 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Surveillance of Vibrio cholerae in a non-sewered sanitation refugee camp setting using culture methods: Dzaleka camp, Malawi
Brandie Banner Shackelford *ab,
Petros Chigwechokhac,
Ernest Chilalikaa,
Lucious Zibad,
Christopher Misomalid,
Mphatso Kanjirud,
Patrick Buleyad,
Ruth Lusungu Nyirendac,
Marlene K. Wolfeb and
Rochelle H. Holm
*ab,
Petros Chigwechokhac,
Ernest Chilalikaa,
Lucious Zibad,
Christopher Misomalid,
Mphatso Kanjirud,
Patrick Buleyad,
Ruth Lusungu Nyirendac,
Marlene K. Wolfeb and
Rochelle H. Holm *e
*e
aThe United Nations High Commissioner for Refugees, Chilanga Drive, Area 10, Plot 441/442, P.O. Box 30230, Lilongwe, Malawi
bThe Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, 1518 Clifton Rd. NE, Atlanta, GA 30322, USA. E-mail: brandie.shackelford@emory.edu
cDepartment of Biological Sciences, Malawi University of Science and Technology, P.O. Box 5196, Limbe, Malawi
dThe Public Health Institute of Malawi, P/Bag 65, Lilongwe, Malawi
eCenter for Healthy Air Water and Soil, Christina Lee Brown Envirome Institute, School of Medicine, University of Louisville, 302 E. Muhammad Ali Blvd, Louisville, KY 40202, USA. E-mail: rochelle.holm@louisville.edu
First published on 30th January 2026
Abstract
Refugees living in camps may be particularly vulnerable to infectious disease outbreaks because of factors such as overcrowding, inconsistent preventive healthcare, and limited water, sanitation, and hygiene services. Due to the endemicity of cholera, Malawi provides a suitable setting for a wastewater and environmental surveillance feasibility study in a refugee camp. We conducted a study in the Dzaleka camp in Malawi during a nationwide cholera outbreak. Incentivized refugee volunteers collected samples for 19 weeks from seven high-use public pit latrines and from a vacuum pump truck used to remove fecal sludge. The National Microbiological Reference Laboratory of the Public Health Institute of Malawi used culture methods, with confirmation using VITEK MS or Analytical Profile Index, to detect Vibrio cholerae. Academic partners provided technical input, training, and quality assurance throughout the study. The results were reviewed weekly at partner-coordination meetings. No V. cholerae were detected in regular samples (n = 147), in concordance with clinical data from the same period. The paired pit latrine and pump truck samples also had consistent negative results for V. cholerae. Other noncholera, pathogenic Vibrio species were identified in seven samples, namely, V. alginolyticus, V. parahaemolyticus, and V. metschnikovii. Here, we discuss the unique challenges of conducting wastewater and environmental surveillance in refugee camp settings, including our operational framework. This study provides a framework for scaling wastewater and environmental surveillance efforts in other humanitarian contexts through robust partnerships, clear protocols for non-sewered sanitation systems, and in-country resources.
Water impactWastewater and environmental surveillance for infectious disease outbreaks in non-sewered sanitation systems offers an important public health tool in densely populated refugee camps with stretched water, sanitation, and hygiene infrastructure. |
1. Introduction
Wastewater and environmental surveillance (WES) has been used across a range of geographic scales and infectious diseases, from single buildings to entire communities,1–3 but its application in areas without formal sewer systems, such as refugee camps, has been limited.4 Although environmental surveillance for polio has been practiced for many years in resource-scarce settings,5 efforts are needed to further contextualize WES methodologies to target additional pathogens, adapt existing clinical laboratory capacities, and appropriately sample non-sewered sanitation systems (NSSS)6–8 where a large camp population is not able to leave a geographic area but has monthly new additions. Researchers have demonstrated the feasibility of conducting WES in resource-scarce settings using samples from NSSS and surface water, including using polymerase chain reaction (PCR) to detect SARS-CoV-2,9 Vibrio cholerae,10,11 and other pathogens of public health concern.12–14 There is also evidence that culture methods, which tend to be less expensive and easier to operationalize in resource-scarce settings, can be leveraged with good specificity to monitor V. cholerae.10 The potential for WES to benefit health systems in emergencies was illustrated in Gaza in 2023, where polio was detected in wastewater before any clinical cases were reported.15 Little is known regarding the feasibility of conducting WES in refugee camps with NSSS using in-country analysis methods. WES could complement clinical surveillance efforts, providing valuable infectious disease public health surveillance data during humanitarian crises.Populations living in refugee camps may be particularly vulnerable to infectious diseases because of disruptions in essential water and sanitation services and high population density.16–21 Furthermore, refugees and asylum seekers may come from countries with different public health risks and arrive in host countries after experiencing inadequate transit conditions and inconsistent access to primary healthcare.22 Health clinics in resource-limited humanitarian settings often lack the capacity to confirm and manage infectious disease outbreaks.23,24
Malawi has not fully integrated WES into its public health system, despite its demonstrated potential.6,10 Both Malawians and refugees living in Malawi have strong public support for WES.25 Malawi experienced a nationwide cholera outbreak between 2022 and 2024.26,27 V. cholerae's persistence in the environment under nutrient-rich conditions is well-documented,28 and persistent phenotypes can remain culturable in nutrient-poor environments for more than 700 days.29 Together, the strong local public opinion, ongoing outbreak, and literature evidence of persistence in the environment provide a rationale for pathogen and site selection for a feasibility study of deploying WES in a refugee camp in Malawi. We sampled both high-use pit latrines and a vacuum desludging pump truck in Dzaleka camp, Malawi, and monitored V. cholerae using culture methods. Although established environmental surveillance guidelines for polio exist in low-resource contexts,30 we propose an additional framework for operationalizing WES for other pathogens in resource-scarce refugee camps.
2. Material and methods
2.1. Study area
The study was conducted in Dzaleka Refugee camp, located in Dowa district, Malawi (Fig. 1A). The United Nations High Commissioner for Refugees (UNHCR) reported that Malawi hosted 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 952 refugees and asylum seekers as of July 31, 2024, of whom 16.5% were children under 5 years old. Dzaleka camp is unfenced, enabling individuals from nearby host communities to access healthcare, education, and economic activities, as well as use the available sanitation facilities. However, at the time of the study, the Government of Malawi did not permit refugees and asylum seekers to leave the 224-hectare camp. The camp is serviced solely by NSSS; no homes or institutions are connected to a central wastewater system.
952 refugees and asylum seekers as of July 31, 2024, of whom 16.5% were children under 5 years old. Dzaleka camp is unfenced, enabling individuals from nearby host communities to access healthcare, education, and economic activities, as well as use the available sanitation facilities. However, at the time of the study, the Government of Malawi did not permit refugees and asylum seekers to leave the 224-hectare camp. The camp is serviced solely by NSSS; no homes or institutions are connected to a central wastewater system.
To determine the sampling sites, the research team first identified 65 publicly accessible pit latrines. We then selected “high-use” pit latrines in conjunction with refugee hygiene promoters. Next, an on-site feasibility check was conducted to ensure that the pit latrines had sufficient fecal waste within reach of the sampling device (i.e., waste was within 1 m of the ground surface) and had a roof tall enough for the sampling device (i.e., at least 1.5 m in height). To ensure sample anonymity, the research team verified the presence of many individual contributors by recording the number of users entering the pit latrine (Table S1). Anonymous user counting was conducted at each site for at least 4 h on at least 5 different days, ensuring that counting occurred on distinct days of the week and at various times of day to account for variability (e.g., market and church days). Where applicable, researchers also noted the most used stances to prioritize sampling there (Table S2). Seven pit latrines were selected for sampling within the camp (Fig. 1B), with the number of stances per site ranging from one to 11.
2.2. Wastewater and environmental sample collection
Before sample collection, refugee hygiene promoters, government health surveillance assistants, and refugee community leaders informed camp residents about the project through house visits, community meetings, a local radio program, and the camp notice board. Samples were collected from the seven public pit latrines and a vacuum desludging pump truck that emptied pit latrines of fecal waste, herein referred to as a “pump truck”. Grab samples were collected weekly on Mondays (or on Tuesdays when Monday was a public holiday) from March 5, 2024, through July 9, 2024. This sampling period coincided with a declared national cholera outbreak period26,27 and included parts of the rainy and cold seasons in Malawi. Pump truck sampling began on March 25, 2024, owing to delays in camp contracting (Table S3). The number of pit latrine sample sites started with four in the first week to allow sample collectors and laboratory technicians to become familiar with the procedures and increased to seven weekly sites thereafter. We also collected two samples from two different latrine blocks at one site (PT4) on April 15, 2024.Pit-latrine samples were collected by three incentive-based refugee volunteers who wore full personal protective equipment (PPE; Fig. 2A). The sampling team received relevant vaccinations, including the oral cholera vaccine, and hands-on training before the study began. The training included donning and doffing PPE, maintaining sample integrity, and site cleanup after collection. The camp water, sanitation, and hygiene cluster accompanied sampling for 19 weeks to ensure compliance with sampling protocols.
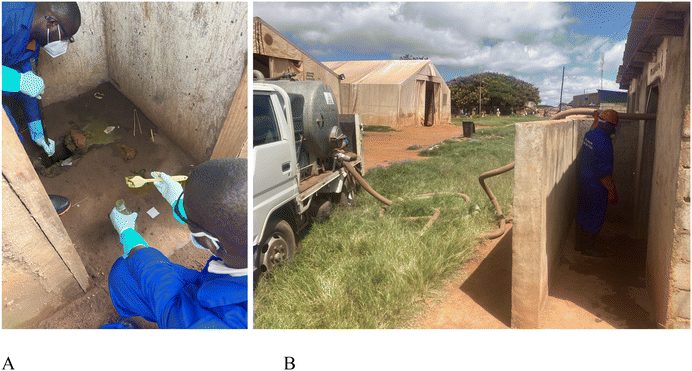 | ||
| Fig. 2 Sample collection, Dzaleka camp. (A) Pit latrine grab sample collection. (B) Pump truck sample collection. | ||
A previous study in Malawi had found that the depth of sampling in pit latrines did not impact the detection of pathogens when using PCR methods.13 The sampling team used a locally fabricated sampling device, a two-meter metal pole welded to a 500 mL metal cup with a spout. Pit latrine fecal sludge was stirred three times using the sampling device to homogenize the freshest material before sample collection. When the fecal sludge was too thick to mix and sample, the sampling team added nonchlorinated water to thin it. A rubber spatula was used to transfer the sample to a collection bottle when necessary (Fig. 2A). A 500 mL sample was collected at each site. Initially, sterile 250 mL glass sample bottles were used; however, the research team switched to disposable, sterile 200 mL plastic bottles and 50 mL plastic conical tubes because sterilizing the glassware was burdensome. A double sample volume was collected from at least one site each week on a rotation, so that each site had quality assurance (laboratory duplicate) samples processed by an independent laboratory throughout the study. The sampling device and spatula were thoroughly washed with water and disinfected with 70% ethanol between samples. The sampling team donned new disposable gloves after each sample.
A pump-truck sample was collected weekly by a private contractor. These samples were collected after the pit latrine grab samples. The commercial pump truck operator extracted 100–200 L of fecal sludge from one of the pit latrine sampling sites into the waste tank and collected the sample directly from the tank using a valve to release the waste directly into the sample bottle (Fig. 2B). The contractor washed the pump truck with water weekly; however, full sterilization of the pump truck was not logistically feasible.
2.3. Clinical data
Dzaleka Health Centre reported de-identified data on the number of diarrhea cases and confirmed cholera cases during the study period through the UNHCR Integrated Refugee Health Information System, as well as the number of cholera vaccinations previously distributed in the camp.2.4. Laboratory analysis
Samples were transported on ice in a cooler box to the Public Health Institute of Malawi (PHIM) National Microbiology Reference Laboratory. The samples were processed using culture methods upon delivery, with an average time of 245 min between sample collection and culture initiation.Clinical culture-based methods are widely used in Malawi; therefore, supplies were relatively easy to procure, and laboratory personnel could be easily trained to adapt existing clinical methods for WES.6 PCR or genomics method supplies would have required a prolonged time for importation during an outbreak period.
We adapted our previously described WES culture methods, which are based on clinical culture methods, for V. cholerae in stool samples.10 Briefly, 10 μL of the sample was inoculated on thiosulfate citrate bile salts sucrose agar plates using disposable, sterile loops, followed by overnight incubation for 18–24 h at 35–37 °C. Presumptive V. cholerae colonies were sub-cultured on nutrient agar for another 18–24 h at 35–37 °C. When oxidase was positive, confirmation analysis (Table S4) was done using VITEK MS (n = 107) with quality control ATCC 8738 or analytical profile index (API) 20E (n = 40) when technical issues prevented the use of the VITEK MS. Both confirmation methods were performed according to the manufacturer's instructions for Enterobacteriaceae and other nonfastidious gram-negative rods, without modifications.
Quality assurance was performed for 15% of the samples (n = 22) at the Malawi University of Science and Technology (MUST), which was conducting other WES research at the time of the study.10 Duplicate volumes were sent in cooler boxes containing ice to MUST, 350 km away from the sampling site, within 24 h of collection, using ground courier services. MUST employed similar techniques for V. cholerae culture as PHIM, but plated 100 μL using spreaders and API was solely used for confirmatory analysis.
Data were manually entered into a password-protected Microsoft Excel version 2301 (Microsoft Corporation, Redmond, WA, USA) file and shared via email with stakeholders who had signed nondisclosure agreements before weekly data-review meetings. Neither laboratory was made aware of the other party's results until the weekly stakeholder results-sharing meeting.
2.5. Ethics
This study was reviewed and approved by the MUST Research Ethics Committee (P.01/2024/117). The University of Louisville Institutional Review Board (23.0969) classified this study as Nonhuman Subject Research.3. Results and discussion
This analysis included 147 samples (131 pit latrine and 16 paired pump truck samples) collected over 19 weeks. All samples cultured by PHIM (n = 147) were negative for V. cholerae after confirmatory analysis (Table S5). Twenty-two additional duplicate samples were independently analyzed by MUST for quality assurance and were confirmed negative, with the exception of one MUST replicate (PT2 on 4/2/2024). The replicate tested positive after biochemical test confirmation, however the other two replicates from that sample and the sample collected from the same latrine at the same time tested by PHIM were all negative. The paired pit latrine and pump truck samples cultured by PHIM had consistent negative results for V. cholerae. This study builds on previous evidence on WES for V. cholerae in NSSS in Malawi10 by demonstrating how this surveillance can be conducted in a camp. While it may be surprising that V. cholerae was only detected in one replicate sample given that cholera is a common issue in Malawi, we note that this is unsurprising given that there was no outbreak or reported cases of cholera in the camp at the time.The presence of other Vibrio species, including V. alginolyticus (n = 3), V. parahaemolyticus (n = 2), and V. metschnikovii (n = 2), was identified in two or more replicates of seven samples using VITEK MS. All these are pathogenic, noncholera Vibrio spp. have been increasingly detected in freshwater in Africa.31 V. parahaemolyticus can be transmitted to humans through ingestion of contaminated water or undercooked aquatic foods and cause gastrointestinal illness. V. alginolyticus can cause ear, eye and wound infections after contact with contaminated water. V. metschnikovii has been associated with both gastrointestinal illness and wound infection, but infrequently causes disease humans.32,33 These Vibrio species are not well documented in previous research in Malawi; additional research is needed to understand transmission routes and develop interventions to protect camp residents.
Based on latrine user counting, we estimate that our samples represented deposits from a total of 310 people per sampling day or approximately 0.6% of the Dzaleka population, assuming 8 h of use. Counting was conducted during the day to ensure researcher safety, and usage may have differed at these sites in the evening. User counting of pit latrines indicated heterogeneity in the use patterns across sites (Table S1). The most used site, PT3, had an average of 11 users per pit latrine stance per h, indicating that more than 85 people use the latrine each day. The least frequently used sites, such as PT1 and PT4, were primarily used by individuals residing in communal tents and buildings nearby. Across sampling sites, the user-counting results indicate that the weekly sampling campaign included a high level of fresh feces, collected from the top layer of the latrine, representing hundreds of people at the camp.
The Dzaleka Health Centre reported no confirmed clinical cholera cases during the study period, consistent with the V. cholerae WES data. According to the Dzaleka Health Centre data, 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640 cholera vaccine doses were administered to camp residents and nearby host community members in 2021; this corresponds to an estimated single-dose vaccination coverage of less than 40% during the study, without considering population changes due to new arrivals, births, deaths, resettlements, and repatriations. The Government of Malawi officially declared the end of the cholera outbreak on July 19, 2024,27 coinciding with the end of our study. Though culture methods via clinical surveillance may be less sensitive than PCR for detecting V. cholerae in stool,34 culture methods for WES have the advantage of selectively detecting viable, culturable V. cholerae, which are more likely to be infectious and clinically relevant.
640 cholera vaccine doses were administered to camp residents and nearby host community members in 2021; this corresponds to an estimated single-dose vaccination coverage of less than 40% during the study, without considering population changes due to new arrivals, births, deaths, resettlements, and repatriations. The Government of Malawi officially declared the end of the cholera outbreak on July 19, 2024,27 coinciding with the end of our study. Though culture methods via clinical surveillance may be less sensitive than PCR for detecting V. cholerae in stool,34 culture methods for WES have the advantage of selectively detecting viable, culturable V. cholerae, which are more likely to be infectious and clinically relevant.
Between February 1, 2024, and July 31, 2024, the Dzaleka Health Centre reported a total of 2269 cases of diarrhea among Dzaleka residents and Malawian patients (Fig. 3). Among those with diarrhea during the study period, 49% (n = 1108) were children under five years of age. Approximately one-third of the total cases (n = 754) involved refugees and asylum seekers, resulting in an average monthly incidence rate of 2.3 cases of diarrhea per 1000 people living in the camp. Previous studies have found a higher incidence of diarrhea in children under five years old living in refugee camps in Africa, with an average of 35.5 cases of diarrhea per 1000 people per month.35
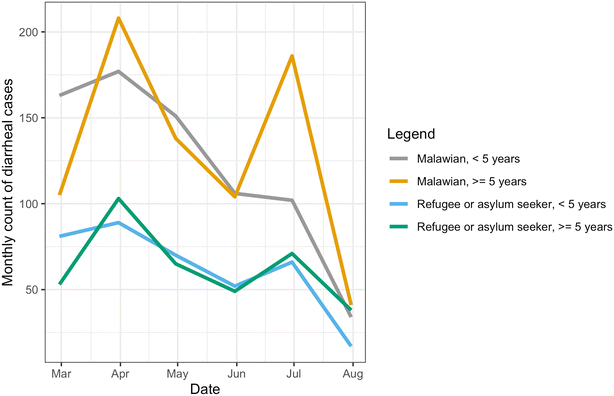 | ||
| Fig. 3 Case count of diarrhea reported by the Dzaleka Health Centre, Malawi, stratified by age and nationality from February to July 2024. | ||
Limitations
Our study has some limitations. First, culture methods limit the range of pathogens that can be surveilled. In clinical surveillance, culture-based methods are less sensitive than PCR for detecting V. cholerae in stool.34 Although our methods adapted guidelines for isolating V. cholerae from stool,10,36 lower V. cholerae abundance is to be expected in environmental samples compared to clinical specimens and could have reduced sensitivity. Further, transporting samples on ice could have induced a viable but nonculturable state in V. cholerae.28The samples in this study may not fully capture the camp population since only certain areas of the camp had high-use pit latrines. Our sampling was limited to areas with the highest use of pit latrines, but these latrines may not be used enough for pathogens with low incidence. Further, symptomatic individuals may have chosen to isolate at home rather than using public latrines. Although we counted latrine users to verify potential individual contributors, usage patterns may have differed in the evening, outside the counting window. Rapid development and a lack of formal road infrastructure at the camp limited the use of other population counting methods, such as GIS or satellite imagery, to understand population size and latrine usage patterns. Composite samplers were not accessible to the research team and were not logistically feasible for use in pit latrines. Finally, not all camps worldwide have established desludging operations, possibly limiting the generalizability of this approach in the future.
Future directions
To increase the number of individuals represented by samples, future studies could prioritize pooled fecal sludge further down the fecal sludge management service chain. We attempted this approach with the pump truck samples; however, these samples had several limitations. First, many pump trucks in Malawi have a capacity of only 1 m3 of fecal sludge, whereas individual pit latrines often contain larger volumes. As a result, pump truck samples may represent fecal sludge from a single pit in this context. Second, pump truck samples could have been cross contaminated between sampling weeks, as there was no practical method to sterilize the desludging truck between sampling weeks.Monitoring bacteriophages via plaque assays may provide a more sensitive alternative method to direct isolation when V. cholerae is at low abundance in WES samples.37 Other methods like colorimetric reverse transcription-loop-mediated isothermal amplification (RT-LAMP) have shown promise for SARS-CoV-2 survelliance in low-resource settings38 and should be considered.
Finally, additional research is needed to understand 1) whether the lower incidence of reported diarrhea in Dzaleka is caused by reduced tendency to seek healthcare in the area, 2) why childhood diarrhea incidence was lower than the initial study by Hershey et al.,35 and 3) how population level WES data can be used alongside clinical diarrheal case rates per 1000 people per month to support refugee health.
Framework for conducting WES in refugee camps
Researchers hoping to apply WES in humanitarian settings with NSSS must consider selecting 1) pathogen targets that can realistically prompt public health action; 2) sampling locations that capture as many users as possible; and 3) engagement by a wide group of stakeholders. Before the start of sample collection in this work, stakeholders agreed to be ready for a range of emergency interventions in the event of a positive sample, including increasing drinking water bucket chlorination campaigns (i.e., point-of-use treatment), providing supplies for disinfecting pit latrines, and advocating for additional vaccinations. Increasing the water supply was not permitted due to groundwater limitations in the camp area. Our framework for implementing WES in Dzaleka (Fig. 4) could support researchers and practitioners to operationalize WES for other pathogens of public health concern in other refugee camps. Given the unique challenges posed by humanitarian contexts for operationalizing WES, we provide detailed suggestions in the SI.4. Conclusion
Vibrio cholerae is a strong candidate for WES in a camp setting during a nationwide cholera outbreak, as early warning of outbreaks allows for prompt response to limit outbreak size and interventions to interrupt transmission are practical across a range of budgets, with some deployable on short notice. We demonstrated that identification and sample collection from high-use pit latrines and pump trucks in an NSSS camp setting was feasible. Quantifying pit-latrine usage in these settings is important to ensure that enough people use the sites to keep site users anonymous and representative of the studied population. Although the culture samples were largely negative for V. cholerae, except for one quality assurance replicate, other pathogenic Vibrio species were identified. Our WES results were consistent with and supported the clinical data reported during the study period. Many methods deployed in global WES rely on PCR or genomic techniques, which are costly, require robust laboratory supply chains, and necessitate extensive staff training. In contrast, culture-based WES may be more practical in humanitarian crises, as it is less expensive and can leverage existing local resources of the ministry of health. Our case study of deploying WES in a refugee camp provides a framework for other governments, academics, intergovernmental organizations, civil society organizations, and community members to leverage WES, thereby enabling them to scale and enhance public health surveillance systems in humanitarian settings.Author contributions
B. B. S., P. C., M. K. W. and R. H. H. conceived and designed research; L. Z., C. M., M. K., P. B. and R. L. N. performed experiments; B. B. S., P. C. and R. H. H. analyzed data; B. B. S. and R. H. H. drafted the manuscript; B. B. S., P. C., E. C., L. Z., C. M., M. K., P. B., R. L. N., M. K. W. and R. H. H. edited and revised manuscript; B. B. S., P. C., E. C., L. Z., C. M., M. K., P. B., R. L. N., M. K. W. and R. H. H. approved final version of manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
Data generated in this study can be found in the published article and its supplementary information (SI) files. Additional data can be requested from The United Nations High Commissioner for Refugees, Chilanga Drive, Area 10, Plot 441/442, P.O. Box 30230, Lilongwe, Malawi.Supplementary information is available. See DOI: https://doi.org/10.1039/d5ew00871a.
Acknowledgements
This work was supported by the UNHCR Innovation Service Data Innovation Fund and grants from the Ellis and Eurofins Foundations. We are grateful for the expertise provided by David Berendes, Jennifer Murphy, Tom Handzel, Ryan Schweitzer, Matthew Kagoli, Joseph B. Bangoh, and S. M. Hasanuzzaman. We also thank the sampling team for their assistance with this study.References
- C. Adams, M. Bias, R. M. Welsh, J. Webb, H. Reese, S. Delgado, J. Person, R. West, S. Shin and A. Kirby, The national wastewater surveillance system (NWSS): from inception to widespread coverage, 2020–2022, United States, Sci. Total Environ., 2024, 924, 171566, DOI:10.1016/j.scitotenv.2024.171566.
- P. Kilaru, D. Hill, K. Anderson, M. B. Collins, H. Green, B. L. Kmush and D. A. Larsen, Wastewater surveillance for infectious disease: a systematic review, Am. J. Epidemiol., 2023, 192(2), 305–322, DOI:10.1093/aje/kwac175.
- N. C. Rondeau, O. J. Rose, E. R. Alt, L. A. Ariyan, A. B. Elikan, J. L. Everard, A. R. Schreier, M. E. Tessler, G. H. Tulinsky, J. R. Vo, C. A. Ray, C. Y. Yang, J. L. Miranda and B. J. Mailloux, Building-level detection threshold of SARS-CoV-2 in wastewater, Microbiol. Spectrum, 2023, 11(2), e0292922, DOI:10.1128/spectrum.02929-22.
- C. S. Truyens, D. M. Berendes, M. E. Cantrell, A. L. Kossik, K. M. McCarthy, A. S. Mehrotra, J. L. Murphy, S. Pillay, S. J. Raj, M. S. Ramaswamy, H. Yakubu and R. H. Holm, Advancing wastewater and environmental surveillance in LMICs for public health response and SDG data gaps, npj Clean Water, 2025, 8(1), 95, DOI:10.1038/s41545-025-00523-w.
- H. Asghar, O. M. Diop, G. Weldegebriel, F. Malik, S. Shetty, L. El Bassioni, A. O. Akande, E. Al Maamoun, S. Zaidi, A. J. Adeniji, C. C. Burns, J. Deshpande, M. S. Oberste and S. A. Lowther, Environmental surveillance for polioviruses in the Global Polio Eradication Initiative, J. Infect. Dis., 2014, 210(Suppl 1), S294–S303, DOI:10.1093/infdis/jiu384.
- R. H. Holm, R. Nyirenda, T. Smith and P. Chigwechokha, Addressing the challenges of establishing quality wastewater or non-sewered sanitation-based surveillance, including laboratory and epidemiological considerations, in Malawi, BMJ Glob. Health., 2023, 8(11), e013307, DOI:10.1136/bmjgh-2023-013307.
- A. Keshaviah, M. B. Diamond, M. J. Wade, S. V. Scarpino and Global Wastewater Action Group, Wastewater monitoring can anchor global disease surveillance systems, Lancet Global Health, 2023, 11(6), e976–e981, DOI:10.1016/S2214-109X(23)00170-5.
- G. Pocock, L. Coetzee, J. Mans, B. Genthe and S. Pillay, Development of a framework for water quality-based COVID-19 epidemiology surveillance for non-sewered communities, Water Research Commission, WRC Report No. 3062/1/22, 2023, https://www.wrc.org.za/wp-content/uploads/mdocs/3062%20final.pdf, (accessed 29 January 2026).
- M. Jakariya, F. Ahmed, M. A. Islam, A. Al Marzan, M. N. Hasan, M. Hossain, T. Ahmed, A. Hossain, H. M. Reza, F. Hossen, T. Nahla, M. M. Rahman, N. M. Bahadur, M. T. Islam, M. Didar-Ul-Alam, N. Mow, H. Jahan, D. Barceló, K. Bibby and P. Bhattacharya, Wastewater-based epidemiological surveillance to monitor the prevalence of SARS-CoV-2 in developing countries with onsite sanitation facilities, Environ. Pollut., 2022, 311, 119679, DOI:10.1016/j.envpol.2022.119679.
- P. Chigwechokha, R. L. Nyirenda, D. Dalitsani, R. L. Namaumbo, Y. Kazembe, T. Smith and R. H. Holm, Vibrio cholerae and Salmonella Typhi culture-based wastewater or non-sewered sanitation surveillance in a resource-limited region, J. Exposure Sci. Environ. Epidemiol., 2024, 34(3), 432–439, DOI:10.1038/s41370-023-00632-z.
- T. Zohra, A. Ikram, M. Salman, A. Amir, A. Saeed, Z. Ashraf and A. Ahad, Wastewater based environmental surveillance of toxigenic Vibrio cholerae in Pakistan, PLoS One, 2021, 16(9), e0257414, DOI:10.1371/journal.pone.02574.
- D. Capone, D. Berendes, O. Cumming, J. Knee, R. Nalá, B. B. Risk, C. Stauber, K. Zhu and J. Brown, Analysis of fecal sludges reveals common enteric pathogens in urban Maputo, Mozambique, Environ. Sci. Technol. Lett., 2020, 7(12), 889–895, DOI:10.1021/acs.estlett.0c00610.
- D. Capone, P. Chigwechokha, F. L. de Los Reyes 3rd, R. H. Holm, B. B. Risk, E. Tilley and J. Brown, Impact of sampling depth on pathogen detection in pit latrines, PLoS Neglected Trop. Dis., 2021, 15(3), e0009176, DOI:10.1371/journal.pntd.0009176.
- E. E. Chukwu, A. Okwuraiwe, C. N. Kunle-Ope, U. T. Igbasi, N. Onyejepu, K. Osuolale, J. O. Shaibu, A. Ojogbede, D. Abuh, E. Afocha, O. Awoderu, K. Obiozor, A. Mustapha and R. Audu, Surveillance of public health pathogens in Lagos wastewater canals: a cross-sectional study, BMC Public Health, 2024, 24, 3590, DOI:10.1186/s12889-024-21157-6.
- Global Polio Eradication Initiative, “Variant Type 2 Poliovirus Isolated from Sewage Samples in Gaza”, 2024, https://polioeradication.org/news/variant-type-2-poliovirus-isolated-from-sewage-samples-in-gaza/, (accessed 29 January 2026).
- N. L. Behnke, R. Cronk, B. B. Shackelford, B. Cooper, R. Tu, L. Heller and J. Bartram, Environmental health conditions in protracted displacement: a systematic scoping review, Sci. Total Environ., 2020, 726, 138234, DOI:10.1016/j.scitotenv.2020.138234.
- B. Cooper, N. L. Behnke, R. Cronk, C. Anthonj, B. B. Shackelford, R. Tu and J. Bartram, Environmental health conditions in the transitional stage of forcible displacement: A systematic scoping review, Sci. Total Environ., 2021, 762, 143136, DOI:10.1016/j.scitotenv.2020.143136.
- J. Cosgrave, Refugee density and dependence: practical implications of camp size, Disasters, 1996, 20(3), 261–270 CrossRef CAS PubMed.
- C. C. Hammer, J. Brainard and P. R. Hunter, Risk factors and risk factor cascades for communicable disease outbreaks in complex humanitarian emergencies: a qualitative systematic review, BMJ Glob. Health, 2018, 3(4), e000647, DOI:10.1136/bmjgh-2017-000647.
- T. Jaber, E. Boelee, J. Bleser and J. K. Bartram, Outbreaks of faecal-orally transmitted diseases in displacement camps: A scoping review of pathogens, risk factors, exposure routes, and drivers of transmission, Global Public Health, 2024, 19(1), 2380847, DOI:10.1080/17441692.2024.2380847.
- B. B. Shackelford, R. Cronk, N. Behnke, B. Cooper, R. Tu, M. D'Souza, J. Bartram, R. Schweitzer and D. Jaff, Environmental health in forced displacement: a systematic scoping review of the emergency phase, Sci. Total Environ., 2020, 714, 136553, DOI:10.1016/j.scitotenv.2020.136553.
- World Health Organization, Common heath needs of refugees and migrants: literature review, WHO, Geneva, 2021, https://iris.who.int/server/api/core/bitstreams/ab9711a7-4894-4605-9400-6ff9027dd3fb/content, (accessed 29 January 2026) Search PubMed.
- A. R. Ario, E. A. Barigye, I. H. Nkonwa, J. Ogwal, D. N. Opio, L. Bulage, D. Kadobera, P. E. Okello, L. W. Kwagonza, S. Kizito, B. Kwesiga and J. Kasozi, Evaluation of public health surveillance systems in refugee settlements in Uganda, 2016–2019: lessons learned, Confl. Health, 2022, 16(1), 15, DOI:10.1186/s13031-022-00449-x.
- M. A. Connolly, M. Gayer, M. J. Ryan, P. Salama, P. Spiegel and D. L. Heymann, Communicable diseases in complex emergencies: impact and challenges, Lancet, 2004, 364(9449), 1974–1983, DOI:10.1016/S0140-6736(04)17481-3.
- D. Jeboda, B. B. Shackelford, P. Chigwechokha, B. A. Chunga, A. Ercumen, C. Workman, J. L. Hart, T. Smith and R. H. Holm, Public opinions from Malawian and Malawi refugee camp residents of wastewater and environmental surveillance, Am. J. Trop. Med. Hyg., 2025, 113(1), 200–213, DOI:10.4269/ajtmh.24-0759.
- C. Chaguza, I. Chibwe, D. Chaima, P. Musicha, L. Ndeketa, W. Kasambara, C. Mhango, U. L. Mseka, J. Bitilinyu-Bangoh, B. Mvula, W. Kipandula, P. Bonongwe, R. J. Munthali, S. Ngwira, C. A. Mwendera, A. Kalizang'oma, K. C. Jambo, D. Kambalame, A. W. Kamng'ona, A. D. Steele, A. Chauma-Mwale, D. Hungerford, M. Kagoli, M. M. Nyaga, Q. Dube, N. French, C. L. Msefula, N. A. Cunliffe and K. C. Jere, Genomic insights into the 2022–2023 Vibrio cholerae outbreak in Malawi, Nat. Commun., 2024, 15(1), 6291, DOI:10.1038/s41467-024-50484-w.
- Republic of Malawi Ministry of Health, 2024, “Press Release: Notice on the End of the Cholera Outbreak in Malawi”.
- C. Lutz, M. Erken, P. Noorian, S. Sun and D. McDougald, Environmental reservoirs and mechanisms of persistence of Vibrio cholerae, Front. Microbiol., 2013, 4, 375, DOI:10.3389/fmicb.2013.00375.
- M. Jubair, J. G. Morris Jr and A. Ali, Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype, PLoS One, 2012, 7(9), e45187, DOI:10.1371/journal.pone.0045187.
- World Health Organization, Guidelines for poliovirus surveillance in the WHO African Region, WHO, Brazzaville, 2025, https://www.afro.who.int/publications/guidelines-poliovirus-surveillance-who-african-region, (accessed 29 January 2026) Search PubMed.
- I. I. Ibangha, D. C. Digwo, C. A. Ozochi, M. C. Enebe, C. N. Ateba and V. N. Chigor, A meta-analysis on the distribution of pathogenic Vibrio Species in water sources and wastewater in Africa, Sci. Total Environ., 2022, 881, 163332, DOI:10.1016/j.scitotenv.2023.163332.
- C. Baker-Austin, J. D. Oliver, M. Alam, A. Ali, M. K. Waldor, F. Qadri and J. Martinez-Urtaza, Vibrio spp. infections, Nat. Rev. Dis. Primers, 2018, 4(1), 1–19, DOI:10.1038/s41572-018-0005-8.
- Y. Konechnyi, Y. Khorkavyi, K. Ivanchuk, I. Kobza, A. Sękowska and O. Korniychuk, Vibrio metschnikovii: Current state of knowledge and discussion of recently identified clinical case, Clin. Case Rep., 2021, 9(4), 2236–2244, DOI:10.1002/ccr3.3999.
- Y. Guillaume, M. Debela, D. Slater, K. Vissieres, R. Ternier, M. F. Franke, J. B. Harris and L. C. Ivers, Poor sensitivity of stool culture compared to polymerase chain reaction in surveillance for Vibrio cholerae in Haiti, 2018–2019, Open Forum Infect. Dis., 2023, 10(6), ofad301, DOI:10.1093/ofid/ofad301.
- C. L. Hershey, S. Doocy, J. Anderson, C. Haskew, P. Spiegel and W. J. Moss, Incidence and risk factors for malaria, pneumonia and diarrhea in children under 5 in UNHCR refugee camps: a retrospective study, Confl. Health, 2011, 5(1), 24, DOI:10.1186/1752-1505-5-24.
- Global Task Force on Cholera Control, Isolation and presumptive identification of Vibrio cholerae O1/O139 from fecal specimens, 2023, https://www.gtfcc.org/wp-content/uploads/2025/03/gtfcc-fact-sheet-isolation-and-identification-of-vibrio-cholerae-from-fecal-specimens-en.pdf, (accessed 29 January 2026).
- M. Akhtar, M. A. Amin, S. N. Hussain, N. N. Nafsi, N. Parvin, F. Khanam, M. T. Islam, M. A. Islam Bhuiyan, R. Afroz, M. G. Firoj, F. Chowdhury, A. I. Khan, M. Jubair, E. T. Ryan, B. J. Shapiro, N. R. Thomson, E. J. Nelson, M. M. Rahman, Y. A. Begum, T. R. Bhuiyan and F. Qadri, Clinical and environmental wastewater-based bacteriophage surveillance for high-impact diarrheal diseases, including cholera, in Bangladesh, MBio, 2025, e0265425, DOI:10.1128/mbio.02654-25.
- J. Akter, W. J. M. Smith, M. Gebrewold, I. Kim, S. L. Simpson, A. Bivins and W. Ahmed, Evaluation of colorimetric RT-LAMP for screening of SARS-CoV-2 in untreated wastewater, Sci. Total Environ., 2024, 907, 167964, DOI:10.1016/j.scitotenv.2023.167964.
| This journal is © The Royal Society of Chemistry 2026 |


