Open Access Article
Open Access ArticleAn AKR1C3-activated kinase inhibitor prodrug
Zhengnian
Li†‡
a,
Michael
Martinez‡
a,
Woong Sub
Byun
a,
Anuradha
Thathireddy
a,
Tian
Qiu
 a,
Yaning
Wang
a,
Leon
Katzengruber
a,
Yaning
Wang
a,
Leon
Katzengruber
 a,
Ani
Chouldjian§
a,
Wenchao
Lu¶
a,
Ani
Chouldjian§
a,
Wenchao
Lu¶
 a,
Wenzhi
Ji
a,
Jianwei
Che
b,
Tinghu
Zhang
a,
Stephen M.
Hinshaw
a,
Wenzhi
Ji
a,
Jianwei
Che
b,
Tinghu
Zhang
a,
Stephen M.
Hinshaw
 *a and
Nathanael S.
Gray
*a
*a and
Nathanael S.
Gray
*a
aDepartment of Chemical and Systems Biology, ChEM-H, and Stanford Cancer Institute, Stanford School of Medicine, Stanford University, Stanford, CA, USA. E-mail: nsgray01@stanford.edu; hinshaw@stanford.edu
bDepartment of Cancer Biology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA
First published on 19th December 2025
Abstract
Enzymatically activated prodrugs can enable context-specific target inhibition. AKR1C3 is an NADPH-dependent aldo-ketoreductase involved in androgen metabolism, prostaglandin synthesis, and cell proliferation that is overexpressed in tumors, making it an ideal candidate for tumor-specific prodrug activation. Reported prodrugs that exploit AKR1C3 catalytic activity release DNA-intercalating toxins or other non-selective poisons upon enzymatic activation. OBI-3424, a prodrug of a DNA alkylating agent, is a prominent example of this strategy. To extend this concept to selective enzymatic inhibitors, we have developed AKR1C3-activated prodrugs of OTS964, a CDK11 inhibitor. We have probed the activities of the compounds with biochemical and cellular assays, finding specific activation of the lead prodrug by AKR1C3. Upon enzymatic conversion, the compound recapitulates the cellular activity of the parent compound. These results demonstrate that the AKR1C3-activated prodrug strategy can be used to convert selective kinase inhibitors into context-dependent prodrugs. Extension of this approach may enable synthesis of prodrugs for targeted therapies that spare normal tissue, further improving their therapeutic windows.
Introduction
Prodrugs are pharmacologically inactive compounds that can be converted to active drugs. Activation mechanisms are either chemical, as for peroxide-cleaved prodrugs,1,2 or enzymatic, as for carboxylesterase substrate prodrugs.3 Enzymatically activated prodrugs are compelling, because they can be targeted to tumor cells that upregulate metabolic enzymes absent from nearby untransformed tissue. This restricts the tissue distribution of the active drugs and improves their toxicity profiles, enabling higher dosage before reaching limiting systemic toxicity.Enzymatic prodrug activation mechanisms that depend on Aldo-ketoreductase 1C3 (AKR1C3) have gained attention in recent years. The enzyme is highly expressed in a wide range of diseases (Fig. S1),4 including hormone-dependent and independent tumors. This makes AKR1C3-mediated conversion a compelling avenue for chemical prodrug activation.5–10 Its endogenous activity is to reduce the sole carbonyl of dehydroepiandrosterone (DHEA) and other signaling metabolites to the corresponding alcohol.11 There are 15 human alpha-ketoreductases, four of which have annotated hydroxysteroid dehydrogenase activity (AKR1C1-4).12 These use nicotinamide adenine dinucleotide phosphate (NADPH) as a hydride donor and, subsequently, a catalytic tyrosine to transfer a proton to the carbonyl oxygen.12,13
Reported AKR1C3-activated prodrugs are cytotoxic DNA intercalating/alkylating agents – effectively caged versions of common chemotherapeutics.14,15 For example, OBI-3424 is a prodrug of a DNA-alkylating nitrogen mustard that has shown early-stage clinical promise and dependence on AKR1C3 for its activation.14,16 Recently reported improvements of this concept that maintain the DNA alkylation activity of OBI-3424 are curative in patient-derived xenograft mouse models of T cell lymphoma and pancreatic cancer.17,18 Curiously, OBI-3424 and other AKR1C3-activated compounds feature a nitrobenzyl group and not a labile ketone, as might be expected due to the known AKR1C3 substrate preference. Indeed, the commonly used AKR1C3-activated protecting groups were initially designed as substrates for nitroreductase enzymes upregulated in the hypoxic tumor microenvironment and were only later discovered to be AKR1C3 substrates under aerobic conditions.15 Thus, activation by AKR1C3 provides access to activated compounds in oxygen-replete tumors. This property serves a second purpose by converting a known chemotherapy liability – drug decomposition by AKR1C319 – into a strength. Despite the existence of a well-defined and apparently chemically transplantable AKR1C3-activated protecting group, selective small molecule inhibitor prodrugs that exploit a similar mechanism have not been described.
One application of the AKR1C3-activated prodrug strategy is to extend the approach to rationally design enzymatically gated selective small molecule inhibitors of disease-specific signaling proteins. Such a strategy would yield chemotherapeutic agents with dual selectivity filters, targeting only tissues that (i) express AKR1C3 and (ii) depend on the targeted protein. To determine the feasibility of this concept, we targeted a kinase due to the wide availability of selective and structurally well-characterized small molecule inhibitors,20 many of which inhibit enzymes with disease-specific relevance. Successful synthesis and validation of AKR1C3-gated kinase inhibitors would provide a conceptually novel advance over existing AKR1C3 prodrug modalities by enabling rational synthesis of targeted prodrug therapeutics at will.
We chose CDK11 (Cyclin-dependent kinase 11) as a first target for AKR1C3-activated kinase inhibitor prodrugs due to the availability and chemical tractability of potent and selective small molecule inhibitors.21,22 CDK11, which is encoded by paralogous genes (CDK11A and CDK11B), regulates transcription and splicing by controlling phosphorylation of RNA polymerase II and by directly phosphorylating its substrate SF3B1 (splicing factor 3b subunit 1).23,24 It is therefore a potential therapeutic target of interest in cancers that depend heavily on RNA splicing,25,26 including high-grade serous ovarian cancer and castrate-resistant prostate cancer. Unfortunately, on-target toxicity from current CDK11 inhibitors disqualifies their clinical use.27 Synthesis of prodrug versions of these active compounds may help ameliorate anticipated on-target toxicity resulting from disruption of transcription in non-cancerous tissues and will provide a template for extension of the prodrug strategy to other kinase inhibitors of therapeutically relevant targets.
Results
Prodrug design and mechanism of AKR1C3-mediated release
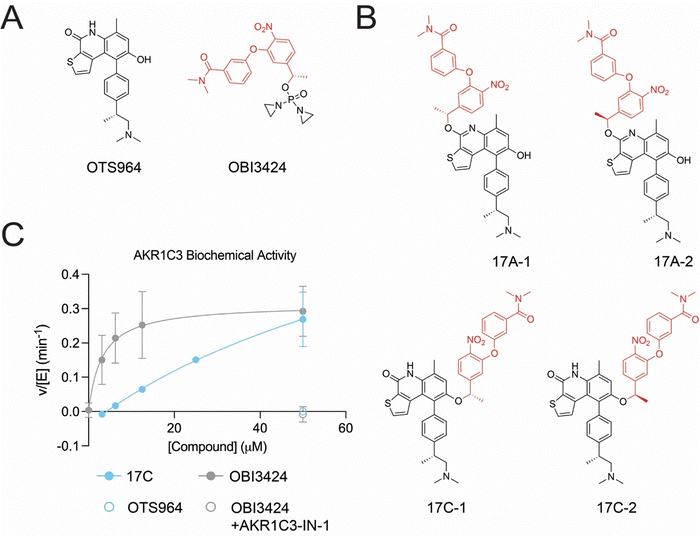
We have synthesized prodrugs that comprise two functional components: (i) a protecting group that serves as a substrate of AKR1C3 and (ii) a selective CDK11 inhibitor, which serves as a leaving group. The two components are linked via a labile ether bond. In the prodrug form, the protecting group disables kinase binding, introducing clashes with either the kinase hinge at the back of the ATP-binding pocket or the sugar binding residues, depending on its attachment point (Fig. 1A). NADPH-dependent reduction of the prodrug nitro group by AKR1C3 generates an unstable phenylhydroxamine intermediate, which collapses rapidly, releasing the active inhibitor and the protecting group. The protecting group eventually resolves to nitrosobenzamine or a phenylhydroxylamine. (Fig. 1B and C).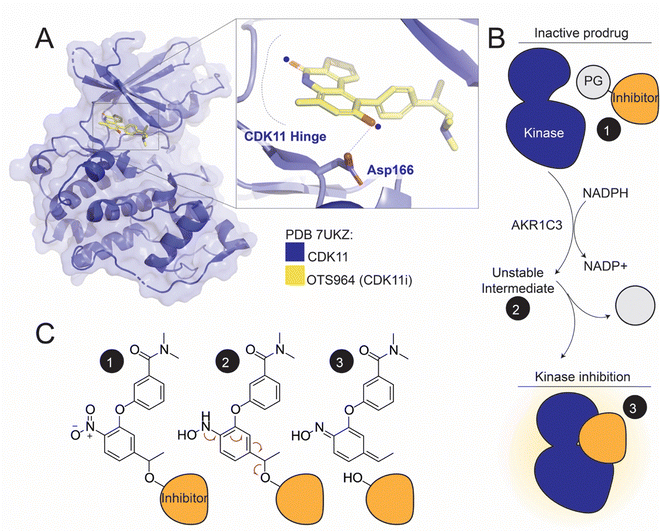 | ||
| Fig. 1 The CDK11 inhibitor OTS964 and the conceptual basis for prodrug activity. (A) Crystal structure of the CDK11-OTS964 complex (PDB 7UKZ).38 The inset shows the attachment points for the prodrug protecting group (17A – top blue dot; 17C – bottom blue dot). A hydrogen bond between OTS964 and CDK11-Asp166 is shown as a dotted line. (B) Schematic showing the stepwise AKR1C3-dependent reduction and activation of a kinase inhibitor prodrug (PG – protecting group). (C) Chemical basis for kinase inhibitor prodrug activation. The black circles match those shown in panel B. The protecting group in state (3) is drawn as the direct product of ester collapse and likely resolves into nitrosobenzene of benzylhydroxylamine (see text). | ||
We synthesized four candidate CDK11-targeted prodrugs to exploit the activation mechanism described above (Fig. 2A). The two pairs of diastereomers differed in the prodrug attachment point: 17A-1 and 17A-2, where the ether bond-linked prodrug protecting group replaces the OTS964 carbonyl group, and 17C-1 and 17C-2, where the protecting group ether linkage replaces the OTS964 hydroxyl group. Both pairs of diastereomers differ in the configuration of the chiral carbon adjacent to the ether oxygen. Because we observed no chirality-dependent activity (see below), we did not determine and do not specify here the absolute chirality of these species.
Enzymatic activation of CDK11-targeting prodrugs
We used an enzymatic assay to detect AKR1C3 activity in vitro. The unmodified kinase inhibitor OTS964 did not activate NADPH oxidation to NADP+ by AKR1C3 (Fig. 2C). By contrast, OBI-3424, an AKR1C3-activated DNA-alkylating prodrug stimulated NADPH oxidation, and the selective AKR1C3 inhibitor, AKR1C3-IN-1,28 abrogated this effect. All newly synthesized prodrug candidates activated NADPH oxidation with similar in vitro potencies (Fig. S2A and B). The apparent KM for OBI-3424 approached or surpassed the lower limit of detection for the assay (KM,app 3.2 µM), and this value was much higher for all kinase inhibitor prodrugs. Nevertheless, the catalytic constants appeared similar for all tested prodrugs, though they could not be measured with high confidence for the newly synthesized compounds. Thus, the same rate-limiting chemical transformation likely gates conversion of all prodrug substrates. The tested diastereomers (17A-1versus17A-2 and 17C-1versus17C-2) were indistinguishable in this assay, and the racemic mixture (17C) was used in the remaining experiments, accordingly. Finally, we further confirmed ketroeductase-mediate transformation of the 17C prodrug by directly monitoring the evolution of reaction products (Fig. S2C and D).Evaluation of cellular CDK11 engagement
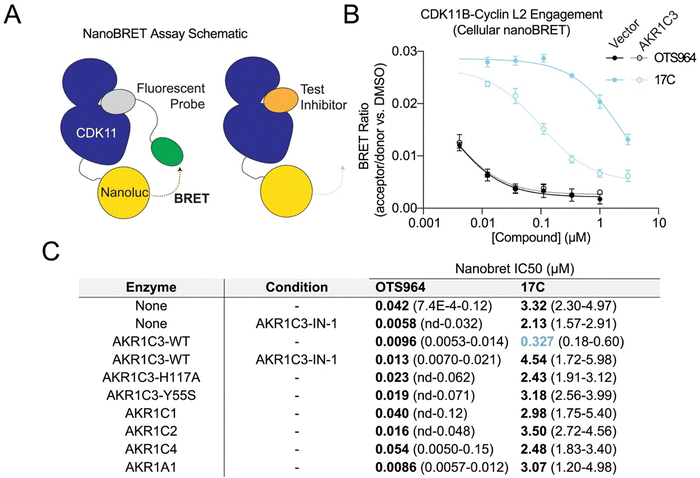
We next evaluated cellular permeation, prodrug activation, and target engagement by the new compounds. To do this, we measured cellular CDK11 binding using a modified NanoBRET assay, which monitors the displacement of a cell-permeable fluorescent probe from CDK11 (CDK11B/Cyclin L2).29 The assay was done in cells overexpressing AKR1C3 or in control cells, which we found to express undetectable levels of AKR1C3 by western blotting (Fig. 3A and Fig. S3A). The 17C prodrug showed CDK11 binding that depended strongly on AKR1C3 expression, consistent with the proposed prodrug activation mechanism (Fig. 3B). Control experiments confirmed that OTS964 is a potent competitive inhibitor of CDK11B/Cyclin L2, and its potency does not depend on AKR1C3 expression.We further investigated the requirement for AKR1C3-dependent prodrug conversion in two ways. First, we measured CDK11 engagement following the chemical inhibition of AKR1C3 using the selective inhibitor AKR1C3-IN-1 (Fig. 3C). Second, we evaluated CDK11 engagement in cells expressing attenuated AKR1C3 mutants (AKR1C3-Y55S and -H117A; Fig. S3B).13 In both experiments, AKR1C3 catalytic activity was required for CDK11 engagement. Thus, AKR1C3 catalyzes the conversion of the kinase inhibitor prodrug into its active drug form.
There are four paralogous enzymes that are closely related in sequence and function to AKR1C3. These are AKR1A1, AKR1C1, AKR1C2, and AKR1C4. Among these, only AKR1C3 is commonly upregulated in cancer.30 The related biochemical activities of the paralogs could, in principle, catalyze undesired prodrug activation in non-cancerous tissues. To assess this possibility, we used the cellular engagement assay to test the abilities of the AKR1C3 paralogs to activate 17C for CDK11 binding. Only AKR1C3 catalyzed prodrug activation; paralogous enzymes produced CDK11 engagement indistinguishable from control conditions (Fig. 3C and Fig. S3A). Thus, the OBI-3424 protecting group, when it is appended to OTS964via an ether linkage, serves as a substrate for AKR1C3 and is not amenable to nitroreduction by paralogous alpha-keto reductases. Collectively, these results demonstrate that 17C is a substrate for AKR1C3 and that AKR1C3-mediated activation releases a potent CDK11 inhibitor.
Cellular activity of 17C and dependence on AKR1C3
We next examined the cellular consequences of AKR1C3-dependent CDK11 prodrug activation. We modelled AKR1C3 dependence using H3122 cells (non-small cell lung cancer), which express high levels of AKR1C3 (Fig. S3A). Unconverted 17C is not a CDK11 inhibitor and is therefore expected to be minimally cytotoxic. Activation by AKR1C3 releases OTS964, inducing cell death. Indeed, OTS964 potently induces cell death in this system irrespective of AKR1C3 activity (IC50 46 nM with DMSO co-treatment versus 44 nM with AKR1C3-IN-1 co-treatment; Fig. 4A). 17C prevented cell proliferation (IC50 202 nM), and this cell killing effect was mitigated by co-treatment with AKR1C3-IN-1 (IC50 ∼2 µM). AKR1C3-IN-1 itself had no effect on cell viability in this cell line. We conclude that AKR1C3-dependent 17C reduction releases OTS964, and resultant CDK11 inhibition is cytotoxic.We next sought to relate this cellular efficacy to our previous observation of selective enzymatic activation by AKR1C3. We assessed the abilities of related alpha ketoreductase enzymes to confer added sensitivity to 17C. H3122 cells transduced to express AKR1A1, AKR1C1, AKR1C2, or AKR1C4 were no more sensitive to 17C than were vector-transduced control cells, whereas those transduced to express additional AKR1C3 were significantly sensitized to the compound compared with untransduced and vector-transduced controls (Fig. 4B). Thus, 17C activation is an exclusive activity of AKR1C3, and related alpha-ketoreductases do not contribute. This predicts that cancer cells overexpressing AKR1C3 will be preferentially susceptible to CDK11 inhibition when challenged with kinase inhibitor prodrugs like 17C.
To further evaluate the dependence of 17C activation and consequent cell cytotoxicity on AKR1C3 expression, we carried out cytotoxicity assays in multiple cancer cell lines. We tested 17C and the nitrogen mustard prodrug OB-3424 in 72-hour cell viability assays (Fig. 4C and D). We found that cells expressing high levels of AKR1C3 (pTPM > ∼15)31 showed 17C cytotoxicity that could be rescued by 2 µM AKR1C3-IN-1. The degree of rescue correlated with AKR1C3 expression level. By contrast, even low levels of AKR1C3 expression were sufficient to confer a differential response to OBI-3424 (±AKR1C3-IN-1), and correlation between AKR1C3 expression and OBI-3424-dependent cell cytotoxicity was poor. We discuss the implications of these observations below.
Cellular kinase inhibition by an AKR1C3-activated prodrug
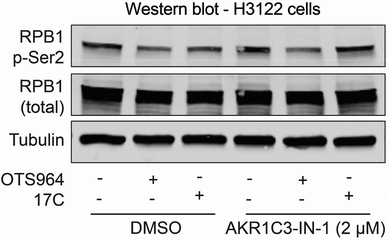
The foregoing data supports a prodrug activation mechanism in which AKR1C3 releases a potent CDK11 inhibitor. To evaluate this idea, we monitored phosphorylation of the RNA polymerase II C-terminal domain (CTD), which depends on CDK11 activity.24 H3122 cells were treated with OTS964 or 17C in the presence and absence of AKR1C3-IN-1 for four hours, and the resultant cell lysates were analyzed by western blotting. OTS964 treatment depleted CTD phosphorylation, irrespective of AKR1C3-IN-1 treatment. By contrast, 17C treatment resulted in diminished CTD phosphorylation, and this effect was rescued by AKR1C3-IN-1 (Fig. 5). Thus, 17C activation produces a cellular effect consistent with CDK11 inhibition, and this depends on AKR1C3 catalytic activity.We carried out initial in vitro pharmacokinetic experiments to determine whether 17C is stable enough to enable similar pharmacodynamic measurements in vivo. In mouse liver microsome stability assay, 17C had a half-life (T1/2) of 3.6 minutes and a Clint of 194 µL min−1 mg−1, demonstrating rapid clearance (Fig. S5A). To determine how the compound is degraded in this assay, we carried out metabolic tracking and identified demethylation of the tertiary amine substituent of OTS964 (Fig. S5B). It is at present unclear if this is a specific property of the prodrug for of the CDK11 inhibitor we have used.
Discussion
We have synthesized and tested a CDK11 inhibitor prodrug that is enzymatically activated by AKR1C3. We verified that CDK11 binding and consequent inhibition require the catalytic activity of AKR1C3. Conditional inhibitor activation via a predefined chemical moiety indicates that this strategy may be broadly applicable to kinase inhibitors.There are multiple reasons to suspect precise enzymatic activation of kinase inhibitors may be useful. First, many kinase inhibitors, including several developed by our laboratory,20,32–36 demonstrate clinically disqualifying on-target toxicity due to the broad essentiality of their targets in proliferating cells. This class includes inhibitors of CDK1, CDK9, PLK1/2/3, AURKA/B, and MPS1. Limiting kinase inhibition to AKR1C3-high cells may enhance the therapeutic windows for these agents. Second, we anticipate it will be possible, by studying the structural basis for AKR1C3-prodrug recognition, to synthesize so-called “bump-hole” ketoreductase-prodrug systems that provide tight enzymatic gating of cell cytotoxicity in experimental systems, providing a novel bio-orthogonal system for kinase inhibition.
The prodrug we describe targets a broadly essential kinase upon its activation. We found that cell cytotoxicity was far more reliant on AKR1C3 expression levels for 17C than it was for the nitrogen mustard prodrug OBI-3424, from which we borrowed the chemical protecting group. We hypothesize two reasons for this. First, OBI-3424 is a better AKR1C3 substrate (superior KM), resulting in appreciable prodrug conversion even in cells expressing very low levels of AKR1C3 (see Molt4 cells, Fig. 4D). Second, cancers of different types have highly variable sensitivities to DNA damage,37 while all cells likely have a similar dependence on CDK11. The latter is admittedly difficult to test in forward genetic experiments due to the functionally equivalent paralogs CDK11A and CDK11B. A prodrug strategy with a more stringent requirement for high AKR1C3 activity (i.e.one with a higher KM for AKR1C3) may provide an unexpected therapeutic benefit by restricting cytotoxicity to cells with exceedingly high AKR1C3 levels. Further, our initial cell line sensitivity data suggests a threshold effect rather than a linear relationship between enzyme expression and cytotoxicity.
There are significant limitations associated with the kinase prodrug approach we have described. The most important unresolved issue is currently the poor pharmacokinetic stability of 17C. Initial metabolite identification experiments suggests pathways to improve this property. It may also be true that AKR1C3-dependent prodrugs for other kinases are more stable. Beyond this first-order concern, a second drawback is likely toxicity due to collateral activation of the prodrug in AKR1C3-high non-cancerous tissues. Tissue versus cancer AKR1C3 expression measurements (Fig. S1)4 indicate the importance of eventual patient stratification on this metric, a point further emphasized by our analysis of AKR1C3 expression versus 17C cytotoxicity in vitro. Though it was generally observed to be well-tolerated, dose-dependent anemia was observed in the Phase I trial of OBI-3424,16 and it remains unclear whether a similar toxicity profile will plague AKR1C3-gated kinase inhibitor prodrugs. Eventually, it may be possible to co-administer AKR1C3-activated kinase inhibitors with tissue-selective AKR1C3 inhibitors to further limit prodrug activation to afflicted tissues.
Methods
Small molecule chemistry
See SI section for complete chemical synthesis methods. The following small molecules were purchased for this work: OBI-3424 (Seleckchem), AKR1C3-IN-1 (Seleckchem), OTS964 (MCE), and NADPH (Sigma Aldrich).Plasmid construction
AKR1A1, AKR1C1, AKR1C2, AKR1C3, and AKR1C4 coding sequences were purchased as codon-optimized gBlocks (IDT Technologies) and cloned by Gibson assembly cloning (NEB HiFi) into vector pN103 or pN144 (PuroR and HygroR, respectively; generous gifts from Sai Gourisankar and Jerry Crabtree, Stanford University) with N-terminal FLAG tags. The resulting plasmids are pNSG273, pNSG304-307. AKR1C3-Y55S and -H117A mutants were subcloned by overlapping PCR extension and ligation independent cloning. Vectors coding for CDK11A- and CDK11B-NanoLuc fusion proteins and for Cyclin L2 were purchased from Promega (NF4601, NF4611, NV2671). The AKR1C3 plasmid for protein production in Escherichia coli was made by PCR-amplification of the codon-optimized AKR1C3 gene from pNSG273 and insertion by ligation-independent cloning into an N-terminal His6-SUMO-TEV fusion vector (Addgene #29711), a generous gift from Dr Scott Gradia.Mammalian cell culture
Mammalian cells were grown at 37 °C with 5% ambient CO2 and high humidity in the following media: DMEM (HEK 293T, A431, A375, and DU-145), RPMI (H3122, Jurkat, MOLT-4, SU-DHL-8, and PC-3). All media were supplemented with 10% FBS and 1% penicillin/streptomycin. Mycoplasma testing was performed monthly using the MycoAlert mycoplasma detection kit (Lonza, Basel, Switzerland), and all lines were negative.Lentivirus production and transduction
Lentiviral supernatant was collected from HEK 293T cells transfected with pVSV-G, psPAX-2, and the indicated transfer plasmids. Virus supernatants were collected ∼48 hours after transfection and stored at −80 °C until use. H3122 cells were infected in 6-well plates by spinfection (one hour, 2000 rpm, 30 °C) in a final volume of 1 ml virus with 10 µg mL−1 polybrene. After infection, viral supernatant was replaced by growth medium. ∼48 hours after transduction, the cells were expanded into flasks and grown in medium supplemented with 1 µg mL−1 puromycin. After uninfected cells were eliminated as judged by a control culture, the resulting transductants were used for cell proliferation assays as described.AKR1C3 protein purification
SoluBL21 cells were transformed with pNSG273 and grown to A600 ∼1 before induction with 0.4 mM IPTG. The total culture volume at induction was 8 L. The temperature was shifted to 18 °C, and the culture was incubated overnight at 18 °C with continued shaking. Cells were pelleted and resuspended in buffer D800 (800 mM NaCl, 20 mM HEPES, pH 7.5, 10 mM imidazole, pH 8.0, 2 mM 2-mercaptoethanol, and 10% glycerol by volume) supplemented with 10 µg mL−1 each of aprotinin, leupeptin, and pepstatin, 1 mM PMSF, and 1 mM benzamidine. The resuspended cell pellets were stored at −80 °C until use. Upon thawing, cells were lysed by sonication and addition of lysozyme, and insoluble material was removed by centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130×g). AKR1C3 was purified by cobalt affinity and eluted in buffer C50 (D800 with 50 mM NaCl and 400 mM imidazole). The eluate was applied directly to an anion exchange cartridge (Cytiva HiTrap SP HP, 5 mL), and the bound protein was separated by gradient elution from B50 (D800 with 50 mM NaCl) to D800. Peak fractions were pooled and cleaved with TEV protease before further purification by nickel affinity chromatography. Unbound material from this step (cleaved AKR1C3) was concentrated and applied to a gel filtration column (Cytiva Superdex 200 Increase 10/300 GL) charged in buffer GF150 (150 mM NaCl, 20 mM Trizma-HCl, pH 8.5, 1 mM TCEP). The peak fractions from this step were concentrated to ∼43 mg mL−1, supplemented to ∼5% glycerol (final, by volume), aliquoted, and stored at −80 °C until use.
130×g). AKR1C3 was purified by cobalt affinity and eluted in buffer C50 (D800 with 50 mM NaCl and 400 mM imidazole). The eluate was applied directly to an anion exchange cartridge (Cytiva HiTrap SP HP, 5 mL), and the bound protein was separated by gradient elution from B50 (D800 with 50 mM NaCl) to D800. Peak fractions were pooled and cleaved with TEV protease before further purification by nickel affinity chromatography. Unbound material from this step (cleaved AKR1C3) was concentrated and applied to a gel filtration column (Cytiva Superdex 200 Increase 10/300 GL) charged in buffer GF150 (150 mM NaCl, 20 mM Trizma-HCl, pH 8.5, 1 mM TCEP). The peak fractions from this step were concentrated to ∼43 mg mL−1, supplemented to ∼5% glycerol (final, by volume), aliquoted, and stored at −80 °C until use.
For detection of reaction progression by column chromatography (Fig. S2C), His6-SUMO-AKR1C3 was purified as above. Uncleaved AKR1C3 was pooled following ion exchange chromatography, concentrated to 10.7 mg mL−1, and frozen in aliquots at −80 °C until use.
AKR1C3 biochemical activity assay
Biochemical assays were carried out according to previous reports. An assay mix containing 4.8 µM recombinant AKR1C3 and 100 µM NADPH in phosphate buffered saline (PBS) was prepared and distributed to a clear 96-well plate. The reactions were initiated by serially diluting the indicated compounds. NADPH absorbance (A340) was measured in kinetics mode on a PHERAStar FSX plate reader at room temperature. Each well was read every 42 or 52 seconds, depending on the experiment. The rate was confirmed to be linear for all measurements, and slopes were extracted from datapoints covering at least 15 minutes for each individual experiment. These slopes were then converted to velocities for each well using a standard dilution curve of NADPH measured equivalent assay conditions. Data shown represent technical triplicates and were repeated at least twice for each condition shown.For detection of reaction progression by column chromatography (Fig. S2C), biochemical reactions were set up with the following components in PBS: 2 µM His6-SUMO-AKR1C3, 1 mM NADH, and 100 µM 17C. Reactions were started by enzyme addition, and 10 µL reaction aliquots were injected every 14 minutes to track reaction progress. Reactants and products were separated by reverse phase chromatography (Shimadzu Nexcol C18 5 µM) with a 10-minute linear gradient (stationary phase – water, mobile phase – acetonitrile, both with 0.1% formic acid). Standard curves (OTS964 and 17C) were used to determine the linear detection range.
NanoBRET assay
HEK 293T cells were plated in 6-well plates and co-transfected on the following day using Lipofectamine 2000 according to the manufacturer's instructions. For each well, the transfection included 0.2 µg Cyclin L2 plasmid, 0.2 µg CDK11B-NanoLuc plasmid, and 2 µg FLAG-AKR1C3 (pN144) or control plasmid. 24 hours after transfection cells were detached and replated in white 384-well plates (Corning, #3570) at 5000 cells per well in 40 µL of fluorobrite DMEM with 10% FBS. NanoBRET and 50 nM K-12 tracer (Promega, #NF1011). AKR1C3-IN-1 or DMSO was then added (1.25 µM final concentration) for 1 hour. Cells were treated with DMSO or test compounds titrated from 10 µM to 50 nM in serial dilutions. After one hour, 20 µL of 3X NanoBRET NanoGlo substrate (Promega, #N1571) and extracellular NanoLuc inhibitor (Promega, #N2162) were added to each well. BRET signal was measured using a PHERAStar FSX plate reader, and BRET ratios were calculated according to the manufacturer's instructions. The results were normalized to DMSO-treated controls. Reported IC50 values were calculated using GraphPad Prism software.Cell proliferation assay
The indicated cells were plated in white 384-well plates at a starting density of ∼500 cells per well in 40 or 50 µL of the indicated growth media. Compounds were added to the final concentrations indicated in the manuscript using a D300 digital drug dispenser (Tecan). DMSO was added to maintain a final volume for all tested wells in each experiment. Triplicate or quadruplicate wells were treated for each concentration and compound. Reported growth IC50 and confidence interval values were determined in Graphpad Prism software (Prism v10, inhibitor versus effect, three-parameter model). For IC50 determination, the minimum viability was constrained (value > 0).Western blotting and antibodies used
Cell lysates were prepared in 2× sample loading buffer (250 mM Tris–HCl, pH 6.8; 4% sodium dodecyl sulfate, 10% glycerol, 0.006% bromophenol blue, 2% β-mercaptoethanol, 50 mM sodium fluoride, and 5 mM sodium orthovanadate). Samples were heated to 95 °C for 5 minutes. Protein concentrations were measured using a BCA Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein were subjected to 4–20% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to 0.45 µm nitrocellulose membranes (Bio-Rad). The membranes were blocked using Intercept® (TBS) Blocking Buffer (LI-COR Biosciences) and subsequently probed with appropriate primary antibodies: anti-phospho-Rpb1 CTD Ser2 (#04-1571 Sigma-Aldrich), anti-Rpb1 CTD (#2629 Cell Signaling Technology), anti-α-Tubulin (#3873 Cell Signaling Technology). After overnight incubation at 4 °C, membranes were washed and incubated with secondary antibodies: IRDye 800-labeled goat anti-rabbit IgG or or IRDye 680RD goat anti-Mouse IgG (#926-32211 and #926-68070, LI-COR Biosciences). After 1 hour at room temperature and PBS washes for 30 min, the membranes were scanned on a LI-COR Odyssey CLx system.Gene expression analysis by MERAV
AKR1C3 gene expression data for normal tissue and primary tumors was downloaded from the Metabolic Gene Rapid Visualizer (MERAV) database.4 We sorted the data according to tissue type and used multiple Kolmogorov-Smirnov tests to calculate P values in Graphpad Prism software (Prism v10).In vitro pharmacokinetics and metabolite identification
17C metabolites were evaluated in mouse liver microsomes. Microsomal incubations were done using 1 mg mL−1 microsomal protein in 0.1 M Tris–HCl buffer pH 7.4, with 1 µM 17C with and without 1 mM NADPH at 37 °C. Aliquots were taken at time points 0, 5, 10, 20, 40, and 60 minutes and the reaction was terminated by the addition of 3× volume of acetonitrile/methanol (ACN/MeOH, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v
1, v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v) containing internal standard (diclofenac). The samples were vortexed for 0.5 min before centrifugation at 15
v) containing internal standard (diclofenac). The samples were vortexed for 0.5 min before centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10 minutes at 4 °C. The supernatant was directly analyzed using a Thermo Scientific Q Exactive hybrid quadrupole-Orbitrap mass spectrometer in positive ion full scan ddMS2 mode. The obtained MS raw data files were submitted to Compound Discoverer 3.3 software for metabolite identification using targeted and non-targeted methods. The exact mass and fragmentation pattern were used to assign the location of individual modifications.
000×g for 10 minutes at 4 °C. The supernatant was directly analyzed using a Thermo Scientific Q Exactive hybrid quadrupole-Orbitrap mass spectrometer in positive ion full scan ddMS2 mode. The obtained MS raw data files were submitted to Compound Discoverer 3.3 software for metabolite identification using targeted and non-targeted methods. The exact mass and fragmentation pattern were used to assign the location of individual modifications.
Author contributions
Z. N. L. synthesized compounds and helped plan the experiments with S. M. H. and M. M. W. S. B., A. T., T. Q., Y. W., M. M, and S. M. H. performed biological and biochemical experiments. L. K. planned compound stability experiments. A. C., W. L., and W. J. contributed reagents and synthesized and tested early iterations of the compounds. J. C., T. H. Z., S. M. H., and N. S. G. planned and administered the project. S. M. H. wrote the manuscript with input from all the authors.Conflicts of interest
N. S. G. is a founder, science advisory board member (SAB), and equity holder in Syros, C4, Allorion, Lighthorse, Matchpoint, Shenandoah (board member), Larkspur (board member), and Soltego (board member). The Gray laboratory receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Jansen, Kinogen, Arbella, Deerfield, Springworks, Interline, and Sanofi. T. Z. is a scientific founder, equity holder, and consultant of Matchpoint, equity holder of Shenandoah.Data availability
The data supporting this article are included in the main text (cell biology and biochemistry) or are included in the supplemental file describing chemical synthesis. Publicly available data was used to determine AKR1C3 expression in cancerous and untransformed tissues. This is described in the Methods section and available for free public download at MERAV (https://merav.wi.mit.edu/). The data used to generate AKR1C3 expression levels shown in Fig. 4C-D is available online at the human protein atlas website: https://www.proteinatlas.org/humanproteome/tissue.Supplementary information (SI) is available. See DOI: https://doi.org/10.1039/d5cb00219b.
Acknowledgements
We thank Dr Michael Cameron and team at the Herbert Wertheim UF Scripps Institute for Biomedical Innovation & Technology for performing in vitro pharmacokinetics and metabolite identification. We thank Esther Elonga and Erhan Kelles for early contributions to this project. We thank all members of the Gray laboratory for helpful discussions. This work was supported by R01 CA218278 from the NIH and departmental funds from Stanford Chemical and Systems Biology and Stanford Cancer Institute to N.S.G.References
- M. Lu, X. Zhang, J. Zhao, Q. You and Z. Jiang, A hydrogen peroxide responsive prodrug of Keap1-Nrf2 inhibitor for improving oral absorption and selective activation in inflammatory conditions, Redox Biol., 2020, 34, 101565, DOI:10.1016/j.redox.2020.101565.
- E. Saxon and X. Peng, Recent Advances in Hydrogen Peroxide Responsive Organoborons for Biological and Biomedical Applications, ChemBioChem, 2022, 23(3), e202100366, DOI:10.1002/cbic.202100366.
- L. D. Lavis, Ester bonds in prodrugs, ACS Chem. Biol., 2008, 3(4), 203–206, DOI:10.1021/cb800065s.
- Y. D. Shaul, B. Yuan, P. Thiru, A. Nutter-Upham, S. McCallum, C. Lanzkron, G. W. Bell and D. M. Sabatini, MERAV: a tool for comparing gene expression across human tissues and cell types, Nucleic Acids Res., 2016, 44(D1), D560–566, DOI:10.1093/nar/gkv1337.
- A. O. Adeniji, M. Chen and T. M. Penning, AKR1C3 as a Target in Castrate Resistant Prostate Cancer, J. Steroid Biochem. Mol. Biol., 2013, 137, 136–149, DOI:10.1016/j.jsbmb.2013.05.012.
- A. Mantel, A. Carpenter-Mendini, J. VanBuskirk and A. P. Pentland, Aldo-keto reductase 1C3 is overexpressed in skin squamous cell carcinoma (SCC) and affects SCC growth via prostaglandin metabolism, Exp. Dermatol., 2014, 23(8), 573–578, DOI:10.1111/exd.12468.
- V. L. Miller, H. K. Lin, P. Murugan, M. Fan, T. M. Penning, L. S. Brame, Q. Yang and K. M. Fung, Aldo-keto reductase family 1 member C3 (AKR1C3) is expressed in adenocarcinoma and squamous cell carcinoma but not small cell carcinoma, Int. J. Clin. Exp. Pathol., 2012, 5(4), 278–289 CAS.
- D. Reddi, B. W. Seaton, D. Woolston, L. Aicher, L. D. Monroe, Z. J. Mao, J. C. Harrell, J. P. Radich, A. Advani and N. Papadantonakis, et al., AKR1C3 expression in T acute lymphoblastic leukemia/lymphoma for clinical use as a biomarker, Sci. Rep., 2022, 12(1), 5809, DOI:10.1038/s41598-022-09697-6.
- Y. Shao, X. Yu, K. Shan, J. Yan and G. Ye, Defining the biological functions and clinical significance of AKR1C3 in gastric carcinogenesis through multiomics functional analysis and immune infiltration analysis, J. Cancer, 2024, 15(9), 2646–2658, DOI:10.7150/jca.94228.
- P. Zhu, R. Feng, X. Lu, Y. Liao, Z. Du, W. Zhai and K. Chen, Diagnostic and prognostic values of AKR1C3 and AKR1D1 in hepatocellular carcinoma, Aging, 2021, 13(3), 4138–4156, DOI:10.18632/aging.202380.
- T. M. Penning, M. E. Burczynski, J. M. Jez, C. F. Hung, H. K. Lin, H. Ma, M. Moore, N. Palackal and K. Ratnam, Human 3alpha-hydroxysteroid dehydrogenase isoforms (AKR1C1-AKR1C4) of the aldo-keto reductase superfamily: functional plasticity and tissue distribution reveals roles in the inactivation and formation of male and female sex hormones, Biochem. J., 2000, 351(Pt 1), 67–77, DOI:10.1042/0264-6021:3510067.
- T. M. Penning, The aldo-keto reductases (AKRs): Overview, Chem. – Biol. Interact., 2015, 234, 236–246, DOI:10.1016/j.cbi.2014.09.024.
- B. P. Schlegel, K. Ratnam and T. M. Penning, Retention of NADPH-linked quinone reductase activity in an aldo-keto reductase following mutation of the catalytic tyrosine, Biochemistry, 1998, 37(31), 11003–11011, DOI:10.1021/bi980475r.
- K. Evans, J. Duan, T. Pritchard, C. D. Jones, L. McDermott, Z. Gu, C. E. Toscan, N. El-Zein, C. Mayoh and S. W. Erickson, et al., OBI-3424, a Novel AKR1C3-Activated Prodrug, Exhibits Potent Efficacy against Preclinical Models of T-ALL, Clin. Cancer Res., 2019, 25(14), 4493–4503, DOI:10.1158/1078-0432.CCR-19-0551.
- C. P. Guise, M. R. Abbattista, R. S. Singleton, S. D. Holford, J. Connolly, G. U. Dachs, S. B. Fox, R. Pollock, J. Harvey and P. Guilford, et al., The bioreductive prodrug PR-104A is activated under aerobic conditions by human aldo-keto reductase 1C3, Cancer Res., 2010, 70(4), 1573–1584, DOI:10.1158/0008-5472.CAN-09-3237.
- A. M. Tsimberidou, C. F. Verschraegen, R. Wesolowski, C. S. Shia, P. Hsu and T. E. Pearce, Phase 1 dose-escalation study evaluating the safety, pharmacokinetics, and clinical activity of OBI-3424 in patients with advanced or metastatic solid tumors, Br. J. Cancer, 2023, 129(2), 266–274, DOI:10.1038/s41416-023-02280-4.
- F. Meng, T. Qi, X. Liu, Y. Wang, J. Yu, Z. Lu, X. Cai, A. Li, D. Jung and J. Duan, Enhanced pharmacological activities of AKR1C3-activated prodrug AST-3424 in cancer cells with defective DNA repair, Int. J. Cancer, 2025, 156(2), 417–430, DOI:10.1002/ijc.35170.
- C. E. Toscan, H. McCalmont, A. Ashoorzadeh, X. Lin, Z. Fu, L. Doculara, H. J. Kosasih, R. Cadiz, A. Zhou and S. Williams, et al., The third generation AKR1C3-activated prodrug, ACHM-025, eradicates disease in preclinical models of aggressive T-cell acute lymphoblastic leukemia, Blood Cancer J., 2024, 14(1), 192, DOI:10.1038/s41408-024-01180-x.
- R. Novotna, V. Wsol, G. Xiong and E. Maser, Inactivation of the anticancer drugs doxorubicin and oracin by aldo-keto reductase (AKR) 1C3, Toxicol. Lett., 2008, 181(1), 1–6, DOI:10.1016/j.toxlet.2008.06.858.
- F. M. Ferguson and N. S. Gray, Kinase inhibitors: the road ahead, Nat. Rev. Drug Discovery, 2018, 17(5), 353–377, DOI:10.1038/nrd.2018.21.
- Z. Li, R. Ishida, Y. Liu, J. Wang, Y. Li, Y. Gao, J. Jiang, J. Che, J. M. Sheltzer and M. B. Robers, et al., Synthesis and Structure-Activity relationships of cyclin-dependent kinase 11 inhibitors based on a diaminothiazole scaffold, Eur. J. Med. Chem., 2022, 238, 114433, DOI:10.1016/j.ejmech.2022.114433.
- A. Lin, C. J. Giuliano, A. Palladino, K. M. John, C. Abramowicz, M. L. Yuan, E. L. Sausville, D. A. Lukow, L. Liu and A. R. Chait, et al., Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials, Sci. Transl. Med., 2019, 11(509) DOI:10.1126/scitranslmed.aaw8412.
- M. Hluchy, P. Gajduskova, I. Ruiz de Los Mozos, M. Rajecky, M. Kluge, B. T. Berger, Z. Slaba, D. Potesil, E. Weiss and J. Ule, et al., CDK11 regulates pre-mRNA splicing by phosphorylation of SF3B1, Nature, 2022, 609(7928), 829–834, DOI:10.1038/s41586-022-05204-z.
- P. Loyer and J. H. Trembley, Roles of CDK/Cyclin complexes in transcription and pre-mRNA splicing: Cyclins L and CDK11 at the cross-roads of cell cycle and regulation of gene expression, Semin. Cell Dev. Biol., 2020, 107, 36–45, DOI:10.1016/j.semcdb.2020.04.016.
- X. Liu, Y. Gao, J. Shen, W. Yang, E. Choy, H. Mankin, F. J. Hornicek and Z. Duan, Cyclin-Dependent Kinase 11 (CDK11) Is Required for Ovarian Cancer Cell Growth In Vitro and In Vivo, and Its Inhibition Causes Apoptosis and Sensitizes Cells to Paclitaxel, Mol. Cancer Ther., 2016, 15(7), 1691–1701, DOI:10.1158/1535-7163.MCT-16-0032.
- Y. Zhou, J. K. Shen, F. J. Hornicek, Q. Kan and Z. Duan, The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer, Oncotarget, 2016, 7(26), 40846–40859, DOI:10.18632/oncotarget.8519.
- L. Julian, L. Crozier, D. Lukow, S. Mishra, A. Swamy, R. A. Hagenson, P. Sennhenn, E. L. Sausville, B. Mendelson and C. Chuaqui, et al., On-target toxicity limits the efficacy of CDK11 inhibition against cancers with 1p36 deletions, bioRxiv, 2025 DOI:10.1101/2025.08.03.668359.
- S. M. Jamieson, D. G. Brooke, D. Heinrich, G. J. Atwell, S. Silva, E. J. Hamilton, A. P. Turnbull, L. J. Rigoreau, E. Trivier and C. Soudy, et al., 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic Acids: highly potent and selective inhibitors of the type 5 17-beta-hydroxysteroid dehydrogenase AKR1C3, J. Med. Chem., 2012, 55(17), 7746–7758, DOI:10.1021/jm3007867.
- T. Machleidt, C. C. Woodroofe, M. K. Schwinn, J. Mendez, M. B. Robers, K. Zimmerman, P. Otto, D. L. Daniels, T. A. Kirkland and K. V. Wood, NanoBRET-A Novel BRET Platform for the Analysis of Protein-Protein Interactions, ACS Chem. Biol., 2015, 10(8), 1797–1804, DOI:10.1021/acschembio.5b00143.
- M. Li, L. Zhang, J. Yu, X. Wang, L. Cheng, Z. Ma, X. Chen, L. Wang and B. C. Goh, AKR1C3 in carcinomas: from multifaceted roles to therapeutic strategies, Front. Pharmacol., 2024, 15, 1378292, DOI:10.3389/fphar.2024.1378292.
- M. Uhlen, L. Fagerberg, B. M. Hallstrom, C. Lindskog, P. Oksvold, A. Mardinoglu, A. Sivertsson, C. Kampf, E. Sjostedt and A. Asplund, et al., Proteomics. Tissue-based map of the human proteome, Science, 2015, 347(6220), 1260419, DOI:10.1126/science.1260419.
- F. M. Ferguson, Z. M. Doctor, A. Chaikuad, T. Sim, N. D. Kim, S. Knapp and N. S. Gray, Characterization of a highly selective inhibitor of the Aurora kinases, Bioorg. Med. Chem. Lett., 2017, 27(18), 4405–4408, DOI:10.1016/j.bmcl.2017.08.016.
- N. S. Gray, L. Wodicka, A. M. Thunnissen, T. C. Norman, S. Kwon, F. H. Espinoza, D. O. Morgan, G. Barnes, S. LeClerc and L. Meijer, et al., Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors, Science, 1998, 281(5376), 533–538, DOI:10.1126/science.281.5376.533.
- N. Kwiatkowski, N. Jelluma, P. Filippakopoulos, M. Soundararajan, M. S. Manak, M. Kwon, H. G. Choi, T. Sim, Q. L. Deveraux and S. Rottmann, et al., Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function, Nat. Chem. Biol., 2010, 6(5), 359–368, DOI:10.1038/nchembio.345.
- C. V. Miduturu, X. Deng, N. Kwiatkowski, W. Yang, L. Brault, P. Filippakopoulos, E. Chung, Q. Yang, J. Schwaller and S. Knapp, et al., High-throughput kinase profiling: a more efficient approach toward the discovery of new kinase inhibitors, Chem. Biol., 2011, 18(7), 868–879, DOI:10.1016/j.chembiol.2011.05.010.
- C. M. Olson, B. Jiang, M. A. Erb, Y. Liang, Z. M. Doctor, Z. Zhang, T. Zhang, N. Kwiatkowski, M. Boukhali and J. L. Green, et al., Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation, Nat. Chem. Biol., 2018, 14(2), 163–170, DOI:10.1038/nchembio.2538.
- B. D. Yard, D. J. Adams, E. K. Chie, P. Tamayo, J. S. Battaglia, P. Gopal, K. Rogacki, B. E. Pearson, J. Phillips and D. P. Raymond, et al., A genetic basis for the variation in the vulnerability of cancer to DNA damage, Nat. Commun., 2016, 7, 11428, DOI:10.1038/ncomms11428.
- S. Kelso, S. O'Brien, I. Kurinov, S. Angers and F. Sicheri, Crystal structure of the CDK11 kinase domain bound to the small-molecule inhibitor OTS964, Structure, 2022, 30(12), 1615–1625, DOI:10.1016/j.str.2022.10.003.
Footnotes |
| † Current address: Septerna, Inc. South San Francisco, CA, USA. |
| ‡ These authors contributed equally |
| § Current address: Department of Plant and Microbial Biology, UC Berkeley, Berkeley, CA, USA. |
| ¶ Current address: Lingang Laboratory, Shanghai, China. |
| This journal is © The Royal Society of Chemistry 2026 |