Open Access Article
Open Access ArticleAnalytical applications of gas-phase ion chemistry enabled by mass spectrometry
Woo-Young Kang and
Boone M. Prentice *
*
Department of Chemistry, University of Florida, 214 Leigh Hall, PO Box 117200, Gainesville, FL 32611, USA. E-mail: booneprentice@chem.ufl.edu; Fax: +1 352 392-4651; Tel: +1 352 392-0556
First published on 6th January 2026
Abstract
Analytical mass spectrometry (MS) has been employed to study a wide variety of analytes, including metabolites, lipids, pharmaceutical compounds, pesticides, petroleum, peptides, proteins, protein complexes, nucleic acids, and glycans. The field of gas-phase ion chemistry in mass spectrometry studies the impacts of the chemical behaviors and properties of ions produced from these samples, along with instrumental and methodological parameters of the MS experiment, in determining the appearance of the resulting mass spectra. This subfield includes the dynamic interactions of the ions with neutral molecules, electrons, photons, and other ions. These interactions are particularly useful in tandem mass spectrometry (MS/MS or MSn) experiments, which provide high specificity and enable analyte structural characterization by fragmenting a compound of interest and then analyzing the product ions. These bimolecular interactions can also result in non-dissociative processes, leading to ion transformation, charge alteration, or the formation of ion/molecule and ion/ion complexes. Such reactions offer valuable insights into chemical behavior across various reaction environments by simulating those conditions within the mass spectrometer. This information can then be used to make novel inferences about the sample and promises to inform MS studies in areas such as metabolomics, lipidomics, drug pharmacology, exposomics, proteomics, glycomics, genomics, and environmental, supramolecular, and interstellar chemistry. In this review, we highlight novel applications of tandem mass spectrometry that have been published in the past ten years, focusing on reactions that take place in the gas phase in a reduced-pressure environment (i.e., ion/neutral atom/molecule, ion/electron, ion/photon, and ion/ion reactions), largely after the ionization step.
1. Introduction
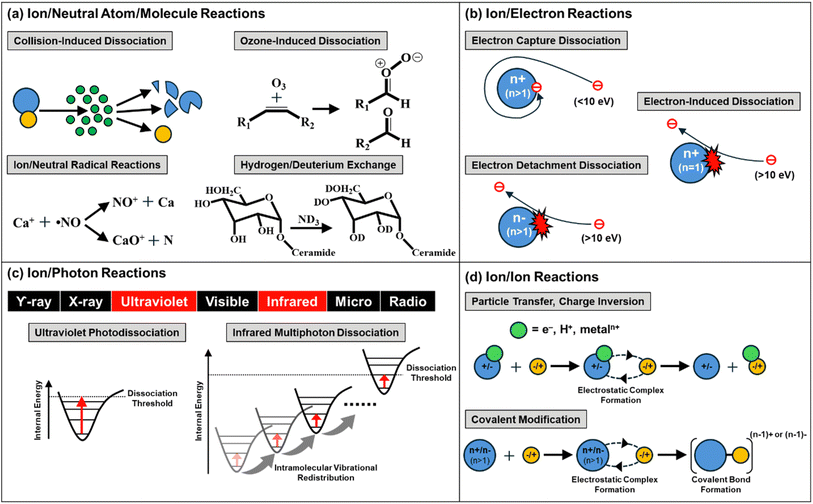
Gas-phase ion chemistry is important in determining the spectra produced from tandem mass spectrometry (MS/MS or MSn) experiments. Gas-phase interactions between ions and neutral molecules, electrons, photons, or oppositely charged ions can cause chemical and physical changes to the analyte ion, such as altering the charge state and breaking bonds, which can produce structural information on the analyte ion. A wide variety of gas-phase reactions have been reported in recent years, leveraging a number of advantages, including proceeding faster than equivalent reactions in the condensed phase and enabling high reaction purities via precise computer-controlled timing and admission of the reactants. Herein, we briefly discuss the fundamental principles and mass spectrometry instrumentation of these gas-phase reactions before focusing on recent novel applications. More comprehensive reviews on reaction mechanisms can be found elsewhere.1–5 This introduction of gas-phase reactions is not intended to be an exhaustive description of these fields. Instead, this prologue is meant to introduce these reactions and provide a context for understanding the recent applications of this technology to analytical challenges highlighted in later sections. Readers are directed to other review articles for more in-depth discussions of ion/neutral atom/molecule reactions,6–8 ion/electron reactions,9 ion/photon reactions,10–13 and ion/ion reactions,14–17 as well as the specific gas-phase ion chemistry of lipids,18 glycans,19 and proteins.4 We aim to describe a wide range of gas-phase chemistry applications to demonstrate the versatility of the reactions and highlight the potential for broader adoption. The types of reactions discussed in this review are summarized in Fig. 1.2. Applications of ion/neutral atom/molecule reactions
Ion/neutral atom/molecule reactions occur between analyte ions and inert or reactive neutral gas molecules.20 In this review, we discuss the recent applications of four different dissociative or non-dissociative ion/neutral atom/molecule reactions, each reaction presenting unique gas-phase chemistries and requiring specific instrumentation: collision-induced dissociation (CID), ozone-induced dissociation (OzID), ion/neutral radical reactions, and hydrogen/deuterium exchange (HDX). CID is the most commonly used form of ion/neutral atom/molecule reaction in mass spectrometry and is available on most MS instrumentation, representing the most widely accessible dissociation method in MS/MS for structural identification of a variety of samples.21 CID generally involves collisions of accelerated ions with inert buffer gas atoms and molecules, such as He, Ar, or N2, which converts the relative translational energy into the internal energy of the ions, resulting in fragmentation from vibrationally excited states of the electronic ground state.22 Another dissociation method, OzID, is increasingly utilized to induce oxidative dissociation using reactive neutral reagents (i.e., ozone). OzID is mainly used to locate carbon–carbon (C–C) double bonds in a wide variety of samples, including unsaturated fatty acids (FAs),23 glycerolipids,24 glycerophospholipids,24,25 cholesteryl esters,26 and sphingolipids,27,28 as well as in amphiphilic small metabolites containing olefinic hydrocarbon chains29 and isoprene chains.30 Ion/neutral radical reactions also involve using reactive neutral reagents (e.g., radical molecules) to facilitate dissociative or non-dissociative reactions. These reactions, for example, can be used to study chemistry in the atmosphere and interstellar space, where naturally occurring free radicals are common due to processes such as ultraviolet (UV) photolysis.31,32 HDX is a non-dissociative, deuterium labelling method that provides structural information for various analytes, including carbohydrates,33–35 small molecules,34,36 and proteins.37–39 Ion/neutral atom/molecule reactions using a variety of other neutral reagents, including hydrocarbons,40 non-radical reactive oxygen/nitrogen species,41 organic acids,42 and aryl radicals43 are discussed elsewhere.2.1. Collision-induced dissociation
The type of instrument determines the range of accessible energies and timescales of CID experiments, which in turn determines the types of fragment ions that can be generated. For example, quadrupole ion traps and Fourier transform ion cyclotron resonance (FT-ICR) traps are typically operated under multiple collision conditions at low energies ranging from <1 to 100 eV and timescales of tens of milliseconds to seconds.44| [M + nH]n+ + N (gas) → [M + nH]n+* + N* → fragmentation | (1) |
The internal energies generated from these collisions progressively accumulate with each collision, eventually exceeding a dissociation threshold within the ion and resulting in fragmentation at the weakest bonds via the lowest energy paths.22 Conversely, sector and time-of-flight instruments are generally operated at keV collision energies under single collision conditions, resulting in less selective dissociation of analytes.21 CID employing keV collisions is less common in recent years and is thus beyond the scope of this discussion.45 In this review, we instead briefly highlight applications of low-energy CID for covalent and noncovalent bond dissociation in the analysis of lipids, peptides, and proteins.
Low-energy CID of lipid ions generated by so-called soft-ionization methods (i.e., those that tend to produce intact ions with minimal fragmentation) such as electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI) preferentially breaks the most labile bonds. For example, this process results in head group loss from protonated glycerophospholipids and ester bond cleavage in deprotonated glycerophospholipids, enabling the identification of lipid subclasses and fatty acyl chains, respectively.18 However, this approach generally does not provide information on intrachain modifications, such as the sites of C–C double bonds,46,47 which necessitates changing the ion type. Chemical derivatization of C–C double bonds via Paternò–Büchi reaction,48 epoxidation,49 or oxidation50 before ionization allows unsaturation site-specific cleavages upon CID and produces diagnostic fragment ions. Fragmentation of unsaturated fatty acyl chains via radical-directed dissociation can also aid in unambiguously identifying C–C double bonds. For example, lipid ions noncovalently modified with bicarbonate during ESI46,51 or covalently derivatized with 2,2,6,6-tetramethylpiperidine-1-oxyl-benzyl (TEMPO) reagents before ionization52 generate radical ions upon CID, which undergo nonselective dissociation along fatty acid chains. TEMPO-assisted radical-directed dissociation of phosphatidylethanolamines (PEs) produces extensive FA chain fragment ions by radical formation via the homolysis of the labile bond between the benzylmethyl carbon and the piperidine oxygen in the TEMPO reagent during CID.52 The presence of doublet fragment ions differing by 2 m/z units following radical-directed dissociation suggests that the loss of two hydrogen atoms occurs at C–C double bonds during their cleavage, thus pinpointing the locations of these unsaturation sites. This method effectively differentiates phosphatidylethanolamines with various combinations of intrachain modifications from an E. coli lipid extract (Fig. 2).52 Overall, this demonstrates the importance of the ion type in determining the type and extent of fragmentation observed during CID.
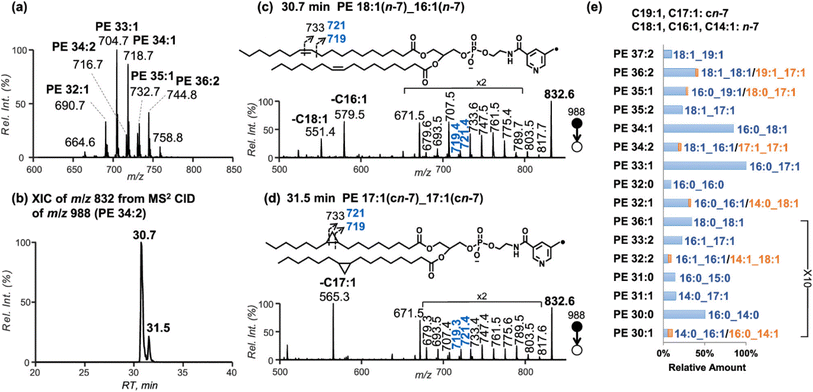 | ||
Fig. 2 (a) The PE profile of E. coli via neutral loss scan of 141 Da in positive-ion mode. (b) Extracted-ion chromatogram of m/z 832 from MS2 CID of [PETPN 34:2 − H]− (m/z 988) and the corresponding spectra for the peaks eluted at (c) 30.7 and (d) 31.5 min, respectively. (e) Identified PEs from E. coli with relative ion abundances normalized to PE 33:1. The locations of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and cyclopropane in fatty acyls are indicated at the top of (e). TPN: 3-(2,2,6,6-tetramethylpiperidin-1-yloxymethyl)-picolinic acid 2,5-dioxopyrrolidin-1-yl ester. Adapted/reproduced from Journal of American Society for Mass Spectrometry, 2022, 33, 714–721 (ref. 52) with permission from the American Society for Mass Spectrometry, Copyright 2022. C and cyclopropane in fatty acyls are indicated at the top of (e). TPN: 3-(2,2,6,6-tetramethylpiperidin-1-yloxymethyl)-picolinic acid 2,5-dioxopyrrolidin-1-yl ester. Adapted/reproduced from Journal of American Society for Mass Spectrometry, 2022, 33, 714–721 (ref. 52) with permission from the American Society for Mass Spectrometry, Copyright 2022. | ||
Low-energy CID of peptide ions generated via ESI leads to amide bond fragmentation, producing b-/y-type fragment ions along the backbone.53 Labile post-translational modifications (PTMs), such as sulfation and phosphorylation,54,55 are generally not retained due to the slow-heating nature of multiple low-energy collisions. However, CID of peptide ions derivatized with TEMPO reagents leads to homolytic bond cleavage in the reagent and produces radical ions. Subsequent radical-directed dissociation results in the production of a˙-/b-/c-/x-/y-/z˙-type fragment ions while preserving PTMs.54,55 Additionally, charge-remote fragmentation via radical-directed dissociation results in more extensive fragmentation across the peptide backbones56–58 than traditional charge-directed cleavages via mobile protons in CID.53,59 Charge-remote fragmentation is particularly useful in enhancing the sequence coverage of large native proteins and protein complexes with relatively low charge states and, thus, low CID efficiency.60 For example, TEMPO-assisted radical-directed dissociation offers approximately 15% greater sequence coverage compared to traditional CID for bovine serum albumin (66 kDa), about 30% more sequence coverage for Concanavalin A tetramer (106 kDa), and a nearly 50% increase for β-lactoglobulin A dimer (36 kDa).60 TEMPO-assisted radical-directed dissociation also enables 3D protein structural analysis by facilitating through-space hydrogen atom radical transfer from the radical site to distant residues in sequence that are close in space. For example, the loss of the TEMPO moiety from the N-terminal benzyl succinic acid conjugate during CID results in the formation of a benzyl radical (Fig. 3), which subsequently induces radical-directed dissociation at C-terminal residues (e.g., E64, S65, and L67) in denatured 5+ ubiquitin, indicating a spatial proximity between the N- and C-terminal regions.61 However, higher ubiquitin charge states do not show fragmentation in this region, suggesting a larger spatial distance between the two termini and a more unfolded conformation compared to 5+ ubiquitin.61 Overall, the use of free radical initiator-assisted radical-directed dissociation for analyzing lipids, peptides, and proteins highlights the versatility of CID and demonstrates that limitations to conventional CID can be addressed without necessitating modifications to the instrument. The TEMPO experiments also do not require MS3 in an ion trap and can effectively utilize beam-type CID at the MS2 level,56 allowing this approach to be implemented in various commercial instruments.
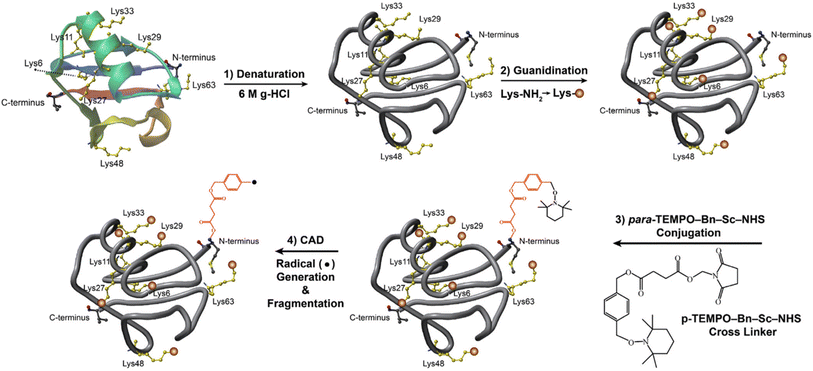 | ||
| Fig. 3 (1–3): p-TEMPO–Bn–Sc–NHS conjugation to guanidinated ubiquitin and (4) FRIPS MS. The ubiquitin protein solutions were first denatured (1), and seven lysines were blocked by guanidination reactions (2), where orange circles denote guanidinated lysines. The only remaining primary amine site, N-terminus, was conjugated with p-TEMPO–Bn–Sc–NHS (3). The resulting guanidinated p-TEMPO–Bn–Sc–ubiquitin was then thermally activated, and a free radical is generated by TEMPO radical loss (4). The loss of the TEMPO radical and the generation of the benzyl radical are represented by the loss of a NHS group. Note that the denatured structure in the figure is not the actual one. Adapted/reproduced from Journal of the American Society for Mass Spectrometry, 2022, 33, 471–481 (ref. 61) with permission from the American Society for Mass Spectrometry, Copyright 2022. | ||
While CID is primarily used for breaking covalent bonds, collisional activation is also increasingly employed to study the bonding dynamics of noncovalent interactions. For example, collision-induced unfolding (CIU) of higher-order protein structures entails the breakdown of noncovalent interactions within the protein, which is determined by measuring collision cross-sections at a range of collisional activation energies using ion mobility mass spectrometry (IM-MS).62–64 CIU50 denotes the acceleration voltage at which 50% of a protein conformer undergoes conformational transitions.65 Determination of CIU50 values for specific ligands and proteins can serve as measures of ligand affinity to binding sites66 and protein conformational stability upon binding67 in protein–ligand complexes. For example, the shift in CIU50 of the pikromycin PKS module 6 (PikAIV) from 45.3 ± 0.0 V to 52.0 ± 0.8 V when bound to the ligand hexaketide indicates an increased conformational stability of the protein–ligand complex relative to other ligands.68 Collisional activation can also provide information on the noncovalent interactions in supramolecular chemistry between a cage-like, synthetic host molecule (H) and its guest molecule (G) encapsulated in the cavity.69,70 Similar to CIU of protein–ligand complexes, CID of H–G complexes obtains survival yield of the complex as a function of collisional activation time and measures the selectivity and affinity of the noncovalent bonds.71–73 The gas-phase dissociation kinetics of H–G complexes allows for studying artificial enzyme and self-assembly models that mimic those operating in nature, while facilitating a better understanding of the intricate characteristics of the relevant noncovalent interactions in a simpler and more controlled environment.69,70 This gas-phase approach also provides more accurate measurements of intrinsic binding energies than the conventional condensed-phase counterpart, as it eliminates interferences from solvation and counterions present in the solution.69,70 Overall, the use of CID for the selective dissociation of noncovalent interactions in protein–ligand and H–G complexes highlights the tunability and versatility of collisional activation.
2.2. Ozone-induced dissociation
OzID involves introducing gaseous ozone into various parts of commercial MS instruments, such as ion sources,74,75 ion traps,76,77 or ion mobility cells.78 External ozone generators produce ozone for injection into MS instruments via a gas inlet system, which requires relatively simple hardware modifications.7,24 The OzID mechanism proceeds through two competitive oxidative cleavage pathways.79 The first is the concerted cycloaddition of an ozone molecule to a C–C double bond to generate an unstable ozonide, which spontaneously dissociates into diagnostic Criegee and aldehyde ions 16 Da apart:80
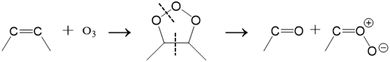 | (2) |
The second, non-classical DeMore mechanism is a biradical stepwise addition of ozone to the C–C double bond that results in a diagnostic ketone and epoxide ([M + 16]+) ions.81
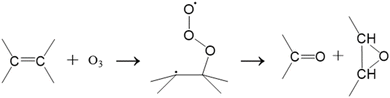 | (3) |
Identifying these diagnostic fragment ions reveals the locations of C–C double bonds in lipids, providing insights into the relationship between unsaturation levels and various physiological conditions, such as cell death82–84 and diseases.85–88 In this review, we discuss the applications of OzID, highlighting uses in MALDI imaging mass spectrometry and shotgun lipidomics.
Imaging mass spectrometry allows for the spatial mapping of analytes sampled directly from tissue surfaces.89,90 Due to the lack of orthogonal separation techniques (e.g., gas or liquid chromatography) compatible with pixel-by-pixel, millisecond timescale acquisition, it can be challenging to identify analyte isomers without MS/MS.91 OzID has been performed on unsaturated glycerophospholipids from biological tissues,25 such as human prostate92 and mouse brain,93 to reveal the spatial distributions of olefinic C–C double bond positional isomers. Sequential CID (MS2) and OzID (MS3) provides additional information on sn-positional isomers of glycerophospholipids by producing structurally specific fragment ions containing sn-1 and sn-2 fatty acyl chains.25,47,78,94,95 For example, CID/OzID of lipids sampled from rat brain tissue provides for the identification of four phosphatidylcholine (PC) 34:1 isomers, PC 16:0/18:1(n-7), PC 16:0/18:1(n-9), PC 18:1(n-7)/16:0, and PC 18:1(n-9)/16:0, with the varying fractional distributions across different subsections of the tissue.78 The use of sequential CID/OzID experiments in imaging mass spectrometry highlights the versatility of OzID in enabling high-throughput lipid isomer identification with spatial context in tissues.
OzID can also be coupled with chromatographic approaches96 or integrated into shotgun lipidomics26 for data-independent analysis (DIA) of the lipidome extracted from various samples, including blood plasma and cell cultures. In this context, the goal is primarily to resolve C–C double bond isomers of fatty acids that are often obscured in sum composition assignments in traditional shotgun workflows. For example, the analysis of the human plasma lipidome using combined DIA and OzID reveals different abundances of double bond positions in different lipid classes. Triacylglycerols (TGs) containing 50–52 carbon atoms have a high abundance of n-9, n-7, and n-6 double bonds, while phosphatidylcholines with 34–38 carbon atoms show a relatively low abundance of these double bonds. Additionally, n-4 double bonds are exclusively found in sphingomyelins.26 The use of OzID in both imaging mass spectrometry and shotgun lipidomics highlights the ability of OzID to accurately identify C–C double bond locations in alkenes, providing additional molecular specificity that can be useful for understanding diseases.
2.3. Ion/neutral radical reactions
Ion/neutral radical reactions often involve exothermic charge transfers via the movement of electrons from neutral radicals with a low ionization potential to cations with a high electron affinity (A+ + B˙ → A˙ + B+).97–102 Various free radical molecules (e.g., allyl,101 organic peroxy (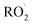 , R = organic functional group),102 hydrofluorocarbon,97 methyl (˙CH3), and nitric oxide (˙NO)) have been used for ion/neutral radical reactions. Free radical molecules can be generated in a laboratory through a variety of methods, including electrical discharges,97,98 laser photolysis,103 and pyrolysis,99,101,104 though it is difficult to produce a sufficient amount with high purity due to their high reactivities.100,101,105 The short lifespan of free radicals (i.e., a few microseconds106) limits the efficiency of ion/neutral radical reactions, which can be improved by extending the reaction time with a continual supply of the radical reactants. For example, flow tube reactors with integrated ion and radical sources97,101,102,107 and quadrupole ion traps108 can serve as reaction vessels with extended ion/neutral radical reaction times (i.e., a few seconds and a few minutes, respectively).
, R = organic functional group),102 hydrofluorocarbon,97 methyl (˙CH3), and nitric oxide (˙NO)) have been used for ion/neutral radical reactions. Free radical molecules can be generated in a laboratory through a variety of methods, including electrical discharges,97,98 laser photolysis,103 and pyrolysis,99,101,104 though it is difficult to produce a sufficient amount with high purity due to their high reactivities.100,101,105 The short lifespan of free radicals (i.e., a few microseconds106) limits the efficiency of ion/neutral radical reactions, which can be improved by extending the reaction time with a continual supply of the radical reactants. For example, flow tube reactors with integrated ion and radical sources97,101,102,107 and quadrupole ion traps108 can serve as reaction vessels with extended ion/neutral radical reaction times (i.e., a few seconds and a few minutes, respectively).
Ion/neutral radical reactions performed under atmospheric conditions in mass spectrometry experiments are commonly used in environmental science for detecting air pollutants arising from various sources.109 Chemical ionization mass spectrometry is often used to indirectly measure the concentration of pollutants in the air.109 Reagent gases, such as methane or ammonia, are ionized through electron ionization to produce cations, which can then react with neutral radicals via protonation of the analyte110 or adduct formation between the two reactants.111 For example, chemical ionization mass spectrometry provides a laboratory setting that enables the mechanistic study of the atmospheric autoxidation of α-pinene emitted from coniferous trees by ionizing the alkyl radical intermediates through NH4+ adduction. This terpenoid generates a series of peroxy radical products (e.g., O,O–C10H15(O2)xO2 with x = 0, 1, 2, 3) via ozonolysis in a flow tube, which then interacts with the NH4+ in a reaction chamber to form ammonium adducts that can be detected using MS.112 The quantitative measurements of these radicals are critical to understanding the self- and cross-reactions of intermediates that lead to the formation of secondary organic aerosols in the atmosphere,112 which can cause health issues in humans, including respiratory diseases.113,114
These types of barrier-free and spontaneous ion/neutral radical reactions under atmospheric conditions100,101,105 do not occur in an interstellar medium, which consists of very low matter density and temperatures (<200 K).115 In this extreme environment, the reactant ions exist in their ground state and require excitation to certain quantum levels through cosmic radiation in order for ion/neutral radical reactions to proceed.116–118 This interstellar chemistry produces a variety of chemicals that are considered the building blocks of earthbound molecules.42 Reactions between simple particles abundant in an interstellar medium, such as calcium ions (Ca+)119 and nitric oxide radicals (˙NO),120 can be studied in laboratory environments that mimic astronomical environments to understand the formation mechanisms of extraterrestrial chemicals. For example, sympathetic cooling of ˙NO radicals using laser-cooled Ca+ in an ion trap117,118,121,122 brings the temperature of the mixture to approximately 180 K. The reaction between Ca+ and ˙NO produces CaO+ (known as lime123 when in neutral form) and N only when Ca+ is repumped from the ground state (S1/2) to the excited state (P1/2) through laser irradiation (Fig. 4).105 This application demonstrates the impact of quantum effects on collisions between cold ions and neutral radicals, which could allow for the study of additional interstellar ion/neutral radical reactions under laboratory conditions.105,124
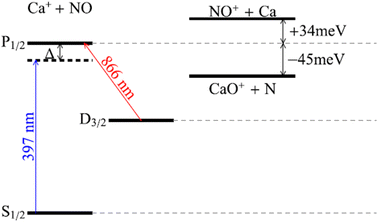 | ||
| Fig. 4 Comparison of reactant and product energies for the Ca+ + NO system. The exothermicity of the reaction depends on the quantum state of Ca+. Excited states of Ca+ can be populated by excitation on the two cooling laser transitions at 397 and 866 nm. To achieve cooling, the 397 nm laser is red-detuned from resonance by an amount Δ. Only Ca+ in the P1/2 state is energetically allowed to react. The charge exchange product channel barrier is overcome by the thermal energy of neutral NO at 180 K. (Energies not to scale.) Adapted/reproduced from Physical Review A, 2018, 98, 032702 (ref. 105) with permission from the American Physical Society, Copyright 2018. | ||
2.4. Hydrogen/deuterium exchange
Gas-phase HDX-MS (gHDX-MS) involves the exchange of labile hydrogen atoms of amides, amines, carboxylic acids, hydroxyls, and thiols with deuterium via reactions with deuterated reagents.33| [M − HL + nH]n+ + RD → [M − D + nH]n+ + RHL (HL: labile hydrogen) | (4) |
The difference in gas-phase proton affinities between analytes and deuterated reagents determines the rate of HDX and the extent of deuterium incorporation.125–127 gHDX-MS often requires relatively simple hardware modifications and can be achieved by flowing a carrier gas, such as N2 or He, through deuterated solvents (e.g., D2O, CD3OD, CD3COOD, ND3) prior to introduction to the instrument via a gas inlet system. Deuterated vapors are then injected via leak valves into ion funnel regions,128 electrodynamic ion traps,129–131 FT-ICR traps,132,133 ion mobility cells,36 and ion guides,35,134 where gHDX reactions occur. In this review, we highlight two gHDX applications: a protein structural biology approach and MALDI imaging mass spectrometry for lipid spatial mapping.
Accurately assessing gas-phase protein conformation is crucial for mechanistic understanding of the charge-state-dependent ESI processes135 and protein gas-phase folding dynamics.136–142 Incorporating deuterium into the labile hydrogen atoms of protein ions through gHDX provides insights into gas-phase surface accessibility and 3-D conformation.131 A recent study of ubiquitin ions generated via ESI from native and denaturing solutions shows the sensitivity of gHDX in detecting local conformational differences.143 In this study, the 6+ charge state of ubiquitin generated from an aqueous solution and exposed to gaseous ND3 shows a significant decrease in deuterium uptake in specific local secondary structures (e.g., β2 and β3 strands and α- and 3.5-helices) compared to the rest of the protein, indicating a tightly folded conformation. By contrast, alcohol-denatured 11+ ubiquitin exhibits extensive deuteration across the entire protein, indicating a large extent of unfolding.143 These results demonstrate the utility of gHDX-MS in detecting gas-phase protein unfolding at varying charge states.
Accurately mapping lipid spatial distributions in MALDI imaging mass spectrometry requires differentiating isobaric compounds (i.e., compounds of the same nominal, but different fractional, m/z value), which is particularly challenging at low mass resolving power.91 However, replacing labile hydrogen atoms with deuterium after ionization can alter the chemical profile of isobaric lipid ions and provide structural information specific to different lipid classes. The MALDI plume, where active chemical reactions occur within the expanding gas cloud of laser-ablated ions and neutral molecules, provides an excellent environment for gHDX.144 For example, an ion mapped in mouse brain tissue with a nominal mass of 838 Da is likely comprised of multiple isobaric lipids, including phosphatidylcholines, phosphatidylethanolamines, and glycolipids, each having a unique number of labile hydrogens (i.e., one, three, and seven, respectively). An online database search based on the isotopic distribution alone cannot differentiate these isobaric lipids.128 However, the incorporation of seven deuterium atoms into this analyte via in-plume gHDX identifies the lipid as a potassium-cationized glycolipid, [GalCer 22:0 (OH)/d18:1 + K]+ (<5 ppm). The seven labile hydrogens correspond to three ceramide and four galactosyl hydroxyl groups, demonstrating improved analyte identification using gHDX in MALDI imaging mass spectrometry without the need for MS/MS.128
3. Applications of ion/electron reactions
Ion/electron reactions occur between analyte ions and energetic electrons, forming radical product ions that undergo radical-directed dissociation.9,145 The energy regime of electron irradiation and the mechanism of odd-electron radical formation determine the types of electron-based dissociation (ExD) methods, though the energy ranges discussed below are not clear-cut boundaries. Electron-capture dissociation (ECD) is widely used for the sequencing and analysis of noncovalent interactions and labile PTMs of multiply charged peptide and protein cations by capturing low-energy (<2 eV) electrons.146,147 Irradiation with electron of energies 3 to 13 eV, a process known as hot ECD (HECD),9,145–147 is an extension of ECD that obtains additional information on amino acid stereochemistry and enhances the dissociation efficiency.146,148 However, the ECD of peptides with low charge states is generally less efficient due to the small electron capture cross-section, and singly charged cations cannot produce detectable fragment ions due to neutralization following the electron capture.149 Singly or doubly charged peptide anions can also capture electrons with 4 to 7 eV and dissociate in a process known as negative-ion ECD (niECD).150 Electron-induced dissociation (EID) is an alternative to ECD, which involves bombarding analyte cations with high-energy (>10 eV) electrons, which results in excitation instead of capture.149,151,152 Multiple terms are used to describe ion activation in this energy regime, including electron-excitation dissociation (EED)152 and electron-ionization dissociation (EIoD).149 The application of electrons with kinetic energies greater than 10 eV can be extended to the analyses of multiply charged analyte anions, referred to as electron detachment dissociation (EDD).9,145ExD instruments enable facile control of electron energies, facilitating a smooth transition between ExD regimes based on analytical requirements. All ExD experiments require sufficient spatial overlap between the analyte ions and energetic electrons, typically using one of three distinctive instrument configurations with magnetic fields to confine both reactants. In one setup, a focused electron beam generated from an external cathode irradiates ions trapped within the electric and magnetic fields of a Fourier-transform ion cyclotron resonance (FT-ICR) instrument.153,154 In other instruments, a focused ion beam can be directed through a dense electron cloud confined within an electromagnetostatic (EMS) cell,4,9,145,155–157 or the ion and electron beams can flow through one another.158
3.1. Electron-capture dissociation
The initial step in ECD of peptides and proteins involves the formation of charge-reduced radical cations following the capture of low-energy electrons. These radical cations undergo radical-directed dissociation and produce c-/z˙-type fragment ions through non-ergodic N-Cα backbone cleavages,9,147 as well as b-/y-type fragment ions generated via vibrational excitation:159| [M + nH]n+ + slow e− → [M + nH]˙(n−1)+* → fragmentation | (5) |
However, the thermodynamically labile nature of N–Cα bonds in the aminoketyl radical intermediates9 directly contributes to rapid dissociation (i.e., within microseconds)160 and the preservation of labile noncovalent interactions161,162 and PTMs163,164 without necessarily invoking non-ergodicity.165–167 Since the slow heating of CID does not maintain PTMs and labile bonds, ECD is commonly used as an alternative to maintain and localize these interactions and modifications to specific amino acid residues in the peptide sequence.168 For example, ECD can locate hydrogen bonds within endocrine hormone islet amyloid polypeptide (hIAPP) oligomers through the detection of noncovalent complexes composed of hIAPP units and c-/z˙-type fragment ions from the neighboring unit.169 Fragment ions [M + c28/29]5+ and [M + z3]3+ from 7+ dimers and [2M + c34]5+ and [2M + z5]4+ from 8+ trimers indicate the protection of C-termini from ECD, localizing the aggregate binding site at S28/S29 in one monomer and N35 in the next, resulting in staggered orientation of the oligomerization.169 While a clear consensus on the mechanism of cellular aggregation has not been reached,170 this result contributes to a better understanding of how these oligomers aggregate in amyloid diseases, and can potentially inform targeted drug development.169 Another noncovalent interaction ECD can explore is a salt bridge network within a phosphopeptide.171 In this application, ECD of VVGARRS(pS)WRVVSSI (pS indicates phosphorylation at S8) reveals strong salt bridge formations between pS8 and R5 or R10 within the binding motif RRS(pS)WR.171 The 3D conformation modelled based on these salt bridges171 aligns well with crystal structures,172 demonstrating ECD as an effective tool to localize noncovalent binding and PTMs simultaneously.
Localization of noncovalent binding sites in protein multimers using ECD is similar to that in peptide oligomers. ECD of α-synuclein proteins, which undergo neurotoxic aggregation in neurodegenerative diseases, results in extensive backbone cleavages across the N-terminal halves of the α-synuclein dimers. However, the C-terminal halves remain intact, indicating the presence of noncovalent binding sites within the intact regions that block ECD from occurring.162 Comparable ECD profiles obtained from wildtype and mutant α-synuclein dimers suggest the presence of natural, non-toxic oligomers in the brain.162 Similar to peptide phosphorylation, ECD can also be used to localize sites of protein glycosylation, such as in glycoforms of α-synuclein with four modification sites (T72, T75, T81, and S87) that prevent toxic aggregation (Fig. 5).164
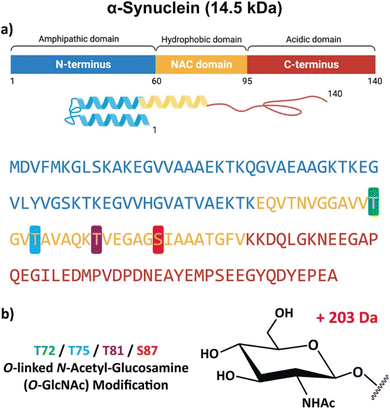 | ||
| Fig. 5 (a) Schematic, tertiary structure, and sequence of the α-synuclein monomer, showing the N-terminal amphipathic (blue), hydrophobic nonamyloid β-component of plaque (NAC, yellow), and acidic unstructured C-terminal (red) domains. (b) The structure and position of the single O-GlcNAc modification at T72 (green), T75 (blue), T81 (purple), and S87 (red). Adapted/reproduced from Analytical Chemistry, 2023, 95, 18039–18045 (ref. 164) with permission from the American Chemical Society, Copyright 2023. | ||
HECD and niECD complement conventional ECD by enhancing structural information and broadening sample compatibility for low-charge-state anions, respectively. The higher electron energies delivered via HECD produce additional secondary d-/w-type fragment ions through endothermic cleavage of primary c-/z˙-type fragment ions,146 while still retaining PTMs, such as glycosylation148,173 and methylation.174 These secondary fragment ions enable the differentiation of amino acid isomers in peptides and proteins that are not identified by traditional ECD.175 For example, z˙-type fragment ions containing valine and norvaline lose ˙CH2CH3 (29 Da) and ˙CH3 (15 Da), respectively, to generate diagnostic w-type counterparts. Accurately identifying these isomers is critical in biologics, as the bacteria used for recombinant protein production naturally synthesize norvaline but can mis-incorporate it into proteins instead of leucine, causing toxicity of the therapeutic proteins.176 During niECD, peptide and protein anions with low charge states and zwitterionic structures177 capture 4–7 eV electrons and form charge-increased radical anions that undergo radical-directed dissociation to produce c-/z˙-type fragment ions while preserving PTMs, similar to ECD and HECD.150 niECD is particularly useful for PTM analyses of acidic peptides, such as sulfopeptides178 and phosphopeptides,179 due to their greater ionization efficiency in negative mode. Singly charged anions typically show greater electron capture efficiency than multiply charged counterparts due to decreased coulombic repulsion of the energetic electrons by the analyte anions,177,178 making MALDI an effective ionization source for niECD. Indeed, MALDI-generated, singly deprotonated hirudin sulfopeptide (∼m/z 1500) and β-casein phosphopeptide (∼m/z 2000) subjected to niECD result in near complete sequence coverage and accurate localization of PTMs.179 Furthermore, similar niECD profiles of synthetic peptides with different salt bridge formations, ionized through MALDI or ESI, suggest that conformationally comparable peptide structures can be generated regardless of the MALDI matrix pKa and the ion source, providing valuable insights into the relative ionization mechanisms.179
3.2. Electron-induced dissociation
A generally accepted dissociation mechanism for EID of analyte cations involves the formation of an oxidized radical cation [M + nH]˙(n+1)+ from the precursor ion [M + nH]n+ via ionization (i.e., an increase in the total positive charge of the analyte from +1 to +2). A slow, low-energy electron is ejected during this bombardment process, which is then recaptured by the [M + nH]˙(n+1)+ to form an electronically excited [M + nH]n+* ion type that then undergoes radical-directed dissociation.9,145,149,152| [M + nH]n+ + fast e− → [M + nH]˙(n+1)+ + fast e− + slow e− → [M + nH]n+* → fragmentation | (6) |
Direct electronic excitation of the oxidized radical cation [M + nH]˙(n+1)+* and subsequent dissociation has also been proposed.149,180 EID has been used for the structural elucidation of peptides and proteins of varying charge states149,151,152,181,182 as well as singly-charge glycans,183 lipids,184–188 metabolites,189,190 and other small molecules.191–193 In this review, we limit the discussion of EID to the study of peptides, proteins, and lipids.
Side-chain loss upon EID of singly or doubly protonated peptides can be abundant for specific amino acid residues, which improves sequencing while preserving labile PTMs, such as glycosylation181,182 and sulfation,194 similar to HECD. However, unlike HECD,182 the dissociation efficiency for EID remains unaffected by charge states due to the electronic excitation that occurs through the interactions between analyte ions and high-energy electrons.149 EID also produces more abundant internal fragment ions from large, multiply protonated protein ions compared to (H)ECD, which can enhance structural information.195,196 For example, the efficient internal fragmentation of lysozyme, ribonuclease A, and α-lactalbumin through EID provides a three-fold increase in sequence coverage and localizes more disulfide bonds compared to ECD.197 Overall, EID studies of peptides and proteins highlight the enhanced and charge-independent sequence coverage that can be obtained with this approach.
EID has also been incorporated into an imaging mass spectrometry workflow to explore the spatial distribution of singly charged glycerophospholipids generated directly from tissue via MALDI, providing information on lipid classes, FA chain lengths, and C–C double bond sites.198,199 Similar to the sequential CID/OzID experiments discussed above, prior CID on glycerophospholipids generates dioxolane intermediates that provide sn-positional information upon subsequent EID.200 Unlike OzID, which produced specific and selective cleavage at double bonds, EID generates extensive cleavage across the entire length of fatty acyl chains, with “V”-shaped dips in allylic and vinyl fragment ion intensities around C–C double bonds. This sequential CID/EID method enables comprehensive structural characterization of singly protonated PC 34:1 from five different regions of rat brain tissue, which shows abundances of PC 16:0/18:1 relative to PC 18:1/16:0 ranging from 57.76 ± 2.23% in the granular layer to 75.77 ± 1.94% in the corpus callosum (Fig. 6).200 These results highlight the facile integration of EID into imaging mass spectrometry, enabling quantitative mapping of glycerophospholipids at sn-isomeric levels without prior separation.200
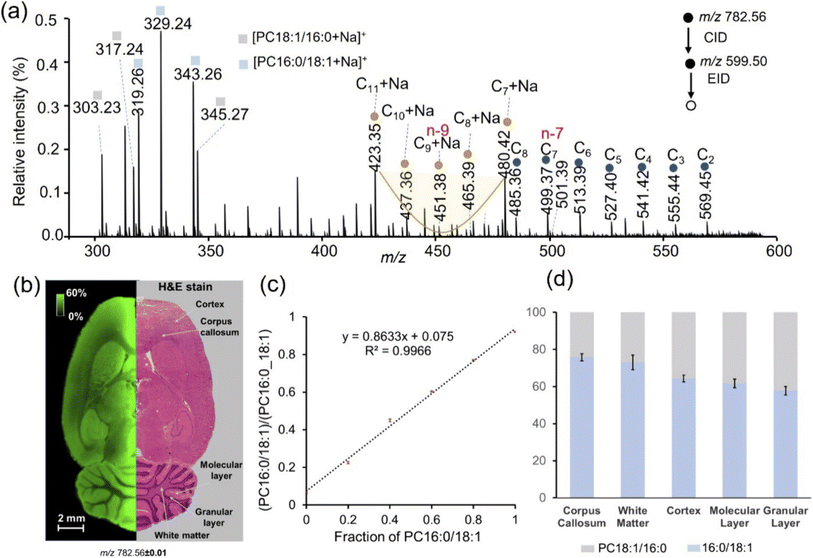 | ||
| Fig. 6 (a) Representative CID/EID spectrum of m/z 782.56 (±1 Da isolation width) from the cortex of rat brain tissue. (b) H&E stain and ion image for m/z 782.5695 ± 0.01 allow for visualization of sodiated PC 34:1 (3.21 ppm) in the tissue. The image is normalized to TIC and acquired using a 75 µm spatial resolution. (c) Calibration curve for the relative quantification of the sn-isomers is constructed using the summed intensities of the three PC 16:0/18:1 diagnostic ions divided by the summed intensities of all diagnostic ions for both isomers (n = 10). (d) Relative percentages of the PC 16:0/18:1 and PC 18:1/16:1 sn-isomers vary in different regions of the rat brain tissue (n = 5). Adapted/reproduced from Analytical Chemistry, 2023, 95, 15707–15715 (ref. 200) with permission from the American Chemical Society, Copyright 2023. | ||
3.3. Electron detachment dissociation
While fewer mechanistic studies have been made for EDD than other ExD methods, a generally accepted mechanism involves bombarding negative ions with high-energy electrons, resulting in subsequent electron detachment.201 Charge-reduced radical ions [M − nH]˙(n−1)− generated via the electron detachment undergo radical-directed dissociation:201| [M − nH]n− + fast e− → [M − nH]˙(n−1)−* + 2e− → fragmentation | (7) |
The charge reduction process requires multiply charged analyte anions to avoid neutralization. A variety of sample types, including glycans,202,203 nucleotides,204,205 and peptides,206,207 can undergo EDD for structural determinations. Here, we discuss recent applications of EDD to differentiate peptide and glycan isomers.
EDD of multiply charged peptide anions leads to cleavages at N–Cα and Cα–C bonds, resulting in the production of a˙-/c-/x-/z˙-type fragment ions, similar to ECD.145 In a recent study, EDD of [M − 3H]3− of antibacterial 21-mer lasso peptides from E. coli (Microcin J25) produces diagnostic bi-peptidic fragment ions that signify the lasso topology, such as 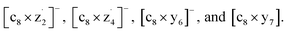 207 c8− represents the intact N-terminal macrolactam ring, and × denotes covalent or noncovalent attachment with z˙- or y-type fragment ions dissociated from the C-terminal tail sterically entangled with the ring.207 By contrast, CID of the same topoisomeric anions predominantly results in side-chain loss from T15 residues and fails to provide structurally meaningful information, highlighting the sensitivity of EDD to the complex 3D topology of peptides.207
207 c8− represents the intact N-terminal macrolactam ring, and × denotes covalent or noncovalent attachment with z˙- or y-type fragment ions dissociated from the C-terminal tail sterically entangled with the ring.207 By contrast, CID of the same topoisomeric anions predominantly results in side-chain loss from T15 residues and fails to provide structurally meaningful information, highlighting the sensitivity of EDD to the complex 3D topology of peptides.207
EDD also enables structural differentiation of glycosaminoglycans isomers, which is typically challenging due to the subtle variation in stereochemistry. For example, EDD of epimeric heparan sulfate tetrasaccharide anions possessing different stereochemistry at C5 of ring-3 produces diagnostic fragment ions that distinguish glucuronic acid (GlcA) from iduronic acid (IdoA) substitution.203 Cleavage at the glycosidic bond between ring-3 and ring-4 (fragment ion 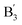 ), in combination with loss of CO2
), in combination with loss of CO2 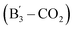 , indicates the presence of a GlcA epimer at the ring-3 position, which is lost with the substitution of IdoA for GlcA.203 The detailed glycosaminoglycans stereochemical information provided by EDD is critical to understanding their binding specificities toward on glycosaminoglycans-binding proteins.
, indicates the presence of a GlcA epimer at the ring-3 position, which is lost with the substitution of IdoA for GlcA.203 The detailed glycosaminoglycans stereochemical information provided by EDD is critical to understanding their binding specificities toward on glycosaminoglycans-binding proteins.
4. Applications of ion/photon reactions
Ion/photon interactions occur when analyte ions absorb photon energies of specific wavelengths.2 Here, we highlight ultraviolet photodissociation (UVPD)11–13 and infrared multiphoton dissociation (IRMPD),208 in which nonselective or selective absorbance of photons of specific wavelengths by analyte ions can result in fragmentation once the ion internal energy exceeds a dissociation threshold.11| [M + nH]n+ + nhν → [M + nH]n+* → fragmentation | (8) |
F2 excimer lasers (157 nm, 7.9 eV per photon), ArF excimer lasers (193 nm, 6.4 eV per photon), and Nd:YAG lasers operated using the 5th (213 nm, 5.8 eV per photon), 4th (266 nm, 4.7 eV per photon), and 3rd (355 nm, 3.5 eV per photon) harmonics are the most commonly employed wavelengths for UVPD, while CO2 lasers (10.6 µm, 0.12 eV per photon) are often used for IRMPD. However, various chromophores and diverse intrinsic electron densities around chromophores dictate the absorption selectivity and efficiencies, and thus the division based on wavelength alone is not clearcut.11 These photodissociation experiments in mass spectrometry require sufficient spatial overlap between the analyte ions and laser beams for efficient dissociation, similar to the ExD experiments discussed above. This can allow for prevalent secondary (i.e., consecutive) dissociation of first-generation fragment ions, as primary fragment ions remain in the laser path and continue to undergo irradiation. Alternatively, some instruments can employ “protective” mass-selective resonant excitation of primary fragment ions to displace them from the laser path and minimize consecutive fragmentation.209–211 RF-ion traps,212,213 RF-ion guides,214 and FT-ICR cells215,216 are commonly used to achieve optimal photoabsorption cross-sections by fine-tuning the ion trajectories, while laser beams are introduced into instrument vacuum system through windows transmitting light at the specified wavelength.11,21,208
4.1. Nonselective UV photodissociation
UVPD of peptide and protein ions at 157, 193, or 213 nm generally involves the selective absorption of single energetic photons via chromophores that are evenly distributed along the backbones, such as π orbitals of amido groups leading to π-to-π* excitation or nonbonding electrons of carbonyl oxygens resulting in n-to-π* excitation.12,13 This high-energy deposition exceeds the energy of peptide bonds (i.e., 3–4 eV) and leads to nonselective dissociation directly from the excited π* electronic states, causing extensive backbone cleavage into a-/b-/c-/x˙-/y-/z˙-type fragment ions.12,13,217 The non-ergodic cleavages at N–Cα and Cα–C bonds enable the study of preserved noncovalent interactions218–220 and labile PTMs, including acetylation,221 methylation,221,222 glycosylation,223 phosphorylation,224 and sulfation.225 Our discussion centers on 193 nm UVPD, as it is the most widely adopted wavelength.The nonselective fragmentation channels and charge-independent dissociation efficiency make 193 nm UVPD particularly useful for low-charge-state peptides and proteins.226 By comparison, CID,227 higher-energy collisional dissociation (HCD, <100 eV),226,228,229 ECD,147 and 213 nm UVPD230 all exhibit strong charge state dependency in fragmentation efficiency, though 213 nm UVPD still provides greater sequence coverage than CID and HCD.231 For example, HCD of ESI-generated 5+ ubiquitin (8.6 kDa), 8+ Staph nuclease (16.1 kDa), and 7+ calmodulin (16.6 kDa) shows limited sequence coverages of 84%, 53%, and 43%, respectively, due to the reduced proton mobility responsible for charge-directed fragmentation.232 However, 193 nm UVPD achieves significant improvements in sequence coverage, up to 98%, 79%, and 66% for each protein, respectively.232 HCD efficiency is even more limited with singly charged proteins commonly produced by MALDI,226,233 as shown by 8% sequence coverage for the 1+ charge state of ubiquitin, which is significantly enhanced to 87% by using 193 nm UVPD.234 These results demonstrate that 193 nm UVPD provides better sequence coverage than conventional HCD at low charge states, and suggest that the MALDI applications, including MALDI imaging mass spectrometry, can potentially adopt UVPD for top-down protein identification of singly charged proteins directly from tissue.233,234
Noncovalent interaction sites in peptide235 and protein complexes236–238 can be explored using 193 nm UVPD, similar to topological mapping studies enabled by ECD. For example, paratopes on engineered biotherapeutics, such as in a complex of a Gnb nanobody and its green fluorescent protein (GFP) antigen, can be deduced from a significant suppression in UVPD fragmentation adjacent to the protein–protein interfaces (Fig. 7).239 Additionally, weighted average charges acquired from a-/x-type fragment ions240 aid in determining sites of inter-subunit salt-bridge formation at Gnb (R36)–GFP (E142) and Gnb (R58)–GFP (E172/D173), which is supported by crystal structures.239 This application highlights the ability of 193 nm UVPD to locate different types of noncovalent interactions and map protein quaternary structures.239
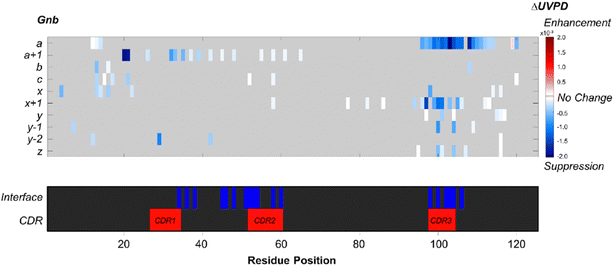 | ||
| Fig. 7 Suppression and enhancement of backbone cleavage sites based on abundances of UVPD fragment ions induced for Gnb by antigen binding. Heat plots display significant differences (p < 0.05, n = 5) for the abundances of each UVPD fragment type between the free and bound states. Blue and red indicate suppression and enhancement of fragment abundances, respectively, for the nanobody upon complexation. Positions that display no significant change are shown in grey. Color maps highlighting interface residues and CDRs are also included. ΔUVPD values correspond to the fragment abundance per residue for the bound state minus the fragment abundance per residue for the free state. Adapted/reproduced from Chemical Science, 2022, 13, 6610–6618 (ref. 239) with permission from the Royal Society of Chemistry, Copyright 2022 The Authors. | ||
4.2. Selective UV photodissociation
UV photodissociation at longer wavelengths, 266 and 355 nm,241 is not as widely utilized in mass spectrometry. However, the high absorption selectivity of these wavelengths by certain chemical bonds provides a unique utility for the application of peptide and protein ions. For example, selective 266 nm UVPD involves modifying peptide ions prior to ionization by introducing carbon–iodine (C–I) bonds via derivatization with chloramine-T242,243 or iodobenzoic acids.244 Irradiation of these C–I bonds with 266 nm light results in selective homolytic cleavage and generates stable free radical ions.242,243 Subsequent CID of the radical cation initiates radical-directed dissociation sensitive to the stereochemistry of neighboring amino acid residues, including Asp, Ser, His, and Glu, via structure-dependent radical migration pathways.245 For example, L-aspartic acid and L-isoaspartic acid in the iodinated synthetic peptide GISEVRSDG are differentiated by assessing the relative abundances of a8+ fragment ions and CO2 loss.244,245 Sequential 266 nm UVPD/CID can also provide information on protein topology.242 Another example of selective UV photoabsorption utilizes a chromogenic diazirine-tagged peptide246,247 for cross-linking mass spectrometry, where distance restraints imposed by cross-linkers provide low-resolution 3-D structural information for peptides and proteins.248 The diazirine group selectively absorbs a 355 nm photon and converts to a reactive carbene intermediate, which then forms a covalent bond with the immediately neighboring amine or carboxylic acid group, signified by the loss of the N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N moiety.249,250 This zero-length chromogenic cross-linker does not introduce any physical distance between the covalently linked molecules, allowing it to probe protein–ligand or protein–protein interfaces via carbene insertion into X–H bonds (X = C, N, or O).249,250
N moiety.249,250 This zero-length chromogenic cross-linker does not introduce any physical distance between the covalently linked molecules, allowing it to probe protein–ligand or protein–protein interfaces via carbene insertion into X–H bonds (X = C, N, or O).249,250
Iodinated lipid ions can also form radical cations by selectively absorbing 266 nm photons at C–I bonds, leading to iodine atom cleavage due to UV irradiation. Subsequent radical-directed dissociation facilitated by CID results in allylic cleavage that unambiguously identifies C–C double bond sites251–255 Alternatively, absorption of 193 and 213 nm light by lipid ions with isolated, non-conjugated C–C double bonds occurs by the entire molecule acting as a chromophore, due to high-energy σ → σ* and n → π* transitions, similar to the universal absorption by peptide and protein backbones.256–260 Following photoexcitation, the molecule redistributes energy internally and fragments preferentially at labile sites, especially near double bonds, producing diagnostic allylic and vinylic fragment ions that localize the unsaturation sites.253,256–262 Sequential HCD/UVPD can be used to identify sn-positional isomers of glycerophospholipid261 and cardiolipin (CL) ions263 by generating dioxolane intermediates through thermal dissociation enabled by HCD, and then subsequent cross-ring cleavages via selective UVPD, similar to the CID/OzID approach discussed above.261,263 For example, HCD/UVPD enables the identification of the tumor-specific and unusual prevalences of phosphatidic acid (PA) 18:2/18:1 (76.5%) over PA 18:1/18:2 (23.5%) and PC 18:1/18:2 (70.2%) over PC 18:2/18:1 (29.8%) in CL 72:6 in a total lipid extract from papillary thyroid carcinoma tissue.263 In another study, HCD/UVPD can be used to identify ester bond locations in isomeric palmitic acid esters of hydroxystearic acids by creating a new C–C double bond specifically at the ester site during the HCD process, further demonstrating selective UVPD for lipid identification.264
4.3. IR photodissociation
Protein molecules have IR-active functional groups that enable the broad absorption of 10.6 µm CO2 laser light, including C–C, C–N, C–O, and P–O stretching and O–H bending vibrational modes.265 However, the degree of energy deposition is highly tunable due to the multi-step absorption process of low-energy photons, resulting in slow heating via intramolecular vibrational energy redistribution.266 This process contributes to the selective fragmentation of labile PTMs, amide bonds, and noncovalent bonds via low-critical-energy pathways.208 For example, step-wise selective dissociation of membrane protein–detergent complex ions is demonstrated using a low-power CO2 laser beam (typically <10 W).265,267 The detergents stabilize the hydrophobic proteins in solution prior to ionization, but should be removed in the gas phase prior to mass analysis of the intact proteins, as the bound detergents complicate spectral interpretation. A low amount of laser power and irradiation time liberates trimeric ammonia channel AmtB protein ions from the protein–detergent complex ions, while additional energy deposition dissociates trimers into monomer subunits (Fig. 8).265 Collisional activation methods can also remove detergents268–270 though with more limited and less efficient energy deposition that leads to incomplete removal of detergents and compromised protein sequence coverage.265 IR irradiation allows for more precise and effective tuning of the energy deposition, decreasing the chemical interferences and enhancing protein sequence coverage.265,267 For example, step-wise IRMPD successfully liberated a class A G protein-coupled receptor (GPCR), beta-1-adrenergic receptor (β1AR), from the detergent and achieved 28% sequence coverage, while an analogous experiment performed using HCD only achieved 17% sequence coverage.265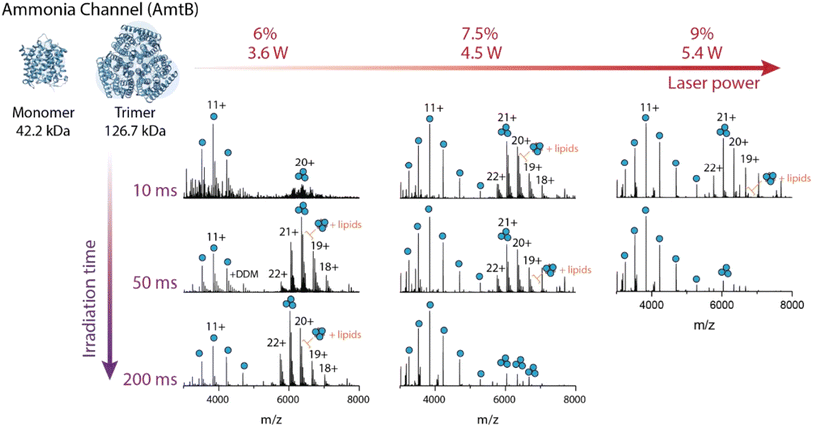 | ||
| Fig. 8 The effect of modulating laser output power and irradiation time for AmtB solubilized in DDM detergent. At low laser output power and short irradiation times, the peaks corresponding to AmtB trimer appear at low abundance due to signal dilution across many adduct peaks. With increasing irradiation time, peaks corresponding to lipid-bound trimers are observed as the protein is liberated from DDM detergent micelles. These peaks dominate the mass spectrum ∼6000 m/z. At intermediate laser output powers from short to long irradiation times, the relative abundances of peaks corresponding to trimer and monomer can be modulated. Finally, at high laser output power and short irradiation times, peaks corresponding to monomer and trimer are approximately equal in abundance, while at long irradiation times, the peaks corresponding to trimer are absent as the protein complex has expectedly dissociated into monomeric subunits (peaks <5000 m/z). Adapted/reproduced from Angewandte Chemie International Edition, 2023, 62, e202305694 (ref. 265) with permission from Wiley-VCH GmbH, Copyright 2023 The Authors. | ||
Lipids also have universal chromophores for nonselective absorption of IR laser light and can undergo selective dissociation via the lowest energy pathway, such as the loss of fatty acyl moieties from glycerophospholipid anions via fragmentation at labile ester bonds.271,272 This feature can be used in MS/MS imaging mass spectrometry to study the spatial distribution of glycerophospholipids from tissue at the sn-positional isomer level.273 In this study, IRMPD of singly charged PC 34:1 anions produces varying abundances of fatty acyl moieties depending on the sn-positions, revealing differential isomer expression in different rat brain regions.273 MS/MS performed at every pixel in this experiment allowed mapping the relative distribution of PC 16:0/18:1 to PC 18:1/16:0 in regions such as the granular layer (64.5 ± 2.5%) and white matter (78.1 ± 2.2%).273 Though not discussed in detail in this review, blackbody infrared radiative dissociation (BIRD) is another photodissociation technique that leverages nonselective photoabsorption of a range of blackbody IR wavelengths emitted from heated ICR cell walls.10 Selective bond dissociation via BIRD is often used to study dissociation kinetics of noncovalent host–guest binding,274–277 similar to CID-based approaches discussed above.
4.4. Action spectroscopy
Spectroscopic analysis of gas-phase ion chemistry in mass spectrometers is known as action spectroscopy,278 in which light absorption is measured indirectly (i.e., by monitoring photodissociation) due to the low ion concentrations that make traditional absorption spectroscopy impractical.279–284 The photodissociation yield can be plotted as a function of wavelength, providing information on the thermodynamic threshold for the photodissociation and electronic transition dynamics of the precursor ion.285–291 Light absorption by gas-phase trapped ions can also be indirectly measured through sensitive fluorescence detection,292,293 facilitating advanced applications such as action – Förster resonance energy transfer (FRET) spectroscopy for protein tertiary structure analysis294 and quantum computing utilizing quantum state-specific fluorescent ions to convert superposition states into classical 0/1 outcomes.295 A detailed discussion of action spectroscopy fundamentals and applications can be found elsewhere,279,281,282,284,286–290,296 as it is beyond the scope of this review.5. Applications of ion/ion reactions
Ion/ion reactions occur when analyte and reagent ions of opposing charge interact due to coulombic attraction, with reaction rates proportional to the square of the charges.14 The coulombic interactions facilitate a variety of changes in the conformational, electrochemical, and physical properties of analyte ions via the transfer of particles (i.e., electrons, protons, and metal ions) and covalent modifications.297,298 Ion/ion reactions can be performed outside mass spectrometers in atmospheric environments, such as with ambient bipolar dual spray, where overlapping streams of positive and negative ions enable ion/ion interactions.299 However, in most ion/ion reactions under reduced pressure environments inside mass spectrometers, it is crucial to bring together ions with opposite charges close enough in space to form stable orbiting electrostatic complexes.297 Therefore, the critical step is securely and mutually trapping these ions in an electrodynamic trap.298,300 Many types of electrodynamic traps can facilitate ion/ion reactions via simultaneous confinement of ions of opposite charges.298,300 Electron transfer dissociation (ETD) is the most widely used ion/ion reaction, and has been commercialized on many instrument platforms.301–303 A broad array of gas phase ion/ion reactions, including proton transfer charge reduction,304,305 charge inversion,306 metal ion transfer,307 and covalent modification308 chemistries can be enabled on commercial instruments already equipped with ETD,301–303 or these reactions can be enabled via instrument modification or construction.15,298,300,3095.1. Electron transfer dissociation
ETD was first developed to enable ECD-like peptide and protein sequencing in electrodynamic ion traps without requiring magnetic fields and thermal electrons.309 ETD involves transferring electrons from singly charged radical reagent anions to multiply charged protein cations during the ion/ion reaction, which electronically excites the analytes. The odd-electron radical protein cations produced undergo cleavages at N-Cα bonds to produce c-/z˙-type fragment ions while preserving labile PTMs, similar to ECD.9,17,145,310,311| [M + nH]n+ + A˙− → [M + nH]˙(n−1)+ + A | (9) |
ETD is widely used for PTM analysis as an alternative to slow-heating collisional activation methods,312 similar to ECD. For example, ETD of U1 small nuclear ribonucleoprotein 70 kDa (U1-70K) enables the identification of 29 PTM sites from 55 peptides, while only 19 PTM sites from 13 peptides are identified via HCD.312 Identifying these PTM sites is crucial for understanding protein–protein interaction sites that can lead to aggregation in diseased individuals.312 However, ETD is often combined with HCD (i.e., EThcD)313,314 to enhance sequence coverage by thermally dissociating ETD fragment ions that have undergone N–Cα bond cleavage, but that are still held together by noncovalent interactions.315–319 For example, EThcD of trypsin-digested peptides from HeLa cell lysates shows abundant c-/z˙-type fragment ions for peptide sequencing and allows for the detection of roughly 1.7-fold more ADP-ribosylation PTM sites320 than conventional ETD.321 ADP-ribosylation is a growing area of research related to cell signaling and DNA damage repair processes, and identifying this PTM enhances the understanding of associated enzyme activities.321 Another benefit of EThcD compared to HCD or ETD alone is the ability to cleave disulfide bonds. For example, EThcD breaks disulfide bonds in the C-terminal cyclic structures of frog skin peptides, leading to enhanced sequencing of the cyclic region, which is typically challenging to sequence when the structures remain closed as with using ETD or HCD.322,323 EThcD also facilitates differentiation of amino acid isomers, such as leucine and isoleucine, through different secondary w-type ions generated from the primary z˙-type fragment ions and provides near-complete sequencing of the amphibian peptides.322,323
EDD-like reactions in electrodynamic ion traps can also be achieved via negative electron transfer dissociation (NETD), where singly charged reagent radical cations abstract single electrons from multiply charged analyte anions, producing charge-reduced, odd-electron radical anion analytes that undergo radical-directed dissociation.324
| [M − nH]n− + B˙+ → [M − nH]˙(n−1)− + B | (10) |
NETD can be used to analyze various sample types, such as peptides,324–326 proteins,327 glycans,328,329 and nucleotides.330 For example, NETD of glycosaminoglycans produces extensive glycosidic (B/Y and C/Z) and cross-ring (A/X) fragment ions, while preserving labile sulfate modifications. The intricate combinations of these fragment ions allow for the distinction of GlcNS3S (NS: 2-N-sulfation, 3S: 3-O-sulfation) and GlcNS3S6S (6S: 6-O-sulfation) in tetrasaccharide structural isomers, which differ only by sulfation on the glucosamine (GclN) moieties at the reducing ends.331 Unambiguously identifying the 3-O- and 6-O-sulfation sites is critical for understanding the biological functions of glycosaminoglycans, as these modification sites play essential roles in the anticoagulant properties of heparan sulfate by binding to antithrombin, which inhibits enzymes responsible for blood clotting.331–333
The transfer of electrons from radical reagent ions to analyte ions does not always result in dissociation.334–336 Still, oxidation or charge reduction resulting from electron transfer can enable charge manipulation to deconvolute crowded spectra of multiply charged protein ions or to explore charge-dependent gas-phase protein conformational changes.337,338 For example, the gas-phase structures of ubiquitin charge conformers, formed either directly from the ESI source or via gas-phase charge manipulation of ESI-generated ions, can be examined. Specifically, the alcohol-denatured 6+ charge state of ubiquitin is compared to 6+ ubiquitin produced via non-dissociative electron transfer (i.e., charge reduction) of alcohol-denatured 8+ ubiquitin (8+ → 6+) in an ion trap.335 Incremental collisional activation significantly alters the collision cross-sections of 6+ ions produced directly from the ESI source, but not those generated via electron transfer charge reduction (Fig. 9). This result indicates that charge-reduced ions (8+ → 6+) retain the conformational memory of the original 8+ ubiquitin, which differentiates them from the 6+ ions formed directly from the ESI process.335 Conformational changes, similar to gas-phase protein folding via non-dissociative electron transfer, can initiate directional motion in molecular switches. High charge states elongate the molecule into a rod shape due to Coulomb repulsion, and a folded globule forms as the charge reduces through stepwise electron transfer, showing the mechanical control of the switch via redox chemistry.339 Gaining insights into the reversible structural changes of molecular switches in response to an external stimulus aids in designing precise control mechanisms at the molecular level for applications such as molecular electronics.339
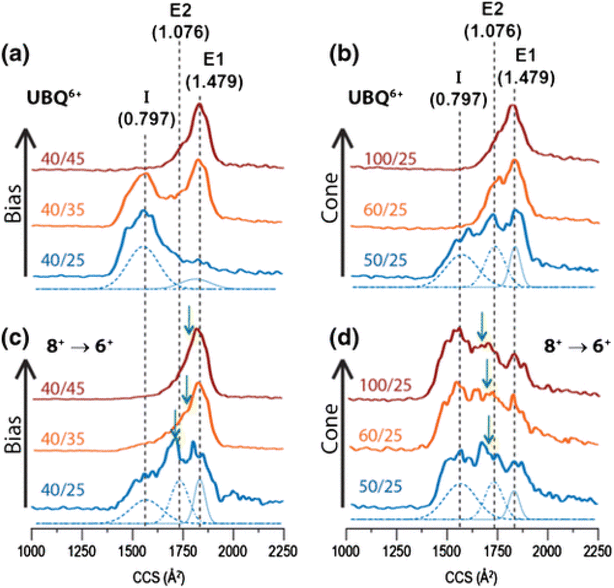 | ||
| Fig. 9 Collision cross sections for 6+ ubiquitin formed by either (a and b) electrospray ionization, or (c and d) charge reduction of ESI-generated [M + 8H]8+ by using ETD. Time axes of arrival time distributions were converted to CCS as described in ref. 340. Data shown in panels (a) and (c) were acquired using a sampling cone voltage of 40 V and a trap DC bias of either 25 (blue), 35 (orange), or 45 V (red). In panels (b) and (d), the bias was kept constant at 25 V and a sampling cone voltage of 50 (blue), 60 (orange), or 100 V (red) was applied. Colored text in panels indicates (cone voltage)/(bias voltage) for each trace. Gaussian components used in deconvolution of the lowest-energy (blue) trace in each panel are shown at the bottom at half-intensity. Values in parentheses are optimized apparent PTR/ETnoD ratios for each of these components, as described in the text. Weighted average CCS values (used to generate Fig. 2) are indicated with green arrows in panels (c) and (d). Adapted/reproduced from Journal of the American Society for Mass Spectrometry, 2017, 28, 69–76 (ref. 335) with permission from the American Chemical Society, Copyright 2017. | ||
5.2. Proton transfer charge reduction
Proton transfer charge reduction features proton transfer from multiply protonated analyte cations to reagent anions with high gas-phase basicity via a single collision between the two reactants.341,342| [M + nH]n+ + A− → [M + (n – 1)H](n−1)+ + AH | (11) |
Proton transfer charge reduction is commonly employed to reduce the charge states of multiply protonated precursor or fragment ions that are present in an m/z range with significant spectral overlap among various ions of different charge states.298 Proton transfer charge reduction decongests complicated mass spectra and can reduce signal dilution from distribution across multiple charge states for a single analyte, which results in a improved signal-to-noise ratio343,344 and sequence coverage345–347 in complex mixtures.341 For example, proton transfer charge reduction streamlines complex spectra from top-down ETD analysis of the antibody–drug conjugate, trastuzumab deruxtecan (T-DXd), enhancing fragment ion detection and resulting in a ∼10% increase in sequence coverage compared to ETD alone.348 The separation of overlapping fragment ions via proton transfer charge reduction also enables a more confident determination of the drug conjugation sites of topoisomerase I inhibitor payload (DXd), such as Cys214 on the light chain and Cys223, Cys229, and Cys232 on the Fd subunit of the heavy chain.348 Proton transfer charge reduction of complex UVPD MS/MS spectra generated from large proteins (>30 kDa) with high charges (>30+) also shows a significant increase in sequence coverage.349 Overall, proton transfer charge reduction demonstrates utility in simplifying MS/MS spectra of multiply charged proteins that commonly produce a large number of fragment ions with overlapping m/z values.
In addition to sequencing, proton transfer charge reduction is utilized to explore the 3D structures of proteins in the gas phase. IM-MS of product ions resulting from sequential proton transfer charge reduction of multiply protonated denatured proteins of various sizes, such as ubiquitin,138 cytochrome c,139 lysozyme,137 and serum albumin,136 reveals collisional cross-sections indicative of a gradual gas-phase folding of the protein ions as the charge states decrease. However, the collisional cross-sections derived from the proton transfer charge reduction products of protein ions initially formed under native solution conditions show overall consistent values across varying charge states, indicating compact conformations with less charge dependence.136–139 The collisional cross-sections of low-charge proton transfer charge reduction products of denatured proteins are generally larger than those of native counterparts, suggesting the proton transfer charge reduction product protein ions retain some of their initial structure.136–139 This trend suggests that the formation of a noncovalent bonding network differs in vacuo from in solution during protein folding,140 providing insights into protein folding dynamics.
Proton transfer charge reduction can also be applied to lipid anions, such as in the charge reduction of [CL − 2H]2− dianions to [CL − H]− monoanions using singly charged proton-donor cations.350 Proton transfer charge reduction improves the signal-to-noise ratio of the dianions by concentrating signals into monoanions, alleviating the challenge of identifying CL dianions in congested m/z ranges dominated by phospholipid monoanions in the complex lipidome.350 This application also demonstrates that proton transfer charge reduction can be effected in either ion polarity.
5.3. Charge inversion reactions
Transferring multiple protons between reactant ions with opposite polarity can result in charge inversion of analyte ions rather than reduction.| [M + H]+ + [N − nH]n− → [M − H]− + [N − (n − 2)H](n−2)− | (12) |
| [M − H]− + [R + nH]n+ → [M + H]+ + [R + (n − 2)H](n−2)+ | (13) |
Ion/ion charge inversion reactions can be used to access new and complementary gas-phase dissociation chemistries by altering the analyte ion type prior to CID. For example, multiply deprotonated phosphopeptides are charge inverted to multiply protonated cations by a reaction with multiply protonated polyaminoamine dendrimers.351 The ionization of the phosphopeptides in negative ion mode allows for their efficient formation without ionization suppression, and the multiply protonated ion/ion reaction product phosphopeptide allows for the use of ECD for the localization of phosphorylation sites.351 Multiply protonated proteins generated in positive ion mode can also be charge inverted to deprotonated anions via reaction with multiply deprotonated hyaluronic acid.352 CID of the protein anion then provides for complementary dissociative channels for PTM localization.353,354
Lipids exhibit different ionization efficiencies in positive and negative ion modes. For example, phosphatidylcholines have higher efficiency in positive mode due to the permanent positive charge embedded in the choline moiety. However, low-energy CID of protonated phosphatidylcholines generally only allows for the characterization of the lipid head group. Conversely, singly protonated phosphatidylcholines can be charge-inverted via an ion/ion reaction with 1,4-phenylenedipropionic acid (PDPA) dianions. Subsequent CID on the resulting demethylated phosphatidylcholine anion provides fragmentation indicative of the fatty acyl chains in the lipid.355,356 The relative fractions of PC 16:0/18:1 compared to PC 18:1/16:0 in different rat brain substructures can be mapped via imaging mass spectrometry combined with this PDPA ion/ion charge inversion reactions, and range from 50.1 ± 1.9% in the granular layer to 76.5 ± 2.1% in the corpus callosum.306 Gas-phase ion/ion reactions can also be combined with other derivatization approaches. For example, a four-membered cyclic ether, incorporating two carbon atoms into C–C double bonds, can be derivatized by a condensed-phase Paternò–Büchi reaction at the double bond sites prior to ionization. When combined with the PDPA charge inversion reaction, subsequent CID provides information on C–C double bond positions within the glycerophospholipids.357 Overall, gas-phase charge inversion reactions resolve analytical challenges arising from different ionization and MS/MS efficiencies by decoupling ion formation from the ion type ultimately subjected to MS/MS. These reactions enable the formation of new ion types in the gas phase, eliminating the need for condensed-phase analyte derivatization.
5.4. Metal ion transfer reactions
A variety of metal ions (i.e., alkali,199,358–360 alkali earth,361–363 and transition363–365) can form adducts with analytes during ionization,| [M + nH]n+ + MetxLy− → [M + nH + MetxLy](n−1)+ → [M + (n − y)H + xMet](n−1)+ + yHL | (14) |
Facilitating novel gas-phase dissociation chemistries upon MS/MS compared to common protonated and deprotonated ion types. Condensed-phase metal ion adduction can also be brought to the gas phase via ion/ion reactions, enabling control over the reactant ion types and their reaction kinetics.366–368 This can be used to obtain enhanced structural information on peptide and lipid analytes.
Cation switching of peptide ions can be achieved by reacting [AuCl2]− anions with [M + 2H]2+ peptide cations to form the electrostatic complexes [M + 2H + AuCl2]+, which lose 2HCl during subsequent low-energy CID and produce aurated peptide ions [M + Au]+.369 Subsequent CID of the mass-selected aurated peptide ion produces a protonated six-membered cyclic iminium ion via HAuNH3 loss facilitated by a lysine residue. This gives in an [M − H − NH3]+ ion that results in preferential cleavage N-terminal to the lysine residue.369 This gold cation-mediated selective dissociation enables site-specific opening of lysine-containing cyclic peptide ions and produces fragment ions with common N- and C-termini, resulting in more unambiguous sequencing than protonated counterparts that produce non-specific CID fragmentation.370 This method can aid in the development of drugs and pesticides using cyclic peptides371–373 by accurately sequencing peptide structures.
Gas-phase charge inversion reactions can also facilitate metal ion transfer with lipids. For example, the ion/ion reaction between FAs ([FA − H]−) and magnesium tris-phenanthroline ([Mg(Phen)3]2+) transfers Mg2+ to fatty acid ions via formation of an electrostatic complex [FA − H + MgPhen]+.368 Mg2+ restricts the positive charge to the carboxylic acid group on the fatty acid ion via strong non-covalent interactions, which unlocks charge-remote fragmentation of the hydrocarbon chain during CID of [FA − H + MgPhen]+ and facilitates unambiguous localization of C–C double bonds.307,374–376 By comparison, low-energy CID of the original [FA − H]− results in uninformative loss of CO2 from the carboxylic acid group as the primary dissociative channel.368 This charge-switching cationization method can also be applied to glycosphingolipids. The positive charge is restricted at the amide oxygen, enabling charge-remote fragmentation upon CID that allows localization of C–C double bond positions on fatty acid ions released from the glycosphingolipids.377,378 In some cases, differences in coordination of the transferred metal allows for differentiation of highly similar compounds in formed ligand–metal–lipid complexes.377,378 For example, sulfatides with either an α-glycosidic or β-glycosidic bond can be separated via a reaction with strontium tris-phenanthroline [Sr(Phen)3]2+, as the diastereomers form complexes containing different numbers of phenanthroline ligands.379 This workflow can be implemented in shotgun lipidomics for differentiating multiple glycosphingolipid isomers in complex lipidomes without the need for chromatographic separation.377,380,381 These ion/ion reactions have also been used to spatially map glycosphingolipid isomers during imaging mass spectrometry analyses of brain tissue.379
5.5. Covalent modifications
Numerous gas-phase ion/ion covalent modifications can be achieved using bioconjugation reagents, such as oxidation by periodate,382 the formation of Schiff bases by aldehyde reagents,383 and the formation of amide384,385 or anhydride386 bonds by ester reagents. The reagent ions are designed to have an electrostatically “sticky” functional group, such as sulf(on)ate and phosph(on)ate groups,387 to aid in forming stable, long-lived electrostatic complexes to allow sufficient time and proximity for covalent chemistry to occur.298 The exothermicity of the ion/ion reaction often drives the spontaneous formation of covalent bonds during the encounter,308,388 signified by the neutral loss of specific byproducts as a result of the reaction or upon subsequent slow-heating activation, such as CID383,389–391 and IRMPD:392| [M + nH]n+ + [R − H]− → [M + (n − 1)H + R](n−1)+ → [M + (n − 1)H + ▲](n−1)+ + (R − ▲) (▲: covalent adduct) | (15) |
The formation of amide bonds can be used for gas-phase cross-linking mass spectrometry via ion/ion reactions between multiply charged protein cations and singly charged ethylene glycol bis(sulfosuccinimidyl succinate) (sulfo-EGS) cross-linker anions. Mutual storage of these oppositely charged reactants leads to the formation of charge-reduced electrostatic complex cations [M + nH + (sulfo-EGS − H)]n−1, followed by spontaneous covalent cross-linking at neutral basic residues, which is signified by the neutral loss of N-hydroxysulfosuccinimide (sulfo-NHS) upon activation.389 The distance restraints imposed by sulfo-EGS provide low-resolution protein conformational information, similar to the cross-linking mass spectrometry studies discussed earlier.249,250 Substituting [sulfo-EGS − 2H + Na]− for [sulfo-EGS − H]− extends cross-linking sites to protonated basic residues by increasing the nucleophilicity of nitrogen atoms in ammonium and guanidinium groups via gas-phase sodium–proton exchange during the ion/ion reactions.392 This enhanced cross-linking reactivity using the linker with sodium allows for differentiation between two gas-phase conformers of alcohol-denatured 8+ ubiquitin, which exhibit subtle conformational differences in the N-terminal region, by detecting additional modification sites compared to using the linker without sodium.392
A Schiff base can be formed in the gas phase during charge inversion ion/ion reactions between analyte ions containing an amine functional group and reagent ions containing an aldehyde functional group.383,391,394 This process is a spontaneous covalent reaction, signified by neutral loss of water during CID of the ion/ion complex formed by the two reactants.383,391,394 For example, mutual storage of a singly deprotonated phosphatidylserine (PS) anion and a doubly charged aliphatic reagent cation, N,N,N′,N′-tetramethyl-N,N′-bis(6-oxohexyl) hexane-1,6-diaminium (TMODA), leads to the formation of a charge-inverted [PS − H + TMODA]+ complex ion.393 Abstraction of a mobile proton from the carboxylic acid group in the serine headgroup is a critical step in the Schiff base condensation reaction, suggesting that phosphatidylserines can be distinguished from other lipid subclasses with different headgroup structures by this reaction.393 Indeed, isobaric PS 40:6 and SHexCer d38:1 are indistinguishable at nominal mass m/z 834 at low mass resolving power, but are readily differentiated via the Schiff base ion/ion reaction by the resulting 18 Da difference in the [PS 40:6 − H + TMODA − H2O]+ and [SHexCer d38:1 − H + TMODA]+ product ions. The use of this ion/ion reaction in an imaging mass spectrometry experiment enables differentiation of these lipids in rat brain tissue, with the phosphatidylserine found localized to the cerebral cortex and the SHexCer localized to the corpus callosum and hippocampus (Fig. 10).393
 | ||
| Fig. 10 (a) A full scan imaging mass spectrometry experiment enables the (b) observation of a single peak at m/z 834.523 and (c) visualization of the H&E staining image and its corresponding spatial distribution. (d) An ion/ion reaction of the ion population at nominal m/z 834 using the TMODA reagent produces (e) a charge inverted product ion complex containing TMODA and SHexCer d38:1 as well as spontaneous water loss signifying Schiff base formation for PS 40:6, which are readily separated and (f) produce distinct spatial localizations where PS 40:6 are localized to the cerebral cortex and SHexCer d38:1 localized to the corpus collosum and hippocampus in the brain. (g) A CASI experiment improves the mass resolving power in the absence of the ion/ion reaction and (h) allows for the separation of SHexCer d38:1 and PS 40:6 using high mass resolving power and (i) validation of the distinct lipid spatial distributions. The analysis times of the ion/ion reaction (565 pixels) and CASI (558 pixels) imaging mass spectrometry experiments were roughly 45 min and 15 min, respectively. Adapted/reproduced from Analytical and Bioanalytical Chemistry, 2023, 415, 4319–4331 (ref. 393) with permission from Springer Nature, Copyright 2023. | ||
The gas-phase ion/ion covalent modifications described here exhibit high reaction efficiency due to the coulombic attraction between oppositely charged ions and the high selectivity of analyte and reagent ions achieved through mass-selective ion isolation prior to the mutual storage for reaction. These features make ion/ion reactions an attractive alternative to their condensed-phase counterparts, which are often slower due to the necessary diffusion of reactants in solvent and can result in low purity because of incomplete and side reactions.298
6. Concluding remarks
The gas-phase reactions in mass spectrometry discussed in this review offer a multitude of approaches to address analytical challenges in a wide variety of sample types, enabling elegant experiments that reveal analyte identity and structure. Gas-phase reactions are generally faster and easier to control than in solution due to the hydrophobic vacuum environment, where analytes do not need to overcome energy barriers involving diffusion, competition between multiple reactions due to the presence of impurities, or difficulty in manipulating equilibrium. The utility of gas-phase chemistries in mass spectrometry, which offer superb control over reactant purity, ion concentration, and reaction kinetics by fine-tuning electronic parameters controlling ion sources, ion optics, and ion traps, is increasingly appreciated by researchers, leading to their widespread adoption in diverse fields of research. However, each technique discussed here also poses unique analytical challenges, some of which can be mitigated by a current trend of combining various complementary methods (e.g., HDX/ECD, CID/EID, CID/UVPD, EThcD) for more comprehensive information on the analytes. Some of these techniques are highly specialized with niche applications, while others hold promise for broad adoption. Continuous efforts to democratize novel approaches are essential for integrating different gas-phase methods, as this process may incur significant costs due to the potential requirement for expert knowledge, hardware and/or software modifications, or the acquisition of a brand-new instrument. An instrument that is flexible enough to perform most of the techniques highlighted here but that is still user-friendly and cost-effective would be ideal.Despite the abundance of novel methods, there is still substantial room for future development and potential new applications through innovation in experimental and instrumental designs. Improvements to reaction and dissociation efficiencies will enable improvements in the limits of detection and sensitivity. Insights from adjacent chemical fields (e.g., organic and inorganic chemistry) can inspire the development and application of novel analytical mass spectrometry techniques, designed to elucidate challenging chemical structures (e.g., stereoisomers) and enhance our understanding of a broader range of analytes across various -omics disciplines. Cross-functional collaborations can lead to technological leaps in different fields, such as the development of quantum computers that use ion traps to confine Ca+ ions and wavelength-specific laser light to precisely manipulate the quantum states of trapped ions, which is essential for quantum gating.395,396 Ion traps provide exceptional control over ion purity, long-term trapping stability, and sustained quantum coherence in well-isolated systems, contributing to their widespread use in quantum computing.295,397 However, rapidly producing and accumulating multifaceted data on a large scale is inevitable considering the current and projected trends in multidisciplinary and multimethod research and automation. This trend requires the construction of powerful computational and software infrastructure to process and utilize big data more effectively and efficiently. Overall, we aim for this exploration of innovative applications and potential future directions in gas-phase ion chemistry to inspire creative solutions for analytical mass spectrometry challenges and to broaden the scope of gas-phase ion chemistry.
Conflicts of interest
There are no conflicts to declare.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Acknowledgements
This work was supported by the National Institutes of Health (NIH) under Award R01 GM138660 (National Institute of General Medical Sciences (NIGMS)). WYK was supported by a Graduate Student Funding Award (GSFA) Fellowship from the University of Florida.References
- J. Laskin and J. H. Futrell, Activation of large lons in FT-ICR mass spectrometry, Mass Spectrom. Rev., 2005, 24(2), 135–167 CrossRef PubMed.
- S. A. McLuckey and M. Mentinova, Ion/neutral, ion/electron, ion/photon, and ion/ion interactions in tandem mass spectrometry: do we need them all? Are they enough?, J. Am. Soc. Mass Spectrom., 2011, 22(1), 3–12 CrossRef PubMed.
- F. Tureček and R. R. Julian, Peptide Radicals and Cation Radicals in the Gas Phase, Chem. Rev., 2013, 113(8), 6691–6733 CrossRef PubMed.
- L. A. Macias, I. C. Santos and J. S. Brodbelt, Ion Activation Methods for Peptides and Proteins, Anal. Chem., 2020, 92(1), 227–251 CrossRef PubMed.
- P. Bayat, D. Lesage and R. B. Cole, Tutorial: Ion Activation In Tandem Mass Spectrometry Using Ultra-High Resolution Instrumentation, Mass Spectrom. Rev., 2020, 39(5–6), 680–702 CrossRef PubMed.
- J. S. Brodbelt, Analytical applications of ion-molecule reactions, Mass Spectrom. Rev., 1997, 16(2), 91–110 CrossRef.
- S. Osburn and V. Ryzhov, Ion–Molecule Reactions: Analytical and Structural Tool, Anal. Chem., 2013, 85(2), 769–778 CrossRef PubMed.
- N. J. Demarais, Ion-atom reactions in bioanalytical mass spectrometry, Int. J. Mass Spectrom., 2021, 468, 116650 CrossRef CAS.
- F. Lermyte, D. Valkenborg, J. A. Loo and F. Sobott, Radical solutions: Principles and application of electron-based dissociation in mass spectrometry-based analysis of protein structure, Mass Spectrom. Rev., 2018, 37(6), 750–771 CrossRef CAS PubMed.
- R. C. Dunbar, BIRD (blackbody infrared radiative dissociation): Evolution, principles, and applications, Mass Spectrom. Rev., 2004, 23(2), 127–158 CrossRef CAS PubMed.
- J. S. Brodbelt, Shedding light on the frontier of photodissociation, J. Am. Soc. Mass Spectrom., 2011, 22(2), 197–206 CrossRef CAS PubMed.
- J. S. Brodbelt, L. J. Morrison and I. Santos, Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules, Chem. Rev., 2020, 120(7), 3328–3380 CrossRef CAS PubMed.
- J. P. Reilly, Ultraviolet photofragmentation of biomolecular ions, Mass Spectrom. Rev., 2009, 28(3), 425–447 CrossRef CAS PubMed.
- S. A. McLuckey and J. L. Stephenson, Ion/ion chemistry of high-mass multiply charged ions, Mass Spectrom. Rev., 1998, 17(6), 369–407 CrossRef CAS.
- S. J. Pitteri and S. A. McLuckey, Recent developments in the ion/ion chemistry of high-mass multiply charged ions, Mass Spectrom. Rev., 2005, 24(6), 931–958 CrossRef CAS PubMed.
- B. M. Prentice and S. A. McLuckey, Gas-phase ion/ion reactions of peptides and proteins: acid/base, redox, and covalent chemistries, Chem. Commun., 2013, 49(10), 947–965 RSC.
- N. M. Riley and J. J. Coon, The Role of Electron Transfer Dissociation in Modern Proteomics, Anal. Chem., 2018, 90(1), 40–64 CrossRef CAS.
- J. R. Bonney and B. M. Prentice, Perspective on Emerging Mass Spectrometry Technologies for Comprehensive Lipid Structural Elucidation, Anal. Chem., 2021, 93(16), 6311–6322 CrossRef CAS PubMed.
- M. Grabarics, M. Lettow, C. Kirschbaum, K. Greis, C. Manz and K. Pagel, Mass Spectrometry-Based Techniques to Elucidate the Sugar Code, Chem. Rev., 2022, 122(8), 7840–7908 CrossRef CAS PubMed.
- P. M. Mayer and C. Poon, The mechanisms of collisional activation of ions in mass spectrometry, Mass Spectrom. Rev., 2009, 28(4), 608–639 CrossRef CAS PubMed.
- L. Sleno and D. A. Volmer, Ion activation methods for tandem mass spectrometry, J. Mass Spectrom., 2004, 39(10), 1091–1112 CrossRef CAS PubMed.
- S. A. McLuckey, Principles of collisional activation in analytical mass spectrometry, J. Am. Soc. Mass Spectrom., 1992, 3(6), 599–614 CrossRef CAS PubMed.
- B. L. J. Poad, D. L. Marshall, E. Harazim, R. Gupta, V. R. Narreddula, R. S. E. Young, E. Duchoslav, J. L. Campbell, J. A. Broadbent, J. Cvačka, T. W. Mitchell and S. J. Blanksby, Combining Charge-Switch Derivatization with Ozone-Induced Dissociation for Fatty Acid Analysis, J. Am. Soc. Mass Spectrom., 2019, 30(10), 2135–2143 CrossRef PubMed.
- F. Berthias, B. L. J. Poad, H. A. Thurman, A. P. Bowman, S. J. Blanksby and A. A. Shvartsburg, Disentangling Lipid Isomers by High-Resolution Differential Ion Mobility Spectrometry/Ozone-Induced Dissociation of Metalated Species, J. Am. Soc. Mass Spectrom., 2021, 32(12), 2827–2836 CrossRef PubMed.
- S. H. J. Brown, T. W. Mitchell and S. J. Blanksby, Analysis of unsaturated lipids by ozone-induced dissociation, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2011, 1811(11), 807–817 CrossRef PubMed.
- D. L. Marshall, A. Criscuolo, R. S. E. Young, B. L. J. Poad, M. Zeller, G. E. Reid, T. W. Mitchell and S. J. Blanksby, Mapping Unsaturation in Human Plasma Lipids by Data-Independent Ozone-Induced Dissociation, J. Am. Soc. Mass Spectrom., 2019, 30(9), 1621–1630 Search PubMed.
- R. C. Barrientos and Q. Zhang, Fragmentation Behavior and Gas-Phase Structures of Cationized Glycosphingolipids in Ozone-Induced Dissociation Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2019, 30(9), 1609–1620 CrossRef PubMed.
- D. H. Ross, J.-Y. Lee, A. Bilbao, D. J. Orton, J. G. Eder, M. C. Burnet, B. L. Deatherage Kaiser, J. E. Kyle and X. Zheng, LipidOz enables automated elucidation of lipid carbon–carbon double bond positions from ozone-induced dissociation mass spectrometry data, Commun. Chem., 2023, 6(1), 74 CrossRef PubMed.
- C. Auranwiwat, A. T. Maccarone, A. W. Carroll, R. Rattanajak, S. Kamchonwongpaisan, S. J. Blanksby, S. G. Pyne and T. Limtharakul, Structure elucidation of cyclohexene (9Z)-octadec-9-enyl ethers isolated from the leaves of Uvaria cherrevensis (Annonaceae), Tetrahedron, 2019, 75(15), 2336–2342 CrossRef.
- S. L. Knowles, N. Vu, D. A. Todd, H. A. Raja, A. Rokas, Q. Zhang and N. H. Oberlies, Orthogonal Method for Double-Bond Placement via Ozone-Induced Dissociation Mass Spectrometry (OzID-MS), J. Nat. Prod., 2019, 82(12), 3421–3431 CrossRef PubMed.
- J. M. Greenberg, Radical formation, chemical processing, and explosion of interstellar grains, Astrophys. Space Sci., 1976, 39(1), 9–18 CrossRef.
- B. F. E. Curchod and A. J. Orr-Ewing, Perspective on Theoretical and Experimental Advances in Atmospheric Photochemistry, J. Phys. Chem. A, 2024, 128(32), 6613–6635 Search PubMed.
- J. B. Hatvany and E. S. Gallagher, Hydrogen/deuterium exchange for the analysis of carbohydrates, Carbohydr. Res., 2023, 530, 108859 CrossRef PubMed.
- Y. Kostyukevich, T. Acter, A. Zherebker, A. Ahmed, S. Kim and E. Nikolaev, Hydrogen/deuterium exchange in mass spectrometry, Mass Spectrom. Rev., 2018, 37(6), 811–853 CrossRef PubMed.
- S. S. Uppal, S. E. Beasley, M. Scian and M. Guttman, Gas-Phase Hydrogen/Deuterium Exchange for Distinguishing Isomeric Carbohydrate Ions, Anal. Chem., 2017, 89(8), 4737–4742 CrossRef PubMed.
- K. Kelly, S. Bell, H. Maleki and S. Valentine, Synthetic Small Molecule Characterization and Isomer Discrimination Using Gas-Phase Hydrogen–Deuterium Exchange IMS-MS, Anal. Chem., 2019, 91(9), 6259–6265 CrossRef PubMed.
- Y. Hamuro, Tutorial: Chemistry of Hydrogen/Deuterium Exchange Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2021, 32(1), 133–151 CrossRef PubMed.
- E. I. James, T. A. Murphree, C. Vorauer, J. R. Engen and M. Guttman, Advances in Hydrogen/Deuterium Exchange Mass Spectrometry and the Pursuit of Challenging Biological Systems, Chem. Rev., 2022, 122(8), 7562–7623 CrossRef PubMed.
- M. K. Green and C. B. Lebrilla, Ion-molecule reactions as probes of gas-phase structures of peptides and proteins, Mass Spectrom. Rev., 1997, 16(2), 53–71 CrossRef PubMed.
- K. Eller and H. Schwarz, Organometallic chemistry in the gas phase, Chem. Rev., 1991, 91(6), 1121–1177 CrossRef.
- B. G. Heikes, V. Treadaway, A. S. McNeill, I. K. C. Silwal and D. W. O'Sullivan, An ion-neutral model to investigate chemical ionization mass spectrometry analysis of atmospheric molecules – application to a mixed reagent ion system for hydroperoxides and organic acids, Atmos. Meas. Tech., 2018, 11(4), 1851–1881 CrossRef.
- K. Parker, N. E. Bollis and V. Ryzhov, Ion-molecule reactions of mass-selected ions, Mass Spectrom. Rev., 2024, 43(1), 47–89 CrossRef PubMed.
- P. E. Williams, B. J. Jankiewicz, L. Yang and H. I. Kenttämaa, Properties and Reactivity of Gaseous Distonic Radical Ions with Aryl Radical Sites, Chem. Rev., 2013, 113(9), 6949–6985 CrossRef PubMed.
- S. A. McLuckey and D. E. Goeringer, SPECIAL FEATURE: TUTORIAL Slow Heating Methods in Tandem Mass Spectrometry, J. Mass Spectrom., 1997, 32(5), 461–474 CrossRef.
- J. Mitchell Wells and S. A. McLuckey, Collision-Induced Dissociation (CID) of Peptides and Proteins, in Methods Enzymol., Academic Press, 2005, vol. 402, pp. 148–185 Search PubMed.
- X. Zhao, W. Zhang, D. Zhang, X. Liu, W. Cao, Q. Chen, Z. Ouyang and Y. Xia, A lipidomic workflow capable of resolving sn- and C=C location isomers of phosphatidylcholines, Chem. Sci., 2019, 10(46), 10740–10748 RSC.
- W. Zhang, R. Jian, J. Zhao, Y. Liu and Y. Xia, Deep-lipidotyping by mass spectrometry: recent technical advances and applications, J. Lipid Res., 2022, 63(7), 100219 CrossRef PubMed.
- X. Ma and Y. Xia, Pinpointing Double Bonds in Lipids by Paternò-Büchi Reactions and Mass Spectrometry, Angew. Chem., Int. Ed., 2014, 53(10), 2592–2596 CrossRef PubMed.
- Y. Feng, B. Chen, Q. Yu and L. Li, Identification of Double Bond Position Isomers in Unsaturated Lipids by m-CPBA Epoxidation and Mass Spectrometry Fragmentation, Anal. Chem., 2019, 91(3), 1791–1795 CrossRef.
- D. Unsihuay, P. Su, H. Hu, J. Qiu, S. Kuang, Y. Li, X. Sun, S. K. Dey and J. Laskin, Imaging and Analysis of Isomeric Unsaturated Lipids through Online Photochemical Derivatization of Carbon–Carbon Double Bonds, Angew. Chem., Int. Ed., 2021, 60(14), 7559–7563 CrossRef PubMed.
- X. Zhao and Y. Xia, Characterization of Fatty Acyl Modifications in Phosphatidylcholines and Lysophosphatidylcholines via Radical-Directed Dissociation, J. Am. Soc. Mass Spectrom., 2021, 32(2), 560–568 CrossRef PubMed.
- Q. Lin, P. Li, R. Jian and Y. Xia, Localization of Intrachain Modifications in Bacterial Lipids Via Radical-Directed Dissociation, J. Am. Soc. Mass Spectrom., 2022, 33(4), 714–721 CrossRef PubMed.
- A. R. Dongré, J. L. Jones, Á. Somogyi and V. H. Wysocki, Influence of Peptide Composition, Gas-Phase Basicity, and Chemical Modification on Fragmentation Efficiency: Evidence for the Mobile Proton Model, J. Am. Chem. Soc., 1996, 118(35), 8365–8374 CrossRef.
- I. Jang, A. Jeon, S. G. Lim, D. K. Hong, M. S. Kim, J. H. Jo, S. T. Lee, B. Moon and H. B. Oh, Free Radical–Initiated Peptide Sequencing Mass Spectrometry for Phosphopeptide Post-translational Modification Analysis, J. Am. Soc. Mass Spectrom., 2019, 30(3), 538–547 CrossRef PubMed.
- N. B. Borotto, K. M. Ileka, C. A. T. M. B. Tom, B. R. Martin and K. Håkansson, Free Radical Initiated Peptide Sequencing for Direct Site Localization of Sulfation and Phosphorylation with Negative Ion Mode Mass Spectrometry, Anal. Chem., 2018, 90(16), 9682–9686 CrossRef PubMed.
- S. T. Lee, H. Park, I. Jang, C. S. Lee, B. Moon and H. B. Oh, New free radical-initiated peptide sequencing (FRIPS) mass spectrometry reagent with high conjugation efficiency enabling single-step peptide sequencing, Sci. Rep., 2022, 12(1), 9494 CrossRef PubMed.
- M. Lee, M. Kang, B. Moon and H. B. Oh, Gas-phase peptide sequencing by TEMPO-mediated radical generation, Analyst, 2009, 134(8), 1706–1712 RSC.
- K. E. Osho, K. A. Wasik, L. M. Geary and N. B. Borotto, Improved Performance of Positive-Ion Mode Free Radical-Initiated Peptide Sequencing with p-TEMPO-Bz, J. Am. Soc. Mass Spectrom., 2023, 34(4), 579–585 CrossRef PubMed.
- V. H. Wysocki, G. Tsaprailis, L. L. Smith and L. A. Breci, Mobile and localized protons: a framework for understanding peptide dissociation, J. Mass Spectrom., 2000, 35(12), 1399–1406 CrossRef.
- C. Rojas Ramírez, R. Murtada, J. Gao and B. T. Ruotolo, Free Radical-Based Sequencing for Native Top-Down Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2022, 33(12), 2283–2290 CrossRef PubMed.
- J.-u. Lee, S. T. Lee, C. R. Park, B. Moon, H. I. Kim and H. B. Oh, TEMPO-Assisted Free-Radical-Initiated Peptide Sequencing Mass Spectrometry for Ubiquitin Ions: An Insight on the Gas-Phase Conformations, J. Am. Soc. Mass Spectrom., 2022, 33(3), 471–481 CrossRef PubMed.
- S. M. Dixit, D. A. Polasky and B. T. Ruotolo, Collision induced unfolding of isolated proteins in the gas phase: past, present, and future, Curr. Opin. Chem. Biol., 2018, 42, 93–100 CrossRef PubMed.
- D. D. Vallejo, C. Rojas Ramírez, K. F. Parson, Y. Han, V. V. Gadkari and B. T. Ruotolo, Mass Spectrometry Methods for Measuring Protein Stability, Chem. Rev., 2022, 122(8), 7690–7719 CrossRef PubMed.
- R. Matney and V. V. Gadkari, Recent advances in gas phase unfolding: Instrumentation and applications, J. Mass Spectrom., 2024, 59(7), e5059 CrossRef.
- A. Phetsanthad, G. Li, C. K. Jeon, B. T. Ruotolo and L. Li, Comparing Selected-Ion Collision Induced Unfolding with All Ion Unfolding Methods for Comprehensive Protein Conformational Characterization, J. Am. Soc. Mass Spectrom., 2022, 33(6), 944–951 CrossRef PubMed.
- S. M. Fantin, K. F. Parson, S. Niu, J. Liu, D. A. Polasky, S. M. Dixit, S. M. Ferguson-Miller and B. T. Ruotolo, Collision Induced Unfolding Classifies Ligands Bound to the Integral Membrane Translocator Protein, Anal. Chem., 2019, 91(24), 15469–15476 CrossRef PubMed.
- Y. Han, A. A. Desai, J. M. Zupancic, M. D. Smith, P. M. Tessier and B. T. Ruotolo, Native ion mobility-mass spectrometry reveals the binding mechanisms of anti-amyloid therapeutic antibodies, Protein Sci., 2024, 33(6), e5008 CrossRef PubMed.
- C. Zhao, N. B. Borotto, J. Schmidt, K. Srivastava, A. Lowell, K. Hakansson, D. H. Sherman and B. T. Ruotolo, Gas-Phase Unfolding Reveals Stability Shifts Associated with Substrate Binding in Modular Polyketide Synthases, J. Am. Soc. Mass Spectrom., 2025, 36(2), 241–246 CrossRef PubMed.
- C. A. Schalley, Supramolecular chemistry goes gas phase: the mass spectrometric examination of noncovalent interactions in host–guest chemistry and molecular recognition, Int. J. Mass Spectrom., 2000, 194(1), 11–39 CrossRef CAS.
- C. A. Schalley, Molecular recognition and supramolecular chemistry in the gas phase, Mass Spectrom. Rev., 2001, 20(5), 253–309 CrossRef CAS PubMed.
- P. Bayat, D. Gatineau, D. Lesage, S. Marhabaie, A. Martinez and R. B. Cole, Investigation of activation energies for dissociation of host-guest complexes in the gas phase using low-energy collision induced dissociation, J. Mass Spectrom., 2019, 54(5), 437–448 CrossRef CAS PubMed.
- P. Bayat, D. Gatineau, D. Lesage, A. Martinez and R. B. Cole, Benchmarking higher-energy collision dissociation (HCD) by investigation of binding energies of gas-phase host-guest complexes of hemicryptophane cages, J. Mass Spectrom., 2022, e4879 CrossRef CAS.
- T. Heravi, J. Shen, S. Johnson, M. C. Asplund and D. V. Dearden, Halide Size-Selective Binding by Cucurbit[5]uril–Alkali Cation Complexes in the Gas Phase, J. Phys. Chem. A, 2021, 125(36), 7803–7812 CrossRef CAS PubMed.
- M. C. Thomas, T. W. Mitchell and S. J. Blanksby, Ozonolysis of Phospholipid Double Bonds during Electrospray Ionization: A New Tool for Structure Determination, J. Am. Chem. Soc., 2006, 128(1), 58–59 CrossRef CAS PubMed.
- M. C. Thomas, T. W. Mitchell, D. G. Harman, J. M. Deeley, R. C. Murphy and S. J. Blanksby, Elucidation of Double Bond Position in Unsaturated Lipids by Ozone Electrospray Ionization Mass Spectrometry, Anal. Chem., 2007, 79(13), 5013–5022 CrossRef CAS PubMed.
- M. C. Thomas, T. W. Mitchell, D. G. Harman, J. M. Deeley, J. R. Nealon and S. J. Blanksby, Ozone-Induced Dissociation: Elucidation of Double Bond Position within Mass-Selected Lipid Ions, Anal. Chem., 2008, 80(1), 303–311 CrossRef CAS PubMed.
- X. Liu, B. Jiao, W. Cao, X. Ma, Y. Xia, S. J. Blanksby, W. Zhang and Z. Ouyang, Development of a Miniature Mass Spectrometry System for Point-of-Care Analysis of Lipid Isomers Based on Ozone-Induced Dissociation, Anal. Chem., 2022, 94(40), 13944–13950 CrossRef CAS PubMed.
- B. S. R. Claes, A. P. Bowman, B. L. J. Poad, R. S. E. Young, R. M. A. Heeren, S. J. Blanksby and S. R. Ellis, Mass Spectrometry Imaging of Lipids with Isomer Resolution Using High-Pressure Ozone-Induced Dissociation, Anal. Chem., 2021, 93(28), 9826–9834 CrossRef CAS PubMed.
- O. B. Gadzhiev, S. K. Ignatov, B. E. Krisyuk, A. V. Maiorov, S. Gangopadhyay and A. E. Masunov, Quantum Chemical Study of the Initial Step of Ozone Addition to the Double Bond of Ethylene, J. Phys. Chem. A, 2012, 116(42), 10420–10434 CrossRef CAS PubMed.
- R. Criegee, Mechanism of Ozonolysis, Angew Chem. Int. Ed. Engl., 1975, 14(11), 745–752 CrossRef.
- B. L. J. Poad, R. S. E. Young, D. L. Marshall, A. J. Trevitt and S. J. Blanksby, Accelerating Ozonolysis Reactions Using Supplemental RF-Activation of Ions in a Linear Ion Trap Mass Spectrometer, Anal. Chem., 2022, 94(9), 3897–3903 CrossRef CAS PubMed.
- L. Tian, Q. Liu, X. Wang, S. Chen and Y. Li, Fighting ferroptosis: Protective effects of dexmedetomidine on vital organ injuries, Life Sci., 2024, 354, 122949 CrossRef CAS PubMed.
- Y. Zou and S. L. Schreiber, Progress in Understanding Ferroptosis and Challenges in Its Targeting for Therapeutic Benefit, Cell Chem. Biol., 2020, 27(4), 463–471 CrossRef CAS PubMed.
- K. S. Chadaideh and R. N. Carmody, Host-microbial interactions in the metabolism of different dietary fats, Cell Metab., 2021, 33(5), 857–872 CrossRef CAS PubMed.
- V. I. Bunik, J. V. Schloss, J. T. Pinto, G. E. Gibson and A. J. L. Cooper, Enzyme-Catalyzed Side Reactions with Molecular Oxygen may Contribute to Cell Signaling and Neurodegenerative Diseases, Neurochem. Res., 2007, 32(4), 871–891 CrossRef CAS.
- B. K. Smith, L. E. Robinson, R. Nam and D. W. L. Ma, Trans-fatty acids and cancer: a mini-review, Br. J. Nutr., 2009, 102(9), 1254–1266 CrossRef CAS PubMed.
- C. Chatgilialoglu and C. Ferreri, Trans Lipids: The Free Radical Path, Acc. Chem. Res., 2005, 38(6), 441–448 CrossRef CAS PubMed.
- L. Yang, L.-m. Cao, X.-j. Zhang and B. Chu, Targeting ferroptosis as a vulnerability in pulmonary diseases, Cell Death Dis., 2022, 13(7), 649 CrossRef CAS PubMed.
- D. S. Cornett, M. L. Reyzer, P. Chaurand and R. M. Caprioli, MALDI imaging mass spectrometry: molecular snapshots of biochemical systems, Nat. Methods, 2007, 4(10), 828–833 Search PubMed.
- K. Chughtai and R. M. A. Heeren, Mass spectrometric imaging for biomedical tissue analysis, Chem. Rev., 2010, 110(5), 3237–3277 Search PubMed.
- B. M. Prentice, An analytical evaluation of tools for lipid isomer differentiation in imaging mass spectrometry, Int. J. Mass Spectrom., 2024, 502, 117268 Search PubMed.
- R. S. E. Young, B. S. R. Claes, A. P. Bowman, E. D. Williams, B. Shepherd, A. Perren, B. L. J. Poad, S. R. Ellis, R. M. A. Heeren, M. C. Sadowski and S. J. Blanksby, Isomer-Resolved Imaging of Prostate Cancer Tissues Reveals Specific Lipid Unsaturation Profiles Associated With Lymphocytes and Abnormal Prostate Epithelia, Front. Endocrinol., 2021, 12, 689600 CrossRef PubMed.
- M. R. L. Paine, B. L. J. Poad, G. B. Eijkel, D. L. Marshall, S. J. Blanksby, R. M. A. Heeren and S. R. Ellis, Mass Spectrometry Imaging with Isomeric Resolution Enabled by Ozone-Induced Dissociation, Angew. Chem., Int. Ed., 2018, 57(33), 10530–10534 CrossRef CAS PubMed.
- H. T. Pham, A. T. Maccarone, M. C. Thomas, J. L. Campbell, T. W. Mitchell and S. J. Blanksby, Structural characterization of glycerophospholipids by combinations of ozone- and collision-induced dissociation mass spectrometry: the next step towards “top-down” lipidomics, Analyst, 2014, 139(1), 204–214 RSC.
- A. M. Batarseh, S. K. Abbott, E. Duchoslav, A. Alqarni, S. J. Blanksby and T. W. Mitchell, Discrimination of isobaric and isomeric lipids in complex mixtures by combining ultra-high pressure liquid chromatography with collision and ozone-induced dissociation, Int. J. Mass Spectrom., 2018, 431, 27–36 CrossRef CAS.
- J. P. Menzel, R. S. E. Young, A. H. Benfield, J. S. Scott, P. Wongsomboon, L. Cudlman, J. Cvačka, L. M. Butler, S. T. Henriques, B. L. J. Poad and S. J. Blanksby, Ozone-enabled fatty acid discovery reveals unexpected diversity in the human lipidome, Nat. Commun., 2023, 14(1), 3940 CrossRef CAS PubMed.
- J. P. Wiens, O. Martinez Jr, S. G. Ard, B. C. Sweeny, A. A. Viggiano and N. S. Shuman, Kinetics of Cations with C2 Hydrofluorocarbon Radicals, J. Phys. Chem. A, 2017, 121(42), 8061–8068 CrossRef CAS PubMed.
- J. C. Sawyer, N. S. Shuman, J. P. Wiens and A. A. Viggiano, Kinetics and Product Branching Fractions of Reactions between a Cation and a Radical: Ar+ + CH3 and O2+ + CH3, J. Phys. Chem. A, 2015, 119(6), 952–958 CrossRef CAS PubMed.
- L. Pei, E. Carrascosa, N. Yang, S. Falcinelli and J. M. Farrar, Velocity Map Imaging Study of Charge-Transfer and Proton-Transfer Reactions of CH3 Radicals with H3+, J. Phys. Chem. Lett., 2015, 6(9), 1684–1689 CrossRef CAS PubMed.
- V. Zhelyazkova, F. B. V. Martins, S. Schilling and F. Merkt, Reaction of an Ion and a Free Radical near 0 K: He+ + NO → He + N+ + O, J. Phys. Chem. A, 2023, 127(6), 1458–1468 CrossRef CAS PubMed.
- X. Zhang, V. M. Bierbaum, G. B. Ellison and S. Kato, Gas-phase reactions of organic radicals and diradicals with ions, J. Chem. Phys., 2004, 120(8), 3531–3534 Search PubMed.
- D. Hanson, J. Orlando, B. Noziere and E. Kosciuch, Proton transfer mass spectrometry studies of peroxy radicals, Int. J. Mass Spectrom., 2004, 239(2), 147–159 CrossRef CAS.
- H. Kersten, V. Funcke, M. Lorenz, K. J. Brockmann, T. Benter and R. O'Brien, Evidence of neutral radical induced analyte ion transformations in APPI and Near-VUV APLI, J. Am. Soc. Mass Spectrom., 2009, 20(10), 1868–1880 CrossRef CAS PubMed.
- X. Zhang, S. Kato, V. M. Bierbaum, M. R. Nimlos and G. B. Ellison, Use of a Flowing Afterglow SIFT Apparatus To Study the Reactions of Ions with Organic Radicals, J. Phys. Chem. A, 2004, 108(45), 9733–9741 CrossRef CAS.
- J. Greenberg, P. C. Schmid, M. Miller, J. F. Stanton and H. J. Lewandowski, Quantum-state-controlled reactions between molecular radicals and ions, Phys. Rev. A, 2018, 98(3), 032702 CrossRef CAS.
- M. Sablier and T. Fujii, Mass Spectrometry of Free Radicals, Chem. Rev., 2002, 102(9), 2855–2924 CrossRef CAS PubMed.
- D. Smith and P. Španěl, The SIFT and FALP techniques; applications to ionic and electronic reactions studies and their evolution to the SIFT-MS and FA-MS analytical methods, Int. J. Mass Spectrom., 2015, 377, 467–478 CrossRef CAS.
- P. C. Schmid, J. Greenberg, M. I. Miller, K. Loeffler and H. J. Lewandowski, An ion trap time-of-flight mass spectrometer with high mass resolution for cold trapped ion experiments, Rev. Sci. Instrum., 2017, 88(12), 123107 CrossRef CAS PubMed.
- W. Zhang, L. Xu and H. Zhang, Recent advances in mass spectrometry techniques for atmospheric chemistry research on molecular-level, Mass Spectrom. Rev., 2024, 43(5), 1091–1134 CrossRef CAS PubMed.
- B. Yuan, A. R. Koss, C. Warneke, M. Coggon, K. Sekimoto and J. A. de Gouw, Proton-Transfer-Reaction Mass Spectrometry: Applications in Atmospheric Sciences, Chem. Rev., 2017, 117(21), 13187–13229 Search PubMed.
- A. Hansel, W. Scholz, B. Mentler, L. Fischer and T. Berndt, Detection of RO2 radicals and other products from cyclohexene ozonolysis with NH4+ and acetate chemical ionization mass spectrometry, Atmos. Environ., 2018, 186, 248–255 CrossRef CAS.
- T. Berndt, B. Mentler, W. Scholz, L. Fischer, H. Herrmann, M. Kulmala and A. Hansel, Accretion Product Formation from Ozonolysis and OH Radical Reaction of α-Pinene: Mechanistic Insight and the Influence of Isoprene and Ethylene, Environ. Sci. Technol., 2018, 52(19), 11069–11077 CrossRef CAS PubMed.
- H. O. T. Pye, C. K. Ward-Caviness, B. N. Murphy, K. W. Appel and K. M. Seltzer, Secondary organic aerosol association with cardiorespiratory disease mortality in the United States, Nat. Commun., 2021, 12(1), 7215 Search PubMed.
- L. A. Eaves, L. Smeester, H. J. Hartwell, Y.-H. Lin, M. Arashiro, Z. Zhang, A. Gold, J. D. Surratt and R. C. Fry, Isoprene-Derived Secondary Organic Aerosol Induces the Expression of MicroRNAs Associated with Inflammatory/Oxidative Stress Response in Lung Cells, Chem. Res. Toxicol., 2020, 33(2), 381–387 Search PubMed.
- Y. Lin, F. Wyrowski, H. B. Liu, A. F. Izquierdo, T. Csengeri, S. Leurini and K. M. Menten, The evolution of temperature and density structures of OB cluster-forming molecular clumps, Astron. Astrophys., 2022, 658(A128), 1–46 Search PubMed.
- M. T. Bell, P. Softley and T., Ultracold molecules and ultracold chemistry, Mol. Phys., 2009, 107(2), 99–132 CrossRef CAS.
- T. P. Softley, Cold and ultracold molecules in the twenties, Proc. R. Soc. A, 2023, 479(2274), 20220806 CrossRef.
- C. A. Perera, C. Amarasinghe, H. Guo and A. G. Suits, Cold collisions of hot molecules, Phys. Chem. Chem. Phys., 2023, 25(34), 22595–22606 Search PubMed.
- Y. Wang, A. Owens, J. Tennyson and S. N. Yurchenko, MARVEL Analysis of the Measured High-resolution Rovibronic Spectra of the Calcium Monohydroxide Radical (CaOH), Astrophys. J., Suppl. Ser., 2020, 248(1), 9 CrossRef CAS.
- R. Dupuy, G. Féraud, M. Bertin, X. Michaut, T. Putaud, P. Jeseck, L. Philippe, C. Romanzin, V. Baglin, R. Cimino and J. H. Fillion, The efficient photodesorption of nitric oxide (NO) ices, Astron. Astrophys., 2017, 606, L9 Search PubMed.
- R. I. Kaiser, Experimental Investigation on the Formation of Carbon-Bearing Molecules in the Interstellar Medium via Neutral−Neutral Reactions, Chem. Rev., 2002, 102(5), 1309–1358 Search PubMed.
- E. Herbst, Unusual Chemical Processes in Interstellar Chemistry: Past and Present, Front. Astron. Space Sci., 2021, 8, 776942 CrossRef.
- A. Bouibes and A. Zaoui, A route to possible civil engineering materials: the case of high-pressure phases of lime, Sci. Rep., 2015, 5(1), 12330 Search PubMed.
- O. A. Krohn and H. J. Lewandowski, Cold Ion–Molecule Reactions in the Extreme Environment of a Coulomb Crystal, J. Phys. Chem. A, 2024, 128(10), 1737–1752 CrossRef CAS PubMed.
- U. H. Mistarz, S. A. Chandler, J. M. Brown, J. L. P. Benesch and K. D. Rand, Probing the Dissociation of Protein Complexes by Means of Gas-Phase H/D Exchange Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2019, 30(1), 45–57 CrossRef CAS PubMed.
- F. Wu, Y. Wang, Y. Chen, Z. Li and C.-F. Ding, Alkali metal ion-induced conformation changes of methionine- and leucine enkephalin investigated by gas-phase hydrogen/deuterium exchange combined with theoretical calculations, J. Mol. Struct., 2021, 1233, 130113 CrossRef CAS.
- H. M. Schramm, T. Tamadate, C. J. Hogan and B. H. Clowers, Ion-neutral clustering alters gas-phase hydrogen–deuterium exchange rates, Phys. Chem. Chem. Phys., 2023, 25(6), 4959–4968 Search PubMed.
- Y. Kostyukevich, G. Vladimirov, E. Stekolschikova, D. Ivanov, A. Yablokov, A. Zherebker, S. Sosnin, A. Orlov, M. Fedorov, P. Khaitovich and E. Nikolaev, Hydrogen/Deuterium Exchange Aiding Compound Identification for LC-MS and MALDI Imaging Lipidomics, Anal. Chem., 2019, 91(21), 13465–13474 CrossRef CAS PubMed.
- B. A. Collings, J. M. Campbell, D. Mao and D. J. Douglas, A combined linear ion trap time-of-flight system with improved performance and MSn capabilities, Rapid Commun. Mass Spectrom., 2001, 15(19), 1777–1795 Search PubMed.
- R. N. Straus and R. A. Jockusch, Hydrogen-Deuterium Exchange and Electron Capture Dissociation to Interrogate the Conformation of Gaseous Melittin Ions, J. Am. Soc. Mass Spectrom., 2019, 30(5), 864–875 Search PubMed.
- M. F. Czar, A. Marchand and R. Zenobi, A Modified Traveling Wave Ion Mobility Mass Spectrometer as a Versatile Platform for Gas-Phase Ion–Molecule Reactions, Anal. Chem., 2019, 91(10), 6624–6631 CrossRef CAS PubMed.
- K. B. Green-Church, P. A. Limbach, M. A. Freitas and A. G. Marshall, Gas-phase hydrogen/deuterium exchange of positively charged mononucleotides by use of Fourier-transform ion cyclotron resonance mass spectrometry, J. Am. Soc. Mass Spectrom., 2001, 12(3), 268–277 CrossRef CAS PubMed.
- M. Miladi, A. D. Olaitan, B. Zekavat and T. Solouki, Competing Noncovalent Host-guest Interactions and H/D Exchange: Reactions of Benzyloxycarbonyl-Proline Glycine Dipeptide Variants with ND3, J. Am. Soc. Mass Spectrom., 2015, 26(11), 1938–1949 CrossRef CAS PubMed.
- A. Mookherjee, S. S. Uppal, T. A. Murphree and M. Guttman, Linkage Memory in Underivatized Protonated Carbohydrates, J. Am. Soc. Mass Spectrom., 2021, 32(2), 581–589 CrossRef CAS PubMed.
- L. Konermann, E. Ahadi, A. D. Rodriguez and S. Vahidi, Unraveling the mechanism of electrospray ionization, Anal. Chem., 2013, 85(1), 2–9 Search PubMed.
- M. M. Gadzuk-Shea and M. F. Bush, Effects of Charge State on the Structures of Serum Albumin Ions in the Gas Phase: Insights from Cation-to-Anion Proton-Transfer Reactions, Ion Mobility, and Mass Spectrometry, J. Phys. Chem. B, 2018, 122(43), 9947–9955 CrossRef CAS PubMed.
- K. J. Laszlo, E. B. Munger and M. F. Bush, Effects of Solution Structure on the Folding of Lysozyme Ions in the Gas Phase, J. Phys. Chem. B, 2017, 121(13), 2759–2766 Search PubMed.
- K. J. Laszlo, E. B. Munger and M. F. Bush, Folding of Protein Ions in the Gas Phase after Cation-to-Anion Proton-Transfer Reactions, J. Am. Chem. Soc., 2016, 138(30), 9581–9588 CrossRef CAS PubMed.
- K. J. Laszlo, J. H. Buckner, E. B. Munger and M. F. Bush, Native-Like and Denatured Cytochrome c Ions Yield Cation-to-Anion Proton Transfer Reaction Products with Similar Collision Cross-Sections, J. Am. Soc. Mass Spectrom., 2017, 28(7), 1382–1391 CrossRef CAS PubMed.
- C. Bleiholder and F. C. Liu, Structure Relaxation Approximation (SRA) for Elucidation of Protein Structures from Ion Mobility Measurements, J. Phys. Chem. B, 2019, 123(13), 2756–2769 Search PubMed.
- S. A. Raab, T. J. El-Baba, A. Laganowsky, D. H. Russell, S. J. Valentine and D. E. Clemmer, Protons Are Fast and Smart; Proteins Are Slow and Dumb: On the Relationship of Electrospray Ionization Charge States and Conformations, J. Am. Soc. Mass Spectrom., 2021, 32(7), 1553–1561 Search PubMed.
- S. L. Koeniger and D. E. Clemmer, Resolution and structural transitions of elongated states of ubiquitin, J. Am. Soc. Mass Spectrom., 2007, 18(2), 322–331 CrossRef CAS PubMed.
- R. Chaturvedi and I. K. Webb, Multiplexed Conformationally Selective, Localized Gas-Phase Hydrogen Deuterium Exchange of Protein Ions Enabled by Transmission-Mode Electron Capture Dissociation, Anal. Chem., 2022, 94(25), 8975–8982 CrossRef CAS PubMed.
- M. F. Mirabelli and R. Zenobi, Observing Proton Transfer Reactions Inside the MALDI Plume: Experimental and Theoretical Insight into MALDI Gas-Phase Reactions, J. Am. Soc. Mass Spectrom., 2017, 28(8), 1676–1686 CrossRef CAS PubMed.
- X. Chen, Z. Wang, Y. L. E. Wong, R. Wu, F. Zhang and T. W. D. Chan, Electron-ion reaction-based dissociation: A powerful ion activation method for the elucidation of natural product structures, Mass Spectrom. Rev., 2018, 37(6), 793–810 CrossRef CAS PubMed.
- F. Kjeldsen, K. F. Haselmann, B. A. Budnik, F. Jensen and R. A. Zubarev, Dissociative capture of hot (3–13 eV) electrons by polypeptide polycations: an efficient process accompanied by secondary fragmentation, Chem. Phys. Lett., 2002, 356(3), 201–206 CrossRef CAS.
- R. A. Zubarev, K. F. Haselmann, B. Budnik, F. Kjeldsen and F. Jensen, Towards An Understanding of the Mechanism of Electron-Capture Dissociation: A Historical Perspective and Modern Ideas, Eur. J. Mass Spectrom., 2002, 8(5), 337–349 Search PubMed.
- N. Manri, H. Satake, A. Kaneko, A. Hirabayashi, T. Baba and T. Sakamoto, Glycopeptide Identification Using Liquid-Chromatography-Compatible Hot Electron Capture Dissociation in a Radio-Frequency-Quadrupole Ion Trap, Anal. Chem., 2013, 85(4), 2056–2063 CrossRef CAS PubMed.
- Y. M. E. Fung, C. M. Adams and R. A. Zubarev, Electron Ionization Dissociation of Singly and Multiply Charged Peptides, J. Am. Chem. Soc., 2009, 131(29), 9977–9985 CrossRef CAS PubMed.
- H. J. Yoo, N. Wang, S. Zhuang, H. Song and K. Håkansson, Negative-Ion Electron Capture Dissociation: Radical-Driven Fragmentation of Charge-Increased Gaseous Peptide Anions, J. Am. Chem. Soc., 2011, 133(42), 16790–16793 CrossRef CAS.
- A. Kalli, G. Grigorean and K. Håkansson, Electron Induced Dissociation of Singly Deprotonated Peptides, J. Am. Soc. Mass Spectrom., 2011, 22(12), 2209–2221 CrossRef CAS PubMed.
- M. L. Nielsen, B. A. Budnik, K. F. Haselmann and R. A. Zubarev, Tandem MALDI/EI ionization for tandem Fourier transform ion cyclotron resonance mass spectrometry of polypeptides, Int. J. Mass Spectrom., 2003, 226(1), 181–187 Search PubMed.
- Y. O. Tsybin, P. Håkansson, B. A. Budnik, K. F. Haselmann, F. Kjeldsen, M. Gorshkov and R. A. Zubarev, Improved low-energy electron injection systems for high rate electron capture dissociation in Fourier transform ion cyclotron resonance mass spectrometry, Rapid Commun. Mass Spectrom., 2001, 15(19), 1849–1854 CrossRef CAS PubMed.
- Y. O. Tsybin, J. P. Quinn, O. Y. Tsybin, C. L. Hendrickson and A. G. Marshall, Electron Capture Dissociation Implementation Progress in Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2008, 19(6), 762–771 CrossRef CAS PubMed.
- J. P. Williams, L. J. Morrison, J. M. Brown, J. S. Beckman, V. G. Voinov and F. Lermyte, Top-Down Characterization of Denatured Proteins and Native Protein Complexes Using Electron Capture Dissociation Implemented within a Modified Ion Mobility-Mass Spectrometer, Anal. Chem., 2020, 92(5), 3674–3681 Search PubMed.
- V. G. Voinov, M. L. Deinzer and D. F. Barofsky, Radio-Frequency-Free Cell for Electron Capture Dissociation in Tandem Mass Spectrometry, Anal. Chem., 2009, 81(3), 1238–1243 CrossRef CAS PubMed.
- V. G. Voinov, M. L. Deinzer, J. S. Beckman and D. F. Barofsky, Electron Capture, Collision-Induced, and Electron Capture-Collision Induced Dissociation in Q-TOF, J. Am. Soc. Mass Spectrom., 2011, 22(4), 607–611 CrossRef CAS PubMed.
- T. Baba, J. L. Campbell, J. C. Y. Le Blanc, J. W. Hager and B. A. Thomson, Electron Capture Dissociation in a Branched Radio-Frequency Ion Trap, Anal. Chem., 2015, 87(1), 785–792 Search PubMed.
- H. Liu and K. Håkansson, Abundant b-type ions produced in electron capture dissociation of peptides without basic amino acid residues, J. Am. Soc. Mass Spectrom., 2007, 18(11), 2007–2013 CrossRef CAS PubMed.
- V. Bakken, T. Helgaker and E. Uggerud, Models of Fragmentations Induced by Electron Attachment to Protonated Peptides, Eur. J. Mass Spectrom., 2004, 10(5), 625–638 CrossRef CAS PubMed.
- H. Zhang, W. Cui and M. L. Gross, Native electrospray ionization and electron-capture dissociation for comparison of protein structure in solution and the gas phase, Int. J. Mass Spectrom., 2013, 354–355, 288–291 Search PubMed.
- K. Jeacock, A. Chappard, K. J. Gallagher, C. L. Mackay, D. P. A. Kilgour, M. H. Horrocks, T. Kunath and D. J. Clarke, Determining the Location of the α-Synuclein Dimer Interface Using Native Top-Down Fragmentation and Isotope Depletion-Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2023, 34(5), 847–856 CrossRef CAS PubMed.
- S. M. M. Sweet, C. M. Bailey, D. L. Cunningham, J. K. Heath and H. J. Cooper, Large Scale Localization of Protein Phosphorylation by Use of Electron Capture Dissociation Mass Spectrometry *S, Mol. Cell. Proteomics, 2009, 8(5), 904–912 CrossRef CAS PubMed.
- S. A. Miller, K. Jeanne Dit Fouque, E. R. Hard, A. T. Balana, D. Kaplan, V. G. Voinov, M. E. Ridgeway, M. A. Park, G. A. Anderson, M. R. Pratt and F. Fernandez-Lima, Top/Middle-Down Characterization of α-Synuclein Glycoforms, Anal. Chem., 2023, 95(49), 18039–18045 CrossRef CAS.
- J. Laskin, J. H. Futrell and I. K. Chu, Is Dissociation of Peptide Radical Cations an Ergodic Process?, J. Am. Chem. Soc., 2007, 129(31), 9598–9599 CrossRef CAS PubMed.
- F. Tureček, NCα Bond Dissociation Energies and Kinetics in Amide and Peptide Radicals. Is the Dissociation a Non-ergodic Process?, J. Am. Chem. Soc., 2003, 125(19), 5954–5963 CrossRef PubMed.
- T. W. Chung and F. Tureček, Proper and improper aminoketyl radicals in electron-based peptide dissociations, Int. J. Mass Spectrom., 2011, 301(1), 55–61 Search PubMed.
- M. Girod, D. Arquier, A. Helms, K. Juetten, J. S. Brodbelt, J. Lemoine and L. MacAleese, Characterization of Phosphorylated Peptides by Electron-Activated and Ultraviolet Dissociation Mass Spectrometry: A Comparative Study with Collision-Induced Dissociation, J. Am. Soc. Mass Spectrom., 2024, 35(5), 1040–1054 CrossRef CAS PubMed.
- Y. P. Y. Lam, C. A. Wootton, I. Hands-Portman, J. Wei, C. K. C. Chiu, I. Romero-Canelon, F. Lermyte, M. P. Barrow and P. B. O'Connor, Determination of the Aggregate Binding Site of Amyloid Protofibrils Using Electron Capture Dissociation Tandem Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2020, 31(2), 267–276 CrossRef CAS PubMed.
- R. Akter, P. Cao, H. Noor, Z. Ridgway, L.-H. Tu, H. Wang, A. G. Wong, X. Zhang, A. Abedini, A. M. Schmidt and D. P. Raleigh, Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology, J. Diabetes Res., 2016, 2016(1), 2798269 Search PubMed.
- A. L. Simmonds, A. F. Lopez-Clavijo, P. J. Winn, D. H. Russell, I. B. Styles and H. J. Cooper, Structural Analysis of 14-3-3-ζ-Derived Phosphopeptides Using Electron Capture Dissociation Mass Spectrometry, Traveling Wave Ion Mobility Spectrometry, and Molecular Modeling, J. Phys. Chem. B, 2020, 124(3), 461–469 CrossRef CAS PubMed.
- K. Rittinger, J. Budman, J. Xu, S. Volinia, L. C. Cantley, S. J. Smerdon, S. J. Gamblin and M. B. Yaffe, Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding, Mol. Cell, 1999, 4(2), 153–166 CrossRef CAS PubMed.
- T. Baba, Z. Zhang, S. Liu, L. Burton, P. Ryumin and J. C. Y. Le Blanc, Localization of Multiple O-Linked Glycans Exhibited in Isomeric Glycopeptides by Hot Electron Capture Dissociation, J. Proteome Res., 2022, 21(10), 2462–2471 CrossRef CAS PubMed.
- F. Berthias, D. A. Cooper-Shepherd, F. H. V. Holck, J. I. Langridge and O. N. Jensen, Full Separation and Sequencing of Isomeric Proteoforms in the Middle-Down Mass Range Using Cyclic Ion Mobility and Electron Capture Dissociation, Anal. Chem., 2023, 95(29), 11141–11148 CrossRef CAS PubMed.
- J. B. Shaw, N. Malhan, Y. V. Vasil’ev, N. I. Lopez, A. Makarov, J. S. Beckman and V. G. Voinov, Sequencing Grade Tandem Mass Spectrometry for Top–Down Proteomics Using Hybrid Electron Capture Dissociation Methods in a Benchtop Orbitrap Mass Spectrometer, Anal. Chem., 2018, 90(18), 10819–10827 CrossRef CAS PubMed.
- Z. Liang, X. Yu and W. Zhong, Peptide Sequence Influence on the Differentiation of Valine and Norvaline by Hot Electron Capture Dissociation, Anal. Chem., 2019, 91(7), 4381–4387 CrossRef CAS PubMed.
- N. Wang, S. M. Dixit, T. Lee, S. A. DeFiglia, B. T. Ruotolo and K. Håkansson, Salt-Bridged Peptide Anion Gaseous Structures Enable Efficient Negative Ion Electron Capture Dissociation, J. Am. Soc. Mass Spectrom., 2024, 35(4), 784–792 CrossRef CAS PubMed.
- K. E. Hersberger and K. Håkansson, Characterization of O-Sulfopeptides by Negative Ion Mode Tandem Mass Spectrometry: Superior Performance of Negative Ion Electron Capture Dissociation, Anal. Chem., 2012, 84(15), 6370–6377 CrossRef CAS PubMed.
- S. A. DeFiglia, C. W. Szot and K. Håkansson, Negative-Ion Electron Capture Dissociation of MALDI-Generated Peptide Anions, Anal. Chem., 2024, 96(21), 8800–8806 CrossRef CAS PubMed.
- Y. Huang, Y. Pu, X. Yu, C. E. Costello and C. Lin, Mechanistic Study on Electronic Excitation Dissociation of the Cellobiose-Na+ Complex, J. Am. Soc. Mass Spectrom., 2016, 27(2), 319–328 CrossRef CAS PubMed.
- R. Li, C. Xia, S. Wu, M. J. Downs, H. Tong, N. Tursumamat, J. Zaia, C. E. Costello, C. Lin and J. Wei, Direct and Detailed Site-Specific Glycopeptide Characterization by Higher-Energy Electron-Activated Dissociation Tandem Mass Spectrometry, Anal. Chem., 2024, 96(3), 1251–1258 CrossRef CAS PubMed.
- Y. L. E. Wong, X. Chen, R. Wu, Y. L. W. Hung and T. W. D. Chan, Structural Characterization of Intact Glycoconjugates by Tandem Mass Spectrometry Using Electron-Induced Dissociation, Anal. Chem., 2017, 89(18), 10111–10117 CrossRef CAS.
- Y. Tang, J. Wei, C. E. Costello and C. Lin, Characterization of Isomeric Glycans by Reversed Phase Liquid Chromatography-Electronic Excitation Dissociation Tandem Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2018, 29(6), 1295–1307 CrossRef CAS PubMed.
- J. W. Jones, C. J. Thompson, C. L. Carter and M. A. Kane, Electron-induced dissociation (EID) for structure characterization of glycerophosphatidylcholine: determination of double-bond positions and localization of acyl chains, J. Mass Spectrom., 2015, 50(12), 1327–1339 CrossRef CAS PubMed.
- J. L. Campbell and T. Baba, Near-Complete Structural Characterization of Phosphatidylcholines Using Electron Impact Excitation of Ions from Organics, Anal. Chem., 2015, 87(11), 5837–5845 CrossRef CAS PubMed.
- T. Baba, J. L. Campbell, J. C. Y. Le Blanc and P. R. S. Baker, Structural identification of triacylglycerol isomers using electron impact excitation of ions from organics (EIEIO)[S], J. Lipid Res., 2016, 57(11), 2015–2027 CrossRef CAS.
- T. Baba, J. L. Campbell, J. C. Y. Le Blanc and P. R. S. Baker, In-depth sphingomyelin characterization using electron impact excitation of ions from organics and mass spectrometry[S], J. Lipid Res., 2016, 57(5), 858–867 CrossRef CAS PubMed.
- H. J. Yoo and K. Håkansson, Determination of Double Bond Location in Fatty Acids by Manganese Adduction and Electron Induced Dissociation, Anal. Chem., 2010, 82(16), 6940–6946 CrossRef CAS.
- A. O. Ducati, D. Ruskic, P. Sosnowski, T. Baba, R. Bonner and G. Hopfgartner, Improved metabolite characterization by liquid chromatography – Tandem mass spectrometry through electron impact type fragments from adduct ions, Anal. Chim. Acta, 2021, 1150, 338207 CrossRef CAS PubMed.
- P. Che, J. T. Davidson, J. Kool and I. Kohler, Electron activated dissociation - a complementary fragmentation technique to collision-induced dissociation for metabolite identification of synthetic cathinone positional isomers, Anal. Chim. Acta, 2023, 1283, 341962 CrossRef CAS PubMed.
- Y. L. E. Wong, X. Chen, W. Li, Z. Wang, Y. L. W. Hung, R. Wu and T. W. D. Chan, Differentiation of Isomeric Ginsenosides by Using Electron-Induced Dissociation Mass Spectrometry, Anal. Chem., 2016, 88(11), 5590–5594 CrossRef CAS PubMed.
- B. P. Marzullo, T. E. Morgan, C. A. Wootton, M. Li, S. J. Perry, M. Saeed, M. P. Barrow and P. B. O'Connor, Comparison of Fragmentation Techniques for the Structural Characterization of Singly Charged Agrochemicals, Anal. Chem., 2020, 92(4), 3143–3151 CrossRef CAS PubMed.
- A. F. Lopez-Clavijo, R. L. Griffiths, R. J. A. Goodwin and H. J. Cooper, Liquid Extraction Surface Analysis (LESA) Electron-Induced Dissociation and Collision-Induced Dissociation Mass Spectrometry of Small Molecule Drug Compounds, J. Am. Soc. Mass Spectrom., 2018, 29(11), 2218–2226 CrossRef CAS PubMed.
- T. Baba, K. Rajabi, S. Liu, P. Ryumin, Z. Zhang, K. Pohl, J. Causon, J. C. Y. Le Blanc and M. Kurogochi, Electron Impact Excitation of Ions from Organics on Singly Protonated Peptides with and without Post-Translational Modifications, J. Am. Soc. Mass Spectrom., 2022, 33(9), 1723–1732 CrossRef CAS PubMed.
- N. N. Mikawy, C. Rojas Ramírez, S. A. DeFiglia, C. W. Szot, J. Le, C. Lantz, B. Wei, M. A. Zenaidee, G. T. Blakney, A. I. Nesvizhskii, J. A. Loo, B. T. Ruotolo, J. Shabanowitz, L. C. Anderson and K. Håkansson, Are Internal Fragments Observable in Electron Based Top-Down Mass Spectrometry?, Mol. Cell. Proteomics, 2024, 23(9), 100814 CrossRef PubMed.
- M. A. Zenaidee, C. Lantz, T. Perkins, W. Jung, R. R. O. Loo and J. A. Loo, Internal Fragments Generated by Electron Ionization Dissociation Enhance Protein Top-Down Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2020, 31(9), 1896–1902 CrossRef PubMed.
- B. Wei, M. A. Zenaidee, C. Lantz, B. J. Williams, S. Totten, R. R. Ogorzalek Loo and J. A. Loo, Top-down mass spectrometry and assigning internal fragments for determining disulfide bond positions in proteins, Analyst, 2023, 148, 26–37 RSC.
- M.-E. N. Born and B. M. Prentice, Structural elucidation of phosphatidylcholines from tissue using electron induced dissociation, Int. J. Mass Spectrom., 2020, 452, 116338 CrossRef.
- T. Yan, M.-E. N. Born and B. M. Prentice, Structural elucidation and relative quantification of sodium- and potassium-cationized phosphatidylcholine regioisomers directly from tissue using electron induced dissociation, Int. J. Mass Spectrom., 2023, 485, 116998 CrossRef PubMed.
- T. Yan, Z. Liang and B. M. Prentice, Imaging and Structural Characterization of Phosphatidylcholine Isomers from Rat Brain Tissue Using Sequential Collision-Induced Dissociation/Electron-Induced Dissociation, Anal. Chem., 2023, 95(42), 15707–15715 CrossRef PubMed.
- B. A. Budnik, K. F. Haselmann and R. A. Zubarev, Electron detachment dissociation of peptide di-anions: an electron–hole recombination phenomenon, Chem. Phys. Lett., 2001, 342(3), 299–302 CrossRef.
- A. Zappe, R. L. Miller, W. B. Struwe and K. Pagel, State-of-the-art glycosaminoglycan characterization, Mass Spectrom. Rev., 2022, 41(6), 1040–1071 CrossRef PubMed.
- I. Agyekum, C. Zong, G.-J. Boons and I. J. Amster, Single Stage Tandem Mass Spectrometry Assignment of the C-5 Uronic Acid Stereochemistry in Heparan Sulfate Tetrasaccharides using Electron Detachment Dissociation, J. Am. Soc. Mass Spectrom., 2017, 28(9), 1741–1750 CrossRef PubMed.
- K. Karasawa, E. Duchoslav and T. Baba, Fast Electron Detachment Dissociation of Oligonucleotides in Electron-Nitrogen Plasma Stored in Magneto Radio-Frequency Ion Traps, Anal. Chem., 2022, 94(44), 15510–15517 CrossRef PubMed.
- K. Breuker, Characterization of Ribonucleic Acids and Their Modifications by Fourier Transform Ion Cyclotron Resonance Mass Spectrometry, in Nucleic Acids in the Gas Phase, ed. V. Gabelica, Springer Berlin Heidelberg, Berlin, Heidelberg, 2014, pp. 185–202 Search PubMed.
- F. Baquero, K. Beis, D. J. Craik, Y. Li, A. J. Link, S. Rebuffat, R. Salomón, K. Severinov, S. Zirah and J. D. Hegemann, The pearl jubilee of microcin J25: thirty years of research on an exceptional lasso peptide, Nat. Prod. Rep., 2024, 41(3), 469–511 RSC.
- M. Pérot-Taillandier, C. Afonso, Q. Enjalbert, R. Antoine, P. Dugourd, R. B. Cole, J.-C. Tabet, S. Rebuffat and S. Zirah, Electron detachment/photodetachment dissociation of lasso peptides, Int. J. Mass Spectrom., 2015, 390, 91–100 CrossRef.
- J. S. Brodbelt and J. J. Wilson, Infrared multiphoton dissociation in quadrupole ion traps, Mass Spectrom. Rev., 2009, 28(3), 390–424 CrossRef PubMed.
- S. A. Hofstadler, K. A. Sannes-Lowery and R. H. Griffey, m/z-Selective infrared multiphoton dissociation in a Penning trap using sidekick trapping and an rf-tickle pulse, Rapid Commun. Mass Spectrom., 2001, 15(12), 945–951 CrossRef.
- A. H. Payne and G. L. Glish, Thermally Assisted Infrared Multiphoton Photodissociation in a Quadrupole Ion Trap, Anal. Chem., 2001, 73(15), 3542–3548 CrossRef PubMed.
- S. D. Dunham, J. D. Sanders, D. D. Holden and J. S. Brodbelt, Improving the Center Section Sequence Coverage of Large Proteins Using Stepped-Fragment Ion Protection Ultraviolet Photodissociation, J. Am. Soc. Mass Spectrom., 2022, 33(3), 446–456 CrossRef PubMed.
- F. C. Liu, M. E. Ridgeway, J. S. R. V. Winfred, N. C. Polfer, J. Lee, A. Theisen, C. A. Wootton, M. A. Park and C. Bleiholder, Tandem-trapped ion mobility spectrometry/mass spectrometry coupled with ultraviolet photodissociation, Rapid Commun. Mass Spectrom., 2021, 35(22), e9192 CrossRef PubMed.
- F. C. Liu, M. E. Ridgeway, M. A. Park and C. Bleiholder, Tandem-trapped ion mobility spectrometry/mass spectrometry (tTIMS/MS): a promising analytical method for investigating heterogenous samples, Analyst, 2022, 147(11), 2317–2337 RSC.
- A. Q. Stiving, S. R. Harvey, B. J. Jones, B. Bellina, J. M. Brown, P. E. Barran and V. H. Wysocki, Coupling 193 nm Ultraviolet Photodissociation and Ion Mobility for Sequence Characterization of Conformationally-Selected Peptides, J. Am. Soc. Mass Spectrom., 2020, 31(11), 2313–2320 CrossRef PubMed.
- D. P. Little, J. P. Speir, M. W. Senko, P. B. O'Connor and F. W. McLafferty, Infrared Multiphoton Dissociation of Large Multiply Charged Ions for Biomolecule Sequencing, Anal. Chem., 1994, 66(18), 2809–2815 CrossRef PubMed.
- M. A. van Agthoven, M.-A. Delsuc and C. Rolando, Two-dimensional FT-ICR/MS with IRMPD as fragmentation mode, Int. J. Mass Spectrom., 2011, 306(2), 196–203 CrossRef.
- M. Lanzillotti and J. S. Brodbelt, Comparison of Top-Down Protein Fragmentation Induced by 213 and 193 nm UVPD, J. Am. Soc. Mass Spectrom., 2023, 34(2), 279–285 CrossRef PubMed.
- E. E. Escobar, W. Yang, M. B. Lanzillotti, K. J. Juetten, S. Shields, D. Siegel, Y. J. Zhang and J. S. Brodbelt, Tracking Inhibition of Human Small C-Terminal Domain Phosphatase 1 Using 193 nm Ultraviolet Photodissociation Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2024, 35(6), 1330–1341 CrossRef PubMed.
- Y. Shi, M. Zhou, K. Zhang, L. Ma and X. Kong, Chiral Differentiation of Non-Covalent Diastereomers Based on Multichannel Dissociation Induced by 213-nm Ultraviolet Photodissociation, J. Am. Soc. Mass Spectrom., 2019, 30(11), 2297–2305 CrossRef PubMed.
- A. Theisen, R. Black, D. Corinti, J. M. Brown, B. Bellina and P. E. Barran, Initial Protein Unfolding Events in Ubiquitin, Cytochrome c and Myoglobin Are Revealed with the Use of 213 nm UVPD Coupled to IM-MS, J. Am. Soc. Mass Spectrom., 2019, 30(1), 24–33 CrossRef PubMed.
- S. A. Miller, K. Jeanne Dit Fouque, M. E. Ridgeway, M. A. Park and F. Fernandez-Lima, Trapped Ion Mobility Spectrometry, Ultraviolet Photodissociation, and Time-of-Flight Mass Spectrometry for Gas-Phase Peptide Isobars/Isomers/Conformers Discrimination, J. Am. Soc. Mass Spectrom., 2022, 33(7), 1267–1275 CrossRef PubMed.
- S. M. Greer, S. Sidoli, M. Coradin, M. Schack Jespersen, V. Schwämmle, O. N. Jensen, B. A. Garcia and J. S. Brodbelt, Extensive Characterization of Heavily Modified Histone Tails by 193 nm Ultraviolet Photodissociation Mass Spectrometry via a Middle–Down Strategy, Anal. Chem., 2018, 90(17), 10425–10433 CrossRef PubMed.
- B. J. Ko and J. S. Brodbelt, Comparison of glycopeptide fragmentation by collision induced dissociation and ultraviolet photodissociation, Int. J. Mass Spectrom., 2015, 377, 385–392 CrossRef PubMed.
- E. E. Escobar, M. K. Venkat Ramani, Y. Zhang and J. S. Brodbelt, Evaluating Spatiotemporal Dynamics of Phosphorylation of RNA Polymerase II Carboxy-Terminal Domain by Ultraviolet Photodissociation Mass Spectrometry, J. Am. Chem. Soc., 2021, 143(22), 8488–8498 CrossRef PubMed.
- M. R. Robinson, K. L. Moore and J. S. Brodbelt, Direct Identification of Tyrosine Sulfation by using Ultraviolet Photodissociation Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2014, 25(8), 1461–1471 CrossRef PubMed.
- J. Hellinger and J. S. Brodbelt, Impact of Charge State on Characterization of Large Middle-Down Sized Peptides by Tandem Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2024, 35(8), 1647–1656 CrossRef PubMed.
- G. E. Reid, J. Wu, P. A. Chrisman, J. M. Wells and S. A. McLuckey, Charge-State-Dependent Sequence Analysis of Protonated Ubiquitin Ions via Ion Trap Tandem Mass Spectrometry, Anal. Chem., 2001, 73(14), 3274–3281 CrossRef PubMed.
- J. V. Olsen, B. Macek, O. Lange, A. Makarov, S. Horning and M. Mann, Higher-energy C-trap dissociation for peptide modification analysis, Nat. Methods, 2007, 4(9), 709–712 CrossRef PubMed.
- C.-C. Chou, B.-Y. Chiang, J. C.-Y. Lin, K.-T. Pan, C.-H. Lin and K.-H. Khoo, Characteristic Tandem Mass Spectral Features Under Various Collision Chemistries for Site-Specific Identification of Protein S-Glutathionylation, J. Am. Soc. Mass Spectrom., 2015, 26(1), 120–132 CrossRef PubMed.
- S. Becher, H. Wang, M. G. Leeming, W. A. Donald and S. Heiles, Influence of protein ion charge state on 213 nm top-down UVPD, Analyst, 2021, 146(12), 3977–3987 RSC.
- L. Fornelli, K. Srzentić, T. K. Toby, P. F. Doubleday, R. Huguet, C. Mullen, R. D. Melani, H. dos Santos Seckler, C. J. DeHart, C. R. Weisbrod, K. R. Durbin, J. B. Greer, B. P. Early, R. T. Fellers, V. Zabrouskov, P. M. Thomas, P. D. Compton and N. L. Kelleher, Thorough Performance Evaluation of 213 nm Ultraviolet Photodissociation for Top-down Proteomics, Mol. Cell. Proteomics, 2020, 19(2), 405–420 CrossRef PubMed.
- A. Bashyal, J. D. Sanders, D. D. Holden and J. S. Brodbelt, Top-Down Analysis of Proteins in Low Charge States, J. Am. Soc. Mass Spectrom., 2019, 30(4), 704–717 CrossRef PubMed.
- M. Dilillo, E. L. de Graaf, A. Yadav, M. E. Belov and L. A. McDonnell, Ultraviolet Photodissociation of ESI- and MALDI-Generated Protein Ions on a Q-Exactive Mass Spectrometer, J. Proteome Res., 2019, 18(1), 557–564 Search PubMed.
- K. J. Zemaitis, M. Zhou, W. Kew and L. Paša-Tolić, 193 nm Ultraviolet Photodissociation for the Characterization of Singly Charged Proteoforms Generated by MALDI, J. Am. Soc. Mass Spectrom., 2023, 34(2), 328–332 CrossRef PubMed.
- R. M. Fleeman, L. A. Macias, J. S. Brodbelt and B. W. Davies, Defining principles that influence antimicrobial peptide activity against capsulated Klebsiella pneumoniae, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(44), 27620–27626 CrossRef PubMed.
- M. S. Blevins, J. N. Walker, J. M. Schaub, I. J. Finkelstein and J. S. Brodbelt, Characterization of the T4 gp32–ssDNA complex by native, cross-linking, and ultraviolet photodissociation mass spectrometry, Chem. Sci., 2021, 12(41), 13764–13776 RSC.
- S. N. Sipe, K. Jeanne Dit Fouque, A. Garabedian, F. Leng, F. Fernandez-Lima and J. S. Brodbelt, Exploring the Conformations and Binding Location of HMGA2·DNA Complexes Using Ion Mobility Spectrometry and 193 nm Ultraviolet Photodissociation Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2022, 33(7), 1092–1102 CrossRef PubMed.
- I. C. Santos and J. S. Brodbelt, Structural Characterization of Carbonic Anhydrase–Arylsulfonamide Complexes Using Ultraviolet Photodissociation Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2021, 32(6), 1370–1379 CrossRef PubMed.
- L. A. Macias, X. Wang, B. W. Davies and J. S. Brodbelt, Mapping paratopes of nanobodies using native mass spectrometry and ultraviolet photodissociation, Chem. Sci., 2022, 13(22), 6610–6618 RSC.
- L. J. Morrison and J. S. Brodbelt, Charge site assignment in native proteins by ultraviolet photodissociation (UVPD) mass spectrometry, Analyst, 2016, 141(1), 166–176 RSC.
- B. M. Walsh, Nonlinear mixing of Nd:YAG lasers; harmonic and sum frequency generation, Opt. Mater., 2017, 65, 2–7 CrossRef CAS.
- T. Ly and R. R. Julian, Elucidating the Tertiary Structure of Protein Ions in Vacuo with Site Specific Photoinitiated Radical Reactions, J. Am. Chem. Soc., 2010, 132(25), 8602–8609 CrossRef CAS PubMed.
- T. Ly and R. R. Julian, Residue-specific radical-directed dissociation of whole proteins in the gas phase, J. Am. Chem. Soc., 2008, 130(1), 351–358 CrossRef CAS PubMed.
- D. L. Riggs, J. W. Silzel, Y. A. Lyon, A. S. Kang and R. R. Julian, Analysis of Glutamine Deamidation: Products, Pathways, and Kinetics, Anal. Chem., 2019, 91(20), 13032–13038 CrossRef CAS PubMed.
- T. R. Lambeth and R. R. Julian, Efficient Isothiocyanate Modification of Peptides Facilitates Structural Analysis by Radical-Directed Dissociation, J. Am. Soc. Mass Spectrom., 2022, 33(8), 1338–1345 CrossRef CAS PubMed.
- C. J. Shaffer, P. C. Andrikopoulos, J. Řezáč, L. Rulíšek and F. Tureček, Efficient Covalent Bond Formation in Gas-Phase Peptide–Peptide Ion Complexes with the Photoleucine Stapler, J. Am. Soc. Mass Spectrom., 2016, 27(4), 633–645 CrossRef CAS PubMed.
- R. Pepin, C. J. Shaffer and F. Tureček, Position-tunable diazirine tags for peptide–peptide ion cross-linking in the gas phase, J. Mass Spectrom., 2017, 52(8), 557–560 CrossRef CAS PubMed.
- L. Piersimoni, P. L. Kastritis, C. Arlt and A. Sinz, Cross-Linking Mass Spectrometry for Investigating Protein Conformations and Protein–Protein Interactions-A Method for All Seasons, Chem. Rev., 2022, 122(8), 7500–7531 CrossRef CAS PubMed.
- H. Zhu, V. Zima, E. R. Ding and F. Tureček, Carbene Cross-Linking in Gas-Phase Peptide Ion Scaffolds, J. Am. Soc. Mass Spectrom., 2023, 34(4), 763–774 CrossRef CAS PubMed.
- Y. Liu and F. Tureček, Photodissociative Cross-Linking of Diazirine-Tagged Peptides with DNA Dinucleotides in the Gas Phase, J. Am. Soc. Mass Spectrom., 2019, 30(10), 1992–2006 CrossRef CAS PubMed.
- V. R. Narreddula, B. I. McKinnon, S. J. P. Marlton, D. L. Marshall, N. R. B. Boase, B. L. J. Poad, A. J. Trevitt, T. W. Mitchell and S. J. Blanksby, Next-generation derivatization reagents optimized for enhanced product ion formation in photodissociation-mass spectrometry of fatty acids, Analyst, 2021, 146(1), 156–169 RSC.
- H. T. Pham and R. R. Julian, Characterization of glycosphingolipid epimers by radical-directed dissociation mass spectrometry, Analyst, 2016, 141(4), 1273–1278 RSC.
- V. R. Narreddula, N. R. Boase, R. Ailuri, D. L. Marshall, B. L. J. Poad, M. J. Kelso, A. J. Trevitt, T. W. Mitchell and S. J. Blanksby, Introduction of a Fixed-Charge, Photolabile Derivative for Enhanced Structural Elucidation of Fatty Acids, Anal. Chem., 2019, 91(15), 9901–9909 CrossRef CAS PubMed.
- H. T. Pham, T. Ly, A. J. Trevitt, T. W. Mitchell and S. J. Blanksby, Differentiation of Complex Lipid Isomers by Radical-Directed Dissociation Mass Spectrometry, Anal. Chem., 2012, 84(17), 7525–7532 CrossRef CAS PubMed.
- V. R. Narreddula, P. Sadowski, N. R. B. Boase, D. L. Marshall, B. L. J. Poad, A. J. Trevitt, T. W. Mitchell and S. J. Blanksby, Structural elucidation of hydroxy fatty acids by photodissociation mass spectrometry with photolabile derivatives, Rapid Commun. Mass Spectrom., 2020, 34(9), e8741 CrossRef CAS PubMed.
- L. A. Macias, K. Y. Garza, C. L. Feider, L. S. Eberlin and J. S. Brodbelt, Relative Quantitation of Unsaturated Phosphatidylcholines Using 193 nm Ultraviolet Photodissociation Parallel Reaction Monitoring Mass Spectrometry, J. Am. Chem. Soc., 2021, 143(36), 14622–14634 CrossRef CAS PubMed.
- M. S. Blevins, S. W. J. Shields, W. Cui, W. Fallatah, A. B. Moser, N. E. Braverman and J. S. Brodbelt, Structural Characterization and Quantitation of Ether-Linked Glycerophospholipids in Peroxisome Biogenesis Disorder Tissue by Ultraviolet Photodissociation Mass Spectrometry, Anal. Chem., 2022, 94(37), 12621–12629 CrossRef CAS PubMed.
- T. Kralj, M. Nuske, V. Hofferek, M.-A. Sani, T.-H. Lee, F. Separovic, M.-I. Aguilar and G. E. Reid, Multi-Omic Analysis to Characterize Metabolic Adaptation of the E. coli Lipidome in Response to Environmental Stress Metabolites [Online], 2022, p. 171 Search PubMed.
- D. R. Klein, C. L. Feider, K. Y. Garza, J. Q. Lin, L. S. Eberlin and J. S. Brodbelt, Desorption electrospray ionization coupled with ultraviolet photodissociation for characterization of phospholipid isomers in tissue sections, Anal. Chem., 2018, 90(17), 10100–10104 CrossRef CAS PubMed.
- C. L. Feider, L. A. Macias, J. S. Brodbelt and L. S. Eberlin, Double Bond Characterization of Free Fatty Acids Directly from Biological Tissues by Ultraviolet Photodissociation, Anal. Chem., 2020, 92(12), 8386–8395 CrossRef CAS PubMed.
- P. E. Williams, D. R. Klein, S. M. Greer and J. S. Brodbelt, Pinpointing Double Bond and sn-Positions in Glycerophospholipids via Hybrid 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry, J. Am. Chem. Soc., 2017, 139(44), 15681–15690 CrossRef CAS PubMed.
- M. S. Blevins, V. K. James, C. M. Herrera, A. B. Purcell, M. S. Trent and J. S. Brodbelt, Unsaturation Elements and Other Modifications of Phospholipids in Bacteria: New Insight from Ultraviolet Photodissociation Mass Spectrometry, Anal. Chem., 2020, 92(13), 9146–9155 CrossRef CAS PubMed.
- L. A. Macias and J. S. Brodbelt, Enhanced Characterization of Cardiolipins via Hybrid 193 nm Ultraviolet Photodissociation Mass Spectrometry, Anal. Chem., 2022, 94(7), 3268–3277 CrossRef CAS PubMed.
- E. W. Buenger and G. E. Reid, Shedding light on isomeric FAHFA lipid structures using 213 nm ultraviolet photodissociation mass spectrometry, Eur. J. Mass Spectrom., 2020, 26(5), 311–323 CrossRef CAS PubMed.
- C. A. Lutomski, T. J. El-Baba, J. D. Hinkle, I. Liko, J. L. Bennett, N. V. Kalmankar, A. Dolan, C. Kirschbaum, K. Greis, L. H. Urner, P. Kapoor, H.-Y. Yen, K. Pagel, C. Mullen, J. E. P. Syka and C. V. Robinson, Infrared Multiphoton Dissociation Enables Top-Down Characterization of Membrane Protein Complexes and G Protein-Coupled Receptors, Angew. Chem., Int. Ed., 2023, 62(36), e202305694 CrossRef CAS PubMed.
- N. C. Polfer and J. Oomens, Vibrational spectroscopy of bare and solvated ionic complexes of biological relevance, Mass Spectrom. Rev., 2009, 28(3), 468–494 CrossRef CAS PubMed.
- B. R. Juliano, J. W. Keating and B. T. Ruotolo, Infrared Photoactivation Enables Improved Native Top-Down Mass Spectrometry of Transmembrane Proteins, Anal. Chem., 2023, 95(35), 13361–13367 CrossRef CAS PubMed.
- J. Gault, I. Liko, M. Landreh, D. Shutin, J. R. Bolla, D. Jefferies, M. Agasid, H.-Y. Yen, M. J. G. W. Ladds, D. P. Lane, S. Khalid, C. Mullen, P. M. Remes, R. Huguet, G. McAlister, M. Goodwin, R. Viner, J. E. P. Syka and C. V. Robinson, Combining native and ‘omics’ mass spectrometry to identify endogenous ligands bound to membrane proteins, Nat. Methods, 2020, 17(5), 505–508 CrossRef CAS PubMed.
- A. Laganowsky, E. Reading, J. T. S. Hopper and C. V. Robinson, Mass spectrometry of intact membrane protein complexes, Nat. Protoc., 2013, 8(4), 639–651 CrossRef CAS PubMed.
- K. Sokratous, D. A. Cooper-Shepherd, J. Ujma, F. Qu, K. Giles, A. Ben-Younis, M. Hensen, J. I. Langridge, J. Gault, A. Jazayeri, I. Liko and J. T. S. Hopper, Enhanced Declustering Enables Native Top-Down Analysis of Membrane Protein Complexes using Ion-Mobility Time-Aligned Fragmentation, J. Am. Soc. Mass Spectrom., 2024, 35(8), 1891–1901 CrossRef CAS PubMed.
- M. Gilleron, B. Lindner and G. Puzo, MS/MS Approach for Characterization of the Fatty Acid Distribution on Mycobacterial Phosphatidyl-myo-inositol Mannosides, Anal. Chem., 2006, 78(24), 8543–8548 CrossRef CAS PubMed.
- N. Zehethofer, T. Scior and B. Lindner, Elucidation of the fragmentation pathways of different phosphatidylinositol phosphate species (PIPx) using IRMPD implemented on a FT-ICR MS, Anal. Bioanal. Chem., 2010, 398(7), 2843–2851 CrossRef CAS PubMed.
- J. R. Bonney, W.-Y. Kang, J. T. Specker, Z. Liang, T. R. I. V. Scoggins and B. M. Prentice, Relative Quantification of Lipid Isomers in Imaging Mass Spectrometry Using Gas-Phase Charge Inversion Ion/Ion Reactions and Infrared Multiphoton Dissociation, Anal. Chem., 2023, 95(48), 17766–17775 CrossRef CAS PubMed.
- Y. Jami-Alahmadi and T. D. Fridgen, Distinguishing complexes of isomeric peptides: Structures, energetic, and reactions of sodium cation-coordinated ProLeu or LeuPro trimers in the gas phase, Int. J. Mass Spectrom., 2018, 429, 136–141 CrossRef CAS.
- D. Gatineau, H. Dossmann, H. Clavier, A. Memboeuf, L. Drahos, Y. Gimbert and D. Lesage, Ligand effects in gold-carbonyl complexes: Evaluation of the bond dissociation energies using blackbody infrared radiative dissociation, Int. J. Mass Spectrom., 2021, 463, 116545 CrossRef CAS.
- M. Azargun, P. J. Meister, J. W. Gauld and T. D. Fridgen, The K2(9-ethylguanine)122+ quadruplex is more stable to unimolecular dissociation than the K(9-ethylguanine)8+ quadruplex in the gas phase: a BIRD, energy resolved SORI-CID, IRMPD spectroscopic, and computational study, Phys. Chem. Chem. Phys., 2019, 21(28), 15319–15326 RSC.
- Y. Chen, P. G. Ghasemabadi, G. J. Bodwell, M. Demireva and T. D. Fridgen, Glycine in a basket: protonated complexes of 1,1,n,n-tetramethyl[n](2,11)teropyrenophane (n = 7, 8, 9) with glycine in the gas-phase, Phys. Chem. Chem. Phys., 2023, 25(24), 16597–16612 RSC.
- A. Pereverzev and J. Roithová, Experimental techniques and terminology in gas-phase ion spectroscopy, J. Mass Spectrom., 2022, 57(5), e4826 CrossRef CAS PubMed.
- T. Baer and R. C. Dunbar, Ion spectroscopy: Where did it come from; where is it now; and where is it going?, J. Am. Soc. Mass Spectrom., 2010, 21(5), 681–693 CrossRef CAS PubMed.
- C. P. Harrilal, A. F. DeBlase, S. A. McLuckey and T. S. Zwier, Two-Color IRMPD Applied to Conformationally Complex Ions: Probing Cold Ion Structure and Hot Ion Unfolding, J. Phys. Chem. A, 2021, 125(42), 9394–9404 CrossRef CAS PubMed.
- J. Roithová and J. M. Bakker, Ion spectroscopy in methane activation, Mass Spectrom. Rev., 2022, 41(4), 513–528 CrossRef PubMed.
- K. Greis, C. Kirschbaum, G. von Helden and K. Pagel, Gas-phase infrared spectroscopy of glycans and glycoconjugates, Curr. Opin. Struct. Biol., 2022, 72, 194–202 CrossRef CAS PubMed.
- J. Martens, R. E. van Outersterp, R. J. Vreeken, F. Cuyckens, K. L. M. Coene, U. F. Engelke, L. A. J. Kluijtmans, R. A. Wevers, L. M. C. Buydens, B. Redlich, G. Berden and J. Oomens, Infrared ion spectroscopy: New opportunities for small-molecule identification in mass spectrometry - A tutorial perspective, Anal. Chim. Acta, 2020, 1093, 1–15 CrossRef CAS PubMed.
- S. Bakels, M.-P. Gaigeot and A. M. Rijs, Gas-Phase Infrared Spectroscopy of Neutral Peptides: Insights from the Far-IR and THz Domain, Chem. Rev., 2020, 120(7), 3233–3260 CrossRef CAS PubMed.
- A. Dang, J. A. Korn, J. Gladden, B. Mozzone and F. Tureček, UV–Vis Photodissociation Action Spectroscopy on Thermo LTQ-XL ETD and Bruker amaZon Ion Trap Mass Spectrometers: a Practical Guide, J. Am. Soc. Mass Spectrom., 2019, 30(9), 1558–1564 CrossRef CAS PubMed.
- O. Asvany and S. Schlemmer, Rotational action spectroscopy of trapped molecular ions, Phys. Chem. Chem. Phys., 2021, 23(47), 26602–26622 RSC.
- L. Polewski, A. Springer, K. Pagel and C. A. Schalley, Gas-Phase Structural Analysis of Supramolecular Assemblies, Acc. Chem. Res., 2021, 54(10), 2445–2456 CrossRef CAS PubMed.
- S. J. P. Marlton and A. J. Trevitt, The combination of laser photodissociation, action spectroscopy, and mass spectrometry to identify and separate isomers, Chem. Commun., 2022, 58(68), 9451–9467 RSC.
- C. J. Gray, I. Compagnon and S. L. Flitsch, Mass spectrometry hybridized with gas-phase InfraRed spectroscopy for glycan sequencing, Curr. Opin. Struct. Biol., 2020, 62, 121–131 CrossRef CAS PubMed.
- P. Maitre, D. Scuderi, D. Corinti, B. Chiavarino, M. E. Crestoni and S. Fornarini, Applications of Infrared Multiple Photon Dissociation (IRMPD) to the Detection of Posttranslational Modifications, Chem. Rev., 2020, 120(7), 3261–3295 CrossRef CAS PubMed.
- F. Tureček, UV-vis spectroscopy of gas-phase ions, Mass Spectrom. Rev., 2023, 42(1), 206–226 CrossRef PubMed.
- I. Djavani-Tabrizi, T. T. Lindkvist, J. Langeland, C. Kjær, M. Graham, H. G. Kjaergaard and S. B. Nielsen, Tautomer-Selective Fluorescence Spectroscopy of Oxyluciferin Anions, J. Am. Chem. Soc., 2024, 146(39), 26975–26982 CrossRef CAS PubMed.
- J. Langeland, T. T. Lindkvist, C. Kjær and S. B. Nielsen, Gas-phase Förster resonance energy transfer in mass-selected and trapped ions, Mass Spectrom. Rev., 2024, 43(3), 477–499 CrossRef CAS PubMed.
- S. Daly, G. Knight, M. A. Halim, A. Kulesza, C. M. Choi, F. Chirot, L. MacAleese, R. Antoine and P. Dugourd, Action-FRET of a Gaseous Protein, J. Am. Soc. Mass Spectrom., 2017, 28(1), 38–49 CrossRef CAS PubMed.
- T. Manovitz, Y. Shapira, L. Gazit, N. Akerman and R. Ozeri, Trapped-Ion Quantum Computer with Robust Entangling Gates and Quantum Coherent Feedback, PRX Quantum, 2022, 3(1), 010347 CrossRef.
- F. Tureček, UV-vis spectroscopy of gas-phase ions, Mass Spectrom. Rev., 2021, 42(1), 206–226 CrossRef PubMed.
- S. A. McLuckey and T.-Y. Huang, Ion/Ion Reactions: New Chemistry for Analytical MS, Anal. Chem., 2009, 81(21), 8669–8676 CrossRef CAS PubMed.
- D. J. Foreman and S. A. McLuckey, Recent Developments in Gas-Phase Ion/Ion Reactions for Analytical Mass Spectrometry, Anal. Chem., 2020, 92(1), 252–266 CrossRef CAS PubMed.
- V. C. Cotham, J. B. Shaw and J. S. Brodbelt, High-Throughput Bioconjugation for Enhanced 193 nm Photodissociation via Droplet Phase Initiated Ion/Ion Chemistry Using a Front-End Dual Spray Reactor, Anal. Chem., 2015, 87(18), 9396–9402 CrossRef CAS PubMed.
- Y. Xia and S. A. McLuckey, Evolution of instrumentation for the study of gas-phase ion/ion chemistry via mass spectrometry, J. Am. Soc. Mass Spectrom., 2008, 19(2), 173–189 CrossRef CAS PubMed.
- D. A. Kaplan, R. Hartmer, J. P. Speir, C. Stoermer, D. Gumerov, M. L. Easterling, A. Brekenfeld, T. Kim, F. Laukien and M. A. Park, Electron transfer dissociation in the hexapole collision cell of a hybrid quadrupole-hexapole Fourier transform ion cyclotron resonance mass spectrometer, Rapid Commun. Mass Spectrom., 2008, 22(3), 271–278 CrossRef CAS PubMed.
- G. C. McAlister, D. Phanstiel, D. M. Good, W. T. Berggren and J. J. Coon, Implementation of Electron-Transfer Dissociation on a Hybrid Linear Ion Trap−Orbitrap Mass Spectrometer, Anal. Chem., 2007, 79(10), 3525–3534 CrossRef CAS PubMed.
- J. P. Williams, J. M. Brown, I. Campuzano and P. J. Sadler, Identifying drug metallation sites on peptides using electron transfer dissociation (ETD), collision induced dissociation (CID) and ion mobility-mass spectrometry (IM-MS), Chem. Commun., 2010, 46(30), 5458–5460 RSC.
- C. D. Wenger, M. V. Lee, A. S. Hebert, G. C. McAlister, D. H. Phanstiel, M. S. Westphall and J. J. Coon, Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging, Nat. Methods, 2011, 8(11), 933–935 CrossRef CAS PubMed.
- D. D. Holden, W. M. McGee and J. S. Brodbelt, Integration of Ultraviolet Photodissociation with Proton Transfer Reactions and Ion Parking for Analysis of Intact Proteins, Anal. Chem., 2016, 88(1), 1008–1016 CrossRef CAS PubMed.
- J. T. Specker, S. L. Van Orden, M. E. Ridgeway and B. M. Prentice, Identification of Phosphatidylcholine Isomers in Imaging Mass Spectrometry Using Gas-Phase Charge Inversion Ion/Ion Reactions, Anal. Chem., 2020, 92(19), 13192–13201 CrossRef CAS PubMed.
- J. R. Bonney and B. M. Prentice, Structural Elucidation and Relative Quantification of Fatty Acid Double Bond Positional Isomers in Biological Tissues Enabled by Gas-Phase Charge Inversion Ion/Ion Reactions, Analysis Sensing, 2023, e202300063 Search PubMed.
- M. Cheung See Kit, S. O. Shepherd, J. S. Prell and I. K. Webb, Experimental Determination of Activation Energies for Covalent Bond Formation via Ion/Ion Reactions and Competing Processes, J. Am. Soc. Mass Spectrom., 2021, 32(9), 2313–2321 CrossRef CAS PubMed.
- J. E. P. Syka, J. J. Coon, M. J. Schroeder, J. Shabanowitz and D. F. Hunt, Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry, Proc. Natl. Acad. Sci. U. S. A., 2004, 101(26), 9528–9533 CrossRef CAS PubMed.
- F. Lermyte, Chapter 2 Mechanism and Implementation of Electron Capture and Electron Transfer Dissociation, in Advanced Fragmentation Methods in Biomolecular Mass Spectrometry: Probing Primary and Higher Order Structure with Electrons, Photons and Surfaces, The Royal Society of Chemistry, 2021, pp. 15–32 Search PubMed.
- S. D. Robinson, L. Kambanis, D. Clayton, H. Hinneburg, L. Corcilius, A. Mueller, A. A. Walker, A. Keramidas, S. S. Kulkarni, A. Jones, I. Vetter, M. Thaysen-Andersen, R. J. Payne, G. F. King and E. A. B. Undheim, A pain-causing and paralytic ant venom glycopeptide, iScience, 2021, 24(10), 103175 CrossRef CAS PubMed.
- S. R. Kundinger, I. Bishof, E. B. Dammer, D. M. Duong and N. T. Seyfried, Middle-Down Proteomics Reveals Dense Sites of Methylation and Phosphorylation in Arginine-Rich RNA-Binding Proteins, J. Proteome Res., 2020, 19(4), 1574–1591 CrossRef CAS PubMed.
- C. K. Frese, A. F. M. Altelaar, H. van den Toorn, D. Nolting, J. Griep-Raming, A. J. R. Heck and S. Mohammed, Toward Full Peptide Sequence Coverage by Dual Fragmentation Combining Electron-Transfer and Higher-Energy Collision Dissociation Tandem Mass Spectrometry, Anal. Chem., 2012, 84(22), 9668–9673 CrossRef CAS PubMed.
- D. L. Swaney, G. C. McAlister, M. Wirtala, J. C. Schwartz, J. E. P. Syka and J. J. Coon, Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors, Anal. Chem., 2007, 79(2), 477–485 CrossRef CAS PubMed.
- D. Wang, J. Baudys, J. L. Bundy, M. Solano, T. Keppel and J. R. Barr, Comprehensive Analysis of the Glycan Complement of SARS-CoV-2 Spike Proteins Using Signature Ions-Triggered Electron-Transfer/Higher-Energy Collisional Dissociation (EThcD) Mass Spectrometry, Anal. Chem., 2020, 92(21), 14730–14739 CrossRef CAS PubMed.
- X. Li, R. Wilmanowski, X. Gao, Z. L. VanAernum, D. P. Donnelly, B. Kochert, H. A. Schuessler and D. Richardson, Precise O-Glycosylation Site Localization of CD24Fc by LC-MS Workflows, Anal. Chem., 2022, 94(23), 8416–8425 CrossRef CAS PubMed.
- M. Li, X. Zhong, Y. Feng and L. Li, Novel Isobaric Tagging Reagent Enabled Multiplex Quantitative Glycoproteomics via Electron-Transfer/Higher-Energy Collisional Dissociation (EThcD) Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2022, 33(10), 1874–1882 CrossRef CAS PubMed.
- M. Penkert, A. Hauser, R. Harmel, D. Fiedler, C. P. R. Hackenberger and E. Krause, Electron Transfer/Higher Energy Collisional Dissociation of Doubly Charged Peptide Ions: Identification of Labile Protein Phosphorylations, J. Am. Soc. Mass Spectrom., 2019, 30(9), 1578–1585 CrossRef CAS PubMed.
- J. A. M. Morgan, A. Singh, L. Kurz, M. Nadler-Holly, M. Ruwolt, S. Ganguli, S. Sharma, M. Penkert, E. Krause, F. Liu, R. Bhandari and D. Fiedler, Extensive protein pyrophosphorylation revealed in human cell lines, Nat. Chem. Biol., 2024, 20, 1305–1316 CrossRef CAS PubMed.
- I. A. Hendriks, S. C. Larsen and M. L. Nielsen, An Advanced Strategy for Comprehensive Profiling of ADP-ribosylation Sites Using Mass Spectrometry-based Proteomics, Mol. Cell. Proteomics, 2019, 18(5), 1010–1026 CrossRef CAS PubMed.
- S. C. Larsen, I. A. Hendriks, D. Lyon, L. J. Jensen and M. L. Nielsen, Systems-wide Analysis of Serine ADP-Ribosylation Reveals Widespread Occurrence and Site-Specific Overlap with Phosphorylation, Cell Rep., 2018, 24(9), 2493–2505 CrossRef CAS PubMed.
- T. Y. Samgina, I. D. Vasileva, R. A. Zubarev and A. T. Lebedev, EThcD as a Unique Tool for the Top-Down De Novo Sequencing of Intact Natural Ranid Amphibian Peptides, Anal. Chem., 2024, 96(29), 12057–12064 CrossRef CAS PubMed.
- I. D. Vasileva, T. Y. Samgina, Z. Meng, R. A. Zubarev and A. T. Lebedev, EThcD Benefits for the Sequencing Inside Intramolecular Disulfide Cycles of Amphibian Intact Peptides, J. Am. Soc. Mass Spectrom., 2023, 34(9), 1979–1988 CrossRef CAS PubMed.
- J. J. Coon, J. Shabanowitz, D. F. Hunt and J. E. P. Syka, Electron transfer dissociation of peptide anions, J. Am. Soc. Mass Spectrom., 2005, 16(6), 880–882 CrossRef CAS PubMed.
- N. G. Rumachik, G. C. McAlister, J. D. Russell, D. J. Bailey, C. D. Wenger and J. J. Coon, Characterizing Peptide Neutral Losses Induced by Negative Electron-Transfer Dissociation (NETD), J. Am. Soc. Mass Spectrom., 2012, 23(4), 718–727 CrossRef CAS PubMed.
- M. J. P. Rush, N. M. Riley, M. S. Westphall, J. E. P. Syka and J. J. Coon, Sulfur Pentafluoride is a Preferred Reagent Cation for Negative Electron Transfer Dissociation, J. Am. Soc. Mass Spectrom., 2017, 28(7), 1324–1332 Search PubMed.
- G. C. McAlister, J. D. Russell, N. G. Rumachik, A. S. Hebert, J. E. P. Syka, L. Y. Geer, M. S. Westphall, D. J. Pagliarini and J. J. Coon, Analysis of the acidic proteome with negative electron-transfer dissociation mass spectrometry, Anal. Chem., 2012, 84(6), 2875–2882 CrossRef CAS PubMed.
- J. J. Wolff, F. E. Leach III, T. N. Laremore, D. A. Kaplan, M. L. Easterling, R. J. Linhardt and I. J. Amster, Negative Electron Transfer Dissociation of Glycosaminoglycans, Anal. Chem., 2010, 82(9), 3460–3466 CrossRef CAS PubMed.
- Y. Huang, X. Yu, Y. Mao, C. E. Costello, J. Zaia and C. Lin, De Novo Sequencing of Heparan Sulfate Oligosaccharides by Electron-Activated Dissociation, Anal. Chem., 2013, 85(24), 11979–11986 CrossRef CAS PubMed.
- T. M. Peters-Clarke, Q. Quan, D. R. Brademan, A. S. Hebert, M. S. Westphall and J. J. Coon, Ribonucleic Acid Sequence Characterization by Negative Electron Transfer Dissociation Mass Spectrometry, Anal. Chem., 2020, 92(6), 4436–4444 CrossRef CAS PubMed.
- J. Wu, J. Wei, J. D. Hogan, P. Chopra, A. Joshi, W. Lu, J. Klein, G.-J. Boons, C. Lin and J. Zaia, Negative Electron Transfer Dissociation Sequencing of 3-O-Sulfation-Containing Heparan Sulfate Oligosaccharides, J. Am. Soc. Mass Spectrom., 2018, 29(6), 1262–1272 CrossRef CAS PubMed.
- E. H. Knelson, J. C. Nee and G. C. Blobe, Heparan sulfate signaling in cancer, Trends Biochem. Sci., 2014, 39(6), 277–288 CrossRef CAS PubMed.
- R. Karlsson, P. Chopra, A. Joshi, Z. Yang, S. Y. Vakhrushev, T. M. Clausen, C. D. Painter, G. P. Szekeres, Y.-H. Chen, D. R. Sandoval, L. Hansen, J. D. Esko, K. Pagel, D. P. Dyer, J. E. Turnbull, H. Clausen, G.-J. Boons and R. L. Miller, Dissecting structure-function of 3-O-sulfated heparin and engineered heparan sulfates, Sci. Adv., 2021, 7(52), eabl6026 CrossRef CAS PubMed.
- Y. Liu, C. Ma, C. J. A. Leonen, C. Chatterjee, G. Nováková, A. Marek and F. Tureček, Tackling a Curious Case: Generation of Charge-Tagged Guanosine Radicals by Gas-Phase Electron Transfer and Their Characterization by UV–vis Photodissociation Action Spectroscopy and Theory, J. Am. Soc. Mass Spectrom., 2021, 32(3), 772–785 CrossRef CAS PubMed.
- F. Lermyte, M. K. Łącki, D. Valkenborg, A. Gambin and F. Sobott, Conformational Space and Stability of ETD Charge Reduction Products of Ubiquitin, J. Am. Soc. Mass Spectrom., 2017, 28(1), 69–76 CrossRef CAS PubMed.
- N. Geue, R. E. P. Winpenny and P. E. Barran, Ion Mobility Mass Spectrometry for Large Synthetic Molecules: Expanding the Analytical Toolbox, J. Am. Chem. Soc., 2024, 146(13), 8800–8819 CrossRef CAS PubMed.
- Y. Zhao, R. R. Abzalimov and I. A. Kaltashov, Interactions of Intact Unfractionated Heparin with Its Client Proteins Can Be Probed Directly Using Native Electrospray Ionization Mass Spectrometry, Anal. Chem., 2016, 88(3), 1711–1718 CrossRef CAS PubMed.
- R. R. Abzalimov and I. A. Kaltashov, Electrospray Ionization Mass Spectrometry of Highly Heterogeneous Protein Systems: Protein Ion Charge State Assignment via Incomplete Charge Reduction, Anal. Chem., 2010, 82(18), 7523–7526 CrossRef CAS PubMed.
- E. Hanozin, B. Mignolet, J. Martens, G. Berden, D. Sluysmans, A.-S. Duwez, J. F. Stoddart, G. Eppe, J. Oomens, E. De Pauw and D. Morsa, Radical-Pairing Interactions in a Molecular Switch Evidenced by Ion Mobility Spectrometry and Infrared Ion Spectroscopy, Angew. Chem., Int. Ed., 2021, 60(18), 10049–10055 CrossRef CAS PubMed.
- B. T. Ruotolo, J. L. P. Benesch, A. M. Sandercock, S.-J. Hyung and C. V. Robinson, Ion mobility–mass spectrometry analysis of large protein complexes, Nat. Protoc., 2008, 3(7), 1139–1152 CrossRef CAS PubMed.
- J. L. Stephenson and S. A. McLuckey, Ion/Ion Reactions in the Gas Phase: Proton Transfer Reactions Involving Multiply-Charged Proteins, J. Am. Chem. Soc., 1996, 118(31), 7390–7397 CrossRef CAS.
- J. L. Stephenson and S. A. McLuckey, Ion-ion proton transfer reactions of multiply-charged oligonucleotide cations, Int. J. Mass Spectrom. Ion Processes, 1997, 165–166, 419–431 CrossRef.
- A. Makarov and E. Denisov, Dynamics of ions of intact proteins in the Orbitrap mass analyzer, J. Am. Soc. Mass Spectrom., 2009, 20(8), 1486–1495 CrossRef CAS PubMed.
- A. O. Bailey, R. Huguet, C. Mullen, J. E. P. Syka and W. K. Russell, Ion–Ion Charge Reduction Addresses Multiple Challenges Common to Denaturing Intact Mass Analysis, Anal. Chem., 2022, 94(9), 3930–3938 CrossRef CAS PubMed.
- J. T. Kline, C. Mullen, K. R. Durbin, R. N. Oates, R. Huguet, J. E. P. Syka and L. Fornelli, Sequential Ion–Ion Reactions for Enhanced Gas-Phase Sequencing of Large Intact Proteins in a Tribrid Orbitrap Mass Spectrometer, J. Am. Soc. Mass Spectrom., 2021, 32(9), 2334–2345 CrossRef CAS PubMed.
- L. Fornelli and T. K. Toby, Characterization of large intact protein ions by mass spectrometry: What directions should we follow?, Biochim. Biophys. Acta, 2022, 1870(4), 140758 CrossRef CAS PubMed.
- J. L. Stephenson and S. A. McLuckey, Simplification of Product Ion Spectra Derived from Multiply Charged Parent Ions via Ion/Ion Chemistry, Anal. Chem., 1998, 70(17), 3533–3544 CrossRef CAS PubMed.
- C. Beaumal, E. Deslignière, H. Diemer, C. Carapito, S. Cianférani and O. Hernandez-Alba, Improved characterization of trastuzumab deruxtecan with PTCR and internal fragments implemented in middle-down MS workflows, Anal. Bioanal. Chem., 2024, 416(2), 519–532 CrossRef CAS PubMed.
- S. D. Dunham and J. S. Brodbelt, Enhancing Top-Down Analysis of Proteins by Combining Ultraviolet Photodissociation (UVPD), Proton-Transfer Charge Reduction (PTCR), and Gas-Phase Fractionation to Alleviate the Impact of Nondissociated Precursor Ions, J. Am. Soc. Mass Spectrom., 2024, 35(2), 255–265 CrossRef CAS PubMed.
- C. E. Randolph, K. C. Fabijanczuk, S. J. Blanksby and S. A. McLuckey, Proton transfer reactions for the gas-phase separation, concentration, and identification of cardiolipins, Anal. Chem., 2020, 92(15), 10847–10855 CrossRef CAS PubMed.
- D. V. Donndelinger, T. Yan, T. R. I. V. Scoggins, J. T. Specker and B. M. Prentice, Sequencing of Phosphopeptides Using a Sequential Charge Inversion Ion/Ion Reaction and Electron Capture Dissociation Workflow, J. Am. Soc. Mass Spectrom., 2024, 35(7), 1556–1566 CrossRef CAS PubMed.
- H.-C. Chao, M. Shih and S. A. McLuckey, Generation of Multiply Charged Protein Anions from Multiply Charged Protein Cations via Gas-Phase Ion/Ion Reactions, J. Am. Soc. Mass Spectrom., 2020, 31(7), 1509–1517 CrossRef CAS PubMed.
- T. Wang, T. T. Nha Tran, H. J. Andreazza, D. Bilusich, C. S. Brinkworth and J. H. Bowie, Negative ion cleavages of (M–H)− anions of peptides. Part 3. Post-translational modifications, Mass Spectrom. Rev., 2018, 37(1), 3–21 CrossRef CAS PubMed.
- D. Bilusich and J. H. Bowie, Fragmentations of (M–H)− anions of underivatised peptides. Part 2: Characteristic cleavages of Ser and Cys and of disulfides and other post-translational modifications, together with some unusual internal processes, Mass Spectrom. Rev., 2009, 28(1), 20–34 CrossRef CAS PubMed.
- J. R. Stutzman, S. J. Blanksby and S. A. McLuckey, Gas-Phase Transformation of Phosphatidylcholine Cations to Structurally Informative Anions via Ion/Ion Chemistry, Anal. Chem., 2013, 85(7), 3752–3757 CrossRef CAS PubMed.
- S. Rojas-Betancourt, J. R. Stutzman, F. A. Londry, S. J. Blanksby and S. A. McLuckey, Gas-Phase Chemical Separation of Phosphatidylcholine and Phosphatidylethanolamine Cations via Charge Inversion Ion/Ion Chemistry, Anal. Chem., 2015, 87(22), 11255–11262 CrossRef CAS PubMed.
- E. T. Franklin, S. W. J. Shields, J. M. Manthorpe, J. C. Smith, Y. Xia and S. A. McLuckey, Coupling Headgroup and Alkene Specific Solution Modifications with Gas-Phase Ion/Ion Reactions for Sensitive Glycerophospholipid Identification and Characterization, J. Am. Soc. Mass Spectrom., 2020, 31(4), 938–945 CrossRef CAS PubMed.
- R. C. Murphy, J. Fiedler and J. Hevko, Analysis of Nonvolatile Lipids by Mass Spectrometry, Chem. Rev., 2001, 101(2), 479–526 CrossRef PubMed.
- T. Fujii, Alkali-metal ion/molecule association reactions and their applications to mass spectrometry, Mass Spectrom. Rev., 2000, 19(3), 111–138 CrossRef PubMed.
- M. T. Cancilla, S. G. Penn, J. A. Carroll and C. B. Lebrilla, Coordination of Alkali Metals to Oligosaccharides Dictates Fragmentation Behavior in Matrix Assisted Laser Desorption Ionization/Fourier Transform Mass Spectrometry, J. Am. Chem. Soc., 1996, 118(28), 6736–6745 CrossRef.
- I. Aloui, V. Legros and W. Buchmann, Study of the gas-phase decomposition pathways of poly(ethylene glycol) by Electron Transfer Dissociation, Int. J. Mass Spectrom., 2023, 487, 117028 CrossRef.
- M. Frańska, Interactions of Nucleobases with Alkali Earth Metal Cations—Electrospray Ionization Mass Spectrometric Study, Eur. J. Mass Spectrom., 2007, 13(5), 339–346 CrossRef PubMed.
- D. D. Carlton and K. A. Schug, A review on the interrogation of peptide–metal interactions using electrospray ionization-mass spectrometry, Anal. Chim. Acta, 2011, 686(1), 19–39 CrossRef PubMed.
- S. H. Toma, A. D. P. Alexiou, H. E. Toma, K. Araki and M. N. Eberlin, Can mass dissociation patterns of transition-metal complexes be predicted from electrochemical data?, J. Mass Spectrom., 2009, 44(3), 361–367 CrossRef PubMed.
- X. Chen, Y. M. E. Fung, W. Y. K. Chan, P. S. Wong, H. S. Yeung and T. W. D. Chan, Transition Metal Ions: Charge Carriers that Mediate the Electron Capture Dissociation Pathways of Peptides, J. Am. Soc. Mass Spectrom., 2011, 22(12), 2232–2245 CrossRef PubMed.
- K. A. Newton and S. A. McLuckey, Gas-phase peptide/protein cationizing agent switching via ion/ion reactions, J. Am. Chem. Soc., 2003, 125(41), 12404–12405 CrossRef PubMed.
- B. M. Prentice, W. M. McGee, J. R. Stutzman and S. A. McLuckey, Strategies for the gas phase modification of cationized arginine via ion/ion reactions, Int. J. Mass Spectrom., 2013, 354–355, 211–218 CrossRef PubMed.
- C. E. Randolph, D. J. Foreman, S. K. Betancourt, S. J. Blanksby and S. A. McLuckey, Gas-phase ion/ion reactions involving tris-phenanthroline alkaline earth metal complexes as charge inversion reagents for the identification of fatty acids, Anal. Chem., 2018, 90(21), 12861–12869 CrossRef PubMed.
- D. J. Foreman, S. K. Betancourt, A. L. Pilo and S. A. McLuckey, Novel peptide ion chemistry associated with gold (I) cationization: Preferential cleavage at lysine residues, Int. J. Mass Spectrom., 2018, 427, 114–122 CrossRef PubMed.
- D. J. Foreman, J. T. Lawler, M. L. Niedrauer, M. A. Hostetler and S. A. McLuckey, Gold(I) Cationization Promotes Ring Opening in Lysine-Containing Cyclic Peptides, J. Am. Soc. Mass Spectrom., 2019, 30(10), 1914–1922 CrossRef PubMed.
- C. Lu, S. Nelson, G. Coy, C. Neumann, E. I. Parkinson and C. A. Rice, Cyclic Peptide Natural Product Inspired Inhibitors of the Free-Living Amoeba Balamuthia mandrillaris, J. Nat. Prod., 2025, 88(2), 274–281 CrossRef PubMed.
- N. Chen, R. Huang, H. Lan, J. Jokela, H. Wei, S. Zhi and L. Liu, Cyclic peptides from cyanobacteria: Structural insights, biological functions, and mechanisms of cyclization, Algal Res., 2025, 86, 103925 CrossRef.
- S. Zhang, S. Fan, H. He, J. Zhu, L. Murray, G. Liang, S. Ran, Y. Z. Zhu, M. J. Cryle, H.-Y. He and Y. Zhang, Cyclic natural product oligomers: diversity and (bio)synthesis of macrocycles, Chem. Soc. Rev., 2025, 54(1), 396–464 RSC.
- C. E. Randolph, D. J. Foreman, S. J. Blanksby and S. A. McLuckey, Generating Fatty Acid Profiles in the Gas Phase: Fatty Acid Identification and Relative Quantitation Using Ion/Ion Charge Inversion Chemistry, Anal. Chem., 2019, 91(14), 9032–9040 CrossRef PubMed.
- C. E. Randolph, D. L. Marshall, S. J. Blanksby and S. A. McLuckey, Charge-switch derivatization of fatty acid esters of hydroxy fatty acids via gas-phase ion/ion reactions, Anal. Chim. Acta, 2020, 1129, 31–39 CrossRef PubMed.
- C. E. Randolph, D. S. M. Shenault, S. J. Blanksby and S. A. McLuckey, Structural Elucidation of Ether Glycerophospholipids Using Gas-Phase Ion/Ion Charge Inversion Chemistry, J. Am. Soc. Mass Spectrom., 2020, 31(5), 1093–1103 CrossRef PubMed.
- H.-C. Chao and S. A. McLuckey, In-Depth Structural Characterization and Quantification of Cerebrosides and Glycosphingosines with Gas-Phase Ion Chemistry, Anal. Chem., 2021, 93(19), 7332–7340 CrossRef PubMed.
- H.-C. Chao and S. A. McLuckey, Differentiation and Quantification of Diastereomeric Pairs of Glycosphingolipids Using Gas-Phase Ion Chemistry, Anal. Chem., 2020, 92(19), 13387–13395 CrossRef PubMed.
- J. R. Bonney, A. E. Stratton, Y. Guo, C. B. Eades and B. M. Prentice, Imaging Mass Spectrometry of Sulfatide Isomers from Rat Brain Tissue Using Gas-Phase Charge Inversion Ion/Ion Reactions, J. Am. Soc. Mass Spectrom., 2025, 36(1), 119–126 CrossRef PubMed.
- R. C. Barrientos and Q. Zhang, Recent advances in the mass spectrometric analysis of glycosphingolipidome – A review, Anal. Chim. Acta, 2020, 1132, 134–155 CrossRef PubMed.
- K. Hořejší, D. Kolářová, R. Jirásko and M. Holčapek, Recent advances, challenges, and future directions in the mass spectrometry analysis of glycosphingolipids in biological samples, TrAC, Trends Anal. Chem., 2024, 178, 117827 CrossRef.
- A. L. Pilo, F. Zhao and S. A. McLuckey, Gas-Phase Oxidation via Ion/Ion Reactions: Pathways and Applications, J. Am. Soc. Mass Spectrom., 2017, 28(6), 991–1004 CrossRef PubMed.
- H. Han and S. A. McLuckey, Selective covalent bond formation in polypeptide ions via gas-phase ion/ion reaction chemistry, J. Am. Chem. Soc., 2009, 131(36), 12884–12885 CrossRef PubMed.
- M. Mentinova and S. A. McLuckey, Covalent modification of gaseous peptide ions with N-hydroxysuccinimide ester reagent ions, J. Am. Chem. Soc., 2010, 132(51), 18248–18257 CrossRef PubMed.
- J. Bu, Z. Peng, F. Zhao and S. A. McLuckey, Enhanced Reactivity in Nucleophilic Acyl Substitution Ion/Ion Reactions Using Triazole-Ester Reagents, J. Am. Soc. Mass Spectrom., 2017, 28(7), 1254–1261 CrossRef PubMed.
- A. M. Pitts-McCoy, C. P. Harrilal and S. A. McLuckey, Gas-Phase Ion/Ion Chemistry as a Probe for the Presence of Carboxylate Groups in Polypeptide Cations, J. Am. Soc. Mass Spectrom., 2019, 30(2), 329–338 CrossRef PubMed.
- M. Shih and S. A. McLuckey, Ion/ion charge inversion/attachment in conjunction with dipolar DC collisional activation as a selective screen for sulfo- and phosphopeptides, Int. J. Mass Spectrom., 2019, 444, 116181 CrossRef PubMed.
- J. Bu, C. M. Fisher, J. D. Gilbert, B. M. Prentice and S. A. McLuckey, Selective Covalent Chemistry via Gas-Phase Ion/ion Reactions: An Exploration of the Energy Surfaces Associated with N-Hydroxysuccinimide Ester Reagents and Primary Amines and Guanidine Groups, J. Am. Soc. Mass Spectrom., 2016, 27(6), 1089–1098 CrossRef PubMed.
- I. K. Webb, M. Mentinova, W. M. McGee and S. A. McLuckey, Gas-Phase Intramolecular Protein Crosslinking via Ion/Ion Reactions: Ubiquitin and a Homobifunctional sulfo-NHS Ester, J. Am. Soc. Mass Spectrom., 2013, 24(5), 733–743 CrossRef PubMed.
- Y. Xia, B. A. Thomson and S. A. McLuckey, Bidirectional Ion Transfer between Quadrupole Arrays: MSn Ion/Ion Reaction Experiments on a Quadrupole/Time-of-Flight Tandem Mass Spectrometer, Anal. Chem., 2007, 79(21), 8199–8206 Search PubMed.
- N. Wang, A. L. Pilo, F. Zhao, J. Bu and S. A. McLuckey, Gas-phase rearrangement reaction of Schiff-base-modified peptide ions, Rapid Commun. Mass Spectrom., 2018, 32(24), 2166–2173 CrossRef PubMed.
- W.-Y. Kang, A. Mondal, J. R. Bonney, A. Perez and B. M. Prentice, Structural Elucidation of Ubiquitin via Gas-Phase Ion/Ion Cross-Linking Reactions Using Sodium-Cationized Reagents Coupled with Infrared Multiphoton Dissociation, Anal. Chem., 2024, 96(21), 8518–8527 CrossRef PubMed.
- X. Diao, N. R. Ellin and B. M. Prentice, Selective Schiff base formation via gas-phase ion/ion reactions to enable differentiation of isobaric lipids in imaging mass spectrometry, Anal. Bioanal. Chem., 2023, 415(18), 4319–4331 CrossRef PubMed.
- J. R. Stutzman, K. M. Hassell and S. A. McLuckey, Dissociation behavior of tryptic and intramolecular disulfide-linked peptide ions modified in the gas phase via ion/ion reactions, Int. J. Mass Spectrom., 2012, 312, 195–200 CrossRef PubMed.
- C. J. Ballance, T. P. Harty, N. M. Linke, M. A. Sepiol and D. M. Lucas, High-Fidelity Quantum Logic Gates Using Trapped-Ion Hyperfine Qubits, Phys. Rev. Lett., 2016, 117(6), 060504 CrossRef PubMed.
- M. F. Brandl, M. W. van Mourik, L. Postler, A. Nolf, K. Lakhmanskiy, R. R. Paiva, S. Möller, N. Daniilidis, H. Häffner, V. Kaushal, T. Ruster, C. Warschburger, H. Kaufmann, U. G. Poschinger, F. Schmidt-Kaler, P. Schindler, T. Monz and R. Blatt, Cryogenic setup for trapped ion quantum computing, Rev. Sci. Instrum., 2016, 87(11), 113103 CrossRef PubMed.
- C. D. Bruzewicz, J. Chiaverini, R. McConnell and J. M. Sage, Trapped-ion quantum computing: Progress and challenges, Appl. Phys. Rev., 2019, 6(2), 021314 Search PubMed.
| This journal is © The Royal Society of Chemistry 2026 |