Open Access Article
Open Access ArticleSelective removal of green pigments and associated proteins from clover-grass protein concentrates: molecular insights into a non-destructive, two-stage membrane-based biorefinery concept for high-quality food protein production
Simon Gregersen Echers *a,
Naim Abdul-Khalek
*a,
Naim Abdul-Khalek a,
Nete Hassing Jensena,
Anders Kjær Jørgensen
a,
Nete Hassing Jensena,
Anders Kjær Jørgensen a,
Tuve Mattssona,
Mads Koustrup Jørgensen
a,
Tuve Mattssona,
Mads Koustrup Jørgensen a,
Peter Stephensen Lübeck
a,
Peter Stephensen Lübeck ab and
Mette Lübeck
ab and
Mette Lübeck a
a
aDepartment of Chemistry and Bioscience, Aalborg University, Fredrik Bajers Vej 7H, 9220 Aalborg, Denmark. E-mail: sgr@bio.aau.dk
bBiomassProtein ApS, Ulvevej 26, DK-7800 Skive, Denmark
First published on 1st December 2025
Abstract
Green leaves are gaining traction as an emerging protein source and a sustainable alternative to animal-based proteins as leaf proteins often possess good nutritional and functional properties. However, the current methods for producing protein isolates from leaves involve partially or fully denaturing conditions, diminishing the protein solubility and functionality. Here, we characterize the performance of a multi-stage, membrane-based green biorefinery concept capable of producing a native protein product from clover grass devoid of attributes such as green color and grassy smell/taste. By sampling at each step along the process, we obtain insights into the fate of the proteins and pigments over each unit operation. This process efficiently removes green pigments (>99.9%), when comparing the product stream with the initial feed stream, based on UV/Vis analysis. Using mass spectrometry-based proteomics, two complementary quantification strategies, and subsequent bioinformatic analysis, it is found that this can particularly be ascribed to the very selective retention (>99%) of unwanted membrane-associated and pigment-binding proteins in the first-stage filtration. In the second-stage filtration and subsequent diafiltration stage, residual unwanted proteins, fragments, and pigments are efficiently washed out while retaining the overall protein composition. The product maintains the high RuBisCO content of green juice and is enriched in proteins with known antioxidant properties while depleted in known food allergens. This work presents an in-depth understanding of the protein-level selectivity in a membrane-based green biorefinery and can help guiding in the process optimization towards improved yields and quality.
Sustainability spotlightGreen biomasses, particularly mixtures of clovers and grasses, are considered highly beneficial in crop rotation schemes as they improve soil health. Moreover, such perennial mixtures require little or no use of fertilizers and pesticides, absorb atmospheric CO2, and allow frequent harvests, ultimately making their cultivation highly sustainable. However, converting these crops into a food grade protein ingredient has been considered mostly a future possibility until recently. But the recent development of a gentle, membrane-based processing method has made it feasible in the near future. Here, we characterize this process from the molecular level to understand why and how it converts green juice into a high-quality and sustainable food-grade protein ingredient for the future. |
1 Introduction
In light of the significant environmental impact imposed by conventional livestock and arable land farming, the increasing focus on the need for sustainable food and protein production is becoming evident on a global scale.1,2 As part of the green transition of society, plants are becoming increasingly important as sources of dietary protein.3,4 The grand challenge associated with plant protein-based ingredients in foods is their subpar nutritional value and functional properties compared with conventional animal-based protein ingredients. While soy protein addresses these needs to some extent, and currently has the largest market share among plant proteins, soy is affiliated with sustainability concerns, emphasizing the need for locally sourced, sustainable alternatives.5 Among them, leaf protein is generating traction as a future source of food protein not just scientifically but also politically–particularly in the Northern Europe.6–9 Perennial grasses and legumes such as clover and alfalfa are garnering increasing attention as emerging sources. Not only are they easily cultivated on a global scale but also possess multiple benefits such as reduced need for fertilizers and pesticides, drought resistance, improved soil quality, high biomass yields, repeated harvest, and a high protein content (up to 30% dry matter).10–16 The utilization of green leaves may also facilitate a reduction in food waste, as leafy parts of food crops, often constituting 75% or more of the biomass of broccoli and kale, are typically discarded.17In photosynthetically active green leaves of plants, such as perennial forage crops like grasses and clovers, the most abundant protein is the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), constituting 50% or more of the soluble leaf protein.18,19 RuBisCO is a hexadecameric globular protein of approximately 550 kDa consisting of eight large (rbcL) and eight small (rbcS) subunits.20 Because of its high content, not only in leaves but also in marine plants, photosynthetic bacteria, and eukaryotic algae, RuBisCO is considered the most abundant protein on Earth.20 In addition to the prevalence of this protein, RuBisCO has several advantageous properties in a food context. Its amino acid composition makes it highly suitable for human nutrition, and furthermore RuBisCO has been shown to possess very promising attributes as a protein-based food ingredient, e.g., excellent solubility as well as strong foaming and gelling properties.21–26 Therefore, obtaining protein isolates and concentrates with a high content of RuBisCO is considered desirable for producing sustainable protein ingredients for foods.
A major challenge in using leafy greens for protein production is the presence of unwanted constituents, posing not only a problem for functionality but also for sensory attributes and consumer acceptance. One group of the unwanted constituents in protein products is pigments such as chlorophylls and carotenoids. Chlorophylls are highly light sensitive and function as photosensitizers, which in turn may lead to undesired sensory attributes when incorporated in foods.27,28 Alternatively, although carotenoids are frequently considered beneficial bioactives and used as health-promoting food additives,29 they typically co-localize with other undesired constituents such as chlorophylls.30 Moreover, other small molecule phytochemicals such as quercetin, a flavonoid polyphenol, and phenolic acids, are associated with lower sensory quality such as bitterness and astringency.31–33 Avoiding these undesirable compounds in a protein product requires the development of strategies for refining plant leaves in a selective manner.
Most of the scalable green biorefinery concepts developed thus far are focused on producing feed protein.34–37 This constitutes a major opportunity for biomass valorization, as functional food protein is generally of much higher value than feed protein.38 The challenge is developing a cost-effective and scalable process.39 On a developmental and experimental scale, conventional green biorefinery concepts for producing “white” food-grade protein from green biomass include initial wet fractionation (typically by screw pressing) to obtain a green juice, followed by a mild heating step (typically around 50 °C) to produce a clarified juice and a “green protein” precipitate. Then, the clarified juice is further processed by precipitating the “white” protein via heating at higher temperatures (typically around 80 °C) or isoelectric precipitation under acidic conditions (typically around pH 4).40 Unfortunately, mild heat treatment causes protein loss and partial denaturation, while the final precipitation step may result in full protein denaturation, and hence loss of its solubility and functionality. Within the last decade, several improvements to the conventional refinery process have been introduced. This includes the addition of pH-stabilizing buffer components41 or sulphites to reduce oxidative enzymatic activity.42–44 Moreover, membrane-based approaches are beginning to emerge as milder and non-destructive biorefinery approaches compared to the conventional ones.43–45
Although membrane-based approaches also have high theoretical potential in green biorefining,46,47 they typically have challenges especially fouling, limiting protein transmission, and thereby reducing the efficiency and yields.15,48,49 In this case, pre-treatments such as mild heat treatment combined with centrifugation have been proposed, albeit at the cost of protein loss and partially impaired functional properties.12,44,48,50 Recently, a green biorefinery concept based on multi-stage cross-flow membrane filtration was described, where the combination of multiple filtration operations and the use of cross-flow membrane modules under carefully selected parameters mitigates the challenges of membrane fouling to a very large extent.51,52 This process does not include the use of any heating, centrifugation, or acid/alkaline treatments, thereby facilitating the production of a fully native and highly functional protein product.52,53 However, producing native protein from leaves is still a challenge due to the very dynamic environment generated immediately after wet fractionation. In green juice, proteins and phytochemicals that are not co-compartmentalized in vivo become mixed. This facilitates a plethora of potential reactions of both spontaneous and enzymatic nature. For instance, plant proteases gain access to new protein substrates, initiating proteolytic degradation,16,54,55 while plant phytochemicals such as polyphenols may interact with proteins. These interactions may cause modifications that hamper functionality, digestibility, and sensory attributes as well as potentially facilitating protein cross-linking, which may ultimately lead to aggregation, precipitation, and protein loss.56–58 Thus, controlling these potentially detrimental reactions is key for producing a high-quality native protein product, as proteins and enzymes are not immediately denatured and isolated, but remain functional and active throughout the process.59
Thus, far, most research has, with good reason, focused on RuBisCO as the main constituent in protein isolates from green leaves, and very little attention has been given to quantitative composition of the remaining ∼50% of the proteome in green juice,60–62 together with downstream refined protein products. In contrast, on the plant level, perennial forage crops such as grasses, clovers, and alfalfa have been extensively studied for their protein-level composition. However, this research has predominantly focused on understanding the plant physiology, metabolism, and response to external stress factors.63–70 Similarly, other leafy greens like sugar beet leaves,71 orange leaves,72 and spinach73 have been characterized for their protein composition, albeit a protein-level understanding of their processing is lacking. Although the high content of RuBisCO makes it a key factor in understanding the bulk functionality of leaf protein as a food ingredient, the remaining proteome, and how it is affected by applied processing and refinement methods will have a large impact on its bulk properties.
The aim of this work was to address this knowledge gap by in-depth characterization across unit operations within a membrane-based green biorefinery concept.52 In this process, macerated green biomass, here a clover and grass blend, is initially mixed with a stabilizing buffer and wet fractionated in a screw press. Then, the resulting green juice is refined via multi-stage cross-flow membrane filtration. Firstly, green juice is separated in the first filtration stage, wherein the retentate represents a “green protein” suitable for feed. The permeate is further separated in the second filtration stage, and subsequent diafiltration removes small molecule contaminants such as free phytochemicals as well as stabilizers and salts from the added buffer. This produces a concentrated product stream with a high protein content and void of green color and grassy smell and taste.52 As the temperature is kept relatively low (5–35 °C) throughout the process and pH is maintained at a near-neutral level, proteins are retained in their native form, thereby rendering the process non-destructive. Consequently, this ensures high solubility across a wide pH range as well as retaining the endogenous functionality of the proteins.53 In this work, mass spectrometry-based bottom-up proteomics was applied to perform an in-depth and quantitative characterization of the proteome across all stages and unit operations within the green biorefinery concept. This approach enables a deeper understanding of the proteome composition and potential, as well as insights into the protein-level selectivity throughout the membrane-based processes. Furthermore, the quantitative characterization enables coupling between the protein-level composition and bulk attributes of the protein extract/isolate. The pigment content was quantified to support the interpretation and enhance the understanding of the processes. Overall, this study provides a foundation for advancing green biorefining by identifying specific requirements and possibilities for optimization of the concept, ultimately improving the yields and quality of the protein products as food ingredients.
2 Materials and methods
2.1 Biomass production and processing
Clover grass biomass was harvested on August 15th 2022 in Denmark (location: 57°02′16.5″N 9°58′42.4″E) and corresponds to the previously described “W33” experiment.52 Briefly, the biomass originated from the commercial blend “Seed Mix 35” (DLF Seeds A/S, Denmark) consisting of 13% white clover (Trifolium repens) and 87% ryegrass (Lolium perenne) of different cultivars, but red clover (Trifolium pratense) was also observed in the field. Biomass was harvested using an Agillo PXC Solo bush trimmer (Einhell, Germany) pre-bloom. Harvest was performed in the morning, and the leaves (15 kg crude mass, 2.1 kg DM) were transported for further processing within one hour. The biomass was mixed with a stabilizing solution51,52 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio (w/w) and shredded for a minimum of five minutes using an HMI012 Blender (Hamilton Beach Commercial, USA) at 20 °C and maximum power. Thereafter, the suspension was immediately transferred to a KP6 screw press (Vincent, USA) for wet fractionation at 3.3 rpm and 4 bar counter pressure.
2 ratio (w/w) and shredded for a minimum of five minutes using an HMI012 Blender (Hamilton Beach Commercial, USA) at 20 °C and maximum power. Thereafter, the suspension was immediately transferred to a KP6 screw press (Vincent, USA) for wet fractionation at 3.3 rpm and 4 bar counter pressure.
Then, the resulting green juice was subjected to a multi-stage filtration process, comprising initial removal of fiber and particles by passing the juice through a stainless-steel strainer (1 × 1 mm square holes), followed by sequential bag filtration using BP-420-x series polypropylene bag filters (Spectrum Filtration, India) with nominal pore sizes of 50 and 1 µm. Then, the permeate was transferred to the filtration unit equipped with a ceramic HTM membrane (60 nm nominal pore size. LiqTech, Denmark) and filtered by cross-flow filtration using a 3 m s−1 cross-flow velocity and a quasi-constant trans-membrane pressure (TMP) of 0.3–0.4 bar until a volume concentration factor (VCF) of 4.7 was reached. Then, the permeate was subjected to second-stage cross-flow filtration using an ST Sanitary 10 kDa polymeric PES membrane (Synder Filtration, USA) at 2 bar TMP until a VCF of 4.6 was reached. In the same rig, the retentate was washed by continuous diafiltration (DF) using a diafactor (i.e. the volume of wash water to permeate volume) of 9.8. The volumetric cross-flow during the second-stage cross-flow filtration and subsequent DF was maintained at ≈2.9 m3 h−1. For further details on the filtration method and setup, pleas1e refer to 51,52.
To characterize the process-induced changes in the protein composition, sample aliquots were collected at all possible sampling points during the process and frozen at −18 °C until analysis. The sampling points represent the following:
• The unprocessed (lyophilized and cryo-ground) clover grass (biomass).
• The green juice immediately after wet fractionation (raw juice).
• The green juice (after bag filtering) used as feed for the first-stage membrane filtration (S1 feed).
• The retentate from the first-stage membrane filtration (S1 ret).
• The permeate from the first-stage membrane filtration (S1 perm), also representing the feed solution for the second-stage membrane filtration.
• The retentate from the second-stage membrane filtration (S2 ret).
• The retentate from the diafiltration operation, representing the final diafiltered concentrate stream (DF C), i.e. the food product stream.
• The permeate from the second-stage membrane filtration (S2 perm).
• The permeate from the diafiltration operation (DF perm).
A replicate of the final product stream (DF C) was included as validation of the protein-level consistency. This sample was extracted from a different container (but from the same batch) as the DF C sample and labelled as “DF C_2”.
2.2 Initial characterization of process streams
All streams were initially characterized to determine the dry matter, crude protein content, optical characteristics (including pigment analysis), and the protein composition analyzed by SDS-PAGE.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. After boiling, the full volume (avoiding any potentially sedimented solids) was loaded to ensure a theoretically equal protein load. 5 µL PIERCE Unstained Protein MW Marker was used for size estimation and electrophoresis was carried out at 150 V for 40–50 min, staining with Coomassie blue and imaging using a ChemDoc MP imaging System (BioRad, USA).
1 ratio. After boiling, the full volume (avoiding any potentially sedimented solids) was loaded to ensure a theoretically equal protein load. 5 µL PIERCE Unstained Protein MW Marker was used for size estimation and electrophoresis was carried out at 150 V for 40–50 min, staining with Coomassie blue and imaging using a ChemDoc MP imaging System (BioRad, USA).Additional UV/Vis analysis was performed for all samples to estimate the content of chlorophyll (a and b) and total carotenoids (xanthophylls and carotenes) by aq. acetone extraction according to74,75 with minor modifications. In the case of the dry (biomass) samples, 30 mg was directly extracted in 300 µL cold 80% aq. acetone (VWR, Denmark) by grinding in a chilled mortar with the addition of 10 mg MgO (Sigma Aldrich, Germany) to prevent pheophytin formation. The slurry was transferred to a centrifuge tube, whereafter the mortar was washed with an additional 100 µL 80% aq. acetone, which was subsequently pooled with the extract. This procedure was repeated twice to minimize losses. In the case of liquid samples, 20 µL was mixed with 180 µL cold 90% aq. acetone, vortexed and incubated on ice for 20 min for extraction. Following pigment extraction, all samples were centrifuged at 13.4k rcf for 10 min at 4 °C to clear residual plant debris. Pigment extracts were analyzed by UV/Vis absorbance measured in the range of 300–750 nm using an SDS-11 FX microvolume spectrophotometer. The conventional absorbance range restriction (0.3–0.85 at 663.2 nm, 646.8 nm, and 470.0 nm74) was not considered due to the use of microvolume analysis. All streams were analyzed in triplicate.
The content of chlorophyll a (eqn. (1)), chlorophyll b (eqn. (2)), and carotenoids (eqn (3)) was calculated according to74 as follows:
| ca = 12.25 × A663 − 2.79 × A647 | (1) |
| cb = 21.50 × A647 − 5.10 × A663 | (2) |
| c(x+c) = (1000 × A470 − 1.82 × ca − 85.02 × cb)/198 | (3) |
2.3 Proteomics analysis by bottom-up-proteomics
All streams were subjected to in-depth qualitative and quantitative analysis using mass spectrometry (MS)-based bottom-up proteomics (BUP). Prior to triplicate analysis, the protocol for protein extraction and sample preparation was optimized to the different biomass streams.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and made up to 100 µL by adding 1× Lyse buffer. After protein extraction, reduction and alkylation, as described above, the protein extract was mixed with resuspended Trypsin/LysC and digested for three hours on a thermomixer at 37 °C and 500 rpm. Subsequently, the digest was transferred to iST cartridges for desalting and cleanup, as previously described.76 The desalted digest was dried in a SpeedVac (Thermo-Fisher Scientific, USA), and subsequently resuspended in LC Load and the digest concentration was estimated using microvolume UV-spectroscopy (A280) on an SDS-11 FX (Denovix, USA) with the standard settings. The samples were diluted as required prior to proteomics analysis. All samples were prepared in triplicate and stored at −18 °C until analysis.
1 ratio and made up to 100 µL by adding 1× Lyse buffer. After protein extraction, reduction and alkylation, as described above, the protein extract was mixed with resuspended Trypsin/LysC and digested for three hours on a thermomixer at 37 °C and 500 rpm. Subsequently, the digest was transferred to iST cartridges for desalting and cleanup, as previously described.76 The desalted digest was dried in a SpeedVac (Thermo-Fisher Scientific, USA), and subsequently resuspended in LC Load and the digest concentration was estimated using microvolume UV-spectroscopy (A280) on an SDS-11 FX (Denovix, USA) with the standard settings. The samples were diluted as required prior to proteomics analysis. All samples were prepared in triplicate and stored at −18 °C until analysis.LC-MS/MS analysis was performed as shotgun bottom-up proteomics using an EASY nLC-1200 ultra-high-performance liquid chromatography system (Thermo) with ESI coupled to a Q Exactive HF tandem mass spectrometer (Thermo). 1 µg digest was loaded onto a PEPMAP trap column (75 µm × 2 cm, C18, 3 µm, 100 Å) followed by separation on a reversed-phase PEPMAP analytical column (75 µm × 50 cm, C18, 2 µm, 100 Å). Solvent A (0.1% (V/V) formic acid) and solvent B (80% (V/V) acetonitrile, and 0.1% (V/V) formic acid), were introduced through a stepwise, 60 min gradient from 5% to 100% solvent B. Data was acquired in full MS/ddMS2 Top20 data-dependent positive mode, employing an MS1 range of 300–1600 m/z. The MS1 and dd-MS2 resolution was set to 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 15
000 and 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, respectively. The automatic gain control (AGC) target and maximum injection time were set to 1 × 105 and 45 ms, respectively, while a 1.2 m/z isolation window and a collisional energy of 28 eV were employed. The dynamic exclusion window was set to 20.0 s, the peptide match was set as “preferred”, and “exclude isotopes” was enabled.
000, respectively. The automatic gain control (AGC) target and maximum injection time were set to 1 × 105 and 45 ms, respectively, while a 1.2 m/z isolation window and a collisional energy of 28 eV were employed. The dynamic exclusion window was set to 20.0 s, the peptide match was set as “preferred”, and “exclude isotopes” was enabled.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 146 entries) were used as reference protein databases for protein identification. Given that L. perenne and T. repens, the main constituents in the biomass, do not have full proteomes, the reference proteomes for the related Brachypodium distachyon (purple false brome, UP000008810, 44
146 entries) were used as reference protein databases for protein identification. Given that L. perenne and T. repens, the main constituents in the biomass, do not have full proteomes, the reference proteomes for the related Brachypodium distachyon (purple false brome, UP000008810, 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 786 entries) and Trifolium subterraneum (subterranean clover, UP000242715, 36
786 entries) and Trifolium subterraneum (subterranean clover, UP000242715, 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 725 entries) were used to increase the depth of the analysis. Fescues are often found in Danish clover grass fields,79 and thus all entries from the Festuca genus (TaxId: 4605, 1498 entries) were also included. All protein lists were downloaded from UniProt80 on December 2nd 2024.
725 entries) were used to increase the depth of the analysis. Fescues are often found in Danish clover grass fields,79 and thus all entries from the Festuca genus (TaxId: 4605, 1498 entries) were also included. All protein lists were downloaded from UniProt80 on December 2nd 2024.In MaxQuant, oxidation of methionine and protein N-terminal methylation were included as variable modifications, while carbamidomethylation of cysteine was defined as a fixed modification. Protein identification was performed using in silico tryptic digest, allowing up to two missed cleavages. Peptide size restrictions were defined using a minimal length of seven amino acids and a maximum mass of 4600 Da. On both the protein- and peptide-level, quality-based filtering was performed using a 1% false discovery rate. To boost the identification rates, matching between runs and dependent peptides was enabled. Protein quantification was done using both label-free quantification (MaxLFQ81) and intensity-based absolute quantification (iBAQ82). The mass spectrometry proteomics data have been deposited into the ProteomeXchange Consortium via the PRIDE83 partner repository with the dataset identifiers PXD069787 and 10.6019/PXD069787.
2.4 Downstream analysis of proteomic data
The MaxQuant output data was analyzed with different complementary workflows to obtain both inter- and intra-sample comparative insights. Prior to any downstream analysis, potential contaminant proteins and false positive (reverted sequences) were removed by categorical column filtering. The MaxQuant output data (proteinGroup) including additional downstream processing and appended information can be found in the SI (Table S1). Proteins identified in at least two of three replicates were considered reproducible and were the basis for a qualitative assessment of overlap between streams. Qualitative assessment was visualized in Venn diagrams using Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/index.html).![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(FC) > 1 or log
2(FC) > 1 or log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(FC) < −1). The built-in Mass Dynamics ANOVA analysis across all stream replicates was used to find globally differential proteins and data was visualized in a heatmap with protein- and replicate-level clustering using a Euclidian cluster distance of seven. Heatmap representations were done both with and without the inclusion of unutilized, low-protein side streams (S2 perm and DF perm).
2(FC) < −1). The built-in Mass Dynamics ANOVA analysis across all stream replicates was used to find globally differential proteins and data was visualized in a heatmap with protein- and replicate-level clustering using a Euclidian cluster distance of seven. Heatmap representations were done both with and without the inclusion of unutilized, low-protein side streams (S2 perm and DF perm).Using both individually identified protein groups within the abundant subset and the defined groups, a fold change (FC) analysis was performed based on riBAQ abundance. This analysis was performed to investigate the protein-level and group-level enrichments across the two filtration stages. In the first filtration stage, the FC was computed by pair-wise comparing S1 feed, S1 ret, and S1 perm. In the second filtration and DF stages, the FC was calculated by pair-wise comparing S1 perm, S2 ret, and DF C. Permeate streams from S2/DF were not considered due to their low protein content, and hence unreliable quantification. The FC between two streams (A and B) was determined as the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 transform of the ratio between riBAQs (B/A). Negative values represent enrichment in stream A, while positive values represent enrichment in stream B. Based on the log
2 transform of the ratio between riBAQs (B/A). Negative values represent enrichment in stream A, while positive values represent enrichment in stream B. Based on the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 transformation, FC < −1 and FC > 1 represent a relative (i.e. part of the whole) two-fold enrichment in the respective stream and a 50% reduction in the other. For proteins not detected in a particular stream, log
2 transformation, FC < −1 and FC > 1 represent a relative (i.e. part of the whole) two-fold enrichment in the respective stream and a 50% reduction in the other. For proteins not detected in a particular stream, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC was set to ±10 based on the maximum values observed in the analysis. If a protein was not found or had a riBAQ < 0.01% in both streams, log
2FC was set to ±10 based on the maximum values observed in the analysis. If a protein was not found or had a riBAQ < 0.01% in both streams, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC was set to zero.
2FC was set to zero.
The dataset initially contained 3580 protein identifications. A preliminary filtering step removed 70 entries that included non-standard amino acids, resulting in a final dataset of 3510 protein groups. In the groups containing multiple protein IDs, only the first ID was retained to represent that unique identification. Protein sequences corresponding to these IDs were retrieved from the respective FASTA files. In each stream, riBAQ values across three replicates were averaged to compute the mean riBAQ, which was subsequently used to determine protein presence per stream.
2.4.4.1 Analysis of physicochemical properties. A range of sequence-derived features was computed using the ProteinAnalysis tool from Biopython (v1.78). These included molecular weight, aromaticity,95 instability index,96 GRAVY score,97 and isoelectric point. Additionally, the predicted fractions of amino acids forming secondary structure elements (helices, turns, and sheets) were calculated. For clarity, all these descriptors, whether purely chemical or structural predictions, are collectively referred to as physicochemical properties, or simply “properties” in this work.
2.4.4.2 Unweighted analysis. Each protein was treated as an independent data point and the distribution of physicochemical properties across streams was initially explored using boxplots. To evaluate if there were significant differences between streams properties, one-way ANOVA was performed using SciPy (v1.10.1). Subsequently, dimensionality reduction was applied using Truncated Singular Value Decomposition (TruncatedSVD) from scikit-learn (v1.3.0), allowing the data to be represented in a two-dimensional space for visualization. Then, clustering analysis was performed on the normalized properties. The optimal number of clusters was estimated using the elbow method and the Calinski–Harabasz index. Clusters were defined using the k-means algorithm. The 2D representation of the data was then visualized to highlight groupings based on clusters, stream assignments, and predicted subcellular localizations.
2.4.4.3 Abundance-weighted analysis. In this analysis, the proteins identified in each stream and replicate were considered as independent observations. To incorporate protein abundance, the physicochemical properties of each protein were multiplied by its corresponding riBAQ value. Then, the resulting weighted values were summed across all proteins within each stream and replicate, yielding 30 aggregated data points (10 streams × 3 replicates), each represented by a vector of weighted physicochemical properties. As in the unweighted analysis, the distribution of these weighted properties across streams was visualized using boxplots and statistically evaluated using one-way ANOVA. Dimensionality reduction was applied to the normalized weighted data, followed by clustering analysis. The results were projected into a two-dimensional space for visualization. Additionally, the distribution of physicochemical properties within each cluster was examined using boxplots to reveal cluster-specific trends and potential biological/process-induced relevance.
2.4.4.4 Analysis of subcellular localization and transmembrane regions. Protein subcellular localization was predicted for each experimental stream, and for all 3510 proteins using DeepLoc v1.0.85 A protein was considered present in a specific stream if it was identified in at least one of the three replicates. Based on this, the number of proteins associated with each subcellular compartment was quantified per stream. This yielded a matrix representing 10 streams, each characterized by the distribution of proteins across the ten compartments. To explore underlying patterns, dimensionality reduction was applied to visualize the data in two dimensions. Subsequently, clustering was performed on the complete dataset, and the resulting groups were visualized within the same 2D space to facilitate interpretation of stream-specific localization trends.
Probable membrane-associated proteins were predicted across all identified lead proteins (3510 after filtering) using TOPCONS2,86 as described above. The abundance of protein-based subcellular localization and membrane association was subsequently computed using the riBAQ values within each stream.
2.5 Statistical analysis
The data from the dry matter, crude protein, pigment analysis and number of protein identifications was analyzed to identify statistical differences using GraphPad Prism (v.10.0.2, build 232). The comparison of means from the different streams was performed as ordinary one-way ANOVA and Tukey's multiple comparison testing using a 95% confidence interval and assuming a Gaussian distribution of residuals and equal standard deviations between groups of unmatched replicates. All p-values are reported as adjusted p-values to account for multiple comparisons.3 Results and discussion
During the entire process, a variety of streams were sampled for investigation. Initially, dry matter (DM) and crude protein (CP) content was determined (Table 1) as the basis for additional analysis and to investigate the fate of the bulk protein and dry matter over the process.| Stream | Description | Dry matter (%) | Crude protein (%DM) |
|---|---|---|---|
| a Dry matter and crude protein were not recorded for the permeate streams from the second filtration and diafiltration stages. | |||
| Biomass | Unprocessed biomass from harvest | 14.20 ± 2.15 a | 24.97 ± 0.74 c |
| Raw juice | Green juice after wet fractionation | 3.42 ± 0.02 c,d | 20.62 ± 0.36 d |
| S1 feed | Green juice after bag filtering (feed for first filtration stage) | 3.36 ± 0.02 c,d | 20.46 ± 0.05 d |
| S1 ret | Retentate from first filtration stage (animal feed protein stream) | 5.48 ± 0.01 b | 38.73 ± 0.67 b |
| S1 perm | Permeate from first filtration stage (feed for second filtration stage) | 2.37 ± 0.35 d,e | 10.33 ± 0.20 e |
| S2 ret | Retentate from second filtration stage (“feed” for diafiltration) | 2.14 ± 0.00 e | 21.05 ± 0.87 d |
| S2 perm a | Permeate from second filtration stage (side stream) | N/A | N/A |
| DF perm a | Permeate from diafiltration (additional side stream) | N/A | N/A |
| DF C | Concentrate from diafiltration (final food protein stream) | 0.48 ± 0.00 f | 57.70 ± 0.86 a |
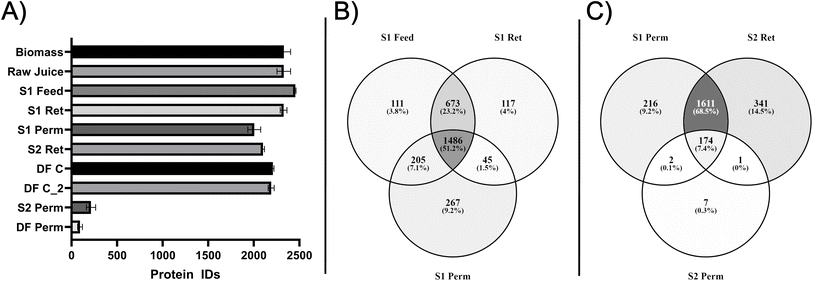
A substantial and highly significant (p < 0.0001) loss of DM is observed in all streams compared to the unprocessed biomass (Table 1 and Fig. S1A). Moreover, a significant (p < 0.0032) accumulation of DM in the S1 retentate is observed compared to both the upstream (raw juice and S1 feed) and downstream sampling points. This can be attributed to the retention of particulate matter and aggregates permeating the initial police filtering stage between raw juice and S1 feed, where no significant (p > 0.99) DM loss was observed. Although notable DM loss was observed between S1 feed and S1 perm due to the retention of larger components in S1 ret, the difference was not significant (p = 0.11), which is likely due to the low number of sample replicates (n = 2). Nevertheless, compared to S1 feed, an additional and significant decrease in DM content was observed moving further downstream to S2 ret (p = 0.047), and finally the DF C stream (p = 0.004). These findings are in good agreement with the general trends observed across a full season of membrane-based biorefining.52 This process produces a native and fully soluble product with CP ranging from 57% to 73% (DM basis). Although the experiment presented here represents a lower purity concentrate compared to other runs within the same season, the protein composition (Fig. 1A) is comparable to concentrates obtained under similar conditions based on previous SDS-PAGE analysis of the final DF C stream.52
 | ||
Fig. 1 Crude protein and pigment/color characterization. (A) Reducing SDS-PAGE analysis of the different streams represented by unprocessed biomass,2 raw juice,3 S1 feed,4 S1 ret,5 S1 perm,6 S2 ret,7 S2 perm,8 DF perm,9 DF C,10 and DF C_2.11 MW marker is loaded in wells 1 and 12 and protein MW (kDa) annotated. (B) Quantification of pigment content in the different streams by UV/Vis analysis following acetone extraction. For each stream, the sum of pigments (chlorophyll a + b and carotenoids (xanthophylls and carotenes)) relative to the stream dry matter content is shown using a log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 scale to accommodate the large dynamic range of values. Quantification of the individual pigments is presented in the SI Materials (Fig. S2). (C) Absorbance spectra of liquid streams in the range of 220–450 nm. (D) Absorbance spectra of liquid streams in the range of 450–750 nm. Absorbance spectra (C and D) are presented in two separate plots due to high UV absorbance in the streams. 10 scale to accommodate the large dynamic range of values. Quantification of the individual pigments is presented in the SI Materials (Fig. S2). (C) Absorbance spectra of liquid streams in the range of 220–450 nm. (D) Absorbance spectra of liquid streams in the range of 450–750 nm. Absorbance spectra (C and D) are presented in two separate plots due to high UV absorbance in the streams. | ||
Although the DF C stream has a significantly higher CP content (and thereby higher protein purity) compared to all the other streams (Table 1 and Fig. S1B), it also contains lower DM compared to all other upstream sampling points, which can be attributed mainly to two aspects. Firstly, the biorefinery concept uses a stabilizing buffer (1.7% DM) to ensure a more consistent process over time. The stabilizing components, such as buffer salts, are washed out during the second filtration and subsequent diafiltration (DF) stages, thereby directly reducing DM.52 Secondly, a substantial loss of protein can be observed over each of the unit operations starting already from the wet fractionation, where approximately 60% of the protein (by CP) was retained in the fiber fraction (pulp).52 Moreover, a substantial loss of nitrogen (approximately 25%) was observed in the downstream permeates (S2 perm and DF perm). This is ascribed to the loss of very small proteins, peptides and partially degraded protein, and particularly non-protein nitrogen of both organic and inorganic origin. Ultimately, around 7% of the initial protein (by CP) ended up in the final product stream. This yield was in fact the lowest of the production season, where the CP yields in DF C range from approximately 7% to 15%,52 but despite its low yield and purity, this specific batch was still chosen for in-depth characterization based on the representative parameters used during the membrane-based biorefining process and the availability of subsamples from all process streams. Although other methods for the estimation of protein content in the streams could have been explored, previous studies have shown that they are incompatible with the workflow.60 For instance, optical methods such as BCA, A280, and the fluorescence-based Qubit assay are impaired by the high level of interference from pigments and other UV-active species present in many of the analyzed streams, leading to inaccurate protein quantification.87,98,99 Their successful application would require initial and selective protein isolation to remove interferences, which may introduce additional bias and/or result in partial protein denaturation and precipitation. As such, only N-based protein estimation was used in this study. While using the Jones N-to-protein conversion factor of 6.25,100 has been deemed inaccurate in plant samples including grass species,101–103 stream-specific factors have not yet been determined. Thus, the Jones factor was employed for methodological consistency.
3.1 Efficient pigment removal and apparent changes to protein composition
Although the overall protein profile appeared largely conserved along the process, based on SDS-PAGE analysis (Fig. 1A), distinct changes were observed. The profile of the two upstream samples (raw juice and S1 feed) as well as S1 ret resembled that of the unprocessed biomass (lanes 2–5). Downstream of the first filtration stage in S1 perm (lane 6), high MW proteins (>∼100 kDa) appeared largely depleted. Several bands became visible again in the downstream samples, potentially due to the differences in salt concentrations. In contrast, apparent high-MW aggregates, observed at the top of the lanes in the upstream samples, appeared selectively depleted. This observation underlines the size-based selectivity of the first stage filtration. Interestingly, the distinct band at ∼25 kDa seen in the upstream samples appears to be retained in the first stage filtration, as this is absent in all the samples downstream (lanes 5–11). This indicates additional selectivity in the first filtration operation that cannot be simplified to monomeric protein MW. This observation is in agreement with an earlier study, where removal of this particular band was also observed.52 The removal of a 25 kDa protein using a 60 nm membrane in the first stage filtration indicates that this particular protein is likely found within larger supramolecular complexes or in aggregated form in the green juice. Overall, the protein profile in the streams resembles that obtained in previous studies on green biomass from not only grass but also other leafy greens.53,104–106 The different appearances of the two DF C samples can be attributed to their differences in protein load.During SDS-PAGE analysis, a green hue was also observed below the lanes containing the samples upstream from the first filtration stage and in S1 ret. Although this observation is not reflected post-imaging of the Coomassie-stained gels, it agrees with the distinct color change in the liquid streams, which is likely ascribed to chlorophyll depletion. To further investigate this phenomenon, we performed UV/Vis analysis of the streams to both characterize their optical properties and specifically quantify the pigments. In the three quantified pigments chlorophyll a (chl a), chlorophyll b (chl b), and total carotenoids (cart) (i.e. xanthophylls and carotenes), similar trends were observed (Fig. S2A–C), respectively, and also reflected in their sum (Fig. 1B). The unprocessed biomass was found to have a relatively high content of chl a (140 µg g−1 DM), chl b (230 µg g−1 DM), and cart (68 µg g−1 DM) for a total pigment content of 435 µg g−1 DM. This also corresponds to a total chlorophyll content of 370 µg g−1 DM or 2.6 mg g−1 fresh biomass (FM). This value is in agreement with previous studies, with total chlorophyll typically reported to be 2–4 mg g−1 FM107,108 in cultivated ryegrass. Further comparisons with the literature are complicated by the differences in analytical methods, as many apply SPAD-meters and report in SPAD units.104,109 The green juice (raw juice) has a significantly lower (p < 0.0001) content of all individual pigments and their sum, albeit still at a total level (7.7 µg g−1 DM) sufficiently high to produce an intense green color in the juice.
The incomplete pigment extraction during wet fractionation by screw pressing agrees with earlier studies, where pigments could be extracted continuously through multiple passes of alfalfa biomass in a screw press.104 The police filter applied prior to first stage filtration had no significant (p > 0.9999) effect on the pigment levels. Over the first filtration stage, a 10-fold enrichment (p < 0.0001) of pigment was observed in S1 ret (77 µg g−1 DM) while a 96% depletion (p < 0.0001) was found in S1 perm (0.27 µg g−1 DM). This shows highly efficient pigment retention in the first filtration stage. The pigment levels were further (27%) reduced during the second filtration stage (S2 ret: 0.20 µg g−1 DM) albeit with no statistical significance. However, pigments were continuously removed during DF (DF C: 0.009 µg g−1 DM), representing a significant (p = 0.046) pigment depletion of 55% from the S1 perm stream. Overall, this illustrates the highly efficient removal of 99.9% of chlorophylls and carotenoids from the green juice (S1 feed) to the final product stream (DF C). These trends were also observed when the pigments were quantified relative to CP (Fig. S2D–F), respectively.
The efficient pigment removal in the first filtration stage is also directly reflected in the UV/Vis spectra of the streams (Fig. 1C and D). It is noteworthy that a distinct peak at ∼670 nm is evident in the upstream samples and in S1 ret (Fig. 1D). This corresponds well with the known absorbance of chl a in this range.109 A distinct peak for chl b and cart was not evident due to the generally high sample absorbance, which increased towards lower wavelengths. The chl a peak is absent downstream of the first filtration stage, further highlighting its efficient removal in this unit operation. In addition to phytopigments, the UV absorbance (Fig. 1C) is also very high in the upstream samples, indicating a high content of UV-active and aromatic compounds. Interestingly, the overall UV absorbance also generally appears to decrease during DF, despite an increasing proportion of CP in the downstream samples. More specifically, the compounds responsible for the absorbance around 230–255 nm and 310–350 nm are washed out initially in S2 perm, and subsequently in DF perm. The absorbance at 250 nm has previously been ascribed to flavonoids like quercetin and rutin, while the absorbance at 325 nm has been ascribed to phenolic acid and chlorogenic acid.110 In contrast, the emergence of a peak around 280 nm, indicative of protein UV absorbance, can be seen in the final product streams (DF C and DF C_2). This coincides with the substantial concentration of CP during the second filtration and DF stages from 10% in S1 perm, over 21% in S2 ret to 58% in DF C. This further substantiates protein enrichment in the product stream, in addition to removal of other and undesired UV-active species such as quercetin and phenolic acids, which are associated with lower sensory quality such as bitterness and astringency.31–33 However, this warrants further investigation.
3.2 Optimization of sample preparation for bottom-up proteomics
Prior to in-depth analysis of the different process streams, the protein extraction protocol was optimized by combining focused ultrasonication (AFA) with the PreOmics iST plant tissue kit. To evaluate the effect of ultrasonication, a varying number of AFA extraction cycles (0, 1, 2, and 4) was applied, combined with the thermochemical extraction from the kit. For this purpose, four different upstream samples (biomass, raw juice, S1 feed, and S1 ret) were selected, while downstream samples were considered fully soluble. According to the SDS-PAGE analysis (Fig. S3A and B), there appeared to be no substantial impact of applying AFA ultrasonication as part of the protein extraction workflow, regardless of the stream and the amount of solids and residual plant material. In fact, by considering the number of identified peptides as a function of AFA cycles (Fig. S3C), a general decreasing trend was observed with an increase in the number of cycles, regardless of considering peptide IDs by MS/MS only or by including matching between runs. Although the origin of this effect was not investigated further, it may reflect partial protein damage (e.g. by degradation or denaturation-induced aggregation and precipitation), as previously reported.111–114 These data suggested that the use of AFA did not appear to have a positive effect on this biomass or resulting streams, underlining the importance of optimizing the workflow every time a new biomass is investigated.76 Although earlier studies on green biomass have indeed included AFA as part of the protein extraction workflow,60 AFA was not included in the protein extraction for the full characterization of process streams in this work.3.3 Process-induced qualitative changes in protein composition
Across all samples, a total of 3580 protein groups, representing 8766 potential proteins, was identified after filtering contaminants and false positives. The number of protein IDs is substantially (∼3 fold) higher than in a recent study on biomass and green juice from lab-cultivated L. perenne.60 This increase can be attributed to the biomass in this work originating from a field cultivated with a mixture of perennial grass and clovers. With the exception of the two “side streams” (S2 perm and DF perm), >2000 protein groups were identified in all streams (Fig. 2A). The lower number of protein IDs in the S2 and DF permeates was expected, as they represent what is transmitted through the second-stage filtration using a 10 kDa membrane. Moreover, a lower number of protein/peptide IDs in the DF permeate is expected as this represents continued washing after second stage filtration. The fact that proteins were identified in these streams can be ascribed to primarily partially degraded protein (i.e. peptides), as no visible protein was observed by SDS-PAGE (Fig. 1A). Interestingly, a distinct drop and significant (p < 0.0001) decrease in the number of protein IDs across the first-stage filtration were found. This observation is consistent regardless of considering individual sample replicates (Fig. 2A) or the number of reproducibly quantified (i.e. found in at least two of three replicates) proteins (Fig. S4A). In all samples upstream of the first stage filtration, >2200 proteins were identified (up to 2575 reproducible IDs in the S1 feed), while the number of protein IDs drops to ∼2000 in S1 perm. This indicated selectivity in the membrane operation. Interestingly, a downstream increase in protein IDs was observed, which continued to increase further downstream in the purification and concentration process. Compared to S1 perm (2004 reproducible IDs), the increase in protein IDs went from being insignificant (p = 0.337) in S2 ret (2127) to being significant (p < 0.005) in the two replicates of DF C (2232 and 2202). This increase could be ascribed to the increasing protein purity, thereby minimizing interference and noise.The selectivity across the first stage filtration was also reflected in the qualitative data when inspecting the overlap among the reproducibly (i.e. in at least two of three replicates) identified proteins for S1 feed, S1 ret, and S1 perm (Fig. 2B). From the ∼2900 protein IDs found in any of these streams, only 51% (1486) were found in all three streams. 27% (790) of the protein IDs were found in S1 feed and S1 ret or only exclusive in S1 ret, indicating that a substantial number of proteins did not appear to cross the first membrane. Furthermore, 16% (472) were found either in both S1 feed and S1 perm or exclusively in S1 perm, further substantiating selective transmittance across the first membrane. In comparison, the second stage filtration (Fig. 1C) displayed almost full protein retention by the second stage membrane based on the number of protein IDs. From the 2352 protein IDs identified across the three streams, less than 0.5%9 proteins were found in the intersect between S1 perm and S2 perm or exclusively in S2 perm. The identification of exclusive proteins in this intersect or in S2 perm can be ascribed to the instrument sensitivity, as all these proteins were of low abundance. In a higher complexity mixture of proteins (i.e. feed side of the second stage membrane), low abundance proteins (i.e. their tryptic peptides) may not be detected due to factors such as ionization competition, co-elution, and ion suppression,115,116 but this does not mean that they are not present on the feed side and suddenly appear on the permeate side. 174 proteins (7%) were found in all three streams, indicating partial transmission or potentially partial degradation and subsequent transmission of peptide products. The remaining proteins were identified in either S1 perm or S2 ret or both representing the majority of protein IDs (69%).
3.4 Distinct and quantitative process-induced enrichment and depletion on the protein level
Using label-free quantification (MaxLFQ), the quantitative proteome changes across the different stages of the membrane-based process were investigated. Based on the normalized MaxLFQ data, a clear differentiation in the quantitative protein composition was evident, when considering differentially abundant proteins by ANOVA (Fig. 3A). This was further reflected in the replicate-level Pearson correlation (Fig. S4B) and principal component analysis (Fig. S4C). The upstream samples (biomass, raw juice, S1 feed, and S1 ret) all display quite comparable qualitative and quantitative compositions, also resulting in a clear sample clustering in PCA (Fig. S4C). Similarly, the downstream samples (S1 perm, S2 ret, DF C, and DF C_2) are highly comparable and cluster, while the residual side streams from the second filtration and DF stages (S2 perm and DF perm) represent a third and distinct cluster. The similarity within the clusters is also noticeable when performing hierarchical clustering on the replicate level. When including the two residual side streams (Fig. S4D), clustering of the S2 perm and DF perm replicates overlap. When omitting these streams (Fig. 3A), DF C and DF C_2 co-cluster. As these are technical replicates but represent the same product batch, this also underlines batch consistency. Similarly, raw juice and S1 feed co-cluster, indicating that the pre-filtering step had very little impact on the quantitative protein composition. Nevertheless, two very distinct clusters are observed at the protein level., i.e. one group of proteins enriched in the upstream samples and one group enriched in the downstream samples (Fig. 3A). This further substantiates the finding by analysis of protein IDs alone that particularly the first membrane filtration represents a highly selective process. This is also reflected in the optical properties of the samples (Fig. 1) and is in agreement with the observations during processing across the whole season.52To obtain further insight into the selectivity in this step, pair-wise analysis of S1 feed and S1 perm as well as S1 ret and S1 perm was performed. In the comparison of S1 feed and S1 perm (Fig. 3B), 190 proteins were found significantly (adjusted p-value < 0.05) and substantially (fold change ≥ 2) enriched in the permeate, while 454 proteins were depleted. Comparing S1 ret and S1 perm (Fig. 3C), 353 proteins were enriched in the permeate, while 484 proteins were depleted. This corroborates that the biggest differences originating from the protein-level selectivity in the first stage filtration come from the comparative analysis of the two products more than by comparison with the feed. A substantial overlap between the enriched or depleted proteins in the S1 feed vs. S1 perm comparison was also found enriched or depleted when comparing S1 ret and S1 perm (Fig. S5), further illustrating the overall comparability of S1 feed and S1 ret, respectively. Ultimately, lists of the 837 differential proteins from the S1 ret and S1 perm pair-wise comparison were exported for further analysis (Table S3).
Subcellular localization prediction with DeepLoc117 was used to investigate if the enriched proteins in the respective streams were biased toward certain subcellular compartments (Fig. 3E). In S1 perm, a larger proportion of extracellular (6.2-fold) and peroxisome (5.5-fold) proteins was enriched. However, the low number of enriched peroxisome proteins (10 of 834) makes this compartment associated with larger uncertainty in terms of relative difference in enrichment. In contrast, S1 ret had a higher proportion of proteins localized in the cell membrane (3.5-fold), endoplasmic reticulum (3.3-fold), and mitochondria (2.6-fold). Plastid and cytoplasmic proteins showed less than 1.05-fold difference, while modest enrichment (1.2–1.7-fold) in S1 ret was seen for the remaining compartments. These observations indicate that particularly extracellular, and thereby presumably soluble proteins, are more easily transmitted through the first stage filtration membrane, while cell membrane proteins are retained, as they are potentially still bound to the cell membrane fragments or prone to aggregation. To test this hypothesis further, we predicted the transmembrane regions for all proteins, as some proteins localized to highly represented compartments such as cytoplasm and plastid may still be membrane-associated. Based on the prediction of all membrane-associated proteins, it was evident that the proteins with predicted transmembrane regions were 4.2-fold more enriched in S1 ret (21.3%) compared to those enriched in S1 perm (5.1%), substantiating a high degree of selective retention of membrane-associated proteins in the first stage filtration (Fig. 3F).
The most highly enriched in S1 perm, and therefore also likely enriched in the final product, were nodulin-related protein 1 (A0A2K3JMR2, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 6.6), non-specific lipid-transfer protein (A0A2K3NE56, log
2FC = 6.6), non-specific lipid-transfer protein (A0A2K3NE56, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 5.5), and reticuline oxidase-like protein (A0A2K3N926, log
2FC = 5.5), and reticuline oxidase-like protein (A0A2K3N926, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 5.4); all from T. pratense. Nodulins are proteins upregulated in legumes because of the symbiotic nitrogen fixation process with Rhizobium bacteria118 but there is no further knowledge on their functionality in a food context or safety issues related to the allergenicity of nodulins. In contrast, non-specific lipid-transfer proteins (nsLTPs) are well-known food allergens with sensitization capabilities. Reticuline oxidase belongs to the class of berberine bridge enzymes (BBE). BBE-like enzymes have also previously been reported to cause allergenic responses in people with pollen allergies.119 The presence of these proteins in a food product may entail certain precautions in terms of proper labelling.
2FC = 5.4); all from T. pratense. Nodulins are proteins upregulated in legumes because of the symbiotic nitrogen fixation process with Rhizobium bacteria118 but there is no further knowledge on their functionality in a food context or safety issues related to the allergenicity of nodulins. In contrast, non-specific lipid-transfer proteins (nsLTPs) are well-known food allergens with sensitization capabilities. Reticuline oxidase belongs to the class of berberine bridge enzymes (BBE). BBE-like enzymes have also previously been reported to cause allergenic responses in people with pollen allergies.119 The presence of these proteins in a food product may entail certain precautions in terms of proper labelling.
In S1 ret, a very different subset of proteins was found highly enriched. The most enriched proteins were chlorophyll a–b binding protein from T. subterraneum (A0A2Z6NV37, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 10.4), photosystem II protein D1 from L. perenne (Q95GL2, log
2FC = 10.4), photosystem II protein D1 from L. perenne (Q95GL2, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 9.1), photosystem II CP47 reaction center protein from T. repens (A0A023HPN2, log
2FC = 9.1), photosystem II CP47 reaction center protein from T. repens (A0A023HPN2, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 9.1), and photosystem II D2 protein from T. repens (A0A023HQ62, log
2FC = 9.1), and photosystem II D2 protein from T. repens (A0A023HQ62, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 8.9). As indicated by the name, chlorophyll a–b binding protein (CBP) directly binds chl a/b as part of plant photosynthesis.120 Similarly, photosystem II proteins, in particular D1/D2 and reaction center proteins, are known for their involvement in binding of pigments such as chl and cart.121–123 As such, this substantial and significant retention in the S1 ret can explain the visible color change and the significant drop in pigment levels (Fig. 1B) obtained in the first stage filtration. In fact, none of the proteins were reproducibly identified in S1 perm, which is why FC is based on the imputed values, further substantiating the very high level of selective retention in the first filtration stage.
2FC = 8.9). As indicated by the name, chlorophyll a–b binding protein (CBP) directly binds chl a/b as part of plant photosynthesis.120 Similarly, photosystem II proteins, in particular D1/D2 and reaction center proteins, are known for their involvement in binding of pigments such as chl and cart.121–123 As such, this substantial and significant retention in the S1 ret can explain the visible color change and the significant drop in pigment levels (Fig. 1B) obtained in the first stage filtration. In fact, none of the proteins were reproducibly identified in S1 perm, which is why FC is based on the imputed values, further substantiating the very high level of selective retention in the first filtration stage.
Performing a similar pairwise comparison across the second filtration and subsequent diafiltration stages (S1 perm vs. DF C), a substantially lower number of differential proteins was found (Fig. 3D). Differential analysis revealed that 84 proteins were significantly enriched in DF C, while 70 proteins were depleted. This indicates a considerably lower effect on protein composition by the second filtration and DF stages compared to the first stage filtration. Surprisingly, T. pratense nsLTP (A0A2K3NE56), which was enriched over the first filtration stage, is significantly depleted in DF C (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 6.1), and in fact, not identified at all. Similarly, T. subterraneum nsLPT (A0A2Z6P2S8) is significantly depleted (log
2FC = 6.1), and in fact, not identified at all. Similarly, T. subterraneum nsLPT (A0A2Z6P2S8) is significantly depleted (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 4.4), indicating the lower allergenic risk of the final product compared to the intermediate product represented by S1 perm. In contrast, T. pratense glutathione S-transferase (GST), a known antioxidant protein,60 and several lactoylglutathione lyase isoforms (A0A2K3JKA7 and A0A2K3L521), known to participate in biosynthesis of the antioxidant glutathione,124 were enriched in the product stream, potentially improving not only the oxidative stability of the product125,126 but also its health-promoting effects.127
2FC = 4.4), indicating the lower allergenic risk of the final product compared to the intermediate product represented by S1 perm. In contrast, T. pratense glutathione S-transferase (GST), a known antioxidant protein,60 and several lactoylglutathione lyase isoforms (A0A2K3JKA7 and A0A2K3L521), known to participate in biosynthesis of the antioxidant glutathione,124 were enriched in the product stream, potentially improving not only the oxidative stability of the product125,126 but also its health-promoting effects.127
Comparing the retentates and permeates from the second filtration stage (Fig. S6A) and DF (Fig. S6B), almost no enrichment in the permeate streams (2 and 4 proteins, respectively) was found. In this context, it is worth considering the underlying data normalization algorithms and missing value imputation included in the LFQ analysis. This can make the direct comparison of two very different samples (e.g. the protein-rich retentates/concentrates and the dilute and protein-depleted permeates compared here) biased and may distort the actual differences between samples. In contrast, a large number of proteins was enriched in the retentates (1116 and 1148 proteins, in S2 ret and DF C, respectively). Among the noteworthy proteins, the RuBisCO small chain from T. repens (P17673, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 8.4 and 9.7), RuBisCO large chain from T. subterraneum (A0A2Z6P9S4, log
2FC = 8.4 and 9.7), RuBisCO large chain from T. subterraneum (A0A2Z6P9S4, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 9.2 and 9.3), superoxide dismutase from T. Pratense (A0A2K3NXG8, log
2FC = 9.2 and 9.3), superoxide dismutase from T. Pratense (A0A2K3NXG8, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 3.5 and 9.5), thioredoxin M (A0A2K3P1S1, log
2FC = 3.5 and 9.5), thioredoxin M (A0A2K3P1S1, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 7.4 and 8.2), and thioredoxin H (A0A2K3NRX5, log
2FC = 7.4 and 8.2), and thioredoxin H (A0A2K3NRX5, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC = 7.1 and 6.5) were highly enriched in the product streams. The enrichment of RuBisCO in the product stream is highly desirable, as RuBisCO is well acknowledged for its good nutritional and functional properties as a food protein ingredient.21–25,53 Moreover, thioredoxins and superoxide dismutase (SOD) are known antioxidant proteins and have previously been shown to be enriched in the green juice during the wet fractionation of L. perenne, and associated with in vitro antioxidant capacity of the protein extract.60
2FC = 7.1 and 6.5) were highly enriched in the product streams. The enrichment of RuBisCO in the product stream is highly desirable, as RuBisCO is well acknowledged for its good nutritional and functional properties as a food protein ingredient.21–25,53 Moreover, thioredoxins and superoxide dismutase (SOD) are known antioxidant proteins and have previously been shown to be enriched in the green juice during the wet fractionation of L. perenne, and associated with in vitro antioxidant capacity of the protein extract.60
Overall, these observations indicate that the product stream is favorable as a protein ingredient for food. Pigment-binding proteins are retained in the first filtration stage, improving the sensory attributes of the bulk protein. Moreover, proteins with desired properties appear enriched in the product stream, while minimal proteins are washed out during the second filtration stage and DF. However, as the MaxLFQ analysis uses normalized data and compares merely levels between samples, it is not evident if a differential protein is of a substantial abundance to be relevant on the bulk scale. To further understand the overall changes induced by the processing, we next performed relative in-sample quantification using relative intensity-based absolute quantification (riBAQ) to estimate the molar distribution of proteins within each stream.
3.5 Process-induced effects for the most abundant proteins in the streams
To identify the most abundant proteins in the respective streams and obtain deeper insight into the unit operation selectivity, we filtered the 3580 protein groups to produce a subset only containing the proteins reproducibly identified (i.e. at least in two of three replicates) and with a mean relative molar abundance of at least 0.5% (by riBAQ) in any stream, excluding the unutilized side streams S2 perm and DF perm. Among the 3347 reproducibly identified protein groups in any stream, 63 protein groups were found above the 0.5% riBAQ threshold (Table S4). Not surprisingly, RuBisCO (rbcL and rbcS) isoforms from different species were the utmost abundant proteins by relative molar quantification using riBAQ (Fig. 4A). The high RuBisCO (rbc) levels were also reflected when summarizing all identified rbc isoforms (including those below the 0.5% threshold), where RuBisCO was found to constitute around 20% of the biomass protein (Fig. 4B). High rbc levels were found in all samples upstream of DF, indicating no rbc selectivity in the first filtration stage but a high retention in the second filtration and DF stage. The high riBAQ of rbc, and particularly one rbcL isoform, in DF perm reflect the identification of a partially degraded protein, as these streams had very low CP and no rbc bands were detected by SDS-PAGE (Fig. 1A). Thus, the contents in both S2 perm and DF perm were not considered representative of the protein content and omitted in further analysis. An rbc content of ∼20% of the total protein in the unprocessed biomass, increasing to ∼26% in Raw Juice, is somewhat lower than expected, as rbc is reported to constitute 50% or more of the soluble leaf protein.18,19 In a previous study, rbc was found to constitute 35–40% of the protein in fresh green juice from L. perenne.60 The slightly lower rbc content may be reflected by the somewhat lower protein yield on the analyzed batch (compared to the rest of the season) or that this particular batch had an extended (up to double the normal value due to technical issues on-site) storage time (i.e. period of time from harvest to mixing with stabilizing buffer), which may facilitate the partial proteolytic degradation of rbc in the green juice. The preliminary data from other batches obtained within the same season indicate that the rbc content in the green juice was typically 30–50% (manuscript in preparation). Besides the high rbc levels, some general tendencies were observed among the abundant proteins across the different unit operations (Fig. 4A). Although some proteins, such as peptidyl-prolyl cis–trans isomerase (PPIase), thioredoxin-M (TRX-M), and superoxide dismutase (SOD), appear enriched on the permeate side, others such as ATP synthase subunit β (ATPs-β), photosystem II protein D1 (PII-D1), and several chlorophyll a–b binding proteins (CBPs), become depleted. The selective depletion of particularly CBPs also correlate well with the disappearance of the ∼25 kDa band in the SDS-PAGE analysis (Fig. 1A). Conversely, the opposite is observed in S1 ret, where the same proteins are depleted and enriched, respectively.To delve further into these observations, we computed the total relative abundance of all identified proteins in a grouped manner, similar to rbc. According to this analysis, the selectivity of the first stage filtration became even more evident (Fig. 4B). In the case of certain types of proteins, such as CBPs, which are proteins associated with photosystem I and II (PI/II), histones (His), and ATP synthases (ATPs), a high level of selective retention was observed. Next, we performed fold-change (FC) analysis across the first filtration stage, where S1 feed, S1 ret, and S1 perm were compared in pair-wise FC analysis. In this analysis, the riBAQ of both individual proteins (Fig. 4C) and grouped proteins (Fig. 4D) was used. The FC analysis further substantiates the depletion of the aforementioned proteins and groups across the first stage filtration, and in fact CBPs were depleted from 5.9% in S1 feed to <0.03% in S1 perm, while PI/II were depleted from 7.6% to 0.1%. The specific depletion of these proteins is considered crucial for removing the undesired color due to their ability to bind pigments such as chlorophylls and carotenoids.128–130 In contrast, proteins with known antioxidant properties such as thioredoxins (TRX), SODs and ferredoxins (FNR) become enriched following first stage filtration from 1.7%, 0.7% and 0.8% in S1 feed (Fig. 4B) to 4.6%, 1.8%, and 1.6% in S1 perm, representing a 2.8, 2.6, and 1.9-fold enrichment, respectively (Fig. 4D). Similarly, nsLTPs were enriched by 3.8-fold, while the oxygen evolving proteins of photosystem II (PII-OES), in contrast to pigment binding PI/II proteins, were also enriched by 2.5-fold.
A much less pronounced effect on the quantitative protein composition was observed across the second filtration and DF stages (Fig. 4E). Most notably, plastocyanin (PC) and some rbcL isoform were enriched while other rbcL, rbcS, and rbcA isoforms were depleted. Several nsLPT isoforms, which were enriched in the permeate from the first stage filtration (S1 perm), were depleted substantially (8- to 20-fold), while any residual CBPs were also depleted. Using the protein groupings (Fig. 4F), CBPs, which were substantially depleted in the first stage filtration, were a further 14-fold depleted to less than 0.002% in DF C from an initial level of 5.9% in S1 feed. This represents a reduction of >99.9% of CBPs across the entire process. Similarly, PI/II proteins were further depleted over the second filtration and DF stages, resulting in a final abundance of 0.06% in DF C from an initial abundance of 7.6% in S1 feed, thereby representing a removal of >99% across the whole process. The total abundance of nsLTPs was depleted to a final abundance of 0.21% from 2.15% in S1 perm, thereby representing a removal of >90% of the potential allergen, which was enriched in the first filtration stage. Based on this observation, it is not unlikely that nsLTPs can be further depleted by extending the DF stage to allow more washing of the protein. Reticuline oxidase-like protein (A0A2K3N926), which was found enriched in S1 perm by LFQ-based analysis, was generally found of low abundance across all streams (<0.1%) and found in very low abundance (<0.004%) in the final product (DF C). As such, this allergen is not considered of particular relevance at the bulk level.
3.6 Process-induced changes cause notable changes to mean protein properties
Next, the mean physicochemical properties of the process stream proteins were investigated using in silico analysis. An unweighted analysis was initially performed based only on reproducibly identified proteins in the different process streams. For this, a range of physicochemical, allergenic, subcellular, and in silico structural properties were computed for all identified proteins. The mean property within each stream was determined by computing the mean for all proteins identified in that process stream. This facilitated investigation of any enrichment or protein-level clustering based on membrane selectivity in the different unit operations. Using computed physicochemical and structural properties as descriptors, no substantial differences between the different streams were observed (Fig. S7A–H) despite the fact that the isoelectric point (pI) and structural features were found to be significant (p < 0.05) in explaining the data variability by ANOVA (Table S5). No significant clustering was observed based on either the protein occurrence in specific streams (Fig. S8A) or by their subcellular localization (Fig. S8B) using TruncatedSVD-based dimensionality reduction.Given that unweighted analysis did not provide any noteworthy insights, the protein-level abundances by riBAQ were used as weights for calculating the mean properties within each process stream. This approach proved to be much more effective for describing the differential mean properties across the different process streams. For instance, distinct differences in mean properties were found between the different process streams (Fig. S9A–H). In fact, all properties were found to be highly significant (p < 1 × 10−6) for describing the data variability by one-way ANOVA (Table S6). These differences were also reflected by distinct clustering using TruncatedSVD-based dimensionality reduction from computed physicochemical and structural properties (Fig. S10). Based on physicochemical and structural properties, the samples upstream of the first filtration stage (including S1 ret) and samples downstream clustered nicely, while the two unutilized side streams (S2 perm and DF perm) were substantially more scattered. Due to the low CP and number of protein IDs, these streams were omitted from subsequent analysis. As a result, two highly distinct clusters were formed (Fig. 5A), an “upstream cluster” and a “downstream cluster” based on their origin relative to the first filtration stage. The same clustering of streams was observed using predicted subcellular localization (Fig. 5B), highlighting the distinct differences in quantitative protein composition.
To identify which properties are important for the observed clustering, the distribution of each property across clusters was visualized using boxplots of the eight descriptive properties individually (Fig. 6A–H). Across all eight properties, distinct and significant (p < 0.05) differences were found between the upstream cluster (cluster 0) and the downstream cluster (cluster 1). Upstream samples were found to have a larger median molecular weight, aromaticity, instability index, GRAVY, and pI. In general, the upstream samples were also found to have a larger proportion of well-defined secondary structures with a higher content of both α-helix and β-sheet, while the proportion of turns decreased.
The higher abundance of membrane-associated proteins in the upstream samples, as indicated by the pair-wise comparison of S1 ret and S1 perm using the LFQ data (Fig. 3E and F), correlates well with the differences in physicochemical properties between the clusters. Plant membrane proteins are similarly reported to generally have a higher MW and more basic pI compared to soluble proteins,131 which correlates with the higher median MW and pI found in the upstream cluster (Fig. 6C and D), respectively. Membrane proteins contain transmembrane domains and often are responsible for pore formation,132 which is why a closer to zero GRAVY in the upstream cluster (Fig. 6B) reflects their more amphiphilic nature.133 Although, reduced amphiphilicity may reduce the ability of the final protein product to form stable oil-in-water emulsions,134 a larger negative GRAVY also indicates its better solubility.135 These results agree with recent work on the functionality of a protein product produced using the presented membrane-based biorefinery concept, where high solubility was observed, while less impressive emulsifying properties were seen.53 Given that membrane proteins require a highly ordered structure, particularly in membrane-spanning regions,132,133 this also explains the higher proportion of defined secondary structure elements (Fig. 6F–H). Furthermore, membrane proteins are reported to have lower stability,132 correlating well with the higher instability index in the upstream cluster (Fig. 6E), which is particularly prominent in the TM-enriched S1 ret (Fig. 3F, 6I and S9E).
Considering the differences in the subcellular localization of proteins, subtle but still distinct differences were observed when performing the unweighted analysis (Tables S7 and S8). A larger proportion of membrane proteins was found in the upstream samples, particularly in S1 ret, in agreement with the pairwise comparison of S1 ret and S1 perm (Fig. 3E). Similarly, the upstream samples generally contained higher proportions of proteins localized in the ER, Golgi apparatus, and mitochondria, while downstream proteins contained a higher proportion of cytoplasmic and extracellular proteins. These differences were more pronounced when factoring the relative protein abundances (riBAQ) in the weighted analysis (Fig. 6J). Here, the selective retention of cell membrane and ER (the site for transmembrane protein synthesis136) proteins was observed across the first filtration stage. Cell membrane proteins constituted 3.7% of the total protein in S1 feed but were depleted to 0.8% in S1 perm. In contrast, the proportion of cytoplasmic and extracellular protein increased. Only subtle differences were observed when comparing S1 perm, S2 ret and the final product DF C. The selective retention of membrane-associated proteins is further substantiated when considering the distribution of all proteins predicted to comprise transmembrane regions (Fig. 6I). Here, the abundance of TM proteins in S1 feed (18%) was substantially depleted over the first membrane into S1 perm (2.7%) and slightly enriched in S1 ret (20%). These results corroborated the findings of the LFQ-based pair-wise comparison between S1 ret and S1 perm (Fig. 3E and F), respectively. In the final product stream (DF C), the low level of membrane-associated proteins was at a similar level as in S1 perm (2.5%), representing a 7.2-fold depletion of membrane-associated proteins compared to S1 feed. Ultimately, our findings show a high degree of selectivity towards particularly membrane-associated and known pigment-binding proteins in the first filtration stage of the biorefinery process, being responsible for significant changes in the optical properties and sensory attributes of the different streams. Meanwhile, the second filtration and DF stages, using a 10 kDa cutoff membrane, merely retains and concentrates proteins in a non-discriminatory manner, while washing out small molecule contaminants, phytochemicals, and residual pigments.
4 Conclusions
Recently, a nondestructive two-stage membrane-based biorefinery concept was developed to produce native leaf protein devoid of grassy attributes. However, the underlying reasons why this process works remain unexplored. In this study, process streams from each unit operation within the two-stage membrane-process were characterized to understand the selectivity and driving forces from a molecular perspective. This process efficiently removes >99.9% of investigated pigments (chlorophylls and carotenoids), with the majority being retained in the first stage filtration. Using mass spectrometry-based proteomics with two complementary quantification strategies and subsequent bioinformatic analysis, this was found to be correlated with the selective retention of membrane-associated and pigment-binding proteins, particularly in the first filtration stage. Over the full process, >99.9% of chlorophyll-binding proteins and other pigment-binding proteins from photosystems I and II were removed from the final protein concentrate obtained following a second filtration and subsequent diafiltration (DF) stage. In contrast, proteins with reported antioxidant properties were enriched in the final protein product intended for use as a food ingredient, which may not only improve the stability of the product but also exhibit potential health benefits from ingestion. Moreover, proteins with known allergenic properties were found to be depleted by the biorefinery concept. Importantly, a high abundance of RuBisCO was found to be retained throughout the process, explaining the previously described promising properties of the protein concentrate.This study clearly demonstrates that the larger pore membrane used in first-stage filtration displays a much higher degree of differential protein retention compared to the 10 kDa membrane used in the second filtration and DF stages. This also reflects why both stages were essential for a successful process, as the first stage initially retains undesired large components. Subsequently, the second stage and DF facilitate permeation of residual small molecule contaminants (e.g. free pigments and other phytochemicals) as well as buffer components and salts from the stabilizing solution employed in the process. This stage facilitates concentrating the protein to a higher purity, while retaining the quantitative composition, and ultimately producing the final product stream for subsequent drying. Overall, this study provides a molecular perspective and protein-level insights into membrane selectivity in green biorefineries and highlights what proteins should be selectively removed from green juice to improve the quality and functionality of a protein product.
Author contributions
SGE: conceptualization, methodology, validation, formal analysis, investigation, visualization, funding acquisition, project administration, supervision, writing – original draft preparation, writing – review and editing. NAK: methodology, formal analysis, investigation, visualization, writing – review and editing. NHJ: formal analysis, investigation, writing – review and editing. AKJ: resources, formal analysis, investigation, writing – review and editing. TM: resources, investigation, writing – review and editing. MKJ: resources, investigation, writing – review and editing. PSL: conceptualization, funding acquisition, project administration, writing – review and editing. ML: conceptualization, funding acquisition, project administration, supervision, writing – review and editing.Conflicts of interest
Peter Stephensen Lübeck reports a relationship with BiomassProtein ApS that includes board membership. Simon Gregersen Echers, Peter Stephensen Lübeck, Tuve Mattsson, Anders K Jørgensen, Mads K Jørgensen, and Mette Lübeck have the patent #WO2025/133209: Method for producing a food-grade protein product and/or feed protein product from plant material. Remaining authors report no competing interests.Data availability
The majority of data is available in the supplementary information (SI). Supplementary information: supplementary tables: Table S1: MaxQuant output data and additional downstream processing hereof (table can be found in appended .xlsx file). Table S2: Uniprot AC#s for protein families/types/groups (table can be found in appended .xlsx file). Table S3: Differential proteins from LFQ-based pair-wise comparison of MF Ret and MF Perm (table can be found in appended .xlsx file). Table S4: Abundant proteins (riBAQ > 0.5%) across all streams (table can be found in appended .xlsx file). Table S5: One-Way ANOVA for unweighted analysis using computed physicochemical properties. Table S6: One-Way ANOVA for riBAQ-weighted analysis using computed physicochemical properties. Table S7: Distribution of protein subcellular localization as predicted by DeepLoc (by count). Table S8: Distribution of protein subcellular localization as predicted by DeepLoc (by %). Supplementary figures: Fig. S1: One-way ANOVA of dry matter and crude protein across streams. Fig. S2: Quantification of individual pigments relative to dry matter and crude protein. Fig. S3: Optimization of protein extraction. Fig. S4: Distribution of protein ID's and sample similarity analysis (replicate level). Fig. S5: Pair-wise differential analysis of MF Ret vs. MF Perm and overlap with differential proteins from pair-wise comparison of MF Feed vs. MF Perm. Fig. S6: Pair-wise differential analysis of retentates and permeates from UF and DF. Fig. S7: Boxplots showing the distribution of computed physicochemical properties by sample based on unweighted analysis. Fig. S8: Two-dimensional representation of proteins obtained via TruncatedSVD from unweighted computed physicochemical properties by sample and predicted subcellular localization. Fig. S9: Boxplots showing the distribution of computed physicochemical properties by sample based on riBAQ-weighted analysis. Fig. S10: Two-dimensional representation of protein properties from each stream (replicate-level) obtained via TruncatedSVD on riBAQ-weighted computed physicochemical properties. Note to the reader: in the raw data supplied (e.g. LC-MS/MS) a different nomenclature is applied to reflect the internal, daily naming of the process steps. As such, samples from first stage filtration (S1) are referred to as microfiltration (MF) while the use of a 60 nm membrane does not qualify as microfiltration per definition but is in principle ultrafiltration. Similarly, the second stage filtration (S2) is referred to as ultrafiltration (UF) despite both stages in principle represent ultrafiltration. The final product stream (DF C) may be referred to as “DC” while its replicate (DF C_2) may be referred to as “DF Ret”. This discrepancy in sample naming is reflected in some plots within the SI. See DOI: https://doi.org/10.1039/d5fb00771b.The mass spectrometry proteomics data have been deposited into the ProteomeXchange Consortium via the PRIDE83 partner repository with the dataset identifier PXD069787 and https://doi.org/10.6019/PXD069787. All other data can be made available upon request.
Acknowledgements
This work was supported by the Danish Green Development and Demonstration Program (GUDP) under the “Grass4Food” project [grant number 34009-20-1657] and the Independent Research Foundation Denmark (DFF) under the “PhyPro” project [grant number DFF 1127-00245B]. The authors would like to acknowledge the laboratory assistance provided by laboratory technician Sofie Albrekt Hansen.References
- H. Lynch, C. Johnston and C. Wharton, Plant-Based Diets: Considerations for Environmental Impact, Protein Quality, and Exercise Performance, Nutrients, 2018, 10(12), 1841 CrossRef PubMed , https://www.mdpi.com/2072-6643/10/12/1841/htm.
- J. Sadhukhan, T. I. J. Dugmore, A. Matharu, E. Martinez-Hernandez, J. Aburto and P. K. S. M. Rahman, et al., Perspectives on “Game Changer” Global Challenges for Sustainable 21st Century: Plant-Based Diet, Unavoidable Food Waste Biorefining, and Circular Economy, Sustainability, 2020, 12(5) CrossRef , https://www.mdpi.com/2071-1050/12/5/1976/htm.
- S. Prasanna, P. Verma and S. Bodh, The role of food industries in sustainability transition: a review, Environ. Dev. Sustain., 2025, 27(7), 15113–15133 CrossRef , https://link.springer.com/article/10.1007/s10668-024-04642-1.
- A. G. A. Sá, Y. M. F. Moreno and B. A. M. Carciofi, Plant proteins as high-quality nutritional source for human diet, Trends Food Sci. Technol., 2020, 97, 170–184 CrossRef.
- N. Sözer and K. S. Poutanen, Plant protein ingredients for future foods, Agro Food Ind. Hi-Tech, 2015, 26(5), 56–59 Search PubMed , https://cris.vtt.fi/en/publications/plant-protein-ingredients-for-future-foods.
- H. M. Meltzer, H. Eneroth, M. Erkkola, E. Trolle, P. Fantke and J. Helenius, et al., Challenges and opportunities when moving food production and consumption toward sustainable diets in the Nordics: a scoping review for Nordic Nutrition Recommendations 2023, Food Nutr. Res., 2024, 68 DOI:10.29219/fnr.v68.10489 , https://pmc.ncbi.nlm.nih.gov/articles/PMC11075468/.
- M. V. Odgaard, M. T. Knudsen, J. E. Hermansen and T. Dalgaard, Targeted grassland production – A Danish case study on multiple benefits from converting cereal to grasslands for green biorefinery, J. Cleaner Prod., 2019, 223, 917–927 CrossRef , https://www.sciencedirect.com/science/article/pii/S0959652619307632.
- T. P. Thomsen, M. B. Karlsson and A. Kamp, Integration of Green Biorefineries and Pyrolysis: Climate Footprint Assessment of Co-Management of Grass Pulp and Cattle Manure in Denmark, Waste Biomass Valorization, 2025, 1–19 Search PubMed , https://link.springer.com/article/10.1007/s12649-025-03249-5.
- S. K. Soni and R. Soni, Challenges and Opportunities in Green Biorefinery, Clean Energy Production Technologies, 2025 pp. 399–433, https://link.springer.com/chapter/10.1007/978-981-96-9013-8_12 Search PubMed.
- J. Gaffey, G. Rajauria, H. McMahon, R. Ravindran, C. Dominguez and M. Ambye-Jensen, et al., Green Biorefinery systems for the production of climate-smart sustainable products from grasses, legumes and green crop residues, Biotechnol. Adv., 2023, 66, 108168 CrossRef PubMed.
- R. Ravindran, S. Koopmans, J. P. M. Sanders, H. McMahon and J. Gaffey, Production of Green Biorefinery Protein Concentrate Derived from Perennial Ryegrass as an Alternative Feed for Pigs, Clean Technol., 2021, 3(3), 656–669 CrossRef , https://www.mdpi.com/2571-8797/3/3/39/htm.
- V. K. Damborg, S. K. Jensen, M. R. Weisbjerg, A. P. Adamsen and L. Stødkilde, Screw-pressed fractions from green forages as animal feed: Chemical composition and mass balances, Anim. Feed Sci. Technol., 2020, 261, 114401 CrossRef.
- Z. Solati, K. Manevski, U. Jørgensen, R. Labouriau, S. Shahbazi and P. E. Lærke, Crude protein yield and theoretical extractable true protein of potential biorefinery feedstocks, Ind. Crops Prod., 2018, 115, 214–226 CrossRef.
- M. Humphreys, U. Feuerstein, M. Vandewalle and J. Baert, Ryegrasses, Fodder Crops and Amenity Grasses, 2010, pp. 211–260, https://link.springer.com/chapter/10.1007/978-1-4419-0760-8_10 Search PubMed.
- U. Jørgensen, S. K. Jensen and M. Ambye-Jensen, Coupling the benefits of grassland crops and green biorefining to produce protein, materials and services for the green transition, Grass Forage Sci., 2022, 77(4), 295–306 CrossRef , https://onlinelibrary.wiley.com/doi/full/10.1111/gfs.12594.
- W. Koshchuh, G. Povoden, V. H. Thang, S. Kromus, K. D. Kulbe and S. Novalin, et al., Production of leaf protein concentrate from ryegrass (Lolium perenne x multiflorum) and alfalfa (Medicago sauva subsp. sativa). Comparison between heat coagulation/centrifugation and ultrafiltration, Desalination, 2004, 163(1–3), 253–259 CrossRef , https://www.sciencedirect.com/science/article/pii/S001191640490197X?via%3Dihub.
- T. S. Shinali, Y. Zhang, M. Altaf, A. Nsabiyeze, Z. Han and S. Shi, et al., The Valorization of Wastes and Byproducts from Cruciferous Vegetables: A Review on the Potential Utilization of Cabbage, Cauliflower, and Broccoli Byproducts, Foods, 2024, 13(8), 1163 CrossRef PubMed , https://www.mdpi.com/2304-8158/13/8/1163/htm.
- I. Andersson and A. Backlund, Structure and function of Rubisco, Plant Physiol. Biochem., 2008, 46(3), 275–291 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/S0981942808000041.
- M. T. Lin, A. Occhialini, P. J. Andralojc, M. A. J. Parry and M. R. Hanson, A faster Rubisco with potential to increase photosynthesis in crops, Nature, 2014, 513(7519), 547–550 CrossRef PubMed , https://www.nature.com/articles/nature13776.
- E. Di Stefano, D. Agyei, E. N. Njoku and C. C. Udenigwe, Plant RuBisCo: An Underutilized Protein for Food Applications, J. Am. Oil Chem. Soc., 2018, 95(8), 1063–1074 CrossRef , https://onlinelibrary.wiley.com/doi/full/10.1002/aocs.12104.
- X. Ma, M. Habibi, J. Landman, L. M. C. Sagis and P. Shen, Rubisco at interfaces II: Structural reassembly enhances oil-water interface and emulsion stabilization, Food Hydrocoll., 2025, 160, 110820 CrossRef , https://www.sciencedirect.com/science/article/pii/S0268005X24010944.
- X. Ma, T. van Polen, M. Habibi, J. Landman, L. M. C. Sagis and P. Shen, Rubisco at interfaces I: Conformational flexibility enhances air-water interface and foam stabilization, Food Hydrocoll., 2025, 160, 110783 CrossRef , https://www.sciencedirect.com/science/article/pii/S0268005X24010579.
- H. Zhou, G. Vu and D. J. McClements, Formulation and characterization of plant-based egg white analogs using RuBisCO protein, Food Chem., 2022, 397, 133808 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/S0308814622017708.
- M. Grácio, S. Oliveira, A. Lima and R. Boavida Ferreira, RuBisCO as a protein source for potential food applications: A review, Food Chem., 2023, 419, 135993 CrossRef PubMed . https://www.sciencedirect.com/science/article/pii/S0308814623006106.
- E. Di Stefano, D. Agyei, E. N. Njoku and C. C. Udenigwe, Plant RuBisCo: An Underutilized Protein for Food Applications, J. Am. Oil Chem. Soc., 2018, 95(8), 1063–1074 CrossRef , https://aocs.onlinelibrary.wiley.com/doi/10.1002/aocs.12104.
- E. Echeverria-Jaramillo, A. K. Jørgensen, M. K. Jørgensen, T. Mattsson, S. G. Echers and P. S. Lübeck, et al., Functional properties of protein fractions from gentle membrane separation of green biomass (legume grass) compared to traditional animal- and plant-based proteins, Future Foods, 2025, 12, 100740 CrossRef CAS , https://www.sciencedirect.com/science/article/pii/S2666833525001996.
- J. P. Wold, A. Veberg, A. Nilsen, V. Iani, P. Juzenas and J. Moan, The role of naturally occurring chlorophyll and porphyrins in light-induced oxidation of dairy products. A study based on fluorescence spectroscopy and sensory analysis, Int. Dairy J., 2005, 15(4), 343–353 CrossRef CAS , https://www.sciencedirect.com/science/article/pii/S0958694604002080.
- M. Suman, R. De Maria and D. Catellani, Chromatographic evaluation of chlorophyll derivatives in pasta-based food products: Effects of pasteurization treatments and correlation with sensory profiles, J. Sci. Food Agric., 2008, 88(3), 471–478 CrossRef CAS , https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/pca.3109.
- A. J. Meléndez-Martínez, A. I. Mandić, F. Bantis, V. Böhm, G. I. A. Borge and M. Brnčić, et al., A comprehensive review on carotenoids in foods and feeds: status quo, applications, patents, and research needs, Crit. Rev. Food Sci. Nutr., 2022, 62(8), 1999–2049 CrossRef , https://www.tandfonline.com/doi/pdf/10.1080/10408398.2020.1867959.
- P. Adadi, N. V. Barakova and E. F. Krivoshapkina, Selected Methods of Extracting Carotenoids, Characterization, and Health Concerns: A Review, J. Agric. Food Chem., 2018, 66(24), 5925–5947 CrossRef CAS , https://www./doi/pdf/10.1021/acs.jafc.8b01407?ref=article_openPDF.
- A. A. Anoop, P. K. S. Pillai, M. Nickerson and K. V. Ragavan, Plant leaf proteins for food applications: Opportunities and challenges, Compr. Rev. Food Sci. Food Saf., 2023, 22(1), 473–501 CrossRef CAS PubMed , https://www./doi/pdf/10.1111/1541-4337.13079.
- Y. H. Chen, Y. H. Zhang, G. S. Chen, J. F. Yin, J. X. Chen and F. Wang, et al., Effects of phenolic acids and quercetin-3-O-rutinoside on the bitterness and astringency of green tea infusion, npj Sci. Food, 2022, 6(1), 1–8 CrossRef CAS , https://www.nature.com/articles/s41538-022-00124-8.
- B. Wan, Q. Xiao, M. Huang and R. Ying, Masking the bitter taste of quercetin by liposomes modified with whey protein isolate: Better to be coated or inserted?, Food Hydrocoll., 2024, 149, 109540 CrossRef CAS , https://www.sciencedirect.com/science/article/pii/S0268005X2301086X.
- R. Ravindran, S. Koopmans, J. P. M. Sanders, H. McMahon and J. Gaffey, Production of Green Biorefinery Protein Concentrate Derived from Perennial Ryegrass as an Alternative Feed for Pigs, Clean Technol., 2021, 3(3), 656–669 CrossRef , https://www.mdpi.com/2571-8797/3/3/39/htm.
- T. A. Andrade and M. Ambye-Jensen, Process Integration and Techno-Economic Assessment of a Green Biorefinery Demonstration Scale Platform for Leaf Protein Production, Comput.-Aided Chem. Eng., 2022, 51, 877–882 CAS , https://www.sciencedirect.com/science/article/abs/pii/B9780323958790501478.
- M. Santamaría-Fernández and M. Lübeck, Production of leaf protein concentrates in green biorefineries as alternative feed for monogastric animals, Anim. Feed Sci. Technol., 2020, 268, 114605 CrossRef.
- M. Santamaria-Fernandez, M. Ambye-Jensen, V. K. Damborg and M. Lübeck, Demonstration-scale protein recovery by lactic acid fermentation from grass clover – a single case of the production of protein concentrate and press cake silage for animal feeding trials, Biofuels, Bioprod. Biorefin., 2019, 13(3), 502–513 CrossRef , https://www./doi/pdf/10.1002/bbb.1957.
- P. Thornton, H. Gurney-Smith and E. Wollenberg, Alternative sources of protein for food and feed, Curr. Opin. Environ. Sustain., 2023, 62, 101277 CrossRef , https://www.sciencedirect.com/science/article/pii/S1877343523000246.
- A. H. Møller, M. Hammershøj, N. H. M. Dos Passos, H. Tanambell, L. Stødkilde and M. Ambye-Jensen, et al., Biorefinery of Green Biomass-How to Extract and Evaluate High Quality Leaf Protein for Food?, J. Agric. Food Chem., 2021, 69(48), 14341–14357 CrossRef PubMed , https://www./doi/pdf/10.1021/acs.jafc.1c04289?ref=article_openPDF.
- A. L. Nynäs, W. R. Newson and E. Johansson, Protein fractionation of green leaves as an underutilized food source—protein yield and the effect of process parameters, Foods, 2021, 10(11), 2533 CrossRef , https://www.mdpi.com/2304-8158/10/11/2533/htm.
- R. J. B. M. Delahaije, A. Kiskini and P. A. Wierenga, Towards predicting the emulsion properties of plant protein extracts from sugar beet (Beta vulgaris L.) leaf and soybean (Glycine max), Colloids Surf., A, 2022, 646, 128950 CrossRef , https://www.sciencedirect.com/science/article/pii/S0927775722007051.
- K. A. Furia, P. J. Torley, M. Majzoobi, C. Balfany and A. Farahnaky, Leaf protein concentrate from cauliflower leaves: Effect of filtration and stabilizing solution on extraction and physio-chemical properties, Innovative Food Sci. Emerging Technol., 2025, 103, 104037 CrossRef , https://www.sciencedirect.com/science/article/pii/S1466856425001213.
- A. H. Martin, O. Castellani, G. A. H. de Jong, L. Bovetto and C. Schmitt, Comparison of the functional properties of RuBisCO protein isolate extracted from sugar beet leaves with commercial whey protein and soy protein isolates, J. Sci. Food Agric., 2019, 99(4), 1568–1576 CrossRef PubMed , https://www./doi/pdf/10.1002/jsfa.9335.
- H. Tanambell, A. H. Møller, L. Roman, M. Corredig and T. K. Dalsgaard, Supramolecular structure modification of RuBisCO from alfalfa during removal of chloroplastic materials, Innovative Food Sci. Emerging Technol., 2023, 87, 103408 CrossRef.
- M. Nieuwland, P. Geerdink, N. P. E. Engelen-Smit, I. M. van der Meer, A. H. P. America and J. J. Mes, et al., Isolation and Gelling Properties of Duckweed Protein Concentrate, ACS Food Sci. Technol., 2021, 1(5), 908–916 CrossRef , https://www./doi/pdf/10.1021/acsfoodscitech.1c00009?ref=article_openPDF.
- I. Gifuni, L. Lavenant, J. Pruvost and A. Masse, Recovery of microalgal protein by three-steps membrane filtration: Advancements and feasibility, Algal Res., 2020, 51, 102082 CrossRef , https://www.sciencedirect.com/science/article/pii/S2211926420305415.
- A. Corona, M. Ambye-Jensen, G. C. Vega, M. Z. Hauschild and M. Birkved, Techno-environmental assessment of the green biorefinery concept: Combining process simulation and life cycle assessment at an early design stage, Sci. Total Environ., 2018, 635, 100–111 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/S0048969718311173.
- A. H. Møller, M. Hammershøj, N. H. M. Dos Passos, H. Tanambell, L. Stødkilde and M. Ambye-Jensen, et al., Biorefinery of Green Biomass-How to Extract and Evaluate High Quality Leaf Protein for Food?, J. Agric. Food Chem., 2021, 69(48), 14341–14357 CrossRef PubMed , https://pubs.acs.org/doi/full/10.1021/acs.jafc.1c04289.
- F. Bazzarelli, R. Mazzei, E. Papaioannou, V. Giannakopoulos, M. R. Roberts and L. Giorno, Biorefinery of Tomato Leaves by Integrated Extraction and Membrane Processes to Obtain Fractions That Enhance Induced Resistance against Pseudomonas syringae Infection, Membranes, 2022, 12(6), 585 CrossRef PubMed , https://www.mdpi.com/2077-0375/12/6/585/htm.
- A. L. Nynäs, E. Berndtsson, W. R. Newson, H. P. Hovmalm and E. Johansson, Protein Fractionation of Leafy Green Biomass at the Pilot Scale: Partitioning and Type of Nitrogen in the Fractions and Their Usefulness for Food and Feed, ACS Food Sci. Technol., 2024, 4(1), 126–138 CrossRef , https://www./doi/pdf/10.1021/acsfoodscitech.3c00426?ref=article_openPDF.
- P. S. Lübeck, T. Mattsson, A. K. Jørgensen, S. Gregersen Echers, M. K. Jørgensen and M. Olsen, et al., Method for Producing A Food Grade Protein Product And/Or A Feed Protein Product from Plant Material, Denmark, WO2025133209, World Intellectual Property Organization, 2025 Search PubMed.
- T. Mattsson, A. K. Jørgensen, S. Gregersen Echers, M. Olsen, N. Badfar and S. U. Heiske, et al., Green Leaf Biorefining – A Demonstrator for Production of High-Quality Food-Grade Protein, Future Foods, 2025, 100846, DOI:10.1016/j.fufo.2025.100846.
- E. Echeverria-Jaramillo, A. K. Jørgensen, M. K. Jørgensen, T. Mattsson, S. G. Echers and P. S. Lübeck, et al., Functional properties of protein fractions from gentle membrane separation of green biomass (legume grass) compared to traditional animal- and plant-based proteins, Future Foods, 2025, 12, 100740 CrossRef , https://www.sciencedirect.com/science/article/pii/S2666833525001996.
- X. Li, J. Tian, Q. Zhang, Y. Jiang, Z. Wu and Z. Yu, Effects of mixing red clover with alfalfa at different ratios on dynamics of proteolysis and protease activities during ensiling, J. Dairy Sci., 2018, 101(10), 8954–8964 CrossRef , https://www.sciencedirect.com/science/article/pii/S0022030218306684.
- Y. Yu, M. Kleuter, L. M. Trindade and A. J. van der Goot, Tomato (Solanum lycopersicum) leaf juice as enzyme source: A study on the impact of endogenous proteases on plant proteins, Food Hydrocoll., 2024, 155, 110245 CrossRef , https://www.sciencedirect.com/science/article/pii/S0268005X24005198.
- J. K. Keppler, K. Schwarz and A. J. van der Goot, Covalent modification of food proteins by plant-based ingredients (polyphenols and organosulphur compounds): A commonplace reaction with novel utilization potential, Trends Food Sci. Technol., 2020, 101, 38–49 CrossRef CAS.
- M. Schneider, D. Esposito, M. A. Lila and E. A. Foegeding, Formation of whey protein–polyphenol meso-structures as a natural means of creating functional particles, Food Funct., 2016, 7(3), 1306–1318 RSC , https://pubs.rsc.org/en/content/articlehtml/2016/fo/c5fo01499a.
- S. Dai, T. Xu, Y. Yuan, Q. Fang, Z. Lian and T. Tian, et al., Combination and precipitation mechanism of soy protein and tea polyphenols, Food Hydrocoll., 2024, 146, 109197 CrossRef CAS , https://www.sciencedirect.com/science/article/pii/S0268005X23007439.
- B. Amer, L. Juul, A. H. Møller, H. S. Møller and T. K. Dalsgaard, Improved solubility of proteins from white and red clover – inhibition of redox enzymes, Int. J. Food Sci. Technol., 2021, 56(1), 302–311 CrossRef CAS , https://publons.com/publon/10.1111/ijfs.14632.
- K. Danner Aakjaer Pedersen, L. Thopholm Andersen, M. Heiselberg, C. Agerskov Brigsted, F. Lyngs Støvring and L. Mailund Mikkelsen, et al., Identifying Endogenous Proteins of Perennial Ryegrass (Lolium perenne) with Ex Vivo Antioxidant Activity, Proteomes, 2025, 13(1), 8 CrossRef , https://www.mdpi.com/2227-7382/13/1/8/htm.
- A. Incamps, F. Hély-Joly, P. Chagvardieff, J. C. Rambourg, A. Dedieu and E. Linares, et al., Industrial process proteomics: Alfalfa protein patterns during wet fractionation processing, Biotechnol. Bioeng., 2005, 91(4), 447–459 CrossRef CAS PubMed , https://www./doi/pdf/10.1002/bit.20520.
- S. Pérez-Vila, F. Acevedo, A. Brodkorb, N. Baptista Dias, M. A. Fenelon and J. A. O'Mahony, et al., Quinoa leaves as a source of soluble proteins: Extraction and proteomic characterization from four different varieties of Chenopodium quinoa, Food Chem., 2025, 468, 142411 CrossRef , https://www.sciencedirect.com/science/article/pii/S0308814624040615.
- J. Liu, X. Hao, Y. Guo, M. Wang and F. Huang, Mechanism of nodulation and nitrogen fixation in Caucasian clover (Trifolium ambiguum Bieb.) based on transcriptomics and proteomics analyses, Front. Genet., 2025, 16, 1600377 CrossRef.
- K. A. Wilson, M. T. McManus, M. E. Gordon and T. W. Jordan, The proteomics of senescence in leaves of white clover, Trifolium repens (L.), Proteomics, 2002, 2(9), 1114–1122 CrossRef , https://www./doi/pdf/10.1002/1615-9861%28200209%292%3A9%3C1114%3A%3AAID-PROT1114%3E3.0.CO%3B2-O.
- A. Bertrand, M. Bipfubusa, Y. Castonguay, S. Rocher, A. Szopinska-Morawska and Y. Papadopoulos, et al., A proteome analysis of freezing tolerance in red clover (Trifolium pratense L.), BMC Plant Biol., 2016, 16(1), 1–18 CrossRef PubMed , https://link.springer.com/articles/10.1186/s12870-016-0751-2.
- J. Yu, N. Fan, R. Li, L. Zhuang, Q. Xu and B. Huang, Proteomic Profiling for Metabolic Pathways Involved in Interactive Effects of Elevated Carbon Dioxide and Nitrogen on Leaf Growth in a Perennial Grass Species, J. Proteome Res., 2019, 18(6), 2446–2457 CrossRef , https://www./doi/pdf/10.1021/acs.jproteome.8b00951?ref=article_openPDF.
- L. Hou, M. Wang, H. Wang, W. H. Zhang and P. Mao, Physiological and proteomic analyses for seed dormancy and release in the perennial grass of Leymus chinensis, Environ. Exp. Bot., 2019, 162, 95–102 CrossRef , https://www.sciencedirect.com/science/article/pii/S0098847218316356.
- X. Peng, D. Yu, J. Yan, N. Zhang, J. Lin and J. Wang, Physiological and Proteomic Analyses Reveal Adaptive Mechanisms of Ryegrass (Annual vs. Perennial) Seedlings to Salt Stress, Agronomy, 2019, 9(12), 843 CrossRef , https://www.mdpi.com/2073-4395/9/12/843/htm.
- J. Xiong, Y. Sun, Q. Yang, H. Tian, H. Zhang and Y. Liu, et al., Proteomic analysis of early salt stress responsive proteins in alfalfa roots and shoots, Proteome Sci., 2017, 15(1), 1–19 CrossRef , https://link.springer.com/articles/10.1186/s12953-017-0127-z.
- W. Li, Z. Wei, Z. Qiao, Z. Wu, L. Cheng and Y. Wang, Proteomics Analysis of Alfalfa Response to Heat Stress, PLoS One, 2013, 8(12), e82725 CrossRef PubMed , https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0082725.
- E. Goktayoglu, M. H. Oztop and S. Ozcan, Proteomics Approach to Differentiate Protein Extraction Methods in Sugar Beet Leaves, J. Agric. Food Chem., 2023, 71(23), 9157–9163 CrossRef , https://www./doi/pdf/10.1021/acs.jafc.2c09190?ref=article_openPDF.
- D. Aiello, C. Siciliano, F. Mazzotti, L. Di Donna, R. Risoluti and A. Napoli, Protein Extraction, Enrichment and MALDI MS and MS/MS Analysis from Bitter Orange Leaves (Citrus aurantium), Molecules, 2020, 25(7), 1485 CrossRef PubMed , https://www.mdpi.com/1420-3049/25/7/1485/htm.
- E. Fasoli, A. D'Amato, A. V. Kravchuk, E. Boschetti, A. Bachi and P. G. Righetti, Popeye strikes again: The deep proteome of spinach leaves, J. Proteomics, 2011, 74(1), 127–136 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/S1874391910002976.
- H. K. Lichtenthaler and C. Buschmann, Chlorophylls and Carotenoids: Measurement and Characterization by UV-VIS Spectroscopy, Current Protocols in Food Analytical Chemistry, 2001, vol. 1, 1, pp. F431–F438, https://onlinelibrary.wiley.com/doi/full/10.1002/0471142913.faf0403s01 Search PubMed.
- H. K. Lichtenthaler and C. Buschmann, Extraction of Phtosynthetic Tissues:Chlorophylls and Carotenoids, Current Protocols in Food Analytical Chemistry, 2001, vol. 1, 1, pp. F421–F426, https://onlinelibrary.wiley.com/doi/full/10.1002/0471142913.faf0402s01 Search PubMed.
- E. S. Gregersen, R. K. Mikkelsen, N. Abdul-Khalek, L. S. Queiroz, T. J. Hobley and B. L. Schulz, et al., Residual barley proteins in brewers' spent grains: Quantitative composition and implications for food ingredient applications, Innovative Food Sci. Emerging Technol., 2025, 106, 104277 CrossRef , https://www.sciencedirect.com/science/article/pii/S1466856425003613.
- S. Tyanova, T. Temu and J. Cox, The MaxQuant computational platform for mass spectrometry-based shotgun proteomics, Nat. Protoc., 2016, 11(12), 2301–2319, DOI:10.1038/nprot.2016.136.
- J. Cox and M. Mann, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification, Nat. Biotechnol., 2008, 26(12), 1367–1372 CrossRef PubMed.
- S. U. Larsen, H. Jørgensen, C. Bukh and J. K. Schjoerring, Green biorefining: Effect of nitrogen fertilization on protein yield, protein extractability and amino acid composition of tall fescue biomass, Ind. Crops Prod., 2019, 130, 642–652 CrossRef , https://www.sciencedirect.com/science/article/pii/S0926669019300238.
- The UniProt Consortium, UniProt: the universal protein knowledgebase, Nucleic Acids Res., 201745(D1), D158–D169 Search PubMed.
- J. Cox, M. Y. Hein, C. A. Luber, I. Paron, N. Nagaraj and M. Mann, Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ, Mol. Cell. Proteomics, 2014, 13(9), 2513–2526 CrossRef , http://www.mcponline.org/article/S1535947620333107/fulltext.
- B. Schwanhüusser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt and J. Wolf, et al., Global quantification of mammalian gene expression control, Nature, 2011, 473(7347), 337–342 CrossRef PubMed , https://www.nature.com/articles/nature10098.
- Y. Perez-Riverol, J. Bai, C. Bandla, D. García-Seisdedos, S. Hewapathirana and S. Kamatchinathan, et al., The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences, Nucleic Acids Res., 2022, 50(D1), D543–D552 CrossRef PubMed , https://academic.oup.com/nar/article/50/D1/D543/6415112.
- A. Quaglieri, J. Bloom, A. Triantafyllidis, B. Green, M. R. Condina and P. B. Ngov, et al., Mass Dynamics 2.0: An improved modular web-based platform for accelerated proteomics insight generation and decision making, bioRxiv, 2022, 517480 Search PubMed , https://www.biorxiv.org/content/10.1101/2022.12.12.517480v1.
- A. J. J. Almagro, C. K. Sønderby, S. K. Sønderby, H. Nielsen and O. Winther, DeepLoc: prediction of protein subcellular localization using deep learning, in Bioinformatics, ed. Hancock J., 2017, vol. 33, 21, pp. 3387–3395, http://www.cbs.dtu.dk/services/DeepLoc.Examplecodeisavailableathttps://github.com/JJAlmagro/subcellular_localization.Thedatasetisavailableathttp://www.cbs.dtu.dk/services/DeepLoc/data.php Search PubMed.
- K. D. Tsirigos, C. Peters, N. Shu, L. Käll and A. Elofsson, The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides, Nucleic Acids Res., 2015, 43(W1), W401–W407, DOI:10.1093/nar/gkv485.
- S. Gregersen Echers, N. Abdul-Khalek, R. K. Mikkelsen, S. L. Holdt, C. Jacobsen and E. B. Hansen, et al., Is Gigartina a potential source of food protein and functional peptide-based ingredients? Evaluating an industrial, pilot-scale extract by proteomics and bioinformatics, Future Foods, 2022, 6, 100189, DOI:10.1016/j.fufo.2022.100189.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion and O. Grisel, et al., Scikit-learn: Machine Learning in Python, J. Mach. Learn. Res., 2012, 12, 2825–2830 Search PubMed , https://arxiv.org/pdf/1201.0490.
- P. J. A. Cock, T. Antao, J. T. Chang, B. A. Chapman, C. J. Cox and A. Dalke, et al., Biopython: freely available Python tools for computational molecular biology and bioinformatics, Bioinformatics, 2009, 25(11), 1422–1423, DOI:10.1093/bioinformatics/btp163.
- W. McKinney, Data Structures for Statistical Computing in Python, SciPy Proceedings, 2010, pp. 56–61, https://proceedings.scipy.org/articles/Majora-92bf1922-00a Search PubMed.
- J. D. Hunter, Matplotlib: A 2D graphics environment, Comput. Sci. Eng., 2007, 9(3), 90–95 Search PubMed.
- M. L. Waskom, seaborn: statistical data visualization, J. Open Source Softw., 2021, 6(60), 3021 CrossRef , https://joss.theoj.org/papers/10.21105/joss.03021.
- P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy and D. Cournapeau, et al., SciPy 1.0: fundamental algorithms for scientific computing in Python, Nat. Methods, 2020, 17(3), 261–272 CrossRef PubMed , https://www.nature.com/articles/s41592-019-0686-2.
- C. R. Harris, K. J. Millman, S. J. van der Walt, R. Gommers, P. Virtanen and D. Cournapeau, et al., Array programming with NumPy, Nature, 2020, 585(7825), 357–362 CrossRef PubMed , https://www.nature.com/articles/s41586-020-2649-2.
- J. R. Lobry and C. Gautier, Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes, Nucleic Acids Res., 1994, 22(15), 3174–3180, DOI:10.1093/nar/22.15.3174.
- K. Guruprasad, B. V. B. Reddy and M. W. Pandit, Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence, Protein Eng., 1990, 4(2), 155–161, DOI:10.1093/protein/4.2.155.
- J. Kyte and R. F. Doolittle, A simple method for displaying the hydropathic character of a protein, J. Mol. Biol., 1982, 157(1), 105–132 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/0022283682905150.
- S. Gregersen, A. S. H. Kongsted, R. B. Nielsen, S. S. Hansen, F. A. Lau and J. B. Rasmussen, et al., Enzymatic extraction improves intracellular protein recovery from the industrial carrageenan seaweed Eucheuma denticulatum revealed by quantitative, subcellular protein profiling: A high potential source of functional food ingredients, Food Chem.: X, 2021, 12, 100137 Search PubMed.
- S. Gregersen, M. Pertseva, P. Marcatili, S. L. Holdt, C. Jacobsen and P. J. Garcia-Moreno, et al., Proteomic characterization of pilot scale hot-water extracts from the industrial carrageenan red seaweed Eucheuma denticulatum, Algal Res., 2022, 62, 102619 CrossRef.
- D. B. Jones and US Department of Agriculture, Factors for Converting Percentages of Nitrogen in Foods and Feeds Into percentages of protein, 1931.
- F. Mariotti, D. Tomé and P. P. Mirand, Converting Nitrogen into Protein—Beyond 6.25 and Jones' Factors, Crit. Rev. Food Sci. Nutr., 2008, 48(2), 177–184 CrossRef PubMed , https://www.tandfonline.com/doi/abs/10.1080/10408390701279749.
- Q. Zhang, D. Qi, X. Dong, X. Li, L. Cheng and H. Liu, et al., Amino acid composition, protein content and accurate nitrogen-to-protein conversion factor for sheepgrass (Leymus chinensis), Botany, 98(3), 137–146 CrossRef , https://www./doi/pdf/10.1139/cjb-2019-0082?download=true.
- H. H. Yeoh and Y. C. Wee, Leaf protein contents and nitrogen-to-protein conversion factors for 90 plant species, Food Chem., 1994, 49(3), 245–250 CrossRef , https://www.sciencedirect.com/science/article/pii/0308814694901678.
- M. Hansen, C. A. Andersen, P. R. Jensen and T. J. Hobley, Scale-Up of Alfalfa (Medicago sativa) Protein Recovery Using Screw Presses, Foods, 2022, 11(20), 3229 CrossRef , https://pmc.ncbi.nlm.nih.gov/articles/PMC9602060/.
- A. Tamayo Tenorio, J. Gieteling, G. A. H. De Jong, R. M. Boom and A. J. Van Der Goot, Recovery of protein from green leaves: Overview of crucial steps for utilisation, Food Chem., 2016, 203, 402–408 CrossRef , https://www.sciencedirect.com/science/article/pii/S0308814616302643.
- R. la Cour, J. K. Schjoerring and H. Jørgensen, Enhancing Protein Recovery in Green Biorefineries by Lignosulfonate-Assisted Precipitation, Front. Sustain. Food Syst., 2019, 3, 503165 Search PubMed , https://www.frontiersin.org/.
- C. Inostroza-Blancheteau, M. Reyes-Díaz, G. Berríos, A. Rodrigues-Salvador, A. Nunes-Nesi and M. Deppe, et al., Physiological and biochemical responses to manganese toxicity in ryegrass (Lolium perenne L.) genotypes, Plant Physiol. Biochem., 2017, 113, 89–97 CrossRef , https://www.sciencedirect.com/science/article/pii/S0981942817300529.
- A. Nikbakht, M. Pessarakli, N. Daneshvar-Hakimi-Maibodi and M. Kafi, Perennial ryegrass growth responses to mycorrhizal infection and humic acid treatments, Agron. J., 2014, 106(2), 585–595 CrossRef , https://www./doi/pdf/10.2134/agronj2013.0275.
- H. Acuña, M. P. Sánchez and L. Inostroza, Morpho-physiological changes induced by soil environment modulate the compatibility of perennial ryegrass and white clover cultivars, Crop Sci., 2021, 61(5), 3775–3786 CrossRef . https://www./doi/pdf/10.1002/csc2.20509.
- A. Solovchenko. Screening Pigments: General Questions. 2010;9–31, https://link.springer.com/chapter/10.1007/978-3-642-13887-4_2.
- J. Su and A. Cavaco-Paulo, Effect of ultrasound on protein functionality, Ultrason. Sonochem., 2021, 76, 105653 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/S1350417721001954.
- L. Huang, X. Ding, Y. Li and H. Ma, The aggregation, structures and emulsifying properties of soybean protein isolate induced by ultrasound and acid, Food Chem., 2019, 279, 114–119 CrossRef , https://www.sciencedirect.com/science/article/pii/S0308814618320971.
- R. Pathak, S. K. Bhangu, G. J. O. Martin, F. Separovic and M. Ashokkumar, Ultrasound-induced protein restructuring and ordered aggregation to form amyloid crystals, Eur. Biophys. J., 2022, 51(4–5), 335–352 CrossRef PubMed , https://link.springer.com/article/10.1007/s00249-022-01601-4.
- H. de F. M. Justino, I. F. dos Santos, R. C. N. de Souza, E. A. Sanches, J. de A. Bezerra and C. V. Lamarão, et al., Exploring ultrasound-assisted technique for enhancing techno-functional properties of plant proteins: a comprehensive review, Int. J. Food Sci. Technol., 2023, 59(1), 498–511, DOI:10.1111/ijfs.16673.
- N. Abdul-Khalek, R. Wimmer, M. T. Overgaard and E. S. Gregersen, Insight on physicochemical properties governing peptide MS1 response in HPLC-ESI-MS/MS: A deep learning approach, Comput. Struct. Biotechnol. J., 2023, 21, 3715–3727 CrossRef PubMed.
- N. Abdul-Khalek, M. Picciani, O. Shouman, R. Wimmer, M. T. Overgaard and M. Wilhelm, et al., To Fly, or Not to Fly, That Is the Question: A Deep Learning Model for Peptide Detectability Prediction in Mass Spectrometry, J. Proteome Res., 2025 Search PubMed , https://www./doi/pdf/10.1021/acs.jproteome.4c00973.
- V. Thumuluri, J. J. Almagro Armenteros, A. R. Johansen, H. Nielsen and O. Winther, DeepLoc 2.0: multi-label subcellular localization prediction using protein language models, Nucleic Acids Res., 2022, 50(W1), W228–W234, DOI:10.1093/nar/gkac278.
- N. Denancé, B. Szurek and L. D. Noël, Emerging Functions of Nodulin-Like Proteins in Non-Nodulating Plant Species, Plant Cell Physiol., 2014, 55(3), 469–474, DOI:10.1093/pcp/pct198.
- B. Daniel, B. Konrad, M. Toplak, M. Lahham, J. Messenlehner and A. Winkler, et al., The family of berberine bridge enzyme-like enzymes: A treasure-trove of oxidative reactions, Arch. Biochem. Biophys., 2017, 632, 88–103 CrossRef CAS , https://www.sciencedirect.com/science/article/pii/S0003986117302552.
- S. Jansson, The light-harvesting chlorophyll ab-binding proteins, Biochim. Biophys. Acta Bioenerg., 1994, 1184(1), 1–19 CrossRef CAS PubMed , https://www.sciencedirect.com/science/article/pii/0005272894901481.
- J. Knoppová, R. Sobotka, J. Yu, M. Bečková, J. Pilný and J. P. Trinugroho, et al., Assembly of D1/D2 complexes of photosystem II: Binding of pigments and a network of auxiliary proteins, Plant Physiol., 2022, 189(2), 790–804, DOI:10.1093/plphys/kiac045.
- R. Bassi, B. Pineau, P. Dainese and J. Marquardt, Carotenoid-binding proteins of photosystem II, Eur. J. Biochem., 1993, 212(2), 297–303 CrossRef PubMed , https://www./doi/pdf/10.1111/j.1432-1033.1993.tb17662.x.
- A. K. Mattoo, M. T. Giardi, A. Raskind and M. Edelman, Dynamic metabolism of photosystem II reaction center proteins and pigments, Physiol. Plant., 1999, 107(4), 454–461 CrossRef , https://www./doi/pdf/10.1034/j.1399-3054.1999.100412.x.
- B. Bhowal, S. L. Singla-Pareek, S. K. Sopory and C. Kaur, From methylglyoxal to pyruvate: a genome-wide study for the identification of glyoxalases and D-lactate dehydrogenases in Sorghum bicolor, BMC Genom., 2020, 21(1), 1–19 CrossRef PubMed , https://link.springer.com/articles/10.1186/s12864-020-6547-7.
- E. A. Decker, Strategies for manipulating the prooxidative/antioxidative balance of foods to maximize oxidative stability, Trends Food Sci. Technol., 1998, 9(6), 241–248 CrossRef , https://www.sciencedirect.com/science/article/pii/S0924224498000454.
- S. Pal, A. Das, B. Sarkar, M. Hasanuzzaman and M. K. Adak, Modulation of secondary metabolism and redox regulation by exogenously applied glutathione improves the shelf life of Capsicum annuum L. fruit, Plant Physiol. Biochem., 2024, 212, 108789 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/S0981942824004571.
- G. Wu, Y. Z. Fang, S. Yang, J. R. Lupton and N. D. Turner, Glutathione Metabolism and Its Implications for Health, J. Nutr., 2004, 134(3), 489–492 CrossRef PubMed , https://www.sciencedirect.com/science/article/pii/S0022316623026639.
- R. BASSI, B. PINEAU, P. DAINESE and J. MARQUARDT, Carotenoid-binding proteins of photosystem II, Eur. J. Biochem., 1993, 212(2), 297–303 CrossRef PubMed , https://www./doi/pdf/10.1111/j.1432-1033.1993.tb17662.x.
- X. P. Li, O. Björkman, C. Shih, A. R. Grossman, M. Rosenquist and S. Jansson, et al., A pigment-binding protein essential for regulation of photosynthetic light harvesting, Nature, 2000, 403(6768), 391–395 CrossRef PubMed , https://www.nature.com/articles/35000131.
- M. Król, M. D. Spangfort, N. P. A. Huner, G. Öquist, P. Gustafsson and S. Jansson, Chlorophyll a/b-Binding Proteins, Pigment Conversions, and Early Light-Induced Proteins in a Chlorophyll b-less Barley Mutant, Plant Physiol., 1995, 107(3), 873–883, DOI:10.1104/pp.107.3.873.
- J. Kiraga, P. Mackiewicz, D. Mackiewicz, M. Kowalczuk, P. Biecek and N. Polak, et al., The relationships between the isoelectric point and: Length of proteins, taxonomy and ecology of organisms, BMC Genom., 2007, 8(1), 1–16 CrossRef PubMed , https://link.springer.com/articles/10.1186/1471-2164-8-163.
- K. R. Vinothkumar and R. Henderson, Structures of membrane proteins, Q. Rev. Biophys., 2010, 43(1), 65–158 CrossRef PubMed , https://www.cambridge.org/core/journals/quarterly-reviews-of-biophysics/article/structures-of-membrane-proteins/C8D6D3015C519307865E18992F94B4F8.
- L. E. Hedin, K. Illergård and A. Elofsson, An introduction to membrane proteins, J. Proteome Res., 2011, 10(8), 3324–3331 CrossRef PubMed , https://www./doi/pdf/10.1021/pr200145a?ref=article_openPDF.
- X. Zhang, Q. Wang, Z. Liu, L. Zhi, B. Jiao and H. Hu, et al., Plant protein-based emulsifiers: Mechanisms, techniques for emulsification enhancement and applications, Food Hydrocoll., 2023, 144, 109008 CrossRef , https://www.sciencedirect.com/science/article/pii/S0268005X23005544.
- P. Smialowski, A. J. Martin-Galiano, A. Mikolajka, T. Girschick, T. A. Holak and D. Frishman, Protein solubility: sequence based prediction and experimental verification, Bioinformatics, 2007, 23(19), 2536–2542, DOI:10.1093/bioinformatics/btl623.
- S. L. Hanton, L. Renna, L. E. Bortolotti, L. Chatre, G. Stefano and F. Brandizzi, Diacidic Motifs Influence the Export of Transmembrane Proteins from the Endoplasmic Reticulum in Plant Cells, Plant Cell, 2005, 17(11), 3081–3093, DOI:10.1105/tpc.105.034900.
| This journal is © The Royal Society of Chemistry 2026 |