Open Access Article
Open Access ArticleDual spatio-functional control of a fission yeast-based bioprocessor upon chemical induction
Stavroula Melina
Sakellakou
a,
Valérie
Migeot
e,
Laure-Elie
Carloni
b,
Elisa
Martino
 c,
André Oliveira
Sequeira
a,
Terézia
Morávková
c,
André Oliveira
Sequeira
a,
Terézia
Morávková
 a,
Lorenzo
Riccio
a,
Lorenzo
Riccio
 a,
Sorin
Melinte
a,
Sorin
Melinte
 d,
Laura
Maggini
d,
Laura
Maggini
 *a,
Damien
Hermand
*ef and
Davide
Bonifazi
*a,
Damien
Hermand
*ef and
Davide
Bonifazi
 *a
*a
aInstitute of Organic Chemistry, Faculty of Chemistry, University of Vienna, Währinger Straße 38, 1090, Vienna, Austria. E-mail: Laura.Maggini@univie.ac.at; Davide.Bonifazi@univie.ac.at
bDepartment of Chemistry, The University of Namur, rue de Bruxelles, 61, Namur 5000, Belgium
cSchool of Chemistry, Cardiff University, Park Place, CF10 3AT, Cardiff, UK
dInstitute of Information and Communication Technologies, Electronics and Applied Mathematics, Université catholique de Louvain, 1348 Louvain-la-Neuve, Belgium
eURPHYM-GEMO, The University of Namur, rue de Bruxelles, 61, Namur 5000, Belgium. E-mail: Damien.Hermand@crick.ac.uk
fThe Francis Crick Institute, 1 Midland Road, London, NW11AT, UK
First published on 17th November 2025
Abstract
Next-generation therapies are advancing beyond small molecules and proteins toward engineered living microorganisms that interact symbiotically with their host and respond to signals precisely when and where needed. Despite progress in the field, engineering cells to both produce biopharmaceuticals and achieve site-specific recruitment remains a challenge. In this work, we genetically engineered the mating pathway of S. pombe to create a “bioprocessor” that responds to a chemical trigger, an artificial replica of the sexual pheromone of the yeast cells, the P-factor, enabling functional control over the production of Albulin as a proof-of-concept biopharmaceutical. This activation simultaneously induces the expression of hydrophobic agglutinins on the cell surface, modifying surface chemistry and adhesion properties. Exploiting this modification, we could simultaneously implement spatial control, allowing selective adhesion to a hydrophobic target surface. Adhesion control tests confirmed the fundamental role of hydrophobic interactions in this adhesion process, enabling selective cell adherence only after activation with P-factor and expression of the agglutinins, even in presence of potentially interfering cells. This approach represents an important milestone in the development of a straightforward chemically-activated multi-control mechanisms, which enable precise and programmable responses in engineered cells. Such advancements pave the way for a new generation of bio-responsive materials and therapeutic devices, including functional implants and targeted delivery systems, where engineered cells can operate in synergy with host tissues, responding to specific environmental cues to produce therapeutic agents exactly when and where they are needed.
Introduction
Next-generation therapies are advancing beyond small molecule and protein approaches towards utilizing whole cells.1 Much of the ongoing enthusiasm for cell-based therapies stems from the redirection of cells’ inherent functions to enable therapeutic performance and efficacy profiles that exceed those of small molecule and protein drugs. In particular, cell-based therapeutic agents have the ability to adapt to different environments without losing their therapeutic potency2 and can harness recognition capabilities with biological macromolecules increasing the specificity of their actions.3 Today only a handful of cell-based therapies have achieved commercial use, with the most successful treatments being Chimeric Antigen Receptor (CAR) T cell-based therapies.4 These involve genetically modifying a patient's T cells (e.g., immune system cells) in the laboratory to express protein receptors. These receptors enable the recognition of malignant cells, facilitating their destruction once the modified cells are re-injected into the patient. Thanks to these technologies, a broad spectrum of diseases can be targeted such as leukemia,5 lymphoma,6 and Crohn's disease.7 However, despite the great advancements in this field, our ability to engineer cells is generally limited to introducing proteins onto the cell surface, while modulating large-scale traits and/or structures of cellular ‘machines’ able to respond to pathogens when needed in vivo still remains elusive.To achieve a responsive cellular therapeutic system, it is crucial to simultaneously gain remote control over both cellular positioning and function (e.g., in situ production of biopharmaceutical), mimicking the in vivo behaviour of cells responding to a signal. One primary consideration is the cell's ability to navigate and adhere to the site where therapeutic action is required. In vivo, cells are embedded within a complex and dynamic microenvironment consisting of the surrounding extracellular matrix (ECM), growth factors, and cytokines, as well as neighbouring cells. Cell adhesion to the ECM scaffolding occurs when required (e.g., upon a chemical stimulus) at specific sites involving physical connection to the microenvironment through receptors present on the cell surface. These adhesive processes in turn trigger of a cascade of intracellular signalling events that can lead to the activation of a cellular function, such as growth, migration, differentiation, and activation of defence mechanisms.8
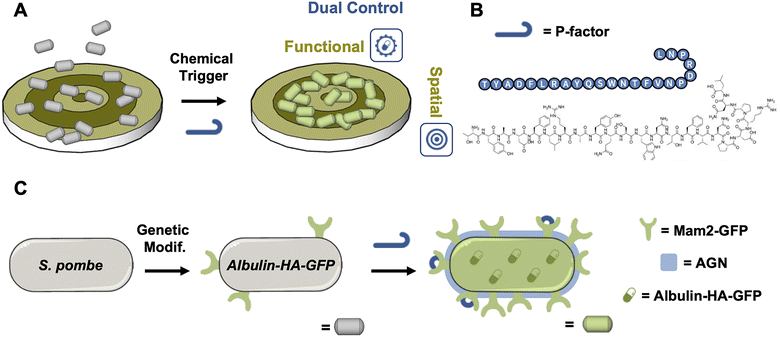
To this goal, one could envision to highjack and rewire a natural regulatory mechanism of a microorganism (e.g., the mating process), exploiting the available genetic modifications toolset, to respond to a chemical stimulus by concomitantly activating adhesion onto a targeted surface and a function of interest (i.e., the production of a biopharmaceutical). This system could be triggered by a specific artificial biomimetic molecular stimulus able to interact exclusively with the therapeutic cells, activating their adhesion and function (e.g., production of a biopharmaceutical) in the adhered two-dimensional (2D) supracellular assembly, advancing the state of the art towards the development of life-like “interactive functional cellular assemblies” (Fig. 1A).
A variety of organisms could be employed in such an application, ranging from prokaryotes9 to eukaryotes, such as yeast10 and mammalian10 cell lines. Therapeutic protein expression is more advantageous in simple eukaryotic hosts since their overall structure is much closer to human cells. In particular, yeast is a very attractive option as it combines the highest genetic engineering capacity, fast growth and affordable manipulations.11 Genetically-engineered yeast cells are industrially employed for the cost-effective production of biotherapeutics12–14 and have recently been used living or dead, intact, permeabilized, or even emptied of all their original cytoplasmic contents, as cell-based microcapsules for drug delivery demonstrating excellent biocompatibility15,16in vivo and no cytotoxicity.
Haploid yeast cells are capable of sexual reproduction implying the recognition of a sexual partner followed by mating to form a diploid cell.17–20 This pathway relies on a pheromone-mediated signal transduction and constitutes an important model system for cell–cell interactions. The fission yeast Schizosaccharomyces pombe (S. pombe) is a well-established, genetically tractable model organism. The recognition between cells of opposite mating type (sex) relies on a simple binary system where h+ cells (called P-cells) produce the P-factor pheromone (Fig. 1B) and express the membrane-associated receptor Map3 that binds the opposite pheromone.21 Conversely, the h− cells (called M-cells) produce the M-factor pheromone and express the Mam2 receptor that binds P-factor.22,23 The M-factor is a nonapeptide in which the C-terminal cysteine is carboxy-methylated and S-alkylated while the P-factor is a 23 amino-acids peptide. Both are encoded in the genome as a repeated polypeptide precursor that needs to be cleaved to generate the functional factors. When nutrients, particularly a nitrogen source, are available the mating system is turned off and fission yeast grow vegetatively.24 Upon starvation, integrated signal transduction pathways activate the expression of the pheromones and their corresponding receptors, leading to mating and sexual development referred to as gametogenesis.17 Large-scale analyses revealed that a set of approximately 100 genes is induced, including the mam3 gene that encodes a cell surface adhesion protein (agglutinin, AGN).25 The simplicity of this communication system offers the possibility to manipulate and control cell interactions exploiting the hydrophobic AGNs, cell wall proteins presenting hydrophobic/aromatic surfaces across all three of their domains.26–28 Furthermore, with regard to biopharmaceuticals production, the baker yeast Sacharomyces cerevisiae has been used to produce dozens of recombinant proteins, including insulin and the α-factor pheromone leader sequence was also used to secrete some of them extracellularly.29 The novelty of our work stands in the fact that we did not use individual components of a pre-existing system but rather completely rewired the pheromone-induced pathway regulating gene expression to reach two goals: set up the fully programmable expression of albulin (functional control) and drive precise spatial arrangement (spatial control) of cells, both of these complex processes being released by the sole addition of a peptide. To the best of our knowledge, this is the first instance of coupling these unrelated phenomena in yeast.
It can thus be postulated that the mating system of S. pombe cells could be genetically tailored to respond to a single chemical trigger enabling both the on-demand production of a biopharmaceutical while also enabling the expression of agglutinins (AGNs) for specific adhesion allowing for a double “functional/spatial” control to take place. This modification would streamline the necessary adjustments to achieve dual control over both the functional activity and spatial positioning of the cells, effectively leveraging the inherent biological machinery and surface properties of S. pombe, without requiring expression or integration of orthogonal ligands either on the cell or the substrate.
In this study, we demonstrate the successful achievement of parallel dual control over both the functional activity and spatial positioning of genetically modified S. pombe cells by engineering their mating pathway (Table 1) to respond to synthetic P-factor. This response triggers the production of the insulin derivative Albulin and simultaneously the expression of hydrophobic AGNs (Mam2-HA-GFP, Table 1 and Fig. 1A and C). For functional control, production of Albulin (HA, a tagged hemagglutinin version, Albulin-HA-GFP), a long-acting insulin analogue that fuses single-chain human insulin with human serum albumin (HSA),30 was selected due to its therapeutic potential for treating insulin-dependent diabetes. To achieve spatial control, we fabricated micrometre-wide striped Au/SiO2 surfaces, functionalizing the Au stripes with aliphatic self-assembled monolayers to enhance hydrophobicity. This setup enabled selective adhesion of genetically modified S. pombe cells exclusively after activation by P-factor and AGN expression. Adhesion specificity was verified using a strain of S. pombe lacking AGNs (noAGN, Table 1), which demonstrated no adhesion in response to P-factor activation, confirming AGNs' essential role in surface attachment. Moreover, selective adhesion persisted in mixed populations containing both adhering and non-adhering cells, further underscoring the system's robustness. This approach establishes a dual-control system that leverages the cells' intrinsic biological machinery in concert with tailored chemical triggers and substrates, creating a streamlined multi-level control strategy. By avoiding unnecessary complexity, this method provides the seminal milestone of an efficient S. pombe model for advancing biotechnological applications, in the framework of living pharmacies, where precise spatial and on-demand activation of cells is required for localized and controlled therapeutic production.
Results and discussion
Construction and optimization of fission yeast strains responding to synthetic P-factor
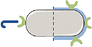
The pheromone/mating yeast system is activated upon starvation to allow the temporary interaction of cells from opposite sexes, called mating types in yeast.31 To use synthetic P-factor as a chemical inducer of a dual spatio-functional control of fission yeast, we first generated a strain where the sxa2 and cyr1 genes were deleted (sxa2Δ cyr1Δ). The Sxa2 protein is a serine carboxypeptidase that degrades extracellular P-factor in fission yeast.32 To avoid desensitization resulting from the degradation of the synthetic P-factor inducer, the sxa2 gene was replaced by the kanR selection marker. The cyr1 gene encodes the adenylate cyclase generating cyclic adenosine monophosphate (cAMP) in the presence of high glucose concentration. The replacement of cyr1 by the hphR selection marker mimics glucose starvation, which allows sexual differentiation even in rich medium, therefore uncoupling mating from the requirement of starvation,33 and greatly facilitating further developments.We next tested if the exposure of M-cells to synthetic P-factor (SI, Fig. S1) would result in the induction of the expression of the Mam2 receptor. We used genome engineering to construct a strain expressing Mam2-Green Fluorescent Protein (Mam2-GFP, Table 1) from the endogenous locus and a control strain where the mam2 gene was deleted (mam2Δ) and replaced by the natR selection marker. Upon addition of synthetic P-factor, the production of the Mam2-GFP mRNA as determined by qRT-PCR was strongly induced within 2 hours and the Mam2-GFP protein accumulated concomitantly (Fig. 2A and B). These data show that the sxa2Δ cyr1Δ cells respond to P-factor independently of starvation by expressing the GFP-tagged P-factor receptor Mam2.
Expression of albulin in response to chemical induction
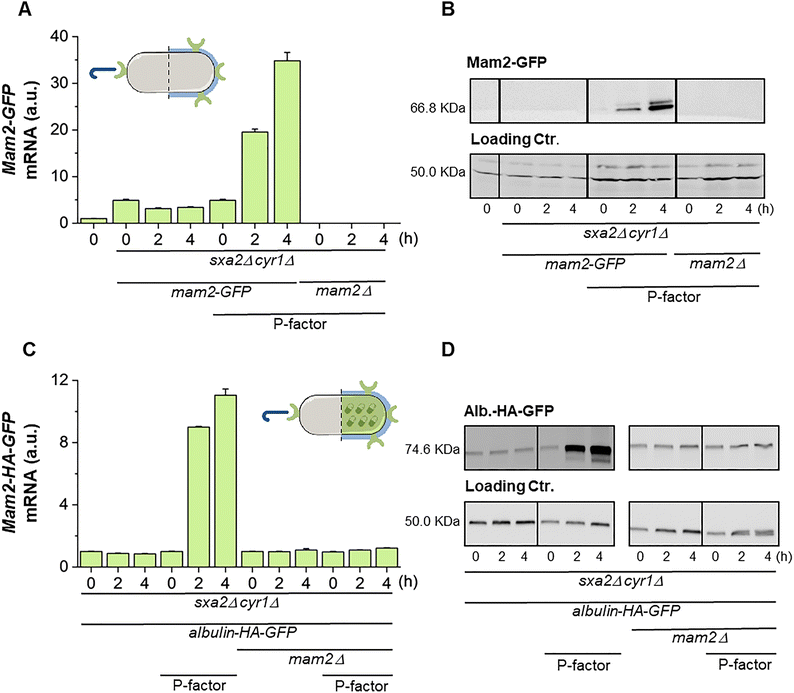
We next designed a plasmid-borne expression system to express HA (Albulin-HA-GFP) in response to the exposure to P-factor. Based on previous works, we choose the SPBC4.01 promoter that is strongly induced by pheromone signalling and regulates the expression of the tetraspan protein claudin Dni2.26,34 The coding sequence of Albulin consists of the B- and A-chains of human insulin (100% identity to human insulin) linked together by a dodecapeptide linker and fused to the NH2-terminus of native HSA.30 The exposure to P-factor of the sxa2Δ cyr1Δ strain transformed with the expression plasmid (Mam2-HA-GFP) resulted in a strong induction of the expression of the mRNA and concomitant accumulation of Albulin-HA-GFP protein (Fig. 2C and D). Importantly, while the induction of Mam2-GFP and Albulin-HA-GFP were concomitant, Albulin-HA-GFP was fully dependent on the presence of the Mam2 receptor, which confirms that it is the binding of the P-factor to its receptor that induces a signal transduction leading to expression of Albulin (Fig. 2C and D).To visualize the expression of both the GFP-tagged Albulin and Mam2 proteins (Fig. 3A) over a longer time frame and to determine the optimal P-factor incubation time for their expression, we monitored the GFP emission levels qualitatively using fluorescence microscopy and quantitatively employing a plate reader. Scoped incubation times varied from 0 to 8 hours with 2 hours intervals (see Materials and methods). Considering as a proxy that GFP expression is linearly related to the P-factor interaction with the receptor, monitoring the GFP levels allows evaluating the efficiency of the synthetic peptide in triggering the fluorescent protein production and verifying its potency. The images reported in Fig. 3A and plate reader measurements plotted in Fig. 3B revealed that the induction occurred within 2 hours, confirming the western blot results (Fig. 2), and was maintained for at least 8 hours even if a slight decrease was observed at that time. Cell viability was determined simultaneously and shown to be constantly above 90% (Fig. 3C). Taken together, these data show that we have created a fission yeast bioprocessor that quickly and specifically responds to the chemical induction of P-factor independently on nitrogen availability by expressing Albulin and high levels of Mam2 receptor.
Activation of surface-specific adhesion by chemical induction
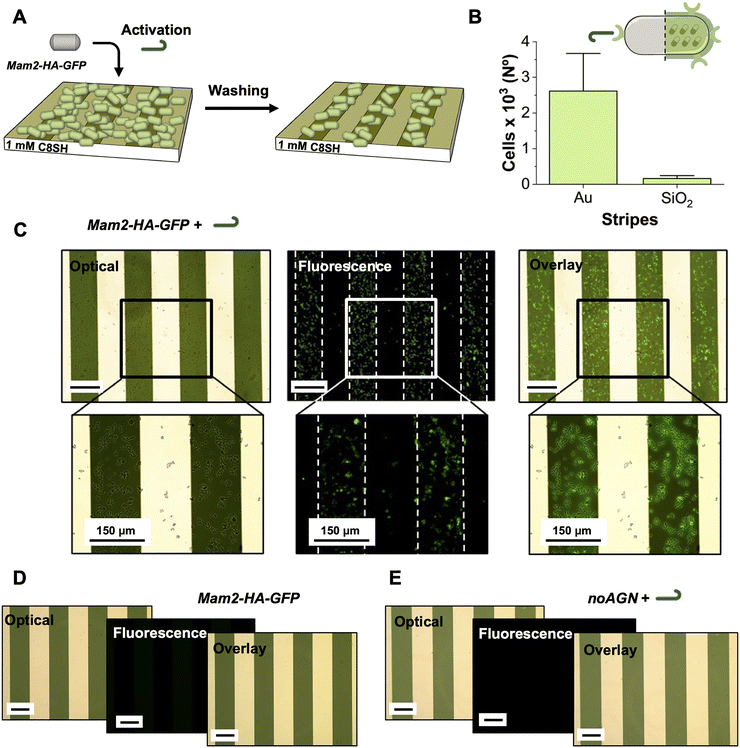
The previous experiments were performed in liquid cultures, where cells randomly interact without following any specific pattern or assembly. We next anticipated the possibility to link the production of Albulin to the formation of surface-specific, self-assembled monolayers (SAMs) of the HA-producing cells. Mating cells are known to express the Mam3 agglutinin (AGN) to increase their capacity to stably interact. Thus, to test the hydrophobicity-driven spatial control of Mam2-HA-GFP, we aimed to exploit the AGNs expressed in response to P-factor activation to drive these cells onto the metallic (Au) portions of striped Au/SiO2 surfaces, selected as an exemplary target location (Scanning Electron Microscopy images of the surfaces SI, Fig. S4).The Au stripes offer a platform, which can easily and orthogonally to SiO2, be functionalized with non-toxic and commercially available hydrophobic alkylthiol, 1-octanethiol (C8-SH),35,36 enhancing the hydrophobic character of these segments facilitating the selective adhesion of the S. pombe expressing AGNs. On the other hand, the SiO2 stripes act as an in situ hydrophilic negative control surface, highlighting the difference in cellular adhesion between functionalized (with alkyl thiols) and non-functionalized area. Hydrophilic SAMs were also prepared for reference, with triethylene glycol mono-11-mercaptoundecyl ether (P-SH), to suppress completely the adhesion of the cells further stressing the importance of the hydrophobic effect in the adhesion process.
Gold stripes 150 µm in width (e.g., enough to accommodate comfortably S. pombe cells approximately 10–14 µm long)37 were patterned onto 1 × 1 cm2 glass surfaces, through standard lithography and lift-off processes. After cleansing to remove the protective polymer and impurities (see Material and methods), the patterned substrates were immersed in EtOH solutions of the given thiol and allowed to react. Different increasing thiol concentrations were tested (10 µM, 100 µM and 1 mM) to tune the polarity of the surface and consequently optimize cellular adhesion. Water contact angle (WCA) measurements for the hydrophobic surfaces, resulted 57.2°, 74.9° and 85.7°, for 10 µM, 100 µM and 1 mM, respectively (SI, Fig. S5).
We next investigated the ability of AGN-expressing Mam2-HA-GFPS. pombe cells to preferentially adhere onto hydrophobic surfaces after 4 h incubation with P-factor, a time corresponding to high induction of Mam2 and Albulin (Fig. 4A). After overnight growing, and a sequence of centrifugation/washing cycles to remove residues of P-factor (see Materials and methods), the cells were suspended in water (5 mL; 0.2 OD595nm corresponding to 2.2 × 106 cells per mL). 2 mL of this suspension was hence placed onto a functionalized surface positioned inside a 24-well plate and further incubated for 3 hours. The surface was visualized by fluorescence microscopy and repeatedly washed to remove unadsorbed cells until the number of the adhered ones remained stable. As shown in Fig. 4B, C and SI, Fig. S6, the cells displayed an increased adhesion preference onto the functionalized hydrophobic Au stripes of the surfaces upon increase of their hydrophobicity, concentrating exclusively on the hydrophobic SAMs. Preferential adhesion was observed onto the 1 mM C8-SH surface, with an average of 2192 cells counted on the Au stripes, compared to 259 cells on the SiO2 part (Fig. 4B). Decreasing the hydrophobicity of the surface, fewer yet detectable adhesion was observed for the 100 µM C8-SH sample (SI, Fig. S6A). An average of 410 cells were counted on the Au patterns, whereas the SiO2 contained only the minimal amount of 52 cells. No adhesion was detected onto the 10 µM sample (SI, Fig. S6B), for which the cells were efficiently removed from the surface during the washing steps. Notably, adhesion was fully dependent upon pre-treatment with P-factor (Fig. 4D) and no adhesion was observed onto the hydrophilic SAMs (SI, Fig. S7 and S8).
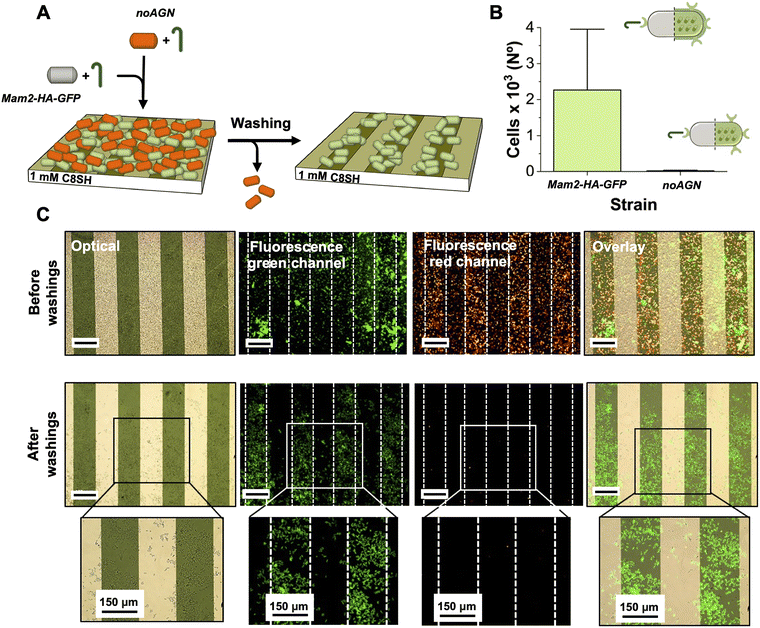
To confirm that the Mam3 AGNs expressed upon addition of P-factor are responsible for the hydrophobic interaction with the surface, we replaced the mam3 gene by the natR selection marker (noAGN). Blocking the expression of AGNs, the deletion of mam3 would completely annul the hydrophobic-driven adhesion. Indeed, as shown in Fig. 4E (SI, Fig. S8), the noAGN strain demonstrated unable to adhere to the substrate even after activation with the P-factor. Taken together, the above data suggests that in addition to activating a function, the chemical induction by the P-factor can direct specific, surface-dependent adhesion and ordering.
We next evaluated the potency of the hydrophobic surface to selectively interact with the Mam3 AGN-expressing cells by implementing a competitive experiment between cells expressing (Mam2-HA-GFP) and not (noAGN) AGNs upon activation with P-factor. These strains were induced with P-factor for 4 hours, mixed and added on the functionalized surface inside a 24 well-plate for 3 hours. They were visualized in two different colours: green for the Mam2-HA-GFP visualizing the GFP expression, and red for the noAGN stained with Rhodamine B (Rho B; see Material and methods), to provide an emission signal which could be differentiated from that of Mam2-HA-GFP. This dye was selected due to its known ability to permeate yeast cell membranes and accumulate within mitochondria without causing cell lysis.38 As shown in Fig. 5, before the washing procedure the surface was saturated with both cells expressing (green) or not (red) the Mam3 agglutinin. After the final washing step, noAGN cells (red channel) were entirely removed from the surface, demonstrating adhesion behaviour consistent with prior tests conducted in isolation (Fig. 4E). This result confirmed that Rho B staining did not alter their adhesion properties. Similarly, Mam2-HA-GFP cells (green channel) demonstrated stable retention on the hydrophobic stripes of the surface, unaffected by the presence of the non-adhering cells. This data shows that the presence of the Mam3 AGNs at the cell surface is necessary to efficiently compete out the cells lacking Mam3 for binding the functionalized hydrophobic surfaces.
Conclusions
In conclusion, this work provides biochemical and nanotechnological insights on the construction of a smart assembly that can produce Albulin on-demand and with high spatial specificity. Here, we have limited our studies to intracellular production of Albulin, which allows easy quantification, in the future we envision to secrete Albulin from the cells using classical secretion signals from yeast39 and to then validate its biological activity. We also plan to optimize the selective attachment to several substrates from varying materials and morphologies40 and control the cell behaviour over time, i.e., control possible mutation of yeast cells that could overwrite their desired characteristics41–43 and genetically encoded information.44–46 Importantly, the presence of P-factor also results in a cell cycle arrest before DNA replication onset. While this feature will impede uncontrolled increase in cell population, it could be modulated by exploiting our broad knowledge of fission yeast cell cycle regulations. Thanks to its integrated approach combining biology, chemistry, and materials science, this work represents an important first step toward the creation of bionanotechnological therapeutic platforms that function as micro bioprocessors for on-demand, locally precise biopharmaceutical production. As next steps, we envisage this dual-control system, which leverages the cells' intrinsic biological machinery alongside carefully designed chemical triggers and substrates, to evolve to incorporate external activation control systems, enabling remote switching of activity. Additionally, further genetic modifications will be introduced to regulate the secretion of the biopharmaceutical. This will create a streamlined, multi-level remotely controlled strategy, enhancing the applicability of this technology for real-world applications.Author contributions
The conceptualization of the study was led by DB and DH. SMS, VM, LEC, EM, AOS, TM, and LR contributed to the investigation, formal analysis, and curation of the data presented in the manuscript. DB, DH, LM, and SM provided supervision. DB, DH, and LM were responsible for project administration and funding acquisition. SMS, AOS, TM, and LM contributed to the writing of the manuscript, while DB, DH, LM, and SM were involved in its review and editing. All authors approved the final version.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supporting information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cb00147aAcknowledgements
DB gratefully acknowledges Cardiff University and University of Vienna for generous financial support. LEC thanks the F.R.S.-FNRS for her F.R.I.A. fellowship. This work was supported by FNRS grant EQP U.N.027.18 and FNRS grant CDR J.0066.16 to DH. We thank Dr G. Sandu and Dr R. Marega (previously at the University of Namur and currently at the CER Groupe) for the initial contribution and the discussions.Notes and references
- C. J. Bashor, I. B. Hilton, H. Bandukwala, D. M. Smith and O. Veiseh, Nat. Rev. Drug Discovery, 2022, 21, 655 CrossRef CAS PubMed.
- M. A. Fischbach, J. A. Bluestone and W. A. Lim, Sci. Transl. Med., 2013, 5, 1 Search PubMed.
- Z. Zhao, A. Ukidve, J. Kim and S. Mitragotri, Cell, 2020, 181, 151 CrossRef CAS.
- H. J. Jackson, S. Rafiq and R. J. Brentjens, Nat. Rev. Clin. Oncol., 2016, 13, 370 CrossRef CAS PubMed.
- M. Sheykhhasan, H. Manoochehri and P. Dama, Cancer Gene Ther., 2022, 29, 1080 CrossRef CAS PubMed.
- A. R. Ramos, E. H. Heslop and K. M. Brenner, Annu. Rev. Med., 2016, 67, 165 CrossRef PubMed.
- N. Hossein-khannazer, S. Torabi, R. Hosseinzadeh, S. Shahrokh, H. Asadzadeh Aghdaei, A. Memarnejadian, N. Kadri and M. Vosough, Human Cell, 2021, 34, 1289 CrossRef.
- B. Alberts, A. Johnson, J. Lewis, M. Raff, K. Roberts and P. Walter, Molecular Biology of the Cell, Garland Science, New York, 2008 Search PubMed.
- T. W. Overton, Drug Discovery Today, 2014, 19, 590 CrossRef CAS.
- P. Balbas and A. Lorence, Recombinant Gene Expression, 2004 Search PubMed.
- C. Trivellin, L. Olsson and P. Rugbjerg, ACS Synth. Biol., 2021, 11, 1686 CrossRef PubMed.
- N. Kulagina, S. Besseau, C. Godon, G. H. Goldman, N. Papon and V. Courdavault, Front. Fungal Biol., 2021, 2, 1 Search PubMed.
- J. Nielsen, Bioengineered, 2013, 4, 207 CrossRef.
- S. Ostegaard, L. Olsson and J. Nielsen, Microbiol. Mol. Biol., 2000, 64, 34 CrossRef.
- G. Coradello and N. Tirelli, Molecules, 2021, 26, 3123 CrossRef CAS.
- C. Sabu, D. Raghav, U. S. Jijith, P. Mufeedha, P. P. Naseef, K. Rathinasamy and K. Pramod, Mater. Sci. Eng., C, 2019, 103, 109753 CrossRef CAS.
- Y. Otsubo and M. Yamamoto, J. Cell Sci., 2012, 125, 2789 CrossRef CAS.
- B. Sieber, J. M. Coronas-Serna and S. G. Martin, Semin. Cell Dev. Biol., 2023, 133, 83 CrossRef CAS.
- H. Zhao, Z. Shen, P. C. Kahn and P. Lipke, J. Bacteriol., 2001, 183, 2874 CrossRef CAS.
- M. Chen, Z. Shen, S. Bobin, P. C. Kahn and P. N. Lipke, J. Biol. Chem., 1995, 270, 26168 CrossRef CAS.
- K. Tanaka, J. Davey, Y. Imai and M. Yamamoto, Mol. Cell. Biol., 1993, 13, 80 CAS.
- Y. Imai and M. Yamamoto, Genes Dev., 1994, 8, 328 CrossRef CAS.
- K. Kitamura and C. Shimoda, EMBO J., 1991, 10, 3743 CrossRef CAS PubMed.
- T. Seike and H. Niki, Microbiol. Mol. Biol. Rev., 2022, 86, 1 CrossRef CAS PubMed.
- Y. Xue-Franzén, S. Kjærulff, C. Holmberg, A. Wright and O. Nielsen, BMC Genomics, 2006, 7, 1 CrossRef PubMed.
- A. Grigorescu, M. Chen, H. Zhao, P. Kahn and P. N. Lipke, IUBMB Life, 2000, 50, 105 CAS.
- P. N. Lipke and J. Kurjan, Microbiol. Rev., 1992, 56, 180 CrossRef CAS.
- H. Zhao, Biochemical studies on a-agglutinin, PhD dissertation, City University of New York, 2000 Search PubMed.
- J. Hou, K. E. J. Tyo, Z. Liu, D. Petranovic and J. Nielsen, Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae, FEMS Yeast Res., 2012, 12, 491–510 CrossRef CAS PubMed.
- A. Duttaroy, P. Kanakaraj, B. L. Osborn, H. Schneider, O. K. Pickeral, C. Chen, G. Zhang, S. Kaithamana, M. Singh, R. Schulingkamp, D. Crossan, J. Bock, T. E. Kaufman, P. Keavey, M. Carey-Barber, S. K. Krishnan, A. Garcia, K. Murphy, J. K. Siskind, M. A. McLean, S. Cheng, S. Ruben, C. E. Birse and O. Blondel, Diabetes, 2005, 54, 251 CrossRef CAS PubMed.
- T. Seike, C. Shimoda and H. Niki, PLoS Biol., 2019, 17, e3000101 CrossRef.
- G. Ladds and J. Davey, Mol. Microbiol., 2000, 36, 377 CrossRef CAS.
- T. Maeda, N. Mochizuki and M. Yamamoto, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 7814 CrossRef CAS.
- M. Á. Curto, S. Moro, F. Yanguas, C. Gutiérrez-González and M. H. Valdivieso, Cell. Mol. Life Sci., 2018, 75, 1687 CrossRef CAS PubMed.
- J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103 CrossRef CAS PubMed.
- S. W. Tarn-Chang, H. A. Biebuyck, G. M. Whitesides, N. Jeon and R. G. Nuzzo, Langmuir, 1995, 11, 4371 CrossRef.
- E. Wood and P. Nurse, Annu. Rev. Cell Dev. Biol., 2015, 31, 11 CrossRef CAS PubMed.
- K. Nowosad, M. Sujka, U. Pankiewicz, D. Miklavčič and M. Arczewska, Biomolecules, 2021, 11, 850 CrossRef CAS PubMed.
- S. Kjærulff and M. R. Jensen, Biochem. Biophys. Res. Commun., 2005, 336, 974 CrossRef.
- V. Brunetti, G. Maiorano, L. Rizzello, B. Sorce, S. Sabella, R. Cingolani and P. P. Pompa, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 6264 CrossRef CAS PubMed.
- S. Labbé, T. Mourer, A. Brault and T. Vahsen, Curr. Genet., 2020, 66, 703 CrossRef.
- T. Mourer, V. Normant and S. Labbé, J. Biol. Chem., 2017, 292, 4898 CrossRef CAS PubMed.
- T. Mourer, J. F. Jacques, A. Brault, M. Bisaillon and S. Labbé, J. Biol. Chem., 2015, 290, 10176 CrossRef CAS PubMed.
- L. Gou, J. S. Bloom and L. Kruglyak, Genetics, 2019, 211, 731 CrossRef CAS PubMed.
- J. Steensels, T. Snoek, E. Meersman, M. P. Nicolino, K. Voordeckers and K. J. Verstrepen, FEMS Microbiol. Rev., 2014, 38, 947 CrossRef CAS PubMed.
- J. Bähler, J. Wu, M. S. Longtine, N. G. Shah, A. Mckenzie III, A. B. Steever, A. Wach, P. Philippsen and J. R. Pringle, Yeast, 1998, 14, 943 CrossRef.
| This journal is © The Royal Society of Chemistry 2026 |