Fractal-shaped droplet microfluidics for highly scalable cell mechanoporation
Myungsuk
Sung
 ab,
Dalei
Jing
cd,
Byeongju
Joo
ab,
Dalei
Jing
cd,
Byeongju
Joo
 ae,
Sungbin
Im
e,
You-Jeong
Kim
ae,
Sungbin
Im
e,
You-Jeong
Kim
 ab,
Yi
Sui
ab,
Yi
Sui
 d and
Aram J.
Chung
d and
Aram J.
Chung
 *abef
*abef
aDepartment of Bioengineering, Korea University, 02841 Seoul, Republic of Korea
bInterdisciplinary Program in Precision Public Health (PPH), Korea University, 02841 Seoul, Republic of Korea
cSchool of Mechanical Engineering, University of Shanghai for Science and Technology, 200240 Shanghai, China
dSchool of Engineering and Materials Science, Queen Mary University of London, E1 4NS London, UK
eMxT Biotech, 04785 Seoul, Republic of Korea
fSchool of Biomedical Engineering, Korea University, 02841 Seoul, Republic of Korea. E-mail: ac467@korea.ac.kr
First published on 10th December 2025
Abstract
Emerging non-viral gene delivery platforms provide alternatives to viral methods. However, they remain limited in scalability and efficiency for clinical translation. We present a fractal-shaped droplet microfluidic system that achieves approximately 98% efficiency and 80% viability at throughputs exceeding 107 cells per min, enabling efficient, large-scale, and clinically relevant cell engineering.
Introduction
Cell-based therapies are rapidly advancing in medicine, with curative potential for treating hematologic malignancies, genetic disorders, and autoimmune diseases.1,2 Among these therapies, chimeric antigen receptor (CAR) T cell therapy presents the clinical impact of genetically engineered immune cells, demonstrating antigen-specific cytotoxicity against hematological cancers.3 Moreover, CAR engineering has been extended to natural killer (NK) cells, macrophages, and stem cell-derived effectors. These applications indicate the broad therapeutic scope and additional demands for scalable and efficient gene delivery systems.4–6 Central to such approaches are intracellular delivery strategies that can introduce functional nucleic acids with both high efficiency and throughput.Various transfection approaches have been developed, each with inherent trade-offs. Viral vectors, such as lentivirus and retrovirus, achieve high transduction efficiencies and stable integrations and are widely used in research and clinical applications. However, their development is limited by immunogenicity, stringent regulatory hurdles, and high manufacturing costs.7–9 Non-viral methods, including electroporation, lipid-mediated delivery, and polymer-based carriers, offer safer and more flexible alternatives. In addition, they have been successfully implemented in mRNA vaccines and CRISPR-based editing. Nevertheless, these approaches suffer from frequent reduced cell viability and inconsistent transfection efficiency.10–12 To overcome these drawbacks, microfluidics-based intracellular delivery platforms have emerged as promising alternatives that combine high delivery efficiency with preservation of cellular integrity.13–17 However, most existing microfluidic systems remain limited in throughput, falling short of the clinical requirement to process several hundred million cells per therapeutic dose.18 Therefore, to meet clinical-scale manufacturing demands, a stable throughput of at least 106 cells per min is typically required.
Within this domain, droplet-based mechanoporation has attracted considerable attention due to its potential to overcome the above-mentioned barriers.19,20 In this technique, cells and cargos are first co-encapsulated in water-in-oil droplets and mechanically deformed as they traverse narrow constrictions. Similar to conventional bulk cell squeezing,21 this process induces reversible membrane disruptions that permit cargo entry.14,15,17,22–27 However, droplet-based systems leverage intrinsic secondary flows inside droplets, enhancing convective and diffusive transport and thereby improving overall delivery efficiency while maintaining high cell viability.19,20,28 Despite these advantages, implementation relies on single-channel architectures that process droplets sequentially, thereby constraining throughput. While increasing the droplet generation frequency may appear simple, it is intrinsically limited by the physical dynamics of droplet formation and device operation. To address this challenge, parallelization of droplet generators has been explored,29–31 but massively parallel droplet arrays require complex and costly fabrication processes.32–34 These drawbacks remain a major obstacle to the translation of droplet-based mechanoporation for large-scale therapeutic applications.
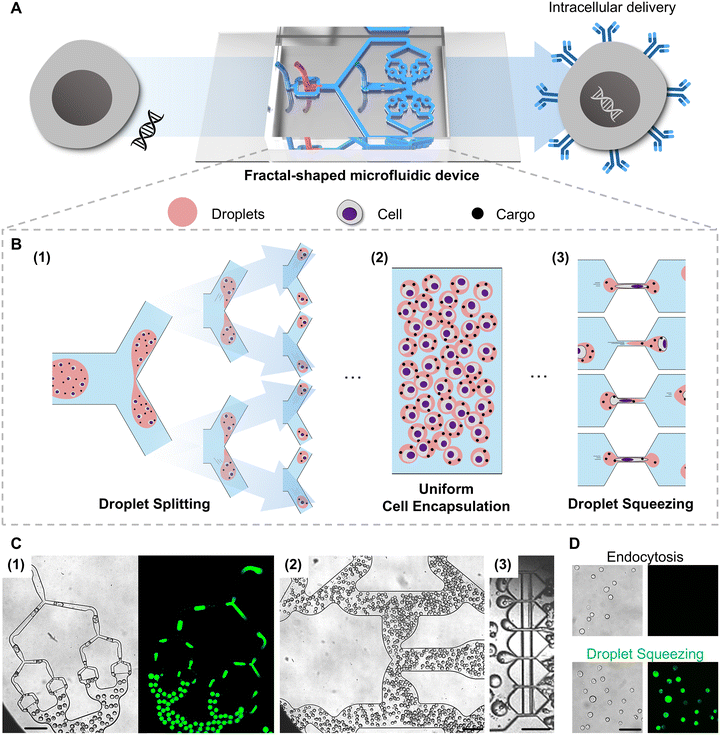
In this study, we present a novel droplet-based highly efficient intracellular delivery platform designed to achieve clinical-scale throughput while maintaining high delivery performance. The system utilizes a fractal-shaped droplet microfluidic architecture, in which a conventional flow-focusing junction35 generates droplets that are subsequently split via a hierarchical network of Y-junctions. The resulting small droplets, typically encapsulating one or two cells, are subsequently directed through constriction regions where mechanical cell deformation induces membrane permeabilization and facilitates cargo entry. This sequential workflow is presented in the schematic overview (Fig. 1A and B), which was visualized using high-speed microscopy (Fig. 1C and Movie S1) and confirmed using fluorescence imaging (Fig. 1D). Collectively, efficient droplet splitting, cell progression, and intracellular delivery of fluorescent cargos are illustrated. We demonstrate that the proposed platform consistently achieves a mean delivery efficiency of approximately 99% and a mean cell viability of approximately 80% at clinically relevant scales exceeding 10 million cells per min, thereby offering a robust and scalable solution for therapeutic cell manufacturing.
Results and discussion
Sequential droplet generation, splitting, and squeezing
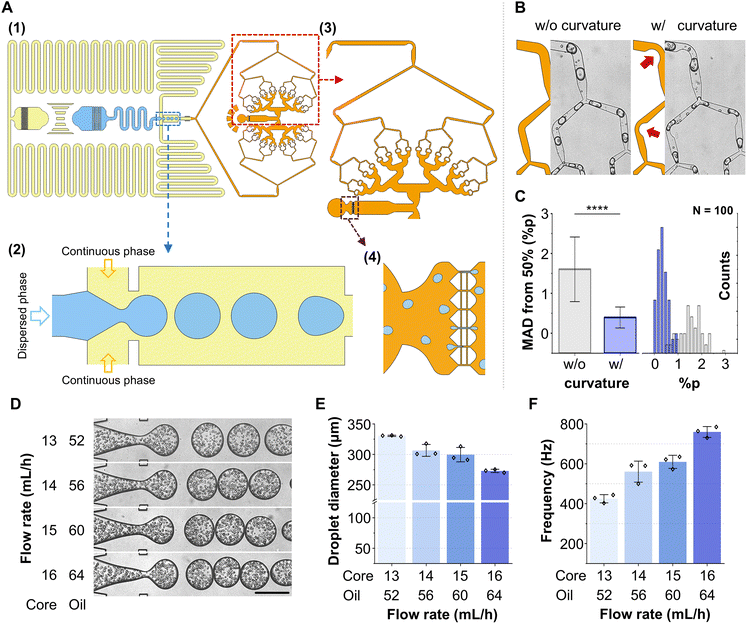
To achieve high scalability while maintaining high delivery efficiency in droplet cell mechanoporation, we hypothesize that stable generation of numerous small droplets upstream, followed by their passage through narrow constrictions for cargo delivery into cells, is crucial. We designed a modular platform comprising droplet generation, geometry-guided splitting, and membrane deformation. The droplet generation module (Fig. 2A(1)) was engineered to produce large, monodisperse water-in-oil droplets under precisely regulated interfacial pressure conditions. This was achieved by a combination of hydrodynamic resistance control with an optimized flow rate ratio.36–39 Pressure modulation through channel resistance, together with regulation of aqueous-to-oil flow rates, enabled stable droplet pinch-off at the flow-focusing junction. The volumetric flow rate ratio between the aqueous core and oil phases was fixed at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, which falls within the reported operating windows of 1
4, which falls within the reported operating windows of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3–1
3–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 for similar microfluidic systems.36–38 A downstream wide orifice was further integrated following the flow-focusing region to stabilize the fluid interface and ensure droplet formation suitable for subsequent splitting and delivery (Fig. 2A(2)).40,41
5 for similar microfluidic systems.36–38 A downstream wide orifice was further integrated following the flow-focusing region to stabilize the fluid interface and ensure droplet formation suitable for subsequent splitting and delivery (Fig. 2A(2)).40,41
The generated droplets were directed into the central splitting module (Fig. 2A(3)), geometrically inspired by the hierarchical branching of pulmonary airways, where recursive bifurcation facilitates uniform flow distribution.42,43 Mirroring this architecture, the device employs a fractal-shaped network to divide large droplets into smaller daughter droplets through a series of 120° Y-junctions. This geometry has been shown to achieve symmetric partitioning while minimizing flow resistance under laminar conditions.44,45 The junctions were radially arranged along concentric circles (Fig. S1). This ensured uniform droplet size and pressure balance by equalizing downstream path lengths. Moreover, this unique fractal splitting structure enabled stepwise isolation of single or a few cells into discrete droplets. Consequently, consistent conditions were established for subsequent membrane permeabilization and molecular delivery (Fig. 2A(3)). By uniformly partitioning the input flow, the system provided each cell with an equivalent microenvironment and identical exposure to cargo.
Curvature was introduced along the inner walls of the branching turns (Fig. 2B) to further enhance uniformity in cell distribution across daughter droplets.46 This modification promoted symmetric flow bifurcation and while suppressing flow dispersion and Dean flow effects, yielding more balanced partitioning. To validate this design principle, we conducted droplet-splitting experiments and quantified the volume ratios of 100 daughter-droplet pairs under each condition (Fig. 2C). Devices without curvature exhibited a mean absolute deviation (MAD) of 1.60%, whereas incorporation of curvature reduced the MAD to 0.40%. This highly significant improvement (p < 0.0001) confirms that the curvature structure substantially enhances droplet-splitting fidelity and ensures reproducible encapsulation performance.
Next, we characterized the flow-rate-dependent behavior of droplet formation by varying the absolute flow rates of the aqueous and oil phases while maintaining a constant core-to-oil ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. Increasing the total flow rate decreased the initial droplet diameter (Fig. 2D and E), which was consistent with shear-driven breakup in microfluidic droplet systems. This inverse correlation enables tunable control of droplet volume and, consequently, encapsulation capacity. At a core flow rate of 16 mL h−1, droplets averaged 272.8 μm in diameter, which was insufficient for complete splitting at the final bifurcation stage. This resulted in frequent incomplete divisions (Fig. S2). Concurrently, the droplet generation frequency increased with flow rate, enhancing throughput (Fig. 2D and F). Under optimized conditions (a core flow rate of 15 mL h−1), droplets with an average diameter of 299.5 μm were generated at a stable frequency of 609.6 Hz. Both droplet diameter and generation frequency exhibited coefficients of variation below 5%, highlighting the robustness and reproducibility of the platform.
4. Increasing the total flow rate decreased the initial droplet diameter (Fig. 2D and E), which was consistent with shear-driven breakup in microfluidic droplet systems. This inverse correlation enables tunable control of droplet volume and, consequently, encapsulation capacity. At a core flow rate of 16 mL h−1, droplets averaged 272.8 μm in diameter, which was insufficient for complete splitting at the final bifurcation stage. This resulted in frequent incomplete divisions (Fig. S2). Concurrently, the droplet generation frequency increased with flow rate, enhancing throughput (Fig. 2D and F). Under optimized conditions (a core flow rate of 15 mL h−1), droplets with an average diameter of 299.5 μm were generated at a stable frequency of 609.6 Hz. Both droplet diameter and generation frequency exhibited coefficients of variation below 5%, highlighting the robustness and reproducibility of the platform.
The final module was designed to mechanically deform cells through constrictions, facilitating transient membrane permeabilization and cargo delivery (Fig. 2A(4)). This region incorporated six squeezing gaps of 6 or 8 μm width (depending on the cell type, optimized in prior work20,47) with a fixed gap length of 70 μm to ensure adequate deformation. To prevent clogging under high-flow-rate conditions, a 6 μm-wide bypass channel was integrated perpendicular to the main gap. This auxiliary flow path acts as a pressure equalizer, enabling continuous droplet progression and enhancing operational robustness.47
Flow rate optimization and intracellular delivery
To maximize throughput, flow rate optimization was performed using K562 cells under four core flow conditions (13–16 mL h−1), while maintaining a constant 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 core-to-oil flow ratio. K562 was chosen due to its suspension phenotype and well-documented resistance to transfection, making it a stringent evaluation model.48 Cells were suspended at 2.5 × 107 cells per mL, and 2000 kDa fluorescein isothiocyanate (FITC)–dextran was used as a model cargo at 0.3 mg mL−1. Throughput was calculated by multiplying the cell concentration [cells per mL] by the core flow rate [mL h−1] and converting the result to cells per minute [cells per min], providing a quantitative measure of the platform's processing capacity.
4 core-to-oil flow ratio. K562 was chosen due to its suspension phenotype and well-documented resistance to transfection, making it a stringent evaluation model.48 Cells were suspended at 2.5 × 107 cells per mL, and 2000 kDa fluorescein isothiocyanate (FITC)–dextran was used as a model cargo at 0.3 mg mL−1. Throughput was calculated by multiplying the cell concentration [cells per mL] by the core flow rate [mL h−1] and converting the result to cells per minute [cells per min], providing a quantitative measure of the platform's processing capacity.
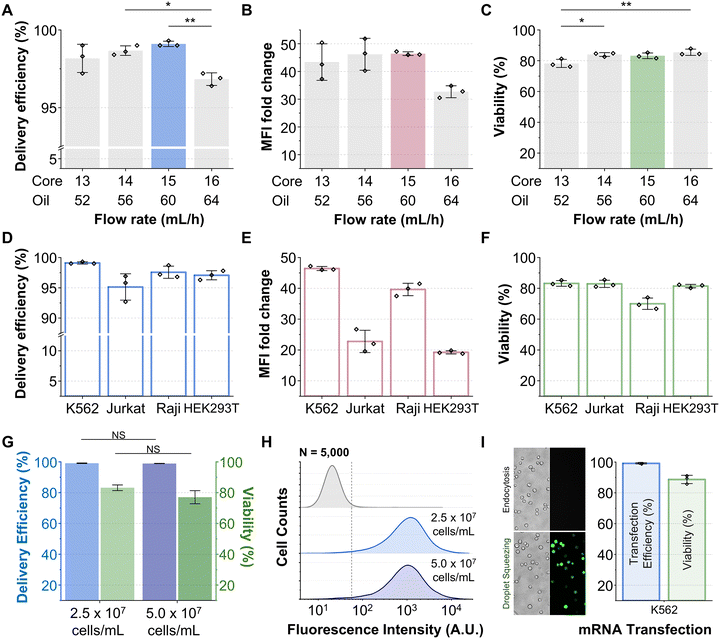
Delivery efficiency, which was measured as previously reported,20,49 increased monotonically from 13 to 15 mL h−1, surpassing 98% at both 14 and 15 mL h−1 but decreased at 16 mL h−1 (Fig. 3A). Mean fluorescence intensity (MFI) fold change exhibited a similar trend, reaching 46.2 and 46.4 at 14 and 15 mL h−1, respectively, before decreasing at 16 mL h−1 (Fig. 3B). These results suggest that increasing the flow rate enhances intracellular delivery up to an optimal threshold, beyond which the performance deteriorates due to bulging of the flexible polydimethylsiloxane (PDMS) squeezing gap under excessive pressure (Fig. S3).
Cell viability exhibited a nonlinear dependence on the flow rate (Fig. 3C). The lowest viability was observed at 13 mL h−1, whereas the other three tested conditions consistently exceeded 80%. Reduced viability at 13 mL h−1 was attributed to flow-dependent clogging in the squeezing gap. Moreover, the infrequent generation of oversized, multiple-cell droplets caused congestion, intensified inter-droplet interactions, and droplet rupture at constrictions.50 Therefore, multiple cells were released simultaneously, inducing blockage (Fig. S4A). To test whether the abrupt release of multiple cells caused clogging, we repeated experiments at 13 mL h−1 and a reduced cell density (1 × 107 cells per mL). Droplet size and frequency remained unchanged (Fig. S5A); however, clogging was substantially alleviated (Fig. S5B), and viability was above 80% without any loss of delivery level (Fig. S5C–E).
At 16 mL h−1, the droplets were smaller and contained fewer cells individually; yet frequent collisions and accumulation at the constriction again caused obstruction (Fig. S4). However, unlike at 13 mL h−1, viability was preserved (Fig. 3C), likely due to the elastic expansion of the PDMS constriction (Fig. S3). To further examine the hydrodynamic influence of the flow rate on cellular responses, three-dimensional two-phase flow simulations were performed using COMSOL Multiphysics®. The numerical results revealed that even under identical inlet flow rates, the presence of droplets locally elevated the shear stress and maximum strain rate within the constriction compared to single-phase flow (Fig. S6; Tables S1 and S2). This indicates that interfacial interactions and geometric confinement amplify the local deformation field around the droplet. Moreover, as the gap width increased, both the shear stress and the maximum strain rate markedly decreased, which is consistent with the experimental observation that cell viability improved under sufficiently high flow conditions (Fig. 3C, S3 and Tables S1–S3).
Finally, both 14 and 15 mL h−1 achieved high delivery efficiencies (>98%) and comparable MFI values, with no significant differences. Given the higher cell processing rate, the optimized condition was selected as 15 mL h−1. These findings suggest that clogging arises from condition-dependent factors rather than structural limitations and can be mitigated through parameter optimization. Importantly, the combination of delivery efficiency and viability above 98% and greater than 80%, respectively, matched that of single-channel-based droplet intracellular delivery systems (Table S4).19,20 Moreover, a clinical-scale throughput previously unattainable was achieved using this method.
Delivery across diverse cell types
To assess generalizability, we tested the platform across four representative cell lines under the optimized condition (core/oil: 15/60 mL h−1): K562 (chronic myelogenous leukemia), Jurkat (acute T cell leukemia), Raji (B lymphocyte), and HEK293T (epithelial, adherent). These represent hematopoietic and non-hematopoietic origins, spanning suspension and adherent phenotypes. All samples were processed under identical conditions (2.5 × 107 cells per mL, 0.3 mg mL−1 2000 kDa FITC-dextran, 15 mL h−1 core flow). Delivery efficiency exceeded 95% in all cases (Fig. 3D), demonstrating robust performance across lineages. MFI fold change values (Fig. 3E) exhibited modest variation, with Jurkat and Raji displaying slightly reduced fluorescence intensity, which may reflect differences in mechanical properties.23 All cell types maintained a viability above 70% (Fig. 3F), including adherent HEK293T, confirming the broad tolerability of mechanical perturbation. These results demonstrate the versatility of the platform for diverse cell engineering applications.System robustness across cell density and cargo types
To evaluate the scalability of the system under increased cellular loads, the concentration of K562 cells increased while maintaining the optimized flow condition. As shown in Fig. 3G, both delivery efficiency and cell viability exhibit slight declines at higher cell density but without statistical significance (delivery efficiency: 99.1% vs. 98.9%; cell viability: 83.2% vs. 77.0%). Fluorescence analysis revealed reduced MFI with higher density (Fig. 3H). This is likely due to hindered deformation or competition for limited cargo. Despite these minor reductions, the platform remained robust, achieving throughputs up to 1.25 × 107 cells per min and enabling engineering of 108 cells within 10 minutes, which is sufficient for clinical dosing.18 Collectively, these results demonstrate a system-level advancement that overcomes scalability barriers beyond method-level novelty.Finally, to confirm functional nucleic acid delivery, EGFP mRNA transfection was performed at the optimized cell concentration (2.5 × 107 cells per mL, 20 μg mL−1, core/oil: 15/60 mL h−1). Robust EGFP expression was observed, with approximately 99% transfection efficiency and above 85% viability (Fig. 3I), comparable to conventional single-channel droplet platforms.19,20 The corresponding mean fluorescence intensity profiles are presented in Fig. S7. These results extend the system's capability beyond inert cargos to functional nucleic acids, underscoring its potential for therapeutic gene delivery.
Conclusions
This study introduced a fractal-structured droplet microfluidic platform that achieved high-throughput intracellular delivery by integrating recursive droplet splitting with geometry-guided membrane deformation. Inspired by pulmonary airway branching, the fractal droplet network enabled uniform cell encapsulation without the need for complex parallelization. Incorporating six constriction gaps and multiple bypasses effectively overcomes the structural limitations of single-gap squeezing channels. Using this platform, the system achieved delivery efficiencies of approximately 98% with cell viabilities of approximately 80% across both suspension (K562, Jurkat, Raji) and adherent (HEK293T) cell types for 2000 kDa FITC-dextran. Importantly, this performance was maintained under elevated cell concentrations, demonstrating the robustness and scalability of the proposed method. Moreover, throughputs exceeded 10 million cells per min, which is well beyond clinical-scale manufacturing requirements.18 Functional delivery was further validated by successful EGFP mRNA transfection, confirming the applicability of the proposed method to nucleic acid-based therapeutics.A limitation of the proposed platform is its dependence on the oil phase, which necessitates an additional recovery step for processed cells. For convenience, perfluorooctanol (PFO) was employed in this study as one of the most commonly used de-emulsifiers;20,51,52 however, we confirmed that nearly complete droplet breakup could also be achieved via electrostatic coalescence53 (Fig. S8). Moreover, because the system operates via tubing and syringe pumps, a certain dead volume is unavoidable, making experiments with very small sample volumes challenging and increasing the required number of cells and cargos such as mRNA. Nevertheless, these drawbacks are outweighed by key advantages, including high-throughput operation, consistent cell encapsulation, and high delivery efficiency across diverse cell types.
Regarding versatility in delivery cargo types, given that the delivery of cargos such as pDNA and CRISPR-Cas9 complexes has already been validated in a previous study,20 this platform is expected to be applicable to these cargos and further extendable to other large therapeutic molecules, including those targeting clinically relevant primary human cells. In summary, the fractal-structured droplet microfluidic system offers a scalable, efficient, and broadly applicable strategy for intracellular delivery, providing significant potential for next-generation cell-based engineering and therapeutic development.
Materials and methods
Microfabrication of microfluidic devices
In this study, the microfluidic channels were designed using AutoCAD (Autodesk, USA) and patterned onto 4-inch silicon-on-insulator (SOI) wafers via deep reactive-ion etching (EPG, Republic of Korea). PDMS channels were fabricated by soft lithography using a Sylgard 184 elastomer kit (Dow-Corning, USA), and inlet/outlet holes were punched with a pin vise. The PDMS layer and glass slide (Marienfeld, Germany) were treated with oxygen plasma (CUTE, Femto Science, Republic of Korea) and bonded immediately. Fluorinated oil (909-FluoroCoat-1ML, RAN Biotech, USA) was introduced via the inlet to passivate the channel walls. Assembled devices were incubated at 75 °C overnight to reinforce bonding strength.Cell culture and preparation
K562 (KCLB no. 10243) and Jurkat (KCLB no. 40152) cell lines were obtained from the Korean Cell Line Bank (KCLB, Republic of Korea), whereas Raji (ATCC no. CCL-86) and HEK293T (ATCC no. CRL-3216) were obtained from the American Type Culture Collection (ATCC, USA). K562, Jurkat, and Raji suspension cultures were maintained in an RPMI-1640 medium (Corning, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin–streptomycin (Gibco, USA). HEK293T adherent cells were cultured in DMEM (Cytiva, USA) with identical supplementation. All cultures were maintained at 37 °C in 5% CO2, and subcultured every 72 h.For device loading, cells were washed twice with Dulbecco's phosphate-buffered saline (DPBS, Cytiva, USA) and sequentially filtered through a 40 μm cell strainer (SPL Life Sciences, Republic of Korea). The delivery medium was prepared by mixing 2000 kDa FITC-dextran (Sigma-Aldrich, USA) and 997-nt EGFP-mRNA (TriLink Biotechnologies, USA) in Opti-MEM (Gibco, USA). Subsequently, the cells were resuspended in this medium at concentrations ranging from 1 × 107 to 5 × 107 cells per mL and injected through the device core inlet.
Droplet generation and recovery
Monodisperse water-in-oil droplets were generated using commercial droplet generation oil (Bio-Rad, USA) filtered sequentially through 0.2 μm hydrophilic and hydrophobic polytetrafluoroethylene (PTFE) syringe filters (Advantec, Japan). Filtered oil and aqueous phases were loaded into Luer-Lok tip syringes (BD, USA), connected to 1/32″ OD × 0.020″ ID PEEK tubing (IDEX, USA), and infused into the microfluidic device using a syringe pump (Harvard Apparatus, USA). Emulsions were collected and destabilized with 1H,1H,2H,2H-perfluoro-1-octanol (Thermo Fisher Scientific, USA). The aqueous phase was recovered, washed twice with DPBS, and incubated at 37 °C in 5% CO2 for either 18 or 24 h, depending on the experiment. Cell viability was assessed by trypan blue exclusion (Lonza, Swiss) using a hemocytometer.Flow cytometry analysis
Following incubation, control and treated samples were washed with phosphate-buffered saline (PBS) and resuspended in eBioscience™ flow cytometry staining buffer (Invitrogen, USA) containing propidium iodide (PI; Invitrogen, USA). Flow cytometry was performed on a Guava® easyCyte™ (Luminex, USA). Intact single cells were gated using forward scatter (FSC) and side scatter (SSC) to exclude debris and dead cells. Live cells were defined as PI-negative, and 5000 events were recorded per sample. Delivery or transfection efficiency was determined by defining the top 5% of fluorescent intensity in the control group as the threshold for endocytosis. The fraction of cells exceeding this threshold in treated samples was reported as efficient. MFI fold change was calculated as the ratio of sample MFI to control MFI.Image acquisition and statistical analysis
High-speed imaging was performed using a Phantom VEO 710 L camera (Vision Research, USA) to visualize droplet formation and cell processing. Quantitative analysis of droplet size, generation frequency, and squeezing dynamics was conducted (NIH, USA). Statistical analyses were conducted in SPSS Statistics v26.0 (IBM, USA): Welch's t-test for two-group comparisons and one-way ANOVA with Tukey's HSD post hoc test for multiple comparisons. Significance was set at p < 0.05. All graphs were generated using OriginPro (OriginLab, USA).Author contributions
All authors contributed to the development of the conceptual framework and its translation into a functional experimental platform. M. S. and Y. K. performed the experiments and data analysis. D. J. and Y. S. conducted the numerical simulations. M. S. and A. J. C. wrote the manuscript with input from all the authors. All authors discussed the results and contributed to the writing of the manuscript.Conflicts of interest
A. J. C. declares the following competing interest: a financial interest in MxT Biotech, which is commercializing the presented technology.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5lc00865d.
Acknowledgements
The authors express their sincere gratitude to all members of the Biomicrofluidics Laboratory at Korea University for their valuable discussions and support. This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the Korean government (MSIT; Ministry of Science and ICT; RS-2023-00242443, and RS-2023-00218543), and the Technology Development Program (RS-2025-25404796) funded by the Ministry of SMEs and Startups (MSS, Korea) awarded to A. J. C. Additional support was provided by a Royal Society Research Grant under the International Exchange Scheme (IES/R1/251157) to A. J. C., Y. S., and D. J. D. J. was further supported by the Newton International Fellowship Follow-on Funding from the Royal Society (AL/24100036).References
- J. Hur and A. J. Chung, Adv. Sci., 2021, 8, e2004595 CrossRef PubMed.
- R. Greco, T. Alexander, N. Del Papa, F. Muller, R. Saccardi, F. Sanchez-Guijo, G. Schett, B. Sharrack, J. A. Snowden, K. Tarte, F. Onida, I. Sanchez-Ortega, J. Burman, C. Castilla Llorente, R. Cervera, F. Ciceri, A. Doria, J. Henes, J. Lindsay, A. Mackensen, P. A. Muraro, E. Ricart, M. Rovira, T. Zuckerman, I. Yakoub-Agha and D. Farge, EClinicalMedicine, 2024, 69, 102476 CrossRef PubMed.
- R. C. Sterner and R. M. Sterner, Blood Cancer J., 2021, 11, 69 CrossRef PubMed.
- M. Daher, L. Melo Garcia, Y. Li and K. Rezvani, Clin. Transl. Immunol., 2021, 10, e1274 CrossRef CAS PubMed.
- K. Pan, H. Farrukh, V. Chittepu, H. Xu, C. X. Pan and Z. Zhu, J. Exp. Clin. Cancer Res., 2022, 41, 119 CrossRef CAS PubMed.
- K. Hadiloo, S. Taremi, M. Heidari and A. Esmaeilzadeh, Biomark. Res., 2023, 11, 103 CrossRef PubMed.
- A. Brave, K. Ljungberg, B. Wahren and M. A. Liu, Mol. Pharmaceutics, 2007, 4, 18–32 CrossRef PubMed.
- Y. Seow and M. J. Wood, Mol. Ther., 2009, 17, 767–777 CrossRef CAS PubMed.
- J. T. Bulcha, Y. Wang, H. Ma, P. W. L. Tai and G. Gao, Signal Transduction Targeted Ther., 2021, 6, 53 CrossRef CAS PubMed.
- J. Gehl, Acta Physiol. Scand., 2003, 177, 437–447 CrossRef CAS PubMed.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Nat. Rev. Mater., 2021, 6, 1078–1094 CrossRef CAS PubMed.
- A. I. S. van den Berg, C. O. Yun, R. M. Schiffelers and W. E. Hennink, J. Controlled Release, 2021, 331, 121–141 CrossRef CAS PubMed.
- J. A. Jarrell, A. A. Twite, K. Lau, M. N. Kashani, A. A. Lievano, J. Acevedo, C. Priest, J. Nieva, D. Gottlieb and R. S. Pawell, Sci. Rep., 2019, 9, 3214 CrossRef PubMed.
- J. Hur, I. Park, K. M. Lim, J. Doh, S. G. Cho and A. J. Chung, ACS Nano, 2020, 14, 15094–15106 CrossRef CAS PubMed.
- G. Kang, D. W. Carlson, T. H. Kang, S. Lee, S. J. Haward, I. Choi, A. Q. Shen and A. J. Chung, ACS Nano, 2020, 14, 3048–3058 CrossRef CAS PubMed.
- P. Chakrabarty, P. Gupta, K. Illath, S. Kar, M. Nagai, F. G. Tseng and T. S. Santra, Mater. Today Bio, 2022, 13, 100193 CrossRef CAS PubMed.
- D. Sevenler and M. Toner, Nat. Commun., 2024, 15, 115 CrossRef CAS PubMed.
- W. Huang, J. Li, M. Z. Liao, S. N. Liu, J. Yu, J. Jing, N. Kotani, L. Kamen, S. Guelman and D. R. Miles, Clin. Pharmacol. Ther., 2022, 112, 968–981 CrossRef CAS PubMed.
- B. Joo, J. Hur, G. B. Kim, S. G. Yun and A. J. Chung, ACS Nano, 2021, 15, 12888–12898 CrossRef CAS PubMed.
- Y. J. Kim, D. Yun, J. K. Lee, C. Jung and A. J. Chung, Nat. Commun., 2024, 15, 8099 CrossRef CAS PubMed.
- G. Zhang, R. Mu, Y. Ma and B. Li, Small Methods, 2025, e2500338 CrossRef PubMed.
- A. Liu, M. Islam, N. Stone, V. Varadarajan, J. Jeong, S. Bowie, P. Qiu, E. K. Waller, A. Alexeev and T. Sulchek, Mater. Today, 2018, 21, 703–712 CrossRef CAS PubMed.
- M. E. Kizer, Y. Deng, G. Kang, P. E. Mikael, X. Wang and A. J. Chung, Lab Chip, 2019, 19, 1747–1754 RSC.
- A. Uvizl, R. Goswami, S. D. Gandhi, M. Augsburg, F. Buchholz, J. Guck, J. Mansfeld and S. Girardo, Lab Chip, 2021, 21, 2437–2452 RSC.
- C. Kwon and A. J. Chung, Lab Chip, 2023, 23, 1758–1767 RSC.
- J. Hur, H. Kim, U. Kim, G. B. Kim, J. Kim, B. Joo, D. Cho, D. S. Lee and A. J. Chung, Nano Lett., 2023, 23, 7341–7349 CrossRef CAS PubMed.
- J. Lim, D. Oh, M. Cheng, U. Chintapula, S. Liu, D. Reynolds, X. Zhang, Y. Zhou, X. Xu and J. Ko, Small, 2025, 21, e2410975 CrossRef PubMed.
- D. Jing, R. Lu, A. Farutin, Z. Guo, F. Wang, W. Wang, C. Misbah and Y. Sui, Commun. Phys., 2024, 7, 310 CrossRef.
- W. Li, J. Greener, D. Voicu and E. Kumacheva, Lab Chip, 2009, 9, 2715–2721 RSC.
- T. Nisisako, T. Ando and T. Hatsuzawa, Lab Chip, 2012, 12, 3426–3435 RSC.
- D. Conchouso, D. Castro, S. A. Khan and I. G. Foulds, Lab Chip, 2014, 14, 3011–3020 RSC.
- M. B. Romanowsky, A. R. Abate, A. Rotem, C. Holtze and D. A. Weitz, Lab Chip, 2012, 12, 802–807 RSC.
- H. H. Jeong, V. R. Yelleswarapu, S. Yadavali, D. Issadore and D. Lee, Lab Chip, 2015, 15, 4387–4392 RSC.
- S. Yadavali, H. H. Jeong, D. Lee and D. Issadore, Nat. Commun., 2018, 9, 1222 CrossRef PubMed.
- S. L. Anna, N. Bontoux and H. A. Stone, Appl. Phys. Lett., 2003, 82, 364–366 CrossRef CAS.
- A. S. Utada, A. Fernandez-Nieves, H. A. Stone and D. A. Weitz, Phys. Rev. Lett., 2007, 99, 094502 CrossRef PubMed.
- G. F. Christopher, N. N. Noharuddin, J. A. Taylor and S. L. Anna, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2008, 78, 036317 CrossRef PubMed.
- M. De Menech, P. Garstecki, F. Jousse and H. A. Stone, J. Fluid Mech., 2008, 595, 141–161 CrossRef.
- W. Zeng, B. Wang, H. Chang and P. Neužil, Phys. Fluids, 2024, 36, 032009 CrossRef CAS.
- Y.-C. Tan, V. Cristini and A. P. Lee, Sens. Actuators, B, 2006, 114, 350–356 CrossRef CAS.
- F. Mardani, S. Falahatian and M. Taghipoor, Results Eng., 2023, 18, 101125 CrossRef CAS.
- R. J. Metzger, O. D. Klein, G. R. Martin and M. A. Krasnow, Nature, 2008, 453, 745–750 CrossRef CAS PubMed.
- D. Iber, Curr. Top. Dev. Biol., 2021, 143, 205–237 Search PubMed.
- D. K. Deka, S. Pati and P. R. Randive, Colloids Surf., A, 2022, 633, 127873 CrossRef CAS.
- S. J. Hymel, H. Lan, H. Fujioka and D. B. Khismatullin, Phys. Fluids, 2019, 31, 082003 CrossRef PubMed.
- S. K. Griffiths and R. H. Nilson, Anal. Chem., 2001, 73, 272–278 CrossRef CAS PubMed.
- Q. Liu and A. J. Chung, Parallelized Droplet Microfluidic Mechanoporation Enables Robust and Clogging-Resistant Intracellular Gene Delivery, 2025, in revision.
- F. Richter, P. Mapfumo, L. Martin, J. I. Solomun, F. Hausig, J. J. Frietsch, T. Ernst, S. Hoeppener, J. C. Brendel and A. Traeger, J. Nanobiotechnol., 2021, 19, 70 CrossRef CAS PubMed.
- H. Kim, M. Lee, B. Han, J. Kim, D. Cho, J. Doh and A. J. Chung, Adv. Sci., 2025, 12, e2412544 CrossRef PubMed.
- N. Bremond, A. R. Thiam and J. Bibette, Phys. Rev. Lett., 2008, 100, 024501 CrossRef PubMed.
- M. S. Chowdhury, W. Zheng, S. Kumari, J. Heyman, X. Zhang, P. Dey, D. A. Weitz and R. Haag, Nat. Commun., 2019, 10, 4546 CrossRef PubMed.
- F. Peng, L. K. Mansson, S. H. Holm, S. Ghosh, G. Carlstrom, J. J. Crassous, P. Schurtenberger and J. O. Tegenfeldt, J. Phys. Chem. B, 2019, 123, 9260–9271 CrossRef CAS PubMed.
- M. Karbaschi, P. Shahi and A. R. Abate, Biomicrofluidics, 2017, 11, 044107 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |