Open Access Article
Open Access ArticleA-type helicity controls efficient nonenzymatic template copying of nucleic acids
Barbara K.
Lech
 *a,
Petr
Jurečka
b,
Marie
Zgarbová
b and
Rafał
Szabla
*a,
Petr
Jurečka
b,
Marie
Zgarbová
b and
Rafał
Szabla
 *ab
*ab
aInstitute of Advanced Materials, Faculty of Chemistry, Wroclaw University of Science and Technology, Wybrzeże Stanisława Wyspiańskiego 27, 50-370, Wrocław, Poland. E-mail: barbara.lech@pwr.edu.pl; rafal.szabla@pwr.edu.pl
bDepartment of Physics, Faculty of Science, University of Ostrava, 30. dubna 22, 70103, Ostrava, Czech Republic
First published on 10th December 2025
Abstract
We performed classical molecular dynamics simulations for DNA self-replication on an RNA template with phosphoroimidazole activation. We demonstrate that the RNA template imposes A-type helicity on the hybrid DNA/RNA helix, as opposed to the B-type helix of pure DNA-based systems. Based on structural analyses, we conclude that helicity type is the primary factor responsible for the feasibility of nonenzymatic primer extension.
Over the past four decades, prebiotic nonenzymatic primer extension has been extensively studied and considered as a crucial aspect of the origin of life on Earth.1 These efforts have primarily focused on RNA self-replication, since RNA is considered a prebiotic predecessor of DNA,1 and the efficiency of the reaction was reported to be significantly higher for RNA than for DNA.2 Generally, such reactions require nucleotide activation, e.g. by the formation of phosphoroimidazolides involving the well-studied 2-aminoimidazole, which facilitates nucleophilic substitution at the phosphate center and enables primer extension.3 Metal cations such as Mg2+ are essential for the process, although their catalytic function is not yet fully understood.4 Elucidating this reaction mechanism is essential to identify the most efficient variants of prebiotic RNA self-replication. However, relatively bad performance of DNA in non-enzymatic self-replication with phosphoroimidazolides is likely the result of the differences in the secondary structure when compared to RNA and the way it affects the reactive conformations in the reaction centre.2,5
The effect of the structural arrangement of the double helix on nonenzymatic RNA self-replication was first reported by Leu and co-workers.6 RNA typically adopts an A-type helical structure, while DNA predominantly forms a B-type double helix.7 These secondary structures can be distinguished by several structural parameters, such as sugar puckering. In A-type helices, the sugar ring favors the north (C3′-endo) conformation, whereas in B-type helices the south (C2′-endo) conformation is more common.8 This affects the spatial positioning of the key atoms involved in the primer extension reaction. That is, the O3′ atom is positioned closer to the incoming 5′ phosphate group of the bound activated nucleotide in A-type helices, which facilitates the formation of a new phosphodiester bond. In contrast, greater distance between these atoms in B-type helices makes the reaction less efficient.9
Despite the low efficiency of nonenzymatic DNA self-replication, it remains an attractive potential alternative method for DNA synthesis. The two most common approaches to DNA synthesis, PCR (polymerase chain reaction)10 and solid-phase synthesis,11 are well established but have several drawbacks. These methods are complex, time-consuming, require costly enzymes and large volumes of solvents, and can produce significant amounts of waste.12 Recently, the Ellis lab demonstrated that the DNA self-replication process can be enhanced by introducing an RNA template.5 Replacing the DNA template with RNA resulted in a 4.5 fold increase in maximum rate and fidelity of primer extension compared to the DNA-only system. The authors suggest that this enhancement is due to a conformational shift in the sugar puckering of nucleotides, from C2′-endo to C3′-endo, specifically at the 3′ terminus of the primer and in the adjacent activated imidazolium-bridged dinucleotide.5
In this work, we employed classical molecular dynamics (MD) simulations to investigate the key structural factors governing nonenzymatic DNA primer extension on an RNA template and to determine the structural properties of the involved nucleotides that can facilitate this process. We also explored the role of Mg2+ cations in the reaction mechanism, particularly their involvement in the deprotonation of the terminal 3′-OH group of the primer, which is believed to be crucial for the primer extension reaction.4
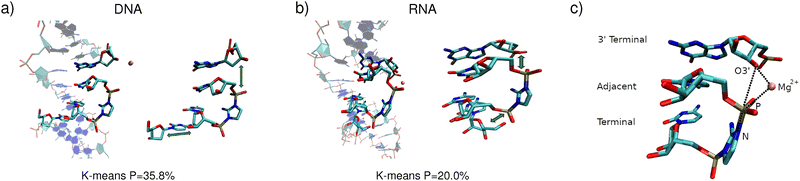
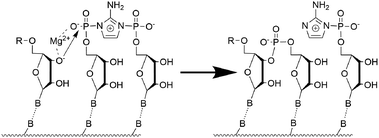
We focused on four systems: pure RNA, pure DNA, and two hybrid structures in which the templating strand is RNA, while the primer, activated dinucleotide, and helper oligonucleotide are composed of DNA. One of these hybrid systems initially adopted an A-type helical conformation (D/RNA A), whereas the MD simulations for the other one were started from B-type helicity (D/RNA B). Each strand consisted of 15 nucleotides, and the activated imidazole-bridged cytidine dinucleotide is positioned after the sixth nucleotide of the primer. These simulations were performed with a single Mg2+ cation placed in between the terminal 3′-OH group of the primer and the neighbouring phosphoroimidazolide group of the activated dinucleotide (see Fig. 1). Since we also investigated the effect of 3′-OH deprotonation on the reactive conformations, we performed eight 3.0 µs-long simulations covering the four systems in total, each with protonated or deprotonated 3′ ends of the primer (see Fig. 2).
We next performed clustering analysis of the MD trajectories. For all investigated systems, the activated cytidine dinucleotide consistently maintains canonical Watson–Crick hydrogen bonding with complementary guanosine nucleotides in the templating strand and is positioned in near proximity to the 3′-terminus of the primer, with the Mg2+ ion located between the 3′ oxygen and the phosphorus atoms. However, a noticeable difference is observed in how the activated dinucleotide fits into A-type versus B-type helices. In RNA (A-type helicity), the imidazolium-bridged dinucleotide is positioned near both the 3′-terminus of the primer and the 5′-terminus of the helper strands (see Fig. 1b)). In contrast, in DNA (B-type helicity), the dinucleotide is located either near the 3′ end of the primer or the 5′ end of the helper, but not both simultaneously (Fig. 1a)). The RMSD analysis (see Section S3 of the SI) of the binding cavity shows that the RNA-activated dinucleotide exhibits relatively small movements on the A-RNA template (average RMSD value of 1.1 Å for the protonated system), whereas the DNA-activated dinucleotide displays considerably larger displacements on the B-DNA template (the average RMSD value of ∼1.6 Å for the protonated system). In other words, the cavity for the binding of the activated dinucleotide is tighter in the A-type double helix, which may contribute to more efficient SN2 reaction and primer extension.
The main structural parameters determining the efficiency of the phosphate-centered SN2 reaction leading to a phosphodiester bond are the distance between the 3′-oxygen and the phosphate atom, as well as the angle of nucleophilic attack.4,13 The proposed reaction mechanism is schematically shown in Fig. 2. The first step involves deprotonation of the 3′-OH group of the primer, and the negatively charged O3′ atom can be coordinated by the Mg2+ cation. This is followed by nucleophilic attack of O3′− on the phosphate group of the adjacent activated nucleotide. Throughout the simulations, we monitored the associated reactive O3′⋯P distance. Fig. 3 shows the probability distribution of this distance for the protonated structures, ranging from 3.1 Å to above 7.0 Å. For RNA, the highest populated distance is ∼3.7 Å, with a median value of 4.1 Å. This demonstrates that the reactive substrate complex for the primer extension of RNA is stable and highly populated, which explains the high yield and efficiency of the self-replication process. In contrast, pure DNA exhibits a different pattern, with the highest populated distance of ∼5.1 Å and a median of 5.2 Å, which does not favor an efficient replication process. The hybrid D/RNA A system behaves similarly to pure RNA, showing the highest peak at 3.6 Å and a median of 3.9 Å, while D/RNA B displays a wide range of populated distances, with a median of 5.5 Å. These results indicate that the system starting from a B-form helical structure exhibits worse results than the system starting from the A-form.
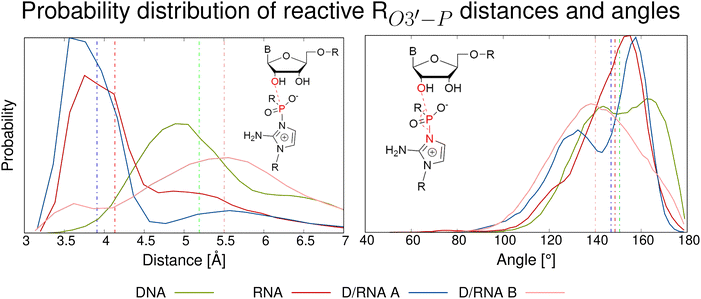 | ||
| Fig. 3 Probability distribution of O3′–P distance and O–P–N angle (attack angle) for protonated systems. Dashed lines represent the median values. | ||
Another parameter characterizing the SN2 reaction is the attack angle, defined by the O3′, P, and N atoms (see Fig. 1c)). As shown in Fig. 3, the protonated systems exhibit attack angles ranging from 130° to 180°, with median values of 151° (DNA), 149° (RNA), 147° (D/RNA A), and 140° (D/RNA B). DNA and D/RNA B exhibit a wide range of attack angles, whereas RNA and D/RNA A display a distinct peak around 150°–160°, indicating the highest population within that angle range. A similar analysis was conducted for the deprotonated structures (see Fig. S15). However, we believe that following the deprotonation, the SN2 reaction proceeds rapidly, and the O–P–N angle becomes less significant compared to its role prior to the deprotonation step.
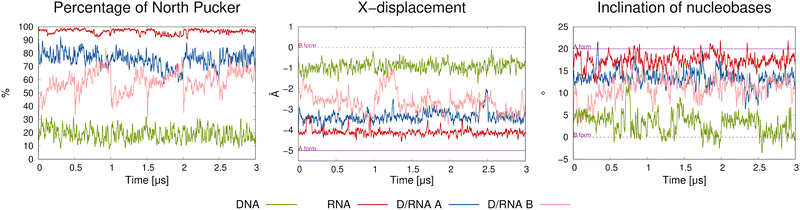
To determine the type of helix, we analysed three parameters: the overall percentage of the north pucker conformation of the sugar rings, the X-displacement, which reflects the size of the central cavity of the double helix relative to its central axis, and the inclination of the nucleobases. The A-form is characterized by a nucleobase inclination of approximately 20°, an X-displacement of about −5 Å, and north-type puckering of the sugar ring. In contrast, the B-form typically exhibits south-type sugar puckering, minimal X-displacement, and an inclination close to 0°.8 As shown in Fig. 4 and Fig. S17, the DNA structure (green line) exhibits a low proportion of north-type puckering (10–30%), X-displacement values fluctuating around −1 Å, and inclination angles ranging from −5° to ∼10°, all of which are indicative of B-type helicity. In contrast, RNA structures (red lines) exhibit the opposite behaviour: the inclination ranges from 10° to 20°, the X-displacement is approximately −4 Å, and the north-type puckering dominates throughout the simulations. These are clear indicators of the A-form helix. The hybrid D/RNA A systems (blue lines) exhibit A-type helicity, as their sugar rings are predominantly in the north conformation (70–80%), the inclination fluctuates around 13°, and the X-displacement remains below −3 Å. In contrast, the D/RNA B systems (pink lines) undergo a rapid transition from B-type to A-type helicity at the beginning of each simulated MD trajectory. This is evidenced by an increased prevalence of north puckering (from 40% up to 60–85%), a sharp decrease in X-displacement to approximately −3 Å, and a rise in inclination to around 15°. However, these parameters do not reach the range observed for pure RNA or D/RNA A, suggesting that more time is required for a full transition between B and A helicities. Overall, these observations confirm that the RNA template promotes A-type helicity in the hybrid duplex.
A more detailed investigation of sugar puckering in two key nucleotides: the 3′-terminal and adjacent residue of the activated dinucleotide, revealed a surprising pattern. Deprotonation of the 3′ end strongly promotes north-type puckering of the 3′-terminal sugar, while adjacent nucleotides predominantly adopt the south conformation, except for D/RNA B (see Table 1). This effect is particularly evident in RNA structures: in the protonated state, the 3′-terminal and adjacent nucleotides display north puckering in 85.39% and 79.50% of frames, respectively, whereas after deprotonation these values shift to 99.99% and 7.12%. This shift correlates with the shortening of the O3′–P distance upon deprotonation and suggests that deprotonation of the 3' OH group is an important step in the reaction mechanism (see Fig. S18–S21). However, the exact cause of this conformational shift remains uncertain. More intriguing is the fact that the 3′ terminal nucleotide of the pure DNA structure favours the north geometry of the sugar ring (82.21%). These results challenge the assumption that efficient primer extension requires both the 3′ terminal and adjacent nucleotides to adopt the north conformation.5 While this hypothesis may be valid for the 3′-terminal nucleotide, our MD simulations show that the pure DNA system predominantly adopts a north conformation of the 3′-terminal nucleotide, which is insufficient for an efficient primer extension reaction to occur.
| Protonated | ||||
|---|---|---|---|---|
| Nucleotide | DNA (%) | RNA (%) | D/RNA A (%) | D/RNA B (%) |
| 3′ Terminal | 82.21 | 85.39 | 77.35 | 35.14 |
| Adjacent | 28.62 | 79.50 | 17.08 | 27.94 |
| Deprotonated | ||||
|---|---|---|---|---|
| Nucleotide | DNA (%) | RNA (%) | D/RNA A (%) | D/RNA B (%) |
| 3′ Terminal | 99.38 | 99.99 | 99.84 | 98.60 |
| Adjacent | 28.68 | 7.12 | 12.31 | 56.93 |
Our results demonstrate that introducing an RNA template for DNA self-replication induces A-type helicity. Moreover, the parameters describing the SN2 reaction indicate that A-type helicity clearly facilitates nonenzymatic primer extension with phosphoroimidazole activation. In both hybrid D/RNA systems, the helix consistently maintained A-type characteristics throughout the simulation. Moreover, the 3′-terminal nucleotide exhibits a strong preference for the north-type sugar pucker, particularly in deprotonated structures. This is associated with a shorter distance between the reactive atoms (e.g., O3′ and P), which could facilitate primer extension. However, the geometries of the 3' -terminal and adjacent sugar rings do not support the hypothesis that the C3′-endo conformation is strictly responsible for efficient self-replication. The DNA exhibits north-type sugar puckers of the 3′-terminal nucleotide even in the absence of an RNA template. Overall, the results indicate that the stability of the A-type helical conformation is the primary structural factor enabling efficient nonenzymatic self-replication of nucleic acids.
Conceptualization: B. K. L. and R. S.; investigation: B. K. L. (major), M. Z., and P. J.; formal analysis: B. K. L.; visualization and writing of the original draft: B. K. L.; funding acquisition: R. S. and P. J.; resources: R. S.; writing – review and editing: R. S.; supervision: R. S. and P. J.
Conflicts of interest
There are no conflicts to declare.Data availability
Data for this article, including trajectories and clustering results are available at Figshare at https://doi.org/10.6084/m9.figshare.29625755.v2.The data supporting this article have been included in part of the supplementary information (SI). The SI contains computational methods and additional analyses of the trajectories. See DOI: https://doi.org/10.1039/d5cc05343a.
This work was supported by a grant from the National Science Centre, Poland (2022/46/E/ST4/00377 to R. S.). This article has been produced with the financial support of the European Union under the LERCO project number (CZ.10.03.01/00/22_003/0000003) via the Operational Programme Just Transition. The simulations were performed in the high-performance Infrastructure PLGrid (grant no. PLG/2024/017359).
References
- (a) M. P. Robertson and G. F. Joyce, Cold Spring Harbor Perspect. Biol., 2012, 4, a003608 CrossRef PubMed; (b) J. Xu, V. Chmela, N. J. Green, D. A. Russell, M. J. Janicki, R. W. Góra, R. Szabla, A. D. Bond and J. D. Sutherland, Nature, 2020, 582, 60–66 CrossRef CAS PubMed; (c) G. Joyce, T. Inoue and L. Orgel, J. Mol. Biol., 1984, 176, 279–306 CrossRef CAS PubMed; (d) F. H. Crick, J. Mol. Biol., 1968, 38, 367–379 CrossRef CAS PubMed.
- (a) T. Walton, W. Zhang, L. Li, C. P. Tam and J. W. Szostak, Angew. Chem., Int. Ed., 2019, 58, 10812–10819 CrossRef CAS PubMed; (b) J. P. Schrum, A. Ricardo, M. Krishnamurthy, J. C. Blain and J. W. Szostak, J. Am. Chem. Soc., 2009, 131, 14560–14570 CrossRef CAS PubMed; (c) C. Deck, M. Jauker and C. Richert, Nat. Chem., 2011, 3, 603–608 CrossRef CAS PubMed; (d) D. Ding, L. Zhou, C. Giurgiu and J. W. Szostak, Nucleic Acids Res., 2022, 50, 35–45 CrossRef CAS PubMed.
- (a) O. E. Leslie, Crit. Rev. Biochem. Mol. Biol., 2004, 39, 99–123 CrossRef PubMed; (b) W. Zhang, T. Walton, L. Li and J. W. Szostak, eLife, 2018, 7, e36422 CrossRef PubMed; (c) W. Zhang, C. P. Tam, L. Zhou, S. S. Oh, J. Wang and J. W. Szostak, J. Am. Chem. Soc., 2018, 140, 2829–2840 CrossRef CAS PubMed; (d) L. Li, N. Prywes, C. P. Tam, D. K. O'Flaherty, V. S. Lelyveld, E. C. Izgu, A. Pal and J. W. Szostak, J. Am. Chem. Soc., 2017, 139, 1810–1813 CrossRef CAS PubMed; (e) J. P. Ferris and G. Ertem, Science, 1992, 257, 1387–1389 CrossRef CAS PubMed; (f) J. Sulston, R. Lohrmann, L. Orgel and H. T. Miles, Proc. Natl. Acad. Sci. U. S. A., 1968, 59, 726–733 CrossRef CAS PubMed.
- (a) S. Mittal, C. Nisler and J. W. Szostak, Biophys. J., 2024, 123, 1579–1591 CrossRef CAS PubMed; (b) R. Lohrmann, P. K. Bridson and L. E. Orgel, Science, 1980, 208, 1464–1465 CrossRef CAS PubMed; (c) T. Inoue and L. E. Orgel, J. Am. Chem. Soc., 1981, 103, 7666–7667 CrossRef CAS.
- S. J. Park, K. L. Callaghan and A. V. Ellis, Org. Biomol. Chem., 2023, 21, 6702–6706 RSC.
- K. Leu, E. Kervio, B. Obermayer, R. M. Turk-MacLeod, C. Yuan, J.-M. J. Luevano, E. Chen, U. Gerland, C. Richert and I. A. Chen, J. Am. Chem. Soc., 2013, 135, 354–366 CrossRef CAS PubMed.
- (a) A. Rich, Nat. Struct. Mol. Biol., 2003, 10, 247–249 CrossRef CAS PubMed; (b) W. Saenger, Principles of Nucleic Acid Structure, Springer Science & Business Media, 2013 Search PubMed; (c) M. Zgarbová, J. Sponer and P. Jurecka, J. Chem. Theory Comput., 2025, 21, 833–846 CrossRef PubMed.
- M. Zgarbová, P. Jurečka, J. Šponer and M. Otyepka, J. Chem. Theory Comput., 2018, 14, 319–328 CrossRef PubMed.
- (a) V. Tereshko, S. Gryaznov and M. Egli, J. Am. Chem. Soc., 1998, 120, 269–283 CrossRef CAS; (b) D. Ding, S. M. Gryaznov and W. D. Wilson, Biochemistry, 1998, 37, 12082–12093 CrossRef CAS PubMed.
- K. Mullis, F. Faloona, S. Scharf, R. Saiki, G. Horn and H. Erlich, Cold Spring Harbor Symp. Quant. Biol., 1986, 51, 263–273 CrossRef CAS PubMed.
- B. Merrifield, Science, 1986, 232, 341–347 CrossRef CAS PubMed.
- B. I. Andrews, F. D. Antia, S. B. Brueggemeier, L. J. Diorazio, S. G. Koenig, M. E. Kopach, H. Lee, M. Olbrich and A. L. Watson, J. Org. Chem., 2021, 86, 49–61 CrossRef CAS PubMed.
- B. K. Lech, B. B. Ogunnaiya, E. F. Petrusevich and R. Szabla, ChemSystemsChem, 2025, 7, e202400086 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2026 |