 Open Access Article
Open Access ArticleLymphatics-on-a-chip microphysiological system: engineering lymphatic structure and function in vitro
Yansong Peng and
Esak Lee
and
Esak Lee *
*
Nancy E. and Peter C. Meinig School of Biomedical Engineering, Cornell University, Ithaca, NY 14853, USA. E-mail: el767@cornell.edu
First published on 7th January 2026
Abstract
The lymphatic system—integral to fluid balance, immune surveillance, and lipid absorption—is frequently overlooked despite its vital roles. Traditional research modalities, including static two-dimensional cultures and animal models, have illuminated key molecular and cellular features but fall short in recapitulating human lymphatic function, due to limited physiological relevance, throughput, and mechanobiological complexity. Recent advances in microfluidic organ-on-a-chip systems offer biomimetic platforms that integrate three-dimensional architecture, fluid flow, and biomechanical stimuli alongside human lymphatic endothelial and supporting cells. These lymphatics-on-a-chip constructs faithfully reproduce dynamic behaviors such as fluid drainage, junction remodeling, and cell trafficking under physiological and pathological responses. This review highlights the foundational lymphatic biology and engineering principles behind these devices, their capacity for disease modeling and drug testing, and their potential to drive future innovation through induced pluripotent stem cell integration, organ-specific customization, and computational modeling. Merging bioengineering, cell biology, and machine learning, lymphatic microphysiological systems stand poised to significantly expand our understanding and treatment of lymphatic-related disorders.
1 Introduction
Lymphatic vessels play critical roles in maintaining fluid balance, transporting dietary fats, and orchestrating immune cell trafficking.1,2 Unlike the blood vasculature, which historically received more attention, the lymphatic system's roles in health and disease have only recently come to the fore. Indeed, an NIH workshop in 2022 highlighted lymphatics as “yet to be charted” and identified critical gaps in our understanding of lymphatic development, function, and pathology.2 These thin-walled vessels were once viewed mainly as passive drains, but modern research reveals them as active regulators at the interface of tissue homeostasis, immune surveillance, and disease progression.3,4 It is now recognized that lymphatic dysfunction underlies a broad spectrum of diseases beyond classical lymphedema—including autoimmune diseases,5–9 obesity,10–14 cardiovascular diseases,15–17 neurological disorders,18–20 and cancer.21,22Existing experimental models for lymphatic biology include traditional two-dimensional (2D) LEC monolayers and in vivo animal systems such as mouse or zebrafish models. While these platforms have revealed many aspects of lymphatic development and physiology, they each carry limitations: 2D cultures lack vessel-level architecture, fluid flow, and matrix anchoring; animal models, though physiologically complete, are species-specific, lower-throughput, and less amenable to mechanical or biochemical control. These limitations motivate the development of organ-on-a-chip models of the lymphatic system, which can recapitulate the unique structure and biomechanical context of lymphatic vessels in vitro.23 Traditional cell culture often falls short in capturing the biophysical environment and discontinuous cell–cell junctions characteristic of lymphatics. By leveraging microfluidic technology and biomaterials, lymphatics-on-a-chip platforms aim to provide more physiologically relevant models for basic research and translational applications. Such systems offer precise control over fluid flow, pressure, and biochemical gradients, enabling researchers to probe lymphatic functions under well-defined conditions.24 Moreover, integrating lymphatic vessels on chip with other tissue compartments or immune components opens new avenues to study interstitial transport, immunology, and cancer metastasis in a controlled setting. Ultimately, lymphatic microphysiological models are poised to accelerate the discovery of lymphatic-specific therapeutics and personalized medicine approaches for lymphatic-related pathologies.
This review provides a comprehensive overview of the emerging field of lymphatics-on-a-chip. We first summarize foundational principles of lymphatic biology (i.e., developmental origins, unique structural features, and mechanobiological hallmarks). We then discuss microfluidic and biomimetic engineering strategies to model lymphatics in vitro, including device designs, biomaterials, and flow control. Next, we survey applications of lymphatic chips in modeling diseases such as lymphedema, cancer, autoimmune diseases, and neurological disorders. Finally, we outline future directions, including induced pluripotent stem cell (iPSC)-derived lymphatic organoids, multi-scale coupling of lymphatics with other organ systems, and AI-driven design and analysis of lymphatic networks. In this review, we synthesize findings across studies to provide a cohesive understanding of how lymphatics-on-a-chip technology advances lymphatic research.
2 Foundational principles of lymphatic biology
Seminal discoveries over the past two decades have redefined our understanding of lymphatic biology. In the early 2000s, a series of studies demonstrated that tumors exploit lymphangiogenesis to facilitate metastasis: overexpression of vascular endothelial growth factor C/D (VEGF-C/D) in mouse models induced new lymphatic growth and increased tumor spread to lymph nodes.25–28 Clinical data support this link in humans, showing that higher tumor-associated lymphatic vessel density (LVD) correlates with increased lymph node metastasis and poorer survival in solid tumors.29–32 Blocking VEGF-C or vascular endothelial growth factor receptor 3 (VEGFR-3) could suppress lymphatic metastasis in mice, establishing these pathways as promising therapeutic targets. Such findings, along with the cloning of prospero homeobox 1 (PROX1) and identifying the first lymphatic-specific markers—LYVE-1, podoplanin (PDPN), and VEGFR-3 (FLT4)—enabled definitive discrimination between lymphatic and blood endothelial cells. These breakthroughs transformed lymphatic research from descriptive anatomy to molecular and functional biology, paving the way for mechanistic and engineering studies. Lymphatics-on-a-chip technology arises from this foundation, aiming to integrate these molecular pathways and cellular behaviors into microengineered platforms that enable controlled, quantitative exploration of lymphatic development, transport, and disease.2.1 Developmental origins and tissue-specific heterogeneity of lymphatic vessels
Lymphatic endothelial cells (LECs) share core endothelial markers such as VE-cadherin and CD31, yet express distinct molecules including LYVE-1, podoplanin, and the transcription factor PROX1, which together define lymphatic identity.33 Classical studies established that mammalian lymphatics arise primarily from PROX1+ endothelial cells sprouting from the cardinal veins.28 More recent lineage-tracing and single-cell analyses, however, reveal additional non-venous sources, including mesenchymal angioblasts in the dermomyotome, mesentery, heart, and kidney, that differentiate directly into LECs through VEGF-C/VEGFR-3 and SOX18/COUP-TFII signaling.33 These findings support a model in which lymphatic vasculogenesis complements venous lymphangiogenesis, contributing to regional diversity. Regardless of origin, PROX1 acts as the master regulator of LEC fate, activating downstream genes (VEGFR-3, LYVE-1, podoplanin) and suppressing the blood endothelial program.34 In mice, loss of Prox1 abolishes lymphatic formation, while mutations in VEGF-C/VEGFR-3 cause primary lymphedema in humans.3,35Recognizing this developmental heterogeneity is important for organ-specific lymphatic modeling on chip, where different progenitor programs and signaling environments may need to be recapitulated to mimic lymphatics across tissues.36 For example, dermal lymphatics form dense, blind-ended capillary networks with permeable button-like junctions that facilitate immune cell trafficking and interstitial fluid drainage.37 Intestinal lacteals located within villi, on the other hand, exhibit dynamic zipper-to-button transitions during lipid absorption, a process tightly regulated by VEGF-A/VEGFR-2 and Notch signaling.38,39 Meningeal lymphatics, which develop postnatally along the dural sinuses and skull base, connect to deep cervical lymph nodes and the glymphatic pathway to drain cerebrospinal fluid and metabolic waste from the brain, linking the central nervous system to systemic immunity.40,41 These diverse architectures underscore how developmental origin and local microenvironment shape lymphatic structure and function. Reproducing such regional diversity, through appropriate matrix stiffness, junctional morphology, and flow regimes, is central to engineering organ-specific lymphatics-on-a-chip that accurately model physiological and pathological lymphatic functions.
2.2 Architectural and functional specialization of lymphatic vessels
The lymphatic vasculature forms a one-way transport network that drains interstitial fluid and cells from tissues back into the blood circulation (Fig. 1).3 The system begins with blind-ended initial lymphatic capillaries, which are composed of a single layer of oak leaf-shaped LECs connected by discontinuous, “button-like” junctions.42,43 These button junctions and overlapping endothelial flaps act as primary valves that allow fluid and immune cell entry from the interstitium.43 Anchoring filaments tether LECs to the extracellular matrix; when interstitial pressure rises, the filaments pull the flaps open, thereby enhancing fluid uptake.3 As these capillaries converge into larger collecting lymphatic vessels, the endothelial junctions transition to continuous “zipper-like” junctions, with fewer gaps that reduce permeability.3,42 Collecting lymphatics also feature intraluminal valves and are ensheathed by lymphatic muscle cells (LMCs), which generate rhythmic contractions to propel lymph forward. This hierarchical organization ensures efficient absorption from tissues and unidirectional transport through lymph nodes—where immune filtering occurs—before lymph empties into the subclavian veins.3 | ||
| Fig. 1 Architectural organization and molecular signatures of lymphatic vessels. The lymphatic vasculature is organized into blind-ended initial lymphatic capillaries, transitional pre-collecting vessels, and contractile collecting lymphatic vessels with intraluminal valves. Initial lymphatics consist of oak leaf-shaped LECs connected by discontinuous button-like junctions (blue: LYVE-1; red: VE-cad) that permit uptake of interstitial fluid, macromolecules, lipids, and immune cells. Anchoring filaments tether LECs to the extracellular matrix, enabling pressure-driven flap opening. Pre-collecting and collecting vessels progressively acquire continuous zipper-like junctions (blue: LYVE-1; red: VE-cad), basement membrane, smooth muscle coverage, and valves that maintain unidirectional flow. Marker expression distinguishes vessel compartments: LYVE-1, VEGFR3, and CCL21 highlight initial lymphatics, while FOXC2 and integrin-α9 are enriched in collecting vessels and valve-forming LECs. This compartmentalization underlies the dual role of lymphatics in tissue fluid absorption and long-distance lymph transport. Adapted with permission from Yijie Hua. Inset panels adapted with permission from ref. 47, copyright 2025 Springer Nature. Created with https://BioRender.com. | ||
This structural design underpins the diverse physiological functions of the lymphatic system. First, lymphatics serve as the body's fluid homeostasis system, continuously returning plasma ultrafiltrate that leaks from blood capillaries. In humans, this amounts to an estimated ∼2–4 liters of fluid per day being returned to the venous circulation under steady-state conditions, preventing edema.1,44 Second, lymphatics provide a specialized route for lipid absorption. In the intestine, lacteals (i.e., specialized lymphatic capillaries in the villi) absorb chylomicrons (i.e., dietary lipid-laden lipoproteins) and transport them via the thoracic duct to the bloodstream.14 In addition, recent studies highlight a role for lymphatics in cholesterol clearance, where interstitial high-density lipoproteins (HDLs) are ferried back to the circulation, reducing atherosclerotic risk.15
Beyond fluid and lipid transport, lymphatic vessels are increasingly recognized as active players in immunity. LECs constitutively secrete CC chemokine ligand 21 (CCL21), which guides CC Chemokine receptor 7-positive (CCR7+) immune cells, including dendritic cells and T cells, into initial lymphatics through button junction gaps.45 Antigens, cytokines, and immune cells entering lymphatics are carried to lymph nodes, where adaptive immune responses are initiated. Moreover, LECs themselves can regulate immunity: they can present antigens directly, modulate their access to lymph nodes, or express checkpoint molecules (e.g., programmed death-ligand 1 (PD-L1)) that influence T-cell activation and tolerance.4,46 Thus, lymphatic vessels are far more than passive conduits—they actively coordinate immune surveillance, tolerance, and inflammation.
2.3 Mechanobiological regulation of lymphatic junctions and valve development
Lymphatic vessels are exquisitely sensitive to mechanical forces within their environment. Fluid shear stress from lymph flow, cyclic stretch caused by vessel pumping, and matrix tethering forces collectively shape LEC phenotypes (Fig. 2). A key outcome of these stimuli is the regulation of intercellular junctions in initial lymphatics, which control permeability and determine whether vessels adopt specialized button- or zipper-like configurations, as well as valvulogenesis in collecting lymphatics, which guide the unidirectional lymph flow. Understanding these mechanobiological mechanisms is essential to decipher how lymphatics maintain homeostasis and how perturbations such as inflammation, hypertension, or exercise drive lymphatic remodeling or dysfunction.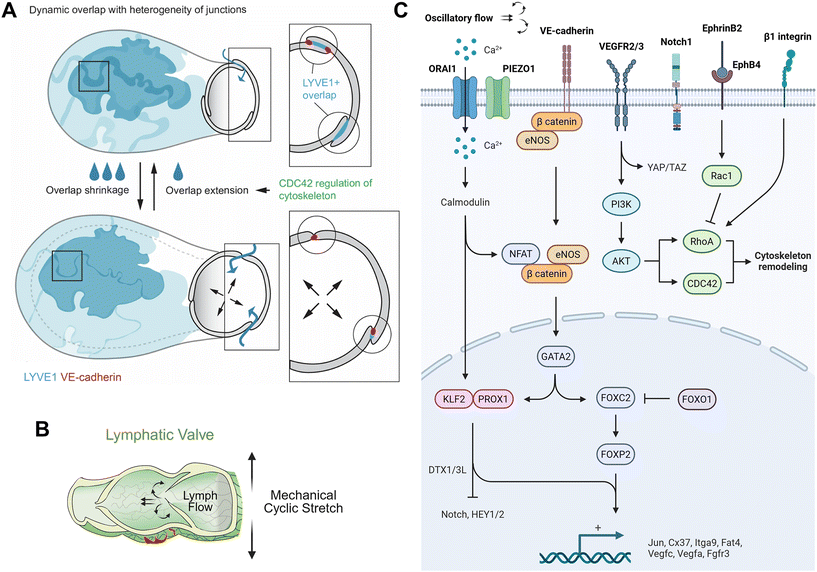 | ||
| Fig. 2 Mechanobiological regulation of lymphatic junction dynamics, shear sensing, and valve development. (A) Proposed model of lymphatic capillary function driven by dynamic remodeling of LEC overlaps. Puzzle-shaped capillary LECs exhibit LYVE1+ cellular overlaps with diverse junctional configurations. Increased interstitial fluid shortens overlaps to permit lumen expansion during uptake, countered by CDC42-mediated actin remodeling that extends lamellipodia-like overlaps to maintain monolayer integrity and barrier strength. This supports a bellows-like mechanism in which overlap extension compresses the vessel and aids fluid propulsion. Adapted with permission from ref. 47, copyright 2025 Springer Nature. (B) Lymph flow maintains expression of mechanosensitive transcription factors FOXC2, GATA2, and NFATc1 and preserves collecting vessel and valve function. Adapted with permission from ref. 40, copyright 2020 American Association for the Advancement of Science. (C) Mechanosensory pathways regulating junction remodeling and valve specification. LECs sense shear stress through a junctional mechanosensory complex (VE-cadherin, VEGFR2, VEGFR3) that activates PI3K-Akt, cytoskeletal remodeling, and YAP/TAZ signaling; and the stretch-activated ion channel PIEZO1, which engages the calcium channel ORAI1. The resulting calcium influx activates calmodulin, which complexes with KLF2 to regulate transcriptional programs. Together, these pathways control key regulators of lymphatic identity and valve morphogenesis, including PROX1, FOXC2, GATA2, NFATc1, Cx37, integrin-α9, FAT4, and E3 ubiquitin ligases DTX1/3L. Adapted with permission from ref. 63, copyright 2023 Springer Nature. Created with https://BioRender.com. | ||
Developmental specialization is reinforced by multiple signaling pathways that stabilize junction architecture. Jun proto-oncogene (Jun) and EphrinB2-EphB4 signaling provide tonic junctional control in lymphatic capillaries and collectors, respectively.48,49 Fu et al. (2023) demonstrated that mechanical pressure at birth induces c-JUN expression in LECs, driving actin cytoskeletal remodeling that enables the opening of neonatal lung lymphatics; loss of c-JUN prevents vessel opening, causes fluid retention, and results in neonatal lethality.48 Likewise, Frye et al. (2020) showed that disruption of EphrinB2-EphB4 signaling destabilizes VE-cadherin junctions, increases actomyosin contractility, and induces lymphatic leakage, effects not observed in blood vessels, while EphrinB2-Fc supplementation restores junctional integrity.49
Beyond developmental regulation, junctions in initial lymphatic capillaries exhibit pronounced mechanical plasticity. These vessels undergo continuous distension and recoil as they fill and empty. Schoofs et al. (2025) used intravital microscopy and on-chip stretch assays to show that cyclic strain drives reversible changes in LEC overlap. During vessel expansion, overlapping flaps partially retract but are reinforced by Cell division control protein 42 homolog (CDC42)-dependent actin protrusions reminiscent of “puzzle-piece” plant cells. On-chip cyclic stretch (∼12% strain at 0.01 Hz) reproduced these dynamics, whereas CDC42 inhibition yielded fragile, leaky junctions in vivo and in vitro. These observations support a “bellows” model in which LEC overlaps narrow during filling and re-expand during emptying, enabling a barrier that is both resilient and permissive to fluid entry (Fig. 2A).47
Junctional plasticity is also shaped by inflammatory and growth factor signaling. Cromer et al. (2014) reported that cytokines such as TNF-α, IL-6, IL-1β, and IFN-γ substantially increase LEC permeability, largely through nitric oxide-dependent VE-cadherin destabilization and actomyosin contraction.50 With prolonged inflammation, these signals can drive reversible transitions between button-like and zipper-like configurations. Seminal work by Baluk et al. (2007) and Yao et al. (2012) demonstrated that button junctions in initial lymphatics can convert to zipper-like junctions, reducing fluid uptake capacity, but that dexamethasone treatment restores button prevalence, highlighting the reversibility of this process.51,52
Together, these findings indicate that lymphatic junction organization is defined by both developmental specialization and post-developmental plasticity, with mechanical forces and inflammatory cues acting synergistically to remodel junctions—often at the expense of drainage efficiency.
Mechanobiology studies collectively demonstrate that lymphatic junctions are dynamic, mechano-responsive structures. Shear stress activates PROX1–KLF2 pathways to downregulate Notch and promote sprouting, stretch engages CDC42-driven actin remodeling to preserve junctional overlap, and inflammatory cues synergize with mechanical load to remodel button/zipper junctions. At valves, piezo1 and FOXC2 translate oscillatory flow into structural stability. These insights highlight multiple druggable targets to modulate lymphatic function and underscore the value of lymphatics-on-a-chip platforms for applying controlled mechanical environments. By bridging in vivo observations with tunable microengineered models, the field is now positioned to dissect mechanotransduction pathways with unprecedented precision. Table 1 summarizes some key physiological values in establishing lymphatic vasculature. It is important to emphasize that most of the available data derives from animal or ex vivo systems. Quantitative, in vivo measurements in human lymphatic vessels remain extremely limited, so translation to human-on-a-chip models should rely on dimensionless analysis and scaling rather than assuming direct equivalence. Future work is required to establish normative values in human lymphatics and validate mechanobiological findings in human-relevant systems.
| Parameter | In vivo values | Species/tissues | Ref. |
|---|---|---|---|
| Interstitial fluid pressure | Subatmospheric (−1 to −3 mmHg) in loose tissues | Various species and tissues | 66–68 |
| Interstitial fluid flow velocity | 0.1 to 1.0 μm s−1 | Rabbit dermis | 69, 70 |
| Hydraulic pressure (initial lymphatics) | Similar to interstitial pressure (−6 to 4 mmHg) | Rabbit and rat diaphragm, cat mesentery | 71–74 |
| Hydraulic pressure (collecting lymphatics) | 5 to 10 cmH2O (4 to 7 mmHg) diastolic pressure during relaxation | Rat mesentery | 75, 76 |
| Interstitial colloid osmotic pressure | 8 to 17 mmHg | Human and pig various tissues | 77, 78 |
| Lymphatic colloid osmotic pressure | 5 to 15 mmHg | Rat tail and dog hindleg prenodal lymph | 79, 80 |
| Lymph flow velocity (initial lymphatics) | 0 to 29 μm s−1 | Mouse and human dermal tissues | 81, 82 |
| Lymph flow velocity (collecting lymphatics) | 40 to 70 μm s−1 in mouse; 0.7 to 1 mm s−1 in rat; up to 9 mm s−1 during phasic contractions in rat | Mouse hindlimb, rat mesentery | 83, 84 |
| Volumetric lymph flow rate (single vessel) | 0.5 to 2.5 μL h−1 in mouse; 9 to 19 μL h−1 in rat | Mouse hindlimb, rat mesentery | 83, 84 |
| Volumetric lymph flow rate (under stress) | Increases ∼6× under acute fluid load: ∼0.1 μL min−1 baseline to ∼0.6 μL min−1 | Rat mesentery (acute edema state) | 85 |
| Total lymph flow (whole body) | 3 to 4 L per day in an average adult at rest; can increase several-fold with exercise | Pig thoracic duct output | 86 |
| Wall shear stress (initial lymphatics) | Extremely low, 0.01 to 0.06 dyne per cm2 | Mouse dermis | 87, 88 |
| Wall shear stress (collecting lymphatics) | 0.5 to 0.8 dyne per cm2 during steady flow; pulsatile peaks 4 to 12 dyne per cm2 due to lymphatic contractions | Rat mesentery | 83 |
| Wall shear stress (edema) | Time averaged 1.5 dyne per cm2; 5 to 40 dyne per cm2 during strong flow pulses | Rat mesentery (acute edema state) | 85 |
| Intrinsic pumping frequency | 0.08 to 0.27 Hz (5 to 19 contractions per minute) | Rat various tissues | 89, 90 |
| Maximal pumping pressure (occlusion pressure) | 80 to 100 mmHg | Human dermis | 91 |
| Lymphatic junction overlap (buttons) | 0.5 to 3.5 μm (width per flap); 70 to 175 μm2 (area between flaps) | Mouse ear and trachea | 47, 51 |
| Lymphatic capillary diameter | 30 to 70 μm | Rat mesentery and human dermis | 82, 92 |
| Lymphatic collector diameter | 60 to 120 μm | Rat mesentery | 89, 93, 94 |
| Hydraulic conductivity | 17.3 × 10−7 cm s−1 cmH2O−1 | Mouse mesentery | 95 |
3 Microfluidic and biomimetic engineering strategies for lymphatic systems
Early efforts in this field have drawn inspiration from well-established blood vessel-on-chip models, adapting them to the unique features of lymphatics.96,97 However, lymphatic vessels differ from blood vessels in essential ways, including their developmental origins, ultrastructure (e.g., specialized cell–cell junctions and valves), lower shear stress environments, and function as unidirectional absorbent conduits.1,98 Over the past decade, researchers have made significant strides in capturing these distinctions (Fig. 3), including (1) engineering lymphatic capillary networks in vitro via lymphangiogenesis99–101 (i.e., sprouting from pre-existing vessels) or vasculogenesis102–105 (i.e., self-assembly from dispersed cells); (2) reproducing lymphatic vessel structure106–110—notably the unique discontinuous “button-like” junctions of initial lymphatics and mural cell coverage of collecting lymphatics; and (3) applying biomechanical forces (e.g., interstitial flow, shear, stretch) to mimic lymphatic fluid dynamics.108,111–113 | ||
| Fig. 3 Microfluidic and biomimetic engineering strategies for reconstituting lymphatic structure and function in vitro. (A) Microfluidic device for reconstitution of 3D sprouting lymphangiogenesis in vitro. Adapted with permission from ref. 99, copyright 2016 Elsevier. (B) Schematic illustration and fabrication process of engineered lymphatic and vascular structures to study cooperative effects of vascular angiogenesis and lymphangiogenesis. Adapted with permission from ref. 100, copyright 2018 Springer Nature. (C) Graphical representation of granular hydrogel composites fabrication and lymphatic sprouting assays, with 3D rendering of V180s gel (green—FITC) exhibiting lymphatic capillary network formation (red—F-actin). Adapted with permission from ref. 105, copyright 2025 Wiley-VCH. (D) An engineering drawing of the lymphangion-chip and a representative confocal image set of on-chip LMCs (green: F-actin) and LECs (green: VE-cadherin) under co-culture conditions. Scale bar: 50 μm. Adapted with permission from ref. 107, copyright 2022 Royal Society of Chemistry. (E) Mμlti-flow platform design and device structure. Adapted with permission from ref. 112, copyright 2022 IOP Publishing. (F) An engineered 3D lymphatic vessel-on-chip model to investigate luminal and interstitial flow effects with VEGF-A or VEGF-C. Adapted with permission from ref. 108, copyright 2023 Springer Nature. | ||
3.1 Lymphangiogenesis-based engineering of perfusable lymphatic capillaries
Noo Li Jeon and colleagues pioneered a biomimetic lymphangiogenesis-on-chip model that closely mimics in vivo sprouting. Kim et al. (2016)99 described a 3D microfluidic platform integrating key elements of the lymphatic niche—biochemical gradients, interstitial flow, and stromal cell interactions—to induce robust sprouting of LECs into a fibrin matrix (Fig. 3A). In their device, a lumen lined by LECs was formed adjacent to a tissue chamber containing fibroblasts, with a gradient of pro-lymphangiogenic factors (e.g., VEGF-C) established across the matrix. Under these conditions, LECs sprouted from the vessel and invaded the matrix, forming multicellular capillary projections. Notably, a slow interstitial flow was applied from the tissue side toward the lymphatic channel to emulate fluid drainage. This interstitial flow proved a crucial regulator: it significantly enhanced the initiation and directional bias of lymphatic sprouting. LEC sprouts preferentially extended upstream against the flow, consistent with how tissue fluid flow in vivo can guide lymphangiogenesis towards the source of drainage. With the right synergy of flow and growth factors, the in vitro sprouts displayed hallmarks of native lymphatic vessels, including migratory tip cells and lumen formation, and responded appropriately to pro- and anti-lymphatic stimuli. This model demonstrated for the first time the feasibility of reconstituting sprouting lymphangiogenesis in vitro in a controlled manner, highlighting that a combination of VEGF-C and interstitial flow is key to inducing robust, directional lymphatic sprouting.99 Osaki et al. (2018)100 extended this synergy concept to a dual vascular model (Fig. 3B). Using sacrificial molding, they created a microdevice embedding both lymphatic vessels and blood vessels—600 μm channels seeded with human LECs and HUVECs, respectively—and induced angiogenesis and lymphangiogenesis via perfusion combined with VEGF-A or VEGF-C and phorbol 12-myristate 13-acetate (PMA). Remarkably, they observed that vascular angiogenesis facilitated lymphatic sprouting through local matrix metalloproteinase (MMP) secretion by endothelial cells. Specifically, a VEGFR-3 inhibitor blocked lymphangiogenesis in the absence of BVs, but not when BVs were present, indicating that blood-vascular-derived MMPs can compensate for lymphatic angiogenic signaling—highlighting crosstalk between the two vascular beds.100 Building on this, Jain and colleagues (2024) recently reported a lymphangiogenesis-on-chip system examining the independent and combined effects of biochemical gradients and interstitial flow. In their study, LECs were subjected to varying conditions: VEGF-C gradients alone, interstitial flow alone, or both together. They found that each factor could stimulate some degree of sprouting, but the combination produced the most extensive and physiologically oriented networks (with flow directing the sprouts).101 This confirms a synergistic paradigm: slow steady flow (∼1–5 μm s−1) synergizes with VEGF-C signaling to amplify lymphatic sprouting and helps define the polarity of growth (i.e., which way sprouts grow). Mechanistically, interstitial flow appears to increase the availability and cell uptake of matrix-bound VEGF-C, as well as to upregulate LEC responsiveness to growth factors. Moreover, flow can work in concert with other biochemical cues; for example, gradients of sphingosine-1-phosphate (S1P)—absent in interstitium but present in lymph—were shown to cooperate with flow in guiding LEC migration and sprouting.63,101 These findings underscore that engineered lymphatic sprouting requires orchestrating multiple biochemical and biophysical cues, much like during developmental lymphangiogenesis.3.2 Matrix and scaffold engineering for lymphatic vasculogenesis
The choice of extracellular matrix (ECM) environment is another critical design parameter. LECs generally require a 3D matrix (e.g., collagen or fibrin) to form tubular structures. Matrix stiffness, porosity, and adhesive ligand composition all influence lymphatic morphogenesis.114,115 Hanjaya-Putra's group has conducted detailed studies on how matrix stiffness “primes” lymphatic capillary formation. In a 2D culture, they observed that softer substrates promoted LEC elongation and sprout-like behavior in response to VEGF-C, whereas stiffer substrates did not—hinting that compliance might be necessary.116 They then moved to 3D hydrogels: Alderfer et al. (2021) showed that a relatively soft hydrogel (with shear modulus on the order of a few hundred Pascals) was optimal for LEC tube formation when VEGF-C was provided, whereas higher stiffness (>1 kPa) markedly impaired sprouting.116 In a follow-up study, Alderfer et al. (2024) developed a synthetic modular hydrogel (i.e., norbornene-functionalized hyaluronic acid) where they could tune stiffness and integrin-binding content independently. They found that a mid-range stiffness (∼500 Pa), including fibronectin-mimetic RGD peptides, yielded robust formation of lumenized lymphatic capillaries in 3D. Softer gels (∼100 Pa) allowed initial capillary assembly but were too weak to support network stability beyond a few days, while stiffer gels (∼1–2 kPa) completely inhibited LEC elongation into tubes. Interestingly, LECs in the stiff hydrogels upregulated matrix-degrading enzymes (MMP-2, MMP-14) and VEGFR3 expression, as if attempting to compensate for the non-permissive environment. However, no stable tubes were formed in the stiff condition even over 3 weeks.104 These results indicate that matrix mechanics can control lymphatic morphogenesis: a compliant matrix “primes” LECs to respond to VEGF-C by forming cords or tubes, whereas a rigid matrix triggers a phenotypic response (i.e., upregulating proteases and VEGFR3) but prevents actual sprout penetration. From an engineering perspective, tuning scaffold stiffness is essential for building lymphatic networks on-chip. It also suggests that the in vivo fibrotic or tumor stroma (often stiffer than healthy tissues) might inherently resist lymphangiogenesis. Adhesive cues are another factor required for LECs to form 3D structures. Alderfer et al. (2024) showed that adding an RGD peptide (which binds integrins α5β1 and αvβ3) to a hydrogel markedly improved LEC sprouting and branch formation. Without RGD, LECs remained mostly round in 3D culture; with an optimal concentration (∼5 mM) of RGD, they formed extensive tube networks. This mirrors earlier observations that fibronectin (an RGD-containing ECM protein) promotes lymphatic endothelial proliferation and tube formation. Moreover, certain integrins like α9β1 are known to be essential for lymphatic valve formation and LEC differentiation, hinting that integrin-specific ECM signals could push LECs toward a lymphatic phenotype.104 In practice, incorporating the right matrix ligand (fibronectin, collagen, laminin, etc.) is vital for successful lymphatic vessel engineering.Granular hydrogels have recently been explored as scaffolds for lymphatic vasculogenesis. Montes et al. (2025) generated granular hydrogels (i.e., jammed microgel suspensions) with tunable porosities to support 3D LEC growth (Fig. 3C). These novel granular hydrogel composites allowed LECs to form capillary-like networks and sprouts in the absence of supporting feeder cells, effectively mimicking the initial lymphatic plexus formation in a synthetic matrix. Interestingly, the microgel fabrication method (pipetting vs. vortexing) produced different pore size distributions that affected lymphatic sprout connectivity and maturation. Vortexed gels, which had tighter packing and smaller pores, led to earlier vessel development but slightly less interconnected networks; by contrast, pipetted gels yielded highly connected lymphatic networks, albeit with delayed sprouting.105 Such materials-driven approaches demonstrate how matrix morphology and stiffness influence lymphatic architecture—insights that can guide tissue engineering of lymphatics and the design of pro-lymphangiogenic/vasculogenic biomaterials.
3.3 Engineering junctional phenotypes and mural cell coverage in lymphatic vessels
Understanding how LECs regulate their intercellular junctions in response to extracellular cues, and how mural cells contribute to this process, is critical for modeling both initial and collecting lymphatic functions. Henderson et al. (2021) pioneered an initial lymphatic vessel model, which allows direct investigation of how ECM composition influences endothelial barrier properties. In this system, human dermal LECs were seeded within a hollow microchannel encased in a 3D collagen hydrogel and cultured under luminal flow. When embedded in pure collagen I, LECs formed relatively permeable junctions, permitting the passage of macromolecules. However, when fibronectin was added to the collagen scaffold, LEC junctions tightened markedly, reducing permeability. Mechanistically, this barrier enhancement was mediated by the activation of integrin α5, either via fibronectin engagement or direct activation of the integrin, highlighting integrin α5 as a key regulator of lymphatic barrier integrity in response to matrix composition.106Building on the theme of multicellular interaction, Selahi et al. (2022) introduced a more structurally faithful model of lymphatic segments—namely, the lymphangion-chip—which expands on barrier regulation by incorporating lymphatic muscle cells (LMCs) alongside LECs (Fig. 3D). Using a novel gravitational lumen patterning (GLP) technique, the researchers fabricated a microfluidic architecture comprising an endothelial channel encased by uniformly thick layers of muscle cells, closely mirroring the cylindrical structure of in vivo lymphangion. Under flow conditions, LECs aligned along the vessel axis while LMCs oriented circumferentially, recapitulating the native organizational pattern viewed in vivo. Crucially, this setup enabled real-time assessment of endothelial-mural cell communication: exposure to pro-inflammatory cytokines produced dynamic changes in LEC barrier function, mediated through LMC interactions.107 The lymphangion-chip thus provides a versatile testbed for investigating lymphatic contractility, inflammation, and barrier dynamics in a physiologically relevant modular format.
3.4 Microfluidic control of shear stress, interstitial flow, and mechanical loading in lymphatics-on-a-chip
Careful modulation of fluid dynamics is a defining feature of lymphatics-on-a-chip systems, since LECs experience markedly lower shear stresses compared to BECs: ∼0.1–2 dyne per cm2 within initial lymphatics, versus 10–20 dyne per cm2 in arterial vessels.57 These cells are also exposed to distinct flow environments—absorbing interstitial fluid from surrounding tissue while simultaneously transporting fluid through the lymphatic lumen. Recapitulating these different shear contexts is therefore essential for physiologically relevant lymphatic models.98 To address this challenge, Lee et al. (2022) developed a multilayered microfluidic device (MμLTI-Flow) that allows simultaneous control of blood, lymphatic, and interstitial fluid pressures to model complex fluid dynamics in tissues (Fig. 3E). The system integrates engineered blood and lymphatic vessels with tunable interstitial flow, enabling direct measurement of fluid velocities and pressure-dependent transport. The authors validate the platform with computational fluid dynamics (CFD) simulations and optical microscopy, showing how luminal, transmural, and interstitial flows are interdependent rather than isolated phenomena.112 Building on this, Ilan et al. (2023) developed a dual-channel lymphatic vessel-on-chip that enables precise and independent control of luminal flow, interstitial flow, both in combination, or static conditions (Fig. 3F). The device was fabricated by embedding two parallel microchannels within a 3D collagen I matrix, with one channel seeded with LECs and the adjacent acellular channel used to pressurize fluid into the surrounding hydrogel. Using this system, the authors demonstrated that interstitial flow in the presence of VEGF-C maintained discontinuous, button-like junctions and minimal sprouting, reflecting the phenotype of quiescent, highly permeable initial lymphatics. In contrast, luminal flow or combined flows in the presence of VEGF-A induced robust sprouting and more continuous, zipper-like junctions, mimicking actively remodeling collecting vessels.108 These results highlight how LECs integrate flow cues differently depending on whether they sense fluid shear from within the lumen or across the vessel wall—an insight uniquely enabled by the two-channel design. It should be noted that VEGF isoform and flow type were varied concurrently in this design; thus, the results reflect the cooperative influence of mechanical and biochemical cues rather than an isolated causal effect of either factor. Future studies that decouple VEGF-A/-C signaling from specific flow regimens will help delineate the relative contribution of VEGFR-mediated signaling versus shear-dependent mechanotransduction in regulating lymphatic junction plasticity.Complementary approaches have sought to reproduce the rhythmic dynamics of collecting lymphatics. Fathi et al. (2020) introduced a pumpless, gravity-driven chip in which periodic tilting of the device generated oscillatory flow through the LEC-lined channel. This design produced physiologically relevant shear stresses on the order of ∼0.9 dyne per cm2 under healthy conditions, while higher tilting rates increased shear to 6–7 dyne per cm2, modeling pathological stress levels. Under low, physiological shear, LECs formed a stable, quiescent endothelium, whereas static conditions triggered pro-inflammatory responses, including elevated TNF-α and IL-8 secretion. Importantly, the cyclical tilting approach also simulated the intrinsic pumping activity of collecting lymphatics, generating 10 second flow pulses multiple times per minute.113 Such platforms reveal how steady versus unsteady flows may elicit divergent effects on LEC biology—a subject of active investigation.
3.5 Engineering limitations and physiological gaps in current lymphatics-on-a-chip models
Although significant progress has been made in engineering lymphatic structure and function in vitro, several defining anatomical and physiological features of native lymphatic vessels remain difficult to reproduce. These gaps highlight the need for continued refinement of lymphatics-on-a-chip microphysiological platforms.Together, these limitations underscore the need for next-generation lymphatic microphysiological systems capable of integrating blind-ended geometry, valve morphogenesis, and authentic junctional specialization. Continued convergence of developmental biology, mechanobiology, and microengineering will be crucial for closing these gaps. Table 2 summarizes the engineering approaches discussed in this section along with their representative features, advantages, and limitations.
| Approach | Representative features | Advantages | Limitations/challenges | Ref. |
|---|---|---|---|---|
| Lymphangiogenesis/sprouting | Guided LEC invasion from preformed vessels into 3D ECM (e.g., collagen, fibrin) driven by interstitial flow or chemokine gradients; often mimics developmental lymphangiogenesis | • Capture early lymphangiogenic morphogenesis • Reconstitute directional growth and tip/stalk cell dynamics • Enable quantification of sprout length and branching • Allow flow conditioning |
• Sprout form continuous, perfusable lumens rather than blunt-ended capillaries • Limited ability to model pressure-driven fluid entry • Require precise biochemical gradient control and matrix tuning • Typically limited to short-term culture (<1 week) |
99, 101 |
| Vasculogenesis/self-assembly | Dispersed LECs seeded within hydrogels self-organize into interconnected networks; may include stromal or perivascular support cells | • Models multicellular assembly • Support heterotypic cell interactions and spontaneous network and lumen formation • Compatible with long-term culture • Flexible for organoid co-culture |
• Self-assembled networks rarely develop blind-ended termini • Authentic “oak-leaf” morphology and alternating VE-cadherin/LYVE-1 segments not reliably reproduced • Lymphangion- and valve-level organization absent • Less geometric and directional control • High heterogeneity in vessel diameter and connectivity |
104, 105, 116 |
| Flow-controlled/perfusable microchannels | LECs line predefined microchannels exposed to luminal or interstitial flow; independent control of flow direction and shear stress | • Enable direct interrogation of mechanobiology (junction remodeling, permeability, valve-like behavior) • Tunable flow environments • Support dynamic perfusion and real-time imaging |
• Cannot induce morphogenesis of valve-like structures • Button-like junction markers only partially reproduced • Complex fabrication and device variability • Often lack early morphogenetic events |
106, 108, 112, 113, 117 |
| Hybrid co-culture systems | Combine lymphangiogenic sprouting with flow conditioning or stromal/immune components | • Capture multicellular crosstalk and adaptive remodeling under physiological cues • Enable modeling of lymphangiocrine signaling • Improved tissue specificity • Suitable for disease modeling |
• Organ-specific lymphatic phenotypes remain incomplete • Greater experimental complexity and variability • Junctional phenotype sensitive to LEC passage number and source • Require advanced imaging and analysis workflows |
100, 107, 118 |
In summary, an expanding toolkit of microengineering strategies—ranging from 3D co-culture approaches to precisely controlled low-shear perfusion—has been integrated to construct lymphatic microphysiological systems. Each technique addresses a distinct facet of lymphatic biology, and in many cases, multiple strategies are combined within a single platform to enhance physiological relevance. Over the past decade, the field has progressed rapidly: what began as simple 2D LEC monolayers exposed to flow88,97,119 has evolved into sophisticated 3D vessel models that closely approximate native lymphatic architecture and function for preclinical drug screening.117,118 As the following sections will explore, these biomimetic lymphatics-on-a-chip platforms not only recapitulate fundamental aspects of lymphatic physiology but also serve as versatile tools for probing disease mechanisms and exploring therapeutics.
4 Disease applications of lymphatics-on-a-chip
Lymphatic dysfunction is implicated in a wide array of diseases, from lymphedema and inflammatory disorders to cancer metastasis and metabolic syndromes. By recreating key aspects of lymphatic physiology in vitro, lymphatics-on-a-chip devices have emerged as powerful tools to model these pathologies and test potential interventions. In this section, we highlight how microfluidic lymphatic models have been used to simulate disease conditions, uncover disease mechanisms, and evaluate therapeutics (Fig. 4). | ||
| Fig. 4 Lymphatics-on-a-chip platforms for modeling lymphatic pathophysiology. (A) A schematic of the lymphatic drainage-on-chip platform. Immunostaining of lymphatic vessels and blood vessels with an adherens junction marker, VE-cadherin (VE-cad), and CD31. Red and yellow arrows indicate exclusive expression of VE-cad and CD31, respectively, showing interdigitated, discontinuous expression of VE-cad in LECs. Scale bars, 100 μm. Adapted with permission from ref. 110, copyright 2023 National Academy of Sciences. (B) TME-on-a-chip reveals that the CCL21(T)-B3 fusion construct effectively targets the tumor cells. Scale bar: 10 μm. Adapted with permission from ref. 125, copyright 2019 American Chemical Society. (C) A schematic of the breast tumor–lymphatic microfluidic device. Adapted with permission from ref. 126, copyright 2020 Wiley-VCH. (D) Illustration of an injection-molded microfluidic device designed in a 384-well plate format. Each unit contains a central channel (C) and two side channels (S1 and S2), enabling 3D LV formation and real-time tracking of DC migration. Adapted with permission from ref. 130, copyright 2025 Springer Nature. (E) Human glymphatics-on-chip model recapitulates 3D neurovascular architecture with astrocytic reactivity to tau. Scale bars: 200 μm. Adapted with permission from ref. 139, copyright 2025 American Institute of Physics. | ||
4.1 Modeling lymphedema pathophysiology
Lymphedema, characterized by tissue swelling due to impaired lymphatic drainage, can arise either from developmental defects (primary lymphedema) or from damage or obstruction of lymphatics (secondary lymphedema), for instance, after lymph node dissection. A hallmark feature is impaired lymph uptake and chronic inflammation in the affected tissues. Microphysiological systems have recently been leveraged to model this pathology and uncover mechanistic drivers of lymphatic dysfunction. Lee et al. (2023) developed a 3D lymphatic vessel-on-chip system with two parallel channels in a collagen hydrogel: one lined with LECs to mimic an initial lymphatic capillary, and the other serving as an interstitial channel for fluorescent dextran infusion (Fig. 4A). In this setup, LECs recapitulated button-like VE-cadherin junctions and supported fluid uptake akin to native initial lymphatics. However, upon exposure to inflammatory cytokines (e.g., IL-2, GM-CSF, G-CSF), the chip revealed pathological junction tightening mediated by a novel ROCK2-JAM-A complex at LEC borders. This hyper-stabilization of VE-cadherin impeded fluid and protein entry, sharply reducing drainage. Importantly, ROCK inhibition restored fluid uptake, identifying ROCK2 as a potential therapeutic target for lymphedema by reversing the “blood-vessel-like” tight junction state induced by inflammation.110 Kraus et al. (2023) established a human initial lymphatic primary-valve-on-a-chip in which interstitial and luminal pressures could be precisely modulated. They found that acute inflammation (i.e., brief high-dose TNF-α exposure) degraded fibrillin fibers and impaired fluid uptake, but corticosteroid treatment reversed the effect. In contrast, chronic low-grade TNF-α exposure caused LEC apoptosis and persistent junctional disruption, leading to valve failure and excessive leakiness—conditions resembling advanced lymphedema.109 This platform underscores that while acute inflammatory injury may be reversible, chronic inflammation drives irreversible valve damage, indicating the need for early anti-inflammatory intervention.4.2 Tumor–lymphatic interactions and metastasis
The lymphatic vasculature plays a central role in metastasis, particularly as tumors exploit lymphangiogenesis to spread to regional lymph nodes. Tumors secrete growth factors such as VEGF-C and VEGF-D, which expand peritumoral lymphatics and remodel draining lymph nodes.28 These changes facilitate tumor cell intravasation and survival in lymphatic niches.119 Lymphatics-on-a-chip offer a unique means to study these processes under controlled conditions. The early coculture model by Pisano et al. (2015) showed that fluid drainage enhanced breast cancer cell invasion toward the lymphatic vessel, mediated by CCR7-CCL21 signaling, supporting the concept of autologous chemotaxis.88 Chung et al. (2017) advanced this concept with a biomimetic tumor microenvironment-on-a-chip containing tumor cells, fibroblasts, and blood and lymphatic vessels. Interstitial flow and fibroblast presence promoted invasive tumor behavior and lymphatic remodeling, underscoring how stromal interactions condition lymphatic endothelium.120 Using a one-channel microfluidic device, Gong et al. (2019) and Lugo-Cintrón et al. (2020) demonstrated that breast cancer-associated fibroblast (CAF) disrupted lymphatic vessel barrier function. Coculture with aggressive breast cancer cells exacerbated lymphatic vessel leakiness and triggered IL-6 secretion, a possible pro-metastatic mechanism of lymphatic metastasis. Blocking IL-6 signaling partially restored lymphatic barrier function, suggesting potential for vessel normalization strategies.121,122 In a follow-up study, Lugo-Cintrón et al. (2021) found that tumor-derived fibroblasts dramatically increased lymphatic sprouting and network density through VEGF-C/D secretion and matrix remodeling.123 Building on this platform, Yada et al. (2023) revealed that LECs secrete macrophage migration inhibitory factor (MIF), which promotes directional tumor cell migration and metabolic reprogramming toward a glycolytic phenotype. Pharmacologic inhibition of MIF reduced cancer cell motility, demonstrating that lymphatic vessels can actively drive tumor invasion through paracrine signaling rather than serving solely as passive conduits.124Beyond modeling invasion, tumor–lymphatic chips are increasingly applied to therapeutic testing. Using a vascularized melanoma model, Fang et al. (2019) evaluated a chemokine/anti-PD-L1 nanobody fusion protein that simultaneously reshaped chemokine gradients and relieved checkpoint inhibition (Fig. 4B). This strategy enhanced T cell infiltration and anti-tumor immunity, illustrating how lymphatic chips can be harnessed to screen innovative immunotherapies.125 Similarly, Ayuso et al. (2020) developed a breast cancer–lymphatic coculture platform pairing perfused LEC channels with estrogen receptor-positive (MCF-7) or triple-negative (MDA-MB-231) breast tumors (Fig. 4C). They showed that aggressive MDA-MB-231 cells reprogrammed LECs into a more inflammatory, leaky, and pro-angiogenic state than MCF-7, mirroring clinical subtype differences.126 Frenkel et al. (2021) introduced immortalized human lymphatic endothelial cells (imLECs) into a membrane-free, three-lane OrganoPlate supporting continuous perfusion on a rocker platform (7° tilt, 8 min cycle) and stable biochemical gradients. The system maintained functional lymphatic microvessels for at least 18 days and enabled co-culture with colon-cancer organoids for 9 days, during which tumors induced VEGF-driven lymphatic sprouting and intravasation of cancer cells into lymphatic lumens, both effects abrogated by VEGFR-3 inhibition.127 The platform's long-term perfusion stability and sustained lymphangiogenic activity make it a robust assay for anti-lymphangiogenic drug testing and real-time analysis of tumor intravasation.
Together, tumor–lymphatic chips now recapitulate key metastatic steps—including flow-guided invasion, stromal and matrix conditioning, subtype-specific endothelial remodeling, and tumor intravasation—and are increasingly used to evaluate therapeutic interventions. From blocking CCR7 or IL-6 signaling to normalizing lymphatic vessels or enhancing checkpoint immunotherapy, these platforms offer powerful translational tools for dissecting and targeting the lymphatic route of cancer metastasis.
4.3 Autoimmune and inflammatory diseases
Lymphatic vessels are pivotal components of the immune system, facilitating dendritic cell (DC) and macrophage trafficking from peripheral tissues to lymph nodes, thereby priming adaptive immune responses.128 In many chronic inflammatory conditions, such as rheumatoid arthritis and inflammatory bowel disease (IBD), lymphatic vascular remodeling is a hallmark feature. For example, in IBD, mesenteric lymphatics may undergo proliferation and contribute to the formation of tertiary lymphoid structures, which in turn hinder normal lymph flow.2 Advances in lymphatics-on-a-chip models have propelled our understanding of immune cell migration under inflammatory conditions. Serrano et al. (2023) developed a microfluidic platform that recreates interstitial flow, lymphatic drainage, and immune cell recruitment on-chip. The engineered vessels achieved physiologically relevant drainage of interstitial proteins and solutes, while faithfully reproducing lymphatic-immune interactions. When peripheral blood mononuclear cells (PBMCs) were introduced under TNF-α-induced inflammatory conditions, the lymphatic chip actively recruited these immune cells; notably, blocking the chemokine receptors C–X–C motif chemokine receptor 4 (CXCR4) and CCR7 markedly reduced PBMC infiltration, demonstrating the robust modeling of chemokine-driven immune trafficking in vitro.129 Complementing this, Jo et al. (2025) introduced a high-throughput microfluidic platform that faithfully recapitulates the structure and function of initial lymphatic vessels in a 3D environment, including DC migration governed by a CCR7-CCL21 chemokine gradient (Fig. 4D). Under conditions of elevated CCR7 expression in DCs or following TNF-α preconditioning of LECs, which induces CCL21 secretion, DCs exhibited enhanced motility and directional migration toward the lymphatic channel. However, chronic inflammation led to DC overshooting their entry, indicative of desensitization or barrier disruption.130 These findings mirror in vivo observations, where acute inflammation may transiently enhance clearance, but chronic inflammation impairs trafficking efficiency.131,132 Such systems hold promise for therapeutic screening, including TNF-α inhibition to restore DC entry dynamics and preserve LEC junctional integrity in diseases like IBD.4.4 Neurological and glymphatic disorders
In the realm of aging and neurological disease, researchers are increasingly focusing on meningeal lymphatics20 and the glymphatic system,133 which together mediate cerebrospinal fluid (CSF) and interstitial solute clearance from the brain. The glymphatic pathway comprises periarterial CSF influx, astrocytic aquaporin-4 (AQP4)-mediated interstitial exchange, and perivenous efflux, functioning as a glial-dependent analogue of classical lymphatic drainage.134 Its activity is coupled to meningeal lymphatic vessels lining the dura mater, which channel glymphatically drained CSF and solutes to the deep cervical lymph nodes.135,136 Disruption of either pathway impairs metabolic waste clearance and has been implicated in Alzheimer's disease, traumatic brain injury, and age-related neuroinflammation.94 Soden et al. (2022) developed a gliovascular unit (GVU) on-a-chip: a microfluidic platform comprised of two parallel channels embedded in a 3D hydrogel populated with human astrocytes forming AQP4-polarized endfeet with the channel lined with human brain microvascular endothelial cells. Under inflammatory stimuli (with LPS, Aβ(1–42) oligomers, or an AQP4 inhibitor), the system showed impaired clearance of fluid and Aβ(1–40) tracer introduced from the other acellular channel; drainage deficits persisted even post-cell lysis, implying roles for both cell-dependent (AQP4-related) and ECM-mediated fluid transport. Notably, inflammation depolarized AQP4, highlighting how altered localization of this channel compromises glymphatic function.137 Building upon Soden's platform, Yslas et al. (2024) found that amyloid-β oligomers, but not monomers, caused pronounced astrocytic calcium signaling disruption and mislocalization of AQP4, impairing fluid clearance.138 Park et al. (2025) demonstrated that hyperphosphorylated tau induced astrocytes to adopt a hypertrophic, hypercontractile phenotype, leading to vasoconstriction of the engineered vessels and reduced glymphatic clearance (Fig. 4E). Critically, pharmacological inhibition of non-muscle myosin II using blebbistatin reversed astrocytic hypercontractility, restored vessel diameter, and rescued fluid transport function.139 By recapitulating the neural clearance pathway in vitro, these glymphatics-on-a-chip platforms advance our understanding of how chronic inflammation disrupts critical glymphatic functions and provide scalable systems for screening novel interventions for neurodegenerative diseases.In summary, lymphatics-on-a-chip platforms rapidly prove their value across a broad spectrum of disease contexts. These microfluidic systems enable the dissection of complex in vivo phenomena—including the physical forces that impair lymph flow in lymphedema, the cellular dialogues that drive cancer dissemination through lymphatic routes, and the nuances of immune cell crosstalk and trafficking—within a controlled, tunable in vitro milieu. Serving both as reductionist models for mechanistic exploration and as preclinical screening platforms, these chips allow researchers to ask focused questions and test interventions with enhanced physiological relevance.
5 Future directions: toward multiscale and human-relevant lymphatic microphysiological systems
The field of lymphatics-on-a-chip is rapidly advancing but remains in an early growth phase, offering significant potential for innovation. Several promising avenues are emerging to enhance the physiological relevance, scalability, and analytical power of these systems. In this section, we explore these future directions and discuss how they can address current limitations and open new avenues in lymphatic research and therapy (Fig. 5). | ||
| Fig. 5 Advanced engineering strategies for next-generation lymphatic microphysiological systems. (A) Schematic of deriving LECs from hiPSCs and capillary-like network formation of LEC stained by cell tracker green. Scale bar, 320 μm. Adapted with permission from ref. 34, copyright 2024 S. Karger AG. (B) The multicellular human ocular fluid outflow on-chip recapitulates steroid-induced glaucoma in vitro. Scale bar: 50 μm. Adapted with permission from ref. 149, copyright 2025 Springer Nature. (C) Chained neural network analysis of a cell transplant preclinical model with vascularized islet-chips designed using the VNQI. Adapted with permission from ref. 154, copyright 2023 American Institute of Chemical Engineers. | ||
5.1 iPSC-derived lymphatic organoids and synthetic developmental programs
A major constraint in current lymphatics-on-a-chip models is the reliance on primary LECs, typically isolated from neonatal or adult skin, which may not capture the molecular and functional heterogeneity among lymphatics across different tissues (e.g., gut, lung, skin, meninges, and heart). In addition to these tissue-specific constraints, commercially available human LECs also pose important demographic limitations. Neonatal or juvenile LECs are generally derived from foreskin and are therefore of male origin, whereas adult LECs are most often obtained from discarded skin collected during cosmetic procedures, which predominantly come from female donors. For researchers investigating sex-dependent or age-dependent differences in endothelial phenotypes, obtaining primary human LECs that are appropriately matched to experimental needs remains challenging. These sourcing limitations further underscore the appeal of iPSC-derived LECs as a more flexible and controllable alternative.iPSCs offer a renewable, patient-specific source of LECs and lymphatic organoids.140 Saha et al. (2024) developed a high-efficiency protocol using the transcription factors ETV2 and ETS2, with precisely timed PROX1 activation, to differentiate human iPSCs (hiPSCs) into LECs expressing VEGFR3, LYVE-1, and podoplanin at levels comparable to mature cells (Fig. 5A). These iPSC-derived LECs can assemble into stable lymphatic networks and secrete lymphangiocrine factors such as reelin, supporting their utility for in vitro modeling.34 However, despite expressing key markers, iPSC-derived LECs often exhibit developmentally immature profiles, with reduced expression of valve and collector related genes (e.g., FOXC2, GATA2, GJA4), incomplete junctional organization, and diminished responsiveness to mechanical cues.34,141 These findings indicate partial functional maturity and highlight the risk of phenotypic drift over prolonged culture. Perfusion and biomechanical conditioning have shown to markedly improve the structural and metabolic maturation of iPSC-derived vascular cells.142 Accordingly, incorporating biophysical stimuli interstitial or oscillatory flow, cyclic stretch, and matrix stiffness gradients during differentiation may help stabilize LEC identity and promote adult-like functionality.
Beyond iPSC sources, organ-specific primary LECs display distinct transcriptional and barrier properties shaped by their local microenvironments. Integrating these specialized LECs into microfluidic platforms can enhance physiological fidelity and enable comparative mechanobiological studies of regional lymphatic function. In parallel, emerging efforts to develop self-assembled vascular organoids, where ECs co-cultured with organotypic spheroids form interconnected vessel-like networks within 3D matrices, provide new avenues for modeling tissue-specific lymphatic morphogenesis.143–145 Combining the scalability of iPSC-derived LECs with organotypic cues, co-cultures, and lymphatic organoid complexity within perfusable microfluidic systems could yield next-generation mesofluidic models that unite tissue specificity with dynamic control and real-time visualization.146
5.2 Organ-specific lymphatic models
Lymphatic vessels are increasingly recognized not only as conduits for fluid clearance and immune cell transport but also as active regulators of organ development, homeostasis, and tissue repair through lymphangiocrine signaling. LECs secrete a wide range of tissue-interacting factors, including metabolic cues, morphogens, chemokines, and extracellular matrix regulators, that shape the identity and function of surrounding stromal, epithelial, immune, and vascular cells.147 Modeling these context-dependent interactions in vitro is therefore essential for recapitulating organ-specific lymphatic phenotypes and enhancing the physiological fidelity of organ-on-a-chip systems. By integrating lymphatic structures directly into organ-specific microphysiological platforms, researchers can interrogate how LECs influence and respond to their tissue environment. This includes immune-cell trafficking through lymphatic channels, stromal-derived guidance cues that pattern lymphatic networks, and reciprocal signaling between lymphatics and blood vessels that coordinates barrier function and antigen transport. Such systems are increasingly enabling body-on-a-chip frameworks capable of simulating drug pharmacokinetics, immune priming, and inter-organ communication.148 A leading example is the human ocular fluid-outflow chip developed by Lu et al. (2025), which reconstructs the trabecular meshwork (TM) and the lymphatic-like Schlemm's canal (SC) using a dual-layer microfluidic architecture (Fig. 5B).149 This platform recapitulates human aqueous humor drainage by channeling interstitial flow through an SC endothelial compartment enveloped by TM. Under steroid exposure, the chip reproduces hallmark features of steroid-induced glaucoma, including impaired fluid egress and strengthened SC junctions, mediated by an ALK5-VEGFC signaling axis between TM and SC cells. This work illustrates how emergent disease phenotypes require heterotypic interactions, which are not captured in monoculture systems, and positions the platform for incorporation into multi-organ models.Beyond the ocular niche, lymph node-on-a-chip technologies have emerged as powerful models for studying lymphatic-immune communication within a structured lymphoid microenvironment.150–152 These platforms recreate subcapsular sinus flow, fibroblastic reticular cell networks, and LEC-immune cell interactions, enabling precise examination of antigen capture, dendritic cell transit, and flow-dependent lymphocyte activation. Multi-compartment organ-on-chip frameworks can further couple peripheral tissue modules to downstream lymph node chambers via perfused lymphatic channels, thereby recreating antigen drainage, cytokine transport, and immune priming in a human-relevant context. Such integration will be crucial for mechanistic studies of how local lymphatic function shapes systemic immunity and immunotherapy responses.
Several additional organ systems present compelling opportunities for lymphatic modeling. Intestinal lacteals coordinate with enterocytes and immune cells to mediate chylomicron uptake and are susceptible to VEGF-A-driven junction zippering in obesity.2,14,38 Adipose lymphatics regulate lipid mobilization and inflammatory tone; their dysfunction, as seen with CD36 loss, promotes visceral inflammation and insulin resistance mirroring type 2 diabetes.12 Microfluidic co-cultures incorporating LECs with adipocytes, enterocytes, renal epithelial cells, or meningeal stromal cells could recapitulate these organ-specific pathophysiologies. Renal and meningeal lymphatic-epithelial chips may further model fibrotic fluid imbalance, intracranial pressure dynamics, and CSF clearance defects relevant to neurodegenerative disease.
Together, these emerging organ-specific and immune-linked lymphatics-on-a-chip platforms illustrate the importance of heterotypic, lymphangiocrine interactions in shaping lymphatic structure and function across tissues. Coupling patient-derived cells, autologous immune components, or disease-relevant organoids with perfusable lymphatic channels will enable personalized mechanistic studies and therapeutic testing across diverse organ systems—from lymph nodes and the eye to intestine, adipose tissue, kidney, brain, and beyond.
5.3 Computational and AI-assisted design of lymphatic networks
As lymphatics-on-a-chip systems grow increasingly sophisticated, they generate rich, high-dimensional datasets that are challenging to interpret using conventional analysis. This complexity presents a compelling opportunity for AI to bridge empirical experimentation and mechanistic understanding. AI offers powerful tools for handling multivariate perturbation experiments typical of lymphatic microfluidic studies. For instance, microfluidics methods have already used evolutionary algorithms to engineer channel geometries optimized for uniform shear stress or physiologically realistic flow distribution.153–155 Such methods could be adapted to lymphatic chips to tailor microchannel networks that mimic native interstitial flow or lymph propulsion dynamics. A landmark advance in applying AI to vascular—and by extension lymphatic—networks comes from the work of Tronolone et al. (2023), who developed a Vascular Network Quality Index (VNQI) using supervised machine learning and a chained neural network architecture (Fig. 5C). This index compresses complex vascular morphology into a single predictive metric that correlates strongly with functional oxygen transport—outperforming simpler morphological measures alone.154 Applied to lymphatics, a similar index could quantify network efficiency in terms of fluid clearance, immune cell trafficking, or barrier integrity, based on features like valve density, contractile behavior, and junctional permeability.Looking ahead, AI-enabled systems could evolve toward adaptive experimental frameworks, where AI “agents” monitor real-time readouts (e.g., flow dynamics, cell migration) and actively modulate stimuli (e.g., growth factors, mechanical stress) to optimize outcomes. Such closed-loop experimentation would dramatically accelerate optimization of drug delivery conditions or personalized therapeutic regimens on a chip. Currently, lymphatics-on-a-chip studies are often proof-of-concept or limited in throughput. Integrating AI and machine learning, coupled with high-throughput microfluidic platforms,156 3D bioprinting,157,158 and advanced microfabrication,159 promises to scale these systems—transforming them from single-use demonstrations into robust screening tools that accelerate drug discovery and mechanistic exploration.
5.4 Standards for validation of human lymphatic microphysiological platforms
Establishing robust validation criteria is essential to ensure that human lymphatics-on-a-chip accurately recapitulate native vessel identity, structure, and function (Fig. 6). Validation should begin with confirming molecular identity and maturity, including expression of canonical lymphatic markers (PROX1, VEGFR-3, PDPN, LYVE-1) and junctional proteins (VE-cadherin, CLDN5), together with valve-associated regulators such as FOXC2 and GATA2 that signify developmental progression. Structural fidelity can be assessed through imaging of button- versus zipper-like junctions, oak-leaf LEC overlaps, and anchoring-filament analogs that maintain tissue attachment. Functional validation should demonstrate physiologically relevant solute and macromolecule transport, quantified by tracer uptake and size-selective permeability assays. | ||
| Fig. 6 Multilevel validation framework for human lymphatic microphysiological systems. Schematic illustrating a multilevel validation framework for human lymphatics-on-a-chip systems, encompassing molecular, structural, functional, and clinical benchmarks. Adapted with permission from ref. 160, copyright 2024 Elsevier. Created with https://BioRender.com. | ||
Mechanobiological validation involves confirming flow-dependent behaviors, such as junctional remodeling, nitric-oxide signaling, and cyclic-stretch responses, that mimic pumping and valve dynamics in collecting vessels. Immune relevance can be established by assessing CCR7-dependent DC and T cell transmigration toward CCL21 gradients, and by testing cytokine-induced barrier modulation under TNF-α or IL-1β stimulation. Pharmacological perturbations provide further functional benchmarks, including expected changes in contractility or permeability following VEGF-A/C or VEGFR-3 blockade, RhoA/ROCK inhibition, or glucocorticoid treatment.
Finally, quantitative benchmarking against human data, encompassing histological architecture, lymphangiography or intralymphatic imaging, and tracer-clearance kinetics, should be reported with clear effect sizes and scaling assumptions to facilitate reproducibility and cross-platform comparison. Collectively, these integrated strategies provide a rigorous foundation for validating human lymphatic microphysiological systems and advancing them as reproducible, predictive tools for both disease modeling and therapeutic testing.
6 Conclusion
In closing, the future of lymphatics-on-a-chip is auspicious, poised to accelerate therapeutic innovation across domains—from lymphedema interventions and immunotherapies to vaccines leveraging lymphatic transport to lymph nodes. These microphysiological systems offer unprecedented potential to accurately replicate human lymphatic physiology and disease, complementing and even replacing traditional animal models, which suffer from translational limitations and rising regulatory and ethical scrutiny. As integrated platforms—combining lymphatic, immune, and multi-organ functionality—continue to materialize, they align tightly with advances in personalized medicine, high-throughput screening, and translational science, drawing on multidisciplinary collaboration across engineering, biology, and computation. For a system as historically underappreciated and complex as the lymphatic vasculature, these advances come at a pivotal time, ensuring that the next generations of lymphatics-on-a-chip models are ever more physiological, predictive, and useful in unraveling the mysteries of the lymphatic system.Author contributions
Writing – original draft: Y. P. Writing – review & editing: Y. P., E. L.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was supported by NIH grants (CA279560, HL165135). Y. P. was supported by the International Foundation for Ethical Research (IFER) Fellowship.References
- L. Alderfer, A. Wei and D. Hanjaya-Putra, J. Biol. Eng., 2018, 12, 32 CrossRef PubMed.
- B. J. Mehrara, A. J. Radtke, G. J. Randolph, B. T. Wachter, P. Greenwel, I. I. Rovira, Z. S. Galis and S. C. Muratoglu, J. Clin. Invest., 2023, 133, e171582 CrossRef PubMed.
- G. Oliver, J. Kipnis, G. J. Randolph and N. L. Harvey, Cell, 2020, 182, 270–296 CrossRef CAS PubMed.
- T. Karakousi, T. Mudianto and A. W. Lund, Nat. Rev. Cancer, 2024, 24, 363–381 CrossRef CAS PubMed.
- N. Schwartz, M. L. S. Chalasani, T. M. Li, Z. Feng, W. D. Shipman and T. T. Lu, Front. Immunol., 2019, 10, 519 CrossRef CAS.
- R. Hokari and A. Tomioka, Inflammation Regener., 2021, 41, 25 CrossRef.
- L. Zhang, D. K. W. Ocansey, L. Liu, C. V. Olovo, X. Zhang, H. Qian, W. Xu and F. Mao, Biomed. Pharmacother., 2021, 140, 111752 CrossRef CAS.
- D. Nikolakis, F. A. E. De Voogd, M. J. Pruijt, J. Grootjans, M. G. Van De Sande and G. R. D'Haens, IJMS, 2022, 23, 1854 CrossRef CAS PubMed.
- B. N. K. Ruliffson and C. F. Whittington, Adv. Biol., 2023, e2200158, DOI:10.1002/adbi.202200158.
- N. Escobedo and G. Oliver, Cell Metab., 2017, 26, 598–609 CrossRef CAS PubMed.
- R. P. Kataru, H. J. Park, J. E. Baik, C. Li, J. Shin and B. J. Mehrara, Front. Physiol., 2020, 11, 459 CrossRef PubMed.
- V. Cifarelli, S. Appak-Baskoy, V. S. Peche, A. Kluzak, T. Shew, R. Narendran, K. M. Pietka, M. Cella, C. W. Walls, R. Czepielewski, S. Ivanov, G. J. Randolph, H. G. Augustin and N. A. Abumrad, Nat. Commun., 2021, 12, 3350 CrossRef CAS PubMed.
- E. Cao, M. J. Watt, C. J. Nowell, T. Quach, J. S. Simpson, V. De Melo Ferreira, S. Agarwal, H. Chu, A. Srivastava, D. Anderson, G. Gracia, A. Lam, G. Segal, J. Hong, L. Hu, K. L. Phang, A. B. J. Escott, J. A. Windsor, A. R. J. Phillips, D. J. Creek, N. L. Harvey, C. J. H. Porter and N. L. Trevaskis, Nat. Metab., 2021, 3, 1175–1188 CrossRef CAS PubMed.
- G. Zarkada, X. Chen, X. Zhou, M. Lange, L. Zeng, W. Lv, X. Zhang, Y. Li, W. Zhou, K. Liu, D. Chen, N. Ricard, J. Liao, Y.-B. Kim, R. Benedito, L. Claesson-Welsh, K. Alitalo, M. Simons, R. Ju, X. Li, A. Eichmann and F. Zhang, Circ. Res., 2023, 133, 333–349 CrossRef CAS PubMed.
- E. Brakenhielm and K. Alitalo, Nat. Rev. Cardiol., 2019, 16, 56–68 CrossRef CAS PubMed.
- K. Klaourakis, J. M. Vieira and P. R. Riley, Nat. Rev. Cardiol., 2021, 18, 368–379 CrossRef.
- X. Liu and G. Oliver, Circ. Res., 2023, 132, 1246–1253 CrossRef CAS.
- A. Louveau, S. Da Mesquita and J. Kipnis, Neuron, 2016, 91, 957–973 CrossRef CAS.
- B.-L. Sun, L.-h. Wang, T. Yang, J.-y. Sun, L.-l. Mao, M.-f. Yang, H. Yuan, R. A. Colvin and X.-y. Yang, Prog. Neurobiol., 2018, 163–164, 118–143 CrossRef PubMed.
- Q. Zhang, Y. Niu, Y. Li, C. Xia, Z. Chen, Y. Chen and H. Feng, Signal Transduction Targeted Ther., 2025, 10, 142 CrossRef.
- L. Gutierrez-Miranda and K. Yaniv, Front. Physiol., 2020, 11, 577584 CrossRef.
- M. Sun, J. Angelillo and S. Hugues, J. Exp. Med., 2025, 222, e20231954 CrossRef CAS.
- W. J. Polacheck, J. B. Dixon and W. Y. Aw, Annu. Rev. Biomed. Eng., 2025, 27, 73–100 CrossRef CAS PubMed.
- C. Ma, Y. Peng, H. Li and W. Chen, Trends Pharmacol. Sci., 2020, 42, 119–133 CrossRef PubMed.
- M. Skobe, T. Hawighorst, D. G. Jackson, R. Prevo, L. Janes, P. Velasco, L. Riccardi, K. Alitalo, K. Claffey and M. Detmar, Nat. Med., 2001, 7, 192–198 CrossRef CAS PubMed.
- T. Karpanen, M. Egeblad, M. J. Karkkainen, H. Kubo, S. Ylä-Herttuala, M. Jäättelä and K. Alitalo, Cancer Res., 2001, 61, 1786–1790 CAS.
- J. Krishnan, V. Kirkin, A. Steffen, M. Hegen, D. Weih, S. Tomarev, J. r. Wilting and J. P. Sleeman, Cancer Res., 2003, 63, 713–722 CAS.
- K. Alitalo, T. Tammela and T. V. Petrova, Nature, 2005, 438, 946–953 CrossRef CAS PubMed.
- S. Zhang, S. Yi, D. Zhang, M. Gong, Y. Cai and L. Zou, Sci. Rep., 2017, 7, 40364 CrossRef CAS.
- J. Li, Y.-B. Liang, Q.-B. Wang, Y.-K. Li, X.-M. Chen, W.-L. Luo, Y. Lakang, Z.-S. Yang, Y. Wang, Z.-W. Li and Y. Ke, Front. Immunol., 2025, 15, 1519999 CrossRef.
- I. Van Der Auwera, Y. Cao, J. C. Tille, M. S. Pepper, D. G. Jackson, S. B. Fox, A. L. Harris, L. Y. Dirix and P. B. Vermeulen, Br. J. Cancer, 2006, 95, 1611–1625 CrossRef CAS PubMed.
- N. Audet, N. J. Beasley, C. Macmillan, D. G. Jackson, P. J. Gullane and S. Kamel-Reid, Arch. Otolaryngol., Head Neck Surg., 2005, 131, 1065 CrossRef PubMed.
- I.-E. Lupu, D. E. Grainger, N. Kirschnick, S. Weischer, E. Zhao, I. Martinez-Corral, H. Schoofs, M. Vanhollebeke, G. Jones, J. Godwin, A. Forrow, I. Lahmann, P. R. Riley, T. Zobel, K. Alitalo, T. Mäkinen, F. Kiefer and O. A. Stone, Nat. Cardiovasc. Res., 2025, 4, 45–63 CrossRef CAS PubMed.
- S. Saha, F. Graham, J. Knopp, C. Patzke and D. Hanjaya-Putra, Cells Tissues Organs, 2024, 1–11, DOI:10.1159/000539699.
- M. Jannaway, D. Iyer, D. M. Mastrogiacomo, K. Li, D. C. Sung, Y. Yang, M. L. Kahn and J. P. Scallan, Cell Rep., 2023, 42, 112777 CrossRef CAS PubMed.
- H. Schoofs and T. Mäkinen, Int. J. Dev. Biol., 2024, 68, 189–198 CrossRef CAS PubMed.
- I. Martinez-Corral, M. H. Ulvmar, L. Stanczuk, F. Tatin, K. Kizhatil, S. W. M. John, K. Alitalo, S. Ortega and T. Makinen, Circ. Res., 2015, 116, 1649–1654 CrossRef.
- F. Zhang, G. Zarkada, J. Han, J. Li, A. Dubrac, R. Ola, G. Genet, K. Boyé, P. Michon, S. E. Künzel, J. P. Camporez, A. K. Singh, G.-H. Fong, M. Simons, P. Tso, C. Fernández-Hernando, G. I. Shulman, W. C. Sessa and A. Eichmann, Science, 2018, 361, 599–603 CrossRef PubMed.
- V. Cifarelli and A. Eichmann, Cell. Mol. Gastroenterol. Hepatol., 2019, 7, 503–513 CrossRef PubMed.
- T. V. Petrova and G. Y. Koh, Science, 2020, 369, eaax4063 CrossRef PubMed.
- A. F. M. Salvador, N. Abduljawad and J. Kipnis, Annu. Rev. Neurosci., 2024, 47, 323–344 CrossRef PubMed.
- P. R. Norden and T. Kume, Front. Cell Dev. Biol., 2020, 8, 627647 CrossRef PubMed.
- F. Zhang, G. Zarkada, S. Yi and A. Eichmann, Front. Physiol., 2020, 11, 509 CrossRef PubMed.
- J. E. Moore, Jr. and C. D. Bertram, Annu. Rev. Fluid Mech., 2018, 50, 459–482 CrossRef.
- M. A. Swartz and A. W. Lund, Nat. Rev. Cancer, 2012, 12, 210–219 CrossRef.
- J. Amersfoort, G. Eelen and P. Carmeliet, Nat. Rev. Immunol., 2022, 22, 576–588 CrossRef PubMed.
- H. Schoofs, N. Daubel, S. Schnabellehner, M. L. B. Gronloh, S. Palacios Martinez, A. Halme, A. M. Marks, M. Jeansson, S. Barcos, C. Brakebusch, R. Benedito, B. Engelhardt, D. Vestweber, K. Gaengel, F. Linsenmeier, S. Schurmann, P. Saharinen, J. D. van Buul, O. Friedrich, R. S. Smith, M. Majda and T. Makinen, Nature, 2025, 641, 465–475 CrossRef PubMed.
- S. Fu, Y. Wang, E. Bin, H. Huang, F. Wang and N. Tang, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2215449120 CrossRef PubMed.
- M. Frye, S. Stritt, H. Ortsäter, M. Hernandez Vasquez, M. Kaakinen, A. Vicente, J. Wiseman, L. Eklund, J. L. Martínez-Torrecuadrada, D. Vestweber and T. Mäkinen, eLife, 2020, 9, e57732 CrossRef.
- W. E. Cromer, S. D. Zawieja, B. Tharakan, E. W. Childs, M. K. Newell and D. C. Zawieja, Angiogenesis, 2014, 17, 395–406 CrossRef PubMed.
- P. Baluk, J. Fuxe, H. Hashizume, T. Romano, E. Lashnits, S. Butz, D. Vestweber, M. Corada, C. Molendini, E. Dejana and D. M. McDonald, J. Exp. Med, 2007, 204, 2349–2362 CrossRef PubMed.
- L.-C. Yao, P. Baluk, R. S. Srinivasan, G. Oliver and D. M. Mcdonald, Am. J. Pathol., 2012, 180, 2561–2575 CrossRef PubMed.
- M. J. Davis, S. D. Zawieja and Y. Yang, Front. Cell Dev. Biol., 2024, 12, 1331291 CrossRef PubMed.
- D. Iyer, D. M. Mastrogiacomo, K. Li, R. Banerjee, Y. Yang and J. P. Scallan, Arterioscler., Thromb., Vasc. Biol., 2023, 43, 2197–2212 CrossRef PubMed.
- T. V. Petrova, T. Karpanen, C. Norrmén, R. Mellor, T. Tamakoshi, D. Finegold, R. Ferrell, D. Kerjaschki, P. Mortimer, S. Ylä-Herttuala, N. Miura and K. Alitalo, Nat. Med., 2004, 10, 974–981 CrossRef PubMed.
- A. Sabine, Y. Agalarov, H. M.-E. Hajjami, M. Jaquet, R. Hägerling, C. Pollmann, D. Bebber, A. Pfenniger, N. Miura, O. Dormond, J.-M. Calmes, R. H. Adams, T. Mäkinen, F. Kiefer, B. R. Kwak and T. V. Petrova, Dev. Cell, 2012, 22, 430–445 CrossRef.
- D. Choi, E. Park, E. Jung, Y. J. Seong, J. Yoo, E. Lee, M. Hong, S. Lee, H. Ishida, J. Burford, J. Peti-Peterdi, R. H. Adams, S. Srikanth, Y. Gwack, C. S. Chen, H. J. Vogel, C. J. Koh, A. K. Wong and Y.-K. Hong, J. Clin. Invest., 2017, 127, 1225–1240 CrossRef PubMed.
- M. N. Hernández Vásquez, M. H. Ulvmar, A. González-Loyola, I. Kritikos, Y. Sun, L. He, C. Halin, T. V. Petrova and T. Mäkinen, EMBO J., 2021, 40, EMBJ2020107192 CrossRef PubMed.
- K. Nonomura, V. Lukacs, D. T. Sweet, L. M. Goddard, A. Kanie, T. Whitwam, S. S. Ranade, T. Fujimori, M. L. Kahn and A. Patapoutian, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 12817–12822 CrossRef.
- D. Choi, E. Park, J. Choi, R. Lu, J. S. Yu, C. Kim, L. Zhao, J. Yu, B. Nakashima, S. Lee, D. Singhal, J. P. Scallan, B. Zhou, C. J. Koh, E. Lee and Y.-K. Hong, Nat. Neurosci., 2024, 27, 913–926 CrossRef PubMed.
- L. Planas-Paz, B. Strilić, A. Goedecke, G. Breier, R. Fässler and E. Lammert, EMBO J., 2012, 31, 788–804 CrossRef PubMed.
- N. Baeyens, S. Nicoli, B. G. Coon, T. D. Ross, K. Van den Dries, J. Han, H. M. Lauridsen, C. O. Mejean, A. Eichmann, J.-L. Thomas, J. D. Humphrey and M. A. Schwartz, eLife, 2015, 4, e04645 CrossRef.
- V. Angeli and H. Y. Lim, Cell. Mol. Immunol., 2023, 20, 1051–1062 CrossRef.
- C. Bonvin, J. Overney, A. C. Shieh, J. B. Dixon and M. A. Swartz, Biotechnol. Bioeng., 2010, 105, 982–991 CrossRef.
- D. Choi, E. Park, E. Jung, Y. J. Seong, M. Hong, S. Lee, J. Burford, G. Gyarmati, J. Peti-Peterdi, S. Srikanth, Y. Gwack, C. J. Koh, E. Boriushkin, A. Hamik, A. K. Wong and Y.-K. Hong, Circ. Res., 2017, 120, 1426–1439 CrossRef.
- H. Wiig and H. Noddeland, Scand. J. Clin. Lab. Invest., 1983, 43, 255–260 CrossRef CAS.
- E. Stranden and H. O. Myhre, Microvasc. Res., 1982, 24, 241–248 CrossRef CAS PubMed.
- S. Jamalian, M. Jafarnejad, S. D. Zawieja, C. D. Bertram, A. A. Gashev, D. C. Zawieja, M. J. Davis and J. E. Moore, Sci. Rep., 2017, 7, 12080 CrossRef PubMed.
- Y. L. Huang, J. E. Segall and M. Wu, Lab Chip, 2017, 17, 3221–3233 RSC.
- S. R. Chary and R. K. Jain, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 5385–5389 CrossRef CAS.
- D. Negrini and M. Del Fabbro, J. Physiol., 1999, 520, 761–769 CrossRef CAS.
- D. Negrini, A. Moriondo and S. Mukenge, Lymphatic Res. Biol., 2004, 2, 69–81 CrossRef.
- A. Moriondo, S. Mukenge and D. Negrini, Am. J. Physiol., 2005, 289, H263–H269 CAS.
- G. Clough and L. H. Smaje, J. Physiol., 1978, 283, 457–468 CrossRef CAS PubMed.
- E. M. Bouta, R. W. Wood, E. B. Brown, H. Rahimi, C. T. Ritchlin and E. M. Schwarz, J. Physiol., 2014, 592, 1213–1223 CrossRef CAS.
- J. P. Scallan and V. H. Huxley, J. Physiol., 2010, 588, 243–254 CrossRef CAS PubMed.
- H. J. T. Guthe, M. Indrebø, T. Nedrebø, G. Norgård, H. Wiig and A. Berg, PLoS One, 2015, 10, e0122779 CrossRef.
- J. K. Heltne, P. Husby, M. E. Koller and T. Lund, Lab. Anim., 1998, 32, 439–445 CrossRef CAS.
- R. D. Manning, Am. J. Physiol., 1998, 275, R135–R140 CAS.
- K. Aukland, G. C. Kramer and E. M. Renkin, Am. J. Physiol., 1984, 247, H74–H79 CrossRef CAS.
- D. A. Berk, M. A. Swartz, A. J. Leu and R. K. Jain, Am. J. Physiol., 1996, 270, H330–H337 CAS.
- M. Fischer, U. K. Franzeck, I. Herrig, U. Costanzo, S. Wen, M. Schiesser, U. Hoffmann and A. Bollinger, Am. J. Physiol., 1996, 270, H358–H363 CAS.
- J. B. Dixon, S. T. Greiner, A. A. Gashev, G. L. Cote, J. E. Moore and D. C. Zawieja, Microcirculation, 2006, 13, 597–610 CrossRef.
- C. Blatter, E. F. J. Meijer, A. S. Nam, D. Jones, B. E. Bouma, T. P. Padera and B. J. Vakoc, Sci. Rep., 2016, 6, 29035 CrossRef CAS PubMed.
- E. Rahbar, T. Akl, G. L. Coté, J. E. Moore and D. C. Zawieja, Microcirculation, 2014, 21, 359–367 CrossRef CAS PubMed.
- S. Molloi, A. R. Polivka, Y. Zhao, J. Redmond, M. Itkin, I. Antunes and Z. Yu, Radiology, 2023, 309, e230959 CrossRef PubMed.
- M. Jafarnejad, W. E. Cromer, R. R. Kaunas, S. L. Zhang, D. C. Zawieja and J. E. Moore, Jr., Am. J. Physiol., 2015, 308, H697–H706 CrossRef CAS.
- M. Pisano, V. Triacca, K. A. Barbee and M. A. Swartz, Integr. Biol., 2015, 7, 525–533 CrossRef CAS.
- A. A. Gashev, M. J. Davis, M. D. Delp and D. C. Zawieja, Microcirculation, 2004, 11, 477–492 CrossRef CAS PubMed.
- H. Cheon, S. H. Lee, S. A. Kim, B. Kim, H. P. Suh and J. Y. Jeon, Arterioscler., Thromb., Vasc. Biol., 2023, 43, 2008–2022 CrossRef CAS PubMed.
- J.-P. Belgrado, L. Vandermeeren, S. Vankerckhove, J.-B. Valsamis, J. Malloizel-Delaunay, J.-J. Moraine and F. Liebens, Lymphatic Res. Biol., 2016, 14, 70–77 CrossRef.
- J. W. Breslin, Microvasc. Res., 2014, 96, 46–54 CrossRef.
- J. P. Scallan, S. D. Zawieja, J. A. Castorena-Gonzalez and M. J. Davis, J. Physiol., 2016, 594, 5749–5768 CrossRef CAS PubMed.
- J. Hagendoorn, T. P. Padera, S. Kashiwagi, N. Isaka, F. Noda, M. I. Lin, P. L. Huang, W. C. Sessa, D. Fukumura and R. K. Jain, Circ. Res., 2004, 95, 204–209 CrossRef CAS PubMed.
- M. Jannaway and J. P. Scallan, Front. Physiol., 2021, 12, 687563 CrossRef PubMed.
- K. H. K. Wong, J. G. Truslow, A. H. Khankhel, K. L. S. Chan and J. Tien, J. Biomed. Mater. Res., Part A, 2013, 101, 2181–2190 CrossRef PubMed.
- M. Sato, N. Sasaki, M. Ato, S. Hirakawa, K. Sato and K. Sato, PLoS One, 2015, 10, e0137301 CrossRef.
- E. Hall, K. Mendiola, N. K. Lightsey and D. Hanjaya-Putra, Biomicrofluidics, 2024, 18, 031502 CrossRef CAS.
- S. Kim, M. Chung and N. L. Jeon, Biomaterials, 2016, 78, 115–128 CrossRef.
- T. Osaki, J. C. Serrano and R. D. Kamm, Regener. Eng. Transl. Med., 2018, 4, 120–132 CrossRef PubMed.
- J. J. Tronolone, N. Mohamed and A. Jain, Adv. Biol., 2024, 8, e2400031 Search PubMed.
- L. Gibot, T. Galbraith, J. Bourland, A. Rogic, M. Skobe and F. A. Auger, Nat. Protoc., 2017, 12, 1077–1088 CrossRef PubMed.
- F. Fan, B. Su, A. Kolodychak, E. Ekwueme, L. Alderfer, S. Saha, M. J. Webber and D. Hanjaya-Putra, ACS Appl. Mater. Interfaces, 2023, 15, 58181–58195 CrossRef PubMed.
- L. Alderfer, S. Saha, F. Fan, J. Wu, L. E. Littlepage and D. Hanjaya-Putra, Commun. Biol., 2024, 7, 1262 CrossRef PubMed.
- D. Montes, S. Saha, A. Taglione, D. P. Jeong, L. Chen, F. Fan, H. C. Chang and D. Hanjaya-Putra, Adv. Mater. Interfaces, 2025, 12, 2401037 CrossRef.
- A. R. Henderson, I. S. Ilan and E. Lee, Microcirculation, 2021, 28, e12730 CrossRef PubMed.
- A. Selahi, T. Fernando, S. Chakraborty, M. Muthuchamy, D. C. Zawieja and A. Jain, Lab Chip, 2022, 22, 121–135 RSC.
- I. S. Ilan, A. R. Yslas, Y. Peng, R. Lu and E. Lee, Cell. Mol. Bioeng., 2023, 16, 325–339 CrossRef PubMed.
- S. Kraus and E. Lee, Lab Chip, 2023, 23, 5180–5194 RSC.
- E. Lee, S.-L. Chan, Y. Lee, W. J. Polacheck, S. Kwak, A. Wen, D.-H. T. Nguyen, M. L. Kutys, S. Alimperti, A. M. Kolarzyk, T. J. Kwak, J. Eyckmans, D. R. Bielenberg, H. Chen and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2308941120 CrossRef PubMed.
- A. Selahi, S. Chakraborty, M. Muthuchamy, D. C. Zawieja and A. Jain, Analyst, 2022, 147, 2953–2965 Search PubMed.
- G. H. Lee, S. A. Huang, W. Y. Aw, M. L. Rathod, C. Cho, F. S. Ligler and W. J. Polacheck, Biofabrication, 2022, 14, 025007 CrossRef PubMed.
- P. Fathi, G. Holland, D. Pan and M. B. Esch, ACS Appl. Bio Mater., 2020, 3, 6697–6707 CrossRef PubMed.
- M. Frye, A. Taddei, C. Dierkes, I. Martinez-Corral, M. Fielden, H. Ortsäter, J. Kazenwadel, D. P. Calado, P. Ostergaard, M. Salminen, L. He, N. L. Harvey, F. Kiefer and T. Mäkinen, Nat. Commun., 2018, 9, 1511 CrossRef PubMed.
- E. Gordon, L. Schimmel and M. Frye, Front. Physiol., 2020, 11, 684 CrossRef PubMed.
- L. Alderfer, E. Russo, A. Archilla, B. Coe and D. Hanjaya-Putra, FASEB J., 2021, 35, e21498 CrossRef.
- R. Lu, B. J. Lee and E. Lee, ACS Biomater. Sci. Eng., 2024, 10, 5752–5763 Search PubMed.
- A. M. Ledo, G. Misiewicz, T. Dimke, W. R. Tschantz, J. Handel, R. Pelis, G. Bruin, K. Bechtold-Peters, M. Sanchez-Felix, S. Deshmukh, S. Kim, M. Proestaki and R. D. Kamm, Lab Chip, 2025, 25, 4660–4676 Search PubMed.
- J. D. Shields, M. E. Fleury, C. Yong, A. A. Tomei, G. J. Randolph and M. A. Swartz, Cancer Cell, 2007, 11, 526–538 CrossRef PubMed.
- M. Chung, J. Ahn, K. Son, S. Kim and N. L. Jeon, Adv. Healthcare Mater., 2017, 6, 1700196 Search PubMed.
- M. M. Gong, K. M. Lugo-Cintron, B. R. White, S. C. Kerr, P. M. Harari and D. J. Beebe, Biomaterials, 2019, 214, 119225 Search PubMed.
- K. M. Lugo-Cintrón, J. M. Ayuso, B. R. White, P. M. Harari, S. M. Ponik, D. J. Beebe, M. M. Gong and M. Virumbrales-Muñoz, Lab Chip, 2020, 20, 1586–1600 Search PubMed.
- K. M. Lugo-Cintrón, J. M. Ayuso, M. Humayun, M. M. Gong, S. C. Kerr, S. M. Ponik, P. M. Harari, M. Virumbrales-Muñoz and D. J. Beebe, EBioMedicine, 2021, 73, 103634 CrossRef.
- R. C. Yada, D. E. Desa, A. A. Gillette, E. Bartels, P. M. Harari, M. C. Skala, D. J. Beebe and S. C. Kerr, Biomaterials, 2023, 298, 122136 CrossRef.
- T. Fang, R. Li, Z. Li, J. Cho, J. S. Guzman, R. D. Kamm and H. L. Ploegh, Mol. Pharmaceutics, 2019, 16, 2838–2844 Search PubMed.
- J. M. Ayuso, M. M. Gong, M. C. Skala, P. M. Harari and D. J. Beebe, Adv. Healthcare Mater., 2020, 9, 1900925 CrossRef PubMed.
- N. Frenkel, S. Poghosyan, C. R. Alarcon, S. B. Garcia, K. Queiroz, L. van den Bent, J. Laoukili, I. B. Rinkes, P. Vulto, O. Kranenburg and J. Hagendoorn, ACS Biomater. Sci. Eng., 2021, 7, 3030–3042 Search PubMed.
- C. Viúdez-Pareja, E. Kreft and M. García-Caballero, Front. Immunol., 2023, 14, 1235812 CrossRef.
- J. C. Serrano, M. R. Gillrie, R. Li, S. H. Ishamuddin, E. Moeendarbary and R. D. Kamm, Adv. Sci., 2023, 11, 2302903 CrossRef.
- H. Jo, S. Lee, I. Park, M. Shim, J. Yu, Y. S. Oh, J. Doh, Y.-K. Hong and N. L. Jeon, BioChip J., 2025, 19, 227 Search PubMed.
- V. Angeli and G. J. Randolph, Lymphatic Res. Biol., 2006, 4, 217–228 CrossRef.
- A. J. Christiansen, L. C. Dieterich, I. Ohs, S. B. Bachmann, R. Bianchi, S. T. Proulx, M. Hollmén, D. Aebischer and M. Detmar, Oncotarget, 2016, 7, 39421–39435 CrossRef.
- K. Ayyappan, L. Unger, P. Kitchen, R. M. Bill and M. M. Salman, Neural Regener. Res., 2026, 21, 534–541 Search PubMed.
- J. J. Iliff, M. Wang, Y. Liao, B. A. Plogg, W. Peng, G. A. Gundersen, H. Benveniste, G. E. Vates, R. Deane, S. A. Goldman, E. A. Nagelhus and M. Nedergaard, Sci. Transl. Med., 2012, 4, 147ra111–147ra141 Search PubMed.
- A. Aspelund, S. Antila, S. T. Proulx, T. V. Karlsen, S. Karaman, M. Detmar, H. Wiig and K. Alitalo, J. Exp. Med., 2015, 212, 991–999 CrossRef PubMed.
- A. Louveau, I. Smirnov, T. J. Keyes, J. D. Eccles, S. J. Rouhani, J. D. Peske, N. C. Derecki, D. Castle, J. W. Mandell, K. S. Lee, T. H. Harris and J. Kipnis, Nature, 2015, 523, 337–341 CrossRef.
- P. A. Soden, A. R. Henderson and E. Lee, Adv. Biol., 2022, 6, 2200027 CrossRef.
- A. R. Yslas, R. Park, N. Nishimura and E. Lee, Lab Chip, 2024, 24, 3826–3839 RSC.
- R. Park, Y. Peng, A. R. Yslas and E. Lee, APL Bioeng., 2025, 9, 026126 CrossRef.
- P. N. Ingram, L. E. Hind, J. A. Jiminez-Torres, A. Huttenlocher and D. J. Beebe, Adv. Healthcare Mater., 2018, 7, 1700497 CrossRef.
- M. P. White, A. J. Rufaihah, L. Liu, Y. T. Ghebremariam, K. N. Ivey, J. P. Cooke and D. Srivastava, Stem Cells, 2013, 31, 92–103 Search PubMed.
- A. Trillhaase, M. Maertens, Z. Aherrahrou and J. Erdmann, Stem Cell Rev. Rep., 2021, 17, 1741–1753 CrossRef.
- C. M. Abreu, A. C. Lima, N. M. Neves, S. C. Kundu, R. L. Reis and D. Caballero, Adv. Mater. Technol., 2025, 10, 2400883 CrossRef.
- S. Zhang, Z. Wan and R. D. Kamm, Lab Chip, 2021, 21, 473–488 Search PubMed.
- J. J. Tronolone, N. Mohamed, C. P. Chaftari, Y. Sun, T. Mathur and A. Jain, Curr. Protoc., 2024, 4, e70058 Search PubMed.
- Z. Li, D. Yu, C. Zhou, F. Wang, K. Lu, Y. Liu, J. Xu, L. Xuan and X. Wang, Biomater. Transl., 2024, 5, 21–32 CrossRef PubMed.
- T. V. Petrova and G. Y. Koh, J. Exp. Med., 2018, 215, 35–49 Search PubMed.
- Y. Peng and E. Lee, Adv. Biol., 2023, e2300077, DOI:10.1002/adbi.202300077.
- R. Lu, A. M. Kolarzyk, W. D. Stamer and E. Lee, Nat. Cardiovasc. Res., 2025, 4, 1066–1076 CrossRef PubMed.
- S. R. Cook, A. G. Ball, A. Mohammad and R. R. Pompano, Lab Chip, 2025, 25, 155–174 RSC.
- K. G. Birmingham, M. J. O'Melia, S. Bordy, D. Reyes Aguilar, B. El-Reyas, G. Lesinski and S. N. Thomas, iScience, 2020, 23, 101751 CrossRef.
- G. Goyal, P. Prabhala, G. Mahajan, B. Bausk, T. Gilboa, L. Xie, Y. Zhai, R. Lazarovits, A. Mansour, M. S. Kim, A. Patil, D. Curran, J. M. Long, S. Sharma, A. Junaid, L. Cohen, T. C. Ferrante, O. Levy, R. Prantil-Baun, D. R. Walt and D. E. Ingber, Adv. Sci., 2022, 2103241, DOI:10.1002/advs.202103241.
- J. J. Tronolone, T. Mathur, C. P. Chaftari and A. Jain, Ann. Biomed. Eng., 2023, 51, 1723–1737 CrossRef.
- J. J. Tronolone, T. Mathur, C. P. Chaftari, Y. Sun and A. Jain, Bioeng. Transl. Med., 2023, 8, e10582 CrossRef PubMed.
- T. Mathur, J. J. Tronolone and A. Jain, PLoS One, 2024, 19, e0299160 CrossRef PubMed.
- M. Kim, S. Choi, D.-H. Choi, J. Ahn, D. Lee, E. Song, H. S. Kim, M. Kim, S. Choi, S. Oh, M. Kim, S. Chung and P. J. Park, NPG Asia Mater., 2024, 16, 7 CrossRef.
- X. Cao, R. Ashfaq, F. Cheng, S. Maharjan, J. Li, G. Ying, S. Hassan, H. Xiao, K. Yue and Y. S. Zhang, Adv. Funct. Mater., 2019, 29, 1807173 CrossRef PubMed.
- K. A. Deo, A. Murali, J. J. Tronolone, C. Mandrona, H. P. Lee, S. Rajput, S. E. Hargett, A. Selahi, Y. Sun, D. L. Alge, A. Jain and A. K. Gaharwar, Adv. Healthcare Mater., 2024, 13, 2303810 CrossRef.
- S. Sundaram, J. H. Lee, I. M. Bjørge, C. Michas, S. Kim, A. Lammers, J. F. Mano, J. Eyckmans, A. E. White and C. S. Chen, Nature, 2024, 636, 361–367 CrossRef PubMed.
- S. Cometta, D. W. Hutmacher and L. Chai, Biomaterials, 2024, 309, 122578 CrossRef.
| This journal is © The Royal Society of Chemistry 2026 |
