Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Gut health monitoring via intestinal barrier function screening using a transepidermal microneedle-based sensor
Nicolas
Maïno
 ad,
Sihui
Xu
ad,
Petter
Brodin
ad,
Sihui
Xu
ad,
Petter
Brodin
 b and
Onur
Parlak
b and
Onur
Parlak
 *acd
*acd
aDepartment of Medicine, Solna, Division of Dermatology and Venereology, Karolinska Institutet, 171 77, Stockholm, Sweden. E-mail: onur.parlak@ki.se
bDepartment of Women's and Children's Health, Unit for Clinical Pediatrics, Karolinska Institutet, Solna 171 65, Sweden
cCenter for the Advancement of Integrated Medical and Engineering Sciences, Karolinska Institutet and KTH Royal Institute of Technology, 171 77, Stockholm, Sweden
dCentre for Molecular Medicine, Karolinska University Hospital, 171 64, Stockholm, Sweden
First published on 19th December 2025
Abstract
The growing prevalence of chronic digestive disorders, such as inflammatory bowel disease, underscores the urgent need for innovative solutions that enable longitudinal monitoring of disease progression and treatment efficacy. Addressing this challenge, we present a novel microneedle-based sensor designed for rapid, point-of-care assessment of intestinal barrier integrity. Through transient application to the skin, the device samples intestinal fatty acid binding protein (IFABP) from systemic circulation, offering a minimally invasive alternative to conventional diagnostics. We demonstrate a versatile, affinity-based electrochemical sensing mechanism integrated into low-cost and clean room-free microneedles. The resulting device is validated in a biomimetic skin-like hydrogel in which it achieves good linearity, a limit of detection of 1.5 ng mL−1 and highly specific response in a short assay format of one hour including the sampling phase. Furthermore, we validate the sensor's biocompatibility, penetration efficiency, and sensing capability in ex vivo human skin, establishing a critical foundation for future clinical applications. This breakthrough technology holds significant promise for transforming the management of gastrointestinal diseases through frequent, patient-friendly monitoring.
1. Introduction
The declining incidence of infectious diseases has coincided with a rising global prevalence of inflammatory and autoimmune disorders.1 This trend is widely attributed to perturbations in early-life immune–microbe interactions, which are critical for immune development.2 A paradigm for this phenomenon is inflammatory bowel disease (IBD), where a loss of immune tolerance to commensal gut microbiota is a central driver of chronic intestinal inflammation and disease pathogenesis.The contribution of intestinal barrier dysfunction to IBD has long been recognized,3,4 and contributing factors such as dysregulation of the gut microbiome are also known to play a major role in disease progression.5,6 Despite the recognized importance of barrier integrity, current point-of-care methodologies for its clinical assessment are hampered by significant practical limitations. Traditional approaches, such as urinary excretion tests using orally administered probes, are cumbersome for patients, requiring prolonged fasting and extended collection periods.7 Similarly, quantification of blood-derived surrogate markers is costly and requires blood draw associated with long sample preparation and assaying time. Consequently, these methods impose a substantial patient burden and are not suited well for frequent, longitudinal monitoring. Parallel challenges exist in microbiome analysis, where 16S ribosomal RNA sequencing of stool samples involves complex processing and is limited by the different composition of the luminal versus fecal microbiota.8,9 Collectively, these limitations underscore an unmet need for rapid analytical methods allowing the longitudinal assessment of intestinal barrier function.
In this context of growing diagnostic challenges, biosensors have emerged as a transformative solution, offering cost-effective, rapid and minimally invasive platforms for both diagnosis10–12 and disease monitoring.13–15 A significant advantage of biosensors lies in their potential for miniaturization and integration into wearable formats such as smartwatches, skin patches or other discreet devices. A particularly relevant innovation is the use of microneedles for sampling dermal interstitial fluids16,17 or capillary blood.18 Microneedles have demonstrated reduced pain and better acceptability compared to standard hypodermic needles.19 Such approaches are particularly interesting for applications where current methods rely on invasive biopsy sampling as for intestinal mucosal or luminal biopsy sampling through endoscopy. Given that ISF shares approximately 90% of its protein composition with plasma and serum,20,21 microneedle-based sampling offers a viable alternative for monitoring systemic biomarkers of intestinal health without the need for blood draws.
Herein, we report a novel microneedle-based biosensor for rapid, point-of-care assessment of intestinal barrier integrity through the detection of intestinal fatty acid binding protein (IFABP), a biomarker released into systemic circulation during enterocyte damage (Fig. 1a). Following the 40 minute transdermal sampling phase, the sensor is removed and interrogated ex situ. Quantification is achieved through a highly specific sandwich immunoassay with electrochemical endpoint detection, completing the entire analysis within 30 minutes (Fig. 1b, i–iii). Owing to an in-depth model of our sensor's mechanism of action, we were successful in optimizing its workflow to deliver adequate performance in this short time frame. We validated sensor performance using a biomimetic hydrogel designed to simulate the skin microenvironment, demonstrating excellent linearity, a low detection limit of 1.5 ng mL−1, and negligible nonspecific binding. Furthermore, we confirmed the device's biocompatibility and its capability for successful IFABP detection in an ex vivo human skin model, thereby establishing a strong foundation for future in vivo applications.
The sensor is fabricated from commercially available components and batch-functionalized, yielding a remarkably low unit cost of $2.59 (Table S4). This cost-effectiveness, combined with the rapid analysis time, positions the technology for practical deployment in longitudinal disease monitoring via single-use devices. In this paradigm, a new sensor is applied for each discrete measurement over a defined observation period. This strategy minimizes patient/device contact time compared to continuous monitors while remaining capable of capturing clinically relevant fluctuations in intestinal permeability that occur over several hours.22,23
The target biomarker, intestinal fatty acid binding protein (IFABP or FABP2), has been extensively studied in the context of intestinal barrier dysfunction. Clinical studies have reported seemingly contradictory concentration trends, a discrepancy attributed to variations in sampling timepoints and the biofluid analyzed (e.g., plasma versus urine).24–27 Furthermore, while existing biosensors for intestinal barrier or microbiome dysregulation exist, they typically rely on complex, microfabricated systems or are hampered by prolonged assay times; consequently, a truly rapid and cost-effective point-of-care solution has remained elusive.28,29
The microneedle sensor presented herein directly addresses these limitations. By enabling facile, high-resolution temporal assessment of intestinal barrier dynamics, this technology provides a powerful new tool to elucidate disease mechanisms and optimize therapeutic interventions. We anticipate that this approach will ultimately pave the way for more objective and personalized management of gastrointestinal disorders.
2. Results and discussion
2.1. Sensor assembly and functionalization
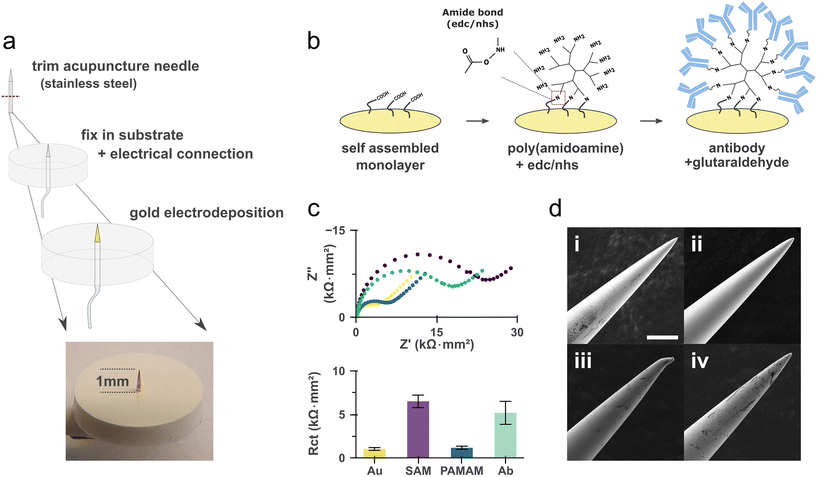
We devised our microneedle sensor with cost-effective and easy to implement fabrication methods in mind while aiming for a small footprint and being uncompromising on the robustness of the microneedles. To achieve this, we utilized commercially available stainless-steel needles with a diameter of 300 μm, which provide superior toughness and a naturally sharp profile for efficient skin penetration (hereafter referred to as microneedles). Each device was assembled by inserting a single trimmed microneedle into an 8 mm diameter polytetrafluoroethylene/silicone substrate. This configuration provided an exposed needle length of 1.04 mm for functionalization and insertion, while the opposite end was reserved for electrical connection (Fig. 2a). The selected insertion length was chosen to specifically target the upper dermis, in accordance with established literature.30–32 To facilitate subsequent biofunctionalization and ensure consistent electrochemical performance, the sensing portion of the microneedle was coated with a gold layer via electrodeposition (Fig. S1 and S2).The sensing strategy employed antibodies as affinity reagents, which were immobilized onto the gold-coated sensor surface via carbodiimide and glutaraldehyde crosslinking chemistries (Fig. 2b). Starting from a self-assembled monolayer with a carboxylic acid end group, an amine-terminated dendrimer is first introduced before conjugation of the antibody. Although direct antibody immobilization is feasible, this branched dendritic architecture was adopted to maximize antibody loading capacity in the subsequent step, thereby enhancing sensor sensitivity (Fig. S3).
We characterized each stage of the functionalization process using electrochemical impedance spectroscopy (EIS) and scanning electron microscopy (SEM) (Fig. 2c and d). The EIS results confirmed that the formation of a self-assembled monolayer can be inferred from an increase in interfacial charge transfer resistance while amine-terminated dendrimer conjugation decreases it because of electrostatic interaction with the redox mediator used.33 Finally, the immobilization of antibodies and blocking with bovine serum albumin (BSA) induced a substantial increase in Rct, consistent with increased steric hindrance that impedes the diffusion of the redox probe to the electrode surface.
SEM imaging confirmed the preservation of the microneedle's structural integrity and conductivity throughout functionalization, revealing a smooth and defect-free gold coating (Fig. 2d and S1d). The mechanical resilience of the coating was qualitatively assessed by insertion through a Parafilm® membrane. We determined that the use of a specialized “hard gold” electroplating bath was critical for achieving a mechanically stable gold film on the stainless-steel substrate. In contrast, gold films electrodeposited from an aqueous chloroauric acid solution consistently delaminated after a single insertion.
2.2. Modeling and optimization of the IFABP detection method
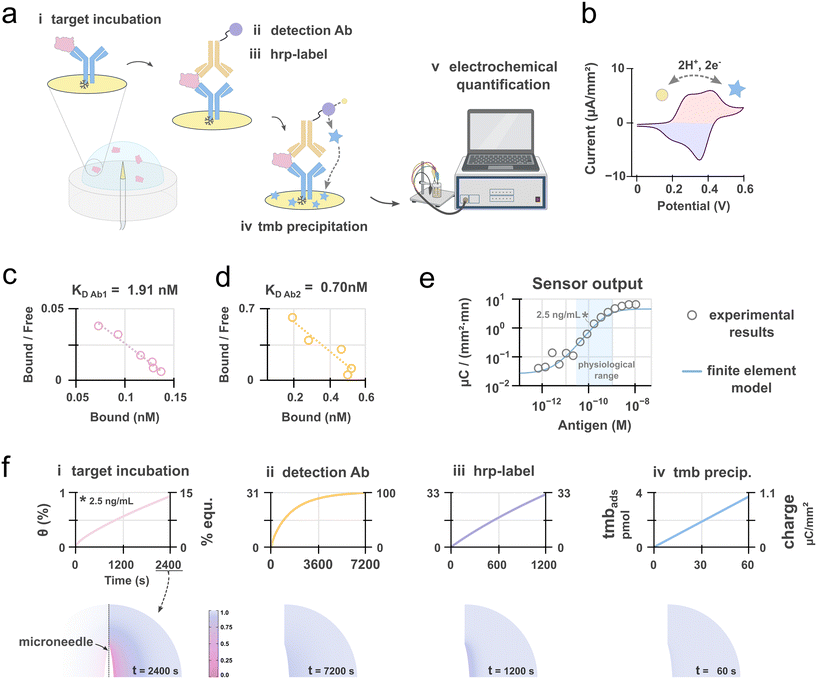
The detection of IFABP was achieved via an electrochemical sandwich immunoassay, directly implemented on the functionalized microneedle platform. The assay protocol consists of five sequential steps following sample introduction: (i) antigen capture from the sample matrix; (ii) binding of a detection antibody; (iii) conjugation of a horseradish peroxidase (HRP) label; (iv) enzymatic reaction upon addition of an HRP substrate; and (v) amperometric detection of the electroactive enzymatic product (Fig. 3a, i–v).In the third enzymatic step, horseradish peroxidase (HRP) catalyzes the oxidation of the chromogenic substrate tetramethylbenzidine (TMB) in the presence of hydrogen peroxide (H2O2), generating an insoluble, blue-colored precipitate. This product becomes adsorbed on the substrate (i.e. the electrode) where it can be electrochemically reduced/re-oxidized through a two electron, two proton reaction corresponding to the dissolution/formation of a charge transfer complex (Fig. 3b).34,35 This electrochemical readout provides a quantitative signal proportional to the target analyte concentration. The HRP/TMB system is widely favored for such applications due to its excellent signal amplification and linear response.36 The assay is performed as an endpoint measurement: the enzymatic reaction is allowed to proceed for a defined period to accumulate sufficient product on the electrode, after which the reaction is quenched, and the electrochemical measurement is immediately performed.
To rationalize the performance of the electrochemical assay, we developed a theoretical model that integrates the affinity-based sensing mechanism with the electrochemical transduction methodology using finite element analysis (FEA). The application of FEA to obtain numerical solutions for systems involving coupled mass transport and surface reactions is well-established; it has been successfully employed to model antibody–antigen binding kinetics, such as those derived from surface plasmon resonance, with high fidelity.37 We have used a similar approach where purely diffusive mass transport within a finite volume is coupled to the surface affinity or catalytic reaction taking place at the microneedle surface (Fig. S4 and Table S1). For model simplification, the wash steps from the experimental protocol were neglected. The simulation was designed to replicate the in vitro testing conditions, wherein a 25 μL droplet containing a defined concentration of the target analyte is dispensed directly onto the microneedle sensor (Fig. 3a).
With this model and experimental affinity data of the antibodies used in our sensor (Fig. 3c and d and S5), we calculated expected sensor outputs for a range of IFABP concentration and compared them to experimental in vitro results (Fig. 3e). We observed good qualitative agreement between the model and the experimental results, with predicted values falling within the same order of magnitude. Notably, within the physiological concentration range of IFABP, the model predictions were within 25% of the measured values. This close correspondence suggests that the model accurately captures the fundamental mechanistic principles governing the sensor's operation.
The FEA model provides critical insights by calculating key performance metrics, including receptor binding site occupancy, the percentage of equilibrium achieved, and the spatiotemporal concentration profiles of targets and reagents during incubation (Fig. 3f, top and bottom rows). These parameters are instrumental for rational assay optimization. We have accordingly used this model to simulate sensor outputs over a wide range of parameters to identify an optimal protocol (Fig. S6). In simulated data, the optimized assay improves the limit-of-detection (LOD) by threefold from 1.15 ng mL−1 to 0.39 ng mL−1. More importantly, the model enabled a drastic reduction in the total assay duration by an order of magnitude, decreasing it from over 300 minutes in a well-plate format to just 25 minutes. This optimized protocol was adopted for all subsequent experiments. The experimental results validated the model's predictions, showing good qualitative agreement; the measured LOD for IFABP was 0.63 ng mL−1, closely aligning with the simulated value of 0.39 ng mL−1 (Fig. S7a).
A further application of the FEA model was to evaluate the feasibility of alternative detection modalities, such as direct, label-free detection via electrochemical impedance spectroscopy (EIS).38 For physiological concentrations of IFABP (0–10 ng mL−1, approximately 50–500 pM), the model calculated that the fractional occupancy of a surface-immobilized antibody with a typical affinity (e.g., Kd = 1.91 nM, as used in our sensor; Fig. S5) increases by a mere 3% across the target range (Fig. S4d).
As the signal magnitude in a label-free method is directly proportional to receptor occupancy, the resultant change in impedance would be negligible and preclude reliable quantification. This theoretical prediction was experimentally validated using a microneedle sensor targeting IL-1β. Although the antibody employed has demonstrated a detection limit of 4 pg mL−1 in a conventional well-plate ELISA, it failed to generate a signal significantly above background on the microneedle platform, even at a high concentration of 10 ng mL−1 (Fig. S8, Table S2). This result confirms the model's assertion that label-free EIS lacks the required sensitivity for low-abundance biomarkers on this platform (Fig. S8 and Table S2).
Conventional approaches to this sensitivity challenge have typically relied on decoupling the sampling and sensing elements, ultimately resorting to complex, laboratory-bound analytical methods that are incompatible with point-of-care use.31,39 Alternative strategies employing microfabricated sensors, while integrated, remain significantly costly, as demonstrated by a previously reported interface for IFABP detection in urine.28 In contrast, the enzyme-linked immunosorbent assay (ELISA) format implemented in our study harnesses the powerful signal amplification inherent to the enzymatic catalytic step. This enables the generation of a well-resolvable electrochemical signal even at low biomarker concentrations and minimal receptor occupancy.40,41 Herein, we demonstrate for the first time the translation of this robust detection principle into a rapid, affordable, and fully integrated microneedle biosensor for the point-of-care quantification of IFABP. This work effectively bridges a critical technological gap, moving beyond costly and complex prototypes to a practical diagnostic tool.
2.3. Analytical performance of microneedle-based sensors in hydrogels
To evaluate the performance of the optimized microneedle sensor under more physiologically relevant conditions, we employed a biomimetic matrix designed to simulate key properties of human skin. This model, adapted from previous reports, utilizes a bilayer structure of Parafilm® and a hydrogel to emulate the mechanical barrier and diffusive environment of the dermis.42–44We significantly improved upon existing models by formulating a hydrogel composed of collagen – a more biologically relevant proxy for the native extracellular matrix – hydrated with artificial interstitial fluid (aISF) and spiked with a defined concentration of the target analyte (Fig. 4a). We term this construct, comprising a spiked collagen/aISF hydrogel beneath a Parafilm® layer, a “skin-like hydrogel”. This collagen-based model presents a substantial advantage over commonly used agarose hydrogels. Crucially, this allows for the precise and homogeneous incorporation of the target analyte via mixing prior to gelation, ensuring a uniform concentration throughout the matrix (Table S3). In contrast, methods relying on topical application or post-gelation soaking often will result in undefined concentration gradients, particularly for large macromolecules like proteins, which exhibit hindered diffusion.45 We therefore anticipate that sensor calibration performed in this refined, more realistic ex vivo system will provide a superior correlation to anticipated in vivo sensor performance.
The functionalized sensors were interrogated upon assay completion using two complementary electrochemical techniques: amperometry and cyclic voltammetry. The amperometric detection results in a more accurate assessment of the total precipitated TMB (from the area under the curve corresponding to the reduction charge, Fig. 4a). The cyclic voltammogram on the other hand has the advantage of displaying the characteristic oxidation/reduction waves of the TMB2+/TMB couple which can be useful to rule out the adsorption of interferent electroactive species on the sensor surface.
Amperometric measurements performed in the spiked skin-like hydrogel were used to construct a calibration curve for the microneedle sensor. The sensor exhibited excellent performance, with a strong linear response (R2 = 0.981) and a calculated limit of detection (LOD) of 1.46 ng mL−1 (Fig. 4b). This sensitivity is well-suited for physiological application, as it resides at the lower end of the reported physiological range for IFABP.46 Future work could focus on achieving even greater sensitivity through strategies such as employing enzymatic labels with higher specific activity or implementing advanced background suppression techniques.47 We observed that the LOD in the skin-like hydrogel is slightly higher than that measured in a simple buffer solution without a penetration step (0.63 ng mL−1, Fig. S7a). This difference is likely attributable to the additional mechanical challenges of insertion into a solid matrix. Importantly, the sensor's performance in this demanding environment remains robust and fully adequate for quantifying physiological IFABP levels. These results confirm that calibration within a biomimetic hydrogel provides a more realistic and transparent assessment of actual sensor performance. This in turns highlights the importance of testing microneedle sensors in a realistic scenario that emulates disruption of the sensor, which may arise in real life application.48
Notably, our analysis revealed that increasing the sensor's electroactive surface area—through longer microneedles, surface roughening, or the use of an array—does not enhance the limit of detection (LOD). This is because the non-specific adsorption of detection antibodies and streptavidin-HRP conjugates scales proportionally with the available surface area, thereby elevating the background signal (Fig. S7b and c). Consequently, the compact, single-needle design of our device presents a distinct advantage, as it minimizes reagent consumption and significantly reduces the associated cost per test (Table S4, 2.59$ per device including reagents).
We further evaluated the sensor's specificity within the skin-like hydrogel by spiking it with a high concentration (10 ng mL−1) of epidermal fatty acid binding protein (eFABP or FABP5), a structurally similar protein. Additionally, a technical control was performed using a sensor fabricated without the capture antibody. In both cases, the measured non-specific response was not significantly different from the blank control and was lower than the signal generated by the specific target (Fig. 4c).
In summary, the microneedle sensor developed herein enables the quantitative and highly specific detection of IFABP within a physiologically relevant environment. The achieved LOD of 1.46 ng mL−1 is not only clinically relevant but also competitive with those reported for more complex and costly systems.28 Furthermore, the entire assay is completed in a fraction of the time required by standard ELISA protocols, representing a significant advancement toward rapid, affordable, and point-of-care diagnostic monitoring.
2.4. Biocompatibility testing of microneedle-based sensors
We investigated our device biocompatibility in accordance with existing guidelines.49 We used two assessment methods to establish the degree of cytotoxicity of our device: proliferation rate and viability staining with a chromogenic dye. To prepare test extracts, devices were immersed in serum-supplemented cell culture medium and agitated for 24 hours at 37 °C, following recommended protocols to isolate potential leachable compounds. The resulting liquid phase was then applied to sub-confluent 2D cultures of N/TERT-2G keratinocytes for 48 hours. Cellular confluence, serving as a metric for proliferation, was monitored optically at two-hour intervals and compared against a negative control (plain serum-supplemented medium) and a positive cytotoxicity control (0.1 mg mL−1 sodium dodecyl sulfate) (Fig. 5a).Representative images captured at the 24 and 48 hour time points reveal no qualitative differences in cell morphology or confluence between keratinocytes treated with device extracts and those in the blank control. The proliferation rate normalized to the blank was not found to be significantly different (Fig. 5c, 100.0 ± 6.0% and 98.2 ± 6.3% respectively). Furthermore, cell viability was assessed after the 48 hour exposure period using resazurin, a cell-permeant chromogenic dye that is reduced by metabolically active cells. This assay also indicated no significant cytotoxic effect, with viability values of 100.0 ± 3.9% for the control and 102.0 ± 2.3% for the extract-treated cells (Fig. 5d). For context, current guidelines for medical devices define the threshold for acceptable cytotoxicity at ≥70% viability. The positive control, as expected, resulted in drastically reduced values of 38.7 ± 1.8% for normalized proliferation and 11.8 ± 0.3% for viability. Collectively, these data from multiple complementary assays demonstrate the excellent biocompatibility of the microneedle device, supporting its potential for translational application.
2.5. Penetration capability in ex vivo human skin
We evaluated the penetration capability and depth of our microneedle sensor in live, ex vivo human skin via histological analysis. The measured insertion depth was 190 ± 50.9 μm, which is less than the total microneedle length of 1040 μm (Fig. 6a). Scanning electron microscopy (SEM) imaging of the microneedle post-insertion confirmed the structural integrity of the device, showing no evidence of bending or damage (Fig. 6b). Consequently, the observed difference between the needle length and penetration depth is attributed to the elastic deformation of the skin tissue during application, a phenomenon consistent with previous reports.31 Critically, and in accordance with our design objective, all insertion sites successfully reached the upper dermis (Fig. 6b).The achieved penetration depth, while sufficient to reach the target upper dermis, is more shallow than those reported for longer microneedles.30 Future in vivo characterization via techniques such as optical coherence tomography would be valuable to eliminate potential artifacts from tissue fixation and to refine the insertion protocol for optimal consistency.50 Notably, a controlled, shallow penetration depth can be considered a beneficial design feature. By consistently targeting the upper dermis while minimizing deeper tissue penetration, the device inherently reduces the risk of pain and bleeding associated with contact with deeper vascularized layers, thereby enhancing patient comfort and compliance.51
2.6. IFABP detection in spiked ex vivo human skin
We further validated the performance of our microneedle sensor in ex vivo human skin. To account for the expected decay of endogenous IFABP following biopsy collection, skin tissues were infused with a 10 ng mL−1 solution of the target analyte in the lower dermis and allowed to equilibrate overnight. The microneedle sensor was then applied using the standard protocol (40 minute insertion followed by ex situ assay completion within one hour). The response from four individual sensors was compared to control measurements obtained in the blank skin-like hydrogel (Fig. 6e). The average sensor response demonstrated a clear positive trend, with a higher signal in spiked biopsies compared to blank biopsies (−3.93 ± 1.68 μC mm−2vs. −0.46 ± 0.26 μC mm−2, respectively). The corresponding calculated IFABP concentration was 1.28 ± 0.20 ng mL−1, corresponding to a coefficient of variation of 15.62% (Fig. 6f). Overall, the sensor readout for ex vivo biopsies was lower compared to the skin-like hydrogel. We attribute this difference to a difference in insertion depth and to the different distribution of the target analyte within the sample. In particular, we hypothesized that analyte redistribution from the injection site (in the lower dermis) results in a gradient of concentration with lower target analyte concentration in the superficial dermis.To verify this hypothesis, we performed complementary blister fluid collection from the same spiked biopsies following microneedle sampling (Fig. S9). As blister formation draws interstitial fluid from deeper tissue layers, often evidenced by slight blood tinting,52,53 it was anticipated that IFABP levels would be higher in this fluid compared to the microneedle readout. This was confirmed experimentally: ELISA analysis of the blister fluid measured a significantly higher IFABP concentration of 3.13 ± 0.35 ng mL−1.
This result provides clear evidence that the microneedle samples ISF from the upper dermis which is reminiscent of glucose measurement from ISF within the dermis as opposed to capillary blood sampled by pricking.54 In such cases, the analyte is said to be measured within a compartment in dynamic equilibrium with the systemic circulation. This measurement modality has shown great clinical value in the management of diabetes through closed loop systems thereby demonstrating the potential of relative analyte measurement from minimally invasive devices.55 Similarly, the IFABP microneedle sensor may prove appropriate to resolve fluctuations of indicators of gut barrier integrity, regardless of whether the recorded sensor output is absolute or relative with respect to blood levels.
In summary, this mock sensing trial in ex vivo skin demonstrates that the microneedle sensor is capable of detecting minute levels of IFABP within the superficial dermis and motivates further testing in live animal models.
3. Conclusion
In this work, we have presented a novel microneedle-based biosensor for the rapid quantification of IFAP from skin interstitial fluid. Our sensor design is underpinned by a theoretical model of the underlying transport, affinity binding and catalytic processes, which enabled excellent analytical performance within a minimal form factor of 0.5 cm2 per device. While future investigation should focus on verifying these results in live animal models, the microneedle-based IFABP sensor has demonstrated exceptional biocompatibility and applicability to ex vivo human skin establishing a strong foundation for future in vivo testing and clinical translation.At a price point of 2.59$ per device (Table S4) including all device components and assay reagents, this minimalist IFABP sensor has great potential as a single-use device for longitudinal assessment of intestinal barrier permeability.56 This could in turn prove extremely useful in developing therapeutics targeting inflammatory-dependent disorders of the digestive system like the irritable bowel disease.57
Another strength of our sensor is its generalizability owing to the use of antibodies as affinity reagents. Unlike scarce catalytic and synthetic biorecognition elements,58 antibodies are available for virtually any previously studied protein biomarker. Coupled with the finite element model we developed to streamline sensor design and optimization, this generalizability creates a versatile diagnostic framework. The combination of low cost and modular design makes our device exceptionally attractive for the future development of multiplexed panels, enabling the simultaneous assessment of multiple biomarkers relevant to gut health.
While continuous monitoring is ideal for rapidly fluctuating metabolites like glucose, many biological processes, including intestinal barrier disruption and bacterial translocation, evolve over hours to days.23 For these applications, our discrete sampling methodology offers significant advantages: it minimizes device–skin contact time, reduces invasiveness, and enhances practicality for at-risk patients. The consistent, short penetration depth of our microneedle is designed to further promote patient comfort and compliance by minimizing pain and anxiety.
Although the current device is tailored for point-of-care testing, its minimal reagent handling requirements and compatibility with simple potentiostats position it ideally for decentralized diagnostics. A compelling future direction involves the full integration of reagent handling and readout into a single, automated device, such as a slip-chip.59–61 Such a platform would deliver the quantitative accuracy of our assay while retaining the speed and portability of lateral flow tests, ultimately bridging a critical gap in accessible quantitative diagnostics.
We envision that the longitudinal monitoring of intestinal permeability enabled by this technology will open new research directions for fundamental and clinical research. It offers a unique opportunity to investigate outstanding questions on the impact of circadian rhythms, infectious diseases, antibiotic courses, and other exposures on intestinal barrier function, paving the way for more personalized and proactive management of gastrointestinal health.
4. Experimental section/methods
Assembly of microneedle devices
Stainless-steel acupuncture needles (Xeno 0.30 mm × 30 mm, 304 stainless-steel, HEGU) were cleaned by successive sonication in deionized water (DIW) and absolute ethanol for 5 min each. After drying, single needles were pushed through polytetrafluoroethylene (PTFE)/silicone septa (27095-U, 8 mm × 1.524 mm diameter/thickness, Merck) and through 8 layers of parafilm (Parafilm® M, Merck) to leave the tip of the needle protruding by 1.04 mm on the silicone side. For electrical interfacing, the resulting microneedle was either connected with an alligator clamp or trimmed on the back PTFE side and a cable attached with a silver conductive epoxy (EPO-TEK® EJ2189, Epoxy Technology Inc.).The microneedles were electrodeposited with gold by galvanostatic deposition at room temperature (RT) from a solution of gold(III) cyanide salt (JE285 pe-gold bath, Jentner) stirred at 100 rpm by applying a current density of −1.86 mA mm−2 for 5 min to the microneedles and using a platinum anode (i.e. a 2-electrode cell with counter and reference electrodes shorted). Bubbles clinging to the surface of the microneedles were occasionally removed along the deposition process by gentle flicking. The gold coated microneedles were then cleaned electrochemically by cyclic voltammetry 10 times in an aqueous solution containing 50 mM of H2SO4 between −0.2 V and 1.4 V against a saturated KCl Ag/AgCl reference electrode. The electroactive surface area of each microneedle was estimated from the last cycle by integration of the cathodic peak corresponding to the reduction of gold oxide and dividing the obtained charge by 410 μC cm−2.62
Sensor functionalization
The functionalization of the biosensor was carried out on freshly cleaned gold coated microneedles in two main steps: i) dendrimer immobilization to increase the number of binding site and ii) antibody immobilization. Electrochemical characterization was carried out between each step as described below. First, the microneedles were incubated for 16 h at RT in 1 mM ethanolic solution of 3-mercaptopropionic acid (M5801, Merck) to obtain a self-assembled monolayer with carboxylic end groups. The carboxylic acid groups were then activated for 30 min in a solution of 0.2/0.05 M of EDC/sNHS (PG82079, PG82071, ThermoFisher) in 25 mM MES buffer at pH 5.5 (1.06126, Merck). After rinsing once with MES buffer, 25 μL of 3rd generation poly(amidoamine) (412422, Merck) at a concentration of 2 mg mL−1 in MES buffer was incubated on the microneedles for 3 hours at room temperature in the dark with gentle shaking to form amide bonds. The microneedles were rinsed once in 15 mM pH 7.4 phosphate buffer (PB: 71643 and P5655, Merck) and any remaining NHS-ester was then quenched by incubation in 0.5 M ethanolamine pH 8.5 (E6133, Merck) for 1 h followed by PB rinsing again. The dendrimer primary amines were then activated with an aqueous solution of 2.5% (w/w) glutaraldehyde (G5882, Merck) with 50 mM phosphate buffer at pH 7.4 for 1 h 30 at RT under gentle shaking in the dark. The microneedles were washed once with PB and then incubated with 25 μL of capture antibody at 5.4 μg mL−1 in 50 mM phosphate buffer at pH 7.4, 4 °C overnight (DY3078, R&D systems for IFABP sensors; DY201, R&D systems for IL1-b in the SI). The microneedles were washed with PB and unreacted sites were blocked with ethanolamine as described above. Non-specifically adsorbed antibodies were washed away with 0.05% Tween 20 in PBS and the surface was blocked for 1 h 30 min with 1% BSA in PBS (DY008, R&D systems). Finally, the devices were kept in assay buffer (0.05% Tween 20 and 1% BSA in PBS) until use.There are a few noteworthy points to ensure successful functionalization (EDC quality, ligand conjugation time, and buffer composition), which are highlighted in the SI (S2).
Electrochemical characterization and measurements
All measurements were made with an Interface 1010E potentiostat (Gamry). Unless explicitly stated, all electrochemical measurements were performed with a platinum wire counter electrode and a saturated KCl Ag/AgCl reference electrode. Electrochemical characterization was performed in an aqueous solution of 0.5 M KNO3, 15 mM phosphate buffer pH 7.4 and 1 mM equimolar ferro/ferricyanide (K3Fe(CN)6 (III), 702587, and K4Fe(CN)6 (II), P3289, Merck). In cyclic voltammetry, the potential was scanned from −0.2 V to 0.6 V and back at 100 mV s−1. In electrochemical impedance spectroscopy (EIS), VDC, the offset voltage, was set to the open circuit potential ∼+240 mV, VAC the probing voltage was 7.07 mVrms and the frequency range investigated was from 100 kHz down to 1 Hz. The resulting spectrum was fitted to a Randles cell, including the diffusional impedance, in series with the spreading resistance in order to report the charge transfer resistance evolution along functionalization steps.63To obtain the readout signal in sensing experiments, the microneedle was immersed in PBS (135 NaCl, 7 mM KCl, 10 mM phosphate pH 7.4). The amount of precipitated TMB was quantified from the reduction charge obtained by amperometry or cyclic voltammetry. In amperometry, the microneedle potential was stepped from open circuit potential (∼+300 mV) to 0 V for 60 s. In cyclic voltammetry, the potential was scanned from 0 to 0.6 V and back at 1 V s−1. In either case, the charge was calculated by integration of the cathodic current trace and is displayed in the results in μC normalized to the electrode surface area and time of enzymatic substrate incubation.
Protocol of the electrochemical immunoassay on the microneedle
Unless specified, the detection of IFABP with the microneedle was performed according to the following optimized protocol. After incubation in the relevant sample (see Finite element model of the microneedle electrochemical assay and experimental comparison, Sensor calibration in skin-like hydrogel medium and Sensing in ex vivo skin), the microneedle sensor was washed twice with assay buffer (0.05% Tween 20 and 1% BSA in PBS DY008, R&D systems) and a 25 μL droplet of biotinylated detection antibody (0.5 μg mL−1, DY3078, R&D systems) was incubated for 10 min on top of the microneedle under 200 rpm agitation on an orbital shaker (Dual-Action Shaker KL 2, Edmund Bühler). The microneedle was washed twice with assay buffer and incubated with 25 μL of labeling reagent (0.5 μg mL−1, poly-HRP-streptavidin, N200 ThermoFisher) for 5 min at 200 rpm and washed again. Finally, 60 μL of precipitating TMB solution was incubated on the microneedle (T9455, Merck) for 10 min at 200 rpm resulting in a blue-colored catalytic product adsorbing on the microneedle surface. The reaction was terminated by rinsing with DIW and the product was used to quantify the amount of IFABP (if any) in the original sample by electrochemical reduction as described above and comparing the total charge to a calibration curve.In other experiments, the raw charge was used for comparison to a finite element model (see Finite element model of the microneedle electrochemical assay and experimental comparison below). In these experiments, the concentration and incubation time of reagents were 50 ng mL−1 of detection antibody incubated for 2 h and 100 ng mL−1 of HRP-label incubated for 20 mn.
Measurement of the antibody affinity constant by indirect ELISA
The affinity constant of the antibodies used in our IFABP sensor was measured by indirect ELISA as reported previously.64 Unless specified otherwise, all reagents in these experiments were obtained from the same supplier (DY008, R&D systems), including the antibodies and antigen under investigation (DY3078, 2694-CL-100/CF, R&D systems). Briefly, the antibody under study was mixed with increasing concentration of the IFABP antigen in a 1% BSA solution in PBS and the resulting solution was incubated for 16 h at RT to allow equilibrium to be reached. The concentration of antibody was i) 0.2 nM for the capture antibody and ii) 0.5 nM for the detection antibody. The concentrations of antigen ranged from i) 2 to 22 nM and ii) 0.5 to 10 nM, respectively. At the same time, a 96-well plate was coated by 16 h incubation at RT of 100 μL from an IFABP solution in PBS at: i) 1 μg mL−1 for the capture antibody and ii) 0.022 μg mL−1 for the detection antibody. After washing the plate 3 times (0.05% Tween 20 in PBS) and blocking for 2 h with 1% BSA solution in PBS, 100 μL of each mix of antibody/antigen at equilibrium was transferred to the plate and incubated for 15 min before washing and incubating for 15 min with a labeling reagent. The labeling reagent was i) a polyclonal HRP-conjugated goat anti-mouse IgG antibody (A16084, ThermoFisher) diluted to 1.25 μg mL−1 and ii) poly-HRP-streptavidin at 0.1 μg mL−1 (N200, ThermoFisher) both in 1% BSA PBS. The plate was washed again three times and 100 μL of a peroxidase substrate solution was incubated in the wells for 5 min before being stopped with 50 μL of 2 N sulfuric acid solution for color development, and absorbance was measured at 450 nm (SpectraMax iD3, Molecular Devices). The concentrations of antibody and antigen and incubation in the plate were determined from preliminary experiments ensuring that the fraction of antibody eventually binding to the plate remained below 10% to avoid displacing the reaction equilibrium.In order to use the absorbance values to calculate the concentrations of antigen-bound and free antibody in the original mixes, a calibration of absorbance vs. free antibody was performed in parallel by directly incubating solutions of increasing antibody concentration in antigen-coated wells and labeling with HRP-conjugated reagents as described above. The raw results (Fig. S5a and b and d and e) show good linearity of the calibration and nearly null values of absorbances for the antibody mixed with the largest antigen concentration indicating full binding of the antibody (saturation). The bound and free fraction hence determined allowed the determination of the antibody affinity constant from a Scatchard plot (Fig. S5c and f).
Finite element model of the microneedle electrochemical assay and experimental comparison
We implemented the finite element model in the COMSOL Multiphysics (version 6.2) software as a 2D axisymmetric, time-dependent model representing half a cross section of the microneedle sensor covered by a hemispherical droplet of 25 μL (Fig. S4a). Mass transport within the liquid phase was defined to occur solely by diffusion while surface reactions were defined kinetically from the relevant reaction's rate constants (for governing equations see SI S1).The time-dependent model was solved sequentially for each step of the assay: target incubation, detection antibody, HRP-label and enzyme substrate. At the end of each step involving an affinity reaction, the concentration of bound species was used to define the surface concentration of binding sites available in the next step. For simplicity, we assumed the unbounding of the surface bound species during wash steps to be negligible and have hence not modeled the wash steps of the actual protocol. In the final catalytic step, the catalysis of TMB by the HRP enzyme was assumed to follow the Michaelis–Menten kinetics and the resulting amount of product was converted to reduction charge (i.e. the sensor output signal) through Faraday's law of electrolysis. This charge was normalized with respect to electrode area and TMB incubation time and added to a background value of 0.11 μC mm−2 mn−1 before being compared to experimental results. In vitro experimental results were obtained by running the assay according to the naïve (non-optimized) supplier recommendation as described above (Protocol of the electrochemical immunoassay on microneedle).
Sensor calibration in skin-like hydrogel medium
The skin-like hydrogels were prepared by mixing (in this order) 22.5 μL collagen type I (08-115, lot#4142896, 4.08 mg ml−1, Merck), 9 μL of 62 mM NaOH, 4.5 μL of concentrated artificial interstitial fluid and 13.5 μL of a solution of concentrated target (IFABP, DY3078, R&D systems) or control molecule (FABP5, OPPA01328, Aviva Systems Biology) where applicable. The mixing was performed on ice at 4 °C to prevent gelation and allow thorough mixing to ensure homogeneous analyte distribution. The pH of the mix was controlled with a micro-pH reader (PH-50, Unisense) to be within pH 7 to 7.4. The composition of the aISF (final concentration after mixing) was 2.5 mM CaCl2, 5.5 mM glucose, 10 mM Na2HPO4, 3.5 mM KCl, 0.7 mM MgSO4, 123 mM NaCl, 1.8 mM KH2PO4, 7.4 mM saccharose, and 188 μM of BSA and the pH was adjusted to pH 7.4.65,66 The mixture was set in a custom printed well and allowed to gel for 30 min at room temperature in a humidified container before being capped with a single layer of parafilm M and used immediately.The sensor calibration was performed with individual devices for each concentration, 4 sensors for each concentration, blanks and negative control hence 24 individual sensors for the results displayed in Fig. 4b for example. Here and throughout the text, error bars correspond to one time the standard deviation of the sample. Unless otherwise stated, the limit of detection (LOD) was calculated according to:
 | (1) |
Cytotoxicity assay
We performed a cytotoxicity assessment of our device according to existing guidelines (ISO 10993-5:2009 and 10993-12:2021). We used N/TERT 2G keratinocytes67 at passage 10 for all experiments. The keratinocytes were maintained in culture by regular passaging before reaching confluence. On the day prior to the cytotoxicity experiments, the keratinocytes were plated in a 96 well-plate and the devices under test were subjected to leachable extraction. First, sub-confluent keratinocytes were detached by trypsinization and re-suspended in fresh culture medium (EpiLife medium, MEPI500CA, supplemented with human keratinocyte growth supplement, S0015, 10% fetal bovine serum, A5256701, and 1% penicillin/streptomycin, 15140122, Gibco). 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 keratinocytes were seeded per well in a 96-well plate in 8 replicates per condition: blank, positive control and leachables from the device under test. The well-plate was placed in an incubator/imaging system (Incucyte, Sartorius) at 37 °C with 5% CO2. The confluence of each well was monitored thereon every second hour and the keratinocytes were allowed to grow for 24 h before the challenge.
000 keratinocytes were seeded per well in a 96-well plate in 8 replicates per condition: blank, positive control and leachables from the device under test. The well-plate was placed in an incubator/imaging system (Incucyte, Sartorius) at 37 °C with 5% CO2. The confluence of each well was monitored thereon every second hour and the keratinocytes were allowed to grow for 24 h before the challenge.
Meanwhile, single microneedle devices placed inside Eppendorf tubes and immersed in 816 μL of supplemented cell culture medium inside acting as an extraction vehicle (equivalent to 1.25 cm2 ml−1). The supplemented cell culture medium was used as a blank and 0.1 mg mL−1 sodium dodecyl sulfate in supplemented cell culture medium was used as a positive control of cytotoxicity. The leachable extraction was carried out for 24 h at 37 °C under agitation (100 rpm) for all three conditions.
24 h after plating, the sub-confluent keratinocytes (confluence ∼22%) were challenged with the leachable extract as described above. The challenge lasted 48 h; meanwhile the confluence was monitored and used to calculate the proliferation rate as % of confluence gained over 48 h. After 48 h, the keratinocyte viability was tested with resazurin (PrestoBlue, A13261, Invitrogen) according to the manufacturer protocol. Briefly, 10 μL of pre-warmed resazurin reagent was added to each well and the plate was incubated in the dark at 37 °C for 15 min. The plate was then immediately placed on ice and 560/590 nm fluorescence values for each well were acquired on a plate reader (SpectraMax iD3, Molecular Devices). The viability was calculated by normalizing the raw fluorescence readout to the average of the blank conditions.
Characterization of the microneedle insertion depth
Complete microneedle sensors were inserted in live ex vivo human skin obtained commercially (NativeSkin access 20 mm, Genoskin). The skin biopsy was obtained in accordance with the local French CPP ethical and CSP health regulations from a 37 year-old consenting female donor with a body mass index of 21. The biopsy was sampled from abdominal skin and maintained in a matrix allowing basal hydration and nutrient delivery until use the following day (less than 36 hours post-sampling). Upon receiving the biopsy, it was first kept for 1 h in a 37 °C incubator before performing insertion in a RT environment. During insertion, the microneedle device was pressed with mild pressure from the thumb and kept in place for 10 seconds before being removed. In another experiment (Sensing in ex vivo skin), the microneedles were kept in place for 40 min but no differences were found in the final insertion site morphology. The skin biopsy was then fixed for 12 h at 4 °C (formalin 10%, HT501128, Merck), dehydrated in ethanol series and embedded in paraffin before being sectioned on microtome (HM360, Rikard). Several 10 μm thick contiguous sections were obtained from each insertion site to identify the deepest penetration depth reached. After transferring to a microscope slide, the sections were stained with hematoxylin and eosin before imaging on an optical microscope. The depth of the insertion indent was measured with ImageJ.Sensing in ex vivo skin
An additional skin biopsy from the same donor (also from the abdominal region) was used for the ex vivo sensing trial (Fig. S9). After allowing the biopsy to equilibrate for 1 h at 37 °C, a solution of 10 ng mL−1 IFABP in aISF with a protease inhibitor (78425, ThermoFisher) was injected from the topical side in the lower dermis (260 μL cm−2) and the biopsy was allowed to equilibrate overnight at 37 °C. This spiked biopsy was then used for interrogation by the microneedle sensors and blister fluid collection. Microneedle sensors were applied as described above (insertion, 40 min incubation, ex situ immunoassay completion; one hour total) and the concentration of IFABP was calculated from the calibration curve obtained in skin-like hydrogel (Fig. 4).The blister fluid was obtained by applying incremental negative pressure: 200, 400 and 600 mmHg for 5 min each followed by 15 min at 600 mmHg (30 min total) using a 3D printed suction cup with 3 blister forming sites of 4 mm diameter each. Interstitial fluid was sampled from the resulting blisters using a hypodermic needle and diluted 13× for assaying with an IFABP ELISA kit according to the manufacturer's protocol (DY3078, R&D systems). Briefly, a 96-well plate was coated with 300 ng (3 μg mL−1) of capture antibody per well in PBS overnight at RT. After successive washing, blocking and washing steps (wash buffer: 0.05% Tween 20 in PBS and blocking buffer: 1% BSA in PBS, DY008, R&D systems), a serial dilution of standards was assayed in duplicates: starting with 2 hours of target incubation, washing, detection antibody incubation for 2 hours, washing, streptavidin-HRP incubation for 20 min, washing and substrate incubation (DY999, R&D systems) for 20 min before acid stop. The absorbance at 450 nm was read with a plate reader (SpectraMax iD3, Molecular Devices) and the raw values were subtracted for absorbance at 540 nm.
Scanning electron microscopy
Scanning electron microscope images of the microneedles at different stages of the sensor functionalization process were acquired on a Zeiss Ultra 55 with an electron extraction voltage of 3 kV using the secondary electron detector. The microneedles were imaged as is: without any conductive coating added.Statistics and results reporting
All values reported in this study correspond to average ± standard deviation. Statistics as annotated in the figures refer to independent-sample double-sided t-tests with an α = 0.05 confidence level. Where significant, the p-values are reported in the figure's legend.Author contributions
N. M., P. B. and O. P. conceived the idea and designed the research. P. B. and O. P. supervised the project. S. X. performed the histological processing of ex vivo human skin and N/TERT-2G cell culture. N. M. performed all other fabrication, experiment and analysis. N. M. and O. P. wrote the manuscript. All the authors discussed the results and commented on the original manuscript and revisions.Conflicts of interest
The authors declare no conflict of interest.Data availability
The data that supports the findings of this study are available from the corresponding author upon reasonable request.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5lc01004g.
Acknowledgements
The Karolinska Institute Research Incubator Fellowship program (2022-02536) & Karolinska Institutet (2-1534/2020) funded this work. The authors acknowledge the help of the EMil electron microscopy facility for their flawless support, Emma Mondoc from the Histocore facility for help on tissue sectioning, Po-Han Huang for early microneedle prototyping, Birce Akpinar for discussion on antibody conjugation, Begüm Kubra Tokyay for discussion on dendrimer chemistries, Matthew Hunt for help with Incucyte experiments and Morteza Hassanpouramiri for his help on printing/milling custom fixtures. The N/TERT-2G keratinocyte cell lines were generously supplied by Jos P. H. Smits and Ellen H. van den Bogaard (Department of Dermatology, Radboud Research Institute for Medical Innovation, Radboud University, Nijmegen, The Netherlands) on behalf of James Rheinwald (Department of Dermatology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA). O. P. and P. B. are thankful for financial support from the Karolinska Institute Research Incubator (KIRI) Fellowship program (2022-02536). O. P. gratefully acknowledges Karolinska Institutet (2-1534/2020) and Swedish Research Council to O. P. (2022-01436) for financial support.References
- J.-F. Bach and N. Engl, J. Med., 2002, 347, 911 Search PubMed.
- P. Brodin, Science, 2022, 376, 945 CrossRef PubMed.
- M. G. Laukoetter, P. Nava and A. Nusrat, World J. Gastroenterol., 2008, 14, 401 CrossRef.
- S. Y. Salim and J. D. Söderholm, Inflammatory Bowel Dis., 2011, 17, 362 CrossRef.
- C. Stolfi, C. Maresca, G. Monteleone and F. Laudisi, Biomedicines, 2022, 10, 289 CrossRef.
- A. Kovaleva, E. Poluektova, R. Maslennikov, A. Karchevskaya, O. Shifrin, A. Kiryukhin, A. Tertychnyy, L. Kovalev, M. Kovaleva, O. Lobanova, A. Kudryavtseva, G. Krasnov and V. Ivashkin, Dig. Dis. Sci., 2023, 68, 4166 CrossRef.
- I. Schoultz and Å. V. Keita, Cells, 2020, 9, 1909 CrossRef.
- J. S. Bajaj, P. B. Hylemon, J. M. Ridlon, D. M. Heuman, K. Daita, M. B. White, P. Monteith, N. A. Noble, M. Sikaroodi and P. M. Gillevet, Am. J. Physiol., 2012, 303, G675 Search PubMed.
- C. Nowicki, L. Ray, P. Engen, A. Madrigrano, T. Witt, T. Lad, M. Cobleigh and E. A. Mutlu, Front. Microbiol., 2023, 14 DOI:10.3389/fmicb.2023.1148097.
- J. Min, M. Nothing, B. Coble, H. Zheng, J. Park, H. Im, G. F. Weber, C. M. Castro, F. K. Swirski, R. Weissleder and H. Lee, ACS Nano, 2018, 12(4), 3378–3384 CrossRef.
- L. Syedmoradi, M. L. Norton and K. Omidfar, Talanta, 2021, 225, 122002 CrossRef.
- J. R. Choi, Front. Chem., 2020, 8 DOI:10.3389/fchem.2020.00517.
- D. C. Klonoff, D. Ahn and A. Drincic, Diabetes Res. Clin. Pract., 2017, 133, 178 CrossRef PubMed.
- O. Parlak, Sens. Actuator Rep., 2021, 3, 100036 Search PubMed.
- H. Chen, M. Xue, Z. Mei, S. Bambang Oetomo and W. Chen, Sensors, 2016, 16, 2134 CrossRef PubMed.
- R. F. Donnelly, T. Raghu Raj Singh and A. D. Woolfson, Drug Delivery, 2010, 17, 187 CrossRef CAS PubMed.
- M. Friedel, I. A. P. Thompson, G. Kasting, R. Polsky, D. Cunningham, H. T. Soh and J. Heikenfeld, Nat. Biomed. Eng., 2023, 1 CAS.
- B. U. W. Lei and T. W. Prow, Biomed. Microdevices, 2019, 21, 81 CrossRef.
- H.-R. Jeong, H.-S. Lee, I.-J. Choi and J.-H. Park, J. Drug Targeting, 2017, 25, 29 CrossRef CAS.
- A. C. Müller, F. P. Breitwieser, H. Fischer, C. Schuster, O. Brandt, J. Colinge, G. Superti-Furga, G. Stingl, A. Elbe-Bürger and K. L. Bennett, J. Proteome Res., 2012, 11, 3715 CrossRef.
- B. Q. Tran, P. R. Miller, R. M. Taylor, G. Boyd, P. M. Mach, C. N. Rosenzweig, J. T. Baca, R. Polsky and T. Glaros, J. Proteome Res., 2018, 17, 479 CrossRef CAS PubMed.
- P. Gallay, D. Heumann, D. Le Roy, C. Barras and M. P. Glauser, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 9935 CrossRef CAS.
- F. Laugerette, C. Vors, M. Alligier, G. Pineau, J. Drai, C. Knibbe, B. Morio, S. Lambert-Porcheron, M. Laville, H. Vidal and M.-C. Michalski, Nutrients, 2020, 12, 1820 CrossRef CAS.
- A. Wiercinska-Drapalo, J. Jaroszewicz, E. Siwak, J. Pogorzelska and D. Prokopowicz, Regul. Pept., 2008, 147, 25 CrossRef CAS.
- M. Logan, M. MacKinder, C. M. Clark, A. Kountouri, M. Jere, U. Z. Ijaz, R. Hansen, P. McGrogan, R. K. Russell and K. Gerasimidis, BMC Gastroenterol., 2022, 22, 260 CrossRef CAS.
- A. G. L. Bodelier, M. J. Pierik, K. Lenaerts, E. de Boer, S. W. Olde Damink, W. M. Hameeteman, A. A. M. Masclee and D. M. Jonkers, Eur. J. Gastroenterol. Hepatol., 2016, 28, 807 CrossRef CAS.
- T. Mogilevski, A. L. Nguyen, M. Ajamian, R. Smith, S. Rosella, M. P. Sparrow, G. T. Moore and P. R. Gibson, Eur. J. Gastroenterol. Hepatol., 2024, 36, 271 CrossRef CAS PubMed.
- G. N. Abdelrasoul, S. MacKay, S. Y. Salim, K. P. Ismond, M. Tamura, C. Khalifa, E. Mannan, D. Lin, T. Mandal, R. R. Montgomery, D. S. Wishart, J. Chen and R. G. Khadaroo, ACS Sens., 2018, 3, 2296 CrossRef CAS PubMed.
- E. Sánchez-Tirado, L. Agüí, A. González-Cortés, S. Campuzano, P. Yáñez-Sedeño and J. M. Pingarrón, Sensors, 2023, 23, 837 CrossRef.
- E. Larrañeta, J. Moore, E. M. Vicente-Pérez, P. González-Vázquez, R. Lutton, A. D. Woolfson and R. F. Donnelly, Int. J. Pharm., 2014, 472, 65 CrossRef.
- F. Ribet, A. Bendes, C. Fredolini, M. Dobielewski, M. Böttcher, O. Beck, J. M. Schwenk, G. Stemme and N. Roxhed, Adv. Healthcare Mater., 2023, 12, 2202564 CrossRef PubMed.
- J. Xu, B. Yang, J. Kong, Y. Zhang and X. Fang, Adv. Healthcare Mater., 2023, 12, 2203133 CrossRef.
- B. L. Garrote, A. Santos and P. R. Bueno, ACS Sens., 2019, 4, 2216 CrossRef PubMed.
- P. D. Josephy, Environ. Health Perspect., 1985, 64, 171 CrossRef PubMed.
- M. Liu, Y. Zhang, Y. Chen, Q. Xie and S. Yao, J. Electroanal. Chem., 2008, 622, 184 CrossRef.
- J. Sabaté del Río, O. Y. F. Henry, P. Jolly and D. E. Ingber, Nat. Nanotechnol., 2019, 14, 1143 CrossRef PubMed.
- D. G. Myszka, X. He, M. Dembo, T. A. Morton and B. Goldstein, Biophys. J., 1998, 75, 583 CrossRef PubMed.
- J. S. Daniels and N. Pourmand, Electroanalysis, 2007, 19, 1239 CrossRef.
- P. R. Miller, R. M. Taylor, B. Q. Tran, G. Boyd, T. Glaros, V. H. Chavez, R. Krishnakumar, A. Sinha, K. Poorey, K. P. Williams, S. S. Branda, J. T. Baca and R. Polsky, Commun. Biol., 2018, 1, 1 CrossRef.
- Z. Wang, J. Luan, A. Seth, L. Liu, M. You, P. Gupta, P. Rathi, Y. Wang, S. Cao, Q. Jiang, X. Zhang, R. Gupta, Q. Zhou, J. J. Morrissey, E. L. Scheller, J. S. Rudra and S. Singamaneni, Nat. Biomed. Eng., 2021, 5, 64 Search PubMed.
- G. R. Yang, W. Kim and J. H. Jung, Biosens. Bioelectron., 2024, 263, 116590 Search PubMed.
- K. Y. Goud, C. Moonla, R. K. Mishra, C. Yu, R. Narayan, I. Litvan and J. Wang, ACS Sens., 2019, 4, 2196 CrossRef PubMed.
- X. Huang, S. Zheng, B. Liang, M. He, F. Wu, J. Yang, H. Chen and X. Xie, Microsyst. Nanoeng., 2023, 9, 1 CrossRef PubMed.
- Q. K. Anjani, A. D. C. Nainggolan, H. Li, A. Miatmoko, E. Larrañeta and R. F. Donnelly, Int. J. Pharm., 2024, 655, 124071 CrossRef PubMed.
- S. Li, J. Dai, M. Zhu, N. Arroyo-Currás, H. Li, Y. Wang, Q. Wang, X. Lou, T. E. Kippin, S. Wang, K. W. Plaxco, H. Li and F. Xia, ACS Nano, 2023, 17, 18525 CrossRef PubMed.
- O. Gawlik-Kotelnicka, K. Czarnecka-Chrebelska, A. Margulska, E. Pikus, J. Wasiak, A. Skowrońska, E. Brzeziańska-Lasota and D. Strzelecki, Prog. Neuropsychopharmacol. Biol. Psychiatry, 2025, 136, 111170 CrossRef.
- B. D. Wilson and H. T. Soh, Trends Biochem. Sci., 2020, 45, 639 CrossRef PubMed.
- J. J. García-Guzmán, C. Pérez-Ràfols, M. Cuartero and G. A. Crespo, TrAC, Trends Anal. Chem., 2021, 135, 116148 CrossRef.
- “ISO 10993-5:2009”, can be found under https://www.iso.org/standard/36406.html.
- R. F. Donnelly, M. J. Garland, D. I. J. Morrow, K. Migalska, T. R. R. Singh, R. Majithiya and A. D. Woolfson, J. Controlled Release, 2010, 147, 333 CrossRef.
- H. S. Gill, D. D. Denson, B. A. Burris and M. R. Prausnitz, Clin. J. Pain, 2008, 24, 585 CrossRef.
- J. Kool, L. Reubsaet, F. Wesseldijk, R. T. Maravilha, M. W. Pinkse, C. S. D'Santos, J. J. van Hilten, F. J. Zijlstra and A. J. R. Heck, Proteomics, 2007, 7, 3638 CrossRef.
- E. A. MacDonald, E. L. Katz, T. F. Pearson and J. E. Harris, Curr. Protoc., 2024, 4, e1073 CrossRef PubMed.
- E. Cengiz and W. V. Tamborlane, Diabetes Technol. Ther., 2009, 11, S11–S16 CrossRef PubMed.
- J. Zhang, J. Xu, J. Lim, J. K. Nolan, H. Lee and C. H. Lee, Adv. Healthcare Mater., 2021, 10, 2100194 CrossRef CAS PubMed.
- I. Schoultz and Å. V. Keita, Cells, 2020, 9, 1909 CrossRef CAS PubMed.
- M. Iacucci, G. Santacroce, S. Majumder, J. Morael, I. Zammarchi, Y. Maeda, D. Ryan, A. D. Sabatino, M. Rescigno, M. R. Aburto, J. F. Cryan and S. Ghosh, Gut, 2024, 73, 1749 CrossRef CAS PubMed.
- N. Arroyo-Currás, P. Dauphin-Ducharme, K. Scida and J. L. Chávez, Anal. Methods, 2020, 12, 1288 RSC.
- X. Li, K. Scida and R. M. Crooks, Anal. Chem., 2015, 87, 9009 CrossRef CAS PubMed.
- I. Banerjee, T. Salih, H. Ramachandraiah, J. Erlandsson, T. Pettersson, A. C. Araújo, M. Karlsson and A. Russom, RSC Adv., 2017, 7, 35048 RSC.
- J. Zhang, Z. Dong, L. Xu, X. Han, Z. Sheng, W. Chen, J. Zheng, D. Lai and F. Shen, Adv. Sci., 2024, 2406367 CrossRef CAS PubMed.
- S. Trasatti and O. A. Petrii, J. Electroanal. Chem., 1992, 327, 353 CrossRef CAS.
- W. Franks, I. Schenker, P. Schmutz and A. Hierlemann, IEEE Trans. Biomed. Eng., 2005, 52, 1295 CrossRef PubMed.
- B. Friguet, A. F. Chaffotte, L. Djavadi-Ohaniance and M. E. Goldberg, J. Immunol. Methods, 1985, 77, 305 CrossRef CAS.
- N. Fogh-Andersen, B. M. Altura, B. T. Altura and O. Siggaard-Andersen, Clin. Chem., 1995, 41, 1522 CrossRef CAS.
- P. Bollella, S. Sharma, A. E. G. Cass and R. Antiochia, Biosens. Bioelectron., 2019, 123, 152 CrossRef CAS.
- M. A. Dickson, W. C. Hahn, Y. Ino, V. Ronfard, J. Y. Wu, R. A. Weinberg, D. N. Louis, F. P. Li and J. G. Rheinwald, Mol. Cell. Biol., 2000, 20, 1436 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |