Open Access Article
Open Access ArticleRNA-binding fluorogenic probes: G-clamp conjugated with a thiazole orange derivative for screening RNA-binding small molecules
Ryosuke
Nagasawa
 ab,
Kazumitsu
Onizuka
ab,
Kazumitsu
Onizuka
 *abc,
Ryohei
Iwata
ab,
Kosuke
Tsuzuki
ab,
Kaoru R.
Komatsu
*abc,
Ryohei
Iwata
ab,
Kosuke
Tsuzuki
ab,
Kaoru R.
Komatsu
 de,
Emi
Miyashita
de,
Sayaka
Dantsuji
e,
Hirotaka
Murase
de,
Emi
Miyashita
de,
Sayaka
Dantsuji
e,
Hirotaka
Murase
 a,
Hirohide
Saito
a,
Hirohide
Saito
 df and
Fumi
Nagatsugi
df and
Fumi
Nagatsugi
 *ab
*ab
aInstitute of Multidisciplinary Research for Advanced Materials, Tohoku University, Miyagi 980-8577, Japan. E-mail: onizuka@tohoku.ac.jp; nagatugi@tohoku.ac.jp
bDepartment of Chemistry, Graduate School of Science, Tohoku University, Miyagi 980-8578, Japan
cDivision for the Establishment of Frontier Sciences of Organization for Advanced Studies, Tohoku University, Miyagi 980-8577, Japan
dCenter for iPS Cell Research and Application (CiRA), Kyoto University, Kyoto 606-8507, Japan
exFOREST Therapeutics, Kyoto 602-0841, Japan
fInstitute for Quantitative Biosciences, The University of Tokyo, Tokyo 113-0032, Japan
First published on 3rd November 2025
Abstract
RNA has emerged as an attractive target for drug discovery, increasing the importance of methods for identifying RNA-binding small molecules. Fluorescent indicator displacement (FID) assays are commonly used for screening such molecules. However, because fluorescent indicators detect hit compounds through competitive binding, developing a diverse range of indicators is essential to avoid missing potential hits. Here, we introduce novel RNA-binding fluorogenic molecular probes for FID assays by conjugating thiazole orange (TO) derivatives to the unique RNA-binding molecule G-clamp, resulting in TO-G-clamp. G-clamp was chosen for its distinct RNA-binding mode compared to TO derivatives, as demonstrated by their large-scale RNA-binding profiles. Four TO-G-clamp analogs were synthesized and evaluated, all retaining the broad RNA-binding selectivity of G-clamp. Among them, TO-G-clamp-Bn, which features a benzyl substituent on the TO moiety, exhibited the highest consistency with the RNA-binding selectivity of G-clamp. FID assays using TO-G-clamp-Bn identified unique hit compounds that were insensitive to the well-known indicator TO-PRO-1. Furthermore, SHAPE-MaP analysis revealed the RNA-binding sites of TO-G-clamp, TO-PRO-1, and one of the hit compounds, AZ191, showing that the binding sites of TO-G-clamp were in close proximity to those of AZ191.
Introduction
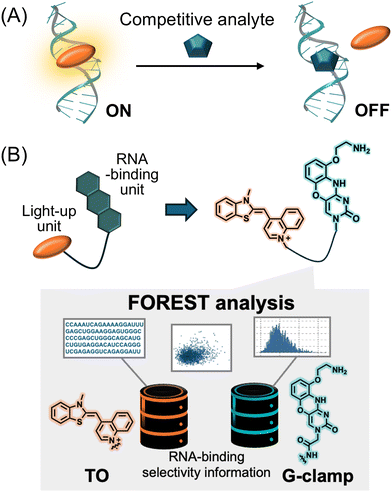
The approval of risdiplam, an RNA-targeting drug, by the Food and Drug Administration in 2020 has fueled increasing interest in RNA-targeting drug discovery.1,2 Despite extensive research efforts, the number of identified RNA-binding small molecules remains limited. As of 2025, the R-BIND database,3 which catalogs RNA-binding small molecules, contains fewer than 200 drug-like compounds. This highlights the need for new methods of identifying novel RNA-binding small molecules.One widely used screening method for discovering RNA-binding small molecules is the fluorescent indicator displacement (FID) assay.4–11 This assay uses an indicator that exhibits a fluorescence change—either Turn-ON or Turn-OFF—upon binding to a target RNA (Fig. 1A). When a competitive analyte binds to the target RNA and displaces the indicator bound to the RNA, the fluorescence intensity of the indicator changes accordingly. This fluorescence intensity change allows for the high-throughput evaluation of the RNA-binding capability of analytes. As a key advantage, the FID assay is label-free, unlike other screening techniques such as microarray-based12–15 or surface plasmon resonance (SPR)-based approaches,16 chemical cross-linking and isolation by pull-down (Chem-CLIP)-based screenings,17 and DNA-encoded library screening,18–20 where either the small molecule or the RNA must be labeled.
However, as a key limitation, the binding site of the fluorescent indicator used in the FID assay must be in close proximity to that of the RNA binder for effective displacement.21 Therefore, developing diverse fluorescent indicators with distinct binding sites and/or modes is essential. Despite this need, the number of reported fluorescent indicators remains limited.22 Designing such indicators is challenging because the molecules must not only emit fluorescence but also undergo fluorescence modulation upon binding and dissociation. Consequently, the development of structurally diverse fluorescent indicators remains a complex task.
Conjugating a light-up unit with an RNA-binding moiety is common strategy for developing RNA-binding fluorogenic molecular probes.23–26 Thiazole orange (TO) is frequently chosen as the light-up unit owing to its ability to emit bright fluorescence upon binding to DNA or RNA, offering a high signal-to-noise ratio.27,28 Several TO-conjugated molecular probes have been developed on this basis.29–35 For example, the conjugation of TO with GMP and AMP yielded TO-GMP and TO-AMP, respectively, where both probes retained the aptamer-binding abilities of GMP and AMP.29 Furthermore, Disney et al. conjugated an r(G4C2)exp-binding molecule to TO and used the resulting probe in FID assays with their in-house library containing compounds structurally similar to the r(G4C2)exp binder.31 A more selective r(G4C2)exp binder was ultimately identified. This highlights the advantage of conjugating TO with an RNA-binding unit possessing binding sites and/or modes distinct from those of TO.
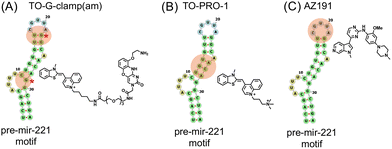
To rationally select an RNA-binding unit, large-scale RNA-binding profiles using folded RNA element profiling with structure library (FOREST) are highly useful.36,37 A previous study using this method ranked the RNA affinities of TO analogs and G-clamp across 3000 and 1824 structured RNA motifs, respectively. G-clamp is an RNA-binding small molecule that specifically recognizes single-stranded guanine in the hairpin and internal loops of RNA.37–39 The study revealed that TO and G-clamp have distinct RNA-binding selectivity.37 Therefore, conjugating TO with G-clamp was expected to confer the light-up fluorogenic function of TO to G-clamp, resulting in novel RNA-binding fluorogenic probes.
In this study, we designed and synthesized molecules that impart fluorescent light-up properties to G-clamp by conjugation with TO derivatives, termed TO-G-clamp analogs (Fig. 1B). FID assays using the TO-G-clamp analog with the closest selectivity to that of G-clamp alongside the commercially available fluorescent indicator TO-PRO-17,40 were performed to compare the hit compounds identified. Furthermore, the binding sites of the fluorescent indicators and the hit compound detected in the FID assay were determined (Fig. S1).
Results and discussion
Molecular design and synthesis of TO-G-clamp analogs
Based on the distinct RNA-binding selectivity of TO analogs and G-clamp, as analyzed by FOREST,37 TO-G-clamp derivatives that integrate a TO derivative and G-clamp via a linker were designed (Fig. 2). Four TO-G-clamp analogs were synthesized: one amide-linked molecule with TO(Me), termed TO-G-clamp(am) (Fig. 2A), and three triazole-linked molecules with TO(R), where R represents a methyl (Me), benzyl (Bn), or phenethyl (Phe) group, denoted as TO-G-clamp-R (R = Me, Bn, or Phe) (Fig. 2B). TO-G-clamp-R derivatives were designed to better retain the RNA-binding selectivity of G-clamp by the substituents that prevent RNA-binding of the TO moiety. (Fig. 2B). The substituents were chosen based on a previous study, which reported that bulky benzyl and phenethyl groups on TO suppressed non-specific interactions resulting from the planarity of TO.41 The modification of TO was expected to reduce its planarity and suppress its interaction with RNA.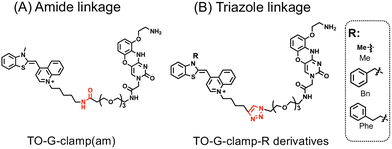 | ||
| Fig. 2 Molecular design of (A) TO-G-clamp(am) and (B) TO-G-clamp-R derivatives (R = Me, Bn, or Phe). | ||
TO-G-clamp(am) was synthesized through amide coupling to conjugate the two moieties (Scheme S1). The TO-G-clamp-R derivatives were synthesized using a copper-catalyzed click reaction to simplify the synthesis (Scheme S2).
Evaluation of fluorescent light-up properties and RNA-binding selectivity of TO-G-clamp analogs
The fluorescent light-up property of TO-G-clamp(am) was assessed using fluorometric binding assays with the pre-mir-4520-1 motif (mir-4520-1) as the target RNA (Fig. 3A), which exhibits the highest affinity for G-clamp, as demonstrated by FOREST.37 The fluorescence intensity of G-clamp decreased to 0.39 times its original intensity upon adding 1 μM of mir-4520-1, whereas the fluorescence intensity of TO-G-clamp(am) increased 77-fold owing to the light-up property of the TO moiety (Fig. 3B).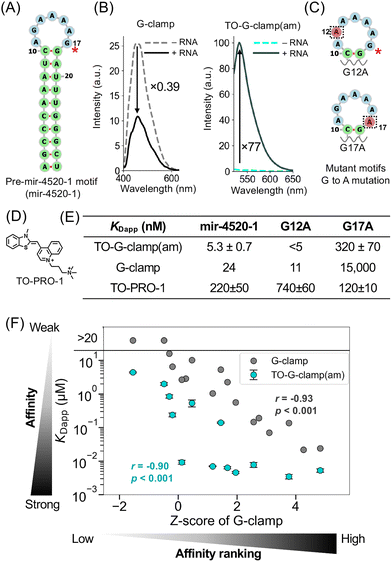 | ||
| Fig. 3 Fluorescence binding assays with TO-G-clamp(am). (A) The secondary structure of the pre-mir-4520-1 motif exhibiting the highest affinity for G-clamp. (B) Fluorescence spectra of TO-G-clamp(am) (5 nM) (left) and G-clamp (10 nM) (right) upon the addition of the pre-mir-4520-1 motif (1 μM) in phosphate buffer (pH 7.0) containing 5% DMSO. (C) The extracted secondary structure of pre-mir-4520-1 mutants, G12A and G17A, predicted by RNAfold. (D) The chemical structure of TO-PRO-1. (E) Apparent dissociation constants (KDapp) of TO-G-clamp(am), G-clamp,37 and TO-PRO-1 in mutation experiments with pre-mir-4520-1 analogs. (F) Correlation graph between the large-scale affinity score, expressed as a Z-score, of G-clamp and KDapp of G-clamp (grey)37 and TO-G-clamp(am) (cyan). Data for G-clamp and TO-G-clamp(am) are presented as mean (n = 2) and mean ± standard error (n = 3), respectively. r represents Spearman's correlation coefficient. Tests of no correlation were performed to calculate p values. One outlier (CUG repeat) was eliminated from this graph. | ||
The apparent dissociation constants (KDapp) for the interaction of TO-G-clamp(am) with mir-4520-1 and its mutants were determined through fluorescence titration assays (Fig. 3C–E). The binding affinity of TO-G-clamp(am) for mir-4520-1 was 4.5 times higher than that of G-clamp (KDapp = 5.3 ± 0.7 nM for TO-G-clamp(am) vs. KDapp = 24 nM for G-clamp37) (Fig. 3E and Fig. S2). This difference suggests that the TO moiety in TO-G-clamp(am) led to stronger binding owing to the anchor effect, as observed with other TO-conjugated probes.24,25,29,32,34 Because G-clamp binds to G17 in mir-4520-1, as demonstrated previously,37 the binding affinity of TO-G-clamp(am) for two mutants, G12A and G17A, was assessed to confirm that the binding of TO-G-clamp(am) to mir-4520-1 was driven by G-clamp (Fig. 3C and Fig. S3). As expected, G12A maintained high affinity for TO-G-clamp(am), with KDapp of <5 nM, whereas the binding affinity of G17A was substantially reduced, with KDapp of 320 ± 70 nM (Fig. 3E). As a control, KDapp was also determined for TO-PRO-1, a TO analog (Fig. 3D) in relation to mir-4520-1 and the mutants (Fig. 3E and Fig. S4). The binding preference of TO-PRO-1 was inverse to that of TO-G-clamp(am). These results confirm that TO-G-clamp(am) maintained the guanine recognition capability of G-clamp and that the binding is G-clamp-driven.
To confirm the binding selectivity, in addition to mir-4520-1, the RNA-binding affinity of TO-G-clamp(am) was further assessed with a total of 13 selected RNAs with various affinities for G-clamp (Table S1 and Fig. S2). The KDapp values for the interaction of G-clamp and TO-G-clamp(am) with the tested RNAs are summarized in Fig. S5A, excluding the outlier, CUG repetitive RNA. As expected, the KDapp values of G-clamp and TO-G-clamp(am) were highly correlated, with a Spearman's correlation coefficient (r) of 0.94. This suggests that TO-G-clamp(am) retained the RNA-binding selectivity even after conjugation with the TO moiety.
The correlation between the large-scale affinity scores, previously obtained using FOREST,37 and the KDapp values of TO-G-clamp(am) were calculated to assess the applicability of large-scale affinity scores as metrics for predicting the RNA-binding affinities of TO-G-clamp(am) (Fig. 3F). The large-scale affinity score, expressed as a Z-score, represents the relative strength of affinity for 1824 RNAs. The Z-score of G-clamp and the KDapp values of TO-G-clamp(am) were highly correlated, with r of −0.90, which is comparable to that of the original binder, G-clamp (r = −0.93). These results suggest that the binding affinities of TO-G-clamp(am) for RNAs in the library can be predicted on the basis of the Z-scores. On the other hand, TO-G-clamp(am) did not follow the binding of the TO moiety, where the RNA-binding selectivity of TO-G-clamp(am) showed poor correlation with the Z-score of the TO moiety (r = −0.57) (Fig. S6A) and KDapp of TO-PRO-1 (r = 0.2) (Fig. S5C), supporting that TO-G-clamp(am) retains the binding selectivity of G-clamp rather than that of TO.
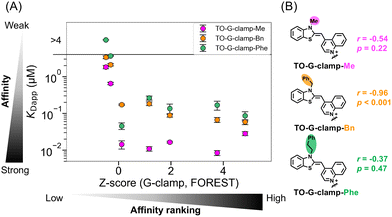
The RNA-binding affinity and selectivity of TO-G-clamp-R derivatives, TO-G-clamp-Me, -Bn, and -Phe were also assessed through fluorescence titration assays using seven RNAs (Fig. 4 and Fig. S7–S9 and Table S1). Notably, TO-G-clamp-Bn and -Phe bound to the all RNAs more weakly than TO-G-clamp-Me, indicating that modification of the TO moiety diminished the anchoring effect of TO. The correlation between KDapp for the TO-G-clamp-R derivatives and the Z-score of G-clamp was investigated to evaluate the extent to which the RNA-binding selectivity is altered by modification of the TO moiety. The KDapp of TO-G-clamp-Me showed a moderate correlation with the Z-score of G-clamp (r = −0.54) (Fig. 4). Remarkably, the RNA selectivity of G-clamp was best retained by TO-G-clamp-Bn (r = −0.96), whereas TO-G-clamp-Phe displayed the weakest correlation (r = −0.37) (Fig. 4). Furthermore, direct comparison of the KDapp of G-clamp and the TO-G-clamp-R derivatives indicated the same degree of correlation as the Z-score of G-clamp (Fig. S5B). These results suggest that TO-Bn reduced TO's RNA-binding and did not disrupt the RNA-binding selectivity of the G-clamp moiety.
To ensure that TO-Bn has less effect on the RNA-binding selectivity of TO-G-clamp-Bn, the RNA-binding selectivity of TO-Bn was analyzed using FOREST and the selectivity between TO-Bn and TO-G-clamp-Bn was compared (Fig. S6B). There was no correlation between the KDapp of TO-G-clamp-Bn and the Z-score of TO-Bn (r = −0.36), whereas the correlation between the KDapp of TO-G-clamp(am) and the Z-score of the TO moiety was slightly stronger (r = −0.57) (Fig. S6A). This suggests that TO-Bn has less influence on the RNA-binding selectivity of the G-clamp moiety of TO-G-clamp-Bn than the TO moiety of TO-G-clamp(am). Among the TO-G-clamp analogs, TO-G-clamp-Bn was selected as the indicator for the FID assays because it most effectively retained the RNA selectivity of G-clamp.
Assessment of TO-G-clamp-Bn as FID probe
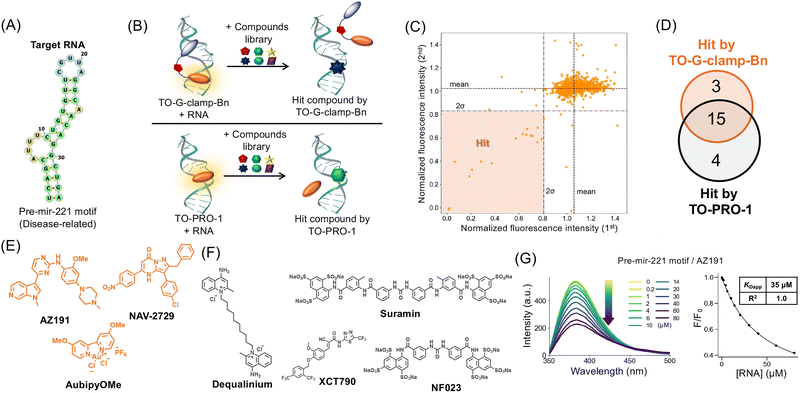
The pre-mir-221 motif was chosen as the target RNA for the FID assays because mature mir-221 is overexpressed in various cancer cells and is promising as a therapeutic target42 (Fig. 5A). Furthermore, reducing mir-221 expression levels in colorectal cancer stem cells has been shown to alleviate tumorigenic functions in mouse models.43 TO-G-clamp-Bn was selected as the fluorescent indicator in the FID assays, as noted above, and its molecular stability was evaluated. TO-G-clamp-Bn was sufficiently stable under photoirradiation and even in cell lysate, indicating its suitability for screening and potential application in living cells (Fig. S10). Additionally, the binding of TO-G-clamp-Bn to the pre-mir-221 motif was validated by SPR, providing a KD of 0.32 ± 0.1 μM, which is similar to that obtained by the fluorescence titration experiments (0.17 ± 0.006 μM) (Fig. S11). The Z′-factor, a measure of the screening quality,44 calculated using TO-G-clamp-Bn, was found to be 0.86, indicating that the probe is amenable to screening (Fig. S12A).To assess if the displacement capability of TO-G-clamp-Bn is distinct from that of TO-PRO-1, both were individually utilized for FID assays with five aminoglycosides, which are known to be less specific RNA binders.45,46 The pre-mir-4520-1 and 221 motifs were selected as target RNAs. For the pre-mir-4520-1 motif, both indicators were sensitive to several aminoglycosides (Fig. S13), suggesting that TO-G-clamp-Bn can exhibit fluorescence changes owing to displacement. In contrast, for the pre-mir-221 motif, TO-PRO-1 detected many of the aminoglycosides, whereas TO-G-clamp-Bn was inert to all of the aminoglycosides (Fig. S13). These observations suggest that the binding site of TO-G-clamp-Bn on the pre-mir-221 motif is distinct from that of TO-PRO-1.
FID assays with TO-G-clamp-Bn and TO-PRO-1 using chemical library
The Sigma-Aldrich 1280 LOPAC chemical library, consisting of 1280 compounds, was used in the assays. The hit threshold was defined as the mean minus twice the standard deviation (mean − 2σ). The assays were performed in duplicate, and hit compounds were those that met the hit threshold in both assays.Ultimately, 18 hit compounds were identified (hit rate: 1.4%) using TO-G-clamp-Bn (Fig. 5B–D). As a control, TO-PRO-1 was used in the FID assays under identical conditions, where 19 hit compounds were detected (hit rate: 1.5%) (Fig. S12B and S14). Fifteen hit compounds were common to both screens (Fig. S14). The common hit compounds are mainly classified into cationic, anionic, and planar compounds. The cationic and planar molecules were detected by both probes, possibly because of non-specific binding of the probes to RNA via electrostatic and pi–pi stacking interactions, respectively.46,47 The anionic compounds seem to interact with the positively charged probes rather than the anionic RNA, thereby inhibiting the binding of the probes to RNA, resulting in a decrease in the fluorescence.22 Myricetin, known to bind to the pre-mir-221 motif with KDapp of 20 ± 2 μM,37 was included in the common hit compounds, further demonstrating the robustness of these screenings. Notably, among the hit compounds, three (AZ191, NAV-2729, and AubipyOMe) were identified only by TO-G-clamp-Bn, whereas four others (Dequalinium, Suramin, NF023, and XCT790) were identified only by TO-PRO-1 (Fig. 5E and F). Dequalinium is known to be an RNA binder48 and is listed in the RNA binder database, R-BIND.3
To validate the screening results, the concentration-dependent fluorescence changes were measured for the seven unique hit compounds. Although AubipyOMe did not show concentration dependence, two of the three compounds identified by TO-G-clamp-Bn (AZ191 and NAV-2729) exhibited concentration dependence, with half-maximal inhibitory concentrations (IC50) of 4.8 ± 0.6 and 11 ± 0.9 μM, respectively (Fig. S15). These IC50 values were statistically significantly lower than those of TO-PRO-1 (14 ± 0.8 and 32 ± 11 μM, respectively), consistent with the data from the FID assay. Thus, TO-G-clamp-Bn was more sensitive for detecting potential hit compounds than TO-PRO-1. Conversely, two of the four compounds (Dequalinium and NF023) uniquely identified by TO-PRO-1 exhibited concentration dependence, with IC50 values of 3.7 ± 1 and 1.1 ± 0.3 μM, respectively. These values were statistically significantly lower than those for TO-G-clamp-Bn (not determined and 97 ± 4 μM, respectively), consistent with the data from the FID assay. The binding of the hit compounds was further validated by selecting four compounds exhibiting dose-dependency (AZ191, NAV-2729, Dequalinium, and NF023) for additional study.
Validation of binding of hit compounds to pre-mir-221 motif
To validate the binding of four selected hit compounds (AZ191, NAV-2729, Dequalinium, NF023) found by each indicator, the KDapp values were calculated based on SPR analysis (Fig. S16). Although AZ191 and Dequalinium showed positive responses, indicating an interaction between the pre-mir-221 motif and the hit compounds, the KDapp values could not be calculated owing to the poor fitting scores, which may be attributed to the occurrence of multiple binding events as the concentration of the compounds increased. The other two compounds (NAV-2729 and NF023) did not show positive responses, suggesting that they did not bind to the RNA and may be false positives.The KDapp values of the compounds were determined through fluorescence titration. Because the intrinsic fluorescence of AZ191 and Dequalinium exhibited changes upon binding to the pre-mir-221 motif, the KDapp values for these two compounds were calculated as 34 ± 1 μM and 2.4 ± 0.3 μM, respectively (Fig. 5G and Fig. S17). These results confirm the binding of the hit compounds (AZ191 and Dequalinium) and highlight the ability of TO-G-clamp-Bn to detect a unique hit compound.
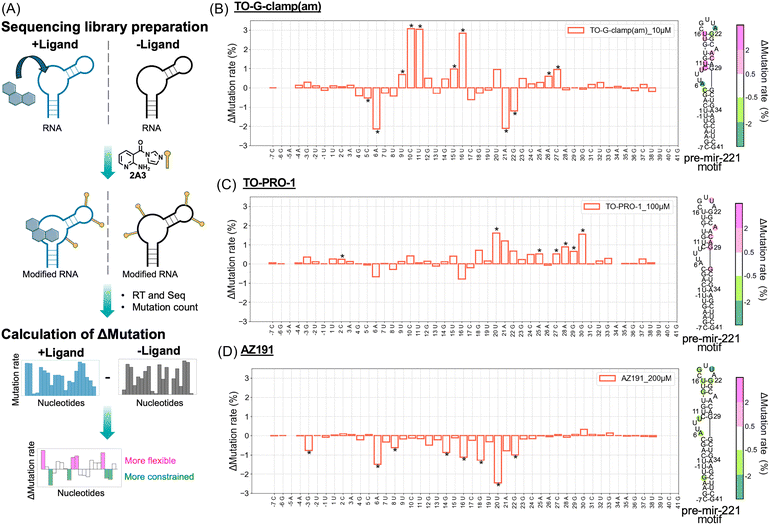
Determination of the binding sites of TO-G-clamp(am), TO-PRO-1, and AZ191
To identify the binding sites of TO-G-clamp(am), TO-PRO-1, and AZ191, we used selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP),49 a technique used to determine the binding sites of small molecules on target RNAs50 (Fig. 6A). Instead of using TO-G-clamp-Bn, TO-G-clamp(am) was selected owing to its higher affinity. The state-of-the-art SHAPE reagent 2A3 was employed for RNA acylation.51 As a positive control, the complex of thiamine pyrophosphate (TPP) and TPP riboswitch, which exhibits detectable binding,52 was applied to SHAPE-MaP. Consequently, TPP binding was clearly observed by SHAPE-MaP (Fig. S18).To further validate the accuracy of this method, the complex of mir-4520-1 and TO-G-clamp(am) (Fig. 3A–C) was used as a model for evaluation using SHAPE-MaP analysis. The mutational profile of mir-4520-1 obtained from the SHAPE-MaP analysis revealed a decrease in the mutation rate at the bound guanine and an increase in the mutation rate at the complementary base to the bound guanine (Fig. S19A). These findings suggest that the binding sites of TO-G-clamp(am) can be identified using SHAPE-MaP given that the flexibility of the bound guanine in mir-4520-1 decreased, whereas the flexibility of the complementary base increased. As a negative control, a complex of TO-G-clamp(am) with a weakly interacting RNA, the pre-mir-4282 motif, was analyzed, indicating no considerable changes in the mutation rates (Fig. S19B). This further supports the specificity of the methodologies.
The mutational profile of the pre-mir-221 motif was determined to identify its binding site. In the mutational profile of TO-G-clamp(am) and the pre-mir-221 motif, the regions surrounding the two guanines (G22 and G29), which form the closing base pairs near the hairpin loop, as well as the four nucleotide (nt) bulge loop, were considerably affected by the addition of 10 μM of TO-G-clamp(am) (Fig. 6B). These results indicate that TO-G-clamp(am) binds to both of these regions near the guanines (G22 and G29). Additionally, SHAPE-guided prediction of the secondary RNA structure in the presence and absence of TO-G-clamp(am) revealed that binding of TO-G-clamp(am) did not change the secondary structure, suggesting that its binding changed only the flexibility of the local structures (Fig. S20).
To further confirm the binding sites, the pre-mir-221 motif was mutated and the KDapp values of the mutants were determined using fluorescence binding assays (Fig. S3 and S20). The mutants were designed based on the predicted secondary structures to ensure that they maintained the structure of the pre-mir-221 motif (Fig. S21A). When the 4-nt bulge was removed from the original sequence (w/o AUUU), the binding affinity of TO-G-clamp(am) decreased considerably (KDapp: 9.3 ± 1 nM for wild type; KDapp: 22 ± 3 nM for w/o AUUU). This suggests that TO-G-clamp(am) binds to the G29 near the 4-nt bulge. Although the affinity of the mutant (G2223A), where G22 and G23 were mutated in the original RNA motif, for TO-G-clamp(am) was not significantly reduced (KDapp: <5 nM), the affinity decreased further when both guanines (G22 and G23) near the hairpin loop were mutated in the w/o AUUU, resulting in the G2223A-w/o AUUU mutant (KDapp: 120 ± 20 nM). This suggests that TO-G-clamp(am) also binds to G22 and G23 near the hairpin loop. Similar results were observed with G-clamp (Fig. S21 and S22).
Similar to TO-G-clamp(am), the binding sites of TO-PRO-1 and AZ191 were identified using SHAPE-MaP (Fig. 6C and D) and mutation experiments (Fig. S23–S25). In the SHAPE-MaP analysis, the mutational profile of TO-PRO-1 and the pre-mir-221 motif showed changes in the mutation rate at the stem region near the 4-nt bulge loop (C27-G30), although these changes were less pronounced compared to those observed with TO-G-clamp(am) (Fig. 6C). For AZ191, the mutation rates at the hairpin loop decreased considerably (Fig. 6D). Overall, the SHAPE-MaP results suggest that TO-PRO-1 binds to the stem region, whereas AZ191 binds to the hairpin loop.
Mutation experiments with TO-PRO-1 and AZ191 further support the findings of the SHAPE-MaP analysis, although the differences in the affinity among the mutants were relatively small (Fig. S23–S25). TO-PRO-1 exhibited weaker affinity for the w/o A and w/o hairpin loop mutants, which lack the stem with the A-bulge, with KDapp values of 0.62 ± 0.05 μM and 1.1 ± 0.1 μM, respectively (compared to 0.44 ± 0.09 μM for the wild type). This suggests that the A-bulge forms the stem, facilitating binding of TO-PRO-1. For AZ191, the w/o hairpin loop mutant showed the lowest affinity, with KDapp = 105 ± 8 μM (compared to 34 ± 1 μM for the wild type), indicating that AZ191 binds to the hairpin loop.
In summary, the TO-G-clamp analog more sensitively detects AZ191 binding than TO-PRO-1, plausibly because the binding site of AZ191 is located near G22, one of the binding sites of the TO-G-clamp analog (Fig. 7). In contrast, the binding site of TO-PRO-1 is farther from the hairpin loop, making it less effective at detecting AZ191 binding.
Conclusions
In this study, we developed fluorogenic molecular probes, TO-G-clamp analogs, employing G-clamp as the RNA-binding unit and TO as the light-up unit for FID assays. We leveraged the distinct large-scale RNA binding profiles of G-clamp and TO by FOREST to design indicators with unique binding modes.37 Four TO-G-clamp analogs were designed and synthesized, and their RNA-binding selectivity was evaluated. The evaluation confirmed that the RNA-binding selectivity of G-clamp was successfully retained. Specifically, TO-G-clamp-Bn, featuring a benzyl substituent on the TO moiety, minimized TO's non-specific binding and best maintained the selectivity of G-clamp. These results suggest that modifying the TO moiety with the Bn group is an advantageous strategy for designing TO-containing RNA-binding fluorogenic probes that minimize the intrinsic RNA-binding properties of the TO unit. Furthermore, our results highlight the utility of large-scale RNA binding profiles in distinguishing selectivity differences between RNA binders and predicting the RNA-binding selectivity of TO-conjugated compounds.FID assays were used to screen 1280 compounds with TO-G-clamp-Bn and the pre-mir-221 motif, a promising therapeutic target RNA. TO-G-clamp-Bn identified unique hit compounds that were insensitive by the well-known indicator, TO-PRO-1. Several binding assays confirmed that one of the three hit compounds, AZ191, specifically bound to the pre-mir-221 motif. SHAPE-MaP and mutation experiments were used to identify the binding sites of the compounds on the pre-mir-221 motif, revealing that the differences in the hit compounds identified by TO-G-clamp-Bn and TO-PRO-1 were plausibly owing to the distinct binding sites on the pre-mir-221 motif.
The fluorescent indicators developed in this study and their molecular design concept are expected to enhance the potential of FID assays and contribute to the discovery of novel RNA-binding molecules. Because this study disclosed that the attachment of the RNA-binding unit to TO changed the hit compounds in FID assays, exploration of other RNA-binding units is crucial and is under investigation in our laboratory. Although pre-mir-221 was used as the target for FID in this study, it is possible to select other disease-related RNAs from the FOREST data, and screening with different targets is also underway. Additionally, the findings of this study are crucial for the design of TO-conjugated molecular probes, not only for FID assays but also for other applications, such as intracellular RNA imaging.
Author contributions
R. N. and K. O. designed the experiments. K. O. and F. N. mentored the research. R. N., R. I., and H.M. synthesized the compounds. R. N., R. I., T. K., K. R. K., E. M., and S. D. performed analytical experiments. R. N., K. O., R. I., T. K., K. R. K., and E. M. analyzed the results. R. N. and K. O. mainly wrote the manuscript. All authors discussed the results and provided feedback on the study and manuscript.Conflicts of interest
K. R. K. and H. S. own shares of xFOREST Therapeutics Co., Ltd. All other authors declare no competing financial interests.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5cb00078e.Acknowledgements
We express our gratitude to Yuko Higashijima for her assistance with FID assays and Miyuki Aihara for her assistance with KDapp calculation. We thank the Tagen Central Analytical Facility for supporting NMR and ESI-HRMS measurements, Research equipment sharing system, Tohoku University (043 Biacore T200) for supporting SPR measurements, and the support program for young researchers from Tohoku University Technical Support Center. This work was supported in part by the Japan Science and Technology Agency (JST) FOREST program (No. JPMJFR2002 to K. O.) and SPRING program (No. JPMJSP2114 to R. N.); Grant-in-Aid for Scientific Research (B) (No. JP24K01641 to K. O. and No. JP23H02076 to F. N.), and Scientific Research on Transformative Research Areas (A) “Biophysical Chemistry for Material Symbiosis” (No. JP23H04051 to K. O.) from the Japan Society for the Promotion of Science; the Uehara Memorial Foundation (K. O.); and the research program of the Crossover Alliance to Create the Future with People, Intelligence, and Materials from MEXT, Japan.References
- A. K. Cheung, B. Hurley, R. Kerrigan, L. Shu, D. N. Chin, Y. Shen, G. O’Brien, M. J. Sung, Y. Hou, J. Axford, E. Cody, R. Sun, A. Fazal, C. Fridrich, C. C. Sanchez, R. C. Tomlinson, M. Jain, L. Deng, K. Hoffmaster, C. Song, M. Van Hoosear, Y. Shin, R. Servais, C. Towler, M. Hild, D. Curtis, W. F. Dietrich, L. G. Hamann, K. Briner, K. S. Chen, D. Kobayashi, R. Sivasankaran and N. A. Dales, J. Med. Chem., 2018, 61, 11021–11036 CrossRef CAS PubMed.
- S. Sturm, A. Günther, B. Jaber, P. Jordan, N. Al Kotbi, N. Parkar, Y. Cleary, N. Frances, T. Bergauer, K. Heinig, H. Kletzl, A. Marquet, H. Ratni, A. Poirier, L. Müller, C. Czech and O. Khwaja, Br. J. Clin. Pharmacol., 2019, 85, 181–193 CrossRef CAS PubMed.
- A. Donlic, E. G. Swanson, L. Y. Chiu, S. L. Wicks, A. U. Juru, Z. Cai, K. Kassam, C. Laudeman, B. G. Sanaba, A. Sugarman, E. Han, B. S. Tolbert and A. E. Hargrove, ACS Chem. Biol., 2022, 17, 1556–1566 CrossRef CAS PubMed.
- R. Pei and M. N. Stojanovic, Anal. Bioanal. Chem., 2008, 390, 1093–1099 CrossRef CAS PubMed.
- M. Krishnamurthy, N. T. Schirle and P. A. Beal, Bioorg. Med. Chem., 2008, 16, 8914–8921 CrossRef CAS PubMed.
- J. Zhang, S. Umemoto and K. Nakatani, J. Am. Chem. Soc., 2010, 132, 3660–3661 CrossRef CAS PubMed.
- T. Tran and M. D. Disney, Nat. Commun., 2012, 3, 1125–1133 CrossRef PubMed.
- N. N. Patwardhan, L. R. Ganser, G. J. Kapral, C. S. Eubanks, J. Lee, B. Sathyamoorthy, H. M. Al-Hashimi and A. E. Hargrove, MedChemComm, 2017, 8, 1022–1036 RSC.
- L. M. Gómez Ramos, N. N. Degtyareva, N. A. Kovacs, S. Y. Holguin, L. Jiang, A. S. Petrov, M. Biesiada, M. Y. Hu, K. J. Purzycka, D. P. Arya and L. D. Williams, Biochemistry, 2017, 56, 5288–5299 CrossRef PubMed.
- Y. Sato, S. Yajima, A. Taguchi, K. Baba, M. Nakagomi, Y. Aiba and S. Nishizawa, Chem. Commun., 2019, 55, 3183–3186 RSC.
- B. Das, A. Murata and K. Nakatani, Nucleic Acids Res., 2021, 49, 8462–8470 CrossRef CAS PubMed.
- J. L. Childs-Disney, M. Wu, A. Pushechnikov, O. Aminova and M. D. Disney, ACS Chem. Biol., 2007, 2, 745–754 CrossRef CAS PubMed.
- C. M. Connelly, R. E. Boer, M. H. Moon, P. Gareiss and J. S. Schneekloth, ACS Chem. Biol., 2017, 12, 435–443 CrossRef CAS PubMed.
- C. M. Connelly, F. A. Abulwerdi and J. S. Schneekloth, Methods Mol. Biol., 2017, 1518, 157–175 CrossRef CAS PubMed.
- F. A. Abulwerdi, W. Xu, A. A. Ageeli, M. J. Yonkunas, G. Arun, H. Nam, J. S. Schneekloth, T. K. Dayie, D. Spector, N. Baird and S. F. J. Le Grice, ACS Chem. Biol., 2019, 14, 223–235 CrossRef CAS PubMed.
- T. Fukuzumi, A. Murata, H. Aikawa, Y. Harada and K. Nakatani, Chem. – Eur. J., 2015, 21, 16859–16867 CrossRef CAS PubMed.
- B. M. Suresh, W. Li, P. Zhang, K. W. Wang, I. Yildirim, C. G. Parker and M. D. Disney, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 33197–33203 CrossRef CAS PubMed.
- Q. M. R. Gibaut, Y. Akahori, J. A. Bush, A. Taghavi, T. Tanaka, H. Aikawa, L. S. Ryan, B. M. Paegel and M. D. Disney, J. Am. Chem. Soc., 2022, 144, 21972–21979 CrossRef CAS PubMed.
- R. I. Benhamou, B. M. Suresh, Y. Tong, W. G. Cochrane, V. Cavett, S. Vezina-Dawod, D. Abegg, J. L. Childs-Disney, A. Adibekian, B. M. Paegel, M. D. Disney and J. Puglisi, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, 1–8 CrossRef PubMed.
- Q. Chen, Y. Li, C. Lin, L. Chen, H. Luo, S. Xia, C. Liu, X. Cheng, C. Liu, J. Li and D. Dou, Nucleic Acids Res., 2022, 50, E67 CrossRef CAS PubMed.
- P. N. Asare-Okai and C. S. Chow, Anal. Biochem., 2011, 408, 269–276 CrossRef CAS PubMed.
- S. L. Wicks and A. E. Hargrove, Methods, 2019, 167, 3–14 CrossRef CAS PubMed.
- J. B.-H. Tok, J. Cho and R. R. Rando, Tetrahedron, 1999, 55, 5741–5758 CrossRef CAS.
- Y. Sato, M. Kudo, Y. Toriyabe, S. Kuchitsu, C. X. Wang, S. Nishizawa and N. Teramae, Chem. Commun., 2014, 50, 515–517 RSC.
- Y. Sato, H. Saito, D. Aoki, N. Teramae and S. Nishizawa, Chem. Commun., 2016, 52, 14446–14449 RSC.
- M. H. Hu, R. J. Guo, S. Bin Chen, Z. S. Huang and J. H. Tan, Dyes Pigm., 2017, 137, 191–199 CrossRef CAS.
- B. A. Armitage, in Heterocyclic Polymethine Dyes: Synthesis, Properties and Applications, ed. L. Strekowski, Springer, Berlin, Heidelberg, Berlin, Heidelberg, 2008, pp. 11–29 Search PubMed.
- O. Suss, L. Motiei and D. Margulies, Molecules, 2021, 26, 2828 CrossRef CAS PubMed.
- R. Pei, J. Rothman, Y. Xie and M. N. Stojanovic, Nucleic Acids Res., 2009, 37, e59 CrossRef PubMed.
- D. Watkins, N. Ranjan, S. Kumar, C. Gong and D. P. Arya, Bioorg. Med. Chem. Lett., 2013, 23, 6695–6699 CrossRef CAS PubMed.
- Z. F. Wang, A. Ursu, J. L. Childs-Disney, R. Guertler, W. Y. Yang, V. Bernat, S. G. Rzuczek, R. Fuerst, Y. J. Zhang, T. F. Gendron, I. Yildirim, B. G. Dwyer, J. E. Rice, L. Petrucelli and M. D. Disney, Cell Chem. Biol., 2019, 26, 179–190 CrossRef CAS PubMed.
- E. T. T. Lee, Y. Sato and S. Nishizawa, Chem. Commun., 2020, 56, 14976–14979 RSC.
- Y. Liang, S. Miao, J. Mao, S. Devari, M. Gonzalez and D. Bong, Chem. – Eur. J., 2022, 28, e202103616 CrossRef CAS PubMed.
- E. T. T. Lee, Y. Sato, A. F. Ujuagu and S. Nishizawa, Analyst, 2024, 149, 4179–4186 RSC.
- A. W. McFarland, L. P. Fernando, P. Kellish, S. P. Story, G. B. Schober, S. Kumar, C. Gong, A. King, X. Gong, A. S. Leutou and D. P. Arya, ChemistryOpen, 2025, 14, e202400189 CrossRef CAS PubMed.
- K. R. Komatsu, T. Taya, S. Matsumoto, E. Miyashita, S. Kashida and H. Saito, Nat. Commun., 2020, 11, 6275–6288 CrossRef CAS PubMed.
- R. Nagasawa, K. Onizuka, K. R. Komatsu, E. Miyashita, H. Murase, K. Ojima, S. Ishikawa, M. Ozawa, H. Saito and F. Nagatsugi, Commun. Chem., 2024, 7, 98 CrossRef CAS PubMed.
- H. Murase and F. Nagatsugi, Bioorg. Med. Chem. Lett., 2019, 29, 1320–1324 CrossRef CAS PubMed.
- R. Nagasawa, K. Onizuka, K. Kawamura, K. Tsuzuki, H. Murase, K. R. Komatsu, E. Miyashita, H. Saito, J. Kondo and F. Nagatsugi, Chem. Commun., 2025, 61, 1120–1123 RSC.
- H. S. Haniff, A. Graves and M. D. Disney, ACS Comb. Sci., 2018, 20, 482–491 CrossRef CAS PubMed.
- C. Y. Zhou, S. C. Alexander and N. K. Devaraj, Chem. Sci., 2017, 8, 7169–7173 RSC.
- M. Garofalo, C. Quintavalle, G. Romano, C. M. Croce and G. Condorelli, Curr. Mol. Med., 2012, 27–33 CrossRef CAS PubMed.
- J. Mukohyama, T. Isobe, Q. Hu, T. Hayashi, T. Watanabe, M. Maeda, H. Yanagi, X. Qian, K. Yamashita, H. Minami, K. Mimori, D. Sahoo, Y. Kakeji, A. Suzuki, P. Dalerba and Y. Shimono, Cancer Res., 2019, 79, 5151–5158 CrossRef CAS PubMed.
- J. H. Zhang, T. D. Y. Chung and K. R. Oldenburg, SLAS Discovery, 1999, 4, 67–73 CrossRef PubMed.
- K. F. Blount, F. Zhao, T. Hermann and Y. Tor, J. Am. Chem. Soc., 2005, 127, 9818–9829 CrossRef CAS PubMed.
- M. L. Kelly, C. C. Chu, H. Shi, L. R. Ganser, H. P. Bogerd, K. Huynh, Y. Hou, B. R. Cullen and H. M. Al-Hashimi, RNA, 2021, 27, 12–26 CrossRef CAS PubMed.
- A. Taghavi, N. A. Springer, P. R. A. Zanon, Y. Li, C. Li, J. L. Childs-Disney and M. D. Disney, RSC Chem. Biol., 2025, 6, 510–527 RSC.
- S. Li, X. Y. Hwang, S. Stav and R. R. Breaker, RNA, 2016, 22, 530–541 CrossRef CAS PubMed.
- N. A. Siegfried, S. Busan, G. M. Rice, J. A. E. Nelson and K. M. Weeks, Nat. Methods, 2014, 11, 959–965 CrossRef CAS PubMed.
- S. Martin, C. Blankenship, J. W. Rausch and J. Sztuba-Solinska, Methods, 2019, 167, 105–116 CrossRef CAS PubMed.
- T. Marinus, A. B. Fessler, C. A. Ogle and D. Incarnato, Nucleic Acids Res., 2021, 49, E34–E34 CrossRef CAS PubMed.
- M. J. Smola, G. M. Rice, S. Busan, N. A. Siegfried and K. M. Weeks, Nat. Protoc., 2015, 10, 1643–1669 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |