Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mimicking a solvent interface at the substrate access channel of nylonase accelerates nylon degradation
Xiaodi
Li
ab,
Yu
Ji
*ab,
Guohua
Li
 b,
Shuaiqi
Meng
cd,
Peng
Zhang
e,
Yiheng
Liu
b,
Shuaiqi
Meng
cd,
Peng
Zhang
e,
Yiheng
Liu
 ac,
Enguang
Ding
a,
Kun
Sun
a,
Tianwei
Tan
*a and
Ulrich
Schwaneberg
ac,
Enguang
Ding
a,
Kun
Sun
a,
Tianwei
Tan
*a and
Ulrich
Schwaneberg
 *bc
*bc
aState Key Laboratory of Green Biomanufacturing, National Energy R&D Center for Biorefinery, Beijing Key Lab of Bioprocess, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing 100029, China. E-mail: twtan@mail.buct.edu.cn; yuji@buct.edu.cn
bBeijing Advanced Innovation Center for Soft Matter Science and Engineering, Beijing University of Chemical Technology, Beijing 100029, China. E-mail: u.schwaneberg@biotec.rwth-aachen.de
cInstitute of Biotechnology, RWTH Aachen University, Worringerweg 3, 52074 Aachen, Germany
dCollege of Life Sciences, Nanjing Normal University, Nanjing 210097, China
eInstitute of Microbial Technology, Shandong University, Qingdao 266237, China
First published on 23rd December 2025
Abstract
The depolymerization of plastics (e.g., PET and nylon) is of great significance to the polymer circular economy. Organic solvents (OSs) often play an important role in plastic recycling by swelling and dissolving polymers. In this study, we mimicked the swelling effect of OSs by redesigning the substrate access channel of nylonase to accelerate the depolymerization of nylon. In detail, eight OSs were evaluated and toluene showed the best swelling effect on nylon 6 (PA6). The key residues in the substrate access channel (loop88–93, loop219–222, and loop300–305) of nylonase were selected and substituted with aromatic amino acids. Multiplex PCR was used for smart library generation. After screening and rescreening, the final variant NylC-V3 (A91W/P220W/D304Y) with improved specific activity (18.7-fold) and thermostability (Tm value: increased by 3.9 °C) was obtained compared to the wild type (WT). Molecular dynamics simulations provided a mechanistic explanation for the enhanced performance of the NylC-V3 variant. The strategic incorporation of aromatic residues strengthened the interactions between adjacent subunits, increasing the structural rigidity of the enzyme. The D304Y substitution induced a steric rearrangement that widened the substrate access channel, thereby facilitating deeper substrate binding. Concurrently, the A91W substitution functioned as a molecular clasp utilizing hydrophobic forces and cation–π interactions from its indole ring to securely lock the substrate in a catalytically optimal conformation. This study provides proof of principle that mimicking the functional groups of OSs in nylon degrading enzymes accelerates depolymerization, providing a biocatalytic solution for developing environmentally friendly plastic recycling technology.
Green foundation1. This study advances green chemistry by engineering an enzyme (nylonase) to mimic the polymer-swelling function of organic solvents, enabling efficient enzymatic depolymerization of nylon 6 without requiring environmentally harmful solvents.2. We quantitatively improved the specific activity of the enzyme by 18.7-fold and increased its thermal stability (Tm+3.9 °C), enhancing its efficiency and robustness for biocatalytic nylon recycling under mild conditions. 3. Future research could further green this process by extending this biomimetic solvent-free strategy to other polyesters/polyamides, optimizing enzyme performance under industrial conditions, and integrating the process with renewable energy inputs. |
1. Introduction
With a global annual production of nearly 10 million tons, polyamide (PA) is a critically important synthetic polymer.1 As a dominant synthetic polyamide material, nylon has broad applications in the automotive industry, textile industry, and medical equipment with excellent mechanical properties and chemical stability.2 Currently, the recycling of nylon still remains at a low level, most nylon being recycled by mechanical or chemical methods. For example, the depolymerization of nylon can be achieved by strong acids, bases, and organic solvents at elevated temperatures by breaking amide bonds, which needs harsh conditions and toxic chemicals.3–5 Developing an environmentally friendly strategy to recycle nylon has attracted great interest.6 An alternative to these traditional processes is the enzymatic polymer recycling process, which has been demonstrated by the PET recycling commercialized by Carbios.7In the case of nylon depolymerization, only a few enzymes have been reported.8–14 Negoro et al.15 found three classes of nylon hydrolases in Arthrobacter sp. KI72, including 6-aminohexanoic acid (6-AHA) cyclic dimer hydrolase (NylA, EC3.5.2.12), 6-AHA dimer hydrolase (NylB, EC3.5.1.46), and 6-AHA oligomer endonuclease (NylC, EC3.5.-.-). Among them, NylC has the best depolymerization performance toward nylon compared to NylA and NylB. The activity of NylCp2 from Arthrobacter plasmid pOAD2 is 7.1 U mg−1.10,11 Beckham et al.13 screened 40 potential natural and engineered nylon-hydrolysing enzymes and NylCK-TS (NylCK-S111G/A137L) achieved the hydrolysis of 0.67 wt% of the PA6 film. Yoshiki Higuchi et al.14 substituted four positions in NylCp2 and obtained NylCp2-TS (NylCp2-D122G/H130Y/D36A/E263Q) with high thermal stability (Tm = 88 °C). We also obtained a variant NylC-HP (NylCp2-TS-F134W/D304M/R330A) through directed evolution, which exhibited a 6.9-fold increase in specific activity (520 ± 19 µmol6-AHAeq. h−1 mgenzyme−1) and enhanced thermal resistance (Tm = 90 °C).16,17 To the best of our knowledge, the degradation performance of even the most advanced nylonase variants is still below 1.3 wt%.6,18
Polymer recycling using organic solvents is a promising alternative for conducting reactions in homogeneous solutions and thereby achieving efficient polymer swelling and degradation. Jones et al.19 achieved 75% conversion of polylactic acid using CH2Cl2 and MeOH at room temperature within 24 h. Another attractive strategy is combining the advantages of organic solvents and the high specificity of enzymes to further improve the degradation efficiency of polymers. Jiang et al.20 used a two-stage process including pretreatment of polyester-cotton blended textiles with an organic solvent, followed by enzymatic depolymerization, achieving 86.8% depolymerization of PET nanoparticles within 2 h. Mimicking the swelling effect of organic solvents on enzymes to boost polymer degradation has not yet been reported.
Inspired by the swelling effect of organic solvents, we first investigated the dissolution properties of eight organic solvents for nylon. Toluene, which efficiently swells PA6, was selected as the favored solvent. To mimic this effect, aromatic amino acids (Phe, Tyr, and Trp) were introduced into the substrate access channel of nylonase, and the degradation performance of PA6 was investigated. In detail, 16 key positions in the substrate access channel and close to the active center of nylonase were selected and simultaneously substituted with F, Y, and W. After screening of 600 variants, the best variant NylC-V3(A91W/P220W/D304Y) was obtained with 18.7-fold improved catalytic activity and the Tm value was increased by 3.9 °C (compared to WT). Molecular dynamics (MD) simulations revealed that the flexibility of the key loop regions was reduced, and the aromatic residue A91W stabilized the conformation of the substrate. This study validates the principle that mimicking the organic solvent can improve the degradation performance of nylonase (NylCp2-TS).
2. Results and discussion
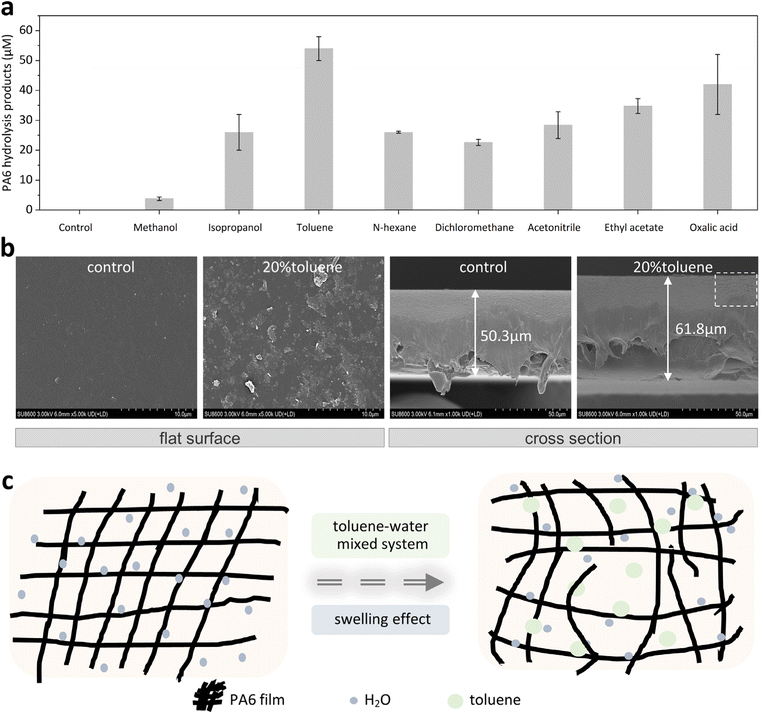
The organic solvent mimicking strategy to improve the catalytic efficiency of nylonase was implemented in four parts. In the first part, we found that toluene was effective in the swelling and depolymerization of the PA6 film. In the second part, key residues near the substrate access channel of nylonase were substituted with aromatic residues by imitating the structure of toluene, and a smart library was generated by multiplex PCR technology and screened. In the third part, the best variant NylC-V3 with improved activity and thermostability was characterized. In the last part, the molecular dynamics analysis was conducted to reveal molecular insights into the improved catalytic performance of NylC-V3.2.1 Screening of organic solvents for depolymerization of PA6
Organic solvents often have a strong swelling effect on polymers at high temperatures (>300 °C).21 However, to explore the mimicking effect of OSs on enzymatic depolymerization under milder conditions, eight structurally different OSs were selected based on the literature, including alcohols (methanol and isopropanol), alkanes (N-hexane and dichloromethane), aromatics (toluene), organic acids (oxalic acid), esters (ethyl acetate), and nitriles (acetonitrile). Then, the swelling effect of OSs on PA6 was investigated and toluene (20% v/v) showed the best swelling effect at 60 °C for 2 h (Fig. 1a). Field-emission scanning electron microscopy (FESEM) analysis revealed significant morphological changes in the toluene-treated PA6 film, including a markedly roughened surface and an increase in thickness of 11.5 µm (Fig. 1b).Toluene showed the most significant swelling effect among all selected solvents. The reason could be its unique molecular structure. Toluene as a hydrophobic aromatic solvent has a nonpolar methyl group with an electron-rich benzene ring capable of extensive π–π delocalization. Therefore, it can enable synergistic interactions with both aliphatic segments and amide groups in PA6 chains via hydrophobic effects and CH–π interactions. Furthermore, the swelling mechanism is governed by the similarity in solubility parameters between toluene and PA6, which enhances miscibility and facilitates solvent diffusion into the polymer matrix.22 This process disrupts intermolecular hydrogen bonding and increases chain mobility, thereby effectively promoting swelling (Fig. 1c).
2.2 Engineering of nylonase by mimicking the toluene swelling effect
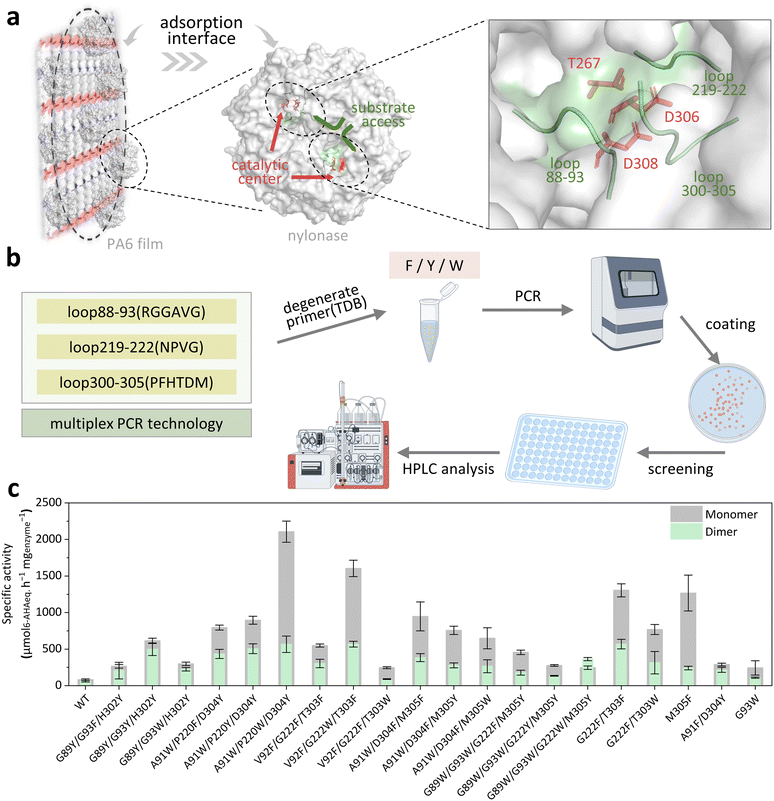
To establish a green and efficient enzymatic alternative to toluene swelling, we would like to introduce aromatic amino acids (Phe, Tyr, and Trp) in nylonase (NylCp2-TS). These aromatic amino acids can mimic the properties of toluene by virtue of their hydrophobic side chains and π-electron-rich characteristics.By visualization of the structure of NylCp2-TS (WT, PDB ID: 5Y0M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24), we identified 16 positions located in flexible loop regions (loop88–93, loop219–222, and loop300–305; Fig. 2a) that are closely associated with substrate binding and are adjacent to the catalytic triad (T267/D306/D308). To explore the synergistic effects, a co-evolution engineering strategy was utilized through simultaneous site-specific mutation mediated by multiplex PCR with degenerate primers (TDB, substituted with aromatic residues F/Y/W, Fig. 2b). A library with 600 clones was generated and its quality was verified by partial sequencing (Table S1). The top 20 variants with activity were used for shake flask fermentation and eleven of them showed more than 5-fold improved specific activity compared with the WT, among which the specific activity of the best variant NylC-V3(A91W/P220W/D304Y) is 2652.1 µM6-AHAeq. h−1 mgenzyme−1 (18.7-fold improvement compared to the WT, Fig. 2c and Fig. S1).
24), we identified 16 positions located in flexible loop regions (loop88–93, loop219–222, and loop300–305; Fig. 2a) that are closely associated with substrate binding and are adjacent to the catalytic triad (T267/D306/D308). To explore the synergistic effects, a co-evolution engineering strategy was utilized through simultaneous site-specific mutation mediated by multiplex PCR with degenerate primers (TDB, substituted with aromatic residues F/Y/W, Fig. 2b). A library with 600 clones was generated and its quality was verified by partial sequencing (Table S1). The top 20 variants with activity were used for shake flask fermentation and eleven of them showed more than 5-fold improved specific activity compared with the WT, among which the specific activity of the best variant NylC-V3(A91W/P220W/D304Y) is 2652.1 µM6-AHAeq. h−1 mgenzyme−1 (18.7-fold improvement compared to the WT, Fig. 2c and Fig. S1).
2.3 Characterization of nylonase NylC-V3 and WT
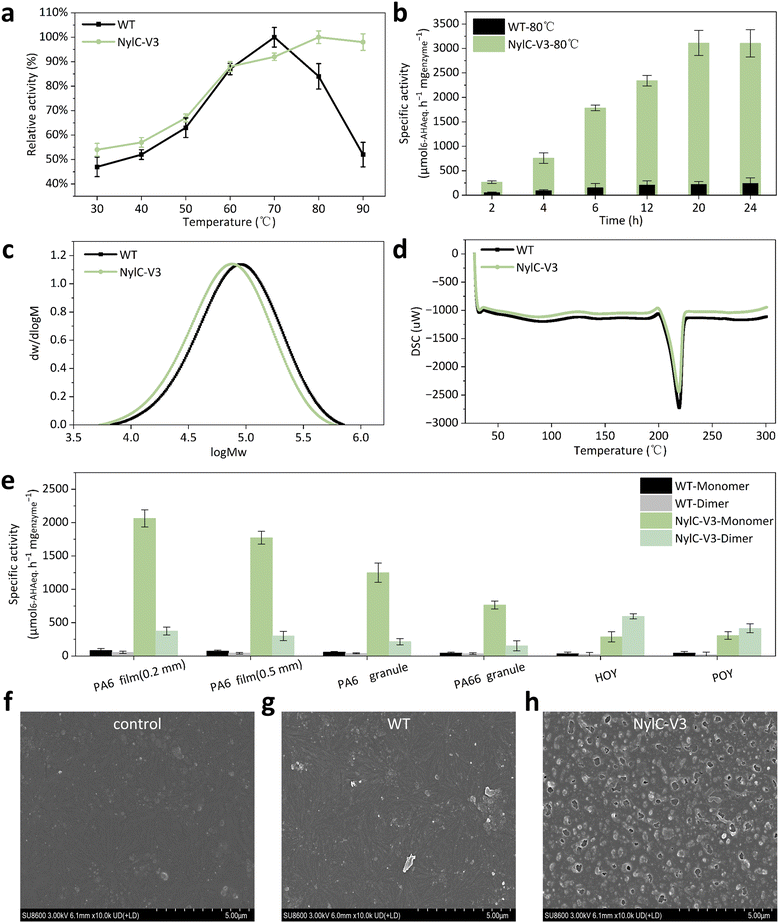
To explore the most suitable reaction temperature of the NylC-V3 variant, we performed the enzymatic hydrolysis of the PA6 film ranging from 30 °C to 90 °C for 2 h. The optimum temperature of NylC-V3 is 80 °C, while that of the WT is 70 °C, which is similar to that reported.16 Surprisingly, the relative activity of NylC-V3 still retains 98.0% at 90 °C while the WT has a sharp decrease (Fig. 3a).Kinetic parameters of the enzymatic reaction are the core indices to evaluate the performance of the enzyme.25 NylC-V3 showed a lower Km value (reduced from 1.7 g L−1 to 1.0 g L−1) compared to the WT, indicating that the enzyme–substrate affinity is enhanced. Meanwhile, kcat increased from 7.6 to 25.8 s−1 (3.4-fold improvement) and kcat/Km increased by 5.7-fold (Table 1). In addition, the NylC-V3 variant is more thermally stable than the WT (the Tm value increased from 77.1 °C to 81.0 °C, Table 1 and Fig. S2). In addition, we also analyzed the kinetic parameters by changing the enzyme concentration, which still showed improved properties (Table S2).
| Enzyme | K m (g L−1) | k cat (s−1) | k cat/Km (L g−1 s−1) | T m (°C) |
|---|---|---|---|---|
| WT | 1.7 ± 0.3 | 7.6 ± 0.2 | 4.5 ± 0.3 | 77.1 ± 0.2 |
| NylC-V3 | 1.0 ± 0.4 | 25.8 ± 1.0 | 25.8 ± 0.6 | 81.0 ± 0.1 |
To further analyze the time scale of PA6 degradation by NylC-V3 at the optimal temperature, we monitored the reaction for 24 h. At 80 °C, NylC-V3 reached a plateau at 20 h (3112.4 µM6-AHAeq. h−1 mgenzyme−1, 12.1-fold compared to the WT), while the WT plateaued at 12 h (Fig. 3b). In addition, the time scale at other temperatures (60 °C and 70 °C) was also analyzed. The degradation performance of NylC-V3 is vastly superior to that of the WT over 2 to 24 h. Notably, for NylC-V3, the activities continuously increased throughout the 24 h period at both temperatures, while the WT reached the platform at 12 h (Fig. S3a and S3b). The degradation performance for a longer time was also analyzed. In order to compare with the reported performance parameters of nylonases,13,16,18 the same experimental conditions (60 °C, 7 days) were selected for characterization. After degradation at 60 °C for 7 days, the 4.3 mM 6-AHA monomer and the 2.6 mM 6-AHA dimer were released from PA6. Based on the total released products, this equates to a hydrolysis percentage of 3.2 wt% of the PA6 film treated with NylC-V3 (Fig. S4a). Operating at its optimal temperature of 80 °C, NylC-V3 reached the 7-day degradation level of 60 °C in only 3 days and achieved a final efficiency of 3.3 wt% after a 7-day reaction (Fig. S4b). This performance surpasses that of the reported nylonase TvgC, which showed only 1.3 wt% conversion of PA6 after 8 days at 60 °C.18 NylC-V3 demonstrates not only higher catalytic efficiency but also superior thermostability (the Tm value increased by 3.9 °C). Due to notable evaporation during prolonged reactions at 80 °C, 60 °C was selected for all subsequent characterization studies.
