Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Effect of protective mutation on structure and dynamics of APOE: a molecular dynamics simulation study
Newton A.
Ihoeghian
 ,
Usman L.
Abass
,
Ibrahim
Imam
and
Qing
Shao
,
Usman L.
Abass
,
Ibrahim
Imam
and
Qing
Shao
 *
*
Department of Chemical & Material Engineering, University of Kentucky, Lexington, KY 40506, USA. E-mail: qshao@uky.edu
First published on 24th December 2025
Abstract
Apolipoprotein E (APOE) plays a significant role in determining the risk of Alzheimer's disease (AD). Three mutations—APOE3–R136S, APOE3–V236E, and APOE4–R251G—have been reported to reduce the risk of AD. Unveiling the molecular mechanism behind this reduction could lay a foundation for developing therapeutics for AD. To shed light on this subject, we investigate the mutation-induced variation in structural and dynamic properties of APOE3–R136S, APOE3–V236E, and APOE4–R251G in explicit solvent using molecular dynamics simulations. The APOE2, APOE3, and APOE4 were used as the reference. The analysis unveiled that the three protective mutations may exert protection through different mechanisms. The R215G mutation makes the flexibility of APOE4 proteins more similar to that of APOE2 and APOE3. In addition, this mutation reduced the exposure area of the oligomerization region by 5–16%. Such a reduction could alleviate the aggregation tendencies of the APOE proteins with amyloid-forming peptides. On the other hand, the R136S and V236 mutations alter the exposure area of the hydrophobic amino acid residues in the lipidation region. Their protective mechanisms may be due to the alteration in the lipidation capability of the APOE3 protein.
1. Introduction
Apolipoprotein E (APOE) transports lipids and maintains lipid balance. It is involved in neurodegenerative diseases, especially Alzheimer's disease (AD).1 In humans, APOE exists as three common isoforms: APOE2 (ε2), APOE3 (ε3), and APOE4 (ε4). These isoforms differ by single amino acid substitutions at positions 112 and 158.1–3 APOE3, the most common isoform, serves as the reference for comparison and has a cysteine (Cys) at position 112 and an arginine (Arg) at position 158. ε2 differs from ε3 by having Cys at position 158. ε4, the strongest genetic risk factor for late-onset AD, differs from ε3 by having an arginine (Arg) at position 112.1,2 Several studies have established an association between ε4 and increased AD risk: APOE4 carriers have a 2–4 fold higher risk compared to non-carriers.1,4,5 ε2 carriers have a lower risk of developing AD.1,4,5Studies have identified rare protective APOE mutations that could reduce the risk of AD.6,7 The APOE3-Jacksonville (V236E), APOE3-Christchurch (R136S), and APOE4–R251G variants are among the discovered protective mutations.6,8–10 Large-scale genetic studies have confirmed that V236E and R251G are associated with a 2- to 3-fold decrease in AD risk.6 The V236E carriers show a 10.5-year delay in AD onset, and R251G carriers exhibit a 6-year delay.6 Notably, R251G is always inherited with APOE4, while V236E is always inherited with APOE3, suggesting that their protective effects may stem from isoform-specific conformational modifications.8
Unveiling the mechanism behind these protective mutations is crucial for understanding their protective. Furthermore, the unveiled mechanism could guide the development of therapeutics mimicking these mutations. Protein functionality depends on its structure and dynamics.2,11,12 Thus, mutation-induced variations in protein structure and dynamics of APOE could be responsible for protection. One APOE protein consists of three domains: an N-terminal domain (RES 1–167), which mediates LDLR binding; a hinge region (RES 168–205), which provides flexibility; and a C-terminal domain (RES 206–299), which facilitates lipid binding.2,11 Nuclear magnetic resonance (NMR) studies show that the C-terminal domain folds against the N-terminal domain in lipid-free APOE, influencing its structural stability and receptor interactions.13 APOE4 adopts a conformation that enhances interdomain interactions, which may increase its aggregation propensity and impair lipid transport.5,14
Knowledge gaps still exist regarding the three protective variants APOE3-Jacksonville (V236E), APOE3-Christchurch (R136S), and APOE4–R251G, particularly in terms of their conformational flexibility and domain interactions with APOE.1,6,9,10,15–19 Molecular dynamics (MD) simulations are a powerful tool for understanding the conformation and dynamics of proteins and their function at the atomic level. MD simulations have provided valuable insights into the molecular mechanisms underlying APOE functions, including lipid binding20 and isoform-dependent effects on AD risk.21 Most of these simulations focus on APOE2, APOE3, and APOE4, three widely occurring variants. The MD simulations of Williams et al.22 revealed an APOE4-specific misfolded intermediate that may disrupt lipid transport and protein aggregation in AD. Henry et al.23 used cross-linking mass spectrometry and MD simulations to model lipidated APOE4. Their findings suggest an activation mechanism involving structural rearrangements for receptor recognition. The computer simulations of Gursky and his team24 showed that APOE undergoes conformational changes to bind amyloid-β fibrils. Mamchur et al.25 used MD simulations to show that the APOE2 isoform retained C-terminal structure while APOE4 showed unwinding at RES 260–266. Stuchell-Brereton et al.26 combined single-molecule spectroscopy and MD simulations to study monomeric APOE4. They found that APOE4 exhibits more disordered structures than previously known, maintaining conformational heterogeneity even when lipid-bound. These studies iterate that a small change in amino acid sequence may result in significant variation in the structure and dynamics of the APOE protein, which could alter its functions. Thus, it is crucial to unveil how the conformation and dynamics of APOE proteins may alter upon the three protective mutations.
The goal of this work is to explore the variation in APOE structure and dynamics induced by the three protective mutations: APOE3–R136S, APOE3–V236E and APOE4–R251G mutations using MD simulations. The three widely occurring isoforms, APOE2, APOE3 and APOE4 will be used as the reference. The analysis will focus on the lipidation and aggregation (oligomerization) domains of the APOE protein. The lipidation domain regulates the ability of APOE to bind to lipid nanoparticles and perform its function to deliver lipids to the desired place. The aggregation domain regulates the tendency of APOE to self-assemble into a dimer. This work will not consider the properties of the receptor binding domain because the receptor binding usually occurs after the APOE protein binds to the lipid nanoparticle. The rest of the paper will be organized as follows: Section 2 will detail the deployed model and simulation procedure. Section 3 will focus on the results and discussions. Section 4 will present the conclusion based on the results and discussions.
2. Molecular model and simulations
2.1 Molecular model
The initial 3D structure of APOE3 (299 amino acid residues) was obtained from the AlphaFold2 database.27 The signal region (first 18 amino acids) was removed. APOE3 has an experimentally determined NMR structure.2 The relaxed conformation from the Alphafold structure was aligned with the 2L7B NMR structure with an RMSD of 0.23 nm, indicating that the AlphaFold2 predicted structure consistent with the NMR structure. The validated AlphaFold2 structure was used as template to create the APOE variants (APOE2, APOE4, APOE3–R136S, APOE3–V236E, and APOE4–R251G), specific mutations were introduced using the Dunbrack 2010 rotamer library28 within the UCSF Chimera software.29The lipidation region (RES 192–272)30,31 is responsible for lipid binding. The amphipathic helices of APOE interact with lipid surfaces.32 Efficient lipid redistribution is vital for the metabolic function of APOE in both circulation and the brain. The oligomerization region (RES 248–299)30,33 is responsible for APOE to oligomerize (aggregate). The lipidation and oligomerization regions overlap on RES 248–272.12 To facilitate the analysis, we divide RES 192–299 into three regions: lipidation-only: RES 192–247; overlapping: RES 248–272, the oligomerization-only: RES 273–299 (Fig. 1a).
The simulation system was built by surrounding the selected APOE protein with a 3.0-nm water molecule shell. This gave a water-shelled protein of volume 9877.47 nm3 and dimensions x − 26.90 nm, y − 19.30 nm, and z − 19.03 nm when placed in a box (Fig. 1b). This water-molecule shell is a balance between the accuracy and efficiency for simulating protein systems. First, it ensures that the APOE protein is immersed in an aqueous reservoir, a widely accepted model setting for proteins in the human body. Second, this water-molecule shell helps accelerate the MD simulation by reducing the number of water molecules in the system. The TIP3P water model34 was used to represent water molecules. Appropriate counterions Na+ ions were introduced to neutralize the net charge of the simulation box.35
The AMBER14 force field with the OL15 modification36 was used to describe the nonbond and bond interactions in the simulation systems. This field has been validated for accurately reproducing the structural and dynamic properties of proteins.37 Non-bonded interactions were calculated using a combination of short-range Lennard-Jones 12-6 potential for van der Waals interactions and long-range Coulombic potential for electrostatic interactions, as represented in eqn (1):
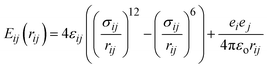 | (1) |
2.2 Simulation detail
The simulations for each system were conducted in three stages: (a) energy minimization, (b) equilibration, and (c) production. Energy minimization utilized the steepest descent to remove unfavorable atomic contacts. The equilibration phase consisted of a 300 ns canonical (NVT) ensemble MD simulation at 310 K and 1 atm with a 2-fs time step, using the velocity-rescaling thermostat37 for temperature control. The production phase involved a 1000 ns canonical (NVT) ensemble MD simulation at 310 K with a 2-fs time step. The temperature was controlled using the same algorithm as for the equilibrium stage. Coordinates of the APOE protein, water molecules, and ions were saved every 500 ps in the production stage for further analysis. Periodic boundary conditions were applied in all simulations. A 1.2 nm cutoff was used for Lennard-Jones interactions, while the particle-mesh Ewald (PME) method38 handled long-range electrostatic interactions. The LINCS algorithm39 constrained all bonds involving hydrogen atoms. The GROMACS 202340 was employed for both energy minimization and MD simulations. To enhance the reliability of our findings, four independent replicates were simulated for each APOE variant.APOE variants were parameterized with the AMBER ff14SB protein force field (as distributed in amber14sb_OL15.ff in GROMACS). ff14SB provides updated protein backbone and side-chain torsions relative to ff99SB and is widely used for protein simulations.36 Each protein was centered in a cubic periodic box and solvated with a spherical water shell of 3.0 nm thickness. The solvation step produces a droplet of explicit water around the protein within a larger periodic box; periodic boundaries were applied in x, y, and z directions for all runs. Using a solvent shell rather than filling the entire box minimizes system size while preserving the first hydration layers of the solute; this approach and related stochastic/spherical boundary methods are established in the literature for biomolecular MD when bulk solvent is not essential,41,42 and more recent implementations in QM/MM shells.43 Water molecules were taken from the AMBER-compatible TIP3P model for consistency with ff14SB (Amber water models overview). The systems were neutralized by replacing solvent with counterions, which adds the minimum number of ions needed for electroneutrality. Before dynamics, each system was minimized by steepest descent (tolerance emtol = 10 kJ mol−1 nm−1, step size emstep = 0.01 nm) to remove bad contacts. All dynamics were conducted in the canonical (NVT) ensemble because the systems are solvent droplets in vacuum. Temperature was controlled with the stochastic velocity-rescaling thermostat (v-rescale) at 310 K with a relaxation time of 1.0 ps.44 After minimization, we performed a 300 ns NVT equilibration to ensure the system was well equilibrated. This was followed by NVT production (up to 1 µs per replica).45–49 Coordinates were saved every 1 ns for analysis (nstxtcout = 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at dt = 0.002 ps).
000 at dt = 0.002 ps).
We used a 2 fs timestep (dt = 0.002 ps) with all covalent bonds involving hydrogen atoms were constrained. The stability and accuracy of v-rescale constraints are established Bussi et al.44 and LINCS efficiently maintains constraints at this timestep.50 Recent protein simulations at 310 K commonly use 2 fs with constraints—for instance, AD-relevant TREM2/ApoE systems (neighbor cutoff 1.4 nm; 2-fs timestep).45 Short-range Lennard-Jones (LJ) and real-space Coulomb interactions used 1.4 nm cutoffs with the Verlet cutoff scheme. The neighbor list was updated every 40 steps (80 fs). A long-range analytical dispersion correction was applied to account for truncated LJ tails, per GROMACS recommendations (long-range vdW corrections). The PME method was used for long-range electrostatics with 4th-order B-spline interpolation (pme-order = 4) and grid spacing 0.12 nm, consistent with standard, well-validated PME settings51 and SPME refinements.52 The 1.4-nm neighbor cutoff and 2-fs timestep are also common in recent AD-related protein MD.45
2.3 Statistical analysis
The statistical analyses utilized in this study include the Kolmogorov–Smirnov (KS) and Cohen's d tests. The KS test was used to assess whether two distributions of solvent accessible surface area (SASA) and hydrogen bonding (HB) values differed significantly in distribution. A large KS statistic, which leads to a low p-value, indicates that the two distributions are likely different, and we reject the null hypothesis. The null hypothesis (H0) assumed identical distributions, while the alternative hypothesis (H1) suggests significant difference. The KS statistic >0.3 reject the null hypothesis (H0) of identical distributions. Cohen's d is a measure of the magnitude of the difference between two group means, standardized by the standard deviation. Cohen's d calculated the effect size of the difference between distributions, with interpretations based on the following ranges: |d| < 0.2 for negligible, 0.2 ≤ |d| < 0.5 for small, 0.5 ≤ |d| < 0.8 for medium, and |d| ≥ 0.8 for large effect sizes.53,54 These methods were applied to compare SASA distributions and the hydrogen bonding distributions between the wild-type APOE isoforms and their mutants. We interpreted the RMSF correlation strength between the mutants and APOE3 or APOE2 using established statistical guidelines. Values near ±1 indicate strong linear or monotonic relationships. Values near 0 indicate weak relationships. We applied Cohen's effect-size thresholds55 and the correlation scale of Hinkle et al.56 All analyses were conducted using Python code developed based on the NumPy, Matplotlib, Seaborn, SciPy, and Pandas modules.3. Results and discussion
3.1 Flexibility
We utilize root mean square fluctuation (RMSF) of the backbone Cα atoms to analyze the flexibility of the APOE protein. Fig. 2 shows the RMSF curves of the backbone Cα atoms for the lipidation-only (RES 192–247), overlapping (RES 248–272), and oligomerization-only (RES 273–299) regions of APOE2, APOE3, APOE4, and APOE3–R136S, APOE3–V236E, and APOE4–R251G. We quantified mutation-induced flexibility changes using Pearson correlation coefficients (rp), Spearman correlation coefficients (rs), mean flexibility deviations (Δμ), percent changes (%Δ), and root mean square deviations (RMSD) to assess dynamic similarities and differences between isoforms and variants. These values are presented in Table S1 of the SI. The discussion will first focus on the lipidation-only and oligomerization-only regions, and then the overlapping regions. | ||
| Fig. 2 RMSF of the backbone Cα atoms in the lipidation-only (RES 192–247), overlapping (RES 248–272), and oligomerization-only (RES 273–299) regions. The APOE4-specific peak at 248–260 lies within the Mahley et al. canonical lipid-binding segment (244–272).57 Peak attenuation by V236E and R251G mirrors reported increases in lipidation and reduced self-aggregation.6,17 | ||
The R136S, V236E and R251G mutations of the APOE proteins attenuate the differences from APOE2 and APOE3 in the lipidation-only region. The lipidation-only region shows two differences in flexibility among APOE2, APOE3 and APOE4. The first is around RES 192–202: both APOE2 and APOE3 RMSF curves exhibit a peak with a height of 0.5–0.6 nm, while the APOE4 curve presents a peak of 0.4 nm. Frieden et al. pointed that APOE3 (as determined by Chen et al.20) presents large intrinsic disordered region (IDRs) around RES 210–223 (particularly 199–209 and 224–239).2 From Fig. 2, we identify that APOE3 presents a flexibility up to 0.57 nm compared to 0.4 nm of APOE4 around RES 192–202. The APOE4 flexibility difference (0.17 nm) relative to APOE3 amount to 26.5% reduction (RMSD = 0.129). This implies that APOE4 may possess fewer IDRs within this segment. Frieden et al. highlighted the importance of IDRs. IDRs allow helices and structured portions of proteins to move relative to one another. The extent of IDR flexibility determines protein function.20 The APOE3 mutations R136S and V236E exhibit RMSF peaks of 0.44 nm and 0.64 nm, respectively. APOE3–R136S reduces the flexibility of APOE3 (12.72% reduction) and the APOE3–V236E variant shows a pronounced increase in flexibility (12.11% increase) around RES 192–202, reaching levels similar to APOE2 with rs (rp) of 0.910 (0.964). The APOE4 R251G mutation shows a flexibility peak of ∼0.52 nm. APOE4–R251G exhibits disrupted correlation with APOE4 (rp = 0.004, rs = −0.082) and introduces a mean flexibility increase of 0.114 nm (33.0% increase, RMSD = 0.139). The mutation shifts the flexibility of APOE4 toward APOE3 levels within this segment. Limited flexibility in APOE4 aligns with stronger inter-domain coupling. APOE4 coupling restricts motions near the IDR-modulated interface. The interface enables lipid-induced domain opening. Restoring flexibility toward APOE2/APOE3 levels by mutation supports the established domain-opening mechanism for lipid binding.12,20 The second is around RES 240–247: both APOE2 and APOE3 present RMSF values around 0.20–0.28 nm, while the RMSF of APOE4 presents a value around 0.36 nm. APOE4 shows moderate negative correlation with APOE3 (rp = −0.416, rs = −0.31) with a mean flexibility increase of 0.064 nm (24.2% increase, RMSD = 0.071). The APOE4 is more flexible than APOE2 and APOE3 in RES 240–247 with a mean flexibility increase of 0.119 nm (56.66% increase, RMSD = 0.119) and 0.064 nm (24.15% increase, RMSD = 0.071) respectively. The APOE3–R136S and APOE3–V235E curves exhibit distinct RMSF. R136S shows strong negative correlation with APOE3 (rp (rs) = −0.858 (−0.833) with a mean flexibility increase of 0.105 nm (39.7% increase, RMSD = 0.113). R136S shows similarity to APOE4 (0.32–0.38 nm) flexibility, while V236E exhibits a similar flexibility range (0.20–0.28 nm) as APOE2 and APOE3. V236E exhibits moderate positive correlation with APOE3 (rp (rs) = 0.436 (0.619)) with a mean flexibility decrease of 0.021 nm (8.0% reduction, RMSD = 0.03). A similar effect was observed with R251G mutation of APOE4. APOE4–R251G shows weak negative correlation with APOE4 (rp = −0.029, rs = −0.071) with a mean flexibility decrease of 0.077 nm (23.5% reduction, RMSD = 0.079). The RMSF curves exhibit 0.20–0.25 nm values. Thus, the V236E and R251G mutations make these proteins more rigid as APOE2 and APOE3 protein around RES 240–247.
The R251G mutation in APOE4 does not alter protein flexibility in the oligomerization-only region (RES 273–299). The APOE2, APOE3, and APOE4 proteins present three distinct peaks around RES 290: 0.28–0.35 nm for APOE2, 0.60 nm for APOE3, and 0.50 for APOE4. Both mutations (R136S and V236E) slightly reduce the flexibility of APOE3 (9.69% and 9.99% reduction respectively), particularly around RES 282–295. The R251G mutation introduce only minimal changes to APOE4 flexibility in the oligomerization-only region (with a mean flexibility increase of 0.022 nm accounting for only 5.8% increase). Chou et al. showed that the C-terminal domain (CTD) (192–271) promotes APOE self-association without marked isoform preference.58 Though they reported higher aggregation in APOE4, our flexibility plot do not provide sufficient information on distinct aggregation tendencies.
The R251G mutation of the APOE4 protein reduces the differences from APOE2 and APOE3 in the overlapping region. The APOE4 presents a 0.5-nm peak around RES 248–260, while APOE2 and APOE3 do not present such a peak in their RMSF curves (this APOE4 peak account for mean 72.81% and 75.94% increase over APOE2 and APOE3 respectively). This difference in RMSF curves illustrates that the APOE4 protein is more flexible than the APOE2 and APOE3 proteins for RES 248–260. The APOE3–V236E and APOE4–R251G RSMF curves do not present a peak similar to the APOE4 one. Instead, their RMSF curves show patterns similar to those of APOE2 and APOE3 (with rp (rs) of 0.958 (0.956) for APOE3_V236E vs. APOE2, 0.969 (0.918) for APOE3_V236E vs. APOE3 and 0.797 (0.736) for APOE4_R251G vs. APOE2 0.888 (0.824) for APOE4_R251G vs. APOE3. APOE4 peak flexibility for RES 248–260 falls within the canonical lipid-binding segment (244–272) defined by Huang and co-workers.57 Helices in this region mediate lipid binding across apolipoproteins.57 Bu had earlier postulated that a structural change in APOE4 by R251G mutation reduces its harmful effect or enhances physiological functions.11 Insights from our study support this position of structural change in APOE4 by R251G and its consequent impact on the binding tendencies, which is more physiological in function. Secondly, the RMSF curves of the APOE3–R136S and APOE3–V236E exhibit distinct peaks of 0.53 nm and 0.7 nm around RES 265. This variation in the flexibility within the overlapping region could imply a less rigid structure. Whereas the APOE2 protein and R251G mutation maintained a similar flexibility within this segment of the overlapping region. This observation is supported with rp (rs) of 0.784 (0.762) and Δμ of 0.028 for APOE4_R251G vs. APOE2.
Several reports have identified RES 261–272 as critical lipid-binding sites.2,12,59 The overlapping-region also participates in APOE oligomerization.60 IDR or high flexibility within this region could favor self-aggregation. Fig. 2 shows that from RES 240–260, APOE4 maintain a higher flexibility than APOE2/3. Higher APOE4 flexibility supports increased aggregation tendencies. On the reverse, APOE2/3 present a gradual flexibility increases from RES 261–272. RES 261–272 represents a segment critical to lipid-binding.2,12,59 Also, higher flexibility could encourage exposure of hydrophobic surfaces, thus improved solubilization.23,57,61,62 The observed trend (high flexibility at RES 262–272) could explain why V236E mutation of APOE3 reduces self-aggregation as reported by Bu and colleagues.17
APOE3–R136S maintained a similar flexibility with APOE4 from RES 238–260; however, it presents higher flexibility than APOE2/3/V236E/R251G within the critical lipid-binding segment. Only comparable to V236E flexibility. This observation still supports enhanced lipid-binding despite earlier similar flexibility trend to APOE4.
Flexibility differences between APOE2, APOE3 and APOE4 concentrate in the lipidation and overlapping regions. Specific mutations, particularly R251G, can restore APOE4 flexibility toward a protective APOE2/3-like state. The lipidation and overlapping regions play a central role in APOE isoform functional differences.
3.2 Secondary structure
The R136S, V236E and R251G mutations change the secondary structure of residues on the lipidation-only region of APOE3 and APOE4. Fig. 3 presents the secondary structure distribution for APOE2, APOE3, APOE4, APOE3–R136S, APOE3–V236E and APOE4–R251G. Two segments of this region are unique: RES 195–202 and RES 232–240. In the region around RES 195–202, APOE4 contains an extended random-coil segment compared to APOE2 and APOE3 [see Fig. 3(a)–(c)]. Fig. 3(f) shows that the R251G mutation reduce the random-coil content in APOE4. This shift toward a more structured form in the lipidation-only region is also illustrated in Fig. 3(d) and (e) for the APOE3 mutations. This makes these mutations more similar to APOE2 and APOE3 around this RES 195–202. RES 195–202 sits inside the hinge region (165–215). The hinge region couples the N-terminal four-helix bundle to the C-terminal lipidation domain.11,13,20 The salt bridge between Arg-61 and Glu-255 drives APOE4 preference toward VLDL.63 Frieden et al. proposed that multiple interactions occur for other APOE isoforms. Different salt bridges influence other isoforms, specifically Lys95 and Glu255 for APOE3.20 Ordered/helical structure in the hinge dampens domain interaction unique to APOE4. Ordered structure reduces N–C proximity and flexing that promotes the salt bridge contact. From Fig. 3, R136S, V236E and R251G mutations reduce random coil in RES 195–202. The coil reduction would weaken APOE4-specific domain interaction feature and trend toward E2/E3-like behavior. | ||
| Fig. 3 Residue-wise secondary-structure fractions for lipidation-only region for the six proteins (a) APOE2, (b) APOE3, (c) APOE4, (d) APOE3–R136S, (e) APOE3–V236E (f) APOE4–R251G. | ||
Secondly, RES 232–240 exhibit additional differences. APOE4 maintains a random-coil conformation in this region, whereas R136E, V236E, and R251G mutants adopt helical structures. This pattern is similar to the helices observed in APOE2. The change from a random coil to helices indicates that these mutations partially restore structural features found in the other common isoforms. This coil to helix structural shift observed with the mutations could improve helical integrity and weaken self-aggregation.64 The mutations alter the secondary structure of APOE in the lipidation-only region.
The R136S, V236E, and R251G mutations cause changes in the secondary structure of RES 259–265 in the overlapping region of APOE3 and APOE4. Fig. 4 shows the secondary structure of the overlap region for APOE2, APOE3, APOE4, and the mutants APOE3–R136S, APOE3–V236E, and APOE4–R251G. In wild-type APOE4, residues in this segment predominantly adopt an α-helical structure, similar to APOE2 and APOE3. However, the mutations introduce structural changes in both APOE3 and APOE4 (see Fig. 4(d)–(f)). R136S mutation makes the helices adopt a coil around RES 252. RES 252 lies next to Glu255. Glu255 participates in N/C-terminal domain interaction that alters lipoprotein preference in APOE4.30,65 Structural changes near RES 252 may disrupt long-range domain coupling. Disrupted coupling could shift APOE toward lipidated states. In APOE3–V236E, residues around 264 and 270 transition from helices to turns and bends. Additionally, APOE4–R251G shifts residue 264 from a helix to a random coil. R251G mutation disrupts the stable helical conformation observed in APOE4. It increases local flexible structures in the overlapping region, shifting helices to coils. RES 261–272 has been reported to be critical to APOE lipid-binding sites.2,12,59 The structural changes from helices to coils presented by these mutation within these sites support higher flexibility in Fig. 2. The consequent effect of this higher flexibility is proposed to improve lipid-binding tendencies within this segment.12,20
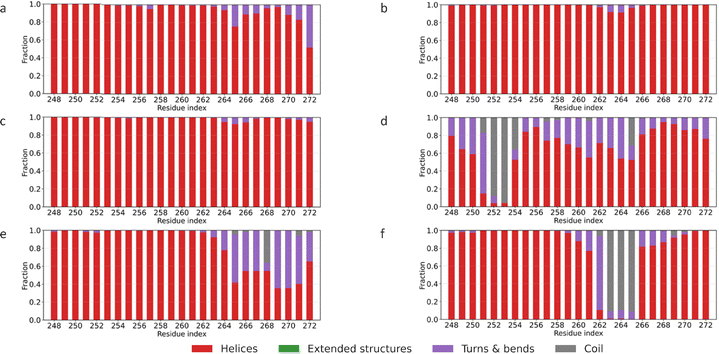 | ||
| Fig. 4 Residue-wise secondary-structure fractions for overlapping region for the six proteins (a) APOE2, (b) APOE03, (c) APOE4, (d) APOE3–R136S, (e) APOE3–V236E (f) APOE4–R251G. | ||
The R136S, V236E and R251G mutations introduce transitions in structures of the oligomerization-only region in APOE3 and APOE4. Fig. 5 presents the secondary structure of the oligomerization-only region for APOE2, APOE3, and APOE4, and APOE3–R136S, APOE3–V236E, and APOE4–R251G mutants. This segment is central to oligomerization. Truncating 273–299 yields monomeric APOE4.66,67 APOE4 shows a wider random-coil region around RES 288–299 compared to APOE2 and APOE3. This implies more flexibility compared to APOE2 and APOE3.12,26,58 As seen in Fig. 5(d) and (e), the APOE3 mutations (R136S and V236E) introduced more coils, turns and bends to APOE3 (RES 273–280). This suggests that the mutations alter the ordered structures of APOE3 to be more like APOE2 in the oligomerization-only region. In APOE4–R251G [Fig. 5(f)], RES 293–299 shift from a predominantly random-coil conformation to a mix of helices and extended structures. This structural change is particularly noticeable as it stabilizes into more ordered forms. These changes suggest the R251G mutation enhances more ordered structures in the oligomerization-only region of the APOE4 protein. It shifts random-coil to helices and extended forms.
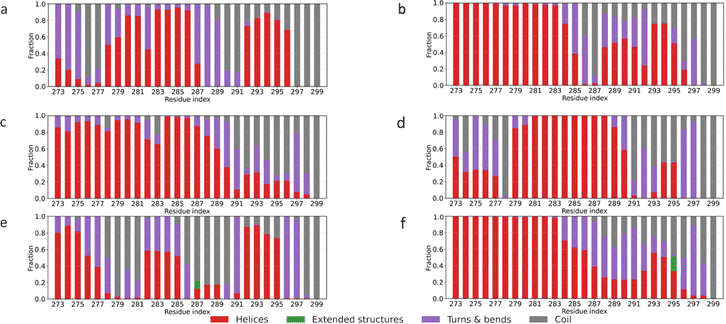 | ||
| Fig. 5 Residue-wise secondary-structure fractions for oligomerization-only region for the six proteins (a) APOE2, (b) APOE3, (c) APOE4, (d) APOE3–R136S, (e) APOE3–V236E (f) APOE4–R251G. | ||
3.3 Solvent accessible surface area
We utilize solvent-accessible surface area to analyze the exposure area of the lipidation and oligomerization of APOE proteins. Fig. 6 shows the cumulative probability of SASA of the lipidation region for the APOE2, APOE3, APOE4, APOE3–R136S, APOE3–V236E, and APOE4–R251G. The lipidation region includes both lipidation-only and overlapping regions. We first analyze whether the three mutations would alter the whole area of the lipidation region, then the hydrophobic and hydrophilic segments of the region. | ||
| Fig. 6 Probability and cumulative distributions of SASA in the lipidation region. (a) Total residues (b) hydrophobic residues (c) hydrophilic residues. | ||
The two protective mutations on APOE3 would not alter the total area of the lipidation domain on APOE3. As shown in Fig. 6(a), APOE2, APOE3, and APOE4 present distinct ranges for the lipidation region SASA: the APOE4 ranges from 52 to 67 nm2, a similar SASA distribution range to APOE2 (52–65 nm2) and APOE3 (55–62 nm2). The values of KS (0.21) and Cohen's d (0.05) indicate that the distributions of SASA of APOE3 and APOE4 are similar with a negligible effect size. APOE2 compared to APOE4 have a KS value of 0.27 and a Cohen's d of 0.43. The KS value shows the distributions are similar. The Cohen's d value indicates a moderate difference between the groups. This implies the distributions have significant overlap and variability within each group. In a surface plasmon resonance (SPR) experiment, Lund-Katz and co-workers demonstrated that APOE4 would need greater interfacial area for lipid binding that APOE3.68 Our study shows that APOE4 presents a similar SASA distribution to APOE3. This could mean, increasing the SASA of APOE4 may improve its lipidation. Furthermore, in Fig. 6(a), the lipidation domain on both APOE3–R136S and APOE3–V236E presents SASA in the range of 53–63 nm2, similar to that of APOE3. The values of KS (0.17 and 0.2) indicate that the lipidation domain on the three proteins also possesses similar distributions. The APOE4–R251G mutation reduce the SASA of APOE4 to the range of 52–62 nm2. The reduction by R251G is moderate as demonstrated by the KS (0.20) and Cohen's d (0.57) values (see S1 of the SI for a complete matrix of these statistical values). The density curves show the leftward shift for APOE4–R251G relative to APOE4, with no widening. This distribution agrees with hydrogen-exchange measurements that report lower helical stability and higher solvent accessibility in lipid-free APOE4 relative to APOE3.13 Furthermore, Stuchell-Brereton et al. reported that APOE4 presents a more disordered structure than initially reported.26 This implies larger, variable exposed surface than a single compact fold would.
The mutation effect on SASA emerges as we divide the functional regions (lipidation and oligomerization) into hydrophobic and hydrophilic segments. The hydrophobic segment is composed of ALA, GLY, ILE, LEU, MET, PHE, PRO, TRP, VAL, and TYR, and the hydrophilic segment contains HIS, TYR, TRP, LYS, ASP, GLU, ARG, CYS, GLN, SER, and THR. Soranno and his team identified isoform dependent solvent accessibility using hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments.26 The APOE3–R136S and APOE3–V236E extends the hydrophobic SASA range of APOE3. Fig. 6(b) presents the exposure of hydrophobic residues in the lipidation region. In the figure, APOE4 presents a SASA range of 17–26 nm2, falling between APOE3 (15.5–25 nm2) and APOE2 (17–30 nm2). The KS test values (0.30 and 0.58) for APOE4 compared to APOE3 and APOE2, respectively, show moderate to greater differences. The Cohen's d values of 0.57 and 1.05 suggest medium to large effect sizes for APOE4 vs. APOE3 and APOE4 vs. APOE2. The APOE3–R136S (15–33 nm2) and APOE3–V236E (17–28 nm2) mutations extend the hydrophobic SASA range of APOE3. For APOE3–V236E, the KS value of 0.48 and Cohen's d of 0.34 indicate moderate difference with medium effect on APOE3. APOE3–R136S, with KS of 0.50 and Cohen's d of 0.73, shows significant difference and a medium-large effect size on APOE3 SASA distribution. These observations imply that APOE3–R136S mutation increased the hydrophobic SASA of APOE3 more compared to V236E mutation.
APOE4–R251G shifts the SASA range of APOE4 minimally, from 17–26 nm2 to 17–29 nm2. As seen in Fig. 6(b), this minimal shift is statistically supported by the KS values (0.21) and small effect size (Cohen's d = 0.28). These results suggest that R251G has minimal impact on the hydrophobic segment of APOE4. The analysis shows that, while APOE3–R136S and APOE3–V236E mutations lead to more hydrophobic exposure of APOE3, APOE–R251G does not for APOE4.
APOE4–R251G mutation reduces the scope of hydrophilic exposure in APOE4. Fig. 6(c) presents the surface exposure of hydrophilic residues in the lipidation region. APOE4 presents a hydrophilic SASA distribution 30–42 nm2, APOE2 32–38 nm2, and APOE3 33–40 nm2. The KS values of APOE4 against APOE2 (0.22) and APOE3 (0.29) indicate only small differences between their distributions. Cohen's d value of 0.15 and 0.57 supports APOE4 presents a small effect size with APOE2 and a moderate effect size with APOE3 on their SASA distributions. The APOE3–R136S and APOE3–V236E mutations do not significantly alter the hydrophilic SASA distribution (KS = 0.17 and 0.25) of APOE3. APOE3–V236E has a greater effect on APOE3 SASA distribution than APOE3–R136S. This is shown by their respective Cohen's d values of 0.57 and 0.19. Meanwhile, the APOE4–R251G mutation narrows APOE4's hydrophilic SASA to 30–35 nm2. The KS value (0.53) and Cohen's d of 1.11 indicate moderate difference and significant large effect size. These indicate that mutations alter common occurring APOE hydrophilic exposure to different degrees.
The evidence suggest that APOE3 mutations moderately increase hydrophobic SASA in the lipidation region. They do not significantly change total or hydrophilic SASA. APOE4–R251G reduces total and hydrophilic SASA. The complete statistic for these KS cumulative distribution plots and the timeseries plot of SASA for lipidation region of the six APOE proteins can be found in S1 and S5 of the SI.
Fig. 7 shows the cumulative distributions for the SASA of the oligomerization region in APOE2, APOE3, APOE4, APOE3–R136S, APOE3–V236E, and APOE4–R251G. The total cumulative SASA distribution is presented in Fig. 7(a), the hydrophobic in Fig. 7(b), and hydrophilic in Fig. 7(c).
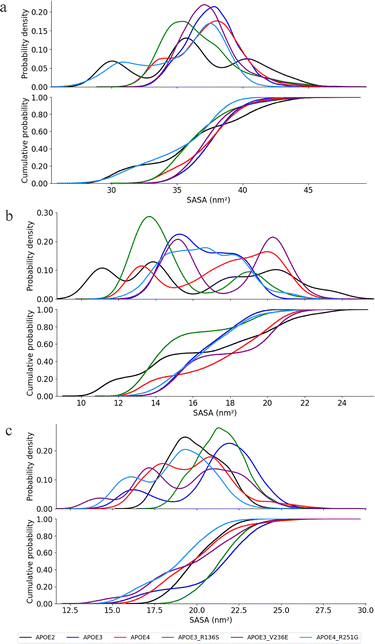 | ||
| Fig. 7 Probability and cumulative distributions of SASA in the oligomerization region. (a) Total residues (b) hydrophobic residues (c) hydrophilic residues. | ||
The two mutations of APOE3 introduce distinct changes in exposure area of APOE3 oligomerization region. As presented in Fig. 7(a), the SASA distributions for APOE4 and APOE3 largely overlap within the 32–42 nm2 range. The KS test shows a value of 0.10. This indicates a minimal statistical difference. Cohen's d (0.13) confirms the effect size is negligible. APOE3–R136S mutation introduces moderate changes to the SASA of APOE3. The KS test shows a value of 0.29 and Cohen's d is 0.46. This indicates a small-to-medium effect for APOE3–R136S on APOE3. APOE3–V236E does not change the SASA of APOE3. The KS test shows a value of 0.10 with Cohen's d of 0.10. This supports the null hypothesis that the distributions are identical.
The APOE4–R251G mutation reduce the exposure area of the APOE4 oligomerization region. APOE4–R251G shifts the APOE4 SASA exposure range for the oligomerization region to 27–40 nm2. The KS test (0.22) and Cohen's d of 0.71 suggest some similarity with a medium-to-large effect size. This effect size confirms the observed shift. The APOE4–R251G exposure range of 27–40 nm2 partially overlaps with the APOE2 SASA distribution of 27–44 nm2. The KS test shows a value of 0.22 and Cohen's d is 0.26. These statistics support the distributions overlaps and show statistical similarities. Generally, APOE4–R251G notably alters the APOE4 exposure with an approximate 5–16% reduction in total SASA distribution.
The APOE mutations demonstrate distinct SASA distribution effects on the hydrophobic segment of APOE's oligomerization region. Fig. 7(b) shows the SASA distributions for APOE2 (9–23 nm2), APOE3 (12–18 nm2), APOE4 (13–22 nm2), and the three mutations. Stuchell-Brereton et al. proposed that CTD of APOE contain α-helices that determines its aggregation in monomer-tetramer equilibrium, lipidation, and interaction with amyloid-β.26 APOE4 shows a different hydrophobic SASA distribution compared to APOE2 and APOE3.26 APOE3–R136S shows a range of 12–21 nm2. APOE3–V236E shows a range of 13–20 nm2. Both mutations maintain the hydrophobic SASA scope of APOE3 but present distinct distributions. The KS test shows values of 0.45 for R136S and 0.39 for V236E. Cohen's d is 0.63 for R136S and 0.57 for V236E. These values confirm medium distributional differences between the mutated variants and APOE3.
APOE4–R251G shows a hydrophobic SASA range similar to APOE4. The KS test shows a value of 0.28. Cohen's d is 0.41. These statistics confirm that APOE4–R251G has a distinct distribution with a marginal difference and medium effect size. These observations show that the mutations increase the hydrophobic exposure of APOE surface in the oligomerization region. The degree of increase differs between the mutations.
The APOE3–R136S, APOE3–V236E, and APOE4–R251G mutations do not alter the hydrophilic segment exposure of APOE protein. Fig. 7(c) presents the hydrophilic SASA distributions of the APOE isoforms APOE2, APOE3, and APOE4, and the mutants APOE3–R136S, APOE3–V236E and APOE4–R251G within the oligomerization region. APOE4 presents a similar SASA scope of 16–27 nm2. This range is similar to APOE3 but wider than APOE2, which shows 17.5–23 nm2. The APOE3–R136S and APOE3–V236E mutations maintain a similar hydrophilic SASA range to APOE3. The KS test shows values of 0.16 for APOE3–R136S and 0.28 for APOE3–V236E. These values support the null hypothesis of similar distributions. Cohen's d is 0.14 for APOE3–R136S and 0.56 for APOE3–V236E. These values indicate a negligible effect size for APOE3–R136S and a moderate effect for APOE3–V236E. These findings show that APOE3–R136S and APOE3–V236E do not significantly alter the distribution of the hydrophilic segment SASA in the oligomerization region. APOE4–R251G mutation introduces a small leftward shift in APOE4's SASA distribution range. APOE4–R251G maintains a distribution similar to APOE4. The KS test shows a value of 0.18. This confirms no statistically significant difference in hydrophilic SASA. Cohen's d test of 0.48 indicates a medium effect size on the distribution. The test suggests that the two distributions are somewhat separated, still there exists significant overlap and variability within each group. These results confirm that the mutation does not significantly alter the hydrophilic exposure of APOE4 in the oligomerization region. The complete statistic for these KS cumulative distribution plots and the timeseries plot of SASA for oligomerization region of the six APOE proteins can be found in S2 and S6 of the SI.
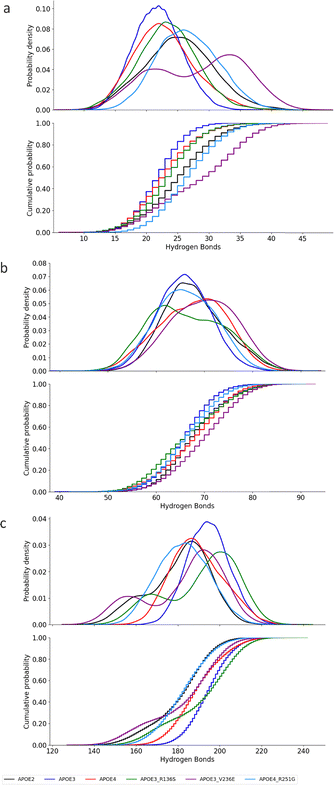
3.4 Hydrogen bond
Experimental and modeling works on lipid-free APOE proteins report hydrogen bonds and salt bridges between N- and C-terminal domains.3,12,13,63 Further study could help explain shielding of the receptor-binding helix in the lipid-free state.We analyze the solvation of the lipidation region of APOE protein based on the hydrogen bonds. First, we analyze hydrogen bonding of lipidation region residues and other residues. Then, between residues within the region, and between lipidation residues and water molecules. Previous reports indicate that the C-terminal domain contains three α-helices which interact with those in the N-terminal helix bundle domain through hydrogen bonds and salt-bridges.13,20,23,59,60,69,70 These interactions need be broken before helix opening occurs. Helix opening exposes hydrophobic surfaces necessary for binding to lipids.13,20,60–62 We define hydrogen bonds using geometric criteria from Chandler et al.71 The definition requires two specific parameters. The first parameter measures the distance between donor and acceptor heavy atoms. This distance must be less than or equal to 0.35 nm. The second parameter measures the donor–hydrogen–acceptor angle. This angle must be less than or equal to 30° from linearity. The absolute angle value must be greater than or equal to 150°. Both criteria must be satisfied simultaneously. We assume hydrogen bond exists only when both conditions are met.71
The APOE3–V236E mutation increases the number of APOE3's hydrogen bonds between the lipidation region and other residues by ∼42%. Fig. 8(a) presents the number of hydrogen bonds between the lipidation region and the rest of each protein. APOE4 hydrogen bonding ranges from 15 to 35. APOE2 and APOE3 show bonding range of 15–38 and 15–28, respectively. For the HB between APOE4 and APOE2, KS of 0.3 suggest marginal difference between their distribution and the Cohen's d of 0.72 reveals a moderate-to-large effect size. A smaller but still moderate effect is observed between APOE4 and APOE3, with Cohen's d of 0.33. The Cohen's value suggest moderate standardized deviations in the HB means of the two proteins.
APOE3–R136S mutation do not introduce significant changes to the HB of APOE3. In Fig. 8(a), APOE3–R136S presents HB range 15–28 similar to APOE3. The KS statistic of 0.0075 and Cohen's d of 0.12 indicate a negligible effect, and a minimal deviation from APOE3. The APOE3–V236E mutation increases HB within the lipidation region from approximately 33 in APOE3 to 44. The KS test (0.22) indicates this increase is not statistically significant. This is supported by the Cohen's d test (0.35) which shows only a moderate deviation and overlap in the standardized mean HB distribution of APOE3–V236E from APOE3. In addition, the APOE4–R251G mutation shifts the HB range from 15–35 in APOE4 to 15–38. The R251G mutation induces a moderate-to-large effect in HB of APOE4 within the lipidation region. The Cohen's d value is 0.73. The KS statistic of 0.3 indicates a marginal difference.
APOE3–R136S and APOE3–V236E mutations would not cause significant change in APOE3's hydrogen bonding within the lipidation region. Fig. 8(b) presents the hydrogen bonding number within the lipidation region for the APOE isoforms and the mutations. APOE4 presents range of 52–82 hydrogen bonding number within lipidation region. Hydrogen bond number range distribution presented by APOE2 (53–80) is similar to but wider than APOE3. The statistic between APOE4 vs. APOE2 (KS = 0.15, and Cohen's d is 0.35) and APOE4 vs. APOE3 (KS = 0.054, and Cohen's d is 0.16), indicates similarity between the distributions with a small effect for APOE2 and negligible effect for APOE3. The APOE3–R136S and APOE3–V236E mutations cause minimal shifts in APOE3's HB range to 55–82. As presented in Fig. 8(b), APOE3–R136S and APOE3–V236E do not alter the distributions of HB number (KS 0.094 and 0.29 with Cohen's d of 0.2 and 0.77). While APOE3–R136S only presents small effect in APOE3, APOE3–V236E shows a medium to large effect size on the number of hydrogen bonding within the lipidation region.
The APOE4–R251G alters the hydrogen bonding within the lipidation region of APOE4. Furthermore, in Fig. 8(b), APOE4–R251G presents the KS statistic 0.4. This indicates a moderate change in HB distribution of APOE4. The Cohen's d value of 1.1 suggests this mutation-induced change has large effect size on hydrogen bond numbers in the lipidation region of APOE4. To sum up, we see that APOE isoforms exhibit distinct hydrogen bonding patterns in the lipidation region.
APOE3 mutations do not introduce significant change in the hydrogen bonding between lipidation residues and water molecules in the lipidation region. Fig. 8(c) presents hydrogen bonding number of lipidation water molecules. From the figure, APOE4 (160–210) shows hydrogen bonding within the range of APOE2 (140–210) and APOE3 (170–220). The two protective mutations of APOE3 only slightly widens the scope of hydrogen bonding in APOE3 to 145–220. The KS statistics (0.11 and 0.25) supports accepting the null hypothesis that these distributions are from similar populations. The standardized mean deviations are however distinct as seen in the Cohen's d (0.1 and 0.65) statistic. These suggest negligible effect by APOE3–R136S and a small to medium effect by APOE3–V236E.
APOE4–R251G changes APOE4 lipidation water molecules hydrogen bonding to resemble APOE2. The APOE4–R251G mutation reduces the number of hydrogen bonds between the lipidation region and water molecules to 155–195. The observed marginal reduction is supported by the KS statistic of 0.31 and a large Cohen's d effect size of 0.85. APOE4–R251G shows similarity in bonding with lipidation water molecules to APOE2. The KS test shows a value of 0.19 and Cohen's d is 0.47. This indicates a medium effect size on their bonding distributions. These results confirm that the APOE4–R251G causes notable changes in HB between APOE4 and lipidation water molecules.
Overall, APOE3–V236E increases intra-lipidation hydrogen bonding with a moderate effect size. APOE4–R251G induces large shifts in APOE4 lipidation region bonding. It also alters bonding with water molecules. The effect sizes are moderate to large, and the distribution changes are marginal. The complete statistic for these KS cumulative distribution and the timeseries plot of number of hydrogen bonds for the lipidation region can be found in S3 and S7 of the SI.
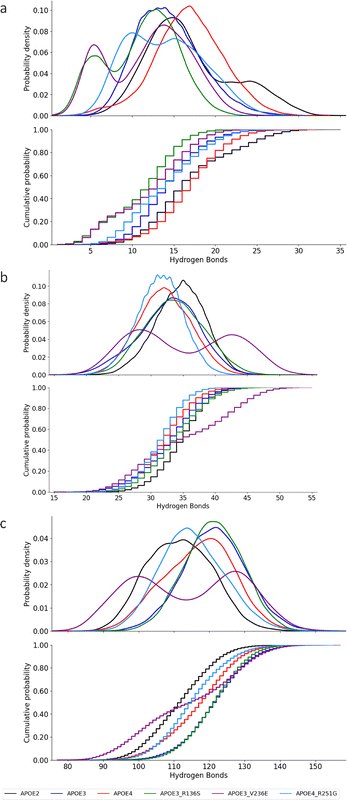
Fig. 9 shows the number of hydrogen bonds of the oligomerization region for the APOE2, APOE3 and APOE4, and APOE3–R136S, APOE3–V236E, and APOE4–R251G. Fig. 9(a) presents hydrogen bonding between the oligomerization region and other resides. Fig. 9(b) shows the hydrogen bonds within the oligomerization region. Fig. 9(c) shows the hydrogen bonds between oligomerization region and oligomers-water molecules.
APOE4–R251G mutation do not change the scope of the hydrogen bonds between the oligomerization region and other residues in APOE4. In Fig. 9(a), APOE4's hydrogen bond range (10–25) falls between that of APOE2 (10–30) and APOE3 (8–23). Statistical comparisons show no significant differences, with negligible effect sizes for APOE2 vs. APOE4 (0.059, Cohen's d 0.023) and APOE3 vs. APOE4 (0.10, Cohen's d 0.17). The APOE3–R136S and APOE3–V236E mutations shift APOE3's hydrogen bond range with other residues from 7–23 to 5–18. APOE3–R136S shows a marginal difference compared to APOE3 (KS 0.4) with a large effect size (Cohen's d 0.97). APOE3–V236E shows some statistical similarity (KS 0.27) with a medium to large effect size (Cohen's d 0.71). The APOE4–R251G mutation does not cause significant changes to APOE4 oligomerization region hydrogen bonding with other residues. The KS statistic of 0.053 and Cohen's d of 0.064 confirm that APOE4–R251G presents no significant shift and a negligible effect size.
APOE3 mutations do not cause significant shift in the hydrogen bonding distribution of APOE3. The distribution of hydrogen bonding within oligomerization region is presented in Fig. 9(b). APOE4 presents a hydrogen bonding range (25–42) similar to APOE2 and APOE3. The KS statistic for APOE2 vs. APOE4 (KS of 0.16, and Cohen's d of 0.34) indicates a no significant difference with a small effect size. In contrast, APOE3 vs. APOE4 show good statistical similarity with a KS of 0.073, and a negligible effect size (Cohen's d 0.094). The APOE3–R136S (25–44) and APOE3–V236E (25–38) mutations do not cause a significant shift in the hydrogen bonding distribution of APOE3. The statistics of APOE3–R136S vs. APOE3 (KS of 0.11 and 0.12, and Cohen's d of 0.22 and 0.028) indicate a no significant difference and a small to negligible effect size.
APOE4–R251G induce moderate reduction of hydrogen bonding within the oligomerization region of APOE4. As presented in Fig. 9(c), APOE4–R251G mutation (25–38) introduces a shift in APOE4's hydrogen bonding distribution. The statistics show KS value of 0.35 and Cohen's d of 0.77. These statistics indicate a marginal shift with a medium to large effect size on APOE4–R251G hydrogen bonding distribution. These statistics confirm that APOE4–R251G alters APOE4's hydrogen bonding within the oligomerization region.
APOE mutations do not alter the hydrogen bonding of oligomers water molecules. In Fig. 9(c), APOE4 presents a HB range 100–135, APOE2 95–130, and APOE3 105–140. The KS statistic for APOE4 vs. APOE2 is 0.079. For APOE4 vs. APOE3, it is 0.13. These values support the similarity between these pairs. They confirm no significant difference in APOE4's hydrogen bonding compared to APOE2. The APOE3–R136S mutation (105–140) shows a KS statistic of 0.032. This supports the null hypothesis. There is no significant difference from APOE3's distribution. The effect size is negligible (Cohen's d 0.038). Similarly, the V236E mutation (105–140) shows no significant change in APOE3 hydrogen bonding with oligomer-water molecules (KS 0.24). However, the effect size is medium. In addition, the APOE4–R251G mutation (100–135) induces a moderate leftward shift in APOE4's hydrogen bonding. However, the distribution remains similar. The KS statistic is 0.24, supporting this similarity. The effect size is medium (Cohen's d 0.56). The complete statistic for these KS cumulative distribution and the timeseries plot of number of hydrogen bonds for the oligomerization region can be found in S4 and S8 of the SI.
Collectively, APOE3 mutations show distinct impacts on APOE3's hydrogen bonding within the oligomerization region. APOE4–R251G marginally alters the hydrogen bonding in APOE4's oligomerization region but does not significantly affect oligomer-water molecule interactions.
4. Conclusion
This study investigates the mutation-induced variation in the structural and dynamic properties of APOE3–R136S, APOE3–V236E, and APOE4–R251G using MD simulations. We examine their flexibility, secondary structure, surface exposure, and hydrogen bonding within lipidation and oligomerization functional regions. The flexibility profiles show region-specific changes. The mutations reduce the difference in flexibility of the lipidation region relative to APOE2 and APOE3. The R251G mutation reduces APOE4-specific flexibility in the overlapping region. These shifts limit disorder that promotes hydrophobic exposure and aggregation. All three mutations do not alter flexibility in the oligomerization-only region (RES 273–299). The mutations change the secondary structures of all three common isoforms. They introduce transitions in the structures of the lipidation-only and oligomerization-only regions. These changes stabilize local structures and reduce aggregation-prone conformations. They disrupt helices in the overlapping region and introduce more flexible structures. SASA analysis shows distinct effects across regions. APOE4–R251G reduces the scope of SASA in both the lipidation and oligomerization regions of APOE4. APOE3–R136S and APOE3–V236E increase hydrophobic SASA in the lipidation region but do not change the total SASA of APOE3. In the hydrophilic segment of the lipidation region, APOE3–R136S does not significantly alter exposure. APOE3–V236E and APOE4–R251G reduce hydrophilic exposure with moderate to large effect sizes. Hydrogen-bond analysis reveals further stabilization. APOE3–R136S and APOE3–V236E increase hydrogen bonding in the lipidation region. They do not alter hydrogen bonding in the oligomerization region or with water molecules. These changes strengthen APOE3 and reduce its aggregation tendency. APOE4–R251G introduces distinct changes in hydrogen bonding in both the lipidation and oligomerization regions. It reduces oligomerization region bonding and modifies lipidation region bonding and water interactions. These findings indicate that the mutations cause APOE4 to resemble APOE2 and APOE3 in terms of lipidation and overlapping regions. These effects weaken APOE4-specific self-association in R251G. This study provides insights into the structure and dynamics of protective APOE mutations. These findings show that APOE3–R136S, APOE3–V236E, and APOE4–R251G reduce aggregation propensity. These findings may support future studies for developing therapeutic strategies for AD.Author contributions
N. A. Ihoeghian performed the molecular dynamics simulations, conducted data analysis, and wrote the manuscript. U. Abass developed the initial simulation protocols. I. Imam created the analysis scripts and computational tools. Q. Shao conceptualized the study, supervised the research, and provided critical manuscript revisions.Conflicts of interest
There are no conflicts to declare.Data availability
Data will be made available upon request.The simulation data will be made availibale upon request. Statistical result data has been made available in the supplementary information (SI). Supplementary information: Fig. S1.1: statistics for total SASA of lipidation region – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S1.2: statistics for hydrophobic SASA of lipidation region – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S1.3: statistics for hydrophilic SASA of lipidation region – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S2.1: statistics for total SASA of oligomerization region – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S2.2: statistics for hydrophobic SASA of oligomerization region – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S2.3: statistics for hydrophilic SASA of oligomerization region – (i) KS test matrix (ii) Cohen’s d matrix. Fig. S3.1: statistics for hydrogen bonding of lipidation with other residues – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S3.2: statistics for hydrogen bonding within lipidation region – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S3.3: statistics for hydrogen bonding of lipidation region with water molecules – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S4.1: statistics for hydrogen bonding of oligomers with other residues – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S4.2: statistics for hydrogen bonding of residues within oligomerization region – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S4.3: statistics for hydrogen bonding between oligomerization region with water molecules – (i) KS test matrix and (ii) Cohen’s d matrix. Fig. S5: time-series plot of SASA for lipidation region of the six APOE proteins – (a) total residues, (b) hydrophobic residues and (c) hydrophilic residues. Fig. S6: time-series plot of SASA for oligomerization region of the six APOE proteins – (a) total residues, (b) hydrophobic residues and (c) hydrophilic residues. Fig. S7: time-series plot of number of hydrogen bonds for the lipidation region for (a) lipidation with other residues, (b) within lipidation region, and (c) lipidation region with water molecules. Fig. S8: time-series plot of number of hydrogen bonds for the oligomerization region for (a) oligomers with other residues, (b) within oligomerization region, and (c) oligomers with water molecules. Table S1: correlation analysis and flexibility metrics for APOE isoform and variant comparisons by structural region. See DOI: https://doi.org/10.1039/d5cp03829d.
Acknowledgements
This study was supported by the Alzheimer's Association AARG-23-1144638. We would thank the University of Kentucky Center for Computational Sciences and Information Technology Services Research Computing for their support and use of the Lipscomb Compute Cluster (LCC) and associated research computing resources. We acknowledge Dr Dave Fardo for providing suggestions on the selection of statistical analysis methods.References
- Y. Yamazaki, N. Zhao, T. R. Caulfield, C. C. Liu and G. Bu, Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies, Nature Publishing Group, 2019, preprint DOI:10.1038/s41582-019-0228-7.
- J. Chen, Q. Li, J. Wang and C. Frieden, Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions, Proc. Natl. Acad. Sci. U. S. A., 2011, 18, 14813–14818 CrossRef PubMed.
- C. Frieden and K. Garai, Concerning the structure of apoE, Protein Sci., 2013, 22, 1820–1825 CrossRef CAS PubMed.
- J. W. Blanchard, L. A. Akay, J. Davila-Velderrain, D. von Maydell, H. Mathys, S. M. Davidson, A. Effenberger, C. Y. Chen, K. Maner-Smith, I. Hajjar, E. A. Ortlund, M. Bula, E. Agbas, A. Ng, X. Jiang, M. Kahn, C. Blanco-Duque, N. Lavoie, L. Liu, R. Reyes, Y. T. Lin, T. Ko, L. R’Bibo, W. T. Ralvenius, D. A. Bennett, H. P. Cam, M. Kellis and L. H. Tsai, APOE4 impairs myelination via cholesterol dysregulation in oligodendrocytes, Nature, 2022, 611, 769–779 CrossRef CAS PubMed.
- Y. A. Martens, N. Zhao, C. C. Liu, T. Kanekiyo, A. J. Yang, A. M. Goate, D. M. Holtzman and G. Bu, ApoE Cascade Hypothesis in the pathogenesis of Alzheimer's disease and related dementias, Cell Press, 2022, preprint DOI:10.1016/j.neuron.2022.03.004.
- Y. Le Guen, M. E. Belloy, B. Grenier-Boley, I. De Rojas, A. Castillo-Morales, I. Jansen, A. Nicolas, C. Bellenguez, C. Dalmasso, F. Küçükali, S. J. Eger, K. L. Rasmussen, J. Q. Thomassen, J. F. Deleuze, Z. He, V. Napolioni, P. Amouyel, F. Jessen, P. G. Kehoe, C. Van Duijn, M. Tsolaki, P. Sánchez-Juan, K. Sleegers, M. Ingelsson, G. Rossi, M. Hiltunen, R. Sims, W. M. Van Der Flier, A. Ramirez, O. A. Andreassen, R. Frikke-Schmidt, J. Williams, A. Ruiz, J. C. Lambert, M. D. Greicius, B. Arosio, L. Benussi, A. Boland, B. Borroni, P. Caffarra, D. Daian, A. Daniele, S. Debette, C. Dufouil, E. Düzel, D. Galimberti, V. Giedraitis, T. Grimmer, C. Graff, E. Grünblatt, O. Hanon, L. Hausner, S. Heilmann-Heimbach, H. Holstege, J. Hort, D. Jürgen, T. Kuulasmaa, A. Van Der Lugt, C. Masullo, P. Mecocci, S. Mehrabian, A. De Mendonça, S. Moebus, B. Nacmias, G. Nicolas, R. Olaso, G. Papenberg, L. Parnetti, F. Pasquier, O. Peters, Y. A. L. Pijnenburg, J. Popp, I. Rainero, I. Ramakers, S. Riedel-Heller, N. Scarmeas, P. Scheltens, N. Scherbaum, A. Schneider, D. Seripa, H. Soininen, V. Solfrizzi, G. Spalletta, A. Squassina, J. Van Swieten, T. J. Tegos, L. Tremolizzo, F. Verhey, M. Vyhnalek, J. Wiltfang, M. Boada, P. García-González, R. Puerta, L. M. Real, V. Álvarez, M. J. Bullido, J. Clarimon, J. M. García-Alberca, P. Mir, F. Moreno, P. Pastor, G. Piñol-Ripoll, L. Molina-Porcel, J. Pérez-Tur, E. Rodríguez-Rodríguez, J. L. Royo, R. Sánchez-Valle, M. Dichgans and D. Rujescu, Association of Rare APOE Missense Variants V236E and R251G with Risk of Alzheimer Disease, JAMA Neurol, 2022, 79, 652–663 CrossRef PubMed.
- G. D. Rabinovici and D. B. Dubal, Rare APOE Missense Variants - Can We Overcome APOE ε4 and Alzheimer Disease Risk?, American Medical Association, 2022, preprint DOI:10.1001/jamaneurol.2022.0854.
- Y. Le Guen, M. E. Belloy, B. Grenier-Boley, I. de Rojas, A. Castillo, I. E. Jansen, A. Nicolas, C. Bellenguez, C. Dalmasso, F. Küçükali, S. J. Eger, V. Álvarez-Martínez, B. Arosio, L. Benussi, A. Boland, B. Borroni, M. J. Bullido, P. Caffarra, J. Clarimón, D. Daian, A. Daniele, S. Debette, J. Deleuze, M. Dichgans, C. Dufouil, E. Duzel, D. Galimberti, J. M. García-Alberca, P. García-González, V. Giedraitis, T. Grimmer, C. Graff, E. Grunblatt, O. Hanon, L. Hausner, S. Heilmann-Heimbach, H. Holstege, J. Hort, D. Jurgen, T. Kuulasmaa, A. van der Lugt, C. Masullo, P. Mecocci, S. Mehrabian, A. de Mendonça, M. Boada, P. Mir, S. Moebus, F. Moreno, B. Nacmias, G. Nicolas, G. Papenberg, L. Parnetti, F. Pasquier, P. Pastor, O. Peters, Y. A. L. Pijnenburg, G. Piñol-Ripoll, J. Popp, L. Molina, R. Puerta, J. Pérez-Tur, I. Rainero, I. H. G. B. Ramakers, K. L. Rasmussen, L. M. Real, S. G. Riedel-Heller, E. R. Rodríguez, J. L. Royo, D. Rujescu, N. Scarmeas, P. Scheltens, N. Scherbaum, A. Schneider, D. Seripa, H. Soininen, V. Solfrizzi, G. Spalletta, A. Squassina, J. C. van Swieten, R. Sanchez-Valle, T. Tegos, J. Q. Thomassen, L. Tremolizzo, F. R. J. Verhey, M. Vyhnalek, J. Wiltfang, Z. He, V. Napolioni, P. Amouyel, F. Jessen, P. G. Kehoe, C. M. van Duijn, M. Tsolaki, P. Sanchez-Juan, K. Sleegers, M. Ingelsson, G. Rossi, M. Hiltunen, R. Sims, W. M. van der Flier, A. Ramirez, O. Andreassen, R. Frikke-Schmidt, J. Williams, A. Ruiz, J. Lambert and M. D. Greicius, Rare missense variant (R251G) on APOE counterbalances the Alzheimer’s disease risk associated with APOE-ε4, Alzheimer’s Dementia, 2022, e060114 CrossRef.
- G. Chen, M. Wang, Z. Zhang, D. K. Hong, E. H. Ahn, X. Liu, S. S. Kang and K. Ye, ApoE3 R136S binds to Tau and blocks its propagation, suppressing neurodegeneration in mice with Alzheimer’s disease, Neuron, 2025, 719–736 CrossRef CAS PubMed.
- M. R. Nelson, P. Liu, A. Agrawal, O. Yip, J. Blumenfeld, M. Traglia, M. J. Kim, N. Koutsodendris, A. Rao, B. Grone, Y. Hao, S. Y. Yoon, Q. Xu, S. De Leon, T. Choenyi, R. Thomas, F. Lopera, Y. T. Quiroz, J. F. Arboleda-Velasquez, E. M. Reiman, R. W. Mahley and Y. Huang, The APOE-R136S mutation protects against APOE4-driven Tau pathology, neurodegeneration and neuroinflammation, Nat. Neurosci., 2023, 26, 2104–2121 CrossRef CAS PubMed.
- G. Bu, APOE targeting strategy in Alzheimer’s disease: lessons learned from protective variants, Mol. Neurodegener., 2022, 15, 51 CrossRef PubMed.
- C. Frieden and K. Garai, Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer's disease, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8913–8918 CrossRef CAS PubMed.
- P. S. Chetty, L. Mayne, S. Lund-Katz, S. W. Englander and M. C. Phillips, Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 968–973 CrossRef CAS PubMed.
- Z. Li, F. Shue, N. Zhao, M. Shinohara and G. Bu, APOE2: protective mechanism and therapeutic implications for Alzheimer's disease, BioMed Central Ltd, 2020, preprint DOI:10.1186/s13024-020-00413-4.
- J. F. Arboleda-Velasquez, F. Lopera, M. O’Hare, S. Delgado-Tirado, C. Marino, N. Chmielewska, K. L. Saez-Torres, D. Amarnani, A. P. Schultz, R. A. Sperling, D. Leyton-Cifuentes, K. Chen, A. Baena, D. Aguillon, S. Rios-Romenets, M. Giraldo, E. Guzmán-Vélez, D. J. Norton, E. Pardilla-Delgado, A. Artola, J. S. Sanchez, J. Acosta-Uribe, M. Lalli, K. S. Kosik, M. J. Huentelman, H. Zetterberg, K. Blennow, R. A. Reiman, J. Luo, Y. Chen, P. Thiyyagura, Y. Su, G. R. Jun, M. Naymik, X. Gai, M. Bootwalla, J. Ji, L. Shen, J. B. Miller, L. A. Kim, P. N. Tariot, K. A. Johnson, E. M. Reiman and Y. T. Quiroz, Resistance to autosomal dominant Alzheimer's disease in an APOE3 Christchurch homozygote: a case report, Nat. Med., 2019, 25, 1680–1683 CrossRef CAS PubMed.
- C. Frieden and K. Garai, Structural differences between apoE3 and apoE4 may be useful in developing therapeutic agents for Alzheimer's disease, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 8913–8918 CrossRef CAS PubMed.
- C. C. Liu, M. E. Murray, X. Li, N. Zhao, N. Wang, M. G. Heckman, F. Shue, Y. Martens, Y. Li, A. C. Raulin, C. L. Rosenberg, S. V. Doss, J. Zhao, M. C. Wren, L. Jia, Y. Ren, T. C. Ikezu, W. Lu, Y. Fu, T. Caulfield, Z. A. Trottier, J. Knight, Y. Chen, C. Linares, X. Wang, A. Kurti, Y. W. Asmann, Z. K. Wszolek, G. E. Smith, P. Vemuri, K. Kantarci, D. S. Knopman, V. J. Lowe, C. R. Jack, J. E. Parisi, T. J. Ferman, B. F. Boeve, N. R. Graff-Radford, R. C. Petersen, S. G. Younkin, J. D. Fryer, H. Wang, X. Han, C. Frieden, D. W. Dickson, O. A. Ross and G. Bu, APOE3-Jacksonville (V236E) variant reduces self-aggregation and risk of dementia, Sci. Transl. Med., 2021, eabc9375 CrossRef CAS PubMed.
- S. Naguib, E. R. Torres, C. Lopez-Lee, L. Fan, M. Bhagwat, K. Norman, S.-I. Lee, J. Zhu, P. Ye, M. Y. Wong, T. Patel, S.-A. Mok, W. Luo, S. Sinha, M. Zhao, S. Gong and L. Gan, APOE3-R136S mutation confers resilience against tau pathology via cGAS-STING-IFN inhibition, bioRxiv, 2025, preprint DOI:10.1101/2024.04.25.591140.
- C. W. Medway, S. Abdul-Hay, T. Mims, L. Ma, G. Bisceglio, F. Zou, S. Pankratz, S. B. Sando, J. O. Aasly, M. Barcikowska, J. Siuda, Z. K. Wszolek, O. A. Ross, M. Carrasquillo, D. W. Dickson, N. Graff-Radford, R. C. Petersen, N. Ertekin-Taner, K. Morgan, G. Bu and S. G. Younkin, ApoE variant p.V236E is associated with markedly reduced risk of Alzheimer’s disease, Mol. Neurodegener., 2014, 9, 11 CrossRef PubMed.
- C. Frieden, H. Wang and C. M. W. Ho, A mechanism for lipid binding to apoE and the role of intrinsically disordered regions coupled to domain-domain interactions, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6292–6297 CrossRef CAS PubMed.
- E. Lewkowicz, M. N. Nakamura, M. J. Rynkiewicz and O. Gursky, Molecular modeling of apoE in complexes with Alzheimer’s amyloid-β fibrils from human brain suggests a structural basis for apolipoprotein co-deposition with amyloids, Cell. Mol. Life Sci., 2023, 80, 376 CrossRef CAS PubMed.
- B. Williams, M. Convertino, J. Das and N. V. Dokholyan, ApoE4-specific Misfolded Intermediate Identified by Molecular Dynamics Simulations, PLoS Comput. Biol., 2015, e1004359 CrossRef PubMed.
- N. Henry, E.-M. Krammer, F. Stengel, Q. Adams, F. Van Liefferinge, E. Hubin, R. Chaves, R. Efremov, R. Aebersold, G. Vandenbussche, M. Prévost, V. Raussens and S. Deroo, Lipidated apolipoprotein E4 structure and its receptor binding mechanism determined by a combined cross-linking coupled to mass spectrometry and molecular dynamics approach, PLoS Comput. Biol., 2018, 14, e1006165 CrossRef PubMed.
- E. Lewkowicz, M. N. Nakamura, M. J. Rynkiewicz and O. Gursky, Molecular modeling of apoE in complexes with Alzheimer’s amyloid-β fibrils from human brain suggests a structural basis for apolipoprotein co-deposition with amyloids, Cell. Mol. Life Sci., 2023, 80, 376 CrossRef CAS PubMed.
- A. A. Mamchur, V. V. Erema, D. A. Kashtanova, M. V. Ivanov, V. S. Yudin, A. A. Keskinov, S. A. Kraevoy and S. M. Yudin, Conformational mobility studies lipid-binding site of apolipoprotein E isoforms ε2, ε3, ε4 molecular dynamics method, Vestn. Mosk. Univ., Ser. 16:Biol., 2023, 78, 70–77 Search PubMed.
- M. D. Stuchell-Brereton, M. I. Zimmerman, J. J. Miller, U. L. Mallimadugula, J. J. Incicco, D. Roy, L. G. Smith, J. Cubuk, B. Baban, G. T. DeKoster, C. Frieden, G. R. Bowman and A. Soranno, Apolipoprotein E4 has extensive conformational heterogeneity in lipid-free and lipid-bound forms, Proc. Natl. Acad. Sci. U. S. A., 2023, e2215371120 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- R. L. Dunbrack, Rotamer Libraries in the 21st Century, Curr. Opin. Struct. Biol., 2002, 431–440 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, UCSF Chimera—A Visualization System for ExploratoryResearch and Analysis, J. Comput. Chem., 2004, 13, 1605–1612 CrossRef PubMed.
- L.-M. Dong, C. Wilson, M. R. Wardellsii, T. Simmons, R. W. Mahleys, K. H. Weisgrabers and D. A. Agardo, Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms, J. Biol. Chem., 1994, 269, 22358–22365 CrossRef CAS PubMed.
- K. H. Weisgraber, Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112, 1990, vol. 31 Search PubMed.
- P. S. Chetty, L. Mayne, S. Lund-Katz, S. W. Englander and M. C. Phillips, Helical structure, stability, and dynamics in human apolipoprotein E3 and E4 by hydrogen exchange and mass spectrometry, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 968–973 CrossRef CAS PubMed.
- J. A. Westerlundz and K. H. Weisgraberg, Discrete Carboxyl-terminal Segments of Apolipoprotein E Mediate Lipoprotein Association and Protein Oligomerization*, 1993, vol. 268 Search PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- C. T. Aiken, R. M. Kaake, X. Wang and L. Huang, Oxidative stress-mediated regulation of proteasome complexes, 2011, preprint DOI:10.1074/mcp.R110.006924.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- G. Bussi, A. Laio and M. Parrinello, Equilibrium free energies from nonequilibrium metadynamics, Phys. Rev. Lett., 2006, 090601 CrossRef PubMed.
- T. Darden, D. York and L. Pedersen, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, LINCS: A Linear Constraint Solver for molecular simulations, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindah, Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- C. L. Brooks, A. Brunger and M. Karplus, Active Site Dynamics in Protein Molecules: A Stochastic Boundary Molecular-Dynamics Approach, Biopolymers, 1985, 24, 843–865 CrossRef CAS PubMed.
- A. Warshel and G. King, Polarization constraints in molecular dynamics simulation of aqueous solutions: The surface constraint all atom solvent (SCAAS) model, Chem. Phys. Lett., 1985, 121, 124–129 CrossRef CAS.
- T. Kubař, M. Elstner and Q. Cui, Hybrid Quantum Mechanical/Molecular Mechanical Methods For Studying Energy Transduction in Biomolecular Machines, Annu. Rev. Biophys., 2025, 238, 19 Search PubMed.
- G. Bussi, D. Donadio and M. Parrinello, Canonical sampling through velocity rescaling, J. Chem. Phys., 2007, 014101 CrossRef PubMed.
- E. E. Lietzke, D. Saeb, E. C. Aldrich, K. D. Bruce and K. G. Sprenger, Synergistic reduction in interfacial flexibility of TREM2R47H and ApoE4 may underlie AD pathology, Alzheimer’s Dementia, 2025, e70120 CrossRef CAS PubMed.
- D. E. Shaw, P. Maragakis, K. Lindorff-Larsen, S. Piana, R. O. Dror, M. P. Eastwood, J. A. Bank, J. M. Jumper, J. K. Salmon, Y. Shan and W. Wriggers, Atomic-Level Characterization of the Structural Dynamics of Proteins, Science, 2010, 330, 341–346 CrossRef CAS PubMed.
- K. Lindorff-Larsen, S. Piana, R. O. Dror and D. E. Shaw, How fast-folding proteins fold, Science, 2011, 334, 517–520 CrossRef CAS PubMed.
- K. Demirtaş, B. Erman and T. Haliloğlu, Dynamic correlations: exact and approximate methods for mutual information, Bioinformatics, 2024, btae076 CrossRef PubMed.
- A. Ray, N. Ahalawat and J. Mondal, Atomistic Insights into Structural Differences between E3 and E4 Isoforms of Apolipoprotein E, Biophys. J., 2017, 113, 2682–2694 CrossRef CAS PubMed.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, 3 LINCS: a linear constraint solver for molecular simulations, 1997, vol. 18 Search PubMed.
- T. Darden, D. York and L. Pedersen, Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, A smooth particle mesh Ewald method, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- S. S. Sawilowsky, Very large and huge effect sizes, J. Mod. Appl. Stat. Methods, 2009, 8, 597–599 CrossRef.
- G. M. Sullivan and R. Feinn, Using Effect Size—or Why the P Value Is Not Enough, J. Grad. Med. Educ., 2012, 4, 279–282 Search PubMed.
- J. Cohen, Statistical Power Analysis for the Behavioral Sciences Second Edition, Lawrence Erlbaum Associates, Publishers, New York, 2nd edn, 1988 Search PubMed.
- D. E. Hinkle, W. Wiersma and S. G. Jurs, pdfcoffee.com_applied-statistics-for-the-behavioral-sciences-by-dennis-e-hinkle-william-wiersma-stephen-g-jurs-pdf-free, Houghton Mifflin, 5th edn, 2002 Search PubMed.
- R. W. Mahley, K. H. Weisgraber and Y. Huang, Apolipoprotein E: Structure determines function, from atherosclerosis to Alzheimer's disease to AIDS, 2009, preprint DOI:10.1194/jlr.R800069-JLR200.
- C. Y. Chou, Y. L. Lin, Y. C. Huang, S. Y. Sheu, T. H. Lin, H. J. Tsay, G. G. Chang and M. S. Shiao, Structural variation in human apolipoprotein E3 and E4: Secondary structure, tertiary structure, and size distribution, Biophys. J., 2005, 88, 455–466 CrossRef CAS PubMed.
- D. M. Hatters, C. A. Peters-Libeu and K. H. Weisgraber, Apolipoprotein E structure: insights into function, 2006, preprint DOI:10.1016/j.tibs.2006.06.008.
- A. B. Chai, H. H. J. Lam, M. Kockx and I. C. Gelissen, Apolipoprotein E isoform-dependent effects on the processing of Alzheimer's amyloid-β, Elsevier B.V., 2021, preprint DOI:10.1016/j.bbalip.2021.158980.
- B. Lu, J. A. Morrow and K. H. Weisgraber, Conformational reorganization of the four-helix bundle of human apolipoprotein E in binding to phospholipid, J. Biol. Chem., 2000, 275, 20775–20781 CrossRef CAS PubMed.
- P. M. M. Weers, V. Narayanaswami, N. Choy, R. Luty, L. Hicks, C. M. Kay and R. O. Ryan, Lipid binding ability of human apolipoprotein E N-terminal domain isoforms: correlation with protein stability?, 2003, vol. 100 Search PubMed.
- L.-M. Dong and K. H. Weisgraber, Human Apolipoprotein E4 Domain Interaction, J. Biol. Chem., 1996, 271, 19053–19057 CrossRef CAS PubMed.
- N. Choy, V. Raussens and V. Narayanaswami, Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain, J. Mol. Biol., 2003, 334, 527–539 CrossRef CAS PubMed.
- R. L. Raffaı, L.-M. Dong, R. V. Farese, K. H. Weisgraber and D. Steinberg, Introduction of human apolipoprotein E4 “domain interaction” into mouse apolipoprotein E Search PubMed.
- T. Nakamura, A. Watanabe, T. Fujino, T. Hosono and M. Michikawa, Apolipoprotein E4 (1-272) fragment is associated with mitochondrial proteins and affects mitochondrial function in neuronal cells, Mol. Neurodegener., 2009, 35 CrossRef PubMed.
- J. V. C. Horn, L. M. Kakutani, V. Narayanaswami and P. M. M. Weers, Insights into the C-terminal domain of apolipoprotein E from chimera studies with apolipophorin III, Mol. Cell. Biochem., 2023, 478, 173–183 CrossRef CAS PubMed.
- D. Nguyen, P. Dhanasekaran, M. C. Phillips and S. Lund-Katz, Molecular mechanism of apolipoprotein E binding to lipoprotein particles, Biochemistry, 2009, 48, 3025–3032 CrossRef CAS PubMed.
- M. C. Phillips, Apolipoprotein e isoforms and lipoprotein metabolism, Blackwell Publishing Ltd, 2014, preprint DOI:10.1002/iub.1314.
- G. Drin and B. Antonny, Amphipathic helices and membrane curvature, 2010, preprint DOI:10.1016/j.febslet.2009.10.022.
- A. Luzar and D. Chandler, Structure and hydrogen bond dynamics of water-dimethyl sulfoxide mixtures by computer simulations, J. Chem. Phys., 1993, 98, 8160–8173 CrossRef CAS.
| This journal is © the Owner Societies 2026 |