Engineering microglial exosome-mediated microRNA-124-3p delivery for Alzheimer's disease combinational therapy
Jia
Ke
ab,
Jing
Ding
a,
Yichong
Xu
a,
Caini
Yu
a,
Yiling
Hong
a,
Sufen
Li
a,
Tingting
Meng
ac,
Yuan
Ping
 ab,
Hong
Yuan
ab,
Hong
Yuan
 ac and
Fuqiang
Hu
ac and
Fuqiang
Hu
 *ac
*ac
aCollege of Pharmaceutical Science, Zhejiang University, Hangzhou, 310058, China. E-mail: hufq@zju.edu.cn
bLiangzhu Laboratory, Zhejiang University Medical Center, Hangzhou, 311121, China
cNational Key Laboratory of Advanced Drug Delivery and Release Systems, Zhejiang University, Hangzhou 310058, China
First published on 15th October 2025
Abstract
Currently, single-target therapy and difficulty in brain drug delivery gravely impede the treatment of Alzheimer's disease (AD). The promising development of microRNA-124-3p (miR-124-3p) serves as a possibility for multiple therapeutic approaches for AD. However, the effective delivery of miR-124-3p to AD-affected brain regions remains a major challenge, primarily due to the blood-brain barrier (BBB) and the inherent instability of therapeutic miR-124-3p. Herein, we engineered miR-124-3p-enriched microglial exosomes (Exo-124-3p) as a biomimetic nanomedicine for the multifunctional treatment of AD. Exo-124-3p can traverse the BBB and facilitate activated-microglia targeting. Subsequently, the on-demand release of miR-124-3p from Exo-124-3p decreased the aggregation of β-amyloid (Aβ) plaques, attenuated the activation of microglia/astrocytes, and exhibited a valuable neuroprotective effect, thereby remolding the AD focal microenvironment. Notably, the in vivo results demonstrated that Exo-124-3p significantly improved the cognitive function in an AD mouse model. Mechanistically, it was elucidated that Exo-124-3p can bind to the 3′UTR region of MEKK3, ultimately inhibiting the MEKK3/NF-κB signaling pathway, thereby ameliorating AD neuroinflammation. Consequently, this study not only provides a promising therapeutic approach for AD combinational therapy, but also advances the development of miRNA delivery in other brain diseases.
1. Introduction
Alzheimer's disease (AD), characterized by progressive cognitive dysfunction and memory loss, is the most common neurodegenerative disorder, and clinically, there is no effective therapy available.1,2 Currently, several hypotheses, such as extracellular Aβ deposition, intracellular neurofibrillary tangles (NFTs), neuroinflammation, synaptic dysfunction, and BBB disruption, have been implicated in the onset of AD.3–7 As the central pathogenesis of AD, the amyloidogenic pathway is believed to be the main intervention target for AD treatment. Research has revealed that the accumulation of extracellular Aβ has been intimately implicated in microglial activation, neuronal apoptosis, synaptic dysfunction and neurogenesis, thereby exacerbating AD progression.8 Meanwhile, activated microglia were reported to generate inflammatory cytokines during the pre-Aβ plaque stage.9,10 Microglial activation triggers excessive pro-inflammatory cytokines, worsening neurogenesis impairment, neuroinflammation and extracellular Aβ accumulation.11 Accordingly, Aβ plaques around activated microglia and neuroinflammation caused a pernicious cycle that substantially contributed to cognitive dysfunction in AD.12,13 Many efforts have been proposed to reduce Aβ production or alleviate neuroinflammation;14–17 however, these strategies have been largely discontinued due to the simple therapeutic target and the lack of efficient drug delivery.18,19 As a result, efficacious brain drug delivery that concurrently eliminates extracellular Aβ deposits and restores normal crosstalk between neurons and microglia may provide a promising strategy for AD treatment.Recent studies have highlighted that microRNAs (miRNAs) have emerged as a promising therapeutic candidate for AD.20–22 In particular, as one of the dominant miRNAs, the level of microRNA-124-3p (miR-124-3p) was significantly decreased in AD brain regions. The stereotactic injection of miR-124-3p has been shown to temper its efficacy in alleviating AD syndromes.23–25 However, due to the autologous characteristic of miR-124-3p, which makes it vulnerable to in vivo degradation with a short half-life, inferior cell membrane permeability and inefficient brain-targeting ability, the effectiveness of miR-124-3p delivery in the AD brain remains challenging. To date, several approaches, such as receptor-mediated transcytosis,26 liposomal nanoparticles27 or polymer nanoparticles,28 have been developed to deliver miR-124-3p in vivo; however, their biocompatibility, safety and complexity greatly limit their biomedical application in the AD brain. Exosomes, a single-membrane lipid bilayer vesicle can be secreted by all cell types, exhibited prolonged tissue retention and great biocompatibility with minimal adverse immune reaction.29,30 Particularly, exosomes generated from microglial cells possess inherent surface proteins and ligands, which endowed the homing and targeting ability with superior BBB penetrability.31,32 These versatile features enable microglial exosomes to mediate efficient brain-targeted delivery of bioactive macromolecules, including proteins, nucleic acids, and miRNAs.
Microglia, the resident immune cells of the central nervous system (CNS), play a pivotal role in neuroinflammation and have emerged as promising therapeutic targets for various CNS diseases. Their unique immunomodulatory properties and innate capability to target pathological CNS environments make them particularly promising therapeutic candidates.33–35 It is important to note that classically activated microglia (M1 phenotype) release pro-inflammatory cytokines and neurotoxic substances that can be detrimental to neuronal health. In contrast, alternatively activated microglia (M2 phenotype) exert neuroprotective effects by clearing protein aggregates and facilitating tissue repair. Therefore, facilitating the transition of microglia from the M1 to the M2 phenotype represents a key therapeutic strategy for modulating the neuroimmune environment and ultimately supporting neuronal recovery in the CNS.
Herein, we fabricated the overexpressed miR-124-3p of engineered M2 microglial exosomes (Exo-124-3p) as a biomimetic and safe nanomedicine for AD combinational therapy. As depicted in Scheme 1, Exo-124-3p exerted good BBB-targeting and -penetrating capabilities, accompanied by sufficient enrichment in the brain. Our results demonstrated that Exo-124-3p exhibited synergistic effects, including diminishing the generation of Aβ1–42, reducing microglial activation and anti-apoptosis and alleviating neuroinflammation. This effective miR-124-3p delivery of Exo-124-3p allowed for cognitive function recovery in AD mice compared with monotherapy. Crucially, it was novel that Exo-124-3p significantly ameliorated neuroinflammation by inhibiting the MEKK3/NF-κB signaling pathway in the AD microenvironment. Altogether, this study reveals the potential of Exo-124-3p employed as an efficient therapeutic possibility for AD combinational treatment and sheds light on a biomimetic approach for miRNA delivery in brain disease therapy.
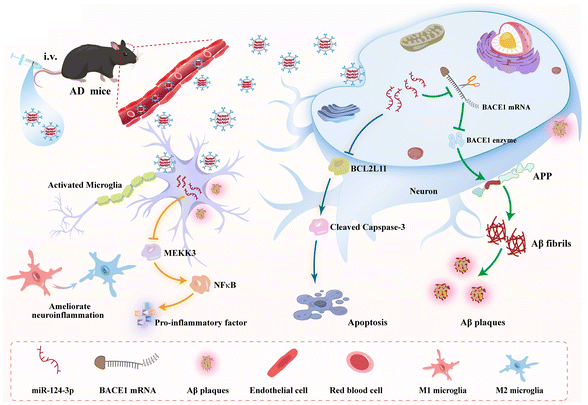 | ||
| Scheme 1 Schematic representation of the underlying therapeutic mechanism of Exo-124-3p for Alzheimer's disease combination therapy. | ||
2. Experimental sections
2.1. Fabrication of exosomes
M2-polarized BV2 cells (M2BV2) were obtained by treating BV2 cells with interleukin-4 at a concentration of 40 ng mL−1. Next, M2BV2 cells were infected with Negative Control (NC) lentivirus and miR-124-3p lentivirus to obtain NC M2BV2 cells and miR-124-3p overexpressed M2BV2 cells. First, the supernatant was collected and then centrifuged at 2000g for 20 min at 4 °C to remove the cells. The resulting supernatant was filtered through a 0.22 μm membrane and then ultracentrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 30 min at 4 °C. The final ultracentrifugation was performed at 100
000g for 30 min at 4 °C. The final ultracentrifugation was performed at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 90 min at 4 °C. The resulting pellet was resuspended in prechilled PBS to obtain purified exosomes, which were stored at −80 °C for further characterization. The particle size was determined using a particle analyzer (Litesizer 500, Anton Paar, AT). The concentration was determined through nanoparticle tracking analysis (Nanosight NS300, Malvern Instruments, UK). Transmission electron microscopy (Tecnai G2 Spirit, ThermoFisher, USA) was used to observe the morphology of the exosomes. The total protein content of the exosomes was measured using a BCA protein assay kit (P0010, Beyotime). Western blotting (WB) was used to detect the exosome markers CD63 and CD9 using the corresponding cell lysate as a control.
000g for 90 min at 4 °C. The resulting pellet was resuspended in prechilled PBS to obtain purified exosomes, which were stored at −80 °C for further characterization. The particle size was determined using a particle analyzer (Litesizer 500, Anton Paar, AT). The concentration was determined through nanoparticle tracking analysis (Nanosight NS300, Malvern Instruments, UK). Transmission electron microscopy (Tecnai G2 Spirit, ThermoFisher, USA) was used to observe the morphology of the exosomes. The total protein content of the exosomes was measured using a BCA protein assay kit (P0010, Beyotime). Western blotting (WB) was used to detect the exosome markers CD63 and CD9 using the corresponding cell lysate as a control.
2.2. Western blotting analysis
Western blotting was performed to detect M2 microglial markers. Briefly, BV2 cells were seeded and allowed to incubate at 70%–80% density; then, interleukin-4 (20 ng ml−1 and 40 ng ml−1) was added to the medium to differentiate primary macrophages into M2-type microglia. After incubation for 24 h, the cells were collected by scraping and lysed with RIPA lysis solution. Protein concentration was determined using a BCA protein quantification kit (Beyotime Institute of Biotechnology, China). In total, 30 μg proteins were separated by 6% SDS-PAGE and transferred onto PVDF membranes (Mini-PEOTEAN, Bio-Rad). The membranes were blocked with 5% BSA and probed with anti-CD206 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and anti-β-actin (1
1000) and anti-β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000) at 4 °C overnight, followed by incubation with HRP-conjugated secondary antibody at room temperature for 1 h. Bands were detected using ECL reagents and analyzed using Image Lab (Bio-Rad).
3000) at 4 °C overnight, followed by incubation with HRP-conjugated secondary antibody at room temperature for 1 h. Bands were detected using ECL reagents and analyzed using Image Lab (Bio-Rad).
2.3. Quantitative real-time PCR
The miR-124-3p, BACE1, TNF-α and IL-1β mRNA expression of PC-12 cells was evaluated by a quantitative real-time polymerase chain reaction (qRT-PCR). Briefly, PC-12 cells were seeded at a density of 2 × 104 cells per well in 6-well plates, as described above. After different treatments for 24 h, total RNA was extracted from the transfected cells using TRIzol (Tiandz Gene Technology Co., Ltd, Beijing) according to the standard protocol, and total RNA concentrations were measured using a Nanodrop spectrophotometer. Reverse transcription was conducted using the PrimeScrit™ RT Reagent Kit (TaKaRa, Shiga, Japan) in a 20 μL SYBR® Green assay. Real-time PCR (StepOne, Applied Biosystems) was used to perform amplification reactions.2.4. Investigation of inhibiting Aβ aggregation
A dot blot assay was used to investigate the aggregation of Aβ oligomers in the hippocampus, which was performed using an oligomer-specific (anti-oligomer A11, Invitrogen) antibody. After 1 week of treatment, the hippocampus of brain tissues was harvested immediately after myocardial perfusion and lysed with RIPA lysis solution. The protein concentration in the samples was determined using the Bradford protein quantification kit according to the manufacturer's instructions. Samples (3 μL, 20 μg) were blotted onto nitrocellulose membranes and air-dried. The membranes were blocked with 5% BSA for 1 h and incubated with primary antibodies (A11, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution) at 4 °C overnight. Then, the membranes were washed three times with TBST and incubated with HRP-conjugated secondary anti-rabbit antibody for 1 h at room temperature. After washing five times, the dots were detected using a ChemiDoc Touch Imaging System (Bio-Rad, USA).
500 dilution) at 4 °C overnight. Then, the membranes were washed three times with TBST and incubated with HRP-conjugated secondary anti-rabbit antibody for 1 h at room temperature. After washing five times, the dots were detected using a ChemiDoc Touch Imaging System (Bio-Rad, USA).
2.5. Evaluation of microglial polarization
The polarization of microglia was evaluated by WB, and BV2 cells were seeded in a 6-well plate at a density of 20 × 104 cells per well and incubated until 90% confluence was achieved. Cells were treated as Exo, Exo-NC and Exo-124-3p. The cells were then collected by scraping and lysed with RIPA lysis solution. In total, 30 μg proteins were separated by 10% and 6% SDS-PAGE and transferred onto PVDF membranes (Mini-PEOTEAN, Bio-Rad). The membranes were blocked with bovine serum albumin, incubated with the primary antibody overnight at 4 °C, and incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. The variations in protein loadings were normalized with an internal control (β-actin). The primary antibodies used were CD16/32 (ab228971, Abcam, UK) and CD206 (ab64693, Abcam, UK).2.6. Measurement of inflammatory cytokines
Briefly, BV2 cells were seeded at a density of 20 × 104 cells per well in a 6-well plate and incubated overnight. Cells were treated with Exo, Exo-NC and Exo-124-3p for 24 h. The supernatant was collected, and the levels of the inflammatory cytokines TNF-α and IL-1β were determined using corresponding Enzyme-Linked Immunosorbent Assay (ELISA) kits (Absin Biotechnology Co., Ltd, Shanghai, China).2.7. Immunofluorescence
After treatment, the mice were anaesthetized and perfused transcardially with saline and 4% paraformaldehyde. Their brains were immediately removed and further fixed for 24 h to perform immunostaining assays. The brains were first dehydrated in 15% sucrose and 30% sucrose at 4 °C; then, 20 μm frozen coronal sections were prepared and processed for immunofluorescence staining using primary antibodies: Iba-1 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), NeuN antibody (1
200), NeuN antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), GFAP antibody (1
200), GFAP antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) and CD206 antibody (1
200) and CD206 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200). Secondary antibodies, including Goat anti-mouse IgG Alexa Flour 488 and Goat anti-mouse IgG Alexa Flour 549, were applied for fluorescence microscope imaging (Olympus VS200, Japan).
200). Secondary antibodies, including Goat anti-mouse IgG Alexa Flour 488 and Goat anti-mouse IgG Alexa Flour 549, were applied for fluorescence microscope imaging (Olympus VS200, Japan).
2.8. Immunobiological staining study
The major organs (heart, liver, spleen, lung, and kidney) of the mice were removed along with their brains and fixed in 4% paraformaldehyde for 24 h. The main organs and brains were embedded in paraffin, and sections of 5 μm were prepared for hematoxylin and eosin (H and E) staining and Nissl staining.2.9. Establishing an AD animal model
Briefly, male C57BL/6 mice (12–14 weeks) were anesthetized and placed in a stereotaxic apparatus (RWD Life Science Co., Ltd, Shenzhen, China). Aβ1–42 oligomers (oAβ1–42) were microinjected stereotaxically into the right dorsal hippocampus (−1.8 mm lateral to the midline, 2.3 mm posterior to the bregma and 2.0 mm ventral to the skull surface) within 5 min. Subsequently, the needle was held in place for an additional 5 min to allow for diffusion and then slowly removed over 5 min. Finally, the mice were intraperitoneally administered 80 units of penicillin for 3 consecutive days to prevent infection. All animal experiments were bred and maintained in a specific pathogen free barrier facility.2.10. Morris water maze behavioral test
The Morris water maze experiment was conducted in a round pool (120 cm in diameter and 50 cm in height). At the pool midpoint, two imaginary perpendicular lines crossed to divide the pool into four quadrants, and a white platform (10 cm in diameter) was submerged 2 cm below the water surface in the middle of the northwest quadrant. The water was painted white, and the water temperature was adjusted to 21 °C ± 2 °C. The maze contained a mass of fixed visual cues on the walls. The behavioral data were acquired and analyzed using a computerized video tracking system.The AD model mice were randomly divided into five groups (n = 10). Normal C57BL/6 mice were used as healthy controls. After 1 week of treatment, as described, the spatial learning and memory ability of mice were evaluated through the Morris water maze test. If the mice found the platform within 60 s, they would stay on the platform for 10 s for memory. If the mice did not find it, they would be guided to the platform for 10 s rest. The mice were trained for 5 days. After training, the platform was removed and the spatial probe test was carried out. They were allowed to swim freely for 60 s. The swimming path and the latencies to find the platform were recorded. Average data from daily tests were used for the statistical analysis.
2.11. Luciferase reporter assay
Through the Targetscan database, we found the miR-124-3p binding site in MEKK3, and the downstream target gene is NF-κB. To construct luciferase reporter vectors, pGL3-MEKK3-3′UTR WT and pGL3-MEKK3-3′UTR MUT were purchased from Genepharma (Shanghai, China). The WT and MUT plasmids were co-transfected into PC-12 cells along with the negative control and miR-124-3p mimics, respectively. After transfection for 48 h, the cells were lysed and the relative luciferase activity was measured with the Dual-Luciferase Reporter Assay System. Renilla luciferase activity was normalized to that of firefly luciferase.2.12. Statistical analysis
Data are expressed as the mean ± SD of at least three experiments performed in triplicate. All statistical analyses were performed using GraphPad Prism 9.5 software. Statistical significance was performed using one-way ANOVA, followed by Tukey's multiple comparisons test, with the following criteria: ns: no significant difference; *p < 0.05; **p < 0.01; ***p < 0.001.3. Results and discussion
3.1. Preparation, characterization and evaluation of Exo-124-3p
First, BV2 cells were directionally polarized to M2 phenotype BV2 (M2BV2) cells by 40 ng mL−1 interleukin-4 (Fig. S1). Subsequently, M2BV2 cells were infected with the corresponding amounts of Negative Control (NC) lentivirus and miR-124-3p lentivirus (Fig. S2). Then, exosome (Exo), exosome-negative control (Exo-NC) and Exo-124-3p were harvested from M2BV2 cells, NC M2BV2 cells and miR-124-3p over-expressed M2BV2 cells using a differential ultracentrifugation method, respectively (Fig. 1A). After that, the morphology, size and related biomarkers were fully evaluated. As shown in the TEM images of Fig. 1B, the structure of Exo-124-3p was found to be canonical vesicles with a double membrane and a clearly observable cup shape, which was similar to Exo and Exo-NC. In addition, western blot assays in Fig. 1C displayed the characteristic exosome markers such as CD9 and CD63 of Exo, Exo-NC and Exo-124-3p. Subsequently, the mean diameter, concentration and surface charge of Exo, Exo-NC and Exo-124-3p were determined through nanoparticle tracking analysis (NTA) and dynamic light scattering. As depicted in Fig. 1D and S3, the particle sizes of Exo, Exo-NC and Exo-124-3p were 73.2 nm, 88.9 nm and 94.8 nm, respectively, while their concentrations were 1.07 × 1012, 4.40 × 1011 and 4.33 × 1011 particles per mL−1, respectively. Besides, both of them exhibited negative zeta potentials of −8.6 mV, −8.7 mV and −8.6 mV, respectively. Importantly, the mRNA levels of miR-124-3p in each group were determined by qRT-PCR. As depicted in Fig. 1F, the levels of miR-124-3p in Exo-124-3p were significantly increased compared with Exo and Exo-NC, suggesting that the applicable miR-124-3p transfected strategy in Exo-124-3p. Overall, these results indicate the successful preparation of engineered exosomes-miR-124-3p (Exo-124-3p).3.2. BBB penetration and brain accumulation of Exo-124-3p
Prior to applying Exo-124-3p in cell assays, we first assessed its biosafety. As shown in the MTT results in Fig. S4, Exo-124-3p exhibited low toxicity and good cytocompatibility. Then, the endocytosis study of PKH67-labeled Exo, Exo-NC and Exo-124-3p was further examined by confocal laser scanning microscopy (CLSM) and FCM. As illustrated in Fig. 2A and S5, after incubation for 6 h, Exo-124-3p exhibited a significantly higher green fluorescence intensity in PC-12 cells, with 1.27-fold cellular uptake compared with Exo, hinting at the superior cell uptake rate of Exo-124-3p (Fig. 2B). After that, the BBB penetrability of Exo-124-3p was evaluated using an in vitro BBB model (Fig. 2C). As presented in Fig. 2D, Exo-124-3p possessed the most prominent transport capability (13.39%) compared with Exo-NC (4.41%) and Exo (7.60%), which demonstrated the superiority carrier of Exo-124-3p for promoting the BBB penetrability and brain accumulation of miR-124-3p. Encouraged by the above promising performance of Exo-124-3p, the biodistribution of Exo-124-3p was further investigated. First, an AD mouse model was successfully established by injecting the oAβ1–42 into the hippocampus according to our previous work.36 Subsequently, DiR-labeled Exo, Exo-NC and Exo-124-3p were intravenously injected into AD mice; then, the fluorescent images were collected and analyzed. As illustrated in Fig. 2E and F, compared with the Exo and Exo-NC groups, a gradually enhanced DiR fluorescence signal was observed in the AD mice brain within 8 h after Exo-124-3p administration, implying the laudable ability of Exo-124-3p to penetrate the BBB and enter the brain. Meanwhile, the distribution of Exo, Exo-NC and Exo-124-3p in the main organs and brains suggested the primary enrichment of Exo-124-3p in the brain (Fig. 2G and H). Therefore, we proposed that M2 microglia-derived exosomes possess activated-microglia targeting capability, as these exosomes inherited the lymphocyte function-associated antigen 1 (LFA-1) and very late antigen-4 (VLA-4) from their parental cells (Fig. S6). These crucial proteins interact with endothelial intercellular adhesion molecule 1 (ICAM-1) and vascular cell adhesion molecule 1 (VCAM-1), thereby mediating the lateral migration and transendothelial diapedesis of exosomes across the BBB. Thus, these results proved that Exo-124-3p can function as an effective delivery carrier for endowing miR-124-3p with satisfactory BBB penetrability, high brain accumulation and retention.3.3. Exo-124-3p mitigates Aβ production
Since abnormal Aβ aggregation is a key hallmark of AD, inhibiting Aβ production and aggregation has been recognized as a potential therapeutic strategy to intervene in AD progression. The β-site APP-cleaving enzyme 1 (BACE1) is a major protease that cleaves amyloid precursor protein (APP) and generates Aβ.37,38 To validate the mechanism of Exo-124-3p for eliminating Aβ, we first investigated the BACE1 gene expression in PC-12 cells by qRT-PCR. As displayed in Fig. 3A, compared with the oAβ1–42 intervened group, both the BACE1 mRNA expression in the Exo and Exo-NC groups independently descended. In particular, the BACE1 mRNA exhibited a significant reduction in the Exo-124-3p groups (44.3% ± 0.4%). In addition, the levels of BACE1 proteins were significantly reduced after Exo-124-3p treatment in oAβ1–42-intervened PC-12 cells and AD mice (Fig. 3B–D and Fig. S7), indicating that Exo-124-3p directly downgraded the expression of BACE1 mRNA in AD focal cells. Subsequently, the contents of Aβ oligomer in the hippocampus of AD mice were also assessed by a dot blot assay in different treatments. As shown in Fig. 3E and F, the aggregation of the Aβ oligomer was strongly inhibited by Exo-124-3p in comparison with the Exo and Exo-NC groups. Overall, these results confirmed that Exo-124-3p could target BACE1 and alleviate Aβ production both in vitro and in vivo in AD.3.4. Exo-124-3p ameliorates neuroinflammation
During AD progression, chronic neuroinflammation is recognized as a significant driver of the disease.39,40 The phenotypic transformation and release of pro-inflammatory cytokines of microglia and astrocytes are involved in neuroinflammation. Thus, we investigated the M1 to M2 phenotypic repolarization of BV-2 cells under various treatments by measuring the expression of the M1 marker CD16/32 and the M2 marker CD206. As illustrated in Fig. 4A, the CD206-to-CD16/32 ratio was significantly decreased in the oAβ1–42-treated group, but pro-inflammatory status was evidently counteracted by the treatment of Exo-124-3p, in contrast to the Exo and Exo-NC groups (Fig. 4B). In addition, several pro-inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β), were determined by ELISA assay. As presented in Fig. 4C and D, the levels of TNF-α and IL-1β were sharply increased under the oAβ1–42 burden compared with the control group. In particular, incubation with Exo-124-3p significantly decreased the expression of pro-inflammatory cytokines. Furthermore, the phenotype of microglia in the AD model mice was examined by immunofluorescence staining. As shown in Fig. 4E and S8, the AD mice presented a major difference in the microglial activation state, characterized by the significant enhancement of ionized calcium-binding adapter molecule 1 (Iba1). In contrast, the level of anti-inflammatory M2-type microglia in the hippocampus of AD mice increased after treatment with Exo-124-3p. Meanwhile, compared with the substantial amount of glial fibrillary acidic protein (GFAP) red fluorescence in the AD hippocampus (Fig. 4F and S9), the Exo-124-3p-treated group presented a significantly decreased GFAP fluorescence signal, indicating lessened astrocyte activation. Additionally, quantitative analysis revealed a significant reduction in the mRNA expression of IL-1β and TNF-α, accompanied by increased expression of the anti-inflammatory cytokine interleukin 10 (IL-10) in AD brains after Exo-124-3p treatment (Fig. 4G–I). Correspondingly, the above results demonstrate that Exo-124-3p effectively remodels the AD microenvironment by alleviating glial cell activation and neuroinflammation.3.5. Exo-124-3p promotes neuroprotective effects
There is growing evidence for the tight connection between neuronal apoptosis and AD degeneration.41,42 To substantiate the neuroprotective effects of Exo-124-3p, its impact on neuronal apoptosis was then investigated by assessing several apoptotic indices. As presented in Fig. 5A–D, the expression of anti-apoptosis protein Bcl-2 in the oAβ1–42-intervened group was significantly reduced. In contrast, the administration of Exo-124-3p observably resulted in enhanced Bcl-2 expression. Additionally, compared to the oAβ1–42 group, the pro-apoptosis protein, including Bcl2-like protein 11 (BCL2L11) and Bax expression, was markedly decreased when treated with Exo-124-3p. Moreover, Annexin-V/PI staining was used for the quantitative determination of the Exo-124-3p anti-apoptotic ability (Fig. 5E). After intervention with oAβ1–42, the apoptotic positive ratio of PC-12 cells was 22.26%. On the contrary, the apoptotic positive ratio diminished to 13.30% under the treatment of Exo-124-3p (Fig. 5F), indicating that Exo-124-3p effectively alleviated neuronal apoptosis in AD.Synaptic plasticity plays a pivotal role in the learning and memory processes of AD. To evaluate the therapeutic efficacy of Exo-124-3p for neuronal plasticity, the neuronal biomarker Neuronal Nuclei (NeuN) in the hippocampus region were determined. As depicted in Fig. 5G and S10, the levels of NeuN-positive cells were significantly decreased in the saline group, implying that Aβ deposition induces the greatest destruction of neurons. Nevertheless, the treatment of Exo-124-3p resulted in a striking increase in NeuN, indicating that Exo-124-3p effectively alleviated neuronal loss and apoptosis in AD mice. Additionally, we further used the Nissl staining assay to assess the neurogenesis impact of Exo-124-3p. As shown in Fig. 5H, the number of Nissl bodies in the DG and cortex of AD mice significantly decreased. On the contrary, the administration of Exo-124-3p noticeably recovered the Nissl bodies and showed no significant difference compared with the normal group, highlighting the efficacy and function of Exo-124-3p for brain neurogenesis restoration. Collectively, these results suggested the neuroprotective effect of Exo-124-3p by preventing neuronal apoptosis, remodeling synaptic plasticity and neurogenesis in AD brains.
3.6. Exo-124-3p improved cognitive functions in AD models
Next, the therapeutic efficacy of Exo-124-3p in improving cognitive deficits in AD model mice was investigated using the Morris water maze (MWM) examination. As illustrated in Fig. 6A, after establishing AD model mice, diverse formulations were daily administered intravenously for one week, and saline was used as the negative control. As shown in Fig. 6B, AD mice treated with Exo-124-3p exhibited improved learning ability during the training process, similar to that of WT mice. In the final test, AD mice displayed prolonged escape latency (55.4 ± 4.5 s) and the least frequency across the target platform (0.4 ± 0.5 times) compared to the normal group, indicating impaired spatial learning and memory. In contrast, the treatment of Exo (47.8 ± 4.2 s) and Exo-NC (43.7 ± 6.1 s) slightly reduced escape latency. It was particularly noteworthy that the Exo-124-3p-treated group exhibited significant improvements in learning and memory function with escape latency (28.1 ± 3.0 s) and frequency across the target platform (2.0 ± 1.0 times), which was close to normal mice (19.09 ± 4.1 s), strongly highlighting the therapeutic core of miR-124-3p in Exo-124-3p in AD brain lesions (Fig. 6C–E).Finally, serum was collected from the mice in each group for subsequent analysis. The results showed that all measured biochemical markers, including alanine transaminase (ALT), aspartate transaminase (AST), urea, creatinine (CREA), creatine kinase (CK), and lactate dehydrogenase (LDH), remained within normal physiological ranges, indicating no significant toxicity in the liver, kidneys, or heart (Fig. S11). In addition, the CD9 content was quantitatively determined by ELISA assay. As shown in Fig. S12, there were no significant differences in CD9 expression among the groups, suggesting that exosomes possess low immunogenicity. Moreover, H and E staining of the major organs shows no visible damage (Fig. S13). These findings confirm that all exosomes are safe for i.v. administration. Collectively, these findings assuredly demonstrated that Exo-124-3p could markedly ameliorate cognitive function in AD model mice and represent a promising therapeutic strategy for AD.
3.7. Anti-inflammatory molecular mechanism of Exo-124-3p
Neuroinflammation has been increasingly recognized as a vital contributor to AD pathogenesis and progression.43,44 In light of the anti-inflammatory performance of Exo-124-3p, we adequately investigated the potential target genes of miR-124-3p in AD neuroinflammation. The results of the online bioinformatics tool TargetScan database indicated that miR-124-3p might target the 3′-UTR of mitogen-activated protein kinase kinase kinase 3 (MEKK3) (Fig. 7A). To verify this, a luciferase reporter was used to determine whether miR-124-3p specifically targets the MEKK3 mRNA. As illustrated in Fig. 7B, the relative luciferase activity of neurons co-transfected with pGL3-MEKK3-3′UTR WT and miR-124-3p was significantly reduced in comparison to the pGL3-MEKK3-3′UTR MUT group, which suggests that miR-124-3p binds to the 3′UTR region of MEKK3, thereby suppressing MEKK3 expression. It has been reported that the knock-down of MEKK3 suppressed the activation of nuclear factor kappa B (NF-κB) and the secretion of pro-inflammatory cytokines in BV2 cells.45,46 Thus, we examined the regulated interplay between MEKK3 and NF-κB in PC-12 cells. It can be observed that the mRNA expression of MEKK3 and NF-κB was overtly suppressed by MEKK3 siRNA (Fig. 7C and D), indicating the upstream action role of MEKK3 to NF-κB. Based on this, we further investigated the potential regulated relationship of miR-124-3p with MEKK3 and NF-κB expression in the AD microenvironment for modulating inflammatory pathogenesis. As displayed in Fig. 7E–G, the upregulation of miR-124-3p can give rise to a downregulated expression of MEKK3 and NF-κB. In addition, the level of related proteins was determined by WB assays. As shown in Fig. 7H, the up-regulated levels of MEKK3 and pP65, a critical biomarker protein of the NF-κB pathway, could be evidently observed after oAβ1–42 intervention. However, the treatment with Exo-124-3p significantly decreased MEKK3 and pP65 expression in comparison with the Exo and Exo-NC groups (Fig. 7I and J), suggesting that miR-124-3p was an initiating signal unit in regulating MEKK3 and NF-κB. Overall, these results convincingly verified that the efficient delivery of miR-124-3p in Exo-124-3p could attenuate neuroinflammation by targeting MEKK3/NF-κB pathway in the AD focal microenvironment (Fig. 7K).4. Conclusions
In summary, to address the dilemma of miR-124-3p delivery in the brain, we successfully developed an engineered miR-124-3p-enriched microglial exosome (Exo-124-3p) and evaluated multiple curative effects for AD therapy. The Exo-124-3p can effectively transport through the BBB, decrease the production of Aβ and exert anti-inflammatory effects in AD focal cells. Furthermore, it has been proven that Exo-124-3p exhibits a superior neuroprotective effect and ameliorates cognitive performance in AD mice. Ultimately, the molecular mechanisms of miR-124-3p mediating the downstream MEKK3/NF-κB signaling pathway in remodeling the AD inflammatory microenvironment were discovered. We envisage that our findings would provide an efficient therapeutic approach to halt AD pathogenesis progression and open new perspectives for the delivery of miRNAs for other neurodegenerative diseases.Author contributions
Jia Ke: investigation, methodology, validation, data curation, formal analysis, writing – original draft. Jing Ding, Yichong Xu, Caini Yu, Yiling Hong and Sufen Li: methodology, data curation, formal analysis. Tingting Meng: methodology, validation. Yuan Ping: supervision, validation. Hong Yuan: review, validation. Fuqiang Hu: conceptualization, supervision, review and editing, funding acquisition, project administration.Conflicts of interest
There are no conflicts to declare.Ethical statement
C57BL/6 mice (male, 12–14 weeks) were purchased from the Ziyuan Laboratory Animal Science and Technology Co., Ltd (Hangzhou, China). All animal experiments and procedures were strictly conducted according to the guidelines of the Laboratory Animal Ethics Committee of Zhejiang University (ZJU20250124).Data availability
Data will be made available on request.Supplementary information (SI): Experimental materials, methods and supplemental figures are available. See DOI: https://doi.org/10.1039/d5bm01080b.
Acknowledgements
The authors gratefully acknowledge the National Natural Science Foundation of China (Grant No. 82473857). The authors would like to thank Wei Yin from the Core Facilities, Zhejiang University School of Medicine, for her technical support on laser confocal analysis.References
- J. Zhang, Y. Zhang, J. Wang, Y. Xia, J. Zhang and L. Chen, Signal Transduction Targeted Ther., 2024, 9, 211 CrossRef CAS.
- S. Oveisgharan, R. S. Wilson, L. Yu, J. A. Schneider and D. A. Bennett, JAMA Neurol., 2020, 77, 1217–1224 CrossRef PubMed.
- P. Griffin, L. Apostolova, B. C. Dickerson, G. Rabinovici, S. Salloway, K. Brandt, J. Masdeu, D. Hammers, S. Raghuram, S. Hall and M. C. Carrillo, Alzheimer's Dementia, 2023, 19, 126–131 CrossRef.
- M. Tzioras, R. I. McGeachan, C. S. Durrant and T. L. Spires-Jones, Nat. Rev. Neurol., 2023, 19, 19–38 CrossRef PubMed.
- E. E. Congdon and E. M. Sigurdsson, Nat. Rev. Neurol., 2018, 14, 399–415 CrossRef CAS.
- M. Cortes-Canteli and C. Iadecola, J. Am. Coll. Cardiol., 2020, 75, 942–951 CrossRef CAS.
- T. K. Ulland and M. Colonna, Nat. Rev. Neurol., 2018, 14, 667–675 CrossRef CAS PubMed.
- F. Panza, M. Lozupone, G. Logroscino and B. P. Imbimbo, Nat. Rev. Neurol., 2019, 15, 73–88 CrossRef.
- V. Calsolaro and P. Edison, Alzheimer's Dementia, 2016, 12, 719–732 CrossRef.
- C. E. Hanzel, A. Pichet-Binette, L. S. B. Pimentel, M. F. Iulita, S. Allard, A. Ducatenzeiler, S. Do Carmo and A. C. Cuello, Neurobiol. Aging, 2014, 35, 2249–2262 CrossRef CAS PubMed.
- M. Lyman, D. G. Lloyd, X. Ji, M. P. Vizcaychipi and D. Ma, Neurosci. Res., 2014, 79, 1–12 CrossRef CAS PubMed.
- V. Bhardwaj, S. Kumari, R. Dhapola, P. Sharma, S. K. Beura, S. K. Singh, B. Vellingiri and D. HariKrishnaReddy, Inflammopharmacology, 2025, 33, 679–702 CrossRef PubMed.
- R. Ebrahimi, S. Shahrokhi Nejad, M. Falah Tafti, Z. Karimi, S. R. Sadr, D. Ramadhan Hussein, N. Talebian and K. Esmaeilpour, Metab. Brain Dis., 2025, 40, 207 CrossRef CAS PubMed.
- H. van Dyck Christopher, C. J. Swanson, P. Aisen, R. J. Bateman, C. Chen, M. Gee, M. Kanekiyo, D. Li, L. Reyderman, S. Cohen, L. Froelich, S. Katayama, M. Sabbagh, B. Vellas, D. Watson, S. Dhadda, M. Irizarry, L. D. Kramer and T. Iwatsubo, N. Engl. J. Med., 2023, 388, 9–21 CrossRef CAS.
- G. D. Rabinovici and R. La Joie, J. Am. Med. Assoc., 2023, 330, 507–509 CrossRef.
- M. T. Heneka, W. M. van der Flier, F. Jessen, J. Hoozemanns, D. R. Thal, D. Boche, F. Brosseron, C. Teunissen, H. Zetterberg, A. H. Jacobs, P. Edison, A. Ramirez, C. Cruchaga, J.-C. Lambert, A. R. Laza, J. V. Sanchez-Mut, A. Fischer, S. Castro-Gomez, T. D. Stein, L. Kleineidam, M. Wagner, J. J. Neher, C. Cunningham, S. K. Singhrao, M. Prinz, C. K. Glass, J. C. M. Schlachetzki, O. Butovsky, K. Kleemann, P. L. De Jaeger, H. Scheiblich, G. C. Brown, G. Landreth, M. Moutinho, J. Grutzendler, D. Gomez-Nicola, R. M. McManus, K. Andreasson, C. Ising, D. Karabag, D. J. Baker, S. A. Liddelow, A. Verkhratsky, M. Tansey, A. Monsonego, L. Aigner, G. Dorothée, K.-A. Nave, M. Simons, G. Constantin, N. Rosenzweig, A. Pascual, G. C. Petzold, J. Kipnis, C. Venegas, M. Colonna, J. Walter, A. J. Tenner, M. K. O'Banion, J. R. Steinert, D. L. Feinstein, M. Sastre, K. Bhaskar, S. Hong, D. P. Schafer, T. Golde, R. M. Ransohoff, D. Morgan, J. Breitner, R. Mancuso and S.-P. Riechers, Nat. Rev. Immunol., 2025, 25, 321–352 CrossRef CAS.
- V. Haage and P. L. De Jager, Mol. Psychiatry, 2022, 27, 3164–3181 CrossRef PubMed.
- Y. Qiu and F. Cheng, Curr. Opin. Struct. Biol., 2024, 85, 102776 CrossRef CAS PubMed.
- D. Shekho, R. Mishra, R. Kamal, R. Bhatia and A. Awasthi, AAPS PharmSciTech, 2024, 25, 207 CrossRef.
- D. Bhatnagar, S. Ladhe and D. Kumar, Mol. Neurobiol., 2023, 60, 5954–5974 CrossRef CAS PubMed.
- E. Lauretti, K. Dabrowski and D. Praticò, Ageing Res. Rev., 2021, 71, 101425 CrossRef CAS PubMed.
- O. Polzer, R. E. Randoe, S. Snoeck, J. Verhaagen, P. J. Lucassen, E. Salta and C. Fitzsimons, Alzheimer's Dementia, 2023, 19, 079625 Search PubMed.
- A. Évora, G. Garcia, A. Rubi, E. De Vitis, A. T. Matos, A. R. Vaz, F. Gervaso, G. Gigli, A. Polini and D. Brites, Front. Pharmacol., 2025, 16, 1474012 CrossRef.
- W. Wuli, S.-Z. Lin, S.-P. Chen, B. A. Tannous, W.-S. Huang, P. Y. Woon, Y.-C. Wu, H.-H. Yang, Y.-C. Chen, R. L. Fleming, J. T. Rogers, C. M. Cahill, T.-J. Ho, T.-W. Chiou and H.-J. Harn, Int. J. Mol. Sci., 2022, 23, 10554 CrossRef CAS.
- Q. Ouyang, K. Liu, Q. Zhu, H. Deng, Y. Le, W. Ouyang, X. Yan, W. Zhou and J. Tong, Small, 2022, 18, 2107534 CrossRef CAS PubMed.
- Y. Liu, M. Zheng, M. Jiao, C. Yan, S. Xu, Q. Du, M. Morsch, J. Yin and B. Shi, Biomaterials, 2021, 276, 121036 CrossRef CAS PubMed.
- C. Saraiva, J. Paiva, T. Santos, L. Ferreira and L. Bernardino, J. Controlled Release, 2016, 235, 291–305 CrossRef CAS.
- H. Yang, M. Han, J. Li, H. Ke, Y. Kong, W. Wang, L. Wang, W. Ma, J. Qiu, X. Wang, T. Xin and H. Liu, ACS Nano, 2022, 16, 14503–14516 CrossRef CAS.
- Nat. Biotechnol., 2020, 38, 1150–1150.
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, 6977 CrossRef.
- T. Wu, Y. Liu, Y. Cao and Z. Liu, Adv. Mater., 2022, 34, 2110364 CrossRef CAS.
- H. Wang, H. Sui, Y. Zheng, Y. Jiang, Y. Shi, J. Liang and L. Zhao, Nanoscale, 2019, 11, 7481–7496 RSC.
- C. Jiang, X. Yang, Q. Huang, T. Lei, H. Luo, D. Wu, Z. Yang, Y. Xu, Y. Dou, X. Ma and H. Gao, Adv. Mater., 2025, 37, 2417869 CrossRef CAS PubMed.
- G. Cheng, A. Xie, Z. Yan, X. Zhu, Y. Song and T. Chen, Brain-X, 2023, 1, e27 CrossRef.
- Y. Wei, X. Xia, X. Wang, W. Yang, S. He, L. Wang, Y. Chen, Y. Zhou, F. Chen, H. Li, F. Peng, G. Li, Z. Xu, J. Fu and H. Gao, Acta Pharm. Sin. B, 2025, 15, 1098–1111 CrossRef CAS PubMed.
- J. Ke, C. Yu, S. Li, Y. Hong, Y. Xu, K. Wang, T. Meng, Y. Ping, Q. Fu, H. Yuan and F. Hu, Adv. Healthcare Mater., 2024, 13, 2302939 CrossRef CAS.
- X. Gu, X. Xu, C. Jia, J. Wang, J. Zhang, Q. Gao and J. Chen, Mol. Neurobiol., 2023, 60, 3569–3583 CrossRef CAS PubMed.
- J. Huang, D. Chen, X. Lin, C. Yang and X. Lin, Hippocampus, 2023, 33, 96–111 CrossRef CAS.
- V. D. Zingale, A. Gugliandolo and E. Mazzon, Int. J. Mol. Sci., 2022, 23, 90 CrossRef CAS.
- C. Gao, J. Jiang, Y. Tan and S. Chen, Signal Transduction Targeted Ther., 2023, 8, 359 CrossRef PubMed.
- G. Perry, A. Nunomura, P. Lucassen, H. Lassmann and M. A. Smith, Science, 1998, 282, 1265–1265 CrossRef PubMed.
- S. Söllvander, E. Nikitidou, R. Brolin, L. Söderberg, D. Sehlin, L. Lannfelt and A. Erlandsson, Mol. Neurodegener., 2016, 11, 38 CrossRef PubMed.
- Y. Liu, Y. Tan, G. Cheng, Y. Ni, A. Xie, X. Zhu, C. Yin, Y. Zhang and T. Chen, Adv. Mater., 2024, 36, 2307081 CrossRef CAS.
- G. C. Tracy, K.-Y. Huang, Y.-T. Hong, S. Ding, H. A. Noblet, K. H. Lim, E. C. Kim, H. J. Chung and H. Kong, Nano Lett., 2023, 23, 10971–10982 CrossRef CAS.
- X. He, X. Wang, L. Yang, Z. Yang, W. Yu, Y. Wang, R. Liu, M. Chen and H. Gao, Acta Pharm. Sin. B, 2022, 12, 1987–1999 CrossRef CAS PubMed.
- J. Sun, Y. Zhang, Y. Kong, T. Ye, Q. Yu, S. Kumaran Satyanarayanan, K.-P. Su and J. Liu, Brain, Behav., Immun., 2022, 106, 76–88 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2026 |







