Open Access Article
Open Access ArticleDeveloping an intelligent data-driven framework for organic photovoltaic research
Yu Cui,
Wei Ma and
Han Yan
and
Han Yan *
*
State Key Laboratory for Mechanical Behavior of Materials, School of Materials Science and Engineering, Xi'an Jiaotong University, Xi'an 710049, China. E-mail: mseyanhan@xjtu.edu.cn
First published on 12th December 2025
Abstract
The current trial-and-error research paradigm is inherently inappropriate for organic photovoltaic (OPV) development because of the diversity in the properties of photovoltaic materials and sophisticated device fabrication conditions. A data-driven paradigm is therefore particularly well-suited to address these challenges. The data-driven research paradigm requires big-data extraction from the literature and high-throughput quantum chemical calculation according to chemical structures and quantitative molecular structure–property relationship (QMSPR). Accordingly, we develop an intelligent data-driven framework (IDDF), leveraging large language models (LLMs), a high-throughput quantum chemistry calculation platform (HQCCP), and explainable machine learning (ML). With the aid of IDDF, we extract structured data from 615 peer-reviewed articles and compute 50 molecular descriptors for 125 Y-series acceptors, forming a QMSPR database linking molecular features to device performance. Using the eXtreme Gradient Boosting (XGBoost)-SHapley Additive exPlanations (SHAP) ML model, IDDF maps molecular substructures to key descriptors and device power conversion efficiency (PCE). The results quantitatively demonstrate the influence of each building block of the Y-series acceptors on the final PCE values with explicit quantum chemical explanations. Our framework shifts OPV research from intuition-based design toward a knowledge-guided, predictive mode, providing a foundational step toward autonomous material discovery and enhancing the competitiveness of OPVs among emerging photovoltaic technologies.
Broader contextThe development of organic photovoltaics (OPVs) has long been relying on iterative, experience-based experimentation, limiting the speed and scalability of their innovation. Despite impressive advances in power conversion efficiencies (PCEs), the discovery of new materials and optimization of device performance remain hindered by fragmented data and a lack of systematic knowledge integration. Here, we introduce an intelligent data-driven framework (IDDF) that unifies large language models, high-throughput quantum chemical calculations, and explainable machine learning to transform OPV research into a data-driven discipline. By extracting structured knowledge from 615 scientific papers and combining it with a comprehensive quantum-chemical dataset, IDDF establishes quantitative structure–performance relationships (QMSPRs) that guide molecular design with unprecedented interpretability and accuracy. This work represents a paradigm shift—not only for OPVs but for materials science at large—by demonstrating how artificial intelligence can automate knowledge synthesis and predictive modeling in complex scientific domains. As a fully integrated AI system tailored for OPV, our approach accelerates discovery, reduces reliance on trial-and-error, and unlocks the hidden value of decades of published research. It exemplifies the future of intelligent scientific infrastructure, where AI acts as a co-researcher, enabling sustainable energy technologies to advance with greater speed, transparency, and reproducibility. |
1. Introduction
The advent of Y6 marked a breakthrough in the power conversion efficiency (PCE) of organic photovoltaics (OPVs).1 Inspired by its molecular design, improved non-fullerene acceptors (NFAs) vigorously elevated the PCE from 15.7% to 21.0%.1–7 The remarkable progress in PCE has sparked global interest in OPVs as a promising candidate for next-generation photovoltaic technology.8–10 In the past ten years, the annual number of OPV research papers has grown from 3532 to 6209 according to the Web of Science search results. However, a noticeable trend is that the pace of PCE improvement has gradually slowed since 2019, owing to the absence of breakthrough materials. The disconnect between research outputs and PCE growth is, to some extent, due to the trial-and-error experimentation research paradigm. As OPV molecular design has become increasingly complex, this traditional paradigm has become unsustainable, limiting both the efficacy of material discovery and its scalability. The sluggish PCE growth erodes the competitive advantage of OPVs against other emerging photovoltaic techniques, such as the perovskite solar cells, even in specialized applications requiring semi-transparent and mechanically flexible properties.9–16To overcome this impasse, a fundamental paradigm shift from empirical exploration to data-driven innovation is urgently needed. Over the past decade, the OPV field has accumulated an extensive body of literature encompassing thousands of photovoltaic materials and their corresponding device performances, providing a solid foundation for the transformation towards data-driven approaches.10,17 However, efforts to fully unlock this knowledge potential have long been constrained due to the lack of an efficient and accurate structured data extraction method. Traditional approaches relying on manual curation or rule-based information extraction are time-consuming, error-prone, and ill-suited to address the prevalent linguistic diversity and unstructured formats in academic literature.18–20 Recent advances in LLMs have demonstrated remarkable capabilities in scientific text comprehension, enabling high-precision, context-aware information extraction, and automated data mining.21,22 By further integrating with optimized prompting strategies, it is feasible to extract high-quality structured device data from the literature.23–26
The next challenge for data-driven research lies in establishing a quantitative molecular structure–property relationship (QMSPR), particularly for Y-series NFAs that dominate the highly performing OPVs.6,7 ML offers a powerful tool for uncovering hidden patterns, directly correlating molecular fingerprints or Simplified Molecular Input Line Entry System (SMILES) strings with PCE, often yielding scientifically limited black-box models, which are insufficient to construct QMSPR in OPVs.27–31 Crucially, PCE is not an intrinsic molecular property but rather an outcome of multiple synergistic photophysical processes dominated by molecular properties. The significant PCE variations observed among structurally similar Y-series NFAs demonstrate how subtle molecular structure tuning can govern device performance by modulating photoelectric properties.9,10,32,33 Thus, an effective modelling framework must decouple the complex structure–property relationship into physically meaningful molecular properties. Central to this approach is a well-defined set of molecular optoelectronic descriptors that serve as physical bridges between the chemical structures of photovoltaic materials and device PCEs. To enable reliable and large-scale acquisition of these descriptors, high-throughput quantum chemical calculations with low costs are essential. After that, using standardized datasets of Y-series molecular properties, a two-stage ML pipeline can be constructed: first, correlating computationally derived optoelectronic descriptors with experimentally extracted device PCEs to identify dominant physical parameters; second, linking molecular structures to these key descriptors to reveal how specific substructures determine optoelectronic descriptors. This hierarchical modelling strategy guarantees data-driven innovation in OPVs.
By integrating these components, we establish an Intelligent Data-Driven Framework (IDDF) for OPVs that links molecular structure to device performance through quantifiable optoelectronic descriptors. Our approach combines LLM-assisted literature mining with a standardized high-throughput quantum chemistry computation platform (HQCCP) to construct a comprehensive structure–property–performance dataset for Y-series NFAs. This dataset includes 32 device-level parameters extracted from 615 peer-reviewed publications and 50 computed molecular descriptors for 125 representative molecules. By leveraging explainable machine learning, we quantitatively disentangle the influence of specific molecular substructures, including central electron-accepting cores, end groups, donor units, and side chains, on fundamental optoelectronic properties and, ultimately, on PCE. This analysis reveals the physical mechanisms by which molecular design dictates device performance. Our framework represents a modest yet significant stride in advancing organic photovoltaic (OPV) molecular design from empirical intuition towards a predictable and interpretable scientific domain, providing a design reference for future Y-series acceptors. It represents a critical step in shifting OPV research away from trial-and-error exploration toward knowledge-driven innovation, thereby helping to accelerate R&D cycles and enhance competitiveness among emerging photovoltaic technologies.
2. Results and discussion
2.1. Overall workflow of IDDF
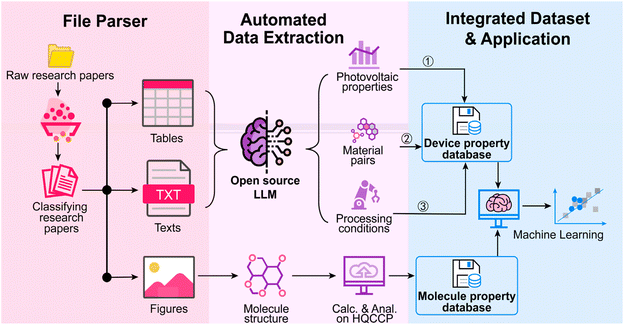
Big-data collection is a prerequisite for OPV's data-driven innovation. By taking advantage of LLM, the structured information on material pairs, device processing conditions, and photovoltaic parameters in texts and tables can be directly converted into metadata.26,34 To obtain missing molecular properties, we developed HQCCP to transform molecular images into strings as inputs and conducted high-throughput quantum chemical computation. Based on the two implementations, IDDF, composed of file parser, automated data extraction, and integrated dataset and application, is proposed to construct the QMSPR in OPVs (Fig. 1). The “File Parser” module first classifies the literature and then extracts texts, tables, and images from the classified literature for downstream data processing. The “Automated Data Extraction” module collects material pairs, photovoltaic parameters, and processing conditions from the contents. Additionally, it incorporates image recognition tools to convert molecular structure images into SMILES strings for subsequent calculations. The “Integrated Dataset and Application” module performs data cleaning, validation, and alignment to merge device performance with molecular electronic properties. The integrated dataset supports explainable ML analysis by applying the XGBoost-SHAP method. This end-to-end automatic workflow enables an efficient transformation from raw literature to structured knowledge, providing reliable data support and intelligent analytical tools for OPV research.2.2. Literature classification
OPV research exhibits interdisciplinary characteristics due to its diverse material systems, complex device architectures, and multiscale physical mechanisms,10,17,34,35 which lead to substantial variations in research focus and evaluation criteria across subfields. The uniform processing of literature data without proper differentiation inevitably introduces interference. Therefore, we conducted a literature classification to select studies directly relevant to molecular design while excluding confounding variables. This classification ensures the dimensional consistency and semantic alignment of the extracted data. As shown in Fig. 2(a), the literature classification comprises two key stages: abstract extraction from literature images using multimodal large models (Fig. S1 in SI), and reliable zero-shot classification through domain knowledge-driven prompt engineering (PE). In the second stage, we designed a structured prompt template incorporating triple constraint mechanisms: (1) establishing a domain-specific classification system, (2) defining deterministic logic for classification decisions, and (3) enforcing a standardized JSON output format to enhance classification reliability (Fig. S2 in SI).We constructed an OPV literature classification framework encompassing 7 core dimensions and 29 subcategories (Fig. 2(a)). The 7 core categories cover the entire innovation chain from molecular design to commercial applications, which strictly follows the “domain-technology” two-dimensional structure. The top-level dimensions classify different “domains” on key scientific and technological challenges; for instance, material development and device physics address efficiency bottlenecks, while stability studies and application technologies target commercialization barriers. At the second level, research directions are categorized based on their technical relevance. Although each “technology” subfield is relatively independent, there are still interconnections among them. Thus, our classification system employs cross-references to reflect these synergistic relationships. This OPV literature classification framework establishes an interdisciplinary knowledge architecture for OPV and demonstrates outstanding adaptability for seamlessly incorporating future research progress.
Building on the classification framework, we analysed the recent landscape of OPV research through a data-driven literature survey. We searched OPV research articles from Web of Science using the querying keywords “organic photovoltaics” and “organic solar cells” during 2019–2025 (starting from the landmark year of Y6). We initially randomly collected 615 peer-reviewed papers from multiple publishers, including Wiley-VCH, the American Chemical Society (ACS), the Royal Society of Chemistry (RSC), and others (Fig. S3 in the SI). We deployed the LLM Qwen2.5-VL-72b for structured abstract information extraction before utilizing DeepSeek-V3 for automatic literature classification. We visualized the literature catalogues using a Sankey diagram (Fig. 2(b)), which effectively represents the flow relationships between hierarchical levels. The line width between nodes intuitively indicates the magnitude of the flow. Sankey visualization reveals that our classification framework achieves broad coverage across all seven “core dimensions” and 25 out of 29 “subcategories”. Nevertheless, at the higher levels of the hierarchy, all 615 papers are fully accounted for within the “core dimensions” and “subcategories,” demonstrating the robustness, comprehensiveness, and systematic design of our classification framework. Within this well-structured catalogue, statistical analysis shows that 57.2% (358/615) of the publications focus on “Material development”, making it the largest node among the core research dimensions. This category typically contains rich information on molecular design, physicochemical characterization, and structure–performance relationships. We thus prioritize an in-depth analysis of this literature subset to systematically establish QMSPR for OPVs. Delving deeper, “Acceptor materials” emerges as the primary output pathway within “Material development,” accounting for 53.1% (190/358) of publications, which highlights its pivotal role in advancing photovoltaic materials. The code for the framework is available at GitHub (https://github.com/limitedcommunication/Data-extraction).
2.3. Text mining
After literature classification, we utilized open-source LLM to extract the material and device data. The DeepSeek-V3 model was selected for two reasons: first, it enables localized data processing; second, it supports targeted model optimization. As illustrated in Fig. 3(a), the corresponding data extraction includes two steps: preliminary screening and data structuring. In the first step, domain knowledge-driven prompts help filter text paragraphs to effectively exclude irrelevant content. Then, multi-round progressive PE precisely extracts the target data from the filtered paragraphs (Fig. S4 in SI). Following the framework, the DeepSeek-V3 model first identifies each set of the four critical photovoltaic parameters reported in the literature (open-circuit voltage (Voc), short-circuit current density (Jsc), fill factor (FF), and PCE) and then assigns a unique condition ID to each photovoltaic parameter set. Subsequently, the model associates each condition ID with the corresponding material pairs and device processing conditions, gradually constructing a complete experimental database. Benefiting from the standardized reporting conventions in OPV research (where most studies consistently report these four photovoltaic parameters), the risk of model-generated hallucinations is substantially reduced. To ensure data reliability, we implemented a verification-feedback mechanism, where the LLM rechecks the original texts to confirm the existence of parameters and automatically triggers condition ID rematching upon detecting omissions. Notably, verification steps employ binary judgments (YES/NO) to control output length and enhance parsing efficiency. All outputs adhere to a predefined JSON schema, preserving both parameter values/units and hierarchical data structures (upper right panel of Fig. 3(a)).To quantitatively evaluate the efficacy of structured data extraction, we established a benchmark dataset through triple cross-validation. 30 representative papers were randomly selected from the original literature corpus and independently annotated by domain experts to create a manual validation set. The evaluation employed a standardized framework to categorize each parameter extraction result into three types: true positive (TP, the model correctly identifies a parameter present in the manual annotation), false positive (FP, the model incorrectly identifies a parameter not present), and false negative (FN, the model fails to extract a reported parameter). The assessment strictly adhered to the data presence principle, only evaluating explicitly documented parameters to ensure unbiased results. As shown in Fig. 3(b), we calculated precision, recall, and F1 scores for 17 kinds of parameters (see the Methods section in the SI for details). The system achieved overall metrics of precision 97.2% ± 1.4%, recall 95.9% ± 0.8%, and F1 score 96.6% ± 1.1%. Collectively, these evaluation metrics highlight the effectiveness of the high-precision text mining methodology, demonstrating its capability to reliably transform unstructured scientific text into structured analysable data. Concurrently, we constructed a complete metadata framework containing literature identifiers, including titles, digital object identifiers (DOIs), author information, abstracts, and publication years. The resulting OPV performance database ultimately incorporated over 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structured data entries, providing a high signal-to-noise ratio foundation for subsequent analysis.
000 structured data entries, providing a high signal-to-noise ratio foundation for subsequent analysis.
Combining the literature classification and the device dataset, we gained clear insights into OPV research trends and performance evolution. Compared to traditional literature review approaches, this data-driven analytical paradigm provides a more objective and comprehensive depiction of the field's developmental trajectory. As the performance data statistics from 2019 to 2025 (Fig. S5 in SI), the reported champion PCE of binary systems increased from 16.5% to 20.8% accompanied by the average value markedly increasing from 11.2% to 17.4%.1,5 The ternary material systems exhibited similar average PCE growth from 14.1% to 17.2% and champion value to the binary counterparts. The statistical results seem somewhat contradictory to the traditional view that OPV devices based on ternary material systems have higher PCE values. Crucially, all high-performance devices—whether binary or ternary—relied to the incorporation of Y6 derivatives. The statistics unequivocally demonstrate the pivotal role of Y6 molecular scaffold innovation in driving OPV performance breakthroughs.
2.4. QMSPR construction
Building upon these analyses, we further elucidate the QMSPR between Y-series acceptors and their photovoltaic performances in OPV devices. A systematic understanding of this underlying correlation necessitates the comprehensive and accurate extraction of electronic characteristics from molecular structures. We therefore developed an analytical framework incorporating HQCCP and explainable ML (Fig. 4(a)). The HQCCP integrates key components of molecular structure generation, input file preparation, job scheduling, output parsing, and data storage into a unified workflow, which implements multi-level high-throughput quantum chemical calculations (Fig. S6 in SI; HQCCP software is available on GitHub: https://github.com/limitedcommunication/HQCCP). We employed the DECIMER package to convert 2D chemical structure images from the literature into SMILES strings, which were subsequently processed by HQCCP. The platform automatically analyses molecular structures and generates standardized input files using template engines. To balance computational efficiency with accuracy, HQCCP adopts a hierarchical calculation strategy: initial geometry optimization is performed using semi-empirical tight-binding (SE-TB) methods, followed by higher-level density functional theory (DFT) geometry optimization and single-point energy corrections based on pre-optimized structures. All quantum chemical calculations employ consistent parameter settings (including functionals and basis sets) to ensure comparability across different Y-series acceptors. The platform supports job submissions to local high-performance computing clusters with automated queue generation, featuring real-time monitoring, failure recovery, and error logging to significantly enhance computational stability and reliability. Upon completion, HQCCP automatically parses quantum chemical output files using Multiwfn, extracts key molecular descriptors, and stores them in standardized database formats (Fig. S6b in SI). The HQCCP systematically transforms implicit structural knowledge into 50 molecular descriptors, organizes them into five main categories: electrostatics and polarity, frontier orbitals, orbital delocalization, molecular geometry, and reactivity indices, and further subdivides these into 13 physically meaningful sub-categories (see Table S2 in SI for details). These descriptors comprehensively characterize the physicochemical properties of small molecule acceptors, which indicate OPV device performance. We selected 125 representative Y-series acceptors from the literature on the “Materials development” catalogue obtained by the afore-mentioned classification (Table S3 in SI), spanning critical molecular engineering aspects. After computational analysis by HQCCP, we established the most comprehensive database of molecular descriptors for Y-series acceptors to date.For each of these acceptors, we employed our HQCCP to compute a comprehensive set of 50 quantum-chemically derived molecular descriptors. To ensure the quality and independence of the input features, we conducted feature engineering with particular emphasis on redundancy analysis and dimensionality reduction among the 50 molecular descriptors. We first calculated the Pearson correlation coefficient matrix across all descriptors to identify the highly correlated feature pairs (Fig. S7 in the SI). If the absolute correlation coefficient between the same two sub-category descriptors exceeds 0.9, they are considered to convey redundant information. After screening, we selected 27 molecular descriptors that exhibited low multicollinearity and broad molecular informativeness (Table S2 in the SI). The database was further integrated with the corresponding photovoltaic properties (PCE parameter), yielding a robust QMSPR dataset comprising a total of 125 × (27 descriptors + 1 PCE label) = 3500 distinct data points (The cleaned database is available on GitHub: https://github.com/limitedcommunication/Cleaned-dataset). Importantly, all machine learning tasks treat each molecule as a single sample represented by a 27-dimensional feature vector paired with one PCE value; thus, the effective sample size is 125. The 125 PCE values span a range of 12.7–19.78%, with a mean of 16.7% and a standard deviation of 1.9% (Fig. S8). Notably, approximately 28% of the entries exhibit PCE values below 15.7%—the benchmark efficiency reported for the Y6 molecule in its original publication. The PCE distribution demonstrates that the dataset encompasses not only state-of-the-art high-efficiency devices but also representative moderate- and lower-performance systems. This balanced distribution significantly enhances the reliability and generalizability of the subsequent ML analysis. However, it should be emphasized that this dataset, despite its performance spread, consists exclusively of published results and therefore reflects only the “survivor” molecules demonstrating sufficient photovoltaic activity to warrant reporting. It does not include truly non-viable candidates that failed during synthesis, processing, or basic device operation. Consequently, the model is best suited for guiding molecular refinement and optimization within the established chemical space of Y-series non-fullerene acceptors, rather than for screening arbitrary structures or identifying fundamentally non-functional materials.
Based on the constructed QMSPR dataset, we developed an ML model for predicting OPV device performance using the XGBoost algorithm.36,37 The model was integrated with the SHAP framework—a cooperative game theory-based interpretability tool—to enable transparent and interpretable predictions (Fig. 4(a)).38,39 The complete dataset was partitioned into training and independent test sets at a 90–10% ratio. To optimize model parameters and ensure predictive performance, we conducted an exhaustive grid search of key XGBoost hyperparameters through 5-fold cross-validation within the training set. The optimized model achieved a root mean square error (RMSE) of 0.26 and a coefficient of determination (r2) of 0.91 on the independent test set, demonstrating robust prediction accuracy (Fig. 4(b)). The comparable performance on the training set (RMSE = 0.24 and r2 = 0.95) indicates that the model effectively learned the underlying structure of the data without significant overfitting (XGBoost framework code is available on https://github.com/limitedcommunication/OPV_analyzer). To rigorously evaluate the generalization capability of our model, we performed a prospective validation on L8-BO-X, a Y-series NFA not included in the original 125-molecule set. For the binary blend PM6:L8-BO-X (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2), the experimentally measured PCE is 17.56% (Fig. S9, validation code is available on: https://github.com/limitedcommunication/L8-BO-X_validation).40 Using our HQCCP, we computed the same 27 quantum-chemical descriptors used in model training and standardized them with the training-set scaler. When input into the frozen XGBoost model, L8-BO-X yielded a predicted PCE of 17.55%, corresponding to an absolute error of just 0.01%. This result further supports the generalizability and predictive reliability of our model. Building upon this well-trained model, we implemented the game theory-based SHAP analysis to quantitatively assess each feature's contribution. This approach provides a unified metric that ranks feature importance and reveals their influences on predictions. It is important to note that the predicted PCEs represent an upper-bound estimate of a molecule's performance potential, contingent upon processing conditions. Consequently, the molecular design rules derived from our model should be interpreted as guidelines for molecular engineering within high-performance OPV systems rather than universal guarantees of device efficiency under arbitrary fabrication protocols. This distinction underscores that our framework captures the interplay between molecular structure and achievable performance under ideal processing, not intrinsic performance independent of processing. It should be noted that the current evaluation relies on a random train-test split, which may inadvertently allow structurally similar molecules to appear in both sets, potentially overestimating predictive performance. Although scaffold- or time-based splitting strategies would provide a more rigorous assessment of generalization to truly novel chemistries, the limited size and scope of the present dataset constrains the feasibility of such approaches.
1.2), the experimentally measured PCE is 17.56% (Fig. S9, validation code is available on: https://github.com/limitedcommunication/L8-BO-X_validation).40 Using our HQCCP, we computed the same 27 quantum-chemical descriptors used in model training and standardized them with the training-set scaler. When input into the frozen XGBoost model, L8-BO-X yielded a predicted PCE of 17.55%, corresponding to an absolute error of just 0.01%. This result further supports the generalizability and predictive reliability of our model. Building upon this well-trained model, we implemented the game theory-based SHAP analysis to quantitatively assess each feature's contribution. This approach provides a unified metric that ranks feature importance and reveals their influences on predictions. It is important to note that the predicted PCEs represent an upper-bound estimate of a molecule's performance potential, contingent upon processing conditions. Consequently, the molecular design rules derived from our model should be interpreted as guidelines for molecular engineering within high-performance OPV systems rather than universal guarantees of device efficiency under arbitrary fabrication protocols. This distinction underscores that our framework captures the interplay between molecular structure and achievable performance under ideal processing, not intrinsic performance independent of processing. It should be noted that the current evaluation relies on a random train-test split, which may inadvertently allow structurally similar molecules to appear in both sets, potentially overestimating predictive performance. Although scaffold- or time-based splitting strategies would provide a more rigorous assessment of generalization to truly novel chemistries, the limited size and scope of the present dataset constrains the feasibility of such approaches.
SHAP-based visualization reveals key predictive features for OPV performance and their underlying mechanisms (Fig. 4(c) and S10 in SI). Quantitative feature importance analysis reveals that the “Electrostatic and polarity” descriptors dominate PCE prediction, accounting for a cumulative contribution of 63.1%. Among these, Pos_average (average positive electrostatic potential) exhibits the highest contribution (12.6%), followed by ESPmin (minimum electrostatic potential, 11.0%), Polar_area (polar surface area, 8.8%), and the MPI (molecular polarity index, 7.0%). SHAP value analysis also confirms that increased values of these descriptors correlate strongly with higher PCE, suggesting that enhancing the four “Electrostatic and polarity” descriptors above improves device performance. Notably, the ZZ component of the molecular quadrupole moment ranks as the fifth most important feature, with a contribution of 6.7%.
For deeper insight into the model's decision-making at the individual molecule level, we visualized SHAP values in a heatmap format (Fig. 5(a)): 125 columns represent 125 distinct molecules in the dataset, and each row corresponds to a molecular descriptor. This representation reveals the direction and magnitude of each feature's contribution to the prediction of individual molecules. Notably, although some molecules exhibit strong and consistent contributions from specific descriptors, others display more complex, multi-feature interaction patterns, highlighting heterogeneity in molecular behaviour. This per-molecule analysis provides a transparent and interpretable view of the model's internal logic. This demonstrates that the model does not rely on uniform feature importance, but its reasoning is based on the unique chemical profile of each acceptor.
To further elucidate the structural origins of key molecular descriptors, we attempted to associate these descriptors with specific molecular blocks to build a QMSPR for Y-series acceptors. We utilize Extended-Connectivity Fingerprints (ECFP) (see calculation details in Method and Fig. S11 in SI) to include the chemical information of the local environment in molecular strings.6,30,31,41 ECFP does not treat molecules as simple strings; instead, it systematically identifies circular topological neighbourhoods around each atom and hashes them into a fixed-length binary vector. Each bit represents the presence or absence of a specific substructural motif. This representation allows ML models to capture complex functional groups and their spatial contexts in a numerically tractable format. By recognizing topological neighbourhoods around each atom and encoding them into fixed-length binary vectors, ECFP efficiently captures information on functional groups and their spatial arrangements. These ECFP fingerprints are subsequently used to map molecular structures to five target descriptors: Pos_average, ESPmin, Polar_area, MPI, and ZZ. Using the XGBoost, we construct high-performance predictive models, achieving R2 of 0.82, 0.95, 0.81, 0.79, and 0.83 on the independent test set, respectively (Fig. S12 in SI). These results indicate a reliable relationship between the ECFP fingerprints and key molecular descriptors. To dissect the specific contributions of molecular substructures to each descriptor, we apply SHAP analysis to all five models (Fig. 5(b–g) and Table S4 in SI). SHAP analysis reveals a positive correlation between the end groups and all five molecular descriptors. Specifically, as the electron-withdrawing capacity of the end groups increases, all molecular descriptors exhibit an upward trend. Through quantitative model evaluation, we conclusively demonstrate that the end groups exert a net positive influence on device PCE, with a weighted contribution of 11.3%. This result indicates that enhancing the electron-withdrawing ability of the end groups can effectively improve the device's PCE. Further analysis reveals that the benzothiadiazole (BTD) core in Y-series acceptors exhibits a significant negative correlation with two key electrostatic potential descriptors (Pos_average and ESPmin). Ultimately, the electron-withdrawing capacity of the BTD core negatively correlates with PCE, with a contribution of −8.7%. Additionally, the donor (D) units in these molecules exhibit a somewhat contradictory influence on electrostatic potential-related descriptors (ESPmin and MPI), suggesting the presence of a nonlinear regulatory mechanism affecting local charge distribution (Fig. 5(g)). Quantitative analysis further reveals that enhancing the electron-donating ability of the D units contributes to improving the device's power conversion efficiency (PCE), with a quantified contribution of 0.1%. Notably, side-chain engineering results demonstrate that increasing the bulkiness of the outer alkyl chains and introducing functionalized substituents on the inner side can enhance device PCE, with an overall contribution of 2.7% (Fig. 5(g)). As depicted in Fig. S13, this confirms the structural consistency of this motif across diverse molecular scaffolds and reveals its atomic-level specificity: the bit is activated by multiple central atoms within the same fluorinated aromatic ring, each contributing to the overall positive SHAP value. The slight variation in SHAP magnitude (+0.4017 vs. +0.3917) between molecules suggests that although the core substructure is universally beneficial, its exact chemical environment can modulate its impact on PCE. This level of granularity links a single fingerprint bit to specific atom environments and quantifies their individual contributions, which is precisely what enables our model to provide actionable, chemically interpretable design rules.
Based on these findings, we propose the following design guidelines for Y-series acceptors. First, strong electron-withdrawing end groups are prioritized to significantly improve the PCE. Second, while maintaining the conjugated core framework, excessive electron-withdrawing strength at the central A′ core should be avoided. Third, D units should be designed with a moderate electron-donating capability to prevent PCE reduction from overly strong donating effects. Finally, side-chain engineering requires precise spatial control for optimal device performance. Collectively, high-performance acceptors should adopt a “strong ends, stable core, moderate donor, controlled side chains” optimization strategy to achieve systematic device improvement through synergistic molecular design. All molecular design guidelines derived from SHAP analysis should be interpreted as testable hypotheses generated from statistical patterns in the data, not as established physical laws. Experimental synthesis and device characterization remain essential to confirm causality and assess practical viability. The primary utility of our framework lies in high-throughput virtual screening and the relative ranking of candidate molecules within the chemical space spanned by known Y-series acceptors, rather than in predicting absolute PCEs for unprecedented, record-breaking materials. The model excels at identifying promising structural motifs and filtering out low-performing candidates, which are tasks that significantly accelerate early-stage discovery. However, absolute PCE predictions for top-tier performers should be treated with caution.
2.5. Applicability domain and generalization potential
The machine learning framework presented in this work is trained and validated exclusively on a dataset of 125 Y-series non-fullerene acceptors, which share a common molecular architecture featuring a fused-ring electron-donating core, electron-withdrawing end groups, and solubilizing side chains. Consequently, the derived structure–property relationships are specific to this chemical family and should not be extrapolated to structurally distinct acceptor classes without validation. The applicability domain of our model is thus defined by its chemical and topological similarities to known Y-series scaffolds. It is well-suited for virtual screening and molecular optimization within this established space, but it cannot reliably predict the performance of molecules that deviate significantly in core topology, conjugation length, or electronic motif.To extend this framework to other photovoltaic material classes, several key steps are required:
(1) construct category-specific datasets with unified device structures and champion PCE values;
(2) develop or adapt molecular descriptors to capture the key physical properties of the new material systems;
(3) retrain or fine-tune the machine learning model using the new data.
Major challenges include the scarcity of standardized data for emerging material classes and the need for descriptors that generalize across diverse chemical motifs. Nevertheless, the modular architecture of our framework combines LLM-based literature mining, quantum-chemical computation, and explainable ML. This provides a scalable foundation for such extensions. Future work will focus on adapting this pipeline to polymer donors, all-polymer systems, and tandem cell subcells, thereby advancing toward a universal data-driven platform for organic photovoltaics.
3. Conclusions
In summary, this study establishes a robust high-throughput computational and analytical pipeline tailored to the OPV field and advances the understanding of the structure–property relationships in Y-series NFA molecules. We developed an IDDF that combines LLM-powered knowledge extraction, HQCCP, and explainable ML. This framework adopts a data-driven approach to accelerate materials innovation. We curated a comprehensive dataset comprising 615 peer-reviewed articles and calculated 27 molecular descriptors for 125 Y-series acceptors. This enabled the construction of a robust QMSPR database that links molecular substructures with multiple descriptors to device performances, establishing a solid data foundation for model training and analysis. On this basis, our explainable XGBoost-SHAP ML model achieves accurate predictions of device performance and quantifies the impact of molecular substructures. The framework represents a step toward replacing the slow and heuristic development of OPV materials with a systematic and computationally efficient approach. The methodology also provides a scalable pathway for advancing high-performance and sustainable energy materials.Author contributions
H. Y. proposed and supervised the project. Y. C. designed and implemented the calculation platform, developed the explainable machine learning models, and constructed the large language model pipeline for literature data extraction. Y. C. and H. Y. co-authored the paper. All authors discussed the results and participated in the analysis.Conflicts of interest
There are no conflicts to declare.Data availability
Data are available within the article or its supplementary information (SI). Supplementary information: includes additional figures, tables, and computational details supporting the main findings. See DOI: https://doi.org/10.1039/d5el00160a.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22475164) and the S&T Program of Energy Shaanxi Laboratory, Grant No. ESLB202440. This work was also supported by the following artificial intelligence technologies: Qwen2.5-72B, Qwen2.5-VL-72B, and DeepSeek-V3. These open-source large language models were employed to analyse textual content, tables, and figures for information extraction and literature classification.Notes and references
- J. Yuan, Y. Zhang, L. Zhou, G. Zhang, H.-L. Yip, T.-K. Lau, X. Lu, C. Zhu, H. Peng, P. A. Johnson, M. Leclerc, Y. Cao, J. Ulanski, Y. Li and Y. Zou, Joule, 2019, 3, 1140–1151 CrossRef CAS.
- Y. Jiang, S. Sun, R. Xu, F. Liu, X. Miao, G. Ran, K. Liu, Y. Yi, W. Zhang and X. Zhu, Nat. Energy, 2024, 9, 975–986 CrossRef CAS.
- C. Chen, L. Wang, W. Xia, K. Qiu, C. Guo, Z. Gan, J. Zhou, Y. Sun, D. Liu, W. Li and T. Wang, Nat. Commun., 2024, 15, 6865 CrossRef CAS.
- J. Fu, Q. Yang, P. Huang, S. Chung, K. Cho, Z. Kan, H. Liu, X. Lu, Y. Lang, H. Lai, F. He, P. W. K. Fong, S. Lu, Y. Yang, Z. Xiao and G. Li, Nat. Commun., 2024, 15, 1830 CrossRef CAS PubMed.
- C. Li, Y. Cai, P. Hu, T. Liu, L. Zhu, R. Zeng, F. Han, M. Zhang, M. Zhang, J. Lv, Y. Ma, D. Han, M. Zhang, Q. Lin, J. Xu, N. Yu, J. Qiao, J. Wang, X. Zhang, J. Xia, Z. Tang, L. Ye, X. Li, Z. Xu, X. Hao, Q. Peng, F. Liu, L. Guo and H. Huang, Nat. Mater., 2025, 24, 1626–1634 CrossRef CAS PubMed.
- C. Li, J. Song, H. Lai, H. Zhang, R. Zhou, J. Xu, H. Huang, L. Liu, J. Gao, Y. Li, M. H. Jee, Z. Zheng, S. Liu, J. Yan, X.-K. Chen, Z. Tang, C. Zhang, H. Y. Woo, F. He, F. Gao, H. Yan and Y. Sun, Nat. Mater., 2025, 24, 433–443 CrossRef.
- J. Ding, H. Mou, H. Chen, J. Xu, W. Sun, J. Zhu, Y. Wang, Y. Huang, Y. Li and Y. Li, Adv. Mater., 2025, 37, 20240439 Search PubMed.
- H. Liang, X. Bi, H. Chen, T. He, Y. Lin, Y. Zhang, K. Ma, W. Feng, Z. Ma, G. Long, C. Li, B. Kan, H. Zhang, O. A. Rakitin, X. Wan, Z. Yao and Y. Chen, Nat. Commun., 2023, 14, 4707 CrossRef PubMed.
- J. Yi, G. Zhang, H. Yu and H. Yan, Nat. Rev. Mater., 2023, 9, 46–62 CrossRef.
- N. Yang, S. Zhang, Y. Cui, J. Wang, S. Cheng and J. Hou, Nat. Rev. Mater., 2025, 10, 404–424 CrossRef.
- Y. Tang, Y. Cui, R. Zhang, W. Xue, W. Ma and H. Yan, Adv. Energy Mater., 2024, 14, 2303799 CrossRef.
- F. Xue, Y. Xie, Y. Cui, D. Y. Paraschuk, W. Ma and H. Yan, Adv. Funct. Mater., 2024, 35, 2415617 CrossRef.
- K. Zhou, D. Han, K. Xian, S. Li, M. Gao, K. Zhang, B. Zhao, X. Li, Y. Chen, Y. Geng and L. Ye, Energy Environ. Sci., 2024, 17, 5950–5961 RSC.
- Z. Wang, D. Zhang, L. Yang, O. Allam, Y. Gao, Y. Su, M. Xu, S. Mo, Q. Wu, Z. Wang, J. Liu, J. He, R. Li, X. Jia, Z. Li, L. Yang, M. D. Weber, Y. Yu, X. Zhang, T. J. Marks, N. Stingelin, J. Kacher, S. S. Jang, A. Facchetti and M. Shao, Science, 2025, 387, 381–387 CrossRef PubMed.
- S. Lee, S. Oh, S. Han, D. Lee, J. Lee, Y. Kim, H.-Y. Jeong, J.-W. Lee, M.-H. Lee, W. B. Ying, S. Jeong, S. Lee, J. Kim, Y. H. Kim, B. J. Kim, E.-c. Jeon, T.-S. Kim, S. Cho and J.-Y. Lee, Energy Environ. Sci., 2024, 17, 8915–8925 RSC.
- J.-W. Lee, H.-G. Lee, E. S. Oh, S. W. Lee, T. N.-L. Phan, S. Li, T.-S. Kim and B. J. Kim, Joule, 2024, 8, 204–223 CrossRef.
- J. Wang, Y. Xie, K. Chen, H. Wu, J. M. Hodgkiss and X. Zhan, Nat. Rev. Phys., 2024, 6, 365–381 CrossRef.
- X. Li, M. Cui, J. Li, R. Bai, Z. Lu and U. Aickelin, Neurocomputing, 2021, 443, 345–355 CrossRef.
- T. Gupta, M. Zaki, N. M. A. Krishnan and Mausam, npj Comput. Mater., 2022, 8, 102 CrossRef.
- H. Huang, R. Long, H. Chen, K. Sun and Q. Li, Sustain. Prod. Consum., 2022, 30, 674–685 CrossRef.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry, A. Askell, S. Agarwal, A. Herbert-Voss, G. Krueger, T. Henighan, R. Child, A. Ramesh, D. M. Ziegler, J. Wu, C. Winter, C. Hesse, M. Chen, E. Sigler, M. Litwin, S. Gray, B. Chess, J. Clark, C. Berner, S. McCandlish, A. Radford, I. Sutskever and D. Amodei, Adv. Neural. Inf. Process. Syst., 2020, 33, 1877 Search PubMed.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, Ł. Kaiser and I. Polosukhin, Adv. Neural. Inf. Process. Syst., 2017, 5998–6008 Search PubMed.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef PubMed.
- Y. Kang and J. Kim, Nat. Commun., 2024, 15, 4705 CrossRef.
- M. P. Polak and D. Morgan, Nat. Commun., 2024, 15, 1569 CrossRef PubMed.
- Y. Kang, W. Lee, T. Bae, S. Han, H. Jang and J. Kim, J. Am. Chem. Soc., 2025, 147, 3943–3958 CrossRef.
- J. Ross, B. Belgodere, V. Chenthamarakshan, I. Padhi, Y. Mroueh and P. Das, Nat. Mach. Intell., 2022, 4, 1256–1264 CrossRef.
- X. Zeng, H. Xiang, L. Yu, J. Wang, K. Li, R. Nussinov and F. Cheng, Nat. Mach. Intell., 2022, 4, 1004–1016 CrossRef.
- P. Bai, F. Miljković, B. John and H. Lu, Nat. Mach. Intell., 2023, 5, 126–136 CrossRef.
- G. Han and Y. Yi, Angew. Chem., Int. Ed., 2022, 61, e202213953 CrossRef.
- S. Liu, D. Zhang, H.-J. Egelhaaf, G. Wang, X. Li, T. Heumüller, C. J. Brabec and N. Li, J. Mater. Chem. A, 2024, 12, 14688–14697 RSC.
- L. Zhu, M. Huang, G. Han, Z. Wei and Y. Yi, Angew. Chem., Int. Ed., 2025, 64, e202413913 CrossRef PubMed.
- S. Shahzadi, T. Shahzadi, Z. Shafiq and M. Janjua, High Energy Chem., 2024, 58, 583–603 CrossRef.
- J. Dagdelen, A. Dunn, S. Lee, N. Walker, A. S. Rosen, G. Ceder, K. A. Persson and A. Jain, Nat. Commun., 2024, 15, 1418 CrossRef.
- J. Luke, E. J. Yang, C. Labanti, S. Y. Park and J.-S. Kim, Nat. Rev. Mater., 2023, 8, 839–852 CrossRef.
- T. Chen and C. Guestrin, in Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Association For Computing Machinery, San Francisco 2016 Search PubMed.
- Y. Cui, Q. Fan, H. Feng, T. Li, D. Y. Paraschuk, W. Ma and H. Yan, Energy Environ. Sci., 2024, 17, 8954–8965 RSC.
- S. M. Lundberg, B. Nair, M. S. Vavilala, M. Horibe, M. J. Eisses, T. Adams, D. E. Liston, D. K.-W. Low, S.-F. Newman, J. Kim and S.-I. Lee, Nat. Biomed. Eng., 2018, 2, 749–760 CrossRef PubMed.
- S. M. Lundberg, G. Erion, H. Chen, A. DeGrave, J. M. Prutkin, B. Nair, R. Katz, J. Himmelfarb, N. Bansal and S.-I. Lee, Nat. Mach. Intell., 2020, 2, 56–67 CrossRef PubMed.
- J. Song, C. Zhang, C. Li, J. Qiao, J. Yu, J. Gao, X. Wang, X. Hao, Z. Tang, G. Lu, R. Yang, H. Yan and Y. Sun, Angew. Chem., Int. Ed., 2024, 63, e202404297 CrossRef.
- D. Rogers and M. Hahn, J. Chem. Inf. Model., 2010, 50, 742 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |