Open Access Article
Open Access ArticleIntegrated DEP presorting and wireless electrode array for high-throughput selective single-cell isolation
Thilini N. Rathnaweera,
Dhatchayani Rajkumar and
Robbyn K. Anand *
*
Department of Chemistry, Iowa State University, Ames, Iowa 50011, USA. E-mail: rkanand@iastate.edu
First published on 7th January 2026
Abstract
Rare cell heterogeneity significantly impacts diagnosis, prognosis, therapeutic options and responses, particularly in diverse diseases like cancer. While single-cell analysis is the most effective route, isolating cells individually with high selectivity, purity, efficiency and throughput remains a major challenge. Thus, we present a unified platform coined “SC-DEPOT” to perform all analytical steps from selective isolation from a mixture of cells to parallel single-cell analysis. The platform integrates three sequential modules – one hydrodynamic and two DEP-based – to independently execute distinct and complementary functions. First, the hydrodynamic module focuses all cells towards the channel centerline. Then by DEP, slanted interdigitated electrodes selectively redirect target cells to the channel walls, where they are finally captured in cell-sized micropockets by insulator-based DEP (iDEP). This final stage builds on our previously reported iDEP device, which isolates cells in nanoliter-scale chambers – which are addressed by “wireless” bipolar electrodes (BPEs) – to facilitate individual analysis. The added two preceding steps enhance sample purity to 96% and enable an eightfold increase in channel width compared to a previous limitation of 100 μm. This result is important because it yields an eight-fold to sixteen-fold enhancement in volumetric throughput for samples comprising a mixture of cell types or only one cell type, respectively. The final iDEP module isolates single cells at 94% efficiency and transfers them into the sealable chambers at 92% efficiency. This combination of high throughput and gentle, extended capture from highly concentrated backgrounds expands the utility of the SC-DEPOT device in clinical workflows.
Introduction
Selective isolation and retention of single target cells is a key requirement to parse cellular heterogeneity, specifically in relation to diagnostics,1 therapeutics,2,3 immunological studies4,5 and personalized medicine.3,6 However, if the target cells are a rare population present in a pool of abundant background cells, their isolation becomes particularly challenging. From a clinical perspective, rare cells are low abundance populations, typically ranging from 1 to 1000 cells in a milliliter of blood7,8 and includes types such as circulating tumor cells (CTCs), adult stem cells and antigen-specific immune cells. These rare cell populations occur in peripheral blood, which comprises mononucleated cells (PBMCs) at 106–107 cells per mL and over 109 red blood cells per mL.Generally, for isolation of rare cells such as CTCs, the volumetric throughput has been a prime focus during platform development. The ensemble-decision aliquot ranking (eDAR) technique introduced by the Chiu group processes blood at 50 μL min−1 flow. eDAR utilizes surface makers to identify nanoliter aliquots containing CTCs and recovers CTCs at a rate of 93%. However, all CTCs are collected in ensemble, thus requiring a secondary platform for single-cell analysis.9 The CTC-chip introduced by the Toner group uses anti-EpCAM antibodies (epithelial cell adhesion molecules) coated on microposts to select CTCs at 50% purity, at an impressive volumetric throughput of 1–2 mL h−1.8 The Herringbone-chip works on a similar principle, but introduces passive mixing to enhance the interaction of CTCs with the antibody-coated surface.10 Nevertheless, targeting a single surface marker such as EpCAM or cytokeratins (CKs) can be disadvantageous, given that EpCAM is downregulated during the disease progression and both markers are limited to cells of epithelial origin.11 Further, specific markers should be pre-identified for their utilization. In addition to the dependence on surface markers, the platforms also lack the single-cell isolation and individual analysis capabilities, thus leading to potential loss of rare cells during transfer to a secondary platform.
Certain other platforms have demonstrated the successful combination of high-throughput aspects with on-chip single-cell isolation in microwells. The Han group has presented a high-throughput platform that utilizes photothermal direction of cells into an array of optimally-sized wells for immunoaffinity-based capturing of exosomes from single cells.12 This platform isolates ∼37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 single cells in 5 min using a temperature gradient to drive cells into cell-sized wells (25 μm diameter for on average 16 μm-diameter cells). While yielding a much higher single-cell isolation rate than gravity-driven microwell loading, the photothermal technique lacks cell-type specificity. In a separate report, a single-cell diagnostic device for lung cancer uses size-based separation of tumor cells from sputum and analyzes them in a single-cell microwell array.13 Since there is substantial overlap in size between many CTC types and background leukocytes,14 cell isolation based on size alone could be disadvantageous in clinical applications. Finally, high-throughput techniques like flow cytometry accompanied by seeding of cells into microwells has also been demonstrated for cell cultivation purposes,15 yet, since flow cytometry often results in the loss of cells,16 these platforms can have detrimental impacts in rare-cell studies.
000 single cells in 5 min using a temperature gradient to drive cells into cell-sized wells (25 μm diameter for on average 16 μm-diameter cells). While yielding a much higher single-cell isolation rate than gravity-driven microwell loading, the photothermal technique lacks cell-type specificity. In a separate report, a single-cell diagnostic device for lung cancer uses size-based separation of tumor cells from sputum and analyzes them in a single-cell microwell array.13 Since there is substantial overlap in size between many CTC types and background leukocytes,14 cell isolation based on size alone could be disadvantageous in clinical applications. Finally, high-throughput techniques like flow cytometry accompanied by seeding of cells into microwells has also been demonstrated for cell cultivation purposes,15 yet, since flow cytometry often results in the loss of cells,16 these platforms can have detrimental impacts in rare-cell studies.
Droplet microfluidics offer excellent capabilities to simultaneously analyze thousands of single cells in fluidically-isolated volumes.17,18 Nonetheless, cell encapsulation into droplets is not selective and individual encapsulation is often limited by Poisson statistics, resulting in a high proportion of empty and multi-cell containing droplets. Therefore, a post-droplet sorting module to remove empty and multi-cell droplets19 is required along with a selective pre-cell sorting module, to tailor droplet microfluidics for efficient single rare-cell analysis.
In contrast to most cell selection strategies, dielectrophoresis (DEP) offers exceptional selectivity. DEP is a label-free technique and leverages the unique intrinsic biophysical properties of individual cells such as membrane and cytoplasmic permittivity and conductivity20 for their isolation. These biophysical properties are determined by various factors such as the nuclear-to-cytoplasmic ratio, the degree of glycosylation21 and membrane folding.22 DEP is sensitive to subtle differences in these properties, thereby enabling fine parsing of closely related cell populations.23 DEP-based cell selection is accomplished by tuning the frequency of the applied electric field, thus modulating the extent of electrical polarization (given by Clausius–Mossotti or CM factor) in cells.
DEP-based sorting of cells has been reported at high throughputs. Lee et al. has reported a nDEP-based cell sorter that contains a macro-sized channel and cantilever-type electrode arrays to separate live K562 cells from nontarget dead cells at ultrahigh throughputs up to 15 μL min−1 at 95% efficiency.24 Similarly, the dielectrophoresis activated cell sorter (DACS) sorts target cells at an efficiency of 94% at a rate of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per min.25 DEP-based cell sorting has also been reported via guided transport of cells along Ag-doped conductive PDMS up to a flowrate of 0.5 mL h−1.26 Further, variants of DEP such as optical DEP (ODEP) has been used to recover 61–83% of CTCs at 64–82% purity from background leukocytes.11 The Jiang group has reported using an array of actuation electrodes across a wide fluidic channel to separate particles (yeast and polystyrene microbeads) into distinct streams, followed by trapping cells from a single stream in a designated area.27 Further, a continuous-flow DEP-based platform for sorting rare stem cells has been reported, using interdigitated electrodes aligned along a millifluidic channel to redirect pDEP-responsive cells to a separate outlet for collection.28 While most of these platforms exhibit exceptional efficiencies and throughputs, a common limitation is that they lack enclosures in which to perform downstream biomolecular assays for the isolated cells. In contrast, the Fujii group has proposed an integrated platform that isolates single-cells into microwells using an underlying interdigitated electrode array and has the capability to conduct on-chip single-cell analysis. This electroactive double well array (EdWA) shows 96% isolation efficiency and works at a flowrate of 2 μL min−1.29 However, this platform relies on collapsing the overlying PDMS membrane to seal the microwells and to mechanically lyse the cells, thus complicating the device architecture and function.
000 cells per min.25 DEP-based cell sorting has also been reported via guided transport of cells along Ag-doped conductive PDMS up to a flowrate of 0.5 mL h−1.26 Further, variants of DEP such as optical DEP (ODEP) has been used to recover 61–83% of CTCs at 64–82% purity from background leukocytes.11 The Jiang group has reported using an array of actuation electrodes across a wide fluidic channel to separate particles (yeast and polystyrene microbeads) into distinct streams, followed by trapping cells from a single stream in a designated area.27 Further, a continuous-flow DEP-based platform for sorting rare stem cells has been reported, using interdigitated electrodes aligned along a millifluidic channel to redirect pDEP-responsive cells to a separate outlet for collection.28 While most of these platforms exhibit exceptional efficiencies and throughputs, a common limitation is that they lack enclosures in which to perform downstream biomolecular assays for the isolated cells. In contrast, the Fujii group has proposed an integrated platform that isolates single-cells into microwells using an underlying interdigitated electrode array and has the capability to conduct on-chip single-cell analysis. This electroactive double well array (EdWA) shows 96% isolation efficiency and works at a flowrate of 2 μL min−1.29 However, this platform relies on collapsing the overlying PDMS membrane to seal the microwells and to mechanically lyse the cells, thus complicating the device architecture and function.
To address limitations in rare-cell isolation platforms, we previously reported a microfluidic system that selectively captures cells at BPEs from a flowing suspension,30 transfers them hydrodynamically into microchambers, and enables downstream analysis of retained cells through assays of both live31 and lysed cells.32 We also demonstrated the clinical applicability of the DEP-BPE platform.14 Further, to ensure the independent analysis of the isolated cells, the chambers were sealed-off by making a polymer plug via electropolymerization of a hydrophobic ionic liquid.33 To address loss of functionality in misaligned devices, an insulator-based DEP approach was also introduced to dramatically increase alignment tolerance.34 However, the volumetric throughput of the DEP- and iDEP-BPE platforms are limited to 100 nL min−1 for a four-channel device. Expansion to thirty-two parallel channels has yielded volumetric throughputs of up to 1600 nL min−1. However, at a fundamental level, these additional channels are present merely to comb-through an overabundance of non-target cells. A more elegant strategy to further increase throughput is to increase the channel width beyond 100 μm by overcoming the limited distance over which DEP force can be exerted.
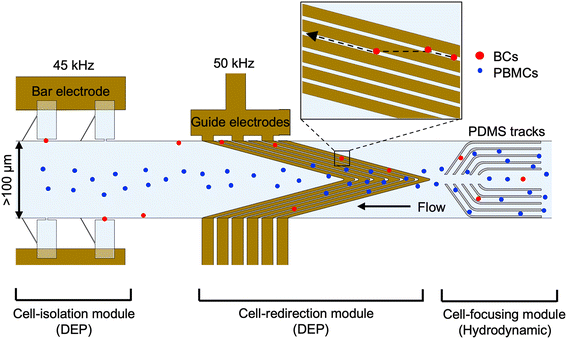
Herein, we report an integrated microfluidic platform coined “Single Cell-DEP on Tracks” or SC-DEPOT (Fig. 1) to isolate single rare cells from a complex leukocyte background with high selectivity and volumetric throughput. Within the SC-DEPOT device, three modules; one hydrodynamic and two DEP-based modules are integrated to enhance the selectivity, purity and throughput of the analysis. The hydrodynamic module focuses all cells toward the center of the channel, after which interdigitated electrodes redirect selected (target) cells toward the channel walls, positioning them for capture and transfer into microchambers. This approach enriches target cells in DEP-active regions near the channel walls, while non-target cells continue along a DEP-inactive ‘superhighway’ at the centerline. Our results demonstrate that by optimizing the spatial configuration of guide electrodes, target cells can be selectively redirected and that the redirection strategy is scalable with 95 ± 5% redirection of target cells in an 800 μm-wide flow channel. Furthermore, when selective frequencies were employed at both DEP modules, rare-target cells were singly captured at micropockets with 94% efficiency and 96% purity. The cells captured at micropockets were also transferred into the microchambers at an efficiency of 92%, thus potentiating evaluation of the heterogeneity of these rare cells. We also demonstrate that the single-cell isolation efficiency within the SC-DEPOT device is unimpacted by the background leukocyte concentration, consistently achieving efficiencies above 90% and purities exceeding 95% at background cell concentrations ranging from 105 to 108 cells per mL. The SC-DEPOT device offers significant improvement to the volumetric throughput of the DEP-BPE platforms while preserving its excellent single-cell isolation performance. The SC-DEPOT platform integrates key capabilities – high selectivity, efficient isolation, robust throughput, and strong cell retention – into a unified system that enables precise and reliable single-cell analysis.
Materials and methods
Chemicals
The silicone elastomer and curing agent (Sylgard 184) was purchased from Ellsworth adhesives. Bovine serum albumin (BSA) (Biotech grade) and 0.25% Trypsin–EDTA (1×) were purchased from Fisher Scientific (Thermo Fisher Scientific, Inc., Waltham, MA). Pluronic® F-127, dextrose (D-glucose), Tris·HCl stock and DMEM/F12 culture medium were obtained from Sigma-Aldrich, Inc. (St. Louis, MO). Fetal bovine serum (FBS) was acquired from ATCC. Sucrose and Ficoll Paque were purchased from Millipore Sigma (Milwaukee, WI). Cell labeling reagents, phycoerythrin anti-human CD-45 and Alexa Fluor 488 anti-human EGFR antibodies were purchased from BioLegend (San Diego, CA). All solutions were prepared in type 1 water (18.2 Ω cm). The “DEP buffer” was prepared with 8.0% sucrose, 0.3% dextrose and 0.1% BSA in 1.0 mM Tris buffer (conductivity: 60–65 μS cm−1).Cell culture and blood fractionation
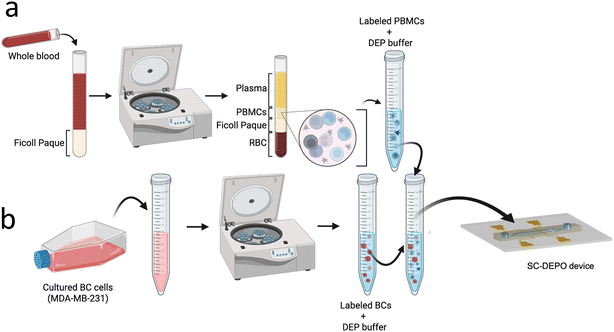
MDA-MB-231 breast-cancer cells were purchased from ATCC and were used as model CTCs. The cells were cultured in DMEM/F12 medium supplemented with 10% FBS. Cells were subcultured every 2 days and the culture was maintained under 80% confluency. To prepare cells for DEP experiments, cells were first detached from the flask using 0.25% Trypsin–EDTA, then pelleted by centrifugation at 1100×g for 5 min. Following two washing steps with the culture medium, the cells were resuspended in freshly prepared DEP buffer. The cell pellet was washed twice with the DEP buffer and the final cell concentration was adjusted to approx. 1 × 104 cells per mL based on the measured concentration and cell viability using Countess™. This concentration was used for baseline experiments (no spiking into PBMCs).To mimic rare cells in background leukocytes, cultured MDA-MB-231 cells were spiked into healthy donor-derived PBMCs. Blood from healthy-volunteer donors were obtained from HCCC-tissue procurement center at the University of Iowa. The whole blood was fractionated to separate the PBMCs. Briefly, 4.0 mL of whole blood was diluted with 4.0 mL of DPBS supplemented with 2% FBS. From the diluted blood, 4.0 mL was carefully layered on top of 3.2 mL of Ficoll Paque without disturbing the layers. The layered sample was centrifuged for 40 min at 0.4 rcf, to separate the layers. Following centrifugation, the plasma layer on top was removed and the layer of PBMCs were carefully pipetted out and was resuspended in 6 mL of DPBS with 2% FBS. The mixture was centrifuged for 15 min at 0.4 rcf, to pellet down the PBMCs. Then the pellet was resuspended in DEP buffer and was washed twice to prepare the working PBMC solution with a concentration of 105–107 cells per mL. As noted in each experiment in the Results and discussion section, either MDA-MB-231 cells were spiked into PBMCs, or one of the two cell types was used alone.
Fluorescence labeling
In spike-in experiments, both cell types, MDA-MB-231 and PBMCs were fluorescently labeled for ease of visualization. MDA-MB-231 cells and PBMCs were labeled with Alexa Fluor 488 anti-human EGFR and phycoerythrin anti-human CD-45 antibodies, respectively. For labeling, in brief, after a wash with either the culture medium or PBS with 2% FBS, cells were washed once with 1× PBS with 10% FBS. The antibody solution was vortexed for 1 min and a cell-labeling buffer was prepared by diluting 5.0 μL of the appropriate antibody solution to 100 μL in 1× PBS with 10% FBS. The diluted antibody solution was centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g (4.0 °C, 10 min). 100 μL of supernatant was added to approx. 1 × 106 cells. The cell solution was mixed well and then placed on a rocker for 1 h at room temperature. Next, the labeled cells were washed once with DEP buffer and the cell viability and concentration were remeasured. Based on the measured concentration, a specific volume of MDA-MB-231 cells was withdrawn to obtain ∼2500 cells. This fraction was washed twice more with DEP buffer, resuspended in a minimum volume of DEP buffer, and spiked-into 1.0 mL containing ∼105 to 109 PBMCs as indicated in the Results and discussion. The overabundance of spiked-in BCs above typical CTC concentrations was used to shorten the experiment time. Further, as approximately 10% of the cells are lost during each washing step and the cell concentration cannot be measured accurately using Countess™ at such low concentrations, prior to each cell capture experiment, the number of BCs that traversed the entire device was counted and averaged to make sure that the maximum number of BCs flowing was less than or equal to 40 cells (because the total capture sites is 40) within 20 min. This value was employed in the calculation of capture efficiency.
000×g (4.0 °C, 10 min). 100 μL of supernatant was added to approx. 1 × 106 cells. The cell solution was mixed well and then placed on a rocker for 1 h at room temperature. Next, the labeled cells were washed once with DEP buffer and the cell viability and concentration were remeasured. Based on the measured concentration, a specific volume of MDA-MB-231 cells was withdrawn to obtain ∼2500 cells. This fraction was washed twice more with DEP buffer, resuspended in a minimum volume of DEP buffer, and spiked-into 1.0 mL containing ∼105 to 109 PBMCs as indicated in the Results and discussion. The overabundance of spiked-in BCs above typical CTC concentrations was used to shorten the experiment time. Further, as approximately 10% of the cells are lost during each washing step and the cell concentration cannot be measured accurately using Countess™ at such low concentrations, prior to each cell capture experiment, the number of BCs that traversed the entire device was counted and averaged to make sure that the maximum number of BCs flowing was less than or equal to 40 cells (because the total capture sites is 40) within 20 min. This value was employed in the calculation of capture efficiency.
3DEP measurements
In this study, the commercial 3DEP instrument (DepTech, Uckfield, U.K.) was used to acquire crossover frequencies and electrical properties (i.e., cytoplasmic conductivity, relative cytoplasmic permittivity, specific membrane conductance and capacitance) of MDA-MB-231 cells and fractionated PBMCs under no-flow conditions. The 3DEP uses a specific chip that contains an array of twenty cylindrical microwells where each well contains several ring-shaped electrodes down the inner wall. This electrode array is designed in a way so that each well receives a specific frequency, typically between 1 kHz to 45 MHz. Upon pipetting the cell suspension into the microwell array, the cells redistribute in response to the electric field. The redistribution occurs such that at frequencies higher than the cell's crossover frequency (pDEP), the cells are drawn towards the walls and at lower frequencies (nDEP), the cells focus to the center of the well. This redistribution is monitored using brightfield imaging and the analysis software maps this redistribution to Clausius–Mossotti polarizability factor and calculates the electrical properties of the cells.Prior to cell loading, the chip was hydrated with 80–100 μL of the conductivity adjusted DEP buffer, removed any trapped air bubbles and was left for 15 min. After hydration the buffer was pipetted out and the cell suspensions with concentrations 1 × 106 cells per mL was loaded and trapped air bubbles were removed. The wells were covered with a glass cover slip and was loaded into the instrument. The boundary conditions were set for cell radius (measured with Countess™ hematocytometer) and buffer conductivity. The full frequency range from 1 kHz to 45 MHz was used to obtain both first and second crossover frequencies. The experiments were triplicated, and the obtained light intensity representative of relative DEP force was plotted against the frequency to estimate the crossover frequency of each cell type.
DEP-based cell capture
Prior to DEP experiments, all devices were treated by filling the channels with 3.0 μM Pluronic® F-127 and incubating overnight at 4 °C. The devices were then flushed with DEP buffer for at least 15 min before introducing the cell sample. Then, the cell solution was introduced, and the flow rate was allowed to stabilize for 2–3 min. The flow was established using a Pico Plus Elite syringe pump (Harvard Apparatus, Holliston, MA) coupled to a 500 μL glass syringe (Hamilton Company, Reno, NV) under withdrawal mode. The volumetric flow rate, AC frequency and the magnitude of the applied AC voltage employed for cell capture are reported for each experiment described in the Results and discussion section. The driving electrodes were energized at reported conditions using a Tektronix AFG3011C waveform generator (Tektronix, Beaverton, OR) paired to Trek Model 2205 amplifier (Trek, Lockport, NY). In preliminary experiments, the cell concentration was adjusted to about 1 × 104 cells per mL and the redirection efficiency was calculated out of the first 30 MDA-MB-231 cells that entered the channel, taking the cell viability into account (dead cells do not experience pDEP). In experiments that quantified the capture efficiency of PBMCs at the employed frequency and voltage for MDA-MB-231 cells, the flow time was adjusted to introduce ∼2 × 104 cells based on the concentration of the stock-PBMC sample, and the capture efficiency was quantified out of this average number of cells that traversed the channels. In spiked-in experiments, a mixture of fluorescently labeled MDA-MB-231 and PBMCs was introduced into the device and was flowed for 20 min under an applied voltage to yield cell capture. Following capture, the AC supply was disconnected to hydrodynamically transfer the cells into the chambers at the same flow rate. In all spiked-cell experiments, the sample mixture was infused at 400 nL min−1 for a 20 min capture interval, corresponding to a total processed volume of ∼8 μL per experiment. All experiments involving unlabeled cells were performed under an AZ-100 microscope (Nikon, Tokyo, Japan) and labeled cells were imaged using a Nikon Eclipse Ti inverted microscope (Nikon, Tokyo, Japan).Results and discussion
Microfluidic platforms capable of isolating selected subpopulation of cells individually is a key requirement for understanding the cellular heterogeneity associated with heterogeneous diseases like cancer. For many applications, the rarity of the subpopulation poses challenges in the efficiency of cell isolation, thus hindering clinical implementation. In this work, we present an integrated platform coined “Single-cell DEP on tracks” or “SC-DEPOT” to perform the key functions of rare cell analysis – label-free cell selection and retention in isolated fluid volumes – at a scaled-up volumetric throughput.The SC-DEPOT device comprises two parallel channels with ten microchambers along either side of the channel leading to a total of forty chambers per device (see SI for device geometry, dimensions and fabrication) the channels are connected to a single inlet and an outlet via bifurcated channels. Each parallel channel comprises three modules shown in Fig. 1.
First, the hydrodynamic cell-focusing module features parallel fluidic tracks arranged across the channel width, with their front edges curved inwards to focus both background peripheral blood mononucleated cells (PBMCs) and MDA-MB-231 breast cancer cells (BCs) towards the centerline. Second, the cell-redirection module incorporates interdigitated wired electrodes termed “guide electrodes” operating at an optimized frequency and voltage to selectively redirect the pDEP-responsive BCs towards the periphery of the channel, while the PBMCs exhibiting nDEP continue to flow along the centerline. Third, the cell isolation module comprises a bar-BPE array aligned to an array of microchambers. The microchambers consist of a leak channel – a 30°-inclined channel on the vertical wall of the chamber which connects the chamber to the main channel and a chamber opening (micropocket) that capture the pDEP-responsive BCs in the presence of an applied electric field. This module is operated at a lower frequency than the guide electrodes to enhance the selectivity of cell capture. Following capture, when the voltage is removed, cells are passively hydrodynamically transferred into the microchambers due to the drag force created by the leak channel, thereby enabling their subsequent analysis. Note that the leak channel dimensions have been previously optimized to provide the necessary drag force.30
As detailed in the following subsections, these modules were independently optimized using BCs at low concentrations before introducing cell mixtures.
The efficiency of guide electrodes in selective redirection of cells
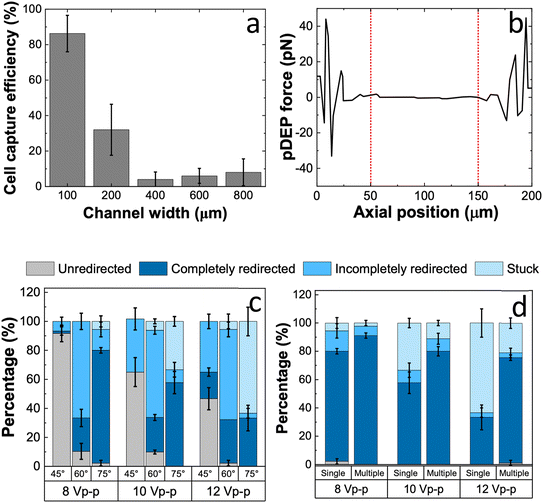
We have previously reported single-cell isolation platforms that utilize electrode-based14,30,32 or insulator-based DEP34 driven by an array of BPEs. Both these platforms enable the target cells flowing in a microfluidic channel to be selectively captured at the micropockets aligned on either side of the channel via DEP. The iDEP-BPE platform, utilized here is an advancement of the original DEP-BPE platform to add both vertical and horizontal alignment tolerances thereby making the device fabrication easier. However, both platforms had a limited volumetric throughput. This limited throughput arises from the short-range nature of the DEP force, which is effective only within 50 μm of each micropocket, thereby constraining the channel width to a maximum of 100 μm.Fig. 2a shows the quantified capture efficiency (i.e., the percentage of cells captured out of the number of cells flowed in) of BCs in response to the increasing channel width. Table S1 shows the design parameters, flow rates, and applied voltages. A key point is that the flow rate and voltage were increased along with the channel width to maintain matched average linear velocities in the channels and average electric field strengths in the micropockets. Accordingly, it is evident that the capture efficiency drastically drops from 86 ± 10% to 32 ± 14%, simply by doubling the channel width to 200 μm. Channel widths from 400 to 800 μm showed a slight increase in the capture efficiency facilitated by the increasing number of cells rolling along the channel wall at higher channel widths.
Numerical simulations demonstrated that, while the magnitude of the pDEP force was not compromised with increasing channel width, the force was constrained to a smaller fraction of the channel width. Fig. 2b shows the simulated pDEP force along a cutline across a 200 μm-wide channel, generated using COMSOL Multiphysics. The simulation clearly indicates that only cells flowing near the channel walls (within 50 μm) experience sufficient pDEP force for capture, while those in the central 100 μm region remain unaffected. Therefore, the channel width and in turn, the volumetric throughput of the device is limited given that a strict balance is required between the flow and the electric field strength to capture cells using DEP. Hence, additional strategies are needed to enhance the functionality of the DEP-BPE platforms at increased channel widths and to make it more compliant with rapid analysis of clinical samples.
To address this limitation, we integrated guide electrodes to redirect cells from the DEP-unresponsive region to the DEP-responsive region of the device. Guide electrodes are a secondary set of Au electrodes that are shaped like “chevrons” and positioned upstream of the BPE array so that the tip of the chevron lies at the central axis of the parallel channel. Importantly, the guide electrodes operate independently from the BPE array, supplied by a secondary voltage source. First, we explored the performance of guide electrodes aligned to a 200 μm-wide channel evaluating their angle of inclination and the number of guiding tracks. Fig. 2c shows percentages of cells classified as unredirected, completely and incompletely redirected, and “stuck” (immobilized on the electrodes), as a function of the inclination of guides to the vertical plane (y-axis) and the applied voltage. At each voltage, the increasing inclination resulted in an increase in the completely redirected fraction. This result is attributed to two aspects of the increasing inclination. At a fixed channel width, increasing the inclination has two effects. First, it lengthens the guiding track, allowing the cell to interact with the electric field for a longer period. Second, it reduces the vertical lift across the channel width, thereby lowering the electric field strength required to guide the cell to the end of the electrode. Aided by these two factors, a single-track guide electrode inclined at 75° from vertical showed 78 ± 2% complete redirection of cells at an applied voltage of 8.0 Vp–p at 100 kHz. The angle of inclination was limited to 75° because the increasing inclination also makes the horizontal length (from tip to end of chevron) wider, demanding an extensively long channel segment.
To further enhance the fraction of redirected cells, parallel guiding tracks (i.e., interelectrode gaps) were added to have a total of five tracks. Accordingly, the completely redirected fraction increased from 78 ± 2% to 91 ± 2% at 8.0 Vp–p, as shown in Fig. 2d. This result is significant because the guide electrodes demonstrate effective redirection efficiencies, potentiating single-cell isolation in channels that are twice as wide as before.
Scalability of the cell redirection strategy
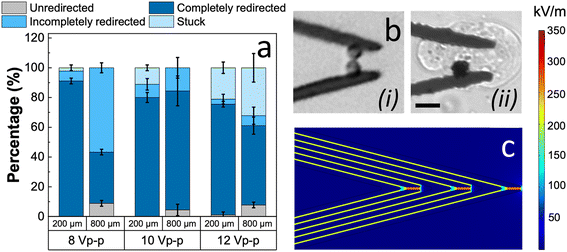
We explored the scalability of the cell redirection strategy by guide electrodes beyond 200 μm widths. Fig. 3a shows a comparison of unredirected, completely and incompletely redirected, and stuck fractions of MDA-MB-231 cells in 200 and 800 μm-wide channels as a function of the applied voltage. Accordingly, a slightly higher voltage (10 Vp–p) was required in 800 μm-wide channels to achieve similar fractions of completely redirected cells as observed in 200 μm-wide channels. This can be attributed to the greater vertical translation required for 800 μm-wide channels before the cells reach the wall at the end of the guiding slit.While the performance of guide electrodes was previously demonstrated using a BPE architecture (in this context, BPEs are electrodes with floating potential that lack direct contact to the power supply), a notable setback was observed: the opening between the wired lead and the BPE led to unintended cell capture and lysis, as shown in Fig. 3b. As simulated using COMSOL, these openings showed a strong localization of electric field at an applied voltage of 10 Vp–p, as depicted in Fig. 3c. As the guide electrodes are intended to selectively redirect rare cells, the loss of a single rare cell can have significant consequences. Therefore, the architecture of guide electrodes was changed from the BPEs to interdigitated wired electrodes while maintaining the track width, the number of tracks and the angle of inclination at optimized values.
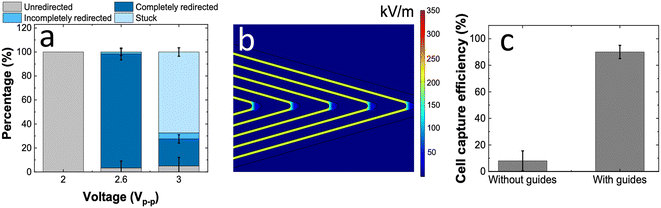
Fig. 4a illustrates the re-optimization of wired interdigitated electrodes to obtain complete redirection of cells up to 95 ± 5% at an applied voltage of only 2.6 Vp–p. While the change in the electrode architecture completely avoids cell lysis, at higher applied voltages, a significant fraction of cells was found to be stuck in the guiding tracks. Additionally, this change also lowers the energy requirement for the functioning of the cell-redirection module. Fig. 4b demonstrates the electric field distribution and its uniformity within the interdigitated guide electrodes. Notably, an applied voltage of 2.6 Vp–p produced an electric field strength comparable to that of the BPE guides operated at 10 Vp–p.
Finally, the efficiency of single-cell capture at the micropockets in an 800 μm-wide channel was quantified in the absence and presence of these optimized guide electrodes (Fig. 4c). Notably, with the introduction of guide electrodes the capture efficiency was increased from 8 ± 7% to 90 ± 5%. These results are significant as they clearly demonstrate the potential of guide electrodes to be used as a strategy to maintain the functionality of the DEP-BPE platforms while widening the flow channels to improve the volumetric throughput. A key point is that non-target cells are expected to undergo nDEP and flow over the guide electrodes without being redirected and remain in the “DEP inactive” middle of the channel width.
Performance of the hydrodynamic cell-focusing module
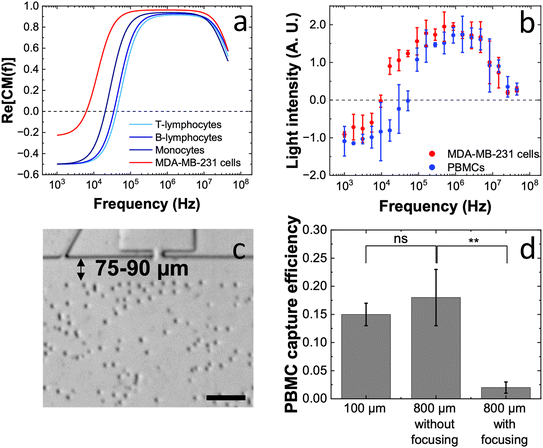
It is important to note that the 90 ± 5% cell capture efficiency observed in the 800 μm-wide channels with guide electrodes includes the ∼8% of cells that were captured even in the absence of guide electrodes. This small fraction corresponds in magnitude to the proportion of cells observed rolling along the channel walls. While this is an added advantage in pure samples of target cells, it can lead to contamination by background cells when working with complex samples, particularly, if the background cells have Clausius Mossotti (CM) factors close to those of the target cells. In the realm of clinical applications, circulating cancer cells (CTCs) occur as a rare population in a highly concentrated background of PBMCs including monocytes and lymphocytes, which possess low crossover frequencies.Fig. 5a shows predicted DEP spectra for MDA-MB-231 cells and various PBMC subtypes, simulated using MyDEP software. The difference in crossover frequencies between MDA-MB-231 cells and PBMCs is sufficient to enable their separation; however, monocytes may still be co-captured with the target breast cancer cells due to overlap in electrical properties – particularly given the heterogeneity among PBMC subtypes and between patients. To circumvent this obstacle, cell isolation kits such as RosetteSep™ monocyte depleting cocktail can be used. However, these add processing steps and cost, and the potential effects of such reagents on the DEP response of both target and non-target cells must be carefully evaluated. To experimentally determine the crossover frequencies of untreated MDA-MB-231 cells and PBMCs under static conditions (no fluid flow), the 3DEP instrument was used. The operating principle and the experimental procedure can be found in the SI. As shown in Fig. 5b, the measured separation in crossover frequencies was greater than predicted. Additionally, the relatively large errors in measured light intensity support the presence of heterogeneous PBMC populations. Cell properties derived from 3DEP measurements are provided in Table S2.
Previous experiments with pure BCs were conducted at 100 kHz to ensure a strong pDEP response. However, in complex mixtures containing PBMCs, this high frequency led to unwanted capture of non-target cells. To address this issue, the frequency was re-optimized using a 100 μm-wide channel prior to BC isolation experiments in complex samples. Since low capture efficiencies were observed for BCs at lower frequencies under a flow velocity of 660 μm s−1, the flow rate was halved to 330 μm s−1 to restore performance. Table 1 summarizes the capture efficiencies of BCs and PBMCs under various frequencies and voltages at this adjusted flow rate. At 75 Vp–p and 45 kHz, BCs exhibited a capture efficiency of 92 ± 5%, whereas only 0.15 ± 0.02% of PBMCs were captured. Further, at the same voltage but at 50 kHz, 100% of BCs were captured. Therefore, 75 Vp–p and 45 kHz were selected for cell capture, while 50 kHz was chosen for the guide electrodes to ensure that all target cells are effectively redirected. The guide electrodes still demonstrated efficient redirection at 50 kHz under an applied voltage of 2.6 Vp–p, and therefore, this voltage was maintained. Notably, in spiked-cell experiments, we did not observe any redirection of PBMCs by guide electrodes operated at 50 kHz.
| Frequency (kHz) | Voltage (Vp–p) | BCs | PBMCs | ||
|---|---|---|---|---|---|
| Capture efficiency (%) | SD | Capture efficiency (%) | SD | ||
| 40 | 60 | 43 | 2.6 | 0.02 | 0.003 |
| 70 | 79 | 8.0 | 0.05 | 0.010 | |
| 75 | 85 | 5.4 | 0.06 | 0.030 | |
| 45 | 60 | 47 | 11 | 0.09 | 0.003 |
| 70 | 87 | 5.8 | 0.14 | 0.010 | |
| 75 | 92 | 5.4 | 0.15 | 0.020 | |
| 50 | 60 | 62 | 2.3 | 0.16 | 0.015 |
| 70 | 96 | 4.0 | 0.18 | 0.020 | |
| 75 | 100 | 0 | 0.14 | 0.070 | |
Note that while the higher frequency applied to the guide electrodes redirects PBMCs with the lowest crossover frequencies – those most likely to be unintentionally captured in the micropockets – toward the channel walls, this redirection is not binary. Unlike the capture mechanism at the micropockets, which either traps a cell or does not, the slanted guide electrodes exhibit a continuous separation effect. For example, a PBMC with a weak pDEP response at the applied frequency may be partially redirected by the guide electrodes without reaching the wall and thus may avoid capture. The same PBMC, if already rolling near the wall, might still be ‘just barely’ captured due to its marginal pDEP responsiveness. Therefore, by directing PBMCs away from the walls upstream of the guide electrodes, their potential capture can be avoided – even if their CM factors are closer to those of BCs. Thus, we introduced a cell focusing module that features parallel tracks defined by narrow walls with inwardly curved front edges fabricated along the width of the 800 μm-wide channel. This module was placed before the guide electrodes to focus all cells flowing in the channel towards the center. Fig. 5c depicts the redirected cells just downstream of the module. While hydrodynamic effects cause considerable divergence of cells after the module, the displaced stream of cells remains 75–90 μm away from the channel wall, effectively bypassing the pDEP-responsive region.
While the flow dynamics in the SC-DEPOT device do not show classical inertial focusing seen at high Reynolds numbers,35,36 it uses geometric flow constriction together with inertially assisted hydrodynamic focusing. In theory, hydrodynamic focusing refers to constricting a sample fluid either by sheath flow or by sheathless focusing using an external or internally induced force, respectively.37 In the SC-DEPOT device, the PDMS tracts act as ‘hydrodynamic rails’ that guide and confine cells into narrow flow lanes and prevent lateral drift. Within the constriction, wall-lift forces push cells away from the PDMS surfaces, and shear-gradient lift drives cells toward a stable off-center streamline. The curved front edge ensures gradual turning of the flow toward the center, further minimizing lateral mixing. As the streamlines merge into the narrow opening at the front edge (approximately one-eighth of the initial channel width), the fluid accelerates through the orifice, creating a Bernoulli or ‘nozzle’ effect and a brief divergence in the flow. Although the diverged flow is not fully developed immediately after the exit, it stabilizes after a short travel distance (the entrance length). Previous studies have also reported this stable expansion of a focused flow.38–40 After exiting the constriction, the channel topology, particle size, wall interactions, rheological properties of the medium, flow velocity, and hydrodynamic interactions among neighboring particles continue to steer them toward the lateral equilibrium positions they maintain downstream. Segrè and Silberberg (1962) first reported that particles traveling through confined flows migrate laterally toward characteristic off-center equilibrium positions, and this behavior has also been observed for soft particles such as red blood cells.41
As anticipated, the cell-focusing module was successful in reducing the capture of PBMCs in the micropockets. Fig. 5d compares the occurrence of unwanted PBMC capture observed in 100 μm- and 800 μm-wide channels in the presence and absence of cell focusing (45 kHz). Note that the electric field strength at the micropockets were matched for the two distinct channel widths using numerical simulations (Table S3). While a slightly higher PBMC capture efficiency was observed in 800 μm-wide channels (0.18 ± 0.05%) than in 100 μm-wide channels (0.15 ± 0.02%), statistical analysis showed that the difference is not significant. However, in the presence of the cell-focusing module, the PBMC capture efficiency was significantly reduced in an 800 μm-wide channel (0.02 ± 0.02%). These results demonstrate this strategy of focusing and redirection successfully separates target and non-target cells into distinct fluid laminae, directly improving the purity of captured cells.
SC-DEPOT device for rare-cell isolation from complex samples
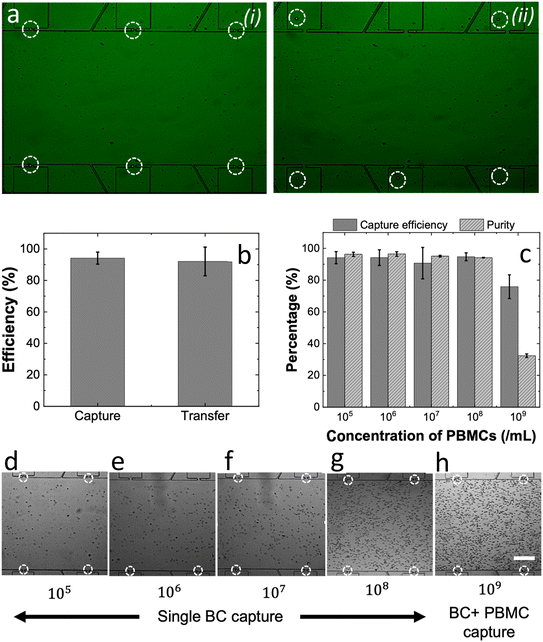
After individual optimization of each device module, we assessed the performance of the integrated SC-DEPOT device in the isolation of target BCs from complex samples. Fig. 6 shows the experimental workflow for separation of PBMCs from healthy blood and spike-in of cultured BCs. The cells were fluorescently labeled for ease of discrimination. The viability of both types of labeled cells were measured using a Countess™ hemacytometer. Table S4 shows the concentration, viability and average diameter measured in three independent experiments.Fig. 7a is a fluorescence micrograph that depicts a section of the cell-isolation module of the SC-DEPOT device. Under an applied voltage of 52 Vp–p at 45 kHz, BCs were singly captured at the micropockets and upon turning off the voltage supply, the captured cells were hydrodynamically transferred into the chambers. As shown in Fig. 7b, a 94 ± 4% capture efficiency was observed for BCs from a complex matrix containing 105 PBMCs per mL at 96 ± 3% purity. And importantly, out of the captured BCs 92 ± 9% were successfully transferred thereby enabling their downstream analysis.
We have previously demonstrated the viability of the cells captured, held in micropockets for 20 min and post transfer at an average electric field of 92 kV m−1.34 However, since a stronger electric field (110 kV m−1) was required to isolate cells at the reduced frequency of 45 kHz, we assessed the viability of cells at these new capture conditions. Fig. S1a depicts the fluorescence of live cells stained with calcein AM, before, during and after redirection by the guide electrodes confirming that the cells do not undergo electroporation (which causes loss of dye) during the cell redirection process. Fig. S1b shows that captured cells remain undamaged after being held in micropockets for 20 min at 110 kV m−1, as indicated by their fluorescence.
Because only cells with intact membranes respond to DEP, repeated capture and release cycles were used to further confirm that viability was maintained (Movie S1). Accordingly, it was evident that the cells remain viable and fully responsive to DEP even after 2.5 h. This result is significant because this extended window for cell analysis, afforded by the iDEP-based approach,34 provides an avenue for processing higher sample volumes.
Assuming an equal number of channels, compared to the previously established DEP-BPE and iDEP-BPE platforms, the SC-DEPOT device offers a 16-fold improvement in volumetric throughput for pure samples and an 8-fold improvement for complex samples, by strategically enabling increased channel width. While the pilot-SC-DEPOT device presented in this work consists only two parallel channels, this architecture can be readily expanded.
Notably, sixteen parallel channels of each 800 μm wide, can be easily fitted to a similar-sized glass substrate, thereby further increasing the volumetric throughput to 6.4 μL min−1 for pure samples and 3.2 μL min−1 for complex samples, while preserving the single-cell isolation characteristics. Although the maximum achievable volume throughput may be suboptimal for clinical applications, its utility can be significantly improved through debulking. For instance, concentrating the buffy coat from a larger blood volume into a small volume of DEP buffer before loading it into the device could render the current flow rate clinically acceptable. To evaluate this approach, we tested the device's single-cell isolation performance at increasing PBMC concentrations. In the results discussed thus far, we demonstrated 94 ± 4% capture of BCs from 105 PBMCs per mL.
To assess the impact of increased “traffic” from PBMCs, we further concentrated the PBMC suspension by 10, 100, 1000 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold and in each case, spiked in the same number of BCs (2500 cells). Fig. 7c shows that even at 108 PBMCs per mL, the capture efficiency of BCs was not depleted, and the purity remained high (>95%). However, at a background PBMC concentration of 109 cells per mL, the capture efficiency for BCs dropped to 75% and the purity drastically dropped to 32%. Thus, it can be established that the SC-DEPOT device can successfully handle concentrated samples up to 108 PBMCs per mL while maintaining purity above 95%. This suggests that the buffy coat from patient blood can be concentrated 10-fold (the normal range for PBMC concentration in peripheral blood is 0.4 to 1.1 × 107 PBMCs per mL) and then introduced into the SC-DEPOT device. Considering a device having sixteen parallel channels and a cell isolation time of 2.5 h, approximately 0.5 mL of concentrated buffy coat can be processed which is equivalent to 4.5 to 12.5 mL of patient blood, while maintaining high purity and cell integrity.
000-fold and in each case, spiked in the same number of BCs (2500 cells). Fig. 7c shows that even at 108 PBMCs per mL, the capture efficiency of BCs was not depleted, and the purity remained high (>95%). However, at a background PBMC concentration of 109 cells per mL, the capture efficiency for BCs dropped to 75% and the purity drastically dropped to 32%. Thus, it can be established that the SC-DEPOT device can successfully handle concentrated samples up to 108 PBMCs per mL while maintaining purity above 95%. This suggests that the buffy coat from patient blood can be concentrated 10-fold (the normal range for PBMC concentration in peripheral blood is 0.4 to 1.1 × 107 PBMCs per mL) and then introduced into the SC-DEPOT device. Considering a device having sixteen parallel channels and a cell isolation time of 2.5 h, approximately 0.5 mL of concentrated buffy coat can be processed which is equivalent to 4.5 to 12.5 mL of patient blood, while maintaining high purity and cell integrity.
Fig. 7d–h, depicts the captured BCs at varying background concentrations of PBMCs. It was observed that even at 109 PBMCs per mL, the 800 μm-wide channel did not exhibit significant congestion or any signs of clogging. Movie S2 shows the focusing, redirection and capture of BCs at a PBMC concentration of 108 cells per mL. This represents a significant improvement over our previous DEP platforms, which featured parallel channels with 100 μm width and demonstrated applicability only within background cell concentrations ranging from 103–106 cells per mL at 100 nL min−1 (0.025–25 cells per min per channel).30,32,34
The performance of the iDEP and SC-DEPOT platforms were compared based on cell capture efficiency and purity at matched throughput in cells per minute. The two highest functional throughputs of the SC-DEPOT device that retained >90% capture efficiency and purity (i.e., 2000 cells per min per channel or 107 cells per mL and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per min per channel or 108 cells per mL at 400 nL min−1) were replicated in the iDEP device. Fig. S2 is a bar graph showing the comparison of cell capture efficiency and purity obtained with iDEP (100 μm-wide channels) and SC-DEPOT (800 μm-wide channels). It is evident that at a throughput of 2000 cells per min per channel, the capture efficiency drops to 49% and the purity depletes to 35% in the 100 μm-wide channel and decreases further at 20
000 cells per min per channel or 108 cells per mL at 400 nL min−1) were replicated in the iDEP device. Fig. S2 is a bar graph showing the comparison of cell capture efficiency and purity obtained with iDEP (100 μm-wide channels) and SC-DEPOT (800 μm-wide channels). It is evident that at a throughput of 2000 cells per min per channel, the capture efficiency drops to 49% and the purity depletes to 35% in the 100 μm-wide channel and decreases further at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per min per channel. In addition, these high concentrations caused significant clogging and flow inconsistencies in the narrow channels, which suggests that the previously established narrow channel-DEP systems must operate at a throughput below 2000 cells pe min per channel while the SC-DEPOT platform can readily attain throughputs up to 20
000 cells per min per channel. In addition, these high concentrations caused significant clogging and flow inconsistencies in the narrow channels, which suggests that the previously established narrow channel-DEP systems must operate at a throughput below 2000 cells pe min per channel while the SC-DEPOT platform can readily attain throughputs up to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per min per channel. This result underscores the applicability of the SC-DEPOT platform to clinical samples.
000 cells per min per channel. This result underscores the applicability of the SC-DEPOT platform to clinical samples.
Conclusion
In this paper, we present an integrated microfluidic platform, SC-DEPOT, for high-throughput, high-purity isolation of individual rare cells from complex biological samples with precise selectivity and on-chip retention for downstream single-cell analysis. The SC-DEPOT device utilizes a combined approach of hydrodynamic focusing and fractionation by DEP-based cell guides prior to cell capture to enhance performance. Optimization of electrode parameters and spatial configuration enabled redirection of 91 ± 2% of target cells to the pDEP-responsive region in 200 μm-wide channels, and scalability demonstrated up to 800 μm wide, achieving 95 ± 5% redirection efficiency. This strategy improved the capture efficiency in wide channels from 8 ± 1% to 90 ± 5%, and resulted in 8-fold higher channel width, yielding up to 16-fold and 8-fold gain in throughput for pure and complex samples, respectively. The hydrodynamic module enhanced the purity of the isolated cell sample to >95% by spatially distancing non-target cells from the capture points. This purity was maintained in the presence of up to 108 cells per mL background cells, which corresponds to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per min per channel. The redirected cells were captured individually via insulator-based DEP at 94% efficiency and remained securely held under the applied electric field for up to 2.5 h, demonstrating the potential for extended cell isolation times when required. Importantly, within this time window, 5.0 mL of patient blood can be processed by a fully expanded SC-DEPOT device. Finally, the captured cells were hydrodynamically transferred into the chambers at 92% efficiency, enabling reliable single-cell analysis. This robust, label-free approach offers a scalable solution for isolating rare cells at clinically relevant throughput and purity, supporting high-resolution studies of cellular heterogeneity.
000 cells per min per channel. The redirected cells were captured individually via insulator-based DEP at 94% efficiency and remained securely held under the applied electric field for up to 2.5 h, demonstrating the potential for extended cell isolation times when required. Importantly, within this time window, 5.0 mL of patient blood can be processed by a fully expanded SC-DEPOT device. Finally, the captured cells were hydrodynamically transferred into the chambers at 92% efficiency, enabling reliable single-cell analysis. This robust, label-free approach offers a scalable solution for isolating rare cells at clinically relevant throughput and purity, supporting high-resolution studies of cellular heterogeneity.
Ethics statement
All experiments were performed in accordance with the Guidelines of the U.S. Department of Health and Human Services for the protection of human subjects, and experiments were approved by the Institutional Review Board of the University of Iowa. Healthy donor blood samples were obtained through the Biospecimen Procurement and Molecular Epidemiology Resource (BioMER) core at Holden Comprehensive Cancer Center, University of Iowa Health Care (IRB #200804792). Informed consents were obtained from all human participants in this study.Author contributions
Thilini N. Rathnaweera: conceptualization, methodology, experimentation, data analysis, writing – original draft. Dhatchayani Rajkumar: device fabrication, cell culture and maintenance. Robbyn K. Anand: supervision, conceptualization, writing – review and editing.Conflicts of interest
The authors declare no conflict of interest.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Supplementary information (SI): the SI includes detailed materials and methods, device design specifications, and tabulated dielectric properties for all cell types used in the study. Movie 1 is a series of brightfield micrographs obtained at 0.5, 1.0, 1.5, 2.0, and 2.5 h post-capture at which timepoints cells were released and recaptured by turning the applied voltage on- and off- to demonstrate their continued viability. Movie 2 is a series of brightfield micrographs that demonstrate the hydrodynamic focusing step. See DOI: https://doi.org/10.1039/d5lc00945f.
Acknowledgements
The authors gratefully acknowledge the 3M Corporation for support through a Non-Tenured Faculty Award. We also acknowledge the National Institutes of Health for funding this project through the R21 Trailblazer Award (EB028583-01). We thank Dr. Beatrise Berzina for suggesting the incorporation of hydrodynamic focusing rails to precondition cell trajectories upstream of the DEP guidance module. The authors thank Prof. Michael Henry, Department of Molecular Physiology and Biophysics, University of Iowa Roy J. and Lucille A. Carver College of Medicine, for facilitating access to biospecimens used in this study.References
- H. Barbot, D. Causeur, Y. Blum and M. Richard, Multi-Omic Statistical Inference of Cellular Heterogeneity, 2024, pp. 1–10 Search PubMed.
- M. Waas and T. Kislinger, Addressing Cellular Heterogeneity in Cancer through Precision Proteomics, J. Proteome Res., 2020, 19(9), 3607–3619, DOI:10.1021/acs.jproteome.0c00338.
- E. Carrasco-Garcia, M. García-Puga, S. Arevalo and A. Matheu, Towards Precision Medicine: Linking Genetic and Cellular Heterogeneity in Gastric Cancer, Ther. Adv. Med. Oncol., 2018, 10, 1–15, DOI:10.1177/1758835918794628.
- L. Ma, Y. Luan and L. Lu, Analyze the Diversity and Function of Immune Cells in the Tumor Microenvironment From the Perspective of Single-Cell RNA Sequencing, Cancer Med., 2025, 14(5), e70622, DOI:10.1002/cam4.70622.
- Z. He, Q. Chen, K. Wang, J. Lin, Y. Peng, J. Zhang, X. Yan and Y. Jie, Single-Cell Transcriptomics Analysis of Cellular Heterogeneity and Immune Mechanisms in Neurodegenerative Diseases, Eur. J. Neurosci., 2024, 59(3), 333–357, DOI:10.1111/ejn.16242.
- I. K. Herrmann and M. Rösslein, Personalized Medicine: The Enabling Role of Nanotechnology, Nanomedicine, 2016, 11(1), 1–3, DOI:10.2217/nnm.15.152.
- A. Salmanzadeh, M. B. Sano, H. Shafiee, M. A. Stremler and R. V. Davalos, Isolation of Rare Cancer Cells from Blood Cells Using Dielectrophoresis, Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, 2012, pp. 590–593, DOI:10.1109/EMBC.2012.6346000.
- S. Nagrath, L. V. Sequist, S. Maheswaran, D. W. Bell, D. Irimia, L. Ulkus, M. R. Smith, E. L. Kwak, S. Digumarthy, A. Muzikansky, P. Ryan, U. J. Balis, R. G. Tompkins, D. A. Haber and M. Toner, Isolation of Rare Circulating Tumour Cells in Cancer Patients by Microchip Technology, Nature, 2007, 450(7173), 1235–1239, DOI:10.1038/nature06385.
- P. G. Schiro, M. Zhao, J. S. Kuo, K. M. Koehler, D. E. Sabath and D. T. Chiu, Sensitive and High-Throughput Isolation of Rare Cells from Peripheral Blood with Ensemble-Decision Aliquot Ranking, Angew. Chem., Int. Ed., 2012, 51(19), 4618–4622, DOI:10.1002/anie.201108695.
- S. L. Stott, C. H. Hsu, D. I. Tsukrov, M. Yu, D. T. Miyamoto, B. A. Waltman, S. Michael Rothenberg, A. M. Shah, M. E. Smas, G. K. Korir, F. P. Floyd, A. J. Gilman, J. B. Lord, D. Winokur, S. Springer, D. Irimia, S. Nagrath, L. V. Sequist, R. J. Lee, K. J. Isselbacher, S. Maheswaran, D. A. Haber and M. Toner, Isolation of Circulating Tumor Cells Using a Microvortex-Generating Herringbone-Chip, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(43), 18392–18397, DOI:10.1073/pnas.1012539107.
- S. B. Huang, M. H. Wu, Y. H. Lin, C. H. Hsieh, C. L. Yang, H. C. Lin, C. P. Tseng and G. B. Lee, High-Purity and Label-Free Isolation of Circulating Tumor Cells (CTCs) in a Microfluidic Platform by Using Optically-Induced-Dielectrophoretic (ODEP) Force, Lab Chip, 2013, 13(7), 1371–1383, 10.1039/c3lc41256c.
- C. Wang, Y. Zhang, J. Wang, Y. Han, Y. Wang, M. Sun, Y. Liang, M. Huang, Y. Yu, H. Hu, H. Liu and L. Han, Single-Cell Isolation Chip Integrated with Multicolor Barcode Array for High-Throughput Single-Cell Exosome Profiling in Tissue Samples, Adv. Mater., 2024, 2411259, 1–215, DOI:10.1002/adma.202411259.
- H. Si, D. Du, W. Li, Q. Li, J. Li, D. Zhao, L. Li and B. Tang, Sputum-Based Tumor Fluid Biopsy: Isolation and High-Throughput Single-Cell Analysis of Exfoliated Tumor Cells for Lung Cancer Diagnosis, Anal. Chem., 2021, 93(30), 10477–10486, DOI:10.1021/acs.analchem.1c00833.
- H. Chen, S. Y. Osman, D. L. Moose, M. Vanneste, J. L. Anderson, M. D. Henry and R. K. Anand, Quantification of Capture Efficiency, Purity, and Single-Cell Isolation in the Recovery of Circulating Melanoma Cells from Peripheral Blood by Dielectrophoresis, Lab Chip, 2023, 23(11), 2586–2600, 10.1039/d2lc01113a.
- S. Lindström, R. Larsson and H. A. Svahn, Towards High-Throughput Single Cell/Clone Cultivation and Analysis, Electrophoresis, 2008, 29(6), 1219–1227, DOI:10.1002/elps.200700536.
- B. D. Hedley and M. Keeney, Technical Issues: Flow Cytometry and Rare Event Analysis, Int. J. Lab. Hematol., 2013, 35(3), 344–350, DOI:10.1111/ijlh.12068.
- X. Yang, W. Liu, D. C. H. Chan, S. U. Ahmed, H. Wang, Z. Wang, C. R. Nemr and S. O. Kelley, Fluorescent Droplet Cytometry for On-Cell Phenotype Tracking, J. Am. Chem. Soc., 2020, 142(35), 14805–14809, DOI:10.1021/jacs.0c05276.
- R. Dimatteo and D. Di Carlo, IL-2 Secretion-Based Sorting of Single T Cells Using High-Throughput Microfluidic on-Cell Cytokine Capture, Lab Chip, 2022, 22(8), 1576–1583, 10.1039/d1lc01098k.
- J. Zhou, A. Wei, A. Bertsch and P. Renaud, High Precision, High Throughput Generation of Droplets Containing Single Cells, Lab Chip, 2022, 22(24), 4841–4848, 10.1039/d2lc00841f.
- A. T. Giduthuri, S. K. Theodossiou, N. R. Schiele and S. K. Srivastava, Dielectrophoresis as a Tool for Electrophysiological Characterization of Stem Cells, Biophys. Rev., 2020, 1(1), 1–15, DOI:10.1063/5.0025056.
- A. R. Yale, J. L. Nourse, K. R. Lee, S. N. Ahmed, J. Arulmoli, A. Y. L. Jiang, L. P. McDonnell, G. A. Botten, A. P. Lee, E. S. Monuki, M. Demetriou and L. A. Flanagan, Cell Surface N-Glycans Influence Electrophysiological Properties and Fate Potential of Neural Stem Cells, Stem Cell Rep., 2018, 11(4), 869–882, DOI:10.1016/j.stemcr.2018.08.011.
- P. R. C. Gascoyne and S. Shim, Isolation of Circulating Tumor Cells by Dielectrophoresis, Cancers, 2014, 6(1), 545–579, DOI:10.3390/cancers6010545.
- A. Y. L. Jiang, A. R. Yale, M. Aghaamoo, D. H. Lee, A. P. Lee, T. N. G. Adams and L. A. Flanagan, High-Throughput Continuous Dielectrophoretic Separation of Neural Stem Cells, Biomicrofluidics, 2019, 13(6), 064111, DOI:10.1063/1.5128797.
- D. Lee, D. Kim, Y. Kim, K. Park, E. Oh, Y. Kim and B. Kim, A Negative Dielectrophoresis and Gravity-Driven Flow-Based High-Throughput and High-Efficiency Cell-Sorting System, J. Lab. Autom., 2014, 19(1), 60–74, DOI:10.1177/2211068213498385.
- D. Lee, B. Hwang, Y. Choi and B. Kim, Sensors and Actuators A : Physical A Novel Dielectrophoresis Activated Cell Sorter (DACS) to Evaluate the Apoptotic Rate of K562 Cells Treated with Arsenic Trioxide (As2O3), Sens. Actuators, A, 2016, 242, 1–8, DOI:10.1016/j.sna.2016.02.034.
- X. Nie, Y. Luo, P. Shen, C. Han, D. Yu and X. Xing, High-Throughput Dielectrophoretic Cell Sorting Assisted by Cell Sliding on Scalable Electrode Tracks Made of Conducting-PDMS, Sens. Actuators, B, 2021, 327, 128873, DOI:10.1016/j.snb.2020.128873.
- Y. Wu, Y. Ren, Y. Tao, L. Hou and H. Jiang, High-Throughput Separation, Trapping, and Manipulation of Single Cells and Particles by Combined Dielectrophoresis at a Bipolar Electrode Array, Anal. Chem., 2018, 90(19), 11461–11469, DOI:10.1021/acs.analchem.8b02628.
- H. Song, J. M. Rosano, Y. Wang, C. J. Garson, B. Prabhakarpandian, K. Pant, G. J. Klarmann, A. Perantoni, L. M. Alvarez and E. Lai, Continuous-Flow Sorting of Stem Cells and Differentiation Products Based on Dielectrophoresis, Lab Chip, 2015, 15(5), 1320–1328, 10.1039/c4lc01253d.
- S. H. Kim and T. Fujii, Efficient Analysis of a Small Number of Cancer Cells at the Single-Cell Level Using an Electroactive Double-Well Array, Lab Chip, 2016, 16(13), 2440–2449, 10.1039/c6lc00241b.
- M. Li and R. K. Anand, Integration of Marker-Free Selection of Single Cells at a Wireless Electrode Array with Parallel Fluidic Isolation and Electrical Lysis, Chem. Sci., 2019, 10(5), 1506–1513, 10.1039/c8sc04804e.
- M. J. Clark, H. J. Moser and R. K. Anand, Dielectrophoretic Capture and Electrochemical Enzyme-Linked Immunosorbent Assay of Single Melanoma Cells at an Array of Interlocked Spiral Bipolar Electrodes, ChemElectroChem, 2024, 11(15), e202400182, DOI:10.1002/celc.202400182.
- J. T. Banovetz, S. Manimaran, B. T. Schelske and R. K. Anand, Parallel Dielectrophoretic Capture, Isolation, and Electrical Lysis of Individual Breast Cancer Cells to Assess Variability in Enzymatic Activity, Anal. Chem., 2023, 95(20), 7880–7887, DOI:10.1021/acs.analchem.3c00078.
- H. Chen, J. L. Anderson and R. K. Anand, Electropolymerization of Pyrrole-Based Ionic Liquids on Selected Wireless Bipolar Electrodes, ACS Appl. Mater. Interfaces, 2022, 14(16), 18087–18096, DOI:10.1021/acsami.1c25230.
- T. N. Rathnaweera and R. K. Anand, IDEP-Based Single-Cell Isolation in a Twodimensional Array of Chambers Addressed by Easyto-Align Wireless Electrodes†, Lab Chip, 2025, 25(Dld), 1600–1610, 10.1039/d4lc00976b.
- A. T. Ciftlik and M. A. M. Gijs, Demonstration of Inertial Focusing in Straight Microfluidic Channels with High Reynolds Numbers up to Turbulence Onset, 2013 Transducers and Eurosensors XXVII: The 17th International Conference on Solid-State Sensors, Actuators and Microsystems, TRANSDUCERS and EUROSENSORS 2013, 2013, No. June, pp. 1468–1471, DOI:10.1109/Transducers.2013.6627057.
- E. Keinan, E. Ezra and Y. Nahmias, Opposing Shear-Induced Forces Dominate Inertial Focusing in Curved Channels and High Reynolds Numbers, Appl. Phys. Lett., 2015, 107(19), 193507, DOI:10.1063/1.4935466.
- X. Xuan, J. Zhu and C. Church, Particle Focusing in Microfluidic Devices, Microfluid. Nanofluid., 2010, 9(1), 1–16, DOI:10.1007/s10404-010-0602-7.
- W. Zhao, W. Wu, D. Yuan, S. Zou, F. Yang, Q. Zhao, K. Mehmood and B. Zhang, Experimental Exploration of Stable Expansion Phenomenon of Sheath Flow in Viscous Microfluidics, Phys. Fluids, 2022, 34(12), 122002, DOI:10.1063/5.0129764.
- H. Dong, L. Huang and L. Zhao, Influence of the Internal Structure of Straight Microchannels on Inertial Transport Behavior of Particles, Heliyon, 2024, 10(8), e29577, DOI:10.1016/j.heliyon.2024.e29577.
- J.-S. Park, S.-H. Song and H.-I. Jung, Continuous Focusing of Microparticles Using Inertial Lift Force and Vorticity via Multi-Orifice Microfluidic Channels, Lab Chip, 2009, 9(7), 939–948, 10.1039/B813952K.
- G. R. Lazaro, A. Hernandez-Machado and I. Pagonabarraga, Rheology of Red Blood Cells under Flow in Highly Confined Microchannels. II. Effect of Focusing and Confinement, Soft Matter, 2014, 10(37), 7207–7217, 10.1039/c4sm01382d.
| This journal is © The Royal Society of Chemistry 2026 |