To evaluate the physical changes of the PA6 film treated with NylC-V3 and the WT (e.g., average molecular weight), GPC and DSC analyses were performed. It was shown that the average molecular weight of the PA6 film treated with NylC-V3 exhibited a 24.1% decrease (Mw decreased from 104![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 851 g mol−1 to 79
851 g mol−1 to 79![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 568 g mol−1, Table S3) compared to the WT, which indicated that NylC-V3 could achieve a higher degree of PA6 degradation (Fig. 3c). In addition, DSC results showed that the melting peak area of the NylC-V3 treated PA6 film decreased from 65.6 mJ mg−1 (WT treated PA6 film) to 58.7 mJ mg−1 (60 °C, 7 days, Table S3), indicating that the energy required to destroy the PA6 film was reduced (Fig. 3d).23
568 g mol−1, Table S3) compared to the WT, which indicated that NylC-V3 could achieve a higher degree of PA6 degradation (Fig. 3c). In addition, DSC results showed that the melting peak area of the NylC-V3 treated PA6 film decreased from 65.6 mJ mg−1 (WT treated PA6 film) to 58.7 mJ mg−1 (60 °C, 7 days, Table S3), indicating that the energy required to destroy the PA6 film was reduced (Fig. 3d).23
Furthermore, we analyzed the performance of NylC-V3 on PA6 with different morphologies, including PA6 films (0.2 mm in thickness, 0.5 mm in thickness), oriented yarn (HOY), and pre-oriented yarn (POY). The PA6 film (0.2 mm in thickness) showed the best degradation (Fig. 3e). As the film thickness increased to 0.5 mm, the degradation efficiency decreased for both NylC-V3 and the WT (decreased by 16.1% and 13.1%, respectively). Notably, NylC-V3 exhibited significantly enhanced degradation activity toward PA66, achieving 11.3-fold higher efficiency than the WT. In contrast, the reference enzyme TvgC showed no detectable activity against PA66.18 NylC-V3 also demonstrated superior performance on two commercially available nylon products, highly oriented yarn (HOY) and pre-oriented yarn (POY), with degradation efficiencies 10.6-fold and 9.2-fold higher than those of the WT, respectively (Fig. 3e). To further visually evaluate the degradation performance of the PA6 film by NylC-V3 and the WT, the morphology of the PA6 films was analyzed using FESEM. After 7 days of treatment at 60 °C, it was observed that PA6 films treated with NylC-V3 and the WT exhibited distinct morphological differences. Notably, the surface of the PA6 film treated with NylC-V3 became markedly rough, with the appearance of numerous holes (average diameter: 314.9 nm, Fig. 3h and S5). In contrast, the surface of the PA6 film treated with the WT was only slightly rough and remained largely intact, showing little difference from the control group (Fig. 3f and g).
The engineered nylonase NylC-V3 demonstrates superior thermostability and catalytic efficiency, enabling efficient nylon depolymerization under mild conditions (60–80 °C, ambient pressure). This enzymatic strategy presents a significant advance for the plastic circular economy, particularly when contrasted with conventional chemical methods, which require extreme temperatures (>200 °C) and pressures (>70 bar), along with their substantial associated environmental burdens and equipment demands.24–27
2.4 MD study of NylC-V3/WT and mechanism analysis
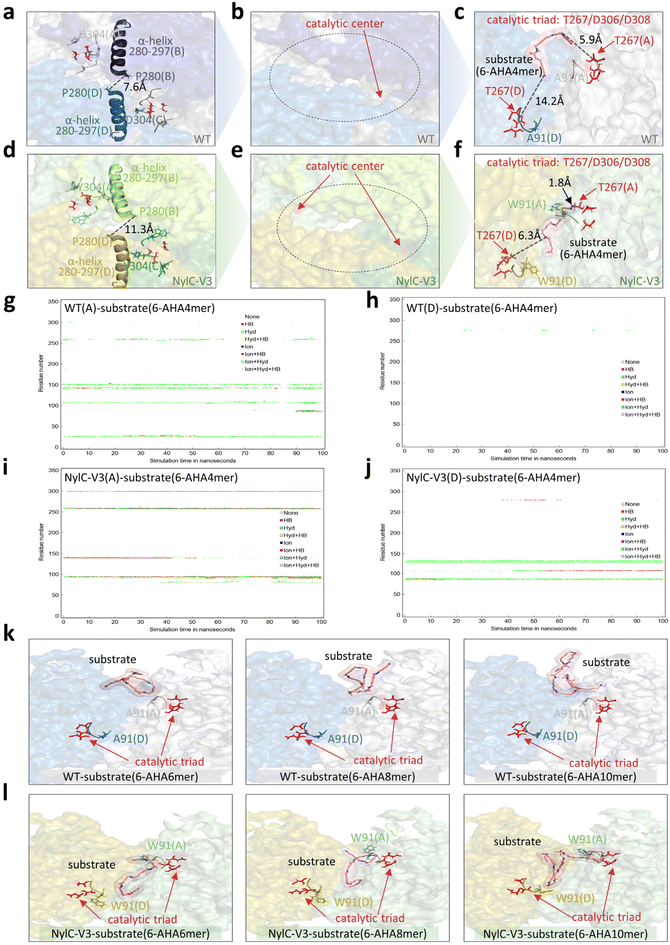
MD simulations demonstrated significantly greater structural rigidity in NylC-V3 than in the WT. RMSD analysis revealed that the average conformational fluctuation of NylC-V3 was 25.0% lower than that of the WT (NylC-V3: average 1.96 Å, WT: average 2.61 Å, Fig. 4a, S6a, and S6b). This increased rigidity was particularly pronounced in three key loop regions. The RMSF value of three key loop regions (loop88–93, loop219–222, and loop300–305) decreased by 47.7%, 29.5%, and 41.7%, respectively (Fig. 4b, S6c, and S6d). In addition, the average number of hydrogen bonds in NylC-V3 is 35 more than that in the WT, which indicates that introduced aromatic acids improve the overall structural rigidity of NylC-V3 (Fig. 4c, S6e, and S6f).We analyzed the substituted sites in NylC-V3 (A91W/P220W/D304Y) and found that both positions 91 and 304 are located at the interfaces between monomers of the homotetrameric enzyme (A/B, B/C, C/D, and D/A, Fig. 4d and e). Substitution of the small alanine at position 91 with the bulky aromatic tryptophan introduced π–π stacking between W91 on monomer D and Y98 on monomer A. Similar interactions are expected across the other three interfacial pairs. Meanwhile, the replacement of negatively charged aspartic acid (D304) with uncharged tyrosine alleviated the electrostatic repulsion between Y304 on monomer B and E282 on monomer A, thereby bringing monomers A and B closer, causing an effect similarly propagated throughout the tetrameric interfaces (Fig. 4d and e). We further examined dynamic inter-subunit interactions by evaluating the dynamic correlation indices involving residues 91 and 304 at the subunit interfaces. A higher correlation indicates stronger inter-subunit communication. The results clearly revealed enhanced dynamic coupling between subunits in NylC-V3 compared to the WT (Fig. 4f). The solvent-accessible surface area (SASA) of NylC-V3 decreased by 108.3 Å2 (WT: 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 877.7 Å2; NylC-V3: 41
877.7 Å2; NylC-V3: 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 769.4 Å2, Fig. S7), indicating a more compact structure. In summary, the increased rigidity and compactness of NylC-V3 contribute to its improved thermostability.
769.4 Å2, Fig. S7), indicating a more compact structure. In summary, the increased rigidity and compactness of NylC-V3 contribute to its improved thermostability.
The local change induced by the D304Y mutation triggered a conformational rearrangement in the NylC-V3 homotetramer. In the WT, the center of the homotetramer adopted a relatively obstructed structure, stabilized by tight interactions among several key amino acids (D99, P280, and V281, Fig. 5a and S8). In NylC-V3, however, the introduction of a bulky aromatic tryptophan residue into the loop300–305 region—specifically the substitution to tyrosine (Y)—displaced adjacent structural elements. The most notable movement occurred in the α-helix spanning residues 280–297 (Fig. 5d). This displacement resulted in the formation of a large central cavity within the homotetramer and increased the distance between the critical gating residue P280 across domains from 7.6 Å to 11.3 Å (Fig. 5a and d). The resulting conformational shift exposed the catalytic center, and the newly formed cavity provided sufficient space for the nylon substrate (6-AHA4mer) to access the catalytic triad (Fig. 5e). It is worth noting that the A91W substitution formed a “molecular clasp” (a stable clamping region between loop88–93 and the catalytic triad), which reduced the distance between the substrate's amide bond and T267 from 5.9 Å to 1.8 Å (Fig. 5c and f). This shortened distance greatly facilitated nucleophilic attack. Consequently, the local and global structural changes enable a novel catalytic mechanism. In NylC-V3, the substrate can shuttle between different monomer domains with both sides of the amide chain inserting into the catalytic centers (monomers A and D), enabling parallel nucleophilic attacks (Fig. 5f). In contrast, in the WT, the substrate remained confined near the catalytic center of monomer A only (Fig. 5c).
Furthermore, the dynamic interactions between the substrate and catalytic triad (T267/D306/D308) across enzyme monomers were analyzed (Fig. 5g–j). In the WT, the substrate shows limited interaction with the catalytic triad in monomer A and virtually no contact with that in monomer D (Fig. 5g and h). By contrast, in NylC-V3, the substrate engages in sustained and strong interactions with the triad in monomer A, along with detectable contact in monomer D (Fig. 5i and j). Accompanying these dynamic changes, a network of intermolecular forces including hydrophobic and ionic interactions mediated by aromatic residues emerged around loop88–93, suggesting a structural role of A91W in reshaping the local binding environment. The enhanced binding was quantitatively corroborated by the calculated binding free energy of −43.5 kcal mol−1 for NylC-V3, which is significantly stronger than the WT (−37.8 kcal mol−1, Table S4). Together, the persistent substrate−catalytic triad contact, the newly formed interaction network near the loop, and the considerably improved binding affinity provide coherent evidence supporting the “molecular clasp” function of A91W in facilitating substrate positioning and stabilization.
Given that the polymerization degree of PA6 is much higher than four units,28 the structural rearrangement in NylC-V3 enables multi-site hydrolysis of a single polymer chain. To validate this, we performed molecular docking with longer substrates (hexamer: 6-AHA6mer, octamer: 6-AHA8mer, and decamer: 6-AHA10mer) into both NylC-V3 and the WT (Fig. 5k and i). The results showed that the tightly packed central structure of the WT sterically hinders substrate binding (Fig. 5k), whereas the central cavity in NylC-V3 accommodates the extended polymer chain, with W91 facilitating substrate recruitment toward the catalytic triad (Fig. 5i).
By mimicking the aromatic functional groups in organic solvents, the binding affinity was enhanced (NylC-V3) by introducing aromatic amino acids in the loop regions of the nylonase surface. Residues on the surface provide the necessary conformational adaptability and substrate accessibility. These introduced aromatic residues via dominant hydrophobic effects and their electron-rich π-system form specific cation–π interactions with the substrate's N-terminal ammonium group, providing directional attraction similar to toluene.
3. Materials and methods
3.1 Gene construction
The gene encoding NylCp2-TS was commercially synthesized by Beijing Tsingke Biotechnology Co., Ltd and cloned into pET 28a (Fig. S9). The nucleotide and amino acid sequences are shown in Table S5. A polyhistidine tag was fused N-terminally to the NylCp2-TS gene for purification. The E. coli BL21(DE3) strain (Addgene_197656) was used for heterologous expression.3.2 Construction of the variant library
NylCp2-TS variants were constructed by multiplex PCR, and the primer sequences are shown in Table S6. The PCR solutions contained DNA polymerase (2 × Phanta Flash Master Mix), forward and reverse primers, template DNA, and ddH2O. The PCR program was 98 °C for 30 s (1 cycle), 98 °C for 10 s/ 55 °C for 5 s/ 72 °C for 36 s (30 cycles), and 72 °C for 10 min (1 cycle). The amplified PCR products were gel-purified, digested with DpnI (37 °C, 1 h), and added cyclase for product formation and transformed into E. coli BL21(DE3) competent cells.3.3 Expression of the variant library
Single colonies were inoculated into 200 μL of LB medium (1% tryptone, 1% sodium chloride and 0.5% yeast extract, containing 50 μg mL−1 kanamycin) and inoculated into 96-well plates. As a positive control, 12 wells were inoculated with NylCp2-TS. The plates were incubated at 37 °C, 70% humidity and 1000 rpm for 16 h; then, 10 μL of preculture were transferred to 130 μL of TB medium containing 50 μg ml−1 of kanamycin and inoculated in 96-well plates. When the cells grew to an OD600 of 0.6–0.8 (37 °C, 70% humidity, 1000 rpm, 2 h), the expression was induced by adding isopropyl-β-D-thiogalactopyranoside (IPTG) with a final concentration of 0.1 mM and continued incubation (1000 rpm, 18 °C, 20 h). Cells were collected by centrifugation (3000 rpm, 4 °C, 15 min), and the cell precipitate was washed with 150 μL of PBS buffer (including 10 mM Na2HPO4, 2 mM NaH2PO4, 135 mM NaCl and 4.7 mM KCl, pH = 7.3) to remove the residual TB medium. Next, cell lysis was completed by resuspending the cells and incubating them in 150 μL of lysis buffer (50 mM Bicine, pH 8.0, 1.5 mg mL−1 lysozyme, 25 μg mL−1 DNase I, 37 °C, 2 h). Finally, it was centrifuged again (3000 rpm, 4 °C, 15 min) to obtain the supernatant.3.4 Selection of variants
Initial screening was performed by following the reported literature.16 By adding 90 mg mL−1 PA6 film (0.2 mm in thickness, 6.4 mm in diameter, AM30-FM-000200, GoodFellow, Germany) to a 96-well plate containing 10 μL of clarified cell lysate and PBS buffer, a reaction system of 200 μL was achieved. After sealing the reaction plate, it was incubated at 60 °C and 800 rpm for 4 h. Subsequently, the reaction plate was centrifuged (4 °C, 3000 rpm, 20 min), and 100 μL of the liquid sample was subjected to further HPLC analysis.The variants with improved product release were selected for sequencing (Table S1), and the top 20 variants were used for shake flask fermentation. The preserved corresponding bacterial liquid was inoculated into 2 mL LB medium with 50 μg mL−1 kanamycin sulfate and grown overnight at 37 °C. The overnight-grown culture (using 1 mL) was then transferred to a 100 mL TB medium supplemented with 50 μg mL−1 kanamycin sulfate. The inoculated TB medium was incubated at 37 °C until the cell density (OD600) reached approximately 0.6–0.8. Proteins were induced by the addition of 0.1 mM of IPTG, and cells were cultured at 18 °C for 20 h. Then, the cells were harvested and ultrasonically broken to obtain the supernatant containing the soluble proteins, which were subsequently purified by Ni-IDA affinity chromatography. Supernatants were loaded onto Ni-IDA columns and equilibrated with binding buffer (20 mM imidazole, 50 mM Tris-HCl, 500 mM NaCl, pH 7.5). Then, the protein was sequentially eluted with wash buffer (50 mM imidazole, 50 mM Tris-HCl, 500 mM NaCl, pH 7.5) and elution buffer (500 mM imidazole, 50 mM Tris-HCl, 500 mM NaCl, pH 7.5). Eluted protein samples with elution buffer were collected and analyzed by 12.5% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (Fig. S10).
3.5 Depolymerization products of PA6 film analysis by HPLC
A reaction system contained 0.1 mM purified WT and the top 20 variants, 90 mg mL−1 PA6 film (0.2 mm in thickness, 6.4 mm in diameter, AM30-FM-000200, Goodfellow, Germany), and PBS buffer in a total volume of 200 μL, and degradation was performed by incubation at 60 °C (500 rpm, 24 h). Afterward, the PA6 film was taken out and protein was removed by heating at 100 °C for 10 min and then the supernatant was collected by centrifugation (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min); the supernatant was further analyzed by HPLC. The depolymerization products were analyzed using a HPLC (Waters, USA) equipped with an AlphaHybrid C18 column (150 Å, 5 µm). The mobile phase was 70% v/v deionized water and 30% v/v acetonitrile. The flow was isocratic and fixed at 0.5 mL min−1. Regarding the UV detection method, this analysis method is based on established literature.16 The absorption observed at 220 nm for underivatized 6-aminohexanoic acid (6-AHA) is primarily due to the electronic transition of its carbonyl group (–C
000 rpm, 10 min); the supernatant was further analyzed by HPLC. The depolymerization products were analyzed using a HPLC (Waters, USA) equipped with an AlphaHybrid C18 column (150 Å, 5 µm). The mobile phase was 70% v/v deionized water and 30% v/v acetonitrile. The flow was isocratic and fixed at 0.5 mL min−1. Regarding the UV detection method, this analysis method is based on established literature.16 The absorption observed at 220 nm for underivatized 6-aminohexanoic acid (6-AHA) is primarily due to the electronic transition of its carbonyl group (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). This wavelength was chosen for detection as it effectively captures this characteristic absorption and is well-suited for routine HPLC-UV analysis. The curves of the monomer and dimer of 6-AHA are shown in Fig. S11. All the experiments were performed in triplicate.
O). This wavelength was chosen for detection as it effectively captures this characteristic absorption and is well-suited for routine HPLC-UV analysis. The curves of the monomer and dimer of 6-AHA are shown in Fig. S11. All the experiments were performed in triplicate.
3.6 Characterization of enzymatic properties
The thermodynamic stability of the WT and variants was evaluated using melting points (Tm) determined using a Prometheus NT.48 (Nano differential scanning fluorimetry, Nano DSF). 10 μL of purified enzymes (0.2 mg mL−1) were filled in capillaries and placed on the sample holder. The device then raised the temperature from 20 °C to 100 °C in increments of 1 °C min−1. The optimum temperature was determined by depolymerizing PA6 films (0.2 mm in thickness) with the WT and variants at different temperatures (30–90 °C) for 2 h, respectively. The depolymerization experiments of the WT and variants with time change were treated at 60–80 °C for 2 h, 4 h, 6 h, 12 h, 20 h, and 24 h, respectively. The flow rate in gel permeation chromatography (GPC) is 1 mL min−1 and the temperature is 35 °C. The temperature range of the differential scanning calorimetry (DSC) reaction is 30–300 °C and the heating rate is 10 °C min−1.The kinetic parameters of the enzyme reactions were measured in PBS buffer (pH 7.3). Various concentrations of the substrate PA6 film (0.2 mm in thickness, 20–320 mg mL−1) were prepared. The 6-AHA monomer and dimer production rates catalyzed by the WT and variants were measured at 60 °C for 2 h (Table 1). In addition, the kinetic parameters were analyzed by varying the enzyme concentration. Various concentrations (0.2 mM, 0.4 mM, 0.6 mM, 0.8 mM, 1.0 mM, 1.5 mM and 2.0 mM) of the enzyme and a constant concentration of the PA6 film (90 mg mL−1) were prepared. The production rates were measured at 60 °C for 2 h (Table S2). To derive the kinetic parameters, nonlinear regression fitting was applied to the collected data using the Origin software.
All the experiments were performed in triplicate to ensure rigor and reliability. Parameter values from three independent analyses were averaged and expressed as mean ± standard deviation.
3.7 Field-emission scanning electron microscopy
The effect of the WT and variants on the surface morphology of the PA6 film (0.2 mm in thickness) was studied by field-emission scanning electron microscopy (FESEM) using a Hitachi Su8600 high-resolution SEM (Hitachi Ltd Corporation, Japan). The PA6 film was incubated (60 °C, 7 days) with 0.1 mM purified WT and variants in reaction buffer (PBS, pH 7.3) and without enzyme for control.3.8 Molecular docking analysis and molecular dynamics (MD) simulations
The X-ray crystallography structure (PDB ID: 5Y0M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29) of the WT downloaded from the protein databank has partial sequence deletion (No. 1–18, 30, and 260–266 amino acids); the whole sequence is modeled based on SWISS-MODEL.30 All variant structures were generated based on the crystal structure utilizing the mutagenesis tool in PyMOL. The structure of the nylon tetramer (6-AHA4mer) was built and energy minimized using Yet Another Scientific Artificial Reality Application (YASARA, version 21.12.19).31 The structure of the WT/variant–6-AHA4mer complex was constructed using AutoDock Vina32 using the default parameters, and the point charges were initially assigned according to the AMBER0333 force field. Only one tetrameric substrate was docked to the vicinity of the catalytic center of monomer A in the homologous tetramers of nylonase, respectively.
29) of the WT downloaded from the protein databank has partial sequence deletion (No. 1–18, 30, and 260–266 amino acids); the whole sequence is modeled based on SWISS-MODEL.30 All variant structures were generated based on the crystal structure utilizing the mutagenesis tool in PyMOL. The structure of the nylon tetramer (6-AHA4mer) was built and energy minimized using Yet Another Scientific Artificial Reality Application (YASARA, version 21.12.19).31 The structure of the WT/variant–6-AHA4mer complex was constructed using AutoDock Vina32 using the default parameters, and the point charges were initially assigned according to the AMBER0333 force field. Only one tetrameric substrate was docked to the vicinity of the catalytic center of monomer A in the homologous tetramers of nylonase, respectively.
All molecular dynamics simulations were performed using YASARA (version 21.12.19).31 The simulation setup included an optimization of the hydrogen bonding network to enhance solute stability and pKa prediction to adjust the protonation states of protein residues at pH 7.4. The system was neutralized by adding Na+ and Cl− ions to achieve a net charge of zero. To match the experimental buffer conditions, NaCl was further added to a physiological concentration of 0.15 mol L−1. The WT–6-AHA4mer complex was centered in a cubic periodic simulation cell with a cube box with a side length of 108 Å.
The AMBER force field was applied to the protein residues, while the GAFF2 force field34 with AM1BCC charges35 was used for the ligand (6-AHA4mer). Water molecules were described using the TIP3P model.36 Short-range van der Waals and electrostatic interactions were treated with a cutoff of 8 Å, and long-range electrostatics were handled using the particle mesh Ewald (PME) method. Bonds involving hydrogen atoms were constrained with the LINCS algorithm, allowing an integration time step of 2 fs.
After initial energy minimization via the steepest descent algorithm followed by simulated annealing to remove steric clashes, the system was equilibrated under the NPT ensemble. The equilibration protocol consisted of restrained heating from 0 to 298 K over 50 ps using the Berendsen thermostat and pressure equilibration at 1 atm for 100 ps using the Berendsen barostat. The thermostat and barostat coupling constants were set to 0.5 ps and 1.0 ps, respectively. Following equilibration, production simulations were conducted for 100 ns at 298 K and 1 atm. Snapshots were saved every 100 ps for subsequent analysis. All simulations were performed in triplicate using different random number seeds. Trajectory analysis was carried out using the YASARA “md_analyze.mcr” macro and PyMOL software.
4. Conclusions
To mimic the swelling effect of toluene, aromatic residues were introduced into the substrate access channel of nylonase. The final variant NylC-V3 (A91W/P220W/D304Y) showed up to an 18.7-fold boosted activity and a 3.9 °C improvement in thermostability when compared to the WT. MD simulations revealed that the D304Y substitution induced a conformational rearrangement that opened a larger central cavity within the homotetramer. This structural expansion enables longer PA6 chains to access and simultaneously engage the catalytic centers of two adjacent monomers, allowing multi-site parallel hydrolysis of a single polymer chain, a mechanistic capability absent in the WT. Concurrently, the A91W substitution formed a “molecular clasp” that significantly shortened the distance between the scissile amide bond of the substrate and the catalytic residue, thereby greatly facilitating nucleophilic attack. Furthermore, the bulky hydrophobic indole ring of tryptophan, with its electron-rich π-system, effectively mimics the solvent-like function of toluene through hydrophobic interactions and cation–π stacking. This study proved to be a highly promising strategy to accelerate the depolymerization of PA6, which is very likely to be expanded to other polymers (e.g., PUR, PET, and PLA).Author contributions
Xiaodi Li: conceptualisation, data curation, formal analysis, investigation, methodology, validation, visualisation, and writing of the original draft. Yu Ji: conceptualisation, data curation, validation, discussion, methodology, supervision, and writing and review of the original draft. Guohua Li: data curation, formal analysis, validation, and software. Shuaiqi Meng: data curation, resources, and software. Peng Zhang: methodology, discussion, and resources. Yiheng Liu: methodology and software. Enguang Ding: methodology and software. Kun Sun: data curation and investigation. Tianwei Tan: discussion and resources. Ulrich Schwaneberg: conceptualisation, data curation, validation, discussion, methodology, supervision, and writing and review of the original draft.Conflicts of interest
Xiaodi Li and Yu Ji have filed a patent application that covers the sequences of the variants described in the paper. The remaining authors declare no competing interests.Data availability
All data supporting this article are included within the supplementary information (SI).Supplementary information: figures and tables. See DOI: https://doi.org/10.1039/d5gc04820f.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2024YFA0920700), the NSFC (22508014), the Fundamental Research Funds for the Central Universities (buctrc202526, JD2528, and buctrc202420) and the Interdisciplinary Research Center of Beijing University of Chemical Technology (No. XK2025-05).References
- Z. Zhang, A. K. Biswal, A. Nandi, K. Frost, J. A. Smith, B. H. Nguyen, S. Patel, A. Vashisth and V. Iyer, Nat. Sustain., 2024, 7, 616–627 CrossRef.
- S. S. Nair and C. Ramesh, Macromolecules, 2005, 38, 454–462 Search PubMed.
- L. Wursthorn, K. Beckett, J. O. Rothbaum, R. M. Cywar, C. Lincoln, Y. Kratish and T. J. Marks, Angew. Chem., Int. Ed., 2022, 62, e202212543 CrossRef PubMed.
- M. Tang, J. Shen, F. Zhang, Y. Zhao, T. Gan, W. Zeng, R. Li, D. Wang, B. Han and Z. Liu, Angew. Chem., Int. Ed., 2024, 64, e202416436 CrossRef.
- A. Kamimura, Y. Shiramatsu and T. Kawamoto, Green Energy Environ., 2019, 4, 166–170 CrossRef.
- S. Sun, Polym. Degrad. Stab., 2025, 238, 111341 CrossRef CAS.
- K. Oda and A. Wlodawer, Biochemistry, 2024, 63, 369–401 CAS.
- S. Negoro, D.-i. Kato, T. Ohki, K. Yasuhira, Y. Kawashima, K. Nagai, M. Takeo, N. Shibata, K. Kamiya and Y. Shigeta, in Enzymatic Plastic Degradation, 2021, vol. 648, pp. 357–389 Search PubMed.
- K. Yasuhira, Y. Tanaka, H. Shibata, Y. Kawashima, A. Ohara, D.-i. Kato, M. Takeo and S. Negoro, Appl. Environ. Microbiol., 2007, 73, 7099–7102 CrossRef CAS PubMed.
- S. Negoro, S. Kakudo, I. Urabe and H. Okada, J. Bacteriol., 1992, 174, 7948–7953 CrossRef CAS PubMed.
- S. Kakudo, S. Negoro, I. Urabe and H. Okada, Appl. Environ. Microbiol., 1993, 59, 3978–3980 CrossRef CAS PubMed.
- S. Negoro, T. Ohki, N. Shibata, K. Sasa, H. Hayashi, H. Nakano, K. Yasuhira, D.-i. Kato, M. Takeo and Y. Higuchi, J. Mol. Biol., 2007, 370, 142–156 CrossRef CAS PubMed.
- E. L. Bell, G. Rosetto, M. A. Ingraham, K. J. Ramirez, C. Lincoln, R. W. Clarke, J. E. Gado, J. L. Lilly, K. H. Kucharzyk, E. Erickson and G. T. Beckham, Nat. Commun., 2024, 15, 1217 CrossRef CAS PubMed.
- S. Negoro, N. Shibata, D. i. Kato, Y. Tanaka, K. Yasuhira, K. Nagai, S. Oshima, Y. Furuno, R. Yokoyama, K. Miyazaki, M. Takeo, K. Hengphasatporn, Y. Shigeta, Y. H. Lee and Y. Higuchi, FEBS J., 2023, 290, 3400–3421 Search PubMed.
- S. Negoro, N. Shibata, Y. Tanaka, K. Yasuhira, H. Shibata, H. Hashimoto, Y.-H. Lee, S. Oshima, R. Santa, S. Oshima, K. Mochiji, Y. Goto, T. Ikegami, K. Nagai, D.-i. Kato, M. Takeo and Y. Higuchi, J. Biol. Chem., 2012, 287, 5079–5090 CrossRef CAS.
- H. Puetz, C. Janknecht, F. Contreras, M. Vorobii, T. Kurkina and U. Schwaneberg, ACS Sustainable Chem. Eng., 2023, 11, 15513–15522 Search PubMed.
- H. Puetz, A. M. Illig, M. Vorobii, C. Janknecht, F. Contreras, F. Flemig and U. Schwaneberg, ChemSusChem, 2025, 18, e202500257 Search PubMed.
- E. R. Hoffman, A. M. M. Rangaswamy, M. E. Cleveland, J. W. Keillor and G. W. Howe, Angew. Chem., Int. Ed., 2024, 64, e202414842 Search PubMed.
- E. L. Whitelaw, M. G. Davidson and M. D. Jones, Chem. Commun., 2011, 47, 7176–7181 Search PubMed.
- J. Lu, W. Wu, W. Wang, B. Wu, J. Deng, J. Zhou, X. Zhou, W. Dong and M. Jiang, Polym. Degrad. Stab., 2025, 240, 111461 Search PubMed.
- G. Tonsi, C. Maesani, S. Alini, M. A. Ortenzia and C. Pirola, Chem. Eng. Trans., 2023, 100, 727–732 Search PubMed.
- G. Porpora, A. Gabriele, R. Pastore and F. Greco, Phys. Rev. Mater., 2023, 7, 115602 Search PubMed.
- R. M. Cywar, N. A. Rorrer, H. B. Mayes, A. K. Maurya, C. J. Tassone, G. T. Beckham and E. Y. X. Chen, J. Am. Chem. Soc., 2022, 144, 5366–5376 CrossRef CAS PubMed.
- A. Kumar, N. von Wolff, M. Rauch, Y.-Q. Zou, G. Shmul, Y. Ben-David, G. Leitus, L. Avram and D. Milstein, J. Am. Chem. Soc., 2020, 142, 14267–14275 Search PubMed.
- T. Kaweetirawatt, T. Yamaguchi, S. Hayashiyama, M. Sumimoto, A. Kamimura and K. Hori, RSC Adv., 2012, 2, 8402–8409 Search PubMed.
- R. Coeck, M. Houbrechts and D. E. De Vos, Chem. Sci., 2023, 14, 7944–7955 RSC.
- U. Češarek, D. Pahovnik and E. Žagar, ACS Sustainable Chem. Eng., 2020, 8, 16274–16282 CrossRef.
- Z. Chen, Q. Zhang, L. Ding, G. Lv, T. Liu, Z. Yang, Y. Jiang, L. Li, W. Li, F. Ding, W. Xu, J. Zhu and B. Zhu, Nat. Commun., 2025, 16, 2009 CrossRef CAS PubMed.
- S. Negoro, N. Shibata, Y.-H. Lee, I. Takehara, R. Kinugasa, K. Nagai, Y. Tanaka, D.-i. Kato, M. Takeo, Y. Goto and Y. Higuchi, Sci. Rep., 2018, 8, 9725 CrossRef PubMed.
- M. Biasini, S. Bienert, A. Waterhouse, K. Arnold, G. Studer, T. Schmidt, F. Kiefer, T. G. Cassarino, M. Bertoni, L. Bordoli and T. Schwede, Nucleic Acids Res., 2014, 42, W252–W258 CrossRef CAS PubMed.
- E. Krieger, G. Koraimann and G. Vriend, Proteins:Struct., Funct., Bioinf., 2010, 47, 393–402 CrossRef PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2009, 31, 455–461 CrossRef PubMed.
- Y. Duan, C. Wu, S. Chowdhury, M. C. Lee, G. Xiong, W. Zhang, R. Yang, P. Cieplak, R. Luo and T. Lee, J. Comput. Chem., 2003, 24, 1999–2012 CrossRef CAS.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS.
- A. Jakalian, D. B. Jack and C. I. Bayly, J. Comput. Chem., 2010, 23, 1623–1641 CrossRef PubMed.
- P. Mark and L. Nilsson, J. Phys. Chem. A, 2001, 105, 9954–9960 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |