Open Access Article
Open Access ArticleLFC-plus: simultaneous multicolour volume cytometry for high-throughput single-cell analysis
Zhi Lingabcd,
Wenhao Liuab,
Kyungduck Yoon abcd,
Zijun Gao
abcd,
Zijun Gao abce,
Keyi Han
abce,
Keyi Han ab,
Mithila Sawantfg,
Aparna Kesarwalafg and
Shu Jia
ab,
Mithila Sawantfg,
Aparna Kesarwalafg and
Shu Jia *abcg
*abcg
aLaboratory for Systems Biophotonics, Georgia Institute of Technology, Atlanta, GA, USA. E-mail: shu.jia@gatech.edu
bWallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, USA
cParker H. Petit Institute for Bioengineering and Biosciences, Georgia Institute of Technology, Atlanta, GA, USA
dGeorge W. Woodruff School of Mechanical Engineering, Georgia Institute of Technology, Atlanta, GA, USA
eSchool of Materials Science and Engineering, Georgia Institute of Technology, Atlanta, GA, USA
fDepartment of Radiation Oncology, Emory University School of Medicine, Atlanta, GA, USA
gWinship Cancer Institute, Emory University, Atlanta, GA, USA
First published on 16th December 2025
Abstract
Imaging flow cytometry demands a careful balance between spatial resolution, spectral multiplexing, throughput, and system complexity. Here, we present LFC-plus, a next-generation light-field cytometry platform that enables multiparametric, simultaneous multi-color, and volumetric single-cell analysis. The system integrates model-based image restoration, custom-designed light-field optics, and spectral aperture partitioning, achieving subcellular resolution in all three dimensions, a near-millimeter-scale imaging cross-section, and an analytical imaging throughput of nearly 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per second. We validate its performance across diverse biological applications, including chemotherapy response profiling, PEG-mediated cell fusion, and stiffness-based flow migration. These results establish LFC-plus as a robust and scalable platform for high-content volumetric cytometry, with broad implications spanning fundamental biology and translational diagnostics.
000 cells per second. We validate its performance across diverse biological applications, including chemotherapy response profiling, PEG-mediated cell fusion, and stiffness-based flow migration. These results establish LFC-plus as a robust and scalable platform for high-content volumetric cytometry, with broad implications spanning fundamental biology and translational diagnostics.
Introduction
Flow cytometry has become an essential tool in biomedical research, enabling high-throughput, high-sensitivity single-cell analysis to characterize and classify heterogeneous cell populations based on their physical and biochemical properties. However, conventional flow cytometry provides no spatial context, lacking the ability to resolve subcellular structures or the intracellular localization of molecular markers. Imaging flow cytometry (IFC) bridges this gap by integrating fluorescence microscopy with flow-based detection, yielding high-resolution images alongside quantitative single-cell measurements.1–3 This combination facilitates cell analysis not only based on antigen expression or fluorescence intensity but also on cellular morphology and spatial marker distribution, enabling extensive research and clinical settings, including cancer biology, immunology, and hematology.4Despite significant promises, a major challenge in IFC lies in achieving high throughput while preserving detection sensitivity and resolution. To accomplish this, existing IFC systems employ strategies, such as high-speed sensors, stroboscopic illumination, and hydrodynamic focusing, to reduce motion blur, maintain cell stream, and ensure image quality.5,6 Meanwhile, single-pixel imaging methods leverage the temporal response of fast photodetectors to reconstruct images from time- or frequency-encoded signals.7–10 For instance, techniques based on optofluidic time-stretch and modulated excitation beams with radiofrequency-tagged emission, or phase-resolved detection combined with corresponding demultiplexing and demodulating strategies, have achieved ultrafast acquisition by temporally encoding spatial information.9–13 Nonetheless, conventional IFC systems remain primarily two-dimensional, lacking depth perception and accurate 3D location, which can lead to signal occlusion and projection artifacts.14
To capture 3D information, IFC systems have adopted advanced microscopy techniques, including light-sheet microscopy,15–20 confocal microscopy,21 beam-shaping illumination,22 and optical tomography.23,24 While these methods offer high-resolution 3D imaging, they typically depend on sequential acquisition through mechanical scanning or multiple exposures, which can constrain temporal resolution. Moreover, their optical complexity, large footprint, and incompatibility with standard epi-fluorescence microscopes pose challenges for scalability and integration into high-throughput workflows.
Light-field cytometry (LFC), in contrast, offers a compelling alternative by capturing both spatial and angular information in a single exposure,25 achieving volumetric, single-shot imaging with a theoretical throughput of over 5000 cells per second under sufficient acquisition speed with near-diffraction-limited resolution (400–600 nm) in all three dimensions. Despite its enhanced 3D resolution and cell throughput, the original LFC implementation possesses a shallow imaging volume (<10 μm focal range), insufficient for multicellular depth, and relies on spectral sequences within each frame for multicolor acquisition. These limitations necessitate stringent control over flow rates and intercellular spacing, thus restricting experimental scalability and hindering broader biological applicability that demands multiplexed fluorescence detection and high-throughput population analysis.
Here, we present LFC-plus, a next-generation cytometric imaging platform that enables simultaneous multicolor acquisition of rapidly flowing cell populations with enhanced volumetry, subcellular resolution, and throughput. In particular, LFC-plus integrates a standard epi-fluorescence microscope with three key system-level configurations. First, LFC-plus leverages a model-based image enhancement scheme to extract biological features from comprehensive noise sources, preserving image quality and analysis, especially during rapid acquisition and under short stroboscopic illumination. Second, LFC-plus implements fully custom-designed light-field optics, enabling an extended imaging flux of up to millimeter scale and spatially resolved 3D visualization of multicellular dynamics and microenvironments. Third, LFC-plus exploits aperture partitioning that allows for instantaneous capture of spectrally distinct 3D volumes without temporal multiplexing or filter switching. We validate the LFC-plus platform across a range of biological assays, including chemotherapeutic profiling, PEG-mediated cell fusion, and stiffness-based spatial sorting in microfluidic channels. These results position LFC-plus as a robust and scalable solution for high-content volume cytometry, thereby broadening the landscape of IFC for biomedical research, translational diagnostics, and therapeutic discovery.
Results
LFC-plus: principle and characterization
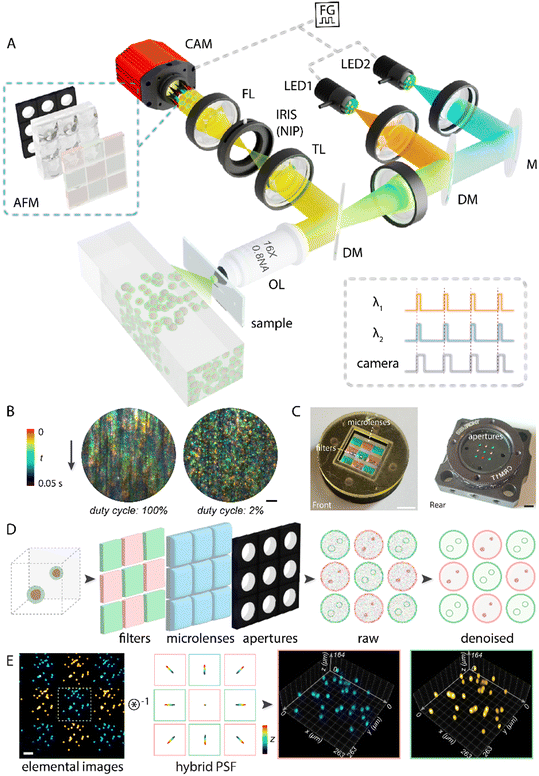
Conventional 3D IFC techniques have been explored to circumvent their inherent trade-offs among throughput, resolution, and multiparametric acquisition that often compromise the field of view (FOV) and imaging speed (Table S1). In this work, LFC-plus realizes simultaneous multicolor capture of volumetric cytometry data, leveraging three key advancements (Materials and methods): (i) a custom-built Fourier light-field microscope26,27 integrated with a microfluidic module (Fig. 1A and S1), which provides a large FOV and extended depth with subcellular resolution for high-throughput single-cell imaging and analysis (Table S2); (ii) stroboscopic, noise-aware acquisition (Fig. 1B) that reduces motion blur and employs physics-informed sparse-filtering to suppress camera-induced noise while preserving delicate structural details;28 and (iii) an integrated array of customized apertures, microfilters, microlenses (Fig. 1C) that partition the pupil plane into segments that distinguish and transport distinct spatial, angular, spectral components into elemental images (Fig. 1D). Unlike previously reported FLFM configurations, where the microlens array collects information from nine different angular perspectives of a single fluorescence channel, our system incorporates two types of emission filters into a microfilter array with distinct bandwidths—ET520/40 m (transmitting fluorescence from 500 to 540 nm) and ET632/60 m (transmitting fluorescence from 600 to 660 nm). The filters are arranged with four along the sides and five at the center and corners (Fig. 1D). This configuration unmixes the overlapping fluorescence at the native image plane into separate spectral components, enabling simultaneous acquisition of multicolor fluorescence signals across different elemental images under dual-color illumination. This implementation underwent wave-optics-based 3D deconvolution with a hybrid point-spread function,26 facilitating accurate volumetric reconstruction of multicolor objects in parallel, calibrated for system deviations while minimizing computational artifacts throughout the entire imaging depth (Fig. 1E and Texts S1 and S2).To characterize the LFC-plus system, we conducted numerical simulations and imaged both phantom and biological samples. In particular, we verified simultaneous subcellular acquisition at 100 frames per second (fps) with a 3D resolution of approximately 4 μm and 8 μm in the lateral and axial dimensions, respectively (Fig. S2–S5), transverse cross-sectional plane of 980 μm × 400 μm, and a flow rate up to approximately 36 mm s−1 under 100 μs illumination, corresponding to an analytical flow-rate estimated cell throughput of nearly 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per s, assuming the sensor is sufficiently fast to capture all passing events without loss, and the theoretical imaging-based throughput of 200
000 cells per s, assuming the sensor is sufficiently fast to capture all passing events without loss, and the theoretical imaging-based throughput of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per s (Fig. S6–S8 and Text S1). These results were obtained without alternating illumination or channel registration, eliminating any speed loss and complex postprocessing, which readies the LFC-plus platform for rapid, multiparametric cytometric analysis.
000 cells per s (Fig. S6–S8 and Text S1). These results were obtained without alternating illumination or channel registration, eliminating any speed loss and complex postprocessing, which readies the LFC-plus platform for rapid, multiparametric cytometric analysis.
Volumetric dual-colour imaging and classification of heterogeneous single-cell populations
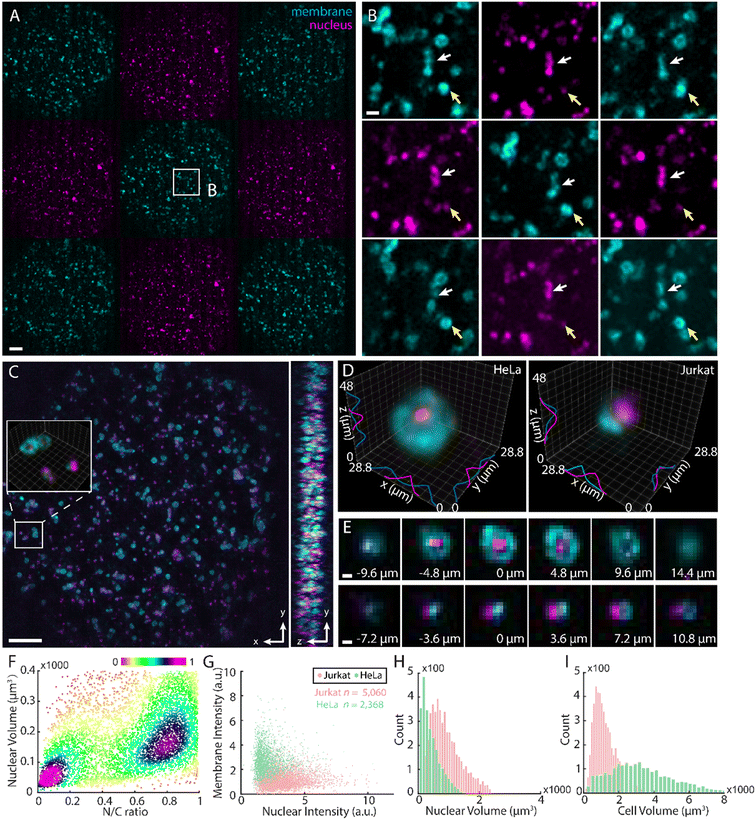
Multicolour information in IFC has provided critical cues for classifying heterogeneous cell populations by linking cellular composition, structure, and morphology to cellular physiology.29 However, compromises in system throughput, resolution, and complexity hamper the accessibility of multicolour 3D-IFC toward broader single-cell investigations. To demonstrate the capabilities of LFC-plus, we first analysed a mixture of HeLa and Jurkat cells, resolving their morphological and fluorescent characteristics at the single-cell level. Both cell types were stained with spectrally distinct fluorophores targeting the nuclei and membranes (Materials and methods). The cell suspension was introduced into the microfluidic system at a flow rate of approximately 80 μL min−1, yielding a moderate throughput of approximately 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per second (Fig. 2A and Movie S1). The images were acquired at 100 fps using stroboscopic illumination (2% duty cycle, 200 μs exposure) to mitigate motion blur.
000 cells per second (Fig. 2A and Movie S1). The images were acquired at 100 fps using stroboscopic illumination (2% duty cycle, 200 μs exposure) to mitigate motion blur.
Meanwhile, given the reduced illumination time, selective noise removal became essential and markedly enhanced the visibility of cellular signals (Fig. 2B). As seen, the reconstructed volume delineated the 3D spatial distribution of both cell types across a 100–200 μm depth range, enabling clear differentiation of their 3D morphological heterogeneity (Fig. 2C and D). Specifically, LFC-plus allowed for 3D measurement of nuclear and cellular volumes of 912.4 μm3 and 1223.9 μm3 for Jurkat cells, and 424.6 μm3 and 3605.7 μm3 for HeLa cells, respectively. The high resolution and 3D capabilities resulted in the distinction of nucleus-to-cell ratios of 0.75 for Jurkat cells and 0.15 for HeLa cells, which facilitated effective cell population classification (Fig. 2E and F) and closely aligned with the known physical profiles of the samples.30,31
Analyzing cell-cycle alterations in chemotherapy-treated lung adenocarcinoma cells
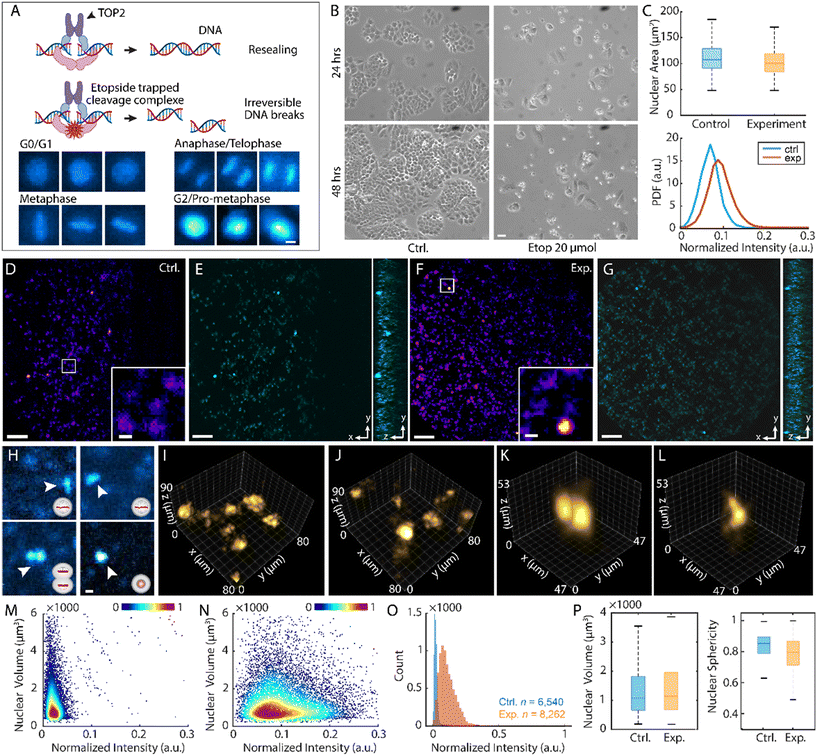
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer diagnoses worldwide.32 Etoposide, a widely used chemotherapeutic agent, functions as a topoisomerase II inhibitor by stabilizing the transient DNA cleavage complex formed during DNA unwinding.33 This inhibition prevents re-ligation of DNA strands, leading to the accumulation of double-strand breaks, which, in turn, trigger apoptosis in rapidly proliferating cancer cells (Fig. 3A and B).34,35 Despite its clinical efficacy, the 3D morphological and biochemical changes associated with etoposide-induced cytotoxicity remain poorly characterized at the single-cell level.Here, we leveraged the high-throughput volumetric capabilities of LFC-plus to extract multiplexed morphological and biochemical features from H460 human NSCLC cells following etoposide treatment. In practice, cells were fixed and stained with propidium iodide (PI) to label the nuclear DNA (Materials and methods). Wide-field images of the floating cells revealed intensified single-cell fluorescent signals post-treatment as determined by the 2D analysis (Fig. 3C), which indicates a reduction in the G1-phase population and an accumulation of cells arrested in the G2/M phase of the cell cycle (Fig. S9).36 With LFC-plus, 3D nuclear morphological changes can be observed and analyzed across a substantially increased population (Fig. 3D–G and Movies S2 and S3). The results enabled further distinction of the G2/M phase into pro-metaphase, metaphase, anaphase, and telophase, details of which were obscured in 2D analysis because chromosome separation may not be clearly visible depending on cellular orientation (Fig. 3H).
Meanwhile, intensity variations indicated the change of DNA content during replication (Fig. 3I and J). Quantitative analysis showed enhanced nuclear fluorescence post-treatment, thus implying elevated single-cell DNA content37 (Fig. 3K–M) and reflecting cell cycle progression toward the mitotic phase during which DNA replication has occurred. Nuclear volumes for both groups remain similar, displaying a medium volume of 1143 μm3 for the control group and 1188 μm3 for the experimental group, given the occurrence of apoptotic shrinkage and the appearance of dividing cells. It was also noted that nuclear sphericity was disturbed, aligning with the elongated morphological characteristic of cells arrested in the G2/M phase (Fig. 3N). These observations agreed with the heterogeneous response in biomass and area to drug exposure suggested by previous studies.38 The results underscore the capability of LFC-plus to capture subtle yet biologically significant 3D phenotypic and molecular alterations at subcellular resolution, enabling precise characterization of drug-induced cellular responses in complex populations.
3D analysis of PEG-mediated fusion in Jurkat cells
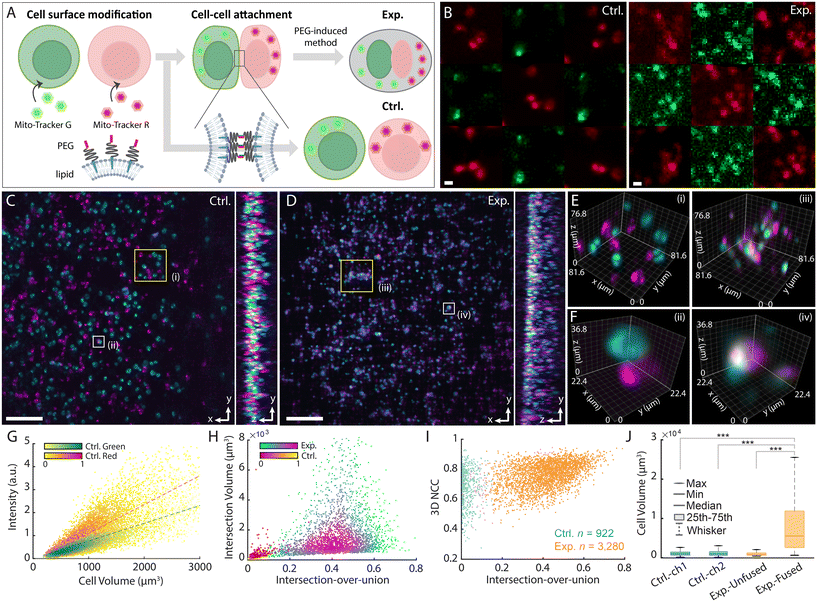
Cell fusion has become an instrumental biotechnology for understanding cell behavior, generating cell lines with desired properties, and developing therapies.39–42 Polyethylene glycol (PEG) is commonly used to induce and control cell fusion in vitro due to its low cytotoxicity, cost-effectiveness, and ease of application.43 As a dehydrating agent, PEG promotes membrane juxtaposition, enhancing lipid bilayer interactions that facilitate hybrid cell formation. Traditional PEG-based fusion assays rely on 2D fluorescence microscopy to quantify fusion efficiency by assessing the co-localization of fluorescent markers in labeled cell populations.44,45Here, we utilized LFC-plus to visualize and quantify the process of cell fusion in Jurkat cells. Jurkat cells were first labelled with spectrally distinct green and red mitochondrial fluorophores and then prepared as untreated mixed controls and PEG-treated fusion groups (Fig. 4A, Materials and methods, Movies S4 and S5). Raw elemental light-field images showed no fused cells or fluorophore transfer in the control group, implying the absence of spontaneous fusion or spectral crosstalk between channels (Fig. 4B). In contrast, PEG-treated cells exhibited dual fluorescence signals within the same cellular structures, which indicated increased proximity and mixing of fluorescent signals indicative of fusion activity (Fig. 4B).
The multicolour platform enabled simultaneous acquisition of the two spectral populations at a flow rate of 15 μL min−1, enabling high morphological alignment and fidelity (Fig. 4C and D). Meanwhile, LFC-plus was capable of observing 3D mitochondrial distributions within individual cells in the control group and cellular clusters containing both labels in the fusion group (Fig. 4E and S10). Notably, LFC-plus allowed for the differentiation of non-associated cells that appeared aligned laterally but separable in depth, preventing their misclassification as fused cells in conventional 2D projections (Fig. 4F). As seen, cellular volumes and fluorescence intensities remain closely relevant and consistent from both channels in the control group (Fig. 4G). Fusion efficiency was quantified by calculating the volumetric intersection between two fluorescent channels relative to the total volume of identified cell clusters. As seen, the controls exhibited an averaged fused cell ratio at 3.7%, while the PEG-treated group showed a significantly elevated ratio of 40.2 ± 1.1% (Fig. 4H). Correspondingly, the experimental group showed higher 3D normalized cross-correlation (NCC) values between the overlapping regions of both channels (0.76 ± 0.08) compared to the control group (0.68 ± 0.12), suggesting a greater extent of true cellular fusion rather than mere structural proximity or clustering (Fig. 4I). Moreover, the cell volume increased markedly after fusion by more than 3 times, from an average of 1123.5 μm3 and 1172.4 μm3 in the green- and red-fluorescence control groups, respectively, to 3873 μm3 in the fusion group (p < 0.001), indicating close membrane contact and cytoplasmic integration with PEG-mediated treatment (Fig. 4J). These results highlighted the capability of LFC-plus to capture and quantify complex cell–cell interactions with volumetric and populational precision, offering a powerful tool for investigating dynamic cellular processes relevant to biomedical research and therapeutic development.
Capturing stiffness-dependent cell flow dynamics in curved channels
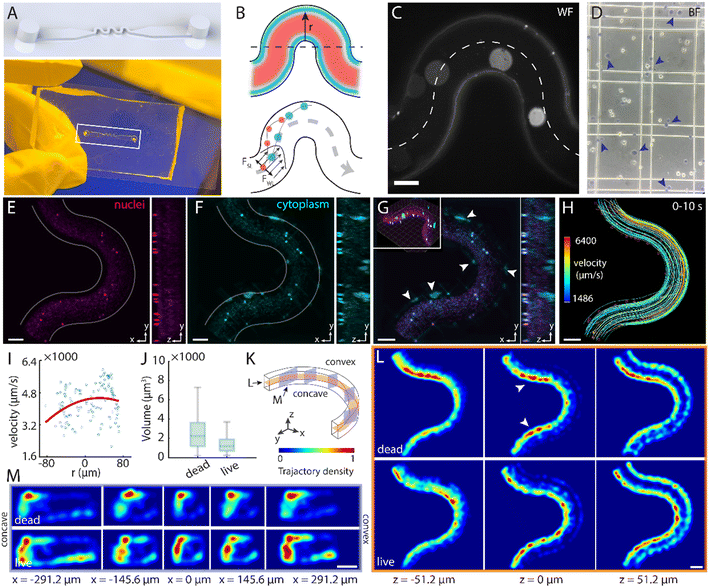
Understanding stiffness-dependent cell distribution in flow is critical for both mechanobiology and clinical diagnostics, as cellular stiffness is closely linked to pathologies such as atherosclerosis, stroke, and cancer progression.46 Curvilinear microfluidic channels have been widely adopted to model vascular geometries and study vasculopathy,47 as well as to enable high-throughput cell sorting via inertial migration and secondary Dean flows.48,49 However, most experimental observations of secondary flow dynamics remain restricted to 2D cross-sections obtained through microscopy50 or simulations.51 Moreover, the influence of cellular mechanical heterogeneity on 3D secondary flow behavior remains underexplored, mainly due to the lack of depth perception of conventional cytometric platforms.Here, we designed and fabricated an S-shaped microfluidic channel with a square cross-section of 200 μm × 200 μm, and a bend length of 800 μm per half-period (Fig. 5A). Numerical simulations predicted that rigid cells experience stronger shear-gradient lift forces and migrate toward the periphery, whereas deformable cells remain nearer the centreline (Fig. 5B).47 To model this stiffness contrast, live (compliant) and heat-treated (stiff) HeLa cells were differentially labelled, mixed, and introduced at a constant pressure of 20 mbar with a flow rate of 15 μL min−1 (Fig. 5C and D and Movie S6).52 For visualization, time-lapse 3D reconstructions averaged 100 time points over 1 second revealed pronounced peripheral accumulation of stiff, dead cells, while live cells preserved central streamlines and showed minimal corner-induced aggregation (Fig. 5E–G). Owing to centrifugal forces, the bulk velocity profile was observed to shift toward the outer wall, producing a slightly higher peak speed with respect to the inner wall (Fig. 5H and I and S11). Furthermore, dead cells displayed enlarged nuclear volumes, consistent with swelling typically associated with cell death (Fig. 5J). This behaviour was noticed in contrast to the more uniform distribution of live cells, as revealed in the axial trajectories (Fig. 5K and L). Longitudinal density plots accumulated over 1000 frames (10 s) exhibited patterns consistent with classic Dean vortices, despite a relatively low Reynolds number of 1.25 and Dean number of 1.77 (Text S3), evidenced by two lateral density peaks corresponding to counter-rotating secondary flows and shifts in trajectory peak positions as the channel curvature evolves (Fig. 5M). Notably, dead stiff cells remained confined to one side with limited lateral displacement, whereas live cells traversed the vortex field, populating a wider cross-sectional distribution. These observations underscore the potential extension of LFC-plus for elucidating how cell mechanics and secondary flows jointly govern cellular transport, with implications for vascular pathology and mechanobiology.53,54
Conclusions
In summary, the LFC-plus platform offers a promising solution to the long-standing trade-offs between throughput, volumetric resolution, and spectral multiplexing that have impeded the adoption of IFC. LFC-plus enables high-throughput, multiparametric single-cell analysis with volumetric capability and subcellular resolution via standard epi-fluorescence settings. The results validated LFC-plus in a diverse range of biological and clinically relevant contexts with rich content and high statistical power. LFC-plus can be further enhanced with novel probes,55–57 optical and instrumental configurations,58–61 super-resolution imaging,62–64 and computational frameworks.65–69 The epi-fluorescence platform adopted by LFC-plus offers feasible integration to support broad biomedical discoveries with scalable model organisms,70 high-throughput screening,71 3D cell culture, organ-on-a-chip,72 and spatial-resolved transcriptomics.73,74 We anticipate that LFC-plus will serve as a powerful paradigm for elucidating the fundamental and translational biological systems beyond current optofluidics limits.Material and methods
LFC-plus setup
The Fourier light-field microscopy system (Fig. S1 and Table S1) was equipped with a water-dipping objective lens (CFI75 LWD 16×/0.8NA W, Nikon) for a long working distance and a large imaging FOV.26 Samples were epi-illuminated by two single-color cold-visible mounted light-emitting diodes (LEDs), peaked at 470 nm (M470L5, Thorlabs) and 590 nm (M590L4, Thorlabs), simultaneously. Two achromatic doublets (AC254-200-A-ML and AC254-400-A-ML, Thorlabs) were inserted between the objective lens and the LED to form Kohler illumination. The fluorescence emitted from the sample was collected by the objective lens and was filtered by the dichroic mirror (ZT 405/488/594rpc-UF2, 50.8 mm × 72 mm × 2 mm, Chroma) and a corresponding emission filter (ZET 488/594 m, 50 mm diam, Chroma), then divided by a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 plate non-polarized beam splitter (#35-947, Edmund optics, diced into 72.5 mm × 50 mm × 1 mm) into two paths: a wide-field path focused by a tube lens (TTL200-A, Thorlabs; fFL = 200 mm) and recorded by an sCMOS camera (Zyla 4.2, Andor; pixel size, 6.5 μm), and a light-field path focused by a 300 mm tube lens (AC508-300-A-ML, Thorlabs) on the native image plane (NIP), then Fourier transformed with a 200 mm Fourier lens (AC508-200-A-ML, Thorlabs) and focused by a customized microlens array (fMLA = 30 mm, pitch size = 3.3 mm, diced from planoconvex lens of 20 mm diameter round by Edmund optics). The signals were filtered by corresponding customized micro filters (3.3 mm × 3.3 mm × 1 mm, ET520/40 m for GFP channel, ET632/60 m for mCherry, Chroma) to form an array of two-color images, which were captured on a second sCMOS camera (ORCA-Flash-4.0 v3, Hamamatsu, pixel size, 6.5 μm) at a full-sensor frame rate of 100 Hz.
50 plate non-polarized beam splitter (#35-947, Edmund optics, diced into 72.5 mm × 50 mm × 1 mm) into two paths: a wide-field path focused by a tube lens (TTL200-A, Thorlabs; fFL = 200 mm) and recorded by an sCMOS camera (Zyla 4.2, Andor; pixel size, 6.5 μm), and a light-field path focused by a 300 mm tube lens (AC508-300-A-ML, Thorlabs) on the native image plane (NIP), then Fourier transformed with a 200 mm Fourier lens (AC508-200-A-ML, Thorlabs) and focused by a customized microlens array (fMLA = 30 mm, pitch size = 3.3 mm, diced from planoconvex lens of 20 mm diameter round by Edmund optics). The signals were filtered by corresponding customized micro filters (3.3 mm × 3.3 mm × 1 mm, ET520/40 m for GFP channel, ET632/60 m for mCherry, Chroma) to form an array of two-color images, which were captured on a second sCMOS camera (ORCA-Flash-4.0 v3, Hamamatsu, pixel size, 6.5 μm) at a full-sensor frame rate of 100 Hz.
Fabrication of customized microfluidic chips
Customized microfluidic chips, including a straight channel (cross-sectional area of 1000 μm × 350 μm) and an S-shaped curved channel, were fabricated. First, the 3D mold was designed in SolidWorks and printed using the BMF microArch S140 3D Microfabrication System in Georgia Tech Biocleanroom. The mold was coated with trichlorosilane (448931, Sigma-Aldrich) via overnight vacuum evaporation. A PDMS base and curing agent mixture (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, Silicone elastomer kit, Sylgard™ 184, 24236-10, VWR) was then prepared and poured onto the coated mold, placed on aluminum foil. Then, a vacuum desiccator (Sigma-Aldrich, Z119008, Scienceware®) was used to remove the bubbles from the PDMS mixture, which was then cured on a hot plate at 80–90 °C for 2–4 hours. The PDMS chip was peeled off the mold after cooling. Holes were punched into the PDMS chip, and the debris was cleaned with an air compressor. Finally, oxygen plasma treatment was applied to activate the surfaces, enabling strong, permanent bonding between the PDMS and the coverslip.
1 ratio, Silicone elastomer kit, Sylgard™ 184, 24236-10, VWR) was then prepared and poured onto the coated mold, placed on aluminum foil. Then, a vacuum desiccator (Sigma-Aldrich, Z119008, Scienceware®) was used to remove the bubbles from the PDMS mixture, which was then cured on a hot plate at 80–90 °C for 2–4 hours. The PDMS chip was peeled off the mold after cooling. Holes were punched into the PDMS chip, and the debris was cleaned with an air compressor. Finally, oxygen plasma treatment was applied to activate the surfaces, enabling strong, permanent bonding between the PDMS and the coverslip.
Flow cytometer and microfluidic preparation
The microfluidic setup (Fig. S1) consists of a single-channel microfluidic flow controller (OB1, MK3+, Elveflow), a microfluidic flow sensor (MFS4, Elveflow), microfluidic chips (10000831, ChipShop), microfluidic reservoirs (LVF-KPT-M-2, Darwin Microfluidics), a syringe (BD-PLSTPK-LL-01, Darwin Microfluidics), and a waste tank. Before the experiments, the reservoirs were filled with deionized (DI) water. During the experiments, Valve 1 (pump side) was opened to flush the chip with DI water, cleaning the channels before measurements. The valve was then closed, and Valve 2 (syringe side) was opened to inject samples into the tubing and microfluidic chips using a syringe. Injection was stopped once the cells were visible within the FOV of the microscope. Finally, Valve 2 was closed, and Valve 1 was reopened, allowing the sample to be pushed with DI water at the rate controlled by the microfluidics controller and the flow sensor.Cytometric imaging acquisition
Time-lapse images were captured at 100 Hz using HClmage Live 4.5.0.0 once the cells appeared in the FOV. Stroboscopic illumination from both LEDs was implemented to minimize the motion blur of cells, with an exposure time as short as 100 μs. The LEDs were synchronized with the sensor to ensure illumination occurred within the camera's exposure time.Model-based correction of sCMOS-related noise
A fluorescence microscopy noise-correction method designed for sCMOS cameras28 was applied to the raw elemental images to enhance the signal-to-noise ratio compromised by the short exposure time of stroboscopic illumination. First, the noisy input image was rescaled using pixel maps of offset and gain. Next, the noise variation was estimated from the intensity distribution of a high-pass-filtered image, with the threshold set at the optical transfer function (OTF) boundary determined by the instrumental parameters. This estimated variation was then used as a parameter for sparse denoising, where each reference patch was grouped with similar patches into a 3D array, and collaborative Wiener filtering was performed using a 3D transform.75 Due to the similarity among patches, the 3D transform provides a sparser representation of the original patches than the 2D transforms. Finally, the aggregation procedure returns the denoised patches to their original locations.Preparation and staining of mixed HeLa and Jurkat cells
HeLa cells (93021013, Sigma-Aldrich) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% Penicillin–Streptomycin (Pen–Strep) at 37 °C in a 5% CO2 atmosphere. On the imaging day, the cells were detached with trypsin–EDTA, washed, and suspended in a solution containing 3 mL modified DMEM, 10 μL SYTO 16, and 150 μL WGA 594 to stain the nucleus and membrane, respectively. After a 30-minute incubation, the cells were washed with 3 mL of phosphate-buffered saline (PBS) by centrifugation at 300 × g for 5 minutes and resuspended in 0.5 mL of PBS. Meanwhile, Jurkat T cells (#88042803, Sigma-Aldrich) were cultured in RPMI supplemented with 10% FBS and 1% Pen–Strep at 37 °C and in a 5% CO2 environment. On the day of imaging, the cultured cells were centrifuged and resuspended in 3 mL RPMI containing 10 μL SYTO 16 and 150 μL WGA 594. The cells were incubated for 30 min, then washed with 3 mL phosphate-buffered saline (PBS) in a centrifuge at 300 × g for 5 min. The cells were resuspended in 0.5 mL PBS and mixed with the HeLa cells at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio for imaging.
1 ratio for imaging.
Cell cycle shift in chemotherapy-treated lung adenocarcinoma cells (H460) and staining
H460 cells (purchased from ATCC) were cultured in RPMI supplemented with 10% FBS and 1% Pen–Strep at 37 °C and in a 5% CO2 environment. The control group was split 48 hours before imaging, while the experimental group received 20 μM etoposide for the same duration. After 48 hours, both groups of cells were rinsed with PBS, detached using trypsin–EDTA, and fixed with 4 mL of 4% paraformaldehyde (PFA) for 10 minutes. The fixed cells were then washed with 3 mL phosphate-buffered saline (PBS) by centrifugation at 300 × g for 5 minutes, and resuspended in 2 mL PBS containing 100 μL PI and 100 μL RNase to stain DNA. The samples were stained at room temperature in the dark for 30 minutes before imaging. High-resolution wide-field images were acquired immediately after light-field cytometric imaging using a 16×, 0.8 NA objective under the same suspension conditions, serving as a reference for comparison (Fig. S9).PEG-mediated fusion of Jurkat cells and staining
On the day of imaging, Jurkat cells were centrifuged, resuspended in 2 mL RPMI containing 2 μL of MitoTracker Red or MitoTracker Green, and incubated for 30 minutes for the experimental group. The cells were then washed twice with 3 mL phosphate-buffered saline (PBS) by centrifugation at 300 × g for 5 minutes. Labeled cells were co-plated at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 red-to-green and treated with 50% PEG 1500/RPMI for 2 minutes to induce cell–cell fusion.76 The treated cells were washed with PBS using the same centrifugation steps. For the control group, Jurkat cells were labeled with MitoTracker Red or MitoTracker Green at the same concentration as the experimental group, washed twice with PBS, and then mixed for imaging.
1 red-to-green and treated with 50% PEG 1500/RPMI for 2 minutes to induce cell–cell fusion.76 The treated cells were washed with PBS using the same centrifugation steps. For the control group, Jurkat cells were labeled with MitoTracker Red or MitoTracker Green at the same concentration as the experimental group, washed twice with PBS, and then mixed for imaging.
Preparation and staining of live and dead HeLa cells in a curved channel
HeLa cells were cultured as described above. On the day of imaging, HeLa cells were rinsed with PBS, detached using trypsin–EDTA, and divided into two parts. One group was subjected to heat treatment at 60 °C for 3 hours to induce necrosis. Live HeLa cells were stained with 10 μL SYTO 16 to label nuclei and 10 μL Live-or-Dye 594 (cell membrane impermeant dyes that selectively stain dead cells) in 2 mL DMEM to label the cytoplasm. The cells were incubated for 30 minutes. Dead HeLa cells were labeled with the same reagents in 2 mL PBS at 4 °C for 30 minutes. After the staining, live and dead HeLa cells were washed with PBS by centrifugation and mixed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio in 1 mL PBS for imaging. 0.5 μL of 0.4% Trypan Blue solution was mixed with 0.5 μL of PBS, then diluted 1
1 ratio in 1 mL PBS for imaging. 0.5 μL of 0.4% Trypan Blue solution was mixed with 0.5 μL of PBS, then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 into 100 μL of cell suspension, loaded into a hemocytometer, and examined under a standard Nikon bright-field microscope to assess cell viability.
100 into 100 μL of cell suspension, loaded into a hemocytometer, and examined under a standard Nikon bright-field microscope to assess cell viability.
Author contributions
Z. L. and S. J. conceived and designed the project. Z. L. and W. L. developed the two-colour Fourier light-field microscopy system. Z. L. constructed the flow cytometer with assistance from K. Y., K. H. Z. G. and Z. L. performed the imaging experiments. Z. L. and M. S. prepared the samples. Z. L. conducted image analysis and generated results. A. K. inspected the research results. S. J. supervised the overall project. Z. L. and S. J. wrote the manuscript with input from all authors.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article are included in the supplementary information (SI). The code for this paper is available as Supplementary Software. The code has been written in MATLAB (MathWorks) and tested with version 2021b. To install the package, unzip the compressed folder and follow the instructions in the file read-me.txt. The most updated version of the software can be found at: https://www.github.com/ShuJiaLab/LFC-plus. Supplementary information is available. See DOI: https://doi.org/10.1039/d5lc00962f.Acknowledgements
We acknowledge the support of the National Science Foundation grants 2145235 and 2503686 (to S. J.), the National Institutes of Health grant R35GM124846 (to S. J.), and the Georgia Institute of Technology Faculty Startup Fund (to S. J.).References
- P. Rees, H. D. Summers, A. Filby, A. E. Carpenter and M. Doan, Nat. Rev. Methods Primers, 2022, 2, 86 CrossRef CAS PubMed.
- M. Doan, I. Vorobjev, P. Rees, A. Filby, O. Wolkenhauer, A. E. Goldfeld, J. Lieberman, N. Barteneva, A. E. Carpenter and H. Hennig, Trends Biotechnol., 2018, 36, 649–652 CrossRef CAS PubMed.
- S. Stavrakis, G. Holzner, J. Choo and A. deMello, Curr. Opin. Biotechnol., 2019, 55, 36–43 CrossRef CAS.
- M. Doan, I. Vorobjev, P. Rees, A. Filby, O. Wolkenhauer, A. E. Goldfeld, J. Lieberman, N. Barteneva, A. E. Carpenter and H. Hennig, Trends Biotechnol., 2018, 36, 649–652 CrossRef CAS.
- H. Mikami, M. Kawaguchi, C. J. Huang, H. Matsumura, T. Sugimura, K. Huang, C. Lei, S. Ueno, T. Miura, T. Ito, K. Nagasawa, T. Maeno, H. Watarai, M. Yamagishi, S. Uemura, S. Ohnuki, Y. Ohya, H. Kurokawa, S. Matsusaka, C. W. Sun, Y. Ozeki and K. Goda, Nat. Commun., 2020, 11, 1162 CrossRef CAS PubMed.
- G. Holzner, B. Mateescu, D. van Leeuwen, G. Cereghetti, R. Dechant, S. Stavrakis and A. deMello, Cell Rep., 2021, 34, 108824 Search PubMed.
- Y. Han, J. Zhao, Z. Chao, K. Liang, C. Zhang, L. Jiang, Z. Jiao, F. Bai, A. Tárnok and Z. You, Device, 2023, 1, 100124 CrossRef.
- S. Ota, R. Horisaki, Y. Kawamura, M. Ugawa, I. Sato, K. Hashimoto, R. Kamesawa, K. Setoyama, S. Yamaguchi, K. Fujiu, K. Waki and H. Noji, Science, 2018, 360, 1246–1251 CrossRef CAS PubMed.
- D. Schraivogel, T. M. Kuhn, B. Rauscher, M. Rodriguez-Martinez, M. Paulsen, K. Owsley, A. Middlebrook, C. Tischer, B. Ramasz, D. Ordonez-Rueda, M. Dees, S. Cuylen-Haering, E. Diebold and L. M. Steinmetz, Science, 2022, 375, 315–320 CrossRef CAS.
- E. D. Diebold, B. Buckley, D. R. Gossett and B. Jalali, Nat. Photonics, 2013, 7, 806–810 CrossRef CAS.
- C. Lei, H. Kobayashi, Y. Wu, M. Li, A. Isozaki, A. Yasumoto, H. Mikami, T. Ito, N. Nitta, T. Sugimura, M. Yamada, Y. Yatomi, D. Di Carlo, Y. Ozeki and K. Goda, Nat. Protoc., 2018, 13, 1603–1631 CrossRef CAS PubMed.
- J. Zhou, L. Mei, M. Yu, X. Ma, D. Hou, Z. Yin, X. Liu, Y. Ding, K. Yang, R. Xiao, X. Yuan, Y. Weng, M. Long, T. Hu, J. Hou, Y. Xu, L. Tao, S. Mei, H. Shen, Y. Yalikun, F. Zhou, L. Wang, D. Wang, S. Liu and C. Lei, Light:Sci. Appl., 2025, 14, 76 CrossRef CAS PubMed.
- H. Kanno, K. Hiramatsu, H. Mikami, A. Nakayashiki, S. Yamashita, A. Nagai, K. Okabe, F. Li, F. Yin, K. Tominaga, O. F. Bicer, R. Noma, B. Kiani, O. Efa, M. Buscher, T. Wazawa, M. Sonoshita, H. Shintaku, T. Nagai, S. Braun, J. P. Houston, S. Rashad, K. Niizuma and K. Goda, Nat. Commun., 2024, 15, 7376 Search PubMed.
- M. Ugawa and S. Ota, Cells, 2024, 13(24), 2073 CrossRef CAS PubMed.
- E. J. Gualda, H. Pereira, G. G. Martins, R. Gardner and N. Moreno, Cytometry, Part A, 2017, 91a, 144–151 CrossRef.
- M. Ugawa and S. Ota, Small Sci., 2022, 2(7), 2100126 CrossRef CAS PubMed.
- M. Ugawa and S. Ota, Biomed. Opt. Express, 2022, 13, 3647–3656 CrossRef CAS.
- J. Son, B. Mandracchia, A. D. Silva Trenkle, G. A. Kwong and S. Jia, Lab Chip, 2023, 23, 624–630 RSC.
- P. Joshi, P. Kumar, A. S. J. M. Varghese and P. P. Mondal, Commun. Phys., 2024, 7, 25 CrossRef CAS.
- P. Paie, G. Calisesi, A. Candeo, A. Comi, F. Sala, F. Ceccarelli, A. De Luigi, P. Veglianese, K. Muhlberger, M. Fokine, G. Valentini, R. Osellame, M. Neil, A. Bassi and F. Bragheri, Lab Chip, 2023, 24, 34–46 RSC.
- S. Quint, A. F. Christ, A. Guckenberger, S. Himbert, L. Kaestner, S. Gekle and C. Wagner, Appl. Phys. Lett., 2017, 111, 103701 CrossRef.
- L. E. Weiss, Y. Shalev Ezra, S. Goldberg, B. Ferdman, O. Adir, A. Schroeder, O. Alalouf and Y. Shechtman, Nat. Nanotechnol., 2020, 15, 500–506 CrossRef CAS PubMed.
- F. Merola, P. Memmolo, L. Miccio, R. Savoia, M. Mugnano, A. Fontana, G. D'Ippolito, A. Sardo, A. Iolascon, A. Gambale and P. Ferraro, Light:Sci. Appl., 2017, 6, e16241 CrossRef CAS.
- D. Pirone, J. Lim, F. Merola, L. Miccio, M. Mugnano, V. Bianco, F. Cimmino, F. Visconte, A. Montella, M. Capasso, A. Iolascon, P. Memmolo, D. Psaltis and P. Ferraro, Nat. Photonics, 2022, 16, 851–859 CrossRef CAS PubMed.
- X. Hua, K. Han, B. Mandracchia, A. Radmand, W. Liu, H. Kim, Z. Yuan, S. M. Ehrlich, K. Li, C. Zheng, J. Son, A. D. Silva Trenkle, G. A. Kwong, C. Zhu, J. E. Dahlman and S. Jia, Nat. Commun., 2024, 15, 1975 Search PubMed.
- W. Liu, G.-A. R. Kim, S. Takayama and S. Jia, Biosens. Bioelectron., 2022, 208, 114201 CrossRef CAS PubMed.
- Z. Ling, W. Liu, K. Yoon, J. Hou, P. Forghani, X. Hua, H. Yoon, M. Bagheri, L. P. Dasi, B. Mandracchia, C. Xu, S. Nie and S. Jia, Nat. Cardiovasc. Res., 2025, 4(5), 637–648 CrossRef.
- B. Mandracchia, X. Hua, C. Guo, J. Son, T. Urner and S. Jia, Nat. Commun., 2020, 11, 94 CrossRef CAS PubMed.
- N. Nitta, T. Sugimura, A. Isozaki, H. Mikami, K. Hiraki, S. Sakuma, T. Iino, F. Arai, T. Endo, Y. Fujiwaki, H. Fukuzawa, M. Hase, T. Hayakawa, K. Hiramatsu, Y. Hoshino, M. Inaba, T. Ito, H. Karakawa, Y. Kasai, K. Koizumi, S. Lee, C. Lei, M. Li, T. Maeno, S. Matsusaka, D. Murakami, A. Nakagawa, Y. Oguchi, M. Oikawa, T. Ota, K. Shiba, H. Shintaku, Y. Shirasaki, K. Suga, Y. Suzuki, N. Suzuki, Y. Tanaka, H. Tezuka, C. Toyokawa, Y. Yalikun, M. Yamada, M. Yamagishi, T. Yamano, A. Yasumoto, Y. Yatomi, M. Yazawa, D. Di Carlo, Y. Hosokawa, S. Uemura, Y. Ozeki and K. Goda, Cell, 2018, 175, 266–276.e213 Search PubMed.
- A. B. Borle, J. Gen. Physiol., 1969, 53, 43–56 CrossRef CAS PubMed.
- U. Schneider, H. U. Schwenk and G. Bornkamm, Int. J. Cancer, 1977, 19, 621–626 Search PubMed.
- J. R. Molina, P. Yang, S. D. Cassivi, S. E. Schild and A. A. Adjei, Mayo Clin. Proc., 2008, 83, 584–594 CrossRef.
- A. Montecucco, F. Zanetta and G. Biamonti, EXCLI J., 2015, 14, 95 Search PubMed.
- R. Santana-Davila, K. Devisetty, A. Szabo, R. Sparapani, C. Arce-Lara, E. M. Gore, A. Moran, C. D. Williams, M. J. Kelley and J. Whittle, J. Clin. Oncol., 2015, 33, 567–574 CrossRef CAS.
- J. Vansteenkiste, D. De Ruysscher, W. E. Eberhardt, E. Lim, S. Senan, E. Felip, S. Peters and E. G. W. Group, Ann. Oncol., 2013, 24(Suppl 6), vi89–vi98 CrossRef.
- P. Olszewska, B. Pazdrak and M. L. Kruzel, Biomedicines, 2022, 10, 2429 CrossRef CAS PubMed.
- M. Tammaro, P. Barr, B. Ricci and H. Yan, PLoS One, 2013, 8, e79202 CrossRef CAS PubMed.
- T. Saleh, L. Tyutyunyk-Massey, G. F. Murray, M. R. Alotaibi, A. S. Kawale, Z. Elsayed, S. C. Henderson, V. Yakovlev, L. W. Elmore, A. Toor, H. Harada, J. Reed, J. W. Landry and D. A. Gewirtz, Biochem. Pharmacol., 2019, 162, 202–212 CrossRef CAS PubMed.
- A. Pedrioli and A. Oxenius, Trends Immunol., 2021, 42, 1143–1158 CrossRef CAS.
- E. Taagen, A. J. Bogdanove and M. E. Sorrells, Trends Plant Sci., 2020, 25, 455–465 CrossRef CAS PubMed.
- H. F. Wang, W. Xiang, B. Z. Xue, Y. H. Wang, D. Y. Yi, X. B. Jiang, H. Y. Zhao and P. Fu, Oncol. Lett., 2021, 22, 530 CrossRef PubMed.
- A. Yoshihara, S. Watanabe, I. Goel, K. Ishihara, K. N. Ekdahl, B. Nilsson and Y. Teramura, Biomaterials, 2020, 253, 120113 CrossRef CAS PubMed.
- C. Villafranca, M. R. Makris, M. J. Garrido Bauerle, R. V. Jensen and W. H. Eyestone, Cytotechnology, 2020, 72, 797–805 CrossRef CAS.
- F. Pedrazzoli, I. Chrysantzas, L. Dezzani, V. Rosti, M. Vincitorio and G. Sitar, Cancer Cell Int., 2011, 11, 32 CrossRef CAS PubMed.
- N. L. Mora, A. L. Boyle, B. J. V. Kolck, A. Rossen, Š. Pokorná, A. Koukalová, R. Šachl, M. Hof and A. Kros, Sci. Rep., 2020, 10, 3087 CrossRef CAS.
- X. Di, X. Gao, L. Peng, J. Ai, X. Jin, S. Qi, H. Li, K. Wang and D. Luo, Signal Transduction Targeted Ther., 2023, 8, 282 CrossRef CAS.
- X. Cheng, C. Caruso, W. A. Lam and M. D. Graham, Sci. Adv., 2023, 9, eadj6423 CrossRef PubMed.
- B. Lombodorj, H. C. Tseng, H. Y. Chang, Y. W. Lu, N. Tumurpurev, C. W. Lee, B. Ganbat, R. G. Wu and F. G. Tseng, Micromachines, 2020, 11, 275 CrossRef PubMed.
- P. B. Howell Jr, D. R. Mott, J. P. Golden and F. S. Ligler, Lab Chip, 2004, 4, 663–669 RSC.
- Y. C. Wong, C. Dai, Q. Xian, Z. Yan, Z. Zhang and W. Wen, Sci. Rep., 2023, 13, 17896 CrossRef CAS.
- Y. Saffar, S. Kashanj, D. S. Nobes and R. Sabbagh, Micromachines, 2023, 14, 2202 CrossRef PubMed.
- M. Islam, R. Mezencev, B. McFarland, H. Brink, B. Campbell, B. Tasadduq, E. K. Waller, W. Lam, A. Alexeev and T. Sulchek, Cell Death Dis., 2018, 9, 239 CrossRef.
- C. A. Dessalles, C. Leclech, A. Castagnino and A. I. Barakat, Commun. Biol., 2021, 4, 764 CrossRef.
- M. L. Jackson, A. R. Bond and S. J. George, Cardiovasc. Drugs Ther., 2023, 37, 997–1010 CrossRef CAS PubMed.
- H. Jeon and J. Han, Annu. Rev. Anal. Chem., 2025, 18, 447–472 CrossRef CAS.
- J. Gala de Pablo, M. Lindley, K. Hiramatsu and K. Goda, Acc. Chem. Res., 2021, 54, 2132–2143 CrossRef CAS PubMed.
- N. Lu, H. M. Tay, C. Petchakup, L. He, L. Gong, K. K. Maw, S. Y. Leong, W. W. Lok, H. B. Ong and R. Guo, Lab Chip, 2023, 23, 1226–1257 RSC.
- S. Daetwyler, H. Mazloom-Farsibaf, F. Y. Zhou, D. Segal, E. Sapoznik, B. Chen, J. M. Westcott, R. A. Brekken, G. Danuser and R. Fiolka, Nat. Methods, 2025, 22, 569–578 CrossRef CAS.
- S. Luro, L. Potvin-Trottier, B. Okumus and J. Paulsson, Nat. Methods, 2020, 17, 93–100 CrossRef CAS.
- J. Mertz, Optica, 2019, 6, 1261–1268 CrossRef CAS.
- Q. Li, J. van de Groep, A. K. White, J.-H. Song, S. A. Longwell, P. M. Fordyce, S. R. Quake, P. G. Kik and M. L. Brongersma, Nat. Nanotechnol., 2022, 17, 1097–1103 CrossRef CAS PubMed.
- B. Mandracchia, J. Son and S. Jia, Lab Chip, 2021, 21, 489–493 RSC.
- K. AbuZineh, L. I. Joudeh, B. Al Alwan, S. M. Hamdan, J. S. Merzaban and S. Habuchi, Sci. Adv., 2018, 4, eaat5304 CrossRef CAS PubMed.
- L. E. Weiss, Y. Shalev Ezra, S. Goldberg, B. Ferdman, O. Adir, A. Schroeder, O. Alalouf and Y. Shechtman, Nat. Nanotechnol., 2020, 15, 500–506 CrossRef CAS PubMed.
- M. Doan, C. Barnes, C. McQuin, J. C. Caicedo, A. Goodman, A. E. Carpenter and P. Rees, Nat. Protoc., 2021, 16, 3572–3595 CrossRef CAS.
- D. M. D. Siu, K. C. M. Lee, B. M. F. Chung, J. S. J. Wong, G. Zheng and K. K. Tsia, Lab Chip, 2023, 23, 1011–1033 RSC.
- K. Huang, H. Matsumura, Y. Zhao, M. Herbig, D. Yuan, Y. Mineharu, J. Harmon, J. Findinier, M. Yamagishi, S. Ohnuki, N. Nitta, A. R. Grossman, Y. Ohya, H. Mikami, A. Isozaki and K. Goda, Lab Chip, 2022, 22, 876–889 RSC.
- J. Riordon, D. Sovilj, S. Sanner, D. Sinton and E. W. K. Young, Trends Biotechnol., 2019, 37, 310–324 CrossRef CAS PubMed.
- K. Han, X. Hua, T. Qi, Z. Gao, X. Wang and S. Jia, Nat. Commun., 2025, 16, 10960 CrossRef.
- N. Frey, U. M. Sönmez, J. Minden and P. LeDuc, Nat. Commun., 2022, 13, 3195 CrossRef PubMed.
- E. M. Payne, D. A. Holland-Moritz, S. Sun and R. T. Kennedy, Lab Chip, 2020, 20, 2247–2262 Search PubMed.
- J. Ko, D. Park, J. Lee, S. Jung, K. Baek, K. E. Sung, J. Lee and N. L. Jeon, Nat. Rev. Bioeng., 2024, 2, 453–469 CrossRef CAS.
- J. Zhu, K. Pang, B. Hu, R. He, N. Wang, Z. Jiang, P. Ji and F. Zhao, Nat. Genet., 2024, 56, 2259–2270 CrossRef CAS PubMed.
- N. V. Menon, J. Lee, T. Tang and C. T. Lim, Lab Chip, 2025, 25(5), 752–763 RSC.
- K. Dabov, A. Foi, V. Katkovnik and K. Egiazarian, IEEE Trans. Image Process., 2007, 16, 2080–2095 Search PubMed.
- A. Nascimento, J. Lannigan and D. Kashatus, Cytometry, Part A, 2016, 89, 708–719 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |