Open Access Article
Open Access ArticleNMR-based conformational analysis of DNA G-quadruplex guides mapping essential structure–function relationship in protein chaperoning
Deepraj Negi†
a,
Zijue Huang† b,
Ahyun Son†
b,
Ahyun Son† b,
Bharathwaj Sathyamoorthy
b,
Bharathwaj Sathyamoorthy *a and
Scott Horowitz
*a and
Scott Horowitz *b
*b
aDepartment of Chemistry, Indian Institute of Science Education and Research Bhopal, Madhya Pradesh 462066, India. E-mail: bharathwaj@iiserb.ac.in
bDepartment of Chemistry & Biochemistry and the Knoebel Institute for Healthy Aging, University of Denver, Denver, CO 80208, USA. E-mail: scott.horowitz@du.edu
First published on 22nd December 2025
Abstract
G-quadruplexes (G4s) are increasingly recognized to chaperone proteins, warranting studies of structure–function relationships. In this study, we apply solution NMR methods to determine the topology and base-level resolution structure of a G-rich DNA sequence with protein chaperoning activity (referred to as Seq576) without chemical shift assignments. Seq576 samples two conformations, in the slow exchange timescale, arising from a G-register shift. Using the structural insights of Seq576, we then perform structure–function studies via mutation and chaperone assays to investigate the G4 properties essential for chaperoning protein aggregation and folding. These studies highlight the possibility of using a construct design to perform in-depth nucleic acid structural biology investigation using inexpensive and fast NMR experiments to obtain and analyze function, such as the residue-level investigation of chaperone activity.
Introduction
A healthy protein folding environment (proteostasis) is crucial to cell health, and defects in proteostasis contribute to multiple protein misfolding diseases.1 Chaperones are a critical part of maintaining proteostasis,1,2 that either prevent protein aggregation (often termed holdases) or help protein folding (often termed foldases).3 A particular nucleic acid structure, the G-quadruplex (G4), is highly efficient at chaperoning proteins, and improper regulation of G4s has been implicated in multiple protein misfolding diseases.4–7 G4s are four-stranded structures of DNA or RNA formed by stacked guanine tetrads and stabilized by central cations.8G4s were previously found to be orders of magnitude more efficient at preventing aggregation than other chaperones in vitro. Furthermore, they were found to be general chaperones, able to prevent the aggregation of a wide variety of different proteins.6,9 Tests of whether G4s could additionally directly affect protein folding found that they rescue partially folded protein intermediates and promote their folding to the native state.10 Further investigation showed that G4s could catalyze protein folding. In these experiments, the protein folded through an on-pathway intermediate while continuously bound to the G4, and with increased rate constants for both folding and unfolding.11 In cells, G4s were found to improve the folding environment to a similar degree to many chaperone proteins and to directly aid the quality of protein folding as their primary effect.9,10
However, our knowledge of how G4s chaperone proteins is far less developed. A previous investigation suggested the roles of overall structural topology, accessibility to the G4 tetrad core, G4 oligomerization state, and structural dynamics in chaperoning proteins.12 Characterizing specific contributions with structure–function studies would identify molecular interactions that facilitate G4s in chaperoning proteins, which are currently not clear.
Performing comprehensive structure–function analyses requires detailed structural information about the binding partners. While solving the structures of any macromolecule is a time-intensive process, it tends to be even more so for nucleic acids than for proteins. Obtaining conformational preferences of nucleic acids with NMR spectroscopy is challenging due to the low proton density and reliance on time and labor-intensive nuclear Overhauser effect (NOE)-based analysis for chemical shift assignments.13 Identification of the G4 topology is further complicated by the range of structural polymorphs, calling for effective and economic methods that provide an alternative to the conventional methodology.14 Recent developments in 13C/1H chemical-shift-based methodologies for DNA G4s enable the rapid and reliable identification of backbone topology and characterization of loop conformations, without the need for expensive isotopic enrichment or complicated NMR experiments/analysis.15,16
In this work, we apply a 13C/1H chemical shifts-based solution-state NMR spectroscopy methodology to analyze the conformations of a G4-forming DNA sequence (referred to as Seq576) that is known to perform protein chaperoning activity, and is found in several organisms, especially fish species. Seq576 is observed to sample two conformations that exchange on the slow timescale (greater than milliseconds).
13C/1H NMR analysis provides topological characterization from a natural isotopic abundance sample without chemical shift assignments, enabling mutational analysis to unravel the conformational dynamics of register shifts. Significantly, the NMR-based topology assignment predictions facilitate structure–function analysis of Seq576, paving the way for understanding atomistic details in the analysis of protein chaperoning performed by nucleic acids.
Results
Preliminary characterization of Seq576
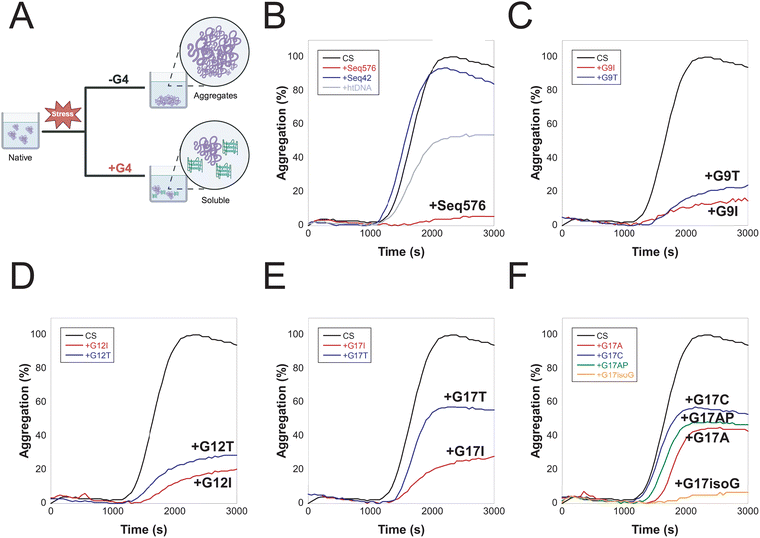
As an initial step in the biophysical characterization of Seq576, circular dichroism (CD) data were acquired to obtain the overall topology of the sequence. CD spectroscopy can distinguish between various topologies of G4s, including parallel (∼264 nm maximum, ∼245 nm minimum), antiparallel (∼295 nm maximum, ∼260 nm minimum), and hybrid (∼295 nm maximum, ∼260 nm maximum, ∼245 nm minimum) structures.17 As previously shown, the CD spectrum obtained for Seq576 suggests the formation of a parallel conformation due to its characteristic ∼264 nm maximum and ∼245 nm minimum (Fig. 1A).9Subsequently, 1D NMR data acquired for Seq576 displayed approximately 24 resonances in the imino region (Fig. 1B), indicating the formation of more than one G4 conformation given a sequence of 20 bases. Such a scenario could arise from two topologies comprising three tetrad planes, each resulting in a doubling of imino resonances in the characteristic Hoogsteen region (10–12 ppm) of the 1H NMR spectrum. This would not be surprising, as Seq576 has fifteen guanine nucleotides, while a three-plane G4 would require a minimum of twelve.
Identifying G-register shifts in Seq576 with single mutant analysis
When surplus guanosines are present, especially in a contiguous manner (as in the regions G9–G12 and G14–G20 for Seq576), G-register shifts are commonly observed that are known to result in structural heterogeneity.18,19 In these cases, the non-tetrad guanosines within the G-tracts can slide with respect to one another, forming multiple conformations. This scenario potentially results in G-register shifts in Seq576, arising from G9–G12 stretching and/or G14–G20 stretching.Generally, extensive polymorphism is reported for G-register shifts with broad and overlapping imino resonances. Seq576 displays sharp and reasonably resolved imino resonances at an almost 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The well-resolved spectral feature of Seq576 is rendered as an amenable target for high-resolution characterization with NMR. It also becomes interesting to determine the two conformational states and to determine whether one or both states contribute to chaperoning activity.
1 ratio. The well-resolved spectral feature of Seq576 is rendered as an amenable target for high-resolution characterization with NMR. It also becomes interesting to determine the two conformational states and to determine whether one or both states contribute to chaperoning activity.
To address this question, we devised a strategy to stabilize each potential G4 conformation of Seq576 individually. If a G-register shift is involved in the case of Seq576, then such a scenario would involve G9–G12 and/or G14–G20 stretches. As a first step, we tested the mutants of the G9–G12 stretch, such that the first G was mutated to thymine (Seq576G9T, Table S1) or inosine (Seq576G9I, Table S1). If G9 were indeed involved in tetrad formation, then one set of resonances belonging to that state would be deleted in the NMR spectrum. Similarly, another set was prepared wherein G12 in Seq576 was mutated to thymine (Seq576G12T, Table S1) or inosine (Seq576G12I, Table S1). In line with our speculations, both Seq576G9T/I and Seq576G12T/I mutations resulted in a single set of imino resonances in the 1H 1D NMR spectrum (Fig. 1B and Fig. S2) that were different from one another. Small variations in the chemical shifts of Seq576 mutants from the wild-type are plausibly due to subtle variations in the structure and/or stabilizing interactions (such as a change in π–π interaction of loop nucleotides proximal to the G-tetrad plane). This experiment effectively separated the two conformations of Seq576.
Now that two sets of NMR resonances of the Seq576 sequence accounted for the parallel topology suggested by CD, we looked to use another mutant to confirm the two conformations. Within the G14–G20 tract, only substitution in G17 may likely allow for two sets of G-stretches (G14–G16 and G18–G20) to be available for three-plane G4 formation. Falling in line, the Seq576G17T mutant displays 24 imino resonances in the 1H NMR spectrum that are almost indistinguishable from those of Seq576 (Fig. 1B), unambiguously showing that G17 is not involved in tetrad formation in either state. Therefore, to allow base-level resolution characterization of Seq576 states, Seq576G9T and Seq576G12T sequences were obtained using 2D NMR spectroscopy.
2D NMR characterization of Seq576
Guanosine bases sampling the anti and syn glycosyl dihedral angle tend to display distinct clusters in the 13C–1H aromatic and sugar spectra.20 As each backbone G4 topology is associated with a unique ratio of anti![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) syn guanines, counting the number in the distinct cluster provides a rapid and reliable way of determining backbone topology before venturing into detailed NOESY analysis.15,16 To confirm the formation of a parallel topology in both states of Seq576, the peaks picked in the 13C–1H HSQC were overlaid on the C8–H8 chemical shift correlations documented previously.15,16 The C8–H8 spectral analysis of Seq576 indicates 24 resonances in the anti region (Fig. S2), confirming that both states (with 12
syn guanines, counting the number in the distinct cluster provides a rapid and reliable way of determining backbone topology before venturing into detailed NOESY analysis.15,16 To confirm the formation of a parallel topology in both states of Seq576, the peaks picked in the 13C–1H HSQC were overlaid on the C8–H8 chemical shift correlations documented previously.15,16 The C8–H8 spectral analysis of Seq576 indicates 24 resonances in the anti region (Fig. S2), confirming that both states (with 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 anti
0 anti![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) syn ratio) sample parallel G4 topology.
syn ratio) sample parallel G4 topology.
The next step was to obtain sequence-specific assignments of Seq576; however, the NOESY analysis is complicated by cross-peaks arising from both states. The single mutants Seq576G9T and Seq576G12T proved helpful, as their 2D HSQC and NOESY spectra arise from a single homogeneous state and are therefore better resolved. Similar to the 1D imino 1H spectra, the 2D spectra of Seq576G9T/G12T agree well with the 2D spectrum of wild-type Seq576, with each mutant displaying 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 anti
0 anti![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) syn resonances in the characteristic C8–H8 aromatic and C1′–H1′ sugar regions of the HSQC. Intranucleotide guanine spin-system 1H resonances (H1′, H2′/H2″ and H8) picked from the NOESY were then subjected to an optimized machine-learning-based methodology, facilitating plane-specific assignments (Table S2).15 The assignments allowed for a rapid and reliable sequence-specific assignment for Seq576G9T/G12T mutants. Resonance assignment obtained for Seq576G9T/G12T (Fig. S3–S5 and Tables S3 and S4) mutants was subsequently transferred to the Seq576 NMR data, allowing complete assignment of chemical shifts (Fig. 1B and 2B and C).
syn resonances in the characteristic C8–H8 aromatic and C1′–H1′ sugar regions of the HSQC. Intranucleotide guanine spin-system 1H resonances (H1′, H2′/H2″ and H8) picked from the NOESY were then subjected to an optimized machine-learning-based methodology, facilitating plane-specific assignments (Table S2).15 The assignments allowed for a rapid and reliable sequence-specific assignment for Seq576G9T/G12T mutants. Resonance assignment obtained for Seq576G9T/G12T (Fig. S3–S5 and Tables S3 and S4) mutants was subsequently transferred to the Seq576 NMR data, allowing complete assignment of chemical shifts (Fig. 1B and 2B and C).
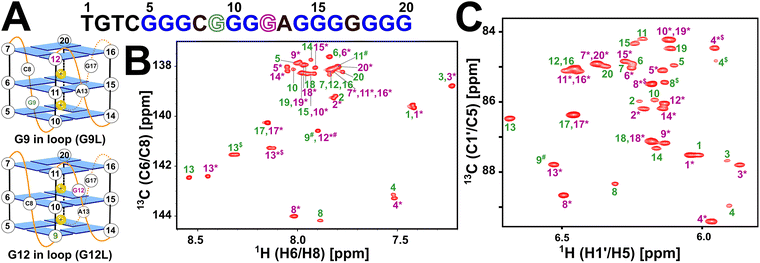 | ||
| Fig. 2 Schematic representation of Seq576 topology and 2D NMR assignments for the two states. (A) Seq576 exchanges across two parallel conformations wherein either G9 (top, G9L) or G12 (bottom, G12L) is involved in G-register shift. All tetrad-forming guanosines sample anti-glycosyl dihedral angle and are represented as blue rectangles. Propeller loops connecting parallel strands are depicted in orange, with labels indicating the nucleotide type and position in the primary sequence. Potassium ions that stabilize tetrad planes are shown as yellow spheres with a positive charge. (B and C) 13C–1H heteronuclear single quantum coherence correlation (HSQC) spectrum obtained for Seq576 observed for (B) aromatic C6/C8–H6/H8 and (C) sugar C1′–H1′ correlation with sequence-specific resonance assignment. NOESY connectivity for sequential walk and 15N–1H imino assignments for Seq576G9T and Seq576G12T are provided in Fig. S3–S5. Nucleotide labels for the state involving G12 in the propeller loop are marked with an asterisk to distinguish it from the state where G9 is involved in loop formation (without an asterisk) to maintain consistency with Fig. 1B. Resonance marked with “$” (adenine C2–H2 for aromatic and cytosine C5–H5 in sugar) indicates aliased peaks arising from folding of the 13C dimension and ambiguous assignment is labeled with “#”. | ||
Combined with the information from single mutant analysis of Seq576, we constructed base-resolution conformations of the two states of Seq576 (Fig. 2A). The sequence adopts two parallel G4 conformations wherein either G9 or G12 is involved in tetrad formation, allowing for G-register shift in the second strand (Fig. 2A), with G10 and G11 involved in tetrad formation in both instances. In both states, as G17 is present in a single-nucleotide propeller loop conformation between strands 3 and 4, only a single resonance is observed in either state (Fig. 2B). In contrast, other states tend to show two resonances.
Furanose 13C–C1' chemical shifts and one-bond 13C–1H scalar coupling (1JC1′–H1′) of tetrad nucleotides shed light on conformational fluctuations observed in parallel topology. Guanines involved in tetrad formation following a single-nucleotide propeller loop present in plane-1 (P1) tend to exhibit a downfield-shifted resonance in the carbon dimension compared to other guanines in plane-2 (P2) and plane-3 (P3).21 For Seq576, when G9/G12 is involved in the loop formation, G14/G9 and G18 are preceded by a single-nucleotide propeller loop that exhibits downfield-shifted 13C–C1′ resonance (Fig. 2B).
Additionally, in both states, the average of 1JC1'-H1' for P1 guanines displays an increased magnitude of ∼4 Hz compared to nucleotides present in plane-2 and plane-3 (P2 and P3) (Fig. S6), in line with the previous results, further confirming common features of parallel topology.21
Conformation-guided chaperoning studies shed light on structure–function relationships
Having confirmed that Seq576 adopts two distinct G4 conformers, we determined whether one or both are functionally responsible for its chaperone activity. To this end, we first used a thermal aggregation assay (Fig. 3A), which monitors the ability of chaperones to suppress heat-induced aggregation of citrate synthase (CS) at 50 °C. We included non-G4 single-stranded DNA, Seq42 (Table S1), and bulk DNA, which serve as controls of basal and medium activity (Fig. 3B), respectively. As previously shown, Seq42 did not prevent aggregation at this concentration, indicating that the holdase activity is not a general property for just any short single-stranded DNA.9 Bulk DNA (∼10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp) showed moderate holdase activity, resulting in approximately 60% aggregation overall, as anticipated.22 In contrast, as previously shown, the G4-forming wild-type Seq576 shows excellent holdase activity, with aggregation below 10% (Fig. 3B).9
000 bp) showed moderate holdase activity, resulting in approximately 60% aggregation overall, as anticipated.22 In contrast, as previously shown, the G4-forming wild-type Seq576 shows excellent holdase activity, with aggregation below 10% (Fig. 3B).9
To dissect the contribution of each conformer to chaperone activity, we introduced inosine and thymine mutations at positions G9 and G12. While structurally analogous to guanine, inosine cannot form the stable Hoogsteen hydrogen bonds required for G-quartet formation. Thymine substitutions at these positions were also intended to prevent specific guanines from engaging in quartet formation, thereby favoring one conformer over the other. Our results showed that inosine (Seq576G9I and Seq576G12I, Table S1) consistently outperformed thymine (Seq576G9T and Seq576G12T, Table S1) at both positions in preventing aggregation, leading to a very modest decrease (Fig. 3C and D). Comparatively, locking the conformation through mutation at G9 (Seq576G9T/I) or G12 (Seq576G12T/I) results in similar activity, with G9 mutants performing slightly better as chaperones at preventing protein aggregation.
Next, we probed how G17 mutants of Seq576 perform in the aggregation assay. NMR and CD data show that the thymine mutation at G17 does not alter tetrad formation or the overall G4 fold (Fig. 1B and Fig. S7, and S8). Interestingly, despite preserving the G4 structure, Seq576G17T substantially reduced holdase activity (∼60% aggregation, Fig. 3E). Yet again, inosine at G17 (Seq576G17I) was comparatively more effective, resulting in ∼30% aggregation (Fig. 3E). These results suggest that the G17 position in the loop is important for activity, meriting additional G17 mutants to explore its role further. Among the mutants at G17, Seq576G17C showed a substantial loss of function, driving the fastest aggregation. Seq576G17T and the abasic mutant (Seq576G17AP) followed nearly identical kinetics (Fig. 3E and F). However, aggregation with G17T continued beyond that of G17AP, leading to higher overall aggregation (Fig. S9), indicating that substituting guanine with thymine is more disruptive than nucleobase removal to chaperone activity. Seq576G17A delayed the aggregation process and marginally decreased aggregation compared to Seq576G17AP (Fig. 3F), with Seq576G17isoG maintaining activity similar to the wild-type (<10% aggregation). Collectively, these results emphasize the importance of G17 present in the propeller conformation in loop-3 modulating the holdase activity.
While aggregation assays demonstrate that G4s can inhibit misfolding, refolding assays are equally essential to determine whether they can promote the restoration of properly folded functional protein. Next, we evaluated the refolding capability of the mutants using the folding biosensor TagRFP675. Chemically denatured TagRFP675 remains unfolded and non-fluorescent at its native excitation/emission wavelength of 598/675 nm. When diluted into refolding conditions, the recovery of native fluorescence over time allows us to assess the effect of the “foldase” activity of Seq576 and its mutants on their ability to assist protein folding (Fig. 4A).
In the presence of non-G4-forming DNA Seq42, TagRFP675 displays no recovery of fluorescence signal, indicating that the control sequence does not aid in refolding (Fig. 4B). Next, G9 and G12 mutants of Seq576 were investigated for foldase activity. Similar to the aggregation activity, inosine mutants at both positions performed slightly better than the thymine mutants in the refolding assays. However, neither position showed a significant increase in fluorescence compared to the wild-type (Fig. 4C and D). The initial folding rate between inosine and thymine was comparable, suggesting that substitutions at these positions do not substantially impair the foldase function (Fig. S10). These findings indicate that the difference in foldase activity between the conformers involving G9 or G12 in the propeller loop is not pronounced.
Given that G17 mutations of Seq576 exhibited strong effects on aggregation despite not disturbing the parallel G4 topology, we investigated its foldase role closely. At G17, consistent with the aggregation results, substitution by inosine continued to outperform thymine (Fig. 4E). The Seq576G17AP mutant shows the worst foldase activity, with lower fluorescence and a prolonged lag phase, indicating a slower and less efficient refolding process. Similarly, Seq576G17T and Seq576G17C also show poor foldase activity, with slower refolding kinetics than the wild-type. Conversely, Seq576G17A and Seq576G17I display nearly comparable refolding activity to the wild-type, suggesting that purine mutations did not substantially impair foldase function (Fig. S10). Seq576G17isoG exhibits rapid folding kinetics, with an exponential phase nearly identical to the wild-type Seq576 (Fig. 4F).
Comparing the activity that reduces aggregation, the G17 mutations can be rank ordered as follows: G17C = G17T < G17AP < G17A < G17I < G17isoG (Fig. 3). While comparing the activity that promotes folding, the mutations can be ranked as follows: G17AP < G17T < G17C < G17A < G17I < G17isoG (Fig. 4). These trends are consistent across the two assays, yet they suggest that the aggregation and folding functions are not entirely dependent on the same features. While pyrimidines were more effective in the G17 position than the absence of a base for folding, this was reversed for the prevention of aggregation. However, purines were more effective in position 17 for chaperone activity in all cases.
AlphaFold3 prediction suggests the interaction of G17 nucleobase with the binding site of TagRFP675
To gain insights into the structural implications of the mutations, especially at G17, we used the machine-learning-based structure prediction tool, AlphaFold3,23 to simulate the G4 structure and its binding site on TagRFP675. While AlphaFold3 predicts that Seq576 will form a parallel G4 topology, the presence of two conformers in Seq576 complicates the modelling of the protein–G4 complex. In the absence of a locking mutation that stabilizes a single conformer, the G4 core appears disordered when bound to the protein (Fig. S11). Thus, the introduction of a locking mutation was necessary to obtain a model of the protein–G4 complex that accurately models the G4 interaction.In the predicted TagRFP675–Seq576G9T and TagRFP675–Seq576G12T complexes, we observed that the loop G17 burrowed into a binding interface formed between the β-sheet (residues 214–217) and a random coil region (residues 150 and 152) (Fig. 5A and B). However, for Seq576G17T/G17AP mutants, the G4 structures move away from the protein surface and no longer form these contacts, indicating a loss of interaction that could contribute to their reduced chaperone function (Fig. 5C and D). These predictions suggest the structural positioning of G17 is essential and mediates binding and chaperone activity.
Role of G4 oligomerization in protein chaperoning
Our previous findings indicated that the G4 chaperone function is influenced by the oligomerization state; it was imperative to examine how these mutations affected G4 self-oligomerization.12 We therefore sought to analyze the oligomerization of Seq576 and its mutants using native gel electrophoresis. To visualize G4 structures, N-methyl mesoporphyrin (NMM), a known quadruplex binder, was used to stain the gels along with general nucleic acid stain SYBR Gold.24 As expected from the CD data (Fig. S8), the appearance of bands in all lanes indicates G4 formation for all mutants, including Seq576G17isoG. Subsequent staining with SYBR Gold revealed that G17C, G17T, and G17AP mutants, which displayed the weakest chaperone activity, existed predominantly in monomeric forms (Fig. 6A). In contrast, Seq576G17isoG was shown to form multiple G4 oligomers, as confirmed by overlaying the NMM- and SYBR-stained gels (Fig. S12–S14).We further quantified the gel to assess the relative oligomerization. Consistent with previous findings, mutants capable of forming higher-order structures largely exhibit stronger holdase activity (Fig. 6B, Pearson's R = −0.90, Spearman's R = −0.92, both p < 0.0001).12 As the relationship between G4 oligomerization and foldase function is unknown, we performed the same comparison for the protein refolding activity.
We observe that mutants with reduced oligomerization showed a slower initial folding phase for foldase function. Based on the ranked foldase performance, Spearman's correlation yielded R = 0.74, indicating that G4 oligomerization is also associated with enhanced foldase activity, but less strongly than aggregation prevention (Fig. 6C and D). Overall, these results suggest that oligomerization is a mechanistic feature that prevents aggregation and facilitates proper protein folding.
Conclusions
In this study, we devised a strategy to solve the base-level resolution of a multi-conformer G4 by NMR without chemical shift assignments. We then investigated the chaperone activity of Seq576, which exists as two parallel G4s, through single-point mutations at three critical positions (G9, G12, and G17). Our results demonstrated that both conformers contributed to both preventing and aiding protein folding, but more stringent changes to chaperone activity occurred due to changes in the loop. These changes in activity are due to changes in G4 oligomerization ability, as well as disruption of a major protein-binding site.The method applied here for characterizing G4 topologies offers a considerably faster approach than traditionally used in NMR. The ability to use unassigned resonances on an unlabeled sample reduces the cost and time to solve an NMR structure considerably. While this technique was used here for G4 structure, a very similar strategy could presumably be employed with other nucleic acid structures.
In terms of chaperone activity, it was interesting that mutations to G17 affected G4 oligomerization so strongly, while it was also crucial to the predicted binding site primarily used for chaperone activity. While unclear how at present, it is possible that these two characteristics are physically linked, and that the ability for G4s to oligomerize uses the same chemical properties as its chaperone function. Regardless, this case emphasizes the importance of considering that chaperoning is not merely binding and that other variables are important to consider in their function.
Author contributions
S. H. and B. S. designed the project and acquired funding. D. N. carried out the NMR experiments, data analysis, Z. H. and A. S. carried out the chaperone assays and related data analysis. All authors wrote and edited the manuscript.Conflicts of interest
There are no conflicts to declare.Data availability
All data supporting this article have been included in the main text and the supplementary information (SI). Supplementary information: chemical shifts for Seq576 mutants have been deposited in Biological Magnetic Resonance Bank (Seq576G9T ID: 53322; Seq576G12T: 53321). See DOI: https://doi.org/10.1039/d5cp03909f.Acknowledgements
B. S. thanks Anusandhan National Research Foundation (ANRF CRG/2022/005088) and IISER Bhopal, and S. H. thanks National Institutes of Health (NIH R35GM142442) for funding. D. N. thanks the University Grants Commission for his fellowship. B. S. thanks the NMR Facility at IISER Bhopal for the infrastructure and maintenance, especially Mr Rajbeer Singh. D. N. and B. S. thanks Dr Rajesh Kumar Reddy Sannapureddi (IISER Bhopal) for insightful discussions during this project.Notes and references
- F. U. Hartl, A. Bracher and M. Hayer-Hartl, Molecular chaperones in protein folding and proteostasis, Nature, 2011, 475, 324–332 CrossRef CAS PubMed.
- B. B. Guzman, A. Son, T. J. Litberg, Z. Huang, D. Dominguez and S. Horowitz, Emerging roles for G-quadruplexes in proteostasis, FEBS J., 2023, 290, 4614–4625 CrossRef CAS.
- B. E. Docter, S. Horowitz, M. J. Gray, U. Jakob and J. C. Bardwell, Do nucleic acids moonlight as molecular chaperones?, Nucleic Acids Res., 2016, 44, 4835–4845 CrossRef CAS PubMed.
- L. Kallweit, E. D. Hamlett, H. Saternos, A. Gilmore, A. C. Granholm and S. Horowitz, Chronic RNA G-quadruplex accumulation in aging and Alzheimer’s disease, eLife, 2025, 14, RP105446 CrossRef.
- K. Matsuo, S. Asamitsu, K. Maeda, H. Suzuki, K. Kawakubo, G. Komiya, K. Kudo, Y. Sakai, K. Hori, S. Ikenoshita, S. Usuki, S. Funahashi, H. Oizumi, A. Takeda, Y. Kawata, T. Mizobata, N. Shioda and Y. Yabuki, RNA G-quadruplexes form scaffolds that promote neuropathological alpha-synuclein aggregation, Cell, 2024, 187, 6835–6848 e6820 CrossRef CAS PubMed.
- B. R. Sahoo, V. Kocman, N. Clark, N. Myers, X. Deng, E. L. Wong, H. J. Yang, A. Kotar, B. B. Guzman, D. Dominguez, J. Plavec and J. C. A. Bardwell, Protein G-quadruplex interactions and their effects on phase transitions and protein aggregation, Nucleic Acids Res., 2024, 52, 4702–4722 CrossRef CAS PubMed.
- E. G. Oldani, K. M. Reynolds Caicedo, M. E. Spaeth Herda, A. H. Sachs, E. G. Chapman, S. Kumar, D. A. Linseman and S. Horowitz, The effect of G-quadruplexes on TDP43 condensation, distribution, and toxicity, Structure, 2025, 8(8), 1294–1303 CrossRef.
- D. Varshney, J. Spiegel, K. Zyner, D. Tannahill and S. Balasubramanian, The regulation and functions of DNA and RNA G-quadruplexes, Nat. Rev. Mol. Cell Biol., 2020, 21, 459–474 CrossRef CAS PubMed.
- A. Begeman, A. Son, T. J. Litberg, T. H. Wroblewski, T. Gehring, V. Huizar Cabral, J. Bourne, Z. Xuan and S. Horowitz, G-Quadruplexes act as sequence-dependent protein chaperones, EMBO Rep., 2020, 21, e49735 CrossRef CAS.
- A. Son, V. Huizar Cabral, Z. Huang, T. J. Litberg and S. Horowitz, G-quadruplexes rescuing protein folding, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2216308120 CrossRef CAS.
- Z. Huang, K. Ghosh, F. Stull and S. Horowitz, G-quadruplexes catalyze protein folding by reshaping the energetic landscape, Proc. Natl. Acad. Sci. U. S. A., 2025, 122, e2414045122 CrossRef PubMed.
- T. J. Litberg, R. K. R. Sannapureddi, Z. Huang, A. Son, B. Sathyamoorthy and S. Horowitz, Why are G-quadruplexes good at preventing protein aggregation?, RNA Biol., 2023, 20, 495–509 CrossRef PubMed.
- B. Sathyamoorthy, R. K. R. Sannapureddi, D. Negi and P. Singh, Conformational characterization of duplex DNA with solution-state NMR spectroscopy, J. Magn. Reson. Open, 2022, 10–11, 100035 CrossRef.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Quadruplex DNA: sequence, topology and structure, Nucleic Acids Res., 2006, 34, 5402–5415 Search PubMed.
- R. K. Reddy Sannapureddi, M. K. Mohanty, A. K. Gautam and B. Sathyamoorthy, Characterization of DNA G-quadruplex Topologies with NMR Chemical Shifts, J. Phys. Chem. Lett., 2020, 11, 10016–10022 Search PubMed.
- R. K. R. Sannapureddi and B. Sathyamoorthy, Loop Nucleotide Chemical Shifts as a Tool to Characterize DNA G-Quadruplexes, ChemPhysChem, 2025, 26, e2401075 CrossRef.
- R. Del Villar-Guerra, J. O. Trent and J. B. Chaires, G-Quadruplex Secondary Structure Obtained from Circular Dichroism Spectroscopy, Angew. Chem., Int. Ed., 2018, 57, 7171–7175 CrossRef PubMed.
- J. T. Grun, C. Hennecker, D. P. Klotzner, R. W. Harkness, I. Bessi, A. Heckel, A. K. Mittermaier and H. Schwalbe, Conformational Dynamics of Strand Register Shifts in DNA G-Quadruplexes, J. Am. Chem. Soc., 2020, 142, 264–273 CrossRef PubMed.
- R. W. T. Harkness and A. K. Mittermaier, G-register exchange dynamics in guanine quadruplexes, Nucleic Acids Res., 2016, 44, 3481–3494 CrossRef PubMed.
- K. L. Greene, Y. Wang and D. Live, Influence of the glycosidic torsion angle on 13C and 15N shifts in guanosine nucleotides: investigations of G-tetrad models with alternating syn and anti bases, J. Biomol. NMR, 1995, 5, 333–338 CrossRef PubMed.
- R. K. R. Sannapureddi, M. K. Mohanty, L. Salmon and B. Sathyamoorthy, Conformational Plasticity of Parallel G-Quadruplex - Implications on Duplex-Quadruplex Motifs, J. Am. Chem. Soc., 2023, 145, 15370–15380 CrossRef PubMed.
- T. J. Litberg, B. Docter, M. P. Hughes, J. Bourne and S. Horowitz, DNA Facilitates Oligomerization and Prevents Aggregation via DNA Networks, Biophys. J., 2020, 118, 162–171 Search PubMed.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritzel, O. Ronneberger, L. Willmore, A. J. Ballard, J. Bambrick, S. W. Bodenstein, D. A. Evans, C. C. Hung, M. O'Neill, D. Reiman, K. Tunyasuvunakool, Z. Wu, A. Zemgulyte, E. Arvaniti, C. Beattie, O. Bertolli, A. Bridgland, A. Cherepanov, M. Congreve, A. I. Cowen-Rivers, A. Cowie, M. Figurnov, F. B. Fuchs, H. Gladman, R. Jain, Y. A. Khan, C. M. R. Low, K. Perlin, A. Potapenko, P. Savy, S. Singh, A. Stecula, A. Thillaisundaram, C. Tong, S. Yakneen, E. D. Zhong, M. Zielinski, A. Zidek, V. Bapst, P. Kohli, M. Jaderberg, D. Hassabis and J. M. Jumper, Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature, 2024, 630, 493–500 CrossRef PubMed.
- J. M. Nicoludis, S. P. Barrett, J. L. Mergny and L. A. Yatsunyk, Interaction of human telomeric DNA with N-methyl mesoporphyrin IX, Nucleic Acids Res., 2012, 40, 5432–5447 CrossRef PubMed.
Footnote |
| † Authors contributed equally to this work. |
| This journal is © the Owner Societies 2026 |