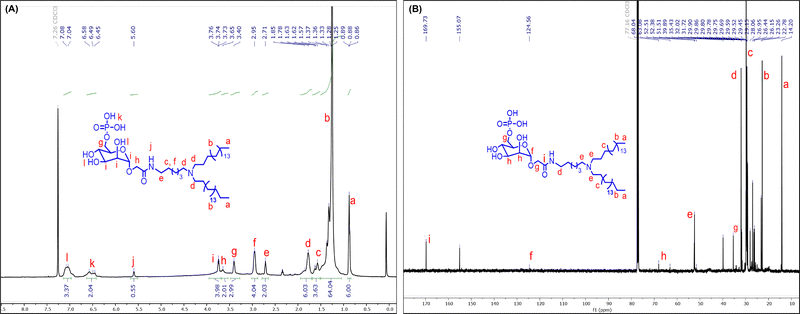 | ||
| Fig. 2 Characterization of M6P functionalized nonester backbone-based glycolipid (M6PGL): (A) 1H NMR, and (B) 13C NMR. | ||
Formulation and characterization of the liposomal nanocarrier
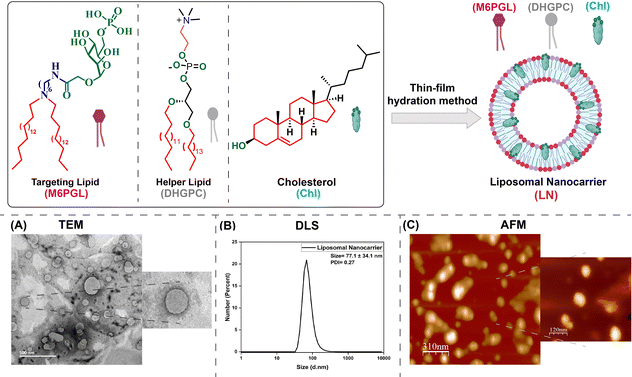
The liposomal nanocarrier (LN) was formulated by following the method described in the experimental section (schematics given in Fig. 3, top). The synthesized M6PGL was used for lysosomal targeting purposes, commercial DHGPC was used as a co-lipid to help in the formulation, and Chl was used to stabilize the bilayer, preventing drug leakage in a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. The formulated normal LN (no dyes/drugs entrapped) solution was filtered using syringe filters and characterized using DLS, TEM, and AFM for its size and morphological analysis. In TEM, a spherical vesicular morphology with a hollow internal core was observed using negative staining, with spheres having an average diameter of ∼116.5 ± 8.32 nm (Fig. 3A). From DLS (Fig. 3B) and AFM (Fig. 3C) analysis, the size value corroborated well with TEM, and the polydispersity index (PDI) value of ∼0.27 from DLS confirmed the monodisperse nature of the LN solution. The 3D image and height profile from the AFM analysis are provided in Fig. S3 of the SI.
2. The formulated normal LN (no dyes/drugs entrapped) solution was filtered using syringe filters and characterized using DLS, TEM, and AFM for its size and morphological analysis. In TEM, a spherical vesicular morphology with a hollow internal core was observed using negative staining, with spheres having an average diameter of ∼116.5 ± 8.32 nm (Fig. 3A). From DLS (Fig. 3B) and AFM (Fig. 3C) analysis, the size value corroborated well with TEM, and the polydispersity index (PDI) value of ∼0.27 from DLS confirmed the monodisperse nature of the LN solution. The 3D image and height profile from the AFM analysis are provided in Fig. S3 of the SI.
Dye/protein encapsulation inside the LN
Liposomes can encapsulate both hydrophilic and hydrophobic cargos simultaneously. Aqueous solutions of hydrophilic dyes (Calcein as a model) are normally added to thin dried lipid films during hydration. After formulation of the LN, the unencapsulated dye was removed using extensive dialysis. Furthermore, the Calcein-loaded liposomal nanocarrier (C-LN) was characterized using UV-vis (Fig. S4(B) in the SI), fluorescence, DLS (Fig. S7(A) in the SI), and TEM (Fig. S8(A) in the SI) techniques. As a model hydrophobic dye, RBOE was encapsulated in the hydrophobic bilayer of the LN. The dual dye encapsulation was confirmed using confocal microscopy (Fig. S5(B) in the SI), where the Calcein and RBOE dyes were localized in the same nanocarrier (CR-LN), which was further confirmed by fluorescence spectroscopy (Fig. S5(A) in the SI). The emission responsible for RBOE is only visible when excited at 540 nm. Still, when the excitation wavelength was set at 470 nm, a sharp peak in the emission spectrum corresponded to Calcein emission, and a broad shoulder region corresponded to RBOE (Fig. S5(A) in the SI). From DLS and TEM analysis, the size of the C-LN and CR-LN was almost comparable to that of the normal LN (Table 2).| S.N. | Liposomes | Enzyme amount in feed (µg) | Enzyme amount encapsulated (µg) | EEE (%) |
|---|---|---|---|---|
| 1 | C-LN | 250 | 145 | 58 |
| 2 | BSA-LN | 500 | 210 | 42 |
| 3 | GUS-LN | 3000 | 1110 | 37 |
To encapsulate proteins into the LN, the protein (BSA, GUS) was hydrated in a 20 mM phosphate buffer (pH ∼7), followed by the protocol described earlier, resulting in the formation of protein-encapsulated liposomal nanocarriers (BSA-LN and GUS-LN) (schematics given in Fig. S6, SI), DLS (Fig. S7(B) and (C)) and TEM (Fig. S8(B) and (C)) analysis revealed a slight increase in the size of the BSA-LN and GUS-LN compared to the normal LN, which can be attributed to the encapsulation of large macromolecular proteins (Table 2). The amount of dye and proteins encapsulated was calculated using the given formula and is provided in Table 1. The amount of Calcein encapsulated in the C-LN is ∼58%, while for the BSA-LN and GUS-LN it is ∼42% and ∼37%, respectively (Table 1).
Time-dependent stability studies of the dye/protein-loaded LN by TEM and DLS
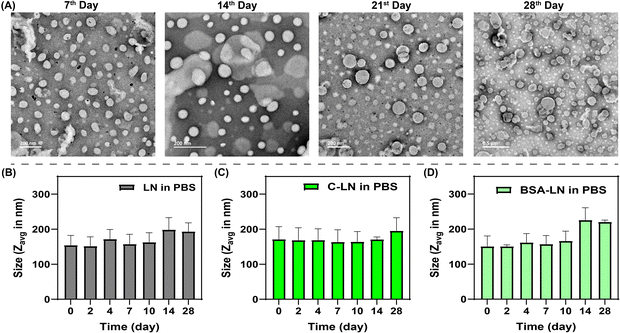
Although the LN offer significant pharmacological advantages, their stability in aqueous media remains a major challenge. Issues such as aggregation, coalescence, flocculation, and drug leakage can compromise the efficacy of this drug delivery system, limiting its practical applications.66 To enhance the stability of the LN, we have employed the lipid composition method by using three different lipids, namely M6PGL, helper lipid DHGPC, and bilayer stabilizer Chl, which reduce the bilayer fluidity by enhancing the rigidity of the liposomal membranes and modulating the release of hydrophilic molecules from the lipid vesicles.54,67,68 To enhance size control and minimize aggregation and coalescence,69,70 we employed extrusion and ultrasonication techniques.After formulating the LN using our established method, we assessed their time-dependent stability by analyzing their morphology through TEM and determining their size distribution using DLS. In TEM, the spherical morphology observed of the LN on the 7th, 14th, 21st, and 28th day (Fig. 4A) was similar to the native morphology of the LNs just after formulation (Fig. 3A). We then investigated the size of the LNs over time using DLS studies and found that the normal LN, C-LN, and BSA-LN were stable in phosphate-buffered saline (100 mM PBS, pH 7.4) (Fig. 4B–D) and in serum (10% FBS) containing DMEM medium (Fig. S9 in the SI). In contrast, the LN solution exhibited a tendency to aggregate in a neutral aqueous medium (Fig. S10 in the SI), suggesting that the negatively charged M6P group remains more stable in a charged environment containing anions. These findings confirm that the LN solution maintains stability under near-physiological conditions for up to 4 weeks.
The rationale behind designing and synthesizing nonester-based M6PGL was to evade degradation from high concentrations of esterase in the blood. To investigate, we incubated the LN in an esterase-rich environment for 48 h. We followed their size and morphology using AFM (Fig. S11(A) in the SI), TEM (Fig. S11(B) in the SI), and DLS (Fig. S11(C) in the SI) analysis at different time points. The nonester-based M6P LN remained quite stable in the esterase-rich environment up to 16 h incubation (Fig. S11 and S12(B) in the SI). Degradation into small and large aggregated structures was observed only after a 48 h period, as observed from TEM images (Fig. S11(B) (iii) and S12(B) in the SI), and very large Z-average values from DLS analysis (Fig. S11(C) in the SI). As a control, we have formulated an ester-based M6P LN, using our previously reported ester-based M6P lipid46 along with DOPC and Chl, and followed its degradation in the presence of esterase (Fig. S12(A) in the SI). Degradation of this ester-based M6P LN occurred only after 16 h of incubation, as observed in both DLS and TEM analyses (Fig. S12(A) in the SI). These findings demonstrate that the degradation of the nonester-based M6P LN is very slow compared to the ester-based M6P LN, highlighting the importance of the novel nonester backbone-based M6PGL. The stability of the formulated LN in physiological medium and an esterase-rich environment demonstrated their potential for biomedical applications, particularly in protein delivery.
In vitro cytotoxicity and lysosome-specific internalization of the dye/protein-loaded LN in HEK293 cells
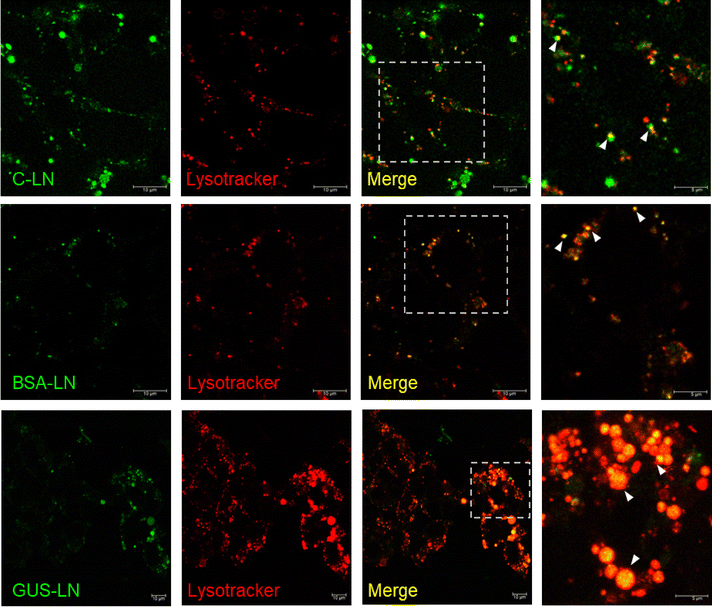
Owing to their stability in physiological conditions, the formulated M6P-functionalized LN were investigated for their cytotoxicity using an MTT assay and cellular uptake using confocal microscopy in mammalian cells. The HEK293 cells treated with different concentrations of LN showed no significant toxicity at any concentration, as determined by the MTT assay (Fig. S13(A) given in the SI). The viability of the cells was compared with that of untreated cells, and more than 75% of the cells remained viable after treatment and incubation of LN at concentrations up to 200 µg mL−1.A model hydrophilic dye-encapsulated C-LN was treated to investigate the delivery location of the cargo inside the cells using LN. After 4 h of incubation, confocal microscopy was performed, and the preliminary results showed a significant uptake of the C-LN (Fig. 5, top panel). Upon staining the lysosomes with lysotracker red dye, it was found that the Calcein fluorescence from the green channel colocalized with the red channel, indicating the lysosome-specific delivery of Calcein dye using the LN. Taking the lead, we investigated the uptake of model protein Fl_BSA loaded in LN (BSA-LN). The green fluorescence signal from fluorescently labelled BSA (Fl_BSA) was also colocalized with the lysotracker (red) positive vesicles, proving the lysosomal delivery. (Fig. 5, middle panel). Finally, the protein of interest, Fl_GUS loaded in LN (GUS-LN), was treated with cells and incubated in a 35 mm glass-bottom confocal dish. The cells were imaged after 4 h of incubation. The GUS delivered using LN was significantly localised in the lysosomes confirmed by the colocalization of the green puncta representing Fl_GUS and red puncta representing the lysotracker positive vesicles (Fig. 5, bottom panel). DIC images of the cells treated with C-LN, Fl_BSA-LN, and Fl_GUS-LN are presented in Fig. S13(B) of the SI, representing the cellular uptake of the dye/protein-loaded LN. These data collectively indicated that LN decorated with M6P can efficiently deliver both small-molecule dyes and large-sized proteins, such as β-GUS, into the lysosomes of mammalian cells.
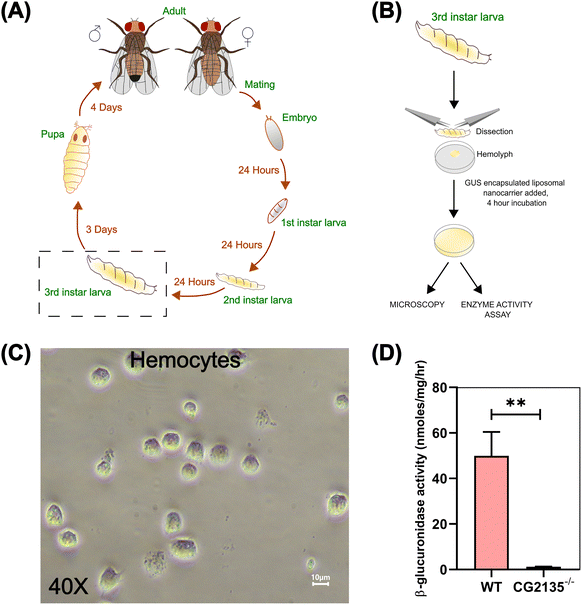
Isolation of the hemocytes from a Drosophila model of MPS VII
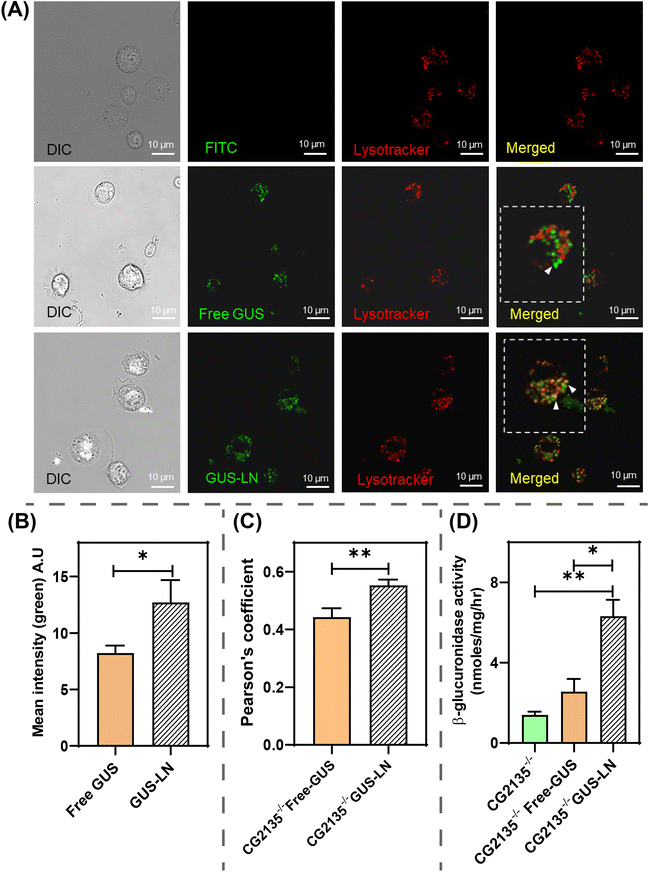
After confirming the cellular uptake and lysosomal delivery of GUS-LN in HEK293 cells, we sought to evaluate the efficacy of this novel delivery system in a disease model of MPS VII. We used the Drosophila model of MPS VII to study GUS-LN uptake assays. We have previously developed a Drosophila model of MPS VII by knocking out the fly β-GUS gene (CG2135).71 The MPS VII fly (CG2135−/−) mimicked the clinical symptoms, such as short lifespan, climbing disability, neurodegeneration, presence of engorged lysosomes, and damaged mitochondria.71,72 For uptake assays, we isolated hemocytes (phagocytic cells)73,74 of Drosophila from the 3rd instar larva CG2135−/− flies. The ease of hemocyte isolation has made it attractive for studies with primary cells of Drosophila. Fig. 6A and B represent the Drosophila lifecycle scheme and schematics of hemocyte isolation, respectively. The bright-field images of the isolated hemocytes revealed an oval-shaped morphology of the primary cells (Fig. 6C). As expected, the isolated hemocytes from CG2135−/− larvae showed 97% less β-GUS activity as compared to wildtype larvae (Fig. 6D). Therefore, we used these cells for studying enzyme delivery.Lysosome-specific internalization of GUS-LN into hemocytes followed by in vitro enzyme activity assay
The isolated hemocytes from CG2135−/− larvae were used for the cellular internalisation study of fluorescein-labelled free-GUS (Fl_GUS) and Fl_GUS-loaded LN (GUS-LN). The CG2135−/− hemocytes treated with Fl_GUS and GUS-LN were studied for lysosome-specific cellular internalization and the in vitro assay of enzyme activity of internalized GUS. To analyze the internalization of GUS-LN and free β-GUS, and their localization with lysosomes, cells were treated for 4 h and stained with lysotracker red. Confocal microscopy indicated the colocalization of the green fluorescence signal from Fl_GUS and red fluorescence signal (lysotracker positive vesicles), proving the lysosome-specific delivery of GUS using the formulated LNs (Fig. 7A, bottom panel).However, the delivery of the free GUS was not specific to the lysosomes, as shown in Fig. 7A (middle panel), where no significant colocalization of green and red fluorescence signals was observed inside the cells. To determine the reason for the low uptake of free-GUS compared to GUS-LN, we analysed the mean intensity of the fluorescence signal (green signal). We observed a significant enhancement in the mean intensity of the green signal for GUS-LN compared to free β-GUS, indicating the effect of targeted delivery using LN (Fig. 7B). Additionally, the efficiency of transport using LN is superior to that of free β-GUS, as indicated by the Pearson's coefficient (Fig. 7C), suggesting a greater degree of colocalization with lysosomes.
Next, the treated cells were assessed for an increase in β-GUS activity by an in vitro enzymatic activity assay. For this, the cells were treated with unlabelled-free-GUS and GUS-LN (unlabelled GUS encapsulated in LN) for 4 h, and untreated cells were kept as controls. Before treatment, we determined the β-GUS activity and used an equal concentration of effective GUS in GUS-LN and free β-GUS at 200 µg mL−1, resulting in an equivalent β-GUS activity of 800 U mg−1 (1 unit equals the production of 4-MU per minute). The model substrate 4-MUG was added to the cell lysate and assessed for the production of 4-MU. We found a significant 6-fold increase in the enzyme activity in GUS-LN-treated cells compared to untreated cells (Fig. 7D). In contrast, the increase in activity in the free GUS-treated cells could not match the increment in activity like that of the GUS-LN (Fig. 7D). The low uptake of free-GUS may result from its instability at physiological temperatures and lack of protection in the cellular environment, in contrast to the LN-protected form. The M6P-functionalized LN successfully enhanced uptake and enabled the targeted delivery of functional enzymes to lysosomes, compared to free GUS, even in Drosophila's MPS VII model cells.
We also examined the time- and dose-dependent internalization of GUS-LN. For the time-dependent uptake, we treated hemocytes with GUS-LN at an effective GUS concentration of 100 µg mL−1 for 2 and 4 hours. After incubation, the enhancement in β-GUS activity was determined and compared with that of untreated cells. The level of activity after 2 h of internalization is nearly that of the untreated case (1.2 nmol mg−1 h−1 for untreated and 1.3 nmol mg−1 h−1 for 2 hours). After 4 hours of incubation, there was a 2-fold increment in the activity level (2.9 nmol mg−1 h−1), indicating a clear time-dependent enhancement in β-GUS activity as shown in Fig. S14(A) in the SI. Furthermore, to evaluate dose dependency, we compared the treatment groups of GUS-LN with effective GUS concentrations of 100 and 200 µg mL−1, incubated for 4 h. Upon determining β-GUS activity after internalization, we found that the enhancement in the activity level for the treatment of 200 µg mL−1 is more than twice that of the 100 µg mL−1 case (2.9 and 6.3 nmol mg−1 h−1 activity for 100 and 200 µg mL−1 treated cells, respectively) (Fig. S14(B) in the SI). This set of experimental data indicated an increase in the enzyme activity with both increasing enzyme concentration and incubation time (Fig. S14 in the SI).
Thus, our data established an efficient system and a novel attempt to deliver the large-sized functional enzyme compared to the existing administration of free-GUS. Also, the usage of Drosophila primary cells, a simple and versatile system, opens a scope of high-throughput studies in the future to explore alternative drug delivery systems.
Conclusions
The development of mannose-6-phosphate (M6P) functionalized liposomal nanocarriers (LN) offered a promising and targeted approach for enzyme replacement therapy (ERT) in Mucopolysaccharidosis type VII (MPS VII), a lysosomal storage disorder caused by β-glucuronidase (GUS) deficiency. Initially, a nonester backbone-based M6P functionalized glycolipid (M6PGL) was designed and synthesized to protect the formulated liposomes from the high concentration of esterases present in the blood. M6PGL, along with DHGPC and Chl, helped us formulate a robust LN, which remained stable for up to 4 weeks. The strategy of incorporating M6P functionality on the surface of the LN was to exploit the cation-independent M6P receptor (CI-MPR) for receptor-mediated endocytosis. In previous reports from our group and others, we have established that M6P-functionalized glycopolypeptides and nanocarriers are internalized into cells via CI-MPR,43,45,47 which demonstrates the CI-MPR-mediated endocytosis of the current M6P-functionalized LN rather than non-specific uptake.Compared to conventional ERT, which often suffers from rapid clearance and suboptimal lysosomal targeting, the M6P-functionalized LN described in this work enhance enzyme stability by protecting it from the blood plasma esterases, improve intracellular enzyme bioavailability, and direct proper organelle-targeted enzyme delivery. The enhanced delivery efficiency and lysosomal targeting capability of these nanocarriers may also help reduce off-target enzyme distribution, minimising adverse effects, and improving overall therapeutic efficacy. Our findings highlight the potential of this nanocarrier system to mitigate the pathological condition of MPS VII. As a next step, demonstrating rescue of disease pathology in the MPS VII Drosophila model would further strengthen the evidence for this ERT. However, delivering M6P-functionalized LN to the entire organism conventionally by injection is technically challenging in Drosophila due to its small body. Future research should focus on designing and evaluating the long-term pharmacokinetics and biodistribution of robust nanocarriers, as well as in vivo MPS VII mouse model studies to validate the translational potential of this approach.
This work paves the way for the next generation of nanocarrier-based systems for targeted ERT strategies, not only for MPS VII but also for other lysosomal storage disorders that require precise intracellular enzyme delivery, keeping the function of the enzymes unaffected. By advancing targeted therapeutic approaches, this research contributes to the ongoing development of more effective and patient-friendly treatments for rare lysosomal storage disorders.
Author contributions
SSG and AP designed the work. SSG and RD supervised the work. AP and AD contributed equally to the work. AP synthesized and characterized all the components, formulated the liposomal nanocarrier, and conducted all the other experiments under the supervision of SSG. AD performed all the in vitro experiments with mammalian cells and Drosophila MPS VII model studies under the supervision of RD. KM contributed to stability studies, and BM contributed to lipid synthesis. The manuscript was written by AP and AD, and edited by RD and SSG. All the authors have approved the final version of the manuscript.Conflicts of interest
The authors declare no competing financial conflicts.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary information (SI). Materials, methods, experimental, and characterization details are provided in the supplementary information and it is available. See DOI: https://doi.org/10.1039/d5ma00526d.Acknowledgements
AP acknowledges the Prime Minister's Research Fellows (PMRF) scheme for the fellowship. AD acknowledges the Indian Council of Medical Research (ICMR) for the senior research fellowship, which provided financial assistance. KM and BM acknowledge the Council of Scientific and Industrial Research (CSIR), New Delhi, for the fellowship. AP acknowledges Mr Nikesh Dewangan for assisting with the extrusion of liposomes. AP acknowledges Mr Gaurab Ghosh and Mr Abhrajyoti Nandi for their assistance with the confocal microscopy analysis of the dual dye-loaded nanocarrier. AP and SSG acknowledge Prof. Arabinda Chaudhuri, IISER Kolkata, for providing valuable insights for liposome formulation, protein encapsulation in the liposomal nanocarrier, and other critical experiments. RD acknowledges ICMR (grant number-6/9-7(318)/2023-ECD-II). SSG acknowledges the DST-Nano mission (grant number DST/NM/NB/2018/16). The TOC and manuscript images were created using BioRender, Adobe Illustrator, and Inkscape. We acknowledge IISER Kolkata for the research environment and facilities.Notes and references
- C. de Duve, B. C. Pressman, R. Gianetto, R. Wattiaux and F. Appelmans, Biochem. J., 1955, 60, 604–617 CrossRef CAS.
- P. Boya, Antioxid. Redox Signaling., 2011, 17, 766–774 CrossRef.
- F. Wang, R. Gómez-Sintes and P. Boya, Traffic, 2018, 19, 918–931 CrossRef CAS PubMed.
- C. Di Malta, D. Siciliano, A. Calcagni, J. Monfregola, S. Punzi, N. Pastore, A. N. Eastes, O. Davis, R. De Cegli, A. Zampelli, L. G. Di Giovannantonio, E. Nusco, N. Platt, A. Guida, M. H. Ogmundsdottir, L. Lanfrancone, R. M. Perera, R. Zoncu, P. G. Pelicci, C. Settembre and A. Ballabio, Science, 2017, 356, 1188–1192 CrossRef CAS PubMed.
- J.-H. Lee, W. H. Yu, A. Kumar, S. Lee, P. S. Mohan, C. M. Peterhoff, D. M. Wolfe, M. Martinez-Vicente, A. C. Massey, G. Sovak, Y. Uchiyama, D. Westaway, A. M. Cuervo and R. A. Nixon, Cell, 2010, 141, 1146–1158 CrossRef CAS.
- X. Ma, H. Liu, S. R. Foyil, R. J. Godar, C. J. Weinheimer, J. A. Hill and A. Diwan, Circulation, 2012, 125, 3170–3181 CrossRef CAS.
- X. Peng, J. Kim, G. Gupta, K. Agaronyan, M. C. Mankowski, A. Korde, S. S. Takyar, H. J. Shin, V. Habet, S. Voth, J. P. Audia, D. Chang, X. Liu, L. Wang, Y. Cai, X. Tian, S. Ishibe, M.-J. Kang, S. Compton, C. B. Wilen, C. S. Dela Cruz and L. Sharma, J. Immunol., 2022, 209, 1314–1322 CrossRef CAS.
- M. Cao, X. Luo, K. Wu and X. He, Signal Transduction Targeted Ther., 2021, 6, 379 CrossRef CAS PubMed.
- R.-M. N. Boustany, Nat. Rev. Neurol., 2013, 9, 583–598 CrossRef CAS PubMed.
- F. M. Platt, A. d’Azzo, B. L. Davidson, E. F. Neufeld and C. J. Tifft, Nat. Rev. Dis. Prim., 2018, 4, 27–51 CrossRef PubMed.
- G. Parenti, D. L. Medina and A. Ballabio, EMBO Mol. Med., 2021, 13, e12836 CrossRef CAS.
- M. T. Fiorenza, E. Moro and R. P. Erickson, Hum. Mol. Genet., 2018, 27, R119–R129 CrossRef CAS.
- W. S. Sly, B. A. Quinton, W. H. McAlister and D. L. Rimoin, J. Pediatr., 1973, 82, 249–257 CrossRef CAS.
- M. I. Hassan, A. Waheed, J. H. Grubb, H. E. Klei, S. Korolev and W. S. Sly, PLoS One, 2013, 8, e79687 CrossRef PubMed.
- A. M. Montaño, N. Lock-Hock, R. D. Steiner, B. H. Graham, M. Szlago, R. Greenstein, M. Pineda, A. Gonzalez-Meneses, M. Çoker, D. Bartholomew, M. S. Sands, R. Wang, R. Giugliani, A. Macaya, G. Pastores, A. K. Ketko, F. Ezgü, A. Tanaka, L. Arash, M. Beck, R. E. Falk, K. Bhattacharya, J. Franco, K. K. White, G. A. Mitchell, L. Cimbalistiene, M. Holtz and W. S. Sly, J. Med. Genet., 2016, 53, 403 CrossRef PubMed.
- M. Beck, Dev. Med. Child Neurol., 2018, 60, 13–18 CrossRef PubMed.
- R. H. Lachmann, Curr. Opin. Pediatr., 2011, 23, 588–593 CrossRef CAS.
- E. Beutler, A. Kay, A. Saven, P. Garver, D. Thurston, A. Dawson and B. Rosenbloom, Blood, 1991, 78, 1183–1189 CrossRef CAS.
- L. D. M. Pena, R. J. Barohn, B. J. Byrne, C. Desnuelle, O. Goker-Alpan, S. Ladha, P. Laforêt, K. E. Mengel, A. Pestronk, J. Pouget, B. Schoser, V. Straub, J. Trivedi, P. Van Damme, J. Vissing, P. Young, K. Kacena, R. Shafi, B. L. Thurberg, K. Culm-Merdek and A. T. van der Ploeg, Neuromuscular Disord., 2019, 29, 167–186 CrossRef.
- M. Beck, Expert Opin. Biol. Ther., 2009, 9, 255–261 CrossRef CAS PubMed.
- E. Jameson, S. Jones and T. Remmington, Cochrane Database Syst. Rev., 2019,(6), CD009354 Search PubMed.
- M.-Y. Chan, A. J. Nelson and L.-H. Ngu, Mol. Genet. Metab. Rep., 2023, 36, 100991 CAS.
- C. J. Hendriksz, R. Parini, M. D. AlSayed, J. Raiman, R. Giugliani, M. L. Solano Villarreal, J. J. Mitchell, B. K. Burton, N. Guelbert, F. Stewart, D. A. Hughes, K. I. Berger, P. Slasor, R. Matousek, E. Jurecki, A. J. Shaywitz and P. R. Harmatz, Mol. Genet. Metab., 2016, 119, 131–143 CrossRef CAS PubMed.
- M. Brunelli, Á. Atallah and E. M. K. da Silva, Cochrane Database Syst. Rev., 2021,(9), CD009806 Search PubMed.
- A. Schulz, N. Specchio, E. de los Reyes, P. Gissen, M. Nickel, M. Trivisano, S. C. Aylward, A. Chakrapani, C. Schwering, E. Wibbeler, L. M. Westermann, D. J. Ballon, J. P. Dyke, A. Cherukuri, S. Bondade, P. Slasor and J. Cohen Pfeffer, Lancet Neurol., 2024, 23, 60–70 CrossRef CAS.
- B. K. Burton, F. Feillet, K. N. Furuya, S. Marulkar and M. Balwani, J. Hepatol., 2022, 76, 577–587 CrossRef CAS.
- J. Cadaoas, G. Boyle, S. Jungles, S. Cullen, M. Vellard, J. H. Grubb, A. Jurecka, W. Sly and E. Kakkis, Mol. Genet. Metab., 2020, 130, 65–76 CrossRef CAS.
- P. Harmatz, C. B. Whitley, R. Y. Wang, M. Bauer, W. Song, C. Haller and E. Kakkis, Mol. Genet. Metab., 2018, 123, 488–494 CrossRef CAS.
- R. Y. Wang, J. F. da Silva Franco, J. López-Valdez, E. Martins, V. R. Sutton, C. B. Whitley, L. Zhang, T. Cimms, D. Marsden, A. Jurecka and P. Harmatz, Mol. Genet. Metab., 2020, 129, 219–227 CrossRef CAS.
- H. A. Lau, D. Viskochil, P. Tanpaiboon, A. G.-M. Lopez, E. Martins, J. Taylor, B. Malkus, L. Zhang, A. Jurecka and D. Marsden, Mol. Genet. Metab., 2022, 136, 28–37 CrossRef CAS PubMed.
- M. Li, Pediatr. Ann., 2018, 47, e191–e197 Search PubMed.
- D. Concolino, F. Deodato and R. Parini, Ital. J. Pediatr., 2018, 44, 120 CrossRef CAS PubMed.
- M. Taylor, S. Khan, M. Stapleton, J. Wang, J. Chen, R. Wynn, H. Yabe, Y. Chinen, J. J. Boelens, R. W. Mason, F. Kubaski, D. D. G. Horovitz, A. L. Barth, M. Serafini, M. E. Bernardo, H. Kobayashi, K. E. Orii, Y. Suzuki, T. Orii and S. Tomatsu, Biol. Blood Marrow Transplant., 2019, 25, e226–e246 Search PubMed.
- C. Vogler, M. S. Sands, B. Levy, N. Galvin, E. H. Birkenmeier and W. S. Sly, Pediatr. Res., 1996, 39, 1050–1054 Search PubMed.
- L. H. O’Connor, L. C. Erway, C. A. Vogler, W. S. Sly, A. Nicholes, J. Grubb, S. W. Holmberg, B. Levy and M. S. Sands, J. Clin. Invest., 1998, 101, 1394–1400 Search PubMed.
- C. Vogler, B. Levy, J. H. Grubb, N. Galvin, Y. Tan, E. Kakkis, N. Pavloff and W. S. Sly, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 14777–14782 CrossRef CAS PubMed.
- H. Xia, Q. Mao and B. L. Davidson, Nat. Biotechnol., 2001, 19, 640–644 CrossRef CAS PubMed.
- K. O. Orii, J. H. Grubb, C. Vogler, B. Levy, Y. Tan, K. Markova, B. L. Davidson, Q. Mao, T. Orii, N. Kondo and W. S. Sly, Mol. Ther., 2005, 12, 345–352 CrossRef CAS PubMed.
- S. X. Lin, W. G. Mallet, A. Y. Huang and F. R. Maxfield, Mol. Biol. Cell, 2003, 15, 721–733 Search PubMed.
- J. Hirst, C. E. Futter and C. R. Hopkins, Mol. Biol. Cell, 1998, 9, 809–816 CrossRef CAS.
- J. Seo and D. B. Oh, Anim. Cells Syst., 2022, 26, 84–91 CrossRef CAS.
- B. Mondal, T. Dutta, A. Padhy, S. Das and S. Sen Gupta, ACS Omega, 2022, 7, 5–16 CrossRef CAS PubMed.
- S. Das, N. Parekh, B. Mondal and S. S. Gupta, ACS Macro Lett., 2016, 5, 809–813 CrossRef CAS PubMed.
- B. Mondal, A. Padhy, S. Maji, A. Gupta and S. Sen Gupta, Biomater. Sci., 2023, 11, 1810–1827 RSC.
- B. Mondal, B. Pandey, N. Parekh, S. Panda, T. Dutta, A. Padhy and S. Sen Gupta, Biomater. Sci., 2020, 8, 6322–6336 RSC.
- B. Mondal, T. Dutta and S. Sen Gupta, Chem. Commun., 2021, 57, 109–112 RSC.
- B. Sevarika, D. Capri, J. Frey, M. C. Dinamarca, D. Häussinger and S. McNeil, Eur. J. Pharm. Biopharm., 2025, 209, 114665 CrossRef CAS PubMed.
- E. Crucianelli, P. Bruni, A. Frontini, L. Massaccesi, M. Pisani, A. Smorlesi and G. Mobbili, RSC Adv., 2014, 4, 58204–58207 Search PubMed.
- A. Padhy, M. Gupta, A. Das, I. Farook, T. Dutta, S. Datta, R. Datta and S. S. Gupta, Bioconjugate Chem., 2025, 36, 383–394 Search PubMed.
- V. P. Torchilin, Nat. Rev. Drug Discovery, 2005, 4, 145–160 CrossRef CAS PubMed.
- N. Grimaldi, F. Andrade, N. Segovia, L. Ferrer-Tasies, S. Sala, J. Veciana and N. Ventosa, Chem. Soc. Rev., 2016, 45, 6520–6545 RSC.
- U. Bulbake, S. Doppalapudi, N. Kommineni and W. Khan, Pharmaceutics, 2017, 9, 12 CrossRef PubMed.
- M. Germain, F. Caputo, S. Metcalfe, G. Tosi, K. Spring, A. K. O. Åslund, A. Pottier, R. Schiffelers, A. Ceccaldi and R. Schmid, J. Controlled Release, 2020, 326, 164–171 CrossRef CAS PubMed.
- H. Nsairat, D. Khater, U. Sayed, F. Odeh, A. Al Bawab and W. Alshaer, Heliyon, 2022, 8, e09394 CrossRef CAS.
- R. Thekkedath, K. Alexander and V. P. Torchilin, Nanomedicine, 2013, 8, 1055–1065 CrossRef CAS.
- J. L. Schneider, R. K. Dingman and S. V. Balu-Iyer, J. Pharm. Sci., 2018, 107, 831–837 CrossRef CAS PubMed.
- D. Jiang, H. Lee and W. M. Pardridge, Sci. Rep., 2020, 10, 13334 CrossRef CAS PubMed.
- D. Wu, Q. Chen, X. Chen, F. Han, Z. Chen and Y. Wang, Signal Transduction Targeted Ther., 2023, 8, 217 Search PubMed.
- A. D. Bangham, M. M. Standish and J. C. Watkins, J. Mol. Biol., 1965, 13, 238 Search PubMed -IN227.
- A. D. Bangham, M. M. Standish and G. Weissmann, J. Mol. Biol., 1965, 13, 253 CrossRef CAS -IN228.
- S. Pal Dhiman, K. Mondal Dipon and R. Datta, Antimicrob. Agents Chemother., 2015, 59, 2144–2152 CrossRef PubMed.
- A. Hiroyasu, D. C. DeWitt and A. G. Goodman, JoVE, 2018, e57077, DOI:10.3791/57077.
- J. H. Glaser and W. S. Sly, J. Lab. Clin. Med., 1973, 82, 969–977 CAS.
- O. Lowry, N. Rosebrough, A. L. Farr and R. Randall, J. Biol. Chem., 1951, 193, 265–275 CrossRef CAS.
- F. M. Williams, Clin. Pharmacokinet., 1985, 10, 392–403 CrossRef CAS.
- M. C. Taira, N. S. Chiaramoni, K. M. Pecuch and S. Alonso-Romanowski, Drug Delivery, 2004, 11, 123–128 CrossRef CAS PubMed.
- S. Kaddah, N. Khreich, F. Kaddah, C. Charcosset and H. Greige-Gerges, Food Chem. Toxicol., 2018, 113, 40–48 CrossRef CAS PubMed.
- A. Jash, A. Ubeyitogullari and S. S. H. Rizvi, J. Mater. Chem. B, 2021, 9, 4773–4792 Search PubMed.
- V. V. S. N. L. Andra, S. V. N. Pammi, L. V. K. P. Bhatraju and L. K. Ruddaraju, Bionanoscience, 2022, 12, 274–291 Search PubMed.
- D. Lombardo and M. A. Kiselev, Pharmaceutics, 2022, 14, 543 Search PubMed.
- S. Bar, M. Prasad and R. Datta, Dis. Models & Mech., 2018, 11, dmm036954 Search PubMed.
- N. Mandal, A. Das and R. Datta, Neurobiol. Dis., 2025, 206, 106825 CrossRef CAS PubMed.
- B. Charroux and J. Royet, Proc. Natl. Acad. Sci., 2009, 106, 9797–9802 Search PubMed.
- H. N. Stephenson, R. Streeck, F. Grüblinger, C. Goosmann and A. Herzig, Development, 2022, 149, dev200286 Search PubMed.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2026 |