Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The effect of microbial community in the self-induced anaerobic fermentation on the aroma precursor development of geothermal coffee
Zikrina Istighfarah a,
C. Hanny Wijaya
a,
C. Hanny Wijaya *a,
Lilis Nuraida
*a,
Lilis Nuraida ab,
Erliza Noor
ab,
Erliza Noor c and
Wisnu Ananta Kusuma
c and
Wisnu Ananta Kusuma d
d
aDepartment of Food Science and Technology, IPB University, IPB Dramaga Campus, Bogor 16680, Indonesia. E-mail: channywijaya@apps.ipb.ac.id
bSoutheast Asian Food and Agricultural Science and Technology (SEAFAST) Center, IPB University, IPB Dramaga Campus, Bogor, Indonesia
cDepartment of Agroindustrial Technology, Faculty of Agricultural Technology, Bogor Agricultural Institute, Bogor 16680, Indonesia
dBioinformatics Study Program, Faculty of Mathematics and Natural Sciences, Bogor Agricultural University, IPB Darmaga, Bogor, West Java 16680, Indonesia
First published on 23rd December 2025
Abstract
Coffee drying is an important stage for character development of green beans, and it is usually conducted in direct sunlight. Thus, this method is very dependent on the weather conditions. Geothermal drying can be a substitute method to address the limitations of conventional drying. However, geothermal drying is very fast, whereas the translocation of chemical compounds into the green bean is not ideal. Process adjustment was established by applying 5 days of Self-Induced Anaerobic Fermentation (SIAF) without adding water before the geothermal drying, to enhance the sensory quality. The whole process was referred to as ‘geothermal coffee processing’. This study aimed to provide knowledge about the role effect of microorganisms involved in the SIAF on the formation of chemical compounds responsible for the geothermal coffee aroma. Samples were extracted to obtain microbial DNA, which was then analyzed using shotgun metagenomics. The species and microbial enzymes were identified by comparing the Open Reading Frames (ORF) with the databases. Furthermore, non-volatile metabolites in fermented green beans were analyzed using LC-MS/MS. The results showed that Tatumella sp. JGM118, Hanseniaspora uvarum, and Leuconostoc pseudomesenteroides were the dominant species. This microbial community secreted enzymes, such as glycoside hydrolases (GH), transaminases, and L-lactate dehydrogenase, as indicated by their ORFs in the samples. Moreover, these enzymes catalyzed the production of non-volatile metabolites in the coffee beans. The non-volatiles were dominated by chlorogenic acid, amino acids, and carboxylic acids, none of which were detected before SIAF. These metabolites were aroma precursors in the geothermal coffee drink after the roasting process. Therefore, it was confirmed that the microbial community in SIAF contributed to the production of geothermal coffee aroma precursors.
Sustainability spotlightThe application of geothermal coffee processing—which involves Self-Induced Anaerobic Fermentation (SIAF) without water addition and geothermal drying—addresses the limitations of conventional coffee processing, including its dependence on weather conditions, time, water use, and labor. Water submersion is not applied during fermentation, resulting in less water being used. The geothermal heat facilitates coffee drying in the dry-house, allowing UV to pass through at higher temperatures and lower relative humidity, creating faster processing times, with less labour required than with direct sunlight drying. Moreover, the microbial community involved in the SIAF enhances the development of aroma precursors in the geothermal coffee drink. |
Introduction
Traditionally, green coffee beans are produced by either a wet or dry process. The wet method involves aerobic and submerged fermentation of pulped coffee to produce high-quality coffee beverages.1–3 Microbial activity eliminates coffee mucilage during fermentation. The fermented coffees are then dried under the sun to obtain green beans. In the dry process, fermentation occurs during drying. Whole coffee cherries are sun-dried on the drying floor, then peeled to obtain green beans.3–5Drying under direct sunlight is a common practice in coffee processing. However, this method has several limitations, including those related to weather conditions. It causes variations in drying times and high workloads in the coffee industry.6 The long rainy season that occurs in tropical areas can pose significant challenges for coffee farmers. The unpredictable weather in Indonesia makes this conventional coffee drying process less effective.7
Mechanical drying using the excess heat from a geothermal installation can be an alternative method that addresses the limitations of conventional drying.7 This excess geothermal energy can be utilized with Heat Pipe Heat Exchangers (HPHE) to increase the temperature in the dry-house, where UV can still penetrate, to 50 °C with RH 30%.8,9 The business evaluation showed a significant surplus for coffee farmers due to the accelerated drying process.9 Conventional coffee drying generally takes up to 16 days, while geothermal drying requires only 2 to 4 days for pulped coffee and 7 to 9 days for whole coffee cherries.
Nevertheless, this acceleration of the drying process results in incomplete translocation of metabolites into coffee beans. Water content evaporates very quickly, with a reduction of more than 40% in the first 3 hours, resulting in a decrease in water activity from 0.88 to 0.62 by the end of the drying process.7,10 This level of water activity is insufficient for the survival of vegetative microorganisms; therefore, fermentation does not occur during geothermal drying.
SIAF is a new coffee fermentation technology that produces CO2 gradually as a result of microbial activities. Whole or pulped coffee is placed in a closed container to maintain anaerobiosis.11,12 This process produces metabolites that impact the quality of the coffee drink. Process adjustments are applied by employing Self-Induced Anaerobic Fermentation (SIAF) before geothermal drying to produce coffee that is favourable to consumers and Q-graders. This combination of SIAF and geothermal drying has not been studied before. Therefore, existing information on the role of microbiota involved in SIAF on the aroma precursor development of geothermal coffee remains limited.
SIAF can improve the sensory quality of coffee beverages by producing favourable chemicals compared to the conventional coffee fermentations.13 Chlorogenic acids, reducing sugars, organic acids, and amino acids are produced throughout coffee fermentation by yeasts and bacteria. These metabolites are precursors for pyrazines, aldehydes, ketones, furans, and esters during the roasting process. Sensory attributes resulting from the mentioned volatiles give roasted, nutty, and fruity notes.14
The microbial community involved in the SIAF was assumed to produce aroma precursors in the fermented green bean before geothermal drying. In this research, we analysed the relative abundance, diversity, and functional properties of the microbial community in the SIAF of Arabica coffee using shotgun metagenomics. LC-MS/MS was used to analyze the non-volatile metabolites resulting from the fermentation. Furthermore, the relations between the microorganisms, their enzymes, and their produced non-volatile metabolites, as the aroma precursors of geothermal coffee, were analyzed by connecting the acquired data from the shotgun metagenomics and LC-MS/MS to the KEGG and NCBI databases. This knowledge aims to provide a theoretical basis for the development and research of Geothermal Coffee Processing.
Materials and methods
Self-induced anaerobic fermentation (SIAF) of arabica coffee
All coffee cherry samples were harvested on the same day from an Arabica coffee plantation in Kamojang, Indonesia. After being washed, 240 kg of whole coffee cherries and 300 kg of pulped coffee, respectively, were placed in three 200-L fermentation barrels (Fig. 1). Every barrel was then tightly closed and equipped with an air lock. The SIAF process was conducted for five days at room temperature. The precise temperature and pH were monitored daily. The geothermal drying was further conducted to obtain the final green coffee beans. However, in this study, we only focus on the effects of the SIAF process on the aroma precursor development of geothermal coffee. | ||
| Fig. 1 Workflow of the experimental process shows that there are two different samples for shotgun metagenomics and non-volatile metabolite analysis using LC-MS/MS. | ||
After five days of fermentation, coffee of each 100 grams and the released water from coffee cherries of each 50 grams, respectively, were collected from the top, middle, and bottom of each barrel and mixed thoroughly. The samples were then submerged with DNA Shield (Zymo Research, California, USA) for shotgun metagenomics analysis. Coffee samples of each 100 grams for non-volatile metabolite analysis were also collected from the top, middle, and bottom of each barrel and mixed thoroughly. The samples were then manually peeled to obtain the green coffee beans and freeze-dried using liquid nitrogen. All composite samples were further transported in a cool box containing ice gel from the coffee processing facility to our laboratory.
Shotgun metagenomic
Shotgun metagenomics was employed to analyse the microbial community. This analysis applied the Next-Generation Sequencing (NGS) method, which provided an overview of the abundance and diversity of various microbial species and enzymes involved in the SIAF of Arabica coffee. Firstly, the samples were thawed, and then DNA extraction was conducted using the CTAB method. The DNA concentration was measured with Qubit dsDNA HS Assay Kits (Thermos Scientific, Massachusetts, USA), and the DNA integrity was measured with 4150 TapeStation (Agilent, California, USA). Further, the NGS library preparation was accomplished using an xGen DNA Library Prep EZ UNI Kit (IDT, Coralville, USA). The DNA concentration and DNA size of the samples were measured again to meet the requirements of the NGS instrument (Illumina NextSeq 2000, San Diego, USA).15The primary process that occurs in the NGS instrument was Surface-Restricted Fluorescence Sequencing (SURF-seq). This process used the principle of sequencing by synthesis. After sequencing was completed, the resulting reads were filtered with fastp (v0.23.2) and quality checked using fastQC (v0.11.9) and MultiQC (v1.13). Further steps of bioinformatics analyses include assembling reads into contigs with de novo assembly using MEGAHIT (v1.2.9), quality control, and visualization of contigs using QUAST (v5.0.2) and Bandage (v0.8.1).
Data transformation from FASTA file to SAM file and deletion of host DNA, coffee tree, was performed using SAMtools (v1.6). Contigs with coverage less than two and/or length shorter than 500 bp were filtered out using BBtools (BBMap v37.62). The complete genome prediction was conducted using MetaGeneMark (v3.38). The predicted ORFs with a length shorter than 100 bp were filtered out using seqkit (v2.5.1). Filtered ORFs from each sample were merged, and the redundant ORFs (percent identity = 100%) were filtered out using BBMap, generating the unique gene catalogues. Other parameters of all the tools were set to the application default.
Taxonomy assignment was performed using Diamond v2.0.15 (blastx)-MEGAN v6.24.24 (LCA, MEGAN-LR) pipeline. The curated NCBI-nr database was used. Taxonomy results were filtered to the species level. Functional binning was performed using various databases, including EC (MEGAN) and Cazy (Diamond). Mapping to gene catalogues from each sample was performed using BWA (v0.7.17-r1188). Read coverage was determined using bedtools (v2.30.0), and a count table (Reads per Kilobases – RPK) was generated using SAMtools. Taxonomy visualization was performed using Krona (v2.8.1). Downstream taxonomy and functional analysis were performed using phyloseq (v1.44.0). Completeness check was performed using BUSCO (v5.4.4).
Moreover, the Cazy database (https://cazy.org/) was used to understand the mechanism of carbohydrate-active enzymes. The detected Enzyme Commission (EC) numbers were compared to the KEGG database (https://www.kegg.jp) to recognize their cellular functions and metabolites. Alpha diversity was calculated with the Shannon-Weaver index and beta diversity with the Sørensen-Dice index. This metagenomic analysis was assisted by PT. Genetika Sains Indonesia.
Non-volatile metabolites analysis
The non-volatile metabolites were analysed using UHPLC Q Orbitrap HRMS (Thermo Fisher Scientific, Bremen, Germany).4,16 Sample preparation was carried out by lyophilizing and then grinding the green coffee beans. The ground green bean (8.25 g) was dissolved in 150 mL of boiling water. The solution was maintained at a temperature of 93 °C while stirring using a magnetic stirrer for 1 minute. The sample solution was then cooled down and filtered using Whatman filter paper number 1. It was stored in a freezer at a temperature of −22 °C before injection.The filtrates of ground green beans were diluted with LC-MS grade methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and filtered through a 0.22-µm PTFE membrane (Merck, KGaA, Darmstadt, Germany). A 5 µL volume of sample was injected onto an Accucore C18+ column (100 × 2
1, v/v) and filtered through a 0.22-µm PTFE membrane (Merck, KGaA, Darmstadt, Germany). A 5 µL volume of sample was injected onto an Accucore C18+ column (100 × 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mm, 1.5 µm) (Thermo Fisher Scientific, Bremen, Germany) at 40 °C. The mobile phases were LC-MS methanol (a) and 0.05% formic acid in LC-MS water (b). The flow rate was set at 0.2 mL min−1, and the gradient elution was 0 min (5% A), 0–17 min (90% A), 17–20 min (90% A), 20–23 min (5% A), and 23–30 min (5% A). Ionization mode was used in positive and negative electrospray ionization (ESI) with Q Orbitrap mass analysis. Analysis was performed at m/z 70–900, accompanied by UV detection at wavelengths of 220, 265, 272, and 320 nm.
1 mm, 1.5 µm) (Thermo Fisher Scientific, Bremen, Germany) at 40 °C. The mobile phases were LC-MS methanol (a) and 0.05% formic acid in LC-MS water (b). The flow rate was set at 0.2 mL min−1, and the gradient elution was 0 min (5% A), 0–17 min (90% A), 17–20 min (90% A), 20–23 min (5% A), and 23–30 min (5% A). Ionization mode was used in positive and negative electrospray ionization (ESI) with Q Orbitrap mass analysis. Analysis was performed at m/z 70–900, accompanied by UV detection at wavelengths of 220, 265, 272, and 320 nm.
Compound identification was performed by analysing raw data (chromatograms) using Compound Discoverer 3.2 software (Thermo Fisher Scientific) and comparing the data to online libraries (FoodDB, HMDB, NIST, Pubchem, and MzCloud). The mass spectra of the identified compounds were analysed using ThermoXcalibur 4.2 (Thermo Fisher Scientific). Identified compounds were screened using Annot (DeltaMass <5 ppm). Compound area data were normalized and displayed as heat maps.
Results and discussion
Microbial profile
The characteristics of reads and contigs in this study are shown in Table 1. Metagenomic datasets usually show 5–23% duplicate reads.17 The whole coffee cherry SIAF sample gave a slightly higher duplication rate, which could be caused by PCR amplification during library preparation. The optimum GC content for metagenomic analysis was 50%. Lower or higher GC content than 50% was caused by lower relative sequencing coverage.18| Sample name (filtered) | Read duplication rate (%) | GC read (%) | Read length (bp) | Reads (M) | Largest contig (bp) | Total length contig (bp) | N50 (bp) | N75 (bp) | L50 (contigs) | L75 (contigs) | GC contig (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a GC read is the guanine and cytosine content in the reads. The N50 and N75 were the lengths of the shortest contigs at 50% and 75% of the total assembly length. GC contig is the guanine and cytosine content in the contigs. The L50 and L75 were the counts of the shortest contigs whose length sum makes up 50% and 75% of the genome size. | |||||||||||
| Whole coffee cherries SIAF | 29.1 | 37 | 138 | 28.4 | 131![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 338 338 |
421![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 774 774![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 401 401 |
1303 | 780 | 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 512 512 |
190![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223 223 |
36.08 |
| Pulped coffee cherries SIAF | 18.3 | 40 | 132 | 26.3 | 290![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 715 715 |
382![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 692 692![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 454 454 |
1175 | 742 | 84![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 334 334 |
188![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 740 740 |
38.39 |
The N50 for metagenomic assembly is usually 20 kbp.19 A low N50 value might be due to low sequencing coverage, high diversity of microorganisms, and high genome complexity. Moreover, the default settings of the tools might not be optimal for the samples. However, a low N50 value was allowed for the diversity study since the contigs were aligned to the databases.
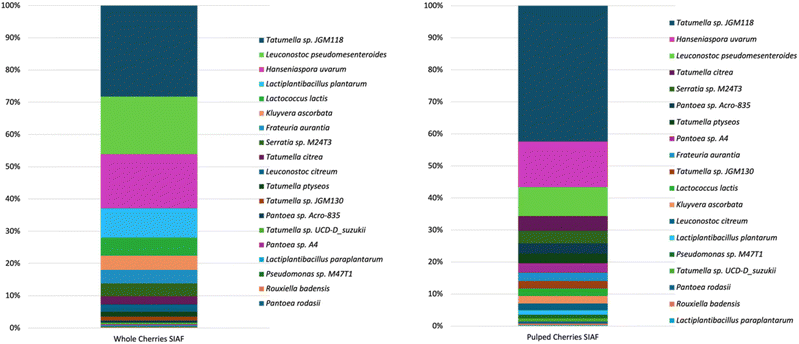
The microbial community of both samples showed low alpha diversity and no overlap for beta diversity. SIAF of the whole coffee was dominated by Tatumella sp. JGM118 (28%), Leuconostoc pseudomesenteroides (18%), and yeast Hanseniaspora uvarum (17%), respectively. The succeeding highest species were Lactiplantibacillus plantarum (9%) and Lactococcus lactis (6%), respectively. The subordinates were species with relative abundance less than 5%, which were Kluyvera ascorbata, Frateuria aurantia, and Serratia sp. M24T3, Tatumella citrea, Leuconostoc citreum, Tatumella ptyseos, Tatumella sp. JGM130, Pantoea sp. Acro-835, Tatumella sp. UCD-D_suzukii, Pantoea sp. A4, Lactiplantibacillus paraplantarum, Pseudomonas sp. M47T1, Rouxiella badensis, and Pantoea rodasii.
Meanwhile, the microbial community in the SIAF of pulped coffee was dominated by Tatumella sp. JGM118 (42%), Hanseniaspora uvarum (14%), and Leuconostoc pseudomesenteroides (9%), respectively. Tatumella citrea (5%) and Serratia sp. M24T3 (4%) were the following two most abundant species. The subordinates were species with relative abundance less than 4%, which were Pantoea sp. Acro-835, Tatumella ptyseos, Pantoea sp. A4, Frateuria aurantia, Tatumella sp. JGM130, Lactococcus lactis, Kluyvera ascorbate, Leuconostoc citreum, Lactiplantibacillus plantarum, Pseudomonas sp. M47T1, Tatumella sp. UCD-D_suzukii, Pantoea rodasii, Rouxiella badensis, and Lactiplantibacillus paraplantarum (Fig. 2).
The final temperature showed 20.2 °C for whole cherries SIAF and 21.2 °C for pulped cherries SIAF (SI Fig. 1). The temperature of whole coffee cherries and pulped coffee cherries SIAF during 5 days increased 1.5 °C and 2.5 °C, respectively. This led to the domination of mesophilic microbiota in the fermentation process. The pH of whole cherries SIAF started at 5.5 and finished at 4.15, while the pH of pulped cherries SIAF gradually decreased from 6.0 to 4.25 on the last day (SI Fig. 2).
The temperature increase is a good sign of proper SIAF process due to the exothermic reaction of sugar conversion to organic acids and alcohols.13 pH monitoring showed that the coffee samples were not over-fermented since pH 4 is the estimated condition of complete mucilage degradation.20,21 SIAF occurs spontaneously due to an epiphytic microbial community. Their fermentative activity is affected by intrinsic and extrinsic factors, which interfere with the quality of final coffee beverages.11
The low-level oxygen available in the SIAF bioreactors was utilized by yeast, Lactic Acid Bacteria (LAB), and facultative anaerobic bacteria like Tatumella. The symbiotic mutualism of yeast and LAB directly affects the aroma development of coffee drinks. Therefore, self-induced anaerobic fermented coffee is associated with improved sensory attributes. LAB delivers an acidic environment that is susceptible to yeast growth. At the same time, yeast provides soluble nitrogen compounds and vitamins that support the bacteria's growth.13
It was also recorded in a study from Brazil that Hanseniaspora uvarum, Lactiplantibacillus plantarum, and Leuconostoc mesenteroides dominated Arabica coffee SIAF at mesophilic conditions.11 The notable difference here was the domination of Tatumella sp. JGM118.Tatumella was included in the family of Erwiniaceae (Fig. 3). This family consists of several plant pathogens. Tatumella ptyesos, one of the species, was found to be the causative agent of pink disease in pineapple.12 However, as a member of the Enterobacterales order, it has been mentioned that Tatumella have the capacity to decompose sucrose under anaerobic conditions during coffee fermentation. The intermediates of sugar degradation were further utilized through glycolysis and the TCA cycle to produce reducing sugars and organic acids, substrates of amino acid metabolism, like fructose and other byproducts.22 These metabolites are the precursors of the Maillard reactions.23
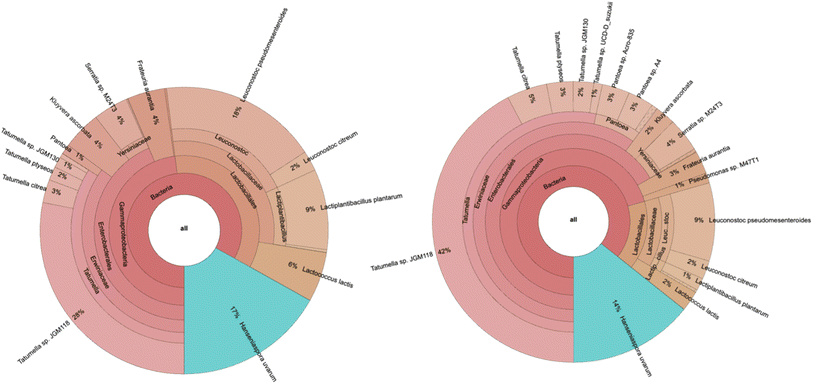 | ||
| Fig. 3 Microbial diversity of whole (left) and pulped (right) coffee cherries SIAF. The taxa showed species, genus, family, order, class and domain of the microbial community. | ||
Yeasts are the key players in coffee fermentation. They hold a crucial role in the development of aroma precursors in the fermented green beans. They hydrolyze macromolecules to produce reducing sugars, citric acid, amino acids, and chlorogenic acids.24 These metabolites were translocated into the beans and affected the coffee drink quality.20
Besides, yeast is a volatile compound producer. The non-Saccharomyces yeasts deliver esters, alcohols, and fatty acids.25 Hanseniaspora uvarum exhibited higher activity of pectinase, amylase, cellulase, and protease compared to other yeast species.26 This yeast prefers fructose and glucose rather than sucrose.21
The LAB was represented by Leuconostocaceae at the beginning of coffee fermentation. Further, the abundance of Lactobacillaceae increases at 48 and 72 h in SIAF of pulped coffee cherries.27 They are notable for being organic acid producers, especially lactic acid. Their activity is responsible for the degradation of coffee mucilage since pectic substances are soluble in organic acids. This acidification causes swelling of the mucilage cell wall.26 The present simple sugars were consumed first by them before polysaccharides, resulting in organic acids via glycolysis and pyruvate metabolism, producing D-lactate, ethanol, and CO2.
Frateuria aurantia28, Serratia sp.,29 and Pseudomonas sp. strain M47T1 (ref. 30) are beneficial for plants as a potassium-solubilizing bacterium, antifungal, and nematotoxic, respectively. While Pantoea rodasii31, Rouxiella badensis32, Pantoea sp. A4 (ref. 33) are plant pathogens, Kluyvera ascorbata is an emerging pathogen and a rare cause of urinary tract infection in immunocompetent patients.34 However, it will not have any effect on consumer health since high-temperature processes are applied before consumption. Aerobic groups of fungi that produce toxins like Aspergillus and Penicillium were not found in this anaerobic environment, although their spores may still survive.25 Temperature and humidity control during coffee drying and storage are therefore needed.
Aroma precursors
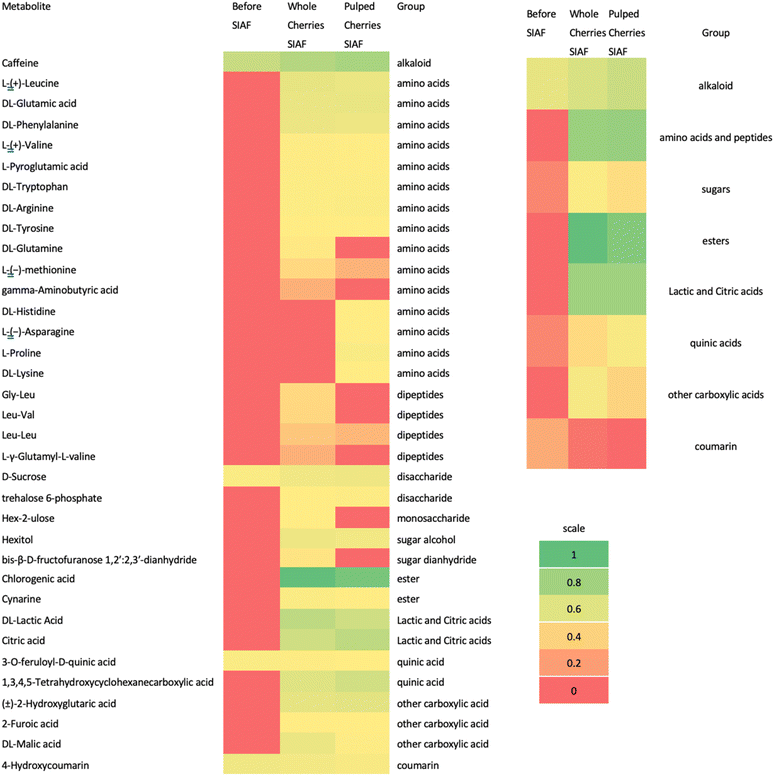
Analysis of the non-volatile metabolites using LC-MS showed that SIAF coffee is characterized by chlorogenic acid, carboxylic acid, and amino acids (Fig. 4). Before SIAF, these metabolites were not detected. Further, its concentration increased 7 to 10-fold after SIAF.Chlorogenic acid (CGA) was the dominant metabolite in the green beans from the SIAF process. CGA is an ester of caffeic acid and quinic acid. This metabolite is produced for plant protection.35 SIAF of whole cherries delivered a higher relative abundance of CGA compared to SIAF of pulped cherries, since CGA was concentrated in the mucilage. During the roasting process, this phenolic compound is degraded back into quinic and caffeic acid, phenol derivatives, and lactones.25
In both coffee SIAF samples, groups of amino acids and peptides, groups of lactic and citric acids, and caffeine from alkaloid groups were the second, third, and fourth highest relative abundance, respectively. Other metabolite groups, such as sugars, quinic acids, and coumarin, were also identified in the fermented green beans. Lactic and citric acids are formed during SIAF. The high abundance of citric acid in the coffee SIAF was due to the microbial community's high yield of Krebs.35
Coffee mucilage is a suitable substrate for microbial growth. It contains sugars like sucrose, as well as organic acids like citric and malic acids.13 During SIAF, microbial pectolytic enzymes catabolize pectin to release sugars. While microbial proteolytic enzymes also release amino acids and peptides.
Malic acid can be converted to lactic acid directly and citric acid through the TCA cycle.25 CO2 influences the anaerobic conditions where glycerol is produced. Glycerol is the precursor of phospholipids, which contribute to the formation of aldehydes and alcohols.13
Steps of the coffee processing interfered with the performance of the microbial community in coffee fermentation, although the calculated beta diversity was zero. Their performance was affected by the substrate and involved enzymes. The pulping process, for instance, changes the substrate composition and abundance. Therefore, the produced metabolites during SIAF may differ from the whole cherry sample.13
Marked by the red colour in the heatmap (Fig. 4), there were several amino acids and peptides that were produced in the pulped cherries SIAF sample but not in the whole cherries, and vice versa. Hex-2-ulose was not found in the pulped cherries SIAF sample. This could be affected by the partial pulp removal and its transformation into Hexitol. Bis-β-D-fructofuranose 1,2′:2,3′-dianhydride might also not be detected due to the pulping process.
Lactic acid in this study was found to be higher in the sample of whole cherries SIAF, compared to the pulped cherries SIAF. Without the pulping process, a high concentration of carbohydrates, the substrate of lactic acid, was still intact in the whole coffee cherries. Lactic acid is mainly produced by LABs, which favor glucose via glycolysis or pyruvate metabolism. It was relevant to the higher relative abundance of Lactobacillales in the whole cherries SIAF sample (35%) compared to the pulped cherries SIAF sample (15%). The microaerophilic condition inside SIAF bioreactors also brought an ideal environment for LAB.
Citric acid is mainly produced by the yeast.13 Pyruvate, as a result of sugar metabolism in the Krebs cycle, is converted into Acetyl-CoA and oxaloacetic acid, the precursors of citric acid. Citric acid was found to be higher in the pulped cherries, SIAF. Although a lower relative abundance of yeast was found in this sample, the relative abundance of Tatumella sp. JGM118 was higher.
Tatumella exhibited strong amylolytic activity.23 The degraded sugars could be used as substrates for the TCA cycle. The elimination of the coffee cherries' outer skin provided direct access for the microbial community to the mucilage, which delivered a variation of citric acid concentrations between the two samples. The pulping process also resulted in microbial enzyme alterations.
Citric acid contributes to sensory attributes of the coffee drink, such as citrus note, fruity, and floral, while lactic acid gives milky, creamy, and tartness.36 The highlighted sensory attributes of SIAF coffee beverages are citrus, honey, caramel, chocolate, and nuts. Malic acid's associated aroma is green fruit, like an apple. Coffee drink from pulped cherries SIAF has an aroma like cereal, nuts, and coconut, corroborating the lactic acid content. While coffee drinks from whole cherries, SIAF deliver notes of rum, watermelon, chocolate, caramel, and brown sugar.36
Proteins, polysaccharides, sugars, and chlorogenic acids are naturally occurring metabolites of coffee pulp.20 These metabolites are coffee aroma precursors. However, fermentation alters the content of these metabolites. The mentioned metabolites are further involved in the formation of volatile compounds such as pyrazines, furans, esters, ketones, aldehydes, and acids during coffee bean drying and roasting via a series of complex reactions. These volatiles eventually generate the aroma of the final coffee beverages.20
The series of complex reactions during roasting includes caramelization, pyrolysis, dehydration, hydrolysis, enolization, cyclization, polymerization, and, mainly, Maillard. Polysaccharides and sugars are the substrates of the caramelization reaction. Phenols and guaiacol are composed via thermal reactions from chlorogenic acids and phenolics. Precursors of Maillard reactions are free amino acids and reducing sugars. Both metabolites interact and generate pyrazines and pyridines, which give the roasted and nutty aroma in the coffee beverages. Pyrazines and pyrroles are also produced via pyrolysis of hydroxyl amino acids like threonine and serine.20
Fermentation also influences the aroma development in the coffee bean through the translocation of microbial metabolites produced in this process into the green beans from the mucilage.20 Chlorogenic acid, citric acid, malic acid, quinic acids, and other acids in the green beans not only contribute to the coffee acidity but also bring masking or synergistic effects to the coffee aroma. They create the cross-modal aroma-taste interaction in coffee through sensory perception.37 Moreover, bitter-tasting sub-qualities of coffee (caffeine, quinic acids, tryptophan, phenylalanine) also influence the retronasal perception of its aroma.38
Microbial enzymes
Coffee mucilage contains 47% wet basis of pectin 39. Carbohydrate content of the mucilage was measured to be sucrose (33.15 mg g−1), glucose (49.24 mg g−1), and fructose (76.26 mg g−1). These numbers are much higher than those for lipid (0.422 mg g−1) and protein (0.996 mg g−1).40 Therefore, carbohydrate-active enzymes were significantly important in the coffee fermentation.Since the coffee plant DNA had been removed using the bioinformatics analysis, only the ORF of microbial enzymes was further observed. Various ORFs of carbohydrate active enzymes (Cazy) were detected. Samples of whole cherries SIAF and pulped cherries SIAF distributed different variations and relative abundance of Cazymes ORF (Fig. 5). Coffee processing alters the microbial enzymes involved in coffee fermentation.
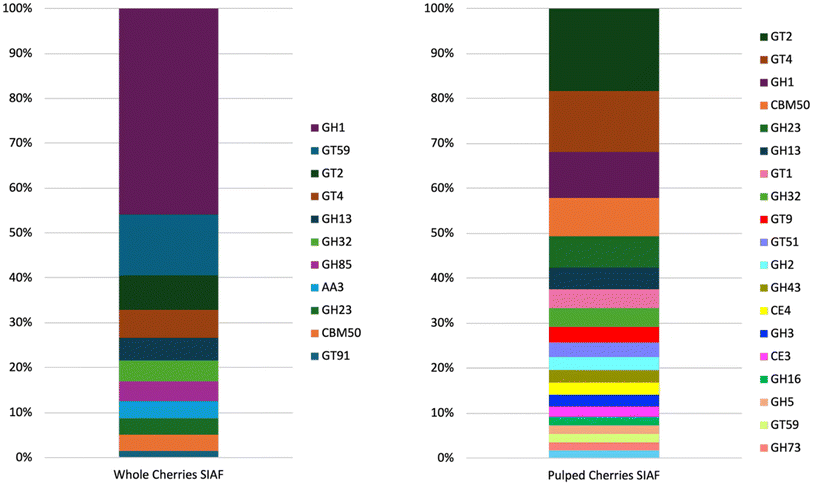 | ||
| Fig. 5 Carbohydrate-active enzymes detected in the whole (left) and pulped (right) coffee cherries SIAF samples. | ||
The main Cazymes that are directly involved in the degradation of carbohydrates are glycoside hydrolases (GH). While glycosyl transferases (GT) are involved in carbohydrate biosynthesis. Carbohydrate esterases (CE) are the ones that remove acyl- or alkyl-group from carbohydrates. Auxiliary Activities (AA) breakdown lignin and non-catalytic carbohydrate-binding modules (CBM) do not have enzyme activity.
In the whole cherry SIAF sample, the ORF of the GH1 family dominated with more than 45% relative abundance. The common enzyme activities of the GH1 family are β-galactosidases and β-glucosidases, which are possessed by Lactococcus lactis, Lactiplantibacillus plantarum, and Hanseniaspora uvarum.41,42 β-galactosidases are one of the pectinases.43 Pectinases break down pectin of the coffee mucilage to produce sugars, organic acids, and alcohols. Hexitol is developed via fructose and mannose metabolism. The activity of this enzyme was confirmed in this study as the sugar content increased in the green bean after SIAF.
The ORF of GH13 was the second most abundant GH family in the whole cherry sample. Based on the Cazy database, there are several Cazymes that are included in this group. Enzyme α-amylase is produced by Lactiplantibacillus plantarum, Pseudomonas sp, and Serratia sp. Amylopullulanase originates from Lactiplantibacillus plantarum. Oligo-α-1,6-glucosidase is associated with Lactiplantibacillus plantarum and yeast. Sucrose isomerase originates from Serratia sp. and sucrose phosphorylase from Leuconostoc mesenteroides. Enzyme α-glucosidase is possessed by yeast.
The most abundant ORFs of the GH families in this sample were GH32, GH85, and G23. Invertase, which breaks down sucrose into glucose and fructose, is included in the GH32 family and is produced by yeast. One of the GH85 family activities is endoglycosidase. An example of the GH23 is peptidoglycan lytic trans glycosylase, which is associated with Pseudomonas sp.
In the pulped cherries SIAF sample, the ORF of GH 1, GH23, GH13, GH32, GH2, GH43, GH3, GH16, GH5, and GH73 families dominated the glycoside hydrolases, respectively. β-galactosidases and β-glucosidases are also included in the GH2, GH3, and GH5 families since both hydrolyse beta-glycosidic bonds. The GH43 family exhibits activity like β-xylosidase or α-L-arabinofuranosidase. Activities of GH16 are reflected by β-porphyranase and β-1,3-glucanase, which are originated from Pseudomonas sp., and 1,6-glucanosyltransferase from yeast. N-acetylglucosaminidase or autolysin from Lactiplantibacillus plantarum and Lactococcus lactis is one of the activities of the GH73 family.
The enzymes that contribute to aroma development detected from the ORF in the coffee SIAF samples are shown in Table 2. The identified EC numbers of enzymes detected from the ORF were compared to the KEGG database to understand their molecular pathways and mechanisms related to coffee aroma development.
| EC number enzymes | Pathways | Metabolites | Reference | Predicted producers |
|---|---|---|---|---|
| a The microbial enzymes were detected from their open reading frames. Pathways were identified from the KEGG database. *Metabolites related to aroma development were detected by LC-MS/MS and matched to the enzymes based on the KEGG database. **Metabolites related to aroma development based on references and not detected by LC-MS/MS, and matched to the enzymes based on the KEGG database. Microbial producers were predicted based on the NCBI database. | ||||
| 2.6.1.42 Branched-chain-amino-acid transaminase | Valine, leucine, isoleucine metabolism | L-(+)-Leucine* | Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Tatumella ptyseos, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii | |
| 1.4.1.13 Glutamate synthase (NADPH) | Alanine, aspartate, and glutamate metabolism; histidine metabolism | DL-Glutamic acid* | Hanseniaspora uvarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Rouxiella badensis, Pantoea rodasii, Pseudomonas sp. | |
| 3.5.3.8 Formimidoylglutamase | Lactococcus lactis | |||
| 3.5.1.68 N-Formylglutamate deformylase | Rouxiella badensis, Pantoea rodasii, Pantoea sp., Serratia sp., Pseudomonas sp. | |||
| 3.5.2.9 5-Oxoprolinase (ATP-hydrolyzing) | Hanseniaspora uvarum | |||
| 3.5.1.2 Glutaminase | Hanseniaspora uvarum, Lactococcus lactis, Kluyvera ascorbata, Tatumella ptyseos, Tatumella sp., Rouxiella badensis, Pantoea rodasii | |||
| 2.6.1.9 Histidinol-phosphate transaminase | Phenylalanine metabolism | DL-Phenylalanine* | Hanseniaspora uvarum, Leuconostoc pseudomesenteroides, Kluyvera ascorbata, Lactiplantibacillus paraplantarum, Rouxiella badensis | |
| 2.6.1 Transaminases | Leuconostoc pseudomesenteroides, Lactiplantibacillus plantarum, Lactococcus lactis, Lactiplantibacillus paraplantarum | |||
| 2.6.1.57 Aromatic-amino-acid transaminase | Hanseniaspora uvarum, Lactococcus lactis, Kluyvera ascorbata, Tatumella sp., Pseudomonas sp., | |||
| 2.6.1.1 Aspartate transaminase | Tatumella sp. JGM118, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Frateuria aurantia, Pseudomonas sp. | |||
| 2.6.1.66 Valine-pyruvate transaminase | Valine, leucine, isoleucine metabolism | L-(+)-Valine* | Tatumella sp. JGM118, Kluyvera ascorbata, Tatumella sp. JGM130, Serratia sp., Pantoea sp. | |
| 2.6.1 Transaminases | Tryptophan metabolism | DL-Tryptophan* | Leuconostoc pseudomesenteroides, Lactiplantibacillus plantarum, Lactococcus lactis, Lactiplantibacillus paraplantarum | |
| 2.6.1.9 Histidinol-phosphate transaminase | Tyrosine metabolism | DL-Tyrosine* | Hanseniaspora uvarum, Leuconostoc pseudomesenteroides, Kluyvera ascorbata, Lactiplantibacillus paraplantarum, Rouxiella badensis | |
| 2.6.1 Transaminases | Leuconostoc pseudomesenteroides, Lactiplantibacillus plantarum, Lactococcus lactis, Lactiplantibacillus paraplantarum | |||
| 2.6.1.57 Aromatic-amino-acid transaminase | Hanseniaspora uvarum, Lactococcus lactis, Kluyvera ascorbata, Tatumella sp., Pseudomonas sp., | |||
| 2.6.1.1 Aspartate transaminase | Tatumella sp. JGM118, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Lactiplantibacillus paraplantarum, Frateuria aurantia, Pseudomonas sp. | |||
| 3.5.1.94 Gamma-glutamyl-gamma-aminobutyrate hydrolase | Alanine, aspartate, and glutamate metabolism; arginine and proline metabolism; butanoate metabolism | Gamma-aminobutyric acid* | Leuconostoc pseudomesenteroides, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Leuconostoc citreum, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii, Serratia sp., Pantoea sp. | |
| 2.6.1.19 4-Aminobutyrate-2-oxoglutarate transaminase | Lactiplantibacillus plantarum, Kluyvera ascorbata, Tatumella ptyseos, Rouxiella badensis, Pantoea rodasii, Pseudomonas sp. | |||
| 1.1.1.23 Histidinol dehydrogenase | Histidine metabolism | DL-Histidine* | Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Tatumella ptyseos, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii | |
| 3.4.13.9 Xaa-Pro dipeptidase | Amino acids metabolism | Proline* | Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii, Serratia sp. | |
| 3.4.13 Dipeptidases | Amino acids metabolism | Gly–Leu* | Tatumella sp. JGM118, Leuconostoc pseudomesenteroides, Lactiplantibacillus plantarum, Lactococcus lactis, Leuconostoc citreum, Tatumella ptyseos, Tatumella sp., Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii | |
| Leu–Val* | ||||
| Leu–Leu* | ||||
| L-γ-Glutamyl-L-valine* | ||||
| 1.1.1.289 Sorbose reductase | Fructose and mannose metabolism | Hexitol* | Hanseniaspora uvarum | |
| 1.1.1.14 L-iditol 2-dehydrogenase | Lactiplantibacillus plantarum, Lactiplantibacillus paraplantarum, Pseudomonas sp. | |||
| 1.1.1.27 L-lactate dehydrogenase | Glycolysis | Lactate* | Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Leuconostoc citreum, Tatumella ptyseos, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii | |
| 1.1.1.40 Malate dehydrogenase | Pyruvate metabolism | DL-Malic acid* | Tatumella sp. JGM118, Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Tatumella sp. JGM130, Rouxiella badensis, Pantoea rodasii, Serratia sp., Pantoea sp. | |
| 2.5.1.18 Glutathione transferase | Glutathione metabolism | Glutathione** | 45 | Tatumella sp. JGM118, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Tatumella ptyseos, Tatumella sp. JGM130, Rouxiella badensis, Pantoea rodasii, Serratia sp., Pantoea sp., Pseudomonas sp. Pantoea sp., Pseudomonas sp. |
| 3.4.11.2 Membrane alanyl aminopeptidase | Glutathione metabolism | Glycine** | 46 | Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Tatumella ptyseos, Lactiplantibacillus paraplantarum |
| 2.3.1.9 Acetyl-CoA C-acetyltransferase | Citrate cycle (TCA cycle) | Acetyl-CoA** | 47 | Hanseniaspora uvarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Pantoea rodasii, Pseudomonas sp. |
| 1.1.1.37 Malate dehydrogenase | Citrate cycle (TCA cycle) | Oxaloacetate** | 47 | Tatumella sp. JGM118, L. pseudomesenteroides, Hanseniaspora uvarum, L. plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Tatumella sp. JGM130, Rouxiella badensis, Pantoea rodasii, Serratia sp., Pantoea sp. |
| 1.1.1 Alcohol dehydrogenases | Glycolysis | Ethanol**, acetaldehyde**, aldehyde**, ketone** | 48,49 | Tatumella sp. JGM118, Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Tatumella sp. JGM130, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii, Serratia sp., Pantoea sp. |
| Pyruvate metabolism | Ethanol**, acetaldehyde** | 48,49 | ||
| 3.5.1.4 Amidase | Phenylalanine metabolism | Phenylacetate** | 50 | Tatumella sp. JGM118, Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Leuconostoc citreum, Tatumella ptyseos, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii, Pseudomonas sp. |
| Aminobenzoate degradation | Benzoate** | 51 | ||
| Styrene degradation | Acrylate** | 51 | ||
| 2.2.1.6 Acetolactate synthase | Butanoate metabolism | Acetolactate** | 52 | Tatumella sp. JGM118, Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Tatumella sp. JGM130, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii, Serratia sp., Pantoea sp., |
| 1.1.1.47 Glucose 1-dehydrogenase | Pentose phosphate pathway | D-glucono-1,5-lactone** | 20 | Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii, Serratia sp., Pantoea sp., Pseudomonas sp. |
| 1.1.1.6 Glycerol dehydrogenase | Glycerolipid metabolism | Glycerol** | 13 | Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Leuconostoc citreum, Tatumella ptyseos, Rouxiella badensis, Pantoea rodasii |
| 3.1.4.46 Glycerophosphodiester phosphodiesterase | Glycerophospholipid metabolism | Glycerol 3-phosphate** | 13 | Leuconostoc pseudomesenteroides, Hanseniaspora uvarum, Lactiplantibacillus plantarum, Lactococcus lactis, Kluyvera ascorbata, Frateuria aurantia, Leuconostoc citreum, Tatumella ptyseos, Tatumella sp. JGM130, Lactiplantibacillus paraplantarum, Rouxiella badensis, Pantoea rodasii, Serratia sp. |
Several of their metabolites were detected in the previous non-volatile metabolite analysis results. While the NCBI database was used to predict the producers of the microbial enzymes. The NCBI database provides information on the genomes and proteins (enzymes) of microorganisms. However, the registered genomes are incomplete genomes that were sequenced by Whole Genome Shotgun (WGS). Therefore, the microbial producers of aroma precursors can only be predicted. It is evident that all detected species in the samples participated in the production of metabolites related to aroma development.
Amino acids are produced via various amino acid metabolisms. Activities of proteolytic enzymes, such as transaminases and dipeptidases, were confirmed, as amino acids were produced in green beans after SIAF. Lactate is composed via glycolysis, and malate via pyruvate metabolism. The detected microbial L-lactate dehydrogenase in the samples explains that microorganisms involved in the SIAF process played an important role in the formation of lactic acid.
As of August 2025, Serratia sp. M24T3, Tatumella citrea, Pantoea sp. Acro-835, and Pseudomonas sp. M47T1 has not yet WGS-sequenced. Their ORF of enzymes is not available. The only available data are ORFs that are common in multiple species of their genus. Therefore, the predicted producers of several metabolites were mentioned as Serratia sp., Tatumella sp., Pantoea sp., and Pseudomonas sp.
The LAB belongs to Lactococcus, which distributes, proteolytic enzymes that enable the hydrolysis of peptides and caseins. This group also provides transaminases that degrade amino acids and transform them into alfa-ketoacids, which are precursors, of aldehydes. Lactiplantibacillus plantarum has tyrosine kinases.44 Moreover, L. lactis subsp. lactis delivers arginine dehydrolase, which releases ammonia from arginine. Hanseniaspora uvarum enhances the abundance of D-lysine, glutamine, isoleucyl, glycyl, prolyl, and alanyl dipeptides. However, they decrease L-tyrosine, L-phenylalanine, Leucyl, and Valyl di- and tripeptides. H. uvarum consumes intermediates of glycolysis and the Krebs cycle.41
Lactate dehydrogenase is possessed by Lactococcus and Lactiplantibacillus plantarum.44 This enzyme converts L-lactate from pyruvate. L. lactis subsp.lactis biovar. Lactobacillus diacetylactis can produce diacetyl from citrate, which brings a buttery aroma.53 Hanseniaspora uvarum possesses alcohol dehydrogenase.41,54
The aforementioned microorganisms were present during SIAF of whole and pulped coffee cherries in relatively high abundance (Fig. 2). The Lactococcus group in this study was predicted to produce transaminase, amino acid hydrolase, and dipeptidase. Lactiplantibacillus plantarum was found to have a transaminase that produces tyrosine. Hanseniaspora uvarum was predicted to have ORFs that produce glutamate synthase, glutaminase, and dipeptidases. Lactate dehydrogenase was indeed predicted to be produced by Lactococcus lactis, Lactiplantibacillus plantarum, and other microbes. Alcohol dehydrogenase was also predicted to be possessed by Hanseniaspora uvarum and other microorganisms. Therefore, LABs and yeasts deliver a significant contribution to the coffee aroma development (Table 2).
Enzymes responsible for producing several metabolites found in fermented green beans, such as caffeine, chlorogenic acid, and citric acid, were not detected. This explains that several metabolites in the coffee were naturally occurring or produced by plant enzymes. As chlorogenic acid was present in the SIAF green beans but not before the fermentation, it was assumed that chlorogenic acid from coffee pulp was not yet translocated to the beans, and the complex matrix was not yet degraded by microbes, so CGA could not be released. Nevertheless, enzymes which generate precursors of citric acid, Acetyl-CoA C-acetyltransferase for Acetyl-CoA and malate dehydrogenase for oxaloacetate, were identified.
Metabolites from the detected enzymes that were not available in the result of non-volatiles analysis could be volatiles, intermediates that had been further transformed during the fermentation process, or had not been translocated from mucilage into the green bean. Ethanol, acetaldehyde, aldehyde, and ketone from alcohol dehydrogenases are volatiles. Therefore, volatile analysis is needed for further investigation.
Glutathione could be further decomposed into glycine by the detected enzyme, aminopeptidase. Other metabolites that were not found in the fermented green beans might be transferred during coffee drying. Thus, metabolite analysis of dried green beans is suggested to gain a more comprehensive understanding.
Conclusions
Geothermal coffee processing, employing SIAF, can be an alternative method that addresses the limitations of conventional coffee processing. This study provided information on the role of microorganisms in the SIAF that affect the aroma development of final coffee beverages. The dominant species in the coffee SIAF samples was found to be Tatumella sp. JGM118, Hanseniaspora uvarum, and Leuconostoc pseudomesenteroides. The concentration of non-volatile metabolites such as chlorogenic acid, amino acids, and carboxylic acids, which were not detected in the green bean before SIAF, increased 7- to 10-fold after SIAF. These metabolites are aroma precursors of coffee drinks. The ORF of microbial enzymes that contribute to the coffee aroma development were identified in the samples, such as glycoside hydrolases (GH), transaminases, and L-lactate dehydrogenase. Therefore, the role of microbial community in SIAF as the producer of geothermal coffee aroma precursors was confirmed. Further, it is recommended to detect volatiles in the self-induced anaerobic fermented green bean samples. Metabolite analyses and sensory evaluation by Q-graders on the geothermal dried green beans are also suggested to get more coherent information.Author contributions
Zikrina Istighfarah, C. Hanny Wijaya, Lilis Nuraida, Erliza Noor, and Wisnu Ananta Kusuma designed the experimentation. Zikrina Istighfarah performed the experiments and wrote the original manuscript. C. Hanny Wijaya, Lilis Nuraida, Erliza Noor, and Wisnu Ananta Kusuma validated and reviewed the final manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting the findings of this study have been included in this article and as part of the supplementary information (SI). No restrictions apply to the availability of these data. Supplementary information is available. See DOI: https://doi.org/10.1039/d5fb00673b.Acknowledgements
This work was funded by the Indonesia Endowment Fund for Education (LPDP). Geothermal Coffee Process (GCP) Group and PT. PGE Area Kamojang is gratefully acknowledged for providing the coffee and research equipment at the coffee processing plant.References
- S. J. Zhang, F. De Bruyn, V. Pothakos, J. Torres, C. Falconi and C. Moccand, et al., Following Coffee Production from Cherries to Cup: Microbiological and Metabolomic Analysis of Wet Processing of Coffea Arabica, Appl. Environ. Microbiol., 2019, 85(6), e02635 CrossRef CAS.
- S. J. Zhang, F. De Bruyn, V. Pothakos, G. F. Contreras, Z. Cai and C. Moccand, et al., Influence of Various Processing Parameters on the Microbial Community Dynamics, Metabolomic Profiles, and Cup Quality During Wet Coffee Processing, Front. Microbiol., 2019, 10, 2621 CrossRef.
- L. H. B. P. de Sousa, J. M. R. da Luz, M. D. C. S. da Silva, A. P. Moreli, T. G. R. Veloso and R. C. Guarçoni, et al., Relationship between sensory and microbial profiles of fermented coffee by dry and washed methods, Food Chem. Adv., 2023, 2, 100259 CrossRef.
- Y. Yulianti, D. R. Adawiyah, D. Herawati, D. Indrasti and N. Andarwulan, Detection of Markers in Green Beans and Roasted Beans of Kalosi-Enrekang Arabica Coffee with Different Postharvest Processing Using LC-MS/MS, Int. J. Food Sci., 2023, 2023, 6696808 Search PubMed.
- F. De Bruyn, S. J. Zhang, V. Pothakos, J. Torres, C. Lambot and A. V. Moroni, et al., Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production, Appl. Environ. Microbiol., 2017, 83(1), e02398 CrossRef CAS PubMed.
- E. Firdissa, A. Mohammed, G. Berecha and W. Garedew, Coffee Drying and Processing Method Influence Quality of Arabica Coffee Varieties (Coffee arabica L.) at Gomma I and Limmu Kossa, Southwest Ethiopia, J. Food Qual., 2022, 2022, 1–8 CrossRef.
- Y. Gunawan, K. T. Margono, R. Rizky, N. Putra, R. A. Faqih and I. I. Hakim, et al., Enhancing the performance of conventional coffee beans drying with low-temperature geothermal energy by applying HPHE: An experimental study, Open Agric., 2021, 6(1), 807–818 CrossRef.
- Y. Gunawan, N. Putra, I. I. Hakim, D. Agustina and T. M. I. Mahlia, Withering of tea leaves using heat pipe heat exchanger by utilizing low-temperature geothermal energy, Int. J. Low-Carbon Technol., 2021, 16(1), 146–155 CrossRef CAS.
- C. U. Farouq, V. A. Brilian, A. N. Fathurrodin, M. I. Pradipta, P. P. Dewi and A. Rahmadi, et al., Technical and Business Evaluation of Geothermal Dryhouse for Coffee in Kamojang, in, IOP Conference Series: Earth and Environmental Science, Institute of Physics, West Java, Indonesia, 2024 Search PubMed.
- P. Pittia, M. C. Nicoli and G. Sacchetti, Effect of moisture and water activity on textural properties of raw and roasted coffee beans, J. Texture Stud., 2007, 38(1), 116–134 CrossRef.
- T. S. Pereira, N. N. Batista, L. P. Santos Pimenta, S. J. Martinez, L. S. Ribeiro and J. A. Oliveira Naves, et al., Self-induced anaerobiosis coffee fermentation: Impact on microbial communities, chemical composition and sensory quality of coffee, Food Microbiol., 2022, 103, 103962 CrossRef CAS.
- V. Marín-Cevada, J. Caballero-Mellado, R. Bustillos-Cristales, J. Muñoz-Rojas, M. A. Mascarúa-Esparza and M. Castañeda-Lucio, et al., Tatumella ptyseos, an unrevealed causative agent of pink disease in pineapple, J. Phytopathol., 2010, 158(2), 93–99 CrossRef.
- M. C. Batista da Mota, N. N. Batista, D. R. Dias and R. F. Schwan, Impact of microbial self-induced anaerobiosis fermentation (SIAF) on coffee quality, Food Biosci., 2022, 47 CAS.
- M. Turan Ayseli and I. Coskun, Analyzing aroma-active compounds of traditional herbal coffee (Mırra) using the headspace-solid phase microextraction method combined with GC–MS/O, Microchem. J., 2024, 205 CrossRef CAS.
- J. J. Schenk, L. E. Becklund, S. J. Carey and P. P. Fabre, What is the “modified” CTAB protocol? Characterizing modifications to the CTAB DNA extraction protocol, Appl. Plant Sci., 2023, 11(3), e11517 CrossRef CAS.
- M. R. Fernández-Alduenda and G. Peter, Coffee Sensory and Cupping Handbook, Speciality Coffee Association, 2021, p. 130 Search PubMed.
- B. Niu, L. Fu, S. Sun and W. Li, Artificial and natural duplicates in pyrosequencing reads of metagenomic data, BMC Bioinf., 2010, 11, 187 Search PubMed.
- P. D. Browne, T. K. Nielsen, W. Kot, A. Aggerholm, M. T. P. Gilbert and L. Puetz, et al., GC bias affects genomic and metagenomic reconstructions, underrepresenting GC-poor organisms, Gigascience, 2020, 9(2), giaa008 CrossRef CAS.
- R. Eisenhofer, J. Nesme, L. Santos-Bay, A. Koziol, S. J. Sørensen and A. Alberdi, et al., A comparison of short-read, HiFi long-read, and hybrid strategies for genome-resolved metagenomics, Microbiol. Spectrum, 2024, 12(4), e03590 CrossRef PubMed.
- H. Elhalis, J. Cox and J. Zhao, Coffee fermentation: Expedition from traditional to controlled process and perspectives for industrialization, Appl. Food Res., 2023, 3, 100253 Search PubMed.
- S. Voce, P. Passaghe and P. Comuzzo, Use of Hansensiaspora uvarum and managing growth conditions increase polysaccharides and antioxidants content in yeast autolysates for winemaking, LWT–Food Sci. Technol., 2024, 203 CrossRef CAS.
- M. Xiao, T. Huang, Y. Xu, Z. Peng, Z. Liu and Q. Guan, et al., Metatranscriptomics reveals the gene functions and metabolic properties of the major microbial community during Chinese Sichuan Paocai fermentation, Food Microbiol., 2021, 98 CAS.
- B. H. Lee, C. H. Huang, T. Y. Liu, J. S. Liou, C. Y. Hou and W. H. Hsu, Microbial Diversity of Anaerobic-Fermented Coffee and Potential for Inhibiting Ochratoxin-Produced Aspergillus niger, Foods, 2023, 12, 2967 Search PubMed.
- C. E. Góngora, L. Holguín-Sterling, B. Pedraza-Claros, R. Pérez-Salinas, A. Ortiz and L. Navarro-Escalante, Metataxonomic Identification of Microorganisms during the Coffee Fermentation Process in Colombian Farms (Cesar Department), Foods, 2024, 13(6), 839 Search PubMed.
- S. J. Martinez, N. N. Batista, A. P. P. Bressani, D. R. Dias and R. F. Schwan, Molecular, Chemical, and Sensory Attributes Fingerprinting of Self-Induced Anaerobic Fermented Coffees from Different Altitudes and Processing Methods, Foods, 2022, 11(24), 3945 CrossRef CAS.
- X. Shen, Q. Wang, H. Wang, G. Fang, Y. Li and J. Zhang, et al., Microbial Characteristics and Functions in Coffee Fermentation: A Review, Fermentation, 2025, 11 Search PubMed.
- A. da Silva Vale, G. Balla, L. R. S. Rodrigues, D. P. de Carvalho Neto, C. R. Soccol and G. V. de Melo Pereira, Understanding the Effects of Self-Induced Anaerobic Fermentation on Coffee Beans Quality: Microbiological, Metabolic, and Sensory Studies, Foods, 2023, 12(1), 37 CrossRef CAS.
- D. V. Subhashini, Growth Promotion and Increased Potassium Uptake of Tobacco by Potassium-Mobilizing Bacterium Frateuria aurantia Grown at Different Potassium Levels in Vertisols, Commun. Soil Sci. Plant Anal., 2015, 46(2), 210–220 CrossRef CAS.
- A. Khanna, M. Khanna and A. Aggarwal, Serratia marcescens- a rare opportunistic nosocomial pathogen and measures to limit its spread in hospitalized patients, J. Clin. Diagn. Res., 2013, 7(2), 243–246 Search PubMed.
- R. K. Aziz, D. Bartels, A. Best, M. DeJongh, T. Disz and R. A. Edwards, et al., The RAST Server: Rapid annotations using subsystems technology, BMC Genomics, 2008, 9, 75 CrossRef PubMed.
- C. L. Brady, I. Cleenwerck, L. van der Westhuizen, S. N. Venter, T. A. Coutinho and P. De Vos, Pantoea rodasii sp. nov., Pantoea rwandensis sp. nov. and Pantoea wallisii sp. nov., isolated from Eucalyptus, Int. J. Syst. Evol. Microbiol., 2012, 62(7), 1457–1464 CrossRef CAS PubMed.
- M. Zhao, C. Tyson, R. Gitaitis, B. Kvitko and B. Dutta, Rouxiella badensis, a new bacterial pathogen of onion causing bulb rot, Front. Microbiol., 2022, 30, 13 Search PubMed.
- K. C. Crosby, M. Rojas, P. Sharma, M. A. Johnson, R. Mazloom and B. H. Kvitko, et al., Genomic delineation and description of species and within-species lineages in the genus Pantoea, Front. Microbiol., 2023, 14, 1254999 CrossRef.
- M. Akiki, R. Ali, A. Jamil, J. Slim and R. Miller, Kluyvera ascorbata: An Unusual Cause of Septic Shock in a Patient With Urothelial Cancer, Cureus, 2023, 15(12), e51057 Search PubMed.
- S. J. Martinez, A. P. P. Bressani, J. B. P. Simão, V. S. Pylro, D. R. Dias and R. F. Schwan, Dominant microbial communities and biochemical profile of pulped natural fermented coffees growing in different altitudes, Food Res. Int., 2022, 1, 159 Search PubMed.
- L. C. F. Silva, P. V. R. Pereira, M. A. D. D. Cruz, G. X. R. Costa, R. A. R. Rocha and P. L. L. Bertarini, et al., Enhancing Sensory Quality of Coffee: The Impact of Fermentation Techniques on Coffea arabica cv. Catiguá MG2, Foods, 2024, 13(5), 653 CrossRef CAS PubMed.
- C. J. B. Rune, D. Giacalone, I. Steen, L. Duelund, M. Münchow and M. P. Clausen, Acids in brewed coffees: Chemical composition and sensory threshold, Curr. Res. Food Sci., 2023, 6, 100485 CrossRef.
- L. Paravisini, A. Soldavini, J. Peterson, C. T. Simons and D. G. Peterson, Impact of bitter tastant sub-qualities on retronasal coffee aroma perception, PLoS One, 2019, 14(10), e0223280 CrossRef.
- M. Haile and W. H. Kang, The Role of Microbes in Coffee Fermentation and Their Impact on Coffee Quality, J. Food Qual., 2019, 2019, 4836709 CrossRef.
- L. J. C. Ferreira, I. N. Casé, P. L. L. Bertarini, L. M. D. Oliveira and L. D. Santos, Impact of immature coffee fruits and water addition during spontaneous fermentation process: Chemical composition and sensory profile, Electron. J. Biotechnol., 2024, 69, 21–29 CrossRef.
- J. Wang, Z. Wang, H. Gao, X. Bai, L. Li and R. Wei, et al., Metabolomics and flavor diversity in Cabernet Sauvignon wines fermented by various origins of Hanseniaspora uvarum in the presence and absence of Saccharomyces cerevisiae, LWT–Food Sci. Technol., 2024, 203 Search PubMed.
- L. Plaza-Vinuesa, A. Sánchez-Arroyo, F. J. Moreno, B. de las Rivas and R. Muñoz, Dual 6Pβ-Galactosidase/6Pβ-Glucosidase GH1 Family for Lactose Metabolism in the Probiotic Bacterium Lactiplantibacillus plantarum WCFS1, J. Agric. Food Chem., 2023, 71(28), 10693–10700 Search PubMed.
- H. A. Ruiz, R. M. Rodríguez-Jasso, A. Hernandez-Almanza, J. C. Contreras-Esquivel and C. N. Aguilar, Pectinolytic Enzymes, in, Current Developments in Biotechnology and Bioengineering: Production, Isolation and Purification of Industrial Products, Elsevier Inc., 2016, pp. 47–71 Search PubMed.
- N. Garcia-Gonzalez, N. Battista, R. Prete and A. Corsetti, Health-promoting role of lactiplantibacillus plantarum isolated from fermented foods, Microorganisms, 2021, 9, 1–30 Search PubMed.
- J. Zhao, T. Wang, J. Xie, Q. Xiao, J. Cheng and F. Chen, et al., Formation mechanism of aroma compounds in a glutathione-glucose reaction with fat or oxidized fat, Food Chem., 2019, 270, 436–444 Search PubMed.
- L. Poisson, I. Blank, A. Dunkel and T. Hofmann, The Chemistry of Roasting-Decoding Flavor Formation, in, The Craft and Science of Coffee, Elsevier Inc., 2017, pp. 273–309 Search PubMed.
- D. K. A. do Rosário, Y. da Silva Mutz, K. M. Vieira, R. F. Schwan and P. C. Bernardes, Effect of self-induced anaerobiosis fermentation (SIAF) in the volatile compounds and sensory quality of coffee, Eur. Food Res. Technol., 2024, 250(2), 667–675 CrossRef.
- G. V. de Melo Pereira, V. T. Soccol, A. Pandey, A. B. P. Medeiros, L. J. M. R. Andrade and A. L. Gollo, et al., Isolation, selection and evaluation of yeasts for use in fermentation of coffee beans by the wet process, Int. J. Food Microbiol., 2014, 188, 60–66 CrossRef PubMed.
- S. R. Evangelista, M. G. da Cruz Pedrozo Miguel, C. de Souza Cordeiro, C. F. Silva, A. C. Marques Pinheiro and R. F. Schwan, Inoculation of starter cultures in a semi-dry coffee (Coffea arabica) fermentation process, Food Microbiol., 2014, 44, 87–95 CrossRef PubMed.
- G. V. de Melo Pereira, E. Neto, V. T. Soccol, A. B. P. Medeiros, A. L. Woiciechowski and C. R. Soccol, Conducting starter culture-controlled fermentations of coffee beans during on-farm wet processing: Growth, metabolic analyses and sensorial effects, Food Res. Int., 2015, 75, 348–356 CrossRef PubMed.
- Y. Feng and A. Zhang, A Floral Fragrance, Methyl Benzoate, is An Efficient Green Pesticide, Scientific Reports, 2017, 7 Search PubMed.
- T. Zhao, Y. Li, S. Yuan, Y. Ye, Z. Peng and R. Zhou, et al., Structure-Based Design of Acetolactate Synthase From Bacillus licheniformis Improved Protein Stability Under Acidic Conditions, Front. Microbiol., 2020, 11 Search PubMed.
- K. Kondrotiene, P. Zavistanaviciute, J. Aksomaitiene, A. Novoslavskij and M. Malakauskas, Lactococcus lactis in Dairy Fermentation—Health-Promoting and Probiotic Properties, Fermentation, 2024, 10 Search PubMed.
- A. K. Langenberg, F. J. Bink, L. Wolff, S. Walter, C. Von Wallbrunn and M. Grossmann, et al., Glycolytic Functions Are Conserved in the Genome of the Wine Yeast Hanseniaspora uvarum, and Pyruvate Kinase Limits Its Capacity for Alcoholic Fermentation, Appl. Environ. Microbiol., 2017, 83(22), e01580 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |