 Open Access Article
Open Access ArticleTuning the efficiency of molecular probes via quinone methide-based in situ labeling
Zachary
Rabinowitz†
 a,
Seyedehalaleh
Anvar†
a,
Seyedehalaleh
Anvar†
 a,
Jun
Liu†
ab,
Zixin
Chen
a,
Jun
Liu†
ab,
Zixin
Chen
 a,
Yuzhao
Zhang
a,
Yuzhao
Zhang
 a,
Chao
Cui
a,
Ashton
Sigler
a,
Chao
Cui
a,
Ashton
Sigler
 b and
Lina
Cui
b and
Lina
Cui
 *ab
*ab
aDepartment of Medicinal Chemistry, College of Pharmacy, University of Florida, Gainesville, FL 32610, USA. E-mail: linacui@cop.ufl.edu
bDepartment of Chemistry and Chemical Biology, University of New Mexico, Albuquerque, NM 87131, USA
First published on 17th December 2025
Abstract
Imaging the activities of hydrolases using molecular imaging probes can reveal underlying molecular mechanisms in the context of cells to organisms and their correlation with different pathological conditions can be used in diagnosis. Due to the nature of hydrolases, substrate-based probes can take advantage of their catalytic cycles to free reporter moieties that can generate amplified signal. This is less ideal in the context of cell- or organism-based detection, as the reporter moiety can easily diffuse away from the target site upon activation. Therefore, spatial resolution is a key factor for probe sensitivity. One strategy to improve the spatial resolution is to form a covalent linkage between the reporter moiety and intracellular proteins upon probe activation by the enzyme via a reactive intermediate. In this work, we tuned the reactivity of the quinone methide intermediate by synthesizing fluorescent probes containing different modifications to the phenol linker for β-galactosidase activation. The labeling efficacy of these probes was evaluated using fluorescence gel electrophoresis, flow cytometry, and fluorescence cell imaging. This study provides insights into the further development of hydrolase-targeting probes for cell- or organism-based imaging with enhanced efficiency via in situ labeling.
Introduction
Molecular imaging probes are important tools to interrogate biological functions, both spatially and temporally, in living cells and organisms.1 Probes can be constructed to detect a myriad of different biological molecules including hydrolases. Hydrolases, enzymes that catalyze hydrolysis of their substrates, are important biomolecules that are involved in a wide range of biological functions. Like other biomolecules, their activities can become enhanced or diminished in diseased states, compared to healthy states. Thus, detecting or imaging the activities of hydrolases can reveal underlying molecular mechanisms in the context of cells-to-organisms, and their correlation with different pathological conditions can be used in diagnosis.2 Due to the nature of hydrolases, substrate-based probes can be activated in their catalytic cycles. Subsequently, hydrolysis of the substrate frees reporter moieties that provide either positive or negative signal output. Substrate-based probes have been widely used to assess enzyme activities in various contexts in vitro or in vivo, taking advantage of their catalytic cycles for amplified signals.3A major limitation of most substrate-based small molecule imaging probes is that they can easily diffuse away from the site of activation, resulting in poor sensitivity and low signal-to-background ratios.4 For test-tube type bulk detection, spatial resolution is not a measure of importance. However, for cell- or organism-based detection or imaging, spatial resolution is a key factor for probe sensitivity that influences signal-to-background ratios.5 Various strategies have been developed to improve spatial resolution.6–8 For example, small molecule probes can be activated by the target enzyme, and due to the change in hydrophobicity before and after enzyme activation, they can self-assemble into nanometer-sized particles or aggregates and retain at the site of activation.9–16 Alternatively, using previously developed strategies for targeted covalent inhibitors,17,18 a covalent bond between the probe and an intracellular, nucleophilic protein can be formed via the formation of an electrophilic moiety generated upon activation of the probe.6,19–22
Quinone methides (QMs) are a reactive, electrophilic species that can be generated in situ from a protected phenol precursor bearing a leaving group on the methyl group at either the para- or ortho-position.23–27 Once formed, QMs are susceptible to nucleophilic attack. Under physiological conditions, QMs can be formed via removal of a “blocking” group, resulting in unmasking of the phenolic oxygen and loss of a leaving group. Enabling bioorthogonal QM chemistry has found numerous applications including developing activity-based probes for various enzymes (e.g., phosphatase,28 glycosidases,29–38 tyrosine phosphatase,39 steroid sulfatase,40 and beta-lactamase41–43), prodrugs,44–46 among others.47–50
Across these applications, fluorine of either the mono- or difluoromethyl group has been frequently used as the leaving group to form the active quinone methide intermediate via spontaneous elimination of a molecule of hydrogen fluoride (HF).26 However, the application of in situ labeling of biological molecules via QM chemistry has been dwarfed by the dilemma that the monofluoromethyl group is more reactive, yet the conjugated products are more stable than its difluoromethyl counterpart.31,41,51 Some molecules containing a monofluoromethyl group suffer from poor stability in aqueous media due to hydrolysis or non-specific reactivity with other nucleophilic molecules before the probe reaches the active site of the target enzyme.52,53 Depending on the properties of the molecules, some monofluoromethyl group-containing probes are reportedly stable,42,54 while many others, including several unpublished probes developed in our lab, are susceptible to hydrolysis.38,52
Identification of self-immolative linkers with better stability in solution, reactivity profiles, and labeling efficiencies remains an important feat towards applying QM-based probes in various applications. Towards solving this issue, the ethyl carbamate moiety has been used as an alternative leaving group to limit hydrolysis and labeling of non-target nucleophilic molecules.52,53,55,56 Furthermore, installation of electron-donating- or electron-withdrawing substituents on QMs has been found to modulate the rate of formation and stability of their adducts in vitro.57 However, thorough investigations of altering the leaving group capability and tuning the aromatic ring electronics to enhance the performance of self-immolative phenol linkers are limited.53 In this paper, we evaluated two different approaches in parallel to provide insights on the design of self-immolative QM-based hydrolase-targeting probes with better stability, reactivity profiles, and labeling efficiencies.
Results and discussion
Design and synthesis of QM-based fluorescent probes
Overall, we designed and synthesized probes each containing three moieties: a β-galactoside moiety, a self-immolative phenol linker, and fluorescein (FITC) (Fig. 1). In addition, to enhance the stability of the linkage between the phenol linker and FITC during synthesis, a previously developed S-methyl-isothiourea linkage was utilized.30 The synthesis of these probes was performed as outlined in Scheme S1.To investigate whether we can improve the performance of QM-based probes, we took two different logical approaches of modifying the self-immolative phenol linker. The first approach is to install a group with better leaving group ability than fluorine. A more stabilized leaving group would theoretically improve the labeling efficiency by facilitating the formation of the QM, which has been shown to be dependent on the type of leaving group.58 Thus, we designed and synthesized BG-FITC-OCONHEt containing an ethyl carbamate leaving group, which is stabilized through electron delocalization and represents a better leaving group than the small, electronegative fluorine atom (Fig. 1). This rationale is further supported by a prior report that the replacement of fluorine with ethyl carbamate led to a more stable molecule in phosphate-buffered saline (PBS).52 The second approach is to adjust the electronics of the aromatic linker. The labeling efficiency and stability of the resulting conjugate has been shown to be tunable by installation of substituents on the self-immobilizing phenol linker.57 To explore how altering the electronics of the aromatic linker affects the labeling efficiency of QM-based probes, electron-donating groups such as methoxy (BG-FITC-OMe-1F) and methyl (BG-FITC-Me-1F) or electron-withdrawing groups such as fluorine (BG-FITC-F-1F) were installed on the aromatic ring in the ortho position to the phenolic oxygen (Fig. 1).
In the presence of β-galactosidase (β-Gal), the β-galactosyl bond is hydrolyzed, allowing the phenol linker to undergo self-immolation to generate the QM in situ upon loss of the leaving group. The QM can then alkylate nearby proteins via their nucleophilic residues. Therefore, we aimed to evaluate and compare the labeling efficiencies (i.e., the amount of FITC-labeled protein) of probes with different modifications in vitro and in living cells to gauge whether we can improve the reactivity profiles and stability of QM-based probes. All probe molecules contained the fluorescent reporter, FITC, in its “on” state, both before and after activation. QM-based probes can also be designed to feature an increase in fluorescence intensity (“turn-on”) after removal of the “blocking group” and nucleophilic attack of the QM, making it more sensitive towards analyte detection.26,38 In this case, without fluorescence change after target activation, our approach provided direct visual comparison of the labeling efficiencies of these molecules both in vitro and in cells.
Evaluation of labeling efficiency in vitro
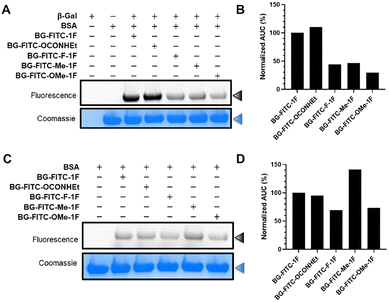
Prior to evaluating the labeling efficiency of these probes, we first confirmed the concentration of all probe stock solutions and assessed their stability in aqueous solution (Fig. S1 and S2). All probes were stable (at least 95% intact probe) after 4-hours of incubation at 37 °C. Therefore, we carried out the labeling efficiency evaluation of all probes using a 4-hour incubation period. With longer incubation times, BG-FITC-1F showed the most degradation, followed by BG-FITC-F-1F, BG-FITC-Me-1F, and BG-FITC-OMe-1F. On the other hand, BG-FITC-OCONHEt exhibited superior stability. LC-MS analysis of the byproducts suggested that hydrolysis of both the monofluoromethyl group and S-methyl-isothiourea linkage are two mechanisms for probe degradation (data not shown).To evaluate the labeling efficiency of these probes in vitro, we incubated bovine serum albumin (BSA, 1 µg µL−1), with 5 µM of each probe with or without Escherichia coli β-Gal (0.4 µg µL−1) for 4-hours in PBS (pH 7.4) at 37 °C. The resulting mixtures were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and the in-gel fluorescence intensities of FITC-labeled BSA were quantified using ImageJ (Fig. 2). As shown in Fig. 2A, in the presence of β-Gal, the probes labeled BSA with FITC via generation of the QM intermediate, followed by alkylation of the protein. BG-FITC-OCONHEt showed a higher labeling efficiency of BSA than BG-FITC-1F (Fig. 2B), suggesting that the superior stability in solution as well as the better leaving group ability of the ethyl carbamate group over fluorine of the monofluoromethyl moiety (Fig. S2) may facilitate the formation and yield of the QM intermediate, leading to more labeling of BSA. Surprisingly, tuning the aromatic ring electronics with either electron-donating- (BG-FITC-Me-1F, BG-FITC-OMe-1F) or electron-withdrawing groups (BG-FITC-F-1F) did not show any improvements in the labeling of BSA, compared to BG-FITC-1F (Fig. 2B). In the absence of β-Gal, the probe cannot generate the electrophilic, QM intermediate, however, proteins can likely still attack the probe to form a covalent bond via nucleophilic substitution reaction, due to the presence of a leaving group. Moreover, the probes can bind to proteins in a non-covalent fashion, leading to non-specific labeling of BSA and poor specificity. To examine non-QM-mediated labeling of BSA, we incubated 5 µM of each probe with BSA (1 µg µL−1) without the addition of Escherichia coli β-Gal (Fig. 2C). To our surprise, in-gel fluorescence revealed that BG-FITC-1F and BG-FITC-OCONHEt exhibited similar non-specific labeling of BSA (Fig. 2D). A previous study observed that molecules containing the less reactive ethyl carbamate leaving group produced less non-specific fluorescence tagging of BSA than fluorine of the monofluoromethyl leaving group.53 On the other hand, BG-FITC-F-1F and BG-FITC-OMe showed lower non-specific labeling of BSA than BG-FITC-1F, whereas BG-FITC-Me-1F exhibited the highest non-specific labeling of BSA (Fig. 2D), suggesting that this probe has more interactions with BSA.
Modification of self-immolative phenol linkers with electron-withdrawing groups (such as ethyl esters) have been shown to result in slow formation of their adducts, however, their adducts are more stable over time.57 On the contrary, electron-donating groups (such as methyl) gave rise to rapid adduct formation, but their adducts are less stable.57 Since this phenomenon is observed to be time-dependent,57 it is likely that the static 4-hour time point did not reflect the true performance of tuning the aromatic ring with various substituents. However, analysis of increasing time-points is limited by the likelihood of probe degradation in aqueous solutions, forbidding accurate time-dependent analyses of these probes. Alternatively, the substituents on the aromatic ring might abrogate the activation of the probe by β-Gal. Therefore, we conducted a molecular docking study to elucidate the binding conformation of our probes with Escherichia coli β-Gal to provide insights into whether these modifications produce a steric hindrance effect towards β-Gal activation.
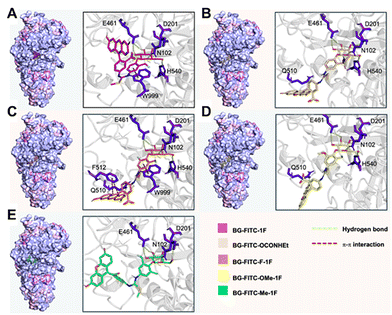
Molecular docking study
To elucidate the binding conformation of our probes with Escherichia coli β-Gal, we performed a molecular docking study within the active site (Fig. 3). To better compare the binding modes across all probes, we selected the binding conformations that adopted a similar binding mode at the β-galactoside moiety. For all probes, the β-galactoside moiety forms polar interactions with Asn102, Asp201, Glu461, and His540. Unlike the other probes, BG-FITC-Me-1F lacks polar interactions with His540. This can be explained by the steric hindrance at the peripheral of the binding pocket caused by the monofluoromethyl group on the self-immolative phenol linker, leading to a small torsion on the β-galactoside moiety. Notably, the catalytic residue Glu461, which serves as the proton donor in the enzyme-catalyzed hydrolysis reaction, is in proximity to the glycosidic bond of all probes. The self-immolative phenol linker moieties of our probes do not frequently interact with the periphery of the binding pocket. However, we observed occasional π–π interactions with Trp999 with both BG-FITC-1F and BG-FITC-F-1F. Across all probes, the FITC moiety did not impart many interactions, and its’ binding conformations were flexible. These observations suggest that the binding conformations of our probes are stabilized primarily by the β-galactoside moiety. Overall, the molecular docking study revealed the suitable binding conformations formed by our probes at the active site of β-Gal. Furthermore, the studies suggests that the self-immolative phenol linker ring did not affect interactions with β-Gal. Therefore, potential steric hinderance of activation by β-Gal caused by the addition of relatively small substituents on the phenol ring (e.g., methyl, methoxy, and fluorine) or replacement of the fluorine leaving group for the ethyl carbamate leaving group likely does not contribute to the observed labeling results.Assessment of cellular performance
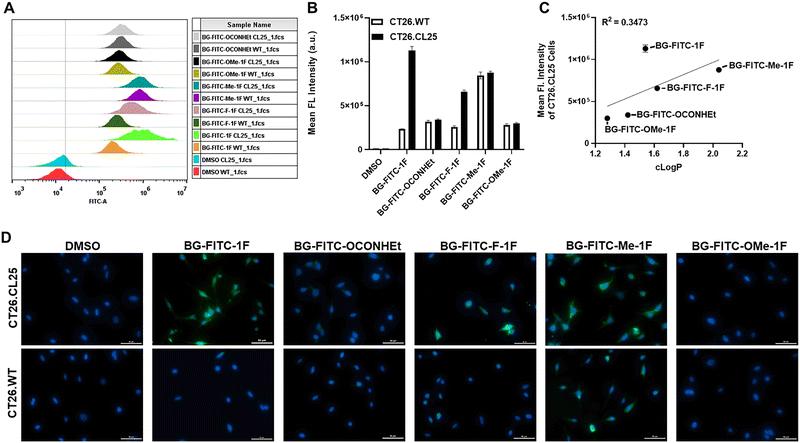
We further characterized the labeling efficiency of these probes in the mouse colon cancer cell lines, CT26.WT and CT26.CL25 (Fig. 4). CT26.CL25 cells, stably transfected with the β-Gal encoding lacZ gene, overexpress β-Gal (Fig. S5). Once the probe enters the cell, β-Gal can recognize and hydrolyze the β-galactoside moiety, triggering the generation of the electrophilic QM intermediate through self-immolation. The QM intermediate, which is linked to the reporter moiety, can then be attacked by intracellular proteins such as β-Gal itself or depart from the active site of β-Gal and react with any other available nucleophilic molecules. Consequently, trapping the reporter moiety in the cell enables improvements in the sensitivity of detecting biological activity as the probe-protein adduct is less likely to efflux from the cell (Fig. 1B). Therefore, we expect a higher fluorescence intensity in CT26.CL25 cells, which overexpress β-Gal and can thus activate the self-immobilizing probe, than CT26.WT cells post-incubation with these probes.Both cell lines were incubated with each probe (5 µM) in serum-free RPMI 1640 medium for 4-hours at 37 °C. The resulting cells were washed once with PBS (pH 7.4) and then analyzed by fluorescence activated cell sorting (FACS) (Fig. 4A). Only BG-FITC-1F and BG-FITC-F-1F showed significant contrast between CT26.CL25 and CT26.WT cells (Fig. 4B) among all probes. Surprisingly, the overall trend observed in the SDS-PAGE analysis (Fig. 2) does not translate to cell-based assays, as the labeling efficiency can be greatly influenced by the probes’ cell permeability. Generally, small molecules can penetrate biological membranes via passive diffusion at a rate related to their lipophilicity.59,60 We calculated the logarithm of the partition coefficient (c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) values of each probe molecule using ChemDraw Professional version 20.0.04.1 (Table S1) and plotted the cLogP for each probe against the mean fluorescence intensities of CT26.CL25 cells after 4-hour incubation (Fig. 4C). Seemingly, there is a positive correlation between c
P) values of each probe molecule using ChemDraw Professional version 20.0.04.1 (Table S1) and plotted the cLogP for each probe against the mean fluorescence intensities of CT26.CL25 cells after 4-hour incubation (Fig. 4C). Seemingly, there is a positive correlation between c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P and the mean fluorescence intensity of CT26.CL25 cells, suggesting that the probes’ lipophilicity and resulting cell penetration capabilities is a factor affecting these probes labeling efficiencies. Subsequently, cross-comparison of self-immobilizing probes with various degrees of lipophilicity in living cells is not straightforward, due to the possible different rates of cell penetration. Interestingly, BG-FITC-Me-1F was the most lipophilic of all the probes (indicated by its high c
P and the mean fluorescence intensity of CT26.CL25 cells, suggesting that the probes’ lipophilicity and resulting cell penetration capabilities is a factor affecting these probes labeling efficiencies. Subsequently, cross-comparison of self-immobilizing probes with various degrees of lipophilicity in living cells is not straightforward, due to the possible different rates of cell penetration. Interestingly, BG-FITC-Me-1F was the most lipophilic of all the probes (indicated by its high c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value), however, it exhibited high fluorescence intensities in both cell lines leading to poor contrast. SDS-PAGE analysis revealed that BG-FITC-Me-1F also exhibited the highest level of non-specific BSA labeling, suggesting that the poor contrast between the two cell lines is largely due to its non-specific interaction. Therefore, optimizing probe hydrophobicity is a critical factor to improve specificity and contrast.
P value), however, it exhibited high fluorescence intensities in both cell lines leading to poor contrast. SDS-PAGE analysis revealed that BG-FITC-Me-1F also exhibited the highest level of non-specific BSA labeling, suggesting that the poor contrast between the two cell lines is largely due to its non-specific interaction. Therefore, optimizing probe hydrophobicity is a critical factor to improve specificity and contrast.
We also performed fluorescence cell imaging of CT26.CL25 and CT26.WT cells after incubation with each probe (5 µM) in serum-free RPMI 1640 medium for 4-hours at 37 °C (Fig. 4D). Cell imaging also demonstrated a similar result as FACS analysis with only BG-FITC-1F and BG-FITC-F-1F exhibiting significant contrast, and BG-FITC-1F had a higher labeling efficiency than BG-FITC-F-1F. It is possible that BG-FITC-F-1F yielded less labeling efficiency than BG-FITC-1F because the rate of formation of QMs bearing an electron-withdrawing substituent such as fluorine is slower, resulting in inferior contrast. This phenomenon has been observed by others previously in buffered conditions.57 Therefore, when developing QM-based probes, we observed that the kinetics of protein adduct formation and the probes stability and lipophilicity are important parameters to be considered.
Conclusions
While QM chemistry is highly desirable in the field of chemical biology, drug discovery, and molecular imaging, its application has been impeded by the difficulty in choosing the proper self-immolative linker to generate the QM species. The monofluoromethyl group is very appealing in many cases for its rapid labeling reaction with proteins and the formation of stable protein-probe conjugates. However, probes containing a monofluoromethyl group tend to be hydrolyzed too fast to be used in most biological applications. In this work, we tuned the self-immolative phenol linker chemistry to investigate the reactivity and stability of the QM that would allow in situ labeling of biological molecules with an imaging reporter via the generation of the QM species. For this purpose, we designed and synthesized various “always-on” fluorescent QM-based probes, each carrying a β-gal-sensitive “blocking” unit, a self-immolative phenol linker, and a FITC reporter. The self-immolative phenol linker featured either different leaving groups (i.e., the widely used monofluoromethy group or ethyl carbamate) or substituents on the phenol linker to investigate their effects on the labeling efficiency of proteins, which depends on both the stability and reactivity of the probe molecule and protein-probe conjugate. The labeling efficiency of these probes were compared in parallel through SDS-PAGE gel analysis, flow cytometry, and fluorescence cell imaging. In addition to its excellent stability in solution, in-gel fluorescence analysis suggested that the ethyl carbamate leaving group, BG-FITC-OCONHEt, provides higher labeling efficiency over the fluorine leaving group of the monofluoromethyl moiety, BG-FITC-1F. In cells, BG-FITC-1F had a superior labeling efficiency producing improved imaging sensitivity, likely due to better cell penetration. Therefore, from this study, we conclude that it is important to factor in the probes molecules stability, kinetics of QM formation, and cell penetration ability towards the successful application of QM-based probes. Moreover, the results from this work can be used to guide the development of QM-based probes for various other applications where FITC can be replaced by the functional moieties of choice.Author contributions
Z. M. R., J. L. and L. C. conceived and designed the project. Z. M. R. and J. L. performed all the chemical synthesis and compound characterization. Z. M. R., S. A., Z. C., C. C., and A. L. S. performed gel electrophoresis, fluorescence microscopy, and flow cytometry. Y. Z. performed docking study. Z. M. R., J. L., Z. C., and L. C. wrote the paper, and all authors commented on the paper.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cb00138b.Acknowledgements
This work is supported by research grants to Prof. L. Cui from the National Institute of General Medical Sciences (R35GM124963, R35GM156219), the National Institute on Aging (R21AG081869, R41AG081007), the University of Florida (Maureen Keller-Wood PROSPER EXCELLENCE Award), and UF Health Cancer Center (Breast Cancer Pilot Award). Z. M. R. was partially supported by UF Graduate School Preeminent Scholarship.Notes and references
- R. Weissleder and U. Mahmood, Molecular Imaging, Radiology, 2001, 219(2), 316–333 CrossRef CAS PubMed.
- C. Yang, Q. Wang and W. Ding, Recent progress in the imaging detection of enzyme activities in vivo, RSC Adv., 2019, 9(44), 25285–25302 RSC.
- S.-D. Li and L. Huang, Pharmacokinetics and Biodistribution of Nanoparticles, Mol. Pharmaceutics, 2008, 5(4), 496–504 CrossRef CAS PubMed.
- I. Johnson, Review: Fluorescent probes for living cells, Histochem. J., 1998, 30(3), 123–140 CrossRef CAS PubMed.
- K. Chen and X. Chen, Design and development of molecular imaging probes, Curr. Top. Med. Chem., 2010, 10(12), 1227–1236 CrossRef CAS PubMed.
- L. E. Edgington, M. Verdoes and M. Bogyo, Functional imaging of proteases: recent advances in the design and application of substrate-based and activity-based probes, Curr. Opin. Chem. Biol., 2011, 15(6), 798–805 CrossRef CAS PubMed.
- G.-B. Qi, Y.-J. Gao, L. Wang and H. Wang, Self-Assembled Peptide-Based Nanomaterials for Biomedical Imaging and Therapy, Adv. Mater., 2018, 30(22), 1703444 CrossRef PubMed.
- Z. Hai and G. Liang, Intracellular Self-Assembly of Nanoprobes for Molecular Imaging, Adv. Biosyst., 2018, 2(8), 1800108 CrossRef.
- Y. Gao, J. Shi, D. Yuan and B. Xu, Imaging enzyme-triggered self-assembly of small molecules inside live cells, Nat. Commun., 2012, 3(1), 1033 CrossRef PubMed.
- G. Liang, H. Ren and J. Rao, A biocompatible condensation reaction for controlled assembly of nanostructures in living cells, Nat. Chem., 2010, 2(1), 54–60 CrossRef CAS PubMed.
- D. Ye, A. J. Shuhendler, L. Cui, L. Tong, S. S. Tee, G. Tikhomirov, D. W. Felsher and J. Rao, Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo, Nat. Chem., 2014, 6(6), 519–526 CrossRef CAS PubMed.
- S. Onogi, H. Shigemitsu, T. Yoshii, T. Tanida, M. Ikeda, R. Kubota and I. Hamachi, In situ real-time imaging of self-sorted supramolecular nanofibres, Nat. Chem., 2016, 8(8), 743–752 CrossRef CAS PubMed.
- J. Li, Y. Gao, Y. Kuang, J. Shi, X. Du, J. Zhou, H. Wang, Z. Yang and B. Xu, Dephosphorylation of d-Peptide Derivatives to Form Biofunctional, Supramolecular Nanofibers/Hydrogels and Their Potential Applications for Intracellular Imaging and Intratumoral Chemotherapy, J. Am. Chem. Soc., 2013, 135(26), 9907–9914 CrossRef CAS PubMed.
- Z. Chen, M. Chen, Y. Cheng, T. Kowada, J. Xie, X. Zheng and J. Rao, Exploring the Condensation Reaction between Aromatic Nitriles and Amino Thiols To Optimize In Situ Nanoparticle Formation for the Imaging of Proteases and Glycosidases in Cells, Angew. Chem., Int. Ed., 2020, 59(8), 3272–3279 CrossRef CAS PubMed.
- Z. Chen, M. Chen, K. Zhou and J. Rao, Pre-targeted Imaging of Protease Activity through In Situ Assembly of Nanoparticles, Angew. Chem., Int. Ed., 2020, 59(20), 7864–7870 CrossRef CAS PubMed.
- K. A. Schleyer, B. D. Datko, B. Burnside, C. Cui, X. Ma, J. K. Grey and L. Cui, Responsive Fluorophore Aggregation Provides Spectral Contrast for Fluorescence Lifetime Imaging, ChemBioChem, 2020, 21(15), 2196–2204 CrossRef CAS PubMed.
- S. De Cesco, J. Kurian, C. Dufresne, A. K. Mittermaier and N. Moitessier, Covalent inhibitors design and discovery, Eur. J. Med. Chem., 2017, 138, 96–114 CrossRef CAS PubMed.
- L. Boike, N. J. Henning and D. K. Nomura, Advances in covalent drug discovery, Nat. Rev. Drug Discovery, 2022, 21(12), 881–898 CrossRef CAS PubMed.
- G. Blum, G. von Degenfeld, M. J. Merchant, H. M. Blau and M. Bogyo, Noninvasive optical imaging of cysteine protease activity using fluorescently quenched activity-based probes, Nat. Chem. Biol., 2007, 3(10), 668–677 CrossRef CAS PubMed.
- G. Blum, S. R. Mullins, K. Keren, M. Fonovič, C. Jedeszko, M. J. Rice, B. F. Sloane and M. Bogyo, Dynamic imaging of protease activity with fluorescently quenched activity-based probes, Nat. Chem. Biol., 2005, 1(4), 203–209 CrossRef CAS PubMed.
- L. E. Edgington, A. B. Berger, G. Blum, V. E. Albrow, M. G. Paulick, N. Lineberry and M. Bogyo, Noninvasive optical imaging of apoptosis by caspase-targeted activity-based probes, Nat. Med., 2009, 15(8), 967–973 CrossRef CAS PubMed.
- D. H. Kwan, H.-M. Chen, K. Ratananikom, S. M. Hancock, Y. Watanabe, P. T. Kongsaeree, A. L. Samuels and S. G. Withers, Self-Immobilizing Fluorogenic Imaging Agents of Enzyme Activity, Angew. Chem., Int. Ed., 2011, 50(1), 300–303 CrossRef CAS PubMed.
- W.-J. Bai, J. G. David, Z.-G. Feng, M. G. Weaver, K.-L. Wu and T. R. R. Pettus, The Domestication of ortho-Quinone Methides, Acc. Chem. Res., 2014, 47(12), 3655–3664 CrossRef CAS PubMed.
- M. S. Singh, A. Nagaraju, N. Anand and S. Chowdhury, ortho-Quinone methide (o-QM): a highly reactive, ephemeral and versatile intermediate in organic synthesis, RSC Adv., 2014, 4(99), 55924–55959 RSC.
- A. Minard, D. Liano, X. Wang and M. Di Antonio, The unexplored potential of quinone methides in chemical biology, Bioorg. Med. Chem., 2019, 27(12), 2298–2305 CrossRef CAS PubMed.
- D. Kern and A. Kormos, Self-Immobilizing Quinone Methides for the Fluorescent Sensing of Enzyme Activity, Chemosensors, 2023, 11, 3 CrossRef.
- A. Abe and M. Kamiya, A versatile toolbox for investigating biological processes based on quinone methide chemistry: From self-immolative linkers to self-immobilizing agents, Bioorg. Med. Chem., 2021, 44 Search PubMed.
- J. R. Betley, S. Cesaro-Tadic, A. Mekhalfia, J. H. Rickard, H. Denham, L. J. Partridge, A. Plückthun and G. M. Blackburn, Direct Screening for Phosphatase Activity by Turnover-Based Capture of Protein Catalysts, Angew. Chem., Int. Ed., 2002, 41(5), 775–777 CrossRef CAS PubMed.
- M. Kurogochi, S.-I. Nishimura and Y. C. Lee, Mechanism-based fluorescent labeling of β-galactosidases. An efficient method in proteomics for glycoside hydrolases, J. Biol. Chem., 2004, 279, 44704–44712 CrossRef CAS PubMed.
- T.-C. Cheng, S. R. Roffler, S.-C. Tzou, K.-H. Chuang, Y.-C. Su, C.-H. Chuang, C.-H. Kao, C.-S. Chen, I. H. Harn, K.-Y. Liu, T.-L. Cheng and Y.-L. Leu, An Activity-Based Near-Infrared Glucuronide Trapping Probe for Imaging β-Glucuronidase Expression in Deep Tissues, J. Am. Chem. Soc., 2012, 134(6), 3103–3110 CrossRef CAS PubMed.
- Y.-L. Hsu, M. Nandakumar, H.-Y. Lai, T.-C. Chou, C.-Y. Chu, C.-H. Lin and L.-C. Lo, Development of Activity-Based Probes for Imaging Human α-l-Fucosidases in Cells, J. Org. Chem., 2015, 80(16), 8458–8463 CrossRef CAS PubMed.
- L. M. Chauvigné-Hines, L. N. Anderson, H. M. Weaver, J. N. Brown, P. K. Koech, C. D. Nicora, B. A. Hofstad, R. D. Smith, M. J. Wilkins, S. J. Callister and A. T. Wright, Suite of Activity-Based Probes for Cellulose-Degrading Enzymes, J. Am. Chem. Soc., 2012, 134(50), 20521–20532 CrossRef PubMed.
- C.-P. Lu, C.-T. Ren, Y.-N. Lai, S.-H. Wu, W.-M. Wang, J.-Y. Chen and L.-C. Lo, Design of a Mechanism-Based Probe for Neuraminidase To Capture Influenza Viruses, Angew. Chem., Int. Ed., 2005, 44(42), 6888–6892 CrossRef CAS PubMed.
- M. A. Lumba, L. M. Willis, S. Santra, R. Rana, L. Schito, S. Rey, B. G. Wouters, M. Nitz and A. β-galactosidase, probe for the detection of cellular senescence by mass cytometry, Org. Biomol. Chem., 2017, 15(30), 6388–6392 RSC.
- C.-S. Tsai, Y.-K. Li and L.-C. Lo, Design and Synthesis of Activity Probes for Glycosidases, Org. Lett., 2002, 4(21), 3607–3610 CrossRef CAS PubMed.
- J. Jiang, Q. Tan, S. Zhao, H. Song, L. Hu and H. Xie, Late-stage difluoromethylation leading to a self-immobilizing fluorogenic probe for the visualization of enzyme activities in live cells, Chem. Commun., 2019, 55(99), 15000–15003 RSC.
- Z. Gao, A. J. Thompson, J. C. Paulson and S. G. Withers, Proximity Ligation-Based Fluorogenic Imaging Agents for Neuraminidases, Angew. Chem., Int. Ed., 2018, 57(41), 13538–13541 CrossRef CAS PubMed.
- J. Liu, X. Ma, C. Cui, Z. Chen, Y. Wang, P. R. Deenik, L. Cui and N. I. R. Noninvasive, Imaging of Senescence via In Situ Labeling, J. Med. Chem., 2021, 64(24), 17969–17978 CrossRef CAS PubMed.
- Y.-Y. Huang, C.-C. Kuo, C.-Y. Chu, Y.-H. Huang, Y.-L. Hu, J.-J. Lin and L.-C. Lo, Development of activity-based probes with tunable specificity for protein tyrosine phosphatase subfamilies, Tetrahedron, 2010, 66(25), 4521–4529 CrossRef CAS.
- C. H. Tai, C. P. Lu, S. H. Wu and L. C. Lo, Synthesis and evaluation of turn-on fluorescent probes for imaging steroid sulfatase activities in cells, Chem. Commun., 2014, 50(46), 6116–6119 RSC.
- W. Mao, L. Xia, Y. Wang, H. Xie and A. Self-Immobilizing, and Fluorogenic Probe for β-Lactamase Detection, Chem. – Asian J., 2016, 11(24), 3493–3497 CrossRef CAS PubMed.
- T. Doura, M. Kamiya, F. Obata, Y. Yamaguchi, T. Y. Hiyama, T. Matsuda, A. Fukamizu, M. Noda, M. Miura and Y. Urano, Detection of LacZ-Positive Cells in Living Tissue with Single-Cell Resolution, Angew. Chem., Int. Ed., 2016, 55(33), 9620–9624 CrossRef CAS PubMed.
- H. Ito, Y. Kawamata, M. Kamiya, K. Tsuda-Sakurai, S. Tanaka, T. Ueno, T. Komatsu, K. Hanaoka, S. Okabe, M. Miura and Y. Urano, Red-Shifted Fluorogenic Substrate for Detection of lacZ-Positive Cells in Living Tissue with Single-Cell Resolution, Angew. Chem., Int. Ed., 2018, 57(48), 15702–15706 CrossRef CAS PubMed.
- M. Chiba, M. Kamiya, K. Tsuda-Sakurai, Y. Fujisawa, H. Kosakamoto, R. Kojima, M. Miura and Y. Urano, Activatable Photosensitizer for Targeted Ablation of lacZ-Positive Cells with Single-Cell Resolution, ACS Cent. Sci., 2019, 5(10), 1676–1681 CrossRef CAS PubMed.
- J. K. Myers and T. S. Widlanski, Mechanism-Based Inactivation of Prostatic Acid Phosphatase, Science, 1993, 262(5138), 1451–1453 CrossRef CAS PubMed.
- K. Haba, M. Popkov, M. Shamis, R. A. Lerner, C. F. Barbas III and D. Shabat, Single-Triggered Trimeric Prodrugs, Angew. Chem., Int. Ed., 2005, 44(5), 716–720 CrossRef CAS PubMed.
- S. Gnaim and D. Shabat, Quinone-Methide Species, A Gateway to Functional Molecular Systems: From Self-Immolative Dendrimers to Long-Wavelength Fluorescent Dyes, Acc. Chem. Res., 2014, 47(10), 2970–2984 CrossRef CAS PubMed.
- K. D. Janda, L.-C. Lo, C.-H. L. Lo, M.-M. Sim, R. Wang, C.-H. Wong and R. A. Lerner, Chemical Selection for Catalysis in Combinatorial Antibody Libraries, Science, 1997, 275(5302), 945–948 CrossRef CAS PubMed.
- S. Cesaro-Tadic, D. Lagos, A. Honegger, J. H. Rickard, L. J. Partridge, G. M. Blackburn and A. Plückthun, Turnover-based in vitro selection and evolution of biocatalysts from a fully synthetic antibody library, Nat. Biotechnol., 2003, 21(6), 679–685 CrossRef CAS PubMed.
- J. Liu, L. Cai, W. Sun, R. Cheng, N. Wang, L. Jin, S. Rozovsky, I. B. Seiple and L. Wang, Photocaged Quinone Methide Crosslinkers for Light-Controlled Chemical Crosslinking of Protein–Protein and Protein–DNA Complexes, Angew. Chem., Int. Ed., 2019, 58(52), 18839–18843 CrossRef CAS PubMed.
- J. Y. Hyun, S.-H. Park, C. W. Park, H. B. Kim, J. W. Cho and I. Shin, Trifunctional Fluorogenic Probes for Fluorescence Imaging and Isolation of Glycosidases in Cells, Org. Lett., 2019, 21(12), 4439–4442 CrossRef CAS PubMed.
- Y. Li, H. Song, C. Xue, Z. Fang, L. Xiong and H. Xie, A self-immobilizing near-infrared fluorogenic probe for sensitive imaging of extracellular enzyme activity in vivo, Chem. Sci., 2020, 11(23), 5889–5894 RSC.
- H. Song, Y. Li, Y. Chen, C. Xue and H. Xie, Highly Efficient Multiple-Labeling Probes for the Visualization of Enzyme Activities, Chem. – Eur. J., 2019, 25(61), 13994–14002 CrossRef CAS PubMed.
- V. Ahmed, Y. Liu and S. D. Taylor, Multiple Pathways for the Irreversible Inhibition of Steroid Sulfatase with Quinone Methide-Generating Suicide Inhibitors, ChemBioChem, 2009, 10(9), 1457–1461 CrossRef CAS PubMed.
- Y. Miao, Z. Q. Yu, S. Xu and M. Yan, Quinone Methide Based Self-Immobilizing Molecular Fluorescent Probes for In Situ Imaging of Enzymes, Chem. Asian J., 2024, 19, e202400189 CrossRef CAS PubMed.
- Y. Li, C. Xue, Z. Fang, W. Xu and H. Xie, In Vivo Visualization of γ-Glutamyl Transpeptidase Activity with an Activatable Self-Immobilizing Near-Infrared Probe, Anal. Chem., 2020, 92(22), 15017–15024 CrossRef CAS PubMed.
- E. E. Weinert, R. Dondi, S. Colloredo-Melz, K. N. Frankenfield, C. H. Mitchell, M. Freccero and S. E. Rokita, Substituents on quinone methides strongly modulate formation and stability of their nucleophilic adducts, J. Am. Chem. Soc., 2006, 128(36), 11940–11947 CrossRef CAS PubMed.
- E. E. Weinert, K. N. Frankenfield and S. E. Rokita, Time-dependent evolution of adducts formed between deoxynucleosides and a model quinone methide, Chem. Res. Toxicol., 2005, 18(9), 1364–1370 Search PubMed.
- P. D. Dobson and D. B. Kell, Carrier-mediated cellular uptake of pharmaceutical drugs: an exception or the rule?, Nat. Rev. Drug Discovery, 2008, 7(3), 205–220 CrossRef CAS PubMed.
- A. K. Ghose, V. N. Viswanadhan and J. J. Wendoloski, Prediction of Hydrophobic (Lipophilic) Properties of Small Organic Molecules Using Fragmental Methods:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) An Analysis of ALOGP and CLOGP Methods, J. Phys. Chem. A, 1998, 102(21), 3762–3772 CrossRef CAS.
An Analysis of ALOGP and CLOGP Methods, J. Phys. Chem. A, 1998, 102(21), 3762–3772 CrossRef CAS.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2026 |


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif)