Lymph node paracortex-inspired on-a-chip recapitulating immunosenescence: a cancer vaccine immunogenicity and antitumoral efficacy screening platform
Surjendu
Maity
 ab,
Alireza
Hassani Najafabadi
ab,
Alireza
Hassani Najafabadi
 ac,
Satoru
Kawakita
a,
Danial
Khorsandi
a,
Can
Yilgor
a,
Christopher
Jewell
a,
Neda
Mohaghegh
a,
Mehmet Remzi
Dokmeci
ac,
Satoru
Kawakita
a,
Danial
Khorsandi
a,
Can
Yilgor
a,
Christopher
Jewell
a,
Neda
Mohaghegh
a,
Mehmet Remzi
Dokmeci
 a,
Ali
Khademhosseini
a,
Ali
Khademhosseini
 a and
Vadim
Jucaud
a and
Vadim
Jucaud
 *a
*a
aTerasaki Institute for Biomedical Innovation, 21100 Erwin St, Woodland Hills, CA 91367, USA. E-mail: vjucaud@terasaki.org
bDepartment of Orthopedic Surgery, Duke University School of Medicine, Duke University, Durham, NC 27710, USA
cDivision of Pharmaceutical Sciences, James L. Winkle College of Pharmacy, University of Cincinnati, Cincinnati, OH 4526, USA
First published on 24th November 2025
Abstract
Immunosenescence dramatically reduces cancer vaccine efficacy in elderly patients, who represent the majority of cancer cases. Despite this clinical reality, the age-related immune decline is rarely considered in preclinical testing. Therefore, novel in vitro models to test cancer vaccine efficacy, considering immunosenescence, are needed. Our novel lymph node paracortex-inspired on-a-chip (LNPoC) platform addresses this gap by recapitulating age-dependent immune responses against cancer vaccines, specifically antigen presentation, antigen-specific T cell activation, and antitumoral responses. Using this platform, we demonstrated that bone marrow-derived antigen-presenting cells (APCs) from young mice (6–7 weeks) displayed significantly enhanced ovalbumin (OVA) peptide presentation compared to APCs from older mice (35–36 weeks). This age-dependent difference translated to significantly greater OVA-specific CD8+ T cell activation and increased cytotoxicity against B16-OVA cancer cells. These age-dependent differences are unique to our LNPoC and undetectable in traditional 2D cultures, confirming that our LNPoC was more effective than 2D cultures at recapitulating immunosenescence-mediated immune responses against cancer vaccines in vitro. The in vivo validation confirms these findings, as young mice demonstrated higher OVA-specific CD8+ T cell responses and smaller tumors than older mice. Our LNPoC is a valuable tool for assessing immunosenescence's impact on cancer vaccines, potentially guiding more effective therapies for older adults.
Introduction
Cancer vaccines are a promising type of immunotherapy that trains the immune system to recognize and attack tumor cells.1,2 They work by immunizing a host against tumor-associated antigens (TAAs) to prevent tumor growth, metastasis, and recurrence.2 Lymph nodes (LNs) play a critical role in this immunization process, as they are the primary site for antigen presentation and T cell activation,2,3 particularly within the paracortex region where the interaction between antigen-presenting cells (APCs) and T cells initiates effective antitumor immune responses. However, the clinical translation of cancer vaccines remains challenging despite their potential and global market expected to reach ∼$15 billion by 2030.4–6The gap in translation can be attributed to the limitations of current preclinical models used to study cancer vaccine efficacy in recapitulating human physiology.7,8 For example, the MAGE-A3 cancer vaccine showed promise in preclinical models yet failed to demonstrate clinical benefit in a phase III trial for non-small cell lung cancer.9,10 Another example is the gp100 melanoma vaccine, which initially showed potent antitumor activity in mice but failed to demonstrate similar efficacy in human clinical trials.11In vivo murine models are the most widely used models for preclinical cancer vaccine testing, but they present significant immunological differences from humans, leading to inaccurate clinical predictions of vaccine efficacy.12,13In vitro models, such as 2D monolayer cultures and simple 3D spheroids, also fail to replicate the complexity of the human immune system.14 These models do not capture the intricate spatial organization and dynamic cellular interactions within the LNs. In addition, they do not simulate in vivo mechanical forces and fluid dynamics, allowing the continuous flow of immune cells and antigens through lymphatic vessels. As a result, in vitro models fail to accurately predict the potential clinical efficacy of cancer vaccines. The limitations of current preclinical models in predicting human immune responses emphasize the need for more clinically relevant platforms for studying cancer vaccines.
With the recent passing of the FDA Modernization Act 2.0, organ-on-a-chip (OoC) technologies have gained tremendous attention due to their capability to represent the human body and generate clinically translatable data.15–17 OoC platforms integrate microfluidic technology with cellular biology and tissue engineering to recreate the structure and function of human organs on a microscale.18–22 Several OoC platforms have been developed to test the efficacy of different immunotherapies;23–29 however, none have been developed for testing cancer vaccine efficacy. In this context, a LN paracortex-inspired on-a-chip (LNPoC) could replicate the native microenvironment of the LN, including the critical APC–T cell interactions. Few LN-on-a-chip platforms can model the paracortical region of the lymph node where dynamic APC–T cell interactions occur, leading to T cell activation; however, none have focused on evaluating cancer vaccine responses (i.e., immunogenicity and antitumoral efficacy).23,30–33 Therefore, there is a need to develop a novel physiologically relevant platform for testing cancer vaccine efficacy in vitro.
Another critical factor often ignored in preclinical testing of cancer vaccine efficacy is immunosenescence, the age-related decline in immune function that affects cancer patients.34,35 Immunosenescence is characterized by a decrease in the population of naïve T cells, reduced T cell receptor (TCR) diversity, and an increase in the number of exhausted or senescent T cells, all of which reduce the immune system's capacity to mount effective antitumor responses.36–42 This is a major hurdle for cancer vaccines, which rely on robust T cell activation and expansion to achieve therapeutic effects. Aging populations, which constitute the majority of cancer patients, are often unaccounted for in preclinical models and excluded from early-phase cancer vaccine trials,43–45 leading to the paucity of clinical data from these populations.43–47 Consequently, cancer vaccines that show efficacy in younger, healthy subjects may fail in elderly populations due to the age-related immune decline or immunosenescence.39,48,49 This gap in preclinical modeling underscores the need for novel in vitro models that can account for the effects of immunosenescence on vaccine efficacy.
The development of a LNPoC is a boon for cancer vaccine research and development if it mimics human immunosenescence. Such technology can expedite the clinical translation of cancer vaccines, particularly for the geriatric population. Therefore, we developed a LNPoC that can recapitulate age-dependent adaptive immune responses against cancer vaccines. We conducted a proof-of-concept study to determine whether our novel LNPoC could recapitulate in vivo adaptive immune responses to cancer vaccines, from immunogenicity to effector functions, better than conventional 2D in vitro models. Using OVA-derived peptides as model antigens and murine immune cells from young or old individuals, we compared antigen presentation, antigen-specific CD8+ T cell generation, and antitumoral efficacy between a 2D static co-culture model, an in vivo B16-OVA mouse model, and our novel LNPoC.
Experimental methods
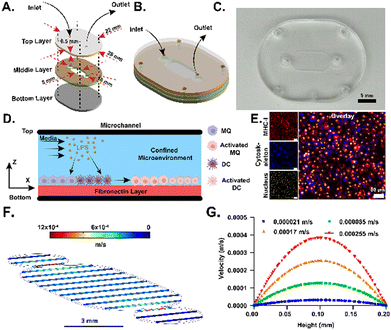
Design and fabrication of the microfluidic device
The 2D computer-aided designs (CADs) for the different layers (top, middle, and bottom) of the microfluidic LNPoC were made in CorelDRAW 2021 (Corel, Canada). PMMA sheets with thicknesses of 3.18 mm (top layer) (McMaster-Carr, USA), 0.2 mm (middle layer) (Astra Products, USA), and 0.2 mm (bottom layer) (Astra Products, USA) were cut using a CO2 laser cutter (Universal Laser Systems, USA) based on the design shown in Fig. 1A. Each PMMA layer was 28 mm long and 22 mm wide. The middle layer's cut-out area consisted of a 12 mm long and 5 mm wide rounded square connected to two 3 mm diameter holes. The top layer had an inlet and outlet port (0.5 mm in diameter). After laser cutting the different PMMA layers, they were cleaned with 100% ethanol (Sigma-Aldrich, USA), rinsed with deionized water, and thoroughly dried before bonding. To bond the PMMA layers and generate the microfluidic LNPoC, the PMMA layers were treated with O2 plasma for 10 minutes, rinsed with 70% isopropyl alcohol (IPA) (Sigma-Aldrich, USA), and held on top of each other for 2 minutes using paper clips. The excess IPA was removed from the microfluidic LNPoC before being placed in a 60 °C oven for 15 minutes to complete the bonding of the different PMMA layers. After bonding, the resulting microchamber was washed with 70% ethanol, followed by 1× phosphate-buffered saline (PBS), and air-dried.Isolation of monocytes from mouse bone marrow
Animals received comprehensive care in compliance with federal, state, and local guidelines. All procedures conducted on animals adhered to the regulations and were approved by the Lundquist Institute Committee on Use and Care of Animals (ICUCA #32706–02). Young (6–7 weeks old) and old (35–36 weeks old) C57BL/6 female mice (The Jackson Laboratory, USA) were housed in a pathogen-free environment for at least 3 days and kept at room temperature with free access to water and a 12 hour light/dark cycle. The C57BL/6 mice were euthanized, and the femurs and tibia were harvested. The bone marrow was flushed out of the femurs and tibia with sterile 1× PBS using a 5 mL syringe with a 30-gauge needle, and the bone marrow was collected on a 40 μm cell strainer (Corning, USA) placed on top of a 50 mL conical tube. The bone marrow was mashed using a syringe's plunger, and then the cell strainer was rinsed with 10 mL of 1× PBS. The resulting bone marrow cell suspension was centrifuged at 400g for 10 minutes at 4 °C. The cells were resuspended in 1× PBS and directly used for subsequent differentiation into MQs and DCs.Macrophages and dendritic cell differentiation
The freshly isolated young or old bone marrow cells were plated in a Petri dish (2 × 106 cells per dish) and cultured for 7 days at 37 °C with 5% CO2, and half of the culture media were replaced with fresh media on days 3 and 5. To generate MQs, the bone marrow cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) + GlutaMax (Fisher Scientific, USA), 1× non-essential amino acid (Thermo Fisher Scientific, USA), 1 mM sodium pyruvate (Cytiva, USA), 100 μM B-mercaptoethanol (Sigma-Aldrich, USA), and mouse macrophage colony-stimulating factor (M-CSF) (5 ng mL−1) (Miltenyi Biotec, USA). To generate DCs, the bone marrow cells were cultured in Roswell Park Memorial Institute (RPMI) medium (Cytiva, USA), 1× non-essential amino acid, 1 mM sodium pyruvate, 100 μM B-mercaptoethanol, and mouse granulocyte-macrophage colony-stimulating factor (GM-CSF) (5 ng mL−1) (GenScript, USA). The differentiation of bone marrow cells into MQs (CD11b+) or DCs (CD11c+) was confirmed with flow cytometry (FACSCanto II flow cytometer, BD Biosciences, USA). Using FlowJo™ software (Becton, Dickinson and Company, USA), flow cytometry data were analyzed using a sequential gating strategy that included singlet discrimination to exclude doublets and time-based gating to ensure acquisition stability. These standard quality control steps were applied consistently across all experimental conditions. Differentiated bone marrow cells were first gated based on size (side-scatter) and granularity (forward-scatter), then MQs (F4/80+/CD11b+) were quantified after staining the cells with an Alexa Fluor® 700-conjugated monoclonal anti-mouse F4/80 antibody (BioLegend, USA) and a Brilliant Violet 711™-conjugated monoclonal anti-mouse CD11b antibody (BioLegend, USA), and DCs (F4/80−/CD11c+) were quantified after staining the cells with a PerCP-conjugated monoclonal anti-mouse F4/80 antibody (BioLegend, USA) and an FITC-conjugated monoclonal anti-mouse CD11c antibody (BioLegend, USA).50,51Stimulation of macrophages and dendritic cells using OVA peptides
Young and old MQs and DCs were collected from Petri dishes and resuspended at a concentration of 1 × 106 cells per mL in their corresponding cell culture media containing 100 ng mL−1 OVA peptide fragment (SIINFEKL) (Anaspec, USA) and 250 ng mL−1 lipopolysaccharides (LPS) (Sigma-Aldrich, USA). For the off-chip stimulation, 100 μL of MQ or DC suspensions were seeded in a flat-bottom 96-well plate and cultured for 7 days at 37 °C with 5% CO2, and half of the culture media were replaced with fresh media on days 3 and 5. For the on-chip stimulation, the LNPoC was first sterilized with 30 minutes of UV treatment, followed by washing with 70% ethanol for 10 minutes. The microfluidic devices were washed 5 times with sterile 1× PBS and then coated with fibronectin (50 μg mL−1) (Thermo Fisher Scientific, USA) for 2 hours at 37 °C with 5% CO2. Finally, 20 μL of young or old cell suspensions were seeded and cultured in the LNPoC for 7 days at 37 °C with 5% CO2, and half of the culture media were replaced with fresh media on days 3 and 5. The presentation of OVA peptides by MHC-I on MQs or DCs was analyzed by flow cytometry using FlowJo™ software. Differentiated bone marrow cells were first gated based on size (side-scatter) and granularity (forward-scatter), and then MQs (OVA+/CD11b+) and DCs (OVA+/CD11c+) were quantified after staining the cells with iTAg Tetramer/PE-H-2 Kb OVA (NIH Core Facility, USA), a Brilliant Violet 711™-conjugated monoclonal anti-mouse CD11b antibody (BioLegend, USA), and an FITC-conjugated monoclonal anti-mouse CD11c antibody (BioLegend, USA).Isolation of naïve CD8+ T cells from mouse spleen
Young (6–7 weeks old) and old (35–36 weeks old) C57BL/6 female mice (The Jackson Laboratory, USA), housed in a pathogen-free environment for at least 3 days and kept at room temperature with free access to water and a 12 hour light/dark cycle, were euthanized under the ICUCA committee of the Lundquist Institute (#32706-02), and spleens were harvested. Under sterile conditions, four spleens were cut into small pieces on a Petri dish using a scalpel and then soaked in T cell media (RPMI, 1× non-essential amino acid, 1 mM sodium pyruvate, and 100 μM B-mercaptoethanol). The solution containing the young or old minced spleens was passed through a 40 μm cell strainer placed on top of a 50 mL conical tube. The spleens were mashed using a syringe's plunger, and then the cell strainer was rinsed with 10 mL of T cell media. The resulting spleen cell suspension was centrifuged at 400g for 10 minutes at 4 °C, and the cell pellet was resuspended in T cell media. Then, the spleen cell suspension was passed through a 40 μm cell strainer to remove clumps or debris. The young and old naïve CD8+ T cells were isolated by negative selection from the purified spleen cell solution using an EasySep™ Mouse CD8+ T Cell Isolation Kit (STEMCELL Technologies, USA). The purification of naïve CD8+ T cells was confirmed by flow cytometry using FCS Express™ software. Purified cells were first gated based on size (side-scatter) and granularity (forward-scatter), and then live naïve CD8+ T cells were quantified after staining the cells with a Zombie Violet™ Fixable Viability Kit (BioLegend, USA) and an APC-conjugated recombinant anti-mouse CD8a antibody (BioLegend, USA).OVA-specific activation of naïve CD8+ T cells
Freshly isolated young or old naïve CD8+ T cells were resuspended in T cell media and then co-cultured with OVA-stimulated MQs and DCs. Equivalent initial cell seeding concentrations of 1 × 106 cells per mL were applied in both the 2D (off-chip) and 3D (on-chip) setups to maintain experimental consistency and enable direct comparison of cellular responses between the two systems. For the off-chip activation of naïve CD8+ T cells, 100 μL of MQ and DC suspensions (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 MQ to DC ratio at a concentration of 1 × 106 cells per mL) were seeded in a flat-bottom 96-well plate and cultured in cell culture media containing 100 ng mL−1 OVA peptides and 250 ng mL−1 LPS for 7 days at 37 °C with 5% CO2, and half of the culture media were replaced with fresh media on days 3 and 5. After 7 days of OVA peptide stimulation, 100 μL of naïve CD8+ T cells (1 × 106 cells per mL) were added to the 96-well plate and co-cultured for 2 days at 37 °C and 5% CO2 with MQs and DCs. After 2 days, activated CD8+ T cells were collected from the 96-well plate. For the on-chip activation of naïve CD8+ T cells, 20 μL of young and old MQ and DC suspensions were seeded and cultured in the LNPoC with cell culture media containing 100 ng mL−1 OVA peptides and 250 ng mL−1 LPS for 7 days at 37 °C with 5% CO2, and half of the culture media were replaced with fresh media on days 3 and 5. After 7 days, 20 μL of naïve CD8+ T cells were seeded into the LNPoC containing OVA-stimulated MQs and DCs and cultured for 2 days at 37 °C with 5% CO2. After 2 days, activated CD8+ T cells were collected from the LNPoC. The generation and proliferation of young and old OVA-specific CD8+ T cells were analyzed by flow cytometry using FlowJo™ software. Activated CD8+ T cells were first gated based on size (side-scatter) and granularity (forward-scatter), and then OVA-specific CD8+ T cells were quantified after staining the cells with a PE-conjugated OVA-tetramer (NIH Core Facility, USA) and an APC-conjugated recombinant anti-mouse CD8a antibody (BioLegend, USA). Activated CD8+ T cells were also stained with CFSE (Thermo Fisher Scientific, USA) to assess the proliferation of OVA-specific CD8+ T cells.
1 MQ to DC ratio at a concentration of 1 × 106 cells per mL) were seeded in a flat-bottom 96-well plate and cultured in cell culture media containing 100 ng mL−1 OVA peptides and 250 ng mL−1 LPS for 7 days at 37 °C with 5% CO2, and half of the culture media were replaced with fresh media on days 3 and 5. After 7 days of OVA peptide stimulation, 100 μL of naïve CD8+ T cells (1 × 106 cells per mL) were added to the 96-well plate and co-cultured for 2 days at 37 °C and 5% CO2 with MQs and DCs. After 2 days, activated CD8+ T cells were collected from the 96-well plate. For the on-chip activation of naïve CD8+ T cells, 20 μL of young and old MQ and DC suspensions were seeded and cultured in the LNPoC with cell culture media containing 100 ng mL−1 OVA peptides and 250 ng mL−1 LPS for 7 days at 37 °C with 5% CO2, and half of the culture media were replaced with fresh media on days 3 and 5. After 7 days, 20 μL of naïve CD8+ T cells were seeded into the LNPoC containing OVA-stimulated MQs and DCs and cultured for 2 days at 37 °C with 5% CO2. After 2 days, activated CD8+ T cells were collected from the LNPoC. The generation and proliferation of young and old OVA-specific CD8+ T cells were analyzed by flow cytometry using FlowJo™ software. Activated CD8+ T cells were first gated based on size (side-scatter) and granularity (forward-scatter), and then OVA-specific CD8+ T cells were quantified after staining the cells with a PE-conjugated OVA-tetramer (NIH Core Facility, USA) and an APC-conjugated recombinant anti-mouse CD8a antibody (BioLegend, USA). Activated CD8+ T cells were also stained with CFSE (Thermo Fisher Scientific, USA) to assess the proliferation of OVA-specific CD8+ T cells.
Simulation of velocity and shear stress distribution in the device
A 3D computational model was developed in COMSOL Multiphysics using the finite element-based method to simulate media flow inside the device (Fig. S1). A single-phase, laminar flow model was used, with the fluid (water) assumed to be isothermal, incompressible, and Newtonian. The governing equations for this simulation are as follows:52| Continuity equation: ∇·u = 0 | (1) |
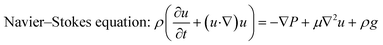 | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 943 triangular elements to ensure a mesh-independent solution. The spatiotemporal distributions of velocity and wall shear stress were obtained and visualized. A cutline (X = 8 mm, Y = 2.5 mm, and Z = 0 to 0.2 mm) was placed across the channel height to extract one-dimensional velocity and shear stress profiles (Fig. S1).
943 triangular elements to ensure a mesh-independent solution. The spatiotemporal distributions of velocity and wall shear stress were obtained and visualized. A cutline (X = 8 mm, Y = 2.5 mm, and Z = 0 to 0.2 mm) was placed across the channel height to extract one-dimensional velocity and shear stress profiles (Fig. S1).
Antitumoral assay
The B16-OVA cells (Sigma-Aldrich, USA) were cultured in DMEM (Fisher Scientific, USA), supplemented with 1% penicillin–streptomycin (Caisson Labs, USA) and 10% fetal bovine serum (FBS) (Life Technologies, USA). The B16-OVA cells were seeded in 6-well plates and grown until confluency (≈1 × 106 cells per well). Then, the young and old activated CD8+ T cells collected from the LNPoC were added to each well (1 × 106 cells per well) and co-cultured with B16-OVA cells. After 2 days, CD8+ T cells and B16-OVA cells were collected and analyzed by flow cytometry using FCS Express™ software. The cells were first gated based on size (side-scatter) and granularity (forward-scatter), and the viability of B16-OVA cells was quantified after staining the cells with a Zombie Violet™ Fixable Viability Kit (BioLegend, USA) and an APC-conjugated recombinant anti-mouse CD8a antibody (BioLegend, USA).In vivo animal study
Animals received comprehensive care in compliance with federal, state, and local guidelines. All procedures conducted on animals adhered to the regulations and were approved by the Lundquist Institute Committee on Use and Care of Animals (ICUCA #32706–02). Young (6–7 weeks old) and old (35–36 weeks old) C57BL/6 female mice (The Jackson Laboratory, USA) were housed in a pathogen-free environment for at least 3 days and were kept at room temperature with free access to water and a 12 hour light/dark cycle. The in vivo study involved C57BL/6 female mice bearing B16-OVA tumors. B16-OVA cells, cultured until 80% confluence and trypsinized with TripleE (Fisher Scientific, USA), were injected (1 × 106 cells in 100 μL per mouse) subcutaneously into the right flank of young and old C57BL/6 mice. After cell injection, the growth of B16-OVA tumors was monitored every other day. Tumor size was measured using a caliper and recorded, and the tumor volume was calculated using the formula: tumor volume (mm3) = length × (width)2 × 0.5. Once the tumors reached 20 mm3, the young and old mice were subcutaneously immunized at the tail base on days 0, 5, and 10 with 100 μL of free OVA peptides (1 mg mL−1) or PBS (negative control). On days 7, 14, and 21 post-first immunization, the mice were anesthetized, and blood samples (100 μl) were collected submandibularly. The animals were euthanized by CO2 asphyxiation when they reached either of the following end-points: (1) day 21 post-first immunization; (2) the tumor mass reached 1.5 cm in any dimension (length or width); or (3) the animals exhibited moribund conditions (>20% weight loss or ulceration). Once euthanized, tumor and LN tissues were harvested and processed. OVA-specific CD8+ T cells in tumors, lymph nodes, and blood samples were quantified by flow cytometry using FCS Express™ software. CD8+ T cells were first gated based on size (side-scatter) and granularity (forward-scatter), and then OVA-specific CD8+ T cells were quantified after staining the cells with a PE-conjugated OVA-tetramer (NIH Core Facility, USA) and an APC-conjugated recombinant anti-mouse CD8a antibody (BioLegend, USA).ELISA
The levels of cytokines in the media collected after 7 days of MQ and DC stimulation using OVA peptides were quantified using IL-6, IL-10, IL-12, TNF-alpha, and IFN-gamma ELISA kits (BioLegend, USA) following the manufacturer's protocol. The absorption measurements used to quantify the concentrations of IL-2, IL-6, IL-10, and IL-12 were acquired with a Varioskan™ LUX multimode microplate reader (Thermo Fisher Scientific, USA).Statistical analysis
All statistical analyses were performed using the GraphPad Prism 9 software. Results are expressed as the mean ± standard deviation from triplicate experiments (N = 3). All data were tested for normality using the Shapiro–Wilk and Shapiro–Francia tests. When applicable, analyses for significance were performed using a parametric or non-parametric analysis of variance (ANOVA) test. Statistical significance was defined by p < 0.05 for two-sided p-values.Results
Microfluidic chip fabrication and characterization
We fabricated a microfluidic LNPoC device using poly(methyl methacrylate) (PMMA). The device comprised a rounded square cell culture microchamber (length: 12 mm; width: 5 mm) connected to two 3 mm diameter holes leading to the inlet and outlet ports (diameter: 0.5 mm) (Fig. 1A–C). The rounded microchamber geometry of the LNPoC was intentionally engineered to promote continuous recirculation of immune cells and to facilitate repeated T cell–APC interactions, effectively emulating the confined flow dynamics of the lymph node paracortex.57 The dimensions of the microchambers were optimized to maximize the frequency of cell–cell contacts while minimizing chamber volume, thereby increasing local cytokine concentrations that support antigen-specific T-cell activation, proliferation, and retention of effector functions. This geometrical configuration ensures the physiological relevance of immune interactions within the platform, while its simplicity allows for reliable and scalable fabrication suitable for high-throughput applications. Once fabricated, the LNPoC device was coated with fibronectin, which promoted macrophage (MQ) and dendritic cell (DC) adhesion on the bottom of the cell culture microchamber and allowed the culture of MQs and DCs within the device (Fig. 1D). This fibronectin coating formed a confined microenvironment appropriate for the cell culture, activation, and antigen presentation by APCs inside the LNPoC device. Successful attachment and distribution of APCs within the device were confirmed through immunofluorescence imaging of MHC-I, cytoskeleton, and nuclei staining (Fig. 1E).LNPoC revealed the age-dependent presentation of OVA peptides
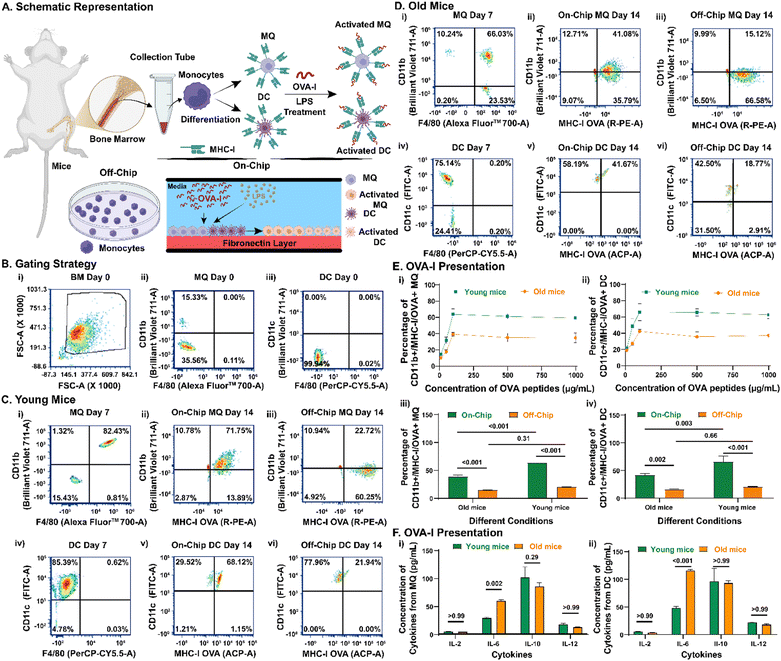
To evaluate the ability of the LNPoC platform to model age-related differences in antigen presentation, we isolated monocytes from the bone marrow of young and old mice and differentiated them into MQs and DCs. A schematic representation of the experimental workflow depicts the process of isolating monocytes, differentiating them into MQs and DCs, and loading OVA peptides either on-chip or off-chip to evaluate antigen presentation capacity (Fig. 2A). We isolated bone marrow cells from the tibia and femur of young (6–7 weeks old) and old (35–36 weeks old) C57BL/6 mice. Prior to initiating the differentiation protocol, we confirmed that the starting cell populations were devoid of macrophages (F4/80+/CD11b+) and dendritic cells (F4/80−/CD11c+), ensuring that only undifferentiated monocytes were present for subsequent experiments (Fig. 2B).58,59 After 7 days of differentiation, we confirmed for young (Fig. 2C) and old (Fig. 2D) mice that the bone marrow cells were differentiated into MQs (F4/80+/CD11b+) and DCs (F4/80−/CD11c+). The flow cytometry plots demonstrate that immune cells derived from young mice exhibit higher activation and antigen presentation levels than those from older mice. Specifically, young mouse-derived macrophages and dendritic cells show increased expression of activation markers and a greater capacity to present OVA peptides. This enhanced antigen presentation leads to more robust activation of OVA-specific CD8+ T cells, critical for effective antitumor immune responses. These findings highlight the age-dependent differences in immune function and underscore the importance of using physiologically relevant models like the LNPoC to study immunosenescence and its impact on cancer vaccine efficacy. After 7 more days of stimulation with OVA peptides, we analyzed the OVA peptide presentation potential of MQs (OVA+/CD11b+) or DCs (OVA+/CD11c+) in young and old cells. We observed that OVA peptides were presented by young and old MQs and DCs, both on-chip and off-chip. On-chip, we observed a dose-dependent OVA peptide presentation response and a plateau of presentation when a 100 μg mL−1 concentration of OVA peptides was used as the immunogen (Fig. 2E). This dosage was used subsequently for all experiments and analyses. For the OVA peptide presentation by MQs, we observed a significant difference in OVA peptide presentation between on-chip and off-chip experiments using cells from young (63.8 ± 4.9% vs. 20.7 ± 0.2%, p < 0.001) and old mice (39.3 ± 2.6% vs. 15.2 ± 0.2%, p < 0.001). In addition, we observed that on-chip OVA peptide presentation was significantly higher with young cells than with old ones (63.8 ± 4.9% vs. 39.3 ± 2.6%, p < 0.001), whereas there was no significant difference in off-chip OVA peptide presentation between young cells and old ones (20.7 ± 0.2% vs. 15.2 ± 0.2%, p = 0.31). The same observations were made for DCs when comparing on-chip and off-chip results using young (65.9 ± 10.1% vs. 21.2 ± 0.5%, p < 0.001) and old cells (42.3 ± 2.0% vs. 16.1 ± 0.9%, p = 0.002), and on-chip OVA peptide presentation with young cells (65.9 ± 10.1% vs. 42.3 ± 2.0%, p = 0.003) and old ones (21.2 ± 0.5% vs. 16.1 ± 0.9%, p = 0.66) (Fig. 2E). The confocal images of the on-chip presentation of the OVA peptides confirmed that the OVA presentation by young MQs and DCs was higher than that by old MQs and DCs (Fig. S2). Lastly, we assessed the cytokine production from MQs and DCs on day 7 after on-chip OVA peptide and lipopolysaccharide (LPS) stimulation, and we observed a significant difference in IL-6 secretion between young and old MQs (p = 0.002) and DCs (p < 0.001). We did not observe a significant difference for IL-2, IL-10, and IL-12 (Fig. 2F).LNPoC revealed the age-dependent generation of OVA-specific CD8+ T cells
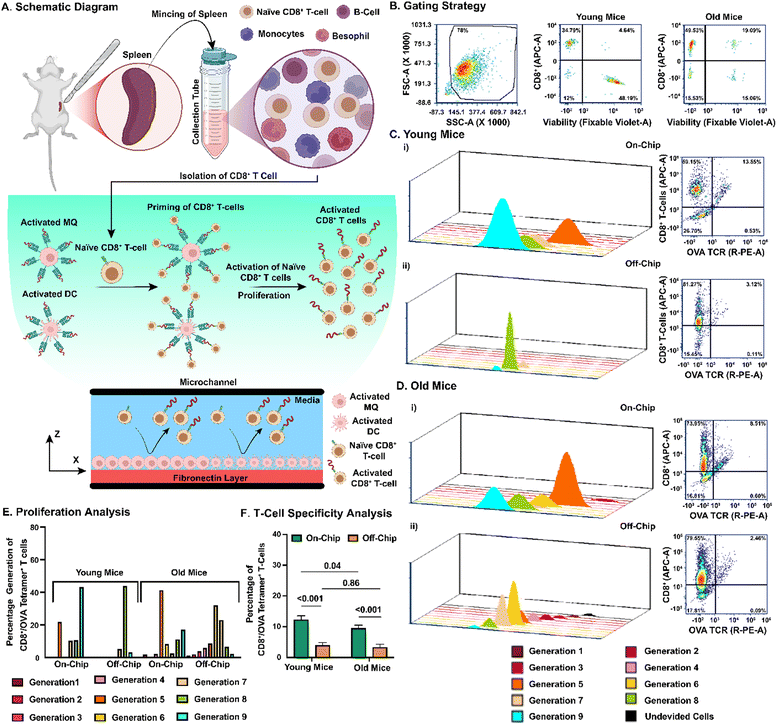
To investigate how aging affects the activation and proliferation of antigen-specific T cells, we isolated CD8+ T cells from the spleens of young and old C57BL/6 mice. We then analyzed their functional responses using the LNPoC platform. A schematic representation of the experimental workflow illustrates the process of isolating CD8+ T cells from mouse splenocytes and co-culturing them with OVA peptide-presenting MQs and DCs within the LNPoC, followed by the subsequent activation and proliferation of OVA-specific CD8+ T cells in a controlled microenvironment (Fig. 3A). We isolated CD8+ T cells through a negative selection process, resulting in a cell fraction containing 34.8% and 49.5% of live CD8+ T cells from young and old mice, respectively (Fig. 3B). Next, we characterized OVA-specific T cell proliferation and OVA peptide specificity in young (Fig. 3C) and old (Fig. 3D) immune cells. We observed that using young and old immune cells, the proliferation of T cells was significantly higher on-chip than off-chip (p < 0.001). Also, we observed a significantly higher on-chip proliferation of young CD8+ T cells than that of old CD8+ T cells (p < 0.001); however, this difference was not observed off-chip (Fig. 3E). Generations 1–9 demonstrate the proliferation of OVA-specific CD8+ T cells, measured by CFSE staining. Young CD8+ T cells show more cells in the later generations (5–9), indicating stronger proliferation. In contrast, old CD8+ T cells mostly remain in the earlier generations (1–4), suggesting impaired proliferation due to immunosenescence. In a similar way, we observed a significantly higher generation of OVA-specific CD8+ T cells on-chip compared to off-chip for young (12.3 ± 1.3% vs. 4.0 ± 0.8%, p < 0.001) and old (9.5 ± 0.9% vs. 3.4 ± 1.0%, p < 0.001) CD8+ T cells (Fig. 3F). We also observed a significantly higher percentage of OVA-specific T cells on-chip using young T cells compared to old ones (12.3 ± 1.3% vs. 9.5 ± 0.9%, p = 0.04), whereas no significant differences were observed between young and old cells (4.00 ± 0.8% vs. 3.36 ± 1.0%, p = 0.86) (Fig. 3F). Confocal images of on-chip activation of OVA-specific CD8+ T cells confirmed higher OVA priming of young CD8+ T cells than that of old CD8+ T cells (Fig. S3).To isolate the activated CD8+ T cells within the device, we employed flow-induced shear stress. We varied the media velocity to optimize the cell isolation process. The spatiotemporal distribution of the isolated CD8+ T cells was assessed within the LNPoC over a time span of 0 to 30 seconds, confirming the minimal optimal velocity (average velocity: 0.00017 m s−1) necessary to separate the activated cells from the total population of CD8+ T cells (Fig. S1D). We also conducted corresponding computational fluid dynamics (CFD) simulations to model shear stress distributions. The CFD simulations indicated the spatial distribution of shear stress (Fig. S1B) across the device at an average velocity of 0.00017 m s−1, highlighting a uniform flow along the microchannel. The shear stress profile was highest near the walls and increased with the inlet velocity, ranging from 0.001 to 0.008 Pa (Fig. S1C). These results validate the controlled microenvironment and physiologically relevant flow conditions within the LNPoC.
Antitumoral efficacy of OVA-I-specific CD8+ T cells
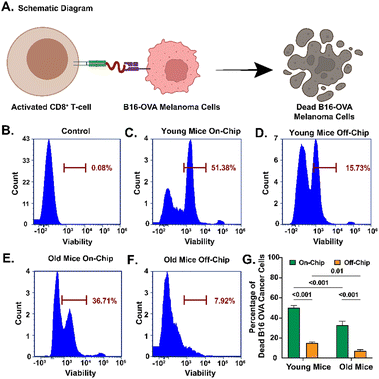
We assessed the antitumoral efficacy of CD8+ T cells stimulated by OVA-presenting MQs and DCs by co-culturing B16-OVA cells with activated CD8+ T cells. A schematic illustration of the experimental workflow displays the co-culture of activated CD8+ T cells with B16-OVA melanoma cells and subsequent assessment of tumor cell death (Fig. 4A). To isolate CD8+ T cells, we used a negative selection process, which removes unwanted blood cell types while leaving CD8+ T cells untouched. This method relies on depleting other immune cells, such as B cells, NK cells, and myeloid cells, resulting in an enriched population of CD8+ T cells. Additionally, our analysis included gating on all live cells rather than exclusively on CD8+ T cells. Because negative selection eliminates many non-T cell populations, the remaining cell suspension contains a higher proportion of CD8+ T cells among all viable cells. This explains why we observed 34.8% live CD8+ T cells from young mice and an even higher percentage of 49.5% from old mice. The increased percentage in older mice may also reflect age-related shifts in immune cell composition. Still, the main factor is the combination of negative selection and the broad gating strategy, which measures CD8+ T cells as a fraction of all remaining live cells rather than just among T cells. Compared to a negative control without OVA-specific CD8+ T cells (Fig. 4B), we assessed the antitumoral efficacy of young and old CD8+ T cells generated on- and off-chip (Fig. 4C–F). We observed a significantly higher percentage of dead B16-OVA cells when OVA-specific CD8+ T cells were generated on-chip compared to off-chip for young (49.9 ± 1.5% vs. 15.1 ± 0.7%, p < 0.001) and old (32.7 ± 4.0% vs. 7.0 ± 0.9%, p < 0.001) immune cells (Fig. 4G). Similarly, we observed a significantly higher percentage of dead B16-OVA cells with on-chip-generated young CD8+ T cells compared to old ones (49.9 ± 1.5% vs. 32.7 ± 4.0%, p < 0.001), and the same trend was observed for OVA-specific CD8+ T cells generated off-chip (15.1 ± 0.7% vs. 7.0 ± 0.9%, p = 0.01). Confocal images also confirmed more dead B16-OVA cells with young OVA-specific CD8+ T cells than with old ones (Fig. S4).In vivo validation of the LNPoC platform
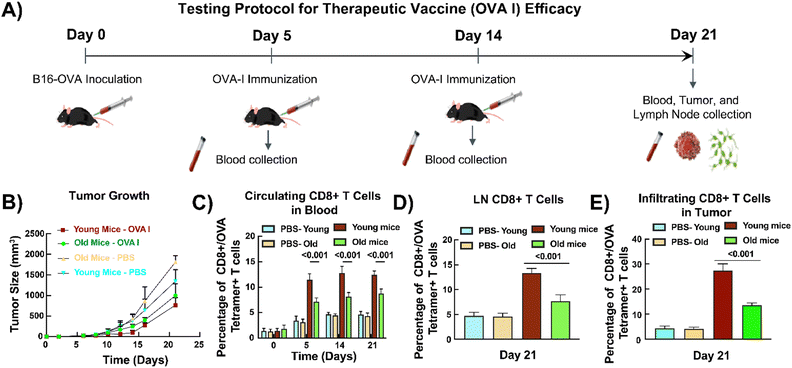
To validate the results from our in vitro platform, we evaluated the immunogenicity and antitumoral efficacy of OVA peptides (without adjuvants) in a subcutaneous B16-OVA murine melanoma model. We inoculated the B16-OVA cells and collected blood on day 0. We immunized the mice with OVA peptides and collected blood one week after inoculation. We also collected blood after two weeks. We sacrificed the mice after three weeks and collected blood, LNs, and B16-OVA cells (Fig. 5A). Although not significant, the tumor growth was lower in young mice compared to old ones (Fig. 5B). The percentage of circulating OVA-specific CD8+ T cells is significantly higher in PBMCs of young mice compared to old mice on days 5 (11.4 ± 1.4% vs. 7.1 ± 1.0%, p < 0.001), 14 (12.8 ± 1.3% vs. 8.2 ± 0.8%, p < 0.001), and 21 (12.4 ± 0.8% vs. 8.8 ± 0.9%, p < 0.001) (Fig. 5C). Similarly, a significantly higher percentage of lymph node-isolated (13.4 ± 0.9% vs. 7.7 ± 2.0%, p < 0.001) and tumor-infiltrating (27.3 ± 2.5% vs. 13.6 ± 1.2%, p < 0.001) OVA-specific CD8+ T cells was observed in young mice compared to old mice on day 21 (Fig. 5D and E). Notably, it was observed that the percentages of OVA-specific CD8+ T cells in young mice present in the circulation and isolated from LNs at day 21 were similar to those of the on-chip young OVA-specific CD8+ T cells (Table S1).Discussion
We developed a LNPoC capable of recapitulating in vivo cancer vaccine immunogenicity in an age-dependent manner, including critical functions of the LN paracortex, such as antigen presentation and antigen-specific T cell activation, proliferation, and effector functions. Our LNPoC platform permitted the flow, capture, processing, and presentation of OVA peptides (as a model antigen) and the migration of T cells over APCs, promoting the interactions necessary for eliciting vaccine-induced immune responses. These functions are integral to mounting an effective antitumor immune response and are known to be compromised with age.36,60 In contrast to conventional 2D co-culture systems, which fail to capture the spatial complexity of lymphoid tissue, the LNPoC demonstrated age-dependent differences in immune functions, demonstrating its potential as a superior model for studying immunosenescence in vitro.61,62 This novel platform provides a robust model to examine immune responses in an age-dependent manner, where older immune cells may be less capable of initiating strong antitumor responses following cancer vaccine immunization than younger cells.The LNPoC platform demonstrated distinct age-related differences in antigen presentation, which were not revealed off-chip using conventional 2D co-culture systems. By comparing immune responses between young and old immune cells, we demonstrated that APCs (MQs and DCs) from young mice presented OVA peptides more effectively than those from older mice on-chip but not off-chip. This significant difference in antigen presentation is consistent with previous findings that aging leads to diminished functions and impaired antigen presentation in MQs and DCs.38,63 Immunosenescent DCs display reduced phagocytosis and the cross-presentation of cell-associated antigens due to the increased oxidative stress associated with aging.63–65 Similarly, MQs have shown a decrease in the ability to activate Toll-like receptors (TLRs), which play a critical role in innate immune responses against cancer vaccines.66–68 Indeed, aging impacts cytokine secretion by APCs, especially on IL-6.69 IL-6 plays an essential role in pro-inflammatory immune responses and age-related chronic inflammations, resulting in altered signaling pathways, increased production of reactive oxygen species, and chronic low-grade inflammation.70–77 Our study confirmed the upregulation of IL-6 in old APCs compared to young ones. The decline in OVA peptide presentation by MQs and DCs from old mice is consistent with established features of immunosenescence, including impaired antigen processing, reduced TLR activation, and altered cytokine secretion profiles in aged APCs.78,79 Although co-stimulatory molecule profiling and mechanistic validation (i.e., TLR/NF-κB pathway analysis) were not conducted, the observed age-dependent OVA peptide presentation and OVA-specific CD8+ T-cell activation provide a robust functional measure of immunological engagement, serving as an integrated outcome of upstream signaling events. While additional cytokine profiling was beyond the scope of the present study, the observed upregulation of IL-6 can be interpreted as an early indicator of the senescence-associated secretory phenotype (SASP), which encompasses key pro-inflammatory mediators such as IL-6, TNF-α, and IL-1β.80 The selective elevation of IL-6 observed in our system is consistent with prior findings that IL-6 dysregulation represents an early event in the development of immunosenescence, preceding broader activation of SASP-related cytokine networks.81 Future iterations of this work will incorporate multiplex cytokine analyses to more comprehensively characterize the SASP profile associated with immune aging within the LNPoC platform.
We also demonstrated a significant reduction in the generation, proliferation, and activation of OVA-specific CD8+ T cells in old cells compared to young ones. These results align with established evidence that aging is associated with a decline in naïve T cell numbers and a reduction in TCR diversity, which collectively reduce the immune system's capacity to respond to novel antigens.82 Indeed, immunosenescence due to aging impairs the priming of T cells and the subsequent proliferation of primed T cells.83–85 The gradual decline in the production of naïve CD8+ T cells with age drives a shift in the population of memory CD8+ T cells toward exhaustion after repeated antigen exposure.44 This state of exhaustion renders T cells less responsive to new antigens and less effective in recognizing APCs.86,87 In addition, the telomer shortening in CD8+ T cells can cause cell senescence by reducing proliferation and functionality.88,89 Moreover, exhausted CD8+ T cells upregulated inhibitory receptors on the surface (i.e., PD-1), which can reduce the capacity of immune cells to recognize PD-L1-expressing cancer cells.90–93 We also demonstrated similar trends for the antitumoral efficacy of OVA-specific CD8+ T cells generated on-chip or off-chip, where the cytotoxicity of LNPoC-generated OVA-specific CD8+ T cells was significantly higher than off-chip. This is consistent with previous findings in vivo, demonstrating that the cytotoxic effect and effector functions of exhausted CD8+ T cells are limited and cannot effectively control the progression of tumors in the later stages.90,94,95
Our results revealed well-known trends in the age-dependent decline in antigen presentation, T cell activation, and antitumoral efficacy characteristic of immunosenescence, which we further validated in vivo using the same species of mice. These age-dependent differences were apparent in our LNPoC but not in conventional 2D co-culture systems, suggesting that our LNPoC can uncover age-related immune function declines that are often masked in simplified culture systems. Unlike 2D models, which lack the spatial architecture and fluidic dynamics of the LN paracortex, the LNPoC facilitates continuous antigen flow and uptake, cell movement, and APC–T cell interactions within the culture microchamber due to the inherently high surface-to-volume ratio of OoC models.96–99 For example, T cell priming relies on dynamic spatial proximity and guided chemokine gradients to effectively interact, which are absent in static 2D cultures.100 In addition, biophysical cues can modulate immune cell functions, including macrophage polarization, T cell selection, and T cell activation.101 The LNPoC's dynamic microenvironment provides physiologically relevant cell–cell interactions, where T cells can engage with APCs to mimic in vivo conditions. This design possibly improves the fidelity of antigen presentation and T cell activation by promoting a consistent flow and concentration gradient of antigens and signaling molecules, a feature that static cultures poorly replicate. Studies have shown that the microfluidic environment enables immune cells to respond to antigen stimuli with enhanced sensitivity and responsiveness by mimicking the natural APC–antigen and APC–T cell interactions that occur in vivo.30 Consequently, our LNPoC model is an accurate platform for studying immunosenescence, as it captures the nuanced cellular interactions and response dynamics that are critical to triggering age-related immune responses. Nevertheless, our LNPoC platform could be used to investigate immunosenescence effects on other vaccines, like influenza. This platform would require integrating additional immune cell types, such as CD4+ T cells, B cells, and plasma cells, to effectively capture germinal center dynamics and antibody production. However, accurately modeling CD8+ T cell priming and effector responses remains crucial in cancer immunotherapy, as these cells are key to tumor clearance after vaccination.
Despite the development of many OoC models recapitulating various functions of the LNs,102 none have focused on recapitulating immunosenescence on-a-chip. Most often, LN on-a-chip models are developed to mimic the architecture, biochemical environment, and fluid dynamics of actual LNs and enable studies on immune cell behavior and interactions. The design of the microfluidic chip intends to capture different key functions of the LN and its complex role in immune responses.30 Several LN on-a-chip models recreate the distinct cellular regions of the LN, where different compartments allow the co-culture and spatial distribution of immune cells to study APC–T cell interactions and T cell activation.103,104 These models can also replicate chemokine gradients to study immune cell chemotaxis, where immune cells migrate in response to controlled gradients of chemokines, similar to their in vivo counterparts.105–107 Several models have been used to test vaccines and to study the immunogenicity of antigens,108,109 including some in an age-dependent manner.110 Indeed, immune cells from young and old human donors demonstrated differential age-associated responses to the influenza vaccine, where old immune cells exhibited reduced IgG production and CD4+ T cell activation upon vaccine stimulation compared to young immune cells.
Vaccine responses modeled on a chip primarily focus on the development of antibodies, with antigen presentation, T cell activation, germinal center formation, and antibody production.111,112 In contrast, our LNPoC platform models CD8+ T cell-specific immune responses, which are the most important to evaluate following cancer vaccination against specific TAAs.113,114 Also, our platform allows the collection of antigen-specific T cells for downstream application, particularly to assess the potency of effector functions downstream. Most importantly, our LNPoC was validated with the same species in vivo, which is a crucial step to ensure that the observed immune responses, particularly the age-dependent declines in antigen presentation, T cell activation, and antitumoral responses, directly correlate with physiological outcomes. These in vivo validations are rarely carried out with the same species.115 This species discrepancy could hinder the translation of cancer vaccines to patients, with potential failures in clinical trials remaining.116 This same-species in vivo validation approach strengthens the translational relevance of our LNPoC model by confirming that it can accurately replicate the species-specific immune dynamics observed in live organisms. Moreover, it supports the LNPoC's reliability in capturing age-related immune function changes and its potential as a predictive tool for evaluating immunosenescence and informing the design of clinical trials to test cancer vaccine efficacy.
Cancer vaccines, particularly those targeting CD8+ T cells, are promising immunotherapies for cancer treatment.117 However, their efficacy weakens in older populations due to immunosenescence.118,119 In this context, our findings have several important implications for cancer vaccine development. First, the LNPoC is a valuable tool for the preclinical screening of cancer vaccines, allowing the testing of multiple candidates in an immune-relevant system before moving to costly and time-consuming clinical trials.120 By providing accurate age-dependent immune responses, our platform can potentially reduce the high failure rates of cancer vaccines in clinical trials and accelerate the development of more effective cancer immunotherapies. Specifically, the ability to model immunosenescence within our in vitro platform addresses a critical gap in the preclinical phase of current vaccine development pipelines.121 For aging populations, constituting the majority of cancer patients, there is an increasing need for effective vaccines;43–45 however, these populations are unaccounted for in preclinical models. Therefore, our LNPoC could provide insights into how vaccines can be optimized for older patients and lead to the development of new vaccine formulations or adjuvants specifically designed to enhance immune responses in geriatric populations. Lastly, our LNPoC platform could be a powerful tool for personalized medicine by enabling the tailored testing of cancer vaccine efficacy for elderly patients from blood-isolated immune cells. Our LNPoC could guide the prediction of older individuals' immune responses to various immunotherapies, which is invaluable for testing and optimizing a broader range of immunotherapies to develop more effective treatments for the aging population.
Despite its strengths, our LNPoC platform has several limitations. We used murine rather than human immune cells, which limits the direct applicability of findings to human systems due to interspecies differences in immune response characteristics.122 Murine cells were selected to maintain species consistency between the in vitro LNPoC experiments and in vivo validation, ensuring biologically coherent comparisons. This same-species validation approach serves as a critical intermediate step that minimizes the need for live animal testing while providing biologically relevant data, consistent with ethical and regulatory priorities outlined in the FDA Modernization Act 2.0.15–17 However, we acknowledge that interspecies differences in T-cell receptor (TCR) diversity and cytokine signaling may influence translational extrapolation.122 Future studies will integrate human PBMCs and iPSC-derived immune cells to enhance translational fidelity and extend the applicability of the LNPoC. The platform will also be optimized with human-specific cytokines, chemokines, and extracellular matrix components to simulate the human lymphoid microenvironment. Overall, our in vivo validation confirms that our platform can reliably recapitulate immune responses in an age-dependent manner.
Furthermore, our LNPoC lacks extracellular matrix (ECM)–cell interactions, which are crucial for accurately mimicking cell migration and positioning, and integral to immune responses in lymphoid tissues.123 Additionally, the simplified tissue architecture of the LNPoC, without stromal or follicular dendritic cells, may overlook critical cell–cell and cell–ECM interactions essential for a fully representative immune microenvironment.124 Our LNPoC is still a simplified model that cannot fully capture the complexity of the immune system. For instance, the platform focuses primarily on the lymph node paracortex and does not include other critical immune responses (i.e., memory or antibody), which also play important roles in vaccine-induced immune responses.125 Like other OoC technologies, our LNPoC represents an isolated environment, excluding systemic interactions with other immune organs, such as the spleen and bone marrow, which are vital for a holistic understanding of immune dynamics after vaccination.126 Also, our model only captures the functional antigen-specific T cell activation processes observed within the lymph node paracortex but does not include the stromal and vascular elements, such as fibroblastic reticular cells and high endothelial venules, which define the native architecture of the LN. While the platform simulates some aspects of immunosenescence, additional research is needed to fully incorporate the range of systemic, cellular, and molecular changes that occur with aging.75
The current study used mice that were 35 to 36 weeks old to model early immunosenescence, capturing the transitional immune decline before late-stage senescence. These middle-aged mice may demonstrate measurable reductions in naïve T-cell populations and IL-2 responsiveness compared to young (6 to 7 weeks old) mice. The goal was to determine whether the LNPoC could detect incipient age-related immune differences that are not visible in 2D cultures. We acknowledge that future studies using mice older than 72 weeks will further validate advanced senescence responses. This will help further validate and expand the applicability of the LNPoC platform in modeling immune dysfunction associated with late-stage aging. We utilized the OVA-I peptide instead of full-length OVA protein for antigen presentation assays. While this peptide allows direct MHC-I-mediated antigen presentation and CD8+ T cell activation, it bypasses natural antigen processing. Future versions of the LNPoC platform will contain full-length proteins or patient-specific tumor antigens to assess better the complete pathways of antigen processing and presentation by APCs. Our in vivo validation demonstrated that young mice had significantly higher OVA-specific CD8+ T cell responses than older mice in blood, lymph nodes, and tumors.
The differences in tumor size between the groups were not statistically significant over the 21 day observation period. This may be due to the aggressive nature of the B16-OVA tumor model, where rapid growth can outpace immune control efforts.127,128 Additionally, immunosuppressive factors in the tumor microenvironment and the short study duration may have suppressed the significant differences in tumor burden, despite notable variations in immune activation.128 Future studies with more extended observation periods (i.e., 30–40 days) or less aggressive tumor models (i.e., MC38-OVA), complemented by Kaplan–Meier survival analysis for a more comprehensive assessment of therapeutic outcomes, could better elucidate age-dependent tumor control dynamics. Although we considered the tumor-killing ability of CD8+ T cells as “cytotoxicity”, our assessment was based on functional effector activity, measured by B16-OVA tumor cell viability assays after co-culture with OVA-specific CD8+ T cells. In this study, we did not directly measure cytotoxic molecules such as granzyme B or perforin. The inclusion of quantitative measurements for effector molecules would further enhance the mechanistic understanding of cytotoxic responses within the LNPoC platform. Although these markers were not assessed in the current study, the cytotoxicity assays performed using B16-OVA demonstrate antigen-specific killing, confirming the functional activation of CD8+ T cells. In future studies, we will incorporate granzyme B staining to more precisely characterize CD8+ T cell cytotoxic effector mechanisms and the corresponding pathways. In addition, future investigation of the LNPoC platform should incorporate clinically relevant neoantigens or influenza antigens to assess broader immunological responses beyond just CD8+ T cells. Another limitation of the current study is the focus on acute antigen-specific CD8+ T-cell activation and effector responses, without extending to the characterization of long-term memory formation. Nevertheless, the platform's architecture is inherently compatible with prolonged co-culture conditions that can support the differentiation and maintenance of memory T cell subsets. Such extended cultures could enable the identification of CD44+/CD62L+ phenotypes and the investigation of memory-like transitions within the engineered lymphoid microenvironment. Future studies will build on this capability to evaluate memory generation and persistence, in line with recent LN on-a-chip models that have demonstrated similar functionality.129 Addressing these limitations by incorporating human cells, ECM components, and multi-organ system integration will be essential for furthering the utility and translational relevance of our LNPoC platform for cancer vaccine development and screening.
Conclusions
Our LNPoC platform represents a significant advancement in studying the impact of immunosenescence on cancer vaccine discovery, development, and translation to the clinic. By replicating age-related declines in immune function, our LNPoC is a valuable tool for understanding the impact of immunosenescence on current cancer vaccines and guiding the development of more effective therapies for older populations. Our LNPoC platform is the first step in improving preclinical cancer vaccine testing by considering age as a variable. It could also be applied beyond cancer vaccines to other areas of immunotherapy, such as immune checkpoint inhibitors or adoptive cell therapies, where LN function plays a crucial role in activating immune cells. Continued research using this platform will be critical for achieving more personalized and age-specific cancer treatment strategies, ultimately improving therapeutic outcomes and quality of life for aging populations.Author contributions
Conceptualization: SM, AHN, VJ; data curation: SM, AHN, SK, DK, CY, CJ, NM, VJ; formal analysis: SM, AHN, VJ; funding acquisition: AK, VJ; investigation: SM, AHN, SK, DK, CY, CJ, NM, VJ; methodology: SM, AHN, SK, DK, CY, CJ, NM, VJ; supervision: MRD, AK, VJ; visualization: SM, DK, VJ; writing – original draft: SM, AHN, VJ; writing – review & editing: SM, AHN, SK, DK, CY, CJ, NM, MRD, AK, VJ.Conflicts of interest
The authors have no conflicts of interest to declare.Data availability
The data for this article are available at Zenodo at https://doi.org/10.5281/zenodo.15550331.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5lc00533g.
Acknowledgements
S. M. and A. H. N. contributed equally to this work. The authors acknowledge funding from the Terasaki Institute for Biomedical Innovation. We thank the NIH Tetramer Core Facility for providing the OVA tetramer.References
- J. Liu, M. Fu, M. Wang, D. Wan, Y. Wei and X. Wei, J. Hematol. Oncol., 2022, 15, 28 CrossRef PubMed
.
- M. Kaczmarek, J. Poznańska, F. Fechner, N. Michalska, S. Paszkowska, A. Napierała and A. Mackiewicz, Cell, 2023, 12(17), 2159 CrossRef CAS PubMed
.
- M. Bilusic and R. A. Madan, Am. J. Ther., 2012, 19, e172–e181 CrossRef PubMed
.
- Cancer Vaccine Market (2024 - 2030), Grand View Research, https://www.grandviewresearch.com/industry-analysis/cancer-vaccine-market-report, (Accessed December 1, 2025) Search PubMed.
- H. Donninger, C. Li, J. W. Eaton and K. Yaddanapudi, Vaccines, 2021, 9, 668 CrossRef CAS PubMed
.
- A. Tojjari, A. Saeed, M. Singh, L. Cavalcante, I. H. Sahin and A. Saeed, Vaccines, 2023, 11(8), 357 Search PubMed
.
- H. Sajjad, S. Imtiaz, T. Noor, Y. H. Siddiqui, A. Sajjad and M. Zia, Anim. Models Exp. Med., 2021, 4, 87–103 CrossRef PubMed
.
- A. Loewa, J. J. Feng and S. Hedtrich, Nat. Rev. Bioeng., 2023, 1, 545–559 CrossRef CAS PubMed
.
- A. D. Martin, X. Wang, M. L. Sandberg, K. R. Negri, M. L. Wu, D. Toledo Warshaviak, G. B. Gabrelow, M. E. McElvain, B. Lee, M. E. Daris, H. Xu and A. Kamb, J. Immunother., 2021, 44, 95–105 CrossRef CAS PubMed
.
- J. F. Vansteenkiste, B. C. Cho, T. Vanakesa, T. De Pas, M. Zielinski, M. S. Kim, J. Jassem, M. Yoshimura, J. Dahabreh, H. Nakayama, L. Havel, H. Kondo, T. Mitsudomi, K. Zarogoulidis, O. A. Gladkov, K. Udud, H. Tada, H. Hoffman, A. Bugge, P. Taylor, E. E. Gonzalez, M. L. Liao, J. He, J. L. Pujol, J. Louahed, M. Debois, V. Brichard, C. Debruyne, P. Therasse and N. Altorki, Lancet Oncol., 2016, 17, 822–835 CrossRef CAS PubMed
.
- D. J. Schwartzentruber, D. H. Lawson, J. M. Richards, R. M. Conry, D. M. Miller, J. Treisman, F. Gailani, L. Riley, K. Conlon, B. Pockaj, K. L. Kendra, R. L. White, R. Gonzalez, T. M. Kuzel, B. Curti, P. D. Leming, E. D. Whitman, J. Balkissoon, D. S. Reintgen, H. Kaufman, F. M. Marincola, M. J. Merino, S. A. Rosenberg, P. Choyke, D. Vena and P. Hwu, N. Engl. J. Med., 2011, 364, 2119–2127 CrossRef CAS PubMed
.
- G. A. Van Norman, JACC: Basic Transl. Sci., 2019, 4, 845–854 Search PubMed
.
- D. Sun, W. Gao, H. Hu and S. Zhou, Acta Pharm. Sin. B., 2022, 12(7), 3049–3062 CrossRef CAS PubMed
.
- O. Urzì, R. Gasparro, E. Costanzo, A. De Luca, G. Giavaresi, S. Fontana and R. Alessandro, Int. J. Mol. Sci., 2023, 24(15), 12046 CrossRef PubMed
.
- C. Ma, Y. Peng, H. Li and W. Chen, Trends Pharmacol. Sci., 2021, 42, 119–133 CrossRef CAS PubMed
.
- J. J. Han, Artif. Organs, 2023, 47(3), 449–450 CrossRef PubMed
.
- A. van Rijt, E. Stefanek and K. Valente, Cancers, 2023, 15(18), 4466 CrossRef CAS PubMed
.
- Y. S. Zhang, J. Aleman, S. R. Shin, T. Kilic, D. Kim, S. A. Mousavi Shaegh, S. Massa, R. Riahi, S. Chae, N. Hu, H. Avci, W. Zhang, A. Silvestri, A. Sanati Nezhad, A. Manbohi, F. De Ferrari, A. Polini, G. Calzone, N. Shaikh, P. Alerasool, E. Budina, J. Kang, N. Bhise, J. Ribas, A. Pourmand, A. Skardal, T. Shupe, C. E. Bishop, M. R. Dokmeci, A. Atala and A. Khademhosseini, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E2293–E2302 CAS
.
- Q. Pi, S. Maharjan, X. Yan, X. Liu, B. Singh, A. M. van Genderen, F. Robledo-Padilla, R. Parra-Saldivar, N. Hu, W. Jia, C. Xu, J. Kang, S. Hassan, H. Cheng, X. Hou, A. Khademhosseini and Y. S. Zhang, Adv. Mater., 2018, 30, e1706913 CrossRef PubMed
.
- N. S. Bhise, J. Ribas, V. Manoharan, Y. S. Zhang, A. Polini, S. Massa, M. R. Dokmeci and A. Khademhosseini, J. Control. Release, 2014, 190, 82–93 CrossRef CAS PubMed
.
- S. R. Shin, T. Kilic, Y. S. Zhang, H. Avci, N. Hu, D. Kim, C. Branco, J. Aleman, S. Massa, A. Silvestri, J. Kang, A. Desalvo, M. A. Hussaini, S.-K. Chae, A. Polini, N. Bhise, M. A. Hussain, H. Lee, M. R. Dokmeci and A. Khademhosseini, Adv. Sci., 2017, 4, 1600522 CrossRef PubMed
.
- S. R. Shin, Y. S. Zhang, D.-J. Kim, A. Manbohi, H. Avci, A. Silvestri, J. Aleman, N. Hu, T. Kilic, W. Keung, M. Righi, P. Assawes, H. A. Alhadrami, R. A. Li, M. R. Dokmeci and A. Khademhosseini, Anal. Chem., 2016, 88, 10019–10027 CrossRef CAS PubMed
.
- M. Chernyavska, M. Masoudnia, T. Valerius and W. Verdurmen, Cancer Immunol., Immunother., 2023, 1–13 Search PubMed
.
- T. I. Maulana, E. Kromidas, L. Wallstabe, M. Cipriano, M. Alb, C. Zaupa, M. Hudecek, B. Fogal and P. Loskill, Adv. Drug Delivery Rev., 2021, 173, 281–305 CrossRef CAS PubMed
.
- A. L. Beckwith, L. F. Velásquez-García and J. T. Borenstein, Adv. Healthcare Mater., 2019, 8, 1900289 CrossRef PubMed
.
- S. Shim, M. C. Belanger, A. R. Harris, J. M. Munson and R. R. Pompano, Lab Chip, 2019, 19, 1013–1026 RSC
.
- X. Cui, C. Ma, V. Vasudevaraja, J. Serrano, J. Tong, Y. Peng, M. Delorenzo, G. Shen, J. Frenster, R.-T. T. Morales, W. Qian, A. Tsirigos, A. S. Chi, R. Jain, S. C. Kurz, E. P. Sulman, D. G. Placantonakis, M. Snuderl and W. Chen, eLife, 2020, 9, e52253 CrossRef CAS PubMed
.
- J. M. Ayuso, S. Rehman, M. Virumbrales-Munoz, P. H. McMinn, P. Geiger, C. Fitzgerald, T. Heaster, M. C. Skala and D. J. Beebe, Sci. Adv., 2021, 7, eabc2331 CrossRef CAS PubMed
.
- Y. Ando, E. L. Siegler, H. P. Ta, G. E. Cinay, H. Zhou, K. A. Gorrell, H. Au, B. M. Jarvis, P. Wang and K. Shen, Adv. Healthcare Mater., 2019, 8, 1900001 CrossRef PubMed
.
- A. Shanti, N. Hallfors, G. A. Petroianu, L. Planelles and C. Stefanini, Front. Pharmacol., 2021, 12, 711307 CrossRef CAS PubMed
.
- C. L. Slingluff, Jr., Cancer J., 2011, 17, 343–350 CrossRef PubMed
.
- M. A. J. Morsink, N. G. A. Willemen, J. Leijten, R. Bansal and S. R. Shin, Micromachines, 2020, 11(9), 849 CrossRef PubMed
.
- A. E. R. Kartikasari, M. D. Prakash, M. Cox, K. Wilson, J. C. Boer, J. A. Cauchi and M. Plebanski, Front. Immunol., 2018, 9, 3109 CrossRef CAS PubMed
.
- L. Donoghue, K. T. Nguyen, C. Graham and P. Sethu, Micromachines, 2021, 12(2), 139 CrossRef PubMed
.
- F. Groell, O. Jordan and G. Borchard, Eur. J. Pharm. Biopharm., 2018, 130, 128–142 CrossRef CAS PubMed
.
- S. N. Crooke, I. G. Ovsyannikova, G. A. Poland and R. B. Kennedy, Immun. Ageing, 2019, 16, 25 CrossRef PubMed
.
- C. A. Chougnet, R. I. Thacker, H. M. Shehata, C. M. Hennies, M. A. Lehn, C. S. Lages and E. M. Janssen, J. Immunol., 2015, 195, 2624–2632 CrossRef CAS PubMed
.
- C. Wong and D. R. Goldstein, Curr. Opin. Immunol., 2013, 25, 535–541 CrossRef CAS PubMed
.
- G. Del Giudice, J. J. Goronzy, B. Grubeck-Loebenstein, P.-H. Lambert, T. Mrkvan, J. J. Stoddard and T. M. Doherty, npj Aging Mech. Dis., 2017, 4, 1 CrossRef PubMed
.
- A. D. Foster, A. Sivarapatna and R. E. Gress, Aging Health, 2011, 7, 707–718 CrossRef CAS PubMed
.
- X. Li, C. Li, W. Zhang, Y. Wang, P. Qian and H. Huang, Signal Transduction Targeted Ther., 2023, 8, 239 CrossRef PubMed
.
- Y. Jiang, Y. Li and B. Zhu, Cell Death Dis., 2015, 6, e1792 CrossRef CAS PubMed
.
- B. Pereira, X.-N. Xu and A. N. Akbar, Front. Immunol., 2020, 11, 583019 CrossRef CAS PubMed
.
- J. Lian, Y. Yue, W. Yu and Y. Zhang, J. Hematol. Oncol., 2020, 13, 151 CrossRef PubMed
.
- H. Donninger, C. Li, J. W. Eaton and K. Yaddanapudi, Vaccines, 2021, 9(6), 668 CrossRef CAS PubMed
.
- A. Agrawal and B. Weinberger, Front. Aging, 2022, 3, 882494 CrossRef PubMed
.
- S. N. Crooke, I. G. Ovsyannikova, G. A. Poland and R. B. Kennedy, Immun. Ageing, 2019, 16, 25 CrossRef PubMed
.
- W. Xu, G. Wong, Y. Y. Hwang and A. Larbi, Semin. Immunopathol., 2020, 42, 559–572 CrossRef PubMed
.
- D. Weiskopf, B. Weinberger and B. Grubeck-Loebenstein, Transplant Int., 2009, 22, 1041–1050 CrossRef CAS PubMed
.
- N. Mohaghegh, A. Ahari, C. Buttles, S. Davani, H. Hoang, Q. Huang, Y. Huang, B. Hosseinpour, R. Abbasgholizadeh, A. L. Cottingham, N. Farhadi, M. Akbari, H. Kang, A. Khademhosseini, V. Jucaud, R. M. Pearson and A. Hassani Najafabadi, ACS Nano, 2024, 18, 27764–27781 CrossRef CAS PubMed
.
- N. Mohaghegh, A. Iyer, E. Wang, N. Z. Balajam, H. Kang, M. Akbari, M. S. Barnhill, A. Khademhosseini, R. M. Pearson and A. Hassani Najafabadi, J. Controlled Release, 2025, 382, 113670 CrossRef CAS PubMed
.
- J. Kim, A. Morgott, Z. Wu, L. Hopaluk, M. Miles, W. Stoner and Q. Li, 2019.
- M. Jafarkhani, Z. Salehi, M. A. Shokrgozar and S. Mashayekhan, Int. J. Numer. Methods Biomed. Eng., 2019, 35, e3154 CrossRef PubMed
.
- S. S. Jensen, H. Jensen, C. Cornett, E. H. Møller and J. Østergaard, J. Pharm. Biomed. Anal., 2014, 92, 203–210 CrossRef CAS PubMed
.
- S. Maity, T. Bhuyan, R. Bhattacharya and D. Bandyopadhyay, ACS Appl. Mater. Interfaces, 2021, 13, 19430–19442 CrossRef CAS PubMed
.
- S. Maity, T. Bhuyan, J. P. Pattanayak, S. S. Ghosh and D. Bandyopadhyay, Nanotechnology, 2021, 32, 505704 CrossRef CAS PubMed
.
- L. E. Harrington, R. D. Hatton, P. R. Mangan, H. Turner, T. L. Murphy, K. M. Murphy and C. T. Weaver, Nat. Immunol., 2005, 6, 1123–1132 CrossRef CAS PubMed
.
- F. Liao, J. Zhang, Y. Hu, A. H. Najafabadi, J. J. Moon, M. S. Wicha, B. Kaspo, J. Whitfield, A. E. Chang and Q. Li, Cancer Immunol., Immunother., 2022, 71, 1959–1973 CrossRef CAS PubMed
.
- A. H. Najafabadi, Z. I. N. Abadi, M. E. Aikins, K. E. Foulds, M. M. Donaldson, W. Yuan, E. B. Okeke, J. Nam, Y. Xu, P. Weerappuli, T. Hetrick, D. Adams, P. A. Lester, A. M. Salazar, D. H. Barouch, A. Schwendeman, R. A. Seder and J. J. Moon, J. Controlled Release, 2021, 337, 168–178 CrossRef CAS PubMed
.
- J. Nikolich-Žugich, Nat. Immunol., 2018, 19, 10–19 CrossRef PubMed
.
- Z. Zhou, Y. Pang, J. Ji, J. He, T. Liu, L. Ouyang, W. Zhang, X.-L. Zhang, Z.-G. Zhang, K. Zhang and W. Sun, Nat. Rev. Immunol., 2024, 24, 18–32 CrossRef CAS PubMed
.
- H. L. Thompson, M. J. Smithey, J. L. Uhrlaub, I. Jeftić, M. Jergović, S. E. White, N. Currier, A. M. Lang, A. Okoye, B. Park, L. J. Picker, C. D. Surh and J. Nikolich-Žugich, Aging Cell, 2019, 18, e12865 CrossRef PubMed
.
- C. Sebastián, C. Herrero, M. Serra, J. Lloberas, M. A. Blasco and A. Celada, J. Immunol., 2009, 183, 2356–2364 CrossRef PubMed
.
- C. A. Chougnet, R. I. Thacker, H. M. Shehata, C. M. Hennies, M. A. Lehn, C. S. Lages and E. M. Janssen, J. Immunol., 2015, 195, 2624–2632 CrossRef PubMed
.
- C. Sebastián, C. Herrero, M. Serra, J. Lloberas, M. A. Blasco and A. Celada, J. Immunol., 2009, 183, 2356–2364 CrossRef PubMed
.
- S. Della Bella, L. Bierti, P. Presicce, R. Arienti, M. Valenti, M. Saresella, C. Vergani and M. L. Villa, Clin. Immunol., 2007, 122, 220–228 CrossRef PubMed
.
- S. X. Leng, Q. L. Xue, J. Tian, Y. Huang, S. H. Yeh and L. P. Fried, Exp. Gerontol., 2009, 44, 511–516 CrossRef PubMed
.
- C. E. Moss, H. Phipps, H. L. Wilson and E. Kiss-Toth, Front. Immunol., 2023, 14, 1222308 CrossRef PubMed
.
- E. Linehan and D. C. Fitzgerald, Eur. J. Microbiol. Immunol., 2015, 5, 14–24 CrossRef PubMed
.
- W. B. Ershler, W. H. Sun, N. Binkley, S. Gravenstein, M. J. Volk, G. Kamoske, R. G. Klopp, E. B. Roecker, R. A. Daynes and R. Weindruch, Lymphokine Cytokine Res., 1993, 12, 225–230 Search PubMed
.
- M. F. Neurath and S. Finotto, Cytokine Growth Factor Rev., 2011, 22, 83–89 CrossRef PubMed
.
- Y. Oishi and I. Manabe, npj Aging Mech. Dis., 2016, 2, 16018 CrossRef PubMed
.
- A. L. Thomas, P. C. Alarcon, S. Divanovic, C. A. Chougnet, D. A. Hildeman and M. E. Moreno-Fernandez, Front. Aging, 2021, 2, 732414 CrossRef PubMed
.
- I. M. Rea, D. S. Gibson, V. McGilligan, S. E. McNerlan, H. D. Alexander and O. A. Ross, Front. Immunol., 2018, 9, 586 CrossRef PubMed
.
- J. Guo, X. Huang, L. Dou, M. Yan, T. Shen, W. Tang and J. Li, Signal Transduction Targeted Ther., 2022, 7, 391 CrossRef PubMed
.
- A. Alberro, A. Iribarren-Lopez, M. Sáenz-Cuesta, A. Matheu, I. Vergara and D. Otaegui, Sci. Rep., 2021, 11, 4358 CrossRef PubMed
.
- N. Unver and F. McAllister, Cytokine Growth Factor Rev., 2018, 41, 10–17 CrossRef PubMed
.
-
A. C. Shaw, in Handbook of Immunosenescence: Basic Understanding and Clinical Implications, ed. T. Fulop, C. Franceschi, K. Hirokawa and G. Pawelec, Springer International Publishing, Cham, 2019, pp. 981–992, DOI:10.1007/978-3-319-99375-1_98
.
- Z. Liu, Q. Liang, Y. Ren, C. Guo, X. Ge, L. Wang, Q. Cheng, P. Luo, Y. Zhang and X. Han, Signal Transduction Targeted Ther., 2023, 8, 200 CrossRef PubMed
.
- J. P. Coppé, C. K. Patil, F. Rodier, Y. Sun, D. P. Muñoz, J. Goldstein, P. S. Nelson, P. Y. Desprez and J. Campisi, PLoS Biol., 2008, 6, 2853–2868 CrossRef PubMed
.
- C. Franceschi, P. Garagnani, P. Parini, C. Giuliani and A. Santoro, Nat. Rev. Endocrinol., 2018, 14, 576–590 CrossRef CAS PubMed
.
- J. J. Goronzy and C. M. Weyand, Nat. Rev. Immunol., 2019, 19, 573–583 CrossRef CAS PubMed
.
- E. Carrasco, M. M. Gómez de las Heras, E. Gabandé-Rodríguez, G. Desdín-Micó, J. F. Aranda and M. Mittelbrunn, Nat. Rev. Immunol., 2022, 22, 97–111 CrossRef CAS PubMed
.
- L. A. Callender, E. C. Carroll, E. A. Bober and S. M. Henson, Ageing Res. Rev., 2018, 47, 24–30 CrossRef CAS PubMed
.
- I. J. Rodriguez, N. Lalinde Ruiz, M. Llano León, L. Martínez Enríquez, M. D. P. Montilla Velásquez, J. P. Ortiz Aguirre, O. M. Rodríguez Bohórquez, E. A. Velandia Vargas, E. D. Hernández and C. A. Parra López, Front. Immunol., 2020, 11, 604591 CrossRef CAS PubMed
.
- M. S. Abdel-Hakeem, S. Manne, J.-C. Beltra, E. Stelekati, Z. Chen, K. Nzingha, M.-A. Ali, J. L. Johnson, J. R. Giles, D. Mathew, A. R. Greenplate, G. Vahedi and E. J. Wherry, Nat. Immunol., 2021, 22, 1008–1019 CrossRef CAS PubMed
.
- P. Tonnerre, D. Wolski, S. Subudhi, J. Aljabban, R. C. Hoogeveen, M. Damasio, H. K. Drescher, L. M. Bartsch, D. C. Tully, D. R. Sen, D. J. Bean, J. Brown, A. Torres-Cornejo, M. Robidoux, D. Kvistad, N. Alatrakchi, A. Cui, D. Lieb, J. A. Cheney, J. Gustafson, L. L. Lewis-Ximenez, L. Massenet-Regad, T. Eisenhaure, J. Aneja, W. N. Haining, R. T. Chung, N. Hacohen, T. M. Allen, A. Y. Kim and G. M. Lauer, Nat. Immunol., 2021, 22, 1030–1041 CrossRef CAS PubMed
.
- M. Bellon and C. Nicot, Viruses, 2017, 9(10), 289 CrossRef PubMed
.
- K. Shirakawa and M. Sano, Cell, 2021, 10, 2435 CrossRef CAS PubMed
.
- W. Jiang, Y. He, W. He, G. Wu, X. Zhou, Q. Sheng, W. Zhong, Y. Lu, Y. Ding, Q. Lu, F. Ye and H. Hua, Front. Immunol., 2020, 11, 622509 CrossRef CAS PubMed
.
- C. Fenwick, V. Joo, P. Jacquier, A. Noto, R. Banga, M. Perreau and G. Pantaleo, Immunol. Rev., 2019, 292, 149–163 CrossRef CAS PubMed
.
- E. J. Wherry and M. Kurachi, Nat. Rev. Immunol., 2015, 15, 486–499 CrossRef CAS PubMed
.
- J. L. Collier, S. A. Weiss, K. E. Pauken, D. R. Sen and A. H. Sharpe, Nat. Immunol., 2021, 22, 809–819 CrossRef CAS PubMed
.
- V. Decman, B. J. Laidlaw, T. A. Doering, J. Leng, H. C. Ertl, D. R. Goldstein and E. J. Wherry, J. Immunol., 2012, 188, 1933–1941 CrossRef CAS PubMed
.
- J. Reiser and A. Banerjee, J. Immunol. Res., 2016, 2016, 8941260 Search PubMed
.
- P. Moura Rosa, N. Gopalakrishnan, H. Ibrahim, M. Haug and Ø. Halaas, Lab Chip, 2016, 16, 3728–3740 RSC
.
- M. Rothbauer, H. Zirath and P. Ertl, Lab Chip, 2018, 18, 249–270 RSC
.
- V. Ortseifen, M. Viefhues, L. Wobbe and A. Grünberger, Front. Bioeng. Biotechnol., 2020, 8, 589074 CrossRef PubMed
.
- A. Polini, L. L. del Mercato, A. Barra, Y. S. Zhang, F. Calabi and G. Gigli, Drug Discovery Today, 2019, 24, 517–525 CrossRef CAS PubMed
.
- K. Poornima, A. P. Francis, M. Hoda, M. A. Eladl, S. Subramanian, V. P. Veeraraghavan, M. El-Sherbiny, S. M. Asseri, A. B. A. Hussamuldin, K. M. Surapaneni, U. Mony and R. Rajagopalan, Front. Oncol., 2022, 12, 891673 CrossRef CAS PubMed
.
- X. Zhang, T. H. Kim, T. J. Thauland, H. Li, F. S. Majedi, C. Ly, Z. Gu, M. J. Butte, A. C. Rowat and S. Li, Curr. Opin. Biotechnol., 2020, 66, 236–245 CrossRef CAS PubMed
.
- Q. Wang, Y. Yang, Z. Chen, B. Li, Y. Niu and X. Li, Pharmaceutics, 2024, 16(5), 666 CrossRef CAS PubMed
.
- A. Shanti, B. Samara, A. Abdullah, N. Hallfors, D. Accoto, J. Sapudom, A. Alatoom, J. Teo, S. Danti and C. Stefanini, Pharmaceutics, 2020, 12(5), 464 CrossRef CAS PubMed
.
- N. Hallfors, A. Shanti, J. Sapudom, J. Teo, G. Petroianu, S. Lee, L. Planelles and C. Stefanini, Bioengineering, 2021, 8(2), 19 CrossRef CAS PubMed
.
- U. Haessler, Y. Kalinin, M. A. Swartz and M. Wu, Biomed. Microdevices, 2009, 11, 827–835 CrossRef CAS PubMed
.
- U. Haessler, M. Pisano, M. Wu and M. A. Swartz, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5614–5619 CrossRef CAS PubMed
.
- B. G. Ricart, B. John, D. Lee, C. A. Hunter and D. A. Hammer, J. Immunol., 2011, 186, 53–61 CrossRef CAS PubMed
.
- L. Radke, G. Sandig, A. Lubitz, U. Schließer, H. H. von Horsten, V. Blanchard, K. Keil, V. Sandig, C. Giese, M. Hummel, S. Hinderlich and M. Frohme, Bioengineering, 2017, 4(3), 70 CrossRef PubMed
.
- T. Kraus, A. Lubitz, U. Schließer, C. Giese, J. Reuschel, R. Brecht, J. Engert and G. Winter, J. Pharm. Sci., 2019, 108, 2358–2366 CrossRef CAS
.
- A. Dauner, P. Agrawal, J. Salvatico, T. Tapia, V. Dhir, S. F. Shaik, D. R. Drake, 3rd and A. M. Byers, Vaccine, 2017, 35, 5487–5494 CrossRef CAS PubMed
.
- A. Purwada, S. B. Shah, W. Béguelin, A. August, A. M. Melnick and A. Singh, Biomaterials, 2019, 198, 27–36 CrossRef CAS PubMed
.
- A. Purwada and A. Singh, Nat. Protoc., 2017, 12(1), 168–182 CrossRef CAS PubMed
.
- J. R. Veatch, N. Singhi, S. Srivastava, J. L. Szeto, B. Jesernig, S. M. Stull, M. Fitzgibbon, M. Sarvothama, S. Yechan-Gunja, S. E. James and S. R. Riddell, J. Clin. Invest., 2021, 131(16), 144195 CrossRef PubMed
.
- Y. Chen, D. Yu, H. Qian, Y. Shi and Z. Tao, J. Transl. Med., 2024, 22, 394 CrossRef PubMed
.
- D. E. Ingber, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed
.
- H. Maeng, M. Terabe and J. A. Berzofsky, Curr. Opin. Immunol., 2018, 51, 111–122 CrossRef CAS PubMed
.
- J. Liu, M. Fu, M. Wang, D. Wan, Y. Wei and X. Wei, J. Hematol. Oncol., 2022, 15, 28 CrossRef PubMed
.
- C. E. DeSantis, K. D. Miller, W. Dale, S. G. Mohile, H. J. Cohen, C. R. Leach, A. Goding Sauer, A. Jemal and R. L. Siegel, CA Cancer J. Clin., 2019, 69, 452–467 Search PubMed
.
- J. Lian, Y. Yue, W. Yu and Y. Zhang, J. Hematol. Oncol., 2020, 13, 151 CrossRef PubMed
.
- C. Ma, Y. Peng, H. Li and W. Chen, Trends Pharmacol. Sci., 2021, 42, 119–133 CrossRef CAS PubMed
.
- T. Fan, M. Zhang, J. Yang, Z. Zhu, W. Cao and C. Dong, Signal Transduction Targeted Ther., 2023, 8, 450 CrossRef CAS
.
- J. Mestas and C. C. Hughes, J. Immunol., 2004, 172, 2731–2738 CrossRef CAS PubMed
.
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123, 4195–4200 CrossRef CAS PubMed
.
- J. G. Cyster and S. R. Schwab, Annu. Rev. Immunol., 2012, 30, 69–94 CrossRef CAS PubMed
.
- F. Sallusto, A. Lanzavecchia, K. Araki and R. Ahmed, Immunity, 2010, 33, 451–463 CrossRef PubMed
.
- D. Huh, G. A. Hamilton and D. E. Ingber, Trends Cell Biol., 2011, 21, 745–754 CrossRef CAS PubMed
.
- J. Oh, A. Magnuson, C. Benoist, M. J. Pittet and R. Weissleder, JCI insight, 2018, 3(21), 122961 CrossRef PubMed
.
- C. Pettan-Brewer, J. Morton, R. Coil, H. Hopkins, S. Fatemie and W. Ladiges, Pathobiol. Aging Age-Relat. Dis., 2012, 2(1), 19182 Search PubMed
.
- G. Goyal, P. Prabhala, G. Mahajan, B. Bausk, T. Gilboa, L. Xie, Y. Zhai, R. Lazarovits, A. Mansour, M. S. Kim, A. Patil, D. Curran, J. M. Long, S. Sharma, A. Junaid, L. Cohen, T. C. Ferrante, O. Levy, R. Prantil-Baun, D. R. Walt and D. E. Ingber, Adv. Sci., 2022, 9, 2103241 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2026 |