Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Minimal-tag polyamines for in situ probing of their intracellular distribution
Zuzanna Sas a,
Maciej Zakrzewski
a,
Maciej Zakrzewski a,
Agnieszka D. Wardak
a,
Agnieszka D. Wardak a,
Przemysław Wanat
a,
Przemysław Wanat b,
Wojciech Czestkowski†
b,
Justyna Zmorzyńska
b,
Wojciech Czestkowski†
b,
Justyna Zmorzyńska a,
Anna A. Marusiak
a,
Anna A. Marusiak *a and
Remigiusz A. Serwa
*a and
Remigiusz A. Serwa *a
*a
aIMol Polish Academy of Sciences, 02-247 Warsaw, Poland. E-mail: a.marusiak@imol.institute; r.serwa@imol.institute
bMolecure SA, 02-089 Warsaw, Poland
First published on 12th December 2025
Abstract
Propargyl polyamines (putrescine, spermidine, and spermine) were synthesized and evaluated as minimal-tag analogs for high-resolution intracellular imaging in mammalian cells and zebrafish. These compounds showed structure-dependent differences in subcellular distribution with unprecedented resolution, discriminating among closely related polyamines.
The polyamines (PAs) putrescine (PUT), spermidine (SPD), and spermine (SPM) are abundant polycations (Fig. 1a) that bind nucleic acids, membranes, and proteins, shaping chromatin and RNA, supporting translation, and tuning organelle function.1 They exist mainly in a biopolymer-bound, functionally engaged pool, with a small freely diffusible fraction. PA homeostasis is maintained through coordinated biosynthesis, catabolism, uptake, secretion, and intracellular trafficking.2 Disturbance of this homeostasis is a hallmark of cancer and neurological disorders, and mutations in genes of the PA pathways (e.g., ATP13A2, SMS) implicate these pathways in disease pathogenesis.3,4
Existing PA detection strategies have complementary strengths but key limitations. Fluorophore-PA conjugates (e.g., BODIPY derivatives)5,6 suit monitoring PA uptake via the PA transport system and have been shown to enable transporter discovery/validation (e.g., ATP13A2),7 yet the bulky dye likely perturbs trafficking and fails to report subcellular localization. Genetically encoded sensors provide single-cell, real-time readouts of intracellular levels of PAs, mainly of SPD/SPM, and can be targeted to cellular compartments, but they quantify free pools rather than specify structure-specific localization patterns.8,9 Methods based on PA-reactive small molecules (e.g., glycine propargyl esters or pyrrolopyrrole aza-BODIPY-based sensors) sensitively assess total cellular PA levels but do not distinguish between PUT, SPD, and SPM.10–12 Finally, chromatographic/mass-spectrometric assays remain ensemble, extraction-based measurements lacking spatial resolution.13
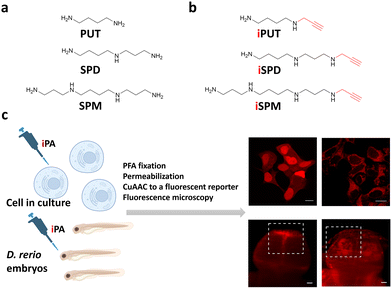
Here, we introduce minimal-tag polyamines (iPAs), specifically propargyl analogs of PUT, SPD, and SPM (iPUT, iSPD, iSPM, respectively; Fig. 1b), that preserve native charge and spacing, and retain the transporter compatibility of the untagged PAs. After paraformaldehyde (PFA) fixation, the alkyne of the propargyl group is labeled by copper-catalyzed azide–alkyne cycloaddition (CuAAC)14,15 with a fluorescent azide, enabling high-resolution intracellular imaging (Fig. 1c). In contrast to conjugation of dyes to PAs, these minimal edits resolve structure-dependent accumulation patterns among closely related PAs in cultured cancer cells and zebrafish embryos. Unlike bulky dye conjugates, the three different iPAs localized to distinct subcellular regions (or to different cell types) and displayed differences in cellular efflux and retention. Complementing real-time genetic sensors that report on free polyamine pools, our polyamine analogs show species-specific uptake and subcellular accumulation while retaining the properties of the corresponding untagged PAs.
iPAs were synthesized from polyamine building blocks previously reported by our group.16 A convergent route introducing a single propargyl tag followed by global deprotection afforded iPUT, iSPD, and iSPM in overall yields of 84%, 68%, and 28%, respectively (Fig. S1).
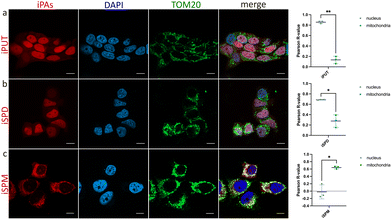
We applied iPAs to the human breast cancer cell line MCF-7, a model in which PA transport has been benchmarked with BODIPY-PA surrogates.17 In parallel, MCF-7 cells were treated with PAs (negative controls) and with commercially available BODIPY-PAs. Imaging revealed differential accumulation of iPAs, iPUT and iSPD localized predominantly to the nucleus, whereas iSPM showed an extranuclear pattern with strong colocalization to the mitochondrial outer membrane marker TOM20 (Fig. 2). As expected, cells treated with native PAs, PFA-fixed and subjected to CuAAC showed no detectable fluorescence (Fig. S2), confirming the specificity of iPA-derived signals. In contrast, all BODIPY-PAs displayed similar extranuclear patterns (Fig. S3), consistent with a previous study utilizing these compounds in MCF-7 cells.17 We then performed an analogous colocalization study using the cervical cancer cell line HeLa and obtained results very similar to those observed using MCF-7 (Fig. S4). The divergent intracellular localization reflected distinct routing—with BODIPY-PAs remaining extranuclear, and specifically localized to the cytoplasm,17 and at least partially accumulated in acidic polyamine-sequestering vesicles,6 but with iPAs entering both cytosolic and nucleoplasmic pools, consistent with protein interactors recovered using spermidine-based photoaffinity probes.16 The prominent colocalization of iSPM with TOM20 may have stemmed from electrostatics and chain length: the higher net charge and increased spacing of amine groups in SPM appeared to have facilitated its multivalent association with anionic mitochondrial membranes, in contrast to the shorter, less charged SPD and PUT.18,19 An additional factor underlying the observed differences in subcellular localization among iPAs may have been the involvement of multiple transporters within the polyamine transport system, which exhibit diverse substrate preferences and organelle-specific expression.2
To verify that intracellular iPAs do not undergo any appreciable metabolic or catabolic transformation that would otherwise yield a mixture of polyamines,1 we developed a mass-spectrometry-based assay. In this assay, cells treated with iPA are subsequently lysed, and solvent-exposed cysteines in cellular proteins react with alkyne-bearing iPAs released from the cells and with azide-PEG3-biotin, added to the lysis buffer, via a copper-catalyzed azide–alkyne–thiol reaction,20 to produce thio-triazole biotinylation sites. Proteins are digested with trypsin, the biotinylated peptides are enriched and analyzed using LC-MS/MS. The assay selectively detects species that retain the bioorthogonal alkyne tag in cells; if the alkyne is metabolically removed, CuAAC reactivity is extinguished and does not confound the microscopy-based steady-state readout of intracellular accumulation. In the current work, the analysis showed the observed cysteine mass shifts overwhelmingly corresponding to incorporation of the intact iPA, with any potential interconversion of these compounds occurring at a minimal level (Fig. S5).
Subsequent time-course experiments showed that intracellular accumulation of iPA increased with incubation time (Fig. S6). iPUT produced a clear signal at 15 min of incubation, whereas iSPD and iSPM required longer incubations, consistent with slower cellular entry. Dose–response studies revealed accumulation proportional to dose (Fig. S7). Importantly, neither changing time nor dose yielded altered intracellular localization patterns.
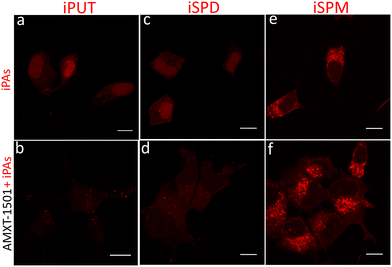
AMXT-1501 is a polyamine transport inhibitor currently under clinical evaluation in oncology.21 After a brief pretreatment of MCF-7 cells with this compound, we observed partial inhibition of intracellular accumulation of iPAs, particularly for iPUT and iSPD, whereas the effect on iSPM was minimal (Fig. 3 and Fig. S8). This substrate profile was found to be consistent with reports of AMXT-1501 limiting P5B-ATPase–dependent polyamine transport, assayed using BODIPY-PAs in HEK293T cells stably expressing polyamine transporter ATP13A3.8 Notably, AMXT-1501 also altered subcellular distribution: both iSPD and iPUT lost their nuclear localization, and iPUT displayed a punctate cytoplasmic pattern, indicating relocalization to the cytoplasm. Together, these observations pointed to PA-species–dependent differences in trafficking that have been difficult to resolve using other analytical methods.
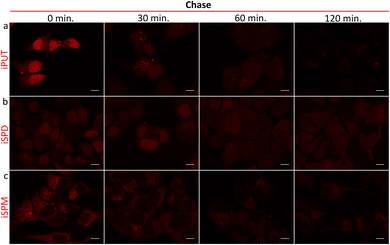
To assess retention of iPAs in MCF-7 cells, we performed pulse-chase assays (incubation followed by washout) and observed distinct, compartment-specific decay kinetics (Fig. 4 and Fig. S9): the nuclear iPUT signal declined rapidly while the cytoplasmic iPUT signal decreased more slowly; for iSPD, nuclear and cytoplasmic signals decayed at similar rates; and the cytoplasmic iSPM signal persisted the longest. To provide evidence for efflux from the cells as being the source of the declining signal, we collected conditioned medium from the iPUT pulse-chase and applied it to naïve cells: this application yielded a robust, iPUT-like nuclear pattern (Fig. S10), consistent with release of import-competent compound and intercellular transfer. These data showed that iPAs can be used to visualize dynamic intercellular transfer of polyamines across cell populations, a phenomenon noted previously but largely only inferred from the presence of secretion/uptake machinery and polyamine-stress transcriptional readouts in heterogeneous tissues rather than having been visualized directly.2 Prior work indicated that mammalian cells actively store and secrete polyamines, and implicated specific solute carrier proteins (SLCs) in these processes: SLC18B1 loads SPD/SPM into secretory vesicles, enabling regulated release,22 and SLC3A2 has been implicated in diamine export to the medium.23 In a separate study, we plan to verify the involvement of these proteins in iPA efflux and use iPAs to investigate polyamine-mediated chemical transmission.
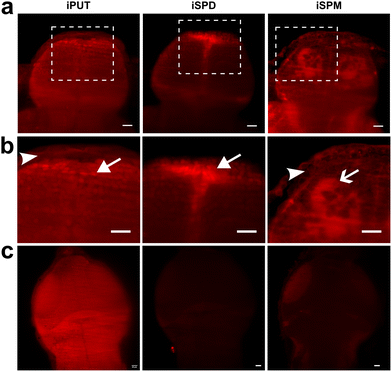
Finally, in the current work, we evaluated the applicability of iPAs to probe the localization of exogenous PAs in vivo in zebrafish larvae (48–120 hpf), with a focus on the developing brain. Polyamines play essential, conserved roles in the brain, including modulation of ion channels and neurotransmitter receptors, regulation of synaptic plasticity, neurodevelopment, and glia-neuron signaling, yet the molecular and spatial mechanisms underlying these roles remain incompletely defined.24 Zebrafish embryos were exposed to iPAs at 48 hpf; and, at 5 dpf, larvae were euthanized, PFA-fixed, permeabilized, and labeled by CuAAC with 5-TAMRA-azide. iPA-dependent fluorescence was clearly above the background in control samples, and accumulation patterns were dependent on iPA structure (Fig. 5 and Fig. S11). Importantly, subcellular localization was tissue specific: brain cells (presumably neurons) showed predominantly nuclear iPUT and iSPD, whereas skin cells exhibited mainly extranuclear iPUT and little to no iSPD (Fig. 5). iSPM localized primarily to the cytoplasm and was observed in both neurons and skin cells. In addition, iSPD was absent from the tectum and cerebellum but was present in the pallium, while iPUT showed nuclear localization and iSPM localized to the cytoplasm of tectal cells, with enrichment in white matter, similar to that in the pallium (Fig. 5). Collectively, the results of our experiments established iPAs as effective tools for visualizing exogenous PA localization in vivo. Importantly, the differential accumulation of iPUT, iSPD, and iSPM across cell types (neurons vs. skin cells) and brain regions (pallium, tectum, cerebellum) demonstrated the ability of iPAs to resolve spatial heterogeneity in PA dynamics. These findings are expected to provide a basis for the future use of iPAs in brain research, including efforts to define how polyamines shape neurodevelopment, synaptic plasticity, and glia-neuron interactions.
In this communication, we present minimally tagged, polyamine analogs for profiling polyamine uptake and intracellular accumulation in cells. We show structure-dependent quantitative and qualitative differences in accumulation and efflux. Using a polyamine transport inhibitor, we demonstrate that iPAs engage the same import pathways as do native polyamines and remain chemically intact under intracellular conditions. We also show that iPAs not only readily enter but also exit cells, enabling future studies on the role of polyamines in intercellular communication. We further demonstrate the applicability of iPAs in an in vivo zebrafish model. Our data indicate that iPA accumulation in zebrafish is iPA-structure dependent and, for a given compound, cell-type dependent. We expect that our minimal-tag polyamines will accelerate mechanistic studies of polyamine biology and enable spatially resolved analyses in disease models in cellulo and in vivo. We anticipate a straightforward adaptation of iPAs to high-throughput formats that interrogate polyamine import and subcellular trafficking. In such settings, these compounds are expected to facilitate screening and quantitative evaluation of therapeutics targeting polyamine pathways.
Author contributions
Conceptualization: RAS. Methodology: RAS, ZS. Investigation: ZS, MZ, PW, WC, ADW. Data analysis: ZS, JZ, AM, RAS. Funding acquisition: RAS. Supervision: RAS, AM, JZ. Writing: RAS with help of ZS, AM, JZ.Conflicts of interest
There are no conflicts to declare.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cc06304c.In addition, cell imaging data and LC-MS/MS data have been uploaded to the public repositories Zenodo (17964427, DOI: 10.5281/zenodo.17529265) and ProteomeXchange25 (PXD070115, https://www.ebi.ac.uk/pride/archive/projects/PXD070115) via PRIDE partner,26 respectively.
Acknowledgements
HRMS and LC-MS/MS measurements were performed at the Proteomics Core Facility, IMol Polish Academy of Sciences. We thank Mrs Viktoriia Lastivka for sample preparation and measurements. This research was funded by the National Science Centre, Poland grant (2020/38/E/ST4/00250). Text editing was performed using the OpenAI ChatGPT-5 environment.References
- A. E. Pegg, J. Biol. Chem., 2016, 291, 14904–14912 CrossRef CAS PubMed.
- M. Azfar, S. van Veen, M. Houdou, N. N. Hamouda, J. Eggermont and P. Vangheluwe, Biochim. Biophys. Acta, Mol. Cell Res., 2022, 1869, 119354 CrossRef CAS PubMed.
- A. Ramirez, A. Heimbach, J. Gründemann, B. Stiller, D. Hampshire, L. P. Cid, I. Goebel, A. F. Mubaidin, A.-L. Wriekat, J. Roeper, A. Al-Din, A. M. Hillmer, M. Karsak, B. Liss, C. G. Woods, M. I. Behrens and C. Kubisch, Nat. Genet., 2006, 38, 1184–1191 CrossRef CAS PubMed.
- C. Li, J. M. Brazill, S. Liu, C. Bello, Y. Zhu, M. Morimoto, L. Cascio, R. Pauly, Z. Diaz-Perez, M. C. V. Malicdan, H. Wang, L. Boccuto, C. E. Schwartz, W. A. Gahl, C. F. Boerkoel and R. G. Zhai, Nat. Commun., 2017, 8, 1257 CrossRef.
- M. Houdou, N. Jacobs, J. Coene, M. Azfar, R. Vanhoutte, C. Van den Haute, J. Eggermont, V. Daniëls, S. H. L. Verhelst and P. Vangheluwe, Biomolecules, 2023, 13, 337 CrossRef CAS.
- D. Soulet, B. Gagnon, S. Rivest, M. Audette and R. Poulin, J. Biol. Chem., 2004, 279, 49355–49366 CrossRef CAS.
- S. van Veen, S. Martin, C. Van den Haute, V. Benoy, J. Lyons, R. Vanhoutte, J. P. Kahler, J.-P. Decuypere, G. Gelders, E. Lambie, J. Zielich, J. V. Swinnen, W. Annaert, P. Agostinis, B. Ghesquière, S. Verhelst, V. Baekelandt, J. Eggermont and P. Vangheluwe, Nature, 2020, 578, 419–424 CrossRef CAS.
- R. Tamura, J. Chen, M. De Jaeger, J. F. Morris, D. A. Scott, P. Vangheluwe and L. L. Looger, 2024 DOI:10.1101/2024.08.21.609037.
- P. Sharma, C. Y. Kim, H. R. Keys, S. Imada, A. B. Joseph, L. Ferro, T. Kunchok, R. Anderson, Y. Sun, Ö. H. Yilmaz, J.-K. Weng and A. Jain, Nat. Commun., 2025, 16, 4921 CrossRef CAS.
- K. K. H. Vong, K. Tsubokura, Y. Nakao, T. Tanei, S. Noguchi, S. Kitazume, N. Taniguchi and K. Tanaka, Chem. Commun., 2017, 53, 8403–8406 RSC.
- W. Li, L. Wang, T. Sun, H. Tang, B. Bui, D. Cao, R. Wang and W. Chen, Commun. Biol., 2021, 4, 803 CrossRef CAS PubMed.
- Y. Gan, B. Lu, J. Zhong, X. Ran, D. Cao and L. Wang, Biosensors, 2025, 15, 542 CrossRef CAS PubMed.
- C. Magnes, A. Fauland, E. Gander, S. Narath, M. Ratzer, T. Eisenberg, F. Madeo, T. Pieber and F. Sinner, J. Chromatogr. A, 2014, 1331, 44–51 CrossRef CAS.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS.
- C. W. Tornøe, C. Christensen and M. Meldal, J. Org. Chem., 2002, 67, 3057–3064 CrossRef.
- M. Zakrzewski, Z. Sas, B. Cocom-Chan, M. E. R. Firdaus, M. Kałek, K. Szczepanowska, P. Gerlach, A. Marusiak and R. A. Serwa, RSC Chem. Biol., 2025, 6, 1787–1796 RSC.
- S. van Veen, A. Kourti, E. Ausloos, J. Van Asselberghs, C. Van den Haute, V. Baekelandt, J. Eggermont and P. Vangheluwe, Biomolecules, 2023, 13, 918 CrossRef CAS PubMed.
- M. W. Yung and C. Green, Biochem. Pharmacol., 1986, 35, 4037–4041 CrossRef CAS PubMed.
- L. Cabrini, B. Tadolini, L. Landi, D. Fiorentini and A. M. Sechi, Comp. Biochem. Physiol. B, 1989, 93, 647–651 CrossRef CAS.
- A. Wiest and P. Kielkowski, J. Am. Chem. Soc., 2024, 146, 2151–2159 CrossRef CAS PubMed.
- S. A. Piha-Paul, A. W. Tolcher, A. L. Vandross, A. I. Spira and M. R. Burns, ESMO Open, 2025, 10, 105576 CrossRef CAS PubMed.
- M. Hiasa, T. Miyaji, Y. Haruna, T. Takeuchi, Y. Harada, S. Moriyama, A. Yamamoto, H. Omote and Y. Moriyama, Sci. Rep., 2014, 4, 6836 CrossRef.
- T. Uemura, H. F. Yerushalmi, G. Tsaprailis, D. E. Stringer, K. E. Pastorian, L. Hawel, C. V. Byus and E. W. Gerner, J. Biol. Chem., 2008, 283, 26428–26435 CrossRef CAS.
- M. G. Makletsova, G. T. Rikhireva, E. Y. Kirichenko, I. Y. Trinitatsky, M. Y. Vakulenko and A. M. Ermakov, Neurochem. J., 2022, 16, 283–294 CrossRef CAS.
- E. W. Deutsch, Nucleic Acids Res., 2023, 51, D1539–D1548 CrossRef PubMed.
- Y. Perez-Rivero, Nucleic Acids Res., 2025, 53, D543–D553 CrossRef.
Footnote |
| † Current address: Ascentia Sp. z o.o., 93-465 Łódź, Poland. |
| This journal is © The Royal Society of Chemistry 2026 |