Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Benchtop NMR signal enhancement of metabolites in urine extract using SABRE
Simon Fleischer *a,
Jing Yang
*a,
Jing Yang a,
Kerti Ausmees
a,
Kerti Ausmees b,
Indrek Reile
b,
Indrek Reile b,
Neil MacKinnon
b,
Neil MacKinnon a,
Jan Gerrit Korvink
a,
Jan Gerrit Korvink a and
Sören Lehmkuhl
a and
Sören Lehmkuhl *a
*a
aInstitute of Microstructure Technology, Karlsruhe Institute of Technology, Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany. E-mail: simon.fleischer@kit.edu; soeren.lehmkuhl@kit.edu
bNational Institute of Chemical Physics and Biophysics, Akadeemia tee 23, 12618 Tallinn, Estonia
First published on 9th January 2026
Abstract
Metabolites in a urine extract are signal-enhanced by SABRE† hyperpolarization and detected using a benchtop NMR spectrometer. Quantification by standard addition is demonstrated for endogenic urinary nicotinamide (vitamin B3). Even higher sensitivity is achieved in an automated setup for multi-scan SABRE experiments. This hyperpolarization scheme is able to expedite biomarker detection and quantification, while maintaining low infrastructure requirements.
Rapid analysis of body fluids has become an important focus in health and metabolomics research. Urine is of particular interest, as it can be sampled non-invasively and can give a comprehensive record of an individual's metabolic state.1,2 The Human Metabolome Database lists over 3000 endogenous low-molecular weight metabolites in urine, only a fraction of which have been quantified.2,3 A range of these metabolites can in principle be detected by NMR, but the low natural concentration of many analytes poses a significant challenge. This is a particular hindrance for benchtop NMR spectrometers, which could offer an affordable way for routine analysis.
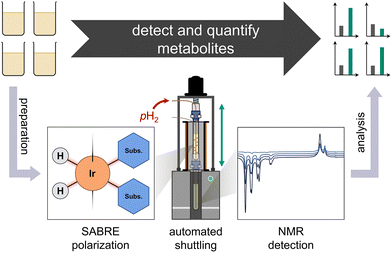
In this work, we increase the concentration sensitivity of benchtop NMR for select metabolites in a urinary extract by Signal Amplification By Reversible Exchange (SABRE) hyperpolarization.4 In contrast to superconducting high-field NMR instruments, benchtop spectrometers require less space, do not require cryogen refills, and their lower magnetic fields eases their integration into less specialized environments while adhering to safety regulations. These lower fields limit both sensitivity in non-hyperpolarized experiments, as well as chemical shift resolution. SABRE generates high polarization levels independent from the magnetic detection field, and allows for multiple polarization cycles and signal averaging, making it particularly attractive for overcoming the lower sensitivity of compact, low-field spectrometers (see Fig. 1).
Over the past decades, several other techniques have been developed to hyperpolarize mixtures as well, including variants of dynamic nuclear polarization (DNP)5 and ParaHydrogen-induced polarization (PHIP).6 Dissolution-DNP was used to identify patients with chronic kidney disease,7 as well as in monitoring metabolic conversions, both in cells8 and ex vivo.9 PHIP was used in several biological studies, e.g. PHIP side arm hydrogenation for spectroscopic imaging of the conversion of pyruvate to lactate in transgenic mice.10 To our knowledge, no PHIP-based routine was reported in natural extract analysis.
The different hyperpolarization variants have specific advantages and disadvantages. Dissolution-DNP is a powerful hyperpolarization technique, but requires radicals to be added to the sample, as well as microwave irradiation at cryogenic temperatures, is limited to single-shot experiments, and often requires considerable preparation times.11,12 Overhauser-DNP is less experimentally demanding and allows for re-hyperpolarization,13 but requires small sample volumes of a few µL or less for in situ polarization, respectively a sub-second shuttling setup for polarization outside of the spectrometer.14 Hydrogenative PHIP allows for large enhancements, but necessarily employs hydrogenation, which makes it inherently single-shot and reduces the scope of target substrates.15 In contrast to that, non-hydrogenative PHIP (nh-PHIP) allows for re-hyperpolarization at a repetition rate similar to regular NMR, and indirect detection of various compounds binding to a transition metal complex.16,17 It has been used for detecting nicotine and its breakdown products in urine,18 and was recently proposed as a broad-scope method for urine metabolome analysis.19 However, hyperpolarized nh-PHIP signals represent analytes bound to the metal complex, with their distinct chemical shifts. Such signals need to be assigned to different species, which is not intuitive and complicates analysis.
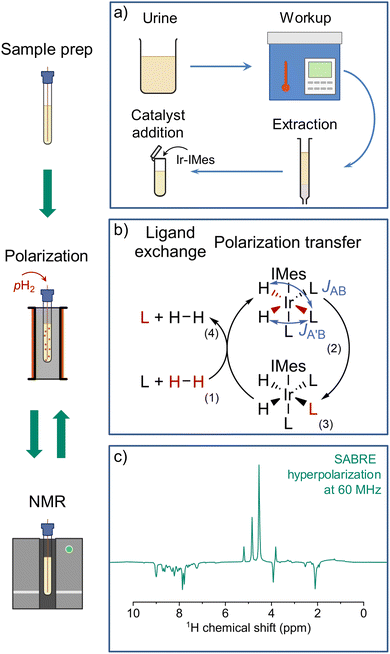
SABRE hyperpolarization, in turn, allows for repeatable, direct polarization of target molecules. Polarization transfer is facilitated by the same class of catalyst as nh-PHIP, and yields hyperpolarized signals at the familiar chemical shifts of the analytes.20,21 However, SABRE hyperpolarization for natural mixture analysis has not yet been published to our knowledge. In this work, we employ a three-step process adapted from Reile et al.22 for urine analysis: urine extract preparation, SABRE hyperpolarization and NMR detection, as illustrated in Fig. 2.
In the preparation step (Fig. 2a), urine is first autoclaved to eliminate potential pathogens. After centrifugation and pH adjustment, a solid-phase extraction (SPE) adapted from published methods,18,22 yields a methanolic solution containing urinary metabolites. These metabolites are in roughly six-fold higher concentration than in the initial urine (see SI for details). SPE reduces the complexity of the metabolic mixture, easing identification on benchtop spectrometers with limited frequency resolution, and provides ideal conditions for SABRE hyperpolarization (see SI). While extraction is not strictly required, raw urine samples would still need to be stripped of unwanted substances, including salts, urea and ammonia, as shown in studies employing nh-PHIP.16,23,24
For the hyperpolarization step (Fig. 2b), the sample is SABRE-hyperpolarized at a magnetic field of 6.5 mT (experimental details see SI) by bubbling 99% enriched parahydrogen (pH2). Applying a magnetic field of 6.5 mT allows for optimum spontaneous polarization transfer to 1H on a broad range of organic compounds,25 particularly for N-heterocycles, including nicotinamide and its derivatives.26
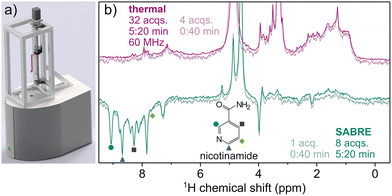
In the NMR detection step (Fig. 2c), the sample is transferred to a 1.5 T benchtop spectrometer, using a linear shuttling setup (Fig. 3a) based on a previously published design, which facilitates sample transfer by a robotic arm.27 Linear shuttling allows for shorter transfer times and higher positional accuracy when moving between polarization transfer field and spectrometer. This ensures consistent polarization build-up and transfer timings, as well as a defined transfer path through the magnetic field, contributing to highly repeatable scan-to-scan performance (experimental details see SI). Consequently, a hyperpolarized 1H NMR spectrum of the urine extract is acquired, displaying enhanced resonances, which stem from endogenous urinary SABRE-compatible analytes.
The resulting hyperpolarized spectrum of urine extract is compared to a thermal reference spectrum recorded at 1.5 T in Fig. 3b). In the SABRE spectrum, the signals of non-hyperpolarized compounds are suppressed compared to the spectrum recorded using thermal equilibrium polarization, as the sample spends only 0.7 s at the detection field before acquisition, allowing for a limited build-up of field-derived polarization. As a result, the SABRE spectrum is significantly less crowded than the spectrum acquired at thermal equilibrium, especially in the range of 0.6–4.8 ppm. In contrast to this, the aromatic region (7–9 ppm) contains various hyperpolarized peaks.
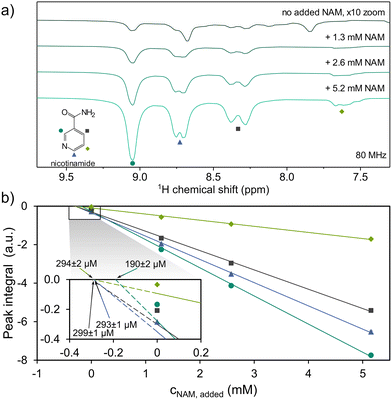
These enhanced signals can be identified by their natural chemical shifts, with assignments verifiable by standard addition. We demonstrate this for nicotinamide (NAM), a form of vitamin B3, which is known to be particularly amenable to SABRE hyperpolarization.28 The presence of NAM was further confirmed, and its concentration estimated, by nh-PHIP (see SI).29 We note that the NAM resonances at 7.6, 8.3 and 8.7 ppm marked in Fig. 3b) overlap with other signals not stemming from NAM. These additional peaks are not baseline-resolved, hindering quantification of NAM based on the respective resonances. A slightly narrower linewidth can be achieved by further optimizing the hydrogen delivery system (details see SI).
In the experiments shown here, the NAM resonance at 9.1 ppm is well isolated and appears to be mostly background-free.
This allows us to quantify NAM in the initial urine by adopting the internal standard spiking approach that has been previously demonstrated for SABRE28 and nh-PHIP,22,29 to a methanolic urine extract (Fig. 4a). The observed relation between added NAM and increase in hyperpolarized 1H signal integrals follows a linear trend (Fig. 4b), despite the complex nature of sample composition and SABRE polarization transfer, which proceeds over several competing equilibrium states with individual rate constants.30 We applied linear regression analysis for all four resonances to determine the concentration of NAM in the extract from each resonance (inset in Fig. 4b).28
Following the NAM resonance integral at 9.1 ppm (dark green circles), we arrive at a concentration of 190 ± 2 µM NAM in the employed sample. For the other resonances, we obtain higher initial concentrations of 294 ± 2 µM (7.6 ppm, bright green diamonds), 293 ± 1 µM (8.3 ppm, blue triangles) and 299 ± 1 µM (8.7 ppm, grey squares), respectively. This difference is attributed to the non-NAM signals found in the chemical shift range of 7.2–8.8 ppm, which cause an offset in integral value for these resonances.
Considering the dilution of extract (400 µL in a sample of 650 µL) and the six-fold concentration increase during extraction, assuming full recovery,22 we arrive at a concentration of 51 µM NAM in the initial urine. This value aligns with the 40 µM estimated by nh-PHIP (details see SI). The limit of quantization in a single scan is equivalent to 1.5 µM NAM in the initial urine. While low, the found value of 51 µM NAM is much higher than the reported normal urinary concentration of roughly 4 µM using HPLC.31 This discrepancy can be traced back to the workup procedure. In contrast to other studies, including work based on nh-PHIP,18,22 the urine samples in this work were sterilized by autoclaving, which can result in thermal degradation of larger NAM-containing molecules, such as nicotinamide adenine dinucleotide, releasing additional free NAM.32,33
SABRE experiments are often carried out with additional co-substrates to ensure an overabundance of coordination partners, which helps to stabilize the active species and fine-tune the exchange rate constants.21,22,28,34 Here, no co-substrates are used. While the individual metabolites are sub-stochiometric with respect to the SABRE catalyst, the sheer number of coordination partners in the urine extract stabilizes the catalyst complex in solution and prevents deterioration over the course of multiple acquisitions. This observation highlights the unexpected robustness and reversible ligand-exchange behavior at the Ir-catalyst in an environment of highly competitive substrate binding, providing insight that could be relevant to other fields, such as in the study of reaction mixtures and catalytic processes. Fitting resonances can improve peak separation in benchtop spectra,35 and increasing chemical shift resolution or applying more advanced pulse schemes, such as pure shift NMR,36 or 2D sequences, as routinely done in nh-PHIP routines,17,37 will aid in identifying and quantifying additional constituents.
Our procedure enables rapid detection of metabolic markers in a complex sample matrix, employing an affordable benchtop spectrometer. The chemoselective nature of SABRE, along with the extraction step, makes the technique useful for targeted analysis.38 After initial calibration by standard addition, conclusive 1H NMR spectra may be produced from just a single scan, requiring less than a minute. This allows adoption in high-throughput laboratories and in clinical settings near point-of-care, requiring fast, targeted identification and quantification of metabolites. While the related nh-PHIP procedure yields more hyperpolarized signals for the same extract (SI), the conceptual simplicity of SABRE and its ability to yield NMR data at the natural chemical shifts of analytes makes it the preferred choice as long as the analyte is amendable to SABRE. Considering that the known list of SABRE hyperpolarizable and metabolically relevant compounds already includes several valuable metabolites39,40 and drugs,41,42 the utility of SABRE in biochemical analysis is improving quickly.
Conceptualization: SL; data curation: SF, KA, IR; formal analysis: SF, KA, IR, NM, SL; funding acquisition: SL, IR, JK; investigation: SF, KA; methodology: SF, JY, IR; project administration: SL; resources: IR, NM, SL, JK; software: SF, JY; supervision: SL, JK; validation: SF, KA; visualization: SF, JY, KA; writing – original draft: SF; writing – review & editing: JY, KA, IR, NM, JK, SL.
Conflicts of interest
There are no conflicts to declare.Data availability
NMR data for this article is available at RADAR4KIT at https://doi.org/10.35097/qfe8pn1s6cc9uzam.Supplementary information: additional experimental details and spectra. See DOI: https://doi.org/10.1039/d5cc06186e.
Acknowledgements
We thank Anna Leismann for her help in sample preparation, and Wenxin Long for prototyping the shuttling setup. 800 MHz NMR spectra were acquired on instrumentation in the Estonian Centre of Analytical Chemistry (AKKI, Taristu24-TK15). The authors acknowledge support from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under contract 454252029 within the context of CRC HyPERiON 1527/1. We further acknowledge support from the Helmholtz Society's program Materials Systems Engineering, in the Research Area Information, and the Karlsruhe Nano Micro Facility (KNMFi), a Helmholtz Research Infrastructure at KIT. JY acknowledges financial support from the DFG under the Grant Agreement No. BR 4175/5-1. KA and IR acknowledge support from the Development Fund of the National Institute of Chemical Physics and Biophysics. SL acknowledges support from the DFG for his Emmy Noether group under contract 528402160.References
- J. Zheng, L. Zhang, M. Johnson, R. Mandal and D. S. Wishart, Anal. Chem., 2020, 92, 10627–10634 CrossRef CAS PubMed.
- S. Bouatra, F. Aziat, R. Mandal, A. C. Guo, M. R. Wilson, C. Knox, T. C. Bjorndahl, R. Krishnamurthy, F. Saleem, P. Liu, Z. T. Dame, J. Poelzer, J. Huynh, F. S. Yallou, N. Psychogios, E. Dong, R. Bogumil, C. Roehring and D. S. Wishart, PLoS One, 2013, 8, e73076 CrossRef CAS PubMed.
- D. S. Wishart, A. Guo, E. Oler, F. Wang, A. Anjum, H. Peters, R. Dizon, Z. Sayeeda, S. Tian, B. L. Lee, M. Berjanskii, R. Mah, M. Yamamoto, J. Jovel, C. Torres-Calzada, M. Hiebert-Giesbrecht, V. W. Lui, D. Varshavi, D. Varshavi, D. Allen, D. Arndt, N. Khetarpal, A. Sivakumaran, K. Harford, S. Sanford, K. Yee, X. Cao, Z. Budinski, J. Liigand, L. Zhang, J. Zheng, R. Mandal, N. Karu, M. Dambrova, H. B. Schiöth, R. Greiner and V. Gautam, Nucleic Acids Res., 2022, 50, D622–D631 CrossRef CAS PubMed.
- R. W. Adams, J. A. Aguilar, K. D. Atkinson, M. J. Cowley, P. I. P. Elliott, S. B. Duckett, G. G. R. Green, I. G. Khazal, J. López-Serrano and D. C. Williamson, Science, 2009, 323, 1708–1711 CrossRef CAS PubMed.
- A. B. Barnes, G. De Paëpe, P. C. A. Van Der Wel, K.-N. Hu, C.-G. Joo, V. S. Bajaj, M. L. Mak-Jurkauskas, J. R. Sirigiri, J. Herzfeld, R. J. Temkin and R. G. Griffin, Appl. Magn. Reson., 2008, 34, 237–263 CrossRef CAS PubMed.
- R. A. Green, R. W. Adams, S. B. Duckett, R. E. Mewis, D. C. Williamson and G. G. R. Green, Prog. Nucl. Magn. Reson. Spectrosc., 2012, 67, 1–48 CrossRef CAS PubMed.
- V. Ribay, B. Charrier, M. Croyal, B. Cariou, S. Hadjadj, J. Boccard, C. Cannet, J.-N. Dumez, M. P. M. Letertre and P. Giraudeau, J. Am. Chem. Soc., 2025, 147, 644–650 CrossRef CAS PubMed.
- P. R. Jensen, F. Sannelli, L. T. Stauning and S. Meier, Chem. Commun., 2021, 57, 10572–10575 RSC.
- C. Kjeldsen, J. H. Ardenkjær-Larsen and J. Ø. Duus, J. Am. Chem. Soc., 2018, 140, 3030–3034 CrossRef CAS PubMed.
- E. Cavallari, C. Carrera, M. Sorge, G. Bonne, A. Muchir, S. Aime and F. Reineri, Sci. Rep., 2018, 8, 8366 CrossRef PubMed.
- A. Dey, B. Charrier, K. Lemaitre, V. Ribay, D. Eshchenko, M. Schnell, R. Melzi, Q. Stern, S. F. Cousin, J. G. Kempf, S. Jannin, J.-N. Dumez and P. Giraudeau, Magn. Reson., 2022, 3, 183–202 CrossRef CAS PubMed.
- V. Ribay, A. Dey, B. Charrier, C. Praud, J. Mandral, J. Dumez, M. P. M. Letertre and P. Giraudeau, Angew. Chem., Int. Ed., 2023, 62, e202302110 CrossRef CAS PubMed.
- R. Kircher, H. Hasse and K. Münnemann, Anal. Chem., 2021, 93, 8897–8905 CrossRef CAS PubMed.
- C. Griesinger, M. Bennati, H. M. Vieth, C. Luchinat, G. Parigi, P. Höfer, F. Engelke, S. J. Glaser, V. Denysenkov and T. F. Prisner, Prog. Nucl. Magn. Reson. Spectrosc., 2012, 64, 4–28 CrossRef CAS PubMed.
- J. Hövener, A. N. Pravdivtsev, B. Kidd, C. R. Bowers, S. Glöggler, K. V. Kovtunov, M. Plaumann, R. Katz-Brull, K. Buckenmaier, A. Jerschow, F. Reineri, T. Theis, R. V. Shchepin, S. Wagner, P. Bhattacharya, N. M. Zacharias and E. Y. Chekmenev, Angew. Chem., Int. Ed., 2018, 57, 11140–11162 CrossRef PubMed.
- R. Fraser, F. P. J. T. Rutjes, M. C. Feiters and M. Tessari, Acc. Chem. Res., 2022, 55, 1832–1844 CrossRef CAS PubMed.
- M. Urbańczyk, K. Kork, K. Ausmees, T. Ratajczyk and I. Reile, Chem. Commun., 2025, 61, 12992–12995 RSC.
- N. Reimets, K. Ausmees, S. Vija and I. Reile, Anal. Chem., 2021, 93, 9480–9485 CrossRef CAS PubMed.
- N. Reimets, K. Ausmees and I. Reile, J. Magn. Reson. Open, 2024, 21, 100171 CrossRef.
- J.-B. Hövener, N. Schwaderlapp, T. Lickert, S. B. Duckett, R. E. Mewis, L. A. R. Highton, S. M. Kenny, G. G. R. Green, D. Leibfritz, J. G. Korvink, J. Hennig and D. Von Elverfeldt, Nat. Commun., 2013, 4, 2946 CrossRef PubMed.
- B. O. Jimmink, M. Negroni, T. B. Posthumus, A. P. M. Kentgens and M. Tessari, Anal. Chem., 2025, 97, 10962–10965 CrossRef CAS PubMed.
- I. Reile, N. Eshuis, N. K. J. Hermkens, B. J. A. Van Weerdenburg, M. C. Feiters, F. P. J. T. Rutjes and M. Tessari, Analyst, 2016, 141, 4001–4005 RSC.
- K. Ausmees, N. Reimets and I. Reile, Chem. Commun., 2022, 58, 463–466 RSC.
- L. Sellies, R. L. E. G. Aspers, M. C. Feiters, F. P. J. T. Rutjes and M. Tessari, Angew. Chem., Int. Ed., 2021, 60, 26954–26959 CrossRef CAS PubMed.
- A. N. Pravdivtsev, A. V. Yurkovskaya, H. Vieth, K. L. Ivanov and R. Kaptein, ChemPhysChem, 2013, 14, 3327–3331 CrossRef CAS PubMed.
- Q. S. Luu, Q. T. Nguyen, H. N. Manh, S. Yun, J. Kim, U. T. Do, K. Jeong, S. U. Lee and Y. Lee, Analyst, 2024, 149, 1068–1073 RSC.
- J. Yang, R. Xin, S. Lehmkuhl, J. G. Korvink and J. J. Brandner, Sci. Rep., 2024, 14, 21022 CrossRef CAS PubMed.
- N. Eshuis, B. J. A. van Weerdenburg, M. C. Feiters, F. P. J. T. Rutjes, S. S. Wijmenga and M. Tessari, Angew. Chem., 2015, 127, 1501–1504 CrossRef.
- N. Reimets, K. Ausmees, S. Vija and I. Reile, Anal. Chem., 2021, 93, 9480–9485 CrossRef CAS PubMed.
- K. Lin, P. TomHon, S. Lehmkuhl, R. Laasner, T. Theis and V. Blum, ChemPhysChem, 2021, 22, 1947–1957 CrossRef CAS PubMed.
- R. Lang, E. F. Yagar, R. Eggers and T. Hofmann, J. Agric. Food Chem., 2008, 56, 11114–11121 CrossRef CAS PubMed.
- S. Hachisuka, T. Sato and H. Atomi, J. Bacteriol., 2017, 119(19) DOI:10.1128/JB.00162-17.
- K. Honda, N. Hara, M. Cheng, A. Nakamura, K. Mandai, K. Okano and H. Ohtake, Metab. Eng., 2016, 35, 114–120 CrossRef CAS PubMed.
- N. Eshuis, N. Hermkens, B. J. A. Van Weerdenburg, M. C. Feiters, F. P. J. T. Rutjes, S. S. Wijmenga and M. Tessari, J. Am. Chem. Soc., 2014, 136, 2695–2698 CrossRef CAS PubMed.
- J. Phuong, B. Salgado, J. Heiß, E. Steimers, P. Nickolaus, L. Keller, U. Fischer, E. Von Harbou, D. J. Holland, F. Jirasek, H. Hasse and K. Münnemann, Food Res. Int., 2025, 203, 115741 CrossRef CAS PubMed.
- D. A. Taylor, L. S. Natrajan, M. Nilsson and R. W. Adams, Magn. Reson. Chem., 2021, 59, 1244–1252 CrossRef CAS PubMed.
- P. M. Richardson, A. J. Parrott, O. Semenova, A. Nordon, S. B. Duckett and M. E. Halse, Analyst, 2018, 143, 3442–3450 RSC.
- I. Reile, N. Eshuis, N. K. J. Hermkens, B. J. A. Van Weerdenburg, M. C. Feiters, F. P. J. T. Rutjes and M. Tessari, Analyst, 2016, 141, 4001–4005 RSC.
- S. J. McBride, M. Pike, E. Curran, A. Zavriyev, B. Adebesin, L. Tucker, J. M. Harzan, I. M. Senanayake, M. Abdulmojeed, F. Theiss, S. Shen, T. Boele, S. B. Duckett, B. M. Goodson, M. S. Rosen, E. Y. Chekmenev, H. Yuan, C. Dedesma, T. Gade, S. Kadlecek, T. Theis and P. TomHon, Angew. Chem., Int. Ed., 2025, 64, e202501231 CrossRef CAS PubMed.
- I. Adelabu, M. R. H. Chowdhury, S. Nantogma, C. Oladun, F. Ahmed, L. Stilgenbauer, M. Sadagurski, T. Theis, B. M. Goodson and E. Y. Chekmenev, Metabolites, 2023, 13, 200 CrossRef CAS PubMed.
- K. MacCulloch, A. Browning, P. TomHon, S. Lehmkuhl, E. Y. Chekmenev and T. Theis, Anal. Chem., 2023, 95, 7822–7829 CrossRef CAS PubMed.
- K. MacCulloch, P. Tomhon, A. Browning, E. Akeroyd, S. Lehmkuhl, E. Y. Chekmenev and T. Theis, Magn. Reson. Chem., 2021, 59, 1225–1235 CrossRef CAS PubMed.
Footnote |
| † SABRE: Signal Amplification By Reversible Exchange. |
| This journal is © The Royal Society of Chemistry 2026 |