Open Access Article
Open Access ArticleTriggering isothermal exponential amplification of microRNA via liposome fusion
Mamiko Tsugane ,
Kosuke Kato,
Keisuke Shinohara,
Tsutomu Okita,
Reiko Sato and
Hiroaki Suzuki
,
Kosuke Kato,
Keisuke Shinohara,
Tsutomu Okita,
Reiko Sato and
Hiroaki Suzuki *
*
Graduate School of Science and Engineering, Chuo University, 112-8551, Japan. E-mail: suzuki.97a@g.chuo-u.ac.jp
First published on 16th December 2025
Abstract
Lipid membrane vesicles serve as essential cellular compartments where diverse biochemical reactions are regulated through dynamic membrane remodeling, including vesicle fusion. Reconstructing these processes in vitro is crucial for advancing artificial nanobiotechnology. Among intracellular metabolic reactions, isothermal nucleic acid amplification is particularly important because it enables sensitive detection of nucleic acid biomarkers. In this study, we focused on exponential amplification reaction (EXPAR), a straightforward isothermal amplification method initiated by microRNA (miRNA), and developed a system, in which fusion of giant unilamellar vesicles (GUVs) triggers the delivery of miRNA into the reaction compartment, thereby initiating amplification. Using our previously reported microdevice with microchambers and high-aspect-ratio electrodes, we monitored both membrane fusion and the subsequent amplification reaction in situ. This platform provides a proof of concept for detecting nucleic acid biomarkers encapsulated in membrane-bound vesicles and offers promising applications in artificial cell systems and in situ analysis of extracellular vesicles such as exosomes.
Introduction
Cellular life fundamentally depends on compartmentalization.1–4 Lipid bilayer vesicles not only form the plasma membrane but also create a complex network of internal organelles that spatially coordinate metabolism, signalling, and genetic information. Eukaryotic cells dynamically generate, transport, and fuse these vesicles to deliver both soluble and membrane-associated cargo with remarkable spatiotemporal precision.5–7 In addition, cells transmit signals to distant targets by producing and fusing extracellular vesicles (EVs) or exosomes that carry signaling molecules such as miRNAs.8–10 Reconstructing these dynamic membrane processes in vitro offers a powerful approach for equipping artificial systems with life-like functionality—from controlled multistep catalysis to signal-responsive molecular processing.11–15Over the past two decades, various synthetic cell-mimetic microcompartments have been developed, including polymer vesicles, colloidosomes, aqueous two-phase coacervates, and other liquid–liquid phase-separated droplets.4,16–28 Each architecture offers distinct advantages, yet lipid vesicles (liposomes) remain the benchmark for cell-mimetic compartments.14,24,29 Their inherent biocompatibility, precisely regulated selective permeability, and ability to host integral membrane proteins make liposomes ideally suited for integrating biological machinery in artificial platforms. In particular, the capacity of liposomes to undergo membrane fusion—a defining feature of cellular trafficking—provides an efficient means to deliver encapsulated reagents between discrete compartments and coordinate sequential biochemical reactions.30,31 In living systems, membrane fusion is mediated by specialized proteins such as SNAREs.32,33 In artificial settings, however, it can be triggered using fusogenic agents34–36 or electrical stimulation.37–39 Because this process enables the transfer of encapsulated materials into reaction compartments, it offers a powerful approach for controlling reaction initiation and detecting membrane-enclosed substances.
Controlled vesicle fusion has been used to initiate chemical and enzymatic reactions. In previous studies, partitioning complementary reagents into separate vesicle populations and triggering their fusion enabled chelation reactions,34,40 enzymatic reactions,41 cell-free protein synthesis,42 and reverse-transcription PCR (RT-PCR).43 Among these, nucleic acid amplification is especially appealing because of its central role in diagnostics, synthetic biology, and information processing. However, PCR requires thermal cycling, which introduces practical challenges: specialized equipment is necessary, and repeated heating cycles can cause membrane leakage or rupture in addition to damage of other biomolecules. In contrast, nature amplifies nucleic acids isothermally, inspiring a range of constant-temperature chemistries—including loop-mediated amplification (LAMP),44 recombinase polymerase amplification (RPA),45 rolling circle amplification (RCA),46 and nucleic acid sequence-based amplification (NASBA47). These methods lower technical requirements and simplify device integration, making them well-suited for point-of-care diagnostics and field-deployable sensors.48,49 Thus, integrating isothermal nucleic acid amplification machinery into the liposome-fusion-mediated reaction system will lead to construction of robust detection/processing system which does not be affected by environmental factors, as demonstrated in nanometer scale vesicles.50 Use of cell-sized giant unilamellar vesicles (GUVs) is expected to provide advantages in uniform encapsulation of complex reaction systems, easy optical-microscope observation, and applicability to local detection/processing.51
Moreover, microRNAs (miRNAs) are short, non-coding RNAs that play essential roles in gene regulation and are widely investigated as biomarkers for various diseases. Their small size and low abundance, however, make detection challenging. Isothermal amplification systems for miRNA have been developed, but many require complex primer designs (LAMP) or prior template circularization (RCA).52,53 To overcome these limitations, several groups introduced the exponential amplification reaction (EXPAR), an isothermal method optimized for short oligonucleotide targets.54–58 EXPAR achieves rapid and sensitive amplification by combining polymerase extension with nicking enzyme cleavage, making it particularly suitable for miRNA detection. Compared with LAMP and RCA, EXPAR offers a simpler design, faster kinetics, and greater compatibility with short RNA inputs.
Here, we demonstrate the feasibility of a liposome-based platform, in which membrane fusion functions as a deterministic on-switch for isothermal amplification of miRNA. Specifically, we adopted the reaction scheme reported by H. Jia et al.55 and aimed to develop a system where miRNA was introduced into EXPAR system-containing vesicles to initiate amplification. In our previous work, we also developed an electrofusion device with chambered groove structures designed to retain GUVs as individual reactors.59 Using this device, we successfully showed that miRNA amplification can be reliably triggered by vesicle membrane fusion.
Materials & methods
EXPAR mixture preparation and thermal conditions
miRNA (let-7a), serving as the trigger oligonucleotide, and the amplification template oligonucleotide were synthesized by FASMAC Co., Ltd (Kanagawa, Japan). Vent (exo−) DNA polymerase and the nicking endonuclease Nt.BstNBI were obtained from New England Biolabs (NEB, Ipswich, MA, USA). RNase inhibitor, dNTP, and SYBR Green I were purchased from Takara Bio Inc. (Shiga, Japan). All solutions were prepared using nuclease-free, ultrapure water (Thermo Fisher Scientific, Waltham, MA, USA). The sequence of the trigger oligonucleotide (let-7a) was 5′-UGAGGUAGUAGGUUGUAUAGUU-3′ and the amplification template sequence was 5′-AACTATACAACCTACTACCTCAAACAGACTCAAACTATACAACCTACTACCTCAA-3′-phosphate. Vent (exo−) DNA polymerase was supplied with ThermoPol buffer (20 mM Tris-HCl; pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, and 0.1% Triton X-100), and Nt.BstNBI was supplied with NEBuffer 3.1 (25 mM Tris-HCl; pH 8.0, 50 mM NaCl, 5 mM MgCl2, and 0.5 mM dithiothreitol). Because the Triton X-100 in ThermoPol buffer could disrupt the lipid bilayer of liposomes, a custom ThermoPol buffer without Triton X-100 was prepared. EXPAR was performed in a total reaction volume of 20 µL containing miRNA at various concentrations, amplification template (0.1 μM), dNTPs (250 μM), Nt.BstNBI (0.4 U μL−1), Vent (exo−) DNA polymerase (0.05 U μL−1), RNase inhibitor (0.8 U μL−1), 2.5× SYBR Green I, 1× custom ThermoPol buffer (without Triton X-100), 0.5× NEBuffer 3.1, and 200 mM sucrose (to match the conditions of in-liposome reactions). The reaction mixture was incubated at 55 °C, and fluorescence intensity was monitored in real time using an Mx3005P real-time PCR system (Agilent Technologies, Santa Clara, CA, USA).Preparation of GUVs containing EXPAR system
1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Cholesterol and liquid paraffin were obtained from Nacalai Tesque (Kyoto, Japan) and Wako Pure Chemical Industries (Osaka, Japan), respectively. Transferrin from human serum, Alexa Fluor 647 Conjugate (TA647) and tetramethylrhodamine (TRITC)-conjugated dextran were purchased from Thermo Fisher Scientific.The compositions of the internal and external aqueous solutions, as well as the lipid solution (oil phase) for the GUVs containing EXPAR, are summarized in Table 1. To create a density difference between the internal and external solutions while maintaining iso-osmotic conditions, sucrose was added to the internal solution, whereas glucose was added to the external solution at identical molar concentrations. To prevent the major electrolyte components of the encapsulated EXPAR buffer—(NH4)2SO4 (10 mM), KCl (10 mM), MgSO4 (2 mM), and NaCl (50 mM)—from leaking out of the GUVs and inhibiting the reaction, these same electrolytes were included in the external solution. MgCl2 contained in NEBuffer 3.1 can cause GUV aggregation; therefore, it was excluded from the external solution. For experiments involving EXPAR triggering via GUV fusion, two distinct GUV populations were prepared: one encapsulating the EXPAR system components and the other encapsulating miRNA (see Tables S1–S3 for detailed compositions). These GUV populations were differentiated by incorporating separate fluorescent markers. The lipid-containing oil phase (liquid paraffin) had the same composition for both populations, with a total lipid concentration of approximately 2 mg mL−1. POPC, POPG, DOPE, and cholesterol were first dissolved in chloroform and mixed at a weight ratio of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. A small amount of DOPE was included to potentially enhance membrane fusion efficiency.60 To enable visualization of the lipid membrane under a microscope, the lipophilic dye DiI (0.05% w/w; Thermo Fisher Scientific) was incorporated into the lipid mixture.
1. A small amount of DOPE was included to potentially enhance membrane fusion efficiency.60 To enable visualization of the lipid membrane under a microscope, the lipophilic dye DiI (0.05% w/w; Thermo Fisher Scientific) was incorporated into the lipid mixture.
| Internal aqueous solution | External aqueous solution | Lipid solution | |||
|---|---|---|---|---|---|
| Sucrose | 200 mM | Glucose | 200 mM | POPC | 1.7 mg mL−1 |
| Custom ThermoPol buffer | 1× | Custom ThermoPol buffer | 1× | POPG | 0.2 mg mL−1 |
| NEBuffer 3.1 | 0.5× | Tris-HCl (pH 8.0) | 25 mM | DOPE | 0.1 mg mL−1 |
| Amplification template | 0.1 μM | NaCl | 50 mM | Cholesterol | 0.1 mg mL−1 |
| SYBR Green I | 2.5× | SYBR Green I | 2.5× | In liquid paraffin | |
| RNase inhibitor | 0.8 unit per μL | ||||
| Nt.BstNBI | 0.4 unit per μL | ||||
| Vent (exo−) | 0.05 unit per μL | ||||
| dNTP | 0.25 mM | ||||
| TA647 | 1 μM | ||||
| miRNA (let-7a) | Various | ||||
Following the method described in our previous publications, we prepared GUVs using the water-in-oil (W/O) emulsion interface transfer method.41,43,59 Briefly, 25 μL of the inner solution was added to the liquid paraffin solution containing lipids and vortexed to form a W/O emulsion. This emulsion was layered on 400 μL of the outer aqueous solution in a test tube and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 20 min, causing the GUVs to precipitate at the bottom of the tube as a pellet. The resulting GUV suspension (∼100 μL) was collected through a small hole pierced in the bottom of the test tube. Next, 400 μL of the outer solution was added, and the suspension was centrifuged again at 15
000 rpm at 4 °C for 20 min, causing the GUVs to precipitate at the bottom of the tube as a pellet. The resulting GUV suspension (∼100 μL) was collected through a small hole pierced in the bottom of the test tube. Next, 400 μL of the outer solution was added, and the suspension was centrifuged again at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C for 10 min. The supernatant was then removed to obtain a concentrated GUV suspension of ∼30 μL.
000 rpm, 4 °C for 10 min. The supernatant was then removed to obtain a concentrated GUV suspension of ∼30 μL.
Small GUVs containing miRNA were prepared using the extrusion method. The GUV suspension was passed through a track-etched membrane with 2 µm pores using the Avanti Mini Extruder (Avanti) according to the instructions of the manufacturer. After three extrusion cycles, the suspension in the syringe was collected. The sample was then centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min, the supernatant was removed, and 30 μL of the outer aqueous solution was added.
000 rpm for 10 min, the supernatant was removed, and 30 μL of the outer aqueous solution was added.
Gel electrophoresis of EXPAR products
EXPAR products were analysed by agarose gel electrophoresis using 4% E-Gel EX Agarose Gels (Thermo Fisher Scientific) and the E-Gel iBase Power System, according to the manufacturer's protocol. A total of 20 µL of each reaction product was mixed with loading dye and loaded into individual chambers. Electrophoresis was performed for 15 min. The E-Gel Ultra Low Range DNA Ladder (Thermo Fisher Scientific) was used as a molecular size marker. DNA bands were visualized using the blue-light transilluminator.Flow cytometry analysis
Quantitative analysis of EXPAR in individual GUVs was performed using an Attune NxT flow cytometer (Thermo Fisher Scientific). The amplification marker (SYBR Green I) and the vesicle volume marker (TA647) were excited with 488 and 638 nm lasers, respectively, to measure their fluorescence intensities. Scatter plots showing the correlation between these intensities were generated after analyzing ∼25 μL of the diluted GUV sample. Flow cytometry data were analyzed using FlowJo software (BD, Franklin Lakes, NJ, USA).Bulk electrofusion
The two GUV suspensions were confirmed to contain approximately equal numbers of vesicles, and they were mixed at a one-to-one volume ratio (40 μL total). This mixture was introduced into an electrofusion cuvette with a 1 mm electrode gap, connected to an electrocell fusion generator (LF201; Nepa Gene Co., Ltd, Chiba, Japan). An alternating current electric field (AC; 15 V cm−1 peak-to-peak, 15 s) was applied to induce pearl-chain alignment, followed by three direct-current pulses (6 kV cm−1, 60 μs each) to trigger membrane fusion.Fabrication of the electrofusion device
The fabrication procedure for the electrofusion device has been described in detail in our previous work.59 Briefly, a low-resistance silicon wafer (resistivity 0.01 Ω cm, thickness 150 µm) served as the electrode material. A positive photoresist (S1818, Shipley, Rohm and Haas, Philadelphia, PA, USA) was patterned to define the microchambers and connecting slits. Using a DRIE apparatus (RIE-400iPB, Samco, Japan), a ∼20 µm deep-etching process created reverse replicas of the microchambers and slits on the silicon wafer. After removing the photoresist, PDMS (Sylgard 184, Dow Corning Midland, Michigan, USA) was spin-coated on cover glass at 3000 rpm for 30 s, and the structured surface of the chamber was gently pressed on top. A second positive photoresist (AZP 4620, MicroChemicals GmbH, Germany) was then patterned to define the electrodes. Deep-etching was repeated using the DRIE apparatus until the trap microchambers were fully exposed. Finally, iron lead wires were attached to the silicon wafer with conductive paste (Polycalm CPT-1211ND, Plascoat Co., Ltd, Japan) and were connected to an electrocell fusion generator for electrofusion.Procedure for the electrofusion in the device
First, two GUV suspensions were confirmed to contain approximately equal numbers of vesicles, and they were mixed at a one-to-one volume ratio (15 μL total). This mixture was placed between the device electrodes and left undisturbed for ∼20 min to allow the GUVs to settle at the bottom. Microscopic images of this pre-fusion state were then acquired. To bring the GUVs in the chamber into contact, an AC electric field of 0.15 kV cm−1 peak-to-peak at 1 MHz was applied for 15 s, followed immediately by two DC pulses of 7.0 kV cm−1 for 60 μs each at 0.2 s intervals. After a 10–20 s pause, post-fusion microscopic images were captured. The device was then incubated for 2 h at 55 °C using a dry block incubator (Biomedical Science Co., Ltd, Tokyo, Japan), and images were collected after the reaction.Image acquisition was performed using a confocal laser scanning microscope (LSM700, Carl Zeiss, Germany) equipped with a 40× water-immersion objective lens (C-Apochromat 40×/1.2 W CorrECS M27). Three-channel fluorescence images were acquired at excitation wavelengths of 488, 555, and 639 nm, along with bright-field (differential interference contrast) images simultaneously.
Results
Experimental scheme
We designed a system, in which two populations of GUVs—one encapsulating the reaction components and the other containing miRNA as the trigger—were fused to initiate nucleic acid amplification. The isothermal amplification method used here was EXPAR developed by H. Jia et al. in 2010 for miRNA detection55 (Fig. 1A). This system is straightforward and primarily consists of DNA polymerase, a nicking enzyme, and a single-stranded DNA (ssDNA) amplification template. The template carries two repeat sequences complementary to the target miRNA (marked blue in Fig. 1A) flanking a recognition site for the nicking enzyme (marked gray). | ||
| Fig. 1 Schematics of the experimental system. (A) Simplified representation of the EXPAR reaction mechanism. (B) Induction of EXPAR within GUVs triggered by membrane electrofusion. | ||
When the target miRNA hybridizes to the 3′ end of the template, DNA polymerase binds and extends the strand by assembling dNTPs. After extension, the nicking enzyme cleaves the newly synthesized strand four base pairs downstream (on the 5′ side). Extension then resumes from the cleavage site, displacing the cleaved ssDNA. Through repeated cycles of extension, cleavage, and strand displacement, amplification proceeds from the target miRNA. Importantly, the displaced ssDNA has the same sequence as the miRNA and can hybridize with additional amplification templates to trigger further replication, enabling exponential amplification. The reaction is monitored by fluorescence using SYBR Green I, which selectively binds to double-stranded DNA (dsDNA).
To initiate this reaction through GUV membrane fusion, we prepared two distinct GUV populations (Fig. 1B): one containing only miRNA and the other containing the amplification template and enzymes. SYBR Green I was basically included in both populations, but does not fluoresce strongly in the presence of ssDNA or miRNA alone. When all components merge within a single compartment via membrane fusion, the EXPAR reaction proceeds, and green fluorescence appears inside the GUVs as the amount of template–miRNA/DNA complex (dsDNA) increases.
EXPAR in GUVs
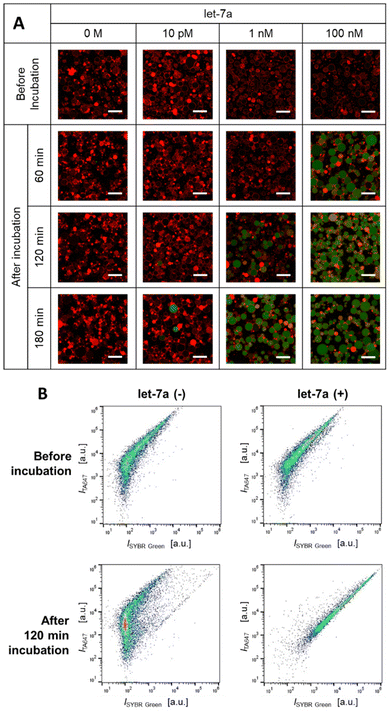
We first validated EXPAR in a test tube to confirm its functionality (Fig. S1a). A 20 μL reaction mixture containing 0.1 μM of the 55 bp template was prepared with initial miRNA concentrations of 0, 10 pM, 1 nM, and 100 nM. The mixtures were incubated at 55 °C, and green fluorescence was monitored using a real-time PCR apparatus. Across this concentration range, samples containing miRNA exhibited markedly earlier fluorescence increases than the blank, with higher miRNA concentrations producing faster amplification onset. At 100 nM miRNA, fluorescence rose sharply approximately 40 min after incubation began. Gel electrophoresis revealed a strong band around 55 bp only in post-reaction samples, corresponding to the amplified cDNA–template complex (Fig. S1b). Together, these results confirmed the successful amplification via miRNA-triggered reactions. Then we confirmed that these post-EXPAR solutions can be encapsulated into GUVs without impeding the production process (Fig. S2).Next, we tested EXPAR within GUVs. Using the W/O emulsion interface transfer method, we encapsulated all components of the EXPAR system with varying miRNA concentrations (0, 10 pM, 1 nM, and 100 nM, Table 1). During incubation at 55 °C, GUVs were sampled and imaged using a fluorescence confocal microscope (Fig. 2A). No fluorescence was detected at 0 M, indicating the absence of nonspecific amplification. At 10 pM, discrete green fluorescence appeared in a few large (>10 μm) GUVs. At 1 nM, green fluorescence became visible in most GUVs after 180 min, although some vesicles remained non-fluorescent. At 100 nM, many GUVs exhibited fluorescence after 60 min, with nearly all fluorescing by 120 min.
On average, vesicles with a diameter of 10 μm should contain ∼3 miRNA molecules at 10 pM and ∼300 molecules at 1 nM. Thus, EXPAR can be initiated with only tens to hundreds of miRNA molecules per GUV. Higher initial miRNA concentrations led to faster fluorescence onset, approaching the kinetics observed in bulk conditions (Fig. S1a) and demonstrating efficient, unimpeded amplification within GUVs. Despite similar sizes, amplification dynamics varied among vesicles, likely reflecting differences in the number of encapsulated molecules or heterogeneity in membrane leakage. Such variability aligned with previous reports on other GUV-based reaction systems.61
At 100 nM miRNA, we further confirmed amplification over time using flow cytometry (Fig. 2B and Fig. S3). After 120 min, dot plots revealed a horizontal shift along the SYBR green fluorescence axis, representing a 140-fold increase in intensity, but only when miRNA (let-7a) was co-encapsulated. These findings indicated that isothermal amplification successfully proceeded while the size and number distribution of the GUVs remained unchanged (Fig. S4 and S5).
EXPAR in GUVs triggered by membrane fusion
Next, we tested electrofusion. As shown in Fig. 1, the EXPAR reaction system and miRNA were encapsulated separately into two distinct populations of GUVs. As an initial experiment, bulk electrofusion tests were performed (Fig. 3). GUVs containing the miRNA were co-loaded with TA647, while GUVs containing EXPAR system were membrane-stained with the red fluorescent dye DiI to distinguish between the two populations. Equal volumes of the two GUV populations were mixed and placed into an electroporation cuvette (1 mm electrode gap) for fusion. Based on previous studies, the electrofusion protocol involved applying an AC field to induce dielectrophoresis and bring the liposomes into contact, followed by three DC pulses to trigger fusion. Note that, as we included POPG, negatively charged lipid in the GUV membrane components, electrostatic repulsion prevented the spontaneous fusion (Fig. S6).62 After applying the electrofusion pulses, a small volume was collected and imaged using confocal microscopy.As a result, GUVs exhibiting both membrane fluorescence and TA647 fluorescence in the lumen were observed, indicating successful fusion of multiple GUVs (Fig. 3A and B). After retrieving the GUV sample from the cuvette into a tube and incubating it at 55 °C for 120 min, increased green fluorescence from EXPAR was detected in multiple GUVs (Fig. 3C). These results confirmed that the miRNA-triggered EXPAR via membrane fusion, as designed in Fig. 1, was successfully implemented in the present system.
EXPAR triggered by membrane fusion within an electrofusion microdevice
Bulk electrofusion, as described in the previous section, does not allow precise control over the number or pairing of GUVs to be fused. Furthermore, dielectrophoresis and unintended electrical convection cause GUV movement, complicating fusion evaluation. To address these limitations, we previously developed a GUV electrofusion device equipped with high-aspect-ratio silicon electrodes and demonstrated its ability to induce and evaluate GUV fusion via a simple chelation reaction.59 This device incorporates PDMS-based microchambers that enable sedimentation and trapping of a defined number of GUVs. Because GUVs remain confined within the microchambers, it becomes possible to visually compare their state before and after fusion as well as monitor reaction progress during incubation. Another advantage of this system is that it requires only a small sample volume (∼10 μL).Using this device, we directly observed changes in GUVs during both fusion and reaction. An overview of the electrofusion device and an electron microscope image of the PDMS microchamber structure are shown in Fig. 4A. The electrodes were fabricated from deep-etched conductive silicon, and PDMS microchambers were formed in the gap between these electrodes. A groove connected the microchambers along the direction of the electric field gradient, promoting dielectrophoresis and fusion. Two types of PDMS microchambers with different geometries were designed and used. One type contained elongated chambers intended to accommodate two adjacent GUVs (each approximately 10 μm in diameter) for fusion. The other type contained circular microchambers sized to trap a single GUV of about 10 μm in diameter. The elongated design was used for fusing two GUV populations with similar size distributions, whereas the circular design was used to simulate detection of miRNA encapsulated in nanoscale biological vesicles (such as exosomes) by attempting fusion between a GUV containing the EXPAR reaction system and ∼1 μm vesicles carrying miRNA.
Before performing fusion experiments, we verified whether the microchamber interfered with the EXPAR reaction inside GUVs. As in the experiment shown in Fig. 2, GUVs containing the complete EXPAR system and 100 nM miRNA were prepared using the W/O emulsion interface transfer method and then sedimented into the microchambers. To prevent drying and unexpected drift flows, a cover glass was placed on top. After incubation at 55 °C for 120 min, all GUVs emitted green fluorescence, confirming that miRNA-triggered nucleic acid amplification proceeded without interference even while GUVs were confined in the microchambers (Fig. S7).
Next, following the experiment shown in Fig. 3, we prepared two GUV populations: one containing the EXPAR reaction components and the other containing miRNA. After mixing them in equal volumes, the mixture was dispensed into the microchamber region of the device. GUVs containing the EXPAR components were labeled with a TA647 volume marker, while GUVs containing miRNA were labeled with a TRITC-conjugated dextran marker to enable visual distinction under a microscope. After dispensing the mixture and allowing 20 min for sedimentation, an AC electric field was applied for 15 s. As a result, multiple microchambers containing both TRITC and TA647 GUVs were observed (because most GUVs were smaller than 10 μm, many microchambers contained multiple GUVs with TRITC and TA647) (Fig. 4B). Note that the edges of PDMS microchambers fluoresced green because of the non-specific adsorption of SYBR green supplemented to the outer solution of GUVs (Table 1). When DC fusion pulses were then applied, GUVs exhibiting both fluorescence signals appeared, confirming that fusion had occurred. Subsequent incubation for 120 min led to an increase of green fluorescence in ∼40% of fused GUVs, indicating progression of the EXPAR reaction (Fig. 4B and Fig. S8). Increase in green fluorescence in fused GUVs was not observed in the control condition without miRNA (Fig. S9). This approach enabled imaging of the same microchamber region at three distinct time points: after GUV seeding, after applying the fusion field, and after incubation. By tile-scanning the microchamber area and comparing these images, we retrospectively verified that green fluorescence in GUVs was triggered by mixing of internal aqueous phases of two different initial GUVs mediated by membrane fusion.
Next, we tested fusion between EXPAR-containing GUVs and smaller GUVs (∼2 μm in diameter) encapsulating miRNA, assuming the detection of nucleic acids inside micro- and nano-extracellular vesicles, which are ubiquitous in the biological world. The miRNA-containing GUVs were first prepared using the W/O emulsion interface transfer method and then downsized by extrusion through a 2 μm pore membrane. The two GUV populations were mixed and introduced into microchambers, followed by application of an AC field for dielectrophoresis and DC pulses for fusion. Similar to the previous experiments, an increase in green fluorescence was observed in multiple GUVs larger than 5 μm in diameter, while the fluorescence increase was not observed in the control condition without miRNA (Fig. 4C and Fig. S10, S11). Under these conditions, the volume of small GUVs is too low, and they are diluted after fusion, so fusion cannot be confirmed by TRITC fluorescence.
These results demonstrate that electrofusion enables the delivery of miRNA encapsulated in small vesicles into compartments containing the EXPAR reaction system, thereby facilitating its detection via isothermal amplification.
Conclusion
In this study, we established the conditions under which EXPAR can be triggered by miRNA within GUVs. We demonstrated that membrane fusion delivers miRNA into compartments containing the EXPAR system, initiating the amplification reaction. Moreover, by using a custom-designed electrofusion device, we monitored the entire process—from membrane fusion to reaction progression—in situ. However, the number of green fluorescence-emitting events as a result of successful fusion and amplification, was insufficient for statistical validation. This low fusion efficiency may be influenced by factors such as the salt composition of the solution deteriorating electrofusion efficiency. Another possibility for the low amplification yield in the fusion experiments is that, when electric pulses were applied, amplification activity of the polymerase (Vent (exo−)) was decreased (Fig. S12). Further challenge remains to effectively fuse small GUVs, as the dielectrophoresis-based vesicle-to-vesicle contact and fusion becomes difficult.63 Despite these limitations, this work provides a proof-of-concept for isothermal detection of miRNA via vesicle fusion and elucidated necessary challenges. Future work will focus on optimizing solution composition, membrane properties, and microdevice architecture to improve fusion efficiency. This platform holds promise for direct detection of nucleic acids encapsulated in membrane-bound compartments, such as exosomes and EVs, without the need for extraction. It also provides a foundation for nucleic acid delivery into artificial cell systems through targeted vesicle fusion.Author contributions
The work was initiated, conceived, and supervised by M. T. and H. S. M. T. designed and performed EXPAR and GUV experiments with assistance of R. S. K. K. performed the electrofusion experiments using a microdevice with guidance of T. O. and K. S. H. S. and M. T. drafted the manuscript and H. S., M. T., and K. K. reviewed the manuscript.Conflicts of interest
The authors declare no conflicts of interest.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information is available. See DOI: https://doi.org/10.1039/d5an01087j.Acknowledgements
This study was supported by JSPS grants 24K08214, 21K04851, 21H05890, 18K04908, and 16J40215 for MS and 24K01320, 24H01155, 20H05935, 19H02576, 19H00901 for HS. This study was also supported by the Institute of Science and Engineering at Chuo University.References
- L. Bar-Peled and N. Kory, Nat. Metab., 2022, 4, 1232–1244 Search PubMed.
- T. Gabaldón and A. A. Pittis, Biochimie, 2015, 119, 262–268 CrossRef PubMed.
- A. Honigmann and A. Pralle, J. Mol. Biol., 2016, 428, 4739–4748 CrossRef CAS PubMed.
- R. J. Wheeler and A. A. Hyman, Philos. Trans. R. Soc., B, 2018, 373, 20170193 CrossRef PubMed.
- S. L. Schmid, Annu. Rev. Biochem., 1997, 66, 511–548 CrossRef CAS PubMed.
- S. D. Conner and S. L. Schmid, Nature, 2003, 422, 37–44 CrossRef CAS PubMed.
- B. S. Glick and A. Nakano, Annu. Rev. Cell Dev. Biol., 2009, 25, 113–132 CrossRef CAS PubMed.
- A. Bobrie, M. Colombo, G. Raposo and C. Théry, Traffic, 2011, 12, 1659–1668 CrossRef CAS PubMed.
- G. van Niel, G. D'Angelo and G. Raposo, Nat. Rev. Mol. Cell Biol., 2018, 19, 213–228 CrossRef CAS PubMed.
- K. Boriachek, M. N. Islam, A. Möller, C. Salomon, N. T. Nguyen, M. S. A. Hossain, Y. Yamauchi and M. J. A. Shiddiky, Small, 2018, 14, 201702153 CrossRef PubMed.
- M. Hagiya, A. Konagaya, S. Kobayashi, H. Saito and S. Murata, Acc. Chem. Res., 2014, 47, 1681–1690 CrossRef CAS PubMed.
- B. A. Grzybowski and W. T. S. Huck, Nat. Nanotechnol., 2016, 11, 584–591 CrossRef PubMed.
- A. Samanta, L. B. Pellejero, M. Masukawa and A. Walther, Nat. Rev. Chem., 2024, 8, 454–470 CrossRef CAS PubMed.
- Z. G. Peng, S. Iwabuchi, K. Izumi, S. Takiguchi, M. Yamaji, S. Fujita, H. Suzuki, F. Kambara, G. Fukasawa, A. Cooney, L. Di Michele, Y. Elani, T. Matsuura and R. Kawano, Lab Chip, 2024, 24, 996–1029 RSC.
- Y. Sato, ChemSystemsChem, 2024, 6, 202400021 CrossRef.
- N. Ichihashi, T. Matsuura, H. Kita, T. Sunami, H. Suzuki and T. Yomo, Cold Spring Harbor Perspect. Biol., 2010, 2, a004945 Search PubMed.
- F. Wu and C. M. Tan, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2014, 6, 369–383 CAS.
- C. Martino and A. J. deMello, Interface Focus, 2016, 6, 20160011 CrossRef PubMed.
- B. C. Buddingh and J. C. M. van Hest, Acc. Chem. Res., 2017, 50, 769–777 CrossRef CAS PubMed.
- E. Rideau, R. Dimova, P. Schwille, F. R. Wurm and K. Landfester, Chem. Soc. Rev., 2018, 47, 8572–8610 RSC.
- Z. Liu, W. Zhou, C. Qi and T. T. Kong, Adv. Mater., 2020, 32, 202002932 Search PubMed.
- X. L. Wang, X. M. Liu and X. Huang, Adv. Mater., 2020, 32, 202001436 Search PubMed.
- M. E. Allen, J. W. Hindley, D. K. Baxani, O. Ces and Y. Elan, Nat. Rev. Chem., 2022, 6, 562–578 CrossRef CAS PubMed.
- Y. Lu, G. Allegri and J. Huskens, Mater. Horiz., 2022, 9, 892–907 RSC.
- X. L. Wang, X. Qiao, H. X. Chen, L. Wang, X. M. Liu and X. Huang, Small Methods, 2023, 7, 2201712 CrossRef PubMed.
- Z. Lin, T. Beneyton, J. C. Baret and N. Martin, Small Methods, 2023, 7, 202300496 Search PubMed.
- V. Maffeis, L. Heuberger, A. Nikoletic, C. A. Schoenenberger and C. G. Palivan, Adv. Sci., 2024, 11, e2305837 CrossRef PubMed.
- A. D. Slootbeek, M. H. van Haren, I. B. A. Smokers and E. Spruijt, Chem. Commun., 2022, 58, 11183–11200 RSC.
- S. S. Mansy, Cold Spring Harbor Perspect. Biol., 2010, 2, a002188 Search PubMed.
- R. Jahn and H. Grubmuller, Curr. Opin. Cell Biol., 2002, 14, 488–495 CrossRef CAS PubMed.
- H. R. Marsden, I. Tomatsu and A. Kros, Chem. Soc. Rev., 2011, 40, 1572–1585 RSC.
- Y. A. Chen and R. H. Scheller, Nat. Rev. Mol. Cell Biol., 2001, 2, 98–106 CrossRef CAS.
- R. Jahn and R. H. Scheller, Nat. Rev. Mol. Cell Biol., 2006, 7, 631–643 CrossRef CAS PubMed.
- D. A. Kendall and R. C. Macdonald, J. Biol. Chem., 1982, 257, 3892–3895 CrossRef.
- B. R. Lentz, Eur. Biophys. J. Biophys. Lett., 2007, 36, 315–326 CrossRef CAS PubMed.
- S. Mondal Roy and M. Sarkar, J. Lipids, 2011, 2011, 528784 Search PubMed.
- N. G. Stoicheva and S. W. Hui, Biochim. Biophys. Acta, Biomembr., 1994, 1195, 31–38 CrossRef CAS PubMed.
- D. T. Chiu, C. F. Wilson, F. Ryttsén, A. Strömberg, C. Farre, A. Karlsson, S. Nordholm, A. Gaggar, B. P. Modi, A. Moscho, R. A. Garza-López, O. Orwar and R. N. Zare, Science, 1999, 283, 1892–1895 CrossRef CAS PubMed.
- R. Dimova, K. A. Riske, S. Aranda, N. Bezlyepkina, R. L. Knorr and R. Lipowsky, Soft Matter, 2007, 3, 817–827 RSC.
- T. Sunami, F. Caschera, Y. Morita, T. Toyota, K. Nishimura, T. Matsuura, H. Suzuki, M. M. Hanczyc and T. Yomo, Langmuir, 2010, 26, 15098–15103 CrossRef CAS PubMed.
- H. Shiomi, S. Tsuda, H. Suzuki and T. Yomo, PLoS One, 2014, 9, 0101820 CrossRef PubMed.
- F. Caschera, T. Sunami, T. Matsuura, H. Suzuki, M. M. Hanczyc and T. Yomo, Langmuir, 2011, 27, 13082–13090 CrossRef CAS PubMed.
- M. Tsugane and H. Suzuki, Sci. Rep., 2018, 8, 9214 CrossRef PubMed.
- L. Becherer, N. Borst, M. Bakheit, S. Frischmann, R. Zengerle and F. von Stetten, Anal. Methods, 2020, 12, 717–746 RSC.
- M. Y. Tan, C. Liao, L. A. Liang, X. L. Yi, Z. H. Zhou and G. J. Wei, Front. Cell. Infect. Microbiol., 2022, 12, 1019071 CrossRef CAS PubMed.
- L. L. Xu, J. X. Duan, J. M. Chen, S. J. Ding and W. Cheng, Anal. Chim. Acta, 2021, 1148, 238187 CrossRef CAS PubMed.
- J. Compton, Nature, 1991, 350, 91–92 CrossRef CAS PubMed.
- Y. X. Zhao, F. Chen, Q. Li, L. H. Wang and C. H. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- P. Srivastava and D. Prasad, 3 Biotech, 2023, 13, 200 CrossRef PubMed.
- B. Ning, Z. Huang, B. M. Youngquist, J. W. Scott, A. Niu, C. M. Bojanowski, K. J. Zwezdaryk, N. S. Saba, J. Fan, X. M. Yin, J. Cao, C. J. Lyon, C. Z. Li, C. J. Roy and T. Y. Hu, Nat. Nanotechnol., 2021, 16, 1039–1044 CrossRef CAS PubMed.
- Y. Sato, K. Komiya, I. Kawamata, S. Murata and S. M. Nomura, Chem. Commun., 2019, 55, 9084–9087 RSC.
- Y. Q. Cheng, L. J. Dong, J. Y. Zhang, Y. Q. Zhao and Z. P. Li, Analyst, 2018, 143, 1758–1774 RSC.
- Z. Cao, X. F. Jiang, G. Z. Xiao, M. C. Xu, H. Liu and S. Cai, Separations, 2021, 8, 166 CrossRef CAS.
- J. Van Ness, L. K. Van Ness and D. J. Galas, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 4504–4509 CrossRef CAS PubMed.
- H. X. Jia, Z. P. Li, C. H. Liu and Y. Q. Cheng, Angew. Chem., Int. Ed., 2010, 49, 5498–5501 CrossRef CAS PubMed.
- C. Shi, Q. Liu, C. P. Ma and W. W. Zhong, Anal. Chem., 2014, 86, 336–339 CrossRef CAS PubMed.
- Q. Zhang, F. Chen, F. Xu, Y. X. Zhao and C. H. Fan, Anal. Chem., 2014, 86, 8098–8105 CrossRef CAS PubMed.
- M. Hiratani, M. Ohara and R. Kawano, Anal. Chem., 2017, 89, 2312–2317 CrossRef CAS PubMed.
- T. Okita, M. Tsugane, K. Kato, K. Shinohara and H. Suzuki, J. Microelectromech. Syst., 2025, 34, 174–183 CAS.
- A. E. Gad, R. Broza and G. D. Eytan, Biochim. Biophys. Acta, 1979, 556, 181–195 CrossRef CAS PubMed.
- G. Rampioni, F. D’Angelo, L. Leoni and P. Stano, Front. Bioeng. Biotechnol., 2019, 7, 1 CrossRef PubMed.
- K. Nishimura, T. Hosoi, T. Sunami, T. Toyota, M. Fujinami, K. Oguma, T. Matsuura, H. Suzuki and T. Yomo, Langmuir, 2009, 25, 10439–10443 CrossRef CAS PubMed.
- L. Rems, M. Ušaj, M. Kandušer, M. Reberšek, D. Miklavčič and G. Pucihar, Sci. Rep., 2013, 3, 3382 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2026 |