Protein–DNA interactions in disease and drug discovery
Richa
Mishra
 and
Shandar
Ahmad
and
Shandar
Ahmad
 *
*
School of Computational and Integrative Sciences, Jawaharlal Nehru University, New Delhi, India. E-mail: shandar@jnu.ac.in
First published on 29th December 2025
Abstract
Protein–DNA interactions (PDIs) are fundamental to all organisms and are often involved in the onset, progression, and severity of diseases or in defence against them. However, their use in drug targeting has remained challenging due to many reasons, including the electrostatic and non-specific interactions of the omnipresent DNA backbone. Nevertheless, PDIs, including regulatory transcriptional events and pathogen-sensing by hosts, have remained critical in drug discovery, and their potential as direct drug targets has been increasingly recognised. In this review article, we survey three key aspects of PDIs in humans, namely transcription, replication-and-repair, and genome organisation, whose misregulation has been implicated in various diseases, thereby highlighting PDIs as viable therapeutic targets. We provide a comprehensive list of targets and drugs used in drug discovery, that have reached the clinical trial or approval stage. We also review the computational methods, including AI-based approaches, that are powering these developments. We observe that, despite the general notion of PDIs being treated as undruggable, the literature shows that the time to use them as effective targets has come, as reflected by the growing number of candidate drugs across these categories.
1 Introduction
Protein–DNA interactions (PDIs) refer to the specific or non-specific binding of proteins to DNA sequences. They can affect a range of cellular functions, including gene expression, DNA replication, DNA repair, chromatin remodelling, and cell cycle regulation.1,2 Disruption of PDIs is a hallmark of many diseases, particularly cancer, where dysregulation of transcription factors (TFs) such as c-Myc, p53, and nuclear factor kappa B (NF-κB), or DNA repair proteins like poly(ADP-ribose) polymerase (PARP) and RAD51, drives tumourigenesis and highlights PDIs as potential therapeutic targets.2–8PDIs have been widely studied using experimental and computational techniques.9 Some of the many questions surrounding PDIs relate to molecular specificity, detailed atomic interactions, identification of amino acid or nucleotide “binding sites”, interaction energetics, and the spatial and temporal order of binding partners.10–16 Identification of binding sites is of particular interest from a drug discovery perspective. In this regard, an early experimental method for the quantitative characterisation of PDIs was DNase I footprinting and titration, developed in the 1980s, which laid the groundwork for mapping DNA–protein binding sites.17 Since then, numerous methods have been developed to study these interactions at multiple levels. For example, electrophoretic mobility shift assays (EMSA) provide evidence of protein–DNA binding in vitro.18 Chromatin immunoprecipitation followed by sequencing (ChIP-seq) enables genome-wide mapping of binding events.19 Surface plasmon resonance (SPR) enables real-time measurement of biomolecular interactions and their kinetics at the ensemble level.20 In contrast, single-molecule nanopore sensing detects PDIs by monitoring ionic current changes as biomolecules interact with or transit through a nanoscale pore, providing direct insight into binding kinetics, conformational dynamics, and molecular heterogeneity that are inaccessible to bulk methods.21–25 More elaborate atomistic insights come from structure-based approaches such as high-resolution X-ray crystallography26 and nuclear magnetic resonance (NMR) spectroscopy, as well as27 molecular assembly-level cryo-electron microscopy (cryo-EM).28 High-throughput experimental methods include sequence-based techniques such as cleavage under targets and tagmentation (CUT&Tag)29 and DNA affinity purification sequencing (DAP-seq),30 as well as optical biosensing methods like bio-layer interferometry (BLI).31
Experimental data generated from these methods for disease-associated systems and their controls have been crucial in advancing our understanding of PDIs and their role in various cellular states, including disease onset, progression, and intervention.6,32–35
However, obtaining such data is often complex for every biological context and cannot be effectively interpreted without a robust infrastructure to store, model, and predict biological meaning from them. Therefore, large biological databases have been developed to compile experimental results on PDIs. The Protein Data Bank (PDB)36,37 contains nearly 5000 human protein–DNA complexes, while Gene Ontology (GO)38,39 annotations list over 14000 nucleic acid-binding proteins (accessed via UniProt40) (GO:0003676), including more than 9000 with DNA-binding activity (GO:0003677) and approximately 3500 with sequence-specific DNA-binding (GO:0043565). Despite this extensive coverage, many interactions lack detailed structural and mechanistic characterisation and therefore depend on computational approaches, which continue to be refined to enhance their predictive accuracy and biological interpretability.
Experimental data and computational analyses reveal that there are three general ways to modify PDIs, e.g. for a therapeutic intent. These include (1) designing molecules that bind to DNA-binding proteins, (2) directly engaging DNA, or (3) interfering with the protein–DNA complex.41–45 Each of these approaches presents unique challenges. For example, DNA-binding proteins often lack well-defined ligand-binding pockets and were therefore long considered non-druggable.46 On the other hand, compounds that interact directly with DNA, including intercalators, groove binders, and alkylating agents, usually display poor sequence selectivity and substantial systemic toxicity.47 Finally, agents that trap transient DNA–protein complexes can cause widespread DNA damage and promote resistance.48 Together, these challenges have contributed to the slow progress in targeting PDIs therapeutically, although the situation is gradually changing. For example, in the case of transcription factors, targeting domains other than the canonical DNA-binding region has been effective, as seen with estrogen and androgen receptor inhibitors such as fulvestrant and enzalutamide.49,50 Targeting via modes other than small molecules has also shown promise; for instance, the mini-protein Omomyc disrupts Myc–Max dimerisation and prevents DNA binding, effectively blocking transcriptional activity,51 while proteolysis targeting chimeras (PROTACs) degraders like bavdegalutamide (ARV-110) promote targeted degradation of DNA-binding proteins.52 The use of small molecules targeting replication or repair proteins through catalytic inhibition, such as olaparib and talazoparib, represents another successful approach.53,54 Chromatin modulators such as vorinostat and tazemetostat provide an epigenetic mode of modulation.55,56 Combination therapies are also being increasingly applied to mitigate off-target effects and enhance therapeutic selectivity.57–61 Thus, PDIs represent an important biomolecular system that holds great promise in solving the next generation of drug discovery problems. In this review, we discuss the current state of knowledge in the field of PDIs that forms the basis of this promise. We highlight the key molecular and structural principles governing protein–DNA recognition (Fig. 1), explore how dysregulation of PDIs contributes to human disease, and examine current and emerging strategies for their therapeutic targeting. We also outline key challenges associated with druggability, specificity, and resistance and discuss future perspectives on how advances in structural biology, chemical biology, and AI-driven modelling may enable more selective and effective modulation of PDIs. Building on this overview, the following section explores the structural features that determine how proteins recognise and bind to DNA.
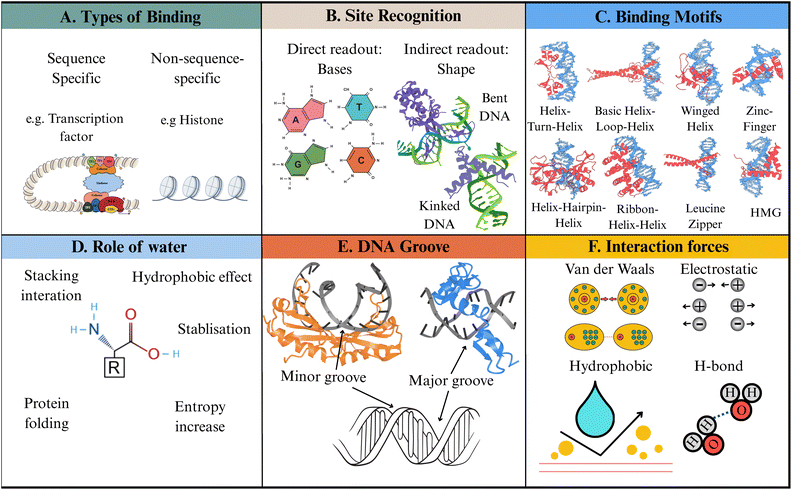 | ||
| Fig. 1 Structural elements of protein–DNA interactions. (A) Types of binding modes. (B) Site recognition: direct readout through base-specific contacts in the major groove, and indirect readout through DNA shape, flexibility, and groove geometry (PDB: 1P51,62 1CKT63). (C) Binding motifs: schematic illustrations of common DNA-binding motifs. (D) Role of water: water-mediated stabilisation via bridging hydrogen bonds, hydrophobic effects, stacking interactions, and entropic contributions. (E) DNA grooves: minor-groove binding (PDB: 1CDW)64 and major-groove binding (PDB: 1AAY).65 (F) Interaction forces: key forces stabilising protein–DNA complexes. Created with icons from BioIcons66 and Servier Medical Art (smart.servier.com), licensed under CC BY 4.0; protein visualisations generated via The Protein Imager67 and Mol*.68 | ||
1.1 Structural features governing protein–DNA recognition
Protein interactions with target DNA molecules can occur either specifically, depending on the DNA sequence and typically involving contacts between protein side chains and nucleotide bases (or sequence-dependent DNA shape features), or non-specifically, through interactions between protein side chains and the sugar–phosphate backbone of the DNA molecule. DNA-binding proteins such as TFs usually exhibit specific binding, whereas histones primarily display non-sequence-specific binding mediated by electrostatic contacts with the DNA backbone.69–71 The DNA replication process also displays a combination of both binding modes: initiator proteins bind specifically to origins of replication (ORI) sequences, while the subsequent machinery, including DNA polymerase and helicase, associates with particular DNA structures (such as the replication fork) rather than specific sequences.72For a protein (typically a TF or DNA-sensing immune response factor) to specifically bind to DNA, it must read the bases paired within the double helix.82 The major groove of DNA is deep, wide, and highly exposed, thereby featuring an easily recognisable pattern of hydrogen-bond donors and acceptors created by the bases. In contrast to the minor groove, this enhances the specificity of binding because hydrogen bonds can be more easily formed between the DNA bases and the protein's side chains. In the minor groove, which is shallow and narrow, binding occurs through electrostatic interactions and sequence-dependent shape complementarity, often influenced by a higher AT content. The high negative potential attracts positively charged arginine and lysine residues on the protein molecule, although arginine is more enriched (60%) compared to lysine (22%) because it forms more hydrogen bond contacts.83 Recent studies show that DNA-binding specificity depends on groove recognition and balanced hydrogen-bonding with both DNA strands.70 Some proteins also show flexibility in groove-binding preference upon post-translational modifications, for example, protamines bind preferentially to the minor groove in their unphosphorylated state, but the binding shifts to the major groove after phosphorylation.84 Given that these binding interactions take place in an aqueous environment, water plays an essential role in PDIs.85 Water molecules in DNA form various arrangements in the major and minor grooves; during binding, some of these water molecules are displaced, contributing to the hydrophobic effect, where the release of ordered water molecules increases the system entropy, making the interaction thermodynamically favourable and stabilising the complex. The specific groove hydration patterns and base-stacking interactions significantly contribute to the stability of different DNA conformations such as A-, B-, Z-DNA or G4. The hydrophobic effect also influences protein folding, as hydrophobic amino acids (such as leucine, isoleucine, and phenylalanine) tend to be located inside the protein core, whereas hydrophilic amino acids generally face outward toward the aqueous environment.82,85,86
Zipper-type motifs have α-helices from two subunits which “zip” together via hydrophobic interactions. In leucine zippers, leucine residues occur at every seventh position in a coiled-coil segment, with the N-terminal basic region extending into the DNA major groove. In contrast, in helix-loop-helix proteins, the DNA-binding and dimerisation helices are separated by a flexible loop.88,96
Histones and high-mobility group (HMG) proteins also use helices for DNA binding and are generally not sequence-specific.88 While the former is primarily involved in DNA packaging, the latter plays a variety of regulatory roles.72 Other motifs, such as the helix-hairpin-helix (involved in base excision and mismatch repair) and the ribbon-helix-helix (found in bacterial Arc repressors), also contribute to DNA recognition during repair and regulation.72,88
DNA-binding motifs are widespread in TFs; for example, the CREB family contains leucine zippers, whereas helix-loop-helix motifs can be found in the mouse and human forms of Max, human USF proteins, and others.72
While structural features enable precise protein–DNA recognition under normal conditions, mutations or alterations in these features can disrupt interactions and lead to disease, as discussed in the following section.
2 Disease-causing dysregulation of PDIs
Mutations in proteins or their target genomic DNA that alter PDIs can disrupt normal gene regulation and genome organisation, leading to detrimental effects on cellular processes such as transcription, replication, and DNA repair. Such disruptions are linked to various diseases.6 For example, missense mutations in DNA-binding proteins have been associated with severe conditions including cancer and neurodevelopmental disorders.97 Studying the effects of these mutations is, therefore, critical for understanding disease mechanisms and developing targeted treatment strategies. The following section explores diseases associated with DNA-binding proteins as well as DNA itself, including non-canonical structures. We have grouped PDI-associated diseases into those involving (1) transcription factors, (2) replication and repair, (3) non-canonical DNA elements, and (4) genome packaging and architectural integrity. Specific examples of disease-causing disruptions in these systems are outlined below.2.1 Diseases linked to transcription factors (TFs)
TFs regulate gene expression by binding to specific DNA sequences and coordinating transcriptional programs essential for cellular identity, development, and environmental responsiveness. Disruption of TF–DNA interactions can have profound pathological consequences, including developmental disorders, inflammatory and metabolic conditions, and cancer.98Developmental TFs are particularly sensitive to perturbations in DNA binding. The paired box (PAX) family of TFs, including PAX3, PAX4, PAX5, PAX6, and PAX7, plays a central role in organogenesis. These transcriptional regulators contain a paired domain (PD) for DNA binding, consisting of an HTH motif that inserts into the major groove of DNA. Some PAX proteins, such as PAX3 and PAX6, also possess an additional homeodomain that provides an extra DNA contact point, increasing binding stability and specificity. PAX homeodomains favour TAAT-containing motifs and can bind paired TAAT sites cooperatively, enabling recognition of regulatory sequences that are usually inaccessible to other homeodomain factors.99 The functional consequences of PAX6 mutations depend on whether they destabilise the paired domain or, instead, introduce more subtle alterations in DNA-binding affinity or specificity. Such disruptions in PAX–DNA interactions can misregulate key gene networks, leading to developmental disorders such as Waardenburg syndrome, renal-coloboma syndrome, and ocular defects, including aniridia and cataracts.2 Aberrant PAX–DNA interactions are also implicated in tumourigenesis, contributing to diseases such as alveolar rhabdomyosarcoma, renal cell carcinoma, and melanoma.2,99
Signal-responsive TFs also represent an important class of disease-associated PDIs. Signal transducer and activator of transcription 3 (STAT3) regulates gene expression by binding to interferon-responsive DNA elements. STAT3 specifically recognises the consensus DNA sequence (TTCNNNGAA), and its DNA-binding affinity is enhanced by cytokine-induced phosphorylation that promotes STAT3 dimerisation.100 Structurally, STAT3 consists of a DNA-binding domain (DBD), an Src homology 2 (SH2) domain, a transactivation domain, and a coiled-coil domain (CCD). As a signalling molecule, dysregulation of STAT3 disrupts multiple cellular processes, including altered chondrocyte metabolism in osteoarthritis, Interleukin-6 (IL-6)-driven inflammation in atherosclerosis, and the dysregulation of fibrosis-related genes in myocardial fibrosis. Because STAT3 activation depends on cytokine signalling, aberrant STAT3 activity is also linked to asthma, autoimmune diseases, Alzheimer's disease, and breast cancer.101
Nuclear hormone receptors constitute another important group of TFs whose disease associations arise from altered DNA binding. Estrogen receptor (ER) and androgen receptor (AR) bind hormone response elements composed of AGGTCA half-sites arranged as inverted or direct repeats. Ligand binding induces conformational changes that enhance DNA-binding affinity and coactivator recruitment.88 IL-6 modulation of the AR coactivator p300 within the AR transcription complex is linked to prostate cancer. Androgens are also implicated in prostate diseases such as benign prostatic hyperplasia, while androgen deficiency is linked to muscular atrophy. ER-mediated transcription through estrogen response elements on DNA is involved in polycystic ovary syndrome, endometriosis, and, most importantly, breast and ovarian cancers. ERα is expressed in approximately 50–80% of breast cancer tissues.102,103
Oncogenic TFs and tumour suppressors frequently drive cancer through dysregulated PDIs. The proto-oncogene c-MYC promotes proliferative gene expression by binding E-box motifs (CACGTG) as a heterodimer with Max, primarily in nucleosome-depleted chromatin regions. Disruption of the c-Myc–Max–DNA interaction has become an important therapeutic strategy in cancers such as breast and prostate cancer.4,5,72 Loss of p53 can worsen c-Myc–driven cancer progression. In breast cancer, combined p53 loss and c-MYC activation increase mitotic gene expression and expand cancer stem cell–like populations. p53 binds DNA as a tetramer through its central DNA-binding domain and recognises the consensus response element RRRCWWGYYY (R = A/G, W = A/T, Y = C/T). p53's ability to activate target genes depends on this sequence and the surrounding chromatin context. Mutations that disrupt the DNA-binding domain or tetramerisation are common in cancer and prevent p53 from regulating its target genes, thereby contributing to tumour initiation and progression.104 SOX2 binds the canonical SOX motif (A/T)TTGT through its HMG domain, inserting into the minor groove and bending DNA. In nucleosomes, SOX2 recognises a degenerate motif (A/T)TTNT, with affinity influenced by local chromatin structure. This enables SOX2 to act as a pioneer factor, maintain open chromatin, and support stem cell self-renewal. In breast cancer, SOX2 promotes cancer stem cell–like properties, tumour initiation, and therapy resistance. Depending on the context, it can function as either an oncogene or a tumour suppressor in cancers such as glioblastoma, lung, and squamous cell carcinoma.105
Activator protein-1 (AP-1) binds to specific DNA sequences called TPA-responsive elements (TREs) via its basic leucine zipper (bZIP) domains, mediating essential protein–DNA interactions for gene regulation.106 The canonical AP-1 binding motif is the TRE consensus sequence 5′-TGAG/CTCA-3′, and DNA-binding affinity and specificity depend on the exact Jun–Fos dimer composition, which positions the basic region in the major groove. Changes in AP-1 subunit makeup, expression, or modifications can alter its DNA-binding behaviour, driving cancers such as breast and lung cancers.107 Disrupted AP-1 binding also affects the regulation of inflammatory cytokines, contributing to diseases such as rheumatoid arthritis and psoriasis.108
In haematological malignancies, fusion transcription factors act as abnormal DNA-binding regulators. In acute myeloid leukaemia (AML), fusion proteins such as AML1/ETO (acute myeloid leukaemia 1–eight-twenty-one), PML/RAR (promyelocytic leukaemia–retinoic acid receptor alpha), and PLZF/RAR (promyelocytic leukaemia zinc finger–retinoic acid receptor alpha) aberrantly regulate transcription by activating stem cell renewal pathways, such as the Notch signalling pathway; while repressing DNA repair genes, including those involved in base excision repair (BER), leading to genomic instability and malignant transformation.109
Finally, hypoxia-inducible factors (HIFs) represent a class of environmentally responsive TFs whose dysregulated DNA binding contributes to disease. The hypoxia pathway works via the stabilisation of the HIF-α subunit (such as HIF-1α, HIF-2α, or HIF-3α), which dimerises with the constitutively expressed aryl hydrocarbon receptor nuclear translocator (HIF-1β, also called ARNT). This complex formation is mediated by the basic helix-loop-helix (bHLH) motif and the Per-ARNT-Sim (PAS) domain, which also facilitates DNA binding. The heterodimer binds DNA at the hypoxia response element (HRE), a consensus sequence 5′-RCGTG-3′ (R = A/G). Its binding affinity and specificity are influenced by the flanking sequence and chromatin accessibility, as well as differences between HIF-1α and HIF-2α in target-gene preference. The stabilised complex then regulates transcription of target genes, mostly upregulating vascular endothelial growth factor (VEGF) and erythropoietin (EPO). Increased HIF levels are related to numerous diseases, such as ischaemic heart disease and heart failure, where HIF exerts a cardioprotective role.110 However, in chronic conditions, persistent HIF activation can lead to diseases such as glioblastoma multiforme (GBM), where HIF promotes tumour growth by activating VEGF in the tumour microenvironment,111 or in von Hippel–Lindau (VHL)–associated cancers like renal cell carcinoma.112 Overexpression of HIF is also linked to chemoresistance, including cisplatin resistance.113
2.2 Diseases linked to replication and repair proteins
Multiple PDIs are also crucial for DNA repair processes in eukaryotes. For example, proteins such as replication protein A (RPA) and xeroderma pigmentosum complementation group A (XPA) must bind to damaged DNA to initiate nucleotide excision repair (NER) by stabilising unwound DNA and recruiting downstream excision factors. Mutations in these proteins disrupt their DNA-binding ability, leading to defective DNA repair, accumulation of DNA lesions, and genomic instability, which can result in diseases such as xeroderma pigmentosum.114RPA is also essential for stabilising ssDNA during replication and repair. Dysfunction or insufficient RPA activity leads to replication stress, impaired homologous recombination (HR), and genomic instability. RPA also assists the activity of DNA polymerases α and θ, helping replication progress smoothly. Defects in RPA are associated with increased susceptibility to cancers such as breast cancer. RPA exhaustion increases cellular sensitivity to DNA-damaging agents.115 For further details on targeting RPA in cancer therapy, see the section on targeting replication/repair protein in this article.
HR repairs double-strand DNA breaks in a template-dependent manner by searching for homologous sequences and performing strand exchange. As described above, RPA stabilises single-stranded DNA and helps recruit RAD51 to form repair filaments. HR works in coordination with interdependent pathways, including the Fanconi anaemia (FA) pathway. BRCA2 mediates RAD51 loading, linking HR with FA pathway repair; mutations in these proteins lead to cancer predisposition.8
In another example under this category of PDI, MSH6, a component of the mismatch repair system (MMR), aids in correcting errors during DNA replication. The MMR corrects base mismatches and small insertion/deletion loops, acting as a crucial proofreading mechanism. MSH6 partners with MSH2 to form the MutSα complex that specifically recognises these errors. The N-terminal PWWP domain of MSH6 contributes to its chromatin localization and mutations that cause pathological effects. For example, the S144I mutation in this protein leads to hereditary non-polyposis colorectal cancer (HNPCC), also known as Lynch syndrome.116 MMR mutations are also commonly found in patients with prostate cancer.117
Base excision repair (BER) is another important pathway which corrects small, non-helix-distorting lesions caused by deamination, oxidation, and alkylation and repairs single-strand breaks (SSBs). If left unrepaired, such lesions can interfere with DNA replication and transcription and increase cytotoxic stress.72 The BER pathway involves several key proteins. Damaged bases are first recognised and excised by DNA glycosylases such as OGG1; defects in OGG1 compromise oxidative damage repair and have been linked to cancer and neurodegeneration. Gap filling is carried out by DNA polymerases such as Pol β (and in some contexts Pol θ). If unrepaired, apurinic/apyrimidinic (AP) sites accumulate, leading to stalled replication forks. The final nick is sealed by DNA ligases (LIG1 or LIG3) that form a complex with XRCC1. Mutations in XRCC1 are associated with neurological disorders due to impaired SSB repair.118,119 Defects in BER contribute to genomic instability, cancer predisposition, and progressive neurodegenerative diseases. For example, mutations in MutY homolog (MYH) increase the risk of colorectal cancer.120 Similarly, defects in PARP-mediated single-strand break repair contribute to genomic instability and cancer predisposition7 (see the section on targeting replication/repair protein).
The FA pathway helps in stabilising the genome. So far, about 23 genes have been recognised as part of this pathway. It is activated when a replication fork stalls due to an inter-strand cross-link (ICL). The FA core complex, including FANCA, FANCB, FANCC, and FANCE, is recruited to the site of the damage. The first event is the monoubiquitination of FANCD2 and FANCI by the core complex. A disruption in this process causes FA, a genetic disorder linked to bone marrow failure and an increased risk of tumours, solid tumours. The FA pathway primarily affects stem cells, such as hematopoietic stem cells, because faulty DNA repair is associated with DNA damage, which in turn activates the p53 protein. It also coordinates with other repair mechanisms, such as HR and NER. It recruits proteins like FANCD1/BRCA2 and FANCR/RAD51 to the site of the lesion, where they protect the DNA from excessive degradation by nucleases, including those involved in the WRN complex. This protection is essential for the later HR repair. The pathway also borrows the XPF-ERCC1 nuclease, a core enzymatic component of NER, to perform the initial incision that “unhooks” the cross-link.121,122
Another interesting example in this group is that of the non-homologous end joining (NHEJ) pathway, which is a faster way to fix double-stranded DNA breaks because it does not need a homologous template. It initiates with the binding of the Ku protein at the breakage site, which then recruits the ATP-dependent DNA-PKcs protein. The DNA-PKcs then facilitates the arrival of other proteins, like Artemis, XRCC4, and DNA Ligase IV, to the site for repair. Defects in this pathway are linked to diseases such as severe combined immunodeficiency (SCID). NHEJ is a very error-prone mechanism. When hyperactive, it can lead to drug resistance by efficiently repairing the damage caused by chemotherapy and radiotherapy.123
Topoisomerases and DNA helicases are some of the additional examples of PDIs involved in replication/repair pathways. Topoisomerases are essential enzymes that regulate the DNA topology. Topoisomerase I (Topo I) relieves supercoiling by introducing transient single-strand breaks, whereas Topoisomerase II (Topo II) introduces ATP-dependent double-strand breaks to allow strand passage and decatenation. Under normal conditions, these reactions preserve genome stability. However, mutations, chemical inhibition, or DNA damage can trap the enzymes as covalent protein–DNA cleavage complexes (TOPccs), leading to persistent DNA breaks and genomic instability.124
Topo IIα contributes to cell proliferation during replication and mitosis, while Topo IIβ regulates transcription and chromatin organization in non-dividing cells.125 Dysregulated topoisomerase activity is implicated in multiple diseases, including cancer,126 neurological disorders such as SCAN1 and ataxia telangiectasia,124 autoimmunity in scleroderma,127 and immunodeficiency in Hoffman syndrome caused by TOP2B mutations.128 Moreover, Topo I has transcriptional roles beyond DNA relaxation, with links to subsets of autism spectrum disorders.127
Finally, DNA helicases are enzymes that use energy from ATP to unwind the DNA double helix. This step is necessary for processes such as DNA replication, repair, and transcription. Helicases also help bring other proteins, such as DNA polymerases, to the replication fork.129 Mutations in the BLM gene, which encodes a member of the RecQ helicase family, cause Bloom syndrome. The BLM helicase works as part of a multiprotein complex that includes helicases, topoisomerases, and DNA repair factors. This Bloom syndrome complex has a central role in HR repair and in maintaining genome stability. Defects in BLM or its partner components impair DNA repair, increasing cancer risk (lymphomas, leukaemias, etc).130
2.3 Diseases linked to non-canonical DNA structures
Non-canonical DNA structures (including hairpin loops, G-quadruplexes (G4), left-handed Z-DNA and triplexes) are often hotspots for DNA breaks and genomic instability. These structures form at characteristic DNA sequences. G4 arise at G-rich runs (such as repeated GGG motifs). Z-DNA forms at alternating purine–pyrimidine sequences, especially (CG)n. Triplex DNA appears at homopurine–homopyrimidine mirror repeats. Hairpin-loops form when inverted repeats or triplet repeats like, CAG or CTG, fold back into a stem–loop shape. These conformations differ from canonical B-DNA in their base pairing, helical geometry, or strand arrangement.131Proteins that normally bind to the B-DNA may not recognise these non-canonical structures or may bind improperly, causing misregulation of gene expression and contributing to various cancers and neurodegenerative diseases. For instance, G4 regulate the transcription of oncogenes such as c-MYC, KRAS, and BCL2. Their stabilisation or misregulation has been linked to cancers, including breast, lung, and colon cancer, while G4 structures have also been implicated in neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS). Autoimmune diseases, particularly Aicardi-Goutières syndrome, are associated with the improper processing of Z-DNA. Cruciform structures have been linked to malignancies such as sarcomas and leukaemia and are commonly found near chromosomal fragile spots. Triplex structures frequently develop at repeating DNA sequences; the expansion of these repeats is a characteristic of hereditary conditions such as myotonic dystrophy, Huntington's disease, and Friedreich's ataxia.132 Hairpin-loop structures slow or block replication fork progression and can impede the activity of all three major replicative polymerases (Polα, Polδ, and Polε), thereby promoting replication stress and repeat-length instability. Hairpin-associated instability contributes to diseases such as spinocerebellar ataxias (SCAs) and other trinucleotide-repeat disorders, in which stable stem–loop formation correlates with pathogenic repeat expansion.133
Some diseases associated with non-B DNA structures appear across more than one category because certain repeat tracts can adopt multiple alternative conformations. For example, CGG repeats can form both hairpins and G-quadruplexes, whereas GAA repeats can form hairpins as well as H-DNA. These overlapping structural possibilities explain why similar disease mechanisms appear under different structural classes. Many of these conditions are collectively recognised as repeat expansion disorders (REDs), including Huntington's disease, myotonic dystrophy, fragile X syndrome and Friedreich's ataxia. These structures are also processed differently by DNA repair pathways, particularly MMR and NER, which can misrecognise or inefficiently resolve them and thereby contribute to disease progression.131,132,134 Research on identifying novel quadruplex structures and addressing their misregulation is in a relatively early stage but holds great promise as these unique structures may provide the missing high specificity targets in PDIs on the DNA side (see Section 3.2, Targeting the DNA).135–140
2.4 Diseases linked to structural and architectural proteins
Structural and architectural proteins shape how DNA is packaged and accessed, playing a key role in regulating gene activity. Histones, CCCTC-binding factor (CTCF), cohesins, high-mobility group (HMG) proteins, alongside other chromatin-associated regulators, collectively maintain genomic organisation and govern the availability of the DNA template.Mutations in core histones can disrupt nucleosome assembly and transcription, leading to neurodevelopmental disorders such as Rahman, Bryant–Li–Bhoj, and Tessadori–Bicknell–van Haaften syndromes. These conditions are marked by developmental delay, intellectual disability, seizures, abnormal growth, craniofacial differences, and problems affecting multiple organs.141
Architectural proteins, such as CTCF and cohesin regulate gene expression via long-distance chromatin interactions. Improper mediation of enhancer–promoter interactions leads to transcriptional dysregulation. Cohesin forms a ring-like complex that mediates chromatin loop extrusion during interphase, while CTCF binds DNA to control topologically associating domain (TAD) boundaries and halt cohesin movement, organising chromatin topology. Mutations in these proteins can lead to TAD boundary disruption and oncogene misregulation, as well as cohesinopathies such as Cornelia de Lange syndrome, and contribute to cancer development by disrupting genome organisation.142
HMG proteins are architectural proteins that bind DNA and induce bends, loops, or other conformational changes, regulating transcription and chromatin accessibility.143 HMGB1, for example, possesses three main domains: the A-box binds damaged DNA and facilitates repair, the B-box has cytokine activity and recruits immune receptors such as Toll-like receptors (TLRs) and RAGE, and the C-terminal tail binds the minor groove of DNA, bending it and helping TF recruitment and repair. During myocardial infarction, HMGB1 is released as a danger associated molecular pattern (DAMP) into the cytoplasm, triggering inflammation and apoptosis; consequently, elevated HMGB1 levels are associated with myocardial damage.144 Similarly, in allergic rhinitis, HMGB1 released by nasal epithelial cells promotes immune responses, activating T cells and inflammatory pathways.145 HMGB1 also plays a role in tumour suppression by facilitating p53 binding to DNA and influencing the NF-κB pathway, which is involved in inflammation and cell survival. Dysregulation or mislocalisation of HMGB1 can promote cancer cell proliferation, induce HIF-1α expression, and support tumour cell survival.146
Proteins like chromatin remodelers and histone modification enzymes regulate how histones interact with other proteins and DNA. Although they do not bind DNA directly, they perform epigenetic regulation, indirectly modulating chromatin accessibility and PDIs critical for transcription, replication, and repair.147 Histone modifications, collectively described as the “histone code”, include methylation, acetylation, phosphorylation, sumoylation and ubiquitination.148 Mutations in these histone modification enzymes (writers and erasers), such as lysine methyltransferases (KMTs), lysine demethylases (KDMs), acetyltransferases (KATs), and deacetylases (HDACs, SIRTs), cause disorders including Kabuki, Wiedemann–Steiner, Sotos, Rubinstein–Taybi, and Luscan–Lumish syndromes. For example, histone deacetylases (HDACs) are involved in transcriptional inactivation of tumour-suppressor genes, leading to cancers, blood disorders, solid tumours, and non-neoplastic disorders.149 Targeting HDAC’s has hence become a significant therapeutic strategy.150 Epigenetic readers recognise chemical modifications (tags) on histones or DNA and mediate interactions between these proteins and DNA, guiding the recruitment of other factors that regulate gene expression and chromatin structure.151 Among them, BRD4 is the most studied and has a role in cell cycle, inflammation, and cancer progression. Likewise, defects in chromatin remodeler complexes such as SWI/SNF, CHD, and INO80, responsible for repositioning nucleosomes, are linked to syndromes including Coffin–Siris, Snijders Blok–Campeau, Sifrim–Hitz–Weiss, and CHARGE. These conditions often share features such as intellectual disability, seizures, skeletal abnormalities, and craniofacial malformations.141
Although this review focuses on human PDIs, it is worth noting that similar recognition mechanisms occur in host–pathogen interactions. For example, Z-DNA binding protein 1 (ZBP1) recognises foreign DNA, and its dysregulation can trigger autoimmune conditions such as Aicardi–Goutieres syndrome.152 While important, these interactions are beyond the scope of this review. With this understanding of dysregulation, the next section addresses therapeutic strategies to target these interactions.
3 Drug targeting strategies for PDIs
Given the central role of PDIs in regulating gene expression, DNA repair, and other cellular processes, therapeutic strategies have been developed to disrupt or modulate these complexes. These strategies include (1) directly targeting DNA-binding proteins, (2) interfering with the DNA molecule itself, or (3) modulating the protein–DNA interface to inhibit or alter binding. Each offers distinct mechanisms and clinical implications as reviewed below.3.1 DNA-binding proteins as drug targets
DNA-binding proteins form a diverse group that includes TFs, replication and repair proteins, histones, and architectural proteins. This section outlines strategies for targeting these proteins to modulate PDIs.| Drug name | Target | Mechanism of action | Approval status | Drug class | Therapeutic area | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: ER, estrogen receptor; AR, androgen receptor; STAT3, signal transducer and activator of transcription 3; Myc, myelocytomatosis oncoprotein; HIF, hypoxia-inducible factor; p53, tumor protein p53; PROTAC, proteolysis-targeting chimera; SERD, selective estrogen receptor degrader; mCRPC, metastatic castration-resistant prostate cancer; PAS-B, Per-ARNT-Sim domain B; SH2, Src-homology 2; DBD, DNA-binding domain; NLS, nuclear localisation signal; LBD, ligand-binding domain; LGL, large granular lymphocyte;T-ALL, T-cell acute lymphoblastic leukaemia; PDAC, pancreatic ductal adenocarcinoma; VHL, von Hippel–Lindau; RCC, renal cell carcinoma; CNS, central nervous system; pNET, pancreatic neuroendocrine tumour; NCT, National Clinical Trial. | ||||||
| WBC100 | Myc | Degrader: Induces proteasomal degradation of Myc by targeting the NLS1–basic–NLS. 2 region | Phase I trial (NCT05100251) | Molecular glue/degrader | Advanced solid tumours | 170 |
| OMO-103 (Omomyc) | Myc | DBD binding: Mini-protein interfering with Myc-Max dimerisation and DNA-binding, blocking transcriptional activity | Phase I/IIa trial (NCT04808362); Phase Ib trial (NCT06059001); Phase II trial (NCT06650514) | Mini protein | Solid tumours, metastatic PDAC, advanced high-grade osteosarcoma | 51 |
| IDP-121 | Myc | Allosteric: Mini-protein interfering with Myc transcriptional activity | Phase I/II trial (NCT05908409) | Mini protein | Relapsed/refractory haematological malignancies | 171 |
| STATTIC | STAT3 | Allosteric: STAT3 SH2-domain inhibitor, prevents dimerisation and DNA-binding | Preclinical | Small-molecule | T-ALL, solid tumours and haematologic malignancies | 159 and 160 |
| TTI-101 | STAT3 | Allosteric: Blocks STAT3 activation at the SH2 domain, preventing dimerisation and transcriptional activity | Phase I trial (NCT03195699); Phase II trial (NCT05440708) | Small-molecule | Advanced/metastatic solid tumours, hepatocellular carcinoma, ovarian cancer, gastric cancer | 161 |
| KT-333 | STAT3 | Degrader: Recruits STAT3 to E3 ubiquitin ligase for proteasomal degradation | Phase I trial (NCT05225584) | Heterobifunctional small-molecule degrader | Refractory lymphomas, LGL leukaemia, solid tumours | 172 |
| Vepdegestrant (ARV-471) | ER | Degrader: Recruits ER to an E3 ubiquitin ligase for proteasomal degradation | Phase III trial (NCT05654623) | PROTAC | ER-positive/HER2-negative advanced or metastatic breast cancer | 61 |
| Fulvestrant | ER | Allosteric: Binds LBD and induces proteasomal degradation | FDA-approved | SERD | Breast cancer | 49 and 162 |
| Enzalutamide | AR | Allosteric: Binds to LBD and prevents nuclear translocation and DNA-binding | FDA-approved | Anti-androgen | Prostate cancer | 50 and 163 |
| ARV-110 | AR | Degrader: Recruits AR to cereblon E3 ligase for proteasomal degradation | Phase I/II trial (NCT03888612) | PROTAC | mCRPC | 52 |
| Eprenetapopt (APR-246) | p53 | DBD binding: Covalently binds to mutant p53, restoring wild-type conformation | Phase III trial (NCT03745716) | Small-molecule | Myelodysplastic syndrome | 157 and 158 |
| Belzutifan | HIF-2α | Allosteric: Binds to the PAS-B domain of HIF-2α and prevents dimerisation with HIF-1β, essential for DNA-binding and transcriptional activity | FDA approved | Small-molecule | VHL-associated tumours: RCC, CNS haemangioblastomas, or pNET | 173 |
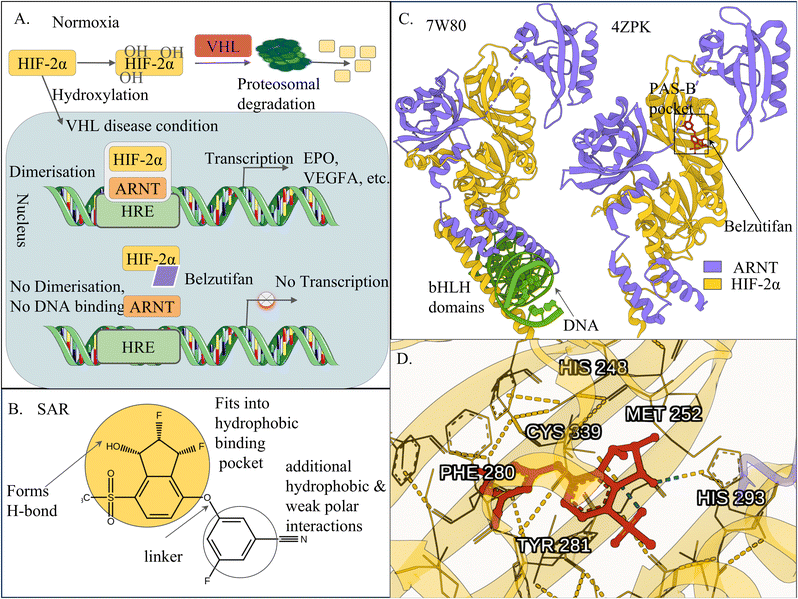 | ||
| Fig. 2 Structural and mechanistic basis of hypoxia-inducible factor 2α (HIF-2α) inhibition by belzutifan. (A) Under normoxic and von Hippel–Lindau (VHL)-proficient conditions, HIF-2α is hydroxylated, recognised by the VHL E3 ubiquitin ligase complex, and degraded. In hypoxia or VHL-deficient states, HIF-2α accumulates, heterodimerises with the aryl hydrocarbon receptor nuclear translocator (ARNT), binds hypoxia-response elements (HREs), and activates transcription. Belzutifan binds the HIF-2α PAS-B domain, disrupting HIF-2α–ARNT dimerisation and preventing DNA binding. (B) Structure–activity relationship (SAR) of belzutifan highlighting chemical features required for selective PAS-B pocket engagement. (C) Structural comparison of DNA-bound HIF-2α–ARNT (PDB: 4ZPK)153 and belzutifan-bound HIF-2α (PDB: 7W80),154 showing that ligand binding stabilises a conformation incompatible with ARNT association and DNA binding. (D) Key interactions of belzutifan within the HIF-2α PAS-B pocket, including Met252, whose side-chain reorientation stabilises a rigid, inactive PAS-B conformation. Created with icons from Servier Medical Art (smart.servier.com), licensed under CC BY 4.0; protein visualisations generated via Mol*,68 MarvinSketch for chemical structures, Chemaxon (https://www.chemaxon.com). | ||
Another approach to target DNA-binding proteins leverages targeted protein degradation, such as PROTACs. For instance, ARV-110 targets the AR, removing the TF entirely rather than merely inhibiting its DNA-binding capability. While these strategies involve direct interaction with the TF, many have not yet achieved full clinical approval across all TF classes.
Due to the difficulties of direct or allosteric targeting of structures, the most clinically feasible method currently is indirect modulation. In this approach, drugs do not physically interact with the TF but instead influence upstream signalling pathways and alter TF phosphorylation, nuclear localisation, or activation, thereby affecting DNA-binding activity. Examples include CDK4/6 inhibitors such as palbociclib, ribociclib, and abemaciclib, which indirectly inhibit E2F TFs,168,169 and AKT inhibitors like capivasertib, which modulate FOXO TFs. These indirect approaches are FDA-approved in various oncology contexts and provide a practical route for therapeutically modulating PDIs.
A summary of key drugs targeting replication and repair proteins is provided in Table 2.
| Drug name | Target | Mechanism of action | Approval status | Drug class | Therapeutic area | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: HR, homologous recombination; DBD, DNA-binding domain; PARP, poly(ADP-ribose) polymerase; SSB, single-strand break; BRCA, breast cancer susceptibility gene; RPA, replication protein A; OB-fold, oligonucleotide/oligosaccharide-binding fold; ssDNA, single-stranded DNA; NHEJ, non-homologous end joining; NSCLC, non-small cell lung cancer; CML, chronic myeloid leukaemia. | ||||||
| IBR2 | RAD51 | Allosteric: Binds at the oligomerisation site, disrupting RAD51 polymer formation, impairing HR repair | Preclinical | Small-molecule | Breast cancer, CML, etc. | 184 |
| B02 | RAD51 | DBD: Binds to the DNA-binding surface, inhibits RAD51 strand invasion preventing HR-mediated repair, also used in combination with cisplatin | Preclinical | Small-molecule | Breast cancer, prostate cancer | 185–187 |
| Olaparib | PARP1/2 | Catalytic: Competitive inhibition of PARP catalytic activity, blocks PARylation, prevents SSB repair | FDA-approved | Small-molecule | BRCA-mutated cancer | 53 and 188 |
| Talazoparib | PARP1/2 | Catalytic: Inhibits and traps PARP, inducing DNA damage, especially in HR-deficient tumours | FDA-approved | Small-molecule | BRCA-mutated HER2-negative breast cancer | 54 and 176 |
| NERx329 | RPA | DBD: Reversibly binds to the ssDNA-binding OB-fold domain of RPA, blocking its interaction with ssDNA and disrupting replication and repair | Preclinical | Small-molecule | NSCLC, BRCA-mutated cancers; potential to overcome platinum resistance | 180 |
| Peposertib (M3814) | DNA-PKcs | Catalytic: ATP-competitive inhibition of kinase activity, blocks NHEJ activation | Phase I trial (NCT04555577, NCT05868174), In combination therapies (NCT05687136) | Small-molecule | Solid tumours, glioblastoma, etc. | 182 and 189 |
The most successful strategy for targeting DNA repair proteins has been the development of PARP inhibitors for BRCA1/2-deficient cancers, exemplified by the FDA-approved drugs olaparib and talazoparib. PARP1 is a protein that detects SSBs in DNA and helps repair them. Olaparib exploits between-pathway synthetic lethality, where in BRCA-mutant cells, the HR pathway is defective and inhibition of PARP blocks the backup repair pathway, leading to cell death (Fig. 3). In contrast, normal cells remain largely unaffected because they retain their primary repair pathway.7 Talazoparib, in addition to catalytic inhibition, is a potent PARP trapper, stabilising PARP–DNA intermediates and preventing their resolution, an example of in-pathway synthetic lethality that enhances cytotoxicity.176 However, resistance to PARP inhibitors in BRCA-deficient cancers has been reported. Some tumour cells restore HR function through secondary mutations that “fix” the defective BRCA1/2 gene or by loss of p53-binding protein 1 (53BP1). For such resistance mechanisms to occur, the cells typically require intact HR-related domains (RING, BRCT, etc).7
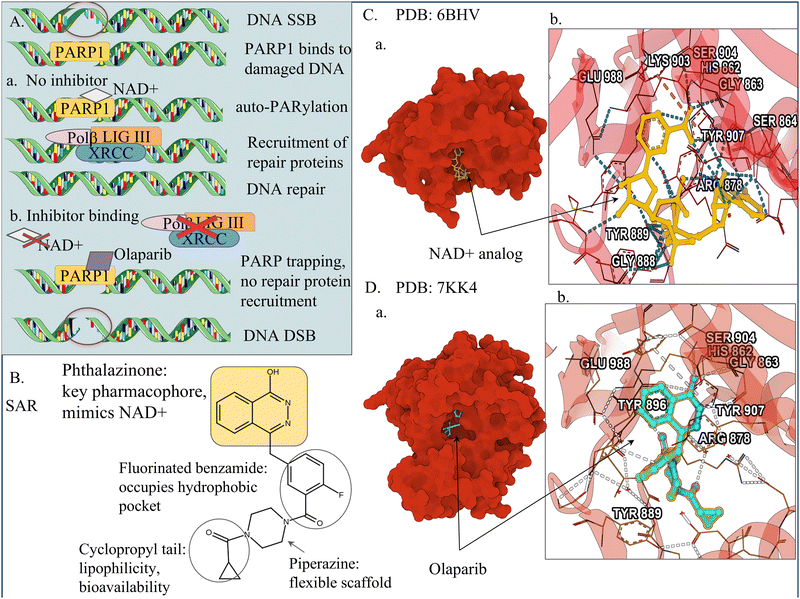 | ||
| Fig. 3 Mechanistic and structural basis of PARP1 inhibition by olaparib. (A) Mechanism of action of olaparib. In the absence of inhibitor, poly(ADP-ribose) polymerase 1 (PARP1) binds to single-strand DNA breaks (SSBs), undergoes auto-poly(ADP-ribosyl)ation using NAD+, recruits DNA repair factors, and facilitates SSB repair. In the presence of olaparib, PARP1 catalytic activity is inhibited and the enzyme becomes trapped on DNA, preventing repair, leading to replication fork collapse, accumulation of double-strand breaks (DSBs), and cell death. (B) Structure–activity relationship (SAR) of olaparib highlighting key chemical features required for high-affinity binding to the PARP1 catalytic domain and efficient PARP trapping. (C) Crystal structure of the PARP1 catalytic domain bound to an NAD+ analogue (PDB: 6BHV)174 showing the overall protein surface, the NAD+-binding pocket, and key ligand–protein interactions. (D) Crystal structure of PARP1 bound to olaparib (PDB: 7KK4)175 showing olaparib (blue) occupying the NAD+-binding site and representative ligand–protein interactions. Created with icons from Servier Medical Art (smart.servier.com), licensed under CC BY 4.0; protein visualisations generated via Mol*;68 MarvinSketch chemical structures, and Chemaxon (https://www.chemaxon.com). | ||
Overexpression of RAD51 contributes to PARP inhibitor resistance by enhancing HR. To target RAD51, small molecules like IBR2 and B02 are being studied, which are currently in preclinical trials. IBR2 disrupts RAD51 oligomerisation and prevents the formation of functional RAD51 filaments, while B02 blocks RAD51 binding to ssDNA and inhibits the strand exchange mediated by it in HR. B02 also makes the tumour cells more sensitive to other therapies like ionising radiation, mitomycin C and cisplatin. It has shown efficacy in triple-negative breast cancer (TNBC) models in mouse xenografts. Combination therapy of B02, PARP inhibitor and p38 inhibitor can overcome resistance in BRCA1/2-deficient cancers, by blocking DNA repair and associated survival pathways.7,177,178 Ongoing studies are exploring new chemical scaffolds inspired by these inhibitors to design next-generation RAD51-targeting molecules.179
For RAD51 to work, it first needs RPA to bind and protect the ssDNA; then, RAD51 replaces RPA to initiate the HR process. RPA is also essential for the NER pathway, thus playing a dual role in DNA replication and repair. RPA can be targeted in two main ways: allosteric inhibition or directly targeting its ssDNA-binding domains, known as OB-folds, to prevent DNA-binding. For example, NERx329 is a small-molecule inhibitor in preclinical trials that has shown promising results by reversibly binding to these domains. This strategy is effective in non-small cell lung cancer (NSCLC). Furthermore, RPA inhibitors are effective in combination with PARP inhibitors for BRCA-mutated cancers and may potentially overcome resistance to platinum-based drugs in lung cancer.180
The FA pathway is another important target in this context. In patients with FA, the pathway is defective, and restoring its function could be a potential therapeutic goal. For example, research is exploring small molecules that can reactivate FANCD2 monoubiquitination thereby bypassing the underlying genetic defect. Conversely, in certain cancers, the FA pathway is often hyperactive, enabling cancer cells to repair DNA damage caused by treatments like chemotherapy. Shutting down this pathway could make these cancer cells more vulnerable. A key step for this therapeutic approach is the targeted inhibition of FANCD2 monoubiquitination. For instance, TAK243, an inhibitor of ubiquitin-activating enzymes (UAE), has been shown to inhibit this crucial step potently. While UAEs have a broad effect, this finding confirms that targeting a key component of the FA pathway is a viable strategy for developing new therapeutic agents. This approach could sensitise cancer cells to existing treatments, while separate efforts could develop activators to treat FA.122
Beyond direct inhibition of repair proteins, another effective strategy is to utilise immunotherapy, particularly for cancers with defective DNA repair mechanisms. Immune checkpoint inhibitors are used as a treatment for cancers with a defective MMR pathway. These tumours have a high number of mutations, a state known as microsatellite instability-high (MSI-H). The presence of these mutations causes the tumour to produce abnormal proteins, which the immune system can recognise. This high mutational load makes these tumours susceptible to immunotherapy. Drugs like pembrolizumab, which is FDA-approved, are used. While not a direct therapeutic for the MMR defect, it causes the immune system to attack tumour cells. It is currently approved for various MSI-H cancers.181
The popular approach to target the NHEJ pathway is to inhibit the DNA-PKcs protein, with drugs like peposertib (currently in clinical trials). These ATP-competitive inhibitors bind to the protein's active site, preventing it from activating other proteins in the pathway. Importantly, these inhibitors block the enzymatic activity of DNA-PKcs after it has bound DNA, rather than preventing DNA-binding itself. This inhibition leaves the DNA damage unrepaired, making the cancer cells more vulnerable.182 In addition to targeting DNA-PKcs, other NHEJ proteins are also being investigated as therapeutic targets. These include inhibitors for the Ku protein (e.g., STL127705) to prevent initial DNA-binding, for Artemis (e.g., Ebselen) to block DNA processing, and for DNA ligase IV (e.g., L189) to stop the final sealing of the DNA break. However, most of these inhibitors are still in the preclinical stage, and further research is needed in this direction.183
| Drug name | Target | Mechanism of action | Approval status | Drug class | Therapeutic area | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: HMGB1, high-mobility group box 1; HDAC, histone deacetylase; CTCL, cutaneous T-cell lymphoma; PTCL, peripheral T-cell lymphoma; EZH2, enhancer of zeste homolog 2; HMT, histone methyltransferase; PRC2, polycomb repressive complex 2; H3K27me3, histone H3 lysine 27 trimethylation; DOT1L, disruptor of telomeric silencing 1-like; H3K79, histone H3 lysine 79; MLL, mixed lineage leukaemia; LSD1 (KDM1A), lysine-specific demethylase 1A; H3K4me1/2, histone H3 lysine 4 mono-/di-methylation; AML, acute myeloid leukaemia; BRD4, bromodomain-containing protein 4; BET, bromodomain and extra-terminal family; BD2, bromodomain 2; NHEJ, non-homologous end joining; CVD, Cardiovascular disease. | ||||||
| Glycyrrhizin | HMGB1 | Direct inhibitor of HMGB1; suppresses tumour proliferation and induces apoptosis via HMGB1-mediated DNA repair (NHEJ) in cancer models | Preclinical | Natural product | Colorectal and cervical cancer | 195 |
| Vorinostat | HDAC1,2,3,6 | Inhibits HDACs, increasing histone acetylation, which loosens chromatin and helps reactivate suppressed genes | FDA-approved | Small-molecule (hydroxamic acid) | Cutaneous T-cell lymphoma | 55 |
| Romidepsin | Class I HDACs | Reduced intracellularly to free thiol which binds zinc ions at the HDAC active site, leading to histone hyperacetylation and chromatin relaxation and apoptosis induction | FDA-approved | Cyclic peptide (prodrug) | CTCL, PTCL | 200 |
| Panobinostat | Pan-HDAC | Binds zinc ions in the HDAC site leading to histone and tubulin acetylation, disrupting chromatin compaction and microtubule function and causing apoptosis | FDA-approved | Small-molecule (hydroxamic acid derivative) | Multiple myeloma (in combination therapy) | 149 and 203 |
| Givinostat | Pan-HDAC | Hydroxamate chelation of zinc ions, preventing histone deacetylation, leading to sustained histone acetylation, reduces fibrosis and inflammation in muscle tissue | FDA-approved | Small-molecule, hydroxamic acid derivative | Duchenne muscular dystrophy | 206 and 213 |
| Tazemetostat | EZH2 (H3K27 methyltransferase, PRC2 complex) | Inhibits EZH2 catalytic activity, preventing trimethylation of H3K27, reduces repressive chromatin marks and reactivates silenced tumour suppressor genes | FDA-approved | Small-molecule (HMT inhibitor) | Epithelioid sarcoma, follicular lymphoma | 56 and 214 |
| Pinometostat | DOT1L (H3K79 methyltransferase) | Selectively blocks DOT1L-mediated H3K79 methylation, essential for maintaining oncogenic transcription programs in MLL-rearranged leukaemias | Phase I/II trial (NCT03701295), Phase I trial (NCT01684150) | Small-molecule (HMT inhibitor) | MLL-rearranged leukaemia | 208 and 209 |
| Iadademstat | LSD1 (KDM1A histone demethylase) | Irreversibly inhibits LSD1, preventing the removal of H3K4me1/2 marks, restoring expression of differentiation genes and promoting maturation of leukemic blasts | Phase IIa trial: EudraCT 2018-000482-36(combination therapy), Phase II trial: EudraCT 2018-000469-35 (combination therapy) | Small-molecule (KDM inhibitor) | AML, SCLC | 207 and 215 |
| Apabetalone | BRD4 | Selectively inhibits the BD2 domain of BET proteins, preventing BRD4 binding to acetylated histones and preventing TF recruitment, leading to gene silencing | Phase III trial (NCT02586155) for type 2 diabetes | Small-molecule | Type 2 diabetes, CVD, long COVID | 212 and 216 |
The design of drugs targeting cohesin and CTCF is still in the conceptual and experimental stage. For example, the first cohesin-inhibiting peptide (CIP), which inhibits the ATPase activity of cohesin by binding to the Smc3 domain, has been developed, suggesting potential applications in cancer therapy. However, challenges remain in using peptides for tumour targeting due to specificity issues.190 There is also potential for treating cohesin-mutated cancers by targeting cohesin components or their regulators, as well as associated transcriptional or signalling events and DNA damage repair systems. Candidates for these strategies include drugs such as glycyrrhizic acid, olaparib, talazoparib, cisplatin, and etoposide.191 CTCF epigenetically regulates gene expression and plays an important role in alternative splicing, with DNA methylation controlling its binding to targeted sequences.192 The development and persistence of CALM-AF10 leukaemia depend on CTCF, which exerts its regulatory effects through histone modifications.193 In pancreatic ductal adenocarcinoma (PDAC), CTCF has been identified as a potential therapeutic target for immunotherapy, and combination of curaxin with gemcitabine has been explored as a treatment strategy.194
Structural proteins such as HMGB1 have also been extensively studied. Glycyrrhizin, a naturally derived compound from licorice, has been identified as a direct inhibitor. Studies in colorectal and cervical cancer models have demonstrated that glycyrrhizin suppresses tumour cell proliferation and induces apoptosis, potentially by modulating HMGB1's involvement in the NHEJ pathway, thereby affecting DNA repair processes.195 In liver disease, glycyrrhizin exerts anti-inflammatory effects by targeting HMGB1's extracellular signalling and has been approved for this use in some countries; this represents a mechanism distinct from its modulation of HMGB1-related DNA interactions in cancer cells.196
Building on these approaches, current efforts increasingly focus on epigenetic regulation as a complementary strategy to target chromatin-associated proteins. Since the activity of structural proteins, like cohesin, CTCF, and HMGB1, is influenced by histone modification and chromatin state, targeting histone-modifying enzymes and chromatin readers offers an indirect yet effective approach.
HDAC inhibitors are well-established drugs that increase histone acetylation, loosening chromatin and reactivating silenced genes. Vorinostat is the first FDA-approved HDAC inhibitor. It is suberoylanilide hydroxamic acid (SAHA) used for the treatment of cutaneous T-cell lymphoma (CTCL). It binds to both Class I and Class II HDACs, with a preference for the former.197 It is currently under trial for other types of tumours, such as head and neck squamous cell carcinoma (HNSCC),198 and breast cancer, both as a monotherapy199 and in combination with drugs like olaparib.60 Romidepsin, a cyclic peptide is used to treat CTCL, peripheral T-cell lymphoma (PTCL) and hepatocellular carcinoma.200–202 Panobinostat, a pan-HDAC inhibitor approved for multiple myeloma,203 can cross the blood–brain barrier, and studies are investigating its use in brain cancers204,205 and other solid tumours.149 The mechanisms of all are explained in Table 3. Givinostat is a recently FDA-approved drug and the first non-steroidal medication for the treatment of Duchenne muscular dystrophy (DMD). DMD is characterised by the loss of dystrophin, which, through a series of processes, affects nitric oxide signalling that controls HDAC activity, leading to its hyperactivation and transcriptional repression of genes such as myogenic microRNAs and follistatin. These genes are important for muscle repair. By inhibiting HDAC2, givinostat is effective in the treatment of DMD.206
Histone methyltransferase inhibitors include tazemetostat, an FDA-approved small-molecule selective inhibitor of EZH2 KMT catalysis. It inhibits EZH2-mediated H3K27 methylation, thereby preventing gene repression. It is used for refractory follicular lymphomas and various solid tumours. Chromatin remodelers, like SWI/SNF, involved in gene transcription, are also linked to EZH2 activity when dysfunctional. Consistent with this, tazemetostat showed clinical activity in SMARCB1/INI1-deficient epithelioid sarcoma, resulting in FDA accelerated approval in 2020.56,207 Pinometostat is another selective inhibitor that targets DOT1L, the H3K79 methyltransferase.208 It has shown activity in early-phase studies of acute leukaemias with MLL rearrangements and has been evaluated in Phase Ib/II trials, both as monotherapy and in combination with chemotherapy or azacitidine; however, the results show limited efficacy.208,209 On the other hand, FDA-approved revumenib, which also downregulates the same oncogenic genes HOXA/MEIS1 as pinometostat, is a menin-KMT2A inhibitor essentially blocking protein–protein interaction and is used in AML therapy too, while pinometostat is also stated to increase the sensitivity to this inhibitor, and trials have been conducted for the combination therapy.209–211 Iadademstat, an LSD1 inhibitor, works opposite to tazemetostat and pinometostat, as it selectively inhibits a histone demethylase and reactivates cancer-suppressing genes. It is still in early clinical trial phases for relapsed AML, both as a single agent and in combination with azacitidine. Iadademstat has also shown activity in SCLC, with ongoing combination trials.207
Emerging approaches target epigenetic readers and chromatin remodelers. Apabetalone is a selective BET inhibitor that binds to the BD2 bromodomain and is currently in Phase III trials for cardiovascular and kidney diseases.212
While the strategies above focus on modulating proteins that bind or organise DNA, another complementary approach is to directly target the DNA molecule itself, either to block protein binding or to modulate its structure, which is discussed in the following section.
3.2 Targeting the DNA
Proteins recognise DNA through either a direct sequence-based readout mechanism, an indirect or shape-based readout mechanism, or a combination of both. Small-molecule drugs work on a similar principle, recognising binding sites through ionic and hydrogen bonding interactions, and indirectly through DNA shape complementarity, depending on the nucleotide sequence. A summary of key drugs targeting DNA instead of proteins for disrupting PDIs is provided in Table 4.| Drug | Target | Mechanism of action | Approval status | Drug class | Therapeutic area | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: Pt, platinum; TBP, TATA-binding protein; NF-Y, nuclear transcription factor Y; RPA, replication protein A; HMGB1, high-mobility group box 1; p53, tumour suppressor protein p53; Bax, BCL-2–associated X protein; c-MYC, cellular myelocytomatosis gene; MutSα, DNA mismatch repair complex MutS alpha; DACH, diaminocyclohexane; bp, base pairs; G-4, G-quadruplex; Topo II, topoisomerase II; DSB, double-strand break; AML, acute myeloid leukaemia; SCLC, small-cell lung cancer; TF, transcription factor. | ||||||
| Cisplatin | N7 of guanines | Forms intra- & interstrand cross links; deforms DNA, blocking transcription and replication. Disrupts TBP, NF-Y, DNA/RNA polymerases, RPA; recruits HMGB1 to adducts; activates p53 in response to damage | FDA-approved | Small-molecule (Pt-based alkylating agent) | Testicular, ovarian, bladder, lung, head and neck cancer | 266 and 267 |
| Oxaliplatin | N7 of guanines | Forms bulky DACH–Pt intra- and interstrand crosslinks (preferentially forms H-bonds at the 3′ G); deforms DNA; blocks DNA synthesis, transcription, and replication. Disrupts TBP, NF-Y, DNA/RNA polymerases, RPA, and repair proteins (MutSα); activates p53 and Bax | FDA-approved | Small-molecule (Pt-based alkylating agent) | Colorectal and oesophageal cancer | 267 and 268 |
| Trabectedin | N2 of guanine | DNA bending and adduct formation, blocks RNA polymerse II binding; inhibits activity of TFs like FUS-CHOP and EWS-FL1 | Phase III trial (NCT02672527, NCT01343277) | Small-molecule (natural product intercalator) | Metastatic liposarcoma, leiomyosarcoma, soft tissue sarcoma | 224, 269 and 270 |
| Melphalan | N7 of guanine and N3 of adenine | DNA crosslinks and adducts causing helix distortion, DNA damage and mutation. Affects DNA polymerase and replication proteins | FDA-approved | Small-molecule (nitrogen mustard, alkylating agent) | Multiple myeloma, ovarian cancer, lymphoma | 271 |
| Actinomycin D | GC-rich minor groove | Intercalates phenoxazone ring between bp, alters DNA structure, prevents binding of RNA polymerase and inhibits transcription; stabilises G4 structures in c-MYC promotor, SOX2 downregulation | FDA-approved | Small-molecule (intercalator) | Rhabdomyosarcoma, choriocarcinoma, Ewing sarcoma, testicular cancer | 243–245 and 272 |
| Doxorubicin | DNA duplex | Intercalates and inhibits Topo II, causing DNA breaks, blocks transcription and replication by evicting histone | FDA-approved | Small-molecule (anthracycline intercalator) | Breast cancer, leukaemia, lymphoma, testicular, bladder, ovarian, lung cancer | 47 and 273 |
| Amsacrine (m-AMSA) | DNA duplex | DNA intercalator, stabilises the Topo II cleavage complex and prevents religation, causing DSBs, leading to apoptosis | Approved (selected countries) | Small-molecule (intercalator) | AML, Hodgkin's and non-Hodgkin's lymphomas | 248 and 249 |
| Voreloxin | DNA duplex, G4 DNA | Intercalates between bp to poison Topo II and induces site-selective DSBs, G2 cell cycle arrest, end-stacks on G-tetrads to suppress c-MYC/BCL-2 expression | ‘Orphan drug’ status, Phase II trials (NCT00408603, NCT00607997, NCT00252382, NCT00298896) | Small-molecule (quinolone-based intercalator) | AML, ovarian cancer, leukaemia, SCLC, solid tumours | 250–252 |
| Mithramycin | GC-rich minor groove | Inhibits TFs Sp1, EWS-FL1, histone modulation | Previously approved now discontinued, currently in trials for cancer | Small-molecule (non-covalent groove binder) | Osteosarcoma, Ewing sarcoma | 237–239 and 274 |
| CX-5461 (Pidnarulex) | G4 DNA | Stabilises G4 structures by π–π stacking on G-quartets, preferentially binding parallel promoter and telomeric G4s (e.g., c-MYC and c-KIT); traps Topo II, induces replication stress, R-loops & DNA damage; synthetic lethality in BRCA1/2- and HR-deficient cancers | Phase I/II trials (NCT04890613); FDA fast track designation | Small-molecule | Breast, ovarian and other BRCA1/2- or HR-deficient cancers | 259 |
Binding can occur in two modes for drug–DNA interactions: covalent binding (alkylating agents), which is irreversible, and non-covalent binding (intercalators and groove binders), which is reversible.74 Direct binding of small molecules is a valuable approach for targeting TFs and topoisomerases.47,48
For example, cisplatin, an alkylating agent, is a square planar platinum(II) complex with two ammine and two chloride ligands.217,218 It forms covalent adducts at the N7 position of guanine, causing DNA crosslinking, bending, and unwinding; this inhibits replication and transcription by blocking the binding of TFs and stalling RNA polymerase.219 While these lesions are normally substrates for the NER pathway, protein shielding of cisplatin adducts can hinder the recruitment of repair factors such as XPA, leading to persistent DNA damage (Fig. 4).220,221 Oxaliplatin, with a bulky diaminocyclohexane (DACH) and oxalate ligands, forms larger adducts and is mainly used in cisplatin-resistant colorectal cancers.218,222 Other new-generation platinum-based drugs, such as Pt nanocluster (NC)-based nanodrugs, are also being investigated.222
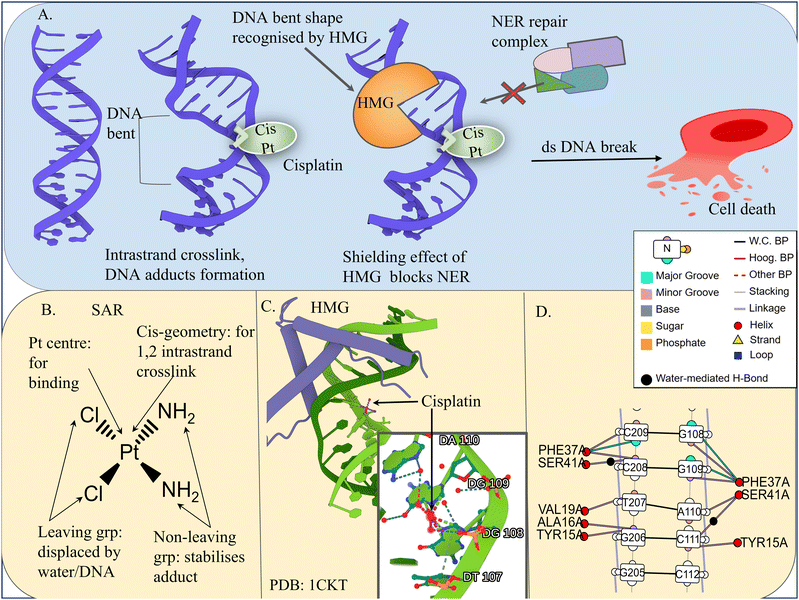 | ||
| Fig. 4 Cisplatin–DNA interactions and structural basis of recognition. (A) Mechanism of action of cisplatin illustrating the formation of intra-crosslinks at the adjacent guanine position, and induction of pronounced DNA bending. The distorted DNA is preferentially recognised and shielded by HMG-domain proteins, thereby limiting access of the nucleotide excision repair (NER) machinery, promoting replication-associated DNA double-strand breaks (DSBs) and activation of apoptotic pathways. (B) Structure–activity relationship (SAR) of cisplatin highlighting the importance of the platinum centre, leaving groups, and coordination geometry for efficient DNA binding and cytotoxic activity. (C) Crystal structure of cisplatin bound to DNA (PDB: 1CKT) showing coordination to adjacent guanine bases dG108 and dG109, with additional stabilising interactions involving dA110 and dT107.63 (D) NUCPLOT representation of the cisplatin–DNA complex highlighting key contacts involving Tyr15, Ala16, Val19, Phe37, and Ser41. Visualisations generated via Mol*,68 DNAproDB93 and Web 3DNA 2.0.223 MarvinSketch was used for drawing chemical structures, Chemaxon (https://www.chemaxon.com). | ||
Apart from platinum drugs, there are also natural product alkylating agents that covalently bind to the DNA minor groove, such as trabectedin, originally isolated from the marine tunicate Ecteinascidia turbinata and now produced synthetically. It is FDA-approved for the treatment of soft-tissue sarcoma, typically as a second-line therapy after doxorubicin failure. Trabectedin disrupts transcription by inhibiting trans-activated TFs (such as Sp1), while also impairing replication and the NER pathway. Homologous recombination-deficient cells are particularly sensitive to trabectedin. In addition to sarcomas, it is under investigation for Ewing sarcoma, myxoid liposarcoma, and in various combination regimens, including immunotherapies.224
Another set of alkylating agents, called nitrogen mustards, also target DNA and are widely used in anticancer therapy.47 However, their disruption of PDIs is generally less selective than that observed with platinum-based drugs, though evidence exists for specific effects, such as inhibition of Sp1 binding sites.225 Melphalan is a classical nitrogen mustard, consisting of an L-phenylalanine moiety linked to a bis(2-chloroethyl)amine group (the “mustard” group, which generates highly reactive aziridinium ions). It is highly reactive and has relatively low selectivity for DNA, as it also reacts with other nucleophiles in the cell such as proteins, leading to DNA–protein crosslinks (DPCs), drug resistance, and genotoxicity.226,227 In contrast, bendamustine is a non-classical nitrogen mustard, incorporating a benzimidazole ring that provides both alkylating activity (via the mustard group) and antimetabolite-like properties (mimicking purine bases).228 It is used in the treatment of non-Hodgkin's lymphoma and multiple myeloma.229
Most small-molecule DNA-binding drugs preferentially target the minor groove, which is narrower and better suited to their small sizes, allowing for strong van der Waals interactions. Major groove binders, mostly intercalators or alkylating agents, also exist, with very few non-covalent binders. Some metal-based small compounds, such as those of platinum, chromium, or rhodium, show this property. Non-covalent minor groove binders can prefer either AT- or GC-rich sites. The minor groove binding drugs include netropsin and distamycin.155 They are pyrrole antibiotics that insert into the minor groove and form hydrogen bonds with donor atoms on adenine (A) or thymine (T) bases.230 These drugs are known to target TFs, such as E2F1/E2F4 and TBP (part of TFIID complex).47 Though they themselves are not very good therapeutics because of binding, toxicity, or specificity issues, they have been the basis for designing many anti-cancer drugs as lead compounds.230
Dervan's polyamides are prime examples of improved sequence-selective minor groove binders based on these. Built from pyrrole-imidazole units, they adopt hairpin-like structures that allow them to recognise predetermined DNA sequences, following Dervan's pairing rules, where Im/Py recognises G·C, Py/Im recognises C·G, and Py/Py recognises both A·T and T·A base pairs.231,232 They have been used to disrupt transcription factor–DNA interactions, such as ERα binding and HIF-responsive elements; in principle, polyamide–peptide conjugates could function as artificial TFs capable of upregulating transcription.233 Although still in preclinical stages, they remain one of the most promising approaches for achieving programmable, sequence-specific targeting of DNA.234
Hoechst 33258 is another minor groove binder,47,155 which is used as a dye for staining DNA and used for fluorescent microscopy, flow cytometry, etc.235
In the case of FDA-approved drugs, mithramycin (also called plicamycin) is a natural product small-molecule inhibitor of TFs such as EWS-FLI1. Earlier, it was used for treating various types of cancers and Paget's disease. It has also been shown to prevent neural cell death in p53-mediated apoptosis by providing neuroprotection, but due to its hepatotoxicity, nephrotoxicity, and bone marrow toxicity, it was discontinued.236 However, it has now seen a resurgence in research and is being used in the therapy of Ewing sarcoma. Unlike most DNA-binding agents, mithramycin acts by directly targeting TFs. It binds in the minor groove of DNA as a Mg2+-coordinated dimer, recognising GC-rich sequences through a two-fold symmetry that complements the DNA site.237 In addition, it serves as a selective Sp1 inhibitor, where it competes for binding at GC-rich motifs, including those within c-MYC promoters.238 Studies have also explored mithramycin analogues (e.g., mithplatin and EC-8042) for overcoming cisplatin resistance. A unique property of mithramycin is its ability to bind non-palindromic GC sequences, which distinguishes it from many other DNA-binding drugs.239,240
Intercalation is a form of non-covalent binding where a flat, aromatic small-molecule inserts itself between the stacked base pairs of the DNA helix. The DNA becomes distorted as a result of this physical insertion, which unwinds the helix (by causing distortions in the sugar-phosphate backbone) and lengthens it (as the drug inserts between bases and increases the spacing). Intercalation also sterically prevents proteins, such as TFs, from binding DNA, following the nearest-neighbour exclusion principle. Similar effects are seen in replication, and many intercalators poison topoisomerase enzymes, causing DNA breakage.241 The unwinding effect is also not limited to that particular base pair, but the torsional strain can extend further along the DNA. The DNA-binding of intercalators is also strongly influenced by π–π stacking interactions between the drug's aromatic rings and the nucleotide base pairs.242
Actinomycin D (dactinomycin) is the first FDA-approved natural DNA intercalator drug, obtained from Streptomyces. Actinomycin D blocks the interaction of RNA polymerase with DNA, thus inhibiting transcription, particularly by halting ribosomal RNA synthesis. Unlike most intercalators, which are cationic (to neutralise DNA's negative charge and reduce repulsion), actinomycin D is largely neutral but possesses a dipole moment. It intercalates in GC-rich regions of the DNA minor groove and is mainly used to treat pediatric tumours, such as Wilms' tumour and rhabdomyosarcoma.243 It also inhibits TFs like Sp1 and Sox2, preventing their binding to DNA.244 In a different mechanism, it stabilises G4 DNA in oncogene promoters like c-MYC, blocking transcription, and reduces mRNA synthesis, effectively “turning off” the oncogene.245
Doxorubicin and daunorubicin are Topo II inhibitors of the anthracycline drug class. They intercalate into DNA and poison Topo II by trapping the enzyme–DNA cleavage complex, causing DSBs. These drugs also impact TFs; for example, they inhibit HIF-1, NF-κB, Sp1, E2F1, and others by blocking their DNA-binding and suppressing gene expression. Doxorubicin is used in breast cancer, sarcoma, and lymphoma therapies, while daunorubicin is used in leukaemias. Their clinical use is limited by cardiotoxicity, among other toxicities. Most clinical intercalators are mono-intercalators (a single aromatic system inserts between bases). In contrast, bis-intercalators like echinomycin have two rings that intercalate at two distinct sites on the DNA. Echinomycin preferentially binds to CpG steps and inhibits transcription. It has been studied as a potential treatment for acute myeloid leukaemia (AML), but toxicity has limited its development.47 For bis-intercalators, the nearest-neighbour exclusion principle does not apply, as they can occupy two sites at once.242
Nogalamycin is another anthracycline-related antibiotic. It uses a “threading intercalation” mechanism, interacting with both the major and minor grooves;246 however, due to severe toxicity, it never advanced beyond trials.247
Amsacrine, though not FDA-approved in the US, is approved in Europe for AML and Hodgkin's and non-Hodgkin's lymphomas. It stabilises the Topo II cleavage complex, causing DSBs.248 Combination therapy with cytarabine has proven effective in AML, especially for patients with cardiac problems, but recent advances focus on improving tolerability and efficacy.249
Voreloxin, a quinolone analogue, functions as a novel intercalator that stabilises G4 structures and selectively induces damage in GC/GG sequences, demonstrating efficacy against AML and solid tumours. It remains in clinical trials, demonstrating sequence-selective cleavage and dual stabilisation of c-MYC and BCL-2 G4s.250–252
In contrast to other drugs, bleomycin chelates metal ions, primarily Fe(II); and when activated by oxygen, it releases free radicals that break DNA strands instead of merely intercalating.253 Cell cycle arrest and apoptosis result from the single- and double-strand breaks caused by this DNA cleavage.254 It has FDA approval for the treatment of squamous cell carcinomas, testicular cancer, and Hodgkin's lymphoma, among other conditions.255 Its efficacy can be enhanced when combined with Ku–DNA binding inhibitors of DNA-PK, which impair the NHEJ pathway essential for repair of bleomycin-induced DSBs.256 Therefore, while bleomycin does not directly interfere with protein–DNA binding, its DNA-cleaving action can lead to indirect effects on these interactions.
Beyond classical intercalators and groove binders, of DNA, non-canonical DNA structures like G4s are emerging targets for anticancer drugs. Camptothecin and indenoisoquinolines, though mainly topoisomerase inhibitors, can also interact with G4s. Camptothecin works synergistically with G4 stabilisers by blocking Topo I-mediated repair, whereas indenoisoquinolines can independently stabilise G4s, such as the c-MYC promoter G4, to suppress oncogene expression.257,258
Several drugs are in development that directly target G4 structures, and CX-5461 (pidnarulex) is a prominent example. Initially characterised as an RNA polymerase I inhibitor, CX-5461 is now known to stabilise G4 DNA and trap Topo II. It exploits synthetic lethality in BRCA1/2-deficient cancers, including tumours resistant to PARP inhibition; its clinical development has been accelerated through FDA Fast Track designation, with early-phase trials in breast and ovarian cancers (see Table 4).259 Concerns regarding its safety arose from an in vitro study reporting extensive collateral mutagenesis in cell lines;260 however, subsequent analyses of clinical samples suggested that these effects may not translate to humans.261 Another G4-targeting compound, QN-302, is in early clinical trials for advanced or metastatic solid tumours.262
Apart from G4s, other non-canonical DNA structures are also being explored as drug targets. Actinomycin D represents a classical model for hairpin-interactive compounds, preferentially binding GC sites within hairpin stems. Its binding affinity is influenced by loop composition and internal mismatches, leading to stabilisation of pathogenic trinucleotide-repeat hairpins.263 More recently, azacryptand ligands, such as tris-acridine and tris-naphthalene derivatives, have demonstrated the ability to bind imperfect DNA hairpins and other folded DNA junctions, supporting a multitargeting strategy that exploits structurally related non-B DNA conformations.264 In parallel, Z-DNA has emerged as an immunologically relevant target. Z-DNA-binding molecules can enhance anti-tumour immune responses and improve the efficacy of anti-PD-1 immunotherapy by unmasking ZBP1-mediated necroptotic signalling normally suppressed by ADAR1.265 In addition to targeting proteins or DNA separately, drugs can also act at the interface between them, directly modulating protein–DNA complexes. This approach is discussed in the next section.
3.3 Targeting the protein–DNA complex
Drugs targeting DNA–protein complexes act at the bound interface between protein and DNA and are often called interfacial inhibitors.45 This strategy addresses the problem of “flat”, otherwise undruggable protein surfaces, since the interface of two molecules creates a pocket for drug binding. The transient complex can be efficiently stabilised by drug binding, making the interaction thermodynamically favourable.275 The clinical applications and specific targets of these inhibitors are summarised in Table 5.Prominent examples include etoposide and mitoxantrone (Topo II–DNA complex poisons), indenoisoquinolines, and camptothecin (Topo I–DNA complex poisons).45 Other agents sometimes discussed in this context are amsacrine and dexrazoxane.
Etoposide traps the Topo II–DNA cleavage complex and stabilises it by blocking DNA religation. Normally, Topo II introduces transient double-strand cleavages to relieve supercoiling. At therapeutic doses, it primarily produces SSBs as only one of the two catalytic sites within the Topo II dimer is trapped (Fig. 5).277 These SSBs remain covalently bound to the enzyme and act as obstacles on the DNA template. When replication or transcription machinery collides with these complexes, they are converted into DSBs or cause fork collapse, which drive cell death. Etoposide targets both Topo IIα and Topo IIβ. Clinically, it is utilised in small-cell lung cancer (SCLC), testicular cancer, soft tissue sarcoma, and other solid tumours. Its main dose-limiting toxicity is bone marrow suppression.45
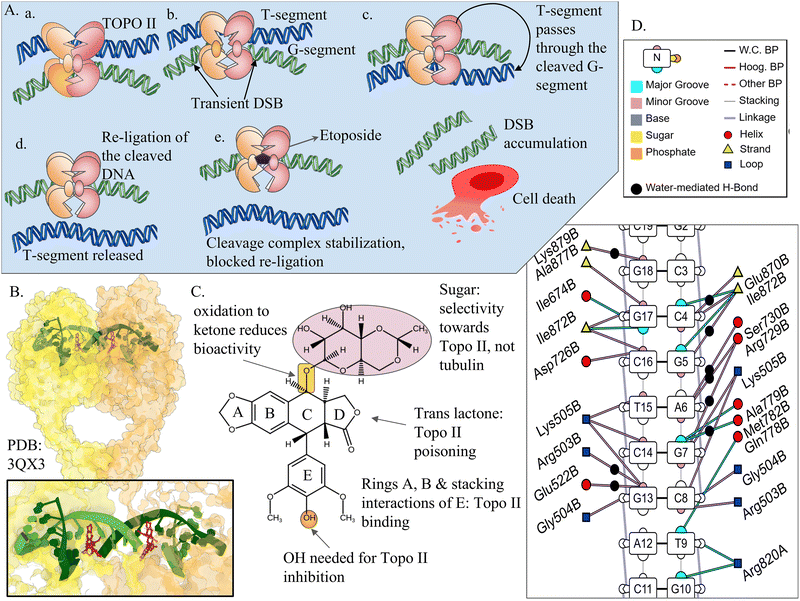 | ||
| Fig. 5 Mechanistic and structural basis of topoisomerase II (Topo II) inhibition by etoposide. (A) Mechanism of action of etoposide showing stabilisation of the transient Topo II–DNA cleavage complex. During the catalytic cycle, Topo II introduces a double-strand break in the gate (G) segment DNA to permit passage of the transported (T) segment. Etoposide intercalates at the protein–DNA interface of the cleaved G segment, inhibits DNA religation, and traps the cleavage complex, leading to accumulation of DNA double-strand breaks, replication stress, and apoptosis. (B) Structure–activity relationship (SAR) of etoposide. (C) Crystal structure of the human Topo IIβ–DNA–etoposide complex (PDB: 3QX3) illustrating the overall surface geometry of the enzyme and the binding of two etoposide molecules at the protein–DNA interface within the cleavage complex.276 (D) NUCPLOT representation of the Topo IIβ–DNA–etoposide complex highlighting representative protein–DNA and protein–drug interactions that stabilise the trapped cleavage intermediate. Visualisations generated via Mol*,68 DNAproDB,93 MarvinSketch for chemical structures, and Chemaxon (https://www.chemaxon.com). | ||
Mitoxantrone acts as both a DNA intercalator and an interfacial inhibitor, stabilising the Topo II–DNA cleavage complex.45,258,278 Unlike anthracyclines such as doxorubicin, mitoxantrone undergoes less efficient redox cycling, generating fewer free radicals. Since anthracycline cardiotoxicity is strongly linked to free radical–mediated mitochondrial damage in the heart, mitoxantrone shows reduced cardiotoxicity in comparison. However, it can still cause dose-dependent cardiac toxicity, mainly through Topo IIβ poisoning in cardiomyocytes.48,258,278,279 Clinically, it is used in AML, acute promyelocytic leukaemia (APL), and multiple sclerosis.278
Camptothecin is a natural product inhibitor that traps the Topo I–DNA cleavage complex, preventing religation of single-strand breaks. This leads to replication fork collapse, S-phase toxicity, and apoptosis. Camptothecin is highly specific, as its only target is Topo I, and shows dose-dependent toxicity. It also halts DNA synthesis. Clinically utilised derivatives include irinotecan and topotecan. Resistance can arise from mutations in Topo I or altered drug efflux.258
Indenoisoquinolines are synthetic non-camptothecin Topo I inhibitors stabilising the Topo I–DNA cleavage complex. They show improved chemical stability and can overcome certain resistance mechanisms associated with camptothecins. Other interfacial inhibitors include raltegravir, elvitegravir, and dolutegravir, which stabilise the HIV integrase–DNA complex, representing microbial applications of this principle.45,280
Some drugs, such as doxorubicin and trabectedin, though primarily classified as DNA intercalators or groove binders, can also act partly as interfacial inhibitors.45 However, as their dominant mechanism is DNA-binding, they are discussed in the preceding section.
Beyond classical small-molecule inhibitors, nanomaterials such as graphene quantum dots have been reported to act as interfacial inhibitors of DNA–protein complexes, including dual inhibition of Topo I and Topo II, with potential to overcome multidrug resistance,281 as discussed in the Section 5.
Apart from the topoisomerase poisons, another type of inhibitor (Polθi) targeting the protein–DNA complex is being explored. It traps the Polθ–Pol–DNA complex in a closed conformation by adding a nucleotide to the primer, thereby blocking the complex transition to the open conformation required for DNA repair. The inhibitor fits into the closed conformation pocket and is stabilised by hydrogen bonding, π-stacking, and hydrophobic interactions. Blocking the N (finger) and O subdomains prevents the polymerase from progressing and renders the complex temporarily resistant to endonuclease cleavage.282
4 Methodological overview: AI and computational approaches in protein–DNA modulator discovery
Rational drug design, supported by computer-aided drug design (CADD) and artificial intelligence (AI), has significantly advanced the discovery of modulators targeting PDIs.284 Molecules identified through AI approaches have reportedly shown higher success rates (up to 80–90%) in early clinical trials compared to conventional candidates.285 Traditional approaches such as structure-based drug design (SBDD) and fragment-based drug design (FBDD), when integrated with molecular modelling, binding site prediction, binding energy estimation and virtual screening, have accelerated the path to the identification of selective and potent inhibitors.284,286 Target identification and prediction represent the first step in drug discovery and have seen a wider application of AI. The integration of multi-omics data and network biology has facilitated the identification of therapeutic targets in complex diseases such as cancer.287 For example, structural modelling of TFs such as c-Myc, facilitated by AlphaFold and refined through molecular dynamics (MD) simulations revealed a druggable conformational switch within its transactivation domain.288 AI-based structural prediction models, including AlphaFold3,289 AlphaFold-Multimer,290 RoseTTAFoldNA,291 and RoseTTAFold All-atom,292 have enabled accurate prediction of novel DNA-binding proteins and protein–DNA complexes.Successful targeting requires detailed information on the binding site. In this direction, both structure-based and sequence-based binding site predictors have been developed. Although the earlier methods were constrained by limited structural data and biological complexity;293–296 recent AI advances have led to powerful binding site predictors such as ATMGBs, which combines protein language models and graph convolutional networks;297 DeepPBS, a geometric deep learning model predicting binding specificity from protein–DNA structures;14 and TransBind, an alignment-free deep learning framework that predicts DNA-binding proteins and residues directly from sequences.298 Tools and databases such as JASPAR,91 AIModules,299 MethMotif90 and MEME Suite300 support motif identification, providing curated TF binding profiles and position weight matrices essential for motif discovery.
The use of curated chemical and structural databases, combined with machine learning (ML) and deep learning (DL) algorithms, enables efficient exploration of chemical space and prioritisation of candidate molecules. Molecular docking tools such as HADDOCK,301 NPDock,302 and MD simulations further refine predicted protein–DNA complex structures, providing insights into binding stability and interaction mechanisms.303 These approaches have been applied to Gli1/DNA interactions as a druggable target for Hedgehog-dependent tumours,304in silico modelling and screening of FOXO TF inhibitors,305 and STAT3 inhibition in cancer therapeutics.3 AI-enhanced MD simulations, incorporating ML force fields, improve the accuracy of studying dynamic PDIs.306,307 From the DNA side, Dictionary-based approaches (DNA shape and Dynaseq) hold great promise through their modelling of large-scale DNA shape and flexibility patterns, revealing conformational signatures crucial for drug binding.308,309 DL methods, such as Deep DNAshape, predict DNA structure.310 High-throughput screening (HTS)311 and DNA-encoded libraries (DELs)312 accelerate the identification of specific inhibitors, including for challenging targets like Myc in cancer,140 while AI-driven virtual screening platforms improve compound selection and druggability assessments.313,314 Chemical databases such as ChEMBL,315 ZINC,316 BindingDB,317 and DrugBank318 provide essential chemical and pharmacological data. Natural language processing (NLP) models extracting drug–target relationships from FDA drug labels further enhance regulatory assessment and drug development.319 QSAR modelling, enhanced by ML, remains vital for predicting biological activity and optimising drug candidates, streamlining drug design effectively.320 Generative AI frameworks, including autoencoders, graph neural networks (GNNs), and reinforcement learning (RL) models, enable de novo molecule generation, opening new frontiers in drug discovery by designing novel compounds with optimised properties.321,322 Additionally, Aloptamer is an AI-driven pipeline that accelerates aptamer optimisation by integrating AI screening, structural modeling, deep learning scoring, and MD simulations, enhancing aptamer discovery.323 Beyond drug design, AI and ML-based frameworks have also advanced predictive oncology, enabling precise prediction of therapeutic response and treatment outcome.324 A recent pan-cancer, pan-treatment model further demonstrated that patient-derived xenograft–driven AI systems can accurately forecast drug responses across cancer types, accelerating preclinical testing and precision drug development.325
While various strategies to target protein–DNA interactions show promise, they face broader challenges related to selectivity, efficacy, and practical application, which are discussed in the next section.
| Drug | Target | Mechanism of action | Approval status | Drug class | Therapeutic area | Ref. |
|---|---|---|---|---|---|---|
| Abbreviations: Topo I, topoisomerase I; Topo IIα, topoisomerase II alpha; DSB, double-strand break; AML, acute myeloid leukaemia | ||||||
| Etoposide | Topo IIα–DNA complex | Stabilises cleavage complex, preventing religation, leading to persistent DSBs during replication | FDA approved | Podophyllotoxin derivative, small-molecule | Leukaemia, lymphoma, testicular/lung cancer | 45 |
| Camptothecin | Topo I–DNA complex | Stabilises cleavage complex, prevents religation leading to replication fork collapse and DSBs | FDA approved | Plant alkaloid, small molecule | Colorectal, ovarian, lung cancers | 45 and 258 |
| Indeno-isoquinolines | Topo I–DNA complex | Stabilises the complex and inhibits the religation reaction of DNA | Phase I trials (NCT03030417) | Small-molecule | Solid tumours | 45 and 283 |
| Mitoxantrone | Topo IIα–DNA complex (also intercalates DNA directly) | Stabilises the cleavage complex, blocking replication and repair | FDA approved | Anthracenedione, small molecule | AML, prostate cancer, breast cancer, multiple sclerosis | 45 and 278 |
5 Challenges and future perspectives
Developing drugs for PDI targets is made complicated by two intersecting challenges: the biological complexity inherent to protein–DNA recognition and interaction surfaces, and technological limitations that restrict our understanding and experimental capacity.11,326,327 Many TFs, for instance, lack well-defined ligand-binding pockets and have historically been considered challenging to target.46,328 Many TFs are intrinsically disordered proteins (IDPs), such as c-Myc with its disordered TAD. As for IDPs, they are highly flexible and dynamic, lacking fixed 3D structures under normal physiological conditions and instead adopting multiple conformations. Their sequences are typically rich in polar residues and depleted of hydrophobic ones, leading to structural instability and a lack of rigid binding sites. Consequently, small-molecule inhibitors that can target them efficiently and specifically are scarce, since these compounds are known to typically bind to stable hydrophobic pockets; the lack of such pockets results in only low-affinity interactions. For example, the p53 binding site for MDM2 lies within a disordered region, making direct inhibition extremely challenging.326 There are attempts at targeting conserved genomic TAD regions belonging to specific targets of TF families, where specificity of binding remains the primary challenge. Even protein-based inhibitors designed to mimic or disrupt these contacts often suffer from rapid degradation and limited stability in vivo.329The intricate nature of PDIs also makes CADD difficult. The key computational technique of MD simulations, hugely successful in other systems, becomes challenging for TFs in PDIs due to their high flexibility and absence of stable structures. Classical force fields often fail to capture IDP behaviour accurately, prompting the use of coarse-grained models or enhanced sampling techniques to improve prediction. One would expect that some of these problems can be solved by using DNA sequences they recognise, instead of new small molecules. However, DNA fragments recognised by TFs are small and this approach may cause off-target effects due to a lack of precision and redundancy of similar DNA sites in the genome. Additionally, there remains a fundamental lack of understanding regarding the binding patterns and mechanisms of these proteins. Developing effective drugs depends critically on identifying their small functional sites and linear motifs. Technologies such as in-cell NMR and AI-driven structure prediction can ensure that compounds bind specifically to their intended targets while avoiding off-target effects.326,330
Proteins involved in DNA repair and replication present their own set of therapeutic challenges. Many DNA repair proteins have broad, flat surfaces, which makes them hard to target. Many of these proteins, such as RAD51, RAD52, and Ku70/80, have large and relatively flat DNA-binding surfaces that are difficult to block selectively. PARP inhibitors, though successful, face resistance through restoration of HR or loss of PARP trapping efficiency.7,331 Synthetic lethality remains a powerful concept, yet maintaining its clinical effectiveness depends on identifying new pathway partners and rational drug combinations. High-throughput CRISPR–Cas9 screening has enabled the identification of novel synthetic lethal interactions outside the BRCA-mutant contexts, such as WRN inhibitors in MSI-H tumours or SMARCA2 degraders in SMARCA4-mutant cancers, providing avenues to circumvent PARP resistance and expand therapeutic options.332 Similarly, inhibitors of DNA-PKcs face challenges related to kinase selectivity and toxicity, since the ATP-binding pocket is highly conserved across the PI3K family. The clinical failure of agents like AZD7648 highlights the unresolved balance between potency, specificity, and safety in targeting DNA repair enzymes.333
Drug resistance is another major phenomenon that complicates therapeutic targeting of PDIs. Defects in the MMR or NER pathways can alter responses to DNA-damaging agents such as cisplatin, temozolomide, and etoposide. Overexpression of repair proteins, such as ERCC1 and MGMT, or mutations in MSH2 and MLH1, contributes to drug resistance by either enhancing lesion removal or impairing DNA damage signalling.334–336 The dynamic crosstalk between repair pathways means that blocking one mechanism often triggers compensatory mechanisms, highlighting the need for rationally designed drug combinations.337
Another inherent challenge in targeting PDIs for drug discovery arises from the limited chemical diversity of DNA itself. DNA is composed of only four bases, yet it encodes thousands of genes, making it challenging to design drugs that selectively target a specific sequence without affecting others. While TFs typically recognise the major groove, small molecules often bind in the minor groove because it provides a snug, shape-complementary site for interaction. In contrast, the major groove is wide, relatively flat, and solvent-exposed, making selective binding of small molecules more difficult.155,234
DNA intercalators remain potent but nonspecific. Bis-intercalators may increase DNA-binding strength but often at the cost of enhanced toxicity and off-target effects.47 Similarly, interfacial inhibitors such as topoisomerase poisons still face limitations in selectivity. For example, lack of isoform-selective inhibitors, particularly those distinguishing Topo IIα from Topo IIβ, results in unwanted side effects such as cardiotoxicity and bone marrow suppression.48
Therefore, enhancing specificity, reducing toxicity, and addressing drug resistance remain key future directions that require substantial scientific effort.209 Solving these challenges depends on precise characterisation of how small molecules interact with DNA. High-resolution analytical platforms, such as single-molecule nanopore sensing, can detect and quantify subtle DNA structural differences at single–base-pair resolution, including canonical and non-canonical hydrogen-bonding states, as well as transient interactions between small molecules and DNA bases.338,339
Combination therapy has shown progress in combating drug resistance. For example, co-inhibition of DOT1L and menin has demonstrated synergistic effects in MLL-rearranged leukaemias,209 while pairing topoisomerase inhibitors with TDP1 or TDP2 blockers may enhance the persistence of DNA damage in tumour cells.48 Advanced drug delivery systems offer another solution. Nanoparticle-encapsulated inhibitors improve tumour targeting and reduce systemic side effects.48 For example, graphene quantum dots (GQDs) enhance drug targeting, improve cellular uptake, and modulate PDIs through π–π stacking and electrostatic interactions. They can self-insert into the DNA major groove sites, enhancing interfacial inhibition of TFs and downregulating cancer stem cell genes, thereby overcoming multidrug resistance. However, optimisation of specificity and biocompatibility is still required.281,340 Liposomal delivery also improves drug safety, bioavailability, and efficacy, as exemplified by lipoplatin, but challenges remain in formulation, stability, and in vitro–in vivo translation. Multi-modal liposomes and liposome–nanoparticle hybrids offer opportunities for the co-delivery of cytotoxic drugs and resistance pathway inhibitors, with potential for targeted and externally triggered release.341
Drug repurposing offers a faster and less risky path to discovering PDI modulators. For example, eltrombopag, an FDA-approved drug for chronic immune thrombocytopenia, has recently been shown to inhibit transcription factor EB (TFEB), enhancing apoptosis inhibition and sensitising tumours to chemo- or radiotherapy.342 Pentamidine, originally FDA-approved for protozoal infections (e.g., PCP, trypanosomiasis, leishmaniasis),343 has been repurposed in cancer therapy.344 It binds to AT-rich regions in the DNA minor groove, thereby interfering with TF–DNA interactions (e.g., STAT3, SOX-2, and CDK4). Studies also show that pentamidine targets the suppression of HIF-1α protein translation and inhibits the interaction between S100P and p53, through protein binding. While it shows activity in prostate and brain cancers and is being tested in combination therapies, challenges such as low bioavailability, delivery issues, and toxicity remain.344
Chemical biology tools, such as PROTACs, decoy PROTACs (D-PROTACs), molecular glues, and molecular glue degrader–antibody conjugates (MACs), offer indirect strategies to degrade, stabilise, or mislocalise PDIs, overcoming the limitations of classical inhibitors. While PROTACs induce targeted protein degradation via the ubiquitin–proteasome system,345 D-PROTACs further extend this strategy by fusing DNA decoys with E3 ligase ligands, enabling sequence-specific recruitment of DNA-binding proteins like STAT3 for targeted degradation and antitumour activity.346 Molecular glues complement these strategies by promoting proximity between E3 ligases and target proteins. Classical examples include thalidomide derivatives that recruit CRBN to degrade IKZF1 and IKZF3, while newer glues such as DKY709 and Helios CELMoD (targeting IKZF2) and WIZ modulators expand this approach to additional TFs.347 Furthermore, c-Myc-degrading MACs have recently demonstrated targeted degradation of the otherwise “undruggable” c-Myc via CRBN recruitment, achieving selective antitumour activity in PSMA-positive prostate cancer models.348 Though there are several limitations of traditional PROTACs, such as solubility, permeability and off-target issues, this has now been addressed using unconventional PROTACs.349
Recent advances in AI-driven structural biology have greatly enhanced our understanding of PDIs and their potential in drug design.290,291,350–352 Despite their promise, these AI-based tools have limitations: motif-specific recognition, non-canonical interactions, and highly dynamic regions can still be mispredicted. Integrating experimental techniques such as ChIP-seq, pull-down assays, or motif discovery algorithms like MEME is essential to refine predicted binding sites.352 Data bias issues, limited representation of disordered/transient complexes and the “black box” of deep learning frameworks limit biological interpretability, suggesting a need for explainable AI (XAI).353,354
Translating the computational findings into clinically viable drugs faces traditional hurdles, including poor stability, bioavailability and efficacy. Data integration across platforms is limited by heterogeneity, computational limitations, and biological interpretability. There is a need for a clear framework to facilitate the transition from in silico findings to in vivo applications, ultimately leading to the development of actual therapeutic drugs in real-life settings.355
Moving forward, a multidisciplinary approach combining structural biology, AI-driven modelling and predictions, chemical biology, nanotechnology, and multi-omics studies will be crucial for rationally designing drugs that precisely target PDIs. At the same time, careful evaluation of safety, specificity, and in vivo efficacy will be crucial for developing effective and clinically relevant therapies.
Conclusion
Transcription, replication, DNA repair, and chromatin regulation are key biological processes mediated by PDIs and provide important opportunities for drug targeting. PDIs have traditionally been considered “undruggable” due to the lack of conventional binding pockets; however a diverse range of targeting strategies have been developed to modulate these interactions. Success stories are evident in FDA-approved drugs, compounds currently under clinical investigation, and emerging modalities such as PROTACs. Targeting approaches span from small molecules that bind directly to DNA or proteins, to interfacial inhibitors that trap the protein–DNA interface in complex. In cases where direct inhibition is unfeasible, such as for histones or architectural proteins, indirect chemical biology strategies are increasingly being explored.This review also highlights key challenges, including selectivity, off-target effects, and drug resistance, that need to be overcome to realise the full potential of PDI targeting. New technologies including AI-based structural predictions, advanced drug delivery systems (e.g., nanoparticles and liposomes), and combination therapies are helping to address these limitations. Rational drug design, guided by structural biology, chemical biology, and computational modelling, will be critical for developing precise, effective, and clinically viable outcomes in this direction.
Author contributions
This article was drafted by RM under the overall guidance of SA. Literature survey and data compilation were done by RM, whereas SA contributed to the overall structure, discussions and editing the draft versions. Both authors read and approved the final version.Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was supported by DBT grants (BIC to JNU, NNP to IIITD, and NNP to CSIR-NEIST) and an ICMR grant on MTB drug-resistance to SA. This was also supported by a Junior Research fellowship to RM from UGC-India.References
- R. A. C. Ferraz, A. L. G. Lopes, J. A. F. da Silva, D. F. V. Moreira, M. J. N. Ferreira and S. V. de Almeida Coimbra, Plant Methods, 2021, 17, 82 CrossRef CAS PubMed
.
- A. Alibes, A. D. Nadra, F. De Masi, M. L. Bulyk, L. Serrano and F. Stricher, Nucleic Acids Res., 2010, 38, 7422–7431 CrossRef CAS PubMed
.
- M. Radaeva, A.-T. Ton, M. Hsing, F. Ban and A. Cherkasov, Drug Discovery Today, 2021, 26, 2660–2679 CrossRef CAS PubMed
.
- C. V. Dang, Cell, 2012, 149, 22–35 CrossRef CAS PubMed
.
- Y. Shiroma, R.-u Takahashi, Y. Yamamoto and H. Tahara, Cancer Sci., 2020, 111, 1058–1064 CrossRef CAS PubMed
.
- M. W. Gonzalez and M. G. Kann, PLoS Comput. Biol., 2012, 8, 1–11 CrossRef PubMed
.
- S. R. Hengel, M. A. Spies and M. Spies, Cell Chem. Biol., 2017, 24, 1101–1119 CrossRef CAS PubMed
.
- X. Li and W.-D. Heyer, Cell Res., 2008, 18, 99–113 CrossRef CAS PubMed
.
- C. Qu and H. Du, Crop Design, 2025, 4, 100082 CrossRef
.
- H. Mousavi and R. Emami, Sci. Rep., 2025, 15, 30565 CrossRef CAS PubMed
.
- A. A. Anashkina, Biophys. Rev., 2023, 15, 1007–1014 CrossRef CAS PubMed
.
- R. Nagarajan, S. Ahmad and M. Michael Gromiha, Nucleic Acids Res., 2013, 41, 7606–7614 CrossRef CAS PubMed
.
- Z. Liu, W.-R. Qiu, Y. Liu, H. Yan, W. Pei, Y.-H. Zhu and J. Qiu, Anal. Biochem., 2025, 115862 CrossRef CAS PubMed
.
- R. Mitra, J. Li, J. M. Sagendorf, Y. Jiang, A. S. Cohen, T.-P. Chiu, C. J. Glasscock and R. Rohs, Nat. Methods, 2024, 21, 1674–1683 CrossRef CAS PubMed
.
- L. R. Genz, S. Nair, A. Sweeney and M. Topf, Curr. Opin. Struct. Biol., 2025, 95, 103165 CrossRef CAS PubMed
.
- Sangeeta, S. K. Mishra and A. Bhattacherjee, JACS Au, 2024, 4, 2640–2655 CrossRef CAS PubMed
.
-
M. Brenowitz, D. F. Senear, M. A. Shea and G. K. Ackers, Methods in enzymology, Elsevier, 1986, vol. 130, pp. 132–181 Search PubMed
.
- M. G. Fried, Electrophoresis, 1989, 10, 366–376 CrossRef CAS PubMed
.
- D. S. Johnson, A. Mortazavi, R. M. Myers and B. Wold, Science, 2007, 316, 1497–1502 CrossRef CAS PubMed
.
- P. Schuck and A. P. Minton, Trends Biochem. Sci., 1996, 21, 458–460 CrossRef CAS PubMed
.
- B. Gyarfas, F. Olasagasti, S. Benner, D. Garalde, K. R. Lieberman and M. Akeson, ACS Nano, 2009, 3, 1457–1466 CrossRef CAS PubMed
.
-
A. H. Squires, T. Gilboa, C. Torfstein, N. Varongchayakul and A. Meller, Methods in enzymology, Elsevier, 2017, vol. 582, pp. 353–385 Search PubMed
.
- J. Li, Y. Wang, L. Wang, Y. Wang, Z. Zhang, S. Liu and L. Wang, ACS Mater. Lett., 2025, 7, 2476–2481 CrossRef CAS
.
- Y.-L. Ying, Z.-L. Hu, S. Zhang, Y. Qing, A. Fragasso, G. Maglia, A. Meller, H. Bayley, C. Dekker and Y.-T. Long, Nat. Nanotechnol., 2022, 17, 1136–1146 CrossRef CAS PubMed
.
- S. Schmid and C. Dekker, Essays Biochem., 2021, 65, 93–107 CrossRef CAS PubMed
.
- V. Bijak, M. Szczygiel, J. Lenkiewicz, M. Gucwa, D. R. Cooper, K. Murzyn and W. Minor, Expert Opin. Drug Discovery, 2023, 18, 1221–1230 CrossRef PubMed
.
- S. Campagne, V. Gervais and A. Milon, J. R. Soc., Interface, 2011, 8, 1065–1078 CrossRef CAS PubMed
.
- M. D. Wilson and A. Costa, Biol. Crystallogr., 2017, 73, 541–548 CAS
.
- H. S. Kaya-Okur, D. H. Janssens, J. G. Henikoff, K. Ahmad and S. Henikoff, Nat. Protoc., 2020, 15, 3264–3283 CrossRef CAS PubMed
.
- A. Bartlett, R. C. O'Malley, S.-S. C. Huang, M. Galli, J. R. Nery, A. Gallavotti and J. R. Ecker, Nat. Protoc., 2017, 12, 1659–1672 CrossRef CAS PubMed
.
- J. Concepcion, K. Witte, C. Wartchow, S. Choo, D. Yao, H. Persson, J. Wei, P. Li, B. Heidecker and W. Ma,
et al.
, Comb. Chem. High Throughput Screening, 2009, 12, 791–800 CrossRef CAS PubMed
.
- M. R. Corces, J. M. Granja, S. Shams, B. H. Louie, J. A. Seoane, W. Zhou, T. C. Silva, C. Groeneveld, C. K. Wong and S. W. Cho,
et al.
, Science, 2018, 362, eaav1898 CrossRef PubMed
.
- C. Sun and Y. Feng, Protein J., 2024, 43, 513–521 CrossRef CAS PubMed
.
- Y. Liu and B. Tian, Briefings Bioinf., 2024, 25, bbad488 CrossRef PubMed
.
- A. Gress, V. Ramensky and O. V. Kalinina, Oncogenesis, 2017, 6, e380–e380 CrossRef CAS PubMed
.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed
.
- H. M. Berman and S. K. Burley, Q. Rev. Biophys., 2025, 58, e9 CrossRef CAS PubMed
.
- G. O. Consortium, Nucleic Acids Res., 2004, 32, D258–D261 CrossRef PubMed
.
- S. A. Aleksander, J. Balhoff, S. Carbon, J. M. Cherry, H. J. Drabkin, D. Ebert, M. Feuermann, P. Gaudet and N. L. Harris,
et al.
, Genetics, 2023, 224, iyad031 CrossRef CAS PubMed
.
- T. U. Consortium, Nucleic Acids Res., 2024, 53, D609–D617 CrossRef PubMed
.
- A. N. Koehler, Curr. Opin. Chem. Biol., 2010, 14, 331–340 CrossRef CAS PubMed
.
- T. Helleday, E. Petermann, C. Lundin, B. Hodgson and R. A. Sharma, Nat. Rev. Cancer, 2008, 8, 193–204 CrossRef CAS PubMed
.
- H. A. Malone and C. W. Roberts, Nat. Rev. Drug Discovery, 2024, 23, 661–681 CrossRef CAS PubMed
.
- Y. S. Raouf, Drug Discovery Today, 2024, 29, 104094 CrossRef CAS PubMed
.
- Y. Pommier and C. Marchand, Nat. Rev. Drug Discovery, 2012, 11, 25–36 CrossRef CAS PubMed
.
- J. H. Bushweller, Nat. Rev. Cancer, 2019, 19, 611–624 CrossRef CAS PubMed
.
- J. Portugal, Biochem. Pharmacol., 2018, 155, 336–345 CrossRef CAS PubMed
.
- Y. Pommier, ACS Chem. Biol., 2013, 8, 82–95 CrossRef CAS PubMed
.
- M. R. Nathan and P. Schmid, Oncol. Ther., 2017, 5, 17–29 CrossRef PubMed
.
- T. M. Beer, A. J. Armstrong, D. E. Rathkopf, Y. Loriot, C. N. Sternberg, C. S. Higano, P. Iversen, S. Bhattacharya, J. Carles and S. Chowdhury,
et al.
, N. Engl. J. Med., 2014, 371, 424–433 CrossRef PubMed
.
- E. Garralda, M.-E. Beaulieu, V. Moreno, S. Casacuberta-Serra, S. Martinez-Martin, L. Foradada, G. Alonso, D. Masso-Valles, S. López-Estévez and T. Jauset,
et al.
, Nat. Med., 2024, 30, 762–771 CrossRef CAS PubMed
.
- L. B. Snyder, T. K. Neklesa, R. R. Willard, D. A. Gordon, J. Pizzano, N. Vitale, K. Robling, M. A. Dorso, W. Moghrabi, S. Landrette, R. Gedrich, S. H. Lee, I. C. Taylor and J. G. Houston, Mol. Cancer Ther., 2025, 24, 511–522 CrossRef CAS PubMed
.
- G. Kim, G. Ison, A. E. McKee, H. Zhang, S. Tang, T. Gwise, R. Sridhara, E. Lee, A. Tzou and R. Philip,
et al.
, Clin. Cancer Res., 2015, 21, 4257–4261 CrossRef CAS PubMed
.
- S. M. Hoy, Drugs, 2018, 78, 1939–1946 CrossRef PubMed
.
- S. Grant, C. Easley and P. Kirkpatrick, Nat. Rev. Drug Discovery, 2007, 6, 21–22 CrossRef CAS PubMed
.
- R. Straining and W. Eighmy, J. Adv. Pract. Oncol., 2022, 13, 158 CrossRef PubMed
.
- A. Bisht, D. Avinash, K. K. Sahu, P. Patel, G. Das Gupta and B. D. Kurmi, Drug Delivery Transl. Res., 2025, 15, 102–133 CrossRef PubMed
.
- D. Tsvetkova and S. Ivanova, Molecules, 2022, 27, 2466 CrossRef CAS PubMed
.
- T. K. Choueiri, T. M. Bauer, D. F. McDermott, E. Arrowsmith, A. Roy, R. F. Perini, D. Vickery and S. S. Tykodi, Phase 2 study of the oral hypoxia-inducible factor 2α (HIF-2α) inhibitor MK-6482 in combination with cabozantinib in patients with advanced clear cell renal cell carcinoma (ccRCC), 2021.
- National Cancer Institute (NCI), Olaparib in Combination with Vorinostat in Patients with Relapsed/Refractory and/or Metastatic Breast Cancer, https://clinicaltrials.gov/study/NCT03742245, 2018, Accessed September 28, 2025.
- S. M. Gough, J. J. Flanagan, J. Teh, M. Andreoli, E. Rousseau, M. Pannone, M. Bookbinder, R. Willard, K. Davenport, E. Bortolon, G. Cadelina, D. Gordon, J. Pizzano, J. Macaluso, L. Soto, J. Corradi, K. Digianantonio, I. Drulyte, A. Morgan, C. Quinn, M. Bekes, C. Ferraro, X. Chen, G. Wang, H. Dong, J. Wang, D. R. Langley, J. Houston, R. Gedrich and I. C. Taylor, Clin. Cancer Res., 2024, 30, 3549–3563 CrossRef CAS PubMed
.
- K. K. Swinger, K. M. Lemberg, Y. Zhang and P. A. Rice, EMBO J., 2003, 22, 3749–3760 CrossRef CAS PubMed
.
- U.-M. Ohndorf, M. A. Rould, Q. He, C. O. Pabo and S. J. Lippard, Nature, 1999, 399, 708–712 CrossRef CAS PubMed
.
- D. B. Nikolov, H. Chen, E. D. Halay, A. Hoffman, R. G. Roeder and S. K. Burley, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 4862–4867 CrossRef CAS PubMed
.
- M. Elrod-Erickson, M. A. Rould, L. Nekludova and C. O. Pabo, Structure, 1996, 4, 1171–1180 CrossRef CAS PubMed
.
- S. Dürr, BioIcons April 2024 Release, https://bioicons.com.
- G. Tomasello, I. Armenia and G. Molla, Bioinformatics, 2020, 36, 2909–2911 CrossRef CAS PubMed
.
- D. Sehnal, S. Bittrich, M. Deshpande, R. Svobodova, K. Berka, V. Bazgier, S. Velankar, S. K. Burley, J. Koca and A. S. Rose, Nucleic Acids Res., 2021, 49, W431–W437 CrossRef CAS PubMed
.
- L. Mariño-Ramrez, M. G. Kann, B. A. Shoemaker and D. Landsman, Expert Rev. Proteomics, 2005, 2, 719–729 CrossRef PubMed
.
- M. Lin and J.-t Guo, Nucleic Acids Res., 2019, 47, 11103–11113 CrossRef CAS PubMed
.
- F. V. Murphy and M. E. Churchill, Structure, 2000, 8, R83–R89 CrossRef CAS PubMed
.
- Z. Zheng and Y. Wang, Biomol. Concepts, 2011, 2, 293–303 CrossRef CAS PubMed
.
- S. Ahmad, H. Kono, M. J. Arauzo-Bravo and A. Sarai, Nucleic Acids Res., 2006, 34, W124–W127 CrossRef CAS PubMed
.
- M. J. Arauzo-Bravo and A. Sarai, Nucleic Acids Res., 2008, 36, 376–386 CrossRef CAS PubMed
.
- A. B. Kolomeisky, Phys. Chem. Chem. Phys., 2011, 13, 2088–2095 RSC
.
- R. Murugan, Biophys. J., 2010, 99, 353–359 CrossRef CAS PubMed
.
- W. K. Olson, A. A. Gorin, X.-J. Lu, L. M. Hock and V. B. Zhurkin, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 11163–11168 CrossRef CAS PubMed
.
- N. M. Luscombe, R. A. Laskowski and J. M. Thornton, Nucleic Acids Res., 2001, 29, 2860–2874 CrossRef CAS PubMed
.
- P. J. Sapienza, T. Niu, M. R. Kurpiewski, A. Grigorescu and L. Jen-Jacobson, J. Mol. Biol., 2014, 426, 84–104 CrossRef CAS PubMed
.
- L.-A. Harris, L. D. Williams and G. B. Koudelka, Nucleic Acids Res., 2014, 42, 14053–14059 CrossRef CAS PubMed
.
- S. Harteis and S. Schneider, Int. J. Mol. Sci., 2014, 15, 12335–12363 CrossRef PubMed
.
- R. Rohs, X. Jin, S. M. West, R. Joshi, B. Honig and R. S. Mann, Annu. Rev. Biochem., 2010, 79, 233–269 CrossRef CAS PubMed
.
- R. Rohs, S. M. West, A. Sosinsky, P. Liu, R. S. Mann and B. Honig, Nature, 2009, 461, 1248–1253 Search PubMed
.
- K. B. Chhetri, Y. H. Jang, Y. Lansac and P. K. Maiti, Phys. Chem. Chem. Phys., 2023, 25, 31335–31345 RSC
.
- J. W. Schwabe, Curr. Opin. Struct. Biol., 1997, 7, 126–134 Search PubMed
.
- R. Satange and M.-H. Hou, RSC Chem. Biol., 2025, 6, 699–720 RSC
.
- C. O. Pabo and R. T. Sauer, Annu. Rev. Biochem., 1992, 61, 1053–1095 CrossRef CAS PubMed
.
- N. M. Luscombe, S. E. Austin, H. M. Berman and J. M. Thornton, Genome Biol., 2000, 1, 001-1 CrossRef PubMed
.
- V. Matys, E. Fricke, R. Geffers, E. Gößling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel and O. V. Kel-Margoulis,
et al.
, Nucleic Acids Res., 2003, 31, 374–378 CrossRef CAS PubMed
.
- M. Dyer, Q. X. X. Lin, S. Shapoval, D. Thieffry and T. Benoukraf, Nucleic Acids Res., 2024, 52, D222–D228 CrossRef CAS PubMed
.
- I. Rauluseviciute, R. Riudavets-Puig, R. Blanc-Mathieu, J. A. Castro-Mondragon, K. Ferenc, V. Kumar, R. B. Lemma, J. Lucas, J. Chèneby and D. Baranasic,
et al.
, Nucleic Acids Res., 2024, 52, D174–D182 CrossRef CAS PubMed
.
- L. Yang, T. Zhou, I. Dror, A. Mathelier, W. W. Wasserman, R. Gordân and R. Rohs, Nucleic Acids Res., 2014, 42, D148–D155 CrossRef CAS PubMed
.
- R. Mitra, A. S. Cohen, J. M. Sagendorf, H. M. Berman and R. Rohs, Nucleic Acids Res., 2025, 53, D396–D402 CrossRef CAS PubMed
.
- R. W. Tak Leung, X. Jiang, K. H. Chu and J. Qin, Nucleic Acids Res., 2019, 47, D322–D329 CrossRef PubMed
.
- Y. Cho, S. Gorina, P. D. Jeffrey and N. P. Pavletich, Science, 1994, 265, 346–355 CrossRef CAS PubMed
.
- A. Emamjomeh, D. Choobineh, B. Hajieghrari, N. MahdiNezhad and A. Khodavirdipour, Mol. Biol. Rep., 2019, 46, 3571–3596 CrossRef CAS PubMed
.
- Z. Yan, F. Ge, Y. Zhang, H. Yan, Y. Liu, Y.-H. Zhu, J. Song and D.-J. Yu, J. Chem. Inf. Model., 2025, 65, 8335–8344 CrossRef CAS PubMed
.
- D. Engelkamp and V. van Heyningen, Curr. Opin. Genet. Dev., 1996, 6, 334–342 CrossRef CAS PubMed
.
- N. Chi and J. A. Epstein, Trends Genet., 2002, 18, 41–47 CrossRef CAS PubMed
.
- J. Husby, A. K. Todd, S. M. Haider, G. Zinzalla, D. E. Thurston and S. Neidle, J. Chem. Inf. Model., 2012, 52, 1179–1192 Search PubMed
.
- M. A. Samad, I. Ahmad, A. Hasan, M. H. Alhashmi, A. Ayub, F. A. Al-Abbasi, A. Kumer and S. Tabrez, MedComm, 2025, 6, e70152 Search PubMed
.
- P. Chen, B. Li and L. Ou-Yang, Front. Endocrinol., 2022, 13, 839005 CrossRef PubMed
.
- G. C. Shukla, A. R. Plaga, E. Shankar and S. Gupta, Andrology, 2016, 4, 366–381 CrossRef CAS PubMed
.
- M. Fischer and M. A. Sammons, Cell Death Differ., 2024, 31, 836–843 CrossRef PubMed
.
- D. W. Hagey, M. Bergsland and J. Muhr, Development, 2022, 149, dev200547 CrossRef CAS PubMed
.
- E. Shaulian and M. Karin, Nat. Cell Biol., 2002, 4, E131–E136 CrossRef CAS PubMed
.
- S. Trop-Steinberg and Y. Azar, Am. J. Med. Sci., 2017, 353, 474–483 CrossRef PubMed
.
- P. B. Bhosale, H. H. Kim, A. Abusaliya, P. Vetrivel, S. E. Ha, M. Y. Park, H. J. Lee and G. S. Kim, J. Evidence-Based Complementary Altern. Med., 2022, 2022, 9797929 Search PubMed
.
- M. Alcalay, N. Meani, V. Gelmetti, A. Fantozzi, M. Fagioli, A. Orleth, D. Riganelli, C. Sebastiani, E. Cappelli and C. Casciari,
et al.
, J. Clin. invest., 2003, 112, 1751–1761 CrossRef CAS PubMed
.
- X. Yuan, W. Ruan, B. Bobrow, P. Carmeliet and H. K. Eltzschig, Nat. Rev. Drug Discovery, 2024, 23, 175–200 CrossRef CAS PubMed
.
- D. P. Vageli, P. G. Doukas, K. Goupou, A. D. BENOS, K. Astara, K. Zacharouli, S. Sotiriou and M. Ioannou, Oncol. Res., 2024, 32, 1239 CrossRef PubMed
.
- G. Roviello, I. De Gennaro, I. Vascotto, G. Venturi, A. D'Angelo, C. Winchler, A. Guarino, S. Cacioppo, M. Modesti and M. M. Mela,
et al.
, Genes, 2024, 16, 6 CrossRef PubMed
.
- J. W. Lee, J. Ko, C. Ju and H. K. Eltzschig, Exp. Mol. Med., 2019, 51, 1–13 Search PubMed
.
- S. Jaiswal, X. Han and H. P. Lu, J. Phys. Chem. B, 2022, 126, 997–1003 Search PubMed
.
- R. Dueva and G. Iliakis, NAR Cancer, 2020, 2, zcaa022 CrossRef PubMed
.
- C. Laguri, I. Duband-Goulet, N. Friedrich, M. Axt, P. Belin, I. Callebaut, B. Gilquin, S. Zinn-Justin and J. Couprie, Biochemistry, 2008, 47, 6199–6207 CrossRef CAS PubMed
.
- D. Jaworski, B. Brzoszczyk and L. Szylberg, Cells, 2023, 12, 1375 CrossRef CAS PubMed
.
- D. M. Wilson III and V. A. Bohr, DNA Repair, 2007, 6, 544–559 CrossRef PubMed
.
- A. Piscone, F. Gorini, S. Ambrosio, A. Noviello, G. Scala, B. Majello and S. Amente, Cells, 2025, 14, 112 CrossRef CAS PubMed
.
- D. C. George, F. E. Bertrand and G. Sigounas, Adv. Biol. Regul., 2024, 91, 101013 CrossRef CAS PubMed
.
- A. Smogorzewska, Chapter 1: The Fanconi Anemia DNA Repair Pathway, https://www.fanconi.org/images/uploads/other/Chapter1.pdf, n.d., Fanconi Anemia Research Fund; Accessed: 2025-09-26.
- M. F. Sharp, V. J. Murphy, S. V. Twest, W. Tan, J. Lui, K. J. Simpson, A. J. Deans and W. Crismani, Sci. Rep., 2020, 10, 7959 CrossRef CAS PubMed
.
- B. Cheng, W. Pan, Y. Xing, Y. Xiao, J. Chen and Z. Xu, Eur. J. Med. Chem., 2022, 230, 114109 CrossRef CAS PubMed
.
- M. Crewe and R. Madabhushi, Front. Aging Neurosci., 2021, 13, 751742 CrossRef CAS PubMed
.
- J. L. Nitiss, Nat. Rev. Cancer, 2009, 9, 327–337 CrossRef CAS PubMed
.
-
Y. Pommier and N. Osheroff, DNA Topoisomerases and Cancer, Springer, 2011, pp. 145–154 Search PubMed
.
- M. Li and Y. Liu, Genomics, Proteomics Bioinf., 2016, 14, 166–171 CrossRef CAS PubMed
.
- L. Broderick, S. Yost, D. Li, M. D. McGeough, L. M. Booshehri, M. Guaderrama, S. D. Brydges, K. Kucharova, N. C. Patel and M. Harr,
et al.
, Nat. Commun., 2019, 10, 3644 CrossRef PubMed
.
- H. Zhang, S.-J. Lee, B. Zhu, N. Q. Tran, S. Tabor and C. C. Richardson, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9372–9377 CrossRef CAS PubMed
.
- R. Bythell-Douglas and A. J. Deans, Structure, 2021, 29, 99–113 CrossRef CAS PubMed
.
- G. Wang and K. M. Vasquez, Nat. Rev. Genet., 2023, 24, 211–234 CrossRef CAS PubMed
.
- A. Bansal, S. Kaushik and S. Kukreti, Front. Genetics, 2022, 13, 959258 CrossRef CAS PubMed
.
- F. Pan, Y. Zhang, P. Xu, V. H. Man, C. Roland, K. Weninger and C. Sagui, Comput. Struct. Biotechnol. J., 2021, 19, 2819–2832 CrossRef CAS PubMed
.
- G. Matos-Rodrigues, J. A. Hisey, A. Nussenzweig and S. M. Mirkin, Mol. Cell, 2023, 83, 3622–3641 CrossRef CAS PubMed
.
- P. Thumpati, S. N. Rai, C. Prajapati, K. Ramakrishna and S. K. Singh, Sci. Pharm., 2025, 93, 6 CrossRef CAS
.
- S.-L. Zhang, H. Fu, Y. Ma, Q. Lin, Y. Xu, Q. Yang, P. He and Z. Wei, Org. Biomol. Chem., 2025, 23, 1112–1119 RSC
.
- A. Sengupta, S. S. Roy and S. Chowdhury, Acc. Chem. Res., 2020, 54, 46–56 CrossRef PubMed
.
- Z.-Y. Sun, X.-N. Wang, S.-Q. Cheng, X.-X. Su and T.-M. Ou, Molecules, 2019, 24, 396 CrossRef PubMed
.
- I. Frasson, V. Pirota, S. N. Richter and F. Doria, Int. J. Biol. Macromol., 2022, 204, 89–102 CrossRef CAS PubMed
.
- J. R. Whitfield and L. Soucek, Nat. Rev. Drug Discovery, 2025, 1–13 Search PubMed
.
- M. Al Ojaimi, B. J. Banimortada, A. Alragheb, R. S. Hajir, C. Alves, D. Walid, A. Raza and A. W. El-Hattab, Hum. Genomics, 2025, 19, 47 CrossRef CAS PubMed
.
- S.-H. Song and T.-Y. Kim, Genomics Bioinf., 2017, 15, 114 Search PubMed
.
- M. Bustin and R. Reeves, Prog. Nucleic Acid Res. Mol. Biol., 1996, 54, 35–100b CAS
.
- S. Date and L. K. Bhatt, Inflammopharmacology, 2025, 33, 767–784 CrossRef CAS PubMed
.
- S. Wu, Y. Yu, Z. Zheng and Q. Cheng, Eur. J. Med. Res., 2023, 28, 430 CrossRef CAS PubMed
.
- B. Wen, Y.-t Wei and K. Zhao, Mol. Cell. Biochem., 2021, 476, 1729–1739 CrossRef CAS PubMed
.
- S. G. Swygert and C. L. Peterson, Biochim. Biophys. Acta, Gene Regul. Mech., 2014, 1839, 728–736 CrossRef CAS PubMed
.
- T. Kouzarides, Cell, 2007, 128, 693–705 CrossRef CAS PubMed
.
- R. Parveen, D. Harihar and B. P. Chatterji, Cancer, 2023, 129, 3372–3380 CrossRef CAS PubMed
.
- Z. Yao, Y. Chen, W. Cao and N. Shyh-Chang, Cell Proliferation, 2020, 53, e12898 CrossRef CAS PubMed
.
- G. A. Holdgate, C. Bardelle, A. Lanne, J. Read, D. H. O'Donovan, J. M. Smith, N. Selmi and R. Sheppard, Drug Discovery Today, 2022, 27, 1088–1098 CrossRef CAS PubMed
.
- A. Jain, S. Mittal, L. P. Tripathi, R. Nussinov and S. Ahmad, Comput. Struct. Biotechnol. J., 2022, 20, 4415–4436 CrossRef CAS PubMed
.
- D. Wu, N. Potluri, J. Lu, Y. Kim and F. Rastinejad, Nature, 2015, 524, 303–308 CrossRef CAS PubMed
.
- X. Ren, X. Diao, J. Zhuang and D. Wu, Mol. Pharmacol., 2022, 102, 240–247 CrossRef CAS PubMed
.
- M. Lambert, S. Jambon, S. Depauw and M.-H. David-Cordonnier, Molecules, 2018, 23, 1479 CrossRef PubMed
.
- J.-j Zhuang, Q. Liu, D.-l Wu and L. Tie, Acta Pharmacol. Sin., 2022, 43, 2474–2481 CrossRef CAS PubMed
.
- D. A. Sallman, Haematologica, 2020, 105, 1470 CrossRef PubMed
.
- G. Garcia-Manero, Lancet Haematol., 2023, 10, e71–e78 CrossRef CAS PubMed
.
- J. Schust, B. Sperl, A. Hollis, T. U. Mayer and T. Berg, Chem. Biol., 2006, 13, 1235–1242 CrossRef CAS PubMed
.
- C.-L. Li, H.-Y. Chen, J.-C. Yen, S.-J. Yu, T.-Y. Chou, S.-W. Yeh, H.-Y. Chuang and F.-L. Huang, Mol. Med. Rep., 2025, 31, 1–11 Search PubMed
.
- M. M. Kasembeli, P. Singhmar, J. Ma, J. Edralin, Y. Tang, C. Adams, C. J. Heijnen, A. Kavelaars and D. J. Tweardy, Biochem. Pharmacol., 2021, 192, 114688 CrossRef CAS PubMed
.
- S. Moverare-Skrtic, A. E. Borjesson, H. H. Farman, K. Sjogren, S. H. Windahl, M. K. Lagerquist, A. Andersson, A. Stubelius, H. Carlsten, J. Ake Gustafsson and C. Ohlsson, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1180–1185 CrossRef CAS PubMed
.
- J. Hoffman-Censits and W. K. Kelly, Clin. Cancer Res., 2013, 19, 1335–1339 CrossRef CAS PubMed
.
- X. Wu, D. Lazris, R. Wong and S. S. Tykodi, Ther. Adv. Med. Oncol., 2025, 17, 17588359251317846 CrossRef CAS PubMed
.
- R. Srinivasan, O. Iliopoulos, K. E. Beckermann, V. Narayan, B. L. Maughan, S. Oudard, T. Else, J. K. Maranchie, A. B. Iversen and J. Cornell,
et al.
, Lancet Oncol., 2025, 26, 571–582 CrossRef CAS PubMed
.
- R. J. Motzer, Y. Liu, R. F. Perini, Y. Zhang and D. Y. C. Heng, Phase III study evaluating efficacy and safety of MK-6482+ lenvatinib versus cabozantinib for second-or third-line therapy in patients with advanced renal cell carcinoma (RCC) who progressed after prior anti-PD-1/L1 therapy, 2021.
- T. K. Choueiri, J. Bedke, J. A. Karam, R. R. McKay, R. J. Motzer, S. M. Pal, C. Suárez, R. Uzzo, H. Liu, J. E. Burgents, et al., Phase 3 LITESPARK-022: Pembrolizumab (pembro) plus hypoxia-inducible factor 2α (HIF-2α) inhibitor belzutifan as adjuvant treatment for clear cell renal cell carcinoma (ccRCC), 2023.
- S. Goel, M. J. DeCristo, S. S. McAllister and J. J. Zhao, Trends Cell Biol., 2018, 28, 911–925 CrossRef CAS PubMed
.
- A. Andrikopoulou, S. Chatzinikolaou, E. Panourgias, M. Kaparelou, M. Liontos, M.-A. Dimopoulos and F. Zagouri, Breast, 2022, 63, 157–167 CrossRef PubMed
.
- Y. Xu, Q. Yu, P. Wang, Z. Wu, L. Zhang, S. Wu, M. Li, B. Wu, H. Li and H. Zhuang,
et al.
, Adv. Sci., 2022, 9, 2104344 CrossRef CAS PubMed
.
- C. Calvis, A. Beier, M. Feichtinger, T. Höfurthner, M. Moreno, R. Messeguer, R. Konrat, S. Esteban and L. Nevola, Cancer Res., 2021, 81, 2471 CrossRef
.
- A. Starodub, A. Gollerkeri, C. De Savi, J. Dey, S. Agarwal, S. Donohue, R. Perea, C. Klaus and J. Gollob, J. Clin. Oncol., 2022, 40, TPS3171 CrossRef
.
- J. Fallah, M. H. Brave, C. Weinstock, G. U. Mehta, D. Bradford, H. Gittleman, E. W. Bloomquist, R. Charlab, S. S. Hamed and C. P. Miller,
et al.
, Clin. Cancer Res., 2022, 28, 4843–4848 CrossRef CAS PubMed
.
- M.-F. Langelier, L. Zandarashvili, P. M. Aguiar, B. E. Black and J. M. Pascal, Nat. Commun., 2018, 9, 844 CrossRef PubMed
.
- K. Ryan, B. Bolanos, M. Smith, P. B. Palde, P. D. Cuenca, T. L. VanArsdale, S. Niessen, L. Zhang, D. Behenna and M. A. Ornelas,
et al.
, J. Biol. Chem., 2021, 296, 100251 CrossRef CAS PubMed
.
- C. J. Lord and A. Ashworth, Science, 2017, 355, 1152–1158 CrossRef CAS PubMed
.
- A. P. Wiegmans, M. Miranda, S. W. Wen, F. Al-Ejeh and A. Möller, Oncotarget, 2016, 7, 60087 CrossRef PubMed
.
- A. Ward, K. K. Khanna and A. P. Wiegmans, Cancer Treat. Rev., 2015, 41, 35–45 CrossRef CAS PubMed
.
- Y. Pan, Q. Zhang, C. Xu, Y. Sun, Q. Zheng, S. Yang and S. Lv, Comput. Biol. Med., 2025, 191, 110105 CrossRef CAS PubMed
.
- P. S. VanderVere-Carozza, N. S. Gavande, S. I. Jalal, K. E. Pollok, E. Ekinci, J. Heyza, S. M. Patrick, A. Masters, J. J. Turchi and K. S. Pawelczak, Front. Oncol., 2022, 12, 826655 CrossRef CAS PubMed
.
- M. T. Roth and S. Das, Expert Rev. Anticancer Ther., 2021, 21, 229–238 CrossRef CAS PubMed
.
- M. T. van Bussel, A. Awada, M. J. de Jonge, M. Mau-Sørensen, D. Nielsen, P. Schöffski, H. M. Verheul, B. Sarholz, K. Berghoff and S. El Bawab,
et al.
, Br. J. Cancer, 2021, 124, 728–735 CrossRef CAS PubMed
.
- J. M. Kelm, A. Samarbakhsh, A. Pillai, P. S. VanderVere-Carozza, H. Aruri, D. S. Pandey, K. S. Pawelczak, J. J. Turchi and N. S. Gavande, Front. Oncol., 2022, 12, 850883 CrossRef CAS PubMed
.
- J. Zhu, L. Zhou, G. Wu, H. Konig, X. Lin, G. Li, X.-L. Qiu, C.-F. Chen, C.-M. Hu and E. Goldblatt,
et al.
, EMBO Mol. Med., 2013, 5, 353–365 CrossRef CAS PubMed
.
- F. Huang, O. M. Mazina, I. J. Zentner, S. Cocklin and A. V. Mazin, J. Med. Chem., 2012, 55, 3011–3020 CrossRef CAS PubMed
.
- F. Huang and A. V. Mazin, PLoS One, 2014, 9, e100993 CrossRef PubMed
.
- F. Huang, N. A. Motlekar, C. M. Burgwin, A. D. Napper, S. L. Diamond and A. V. Mazin, ACS Chem. Biol., 2011, 6, 628–635 CrossRef CAS PubMed
.
- P. C. Fong, D. S. Boss, T. A. Yap, A. Tutt, P. Wu, M. Mergui-Roelvink, P. Mortimer, H. Swaisland, A. Lau and M. J. O'Connor,
et al.
, N. Engl. J. Med., 2009, 361, 123–134 CrossRef CAS PubMed
.
- F. T. Zenke, A. Zimmermann, C. Sirrenberg, H. Dahmen, V. Kirkin, U. Pehl, T. Grombacher, C. Wilm, T. Fuchss and C. Amendt,
et al.
, Mol. Cancer Ther., 2020, 19, 1091–1101 CrossRef CAS PubMed
.
- M. Elias, S. Gani, Y. Lerner, K. Yamin, C. Tor, A. Patel, A. Matityahu, M. Dessau, N. Qvit and I. Onn, iScience, 2023, 26, 107498 CrossRef CAS PubMed
.
- J. Antony, C. V. Chin and J. A. Horsfield, Int. J. Mol. Sci., 2021, 22, 6788 CrossRef CAS PubMed
.
- A. B. Alharbi, U. Schmitz, C. G. Bailey and J. E. Rasko, Nucleic Acids Res., 2021, 49, 7825–7838 CrossRef CAS PubMed
.
- Y. Kuroki, K. Yamagata, Y. Aikawa, Y. Shima, H. Shioneri and I. Kitabayashi, Cancer Sci., 2025, 3016–3026 CrossRef CAS PubMed
.
- Y. Liu, P. Liu, S. Duan, J. Lin, W. Qi, Z. Yu, X. Gao, X. Sun, J. Liu and J. Lin,
et al.
, Cell Death Differ., 2025, 32, 745–762 CrossRef CAS PubMed
.
- Y. Han, W. Sheng, X. Liu, H. Liu, X. Jia, H. Li, C. Wang, B. Wang, T. Hu and Y. Ma, Sci. Rep., 2024, 14, 24948 CrossRef CAS PubMed
.
- G. Vergoten and C. Bailly, Med. Drug Discovery, 2020, 7, 100058 CrossRef
.
- E. San José-Enériz, N. Gimenez-Camino, X. Agirre and F. Prosper, Cancers, 2019, 11, 1794 CrossRef PubMed
.
- T. N. Teknos, J. Grecula, A. Agrawal, M. Old, E. Ozer, R. Carrau, S. Kang, J. Rocco, D. Blakaj and V. Diavolitsis,
et al.
, Invest. New Drugs, 2019, 37, 702–710 CrossRef CAS PubMed
.
- T. H. Luu, R. J. Morgan, L. Leong, D. Lim, M. McNamara, J. Portnow, P. Frankel, D. D. Smith, J. H. Doroshow and D. R. Gandara,
et al.
, Clin. Cancer Res., 2008, 14, 7138–7142 CrossRef CAS PubMed
.
- P. Smolewski and T. Robak, Expert Opin. Drug Discovery, 2017, 12, 859–873 CAS
.
- C. Sequera, M. Grattarola, F. Cannet, A. Dobric, P. Michea Veloso, M. Methia, S. Richelme, A. El Kaoutari, P. Kousteridou and D. Debayle,
et al.
, Nat. Commun., 2025, 16, 7919 CrossRef CAS PubMed
.
- S. M. Horwitz, A. J. Nirmal, J. Rahman, R. Xu, E. Drill, N. Galasso, N. Ganesan, T. Davey, H. Hancock and L. Perez,
et al.
, Nat. Med., 2024, 30, 2517–2527 CrossRef CAS PubMed
.
- J. P. Laubach, P. Moreau, J. F. San-Miguel and P. G. Richardson, Clin. Cancer Res., 2015, 21, 4767–4773 CrossRef CAS PubMed
.
- M. J. Homan, A. Franson, K. Ravi, H. Roberts, M. P. Pai, C. Liu, M. He, A. Matvekas, C. Koschmann and B. L. Marini, Cancer Chemother. Pharmacol., 2021, 88, 555–562 CrossRef CAS PubMed
.
- M. Monje, T. Cooney, J. Glod, J. Huang, C. J. Peer, D. Faury, P. Baxter, K. Kramer, A. Lenzen and N. J. Robison,
et al.
, Neuro-Oncology, 2023, 25, 2262–2272 CrossRef CAS PubMed
.
- A. Aartsma-Rus, Front. Cell Dev. Biol., 2025, 12, 1514898 CrossRef PubMed
.
- F. Perner, T. Berg, D. Sasca, S.-L. Mersiowsky, J. Y. Gadrey, J. Thomas, M. W. Kühn and M. Lübbert, Int. J. Cancer, 2025 Search PubMed
.
- E. M. Stein, G. Garcia-Manero, D. A. Rizzieri, R. Tibes, J. G. Berdeja, M. R. Savona, M. Jongen-Lavrenic, J. K. Altman, B. Thomson and S. J. Blakemore,
et al.
, Blood, 2018, 131, 2661–2669 CrossRef CAS PubMed
.
- A. Schurer, S. G. Glushakow-Smith and K. Gritsman, Stem Cells Transl. Med., 2024, 14, szae089 CrossRef PubMed
.
- S. Zehtabcheh, H. Soleimani Samarkhazan, M. Asadi, M. Zabihi, S. Parkhideh and M. H. Mohammadi, Biomarker Res., 2025, 13, 73 CrossRef PubMed
.
- L. L. Marzochi, C. I. Cuzziol, C. H. V. Do Nascimento Filho, J. A. Dos Santos, M. M. U. Castanhole-Nunes, É. C. Pavarino, E. N. S. Guerra and E. M. Goloni-Bertollo, Eur. J. Pharmacol., 2023, 944, 175590 CrossRef CAS PubMed
.
- H. Dhulkifle, M. I. Diab, M. Algonaiah, H. M. Korashy and Z. H. Maayah, ACS Pharmacol. Transl. Sci., 2024, 7, 546–559 CrossRef CAS PubMed
.
- Y. N. Lamb, Drugs, 2024, 84, 849–856 CrossRef PubMed
.
- S. M. Hoy, Drugs, 2020, 80, 513–521 CrossRef PubMed
.
- T. Maes, C. Mascaró, I. Tirapu, A. Estiarte, F. Ciceri, S. Lunardi, N. Guibourt, A. Perdones, M. M. Lufino and T. C. Somervaille,
et al.
, Cancer Cell, 2018, 33, 495–511 CrossRef CAS PubMed
.
- K. K. Ray, S. J. Nicholls, K. A. Buhr, H. N. Ginsberg, J. O. Johansson, K. Kalantar-Zadeh, E. Kulikowski, P. P. Toth, N. Wong and M. Sweeney,
et al.
, JAMA, 2020, 323, 1565–1573 CrossRef CAS PubMed
.
- A. M. J. Fichtinger-Schepman, J. L. Van der Veer, J. H. Den Hartog, P. H. Lohman and J. Reedijk, Biochemistry, 1985, 24, 707–713 CrossRef CAS PubMed
.
- D. Sahoo, P. Deb, T. Basu, S. Bardhan, S. Patra and P. K. Sukul, Bioorg. Med. Chem., 2024, 112, 117894 CrossRef CAS PubMed
.
- M.-H. Baik, R. A. Friesner and S. J. Lippard, J. Am. Chem. Soc., 2003, 125, 14082–14092 CrossRef CAS PubMed
.
- R. D. Wood, Biochimie, 1999, 81, 39–44 CrossRef CAS PubMed
.
- A. Yimit, O. Adebali, A. Sancar and Y. Jiang, Nat. Commun., 2019, 10, 309 CrossRef PubMed
.
- C. Zhang, C. Xu, X. Gao and Q. Yao, Theranostics, 2022, 12, 2115 CrossRef CAS PubMed
.
- S. Li, W. K. Olson and X.-J. Lu, Nucleic Acids Res., 2019, 47, W26–W34 CrossRef CAS PubMed
.
- A. K. Larsen, C. M. Galmarini and M. D'Incalci, Cancer Chemother. Pharmacol., 2016, 77, 663–671 CrossRef CAS PubMed
.
- X.-M. Chen, C. Cullinane, P. J. Gray and D. R. Phillips, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 1999, 445, 45–54 CrossRef CAS PubMed
.
- M. Shang, M. Ren and C. Zhou, Chem. Res. Toxicol., 2019, 32, 2517–2525 Search PubMed
.
- V. L. Souliotis, M. A. Dimopoulos and P. P. Sfikakis, Clin. Cancer Res., 2003, 9, 4465–4474 CAS
.
- L. M. Leoni, B. Bailey, J. Reifert, H. H. Bendall, R. W. Zeller, J. Corbeil, G. Elliott and C. C. Niemeyer, Clin. Cancer Res., 2008, 14, 309–317 CrossRef CAS PubMed
.
- T. W. Dennie and J. M. Kolesar, Clin. Ther., 2009, 31, 2290–2311 CrossRef CAS PubMed
.
- G. S. Khan, A. Shah and D. Barker,
et al.
, J. Photochem. Photobiol., B, 2012, 115, 105–118 CrossRef CAS PubMed
.
- P. B. Dervan, Bioorg. Med. Chem., 2001, 9, 2215–2235 CrossRef CAS PubMed
.
- L. Pett, J. A. Hartley and K. Kiakos, Curr. Top. Med. Chem., 2015, 15, 1293–1322 CrossRef CAS PubMed
.
- P. B. Dervan and B. S. Edelson, Curr. Opin. Struct. Biol., 2003, 13, 284–299 CrossRef CAS PubMed
.
- J. L. Yap, J. Chauhan, K.-Y. Jung, L. Chen, E. V. Prochownik and S. Fletcher, MedChemComm, 2012, 3, 541–551 RSC
.
- L. H. Fornander, L. Wu, M. Billeter, P. Lincoln and B. Nordén, J. Phys. Chem. B, 2013, 117, 5820–5830 CrossRef CAS PubMed
.
- J. Kormanec, R. Novakova, D. Csolleiova, L. Feckova, B. Rezuchova, B. Sevcikova and D. Homerova, Appl. Microbiol. Biotechnol., 2020, 104, 7701–7721 CrossRef CAS PubMed
.
- C. Hou, S. Weidenbach, K. E. Cano, Z. Wang, P. Mitra, D. N. Ivanov, J. Rohr and O. V. Tsodikov, Nucleic Acids Res., 2016, 44, 8990–9004 CrossRef CAS PubMed
.
- S. F. Sleiman, B. C. Langley, M. Basso, J. Berlin, L. Xia, J. B. Payappilly, M. K. Kharel, H. Guo, J. L. Marsh and L. M. Thompson,
et al.
, J. Neurosci., 2011, 31, 6858–6870 CrossRef CAS PubMed
.
- J. Portugal, Pharmacol. Ther., 2024, 260, 108672 CrossRef CAS PubMed
.
- S. S. Bhosale, A. Mandal, C. Hou, J. R. McCorkle, D. Schweer, K. S. Hill, V. Subramanian, J. M. Kolesar, O. V. Tsodikov and J. Rohr, ChemMedChem, 2023, 18, e202200368 CrossRef CAS PubMed
.
- S. Venugopal, V. Sharma, A. Mehra, I. Singh and G. Singh, Chem. Biol. Drug Des., 2022, 100, 580–598 CrossRef CAS PubMed
.
- F. Gago, Methods, 1998, 14, 277–292 CrossRef CAS PubMed
.
-
C. Avendano and J. C. Menendez, Medicinal Chemistry of Anticancer Drugs, Elsevier, Amsterdam, 2008, pp. 199–228 Search PubMed
.
- J. T. Taylor, S. Ellison, A. Pandele, S. Wood, E. Nathan, G. Forte, H. Parker, E. Zindy, M. Elvin and A. Dickson,
et al.
, Neuro-Oncology, 2020, 22, 1289–1301 CrossRef CAS PubMed
.
- H.-J. Kang and H.-J. Park, Biochemistry, 2009, 48, 7392–7398 CrossRef CAS PubMed
.
- T. Banerjee, S. Banerjee, S. Sett, S. Ghosh, T. Rakshit and R. Mukhopadhyay, PLoS One, 2016, 11, e0154666 CrossRef CAS PubMed
.
-
B. K. Bhuyan and C. G. Smith, in Nogalamycin, ed. A. C. Sartorelli and D. G. Johns, Springer Berlin Heidelberg, Berlin, Heidelberg, 1975, pp. 623–632 Search PubMed
.
- A. C. Ketron, W. A. Denny, D. E. Graves and N. Osheroff, Biochemistry, 2012, 51, 1730–1739 CrossRef CAS PubMed
.
- D. Kuron, A. Pohlmann, L. Angenendt, T. Kessler, R. Mesters, W. E. Berdel, M. Stelljes, G. Lenz, C. Schliemann and J.-H. Mikesch, Ann. Hematol., 2023, 102, 755–760 CrossRef CAS PubMed
.
- R. E. Hawtin, D. E. Stockett, J. A. W. Byl, R. S. McDowell, N. Tan, M. R. Arkin, A. Conroy, W. Yang, N. Osheroff and J. A. Fox, PLoS One, 2010, 5, e10186 CrossRef PubMed
.
- Y. Singh, N. Bhatia, A. Biharee, S. Kulkarni, S. Thareja and V. Monga, Expert Opin. Drug Discovery, 2023, 18, 1151–1167 CrossRef CAS PubMed
.
- J. Yin, P. Jia, X. Qu, Z. Han, L. Yao, S. Wang and J. Gao, Chem. Biol. Drug Des., 2024, 104, e70034 CrossRef CAS PubMed
.
- E. A. Sausville, J. Peisach and S. B. Horwitz, Biochemistry, 1978, 17, 2740–2746 CrossRef CAS PubMed
.
- O. Tounekti, A. Kenani, N. A. Foray, S. Orlowski and L. M. Mir, Br. J. Cancer, 2001, 84, 1272–1279 CrossRef CAS PubMed
.
- J. M. Bennett and S. D. Reich, Ann. Intern. Med., 1979, 90, 945–948 CrossRef CAS PubMed
.
- P. L. Mendoza-Munoz, N. S. Gavande, P. S. VanderVere-Carozza, K. Pawelczak, J. Dynlacht, J. Garrett and J. Turchi, NAR Cancer, 2023, 5, zcad003 CrossRef PubMed
.
- K.-B. Wang, M. S. A. Elsayed, G. Wu, N. Deng, M. Cushman and D. Yang, J. Am. Chem. Soc., 2019, 141, 11059–11070 CrossRef CAS PubMed
.
- A. Ali and S. Bhattacharya, Bioorg. Med. Chem., 2014, 22, 4506–4521 CrossRef CAS PubMed
.
- H.-X. Li, Y.-M. He, J. Fei, M. Guo, C. Zeng, P.-J. Yan, Y. Xu, G. Qin and F.-Y. Teng, J. Transl. Med., 2025, 23, 457 CrossRef CAS PubMed
.
- G. C. C. Koh, S. Boushaki, S. J. Zhao, A. M. Pregnall, F. Sadiyah, C. Badja, Y. Memari, I. Georgakopoulos-Soares and S. Nik-Zainal, Nat. Genet., 2024, 56, 23–26 CrossRef CAS PubMed
.
- S. Kanwal, N. Brajanovski, J. Zhan, J. Chan, R. Rebello, O. Hofmann, S. Grimmond, L. Furic, R. D. Hannan and G. McArthur,
et al.
, MedRxiv, 2025, 2025.09.30.25336525, DOI:10.1101/2025.09.30.25336525
.
- A. A. Ahmed, W. Greenhalf, D. H. Palmer, N. Williams, J. Worthington, T. Arshad, S. Haider, E. Alexandrou, D. Guneri and Z. A. Waller,
et al.
, Molecules, 2023, 28, 2452 CrossRef CAS PubMed
.
- R. M. Wadkins, Curr. Med. Chem., 2000, 7, 1–15 CrossRef CAS PubMed
.
- J. Zell, K. Duskova, L. Chouh, M. Bossaert, N. Chéron, A. Granzhan, S. Britton and D. Monchaud, Nucleic Acids Res., 2021, 49, 10275–10288 CrossRef CAS PubMed
.
- T. Zhang, C. Yin, A. Fedorov, L. Qiao, H. Bao, N. Beknazarov, S. Wang, A. Gautam, R. M. Williams and J. C. Crawford,
et al.
, Nature, 2022, 606, 594–602 CrossRef CAS PubMed
.
- E. A. Elmorsy, S. Saber, R. S. Hamad, M. A. Abdel-Reheim, A. F. El-Kott, M. A. AlShehri, K. Morsy, S. A. Salama and M. E. Youssef, Eur. J. Pharm. Sci., 2024, 203, 106939 CrossRef CAS PubMed
.
- R. C. Todd and S. J. Lippard, Metallomics, 2009, 1, 280–291 CrossRef CAS PubMed
.
- P. D. O'Dowd, D. F. Sutcliffe and D. M. Griffith, Coord. Chem. Rev., 2023, 497, 215439 CrossRef
.
- A. Le Cesne, J.-Y. Blay, D. Cupissol, A. Italiano, C. Delcambre, N. Penel, N. Isambert, C. Chevreau, E. Bompas and F. Bertucci,
et al.
, Ann. Oncol., 2021, 32, 1034–1044 CrossRef CAS PubMed
.
- G. D. Demetri, M. Von Mehren, R. L. Jones, M. L. Hensley, S. M. Schuetze, A. Staddon, M. Milhem, A. Elias, K. Ganjoo and H. Tawbi,
et al.
, J. Clin. Oncol., 2016, 34, 786–793 CrossRef CAS PubMed
.
- M. van Kan, K. E. Burns and N. A. Helsby, Cancer Chemother. Pharmacol., 2021, 88, 755–769 CrossRef CAS PubMed
.
- U.S. Food and Drug Administration, Cosmegen® (dactinomycin for injection) Prescribing Information, 2018, https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/050682s033lbl.pdf, Accessed: 2025-09-26.
- M. Kciuk, A. Gielecinska, S. Mujwar, D. Kolat, Z. Kaluzinska-Kolat, I. Celik and R. Kontek, Cells, 2023, 12, 659 CrossRef CAS PubMed
.
- A. Banerjee, S. Sanyal, K. K. Kulkarni, K. Jana, S. Roy, C. Das and D. Dasgupta, FEBS Open Bio, 2014, 4, 987–995 CrossRef CAS PubMed
.
- Y. Pommier and J. Cherfils, Trends Pharmacol. Sci., 2005, 26, 138–145 CrossRef CAS PubMed
.
- C.-C. Wu, T.-K. Li, L. Farh, L.-Y. Lin, T.-S. Lin, Y.-J. Yu, T.-J. Yen, C.-W. Chiang and N.-L. Chan, Science, 2011, 333, 459–462 CrossRef CAS PubMed
.
- V. Singh, P. Johansson, E. Ekedahl, Y.-L. Lin, O. Hammarsten and F. Westerlund, Biochem. Biophys. Res. Commun., 2022, 594, 57–62 CrossRef CAS PubMed
.
- B. J. Evison, B. E. Sleebs, K. G. Watson, D. R. Phillips and S. M. Cutts, Med. Res. Rev., 2016, 36, 248–299 CrossRef CAS PubMed
.
- D. Cardinale, F. Iacopo and C. M. Cipolla, Front. Cardiovasc. Med., 2020, 7, 26 CrossRef CAS PubMed
.
- G. N. Maertens, A. N. Engelman and P. Cherepanov, Nat. Rev. Microbiol., 2022, 20, 20–34 CrossRef CAS PubMed
.
- B. Geng, J. Hu, P. Li, D. Pan and L. Shen, Carbon, 2022, 187, 365–374 Search PubMed
.
- W. Fried, M. Tyagi, L. Minakhin, G. Chandramouly, T. Tredinnick, M. Ramanjulu, W. Auerbacher, M. Calbert, T. Rusanov and T. Hoang,
et al.
, Nat. Commun., 2024, 15, 2862 CrossRef CAS PubMed
.
- B. Ko, A. P. Chen, S. Kummar, M. Hogu, L. V. Rubinstein, N. Takebe, R. Piekarz, E. Sharon, J. H. Beumer, D. Wilsker, D. Ketchum, K. V. Ferry-Galow, R. E. Parchment, J. L. Holleran, J. J. Deppas, J. M. Collins, B. C. Johnson, S. P. Ivy, J. H. Doroshow and G. H. O'Sullivan Coyne, J. Clin. Oncol., 2023, 41, 3022 CrossRef
.
- H. Gamal, E. M. Shoeib, A. Hajjaj, H. E. A. Abdullah, E. H. Elramy, D. A. Abd Ellah, S. M. El-Sayed and M. F. Khder, RSC Pharm., 2025, 1019–1033 Search PubMed
.
- M. K. Jayatunga, M. Ayers, L. Bruens, D. Jayanth and C. Meier, Drug Discovery Today, 2024, 29, 104009 CrossRef PubMed
.
- H. Wei and J. A. McCammon, npj Drug Discovery, 2024, 1, 1 Search PubMed
.
- Y. You, X. Lai, Y. Pan, H. Zheng, J. Vera, S. Liu, S. Deng and L. Zhang, Signal Transduction Targeted Ther., 2022, 7, 156 Search PubMed
.
- D. Lama, T. Vosselman, C. Sahin, J. Liaño-Pons, C. P. Cerrato, L. Nilsson, K. Teilum, D. P. Lane, M. Landreh and M. Arsenian Henriksson, Nat. Commun., 2024, 15, 1865 CrossRef CAS PubMed
.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritzel, O. Ronneberger, L. Willmore, A. J. Ballard and J. Bambrick,
et al.
, Nature, 2024, 630, 493–500 CrossRef CAS PubMed
.
- A. Omidi, M. H. Møller, N. Malhis, J. M. Bui and J. Gsponer, Proc. Natl. Acad. Sci. U. S. A., 2024, 121, e2406407121 Search PubMed
.
- M. Baek, R. McHugh, I. Anishchenko, H. Jiang, D. Baker and F. DiMaio, Nat. Methods, 2024, 21, 117–121 Search PubMed
.
- R. Krishna, J. Wang, W. Ahern, P. Sturmfels, P. Venkatesh, I. Kalvet, G. R. Lee, F. S. Morey-Burrows, I. Anishchenko and I. R. Humphreys,
et al.
, Science, 2024, 384, eadl2528 CrossRef CAS PubMed
.
- M. Andrabi, K. Mizuguchi, A. Sarai and S. Ahmad, BMC Struct. Biol., 2009, 9, 30 CrossRef PubMed
.
- S. Chauhan and S. Ahmad, Proteins: Struct., Funct., Bioinf., 2020, 88, 15–30 CrossRef CAS PubMed
.
- D. M. Varghese, R. Nussinov and S. Ahmad, NAR: Genomics Bioinf., 2022, 4, lqac091 CrossRef PubMed
.
-
S. Mirza, P. Pandey, S. Hasnain and S. Ahmad, Emerging Topics in Protein–DNA Interactions, in Reference Module in Life Sciences, Elsevier, 2025, pp. 373–393 Search PubMed
.
- X. Li, W. Peng and X. Zhu, Briefings Bioinf., 2025, 26, bbaf457 CrossRef CAS PubMed
.
- M. T. Tahmid, A. M. Hasan and M. S. Bayzid, Commun. Biol., 2025, 8, 568 CrossRef PubMed
.
- M. Aydinli, C. Liang and T. Dandekar, Sci. Rep., 2022, 12, 17588 CrossRef CAS PubMed
.
- S. L. Nystrom and D. J. McKay, PLoS Comput. Biol., 2021, 17, e1008991 CrossRef CAS PubMed
.
- R. V. Honorato, M. E. Trellet, B. Jiménez-Garca, J. J. Schaarschmidt, M. Giulini, V. Reys, P. I. Koukos, J. P. Rodrigues, E. Karaca and G. C. van Zundert,
et al.
, Nat. Protoc., 2024, 19, 3219–3241 CrossRef CAS PubMed
.
- I. Tuszynska, M. Magnus, K. Jonak, W. Dawson and J. M. Bujnicki, Nucleic Acids Res., 2015, 43, W425–W430 CrossRef CAS PubMed
.
- M. M. Gromiha and K. Harini, Curr. Opin. Struct. Biol., 2025, 90, 102955 CrossRef CAS PubMed
.
- P. Infante, M. Mori, R. Alfonsi, F. Ghirga, F. Aiello, S. Toscano, C. Ingallina, M. Siler, D. Cucchi and A. Po,
et al.
, EMBO J., 2015, 34, 200–217 CrossRef CAS PubMed
.
- G. Calissi, E. W.-F. Lam and W. Link, Nat. Rev. Drug Discovery, 2021, 20, 21–38 CrossRef CAS PubMed
.
- Y. Wang, J. M. L. Ribeiro and P. Tiwary, Curr. Opin. Struct. Biol., 2020, 61, 139–145 CrossRef CAS PubMed
.
- O. T. Unke, S. Chmiela, H. E. Sauceda, M. Gastegger, I. Poltavsky, K. T. Schutt, A. Tkatchenko and K.-R. Muller, Chem. Rev., 2021, 121, 10142–10186 CrossRef CAS PubMed
.
- M. Andrabi, A. P. Hutchins, D. Miranda-Saavedra, H. Kono, R. Nussinov, K. Mizuguchi and S. Ahmad, Sci. Rep., 2017, 7, 4071 CrossRef PubMed
.
-
M. Kalsan, A. Jabeen and S. Ahmad, Transcriptome Data Analysis, Springer, 2024, pp. 317–343 Search PubMed
.
- J. Li, T.-P. Chiu and R. Rohs, Nat. Commun., 2024, 15, 1243 CrossRef CAS PubMed
.
- M. J. Wildey, A. Haunso, M. Tudor, M. Webb and J. H. Connick, Annu. Rep. Med. Chem., 2017, 50, 149–195 Search PubMed
.
- A. A. Peterson and D. R. Liu, Nat. Rev. Drug Discovery, 2023, 22, 699–722 Search PubMed
.
- T. A. A. Program, Sci. Rep., 2024, 14, 7526 Search PubMed
.
- R. Qureshi, M. Irfan, T. M. Gondal, S. Khan, J. Wu, M. U. Hadi, J. Heymach, X. Le, H. Yan and T. Alam, Heliyon, 2023, 9, e17575 CrossRef CAS PubMed
.
- B. Zdrazil, E. Felix, F. Hunter, E. J. Manners, J. Blackshaw, S. Corbett, M. De Veij, H. Ioannidis, D. M. Lopez and J. F. Mosquera,
et al.
, Nucleic Acids Res., 2024, 52, D1180–D1192 CrossRef CAS PubMed
.
- J. J. Irwin and B. K. Shoichet, J. Chem. Inf. Model., 2005, 45, 177–182 Search PubMed
.
- T. Liu, L. Hwang, S. K. Burley, C. I. Nitsche, C. Southan, W. P. Walters and M. K. Gilson, Nucleic Acids Res., 2025, 53, D1633–D1644 CrossRef CAS PubMed
.
- C. Knox, M. Wilson, C. M. Klinger, M. Franklin, E. Oler, A. Wilson, A. Pon, J. Cox, N. E. Chin and S. A. Strawbridge,
et al.
, Nucleic Acids Res., 2024, 52, D1265–D1275 CrossRef CAS PubMed
.
- Y. Shi, P. Ren, Y. Zhang, X. Gong, M. Hu and H. Liang, Front. Res. Metrics Anal., 2021, 6, 670006 CrossRef PubMed
.
- A. Cherkasov, E. N. Muratov, D. Fourches, A. Varnek, I. I. Baskin, M. Cronin, J. Dearden, P. Gramatica, Y. C. Martin and R. Todeschini,
et al.
, J. Med. Chem., 2014, 57, 4977–5010 CrossRef CAS PubMed
.
- A. Gangwal, A. Ansari, I. Ahmad, A. K. Azad, V. Kumarasamy, V. Subramaniyan and L. S. Wong, Front. Pharmacol., 2024, 15, 1331062 CrossRef CAS PubMed
.
- D. Crucitti, C. Pérez Mguez, J. Á. Daz Arias, D. B. Fernandez Prada and A. Mosquera Orgueira, Front. Hematol., 2024, 3, 1305741 CrossRef
.
- T. Gupta, P. Sharma, S. Malik and P. Pant, Mol. Pharm., 2025, 4076–4090 CrossRef CAS PubMed
.
- T.-G. Chang, S. Park, A. A. Schäffer, P. Jiang and E. Ruppin, Nat. Cancer, 2025, 1–15 CAS
.
- S. Gupta, V. K. Mohani, G. Ghislat, P. J. Ballester and S. Ahmad, NAR: Genomics Bioinf., 2025, 7, lqaf111 CrossRef CAS PubMed
.
- T. Lazar, A. Connor, C. F. DeLisle, V. Burger and P. Tompa, Nat. Rev. Drug Discovery, 2025, 1–21 Search PubMed
.
- A. van der Vaart, Biochim. Biophys. Acta, Gen. Subj., 2015, 1850, 1091–1098 CrossRef CAS PubMed
.
- J. E. Darnell Jr, Nat. Rev. Cancer, 2002, 2, 740–749 CrossRef PubMed
.
- R. Edaibis, R. Akel and J. A. Shin, Transcription, 2025, 16, 67–85 CrossRef CAS PubMed
.
- Q. Sun, H. Wang, J. Xie, L. Wang, J. Mu, J. Li, Y. Ren and L. Lai, Chem. Rev., 2025, 125(13), 6309–6365 CrossRef CAS PubMed
.
- N. S. Gavande, P. S. VanderVere-Carozza, K. S. Pawelczak, P. Mendoza-Munoz, T. L. Vernon, L. A. Hanakahi, M. Summerlin, J. R. Dynlacht, A. H. Farmer and C. R. Sears,
et al.
, Nucleic Acids Res., 2020, 48, 11536–11550 CrossRef CAS PubMed
.
- N. Y. Ngoi, D. Gallo, C. Torrado, M. Nardo, D. Durocher and T. A. Yap, Nat. Rev. Clin. Oncol., 2025, 22, 46–64 CrossRef PubMed
.
- T. A. Yap, P. LoRusso, R. E. Miller, R. Kristeleit, A. G. Paulovich, S. McMorn, L. Oplustil O'Connor, B. Lombardi, P. Marco-Casanova and E. T. Gangl,
et al.
, Br. J. Cancer, 2025, 1–10 Search PubMed
.
- S.-H. Chen and J.-Y. Chang, Int. J. Mol. Sci., 2019, 20, 4136 CrossRef PubMed
.
- L. Dong, H. Jiang, Z. Kang and M. Guan, Clin. Chim. Acta, 2023, 544, 117338 Search PubMed
.
- N. J. Curtin, L.-Z. Wang, A. Yiakouvaki, S. Kyle, C. A. Arris, S. Canan-Koch, S. E. Webber, B. W. Durkacz, H. A. Calvert and Z. Hostomsky,
et al.
, Clin. Cancer Res., 2004, 10, 881–889 CrossRef CAS PubMed
.
- R. C. van Waardenburg and E. S. Yang, Cancer Drug Res., 2021, 4, 837 Search PubMed
.
- S. Liu, J. Li, Y. Wang, Z. Zhang, Y. Wang, L. Wang and L. Wang, Small Methods, 2025, 9, e01445 CrossRef CAS PubMed
.
- W. Li, J. Li, L. Wang, Y. Wang, Z. Zhang, S. Liu, B. Gong, Y. Wang and L. Wang, Nano Lett., 2025, 25, 1706–1714 CrossRef CAS PubMed
.
- Y. Su, K. Ye, J. Hu, Z. Zhang, Y. Wang, B. Geng, D. Pan and L. Shen, Adv. Healthcare Mater., 2024, 13, 2304648 CrossRef CAS PubMed
.
- M. Stepie, J. Zajda, B. K. Keppler, A. R. Timerbaev and M. Matczuk, Talanta, 2025, 128331 Search PubMed
.
- Y. Lin, Q. Shi, G. Yang, F. Shi, Y. Zhou, T. Wang, P. Xu, P. Li, Z. Liu and H. Sun,
et al.
, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2213670120 CrossRef CAS PubMed
.
- M. Sands, M. A. Kron and R. B. Brown, Rev. Infect. Dis., 1985, 7, 625–6344 CrossRef CAS PubMed
.
- N. Rastegar-Pouyani, M. A. Farzin, J. Zafari, M. Haji Abdolvahab and S. Hassani, J. Transl. Med., 2025, 23, 258 CrossRef CAS PubMed
.
- H. Jung and Y. Lee, Cancers, 2025, 17, 1871 Search PubMed
.
- S. Li, X. Wang, J. Huang, X. Cao, Y. Liu, S. Bai, T. Zeng, Q. Chen, C. Li and C. Lu,
et al.
, Cell Death Dis., 2025, 16, 197 Search PubMed
.
- C. Kang and W. Xu, J. Med. Chem., 2025, 68, 2048–2051 CrossRef CAS PubMed
.
- Y. Tao, Y. Lu, B. Yu and Y. Wang, Trends Pharmacol. Sci., 2025, 520–534 CrossRef CAS PubMed
.
- X. Zhang, Z. Song, X. Zhang, J. Zou, G. Yu, X. Chen and S. Wang, Angew. Chem., Int. Ed., 2025, 64, e202507702 CrossRef CAS PubMed
.
- X. Yang, H. Zhu, L. Shi, T. Song, W. Gong, S. He, S. Shan, C. Xu and Z. Zhou, Nucleic Acids Res., 2025, 53, gkaf735 CrossRef PubMed
.
- L. Gerasimavicius, S. C. Biddie and J. A. Marsh, Nucleic Acids Res., 2026, 54(1), gkaf1417 CrossRef CAS PubMed
.
- P.-Y. Lin, S.-C. Huang, K.-L. Chen, Y.-C. Huang, C.-Y. Liao, G.-J. Lin, H. Lee and P.-Y. Chen, Botanical Studies, 2025, 66, 14 CrossRef PubMed
.
- M. R. Karim, T. Islam, M. Shajalal, O. Beyan, C. Lange, M. Cochez, D. Rebholz-Schuhmann and S. Decker, Briefings Bioinf., 2023, 24, bbad236 CrossRef PubMed
.
- A. Blanco-Gonzalez, A. Cabezon, A. Seco-Gonzalez, D. Conde-Torres, P. Antelo-Riveiro, A. Pineiro and R. Garcia-Fandino, Pharmaceuticals, 2023, 16, 891 CrossRef PubMed
.
- F. J. Ferreira and A. S. Carneiro, ACS Omega, 2025, 10(23), 23889–23903 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2026 |


