Open Access Article
Open Access ArticleEvaluating the strength of molecular interactions in a deep eutectic solvent (DES) by means of ionization mechanisms involved in cold-spray ionization mass spectrometry and by DFT calculations
Emilie Bertranda,
Camille Cousseaub,
Swaroop V. S. Kunapulic,
Thomas Delhayea,
Rachel Schurhammer c,
Alain Chaumont
c,
Alain Chaumont c,
Emmanuelle Limantonb,
Béatrice Legouinb,
Ludovic Paquinb,
Xavier Castela,
Mohammed Himdia and
David Rondeau
c,
Emmanuelle Limantonb,
Béatrice Legouinb,
Ludovic Paquinb,
Xavier Castela,
Mohammed Himdia and
David Rondeau *a
*a
aUniv Rennes, CNRS, IETR – UMR 6164, F-35000 Rennes, France. E-mail: david.rondeau@univ-rennes.fr
bUniv Rennes, CNRS, ISCR – UMR 6226, F-35000 Rennes, France
cUniversité de Strasbourg, CNRS CMC UMR 7140, Laboratoire MSM, 4 rue Blaise Pascal, Strasbourg, F-67000, France
First published on 21st October 2025
Abstract
This paper regroups the results of mass spectrometry analysis of two categories of deep eutectic solvents (DESs) using electrospray (ESI) and cold-spray (CSI) ionizations. Mass spectrometry of octanoic based DES's is first presented by proposing an interpretation of the ESI and CSI mass spectra based on the knowledge of the theoretical principles of ion formation by these two ionization methods. Whereas in the case of the ESI-MS analysis of the menthol/octanoic based DES, only the cationized adducts of the ester derivatives are observed in positive ion mode, the CSI-MS analysis discloses the formation of protonated non-covalent adducts of menthol and ester. By means of the acid properties of the DES precursors such as octanoic acid and thymol, CSI was used in negative ion mode to evidence the formation of deprotonated non-covalent species that are characteristic of an octanoic based DES. The interpretation is based on the main difference between ESI and CSI which is the spray temperature that induces a lowering of the dielectric constant of the solvent and then the ability of the medium to separate and then disperse opposite charges. In this context, the fact that diagnostic ions of DES are observed only in CSI indicates that the charge of the non-covalent species, whether the proton in positive ion mode or the deprotonated group in negative ion mode, is likely localised inside the supramolecular assemblies participating both to the ionization and the network structuration. In the case of the choline-chloride based DESs, the interpretation of their mass spectra obtained in negative CSI and the gas-phase stability studies of their characteristic ions, is supported by DFT calculations. The association of mass spectrometry experiments and computational studies shows that as the size and composition of clusters increase, the DES clusters containing thiourea are more stable in the gas-phase than those associating urea and glycerol, due to a greater orientation of thiourea towards the cationic quaternary ammonium group of choline in the DES.
Introduction
Depending on the field of application and the properties required, finding the right solvent can be a real challenge. According to Prat, there are 53 classical solvents, divided into different chemical families, including alcohols, ketones, esters, ethers, hydrocarbons, halogens, polar aprotics, acids and amines.1 The study that was carried out – according to the CHEM21 methodology, which classifies solvents according to safety, health and environmental scores – highlighted that of these 53 classical solvents, only 14 were recommended. Of these recommended solvents, only four chemical families are represented: 1 ether, 3 esters, 3 ketones, 6 alcohols, and of course water. This leaves few options for choosing a solvent that is safe, environmentally friendly and suitable for the intended application. It therefore became necessary to find alternatives to conventional solvents.Today, there are several alternatives, including ionic liquids (ILs). ILs were first described in the literature in 1914.2,3 Composed of molten salts – an organic cation and an organic or inorganic anion, associated by ionic bonds – and liquid at room temperature, ILs have physicochemical properties that vary according to their composition (viscosity, density, and polarity).4 Applied to extraction, organic synthesis and catalysis, ILs have attracted growing interest in recent decades.3 Although they offer advantages of non-flammability, high thermal stability and low volatility, as well as good extraction capacities for bioactive compounds, ILs are toxic and costly to produce.5 The research on biobased ionic liquids based on choline chloride has led to a new class of solvents: deep eutectic solvents (DESs).6–8 These new solvents emerged in 2003.9 Inexpensive and easy to prepare, DESs are generally composed of 2 or 3 compounds. The association of these compounds by weak interactions generates a “drop” in the real eutectic point, presenting significant negative deviation from ideality.10,11 The mixture is thus fully molten at a temperature well below the initial melting temperature of each of the compounds. Compounds can be quaternary ammoniums, carboxylic acids, alcohols or amides. In 2011, Choi et al. have defined a sub-category of DESs, natural deep eutectic solvents (NaDESs), for eutectic mixtures composed of primary metabolites such as amino acids, organic acids, sugars or polyols.12 Although they are often considered as “green solvents”, the toxicity and biodegradability of DESs depend on their composition. Elsewhere, depending on this composition, these solvents can be designed to meet the requirements of targeted applications. Indeed, certain physicochemical properties of DESs can be adjusted: they can be more or less polar, viscous, ionically conductive or dense. In addition to being easy to prepare, DESs have the advantage of not requiring a purification step. Several preparation methods have been reported, such as freeze-drying, grinding or heating using microwaves, but the most widely reported in the literature simply involves heating the mixture to between 60 and 100 °C.13
Since their discovery, DESs have been used in many fields: in synthesis,14 in biocatalysis,15 for CO2 captation,16 in electrochemistry,17 as extraction solvents,18,19 in medicine,20 and in separation processes.21 However, in 2018, according to Clarke, it was estimated that another 20 million tonnes a year of classical organic solvents were being used by industries, impacting the environment through their toxicity and volatility.22
Although DESs are becoming increasingly popular and finding more and more applications, knowledge and understanding of these mixtures is still limited, particularly concerning the interactions that govern their structure.23 Often described as being formed by hydrogen bonding of compounds, the importance of other weak interactions such as electrostatic interactions in the formation of DESs based on quaternary ammonium compounds has been highlighted. Indeed, the eutectic depth is largely influenced by the nature of the anion and its interaction preferences with the cation or molecular counterpart.8,24–27 DES organization modes have been studied by spectroscopic methods (RMN and IR),28–31 or by interpreting inelastic neutron scattering (INS) spectra in light of the results of ab initio quantum chemical calculations and molecular dynamic simulations.32–37 Some of us have shown that mass spectrometry (MS) can be considered a versatile tool for DES characterization if cold-spray ionization (CSI) is used for the transfer into the gas-phase of their characteristic ionic species and mass spectra are interpreted with DFT calculations.38 CSI is a low-temperature variant of the electrospray ionization (ESI) referenced for identifying labile non-covalent complexes.39,40 In CSI, the coaxial nebulizing gas is cooled by a liquid nitrogen device in order to maintain the temperature of the inlet capillary below −30 °C.41 The interest in CSI-MS has been demonstrated for the characterization of many supramolecular assemblies or non-covalent complexes such as labile inorganic and organometallic compounds,42–44 labile biomolecules,45 encapsulating supramolecules,46 oligomeric coordination species,47–49 large-scale aggregated chain structures,50 and organic molecule clusters.51
The aim of this work was therefore to show how the use of a chemical analysis method like cold-spray ionization mass spectrometry can not only characterize DES's synthesized from different molecular and ionic precursors, but also disclose the strength of the interactions that maintain the structure of such non-covalent assemblies. Toward this purpose, CSI mass spectra are interpreted in light of some theoretical aspects of the ionization mechanisms in ESI and CSI or by the help of DFT calculations. Toward this goal, we studied DESs governed by H-bonds, based on octanoic acid and DESs governed mainly by electrostatic interactions, based on choline chloride. Therefore, in order to better understand the interpretations that can be obtained from the study of CSI mass spectra, a theoretical background section for anticipating DES's response in ESI and CSI, is presented in this manuscript.
Theoretical background for anticipating DES's response in ESI and CSI
Cold-spray ionization is known to be more suitable than electrospray for maintaining in the gas-phase the non-covalent interactions formed in solution and for characterizing the most labile compounds.38 It's often admitted that the main concept difference between the two ionization methods concerns the temperature of the solution infusing the capillary, the one inducing the temperature of the ions emitted into the gas-phase from the last charged micro-droplets.52 Starting from this paradigm, we can review the main factors influencing the responsiveness of molecules in ESI in order to anticipate their behavior when ESI is substituted by CSI.53 In the overall process, the ionization by protonation of the analytes in solution is considered the initial step that will lead to the final formation of gaseous protonated molecules. In positive ion mode, the basicity of the analyte in solution is thus the first factor to consider through the pKa or pKb value. The pH of the solution perfusing the ESI capillary is definitively lower than 7 due to the charge-balancing redox reactions inherent to the electrospray process where the water oxidation is equivalent to the production of strong acid in solution.54–59 It has been shown that the intensity of a [M + H]+ ion produced in ESI by using a pure solution of methanol is not under the influence of the pKb value if this one is less than ∼20.60 Table 1 regroups the properties of the DES precursors used for the present study. These ones can be considered as relevant for understanding the ESI response of the molecules considered for these studies. From the pKa and pKb values of Table 1, whether they are taken from literature data for the former or calculated using the Kebarle approach,61,62 it appears that almost all the selected DES precursors, have a pKb value < 20. Note that there is no available data for thiourea. The cholinium cation is already charged in solution without protonation.| Compound | pKa | pKbb | PAc | GBc | ΔacidH°c | GAc | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pa Pa |
|---|---|---|---|---|---|---|---|
| a pKa values in water from the National Center for Biotechnology Information (2024) https://pubchem.ncbi.nlm.nih.gov.b pKb values in methanol calculated using the Kebarle approach where pKa(MeOH) = pKa(H2O) + 0.6 with the methanol autoprotolysis constant KSH(MeOH) = Ka(MeOH)Kb(MeOH) = 3.2 × 10−17.61,62c Data (kJ mol−1) from the National Institute of Standards and Technology (2024). https://webbook.nist.gov/chemistry/.d Considered for the conjugate acid.e Data taken from that of 1-hexanol.f Data taken from that of propanoic acid.g Data taken from that of phenol.h Data taken from that of propanoic acid methyl ester.i Data taken from that of 1,2-ethandiol. NI: Data are not available for compounds that form no ions in water such as hydroxyl anion or proton. NA: Data are not available. | |||||||
| Water | 7 | 8.9 | 691 | 660 | 1622.1 | 1594.5 | NA |
| Acetonitrile | −4.3 | 20.2 | 779.2 | 748 | 1534 | 1528 | NA |
| Methanol | 15.3 | 0.6 | 754 | 724.5 | 1597 | 1573.3 | NA |
| Menthol | 19.6 | −3.7 | 799e | NA | 1565e | 1537e | 2.75 |
| Octanoic acid | 4.9 | 11.0 | 797f | 766.2f | 1454f | 1424f | 2.43 |
| Thymol | 10.6 | 5.3 | 817.3g | 786.3g | 1462g | 1426g | 3.3 |
| Ester | 18.4 | −2.5 | 821.6h | 790.7h | 1573g | 1528h | 3.72 |
| Choline | NI | NI | NA | NA | NA | NA | NE |
| Urea | 0.1 | 15.8 | 868.4 | 838.7 | 1495 | 1488 | −1.29 |
| Thiourea | NI | NA | NA | NA | NA | NA | −0.71 |
| Glycerol | 14.4d | 1.5 | 874.8i | 820i | 1528i | NAf | −1.33 |
In addition to the acid–base properties in solution, the ESI response is also related to the evolution of the ionized species in the charged droplets. Indeed, a molecule with a non-polar region will be preferentially distributed at the solvent–air interface of the droplets.63,64 This so-called “surface activity” is usually correlated to a partition coefficient between octanol and water (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) and has been demonstrated to influence noticeably the intensity of the [M + H]+ ions detected in ESI.65,66 An analyte with high polarity can be “easily” protonated in solution but can poorly respond in positive ESI-MS due to its very low log
P) and has been demonstrated to influence noticeably the intensity of the [M + H]+ ions detected in ESI.65,66 An analyte with high polarity can be “easily” protonated in solution but can poorly respond in positive ESI-MS due to its very low log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value. By referring to the properties of compounds reported in Table 1, one can expect that if an ester is involved in the DES formation as a precursor, its participation could lead to relatively intense ions as protonated molecules in a supramolecular assembly because its log
P value. By referring to the properties of compounds reported in Table 1, one can expect that if an ester is involved in the DES formation as a precursor, its participation could lead to relatively intense ions as protonated molecules in a supramolecular assembly because its log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value will be relatively high, although it has a low pKb value. Positive ESI-MS could be used for characterizing DES's formed from mixtures of menthol or thymol by referring to their pKb and log
P value will be relatively high, although it has a low pKb value. Positive ESI-MS could be used for characterizing DES's formed from mixtures of menthol or thymol by referring to their pKb and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values (see Table 1).
P values (see Table 1).
When the [M + H]+ ions are emitted from the charged droplets into the gaseous atmosphere of the source, they can undergo gas-phase proton transfer reactions with solvent vapors before being sampled into the mass spectrometer analyzer vacuum.67,68 Such a reaction is governed by the values of the gas-phase basicity (GB) or the proton affinity (PA) of the gaseous analyte compared to that of the solvent molecule. The analytes of Table 1 have PA or GB values greater than that of methanol or acetonitrile.
As a result, the protonated molecule of the analyte [M + H]+ should retain the charge without proton transfer reaction to the surrounding gaseous solvent molecule or during the dissociation of a solvent adduct [M + S + H]+, where S is the solvent molecule. To observe DES assemblies as protonated states, the precursor molecules must thus have high-gas phase basicity in addition to a high basicity in solution. If molecules of Table 1, such as menthol, thymol and esters can lead to DES formation, through their HBD (hydrogen bonding donor) or HBA (hydrogen bonding acceptor) properties, their solution and gas-phase basicities will favor the detection of protonated DES's.69 Note that the position of the proton on a molecule in solution can differ from that of the gaseous ion. The protonation of some groups can indeed occur in solution whereas other sites of the molecule can be preferred during the gas-phase proton transfer reaction.70,71
Other analytes that are nevertheless polar, exhibit no basicity in solution. In this case, the produced ion is due to a cation attachment process leading mainly to the formation of a [M + Na]+ ion and sometimes to a [M + K]+ ion, the sodium adduct formation being considered to be in competition with the protonation in solution.72 Even though the [M + Na]+ ions are observed for low pKb value analytes, they must have the ability to induce cation interaction by ion-dipole attraction. For such compounds, the most important factor determining the responsiveness is the partial charge of their atoms and their chelating ability towards Na+.73 The main interaction is supposed to occur with the oxygen of a carbonyl group where the sodium ion is positioned along the axis of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, the non-covalent bonding strength depending on the dipole moment value.73 It means that octanoic acid and the so-called ester of Table 1, can produce [M + Na]+ ions in positive ESI. By contrast, the nitrogen bases where the partial charges on nitrogen atoms are often less negative than that of oxygen, should undergo protonation reactions rather than sodium adduct formation processes.
O bond, the non-covalent bonding strength depending on the dipole moment value.73 It means that octanoic acid and the so-called ester of Table 1, can produce [M + Na]+ ions in positive ESI. By contrast, the nitrogen bases where the partial charges on nitrogen atoms are often less negative than that of oxygen, should undergo protonation reactions rather than sodium adduct formation processes.
In negative ion electrospray mass spectrometry, the main ionized species that can be detected are [M − H]− and [M + Cl]− ions. The [M − H]− ions are produced in solution by acid–base reaction of the analyte molecule with the hydroxyl anion electrochemically generated from water reduction, the main electrochemical reaction required to counterbalance the loss of same polarity ions producing the charged ES droplets.74 The response of the [M − H]− ions depends first of the ionizability of the analyte with an equilibrium governed by the pKa value in conditions where the water electrolytic reaction leads to a change in the solution pH that can reach a value of 10.75 Molecules with pKa values higher than 7 can thus be theoretically deprotonated in aqueous solution. Molecules with low pKa values will not necessarily yield intense [M − H]− ions if the equilibrium is shifted to the neutral formation owing to the low solubility of the corresponding anion in organic solvent. The negative ions are indeed less solvated in methanol than in water and still less in acetonitrile. In this context, among the DES precursors reported in Table 1, only the octanoic acid and the thymol could be considered as fairly good candidates for detecting ionized DES that will encompass these two precursors. DES precursor analytes with non-polar regions in their structure and high gas-phase acidity such as octanoic acid should lead to relatively intense [M − H]− ions. The emission of the deprotonated molecules into the gas-phase is also under the influence of both their surface activity (molecular structure with a non-polar region) and thermochemical properties (gas-phase acidity).76,77 The [M − H]− ion emitted into the gas-phase is detected on the mass spectra if the proton loss from the analyte molecule (M) is less endothermic than that of the solvent molecule (S). In another words, a molecule will be more acidic in the gas-phase than a solvent if GA(M) < GA(S). Let's however keep in mind that the deprotonated group after emission of the ion in the gas-phase, is not necessarily the more acidic site in solution.78,79 From the GA and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values available in Table 1, it appears that the greater candidates to observe [M − H]− ions from DES are thymol and octanoic acid. When the negative ESI-MS analysis is performed in chlorinated solvents or with chlorinated molecules at the trace level, the reduction cleavage of the carbon-chlorine bond is the main charge-balancing reaction that releases chloride anions.80 The [M + Cl]− adducts are expected to be formed from analytes whom pKa values are between 5 and 28 if one considers that the solution pH should not increase beyond ∼7 in the absence of electrochemical production of hydroxyl ions.81 The gaseous [M + Cl]− adduct is detected in negative mass spectrometry if GA(M) is greater than that of HCl, i.e. GA < ∼1400 kJ mol−1, considering that a gas-phase proton transfer reaction occurs between the analyte M and the chloride anion for yielding a [M − H]− ion.82,83
P values available in Table 1, it appears that the greater candidates to observe [M − H]− ions from DES are thymol and octanoic acid. When the negative ESI-MS analysis is performed in chlorinated solvents or with chlorinated molecules at the trace level, the reduction cleavage of the carbon-chlorine bond is the main charge-balancing reaction that releases chloride anions.80 The [M + Cl]− adducts are expected to be formed from analytes whom pKa values are between 5 and 28 if one considers that the solution pH should not increase beyond ∼7 in the absence of electrochemical production of hydroxyl ions.81 The gaseous [M + Cl]− adduct is detected in negative mass spectrometry if GA(M) is greater than that of HCl, i.e. GA < ∼1400 kJ mol−1, considering that a gas-phase proton transfer reaction occurs between the analyte M and the chloride anion for yielding a [M − H]− ion.82,83
The main difference between CSI and ESI is that the former acts at a temperature of the sprayed solution that is lowered until −30 °C, inducing an increase in the dielectric constant value (εr) of the solvent. The value of εr for methanol is of 32.62 at 278 K and 37.91 at 273 K.84 A dielectric constant of a solvent is related to its ability to disperse the attractive forces between either opposite charges or a charge and a dipole. In spray ionization, a solvent with high dielectric constant is expected to increase the separation degrees between an ionized analyte and either a counter-ion or a dipolar solvent molecule. As previously reported in the literature, the fact to detect supramolecular assemblies involving cholinium cations, chloride anions and urea molecule as dipole as CSI-MS is used, is not only due to the stabilization of brittle species by the use of a low source temperature.38 Such results suggest also that the observed chloride adducts are not related to a simple charge interaction but rather to the organization in a network of the different charged partners. DFT calculations have indicated that such ionic network organization is supported by a maximization of hydrogen bonds of the chlorides not only with the hydroxyl and methyl moieties of the cholinium cations but also with the amino groups of urea.38 Most of the publications devoted to the use of CSI-MS supports this assertion. In positive ion mode, the cationized labile non-covalent complexes that should be observed only in cold-spray, must be considered as ionic aggregates or supramolecular assemblies encapsulating counter-ions or solvent molecules, rather than a cation or anion attachment as in electrospray. This later kind of interaction is very likely dispersed in CSI due to the high dielectric constant of the solvent. From the diverse set of molecular clusters that can be observed when mixtures of cholinium chloride (ChCl) with urea (U), thiourea (ThU) or glycerol (Gly) molecules are analysed in CSI-MS, the possibilities to detect molecular clusters following the formulae [M + ChCl + Cl]−, [2M + ChCl + Cl]− and [M + 2ChCl + Cl]− where M = U, ThU and Gly, suggest that the motivations for the DFT calculations can be related to: (i) the structures of the studied molecular clusters in the gas-phase, giving an idea of the spectrum of cluster geometries possible in the mass-spectrometry, (ii) the evolution of the calculated binding energies of the clusters with the addition or removal of either the HBDs (U, ThU and Gly) or the cholinium chloride (ChCl) entity and (iii) the reasons for the relative stability of the molecular clusters and the facile integration of some molecules into the structure of the charged ion cluster entity as opposed to the destabilizing integration of others.
Results and discussion
Mass spectrometry of octanoic acid-based DES's
The negative ESI-MS and CSI-MS analyses of menthol/octanoic acid-based DESs, have shown the formation of [M − H]− and [2M − H]− ions of the octanoic acid (data not shown). Any criteria other than that of the characterization of the DES precursors are provided from these ESI mass spectra. Positive ESI and CSI mass spectra of the menthol/octanoic acid based DES sample are depicted in Fig. 1 and 2, respectively. The accurate mass measurements performed on the detected ions shown in Fig. 1 and 2 and their comparison with the theoretical masses calculated from elemental compositions, are reported in Table S1 (SI). The chemical structure that we can assume to represent the ions detected in positive ESI and CSI are reported in Schemes 1 and 2, respectively. It appears from Fig. 1 and the interpretations of the measurements regrouped in Table S1 (SI), that the positive ESI-MS analysis characterizes rather the esters produced from the reaction of octanoic acid with menthol rather than all the other menthol/octanoic acid based DES components, as depicted in Scheme 1. Such a behavior has been previously described in the positive ESI-MS analysis of deep eutectic solvents based on cholinium chloride and carboxylic acids where the cationic ester produced between the cholinium cation and the glutaric acid was detected.85 From the “Theoretical background” section, the observation of ester adducts with a sodium cation can be expected if one refers to the surface activity related to their log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value (see Table 1) and the ability for interacting with Na+ of these molecules as they are submitted to ESI analysis. The difference between the two ESI mass spectra of Fig. 1, is due to the raising of the Orifice 1 voltage (OR 1) that activates the dissociations of all the ions produced in the source.
P value (see Table 1) and the ability for interacting with Na+ of these molecules as they are submitted to ESI analysis. The difference between the two ESI mass spectra of Fig. 1, is due to the raising of the Orifice 1 voltage (OR 1) that activates the dissociations of all the ions produced in the source.
 | ||
Fig. 2 Positive CSI mass spectrum of menthol/octanoic acid DES in methanol at Tnebulizing![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) gaz = −30 °C and OR1 = 20 V. Note the signal magnification of 25 times in the m/z 590–1000 range. gaz = −30 °C and OR1 = 20 V. Note the signal magnification of 25 times in the m/z 590–1000 range. | ||
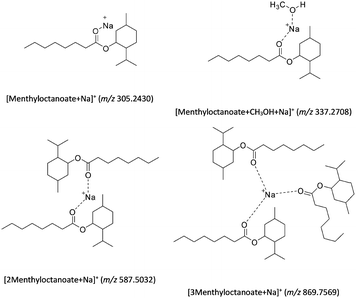 | ||
| Scheme 1 Proposed chemical structures to account for the formation of ions observed in positive ESI-MS (see Table S1 (SI) for the elemental formulae proposed from the exact mass measurements). | ||
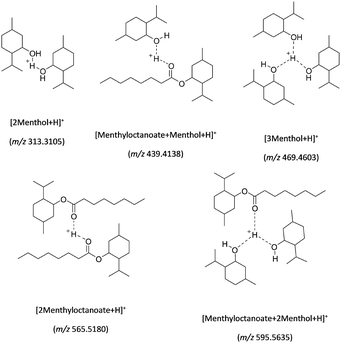 | ||
| Scheme 2 Proposed chemical structures to account for the formation of ions observed in positive CSI-MS (see Table S1 (SI) for the elemental formulae proposed from the exact mass measurements). | ||
The decrease in intensity of the m/z 870 signal in Fig. 1 (lower figure) occurs with the increase of the m/z 587 peak and the appearance of the m/z 305 and 337 ions.
This suggests that the [2Menthyloctanoate + Na]+ (m/z 587) and the [Menthyloctanoate + Na]+ (m/z 305) ions are produced from the [3Menthyloctanoate + Na]+ (m/z 870) (see Scheme 1 and Table S1 (SI)). The detection of the signal at m/z 337 is attributed to the [Menthyloctanoate + CH3OH + Na]+ ion. This ion is due to an interaction of the m/z 305 ion with the gaseous solvent that occurs downstream into the mass spectrometer as previously described for such atmospheric pressure interface devices.86–88 This is not a complex encapsulating a solvent molecule. It is clear that the ESI-MS analysis allows only detection of sodium adducts that are not diagnostic ions of the interaction between HBD and HBA compounds into a DES. By contrast to ESI-MS, the charged species observed in CSI-MS (see Fig. 2) are all protonated adducts (see Table S1 (SI) and Scheme 2). As the solution temperature is dramatically decreased, the sodium/ester interactions are also lowered and are not maintained during the formation of the charged droplets. In CSI, there are the protonated species that participate in turn to the charge excess for the formation of the charged droplets. In this case, the use of cold-spray in positive ion mode appears to be more suitable than ESI for highlighting the participation of hydrogen-bonds in a DES structuration. Indeed the formation of protonated adducts at m/z 469 and 596 in Fig. 2 that involve the participation of more than two molecules, suggests the hydrogen-bond formation for maintaining the structure whatever the localization of the proton (see the proposed structures in Scheme 2). To summarize, if the [M − H]− and [2M − H]− ions of octanoic acid are the only charged species detected in negative ion mode whether in ESI or in CSI, the interactions of ester with menthol are only evidenced in positive ion mode using cold-spray ionization (see Fig. 2). The formation of non-covalent assemblies maintained by hydrogen bonds are not characterized in positive ESI-MS analysis where sodium adducts are observed (see Fig. 1).
For thymol/octanoic acid based DES, the two pKa values (see Table 1) would indicate that the HBA compound is thymol and the HBD compound is octanoic acid. The physicochemical properties listed in Table 1 for evaluating their responses in mass spectrometry, suggest that the negative ion mode can be assumed to be the most suitable in CSI-MS. CSI mass spectra of thymol/octanoic acid based DES are depicted in Fig. 3. The assignment propositions of the signals detected in the mass spectra shown in Fig. 3 are given in Table S2 (SI), after the accurate mass measurements. The assumed chemical structures of the ions are proposed in Scheme 3. In Fig. 3, the ions at m/z 443 and m/z 437 represent the association of two thymol molecules with one octanoic acid for the former and two octanoic acid with a thymol molecule for the later (see Table S2 (SI) and Scheme 3). It's not astonishing to observe such interactions in the gas-phase if one refers to the close values of the gas-phase acidity (GA) of thymol and octanoic acid regrouped in Table 1. Indeed, as previously discussed, the closer they are, the less the non-covalent heterodimer maintained by the proton bond, tends to dissociate. In Fig. 3, the m/z 443 intensities are greater than that of the m/z 437 ion. However, one could expect that for the m/z 437, the non-polar part of the two octanoic acids bestow to this ion a higher response in spray ionization. The [Octanoic acid + 2Thymol-H]− ion seems to be the major species present in solution and characterizing the thymol/octanoic acid-based DES. The change in Fig. 3 between the upper and lower figures, is due to the increase in the orifice 1 voltage value. The intensities of the m/z 443 and m/z 437 ions drop whereas the intensity ratio between the m/z 293 and 287 ions remains constant. The [M − H]− ion of octanoic acid (m/z 143) appears under in-source activation in Fig. 3 (lower figure), but not the thymol [M − H]− ion. This behavior can be understood from the data regrouped in Table 1 and taken from that of phenol and propanoic acid (ΔacidH° and GA values), suggesting that the octanoic acid has a greater acidity in the gas phase than thymol especially since the presence of isopropyl and methyl groups would increase the endothermicity of its deprotonation reaction. One can remark that only the chloride adduct with thymol is detected at m/z 185 (see Table S2 (SI)) with low intensity in the mass spectra of Fig. 3. This can be attributed to the fact that octanoic acid and thymol are higher acids in the gas phase than HCl, i.e. with GA values lower than 1400 kJ mol−1. Nevertheless, the negative CSI-MS analysis of the thymol/octanoic acid-based DES indicates that the most probable non-covalent assembly present in solution is the complex formed by one octanoic acid and two thymol. The CSI-MS results show also that the major species in solution is not the most stable in the gas-phase due to the weakest hydrogen bonds involving the thymol into the DES assemblies.
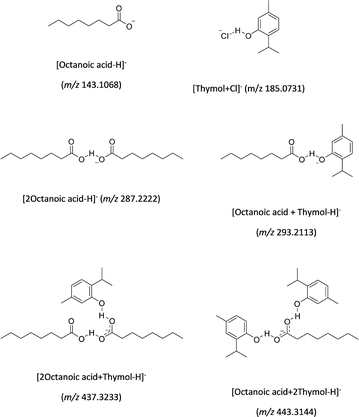 | ||
| Scheme 3 Proposed chemical structures to account for the formation of common ions observed in negative CSI-MS (see Table S2 (SI) for the elemental formulae proposed from the exact mass measurements). | ||
Mass spectrometry of choline-chloride based DESs
According to the works previously published on reline analysis, cold-spray ionization mass spectrometry of choline-chloride based DESs has been performed using the negative ion mode.38Using gentle desolvation conditions, i.e. OR 1 = 10 V, a new negative CSI mass spectrum of the choline chloride:urea (U:ChCl) was obtained in order to compare them to the mass spectra of choline chloride:thiourea (ThU:ChCl) and choline chloride:glycerol (Gly:ChCl) DES. All these spectra are reported in Fig. 4 for U:ChCl (upper), ThU:ChCl (middle) and Gly:ChCl (lower) DESs. The ion elemental compositions proposed by comparison between the measured and theoretical masses are reported in Tables S3–S5 (SI) for the U:ChCl, ThU:ChCl and Gly:ChCl samples, respectively. All the CSI mass spectra of Fig. 4–6 present three chloride adduct populations. The first one encompasses the species for those where the cholinium chloride is in interaction as a HBD compound with the chloride anion as a HBA compound. They are all detected as Cl− adducts, i.e. the [nChCl + Cl]− adducts with 1 < n < 5. Choline chloride is considered as the common HBD compound for the three DES samples. The second series concerns the HBD compound that is specific to each DES sample. This ion population is either urea, thiourea or glycerol that interacts with the chloride anion as a HBA compound, i.e. the [mM + Cl]− adducts with m < 3 in all cases and where M is either urea (Fig. 4 upper), thiourea (Fig. 4 middle) or glycerol (Fig. 4 lower). The last series characterizes the DES ions where the two HBD precursors and the HBA anion are associated in a [mM + nChCl + Cl]− chloride adduct.
From the assignments proposed in Tables S3–S5 (SI), only the CSI mass spectrum of the DES sample prepared with a mixture of cholinium chloride and thiourea (ChCl:ThU) exhibits [mM + nChCl + Cl]− ions with m > 2. Indeed, any non-covalent complex associating more than two urea or glycerol molecules with choline chlorides are assigned to the signals observed on the upper and lower mass spectra of Fig. 4. These behaviors can be interpreted in light of the DES organization model previously reported in the literature.38,89–92 In this model, the cholinium is assumed as interacting by hydrogen bonds with the chloride through its methyl and hydroxyl groups. The presence of urea, thiourea or glycerol would lead to a re-organization of the pre-existent network by means of the maximization of the hydrogen bonds induced by the introduction of their amino or hydroxyl groups. From the interpretations of the mass spectra of Fig. 4 (see Tables S4–S6 (SI)), thiourea would take a larger part in the organization of the non-covalent stable complex assembly since there are more assemblies encompassing thiourea and cholinium chloride in the mass spectrum shown in the middle of Fig. 4, compared with the two others in Fig. 4.
A m/z 680 [3ThU + 3ChCl + Cl]− ion is even observed in Fig. 4 (middle), whereas the equivalent for urea and glycerol is absent in Fig. 4 (upper) and Fig. 4 (lower), respectively. This assertion is reinforced by the fact that, by contrast, some ions characteristic of the only cholinium chloride associations, i.e. the m/z 591 [4ChCl + Cl]− and 730 [4ChCl + Cl]− ions, are detected in the mass spectra shown in Fig. 4 (upper) and Fig. 4 (lower), but are of lower relative intensity in Fig. 4 (middle). The relative stabilities of the different choline-chloride based DES's have been checked by studying the ion behaviors under collision activation. The potential difference between the so-called orifice 1 (OR 1) and orifice 2 (OR 2) lenses of the atmospheric interface of the mass spectrometer has then been increased. It's important to note that the method used for the in-source collision induced dissociation (in-source CID) experiments performed in this work, are different from what was published about the gas-phase stability of reline ions.38 In this previous paper, the variation of the potential differences was considered between the orifice 2 (OR 2) lens and the ion guide (IG) of the same mass spectrometer. For the present work the relative stabilities of the three ionized DESs are studied by raising the OR1 potential for the in-source CID experiments.
Recall also that the in-source CID experiments are not true MS-MS methods since no particular ion is selected to undergo dissociation by collisions. This is rather all the gaseous ion population produced from the ion source that is activated for fragmentation.
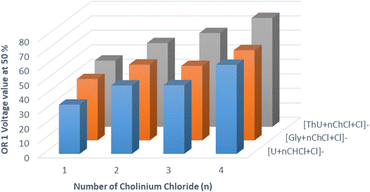
In this context, the intensities of the monoisotopic peaks of the DES ions whose elemental compositions are disclosed in Tables S3–S5 (SI), have been measured for different OR 1 voltage values. These intensities have been summed and the contribution of each peak has been reported as a fraction of the total ion intensity for each DES. The plots representing the evolutions of the fractions of the total ion intensity are displayed in Fig. S1–S3 (SI) for the U:ChCl DES, the Gly:ChCl DES and the ThU:ChCl DES, respectively. The data of Fig. S1–S3 (SI) have been fitted with the well-known half maximal effective concentration (EC50) non-linear regression curve by considering the Orifice 1 voltage value (OR1) for which the fraction of the total ion intensity is half of the intensities between the bottom and top plateaus of the curve. The graphs representing these so-called OR1(50) values reported as a function of the number (n) of cholinium chloride in a [M + nChCl + Cl]− chloride adduct and in a [2M + nChCl + Cl]− chloride adduct, are reported in Fig. 5 and 6, respectively. Hence it appears that independently of the relative intensity of detected ions, the OR1(50) values increase with the number (n) of cholinium chloride in a DES whatever the nature and the number (m = 1 or 2) of urea, thiourea or glycerol, in the DES. In addition, the presence of thiourea in a given DES leads us to observe the highest OR1(50) values as its behavior is compared with that of the others DES's considering the same n and m values and under in-source CID activation. From these CSI-MS experiments, it can be inferred that the presence of thiourea as a HBA compound in a cholinium chloride based DES, tends to stabilise the DES, as well as an increasing number of cholinium chloride. One will note that it is only in the case of thiourea based DESs that ionic species [mM + nChCl + Cl]−, where m = 3, are observed on the CSI mass spectra.
From this diverse set of molecular clusters, whose stoichiometries are disclosed in the CSI mass spectra of Fig. 4 and relatives stabilities are illustrated in Fig. 5 and 6, the ions of molecular clusters following the formulae [M + ChCl + Cl]−, [2M + ChCl + Cl]− and [M + 2ChCl + Cl]− where M = U, ThU and Gly were selected for DFT calculations. Out of 10 conformers sampled for each molecular cluster ion, the structures of the three most stable conformers of [M + ChCl + Cl]−, [2M + ChCl + Cl]− and [M + 2ChCl + Cl]− cluster entities where M = U, ThU and Gly are successively presented below.
These structures and energetics of these stable conformers are the most relevant when relating the experimental trends to the theoretical calculations.
By focussing on the study of the relative stabilities, the results of which are illustrated in Fig. 5 for n = 1, the binding energies, the local energy decomposition (LED) contribution energies of the 10 lowest-lying conformers for the clusters [U + ChCl + Cl]−, [ThioU + ChCl + Cl]− and [Gly + ChCl + Cl]− are calculated and their values are given in Table S6 (SI).
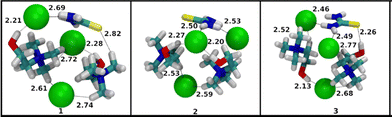
The binding energies of the three cluster entities reveal that even though glycerol molecules have more atoms than urea and thiourea and more polar hydrogens to act as a good HBD, the thiourea clusters exhibit higher relative stability. One explanation comes from the fact that in the unbound state the glycerol molecule exhibits intra molecular hydrogen bonds that have first to be broken before they can interact with the Cl− anion, which is reflected in the geometric and electronic preparation energies of the HBD molecule (Table S6 (SI)). The binding energy difference can further be understood when the structures of these clusters are analysed. Fig. 7, 8 and 9 show the three lowest-energy structures of [U + ChCl + Cl]−, [ThU + ChCl + Cl]− and [Gly + ChCl + Cl]− cluster ions sorted in increasing order of binding energy. The groups of conformers of [U + ChCl + Cl]− in Fig. 7 are populated by structures with hydrogens of urea chelating one chloride ion, the hydroxyl group of choline interacting with the other chloride ion and the interaction of chloride ion with a side of the tetrahedral structure of the cationic quaternary ammonium group of the chloride. The relative binding energies of these three lowest-energy conformers lie within the range of 0.5 kJ mol−1 and show slight differences in the hydrogen bond distances in their structures. The structures of the total set of 10 lowest-lying conformers of [U + ChCl + Cl]− and [ThU + ChCl + Cl]− are presented in Fig. S4 and S5 (SI). The binding energies of the [ThU + ChCl + Cl]− cluster in Table S6 (SI) show much greater degeneracy in structures as all of the starting conformer geometries converged to a group of conformers with similar optimised energy with relative binding energy of the highest-energy conformer only 1.5 kJ mol−1 higher than the lowest-energy conformer. The significant variation in this set of three conformers in Fig. 8 is the orientation of the thiourea group with respect to the choline cation and the associated chloride anion. It is also important to note that while the oxygen of the urea molecule in [U + ChCl + Cl]− (Fig. 7) generally orients away from the choline cation in the optimised geometries, the sulphur of thiourea in [ThU + ChCl + Cl]− orients towards the choline cation and interacts with the quaternary ammonium cationic group. The greater orientation of thiourea towards the cationic quaternary ammonium group in choline coupled with the chelation interaction with chloride ion helps in stabilization of the cluster. The experimental observations noting elevated stability of larger thiourea containing clusters can find origin in this behaviour of the interaction thiourea engages with choline and chloride where the HBD integrates into the ionic complex much better than clusters containing urea and glycerol.
 | ||
| Fig. 7 Structures of the 10 lowest-lying conformers of [U + ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
 | ||
| Fig. 8 Structures of the 10 lowest-lying conformers of [ThU + ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
 | ||
| Fig. 9 Structures of the 10 lowest-lying conformers of [Gly + ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
The set of three lowest-energy conformers of [Gly + ChCl + Cl]− structures in Fig. 9 and the total set of 10 conformers in Fig. S6 (SI) represent a diverse conformational and energetic set of conformers. Similar to the [U + ChCl + Cl]− and [ThU + ChCl + Cl]− structures discussed above, these structures feature interactions between the cationic quaternary ammonium group and hydroxyl group of choline with the chloride ions. The degree of chelation of the chloride ion with the hydrogens of the glycerol groups shown in Fig. S6 (SI) determines the binding energies in Table S6 (SI). In these structures we note the presence of a stabilizing interaction between the hydroxyl group of choline and chloride. The glycerol molecule presents three polar hydrogen atoms for interaction with the chloride ion. The interaction of all three hydrogens of glycerol with the chloride ion comes at the expense of the geometrical preparation energy required to bring the glycerol molecules into the structures in the system.
In structures presented in Fig. 9 we note that all three hydrogens of glycerol tend to orient towards the chloride ion even if they are not directionally interacting with the ion. It should be noted that none of the glycerol groups show interaction with the cationic quaternary ammonium group of choline, this fact combined with the geometric preparation energy expended in orienting the glycerol molecule to facilitate interaction with the chloride ion contributes to the lower stability of [Gly + ChCl + Cl]− relative to [ThU + ChCl + Cl]− in the gas-phase.
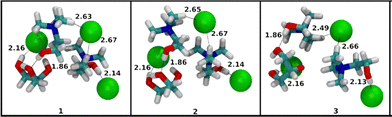
The behaviors of the choline-chloride based DESs depicted in Fig. 5 for n = 2 and m = 1, can be considered as those of the family of clusters that is the largest of the set of clusters presented in this work. The structures of the total set of 10 lowest-lying conformers of these [U + 2ChCl + Cl]−, [ThU + 2ChCl + Cl]−, [Gly + 2ChCl + Cl]− are presented in Fig. S7–S9 (SI), respectively. The binding energies of [U + 2ChCl + Cl]−, [ThU + 2ChCl + Cl]− and [Gly + 2ChCl + Cl]− are presented in Table S7 (SI). We will see further in this paper that the difference in average binding energy between [ThU + 2ChCl + Cl]− and its urea and glycerol counterparts is not as large as observed in the case of the [2M + ChCl + Cl]− family of clusters above. The presence of multiple choline groups opens up the possibility of observation of a new set of interactions. The thiourea-containing clusters set themselves apart from the urea and glycerol-containing clusters in this family of clusters. The set of three lowest-lying conformers of clusters [U + 2ChCl + Cl]− and [Gly + 2ChCl + Cl]− presented in Fig. 10 and 12 feature the replacement of at least one hydroxyl of choline choline-chloride interaction with less energetic hydroxyl of choline-HBD interaction. The same is observed for the majority of structures of [U + 2ChCl + Cl]− and [Gly + 2ChCl + Cl]− in Fig. S7 and S9 (SI) for the total set of 10 conformers of each cluster. Unlike these clusters, thiourea molecules in [ThU + 2ChCl + Cl]− show a lower tendency to replace the more energetic interaction between the hydroxyl hydrogen of choline and chloride with the interaction with the HBD molecule as seen in structures of three conformers in Fig. 11 and total set of conformers in Fig. S8 (SI). This can be illustrated by the strengths of interaction between choline and chloride ions in the three clusters. The LED scheme employed in this work decomposes the binding energy into multiple physically meaningful components. For the sake of brevity, the electrostatic interaction components discussed here and the data of the remaining components are presented in Fig. S10 and S11 (SI). In the LED scheme the electronic preparation energy of molecules is related to the pair-wise electrostatic interaction term between the molecules as the preparation energy is the energy expended to bring the electron cloud of the cluster in its cluster structure to facilitate electrostatic interaction. The magnitude of electronic preparation energy and electrostatic interactions changes with the nature of molecules involved and this feature is discussed here. Fig. S10 (SI) shows the electrostatic interactions between choline and chloride ions in the 10 lowest-energy conformers of the three clusters while Fig. S11 (SI) shows the electronic preparation energy expended by choline ions in these clusters. The Boltzmann-weighted average and standard deviation of electrostatic interaction energy between choline and chloride molecules is −451 ± 3.2, −456 ± 5.1, and −427 ± 2.3 kJ mol−1 for the 10 conformers of molecular clusters [U + 2ChCl + Cl]−, [ThU + 2ChCl + Cl]− and [Gly + 2ChCl + Cl]−, respectively. The electrostatic interaction strength in thiourea-containing clusters is significantly higher than the glycerol-containing clusters because of the facilitation of the interaction between choline and chloride ions by thiourea molecules when glycerol molecules disrupt this interaction. The urea molecules in [U + 2ChCl + Cl]− also disrupt the choline–chloride interactions as seen in Fig. 10 where the oxygen of the urea molecule has an interaction with the hydroxyl hydrogen of choline ions. The corresponding Boltzmann-weighted average and standard deviation of electronic preparation energies expended by choline ions was calculated to be 256 ± 1.4, 238 ± 4.6, 247 ± 1.8 kJ mol−1 molecular clusters [U + 2ChCl + Cl]−, [ThU + 2ChCl + Cl]− and [Gly + 2ChCl + Cl]−, respectively. The [ThU + 2ChCl + Cl]− clusters feature the strongest choline–chloride interaction while expending the least amount of electronic preparation energy by choline ions. The role of urea and glycerol in disrupting the electrostatic interaction network in the charged clusters of choline and chloride ions lead thus to relative reduction of binding energies compared to thiourea-containing clusters. This points to a moderating role thiourea plays in the formation and stabilization of DES opening the possibility for the creation of tailor-made choline-chloride containing DES with HBD that show similar properties to thiourea. The origin of the relative stabilities of molecular clusters containing at least two urea, thiourea or glycerol as HBD compounds, disclosed in CSI-MS in Fig. 6 for n = 1 and m = 2, can be compared to the structures of the total set of 10 lowest-lying conformers of [2U + ChCl + Cl]−, [2ThU + ChCl + Cl]−, and [2Gly + ChCl + Cl]− presented in Fig. S12–S14 (SI), respectively.
 | ||
| Fig. 10 Structures of the 10 lowest-lying conformers of [U + 2ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
 | ||
| Fig. 11 Structures of the 10 lowest-lying conformers of [ThU + 2ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
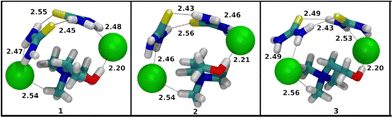
The binding energies of 10 lowest-lying conformers of [2U + ChCl + Cl]−, [2ThU + ChCl + Cl]−, [2Gly + ChCl + Cl]− are presented in Table S8 (SI). The difference in the average binding energy between [2ThU + ChCl + Cl]− and the other two cluster combinations is much larger than in the case of [ThU + ChCl + Cl]− and its counterparts. This corroborates the experimental observation that as size and composition of clusters increase, the clusters containing thiourea are more stable in the gas-phase than their urea and glycerol counterparts. This illustrates the fact that thiourea molecules integrate into the ionic cluster structure much more efficiently than urea or glycerol. With the addition of a urea molecule to [U + ChCl + Cl]−, the complementary interaction between two urea molecules where the oxygen of one urea molecule interacts with the hydrogen of the other urea molecule, is the most noticeable addition to the type of interaction observed in structures of [2U + ChCl + Cl]− as presented in Fig. 13. An analogue interaction was observed in the structures of [2ThU + ChCl + Cl]− clusters in Fig. 14, while this interaction is absent in the [2Gly + ChCl + Cl]− clusters in Fig. 15, which also importantly do not feature any interaction between the hydroxyl hydrogen of choline and the chloride ion being replaced by interaction of the hydroxyl hydrogen of choline with oxygen of glycerol molecules. In choline chloride-containing DESs the interaction between choline and chloride ions is a strong stabilization interaction and its absence contributes to relatively less binding energies observed for [2Gly + ChCl + Cl]− clusters.
 | ||
| Fig. 12 Structures of the 10 lowest-lying conformers of [Gly + 2ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
 | ||
| Fig. 13 Structures of the 10 lowest-lying conformers of [2U + ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
 | ||
| Fig. 14 Structures of the 10 lowest-lying conformers of [2ThU + ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
 | ||
| Fig. 15 Structures of the 10 lowest-lying conformers of [2Gly + ChCl + Cl]− after geometry optimisation. Hydrogen-bond distances in Å. | ||
It's then possible to summarise the interpretations of computational data by mentioning that with the help of binding energy calculations and the local energy decomposition (LED) analysis method, we were able to corroborate the experimental trends of relative gas-phase stability of thiourea containing clusters over urea and glycerol containing clusters as the size of the clusters is increased. When glycerol containing clusters are compared to the thiourea containing clusters, we observe that the destabilizing preparation energy of glycerol reduces the binding energy of glycerol containing clusters. On the other hand, thiourea containing clusters show larger stabilizing interaction energy terms compared to urea containing clusters. Thiourea molecules also facilitate a higher degree of interaction between choline and chloride ions as cluster size increases as compared to urea and glycerol molecules as HBD. This supports the hypothesis from the experimental data that the thiourea molecules integrate into the ionic cluster structure most effectively.
Experimental section
Chemicals
Chemicals used for DES preparation supplied by Sigma-Aldrich with a purity >99%.Preparation
The deep eutectic solvents (DESs) were prepared by heating their components to a temperature between 65 °C to 80 °C for 1 to 4 hours, a duration that depended on the specific mixture. This synthesis yielded viscous liquids with a color ranging from transparent to a pale yellow. Upon completion, the DESs were cooled and stored at room temperature. For NMR analysis, 1 mL of the DES was placed in an NMR tube. A deuterated solvent was then added to a coaxial insert, which was subsequently placed inside the NMR tube. The water content of the eutectic mixtures was determined by mass percentage using a Metrohm Karl Fischer 831 KF Coulometer titrator. A drop of DES was then diluted in 7 mL of methanol before being analyzed by mass spectrometry.Mass spectrometry
All the CSI and ESI mass spectra were acquired on an AccuTOF CS mass spectrometer (JEOL, Tokyo, Japan). Nitrogen was used as a nebulizer and dry gas at flow rates of 1.0 and 2.0 L min−1, respectively. Several source parameters were used depending on both the source set-up and the analyzed sample. They are regrouped in Table S9 (SI). The mass spectra were recorded at a scan rate of one spectra every second in a 50 to 1200 m/z range and with a resolution of 6000 (fwhm definition). The mass drift compensation procedure available on the main program that controls the AccuTOF CS was used for compensating for the m/z drift in the range of m/z 100–1100 and performing the accurate mass measurements.Quantum chemical calculations
To understand the structure and interactions in the DES of interest, the gas-phase structure, energies and properties of the molecular clusters observed in the ESI and CSI experiments of choline-based DES were studied using quantum chemical thermochemistry calculations. The potential energy surface of each molecular cluster was sampled using the CREST software93 with a GFN-FF semiempirical method.94 The configurations were sorted energetically and the 10 lowest-energy ensemble of conformers were optimised with the semi-empirical GFN2-xTB method95 in the CREST software. The resultant geometries of 10 lowest-lying conformers of each molecular cluster were further optimised in the density functional theory (DFT) calculations at the hybrid-functional B3LYP level of theory96 using the augmented 6-311++G(d,p) basis-set from the Pople family of basis-sets. The dispersion interactions were taken into account with the D3 dispersion correction97 and the basis-set superposition error (BSSE) was corrected using the geometric counterpoise (gCP) method98 during the geometry optimisation. This is similar to what has been done in earlier studies of reline clusters.38,91 The resolution of identity (RI) approximation was used to calculate the two-electron integrals in the calculation of the Hartree–Fock (HF) reference electrostatic term (RI-J),99,100 where the Chains-Of-Spheres (COS) approximation was additionally used in the calculation of the two-electron integrals in the reference exchange term (COS-X)101 to speed-up the calculations. The geometry optimisation and the further single-point energy calculation were performed using the quantum chemistry software ORCA 6.0102 which provides a sophisticated method to speed-up the calculation of two-electron integrals described above with the RIJCOSX keyword.103 The single-point energy calculations for the determination of binding energies of the molecular cluster were calculated with coupled-cluster singles and doubles with the perturbative triples (CCSD(T)) method. The Domain-based local pair natural-orbital (DLPNO) method104,105 was used to localise the molecular orbitals on the molecules where they are pre-dominantly situated using the Foster-Boys Scheme.106 The BSSE-corrected binding energies were calculated according to eqn (1) given by the Boys-Bernardi counterpoise method.107| ΔE = [EXYXY(XY) − (EXX(X) + EYY(Y))] − [(EXYX(XY) − EXYX(X)) + (EXYY(XY) − EXYY(Y))] | (1) |
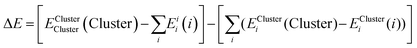 | (2) |
| ΔE = ΔEHFElprep + EHFelstat + EHFexch + EC-CCSDdisp + ΔEC-CCSDnon-disp + ΔEC-(T)int + EGeoprep | (3) |
Conclusions
This work shows that using only electrospray may not be sufficient for characterizing DES ion formation in mass spectrometry. Rather, the additional use of the cold-spray ionization method associated with molecular dynamic simulations can be considered a tool of choice for the chemical characterization of DESs. In this context, it appears from this work that the mass spectrometry of octanoic acid-based DESs where the HBD compound is either menthol or thymol, discloses the ion formation characteristic of supramolecular species. In this case, the charge of the ion interacts most likely with DES precursors and is not dispersed by the high dielectric constant of the cooled solvent. The interpretation of positive CSI mass spectra of the menthol/octanoic acid-based DES indicates that this kind of DES is mainly produced between the menthyloctanoate and the menthol instead of an interaction of the octanoic acid and the menthol. As menthol is substituted by thymol, the use of CSI in negative ion mode is most suitable for the analysis of the corresponding DES, if one refers to the physical and chemical properties of the precursors. In this case the CSI-MS results indicate that the major non-covalent assembly present in solution is a complex formed by octanoic acid and two thymol and where the negative charge is maintained into the DES assemblies through hydrogen bonds with all the DES precursors. CSI mass spectrometry of choline-chloride (ChCl) based DES's allows the state of the ability of the so-called second HBD precursors such as urea (U), thiourea (ThU) and glycerol (Gly), to form DES with cholinium chloride. Indeed, it is through the CSI mass spectra of the choline chloride:thiourea DES that the ions with the higher stoichiometry of each of the precursors are observed, namely through the m/z 680 [3ThU + 3CHCl + Cl]− detected ion. In addition, the studies of the stability in the gas-phase of the DES characteristic ions through in-source CID experiments, disclose that the most stable ions are the [mThU + nCHCl + Cl]− ions (where m = 1 or 2 and n = 1 to 4) with regards to the [mU + nCHCl + Cl]− and [mGly + nCHCl + Cl]− ions (where m = 1 or 2 and n = 1 to 4). The quantum chemical calculations performed on these choline-based DES corroborate these mass spectrometry results, by disclosing that the [mThU + CHCl + Cl]− clusters (where m = 1 or 2) are maintained by larger stabilizing interaction energies than in the [mU + CHCl + Cl]− clusters, whereas the destabilizing steric energy of glycerol reduces the binding energy of the [mGly + CHCl + Cl]−.Author contributions
Emilie Bertrand, Camille Cousseau and Thomas Delhaye have conducted a research and investigation process, specifically performing the experiments, or data/evidence collection in mass spectrometry and chemical synthesis. Swaroop V. S. Kunapuli was involved in the software development, implementation of the computer code and supporting algorithms for the molecular dynamic simulations, and the writing of this part. Rachel Schurhammer and Alain Chaumont had management and coordination responsibilities for the research activity in the molecular dynamic simulations and the writing of this part. Emmanuelle Limanton and Béatrice Legouin developed and designed the methodology for the DES synthesis. Ludovic Paquin had management and coordination responsibilities for the research activity in DES synthesis and the writing. Xavier Castel and Mohammed Himdi managed the coordination between the knowledge of DES properties and the interpretation of the analytical results. The contributions of David Rondeau for this publication were related to the conceptualization, the project administration, the validation of the mass spectrometry results and the writing, the review and the editing of the published work.Conflicts of interest
The authors claim that there are no conflicts of interest to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: all data related to mass spectrometry and molecular simulation dynamics. See DOI: https://doi.org/10.1039/d5cp02383a.Acknowledgements
This work is supported by the European Union through European Regional Development Fund (ERDF), the Ministry of Higher Education and Research, CNRS, Brittany region, Conseils Départementaux d’Ille-et-Vilaine and Côtes d’Armor, Rennes Métropôle and Saint-Brieuc Armor Agglomération, through the CPER Project Mat&Trans.Notes and references
- D. Prat, A. Wells, J. Hayler, H. Sneddon, C. Robert McElroy, S. Abou-Shehada and P. J. Dunn, Green Chem., 2016, 18, 288–296 RSC.
- F. Pena-Pereira and J. Namieśnik, ChemSusChem, 2014, 7, 1784–1800 CrossRef CAS PubMed.
- H. Vanda, Y. Dai, E. G. Wilson, R. Verpoorte and Y. H. Choi, C. R. Chim., 2018, 21, 628–638 CrossRef CAS.
- A. Paiva, R. Craveiro, I. Aroso, M. Martins, R. L. Reis and A. R. C. Duarte, ACS Sustainable Chem. Eng., 2014, 2, 1063–1071 CrossRef CAS.
- M. H. Zainal-Abidin, M. Hayyan, A. Hayyan and N. S. Jayakumar, Anal. Chim. Acta, 2017, 979, 1–23 CrossRef CAS PubMed.
- A. P. Abbott, D. Boothby, G. Capper, D. L. Davies and R. K. Rasheed, J. Am. Chem. Soc., 2004, 126, 9142–9147 CrossRef CAS PubMed.
- E. L. Smith, A. P. Abbott and K. S. Ryder, Chem. Rev., 2014, 114, 11060–11082 CrossRef CAS PubMed.
- A. Sharma, R. Sharma, R. C. Thakur and L. Singh, J. Energy Chem., 2023, 82, 592–626 CrossRef CAS.
- A. P. Abbott, G. Capper, D. L. Davies, R. K. Rasheed and V. Tambyrajah, Chem. Commun., 2003, 70–71 RSC.
- L. J. B. M. Kollau, M. Vis, A. van den Bruinhorst, A. C. C. Esteves and R. Tuinier, Chem. Commun., 2018, 54, 13351–13354 RSC.
- M. A. R. Martins, S. P. Pinho and J. A. P. Coutinho, J. Solution Chem., 2019, 48, 962–982 CrossRef CAS.
- Y. H. Choi, J. van Spronsen, Y. Dai, M. Verberne, F. Hollmann, I. W. C. E. Arends, G.-J. Witkamp and R. Verpoorte, Plant Physiol., 2011, 156, 1701–1705 CrossRef CAS PubMed.
- A. Prabhune and R. Dey, J. Mol. Liq., 2023, 379, 121676 CrossRef CAS.
- S. Handy, in Ionic Liquids – Current State of the Art, ed. S. Handy, InTech, 2015 Search PubMed.
- M. Pätzold, S. Siebenhaller, S. Kara, A. Liese, C. Syldatk and D. Holtmann, Trends Biotechnol., 2019, 37, 943–959 CrossRef PubMed.
- Y. Zhang, X. Ji and X. Lu, Renewable Sustainable Energy Rev., 2018, 97, 436–455 CrossRef CAS.
- A. P. Abbott, Curr. Opin. Green Sustainable Chem., 2022, 36, 100649 CrossRef CAS.
- L. Percevault, E. Limanton, F. Gauffre, C. Lagrost and L. Paquin, in Deep Eutectic Solvents for Medicine, Gas Solubilization and Extraction of Natural Substances, ed. S. Fourmentin, M. Costa Gomes and E. Lichtfouse, Springer International Publishing, Cham, 2021, vol. 56, pp. 241–306 Search PubMed.
- Y. H. Choi and R. Verpoorte, Curr. Opin. Food Sci., 2019, 26, 87–93 CrossRef.
- A. M. Curreri, J. Kim, M. Dunne, P. Angsantikul, M. Goetz, Y. Gao and S. Mitragotri, Adv. Sci., 2023, 10, 2205389 CrossRef CAS PubMed.
- J.-B. Chagnoleau, I. L. Rocha, R. Khedher, J. A. Coutinho, T. Michel, X. Fernandez and N. Papaiconomou, J. Chromatogr. A, 2023, 1691, 463812 CrossRef CAS PubMed.
- C. J. Clarke, W.-C. Tu, O. Levers, A. Bröhl and J. P. Hallett, Chem. Rev., 2018, 118, 747–800 CrossRef CAS PubMed.
- J. Afonso, A. Mezzetta, I. M. Marrucho and L. Guazzelli, Green Chem., 2023, 25, 59–105 RSC.
- O. S. Hammond, D. T. Bowron and K. J. Edler, Green Chem., 2016, 18, 2736–2744 RSC.
- M. Gilmore, L. M. Moura, A. H. Turner, M. Swadźba-Kwaśny, S. K. Callear, J. A. McCune, O. A. Scherman and J. D. Holbrey, J. Chem. Phys., 2018, 148, 193823 CrossRef PubMed.
- M. E. Di Pietro, O. Hammond, A. Van Den Bruinhorst, A. Mannu, A. Padua, A. Mele and M. Costa Gomes, Phys. Chem. Chem. Phys., 2021, 23, 107–111 RSC.
- A. van den Bruinhorst, L. J. B. M. Kollau, M. Vis, M. M. R. M. Hendrix, J. Meuldijk, R. Tuinier and A. C. C. Esteves, J. Chem. Phys., 2021, 155, 014502 CrossRef CAS PubMed.
- O. S. Hammond, D. T. Bowron and K. J. Edler, Angew. Chem., 2017, 129, 9914–9917 CrossRef.
- P. L. Pisano, M. Espino, M. Á. Fernández, M. F. Silva and A. C. Olivieri, Microchem. J., 2018, 143, 252–258 CrossRef CAS.
- I. Delso, C. Lafuente, J. Munoz-Embid and M. Artal, J. Mol. Liq., 2019, 290, 111236–111249 CrossRef CAS.
- S. L. Perkins, P. Painted and C. M. Colina, J. Phys. Chem. B, 2013, 117, 10250–10260 CrossRef CAS PubMed.
- O. S. Hammond, T. D. Bowron and K. J. Edler, Green Chem., 2016, 18, 2736–2744 RSC.
- C. F. Araujo, J. A. P. Coutinho, M. M. Nolasco, S. F. Parker, P. J. A. Ribeiro-Claro, S. Rudic, B. I. G. Soares and P. D. Vaz, Phys. Chem. Chem. Phys., 2017, 19, 17998–18009 RSC.
- S. L. Perkins, P. Painter and C. M. Colina, J. Chem. Eng. Data, 2014, 59, 3652–3662 CrossRef CAS.
- C. R. Ashworth, R. P. Matthews, T. Welton and P. A. Hunt, Phys. Chem. Chem. Phys., 2016, 18, 18145–18160 RSC.
- H. Sun, Y. Li, X. Wu and G. Li, J. Mol. Model., 2013, 19, 2433–2441 CrossRef CAS PubMed.
- G. Garcia, M. Atilhan and S. Aparicio, Chem. Phys. Lett., 2015, 634, 151–155 CrossRef CAS.
- L. Percevault, T. Delhaye, A. Chaumont, R. Schurhammer, L. Paquin and D. Rondeau, J. Mass Spectrom., 2021, 56, e4725 CrossRef CAS PubMed.
- S. Sakamoto, M. Fujita, K. Kim and K. Yamaguchi, Tetrahedron, 2000, 56, 955–964 CrossRef CAS.
- K. Yamaguchi, J. Mass Spectrom., 2003, 38, 473–490 CrossRef CAS PubMed.
- K. Yamaguchi, J. Mass Spectrom. Soc. Japan, 2013, 2, 1–7 Search PubMed.
- S. Sakamoto, T. Imamoto and K. Yamaguchi, Org. Lett., 2001, 3, 1793–1795 CrossRef CAS PubMed.
- E. F. Wilson, H. Abbas, B. J. Duncombe, C. Streb, D. L. Long and L. Cronin, J. Am. Chem. Soc., 2008, 130, 13876–13884 CrossRef CAS PubMed.
- H. N. Miras, E. Wilson and L. Cronin, Chem. Commun., 2009, 1297–1311 RSC.
- S. Sakamoto and K. Yamaguchi, Angew. Chem., Int. Ed., 2003, 4, 905–908 CrossRef PubMed.
- S. Sakamoto, M. Yoshizawa, T. Kusukawa, M. Fujita and K. Yamaguchi, Org. Lett., 2001, 3, 1601–1604 CrossRef CAS PubMed.
- K. Ohara and K. Yamaguchi, Anal. Sci., 2012, 28, 635–637 CrossRef CAS PubMed.
- K. Ohara, M. Tominaga, I. Azumaya and K. Yamaguchi, Anal. Sci., 2013, 29, 773–776 CrossRef CAS PubMed.
- K. Ohara, M. Tominaga, H. Masu, I. Azumaya and K. Yamaguchi, Anal. Sci., 2016, 32, 1347–1352 CrossRef CAS PubMed.
- K. Shikii, S. Sakamoto, H. Seki, H. Utsumi and K. Yamaguchi, Tetrahedron, 2004, 60, 3487–3492 CrossRef CAS.
- X. Xu, J. Qiao, X. Deng, X. N. Na and J. Ouyang, J. Mass Spectrom., 2013, 48, 961–968 CrossRef CAS PubMed.
- E. Bertrand, D. Rondeau, T. Delhaye, X. Castel and M. Himdi, J. Mass Spectrom., 2023, 58, e4977 CrossRef CAS PubMed.
- N. B. Cech and C. G. Enke, Mass Spectrom. Rev., 2001, 20, 362–387 CrossRef CAS PubMed.
- C. L. Gatlin and F. Turecek, Anal. Chem., 1994, 66, 712–718 CrossRef CAS.
- G. J. Van Berkel, F. Zhou and J. T. Aronson, Int. J. Mass Spectrom. Ion Processes, 1997, 162, 55–67 CrossRef CAS.
- J. F. del la Mora, G. J. Van Berkel, C. G. Enke, R. B. Cole, M. Martinez-Sanchez and J. B. Fenn, J. Mass Spectrom., 2000, 35, 939–952 CrossRef.
- S. Zhou, B. S. Prebyl and K. D. Cook, Anal. Chem., 2002, 74, 4885–4888 CrossRef CAS PubMed.
- D. Rondeau, F. Rogalewicz, G. Ohanessian, E. Levillain, F. Odobel and P. Richomme, J. Mass Spectrom., 2005, 40, 628–635 CrossRef CAS PubMed.
- G. J. Van Berkel and V. Kertesz, Anal. Chem., 2007, 79, 5510–5520 CrossRef CAS PubMed.
- B. M. Ehrmann, T. Henriksen and N. B. Cech, J. Am. Soc. Mass Spectrom., 2008, 19, 719–728 CrossRef CAS PubMed.
- M. G. Ikonomou, A. T. Blades and P. Kebarle, Anal. Chem., 1990, 62, 957–967 CrossRef CAS.
- S. Rondinini, P. Longhi, P. R. Mussini and T. Mussini, Pure Appl. Chem., 1987, 59, 1693–1702 CrossRef CAS.
- C. G. Enke, Anal. Chem., 1997, 69, 4885–4893 CrossRef CAS PubMed.
- N. B. Cech and C. G. Enke, Anal. Chem., 2000, 72, 2717–2723 CrossRef CAS PubMed.
- N. B. Cech, J. R. Krone and C. G. Enke, Anal. Chem., 2001, 73, 208–213 CrossRef CAS PubMed.
- J. Hermans, S. Ongay, V. Markov and R. Bischoff, Anal. Chem., 2017, 89, 9159–9166 CrossRef CAS PubMed.
- M. G. Ikonomou, A. T. Blades and P. Kebarle, Anal. Chem., 1990, 62, 957–967 CrossRef CAS.
- M. H. Amad, N. B. Cech, G. S. Jackson and C. G. Enke, J. Mass Spectrom., 2000, 35, 784–789 CrossRef CAS.
- J. Evans, G. Nicol and B. Munson, J. Am. Soc. Mass Spectrom., 2000, 11, 789–796 CrossRef CAS PubMed.
- Z. Tian and S. Kass, Angew. Chem., Int. Ed., 2009, 48, 1321–1323 CrossRef CAS PubMed.
- J. R. Joyce and D. S. Richards, J. Am. Soc. Mass Spectrom., 2011, 22, 360–368 CrossRef CAS PubMed.
- A. Kruve, K. Kaupmees, J. Liigand, M. Oss and I. Leito, J. Mass Spectrom., 2013, 48, 695–702 CrossRef CAS PubMed.
- S. K. Loh, L. Lian and P. B. Armentrout, J. Am. Chem. Soc., 1989, 111, 3167–3176 CrossRef CAS.
- G. J. Van Berkel, J. Am. Soc. Mass Spectrom., 2000, 11, 951–960 CrossRef CAS PubMed.
- T. Henriksen, R. K. Juhler, B. Svensmark and N. B. Cech, J. Am. Soc. Mass Spectrom., 2005, 16, 446–455 CrossRef CAS PubMed.
- T. B. McMahon and P. Kebarle, J. Am. Chem. Soc., 1977, 99, 2222–2230 CrossRef CAS.
- G. Caldwell, R. Renneboog and P. Kebarle, Can. J. Chem., 1989, 67, 611–618 CrossRef CAS.
- Z. Tian and S. R. Kass, J. Am. Chem. Soc., 2008, 130, 10842–10843 CrossRef CAS PubMed.
- Z. Tian, X.-B. Wang, L.-S. Wang and S. R. Kass, J. Am. Chem. Soc., 2009, 131, 1174–1181 CrossRef CAS PubMed.
- G. J. Van Berkel, The Electrolytic Nature of Electrospray, in Electrospray Ionization Mass Spectrometry: Fundamentals, Instrumentation, and Applications, ed. R. B. Cole, Wiley-Interscience, NY, 1997, pp. 65–105 Search PubMed.
- R. B. Cole and J. Zhu, Rapid Commun. Mass Spectrom., 1999, 13, 607–611 CrossRef CAS.
- J. Zhu and R. B. Cole, J. Am. Soc. Mass Spectrom., 2000, 11, 932–941 CrossRef CAS PubMed.
- Y. Cai and R. B. Cole, Anal. Chem., 2002, 74, 985–991 CrossRef CAS PubMed.
- R. D. Bezman, E. F. Casassa and R. L. Kay, J. Mol. Liq., 1997, 73–74, 397–402 CrossRef.
- C. Florindo, F. S. Oliveira, L. P. N. Rebelo, A. M. Fernandes and I. M. Marrucho, ACS Sustainable Chem. Eng., 2014, 2, 2416–2425 CrossRef CAS.
- V. Gabelica, D. Lemaire, O. Laprevote and E. De Pauw, Int. J. Mass Spectrom., 2001, 210–211, 113–119 CrossRef.
- P. Lecchi, J. Zhao, W. S. Wiggins, T. H. Chen, P. F. Yip, B. C. Mansfield and J. M. Peltier, J. Am. Chem. Soc., 2009, 20, 398–410 CAS.
- D. Rondeau, S. Perruchas, N. Avarvari, P. Batail and K. Vékey, J. Mass Spectrom., 2005, 40, 60–65 CrossRef CAS PubMed.
- C. F. Araujo, J. A. P. Coutinho, M. M. Nolasco, S. F. Parker, P. J. A. Ribeiro-Claro, S. Rudić, B. I. G. Soares and P. D. Vaz, Phys. Chem. Chem. Phys., 2017, 19, 17998–18009 RSC.
- S. L. Perkins, P. Painter and C. M. Colina, J. Chem. Eng. Data, 2014, 59, 3652–3662 CrossRef CAS.
- C. R. Ashworth, R. P. Matthews, T. Welton and P. A. Hunt, Phys. Chem. Chem. Phys., 2016, 18, 18145–18160 RSC.
- H. Sun, Y. Li, X. Wu and G. Li, J. Mol. Model., 2013, 19, 2433–2441 CrossRef CAS PubMed.
- P. Pracht, F. Bohle and S. Grimme, Phys. Chem. Chem. Phys., 2020, 22, 7169–7192 RSC.
- S. Spicher and S. Grimme, Angew. Chem., Int. Ed., 2020, 59, 15665–15673 CrossRef CAS PubMed.
- C. Bannwarth, S. Ehlert and S. Grimme, J. Chem. Theory Comput., 2019, 15, 1652–1671 CrossRef CAS PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- S. Grimme, A. Hansen, J. G. Brandenburg and C. Bannwarth, Chem. Rev., 2016, 116, 5105–5154 CrossRef CAS PubMed.
- H. Kruse and S. Grimme, J. Chem. Phys., 2012, 136, 154101 CrossRef PubMed.
- F. Neese, J. Comput. Chem., 2003, 24, 1740–1747 CrossRef CAS PubMed.
- C.-K. Skylaris, L. Gagliardi, N. C. Handy, A. G. Ioannou, S. Spencer and A. Willetts, J. Mol. Struct. THEOCHEM, 2000, 501–502, 229–239 CrossRef CAS.
- F. Neese, F. Wennmohs, A. Hansen and U. Becker, Chem. Phys., 2009, 356, 98–109 CrossRef CAS.
- F. Neese, Software Update: The ORCA Program System—Version 5.0, WIREs Comput. Mol. Sci., 2022, 12, e1606, DOI:10.1002/wcms.1606.
- B. Helmich-Paris, B. De Souza, F. Neese and R. Izsák, J. Chem. Phys., 2021, 155, 104109 CrossRef CAS PubMed.
- M. Gray and J. M. Herbert, J. Chem. Phys., 2024, 161, 054114–1-9 CrossRef CAS PubMed.
- M. Saitow, U. Becker, C. Riplinger, E. F. Valeev and F. A. Neese, J. Chem. Phys., 2017, 146, 164105-1-30 CrossRef CAS PubMed; S. F. Boys, Rev. Mod. Phys., 1960, 32, 296–299 CrossRef.
- S. F. Boys and F. Bernardi, Mol. Phys., 1970, 19(4), 553–566 CrossRef CAS.
- S. F. Boys, Rev. Mod. Phys., 1960, 32(2), 296–299 CrossRef CAS.
- A. Altun, R. Izsák and G. Bistoni, Int. J. Quantum Chem., 2021, 121, e26339 CrossRef CAS.
- A. Altun, I. F. Leach, F. Neese and G. Bistoni, Angew. Chem., Int. Ed., 2025, 64, e202421922 CrossRef CAS PubMed.
- G. Bisto, A. Altun, Z. Wang and F. Neese, Acc. Chem. Res., 2024, 57, 1411–1420 CrossRef PubMed.
| This journal is © the Owner Societies 2026 |