 Open Access Article
Open Access ArticleAffinity-based protein profiling of the antiviral natural product nanchangmycin
Santiago
Leiva
 a,
Chloé
Freyermuth
a,
Chloé
Freyermuth
 a,
Stéphane
Claverol
b,
Daniele
Mantione
a,
Stéphane
Claverol
b,
Daniele
Mantione
 cd and
Emmanuelle
Thinon
cd and
Emmanuelle
Thinon
 *a
*a
aUniv. Bordeaux, CNRS, Bordeaux INP, CBMN, UMR 5248, IECB, F-33600 Pessac, France. E-mail: e.thinon@iecb.u-bordeaux.fr
bUniv. Bordeaux, Bordeaux Proteome, Bordeaux, France
cPOLYMAT, University of the Basque Country UPV/EHU, Joxe Mari Korta Center, Avda. Tolosa 72, 20018 Donostia-San Sebastian, Spain
dIkerbasque, Basque Foundation for Science, 48013 Bilbao, Spain
First published on 21st October 2025
Abstract
Nanchangmycin is a natural product with broad-spectrum activity against various organisms, exhibiting antibiotic, antiviral, anticancer, and antifibrotic effects. Nanchangmycin belongs to the family of polyether ionophores and is proposed to exert its therapeutic effects by altering ion gradients across biological membranes. Although this therapeutic mechanism has been well characterised in cancer models, it does not fully explain how nanchangmycin inhibits Zika virus infection, as recently reported. The specific molecular targets responsible for mediating nanchangmycin's antiviral activity remain unknown. Here, we designed a photoreactive clickable nanchangmycin probe and employed chemical proteomics to identify protein targets of nanchangmycin related to Zika virus infection in human cells. Among the most prominent targets was the protein SEC11A, a key component of the signal peptidase complex, which is essential for cleaving and processing Zika virus proteins. We showed that nanchangmycin blocks the cleavage of a Zika virus polyprotein, suggesting a novel mechanism for nanchangmycin-mediated inhibition of Zika virus infection.
Introduction
Zika virus (ZIKV) is a vector-borne RNA virus belonging to the Flavivirus genus.1 Flaviruses enter cells through endocytosis, and rely on the acidification of the endosome to fuse with the endosomal membrane, releasing their RNA genome into the cytoplasm. The viral proteins are translated as a single polyprotein which is post-translationally cleaved into ten different proteins: three structural proteins – capsid (C), membrane precursor (prM), and envelope (E) – as well as seven nonstructural proteins – NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5.2 The viral protease NS2B/NS3, mediates several of these cleavage events during polyprotein maturation.3 However, host proteases, such as the signal peptidase complex (SPC), are also essential for the complete polyprotein processing, including the cleavage of a short hydrophobic segment called 2K, located immediately upstream of the NS4B protein. Hence, these proteases are critical host factors in the viral life cycle.4 Once processed, viral proteins enable the replication of the viral genome and the assembly of new viral particles, which escape from cells to restart the cycle of infection.ZIKV remains a serious threat to human health, causing severe birth defects and neurological complications, such as Guillain-Barré syndrome.5 In addition, neither a ZIKV vaccine nor an effective drug has been developed, highlighting the urgent need for new ZIKV therapeutics to prevent future ZIKV outbreaks, such as the 2015 epidemic in South America.6
Over the last decade, several high-throughput screenings (HTS) of compound libraries have been performed to identify new ZIKV inhibitors or repurpose FDA-approved drugs.7–11 Interestingly, nanchangmycin (Nan), a natural product isolated from Streptomyces nanchangensis, was identified as an inhibitor of ZIKV and other Flaviviruses in a microscopy-based screening assay.12 Nanchangmycin has a wide spectrum of reported activities, including antibiotic,13 anticancer,14 antiviral,15 and antifibrotic16 effects. Like other ionophores, it has been proposed that its mechanism of action involves ion flux modulation across biological membranes;16–18 however, this alone does not fully explain its diverse biological activities. Therefore, additional mechanisms have been proposed in different models, such as inhibition of the Wnt signaling pathway14 and decreased phosphorylation of proteins like FYN, PTK2, and MAPK1/3.16
It is possible that Nan exerts its anti-ZIKV activity thanks to its ability to change ion gradients in cells. However, salinomycin, another ionophore that modulates ion concentrations across biological membranes, did not inhibit ZIKV infection when it was tested in the same high-throughput screen.10 This suggests that Nan may have a different or additional mode of action regarding its antiviral properties.
Identifying the cellular targets of Nan could help elucidate the precise molecular mechanism through which Nan exerts its antiviral effects, especially against ZIKV. To date, no proteomic approach19 has been applied to elucidate nanchangmycin's interaction partners at the proteome level in living cells in an unbiased manner.
Herein, we report the synthesis and validation of a photoreactive and clickable Nan probe (Nan-DYne). Nan-DYne was applied in human cells to identify the cellular target(s) of Nan, using quantitative mass spectrometry-based proteomics and competition with the natural product Nan. Among the prominent targets identified was the protein SEC11A, a subunit of the SPC. Subsequent experiments confirmed the binding of Nan to SEC11A, and functional studies revealed that Nan blocked the activity of the SPC in human cells. This host–protein complex is essential for ZIKV polyprotein cleavage and processing during ZIKV infection in host human cells. These findings suggest a novel mechanism for the therapeutic effect of Nan in ZIKV infection.
Results and discussion
Synthesis and validation of a Nan probe
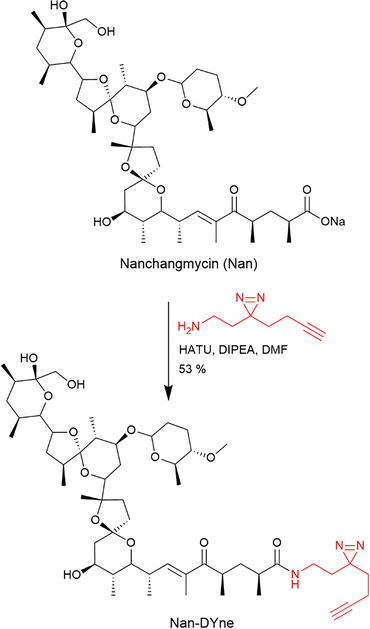
In order to assess the protein targets of Nan, we designed a photoreactive clickable Nan probe by derivatising Nan at the carboxylic acid position. This position was chosen since modification of the same position in other ionophore compounds did not significantly alter their biological activitiy.20 Nan was coupled in one step to a minimalist photoreactive clickable moiety21 to provide Nan-DYne (Scheme 1).Nan-DYne comprises: (1) a diazirine photocrosslinker moiety for UV-induced covalent capture of proteins interacting with Nan-DYne in cells; and (2) a terminal alkyne for subsequent conjugation to an azido-tagged reporter via copper Azide–Alkyne cycloaddition (CuAAC), such as a fluorophore for in-gel fluorescence visualisation or a biotin moiety for enrichment and identification of Nan-DYne binding proteins.
To validate that this modification did not affect the biological activity of Nan, we assessed the antiviral activity of Nan-DYne against ZIKV. A549 human cells were treated with 1 μM Nan, 1 μM Nan-DYne or vehicle (DMSO), and infected with ZIKV at a multiplicity of infection (MOI) of 1.
At 72 hours post-infection, viral RNA levels were measured by RT-qPCR (Fig. 1A) and the number of newly generated mature viral particles was estimated using a TCID50 (50% tissue culture infectious dose) assay (Fig. 1B). In addition, the percentage of infected cells was evaluated by flow cytometry using an anti-flavivirus envelope antibody (Fig. 1C and Fig. S1). Nan and Nan-DYne inhibited ZIKV infection to a similar extent, albeit with a slight decrease in activity for Nan-DYne, which is often observed for probe derivatives.22
A cell cytotoxicity assay was performed over 72 h to assess the cytoxicity of the probe and to rule out nonspecific effects in the antiviral assays and subsequent experiments (Fig. S2). Nan-Dyne appeared to be slightly less toxic, with a half maximal effective concentration (EC50) of 9.1 μM, compared to 4.3 μM for Nan. This difference might be attributed to the weaker ability of Nan-Dyne to chelate ions, a well-known mechanism of toxicity for Nan. Importantly, both Nan-Dyne and Nan showed weak cytotoxicity at the working concentration of 1 μM.
Gel-based and mass spectrometry-based profiling of Nan-Dyne protein targets
With this Nan probe in hand, we assessed the labelling profile of Nan-Dyne in human cells using in-gel fluorescence. Cells were treated with increasing concentrations of Nan-Dyne for 1 hour, irradiated with UV light, lysed, and Nan-Dyne-labelled proteins were coupled to azido-TAMRA-biotin via CuAAC (Fig. 2A). Nan-Dyne showed a concentration-dependent labelling, with little to no labelling observed in control samples that were not UV-irradiated (Fig. S3A). Competition experiments with the natural product Nan were performed to exclude non-specific binding.Competition with Nan resulted in a decrease of labelling in a concentration-dependent manner of a subset of proteins (Fig. S3B and C). A 50-fold excess of Nan was shown to result in the best competition. However, since Nan was toxic to cells at concentrations above 25 μM, a concentration of 0.5 μM of Nan-DYne for the labelling and 25 μM of Nan for the competition (Fig. 2B and Fig. S3C) were chosen in subsequent target identification and quantification experiments.
Next, we employed an activity-based protein profiling (ABPP) approach to identify the protein interaction partners of Nan. Cells were treated with 25 μM Nan or DMSO, followed by 0.5 μM Nan-DYne for 1 hour, irradiated with UV light, lysed, and Nan-DYne-labelled proteins were coupled to azido-TAMRA-biotin via CuAAC. Proteins were enriched on NeutrAvidin beads, enzymatically digested with trypsin, and the peptides were analysed by LC-MS/MS using label-free quantification (LFQ) (Fig. 2C and Table S1).23
Four proteins, TIMM17A, TIMM17B, TMEM126A and SEC11A were found to be significantly labelled and enriched by Nan-DYne and sensitive to a 50-fold competition with Nan (p-value < 0.01 and Log2 fold change > 2). TIMM17A and TIMM17B are components of the TIM23 mitochondrial import inner membrane translocase complex, which imports presequence-containing proteins into the mitochondrial matrix and inner membrane.24
TMEM126A, a transmembrane protein localised on the inner membrane of mitochondria, is essential for the synthesis and quality control of mitochondrial-encoded protein.25 To date, TIMM17A, TIMM17B and TMEM126A have not been associated with viral infection. Interestingly, the protein SEC11A is a subunit of the signal peptidase complex (SPC), known to be essential for the processing of viral polyproteins in flaviviruses infections, including ZIKV infection.10,26,27 Thus, we decided to further investigate the link between Nan, SEC11A and ZIKV.
Confirmation that nanchangmycin binds to SEC11A
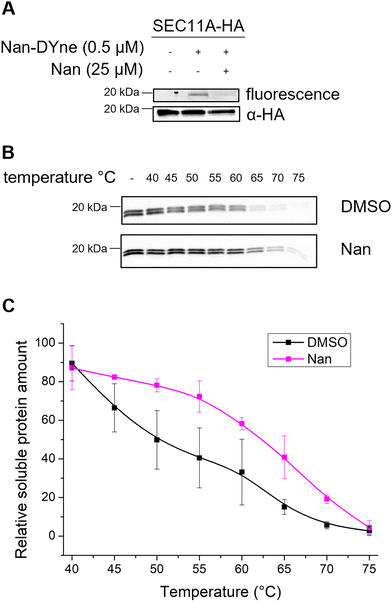
To validate that SEC11A is a target of Nan, we expressed HA-tagged SEC11A in HeLa cells and treated the cells with Nan-DYne with or without Nan competition. Cells were irradiated with UV, lysed, immunoprecipitated using anti-HA beads, and Nan-DYne-labelled proteins were coupled to azido-TAMRA via CuAAC before analysis by in-gel fluorescence and western blotting (Fig. 3A). A fluorescent band was observed in Nan-DYne treated cells, confirming the binding of Nan-DYne to SEC11A. Importantly, Nan-DYne binding and labelling appeared to drastically decrease upon Nan competition, confirming that SEC11A is a specific protein target of Nan.To further assess Nan target engagement and selectivity, we employed a cellular thermal shift assay (CETSA), which detects changes in a protein stability upon binding to a small molecule. Lysates from cells expressing SEC11A-HA were incubated with Nan or DMSO and were subjected to a 3-minute heat treatment, ranging from 40 to 75 °C. At high temperatures, proteins denature and aggregate, allowing soluble proteins to be separated by centrifugation. Soluble SEC11A-HA was then analysed and quantified by western blot analysis (Fig. 3B and C). In lysates treated with Nan, SEC11A-HA was more resistant to thermal degradation.
Taken together, these results suggest that Nan directly binds to SEC11A.
Nanchangmycin inhibits the activity of the signal peptidase complex responsible for cleaving viral polyproteins
We next tested whether Nan could disturb the function of SEC11A. SEC11A is a subunit of the SPC, which is involved in the cleavage of the ZIKV polyprotein.10,26,27 Specifically, the SPC cleaves the ZIKV polyprotein at four sites, at the N-termini of the prM, E, NS1, and between the 2K segment and NS4B proteins.In order to study the effect of Nan on the cleavage of the viral polyprotein by SPC, we transfected human cells with the ZIKV precursor protein 2K-NS4B tagged with V5 (2K-NS4B-V5).28 The blockage of this cleavage alters the maturation of NS4B protein and impairs ZIKV replication.27,29 To monitor only newly synthesised 2K-NS4B-V5, rather than steady-state 2K-NS4B-V5 that may have already been cleaved by the SPC, we metabolically labelled proteins with homopropargylglycine (HPG) for 1 hour.27 Cells were collected 12 hours post HPG-pulse to allow sufficient time for SPC-mediated processing of the HPG-labelled proteins.30 Following cell lysis, samples were labelled with azido-biotin and newly synthesized proteins were affinity-purified using NeutrAvidin beads.
In control cells, the SPC efficiently cleaved 2K-NS4B-V5 to yield NS4B-V5 (Fig. 4A). In contrast, cells treated with 1 μM Nan or 1 μM Nan-DYne exhibited a band shift corresponding to the accumulation of the full-length precursor protein 2K-NS4B-V5, indicating inhibition of the SPC-mediated cleavage. This accumulation of the full-length precursor protein 2K-NS4B-V5 was dose-dependent (Fig. 4B). Taken together, these data demonstrate that Nan impairs the cleavage of ZIKV proteins by SPC, a process that is crucial for the ZIKV life cycle.27,29
To further understand how the binding of Nan to SEC11A could inhibit the SPC function, we performed a molecular docking analysis to model their interaction using the Swissdock web tool which employs the docking algorithm AutoDockVina (Fig. S4).31,32 Ser56, His96, and Asp122 have previously been identified as the catalytic residues of SEC11A.26 Remarkably, the docking analysis revealed that the most favourable binding mode of nanchangmycin was located within the active site of SEC11A (Fig. S4).
Conclusions
In this study, we synthesised a photoreactive clickable probe of nanchangmycin (Nan-DYne) and applied it in activity-based protein profiling to identify its protein interaction partners. Strong enrichment and competition with the natural product were observed for four proteins, including SEC11A, which is a component of the SPC complex. SEC11A has previously been shown to be essential for the activity of the SPC33 and for ZIKV infection.27 Here, we confirmed that Nan directly engages SEC11A, possibly binding within its catalytic site. The SPC activity was abrogated upon treatment with Nan, suggesting a novel mechanism by which Nan inhibits ZIKV infection. Interestingly, another natural product, cavinafungin, was previously found to inhibit ZIKV infection by impairing the SPC complex activity through targeting of SEC11A.27 While additional cellular effects may contribute to the antiviral activity of Nan, this study on cavinafungin supports SPC inhibition, via SEC11A, as one important mechanism contributing to Nan's antiviral effect.The SPC is a membrane-bound complex located in the endoplasmic reticulum (ER), composed of four subunits, SPC22/23, SPC 25, SPC12, and a catalytic subunit SEC11A.26 The SPC removes signal peptides from host secretory pre-proteins but is also highjacked by flaviviruses to cleave the viral polyprotein at the N-termini of four proteins: prM, E, NS1, and NS4B proteins. Both NS1 and NS4B are non-enzymatic proteins that play critical roles in viral replication and immune evasion through interactions with viral and host components,34,35 while prM and E are structural proteins which are necessary for the formation of the viral capsid and envelope.10 Incomplete cleavage of the viral polyprotein disrupts the maturation and function of these proteins, and consequently compromises viral replication and secretion of viral particles.4,10,27,29
Accordingly, the SPC represents a promising drug target for the development of new ZIKV inhibitors, particularly through inhibition of SEC11A, as suggested by our study and previous findings on cavinafungin.27
Since ZIKV and other flaviviruses rely on the same mechanism and host processing machinery to infect cells, and given the high sequence homology of SPC cleavage sites across flaviviruses,27 SEC11A may also represent a viable antiviral target for a broader range of flavivirus infections.
Author contributions
SL: investigation, writing – review and editing. CF: investigation, – review and editing. SC: investigation, resources, writing – review and editing. DM: investigation – review and editing. ET: conceptualization, funding acquisition, investigation, methodology, resources, supervision, writing original draft, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supporting information: Table S1, supplementary figures, NMR and HRMS spectra, and experimental details. See DOI: https://doi.org/10.1039/d5cb00126a.The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the dataset identifier PXD062176. Raw data (gels and western blot analyses, NMR spectra) associated with this study are available through the Zenodo Repository and can be accessed here: https://doi.org/10.5281/zenodo.15482314.
Acknowledgements
This work was supported by research grants from the he Agence Nationale de la Recherche (ANR-20-CE44-0015 to ET), the Conseil Régional de Nouvelle-Aquitaine (AAPR2020-2019-8034510 to ET) and an Idex Univ. Bordeaux Junior chair (OPE 2019-0436 to ET). This work has benefited from the facilities and expertise of IECB NMR facility, CNRS UMR3033, Inserm US001, Univ. Bordeaux. We thank Marie-Line Andreola and Patricia Recordon-Pinson from the UB’L3 facility, TBMCore, University of Bordeaux for their help. We thank Prof. Dr Jan Rehwinkel for kindly providing pcDNA-2K-NS4B-V5. We thank Zoeisha Chinoy, IECB, ISM, Univ. Bordeaux, for her help with the HRMS analysis.References
- M.-D. Fernandez-Garcia, M. Mazzon, M. Jacobs and A. Amara, Cell Host Microbe, 2009, 5, 318–328 CrossRef CAS PubMed.
- W. Hou, R. Cruz-cosme, N. Armstrong, L. A. Obwolo, F. Wen, W. Hu, M.-H. Luo and Q. Tang, Gene, 2017, 628, 117–128 CrossRef CAS PubMed.
- H. Xing, S. Xu, F. Jia, Y. Yang, C. Xu, C. Qin and L. Shi, Virus Res., 2020, 275, 197793 CrossRef CAS PubMed.
- M. Rother and M. Naumann, Virus Res., 2021, 296, 198338 CrossRef CAS.
- T. C. Pierson and M. S. Diamond, Nat. Microbiol., 2020, 5, 796–812 CrossRef CAS.
- E. M. Lackritz, L.-C. Ng, E. T. A. Marques, I. B. Rabe, N. Bourne, J. E. Staples, J. A. Méndez-Rico, E. Harris, A. C. Brault, A. I. Ko, D. W. C. Beasley, T. Leighton, A. Wilder-Smith, J. T. Ostrowsky, A. J. Mehr, A. K. Ulrich, R. Velayudhan, J. P. Golding, P. C. Fay, A. Cehovin, N. M. Moua, K. A. Moore, M. T. Osterholm and A. D. T. Barrett, Lancet Infect. Dis., 2025, 25(7), e390–e401 CrossRef CAS.
- Y. D. Fong and J. J. H. Chu, Med. Res. Rev., 2022, 42, 1739–1780 CrossRef CAS.
- J. Zou and P.-Y. Shi, Curr. Opin. Virol., 2019, 35, 19–26 CrossRef CAS.
- L. Riva, S. Goellner, S. B. Biering, C.-T. Huang, A. N. Rubanov, U. Haselmann, C. M. Warnes, P. D. De Jesus, L. Martin-Sancho, A. V. Terskikh, E. Harris, A. B. Pinkerton, R. Bartenschlager and S. K. Chanda, J. Virol., 2021, 95(22), e00996-21 CrossRef PubMed.
- R. Zhang, J. J. Miner, M. J. Gorman, K. Rausch, H. Ramage, J. P. White, A. Zuiani, P. Zhang, E. Fernandez, Q. Zhang, K. A. Dowd, T. C. Pierson, S. Cherry and M. S. Diamond, Nature, 2016, 535, 164–168 CrossRef CAS PubMed.
- L. Zhou, J. Zhou, T. Chen, X. Chi, X. Liu, S. Pan, W. Chen, T. Wu, T. Lin, X. Zhang, Y.-P. Li and W. Yang, Antiviral Res., 2021, 196, 105210 CrossRef CAS.
- K. Rausch, B. A. Hackett, N. L. Weinbren, S. M. Reeder, Y. Sadovsky, C. A. Hunter, D. C. Schultz, C. B. Coyne and S. Cherry, Cell Rep., 2017, 18, 804–815 CrossRef CAS.
- H. A. Lardy, D. Johnson and W. C. McMurray, Arch. Biochem. Biophys., 1958, 78, 587–597 CrossRef CAS PubMed.
- M. Huang, B. Liu, R. Liu, J. Li, J. Chen, F. Jiang, H. Ding, Z. Deng and T. Liu, ACS Pharmacol. Transl. Sci., 2018, 1, 84–95 CrossRef CAS PubMed.
- J. J. Patten, P. T. Keiser, D. Morselli-Gysi, G. Menichetti, H. Mori, C. J. Donahue, X. Gan, I. do Valle, K. Geoghegan-Barek, M. Anantpadma, R. Boytz, J. L. Berrigan, S. H. Stubbs, T. Ayazika, C. O’Leary, S. Jalloh, F. Wagner, S. Ayehunie, S. J. Elledge, D. Anderson, J. Loscalzo, M. Zitnik, S. Gummuluru, M. N. Namchuk, A.-L. Barabási and R. A. Davey, iScience, 2022, 25, 104925 CrossRef CAS.
- W. Li, J. Y. Chen, C. Sun, R. P. Sparks, L. Pantano, R.-U. Rahman, S. P. Moran, J. V. Pondick, R. Kirchner, D. Wrobel, M. Bieler, S. J. H. Sui, J. F. Doerner, J. F. Rippmann and A. C. Mullen, bioRxiv, 2021, preprint DOI:10.1101/2021.10.08.463221.
- J. Rutkowski and B. Brzezinski, BioMed Res. Int., 2013, 2013, 1–31 CrossRef.
- P. J. Henderson, J. D. McGivan and J. B. Chappell, Biochem. J., 1969, 111, 521–535 CrossRef CAS PubMed.
- M. H. Wright and S. A. Sieber, Nat. Prod. Rep., 2016, 33, 681–708 RSC.
- A. Huczyński, J. Stefańska, P. Przybylski, B. Brzezinski and F. Bartl, Bioorg. Med. Chem. Lett., 2008, 18, 2585–2589 CrossRef.
- Z. Li, P. Hao, L. Li, C. Y. J. Tan, X. Cheng, G. Y. J. Chen, S. K. Sze, H.-M. Shen and S. Q. Yao, Angew. Chem., Int. Ed., 2013, 52, 8551–8556 CrossRef CAS.
- M. E. Bunnage, E. L. P. Chekler and L. H. Jones, Nat. Chem. Biol., 2013, 9, 195–199 CrossRef CAS.
- J. Cox, M. Y. Hein, C. A. Luber, I. Paron, N. Nagaraj and M. Mann, Mol. Cell. Proteomics MCP, 2014, 13, 2513–2526 CrossRef CAS PubMed.
- S. I. Sim, Y. Chen, D. L. Lynch, J. C. Gumbart and E. Park, Nature, 2023, 621, 620–626 CrossRef CAS.
- S. Poerschke, S. Oeljeklaus, L. D. Cruz-Zaragoza, A. Schendzielorz, D. Dahal, H. S. Hillen, H. Das, L. S. Kremer, A. Valpadashi, M. Breuer, J. Sattmann, R. Richter-Dennerlein, B. Warscheid, S. Dennerlein and P. Rehling, Mol. Cell, 2024, 84, 345–358.e5 CrossRef CAS PubMed.
- A. M. Liaci, B. Steigenberger, P. C. Telles de Souza, S. Tamara, M. Gröllers-Mulderij, P. Ogrissek, S. J. Marrink, R. A. Scheltema and F. Förster, Mol. Cell, 2021, 81, 3934–3948.e11 CrossRef CAS.
- D. Estoppey, C. M. Lee, M. Janoschke, B. H. Lee, K. F. Wan, H. Dong, P. Mathys, I. Filipuzzi, T. Schuhmann, R. Riedl, T. Aust, O. Galuba, G. McAllister, C. Russ, M. Spiess, T. Bouwmeester, G. M. C. Bonamy and D. Hoepfner, Cell Rep., 2017, 19, 451–460 CrossRef CAS PubMed.
- J. Hertzog, A. G. Dias Junior, R. E. Rigby, C. L. Donald, A. Mayer, E. Sezgin, C. Song, B. Jin, P. Hublitz, C. Eggeling, A. Kohl and J. Rehwinkel, Eur. J. Immunol., 2018, 48, 1120–1136 CrossRef CAS PubMed.
- D. Kiemel, A.-S. H. Kroell, S. Denolly, U. Haselmann, J.-F. Bonfanti, J. I. Andres, B. Ghosh, P. Geluykens, S. J. F. Kaptein, L. Wilken, P. Scaturro, J. Neyts, M. Van Loock, O. Goethals and R. Bartenschlager, Nat. Commun., 2024, 15, 6080 CrossRef CAS.
- P. Landgraf, E. R. Antileo, E. M. Schuman and D. C. Dieterich, Methods Mol. Biol. Clifton NJ, 2015, 1266, 199–215 CAS.
- M. Bugnon, U. F. Röhrig, M. Goullieux, M. A. S. Perez, A. Daina, O. Michielin and V. Zoete, Nucleic Acids Res., 2024, 52, W324–W332 CrossRef.
- J. Eberhardt, D. Santos-Martins, A. F. Tillack and S. Forli, J. Chem. Inf. Model., 2021, 61, 3891–3898 CrossRef CAS.
- H. Fang, C. Mullins and N. Green, J. Biol. Chem., 1997, 272, 13152–13158 CrossRef CAS.
- D. R. Perera, N. D. Ranadeva, K. Sirisena and K. J. Wijesinghe, ACS Infect. Dis., 2024, 10, 20–56 CrossRef CAS PubMed.
- Y. Wang, X. Xie and P.-Y. Shi, Antiviral Res., 2022, 207, 105423 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ±
± 