Open Access Article
Open Access ArticleIntegrating nucleic acid research and computational strategies for advancing plant food security
Yashikaa,
Leena Aggarwal *a and
Pradeep Pant
*a and
Pradeep Pant *b
*b
aDepartment of Chemistry, Netaji Subhas University of Technology, Delhi-110078, India. E-mail: leena.aggarwal@nsut.ac.in
bDepartment of Biotechnology, Bennett University, Greater Noida, India. E-mail: pradeep.pant25@gmail.com
First published on 11th December 2025
Abstract
The evolution of plant breeding, from traditional techniques to cutting-edge omics-driven approaches (genomics, transcriptomics, proteomics, and metabolomics), has revolutionized crop yield enhancement since the mid-twentieth century. Today, genomics empowers breeders with powerful tools to directly associate genetic variation with phenotypes, accelerating the development of desirable traits. Building on this foundation, nucleic acid-based diagnostic techniques (PCR and LAMP) and gene-editing platforms (ZFNs, TALENs, and CRISPR/Cas), along with RNAi enable precise detection and manipulation of plant gene expression. This review focuses on various genome editing and diagnostic strategies for plant protection that aim to improve stress resilience, productivity, and nutritional value. Additionally, we highlight the latest computational strategies and methodologies that support the precise and rational design of such interventions with robust tools for targeted crop improvement. Looking ahead, these breakthroughs are poised to drive innovation across agriculture, biotechnology, and nucleic acid testing, opening new frontiers in sustainable food production and precision breeding.
Sustainability spotlightEnsuring global food security while facing climate change and population growth demands sustainable innovations in agriculture. This review highlights how nucleic acid-based diagnostics and gene-editing technologies—such as PCR, LAMP, CRISPR/Cas, and RNAi—are revolutionizing plant protection and crop improvement. By enabling precise, efficient, and targeted breeding strategies, these tools support resilient, high-yield crops and reduce dependence on chemical inputs. The integration of computational approaches further enhances the rational design of genetic interventions. This work aligns with UN SDG 2 (Zero Hunger) and SDG 13 (Climate Action) by promoting sustainable agricultural practices and developing climate-resilient crops, thus contributing to global efforts toward sustainable food production and improved nutritional outcomes. |
1. Introduction
Population growth and climate change exert substantial pressure on global food security by inducing physiological and environmental stresses in plants, thereby enhancing their susceptibility to both abiotic stressors (such as drought, salinity, and temperature extremes) and biotic stressors (including pathogens, pests, and weeds).1,2 Biotic stressors contribute significantly to global agricultural losses, with estimated yield reductions ranging from 20% to 40%, leading to substantial economic impacts.3 According to the Food and Agriculture Organization (FAO), plant pests destroy as much as 40% of global crop yields annually, leading to economic losses exceeding USD 220 billion from plant diseases and at least USD 70 billion from invasive insect infestations.4 Plants are pivotal in sustaining life on Earth, as they are primary sources of food and energy, forming the foundation of ecological and biological systems. Ensuring effective crop protection against threats is crucial for sustaining agricultural productivity and meeting the growing population demand.5 According to World Population Prospects 2022,6 the global population reached 8 billion in November 2022, and the UN's medium-variant projection indicates that it will peak at approximately 10.4 billion in 2080s.7 To address these challenges and to attain the second Sustainable Development Goal, i.e., “zero hunger”, substantial efforts are necessary for the transition from conventional high-input agriculture toward more resilient, diversified and technologically supported production approaches.8–10 Over the last few decades, plant breeding and other technologies have made significant contributions toward minimizing hunger and extreme poverty.2,11 Prior to the genomic era, conventional breeding methods rely on controlled hybridisation and selection of desirable traits through natural processes to develop new plant varieties.12 Unlike marker-assisted selection, conventional breeding is slower and less precise, making it insufficient to meet the rising global food demand.13 Innovative strategies to enhance crop yield and stress resistance have been transformed by genetic engineering.2,14 Techniques, such as Novel Plant Breeding Techniques (NPBTs) and Next Generation Sequencing (NGS), collectively accelerate crop improvement by providing genomic insights and enabling targeted manipulation of plant genetic materials. These approaches offer relevant, versatile, cost-effective, and time-efficient strategies that enhance precision in modern plant breeding.2,15 These techniques facilitated the development of crop plants with enhanced agronomic traits, improved nutritional quality, and increased resistance to both biotic and abiotic stresses. These technologies not only address the shortcomings of conventional breeding but also offer flexibility by enabling the use of genomic information from both model and non-model plant species.15 In addition to genetic improvements, early and accurate detection of pathogens plays a crucial role in managing plant diseases and minimizing crop loss. Nucleic acid-based diagnostic techniques such as Polymerase Chain Reaction16 (PCR), quantitative Polymerase Chain Reaction16 (qPCR), and digital Polymerase Chain Reaction17 (dPCR) enable rapid and sensitive detection of plant pathogens even at early infection stages. These techniques detect specific DNA or RNA sequences of pathogens, providing precision and timeliness in field-level diagnostics. This review critically discusses genome editing tools, nucleic acid-based diagnostic techniques, and RNA-based technologies, along with the computational approaches that enhance their precision and applicability. A critical aspect of these technologies is the integration of computational approaches to design, evaluate, and optimize editing tools. For genome editing techniques such as Zinc Finger Nucleases18 (ZFNs) and Transcription Activator-Like Effector Nucleases19 (TALENs), software tools predict target site specificity based on DNA-binding motifs, while for Clustered Regularly Interspaced Short Palindromic Repeats20 (CRISPR/Cas9), several platforms are available such as CRISPR Off-target Sites with Mismatches, Insertions and Deletion21 (COSMID) for designing guide RNA (gRNA) and CHaracterization and OPtimization of CHOPping tools22 (CHOPCHOP) for assisting in identifying suitable target sites and minimizing off-target effects. Likewise, nucleic acid-based diagnostic techniques such as PCR16 and dPCR17 rely on in silico tools for primer/probe design and target validation. These computational approaches enhance the accuracy, efficiency, and scalability of both genome editing and diagnostics, facilitating precision in plant protection and crop improvement, as shown in Fig. 1. Together, these pipelines demonstrate how computational approaches have been developed to support the design and optimization of nucleic acid-based tools, including genome editing nucleases, by enabling the prediction and reduction of off-target effects. These advancements contribute significantly to crop improvement, enhance resistance to biotic and abiotic stresses, and improve the nutritional quality of plants.2. Literature search strategy
A comprehensive literature search was undertaken to identify studies relevant to plant genome-editing platforms, RNA-based technologies, and nucleic acid-based diagnostic approaches. The search was performed across major scientific databases, including PubMed, SciFinder and Google Scholar. Boolean operators and targeted keywords were applied, such as “CRISPR/Cas9 and plant genome editing”, “ZFNs”, “TALENs”, “RNA interference in plants”, “nucleic acid-based diagnostics”, “LAMP”, “NASBA”, “plant pathogen detection”, and “computational genome-editing tools”. Additional terms such as “RNAi delivery”, “siRNA design”, “omics-driven crop improvement”, and “biosensing technologies” were also used. Literature published between 2000 and 2024 was prioritized, with earlier foundational studies incorporated when scientifically justified.3. Genome editing techniques
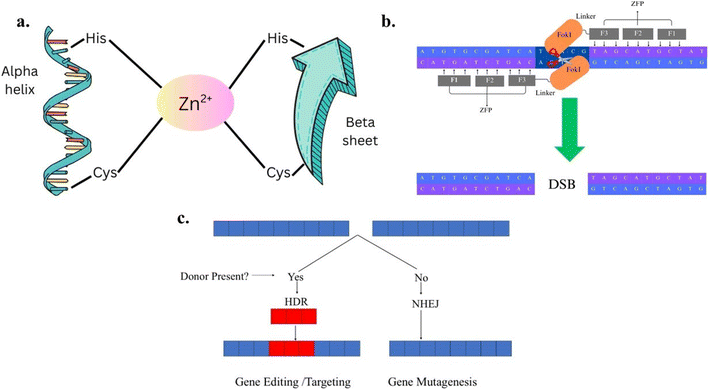
Genetic engineering has been a central area of research for several years in elucidating gene functions. This field encompasses the application of physical and biological mutagenesis, along with the identification of molecular mechanisms to enhance the crop yield.23 It relies on utilization of engineered nucleases, which consist of sequence-specific DNA-binding domains conjugated to a nonspecific DNA cleavage module.24 It includes several methods, such as ZFN,18 TALEN,19 and CRISPR/Cas9.25 These are chimeric nucleases that facilitate highly efficient and precise genome editing by inducing site-specific DNA double-strand breaks (DSBs). These DSBs activate intrinsic cellular DNA repair pathways, including the error-prone Non-Homologous End Joining (NHEJ) and the high-fidelity Homology-Directed Repair (HDR) mechanisms.263.1. Zinc-finger nucleases (ZFNs)
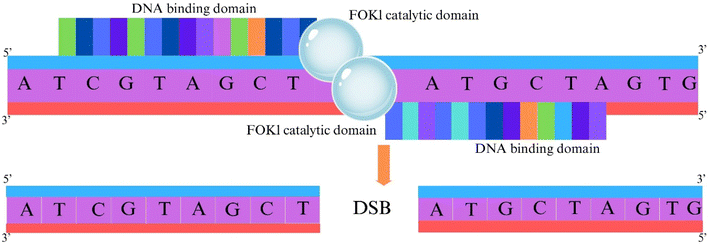
ZFNs are site-specific endonucleases engineered for targeted DNA cleavage, facilitating precise genome modification. It consists of two distinct domains: a zinc finger domain that binds to the target DNA and a nuclease domain derived from the FokI restriction enzyme, which is responsible for generating DSBs. Zinc finger domains are protein motifs that fold around one or more zinc ions and are capable of recognizing specific DNA sequences18,24 as shown in Fig. 2a. Zinc Finger Proteins (ZFPs) contain a tandem array of Cys2-His2 zinc fingers, each of which binds with approximately 3 base pairs (bp) of targeted DNA.27,28 It was reported18,24 that individual ZFNs used three fingers to bind a 9-bp target, which enabled ZFN dimers to specify 18 bp of DNA per cleavage site. More recent advancements18,24 have incorporated more fingers, and a variety of strategies have been described in the literature for designing ZFPs with new, user-chosen binding specificities.18,24 By designing multiple zinc finger domains, ZFNs can be engineered to target a particular DNA sequence with high precision.The nuclease domain of FokI restriction enzyme is crucial for the function of ZFNs, as it facilitates the targeted cleavage of DNA within complex genomes.18,24 It must dimerize for effective cleavage of DNA.29,30 The monomer of FokI is catalytically inactive and its natural dimerization is weak therefore, cleavage can be achieved by constructing two sets directed to DNA sequence and joined to the cleavage domain31 as shown in Fig. 2b. The nuclease domain introduces DSBs in the DNA at the targeted site, and this break activates the repair mechanisms, which occur via NHEJ (absence of donor DNA) or HDR (presence of donor DNA), as shown in Fig. 2c. Targeting or editing of gene at the site of break takes place in the presence of a template donor flanking DNA gene.18,24 Rejoining of the two broken DNA ends takes place in the absence of template donor flanking DNA gene, with some insertion or deletion often causing mutations due to frameshifts and effectively knocking out the targeted gene at the site of break, which leads to disruption of the target gene.18,24
Although ZFNs have been proven as effective technology, it also has certain drawbacks. It is an expensive process of designing protein domains for individual genes and has a risk of off-target DNA cleavage caused by faulty interactions. These challenges prompted the emergence of newer gene-editing tools, such as TALENs and CRISPR/Cas9 which offer easier construction and greater efficiency.32
3.2. Transcription activator-like effector nucleases (TALENs)
In 2011, Nature Methods recognized TALENs as the Method of the Year.19,33 The advancement of the TALEN system technique is linked to research on bacteria of the genus Xanthomonas, which is pathogenic to crop plants such as rice, pepper, and tomato. These bacteria secrete effector proteins (transcription activator-like effectors, TALEs) into the cytoplasm of plant cells, modulating cellular processes and enhancing host susceptibility to infection.32 Later, it was found that these effector proteins are also capable of DNA binding and activate the expression of the target genus.32TALE proteins consist of a central domain responsible for DNA binding and a nuclear localization signal that facilitates their targeting of gene transcription.34 The central domain is composed of repeating monomeric units, each specifically recognizing and binding a single nucleotide within the target sequence. These units are tandem repeats of 34 amino acid residues, and the last tandem repeat in the domain has 20 amino acids.35 Amino acids at positions 12 and 13 are highly variable and thus called Repeat Variable Diresidues30,35 (RVDs). RVDs are responsible for recognition of a specific nucleotide, and the four most common RVDs are histidine–aspartic acid (HD), asparagine–glycine (NG), asparagine–isoleucine (NI), and asparagine–asparagine (NN), accounting for each of the four nucleotides cytosine (C), thymine (T), adenine (A) and guanine (G), respectively.35,36 The first amino acid residue in RVDs, i.e. H or N, is not directly involved in nucleotide binding but plays a crucial role in stabilizing the spatial conformation of the domain. In contrast, the second amino acid residues in RVDs, i.e. D, G, I and N interact directly with the target nucleotide: D and N form hydrogen bonds, while I and G engage via van der Waals forces.35,37 The DNA-binding domain is integrated into a genetic construct containing a half-repeat, the N-terminal domain, and the catalytic domain of FokI.35 It functions in pairs, with binding sites strategically positioned on opposite DNA strands and separated by a short spacer sequence (12–25 bp). Upon nuclear entry, the nucleases recognize and bind to their target sites, facilitating the dimerization of FokI domains at the C-terminal of the chimeric protein. This dimerization induces DSBs within the spacer sequence32 as shown in Fig. 3. DSBs activate two repair pathways in the cell i.e. NHEJ and HDR.38
 | ||
| Fig. 3 Schematic representation of transcription activator-like effector nuclease architecture and double-strand break induction. | ||
Despite the success of TALENs, a significant limitation remains as it recognizes the target site based on DNA–protein interactions due to which it leads to some fundamental structural difference which causes its low off target activity, low specificity/effectiveness, and high production cost.39 These challenges prompted the emergence of a newer gene-editing tool i.e., CRISPR/Cas9.
3.3. Clustered regularly interspaced short palindromic repeats (CRISPR/Cas9)
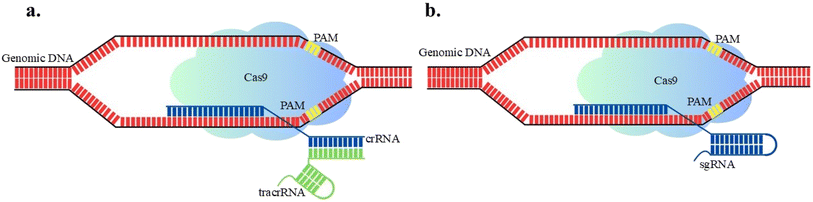
In 1987, Yoshizumi Ishino40 and team first discovered Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) in Escherichia coli, characterizing them as unique DNA repeats separated by spacer sequences. In the early 2000s, Barrangou et al.41 worked on Streptococcus thermophilus and identified that bacteria containing spacer sequences homologous to bacteriophages and viral genomes exhibit immunity against infection. During viral infection, CRISPR spacers are transcribed into CRISPR RNAs (crRNAs) with Cas proteins, target and cleave viral DNA or RNA to block infection.20 Emerging CRISPR-associated nucleases such as Cas12, Cas13, and Cas14 hold significant promise for the development of transgene-free (foreign DNA free) crops.42 These next-generation genome editors enable precise genetic modifications, offering a regulatory advantage that facilitates easier approval and commercialization across various countries.20 It is one of the best state-of-the-art genome editing technologies that is gaining popularity because of its broader applications across different organisms.42–44 The CRISPR–Cas system is mainly classified into two main classes as 1 and 2. Class 1 system includes type I, III and IV multiple effector proteins for the RNA-guided target cleavage, while Class 2 includes type II, V, and VI effector proteins which required only one RNA-guided endonuclease for the DNA sequence cleavage.20 Type I utilizes Cas3, which acts like a molecular motor, helping to unwind and cut DNA or RNA structures. Type II employs the well-known Cas9 protein, which accurately cuts double-stranded DNA at targeted locations. Cas9 contains two domains i.e., HNH domain and RucV-like domain. The HNH domain cuts the complementary strand of crRNA, whereas the RucV-like domain cleaves the opposite strand of double-stranded DNA.45 Type III involves Cas10, which can target single-stranded DNA and contains a specialized region called the palm domain that enables its activity. In Type IV, the system includes a protein known as csf1, which functions in a similar way to Cas8. Type V uses Cas12 (also called Cpf1), a versatile protein capable of cleaving both single and double-stranded DNA, and it also plays a role in DNA repair. Lastly, Type VI features Cas13, which is unique because it targets RNA instead of DNA. It contains the Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domain that allows it to cleave single-stranded RNA in both simple and complex organisms.46 The recent advent of CRISPR derived methodologies is base editing and prime editing, which have substantially expanded the scope and utility of precision genome editing.47,48 This review presents an overview of key developments in CRISPR/Cas technology and its applications in modern horticulture and agriculture.The CRISPR/Cas9 system is a bacterial RNA-guided immune defense, which targets and eliminates foreign DNA from plasmids, bacteriophages and functions as a form of bacterial “immune system”.49 It facilitates precise gene modification by introducing targeted DNA cleavage, followed by endogenous DNA repair mechanisms.43,50 CRISPR/Cas9 technology, first applied in plant genome editing in 2013, has revolutionized the field by enabling precise, efficient, and versatile genetic modifications.51,52 CRISPR/Cas9 requires the Cas9 protein and a Protospacer Adjacent Motif (PAM) sequence for efficient DSB formation in the target DNA. Cas9 utilizes a guide RNA (gRNA) duplex composed of CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) for target recognition as shown in Fig. 4a. The crRNA typically 18–20 nucleotides long, is essential for recognizing and binding to the target DNA sequence.53,54 The tracrRNA ranging from 50 to 150 nucleotides, plays a vital role in the Cas9-mediated DNA cleavage process.53,54 In current genetic engineering techniques, the duplex is combined into a single molecule called single guide RNA (sgRNA) as shown in Fig. 4b. More specifically, sgRNA is generated by linking the 3′ end of crRNA with the 5′ end of tracrRNA using a connector sequence.55 The PAM sequence length varies between two and six nucleotides, such as NGG (any nucleotide (N) followed by two guanine units), NAG (any nucleotide followed by adenine and thymine units), CTT (cytosine followed by two thymine units), and TTTV (three thymine units followed by V, where V is A, C, or G). PAM functions as a key indicator for identifying target sites as it instructs Cas9 to cleave a DNA strand at a precise location.56,57 The seed region adjacent to PAM consisting of 10–12 base pairs determines Cas9 specificity and makes it more essential than other regions in the sgRNA.55 The interaction allows Cas9 to make precise DSBs in the DNA.58 DSBs then activate repair processes, which can occur through one of the two primary mechanisms NHEJ and HDR, that are then utilized to introduce genetic modifications.58,59 Both NHEJ and HDR provide versatility and precision in plant genome editing and enhance resistance to diseases and insect pests60,61 however, HDR remains challenging due to its limitations in supplying sufficient repair templates.62
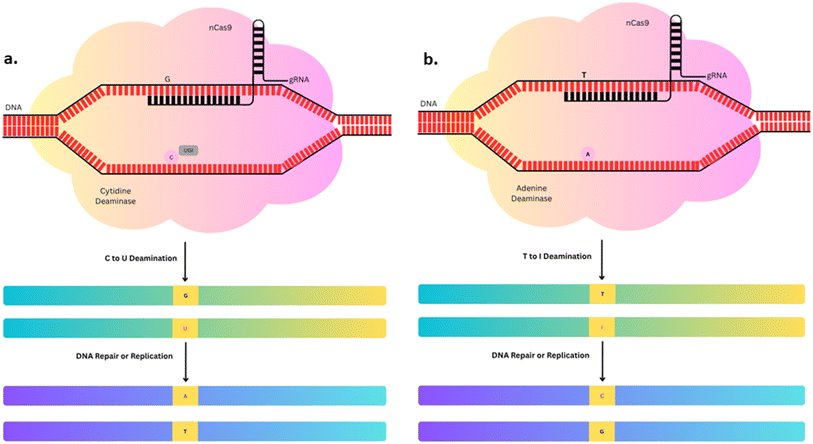
3.3.1.1 Cytosine Base Editors (CBEs). The first-generation CBE (CBE1) was developed by Liu and co-workers in 2016.65 The CRISPR/Cas9-mediated base editing platform incorporating cytosine deaminase facilitates highly precise and efficient single-nucleotide substitutions at defined genomic loci without generating double-stranded DNA breaks. This approach holds substantial potential for targeted gene correction and enhancement of genetic diversity across yeast, plant, mammalian, and human cells.66 It converts C to U (uracil), which is subsequently recognized as T during replication, resulting in a C–G to T–A transition. It has low efficiency because of cellular-mediated repair of the U–G intermediate in DNA by Base Excision Repair (BER). To overcome this, several base-editors such as CBE2 and CBE3 were developed to improve the editing efficiency. CBE3 shows the highest efficiency by six-fold over CBE2. It was developed by restoring the histidine residue at position 840 (H840) within the HNH catalytic domain of Cas9 to produce a base editor utilizing the Cas9 nickase (nCas9) variant. This introduced a single-strand nick in G containing a strand of U–G intermediate, directing the cellular repair machinery to favour conversion of the intermediate to a U–A pair, which is subsequently transformed into a stable T–A base pair during DNA replication64 as shown in Fig. 5a.
 | ||
| Fig. 5 Schematic illustration of the primary class of base editing tools. (a) Cytosine Base Editors (CBEs) and (b) Adenine Base Editors (ABEs). | ||
3.3.1.2 Adenine Base Editors (ABEs). CBEs are restricted to mediating C–G to T–A transitions, which substantially limit their capacity to correct a wider set of pathogenic nucleotide substitutions. Notably, methylated cytosines exhibit a high susceptibility to spontaneous deamination, and it is estimated that nearly half of all pathogenic point mutations could theoretically be corrected through ABEs by converting an A–T base pair back to its original G–C configuration.67 ABEs function through a similar mechanism as CBEs. The ABE–dCas9 fusion complex binds to the target DNA sequence in a guide RNA directed manner, where the deoxyadenosine deaminase domain catalyzes the conversion of adenine to inosine. During DNA replication, inosine is recognized as guanine, leading to the substitution of the original A–T base pair with a G–C base pair at the designated genomic site64 as shown in Fig. 5b.
In comparison to CBEs, ABEs produce markedly cleaner editing outcomes, characterized by an almost complete absence of indels and no reported instances of significant off-target A-to-G substitutions. Alkyladenine DNA Glycosylase (AAG), the enzyme responsible for recognizing and excising inosine from DNA, does not enhance ABE efficiency or product purity relative to wild-type cells.68 CBEs and ABEs are limited to four transition mutations, a constraint that prompted the development of prime editing for broader nucleotide modification.
 | ||
| Fig. 6 Prime editing and its derived techniques. Prime editors expand the scope of DNA editing to not all transition and transversion mutations, as well as small insertion and deletion mutations. | ||
CRISPR/Cas9 leads to these advancements, providing unmatched precision and efficiency in genome editing. This integrative approach will enable the development of crops that are not only resistant to diseases, pests, and weeds but also tailored to thrive under specific environmental conditions. Researchers experienced with CRISPR have helped newcomers by openly sharing methods and resources, unlike the restricted, company-controlled framework of zinc-finger nucleases.45 Several studies have successfully demonstrated the use of ZFNs, TALENs, and CRISPR/Cas9 in various plant species for precise gene editing to improve agronomic traits. These applications are summarized in Table 1, highlighting the practical outcomes of each technology in enhancing crop resistance, yield, and nutritional quality.
| Tool | Researcher(s) | Plant species | Target gene(s)/region | Outcome | References |
|---|---|---|---|---|---|
| ZFNs | Sylvia de Pater | Arabidopsis | PPO | Herbicide-insensitive enzyme due to gene mutation | 70 |
| ZFNs | Vipula K. Shukla | Zea maize | IPK1 | Herbicide tolerance; modified inositol phosphate profile | 71 |
| TALENs | Qiwei Shan | Brachypodium, rice | Multiple genes | Gene editing via NHEJ; large deletions with dual TALENs | 72 |
| TALENs | Yanpeng Wang | Bread wheat | 3 homoeoalleles | Powdery mildew resistance | 73 |
| TALENs | William Haun | Soybean | FAD2-1A and FAD2-1B | Improved oil quality and shelf life | 74 |
| TALENs | Toni Wendt | Barley | Genome-wide | ∼20% transformation efficiency; small deletions via NHEJ | 75 |
| CRISPR | Andrew S. Fister | Cacao | TcNPR3 | Resistance to Phytophthora tropicalis | 76 |
| CRISPR | Valero Pompili | Apple | MdDIPM4 | Potential for improved stress and disease resistance | 77 |
| CRISPR | Jun Li | Rice | EPSPS (via NHEJ) | Glyphosate resistance via targeted substitution (TILLING) | 78 |
| CRISPR | Anning Zhang | Rice | OsRR22 | Enhanced salinity tolerance | 79 |
| CRISPR | Xiaohong Sun | Apple (Malus sp.) | MdMKK9 | Increased anthocyanin accumulation | 80 |
| CRISPR | Akira Endo | Rice | OsOr | Elevated β-carotene levels | 81 |
4. Nucleic acid-based diagnostic techniques
Foodborne illnesses pose a significant threat to public health and the global economy. They impact the consumers, food industry, and regulatory systems. Bacterial and viral pathogens are the primary biotic agents compromising food safety.82 Traditionally, pathogen detection has relied on culture-based methods, but these are often time-consuming, costly, and labour-intensive. Therefore, nucleic acid-based techniques are widely adopted due to their higher sensitivity, speed and specificity. These molecular tools are increasingly replacing conventional approaches in routine food testing.84,85Conventional detection methods for plant and foodborne pathogens are broadly categorized as direct and indirect techniques.83,85 Direct methods include Polymerase Chain Reaction16,86 (PCR), immunoassays,87 and culture-based colony counting.88 Indirect methods are non-invasive and involve technologies such as thermography, gas chromatography, hyperspectral imaging, and fluorescence imaging.89 Despite their accuracy, these standard techniques are often time-consuming, costly, and labour-intensive.90 Initially, nucleic acid-based methods were limited to research laboratories due to their complexity and the requirement for skilled personnel.91 However, with technological advancements these approaches have become more accessible and are replacing culture-based and immunoassay techniques in routine food safety analysis.85,92 Nucleic acid-based technologies also play a critical role in both basic research and applied sciences. They are extensively used in clinical diagnostics, pathogen identification, gene cloning, and industrial quality control.85
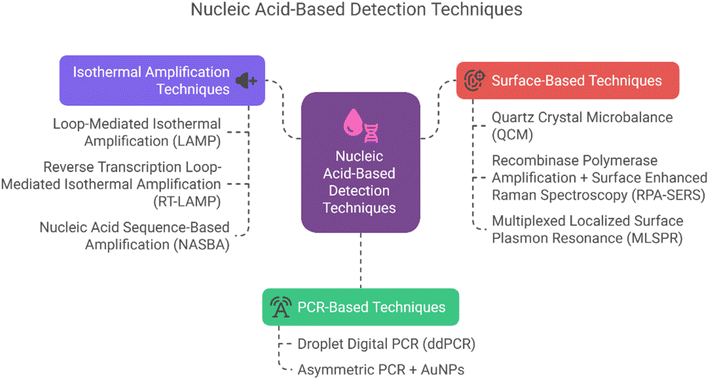
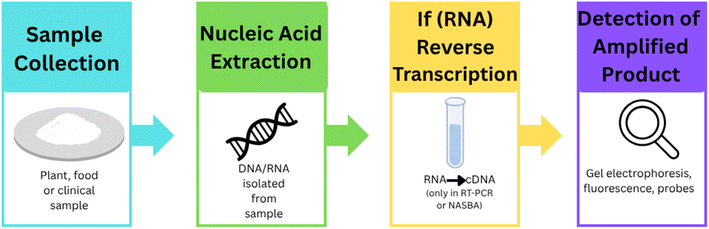
In recent years, various advanced nucleic acid-based diagnostic methods have emerged that overcome the limitations of conventional assays. These include techniques like Quartz Crystal Microbalance90 (QCM) using DNA amplicons, Recombinase Polymerase Amplification Surface-Enhanced Raman Scattering93 (RPA-SERS) and gold nanoparticle-based PCR.94 Additionally, isothermal amplification methods such as Loop-Mediated Isothermal Amplification95 (LAMP), Reverse Transcription Loop-Mediated Isothermal Amplification96 (RT-LAMP), and Nucleic Acid Sequence-Based Amplification97 (NASBA) offer rapid detection without the need for thermal cyclers. Multiplexed Localized Surface Plasmon Resonance98 (LSPR) approaches allow simultaneous detection of multiple DNA targets with high sensitivity. These technologies are not only faster but also suitable for point-of-care and field diagnostics.98 A schematic overview of these diagnostic techniques is presented in Fig. 7. Although these techniques differ in their experimental setups, they largely follow a similar core mechanism.99 All these methods target and amplify specific nucleic acid sequences (DNA or RNA) of the pathogen to enable sensitive detection. The general mechanism involves four key steps: sample preparation, nucleic acid extraction, amplification, and detection.99,100 The process begins with sample collection followed by nucleic acid extraction (DNA or RNA). If the target is RNA, it is usually converted to complementary DNA (cDNA) using reverse transcription. This is followed by amplification of the target nucleic acid, which can be achieved through isothermal techniques. Finally, detection is carried out using various methods including gel electrophoresis, fluorescence analysis, colorimetric changes, nanoparticle-based sensors (AuNPs), or signal-enhancing platforms such as quartz crystal-based biosensors.101,102 A systematic diagram representing this common mechanism is shown in Fig. 8, while the methodological variations among these techniques are summarized in Table 2. Several studies have successfully demonstrated the use of LAMP,95 QCM,90 and NASBA97 in various plant species for detection of plant pathogens to improve agronomic traits. These applications summarized in Table 3, demonstrate the use of nucleic acid-based biosensing tools for sensitive and rapid detection of plant pathogens and genetic markers using integrated amplification and nanotechnology platforms. Several other technologies are also used to detect plant pathogens such as CRISPR-based diagnostics101 that represent a transformative approach in the field of molecular diagnostics, offering rapid, accurate, and sensitive detection capabilities across various applications. These diagnostics leverage the CRISPR/Cas system, originally known for its gene-editing capabilities, to identify specific nucleic acid sequences, making them highly versatile tools for detecting pathogens, genetic mutations, and invasive species. While CRISPR-based diagnostics offer significant advantages, challenges remain in terms of scalability and integration into existing diagnostic frameworks. Future developments may focus on automation and modularization to enhance the accessibility and efficiency of these diagnostics in various settings.102
| Technique | Target | Amplification type | Isothermal? | Key enzymes/features | Detection method | References |
|---|---|---|---|---|---|---|
| ddPCR | DNA | PCR-based (partitioned) | ✗ | DNA polymerase; droplet-based quantification | Fluorescence in droplets | 103 |
| QCM | DNA | PCR (multiplex/conventional PCR) | ✗ | PCR-amplified; DNA detected | Mass-change-based acoustic biosensing using QCM | 90 |
| RPA–SERS | DNA | Recombinase-based | ☑ | Recombinase, DNA polymerase, and SSB | Raman signal of labeled probes | 93 |
| LAMP | DNA | Polymerase-based loop amplification | ☑ | DNA polymerase with loop primers | Turbidity, color, and fluorescence | 95 |
| RT-LAMP | RNA (converted to cDNA) | Isothermal | ☑ | Reverse transcriptase + Bst polymerase | Color change, fluorescence, and turbidity | 96 |
| NASBA | RNA | Transcription-based | ☑ | Reverse transcriptase, RNase H, and T7 RNA polymerase | Fluorescence- or probe-based | 97 |
| Asymmetric PCR + AuNPs | DNA | PCR (with excess primer) | ✗ | DNA polymerase; creates more ssDNA | Gold nanoparticle colour shift | 94 |
| MISPR | DNA | Not always with amplification | Sometimes | Surface plasmon resonance on nanoparticle arrays | Optical signal based on nanoparticle resonance | 104 |
| Tool/technique | Researcher(s) | Plant species/matrix | Target gene(s)/pathogen | Outcomes/findings | References |
|---|---|---|---|---|---|
| LAMP–QCM | Reona Takabatake | Papaya | Cauliflower mosaic virus 35S promoter and papaya endogenous | Detect genetically modified papaya with high specificity and sensitivity (≤0.05%) without requiring PCR instruments | 105 |
| RPA–LFD | Y. Zhou | Tomato | Tomato yellow leaf curl virus (TYLCV) | Rapid DNA detection of the TYLCV | 106 |
| LAMP | M. K. Prasannakumar | Rice | Sarocladium oryzae and Magnaporthe oryzae | Detection of 100 fg of pathogen DNA | 107 |
| RT-PCR | Sunil B. Kokane | Citrus tristiza | Citrus tristeza virus RNA | Real-time RNA detection using RT-PCR with high specificity | 108 |
| Multiplex PCR | Nelly Datukishvili | Maize, Soybean | 35S, NOS, EPSPS and cry1Ab gene | Identification of new DNA markers in GMO | 109 |
| LAMP–QCM | Sirirat Wachiralurpan | Bacterial DNA solution | Listeria monocytogenes | LAMP products monitored by QCM in real-time | 110 |
| LAMP–AuNPs (colorimetric) | Mila Djisalov | Mushroom substrate | Trichoderma spp., tef1 | AuNP-based colorimetric detection for rapid fungal screening | 111 |
5. RNA-based gene silencing
Sustainable production strategies including the development of safer pesticide alternatives are critically required to improve the current cropping system. The RNA interference (RNAi) mechanism has become a promising approach for effectively managing phytophagous pests and combating pathogenic attacks.112 With its inherent capability for the sequence-specific target, this technology is rapidly becoming a major focus of research as an environmentally sustainable and cost-efficient solution for pest management.113 It is a conserved eukaryotic mechanism that regulates gene expression at either the post-transcriptional level (post-transcriptional gene silencing (PTGS)) or the transcriptional level (transcriptional gene silencing (TGS)).114 TGS halts transcription by methylating the 5′-untranslated region (5′ UTR), preventing the binding of transcription factors, while in PTGS methylation of the coding region marks the transcript for degradation.115 In fungi, this PTGS mechanism is referred to as quelling.116,117In agriculture, RNAi has been widely utilized, especially for developing resistance to biotic stressors, bacteria, nematodes, fungi, and viruses.118 This phenomenon was initially discovered in the free-living nematode Caenorhabditis elegans.119 The RNAi mechanism is used to inhibit key growth and developmental genes in targeted phytophagous pests at the post-transcriptional level that helps to mitigate their harmful impact on crop plants. RNAi is activated by double strand RNA (dsRNA) and multidomain enzymes, such as the RNase III classes of enzymes in the Dicer family, which convert dsRNA into small interference RNA (siRNA), a double-strand RNA.120,121 These siRNAs are fused with the RNA induced silencing complex (RISC) having Argonaute (AGO) proteins. This RISC complex and siRNA complement with viral RNA and cleave/degrade the viral RNA122 as shown in Fig. 9.
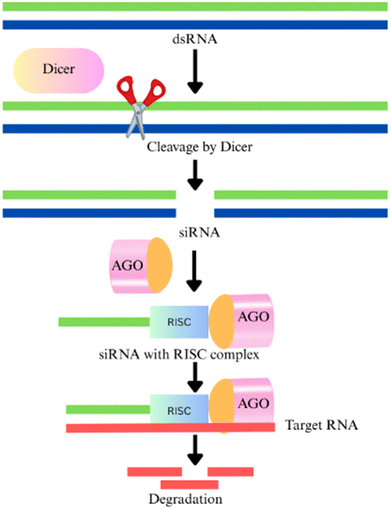 | ||
| Fig. 9 Mechanism of RNA silencing incorporating the Dicer family enzyme, RNA-induced silencing complex (RISC) formation and degradation of viral RNA. | ||
There are two sources of dsRNA i.e. endogenous dsRNA (inside the cell) and exogenous dsRNA (outside the cell).123,124 The introduction of exogenous dsRNA in plants involves various approaches, depending on the specific research objectives and intended applications. These approaches have been widely investigated in plants to examine gene functions via reverse genetics and to attain crop enhancement objectives through genetic modifications.115 The major strategies of dsRNA introduction in plants to protect from pests are Host Induced Gene Silencing125 (HIGS), Spray Induced Gene Silencing125 (SIGS), and a specialized RNAi-based approach – Virus Induced Gene Silencing126 (VIGS).
In HIGS, transgene plants (wheat, barley, rice, and maize) are genetically modified to produce dsRNA, which is taken by pests to silence the insect critical gene and protect plants from degradation125 as shown in Fig. 10a.
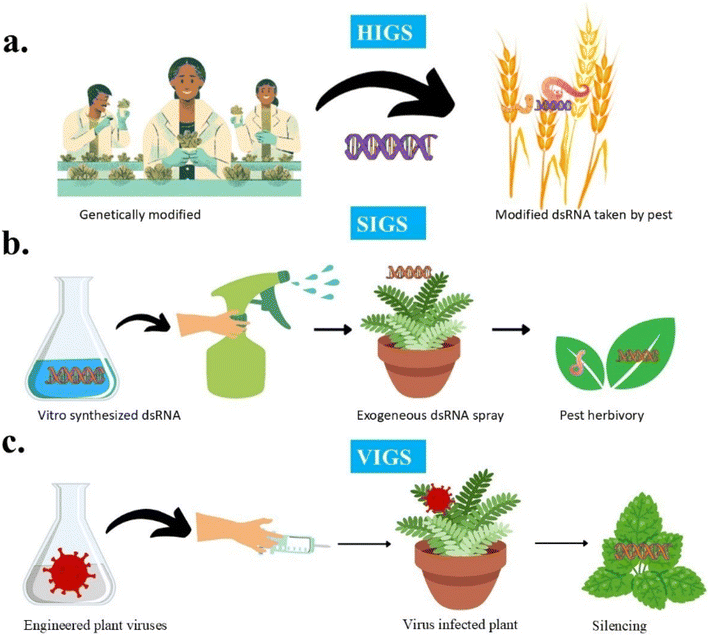 | ||
| Fig. 10 Strategies for dsRNA delivery for pest/pathogen control. (a) Host Induced Gene Silencing (HIGS), (b) Spray Induced Gene Silencing (SIGS) and (c) Virus Induced Gene Silencing (VIGS). | ||
In SIGS, dsRNA is synthesized in vitro which targets the essential gene in pathogens. Synthesized dsRNA is sprayed onto the plant surface (strawberry, lettuce, canola, and barley) by using nanocarriers or protective formulations which are absorbed by the pest or pathogen, either through feeding or direct contact. As a result, silencing of the targeted gene takes place inside the target organism125 as shown in Fig. 10b.
Virus induced gene silencing is a specialized RNAi-based approach that uses engineered plant viruses to assess gene functions in plants and control plant pests. Once the virus infects the plant, it triggers the plant defense mechanism. The plant produces dsRNA as a part of its response to the viral infection, and as a result silencing of gene takes place126 as shown in Fig. 10c. It offers significant advantages for functional genomics as it is rapid, does not require stable transformation, and also allows high-throughput screening of candidate genes involved in stress responses, defense mechanisms, and developmental processes. This makes VIGS particularly valuable for studying gene function in non-model plants or those with long generation times. Xijun Chen127 explored the use of composite nanomaterials as delivery systems for dsRNA to enhance plant protection against viral pathogens.
Artificial dsRNA produces a large population of siRNAs, out of which only a limited subset is functionally active in guiding RISC mediated target cleavage. The remaining, non-functional siRNAs can contribute to off-target effects, thereby reducing the overall efficiency and specificity of RNA silencing.128 However, a major challenge with topical application of naked dsRNA is its instability on the plant surface, leading to limited protection duration. Neena Mitter129 and her colleagues developed a novel nanocarrier system called BioClay, which uses Layered Double Hydroxide (LDH) nanosheets to deliver dsRNA in a stable form. When dsRNA is loaded onto these clay nanosheets, it becomes protected from environmental degradation, including nucleases and rain. Similarly, Marie Knoblich130 and colleagues developed an experimental platform termed ‘eNA screen’, which utilizes lysates from cultured Nicotiana tabacum BY-2 cells (BY-2 lysates). This system enables the identification of ‘effective siRNAs’ (esiRNAs) which are capable of directing efficient AGO/RISC-mediated target RNA cleavage. The application of the eNA screen to cucumber mosaic virus (CMV) allowed the identification of esiRNAs with strong antiviral activity, demonstrating their potential in conferring resistance against CMV infection.
RNAi can be effectively used to enhance the yield of crops, fruits and plants by modifying key agronomic traits such as plant height, branching, and size. Kenneth A. Feldmann131 employed RNAi to reduce the activity of the gene OsDWARF4 of rice that resulted in shorter plants with their leaves growing more upright instead of drooping, thus this increases light interception, leading to higher photosynthesis, and improved yields are observed under dense planting conditions. Hisano et al.132 used RNAi in the downregulation of some lignin genes like cinnamate 4-hydroxylase (C4H), shikimate hydroxycinnamoyl transferase (HCT) and 4-coumarate:CoA ligase (4CL) in plants that resulted in the reduction of lignin content, improved accessibility of cellulases for cellulose degradation, and increased dry matter degradability. Qiao et al.133 employed RNAi to reduce the activity of the enzyme GA 20-oxidase (OsGA20ox2) that resulted in a decrease in the length of rice variety named QX1, which helps in increasing the panicle length, number of seeds per panicle and test weight (1000 grains). Jiao134 and Miura135 observed that the OsSPL16 gene is a positive regulator of cell proliferation with an increase in grain width and yield in over-expressing rice plants. Its overexpression decreases the grain appearance and quality therefore, Jiao134 and Miura135 decreased the expression of OsSPL16 gene and resulted in slender grains with better quality. Davuluri et al.136 suppressed the DET1 gene in tomato that resulted in an increase in the level of flavonoid and carotenoid, which are highly beneficial for human health. Yu et al.137 suppressed the expression of lycopene epsilon cyclase (ε-CYC) gene by using RNAi and resulted in the enhancement of carotenoid content of rapeseed (Brassica napus). Dandekar et al.138 also utilized RNAi in apple to improve the fruit quality by enhanced self-life. Regina et al.139 used RNAi to down-regulate the starch-branching enzyme resulting in high-amylose wheat, which has great potential to improve human health. Gil Humanes et al.140 silenced the expression of specific γ-gliadins in different wheat cultivars and resulted in the enhancement of protein content of transgenic lines.
6. Integration of artificial intelligence with genome editing techniques for enhanced precision
Artificial Intelligence (AI) is being integrated into genome-editing pipelines to overcome major bottlenecks in precision and predictability. Deep learning and machine learning (ML) models are widely used to design tools with high efficiency, generate efficient gRNAs with minimal off-target effects by analyzing sequence performance patterns from extensive data. AI platforms such as DeepCRISPR,141 ZFN-Site,143 CCTop143 and siRNA-Finder144 enable prediction of off-target sites with far greater accuracy than rule-based tools. In base and prime editing ML frameworks help in optimizing the sequence context, thereby reducing byproducts and improving editing fidelity. AI-driven prediction supports the selection of biologically relevant targets before experimental validation. In plants, AI also assists in optimizing delivery constructs, regulatory elements, and tissue specific expression strategies. Collectively, the integration of AI with genome editing substantially enhances edit precision, reduces experimental iterations, and accelerates the development of reliable, high-performance, crop-improvement strategies.7. Computational resources for precision genome editing and nucleic acid-based biosensing
AI models can analyze large genomic datasets to optimize gRNA selection, predict off-target effects, and model protein–DNA/RNA interactions with high accuracy. Platforms such as DeepCRISPR141 and CNN-based gRNA scoring145 systems exemplify how AI is transforming genome engineering into a more predictive, efficient, and scalable process. Over the past few years, various computational technologies utilizing AI and ML have been designed for target identification, predicting/reducing off-target activity of gene editing and gene regulation methodologies. In addition to genome editing, AI/ML approaches are also being employed in nucleic acid-based detection techniques such as droplet digital PCR103 (ddPCR) and LAMP107 where they assist in designing specific primers and probes, enhance signal analysis from biosensor platforms and improve multiplex detection efficiency. Several dedicated tools are now available for designing primers, like PrimerExplorer,146 NEB LAMP Primer146 Design Tool, FastPCR,147 etc., as listed in Table 4. Databases used for providing target sequences in ZFN, TALEN, CRISPR/Cas9 and RNAi are presented in Table 5. Off-target activity encompasses mainly three major categories: software tools and packages, specialized databases, and web-based platforms, each of which exhibits diverse areas of application. Certain resources are tailored for specific genomes, whereas others offer broader compatibility, accommodating any user-defined genomic input. While some tools prioritize computational speed and scalability for larger genomes, others are optimized for precise algorithmic control and customization.148 Common features shown by each tool are off-targets, scoring, ranking and genome specificity. Computational tools and their detailed comparison of off-target feature are presented in Table 6.| Software/Tool | Purpose | References |
|---|---|---|
| PrimerExplorer | A widely used software for designing LAMP primers, offering a user-friendly interface for constructing primers for loop-mediated isothermal amplification | 146 |
| NEB LAMP primer design tool | A tool provided by New England Biolabs for designing LAMP primers, gaining popularity for its ease of use and specificity | 146 |
| FastPCR | An integrated tool for designing primers for various PCR applications, including LAMP, multiplex PCR, and long-distance PCR | 147 |
| ThermoPlex | An automated design tool for target-specific multiplex PCR primers based on DNA thermodynamics, ensuring optimal primer design | 149 |
| PrimerJinn | A tool for designing multiplex PCR primer sets and performing in silico PCR evaluation, particularly for targeted sequencing of pathogens | 150 |
| PMPrimer | A Python-based tool for automated design and evaluation of multiplex PCR primer pairs using diverse templates | 151 |
| PrimerScore2 | A high-throughput primer design tool that uses a piecewise logistic model to score primers for multiple PCR variants | 152 |
| SADDLE | A stochastic algorithm for designing highly multiplex PCR primer sets with minimal primer dimer formation | 153 |
| MOPSO-based primer design | A multiobjective particle swarm optimization (MOPSO) approach for designing primers based on user-specified parameters | 154 |
| Ultiplex | A web-based multiplex PCR primer design tool that supports up to 100-plex multiplicity and includes compatibility checking for primer groups | 155 |
| NGS-PrimerPlex | A command-line application for designing primers for amplicon-based genome target enrichment in multiplex PCR | 156 |
| MRPrimerW | A web-based tool for designing high-quality primers for multiple target qPCR experiments, including homology testing and TaqMan probes | 157 |
| GPrimer | A GPU-based pipeline for rapid primer design, significantly improving computational speed compared to traditional methods | 158 |
| PrimerServer | A high-throughput primer design and specificity-checking platform with web and command-line interfaces for large-scale applications | 159 |
| Database | Purpose | References |
|---|---|---|
| EENdb | Database for ZFNs and TALENs, providing target sequences and efficiency data | 160 |
| PICKLES | Provides data from pooled CRISPR knockout screens | 161 |
| Cas-database | Designs genome-wide guide RNA libraries for CRISPR screens | 162 |
| CHOPCHOP | Designs CRISPR/Cas9 and TALEN constructs | 22 |
| grlD | Guides RNA database with gRNA properties | 163 |
| CRISPRz | Validates CRISPR target sites in zebrafish | 164 |
| CRISPRdirect | Designs target site-specific gRNA sequences | 165 |
| CRISPR-based | |||
|---|---|---|---|
| CRISPR tools | Type | Off-targets | References |
| Cas9 design | Tool and database | ✓ | 173 |
| CasOT | Tool | ✓ | 174 |
| Cas-OFFinder | Tool | ✓ | 175 |
| CRISPR optimal target finder | Tool | ✓ | 176 |
| E-CRISP | Tool | ✓ | 177 |
| CRISPR-P | Tool | ✓ | 178 |
| GT-SCAN | Tool | ✓ | 179 |
| CRISPy | Tool | ✓ | 180 |
| sgRNAcas9 | Package | ✓ | 181 |
| CRISPRseek | Package | ✓ | 182 |
| COSMID | Tool | ✓ | 21 |
| CRISPRdirect | Tool | ✓ | 183 |
| Off-Spotter | Tool | ✓ | 184 |
| CRISPR multitargeter | Tool | ✗ | 185 |
| CCTop | Tool | ✓ | 143 |
| CrisprGE | Database | ✓ | 186 |
| WGE | Package | ✓ | 187 |
| RNAi-based | |||
|---|---|---|---|
| RNAi tools | Type | Applications | References |
| pssRNAit | Tool | Designing siRNAs | 190 |
| siRNA-Finder | Tool | Designing siRNAs | 144 |
| sPARTA | Tool | Analyzing miRNA | 191 |
| AttSiOff | Tool | Off-targets | 192 |
| MIRZA-G | Tool | Off-targets | 193 |
| siRNADesign | Tool | Off-targets | 194 |
| dsRNAEngineer | Tool | Designing dsRNAs | 195 |
| kmerPMTF | Tool | Predicting miRNA | 196 |
| PAREameters | Tool | Identifying miRNA | 197 |
| RNA Degradome | Tool | Predicting sRNA | 198 |
8. Conclusions and future prospects
Gene editing and nucleic acid-based technologies are opening new possibilities in plant biotechnology. These tools offer accurate and sustainable methods to boost crop yield, improve disease resistance, and ensure food security. Among them, genome editing tools like ZFNs, TALENs, and CRISPR/Cas9 have transformed plant breeding. They allow targeted changes in the plant genome by creating DSBs, which are repaired by the plant's natural mechanisms. On the other hand, the base editing and prime editing tools enable precise, programmable nucleotide changes without DSBs. While ZFNs and TALENs provided the early framework for gene editing, CRISPR/Cas9 is now most widely used. Its popularity comes from its simplicity, high precision, and ease of design. The capacity to induce targeted genetic modifications enables the precise introduction or alteration of desirable traits in elite crop varieties utilized for breeding and agricultural production. Moreover, the potential to stack multiple genetic modifications offers opportunities for the development of multi-trait resistance and the controlled expression of pharmaceutically important proteins. However, challenges such as off-target mutations, low editing accuracy, and inefficient delivery methods still need to be addressed. While no system has yet been established as most efficient and safe, ongoing efforts, particularly computational approaches aimed at minimizing off-target effects, hold promise for enhancing the precision and overall efficacy of therapeutic applications.Alongside gene editing, nucleic acid-based diagnostic methods are essential for plant health monitoring. Techniques such as PCR16 and LAMP95 offer fast and sensitive detection of pathogens, even at early stages. These diagnostics are being improved through integration with nanotechnology. These enhancements make them suitable for field-level use and point-of-care testing. This helps in reducing yield loss caused by undetected plant diseases.
In recent years, the global scientific and regulatory communities have been engaged in an ongoing debate regarding whether the application of these genome editing techniques falls within the framework of existing genetically modified organism legislation.199 Regulatory clarity for New Genomic Techniques (NGTs) is still evolving. The European Food Safety Authority (EFSA) concludes that cisgenic and intragenic plants developed through NGTs do not pose new or additional risks compared to conventionally bred plants and recommends risk-proportionate, science-based assessment criteria.200 The US Department of Agriculture decides in a case-by-case manner and has stated on requests that small mutations in corn induced by ZFNs fall outside their scope of regulation.201
RNA interference (RNAi) technologies, including siRNA and miRNA, have become valuable for plant researchers. These techniques are employed not only to explore plant functions but also to engineer plants with enhanced or novel traits through the manipulation of both beneficial and detrimental genes. This technology has been effectively applied in crops to enhance not only food productivity (such as biomass and grain yield) but also their nutritional value, with cereals, fruits, and vegetables being enriched with essential minerals, vitamins, fatty acids, and amino acids. RNAi technology has also been exploited to develop plants with improved resistance against various environmental stresses (especially drought). Although RNAi technology can serve as a potential tool for crop improvement, certain limitations are also associated with it. Altering the expression of a target gene might lead to undesirable changes in plant morphology and development; therefore, transgenic strategies should be designed only after completely understanding the mechanism of its regulation. siRNA-based RNAi strategies might not be suitable for some applications requiring tissue-specific silencing of genes.202
In coming years, ongoing advancements and innovations are expected to yield more robust computational frameworks for genome editing. Computational tools and AI play key roles in advancing these technologies. They help in improving the accuracy, speed, and scalability of genome editing, diagnostics, and RNAi technologies. Tools including grlD,163 CHOPCHOP,22 PrimerExplorer,146 siFi144 and sPARTA172 assist in designing gRNAs, primers, and siRNAs. These tools reduce off-target effects and improve the targeting efficiency. Specialized databases also support gene selection, off-target prediction, and RNA structure analysis. Together, they make molecular tools more reliable and user-friendly. Notably, recent computational studies demonstrated the potential of modified siRNA molecules to effectively inhibit viral RNA-silencing suppressor proteins, such as p19, thereby restoring the RNAi mechanism and improving resistance against plant viruses.203 These emerging tools and algorithms hold the potential to significantly enhance genome editing and nucleic acid-based technologies by contributing to both pre-editing design and post-editing analysis, thereby expanding their applicability.
Looking ahead, combining genome editing, RNA-based methods, diagnostics, and AI could transform agriculture. These integrated systems will help develop crops that are more resilient, high-yielding, and rich in nutrients. These approaches will also allow precision breeding tailored to the local environment and nutritional needs. The integration of genome editing with synthetic biology could pave the way for designing new metabolic pathways and creating biofortified crops to tackle malnutrition. However, the widespread application of these technologies will also require robust regulatory frameworks, ethical considerations, and increased public awareness to ensure their safe and acceptable use. Promoting farmer education and consumer acceptance will be essential for realizing the full potential of nucleic acid research in ensuring sustainable food security for future generations. In future, tools that exhibit substantial potential in terms of their applicability may consequently be envisioned as among the most widely adopted next-generation technologies for both scientific and therapeutic applications.
In conclusion, multiple proof-of-concept studies have already demonstrated the practical success of various technologies. CRISPR-edited rice lines with OsRR22 mutations show enhanced salinity tolerance,79 TALEN-mediated MLO knockouts in wheat confer powdery mildew resistance73 and ZFN-engineered IPK1 maize exhibits herbicide tolerance.71 Likewise, LAMP-QCM assays detect GMO papaya with 0.05% sensitivity.102 Such validated examples collectively confirm that nucleic-acid-based editing, diagnostics, and RNAi technologies are not merely theoretical innovations but proven tools with strong translational potential in agriculture. They offer precise, efficient, and eco-friendly solutions to address global challenges such as food insecurity and climate change. Continued innovation, supportive policies, and responsible use will ensure these technologies become foundational for resilient food systems in the future.
9. Limitations and biosafety considerations
Notwithstanding the accelerating developments in genome editing and RNA-based methodologies for enhancing crop traits, a number of considerable challenges remain unresolved. The efficacy of introducing editing tools is severely compromised in plant species characterized by complex genomic architectures or strong inherent biological barriers. Off-target effects and variable editing efficiency restrict the reproducibility and stability of edited traits. The stability and successful cellular uptake of dsRNA in RNAi-based applications are highly susceptible to ambient environmental parameters.From a biosafety standpoint, the comprehensive assessment of ecological and regulatory implications is imperative. Elevated risks require analysis, specifically concerning unintended genetic dispersal (gene flow), non-specific gene silencing in organisms outside the target scope, and the environmental persistence of the introduced nucleic acids or editing machinery. Risk assessment frameworks and public acceptance also play critical roles in translating laboratory advances into sustainable agricultural solutions.
Conflicts of interest
The authors declare that they have no conflicts of interest.Data availability
New data are not generated as a part of the submitted study.Acknowledgements
Yashika acknowledges Netaji Subhas University of Technology for funding.References
- A. Faizal, S. Nugroho, A. A. Sembada, Y. Theda, T. Komariyah and R. R. Esyanti, Genome Editing in Future Crop Protection: Utilizing CRISPR/Cas9 to Improve Crop Resistance against Diseases, Pests, and Weeds, Discov. Agric., 2024, 2(1), 104 CrossRef.
- S. Fiaz, S. Ahmar, S. Saeed, A. Riaz, F. Mora-Poblete and K. H. Jung, Evolution and application of genome editing techniques for achieving food and nutritional security, Int. J. Mol. Sci., 2021, 22(11), 5585 CrossRef CAS.
- S. He and K. M. Creasey Krainer, Pandemics of People and Plants: Which Is the Greater Threat to Food Security?, Mol. Plant, 2020, 13(7), 933–934 Search PubMed.
- Food and Agriculture Organization (FAO) of the United Nations, Climate Change Fans Spread of Pests and Threatens Plants and Crops, New FAO Study. Rome, 2021 Search PubMed.
- R. N. Strange and P. R. Scott, Plant Disease: A Threat to Global Food Security, Annu. Rev. Phytopathol., 2005, 43, 83–116 Search PubMed.
- UNDESA. World Population Prospects, the 2022 Revision; Population Division, Department of Economic and Social Affairs, United Nations, New York, NY, USA, 2022 Search PubMed.
- E. K. Norrman, World Population Growth: A Once and Future Global Concern, World, 2023, 4, 684–697 CrossRef.
- S. K. Patel, A. Sharma and G. S. Singh, Traditional agricultural practices in India: an approach for environmental sustainability and food security, Energy, Ecol. Environ., 2020, 5(4), 253–271 CrossRef.
- A. O. Adefila, O. O. Ajayi, A. S. Toromade and N. J. Sam-Bulya, Integrating traditional knowledge with modern agricultural practices: A sociocultural framework for sustainable development, World J. Biol. Pharm. Health Sci., 2024, 20(02), 125–135 Search PubMed.
- T. Johns, B. Powell, P. Maundu and P. B. Eyzaguirre, Agricultural biodiversity as a link between traditional food systems and contemporary development, social integrity and ecological health, J. Sci. Food Agric., 2013, 93(14), 3433–3442 Search PubMed.
- H. N. Barman, Z. Sheng, S. Fiaz, M. Zhong, Y. Wu, Y. Cai, W. Wang, G. Jiao, S. Tang, X. Wei and P. Hu, Generation of a New Thermo-Sensitive Genic Male Sterile Rice Line by Targeted Mutagenesis of TMS5 Gene through CRISPR/Cas9 System, BMC Plant Biol., 2019, 19(1), 109 Search PubMed.
- R. ManshardtCrop Improvement by Conventional Breeding or Genetic Engineering: How Different Are They, 2004. http://www.ctahr.hawaii.edu Search PubMed.
- K. P. Voss-Fels, A. Stahl, B. Wittkop, C. Lichthardt, S. Nagler, T. Rose, T. W. Chen, H. Zetzsche, S. Seddig, M. Majid Baig, A. Ballvora, M. Frisch, E. Ross, B. J. Hayes, M. J. Hayden, F. Ordon, J. Leon, H. Kage, W. Friedt, H. Stützel and R. J. Snowdon, Breeding Improves Wheat Productivity under Contrasting Agrochemical Input Levels, Nat. Plants, 2019, 5(7), 706–714 CrossRef.
- M. C. Lallawmkimi, P. Ashoka, D. J. M. S. N. K. S. Veda, A. Yadav, B. Dhivya, M. Kumar and A. Rout, Innovative Approaches in Crop Genetic Engineering for Sustainable Agriculture: A Review, J. Adv. Biol. Biotechnol., 2024, 27(8), 615–631 Search PubMed.
- S. Fiaz, S. Ahmar, S. Saeed, A. Riaz, F. Mora-Poblete and K. H. Jung, Evolution and Application of Genome Editing Techniques for Achieving Food and Nutritional Security, Int. J. Mol. Sci., 2021, 22(11), 5585 CrossRef CAS.
- M. Jalali, J. Zaborowska and M. Jalali, The Polymerase Chain Reaction: PCR, QPCR, and RT-PCR, in Basic Science Methods for Clinical Researchers, 2017, pp. 1–18 Search PubMed.
- S. Lei, S. Chen and Q. Zhong, Digital PCR for Accurate Quantification of Pathogens: Principles, Applications, Challenges and Future Prospects, Int. J. Biol. Macromol., 2021, 184, 750–759 CrossRef CAS PubMed.
- C. T. Scott, The zinc finger nuclease monopoly, Nat. Biotechnol., 2005, 23(8), 915–919 Search PubMed.
- K. Chen and C. Gao, TALENs: Customizable Molecular DNA Scissors for Genome Engineering of Plants, J. Genet. Genomics, 2013, 40(6), 271–279 Search PubMed.
- S. Mishra, S. Nayak, N. Tuteja, S. Poosapati, D. M. Swain, R. K. Sahoo, CRISPR/Cas Mediated Genome Engineering in Plants: Application and Future Prospective, preprint, 2024, DOI:10.20944/preprints202404.0435.v2.
- T. J. Cradick, P. Qiu, C. M. Lee, E. J. Fine and G. Bao, COSMID: A Web-Based Tool for Identifying and Validating CRISPR/Cas off-Target Sites, Mol. Ther.--Nucleic Acids, 2014, 3(12), e214 CrossRef CAS.
- T. G. Montague, J. M. Cruz, J. A. Gagnon, G. M. Church and E. Valen, CHOPCHOP: A CRISPR/Cas9 and TALEN Web Tool for Genome Editing, Nucleic Acids Res., 2014, 42(W1), W401–W407 Search PubMed.
- M. K. Samanta, A. Dey and S. Gayen, CRISPR/Cas9: An Advanced Tool for Editing Plant Genomes, Transgenic Res., 2016, 25, 561–573 CrossRef CAS.
- F. D. Urnov, E. J. Rebar, M. C. Holmes, H. S. Zhang and P. D. Gregory, Genome Editing with Engineered Zinc Finger Nucleases, Nat. Rev. Genet., 2010, 11, 636–646 Search PubMed.
- A. Rasheed, R. A. Gill, M. U. Hassan, A. Mahmood, S. Qari, Q. U. Zaman, M. Ilyas, M. Aamer, M. Batool, H. Li and Z. Wu, A Critical Review: Recent Advancements in the Use of Crispr/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises, Curr. Issues Mol. Biol., 2021, 43(3), 1950–1976 Search PubMed.
- C. Wyman and R. Kanaar, DNA Double-Strand Break Repair: All's Well That Ends Well, Annu. Rev. Genet., 2006, 40, 363–383 Search PubMed.
- T. Gaj, C. A. Gersbach and C. F. Barbas, ZFN, TALEN, and CRISPR/Cas-Based Methods for Genome Engineering, Trends Biotechnol., 2013, 31(7), 397–405 Search PubMed.
- S. A. Wolfe, L. Nekludova and C. O. Pabo, DNA Recognition by Cys 2 His 2Zinc Finger Proteins, Annu. Rev. Biophys., 2000, 29, 183–212 CrossRef CAS.
- É. S. Vanamee, S. Santagata and A. K. Aggarwal, FokI Requires Two Specific DNA Sites for Cleavage, J. Mol. Biol., 2001, 309(1), 69–78 CrossRef.
- J. Bitinaite, D. A. Wah, A. K. Aggarwal and I. Schildkraut, FokI Dimerization Is Required for DNA Cleavage, Biophy. Comput. Biol., 1998, 95(18), 10570–10575 CAS.
- D. Carroll, Genome Engineering with Zinc-Finger Nucleases, Genetics, 2011, 188(4), 773–782 CrossRef CAS.
- A. A. Nemudryi, K. R. Valetdinova, S. P. Medvedev and S. M. Zakian, TALEN and CRISPR/Cas Genome Editing Systems, Tools Discovery, 2014, 6, 19–40 CAS.
- Editorial, Method of the Year 2011, Nat. Methods, 2012, 1 DOI:10.1038/nmeth.1852.
- S. Schornack, A. Meyer, P. Römer, T. Jordan and T. Lahaye, Gene-for-Gene-Mediated Recognition of Nuclear-Targeted AvrBs3-like Bacterial Effector Proteins, J. Plant Physiol., 2006, 163(3), 256–272 CrossRef CAS.
- A. J. Bogdanove and D. F. Voytas, TAL Effectors: Customizable Proteins for DNA Targeting, Science, 2011, 333, 1843–1846 CrossRef CAS PubMed.
- C. Bassi, J. Ho, T. Srikumar, R. J. O. Dowling, C. Gorrini, S. J. Miller, T. W. Mak, B. G. Neel, B. Raught and V. Stambolic, Nuclear PTEN Controls DNA Repair and Sensitivity to Genotoxic Stress, Science, 2013, 341(6144), 395–399 CrossRef CAS.
- L. W. Seymour and A. J. Thrasher, Gene Therapy Matures in the Clinic, Nat. Biotechnol., 2012, 30, 588–593 CrossRef CAS PubMed.
- A. Knoll, F. Fauser and H. Puchta, DNA Recombination in Somatic Plant Cells: Mechanisms and Evolutionary Consequences, Chromosome Res., 2014, 22, 191–201 CrossRef CAS.
- A. Bhardwaj and V. Nain, TALENs—an Indispensable Tool in the Era of CRISPR: A Mini Review, J. Genet. Eng. Biotechnol., 2021, 19(1), 125 CrossRef PubMed.
- Y. Ishino, H. Shinagawa, K. Makino, M. Amemura and A. Nakata, Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product, J. Bacteriol., 1987, 169(12), 5429–5433 CrossRef CAS PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, A. D. Romero and P. Horvath, CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes, Science, 2007, 315(5819), 1709–1712 CrossRef CAS.
- S. Rauf, A. Shahid, M. Faizan, M. N. Khalid and I. Amjad, CRISPER/CAS: A potential tool for genomes editing, Agrobiol. Rec., 2024, 15, 13–23 CrossRef PubMed.
- F. Akram, S. Sahreen, F. Aamir, I. u. Haq, K. Malik, M. Imtiaz, W. Naseem, N. Nasir and H. M. Waheed, An Insight into Modern Targeted Genome-Editing Technologies with a Special Focus on CRISPR/Cas9 and Its Applications, Mol. Biotechnol., 2023, 65, 227–242 CrossRef CAS.
- M. Lyu, Y. Sun, N. Yan, Q. Chen, X. Wang, Z. Wei, Z. Zhang and K. Xu, Efficient CRISPR/Cas9-Mediated Gene Editing in Mammalian Cells by the Novel Selectable Traffic Light Reporters, Int. J. Biol. Macromol., 2023, 243, 124926 CrossRef PubMed.
- S. Fiaz, S. Ahmad, M. A. Noor, X. Wang, A. Younas, A. Riaz, A. Riaz and F. Ali, Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities, Int. J. Mol. Sci., 2019, 20(4), 888 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf, J. Iranzo, S. A. Shmakov, O. S. Alkhnbashi, S. J. J. Brouns, E. Charpentier, D. Cheng, D. H. Haft, P. Horvath, S. Moineau, F. J. M. Mojica, D. Scott, S. A. Shah, V. Siksnys, M. P. Terns, Č. Venclovas, M. F. White, A. F. Yakunin, W. Yan, F. Zhang, R. A. Garrett, R. Backofen, J. van der Oost, R. Barrangou and E. V. Koonin, Evolutionary Classification of CRISPR–Cas Systems: A Burst of Class 2 and Derived Variants, Nat. Rev. Microbiol., 2020, 18, 67–83 CrossRef CAS.
- Y. Xie, S. I. U. Haq, X. Jiang, D. Zheng, N. Feng, W. Wang and Q. S. Qiu, Plant genome editing: CRISPR, base editing, prime editing, and beyond, Grassl. Res., 2022, 1(4), 234–243 CrossRef CAS.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage, Nature, 2017, 551(7681), 464–471 CrossRef CAS.
- Y. Xu and Z. Li, CRISPR-Cas Systems: Overview, Innovations and Applications in Human Disease Research and Gene Therapy, Comput. Struct. Biotechnol. J., 2020, 18, 2401–2415 CrossRef CAS.
- A. Rasheed, R. A. Gill, M. U. Hassan, A. Mahmood, S. Qari, Q. U. Zaman, M. Ilyas, M. Aamer, M. Batool, H. Li and Z. Wu, A Critical Review: Recent Advancements in the Use of Crispr/Cas9 Technology to Enhance Crops and Alleviate Global Food Crises, Curr. Issues Mol. Biol., 2021, 43(3), 1950–1976 CrossRef CAS PubMed.
- Y. Fu, J. A. Foden, C. Khayter, M. L. Maeder, D. Reyon, J. K. Joung and J. D. Sander, High-Frequency off-Target Mutagenesis Induced by CRISPR-Cas Nucleases in Human Cells, Nat. Biotechnol., 2013, 31(9), 822–826 CrossRef CAS.
- Y. Mao, H. Zhang, N. Xu, B. Zhang, F. Gou and J. K. Zhu, Application of the CRISPR-Cas System for Efficient Genome Engineering in Plants, Mol. Plant, 2013, 6(6), 2008–2011 Search PubMed.
- M. A. Nethery and R. Barrangou, Predicting and Visualizing Features of CRISPR–Cas Systems, Methods Enzymol., 2019, 616, 1–25 CAS.
- M. A. Mengstie and B. Z. Wondimu, Mechanism and Applications of Crispr/Cas-9-Mediated Genome Editing, Biol.: Targets Ther., 2021, 15, 353–361 Search PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity, Science, 2012, 337(6096), 816–821 CrossRef CAS.
- G. Gasiunas, R. Barrangou, P. Horvath and V. Siksnys, Cas9-CrRNA Ribonucleoprotein Complex Mediates Specific DNA Cleavage for Adaptive Immunity in Bacteria, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(39), E2579–E2586 CrossRef CAS PubMed.
- O. Rawashdeh, R. Y. Rawashdeh, T. Kebede, D. Kapp and A. Ralescu, Bio-Informatic Analysis of CRISPR Protospacer Adjacent Motifs (PAMs) in T4 Genome, BMC Genomic Data, 2022, 23(1), 40 CrossRef CAS.
- J. A. Doudna and E. Charpentier, The New Frontier of Genome Engineering with CRISPR-Cas9, Science, 2014, 346, 6213 CrossRef PubMed.
- W. S. Ganpatrao and P. M. Baliram, Current and Future Prospects of Plant Breeding with CRISPR/Cas, Curr. J. Appl. Sci. Technol., 2019, 1–17, 2457–1024 Search PubMed.
- A. Macovei, N. R. Sevilla, C. Cantos, G. B. Jonson, I. Slamet-Loedin, T. Čermák, D. F. Voytas, I. R. Choi and P. Chadha-Mohanty, Novel Alleles of Rice EIF4G Generated by CRISPR/Cas9-Targeted Mutagenesis Confer Resistance to Rice Tungro Spherical Virus, Plant Biotechnol. J., 2018, 16(11), 1918–1927 CrossRef CAS PubMed.
- Y. Wang, X. Cheng, Q. Shan, Y. Zhang, J. Liu, C. Gao and J. L. Qiu, Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew, Nat. Biotechnol., 2014, 32(9), 947–951 CrossRef CAS.
- M. Wang, Y. Lu, J. R. Botella, Y. Mao, K. Hua and J. K. Zhu, Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System, Mol. Plant, 2017, 10(7), 1007–1010 CrossRef CAS PubMed.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage, Nature, 2016, 533(7603), 420–424 CrossRef CAS PubMed.
- A. Kantor, M. E. McClements and R. E. MacLaren, CRISPR-Cas9 DNA base-editing and prime-editing, Int. J. Mol. Sci., 2020, 21(17), 6240 CrossRef CAS.
- A. C. Komor, A. H. Badran and D. R. Liu, CRISPR-based technologies for the manipulation of eukaryotic genomes, Cell, 2017, 168(1), 20–36 CrossRef CAS PubMed.
- S. S. Bharat, S. Li, J. Li, L. Yan and L. Xia, Base editing in plants: current status and challenges, Crop J., 2020, 8(3), 384–395 CrossRef.
- L. Alsøe, A. Sarno, S. Carracedo, D. Domanska, F. Dingler, L. Lirussi, T. SenGupta, B. N. Tekin, L. Jobert, B. L. Alexandrov, A. Galashevskaya, C. Rada, K. J. Sandve, T. Rongnes, E. H. Krokan and H. Nilsen, Uracil accumulation and mutagenesis dominated by cytosine deamination in CpG dinucleotides in mice lacking UNG and SMUG1, Sci. Rep., 2017, 7(1), 7199 CrossRef.
- T. P. Huang, K. T. Zhao, S. M. Miller, N. M. Gaudelli, B. L. Oakes, C. Fellmann, F. D. Savage and D. R. Liu, Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors, Nat. Biotechnol., 2019, 37(6), 626–631 CrossRef CAS PubMed.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, J. P. Chen, C. Wilson, A. G. Newby, A. Raguram and D. R. Liu, Search-and-replace genome editing without double-strand breaks or donor DNA, Nature, 2019, 576(7785), 149–157 CrossRef CAS.
- S. De Pater, J. E. Pinas, P. J. J. Hooykaas and B. J. van der Zaal, ZFN-Mediated Gene Targeting of the Arabidopsis Protoporphyrinogen Oxidase Gene through Agrobacterium-Mediated Floral Dip Transformation, Plant Biotechnol. J., 2013, 11(4), 510–515 CrossRef CAS.
- V. K. Shukla, Y. Doyon, J. C. Miller, R. C. Dekelver, E. A. Moehle, S. E. Worden, J. C. Mitchell, N. L. Arnold, S. Gopalan, X. Meng, V. M. Choi, J. M. Rock, Y. Y. Wu, G. E. Katibah, G. Zhifang, D. McCaskill, M. A. Simpson, B. Blakeslee, S. A. Greenwalt, H. J. Butler, S. J. Hinkley, L. Zhang, E. J. Rebar, P. D. Gregory and F. D. Urnov, Precise Genome Modification in the Crop Species Zea Mays Using Zinc-Finger Nucleases, Nature, 2009, 459(7245), 437–441 CrossRef CAS PubMed.
- Y. Zhang, Q. Shan, Y. Wang, K. Chen, Z. Liang, J. Li, Y. Zhang, K. Zhang, J. Liu, D. F. Voytas, X. Zheng and C. Gao, Rapid and Efficient Gene Modification in Rice and Brachypodium Using TALENs, Mol. Plant, 2013, 6(4), 1365–1368 CrossRef.
- Y. Wang, X. Cheng, Q. Shan, Y. Zhang, J. Liu, C. Gao and J. L. Qiu, Simultaneous Editing of Three Homoeoalleles in Hexaploid Bread Wheat Confers Heritable Resistance to Powdery Mildew, Nat. Biotechnol., 2014, 32(9), 947–951 CrossRef CAS.
- W. Haun, A. Coffman, B. M. Clasen, Z. L. Demorest, A. Lowy, E. Ray, A. Retterath, T. Stoddard, A. Juillerat, F. Cedrone, L. Mathis, D. F. Voytas and F. Zhang, Improved Soybean Oil Quality by Targeted Mutagenesis of the Fatty Acid Desaturase 2 Gene Family, Plant Biotechnol. J., 2014, 12(7), 934–940 CrossRef CAS PubMed.
- T. Wendt, P. B. Holm, C. G. Starker, M. Christian, D. F. Voytas, H. Brinch-Pedersen and I. B. Holme, TAL Effector Nucleases Induce Mutations at a Pre-Selected Location in the Genome of Primary Barley Transformants, Plant Mol. Biol., 2013, 83(3), 279–285 CrossRef CAS PubMed.
- A. S. Fister, L. Landherr, S. N. Maximova and M. J. Guiltinan, Transient Expression of CRISPR/Cas9 Machinery Targeting TcNPR3 Enhances Defense Response in Theobroma Cacao, Front. Plant Sci., 2018, 9, 268 CrossRef.
- V. Pompili, L. Dalla Costa, S. Piazza, M. Pindo and M. Malnoy, Reduced Fire Blight Susceptibility in Apple Cultivars Using a High-Efficiency CRISPR/Cas9-FLP/FRT-Based Gene Editing System, Plant Biotechnol. J., 2020, 18(3), 845–858 CrossRef CAS PubMed.
- J. Li, X. Meng, Y. Zong, K. Chen, H. Zhang, J. Liu, J. Li and C. Gao, Gene Replacements and Insertions in Rice by Intron Targeting Using CRISPR-Cas9, Nat. Plants, 2016, 2(10), 16139 CrossRef CAS PubMed.
- A. Zhang, Y. Liu, F. Wang, T. Li, Z. Chen, D. Kong, J. Bi, F. Zhang, X. Luo, J. Wang, J. Tang, X. Yu, G. Liu and L. Luo, Enhanced Rice Salinity Tolerance via CRISPR/Cas9-Targeted Mutagenesis of the OsRR22 Gene, Mol. Breed., 2019, 39(3), 47 CrossRef PubMed.
- X. Sun, X. Li, Y. Wang, J. Xu, S. Jiang and Y. Zhang, MdMKK9-Mediated the Regulation of Anthocyanin Synthesis in Red-Fleshed Apple in Response to Different Nitrogen Signals, Int. J. Mol. Sci., 2022, 23(14), 7755 CrossRef CAS.
- A. Endo, H. Saika, M. Takemura, N. Misawa and S. Toki, A Novel Approach to Carotenoid Accumulation in Rice Callus by Mimicking the Cauliflower Orange Mutation via Genome Editing, Rice, 2019, 12(1), 81 CrossRef PubMed.
- A. Souii, M. B. M’hadheb-Gharbi and J. Gharbi, Nucleic Acid-Based Biotechnologies for Food-Borne Pathogen Detection Using Routine Time-Intensive Culture-Based Methods and Fast Molecular Diagnostics, Food Sci. Biotechnol., 2016, 25, 11–20 CrossRef CAS PubMed.
- K. S. Gracias and J. L. McKillip, A Review of Conventional Detection and Enumeration Methods for Pathogenic Bacteria in Food, Can. J. Microbiol., 2004, 50, 883–890 CrossRef CAS PubMed.
- A. Pathologica, Rapid Detection, Character-Ization and Enumeration of Foodborne Pathogens, J. Pathol. Microbiol. Immunol., 2011, 119(s133), 1–24 Search PubMed.
- R. Patel, B. Mitra, M. Vinchurkar, A. Adami, R. Patkar, F. Giacomozzi, L. Lorenzelli and M. S. Baghini, A Review of Recent Advances in Plant-Pathogen Detection Systems, Heliyon, 2022, 8(12) CrossRef CAS PubMed.
- B. Fraaije and D. Lovell, Septoria epidemics on wheat: combined use of visual assessment and PCR-based diagnostics to identify mechanisms of disease escape, Plant Prot. Sci., 2017, 38, 421–424 CrossRef.
- A. C. Oluwaseun, P. Phazang and N. B. Sarin, Biosensors: A fast-growing technology for pathogen detection in agriculture and food sector, in Biosensing Technologies for the Detection of Pathogens—A Prospective Way for Rapid Analysis, 2018, pp. 37–52 Search PubMed.
- A. K. Gautam and S. Kumar, Techniques for the Detection, Identification, and Diagnosis of Agricultural Pathogens and Diseases, in Natural Remedies for Pest, Disease and Weed Control, Elsevier, 2020, pp. 135–142 Search PubMed.
- Y. Fang and R. P. Ramasamy, Current and Prospective Methods for Plant Disease Detection, Biosensors, 2015, 5(3), 537–561 CrossRef CAS.
- G. Papadakis, N. Skandalis, A. Dimopoulou, P. Glynos and E. Gizeli, Bacteria Murmus; Application of an Acoustic Biosensor for Plant Pathogen Detection, PLoS One, 2015, 10(7), e0132773 CrossRef.
- N. P. Rijpens and L. M. F. Herman, Molecular Methods for Identification and Detection of Bacterial Food Pathogens, J. AOAC Int., 2002, 85(4), 984–995 CrossRef CAS.
- A. Souii, M. B. M’hadheb-Gharbi and J. Gharbi, Nucleic Acid-Based Biotechnologies for Food-Borne Pathogen Detection Using Routine Time-Intensive Culture-Based Methods and Fast Molecular Diagnostics, Food Sci. Biotechnol., 2016, 25, 11–20 CrossRef CAS PubMed.
- H. Y. Lau, H. Wu, E. J. H. Wee, M. Trau and J. R. Botella, Field Demonstration of Multiplexed Point-of-Care Diagnostic Platform for Plant Pathogens, Anal. Chem., 2016, 88(16), 8074–8081 CrossRef CAS.
- F. Zhan, T. Wang, L. Iradukunda and J. Zhan, A Gold Nanoparticle-Based Lateral Flow Biosensor for Sensitive Visual Detection of the Potato Late Blight Pathogen, Phytophthora Infestans, Anal. Chim. Acta, 2018, 1036, 153–161 CrossRef CAS PubMed.
- Z. D. Chen, H. J. Kang, A. L. Chai, Y. X. Shi, X. W. Xie, L. Li and B. J. Li, Development of a Loop-Mediated Isothermal Amplification (LAMP) Assay for Rapid Detection of Pseudomonas Syringae Pv. Tomato in Planta, Eur. J. Plant Pathol., 2020, 156(3), 739–750 CrossRef CAS.
- X. Zong, W. Wang, H. Wei, J. Wang, X. Chen, L. Xu, D. Zhu, Y. Tan and Q. Liu, Rapid Detection of Prunus Necrotic Ringspot Virus Using Magnetic Nanoparticle-Assisted Reverse Transcription Loop-Mediated Isothermal Amplification, J. Virol. Methods, 2014, 208, 85–89 CrossRef CAS.
- Y. Qiu, S. Zhang and A. Tan, et al., Detection of TuMV by a toehold switch sensor coupled with NASBA amplification in Pseudostellaria heterophylla, Plant Methods, 2025, 21, 81 CrossRef CAS PubMed.
- J. A. Ruemmele, W. P. Hall, L. K. Ruvuna and R. P. Van Duyne, A localized surface plasmon resonance imaging instrument for multiplexed biosensing, Anal. Chem., 2013, 85(9), 4560–4566 CrossRef CAS PubMed.
- A. Stals, L. Baert, E. Van Coillie and M. Uyttendaele, Extraction of Food-Borne Viruses from Food Samples: A Review, Int. J. Food Microbiol., 2012, 153, 1–9 CrossRef.
- A. Stals, E. Van Coillie and M. Uyttendaele, Viral Genes Everywhere: Public Health Implications of PCR-Based Testing of Foods, Curr. Opin. Virol., 2013, 3(1), 69–73 CrossRef CAS.
- W. Sun, X. Huang and X. Wang, CRISPR-based molecular diagnostics: a review, Shengwu Gongcheng Xuebao, 2023, 39(1), 60–73 CAS.
- P. R. Shashank, B. M. Parker, S. R. Rananaware, D. Plotkin, C. Couch, L. G. Yang, L. T. Nguyen, N. R. Prasannakumar, W. E. Braswell, P. K. Jain and A. Y. Kawahara, CRISPR-Based Diagnostics Detects Invasive Insect Pests, Mol. Ecol. Resour., 2023, 24(1), e13881 CrossRef.
- B. J. Hindson, K. D. Ness, D. A. Masquelier, P. Belgrader, N. J. Heredia, A. J. Makarewicz, I. J. Bright, M. Y. Lucero, A. L. Hiddessen, T. C. Legler, T. K. Kitano, M. R. Hodel, J. F. Petersen, P. W. Wyatt, E. R. Steenblock, P. H. Shah, L. J. Bousse, C. B. Troup, J. C. Mellen, D. K. Wittmann, N. G. Erndt, T. H. Cauley, R. T. Koehler, A. P. So, S. Dube, K. A. Rose, L. Montesclaros, S. Wang, D. P. Stumbo, S. P. Hodges, S. Romine, F. P. Milanovich, H. E. White, J. F. Regan, G. A. Karlin-Neumann, C. M. Hindson, S. Saxonov and B. W. Colston, High-throughput droplet digital PCR system for absolute quantitation of DNA copy number, Anal. Chem., 2011, 83(22), 8604–8610 CrossRef CAS.
- M. Z. Hossain and C. M. Maragos, Gold Nanoparticle-Enhanced Multiplexed Imaging Surface Plasmon Resonance (ISPR) Detection of Fusarium Mycotoxins in Wheat, Biosens. Bioelectron., 2018, 101, 245–252 CrossRef CAS.
- R. Takabatake, Y. Kagiya, S. Futo, Y. Minegishi, K. Soga, N. Shibata and K. Kondo, Rapid Screening Detection of Genetically Modified Papaya by Loop-Mediated Isothermal Amplification, Biol. Pharm. Bull., 2023, 46(5), 713–717 CrossRef CAS.
- Y. Zhou, H. Y. Zheng, D. M. Jiang, M. Liu, W. Zhang and J. Y. Yan, A Rapid Detection of Tomato Yellow Leaf Curl Virus Using Recombinase Polymerase Amplification-Lateral Flow Dipstick Assay, Lett. Appl. Microbiol., 2022, 74(5), 640–646 CrossRef CAS PubMed.
- M. K. Prasannakumar, P. B. Parivallal, D. Pramesh, H. B. Mahesh and E. Raj, LAMP-based foldable microdevice platform for the rapid detection of Magnaporthe Oryzae and Sarocladium Oryzae in rice seed, Sci. Rep., 2021, 11, 178 CrossRef CAS.
- S. B. Kokane, P. Misra and A. D. Kokane, et al., Development of a real-time RT-PCR method for the detection of Citrus tristeza virus (CTV) and its implication in studying virus distribution in planta, 3 Biotech, 2021, 11, 431 CrossRef PubMed.
- N. Datukishvili, T. Kutateladze, I. Gabriadze, K. Bitskinashvili and B. Vishnepolsky, New multiplex PCR methods for rapid screening of genetically modified organisms in foods, Front. Microbiol., 2015, 6, 757 Search PubMed.
- S. Wachiralurpan, I. Phung-On, N. Chanlek, S. Areekit, K. Chansiri and P. A. Lieberzeit, In-Situ Monitoring of Real-Time Loop-Mediated Isothermal Amplification with Qcm: Detecting Listeria Monocytogenes, Biosensors, 2021, 11(9), 308 CrossRef CAS PubMed.
- M. Djisalov, L. Janjušević, V. Léguillier, L. Šašić Zorić, C. Farre, J. Anba-Mondoloni, J. Vidic and I. Gadjanski, Loop-Mediated Isothermal Amplification (LAMP) Assay Coupled with Gold Nanoparticles for Colorimetric Detection of Trichoderma Spp. in Agaricus Bisporus Cultivation Substrates, Sci. Rep., 2024, 14(1), 15539 CrossRef CAS.
- F. Jacquet, M. H. Jeuffroy, J. Jouan, E. Le Cadre, I. Litrico, T. Malausa, X. Reboud and C. Huyghe, Pesticide-Free Agriculture as a New Paradigm for Research, Agron. Sustainable Dev., 2022, 42, 8 CrossRef.
- J. Zhang, H. Li, X. Zhong, J. Tian, A. Segers, L. Xia and F. Francis, RNA-Interference-Mediated Aphid Control in Crop Plants: A Review, Agriculture, 2022, 12(12), 2108 CrossRef CAS.
- N. G. Bologna and O. Voinnet, The Diversity, Biogenesis, and Activities of Endogenous Silencing Small RNAs in Arabidopsis, Annu. Rev. Plant Biol., 2014, 65, 473–503 CrossRef CAS PubMed.
- M. W. Choudry, P. Nawaz, N. Jahan, R. Riaz, B. Ahmed, M. H. Raza, Z. Fayyaz, K. Malik and S. Afzal, RNA Based Gene Silencing Modalities to Control Insect and Fungal Plant Pests – Challenges and Future Prospects, Physiol. Mol. Plant Pathol., 2024, 130, 102241 CrossRef CAS.
- N. Agrawal, P. V. N. Dasaradhi, A. Mohmmed, P. Malhotra, R. K. Bhatnagar and S. K. Mukherjee, RNA Interference: Biology, Mechanism, and Applications, Microbiol. Mol. Biol. Rev., 2003, 67(4), 657–685 CrossRef CAS.
- C. Cogoni, J. T. Irelan, M. Schumacher, T. J. Schmidhauser, E. U. Selker and G. Macino, Transgene Silencing of the Al-1 Gene in Vegetative Cells of Neurospora Is Mediated by a Cytoplasmic Effector and Does Not Depend on DNA-DNA Interactions or DNA Methylation, EMBO J., 1996, 15(12), 3153–3163 CrossRef CAS.
- F. L. Tan and J. Q. Yin, RNAi, a New Therapeutic Strategy against Viral Infection, Cell Research, 2004, 4, 460–466 CrossRef.
- A. Fire, et al., Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans, Nature, 1998, 391(6669), 806–811 CrossRef CAS.
- I. R. Henderson, X. Zhang, C. Lu, L. Johnson, B. C. Meyers, P. J. Green and S. E. Jacobsen, Dissecting Arabidopsis Thaliana DICER Function in Small RNA Processing, Gene Silencing and DNA Methylation Patterning, Nat. Genet., 2006, 38(6), 721–725 CrossRef CAS.
- E. J. Finnegan and M. A. Matzke, The Small RNA World, J. Cell Sci., 2003, 116(23), 4689–4693 CrossRef CAS.
- M. Ghildiyal and P. D. Zamore, Small Silencing RNAs: An Expanding Universe, Nat. Rev. Genet., 2009, 10, 94–108 CrossRef CAS.
- J. K. W. Lam, M. Y. T. Chow, Y. Zhang and S. W. S. Leung, SiRNA versus MiRNA as Therapeutics for Gene Silencing, Mol. Ther.--Nucleic Acids, 2015, 4, e252 CrossRef CAS.
- F. Betti, M. J. Ladera-Carmona, D. A. Weits, G. Ferri, S. Iacopino, G. Novi, B. Svezia, A. B. Kunkowska, A. Santaniello, A. Piaggesi, E. Loreti and P. Perata, Exogenous MiRNAs Induce Post-Transcriptional Gene Silencing in Plants, Nat. Plants, 2021, 7(10), 1379–1388 CrossRef CAS PubMed.
- H. Sang and J. I. Kim, Advanced Strategies to Control Plant Pathogenic Fungi by Host-Induced Gene Silencing (HIGS) and Spray-Induced Gene Silencing (SIGS), Agricultural Sciences, 2020, 14, 1–8 Search PubMed.
- C. Rössner, D. Lotz and A. Becker, VIGS Goes Viral: How VIGS Transforms Our Understanding of Plant Science, Annu. Rev. Plant Biol., 2025, 73, 703–728 CrossRef.
- X. Chen, T. Shi, T. Tang, C. Chen, Y. Liang and S. Zuo, Nanosheet-Facilitated Spray Delivery of dsRNAs Represents a Potential Tool to Control Rhizoctonia Solani Infection, Int. J. Mol. Sci., 2022, 23(21), 12922 CrossRef CAS.
- A. L. Jackson, J. Burchard, J. Schelter, B. N. Chau, M. Cleary, L. Lim and P. S. Linsley, Widespread SiRNA “off-Target” Transcript Silencing Mediated by Seed Region Sequence Complementarity, RNA, 2006, 12(7), 1179–1187 CrossRef CAS.
- N. Mitter, E. A. Worrall, K. E. Robinson, P. Li, R. G. Jain, C. Taochy, S. J. Fletcher, B. J. Carroll, G. Q. Lu and Z. P. Xu, Clay Nanosheets for Topical Delivery of RNAi for Sustained Protection against Plant Viruses, Nat. Plants, 2017, 3(2), 1–10 Search PubMed.
- M. Knoblich, T. Gursinsky, S. Gago-Zachert, C. Weinholdt, J. Grau and S.-E. Behrens, A New Level of RNA-Based Plant Protection: DsRNAs Designed from Functionally Characterized SiRNAs Highly Effective against Cucumber Mosaic Virus, Nucleic Acids Res., 2025, 53(5), gkaf136 CrossRef CAS.
- K. A. Feldmann, Steroid regulation improves crop yield, Nat. Biotechnol., 2006, 24(1), 46–47 CrossRef CAS.
- H. Hisano, R. Nandakumar and Z.-Y. Wang, Genetic Modification of Lignin Biosynthesis for Improved Biofuel Production, in Biofuels: Global Impact on Renewable Energy, Production Agriculture, and Technological Advancements, 2011, pp. 223–235 Search PubMed.
- F. Qiao, Q. Yang, C. L. Wang, Y. L. Fan, X. F. Wu and K. J. Zhao, Modification of Plant Height via RNAi Suppression of OsGA20ox2 Gene in Rice, Euphytica, 2007, 158(1–2), 35–45 CrossRef CAS.
- Y. Jiao, Y. Wang, D. Xue, J. Wang, M. Yan, G. Liu, G. Dong, D. Zeng, Z. Lu, X. Zhu, Q. Qian and J. Li, Regulation of OsSPL14 by OsmiR156 Defines Ideal Plant Architecture in Rice, Nat. Genet., 2010, 42(6), 541–544 CrossRef CAS.
- K. Miura, M. Ikeda, A. Matsubara, X. J. Song, M. Ito, K. Asano, M. Matsuoka, H. Kitano and M. Ashikari, OsSPL14 Promotes Panicle Branching and Higher Grain Productivity in Rice, Nat. Genet., 2010, 42(6), 545–549 CrossRef CAS PubMed.
- G. R. Davuluri, A. Van Tuinen, P. D. Fraser, A. Manfredonia, R. Newman, D. Burgess, D. A. Brummell, S. R. King, J. Palys, J. Uhlig, P. M. Bramley, H. M. J. Pennings and C. Bowler, Fruit-Specific RNAi-Mediated Suppression of DET1 Enhances Carotenoid and Flavonoid Content in Tomatoes, Nat. Biotechnol., 2005, 23(7), 890–895 CrossRef CAS PubMed.
- B. Yu, D. J. Lydiate, L. W. Young, U. A. Schäfer and A. Hannoufa, Enhancing the Carotenoid Content of Brassica Napus Seeds by Downregulating Lycopene Epsilon Cyclase, Transgenic Res., 2008, 17(4), 573–585 CrossRef CAS PubMed.
- A. M. Dandekar, G. Teo, B. G. Defilippi, S. L. Uratsu, A. J. Passey, A. A. Kader, J. R. Stow, R. J. Colgan and D. J. James, Effect of Down-Regulation of Ethylene Biosynthesis on Fruit Flavor Complex in Apple Fruit, Transgenic Res., 2004, 13, 373–384 CrossRef CAS.
- A. Regina, A. Bird, D. Topping, S. Bowden, J. Freeman, T. Barsby, B. Kosar-Hashemi, Z. Li, S. Rahman and M. Morell, High-Amylose Wheat Generated by RNA Interference Improves Indices of Large-Bowel Health in Rats, Agric. Sci., 2006, 103(10), 3546–3551 CAS.
- J. Gil-Humanes, F. Pistón, C. M. Rosell and F. Barro, Significant Down-Regulation of γ-Gliadins Has Minor Effect on Gluten and Starch Properties of Bread Wheat, J. Cereal Sci., 2012, 56(2), 161–170 CrossRef CAS.
- G. Chuai, H. Ma, J. Yan, M. Chen, N. Hong, D. Xue, C. Zhou, C. Zhu, K. Chen, B. Duan, F. Gu, S. Qu, D. Huang, J. Wei and Q. Liu, DeepCRISPR: Optimized CRISPR Guide RNA Design by Deep Learning, Genome Biol., 2018, 19(1), 18 CrossRef.
- T. J. Cradick, G. Ambrosini, C. Iseli, P. Bucher and A. P. McCaffrey, ZFN-Site Searches Genomes for Zinc Finger Nuclease Target Sites and off-Target Sites, BMC Bioinf., 2011, 12, 152 CrossRef CAS PubMed.
- M. Stemmer, T. Thumberger, M. Del Sol Keyer, J. Wittbrodt and J. L. Mateo, CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool, PLoS One, 2015, 10(4), e0176619 CrossRef.
- S. Lück, T. Kreszies, M. Strickert, P. Schweizer, M. Kuhlmann and D. Douchkov, SiRNA-Finder (Si-Fi) Software for RNAi-Target Design and Off-Target Prediction, Front. Plant Sci., 2019, 10, 1023 CrossRef.
- G. Zhang, Z. Dai and X. Dai, C-RNNCrispr: Prediction of CRISPR/Cas9 SgRNA Activity Using Convolutional and Recurrent Neural Networks, Comput. Struct. Biotechnol. J., 2020, 18, 344–354 CrossRef CAS.
- K. G. Ptitsyn, S. A. Khmeleva, L. K. Kurbatov, O. S. Timoshenko, E. V. Suprun, S. P. Radko and A. V. Lisitsa, LAMP Primer Designing Software: The Overview, Biomed. Chem. Res. Methods, 2024, 7(4), e00226 CrossRef.
- R. Kalendar, A Guide to Using FASTPCR Software for PCR, In Silico PCR, and Oligonucleotide Analysis, in PCR Primer Design, ed. C. Basu, Springer US, New York, NY, 2021, pp. 223–243 Search PubMed.
- V. Periwal, A Comprehensive Overview of Computational Resources to Aid in Precision Genome Editing with Engineered Nucleases, Briefings Bioinf., 2017, 18(4), 698–711 CAS.
- A. Agmata, K. Labrador, J. D. Palermo and M. J. Pante, ThermoPlex: An Automated Design Tool for Target-Specific Multiplex PCR Primers Based on DNA Thermodynamics, Biol. Methods Protoc., 2024, 10(1), bpaf074 CrossRef PubMed.
- J. D. Limberis and J. Z. Metcalfe, PrimerJinn: A Tool for Rationally Designing Multiplex PCR Primer Sets for Amplicon Sequencing and Performing in Silico PCR, BMC Bioinf., 2023, 24(1), 468 CrossRef CAS PubMed.
- L. Yang, F. Ding, Q. Lin, J. Xie, W. Fan, F. Dai, P. Cui and W. Liu, A Tool to Automatically Design Multiplex PCR Primer Pairs for Specific Targets Using Diverse Templates, Sci. Rep., 2023, 13(1), 16451 CrossRef CAS.
- H. Zeng, K. Chen, C. Ma, B. Zhu, J. Chuan, S. Zhang, L. Tang, T. Yang, Z. Sun, X. Yang and Y. Wang, High-Throughput Primer Design by Scoring in Piecewise Logistic Model for Multiple Polymerase Chain Reaction Variants, Sci. Rep., 2022, 12(1), 21136 CrossRef CAS.
- N. G. Xie, M. X. Wang, P. Song, S. Mao, Y. Wang, Y. Yang, J. Luo, S. Ren and D. Y. Zhang, Designing Highly Multiplex PCR Primer Sets with Simulated Annealing Design Using Dimer Likelihood Estimation (SADDLE), Nat. Commun., 2022, 13(1), 1881 CrossRef CAS.
- C. H. Yang, Y. H. Cheng, E. C. Yang, L. Y. Chuang and Y. Da. Lin, Multiobjective Optimization-Driven Primer Design Mechanism: Towards User-Specified Parameters of PCR Primer, Briefings Bioinf., 2022, 23(3), bbac121 CrossRef PubMed.
- J. Yuan, J. Yi, M. Zhan, Q. Xie, T. T. Zhen, J. Zhou, Z. Li and Z. Li, The Web-Based Multiplex PCR Primer Design Software Ultiplex and the Associated Experimental Workflow: Up to 100- Plex Multiplicity, BMC Genomics, 2021, 22(1), 835 CrossRef CAS PubMed.
- A. Kechin, V. Borobova, U. Boyarskikh, E. Khrapov, S. Subbotin and M. Filipenko, NGS-PrimerPlex: High-Throughput Primer Design for Multiplex Polymerase Chain Reactions, PLoS Comput. Biol., 2020, 16, e1008468 CrossRef CAS.
- H. Kim, N. N. Kang, K. H. An, J. H. Koo and M. S. Kim, MRPrimerW: A Tool for Rapid Design of Valid High-Quality Primers for Multiple Target QPCR Experiments, Nucleic Acids Res., 2016, 44(W1), W259–W266 CrossRef CAS.
- J. Bae, H. Jeon and M. S. Kim, GPrimer: A Fast GPU-Based Pipeline for Primer Design for QPCR Experiments, BMC Bioinf., 2021, 22(1), 220 CrossRef CAS.
- T. Zhu, C. Liang, Z. Meng, Y. Li, Y. Wu, S. Guo, R. Zhang, PrimerServer: A High-Throughput Primer Design and Specificity-Checking Platform, bioRxiv, 2017, preprint, DOI:10.1101/181941.
- A. Xiao, Y. Wu, Z. Yang, Y. Hu, W. Wang, Y. Zhang, L. Kong, G. Gao, Z. Zhu, S. Lin and B. Zhang, EENdb: A Database and Knowledge Base of ZFNs and TALENs for Endonuclease Engineering, Nucleic Acids Res., 2013, 41(D1), D415–D422 CrossRef CAS.
- W. F. Lenoir, T. L. Lim and T. Hart, PICKLES: The Database of Pooled in-Vitro CRISPR Knockout Library Essentiality Screens, Nucleic Acids Res., 2018, 46(D1), D776–D780 CrossRef CAS PubMed.
- J. Park, J. S. Kim and S. Bae, Cas-Database: Web-Based Genome-Wide Guide RNA Library Design for Gene Knockout Screens Using CRISPR-Cas9, Bioinformatics, 2016, 32(13), 2017–2023 CrossRef CAS.
- V. Jaskula-Ranga, D. J. Zack, GrID: A CRISPR-Cas9 Guide RNA Database and Resource for Genome-Editing, bioRxiv, 2016, preprint, DOI:10.1101/097352.
- G. K. Varshney, S. Zhang, W. Pei, A. Adomako-Ankomah, J. Fohtung, K. Schaffer, B. Carrington, A. Maskeri, C. Slevin, T. Wolfsberg, J. Ledin, R. Sood and S. M. Burgess, CRISPRz: A Database of Zebrafish Validated SgRNAs, Nucleic Acids Res., 2016, 44(D1), D822–D826 CrossRef CAS PubMed.
- Y. Naito, K. Hino, H. Bono and K. Ui-Tei, CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA with Reduced off-Target Sites, Bioinformatics, 2015, 31(7), 1120–1123 CrossRef CAS.
- J. G. Mandell and C. F. Barbas, Zinc Finger Tools: Custom DNA-Binding Domains for Transcription Factors and Nucleases, Nucleic Acids Res., 2006, 34, W516–W523 CrossRef CAS PubMed.
- M. Jayakanthan, J. Muthukumaran, S. Chandrasekar, K. Chawla, A. Punetha and D. Sundar, ZifBase: A Database of Zinc Finger Proteins and Associated Resources, BMC Genomics, 2009, 10(1), 421 CrossRef.
- J. D. Sander, M. L. Maeder, D. Reyon, D. F. Voytas, J. K. Joung and D. Dobbs, ZiFiT (Zinc Finger Targeter): An Updated Zinc Finger Engineering Tool, Nucleic Acids Res., 2010, 38, W462–W468 CrossRef CAS.
- E. L. Doyle, N. J. Booher, D. S. Standage, D. F. Voytas, V. P. Brendel, J. K. Vandyk and A. J. Bogdanove, TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: Tools for TAL Effector Design and Target Prediction, Nucleic Acids Res., 2012, 40, W117–W122 CrossRef CAS PubMed.
- J. Grau, J. Boch and S. Posch, TALENoffer: Genome-Wide TALEN off-Target Prediction, Bioinformatics, 2013, 29(22), 2931–2932 CrossRef CAS.
- F. Heigwer, G. Kerr, N. Walther, K. Glaeser, O. Pelz, M. Breinig and M. Boutros, E-TALEN: A Web Tool to Design TALENs for Genome Engineering, Nucleic Acids Res., 2013, 41(20), e190 CrossRef CAS PubMed.
- Y. Lin, E. J. Fine, Z. Zheng, C. J. Antico, R. A. Voit, M. H. Porteus, T. J. Cradick and G. Bao, SAPTA: A New Design Tool for Improving TALE Nuclease Activity, Nucleic Acids Res., 2014, 42(6), e47 CrossRef CAS PubMed.
- M. Ma, A. Y. Ye, W. Zheng and L. Kong, A Guide RNA Sequence Design Platform for the CRISPR/Cas9 System for Model Organism Genomes, BioMed Res. Int., 2013, 10, 270805 Search PubMed.
- A. Xiao, Z. Cheng, L. Kong, Z. Zhu, S. Lin, G. Gao and B. Zhang, CasOT: A Genome-Wide Cas9/GRNA off-Target Searching Tool, Bioinformatics, 2014, 30(8), 1180–1182 CrossRef CAS.
- S. Bae, J. Park and J. S. Kim, Cas-OFFinder: A Fast and Versatile Algorithm That Searches for Potential off-Target Sites of Cas9 RNA-Guided Endonucleases, Bioinformatics, 2014, 30(10), 1473–1475 CrossRef CAS.
- S. J. Gratz, F. P. Ukken, C. D. Rubinstein, G. Thiede, L. K. Donohue, A. M. Cummings and K. M. Oconnor-Giles, Highly Specific and Efficient CRISPR/Cas9-Catalyzed Homology-Directed Repair in Drosophila, Genetics, 2014, 196(4), 961–971 CrossRef CAS.
- F. Heigwer, G. Kerr and M. Boutros, E-CRISP: Fast CRISPR Target Site Identification, Nat. Methods, 2014, 11, 122–123 CrossRef CAS PubMed.
- Y. Lei, L. Lu, H. Y. Liu, S. Li, F. Xing and L. L. Chen, CRISPR-P: A Web Tool for Synthetic Single-Guide RNA Design of CRISPR-System in Plants, Mol. Plant, 2014, 7(9), 1494–1496 CrossRef CAS PubMed.
- A. O'Brien and T. L. Bailey, GT-Scan: Identifying Unique Genomic Targets, Bioinformatics, 2014, 30(18), 2673–2675 CrossRef.
- C. Ronda, L. E. Pedersen, H. G. Hansen, T. B. Kallehauge, M. J. Betenbaugh, A. T. Nielsen and H. F. Kildegaard, Accelerating Genome Editing in CHO Cells Using CRISPR Cas9 and CRISPy, a Web-Based Target Finding Tool, Biotechnol. Bioeng., 2014, 111, 1604–1616 CrossRef CAS PubMed.
- S. Xie, B. Shen, C. Zhang, X. Huang and Y. Zhang, SgRNAcas9: A Software Package for Designing CRISPR SgRNA and Evaluating Potential off-Target Cleavage Sites, PLoS One, 2014, 9(6), e100448 CrossRef PubMed.
- L. J. Zhu, B. R. Holmes, N. Aronin and M. H. Brodsky, CRISPRseek: A Bioconductor Package to Identify Target-Specific Guide RNAs for CRISPR-Cas9 Genome-Editing Systems, PLoS One, 2014, 9(9), e108424 CrossRef.
- Y. Naito, K. Hino, H. Bono and K. Ui-Tei, CRISPRdirect: Software for Designing CRISPR/Cas Guide RNA with Reduced off-Target Sites, Bioinformatics, 2015, 31(7), 1120–1123 CrossRef CAS PubMed.
- V. Pliatsika and I. Rigoutsos, “Off-Spotter”: Very Fast and Exhaustive Enumeration of Genomic Lookalikes for Designing CRISPR/Cas Guide RNAs, Biol. Direct, 2015, 10(1), 4 CrossRef PubMed.
- S. V. Prykhozhij, V. Rajan, D. Gaston and J. N. Berman, CRISPR Multitargeter: A Web Tool to Find Common and Unique CRISPR Single Guide RNA Targets in a Set of Similar Sequences, PLoS One, 2015, 10(3), e0138634 CrossRef PubMed.
- K. Kaur, H. Tandon, A. K. Gupta and M. Kumar, CrisprGE: A Central Hub of CRISPR/Cas-Based Genome Editing, Database, 2015, 2015, bav055 CrossRef PubMed.
- A. Hodgkins, A. Farne, S. Perera, T. Grego, D. J. Parry-Smith, W. C. Skarnes and V. Iyer, WGE: A CRISPR Database for Genome Engineering, Bioinformatics, 2015, 31(18), 3078–3080 CrossRef CAS PubMed.
- K. L. Neff, D. P. Argue, A. C. Ma, H. B. Lee, K. J. Clark and S. C. Ekker, Mojo Hand, a TALEN Design Tool for Genome Editing Applications, BMC Bioinf., 2013, 14(1), 1 CrossRef CAS.
- E. J. Fine, T. J. Cradick, C. L. Zhao, Y. Lin and G. Bao, An Online Bioinformatics Tool Predicts Zinc Finger and TALE Nuclease Off-Target Cleavage, Nucleic Acids Res., 2014, 42(6), e42 CrossRef CAS PubMed.
- F. Ahmed, M. Senthil-Kumar, X. Dai, V. S. Ramu, S. Lee, K. S. Mysore and P. X. Zhao, PssRNAit: A Web Server for Designing Effective and Specific Plant Sirnas with Genome-Wide off-Target Assessment1[OPEN], Plant Physiol., 2020, 184(1), 65–81 CrossRef CAS.
- A. Kakrana, R. Hammond, P. Patel, M. Nakano and B. C. Meyers, SPARTA: A Parallelized Pipeline for Integrated Analysis of Plant MiRNA and Cleaved MRNA Data Sets, Including New MiRNA Target-Identification Software, Nucleic Acids Res., 2014, 42(18), e139 CrossRef.
- B. Liu, Y. Yuan, X. Pan, H.-B. Shen and C. Jin, AttSiOff: A Self-Attention-Based Approach on SiRNA Design with Inhibition and off-Target Effect Prediction, Med-X, 2024, 2(1), 5 CrossRef CAS.
- R. Gumienny and M. Zavolan, Accurate Transcriptome-Wide Prediction of MicroRNA Targets and Small Interfering RNA off-Targets with MIRZA-G, Nucleic Acids Res., 2015, 43(3), 1380–1391 CrossRef CAS.
- R. Long, Z. Guo, D. Han, X. Yuan, G. Chen, P.-A. Heng, L. Zhang, SiRNADesign: A Graph Neural Network for SiRNA Efficacy Prediction via Deep RNA Sequence Analysis, bioRxiv, 2024, preprint, DOI:10.1101/2024.04.28.591509.
- Y. Chen, Y. Shi, Z. Wang, X. An, S. Wei, C. Andronis, J. Vontas, J. Wang and J. Niu, DsRNAEngineer: A Web-Based Tool of Comprehensive DsRNA Design for Pest Control, Trends Biotechnol., 2024, 43(4), 969–983 CrossRef.
- W. Zhang, P. Zhang, W. Sun, J. Xu, L. Liao, Y. Cao and Y. Han, Improving Plant MiRNA-Target Prediction with Self-Supervised k-Mer Embedding and Spectral Graph Convolutional Neural Network, PeerJ, 2024, 12(5), e17396 CrossRef.
- J. Thody, V. Moulton, I. Mohorianu, PAREameters: Computational Inference of Plant MicroRNA-MRNA Targeting Rules Using RNA Sequencing Data, bioRxiv, 2019, preprint, DOI:10.1101/710814.
- J. Thody, Computational Methods for Functional Analysis of Plant Small RNAs Using the RNA Degradome, Doctoral thesis, University of East Anglia, 2020, https://ueaeprints.uea.ac.uk/id/eprint/79639. Search PubMed.
- F. Hartung and J. Schiemann, Precise Plant Breeding Using New Genome Editing Techniques: Opportunities, Safety and Regulation in the EU, Plant J., 2014, 78(5), 742–752 CrossRef CAS PubMed.
- Scientific Opinion, Updated scientific opinion on plants developed through cisgenesis and intragenesis, EFSA J., 2022, 20(10), e07621 Search PubMed.
- E. Waltz, Tiptoeing around Transgenics. New techniques for manipulating plant genomes are yielding plants touted as non-transgenic. Will that relieve regulatory burden?, Nat. Biotechnol., 2012, 30(3), 215–217 CrossRef CAS.
- A. Kamthan, A. Chaudhuri, M. Kamthan and A. Datta, Small RNAs in Plants: Recent Development and Application for Crop Improvement, Front. Plant Sci., 2015, 6, 208 Search PubMed.
- P. Pant and F. Leese, Probing the Nucleic Acid Flexibility to Disarm the Viral Counter-Defense Machinery: Design and Characterization of Potent P19 Inhibitors, J. Phys. Chem. B, 2023, 127(41), 8842–8851 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |