Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Serum-free media drive lipid accumulation in porcine satellite cells intended for cultivated meat
Anupam Abraham ,
Marc Auguet-Lara,
Stig Skrivergaard,
Margrethe Therkildsen,
Martin Krøyer Rasmussen
,
Marc Auguet-Lara,
Stig Skrivergaard,
Margrethe Therkildsen,
Martin Krøyer Rasmussen and
Jette Feveile Young
and
Jette Feveile Young *
*
Department of Food Science, Aarhus University, AgroFood Park 48, Aarhus, Denmark. E-mail: jettef.young@food.au.dk
First published on 2nd January 2026
Abstract
Fat is a key contributor to the sensory qualities of meat, particularly flavour. In the context of cultivated meat, enhancing lipid accumulation in muscle satellite cells (SCs) may improve the organoleptic properties of the final product. This study investigated the effects of serum concentration and fatty acid supplementation on lipid accumulation in porcine SCs and differentiated myotubes. Cells cultured in reduced-serum media (2% horse serum or 2% fetal bovine serum) and a custom serum-free medium (SFM) exhibited significantly higher lipid accumulation upon exposure to oleic acid compared to those maintained in standard growth medium (10% fetal bovine serum and 10% horse serum). Notably, cells in SFM retained lipid droplets for extended periods relative to those in serum-containing media. Furthermore, supplementation with unsaturated and polyunsaturated fatty acids such as linolenic, oleic, and erucic acids induced greater lipid droplet formation than saturated or branched-chain fatty acids, including palmitic acid.
Sustainability spotlightCultivated meat presents an opportunity to reduce the environmental footprint of meat production. In conventional meat, fat is essential for attributes such as taste and texture, whereas cultivated meat typically has low lipid levels. Replacing or reducing fetal bovine serum with serum-free media is a crucial step towards making this technology more sustainable. This study shows that such media, combined with specific fatty acid supplementation, can enhance lipid accumulation within cultivated pork muscle cells by inducing lipids directly in muscle tissue. Monounsaturated and polyunsaturated C18 fatty acids were particularly effective in increasing lipid content, helping address the limitation of low lipid levels and supporting the development of products with a composition more comparable to conventional meat. |
1. Introduction
Cultivated meat offers a new and sustainable approach to producing environmentally friendly meat,1 providing essential nutrients similar to those found in traditional meat. This technology relies on the ability of stem cells to multiply in a suitable environment, not only following the myogenic lineage, but also to transform into different cell types, such as osteoblasts, myofibroblasts, or adipocytes.2 While much of the research in cultivated meat has focused on reducing production costs by developing cell lines3,4 and supporting biotechnologies,5,6 it is equally important to address the sensory quality of the final product to meet consumer demands and expectations. Therefore, early-stage research and development should also concentrate on enhancing the taste of cultured meat.7 Pork holds a significant position in the meat industry due to its long-standing history of consumption in various forms, resulting in high demand.8 Despite recent advances in cultivated pork muscle and fat research,9 optimising intramyocellular fat deposition within porcine muscle cells remains underexplored.Consumers associate the quality of meat products with characteristics such as tenderness, juiciness, flavour, and colour.10 Fat plays a significant role in juiciness, texture, flavour and water-holding capacity, and adipocytes in meat can be found in intramuscular or intermuscular depots, with marbling fat located between muscle fibres in a muscle bed,11 in traditional meat, the development of a meaty flavour results from post-mortem metabolic processes such as pH reduction, protein and lipid oxidation, as well as from reactions that occur during cooking.12,13 During cooking, aroma is developed through the interaction between small metabolites, including amino acids, sugars, and fat precursors, via the Maillard reaction, lipid oxidation, thiamine degradation, and the interaction between Maillard reaction products and lipid oxidation products14 This highlights the importance of incorporating fat in cultivated meat products for the flavour of the final product. However, in the case of cultivated meat, the lipid content is low compared to conventional meat.15–17
Various approaches have been proposed to increase the fat content in cultivated meat systems. These include adding separately cultured fat cells to the muscle cell culture product,18 transdifferentiating muscle cells into adipocytes,19–21 co-culturing muscle cells and adipose cells22,23 or adding fat into the final product. While successful isolation and separate culturing of satellite cells (SCs)24,25 and adipocytes26 have been achieved, co-culturing adipocytes and muscle SCs has proven challenging, as shown by Takegahara et al.,27 who reported that co-culture of C2C12 myoblasts with 3T3-L1 adipocytes led to suppressed myotube formation, likely due to interference from adipogenic factors and difficulty in maintaining a stable adipocyte phenotype in the presence of myoblasts. Producing adipocytes from progenitor cells typically requires a continuous supply of free fatty acids and chemicals that are not food-compatible, such as 3-isobutyl-1-methylxanthine (IBMX) and thiazolidinediones like rosiglitazone.19,28
In cultivated meat systems, intramuscular fat can be considered equivalent to intramyocellular lipids (IMCL), contributing to processability and flavour, like marbling fat in traditional meat.29 Previous research has demonstrated that adding fatty acids like oleic or palmitoleic acid to the culture media facilitates the accumulation of lipid droplets in bovine satellite cell cultures and other cell lines.26,30–34 Different types of fatty acids, including monounsaturated32,35 polyunsaturated,34,36,37 and branched-chain fatty acids38 have been used to induce lipid accumulation or adipogenesis in different cell types. However, research investigating the role of different types of fatty acids in inducing lipid droplets in porcine SCs is lacking. Various concentrations and types of serum and basal media have been reported for adipogenic differentiation, including 2–3% fetal bovine serum (FBS),39,40 2% horse serum (HS),19,41 and serum-free medium (SFM).42 Additionally, different basal media have also been employed, most commonly Dulbecco's Modified Eagle Medium (DMEM)40,43–45 and DMEM/F12.40,46–48 However, studies on the optimal media type and serum concentration for lipid accumulation remain scarce.
This study investigated lipid droplet accumulation in porcine SCs at distinct developmental stages: proliferating SCs and differentiated myotubes under varying serum conditions. Specifically, we compared the effects of FBS, HS, and no serum on lipid storage. Additionally, we assessed the influence of different classes of fatty acids, including saturated, unsaturated, long-chain, and short-chain fatty acids, on lipid accumulation in these cells.
2. Experimental procedures
2.1 Satellite cell culture
The SCs were isolated from the muscles of three male pigs (approximately 20 kg body weight, Duroc-Danish Landrace-Yorkshire crossbreed), sacrificed at the Department of Animal and Veterinary Science facility, Aarhus University, Denmark, by authorised personnel. Muscle tissue was removed from the semimembranosus muscle (approximately 100 g) right after slaughter and transported to the cell laboratory, Department of Food Science, Aarhus University, in a 50 mL falcon tube kept on ice in transport solution containing Dulbecco's phosphate buffered saline (DPBS, without Ca2+ and Mg2+, Gibco-14190144) supplemented with 1% w/v D-glucose (Sigma-G7021), gentamicin 0.2 mg mL−1 (Sigma-G1397), penicillin 200 units per mL and streptomycin 0.2 mg mL−1 (Sigma-P4333), and amphotericin B 5 µg mL−1 (Sigma-A2942). SC isolation was initiated within 2 hours post-mortem, as reported by Skrivergaard et al.25 Briefly, five grams of muscle tissue devoid of visible fat and connective tissue was minced using sterile scissors in transport solution and subjected to enzymatic digestion for 1 hour at 37 °C in a 50 mL sterile falcon tube using 20 mL of prewarmed digest solution (427.5 U mL−1 of collagenase type II (Worthington-LS004177), 0.01% DNase (Sigma-DN25), 0.25% v/v Trypsin (Gibco-15090) and 1% w/v D-glucose in DPBS). The tubes were inverted every five minutes, and post-incubation vigorous trituration was performed using 25 mL serological pipettes. Subsequently, the tubes were centrifuged at 100×g for 10 seconds at 4 °C. The supernatant was collected in a new 50 mL Falcon tube containing ice-cold growth medium (GM) (DMEM-Glutamax, Gibco-61965-026, supplemented with 10% v/v FBS (Gibco-A5669801), 10% v/v HS (Gibco-26050088), 1 mM sodium pyruvate (Gibco-11360070), 0.1 mg mL−1 gentamicin, amphotericin B 2.5 µg mL−1, penicillin 100 units per mL and streptomycin 0.1 mg mL−1) on ice. The cell supernatant was centrifuged at 1000×g for 10 minutes at 4 °C, and the pellet was resuspended in ice-cold GM.After centrifugation of the first digestion, the tissue pellet was subjected to a second digestion, following the same procedure. Both cell suspensions were then pooled. The pooled suspension was subjected to sequential straining to remove tissue debris by passing the solution through a 100 µm cell strainer (Corning-431752) followed by a 40 µm cell strainer (Corning-431750). The filtrate was then centrifuged at 1000×g for 10 minutes at 4 °C, and the cell pellet was resuspended in 30 mL of GM. To remove fibroblasts from the cell population, the cell suspension was pre-plated three times for 1 hour at 37 °C in a humidified 5% CO2 environment. The media with unattached SCs was then pelleted by centrifugation at 1000×g for 10 minutes at 4 °C, a modified method from Yoshioka et al.49 The SC pellet was resuspended in 10 mL of GM containing 10% v/v dimethyl sulfoxide (DMSO, Sigma-D2650) and then cryopreserved.
Cryopreserved SCs were thawed quickly and, after dilution with GM, subjected to centrifugation at 1000×g for 5 minutes at 4 °C. The pellet was resuspended in GM and cultured in a 6-well plate (Nunc, Thermo Scientific-140675) coated with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 diluted Matrigel Matrix (Corning, 356237) in DMEM/F-12 (Gibco-11330032). Trypsin–EDTA (0.05% Trypsin–EDTA, Gibco-25300062) solution was used to detach the cells for passaging. SCs were passaged every 3–4 days at 80% confluency. For the experiment, cells were seeded in 96-well (Nunc, ThermoScientific-167008) plates coated with Matrigel Matrix at densities of 2500 cells per well. SC from each biological replicate (n) were seeded in triplicate or duplicate wells (technical replicates).
50 diluted Matrigel Matrix (Corning, 356237) in DMEM/F-12 (Gibco-11330032). Trypsin–EDTA (0.05% Trypsin–EDTA, Gibco-25300062) solution was used to detach the cells for passaging. SCs were passaged every 3–4 days at 80% confluency. For the experiment, cells were seeded in 96-well (Nunc, ThermoScientific-167008) plates coated with Matrigel Matrix at densities of 2500 cells per well. SC from each biological replicate (n) were seeded in triplicate or duplicate wells (technical replicates).
2.2 Satellite cell differentiation and myotube formation
To assess the myogenic potential, SCs at passage 5 were grown in Matrigel-coated 96-well plates in GM. After reaching confluence, the medium was replaced with 2% HS, maintaining the same basal medium and antibiotics as in GM, with only the serum composition altered. The cells were then cultured for 12 days in 96-well plates. Cells in the 96-well plates were stained for F-actin using Phalloidin-AF647 (Thermo Scientific, A22287) and for nuclei using Hoechst 33342 (Invitrogen, H3570, 5 mL), following the method described by Skrivergaard et al. (2023).50 Imaging was performed using a BioTek Cytation 5 imager with Gen 5 (3.11) software. The DAPI filter (excitation ≈ 377 nm, emission ≈ 447 nm) was used to visualise Hoechst-stained nuclei, and the Texas Red filter (excitation ≈ 586 nm, emission ≈ 647 nm) was used to image Phalloidin-AF647-stained F-actin (SI Fig. 1C and D).2.3 Fatty acid conjugations
All fatty acids (FAs; Sigma Aldrich) were solubilised in ethanol (99.8%, Sigma) at a concentration of 50 mM and stored in glass vials at −20 °C. FAs were conjugated with bovine serum albumin (BSA, 10% in DPBS, FA-free, Sigma-A1595) according to Pappas et al., (2002)51 on the day of the experiment. In short, FAs were added to the BSA solution stepwise by adding small volumes at 37 °C (90 °C for saturated FAs) and diluted with deionised water. The final concentration of FA in the FA-BSA conjugate was 5 mM. The FA-BSA conjugate was mixed with the appropriate media used for cell treatment at the final concentration and incubated at 37 °C for 1 hour, with manual shaking at 10 minutes intervals before adding it to SCs.2.4 Quantification of lipid accumulation
We quantified only neutral lipids, which are in cytoplasmic lipid droplets. SCs were washed twice with DPBS, fixed for 15 minutes using 10% neutral buffered formalin, and washed three times with DPBS. The cells were then stained for lipid droplets with BODIPY 493/503 (2 µg mL−1 in DPBS, Invitrogen) and incubated for 30 minutes at room temperature, followed by three washes with DPBS, as described by Grandl and Schmitz.52 Subsequently, nuclei were stained by Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
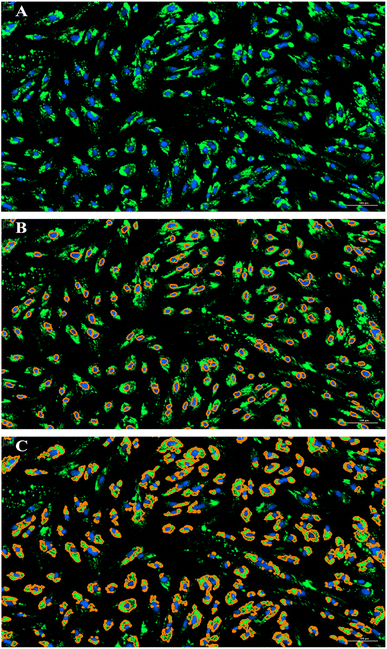
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in DPBS, Invitrogen) and incubated for 15 minutes at room temperature. The cells were imaged (4× and 20× magnification) using a Biotek Cytation 5 imager (Biotek) (SI Fig. 1A and B). Primary and secondary mask analysis of the images (4× objective) was used to quantify lipid droplets using Gen5 3.11 Image Prime software (Biotek). Using a primary mask, nuclei (DAPI channel) were identified, and counted and the secondary mask (GFP channel) was designated as an area up to 30 µm surrounding the nuclei, representing a cytoplasmic region having lipid droplets (Fig. 1). For each experiment, image acquisition settings (including exposure time and gain) for BODIPY fluorescence were optimized at the beginning and then kept constant across all plates within that experiment to ensure comparability. While settings varied slightly between different experiments, the same parameters were applied consistently across all wells and replicates within each experiment series. Mean green fluorescence intensity values (GFP channel) from the secondary mask were used as a proxy for neutral lipid content, with higher values indicating greater lipid droplet accumulation.
1000 in DPBS, Invitrogen) and incubated for 15 minutes at room temperature. The cells were imaged (4× and 20× magnification) using a Biotek Cytation 5 imager (Biotek) (SI Fig. 1A and B). Primary and secondary mask analysis of the images (4× objective) was used to quantify lipid droplets using Gen5 3.11 Image Prime software (Biotek). Using a primary mask, nuclei (DAPI channel) were identified, and counted and the secondary mask (GFP channel) was designated as an area up to 30 µm surrounding the nuclei, representing a cytoplasmic region having lipid droplets (Fig. 1). For each experiment, image acquisition settings (including exposure time and gain) for BODIPY fluorescence were optimized at the beginning and then kept constant across all plates within that experiment to ensure comparability. While settings varied slightly between different experiments, the same parameters were applied consistently across all wells and replicates within each experiment series. Mean green fluorescence intensity values (GFP channel) from the secondary mask were used as a proxy for neutral lipid content, with higher values indicating greater lipid droplet accumulation.
2.5 Serum alteration and source of lipids
SCs (passage number 5) were seeded in 96-well plates and incubated for 48 hours at 37 °C in a humidified 5% CO2 environment in GM. Then, the media was changed to experimental media (100 µL in each well) containing oleic acid at varying concentrations (50 µM, 100 µM, 200 µM and 400 µM). Separate plates were incubated for 24 and 48 hours, respectively, for subsequent analysis of cell count and lipid accumulation, as described in the section on quantification of lipid accumulation.To test the effect of serum, we used different basal media combined with three different serum concentrations (2% FBS, 2% HS, GM or SFM). DMEM-Glutamax was the basal medium used for 2% FBS, 2% HS, and GM. The SFM was prepared according to previous research from our lab (DMEM/F12) with fetuin (600 µg mL−1, Sigma-F2379), BSA (75 µg mL−1, Sigma-A8412), Fibroblast Growth Factor 2 (FGF2, 2 ng mL−1 Gibco-100-18B-50UG), insulin 10 µg mL−1, transferrin 5.5 µg mL−1 and selenium 6.7 ng mL−1 as ITS-1X (Gibco-41400045).5
To assess whether lipid droplets can be sustained within SCs and myotubes, SCs in passage 5 were cultured as described above in GM for 2 days. For SCs (see Fig. 2A), after 2 days in GM, experimental media (2% FBS, 2% HS, GM, or SFM) containing 200 µM oleic acid were added to the cells and incubated for 2 days. Following a 2 days exposure, the media were replenished with the same specific media, but without oleic acid, every two days for up to 8 days. Separate plates, seeded simultaneously under identical conditions, were used for analysis at days 2, 4, and 8 to determine lipid accumulation and cell count. For myotubes (Fig. 2B), to initiate the differentiation of SCs, the media were changed to 2% HS after growing SCs for 2 days in GM. Cells were kept for 20 days, with media replenishment every two days. Afterwards, cells were exposed to experimental media containing 200 µM oleic acid in different serum media (2% FBS, 2% HS, GM, or SFM) for 2 days. After two days, the media were replaced with the same specific media, but without oleic acid, for eight days, with replenishment every two days, as described earlier.
To assess the effect of different types of FAs (myristoleic-M3525, palmitoleic-P9417, oleic-O1383, erucic-E3385, elaidic-E4637, linoleic-L1012, linolenic-L2376, DHA-D2534, EPA-734322, pristanic-P6617, phytanic-P4060, palmitic-P5585, stearic-S4751, and butyrate-B103500, all from Sigma), SCs were seeded in 96-well plate as described previously, and post 48 hours in GM, cells were exposed to BSA conjugated FA in different serum media (2% FBS GM or SFM) for 24 hours. Afterwards, lipid droplets were measured using fluorescent staining and imaging, as described in the section on the quantification of lipid accumulation.
2.6 Effect of serum-free media components, basal medium composition, and oleic acid exposure time on lipid accumulation
To evaluate the effect of individual components of the SFM on lipid accumulation, SCs were seeded in 96-well plates and cultured in GM for 24 hours. Subsequently, cells were exposed for 24 hours to SFM supplemented with 200 µM oleic acid, with the systematic omission of individual components to assess their specific contributions. The tested conditions included SFM without fetuin, without BSA, without FGF2, and without ITS, respectively, and were compared against complete SFM. To further investigate the role of the ITS supplement, cells were exposed to SFM lacking ITS but containing individual or combinations of ITS components: insulin, transferrin, and sodium selenite. The tested combinations included: insulin alone, transferrin alone, sodium selenite alone, insulin + transferrin, insulin + sodium selenite, and transferrin + sodium selenite, all within the ITS-depleted SFM and supplemented with 200 µM oleic acid. Transferrin and sodium selenite were dissolved in Milli-Q water, and the final concentrations in the media were 10 µg mL−1 insulin, 5.5 µg mL−1 transferrin, and 6.7 µg mL−1 sodium selenite, corresponding to the levels present in the ITS 1X used in SFM. Additionally, the impact of basal media composition was assessed by preparing SFM using either DMEM or DMEM/F12. Conditions included SFM prepared in DMEM versus DMEM/F12, DMEM or DMEM/F12 supplemented with 2% FBS, and DMEM or DMEM/F12 with ITS alone. Cells were grown in SFM before treatments. All treatments were carried out in the presence of 200 µM oleic acid for 24 hours.To assess the effect of exposure time on lipid droplet accumulation in porcine SCs, SCs were grown in 96-well plates in GM for 24 hours and then in SFM containing 100 µM oleic acid for another 24 hours. Lipid accumulation was measured at 0.5, 1, 2, 4, 6, 12 and 24 hours using separate wells prepared under identical conditions for each time point.
2.7 Statistical analysis
All statistical analyses and data visualisations were conducted using Prism version 10.1.0 (GraphPad-Dotmatics, USA). Results are expressed as mean ± standard deviation (SD). Statistical tests used for each analysis are specified in the corresponding figure legends. A p-value of less than 0.05 was considered statistically significant.3. Results
3.1 Effect of media on lipid droplet accumulation
To evaluate the effect of media composition on lipid droplet accumulation, we exposed cells to various concentrations of oleic acid using media with either no serum or varying amounts of serum. Following 24 hours of exposure to different concentrations of oleic acid, lipid droplet accumulation was highest (almost two-fold that of the control) in the serum-free media at all concentrations of oleic acid. Moreover, lipid droplet accumulation within serum-containing media was higher in 2% HS media at 200 µM oleic acid and 2% HS and 2% FBS media in 400 µM oleic acid compared to GM (Fig. 2A). After 48 hours of incubation, the cells in SFM accumulated a higher (almost two-fold) lipid content compared to cells in serum-containing media. In contrast, there was no significant difference in lipid droplet accumulation between serum-containing media within a specific oleic acid concentration (Fig. 2B).To examine whether cells can retain the accumulated lipid droplets, cells were exposed to fatty acids for 48 hours, then changed to media without fatty acids for 2, 4 or 8 days. SCs in SFM had a higher lipid accumulation, compared to 2% HS media and GM, at all time points (Fig. 3A). For SCs, there was no significant difference in cell count between serum-containing media at 2 days exposure. However, GM had the highest cell number, followed by SFM (Fig. 3C). After 2 days of exposure to oleic acid, SCs in SFM and 2% FBS media had the highest lipid accumulation. In contrast, SCs in GM had the lowest lipid accumulation. SFM helped maintain the lipid accumulation, which was evident as lipid content was higher in SCs in SFM. SCs in SFM accumulated more lipid by day 4 and then decreased by day 8, whereas lipid accumulation decreased over time for SCs in 2% FBS and 2% HS. Hence, SCs in SFM sustained the lipid droplets for a longer time compared to serum-containing media. GM supported SC proliferation, which was significantly better compared to 2% FBS and 2% HS at both day 4 and day 8. To further assess lipid accumulation during differentiation, we also investigated lipid accumulation in myotubes (SI Fig. 2) under the same serum media conditions with oleic acid. At all time-points, lipid droplet content was substantially higher in myotubes grown in SFM compared to all other media types (Fig. 3B). On day 2, myotubes in the 2% FBS media showed a higher lipid accumulation than 2% HS and GM. As the days progressed, the lipid content decreased in all media types. The cell count was significantly higher in GM (except on day 2) compared to all other media (Fig. 3D). A representative fluorescent image of cells exposed to 400 µM oleic acid in different serum media is shown in SI Fig. 3.
3.2 Effect of fatty acid type on lipid droplet accumulation
In both 2% FBS media (SI Fig. 4A) and SFM (SI Fig. 4B), SCs exposed to monounsaturated fatty acids and C18 polyunsaturated fatty acids had higher lipid content compared to SCs exposed to saturated fatty acids, branched-chain fatty acids, short-chain fatty acids and C20–22 polyunsaturated fatty acids. At 400 µM, lipid accumulation varied depending on the type of fatty acid. In 2% FBS media, all monounsaturated fatty acids, as well as linoleic and linolenic acid, led to a higher accumulation of lipid droplets (almost three-fold) in SCs compared to the other tested fatty acids, except for palmitoleic acid in 2% FBS media, which did not show the same effect (Fig. 4A). However, SCs exposed to eicosapentaenoic acid (EPA), palmitic acid, and butyric acid had a lower cell count (Fig. 4C).In SFM, all monounsaturated fatty acids, linoleic and linolenic acid, resulted in higher lipid droplet accumulation (almost threefold) compared to the other FAs tested in SFM (Fig. 4B). Docosahexaenoic acid (DHA), a C22 omega-3 fatty acid, induced more lipid accumulation in SCs compared to the branched, saturated and short-chain FAs phytanic, palmitic, stearic and butyric acid when cultured in SFM (Fig. 4B). Cells with erucic, myristoleic, palmitic and stearic exposure had a lower cell count compared to the control (Fig. 4D). A representative fluorescent image of cells exposed to 400 µM of different fatty acids in SFM is shown in SI Fig. 5.
3.3 Effect of serum-free media components, basal medium composition, and oleic acid exposure time on lipid accumulation
Removal of individual components from the complete SFM did not affect cell count (Fig. 5A). However, lipid accumulation was significantly reduced upon omission of ITS, BSA, or fetuin compared to the complete SFM control, with the most substantial decrease observed in the ITS-omitted condition, while FGF2 omission had no significant effect (one-way ANOVA with post-hoc comparisons only to SFM; Fig. 5B). Since ITS is a composite of insulin, transferrin, and sodium selenite, the individual and combined effects of these components were assessed in ITS-depleted SFM. Insulin alone or combined with transferrin or sodium selenite did not significantly reduce lipid accumulation compared to ITS, while treatments lacking insulin showed reduced lipid content (Fig. 5C). Across all tested basal medium conditions (SFM, 2% FBS, and ITS-supplemented media), SCs cultured in DMEM/F12 exhibited higher lipid accumulation than those cultured in DMEM alone (Fig. 5D), indicating a superior capacity of DMEM/F12 to support lipid deposition.Lipid droplet accumulation increased progressively over time in proliferating SCs exposed to 100 µM oleic acid in SFM, with a marked increase observed up to 12 hours. No significant difference in lipid content was detected between 12 hours and 24 hours of exposure (Fig. 6).
4. Discussion
In the present study, we tested the effect of different serum concentrations and SFM on lipid accumulation. We demonstrated that both proliferating cells and myotubes cultured in SFM had a higher lipid accumulation than those cultured in serum-containing media. In our study, lower serum media (2% HS or 2% FBS) facilitated higher lipid accumulation compared to GM (10% FBS + 10% HS), which agrees with previous reports,39,40 where in fibroadipogenic precursor cells and preadipocytes accumulated more lipids in a serum free environment or a lower serum environment, compared to 10% or 20% FBS media. Also, Entenmann and Hauner reported that when human adipocyte precursor cells were incubated in serum-free conditions, they had a higher adipogenic differentiation compared to media supplemented with 10% FBS, and they found that replication and differentiation of cells were inversely related.53 Among the sources of fatty acids that can induce lipid accumulation, we identified the polyunsaturated fatty acid linoleic acid, as having a substantial effect on increasing lipid accumulation, along with monounsaturated fatty acids such as oleic acid. In contrast, saturated and short-chain fatty acids had a minor impact on lipid droplet accumulation. Unsaturated fatty acids can act as PPAR γ ligands, with monounsaturated fatty acids, such as oleic acid, serving as weak ligands, and polyunsaturated fatty acids, including linoleic acid, linolenic acid, and arachidonic acid, being more potent.54 Branched-chain fatty acids (pristanic acid and phytanic acid) have been used as adipogenic agents42 but did not increase lipid accumulation compared to the control in the present study. The omega-3 fatty acids DHA and EPA in SFM stimulated less lipid accumulation than monounsaturated and C18 polyunsaturated fatty acids. Ghnaimawi et al. reported that DHA had a greater adipogenic capacity than EPA in transdifferentiating C2C12 muscle cells toward adipogenesis,31 a tendency also observed in the present study. Short-chain fatty acids, such as butyrate, are used to enhance adipogenesis in the presence of insulin, dexamethasone and IBMX in bovine dedifferentiated fat cells;55 however, during short-term exposure (24 hours), butyrate did not induce lipid accumulation in our study. It has been shown in animal models that medium-chain fatty acids do not contribute to intramyocellular lipid accumulation56 In contrast, oleic acid alone induced adipogenesis by upregulating PPAR γ in bovine muscle SCs, although no fully transdifferentiated adipose tissue was observed.32 A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mix of oleic and linoleic acids and insulin increased adipogenic gene expression and lipid filling in chicken embryonic fibroblasts.57 These studies all indicate the possibility of using long-chain unsaturated fatty acids and insulin or ITS to transdifferentiate or induce adipogenesis.
1 mix of oleic and linoleic acids and insulin increased adipogenic gene expression and lipid filling in chicken embryonic fibroblasts.57 These studies all indicate the possibility of using long-chain unsaturated fatty acids and insulin or ITS to transdifferentiate or induce adipogenesis.
Oleic, palmitic, stearic, and linoleic acids are the major fatty acids found in pork;58 however, the saturated fatty acids palmitic and stearic acids have limited lipid droplet induction capacity compared to oleic and linoleic acids, as evident from our study. Here, we quantified the accumulation of neutral lipid droplets, representing the most significant fraction, but not phospholipids, which form part of the cell membrane structure. Kim et al. reported that bovine muscle cells containing intramyocellular lipids exhibit both myogenic and adipose-like characteristics.32 This finding may also apply to porcine muscle cells, as observed in our study. Cells possessing muscle-like and fat-like characteristics could be significant in developing the flavour profile of cultivated meat products, which closely resemble traditional meat products. We were also able to induce lipid droplets using DHA, which could be used to alter the nutritional profile of the cultivated meat product. Future research should also investigate whether the fatty acid composition of cultivated muscle cells can be matched to that of pork by combining different fatty acids. However, high lipid accumulation could result in insulin resistance, thereby reducing glycogen synthase activity.59 Glycogen content is essential for the post-mortem pH decline, subsequent protease activity, and flavour precursor development during muscle to meat conversion.13
The SFM used in this study (referred to as Tribasal 2.0+ by Skrivergaard et al.) has been shown to upregulate lipid metabolism genes such as ACSS2, FASN, SCD, and FADS2 in bovine satellite cells.5 FASN and SCD are key enzymes in de novo lipogenesis, providing saturated and monounsaturated fatty acids essential for triacylglycerol synthesis and lipid droplet formation.60 In our study, supplementation of SFM with mono or polyunsaturated fatty acids was associated with increased intramyocellular lipid accumulation and retention of lipid droplets for at least 8 days.
DMEM/F12 supported greater lipid accumulation than DMEM, likely due to differences in nutrient composition. Although DMEM/F12 glucose concentration is lower than that of high-glucose DMEM, the presence of exogenous fatty acids appeared to offset the influence of glucose on lipid accumulation. Additionally, components such as linoleic acid,61 choline,62 inositol,63 and biotin,64 known for their adipogenic potential, may contribute to the enhanced lipid deposition. Further research is needed to elucidate the specific role of basal media composition in regulating lipid accumulation in muscle cells.
In our study, we observed that FGF2 had a minimal effect on lipid accumulation in SCs, despite previous reports demonstrating its potential for adipogenesis. Prior studies have shown that FGF2 promotes adipogenesis in preadipocytes65,66 and upregulates PPARγ expression in human adipose-derived stem cells, thereby enhancing lipid storage.67 Our findings, however, suggest a limited role for FGF2 in promoting lipid accumulation under the tested conditions.
In contrast, fetuin, an abundant glycoprotein in serum (∼20 mg mL−1 in FBS),68 showed a significant effect. Fetuin functions as a free fatty acid (FFA) carrier and adipokine that enhances lipogenesis and attenuates lipolysis, particularly in monogastric and bovine adipocytes.69 It has been reported to upregulate the lipogenic enzyme AGPAT2, thereby promoting triglyceride synthesis.69 In smooth muscle cell cultures, adding fetuin significantly increased the amount of fatty acid stored as triglycerides, even though the amount of free (unbound) fatty acids available for cellular uptake remained the same.70 Similar effects were observed in human fetal skin fibroblasts using both oleic and arachidonic acids. However, fetuin may also exert complex regulatory effects. For example, it has been shown to downregulate CD36, a major fatty acid transporter in muscle and heart cells, by inhibiting PPARγ phosphorylation, which potentially alters fatty acid uptake dynamics.71
Albumin, especially BSA, is another key modulator of lipid metabolism in vitro. While its primary role is to bind FFAs and increase their solubility in aqueous media, albumin also facilitates lipid accumulation through controlled FFA delivery. Studies using hepatocyte cultures demonstrate that albumin promotes palmitate uptake more efficiently than free fatty acids alone, following a “pseudo facilitation” model, where delivery is governed by unbound FFA concentration.72 At physiological albumin levels (∼200 µM), fatty acids are steadily released, allowing gradual cellular uptake that mimics in vivo transport.73 Moreover, albumin not only buffers FFAs to prevent lipotoxicity but also enhances fatty acid incorporation into triglycerides in cells with high metabolic demand. The direct addition of free fatty acids to media can result in abrupt, cytotoxic spikes, whereas BSA conjugation ensures safe and sustained uptake.72,74 Although free and BSA-bound FFAs enter cells in unbound form, the albumin complex stabilises delivery and prevents detergent-like membrane effects, reducing stress and enhancing lipid storage capacity.
In our study, ITS enhanced lipid accumulation compared to other components of SFM. Insulin had the most pronounced effect on lipid accumulation among the ITS components. Cells cultured without insulin, either alone or combined with transferrin or selenium, exhibited significantly lower lipid accumulation. Insulin has lipogenic effects75 and it increases the translocation of fatty acid transporters such as CD36, FABPpm, FATP1, and FATP4,76,77 glucose transport via GLUT4 (ref. 77) and it has also been reported that insulin inhibits carnitine palmitoyl transferase 1 and 5′ adenosine monophosphate-activated protein kinase, thereby decreasing fatty acid oxidation.78,79 Stout et al.80 reported that a serum-free medium led to increased lipid droplets in bovine satellite cells in the long-term culture without the addition of free fatty acids and suggested the possibility of insulin resistance due to high insulin concentration in the media (20 µg mL−1). However, the relationship between insulin resistance and intramyocellular lipid accumulation is interdependent and quite complex.81
Selenium has been used in adipogenic induction media previously, and Heart et al. reported that selenium increases glucose uptake.82 Similarly, Hassan et al. found that selenium increased adipogenic expression markers and fatty acid uptake in chicken fibroblasts.83 However, in our experiments using SCs, selenium and transferrin, alone or in combination, did not increase lipid accumulation compared to the control.
5. Conclusion
In conclusion, this study demonstrated that serum-free medium facilitates higher lipid accumulation in both proliferating SCs and myotubes compared to serum-containing media. The presence of ITS in SFM likely plays a key role in enhancing lipid deposition, with insulin alone exhibiting a notable lipogenic effect. Among the tested fatty acids, polyunsaturated fatty acids like linoleic acid and monounsaturated fatty acids like oleic acid were the most effective in inducing lipid droplet accumulation. In contrast, saturated and short-chain fatty acids had a minimal impact. Additionally, the ability to modulate lipid composition using specific fatty acids such as DHA presents an opportunity to optimise the nutritional profile of cultivated meat. Future research should focus on understanding the mechanisms of lipid uptake and biosynthesis in muscle cells, as well as exploring whether a fatty acid profile matching that of conventional pork can be achieved to enhance the flavour and texture of cultivated meat.Author contributions
Anupam Abraham: conceptualisation, resources, methodology, data curation, formal analysis, investigation, writing-original draft, review and editing. Marc Auguet-Lara: investigation, methodology, formal analysis, writing-review and editing. Stig Skrivergaard: methodology, resources, writing-review and editing. Margrethe Therkildsen: conceptualization, methodology, formal analysis, writing-review and editing, supervision, resources, funding acquisition, project administration. Martin Krøyer Rasmussen: conceptualization, methodology, resources, formal analysis, writing-review and editing, supervision, funding acquisition, project administration. Jette Feveile Young: conceptualization, methodology, resources, formal analysis, writing-review and editing, supervision, funding acquisition, project administration.Conflicts of interest
The authors declare no competing interests.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5fb00572h.Acknowledgements
This project is supported by AU Aarhus Universitets Forskningsfond (AUFF)/Aarhus University Research Foundation. Data was generated by accessing research infrastructure at Aarhus University, including FOODHAY (Food and Health Open Innovation Laboratory, Danish Roadmap for Research Infrastructure). The authors greatly acknowledge the technical support of Navid Sahebekhtiari (Department of Food Science, Aarhus University) during the investigation and Jens Askov Jensen (Department of Food Science, Aarhus University) during the sampling. The authors acknowledge the use of AI-based tools (Co-Pilot) for language enhancement and editing to improve the clarity, coherence and readability of this manuscript. The AI was not used for data analysis, experimental design, or the generation of original scientific content. The final manuscript was carefully reviewed and edited by the authors to ensure accuracy and compliance with scientific standards.References
- H. L. Tuomisto, EMBO Rep., 2019, 20, e47395 CrossRef.
- M. J. Post, J. Sci. Food Agric., 2014, 94, 1039–1041 CrossRef PubMed.
- M. K. Saad, J. S. K. Yuen, C. M. Joyce, X. Li, T. Lim, T. L. Wolfson, J. Wu, J. Laird, S. Vissapragada, O. P. Calkins, A. Ali and D. L. Kaplan, Sci. Rep., 2023, 13, 5098 CrossRef PubMed.
- A. J. Stout, M. J. Arnett, K. Chai, T. Guo, L. Liao, A. B. Mirliani, M. L. Rittenberg, M. Shub, E. C. White, J. S. K. Yuen Jr., X. Zhang and D. L. Kaplan, ACS Synth. Biol., 2023, 12, 1567–1573 CrossRef PubMed.
- S. Skrivergaard, J. F. Young, N. Sahebekhtiari, C. Semper, M. Venkatesan, A. Savchenko, P. J. Stogios, M. Therkildsen and M. K. Rasmussen, Food Res. Int., 2023, 172, 113194 CrossRef.
- A. J. Stout, X. Zhang, S. M. Letcher, M. L. Rittenberg, M. Shub, K. M. Chai, M. Kaul and D. L. Kaplan, Cell Rep. Sustain., 2024, 1, 100009 Search PubMed.
- I. Fraeye, M. Kratka, H. Vandenburgh and L. Thorrez, Front Nutr., 2020, 7, 35 CrossRef PubMed.
- L. Lin-Schilstra, G. Backus, H. Snoek and D. Mörlein, Meat Sci., 2022, 187, 108736 CrossRef PubMed.
- E. T. Lew, J. S. K. Yuen, K. L. Zhang, K. Fuller, S. C. Frost and D. L. Kaplan, Sci. Rep., 2024, 14, 17643 CrossRef.
- K. G. Grunert, Food Qual. Prefer., 1997, 8, 157–174 CrossRef.
- G. J. Hausman, U. Basu, M. Du, M. Fernyhough-Culver and M. V. Dodson, Adipocyte, 2014, 3, 242–255 CrossRef CAS PubMed.
- M. D. Aaslyng and L. Meinert, Meat Sci., 2017, 132, 112–117 CrossRef.
- D. Dashdorj, T. Amna and I. Hwang, Eur. Food Res. Technol., 2015, 241, 157–171 CrossRef.
- E. G. Vilar, M. G. O'Sullivan, J. P. Kerry and K. N. Kilcawley, Sep. Sci. plus, 2022, 5, 482–512 CrossRef.
- X. Guan, Q. Yan, Z. Ma and J. Zhou, Food Funct., 2023, 14, 3576–3587 RSC.
- M. Lee, S. Park, B. Choi, W. Choi, H. Lee, J. M. Lee, S. T. Lee, K. H. Yoo, D. Han and G. Bang, Nat. Commun., 2024, 15, 77 CrossRef.
- Y. Wang, Z. Zhong, N. Munawar, L. Zan and J. Zhu, Int. J. Biol. Macromol., 2024, 259, 129134 CrossRef PubMed.
- J. S. K. Yuen, Jr., A. J. Stout, N. S. Kawecki, S. M. Letcher, S. K. Theodossiou, J. M. Cohen, B. M. Barrick, M. K. Saad, N. R. Rubio, J. A. Pietropinto, H. DiCindio, S. W. Zhang, A. C. Rowat and D. L. Kaplan, Biomaterials, 2022, 280, 121273 CrossRef.
- N. K. Singh, H. S. Chae, I. H. Hwang, Y. M. Yoo, C. N. Ahn, S. H. Lee, H. J. Lee, H. J. Park and H. Y. Chung, J. Anim. Sci., 2007, 85, 1126–1135 CrossRef.
- N. Singh, Asian-Australas. J. Anim. Sci., 2007, 20, 432–439 CrossRef.
- J. Singh, N. K. Verma, S. M. Kansagra, B. N. Kate and C. S. Dey, Mol. Cell. Biochem., 2007, 294, 163–171 CrossRef PubMed.
- P. Kuppusamy, D. Kim, I. Soundharrajan, I. Hwang and K. C. Choi, Biology, 2021, 10, 6 CrossRef.
- Y. Wang, D. Zhuang, N. Munawar, L. Zan and J. Zhu, Food Chem., 2024, 460, 140696 CrossRef PubMed.
- S. B. Rønning, P. V. Andersen, M. E. Pedersen and K. Hollung, PLoS One, 2017, 12, e0182928 CrossRef PubMed.
- S. Skrivergaard, M. K. Rasmussen, M. Therkildsen and J. F. Young, Int. J. Mol. Sci., 2021, 22, 8376 CrossRef PubMed.
- F. Mehta, R. Theunissen and M. J. Post, Methods Mol. Biol., 2019, 1889, 111–125 CrossRef PubMed.
- Y. Takegahara, K. Yamanouchi, K. Nakamura, S. Nakano and M. Nishihara, Exp. Cell Res., 2014, 324, 105–114 CrossRef.
- S. Benvenuti, I. Cellai, P. Luciani, C. Deledda, S. Baglioni, C. Giuliani, R. Saccardi, B. Mazzanti, S. Dal Pozzo, E. Mannucci, A. Peri and M. Serio, J. Endocrinol. Invest., 2007, 30, Rc26–Rc30 CrossRef PubMed.
- J. F. Young, A. Abraham, M. K. Rasmussen, S. Skrivergaard and M. Therkildsen, Developing cultured meat as a food product, in Advances in cultured meat technology, ed. M. Post, C. Cannon and C. Bryant, Burleigh Dodds Science Publishing, 2023, pp. 299–318 Search PubMed.
- S. Ghnaimawi, S. Shelby, J. Baum and Y. Huang, Anim. Cells Syst., 2019, 23, 355–364 CrossRef PubMed.
- S. Ghnaimawi, L. Rebello, J. Baum and Y. Huang, PLoS One, 2021, 16, e0249438 CrossRef PubMed.
- J. Kim, K. Chung, L. Fuerniss and B. Johnson, Open J. Anim. Sci., 2020, 10, 649–664 CrossRef CAS.
- X. Z. Li, Y. Yan, J. F. Zhang, J. F. Sun, B. Sun, C. G. Yan, S. H. Choi, B. J. Johnson, J. K. Kim and S. B. Smith, J. Anim. Sci., 2019, 97, 4114–4123 CrossRef.
- M.-È. Ouellette, J.-C. Bérubé, J.-M. Bourget, M. Vallée, Y. Bossé and J. Fradette, PLoS One, 2019, 14, e0224228 CrossRef CAS.
- U. Abou-Rjeileh, A. L. Lock and G. A. Contreras, Animal, 2025, 19, 101505 CrossRef CAS.
- S. A. Belal, D. R. Kang, A. S. Sivakumar, H. S. Choe and K. S. Shim, Anim. Biotechnol., 2019, 30, 323–331 CrossRef CAS PubMed.
- S. A. Belal, J. Lee, J. Park, D. Kang and K. Shim, Foods, 2024, 13, 2200 CrossRef CAS PubMed.
- G. Li, W. Yao and H. Jiang, J. Nutr., 2014, 144, 1887–1895 CrossRef CAS PubMed.
- R. Mitić, F. Cantoni, C. S. Börlin, M. J. Post and L. Jackisch, iScience, 2023, 26, 105822 CrossRef.
- J. S. K. Yuen Jr, M. K. Saad, N. Xiang, B. M. Barrick, H. DiCindio, C. Li, S. W. Zhang, M. Rittenberg, E. T. Lew, K. L. Zhang, G. Leung, J. A. Pietropinto and D. L. Kaplan, eLife, 2023, 12, e82120 CrossRef PubMed.
- Z. Redshaw, S. McOrist and P. Loughna, Cell Biochem. Funct., 2010, 28, 403–411 CrossRef CAS.
- L. Pasitka, M. Cohen, A. Ehrlich, B. Gildor, E. Reuveni, M. Ayyash, G. Wissotsky, A. Herscovici, R. Kaminker, A. Niv, R. Bitcover, O. Dadia, A. Rudik, A. Voloschin, M. Shimoni, Y. Cinnamon and Y. Nahmias, Nat. Food, 2023, 4, 35–50 CrossRef CAS PubMed.
- Y. N. Ma, B. Wang, Z. X. Wang, N. A. Gomez, M. J. Zhu and M. Du, Animal, 2018, 12, 2123–2129 CrossRef.
- D. Ozhava, K. Lee, C. Bektas, A. Jackson, K. Patel and Y. Mao, Gels, 2024, 10, 488 CrossRef PubMed.
- S. Yonekura, S. Hirota, Y. Tokutake, M. T. Rose, K. Katoh and H. Aso, Asian-Australas. J. Anim. Sci., 2014, 27, 567–573 CrossRef.
- E. J. Lee, H. J. Lee, M. R. Kamli, S. Pokharel, A. R. Bhat, Y.-H. Lee, B.-H. Choi, T. Chun, S. W. Kang, Y. S. Lee, J. W. Kim, R. D. Schnabel, J. F. Taylor and I. Choi, Genomics, 2012, 100, 195–202 CrossRef.
- R. V. Rajesh, G. N. Heo, M. R. Park, J. S. Nam, N. K. Kim, D. Yoon, T. H. Kim and H. J. Lee, Comp. Biochem. Physiol., Part D:Genomics Proteomics, 2010, 5, 234–244 Search PubMed.
- C. Strieder-Barboza, E. Thompson, K. Thelen and G. A. Contreras, J. Dairy Sci., 2019, 102, 3622–3629 CrossRef PubMed.
- K. Yoshioka, Y. Kitajima, N. Okazaki, K. Chiba, A. Yonekura and Y. Ono, Front. Cell Dev. Biol., 2020, 8, 793 CrossRef PubMed.
- S. Skrivergaard, M. Krøyer Rasmussen, N. Sahebekhtiari, J. Feveile Young and M. Therkildsen, Food Res. Int., 2023, 173, 113217 CrossRef.
- A. Pappas, M. Anthonavage and J. S. Gordon, J. Invest. Dermatol., 2002, 118, 164–171 CrossRef PubMed.
- M. Grandl and G. Schmitz, Cytometry, Part A, 2010, 77, 231–242 CrossRef.
- G. Entenmann and H. Hauner, Am. J. Physiol.: Cell Physiol., 1996, 270, C1011–C1016 CrossRef.
- O. A. Macdougald and M. D. Lane, Curr. Biol., 1995, 5, 618–621 CrossRef.
- Y. Oki, R. Hagiwara, T. Matsumaru and K. Kano, Genes Cells, 2022, 27, 5–13 CrossRef PubMed.
- J. De Vogel-van den Bosch, J. Hoeks, S. Timmers, S. M. Houten, P. J. van Dijk, W. Boon, D. van Beurden, G. Schaart, S. Kersten, P. J. Voshol, R. J. A. Wanders, M. K. Hesselink and P. Schrauwen, Obesity, 2011, 19, 792–799 CrossRef PubMed.
- D.-H. Kim, J. Lee, Y. Suh, M. Cressman, S. S. Lee and K. Lee, Lipids, 2020, 55, 163–171 CrossRef.
- T. T. N. Dinh, K. V. To and M. W. Schilling, Meat Muscle Biol., 2021, 5(34), 31–16 Search PubMed.
- D. I. W. Phillips, S. Caddy, V. Ilic, B. A. Fielding, K. N. Frayn, A. C. Borthwick and R. Taylor, Metabolism, 1996, 45, 947–950 CrossRef.
- N. Koundouros and G. Poulogiannis, Br. J. Cancer, 2020, 122, 4–22 CrossRef PubMed.
- I. Abe, Y. Oguri, A. R. P. Verkerke, L. B. Monteiro, C. M. Knuth, C. Auger, Y. Qiu, G. P. Westcott, S. Cinti, K. Shinoda, M. G. Jeschke and S. Kajimura, Dev. Cell, 2022, 57, 2623–2637.e2628 CrossRef.
- Y. Takeda and P. Dai, Sci. Rep., 2024, 14, 18252 CrossRef.
- M. M. Montt-Guevara, M. Finiguerra, I. Marzi, T. Fidecicchi, A. Ferrari, A. D. Genazzani and T. Simoncini, Front. Endocrinol., 2021, 12, 660815 CrossRef.
- S. Selvam, A. Ramaian Santhaseela, D. Ganesan, S. Rajasekaran and T. Jayavelu, Cell Stress Chaperones, 2019, 24, 343–350 CrossRef PubMed.
- H. Eda, K. Aoki, K. Marumo, K. Fujii and K. Ohkawa, Biochem. Biophys. Res. Commun., 2008, 366, 471–475 CrossRef PubMed.
- Y. Hiraoka, H. Yamashiro, K. Yasuda, Y. Kimura, T. Inamoto and Y. Tabata, Tissue Eng., 2006, 12, 1475–1487 CrossRef.
- N. Kakudo, A. Shimotsuma and K. Kusumoto, Biochem. Biophys. Res. Commun., 2007, 359, 239–244 CrossRef.
- M. N. Kundranda, M. Henderson, K. J. Carter, L. Gorden, A. Binhazim, S. Ray, T. Baptiste, M. Shokrani, M. L. Leite-Browning, W. Jahnen-Dechent, L. M. Matrisian and J. Ochieng, Cancer Res., 2005, 65, 499–506 CrossRef.
- C. Strieder-Barboza and G. A. Contreras, J. Dairy Sci., 2019, 102, 4628–4638 CrossRef.
- A. J. Cayatte, L. Kumbla and M. T. Subbiah, J. Biol. Chem., 1990, 265, 5883–5888 CrossRef PubMed.
- E. Chekol Abebe, Z. Tilahun Muche, T. M. A. Behaile, T. Mengie Ayele, M. Mekonnen Agidew, M. Teshome Azezew, E. Abebe Zewde, T. Asmamaw Dejenie and M. Asmamaw Mengstie, Front. Cell Dev. Biol., 2022, 10, 945287 CrossRef PubMed.
- N. Alsabeeh, B. Chausse, P. A. Kakimoto, A. J. Kowaltowski and O. Shirihai, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2018, 1863, 143–151 CrossRef CAS.
- G. J. van der Vusse, Drug Metab. Pharmacokinet., 2009, 24, 300–307 CrossRef CAS PubMed.
- G. L. Francis, Cytotechnology, 2010, 62, 1–16 CrossRef CAS PubMed.
- D. M. Muoio, G. L. Dohm, E. B. Tapscott and R. A. Coleman, Am. J. Physiol.: Endocrinol. Metab., 1999, 276, E913–E921 CrossRef CAS.
- S. S. Jain, J. J. F. P. Luiken, L. A. Snook, X. X. Han, G. P. Holloway, J. F. C. Glatz and A. Bonen, FEBS Lett., 2015, 589, 2769–2775 CrossRef CAS.
- J. J. F. P. Luiken, D. P. Y. Koonen, J. Willems, A. Zorzano, C. Becker, Y. Fischer, N. N. Tandon, G. J. van der Vusse, A. Bonen and J. F. C. Glatz, Diabetes, 2002, 51, 3113–3119 CrossRef CAS PubMed.
- M. M. Awan and E. D. Saggerson, Biochem. J., 1993, 295(Pt 1), 61–66 CrossRef CAS PubMed.
- J. Gamble and G. D. Lopaschuk, Metabolism, 1997, 46, 1270–1274 CrossRef CAS PubMed.
- A. J. Stout, A. B. Mirliani, M. L. Rittenberg, M. Shub, E. C. White, J. S. K. Yuen and D. L. Kaplan, Commun. Biol., 2022, 5, 466 CrossRef CAS.
- T. S. Varman and G. I. Shulman, Cell, 2012, 148, 852–871 CrossRef PubMed.
- E. Heart and C. K. Sung, J. Cell. Biochem., 2003, 88, 719–731 CrossRef CAS PubMed.
- A. Hassan, J. Ahn, Y. Suh, Y. M. Choi, P. Chen and K. Lee, J. Nutr. Biochem., 2014, 25, 858–867 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |